- Key Laboratory of Marine Biotechnology of Zhejiang Province, School of Marine Sciences, Ningbo University, Ningbo, China
The use of antibiotics to facilitate resistance to pathogens in aquatic animals is a traditional method of pathogen control that is harmful to the environment and human health. RNAi is an emerging technology in which homologous small RNA molecules target specific genes for degradation, and it has already shown success in laboratory experiments. However, further research is needed before it can be applied in aquafarms. Many laboratories inject the dsRNA into aquatic animals for RNAi, which is obviously impractical and very time consuming in aquafarms. Therefore, to enable the use of RNAi on a large scale, the methods used to prepare dsRNA need to be continuously in order to be fast and efficient. At the same time, it is necessary to consider the issue of biological safety. This review summarizes the key harmful genes associated with aquatic pathogens (viruses, bacteria, and parasites) and provides potential targets for the preparation of dsRNA; it also lists some current examples where RNAi technology is used to control aquatic species, as well as how to deliver dsRNA to the target hydrobiont.
1 Introduction
Aquatic animals are very important part of the contemporary food and along with other industries dominate the global economy. Currently, aquatic organisms are severely endangered, with infections by various bacterial, fungal pathogens and invasion by viruses causing a significant decline in aquatic animal populations. For example, infectious myonecrosis virus (IMNV) poses a serious threat to shrimp farming in many countries around the world, especially in Brazil (Andrade et al., 2022). Decapod iridescent virus 1 (DIV1) is an emerging virus that has posed a serious threat to crustacean farming in recent years and has caused significant economic losses (Liao et al., 2022). White spot syndrome virus (WSSV) is considered one of the most devastating diseases for shrimp farming, and there are many ways to prevent the invasion of this virus (Kumar et al., 2022). Much research has been devoted to the safety of aquatic products. For a long time, antibiotics have been used to protect aquatic animals from pathogens (Jones, 1986; Xu et al., 2021), but they have severely affected the marine ecosystem, causing eutrophication of the water and also posing a risk to humans. New ways of aquatic control are thus being sought.
The phenomenon of RNAi (RNA interference) is a molecularly mediated post-transcriptional gene-silencing mechanism, and the molecule is known as double-stranded RNA., which was discovered in plants (Napoli et al., 1990) and in Caenorhabditis elegans in 1998 (Andrew Fire, 1998). Since the existence of RNAi was reported, researchers began to use it to study the functions of certain genes. Also, RNAi can be used for biological control, likely aquaculture and insects. (Katoch et al., 2013). In this review, we summarize the applications of RNAi in some aquatic organisms, including studies targeting viral, bacterial, and parasitic genes. We also explore the latest pathways used to prepare RNAi and look at the future of RNAi.
2 Mechanisms of RNAi
RNAi is a pathway through which gene expression is downregulated, which consists of dsRNA targeting specific mRNAs inside and outside the cell for degradation (Hannon, 2002). RNAi represents an innate immune system (Reshi et al., 2014; Gong and Zhang, 2021), and as an innate immune response, when an exogenous mRNA enters a cell, it is cleaved to inhibit the replication and translation of the mRNA (Hannon, 2002). It is this property that allows RNAi technology to target certain genes for knock out and suppress viral invasion. In the initiation phase of RNAi, dsRNA is cleaved into small segments of 21–23 nucleotides by an RNase-III-like enzyme, specifically an ATP-dependent enzyme called Dicer (Hamilton and Baulcombe, 1999; Zamore et al., 2000; Bernstein et al., 2001). These small nucleotide segments of double-stranded RNA are called siRNA and have approximately 19 bp duplexes and two-base 3′-overhangs(Lingel and Izaurralde, 2004). The siRNA is then involved in the formation of the RNA-induced silencing complex (RISC). RISC is the key role in RNAi technology (Chendrimada et al., 2005; Schuster et al., 2019).
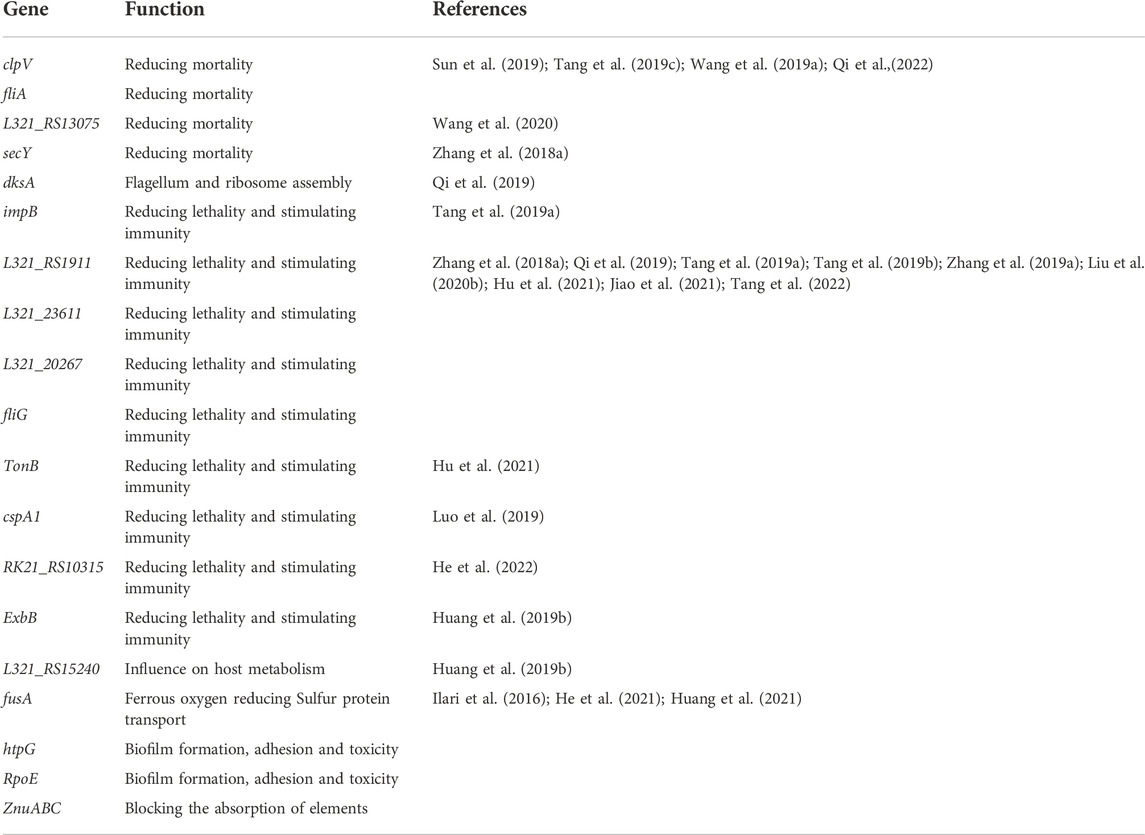
3 RNAi-mediated inhibition of Pseudomonas
Pseudomonas plecoglossicida is a pathogen that can cause significant harm to marine organisms (Zhang et al., 2013) and has caused serious economic losses to the marine economy. Since the isolation of P. plecoglossicida, its pathogenic mechanism has been extensively studied, from which relevant therapeutic targets can be discovered (Table 1). The silencing of genes in the bacterium via RNAi technology can significantly reduce mortality of the host, and these genes share similar functions, namely, toxicity, adhesion, and flagellar movement.
Trace elements such as Mn and Zn are necessary for bacterial growth, and these elements are only available through the host, which uses a pathway to prevent bacterial uptake of the elements (Kehl-Fie and Skaar, 2010; Hood and Skaar, 2012). Zn is a very important element that plays a catalytic role in proteins and is also able to maintain protein functions and partial bacterial toxicity (Hantke, 2005). Many bacteria transport Zn via the ZnuABC transporter (Ilari et al., 2016). It was found that ZnuC is an important protein for Zn uptake in Pseudomonas aeruginosa (Huang et al., 2021). Similarly, the authors found that silencing the znuA gene in this strain could achieve an 89.2% reduction of deaths and that RNAi-treated strains also induced antibody production in grouper (He et al., 2021). We have summarized some genes that we expect to enhance host defense through RNAi technology (Table 1).
4 RNAi-mediated inhibition of Aeromonas hydrophila
Aeromonas hydrophila is a species of Aeromonas capable of causing significant harm to both aquatic organisms and humans, impacting the aquatic industry worldwide (Sha et al., 2002; Sabili et al., 2015). Until the mechanisms of A. hydrophila virulence were understood, antibiotics were used to control related diseases (Samir et al., 2017). With the introduction of a large number of antibiotics, A. hydrophila has developed drug resistance (Mao et al., 2020). People found silencing LuxR by using RNAi technology can partially restore the susceptibility of A. hydrophila to antibiotics (Chang et al., 2010). Therefore, inhibition of this strain with RNAi is a promising approach. Many genes have important roles in A. hydrophila biofilm formation, bacterial motility, and virulence.
The escape of bacteria in macrophages is necessary for a rapid infection process (Qin et al., 2014). In an in vitro invasion assay of A. hydrophila, flgE was found to be an important gene for flagellogenesis and key to the infestation of macrophages (Qin et al., 2014). Further, acuC-RNAi strains were able to reduce mortality in zebrafish (Jiang et al., 2017), which also resulted the expression of hlyA (a key virulence gene of A. hydrophila) decreased (Qian et al., 1995), demonstrating that acuC is an important regulatory protein affecting the survival and pathogenicity of A. hydrophila. Moreover, icmF is thought to be an ATPase capable of stimulating the T6SS secretome of A. hydrophila (Ma et al., 2012). The maximum efficiency of knockdown for icmF using RNAi technology can reach 94.42%, resulting in a decrease in the survival of A. hydrophila from 92.3% to 20.58%, as well as a significant reduction in the probability of escape from macrophages(Wang S. et al., 2019).
Reactive oxygen species (ROS) are an important way for host macrophages to mitigate bacterial invasion (Grayfer et al., 2014). Zhang et al. (Zhang M. et al., 2018) found that katG facilitates the removal of host H2O2 by A. hydrophila and aids in its survival in macrophages and that RNAi-katG could reduce immune escape and fish mortality by 85%; the authors also found that sodA-RNAi and sodB-RNAi were able to reduce A. hydrophila survival in fish macrophages by 91.8% and 74.9% and reduce immune evasion by 32% and 92%, respectively, in addition to restoring ROS content in some macrophages and enhancing host immunity (Zhang M. et al., 2019). These genes are summarized. (Table 2).
5 RNAi to target Vibrio alginolyticus
Vibrio alginolyticus is capable of harming coral polyps, aquatic organisms (Zhenyu et al., 2013; Huang et al., 2019b), and crustaceans. Oxidative phosphorylation is an important pathway for aerobic growth and energy acquisition, suggesting that key oxidative phosphorylation proteins are associated with bacterial adherence. However, silencing the relevant oxidative phosphorylation protein-encoding genes via RNAi technology can result in reduced bacterial adherence and cytochrome C oxidase activity. The adherence in V. alginolyticus is critical in the early stages of pathogenesis, and there have been many studies on targeting adhesion genes using RNAi for Vibrio lysozyme (table below), including some tricarboxylic acid cycle genes (Huang et al., 2016b). These proteins could be potential targets for the RNAi-mediated control of aquatic pathogens. Zhang (Liu et al., 2012) found that luxT promotes the transcription of luxO and luxR, luxO can regulate MviN (an extracellular protein that produces toxicity) (Cao et al., 2010), and luxO promotes the secretion of extracellular substances and the formation of iron carriers in V. alginolyticus (Wang et al., 2007). The pep protein (a protein required for Vibrio lysis motility) is also regulated by luxO (Cao et al., 2011), and luxR regulates extracellular alkaline serine protease A and reduces the production of extracellular sugars and motility in V. alginolyticus (Rui et al., 2008). LuxR and AphA are two of the most important molecules involved in sensing the Vibrio lysogenicus population. One related study found that the AphB (Gao et al., 2017) gene can positively regulate the expression of luxR and the toxin asp (alkaline serine protease), which is expected to reduce Vibrio lysis by repressing AphB, whereas AphA can negatively regulate the asp toxin through luxR (Gu et al., 2016). Further, luxO-luxR can regulate asp production (Rui et al., 2009). ValR, a gene homologous to luxR, affects both cell membrane formation and bacterial motility by regulating flagellar synthesis (Chang et al., 2010). DctP, a protein-transporting subunit, was found to regulate the expression of 22 genes involved in the pathogenesis of Vibrio lysogenicus, without inducing any morphological changes, but with significantly reduced adherence and virulence (Zhang et al., 2022). VqsA (Gao et al., 2018) is a transcription factor that functions in Type VI secretion systems. AcfA is a factor required for Vibrio infection, and in acfA-deficient strains in which DctP (Zhang et al., 2022) and deoD transcript levels were found to be increased, pepD, arA, fla, and ompA genes are repressed (Cai et al., 2018). Both TonB systems are important for virulence in Vibrio lysogenicus, and the absence of TonB diminishes this virulence (Wang et al., 2008). A previous team used pulsed-field gel electrophoresis to isolate a number of Vibrio pathogenesis-related genes for subsequent RNAi technology (Ren et al., 2013). Huang et al. (Huang et al., 2015a; Huang et al., 2016a) used high-throughput sequencing to find a number of non-coding RNA molecules that they believe play a key role in Vibrio infection of the host. Srvg17985, a small RNA that regulates various aspects of stress balance in Vibrio, is involved in adapting to environmental stress, and is expected to be a new target (Deng et al., 2019). Vvrr1 (a non-coding RNA) and pykF interact with each other and are involved in the mechanism of virulence in V. alginolyticus (Zuo et al., 2019). Qrr (Liu H. et al., 2020) is a non-coding small molecule RNA that activates luxR and inhibits aphA.
Micronutrient uptake is necessary for growth and physiological processes in bacteria (Kehl-Fie and Skaar, 2010; Hood and Skaar, 2012). OmpU is a pore protein located on the surface of V. alginolyticus that regulates Fe uptake, and physiological growth is compromised in strains lacking this protein (Lv et al., 2020). Branched-chain amino acid metabolism is also capable of influencing bacterial physiological activity. Deng et al. (Deng et al., 2017) detected 311 upregulated genes and 251 downregulated genes in nitrogen source culture, which provides a foundation for subsequent RNAi applications. Genes which have mentioned are summarized. (Table 3).
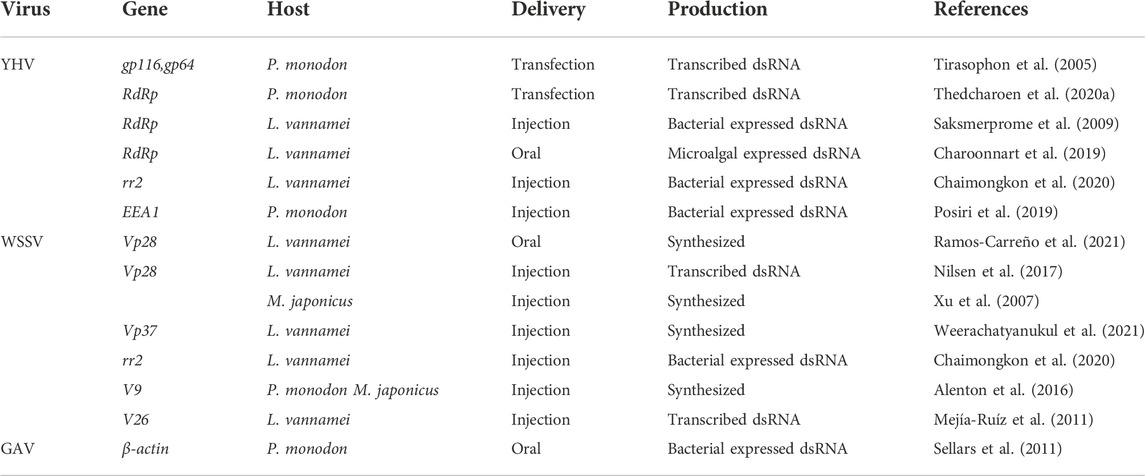
6 Viruses and RNAi
RNAi-mediated long dsRNA or siRNA causes cellular resistance to foreign nucleic acid invasion and is a natural mechanism prevalent in many species (Keene et al., 2005; Wang et al., 2006; Ding and Voinnet, 2007). Aquatic animals can be infected by many types of viruses, and one virus can carry multiple disease-causing genes (Table 4). The injection of these specific viral sequences into shrimp enables the shrimp to produce RNAi to resist the virus. YHV (yellow head virus), is a positive sense, single-stranded RNA virus; WSSV, white spot syndrome virus, is a DNA virus comprised of double-stranded circular DNA, and we list some evidence for RNAi silencing of key viral genes. It has been demonstrated that YHV-specific dsRNA introduced into spotted shrimp can effectively inhibit the replication of YHV (Tirasophon et al., 2005; Yodmuang et al., 2006; Tirasophon et al., 2007).
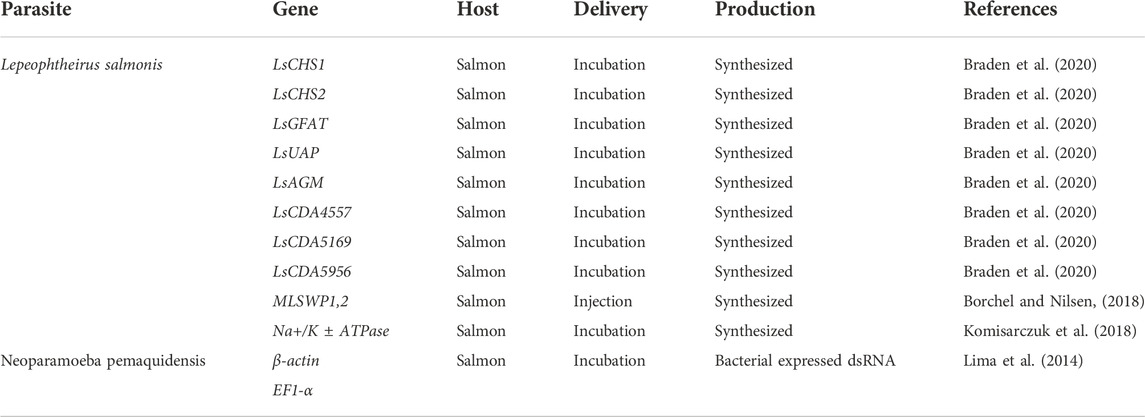
7 Parasites and RNAi
Salmon farming has been largely affect by Lepeophtheirus salmonis (salmon louse) (Brooker et al., 2018). AGD (Amoebic gill disease) is a parasitic disease of salmonids (Crosbie et al., 2012). The use of large amounts of anti-parasitic drugs seems to alleviate the development of the association disease (Carmichael et al., 2013). But as we mentioned before, drug abuse can put our health at risk. We use RNAi to target certain pathway genes, which can also prevent parasitism. For example, inhibition of the titin synthesis pathway in this parasite is a very effective way of targeting it (Liu et al., 2019). We list some examples of key genes in aquatic parasites that have been knocked out using RNAi technology (Table 5).
8 Preparation and uptake of RNAi
Many RNAi molecules can now be injected in the laboratory to control the death of aquatic animals, but this is a time-consuming and impractical approach for aquaculture farms. The uptake of RNAi molecules in cells is hampered by properties, such as the molecular length and volume, charge, and nuclease degradation (Gupta et al., 2019; Sajid et al., 2020). Therefore, to apply RNAi in aquaculture farms, two conditions must be met as follows: 1) more dsRNA production (including the preparation of dsRNA) and 2) the stable presence of dsRNA in aquatic organisms (including ingestion of dsRNA).
8.1 Preparation
8.1.1 Expressing in bacteria
The use of fed-batch fermentation was able to increase the growth of the bacteria and increase the nucleic acids. The fermentation of E. coli HT115 (DE3) was able to increase the growth of the bacteria, increase the amount of nucleic acid and then increase the level of dsRNA molecular, and maximize the expression function of E. coli HT115 (DE3) (Papić et al., 2018). Moreover, in E. coli, p19, an siRNA binding protein, stabilizes the siRNA produced by this bacterium and increases the amount transferred to siRNA (Huang and Lieberman, 2013).
Yarrowia lipolytica is also a harmless bacterium. Álvarez-Sánchez et al. (Álvarez-Sánchez et al., 2018) used Y. lipolytica to produce dsRNA to help L. vannamei resist WSSV, and when Y. lipolytica produced hairpin RNA against WSSV-orf89, injection of the extracted RNA into the muscle of shrimp improved survival after WSSV infection by 25%.
Lactobacillus plantarum is a plant-derived lactic acid bacterium that is harmless to humans. Thammasorn et al. (Thammasorn et al., 2017) modified L. plantarum to produce hairpin RNA by transferring genes targeting YHV specific to this probiotic and were able to enhance resistance to YHV in shrimp consuming the probiotic while retaining the original function of the probiotic, which is to inhibit the development of some diseases. Another probiotic, W2, was found to be non-toxic, containing an active bacteriostatic component, and was able to antagonize seven strains of aquatic pathogens, while also promoting shrimp growth (Wei et al., 2022). W2 appears to be equally useful for RNAi control in the same manner as L. plantarum. Dekham et al. (Dekham et al., 2022) transferred hpRNA targeting WSSV-vp28 into L. plantarum and Lactococcus lactis via pWH1520-VP28 and found that both probiotics were able to protect the host against the virus, with L. lactis not only reducing shrimp mortality due to WSSV but also stimulating RNAi and activating the innate immune system of the shrimp.
8.1.2 Reorganization of the particles
Sarathi et al., (2008) fed Penaeus monodon inactivated bacteria specifically expressing dsRNA from WSSV vp28 and pelleted feed covered with vp28 dsRNA-chitosan composite nanoparticles and invaded P. monodon by WSSV. Romo-Quiñonez et al. (Romo et al., 2020) were able to enhance the antiviral potential of Litopenaeus vannamei after using a silver nanoparticle, Argovit4, administered as feed. Sinnuengnong et al., (2018) co-expressed virus-like particles of pstDNV with dsRNA molecules of YHV-Pro to deliver dsRNA using viral proteins, which was also very effective. Thedcharoen et al., (2020b) used the pLVX-AcGFP1-N1 vector with a gene encoding long hairpin RNA, namely pLVX-lhRdRp, introduced under control of the CMV promoter, to inhibit the RdRp (RNA-dependent RNA polymerase) of YHV with significant effects.
8.1.3 Small creatures
Artemia, a common feed for marine organisms, is able to enrich proteins and other molecules in the gut (Subhadra et al., 2010). Feeding P. monodon with LSNV-enriched dsRNA artemia can effectively inhibit LSNV infection (Thammasorn et al., 2013).
Microalgae possess essential nutrients for aquatic organisms, and many natural antimicrobial substances have become a promising new green and environmentally friendly platform (Charoonnart et al., 2018; Fayyaz et al., 2020). Chlamydomonas reinhardtii is a distinct group of microalgae in which exogenous genes can be recombined into nuclear DNA or chloroplast DNA to express recombinant proteins (Rosales-Mendoza et al., 2012; Shamriz and Ofoghi, 2016). Somchai et al. (Somchai et al., 2016) transferred dsRNA targeting YHV-RdRp into the nuclear genome of C. reinhardtii to produce hairpin RNA. Feeding this to larval shrimp increased their survival by 22% after YHV infection. However, eukaryotic cell nuclei have mechanisms associated with RNAi that target dsRNA for degradation, which are lacking in chloroplasts (Boynton et al., 1988). In chloroplasts, dsRNA can accumulate and produce large amounts of dsRNA. At the same time, gene transformation in chloroplasts is not affected by off-target effects, mutations, and antibiotic screening (Doron et al., 2016). Thus, chloroplast transformation to produce dsRNA is more advantageous than nuclear transformation. Charoonnart et al., (2019) transferred dsRNA targeting YHV-RdRp into chloroplasts of C. reinhardtii, and 8 days after YHV infection, the survival of shrimp consuming dsRNA-expressing microalgae increased by 34% compared to that in controls, although the microalgae produced less dsRNA than that with nuclear transformation. Purton et al. (Kiataramgul et al., 2020) used WSSV-vp28 gene to integrate into C. reinhardtii which is a chloroplast of cell wall-deficient, and the C. reinhardtii was fed to shrimp, and the survival rate of shrimp was 87% in the presence of WSSV invasion. Pham et al., (2021) transferred the vp28 protein of WSSV into the nucleus of C. reinhardtii cells, and after feeding, the shrimp developed an immune response against WSSV. These results indicate that C. reinhardtii is a very promising oral substance that could be used to protect aquatic animals from pathogens.
8.2 Oral delivery is very common
Rattanarojpong et al., (2016) transferred dsRNA targeting the rr2 gene into a recombinant baculovirus expressing vp28 and then injected it into shrimp. Shrimp mortality was greatly reduced and WSSV infection was also prevented. Weerachatyanukul et al., (2021) co-packaged genes for silencing vp28 and vp37 into a virus like particle-IHHNV and injected this particle into shrimp, which not only improved survival but also stimulated the immune response.
The most promising method of RNAi delivery in aquatic waters is oral. Pathogens such as microorganisms and viruses can overcome host immunity and cellular barriers that facilitate the delivery of RNAi molecules (Abo-Al-Ela, 2020). Thammasorn et al. (Ongvarrasopone and Panyim, 2007; Thammasorn et al., 2015) used Escherichia coli to express dsRNA targeting the vp28 and WSSV051 structural protein-encoding genes of WSSV and used oral delivery of dsRNA to shrimp, which was able to reduce the risk of WSSV infection and mortality in shrimp, and this method was less costly than traditional transcriptional techniques.
They found that the numbers of surviving inactivated bacteria consumed were higher than those in shrimp that were orally fed nanoparticles, suggesting that the oral delivery of dsRNA-expressing bacteria improves host survival, as illustrated in another study (Leigh et al., 2015).
9 Outlook
Aquaculture is becoming increasingly important and a major support for the world economy. At the same time, there are many pathogens in aquaculture that are constantly attacking aquatic organisms. People began to use antibiotics in large quantities to address this (Monahan et al., 2021), which has had a considerable impact on the environment and human health. With the continuous research on and maturity of RNAi technology, researchers have shifted their attention from antibiotics to RNAi for the control of aquatic aquaculture. This paper highlights the potential dsRNA target genes in aquatic pathogens that can reduce pathogen infection and improve the survival of aquatic host animals after silencing and also introduces species used as feed for oral-based dsRNA, such as E. coli and microalgae. An increasing number of studies has shown that the use of E. coli is a very inexpensive way to synthesize dsRNA, and E. coli is constantly being modified to obtain high yields (Hashiro and Yasueda, 2022).
However, the long-term effects of the regular use of E. coli cells in shrimp feed on animals and the environment have yet to be studied. Therefore, finding alternative dsRNA production and delivery systems that can be used safely in shrimp farms is a key challenge for future applications of this technology. C. reinhardtii is considered a very safe organism (Enzing et al., 2014) that has great appeal as a cell factory. Probiotics are also very promising as oral feed for delivering dsRNA, while enhancing host immunity.
At the same time, to be able to apply RNAi technology in farms, we have to consider the cost and safety. Since dsRNA molecules are constantly degraded, a large amount of dsRNA is needed for the organisms, and ways to increase the yield while controlling the cost is a problem that needs to be solved (Papić et al., 2018; Huang and Lieberman, 2013). The use of RNAi technology for aquatic control needs to be improved and the threat to biodiversity needs to be further explored. The continuous development of RNAi molecule production and delivery methods targeting pathogen genes to reduce off-target effects and enhance host resistance remains a direction to be pursued.
Author contributions
WN and HZ contributed to the conception and design of the study. WN and HZ wrote the draft of the manuscript. XC, YT, and NX revised the draft of the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (Grant No. 31702061).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abo-Al-Ela, H. G. (2020). Are pathogens completely harmful or useless? ACS Chem. Neurosci. 11 (16), 2388–2390. doi:10.1021/acschemneuro.0c00035
Alenton, R. R., Kondo, H., Hirono, I., and Maningas, M. B. (2016). Gene silencing of VP9 gene impairs WSSV infectivity on Macrobrachium rosenbergii. Virus Res. 214, 65–70. doi:10.1016/j.virusres.2016.01.013
Álvarez-Sánchez, A. R., Romo-Quinones, C., Rosas-Quijano, R., Reyes, A. G., Barraza, A., Magallón-Barajas, F., et al. (2018). Production of specific dsRNA against white spot syndrome virus in the yeast Yarrowia lipolytica. Aquac. Res. 49 (1), 480–491. doi:10.1111/are.13479
Andrade, T. P. D., Cruz-Flores, R., Mai, H. N., and Dhar, A. K. (2022). Novel infectious myonecrosis virus (IMNV) variant is associated with recent disease outbreaks in Penaeus vannamei shrimp in Brazil. Aquaculture 554, 738159. doi:10.1016/j.aquaculture.2022.738159
Andrew Fire, S. X., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by doublestranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Bernstein, E., Caudy, A., Hammond, S., Hannon, G., Bernstein, E., Caudy, A. A., et al. (2001). Role for a bidentate ribonuclease in the initiation step of RNA interference. Nature 409, 363–366. doi:10.1038/35053110
Bontemps-Gallo, S., Fernandez, M., Dewitte, A., Raphaël, E., Gherardini, F. C., Elizabeth, P., et al. (2019). Nutrient depletion may trigger the Yersinia pestis OmpR-EnvZ regulatory system to promote flea-borne plague transmission. Mol. Microbiol. 112 (5), 1471–1482. doi:10.1111/mmi.14372
Borchel, A., and Nilsen, F. (2018). A novel gene-family involved in spermatophore generation in the economically important salmon louse Lepeophtheirus salmonis. Mol. Reprod. Dev. 85 (6), 478–489. doi:10.1002/mrd.22984
Boynton, J., Gillham, N., Harris, E., Hosler, J., Johnson, A., Jones, A., et al. (1988). Chloroplast transformation in chlamydomonas with high velocity microprojectiles. Sci. (New York, N.Y.) 240, 1534–1538. doi:10.1126/science.2897716
Braden, L., Michaud, D., Igboeli, O. O., Dondrup, M., Hamre, L., Dalvin, S., et al. (2020). Identification of critical enzymes in the salmon louse chitin synthesis pathway as revealed by RNA interference-mediated abrogation of infectivity. Int. J. Parasitol. 50 (10-11), 873–889. doi:10.1016/j.ijpara.2020.06.007
Brooker, A., Skern-Mauritzen, R., and Bron, J. (2018). Production, mortality, and infectivity of planktonic larval sea lice, Lepeophtheirus salmonis (Kroyer, 1837): Current knowledge and implications for epidemiological modelling. ICES J. Mar. Sci. 75, 1214–1234. doi:10.1093/icesjms/fsy015
Cai, S., Cheng, H., Pang, H., Jian, J., and Wu, Z. (2018). AcfA is an essential regulator for pathogenesis of fish pathogen Vibrio alginolyticus. Vet. Microbiol. 213, 35–41. doi:10.1016/j.vetmic.2017.11.016
Cao, X., Wang, Q., Liu, Q., Liu, H., He, H., and Zhang, Y. (2010). Vibrio alginolyticus MviN is a LuxO-regulated protein and affects cytotoxicity toward EPC cell. J. Microbiol. Biotechnol. 20 (2), 271–280. doi:10.4014/jmb.0904.04031
Cao, X., Wang, Q., Liu, Q., Rui, H., Liu, H., and Zhang, Y. (2011). Identification of a luxO-regulated extracellular protein Pep and its roles in motility in Vibrio alginolyticus. Microb. Pathog. 50 (2), 123–131. doi:10.1016/j.micpath.2010.12.003
Carmichael, S., Bron, J., Taggart, J., Ireland, J., Bekaert, M., Burgess, S., et al. (2013). Salmon lice (Lepeophtheirus salmonis) showing varying emamectin benzoate susceptibilities differ in neuronal acetylcholine receptor and GABA-gated chloride channel mRNA expression. BMC genomics 14, 408. doi:10.1186/1471-2164-14-408
Chaimongkon, D., Assavalapsakul, W., Panyim, S., and Attasart, P. (2020). A multi-target dsRNA for simultaneous inhibition of yellow head virus and white spot syndrome virus in shrimp. J. Biotechnol. 321, 48–56. doi:10.1016/j.jbiotec.2020.06.022
Chang, C., Jing-Jing, Z., Chun-Hua, R., and Chao-Qun, H. (2010). Deletion of valR, a homolog of Vibrio harveyiś luxR generates an intermediate colony phenotype between opaque/rugose and translucent/smooth in Vibrio alginolyticus. Biofouling 26 (5), 595–601. doi:10.1080/08927014.2010.499511
Charoonnart, P., Purton, S., and Saksmerprome, V. (2018). Applications of microalgal Biotechnology for disease control in aquaculture. Biology 7 (2), 24. doi:10.3390/biology7020024
Charoonnart, P., Worakajit, N., Zedler, J. A. Z., Meetam, M., Robinson, C., and Saksmerprome, V. (2019). Generation of microalga Chlamydomonas reinhardtii expressing shrimp antiviral dsRNA without supplementation of antibiotics. Sci. Rep. 9 (1), 3164. doi:10.1038/s41598-019-39539-x
Chen, Y., Wu, F., Pang, H., Tang, J., Cai, S., and Jian, J. (2019). Superoxide dismutase B (sodB), an important virulence factor of Vibrio alginolyticus, contributes to antioxidative stress and its potential application for live attenuated vaccine. Fish. Shellfish Immunol. 89, 354–360. doi:10.1016/j.fsi.2019.03.061
Chendrimada, T. P., Gregory, R. I., Kumaraswamy, E., Norman, J., Cooch, N., Nishikura, K., et al. (2005). TRBP recruits the Dicer complex to Ago2 for microRNA processing and gene silencing. Nature 436 (7051), 740–744. doi:10.1038/nature03868
Crosbie, P., Bridle, A. R., Cadoret, K., and Nowak, B. F. (2012). In vitro cultured Neoparamoeba perurans causes amoebic gill disease in Atlantic salmon and fulfils Koch's postulates. Int. J. Parasitol. 42, 511–515. doi:10.1016/j.ijpara.2012.04.002
Dekham, K., Jitrakorn, S., Charoonnart, P., Isarangkul, D., Chaturongakul, S., and Saksmerprome, V. (2022). Probiotics expressing double-stranded RNA targeting VP28 efficiently protect shrimps from WSSV infection. Aquac. Rep. 23, 101067. doi:10.1016/j.aqrep.2022.101067
Deng, Y., Ding, X., Huang, X., Yang, Y., and Chen, C. (2017). Metabolism of branched-chain amino acids revealed by transcriptome analysis in Vibrio alginolyticus. Mar. Genomics 35, 23–26. doi:10.1016/j.margen.2017.03.006
Deng, Y., Su, Y., Liu, S., Bei, L., Guo, Z., Li, H., et al. (2019). A novel sRNA srvg17985 identified in Vibrio alginolyticus involving into metabolism and stress response. Microbiol. Res. 229, 126295. doi:10.1016/j.micres.2019.126295
Ding, S.-W., and Voinnet, O. (2007). Antiviral immunity directed by small RNAs. Cell 130, 413–426. doi:10.1016/j.cell.2007.07.039
Doron, L., Segal, N., and Shapira, M. (2016). Transgene expression in microalgae-from tools to applications. Front. Plant Sci. 7, 505. doi:10.3389/fpls.2016.00505
Enzing, C., Ploeg, M., Barbosa, M. J., and Sijtsma, L. (2014). Microalgae-based products for food and feed sector: An outlook for europe. Europe: JCR. doi:10.2791/3339
Fayyaz, M., Chew, K. W., Show, P. L., Ling, T. C., Ng, I. S., and Chang, J.-S. (2020). Genetic engineering of microalgae for enhanced biorefinery capabilities. Biotechnol. Adv. 43, 107554. doi:10.1016/j.biotechadv.2020.107554
Gao, X., Liu, Y., Liu, H., Yang, Z., Liu, Q., Zhang, Y., et al. (2017). Identification of the regulon of AphB and its essential roles in LuxR and exotoxin asp expression in the pathogen Vibrio alginolyticus. J. Bacteriol. 199 (20), 002522-e317. doi:10.1128/jb.00252-17
Gao, X., Wang, X., Mao, Q., Xu, R., Zhou, X., Ma, Y., et al. (2018). VqsA, a novel LysR-type transcriptional regulator, coordinates quorum sensing (QS) and is controlled by QS to regulate virulence in the pathogen Vibrio alginolyticus. Appl. Environ. Microbiol. 84 (12), 004444-e518. doi:10.1128/aem.00444-18
Grayfer, L., Hodgkinson, J. W., and Belosevic, M. (2014). Antimicrobial responses of teleost phagocytes and innate immune evasion strategies of intracellular bacteria. Dev. Comp. Immunol. 43 (2), 223–242. doi:10.1016/j.dci.2013.08.003
Gu, D., Liu, H., Yang, Z., Zhang, Y., and Wang, Q. (2016). Chromatin immunoprecipitation sequencing technology reveals global regulatory roles of low-cell-density quorum-sensing regulator AphA in the pathogen Vibrio alginolyticus. J. Bacteriol. 198 (21), 2985–2999. doi:10.1128/jb.00520-16
Gu, D., Zhang, J., Hao, Y., Xu, R., Zhang, Y., Ma, Y., et al. (2019). Alternative sigma factor RpoX is a part of the RpoE regulon and plays distinct roles in stress responses, motility, biofilm formation, and hemolytic activities in the marine pathogen Vibrio alginolyticus. Appl. Environ. Microbiol. 85 (14), 002344-e319. doi:10.1128/aem.00234-19
Guo, L., Huang, L., Su, Y., Qin, Y., Zhao, L., and Yan, Q. (2018). secA, secD, secF, yajC, and yidC contribute to the adhesion regulation of Vibrio alginolyticus. Microbiologyopen 7 (2), e00551. doi:10.1002/mbo3.551
GuptaRai, D., Jangid, A., Kulhari, D., and Kulhari, H. (2019). Nanomaterials-based siRNA delivery: Routes of administration, hurdles and role of nanocarriers. Nanotechnol. Mod. Animal Biotechnol. 31, 67–114. doi:10.1007/978-981-13-6004-6_3
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in posttranscriptional gene silencing in plants. Science 286 (5441), 950–952. doi:10.1126/science.286.5441.950
Hantke, K. (2005). Bacterial zinc uptake and regulators. Curr. Opin. Microbiol. 8 (2), 196–202. doi:10.1016/j.mib.2005.02.001
Hashiro, S., and Yasueda, H. (2022). RNA interference-based pesticides and antiviral agents: Microbial overproduction systems for double-stranded RNA for applications in agriculture and aquaculture. Appl. Sci. 12 (6), 2954. doi:10.3390/app12062954
He, L., Wang, L., Zhao, L., Zhuang, Z., Wang, X., Huang, H., et al. (2021). Integration of RNA-seq and RNAi reveals the contribution of znuA gene to the pathogenicity of Pseudomonas plecoglossicida and to the immune response of Epinephelus coioides. J. Fish. Dis. 44 (11), 1831–1841. doi:10.1111/jfd.13502
He, R., Wang, J., Lin, M., Tian, J., Wu, B., Tan, X., et al. (2022). Effect of ferredoxin receptor FusA on the virulence mechanism of Pseudomonas plecoglossicida. Front. Cell. Infect. Microbiol. 12, 808800. doi:10.3389/fcimb.2022.808800
Hood, M. I., and Skaar, E. P. (2012). Nutritional immunity: Transition metals at the pathogen-host interface. Nat. Rev. Microbiol. 10 (8), 525–537. doi:10.1038/nrmicro2836
Hu, L., Zhao, L., Zhuang, Z., Wang, X., Fu, Q., Huang, H., et al. (2021). The effect of tonB gene on the virulence of Pseudomonas plecoglossicida and the immune response of Epinephelus coioides. Front. Microbiol. 12, 720967. doi:10.3389/fmicb.2021.720967
Huang, L., Guo, L., Xu, X., Qin, Y., Zhao, L., Su, Y., et al. (2019a). The role of rpoS in the regulation of Vibrio alginolyticus virulence and the response to diverse stresses. J. Fish. Dis. 42 (5), 703–712. doi:10.1111/jfd.12972
Huang, L., Hu, J., Su, Y., Qin, Y., Kong, W., Ma, Y., et al. (2015a). Identification and characterization of three Vibrio alginolyticus non-coding RNAs involved in adhesion, chemotaxis, and motility processes. Front. Cell. Infect. Microbiol. 5, 56. doi:10.3389/fcimb.2015.00056
Huang, L., Hu, J., Su, Y., Qin, Y., Kong, W., Zhao, L., et al. (2016a). Genome-wide detection of predicted non-coding RNAs related to the adhesion process in Vibrio alginolyticus using high-throughput sequencing. Front. Microbiol. 7, 619. doi:10.3389/fmicb.2016.00619
Huang, L., Huang, L., Yan, Q., Qin, Y., Ma, Y., Lin, M., et al. (2016b). The TCA pathway is an important player in the regulatory network governing Vibrio alginolyticus adhesion under adversity. Front. Microbiol. 7, 40. doi:10.3389/fmicb.2016.00040
Huang, L., Huang, L., Zhao, L., Qin, Y., Su, Y., and Yan, Q. (2019b). The regulation of oxidative phosphorylation pathway on Vibrio alginolyticus adhesion under adversities. MicrobiologyOpen 8 (8), e00805. doi:10.1002/mbo3.805
Huang, L., and Lieberman, J. (2013). Production of highly potent recombinant siRNAs in Escherichia coli. Nat. Protoc. 8 (12), 2325–2336. doi:10.1038/nprot.2013.149
Huang, L., Qin, Y., Yan, Q., Lin, G., Huang, L., Huang, B., et al. (2015b). MinD plays an important role in Aeromonas hydrophila adherence to Anguilla japonica mucus. Gene 565 (2), 275–281. doi:10.1016/j.gene.2015.04.031
Huang, L., Wang, L., Lin, X., Su, Y., Qin, Y., Kong, W., et al. (2017). mcp, aer, cheB, and cheV contribute to the regulation of Vibrio alginolyticus (ND-01) adhesion under gradients of environmental factors. Microbiologyopen 6 (6), e00517. doi:10.1002/mbo3.517
Huang, L., Xu, W., Su, Y., Zhao, L., and Yan, Q. (2018). Regulatory role of the RstB-RstA system in adhesion, biofilm production, motility, and hemolysis. Microbiologyopen 7 (5), e00599. doi:10.1002/mbo3.599
Huang, L., Zuo, Y., Qin, Y., Zhao, L., Lin, M., and Yan, Q. (2021). The zinc nutritional immunity of Epinephelus coioides contributes to the importance of znuC during Pseudomonas plecoglossicida infection. Front. Immunol. 12, 678699. doi:10.3389/fimmu.2021.678699
Ilari, A., Pescatori, L., Di Santo, R., Battistoni, A., Ammendola, S., Falconi, M., et al. (2016). Salmonella enterica serovar Typhimurium growth is inhibited by the concomitant binding of Zn(II) and a pyrrolyl-hydroxamate to ZnuA, the soluble component of the ZnuABC transporter. Biochimica Biophysica Acta (BBA) - General Subj. 1860 (3), 534–541. doi:10.1016/j.bbagen.2015.12.006
Jiang, Q., Chen, W., Qin, Y., Huang, L., Xu, X., Zhao, L., et al. (2017). AcuC, a histone deacetylase, contributes to the pathogenicity of Aeromonas hydrophila. MicrobiologyOpen 6 (4), e00468. doi:10.1002/mbo3.468
Jiao, J., Zhao, L., Huang, L., Qin, Y., Su, Y., Zheng, W., et al. (2021). The contributions of fliG gene to the pathogenicity of Pseudomonas plecoglossicida and pathogen-host interactions with Epinephelus coioides. Fish. Shellfish Immunol. 119, 238–248. doi:10.1016/j.fsi.2021.09.032
Jones, J. G. (1986). Antibiotic resistance in aquatic bacteria. J. Antimicrob. Chemother. 18, 149–154. doi:10.1093/jac/18.supplement_c.149
Katoch, R., Sethi, A., Thakur, N., and Murdock, L. L. (2013). RNAi for insect control: Current perspective and future challenges. Appl. Biochem. Biotechnol. 171 (4), 847–873. doi:10.1007/s12010-013-0399-4
Keene, K., Foy, B., Sanchez-Vargas, I., Beaty, B., Blair, C., and Olson, K. (2005). RNA interference acts as a natural antiviral response to O'nyong-nyong virus (Alphavirus; Togaviridae) infection of Anopheles gambiae. Proc. Natl. Acad. Sci. U. S. A. 101, 17240–17245. doi:10.1073/pnas.0406983101
Kehl-Fie, T. E., and Skaar, E. P. (2010). Nutritional immunity beyond iron: A role for manganese and zinc. Curr. Opin. Chem. Biol. 14 (2), 218–224. doi:10.1016/j.cbpa.2009.11.008
Kiataramgul, A., Maeenin, S., Purton, S., Hirono, I., Brocklehurst, T., Unajak, S., et al. (2020). An oral delivery system for controlling white spot syndrome virus infection in shrimp using transgenic microalgae. Aquaculture 521, 735022. doi:10.1016/j.aquaculture.2020.735022
Komisarczuk, A. Z., Kongshaug, H., and Nilsen, F. (2018). Gene silencing reveals multiple functions of Na(+)/K(+)-ATPase in the salmon louse (Lepeophtheirus salmonis). Exp. Parasitol. 185, 79–91. doi:10.1016/j.exppara.2018.01.005
Kumar, S., Verma, A. K., Singh, S. P., and Awasthi, A. (2022). Immunostimulants for shrimp aquaculture: Paving pathway towards shrimp sustainability. Environ. Sci. Pollut. Res. Int.1–19. doi:10.1007/s11356-021-18433-y
Leigh, O., Kelly, C., Praveen, R., and Indrani, K. (2015). Diet-delivery of therapeutic RNA interference in live Escherichia coli against pre-existing Penaeus merguiensis hepandensovirus in Penaeus merguiensis. Aquaculture 437, 360–365. doi:10.1016/j.aquaculture.2014.12.014
Liao, X., He, J., and Li, C. (2022). Decapod iridescent virus 1: An emerging viral pathogen in aquaculture. Rev. Aquac. 14, 1779–1789. doi:10.1111/raq.12672
Lima, P. C., Botwright, N. A., Harris, J. O., and Cook, M. (2014). Development of an in vitro model system for studying bacterially expressed dsRNA-mediated knockdown in Neoparamoeba genus. Mar. Biotechnol. (NY). 16 (4), 447–455. doi:10.1007/s10126-014-9561-4
Lin, G., Chen, W., Su, Y., Qin, Y., Huang, L., and Yan, Q. (2017). Ribose operon repressor (RbsR) contributes to the adhesion of Aeromonas hydrophila to Anguilla japonica mucus. Microbiologyopen 6 (4), e00451. doi:10.1002/mbo3.451
Lingel, A., and Izaurralde, E. (2004). RNAi: Finding the elusive endonuclease. RNA 10 (11), 1675–1679. doi:10.1261/rna.7175704
Liu, H., Gu, D., Cao, X., Liu, Q., Wang, Q., and Zhang, Y. (2012). Characterization of a new quorum sensing regulator luxT and its roles in the extracellular protease production, motility, and virulence in fish pathogen Vibrio alginolyticus. Arch. Microbiol. 194 (6), 439–452. doi:10.1007/s00203-011-0774-x
Liu, H., Liu, W., He, X., Chen, X., Yang, J., Wang, Y., et al. (2020a). Characterization of a cell density-dependent sRNA, Qrr, and its roles in the regulation of the quorum sensing and metabolism in Vibrio alginolyticus. Appl. Microbiol. Biotechnol. 104 (4), 1707–1720. doi:10.1007/s00253-019-10278-3
Liu, H., Wang, Q., Liu, Q., Cao, X., Shi, C., and Zhang, Y. (2011). Roles of Hfq in the stress adaptation and virulence in fish pathogen Vibrio alginolyticus and its potential application as a target for live attenuated vaccine. Appl. Microbiol. Biotechnol. 91 (2), 353–364. doi:10.1007/s00253-011-3286-3
Liu, W., Huang, L., Su, Y., Qin, Y., Zhao, L., and Yan, Q. (2017). Contributions of the oligopeptide permeases in multistep of Vibrio alginolyticus pathogenesis. MicrobiologyOpen 6 (5), e00511. doi:10.1002/mbo3.511
Liu, X., Cooper, A., Yu, Z., Silver, K., Zhang, J., and Zhu, K. Y. (2019). Progress and prospects of arthropod chitin pathways and structures as targets for pest management. Pesticide Biochem. Physiology 161, 33–46. doi:10.1016/j.pestbp.2019.08.002
Liu, Z., Zhao, L., Huang, L., Qin, Y., Zhang, J., Zhang, J., et al. (2020b). Integration of RNA-seq and RNAi provides a novel insight into the immune responses of Epinephelus coioides to the impB gene of Pseudomonas plecoglossicida. Fish. Shellfish Immunol. 105, 135–143. doi:10.1016/j.fsi.2020.06.023
Luo, G., Huang, L., Su, Y., Qin, Y., Xu, X., Zhao, L., et al. (2016). flrA, flrB and flrC regulate adhesion by controlling the expression of critical virulence genes in Vibrio alginolyticus. Emerg. Microbes Infect. 5 (8), 1–11. doi:10.1038/emi.2016.82
Luo, G., Zhao, L., Xu, X., Qin, Y., Huang, L., Su, Y., et al. (2019). Integrated dual RNA-seq and dual iTRAQ of infected tissue reveals the functions of a diguanylate cyclase gene of Pseudomonas plecoglossicida in host-pathogen interactions with Epinephelus coioides. Fish. Shellfish Immunol. 95, 481–490. doi:10.1016/j.fsi.2019.11.008
Lv, T., Dai, F., Zhuang, Q., Zhao, X., Shao, Y., Guo, M., et al. (2020). Outer membrane protein OmpU is related to iron balance in Vibrio alginolyticus. Microbiol. Res. 230, 126350. doi:10.1016/j.micres.2019.126350
Ma, L. S., Narberhaus, F., and Lai, E.-M. (2012). IcmF family protein TssM exhibits ATPase activity and energizes type VI secretion. J. Biol. Chem. 287, 15610–15621. doi:10.1074/jbc.M111.301630
Mao, L., Qin, Y., Kang, J., Wu, B., Huang, L., Wang, S., et al. (2020). Role of LuxR-type regulators in fish pathogenic Aeromonas hydrophila. J. Fish. Dis. 43 (2), 215–225. doi:10.1111/jfd.13114
Mejía-Ruíz, C. H., Vega-Peña, S., Alvarez-Ruiz, P., and Escobedo-Bonilla, C. M. (2011). Double-stranded RNA against white spot syndrome virus (WSSV) vp28 or vp26 reduced susceptibility of Litopenaeus vannamei to WSSV, and survivors exhibited decreased susceptibility in subsequent re-infections. J. Invertebr. Pathol. 107 (1), 65–68. doi:10.1016/j.jip.2011.02.002
Monahan, C., Nag, R., Morris, D., and Cummins, E. (2021). Antibiotic residues in the aquatic environment - current perspective and risk considerations. J. Environ. Sci. Health Part A 56 (7), 733–751. doi:10.1080/10934529.2021.1923311
Napoli, C., Lemieux, C., and Jorgensen, R. (1990). Introduction of a chimeric chalcone synthase gene into petunia results in reversible co-suppression of homologous genes in trans. Plant Cell 2 (4), 279–289. doi:10.1105/tpc.2.4.279
Nilsen, P., Karlsen, M., Sritunyalucksana, K., and Thitamadee, S. (2017). White spot syndrome virus VP28 specific double-stranded RNA provides protection through a highly focused siRNA population. Sci. Rep. 7 (1), 1028. doi:10.1038/s41598-017-01181-w
Ongvarrasopone, C., and Panyim, S. (2007). A simple and cost effective method to generate dsRNA for RNAi studies in invertebrates. ScienceAsia 33, 035. doi:10.2306/scienceasia1513-1874.2007.33.035
Pang, H., Qiu, M., Zhao, J., Hoare, R., Monaghan, S. J., Song, D., et al. (2018). Construction of a Vibrio alginolyticus hopPmaJ (hop) mutant and evaluation of its potential as a live attenuated vaccine in orange-spotted grouper (Epinephelus coioides). Fish. Shellfish Immunol. 76, 93–100. doi:10.1016/j.fsi.2018.02.012
Papić, L., Rivas, J., Toledo, S., and Romero, J. (2018). Double-stranded RNA production and the kinetics of recombinant Escherichia coli HT115 in fed-batch culture. Biotechnol. Rep. 20, e00292. doi:10.1016/j.btre.2018.e00292
Pham, L., Nguyen, H., Duong, B., Hòa, N., Thom, L., Tam, L., et al. (2021). Generation of microalga Chlamydomonas reinhardtii expressing VP28 protein as oral vaccine candidate for shrimps against White Spot Syndrome Virus (WSSV) infection. Aquaculture 540, 736737. doi:10.1016/j.aquaculture.2021.736737
Posiri, P., Thongsuksangcharoen, S., Chaysri, N., Panyim, S., and Ongvarrasopone, C. (2019). PmEEA1, the early endosomal protein is employed by YHV for successful infection in Penaeus monodon. Fish. Shellfish Immunol. 95, 449–455. doi:10.1016/j.fsi.2019.10.054
Qi, W., Gao, Q., Tian, J., Wu, B., Lin, M., Qi, S., et al. (2022). Immune responses and inorganic ion transport regulations of Epinephelus coioides in response to L321_RS13075 gene of Pseudomonas plecoglossicida. Fish. Shellfish Immunol. 120, 599–609. doi:10.1016/j.fsi.2021.12.036
Qi, W., Xu, W., Zhao, L., Xu, X., Luo, Z., Huang, L., et al. (2019). Protection against Pseudomonas plecoglossicida in Epinephelus coioides immunized with a cspA1-knock-down live attenuated vaccine. Fish. Shellfish Immunol. 89, 498–504. doi:10.1016/j.fsi.2019.04.029
Qian, D., Chen, Y., Shen, J., and Shen, Z. (1995). Serogroups, virulence and hemolytic activity of Aeromon as hydrophila which caused fish bacterial septicaemia. Wei Sheng Wu Xue Bao 35 (6), 460–464.
Qin, Y., Lin, G., Chen, W., Huang, B., Huang, W., and Yan, Q. (2014). Flagellar motility contributes to the invasion and survival of Aeromonas hydrophila in Anguilla japonica macrophages. Fish. Shellfish Immunol. 39 (2), 273–279. doi:10.1016/j.fsi.2014.05.016
Ramos-Carreño, S., Giffard-Mena, I., Zamudio-Ocadiz, J. N., Nuñez-Rivera, A., Valencia-Yañez, R., Ruiz-Garcia, J., et al. (2021). Antiviral therapy in shrimp through plant virus VLP containing VP28 dsRNA against WSSV. Beilstein J. Org. Chem. 17, 1360–1373. doi:10.3762/bjoc.17.95
Rattanarojpong, T., Khankaew, S., Khunrae, P., Vanichviriyakit, R., and Poomputsa, K. (2016). Recombinant baculovirus mediates dsRNA specific to rr2 delivery and its protective efficacy against WSSV infection. J. Biotechnol. 229, 44–52. doi:10.1016/j.jbiotec.2016.05.007
Ren, C., Hu, C., Jiang, X., Sun, H., Zhao, Z., Chen, C., et al. (2013). Distribution and pathogenic relationship of virulence associated genes among Vibrio alginolyticus from the mariculture systems. Mol. Cell. Probes 27 (3-4), 164–168. doi:10.1016/j.mcp.2013.01.004
Romo, C., Alvarez Sánchez, A., Alvarez-Ruiz, P., Chavez-Sanchez, C., Bogdanchikova, N., Pestryakov, A., et al. (2020). Evaluation of a new Argovit as an antiviral agent included in feed to protect the shrimp Litopenaeus vannamei against White Spot Syndrome Virus infection. PeerJ 8, e8446. doi:10.7717/peerj.8446
Rosales-Mendoza, S., Paz-Maldonado, L. M. T., and Soria-Guerra, R. E. (2012). Chlamydomonas reinhardtii as a viable platform for the production of recombinant proteins: Current status and perspectives. Plant Cell Rep. 31 (3), 479–494. doi:10.1007/s00299-011-1186-8
Rui, H., Liu, Q., Ma, Y., Wang, Q., and Zhang, Y. (2008). Roles of LuxR in regulating extracellular alkaline serine protease A, extracellular polysaccharide and mobility of Vibrio alginolyticus. FEMS Microbiol. Lett. 285 (2), 155–162. doi:10.1111/j.1574-6968.2008.01185.x
Rui, H., Liu, Q., Wang, Q., Ma, Y., Liu, H., Shi, C., et al. (2009). Role of alkaline serine protease, asp, in vibrio alginolyticus virulence and regulation of its expression by luxO-luxR regulatory system. J. Microbiol. Biotechnol. 19 (5), 431–438. doi:10.4014/jmb.0807.404
Sabili, A., Md Yasin, I., Mohd Daud, H., Mariana, N., and Abdelhadi, Y. (2015). Pathogenicity of Aeromonas hydrophila in giant freshwater prawn; Macrobrachium rosenbergii, cultured in East. Iran. J. Fish. Sci. 14, 232–245.
Sajid, M., Moazzam, M., Kato, S., Cho, K., and Tiwari, R. (2020). Overcoming barriers for siRNA therapeutics: From bench to bedside. Pharm. (Basel) 13, 294. doi:10.3390/ph13100294
Saksmerprome, V., Charoonnart, P., Gangnonngiw, W., and Withyachumnarnkul, B. (2009). A novel and inexpensive application of RNAi technology to protect shrimp from viral disease. J. Virol. Methods 162 (1-2), 213–217. doi:10.1016/j.jviromet.2009.08.010
Samir, F., Abd El-Naby, A., and Monier, M. (2017). Antioxidative and immunostimulatory effect of dietary cinnamon nanoparticles on the performance of Nile tilapia, Oreochromis niloticus (L.) and its susceptibility to hypoxia stress and Aeromonas hydrophila infection. Fish Shellfish Immunol. 74, 19–25. doi:10.1016/j.fsi.2017.12.033
Sarathi, M., Simon, M. C., Venkatesan, C., and Hameed, A. S. S. (2008). Oral administration of bacterially expressed VP28dsRNA to protect Penaeus monodon from white spot syndrome virus. Mar. Biotechnol. (NY). 10 (3), 242–249. doi:10.1007/s10126-007-9057-6
Schuster, S., Miesen, P., and van Rij, R. P. (2019). Antiviral RNAi in insects and mammals: Parallels and differences. Viruses 11 (5), 448. doi:10.3390/v11050448
Sellars, M., Rao, M., Arnold, S., Wade, N., and Cowley, J. (2011). Penaeus monodon is protected against Gill-associated virus by muscle injection but not oral delivery of bacterially expressed dsRNAs. Dis. Aquat. Organ. 95, 19–30. doi:10.3354/dao02343
Sha, J., Kozlova, E., and Chopra, A. K. (2002). Role of various enterotoxins in Aeromonas hydrophila-induced gastroenteritis: Generation of enterotoxin gene-deficient mutants and evaluation of their enterotoxic activity. Infect. Immun. 70, 1924–1935. doi:10.1128/IAI.70.4.1924-1935.2002
Shamriz, S., and Ofoghi, H. (2016). Outlook in the application of Chlamydomonas reinhardtii chloroplast as a platform for recombinant protein production. Biotechnol. Genet. Eng. Rev. 32 (1-2), 92–106. doi:10.1080/02648725.2017.1307673
Sinnuengnong, R., Attasart, P., Smith, D. R., Panyim, S., and Assavalapsakul, W. (2018). Administration of co-expressed Penaeus stylirostris densovirus-like particles and dsRNA-YHV-Pro provide protection against yellow head virus in shrimp. J. Biotechnol. 267, 63–70. doi:10.1016/j.jbiotec.2018.01.002
Somchai, P., Jitrakorn, S., Thitamadee, S., Meetam, M., and Saksmerprome, V. (2016). Use of microalgae Chlamydomonas reinhardtii for production of double-stranded RNA against shrimp virus. Aquac. Rep. 3, 178–183. doi:10.1016/j.aqrep.2016.03.003
Subhadra, B., Hurwitz, I., Fieck, A., Rao, D. V. S., Subba Rao, G., and Durvasula, R. (2010). Development of paratransgenic Artemia as a platform for control of infectious diseases in shrimp mariculture. J. Appl. Microbiol. 108 (3), 831–840. doi:10.1111/j.1365-2672.2009.04479.x
Sun, Y., Nie, P., Zhao, L., Huang, L., Qin, Y., Xu, X., et al. (2019). Dual RNA-seq unveils the role of the Pseudomonas plecoglossicida fliA gene in pathogen-host interaction with larimichthys crocea. Microorganisms 7 (10), 443. doi:10.3390/microorganisms7100443
Tang, R., Luo, G., Zhao, L., Huang, L., Qin, Y., Xu, X., et al. (2019a). The effect of a LysR-type transcriptional regulator gene of Pseudomonas plecoglossicida on the immune responses of Epinephelus coioides. Fish. Shellfish Immunol. 89, 420–427. doi:10.1016/j.fsi.2019.03.051
Tang, R., Zhao, L., Xu, X., Huang, L., Qin, Y., Su, Y., et al. (2019b). Dual RNA-Seq uncovers the function of an ABC transporter gene in the host-pathogen interaction between Epinephelus coioides and Pseudomonas plecoglossicida. Fish. Shellfish Immunol. 92, 45–53. doi:10.1016/j.fsi.2019.05.046
Tang, Y., Jiao, J., Zhao, L., Zhuang, Z., Wang, X., Fu, Q., et al. (2022). The contribution of exbB gene to pathogenicity of Pseudomonas plecoglossicida and its interactions with Epinephelus coioides. Fish. Shellfish Immunol. 120, 610–619. doi:10.1016/j.fsi.2021.12.026
Tang, Y., Sun, Y., Zhao, L., Xu, X., Huang, L., Qin, Y., et al. (2019c). Mechanistic insight into the roles of Pseudomonas plecoglossicida clpV gene in host-pathogen interactions with Larimichthys crocea by dual RNA-seq. Fish Shellfish Immunol. 93, 344–353. doi:10.1016/j.fsi.2019.07.066
Thammasorn, T., Jitrakorn, S., Charoonnart, P., Sirimanakul, S., Rattanarojpong, T., Chaturongakul, S., et al. (2017). Probiotic bacteria (Lactobacillus plantarum) expressing specific double-stranded RNA and its potential for controlling shrimp viral and bacterial diseases. Aquac. Int. 25 (5), 1679–1692. doi:10.1007/s10499-017-0144-z
Thammasorn, T., Sangsuriya, P., Meemetta, W., Senapin, S., Jitrakorn, S., Rattanarojpong, T., et al. (2015). Large-scale production and antiviral efficacy of multi-target double-stranded RNA for the prevention of white spot syndrome virus (WSSV) in shrimp. BMC Biotechnol. 15, 110. doi:10.1186/s12896-015-0226-9
Thammasorn, T., Somchai, P., Laosutthipong, C., Jitrakorn, S., Wongtripop, S., Thitamadee, S., et al. (2013). Therapeutic effect of Artemia enriched with Escherichia coli expressing double-stranded RNA in the black tiger shrimp Penaeus monodon. Antivir. Res. 100 (1), 202–206. doi:10.1016/j.antiviral.2013.08.005
Thedcharoen, P., Pewkliang, Y., Kiem, H. K. T., Nuntakarn, L., Taengchaiyaphum, S., Sritunyalucksana, K., et al. (2020a). Effective suppression of yellow head virus replication in Penaeus monodon hemocytes using constitutive expression vector for long-hairpin RNA (lhRNA). J. Invertebr. Pathol. 175, 107442. doi:10.1016/j.jip.2020.107442
Thedcharoen, P., Pewkliang, Y., tran kiem, H., Nuntakarn, L., Taengchaiyaphum, S., Sritunyalucksana, K., et al. (2020b). Effective suppression of yellow head virus replication in Penaeus monodon hemocytes using constitutive expression vector for long-hairpin RNA (lhRNA). J. Invertebr. Pathology 175, 107442. doi:10.1016/j.jip.2020.107442
Tian, Y., Wang, Q., Liu, Q., Ma, Y., Cao, X., Guan, L., et al. (2008). Involvement of LuxS in the regulation of motility and flagella biogenesis in Vibrio alginolyticus. Biosci. Biotechnol. Biochem. 72 (4), 1063–1071. doi:10.1271/bbb.70812
Tirasophon, W., Roshorm, Y., and Panyim, S. (2005). Silencing of yellow head virus replication in penaeid shrimp cells by dsRNA. Biochem. biophysical Res. Commun. 334, 102–107. doi:10.1016/j.bbrc.2005.06.063
Tirasophon, W., Yodmuang, S., Chinnirunvong, W., Plongthongkum, N., and Panyim, S. (2007). Therapeutic inhibition of yellow head virus multiplication in infected shrimps by YHV-protease dsRNA. Antivir. Res. 74 (2), 150–155. doi:10.1016/j.antiviral.2006.11.002
Wang, L., Sun, Y., Zhao, L., Xu, X., Huang, L., Qin, Y., et al. (2019a). Dual RNA-seq uncovers the immune response of Larimichthys crocea to the secY gene of Pseudomonas plecoglossicida from the perspective of host-pathogen interactions. Fish. Shellfish Immunol. 93, 949–957. doi:10.1016/j.fsi.2019.08.040
Wang, L. Y., Liu, Z. X., Zhao, L. M., Huang, L. X., Qin, Y. X., Su, Y. Q., et al. (2020). Dual RNA-seq provides novel insight into the roles of <i>dksA</i> from <i>Pseudomonas plecoglossicida</i> in pathogen-host interactions with large yellow croakers (<i>Larimichthys crocea</i>). Zool. Res. 41 (4), 410–422. doi:10.24272/j.issn.2095-8137.2020.048
Wang, Q., Liu, Q., Cao, X., Yang, M., and Zhang, Y. (2008). Characterization of two TonB systems in marine fish pathogen Vibrio alginolyticus: Their roles in iron utilization and virulence. Arch. Microbiol. 190 (5), 595–603. doi:10.1007/s00203-008-0407-1
Wang, Q., Liu, Q., Ma, Y., Rui, H., and Zhang, Y. (2007). LuxO controls extracellular protease, haemolytic activities and siderophore production in fish pathogen Vibrio alginolyticus. J. Appl. Microbiol. 103 (5), 1525–1534. doi:10.1111/j.1365-2672.2007.03380.x
Wang, S., Yan, Q., Zhang, M., Huang, L., Mao, L., Zhang, M., et al. (2019b). The role and mechanism of icmF in Aeromonas hydrophila survival in fish macrophages. J. Fish. Dis. 42 (6), 895–904. doi:10.1111/jfd.12991
Wang, X.-H., Aliyari, R., Li, W.-X., Li, H.-W., Kim, K., Carthew, R., et al. (2006). RNA interference directs innate immunity against viruses in adult Drosophila. Sci. (New York, N.Y.) 312, 452–454. doi:10.1126/science.1125694
Weerachatyanukul, W., Chotwiwatthanakun, C., and Jariyapong, P. (2021). Dual VP28 and VP37 dsRNA encapsulation in IHHNV virus-like particles enhances shrimp protection against white spot syndrome virus. Fish Shellfish Immunol. 113, 89–95. doi:10.1016/j.fsi.2021.03.024
Wei, C., Luo, K., Wang, M., Li, Y., Miaojun, P., Xie, Y., et al. (2022). Evaluation of potential probiotic properties of a strain of lactobacillus plantarum for shrimp farming: From beneficial functions to safety assessment. Front. Microbiol. 13, 854131. doi:10.3389/fmicb.2022.854131
Xu, J., Han, F., and Zhang, X. (2007). Silencing shrimp white spot syndrome virus (WSSV) genes by siRNA. Antivir. Res. 73 (2), 126–131. doi:10.1016/j.antiviral.2006.08.007
Xu, S., Jiang, Y., Liu, Y., and Zhang, J. (2021). Antibiotic-accelerated cyanobacterial growth and aquatic community succession towards the formation of cyanobacterial bloom in eutrophic lake water. Environ. Pollut. 290, 118057. doi:10.1016/j.envpol.2021.118057
Yang, Z., Wang, X., Xu, W., Zhou, M., Zhang, Y., Ma, Y., et al. (2018). Phosphorylation of PppA at threonine 253 controls T6SS2 expression and bacterial killing capacity in the marine pathogen Vibrio alginolyticus. Microbiol. Res. 209, 70–78. doi:10.1016/j.micres.2018.02.004
Yodmuang, S., Tirasophon, W., Roshorm, Y., Chinnirunvong, W., and Panyim, S. (2006). YHV-protease dsRNA inhibits YHV replication in Penaeus monodon and prevents mortality. Biochem. biophysical Res. Commun. 341, 351–356. doi:10.1016/j.bbrc.2005.12.186
Zamore, P., Tuschl, T., Sharp, P. A., and Bartel, D. (2000). RNAi: Double-stranded RNA directs the ATP-dependent cleavage of mRNA at 21 to 23 nucleotide intervals. Cell 101, 25–33. doi:10.1016/s0092-8674(00)80620-0
Zeng, L.-R., and Xie, J.-P. (2011). Molecular basis underlying LuxR family transcription factors and function diversity and implications for novel antibiotic drug targets. J. Cell. Biochem. 112, 3079–3084. doi:10.1002/jcb.23262
Zhang, B., Luo, G., Zhao, L., Huang, L., Qin, Y., Su, Y., et al. (2018a). Integration of RNAi and RNA-seq uncovers the immune responses of Epinephelus coioides to L321_RS19110 gene of Pseudomonas plecoglossicida. Fish. Shellfish Immunol. 81, 121–129. doi:10.1016/j.fsi.2018.06.051
Zhang, B., Zhuang, Z., Wang, X., Huang, H., Fu, Q., and Yan, Q. (2019a). Dual RNA-Seq reveals the role of a transcriptional regulator gene in pathogen-host interactions between Pseudomonas plecoglossicida and Epinephelus coioides. Fish. Shellfish Immunol. 87, 778–787. doi:10.1016/j.fsi.2019.02.025
Zhang, J., Zhou, S.-M., An, S., Chen, L., and Wang, G. (2013). Visceral granulomas in farmed large yellow croaker, Larimichthys crocea (Richardson), caused by a bacterial pathogen, Pseudomonas plecoglossicida. J. Fish. Dis. 37, 113–121. doi:10.1111/jfd.12075
Zhang, M., Kang, J., Wu, B., Qin, Y., Huang, L., Zhao, L., et al. (2020). Comparative transcriptome and phenotype analysis revealed the role and mechanism of ompR in the virulence of fish pathogenic Aeromonas hydrophila. MicrobiologyOpen 9 (7), e1041. doi:10.1002/mbo3.1041
Zhang, M., Qin, Y., Huang, L., Yan, Q., Mao, L., Xu, X., et al. (2019b). The role of sodA and sodB in Aeromonas hydrophila resisting oxidative damage to survive in fish macrophages and escape for further infection. Fish. Shellfish Immunol. 88, 489–495. doi:10.1016/j.fsi.2019.03.021
Zhang, M., Yan, Q., Mao, L., Wang, S., Huang, L., Xu, X., et al. (2018b). KatG plays an important role in Aeromonas hydrophila survival in fish macrophages and escape for further infection. Gene 672, 156–164. doi:10.1016/j.gene.2018.06.029
Zhang, Y., Tan, H., Yang, S., Huang, Y., Cai, S., Jian, J., et al. (2022). The role of dctP gene in regulating colonization, adhesion and pathogenicity of Vibrio alginolyticus strain HY9901. J. Fish. Dis. 45 (3), 421–434. doi:10.1111/jfd.13571
Zhenyu, X., Shaowen, K., Chaoqun, H., Zhixiong, Z., Shifeng, W., and Yongcan, Z. (2013). First characterization of bacterial pathogen, Vibrio alginolyticus, for Porites andrewsi White syndrome in the South China Sea. PLoS One 8 (9), e75425. doi:10.1371/journal.pone.0075425
Zhou, Z., Pang, H., Ding, Y., Cai, J., Huang, Y., Jian, J., et al. (2013). VscO, a putative T3SS chaperone escort of Vibrio alginolyticus, contributes to virulence in fish and is a target for vaccine development. Fish. Shellfish Immunol. 35 (5), 1523–1531. doi:10.1016/j.fsi.2013.08.017
Keywords: RNAi, aquacultur, target genes, preparation, uptake
Citation: Nie W, Chen X, Tang Y, Xu N and Zhang H (2022) Potential dsRNAs can be delivered to aquatic for defense pathogens. Front. Bioeng. Biotechnol. 10:1066799. doi: 10.3389/fbioe.2022.1066799
Received: 11 October 2022; Accepted: 03 November 2022;
Published: 17 November 2022.
Edited by:
Ruobing Guan, Henan Agricultural University, ChinaReviewed by:
Shaoru Hu, Shanghai Jiao Tong University School of Medicine, Shanghai, ChinaFengqi Li, Guizhou University, China
Copyright © 2022 Nie, Chen, Tang, Xu and Zhang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hao Zhang, zhanghao@nbu.edu.cn
 Wenhao Nie
Wenhao Nie