- 1Medical Laboratory, Xiamen Humanity Hospital, Fujian Medical University, Xiamen, China
- 2Ultrasonography Department, Women and Children’s Hospital, School of Medicine, Xiamen University, Xiamen, China
CRC poses a significant challenge in the global health domain, with a high number of deaths attributed to this disease annually. If CRC is detected only in its advanced stages, the difficulty of treatment increases significantly. Therefore, biomarkers for the early detection of CRC play a crucial role in improving patient outcomes and increasing survival rates. The development of a reliable biomarker for early detection of CRC is particularly important for timely diagnosis and treatment. However, current methods for CRC detection, such as endoscopic examination, blood, and stool tests, have certain limitations and often only detect cases in the late stages. To overcome these constraints, researchers have turned their attention to molecular biomarkers, which are considered a promising approach to improving CRC detection. Non-invasive methods using biomarkers such as mRNA, circulating cell-free DNA, microRNA, LncRNA, and proteins can provide more reliable diagnostic information. These biomarkers can be found in blood, tissue, stool, and volatile organic compounds. Identifying molecular biomarkers with high sensitivity and specificity for the early and safe, economic, and easily measurable detection of CRC remains a significant challenge for researchers.
Introduction
CRC is a common type of cancer, ranking as the third most prevalent malignant tumor globally and the second leading cause of cancer-related deaths (1, 2). The survival rate of CRC patients is significantly influenced by the stage at which the tumor is detected, with a general 5-year overall survival rate of about 65% (3). Most cases of CRC are diagnosed at an advanced stage due to the lack of unique, early-stage disease-specific symptoms and limitations in early diagnosis methods. The most critical portion of cancer-related deaths is caused by metastatic cancer, with the liver being the most common site of metastasis in CRC. More than half of the patients in stage III, and a quarter in stages II and III, appear to have local recurrence or metastasis (4). CRC is a unique cancer that can be prevented and cured through early identification and removal of high-risk adenomas (5). Thus, implementing early detection screening programs is crucial for reducing the incidence and mortality of the disease. Early detection increases the likelihood of successful treatment and improves patient health outcomes (6). Colonoscopy is a widely accepted and effective screening method for early detection of CRC, though it carries certain risks. Patients may experience bleeding during sampling or polyp removal, and other complications may occur (7). In recent years, advanced molecular techniques have played a significant role in the early diagnosis and treatment of various cancers, including CRC, helping to reveal some of the genetic mechanisms that lead to CRC (8). Therefore, clarifying the molecular mechanisms of colon cancer occurrence is crucial. In recent years, non-coding RNAs (ncRNAs) have been proven to participate in the onset and progression of colon cancer (9, 10). It is well-known that ncRNAs, mostly not translated into proteins, also play significant roles in various cellular and physiological processes (11). LncRNAs are longer than 200 nucleotides, participate in various biological processes including cell proliferation, differentiation, development, apoptosis, and metastasis, often acting as competitive endogenous RNAs (ceRNAs) to regulate the expression of specific miRNAs, then targeting molecules downstream of these miRNAs (12). In fact, lncRNAs can interact with RNA, DNA, and proteins to form RNA-RNA, RNA-DNA, and RNA-protein complexes, regulating gene expression through various mechanisms, including transcriptional regulation, mRNA stability, and translation (13, 14). Numerous studies suggest that lncRNAs may play key roles in the biological processes of cancer, such as apoptosis, cell proliferation, cell invasion, and metastasis (15–17). In this article, we will summarize the functions and mechanisms of lncRNAs in the occurrence and malignant progression of human CRC.
The history of lncRNAs
LncRNAs is a class of RNA molecules longer than 200 nucleotides, first discovered in the 1970s. Initially, scientists primarily focused on mRNA, which encodes proteins, while non-coding RNAs were considered “noise” or “by-products.” However, with technological advancements and deeper research, it gradually became apparent that non-coding RNAs play crucial roles in gene regulation, epigenetics, and disease occurrence. The research journey of LncRNA can be traced back to a series of groundbreaking studies in the late 20th and early 21st centuries. In 2002, researchers first discovered a LncRNA associated with gene silencing on the X chromosome (18). Subsequently, Guttman et al. discovered the LncRNA-HOTAIR, which plays a significant role in gene locus regulation (11). In 2009, Rinn et al. identified an lncRNA (HOTTIP) located in the HOX gene cluster and found its crucial involvement in gene locus regulation (19). It has also been reported that LncRNA plays crucial roles in embryonic development (20). Studies have also indicated the role of lncRNA in tumor initiation and progression, sparking a research frenzy into the role of lncRNA in cancer (12, 21, 22).
Classification of lncRNAs
Based on the genomic database [Ensembl Release 96 (April 2019)], human lncRNAs are classified into several categories, including 3’ overlapping ncRNA, antisense LncRNA, long interspersed ncRNA, retained intron, sense intronic, sense overlapping, and macro lncRNAs.Intronic lncRNA is located within the intron regions of protein-coding genes, transcribed from the introns of these genes, but does not itself participate in encoding proteins (23). Antisense lncRNA overlaps with the antisense strand of coding genes and may affect gene expression by forming double-stranded RNA structures with coding regions through base complementary pairing (24, 25). Intergenic lncRNA is located in the region between two coding genes and may play a role in the regulation of genes in its region (11), sense lncRNAs are overlapped with the sense strand of protein coding genes containing exons (26). Messenger lncRNA can act as a regulatory factor, involved in regulating the expression of specific genes (20). Structural lncRNA may play an important role in the physical structure within cells or the chromosomal architecture within the nucleus (27).
lncRNAs localization and related research techniques
lncRNAs can be located in the cytoplasm (28), nucleus (29), nucleolus (30)s, and other subcellular regions and vesicles (such as nucleoli and exosomes), and their localization is related to their molecular functions (28, 31). Certain sequence motifs in their primary sequences are related to subcellular localization (32). Exploring the localization of lncRNAs is important for understanding their roles in gene regulation, disease development, and cell functions. Techniques for studying the localization of lncRNAs include in situ hybridization (33), RNA immunoprecipitation (34), RNA-seq (35), single-cell RNA sequencing (36), FISH-Flow (37), etc.
The conservation of lncRNAs
Although lncRNAs are crucial in function, most lncRNA sequences exhibit low conservation across different species, meaning the same lncRNA may be difficult to identify in different species through sequence similarity. Low conservation is considered to reflect the diversity and specificity of lncRNA functions, as well as their rapid changes during evolution (38). Despite low sequence conservation, some lncRNAs exhibit a degree of structural and functional conservation across different species. These lncRNAs may maintain similar three-dimensional structures or play roles in the same pathways of gene expression regulation across species (27, 39). Moreover, many lncRNAs exhibit strong species specificity, existing in certain species while absent in others. This species specificity suggests that lncRNAs may play specialized roles in the development and adaptation processes of specific species (38, 40). The conservation level of lncRNA promoters is comparable to that of protein-coding genes (41, 42).
LncRNAs as diagnostic biomarkers for CRC in blood
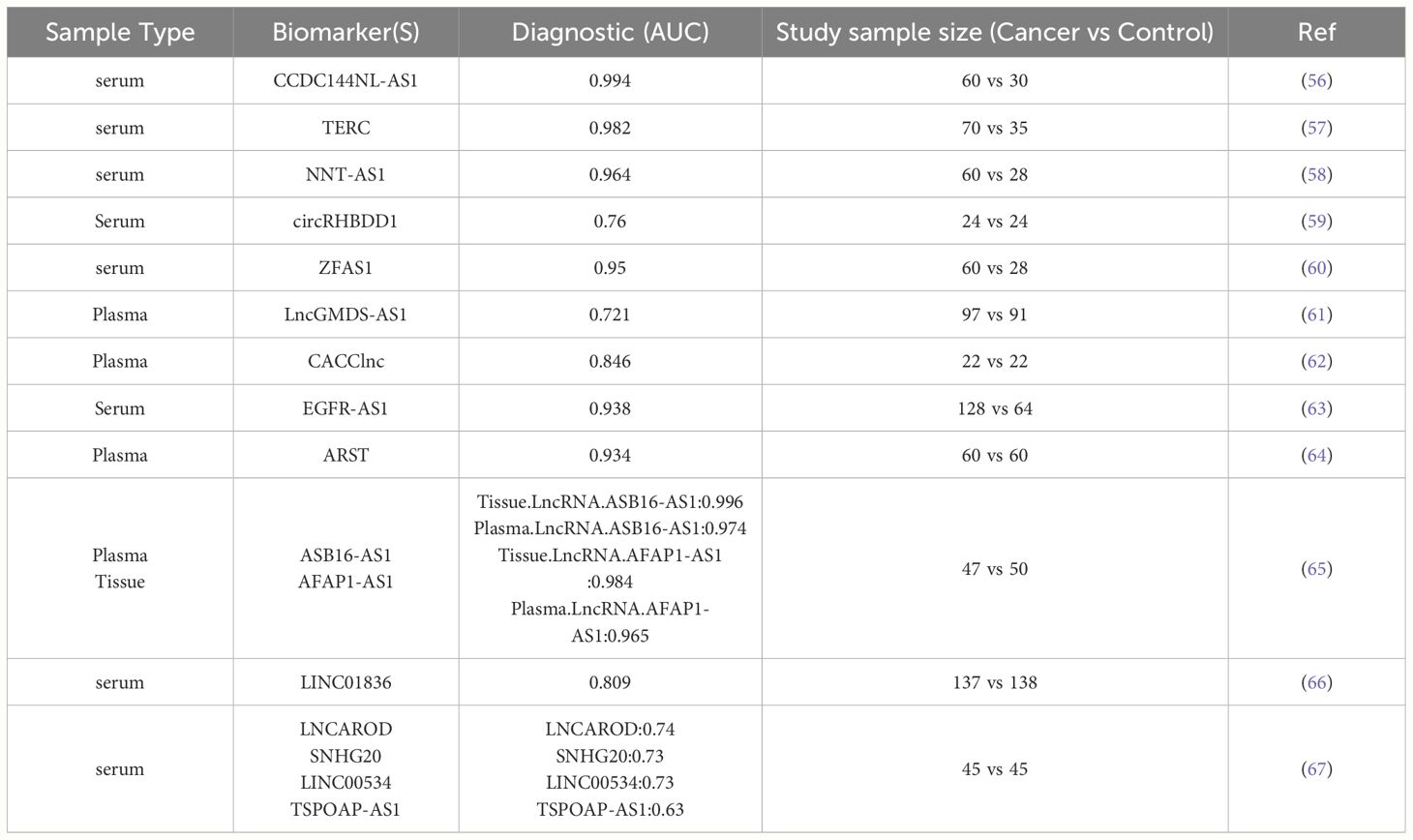
Ease of acquisition and detectability are preferred criteria for diagnostic biomarkers. For potential early-stage CRC patients, undergoing a colonoscopy to obtain tissue samples might be strongly resisted by patients. Therefore, biomarkers that can be detected in blood or other body fluids are ideal for broader clinical use. Over the past decade, numerous studies have demonstrated that lncRNAs are stable in the bloodstream and hold diagnostic potential. This makes lncRNAs promising candidates for non-invasive diagnostic tests in CRC (43–46). LncRNAs are present in different body fluids such as blood and urine because they can cross cellular membranes. This property allows for their detection in non-invasive diagnostic tests (47). LncRNAs in body fluids directly represent the expression levels of certain genes and can differentiate between cancer patients and healthy individuals (48). Additionally, a key characteristic of circulating lncRNAs is their ability to resist degradation by RNase enzymes (49, 50). Apoptotic bodies, microvesicles, and exosomes are vesicles encapsulated by a phospholipid bilayer that contain DNA, RNA, lipids, proteins, polysaccharides, and metabolites. These vesicles are released into the human circulatory system for dissemination, facilitating the transfer of materials between cells (51–53). Quantitative reverse transcription PCR (qRT-PCR) is frequently employed to detect circulating lncRNAs because of its notable sensitivity and specificity (54). CCAT1 and HOTAIR are the first lncRNA markers reported to show significantly higher levels in the plasma of CRC patients compared to healthy individuals (55). Numerous other circulating lncRNAs have also been identified as potential biomarkers for detecting CRC (Table 1).
The role and regulatory axes of LncRNAs in CRC
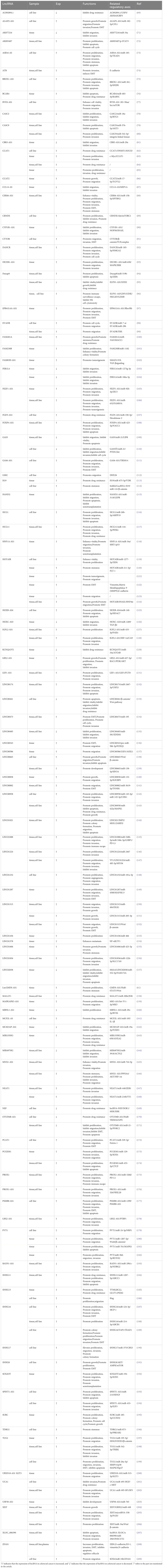
As research progresses, it has been discovered that lncRNAs play a crucial role in the onset and development of CRC (68). LncRNAs play multifaceted roles in CRC, influencing various biological processes including cell cycle control, cell proliferation, epithelial-mesenchymal transition (EMT), migration, invasion, drug resistance, apoptosis, and cellular stemness. This section will elaborate on the specific roles of lncRNAs within CRC, highlighting how they contribute to both the development and progression of the disease. We will also focus on the signaling pathways associated with lncRNAs, enhancing our understanding of their mechanistic impact on the pathophysiology of CRC (Table 2).
Regulation of cell proliferation by LncRNAs
Increasing research evidence suggests that lncRNAs play a crucial role in regulating cellular proliferation. Many studies have shown that abnormal expression of lncRNAs is closely associated with the regulation of proliferation in CRC cells. LncRNAs can promote and inhibit cancer cell proliferation. Generally speaking, lncRNAs that promote cancer cell proliferation are highly expressed in cancer, while lncRNAs that inhibit cancer cell proliferation are low expressed in cancer.
Studies on lncRNAs that promote CRC proliferation are as follows. Guo et al. found that AGAP2-AS1 can be transcriptionally activated by E2F transcription factor 4(E2F4). Downregulating AGAP2-AS1 impairs the proliferation of CRC cells. Mechanistically, AGAP2-AS1 enhances CFL1 expression by competitively binding with miR-182-5p in CRC, providing a possible target for therapeutic intervention (70). When AK093407 is silenced, the proliferation of HCT-15 and HCT-116 cells is reduced (72). ASB16-AS1 promotes the proliferation of CRC cells through the ASB16-AS1-miR-185-5p/TEAD1 axis (73). BBOX1-AS1 promotes the progression of CRC by acting as a sponge for hsa-miR-361-3p and upregulating SH2B1 (75). Silencing CASC9 can not only downregulate AKT3 expression by reducing the competitive binding of CASC9 with miR-576-5p, thereby inhibiting CRC cell proliferation (210), it can also inhibit the proliferation of CRC cells through the miR-542-3p/integrin-linked kinase pathway (80), CCAT2 is highly overexpressed in microsatellite stable CRC and can enhance tumor growth by activating the WNT signaling pathway through the CCAT2/miR-17-5p/TCF7L2 axis (86). Studies have found that by overexpressing or knocking down CCAT2, reducing CCAT2 can increase miR-145, thereby inhibiting the proliferation of cancer cells (211). A study indicated that CCDC144NL-AS1 is upregulated in CRC tissues and cells, and the elevated expression of CCDC144NL-AS1 promotes cell proliferation through the miR-363-3p/GALNT7 axis (212). Another study established cell models with either the gain or loss of function of DANCR. It was discovered that DANCR promotes CRC cell proliferation through the miR-185-5p/HMGA2 axis (92). Research utilizing public datasets and experimental evaluation has explored the function and expression of DICER1-AS1 in CRC. It was found that the upregulation of DICER1-AS1 promotes CRC proliferation in vitro by acting as a sponge for miR-650, thereby activating the MAPK/ERK signaling pathway (92). Further research has revealed the DLGAP1-AS2/CTCF/Myc axis as an oncogenic regulatory pathway in CRC, promoting CRC cell proliferation (213). Multiple studies have shown that ELFN1-AS1 is highly expressed in CRC and promotes CRC proliferation through various regulatory axes (95, 214, 215). Overexpression of FAM230B promotes tumor growth by increasing the levels of immature miR-1182 in CRC cells, while simultaneously inhibiting the expression levels of mature miR-1182 (101). Another study used lentiviral transfection to knock down FEZF1-AS1 in CRC cells and found that FEZF1-AS1 likely promotes CRC proliferation through the miR-92b-3p/ZIC5 axis by activating the PI3K/AKT signaling pathway (105). Further research has discovered that knocking out FEZF1-AS1 may inhibit cell proliferation through the miR-632/FAM83A axis (106). FOXP4-AS1 acts as a molecular sponge for miR-423-5p, with NACC1 being a direct target of miR-423-5p.Overexpression of FOXP4-AS1 promotes the proliferation of HCT116 cells (108). GAS6-AS1 acts as an oncogene by competitively binding with miR-370-3p and miR-1296-5p. This interaction leads to the upregulation of TRIM14, which positively regulates CRC proliferation in vitro (111). GATA2-AS1 is highly expressed in CRC cells, and knocking down GATA2-AS1 impedes the proliferation of CRC cells (216). Guo et al. found that HCG11 enhances cell proliferation by targeting the miR-26b-5p/ARPP19 axis (116). Another found that silencing HCG11 inhibits the proliferation of CRC cells through the miR-144-3p/PDK4 pathway (117). HOTAIR regulates the levels of HNF4α by recruiting SNAIL. Knocking down HOTAIR inhibits the proliferation of CRC cell lines in vitro (124). Deng and colleagues discovered that knocking out HOXB-AS4 inhibits proliferation, and functional restoration experiments further confirmed the critical role of the HOXB-AS4/miR-140-5p/HDAC7 axis in regulating the malignant phenotype of CRC cells (125). Liu and colleagues found that silencing the lncRNA IGFL2-AS1 impedes the malignant proliferation of HCT116 cells and facilitates miR-433-3p-mediated inhibition of PAK4 transcription. Conversely, overexpression of lncRNA IGFL2-AS1 has the opposite effect, promoting cell proliferation by inhibiting miR-433-3p activity and thereby enhancing PAK4 transcription (127). Another study on IGFL2-AS1 also observed that it is highly upregulated in CRC tumor tissues and cells, functionally promoting CRC cell proliferation in vitro. Mechanistic investigations revealed that IGFL2-AS1 elevates CA9 levels by affecting the degradation pathway of HIF-1α (128). Overexpression of LBX2-AS1 promotes the proliferation of CRC cells through the miR-627-5p/RAC1/PI3K/AKT pathway (130). In vitro, LEF1-AS1 mediates the proliferation of CRC cells through the LEF1-AS1/LEF1/FUT8 axis (131). Liang et al. demonstrated that both loss and gain of function of LINC00174 promote CRC cell proliferation in vitro. Mechanistic experiments revealed that LINC00174 binds to miR-2467-3p, enhancing the expression and function of USP21, which in turn affects the proliferation of CRC cells (132). RNA immunoprecipitation (RIP) and RNA pull-down experiments have confirmed that LINC00883 binds with miR-577, and miR-577 interacts with FKBP14. Disrupting the expression of LINC00883 inhibits the proliferation of CRC cells via the miR-577/FKBP14 axis (217). Deficiency of LINC00958 inhibits the proliferation of cancer cells. A study found that LINC00958 functions by interacting with miR-145-3p and modulating the miR-145-3p/CDK1 axis, thereby influencing the proliferation dynamics of the cancer cells (142), Another study found that the pro-proliferative function of LINC00958 is facilitated through the miR-422a/MAPK1 pathway. This interaction contributes to the regulatory mechanism by which LINC00958 enhances cell proliferation (143). Wu et al. confirmed that LINC021 is significantly upregulated in CRC cell lines and clinical tissues, and showed that the oncogenic LINC021 specifically binds with the m(6)A “reader” IMP2 protein, enhancing the mRNA stability of MSX1 and JARID2 through m(6)A regulatory mechanisms during CRC tumorigenesis and pathogenesis (144). Li’s findings suggest that LINC01088 directly targets miR-548b-5p and miR-548c-5p, enhancing the expression of G3BP1 and PD-L1, and thereby promoting the proliferation of CRC cells (145). Silencing LINC01224 inhibits the proliferation of CRC cells. This effect can be reversed by co-transfecting with an miR-2467 inhibitor, indicating that miR-2467 negatively regulates LINC01224. RNA-binding protein immunoprecipitation (RIP) assays further confirm that the inhibition of LINC01224 by miR-2467 is dependent on the RNA-induced silencing complex (RISC) (146) In CRC tissue samples and cell lines, the abundance of LINC01224 is elevated. Knocking down LINC01224 inhibits CRC cell proliferation. Mechanistic studies have shown that YY1-induced LINC01224 regulates CRC proliferation through the miR-485-5p/MYO6 axis (147). It has been observed that LINC1436 can reduce the level of mature miR-466 without affecting its precursors. Proliferation studies indicated that the overexpression of LINC01436 counteracted the reduction in cell proliferation caused by miR-466. Thus, LINC01436, which is found at elevated levels in CRC, enhances cancer cell growth by inhibiting the maturation of miR-466 (153). Ye et al. found that knocking down GMDS-AS1 impairs CRC cell proliferation both in vitro and in vivo. Through RNA sequencing (RNA-seq) and mass spectrometry (MS) studies, it was discovered that in CRC cells, GMDS-AS1 physically interacts with the RNA-binding protein HuR. This interaction protects HuR from polyubiquitination and proteasomal degradation, highlighting a crucial mechanism by which GMDS-AS1 contributes to the stabilization of proteins involved in cancer cell proliferation (61). The lncRNA MNX1-AS1/PPFIA4 activates the downstream AKT/HIF-1α signaling pathway to promote COAD proliferation (166). NEAT1 can regulate the miR-448/ZEB1 axis (167), and also directly acts as a molecular sponge for miR-216b, which in turn activates YY1 to promote CRC proliferation (168). PCGEM1 is elevated in CRC tissues and cells, and it regulates CRC proliferation through the miR-129-5p/SOX4 axis and the miR-433-3p/CTCF axis (173, 174). PVT1 promotes CRC proliferation through a variety of regulatory pathways, these pathways include the PVT1/miR-24-3p/NRP1 axis, PVT1/miR-1207-5p/Wnt6/β-catenin axis,PVT1/miR-761/MAPK1 axis and PVT1/miR-30d-5p/RUNX2 axis (179–182). Huang et al. found that SNHG11 is overexpressed in bevacizumab-resistant CRC tissues and cells. Knocking down SNHG11 inhibits cell proliferation, possibly through the miR-1207-5p/ABCC1 axis (184). SNHG15 promotes CRC cell proliferation, while knocking down SNHG15 inhibits it. RT-qPCR and Western blot analyses confirm that SNHG15 enhances the expression of TYMS, BCL2, GLUT1, and PKM2 in CRC cells (185). Additionally, another study suggests that SNHG15 interacts with the ubiquitin-proteasome system to block the degradation of Slug, thereby inhibiting its breakdown (186). SNHG16 regulates CRC proliferation through multiple pathways, including SNHG16/miR-124-3p/MCP-1 axis, SNHG16/miR-214-3p/ABCB1 axis and SNHG16/YAP1/TEAD1 axis (187–189). Functionally, SNHG17 can promote the proliferation of COAD cells in vitro, and its effect is mediated through the miR-375/CBX3 axis (190). Islam et al. used dicer-substrate siRNA transfection to modulate the expression of SNHG8 in the HCT-116 and SW480 cell lines. The knockdown of SNHG8 induced autophagy and apoptosis pathways through the AKT/AMPK/mTOR axis, significantly reducing the proliferation of CRC cells (191). SOX2OT is an oncogene in human CRC tissues and cell lines. Silencing of SOX2OT in vitro inhibits cell proliferation in CRC cells. Mechanistically, SOX2OT acts as a ceRNA that upregulates SOX5 by sponging miR-194-5p (192). SPINT1-AS1 is upregulated in CRC, and the silencing of SPINT1-AS1 inhibits the proliferation of CRC cells. SPINT1-AS1 mediates the expression of HDGF by targeting miR-214 (193). Another study indicated that knocking down SPINT1-AS1 weakened the proliferation of KRAS-silenced CRC cells. Rescue experiments confirmed the regulatory effect of the SPINT1-AS1/miR-433-3p/E2F3 axis on the proliferation of CRC cells (194). Li et al. found that mutated APC genes can promote the transcription of SURC in CRC by reducing the degradation of β-catenin. Functional assays showed that knockdown of SURC inhibits the proliferation of CRC cells, potentially through the SURC/miR-185-5p/CCND2 axis (195). TUG1 is involved in the regulation of proliferation in CRC through multiple pathways. Xia et al. reported that TUG1, stabilized by IGF2BP2, promotes CRC cell proliferation through the miR-195-5p/HDGF/DDX5/β-catenin axis (197). Liu et al. found that knockdown of TUG1 inhibits tumor growth in vivo, where TUG1 interacts with miR-542-3p, affecting the expression of TRIB2 and inhibiting CRC proliferation through the Wnt/β-catenin pathway (198). Tian et al. reported that TUG1 promotes CRC proliferation both in vivo and in vitro by regulating the miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis (199). Liu et al. transfected pcDNA-UCA1 plasmids to overexpress UCA1 and found that dysregulation of the UCA1/miR-495/SP1/SP3 axis in CRC leads to malignant proliferation of CRC cells (202). XIST is involved in the regulation of CRC proliferation through various pathways. Yan et al. reported that XIST is highly expressed in CRC tissues and cells, and its downregulation in vitro inhibits CRC cell proliferation, which is regulated through the XIST/miR-448/GRHL2 axis (204), Li et al. found that knockdown of XIST inhibits cell proliferation through the miR-338-3p/PAX5 axis (205). Sun et al.’s study indicates that XIST regulates cell proliferation through the miR-34a/Wnt/β-catenin signaling pathway (206). In cells where ZFAS1 is knocked down, an increase in the expression of epithelial markers E-cadherin and ZO-1 is observed, along with a decrease in the expression of mesenchymal markers vimentin and N-cadherin (208).
The research on lncRNAs that inhibit the proliferation of CRC is as follows. CASC2 inhibits the expression of miR-18a-5p by acting as a molecular sponge for miR-18a-5p, positively regulates the expression of BTG3, and thereby achieves the suppression of CRC cell proliferation (78). Experimental studies both in vitro and in vivo have confirmed that upregulating CYP1B1-AS1 markedly inhibits CRC cell proliferation. Additionally, CYP1B1-AS1 directly binds to and negatively regulates NOP58. The effects of CYP1B1-AS1 can be reversed by the overexpression of NOP58 (90). Xiong et al. transfected HCT-116 cells with si-GAS5 to inhibit GAS5 expression and found that this suppression led to an upregulation of miR-21. This increase in miR-21 subsequently affected the PTEN/Akt signaling pathway, promoting cell proliferation (110). Yi et al.found that overexpression of HAND2-AS1 in CRC cell lines using an expression vector, revealed that HAND2-AS1 suppresses CRC cell proliferation by acting as a molecular sponge for miR-3118, which affects the LEPR axis (115). Li et al. found that knocking down LINC00485 in FHC cells promoted cell proliferation. Mechanistic studies revealed that LINC00485 inhibits CRC tumor proliferation by targeting the miR-581/EDEM1 axis (135). In CRC tissues, overexpressing LINC02038 significantly inhibits CRC cell proliferation. It acts as a sponge for miR-552-5p, protecting its target gene, FAM172A, from degradation (157). MBNL1-AS1 primarily inhibits the proliferation of CRC cells by regulating the miR-29c-3p/BVES signaling pathway (160). OTUD6B-AS1 is expressed at lower levels in CRC tissues and cells. Mechanistic studies have shown that OTUD6B-AS1 inhibits CRC proliferation by sponging miR-21-5p and upregulating PNRC2 (171). MBNL1-AS1 inhibits CRC cell proliferation by regulating the miR-29c-3p/BVES signaling pathway (218). Zhang et al. observed that RPL34-AS1 may act as a molecular sponge for miR-3656, inhibiting CRC cell proliferation (219).
LncRNAs regulate cell apoptosis
One of the important reasons for the infinite proliferation of cancer cells is that their apoptosis function is inhibited, thus allowing cells to escape programmed death and continue to proliferate, and lncRNA has the function of regulating apoptosis of cancer cells (220, 221). The following is a summary of research on lncRNA that promotes cell apoptosis.
In the study by Xue et al., it was found that silencing AK093407 increased the apoptosis rate of HCT-15 and HCT-116 cells (72). Yu et al. reported that silencing ASB16-AS1 accelerates apoptosis in CRC cells by regulating the miR-185-5p/TEAD1 axis (73). Another study indicated that knocking down BBOX1-AS1 reduces apoptosis, which may be achieved by upregulating SH2B1 through the sponge effect on hsa-miR-361-3p (75). The study by Li et al. reported that knocking down BCAR4 promotes apoptosis in CRC cells, which can be reversed by overexpressing RAB5C.The related regulatory axis is the BCAR4/miR-483-3p/RAB5C axis (76). Liu’s report indicates that silencing CASC9 can promote apoptosis by downregulating AKT3 expression through reducing the competitive binding of CASC9 with miR-576-5p (222). Liang et al. found that Duxap8 inhibits apoptosis in vitro. Mechanistically, Duxap8 is primarily located in the cytoplasm and acts as a competitive endogenous RNA, affecting apoptosis by upregulating ZNF277 through the sponge effect on miR-519b-3p (94). FOXP4-AS1 was found to act as a molecular sponge for miR-423-5p in HCT116 cells and animal models, with NACC1 being a direct target of miR-423-5p.Overexpression of FOXP4-AS1 inhibits apoptosis in CRC cells (108). Guo et al.found that knocking down HCG11 promoted apoptosis. Through bioinformatics analysis and mechanistic studies, HCG11, primarily located in the cytoplasm, has been shown to competitively bind with miR-26b-5p to regulate the expression of cAMP-regulated phosphoprotein 19 (ARPP19) (116). Deng et al. discovered that knocking out HOXB-AS4 can promote apoptosis.HOXB-AS4 acts as a molecular sponge by binding and adsorbing miR-140-5p to regulate the expression of histone deacetylase 7 (HDAC7) (125). Liang et al. demonstrated through loss and gain of function studies of LINC00174 that it enhances resistance to apoptosis in CRC cells in vitro via the miR-2467-3p/USP21 axis (132). Another study reported that LINC00665 stimulates apoptosis in CRC by activating the Wnt/β-catenin signaling pathway (138). LINC00958 promotes the expression of MAPK1 by targeting miR-422a and inhibits apoptosis (143). Wu et al. found that LINC021 directly recognizes the IMP2 protein, which enhances the mRNA stability of transcripts such as MSX1 and JARID2 by recognizing their m(6)A-modified element RGGAC, thereby reducing apoptosis (144). Xu et al. found that knocking down PCGEM1 inhibits apoptosis in CRC cells (174). Liu et al. believe that knocking down the expression of PVT1 promotes apoptosis in CRC cells through the miR-761/MAPK1 axis (181). Huang et al. found that knocking down SNHG11 promotes apoptosis in bevacizumab-resistant CRC cells, potentially through the miR-1207-5p/ABCC1 axis (184). The silencing of SPINT1-AS1 mediates the expression of HDGF by targeting miR-214, increasing apoptosis in CRC cells (193). Ma et al.’s study reveals that silencing XLOC_006390 inhibits apoptosis in CRC through the regulation of the miR-296/ONECUT2 axis (207). Fang et al. believe that ZFAS1 prevents apoptosis. Knocking down ZFAS1 can reduce the expression of ZEB1 and increase epithelial markers E-cadherin and ZO-1, while decreasing mesenchymal markers vimentin and N-cadherin (208).
The following is a summary of relevant studies on lncRNAs that inhibit cell apoptosis.
A study indicates that GAS5 is significantly reduced in CRC tumor tissues and cells, and knocking down GAS5 can promote apoptosis in cancer cells (109). Wang et al.’s study suggests that LINC00261 may promote apoptosis by downregulating nuclear β-catenin through inhibiting the translocation of β-catenin from the cytoplasm to the nucleus or by promoting the degradation of β-catenin and inhibiting the activation of the Wnt pathway (133). Research has found that the m(6)A/LINC02038/miR-552-5p/FAM172A axis may represent a novel anti-tumor pathway. Overexpression of LINC02038 can accelerate apoptosis in CRC cells through this axis (157). Cai et al. suggest that the overexpression of OTUD6B-AS1 promotes apoptosis through the OTUD6B-AS1/miR-21-5p/PNRC2 axis (171).
LncRNAs regulate invasion and metastasis
Invasion refers to the ability of cancer cells to penetrate from the original tumor site into surrounding normal tissues, while metastasis is the process by which cancer cells spread through the blood or lymphatic system to form new tumors at other sites in the body. The invasion and metastasis of CRC cells are related to a variety of factors, including cellular pathways and molecular mechanisms, epithelial-mesenchymal transition (EMT), and the influence of microorganisms, all of which together promote the aggressive and metastatic characteristics of CRC (223, 224). LncRNAs can also promote and inhibit the migration and invasion of cancer cells. The following is a brief summary of the research on LncRNAs promoting cancer migration and invasion.
The expression of AGAP2-AS1 is significantly elevated in CRC cells. Downregulation of AGAP2-AS1 can attenuate the migration and invasion of CRC cells. Mechanistically, miR-182-5p is a downstream target molecule of AGAP2-AS1, and CFL1 is a target of miR-182-5p (70). Yu et al. found that ASB16-AS1 is highly expressed in CRC cells. Silencing ASB16-AS1 suppressed the proliferation, migration, and invasion of CRC cells. Mechanistic experiments revealed that ASB16-AS1 drives the progression of CRC by regulating the miR-185-5p/TEAD1 axis (73). Yue et al. found that the levels of lncRNA ATB are higher in three highly invasive CRC cell lines compared to three low-invasive cell lines (74). Knockdown of BBOX1-AS1 enhances cancer cell migration and invasion, with its regulatory axis being hsa-miR-361-3p/SH2B1 (75). BVES-AS1 can encode a micropeptide that promotes the migration and invasion of CRC cells in vitro (77). Zhang et al. discovered that the CASC9/miR-542-3p/integrin-linked kinase regulatory axis is involved in the invasion and migration of CRC cells (80). The upregulation of CCAT1 expression promotes cell proliferation and invasion (83). Depletion of CERS6-AS1 inhibits cell migration and invasion through the miR-15b-5p/SPTBN2 axis (88). Knockdown of CYTOR reduces the migration and invasion of colon cancer cells (91). The DANCR/miR-185-5p/HMGA2 axis is involved in the migration and invasion processes of CRC (92). The upregulation of DICER1-AS1 promotes CRC migration and invasion by activating the MAPK/ERK signaling pathway through a sponge-like effect on miR-650 (93). Bin et al. found that EPB41L4A-AS1 promotes the invasion and migration of CRC (97). FEZF1-AS1 can activate the PI3K/AKT signaling pathway to promote the proliferation and invasion of RKO cells (105), it can also promote CRC migration and invasion through the miR-632/FAM83A axis (106). ATF3-activated FOXP4-AS1 enhances CRC migration and invasion by regulating the miR-423-5p/NACC1 axis (108). GAS6-AS1 promotes TRIM14-mediated CRC cell migration and invasion through a ceRNA network and FUS-dependent mechanism (111). Guo et al. reported that upregulated HCG11 in CRC cells can promote cell migration and invasion by targeting the miR-26b-5p/ARPP19 axis (116), Cui reported another regulatory axis, HCG11/miR-144-3p/PDK4 (117). Upregulated HNF1A-AS1 promotes migration and invasion of colon cancer cells in vitro and in vivo (118). IGFL2-AS1 promotes CRC cell migration and invasion in vitro and accelerates the occurrence of CRC in vivo (128). Fang’s research team reported that overexpression of LBX2-AS1 significantly promotes the metastasis of colon cancer cells (130). Liang and others designed loss and gain of function experiments for LINC00174, proving its key role in promoting CRC cell migration and invasion in vitro (132). LINC00473 acts as a ceRNA for miR-195 to promote cell invasion in CRC (134). The expression of LINC00473 accelerates the aggressiveness of CRC (134). Knockdown of LINC00485 promotes migration and invasion in FHC cells, while overexpression weakens these abilities in LoVo cells (135). LINC00665, acting as a sponge for miR-214-3p, upregulates CTNNB1 expression, thereby activating the Wnt/β-catenin signaling pathway and regulating CRC cell migration and invasion (138). LINC00958 directly binds to miR-145-3p, which interacts with downstream effector CDK1, forming the LINC00958 regulatory axis for CRC migration and invasion (142). Knockdown of LINC01088 inhibits proliferation, migration, invasion, and immune evasion of CRC cells. Mechanistically, LINC01088 directly binds to miR-548b-5p and miR-548c-5p, with miR-548c-5p significantly upregulating the expression of Ras GTPase activating protein binding protein 1 (G3BP1) and programmed death ligand 1 (PD-L1), thereby regulating CRC cell migration and invasion (145). LINC01224 can promote CRC invasion through a sponge-like mechanism on miR-2467 (146), and can also regulate migration and invasion through the miR-485-5p/MYO6 axis (147). Knockdown of LINC01287 inhibits colon cancer cell migration and invasion through the LINC01287/miR-4500/MAP3K13 axis (149). Li et al. reported that silencing of LINC01315 regulates CRC cell migration and invasion through the miR-484/DLK1 axis (150), while Liang and others reported another regulatory axis: LINC01485/miR-383-5p/KRT80 (151). Liu’s team used gain and loss of function assays to show that LINC01578 enhances colon cancer cell vitality and migration rate in vitro, and enhances colon cancer liver metastasis in vivo (154). LINC01606 acts as an oncogene promoting colon cancer cell invasion in vitro and in vivo, and is associated with the Wnt/β-catenin signaling pathway (155). Zhou et al. found that knockdown of MIR155HG inhibits CRC cell migration and invasion, regulated through the miR-650/ANXA2 axis (163). Tang et al.’s study suggests that MIR497HG, as a ceRNA regulating the miR-3918/ACTG2 axis, plays a key role in CRC cell migration and invasion (218). MNX1-AS1 regulates miR-744-5p (165) or the PPFIA4/AKT/HIF-1α axis (166), playing a critical role in the invasion and migration of CRC cells.NEAT1 promotes the expression of ZEB1 by targeting miR-448 (167), or enhances the proliferation and invasion of CRC through the miR-216b/YY1 axis (168). Guan et al. found that PCGEM1 mediates the invasion and migration of CRC cells through targeting the miR-129-5p/SOX4 axis (173). Li et al.’s research confirms that PROX1-AS1 can absorb miR-520d to upregulate PD-L1 in CRC (175), while Liu et al. discovered that the transcription factor SP1 activates the PROX1-AS1/miR-32/FBXL20 axis (176), both of which are involved in the regulation of CRC cell migration and invasion. Yu et al.’s results indicate that PVT1 can promote the metastasis of colon cancer by inhibiting the miR-30d-5p/RUNX2 axis (182). Chen et al. found that SNHG16 facilitates CRC cell migration through the miR-124-3p/MCP-1 axis (187). Xiang et al. discovered that SNHG16 acts as a miRNA sponge to sequester miR-195-5p on Ago2, thereby protecting YAP1 from inhibition and subsequently promoting CRC cell migration and invasion (189). Liu et al. demonstrated a new triad involving SNHG17/miR-375/CBX3 that participates in the migration and invasion of COAD (190). SOX2OT,acting as a ceRNA, upregulates SOX5 through a sponge-like effect on miR-194-5p, regulating CRC cell migration and invasion (192). Sui et al. observed that knockdown of SPINT1-AS1 weakens the migration and invasion of CRC cells with silenced KRAS (194). Knockdown of TUG1 not only inhibits CRC migration and invasion through the miR-542-3p/TRIB2 axis (198) but also, by overexpressing it, regulates the miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis in vitro and in vivo to accelerate cancer invasion (199). Liu et al. found that UCA1 is significantly upregulated in CRC tissues. UCA1 enhances the migration and invasion of CRC cell lines (202). Li et al. reported that downregulation of XIST inhibits migration and invasion in CRC cells by regulating the miR-338-3p/PAX5 axis (205). The silencing of XLOC006390 significantly inhibits their viability by inducing apoptosis and limiting the migration and invasion of cancer cells (207). Fang et al.’s study shows that ZFAS1 promotes CRC invasion, and its mechanism is associated with E-cadherin, ZO-1, vimentin, and N-cadherin (208).
The following is an overview of research on LncRNAs inhibiting cancer migration and invasion. Liu et al. found that AK077216 is downregulated in CRA tissues. AK077216 may inhibit the migration and invasion of CRA cells by downregulating miR-34a (71). Overexpression of CASC2 inhibits the migration and invasion capabilities of CRC cells through the miR-18a-5p/BTG3 axis (78). CBR3-AS1 is downregulated in CRC and inhibits miR-29a-mediated cell migration and invasion through a sponge-like mechanism on miR-29a (81). Knockdown of CCL14-AS enhances the invasiveness and lymph node metastatic capability of CRC cells in nude mice, with the CCL14-AS/MEP1A axis playing a key role (87). CYP1B1-AS1 is significantly downregulated in CRC. Experiments conducted in vitro and in vivo have confirmed that upregulation of CYP1B1-AS1 significantly inhibits the migration and invasion of CRC cells (90). Knockdown of both FER1L4 and p73 enhances the migration and invasion of CRC cells. Mechanistic studies have found that FER1L4 promotes the expression of miR-1273g-3p, which in turn increases the expression of PTEN (103). Another study found that restoration of FER1L4 reduces the expression of miR-106a-5p and significantly affects the migration and invasion of colon cancer cells (104). GAS5 can act as a competitive endogenous RNA for miR-21. The downregulation of GAS5 can promote CRC migration and invasion by activating the miR-21/PTEN/Akt signaling pathway (110). Overexpression of HOXC-AS3 reduces cell migration and invasion (126). Wang et al. reported that LINC00261 is downregulated in colon cancer cell lines, tissues, and cisplatin-resistant cells; overexpression of LINC00261 may inhibit cell migration and invasion (133). Knockdown of LINC00485 promotes migration and invasion in FHC cells, and the miR-581/EDEM1 axis is involved (135). Liu et al. reported that m(6)A/LINC02038/miR-552-5p/FAM172A might be a novel anti-tumor axis, closely related to regulating CRC migration and invasion (157). Cai et al. reported that overexpression of OTUD6B-AS1 inhibits the migration and invasion of CRC cells (171).
LncRNAs regulate cell cycle
LncRNAs play a crucial role in the development of CRC by regulating the cell cycle. LncRNAs can intervene in the control of the cell cycle through various mechanisms. Research has found that the levels of AK093407 are higher in the CRC cell lines HCT-15 and HCT-116 compared to normal colonic epithelial NM460 cells. When AK093407 is silenced, the cell cycle of HCT-15 and HCT-116 cells stalls at the G1/S phase (72).Overexpression of DANCR promotes the progression of the CRC cell cycle, with the DANCR/miR-185-5p/HMGA2 axis involved in the regulation (92). Interestingly, during the amplification of the full-length cDNA of EVADR (named EVADR-v1), a new/shorter variant (EVADR-v2) was discovered. When both variants, EVADR-v1 and EVADR-v2, are overexpressed in SW480/HCT116 cells, there is an increase in PI3K activity and upregulation of WNT signaling, thereby promoting cell cycle progression (98). Xiong et al. reported that inhibiting GAS5 can upregulate the expression levels of miR-21, decrease G1 phase cells, and increase S phase cells (110). Reports also suggest that increased expression of LINC00473 accelerates the cell cycle process. Li et al. demonstrated through functional assays that knockdown of SURC inhibits the CRC cell cycle (195).
LncRNAs regulate cancer cell stemness
Yu et al. discovered that ASB16-AS1 is highly expressed in CRC cells, and silencing ASB16-AS1 inhibits the stemness of CRC cells. Mechanistic exploration revealed that ASB16-AS1 drives the progression of CRC by regulating the miR-185-5p/TEAD1 axis (73). Zhao used sphere-formation assays to examine cell stemness and found that depletion of CERS6-AS1 inhibits cell stemness (88). Knockdown of FAM201A inhibits cell stemness, and its regulation is achieved through the miR-3163/MACC1 axis (100). hang et al. demonstrated that Sohlh2 is associated with the inhibition of the LncRNA-H19/miR-141/β-catenin signaling pathway, leading to the suppression of colon CSC (cancer stem cell) stemness (114). Huang et al. used an analysis of stemness-related markers to determine cell stemness, finding that HOTAIR promotes the expression of the miR-211-5p target gene FLT-1, thereby regulating the characteristics of cancer stem cells (CSC) in CRC (120). LINC01606 acts as an oncogene and promotes the stemness of colon cancer cells both in vitro and in vivo. Mechanistically, LINC01606 enhances the expression of SCD1 and regulates the expression of miR-423-5p as a competitive endogenous RNA, subsequently activating the classic Wnt/β-catenin signaling pathway. Additionally, the transcription factor’s binding to IGHM enhancer 3 (TFE3) increases the transcription of LINC01606 after being recruited to the promoter region of LINC01606 (155). Ye et al.’s study indicates that the lncRNA GMDS-AS1 and its direct target HuR constitutively activate the STAT3/Wnt signaling pathway and promote the stemness of CRC tumors (61). Sun et al. reported that PPFIA4 mediates the expression of lncRNA MNX1-AS1 and affects the stemness of COAD cells. The MNX1-AS1/PPFIA4 activates the downstream AKT/HIF-1α signaling pathway to promote the progression of COAD (166). TDRG1 levels are significantly upregulated in 3D non-adherent spheroids derived from parental CRC cells. Further studies indicate that knockdown of TDRG1 inhibits the stemness of CRC cells (196).
LncRNAs regulate EMT
LncRNAs also play an important role in the EMT process in CRC.E2F4-stimulated AGAP2-AS1 exacerbates the EMT process in CRC by regulating the miR-182-5p/CFL1 axis (70). lncRNA-ATB activated by TGF-β may contribute to the development of colon cancer by inhibiting E-cadherin expression and promoting the EMT process (74). Zhao et al. reported that CERS6-AS1 also participates in regulating the EMT process (88). Yue et al.’s experiments show that ectopic expression of CYTOR induces EMT (91).Research indicates that knockdown of EPB41L4A-AS1 inhibits EMT (97). Chen et al. reported that GAS6-AS1 positively regulates the CRC EMT process in vitro through a ceRNA network and FUS-dependent manner (111). Wu et al. consider HOTAIR to be a pleiotropic regulator involved in EMT (122), and Jin et al. demonstrated that HOTAIR regulates HNF4α levels by recruiting SNAIL, thereby regulating the EMT process in CRC cells (124). Li et al. report that LINC00473 regulates EMT progression by modulating miR-195 expression (134). Zheng et al.’s study shows that LINC00543 enhances CRC cell EMT through the pre-miR-506-3p/FOXQ1 axis (136). Research shows that knockdown of LINC00882 impedes the EMT process in CRC cells (141). Fu et al.’s research found that the LINC01287/miR-4500/MAP3K13 axis promotes EMT in colon cancer (149). Studies also suggest that the LINC01315/Wnt/β-catenin signaling pathway provides new insights into regulating CRC EMT (152). Cai et al.’s study found that the OTUD6B-AS1/miR-21-5p/PNRC2 axis is involved in regulating CRC EMT (171). Research finds that SNHG16 promotes CRC cell EMT through the miR-124-3p/MCP-1 axis (187) and SNHG16/YAP1/TEAD1 axis (189). Islam et al. consider SNHG8 as an oncogene in CRC through the EMT pathway (191). Tian et al. believe that TUG1 accelerates cancer EMT by regulating the miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis in vitro and in vivo (199). Fang et al.’s study indicates that ZFAS1 may act as an oncogene by regulating ZEB1 to induce EMT (208).
LncRNAs regulate drug resistance
Drug resistance is a complex process and has always been one of the most unfavorable factors in the process of CRC.Zheng et al.’s research, through microarray screening, identified lncRNAs associated with resistance to oxaliplatin. They experimentally confirmed that the AC092894.1/USP3/AR/RASGRP3 signaling axis is a new option for treating oxaliplatin resistance (69). Li et al. reported that BCAR4 promotes oxaliplatin resistance by inhibiting apoptosis through the BCAR4/miR-483-3p/RAB5C axis (76). Liu found that CCAT1 enhances CRC cell resistance to oxaliplatin by transcriptionally activating CCAT1 through B-MYB, increasing SOCS3 promoter methylation to recruit DNMT1, thereby inhibiting SOCS3 expression and enhancing CRC cell resistance to oxaliplatin (82). Another study found that downregulation of CCAT1 effectively reversed the resistance of HCT 116/5-FU and HT-29/5-FU cells to 5-FU chemotherapy (84). Yang et al. reported that silencing CRNDE promotes apoptosis in CRC cells and enhances sensitivity to cisplatin, possibly through regulation of the Warburg effect mediated by the Akt/mTORC1 pathway (89). Li et al. emphasized the potential of targeting ELFN1-AS1 as a therapeutic agent in cell survival and resistance to oxaliplatin (95). Gao et al., through in vitro and in vivo xenograft experiments, showed that the specific inhibitor erlotinib enhances the anti-tumor toxicity of 5-Fu by targeting the EGFR/FGD5-AS1/miR-330-3p/HK2 pathway (107). Chen et al. found that overexpression of H19 may be one of the mechanisms by which advanced colon cancer develops resistance to treatment with 1,25(OH)2D3 (113). A study revealed the crucial role and molecular mechanism of HCG11-mediated 5-FU resistance in CRC through regulation of the miR-144-3p/PDK4/glucose metabolism pathway (117). Zhang et al. reported that patients carrying allele 10 and having methylation in the KCNQ1OT1 promoter exhibited the greatest reduction in resistance to colon cancer treatment (129). Research has found that LINC00261 reduces cisplatin resistance in colon cancer in vivo, and enhances the anti-colon cancer effects of cisplatin by reducing tumor volume and weight (133). LINC00958 promotes the expression of MAPK1 by targeting miR-422a, inhibiting cell radiosensitivity (143). In Wei’s study on the traditional Chinese medicine Zuojin Wan, it was found that MALAT1 promotes resistance to oxaliplatin in CRC by decreasing the expression of miR-200s. Zuojin Wan may reverse chemotherapy resistance by inhibiting the expression of MALAT1 and regulating the miR-200s/JNK pathway (158). Cai et al. reported that knockdown of MCF2L-AS1 can alleviate the chemoresistance of CRC/OXA cells (161). Zhou et al.’s research reported that MIR155HG promotes CRC progression and enhances CRC cell resistance to oxaliplatin by regulating the miR-650/ANXA2 axis and through M2 macrophage polarization (163). Shi et al.’s work found that lncRNA-NEF plays a key role in mediating oxaliplatin chemotherapy resistance in CRC, providing a promising therapeutic strategy for CRC patients resistant to oxaliplatin (169). Zhang et al. reported that overexpression of OTUD6B-AS1 by binding with HuR stabilizes TRIM16 and increases iron accumulation mediated by GPX4, thus weakening CRC’s radioresistance (170). Another study indicated that exosomal SNHG11 is upregulated in bevacizumab-resistant CRC cells, and SNHG11 contributes to the resistance to bevacizumab in CRC, depending on the regulation of miR-1207-5p and ABCC1 (184). Li et al.’s research suggests that SNHG15 promotes 5-FU chemoresistance in CRC by potentially regulating the expression of TYMS, BCL2, GLUT1, and PKM2 (185). Yuan et al.’s research provides the first evidence that the UCA1-miR-495-HGF/c-MET regulatory network is involved in CRC resistance to cetuximab (201).
Discussion
CRC is one of the major health challenges globally, with a high mortality rate, especially when the disease is diagnosed at a late stage. To improve the success rate of treatments and the survival rates of patients, the development of reliable early detection biomarkers is particularly crucial. In recent years, researchers have begun to focus on the potential role of LncRNAs in CRC, especially as non-invasive molecular biomarkers (225).
LncRNAs have multifaceted functions in CRC, including the regulation of the cell cycle, proliferation, apoptosis, and metastasis. They often function as ceRNAs that affect the expression of specific miRNAs and subsequently the downstream molecules of these miRNAs. Additionally, LncRNAs play an important role in regulating gene expression, including transcriptional regulation, mRNA stability, and translation. Research indicates that LncRNAs might interact with RNA, DNA, and proteins to form complex regulatory networks, thereby affecting the onset and progression of CRC.Given the stability of LncRNAs in blood and their potential in early CRC detection, they are considered a promising tool for non-invasive testing. Furthermore, studies on LncRNAs have also revealed their potential key roles in regulating various pathological processes related to CRC, such as influencing the aggressiveness and metastatic properties of cancer cells through specific regulatory axes.
At present, the main diagnostic methods for CRC include colonoscopy, tissue biopsy, and serum tumor marker detection. Although these methods are widely used, they have some significant limitations. Colonoscopy is an invasive procedure where patients need to prepare their intestines and may experience discomfort or pain during the examination process (226). In addition, although tissue biopsy is the gold standard for diagnosis, the limitations of sampling may lead to missed diagnosis (227). The detection sensitivity and specificity of serum tumor markers such as CEA and CA19-9 are low, and false positive and false negative results are prone to occur, which cannot accurately reflect early carcinogenesis (228). In terms of treatment, surgical resection is the main treatment for early CRC, while chemotherapy and radiotherapy are used as adjuvant treatments for advanced or postoperative conditions. However, these treatment methods also have certain limitations. Although surgery can remove tumors, it is difficult to completely remove early metastases or small lesions (228). Chemotherapy and radiation therapy may cause serious side effects such as nausea, vomiting, and bone marrow suppression, and some patients may develop drug resistance, leading to poor treatment outcomes (229). In addition, although targeted therapy is effective for patients with certain specific gene mutations, its scope of application is limited and the treatment cost is high (227). LncRNA provides a certain possibility to solve the above problems. In the past decade, numerous studies have shown that lncRNA is stable in the bloodstream and has excellent diagnostic potential, making lncRNA a promising candidate for non-invasive diagnostic testing of CRC (43–46). In the treatment of CRC, LncRNA can serve as a novel therapeutic target, affecting tumor growth and metastasis by regulating the expression of oncogenes and tumor suppressor genes. Combining lncRNA with small molecule drugs or gene therapy to develop novel therapies with stronger targeting and fewer side effects may break through the limitations of existing treatments (230, 231). LncRNA also plays an important role in the mechanism of chemotherapy and radiation resistance. By intervening in the function of these lncRNAs, it is expected to reverse drug resistance and improve treatment efficacy (232, 233).
Although lncRNAs have broad application prospects, their application in clinical diagnosis and treatment still faces multiple challenges:1. In the previous chapters, we listed many lncRNAs with a diagnostic AUC value greater than 0.9 in Table 1, but most of the experimental and control groups included in these studies were less than 100 cases, so there may be a large bias in the research results. In addition, the research is also limited by age, race, gender and other factors. In order to obtain more credible results, these studies should be verified with a larger sample size and wider population distribution; 2. Currently, there are various methods for detecting lncRNA, including qRT qPCR, RNA seq, Sanger sequencing, and Northern blot. The standardization and optimization of these methods still need further development to ensure consistency and reproducibility in different laboratory and clinical environments (234, 235). 3.The data analysis process of high-throughput lncRNAs is complex and requires high-level professional knowledge and computing resources (236, 237). 4.Although many lncRNAs have been found to be associated with cancer, research on their specific functions and mechanisms is still in its early stages. The current research lacks a comprehensive understanding of the mechanism of action of lncRNA, which limits its potential in clinical applications (238, 239). 5.Treatment strategies based on lncRNA, such as siRNA and antisense oligonucleotides (ASO), face issues such as low delivery efficiency, poor specificity, and off target effects, which further limit their therapeutic efficacy (240).
Conclusion
In summary, research on LncRNAs provides a new perspective for understanding the molecular mechanisms of CRC and brings hope for the development of new diagnostic and therapeutic strategies. Future studies are needed to further explore the specific functions of these molecules in CRC and their potential clinical applications, with the hope of bringing more effective treatment options and better survival prospects for CRC patients.
Author contributions
YNL: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. WZ: Investigation, Visualization, Writing – review & editing. ZL: Investigation, Writing – review & editing. HX: Investigation, Methodology, Writing – review & editing. YL: Investigation, Software, Writing – review & editing. ZZ: Conceptualization, Data curation, Methodology, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by Xiamen medical and health guidance project(No.3502Z20224ZD1116).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
1. Biller LH, Schrag D. Diagnosis and treatment of metastatic colorectal cancer: A review. JAMA-J Am Med Assoc. (2021) 325:669–85. doi: 10.1001/jama.2021.0106
2. Yamashita R, Long J, Longacre T, Peng L, Berry G, Martin B, et al. Deep learning model for the prediction of microsatellite instability in colorectal cancer: a diagnostic study. Lancet Oncol. (2021) 22:132–41. doi: 10.1016/S1470-2045(20)30535-0
3. Kather JN, Krisam J, Charoentong P, Luedde T, Herpel E, Weis CA, et al. Predicting survival from colorectal cancer histology slides using deep learning: A retrospective multicenter study. PloS Med. (2019) 16:e1002730. doi: 10.1371/journal.pmed.1002730
4. Cancer Genome Atlas Network. Comprehensive molecular characterization of human colon and rectal cancer. Nature. (2012) 487(7407):330–7. doi: 10.1038/nature11252
5. Xi Y, Xu P. Global colorectal cancer burden in 2020 and projections to 2040. Trans Oncol. (2021) 14:101174. doi: 10.1016/j.tranon.2021.101174
6. Brandi G, Ricci AD, Rizzo A, Zanfi C, Tavolari S, Palloni A, et al. Is post-transplant chemotherapy feasible in liver transplantation for colorectal cancer liver metastases? Cancer Commun (London England). (2020) 40:461–4. doi: 10.1002/cac2.12072
7. García-Albéniz X, Hsu J, Bretthauer M, Hernán MA. Effectiveness of screening colonoscopy to prevent colorectal cancer among medicare beneficiaries aged 70 to 79 years: A prospective observational study. Ann Internal Med. (2017) 166:18–26. doi: 10.7326/M16-0758
8. Zarkavelis G, Boussios S, Papadaki A, Katsanos KH, Christodoulou DK, Pentheroudakis G. Current and future biomarkers in colorectal cancer. Ann gastroenterology. (2017) 30:613–21. doi: 10.20524/aog.2017.0191
9. Tang X, Qiao X, Chen C, Liu Y, Zhu J, Liu J. Regulation mechanism of long noncoding RNAs in colon cancer development and progression. Yonsei Med J. (2019) 60:319–25. doi: 10.3349/ymj.2019.60.4.319
10. Okayama H, Schetter AJ, Harris CC. MicroRNAs and inflammation in the pathogenesis and progression of colon cancer. Dig Dis. (2012) 30 Suppl 2(Suppl 2):9–15. doi: 10.1159/000341882
11. Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. (2009) 458(7235):223–7. doi: 10.1038/nature07672
12. Bhan A, Mandal SS. Long noncoding RNAs: emerging stars in gene regulation, epigenetics and human disease. ChemMedChem. (2014) 9(9):1932–56. doi: 10.1002/cmdc.201300534
13. Gil N, Ulitsky I. Regulation of gene expression by cis-acting long non-coding RNAs. Nat Rev Genet. (2020) 21(2):102–17. doi: 10.1038/s41576-019-0184-5
14. Yao RW, Wang Y, Chen LL. Cellular functions of long noncoding RNAs. Nat Cell Biol. (2019) 21(5):542–51. doi: 10.1038/s41556-019-0311-8
15. Castro-Oropeza R, Melendez-Zajgla J, Maldonado V, Vazquez-Santillan K. The emerging role of lncRNAs in the regulation of cancer stem cells. Cell Oncol. (2018) 41:585–603. doi: 10.1007/s13402-018-0406-4
16. Li ZR, Qin XB, Bian W, Li YS, Shan BE, Yao ZM, et al. Exosomal lncRNA ZFAS1 regulates esophageal squamous cell carcinoma cell proliferation, invasion, migration and apoptosis via microRNA-124/STAT3 axis. J Exp Clin Cancer Res. (2019) 38:13. doi: 10.1186/s13046-019-1473-8
17. Zhou YC, Shi HT, Du YQ, Zhao GQ, Wang XX, Li Q, et al. lncRNA DLEU2 modulates cell proliferation and invasion of non-small cell lung cancer by regulating miR-30c-5p/SOX9 axis. Aging-US. (2019) 11:7386–401. doi: 10.18632/aging.v11i18
18. Plath K, Fang J, Mlynarczyk-Evans SK, Cao R, Worringer KA, Wang H, et al. Role of histone H3 lysine 27 methylation in X inactivation. Sci (New York NY). (2003) 300:131–5. doi: 10.1126/science.1084274
19. Rinn JL, Kertesz M, Wang JK, Squazzo SL, Xu X, Brugmann SA, et al. Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell. (2007) 129:1311–23. doi: 10.1016/j.cell.2007.05.022
20. Rinn JL, Chang HY. Genome regulation by long noncoding RNAs. Annu Rev Biochem. (2012) 81:145–66. doi: 10.1146/annurev-biochem-051410-092902
21. Huarte M. The emerging role of lncRNAs in cancer. Nat Med. (2015) 21:1253–61. doi: 10.1038/nm.3981
22. Schmitt AM, Chang HY. Long noncoding RNAs in cancer pathways. Cancer Cell. (2016) 29:452–63. doi: 10.1016/j.ccell.2016.03.010
23. Mattick JS, Makunin IV. Non-coding RNA. Hum Mol Genet. (2006) 15 Spec No 1:R17–29. doi: 10.1093/hmg/ddl046
24. Katayama S, Tomaru Y, Kasukawa T, Waki K, Nakanishi M, Nakamura M, et al. Antisense transcription in the mammalian transcriptome. Sci (New York NY). (2005) 309:1564–6. doi: 10.1126/science.1112009
25. Pelechano V, Steinmetz LM. NON-CODING RNA Gene regulation by antisense transcription. Nat Rev Genet. (2013) 14:880–93. doi: 10.1038/nrg3594
26. Ma LN, Bajic VB, Zhang Z. On the classification of long non-coding RNAs. RNA Biol. (2013) 10:925–34. doi: 10.4161/rna.24604
27. Quinn JJ, Chang HY. Unique features of long non-coding RNA biogenesis and function. Nat Rev Genet. (2016) 17:47–62. doi: 10.1038/nrg.2015.10
28. Cheng S, Jia Y, Wu J, Li J, Cao Y. Helicobacter pylori infection induces gastric cancer cell Malignancy by targeting HOXA-AS2/miR-509-3p/MMD2 axis. Genes Genomics. (2024) 46(6):647–57. doi: 10.1007/s13258-024-01500-2
29. Yan S, Teng L, Du J, Ji L, Xu P, Zhao W, et al. Long non−coding RNA DANCR aggravates breast cancer through the miR−34c/E2F1 feedback loop. Mol Med Rep. (2024) 29(6):93. doi: 10.3892/mmr.2024.13217
30. Wang C, Chen R, Zhu X, Zhang X, Lian N. Long noncoding RNA small nucleolar RNA host gene 5 facilitates neuropathic pain in spinal nerve injury by promoting SCN9A expression via CDK9. Hum Cell. (2024) 37:451–64. doi: 10.1007/s13577-023-01019-w
31. Geisler S, Coller J. RNA in unexpected places: long non-coding RNA functions in diverse cellular contexts. Nat Rev Mol Cell Biol. (2013) 14:699–712. doi: 10.1038/nrm3679
32. Gudenas BL, Wang LJ. Prediction of lncRNA subcellular localization with deep learning from sequence features. Sci Rep. (2018) 8:10. doi: 10.1038/s41598-018-34708-w
33. Gozzetti A, Le Beau MM. Fluorescence in situ hybridization: uses and limitations. Semin hematology. (2000) 37:320–33. doi: 10.1016/S0037-1963(00)90013-1
34. Keene JD, Komisarow JM, Friedersdorf MB. RIP-Chip: the isolation and identification of mRNAs, microRNAs and protein components of ribonucleoprotein complexes from cell extracts. Nat Protoc. (2006) 1:302–7. doi: 10.1038/nprot.2006.47
35. Wang Z, Gerstein M, Snyder M. RNA-Seq: a revolutionary tool for transcriptomics. Nat Rev Genet. (2009) 10:57–63. doi: 10.1038/nrg2484
36. Macosko EZ, Basu A, Satija R, Nemesh J, Shekhar K, Goldman M, et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell. (2015) 161:1202–14. doi: 10.1016/j.cell.2015.05.002
37. Shaffer SM, Dunagin MC, Torborg SR, Torre EA, Emert B, Krepler C, et al. Rare cell variability and drug-induced reprogramming as a mode of cancer drug resistance. Nature. (2017) 546:431–5. doi: 10.1038/nature22794
38. Hezroni H, Koppstein D, Schwartz MG, Avrutin A, Bartel DP, Ulitsky I. Principles of long noncoding RNA evolution derived from direct comparison of transcriptomes in 17 species. Cell Rep. (2015) 11:1110–22. doi: 10.1016/j.celrep.2015.04.023
39. Necsulea A, Soumillon M, Warnefors M, Liechti A, Daish T, Zeller U, et al. The evolution of lncRNA repertoires and expression patterns in tetrapods. Nature. (2014) 505:635–40. doi: 10.1038/nature12943
40. Pang KC, Frith MC, Mattick JS. Rapid evolution of noncoding RNAs: lack of conservation does not mean lack of function. Trends genetics: TIG. (2006) 22:1–5. doi: 10.1016/j.tig.2005.10.003
41. Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, et al. The transcriptional landscape of the mammalian genome. Sci (New York NY). (2005) 309:1559–63. doi: 10.1126/science.1112014
42. Derrien T, Johnson R, Bussotti G, Tanzer A, Djebali S, Tilgner H, et al. The GENCODE v7 catalog of human long noncoding RNAs: analysis of their gene structure, evolution, and expression. Genome Res. (2012) 22:1775–89. doi: 10.1101/gr.132159.111
43. Dong L, Lin W, Qi P, Xu MD, Wu X, Ni S, et al. Circulating long RNAs in serum extracellular vesicles: their characterization and potential application as biomarkers for diagnosis of colorectal cancer. Cancer epidemiology Biomarkers prevention: Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol. (2016) 25:1158–66. doi: 10.1158/1055-9965.EPI-16-0006
44. Ferracin M, Lupini L, Mangolini A, Negrini M. Circulating non-coding RNA as biomarkers in colorectal cancer. Adv Exp Med Biol. (2016) 937:171–81. doi: 10.1007/978-3-319-42059-2_9
45. Ragusa M, Barbagallo C, Statello L, Condorelli AG, Battaglia R, Tamburello L, et al. Non-coding landscapes of colorectal cancer. World J gastroenterology. (2015) 21:11709–39. doi: 10.3748/wjg.v21.i41.11709
46. Saplacan RM, Mircea PA, Balacescu L, Balacescu O. MicroRNAs as non-invasive screening biomarkers of colorectal cancer. Clujul Med (1957). (2015) 88:453–6. doi: 10.15386/cjmed-568
47. Sole C, Arnaiz E, Manterola L, Otaegui D, Lawrie CH. The circulating transcriptome as a source of cancer liquid biopsy biomarkers. Semin Cancer Biol. (2019) 58:100–8. doi: 10.1016/j.semcancer.2019.01.003
48. Lo YM. Circulating nucleic acids in plasma and serum: an overview. Ann New York Acad Sci. (2001) 945:1–7. doi: 10.1111/j.1749-6632.2001.tb03858.x
49. Shi T, Gao G, Cao Y. Long noncoding RNAs as novel biomarkers have a promising future in cancer diagnostics. Dis markers. (2016) 2016:9085195. doi: 10.1155/2016/9085195
50. Qi P, Zhou XY, Du X. Circulating long non-coding RNAs in cancer: current status and future perspectives. Mol cancer. (2016) 15:39. doi: 10.1186/s12943-016-0524-4
51. Xie H, Ma H, Zhou D. Plasma HULC as a promising novel biomarker for the detection of hepatocellular carcinoma. BioMed Res Int. (2013) 2013:136106. doi: 10.1155/2013/136106
52. Revenfeld AL, Bæk R, Nielsen MH, Stensballe A, Varming K, Jørgensen M. Diagnostic and prognostic potential of extracellular vesicles in peripheral blood. Clin Ther. (2014) 36:830–46. doi: 10.1016/j.clinthera.2014.05.008
53. Mathieu M, Martin-Jaular L, Lavieu G, Théry C. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat Cell Biol. (2019) 21:9–17. doi: 10.1038/s41556-018-0250-9
54. Benes V, Castoldi M. Expression profiling of microRNA using real-time quantitative PCR, how to use it and what is available. Methods (San Diego Calif). (2010) 50:244–9. doi: 10.1016/j.ymeth.2010.01.026
55. Zhao W, Song M, Zhang J, Kuerban M, Wang H. Combined identification of long non-coding RNA CCAT1 and HOTAIR in serum as an effective screening for colorectal carcinoma. Int J Clin Exp pathology. (2015) 8(11):14131–40.
56. Abd El Fattah YK, Abulsoud AI, AbdelHamid SG, AbdelHalim S, Hamdy NM. CCDC144NL-AS1/hsa-miR-143-3p/HMGA2 interaction: In-silico and clinically implicated in CRC progression, correlated to tumor stage and size in case-controlled study; step toward ncRNA precision. Int J Biol macromolecules. (2023) 253:126739. doi: 10.1016/j.ijbiomac.2023.126739
57. Bakr M, Abd-Elmawla MA, Elimam H, Gamal El-Din H, Fawzy A, Abulsoud AI, et al. Telomerase RNA component lncRNA as potential diagnostic biomarker promotes CRC cellular migration and apoptosis evasion via modulation of β-catenin protein level. Non-coding RNA Res. (2023) 8:302–14. doi: 10.1016/j.ncrna.2023.03.004
58. El-Sheikh NM, Abulsoud AI, Fawzy A, Wasfey EF, Hamdy NM. LncRNA NNT-AS1/hsa-miR-485-5p/HSP90 axis in-silico and clinical prospect correlated-to histologic grades-based CRC stratification: A step toward ncRNA Precision. Pathology Res practice. (2023) 247:154570. doi: 10.1016/j.prp.2023.154570
59. Long F, Zhong C, Long Q, Zhu K, Wang J, Yu Y, et al. Circular RNA RHBDD1 regulates tumorigenicity and ferroptosis in colorectal cancer by mediating the ELAVL1/SCD mRNA interaction. Cancer Gene Ther. (2024) 31(2):237–49. doi: 10.1038/s41417-023-00698-9
60. Salman IT, Abulsoud AI, Abo-Elmatty DM, Fawzy A, Mesbah NM, Saleh SM. The long non-coding RNA ZFAS1 promotes colorectal cancer progression via miR200b/ZEB1 axis. Pathology Res practice. (2023) 247:154567. doi: 10.1016/j.prp.2023.154567
61. Ye D, Liu H, Zhao G, Chen A, Jiang Y, Hu Y, et al. LncGMDS-AS1 promotes the tumorigenesis of colorectal cancer through HuR-STAT3/Wnt axis. Cell Death disease. (2023) 14:165. doi: 10.1038/s41419-023-05700-8
62. Zhang X, Ma D, Xuan B, Shi D, He J, Yu M, et al. LncRNA CACClnc promotes chemoresistance of colorectal cancer by modulating alternative splicing of RAD51. Oncogene. (2023) 42:1374–91. doi: 10.1038/s41388-023-02657-y
63. Dai W, Wang Z, Liang X, Wang M, Ni W, Yang Y, et al. Circulating lncRNA EGFR-AS1 as a diagnostic biomarker of colorectal cancer and an indicator of tumor burden. J gastrointestinal Oncol. (2022) 13:2439–46. doi: 10.21037/jgo-22-968
64. Dong L, Liu D, Jing D, Xu H, Zhang C, Qi D, et al. LncRNA ARST is a novel prognostic and diagnostic biomarker for colorectal cancer. Cancer Manage Res. (2022) 14:19–24. doi: 10.2147/CMAR.S338997
65. Elabd NS, Soliman SE, Elhamouly MS, Gohar SF, Elgamal A, Alabassy MM, et al. Long non-coding RNAs ASB16-AS1 and AFAP1-AS1: diagnostic, prognostic impact and survival analysis in colorectal cancer. Appl Clin Genet. (2022) 15:97–109. doi: 10.2147/TACG.S370242
66. Shen L, Zong W, Feng W, Chen E, Ma S, Yuan J, et al. Upregulated linc01836 in serum promisingly serving as a diagnostic and prognostic biomarker for colorectal cancer. Front Pharmacol. (2022) 13:840391. doi: 10.3389/fphar.2022.840391
67. Ye B, Li F, Chen M, Weng Y, Qi C, Xie Y, et al. A panel of platelet-associated circulating long non-coding RNAs as potential biomarkers for colorectal cancer. Genomics. (2022) 114:31–7. doi: 10.1016/j.ygeno.2021.11.026
68. Wu Y, Xu X. Long non-coding RNA signature in colorectal cancer: research progression and clinical application. Cancer Cell Int. (2023) 23:28. doi: 10.1186/s12935-023-02867-0
69. Zheng Z, Wu M, Li H, Xu W, Yang M, Pan K, et al. Downregulation of AC092894.1 promotes oxaliplatin resistance in colorectal cancer via the USP3/AR/RASGRP3 axis. BMC Med. (2023) 21:132. doi: 10.1186/s12916-023-02826-6
70. Guo Z, Liu X, Shao H. E2F4-induced AGAP2-AS1 up-regulation accelerates the progression of colorectal cancer via miR-182-5p/CFL1 axis. Digestive liver disease: Off J Ital Soc Gastroenterol Ital Assoc Study Liver. (2022) 54:878–89. doi: 10.1016/j.dld.2021.08.002
71. Liu H, Li Y, Lv Y, Guo Z, Guo S. LncRNA AK077216 affects the survival of colorectal adenocarcinoma patients via miR-34a. Arab J gastroenterology: Off Publ Pan-Arab Assoc Gastroenterology. (2022) 23:65–9. doi: 10.1016/j.ajg.2021.11.001
72. Xuerong Z, Ao S, Jianping W, Xin Z, Duoduo T, Mingjuan W, et al. Effects of long noncoding RNA AK093407 on the biological behavior of colon cancer cells and the underlying mechanism. Combinatorial Chem High throughput screening. (2023) 26:289–300. doi: 10.2174/1386207325666220408092028
73. Yu M, Shi C, Xu D, Lin X, Ji T, Shi Z, et al. LncRNA ASB16-AS1 drives proliferation, migration, and invasion of colorectal cancer cells through regulating miR-185-5p/TEAD1 axis. Cell Cycle (Georgetown Tex). (2022) 21:1–11. doi: 10.1080/15384101.2021.1973700
74. Yue B, Qiu S, Zhao S, Liu C, Zhang D, Yu F, et al. LncRNA-ATB mediated E-cadherin repression promotes the progression of colon cancer and predicts poor prognosis. J Gastroenterol hepatology. (2016) 31:595–603. doi: 10.1111/jgh.13206
75. Liu J, Zhu J, Xiao Z, Wang X, Luo J. BBOX1-AS1 contributes to colorectal cancer progression by sponging hsa-miR-361-3p and targeting SH2B1. FEBS Open bio. (2022) 12:983–92. doi: 10.1002/2211-5463.12802
76. Li X, Chen X, Fu C, Xie M, Ouyang S. Long non-coding RNA BCAR4 promotes oxaliplatin resistance in colorectal cancer by modulating miR-484-3p/RAB5C expression. Chemotherapy. (2023) 68:119–30. doi: 10.1159/000529134
77. Zheng W, Guo Y, Zhang G, Bai J, Song Y, Song X, et al. Peptide encoded by lncRNA BVES-AS1 promotes cell viability, migration, and invasion in colorectal cancer cells via the SRC/mTOR signaling pathway. PloS One. (2023) 18:e0287133. doi: 10.1371/journal.pone.0287133
78. Kang L, Sun J, Liu J, Xu F, Zhu Q, Shi X. Long Non-Coding RNA CASC2 Functions as A Tumor Suppressor in Colorectal Cancer via Modulating The miR-18a-5p/BTG3 Pathway. Cell J. (2022) 24:665–72. doi: 10.22074/cellj.2022.8036
79. Liu HZ, Shan TD, Han Y, Liu XS. Correction to: Silencing long non-coding RNA CASC9 inhibits colorectal cancer cell proliferation by acting as a competing endogenous RNA of miR-576-5p to regulate AKT3. Cell Death discovery. (2021) 7:185. doi: 10.1038/s41420-021-00564-3
80. Zhang H, Wang J, Yu T, Wang J, Lu J, Yu Z. Silencing LncRNA CASC9 inhibits proliferation and invasion of colorectal cancer cells by MiR-542-3p/ILK. PloS One. (2022) 17:e0265901. doi: 10.1371/journal.pone.0265901
81. Yang M, Chen W, Liu H, Yu L, Tang M, Liu Y. Long non-coding RNA CBR3 antisense RNA 1 is downregulated in colorectal cancer and inhibits miR-29a-mediated cell migration and invasion. Mol Biotechnol. (2022) 64:773–9. doi: 10.1007/s12033-021-00444-2
82. Liu F, Wang Y, Cao Y, Wu Z, Ma D, Cai J, et al. Transcription factor B-MYB activates lncRNA CCAT1 and upregulates SOCS3 to promote chemoresistance in colorectal cancer. Chemico-biological interactions. (2023) 374:110412. doi: 10.1016/j.cbi.2023.110412
83. He X, Tan X, Wang X, Jin H, Liu L, Ma L, et al. C-Myc-activated long noncoding RNA CCAT1 promotes colon cancer cell proliferation and invasion. Tumour biology: J Int Soc Oncodevelopmental Biol Med. (2014) 35:12181–8. doi: 10.1007/s13277-014-2526-4
84. Yang C, Pan Y, Deng SP. Downregulation of lncRNA CCAT1 enhances 5-fluorouracil sensitivity in human colon cancer cells. BMC Mol Cell Biol. (2019) 20:9. doi: 10.1186/s12860-019-0188-1
85. Kam Y, Rubinstein A, Naik S, Djavsarov I, Halle D, Ariel I, et al. Detection of a long non-coding RNA (CCAT1) in living cells and human adenocarcinoma of colon tissues using FIT-PNA molecular beacons. Cancer Lett. (2014) 352(1):90–6. doi: 10.1016/j.canlet.2013.02.014
86. Ling H, Spizzo R, Atlasi Y, Nicoloso M, Shimizu M, Redis RS, et al. CCAT2, a novel noncoding RNA mapping to 8q24, underlies metastatic progression and chromosomal instability in colon cancer. Genome Res. (2013) 23(9):1446–61. doi: 10.1101/gr.152942.112
87. Li M, Huang C, Wu Y, Zhu L, Zhang Y, Zhou Y, et al. Long non-coding RNA CCL14-AS suppresses invasiveness and lymph node metastasis of colorectal cancer cells by regulating MEP1A. Cancer Cell Int. (2023) 23:27. doi: 10.1186/s12935-023-02866-1
88. Zhao SY, Wang Z, Wu XB, Zhang S, Chen Q, Wang DD, et al. CERS6-AS1 contributes to the Malignant phenotypes of colorectal cancer cells by interacting with miR-15b-5p to regulate SPTBN2. Kaohsiung J Med Sci. (2022) 38:403–14. doi: 10.1002/kjm2.12503
89. Yang W, Wang Y, Tao C, Li Y, Cao S, Yang X. CRNDE silencing promotes apoptosis and enhances cisplatin sensitivity of colorectal carcinoma cells by inhibiting the Akt/mTORC1-mediated Warburg effect. Oncol letters. (2022) 23:70. doi: 10.3892/ol.2022.13190
90. Wu Z, Cheng F, Yuan L, Li X, Li Z, Huang Z, et al. CYP1B1-AS1 delays the Malignant progression of colorectal cancer by binding with NOP58. Digestive Dis Sci. (2024) 31(2):237–49. doi: 10.1007/s10620-023-08206-7
91. Yue B, Liu C, Sun H, Liu M, Song C, Cui R, et al. A positive feed-forward loop between lncRNA-CYTOR and wnt/β-catenin signaling promotes metastasis of colon cancer. Mol therapy: J Am Soc Gene Ther. (2018) 26:1287–98. doi: 10.1016/j.ymthe.2018.02.024
92. Lu W, Huang Z, Wang J, Liu H. Long non-coding RNA DANCR accelerates colorectal cancer progression via regulating the miR-185-5p/HMGA2 axis. J Biochem. (2022) 171:389–98. doi: 10.1093/jb/mvab011
93. Li W, Ke C, Yang C, Li J, Chen Q, Xia Z, et al. LncRNA DICER1-AS1 promotes colorectal cancer progression by activating the MAPK/ERK signaling pathway through sponging miR-650. Cancer Med. (2023) 12:8351–66. doi: 10.1002/cam4.5550
94. Liang H, Wang J, Zhang P, Yang W, Yang Y, Zhi Y, et al. Long Non-Coding RNA Duxap8 Facilitates Cell Proliferation and Induces Apoptosis in Colorectal Cancer via miR-519b/ZNF277 Axis. OncoTargets Ther. (2021) 14:4693–703. doi: 10.2147/OTT.S301233
95. Li Y, Gan Y, Liu J, Li J, Zhou Z, Tian R, et al. Downregulation of MEIS1 mediated by ELFN1-AS1/EZH2/DNMT3a axis promotes tumorigenesis and oxaliplatin resistance in colorectal cancer. Signal transduction targeted Ther. (2022) 7:87. doi: 10.1038/s41392-022-00902-6
96. Han B, He J, Chen Q, Yuan M, Zeng X, Li Y, et al. ELFN1-AS1 promotes GDF15-mediated immune escape of colorectal cancer from NK cells by facilitating GCN5 and SND1 association. Discover Oncol. (2023) 14:56. doi: 10.1007/s12672-023-00675-6
97. Bin J, Nie S, Tang Z, Kang A, Fu Z, Hu Y, et al. Long noncoding RNA EPB41L4A-AS1 functions as an oncogene by regulating the Rho/ROCK pathway in colorectal cancer. J Cell Physiol. (2021) 236:523–35. doi: 10.1002/jcp.29880
98. Yari M, Soltani BM, Ghaemi Z, Omrani MD. EVADR ceRNA transcript variants upregulate WNT and PI3K signaling pathways in SW480 and HCT116 cells by sponging miR-7 and miR-29b. Biol Chem. (2023) 404:71–83. doi: 10.1515/hsz-2022-0246
99. Lu X, Xu Q, Tong Y, Zhang Z, Dun G, Feng Y, et al. Long non-coding RNA EVADR induced by Fusobacterium nucleatum infection promotes colorectal cancer metastasis. Cell Rep. (2022) 40:111127. doi: 10.1016/j.celrep.2022.111127
100. Zeng L, Luo X, Zhang Z, Wang Z, Qian J. Long non-coding FAM201A accelerates the tumorigenesis and progression of colorectal cancer through miR-3163/MACC1 axis. Trans Oncol. (2022) 25:101490. doi: 10.1016/j.tranon.2022.101490
101. Li N, Zhou C, Yang F. lncRNA FAM230B is highly expressed in colorectal cancer and suppresses the maturation of miR-1182 to increase cell proliferation. Open Med (Warsaw Poland). (2022) 17:1559–67. doi: 10.1515/med-2022-0500
102. Yang L, Cui J, Wang Y, Tan J. FAM83H-AS1 is upregulated and predicts poor prognosis in colon cancer. Biomedicine pharmacotherapy = Biomedecine pharmacotherapie. (2019) 118:109342. doi: 10.1016/j.biopha.2019.109342
103. Uboveja A, Satija YK, Siraj F, Saluja D. p73-regulated FER1L4 lncRNA sponges the oncogenic potential of miR-1273g-3p and aids in the suppression of colorectal cancer metastasis. iScience. (2022) 25:103811. doi: 10.1016/j.isci.2022.103811
104. Yue B, Sun B, Liu C, Zhao S, Zhang D, Yu F, et al. Long non-coding RNA Fer-1-like protein 4 suppresses oncogenesis and exhibits prognostic value by associating with miR-106a-5p in colon cancer. Cancer science. (2015) 106:1323–32. doi: 10.1111/cas.12759
105. Yang X, Wu P, Wang Z, Su X, Wu Z, Ma X, et al. Constructed the ceRNA network and predicted a FEZF1-AS1/miR-92b-3p/ZIC5 axis in colon cancer. Mol Cell Biochem. (2023) 478:1083–97. doi: 10.1007/s11010-022-04578-y
106. Xie R, Liu C, Liu L, Lu X, Tang G. Long non-coding RNA FEZF1-AS1 promotes rectal cancer progression by competitively binding miR-632 with FAM83A. Acta Biochim Biophys Sinica. (2022) 54:452–62. doi: 10.3724/abbs.2022022
107. Gao SJ, Ren SN, Liu YT, Yan HW, Chen XB. Targeting EGFR sensitizes 5-Fu-resistant colon cancer cells through modification of the lncRNA-FGD5-AS1-miR-330-3p-Hexokinase 2 axis. Mol Ther oncolytics. (2021) 23:14–25. doi: 10.1016/j.omto.2021.06.012
108. Cheng Z, Jiang S, Tao R, Ge H, Qin J. Activating transcription factor 3-activated long noncoding RNA forkhead box P4-antisense RNA 1 aggravates colorectal cancer progression by regulating microRNA-423-5p/nucleus accumbens associated 1 axis. Bioengineered. (2022) 13:2114–29. doi: 10.1080/21655979.2021.2023798
109. Xie J, Wang JJ, Li YJ, Wu J, Gu XJ, Yang XR. LncRNA GAS5 suppresses colorectal cancer progress by target miR-21/LIFR axis. Evidence-Based complementary Altern medicine: eCAM. (2022) 2022:3298939. doi: 10.1155/2022/3298939
110. Xiong BH, Li SS, Ren ZY, Zhang Z, Liu YZ, Sun Y, et al. [Inhibition of GAS5 promoted invasion, migration and epithelial-mesenchymal transition of colorectal cancer cells via miR-21/PTEN/Akt axis]. Zhonghua zhong liu za zhi [Chinese J oncology]. (2022) 44:1168–74. doi: 10.3760/cma.j.cn112152-20200321-00243
111. Chen Q, Zhou L, Ma D, Hou J, Lin Y, Wu J, et al. LncRNA GAS6-AS1 facilitates tumorigenesis and metastasis of colorectal cancer by regulating TRIM14 through miR-370-3p/miR-1296-5p and FUS. J Trans Med. (2022) 20:356. doi: 10.1186/s12967-022-03550-0
112. Matsumura K, Kawasaki Y, Miyamoto M, Kamoshida Y, Nakamura J, Negishi L, et al. The novel G-quadruplex-containing long non-coding RNA GSEC antagonizes DHX36 and modulates colon cancer cell migration. Oncogene. (2017) 36:1191–9. doi: 10.1038/onc.2016.282
113. Chen S, Bu D, Ma Y, Zhu J, Chen G, Sun L, et al. H19 overexpression induces resistance to 1,25(OH)2D3 by targeting VDR through miR-675-5p in Colon Cancer cells. Neoplasia (New York NY). (2017) 19(3):226–36. doi: 10.1016/j.neo.2016.10.007
114. Zhang R, Zhang X, Zhang W, Cui W, Xiao Y, Liu L, et al. Sohlh2 regulates the stemness and differentiation of colon cancer stem cells by downregulating lncRNA-H19 transcription. Mol Cancer research: MCR. (2023) 21:115–26. doi: 10.1158/1541-7786.MCR-22-0134
115. Yi C, Zhao C, Peng C. Long non-coding RNA HAND2 antisense RNA 1 inhibits colorectal cancer development by sponging microRNA-3118/leptin receptor axis. Ann Clin Lab science. (2023) 53(1):82–93.
116. Guo S, Song B, Li L, Li H, Yang T, Cao L, et al. lncRNA HCG11 Promotes Colorectal Cancer Cell Malignant Behaviors via Sponging miR-26b-5p. J Immunol Res. (2023) 2023:9011232. doi: 10.1155/2023/9011232
117. Cui Z, Wang Q, Deng MH, Han QL. LncRNA HCG11 promotes 5-FU resistance of colon cancer cells through reprogramming glucose metabolism by targeting the miR-144-3p-PDK4 axis. Cancer biomarkers: section A Dis markers. (2022) 34:41–53. doi: 10.3233/CBM-210212
118. Fang C, Qiu S, Sun F, Li W, Wang Z, Yue B, et al. Long non-coding RNA HNF1A-AS1 mediated repression of miR-34a/SIRT1/p53 feedback loop promotes the metastatic progression of colon cancer by functioning as a competing endogenous RNA. Cancer letters. (2017) 410:50–62. doi: 10.1016/j.canlet.2017.09.012
119. Weng X, Liu H, Ruan J, Du M, Wang L, Mao J, et al. HOTAIR/miR-1277-5p/ZEB1 axis mediates hypoxia-induced oxaliplatin resistance via regulating epithelial-mesenchymal transition in colorectal cancer. Cell Death discovery. (2022) 8:310. doi: 10.1038/s41420-022-01096-0
120. Huang Y, Wang L, Liu D. HOTAIR regulates colorectal cancer stem cell properties and promotes tumorigenicity by sponging miR-211-5p and modulating FLT-1. Cell Cycle (Georgetown Tex). (2021) 20:1999–2009. doi: 10.1080/15384101.2021.1962636
121. Luo ZF, Zhao D, Li XQ, Cui YX, Ma N, Lu CX, et al. Clinical significance of HOTAIR expression in colon cancer. World J gastroenterology. (2016) 22:5254–9. doi: 10.3748/wjg.v22.i22.5254
122. Wu ZH, Wang XL, Tang HM, Jiang T, Chen J, Lu S, et al. Long non-coding RNA HOTAIR is a powerful predictor of metastasis and poor prognosis and is associated with epithelial-mesenchymal transition in colon cancer. Oncol Rep. (2014) 32:395–402. doi: 10.3892/or.2014.3186
123. Tatangelo F, Di Mauro A, Scognamiglio G, Aquino G, Lettiero A, Delrio P, et al. Posterior HOX genes and HOTAIR expression in the proximal and distal colon cancer pathogenesis. J Trans Med. (2018) 16:350. doi: 10.1186/s12967-018-1725-y
124. Jin L, Pan YL, Zhang J, Cao PG. LncRNA HOTAIR recruits SNAIL to inhibit the transcription of HNF4α and promote the viability, migration, invasion and EMT of colorectal cancer. Trans Oncol. (2021) 14:101036. doi: 10.1016/j.tranon.2021.101036
125. Deng Q, Yang J, Chen Y, Chen Z, Li J, Fu Z. LncRNA HOXB-AS4 promotes proliferation and migration of colorectal cancer via the miR-140-5p/hdac7 axis. Biotechnol Genet Eng Rev. (2023) 23:1–19. doi: 10.1080/02648725.2023.2193465
126. Zhang TT, Chen HP, Yu SY, Zhao SP. LncRNA HOXC-AS3 overexpression inhibits TGF-β2-induced colorectal cancer cell migration and invasion by sponging miR-1269. Hum Exp toxicology. (2022) 41:9603271221093630. doi: 10.1177/09603271221093630
127. Liu S, Wang X, Zhang J, Liu Y, Li B. LncRNA IGFL2-AS1 as a ceRNA Promotes HCT116 Cell Malignant Proliferation via the miR-433-3p/PAK4 Axis. Turkish J gastroenterology: Off J Turkish Soc Gastroenterology. (2023) 34:497–507. doi: 10.5152/tjg.2023.21998
128. Qin M, Liu Q, Yang W, Wang Q, Xiang Z. IGFL2-AS1-induced suppression of HIF-1α degradation promotes cell proliferation and invasion in colorectal cancer by upregulating CA9. Cancer Med. (2023) 12:8415–32. doi: 10.1002/cam4.5562
129. Zhang C, Wang Y. KCNQ1OT1 polymorphism rs35622507 and methylation status of KCNQ1OT1 promoter influence the drug resistance to L-OHP. Aging. (2022) 14:1836–47. doi: 10.18632/aging.v14i4
130. Fang J, Yang J, Chen H, Sun W, Xiang L, Feng J. Long non-coding RNA LBX2-AS1 predicts poor survival of colon cancer patients and promotes its progression via regulating miR-627-5p/RAC1/PI3K/AKT pathway. Hum Cell. (2022) 35:1521–34. doi: 10.1007/s13577-022-00745-x
131. Qi Y, Shan Y, Li S, Huang Y, Guo Y, Huang T, et al. LncRNA LEF1-AS1/LEF1/FUT8 axis mediates colorectal cancer progression by regulating α1, 6-fucosylationvia wnt/β-catenin pathway. Digestive Dis Sci. (2022) 67:2182–94. doi: 10.1007/s10620-021-07051-w
132. Liang W, Liu D, Wu J. c-JUN-induced upregulation of LINC00174 contributes to colorectal cancer proliferation and invasion through accelerating USP21 expression. Cell Biol Int. (2023) 47:1782–98. doi: 10.1002/cbin.12069
133. Wang ZK, Yang L, Wu LL, Mao H, Zhou YH, Zhang PF, et al. Long non-coding RNA LINC00261 sensitizes human colon cancer cells to cisplatin therapy. Braz J Med Biol Res = Rev Bras pesquisas medicas e biologicas. (2017) 51:e6793. doi: 10.1590/1414-431x20176793
134. Li S, Lv C, Li J, Xie T, Liu X, Zheng Z, et al. LncRNA LINC00473 promoted colorectal cancer cell proliferation and invasion by targeting miR-195 expression. Am J Trans Res. (2021) 13(6):6066–75.
135. Li C, Pan B, Liu X, Qin J, Wang X, He B, et al. Long intergenic non-coding RNA LINC00485 exerts tumor-suppressive activity by regulating miR-581/EDEM1 axis in colorectal cancer. Aging. (2021) 13:3866–85. doi: 10.18632/aging.v13i3
136. Zheng J, Dou R, Zhang X, Zhong B, Fang C, Xu Q, et al. LINC00543 promotes colorectal cancer metastasis by driving EMT and inducing the M2 polarization of tumor associated macrophages. J Trans Med. (2023) 21:153. doi: 10.1186/s12967-023-04009-6
137. Liu F, Ma X, Bian X, Zhang C, Liu X, Liu Q. LINC00586 represses ASXL1 expression thus inducing epithelial-to-mesenchymal transition of colorectal cancer cells through LSD1-mediated H3K4me2 demethylation. Front Pharmacol. (2022) 13:887822. doi: 10.3389/fphar.2022.887822
138. Han T, Gao M, Wang X, Li W, Zhuo J, Qu Z, et al. LINC00665 activates Wnt/β-catenin signaling pathway to facilitate tumor progression of colorectal cancer via upregulating CTNNB1. Exp Mol pathology. (2021) 120:104639. doi: 10.1016/j.yexmp.2021.104639
139. Nan S, Zhang S, Jin R, Wang J. LINC00665 up-regulates SIN3A expression to modulate the progression of colorectal cancer via sponging miR-138-5p. Cancer Cell Int. (2022) 22:51. doi: 10.1186/s12935-021-02176-4
140. Sun P, Luan Y, Cai X, Liu Q, Ren P, Peng P, et al. LINC00858 facilitates formation of hepatic metastases from colorectal cancer via regulating the miR-132-3p/IGF2BP1 axis. Biol Chem. (2023) 405(2):129–41. doi: 10.1515/hsz-2022-0328
141. Song B, Li H, Guo S, Yang T, Li L, Cao L, et al. LINC00882 plays a tumor-promoter role in colorectal cancer by targeting miR-3619-5p to up-regulate CTNNB1. Arch Med Res. (2022) 53:29–36. doi: 10.1016/j.arcmed.2021.06.001
142. Zhou L, Mu D, Chen Y. LINC00958 targets miR-145-3p/CDK1 axis to aggravate the Malignancy of colon cancer. Ann Clin Lab science. (2022) 52(5):695–706.
143. Liang H, Zhao Q, Zhu Z, Zhang C, Zhang H. Long noncoding RNA LINC00958 suppresses apoptosis and radiosensitivity of colorectal cancer through targeting miR-422a. Cancer Cell Int. (2021) 21:477. doi: 10.1186/s12935-021-02188-0
144. Wu H, Ding X, Hu X, Zhao Q, Chen Q, Sun T, et al. LINC01021 maintains tumorigenicity by enhancing N6-methyladenosine reader IMP2 dependent stabilization of MSX1 and JARID2: implication in colorectal cancer. Oncogene. (2022) 41:1959–73. doi: 10.1038/s41388-022-02189-x
145. Li C, Pan B, Wang X, Liu X, Qin J, Gao T, et al. Upregulated LINC01088 facilitates Malignant phenotypes and immune escape of colorectal cancer by regulating microRNAs/G3BP1/PD-L1 axis. J Cancer Res Clin Oncol. (2022) 148:1965–82. doi: 10.1007/s00432-022-03981-8
146. Chen L, Chen W, Zhao C, Jiang Q. LINC01224 promotes colorectal cancer progression by sponging miR-2467. Cancer Manage Res. (2021) 13:733–42. doi: 10.2147/CMAR.S281625
147. Gu J, Dong L, Wang Y, Nie W, Liu W, Zhao JA. LINC01224 promotes colorectal cancer progression through targeting miR-485-5p/MYO6 axis. World J Surg Oncol. (2021) 19:281. doi: 10.1186/s12957-021-02389-x
148. Yuan Y, Long Z. LncRNA LINC01232 enhances proliferation, angiogenesis, migration and invasion of colon adenocarcinoma cells by downregulating miR-181a-5p. J Microbiol Biotechnol. (2023) 33:398–409. doi: 10.4014/jmb.2206.06032
149. Fu D, Ren Y, Wang C, Yu L, Yu R. LINC01287 facilitates proliferation, migration, invasion and EMT of colon cancer cells via miR-4500/MAP3K13 pathway. BMC cancer. (2021) 21:782. doi: 10.1186/s12885-021-08528-7
150. Li Y, Wang W, Wu M, Zhu P, Zhou Z, Gong Y, et al. LncRNA LINC01315 silencing modulates cancer stem cell properties and epithelial-to-mesenchymal transition in colorectal cancer via miR-484/DLK1 axis. Cell Cycle (Georgetown Tex). (2022) 21:851–73. doi: 10.1080/15384101.2022.2033415
151. Liang R, Zhang J, Zhang RM, Qiu H. LINC01315 silencing inhibits the aggressive phenotypes of colorectal carcinoma by sponging miR-205-3p. Biochem Biophys Res Commun. (2021) 534:1033–9. doi: 10.1016/j.bbrc.2020.10.041
152. Liu Y, Zhou WL. LINC01315 accelerates the growth and epithelial-mesenchymal transition of colorectal cancer cells via activating the Wnt/β-catenin signal. Bioengineered. (2022) 13:8396–406. doi: 10.1080/21655979.2022.2044275
153. Li H, Dong W, Hou J, He D. LINC 01436 is overexpressed in colorectal cancer and promotes cancer cell proliferation by suppressing tumor-suppressive miR-466 maturation. In Vitro Cell Dev Biol Animal. (2022) 58:109–15. doi: 10.1007/s11626-021-00642-x
154. Liu J, Zhan Y, Wang J, Wang J, Guo J, Kong D. Long noncoding RNA LINC01578 drives colon cancer metastasis through a positive feedback loop with the NF-κB/YY1 axis. Mol Oncol. (2020) 14:3211–33. doi: 10.1002/1878-0261.12819
155. Luo Y, Huang S, Wei J, Zhou H, Wang W, Yang J, et al. Long noncoding RNA LINC01606 protects colon cancer cells from ferroptotic cell death and promotes stemness by SCD1-Wnt/β-catenin-TFE3 feedback loop signalling. Clin Trans Med. (2022) 12:e752. doi: 10.1002/ctm2.752
156. Xu Z, Yu Y, Ni H, Sun W, Kuang Y. LINC01836 Promotes Colorectal Cancer Progression and Functions as ceRNA to Target SLC17A9 by Sponging miR-1226-3p. Protein Pept Lett. (2024) 31(1):43–60. doi: 10.2174/0109298665248028231122064831
157. Liu W, Zhang Z, Luo X, Qian K, Huang B, Liang J, et al. m(6)A−mediated LINC02038 inhibits colorectal cancer progression via regulation of the FAM172A/PI3K/AKT pathway via competitive binding with miR−552−5p. Int J Oncol. (2023) 63(1):81. doi: 10.3892/ijo.2023.5529
158. Wei Z, Zhou J, Yu H, Pu Y, Cheng Y, Zhang Y, et al. Zuo jin wan reverses the resistance of colorectal cancer to oxaliplatin by regulating the MALAT1/miR-200s/JNK signaling pathway. Evidence-Based complementary Altern medicine: eCAM. (2022) 2022:3032407. doi: 10.1155/2022/3032407
159. Yang T, Chen WC, Shi PC, Liu MR, Jiang T, Song H, et al. Correction: Long noncoding RNA MAPKAPK5-AS1 promotes colorectal cancer progression by cis-regulating the nearby gene MK5 and acting as a let-7f-1-3p sponge. J Exp Clin Cancer research: CR. (2022) 41:341. doi: 10.1186/s13046-022-02537-5
160. Chen WS, Zhang X, Zhao ZF, Che XM. MBNL1−AS1 attenuates tumor cell proliferation by regulating the miR−29c−3p/BVES signal in colorectal cancer. Oncol Rep. (2023) 50(4):191. doi: 10.3892/or.2023.8628
161. Cai M, Hu W, Huang C, Zhou C, Li J, Chen Y, et al. lncRNA MCF2L-AS1/miR-105/ IL-1β Axis regulates colorectal cancer cell oxaliplatin resistance. Cancer Manage Res. (2021) 13:8685–94. doi: 10.2147/CMAR.S313905
162. Dai W, Zeng W, Lee D. lncRNA MCM3AP-AS1 inhibits the progression of colorectal cancer via the miR-19a-3p/FOXF2 axis. J Gene Med. (2021) 23:e3306. doi: 10.1002/jgm.3306
163. Zhou L, Li J, Liao M, Zhang Q, Yang M. LncRNA MIR155HG induces M2 macrophage polarization and drug resistance of colorectal cancer cells by regulating ANXA2. Cancer immunology immunotherapy: CII. (2022) 71:1075–91. doi: 10.1007/s00262-021-03055-7
164. Tang G, Wu D, Guo M, Li H. Correction to: LncRNA MIR497HG inhibits colorectal cancer progression by the miR-3918/ACTG2 axis. J Genet. (2022) 101:53. doi: 10.1007/s12041-022-01403-9
165. Huang S, Sun Y. Long noncoding RNA MNX1-AS1 functions as a competing endogenous RNA to regulate epithelial-mesenchymal transition by sponging MiR-744-5p in colorectal cancer. Bioscience biotechnology Biochem. (2021) 85:568–78. doi: 10.1093/bbb/zbaa096
166. Sun Q, Gui Z, Zhao Z, Xu W, Zhu J, Gao C, et al. Overexpression of lncRNA MNX1-AS1/PPFIA4 activates AKT/HIF-1α Signal pathway to promote stemness of colorectal adenocarcinoma cells. J Oncol. (2022) 2022:8303409. doi: 10.1155/2022/8303409
167. Wu H, Dong D, Wang J, Yin S, Gong Y, Yang C, et al. LncRNA NEAT1 Promotes the Malignant Progression of Colorectal Cancer by Targeting ZEB1 via miR-448. Technol Cancer Res Treat. (2022) 21:15330338221085348. doi: 10.1177/15330338221085348
168. Zhu Y, Wang X, Zheng L, Li D, Liu Z, Teng L. The lncRNA NEAT1 Inhibits miRNA-216b and Promotes Colorectal Cancer Progression by Indirectly Activating YY1. J Oncol. (2022) 2022:8130132. doi: 10.1155/2022/8130132
169. Shi CJ, Xue ZH, Zeng WQ, Deng LQ, Pang FX, Zhang FW, et al. LncRNA-NEF suppressed oxaliplatin resistance and epithelial-mesenchymal transition in colorectal cancer through epigenetically inactivating MEK/ERK signaling. Cancer Gene Ther. (2023) 30:855–65. doi: 10.1038/s41417-023-00595-1
170. Zhang Z, Ye B, Lin Y, Liu W, Deng J, Ji W. LncRNA OTUD6B-AS1 overexpression promoted GPX4-mediated ferroptosis to suppress radioresistance in colorectal cancer. Clin Trans oncology: Off Publ Fed Spanish Oncol Societies Natl Cancer Institute Mexico. (2023) 25:3217–29. doi: 10.1007/s12094-023-03193-7
171. Cai Y, Li Y, Shi C, Zhang Z, Xu J, Sun B. LncRNA OTUD6B-AS1 inhibits many cellular processes in colorectal cancer by sponging miR-21-5p and regulating PNRC2. Hum Exp toxicology. (2021) 40:1463–73. doi: 10.1177/0960327121997976
172. Fang X, Xu Y, Li K, Liu P, Zhang H, Jiang Y, et al. Exosomal lncRNA PCAT1 Promotes Tumor Circulating Cell-Mediated Colorectal Cancer Liver Metastasis by Regulating the Activity of the miR-329-3p/Netrin-1-CD146 Complex. J Immunol Res. (2022) 2022:9916228. doi: 10.1155/2022/9916228
173. Guan B, Chen F, Wu Z, Wang C, Yang J. lncRNA PCGEM1 Regulates the Progress of Colorectal Cancer through Targeting miR-129-5p/SOX4. Dis markers. (2022) 2022:2876170. doi: 10.1155/2022/2876170
174. Xu W, Wu L, Lu H, Xiang X, Wang F, Li S. LncRNA PCGEM1 promotes colorectal cancer cell proliferation and migration in positive feedback loop through PCGEM1/miR-433-3p/CTCF axis. Pathology Res practice. (2022) 237:154017. doi: 10.1016/j.prp.2022.154017
175. Li JS, Liu TM, Li L, Jiang C. LncRNA PROX1 antisense RNA 1 promotes PD-L1-mediated proliferation, metastasis, and immune escape in colorectal cancer by interacting with miR-520d. Anti-cancer Drugs. (2023) 34:669–79. doi: 10.1097/CAD.0000000000001437
176. Liu J, Zhan W, Chen G, Yan S, Chen W, Li R. SP1-induced PROX1-AS1 contributes to tumor progression by regulating miR-326/FBXL20 axis in colorectal cancer. Cell signalling. (2023) 101:110503. doi: 10.1016/j.cellsig.2022.110503
177. Zhao F, Wang M, Zhang Y, Su R, He C, Gao X, et al. lncRNA PSMB8-AS1 promotes colorectal cancer progression through sponging miR-1299 to upregulate ADAMTS5. Neoplasma. (2022) 69:1138–53. doi: 10.4149/neo_2022_220111N42
178. Zhao A, Wang Y, Lin F, Bai K, Gu C. Long noncoding RNA LBX2-AS1 promotes colorectal cancer progression via binding with PTBP1 and stabilizing KAT2A expression. J Biochem Mol toxicology. (2022) 36:e23020. doi: 10.1002/jbt.23020
179. Yin H, Gu S, Li G, Yu H, Zhang X, Zuo Y. Long noncoding RNA PVT1 predicts poor prognosis and promotes the progression of colorectal cancer through the miR-24-3p/NRP1 axis in zebrafish xenografts. Neoplasma. (2023) 70:500–13. doi: 10.4149/neo_2023_221208N1169
180. Yu P, Zhang J, Zhu A, Kong W, Shen X. LncRNA PVT1 Regulates miR-1207-5p to Affect Colon Cancer Proliferation and Migration via the Wnt6/β-catenin2 Pathway. Genet testing Mol biomarkers. (2022) 26:307–15. doi: 10.1089/gtmb.2021.0259
181. Liu Y, Wu Y, Zhu Z, Gong J, Dou W. Knockdown of lncRNA PVT1 inhibits the proliferation and accelerates the apoptosis of colorectal cancer cells via the miR−761/MAPK1 axis. Mol Med Rep. (2021) 24(5):794. doi: 10.3892/mmr.2021.12434
182. Yu X, Zhao J, He Y. Long non-coding RNA PVT1 functions as an oncogene in human colon cancer through miR-30d-5p/RUNX2 axis. J BUON: Off J Balkan Union Oncol. (2018) 23:48–54.
183. Li C, Wang P, Du J, Chen J, Liu W, Ye K. LncRNA RAD51-AS1/miR-29b/c-3p/NDRG2 crosstalk repressed proliferation, invasion and glycolysis of colorectal cancer. IUBMB Life. (2021) 73:286–98. doi: 10.1002/iub.2427
184. Huang W, Zhang H, Tian Y, Li Y, Li J, Zhong X, et al. LncRNA SNHG11 enhances bevacizumab resistance in colorectal cancer by mediating miR-1207-5p/ABCC1 axis. Anti-cancer Drugs. (2022) 33:575–86. doi: 10.1097/CAD.0000000000001289
185. Li M, Sun S, Bian Z, Yao S, Liu M, You X, et al. SNHG15 promotes chemoresistance and glycolysis in colorectal cancer. Pathology Res practice. (2023) 246:154480. doi: 10.1016/j.prp.2023.154480
186. Jiang H, Li T, Qu Y, Wang X, Li B, Song J, et al. Long non-coding RNA SNHG15 interacts with and stabilizes transcription factor slug and promotes colon cancer progression. Cancer Lett. (2018) 425:78–87. doi: 10.1016/j.canlet.2018.03.038
187. Chen ZY, Wang XY, Yang YM, Wu MH, Yang L, Jiang DT, et al. LncRNA SNHG16 promotes colorectal cancer cell proliferation, migration, and epithelial-mesenchymal transition through miR-124-3p/MCP-1. Gene Ther. (2022) 29:193–205. doi: 10.1038/s41434-020-0176-2
188. Tan P, Xu M, Nie J, Qin J, Liu X, Sun H, et al. LncRNA SNHG16 promotes colorectal cancer proliferation by regulating ABCB1 expression through sponging miR-214-3p. J Biomed Res. (2022) 36:231–41. doi: 10.7555/JBR.36.20220049
189. Xiang Z, Huang G, Wu H, He Q, Yang C, Dou R, et al. SNHG16 upregulation-induced positive feedback loop with YAP1/TEAD1 complex in Colorectal Cancer cell lines facilitates liver metastasis of colorectal cancer by modulating CTCs epithelial-mesenchymal transition. Int J Biol Sci. (2022) 18:5291–308. doi: 10.7150/ijbs.73438
190. Liu J, Zhan Y, Wang J, Guo J, Kong D. lncRNA-SNHG17 promotes colon adenocarcinoma progression and serves as a sponge for miR-375 to regulate CBX3 expression. Am J Trans Res. (2020) 12(9):5283–95.
191. Islam Khan MZ, Law HKW. Suppression of small nucleolar RNA host gene 8 (SNHG8) inhibits the progression of colorectal cancer cells. Non-coding RNA Res. (2023) 8:224–32. doi: 10.1016/j.ncrna.2023.02.003
192. Feng Y, Xu Y, Gao Y, Chen Y, Wang X, Chen Z. A novel lncRNA SOX2OT promotes the Malignancy of human colorectal cancer by interacting with miR-194-5p/SOX5 axis. Cell Death disease. (2021) 12:499. doi: 10.1038/s41419-021-03756-y
193. Fang Y, Yang Q. Specificity protein 1-induced serine peptidase inhibitor, Kunitz Type 1 antisense RNA1 regulates colorectal cancer cell proliferation, migration, invasion and apoptosis through targeting heparin binding growth factor via sponging microRNA-214. Bioengineered. (2022) 13:3309–22. doi: 10.1080/21655979.2022.2026859
194. Sui X, Hu N, Wang P, Wang Y, Meng C, Zhang Z. LncRNA SPINT1-AS1/miR-433-3p/E2F3 positive feedback loop promotes the KRAS-mutant colorectal cancer cell proliferation, migration and invasion. Pathology Res practice. (2022) 239:154064. doi: 10.1016/j.prp.2022.154064
195. Li J, Ji Y, Chen N, Wang H, Fang C, Yin X, et al. A specific upregulated long noncoding RNA in colorectal cancer promotes cancer progression. JCI Insight. (2022) 7(15):e158855. doi: 10.1172/jci.insight.158855
196. Hong Q, Mai P, Wu B, Wang H, Xiao M, You J. Long non-coding RNA TDRG1 aggravates colorectal cancer stemness by binding with miR-873-5p to upregulate PRKAR2. Environ toxicology. (2022) 37:2366–74. doi: 10.1002/tox.23602
197. Xia C, Li Q, Cheng X, Wu T, Gao P, Gu Y. Insulin-like growth factor 2 mRNA-binding protein 2-stabilized long non-coding RNA Taurine up-regulated gene 1 (TUG1) promotes cisplatin-resistance of colorectal cancer via modulating autophagy. Bioengineered. (2022) 13:2450–69. doi: 10.1080/21655979.2021.2012918
198. Liu Q, Zhang W, Luo L, Han K, Liu R, Wei S, et al. Long noncoding RNA TUG1 regulates the progression of colorectal cancer through miR-542-3p/TRIB2 axis and Wnt/β-catenin pathway. Diagn pathology. (2021) 16:47. doi: 10.1186/s13000-021-01101-7
199. Tian L, Zhao ZF, Xie L, Zhu JP. Taurine up-regulated 1 accelerates tumorigenesis of colon cancer by regulating miR-26a-5p/MMP14/p38 MAPK/Hsp27 axis in vitro and in vivo. Life Sci. (2019) 239:117035. doi: 10.1016/j.lfs.2019.117035
200. Tang Y, Cai J, Lv B. LncRNA ubiquitin-binding protein domain protein 10 antisense RNA 1 inhibits colon adenocarcinoma progression via the miR-515-5p/slit guidance ligand 3 axis. Bioengineered. (2022) 13:2308–20. doi: 10.1080/21655979.2021.2024396
201. Yuan HH, Zhang XC, Wei XL, Zhang WJ, Du XX, Huang P, et al. LncRNA UCA1 mediates Cetuximab resistance in Colorectal Cancer via the MiR-495 and HGF/c-MET Pathways. J Cancer. (2022) 13:253–67. doi: 10.7150/jca.65687
202. Liu SJ, Li ZQ, Wang XY, Liu F, Xiao ZM, Zhang DC. lncRNA UCA1 induced by SP1 and SP3 forms a positive feedback loop to facilitate Malignant phenotypes of colorectal cancer via targeting miR-495. Life Sci. (2021) 277:119569. doi: 10.1016/j.lfs.2021.119569
203. Li C, Liang X, Liu Y. lncRNA USP30-AS1 sponges miR-765 and modulates the progression of colon cancer. World J Surg Oncol. (2022) 20:73. doi: 10.1186/s12957-022-02529-x
204. Yan Z, Li J, Guo J, He R, Xing J. LncRNA XIST sponges microRNA-448 to promote Malignant behaviors of colorectal cancer cells via regulating GRHL2. Funct Integr Genomics. (2022) 22:977–88. doi: 10.1007/s10142-022-00873-5
205. Li W, He Y, Cheng Z. Long noncoding RNA XIST knockdown suppresses the growth of colorectal cancer cells via regulating microRNA-338-3p/PAX5 axis. Eur J Cancer prevention: Off J Eur Cancer Prev Organisation (ECP). (2021) 30:132–42. doi: 10.1097/CEJ.0000000000000596
206. Sun N, Zhang G, Liu Y. Long non-coding RNA XIST sponges miR-34a to promotes colon cancer progression via Wnt/β-catenin signaling pathway. Gene. (2018) 665:141–8. doi: 10.1016/j.gene.2018.04.014
207. Ma T, Qiao T, Huang R, Wang M, Hu H, Yuan Z, et al. Long Noncoding XLOC_006390 Regulates the Proliferation and Metastasis of Human Colorectal Cancer via miR-296/ONECUT2 Axis. J Oncol. (2022) 2022:4897201. doi: 10.1155/2022/4897201
208. Fang C, Zan J, Yue B, Liu C, He C, Yan D. Long non-coding ribonucleic acid zinc finger antisense 1 promotes the progression of colonic cancer by modulating ZEB1 expression. J Gastroenterol hepatology. (2017) 32:1204–11. doi: 10.1111/jgh.13646
209. Deng H, Wang M, Xu Q, Yao H. ZFAS1 promotes colorectal cancer metastasis through modulating miR-34b/SOX4 targeting. Cell Biochem biophysics. (2021) 79:387–96. doi: 10.1007/s12013-021-00976-z
210. Wang JJ, Li XM, He L, Zhong SZ, Peng YX, Ji N. Expression and function of long non-coding RNA CASC19 in colorectal cancer. Zhongguo yi xue ke xue yuan xue bao Acta Academiae Medicinae Sinicae. (2017) 39:756–61. doi: 10.3881/j.issn.1000-503X.2017.06.004
211. Yu Y, Nangia-Makker P, Farhana L, Majumdar APN. A novel mechanism of lncRNA and miRNA interaction: CCAT2 regulates miR-145 expression by suppressing its maturation process in colon cancer cells. Mol cancer. (2017) 16:155. doi: 10.1186/s12943-017-0725-5
212. Zhang Y, Peng C, Li J, Zhang D, Zhang C, Jin K, et al. Long non-coding RNA CCDC144NL-AS1 promotes cell proliferation by regulating the miR-363-3p/GALNT7 axis in colorectal cancer. J Cancer. (2022) 13:752–63. doi: 10.7150/jca.65885
213. Zhuang ST, Cai YJ, Gao HC, Qiu JF, Zeng L, Zheng WJ. Study on the function and mechanism of long non-coding RNA DMTF1v4 in the occurrence of colon cancer. Eur Rev Med Pharmacol Sci. (2018) 22:3779–88. doi: 10.26355/eurrev_201806_15260
214. Li C, Hong S, Hu H, Liu T, Yan G, Sun D. MYC-induced upregulation of lncrna ELFN1-AS1 contributes to tumor growth in colorectal cancer via epigenetically silencing TPM1. Mol Cancer research: MCR. (2022) 20:1697–708. doi: 10.1158/1541-7786.MCR-22-0009
215. Wu F, Zhang W, Wei H, Ma H, Leng G, Zhang Y. lncRNA ELFN1-AS1 promotes proliferation, migration and invasion and suppresses apoptosis in colorectal cancer cells by enhancing G6PD activity. Acta Biochim Biophys Sinica. (2023) 55:649–60. doi: 10.3724/abbs.2023010
216. Pan Y, Zhu Y, Zhang J, Jin L, Cao P. A feedback loop between GATA2-AS1 and GATA2 promotes colorectal cancer cell proliferation, invasion, epithelial-mesenchymal transition and stemness via recruiting DDX3X. J Trans Med. (2022) 20:287. doi: 10.1186/s12967-022-03483-8
217. Bai Y, Li L, Zhang Z. Linc00883 affects colorectal cancer through miR-577/FKBP14 axis: a novel mechanism for regulating colorectal cancer cell proliferation, invasion, and migration. Cell Cycle (Georgetown Tex). (2022) 21:2403–16. doi: 10.1080/15384101.2022.2097824
218. Tang G, Wu D, Guo M, Li H. LncRNA MIR497HG inhibits colorectal cancer progression by the miR-3918/ACTG2 axis. J Genet. (2022) 101:53. doi: 10.1007/s12041-022-01367-w
219. Zhang C, Zhang P, Liu J, Song H, Zhou X, Liu X. LncRNA RPL34-AS1 sponges miR-3656 to suppress cell proliferation in colorectal cancer. In Vitro Cell Dev Biol Animal. (2022) 58:462–70. doi: 10.1007/s11626-022-00686-7
221. Carneiro BA, El-Deiry WS. Targeting apoptosis in cancer therapy. Nat Rev Clin Oncol. (2020) 17:395–417. doi: 10.1038/s41571-020-0341-y
222. Liu HZ, Shan TD, Han Y, Liu XS. Silencing long non-coding RNA CASC9 inhibits colorectal cancer cell proliferation by acting as a competing endogenous RNA of miR-576-5p to regulate AKT3. Cell Death discovery. (2020) 6:115. doi: 10.1038/s41420-020-00352-5
223. Malki A, ElRuz RA, Gupta I, Allouch A, Vranic S, Al Moustafa AE. Molecular mechanisms of colon cancer progression and metastasis: recent insights and advancements. Int J Mol Sci. (2020) 22(1):130. doi: 10.3390/ijms22010130
224. Hou S, Zhao Y, Chen J, Lin Y, Qi X. Tumor-associated macrophages in colorectal cancer metastasis: molecular insights and translational perspectives. J Trans Med. (2024) 22:62. doi: 10.1186/s12967-024-04856-x
225. Bresalier RS, Grady WM, Markowitz SD, Nielsen HJ, Batra SK, Lampe PD. Biomarkers for early detection of colorectal cancer: the early detection research network, a framework for clinical translation. Cancer epidemiology Biomarkers prevention: Publ Am Assoc Cancer Research cosponsored by Am Soc Prev Oncol. (2020) 29:2431–40. doi: 10.1158/1055-9965.EPI-20-0234
226. Dohrn N, Klein MF. Colorectal cancer: current management and future perspectives. Br J Surgery. (2023) 110:1256–9. doi: 10.1093/bjs/znad095
227. Shinji S, Yamada T, Matsuda A, Sonoda H, Ohta R, Iwai T, et al. Recent advances in the treatment of colorectal cancer: A review. J Nippon Med Sch. (2022) 89:246–54. doi: 10.1272/jnms.JNMS.2022_89-310
228. Ohishi T, Kaneko MK, Yoshida Y, Takashima A, Kato Y, Kawada M. Current targeted therapy for metastatic colorectal cancer. Int J Mol Sci. (2023) 24(2):1702. doi: 10.3390/ijms24021702
229. Xue WH, Li XW, Ding YQ, Wu N, Pei BB, Ma XY, et al. Efficacy and safety of third-line or later-line targeted treatment for patients with metastatic colorectal cancer: a meta-analysis. Front Oncol. (2023) 13:1165040. doi: 10.3389/fonc.2023.1165040
230. Hossain MS, Karuniawati H, Jairoun AA, Urbi Z, Ooi J, John A, et al. Colorectal cancer: A review of carcinogenesis, global epidemiology, current challenges, risk factors, preventive and treatment strategies. Cancers. (2022) 14(7):1732. doi: 10.3390/cancers14071732
231. Shan C, Zhang Y, Hao X, Gao J, Chen X, Wang K. Biogenesis, functions and clinical significance of circRNAs in gastric cancer. Mol cancer. (2019) 18:136. doi: 10.1186/s12943-019-1069-0
232. Hernandez Dominguez O, Yilmaz S, Steele SR. Stage IV colorectal cancer management and treatment. J Clin Med. (2023) 12(5):2072. doi: 10.3390/jcm12052072
233. Wang B, Tang D, Zhang Z, Wang Z. Identification of aberrantly expressed lncRNA and the associated TF-mRNA network in hepatocellular carcinoma. J Cell Biochem. (2020) 121:1491–503. doi: 10.1002/jcb.29384
234. Kerachian MA, Azghandi M. Identification of long non-coding RNA using single nucleotide epimutation analysis: a novel gene discovery approach. Cancer Cell Int. (2022) 22:337. doi: 10.1186/s12935-022-02752-2
235. Li M, Guo D, Chen X, Lu X, Huang X, Wu Y. Transcriptome profiling and co-expression network analysis of lncRNAs and mRNAs in colorectal cancer by RNA sequencing. BMC cancer. (2022) 22:780. doi: 10.1186/s12885-022-09878-6
236. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. (2012) 9:703–19. doi: 10.4161/rna.20481
237. Statello L, Guo CJ, Chen LL, Huarte M. Gene regulation by long non-coding RNAs and its biological functions. Nat Rev Mol Cell Biol. (2021) 22:96–118. doi: 10.1038/s41580-020-00315-9
238. TaNiue K, Akimitsu N. The functions and unique features of lncRNAs in cancer development and tumorigenesis. Int J Mol Sci. (2021) 22(2):632. doi: 10.3390/ijms22020632
239. Wang L, Bitar M, Lu X, Jacquelin S, Nair S, Sivakumaran H, et al. CRISPR-Cas13d screens identify KILR, a breast cancer risk-associated lncRNA that regulates DNA replication and repair. Mol cancer. (2024) 23:101. doi: 10.1186/s12943-024-02021-y
Keywords: lncRNA, colorectal cancer, biomarker, functions, mechanisms
Citation: Lin Y, Zhao W, Lv Z, Xie H, Li Y and Zhang Z (2024) The functions and mechanisms of long non-coding RNA in colorectal cancer. Front. Oncol. 14:1419972. doi: 10.3389/fonc.2024.1419972
Received: 19 April 2024; Accepted: 18 June 2024;
Published: 04 July 2024.
Edited by:
José Díaz-Chávez, Instituto Nacional de Cancerología (INCAN), MexicoReviewed by:
Giovannino Silvestri, University of Maryland Medical Center, United StatesEswari Dodagatta-Marri, University of California, San Francisco, United States
Copyright © 2024 Lin, Zhao, Lv, Xie, Li and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Zhongying Zhang, zhangzy1121@xmu.edu.cn
 Yuning Lin1
Yuning Lin1