
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 14 November 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1054943
This article is part of the Research Topicds-RNA-based Pesticides: Production, Development, and Application TechnologyView all 9 articles
RNA interference (RNAi) is powerful biotechnology for studying the in vivo functions of key genes. Based on this property, RNAi can also be used for pest control as an effective alternative to chemical pesticides. The management of phloem-sucking pests is a tricky issue in current agricultural and forestry pest control. RNAi can silence key chemoreceptor genes of phloem-sucking pests; thereby regulating the behavior of these pests can be manipulated. So, it is considered to be a promising new type of ecological pest management strategy. In this study, we identified a candidate taste receptor gene, BtGR11, that controls the taste sensitivity to sucrose in the whitefly Bemisia tabaci, which is a serious invasive phloem-sucking pest worldwide. Functional analyses using the Xenopus oocyte expression system and the two-electrode voltage-clamp system revealed that the oocytes expressing BtGR11 responded to sucrose. Furthermore, we found that silencing BtGR11 by RNAi inhibited the function of sensing sucrose in the whitefly. This study reports a key chemoreceptor gene that can be used for the understanding of the gustatory sensing mechanisms of whitefly to deterrent.
RNA interference (RNAi) is a technology that involves post-transcriptional gene regulation, and it is a powerful tool for understanding various aspects of insect physiology (Saleh et al., 2006). Using RNAi for pest management requires the introduction of dsRNA in vivo to knockdown genetic targets of interest, thereby activating the siRNA pathway. By silencing specific genes, the roles of multiple functional genes in the sensing system have been elucidated (Huvenne and Smagghe, 2010; Zhao et al., 2011; Guan et al., 2021; Zhan et al., 2021). RNAi can also be used for pest control, as an effective alternative to chemical pesticides. For the future, RNAi is a promising ecological pest management strategy for phloem-sucking pests, which is a tricky issue in current agricultural and forestry pest control. Ac.
The interaction between phloem-sucking pests and their host plants involves many aspects (Li et al., 2019b; Jiang et al., 2019; Xia et al., 2021). In insects, the taste system not only has a significant impact on food selection, feeding, mating, and oviposition behavior (Isono and Morita, 2010), but it also has key functions in development and metabolism. Taste receptors play a vital role in insects that confer their ability to detect various chemicals and to use this information to make feeding decisions. Taste sensations from the environment are identified and evaluated by multiple gustatory receptors (GRs) and gustatory receptor neurons that are distributed in different tissues, including the antennae, proboscises, maxillary palps, labial palps, tarsi, wings, and ovipositors (Scott et al., 2001; Amrein and Thorne, 2005; Montell, 2009). Most receptors have four taste neurons and one mechanosensory neuron. In the taste neurons, one is a “sugar” neuron that is sensitive to sugars such as sucrose, glucose, and fructose; one is a “salt” neuron that is sensitive to salt; one is a “bitter” neuron that is sensitive to deterrent compounds such as quinine, chloroquine, caffeine, and strychnine; and one is a “CO2” neuron that is CO2 sensitive (Jones et al., 2007; Isono and Morita, 2010; Poudel et al., 2015; Poudel et al., 2017).
Previous studies of genomes and transcriptome analyses in insects such as Drosophila melanogaster, Nilaparvata lugens, Anopheles gambiae, and Linepithema humile have shown that the number of GR genes in a given species ranges from 16 to 96 (Hill et al., 2002; Robertson and Wanner, 2006; Wanner and Robertson, 2008; Smadja et al., 2009; Scott, 2018). To date, most of the studies related to gustatory receptors have been conducted on the model organism Drosophila melanogaster. In Drosophila melanogaster, GR43a (Miyamoto et al., 2012) and GR64f and GR5a (Jiao et al., 2008) respond specifically to D-fructose and trehalose, respectively, while the combination of GR64f and GR64a responds to multiple sugar components (Chyb et al., 2003; Jiao et al., 2007). In the silkworm, Bombyx mori, some gustatory receptors have the function of sensing fructose and inositol (Sato et al., 2011). In Helicoverpa armigera, several gustatory receptors exhibit sensitivity to galactose, maltose, sucrose, and fructose (Xu et al., 2012; Jiang et al., 2015). In Spodoptera litura, SlitGR8 has a specific response to D-fructose (Liu et al., 2019). In the brown planthopper, Nilaparvata lugens, fructose, galactose, and arabinose are ligands of NlGR11. The fecundity of brown planthoppers was affected after the expression of the NlGR11 gene was inhibited (Chen et al., 2019).
The whitefly Bemisia tabaci (Gennadius) Hemiptera: Aleyrodidae) is one of the main pests in tropical and subtropical regions. In recent years, with global warming and frequent economic and trade exchanges between countries, the whitefly has also spread and invaded on a large scale. It is widely distributed in China, India, and other countries throughout the world, and has become a disaster pest worldwide (Chen et al., 2016). In China, there are currently two main biotypes of B. tabaci, B and Q. The list of host plants of the whitefly is extensive, including more than 600 plant species. Although there are many kinds of hosts, there are differences in the feeding and oviposition preferences for different host plants (De Barro et al., 2011; Tian et al., 2021). The success of the whitefly, in colonization, spreading, and even outbreaks in invasive areas is inseparable from its strong host adaptability (Wang et al., 2017). The excessive usage of pesticides is an important factor restricting the quality and safety of vegetables, such as tomatoes and peppers which is related to human health. Thus, explore new type of control technologies for whitefly is required. RNAi technology is a new, effective and promising pest control technology. At present, the control of the whitefly is heavily dependent on chemical pesticides, and there is a lack of effective green pest management technology to date. Develop RNAi techniques will provide technical support for the prevention and management of this insect. The taste sensing system of the whitefly plays an important role in its identification of and adaptation to different host plants. Understanding the taste mechanism can provide a theoretical basis for revealing the adaptive mechanism of the whitefly to host plants and understanding the invasion and rapid expansion mechanism of this invasive species. It will also provide a new potential target gene for whitefly management. At present, our understanding of the host plant adaptation mechanism of the whitefly mainly focuses on the olfactory system (Li et al., 2019a; Li et al., 2021; Zhan et al., 2021; Tian et al., 2022), so our understanding of the taste recognition of host plants by the whitefly is very lacking.
In the current study, we identified and cloned a candidate gustatory receptor gene, BtGR11, based on whitefly biotype B’s genome and transcriptome data. The ligands of BtGR11 were identified using the Xenopus oocyte expression system and two-electrode voltage-clamp recording. We further verified the function of BtGR11 by RNAi and insect behavioral techniques. The identification and functional study of this gene lay an essential foundation for understanding the whitefly’s mechanism of recognition and adaptation to host plants, and will allow for the development of techniques for the behavioral regulation of this insect.
The B biotype of whitefly (Bemisia tabaci) was reared in a greenhouse of the Beijing Academy of Agriculture and Forestry Sciences (Beijing, China), and identified as biotype B by the mitochondrial cytochrome oxidase I (mtCOI) gene. The whitefly population was raised on Phaseolus vulgaris plants in a cage (60 cm × 60 cm × 60 cm) with a 16 h day/8 h night photoperiod, a room temperature of 26°C ± 1°C, and a relative humidity of 65% ± 5%.
RNA sample from different tissues of the whitefly was extracted using the Trizol Kit (Invitrogen, Dalian, China) according to the manufacturer’s instructions. RNA concentration and quality were detected using NanoDrop 2000 (NanoDrop Products, United States) and agarose gel electrophoresis. The extracted RNA was converted into cDNA using the PrimeScript RT reagent Kit with gDNA Eraser (perfect Real Time) (TaKaRa, Japan). The cDNA was stored at −20°C until the gene cloning and real-time PCR experiments.
Based on the genome and previously published transcriptome sequences (SRA numbers are SRX022878, SRA036954 and SRR835757) of the whitefly biotype B (Chen et al., 2016; Tian et al., 2021), one gustatory receptor gene with a complete open reading frame was identified. PCR primers were designed to clone the coding sequences of the BtGR11 gene based on the above predicted sequence (Table 1). Then, 2× TransStart FastPfu Fly PCR SuperMix (TransGen Biotech, China) was used to amplify the full-length coding sequences of the BtGR11 gene. The PCR product was purified using the MiniBEST DNA Fragment Purification Kit (TaKaRa, China), and the purified product was incorporated into the pEASY-Blunt Cloning vector, and then sequenced by TsingKe (Beijing, China). The open reading frame of BtGR11 was predicted using the online tool of ORF Finder (https://www.ncbi.nlm.nih.gov/orffinder/). Multiple sequence alignment was conducted using BLASTp (https://blast.ncbi.nlm. nih.gov/Blast.cgi) and DNAMAN. The transmembrane domains were predicted using TOPO2 (http://www.sacs.ucsf.edu/cgi-bin/open-topo2.py). The phylogenetic tree was built using MEGA-X (Kumar et al., 2018).
The newly emerged whitefly adults (males and females) were dissected separately into head with antennae, head without antennae, thorax and abdomen for RNA extraction and synthesis of the cDNA. qRT-PCR was preformed using the qPCR Master Mix A6002 (Promega, China) system containing 7.2 μl of ddH2O, 10 μl of 2×GoTaq® qPCR Master Mix (Promega, China), 10 μM of each specific primer, 100 ng cDNA. The qPCR program included 95 °C for 10 min followed by 40 cycles of 95°C for 15 s, 60°C for 1min β-actin was used as endogenous control. The amplification of BtGR11 and β-actin by qPCR was confirmed by 1% agarose gel electrophoresis. For standard curve, cDNA was used as template with five 10-fold different concentrations serial dilutions. The amplified fragments were sequenced to confirm the targeted gene sequences. The primer sequences for BtGR11 qRT-PCR are listed in Table 1. Each qRT-PCR experiment was performed with three biological replicates and three technical replicates.
The full-length sequence of BtGR11 was ligated into a pT7Ts expression vector using the ClonExpress II One Step Cloning Kit (Vazyme Biotech Co., Ltd., China) with the specific primers listed in Table 1. The constructed pT7TS vectors were linearized by restriction enzyme Smal I, and the cRNA of BtGR11 was synthesized using the Ambion mMESSAGE mMACHINE T7 Ultra Kit (Ambion, Austin, TX). Mature healthy Xenopus oocytes were treated in washing buffer (5 mM MgCl2, 2 mM KCl, 96 mM NaCl, and 5 mM HEPES [pH = 7.6]) with 2 mg/ml collagenase I for 1 h at room temperature until almost all of them were separated. After culturing overnight in an 18°C incubator, 50 ng BtGR11 cRNA was microinjected into each oocyte.
The injected oocytes were cultured in an incubation medium (1 × Ringer’s buffer, 5% dialyzed horse serum, 50 mg/ml tetracycline, 100 mg/ml streptomycin, and 550 mg/ml sodium pyruvate) at 18°C for about 3 days. Then, the reaction of the cultured oocytes to the ligands was tested using a two-electrode voltage clamp system (OC-725C oocyte clamp, Warner Instruments, Hamden, CT, United States). A total of eight sugars were tested in the experiment, including D-fructose, D-glucose, D-sucrose, D-galactose, D-arabinose, D-maltose, D-mannose, and D-xylose (Supplementary Table S1). These compounds were purchased from Sigma and had a purity of ≥99.5%. The sugars to be measured were dissolved in water to a concentration of 0.1 M. Then, the solution was diluted to 10−4 mol/L using 1× Ringer solution (2 mM KCl, 96 mM NaCl, 5 mM MgCl2, 5 mM HEPES, and 0.8 mM CaCl2, pH 7.6). The data were recorded and analyzed using Digidata 1440 A and PCLAMP 10.2 software (Axon Instruments Inc., Union City, CA, United States).
RNA interference of the whitefly was carried out according to our previously reported method (Li et al., 2021). The dsRNA primers were designed using the E-RNAi web tool (https://www.dkfz.de/signaling/e-rnai3/) (Table 1). The red circle in Figure 1 represents the region of RNAi. The dsRNAs were amplified using primers in Table 1 from the GR11 plasmid Cloning vector cloned into pEASY-Blunt vector. dsRNA was obtained using the TranscriptAid T7 High Yield Transcription Kit (Thermo Fisher Scientific, DE, United States) according to the manufacturer’s instructions. The synthesized dsRNA of BtGR11 and EGFP (Enhanced Green Fluorescent Protein) was diluted up to 500 ng/μl with a 30% glucose water solution. The double-layer film feeding method was performed to interfere with the gene mRNA expression levels of the newly emerging whitefly using a female-to-male adult ratio of 1:1. The interference lasted for 2 days, and then the reduced expression of the mRNA level was evaluated by qRT-PCR. The primers of the qRT-PCR are listed in Table 1. The successfully disturbed whiteflies were chosen for subsequent behavioral and biological studies.
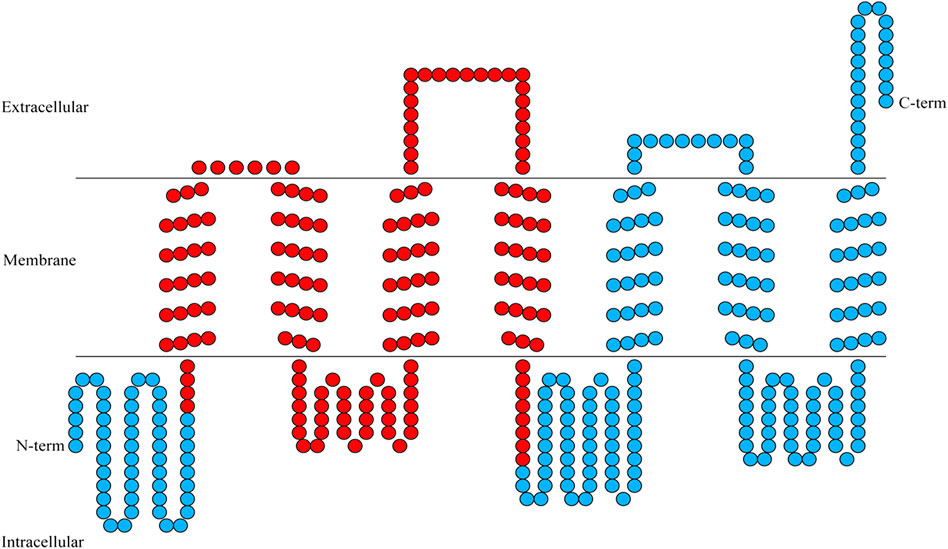
FIGURE 1. Transmembrane helical structures in the amino acid sequence encoding BtGR11. Each circle represents an amino acid, and the red circles represent the region of RNAi.
To test the ability of the whiteflies to detect sucrose, adults were deprived of food for 24 h (only supplied with water). Then, 50–60 adult whiteflies (1:1 female/male ratio) were selected and placed into a glass tube (45 cm length ×3 cm diameter) with a double-layer film at both ends of the tube. The double-layer film at one end contained a sucrose solution colored with red dye (0.2 mg/ml sulforhodamine B, Solarbio,Lot.No.922E021) and the film at the other end contained distilled water colored with blue dye (0.125 mg/ml brilliant blue FCF, Solarbio, Lot.No.917E031). The test was conducted in an appropriate environment conditions of 25 ± 1°C, 75 ± 5% relative humidity and under dark conditions. After 12 h, the abdomen colors of whiteflies were observed using a stereomicroscope. Whiteflies that fed on sucrose had a red color in their abdomens, while whiteflies that fed on distilled water had a green color in their abdomens, and whiteflies that fed on both solutions had a purple color in their abdomens. The preference index (PI) was calculated with the following formula: PI = (N Red +0.5 N Mix)/(N Red + N Blue + N Mix) (Weiss et al., 2011), where N Red is the number of whiteflies with the red color, N Blue is the number of whiteflies with the green color (resulting from the blue dye) and N Mix is the number of whiteflies with the purple color (Figure 5A). The PI is considered to reflect a preference when the value range is between 0.5 and 1, or no preference when the value is below 0.5. The attractiveness was tested using D-fructose with a range of concentrations (0.05 M, 0.10 M, 0.30 M, 0.50 M, 0.80 M, and 1.0 M). The sucrose recognition preference before and after BtGR11 gene silencing was also evaluated by the above method. All experiments were replicated three times.
For the whitefly fecundity experiment, the female adults were transferred to three-week-old common bean plants after 48 h of feeding on dsRNA. Each group of five whiteflies was placed in a clip cage, and the number of eggs in each cage was recorded after 3 days. The plants and whiteflies were maintained in a plant growth chamber under a photoperiod of 16 h light: 8 h dark, at 25°C and 65% relative humidity.
The relative gene expression level was calculated using the relative standard curve method with the help of the QuantStudio 7 Flex Real-Time PCR system (ThermoFisher, United States). The mRNA relative expression levels were analyzed by the 2−ΔΔCT method. and the statistical significance was conducted using the IBM SPSS Statistics 22.0 statistical analysis system.
According to the sequencing results, the ORF of BtGR11 (Genbank ID OP653787) is 1,242 bp (414 amino acids). Transmembrane structure analysis showed that the BtGR11 protein contains seven putative transmembrane domains, located in TM-I = 56–78 amino acids (aa); TM-II = 85–107 aa; TM-III = 142–164 aa; TM-IV = 188–210 aa; TM-V = 271–293 aa; TM-VI = 306–328 aa; and TM-VII = 371–393 aa (Figure 1).
The N-terminus of the BtGR11 protein is located in the cell membrane, and the C-terminus is located outside the cell membrane. The amino acid sequence of BtGR11 was searched for homologous sequences in the NCBI database using BlastP, and the BtGR11 protein was found to be highly similar to the GR sequences of various hemipteran insects. The amino acid sequence similarities between BtGR11 and Rhopalosiphum maidis GR64f-like (XP_026821636.1), Myzus persicae GR64f-like (XP_022180784.1) and Nilaparvata lugens GR 11 (AUD08731.1) were 61.35%, 59.93% and 58.44%, respectively. The results of the protein multiple sequence alignment showed that similar sequences were mostly located in the second half of the protein sequences (Supplementary Figure S1). After building a phylogenetic tree with the amino acid sequences of BtGR11 and the other hemipteran GR genes, BtGR11 was found to cluster into the gustatory receptor GR64f-like subfamily (Figure 2).
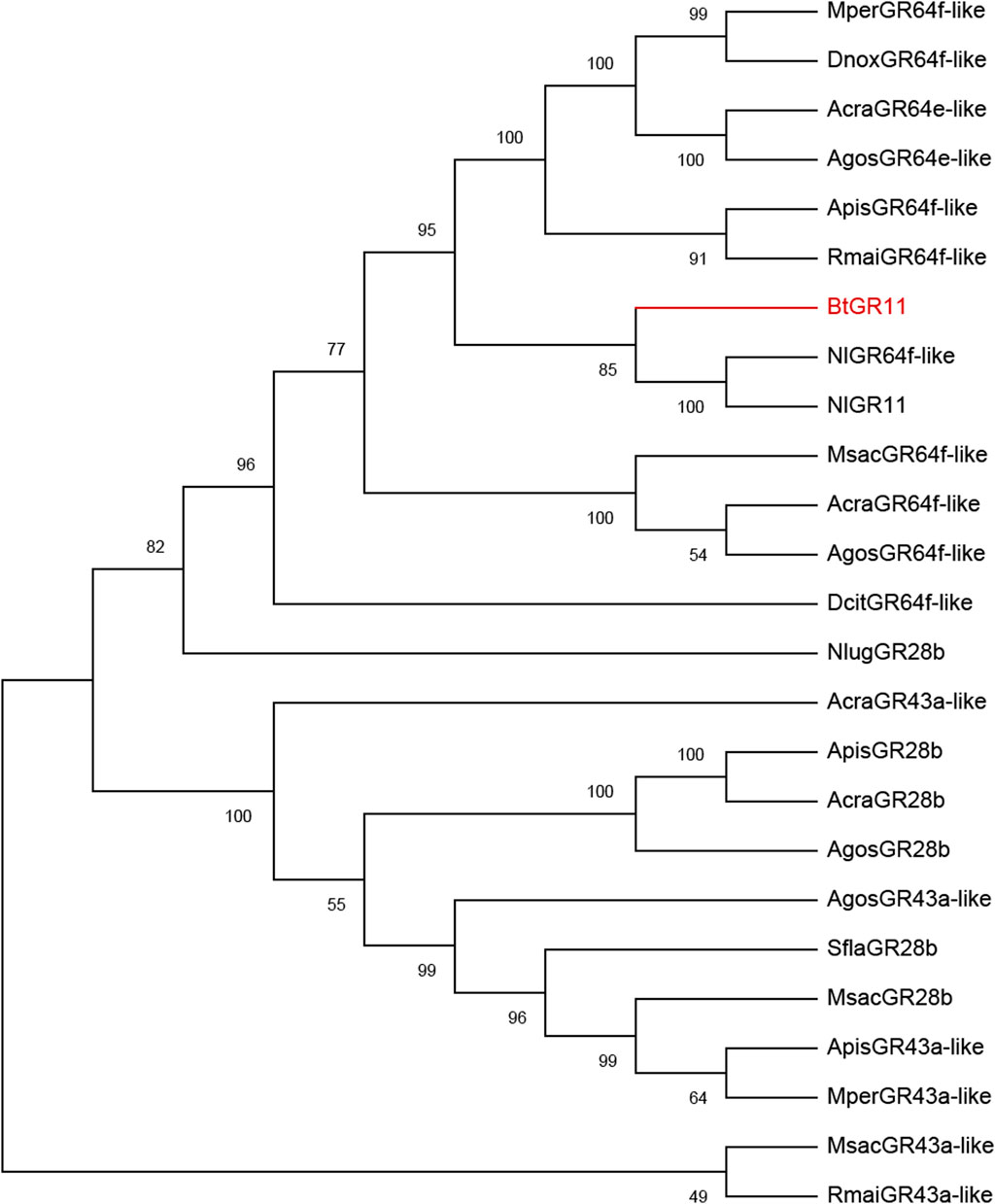
FIGURE 2. Phylogenetic tree of BtGR11 and other hemipteran gustatory receptors based on amino acid sequences. Gene names and GenBank numbers are as follows: ApisGR28b, ApisGR43a-like, ApisGR64f-like: Acyrthosiphon pisum (XP_008178128.1, XP_003244120.4, XP_001942787.2); AcraGR43a-like, AcraGR64e-like, AcraGR64f-like, AcraGR28b: Aphis craccivora (KAF0773920.1, KAF0765871.1, KAF0765661.1, KAF0749339.1); AgosGR28b, AgosGR43a-like, AgosGR64f-like, AgosGR64e-like: Aphis gossypii (XP_027840408.1, XP_027840395.2, XP_027839680.2, XP_027847461.1); DcitGR64f-like: Diaphorina citri (XP_017300309.2); DnoxGR64f-like: Diuraphis noxia (XP_015369418.1); MperGR43a-like, MperGR64f-like: Myzus persicae (XP_022165389.1, XP_022180782.1); MsacGR43a-like, MsacGR64f-like, MsacGR28b: Melanaphis sacchari (XP_025195577.1, XP_025204513.1, XP_025202779.1); NlugGR28b, NlugGR11, NlugGR64f-like: Nilaparvata lugens (XP_039287957.1, AUD08731.1, XP_022186608.2); RmaiGR43a-like, RmaiGR64f-like: Rhopalosiphum maidis (XP_026807230.1, XP_026822156.1); SflaGR28b: Sipha flava (XP_025417386.1)
The mRNA relative expression levels were tested by qRT-PCR using different tissue samples. Standard curve plots with correlation coefficient (R2) values were above 0.99 (Supplementary Figure S2). This indicated the measurement of the relative expression level of the BtGR11 was accurate. The representative pictures of qRT-PCR efficiency checking is shown in Supplementary Figure S2. The qRT-PCR results indicated that the expression of the BtGR11 gene in whitefly female adults was highest in the head with antennae, and relatively high in the head without antennae and thorax. In male adults, the expression of the BtGR11 gene was highest in the head without antennae, with moderately high expression levels mainly in the thorax and head with antennae (Figure 3).
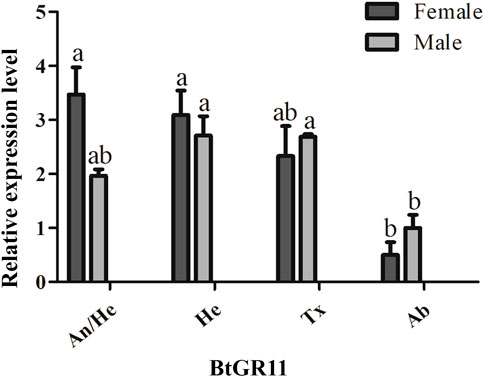
FIGURE 3. Tissue expression profiles of the BtGR11 gene. Female: female adult; Male: male adult; An/He: head with antennae; He: head without antennae; Tx: thorax; Ab: abdomen.
A Xenopus oocyte expression and voltage-clamp recording system was used to test the functionality of BtGR11. Eight sugar ligands were selected for testing. First, BtGR11 was tested using 0.1 M of each ligand to determine its responsiveness, and the results showed that BtGR11 was only responsive to sucrose (Figure 4). Then, a range of sucrose concentrations (0.05–1.1 M) was tested to determine the dose-response relationships. According to the dose-response curve, the sucrose-induced current increased with the concentration from 0.05 M to 1.1 M, and the EC50 value of sucrose was 0.563 M (Figure 4C,D).
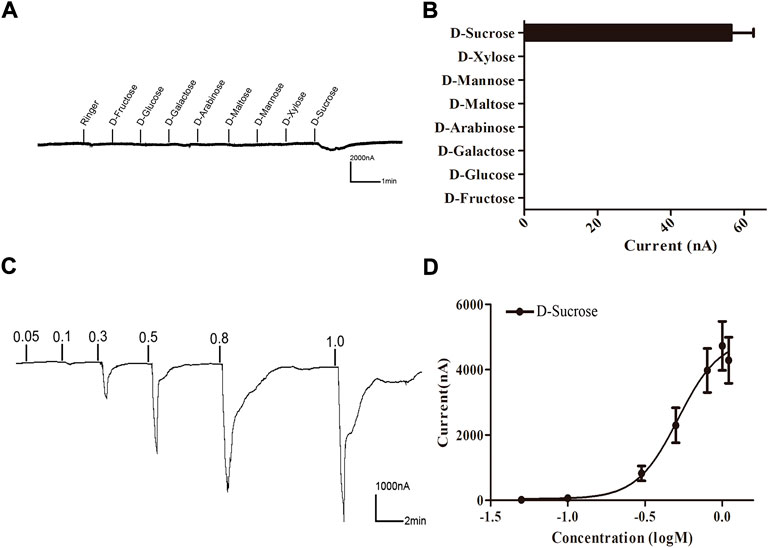
FIGURE 4. Responses of Xenopus oocytes expressing BtGR11 to different sugar ligands. (A) Inward current responses of Xenopus oocytes injected with BtGR11 cRNA to sugar compounds (0.1 M). (B) Response profile of Xenopus oocytes expressing BtGR11. Error bars indicate SEM (n = 4). (C) Response of BtGR11-expressing Xenopus oocytes to D-sucrose at different concentrations. (D) Dose–response curve of BtGR11-expressing Xenopus oocytes to D-sucrose, with EC50 = 0.563 M. Error bars indicate SEM (n = 4).
We conducted a two-way choice test to estimate the preference of whitefly for sucrose. The results indicated that whitefly adults showed a clear preference for sucrose compared with water (Figure 5A). The attraction preference index was increased with increasing sucrose concentrations (Figure 5B). The lowest PI of sucrose was 0.462 at a concentration of 0.05 M, while the highest PI was 0.807 at the 1 M concentration. These data suggest that whitefly adults have a preference for sucrose in a dose-dependent manner.
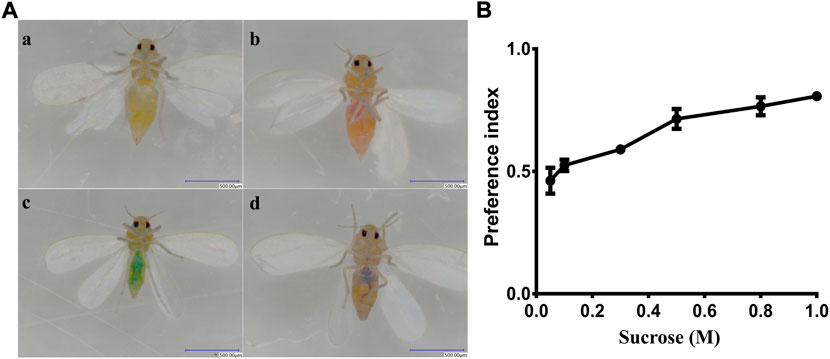
FIGURE 5. Behavioral preference of whitefly to sucrose. (A) (a) whitefly fed nothing; (b) whitefly fed sucrose (with red dye); (c) whitefly fed water (with blue dye); (d) whitefly fed both sucrose and water. (B) PI values for sucrose.
After interfering with the BtGR11 mRNA gene expression, the relative expression of the BtGR11 gene significantly decreased by 47% after 48 h compared with the EGFP control group (student t-test, p < 0.05) (Figure 6A). After interference with dsBtGR11, the PI value decreased by 0.22 compared with the untreated group, and the PI value decreased by 0.18 compared with the EGFP control group. There was no difference in the preference of whitefly for sucrose between the untreated group and the EGFP control group, but there was a significant difference between the experimental group and these two (student t-test, p < 0.05, Figure 6B), indicating that the RNAi of the BtGR11 gene would affect the preference of whitefly adults for sucrose. We further evaluated the effect of BtGR11 interference on whitefly fecundity. After 48 h of interference with dsBtGR11 and dsEGFP, the average numbers of eggs laid by five female adults for 3 days were 149 and 152, respectively. These results showed that the disturbance of BtGR11 expression did not affect the fecundity of whitefly (Supplementary Figure S3).
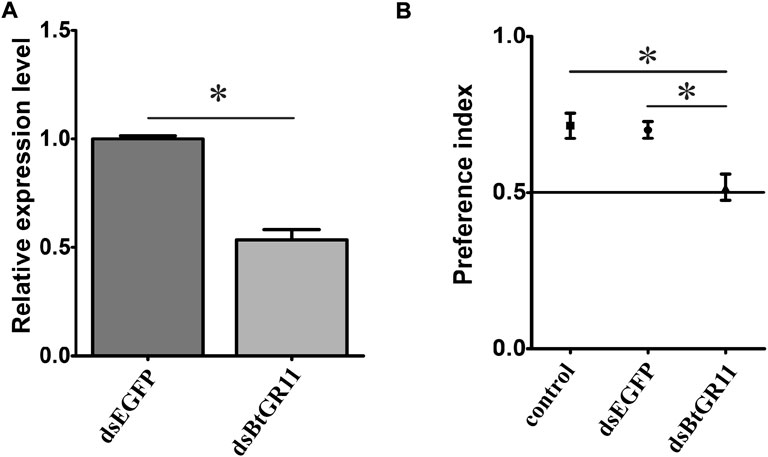
FIGURE 6. Preference index of female whitefly after interference of BtGR11. (A) the mRNA relative expression of the BtGR11 after interfering. (B) Preference index of female whitefly for sucrose, control: untreated group, dsEGFP mean EGFP control group, and dsBtGR11 mean BtGR11 RNAi experimental group. * mean p < 0.05.
The gustatory system plays a crucial role in a variety of insect behaviors, including feeding, mating, and many others (Isono and Morita, 2010). Functional studies of GR genes are important for a better understanding of the mechanisms of gustatory perception, and may also provide potential targets for developing a better strategy for pest control technologies (Weiss et al., 2011). In this study, we cloned a gustatory receptor gene, BtGR11, based on the genome and transcriptome data of whitefly biotype B. BtGR11 has seven transmembrane domains, and the N-terminus of the protein is located in the cell membrane, while the C-terminus is located outside the cell membrane, which is consistent with the previously published structural characteristics of gustatory receptor proteins (Kang et al., 2018).
Through multiple sequence alignment and phylogenetic tree analysis, the BtGR11 gene was found to be in the same clade with the GR64f-like gustatory receptors of other insects, indicating that BtGR11 may belong to the GR64f-like subfamily and have similar functions. A study on GR64f-like in D. melanogaster showed that the DmelGR64f gene is broadly active as a coreceptor for detecting sucrose, maltose, and glucose (Jiao et al., 2008). Currently, there are few studies on the functions of GR64f-like in agricultural pests. One study of the GR64f gene (NlGR11) in N. lugens showed that NlGR11 can recognize fructose, galactose and arabinose, and disturbing NlGR11 affected the fecundity of N. lugens (Chen et al., 2019). The current study found that BtGR11 in whitefly is specifically sensing sucrose. This result is different from the previously reported functions of GR64f-like genes in insects, so the findings in this study expand our understanding of the functions of GR64f-like genes in insects.
Studying the expression of gustatory receptor genes in different tissues of insects is useful for speculating on the possible functions of the receptors (Miyamoto et al., 2012). It has been reported that the GR genes have a wide distribution, and they are distributed in various tissues of insects. Tissue expression profiling showed that the BtGR11 gene was highly expressed in the head and head with antenna. This may be due to the various taste organs, including mouthparts, in the head of whitefly adults. We noticed that BtGR11 was also expressed in the thorax and abdomen. The GR gene is reportedly not only expressed in the taste organs, but also in the antennae, thorax, abdomen, and reproductive organs. For example, the GR gene of Heliconius melpomene is mostly expressed in the labial palps, proboscis, antennae and legs of female and male adults, and the female-specific GR gene is mainly expressed in the legs (Briscoe et al., 2013). In N. lugens, the GR genes are distributed in adult head, leg, midgut, fat body and ovary (Kang et al., 2018). We speculate that whitefly BtGR11 not only acts as a carbohydrate sensor in chemosensory receptors, but is also used in other organs to assess nutrients. In Drosophila adults, GR43a is present in the brain and gut, which is sufficient to assess nutrient carbohydrates and regulate feeding behavior (Miyamoto et al., 2012). In the foregut of the cotton bollworm, HaGR9 acts as a nutrient sensor to guide the digestive process and protect against harmful substances (Xu et al., 2012).
Functional analysis using the Xenopus oocyte expression and voltage-clamp recording system showed that whitefly BtGR11 was specifically responsive to D-sucrose among the eight tested sugar compounds. Sucrose is the most common sugar translocated in plants. Large quantities of sucrose accumulate in the edible parts of some plants, making it the most abundant natural sweetener in food (Salvucci, 2003). A previous study using artificial diets containing radiolabeled carbohydrates showed that the whitefly rapidly hydrolyzes and metabolizes ingested sucrose (Salvucci et al., 1997). In addition to providing nutrients for the whitefly, when the carbon input from sucrose exceeds the metabolic needs, trehalose is synthesized from the excess sucrose for honeydew excretion (Salvucci, 2003). In this study, a series of in vitro and in vivo functional experiments showed that BtGR11 is required for the whitefly’s recognition of sucrose.
Sugar can stimulate feeding behavior in insects (Scott, 2018), most insects rely on sugars for their energy needs, and the insect lifespan and fecundity are limited by the energy ingested and expended (Wäckers, 2001). After the interference of BtGR11, the fecundity of the whitefly did not decrease significantly. There may be other sugar receptors involved in the effects of BtGR11 on fecundity in the whitefly. Although no difference in fecundity was detected after BtGR11 interference, its sensitivity to sucrose was significantly inhibited, which may have various effects on the insect’s lifespan, development and metabolism. These specific effects need to be further investigated in the future. In addition, studies have shown that various sugars of whitefly, including sucrose, have important effects on the detoxification of defensive metabolites in host plants. For example, the whitefly detoxifies the majority of ingested glucosinolates by the stereoselective addition of glucose moieties, which prevents the hydrolytic activation of these defense compounds (Malka et al., 2020). The whitefly has feeding and oviposition preferences for different plants, and even different varieties of the same kind of plant (Li et al., 2016; Li et al., 2021). The whitefly may sense the sucrose content in plants to judge whether they are suitable hosts for feeding and/or oviposition, because sucrose is often the main saccharide in the phloem vasculature, with concentrations ranging from 0.4 to 0.8 M (Rennie and Turgeon, 2009; Malka et al., 2020). Therefore, the identification of BtGR11 will help us to understand the mechanisms of whitefly selection and recognition among different host plants. In the future, by silencing the expression of GR11, the feeding and nutritional supplementation of the whitefly could be inhibited; thereby the host selection, nutrient acquisition and development of this pest can be manipulated, so as to achieve the purpose of controlling the whitefly population.
In conclusion, this study revealed the function of the BtGR11 gene in the whitefly. The full-length cDNA of BtGR11 was cloned. In addition, the tissue expression profiles of BtGR11 in male and female adults indicated that this gene may have specific taste functions in different tissues of both sexes. The specific sensitivity of BtGR11 to D-sucrose was determined using the Xenopus oocyte expression and voltage-clamp recording system. Furthermore, the effect of BtGR11 on sucrose recognition in whitefly was determined by RNAi. These results are crucial for improving the understanding of the role of taste in whitefly host recognition and adaptation, and also provide a potential target gene for the development of new control methods for whitefly.
The sequence of BtGR11 presented in the study is deposited in the NCBI GenBank, accession number OP653787.
CL conceived and designed the experiments. FL, ZD, JT, CQ, SY, and YF conducted the experiments and analyzed the data. FL, ZD, and YD wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by grants from the Funding Program for National Natural Science Foundation of China (Grant No. 31871972 and 31701789).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fbioe.2022.1054943/full#supplementary-material
Amrein, H., and Thorne, N. (2005). Gustatory perception and behavior in Drosophila melanogaster. Curr. Biol. 15, R673–R684. doi:10.1016/j.cub.2005.08.021
Briscoe, A. D., Macias-Muñoz, A., Kozak, K. M., Walters, J. R., Yuan, F., Jamie, G. A., et al. (2013). Female behaviour drives expression and evolution of gustatory receptors in butterflies. PLoS Genet. 9, e1003620. doi:10.1371/journal.pgen.1003620
Chen, W., Hasegawa, D. K., Kaur, N., Kliot, A., Pinheiro, P. V., Luan, J., et al. (2016). The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 14, 110. doi:10.1186/s12915-016-0321-y
Chen, W. W., Kang, K., Yang, P., and Zhang, W. Q. (2019). Identification of a sugar gustatory receptor and its effect on fecundity of the Brown planthopper Nilaparvata lugens. Insect Sci. 26, 441–452. doi:10.1111/1744-7917.12562
Chyb, S., Dahanukar, A., Wickens, A., and Carlson, J. R. (2003). Drosophila Gr5a encodes a taste receptor tuned to trehalose. Proc. Natl. Acad. Sci. U. S. A. 100 (2), 14526–14530. doi:10.1073/pnas.2135339100
De Barro, P. J., Liu, S. S., Boykin, L. M., and Dinsdale, A. B. (2011). Bemisia tabaci: A statement of species status. Annu. Rev. Entomol. 56, 1–19. doi:10.1146/annurev-ento-112408-085504
Guan, R., Chu, D., Han, X., Miao, X., and Li, H. (2021). Advances in the development of microbial double-stranded RNA production systems for application of RNA interference in agricultural pest control. Front. Bioeng. Biotechnol. 9, 753790. doi:10.3389/fbioe.2021.753790
Hill, C. A., Fox, A. N., Pitts, R. J., Kent, L. B., Tan, P. L., Chrystal, M. A., et al. (2002). G protein-coupled receptors in Anopheles gambiae. Science 298, 176–178. doi:10.1126/science.1076196
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiol. 56, 227–235. doi:10.1016/j.jinsphys.2009.10.004
Isono, K., and Morita, H. (2010). Molecular and cellular designs of insect taste receptor system. Front. Cell. Neurosci. 4, 20. doi:10.3389/fncel.2010.00020
Jiao, Y., Moon, S. J., and Montell, C. (2007). A Drosophila gustatory receptor required for the responses to sucrose, glucose, and maltose identified by mRNA tagging. Proc. Natl. Acad. Sci. U. S. A. 104, 14110–14115. doi:10.1073/pnas.0702421104
Jiang, X. J., Ning, C., Guo, H., Jia, Y. Y., Huang, L. Q., Qu, M. J., et al. (2015). A gustatory receptor tuned to D-fructose in antennal sensilla chaetica of Helicoverpa armigera. Insect biochem. Mol. Biol. 60, 39–46. doi:10.1016/j.ibmb.2015.03.002
Jiang, Y., Zhang, C. X., Chen, R., and He, S. Y. (2019). Challenging battles of plants with phloem-feeding insects and prokaryotic pathogens. Proc. Natl. Acad. Sci. U. S. A. 116, 23390–23397. doi:10.1073/pnas.1915396116
Jiao, Y., Moon, S. J., Wang, X., Ren, Q., and Montell, C. (2008). Gr64f is required in combination with other gustatory receptors for sugar detection in Drosophila. Curr. Biol. 18, 1797–1801. doi:10.1016/j.cub.2008.10.009
Jones, W. D., Cayirlioglu, P., Kadow, I. G., and Vosshall, L. B. (2007). Two chemosensory receptors together mediate carbon dioxide detection in Drosophila. Nature 445, 86–90. doi:10.1038/nature05466
Kang, K., Yang, P., Chen, L. E., Pang, R., Yu, L. J., Zhou, W. W., et al. (2018). Identification of putative fecundity-related gustatory receptor genes in the Brown planthopper Nilaparvata lugens. BMC Genomics 19, 970. doi:10.1186/s12864-018-5391-5
Kumar, S., Stecher, G., Li, M., Knyaz, C., and Tamura, K. (2018). Mega X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 35, 1547–1549. doi:10.1093/molbev/msy096
Li, F., Dewer, Y., Li, D., Qu, C., and Luo, C. (2021). Functional and evolutionary characterization of chemosensory proteinCSP2in the whitefly, Bemisia tabaci. Pest Manag. Sci. 77, 378–388. doi:10.1002/ps.6027
Li, F., Li, D., Dewer, Y., Qu, C., Yang, Z., Tian, J., et al. (2019a). Discrimination of oviposition deterrent volatile β-ionone by odorant-binding proteins 1 and 4 in the whitefly Bemisia tabaci. Biomolecules 9, 563. doi:10.3390/biom9100563
Li, F., Wu, C., Gao, M., Jiao, M., Qu, C., Gonzalez-Uriarte, A., et al. (2019b). Transcriptome sequencing, molecular markers, and transcription factor discovery of Platanus acerifolia in the presence of Corythucha ciliata. Sci. Data 6, 128. doi:10.1038/s41597-019-0111-9
Li, J., Zhu, L., Hull, J. J., Liang, S., Daniell, H., Jin, S., et al. (2016). Transcriptome analysis reveals a comprehensive insect resistance response mechanism in cotton to infestation by the phloem feeding insect Bemisia tabaci (whitefly). Plant Biotechnol. J. 14, 1956–1975. doi:10.1111/pbi.12554
Liu, X. L., Yan, Q., Yang, Y. L., Hou, W., Miao, C. L., Peng, Y. C., et al. (2019). A gustatory receptor GR8 tunes specifically to D-fructose in the common cutworm spodoptera litura. Insects 10 (9). doi:10.3390/insects10090272
Malka, O., Easson, M., Paetz, C., Götz, M., Reichelt, M., Stein, B., et al. (2020). Glucosylation prevents plant defense activation in phloem-feeding insects. Nat. Chem. Biol. 16, 1420–1426. doi:10.1038/s41589-020-00658-6
Miyamoto, T., Slone, J., Song, X., and Amrein, H. (2012). A fructose receptor functions as a nutrient sensor in the Drosophila brain. Cell 151, 1113–1125. doi:10.1016/j.cell.2012.10.024
Montell, C. (2009). A taste of the Drosophila gustatory receptors. Curr. Opin. Neurobiol. 19, 345–353. doi:10.1016/j.conb.2009.07.001
Poudel, S., Kim, Y., Gwak, J. S., Jeong, S., and Lee, Y. (2017). Gustatory receptor 22e is essential for sensing chloroquine and strychnine in Drosophila melanogaster. Insect biochem. Mol. Biol. 88, 30–36. doi:10.1016/j.ibmb.2017.07.007
Poudel, S., Kim, Y., Kim, Y. T., and Lee, Y. (2015). Gustatory receptors required for sensing umbelliferone in Drosophila melanogaster. Insect biochem. Mol. Biol. 66, 110–118. doi:10.1016/j.ibmb.2015.10.010
Rennie, E. A., and Turgeon, R. (2009). A comprehensive picture of phloem loading strategies. Proc. Natl. Acad. Sci. U. S. A. 106, 14162–14167. doi:10.1073/pnas.0902279106
Robertson, H. M., and Wanner, K. W. (2006). The chemoreceptor superfamily in the honey bee, Apis mellifera: Expansion of the odorant, but not gustatory, receptor family. Genome Res. 16, 1395–1403. doi:10.1101/gr.5057506
Saleh, M. C., Van Rij, R. P., Hekele, A., Gillis, A., Foley, E., O'farrell, P. H., et al. (2006). The endocytic pathway mediates cell entry of dsRNA to induce RNAi silencing. Nat. Cell Biol. 8, 793–802. doi:10.1038/ncb1439
Salvucci, M. E. (2003). Distinct sucrose isomerases catalyze trehalulose synthesis in whiteflies, Bemisia argentifolii, and Erwinia rhapontici. Comp. Biochem. Physiology Part B Biochem. Mol. Biol. 135, 385–395. doi:10.1016/s1096-4959(03)00092-7
Salvucci, M. E., Wolfe, G. R., and Hendrix, D. L. (1997). Effect of sucrose concentration on carbohydrate metabolism in Bemisia argentifolii: Biochemical mechanism and physiological role for trehalulose synthesis in the silverleaf whitefly. J. Insect Physiology 43, 457–464. doi:10.1016/s0022-1910(96)00124-2
Sato, K., Tanaka, K., and Touhara, K. (2011). Sugar-regulated cation channel formed by an insect gustatory receptor. Proc. Natl. Acad. Sci. U. S. A. 108, 11680–11685. doi:10.1073/pnas.1019622108
Scott, K., Brady, R., Cravchik, A., Morozov, P., Rzhetsky, A., Zuker, C., et al. (2001). A chemosensory gene family encoding candidate gustatory and olfactory receptors in Drosophila. Cell 104, 661–673. doi:10.1016/s0092-8674(01)00263-x
Scott, K. (2018). Gustatory processing in Drosophila melanogaster. Annu. Rev. Entomol. 63, 15–30. doi:10.1146/annurev-ento-020117-043331
Smadja, C., Shi, P., Butlin, R. K., and Robertson, H. M. (2009). Large gene family expansions and adaptive evolution for odorant and gustatory receptors in the pea aphid, Acyrthosiphon pisum. Mol. Biol. Evol. 26, 2073–2086. doi:10.1093/molbev/msp116
Tian, J., Dewer, Y., Hu, H., Li, F., Yang, S., and Luo, C. (2022). Diversity and molecular evolution of odorant receptor in Hemipteran insects. Insects 13, 214. doi:10.3390/insects13020214
Tian, J., Zhan, H., Dewer, Y., Zhang, B., Qu, C., Luo, C., et al. (2021). Whitefly network analysis reveals gene modules involved in host plant selection, development and evolution. Front. Physiol. 12, 656649. doi:10.3389/fphys.2021.656649
Wäckers, F. L. (2001). A comparison of nectar- and honeydew sugars with respect to their utilization by the hymenopteran parasitoid Cotesia glomerata. J. Insect Physiol. 47, 1077–1084. doi:10.1016/s0022-1910(01)00088-9
Wang, X. W., Li, P., and Liu, S. S. (2017). Whitefly interactions with plants. Curr. Opin. Insect Sci. 19, 70–75. doi:10.1016/j.cois.2017.02.001
Wanner, K. W., and Robertson, H. M. (2008). The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol. Biol. 17, 621–629. doi:10.1111/j.1365-2583.2008.00836.x
Weiss, L. A., Dahanukar, A., Kwon, J. Y., Banerjee, D., and Carlson, J. R. (2011). The molecular and cellular basis of bitter taste in Drosophila. Neuron 69, 258–272. doi:10.1016/j.neuron.2011.01.001
Xia, J., Guo, Z., Yang, Z., Han, H., Wang, S., Xu, H., et al. (2021). Whitefly hijacks a plant detoxification gene that neutralizes plant toxins. Cell 184, 3588–1705. e1617. doi:10.1016/j.cell.2021.06.010
Xu, W., Zhang, H. J., and Anderson, A. (2012). A sugar gustatory receptor identified from the foregut of cotton bollworm Helicoverpa armigera. J. Chem. Ecol. 38, 1513–1520. doi:10.1007/s10886-012-0221-8
Zhan, H., Dewer, Y., Zhang, J., Tian, J., Li, D., Qu, C., et al. (2021). Odorant-binding protein 1 plays a crucial role in the olfactory response of Bemisia tabaci to R-curcumene. J. Agric. Food Chem. 69, 12785–12793. doi:10.1021/acs.jafc.1c03825
Keywords: RNAi, phloem-sucking pests, behavioral regulation, sugar receptor, sucrose
Citation: Li F, Di Z, Tian J, Dewer Y, Qu C, Yang S and Luo C (2022) Silencing the gustatory receptor BtGR11 affects the sensing of sucrose in the whitefly Bemisia tabaci. Front. Bioeng. Biotechnol. 10:1054943. doi: 10.3389/fbioe.2022.1054943
Received: 27 September 2022; Accepted: 26 October 2022;
Published: 14 November 2022.
Edited by:
Tong Li, Henan Academy of Agricultural Science, ChinaReviewed by:
Fangzhou Liu, Cornell University, United StatesCopyright © 2022 Li, Di, Tian, Dewer, Qu, Yang and Luo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chen Luo, bHVvY2hlbjEwMTBAMTI2LmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.