
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Cell. Infect. Microbiol. , 17 November 2021
Sec. Clinical Microbiology
Volume 11 - 2021 | https://doi.org/10.3389/fcimb.2021.789919
This article is part of the Research Topic Insights In Clinical Microbiology: 2021 View all 8 articles
 Bing-Yan Wang1*
Bing-Yan Wang1* Tom Lu2*
Tom Lu2* Qiuyin Cai3
Qiuyin Cai3 Meng-Hsuan Ho4
Meng-Hsuan Ho4 Sally Sheng1
Sally Sheng1 Hsiu-Wan Meng1
Hsiu-Wan Meng1 Laura Arsto1
Laura Arsto1 Jianming Hong1
Jianming Hong1 Hua Xie4*
Hua Xie4*Periodontitis disproportionately affects different racial and ethnic populations. In this study, we used qPCR to determine and compare oral microbial profiles in dental plaque samples from 191 periodontitis patients of different ethnic/racial backgrounds. We also obtained the periodontal parameters of these patients retrospectively using axiUm and performed statistical analysis using SAS 9.4. We found that in this patient cohort, neighborhood median incomes were significantly higher among Caucasians Americans (CAs) than among African Americans (AAs) and Hispanic Americans (HAs). Levels of total bacteria and Porphyromonas gingivalis, a keystone periodontal pathogen, were not evenly distributed among the three groups. We confirmed our previous findings that Streptococcus cristatus reduces P. gingivalis virulence potential and likely serves as a beneficial bacterium. We also showed the ratio of S. cristatus to P. gingivalis to be significantly higher in CAs than in HAs and AAs. Our results suggest that higher levels of P. gingivalis and lower ratios of S. cristatus to P. gingivalis may contribute to periodontal health disparities.
Periodontitis is recently defined as a dysbiotic disease resulting from imbalanced oral microbiota (Hajishengallis and Lamont, 2021). The etiology of periodontitis has been developed from microbial accumulation and specific periodontal pathogens to keystone pathogen-associated polymicrobial dysbiosis (Socransky and Haffajee, 1994; Hajishengallis et al., 2012; Hajishengallis and Lamont, 2012). Porphyromonas gingivalis, a gram-negative bacterium, plays a vital role in the development of dysbiotic microbial communities. While a low-abundance species in oral microbial communities, P. gingivalis can act in concert with other microbes to disrupt host-microbial homeostasis and induce uncontrolled inflammatory responses in periodontal tissues (Hajishengallis et al., 2011; Hajishengallis and Lamont, 2021).
The National Health and Nutrition Examination Survey (NHANES) 2009–2014 revealed the prevalence of periodontitis among dentate adults aged 30 years and older to be significantly different among African Americans (AAs), Caucasian Americans (CAs), and Hispanic Americans (HAs) (Eke et al., 2018). AAs and HAs exhibit much a higher incidence of periodontitis than CAs, a phenomenon evident even in populations with severe periodontitis. We observed higher detection rates of P. gingivalis in AAs and HAs than in CAs, diagnosed either as periodontal health or with biofilm-induced gingivitis on an intact periodontium (unpublished data). We also found that most P. gingivalis detected in AAs were of fimA genotypes II and IV that are associated with higher plaque indexes and levels of bleeding on probing in an intact periodontium cohort (unpublished data). Based on these observations, we hypothesized that differential oral microbial profiles exist in periodontitis patients of different racial/ethnic backgrounds. Here, we investigated a potential link between microbial composition, particularly the prevalence of P. gingivalis and Streptococcus cristatus, and periodontitis risks among AAs, CAs, and HAs. Our results suggest that differences in the microbial composition of dental biofilms may influence the initiation of periodontitis, and that individuals susceptible to periodontitis may depend, to some extent, on the microbial composition of early dental biofilm colonizers such as S. cristatus.
The research protocol was approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston (UTHealth at Houston) (IRB number: HSC-DB-17-0636). Candidates were screened during routine dental visits at the clinic of School of Dentistry at UTHealth at Houston between 2018 and 2021. Individuals aged 21–75 with self-reported ethnicity/race of non-Hispanic African American (AA), non-Hispanic Caucasian American (CA), or Hispanic American (HA) were enrolled after initial periodontal examination. The examination documented plaque index (PI), bleeding on probing (BOP), probing depth (PD), clinical attachment level (CAL), furcation involvement, tooth mobility, and keratinized tissue on all teeth (Newman et al., 2018). Radiographs were taken during this screening phase to assess bone loss. The clinical periodontal examinations were performed by trained dental examiners who are faculty members of the School of Dentistry, UTHealth at Houston. All study participants were diagnosed with generalized periodontitis Stage II or III, regardless of their grading, based on the 2017 World Workshop classification (Tonetti et al., 2018; Papapanou et al., 2018a). The enrolled patients also met the following criteria: ≤4 tooth loss due to periodontitis, interdental CAL ≥3mm and PD ≥5 mm at two or more teeth in different quadrants, and radiographic bone loss ≥15%. Other criteria for study participation were 1) no scaling and root planning within the previous year or periodontal surgeries in the previous five years; 2) no antibiotic therapy in the previous six months; 3) not pregnant. The periodontal parameters and diagnosis of participants were abstracted from the Electronic Health Record (axiUm). In addition, medical history and dental history were extracted from axiUm and recorded by a dental student (LA) who is blinded to study design and bacterial data. Cardiovascular diseases (including infective endocarditis, coronary artery disease, heart attach, congestive heart failure, hypertension, cardiac arrhythmia, rheumatic fever, stroke, mitral valve prolapse), respiratory diseases (chronic bronchitis or emphysema, asthma), sleep apnea, renal failure, gastrointestinal diseases (including hepatitis, stomach ulcer, crohn’s disease, colitis, liver disease), endocrine diseases (diabetes, thyroid disorder), hemotologic disorders, arthritis, osteoporosis, depression, autoimmune diseases, cancer, usage of alcohol or tobacco, dry mouth, and habitually clench or grind teeth were recorded as present or abscent. Body mass index (BMI) was recorded as numerical numbers. Neiberhood median incomes of the participants were decided based on their residential zip codes (https://www.incomebyzipcode.com/).
Dental plaque samples were collected by board-certified periodontists using sterile paper points at baseline prior to any dental treatment. The samples were labelled with numbers according to sampling sequence. The paper points were placed in ≥5mm pockets in different quadrant for 1 minute and then immersed immediately in an Eppendorf tube with 0.5 ml of Tris-EDTA (TE) buffer (pH 7.5) (Wang et al., 2009). Oral bacteria were harvested by centrifugation and the bacterial pellets were resuspended in 100 µl TE buffer. Chromosomal DNA was released by two cycles of freezing at -80°C overnight and boiling for 10 minutes.
All bacteria tested were enumerated by qPCR using SYBR Green PCR mix (Bio-Red Laboratories Inc., Redmond, WA, USA) with species-specific primers listed in Table 1. P. gingivalis strains with different fimA genotypes were identified by qPCR using strain-specific primers (Zheng et al., 2011). Levels of total bacteria were determined using primers corresponding to the conserved sequences of cyanobacterial small subunit rRNA genes (Turner et al., 1999) (Table 1). Standard curves used to enumerate bacterial cells were generated by qPCR using the genomic DNA from each bacterial species as previously described (Wang et al., 2009). Bacterial ratios were calculated using the numerical levels of two bacteria from the same sample. Bacterial quantitation by qPCR was performed by a designated technician blinded to subject information.
Continuous variables were analyzed using one-way ANOVA. The chi-squared test or Fisher’s exact test was performed for categorical variables. Levels of periodontitis-associated bacteria were categorized into two groups by their medians and compared among the three study groups. Stratified analyses were conducted to evaluate the differences in levels of periodontitis-associated bacteria between perodontitis stages II and III. Correlations between levels of bacterial species (r) were determined using Pearson correlation coefficient. P-values <0.05 were considered statistically significant. SAS version 9.4 (SAS Institute, Cary, NC, USA) was used to conduct all statistical analyses.
We enrolled 191 periodontitis patients in this study, including 56 AAs, 67 CAs, and 68 HAs with the mean age of 51.66 ± 12.79 years. Diagnosis creteria for the patients were based on the 2017 World Workshop classification (Tonetti et al., 2018; Papapanou et al., 2018b). Among them, 61.78% were diagnosed with periodontitis stage III and 38.22% were diagnosed with stage II. There was no significant difference in the distribution of periodontitis stages among AAs (66.04% with stage III), CAs (54.41% with stage III), and HAs (67.64% with stage III) (p = 0.291). In addition, there was no significant difference among the racial and ethnic groups with regards to gender (p = 0.228). To investigate the possible existence of social inequalities among these racial/ethnic groups, we used the participants’ residential zip codes to obtain median incomes of the neighborhoods in which they reside. Our results showed the neighborhood median income of the CA group to be significantly higher ($73,895 ± $26,906) than that of the AA or HA group ($58,013 ± 22,470 and $58,558 ± $ 21,434, respectively) (p<0.001). Our observation agrees with a previously reported likelihood of an association between neighborhood socioeconomic circumstances and periodontitis (Borrell et al., 2006). However, further studies are needed to elucidate how economic conditions of a neighborhood impact periodontal health of its residents.
We also found significant differences in levels of BOP among the racial/ethnic groups. We observed the highest degree of BOP in the HA group (44.18% ± 25.80%), followed by the AA group (41.91% ± 25.74%) and the CA group (38.39% ± 24.51%) (p = 0.007) (Table 2). However, we did not observe significant differences in the levels of PI among AAs (66.61% ± 32.27%), CAs (61.65% ± 24.57%), and HAs (64.77% ± 27.57%) (p = 0.569). These results suggest that different profiles of microbiota likely led to a more severe inflammatory response found in AA and HA patients compared to CA patients.
To investigate the oral microbial profiles of the study subjects, we used qPCR to measure the distribution and levels of several well-studied oral bacteria in their dental plaque samples, including keystone pathogens, accessory pathogens, and pathobionts (Hajishengallis and Lamont, 2016). Since the data are not evenly distributed, we designated bacterial levels as either higher or lower than their medians. As shown in Table 3, total bacterial levels in the dental plaque samples were determined using primers corresponding to cyanobacterial 16S rRNA as probes (Turner et al., 1999). Approximately 50% of all samples had more than 109 total bacterial cells. However, 71% of samples from the AA patients had more than 109 total bacterial cells, which was much higher than that for the CA patients (47%) and for the HA patients (38%) (p = 0.0009). Using qPCR, we detected P. gingivalis in all 191 samples. Higher levels of P. gingivalis (>106) was detected in 54.41% of samples from the HA patients and in 48.21% of samples from the AA patients, but in only 24.88% of samples from the CA patients (p = 0.0008), which may be linked to higher BOP index observed in the HA and AA patients. We also detected Tannerella forsythia and Fusobacterium nucleatum in all samples, while Treponema denticola was present in 66.49% of the samples. These three species were all evenly distributed among the AA, CA, and HA groups.
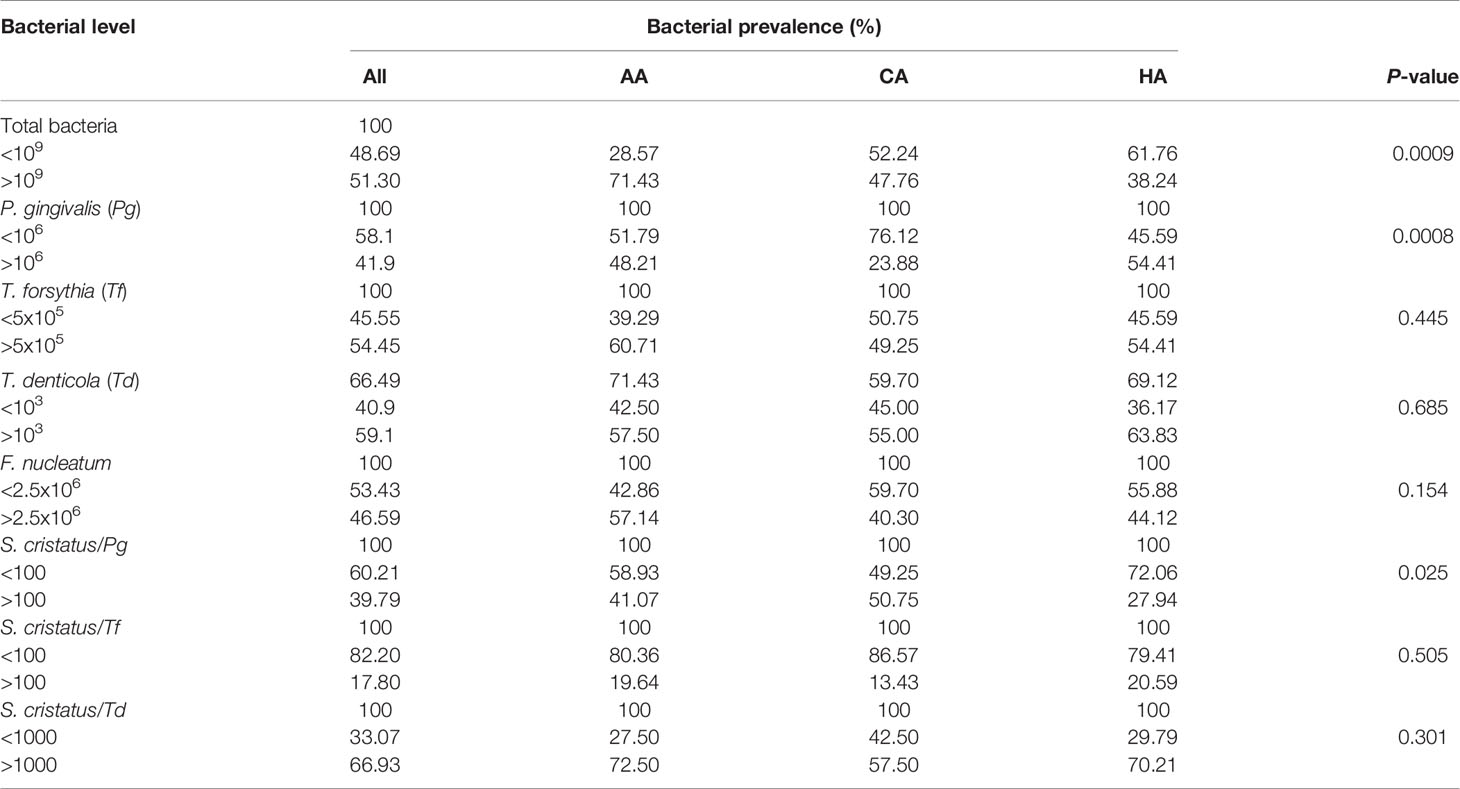
Table 3 Distributions of periodontitis-associated bacteria in samples from patients of different racial/ethnic backgrounds.
Our previous study on 13 subjects with periodontitis demonstrated that there appeared to be an inverse relationship between the number of S. cristatus versus P. gingivalis cells in dental plaque (Wang et al., 2009). Therefore, we postulated that higher levels of S. cristatus, an early colonizer of oral microbial communities, may control and suppress P. gingivalis levels, and that the higher risk of periodontitis in AA and HA populations is, at least in part, dependent on a different microbial composition containing less S. cristatus. Here, we found that 50.75% of dental plaque samples from the CA patients had S. cristatus/P. gingivalis ratios greater than 100, compared to 41.07% of samples from the AA patients and 27.94% of those from the HA patients (p = 0.025) (Table 3). Further analysis of the S. cristatus/P. gingivalis ratios revealed a significant difference in the levels of P. gingivalis and T. denticola between dental plaque samples with higher S. cristatus/P. gingivalis ratios and samples with lower ratios (Table 4). Particularly, levels of P. gingivalis were approximately 700 time higher in samples with relatively lower levels of S. cristatus. We also observed a slight increase in the abundance of total bacteria, T. denticola, and F. nucleatum in samples with lower S. cristatus/P. gingivalis ratios. These results further support our postulation that S. cristatus benefits the host by antagonizing the colonization and accumulation of P. gingivalis (Wang et al., 2009).
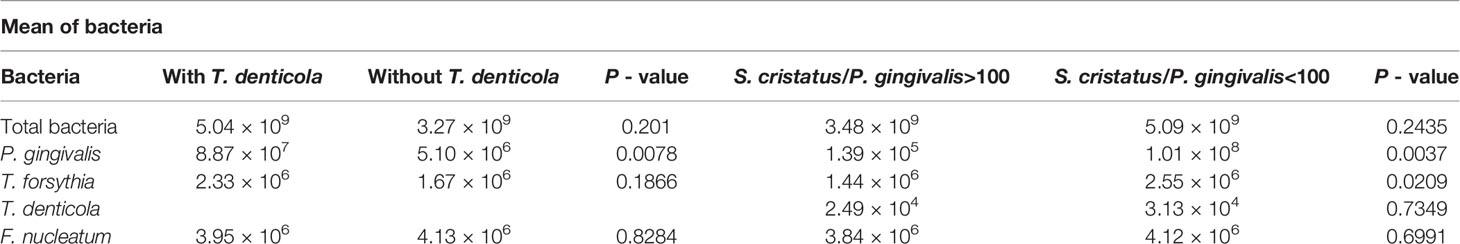
Table 4 Impact of T. denticola and the S. cristatus/P. gingivalis ratios on the abundance of periodontitis-associated species.
In addition, we examined the distribution of P. gingivalis of different fimA genotypes (types I and Ib–V), which are classified based on their nucleotide sequences of the fimA gene. We found P. gingivalis of fimA genotype II to be present in 55.5% of all samples. This is consistent with previous reports stating P. gingivalis type II as the predominant P. gingivalis strain found in periodontitis patients (Amano et al., 1999; Enersen et al., 2008). We also detected the type IV strain in 20.94% of all samples, followed by type III (10.47%), type I (9.94%), type Ib (2.10%), and type V (2.09%) (Table 5). We did not observe any significant difference in the distribution of the different fimA strains among the three racial/ethnic groups and between patients with periodontitis stage II and III.
To examine the relationship between P. gingivalis levels and periodontitis progression as well as the abundance of other bacteria, we sequentially analyzed the keystone and accessory pathogen levels in patients with different periodontitis stages and characteristics. As shown in Table 6, more samples from patients with periodontitis stage III exhibited higher levels of total bacteria (54.17% >109) and P. gingivalis (44.17% >106) than samples from stage II patients, the latter with 46.48% >109 for total bacteria and 38.03% >106 for P. gingivalis. However, these differences are not statistically significant. We also observed a similar trend for the S. cristatus/P. gingivalis ratios. Specifically, 46.48% of samples from stage II patients had ratios greater than 100, but only 35.83% of samples from stage III patients had ratios greater than 100. We observed significantly higher levels of T. denticola in samples from stage III patients than in those from stage II patients (66.28% vs. 43.90% >103, p = 0.016). These results are consistent with our observation that samples with higher T. denticola levels also possessed higher levels of P. gingivalis (mean = 8.86 × 107) than samples with lower T. denticola levels (mean = 5.10 × 106) (p = 0.0078) (Table 4). These findings indicate that co-colonization of P. gingivalis and T. denticola may be associated with periodontitis progression. We also examined correlation between other independent variables, including body mass index, smoking, and diabetes, and severities of periodontitis. We did not found significant influence of these variables on stages of periodontitis in this cohort (Table 6).
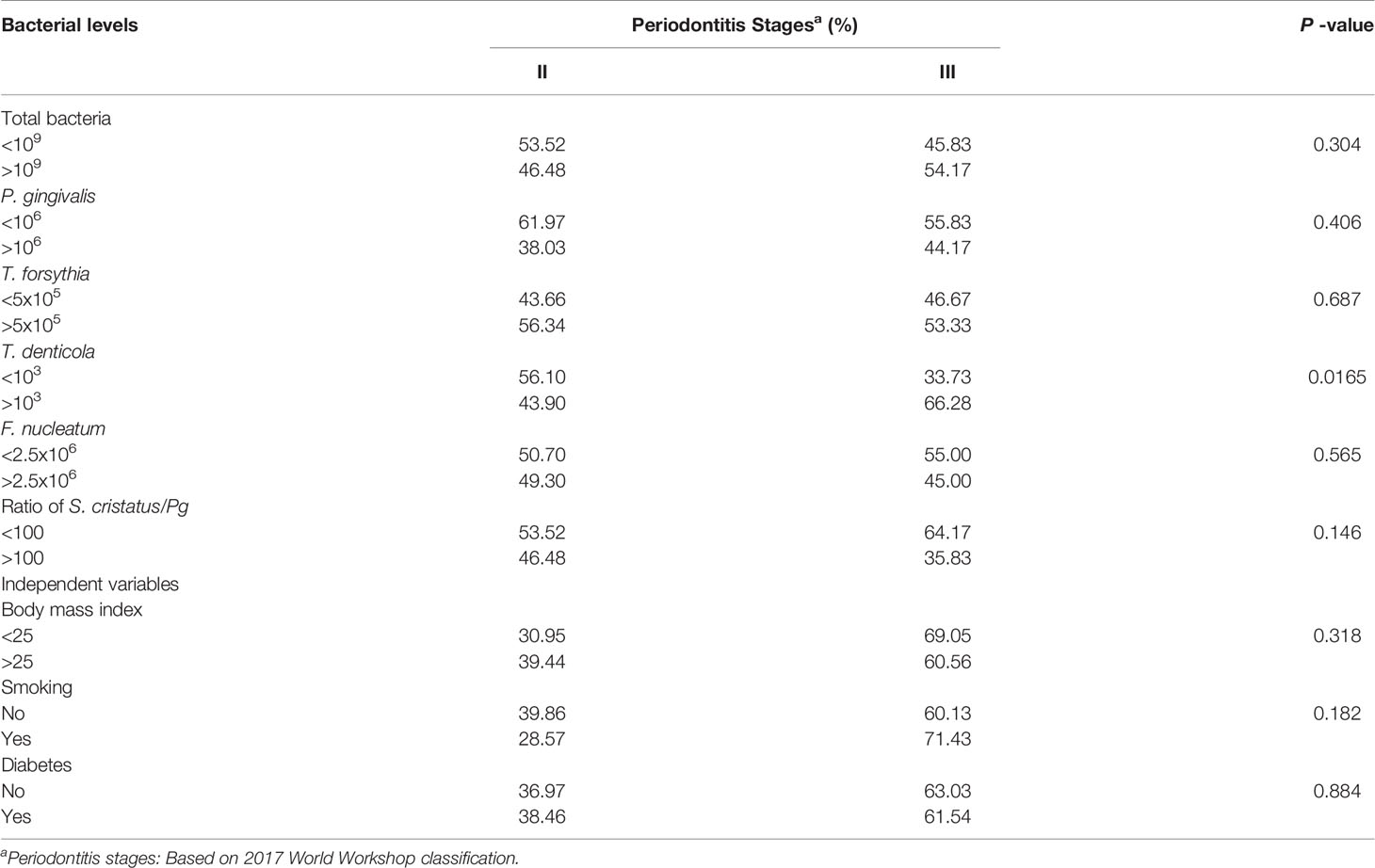
Table 6 Correlation between oral bacterial levels or independent variables and periodontal disease stages.
In addition, we examined the correlations among these oral bacteria. Levels of P. gingivalis, T forsythia, T. denticola, and F. nucleatum in the samples positively correlated with the level of total bacteria to various extents (Table 7). The level of P. gingivalis correlated most strongly with the level of total bacteria (r = 0.66), followed by T. forsythia (r = 0.59), F. nucleatum (r = 0.29), and T. denticola (r = 0.18). Interestingly, T. forsythia exhibited a relatively high degree of correlation with P. gingivalis, T. denticola, and F. nucleatum (r > 0.45), suggesting the importance of T. forsythia in periodontal dysbiosis. In contrast, P. gingivalis showed weaker correlations with T. denticola (r = 0.18) and F. nucleatum (r = 0.15). We did not observe any significant correlation between T. denticola and F. nucleatum.
Previous studies based on the 2009–2010 NHANES demonstrated periodontitis incidence to be significantly higher in AAs (58.6%) and HAs (59.7%) than in CAs (42.6%) (Thornton-Evans et al., 2013). Here, we examined the potential risk factors for periodontal health disparities by comparing the abundance of several well-studied bacteria in dental plaque samples from periodontitis patients of different racial/ethnic backgrounds. We detected a significantly higher bacterial mass in the AA patients than in the CA and HA patients. Additionally, P. gingivalis levels were much higher in the AA and HA patients than in the CA patients, rendering P. gingivalis levels a potential risk factor for periodontitis progression in the AA and HA populations. Our findings also indicate the involvement of S. cristatus in regulating P. gingivalis levels. We previously reported an antagonistic relationship between S. cristatus and P. gingivalis (Xie et al., 2000). We identified arginine deiminase (ArcA), a surface protein of S. cristatus, as the signaling molecule to which P. gingivalis responds by repressing fimA gene expression and protein production (Xie et al, 2007; Wu and Xie, 2010). Our previous study on 13 subjects with periodontitis revealed the possibility of an inverse relationship between the number of S. cristatus cells and that of P. gingivalis cells in dental plaques, suggesting that S. cristatus may benefit the host by antagonizing the colonization and accumulation of P. gingivalis (Wang et al., 2009). This study with larger sample size further revealed significantly lower ratios of S. cristatus to P. gingivalis in samples from the AA and HA patients than in those from the CA patients, thereby establishing the association between the S. cristatus/P. gingivalis ratio and disparity in periodontitis. Consistent with our previous discovery of significantly higher arcA expression in S. cristatus than in Streptococcus gordonii (Lin et al., 2008), we did not find a negative correlation between S. gordonii and P. gingivalis (data not shown). Taken together, our findings suggest that these two streptococcal species play distinct roles in the highly orchestrated dental plaque formation. Furthermore, results from these clinical studies provide a rationale for eliminating P. gingivalis from oral microbial communities using synthesized peptide analogs derived from S. cristatus ArcA.
In this study, T. forsythia appeared to be the most closely correlated to P. gingivalis in dental plaques. Levels of both P. gingivalis and T. forsythia increased when the S. cristatus/P. gingivalis ratio was low. These results agree with our previous observation that levels of P. gingivalis and T. forsythia were correlated in multispecies biofilms using an ex-vivo binding assay (Ho et al., 2018). Several clinical studies also reported a strong co-occurrence relationship among P. gingivalis, T. forsythia, T. denticola, and F. nucleatum in subgingival plaques (Gmur et al., 1989; Wara-aswapati et al., 2009; da Silva-Boghossian et al., 2011). Here, we confirmed the positive correlations among these bacterial species in all 191 dental plaque samples, though with varying degrees of coefficiency. Interestingly, T. forsythia showed higher correlations with P. gingivalis, T. denticola, and F. nucleatum compared to those among other bacterial species. A recent clinical study used immunohistochemistry and qPCR to show that P. gingivalis and T. forsythia aggregated in dental plaques and periodontal tissues from periodontitis patients (Rajakaruna et al., 2018). The interaction between F. nucleatum and T. forsythia is well established; it likely involves the hydrolyzation of β-glucans by T. forsythia β-glucanase (Sharma et al., 2005; Honma et al., 2018). However, direct interaction between T. forsythia and T. denticola has not been revealed. In an in vitro polymicrobial biofilm study using P. gingivalis, T. denticola, and T. forsythia, Zhu et al. found a significant increase in the total biovolume for P. gingivalis and T. denticola in polymicrobial microcolonies compared to single-species biofilms. Conversely, only a few T forsythia cells were detected in the polymicrobial biofilms (Zhu et al., 2013). A study by Hashimoto et al. revealed coaggregation of P. gingivalis and T. denticola mediated by a P. gingivalis fimbrial protein and T. denticola dentilisin (Hashimoto et al., 2003). Here, our results support a recently proposed concept that bacterial properties within oral microbial communities are context-dependent (Hajishengallis and Lamont, 2021). Particularly, we showed that S. cristatus may regulate the levels of P. gingivalis and T. forsythia in dental plaques, and that T. denticola was only detected in dental plaques with relatively high levels of P. gingivalis.
In conclusion, characteristics of periodontal dysbiosis including increase in levels of P. gingivalis and its association with T. forsythia, T. denticola, and F. nucleatum are potential risk factors of disparities in periodontal health and periodontitis severity. Moreover, S. cristatus controls P. gingivalis levels in oral microbiota and plays an important role in regulating the virulence potential of multispecies communities.
The original contributions presented in the study are included in the article/supplementary material. Further inquiries can be directed to the corresponding authors.
The studies involving human participants were reviewed and approved by the Committee for the Protection of Human Subjects of the University of Texas Health Science Center at Houston. The patients/participants provided their written informed consent to participate in this study.
HX and B-YW conceived the study and supervised the project. TL and QC performed and verified the statistical analyses. B-YW, SS, H-WM, and LA enrolled study participants. M-HH and JH helped sample process. All authors discussed the results and contributed to the final manuscript.
The study was supported in part by grant MD007586 from the National Institute on Minority Health and Health Disparities, United States of America. This work was supported by in part by Meharry RCMI MD007593 from NIMHD.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors are grateful to all enrolled subjects for their participation in this research. The authors thank Krishna Kookal for abstracting clinical parameters from the Electronic Health Records at the School of Dentistry, University of Texas Health Science Center at Houston. The authors also thank the Meharry Office of Scientific Editing and Publications (S21MD000104) for editorial assistance.
Amano, A., Nakagawa, I., Kataoka, K., Morisaki, I., Hamada, S. (1999). Distribution of Porphyromonas Gingivalis Strains With fimA Genotypes in Periodontitis Patients. J. Clin. Microbiol. 37 (5), 1426–1430. doi: 10.1128/JCM.37.5.1426-1430.1999
Borrell, L. N., Burt, B. A., Warren, R. C., Neighbors, H. W. (2006). The Role of Individual and Neighborhood Social Factors on Periodontitis: The Third National Health and Nutrition Examination Survey. J. Periodontol. 77 (3), 444–453. doi: 10.1902/jop.2006.050158
da Silva-Boghossian, C. M., do Souto, R. M., Luiz, R. R., Colombo, A. P. (2011). Association of Red Complex, A. Actinomycetemcomitans and Non-Oral Bacteria With Periodontal Diseases. Arch. Oral. Biol. 56 (9), 899–906. doi: 10.1016/j.archoralbio.2011.02.009
Eke, P. I., Thornton-Evans, G. O., Wei, L., Borgnakke, W. S., Dye, B. A., Genco, R. J. (2018). Periodontitis in US Adults: National Health and Nutrition Examination Survey 2009-2014. J. Am. Dent. Assoc. 149 (7), 576–588.e576. doi: 10.1016/j.adaj.2018.04.023
Enersen, M., Olsen, I., Kvalheim, O., Caugant, D. A. (2008). fimA Genotypes and Multilocus Sequence Types of Porphyromonas Gingivalis From Patients With Periodontitis. J. Clin. Microbiol. 46 (1), 31–42. doi: 10.1128/JCM.00986-07
Gmur, R., Strub, J. R., Guggenheim, B. (1989). Prevalence of Bacteroides Forsythus and Bacteroides Gingivalis in Subgingival Plaque of Prosthodontically Treated Patients on Short Recall. J. Periodontal Res. 24 (2), 113–120. doi: 10.1111/j.1600-0765.1989.tb00865.x
Hajishengallis, G., Darveau, R. P., Curtis, M. A. (2012). The Keystone-Pathogen Hypothesis. Nat. Rev. Microbiol. 10 (10), 717–725. doi: 10.1038/nrmicro2873
Hajishengallis, G., Lamont, R. J. (2012). Beyond the Red Complex and Into More Complexity: The Polymicrobial Synergy and Dysbiosis (PSD) Model of Periodontal Disease Etiology. Mol. Oral. Microbiol. 27 (6), 409–419. doi: 10.1111/j.2041-1014.2012.00663.x
Hajishengallis, G., Lamont, R. J. (2016). Dancing With the Stars: How Choreographed Bacterial Interactions Dictate Nososymbiocity and Give Rise to Keystone Pathogens, Accessory Pathogens, and Pathobionts. Trends Microbiol. 24 (6), 477–489. doi: 10.1016/j.tim.2016.02.010
Hajishengallis, G., Lamont, R. J. (2021). Polymicrobial Communities in Periodontal Disease: Their Quasi-Organismal Nature and Dialogue With the Host. Periodontol. 2000 86, 210–30. doi: 10.1111/prd.12371
Hajishengallis, G., Liang, S., Payne, M. A., Hashim, A., Jotwani, R., Eskan, M. A., et al (2011). Low-Abundance Biofilm Species Orchestrates Inflammatory Periodontal Disease Through the Commensal Microbiota and Complement. Cell Host Microbe 10 (5), 497–506. doi: 10.1016/j.chom.2011.10.006
Hashimoto, M., Ogawa, S., Asai, Y., Takai, Y., Ogawa, T. (2003). Binding of Porphyromonas Gingivalis Fimbriae to Treponema Denticola Dentilisin. FEMS Microbiol. Lett. 226 (2), 267–271. doi: 10.1016/S0378-1097(03)00615-3
Ho, M. H., Lamont, R. J., Chazin, W. J., Chen, H., Young, D. F., Kumar, P., et al (2018). Characterization and Development of SAPP as a Specific Peptidic Inhibitor That Targets Porphyromonas Gingivalis. Mol. Oral. Microbiol. 33 (6), 430–439. doi: 10.1111/omi.12246
Honma, K., Ruscitto, A., Sharma, A. (2018). Beta-Glucanase Activity of the Oral Bacterium Tannerella Forsythia Contributes to the Growth of a Partner Species, Fusobacterium Nucleatum, in Cobiofilms. Appl. Environ. Microbiol. 84:e01759–17. doi: 10.1128/AEM.01759-17
Lin, X., Lamont, R. J., Wu, J., Xie, H. (2008). Role of Differential Expression of Streptococcal Arginine Deiminase in Inhibition of fimA Expression in Porphyromonas Gingivalis. J. Bacteriol. 190 (12), 4367–4371. doi: 10.1128/JB.01898-07
Newman, M., Takei, H., Klokkevold, H., Carranza, F. (2018). Newman and Carranza's Clinical Periodontology. Thirteenth Edition. Chapter 32 (Pages 387–389) and Chapter 48 (Page 520.e5).
Papapanou, P. N., Sanz, M., Buduneli, N., Dietrich, T., Feres, M., Fine, D. H., et al (2018a). Periodontitis: Consensus Report of Workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Periodontol. 89 Suppl 1, S173–S182. doi: 10.1002/JPER.17-0721
Papapanou, P. N., Sanz, M., Buduneli, N., Dietrich, T., Feres, M., Fine, D. H., et al (2018b). Periodontitis: Consensus Report of Workgroup 2 of the 2017 World Workshop on the Classification of Periodontal and Peri-Implant Diseases and Conditions. J. Clin. Periodontol. 45 Suppl 20, S162–S170. doi: 10.1111/jcpe.12946
Rajakaruna, G. A., Negi, M., Uchida, K., Sekine, M., Furukawa, A., Ito, T., et al (2018). Localization and Density of Porphyromonas Gingivalis and Tannerella Forsythia in Gingival and Subgingival Granulation Tissues Affected by Chronic or Aggressive Periodontitis. Sci. Rep. 8 (1), 9507. doi: 10.1038/s41598-018-27766-7
Sharma, A., Inagaki, S., Sigurdson, W., Kuramitsu, H. K. (2005). Synergy Between Tannerella Forsythia and Fusobacterium Nucleatum in Biofilm Formation. Oral. Microbiol. Immunol. 20 (1), 39–42. doi: 10.1111/j.1399-302X.2004.00175.x
Socransky, S. S., Haffajee, A. D. (1994). Evidence of Bacterial Etiology: A Historical Perspective. Periodontol. 2000 5, 7–25. doi: 10.1111/j.1600-0757.1994.tb00016.x
Thornton-Evans, G., Eke, P., Wei, L., Palmer, A., Moeti, R., Hutchins, S., et al (2013). Periodontitis Among Adults Aged >/=30 Years - United States 2009-2010. MMWR Suppl. 62 (3), 129–135.
Tonetti, M. S., Greenwell, H., Kornman, K. S. (2018). Staging and Grading of Periodontitis: Framework and Proposal of a New Classification and Case Definition. J. Clin. Periodontol. 45 Suppl 20, S149–S161. doi: 10.1111/jcpe.12945
Turner, S., Pryer, K. M., Miao, V. P., Palmer, J. D. (1999). Investigating Deep Phylogenetic Relationships Among Cyanobacteria and Plastids by Small Subunit rRNA Sequence Analysis. J. Eukaryot. Microbiol. 46 (4), 327–338. doi: 10.1111/j.1550-7408.1999.tb04612.x
Wang, B. Y., Wu, J., Lamont, R. J., Lin, X., Xie, H. (2009). Negative Correlation of Distributions of Streptococcus Cristatus and Porphyromonas Gingivalis in Subgingival Plaque. J. Clin. Microbiol. 47 (12), 3902–3906. doi: 10.1128/JCM.00072-09
Wara-aswapati, N., Pitiphat, W., Chanchaimongkon, L., Taweechaisupapong, S., Boch, J. A., Ishikawa, I. (2009). Red Bacterial Complex Is Associated With the Severity of Chronic Periodontitis in a Thai Population. Oral. Dis. 15 (5), 354–359. doi: 10.1111/j.1601-0825.2009.01562.x
Wu, J., Xie, H. (2010). Role of Arginine Deiminase of Streptococcus Cristatus in Porphyromonas Gingivalis Colonization. Antimicrob. Agents Chemother. 54 (11), 4694–4698. doi: 10.1128/AAC.00284-10
Xie, H., Cook, G. S., Costerton, J. W., Bruce, G., Rose, T. M., Lamont, R. J. (2000). Intergeneric Communication in Dental Plaque Biofilms. J. Bacteriol. 182 (24), 7067–7069.
Xie, H., Lin, X., Wang, B. Y., Wu, J., Lamont, R. J. (2007). Identification of a Signalling Molecule Involved in Bacterial Intergeneric Communication. Microbiology 153 (Pt 10), 3228–3234. doi: 10.1099/mic.0.2007/009050-0
Zheng, C., Wu, J., Xie, H. (2011). Differential Expression and Adherence of Porphyromonas Gingivalis FimA Genotypes. Mol. Oral. Microbiol. 26 (6), 388–395. doi: 10.1111/j.2041-1014.2011.00626.x
Keywords: Porphyromonas gingivalis, periodontitis, racial and ethnic groups, Streptococcus cristatus, microbial profiles
Citation: Wang B-Y, Lu T, Cai Q, Ho M-H, Sheng S, Meng H-W, Arsto L, Hong J and Xie H (2021) Potential Microbiological Risk Factors Associated With Periodontitis and Periodontal Health Disparities. Front. Cell. Infect. Microbiol. 11:789919. doi: 10.3389/fcimb.2021.789919
Received: 05 October 2021; Accepted: 29 October 2021;
Published: 17 November 2021.
Edited by:
Max Maurin, Université Grenoble Alpes, FranceReviewed by:
Vijay Kumar Chava, Narayana Dental College and Hospital, IndiaCopyright © 2021 Wang, Lu, Cai, Ho, Sheng, Meng, Arsto, Hong and Xie. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Bing-Yan Wang, bing-yan.wang@uth.tmc.edu; Tom Lu, t.lu@ttu.edu; Hua Xie, hxie@mmc.edu
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.