- 1Agricultural College, Yanbian University, Yanji, China
- 2Institute of Edible and Medicinal Fungi, Agricultural College, Yanbian University, Yanji, China
Introduction: Fungal secondary metabolites (SMs) have broad application prospects in the food and medicine industries. Co-culturing strategies that simulate natural symbiotic relationships among microorganisms are used to discover and enhance the production of new SMs. We aimed to use the abundant resources of large edible and medicinal fungi to enhance the yield of desired metabolites through co-culture and potentially produce metabolites that cannot be generated in pure cultures.
Methods: We assessed the biomass and intracellular polysaccharide (IPS) content of liquid co-cultures of Sanghuangporus vaninii and Pleurotus sapidus. Subsequently, the effect of the liquid co-culture on fungal intracellular metabolites was studied using UPLC-QTOF.
Results: Co-culturing of S. vaninii with P. sapidus resulted in significantly increased biomass and IPS content; however, P. sapidus had a significant inhibitory effect on the growth of S. vaninii. Metabolomic data further indicated that amino acid, nucleotide, and glycerophospholipid metabolisms were the primary metabolic pathways affected by symbiosis.
Discussion: This study provides insights into fungal interactions and cellular metabolic mechanisms, contributing to the understanding and enhancement of the fungal fermentation potential.
1 Introduction
Secondary metabolites (SMs) are a class of low-molecular-weight compounds produced by microorganisms with various biological activities (Wang Y. et al., 2023). In a continuous interaction state, microorganisms activate complex regulatory mechanisms, specifically for SM production, to induce the biosynthesis of highly symbiotic natural products or novel metabolic compounds (Liu et al., 2022). Rather than passively existing in natural or unnatural growth environments, microorganisms establish physiological and metabolic interactions and adapt to changes in their living environments by regulating their metabolism, particularly through SM production (Singh and Lee, 2018). However, the underlying mechanisms regulating microbial interactions, community composition, dynamics, and functions remain elusive (Bertrand et al., 2013a,b). Fungal SMs have broad application prospects in the food and medicine industries (Guo et al., 2024). Therefore, many studies have been conducted on the co-cultures of microorganisms to increase SM yield (Li et al., 2023; Tan et al., 2024; Xu et al., 2024).
In 1925, Sack first proposed the concept of microbial co-culture (Sun, 2022). Microbial co-culture usually involves the culturing of two or more strains (including fungus–fungus, fungus–bacteria, and bacteria–bacteria) in one culture vessel (Mantravadi et al., 2019) and is considered an excellent strategy to enhance SM yield and variety (Caudal et al., 2022). As microorganisms continually interact, they compete for survival space and nutrients, which activates the expression of gene clusters encoding SMs, affecting the metabolic activities of the strains (Costa et al., 2019). Because microbial co-cultures involve interactions between different species or strains of the same species, effective detection methods for metabolites in co-cultures are particularly essential (Adnani et al., 2015).
Metabolomic analysis has been shown to effectively reveal the metabolic accumulation mechanisms of co-cultured microorganisms (Li et al., 2024) and enable the simultaneous measurement of changes in various cellular small-molecule metabolites to determine all metabolic changes in a biological system before and after stimulation, elucidating the metabolic network (Wang J. et al., 2024). Metabolomics represents a significant tool for studying fungus-induced changes in metabolites during fermentation and has broad applications in fields such as food science, agriculture, and microbiology, with metabolite levels revealing the functional state of the cell (Liu et al., 2022). For example, co-culturing Eurotium amstelodami GZ23 and Bacillus licheniformis GZ241 significantly enhanced the inhibitory effect on Staphylococcus aureus ATCC25923. Non-targeted metabolomics analysis revealed that, intracellularly, the differential metabolites primarily involved the metabolism of amino acids, nucleic acids, and glycerophospholipids (Wang Y. et al., 2023). When Aspergillus oryzae AS3.951 and Zygosaccharomyces rouxii HH18 were co-cultured, A. oryzae significantly inhibited the growth of Z. rouxii. Analysis of intracellular and extracellular metabolites using UPLC-QTOF-MS technology showed that the main pathways affected by co-culturing included histidine metabolism and Phenylalanine, tyrosine, and tryptophan biosynthesis (Liu et al., 2022).
Sanghuangporus spp., belonging to the genus Sanghuangporus, are large medicinal mushrooms with food and medicinal purposes (Gao et al., 2024). Their medicinal value was first recorded in the Chinese herbal classic “Shennong Bencao Jing,” and they are internationally recognized among the best medicinal large fungi with anti-cancer effects (Luan et al., 2022; Wu et al., 2022; Hu et al., 2024). Polysaccharides are among the primary active components of Sanghuang that contribute to its pharmacological effects (Wang et al., 2022) and possess anti-tumor (Boateng et al., 2024), anti-inflammatory (Hou et al., 2020), hepato-protective (Chiu et al., 2023), and immune-modulating properties (Wang F. et al., 2024). The current studies on Sanghuang polysaccharides primarily focus on fruiting bodies and pure liquid cultures, whereas studies on its co-cultures are rare.
In a preliminary study (Lu and Liu, 2024), we screened seven species of large edible and medicinal fungi and found that a co-culture of Sanghuangporus vaninii and Pleurotus sapidus yielded the optimum results. Based on preliminary research findings, it is speculated that under co-culture conditions, S. vaninii and P. sapidus interact to alter the abundance of intracellular metabolites, affecting their functions. In this study, we compared and measured the biomass and intracellular polysaccharide (IPS) content of pure cultures and co-cultures of S. vaninii and P. sapidus through liquid cultures. Additionally, we conducted a differential analysis of the intracellular metabolites of pure cultures and co-cultures of S. vaninii and P. sapidus using non-targeted metabolomics to explore the impact of co-culture on intracellular metabolism. This study primarily aimed to (1) understand the interactions between co-cultures of S. vaninii and P. sapidus; (2) determine and analyze the biomass and IPS content of pure cultures and co-cultures of S. vaninii and P. sapidus; and (3) explore the changes in the abundance of intracellular metabolites after co-culturing S. vaninii and P. sapidus. We aimed to use the abundant resources of large edible and medicinal fungi to enhance the yield of desired metabolites through co-culture and potentially produce metabolites that cannot be generated in pure cultures. This study seeks to contribute theoretical reference materials for the diversification of S. vaninii or P. sapidus products and the sustainable development of the industry.
2 Materials and methods
2.1 Strains
Strains of S. vaninii ST (NMDC Accession: NMDCN0003GMT) and P. sapidus BLZ (NMDC Accession: NMDCN0002T9L) were obtained from the Institute of Edible and Medicinal Fungi, Agricultural College, Yanbian University, Yanji, China.
2.2 Culture medium
PDA medium: potato 200 g/L, glucose 20 g/L, agar 20 g/L, and natural pH.
Seed medium: yeast extract 5 g/L, glucose 20 g/L, KH2PO4 1.5 g/L, MgSO4·7H2O 0.5 g/L, and natural pH.
Co-culture medium: yeast extract 6 g/L, glucose 30 g/L, KH2PO4 1.5 g/L, MgSO4·7H2O 2 g/L, and pH 9.
2.3 Strain activation
Using an inoculating loop, 0.5*0.5 cm mycelial block was transferred from the seed tube into a fresh PDA medium plate and incubated invertedly in the dark at 28°C. The mycelium grew until it covered approximately 90% of the plate, indicating completion of strain activation.
2.4 Liquid co-culture of Sanghuangporus vaninii and Pleurotus sapidus
Cultivate according to the methods in the preliminary research findings (Lu and Liu, 2024).
Five blocks of S. vaninii and one block of P. sapidus, each 9 mm in diameter, were inoculated separately into 250-mL shake flasks containing 100 mL of seed medium. The S. vaninii and P. sapidus were cultured for 7 and 5 days, respectively. After culturing, the seed culture solution was homogenized using an adjustable high-speed homogenizer. At an inoculation volume of 20% (v/v), the seed culture solution was transferred into 250-mL shake flasks containing 80 mL of seed medium. The S. vaninii and P. sapidus were cultured for 2 and 0 days, respectively, to obtain the seed pre-culture solution.
Pure culture of S. vaninii and P. sapidus: With an inoculation volume of 10% (v/v), the seed pre-culture solutions of S. vaninii and P. sapidus, which were cultured for 0 days, were transferred into 250-mL shake flasks containing 95 mL of a co-culture medium, where S. vaninii and P. sapidus were cultured for 7 and 5 days, respectively.
Co-culture of S. vaninii and P. sapidus: With inoculation volumes of 6.9 and 3.1% (v/v), the seed pre-culture solutions of S. vaninii and P. sapidus were transferred into 250-mL shake flasks containing 95 mL of a co-culture medium and cultured for 5 days. All culture conditions were at 28°C and 150 × g.
2.5 Biomass determination and sample preparation
After culturing, all fermentation liquids were centrifuged at 5,000 × g at 4°C for 5 min. The supernatant was discarded, and the mycelia were collected, which were washed with distilled water and divided into two parts. One part was dried in a 60°C constant temperature oven until a constant weight was achieved, cooled, and weighed for biomass measurement. The dried mycelia were crushed and sieved through a 60-mesh standard sieve, and the samples were collected for IPS concentration determination. For the other part, based on the different mycelium colors and morphologies of different strains, the mycelia were isolated from the co-culture to obtain a pure mycelial strain, which was treated with liquid nitrogen and stored at −80°C for metabolomics analysis.
2.6 IPS content determination
We adopted the method of Wang (2021) with slight modifications.
(1) IPS extraction: The tested sample (0.3 g) was placed in a centrifuge tube, followed by the addition of distilled water at a material-to-liquid ratio of 1:30 (g/mL) and extraction at 90°C in a constant temperature water bath for 2 h. Subsequently, centrifugation was performed at 6,000 × g for 10 min, and the supernatant was collected. The extraction was repeated twice, and the supernatants were combined. The supernatant was concentrated to one-fifth of the original volume in a 60°C electric constant temperature drying oven. Four times the volume of anhydrous ethanol was added, and the mixture was allowed to stand for 12 h at 4°C. Next, centrifugation was performed at 8,000 × g for 30 min, and the precipitate was collected, dried at 60°C, and dissolved in distilled water. The volume was made up to obtain the IPS solution for testing.
(2) Glucose standard curve preparation: Glucose standard solution (0.1 mg/mL): Glucose standard (0.1 g) was placed in a 100-mL beaker and dissolved in distilled water (to obtain a volume of 1,000 mL) and stored at 4°C. Pipettes (0, 0.2, 0.4, 0.6, 0.8, and 1.0 mL) of the glucose standard solution were placed into test tubes, followed by the addition of distilled water (to obtain a volume of 1.0 mL) and thorough mixing. Subsequently, 1 mL of 5% phenol and 5 mL of concentrated sulfuric acid were added, mixed thoroughly, placed in a boiling water bath for 15 min, and quickly cooled under running water. After 10 min at room temperature, the absorbance was measured at 490 nm using distilled water as the blank. The glucose standard curve was plotted with glucose content (mg) on the x-axis and absorbance on the y-axis. The linear regression equation for the glucose standard is Y = 10.185X − 0.0453 (R2 = 0.9927).
(3) IPS content determination: We measured according to step (2) and calculated the IPS content thus:
where m1 is the glucose content calculated from the standard curve (mg), V1 is the volume (mL), N is the dilution factor, m is the sample mass (g), and V is the volume of the sample solution used for the measurement (mL).
2.7 Metabolomic analysis
2.7.1 Intracellular metabolite extraction
Frozen samples were mixed into a fine powder in liquid nitrogen. A 50 mg sample and 1,000 μL of extraction liquid (methanol: acetonitrile: deionized water = 2:2:1, v/v) and 20 mg/L internal standard (L-2-chlorophenylalanine) were vortexed for 30 s. The mixture was thoroughly ground using a grinder (45 Hz, with steel balls), followed by ultrasonic extraction for 10 min in an ice-water bath. Subsequently, the samples were left to stand at −20°C for 1 h and centrifuged for 15 min (13,000 × g, 4°C), and 500 μL of the supernatant was collected into a PE tube, where they extracted a vacuum concentrator. To the extract, 160 μL of extraction liquid (acetonitrile: deionized water = 1:1, v/v) was added, followed by vortexing for 30 s, ultrasonication for 10 min (in an ice-water bath), and centrifugation for 15 min (12,000 × g, 4°C). Next, 120 μL of the supernatant was taken into a 2 mL sample vial, with 10 μL from each sample mixed into a QC sample for analysis (Dunn et al., 2011).
2.7.2 Metabolomics detection
The samples were analyzed using Waters Acquity I-Class PLUS ultra-performance liquid chromatography on a Waters Xevo G2-XS QTOF high-resolution mass spectrometer. The chromatographic column was Waters Acquity UPLC HSS T3 (1.8 um, 2.1 mm × 100 mm). The operational parameters were as follows: injection volume 1 μL; flow rate 400 μL/min; mobile phase, 0.1% (v/v) formic acid aqueous solution (A) and formic acid acetonitrile (B). The gradient program was as follows: 2% solvent B (0 min) increased to 98% (10 min) and maintained at 98% for 3 min, reduced to 2% at 13.00–13.10 min and maintained at 2% for 1.90 min; the total runtime was 15 min. Mass charge ratio (m/z) acquisition range 50–1,200, capillary voltage 2,500 V (positive ion mode) or − 2,000 V (negative ion mode), cone voltage 30 V, ion source temperature 100°C, desolvation gas temperature 500°C, cone gas flow rate 50 L/h, desolvation gas flow rate 800 L/h, a low collision energy of 2 V, a high collision energy range of 10–40 V, and the scanning frequency was 0.2 s/spectrum (Wang et al., 2016).
2.8 Data processing and statistical analysis
The determination of biomass and IPS content in pure and co-cultured liquid cultures of S. vaninii and P. sapidus was performed thrice, and metabolomic analyses were performed six times. Data were presented as mean ± standard deviation. Microsoft Excel 2022 was used for the statistical analysis and plotting. Biomass and IPS contents were determined using a one-way ANOVA and Duncan’s test (p < 0.05).
The raw data collected using MassLynx V4.2 were processed using Progenesis QI software for peak extraction, peak alignment, and other data processing operations, based on the Progenesis QI software online METLIN database and Biomark’s self-built library for identification, as well as theoretical fragment identification and mass deviation, which were within 100 ppm (Wang et al., 2016). After normalizing the original peak area information to the total peak area, a follow-up analysis was performed. Principal component analysis (PCA) and Spearman’s correlation analysis were used to assess the repeatability of the samples within the group and the quality control samples. The identified compounds were searched for classification and pathway information in the KEGG, HMDB, and lipid map databases. Based on the grouping information, we calculated and compared the difference multiples, and a t-test was used to calculate the significance p-value of each compound. The R language package ropls was used to perform orthogonal partial least squares discriminant analysis (OPLS-DA) modeling, and permutation tests were performed 200 times to verify the reliability of the model. The variable importance in the projection (VIP) value of the model was calculated using multiple cross-validations. The method of combining the difference multiple, p value, and VIP value of the OPLS-DA model was adopted to screen the differential metabolites. The screening criteria were (VIP) ≥1 and p value (p < 0.05). Differences in metabolites with significant KEGG pathway enrichment were calculated using the hypergeometric distribution test.
3 Results and discussion
3.1 Physical interactions in the co-culture of Sanghuangporus vaninii and Pleurotus sapidus
Extensive interactions between microbes of the same or different species regulate the activities (such as replication, transcription, translation, and metabolism) of this biological community at different levels (Wu and Liu, 2022). We conducted a liquid co-culture study to understand the interactions between S. vaninii and P. sapidus during liquid co-culturing. After the co-culturing, we assessed the mycelial growth rate by comparing the size of the mycelia of the strains with those of the pure cultures. The mycelium morphology (Figures 1A–C) showed that the growth rate of P. sapidus under co-culture conditions did not significantly decline compared with that under pure culture conditions, and the color of the mycelial pellets showed no significant changes. However, under co-culture conditions, S. vaninii was slowed down, the color of the mycelial pellets became lighter, the pellets were smaller, and the mycelia were fewer and shorter, indicating that P. sapidus inhibited S. vaninii growth. Therefore, further studies were conducted on the biomass and IPS content in the co-culture and pure cultures of S. vaninii and P. sapidus to assess the influence of the co-culture on the fungi. The biomass under co-culture conditions was 3.23 and 1.08 times those of S. vaninii and P. Sapidus, respectively, in pure cultures (Figure 1D); the IPS content under co-culture conditions was 5.08 and 1.23 times those of S. vaninii and P. sapidus, respectively, in pure cultures (Figure 1E), indicating that the co-culture of S. vaninii and P. sapidus enhanced the biomass and IPS content in the liquid co-culture system.
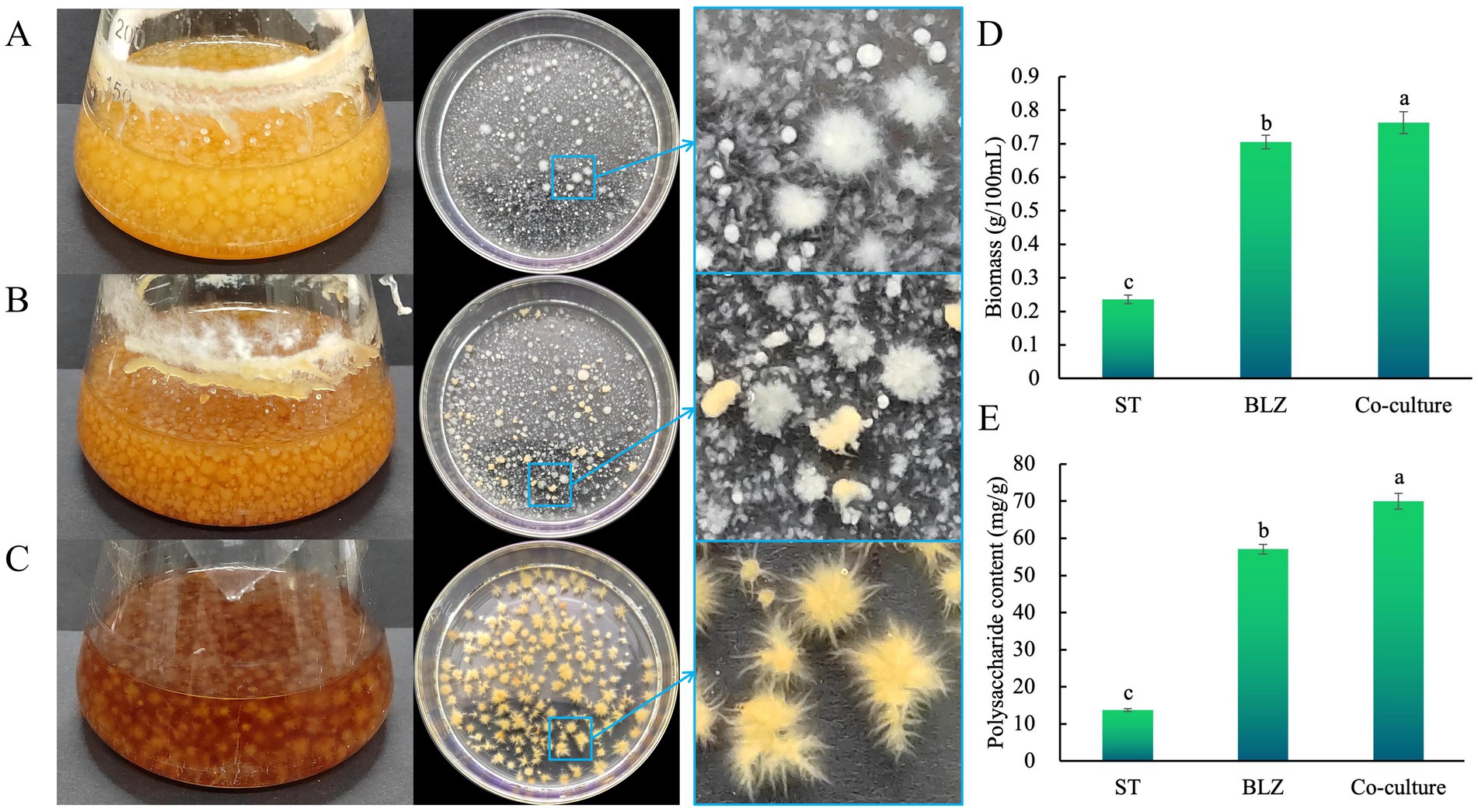
Figure 1. (A) Pure culture of Pleurotus sapidus; (B) co-culture of Sanghuangporus vaninii and P. sapidus; (C) pure culture of S. vaninii; (D) biomass; and (E) polysaccharide content. Lowercase letters indicate significant differences at a p value of <0.05 level.
3.2 Metabolite qualitative and quantitative analysis and data quality assessment
Fungal SMs have several applications in the food and medicine industries. Therefore, the co-culture of multiple microorganisms has a common application in various studies to increase SM yield (Liu et al., 2022). However, studies on the differences in SMs between S. vaninii and P. sapidus under pure culture and co-culture conditions are rare. To determine the changes in related metabolomes after co-culturing S. vaninii and P. sapidus, we conducted an untargeted metabolomic analysis of the mycelia of both species under pure culture and co-culture conditions. The 17,589 peaks were detected in the positive and negative ion modes, annotating 3,188 metabolites. Multivariate statistical analysis was performed to determine changes in metabolite profiles between the different treatments. PCA score plots showed a significant separation trend of metabolites for the pure culture of S. vaninii (S), co-culture of S. vaninii (S_C), pure culture of P. sapidus (P), and co-culture of P. sapidus (P_C) (PC1: 35.11%, PC2: 14.34%) (Figure 2A), indicating significant differences between groups and good consistency within the groups. This trend was also illustrated in the sample correlation graph (Figure 2B), a possible “stress” response during co-culturing.
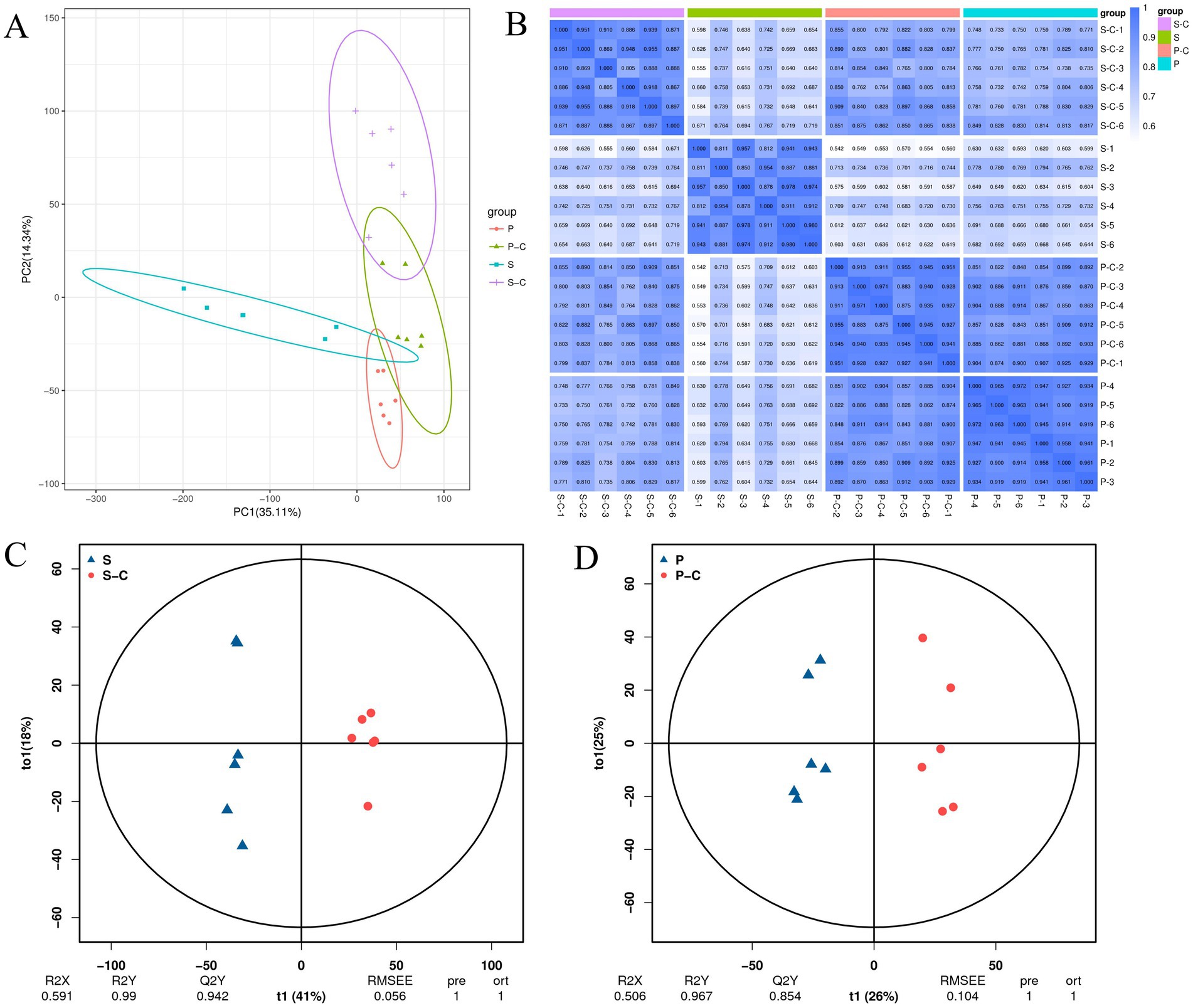
Figure 2. (A) PCA analysis of all samples; (B) correlation map between samples; (C) S-vs-S_C OPLS-DA score plot; (D) P-vs-P_C OPLS-DA score plot. S, Pure culture of Sanghuangporus vaninii; S_C, Co-culture of S. vaninii; P, Pure culture of Pleurotus sapidus; P_C, Co-culture of P. sapidus.
To show a smaller value, we plotted OPLS-DA score graphs for the pure cultures and co-cultures of S. vaninii and P. sapidus. The results showed that Component 1 accounted for 41 and 26% of the variation, whereas Component 2 accounted for 18 and 25% (Figures 2C,D). In the PCA score plot, the overlapping samples (P and P_C) were distinguished in the OPLS-DA score plot. The R2Y values were 0.99 and 0.967, and the Q2Y values were 0.942 and 0.854, respectively. R2Y explains the calibration of the model samples, and Q2Y describes the predictive capability. According to a study, when the R2Y and Q2Y are strikingly similar, the model in the OPLS-DA score plot shows meaningful results (Bevilacqua and Bro, 2020). Thus, this result implies that the model is stable, reliable, and suitable for further screening of differential metabolites and indicates significant reprogramming of the metabolome after co-culturing S. vaninii and P. sapidus.
3.3 Differential metabolite analysis
Based on the OPLS-DA, differential metabolites were determined using VIP ≥ 1 and p < 0.05 as criteria. A total of 1,591 significantly different metabolites were found between the pure cultures and co-cultures of S. vaninii (907 upregulated and 684 downregulated) (Figure 3A). Many upregulated metabolites were annotated as amino acids, peptides, and analogs (Supplementary Table S1), such as D-glutamine, D-proline, glutathione, L-threonine, L-histidine, L-isoleucine, and L-glutamate. Additionally, the contents of carbohydrates and carbohydrate conjugates (Supplementary Table S2) and fatty acids and conjugates, including D-fucose, D-ribulose, L-rhamnose, rutinose, isomaltose, undecylic acid, suberic acid, heptadecanoic acid, and nonadecanoic acid, were enhanced (Supplementary Table S3). However, during co-culturing, the content of certain amino acids, peptides, and analogs (Supplementary Table S4) and fatty acids and conjugates (Supplementary Table S5), such as L-aspartic acid, L-serine, L-asparagine, octanoic acid, azelaic acid, hexadecanedioate, and butanoic acid, were decreased, indicating that S. vaninii requires significant protein and fatty acid metabolism to cope with the competition for space and nutrients in the co-culture. The differentially expressed metabolites were used for functional pathway enrichment in the KEGG database. The results showed that the differential metabolites primarily participate in carotenoid biosynthesis, caffeine metabolism, indole diterpene alkaloid biosynthesis, lysine degradation, and carbon fixation in photosynthetic organisms (Figure 3C).
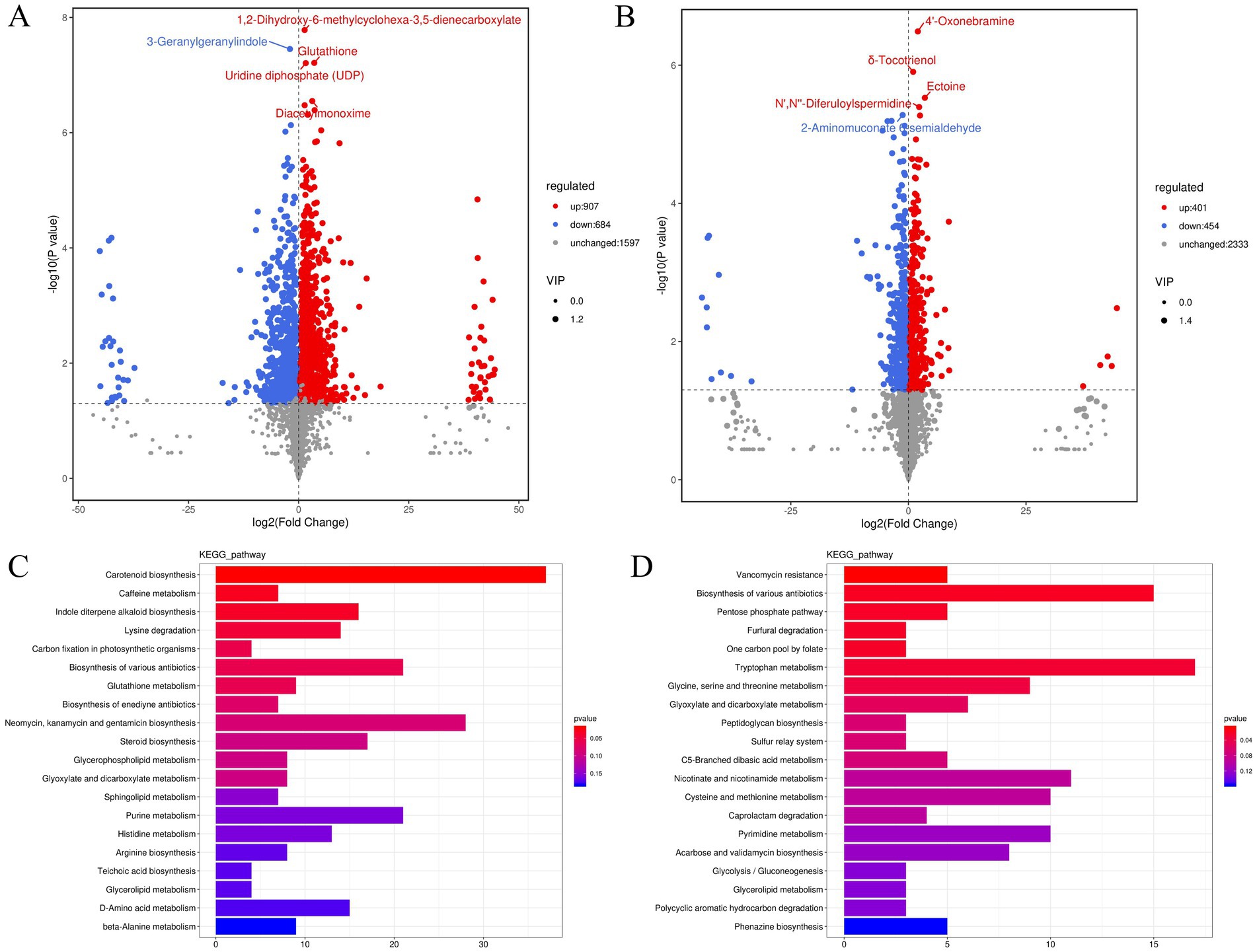
Figure 3. (A) Volcano plot of S-vs-S_C differential metabolites; (B) volcano plot of P-vs-P_C differential metabolites; (C) bar chart of S-vs-S_C differential metabolite pathways; (D) bar chart of P-vs-P_C differential metabolite pathways. S, Pure culture of Sanghuangporus vaninii; S_C, Co-culture of S. vaninii; P, Pure culture of P. sapidus; P_C, Co-culture of P. sapidus.
Between the pure cultures and co-cultures of P. sapidus, 855 significantly different metabolites were screened (401 upregulated and 454 downregulated) (Figure 3B). Compared with those in pure cultures, many upregulated metabolites, such as L-glutamine and tryptophyl-tryptophan, were annotated as amino acids, peptides, and analogs (Supplementary Table S6). Among the downregulated metabolites, L-serine and DL-O-tyrosine were annotated as amino acids, peptides, and analogs (Supplementary Table S7). Additionally, the contents of some fatty acids and conjugates (Supplementary Table S8), such as itaconate, suberic acid, azelaic acid, and hexadecanoic acid, were decreased. Itaconate is a small-molecule metabolite with significant anti-inflammatory activity discovered in macrophages in recent years (Chen et al., 2022), playing a crucial role in antibacterial defense and inflammation inhibition (McGettrick et al., 2024). It was not until 2011 that it was identified as a related metabolite produced by macrophages (Lang and Siddique, 2024). However, some studies have shown that itaconate can be obtained through microbial fermentation processes, including filamentous fungi, yeasts, and bacteria (Diankristanti and Ng, 2023; Blaga et al., 2024). Additionally, in the HMDB data, the route of exposure includes mushroom, such as Chanterelle, Common mushroom, and Shiitake.1 Based on this, we hypothesize that P. sapidus can produce itaconate in co-cultures, and further research is needed to elucidate the molecular mechanisms by which it is produced through co-culture. The above indicates that P. sapidus requires significant protein and fatty acid metabolism to cope with the competition for space and nutrients in the co-culture. The primary KEGG functional pathways enriched with differential metabolites were vancomycin resistance, biosynthesis of various antibiotics, pentose phosphate pathway, furfural degradation, and one-carbon pool by folate (Figure 3D). Vancomycin is a clinically significant glycopeptide antibiotic, isolated in 1956 from Amycolatopsis orientalis (also known as Streptomyces orientalis and Nocardia orientalis) (Chen S. et al., 2024; Pisani et al., 2024). Moussa et al. (2022) summarized that 39 compounds and extracts from edible mushrooms exhibited activity against VRSA (vancomycin-resistant Staphylococcus aureus), with terpenoids showing the strongest antimicrobial effects. Li et al. (2021) summarized the isolation of a tricyclic diterpenoid antibiotic, pleuromutilin, from the species Pleurotus mutilis and Pleurotus passeckeranius, which demonstrated good inhibitory activity against vancomycin-intermediate Staphylococcus aureus (VISA), heterogeneous VISA (hVISA), and vancomycin-resistant Staphylococcus aureus (VRSA). Pleurotus sapidus, Pleurotus mutilis, and Pleurotus passeckeranius all belong to the Pleurotus. In our study, the vancomycin-resistance pathway showed the most significant differences, while reports on vancomycin resistance in large edible and medicinal fungi are rare.
3.4 Analysis of key metabolic pathways
3.4.1 Glycolysis pathway
Glycolysis is an essential metabolic pathway for most organisms to obtain energy through the oxidation of sugars, consisting of a total of 10 enzymatic reactions (Pyrihová et al., 2024). In the seventh and eighth enzymatic reactions, under the action of phosphoglycerate kinase, the high-energy phosphoryl group of 1,3-bisphosphoglycerate is transferred to ADP to form ATP and 3-phosphoglycerate. Subsequently, 3-phosphoglycerate is converted to 2-phosphoglycerate under the catalysis of phosphoglycerate mutase (Chandel, 2021a). Additionally, 3-phosphoglycerate is a key intermediate in several central metabolic pathways, including glycolysis and glucose generation (Jang et al., 2009). During co-culture, the expression of both 3-phosphoglycerate and 2-phosphoglycerate was significantly upregulated in S. vaninii, while both were significantly downregulated in P. sapidus (Supplementary Table S9). This indicates that in the co-culture system, the glycolytic activity of S. vaninii is stronger than that of P. sapidus, providing more energy and starting materials for synthesizing substances in other metabolic pathways.
3.4.2 Tricarboxylic acid cycle
The TCA cycle is the center of glucose, protein, lipid, and energy metabolisms within the cell (Kato et al., 2022). The expression of succinate and fumarate related to the TCA cycle was significantly upregulated in S. vaninii, whereas fumarate was significantly downregulated in P. sapidus (Supplementary Table S10). Succinate plays a crucial role in mitochondrial energy production and is also involved in signaling pathways, reactive oxygen species generation, and other processes (Chen H. et al., 2024). Mitochondrial respiratory complex II (CII) reversibly oxidizes succinate to fumarate, producing electrons that are transported via the Fe–S cluster chain to the ubiquinone membrane pool, thus providing ubiquinol QH2 for the respiratory chain during oxidative phosphorylation (Markevich et al., 2020). In summary, the S. vaninii TCA system in the co-culture system is in an active state, providing an effective energy and material exchange mechanism for its growth and metabolism. Co-culturing S. vaninii with P. sapidus accelerated the energy and material metabolism of S. vaninii, providing a material synthesis and energy foundation for maintaining the stability of S. vaninii in the external environment.
3.4.3 Nucleotide metabolism
Nucleotides are DNA and RNA components that provide energy, activate signaling pathways, and increase cellular biomass in living cells (Ariav et al., 2021; Chandel, 2021b). In the co-culture systems, the expression levels of cytidine and cytosine in cells were significantly downregulated, whereas those of uridine, uracil, guanine, xanthosine, xanthine, and adenine were significantly upregulated (Supplementary Table S11). These results indicate that the interactions in the co-culture play a role in regulating cell division. This further suggests a symbiotic, mutually beneficial stage during the co-culturing process between S. vaninii and P. sapidus.
3.4.4 Amino acid metabolism
As a glutamine precursor, glutamate regulates cellular nitrogen assimilation through the synergistic action of glutamine synthetase/glutamate synthase (GS/GOGAT) (Sanz-Luque et al., 2015). In the co-culture, glutamate expression in S. vaninii was significantly increased along with the constancy of glutamine, and glutamine expression in P. sapidus was also significantly increased, indicating that the symbiotic system effectively utilized the nitrogen source in the medium to survive by maintaining stable levels of these amino acids (León-Vaz et al., 2021). Threonine, which has strong antioxidant properties (Huang et al., 2021), showed significantly increased expression in S. vaninii during co-culturing (Supplementary Table S12), indicating that it participates in the balance of oxidative stress in the system, further enhancing its self-defense capabilities.
3.4.5 Polysaccharide synthesis
The most extensively studied monosaccharides in mushrooms are glucose, mannose, galactose, xylose, arabinose, rhamnose, and fucose (Wang et al., 2017). Previous results showed that the IPS in the co-culture was higher than that in pure cultures of S. vaninii and P. sapidus. The expression levels of mannose, galactose, and glucose were significantly increased in S. vaninii but not in P. sapidus. In the fructose and mannose metabolism pathways, L-rhamnose expression in S. vaninii was significantly increased, with a difference multiple of 16.38 (Supplementary Table S13). These results indicate that the co-culture primarily promoted an increase in monosaccharide expression in S. vaninii cells, while the impact on P. sapidus was relatively small. This significant increase in monosaccharide expression enhanced the stability and resistance of S. vaninii cells to the co-culture environment.
3.4.6 Antioxidant system
The activation of a robust and complex antioxidant system is the reaction mechanism of the symbiotic system to environmental stress, including non-enzymatic antioxidants (glutathione, betaine, proline, and ascorbic acid) and antioxidant enzymes (Wang J. et al., 2023). The expression levels of reduced glutathione, oxidized glutathione, and D-proline were significantly increased in S. vaninii but not in P. sapidus (Supplementary Table S12). Glutathione is the most abundant intracellular antioxidant (Wang et al., 2014) that primarily alleviates intracellular oxidative stress (Mochizuki et al., 2023). Glutathione expression was significantly increased in S. vaninii, indicating its participation in alleviating cellular stress. Proline is a multifunctional amino acid that is essential for protein biosynthesis. Proline plays a significant role in enhancing abiotic stress tolerance (Jurkonienė et al., 2023), acting as an osmoprotectant, stabilizing macromolecules, scavenging free radicals, and maintaining redox balance (Alvarez et al., 2022). This indicates that the co-culture environment imposed survival pressure on S. vaninii, prompting the overexpression of glutathione and proline under unfavorable conditions to maintain cellular homeostasis.
3.4.7 Transport proteins
ABC transport proteins are ubiquitous in prokaryotic (blue-green algae) and eukaryotic (fungi) cells and obtain energy through ATP hydrolysis to mediate active transmembrane transport (Nickerson et al., 2018). In the ATP hydrolysis process, ABC transport proteins transport natural metabolites and various xenobiotics, including antifungal compounds, making certain ABC transporters crucial players in antifungal resistance (Víglaš and Olejníková, 2021). The activation of the ABC transport protein system (Supplementary Table S14) in the co-culture system signifies that the environmental pressure exerted by co-culturing caused the rapid exchange and transport of substances inside and outside the cells to maintain the stability requirements of symbiosis between S. vaninii and P. sapidus in the co-culture system.
3.5 Metabolic molecular mechanism of the co-culture system of Sanghuangporus vaninii and Pleurotus sapidus
The cumulative effects on S. vaninii and P. sapidus owing to co-culturing are attributable to the fact that fungal interactions in the co-culture enhance IPS production within the system, making the production of commercially valuable products feasible. The synthesis of the intermediate product, UDP-Glc, was sufficient to support this conclusion. In S. vaninii, co-culturing leads to the upregulation of substances related to the glycolysis pathway, indicating that the stress produced by co-culturing benefits the cycle. The expression of substances involved in the TCA cycle was upregulated, leading to more SMs entering different pathways for amino acid, fatty acid, transport protein, and nucleotide synthesis (Figure 4). It provides a co-culture system with buffering and supplementation to cope with environmental pressures and nutritional competition.
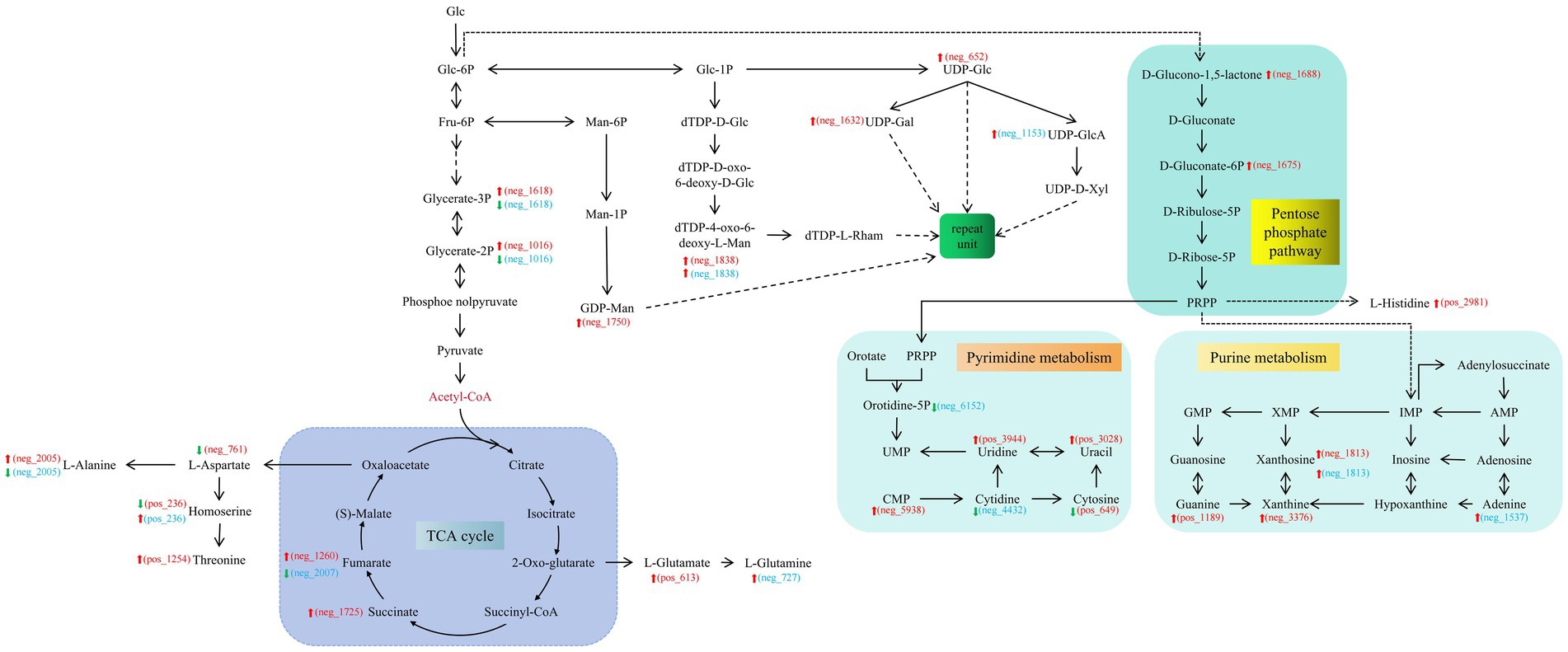
Figure 4. Key metabolic network diagram of the co-culture of Sanghuangporus vaninii and Pleurotus sapidus. The red font ID represents the differential metabolites in S_vs_S-C. The blue font ID represents the differential metabolites in P_vs_P-C. S, Pure culture of S. vaninii; S_C, co-culture of S. vaninii; P, Pure culture of P. sapidus; P_C, Co-culture of P. sapidus.
4 Conclusion
The liquid co-culturing of S. vaninii and P. sapidus significantly enhanced the biomass and IPS content of the co-culture system. Similarly, P. sapidus exerted a noticeable inhibitory effect on S. vaninii growth. The detection of intracellular metabolites indicated that fungal co-culture affects SM production via multiple metabolic pathways. In addition, it elucidates the rapid adaptation to environmental changes in co-cultures, a process revealed to involve the regulation of several metabolic pathways (amino acid metabolism, TCA cycle, glycolysis pathway, polysaccharide metabolism, and nucleotide metabolism). These results provide new insights into the selection of good co-culture combinations and activation of silent gene clusters for microbial SM synthesis. In the future, we will investigate whether new metabolites are produced and explore the molecular mechanisms underlying these changes. This study will help enrich the product development of large edible and medicinal fungi by providing new ideas.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
YL: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft, Writing – review & editing. DL: Funding acquisition, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (Grant No.2017YFD0300104).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1445993/full#supplementary-material
Abbreviations
SMs, Secondary metabolites; IPS, Intracellular polysaccharide; PCA, Principal component analysis; OPLS-DA, Orthogonal partial least squares discriminant analysis; VIP, Variable importance in projection; TCA, Tricarboxylic acid
Footnotes
References
Adnani, N., Vazquez-Rivera, E., Adibhatla, S. N., Ellis, G. A., Braun, D. R., and Bugni, T. S. (2015). Investigation of interspecies interactions within marine micromonosporaceae using an improved co-culture approach. Mar Drugs 13, 6082–6098. doi: 10.3390/md13106082
Alvarez, M. E., Savouré, A., and Szabados, L. (2022). Proline metabolism as regulatory hub. Trends Plant Sci 27, 39–55. doi: 10.1016/j.tplants.2021.07.009
Ariav, Y., Chng, J. H., Christofk, H. R., Ron-Harel, N., and Erez, A. (2021). Targeting nucleotide metabolism as the nexus of viral infections, cancer, and the immune response. Sci Adv 7:eabg6165. doi: 10.1126/sciadv.abg6165
Bertrand, S., Schumpp, O., Bohni, N., Bujard, A., Azzollini, A., Monod, M., et al. (2013a). Detection of metabolite induction in fungal co-cultures on solid media by high-throughput differential ultra-high pressure liquid chromatography–time-of-flight mass spectrometry fingerprinting. J Chromatogr A 1292, 219–228. doi: 10.1016/j.chroma.2013.01.098
Bertrand, S., Schumpp, O., Bohni, N., Monod, M., Gindro, K., and Wolfender, J.-L. (2013b). De novo production of metabolites by fungal co-culture of Trichophyton rubrum and Bionectria ochroleuca. J Nat Prod 76, 1157–1165. doi: 10.1021/np400258f
Bevilacqua, M., and Bro, R. (2020). Can we trust score plots? Meta 10:278. doi: 10.3390/metabo10070278
Blaga, A. C., Kloetzer, L., Cascaval, D., Galaction, A.-I., and Tucaliuc, A. (2024). Studies on reactive extraction of itaconic acid from fermentation broths. PRO 12:725. doi: 10.3390/pr12040725
Boateng, I. D., Guo, Y.-Z., and Yang, X.-M. (2024). Extraction, purification, structural characterization, and antitumor effects of water-soluble intracellular polysaccharide (IPSW-1) from Phellinus igniarius mycelia. J Agric Food Chem 72, 19721–19732. doi: 10.1021/acs.jafc.4c01059
Caudal, F., Tapissier-Bontemps, N., and Edrada-Ebel, R. A. (2022). Impact of co-culture on the metabolism of marine microorganisms. Mar Drugs 20:153. doi: 10.3390/md20020153
Chandel, N. S. (2021b). Nucleotide metabolism. Cold Spring Harb Perspect Biol 13:a040592. doi: 10.1101/cshperspect.a040592
Chen, H., Jin, C., Xie, L., and Wu, J. (2024). Succinate as a signaling molecule in the mediation of liver diseases. Biochim Biophys Acta (BBA) Mol Basis Dis 1870:166935. doi: 10.1016/j.bbadis.2023.166935
Chen, L.-L., Morcelle, C., Cheng, Z.-L., Chen, X., Xu, Y., Gao, Y., et al. (2022). Itaconate inhibits TET DNA dioxygenases to dampen inflammatory responses. Nat Cell Biol 24, 353–363. doi: 10.1038/s41556-022-00853-8
Chen, S., Rao, M., Jin, W., Hu, M., Chen, D., Ge, M., et al. (2024). Metabolomic analysis in Amycolatopsis keratiniphila disrupted the competing ECO0501 pathway for enhancing the accumulation of vancomycin. World J Microbiol Biotechnol 40:297. doi: 10.1007/s11274-024-04105-9
Chiu, C.-H., Chen, M.-Y., Lieu, J.-J., Chen, C.-C., Chang, C.-C., Chyau, C.-C., et al. (2023). Inhibitory effect of styrylpyrone extract of Phellinus linteus on hepatic steatosis in HepG2 cells. Int J Mol Sci 24:3672. doi: 10.3390/ijms24043672
Costa, J. H., Wassano, C. I., Angolini, C. F. F., Scherlach, K., Hertweck, C., and Pacheco Fill, T. (2019). Antifungal potential of secondary metabolites involved in the interaction between citrus pathogens. Sci Rep 9:18647. doi: 10.1038/s41598-019-55204-9
Diankristanti, P. A., and Ng, I.-S. (2023). Microbial itaconic acid bioproduction towards sustainable development: insights, challenges, and prospects. Bioresour Technol 384:129280. doi: 10.1016/j.biortech.2023.129280
Dunn, W. B., Broadhurst, D., Begley, P., Zelena, E., Francis-McIntyre, S., Anderson, N., et al. (2011). Procedures for large-scale metabolic profiling of serum and plasma using gas chromatography and liquid chromatography coupled to mass spectrometry. Nat Protoc 6, 1060–1083. doi: 10.1038/nprot.2011.335
Gao, Y., Li, X., Xu, H., Sun, H., Zhang, J., Wu, X., et al. (2024). The liquid-fermentation formulation of Sanghuangporus sanghuang optimized by response surface methodology and evaluation of biological activity of extracellular polysaccharides. Food Secur 13:1190. doi: 10.3390/foods13081190
Guo, L., Xi, B., and Lu, L. (2024). Strategies to enhance production of metabolites in microbial co-culture systems. Bioresour Technol 406:131049. doi: 10.1016/j.biortech.2024.131049
Hou, C., Chen, L., Yang, L., and Ji, X. (2020). An insight into anti-inflammatory effects of natural polysaccharides. Int J Biol Macromol 153, 248–255. doi: 10.1016/j.ijbiomac.2020.02.315
Hu, X., Ganesan, K., Khan, H., and Xu, B. (2024). Critical reviews on anti-cancer effects of edible and medicinal mushroom Phellinus linteus and its molecular mechanisms. Food Rev Int 40, 1118–1137. doi: 10.1080/87559129.2023.2212036
Huang, L., Fang, Z., Gao, J., Wang, J., Li, Y., Sun, L., et al. (2021). Protective role of l-threonine against cadmium toxicity in Saccharomyces cerevisiae. J Basic Microbiol 61, 339–350. doi: 10.1002/jobm.202100012
Jang, M., Kang, H. J., Lee, S. Y., Chung, S. J., Kang, S., Chi, S. W., et al. (2009). Glyceraldehyde-3-phosphate, a glycolytic intermediate, plays a key role in controlling cell fate via inhibition of caspase activity. Mol Cell 28, 559–564. doi: 10.1007/s10059-009-0151-7
Jurkonienė, S., Mockevičiūtė, R., Gavelienė, V., Šveikauskas, V., Zareyan, M., Jankovska-Bortkevič, E., et al. (2023). Proline enhances resistance and recovery of oilseed rape after a simulated prolonged drought. Plan Theory 12:2718. doi: 10.3390/plants12142718
Kato, Y., Inabe, K., Hidese, R., Kondo, A., and Hasunuma, T. (2022). Metabolomics-based engineering for biofuel and bio-based chemical production in microalgae and cyanobacteria: a review. Bioresour Technol 344:126196. doi: 10.1016/j.biortech.2021.126196
Lang, R., and Siddique, M. N. A. A. (2024). Control of immune cell signaling by the immuno-metabolite itaconate. Front Immunol 15:1352165. doi: 10.3389/fimmu.2024.1352165
León-Vaz, A., Romero, L. C., Gotor, C., León, R., and Vigara, J. (2021). Effect of cadmium in the microalga Chlorella sorokiniana: a proteomic study. Ecotoxicol Environ Saf 207:111301. doi: 10.1016/j.ecoenv.2020.111301
Li, W., Huang, X., Liu, H., Lian, H., Xu, B., Zhang, W., et al. (2023). Improvement in bacterial cellulose production by co-culturing Bacillus cereus and Komagataeibacter xylinus. Carbohydr Polym 313:120892. doi: 10.1016/j.carbpol.2023.120892
Li, X., Li, J., Zhu, Z., Li, T., Zhang, W., Xia, J., et al. (2021). Advances of Lefamulin: a new pleuromutilin antibiotic. Acta Pharm Sin 56, 1006–1015. doi: 10.16438/j.0513-4870.2020-1617
Li, Q., Lin, W., Zhang, X., Wang, M., Zheng, Y., Wang, X., et al. (2024). Transcriptomics integrated with metabolomics reveal the competitive relationship between co-cultured Trichoderma asperellum HG1 and Bacillus subtilis Tpb55. Microbiol Res 280:127598. doi: 10.1016/j.micres.2023.127598
Liu, Z., Kang, B., Duan, X., Hu, Y., Li, W., Wang, C., et al. (2022). Metabolomic profiles of the liquid state fermentation in co-culture of A. oryzae and Z. rouxii. Food Microbiol 103:103966. doi: 10.1016/j.fm.2021.103966
Lu, Y., and Liu, D. (2024). Optimization of polysaccharide conditions and analysis of antioxidant capacity in the co-culture of Sanghuangporus vaninii and Pleurotus sapidus. PeerJ 12:e17571. doi: 10.7717/peerj.17571
Luan, F., Peng, X., Zhao, G., Zeng, J., Zou, J., Rao, Z., et al. (2022). Structural diversity and bioactivity of polysaccharides from medicinal mushroom Phellinus spp.: a review. Food Chem 397:133731. doi: 10.1016/j.foodchem.2022.133731
Mantravadi, P. K., Kalesh, K. A., Dobson, R. C. J., Hudson, A. O., and Parthasarathy, A. (2019). The quest for novel antimicrobial compounds: emerging trends in research, development, and technologies. Antibiotics 8:8. doi: 10.3390/antibiotics8010008
Markevich, N. I., Galimova, M. H., and Markevich, L. N. (2020). Hysteresis and bistability in the succinate-CoQ reductase activity and reactive oxygen species production in the mitochondrial respiratory complex II. Redox Biol 37:101630. doi: 10.1016/j.redox.2020.101630
McGettrick, A. F., Bourner, L. A., Dorsey, F. C., and O’Neill, L. A. J. (2024). Metabolic messengers: itaconate. Nat Metab 6, 1661–1667. doi: 10.1038/s42255-024-01092-x
Mochizuki, R., Kobayashi, A., Takayama, H., Toida, T., and Ogra, Y. (2023). Simultaneous determination of intracellular reduced and oxidized glutathiones by the König reaction. Anal Methods 15, 3426–3431. doi: 10.1039/D3AY00860F
Moussa, A. Y., Fayez, S., Xiao, H., and Xu, B. (2022). New insights into antimicrobial and antibiofilm effects of edible mushrooms. Food Res Int 162:111982. doi: 10.1016/j.foodres.2022.111982
Nickerson, N. N., Jao, C. C., Xu, Y., Quinn, J., Skippington, E., Alexander, M. K., et al. (2018). A novel inhibitor of the LolCDE ABC transporter essential for lipoprotein trafficking in gram-negative bacteria. Antimicrob Agents Chemother 62:16. doi: 10.1128/AAC.02151-17
Pisani, S., Tufail, S., Rosalia, M., Dorati, R., Genta, I., Chiesa, E., et al. (2024). Antibiotic-loaded nano-sized delivery systems: an insight into gentamicin and vancomycin. J Funct Biomater 15:194. doi: 10.3390/jfb15070194
Pyrihová, E., King, M. S., King, A. C., Toleco, M. R., van der Giezen, M., and Kunji, E. R. (2024). A mitochondrial carrier transports glycolytic intermediates to link cytosolic and mitochondrial glycolysis in the human gut parasite Blastocystis. eLife 13:RP94187. doi: 10.7554/eLife.94187
Sanz-Luque, E., Chamizo-Ampudia, A., Llamas, A., Galvan, A., and Fernandez, E. (2015). Understanding nitrate assimilation and its regulation in microalgae. Front Plant Sci 6:899. doi: 10.3389/fpls.2015.00899
Singh, D., and Lee, C. H. (2018). Volatiles mediated interactions between aspergillus oryzae strains modulate morphological transition and exometabolomes. Front Microbiol 9:628. doi: 10.3389/fmicb.2018.00628
Sun, Y. (2022). Induction mechanism and secondary metabolites in co-culture of Aspergillus sydowii. Dalian, Liaoning, China: Dalian University of Technology.
Tan, X., Zhang, Q., Liu, J., Shang, Y., Min, Y., Sun, X., et al. (2024). Enhanced γ-aminobutyric acid production by co-culture fermentation with Enterococcus faecium AB157 and Saccharomyces cerevisiae SC125. LWT 208:116739. doi: 10.1016/j.lwt.2024.116739
Víglaš, J., and Olejníková, P. (2021). An update on ABC transporters of filamentous fungi – from physiological substrates to xenobiotics. Microbiol Res 246:126684. doi: 10.1016/j.micres.2020.126684
Wang, L. (2021). The effect of different light quality on the main active components of Sanghuangporus vaninii mycelium and transcriptomics study. Yanji, Jilin, China: Yanbian University.
Wang, Y., Chen, Y., Xin, J., Chen, X., Xu, T., He, J., et al. (2023). Metabolomic profiles of the liquid state fermentation in co-culture of Eurotium amstelodami and Bacillus licheniformis. Front Microbiol 14:1080743. doi: 10.3389/fmicb.2023.1080743
Wang, F., Li, N., Li, H., Di, Y., Li, F., Jiang, P., et al. (2024). An alkali-extracted neutral heteropolysaccharide from Phellinus nigricans used as an immunopotentiator in immunosuppressed mice by activating macrophages. Carbohydr Polym 335:122110. doi: 10.1016/j.carbpol.2024.122110
Wang, H., Ma, J.-X., Zhou, M., Si, J., and Cui, B.-K. (2022). Current advances and potential trends of the polysaccharides derived from medicinal mushrooms sanghuang. Front Microbiol 13:965934. doi: 10.3389/fmicb.2022.965934
Wang, W., Tang, L., Zhou, W., Yang, Y., Gao, B., Zhao, Y., et al. (2014). Progress in the biosynthesis and metabolism of glutathione. China Biotechnol 34, 89–95. doi: 10.13523/j.cb.20140714
Wang, J., Tian, Q., Kang, J., Zhou, H., Yu, X., Qiu, G., et al. (2024). Metabolomics-based analysis of the interaction effects and stress response mechanisms in the co-culture system of fungi and microalgae under cadmium stress. J Clean Prod 447:141473. doi: 10.1016/j.jclepro.2024.141473
Wang, J., Tian, Q., Zhou, H., Kang, J., Yu, X., and Shen, L. (2023). Key metabolites and regulatory network mechanisms in co-culture of fungi and microalgae based on metabolomics analysis. Bioresour Technol 388:129718. doi: 10.1016/j.biortech.2023.129718
Wang, Q., Wang, F., Xu, Z., and Ding, Z. (2017). Bioactive mushroom polysaccharides: a review on monosaccharide composition, biosynthesis and regulation. Molecules 22:955. doi: 10.3390/molecules22060955
Wang, J., Zhang, T., Shen, X., Liu, J., Zhao, D., Sun, Y., et al. (2016). Serum metabolomics for early diagnosis of esophageal squamous cell carcinoma by UHPLC-QTOF/MS. Metabolomics 12:116. doi: 10.1007/s11306-016-1050-5
Wu, M., and Liu, G. (2022). Co-culture strategy for activating the cryptic gene cluster in microorganisms. Acta Microbiol Sin 62, 4247–4261. doi: 10.13343/j.cnki.wsxb.20220320
Wu, F., Zhou, L.-W., Vlasak, J., and Dai, Y.-C. (2022). Global diversity and systematics of Hymenochaetaceae with poroid hymenophore. Fungal Divers 113, 1–192. doi: 10.1007/s13225-021-00496-4
Keywords: Sanghuangporus vaninii, Pleurotus sapidus, polysaccharide, co-culture, metabolomics profiles
Citation: Lu Y and Liu D (2024) Metabolomics profiles of the liquid co-culture of Sanghuangporus vaninii and Pleurotus sapidus. Front. Sustain. Food Syst. 8:1445993. doi: 10.3389/fsufs.2024.1445993
Edited by:
Reza Rastmanesh, American Physical Society, United StatesReviewed by:
Jing Si, Beijing Forestry University, ChinaAntero Ramos-Fernández, Instituto de Ecología (INECOL), Mexico
Copyright © 2024 Lu and Liu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Di Liu, bGl1ZGlAeWJ1LmVkdS5jbg==
 Yuantian Lu
Yuantian Lu Di Liu
Di Liu