- The Second Department of Gynecological Surgery, The Affiliated Tumor Hospital of Xinjiang Medical University, Urumqi, China
Introduction: The occurrence of cervical cancer may be related to estrogen and estrogen receptors. This study investigated the expression of lnc-CCDC170–4:1, ESR1 (estrogen receptor 1), lncRNA SRA, and CYP19A1 (aromatase) in cervical squamous cell carcinoma tissues, as well as their relationship with the clinical characteristics of patients.
Methods: Whole transcriptome sequencing analysis was performed on cervical squamous cell carcinoma tissues (n=4) and normal tissues (n=4). The expressions of lnc-CCDC170–4:1, ESR1, lncRNA SRA, and CYP19A1 were validated in 26 cases of cervical cancer tissue and 30 cases of normal cervical tissue using qRT-PCR. The relationship of gene expression with the clinical characteristics and 5-year overall survival rates of cervical cancer patients was analyzed.
Results: The expression levels of CYP19A1 and lncRNA SRA were upregulated, while those of ESR1 and lnc-CCDC170–4:1 were downregulated in cervical squamous cell carcinoma tissue. However, their expression was not related to 5-year overall survival rates (p>0.05). Low expression of lnc-CCDC170–4:1 was associated with lymph node metastasis (p=0.030) and Tumor size (p=0.047), Low expression of ESR was associated with FIGO Staging (p=0.041)and Tumor size(p=0.002),High expression of LncSRA was associated with FIGO Staging(p=0.004).
Conclusion: Estrogen and estrogen receptors may play a role in the occurrence and development of cervical squamous cell carcinoma. Low expression of lnc-CCDC170–4:1 and ESR1 are associated with lymph node metastasis and FIGO stage, so it may be a potential biomarker to evaluate the prognosis of cervical cancer.
Introduction
Cervical cancer is the fourth-leading cause of cancer-related death in women (1). Despite the rapid development in prevention and treatment strategies over the past few decades, the prognosis for late-stage or recurrent cervical cancer patients remains poor (2). Cervical squamous cell carcinoma exhibits a notably improved prognosis compared to other types of cervical cancer. Patients diagnosed with advanced clinical stage of cervical squamous cell carcinoma present increased rates of lymph node metastasis and reduced 5-year survival rates, which indicate an unfavorable prognosis. Furthermore, the degree of differentiation significantly impacts the postoperative prognosis of cervical squamous cell carcinoma, with 5-year survival rates of 100%, 75%, and 55% observed for high, moderate, and low differentiation, respectively (3). Therefore, investigating the pathogenesis of cervical cancer, identifying prognosis factors, and improving the prognosis is of great clinical significance (4).
It is currently believed that long-term use of oral contraceptives and multiple pregnancies can increase the risk of cervical cancer, indicating that estrogen and estrogen receptors (ER) are closely related to cervical cancer (5, 6). Estrogen receptor inhibitors play an important role in the treatment of breast cancer. Studies have shown that estrone has a pro-inflammatory effect, can increase the expression of embryonic stem cell transcription factors, and induce the epithelial-mesenchymal transition of ER+ HeLa cells (7, 8). ESR1 (estrogen receptor 1) is the encoding gene for estrogen receptor-α (ER-α) and has been shown to play an important role in mediating the invasion and progression of cervical cancer with ER-α expression loss (9). It has been shown that there are ESR1 mutations in patients with cervical squamous cell carcinoma (10). However, the prognostic properties of ESR1 in cervical cancer have been less studied.
P450 aromatase is encoded by the CYP1A1 (aromatase) gene and participates in the hydroxylation metabolism of estradiol. There is a significant correlation between CYP1A1 and the infiltration level of T cells in cervical cancer (11). In addition, the high expression of CYP1A1 and T cells in cervical cancer is unfavorable for the prognosis of cervical cancer patients. Our previous study found that P450 aromatase expression was increased and ER-β expression was decreased in local cervical tissue, indicating that they may play a role in the occurrence and development of cervical squamous cell carcinoma (12).
Long non-coding RNAs (lncRNAs) play an important role in the occurrence and development of tumors (13). Recently, the abnormal regulation of lncRNAs in cervical cancer cells and tissues has been reported 11. Some of these lncRNAs include homeobox transcript antisense RNA, H19, metastasis-associated lung adenocarcinoma transcript 1 (2), cervical carcinoma high-expressed 1, antisense noncoding RNA in the inhibitors of cyclin-dependent kinase 4, growth arrest special 5, and plasmacytoma variant translocation 1. These lncRNAs have been found to play a role in various disease-related processes such as cell growth, cell proliferation, cell survival, metastasis, invasion, and therapeutic resistance (11). The lncRNA SRA facilitates the migration and invasion of cervical cancer cells via the NOTCH signaling pathway and by upregulating MMP-2, MMP-9, and VEGF (14). Additionally, it could potentially function as a therapeutic target and prognostic marker for cervical cancer. Therefore, lncRNAs may serve as potential targets for cancer treatment.
Herein, this study explores the expression of mRNA and corresponding lncRNA related to estrogen and ER in cervical squamous cell carcinoma and their relationship with clinical pathological features (such as tumor size and lymph node metastasis) of patients with cervical squamous cell carcinoma. Our results may provide new ideas for the prognosis assessment of cervical squamous cell carcinoma.
Materials and methods
Study participants
Four patients with primary cervical squamous cell carcinoma diagnosed by the Pathology Department of the Affiliated Tumor Hospital of Xinjiang Medical University in October 2017 were enrolled, along with four control individuals who underwent simple hysterectomy due to uterine leiomyoma. The cervical cancer tissues and normal cervical tissues were collected for whole transcriptome sequencing. Additionally, 26 patients with primary cervical cancer diagnosed by the same department from October 2017 to March 2018 and 30 control individuals who underwent simple hysterectomy due to uterine leiomyoma were also recruited. The cervical cancer tissues and normal cervical tissues were collected for quantitative real-time PCR (qRT-PCR) validation. The inclusion criteria for cervical cancer patients included: 1) non-pregnant patients with primary cervical cancer; 2) patients without immunodeficiency disease or other malignant tumors; 3) patients who had no prior history of chemotherapy or pelvic radiotherapy; 4) patients with pathologically confirmed cervical squamous cell carcinoma. The exclusion criteria for cervical cancer patients included: 1) Non-cervical squamous cell carcinoma confirmed by cervical biopsy and pathology at the Affiliated Cancer Hospital of Xinjiang Medical University; 2) Patients with other malignant tumors; 3) After the medical staff explained the purpose of the study, they were unwilling to cooperate Investigation of patients. The inclusion criteria for control individuals included: 1) non-pregnant individuals; 2) individuals without immunodeficiency disease or other malignant tumors; 3) individuals with negative cervical cytology results; 4) individuals with uterine leiomyoma. The exclusion criteria for control individuals included: 1) Abnormal uterine cervical cytology; 2) Those who are unwilling to cooperate with the investigation after the medical staff has explained the purpose of the research to Patient; 3) Patients with other malignant tumors. All procedures and studies involving human subjects to have been carried out according to the ethical guidelines outlined in Declaration of Helsinki. This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Xinjiang Medical University (No.: K-2021018). All participants have signed the informed consent form.
Data collection
The basic clinical data were collected, such as patient age, menopausal status, tumor size, lymph node involvement, vascular invasion, FIGO stage, etc.
Whole transcriptome sequencing
Total RNA was extracted from four pairs of frozen cervical cancer and normal cervical tissues using TRIzol reagent (TransGen Biotech, Beijing, China). The purity and quantity of RNA were assessed using a NanoDrop 2000 spectrophotometer (Thermo Scientific, Waltham, MA, USA). RNA integrity was evaluated using an Agilent 2100 bioanalyzer (Agilent Technologies, Santa Clara, CA, USA). After quality control, the RNA underwent fragmentation, followed by the synthesis of a single-stranded cDNA using six-base random primers. A double-stranded cDNA synthesis was then performed, with the dTTP replaced with dUTP. Different adapters were ligated. The cDNA strand containing dUTP was digested with the Uracil DNA N-Glycosylase. The cDNA single strand with adapters was purified using Agencourt AMPure XP (BECKMAN COULTER, CA, USA) and then underwent end-repair, A-tailing, and ligation of the sequencing adapters. Subsequently, PCR was performed. After purifying the PCR products using AMPure XP beads (BECKMAN COULTER), the library was obtained. An Agilent 2100 chip (Agilent Technologies, SantaClara, CA, USA) was used to detect insert fragments, and the effective concentration was quantified using qRT-PCR to ensure library quality. Finally, the library was sequenced on the Illumina HiSeq X Ten platform (Illumina, San Diego, CA, USA).
Sequencing data analysis
The quality of sequencing data was typically above Q30, which met the requirements for subsequent analysis. After removing the adapter information and low-quality bases low-quality bases that are present in the original data, clean reads were obtained. We used HISAT2 v2.1.0 to align clean reads with the specified reference genome, and the aligned reads on the specified reference genome were called Mapped Reads. We used Cufflinks v2.2.1 to splice Mapped Reads into transcripts. Using reference transcripts as the library, we used the sequence similarity alignment method of bowtie2 v2.3.1 and eXpress v1.5.1 software to calculate the expression level of each transcript in each sample.
Identification of differentially expressed genes and gene function analysis
The CuffDiff software was used to identify differentially expressed genes, with the screening criteria of p < 0.05 and a fold change greater than 2. The DAVID tool (https://david.ncifcrf.gov/) was used for Gene Ontology (GO) analysis. KEGG database was used for pathway analysis of differential transcripts.
Validation with qRT-PCR
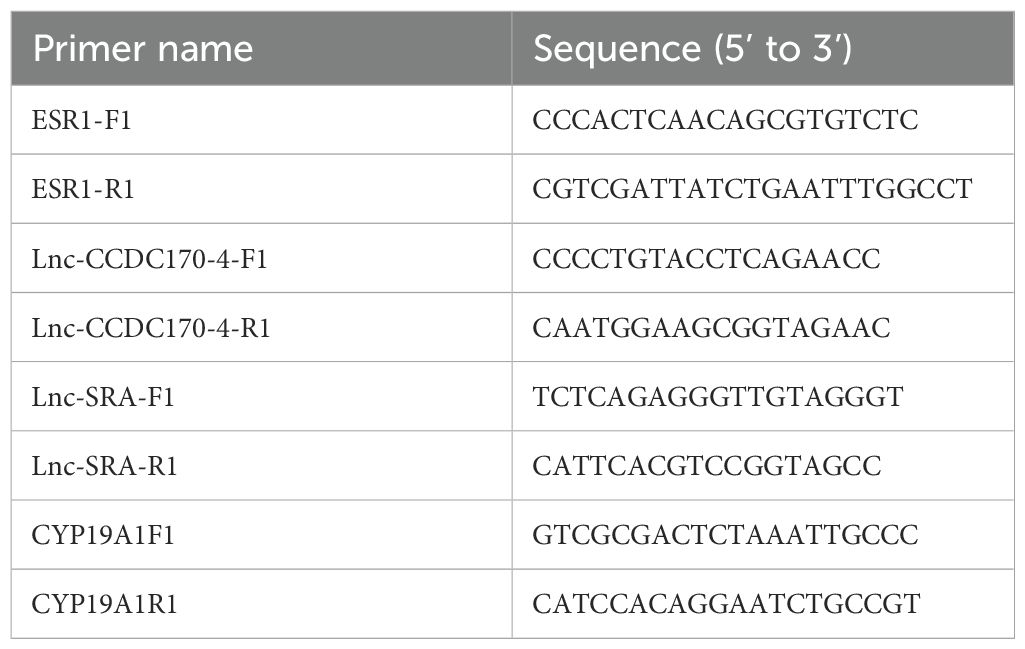
Total RNA was extracted using TRIzol reagent (TransGen Biotech) and reversed transcribed into cDNA using 5X All-In-One RT MasterMix (ABM, Shanghai, China). The qRT-PCR reaction mixture comprised 2×SYBR Green Select Mix (5 μL), cDNA (1 μL), upstream primer (0.7 μL), downstream primer (0.7 μL), and enzyme-free water (10 μL). The PCR reaction conditions were as follows: 95°C for 2 min, followed by 40 cycles of 95°C for 5s and 60°C for 30s. The qRT-PCR experiment was conducted in triplicate, and the relative expression level of the gene was determined using the 2-ΔΔct method. The primers are listed in Table 1.
Follow-up
Patients were followed up by telephone or outpatient visits. The follow-up deadline was March 2023, and the overall survival (OS) was calculated from the surgery until the end of the follow-up period or death.
Statistical analysis
Statistical analysis was performed using SPSS 26.0 software, and all data are represented as mean ± standard deviation. T-test or one-way analysis of variance compared data of normal distribution, while the rank-sum test analyzed data of non-normal distribution. The independent sample t-test was used to evaluate the relationship of lnc-CCDC170–4:1, ESR, lncRNA SRA, and CYP19A1 expression with clinical pathological features. Correlation analysis was conducted with Pearson’s correlation. The Kaplan-Meier method analyzed the OS of patients. P < 0.05 indicates a significant difference.
Results
Results of whole transcriptome high-throughput sequencing
To identify differentially expressed mRNAs and lncRNAs in cervical cancer, we analyzed four cases of cervical cancer tissues and four normal cervical cases through whole transcriptome high-throughput sequencing. The results showed a significant increase in transcripts that were differentially expressed in cervical cancer tissues when compared to normal cervical tissues (P<0.05) (Figures 1A, B), including 3035 significantly up-regulated genes and 3319 significantly down-regulated genes. We analyzed the functions of the differentially expressed transcripts through GO (Figure 1C) and KEGG (Figure 1D) enrichment analysis. We calculated the significance of enrichment for each pathway entry using the hypergeometric distribution test. Finally, we screened mRNA related to estrogen and co-expressed and/or related lncRNAs from lncRNA cis and trans mechanisms, namely CYP19A1, lncRNA SRA, ESR, and lnc-CCDC170–4:1.
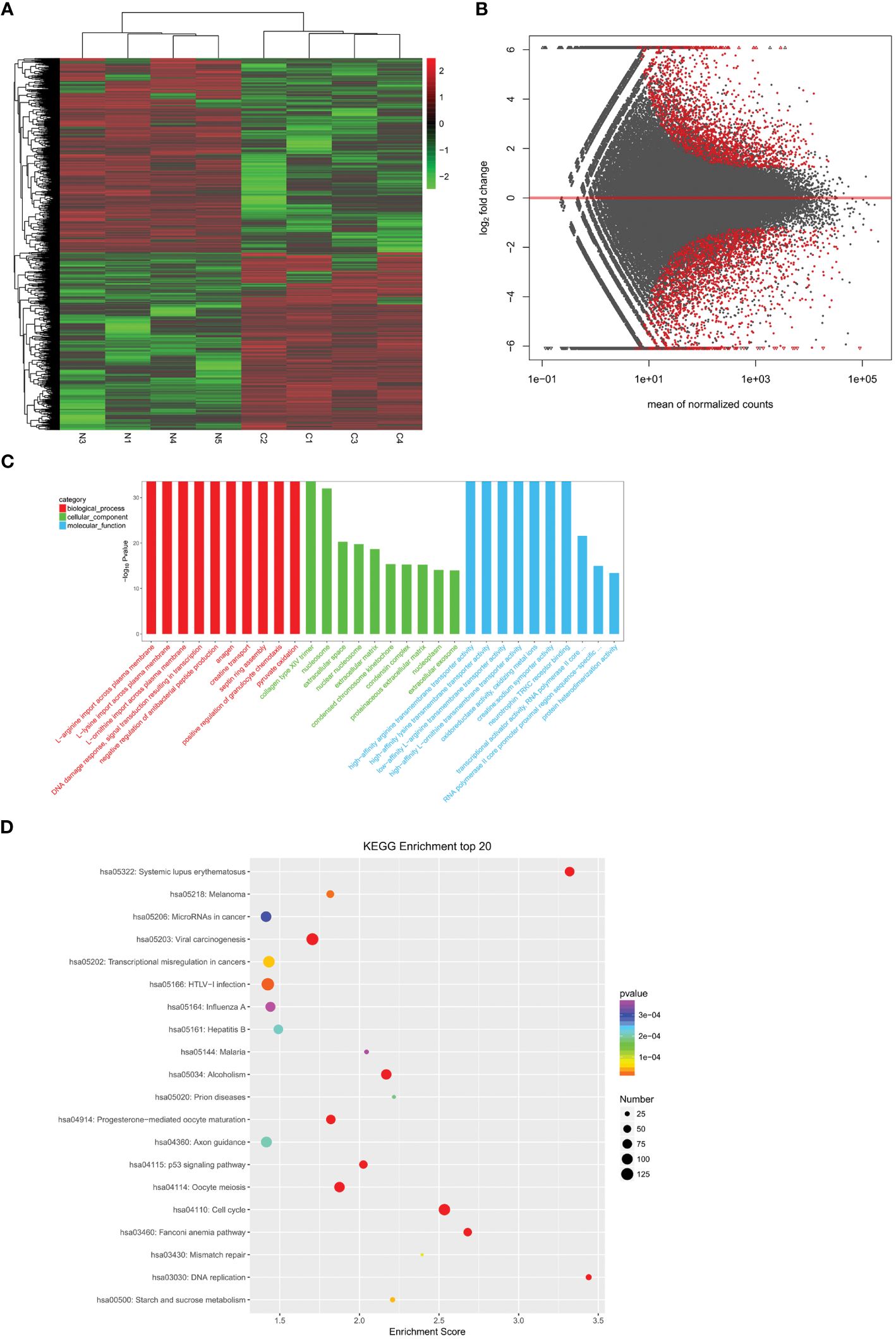
Figure 1. Analysis of differentially expressed genes. (A) A heatmap showing differentially expressed mRNAs between cervical cancer tissues (n =4) and normal cervical tissues (n =4). (B) The volcano map showing the differential transcripts. (C) The bar diagram showing the Top30 enriched GO terms. (D) Example of KEGG enrichment corresponding pathway map.
Validation of lnc-CCDC170–4:1, ESR, lncRNA SRA, and CYP19A1 expression with qRT-PCR
The expression levels of lnc-CCDC170–4:1, ESR, lncRNA SRA, and CYP19A1 were validated using qRT-PCR in 26 cases of cervical cancer tissue and 30 cases of normal cervical tissue. We found that the relative expression levels of lnc-CCDC170–4:1 (Figure 2A) and ESR mRNA (Figure 2B) were significantly lower in cervical cancer tissue than in the control (P<0.05). The relative expression level of lncRNA SRA (Figure 2C) and CYP19A1 mRNA (Figure 2D) were significantly higher in cervical cancer tissue.

Figure 2. Expression levels of lnc-CCDC170–4:1, ESR, lncRNA SRA, and CYP19A1. Their levels were measured with qRT-PCR. (A) Level of lnc-CCDC170–4:1. (B) Level of ESR. (C) Level of lncRNA SRA. (D) Level of CYP19A1. *P < 0.05, **P<0.01.
Correlation analysis
Pearson’s correlation analysis was used to analyze the correlation between the expression levels of lncRNASRA and CYP19A1, as well as lnc-CCDC170–4:1 andESR. The results showed that lnc-CCDC170–4:1 and ESR were positively correlated (r=0.477, P<0.001) (Figure 3A). Similarly, lncRNASRA and CYP19A1 were also positively correlated (r=0.532, P<0.001) (Figure 3B).

Figure 3. Pearson correlation analysis. (A) The positive association between lnc-CCDC170–4:1 and ESR1. (B) The positive association between lncRNASRA and CYP19A1.
The effect of lnc-CCDC170–4:1, ESR, and lncRNA SRA on the prognosis of patients
Kaplan-Meier survival curve analysis was used to evaluate the OS of 26 cervical cancer patients. The survival rate of cervical cancer patients with high expression levels of lncRNA SRA was lower than that of patients with low expression levels of lncRNA SRA (Figure 4A), while the survival rate of cervical cancer patients with low expression levels of lnc-CCDC170–4:1 (Figure 4B) and ESR (Figure 4C) was lower than that of patients with high expression levels of lnc-CCDC170–4:1 and ESR. However, due to the small sample size, the differences were not statistically significant (P>0.05).
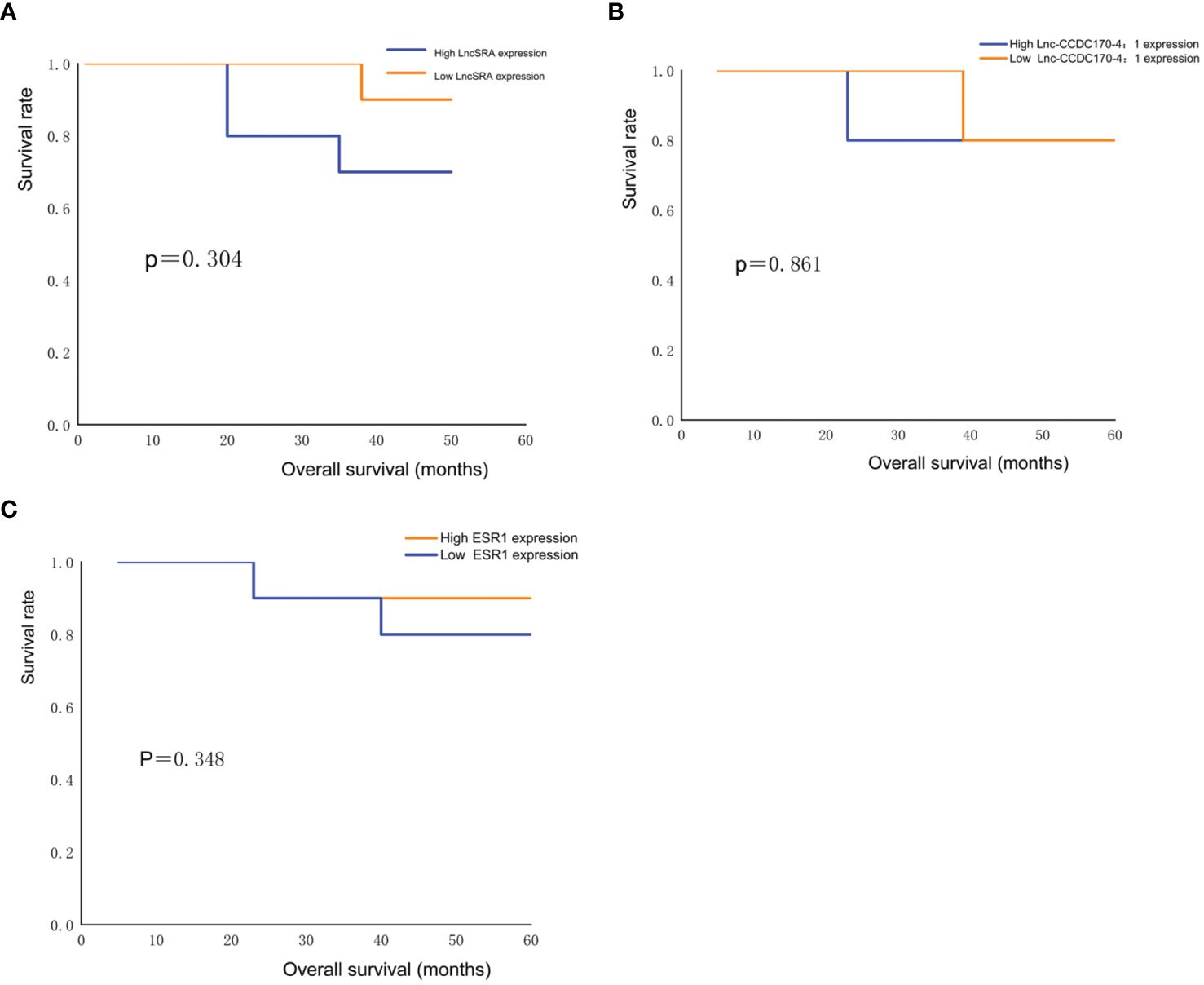
Figure 4. Survival analysis. (A) Comparison of survival rate of cervical cancer patients with low and high lncRNASRA expression. (B) Comparison of survival rate of cervical cancer patients with low and high lnc-CCDC170–4:1 expression. (C) Comparison of survival rate of cervical cancer patients with low and high ESR expression.
The association between the expression levels of lncRNA SRA, CYP19A1, lnc-CCDC170–4:1, and ESR and the clinical-pathological features of patients with cervical cancer
We analyzed the relative expression levels of these lncRNAs and mRNAs in the cancer tissues of 26 patients with cervical cancer and divided the patients into low- and high-expression groups based on the median value. Subsequently, we assessed the relationship between the expression levels of these genes and the clinical-pathological features of cervical cancer patients. The results showed that the expression of lnc-CCDC170–4:1 was associated with lymph node metastasis (p=0.030)and Tumor size(p=0.047), Low expression of ESR was associated with FIGO Staging(p=0.041)and Tumor size(p=0.002) (Table 2). High expression of LncSRA was associated with FIGO Staging(p=0.004) (Table 3).
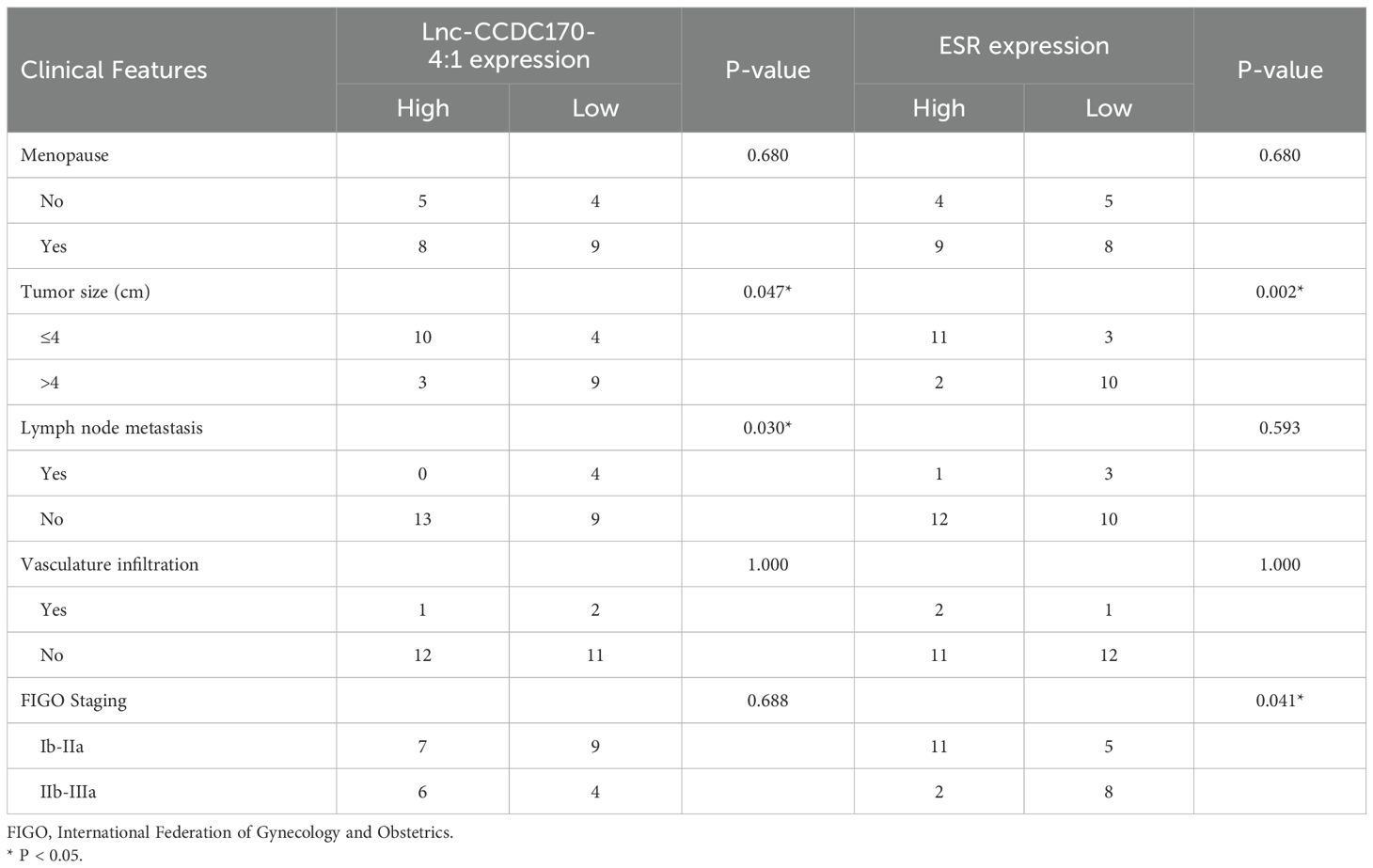
Table 2. Relationship between Lnc-CCDC170-4:1 and ESR expression and clinicopathological characteristics.

Table 3. Relationship between lncRNASRA and CYP19A1 expression and clinicopathological characteristics.
Discussion
Cervical cancer remains one of the main causes of cancer-related death in women worldwide. The malignant proliferation of cervical cancer cells, lymphatic infiltration, and distant organ metastasis are the main reasons for the high mortality rate of cervical cancer patients. More and more studies are focused on prognostic-specific molecular markers in cervical cancer (15, 16). As one of the important female sex hormones, estrogen can regulate gene expression and the function of the female reproductive system, and stimulate the development, proliferation, migration, and survival of target cells by binding with ER (17–19). The cervix is a target organ of estrogen, and most cervical cancers originate from the squamous-columnar junction of the cervix, where the immature metaplastic squamous epithelium is metabolically active and highly hormone-sensitive. Some scholars believe that estrogen may play a carcinogenic role in cervical lesion progression (20). LncRNA in cells regulates the proliferation, invasion, and migration of tumor cells. Thus, lncRNAs related to estrogen and ER may be potential targets for tumor therapy (21).
P450 aromatase is a key enzyme in local estrogen synthesis. Our previous study 12 revealed an increase in P450 aromatase expression in cervical cancer. Due to its involvement in local estrogen production, molecular therapy for breast cancer is based on the inhibition of aromatase to minimize androgen-to-estrogen conversion and attenuate estrogen levels. CYP19A1, a member of the cytochrome P450 superfamily, serves as a monooxygenase during steroidogenesis. It is reported that CYP19A1 was a potential biomarker for gastric cancer prognosis and immune cell infiltration (22). There is evidence that the overexpression of lncRNA SRA can induce EMT, cell proliferation, migration, and invasion in vitro (23). As a result, lncRNA SRA may elevate EMT-associated genes, leading to increased cancer cell aggressiveness and making it a promising biomarker for predicting cervical cancer relapse and prognosis as well as a therapeutic target. In this study, we carried out qRT-PCR to evaluate the lncRNA SRA expression and its corresponding mRNA, CYP19A1, in cervical cancer and control tissues. We found a significant increase in lncRNA SRA expression in cervical cancer tissues compared to the control, while the expression of CYP19A1 did not significantly differ between the two groups. Moreover, we found a positive correlation between the lncRNA SRA and CYP19A1 expression levels, despite the regulatory mechanism still requiring further investigation. Further, they had no significant effect on patient survival. Additionally, we analyzed the relationship between the expression of lncRNA SRA and CYP19A1 and the clinical-pathological characteristics of 26 cervical cancer patients. We found that high expression of LncSRA was associated with FIGO Staging, it may be an important factor for evaluating the prognosis of cervical cancer.
In addition, ESR1 is a nuclear factor that mediates the biological effects of estrogen and is the encoding gene for ERα. Studies (24, 25) have shown that if the ESR1 gene is overexpressed in breast cancer cells, the risk of postmenopausal women developing ER-positive breast cancer increases. Chen et al. (26) found that overexpression of ESR1 partially reversed the effect of miR-944 on the expression levels of E-cadherin, N-cadherin, and Vimentin in cervical cancer cells. Wen et al. (27) revealed that the expression of ERα gradually decreased with the progression of cervical lesions, which may become a diagnostic or staging indicator for cervical cancer. Tumor size and lymph node metastasis are independent survival factors for cervical cancer patients (28). In this study, we investigated the role of ESR1 and its corresponding lnc-CCDC170–4:1 in cervical cancer. Our findings showed that the expression of ESR1 and lnc-CCDC170–4:1 was downregulated in cervical cancer tissues, and the decreased expression of lnc-CCDC170–4:1 was associated with lymph node metastasis and Tumor size, expression of ESR was associated with FIGO Staging and Tumor size, may be an important target for prognosis evaluation in cervical cancer patients.
It is known that estrogen and ER play an important role in the treatment of breast cancer. If ERα plays an important role in the development of cervical cancer, drugs targeting ER may also play a role in the treatment of cervical cancer (29). It has been shown that ERα can be used as a biomarker to evaluate the OS of HPV-positive oropharyngeal cancer patients (30). Therefore, the lnc-CCDC170–4:1 and ESR1 may be an important factor for evaluating the prognosis of cervical cancer, and their specific mechanism of action in cervical cancer is also one of our main research directions in the future.
Conclusions
In conclusion, the expression of lncRNA SRA and CYP19A1 is elevated in cervical cancer, while lnc-CCDC170–4:1 and ESR1 expression is decreased in cervical cancer. Moreover, low expression of lnc-CCDC170–4:1 and ESR1 are associated with lymph node metastasis and FIGO stage, so it may be a potential biomarker to evaluate the prognosis of cervical cancer.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/supplementary material.
Ethics statement
All procedures and studies involving human subjects to have been carried out according to the ethical guidelines outlined in Declaration of Helsinki. This study was approved by the Ethics Committee of the Affiliated Tumor Hospital of Xinjiang Medical University (No.: K-2021018). All participants have signed the written informed consent form.
Author contributions
JY: Data curation, Formal analysis, Writing – original draft. MW: Conceptualization, Methodology, Writing – original draft. AM: Software, Writing – original draft, Supervision. SZ: Validation, Writing – original draft. NJ: Investigation, Writing – review & editing. GS: Funding acquisition, Project administration, Resources, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. The work was supported by Xinjiang Uygur Autonomous Region Natural Science Foundation (No. 2020D01C204). The funding source had no role in the design, practice or analysis of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abbreviations
ER, estrogen receptors; ESR1, estrogen receptor 1; lncRNAs, Long non-coding RNAs; qRT-PCR, quantitative real-time PCR; OS, overall survival.
References
1. Ou Z, Lin S, Qiu J, Ding W, Ren P, Chen D, et al. Single-nucleus RNA sequencing and spatial transcriptomics reveal the immunological microenvironment of cervical squamous cell carcinoma. Adv Sci (Weinh). (2022) 9:e2203040. doi: 10.1002/advs.202203040
2. Hu Y, Li G, Ma Y, Luo G, Wang Q, Zhang S. Effect of exosomal lncRNA MALAT1/miR-370-3p/STAT3 positive feedback loop on PI3K/akt pathway mediating cisplatin resistance in cervical cancer cells. J Oncol. (2023) 2023:6341011. doi: 10.1155/2023/6341011
3. Xiong Y, Liang LL, Liu JH, Zhang F, Liu H. Stratification of prognostic factors for squamous cell carcinoma of the cervix and its clinical significance. Chin J Clinical Oncol. (2007) 34:1048–52.
4. Wang MN, Zhang Y, Wu L, Liu CC. Expression of LncRNA DGCR5 in cervical cancer and its relationship with clinical features and prognosis. Adv Anatomical Sci. (2022) 28:204–8.
5. Bronowicka-Klys DE, Lianeri M, Jagodzinski PP. The role and impact of estrogens and xenoestrogen on the development of cervical cancer. Biomed Pharmacother. (2016) 84:1945–53. doi: 10.1016/j.biopha.2016.11.007
6. Li H, Ren J, Wang W, Hao M. Research progress on the synergistic effects of estrogen and its receptors with HPV in the tumor microenvironment of cervical cancer. J Basic Med Clinical. (2022) 42:1438–42.
7. Diaz-Ruano AB, Martinez-Alarcon N, Perán M, Benabdellah K, Garcia-Martinez MLÁ, Preda O, et al. Estradiol and estrone have different biological functions to induce NF-κB-driven inflammation, EMT and stemness in ER+ Cancer cells. Int J Mol Sci. (2023) 24:1221. doi: 10.3390/ijms24021221
8. Wang D, Bu F, Zhang W. The role of ubiquitination in regulating embryonic stem cell maintenance and cancer development. Int J Mol Sci. (2019) 20:2667. doi: 10.3390/ijms20112667
9. Ding B, Sun W, Han S, Cai Y, Ren M, Shen Y. Cytochrome P450 1A1 gene polymorphisms and cervical cancer risk: A systematic review and meta-analysis. Med (Baltimore). (2018) 97:e0210. doi: 10.1097/MD.0000000000010210
10. Kirn V, Zaharieva I, Heublein S, Thangarajah F, Friese K, Mayr D, et al. ESR1 promoter methylation in squamous cell cervical cancer. Anticancer Res. (2014) 34:723–7.
11. Wang J, Mijiti Y, Chen Y, Liu Z. Aryl hydrocarbon receptor is a prognostic biomarker and is correlated with immune responses in cervical cancer. Bioengineered. (2021) 12:11922–35. doi: 10.1080/21655979.2021.2006953
12. Shen G, Yuan J, Cheng J, Zhang G, Xiong T. Expression of P450arom and ER-β in cervical squamous cell carcinoma and precancerous lesions of Xinjiang Uyghur and Han ethnic groups. J Xinjiang Med University. (2014) 37:1470–1472 + 1476.
13. Liu J, Zhang Q, Yang D, Xie F, Wang Z. The role of long non-coding RNAs in angiogenesis and anti-angiogenic therapy resistance in cancer. Mol Ther Nucleic Acids. (2022) 28:397–407. doi: 10.1016/j.omtn.2022.03.012
14. Eoh KJ, Paek J, Kim SW, Kim HJ, Lee HY, Lee SK, et al. Long non-coding RNA, steroid receptor RNA activator (SRA), induces tumor proliferation and invasion through the NOTCH pathway in cervical cancer cell lines. Oncol Rep. (2017) 38:3481–8. doi: 10.3892/or
15. Zhang Q. Expression and significance of immune checkpoint B7-H3 and B7-H4 in cervical cancer and precancerous lesions. Shandong University. (2021).
16. Volkova LV, Pashov AI, Omelchuk NN. Cervical carcinoma: oncobiology and biomarkers. IntJ Mol Sci. (2021) 22:12571. doi: 10.3390/ijms222212571
17. Chen Y, Geng Y, Huang J, Xi D, Xu G, Gu W, et al. CircNEIL3 promotes cervical cancer cell proliferation by adsorbing miR-137 and upregulating KLF12. Cancer Cell Int. (2021) 21:34. doi: 10.1186/s12935-020-01736-4
18. Fuentes N, Silveyra P. Estrogen receptor signaling mechanisms. Adv Protein Chem Struct Biol. (2019) 116:135–70. doi: 10.1016/bs.apcsb.2019.01.001
19. Wang C, Zhang T, Wang K, Zhang S, Sun Q, Yang X. ER-α36 promotes the Malignant progression of cervical cancer mediated by estrogen via HMGA2. Front Oncol. (2021) 11:712849. doi: 10.3389/fonc.2021.712849
20. Liu C, Wu HT, Zhu N, Cheng H, Zhang LG. Steroid receptor RNA activator: Biologic function and role in disease. Clin Chim Acta. (2016) 459:137–46. doi: 10.1016/j.cca.2016.06.004
21. Liu Y, Li MZ, Wei LH. Research progress of estrogen and its receptor in cervical cancer. Chin Clin J Obstetrics Gynecol. (2019) 20:188–90.
22. Wang N, Huang X, Long Q. Lipid metabolic-related signature CYP19A1 is a potential biomarker for prognosis and immune cell infiltration in gastric cancer. J Inflammation Res. (2022) 15:5075–88. doi: 10.2147/JIR.S378212
23. Kim HJ, Kim LK, Lee SH, Park SA, Eoh KJ, Kim YT. Expression levels of the long noncoding RNA steroid receptor activator promote cell proliferation and invasion and predict patient prognosis in human cervical cancer. Oncol Lett. (2018) 16:5410–8. doi: 10.3892/ol
24. Mou Y, Xiong Y, Qin Y. Mechanisms of endocrine therapy resistance and targeted treatment strategies in estrogen receptor-positive breast cancer. Chin Bull Life Sci. (2022) 34:1559–68.
25. Herzog SK, Fuqua SAW. ESR1 mutations and therapeutic resistance in metastatic breast cancer: progress and remaining challenges. Br J Cancer. (2022) 126:174–86. doi: 10.1038/s41416-021-01564-x
26. Chen S, Chen JZ, Zhang JQ, Chen HX, Qiu FN, Yan ML, et al. Silencing of long noncoding RNA LINC00958 prevents tumor initiation of pancreatic cancer by acting as a sponge of microRNA-330-5p to down-regulate PAX8. Cancer Lett. (2019) 446:49–61. doi: 10.1016/j.canlet.2018.12.017
27. Wen L, Li X, Jia Y. Expression and clinical significance of CHAF1A and ERα in cervical squamous cell carcinoma. Chin J Cell Biol. (2021) 43:561–9.
28. Yang XL, Wang MM, Kou LN, Lai H, Wu DJ. Conditional survival for high-risk early-stage cervical cancer patients with lymph node metastasis after hysterectomy. Curr Probl Cancer. (2021) 45:100756. doi: 10.1016/j.currproblcancer.2021.100756
29. Su F, Kong W. Analysis of the characteristics and trends of cervical cancer incidence in Beijing Maternity Hospital of Capital Medical University in the past 40 years. China Family Plann Obstetrics Gynecol. (2021) 13:60–3.
Keywords: lnc-CCDC170-4:1, ESR, lncRNA SRA, CYP19A1, estrogen
Citation: Yuan J, Wen M, Matnuri A, Zhao S, Jian N and Shen G (2024) The expression of lnc-CCDC170-4:1, ESR1, lncRNA SRA, and CYP19A1 in cervical squamous cell carcinoma and their relationship with the clinical characteristics. Front. Oncol. 14:1430826. doi: 10.3389/fonc.2024.1430826
Received: 10 May 2024; Accepted: 12 July 2024;
Published: 14 August 2024.
Edited by:
Anjali Geethadevi, Medical College of Wisconsin, United StatesReviewed by:
Lavannya Sabharwal, Medical College of Wisconsin, United StatesNeha Nanda, Harvard Medical School, United States
Copyright © 2024 Yuan, Wen, Matnuri, Zhao, Jian and Shen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Guqun Shen, c2dxMTIyOEB0b20uY29t
†These authors have contributed equally to this work
 Jinrui Yuan
Jinrui Yuan Mengke Wen†
Mengke Wen†