
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Bioeng. Biotechnol., 18 November 2022
Sec. Synthetic Biology
Volume 10 - 2022 | https://doi.org/10.3389/fbioe.2022.1023729
This article is part of the Research Topicds-RNA-based Pesticides: Production, Development, and Application TechnologyView all 9 articles
 Xifa Yang1†
Xifa Yang1† Shaokai Liu2†
Shaokai Liu2† Wenhui Lu1
Wenhui Lu1 Mengfang Du1
Mengfang Du1 Zhuangzhuang Qiao1
Zhuangzhuang Qiao1 Zhen Liang1
Zhen Liang1 Yiting An1
Yiting An1 Jing Gao1
Jing Gao1 Xiang Li1*
Xiang Li1*The brown planthopper (BPH; Nilaparvata lugens) is an important pest in rice cultivation, and chemical pesticide over-use and ineffectiveness of existing Bt transgenic rice against piercing-sucking insects make novel control methods necessary. RNA interference (RNAi) biopesticide is a new type of product with high efficiency and specificity and are simple to use. The Notch signaling pathway has extensive and important physiological functions and plays a key role in the development of insects. In this study, two key ligand genes of the Notch signaling pathway, delta (dl) and jagged (jag), were selected and their lethal effects and functional analysis were systematically evaluated using a stable short-winged population (Brachypterous strain) and a long-winged population (Macropterous strain) of BPHs. The full-length coding sequences of Nldl and Nljag comprised 1,863 and 3,837 base pairs, encoding 620 and 1,278 amino acids, respectively. The nucleic acid sequences of Nldl and Nljag were identical between the two strains. The expression levels of Nldl and Nljag were relatively high in the head of the nymphs, followed by those in the abdomen. Through RNAi treatment, we found that injection of BPH nymphs of both strains with dsNldl (10–50 ng/nymph) or dsNljag (100 ng/nymph) produced lethal or teratogenic effects. dsRNA treatment showed excellent inhibitory effects on the expression of target genes on days 1 and 5, suggesting that RNAi rapidly exhibits effects which persist for long periods of time in BPHs. Taken together, our results confirm the potential of Nldl and Nljag as target genes of RNAi biopesticides, and we propose optimized dosages for the control of BPHs.
The brown planthopper (BPH; Nilaparvata lugens, Hemiptera: Delphacidae) is a notorious pest in rice cultivation which can cause devastating damage. Adults and nymphs prefer to gather at the base of the rice plexus and suck the sap from stems and leaves, resulting in the loss of rice nutrients and decreased yield and quality (Alagar et al., 2007; Vanitha et al., 2011). In addition to direct feeding, BPHs are also a vector of pathogens including rice grassy stunt virus, rice ragged stunt virus, and rice wilted stunt virus (Hibino, 1995; Li et al., 2011). Honeydew excreted by BPHs contains large amounts of sugar, which is highly conducive to the propagation of bacteria and fungi, leading to mildew and wilting of rice (Sōgawa, 2003; Wei et al., 2010). Depending on habitat quality, adult BPHs can develop into a highly mobile long-winged morph or a highly fecund short-winged morph (Liu et al., 2020; Li et al., 2021). The differential formation of two wing morphs aggravates the damage caused by BPH and increases the difficulty of controlling its spread. In normal years, BPH infestation can cause a reduction of 10%–20% in rice production, and in severe cases, it can result in crop failure (Heong and Hardy, 2009).
RNA interference (RNAi) is a conserved regulatory mechanism mediated by RNA in eukaryotes. RNAi effectors target genes paired with homologous sequences and effectively suppress gene expression at the transcriptional or post-transcriptional level (Carthew and Sontheimer, 2009; Chen et al., 2010). RNAi biopesticide is a new type of product with high efficiency and specificity and which are easy to use, thus they are expected to constitute a scientific and technological revolution and facilitate sustainable agricultural development (Li et al., 2022). RNAi biopesticides can specifically interfere with the expression of designated genes of agricultural pests, pathogens, and weeds, thereby hindering their growth or preventing them from exerting harmful effects. In 2002, the feasibility of RNAi technology for gene interference in insects was first demonstrated using Hemolin-RNAi in Hyalophora cecropia (Bettencourt et al., 2002). Since then, numerous studies have demonstrated that exogenous synthesized dsRNA applied through feeding, injecting, or spraying can induce RNAi effects in various insects (Kim et al., 2015; Vogel et al., 2019).
Genes that are essential for insect growth and development can serve as targets for RNAi biopesticides. A very wide range of targets available for RNAi biopesticides can decrease the upfront development cost, which constitutes an important advantage of RNAi-based pesticides, compared to chemical pesticides. In a previous study, 290 insect genes were selected and silenced, and most of them led to a lethal effect (some could also cause larval maldevelopment), thus they may serve as insecticidal targets (Baum et al., 2007). Similarly, 100 potential target genes were isolated from random genes of Tribolium castaneum, and dsRNA injection inhibited the expression of these genes at the larval or pupal stage and resulted in >90% mortality (Ulrich et al., 2015).
The Notch protein is a heterodimer formed by a non-covalent bond between an extracellular domain subunit and an intracellular domain subunit (Kopan and Ilagan, 2009). Notch is a highly conserved transmembrane protein of vertebrates and invertebrates that can transmit signals unidirectionally and regulate cell proliferation, differentiation, metabolism, and apoptosis through collateral inhibition (Palomero et al., 2006; Lewis et al., 2007; Joshi et al., 2009). During insect ontogeny, Notch can affect normal development of the embryonic ectoderm, mesoderm, central nervous system, germ cells, eyes, wings, etc. (Bray, 2006; Irles et al., 2016; Hu et al., 2022). If Notch is abnormally expressed in the early stage of embryonic development, it may lead to abnormal embryonic differentiation and death (Kidd and Lieber, 2016).
Notch ligands are transmembrane proteins located on the surface of cells in close proximity to the Notch receptors, and they are also composed of extracellular and intracellular domains (Schroeter et al., 1998). When performing its function, Notch must bind to a ligand protein, i.e., Delta (Dl) or Jagged (Jag), and induce a conformational change to release the intracellular subunit. The modified intracellular subunits are transported into the nucleus and act as transcription factors to regulate the expression of downstream genes (Struhl and Adachi, 1998; Mumm et al., 2000; Mumm and Kopan, 2000; Gordon et al., 2007). Notch has both cis- and trans- regulation modes on its ligands. In addition, Notch exerts cis-inhibition effects on Jag, suggesting complex and diverse functions of Notch and emphasizing the critical role of ligands in signal transduction through the Notch signaling pathway (Matsuda and Chitnis, 2009; Fiuza et al., 2010; Yamamoto et al., 2012; Fleming et al., 2013).
In the current study, the cDNA sequences of the Notch ligand genes Nldl and Nljag were cloned and analyzed. Furthermore, the nucleotide homology and phylogenetic relationships of Nldl/Nljag between BPH and several common agricultural insects were investigated. The spatiotemporal expression profiles of Nldl and Nljag in BPH nymphs were measured by qPCR, and the effects of RNAi-Nldl/Nljag on the development and lethality of BPH were investigated by microinjection method. Taken together, this study evaluated the potential application of Nldl and Nljag as an RNAi biopesticides for BPH control.
BPHs were collected from paddy fields of the Huazhong Agricultural University, Wuhan, China. According to Morooka and Tojo (1992), BPHs were reared in growth chambers containing rice seedlings (variety: Taichung Native 1) and were screened for more than seventy successive generations to cultivate and obtain a stable long-winged population (Macropterous strain, long-winged rate approximately 85% ± 5%) and a short-winged strain (Brachypterous strain, short-winged rate approximately 100%). The rearing conditions included a photoperiod of 14 h, 27 ± 1°C, and 60% ± 10% relative humidity. The BPHs of the Brachypterous and Macropterous strain tested in this study were generously donated by Professor Hongxia Hua from Huazhong Agricultural University.
Total RNA was isolated and extracted from a mixture of BPH adults and nymphs at different instar stages of the Brachypterous or Macropterous strain using TsingZol Reagent (Tsingke Biotech, Beijing, China). cDNA synthesis reactions were performed using 1 μg total RNA and an All-in-one First Strand cDNA Synthesis Kit Ⅲ for qPCR (Sevenbio, Beijing, China), according to the manufacturer’s instructions. High-accuracy sequences were amplified with 2 × KOD PCR MasterMix (Bioman, Beijing, China). The correctness of PCR products was confirmed by sequencing of Tsingke Biotech. The experimental reaction parameters refer to the default parameters given by the kits. Nucleic acid homology analysis was performed in the Blast module (https://blast.ncbi.nlm.nih.gov/Blast.cgi) of the National Center for Biotechnology Information (NCBI) database. Expasy (http://web.expasy.org) was employed to predict the sequence structures of Nldl and Nljag. MEGA 11.0 software was used to perform the sequence alignment and phylogenetic analysis with 1,000 replicates of the bootstrap test. STRING (https://cn.string-db.org/) was used to identify physical interactions and co-expression of the test proteins.
BPH nymphs were divided into three body segments, i.e., the head, thorax, and abdomen, and additional appendages: the legs. qPCR was employed to investigate the expression profiles of Nldl and Nljag in these tissues of second- to fifth-instar BPH nymphs. Each sample contained 50 corresponding nymph tissues of the Macropterous strain or the Brachypterous strain. Total RNA was isolated and extracted from different tissues of BPHs using TRIzol reagent (Sangon Biotech, Shanghai, China). cDNA synthesis was performed using 1 μg total RNA and a TransScript Uni All-in-One First-Strand cDNA Synthesis SuperMix for qPCR (TransGen Biotech, Beijing, China), according to the manufacturer’s instructions. The amplification efficiencies of the qPCR primers of Nldl and Nljag were determined by using more than five concentrations of cDNA templates for amplification on QuantStudio3 Real-time PCR system. Detection of gene expression levels by qPCR was performed on an Applied Biosystems 7,500 Fast Real-time PCR system using the Hieff qPCR SYBR Green Master Mix (Yeasen Biotech, Shanghai, China) according to the manufacturer’s instructions. The thermal cycle conditions for qPCR were 95°C for 5 min, followed by 40 cycles of 95°C for 10 s and 60°C for 30 s. The experimental reaction parameters refer to the default parameters given by the kits. The housekeeping gene Nlactin1 (GenBank accession EU179846.1) was used as a reference. The relative expression level of the test gene was calculated and compared according to Livak and Schmittgen (2001). Signal intensities of the target genes are presented as average values. Each sample was examined using three independent replicates, and each treatment was performed using three biological replicates. qPCR primers are listed in Table 1.
The PCR template (cDNA) for Nldl and Nljag was amplified using gene-specific primers containing T7 polymerase sites and with 2 × Taq Plus PCR Master Mix (Solarbio, Beijing, China). dsRNA was generated using a T7 RNAi Transcription Kit (Vazyme Biotech, Nanjing, China) and was dissolved in diethyl pyrocarbonate-treated. Third-instar nymphs were injected with dsNldl (10, 25, or 50 ng/nymph) or dsNljag (100 ng/nymph) dissolved in 20 nl volume of the nuclease-free water on the second day after ecdysis from the ventral side of the thorax using a Nanoliter 2010 microinjector (WPI, Sarasota, United States). An equivalent amount of dsGFP (GenBank accession U76561) was injected as a control. The survival numbers of injected BPHs were recorded daily until adult emergence. Three replicates were used per treatment, and each replicate comprised 50 nymphs. The whole bodies of five treated-BPH were sampled randomly on the first and fifth day after injection (corresponding to third- and fifth-instar nymphs) for evaluating RNAi delivery efficiency by qPCR. Adult phenotypes were observed and photographed using a stereomicroscope (Olympus szx16, Tokyo, Japan). Primers used for dsRNA synthesis are shown in Table 1.
Statistical analyses were performed using SPSS 20.0 software (IBM, Armonk, United States). Differences between treatments regarding RNAi efficiency and survival responses were compared using Student’s t-tests. Expression profiles of Nldl or Nljag in the same tissue of BPH nymph at different instars were analyzed using one-way analysis of variance followed by Tukey’s test for multiple comparisons. Statistical significance is reported at p < 0.05.
cDNA of dl and jag was amplified using BPHs of the Brachypterous and Macropterous strain, respectively. The full-length coding sequence of Nldl comprised 1,863 base pairs (bp), encoding 620 amino acids, with a molecular weight of 67.25 kDa (Figure 1). The Nljag full-length coding sequence comprised 3,837 bp, encoding 1,278 amino acids; its molecular weight was 137.83 kDa (Figure 2). The nucleic acid sequences of Nldl and Nljag were identical between the two strains.

FIGURE 1. Nucleotide and deduced amino acid sequence of Nldelta. The target sequence of dsNldelta is indicated in grey; the fragment amplified by qPCR is indicated by a box.

FIGURE 2. Nucleotide and deduced amino acid sequences of Nljagged. The target sequence of dsNljagged is indicated in grey; the fragment amplified by qPCR is indicated by a box.
The amino acid sequences of Dl and Jag of 15 agriculturally important insects (including natural enemies and pests) of the Hemiptera, Coleoptera, Diptera, Orthoptera, Hymenoptera, etc. were collected and used for phylogenetic analyses. The evolutionary pattern of NlDl and NlJag did not produce a consistent picture. With regard to Dl, N. lugens appeared to be more closely related to Drosophila melanogaster and Homalodisca vitripennis (Figure 3A). However, the Jag sequence of N. lugens suggested that it was more closely related to Hemipteran insects such as H. vitripennis, Bemisia tabaci, and Cimex lectularius (Figure 3B). Through the nucleic acid homology analysis in the Blast module of the NCBI database, it was found that the nucleotide sequence of dl had no more than 76% homology with other species (mainly insects), while the nucleotide sequence homology of jag and other species (mainly insects) is no more than 71%.
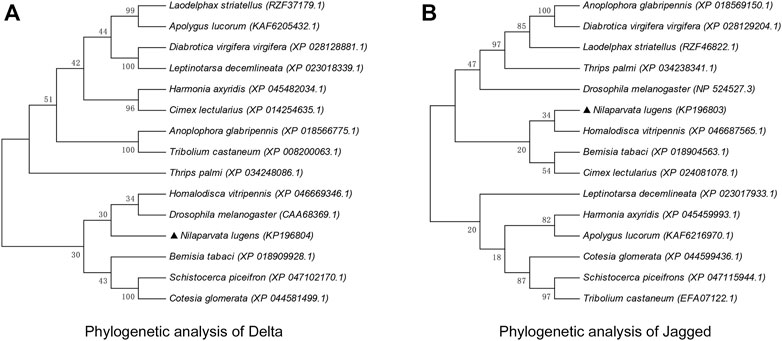
FIGURE 3. Phylogenetic analysis of Delta (A) and Jagged (B). The phylogenetic tree was constructed through neighbor-joining using MEGA11.0 software. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1,000 replicates) is shown above the branches. Amino acid sequences of Dl and Jag (and its homologous protein Ser) were retrieved from the NCBI database; the respective GenBank accession numbers are shown in brackets after the Latin species name.
The amplification specificity and amplification efficiency of the qPCR primers of Nldl and Nljag were first examined. The results showed that the amplification efficiencies of the primers of Nldl and Nljag were 90.65% and 91.44%, respectively, and no non-specific amplification occurred, confirming the availability of these two pairs of qPCR primers. The expression levels of Nldl and Nljag were relatively high in the head, followed by those in the abdomen (Figure 4). Nldl expression was relatively high in the abdomen of fourth- to fifth-instar nymphs (Figures 4A,B). The expression of Nljag in the thorax of fourth-instar nymphs of the Macropterous strain was significantly higher than that in other instars (Figure 4D).
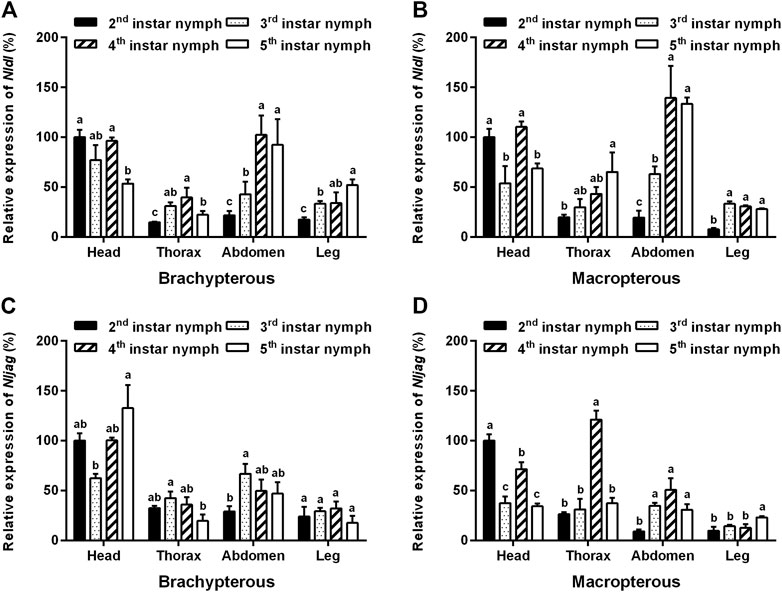
FIGURE 4. Spatiotemporal expression analysis of Nldelta and Nljagged in BPH nymphs by qPCR detection. (A) Expression profile of Nldelta in the Brachypterous strain. (B) Expression profile of Nldelta in the Macropterous strain. (C) Expression profile of Nljagged in the Brachypterous strain. (D) Expression profile of Nljagged in the Macropterous strain. Shown are the means ± standard error of three biological replicates, with 50 respective nymph tissues per replicate. Nlactin1 was used as a housekeeping gene for quantitation of relative expression levels of the test genes. Data represent the mean ± standard error (n = 3). The expression level of the test gene in the head of the second-instar nymph was defined as 100%. Different letters on the top of the columns indicate significant differences in expression levels between instars in the same tissue, as assessed using Tukey’s test (p < 0.05).
Different doses of dsRNA were injected from the ventral side of thorax into the third-instar BPH nymphs of the Brachypterous and Macropterous strain, respectively. dsRNA delivery efficiency was measured on the first and fifth day after treatment. Nldl-RNAi and Nljag-RNAi showed excellent interference efficiency and persistent effects (Figure 5). The expression levels of Nldl and Nljag decreased by 46.65% and 44.48%, respectively, in the Brachypterous strain on day 1 after treatment, and they decreased by 58.67% and 59.79% in the Macropterous strain (Figures 5A,C). On day 5, the expression levels of Nldl and Nljag in the Brachypterous strain were 58.67% and 52.35% of those in the control group, and 46.04% and 49.37% in the Macropterous strain, respectively (Figures 5B,D).
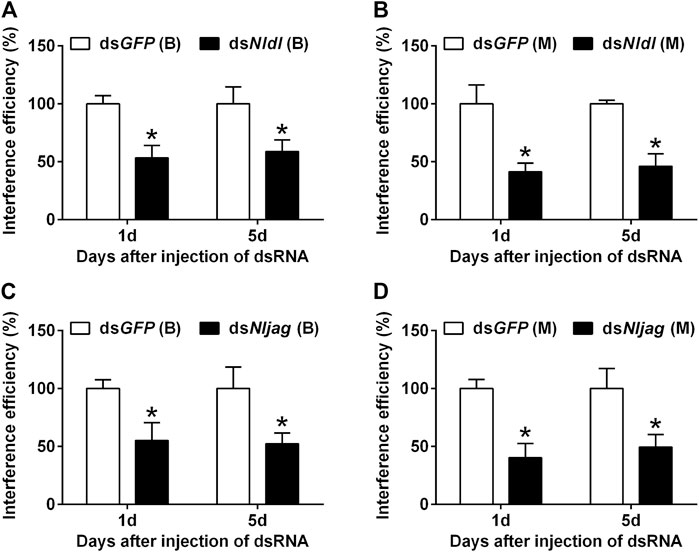
FIGURE 5. Interference efficiency of Nldelta-RNAi and Nljagged-RNAi in BPH nymphs. (A) Interference efficiency of Nldelta in the Brachypterous strain. (B) Interference efficiency of Nldelta in the Macropterous strain. (C) Interference efficiency of Nljagged in the Brachypterous strain. (D) Interference efficiency of Nljagged in the Macropterous strain. Shown are the means ± standard error of three replicates, with ten nymphs per replicate. Third-instar nymphs were injected with 25 ng dsNldelta or 100 ng dsNljagged. Equivalent dsGFP was used as a control. Nlactin1 was used as a housekeeping gene quantitation of relative expression levels of the test genes. Data represent the mean ± standard error (n = 3). Asterisks indicate significant differences in expression levels between dsGFP and dsNldelta/dsNljagged, as assessed using Student’s t-tests (p < 0.05).
Third-instar nymphs treated with 50 ng dsNldl produced very high mortality rates in the current study, and no nymphs reached the adult stage (Figures 6A,B). We thus reduced the injection dose to 25 ng/nymph, and approximately 2% of the Brachypterous and Macropterous strain nymphs successfully emerged (Figures 6C,D). These few survivors of both the long-winged and short-winged morphs produced a phenotype unable to close its wings (Figure 7). At 10 ng/nymph, the survival rate 6 days after treatment was 34.67% in the Brachypterous strain and 18.67% in the Macropterous strain, and 19.33% and 6.67% of the treated nymphs, respectively, successfully emerged and showed deformities rendering them unable to close their wings (Figures 6E,F). This constitutes a reference dosage for the application of dsNldl in the field control of BPH.
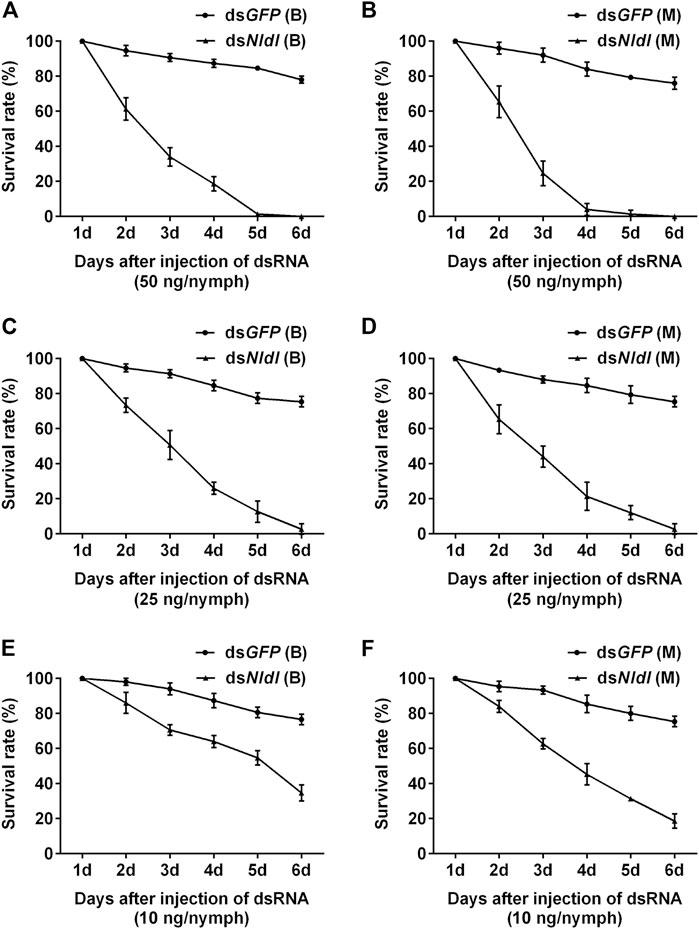
FIGURE 6. Survival rates of BPH developed from nymphs treated with dsNldelta. (A,B): Survival rate of 50 ng dsNldelta injection of the Brachypterous (A) and Macropterous strain (B). (C,D): Survival rate of 25 ng dsNldelta injection of the Brachypterous (C) and Macropterous strain (D). (E,F): Survival rate of 10 ng dsNldelta injection of the Brachypterous (E) and Macropterous strain (F). The nymphs injected with dsRNA were third-instar nymphs on the second day after ecdysis. The test nymphs began transforming to adults on the sixth day, and all surviving nymphs developed into adults on the seventh day. We therefore recorded nymph survival rates on the sixth day after treatment and additionally recorded the emergence rates. Equivalent dsGFP was used as a control. Each value is a mean of three replicates, with 50 nymphs per replicate. Data represent the mean ± standard error (n = 3).

FIGURE 7. Phenotypes of BPH adults developed from nymphs treated with dsNldelta. (A,B): Phenotypes of BPH adults of the Brachypterous strain with dsGFP-treatment (A) or dsNldelta-treatment (B). (C,D): Phenotypes of BPH adults of the Macropterous strain with dsGFP-treatment (C) or dsNldelta-treatment (D). Third-instar nymphs were injected with 25 ng dsNldelta. Equivalent dsGFP was used as a control.
Injection with 100 ng dsNljag caused a significant lethal effect on BPH nymphs. The survival rates after Nljag-RNAi injection in the Brachypterous and Macropterous strains were 22.00% and 18.00%, respectively, on day 6 after treatment, which were significantly lower than those of the control group (77.33%, p = 0.005328 and 74.00%, p = 0.002963, respectively) (Figure 8). On days 6 or 7 after dsNljag-treatment, the surviving nymphs developed into adults, and all of them showed curled wings (Figure 9). Neither dsNldl nor dsNljag injection caused obvious developmental delay.
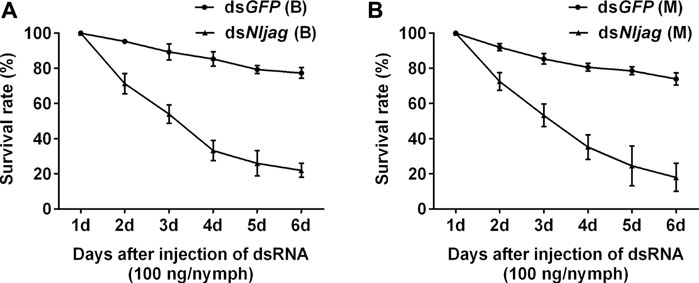
FIGURE 8. Survival rates of BPH developed from nymphs treated with dsNljagged. (A): Survival rate of 100 ng dsNljagged injection of the Brachypterous strain. (B): Survival rate of 100 ng dsNljagged injection of the Macropterous strain. Third-instar nymphs on the second day after ecdysis were injected with dsRNA. Equivalent dsGFP was used as a control. Each value is a mean of three replicates, with 50 nymphs per replicate. Data represent the mean ± standard error (n = 3).

FIGURE 9. Phenotypes of BPH adults developed from nymphs treated with dsNljagged. (A,B): Phenotypes of BPH adults of the Brachypterous strain with dsGFP-treatment (A) or dsNljagged-treatment (B). (C,D): Phenotypes of BPH adults of the Macropterous strain with dsGFP-treatment (C) or dsNljagged-treatment (D). Third-instar nymphs were injected with 100 ng dsNljagged. Equivalent dsGFP was used as a control.
At present, no respective data of BPH is available from public databases; however, the function of the Notch signaling pathway is conserved in most species. Therefore, we chose D. melanogaster as the model insect to predict the partner proteins of Dl and Serrate (Ser, homologous to Jag). Through STRING analysis, both Dl and Ser were found to be closely related to seven proteins including Notch, Mind-bomb, Neuralized, Suppressor of hairless protein, Kuzbanian, Presenilin, and Fringe (Figure 10). Furthermore, Dl interacts specifically with Scabrous and Hairless (Figure 10A), while Ser interacts specifically with Deltex and Mastermind (Figure 10B).
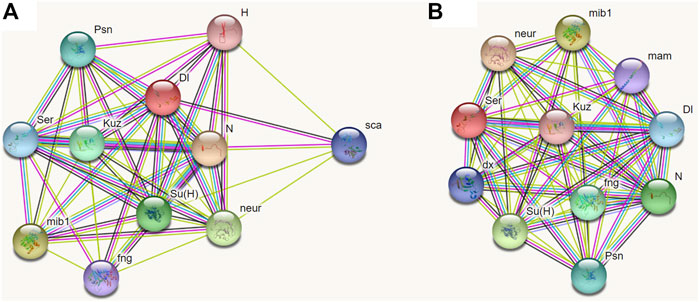
FIGURE 10. Prediction of interacting proteins of Delta and Serrate in Drosophila melanogaster by STRING analyses. (A): Interacting proteins of Delta. (B): Interacting proteins of Serrate. Note: N, Notch; mib1, Mind-bomb; neur, Neuralized; Su(H), Suppressor of hairless protein; Kuz, Kuzbanian; Psn, Presenilin; Ser, Serrate; fng, Fringe; Dl, Delta; sca, Scabrous; H, Hairless; dx, Deltex; mam, Mastermind.
Due to the high accuracy of RNAi treatment compounds, high degradability, and environmental friendliness, RNA biopesticides are considered “the third revolution in the history of agriculture” (Li et al., 2022; Yang et al., 2022), and RNAi biopesticide is a promising novel approach in the control of BPH. In the present study, two key ligand genes (dl and jag) of the Notch signaling pathway were selected and were systematically evaluated regarding their potential as targets for RNAi biopesticides. Short-winged BPHs are more reproductive and destructive, whereas long-winged adults are more migratory (Xu et al., 2015; Zhang et al., 2017). This difference in characteristics may require differentiated control measures. It is almost impossible to distinguish the wing morph of adults before emergence, which limits differentiated research on nymph control. To address this problem, we cultivated a stably long-winged (Macropterous strain) and a short-winged population (Brachypterous strain) of BPH strains through more than 70 generations of continuous cultivation, and we evaluated the RNAi effects of Nldl and Nljag in these two strains. In our observations and some previous reports, the RNAi experiment performance in these two strains was not significantly different from that of wild-type BPH (Li et al., 2019a; 2019b, 2021). These cultivated strains have thus become precious and scarce materials for the research on BPH.
We first cloned the cDNA sequences of dl and jag in both strains and found no nucleic acid sequence mutations, suggesting that there is no need to distinguish strains when designing dsRNA for interference. We selected 15 species for phylogenetic analysis of Dl and Jag respectively, however, the two proteins did not produce congruent results, suggesting evolutionary and functional differences between them. Although dl and jag have high homology with various insects (no more than 76%), we can still select suitable sequences to specifically interfere with BPH. If there are enough nucleic acid databases available in the future, we can even develop dsdl or dsjag to control multiple pests simultaneously. The related genes of the Notch signaling pathway mainly exist in animals, indicating that it should be difficult to interfere with the homologous genes in rice when dsNldl and dsNljag are applied as RNAi biopesticides. Furthermore, the identity of dl and jag with human homologous genes is far lower than those of insects. As long as the dsRNA targeting sequences are properly screened and designed, they will not be harmful to humans.
Through spatiotemporal expression analysis by qPCR, we found that Nldl and Nljag were higher expressed in the head and abdomen, indicating that these two genes were involved more in neural activity and metabolism. In particular, the expression level of Nljag was the highest in the thorax of fourth-instar nymphs of the Macropterous strain, representing the critical period of wing-pad development (Li et al., 2019b; Zhang et al., 2019). We therefore speculate that this gene may be involved in the wing development of BPH, as confirmed by adult wing deformity caused by Nljag-knockdown.
Through RNAi treatment, we found that injection of BPH nymphs of both Brachypterous and Macropterous strain with dsNldl or dsNljag produced the expected lethal effects. dsNldl at only 10 ng/nymph caused failure to emerge in >80% of the nymphs. Even if some treated nymphs can survive to adulthood, wing development deformities caused by dsRNA treatment may also reduce their mobility, fitness, and reproductive success. dsRNA treatment showed excellent inhibitory effects on the expression of target genes on days 1 and 5, which suggested that RNAi works rapidly and its effects are persistent in BPH. We have also tried to increase the injection dosage to detect whether the interference efficiency can be improved, but the change is not obvious. However, this does not prevent us from obtaining the stable adult phenotypes. The phenotypes after dsRNA treatment can obtain 100% consistent phenotypes regardless of whether the dosages were increased or decreased. Our findings support the potential of these two genes as RNAi biopesticide targets for controlling BPH. It should be noted that with the same injection dosage of dsNldl, mortality differed between the Brachypterous and Macropterous strains. Interference efficiency of dsRNA was higher in the Macropterous than in the Brachypterous strain. This may be related to the physiological tolerance of different strains of nymphs. Rice at the yellow-ripe stage (relatively low nutritional value) can stimulate the development of more long-winged BPH, whereas rice at the tillering-stage (relatively high nutritional value) induces development of more short-winged adults (Liu et al., 2020; Li et al., 2021). Therefore, in the future control and when dsNldl is used as an RNAi biopesticide, the dosage should be adjusted according to the different growth periods of rice.
To further explore the functions of Dl and Jag in insects, the associated proteins of these two ligands were predicted using STRING software. However, the database did not include data on N. lugens, nor on Hemiptera. Referring to the phylogenetic analysis of Dl, N. lugens is closely related to D. melanogaster, we thus used D. melanogaster as a model insect to predict the respective partner protein of Dl and Jag/Ser. As expected, most of these co-expressed proteins were associated with signal transduction and functioning of the Notch signaling pathway, and most of the predicted associated proteins of Dl and Jag/Ser were identical. According to our findings, the lethal effect produced by dsNldl injection was stronger than that of dsNljag treatment, and there were also obvious differences in the phenotype after emergence. We propose that these differences may originate from differences in associated proteins. In detail, Dl is specifically associated with Scabrous and Hairless, which may be responsible for the higher lethality. The Deltex and Mastermind associated with Jag/Ser may be the cause of the curling of wings in adults. However, these speculations need further verification.
Our study preliminarily investigated the expression profiles and functions of dl and jag in BPH and confirmed their application potential. This potential was manifested by the outstanding lethal and teratogenic effects of dl and jag on the BPH. We further propose dosages regarding target genes of RNAi biopesticide to control BPH. RNAi biopesticides can be delivered in a variety of ways, mainly divided into resistant plant-based plant-incorporated protectants and non-plant-incorporated protectants. It should be emphasized that studies have demonstrated that transgenic plants expressing dsRNA have poor control effects on a variety of piercing-sucking insects, including the BPH, mainly because these insects are difficult to effectively consume sufficient dsRNA during the feeding process (Kim and Zhang, 2022). Delivery of dsRNA by feeding is also a common method in research and production, especially for Coleoptera and Lepidoptera insects. But for oligophagous BPH, the attraction of food attractants is significantly lower than that of host rice plants in the field. Fortunately, RNAi biopesticides can also follow the application methods of traditional chemical pesticide and are designed to exert a pest control role by the means of spray or root-irrigation, although several problems remain to be addressed before practical application, including risk assessment, cost control, and application methods. No fully commercialized non-PIP RNAi-based biopesticide products have been released to the market thus far (Kunte et al., 2020; Liu et al., 2020). However, RNAi biopesticides bear considerable application value and are expected to become an important means of integrated pest control.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author.
XY and XL contributed to the conception and design of the study. XL and MD wrote and revised the draft of the manuscript. ZQ, ZL, YA, and JG conducted the experiment, SL and WL performed the analysis. All authors have read and agreed to the published version of the manuscript.
This work was supported by the National Natural Science Foundation of China (31970472); and the Henan Agricultural Research System (HARS-22-09-G3).
The brown planthoppers of the Brachypterous and Macropterous strain tested in this study were generously donated by Professor Hongxia Hua from Huazhong Agricultural University.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Alagar, M., Suresh, S., Samiyappan, R., and Saravanakumar, D. (2007). Reaction of resistant and susceptible rice genotypes against Brown planthopper (Nilaparvata lugens). Phytoparasitica 35 (4), 346–356. doi:10.1007/bf02980697
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of Coleopteran insect pests through RNA interference. Nat. Biotechnol. 25 (11), 1322–1326. doi:10.1038/nbt1359
Bettencourt, R., Terenius, O., and Faye, I. (2002). Hemolin gene silencing by ds–RNA injected into Cecropia pupae is lethal to next generation embryos. Insect Mol. Biol. 11 (3), 267–271. doi:10.1046/j.1365–2583.2002.00334.x
Bray, S. J. (2006). Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol. 7 (9), 678–689. doi:10.1038/nrm2009
Carthew, R. W., and Sontheimer, E. J. (2009). Origins and mechanisms of miRNAs and siRNAs. Cell 136 (4), 642–655. doi:10.1016/j.cell.2009.01.035
Chen, H. M., Chen, L. T., Patel, K., Li, Y. H., Baulcombe, D. C., and Wu, S. H. (2010). 22-Nucleotide RNAs trigger secondary siRNA biogenesis in plants. Proc. Natl. Acad. Sci. U. S. A. 107 (34), 15269–15274. doi:10.1073/pnas.1001738107
Fiuza, U. M., Klein, T., Arias, M. A., and Hayward, P. (2010). Mechanisms of ligand–mediated inhibition in notch signaling activity in Drosophila. Dev. Dyn. 239 (3), 798–805. doi:10.1002/dvdy.22207
Fleming, R. J., Hori, K., Sen, A., Filloramo, G. V., Langer, J. M., Obar, R. A., et al. (2013). An extracellular region of serrate is essential for ligand-induced cis-inhibition of Notch signaling. Development 140 (9), 2039–2049. doi:10.1242/dev.087916
Gordon, W. R., Vardar-Ulu, D., Histen, G., Sanchez-Irizarry, C., Aster, J. C., Blacklow, S. C., et al. (2007). Structural basis for autoinhibition of Notch. Nat. Struct. Mol. Biol. 14 (4), 295–300. doi:10.1038/nsmb1227
Heong, K., and Hardy, B. (2009). Planthoppers: New threats to the sustainability of intensive rice production systems in Asia. Los Baños, the Philippines: International Rice Research Institute, 1–460.
Hibino, H. (1995). Biology and epidemiology of rice viruses. Annu. Rev. Phytopathol. 34, 249–274. doi:10.1146/annurev.phyto.34.1.249
Hu, D. B., Xiao, S., Wang, Y., and Hua, H. X. (2022). Notchis an alternative splicinggene in Brown planthopper, Nilaparvata lugens. Arch. Insect Biochem. Physiol. 110, e21894. doi:10.1002/arch.21894
Irles, P., Elshaer, N., and Piulachs, M. D. (2016). The Notch pathway regulates both the proliferation and differentiation of follicular cells in the panoistic ovary of Blattella germanica. Open Biol. 6 (1), 150197. doi:10.1098/rsob.150197
Joshi, I., Minter, L. M., Telfer, J., Demarest, R. M., Capobianco, A. J., Aster, J. C., et al. (2009). Notch signaling mediates G1/S cell-cycle progression in T cells via cyclin D3 and its dependent kinases. Blood 113 (8), 1689–1698. doi:10.1182/blood-2008-03-147967
Kidd, S., and Lieber, T. (2016). Mechanism of Notch pathway activation and its role in the regulation of olfactory plasticity in Drosophila melanogaster. PLoS One 11 (3), e0151279. doi:10.1371/journal.pone.0151279
Kim, D. S., and Zhang, J. (2022). Strategies to improve the efficiency of RNAi-mediated crop protection for pest control. Entomol. Gen. (Early Access). doi:10.1127/entomologia/2022/1638
Kim, Y. H., Issa, M. S., Cooper, A., and Zhu, K. Y. (2015). RNA Interference: Applications and advances in insect toxicology and insect pest management. Pesticide Biochem. Physiology 120, 109–117. doi:10.1016/j.pestbp.2015.01.002
Kopan, R., and Ilagan, M. X. (2009). The canonical Notch signaling pathway: Unfolding the activation mechanism. Cell 137 (2), 216–233. doi:10.1016/j.cell.2009.03.045
Kunte, N., McGraw, E., Bell, S., Held, D., and Avila, L. A. (2020). Prospects, challenges and current status of RNAi through insect feeding. Pest Manag. Sci. 76 (1), 26–41. doi:10.1002/ps.5588
Lewis, H. D., Leveridge, M., Strack, P. R., Haldon, C. D., O’Neil, J., Kim, H., et al. (2007). Apoptosis in T cell acute lymphoblastic leukemia cells after cell cycle arrest induced by pharmacological inhibition of Notch signaling. Chem. Biol. 14 (2), 209–219. doi:10.1016/j.chembiol.2006.12.010
Li, J., Chen, Q., Wang, L., Liu, J., Shang, K., and Hua, H. (2011). Biological effects of rice harbouring Bph14 and Bph15 on Brown planthopper. Pest Manag. Sci. 67 (5), 528–534. doi:10.1002/ps.2089
Li, X., Liu, F., Cai, W., Zhao, J., Hua, H., and Zou, Y. (2019a). The function of spineless in antenna and wing development of the Brown planthopper, Nilaparvata lugens. Insect Mol. Biol. 28 (2), 196–207. doi:10.1111/imb.12538
Li, X., Liu, F., Wu, C., Zhao, J., Cai, W., and Hua, H. (2019b). Decapentaplegic function in wing vein development and wing morph transformation in Brown planthopper, Nilaparvata lugens. Dev. Biol. 449 (2), 143–150. doi:10.1016/j.ydbio.2019.02.016
Li, X., Liu, X., Lu, W., Yin, X., and An, S. (2022). Application progress of plant-mediated RNAi in pest control. Front. Bioeng. Biotechnol. 10, 963026. doi:10.3389/fbioe.2022.963026
Li, X., Zhao, M., Tian, M., Zhao, J., Cai, W., and Hua, H. (2021). An InRs/mir-9a/NlUbx regulatory cascade regulates wing diphenism in Brown planthopoper. Insect Sci. 28, 1300–1313. doi:10.1111/1744-7917.12872
Liu, F., Li, X., Zhao, M., Guo, M., Han, K., Dong, X., et al. (2020). Ultrabithorax is a key regulator for the dimorphism of wings, a main cause for the outbreak of planthoppers in rice. Nat Sci. Rev. 7 (7), 1181–1189. doi:10.1093/nsr/nwaa061
Livak, K. J., and Schmittgen, T. D. (2001). Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods 25 (4), 402–408. doi:10.1006/meth.2001.1262
Matsuda, M., and Chitnis, A. B. (2009). Interaction with notch determines endocytosis of specific delta ligands in zebrafish neural tissue. Development 136 (2), 197–206. doi:10.1242/dev.027938
Morooka, S., and Tojo, S. (1992). Maintenance and selection of strains exhibiting specific wing form and body colour under high density conditions in the Brown planthopper, Nilaparvata lugens (Homoptera: Delphacidae). Appl. Entomol. Zool. 27 (3), 445–454. doi:10.1303/aez.27.445
Mumm, J. S., and Kopan, R. (2000). Notch signaling: From the outside in. Dev. Biol. 228 (2), 151–165. doi:10.1006/dbio.2000.9960
Mumm, J. S., Schroeter, E. H., Saxena, M. T., Griesemer, A., Tian, X., Pan, D. J., et al. (2000). A ligand-induced extracellular cleavage regulates gamma-secretase-like proteolytic activation of Notch1. Mol. Cell 5 (2), 197–206. doi:10.1016/s1097-2765(00)80416-5
Palomero, T., Lim, W. K., Odom, D. T., Sulis, M. L., Real, P. J., Margolin, A., et al. (2006). NOTCH1 directly regulates c-MYC and activates a feed-forward-loop transcriptional net work promoting leukemic cell growth. Proc. Natl. Acad. Sci. U. S. A. 103 (28), 18261–18266. doi:10.1073/pnas.0606108103
Schroeter, E. H., Kisslinger, J. A., and Kopan, R. (1998). Notch-1 signalling requires ligand-induced proteolytic release of intracellular domain. Nature 393 (6683), 382–386. doi:10.1038/30756
Sōgawa, K. (2003). The rice Brown planthopper: Feeding physiology and host plant interactions. Annu. Rev. Entomol. 27 (1), 49–73. doi:10.1146/annurev.en.27.010182.000405
Struhl, G., and Adachi, A. (1998). Nuclear access and action of Notch in vivo. Cell 93 (4), 649–660. doi:10.1016/s0092-8674(00)81193-9
Ulrich, J., Dao, V. A., Majumdar, U., Schmitt-Engel, C., Schwirz, J., Schultheiset, D., et al. (2015). Large scale RNAi screen in tribolium reveals novel target genes for pest control and the proteasome as prime target. BMC Genomics 16 (1), 674. doi:10.1186/s12864-015-1880-y
Vanitha, K., Suresh, S., and Gunathilagaraj, K. (2011). Influence of Brown planthopper Nilaparvava lugens (Stål.) feeding on nutritional biochemistry of rice plant. Oryza 48 (2), 142–146.
Vogel, E., Santos, D., Mingels, L., Verdonckt, T. W., and Broeck, J. V. (2019). RNA interference in insects: Protecting beneficials and controlling pests. Front. Physiol. 9, 1912. doi:10.3389/fphys.2018.01912
Wei, Z., Hu, W., Lin, Q., Cheng, X., Tong, M., Zhu, L., et al. (2010). Understanding rice plant resistance to the Brown planthopper (Nilaparvata lugens): A proteomic approach. Proteomics 9 (10), 2798–2808. doi:10.1002/pmic.200800840
Xu, H. J., Xue, J., Lu, B., Zhang, X. C., Zhuo, J. C., He, S. F., et al. (2015). Two insulin receptors determine alternative wing morphs in planthoppers. Nature 519 (7544), 464–467. doi:10.1038/nature14286
Yamamoto, S., Charng, W. L., Rana, N. A., Kakuda, S., Jaiswal, M., Bayat, V., et al. (2012). A mutation in EGF repeat–8 of Notch discriminates between Serrate/Jagged and Delta family ligands. Science 338 (6111), 1229–1232. doi:10.1126/science.1228745
Yang, E. L., Wang, B. T., Lei, G., Chen, G. C., and Liu, D. H. (2022). Advances in nanocarriers to improve the stability of dsRNA in the environment. Front. Bioeng. Biotechnol. 10, 974646. doi:10.3389/fbioe.2022.974646
Zhang, C. X., Brisson, J. A., and Xu, H. J. (2019). Molecular mechanisms of wing polymorphism in insects. Annu. Rev. Entomol. 64, 297–314. doi:10.1146/annurev-ento-011118-112448
Keywords: Nilaparvata lugens, RNAi biopesticide, Notch signaling pathway, delta, jagged
Citation: Yang X, Liu S, Lu W, Du M, Qiao Z, Liang Z, An Y, Gao J and Li X (2022) Delta and jagged are candidate target genes of RNAi biopesticides for the control of Nilaparvata lugens. Front. Bioeng. Biotechnol. 10:1023729. doi: 10.3389/fbioe.2022.1023729
Received: 20 August 2022; Accepted: 10 November 2022;
Published: 18 November 2022.
Edited by:
Haichao Li, Center for Excellence in Molecular Plant Sciences (CAS), ChinaReviewed by:
Weihua Ma, Huazhong Agricultural University, ChinaCopyright © 2022 Yang, Liu, Lu, Du, Qiao, Liang, An, Gao and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiang Li, bGl4aWFuZzAyMTdAMTI2LmNvbQ==
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.