
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Bioeng. Biotechnol., 10 October 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.980592
This article is part of the Research Topicds-RNA-based Pesticides: Production, Development, and Application TechnologyView all 9 articles
Current crop pest control strategies rely on insecticidal and fungicidal sprays, plant genetic resistance, transgenes and agricultural practices. However, many insects, plant viruses, and fungi have no current means of control or have developed resistance against traditional pesticides. dsRNA is emerging as a novel sustainable method of plant protection as an alternative to traditional chemical pesticides. The successful commercialisation of dsRNA based biocontrols for effective pest management strategies requires the economical production of large quantities of dsRNA combined with suitable delivery methods to ensure RNAi efficacy against the target pest. A number of methods exist for the production and delivery of dsRNA based biocontrols and here we review alternative methods currently employed and emerging new approaches for their production. Additionally, we highlight potential challenges that will need to be addressed prior to widespread adoption of dsRNA biocontrols as novel sustainable alternatives to traditional chemical pesticides.
Plant pests and pathogens are estimated to reduce crop yields by 20–40% each year, with insect pests alone already consuming anywhere from 5–20% of major grain crops (Deutsch et al., 2018), leading to reduced food security at household, national and global levels (Flood, 2010; Cerda et al., 2017; Douglas, 2018). Food demand is currently at its highest and is set to increase with global population, which is predicted to hit 9 billion within four decades (Andreev et al., 2013). The free movement of populations, combined with the effects of global warming are increasing the range of pest species and crop diseases, generating new challenges for current crop protection strategies (Hilder and Boulter, 1999; Bebber et al., 2013). Historically, a range of alternative crop protection agents have been implemented in the agricultural sector (Dubey et al., 2011). However, recently there have been several developments which threaten current global food security. Worldwide, the use of pesticides has been in decline since 2007 - largely due to stricter regulations and public opinion pressures. Whilst providing benefit by protecting crop yields and helping to ensure land use efficiency, issues with pesticide use have arisen from the appearance of resistance to existing products and the growing customer concerns about high-intensity agricultural practices (Hilder and Boulter, 1999; Zhang, 2018). Beyond the use of traditional small molecule pesticides as a “chemical” method of pest control, alternative “biological” methods of pest control have been developed, such as the spray application of toxins derived from Bacillus thuringiensis (Bt) to crops, which has been deployed for more than 60 years (Bravo et al., 2011; Heckel, 2020). However, many important insect pest species are not susceptible to these methods of control, while other previously susceptible species have developed resistance to treatments such as Bt toxins (Gordon and Waterhouse, 2007; Tabashnik et al., 2013; Tabashnik and Carrière, 2017).
There are several major drivers for the development of new classes of pesticides, chief among which is the economic cost of pest damage to agriculture, and particularly the increasing cost due to increasing pesticide resistance (Gould et al., 2018), with pest and pathogen damage resulting in over $100 billion worth of damage annually. Incidences of pesticide resistance have dramatically and relentlessly increased since the 1950s (Whalon et al., 2008), and the spread of the natural range of resistant insects, such as Colorado potato beetle, with climate change threatens to magnify the amount of crop damage done by these insects (Bebber, 2015). Just a one degree Celsius rise in temperatures could increase the total losses of rice, corn and wheat alone by 10–25%, with a two degree Celsius rise resulting in approximately 213 million tons of lost produce (Deutsch et al., 2018).
Current crop pest control strategies rely on insecticidal and fungicidal sprays and plant genetic resistance and/or transgenes. There is a growing demand for innovative, sustainable approaches to crop protection motivated by: an ever increasing population, climate-driven pest range expansion, community and regulatory demands, and pest resistance to traditional agro-chemicals (Savary et al., 2012).
The application of double-stranded RNA (dsRNA) for the sequence specific degradation of targeted mRNA via RNA interference (RNAi) is emerging as an important tool for the development of novel RNA-based sustainable insect management strategies (Katoch et al., 2013; Dalaisón-Fuentes et al., 2022). The advantages and disadvantages of using different RNAi technologies to protect plants from insect pests have been reviewed (Liu et al., 2020). In addition to the regulation of endogenous gene expression, another function of the RNAi pathway in invertebrates and plants is to act as a defence mechanism, providing innate immunity against viruses that produce dsRNA (Paces et al., 2017). It is an endogenous cellular process and is a form of post-transcriptional regulation in which dsRNA directs cleavage of complementary endogenous mRNA (Price and Gatehouse, 2008), resulting in loss of protein production (see Figure 1). RNAi is believed to have evolved as a defence mechanism against viral RNA (Obbard et al., 2009) and to maintain the integrity of genomic DNA (Agrawal et al., 2003).
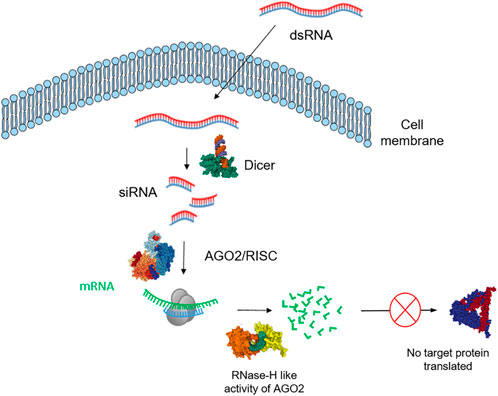
FIGURE 1. Schematic illustration of the exogeneous RNA interference (RNAi) pathway in insects. Dicer-2, cleaves long dsRNAs of either cellular or viral origin into siRNAs that mediate an endogenous or antiviral RNA interference (RNAi) response preferentially sorted to the Argonaute-2 (AGO2) RISC, which mediates sequence-specific target cleavage and degradation.
The 2006 Nobel Prize in Physiology or Medicine was awarded to Andrew Fire and Craig Mello following their pioneering work in 1998, in which this endogenous process was manipulated (Fire et al., 1998). This research monitored phenotypic changes in nematode worms (Caenorhabditis elegans) following the introduction of exogenously produced dsRNA (Fire et al., 1998). This discovery revolutionised functional genomics studies, and further work highlighted the potential for RNAi to play a key role in the agricultural sector for the protection of crops (Baum et al., 2007; Mao et al., 2007). The RNAi mechanism can result in the degradation of target mRNA upon entry of a specific dsRNA into the cell (Zhang, 2018). Therefore, by delivering dsRNA targeting an endogenous mRNA of the intended pest, production of the encoded protein can be reduced at the post-transcriptional level. Thus, through careful selection of an essential target mRNA, this mechanism can lead to insect mortality. The sequence-specific nature of RNAi makes dsRNA an ideal candidate for further application as a species-selective insecticide and it is emerging as an important novel and sustainable insect and fungal management strategy.
Potential uses for triggering RNAi in the agricultural sector include: bio-pesticides (Baum et al., 2007; Mao et al., 2011; Zhang et al., 2017; Meng et al., 2020; Yan et al., 2020), plant virus repression (Viswanath et al., 2000; Wang et al., 2000; Mitter et al., 2017) and prevention of parasites or viral infections of pollinators (Hunter et al., 2010; Paldi et al., 2010; Garbian et al., 2012). There is also scope for using this approach to target insect vectors of human disease, for example mosquitoes or zika flies. A significant advantage of RNA-based bio-controls is the ability to target individual pest species, therefore reducing the use of non-specific pesticides (Zhang et al., 2015).
There are several strategies for the delivery of dsRNA for pest management, all of which have suitable applications and limitations (Figure 2). Firstly, production and delivery of dsRNA can be broadly divided into transformative and non-transformative strategies. Transformative strategies involve the introduction of genes encoding insecticidal dsRNA into plants by genetic engineering, resulting in endogenous production of dsRNA within the plant (Baum et al., 2007). This is typically within plastids (chloroplasts etc.) which lack genes involved in plant post-transcriptional gene silencing (PTGS) such as the dsRNA-specific Dicer-like (DCL) ribonucleases. Therefore, dsRNA is accumulated in plastids within the plant without being degraded by DCL ribonucleases as it would be in the cytoplasm (Cagliari et al., 2019). However, in some commercial dsRNA insecticide products such as MON 87411 maize, the dsRNA expression cassette is chromosomally integrated rather than being within plastids (Naegeli et al., 2018). In the case of MON 87411, this is because it targets corn rootworm, which feed on the roots of the plant which lack chloroplasts. These strategies are also often referred to as plant incorporated protectants (PIPs) (Parker and Sander, 2017).

FIGURE 2. A summary of the delivery methods for endogenous and exogenous dsRNA. Non-transformative strategies which deliver exogenously synthesised dsRNA, include methods where dsRNA is synthesised in vitro, and those where dsRNA is synthesised and/or delivered in vivo in bacteria or fungi. Transformative strategies refer only to methods where the crop plant itself contains a dsRNA transgene, often in chloroplasts or other plastids.
Non-transformative strategies encompass a wider range of delivery methods, though all of these involve the topical application of dsRNA synthesised (exogenously from the target crop plant) either in vitro, or in microorganisms (Wang et al., 2018; Yoon et al., 2018; Hashiro and Yasueda, 2022). dsRNA synthesised in microorganisms can be extracted prior to application, applied within dead microorganisms, or applied within live microorganisms. This latter strategy is referred to as bacterium-mediated or bacterially mediated RNAi, and there are several different sub-strategies encompassed within this term, which will be discussed below. It is worth clarifying that “non-transformative” specifically refers to a strategy where there is no genetic transformation of the crop plant but includes strategies where microorganisms may be genetically transformed to produce dsRNA based biocontrols.
Exogenous (i.e. non-transformative) strategies require the dsRNA to be applied to the crop, and therefore utilise a number of delivery methods. For example, similar to the current application methods used for chemical pesticides, the purified dsRNA or material containing the dsRNA can simply be sprayed directly on the crop (Tian et al., 2009; Huvenne and Smagghe, 2010; Zhu et al., 2011; Petrov and Galabov, 2012; San Miguel and Scott, 2016; Gogoi et al., 2017; Mamta and Rajam, 2017; Davis-Vogel et al., 2018; Niehl et al., 2018) (see Figure 2). The dsRNA can be applied by direct spraying onto the target crop plant, and these methods are collectively referred to as spray-induced gene silencing (SIGS) strategies (Koch et al., 2016), as opposed to methods involving transgene insertion into plants, known as host-induced gene silencing (HIGS) (Koch et al., 2019). Furthermore, additional successful methods of exogenous foliar application include application of formulated dsRNA (Christiaens et al., 2018), liposome encapsulated dsRNA (Castellanos et al., 2019), nanoparticle-bound siRNAs (Thairu et al., 2017), and dsRNA loaded onto clay nanosheets (Mitter et al., 2017; Jain et al., 2022). Another approach for introducing exogenous dsRNA based bio-controls is root absorption, which involves integration of the dsRNA into the irrigation systems of crops (see Figure 2) (Hunter et al., 2012; Ghosh et al., 2017). There are also many examples of successful delivery of insecticidal dsRNA in microorganisms, including delivery in yeast (Murphy et al., 2016), and delivery in E. coli (García et al., 2015; Kim et al., 2015).
With the continued development of new innovative dsRNA production methods, alternative delivery methods are now emerging. For example, the recent development of dsRNA produced in yeast allows for live bait stations to be utilised as a potential method of delivery (Duman-Scheel, 2018) and fungal synthesis of dsRNA enables live fungus to be grown on the exterior of crops, protecting them from feeding insects (Hu and Wu, 2016). In addition to these exogenous approaches, endogenous (transformative) approaches involving modifying the crop to endogenously produce insecticidal dsRNA (see Figure 2) (Baum et al., 2007; Mao et al., 2011) continue to be improved. The methods used and challenges associated with the delivery of dsRNA based biocontrols have been reviewed elsewhere (Terenius et al., 2011; Christiaens et al., 2020). For each of these delivery methods, there are several alternative production methods which are the focus of this review.
Non-transformative methods used to produce exogenous dsRNA include in vitro transcription (IVT), microbial expression in bacteria or fungi, and cell-free synthesis. Conversely, transformative methods require the creation of transgenic plants (GM crops). Both methods have their advantages and limitations, with each of the production methods providing specific modes of action. Within this review the mechanism of each dsRNA production system will be described, with a focus on production yields, scalability and the current implementation status of each.
In vitro transcription (IVT) has been routinely used for the production of RNA for a wide range of molecular biology applications, including the synthesis of dsRNA for laboratory studies of RNAi in insects such as Colorado potato beetle (San Miguel and Scott, 2016), Western corn rootworm and Southern corn rootworm (Baum et al., 2007; Hu et al., 2019), Asian corn borer moth (Wang et al., 2011), spider mites (Yoon et al., 2018), brown planthoppers (Wang et al., 2018), and pea aphids (Yang et al., 2020), among many others. IVT involves the enzymatic synthesis of RNA from a DNA template in a single reaction. Most IVT reactions employ the bacteriophage T7 DNA dependant RNA polymerase (DdRp) (Wang et al., 2018; Yoon et al., 2018; Hu et al., 2019; Dumas et al., 2020; Yan et al., 2020; Yang et al., 2020). Alternative DdRp used include T3 or SP6 (Rajagopal et al., 2002; Alder et al., 2003; Skelly et al., 2003). The RNA polymerase promoter sequence required differs between T3, T7 and SP6 DdRps, as does the optimum number and arrangement of transcriptional promoters and/or terminators (see Figure 3). Alternative in vitro systems have also been utilised including a dual polymerase system using both T7 DdRp and Phi6 (ϕ6) RNA dependant RNA polymerase (RdRp) (see Figure 3C) (Petrov and Galabov, 2012; Levanova and Poranen, 2018; Niehl et al., 2018).
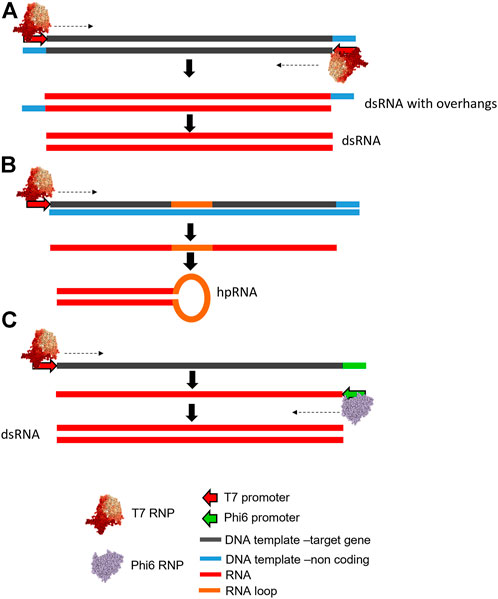
FIGURE 3. Schematic illustration of alternative DNA template designs for in vitro transcription of dsRNA. (A) A DNA template containing convergent T7 DdRp promoters flanking the target sequence is used to generate dsRNA in a single reaction with resulting 3′ overhangs that can be removed using RNase enzymes. (B) A DNA template using a single T7 DdRp promoter, target sequence and a sequence corresponding to an RNA loop is used to generate a long hairpin RNA (lhpRNA). (C) A DNA template containing a single T7 DdRp promoter site, and a ϕ6 promoter region which flank the dsRNA target sequence is used to synthesise the sense ssRNA containing the ϕ6 promoter. ϕ6 RdRP subsequently synthesises the antisense strand resulting in the production of dsRNA with blunt ends.
For the synthesis of dsRNA, run-off transcription is typically employed. It has previously been demonstrated that products of in vitro transcription by T7 DdRp may have up to three additional 5′ G residues, and 3′ overhangs of up to 12 additional nucleotides at the 3′ end (n + 12) (Gholamalipour et al., 2018). The same study also proposed a mechanism by which T7 polymerase switches from the DNA to bind to the end of the RNA product and continues synthesis of further RNA using the initial RNA product as a template, in a process termed primer extension. While deliberate utilisation of this mechanism in the dual polymerase T7-ϕ6 system produces effective insecticidal dsRNA, random primer extension results in double-stranded hairpin regions at the end of single stranded transcripts, which will subsequently affect the binding of complimentary ssRNA transcripts into dsRNA, and how key RNAi pathway enzymes such as Dicer effectively process these dsRNA substrates. IVT reactions can also produce by-products comprised of short products formed by inefficient transcription, often referred to as “shortmers.” Additionally long contaminants or “longmers” may arise, either from promoter-less transcription, extension by priming using RNA-dependant RNA synthesis (Gholamalipour et al., 2018, 2019), or as a result of the formation of multimers or aggregates of dsRNA (Nwokeoji et al., 2019). However, short single-stranded overhangs are easily removed by incubation with a ssRNA-specific ribonucleases such as RNase T1, in order to produce blunt end fragments. This ensures the purity of the product is high and minimal purification steps are required to remove excess NTPs, template DNA and protein contaminants (Losick, 1972; Thermo Fisher Scientific, 2009). This strategy is well characterised, efficient, and robust, making it an ideal method for producing insecticidal dsRNA.
Synthesis of dsRNA by IVT requires a DNA template of the target gene region conforming to one of the designs illustrated in Figure 3. The most widely exploited design (see Figure 3A) consists of the target sequence for the target gene, flanked by two convergent (i.e. facing each other in opposite directions) 5′ RNA polymerase promoter sites (Wang et al., 2018; Yoon et al., 2018; Dumas et al., 2020; Yan et al., 2020). This DNA template transcribes both sense and antisense strands which anneal rapidly within the same reaction vessel to produce dsRNA (Hu et al., 2016). However, sense and anti-sense strand synthesis is not always equal and therefore, dsRNA yield is governed by the least transcribed strand. Inefficient transcription of one strand will produce an excess of one of the component ssRNAs, and subsequently cause inefficient production of dsRNA. Additionally, promoter-less transcription, in which transcribed RNA folds back on itself to prime its own RNA-templated extension (Gholamalipour et al., 2019), creates aberrant transcripts, also reducing the efficiency of dsRNA production.
To circumvent this issue, two separate DNA templates may be generated, each of which contain a single 5′ RNA promoter region upstream of either the sense or anti-sense sequence. Sense and anti-sense ssRNAs are synthesised separately, and subsequently combined in equimolar amounts to generate dsRNA. The separate synthesis eliminates the issue of unequal synthesis of the corresponding ssRNAs. This method has been used to ensure production of high-quality dsRNA for downstream RNAi applications in insects and flatworms, and to produce separate sense and antisense strands individually with both SP6 and T7 promoter systems (Rajagopal et al., 2002; Skelly et al., 2003).
An alternative approach for synthesising dsRNA biocontrols using IVT requires only a single RNA polymerase promoter region at the 5′ end of the DNA template. The DNA template has both sense and antisense target sequences on the same strand separated by a short spacer sequence which forms a hairpin loop post-transcription, thus generating long hairpin RNA (lhpRNA) (see Figure 3B). This DNA template is rarely employed for in vitro synthesis of long dsRNA and is utilised only when hairpin templates are cloned from pre-synthesised plasmid DNA (Alder et al., 2003; Urquhart et al., 2015).
In addition to the IVT methods described, alternative hybrid approaches using two different RNA polymerases have been used. For example, a system has been developed in which the DNA template contains a single T7 RNA polymerase promoter site, and a ϕ6 RNA polymerase promoter (AAAAAAAAGG), which flank the dsRNA target sequence. Following the synthesis of the sense ssRNA by T7 DdRp, the ϕ6 RdRp is subsequently able to recognise the RNA polymerase promoter sequence and synthesises an entire complimentary antisense ssRNA strand in situ, to form the dsRNA (Niehl et al., 2018) (see Figure 3C). The advantage of using the hybrid system over other methods is the high purity of the dsRNA product, which has true blunt ends and lacks the potential overhangs which are produced by other polymerases such as T7 (Makeyev and Bamford, 2000) or other template configurations, as previously described. Notably, convergent promoters (Figure 3A) are the most widely employed templates for IVT synthesis, and have been widely adopted for screening for potential RNAi targets (Wang et al., 2018; Yoon et al., 2018; Hu et al., 2019). This is largely due to the simplicity of construct design and synthesis, which can be achieved by generation of a DNA template using PCR reactions with sequence specific primers typically conjugated to a 5′ T7 RNA polymerase promoter sequence (5′-TAATACGACTCACTATAG-[Target primer]-3′), which following purification can be used to produce dsRNA of the target (Wang et al., 2018; Hu et al., 2019; Dumas et al., 2020; Yan et al., 2020; Yang et al., 2020). This relatively short method produces high purity dsRNA, enabling the rapid analysis of large numbers of potential new targets. Current commercial high yield IVT reactions are widely available and typically generate approximately 50–100 µg of RNA per reaction. Large-scale IVT is limited but commercially available (http://genolution.co.kr/agrorna/service-overview/).
There are several advantages of in vitro transcription over other scalable systems, including the relatively simple purification of the dsRNA product when compared to in vivo production systems. Agricultural products do not necessarily require the high standard of purity required for oligonucleotide/siRNA therapeutics, although some delivery methods may be inhibited by impurities. Many studies have shown the short life span of naked dsRNA in the environment, with improvements in RNAi activity achieved when the dsRNA is encapsulated and therefore protected from environmental nucleases (Aalto et al., 2007; Lundgren and Duan, 2013; Roberts et al., 2015; Ghosh et al., 2017; Leeuwen et al., 2019; Bachman et al., 2020). In laboratory studies, dsRNA produced enzymatically has been proven effective against numerous target species through either injection and/or ingestion, including relevant crop pest insects such as Colorado potato beetle (San Miguel and Scott, 2016; Dumas et al., 2020), Western corn rootworm (Baum et al., 2007; Hu et al., 2019), soybean aphid (Yan et al., 2020), spider mite (Yoon et al., 2018), brown plant hopper (Wang et al., 2018), pea aphid (Yang et al., 2020) and grain aphid (Liu et al., 2021). In addition, larger scale field trials of dsRNA produced enzymatically have also demonstrated RNAi efficacy (Vogel et al., 2019).
Cell-free systems for in vitro protein expression (also referred to as in vitro translation, or cell-free protein expression) have been widely employed to rapidly express and manufacture small amounts of functional proteins (Kigawa et al., 1999; Shimizu et al., 2001; Zimmerman et al., 2014). Cell-free coupled transcription–translation systems utilise cell lysates to transcribe mRNA coupled to protein translation in vitro. The cell-free extracts are optimised to contain most of the cellular cytoplasmic components required for transcription and translation. The first known cell-free extracts capable of supporting translation were made from E. coli prior to the development of eukaryotic in vitro translational systems including lysates prepared from insect embryos (Haley et al., 2003).
Cell free synthesis has several advantages over in vivo production systems. For example, the elimination of the ancillary processes required for cell viability and growth, enables all of the RNA polymerase activity and the entire pool of ribonucleotides in the reaction mixture to be utilised entirely for transcription of the dsRNA product. The absence of a cell wall generates a system that can be actively monitored with reduced sample preparation (Kanter et al., 2007; Carlson et al., 2012; Garamella et al., 2016; Fujiwara et al., 2017). Cell-free systems for the large-scale production of dsRNA focus on in vitro transcription from DNA templates and have been developed and commercialised by GreenLight Biosciences (https://www.greenlightbiosciences.com/in-the-pipeline-colorado-potato-beetle/). Their GreenWorx system is a cell-free platform technology that offers large scale production of dsRNA at low cost. The dsRNA is generated via enzymatic synthesis, with all the components required for RNA synthesis present in the cell-free system. Cell-free systems can generate hpRNA and dsRNA with a variety of potential different polymerases, using linear DNA templates similar to those used in IVT systems (see Figure 3) or alternatively plasmid DNA templates similar to those used in in vivo systems (Figure 4). Cell free systems are scalable and able to generate high yields, with production costs proposed to be as little as $0.5/G (Maxwell et al., 2018). The system also allows for the rapid change of dsRNA targets, as the DNA template is added in the final stage of the system. Potentially, it is possible to produce multiple dsRNA targets within the same reaction vessel. This would allow for the one-step synthesis of a range of dsRNA biocontrols which consist of dsRNAs targeting multiple targets within the same species for increased efficacy, or target multiple species specifically, to deal with multi-pest infestations.
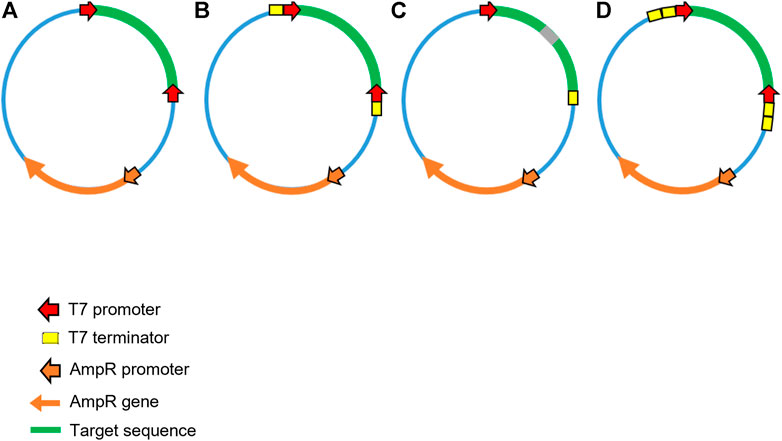
FIGURE 4. Schematic illustration of alternative plasmid DNA template designs for production of dsRNA in microbial cells. A range of alternative plasmids have been utilised for production in microbial cells. (A) Convergent T7 DdRp promoters flanking the target sequence without transcriptional terminators. (B) Convergent T7 DdRp promoters flanking the target with transcriptional terminator sequences outside of the T7 promoters. (C) Single T7 DdRp promoter prior to the target, a sequence corresponding to an RNA loop, the antisense target sequence and finally a T7 terminator sequence used to generate a long hairpin RNA (lhpRNA). (D) Convergent T7 DdRp promoters flanking the target sequence with two transcriptional terminator sequences outside of the T7 RNA polymerase promoters.
Escherichia coli (E. coli) has been the widely adopted “cell factory” for production of recombinant proteins for many years. This well understood organism has high transformation efficiencies and methods of culturing it are relatively simple, inexpensive and scalable, therefore making it an ideal host for the production of biomolecules (Terpe, 2006). In addition, E. coli is amenable to genetic manipulation and has been widely engineered to enhance protein and biochemical production (Sørensen and Mortensen, 2005; Chen et al., 2013). Many of the methods used for recombinant protein production in E. coli have been adopted for the production of dsRNA (Timmons et al., 2001; Nwokeoji et al., 2017). T7 DdRp is one of the most widely utilised expression systems for the production of proteins. It is extremely well characterised, with numerous genetically engineered E. coli strains commercially available to suit the desired recombinant product. This availability reduces optimisation time and cost for small scale laboratory work, and provides a cost effective starting point for many research laboratories to generate larger quantities of dsRNA as an alternative to IVT (Chamberlin et al., 1970; Tunitskaya and Kochetkov, 2002; Terpe, 2006; Tegel et al., 2011).
The production of dsRNA in E. coli has predominantly used the bacterial strain HT115 (DE3) (Hull and Timmons, 2004; Ongvarrasopone et al., 2007; Mao et al., 2011; Posiri et al., 2013; García et al., 2015; Nwokeoji et al., 2016; Ahn et al., 2019; Bento et al., 2020; Meng et al., 2020). The HT115 cell line is of K12 lineage and was generated following the chromosomal deletion of the RNase III gene (rnc) to generate an RNase III deficient strain (Takiff et al., 1989). Further genetic manipulation was performed by lysogenization of the HT115 strain to insert the T7 DdRP within its genome, under the control of an IPTG inducible promoter (Timmons et al., 2001). Several alternative E. coli strains which are RNase III deficient have been developed. In 2009 Yin et al., produced an RNase III deficient strain (M-JM109-YLac) of the common JM109-LacY (DE3) bacteria using Red proteins to knock out the rnc gene (Yin et al., 2009). Production of dsRNA was shown to be increased when compared to HT115 cells containing the same plasmid. It was proposed that the increase in yield was due to the strain harbouring endA1 and recA1 genes which increase exogenous plasmid stability and produce more stable dsRNA respectively (Yin et al., 2009). Recently Ma et al., produced a BL21(DE3) RNase III deficient strain by using a no-SCAR CRISPR system to efficiently knock out rnc. This strain demonstrated an improvement in dsRNA production yield when compared to HT115 cells (Ma et al., 2020).
The workflows adopted for production of dsRNA in E. coli are typical of those used for the production of recombinant proteins. Following the transformation of E. coli with a plasmid harbouring a dsRNA sequence which targets a mRNA of interest under the control of T7 RNA polymerase promoters, cells are then grown to exponential phase and induced for around 4–6 h. Cells are then harvested, lysed and dsRNA subsequently purified. Like IVT, there are a variety of plasmid construct designs which can be utilised to generate varying forms of dsRNA (hpRNA and linear dsRNA), all of which currently utilise T7 DdRp (see Figure 4). Yields from small scale shake-flask production vary but typical reported yields are <20 µg/108 cells (Nwokeoji et al., 2016).
Initial plasmid DNA constructs consisted of two opposing convergent T7 promoters (face to face), flanking the dsRNA sequence (see Figure 4A) as described for in vitro transcription. The L4440 plasmid, which lacks transcriptional terminators, first designed and used by Timmons and Fire in 1998, has been widely employed for microbial expression of dsRNA and was utilised in early RNAi papers for delivery of dsRNA to C. elegans in E. coli (Timmons et al., 2001). Alterations to the initial plasmid design have been adopted. The use of T7 terminators just outside of the T7 promoters suggested that this diminished the effectiveness of initiating RNAi via feeding in C. elegans (Kamath et al., 2000). In contrast, Sturm et al. indicated that the efficiency of RNAi was improved by the incorporation of terminators (Sturm et al., 2018). Alternative plasmid designs have also utilised dual terminators which improve termination efficiency and therefore increase dsRNA yield, and this strategy enabled effective RNAi effects in A. thaliana (Baum et al., 2014). The plasmid constructs described by Chen et al., (Chen et al., 2019) produced hpRNA, using a single T7 promoter, in front of the sense-loop-antisense sequence, subsequently followed by two sequential terminators similar to the arrangement in Figure 4C. These hpRNA-coding plasmid constructs produced twice as much dsRNA compared to a construct expressing dsRNA targeting the same sequence (see Figure 4D). Yields from E. coli HT115 cells transformed with the L4440 plasmid vary dependant on culture method, dsRNA sequence, and length. Fed batch cultures have produced up to 182 mg/L of dsRNA (Papić et al., 2018), making the production cost per mg of dsRNA less expensive when routinely performed, compared to IVT. Other combinations of vector and dsRNA sequence vary in yield when also expressed in the HT115 strain, for example a pET3a-Pro construct yielded 30 mg/L dsRNA (Ongvarrasopone et al., 2007) and a pLitmus28i construct produced 20 mg/L (Ramesh Kumar et al., 2016).
Insecticidal dsRNA expressed in E. coli has been utilised to induce RNAi or mortality as a result of RNAi in various species. This includes purification from E. coli and injection to induce RNAi in Asian lady beetle (Ma et al., 2020), and aquatic feeding of Aedes mosquitoes with E. coli lysate containing insecticidal dsRNA to induce mortality (Lopez et al., 2019). dsRNA expressed in E. coli and then purified prior to application, has also been used to induce RNAi in Arabidopsis plants (Chen et al., 2019).
In addition to E. coli, alternative production systems have been developed. These include a novel bacteriophage-based dsRNA production system, utilising ϕ6 RdRp to synthesise dsRNA from ssRNA templates in Pseudomonas syringae (Aalto et al., 2007). The system involves transfecting P. syringae with three plasmids: one containing a constitutively expressed T7 DdRp; a second expressing the L genes of the bacteriophage ϕ6, which encode for the RdRp and capsid packaging genes; and a final plasmid containing a single stranded RNA sequence downstream of a T7 DdRp promoter, followed by a 3ʹ antisense ϕ6 RdRp promoter region.
This system has similarities to the hybrid T7-ϕ6 IVT system, however, no dsRNA is produced in the cytoplasm of the cell. ssRNA produced by T7 DdRp is transported into capsids produced by the L genes of the ϕ6 bacteriophage on the second plasmid. Housed within the capsid is the ϕ6 RdRp, which recognises the ϕ6 promoter region on the ssRNA, propagating synthesis of the complimentary antisense RNA strand, subsequently forming a dsRNA duplex. This method encapsulates dsRNA within a protective layer against endogenous RNases. dsRNA production in both small batches and in larger scale fermenters was performed, and it was established that the combination of these three plasmids within the host cell produces highly specific dsRNA in quantities of ∼1.6 mg of dsRNA/g of wet cells (Aalto et al., 2007). However, the system was unstable and inefficient in production when numerous RNA sequences were analysed (Niehl et al., 2018). A more stable and efficient system was later developed with the introduction of other ϕ6 genes which helped to stabilise plasmids within the host cells (Niehl et al., 2018). This system increased reproducibility and titre of dsRNA across a wider range of target sequences. Successful viral suppression was also reported when targeting the Tobacco Mosaic Virus (TMV), resulting in propagation suppression when exogenous dsRNA produced by P. syringae was applied topically (Niehl et al., 2018).
The scalability of production, and the ability to protectively encapsulate the dsRNA are key advantages to dsRNA bio-synthesis in P. syringae. However, the dsRNA yields achieved with this system (7 mg/L of cells at a density of 4 × 109 cells/ml) (Niehl et al., 2018) are lower compared to other microbial systems e.g. 182 mg/L was obtained from E. coli fed batch cultures (Papić et al., 2018) In addition, P. syringae cultures require much longer production times, largely due to the requirement for three transfections, and the slower growth rate of this species, which has a doubling time of 1.53 h (Young et al., 1977).
C. glutamicum has recently been studied as a novel model organism for production of insecticidal dsRNA. C. glutamicum is a well characterised microbe capable of recombinantly producing high yields of amino acids such as L-glutamate (Ikeda and Takeno, 2013) and has recently been reported to be capable of producing large quantities of RNA (Hashiro et al., 2019c; 2019b; 2019a). These studies highlight many advantages of the bacterium as a cell factory, including its low-cost culture methods, non-pathogenicity, and it being safe and robust in large scale fermentation. This combination of traits and historical use, make it an ideal candidate for industrial RNAi applications.
An RNase III deficient C. glutamicum strain (2256LΔrnc) was generated via knockout of the rnc gene from strain 2256L (Hashiro et al., 2019a) – which was shown not to be critical to the survival of the organism (Maeda et al., 2016). Recombinant plasmids for production of dsRNA were designed which replicated those seen in E. coli systems. Construct design was synergistic to the L4440 plasmid, with convergent synthetic strong F1 promoters from the corynephage BFK20 replacing the T7 RNA polymerase promoters (Figure 3A). These flanked a 380 bp homologue of the diap1 gene of H. vigintioctopunctata. In a 30-h culture 75 mg/L of target dsRNA was isolated. Heat-sterilised C. glutamicum containing recombinantly produced insecticidal dsRNA were used in H. vigintioctopunctata feeding assays, and ingestion of the dsRNA-containing bacteria by the insects resulted in a significant decrease in leaf consumption and suppression in insect weight gain (Hashiro et al., 2019a).
Recently, higher yields of over 1 g/L of dsRNA targeting the H. vigintioctopunctata gene diap1 were achieved in C. glutamicum using a similar convergent promoter arrangement, though utilising T7 RNA polymerase and the associated promoters, supplied by co-transfection of the pVC7T7pol1 plasmid (Hashiro et al., 2021). Ethanol-sterilised C. glutamicum containing recombinantly produced diap1-targeting dsRNA were again used in H. vigintioctopunctata feeding assays, and once more ingestion of the dsRNA-containing bacteria significantly reduced larval weight gain in a 48-h period. These studies demonstrate great promise for the industrial scale production of RNAi insecticide products in C. glutamicum.
The potential for expression of dsRNA in yeast was demonstrated with the feeding of live genetically modified S. cerevisiae INVSc1 strain to fruit flies (D. suzukii) (Murphy et al., 2016). This study showed the potential of a new and novel application method to be implemented into the agricultural sector–live bait feeding. Previously, bait-fed live yeast has been presented for pest management in berry crops. However, this is the first study to provide a proof of concept for triggering RNAi via fed yeast (Murphy et al., 2016). Plasmids producing hpRNA products targeting mRNAs whose degradation is known to cause lower motility in the target species were constructed. Plasmids were of a relatively simple design and constitutively produced dsRNA, with no induction being necessary. RNAi effect was observed in D. suzukii, but not in other target species. Production of dsRNA in other live fungi–besides yeast–which parasitise insects has also been examined as a combined production/delivery method (see below).
A more recent study by Alvarez-Sanchez, et al., produced dsRNA in the yeast strain Y. lipolytica, targeting white spot syndrome virus of the white legged shrimp Litopenaeus vannamei (Álvarez-Sánchez et al., 2018). dsRNA was isolated and injected into the host species. Yields of dsRNA were <182 ng/L which is low when compared to other in vivo systems, however, yeast has several advantages. Common baker’s yeast (S. cerevisiae) is supplied throughout the world as a dried product therefore, applications in live feeding stations are particularly useful in developing countries with warm climates, where cold transport is required to ensure long-term stability of dsRNA in transit (Murphy et al., 2016) but may not be available. Dried packet yeast is easily transported at a wide range of ambient temperatures, removing the limitation of cold storage availability for application of dsRNA biocontrols in warmer climates.
The fact that yeast lack the RNAi machinery used to process dsRNA is also advantageous (Zhong et al., 2019). However, in contrast to other systems they require little genetic manipulation such as the removal of ribonuclease genes (which can hinder growth rates) (Hofmann and Miller, 1977), therefore, ensuring accumulation of synthesised dsRNA. These factors, and the fact that yeast is biologically safe for human consumption, demonstrate that yeast offers an alternative system for the production and application of dsRNA based biocontrols.
In recent years, there have been a small number of studies which have exploited Bacillus thuringiensis as a host bacterial production system for the expression of dsRNA (Park et al., 2020, 2021; Jiang et al., 2021). Bacillus thuringiensis are soil dwelling bacteria, and are also entomopathogens i.e. pathogenic to insects. In both studies by Park et al. dsRNA was expressed to target sacbrood virus (SBV), a ssRNA virus which threatens Asian Honey Bee populations. Jiang et al. produced dsRNA targeting arginine kinase in Plutella xylostella (diamondback both), a major pest of Brassica crops (Park et al., 2020, 2021). In all cases, dsRNA was expressed using convergent promoters surrounding a target dsRNA. Park et al., used cyt1Aa promoters, whereas Jiang et al. used Pro3α promoters.
The studies by Park et al. involved the extraction of dsRNA from the bacteria after expression in culture. Neither study provides details on the yield of dsRNA so how the efficiency of dsRNA production in Bacillus thuringiensis compares to other microbial systems is unknown. In contrast, Jiang et al. directly applied the live expression culture to the surface of leaves in the lab. It was determined that the intrinsic rate of increase and net reproductive rate of the insects treated with a 9:1 ratio of wildtype bacteria to bacteria expressing dsRNA were lower than those of the population treated with either wildtype bacteria alone or dsRNA-expressing bacteria alone. A similar approach was previously utilised in which Bacillus thuringiensis were delivered alongside an E. coli-expressed dsRNA targeting the immune related Sl102 gene in Spodoptera littoralis (Egyptian cotton leafworm), resulting in enhanced mortality compared to the application of Bacillus thuringiensis alone (Caccia et al., 2020).
The minimal purification and formulation costs associated with this latter method make it an attractive approach for the joint production and delivery of a dsRNA biocontrol. However, no live transgenic Bacillus thuringiensis for insect pest management has been approved by regulatory bodies. The use of Bacillus thuringiensis as a dsRNA biocontrol expression system is in its infancy compared to other more well established systems, and further research into its viability as a practical insect control strategy is required.
The direct application of dsRNA in the agricultural sector has a number of limitations associated with product degradation by UV and environmental RNases which may limit overall efficacy when applied directly without formulation to protect the dsRNA (San Miguel and Scott, 2016; Leeuwen et al., 2019; Bachman et al., 2020). A potential method for protecting ssRNA and dsRNA products is the coexpression of proteins that will bind to and protect the RNA. Similar approaches have been utilised to produce and purify short hairpin dsRNA in E. coli (Huang et al., 2013). Co-expression of a recombinant His-tagged p19 protein and a long hairpin RNA containing sense and antisense sequences of the target mRNA was performed. The p19 binds siRNAs with high affinity once the hpRNA is processed by RNase III into short 21–24 nt siRNAs. The siRNA-protein complex was subsequently purified by Ni-NTA affinity chromatography followed by anion-exchange chromatography (Huang et al., 2013).
Encapsulation of dsRNA has been developed as a method to increase yields of dsRNA in microbial host species, and to protect the product from degradation in the application environment. A platform using bacterial minicells for the production and encapsulation of dsRNA has been developed under the name AgriCell technology by AgroSpheres (www.agrospheres.com). This technology involves non-GMO bioparticles produced during the fermentation of bacterial cells which encapsulate the dsRNA, providing protection from environmental degradation including heat, UV, microbes and nucleases and therefore has potential benefits for the endogenous delivery and stable release of the dsRNA. Using this technology dsRNA was manufactured at a yield of 100 mg/L and shown to trigger an RNAi effect in Botryotinia fuckeliana, and to inhibit grey mould infection in glasshouse-grown strawberries (Islam et al., 2021).
The potential for the use of virus like particles (VLPs) for dsRNA delivery to insect pests, and the challenges associated with this method, have been reviewed by Kolliopoulou et al. (2017) and VLPs have been successfully used in order to protect against yellow head virus in shrimp (Sinnuengnong et al., 2018). The technology utilises VLPs that co-assemble in the expression species, specifically non-infectious viral envelopes co-expressed in E. coli with the dsRNA and protect dsRNA from nucleases and harsh lysis steps, assisting in the isolation, purification and application of crop protecting agents.
The production of dsRNA in microorganisms offers alternative routes for foliar application of the dsRNA. The dsRNA can either be purified from the host microorganism or delivered via direct foliar application of the microorganism containing the dsRNA itself. However, microorganisms transformed with plasmids for production of dsRNA that target insect mRNAs are GM organisms. Therefore prior to application of the microorganism, strategies have been employed to inactivate the microorganisms. For example, E. coli cells expressing insecticidal dsRNA and subsequently heat inactivated, resulted in high mortality of beet armyworm through feeding, and resulted in no detectable bacterial colonies when plated on standard LB agar plates (Kim et al., 2015) demonstrating the organism is not able to reproduce in the environment.
Heat-inactivated yeast expressing a dsRNA insecticide have also been demonstrated to be successful in killing larvae of Anopheles mosquitoes, as a possible measure against this common disease vector (Mysore et al., 2017). dsRNA produced in E. coli which are subsequently heat-inactivated has also been demonstrated to successfully induce RNAi in fish cells (García et al., 2015) and if uptake into live fish could be achieved this technique may have applications outside of insecticides, such as virus control in fish farming.
RNAi has also been induced successfully in Aedes mosquitoes fed E. coli containing insecticidal dsRNA that were inactivated and lysed by chlorhexidine (Lopez et al., 2019). In addition, Hashiro et al., evaluated the use of alcohols as a sterilising agent for C. glutamicum (Hashiro et al., 2019a) and identified this as a preferred method of microbe inactivation, as opposed to heat sterilisation, previously reported (Zhu et al., 2011). Alcohols were shown to permeate cell membranes and inactivate RNases without disruption and degradation of dsRNA. Microbe membranes were noted to still be partially intact, thus protecting active dsRNA from environmental degradation.
An alternative approach, also using microorganisms for production of dsRNA is bacterium-mediated RNA interference. In this system live bacteria expressing dsRNA are applied to and colonise an organism to produce and facilitate the uptake of dsRNA resulting in RNAi effects on the targeted mRNA. Using this approach, an appropriate delivery bacterium is selected in conjunction with a plasmid construct to express the dsRNA similarly to approaches outlined previously. The overall aim is to enable the bacterium to replicate in vivo, synthesise specific dsRNA molecules that can be absorbed by the insect gut and induce systemic RNAi, either following secretion by the bacteria or after bacterial cell death and lysis in insecta.
Although such approaches have been utilised in other systems, their application in crop plant protection is limited. Such approaches have been demonstrated using two different insect species: Rhodnius prolixus, a trypanosome-transmitting assassin bug, and Frankliniella occidentalis, the western flower thrip, an invasive agricultural pest (Whitten et al., 2016). RNase III-deficient, dsRNA-expressing strains of R. rhodnii–a symbiont of these pest species–were created. The production system included an RNase III mutant with a stably integrated dsRNA expression cassette within the R. rhodnii chromosome, and dsRNA expressed from plasmids. In both systems the dsRNA expressing R. rhodnii were able to produce sustained systemic silencing. In F. occidentalis, RNAi reduction of α-tubulin mRNA was achieved and produced significant mortality in both larvae and adults, which was not observed using heat-inactivated bacteria expressing dsRNA (Whitten et al., 2016).
In addition to using live bacteria for dsRNA production, the use of live insect-parasitising fungi for production and delivery of insecticidal dsRNA is also a possibility. Since 1995, there have been 26 licences approved for the use of genetically modified fungi as crop protection agents in the USA (Hokanson et al., 2014). This generates an appealing avenue for RNA based crop protection strategies as approval remains a significant hurdle to overcome.
Dialeurodes citri is a citrus fruit pest which causes soot disease in citrus crops in many locations globally. Currently, only chemical pesticides are used to control D. citri (Yu et al., 2019). L. attenuatum, an entomopathogenic fungus which causes decreased population size in D. citri, has been explored as a natural alternative to chemical pesticides. However, application of the wild type fungi only results in low mortality rates and therefore, it has not been successfully implemented as a biological agent. Different strains of L. attenuatum were developed to produce dsRNA specific to four of the C. citri immune responses, under the control of a constitutive promoter PtrpC (Yu et al., 2019). The fungi produced hpRNA which was subsequently processed by the fungal RNAi machinery into siRNAs. Although siRNAs are not usually effective for RNAi in insects (typically dsRNA >80 bp is required for entry via the midgut (Huvenne and Smagghe, 2010; Yu et al., 2013)), in this case the delivery method is not through ingestion but via a fungal host. RNAi activity was detected by day three of infection with live fungi producing hpRNA, and mortality was slightly increased in two of the targets (Yu et al., 2019).
In a similar study by Hu et al. (Hu and Xia, 2019), virulence of the locust specific fungus Metarhizium acridum was increased by the introduction of dsRNA targeting mRNAs specific to the host’s ATPase subunits F0-F1. Again, the resulting hpRNA was generated under the control of a strong constitutive fungal promoter PmaGDP (Cao et al., 2012; Hu and Xia, 2019). Production of insecticidal dsRNA in live non-parasitic fungi has also been demonstrated, as in the case of production in live yeast (see above). Similarly, genetically modified live algae have also been used to deliver dsRNA to mosquito larvae in an aquatic environment (Kumar et al., 2013). In addition, the green microalgae Chlamydomonas reinhardtii was engineered to produce dsRNA targeting the lethal shrimp yellow head virus (YHV) and is considered as “Generally Recognized As Safe (GRAS)” by the US Food and Drug Administration. The microalgae does not produce any endotoxins and infectious agents, and therefore is unlikely to result in associated health risk or environmental contamination (Dauvillée et al., 2010).
A wide range of studies have shown the feasibility of transgenic crops protecting plants against pests and pathogens (Baum et al., 2007; Mao et al., 2011; Pitino et al., 2011; Zha et al., 2011; Zhang et al., 2015; Malik et al., 2016; Yu et al., 2016; Khalid et al., 2017; Li et al., 2017). Transgenic crop protection has also been reviewed in a number of publications (Talakayala et al., 2020; Chung et al., 2021; Kumari et al., 2022). The main advantage associated with in planta dsRNA synthesis (endogenous dsRNA production) in comparison to alternative production systems, is the elimination of downstream processing, with no purification, formulation and/or delivery of the dsRNA or material containing the dsRNA required, thus reducing labour for agricultural workers and consequently reducing costs to the consumer (Zotti and Smagghe, 2015). Research has been undertaken examining the feasibility of protecting crops against a variety of organisms, with particular success in protection against plant viruses. Crops which produce their own dsRNA are efficient at “self-protection,” a highly useful technique used for cultivated crops. Some of the previously reported crops which have increased resistance to viruses are tobacco against TMV (Kalantidis et al., 2002), potato against SPV (Sano et al., 1997) and tomato which showed an 82% increase in resistance to four tomato specific viruses with a single transgene (Bucher et al., 2006).
Other research has focused on the protection of crops against insect pests, the earliest of which was undertaken by Mao et al. (Baum et al., 2007), in which F1 corn plants were genetically engineered to express a dsRNA homologue of the V-ATPase A mRNA of the Western corn rootworm (WCR). A cassette synthesising a hairpin RNA homologue of the V-ATPase A mRNA was produced, under the control of a constitutive promoter CaMV e35S–a design like that in Figure 4C. The cassette was cloned into a vector which aided in chromosomal insertion of the cassette into the host genome (Baum et al., 2007), resulting in dsRNA generation in all cells of the plant. In the study dsRNA was isolated and imaged, showing the product to be intact hpRNA and Dicer processed siRNAs (21–24 nt in length). As previously mentioned, siRNA is not active for RNAi when ingested by insect pests, and dsRNA of lengths <60 bp have limited uptake (Bolognesi et al., 2012). However, as some hpRNA was available, despite being processed by the host’s RNAi machinery, uptake was achieved by the target pest and RNAi activity was observed, with the insects demonstrating signs of impaired growth and development. Other studies also achieved similar results but none could produce full plant protection or 100% insect mortality (Baum et al., 2007; Mao et al., 2011; Pitino et al., 2011; Zha et al., 2011).
Recently, a number of laboratories have shown that producing dsRNA in a plant’s chloroplast, rather than in its cellular cytoplasm, results in improved delivery of dsRNA (Jin et al., 2015; Zhang et al., 2015; Bally et al., 2016). Plastids are plant organelles formally believed to have been freely living prokaryotic cyanobacteria, and therefore lack RNAi machinery. It was hypothesised that dsRNA may therefore accumulate in plastids, for example in transplastomic potato plants with dsRNA targeting the Colorado potato beetle (CPB) (He et al., 2020). Three forms of dsRNA were produced in tobacco plastids to assess yield and homogeneity: linear dsRNA, hairpin RNA, and a linear RNA with hairpin protective ends. Constructs for the linear and hairpin forms of the dsRNA were identical to those previously described (see Figures 4B, C respectively) except for the substitution of promoter type. The novel hairpin protected dsRNA (hppRNA) was synthesised in a similar manner to linear dsRNA, apart from a small alteration by way of insertion of small self-complimentary inverted repeats at either end of the targeting region. These are predicted to self fold into small hairpins which protect linear dsRNA from RNases.
Accumulation of dsRNA was achieved in all three cases, with linear dsRNA having the highest yields. Zhang et al., subsequently produced transplastomic potato crops that showed dsRNA accumulation levels in leaves of 0.4% of the total cellular RNA (Zhang et al., 2015). This level of expression was enough to provide full plant protection from the CPB larvae when targeting β-actin (an essential cytoskeleton protein) causing 100% mortality after 5 days of insect feeding. These studies also highlight the importance of producing large amounts of long dsRNAs to achieve efficient protection.
The development of GM crops is a long and costly process, and engineered plants may become redundant in a short period of time (Khajuria et al., 2018) with the ever growing issue of pest resistance. Additionally, expression of dsRNA in transgenic plants is less flexible and adaptable than other RNA-based control methods, and hampered by political issues surrounding GM crops (Borel, 2017). There is strong resistance to the introduction of GM crops from public groups and the media, in spite of the ever-growing support from scientific research (Zhang et al., 2015). Legislation also holds back the development of transgenic crops, particularly in Europe where growth of genetically modified crops is restricted (Papademetriou, 2015). A further limitation of genetically engineered crops is well described by the control of the Canadian pine beetle, which threatens the pine forests of Canada. The beetles’ main food source is well-established trees, and thus genetically engineering plants would not prove an effective biocontrol measure (Keeling et al., 2013).
2017 was a landmark year for dsRNA crop protection as the first commercially available GMO crop was approved by the FDA with Monsanto and Dow’s SMARTSTAX PRO maize (Head et al., 2017). The crop produces an active dsRNA targeting the western corn rootworm mRNA of Snf7. Furthermore, studies were also approved to investigate quality enhancement of apple and potato crops which express dsRNA for the regulation of endogenous proteins (Waltz, 2015; Baranski et al., 2019).
The development of dsRNA based biocontrols for effective pest management strategies, requires the production of large quantities of dsRNA combined with suitable delivery methods to ensure RNAi is effectively triggered in the target pest. For large field-scale management of crop pests and pathogens, large quantities of dsRNA would be required (>1000 kg of dsRNA). Previous estimations have suggested up to 10 g of dsRNA per hectare (Zotti et al., 2018), although this amount may vary depending on the target’s sensitivity to trigger RNAi via oral uptake of dsRNA and its capacity for systemic RNAi. Such large scale manufacturing at appropriate cost is challenging and will be difficult to achieve using in vitro dsRNA transcription systems, which exhibit a minimum cost of $100 per g of dsRNA (Zotti et al., 2018). However recent advances in the manufacturing of mRNA vaccines have demonstrated the scalability of IVT for the large-scale manufacturing of RNA using such approaches (Sahin et al., 2020). Large scale microbial fermentation methods offer the potential for more economical large scale biomanufacturing of dsRNA and several industrial companies are currently developing low-cost, large-scale manufacturing platforms for the production of dsRNA typically <$1/g (Zotti et al., 2018).
In vitro transcription is a rapid, versatile method capable of producing high purity dsRNA which is particularly useful for research focussing on the development of new dsRNA based biocontrols and laboratory scale research studies. Large-scale IVT is limited but commercially available and capable of generating Gram to kilogram quantities of 100–800 bp dsRNA (Cagliari et al., 2018). Large scale IVT typically uses convergent T7 DdRp promoters flanking the target sequence. However, limited yields and lengthy production times for industrial scale production potentially limit its agricultural implementation. Cell-free systems offer an alternative scalable production method with the advantages of low cost and high yield, although currently this requires specialist commercial production. Microbial systems offer an alternative approach for production of dsRNA which is versatile and scalable. Moreover E. coli is currently widely used in high cell density fermentation processes for industrial recombinant protein production and is also able to achieve high dsRNA yields. In addition, alternative new microbial hosts are emerging as alternative production systems which will provide new opportunities and strategies for the in vivo production of exogenous dsRNA based biocontrols economically at scale, for example C. glutamicum which recently achieved yields of over 1 g/L dsRNA (Hashiro et al., 2021).
Bacterially mediated RNAi could provide an alternative approach for the production and delivery of dsRNA, enabling a semi-permanent RNAi silencing effect to be induced by exogenous application. Use of a plant symbiote microorganism offers additional benefits compared to topical application of dsRNA, as the symbiote can colonise the host plant, synthesising protective dsRNA in situ. However, such approaches are limited by the availability of an appropriate delivery bacterium for which suitable dsRNA expression cassettes and vectors have been developed. It is also presently unclear how bacteria might transmit silencing RNA to the plant. Furthermore, such approaches will require the release of GM organisms into the environment and raise potential issues of public acceptance.
Transgenic organisms for the production of endogenous dsRNA possess a number of advantages over non-transformative methods as well as a few drawbacks. Fungal production is in its infancy when compared to other methods but with further optimisation–for example, the knockout of RNAi machinery within the fungi–there is potential that transgenic fungi may play a part in the future of bio-pesticides. Additionally, the approval of a number of licences for the use of transgenic fungi by regulatory bodies shows promise for further approval for crop protection. In planta production, again shows promise for crop protection with a number for studies highlighting its key benefits. Expression in transgenic plants works for commercial crops such as corn. However, such approaches are expensive, time consuming, less flexible and hampered by issues surrounding GM crops.
Beyond the challenges associated with the production method, there are also challenges surrounding the delivery of dsRNA biocontrols and the variability of RNAi efficacy between different target species. Different insect orders and species show variation in RNAi efficacy, with orders such as Coleoptera and Hemipterea generally showing good RNAi efficacy. In contrast, many Lepidoptera demonstrate poor RNAi efficacy. This variation is due to factors such as insect nuclease potency and upregulation (Guan et al., 2018), physiological pH prone to causing RNA hydrolysis (Shukla et al., 2016), and differences in dsRNA uptake and subsequent intracellular transport (Shukla et al., 2016). As these factors can often be overcome by varying the dsRNA delivery method, and certain delivery methods are also more or less amenable to particular production methods, both production and delivery will have to be dovetailed in order to achieve efficient dsRNA biocontrols against certain species of pest insect.
Whilst research in production of dsRNA biocontrols has resulted in many publications few products have been commercialised. The plant-incorporated protectant product MON-87411 is a genetically engineered maize that among other transgenes produces a dsRNA matching the sequence of part of the snf7 essential gene from Western corn rootworm (Diabrotica virgifera virgifera). MON-87411 has been registered for cultivation in the US and elsewhere since 2015 (Firko, 2015). The grain product of this crop has been registered for food and feed uses within the European Union (Naegeli et al., 2019). To date, no other dsRNA-based GM crop has been commercialized.
No spray applied RNA-based biocontrols have been commercialized. However, Greenlight Biosciences indicates that it plans to register a treatment for Colorado potato beetle during 2022 with first sales planned in the US during 2023. One RNA-based biocontrol has been given initial registration to protect honeybees from Israeli acute paralysis virus (IAPV). The registration holder, Beeologics was acquired by Monsanto, subsequently acquired by Bayer who sold the technology to GreenLight. Research data suggested that this technology was effective in the controlling IAPV in commercial beehives (Hunter et al., 2010).
In conclusion, dsRNA based biocontrols have the potential to provide a species-selective and sustainable insect management strategy that overcomes many current issues associated with chemical pesticides. However, research on the large-scale manufacturing and mass delivery of dsRNA insecticides is in its infancy, and all potential methods present challenges. Different methods may also be more suited to particular target insects or application environments, and have additional challenges related to product characterisation, quantification, quality control and regulatory approval that need to be addressed. Furthermore, there is unlikely to be a universal method that is effective for control of all conceivable target species, and production and delivery methods will have to be tailored accordingly. However, a number of methods presented here demonstrate potential for large scale commercial production of dsRNA based biocontrols as sustainable alternatives to chemical pesticides.
Conceptulization MD, PK, SB, JoH, and JaH. Manuscript writing JoH, JaH, MD, and PK. Manuscript editing and proofing MD and PK.
JoH was funded on a Biotechnology and Biological Science Research Council iCASE studentship in collaboration with Syngenta (BB/N504099/1). MD acknowledges further support from the Biotechnology and Biological Science Research Council (BB/M012166/1). For the purpose of open access, the author has applied a Creative Commons Attribution (CC BY) licence to any Author Accepted Manuscript version arising.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aalto, A. P., Sarin, L. P., Van Dijk, A. A., Saarma, M., Poranen, M. M., Arumäe, U., et al. (2007). Large-scale production of dsRNA and siRNA pools for RNA interference utilizing bacteriophage φ6 RNA-dependent RNA polymerase. RNA 13, 422–429. doi:10.1261/rna.348307
Agrawal, N., Dasaradhi, P. V. N., Mohmmed, A., Malhotra, P., Bhatnagar, R. K., and Mukherjee, S. K. (2003). RNA interference: Biology, mechanism, and applications. Microbiol. Mol. Biol. Rev. 67, 657–685. doi:10.1128/mmbr.67.4.657-685.2003
Ahn, S.-J., Donahue, K., Koh, Y., Martin, R. R., and Choi, M.-Y. (2019). Microbial-based double-stranded RNA production to develop cost-effective RNA interference application for insect pest management. Int. J. Insect Sci. 11, 117954331984032. doi:10.1177/1179543319840323
Alder, M. N., Dames, S., Gaudet, J., and Mango, S. E. (2003). Gene silencing in Caenorhabditis elegans by transitive RNA interference. RNA 9, 25–32. doi:10.1261/rna.2650903
Andreev, K., Kantorová, V., and Bongaarts, J. (2013). Demographic components of future population growth. New York: United Nations, Department of Economic and Social Affairs, Population Division. Available at: https://kirillandreev.com/2013_dcpg/Andreev-etal_2013_Demographic-Components-of-Future-Population-Growth.pdf.
Álvarez-Sánchez, A. R., Romo-Quinones, C., Rosas-Quijano, R., Reyes, A. G., Barraza, A., Magallón-Barajas, F., et al. (2018). Production of specific dsRNA against white spot syndrome virus in the yeast Yarrowia lipolytica. Aquac. Res. 49, 480–491. doi:10.1111/are.13479
Bachman, P., Fischer, J., Song, Z., Urbanczyk-Wochniak, E., and Watson, G. (2020). Environmental fate and dissipation of applied dsRNA in soil, aquatic systems, and plants. Front. Plant Sci. 11, 21. doi:10.3389/fpls.2020.00021
Bally, J., McIntyre, G. J., Doran, R. L., Lee, K., Perez, A., Jung, H., et al. (2016). In-plant protection against helicoverpa armigera by production of long hpRNA in chloroplasts. Front. Plant Sci. 7, 1453. doi:10.3389/fpls.2016.01453
Baranski, R., Klimek-Chodacka, M., and Lukasiewicz, A. (2019). Approved genetically modified (GM) horticultural plants: A 25-year perspective. Folia Hortic. 31, 3–49. doi:10.2478/fhort-2019-0001
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi:10.1038/nbt1359
Baum, J. A., Christian, A. T., Evdokimov, A., Moshiri, F., Weaver, L. M., and Zhang, H. (2014). Compositions and methods for the improved production and delivery of RNA by efficient transcription termination. Patent no.: US9814243B2, Alexandria, Virginia: US Patent & Trademark Office.
Bebber, D. P., Ramotowski, M. A. T., and Gurr, S. J. (2013). Crop pests and pathogens move polewards in a warming world. Nat. Clim. Chang. 3, 985–988. doi:10.1038/nclimate1990
Bebber, D. P. (2015). Range-expanding pests and pathogens in a warming world. Annu. Rev. Phytopathol. 53, 335–356. doi:10.1146/annurev-phyto-080614-120207
Bento, F. M., Marques, R. N., Campana, F. B., Demétrio, C. G., Leandro, R. A., Parra, J. R. P., et al. (2020). Gene silencing by RNAi via oral delivery of dsRNA by bacteria in the South American tomato pinworm, Tuta absoluta. Pest Manag. Sci. 76, 287–295. doi:10.1002/ps.5513
Bolognesi, R., Ramaseshadri, P., Anderson, J., Bachman, P., Clinton, W., Flannagan, R., et al. (2012). Characterizing the mechanism of action of double-stranded RNA activity against western corn rootworm (diabrotica virgifera virgifera LeConte). PLoS ONE 7, e47534. doi:10.1371/journal.pone.0047534
Borel, B. (2017). CRISPR, microbes and more are joining the war against crop killers. Nature 543, 302–304. doi:10.1038/543302a
Bravo, A., Likitvivatanavong, S., Gill, S. S., and Soberón, M. (2011). Bacillus thuringiensis: A story of a successful bioinsecticide. Insect Biochem. Mol. Biol. 41, 423–431. doi:10.1016/J.IBMB.2011.02.006
Bucher, E., Lohuis, D., van Poppel, P. M. J. A., Geerts-Dimitriadou, C., Goldbach, R., and Prins, M. (2006). Multiple virus resistance at a high frequency using a single transgene construct. J. Gen. Virol. 87, 3697–3701. doi:10.1099/vir.0.82276-0
Caccia, S., Astarita, F., Barra, E., Di Lelio, I., Varricchio, P., et al. (2020). Enhancement of Bacillus thuringiensis toxicity by feeding Spodoptera littoralis larvae with bacteria expressing immune suppressive dsRNA. J. Pest Sci. 93, 303–314. doi:10.1007/s10340-019-01140-6
Cagliari, D., Dias, N. P., Galdeano, D. M., dos Santos, E. Á., Smagghe, G., and Zotti, M. J. (2019). Management of pest insects and plant diseases by non-transformative RNAi. Front. Plant Sci. 10, 1319. doi:10.3389/fpls.2019.01319
Cagliari, D., Santosdos, E. A., Dias, N., Smagghe, G., and Zotti, M. (2018). “Nontransformative strategies for RNAi in crop protection,” in Modulating gene expression - abridging the RNAi and CRISPR-Cas9 technologies. Editor A. Singh, and M. W. Khan (London: IntechOpen). doi:10.5772/intechopen.80874
Cao, Y., Jiao, R., and Xia, Y. (2012). A strong promoter, PMagpd, provides a tool for high gene expression in entomopathogenic fungus, Metarhizium acridum. Biotechnol. Lett. 34, 557–562. doi:10.1007/s10529-011-0805-3
Carlson, E. D., Gan, R., Hodgman, C. E., and Jewett, M. C. (2012). Cell-free protein synthesis: Applications come of age. Biotechnol. Adv. 30, 1185–1194. doi:10.1016/j.biotechadv.2011.09.016
Castellanos, N. L., Smagghe, G., Sharma, R., Oliveira, E. E., and Christiaens, O. (2019). Liposome encapsulation and EDTA formulation of dsRNA targeting essential genes increase oral RNAi-caused mortality in the Neotropical stink bug Euschistus heros. Pest Manag. Sci. 75, 537–548. doi:10.1002/ps.5167
Cerda, R., Avelino, J., Gary, C., Tixier, P., Lechevallier, E., and Allinne, C. (2017). Primary and secondary yield losses caused by pests and diseases: Assessment and modeling in coffee. PLOS ONE 12, e0169133. doi:10.1371/journal.pone.0169133
Chamberlin, M., Mcgrath, J., and Waskell, L. (1970). New RNA polymerase from escherichia coli infected with bacteriophage T7. Nature 228, 227–231. doi:10.1038/228227a0
Chen, X., Zhou, L., Tian, K., Kumar, A., Singh, S., Prior, B. A., et al. (2013). Metabolic engineering of Escherichia coli: A sustainable industrial platform for bio-based chemical production. Biotechnol. Adv. 31, 1200–1223. doi:10.1016/j.biotechadv.2013.02.009
Chen, Z., He, J., Luo, P., Li, X., and Gao, Y. (2019). Production of functional double-stranded RNA using a prokaryotic expression system in Escherichia coli. Microbiologyopen 8, e00787. doi:10.1002/mbo3.787
Christiaens, O., Tardajos, M. G., Martinez Reyna, Z. L., Dash, M., Dubruel, P., and Smagghe, G. (2018). Increased RNAi efficacy in Spodoptera exigua via the formulation of dsRNA with guanylated polymers. Front. Physiol. 9, 316. doi:10.3389/fphys.2018.00316
Christiaens, O., Whyard, S., Vélez, A. M., and Smagghe, G. (2020). Double-stranded RNA technology to control insect pests: Current status and challenges. Front. Plant Sci. 11, 451. doi:10.3389/fpls.2020.00451
Chung, S. H., Feng, H., and Jander, G. (2021). Engineering pest tolerance through plant-mediated RNA interference. Curr. Opin. Plant Biol. 60, 102029. doi:10.1016/J.PBI.2021.102029
Dalaisón-Fuentes, L. I., Pascual, A., Gazza, E., Welchen, E., Rivera-Pomar, R., and Inés Catalano, M. (2022). Development of efficient RNAi methods in the corn leafhopper Dalbulus maidis, a promising application for pest control. Pest Manag. Sci. 78 (7), 3108–3116. doi:10.1002/ps.6937
Dauvillée, D., Delhaye, S., Gruyer, S., Slomianny, C., Moretz, S. E., d’Hulst, C., et al. (2010). Engineering the chloroplast targeted malarial vaccine antigens in Chlamydomonas starch granules. PLoS ONE 5, e15424. doi:10.1371/journal.pone.0015424
Davis-Vogel, C., Ortiz, A., Procyk, L., Robeson, J., Kassa, A., Wang, Y., et al. (2018). Knockdown of RNA interference pathway genes impacts the fitness of Western corn rootworm. Sci. Rep. 8, 7858. doi:10.1038/s41598-018-26129-6
Deutsch, C. A., Tewksbury, J. J., Tigchelaar, M., Battisti, D. S., Merrill, S. C., Huey, R. B., et al. (2018). Increase in crop losses to insect pests in a warming climate. Science 361, 916–919. doi:10.1126/science.aat3466
Douglas, A. E. (2018). Strategies for enhanced crop resistance to insect pests. Annu. Rev. Plant Biol. 69, 637–660. doi:10.1146/annurev-arplant-042817-040248
Dubey, S., Jhelum, V., and Patanjali, P. K. (2011). Controlled release agrochemicals formulations: A review. J. Sci. Ind. Res. (India). 70, 105–112.
Duman-Scheel, M. (2018). Saccharomyces cerevisiae (Baker’s Yeast) as an Interfering RNA Expression and Delivery System. Curr. Drug Targets 20, 942–952. doi:10.2174/1389450120666181126123538
Dumas, P., Sambou, M., Gaudet, J. D., Morin, M. D., Moffat, C. E., Boquel, S., et al. (2020). Differential expression of transcripts with potential relevance to chlorantraniliprole response in the Colorado potato beetle, Leptinotarsa decemlineata. Arch. Insect Biochem. Physiol. 103, e21642. doi:10.1002/arch.21642
Fire, A., Xu, S., Montgomery, M. K., Kostas, S. A., Driver, S. E., and Mello, C. C. (1998). Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature 391, 806–811. doi:10.1038/35888
Firko, M. J. (2015). Determination of Nonregulated Status for MON 87411 corn. Available at: https://www.aphis.usda.gov/brs/aphisdocs/13_29001p_det.pdf (Accessed August 15, 2022).
Flood, J. (2010). The importance of plant health to food security. Food Secur. 2, 215–231. doi:10.1007/s12571-010-0072-5
Fujiwara, K., Sawamura, T., Niwa, T., Deyama, T., Nomura, S. I. M., Taguchi, H., et al. (2017). In vitro transcription-Translation using bacterial genome as a template to reconstitute intracellular profile. Nucleic Acids Res. 45, 11449–11458. doi:10.1093/nar/gkx776
Garamella, J., Marshall, R., Rustad, M., and Noireaux, V. (2016). The All E. coli TX-TL Toolbox 2.0: A Platform for Cell-Free Synthetic Biology. ACS Synth. Biol. 5, 344–355. doi:10.1021/acssynbio.5b00296
Garbian, Y., Maori, E., Kalev, H., Shafir, S., and Sela, I. (2012). Bidirectional Transfer of RNAi between Honey Bee and Varroa destructor: Varroa Gene Silencing Reduces Varroa Population. PLoS Pathog. 8, e1003035. doi:10.1371/journal.ppat.1003035
García, K., Ramírez-Araya, S., Díaz, Á., Reyes-Cerpa, S., Espejo, R., Higuera, G., et al. (2015). Inactivated E. coli transformed with plasmids that produce dsRNA against infectious salmon anemia virus hemagglutinin show antiviral activity when added to infected ASK cells. Front. Microbiol. 6, 300–311. doi:10.3389/fmicb.2015.00300
Gholamalipour, Y., Johnson, W. C., and Martin, C. T. (2019). Efficient inhibition of RNA self-primed extension by addition of competing 3′-capture DNA-improved RNA synthesis by T7 RNA polymerase. Nucleic Acids Res. 47, e118. doi:10.1093/NAR/GKZ700
Gholamalipour, Y., Karunanayake Mudiyanselage, A., and Martin, C. T. (2018). 3′ end additions by T7 RNA polymerase are RNA self-templated, distributive and diverse in character—RNA-Seq analyses. Nucleic Acids Res. 46, 9253–9263. doi:10.1093/nar/gky796
Ghosh, S. K. B., Hunter, W. B., Park, A. L., and Gundersen-Rindal, D. E. (2017). Double strand RNA delivery system for plant-sap-feeding insects. PLOS ONE 12, e0171861. doi:10.1371/journal.pone.0171861
Gogoi, A., Sarmah, N., Kaldis, A., Perdikis, D., and Voloudakis, A. (2017). Plant insects and mites uptake double-stranded RNA upon its exogenous application on tomato leaves. Planta 246, 1233–1241. doi:10.1007/s00425-017-2776-7
Gordon, K. H. J., and Waterhouse, P. M. (2007). RNAi for insect-proof plants. Nat. Biotechnol. 25, 1231–1232. doi:10.1038/nbt1107-1231
Gould, F., Brown, Z. S., and Kuzma, J. (2018). Wicked evolution: Can we address the sociobiological dilemma of pesticide resistance? Science 360, 728–732. doi:10.1126/science.aar3780
Guan, R.-B., Li, H.-C., Fan, Y.-J., Hu, S.-R., Christiaens, O., Smagghe, G., et al. (2018). A nuclease specific to lepidopteran insects suppresses RNAi. J. Biol. Chem. 293, 6011–6021. doi:10.1074/jbc.RA117.001553
Haley, B., Tang, G., and Zamore, P. D. (2003). In vitro analysis of RNA interference in Drosophila melanogaster. Methods 30, 330–336. doi:10.1016/S1046-2023(03)00052-5
Hashiro, S., Chikami, Y., Kawaguchi, H., Krylov, A. A., Niimi, T., and Yasueda, H. (2021). Efficient production of long double-stranded RNAs applicable to agricultural pest control by Corynebacterium glutamicum equipped with coliphage T7-expression system. Appl. Microbiol. Biotechnol. 105, 4987–5000. doi:10.1007/s00253-021-11324-9/
Hashiro, S., Mitsuhashi, M., Chikami, Y., Kawaguchi, H., Niimi, T., and Yasueda, H. (2019a). Construction of Corynebacterium glutamicum cells as containers encapsulating dsRNA overexpressed for agricultural pest control. Appl. Microbiol. Biotechnol. 103, 8485–8496. doi:10.1007/s00253-019-10113-9
Hashiro, S., Mitsuhashi, M., and Yasueda, H. (2019b). High copy number mutants derived from Corynebacterium glutamicum cryptic plasmid pAM330 and copy number control. J. Biosci. Bioeng. 127, 529–538. doi:10.1016/j.jbiosc.2018.10.012
Hashiro, S., Mitsuhashi, M., and Yasueda, H. (2019c). Overexpression system for recombinant RNA in Corynebacterium glutamicum using a strong promoter derived from corynephage BFK20. J. Biosci. Bioeng. 128, 255–263. doi:10.1016/j.jbiosc.2019.03.003
Hashiro, S., and Yasueda, H. (2022). RNA Interference-Based Pesticides and Antiviral Agents: Microbial Overproduction Systems for Double-Stranded RNA for Applications in Agriculture and Aquaculture. Appl. Sci. Switz. 12, 2954. doi:10.3390/app12062954
He, W., Xu, W., Xu, L., Fu, K., Guo, W., Bock, R., et al. (2020). Length-dependent accumulation of double-stranded RNAs in plastids affects RNA interference efficiency in the Colorado potato beetle. J. Exp. Bot. 71, 2670–2677. doi:10.1093/JXB/ERAA001
Head, G. P., Carroll, M. W., Evans, S. P., Rule, D. M., Willse, A. R., Clark, T. L., et al. (2017). Evaluation of SmartStax and SmartStax PRO maize against Western corn rootworm and northern corn rootworm: efficacy and resistance management. Pest Manag. Sci. 73, 1883–1899. doi:10.1002/ps.4554
Heckel, D. G. (2020). How do toxins from Bacillus thuringiensis kill insects? An evolutionary perspective. Arch. Insect Biochem. Physiol. 104, e21673. doi:10.1002/ARCH.21673
Hilder, V. A., and Boulter, D. (1999). Genetic engineering of crop plants for insect resistance - A critical review. Crop Prot. 18, 177–191. doi:10.1016/S0261-2194(99)00028-9
Hofmann, S., and Miller, O. L. (1977). Visualization of ribosomal ribonucleic acid synthesis in a ribonuclease III deficient strain of Escherichia coli. J. Bacteriol. 132, 718–722. doi:10.1128/jb.132.2.718-722.1977
Hokanson, K. E., Dawson, W. O., Handler, A. M., Schetelig, M. F., and St. Leger, R. J. (2014). Not all GMOs are crop plants: non-plant GMO applications in agriculture. Transgenic Res. 23, 1057–1068. doi:10.1007/s11248-013-9769-5
Hu, J., and Xia, Y. (2019). Increased virulence in the locust-specific fungal pathogen Metarhizium acridum expressing dsRNAs targeting the host F 1F0-ATPase subunit genes. Pest Manag. Sci. 75, 180–186. doi:10.1002/ps.5085
Hu, Q., and Wu, W. (2016). Recombinant fungal entomopathogen RNAi target insect gene. Bioengineered 7, 504–507. doi:10.1080/21655979.2016.1146833
Hu, X., Richtman, N. M., Zhao, J. Z., Duncan, K. E., Niu, X., Procyk, L. A., et al. (2016). Discovery of midgut genes for the RNA interference control of corn rootworm. Sci. Rep. 6, 30542. doi:10.1038/srep30542
Hu, X., Steimel, J. P., Kapka-Kitzman, D. M., Davis-Vogel, C., Richtman, N. M., Mathis, J. P., et al. (2019). Molecular characterization of the insecticidal activity of double-stranded RNA targeting the smooth septate junction of Western corn rootworm (Diabrotica virgifera virgifera). PLOS ONE 14, e0210491. doi:10.1371/journal.pone.0210491
Huang, L., Jin, J., Deighan, P., Kiner, E., McReynolds, L., and Lieberman, J. (2013). Efficient and specific gene knockdown by small interfering RNAs produced in bacteria. Nat. Biotechnol. 31, 350–356. doi:10.1038/nbt.2537
Hull, D., and Timmons, L. (2004). Methods for delivery of double-stranded RNA into Caenorhabditis elegans. Methods Mol. Biol. 265, 23–58. doi:10.1385/1-59259-775-0:023
Hunter, W. B., Glick, E., Paldi, N., and Bextine, B. R. (2012). Advances in RNA interference: dsRNA Treatment in Trees and Grapevines for Insect Pest Suppression. Southwest. Entomol. 37, 85–87. doi:10.3958/059.037.0110
Hunter, W., Ellis, J., vanEngelsdorp, D., Hayes, J., Westervelt, D., Glick, E., et al. (2010). Large-Scale Field Application of RNAi Technology Reducing Israeli Acute Paralysis Virus Disease in Honey Bees (Apis mellifera, Hymenoptera: Apidae). PLoS Pathog. 6, e1001160. doi:10.1371/journal.ppat.1001160
Huvenne, H., and Smagghe, G. (2010). Mechanisms of dsRNA uptake in insects and potential of RNAi for pest control: A review. J. Insect Physiology 56, 227–235. doi:10.1016/j.jinsphys.2009.10.004
Ikeda, M., and Takeno, S. (2013). “Amino Acid Production by Corynebacterium glutamicum,” in Corynebacterium glutamicum: Biology and biotechnology (Berlin: Springer-Verlag), 107–147. doi:10.1007/978-3-642-29857-8_4
Islam, M. T., Davis, Z., Chen, L., Englaender, J., Zomorodi, S., Frank, J., et al. (2021). Minicell-based fungal RNAi delivery for sustainable crop protection. Microb. Biotechnol. 14, 1847–1856. doi:10.1111/1751-7915.13699
Jain, R. G., Fletcher, S. J., Manzie, N., Robinson, K. E., Li, P., Lu, E., et al. (2022). Foliar application of clay-delivered RNA interference for whitefly control. Nat. Plants 8, 535–548. doi:10.1038/s41477-022-01152-8
Jiang, Y. X., Chen, J. Z., Li, M. W., Zha, B. H., Huang, P. R., Chu, X. M., et al. (2021). The Combination of Bacillus Thuringiensis and Its Engineered Strain Expressing dsRNA Increases the Toxicity against Plutella Xylostella. Int. J. Mol. Sci. 23, 444. doi:10.3390/IJMS23010444
Jin, S., Singh, N. D., Li, L., Zhang, X., and Daniell, H. (2015). Engineered chloroplast dsRNA silences cytochrome p450 monooxygenase, V-ATPase and chitin synthase genes in the insect gut and disrupts Helicoverpa armigera larval development and pupation. Plant Biotechnol. J. 13, 435–446. doi:10.1111/pbi.12355
Kalantidis, K., Psaradakis, S., Tabler, M., and Tsagris, M. (2002). The occurrence of CMV-specific short RNAs in transgenic tobacco expressing virus-derived double-stranded RNA is indicative of resistance to the virus. Mol. Plant. Microbe. Interact. 15, 826–833. doi:10.1094/MPMI.2002.15.8.826
Kamath, R. S., Martinez-Campos, M., Zipperlen, P., Fraser, A. G., and Ahringer, J. (2000). Effectiveness of specific RNA-mediated interference through ingested double-stranded RNA in Caenorhabditis elegans. Genome Biol. 2, RESEARCH0002–1. doi:10.1186/GB-2000-2-1-RESEARCH0002
Kanter, G., Yang, J., Voloshin, A., Levy, S., Swartz, J. R., and Levy, R. (2007). Cell-free production of scFv fusion proteins: An efficient approach for personalized lymphoma vaccines. Blood 109, 3393–3399. doi:10.1182/blood-2006-07-030593
Katoch, R., Sethi, A., Thakur, N., and Murdock, L. L. (2013). RNAi for Insect Control: Current Perspective and Future Challenges. Appl. Biochem. Biotechnol. 171, 847–873. doi:10.1007/s12010-013-0399-4
Keeling, C. I., Chiu, C. C., Aw, T., Li, M., Henderson, H., Tittiger, C., et al. (2013). Frontalin pheromone biosynthesis in the mountain pine beetle, Dendroctonus ponderosae, and the role of isoprenyl diphosphate synthases. Proc. Natl. Acad. Sci. U. S. A. 110, 18838–18843. doi:10.1073/pnas.1316498110
Khajuria, C., Ivashuta, S., Wiggins, E., Flagel, L., Moar, W., Pleau, M., et al. (2018). Development and characterization of the first dsRNA-resistant insect population from Western corn rootworm, Diabrotica virgifera virgifera LeConte. PLOS ONE 13, e0197059. doi:10.1371/journal.pone.0197059
Khalid, A., Zhang, Q., Yasir, M., and Li, F. (2017). Small RNA based genetic engineering for plant viral resistance: Application in crop protection. Front. Microbiol. 8, 43. doi:10.3389/fmicb.2017.00043
Kigawa, T., Yabuki, T., Yoshida, Y., Tsutsui, M., Ito, Y., Shibata, T., et al. (1999). Cell-free production and stable-isotope labeling of milligram quantities of proteins. FEBS Lett. 442, 15–19. doi:10.1016/S0014-5793(98)01620-2
Kim, E., Park, Y., Kim, Y., and Ling, E. (2015). A transformed bacterium expressing double-stranded RNA specific to integrin β1 enhances Bt Toxin efficacy against a polyphagous insect pest, Spodoptera exigua. PLoS ONE 10, e0132631–15. doi:10.1371/journal.pone.0132631
Koch, A., Biedenkopf, D., Furch, A., Weber, L., Rossbach, O., Abdellatef, E., et al. (2016). An RNAi-Based Control of Fusarium graminearum Infections Through Spraying of Long dsRNAs Involves a Plant Passage and Is Controlled by the Fungal Silencing Machinery. PLoS Pathog. 12, e1005901. doi:10.1371/JOURNAL.PPAT.1005901
Koch, A., Höfle, L., Werner, B. T., Imani, J., Schmidt, A., Jelonek, L., et al. (2019). SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non‐host plants. Mol. Plant Pathol. 20, 1636–1644. doi:10.1111/MPP.12866
Kolliopoulou, A., Taning, C. N. T., Smagghe, G., and Swevers, L. (2017). Viral Delivery of dsRNA for Control of Insect Agricultural Pests and Vectors of Human Disease: Prospects and Challenges. Front. Physiol. 8, 399. doi:10.3389/fphys.2017.00399
Kumar, A., Wang, S., Ou, R., Samrakandi, M., Beerntsen, B. T., and Sayre, R. T. (2013). Development of an RNAi based microalgal larvicide to control mosquitoes. MalariaWorld J. 4 (6).
Kumari, P., Jasrotia, P., Kumar, D., Kashyap, P. L., Kumar, S., Mishra, C. N., et al. (2022). Biotechnological Approaches for Host Plant Resistance to Insect Pests. Front. Genet. 13, 914029. doi:10.3389/FGENE.2022.914029
Leeuwen, D. M., Lever, M. A., Mateescu, B., and Sander, M. (2019). Environmental Fate of RNA Interference Pesticides: Adsorption and Degradation of Double-Stranded RNA Molecules in Agricultural Soils. Environ. Sci. Technol. 53 (6), 3027–3036. doi:10.1021/acs.est.8b05576
Levanova, A., and Poranen, M. M. (2018). RNA interference as a prospective tool for the control of human viral infections. Front. Microbiol. 9, 2151. doi:10.3389/fmicb.2018.02151
Li, Y., Wang, K., Lu, Q., Du, J., Wang, Z., Wang, D., et al. (2017). Transgenic Nicotiana benthamiana plants expressing a hairpin RNAi construct of a nematode Rs-cps gene exhibit enhanced resistance to Radopholus similis. Sci. Rep. 7, 13126. doi:10.1038/s41598-017-13024-9
Liu, S., Jaouannet, M., Dempsey, D. A., Imani, J., Coustau, C., and Kogel, K. H. (2020). RNA-based technologies for insect control in plant production. Biotechnol. Adv. 39, 107463. doi:10.1016/J.BIOTECHADV.2019.107463
Liu, S., Ladera-Carmona, M. J., Poranen, M. M., van Bel, A. J. E., Kogel, K. H., and Imani, J. (2021). Evaluation of dsRNA delivery methods for targeting macrophage migration inhibitory factor MIF in RNAi-based aphid control. J. Plant Dis. Prot. 128, 1201–1212. doi:10.1007/S41348-021-00464-9
Lopez, S. B. G., Guimarães-Ribeiro, V., Rodriguez, J. V. G., Dorand, F. A. P. S., Salles, T. S., Sá-Guimarães, T. E., et al. (2019). RNAi-based bioinsecticide for Aedes mosquito control. Sci. Rep. 9, 4038. doi:10.1038/s41598-019-39666-5
Losick, R. (1972). In vitro transcription. Annu. Rev. Biochem. 41, 409–446. doi:10.1146/annurev.bi.41.070172.002205
Lundgren, J. G., and Duan, J. J. (2013). RNAi-Based Insecticidal Crops: Potential Effects on Nontarget Species. BioScience 63, 657–665. doi:10.1525/bio.2013.63.8.8
Ma, Z. Z., Zhou, H., Wei, Y. L., Yan, S., and Shen, J. (2020). A novel plasmid–Escherichia coli system produces large batch dsRNAs for insect gene silencing. Pest Manag. Sci. 76, 2505–2512. doi:10.1002/ps.5792
Maeda, T., Tanaka, Y., Takemoto, N., Hamamoto, N., and Inui, M. (2016). RNase III mediated cleavage of the coding region of mraZ mRNA is required for efficient cell division in Corynebacterium glutamicum. Mol. Microbiol. 99, 1149–1166. doi:10.1111/mmi.13295
Makeyev, E. V., and Bamford, D. H. (2000). Replicase activity of purified recombinant protein P2 of double-stranded RNA bacteriophage phi 6. EMBO J. 19, 124–133. doi:10.1093/emboj/19.1.124
Malik, H. J., Raza, A., Amin, I., Scheffler, J. A., Scheffler, B. E., Brown, J. K., et al. (2016). RNAi-mediated mortality of the whitefly through transgenic expression of double-stranded RNA homologous to acetylcholinesterase and ecdysone receptor in tobacco plants. Sci. Rep. 6, 38469. doi:10.1038/srep38469
Mamta, B., and Rajam, M. V. (2017). RNAi technology: a new platform for crop pest control. Physiol. Mol. Biol. Plants 23, 487–501. doi:10.1007/s12298-017-0443-x
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi:10.1038/nbt1352
Mao, Y. B., Tao, X. Y., Xue, X. Y., Wang, L. J., and Chen, X. Y. (2011). Cotton plants expressing CYP6AE14 double-stranded RNA show enhanced resistance to bollworms. Transgenic Res. 20, 665–673. doi:10.1007/s11248-010-9450-1
Maxwell, B., Boyes, D., Tang, J., Rodrigues, T., Desai, S., Cunningham, D., et al. (2018). New pest control MOA Enabling the RNA revolution; Cell-free dsRNA production and control of Colorado potato beetle. Medford, MA: GreenLight Biosciences. Available at: http://www.globalengage.co.uk/pgc/docs/PosterMaxwell.pdf.
Meng, J., Lei, J., Davitt, A., Holt, J. R., Huang, J., Gold, R., et al. (2020). Suppressing tawny crazy ant (Nylanderia fulva) by RNAi technology. Insect Sci. 27, 113–121. doi:10.1111/1744-7917.12604
Mitter, N., Worrall, E. A., Robinson, K. E., Li, P., Jain, R. G., Taochy, C., et al. (2017). Clay nanosheets for topical delivery of RNAi for sustained protection against plant viruses. Nat. Plants 3, 16207. doi:10.1038/nplants.2016.207
Murphy, K. A., Tabuloc, C. A., Cervantes, K. R., and Chiu, J. C. (2016). Ingestion of genetically modified yeast symbiont reduces fitness of an insect pest via RNA interference. Sci. Rep. 6, 22587. doi:10.1038/srep22587
Mysore, K., Hapairai, L. K., Sun, L., Harper, E. I., Chen, Y., Eggleson, K. K., et al. (2017). Yeast interfering RNA larvicides targeting neural genes induce high rates of Anopheles larval mortality. Malar. J. 16, 461. doi:10.1186/s12936-017-2112-5
Naegeli, H., Birch, A. N., Casacuberta, J., De Schrijver, A., Gralak, M. A., Guerche, P., et al. (2018). Assessment of genetically modified maize MON 87411 for food and feed uses, import and processing, under Regulation (EC) No 1829/2003 (application EFSA-GMO-NL-2015-124). EFSA J. 16, e05310. doi:10.2903/J.EFSA.2018.5310
Naegeli, H., Bresson, J. L., Dalmay, T., Dewhurst, I. C., Epstein, M. M., Firbank, L. G., et al. (2019). Assessment of genetically modified maize MON 87427 × MON 89034 × MIR162 × MON 87411 and subcombinations, for food and feed uses, under Regulation (EC) No 1829/2003 (application EFSA-GMO-NL-2017-144). EFSA J. 17, e05848. doi:10.2903/J.EFSA.2019.5848
Niehl, A., Soininen, M., Poranen, M. M., and Heinlein, M. (2018). Synthetic biology approach for plant protection using dsRNA. Plant Biotechnol. J. 16, 1679–1687. doi:10.1111/pbi.12904
Nwokeoji, A. O., Kilby, P. M., Portwood, D. E., and Dickman, M. J. (2016). RNASwift: A rapid, versatile RNA extraction method free from phenol and chloroform. Anal. Biochem. 512, 36–46. doi:10.1016/j.ab.2016.08.001
Nwokeoji, A. O., Kumar, S., Kilby, P. M., Portwood, D. E., Hobbs, J. K., and Dickman, M. J. (2019). Analysis of long dsRNA produced in vitro and in vivo using atomic force microscopy in conjunction with ion-pair reverse-phase HPLC. Analyst 144, 4985–4994. doi:10.1039/c9an00954j
Nwokeoji, A. O., Kung, A.-W., Kilby, P. M., Portwood, D. E., and Dickman, M. J. (2017). Purification and characterisation of dsRNA using ion pair reverse phase chromatography and mass spectrometry. J. Chromatogr. A 1484, 14–25. doi:10.1016/J.CHROMA.2016.12.062
Obbard, D. J., Gordon, K. H. J., Buck, A. H., and Jiggins, F. M. (2009). The evolution of RNAi as a defence against viruses and transposable elements. Phil. Trans. R. Soc. B 364, 99–115. doi:10.1098/rstb.2008.0168
Ongvarrasopone, C., Roshorm, Y., and Panyim, S. (2007). A simple and cost effective method to generate dsRNA for RNAi studies in invertebrates. ScienceAsia 33, 035–039. doi:10.2306/scienceasia1513-1874.2007.33.035
Paces, J., Nic, M., Novotny, T., and Svoboda, P. (2017). Literature review of baseline information to support the risk assessment of RNAi-based GM plants. EFSA Support. Publ. 14, 1246E. doi:10.2903/SP.EFSA.2017.EN-1246
Paldi, N., Glick, E., Oliva, M., Zilberberg, Y., Aubin, L., Pettis, J., et al. (2010). Effective gene silencing in a microsporidian parasite associated with honeybee (Apis mellifera) colony declines. Appl. Environ. Microbiol. 76, 5960–5964. doi:10.1128/AEM.01067-10
Papademetriou, T. (2015). Restrictions on genetically modified organisms. Washington, D.C.: European Union. Library of Congress.
Papić, L., Rivas, J., Toledo, S., and Romero, J. (2018). Double-stranded RNA production and the kinetics of recombinant Escherichia coli HT115 in fed-batch culture. Biotechnol. Rep. 20, e00292. doi:10.1016/j.btre.2018.e00292
Park, M. G., Choi, J. Y., Park, D. H., Wang, M., Kim, H. J., and Je, Y. H. (2021). Simultaneous control of sacbrood virus (SBV) and Galleria mellonella using a Bt strain transformed to produce dsRNA targeting the SBV vp1 gene. Entomol. Gen. 41, 233–242. doi:10.1127/ENTOMOLOGIA/2021/1114
Park, M. G., Kim, W. J., Choi, J. Y., Kim, J. H., Park, D. H., Kim, J. Y., et al. (2020). Development of a Bacillus thuringiensis based dsRNA production platform to control sacbrood virus in Apis cerana. Pest Manag. Sci. 76, 1699–1704. doi:10.1002/PS.5692
Parker, K. M., and Sander, M. (2017). Environmental Fate of Insecticidal Plant-Incorporated Protectants from Genetically Modified Crops: Knowledge Gaps and Research Opportunities. Environ. Sci. Technol. 51, 12049–12057. doi:10.1021/acs.est.7b03456
Petrov, N., and Galabov, A. (2012). Conserved regions from Coxsackievirus genome as targets for gene silencing (RNAi). Sci. Technol. II (1), 32–36.
Pitino, M., Coleman, A. D., Maffei, M. E., Ridout, C. J., and Hogenhout, S. A. (2011). Silencing of Aphid Genes by dsRNA Feeding from Plants. PLoS ONE 6, e25709. doi:10.1371/journal.pone.0025709
Posiri, P., Ongvarrasopone, C., and Panyim, S. (2013). A simple one-step method for producing dsRNA from E. coli to inhibit shrimp virus replication. J. Virological Methods 188, 64–69. doi:10.1016/j.jviromet.2012.11.033
Price, D. R. G., and Gatehouse, J. a. (2008). RNAi-mediated crop protection against insects. Trends Biotechnol. 26, 393–400. doi:10.1016/j.tibtech.2008.04.004
Rajagopal, R., Sivakumar, S., Agrawal, N., Malhotra, P., and Bhatnagar, R. K. (2002). Silencing of midgut aminopeptidase N of Spodoptera litura by double-stranded RNA establishes its role as Bacillus thuringiensis toxin receptor. J. Biol. Chem. 277, 46849–46851. doi:10.1074/jbc.C200523200
Ramesh Kumar, D., Saravana Kumar, P., Gandhi, M. R., Al-Dhabi, N. A., Paulraj, M. G., and Ignacimuthu, S. (2016). Delivery of chitosan/dsRNA nanoparticles for silencing of wing development vestigial (vg) gene in Aedes aegypti mosquitoes. Int. J. Biol. Macromol. 86, 89–95. doi:10.1016/J.IJBIOMAC.2016.01.030
Roberts, A. F., Devos, Y., Lemgo, G. N. Y., and Zhou, X. (2015). Biosafety research for non-target organism risk assessment of RNAi-based GE plants. Front. Plant Sci. 6, 958. doi:10.3389/fpls.2015.00958
Sahin, U., Muik, A., Derhovanessian, E., Vogler, I., Kranz, L. M., Vormehr, M., et al. (2020). COVID-19 vaccine BNT162b1 elicits human antibody and TH1 T cell responses. Nature 586, 594–599. doi:10.1038/s41586-020-2814-7
San Miguel, K., and Scott, J. G. (2016). The next generation of insecticides: DsRNA is stable as a foliar-applied insecticide. Pest Manag. Sci. 72, 801–809. doi:10.1002/ps.4056
Sano, T., Nagayama, A., Ogawa, T., Ishida, I., and Oada, Y. (1997). Transgenic potato expressing a double- stranded RNA-specific ribonuclease is resistant to potato spindle tuber viroid Teruo. Nature 3, 3–8.
Savary, S., Ficke, A., Aubertot, J. N., and Hollier, C. (2012). Crop losses due to diseases and their implications for global food production losses and food security. Food Secur. 4, 519–537. doi:10.1007/s12571-012-0200-5
Shimizu, Y., Inoue, A., Tomari, Y., Suzuki, T., Yokogawa, T., Nishikawa, K., et al. (2001). Cell-free translation reconstituted with purified components. Nat. Biotechnol. 19, 751–755. doi:10.1038/90802
Shukla, J. N., Kalsi, M., Sethi, A., Narva, K. E., Fishilevich, E., Singh, S., et al. (2016). Reduced stability and intracellular transport of dsRNA contribute to poor RNAi response in lepidopteran insects. RNA Biol. 13, 656–669. doi:10.1080/15476286.2016.1191728
Sinnuengnong, R., Attasart, P., Smith, D. R., Panyim, S., and Assavalapsakul, W. (2018). Administration of co-expressed Penaeus stylirostris densovirus-like particles and dsRNA-YHV-Pro provide protection against yellow head virus in shrimp. J. Biotechnol. 267, 63–70. doi:10.1016/J.JBIOTEC.2018.01.002
Skelly, P. J., Da’dara, A., and Harn, D. A. (2003). Suppression of cathepsin B expression in Schistosoma mansoni by RNA interference. Int. J. Parasitol. 33 (4), 363–369. doi:10.1016/S0020-7519(03)00030-4
Sørensen, H. P., and Mortensen, K. K. (2005). Advanced genetic strategies for recombinant protein expression in Escherichia coli. J. Biotechnol. 115, 113–128. doi:10.1016/j.jbiotec.2004.08.004
Sturm, Á., Saskoï, É., Tibor, K., Weinhardt, N., and Vellai, T. (2018). Highly efficient RNAi and Cas9-based auto-cloning systems for C. Elegans research. Nucleic Acids Res. 46, e105. doi:10.1093/nar/gky516
Tabashnik, B. E., Brévault, T., and Carrière, Y. (2013). Insect resistance to Bt crops: lessons from the first billion acres. Nat. Biotechnol. 31, 510–521. doi:10.1038/nbt.2597
Tabashnik, B. E., and Carrière, Y. (2017). Surge in insect resistance to transgenic crops and prospects for sustainability. Nat. Biotechnol. 35, 926–935. doi:10.1038/nbt.3974
Takiff, H. E., Chen, S. M., and Court, D. L. (1989). Genetic analysis of the rnc operon of Escherichia coli. J. Bacteriol. 171, 2581–2590. doi:10.1128/jb.171.5.2581-2590.1989
Talakayala, A., Katta, S., and Garladinne, M. (2020). Genetic engineering of crops for insect resistance: An overview. J. Biosci. 45, 114. doi:10.1007/S12038-020-00081-Y
Tegel, H., Ottosson, J., and Hober, S. (2011). Enhancing the protein production levels in Escherichia coli with a strong promoter. FEBS J. 278, 729–739. doi:10.1111/j.1742-4658.2010.07991.x
Terenius, O., Papanicolaou, A., Garbutt, J. S., Eleftherianos, I., Huvenne, H., Kanginakudru, S., et al. (2011). RNA interference in Lepidoptera: An overview of successful and unsuccessful studies and implications for experimental design. J. Insect Physiology 57, 231–245. doi:10.1016/J.JINSPHYS.2010.11.006
Terpe, K. (2006). Overview of bacterial expression systems for heterologous protein production: From molecular and biochemical fundamentals to commercial systems. Appl. Microbiol. Biotechnol. 72, 211–222. doi:10.1007/s00253-006-0465-8
Thairu, M. W., Skidmore, I. H., Bansal, R., Nováková, E., Hansen, T. E., Li-Byarlay, H., et al. (2017). Efficacy of RNA interference knockdown using aerosolized short interfering RNAs bound to nanoparticles in three diverse aphid species. Insect Mol. Biol. 26, 356–368. doi:10.1111/imb.12301
Tian, H., Peng, H., Yao, Q., Chen, H., Xie, Q., Tang, B., et al. (2009). Developmental Control of a Lepidopteran Pest Spodoptera exigua by Ingestion of Bacteria Expressing dsRNA of a Non-Midgut Gene. PLoS ONE 4, e6225. doi:10.1371/journal.pone.0006225
Timmons, L., Court, D. L., and Fire, A. (2001). Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene 263, 103–112. doi:10.1016/S0378-1119(00)00579-5
Tunitskaya, V. L., and Kochetkov, S. N. (2002). Structural-functional analysis of bacteriophage T7 RNA polymerase. Biochemistry. 67, 1124–1135. doi:10.1023/A:1020911223250
Urquhart, W., Mueller, G. M., Carleton, S., Song, Z., Perez, T., Uffman, J. P., et al. (2015). A novel method of demonstrating the molecular and functional equivalence between in vitro and plant-produced double-stranded RNA. Regul. Toxicol. Pharmacol. 73, 607–612. doi:10.1016/j.yrtph.2015.09.004
Viswanath, V., Story, G. M., Peier, M., Petrus, M. J., Lee, V. M., Hwang, S. W., et al. (2000). Total silencing by intron-spliced hairpin RNAs. Nature 407, 319–320. doi:10.1038/35030305
Vogel, E., Santos, D., Mingels, L., Verdonckt, T.-W., and Broeck, J. V. (2019). RNA Interference in Insects: Protecting Beneficials and Controlling Pests. Front. Physiol. 9, 1912. doi:10.3389/fphys.2018.01912
Waltz, E. (2015). USDA approves next-generation GM potato. Nat. Biotechnol. 33, 12–13. doi:10.1038/nbt0115-12
Wang, M. B., Abbott, D. C., and Waterhouse, P. M. (2000). A single copy of a virus-derived transgene encoding hairpin RNA gives immunity to barley yellow dwarf virus. Mol. Plant Pathol. 1, 347–356. doi:10.1046/j.1364-3703.2000.00038.x
Wang, W., Wan, P., Lai, F., Zhu, T., and Fu, Q. (2018). Double-stranded RNA targeting calmodulin reveals a potential target for pest management of Nilaparvata lugens. Pest Manag. Sci. 74, 1711–1719. doi:10.1002/ps.4865
Wang, Y., Zhang, H., Li, H., and Miao, X. (2011). Second-generation sequencing supply an effective way to screen RNAi targets in large scale for potential application in pest insect control. PLoS ONE 6, 18644. doi:10.1371/journal.pone.0018644
Whalon, M. E., Mota-Sanchez, D., and Hollingworth, R. M. (2008). Global pesticide resistance in arthropods. Wallingford: CABI International.
Whitten, M. M. A., Facey, P. D., Del Sol, R., Fernández-Martínez, L. T., Evans, M. C., Mitchell, J. J., et al. (2016). Symbiont-mediated RNA interference in insects. Proc. R. Soc. B 283, 20160042. doi:10.1098/rspb.2016.0042
Yan, S., Qian, J., Cai, C., Ma, Z., Li, J., Yin, M., et al. (2020). Spray method application of transdermal dsRNA delivery system for efficient gene silencing and pest control on soybean aphid Aphis glycines. J. Pest Sci. 93, 449–459. doi:10.1007/s10340-019-01157-x
Yang, L., Tian, Y., Peng, Y. Y., Niu, J., and Wang, J. J. (2020). Expression dynamics of core RNAi machinery genes in pea aphids upon exposure to artificially synthesized dsRNA and miRNAs. Insects 11, 70. doi:10.3390/insects11020070
Yin, G., Sun, Z., Liu, N., Zhang, L., Song, Y., Zhu, C., et al. (2009). Production of double-stranded RNA for interference with TMV infection utilizing a bacterial prokaryotic expression system. Appl. Microbiol. Biotechnol. 84, 323–333. doi:10.1007/s00253-009-1967-y
Yoon, J. S., Sahoo, D. K., Maiti, I. B., and Palli, S. R. (2018). Identification of target genes for RNAi-mediated control of the Twospotted Spider Mite. Sci. Rep. 8, 14687. doi:10.1038/s41598-018-32742-2
Young, J., Luketina, R., and Marshall, A. M. (1977). The Effects on Temperature on Growth in vitro of Pseudomonas syringae and Xanthomonas pruni. J. Appl. Bacteriol. 42, 345–354. doi:10.1111/j.1365-2672.1977.tb00702.x
Yu, N., Christiaens, O., Liu, J., Niu, J., Cappelle, K., Caccia, S., et al. (2013). Delivery of dsRNA for RNAi in insects: An overview and future directions. Insect Sci. 20, 4–14. doi:10.1111/j.1744-7917.2012.01534.x
Yu, S. J., Pan, Q., Luo, R., Wang, C. L., Cheng, L. Y., Yang, J. S., et al. (2019). Expression of exogenous dsRNA by Lecanicillium attenuatum enhances its virulence to Dialeurodes citri. Pest Manag. Sci. 75, 1014–1023. doi:10.1002/ps.5210
Yu, X. D., Liu, Z. C., Huang, S. L., Chen, Z. Q., Sun, Y. W., Duan, P. F., et al. (2016). RNAi-mediated plant protection against aphids. Pest Manag. Sci. 72, 1090–1098. doi:10.1002/ps.4258
Zha, W., Peng, X., Chen, R., Du, B., Zhu, L., and He, G. (2011). Knockdown of Midgut Genes by dsRNA-Transgenic Plant-Mediated RNA Interference in the Hemipteran Insect Nilaparvata lugens. PLoS ONE 6, e20504. doi:10.1371/journal.pone.0020504
Zhang, J., Khan, S. A., Hasse, C., Ruf, S., Heckel, D. G., and Bock, R. (2015). Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids. Science 347, 991–994. doi:10.1126/science.1261680
Zhang, J., Khan, S. A., Heckel, D. G., and Bock, R. (2017). Next-Generation Insect-Resistant Plants: RNAi-Mediated Crop Protection. Trends Biotechnol. 35, 871–882. doi:10.1016/j.tibtech.2017.04.009
Zhang, W. (2018). Global pesticide use: Profile, trend, cost/benefit and more. Proc. Int. Acad. Ecol. Environ. Sci. 8, 1–27.
Zhong, C., Smith, N. A., Zhang, D., Goodfellow, S., Zhang, R., Shan, W., et al. (2019). Full-Length Hairpin RNA Accumulates at High Levels in Yeast but Not in Bacteria and Plants. Genes 10, 458. doi:10.3390/GENES10060458
Zhu, F., Xu, J., Palli, R., Ferguson, J., and Palli, S. R. (2011). Ingested RNA interference for managing the populations of the Colorado potato beetle, Leptinotarsa decemlineata. Pest Manag. Sci. 67, 175–182. doi:10.1002/ps.2048
Zimmerman, E. S., Heibeck, T. H., Gill, A., Li, X., Murray, C. J., Madlansacay, M. R., et al. (2014). Production of site-specific antibody-drug conjugates using optimized non-natural amino acids in a cell-free expression system. Bioconjug. Chem. 25, 351–361. doi:10.1021/bc400490z
Zotti, M., dos Santos, E. A., Cagliari, D., Christiaens, O., Taning, C. N. T., and Smagghe, G. (2018). RNA interference technology in crop protection against arthropod pests, pathogens and nematodes. Pest Manag. Sci. 74, 1239–1250. doi:10.1002/ps.4813
Keywords: dsRNA, RNAi, RNAi pest control, pest management, biopesticicdes
Citation: Hough J, Howard JD, Brown S, Portwood DE, Kilby PM and Dickman MJ (2022) Strategies for the production of dsRNA biocontrols as alternatives to chemical pesticides. Front. Bioeng. Biotechnol. 10:980592. doi: 10.3389/fbioe.2022.980592
Received: 28 June 2022; Accepted: 23 August 2022;
Published: 10 October 2022.
Edited by:
Ruobing Guan, Henan Agricultural University, ChinaCopyright © 2022 Hough, Howard, Brown, Portwood, Kilby and Dickman. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mark J. Dickman, bS5kaWNrbWFuQHNoZWZmaWVsZC5hYy51aw==
†These authors have contributed equally to this work and share first authorship
‡Present address: John D. Howard, Vector Biology Department, Liverpool School of Tropical Medicine, Liverpool, United Kingdom
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.