- 1Department of Biomedical Science, School of Medicine, Debre Markos University, Debre Markos, Ethiopia
- 2Department of Medical Laboratory Sciences, College of Health Sciences, Debre Markos University, Debre Markos, Ethiopia
- 3Department of Pharmacy, College of Medicine and Health Sciences, Debre Markos University, Debre Markos, Ethiopia
- 4Department of Biomedical Sciences, School of Medicine, Wolaita Sodo University, Wolaita Sodo, Ethiopia
Breast cancer (BC) is a prevalent malignant tumor in women, and its incidence has been steadily increasing in recent years. Compared with other types of cancer, it has the highest mortality and morbidity rates in women. So, it is crucial to investigate the underlying mechanisms of BC development and identify specific therapeutic targets. Pyruvate kinase M2 (PKM2), an important metabolic enzyme in glycolysis, has been found to be highly expressed in BC. It can also move to the nucleus and interact with various transcription factors and proteins, including hypoxia-inducible factor-1α (HIF-1α), signal transducer and activator of transcription 3 (STAT3), β-catenin, cellular-myelocytomatosis oncogene (c-Myc), nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB), and mammalian sterile 20-like kinase 1 (MST1). This interaction leads to non-metabolic functions that control the cell cycle, proliferation, apoptosis, migration, invasion, angiogenesis, and tumor microenvironment in BC. This review provides an overview of the latest advancements in understanding the interactions between PKM2 and different transcription factors and proteins that influence the initiation and progression of BC. It also examined how natural drugs and noncoding RNAs affect various biological processes in BC cells through the regulation of the non-metabolic enzyme functions of PKM2. The findings provide valuable insights for improving the prognosis and developing targeted therapies for BC in the coming years.
Introduction
Breast cancer is the most commonly occurring cancer in women and the most common cancer overall (1). There were more than 2.26 million new cases of BC, and approximately 685 000 women died from the disease in 2020 (2). It can be divided into three main subtypes: luminal, HER2-positive, and triple-negative breast cancer (TNBC) (3). TNBC, in particular, is known for its high invasiveness. Around 30% of individuals with early-stage BC experience metastases, resulting in a 5-year relative survival rate of 25% (4). Nonetheless, the precise molecular mechanisms responsible for BC development across different subtypes remain unclear. Further exploration is needed to identify specific biomarkers that can be targeted to improve the overall prognosis of patients with this disease (5).
The primary distinguishing feature of cancer cells is their metabolic reprogramming. Unlike healthy cells, tumor cells rely on aerobic glycolysis for energy production, even when enough oxygen present in the environment (6, 7). This shift from the typical respiratory pathway to aerobic glycolysis is referred to as the Warburg effect (7, 8). Aerobic glycolysis is a characteristic feature of the Warburg effect and is crucial for the survival of cancer cells (9). Pyruvate kinase (PK) is a key rate-limiting enzyme for glycolysis that catalyzes the phosphorylation of phosphoenolpyruvate (PEP) and adenosine diphosphate (ADP) to produce pyruvate and adenosine triphosphate (ATP), which play critical roles in glycolysis during tumor formation (10). Since PKs are found in various tissues and exhibit distinct catalytic activities, it suggests that there may be different isotypes of this enzyme (11). There are four different subtypes of PKs that are expressed in specific tissues: muscle (M1), liver (L), erythrocyte (R), and ubiquitous (M2) (12). Among these subtypes, PKM2 is frequently overexpressed in cancer cells and has been extensively studied as a subtype specific to tumors (13).
Since Christofk et al. first demonstrated the necessity of PKM2 expression for cancer-specific aerobic glycolysis, known as the Warburg effect, there has been significant interest in its role in cancer development (14). In addition to its role in tumor metabolism, PKM2 plays a role in oncogenic cytokinesis, tumor growth, and metastasis (15–17). Furthermore, PKM2 functions as a protein kinase by phosphorylating its substrates and regulating gene expression (18). Previous studies have highlighted the importance of PKM2 in promoting cancer cell growth and survival (16, 19). Therefore, comprehending the biochemical functions of PKM2 during tumor progression is essential for identifying possible therapeutic targets and developing novel therapies for BC (5). Hence, this review highlights current advancements in understanding how PKM2 interacts with different transcription factors and proteins that influence the initiation and progression of BC. Additionally, it explored the impact of natural products and noncoding RNAs on various biological functions of BC cells by controlling the nonmetabolic functions of PKM2.
Structure and function of PKM2
The PKM gene, which is located in the 15q23 region of the chromosome, has the ability to undergo alternative splicing to produce either PKM1 or PKM2. The PKM gene spans approximately 32 kb and comprises 12 exons and 11 introns. The lengths of exons 9 and 10 are identical, which contributes to the differences in the final transcript. For PKM2, the final messenger ribonucleic acid (mRNA) product includes exon 10 but omits exon 9, a feature unique to PKM1 (20, 21). PKM2 is composed of 531 amino acids and has four domains: A (244 aa), B (102 aa), C (142 aa), and N (43 aa)-terminal domains (22). The catalytic active site is formed by the interface between the A- and B-domains, whereas the C-domain includes the fructose 1,6-bisphosphate (FBP) binding site, which acts as an allosteric activator. Additionally, the C-domain contains a nuclear localization signal sequence (NLS) and an inter-subunit contact domain (ISCD) (Figure 1) (22, 23). Furthermore, the arrangement of the C-domain plays a crucial role in explaining the differences observed in allosteric regulation by FBP among various PKM isoforms (22). The ISCD domain of PKM2 has a protein sequence that differs from that of its alternate splice variant, PKM1, by 23 amino acids, resulting in distinct properties such as allosteric regulation by FBP. These differences also enable PKM2 to interact with specific protein partners, including phosphotyrosine proteins (24).
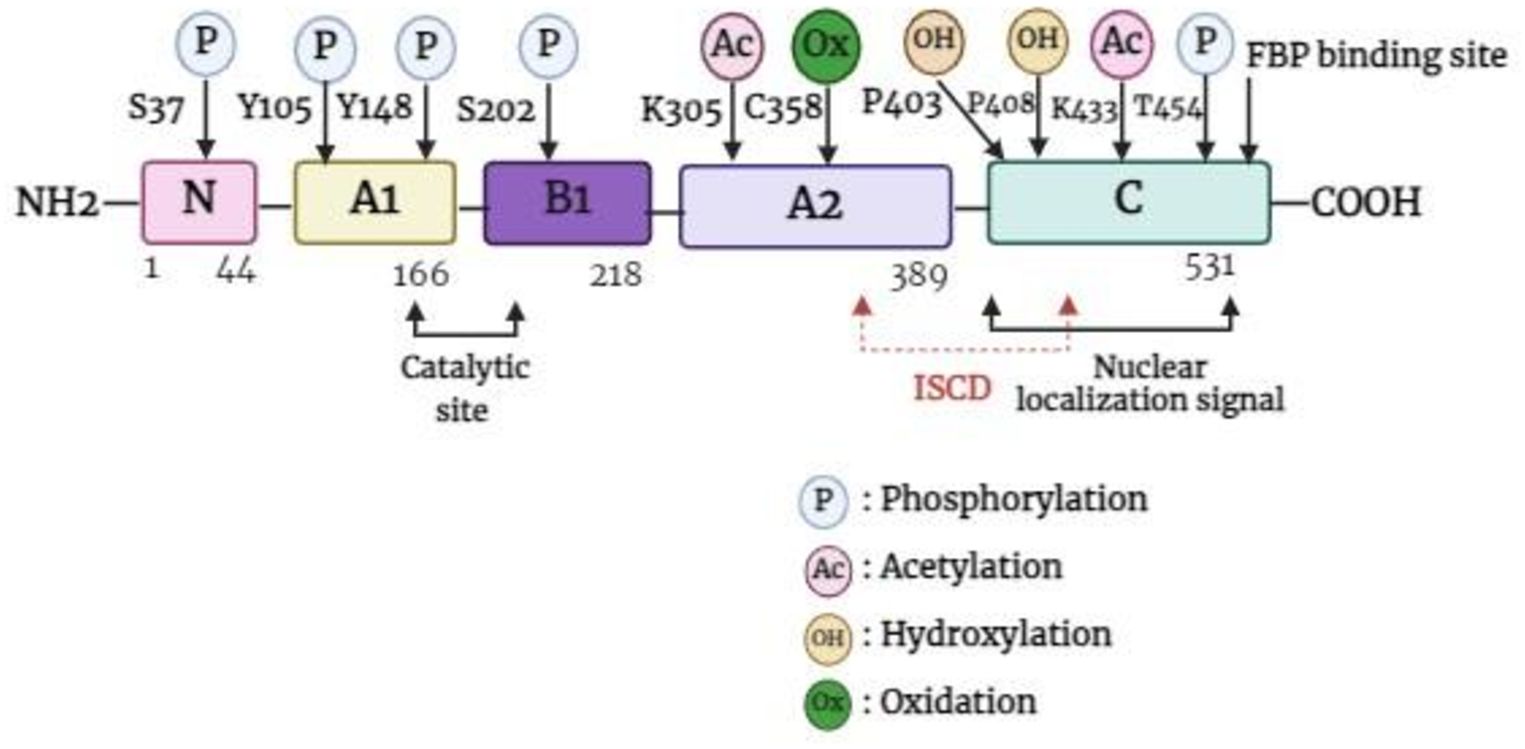
Figure 1. Basic structure of PKM2 monomer. FBP, fructose 1,6-bisphosphate; ISCD, inter-subunit contact domain.
Unlike other PK isoforms, such as PKL, PKR, and PKM1 which are exclusively tetramers, PKM2 is present in both tetrameric and dimeric forms. The A-domain of individual PKM2 units combines to form a dimer, and two such dimers bind at the interface of the ISCD (the C-domain) to create the full PKM2 tetramer (25). The shift between PKM2 dimers and tetramers is controlled by the structural changes caused by endogenous and exogenous activators and inhibitors (26). Fructose-1,6-bisphosphate (FBP) and serine are both potent allosteric activators of PKM2, which directly bind to PKM2 and stabilize it in the active tetramer configuration (27). In addition, when succinyl-5-aminoimidazole-4-carboxamide-1-ribose 5’-phosphate (SAICAR) binds to PKM2, it can trigger both the pyruvate kinase and the protein kinase activity of PKM2 (28). Furthermore, modifications such as phosphorylation, acetylation, and oxidation of PKM2 at the Tyr-105, Lys-305, and Cys-358 sites can prevent FBP from binding to tetrameric PKM2, thereby maintaining it in dimer form (29). PKM2 in tetrameric form is highly active at physiological concentrations of PEP and has a high affinity for PEP (30). In cases where PKM2 exists mainly in its highly active tetrameric form, as is the case in differentiated tissues and most normal proliferating cells, glucose is converted to pyruvate to produce energy (31). Meanwhile, PKM2 dimer is described by weak attraction to its substrate, PEP, and is virtually inactive at normal concentrations of PEP. In this form, PKM2 generates minimal ATP during the conversion of PEP to pyruvate, resulting in no net production of ATP through glycolysis (31, 32). This scenario occurs primarily in tumor cells, where PKM2 predominantly exists in the less active dimeric state. As a consequence, all glycolytic intermediates beyond PK accumulate and are redirected toward various synthetic processes such as nucleic acid production, phospholipid synthesis, and amino acid synthesis (26, 30, 33). Like tumor cells, cells that undergo rapid proliferation require a large amount of essential components such as nucleic acids, phospholipids, and amino acids (34). Importantly, the dimeric form of PKM2 can enter the nucleus and act as a protein kinase (35). The basic functions of the PKM2 dimer and tetramer are described in Figure 2.
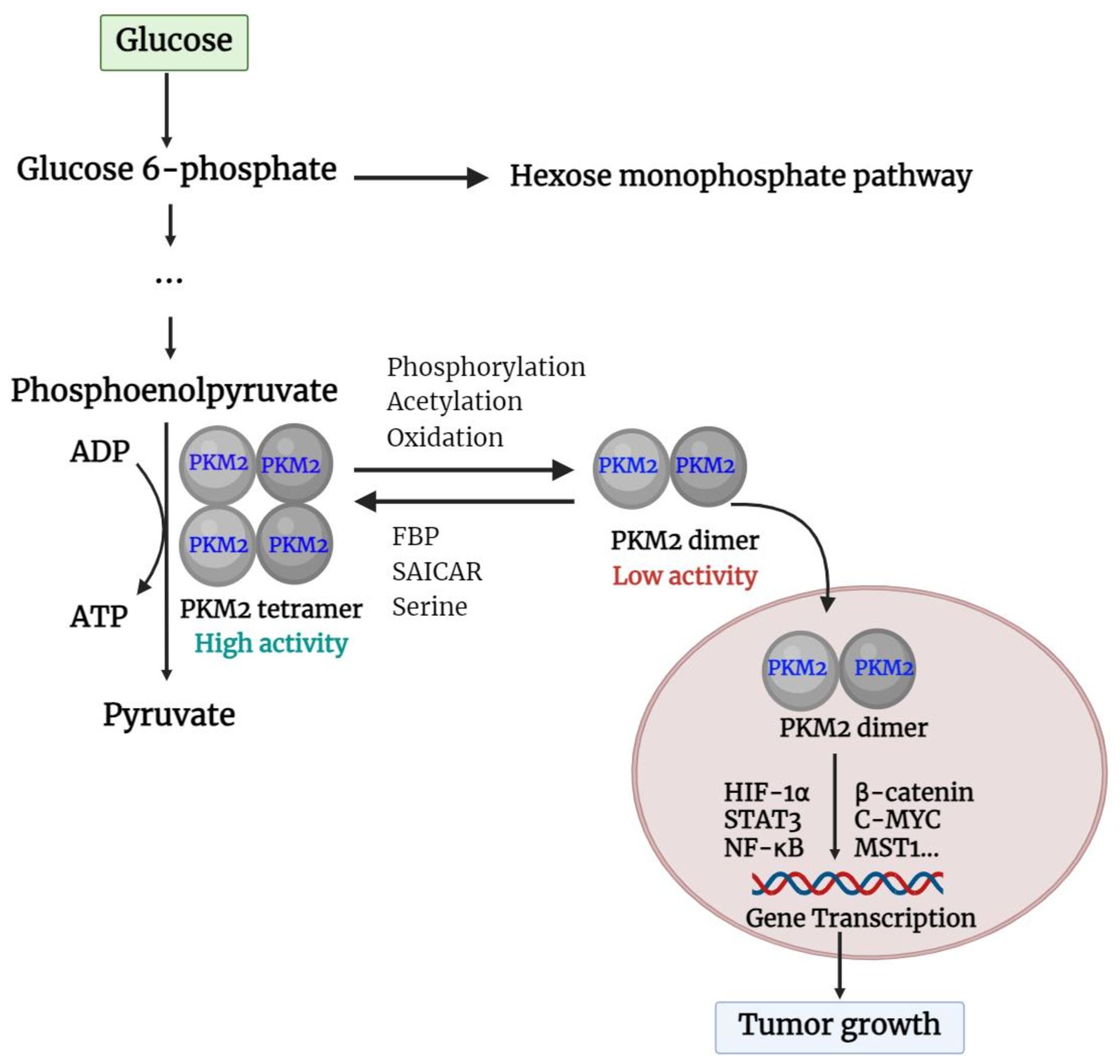
Figure 2. Basic function of PKM2 dimer and tetramer. PKM2, pyruvate kinase M2; FBP, fructose 1,6-bisphosphate; SAICAR, succinyl-5-aminoimidazole-4-carboxamide-1-ribose 5’-Phosphate; HIF-1α, hypoxia-inducible factor-1α; STAT3, Signal transducer and activator of transcription 3; NF-κB, nuclear factor kappa-light-chain enhancer of activated B cells; c-Myc, cellular-myelocytomatosis oncogene; MST1, mammalian sterile 20-like kinase 1.
The effect of PKM2 on breast cancer tumorigenesis and development
A large body of evidence supports the notion that cancers predominantly express PKM2 (14). Immunohistochemical analysis revealed that PKM2 is commonly expressed in BC (5). In BC, the activation of HIF-1α and epidermal growth factor receptor (EGFR) can facilitate the nuclear translocation of PKM2 (36), which is determined by the nuclear localization signal at its C-terminus. The phosphorylation of the PKM2 S37 site by extracellular regulatory protein kinases leads to its transformation from a tetramer to a monomer through the peptidyl-prolyl cis-trans isomerase NIMA-interacting 1 (PIN1), enabling the nuclear localization signal to enter the nucleus (23). In addition, acetylation of PKM2 at Lys433 decreases FBP binding to PKM2 and the conversion of monomers or dimers to tetramers but increases PKM2 nuclear import and protein kinase activity (37). Furthermore, the Jumonji C domain-containing dioxygenase Jumonji domain-containing protein 5 (JMJD5) interacts directly with PKM2, promoting its movement into the nucleus and HIF-1α-mediated transactivation. The interaction between JMJD5 and PKM2 occurs at the intersubunit interface region of PKM2, preventing its tetramerization and inhibiting its kinase activity (38). Once within the nucleus, PKM2 serves as a transcriptional co-activator and stimulates the activation of various transcription factors, such as HIF-1α, β-catenin, STAT3, C-MYC, NF-κB, etc., which influences the expression of their respective downstream target genes. Overall, Figure 3 shows how nuclear PKM2 regulates gene expression in relation to breast cancer development and progression.
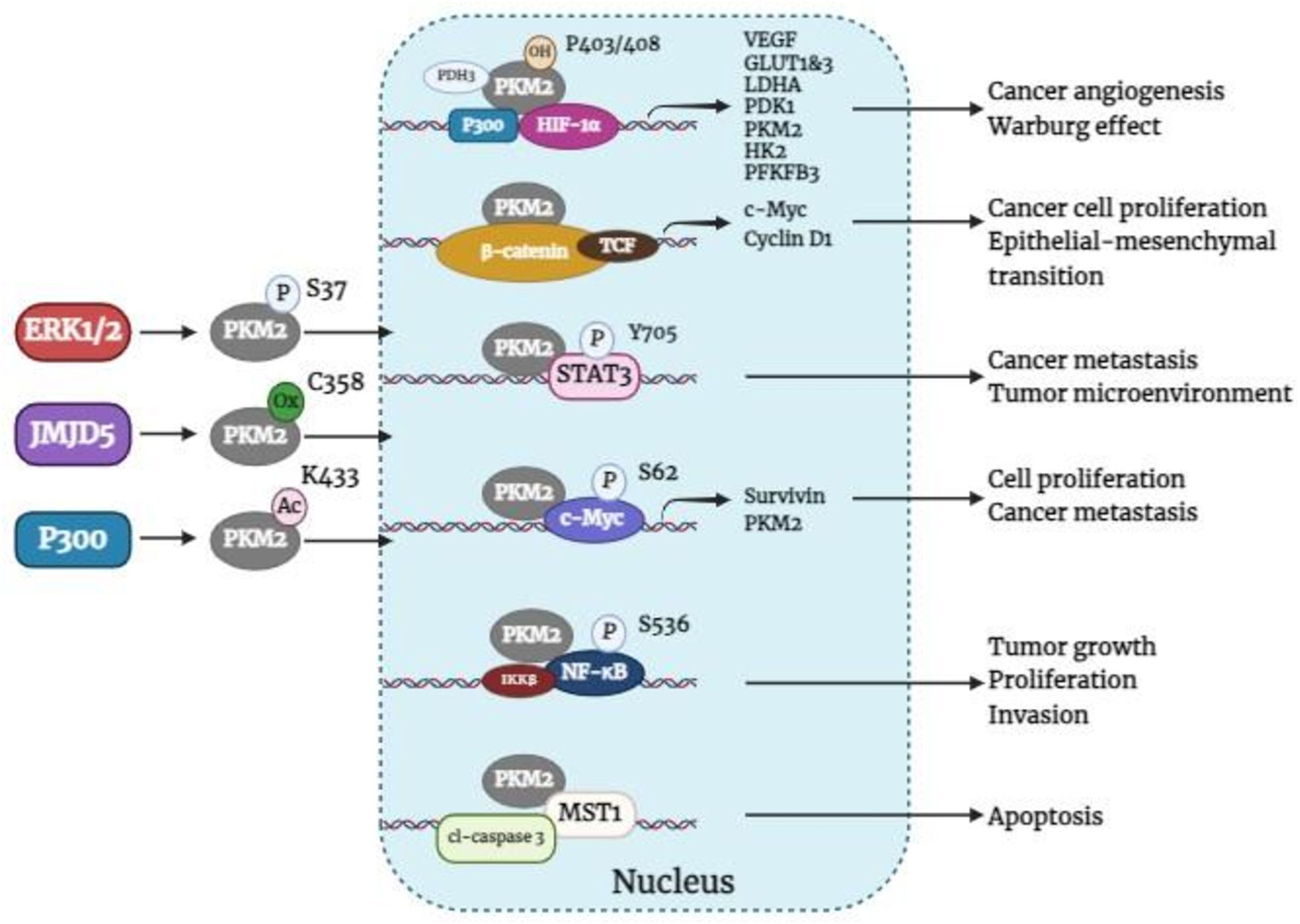
Figure 3. Nuclear PKM2 regulates the expression of related genes affecting BC tumorigenesis and development. ERK1/2, extracellular signal-regulated kinase 1/2; JMJD5, jumonji C domain-containing protein 5; PKM2, pyruvate kinase M2; HIF-1α, hypoxia-inducible factor-1α; PDH3, prolyl hydroxylase domain 3; TCF, T-cell factor; VEGF, vascular endothelial growth factor; GLUT1&3, glucose transporters 1 and 3; LDHA, lactate dehydrogenase A; PDK1, pyruvate dehydrogenase kinase 1, HK2, hexokinase 2; PFKFB3, 6-phosphofructo-2-kinase/fructose-2, 6-bisphosphatase 3; c-Myc, cellular-myelocytomatosis oncogene; MST1, mammalian sterile 20-like kinase 1; cl-caspase 3, cleaved-Caspase-3.
PKM2/HIF-1α
Cancer cells exhibit unregulated cell proliferation, unrestricted cell division, and suppression of autophagy, making them reliant on additional oxygen and nutrients for survival (39). To meet these demands, tumor cells can utilize angiogenesis and metabolic alterations. The transcription factor HIF-1α plays a crucial role in regulating genes related to angiogenesis and the Warburg effect, both of which contribute to tumor formation (40). Previous studies have focused primarily on HIF-1α as a transcription factor that increases the expression of various glycolysis-related genes, such as glucose transporters (GLUT-1 and GLUT3) and enzymes involved in glycolytic pathways [lactate dehydrogenase A, pyruvate dehydrogenase kinase 1, hexokinase 2 (HK2), and PKM2], to promote BC glycolysis and consequently affect the proliferation of BC cells (36). Intriguingly, studies have revealed that PKM2 can influence the expression and activation of HIF-1α (41, 42). The activation of PKM2 in hypoxic BC cells leads to the activation of NF-κB/p65 and HIF-1α, resulting in increased production and secretion of vascular endothelial growth factor (VEGF), which promotes angiogenesis and tumor growth (41). Angiogenesis plays a crucial role in tumor invasion and migration. Notably, studies have shown that vascular endothelial growth factor A (VEGFA) can enhance the self-renewal of cancer stem cells and promote epithelial–mesenchymal transition (EMT), potentially contributing to tumor metastasis in BC cells (43). Furthermore, clinical studies have shown that patients with metastatic BC have increased circulating VEGFA levels (44, 45). Consequently, targeting VEGF through the PKM2/HIF-1α axis in BC is a feasible approach. Chai et al. (46) reported that silencing PKM2 in BC cells resulted in reduced expression of HIF-1α and VEGF, as well as cell migration capabilities. These findings suggest that PKM2 influences the expression of HIF-1α and subsequently affects the onset and progression of various tumors, including BC. Specifically, prolyl hydroxylase domain 3 (PDH3) hydroxylates PKM2 at proline 403/408, leading to its binding to the HIF-1α subunit, which enhances HIF-1α binding to p300. This interaction then recruits p300 to the hypoxia response element, facilitating the transactivation of HIF-1α target genes and promoting PKM2 transcription (47). PKM2 and HIF-1α establish a positive feedback loop. Recent investigations have revealed that in BC cells under hypoxic conditions, nuclear PKM2 recruits HIF-1α and p300, leading to the upregulation of 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase 3 (PFKFB3), an enzyme involved in glycolysis that is associated with cancer progression and aggressiveness (36). These findings suggest that nuclear PKM2 can induce glycolysis by activating HIF-1α, potentially influencing various biological processes in BC. These results underscore the potential of targeting the PKM2/HIF-1α axis as a promising strategy for anti-BC therapies.
PKM2/β-catenin
β-catenin is the central component of the Wnt signaling pathway and plays a key role in the regulation of cell proliferation, differentiation, and apoptosis (48). The Wnt-signaling pathway is an evolutionarily conserved and complex signaling cascade that plays crucial roles in both development and disease (49). The Wnt signaling pathway plays an important role not only in the development and maintenance of healthy breast and mammary glands but also in BC etiology (50). To validate the regulation of β-catenin by PKM2, Zhao and colleagues investigated the levels of β-catenin in MDA-A-231-shPKM2 and BT20-PKM2 cells. The findings demonstrated that β-catenin expression was suppressed in PKM2-silenced cells, whereas both the protein and RNA levels of β-catenin were increased in PKM2-overexpressing cells (51). In addition, several studies have indicated that PKM2 is translocated into the nucleus in cancer cells, where it acts as a transcription factor and controls β-catenin transactivation (52, 53). Once inside the nucleus, PKM2 can interact with the transcription factor T-cell factor (TCF), leading to an increase in the transcription of the c-Myc and cyclin D1 genes. This process ultimately facilitates BC cell proliferation and EMT (51, 54). Notably, the involvement of β-catenin in various cancer cell activities, such as cell migration, invasion, and angiogenesis, is noteworthy. Findings from a clinical investigation indicated a potential correlation between the expression of β-catenin in TNBC tissue and survival prognosis (52). A recent study revealed that cryptotanshinone significantly reduced BC migration and invasion, possibly through the suppression of the PKM2/β-catenin pathway and the overexpression of PKM2, which decreased cryptotanshinone sensitivity in BC cells (54). Consequently, these findings suggest that targeting the PKM2/β-catenin signaling pathway may hold promise for treating BC.
PKM2/STAT3
STAT3 is activated in several types of tumor cells and can promote the malignant transformation of cells and inhibit apoptosis, suggesting that the STAT3 signaling pathway could be a new target for tumor gene therapy (55, 56). The Janus kinase (JAK)/STAT3 pathway is responsible for regulating the gene expression of several enzymes involved in glucose metabolism. Recently, interest in understanding the importance of the PKM2/STAT3 pathway in the advancement of cancer has increased (57). PKM2 overexpression facilitates the nuclear translocation of STAT3, a transcription factor crucial for PKM2-driven metastasis. The protein kinase activity of PKM2 mediates the nuclear translocation and up-regulation of STAT3, which regulates the aggressive progression of colorectal cancer (58). Nuclear PKM2 activates the transcription of MAP kinase kinase 5 (MEK5/ERK5) by phosphorylating STAT3 at Y705, resulting in cell proliferation (18, 59). Some investigations related to the STAT3 oncogene have laid the foundation for the interpretation and treatment of BC formation mechanisms (60). Guan et al. reported that knocking down PKM2 in BC cells resulted in a decrease in the expression of STAT3 and STAT3 (Tyr(P)-705), leading to the suppression of gene transcription and the inhibition of BC cell proliferation (61). In vitro and in vivo studies have confirmed that pY705 modification of STAT3 is necessary for tumor growth, autophagy, and metastasis and suggest that STAT3 is an effective approach for treating cancer (62). Furthermore, clinical studies using STAT3 inhibitors indicate encouraging outcomes in malignant conditions. For example, yuanhuacine (YHC), a daphnane-type diterpenoid as the main active ingredient, may inhibit BC cell proliferation and induce apoptosis in vivo and in vitro by regulating the STAT3 pathway and glycolysis through targeting PKM2 (63). The suppression of STAT3 leads to apoptosis, inhibition of growth, reduced tumor cell invasion, and increased sensitivity to treatment in BC cells (64). These results emphasize the potential significance of targeting PKM2/STAT3 as a therapeutic approach for BC.
PKM2/C-MYC
c-Myc is a frequently activated oncogene that is closely associated with the initiation and progression of cancer in humans (65). It functions as an oncoprotein involved in various cellular processes such as deoxyribonucleic acid (DNA) replication, transcription, and RNA splicing (66). One crucial transcriptional target of c-Myc is survivin, which is an inhibitor of the apoptosis protein family and plays a significant role in tumorigenesis (67–69). Survivin is highly expressed in BC and promotes the proliferation and migration of cancer cells (67, 70). Transcriptional factors play a role in the development and advancement of cancer, such as metastasis and cellular proliferation (71). PKM2’s nuclear translocation enables it to serve as a transcriptional activator for the c-Myc gene (70, 72, 73). Yu et al. (70) revealed that PKM2 interacts with c-Myc and regulates its phosphorylation at Ser-62. This interaction suggests that c-Myc may be a novel substrate of PKM2. Inhibiting PKM2 reduces c-Myc phosphorylation, leading to the downregulation of c-Myc protein expression through the promotion of its degradation in BC cells. The results of c-Myc knockdown indicated decreased survivin protein and mRNA levels, suggesting that PKM2 regulates survivin via c-Myc (70, 74). Thus, a novel approach to inhibit BC cell proliferation and migration could involve focusing on the PKM2-c-Myc-survivin pathway.
PKM2/NF-κB
The nuclear factor kappa-light-chain enhancer of activated B cells (NF-κB) family of transcription factors plays important roles in regulating inflammation, immunological response, cell differentiation, proliferation, and survival (75). NF-κB consists of five subunits: p65/RelA, c-Rel, RelB, p50/NF-κB1, and p52/NF-κB2, which form unique protein complexes that bind to consensus DNA sequences at gene promoter regions to control various biological activities (76, 77). PKM2 plays a role in controlling the NF-κB signaling pathway in cancer cells. For example, Azoitei et al. reported that PKM2 stimulated the release of VEGF-A by activating NF-κB and HIF-1α, which in turn affected tumor angiogenesis in pancreatic cancer (41). In addition, Zheng et al. found that PKM2 facilitates the movement of NF-κB/P65 to the nucleus, resulting in the promotion of ovarian cancer cell migration and invasion (78). In BC, there has also been evidence of positive crosstalk between NF-κB and PKM2 (79, 80). Knockdown of PKM2 in TNBC cells significantly reduces the activity of NF-κB by decreasing the phosphorylation of p65 at serine 536 and suppressing the expression of NF-κB target genes (80, 81). However, the exact mechanism by which PKM regulates the activity of NF-κB through p65 phosphorylation remains unclear. IKKβ is known to play a critical role in phosphorylating RelA/p65 at serine 536 and controlling the activation of NF-κB (81). Dimeric PKM2 acts as an active protein kinase that can phosphorylate certain nuclear proteins (18, 82). Therefore, future investigations will explore whether the localization of PKM2 differs between the cytoplasm and nucleus and whether dimeric PKM2 can phosphorylate the p65 protein at serine 536 or influence IKKβ signaling molecules. These studies may reveal additional mechanisms and metabolic changes in PKM2/NF-κB in TNBC cells.
PKM2/MST1
Mammalian sterile 20-like kinase 1 (MST1), also known as STK4 and Krs-2, is widely present in all human tissues and cell lines and serves as a crucial upstream kinase in the Hippo signaling pathway (83). MST1 plays a vital role in regulating mammalian cell shape, controlling organ size, managing apoptotic responses, and influencing cancer development (83, 84). PKM2 physically interacts with MST1 in vivo. The phosphorylation and proteolysis of MST1 regulated by PKM2 could affect downstream signaling pathways controlled by MST1, which are involved in regulating cell death (85, 86). Treatment with 4-hydroxytamoxifen reduces the levels of PKM2 and disrupts its interaction with MST1, leading to the activation of caspase-3. This further enhances the cleavage of MST1 mediated by caspase-3 in BC cells. The cleaved form of MST1 relocates to the nucleus, where it causes chromatin condensation and ultimately triggers cell apoptosis (85). Silencing PKM2 can activate Caspase-3, which then cleaves its substrate to enhance the apoptotic signaling cascade induced by tamoxifen treatment (85, 87). These findings offer new insight for the development of therapies against BC.
Other transcription factors or signaling pathways affected by nuclear PKM2
Besides, studies have revealed that the activation of PKM2 induces the phosphorylation of adenosine monophosphate-activated protein kinase (AMPK), resulting in the phosphorylation and subsequent inhibition of acetyl-CoA carboxylase. This suggests that heightened PK activity results in an energy deficit (88). AMPK activation is known to hinder mammalian target of rapamycin (mTOR) signaling during low-energy conditions, exerting a cytostatic effect (89). The classic antidiabetic drug metformin, for example, has been extensively studied for its anticancer effects via AMPK signaling (90, 91). Existing studies have demonstrated that PKM2 interacts directly with the histone methyltransferase enhancer of zeste homolog 2 (EZH2) to orchestrate the epigenetic silencing of the carnitine transporter, SLC16A9. Inhibiting PKM2 disrupts the binding of EZH2 to SLC16A9, which de-represses the expression of SLC16A9 and increases intracellular carnitine influx, thus programming TNBC cells toward fatty acid oxidation-dependent and luminal-like cell states (92). According to previous reports, nuclear PKM2 promotes the proliferation of liver, breast, colon, and lung cancer cells by phosphorylating nuclear sterol regulatory element-binding protein (SREBP)-1a T59, which prevents SREBP-1a from being ubiquitinated and degraded and consequently enhances the expression of SREBP-1a’s target genes related to lipid metabolism. This study demonstrated that PKM2 functions as a transcriptional co-activator by promoting SREBP-1a’s transcriptional activation function and thus driving the proliferation of BC cells (93).
The effect of regulating nonmetabolic enzyme function of PKM2 on breast cancer tumorigenesis and development
The above studies revealed that the nuclear localization of dimeric PKM2 can affect a variety of biological processes in BC cells. As a result, controlling the nonmetabolic function of PKM2 is critical for intervening in BC carcinogenesis and progression. Currently, a growing body of research is focused on exploring the potential role of natural products and noncoding RNAs in regulating the nonmetabolic function of PKM2 to affect BC tumorigenesis and development.
Natural products regulate the nonmetabolic function of PKM2
Shikonin
Shikonin, an active compound extracted from Lithospermum erythrorhizon, has exhibited various pharmacological effects and has been found to possess antitumor capabilities in a variety of human cancer types (94, 95). Specifically, it can impede the glycolytic function of PKM2 in BC cells (96, 97), and suppress its nonmetabolic enzyme function to impact the biological processes of these cells (36, 97–99). Shikonin works by binding with the R399/400 residues of PKM2 to hinder its nuclear translocation, hence leading to the disruption of the PKM2-HIFs-PFKFB3 pathway (36). As PFKFB3 facilitates aerobic glycolysis, reducing its levels increases the reliance on OXPHOS to meet ATP needs (100). Cancer cell growth is known to be inhibited by a transition from a glycolysis-dependent state to an OXPHOS-dependent state (101). Pandkar et al. found that shikonin inhibits the hypoxic activation of PFKFB3 by engaging with a putative nuclear localization signal (NLS) on PKM2, thereby blocking its movement into the nucleus during hypoxia. Furthermore, studies in mice carrying WT BBS MCF7 tumors revealed that shikonin treatment led to significantly reduced tumor growth, emphasizing the crucial role of targeting PKM2 signaling to inhibit the progression of tumors (36). Moreover, LEE et al. showed that shikonin is a robust and rapid inducer of late apoptosis in MDA-MB-231 cells, implying that inhibitors of PKM2 may be validated treatment targets for TNBC. Targeting PKM2 in TNBC paves the way for the development of PKM2 inhibitors as potential anti-TNBC agents (98). Therefore, focusing on targeting PKM2 in BC sets the stage for developing promising anti-BC agents in the form of PKM2 inhibitors.
Cryptanshinone
Cryptotanshinone is a liposoluble monomer of tanshinones isolated from the dried roots and rhizomes of Salvia miltiorrhiza (102). Cryptotanshinone has been recognized as a medication with anti-inflammatory and anti-oxidative properties, and it has demonstrated its ability to combat different forms of cancer by impeding cell growth, movement, and infiltration (103). Notably, significant evidence suggests that cryptotanshinone has the capacity to inhibit the metabolism of glucose in ovarian cancer cells, suggesting that it could be an effective anti-cancer agent by controlling glycolysis (104). Zhou et al. showed that cryptotanshinone has an anti-cancer effect on BC cells by inhibiting cell proliferation, migration, and invasion in vitro. In addition, it significantly decreases the expression of glycolysis-related proteins and PKM2/β-catenin signaling in BC cells (54). PKM2 is a crucial regulator that controls the transactivation of β-catenin, which is responsible for numerous critical functions of cancer cells, including invasion, migration, and angiogenesis (105). Cryptotanshinone successfully hindered the migration and invasion of BC, which could be attributed to the suppression of the PKM2/β-catenin pathway (54). Therefore, cryptotanshinone may hold promise for the advancement of novel targeted medications for BC.
Cyclosporine A
Cyclosporin A is an immunosuppressant that is not toxic to cells and was first identified in the 1970s. Initially, it was utilized to suppress immune responses after organ and marrow transplantation (106). Afterwards, it has been utilized in various medical fields where autoimmune or inflammatory processes are involved in pathology (107, 108). Recent studies have highlighted the potential antitumor activity of this compound against different cancer cells, including BC. Jiang et al. found that cyclosporin A inhibited the cell proliferation, cell cycle progression, and G1/S phase transition of BC cell lines (109). Several studies have reported that cyclosporin A hinders the expression of some oncogenes. Cyclosporin A can inhibit the proliferation of cancer cells, possibly by controlling the expression levels of c-Myc, p21, and proliferating cell nuclear antigen (PCNA) through the suppression of calcineurin (CaN)/nuclear factor of activated T cells (NFAT) activity (110). It also suppresses cancer-specific PKM2, which supports the advancement of the tumor (109, 111). The function of the PKM2 enzyme was also suppressed in MCF-7 cells when they were exposed to cyclosporin A. Pyruvate kinase catalyzes the final step in the glycolytic pathway and is responsible for net ATP production; hence, the production of ATP was evaluated in MCF-7 cells treated with cyclosporin A and was found to be significantly reduced (109). This finding shows that cyclosporin A hampers the expression of PKM2, leading to a reduction in intracellular ATP within tumor cells, which ultimately slows down cell growth and may even trigger cell death (109, 111). Thus, cyclosporin A acts as a suppressor of BC growth by targeting tumor-related PKM2.
Lapatinib
Lapatinib is a small-molecule tyrosine kinase inhibitor that targets EGFR and HER2 (112, 113). Clinical studies have revealed that lapatinib is well-tolerated and can be used alone or in combination with other drugs for the treatment of BC (114, 115). The specific molecular mechanism of lapatinib involves regulating PKM2-mediated STAT3 tyrosine phosphorylation (18, 61). Guan et al. (61) demonstrated that lapatinib hinders the proliferation of BC cells by influencing the expression of PKM2, which decreases the levels of STAT3 and phosphorylated STAT3. Downregulation of PKM2 expression by lapatinib-mediated EGFR and HER2 suppression decreases STAT3 and phosphorylated STAT3 expression, resulting in decreased gene transcription and prevention of tumor cell proliferation.
Mangifera indica
M. indica, also known as mango, is a member of the Anacardiaceae flowering family (116). Various sections of the M. indica plant contain different types of phytochemicals, and they have traditionally been used to treat a wide range of health issues such as gastrointestinal, genitourinary, ophthalmic, and respiratory conditions (117, 118). Preclinical research on extracts derived from diverse plant sections has indicated their anti-cancer, anti-inflammatory, antimicrobial, antioxidant, and immunomodulatory properties (119, 120). Specifically, various investigations have reported the anti-cancer properties of the pulp extracts of M. indica in BC. These findings suggest that consuming M. indica fruits could be beneficial for BC treatment (121). M. indica extracts (leaf, bark, and seed coats) were also found to have anticancer activity against TNBC (122, 123). These studies revealed that M. indica extracts could inhibit PKM2, but the exact mechanism underlying these effects is still not fully understood. Further research is also suggested to examine their potential in in vivo studies.
Carpesium abrotanoides (L.)
Carpesium abrotanoides Linn. (CA) is a popular medicinal plant recognized for its anti-inflammatory effects and wide range of therapeutic applications (46). Some studies have demonstrated that CA extracts exhibit promising antitumor effects on various cancer cells in vitro, including non–small cell lung, ovarian, skin, and colon cancer cells (124, 125). As a traditional herbal remedy, it is commonly used as an oral decoction for treating chronic inflammatory illnesses (126). Currently, various compounds have been isolated from the whole plant of CA that display significant cytotoxicity against MCF-7 and MDAMB-231 cells, indicating its potential anti-BC effects (46, 127). In addition, Chai et al. found that CA has dose-dependent antiproliferative effects on both metastatic (MDA-MB-231) and nonmetastatic (MCF-7) BC cells while inhibiting their migration ability. Furthermore, CA downregulates the expression of glycolysis-related proteins (PKM2, LDHA, HK2, and GLUT1) and suppresses the PKM2/HIF-1α/VEGF signaling pathway to exert its anticancer effects (46). In cancer cells, PKM2 activates the transcription of HIF-1α and its target gene VEGF in the nucleus; this stimulates the secretion of VEGF to promote angiogenesis and enhance tumor growth (41). Angiogenesis is a key event in tumor invasion and migration. Moreover, VEGFA has been linked to promoting cancer stem cell self-renewal and EMT while contributing to tumor metastasis in BC cells (43). Clinical studies have also revealed that patients with metastatic BC tend to have higher circulating levels of VEGFA (45). Consequently, targeting the PKM2/HIF-1α axis holds promise as a feasible approach for regulating VEGF in BC.
Yuanhuacine
Yuanhuacine, a daphnane-type diterpenoid as the main active ingredient, has been previously documented to exhibit cytotoxic effects in vitro against a wide range of human cancer cell lines (128–130). For instance, yuanhuacine has notable inhibitory effects on human lung cancer cells (131). Recent studies have revealed that yuanhuacine significantly inhibits effect BC cell growth (63, 132). Yuanhuacine reduced the growth of BC cells and caused apoptosis both in vivo and in vitro. The mechanism of action has been linked to yuanhuacine, which has been hypothesized to potentially disrupt the interactions of PKM2 and STAT3, hence inhibiting downstream proteins. Besides, yuanhuacine suppresses BC cells by targeting PKM2, which controls the STAT3 pathway and glycolysis. The combination of yuanhuacine and PKM2 siRNA treatment significantly hindered the phosphorylation of STAT3 (Y705) and its downstream effects, in contrast to yuanhuacine treatment alone (63). This finding could also indicate a synergistic effect of yuanhuacine when it is used with other cancer-specific treatments.
Cantharidin
Cantharidin, a sesquiterpenoid bioactive component, is among the active ingredients found in mylabris. Many in vitro studies have investigated cantharidin’s antitumor activity, which includes hindering migration, inducing apoptosis, halting the G2/M transition, and suppressing invasion (133). Yang et al. showed that EGFR activation results in the translocation of PKM2 into the nucleus, leading to increased expression of GLUT-1 and LDHA, which work together to enhance aerobic glycolysis in a positive feedback loop (105). This mechanism relies on ERK signaling, which is facilitated by importin α5 (23). Pan et al. (134) investigated the mechanism by which cantharidin influences PKM2 translocation into the nucleus in BC cells. Cantharidin reduces GLUT1 transcription and glucose uptake by blocking the transformation of PKM2 dimers into tetramers and their entry into the nucleus. Most importantly, the inhibitory effect of cantharidin on migration and invasion was significantly reversed when FBP and L-cysteine were introduced (134). These findings suggest that cantharidin and its derivatives have strong potential as anti-metastatic agents, making them highly valuable in clinical applications.
Beta‐elemene
Beta-elemene is a prominent non‐cytotoxic anticancer ingredient extracted from Curcuma zedoary that triggers cancer cell death, halts cell cycle progression, and improves radiotherapy and chemotherapy sensitivity without causing myelosuppression or notable harm to the liver or kidney (135–137). In cancer cells, the balance between tetrameric and dimeric forms of PKM2 regulates the glucose metabolic pathway, influencing energy production and the synthesis of anabolic precursors. Additionally, this interconversion between dimeric and tetrameric PKM2 helps to maintain a dynamic equilibrium (37, 138). Pan et al. showed that β‐elemene inhibited the spread of BC by impeding aerobic glycolysis. Specifically, β‐elemene hindered the transformation of dimeric and tetrameric forms of PKM2, thus inhibiting its pyruvate kinase activity, resulting in reduced utilization of glucose and generation of pyruvate and lactate. Besides its metabolic function, PKM2 acts as a nonmetabolic protein kinase and transcriptional coactivator for HIF-1α and c-MYC, which are crucial for tumorigenesis induced by the activation of EGFR. Activated EGFR facilitates the translocation of PKM2 to the nucleus through importin α5 (23, 105). β‐elemene inhibited the EGFR‐importin α5‐mediated movement of PKM2 into the nucleus and the expression of GLUT1, monocarboxylate transporter 1 (MCT1), MCT4, and LDHA (139). Collectively, these studies suggest that β‐elemene can inhibit BC metastasis by disrupting PKM2‐mediated metabolic signaling to exert an anti-BC effect.
Noncoding RNAs regulate nonmetabolic function of PKM2 in breast cancer
Current studies are focused on the impact of long noncoding RNA (lncRNA), circular RNA (circRNA), and microRNA (miRNA) on PKM2 in BC tumorigenesis and development (140–142). Noncoding RNA can influence BC cell metabolic reprogramming by targeting PKM2’s metabolic enzyme function as well as its nonmetabolic enzyme function (141, 143). In some cases, miRNAs have been found to regulate PKM2 expression in BC cells. For instance, miR-122 is abundantly released by tumor cells and promotes metastasis by adapting to the metabolic environment in the pre-metastatic niche, whereas down-regulation of PKM2 and GLUT1 limits glucose consumption in BC cells (144). miR-Let-7a inhibited MDA-MB-231 cell proliferation and particularly upregulated the levels of PMK2, oxidative phosphorylation, and reactive oxygen species (ROS) in TNBC and improved the sensitivity of BC cells to the tumor suppressor doxorubicin in BC metastasis (145). CircKIF4A, which is largely found in the cytoplasm, may interact with miR-335, while aldolase A (ALDOA) and octamer-binding transcription factor 4 (OCT4) serve as miR-335’s downstream targets. ALDOA and OCT4 are metabolic proteins that control glycolytic proteins such as HK2 and PKM2 (146–148). Increased miR-335 expression resulted in decreased levels of ALDOA/HK2 and OCT4/PKM2 proteins. As a result, circKIF4A regulates glucose metabolism through the miR-335-ALDOA/OCT4-HK2/PKM2 pathway (149).
Zheng et al. demonstrated the importance of HIF-1α antisense lncRNA (HIFAL) in maintaining and increasing HIF-1α-driven transactivation and glycolysis. Functionally, HIFAL recruits PHD3 to PKM2, causing its prolyl hydroxylation and facilitating the entry of the PKM2/PHD3 complex into the nucleus by interacting with heterogeneous nuclear ribonucleoprotein (hnRNP) F. This promotes HIF-1α transactivation, glucose absorption, and lactate generation in BC cells (150). Besides, Yao et al. revealed that miR-Let-7a-5p suppresses aerobic glycolysis by modulating the STAT3/hnRNP-A1/PKM2 signaling pathway. They showed that hnRNP-A1 controls PKM2 gene selective cleavage, resulting in enhanced PKM2 synthesis. STAT3 may increase hnRNP-A1 transcription, whereas miR-Let-7a-5p suppresses STAT3 and hence reduces PKM2 (141, 151).
A study conducted by Wen et al. revealed that miR-152 suppresses growth and angiogenesis in BC by suppressing both β-catenin and PKM2. β-catenin, which is a downstream target of insulin-like growth factor 1 (IGF-1), plays a role in controlling cell proliferation (53). Recent evidence has shown that miR-148a/152 activation contributes to the suppression of PKM2 and NF-κB p56 expression in TNBC cells. NF-κB p56 interacts with PKM2 to regulate early growth response 1 (EGR1) expression. EGR1 can bind to multiple sites on miRNA gene promoters, thereby controlling miR-148a and miR-152 expression (152).
Conclusion and future perspectives
In this review, we reviewed how PKM2 has emerged as a crucial player in BC; through its role as a transcriptional co-activator and regulator of gene expression, PKM2 has been demonstrated to contribute to the epigenetic regulation of gene transcription. Understanding the diverse functions of PKM2 beyond its metabolic role opens new avenues for targeting this enzyme in BC treatment. The nonmetabolic role of PKM2 controls the expression of cancer-related genes implicated in various aspects of BC progression, such as cell cycle, proliferation, programmed cell death, angiogenesis, migration and invasion, tumor microenvironment, and others. Natural products such as shikonin, cryptanshinone, cyclosporine A, lapatinib, mangifera indica, beta-elemene, yuanhuacine, cantharidin, and others control PKM2’s nonmetabolic function and affect BC progression. In addition, the interaction of PKM2 with noncoding RNAs has garnered increasing interest. Noncoding RNAs regulate PKM2’s nonmetabolic enzyme function and influence BC growth. In the future, it will be imperative to further investigate the nonmetabolic function of PKM2 in BC in order to gain a comprehensive understanding of its involvement in the growth and advancement of tumors. In addition, exploring the interaction between metabolic reprogramming and PKM2-mediated nonmetabolic functions in BC is an intriguing area for further research. This knowledge could potentially lead to the use of PKM2 as a molecular marker for BC diagnosis and prognosis, which could be highly important for the therapeutic targeting of BC.
Author contributions
MJ: Conceptualization, Data curation, Methodology, Validation, Writing – original draft, Writing – review & editing. MW: Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. GA: Data curation, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. BT: Data curation, Formal analysis, Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. TB: Formal analysis, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. EM: Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. EO: Formal analysis, Investigation, Methodology, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NC: Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing. AA: Investigation, Methodology, Validation, Visualization, Writing – original draft, Writing – review & editing.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations/acronyms
CA, Carpesium abrotanoides Linn.; TNBC, triple-negative breast cancer; PK, pyruvate kinase; PEP, phosphoenolpyruvate; PKM2, pyruvate kinase M2; FBP, fructose 1,6-bisphosphate; ISCD, HIF-1α, hypoxia-inducible factor-1α; EGFR, epidermal growth factor receptor; VEGF, vascular endothelial growth factor; BC, breast cancer; STAT3, signal transducer and activator of transcription 3; YHC, yuanhuacine; RNA, ribonucleic acid; DNA, deoxyribonucleic acid; NF-κB, nuclear factor-kappa B; CTS, cryptotanshinone; CsA, cyclosporin A; HER2, human epidermal growth factor receptor 2.
References
1. Sung H, Ferlay J, Siegel RL, Laversanne M, Soerjomataram I, Jemal A, et al. Global cancer statistics 2020: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA: Cancer J Clin. (2021) 71:209–49. doi: 10.3322/caac.21660
2. Anderson BO, Ilbawi AM, Fidarova E, Weiderpass E, Stevens L, Abdel-Wahab M, et al. The Global Breast Cancer Initiative: a strategic collaboration to strengthen health care for non-communicable diseases. Lancet Oncol. (2021) 22:578–81. doi: 10.1016/S1470-2045(21)00071-1
3. Yeo SK, Guan J-L. Breast cancer: multiple subtypes within a tumor? Trends Cancer. (2017) 3:753–60. doi: 10.1016/j.trecan.2017.09.001
4. Bianchini G, Balko JM, Mayer IA, Sanders ME, Gianni L. Triple-negative breast cancer: challenges and opportunities of a heterogeneous disease. Nat Rev Clin Oncol. (2016) 13:674–90. doi: 10.1038/nrclinonc.2016.66
5. Xiao H, Zhang L, Chen Y, Zhou C, Wang X, Wang D, et al. PKM2 promotes breast cancer progression by regulating epithelial mesenchymal transition. Anal Cell Pathol (Amst). (2020) 2020:8396023. doi: 10.1155/2020/8396023
6. Wu H, Yang P, Hu W, Wang Y, Lu Y, Zhang L, et al. Overexpression of PKM2 promotes mitochondrial fusion through attenuated p53 stability. Oncotarget. (2016) 7:78069–82. doi: 10.18632/oncotarget.12942
7. Christofk HR, Vander Heiden MG, Wu N, Asara JM, Cantley LC. Pyruvate kinase M2 is a phosphotyrosine-binding protein. Nature. (2008) 452:181–6. doi: 10.1038/nature06667
8. Gu Z, Xia J, Xu H, Frech I, Tricot G, Zhan F. NEK2 promotes aerobic glycolysis in multiple myeloma through regulating splicing of pyruvate kinase. J Hematol Oncol. (2017) 10:17. doi: 10.1186/s13045-017-0392-4
9. Fu Y, Liu S, Yin S, Niu W, Xiong W, Tan M, et al. The reverse Warburg effect is likely to be an Achilles’ heel of cancer that can be exploited for cancer therapy. Oncotarget. (2017) 8:57813–25. doi: 10.18632/oncotarget.18175
10. Witney TH, James ML, Shen B, Chang E, Pohling C, Arksey N, et al. PET imaging of tumor glycolysis downstream of hexokinase through noninvasive measurement of pyruvate kinase M2. Sci Trans Med. (2015) 7:310ra169–310ra169. doi: 10.1126/scitranslmed.aac6117
11. Tanaka T, Harano Y, Sue F, Morimura H. Crystallization, characterization and metabolic regulation of two types of pyruvate kinase isolated from rat tissues. J Biochem. (1967) 62:71–91. doi: 10.1093/oxfordjournals.jbchem.a128639
12. Tsutsumi H, Tani K, Fujii H, Miwa S. Expression of L- and M-type pyruvate kinase in human tissues. Genomics. (1988) 2:86–9. doi: 10.1016/0888-7543(88)90112-7
13. Goonetilleke KS, Mason JM, Siriwardana P, King NK, France MW, Siriwardena AK. Diagnostic and prognostic value of plasma tumor M2 pyruvate kinase in periampullary cancer: evidence for a novel biological marker of adverse prognosis. Pancreas. (2007) 34:318–24. doi: 10.1097/MPA.0b013e31802ee9c7
14. Christofk HR, Vander Heiden MG, Harris MH, Ramanathan A, Gerszten RE, Wei R, et al. The M2 splice isoform of pyruvate kinase is important for cancer metabolism and tumour growth. Nature. (2008) 452:230–3. doi: 10.1038/nature06734
15. Hamabe A, Konno M, Tanuma N, Shima H, Tsunekuni K, Kawamoto K, et al. Role of pyruvate kinase M2 in transcriptional regulation leading to epithelial-mesenchymal transition. Proc Natl Acad Sci U.S.A. (2014) 111:15526–31. doi: 10.1073/pnas.1407717111
16. Harris I, McCracken S, Mak TW. PKM2: a gatekeeper between growth and survival. Cell Res. (2012) 22:447–9. doi: 10.1038/cr.2011.203
17. Wong N, Ojo D, Yan J, Tang D. PKM2 contributes to cancer metabolism. Cancer Lett. (2015) 356:184–91. doi: 10.1016/j.canlet.2014.01.031
18. Gao X, Wang H, Yang JJ, Liu X, Liu ZR. Pyruvate kinase M2 regulates gene transcription by acting as a protein kinase. Mol Cell. (2012) 45:598–609. doi: 10.1016/j.molcel.2012.01.001
19. Zhu K, Li Y, Deng C, Wang Y, Piao J, Lin Z, et al. Significant association of PKM2 and NQO1 proteins with poor prognosis in breast cancer. Pathol Res Pract. (2020) 216:153173. doi: 10.1016/j.prp.2020.153173
20. Noguchi T, Inoue H, Tanaka T. The M1-and M2-type isozymes of rat pyruvate kinase are produced from the same gene by alternative RNA splicing. J Biol Chem. (1986) 261:13807–12. doi: 10.1016/S0021-9258(18)67091-7
21. Takenaka M, Noguchi T, Sadahiro S, Hirai H, Yamada K, Matsuda T, et al. Isolation and characterization of the human pyruvate kinase M gene. Eur J Biochem. (1991) 198:101–6. doi: 10.1111/j.1432-1033.1991.tb15991.x
22. Dombrauckas JD, Santarsiero BD, Mesecar AD. Structural basis for tumor pyruvate kinase M2 allosteric regulation and catalysis. Biochemistry. (2005) 44:9417–29. doi: 10.1021/bi0474923
23. Yang W, Zheng Y, Xia Y, Ji H, Chen X, Guo F, et al. ERK1/2-dependent phosphorylation and nuclear translocation of PKM2 promotes the Warburg effect. Nat Cell Biol. (2012) 14:1295–304. doi: 10.1038/ncb2629
24. Keller KE, Tan IS, Lee Y-S. SAICAR stimulates pyruvate kinase isoform M2 and promotes cancer cell survival in glucose-limited conditions. Science. (2012) 338:1069–72. doi: 10.1126/science.1224409
25. Mazurek S. Pyruvate kinase type M2: a key regulator of the metabolic budget system in tumor cells. Int J Biochem Cell Biol. (2011) 43:969–80. doi: 10.1016/j.biocel.2010.02.005
26. Zhang Z, Deng X, Liu Y, Liu Y, Sun L, Chen F. PKM2, function and expression and regulation. Cell Biosci. (2019) 9:1–25. doi: 10.1186/s13578-019-0317-8
27. Luo W, Semenza GL. Emerging roles of PKM2 in cell metabolism and cancer progression. Trends Endocrinol Metab. (2012) 23:560–6. doi: 10.1016/j.tem.2012.06.010
28. Keller Kirstie E, Doctor Zainab M, Dwyer Zachary W, Lee Y-S. SAICAR induces protein kinase activity of PKM2 that is necessary for sustained proliferative signaling of cancer cells. Mol Cell. (2014) 53:700–9. doi: 10.1016/j.molcel.2014.02.015
29. Prakasam G, Iqbal MA, Bamezai RN, Mazurek S. Posttranslational modifications of pyruvate kinase M2: tweaks that benefit cancer. Front Oncol. (2018) 8:22. doi: 10.3389/fonc.2018.00022
30. Mazurek S, Boschek CB, Hugo F, Eigenbrodt E. Pyruvate kinase type M2 and its role in tumor growth and spreading. Semin Cancer Biol. (2005) 15(4):300–8. doi: 10.1016/j.semcancer.2005.04.009
31. Verma H, Cholia RP, Kaur S, Dhiman M, Mantha AK. A short review on cross-link between pyruvate kinase (PKM2) and Glioblastoma Multiforme. Metab Brain Dis. (2021) 36:751–65. doi: 10.1007/s11011-021-00690-y
32. Zahra K, Dey T, Ashish Mishra SP, Pandey U. Pyruvate kinase M2 and cancer: the role of PKM2 in promoting tumorigenesis. Front Oncol. (2020) 10:159. doi: 10.3389/fonc.2020.00159
33. Eigenbrodt E, Reinacher M, Scheefers-Borchel U, Scheefers H, Friis R. Double role for pyruvate kinase type M2 in the expansion of phosphometabolite pools found in tumor cells. Crit Rev Oncogenesis. (1992) 3:91–115.
34. Soga T. Cancer metabolism: key players in metabolic reprogramming. Cancer Sci. (2013) 104:275–81. doi: 10.1111/cas.2013.104.issue-3
35. He Y, Gao M, Cao Y, Tang H, Liu S, Tao Y. Nuclear localization of metabolic enzymes in immunity and metastasis. Biochim Biophys Acta Rev Cancer. (2017) 1868:359–71. doi: 10.1016/j.bbcan.2017.07.002
36. Pandkar MR, Raveendran A, Biswas K, Mutnuru SA, Mishra J, Samaiya A, et al. PKM2 dictates the poised chromatin state of PFKFB3 promoter to enhance breast cancer progression. NAR Cancer. (2023) 5. doi: 10.1093/narcan/zcad032
37. Lv L, Xu YP, Zhao D, Li FL, Wang W, Sasaki N, et al. Mitogenic and oncogenic stimulation of K433 acetylation promotes PKM2 protein kinase activity and nuclear localization. Mol Cell. (2013) 52:340–52. doi: 10.1016/j.molcel.2013.09.004
38. Wang HJ, Hsieh YJ, Cheng WC, Lin CP, Lin YS, Yang SF, et al. JMJD5 regulates PKM2 nuclear translocation and reprograms HIF-1α-mediated glucose metabolism. Proc Natl Acad Sci U.S.A. (2014) 111:279–84. doi: 10.1073/pnas.1311249111
39. Gutschner T, Diederichs S. The hallmarks of cancer: a long non-coding RNA point of view. RNA Biol. (2012) 9:703–19. doi: 10.4161/rna.20481
40. Yi X, Qi M, Huang M, Zhou S, Xiong J. Honokiol inhibits HIF-1α-mediated glycolysis to halt breast cancer growth. Front Pharmacol. (2022) 13:796763. doi: 10.3389/fphar.2022.796763
41. Azoitei N, Becher A, Steinestel K, Rouhi A, Diepold K, Genze F, et al. PKM2 promotes tumor angiogenesis by regulating HIF-1α through NF-κB activation. Mol Cancer. (2016) 15:3. doi: 10.1186/s12943-015-0490-2
42. Liu WR, Tian MX, Yang LX, Lin YL, Jin L, Ding ZB, et al. PKM2 promotes metastasis by recruiting myeloid-derived suppressor cells and indicates poor prognosis for hepatocellular carcinoma. Oncotarget. (2015) 6:846–61. doi: 10.18632/oncotarget.2749
43. Kim M, Jang K, Miller P, Picon-Ruiz M, Yeasky TM, El-Ashry D, et al. VEGFA links self-renewal and metastasis by inducing Sox2 to repress miR-452, driving Slug. Oncogene. (2017) 36:5199–211. doi: 10.1038/onc.2017.4
44. Adams J, Carder PJ, Downey S, Forbes MA, MacLennan K, Allgar V, et al. Vascular endothelial growth factor (VEGF) in breast cancer: comparison of plasma, serum, and tissue VEGF and microvessel density and effects of tamoxifen. Cancer Res. (2000) 60:2898–905.
45. Arias-Pulido H, Chaher N, Gong Y, Qualls C, Vargas J, Royce M. Tumor stromal vascular endothelial growth factor A is predictive of poor outcome in inflammatory breast cancer. BMC Cancer. (2012) 12:298. doi: 10.1186/1471-2407-12-298
46. Chai X-X, Le Y-F, Wang J-C, Mei C-X, Feng J-F, Zhao H, et al. Carpesium abrotanoides (L.) root as a potential source of natural anticancer compounds: targeting glucose metabolism and PKM2/HIF-1α Axis of breast cancer cells. J Food Sci. (2019) 84:3825–32. doi: 10.1111/1750-3841.14953
47. Luo W, Hu H, Chang R, Zhong J, Knabel M, O'Meally R, et al. Pyruvate kinase M2 is a PHD3-stimulated coactivator for hypoxia-inducible factor 1. Cell. (2011) 145:732–44. doi: 10.1016/j.cell.2011.03.054
48. Liu J, Xiao Q, Xiao J, Niu C, Li Y, Zhang X, et al. Wnt/β-catenin signalling: function, biological mechanisms, and therapeutic opportunities. Signal Transduct Target Ther. (2022) 7:3. doi: 10.1038/s41392-021-00762-6
49. Ring A, Kim YM, Kahn M. Wnt/catenin signaling in adult stem cell physiology and disease. Stem Cell Rev Rep. (2014) 10:512–25. doi: 10.1007/s12015-014-9515-2
50. Xu X, Zhang M, Xu F, Jiang S. Wnt signaling in breast cancer: biological mechanisms, challenges and opportunities. Mol Cancer. (2020) 19:165. doi: 10.1186/s12943-020-01276-5
51. Zhao Z, Song Z, Liao Z, Liu Z, Sun H, Lei B, et al. PKM2 promotes stemness of breast cancer cell by through Wnt/β-catenin pathway. Tumour Biol. (2016) 37:4223–34. doi: 10.1007/s13277-015-4121-8
52. Wang J, Li M, Chen D, Nie J, Xi Y, Yang X, et al. Expression of C-myc and β-catenin and their correlation in triple negative breast cancer. Minerva Med. (2017) 108:513–7. doi: 10.23736/S0026-4806.17.05213-2
53. Wen Y-Y, Liu W-T, Sun H-R, Ge X, Shi Z-M, Wang M, et al. IGF-1-mediated PKM2/β-catenin/miR-152 regulatory circuit in breast cancer. Sci Rep. (2017) 7:15897. doi: 10.1038/s41598-017-15607-y
54. Zhou J, Su CM, Chen HA, Du S, Li CW, Wu H, et al. Cryptanshinone inhibits the glycolysis and inhibits cell migration through PKM2/β-catenin axis in breast cancer. Onco Targets Ther. (2020) 13:8629–39. doi: 10.2147/OTT.S239134
55. Song L, Turkson J, Karras JG, Jove R, Haura EB. Activation of Stat3 by receptor tyrosine kinases and cytokines regulates survival in human non-small cell carcinoma cells. Oncogene. (2003) 22:4150–65. doi: 10.1038/sj.onc.1206479
56. Tolomeo M, Cascio A. The multifaced role of STAT3 in cancer and its implication for anticancer therapy. Int J Mol Sci. (2021) 22. doi: 10.3390/ijms22020603
57. Demaria M, Poli V. PKM2, STAT3 and HIF-1α: The Warburg’s vicious circle. Jakstat. (2012) 1:194–6. doi: 10.4161/jkst.20662
58. Yang P, Li Z, Fu R, Wu H, Li Z. Pyruvate kinase M2 facilitates colon cancer cell migration via the modulation of STAT3 signalling. Cell Signal. (2014) 26:1853–62. doi: 10.1016/j.cellsig.2014.03.020
59. Li L, Zhang Y, Qiao J, Yang JJ, Liu Z-R. Pyruvate kinase M2 in blood circulation facilitates tumor growth by promoting angiogenesis*. J Biol Chem. (2014) 289:25812–21. doi: 10.1074/jbc.M114.576934
60. Niu G, Wright KL, Huang M, Song L, Haura E, Turkson J, et al. Constitutive Stat3 activity up-regulates VEGF expression and tumor angiogenesis. Oncogene. (2002) 21:2000–8. doi: 10.1038/sj.onc.1205260
61. Guan M, Tong Y, Guan M, Liu X, Wang M, Niu R, et al. Lapatinib inhibits breast cancer cell proliferation by influencing PKM2 expression. Technol Cancer Res Treat. (2018) 17:1533034617749418. doi: 10.1177/1533034617749418
62. Qin W, Tian Y, Zhang J, Liu W, Zhou Q, Hu S, et al. The double inhibition of PDK1 and STAT3-Y705 prevents liver metastasis in colorectal cancer. Sci Rep. (2019) 9(1):12973. doi: 10.1038/s41598-019-49480-8
63. Shang XY, Wang YJ, Hou ZL, Wang XY, Zhang H, Yang CY, et al. A natural PKM2 targeting agent as a potential drug for breast cancer treatment. Clin Transl Med. (2023) 13:e1157. doi: 10.1002/ctm2.v13.1
64. Nalla LV, Dharavath A, Behera SK, Khairnar A. Alpha mangostin inhibits proliferation, migration, and invasion of human breast cancer cells via STAT3 inhibition. Adv Cancer Biol Metastasis. (2023) 7:100089. doi: 10.1016/j.adcanc.2023.100089
65. Chen H, Liu H, Qing G. Targeting oncogenic Myc as a strategy for cancer treatment. Signal Transduct Target Ther. (2018) 3:5. doi: 10.1038/s41392-018-0008-7
66. Dominguez-Sola D, Gautier J. MYC and the control of DNA replication. Cold Spring Harb Perspect Med. (2014) 4. doi: 10.1101/cshperspect.a014423
67. Haque R, Song J, Haque M, Lei F, Sandhu P, Ni B, et al. c-Myc-induced survivin is essential for promoting the notch-dependent T cell differentiation from hematopoietic stem cells. Genes (Basel). (2017) 8(3):97. doi: 10.3390/genes8030097
68. Garg H, Suri P, Gupta JC, Talwar G, Dubey S. Survivin: a unique target for tumor therapy. Cancer Cell Int. (2016) 16:1–14. doi: 10.1186/s12935-016-0326-1
69. Cosgrave N, Hill AD, Young LS. Growth factor-dependent regulation of survivin by c-myc in human breast cancer. J Mol Endocrinol. (2006) 37:377–90. doi: 10.1677/jme.1.02118
70. Yu P, Li AX, Chen XS, Tian M, Wang HY, Wang XL, et al. PKM2-c-myc-survivin cascade regulates the cell proliferation, migration, and tamoxifen resistance in breast cancer. Front Pharmacol. (2020) 11:550469. doi: 10.3389/fphar.2020.550469
71. Rodríguez-Enríquez S, Marín-Hernández Á, Gallardo-Pérez JC, Pacheco-Velázquez SC, Belmont-Díaz JA, Robledo-Cadena DX, et al. Transcriptional regulation of energy metabolism in cancer cells. Cells. (2019) 8. doi: 10.3390/cells8101225
72. Varlı M, Kim SJ, Noh M-G, Kim YG, Ha H-H, Kim KK, et al. KITENIN promotes aerobic glycolysis through PKM2 induction by upregulating the c-Myc/hnRNPs axis in colorectal cancer. Cell Biosci. (2023) 13:146. doi: 10.1186/s13578-023-01089-1
73. Li L, Peng G, Liu X, Zhang Y, Han H, Liu ZR. Pyruvate kinase M2 coordinates metabolism switch between glycolysis and glutaminolysis in cancer cells. iScience. (2020) 23:101684. doi: 10.1016/j.isci.2020.101684
74. Fang ZH, Dong CL, Chen Z, Zhou B, Liu N, Lan HF, et al. Transcriptional regulation of survivin by c-Myc in BCR/ABL-transformed cells: implications in anti-leukaemic strategy. J Cell Mol Med. (2009) 13(8b):2039–52. doi: 10.1111/j.1582-4934.2008.00549.x
75. Hayden MS, Ghosh SJC. Shared principles in NF-κB signaling. Cell. (2008) 132(3):344–62. doi: 10.1016/j.cell.2008.01.020
76. Hoffmann A, Baltimore D. Circuitry of nuclear factor κB signaling. Immunol Rev. (2006) 210:171–86. doi: 10.1111/j.0105-2896.2006.00375.x
77. Chariot A. 20 years of NF-kappa B. Biochem Pharmacol. (2006) 72:1051–3. doi: 10.1016/j.bcp.2006.08.023
78. Zheng B, Geng L, Zeng L, Liu F, Huang Q. AKT2 contributes to increase ovarian cancer cell migration and invasion through the AKT2-PKM2-STAT3/NF-κB axis. Cell Signal. (2018) 45:122–31. doi: 10.1016/j.cellsig.2018.01.021
79. Han D, Wei W, Chen X, Zhang Y, Wang Y, Zhang J, et al. NF-κB/RelA-PKM2 mediates inhibition of glycolysis by fenofibrate in glioblastoma cells. Oncotarget. (2015) 6(28):26119. doi: 10.18632/oncotarget.v6i28
80. Ma C, Zu X, Liu K, Bode AM, Dong Z, Liu Z, et al. Knockdown of pyruvate kinase M inhibits cell growth and migration by reducing NF-kB activity in triple-negative breast cancer cells. Mol Cells. (2019) 42:628–36. doi: 10.14348/molcells.2019.0038
81. Israël A. The IKK complex, a central regulator of NF-κB activation. Cold Spring Harb Perspect Biol. (2010) 2:a000158. doi: 10.1101/cshperspect.a000158
82. Yang W, Xia Y, Hawke D, Li X, Liang J, Xing D, et al. PKM2 phosphorylates histone H3 and promotes gene transcription and tumorigenesis. Cell. (2012) 150(4):685–96. doi: 10.1016/j.cell.2012.07.018
83. Pan D. The hippo signaling pathway in development and cancer. Dev Cell. (2010) 19:491–505. doi: 10.1016/j.devcel.2010.09.011
84. Song H, Mak KK, Topol L, Yun K, Hu J, Garrett L, et al. Mammalian Mst1 and Mst2 kinases play essential roles in organ size control and tumor suppression. Proc Natl Acad Sci U.S.A. (2010) 107:1431–6.
85. Ji F, Guo B, Wang N, Zhong C, Huang L, Huang Y, et al. Pyruvate kinase M2 interacts with mammalian sterile 20-like kinase 1 and inhibits tamoxifen-induced apoptosis in human breast cancer cells. Tumour Biol. (2017) 39(6):1010428317692251. doi: 10.1177/1010428317692251
86. Singh K, Pruski MA, Polireddy K, Jones NC, Chen Q, Yao J, et al. Mst1/2 kinases restrain transformation in a novel transgenic model of Ras driven non-small cell lung cancer. Oncogene. (2020) 39:1152+. doi: 10.1038/s41388-019-1031-z
87. Mandlekar S, Yu R, Tan TH, Kong AN. Activation of caspase-3 and c-Jun NH2-terminal kinase-1 signaling pathways in tamoxifen-induced apoptosis of human breast cancer cells. Cancer Res. (2000) 60:5995–6000.
88. Almouhanna F, Blagojevic B, Can S, Ghanem A, Wölfl S. Pharmacological activation of pyruvate kinase M2 reprograms glycolysis leading to TXNIP depletion and AMPK activation in breast cancer cells. Cancer Metab. (2021) 9:5. doi: 10.1186/s40170-021-00239-8
89. Liang J, Mills GB. AMPK: a contextual oncogene or tumor suppressor? Cancer Res. (2013) 73:2929–35. doi: 10.1158/0008-5472.CAN-12-3876
90. Rocha GZ, Dias MM, Ropelle ER, Osório-Costa F, Rossato FA, Vercesi AE, et al. Metformin amplifies chemotherapy-induced AMPK activation and antitumoral growth. Clin Cancer Res. (2011) 17(12):3993–4005. doi: 10.1158/1078-0432.CCR-10-2243
91. Zhang Y, Wang Y, Bao C, Xu Y, Shen H, Chen J, et al. Metformin interacts with AMPK through binding to γ subunit. Mol Cell Biochem. (2012) 368:69–76. doi: 10.1007/s11010-012-1344-5
92. Zhang Y, Wu M-J, Lu W-C, Li Y-C, Chang CJ, Yang J-Y. Metabolic switch regulates lineage plasticity and induces synthetic lethality in triple-negative breast cancer. Cell Metab. (2024) 36:193–208.e8. doi: 10.1016/j.cmet.2023.12.003
93. Zhao X, Zhao L, Yang H, Li J, Min X, Yang F, et al. Pyruvate kinase M2 interacts with nuclear sterol regulatory element–binding protein 1a and thereby activates lipogenesis and cell proliferation in hepatocellular carcinoma. J Biol Chem. (2018) 293(17):6623–34. doi: 10.1074/jbc.RA117.000100
94. Chen C, Xiao W, Huang L, Yu G, Ni J, Yang L, et al. Shikonin induces apoptosis and necroptosis in pancreatic cancer via regulating the expression of RIP1/RIP3 and synergizes the activity of gemcitabine. Am J Transl Res. (2017) 9:5507–17.
95. Lin TJ, Lin HT, Chang WT, Mitapalli SP, Hsiao PW, Yin SY, et al. Shikonin-enhanced cell immunogenicity of tumor vaccine is mediated by the differential effects of DAMP components. Mol Cancer. (2015) 14:174. doi: 10.1186/s12943-015-0435-9
96. Dai Y, Liu Y, Li J, Jin M, Yang H, Huang G. Shikonin inhibited glycolysis and sensitized cisplatin treatment in non-small cell lung cancer cells via the exosomal pyruvate kinase M2 pathway. Bioengineered. (2022) 13:13906–18. doi: 10.1080/21655979.2022.2086378
97. Chen X, Yang L, Oppenheim JJ, Howard MZ. Cellular pharmacology studies of shikonin derivatives. Phytother Res. (2002) 16:199–209. doi: 10.1002/ptr.v16:3
98. LEE JS, Oh Y, Lee J-S, Park JH, Shin J-K, Han J-H, et al. Combination treatment using pyruvate kinase M2 inhibitors for the sensitization of high density triple-negative breast cancer cells. In Vivo. (2022) 36:2105–15. doi: 10.21873/invivo.12936
99. Zhang Q, Liu Q, Zheng S, Liu T, Yang L, Han X, et al. Shikonin Inhibits Tumor Growth of ESCC by suppressing PKM2 mediated Aerobic Glycolysis and STAT3 Phosphorylation. J Cancer. (2021) 12:4830–40. doi: 10.7150/jca.58494
100. Lu L, Chen Y, Zhu Y. The molecular basis of targeting PFKFB3 as a therapeutic strategy against cancer. Oncotarget. (2017) 8:62793–802. doi: 10.18632/oncotarget.19513
101. Luengo A, Li Z, Gui DY, Sullivan LB, Zagorulya M, Do BT, et al. Increased demand for NAD(+) relative to ATP drives aerobic glycolysis. Mol Cell. (2021) 81:691–707.e6. doi: 10.1016/j.molcel.2020.12.012
102. Wang Z, Wan H, Tong X, He Y, Yang J, Zhang L, et al. An integrative strategy for discovery of functional compound combination from Traditional Chinese Medicine: Danhong Injection as a model. Biomed Pharmacother. (2021) 138:111451. doi: 10.1016/j.biopha.2021.111451
103. Feng Z, Zheng W, Li X, Lin J, Xie C, Li H, et al. Cryptotanshinone protects against IL-1β-induced inflammation in human osteoarthritis chondrocytes and ameliorates the progression of osteoarthritis in mice. Int Immunopharmacol. (2017) 50:161–7. doi: 10.1016/j.intimp.2017.06.017
104. Yang Y, Cao Y, Chen L, Liu F, Qi Z, Cheng X, et al. Cryptotanshinone suppresses cell proliferation and glucose metabolism via STAT3/SIRT3 signaling pathway in ovarian cancer cells. Cancer Med. (2018) 7:4610–8. doi: 10.1002/cam4.2018.7.issue-9
105. Yang W, Xia Y, Ji H, Zheng Y, Liang J, Huang W, et al. Nuclear PKM2 regulates β-catenin transactivation upon EGFR activation. Nature. (2011) 480:118–22. doi: 10.1038/nature10598
106. Ferns G, Reidy M, Ross R. Vascular effects of cyclosporine A in vivo and in vitro. Am J Pathol. (1990) 137:403.
107. Elliott JF, Lin Y, Mizel SB, Bleackley RC, Harnish DG, Paetkau V. Induction of interleukin 2 messenger RNA inhibited by cyclosporin A. Science. (1984) 226:1439–41. doi: 10.1126/science.6334364
108. Lim KK, Su WD, Schroeter AL, Sabers CJ, Abraham RT, Pittelkow MR. Cyclosporine in the treatment of dermatologic disease: an update. Mayo Clinic Proc. (1996) 71(12):1182–91. doi: 10.4065/71.12.1182
109. Jiang K, He B, Lai L, Chen Q, Liu Y, Guo Q, et al. Cyclosporine A inhibits breast cancer cell growth by downregulating the expression of pyruvate kinase subtype M2. Int J Mol Med. (2012) 30:302–8. doi: 10.3892/ijmm.2012.989
110. Masuo T, Okamura S, Zhang Y, Mori M. Cyclosporine A inhibits colorectal cancer proliferation probably by regulating expression levels of c-Myc, p21(WAF1/CIP1) and proliferating cell nuclear antigen. Cancer Lett. (2009) 285:66–72. doi: 10.1016/j.canlet.2009.05.001
111. Trombino S, Curcio F, Poerio T, Pellegrino M, Russo R, Cassano R. Chitosan membranes filled with cyclosporine A as possible devices for local administration of drugs in the treatment of breast cancer. Molecules. (2021) 26:1889. doi: 10.3390/molecules26071889
112. Ito Y, Tokudome N, Sugihara T, Takahashi S, Hatake K. Does lapatinib, a small-molecule tyrosine kinase inhibitor, constitute a breakthrough in the treatment of breast cancer? Breast Cancer. (2007) 14:156–62. doi: 10.2325/jbcs.971
113. Gilmer TM, Cable L, Alligood K, Rusnak D, Spehar G, Gallagher KT, et al. Impact of common epidermal growth factor receptor and HER2 variants on receptor activity and inhibition by lapatinib. Cancer Res. (2008) 68:571–9. doi: 10.1158/0008-5472.CAN-07-2404
114. Gui X, Li H, Yan Y, Zhang R. Efficacy of lapatinib combined with capecitabine in patients with HER2−positive metastatic breast cancer in a real−world study. Oncol Lett. (2020) 20:1–1. doi: 10.3892/ol.2020.12241
115. Elwaie TA, Abbas SE, Aly EI, George RF, Ali H, Kraiouchkine N, et al. HER2 kinase-targeted breast cancer therapy: Design, synthesis, and in vitro and in vivo evaluation of novel lapatinib congeners as selective and potent HER2 inhibitors with favorable metabolic stability. J Medicinal Chem. (2020) 63:15906–45. doi: 10.1021/acs.jmedchem.0c01647
116. Hanif MA, Nawaz H, Khan M, Byrne H. Medicinal Plants of South Asia. Amsterdam: Susan Dennis (2020).
117. Masibo M, He Q. Major mango polyphenols and their potential significance to human health. Compr Rev Food Sci Food Saf. (2008) 7:309–19. doi: 10.1111/j.1541-4337.2008.00047.x
118. Derese S, Guantai E, Souaibou Y, Kuete V. Mangifera indica L.(anacardiaceae). In: Medicinal Spices and Vegetables from Africa. Elsevier (2017). p. 451–83.
119. El-Gied AAA, Joseph MR, Mahmoud IM, Abdelkareem AM, Al Hakami AM, Hamid ME. Antimicrobial activities of seed extracts of mango (Mangifera indica L.). Adv Microbiol. (2012) 2:571–6. doi: 10.4236/aim.2012.24074
120. Parham S, Kharazi AZ, Bakhsheshi-Rad HR, Nur H, Ismail AF, Sharif S, et al. Antioxidant, antimicrobial and antiviral properties of herbal materials. Antioxid (Basel). (2020) 9. doi: 10.3390/antiox9121309
121. Banerjee N, Kim H, Krenek K, Talcott ST, Mertens-Talcott SU. Mango polyphenolics suppressed tumor growth in breast cancer xenografts in mice: Role of the PI3K/AKT pathway and associated microRNAs. Nutr Res. (2015) 35:744–51. doi: 10.1016/j.nutres.2015.06.002
122. Rasul A, Riaz A, Wei W, Sarfraz I, Hassan M, Li J, et al. Mangifera indica extracts as novel PKM2 inhibitors for treatment of triple negative breast cancer. BioMed Res Int. (2021) 2021:5514669. doi: 10.1155/2021/5514669
123. Yap KM, Sekar M, Seow LJ, Gan SH, Bonam SR, Mat Rani NNI, et al. Mangifera indica (Mango): A promising medicinal plant for breast cancer therapy and understanding its potential mechanisms of action. Breast Cancer (Dove Med Press). (2021) 13:471–503. doi: 10.2147/BCTT.S316667
124. Li X-W, Weng L, Gao X, Zhao Y, Pang F, Liu J-H, et al. Antiproliferative and apoptotic sesquiterpene lactones from Carpesium faberi. Bioorg Medicinal Chem Lett. (2011) 21:366–72. doi: 10.1016/j.bmcl.2010.10.138
125. Ibrahim SRM, Fadil SA, Fadil HA, Hareeri RH, Abdallah HM, Mohamed GA. Ethnobotanical uses, phytochemical composition, biosynthesis, and pharmacological activities of Carpesium abrotanoides L. (Asteraceae). Plants (Basel). (2022) 11. doi: 10.3390/plants11121598
126. Liberti MV, Locasale JW. The Warburg effect: how does it benefit cancer cells? Trends Biochem Sci. (2016) 41:211–8. doi: 10.1016/j.tibs.2015.12.001
127. Wang L, Qin W, Tian L, Zhang X-X, Lin F, Cheng F, et al. Caroguaianolide A–E, five new cytotoxic sesquiterpene lactones from Carpesium abrotanoides L. Fitoterapia. (2018) 127:349–55. doi: 10.1016/j.fitote.2018.03.015
128. Kang JI, Hong J-Y, Lee H-J, Bae SY, Jung C, Park HJ, et al. Anti-tumor activity of yuanhuacine by regulating AMPK/mTOR signaling pathway and actin cytoskeleton organization in non-small cell lung cancer cells. PloS One. (2015) 10:e0144368. doi: 10.1371/journal.pone.0144368
129. Pan R-R, Zhang C-Y, Li Y, Zhang B-B, Zhao L, Ye Y, et al. Daphnane diterpenoids from Daphne genkwa inhibit PI3K/Akt/mTOR signaling and induce cell cycle arrest and apoptosis in human colon cancer cells. J Natural Prod. (2020) 83:1238–48. doi: 10.1021/acs.jnatprod.0c00003
130. Li F, Sun Q, Hong L, Li L, Wu Y, Xia M, et al. Daphnane-type diterpenes with inhibitory activities against human cancer cell lines from Daphne genkwa. Bioorg Medicinal Chem Lett. (2013) 23:2500–4. doi: 10.1016/j.bmcl.2013.03.025
131. Hong J-Y, Nam J-W, Seo E-K, Lee SKJC, Bulletin P. Daphnane diterpene esters with anti-proliferative activities against human lung cancer cells from Daphne genkwa. Chem Pharm Bull (Tokyo). (2010) 58(2):234–7. doi: 10.1248/cpb.58.234
132. Hou ZL, Yao GD, Song SJ. Daphnane-type diterpenes from genus Daphne and their anti-tumor activity. Chin Herb Med. (2021) 13:145–56. doi: 10.1016/j.chmed.2020.09.006
133. Xu M-D, Liu L, Wu M-Y, Jiang M, Shou L-M, Wang W-J, et al. The combination of cantharidin and antiangiogenic therapeutics presents additive antitumor effects against pancreatic cancer. Oncogenesis. (2018) 7:94. doi: 10.1038/s41389-018-0102-2
134. Pan Y, Zheng Q, Ni W, Wei Z, Yu S, Jia Q, et al. Breaking glucose transporter 1/pyruvate kinase M2 glycolytic loop is required for cantharidin inhibition of metastasis in highly metastatic breast cancer. Front Pharmacol. (2019) 10:590. doi: 10.3389/fphar.2019.00590
135. Mu L, Wang T, Chen Y, Tang X, Yuan Y, Zhao Y. [amp]]beta;-Elemene enhances the efficacy of gefitinib on glioblastoma multiforme cells through the inhibition of the EGFR signaling pathway. Int J Oncol. (2016) 49:1427–36. doi: 10.3892/ijo.2016.3626
136. Liu S, Zhou L, Zhao Y, Yuan Y. [amp]]beta;-elemene enhances both radiosensitivity and chemosensitivity of glioblastoma cells through the inhibition of the ATM signaling pathway. Oncol Rep. (2015) 34:943–51. doi: 10.3892/or.2015.4050
137. Zhang Y, Mu XD, Li EZ, Luo Y, Song N, Qu XJ, et al. The role of E3 ubiquitin ligase Cbl proteins in β-elemene reversing multi-drug resistance of human gastric adenocarcinoma cells. Int J Mol Sci. (2013) 14:10075–89. doi: 10.3390/ijms140510075
138. Chaneton B, Gottlieb E. Rocking cell metabolism: revised functions of the key glycolytic regulator PKM2 in cancer. Trends Biochem Sci. (2012) 37:309–16. doi: 10.1016/j.tibs.2012.04.003
139. Pan Y, Wang W, Huang S, Ni W, Wei Z, Cao Y, et al. Beta-elemene inhibits breast cancer metastasis through blocking pyruvate kinase M2 dimerization and nuclear translocation. J Cell Mol Med. (2019) 23:6846–58. doi: 10.1111/jcmm.v23.10
140. Palcau AC, Brandi R, Mehterov NH, Botti C, Blandino G, Pulito C. Exploiting long non-coding RNAs and circular RNAs as pharmacological targets in triple-negative breast cancer treatment. Cancers (Basel). (2023) 15. doi: 10.3390/cancers15164181
141. Yao A, Xiang Y, Si YR, Fan LJ, Li JP, Li H, et al. PKM2 promotes glucose metabolism through a let-7a-5p/Stat3/hnRNP-A1 regulatory feedback loop in breast cancer cells. J Cell Biochem. (2019) 120:6542–54. doi: 10.1002/jcb.v120.4
142. Liang J, Ye C, Chen K, Gao Z, Lu F, Wei K. Non-coding RNAs in breast cancer: with a focus on glucose metabolism reprogramming. Discovery Oncol. (2023) 14:72. doi: 10.1007/s12672-023-00687-2
143. Xu S, Wang L, Zhao Y, Mo T, Wang B, Lin J, et al. Metabolism-regulating non-coding RNAs in breast cancer: roles, mechanisms and clinical applications. J BioMed Sci. (2024) 31:25. doi: 10.1186/s12929-024-01013-w
144. Fong MY, Zhou W, Liu L, Alontaga AY, Chandra M, Ashby J, et al. Breast-cancer-secreted miR-122 reprograms glucose metabolism in premetastatic niche to promote metastasis. Nat Cell Biol. (2015) 17(2):183–94. doi: 10.1038/ncb3094
145. Serguienko A, Grad I, Wennerstrøm AB, Meza-Zepeda LA, Thiede B, Stratford EW, et al. Metabolic reprogramming of metastatic breast cancer and melanoma by let-7a microRNA. Oncotarget. (2015) 6(4):2451. doi: 10.18632/oncotarget.3235
146. Marsboom G, Zhang G-F, Pohl-Avila N, Zhang Y, Yuan Y, Kang H, et al. Glutamine metabolism regulates the pluripotency transcription factor OCT4. Cell Rep. (2016) 16(2):323–32. doi: 10.1016/j.celrep.2016.05.089
147. Shen Y, Xu J, Pan X, Zhang Y, Weng Y, Zhou D, et al. LncRNA KCNQ1OT1 sponges miR-34c-5p to promote osteosarcoma growth via ALDOA enhanced aerobic glycolysis. Cell Death Dis. (2020) 11(4):278. doi: 10.1038/s41419-020-2485-1
148. Morfouace M, Lalier L, Oliver L, Cheray M, Pecqueur C, Cartron P-F, et al. Control of glioma cell death and differentiation by PKM2–Oct4 interaction. Cell Death Dis. (2014) 5(1):e1036–6. doi: 10.1038/cddis.2013.561
149. Huang J, Deng X, Chen X, Chang Z, Lu Q, Tang A, et al. Circular RNA KIF4A promotes liver metastasis of breast cancer by reprogramming glucose metabolism. J Oncol. (2022) 2022:. doi: 10.1155/2022/8035083
150. Zheng F, Chen J, Zhang X, Wang Z, Chen J, Lin X, et al. The HIF-1α antisense long non-coding RNA drives a positive feedback loop of HIF-1α mediated transactivation and glycolysis. Nat Commun. (2021) 12(1):1341. doi: 10.1038/s41467-021-21535-3
151. David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. (2010) 463(7279):364–8. doi: 10.1038/nature08697
Keywords: breast cancer, non-metabolic enzyme function, pyruvate kinase M2, noncoding RNA, tumorigenesis
Citation: Jemal M, Getinet M, Amare GA, Tegegne BA, Baylie T, Mengistu EF, Osman EE, Chura Waritu N and Adugna A (2024) Non-metabolic enzyme function of pyruvate kinase M2 in breast cancer. Front. Oncol. 14:1450325. doi: 10.3389/fonc.2024.1450325
Received: 17 June 2024; Accepted: 12 September 2024;
Published: 01 October 2024.
Edited by:
Wenwen Zhang, Nanjing Medical University, ChinaReviewed by:
Yu Xiao, Shenzhen Second People’s Hospital, ChinaIndranil Chakrabarti, All India Institute of Medical Sciences, India
Copyright © 2024 Jemal, Getinet, Amare, Tegegne, Baylie, Mengistu, Osman, Chura Waritu and Adugna. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mohammed Jemal, mohajem9801@gmail.com
 Mohammed Jemal
Mohammed Jemal