- Department of Biology, Institute of Molecular Plant Biology, ETH Zurich (Swiss Federal Institute of Technology), Zurich, Switzerland
Iron deficiency leads to severe chlorosis in crop plants, including wheat, thereby reducing total yield and quality. Furthermore, grains of most bread wheat varieties are poor source of iron, which is vital for human nutrition. Despite the significance, iron uptake and translocation mechanisms in bread wheat have not been studied in detail, particularly under iron limited growth conditions. In this study, bread wheat plants were grown under iron deficiency stress until maturity. Data were collected at three distinct developmental time points during grain-filling. The plants experiencing low iron availability exhibited significantly lower chlorophyll content as well as low iron concentration in leaves and grains. The expression levels of bread wheat genes homologous to iron deficiency responsive genes of rice, barley, and Arabidopsis were significantly changed under iron deficiency stress. The wheat homologs of genes involved in phytosiderophore (PS) synthesis and transport were significantly up-regulated in the iron-deficient roots through all development stages, confirming an important role of deoxymugineic acid (DMA) in iron acquisition. The up-regulation of NICOTIANAMINE SYNTHASE (NAS) and DEOXYMUGINEIC ACID SYNTHASE (DMAS) in flag leaves and grains suggested the involvement of nicotianamine (NA) and DMA in iron chelation and translocation in wheat, particularly at the commencement of grain-filling. In line with this, the homolog of gene encoding TRANSPORTER OF MUGINEIC ACID (TOM) was up-regulated in the wheat roots under iron deficiency. Additionally, genes encoding long-distance iron transporter YELLOW STRIPE-LIKE (YSL), the vacuolar transporter NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN (NRAMP), and the transcription factor BASIC HELIX-LOOP-HELIX (bHLH), were also up-regulated upon iron starvation. A tissue specific and growth stage specific gene expression differences in response to iron deficiency stress were observed, providing new insights into iron translocation, storage and regulation in bread wheat.
Introduction
Iron is an essential micronutrient for all living organisms. In plants, iron functions as redox-active metal in many important reactions of metabolic processes such as photosynthesis, mitochondrial respiration, nitrogen assimilation, hormone biosynthesis, and the production and scavenging of reactive oxygen species (Hansch and Mendel, 2009). Although iron ranks as the fourth most abundant element in the earth's crust (Buckhout and Schmidt, 2013), plants often suffer from iron deficiency due to the low bioavailability of iron in aerobic, calcareous and/or high pH soils (Morrissey and Guerinot, 2009). Iron deficiency causes interveinal chlorosis because of insufficient chlorophyll production that is often identified as alternate yellow and green stripes in younger leaves of most cereals (Barker and Stratton, 2015). As one of the key agricultural problems, iron deficiency restrains plant growth, ultimately leading to yield losses. Additionally, the grain iron content in cereal crops affected by iron deficiency is further reduced, thereby impacting human nutrition. Several studies investigated plant responses to iron deficiency stress and its effect on iron uptake, translocation and utilization (Thimm et al., 2001; Kobayashi et al., 2005; Buckhout et al., 2009; Yang et al., 2010; Rodriguez-Celma et al., 2013) but such information is rather limited for wheat (Bonneau et al., 2016; Beasley et al., 2017; Connorton et al., 2017; Garnica et al., 2018; Kumar et al., 2018; Mathpal et al., 2018).
Since excess iron can also be reactive and toxic via the Fenton reaction, plants tightly control and maintain iron homeostasis. Non-graminaceous and graminaceous plants utilize different strategies for iron acquisition from soil. Non-graminaceous plants acquire iron through the reduction strategy (strategy I), in which protons are first released into the rhizosphere by H+-ATPases to increase the solubility of iron (Santi and Schmidt, 2009). FERRIC CHELATE REDUCTASES (FRO) reduce ferric iron [Fe(III)] to ferrous iron [Fe(II)] (Robinson et al., 1999; Waters et al., 2002). IRON REGULATED TRANSPORTER 1 (IRT1), which is a divalent metal ion transporter localized on the plasma membrane, subsequently transports Fe(II) into the plant roots (Vert et al., 2002). Graminaceous plants use the chelation strategy (strategy II) for iron uptake from the soil. The production of iron chelators, i.e., the mugineic acid (MA) family phytosiderophores (PSs), is the key step. The PSs are synthesized from S-adenosyl-L-methionine, through a series of conserved reactions catalyzed by NICOTIANAMINE SYNTHASE (NAS), NICOTIANAMINE AMINOTRANSFERASE (NAAT) and DEOXYMUGINEIC ACID SYNTHASE (DMAS) (Kobayashi et al., 2010). In addition to being a precursor for DMA production, NA also chelates iron forming Fe(II)-NA and Fe(III)-NA complexes, and transports iron within the plants (von Wiren et al., 1999; Takahashi et al., 2003). The DMAs are either directly exported to the soil as chelators for iron acquisition, for example in rice, or they are further converted into other MAs as is the case in barley (Kobayashi et al., 2010). In wheat, 21 NAS genes were identified and classified into two distinct clades (Bonneau et al., 2016). Under iron deficiency condition, the clade I NAS genes were up-regulated in root tissues (Bonneau et al., 2016). In addition, six TaNAAT homeologs and three TaDMAS1 homeologs have been identified, and these are most closely related to the barley HvNAAT and HvDMAS1 (Beasley et al., 2017). The TaNAAT1, TaNAAT2, and TaDMAS1 genes were also significantly upregulated in root tissues under Fe deficiency treatment (Beasley et al., 2017). In rice, the PSs are transported to the rhizosphere by the phytosiderophore efflux transporter TRANSPORTER OF MUGINEIC ACID 1 (TOM1) (Nozoye et al., 2011). The released PSs chelate Fe(III) and the resulting Fe(III)-PS complexes are transported back into roots cells by the YELLOW STRIPE-LIKE (YSL) family of transporters (Kobayashi and Nishizawa, 2012). OsYSL15 in rice encodes a Fe(III)-DMA and Fe(II)-NA transporter and is induced in the root epidermis under iron deficiency (Inoue et al., 2008; Lee et al., 2009). The YSL family of transporters is also suggested to facilitate long-distance iron transport through the phloem (Inoue et al., 2009; Lee et al., 2009; Ishimaru et al., 2010; Kumar et al., 2018). The function of YSL transporters in iron translocation is well-established in several crop plants, including maize and barley (Curie et al., 2001; Koike et al., 2004; Murata et al., 2006; Aoyama et al., 2009; Ishimaru et al., 2010; Kakei et al., 2012). Recently, 26 wheat YSL genes have been identified and comprise the largest plant YSL gene family known so far. These wheat YSL genes are differently regulated in wheat roots and shoots in response to iron-deficient stress (Kumar et al., 2018).
Specific transporters control iron translocation between the subcellular compartments. Iron in plants is often stored in vacuoles and iron import and export between vacuoles and the cytoplasm is dynamically regulated by the plant iron status. Some NATURAL RESISTANCE ASSOCIATED MACROPHAGE PROTEIN (NRAMP) such as AtNRAMP3 and AtNRAMP4 function in transporting iron out of vacuoles, whereas VACUOLAR IRON TRANSPORTER (VIT) has the opposite function (Lanquar et al., 2005; Zhang et al., 2012). Most of the iron in plants is found in leaf chloroplasts (Buckhout and Schmidt, 2013). PERMEASE IN CHLOROPLASTS 1 (PIC1) is associated with iron transport across the inner chloroplast envelope in Arabidopsis and tobacco (Duy et al., 2007, 2011; Gong et al., 2015), and Arabidopsis AtYSL4 and AtYSL6 export iron out of chloroplasts (Divol et al., 2013). The MITOCHONDRIAL IRON TRANSPORTER (MIT) (Bashir et al., 2011) is involved in iron transport into mitochondria.
In addition to the storage in vacuoles, iron is also stored in plastids where it is bound to FERRITIN (Briat et al., 2010). The FERRITIN globular protein complex can harbor up to 4,500 iron atoms (Harrison and Arosio, 1996) and therefore is an important iron storage protein in plants. Because of this iron storage capacity, targeted expression of FERRITIN in the endosperm is often used in iron biofortification strategies (Wirth et al., 2009; Boonyaves et al., 2016, 2017; Trijatmiko et al., 2016; Wu et al., 2018).
Additionally, several transcription factor families such as the BASIC HELIX-LOOP-HELIX (bHLH), WRKY, and NO APICAL MERISTEM (NAM)/ARABIDOPSIS TRANSCRIPTION ACTIVATION FACTOR (ATAF)/CUP-SHAPED COTYLEDON (CUC) (NAC) families are known to regulate the iron deficiency stress response (Ogo et al., 2006; Uauy et al., 2006; Sperotto et al., 2008; Wang et al., 2013). Transcription factors from the bHLH family, such as Fe-DEFICIENCY-INDUCIBLE BASIC HELIX-LOOP-HELIX TRANSCRIPTION FACTOR 2 (OsIRO2) and OsbHLH133 in rice, are known to regulate iron homeostasis-related genes in many plants (Ogo et al., 2006; Wang et al., 2013). Studies have shown that bHLH proteins are also involved in the regulation of iron uptake in chrysanthemum (Zhao et al., 2014) and Populus plants (Huang and Dai, 2015).
Wheat is the most widely cultivated commercial crop in the world and feeds 40% of the population and provides 20% of the energy for human nutrition (Gupta et al., 2005). Although the knowledge pool concerning iron homeostasis in bread wheat is slowly advancing, the information is very limited when compared to model plants such as Arabidopsis and rice (Uauy et al., 2006; Waters et al., 2009; Bonneau et al., 2016; Beasley et al., 2017; Garnica et al., 2018; Kumar et al., 2018; Mathpal et al., 2018). In particular, the molecular response of bread wheat to iron deficiency stress is unknown but could help breeders to increase grain iron content and growers to optimize agricultural management. We investigated wheat plants grown on iron deficiency condition and observed their response. The chlorophyll content was significantly lower in plants experiencing iron deficiency condition. The expression of endogenous iron homeostasis-related genes in roots, flag leaves and grains of iron deficient wheat plants indicated tissue- and growth stage-specific roles for iron chelators, transporters and transcription factors.
Materials and Methods
Plant Material and Growth Conditions
Bread wheat (Triticum aestivum cv. Bobwhite S26) seeds were germinated on wet filter paper for 1 week. The seedlings were then transferred to hydroponic nutrient solutions (pH 6.0): 0.88 mM K2SO4, 2 mM Ca(NO3)2, 0.2 mM KH2PO4, 1.0 mM MgSO4, 0.1 mM KCl, 1.0 μM H3BO3, 1.0 μM MnSO4, 0.2 μM CuSO4, 0.02 μM (NH4)6Mo7O24, 1.0 μM ZnSO4, and 100.0 μM Fe(III)-EDTA for maintenance of growth (control condition) (Durmaz et al., 2011). After 1 week, half of seedlings were transferred to hydroponic nutrient solutions with 10.0 μM Fe(III)-EDTA (for iron deficiency stress treatment) under greenhouse conditions (22°C/18°C with 16-h light/8-h dark cycle, 60% humidity). Air was continuously pumped into the hydroponic system for better circulation. The hydroponic medium was changed once per week and the solution was neutralized to pH 6.0 twice per week. Roots, flag leaves and grains were harvested at three different development stages, i.e., 8–10 DPA - days post anthesis (stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3). The three sampling stages correspond to 96, 113, 121 days after transplantation to the hydroponics solution, respectively. Three biological replicates were collected for each sample.
Chlorophyll Content Measurement
Twenty-five (25) mg of ground flag leaves samples were incubated in 15 mL of 95% ethanol solution for 24 h in dark at room temperature. Extraction at room temperature for 24 h is suggested to have better efficiency than in the refrigerator or for shorter duration (Arvola, 1981). Three biological replicates and two technical replicates were analyzed. The ethanol extract was subjected to Ultrospec 2100 UV/Visible spectrometer measurements (Pharmacia Biotech, Sweden) at A649 and A664. The quantification of chlorophyll a and chlorophyll b were calculated according to the protocol by Lichtenthaler and Buschmann (2001). The sample used for chlorophyll measurement was standardized at 25 mg fresh weight (FW) and chlorophyll content/g FW was calculated accordingly.
Metal Measurements
Roots, flag leaves and grain samples were ground and digested with 4 mL 32.5% HNO3 in a single reaction chamber microwave system (turboWave, MLS GmbH, Switzerland). After the digestion, samples were volumed up to 10 mL with water, and this solution was further diluted 10 times to be analyzed in ICP-OES (ICPE 9820, Shimadzu, Japan). The wavelength used for iron, zinc, copper and manganese was 259.940, 202.548, 324.754, and 257.610, respectively.
Total RNA Extraction and cDNA Synthesis
Isolation of total RNA from the roots and flag leaves was carried out using the Isol- RNA Lysis Reagent (5 PRIME, USA). RNA extraction from wheat grains was carried out following the protocol described by Singh and colleagues (Singh et al., 2003). Total RNA used for real-time quantitative PCR was treated with DNase I (Thermo Fisher Scientific, USA). The RevertAid™ first strand cDNA synthesis kit (Thermo Fisher Scientific, USA) was used for cDNA synthesis. All steps were carried out following the manufacturers' instructions.
Gene Expression and Statistical Analysis
Gene expression analysis was carried out using qRT-PCR performed with the LightCycler® 480 Instrument II Real Time PCR system (Roche, Switzerland) using Taqman hydrolysis probes (Roche, Switzerland). The total reaction volume of each sample was set as 10 μL, comprising 5 μL of mastermix (Applied Biosystems, USA), 3 μL of 10× diluted cDNA, 0.9 μL of forward primer, 0.9 μL of reverse primer, 0.1 μL of the probe and 0.1 μL of H2O.
The primers were designed based on the Roche universal probe library assay design center website (https://lifescience.roche.com/shop/CategoryDisplay?catalogId=10001&tab=&identifier=Universal±Probe±Library&langId=-1&storeId=15006). The corresponding probes were also selected using the same webportal. The primer sequences and probe numbers are provided in Supplementary Table S1. Primer efficiency was determined using a standard curve calculated with the following formula: E = 10∧(−1/slope) (Supplementary Table S1). Genes for the study were selected based on % sequence identity to known homologs in other plant species using NCBI UniGene database as well as on published reports on wheat specific genes. The 19 studied wheat genes included phytosiderophore biosynthesis-related genes as well as several genes encoding different iron transporters, transcription factors and the storage protein FERRITIN (Table 1). Amplicons were searched against IWGSC (International Wheat Genome Sequencing Consortium) chromosome-based wheat genome sequencing (RefSeq assembly v1.0) in Ensembl database (Supplementary Table S2). For the already characterized wheat genes, same gene name as in the original publication are used in this study (Borg et al., 2012; Bonneau et al., 2016; Beasley et al., 2017; Connorton et al., 2017; Guo and Wang, 2017; Kumar et al., 2018). The qRT-PCR reaction was carried out using the following steps: incubation at 50°C for 2 min, initialization at 95°C for 10 min, followed by denaturation at 95°C for 15 s, and annealing and extension at 60°C for 1 min (these two steps were repeated for 40 cycles).
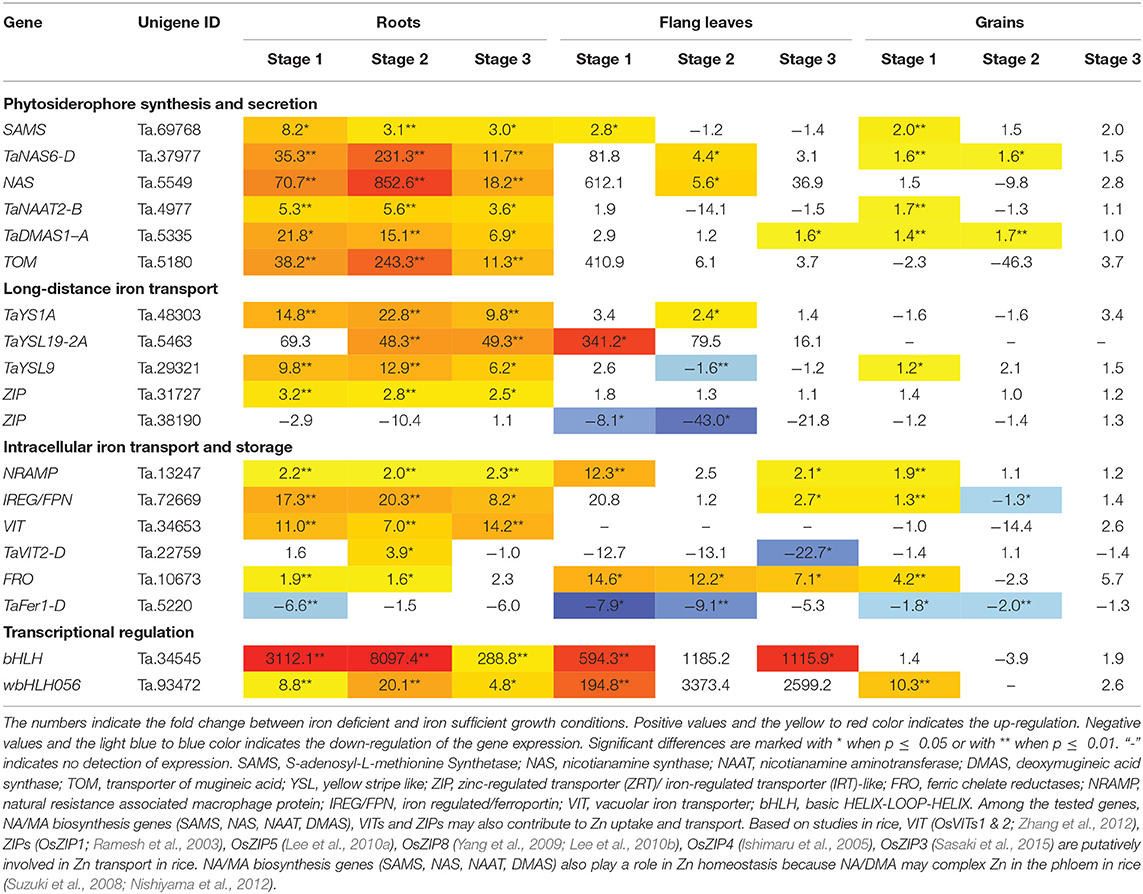
Table 1. Gene expression fold change between iron deficient and sufficient condition in bread wheat.
The reference genes for qRT-PCR normalization were chosen from 16 candidate genes that were demonstrated as stably expressed genes in the Genevestigator™ database and the literature (Zimmermann et al., 2005; Paolacci et al., 2009). Among these 16 tested genes, Ta.55681 (casein kinase II subunit beta-like), Ta.40026 (BTB/POZ and MATH domain-containing protein 4-like) and Ta.22845 (26S proteasome regulatory subunit) were found to be the most stable reference genes across all of the tested samples (unpublished data). Ta.55681 was used for the normalization of qRT-PCR data on genes of interest. Data normalization was performed as described by Schefe and colleagues (Schefe et al., 2006). Statistical analysis (one-way ANOVA) was carried out using RStudio (Version 1.1.456) and Excel. Figures were prepared in RStudio using the R plotting packages (ggplot2, pheatmap).
Results
Iron Deficiency Stressed Plants Have Lower Tissue Iron Concentration and Lower Chlorophyll Content in Flag Leaves
The chlorophyll content in flag leaves and metal concentration in roots, flag leaves and grains were measured in the iron deficiency stressed plants in comparison to the plants grown on normal condition. For the chlorophyll measurement, the samples collected at three development stages during grain filling, i.e., 8–10 days post anthesis (DPA; stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3) were analyzed. In growth stage 1, the content of chlorophyll a and chlorophyll b, together with the total chlorophyll content (chlorophyll a+b), decreased around 70% in the wheat plants experiencing iron deficiency (Figure 1). In stage 2, the total chlorophyll content and chlorophyll a content decreased nearly 50%. In stage 3, 50% decrease in chlorophyll a content could be observed in iron-deficient wheat flag leaves. For the metal concentration measurement, the samples collected at 25–28 DPA (stage 2) were analyzed. The iron concentration in flag leaves and grains was significantly lower in plants under iron deficiency stress (Figure 1). On the other hand, the concentration of copper and zinc in all tissues and the manganese concentration in roots and flag leaves was higher in plants treated with low iron (Supplementary Figure S1). Further, plant height and panicle number were significantly reduced under iron deficiency stress (Supplementary Figure S2). Together, these observations indicate strong iron stress in the plants (Figure 1). We explored the expression differences of iron homeostasis related genes under iron deficiency stress (results below).
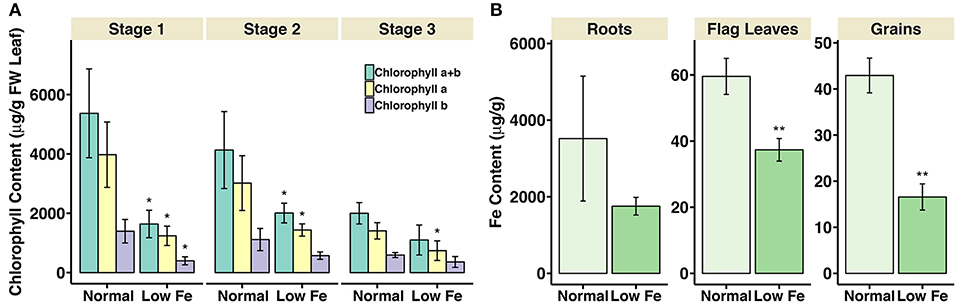
Figure 1. Chlorophyll content in flag leaves and iron concentration in roots, flag leaves and grains of iron deficiency stressed plants. (A) Total chlorophyll content (chlorophyll a+b), chlorophyll a, and chlorophyll b content are presented. Values are the mean ± standard deviation of the three biological replicates (*p ≤ 0.05). (B) Iron concentration in roots, flag leaves and grains of samples collected at 25-28DPA is presented. Values are the average of three biological replicates. Asterisks indicate statistically significantly higher values calculated using Student's t-test as compared to the control (*p < 0.05, **p < 0.01).
Iron Deficiency Induces Expression of Genes Involved in the Phytosiderophore Synthesis and Secretion
The relative mRNA levels of the wheat homologous genes SAMS (Ta.69768), TaNAS6-D (Ta.37977), NAS (Ta.5549), TaNAAT2-B (Ta.4977), and TaDMAS1-A (Ta.5335) that are related to PS biosynthesis were significantly increased in response to iron-deficient conditions, particularly in the roots (Table 1, Figure 2). In the iron-sufficient growth condition, their expression was similar in all tissues and development stages (Figure 2). Table 1 summarizes the fold change difference between iron sufficient and deficient condition, while relative gene expression level differences can be observed in Figure 2, Supplementary Table S3. During iron deficient growth, the expression of SAMS (Ta.69768) increased in roots by 8.2-fold at stage 1, and around 3-fold at stages 2 and 3 (Table 1). The NAS homologs were up-regulated in roots at least 11-fold at all three stages, but in stage 2 their mRNA levels increased more than 200-fold, indicating a very strong but transient up-regulation of the genes during the peak grain filling period (Table 1). Likewise, the expression of TaNAAT2-B (Ta.4977) increased in roots by 5-fold at stage 1 and 2, and by 3.6-fold at stage 3. The expression of TaDMAS1-A (Ta.5335) was highest at stage 1 (21.8- fold up-regulated in roots), followed by stage 2 (15.1-fold) and 3 (6.9-fold). In line with the increased expression of genes involved in phytosiderophore synthesis, the expression of the phytosiderophore efflux transporter TOM homolog Ta.5180 was also significantly up-regulated in iron-deficient roots at all the development stages, with highest expression at stage 2 (243.3-fold) (Table 1, Figure 2).
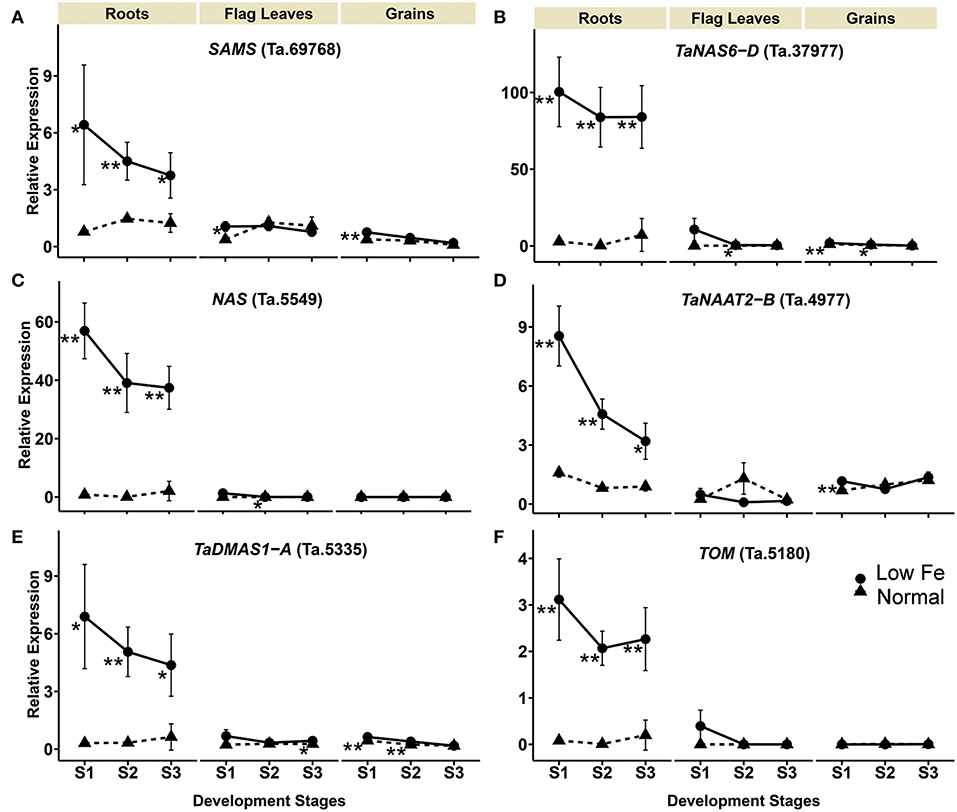
Figure 2. Iron deficiency triggered differences in expression levels of wheat homologs of phytosiderophore synthesis related genes and the TRANSPORTER OF MUGINEIC ACID (TOM). The data collected at three different development stages, i.e., 8–10 DPA - days post anthesis (stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3) are presented. Values are the mean ± standard deviation of the three biological replicates (*p ≤ 0.05; **p ≤ 0.01). Note the differences in scale at Y-axis for relative expression levels of tested genes. (A) SAMS (Ta.69768), (B) TaNAS6-D (Ta.37977), (C) NAS (Ta.5549), (D) TaNAAT2-B (Ta.4977), (E) TaDMAS1-A (Ta.5335), (F) TOM (Ta.5180).
The expression of phytosiderophore biosynthesis related genes was also increased in the flag leaves in iron-deficient conditions, including SAMS (Ta.69768) at stage 1, TaNAS6-D (Ta.37977), NAS (Ta.5549) at stage 2 and TaDMAS1-A (Ta.5335) at stage 3, but generally the increase of their expression was lower than in roots. In addition to roots and the flag leaves, iron deficiency also increased gene expression in developing wheat grains. With a few exceptions, most of the expression changes in grains were restricted to growth stage 1 (Table 1, Figure 2). The increased expression of the phytosiderophore biosynthesis and iron acquisition-related genes indicate that NA and DMA production was enhanced in response to iron deficiency, especially in the roots. The stronger increase in expression in roots as compared with the flag leaves and grains is consistent with the required increase in phytosiderophore production by the roots and their export into the rhizosphere for iron chelation. Furthermore, the up-regulation of NAS and DMAS in the flag leaves and grains suggests the involvement of NA and DMA in long-distance iron translocation within the plants, and also indicates an active transport of iron into the grains at growth stage 1 (Figure 2).
Expression of Genes Encoding Long-Distance Iron Transporters
The transcript levels of TaYS1A (Ta.48303), TaYSL19-2A (Ta.5463), TaYSL9 (Ta.29321) encoding the wheat homologs of the metal-nicotianamine transporter YSL increased significantly in the iron deficient condition (Table 1). The expression of TaYS1A (Ta.48303) was up-regulated more than 9-fold in roots at all stages, and by 2.4-fold in flag leaves at stage 2 (Table 1, Figure 3). The expression of TaYSL19-2A (Ta.5463) in roots increased by 48.3- and 49.3-fold at stages 2 and 3, respectively, and in the flag leaves by 341.2-fold at stage 1. Expression of the TaYSL9 (Ta.29321) was also significantly increased in roots at all the growth stages (Table 1, Figure 3). While a small increase in the expression of TaYSL9 was detected in the grains at stage 1, the gene was down-regulated in the flag leaves collected at stage 2, suggesting that the wheat genes encoding the YSL homologs are differentially regulated.
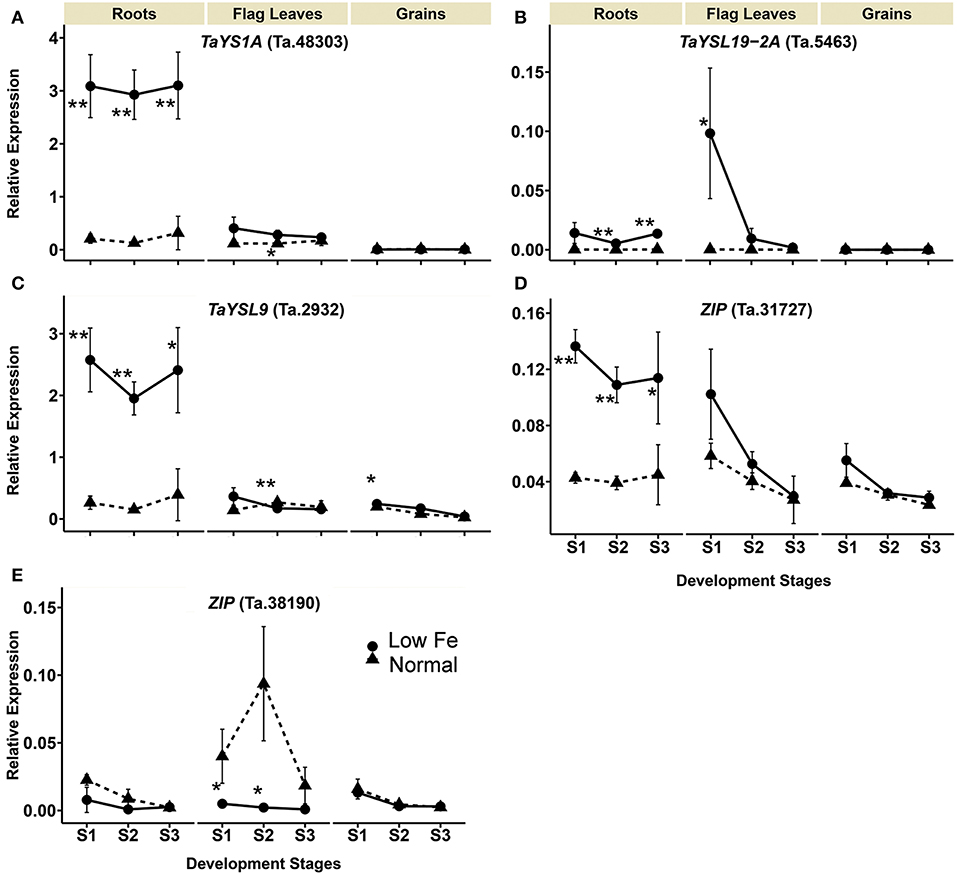
Figure 3. Iron deficiency triggered expression differences of wheat homologs of long-distance iron transporters encoding genes. The data collected at three different development stages, i.e., 8–10 DPA (stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3) are presented. Values are the mean ± standard deviation of the three biological replicates (*p ≤ 0.05; **p ≤ 0.01). Note the differences in scale at Y-axis for relative expression levels of tested genes. (A) TaYS1A (Ta.48303), (B) TaYSL19-2A (Ta.5463), (C) TaYSL9 (Ta.29321), (D) ZIP (Ta.31727), (E) ZIP (Ta.38190).
The expression of two genes from the ZIP family (Ta.31727, Ta.38190) differed in the iron-deficient condition (Table 1, Figure 3). Expression of the ZIP homolog (Ta.31727) in roots was increased more than 2.5-fold at all development stages but was not altered in the flag leaves and grains. On the contrary, Ta.38190 showed a significant decrease in expression in the flag leaves at stages 1 and 2.
Differential Expression of Genes Encoding Intracellular Iron Transporters and Storage Proteins
Expression of the NRAMP homolog Ta.13247 increased in the flag leaves by 12- and 2- fold in stages 1 and 3, respectively. In the roots, the Ta.13247 expression increased approximately 2-fold in all of the development stages. In the grains, the gene was up-regulated in the stages 1 with 1.9-fold increase (Table 1, Figure 4). IRON REGULATED PROTEIN/ FERROPORTIN (IREG/FPN) homolog (Ta.72669) showed increased expression in the roots during all stages. In addition, the expression of Ta.72669 also increased by 2.7- and 1.3-fold in stage 3 of flag leaves, and in stage 1 of the grains, respectively. However, a 1.3-fold decrease in expression of Ta.72669 was detected in the grains in stage 2 (Table 1, Figure 4). The expression of VIT homolog (Ta.34653) could not be detected in the flag leaves regardless of the growth conditions and stage of development. However, a significant increase in gene expression was observed in the roots, ranging from 7- to 14.2-fold changes (Table 1). The TaVIT2-D (Ta.22759) was only up-regulated in the roots in stage 2, but a significant decrease (22.7-fold) in gene expression was observed in the flag leaves in stage 3 (Table 1, Figure 4). The dynamic modulation of vacuolar iron transporters suggest that plant cells tightly maintain the balance between iron storage and usage, and actively re-distribute iron to the cell organs based on the iron status and demand within the plants. Iron deficiency also caused an increase in the expression of the FRO homolog Ta.10673 in roots at stages 1 and 2 (1.9- and 1.6-fold, respectively) (Table 1, Figure 4). FRO expression was also highly up-regulated in the flag leaves at all development stages (14.6-, 12.2-, and 7.1-fold) and also 4.2-fold in the grains during stage 1.
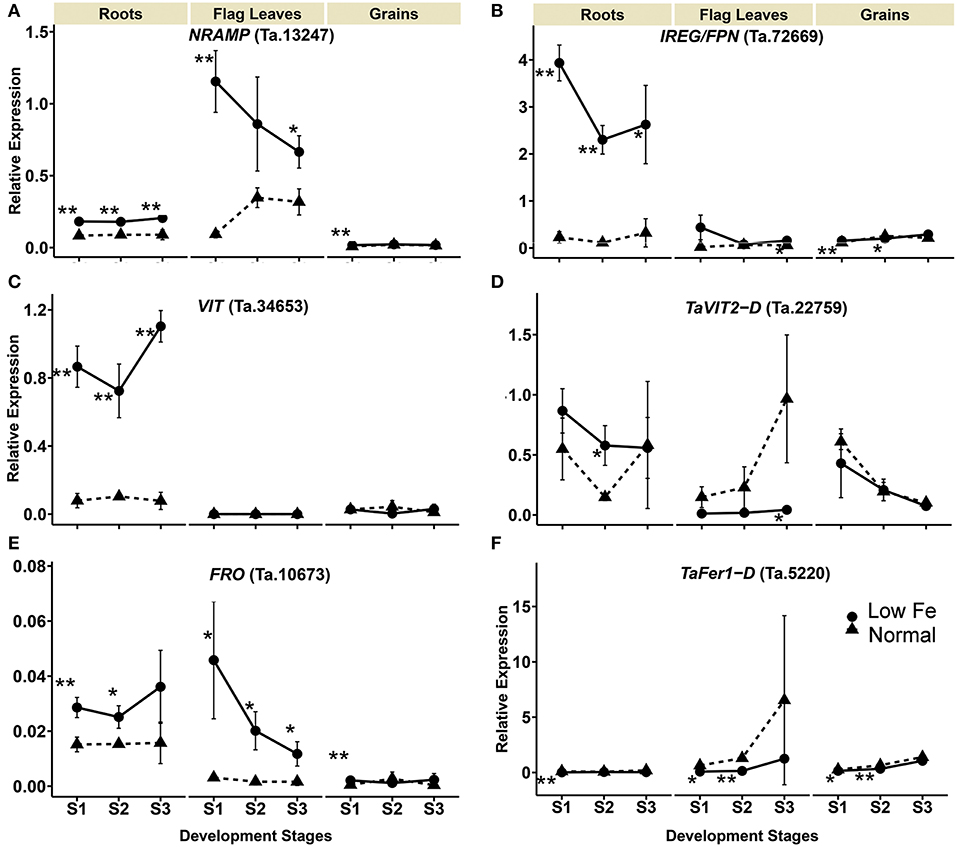
Figure 4. Differentially expressed wheat homologs of intracellular iron transporters and storage proteins encoding genes under iron deficiency. The data collected at three different development stages, i.e., 8–10 DPA (stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3) are presented. Values are the mean ± standard deviation of the three biological replicates (*p ≤ 0.05; **p ≤ 0.01). Note the differences in scale at Y-axis for relative expression levels of tested genes. (A) NRAMP (Ta.13247), (B) IREG/FPN (Ta.72669), (C) VIT (Ta.34653), (D) TaVIT2-D (Ta.22759), (E) FRO (Ta.10673), (F) TaFer1-D (Ta.5220).
The expression of FERRITIN homolog (TaFer1-D, Ta.5220) was down-regulated in the roots, flag leaves and grains due to iron starvation. A more than 6-fold decrease in the expression of TaFer1-D was observed in the roots during stage 1. In the flag leaves and grains, the gene was down-regulated in stages 1 and 2 by more than 7.9- and 1.8-fold, respectively (Table 1, Figure 4). The results indicate that the plants reduce iron storage under low iron availability.
Expression Modulation of bHLH Transcription Factor Encoding Genes
Upon iron deficiency stress, the expression of bHLH homolog Ta.34545 was significantly up-regulated by 3112.1-, 8097.4-, and 288.8-fold in the roots in stage 1, 2, and 3, respectively. Similar increases were observed for the flag leaves, particularly in stage 1 and 3, with at least a 594.3-fold increase in gene expression. No significant difference in the expression was observed in the grains. Additionally, the expression of the IRO2 homolog (wbHLH056, Ta.93472), which is also a bHLH member, was up-regulated by 4.8- to 20.1-fold in the roots when plants experienced iron-deficiency (Table 1, Figure 5). Increased expression of wbHLH056 was observed in flag leaves (195-fold) and grains (10-fold), particularly during stage 1 (Table 1, Figure 5).
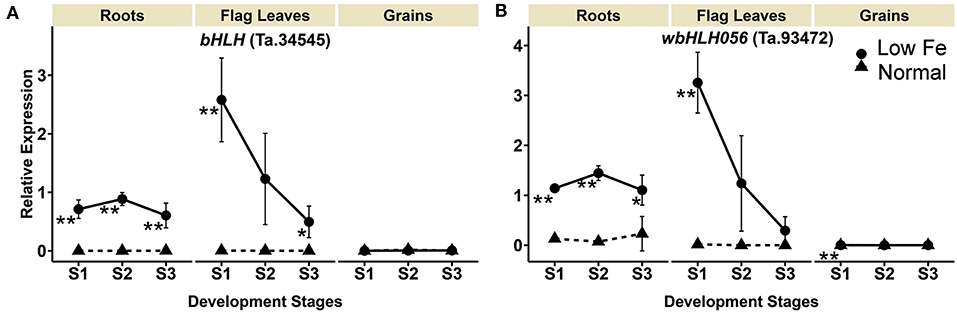
Figure 5. Iron deficiency induced differences in expression levels of wheat homologs of genes encoding transcription factors. The data collected at three different development stages, i.e., 8–10 DPA (stage 1), 25–28 DPA (stage 2), and 33–35 DPA (stage 3) are presented. Values are the mean ± standard deviation of the three biological replicates (*p ≤ 0.05; **p ≤ 0.01). Note the differences in scale at Y-axis for relative expression levels of tested genes. (A) bHLH (Ta.34545), (B) wbHLH056 (Ta.93472).
Discussion
It has been suggested that PS production increases during iron deficiency in strategy II plants, including barley, rice, maize, sorghum, red fescue, and wheat (Kanazawa et al., 1994; Ma et al., 2003; Ishimaru et al., 2006; Tsednee et al., 2012; Garnica et al., 2018). Several reports have shown that iron deficiency in rice, barley and maize increases the expression of genes involved in NA and DMA synthesis (Negishi et al., 2002; Inoue et al., 2003, 2008; Kobayashi et al., 2005; Bashir et al., 2006; Zheng et al., 2009; Zhou et al., 2013). Similarly, an increased expression of TaNAS genes, TaNAAT genes, and TaDMAS1 in wheat roots in response to iron-limited conditions during the vegetative growth stage have been reported (Bonneau et al., 2016; Beasley et al., 2017). In this study, we find that the expression of SAMS, NAS, NAAT, and DMAS homologs is increased in iron-deficient conditions, particularly in roots, suggesting increased production of NA and DMA for iron acquisition and translocation (Figure 6). NA chelates iron for long-distance and intracellular transport, and also is an intermediate in DMA production (Takahashi et al., 2003; Schuler et al., 2012). Recent evidence suggests that in addition to its well-known role in iron uptake from the rhizosphere, DMA also chelates iron in the xylem and phloem for long-distance transport in rice (Kakei et al., 2009; Nishiyama et al., 2012). The increased expression of NAS and DMAS genes in the flag leaves and grains suggest similar roles for DMA and NA in long-distance iron translocation in wheat. In rice, contrary to the other two NAS gens, OsNAS3 is not induced under low iron stress in shoots and it is likely to be rather involved in mitigation of excessive iron stress (Inoue et al., 2003; Suzuki et al., 2008; Aung et al., 2018, 2019). Of the wheat NAS genes investigated in the present study, no NAS gene showed a similar expression pattern as OsNAS3, which is in line with findings by Bonneau et al. (2016).
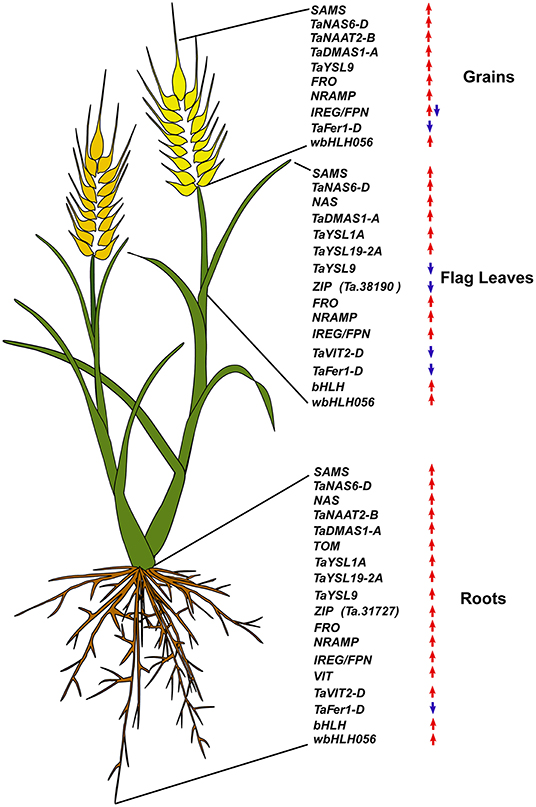
Figure 6. Summary of the expression modulation of wheat iron homeostasis related genes. Upregulated genes are indicated by an upward red arrow and the downregulated ones are indicated by a downward blue arrow.
Parallel to the increased PS production, the expression of genes encoding PS transporters is also positively modulated by iron deficiency to facilitate efficient iron translocation. TOM1 and TOM2 transporters facilitate DMA secretion into the rhizosphere and vascular loading, respectively (Nozoye et al., 2011, 2015). The wheat TOM homolog Ta.5180 has high DNA sequence similarity with the barley and rice TOM1 genes and was significantly increased in wheat roots during iron deficiency, suggesting a potential role of the transporter in DMA efflux in wheat roots. The YSL transporters in non-grass plants such as Arabidopsis have been studied in detail for their function in iron transport and homeostasis. For example, YSL2 is proposed to facilitate lateral movement of metals in the vasculature (DiDonato et al., 2004), and YSL4 and YSL6 control iron release from chloroplasts (Divol et al., 2013). Detailed functions of the wheat YSL homologs in the iron deficiency response and maintenance of homeostasis remain unclear, but their increased expression in roots at all development stages suggest a primary role in iron uptake as well (Kumar et al., 2018; this study).
In rice, the transcription factor OsIRO2 regulates the expression of genes involved in phytosiderophore synthesis (OsNAS1, OsNAS2, OsNAAT1, OsDMAS1), and genes encoding the PS transporter TOM1 as well as other iron transporters such as OsYSL15 (Ogo et al., 2006, 2007, 2011). Under iron deficiency, OsIRO2 expression is positively regulated by the transcription factor IDE BINDING FACTOR (OsIDEF1) (Kobayashi and Nishizawa, 2012), which together with the NAC family transcription factor IDEF2 also regulates some of the iron deficiency-responsive genes in rice (Ogo et al., 2008). The Arabidopsis genes encoding the Ib subgroup bHLH transcription factors, including bHLH38, bHLH39, bHLH100, and bHLH101, have a closer phylogenetic relationship with the rice OsIRO2 gene (Hindt and Guerinot, 2012). These bHLH transcription factors function in Arabidopsis iron homeostasis and are up-regulated under iron starvation (Wang et al., 2007). bHLH38 and bHLH39 can form heterodimers with the FER-Like iron deficiency-induced transcription factor (FIT) and they regulate the expression of FRO2 and IRT1 in Arabidopsis (Yuan et al., 2008). In contrast, the bHLH100 and bHLH101 function in a FIT-independent manner (Sivitz et al., 2012). The wheat homologs of IRO2 were significantly up-regulated in roots and the flag leaves during iron deficiency, suggesting that they have a significant function in iron homeostasis and regulation of iron deficiency-responsive genes in wheat. Similar to reports from other plants (Ogo et al., 2006, 2007; Wang et al., 2007; Sivitz et al., 2012), the increased expression of PS-related genes and genes encoding iron transporters parallels the up-regulation of IRO2 during iron deficiency in wheat.
The expression of iron homeostasis-related genes often depends on the external and/or internal iron status of plants. This is also relevant for the gene encoding important iron storage protein FERRITIN. Similar to the Arabidopsis AtFERRITIN genes (Buckhout et al., 2009; Rodriguez-Celma et al., 2013), the TaFer1-D gene in bread wheat was down-regulated under the iron-deficient conditions. Excess iron induces the FERRITIN transcript levels (Briat et al., 2010), supporting the storage function of FERRITIN in wheat. Further, we observed that the expression level of the TaFer1-D gene in the grains was also growth stage dependent, since a gradual increase in the expression of TaFer1-D was observed during the three development stages (Supplementary Figure S3, Supplementary Table S3).
Localizing on the vacuolar membrane, AtNRAMP3 and AtNRAMP4 mediate iron efflux from the vacuoles in Arabidopsis (Lanquar et al., 2005). The VIT and IRON REGULATED PROTEIN2/FERROPORTIN2 (IREG2/NPN2) perform the opposite function (Lanquar et al., 2005; Morrissey et al., 2009; Zhang et al., 2012). The expression of their homologs in wheat roots was significantly increased upon iron deficiency, which is in agreement with the expression modulation of AtNRAMP3, AtNRAMP4 and IREG2/FPN2 in Arabidopsis but conflict with the OsVIT2 expression in rice (Lanquar et al., 2005; Morrissey et al., 2009; Zhang et al., 2012). This induction of VITs in roots can be due to the accumulation of metals other than iron, such as zinc and manganese, in the roots of plants under iron deficiency. It is known that VITs can transport not only iron but also zinc and manganese (Zhang et al., 2012; Connorton et al., 2017). In the roots of iron-deficient plants, we detected higher concentration of zinc and manganese, which can be toxic when present in excess (Marschner, 1995). The plants under low iron treatment potentially up-regulated the VIT genes in order to maintain the cytoplasmic zinc and manganese concentration low. In contrast to the roots, the VIT expression was either unaltered or reduced in the flag leaves and grains.
Localized on the root epidermal cells, FRO2 has been reported to function in iron reduction in strategy I plants (Robinson et al., 1999; Waters et al., 2002). Furthermore, leaf mesophyll cells (Bruggemann et al., 1993) and chloroplasts (Jeong et al., 2008) also rely on FROs for iron uptake. Recently, in rice, it has been suggested that OsFRO1 localizes on tonoplast and play a role in making more iron available in the cytoplasm (Li et al., 2019). The induction of FRO expression in bread wheat roots, flag leaves and grains under iron limited conditions suggested their role being similar to their homolog in rice. However, whether FROs in wheat are involved in iron uptake from the soil into root cells needs to be verified by further experimentation.
In summary, iron homeostasis in bread wheat is maintained via a comprehensive modulation of various genes and gene families. Under iron-limited conditions, overall iron uptake and transport appears to be regulated by iron deficiency inducible transcription factors, such as those from the bHLH gene family (Figure 6). In order to cope with reduced iron supply during iron deficiency stress, the genes related to the synthesis of iron chelators (NA and DMA) as well as those encoding various iron translocation related transporters are up-regulated throughout the grain filling period. This suggests that iron uptake by the root and/or iron translocation from the roots to different plants parts lasted until the maturity in iron-deficient plants. Consistently, the root iron concentration was not significantly different between normal and iron-deficient plants. On the other hand, in the flag leaf and the grains, such genes involved in iron homeostasis were more highly expressed in iron-deficient plants than in normal plants at Stage 1, but from Stage 2 onwards, the gene expression difference between the iron treatments became insignificant or the expression level became even higher in normal plants than in iron-deficient plants. Given that YSLs play a role in iron remobilization in rice (Aoyama et al., 2009; Ishimaru et al., 2010), these results indicate that low external iron availability prompted both the onset and the end of iron remobilization from the flag leaf to the grains. The upregulation of genes involved in NA/PS synthesis in grains under iron deficiency implies a promotional role of NA/DMA in grain iron storage, which is in line with the study in rice (Lee et al., 2009). In rice, OsYSL9 is involved in iron export from the endosperm to embryo (Senoura et al., 2017). The up-regulation of TaYSL9 in grains in iron-deficient plants may indicate that this gene plays a similar role as OsYSL9.
Overall, zinc, copper and manganese accumulated in tissues in an opposite way than iron. In other graminaceous species, it is known that YSLs can transport other metals than iron (Schaaf et al., 2004; Zheng et al., 2012; Zhang et al., 2018). Therefore, it is likely that at least some of the YS/YSL genes investigated in this study can transport both iron and other metals and that increased concentration of these metals is a consequence of the up-regulation of some YS/YSL genes.
A comprehensive study comprising of transcriptome profiling of the iron deficiency wheat plants could provide further understanding of the coordinated roles of different transporters in combating iron deficiency stress. The insight into the wheat molecular iron homeostasis provided in this study may help researchers and breeders design well-tuned and more diversified biofortification studies (Figure 6). Experience from rice biofortification efforts suggest that the same biofortification strategies work differently in different genetic backgrounds (Masuda et al., 2012; Aung et al., 2013; Trijatmiko et al., 2016), and therefore it is necessary to have a wide range of biofortification strategies available. To date, however, there are only a limited number of wheat iron biofortification strategies reported (Borg et al., 2012; Connorton et al., 2017; Singh et al., 2017). In the present study, we found some endogenous transporters and transcription factors putatively involved in wheat iron homeostasis, whose use has not yet been explored in biofortification efforts. In particular, combining boost in NA/PS synthesis and iron remobilization from leaves to grains by tissue-specific expression of YSL genes could be an effective approach. Future biofortification studies may capitalize on this information to design and test novel biofortification strategies.
Data Availability
All datasets generated for this study are included in the manuscript and/or the Supplementary Files.
Author Contributions
NB conceived the experiments. MW and YK carried out the experiments. MW and NB interpreted the data and wrote the manuscript. All authors revised the final manuscript.
Funding
This research was supported by research grant from the State Secretariat for Education, Research and Innovation (SERI) within the COST FA0905 program to NB.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank Prof. Gruissem for providing access to Lab infrastructure and scientific discussions. We thank Irene Zurkirchen and Katja Bärenfaller for the technical support in the greenhouse and in data analysis, respectively. We thank Dr. Laurie Paule Schönholzer for the support in ICP-OES measurement.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2019.00067/full#supplementary-material
References
Aoyama, T., Kobayashi, T., Takahashi, M., Nagasaka, S., Usuda, K., Kakei, Y., et al. (2009). OsYSL18 is a rice iron(III)-deoxymugineic acid transporter specifically expressed in reproductive organs and phloem of lamina joints. Plant Mol. Biol. 70, 681–692. doi: 10.1007/s11103-009-9500-3
Arvola, L. (1981). Spectrophotometric determination of chlorophyll-alpha and phaeopigments in ethanol extractions. Annal. Bot. Fenn. 18, 221–227.
Aung, M. S., Masuda, H., Kobayashi, T., Nakanishi, H., Yamakawa, T., and Nishizawa, N. K. (2013). Iron biofortification of Myanmar rice. Front. Plant Sci. 4:158. doi: 10.3389/fpls.2013.00158
Aung, M. S., Masuda, H., Kobayashi, T., and Nishizawa, N. K. (2018). Physiological and transcriptomic analysis of responses to different levels of iron excess stress in various rice tissues. J. Soil Sci. Plant Nutr 64, 370–385. doi: 10.1080/00380768.2018.1443754
Aung, M. S., Masuda, H., Nozoye, T., Kobayashi, T., Jeon, J.-S., An, G., et al. (2019). Nicotianamine synthesis by OsNAS3 is important for mitigating iron excess stress in rice. Front. Plant Sci. 10:660. doi: 10.3389/fpls.2019.00660
Barker, A. V., and Stratton, M. L. (2015). “Iron,” in Handbook of Plant Nutrition, 2nd Edn, eds A. V. Barker and D. J. Pilbeam (Boca Raton, FL: CRC Press), 399–426.
Bashir, K., Inoue, H., Nagasaka, S., Takahashi, M., Nakanishi, H., Mori, S., et al. (2006). Cloning and characterization of deoxymugineic acid synthase genes from graminaceous plants. J. Biol. Chem. 281, 32395–32402. doi: 10.1074/jbc.M604133200
Bashir, K., Ishimaru, Y., Shimo, H., Nagasaka, S., Fujimoto, M., Takanashi, H., et al. (2011). The rice mitochondrial iron transporter is essential for plant growth. Nat. Commun. 2:322. doi: 10.1038/ncomms1326
Beasley, J. T., Bonneau, J. P., and Johnson, A. A. T. (2017). Characterisation of the nicotianamine aminotransferase and deoxymugineic acid synthase genes essential to strategy II iron uptake in bread wheat (Triticum aestivum L.). PLoS ONE 12:e0177061. doi: 10.1371/journal.pone.0177061
Bonneau, J., Baumann, U., Beasley, J., Li, Y., and Johnson, A. A. T. (2016). Identification and molecular characterization of the nicotianamine synthase gene family in bread wheat. Plant Biotechnol. J. 14, 2228–2239. doi: 10.1111/pbi.12577
Boonyaves, K., Gruissem, W., and Bhullar, N. K. (2016). NOD promoter-controlled AtIRT1 expression functions synergistically with NAS and FERRITIN genes to increase iron in rice grains. Plant Mol. Biol. 90, 207–215. doi: 10.1007/s11103-015-0404-0
Boonyaves, K., Wu, T. Y., Gruissem, W., and Bhullar, N. K. (2017). Enhanced grain iron levels in rice expressing an iron-regulated metal transporter, nicotianamine synthase, and ferritin Gene Cassette. Front. Plant Sci. 8:130. doi: 10.3389/fpls.2017.00130
Borg, S., Brinch-Pedersen, H., Tauris, B., Madsen, L. H., Darbani, B., Noeparvar, S., et al. (2012). Wheat ferritins: improving the iron content of the wheat grain. J. Cereal Sci. 56, 204–213. doi: 10.1016/j.jcs.2012.03.005
Briat, J. F., Duc, C., Ravet, K., and Gaymard, F. (2010). Ferritins and iron storage in plants. Biochim. Biophys. Acta 1800, 806–814. doi: 10.1016/j.bbagen.2009.12.003
Bruggemann, W., Maaskantel, K., and Moog, P. R. (1993). Iron uptake by leaf mesophyll-cells—the role of the plasma membrane-bound ferric-chelate reductase. Planta 190, 151–155. doi: 10.1007/BF00196606
Buckhout, T. J., and Schmidt, W. (2013). “Iron in Plants,” in eLS, ed. A. M. Hetherington (Chichester: John Wiley & Sons Ltd), 1–9
Buckhout, T. J., Yang, T. J., and Schmidt, W. (2009). Early iron-deficiency-induced transcriptional changes in arabidopsis roots as revealed by microarray analyses. BMC Genom. 10:147. doi: 10.1186/1471-2164-10-147
Connorton, J. M., Jones, E. R., Rodriguez-Ramiro, I., Fairweather-Tait, S., Uauy, C., and Balk, J. (2017). Wheat vacuolar iron transporter TaVIT2 transports Fe and Mn and is effective for biofortification. Plant Physiol. 174, 2434–2444. doi: 10.1104/pp.17.00672
Curie, C., Panaviene, Z., Loulergue, C., Dellaporta, S. L., Briat, J. F., and Walker, E. L. (2001). Maize yellow stripe1 encodes a membrane protein directly involved in Fe(III) uptake. Nature 409, 346–349. doi: 10.1038/35053080
DiDonato, R. J. Jr., Roberts, L. A., Sanderson, T., Eisley, R. B., and Walker, E. L. (2004). Arabidopsis yellow stripe-like2 (YSL2): a metal-regulated gene encoding a plasma membrane transporter of nicotianamine-metal complexes. Plant J. 39, 403–414. doi: 10.1111/j.1365-313X.2004.02128.x
Divol, F., Couch, D., Conejero, G., Roschzttardtz, H., Mari, S., and Curie, C. (2013). The Arabidopsis YELLOW STRIPE LIKE4 and 6 transporters control iron release from the chloroplast. Plant Cell 25, 1040–1055. doi: 10.1105/tpc.112.107672
Durmaz, E., Coruh, C., Dinler, G., Grusak, M. A., Peleg, Z., Saranga, Y., et al. (2011). Expression and cellular localization of ZIP1 transporter under zinc deficiency in wild emmer wheat. Plant Mol. Biol. Rep. 29, 582–596. doi: 10.1007/s11105-010-0264-3
Duy, D., Stube, R., Wanner, G., and Philippar, K. (2011). The chloroplast permease PIC1 regulates plant growth and development by directing homeostasis and transport of iron. Plant Physiol. 155, 1709–1722. doi: 10.1104/pp.110.170233
Duy, D., Wanner, G., Meda, A. R., von Wiren, N., Soll, J., and Philippar, K. (2007). PIC1, an ancient permease in Arabidopsis chloroplasts, mediates iron transport. Plant Cell 19, 986–1006. doi: 10.1105/tpc.106.047407
Garnica, M., Bacaicoa, E., Mora, V., San Francisco, S., Baigorri, R., Zamarreno, A. M., et al. (2018). Shoot iron status and auxin are involved in iron deficiency-induced phytosiderophores release in wheat. BMC Plant Biol. 18:105. doi: 10.1186/s12870-018-1324-3
Gong, X., Guo, C. H., Terachi, T., Cai, H. S., and Yu, D. S. (2015). Tobacco PIC1 mediates iron transport and regulates chloroplast development. Plant Mol. Biol. Rep. 33, 401–413. doi: 10.1007/s11105-014-0758-5
Guo, X. J., and Wang, J. R. (2017). Global identification, structural analysis and expression characterization of bHLH transcription factors in wheat. BMC Plant Biol.17:90. doi: 10.1186/s12870-017-1038-y
Gupta, P. K., Kulwal, P. L., and Rustgi, S. (2005). Wheat cytogenetics in the genomics era and its relevance to breeding. Cytogenet. Genome Res. 109, 315–327. doi: 10.1159/000082415
Hansch, R., and Mendel, R. R. (2009). Physiological functions of mineral micronutrients (Cu, Zn, Mn, Fe, Ni, Mo, B, Cl). Curr. Opin. Plant Biol. 12, 259–266. doi: 10.1016/j.pbi.2009.05.006
Harrison, P. M., and Arosio, P. (1996). Ferritins: Molecular properties, iron storage function and cellular regulation. Biochim. Biophys. Acta Bioenerg. 1275, 161–203. doi: 10.1016/0005-2728(96)00022-9
Hindt, M. N., and Guerinot, M. L. (2012). Getting a sense for signals: Regulation of the plant iron deficiency response. Biochim. Biophys. Acta Mol. Cell Res. 1823, 1521–1530. doi: 10.1016/j.bbamcr.2012.03.010
Huang, D. Q., and Dai, W. H. (2015). Molecular characterization of the basic helix-loop-helix (bHLH) genes that are differentially expressed and induced by iron deficiency in Populus. Plant Cell Rep. 34, 1211–1224. doi: 10.1007/s00299-015-1779-8
Inoue, H., Higuchi, K., Takahashi, M., Nakanishi, H., Mori, S., and Nishizawa, N. K. (2003). Three rice nicotianamine synthase genes, OsNAS1, OsNAS2, and OsNAS3 are expressed in cells involved in long-distance transport of iron and differentially regulated by iron. Plant J. 36, 366–381. doi: 10.1046/j.1365-313X.2003.01878.x
Inoue, H., Kobayashi, T., Nozoye, T., Takahashi, M., Kakei, Y., Suzuki, K., et al. (2009). Rice OsYSL15 is an iron-regulated iron(III)-deoxymugineic acid transporter expressed in the roots and is essential for iron uptake in early growth of the seedlings. J. Biol. Chem. 284, 3470–3479. doi: 10.1074/jbc.M806042200
Inoue, H., Takahashi, M., Kobayashi, T., Suzuki, M., Nakanishi, H., Mori, S., et al. (2008). Identification and localisation of the rice nicotianamine aminotransferase gene OsNAAT1 expression suggests the site of phytosiderophore synthesis in rice. Plant Mol. Biol. 66, 193–203. doi: 10.1007/s11103-007-9262-8
Ishimaru, Y., Masuda, H., Bashir, K., Inoue, H., Tsukamoto, T., Takahashi, M., et al. (2010). Rice metal-nicotianamine transporter, OsYSL2, is required for the long-distance transport of iron and manganese. Plant J. 62, 379–390. doi: 10.1111/j.1365-313X.2010.04158.x
Ishimaru, Y., Suzuki, M., Kobayashi, T., Takahashi, M., Nakanishi, H., Mori, S., et al. (2005). OsZIP4, a novel zinc-regulated zinc transporter in rice. J. Exp. Bot. 56, 3207–3214. doi: 10.1093/jxb/eri317
Ishimaru, Y., Suzuki, M., Tsukamoto, T., Suzuki, K., Nakazono, M., Kobayashi, T., et al. (2006). Rice plants take up iron as an Fe3+-phytosiderophore and as Fe2+. Plant J. 45, 335–346. doi: 10.1111/j.1365-313X.2005.02624.x
Jeong, J., Cohu, C., Kerkeb, L., Pilon, M., Connolly, E. L., and Guerinot, M. L. (2008). Chloroplast Fe(III) chelate reductase activity is essential for seedling viability under iron limiting conditions. Proc. Natl. Acad. Sci. U.S.A. 105, 10619–10624. doi: 10.1073/pnas.0708367105
Kakei, Y., Ishimaru, Y., Kobayashi, T., Yamakawa, T., Nakanishi, H., and Nishizawa, N. K. (2012). OsYSL16 plays a role in the allocation of iron. Plant Mol. Biol. 79, 583–594. doi: 10.1007/s11103-012-9930-1
Kakei, Y., Yamaguchi, I., Kobayashi, T., Takahashi, M., Nakanishi, H., Yamakawa, T., et al. (2009). A highly sensitive, quick and simple quantification method for nicotianamine and 2-deoxymugineic acid from minimum samples using LC/ESI-TOF-MS achieves functional analysis of these components in plants. Plant Cell Physiol. 50, 1988–1993. doi: 10.1093/pcp/pcp141
Kanazawa, K., Higuchi, K., Nishizawa, N. K., Fushiya, S., Chino, M., and Mori, S. (1994). Nicotianamine aminotransferase activities are correlated to the phytosiderophore secretions under Fe-deficient conditions in gramineae. J. Exp. Bot. 45, 1903–1906. doi: 10.1093/jxb/45.12.1903
Kobayashi, T., Nakanishi, H., and Nishizawa, N. K. (2010). Recent insights into iron homeostasis and their application in graminaceous crops. Proc. Jpn. Acad. Ser. B Phys. Biol. Sci. 86, 900–913. doi: 10.2183/pjab.86.900
Kobayashi, T., and Nishizawa, N. K. (2012). Iron uptake, translocation, and regulation in higher plants. Annu. Rev. Plant Biol. 63, 131–152. doi: 10.1146/annurev-arplant-042811-105522
Kobayashi, T., Suzuki, M., Inoue, H., Itai, R. N., Takahashi, M., Nakanishi, H., et al. (2005). Expression of iron-acquisition-related genes in iron-deficient rice is co-ordinately induced by partially conserved iron-deficiency-responsive elements. J. Exp. Bot. 56, 1305–1316. doi: 10.1093/jxb/eri131
Koike, S., Inoue, H., Mizuno, D., Takahashi, M., Nakanishi, H., Mori, S., et al. (2004). OsYSL2 is a rice metal-nicotianamine transporter that is regulated by iron and expressed in the phloem. Plant J. 39, 415–424. doi: 10.1111/j.1365-313X.2004.02146.x
Kumar, A., Kaur, G., Goel, P., Bhati, K. K., Kaur, M., Shukla, V., et al. (2018). Genome-wide analysis of oligopeptide transporters and detailed characterization of yellow stripe transporter genes in hexaploid wheat. Funct. Integr. Genomics. 19, 75-90 doi: 10.1007/s10142-018-0629-5
Lanquar, V., Lelievre, F., Bolte, S., Hames, C., Alcon, C., Neumann, D., et al. (2005). Mobilization of vacuolar iron by AtNRAMP3 and AtNRAMP4 is essential for seed germination on low iron. EMBO J 24, 4041–4051. doi: 10.1038/sj.emboj.7600864
Lee, S., Chiecko, J. C., Kim, S. A., Walker, E. L., Lee, Y., Guerinot, M. L., et al. (2009). Disruption of OsYSL15 leads to iron inefficiency in rice plants. Plant Physiol. 150, 786–800. doi: 10.1104/pp.109.135418
Lee, S., Jeong, H. J., Kim, S. A., Lee, J., Guerinot, M. L., and An, G. (2010a). OsZIP5 is a plasma membrane zinc transporter in rice. Plant Mol. Biol. 73, 507–517. doi: 10.1007/s11103-010-9637-0
Lee, S., Kim, S. A., Lee, J., Guerinot, M. L., and An, G. (2010b). Zinc deficiency-inducible OsZIP8 encodes a plasma membrane-localized zinc transporter in rice. Mol. Cells 29, 551–558. doi: 10.1007/s10059-010-0069-0
Li, L., Ye, L., Kong, Q., and Shou, H. (2019). A vacuolar membrane ferric-chelate reductase, OsFRO1, alleviates Fe toxicity in rice (Oryza sativa L.). Front. Plant Sci. 10:700. doi: 10.3389/fpls.2019.00700
Lichtenthaler, H. K., and Buschmann, C. (2001). Chlorophylls and carotenoids: measurement and characterization by UV-VIS spectroscopy. Curr. Protoc. Food Anal. Chem. 1, F4.3.1–F4.3.8. doi: 10.1002/0471142913.faf0403s01
Ma, J. F., Ueno, H., Ueno, D., Rombol,à, A. D., and Iwashita, T. (2003). Characterization of phytosiderophore secretion under Fe deficiency stress in Festuca rubra. Plant Soil 256, 131–137. doi: 10.1023/a:1026285813248
Masuda, H., Ishimaru, Y., Aung, M. S., Kobayashi, T., Kakei, Y., Takahashi, M., et al. (2012). Iron biofortification in rice by the introduction of multiple genes involved in iron nutrition. Sci. Rep. 2:543. doi: 10.1038/srep00543
Mathpal, P., Kumar, U., Kumar, A., Kumar, S., Malik, S., Kumar, N., et al. (2018). Identification, expression analysis, and molecular modeling of Iron-deficiency-specific clone 3 (Ids3)-like gene in hexaploid wheat. Biotech 8:219. doi: 10.1007/s13205-018-1230-2
Morrissey, J., Baxter, I. R., Lee, J., Li, L. T., Lahner, B., Grotz, N., et al. (2009). The ferroportin metal efflux proteins function in iron and cobalt homeostasis in Arabidopsis. Plant Cell 21, 3326–3338. doi: 10.1105/tpc.109.069401
Morrissey, J., and Guerinot, M. L. (2009). Iron uptake and transport in plants: the good, the bad, and the ionome. Chem. Rev. 109, 4553–4567. doi: 10.1021/cr900112r
Murata, Y., Ma, J. F., Yamaji, N., Ueno, D., Nomoto, K., and Iwashita, T. (2006). A specific transporter for iron(III)-phytosiderophore in barley roots. Plant J. 46, 563–572. doi: 10.1111/j.1365-313X.2006.02714.x
Negishi, T., Nakanishi, H., Yazaki, J., Kishimoto, N., Fujii, F., Shimbo, K., et al. (2002). cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J. 30, 83–94. doi: 10.1046/J.1365-313x.2002.01270.X
Nishiyama, R., Kato, M., Nagata, S., Yanagisawa, S., and Yoneyama, T. (2012). Identification of Zn-nicotianamine and Fe-2'-deoxymugineic acid in the phloem sap from rice plants (Oryza sativa L.). Plant Cell Physiol. 53, 381–390. doi: 10.1093/pcp/pcr188
Nozoye, T., Nagasaka, S., Kobayashi, T., Sato, Y., Uozumi, N., Nakanishi, H., et al. (2015). The phytosiderophore efflux transporter TOM2 is involved in metal transport in rice. J. Biol. Chem. 290, 27688–27699. doi: 10.1074/jbc.M114.635193
Nozoye, T., Nagasaka, S., Kobayashi, T., Takahashi, M., Sato, Y., Sato, Y., et al. (2011). Phytosiderophore efflux transporters are crucial for iron acquisition in graminaceous plants. J. Biol. Chem. 286, 5446–5454. doi: 10.1074/jbc.M110.180026
Ogo, Y., Itai, R. N., Inoue, H., Kobayashi, T., Suzuki, M., Takahashi, M., et al. (2006). Isolation and characterization of IRO2, a novel iron-regulated bHLH transcription factor in graminaceous plants. J. Exp. Bot. 57, 2867–2878. doi: 10.1093/jxb/erl054
Ogo, Y., Itai, R. N., Kobayashi, T., Aung, M. S., Nakanishi, H., and Nishizawa, N. K. (2011). OsIRO2 is responsible for iron utilization in rice and improves growth and yield in calcareous soil. Plant Mol. Biol. 75, 593–605. doi: 10.1007/s11103-011-9752-6
Ogo, Y., Itai, R. N., Nakanishi, H., Kobayashi, T., Takahashi, M., Mori, S., et al. (2007). The rice bHLH protein OsIRO2 is an essential regulator of the genes involved in Fe uptake under Fe-deficient conditions. Plant J. 51, 366–377. doi: 10.1111/j.1365-313X.2007.03149.x
Ogo, Y., Kobayashi, T., Itai, R. N., Nakanishi, H., Kakei, Y., Takahashi, M., et al. (2008). A novel NAC transcription factor, IDEF2, that recognizes the iron deficiency-responsive element 2 regulates the genes involved in iron homeostasis in plants. J. Biol. Chem. 283, 13407–13417. doi: 10.1074/jbc.M708732200
Paolacci, A. R., Tanzarella, O. A., Porceddu, E., and Ciaffi, M. (2009). Identification and validation of reference genes for quantitative RT-PCR normalization in wheat. BMC Mol. Biol. 10:11. doi: 10.1186/1471-2199-10-11
Ramesh, S. A., Shin, R., Eide, D. J., and Schachtman, D. P. (2003). Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 133, 126–134. doi: 10.1104/pp.103.026815
Robinson, N. J., Procter, C. M., Connolly, E. L., and Guerinot, M. L. (1999). A ferric-chelate reductase for iron uptake from soils. Nature 397, 694–697. doi: 10.1038/17800
Rodriguez-Celma, J., Pan, I. C., Li, W., Lan, P., Buckhout, T. J., and Schmidt, W. (2013). The transcriptional response of Arabidopsis leaves to Fe deficiency. Front. Plant. Sci. 4, 276. doi: 10.3389/fpls.2013.00276
Santi, S., and Schmidt, W. (2009). Dissecting iron deficiency-induced proton extrusion in Arabidopsis roots. New Phytol. 183, 1072–1084. doi: 10.1111/j.1469-8137.2009.02908.x
Sasaki, A., Yamaji, N., Mitani-Ueno, N., Kashino, M., and Ma, J. F. (2015). A node-localized transporter OsZIP3 is responsible for the preferential distribution of Zn to developing tissues in rice. Plant J. 84, 374–384. doi: 10.1111/tpj.13005
Schaaf, G., Ludewig, U., Erenoglu, B. E., Mori, S., Kitahara, T., and von Wiren, N. (2004). ZmYS1 functions as a proton-coupled symporter for phytosiderophore- and nicotianamine-chelated metals. J. Biol. Chem. 279, 9091–9096. doi: 10.1074/jbc.M311799200
Schefe, J. H., Lehmann, K. E., Buschmann, I. R., Unger, T., and Funke-Kaiser, H. (2006). Quantitative real-time RT-PCR data analysis: current concepts and the novel “gene expression's CT difference” formula. J. Mol. Med. 84, 901–910. doi: 10.1007/s00109-006-0097-6
Schuler, M., Rellan-Alvarez, R., Fink-Straube, C., Abadia, J., and Bauer, P. (2012). Nicotianamine functions in the phloem-based transport of iron to sink organs, in pollen development and pollen tube growth in Arabidopsis. Plant Cell 24, 2380–2400. doi: 10.1105/tpc.112.099077
Senoura, T., Sakashita, E., Kobayashi, T., Takahashi, M., Aung, M. S., Masuda, H., et al. (2017). The iron-chelate transporter OsYSL9 plays a role in iron distribution in developing rice grains. Plant Mol. Biol. 95, 375–387. doi: 10.1007/s11103-017-0656-y
Singh, G., Kumar, S., and Singh, P. (2003). A quick method to isolate RNA from wheat and other carbohydrate-rich seeds. Plant Mol. Biol. Rep. 21, 93–93. doi: 10.1007/bf02773401
Singh, S. P., Keller, B., Gruissem, W., and Bhullar, N. K. (2017). Rice NICOTIANAMINE SYNTHASE 2 expression improves dietary iron and zinc levels in wheat. Theor. Appl. Genet. 130, 283–292. doi: 10.1007/s00122-016-2808-x
Sivitz, A. B., Hermand, V., Curie, C., and Vert, G. (2012). Arabidopsis bHLH100 and bHLH101 control iron homeostasis via a FIT-independent pathway. PLoS ONE 7:e44843. doi: 10.1371/journal.pone.0044843
Sperotto, R. A., Boff, T., Duarte, G. L., and Fett, J. P. (2008). Increased senescence-associated gene expression and lipid peroxidation induced by iron deficiency in rice roots. Plant Cell Rep. 27, 183–195. doi: 10.1007/s00299-007-0432-6
Suzuki, M., Tsukamoto, T., Inoue, H., Watanabe, S., Matsuhashi, S., Takahashi, M., et al. (2008). Deoxymugineic acid increases Zn translocation in Zn-deficient rice plants. Plant Mol. Biol. 66, 609–617. doi: 10.1007/s11103-008-9292-x
Takahashi, M., Terada, Y., Nakai, I., Nakanishi, H., Yoshimura, E., Mori, S., et al. (2003). Role of nicotianamine in the intracellular delivery of metals and plant reproductive development. Plant Cell 15, 1263–1280. doi: 10.1105/tpc.010256
Thimm, O., Essigmann, B., Kloska, S., Altmann, T., and Buckhout, T. J. (2001). Response of arabidopsis to iron deficiency stress as revealed by microarray analysis. Plant Physiol. 127, 1030–1043. doi: 10.1104/Pp.010191
Trijatmiko, K. R., Duenas, C., Tsakirpaloglou, N., Torrizo, L., Arines, F. M., Adeva, C., et al. (2016). Biofortified indica rice attains iron and zinc nutrition dietary targets in the field. Sci. Rep. 6, 19792. doi: 10.1038/srep19792
Tsednee, M., Mak, Y. W., Chen, Y. R., and Yeh, K. C. (2012). A sensitive LC-ESI-Q-TOF-MS method reveals novel phytosiderophores and phytosiderophore-iron complexes in barley. New Phytol. 195, 951–961. doi: 10.1111/j.1469-8137.2012.04206.x
Uauy, C., Distelfeld, A., Fahima, T., Blechl, A., and Dubcovsky, J. (2006). A NAC gene regulating senescence improves grain protein, zinc, and iron content in wheat. Science 314, 1298–1301. doi: 10.1126/science.1133649
Vert, G., Grotz, N., Dedaldechamp, F., Gaymard, F., Guerinot, M. L., Briat, J. F., et al. (2002). IRT1, an Arabidopsis transporter essential for iron uptake from the soil and for plant growth. Plant Cell 14, 1223–1233. doi: 10.1105/tpc.00138
von Wiren, N., Klair, S., Bansal, S., Briat, J. F., Khodr, H., Shioiri, T., et al. (1999). Nicotianamine chelates both FeIII and FeII. Implications for metal transport in plants. Plant Physiol. 119, 1107–1114. doi: 10.1104/pp.119.3.1107
Wang, H. Y., Klatte, M., Jakoby, M., Baumlein, H., Weisshaar, B., and Bauer, P. (2007). Iron deficiency-mediated stress regulation of four subgroup Ib BHLH genes in Arabidopsis thaliana. Planta 226, 897–908. doi: 10.1007/s00425-007-0535-x
Wang, L., Ying, Y., Narsai, R., Ye, L., Zheng, L., Tian, J., et al. (2013). Identification of OsbHLH133 as a regulator of iron distribution between roots and shoots in Oryza sativa. Plant Cell Environ. 36, 224–236. doi: 10.1111/j.1365-3040.2012.02569.x
Waters, B. M., Blevins, D. G., and Eide, D. J. (2002). Characterization of FRO1, a pea ferric-chelate reductase involved in root iron acquisition. Plant Physiol. 129, 85–94. doi: 10.1104/pp.010829
Waters, B. M., Uauy, C., Dubcovsky, J., and Grusak, M. A. (2009). Wheat (Triticum aestivum) NAM proteins regulate the translocation of iron, zinc, and nitrogen compounds from vegetative tissues to grain. J. Exp. Bot. 60, 4263–4274. doi: 10.1093/jxb/erp257
Wirth, J., Poletti, S., Aeschlimann, B., Yakandawala, N., Drosse, B., Osorio, S., et al. (2009). Rice endosperm iron biofortification by targeted and synergistic action of nicotianamine synthase and ferritin. Plant Biotechnol. J. 7, 631–644. doi: 10.1111/j.1467-7652.2009.00430.x
Wu, T. Y., Gruissem, W., and Bhullar, N. K. (2018). Facilitated citrate-dependent iron translocation increases rice endosperm iron and zinc concentrations. Plant Sci. 270, 13–22. doi: 10.1016/j.plantsci.2018.02.002
Yang, T. J. W., Lin, W. D., and Schmidt, W. (2010). Transcriptional profiling of the Arabidopsis iron deficiency response reveals conserved transition metal homeostasis networks. Plant Physiol. 152, 2130–2141. doi: 10.1104/pp.109.152728
Yang, X., Huang, J., Jiang, Y., and Zhang, H.-S. (2009). Cloning and functional identification of two members of the ZIP (Zrt, Irt-like protein) gene family in rice (Oryza sativa L.). Mol. Biol. Rep.36, 281–287. doi: 10.1007/s11033-007-9177-0
Yuan, Y. X., Wu, H. L., Wang, N., Li, J., Zhao, W. N., Du, J., et al. (2008). FIT interacts with AtbHLH38 and AtbHLH39 in regulating iron uptake gene expression for iron homeostasis in Arabidopsis. Cell Res.18, 385–397. doi: 10.1038/cr.2008.26
Zhang, C., Lu, W. H., Yang, Y., Shen, Z. G., Ma, J. F., and Zheng, L. Q. (2018). OsYSL16 is required for preferential Cu distribution to floral organs in rice. Plant Cell Physiol. 59, 2039–2051. doi: 10.1093/pcp/pcy124
Zhang, Y., Xu, Y. H., Yi, H. Y., and Gong, J. M. (2012). Vacuolar membrane transporters OsVIT1 and OsVIT2 modulate iron translocation between flag leaves and seeds in rice. Plant J. 72, 400–410. doi: 10.1111/j.1365-313X.2012.05088.x
Zhao, M., Song, A. P., Li, P. L., Chen, S. M., Jiang, J. F., and Chen, F. D. (2014). A bHLH transcription factor regulates iron intake under Fe deficiency in chrysanthemum. Sci.Rep. 4:6649. doi: 10.1038/srep06694
Zheng, L. Q., Huang, F. L., Narsai, R., Wu, J. J., Giraud, E., He, F., et al. (2009). Physiological and transcriptome analysis of iron and phosphorus interaction in rice seedlings. Plant Physiol. 151, 262–274. doi: 10.1104/pp.109.141051
Zheng, L. Q., Yamaji, N., Yokosho, K., and Ma, J. F. (2012). YSL16 is a phloem-localized transporter of the copper-nicotianamine complex that is responsible for copper distribution in rice. Plant Cell 24, 3767–3782. doi: 10.1105/tpc.112.103820
Zhou, X. J., Li, S. Z., Zhao, Q. Q., Liu, X. Q., Zhang, S. J., Sun, C., et al. (2013). Genome-wide identification, classification and expression profiling of nicotianamine synthase (NAS) gene family in maize. BMC Genomics 14:238. doi: 10.1186/1471-2164-14-238
Keywords: iron deficiency, gene expression, wheat, phytosiderophore synthesis, transcription factors, iron transporters
Citation: Wang M, Kawakami Y and Bhullar NK (2019) Molecular Analysis of Iron Deficiency Response in Hexaploid Wheat. Front. Sustain. Food Syst. 3:67. doi: 10.3389/fsufs.2019.00067
Received: 02 April 2019; Accepted: 07 August 2019;
Published: 04 September 2019.
Edited by:
Avtar Krishan Handa, Purdue University, United StatesReviewed by:
Jorge Rodriguez-Celma, John Innes Centre (JIC), United KingdomHiroshi Masuda, Akita Prefectural University, Japan
Copyright © 2019 Wang, Kawakami and Bhullar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Navreet K. Bhullar, Ymh1bGxhcm5AZXRoei5jaA==; orcid.org/0000-0003-1657-0422
†Present address: Meng Wang, School of Life Sciences, University of Science and Technology of China, Hefei, China
 Meng Wang
Meng Wang Yuta Kawakami
Yuta Kawakami Navreet K. Bhullar
Navreet K. Bhullar