
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 06 March 2025
Sec. Land, Livelihoods and Food Security
Volume 9 - 2025 | https://doi.org/10.3389/fsufs.2025.1533423
This article is part of the Research Topic Technologies To Assess Soil Quality Towards Sustaining Food Security View all 7 articles
 Olusegun Folorunso1*
Olusegun Folorunso1* Oluwafolake Ojo2
Oluwafolake Ojo2 Mutiu Busari3
Mutiu Busari3 Muftau Adebayo4
Muftau Adebayo4 Joshua Adejumobi1
Joshua Adejumobi1 Daniel Folorunso1
Daniel Folorunso1 Femi Ayo5
Femi Ayo5 Orobosade Alabi1
Orobosade Alabi1 Olusola Olabanjo1
Olusola Olabanjo1Introduction: Most farmers in Nigeria lack knowledge of their farmland’s nutrient content, often relying on intuition for crop cultivation. Even when aware, they struggle to interpret soil information, leading to improper fertilizer application, which can degrade soil and ground water quality. Traditional soil nutrient analysis requires field sample collection and laboratory analysis; a tedious and time-consuming process. Digital Soil Mapping (DSM) leverages Machine Learning (ML) to create detailed soil maps, helping mitigate nutrient depletion. Despite its growing use, existing DSM-based ML methods face challenges in prediction accuracy and data representation.
Aim: This study presents GeaGrow, an innovative mobile app that enhances agricultural productivity by predicting soil properties and providing tailored fertilizer recommendations for yam, maize, cassava, upland rice, and lowland rice in southwest Nigeria using Artificial Neural Networks (ANN).
Materials and methods: The presented method involved the collection of soil samples from six states in southwest Nigeria which were analysed in the laboratory to compile the primary dataset mapped to the coordinates. A secondary dataset was compiled using iSDAsoil’s API for data augmentation and validation. The two sets of data were pre-processed and normalized using Python, and an ANN was employed to predict soil properties such as NPK, Organic Carbon, Soil Textural Composition and pH levels through regressive analysis while building a composite model for Soil Texture Classification based on the predicted soil composition. The model’s performance yielded a Mean Absolute Error (MAE) of 1.9750 for NPK and Organic Carbon prediction, 3.5461 for Soil Textural Composition prediction, and 0.1029 for pH prediction. For the classification of the soil texture, the results showed a high accuracy value of 99.9585%.
Results: The results highlight the effectiveness of combining soil texture with water retention, NPK, and Organic Carbon to predict pH and optimize fertilizer application. The GeaGrow app provides farmers with accessible, location-based soil insights and personalized crop recommendations, marking a significant advancement in agricultural technology. The GeaGrow app also provides smallholder farmers with scalable, ease of adoption and use of the developed mobile application.
Conclusion: This research demonstrates the potential of ML to transform soil nutrient management and improve crop yields, contributing to sustainable farming practices in Nigeria.
Digital Soil Mapping (DSM) is a recent method that leverages Machine Learning (ML) algorithms to produce detailed maps of soil features over extensive land areas, offering accurate spatial information on soil properties such as nutrient concentrations (Buthelezi et al., 2024; Araujo-Carrillo et al., 2021). By integrating various ML techniques, statistical models, and data sources—including satellite imagery, topographic features, and soil samples—DSM provides a data-driven solution to soil nutrient depletion, a pressing issue in agriculture (Esmaeilizad et al., 2024). The adoption of DSM, enhanced by ML algorithms, has revolutionized soil analysis, enabling precise predictions of nutrient status through trained models that incorporate environmental factors (Barathkumar et al., 2024; Júnior et al., 2024). This aligns with broader findings that highlight the importance of advanced modeling tools in mitigating climate variability and improving resource management in farming systems (Mkuhlani et al., 2024). One subset of ML, Artificial Neural Networks (ANN), models interconnected inputs through weighted connections, producing highly accurate predictions (Ibrahim et al., 2024; Kurani et al., 2023). In developing nations, farmers struggle with declining soil nutrients due to continuous cultivation, exacerbated by limited access to reliable laboratory soil testing. Although advanced practices such as water-fertilizer coupling improve soil health, smallholder farmers often lack the technological resources for comprehensive soil nutrient analysis, relying instead on observational methods that can lead to suboptimal fertilization (Xing et al., 2024). Given the labor-intensive and costly nature of traditional soil analysis, new approaches such as DSM and ANN-based models have become essential for efficient soil mapping and nutrient management.
Data on land suitability for agricultural production, soil nutrients, trace elements, moisture content, soil classifications, soil color, soil maturity, soil texture, and meteorological information are all included in soil databases. The most common soil nutrients, trace elements, and their descriptions are shown in Table 1. Establishing relationships between different environmental elements and soil qualities is made easier by using covariate environmental data. The formation of soil and its properties can be attributed to different factors some of which are but not limited to parent material’s nature, topographical features, vegetation cover, land use, and climate. To have a detailed insight into the relationship between soil and its environment, the inclusion of covariate data can boost the effectiveness of the soil prediction model which is important when predicting soil as it improves our understanding of the correlations between soil and its environment, capture spatial heterogeneity, provide insights into basic mechanisms, enable data fusion, and help make well-informed land management decisions. The application of Covariate data increases the accuracy of forecasting the accuracy of soil models and their use in different industries like land use management, environmental governance and agriculture (Tziachris et al., 2020; Zeraatpisheh et al., 2022).

Table 1. Soil nutrients and trace elements (Hudson, 1992).
In the process of trying to improve on difficult challenges in the field of agricultural and environmental management, the use of machine learning and Data Analytics are on the rise as both technologies offer creative solutions for enhancing productivity, resource efficiency, and sustainable practices. When it comes to predicting soil and crop health, mapping vulnerabilities, and optimizing management strategies, machine learning and Data analytics are great tools for smart farming. Also, in mapping soil vulnerabilities and predicting environmental risks machine learning serves as a great tool. This major difference is further exemplified in studies that have developed models to assess soil erosion risks (Sarkar and Mishra, 2018), while other research has used ML techniques to map areas prone to gully erosion (Garosi et al., 2019). Different satellite imagery, GPS and soil sensors have been combined using this model to help have a better understanding of specific measures such as erosion control and soil health improvements. ML plays a pivotal role in the management and prediction of crop disease outbreaks and it also helps in predicting crop real-time nutrient needs. An example is found in the study of Qiu et al. (2021) where ML algorithms and drone imagery were used to assess nitrogen levels in rice fields quickly, enabling data-driven fertilization decisions. The study produces high computational complexity and lacks interpretability. Another study by Akhter and Sofi (2022) made use of ML and IoT sensors to help farmers improve yield production and manage Apple diseases in Kashmir’s orchards. The study also discusses the difficulty faced in the deployment of the proposed technology into traditional farming activities. The study did not cover a wide range of parameters that can enhance Apple disease prediction and management. The studies of Landeta-Escamilla et al. (2023) and Ahmad et al. (2024) developed predictive models to assist in allocating resources like fertilizers and water more efficiently by identifying the most crucial factors affecting crop health and productivity. The study in Landeta-Escamilla et al. (2023) applied machine learning methods to predict the soil parameters that will be suitable for sugarcane higher productivity but did not cover a wide range of parameters that can enhance the prediction of soil suitability for sugarcane production. Similarly, the study in Ahmad et al. (2024) applied machine learning to predict the relationship between soil features and pathogen occurrences and classification but the limitation is that the size of their dataset is limited. Guo et al. (2024) developed a machine learning model to analyse the pollution features and health risks of cadmium in paddy soils across Hainan Island based on multiple environmental data. The study demonstrated how ML models could identify pollution hotspots and assess potential health impacts, guiding regulatory actions and remediation efforts. Machine learning is improving farming activities day by day by providing data-driven information that guides farmers’ decisions. This approach of digital farming helps improve agricultural efficiency and sustainability in the process tackling environmental challenges like soil degradation, pollution, and climate change. In all, digital farming improves food security and land management sustainability.
In the process of assessing environmental pollutants like cadmium in paddy soils and their related health risk blending the use of ML with geospatial data and advanced statistical methods helps improve the assessment of environmental pollutants. The work used fewer soil properties and environmental variables leading to less accurate predictions. Makungwe et al. (2021) proposed linear mixed models and random forests for the prediction of soil pH for rice production using environmental variables. The study used machine learning methods in the field of soil quality assessment, environmental monitoring and agricultural forecasting relating to soil pH predictions for optimal agricultural productivity. The limitation of the study is related to the selection of the environmental variables and the predictive models which lead to an overfitting problem. Martinho (2024) proposed predictive machine learning models for the prediction of agricultural outputs. The research highlights the potential of machine learning to provide innovative solutions to critical issues in agriculture and environmental management, ultimately contributing to more informed and effective strategies for ensuring food security and environmental sustainability. The research identified the most suitable model for the prediction of agricultural outputs using farm environmental data. The validation of the proposed models was limited to the availability of a few farm datasets. Kumar et al. (2023) presented an artificial intelligence solution for optimizing irrigation and nutrient management in Agriculture. The study examined different studies involving the broad applications of AI for optimal nutrient and irrigation management that enhances agricultural productivity. The study also discusses the bad and good associated with the application of AI in agriculture which include model interpretability, data quality and adoption of AI technologies by farmers. Reddy et al. (2024) presented machine learning algorithms for real-time analysis and recommendation of optimal soil nutrients for higher agricultural productivity. The study provided a user-friendly mobile app that provided timely accessibility to environmental parameters for the prediction of insufficient soil nutrients and recommendations for best alternatives. The presented method can suffer from high complexity and computational time. Khanna et al. (2024) presented a theory on the application of machine learning algorithms on environmental parameters for the prediction of medicinal plant cultivation. The study presented a machine learning method for the recommendation of the right amount of soil nutrients required for the cultivation of optimal medicinal plants. The presented theory was validated against three different cases to prove its effectiveness for the production of high-quality medicinal plants. However, the study only provided a hypothesis without any real implementation. Purohit et al. (2024) proposed a stack regressor learning algorithm for balancing soil nutrients in the right proportions for enhancing crop production. The proposed algorithm takes into consideration the joint effect of some environmental factors to predict the optimal nutrient requirements for a particular type of crop and agricultural soil. The results showed that the proposed stack regressor learning algorithm performed better in prediction accuracy than similar methods for improved crop productivity. However, a broad comparative analysis showed that the accuracy of the proposed method is slightly lower than that of the decision tree and random forest models. The simulation results of the proposed method also showed higher complexity and computational time. Padhiary et al. (2025) presented a smart recommender farming system using artificial intelligence for automated decision-making for farm operations to enhance productivity and sustainability in farming practices. The study only adopted real-world case studies for the validation of the presented method and discusses drawbacks in the application of smart agriculture but lacks actual implementation results for the evaluation of the presented method.
Recent years have seen increasing research interest in applying ML to DSM, yet current approaches still face challenges related to prediction accuracy and data representation (Esmaeilizad et al., 2024; Lamichhane et al., 2019; Wadoux et al., 2020). Additionally, there is a lack of mobile technology solutions in countries like Nigeria for real-time soil nutrient prediction, particularly for nitrogen, phosphorus, and potassium. In response, this study introduces GeaGrow, an innovative mobile application developed by the SmartSoil Team, which provides precise soil property predictions, including organic carbon, nitrogen, phosphorus, potassium, and pH levels. GeaGrow also recommends suitable crops—such as yam, maize, cassava, and rice—and offers tailored fertilizer advice to improve agricultural productivity. The application integrates ML for soil nutrient prediction and farm management, bridging the technological gap for smallholder farmers. This study developed a mobile DSM application that dynamically predicts soil properties and optimizes fertilizer applications using ANN, improving upon traditional DSM methods that provide static soil maps. The methodology involved data collection from six Nigerian states and the iSDAsoil API, preprocessing with max-min normalization, and ANN-based prediction of soil texture class. By integrating ML into soil nutrient mapping, this research advances precision agriculture and supports sustainable farming practices. Despite challenges related to static datasets and real-time data integration, GeaGrow represents a significant step in empowering smallholder farmers and enhancing soil fertility management through AI-driven insights.
The study aims to answer the following research questions: (1) How to develop a friendly application that enhances agricultural productivity by predicting soil properties; (2) How to provide tailored fertilizer recommendations for crops through the application of artificial intelligence; (3) Can the developed application provide farmers with accessible, location-based soil insights and personalized crop recommendations; and (4) Can the developed application provides smallholder farmers with scalable, ease of adoption and use. The rest of this study is designed as follows: Section 2 presents related work. Materials and methods are presented in Section 3. The results and discussion are presented in Section 4. Section 5 presents the conclusion and future directions for the study.
This research employed an Artificial Neural Network to develop a mobile digital soil mapping application for assessing soil nutrients and optimizing fertilizer application for specific crops. The approach involved data collection, feature extraction, prediction, and recommendation. A total of 710 soil samples were collected from six states in southwest Nigeria, supplemented by a secondary dataset of over 2,500 samples from the iSDAsoil API, using a 5 km sampling interval to prevent data redundancy. Preprocessing in Python included handling missing values, eliminating incomplete rows, and random sampling to mitigate class imbalance. Feature extraction used a sequential model, and values were normalized via max-min scaling. The ANN model predicted soil texture class, forming the basis for a recommendation system that suggests optimal fertilizer quantities to enhance soil conditions for specific crops. Figure 1 shows the flow diagram of the developed mobile digital soil mapping application. This study significantly advances agricultural science by integrating machine learning into soil nutrient mapping and precision farming.
The study area is located in the Southwest of Nigeria covering Oyo, Osun, Ekiti, Ogun, Ondo and Lagos states. Figure 2 shows the map of the study area. The justification for the choice of the Southwest regions is based on their economic contribution, popularity, and ecological and agricultural diversity to the Nation. Lagos and Ibadan are recognized as one of the largest economic contributors to Nigeria. The South-west zone alone accounts for over 50 million people, which is more than 22% of the total population of the country. Lagos is the largest urban centre in the Southwest and is the largest city in Nigeria and among the largest in Africa.
The study used both secondary and primary datasets. The primary datasets were derived from six (6) states in southwest, Nigeria with 710 samples to predict the soil properties (NPK, Organic Carbon) and soil composition (Sand, Silt, Clay) by analysing soil samples in the laboratory with an Atomic Absorption Spectrometry (AAS) machine and other standard laboratory procedures for soil composition analysis. The sampling frequency for the primary include Ekiti (41%), Ondo (116%), Osun (70%), Oyo (321%), Lagos (21%) and Ogun (141%). Similarly, all six states were covered in the secondary dataset. The secondary dataset utilized data obtained from the iSDAsoil website in predicting soil texture. The ISDASoil data was used to develop the initial model and the implementation was transferred to the training of the primary dataset to validate the iSDAsoil’s API data against the primary datasets. The iSDAsoil is the first field-level soil map for Africa, offering predictions on over 20 soil properties at a 30 m resolution across the continent. The data driving their soil map was collected by analysing over 130,000 soil samples across Africa. This wealth of data, accessible via an open-source API, supports advanced agricultural and environmental analysis, underscoring its utility for precise, localized soil health and nutrient assessments. This study extracted data for over 20 different soil properties at 2 different depths (0–20 and 20-50 cm) within the Southwest region of Nigeria. Utilizing coordinates to interface with the iSDAsoil’s API, the study collaborated with a GIS expert who employed Aeronautical Reconnaissance Coverage Geographic Information System (ArcGIS) software to map out 2,762 specific locations in the southwest every 5 km, avoiding aquatic and built-up areas to ensure the integrity of the extracted data. Initially, the extracted data was obtained in JSON format and later converted into a CSV file for streamlined analysis and incorporation into the developed model. The extracted data consists of 2,761 rows and 21 columns, the columns include geographical coordinates, land and crop cover types, and slope metrics. These soil properties encompass vital macronutrients such as NPK, along with organic carbon content and texture. Further information on the dataset can be found at https://www.isda-africa.com/isdasoil/.
In this study, for effective soil sampling method and analysis, a 30 m resolution land use and land cover map of the study area from 1st January 2022 to 31st January 2023 was downloaded from the USGS website. The download was classified by supervised learning using a maximum likelihood algorithm. The stratified sampling method at a 10 km sampling interval within each stratum was adopted. Soil sample points were geo-referenced using ArcGIS and the samples were taken at 0–20 cm and 20–50 cm soil depth. The soil samples were properly labeled and transferred to the laboratory for processing. In the laboratory, the samples were air-dried and passed through a 2.0 mm sieve. Particle size analysis was carried out using the hydrometer method (Bouyoucos, 1962). The pH (in water) was determined using a glass electrode pH meter in a soil: water ratio of 1:1. Organic carbon content of the soils was determined by the Walkley-Black chromic acid wet oxidation method (Nelson and Sommers, 1982). Soil Total N was determined using the micro Kjeldhal method (Bremmer and Mulvaney, 1982). Available phosphorus (Avail P.) was analysed using Bray-1 P extractant and determined colorimetrically by the molybdenum blue procedure. Exchangeable cations were extracted using 1 M Ammonium Acetate pH 7.0 and the potassium and calcium in the extract were determined using an atomic absorption spectrophotometer (AAS).
The CSV format of the dataset was pre-processed using Python. The pandas’ package was used with the “drop” function to remove rows that match the “No data” value. Preprocessing entails changing the “No data” value to a “0” value, dropping rows where at least one column is not available and random sampling of the entire dataset to prevent class imbalance problems. The dropout layer was used in the developed ANN model to reduce overfitting by setting a few neurons to 0 and reducing computation in the training process. Furthermore, the features that exist in both the primary and secondary data were standardized to ensure consistency across the board. The units of the primary dataset were converted to the units of the secondary dataset and vice-versa.
The SCORPAN model which is a conceptual framework for predicting soil properties and mapping soil classes was used to extract predictors that are possibly predictive of the outcome of this study from the primary dataset. It integrates various environmental covariates to understand and predict soil variability. The acronym SCORPAN stands for:
• S: Soil properties (existing soil data)
• C: Climate (temperature, precipitation, etc.)
• O: Organisms (vegetation, fauna, human activity)
• R: Relief (topography, slope)
• P: Parent material (geological material from which soil develops)
• A: Age (time, soil formation processes)
• N: Spatial position (geographic location, longitude, latitude)
The parameters extracted for this study were derived from terrain attributes, climate and hydrology variables, soil and geological characteristics, and additional land-use factors. Terrain attributes such as elevation, slope, aspect, curvature, and wetness indices were generated from digital elevation models. Climate and hydrology variables, including temperature, precipitation, evapotranspiration, and soil moisture, were sourced from global climate datasets. Soil properties and geological information were obtained from existing soil maps and surveys, providing insight into soil type, texture, and underlying rock formations. Additionally, land-use patterns and proximity to water bodies were analyzed to assess environmental influences on soil properties. By integrating these diverse parameters, the study effectively applied SCORPAN for digital soil mapping, improving soil property predictions and supporting more precise agricultural decision-making. Table 2 describes the SCORPAN parameters used in this study.
All SCORPAN factors are inherently tied to a specific spatial position (n), making longitude and latitude effective covariates. The neural network clustering approach further ensures coverage for unmapped areas, justifying the decision to use only spatial position.
These parameters are then used as inputs for the SCORPAN model to predict soil properties, such as soil type, texture, and fertility, and to generate digital soil maps. Furthermore, feature extraction was performed on the dataset in CSV format, the feature extraction process used a sequential model in Python to extract needed columns from the pre-processed datasets. The value obtained from the features was then normalized to a common scale using the max-min normalization method as in Equation 1:
where and are the feature’s maximum and minimum values, respectively. The numerator will be zero and will be zero when the value of is the lowest in the column. In contrast, the value of equals 1 when it is the largest value in the column since the numerator and denominator are equal. The value of is between 0 and 1 if it falls between the lowest and maximum values.
The ANN model was used purely for regressive and predictive analysis of both primary and secondary data. The developed ANN model for prediction is based on the sequential model object in Keras which allows easy creation of a linear stack of layers. The rationale for selecting the sequential ANN model in Keras is to allow for the easy addition of a linear stack of layers for different distinct functions and the choice of the ReLU activation function is to increase the learning rate of the network to overcome the vanishing gradient problem of ANN models. The hyperparameter tuning of the ANN model was done on the results from each iteration of the network. The developed sequential model consists of ANN with a combination of dense layer, Rectified Linear Unit (ReLU) activation functions, and downsampling layer to train the dataset.
Dense layer: a dense layer is a regular neural network layer that links every neuron in the previous layer to every neuron in the defined layer. The dense layer is also based on the ReLU activation function for the final prediction task. In this study, the customization ability of the Keras API to specify any given number of layers was utilized. The developed ANN with the sequential model was used to specify the number of neurons for the layers, the activation type, the type initialization for kernel and bias, and the training method. This study used the number of neurons for the layers and activation function parameters while keeping other parameters at default settings for simplicity. The output y is computed using the ReLU activation function. The ANN-based model combines an input x in the training data and some randomly generated weights w to get the output y as in Equation 2.
Where is the number of instances.
Dropout layer: the dropout layer was used in the developed ANN based on the sequential model to reduce overfitting by setting a few neurons to 0 and reducing computation in the training process. The justification for this is based on the fact that randomly dropping neurons can greatly reduce overfitting for the prediction task.
Loss function: when trying to evaluate the learning process of a network, a loss function is used which measures the deviation from the target. This is then used to measure the performance level of ANN for predicting the soil nutrient properties and optimizing fertilizer application for specific crops. This loss function serves as the error rate that the network can use to adjust its weights and become more intelligent. In this research, the Mean Absolute Error (MAE) loss function was used to compute the average absolute error between actual and predicted values (Equation 3). In this study, Stochastic Gradient Descent (SGD) was used as the training method to calculate the loss and update the weight of the network (Equation 4).
where actual is the target value, predicted is the network predictions, and is the total number of instances.
Where the learning rate is a defined parameter in the network architecture.
Optimizer: the Stochastic Gradient Descent-based optimization referred to as Adams helps define momentum and variance of the gradient of the loss and leverage a combined effect to update the weight parameters as in Equation 5. In the course of this research, the Adaptive Moment Estimation (Adam) was used as the optimization technique to compute an adaptive learning rate for each parameter in the network. The momentum and variance together help to effectively improve the learning process.
Where momentum and variance are used to enhance the convergence rate of the Stochastic Gradient Descent-based optimization.
A recommendation system was then developed to suggest the amount of fertilizer needed to put the soil in the perfect condition for optimal crop yield. Based on the results obtained from the prediction stage on the available nutrients in the soil for the specific crops being considered in this paper; Yam, Maize, cassava, Upland and Lowland rice, the amount of fertilizer needed to put the soil in the perfect condition for optimal crop yield is then recommended for that location using a developed recommendation system. The knowledge of experts in the form of recommendation rules was used in the development of the recommendation system.
This section presents the overall algorithm for the developed mobile digital soil mapping application for predicting soil nutrient properties and optimizing fertilizer applications for specific crops using artificial neural networks. The algorithm takes as input the secondary and the primary datasets. The data preprocessing steps were then executed to remove any anomalies in the datasets. The preprocessing involves changing the “No data” value to a “0” value, dropping rows where at least one column is not available and random sampling of the entire dataset to prevent class imbalance problems. In addition, the features in both the primary and secondary datasets were standardized to the same units to ensure consistency. The secondary dataset was converted to the units of the primary dataset. The feature extraction was carried out in Python using the CSV format of the dataset. The feature extraction used the sequential model in Python to extract the needed columns from the pre-processed datasets. This study developed an ANN model with fully connected dense layers stacked together. The value of the features was normalized to a common scale using the max-min normalization. The ANN model was used purely for regressive and predictive analysis of both primary and secondary data. The developed ANN model for prediction is based on the sequential model object in Keras which allows easy creation of a linear stack of layers. The developed sequential model consists of ANN with a combination of dense layers, Rectified Linear Unit (ReLU) activation functions, and a down-sampling layer to train the dataset (Algorithm 1).
The dataset size did not change during the preprocessing, however, to predict the soil texture from the sand, silt, and clay composition, the data was oversampled having 1,956 values for each texture class. Null values were also removed and the primary and secondary datasets were combined. This combination resulted in a final dataset with 3,241 rows and 7 columns. The modeling process was conducted in stages, splitting the dataset into an 80–20 train-test split, with 80% used for training and 20% for testing using the neural networks model.
Table 3 presents the training and validation loss results across four stages of soil property prediction using the developed model. For predicting nitrogen, phosphorus, potassium, and organic carbon, the model attained a training loss of 1.98 and a validation loss of 2.14, with standard deviations of 3.52and 4.06, respectively. These results indicate minimal error between target and predicted values, confirming effective learning and minimal overfitting. Similarly, for soil composition (sand, clay, and silt), the training loss was 3.55, and validation loss was 4.14, with standard deviations of 5.53 and 4.23, respectively, showing a slight increase compared to stage I but maintaining strong predictive accuracy. The prediction of soil texture yielded a significantly lower training loss of 0.04 and validation loss of 0.05, with respective standard deviations of 0.03 and 0.04, indicating improved model efficiency. For soil pH prediction, the training loss was 0.10, while validation loss was 0.28, with standard deviations of 0.12 and 0.46, reflecting a slight increase compared to stage III but still within acceptable margins. Across all stages, the small differences between training and validation loss confirm the model’s robustness, demonstrating its ability to generalize well without overfitting.
The training and validation accuracy were also investigated. For the prediction of nitrogen, phosphorus, potassium, and organic carbon, the model achieved a training accuracy of 98.02% and a validation accuracy of 97.86%, indicating minimal misclassification and effective learning. Similarly, for soil composition (sand, clay, and silt), the model attained 96.45% training accuracy and 95.86% validation accuracy, demonstrating robust generalization. In predicting soil texture, training accuracy reached 99.95% with validation accuracy at 99.95%, showing negligible overfitting. The prediction of soil pH yielded 99.90% training accuracy and 99.72% validation accuracy, confirming strong model performance. Figures 4A–D illustrate the training and validation loss trends, showing consistent reductions in loss across epochs and stabilization over time, further affirming the model’s reliability. Minimal fluctuations and close alignment between training and validation losses suggest that the model effectively mitigates overfitting and generalizes well across soil property predictions. While minor inconsistencies in validation accuracy trends for soil texture prediction suggest slight overfitting, the overall results confirm the model’s precision, stability, and suitability for digital soil mapping applications.
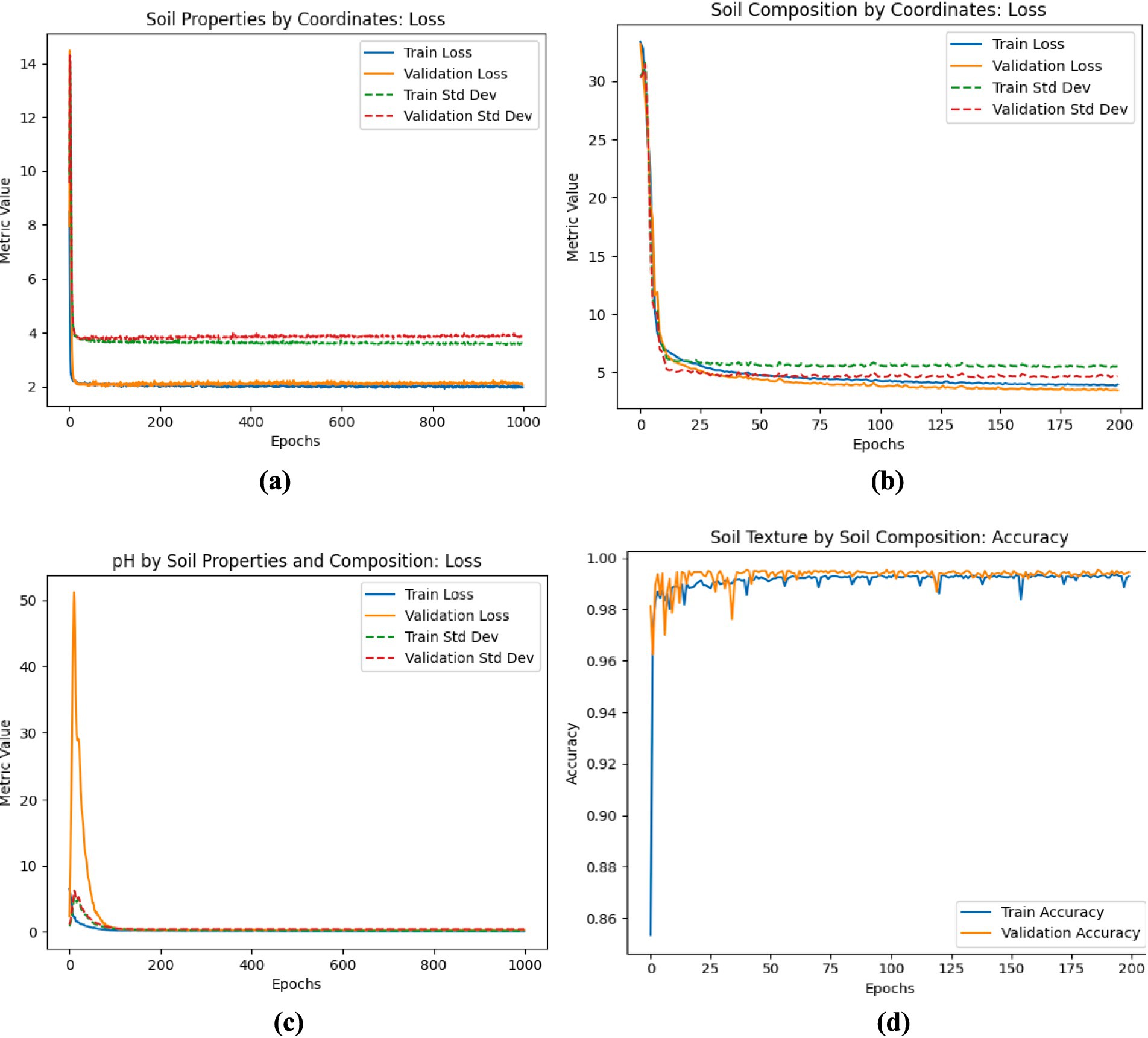
Figure 4. (A) Results of the stage I training and validation loss for the prediction of Nitrogen, Phosphorus, Potassium, and Organic Carbon properties of the soil (B) results of the stage II training and validation loss for the prediction of Sand, clay, and silt composition of the soil (C) results of the stage IV training and validation loss for the prediction of pH level of the soil (D) results of the stage III training and validation accuracy for the prediction of soil texture.
Table 4 shows the comparison of the developed GeaGrow’s performance with other existing ML methods using the same dataset. Most of the ML algorithms on soil texture class prediction obtained an F1-score of at least 80% classification rates. The F1-score of the developed GeaGrow is better at 98.60% compared to AdaboostM1 with the closest score of 97.60%. The results suggest LibSVM as the least ML algorithm for the prediction of soil texture class with an F1-score of 80.20%. The outcomes demonstrated that the optimum predictor for soil texture class is the developed GeaGrow while the worst predictor for soil texture class is LibSVM across the evaluation metrics. The results showed that the developed GeaGrow using the same dataset is an improvement over the traditional Multilayer Perceptron with accuracy, error rate, precision, recall, and F1-score of 98.35, 1.65, 98.50, 97.45, and 98.60%, respectively, when compared to the traditional Multilayer Perceptron with 95.11, 4.89, 97.10, 96.90, and 97.00%, respectively. The results of the developed GeaGrow are a justification that the method can provide better results than the traditional ML algorithms. Generally, the results demonstrated that most of the ML methods on the same dataset performed in a well-balanced manner.
The larger part of the study area (northern part) has soil native total nitrogen below 0.10 gkg−1 while some parts within western and eastern part had total nitrogen between 0.20 and 0.30 gkg−1. The remaining land area had the nutrient considerably between 0.10 and 0.20 gkg−1 (Figure 5A). Most portions of Southwest Nigeria had extractable phosphorus between 23 and 28 mgkg−1 while some areas toward the west and east of the study area had values above 30 mgkg−1 (Figure 5B) while extractable potassium ranged between 0.29 and 0.33 gkg−1 (Figure 5C). The soil organic carbon in most parts of the study areas were <1%, though some areas had organic carbon between 1 and 2% (Figure 5D). The soil pH (H2O) in the southern part of the study area had pH less than or equals 5.50 with the part toward the mid-belt having values of pH between 5.50 and 6.20, while the northern part had values between 6.50 and 7.40 (Figure 5E).
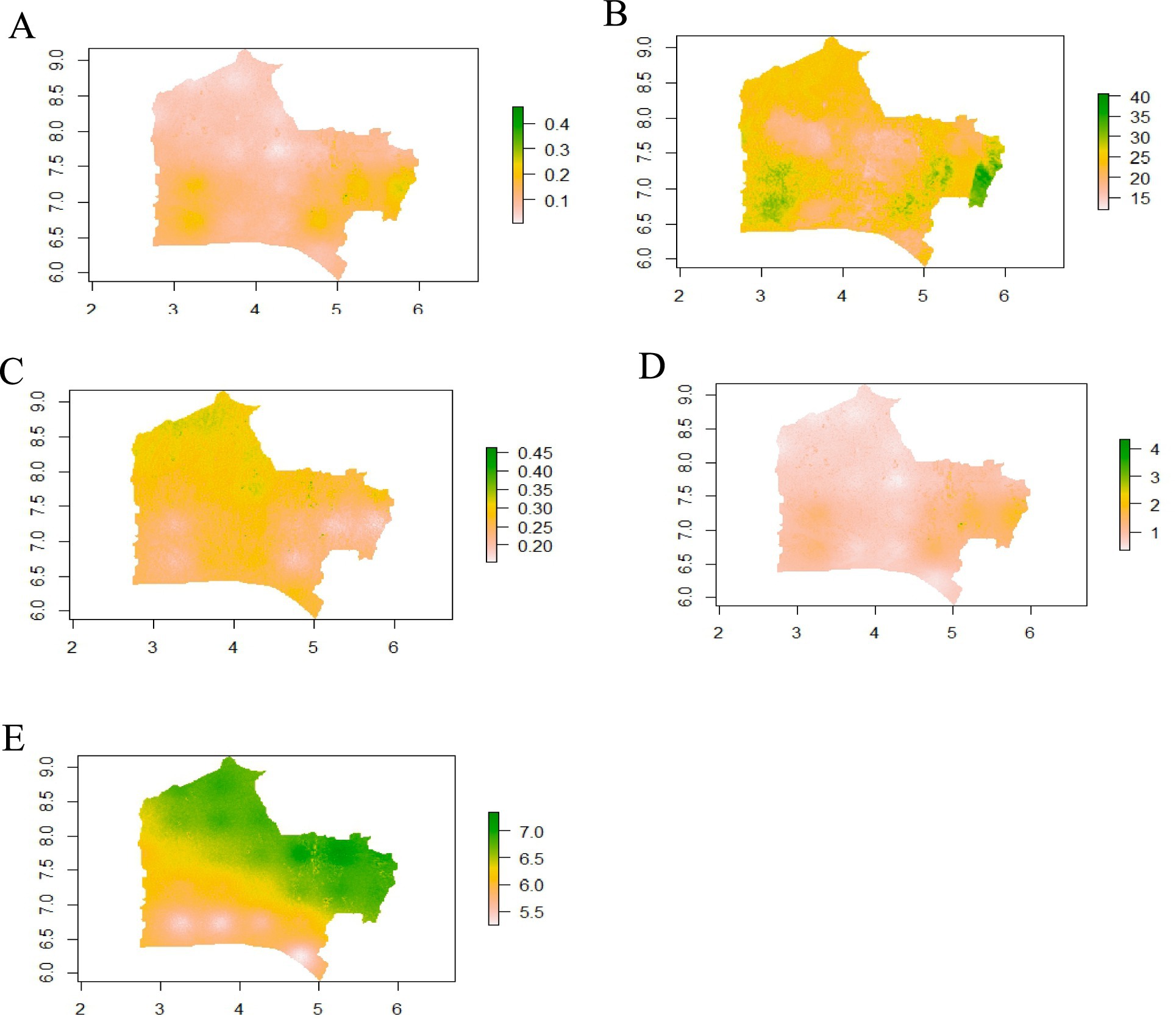
Figure 5. Soil native nutrients generated by multiple linear regression on environmental covariates. (A) Soil total nitrogen (g kg−1); (B) Extractable phosphorus (mg kg−1); (C) Extractable potassium; (D) Organic carbon (g kg−1); E: Soil pH (H2O).
Based on the results obtained from the modeling stage on the available nutrients in the soil for the specific crops being considered in this paper; Yam, Maize, cassava, Upland and Lowland rice, the amount of fertilizer needed to put the soil in the perfect condition for optimal crop yield is then recommended for that location. The information being used in the system was obtained from the soil experts and it is displayed in Tables 5, 6. To utilize the information in Tables 5, 6, a retrieval system called the GeaGrow mobile application was set up.
GeaGrow, a cutting-edge agritech application, derives its name from “Gea,” an alternative form of “Geo,” symbolizing the Earth’s essential role in agriculture, and “grow,” representing the development enabled by fertile soil. The app provides farmers, researchers, and extension agents with detailed soil property predictions, including nitrogen, phosphorus, potassium, and pH levels, directly on their mobile devices. By leveraging advanced predictive analysis, it offers personalized crop recommendations for yam, maize, cassava, upland rice, and lowland rice, alongside tailored fertilizer application advice to optimize soil conditions for each crop. Designed with cross-platform compatibility using Google’s Flutter SDK, the app incorporates Google Maps SDK for spatial functionality, allowing users to analyze individual locations through the “Point Scan” feature or multiple sites via the “Multi Scan” function. Additionally, user-centric features such as reminders, collections, and history enhance efficiency in farm management by synchronizing across devices, providing an adaptable platform for both technologically advanced and traditional farmers. Localization features, including Yoruba language support, further improve accessibility, ensuring broader usability in Nigeria’s Southwest region.
The user interface of GeaGrow (Figure 6) is designed for intuitive navigation, beginning with a login page where existing users can sign in while new users must register before accessing the app’s features. The homepage includes “My Farms,” which displays a list of registered farms and allows users to add new ones by entering farm details such as name, location, crop type, and estimated yield, either manually or through Google Maps integration. The app then provides insights into soil chemical properties and crop suitability, offering recommendations for optimized fertilizer application. Beyond individual farm management, the scalability of GeaGrow ensures the potential for expansion to other regions through the integration of additional environmental data, made possible by the Google Maps SDK and contributions from the SmartSoil Team project. However, adoption challenges remain, particularly due to limited technological literacy and poor internet access in rural areas, which may hinder smallholder farmers’ ability to fully utilize the app. Despite these limitations, GeaGrow presents a transformative tool for precision agriculture, capable of influencing agricultural policy and enhancing farming practices through AI-driven soil analysis and optimization.
In this study, a mobile digital soil mapping application for predicting soil nutrient properties and optimizing fertilizer applications for specific crops using artificial neural networks was developed. The developed method included data collection, data preprocessing, feature extraction, prediction and recommendation. The data collection was done through coordinates interfacing with the iSDAsoil’s API and primary datasets derived from six (6) states in southwest, Nigeria. The preprocessing and feature extraction were carried out in Python using the CSV format of the dataset. The preprocessing involves changing the “No data” value to a “0” value, dropping rows where at least one column is not available and random sampling of the entire dataset to prevent class imbalance problems. The feature extraction used the sequential model to extract the needed columns from the datasets. The value of the features was normalized to a common scale using the max-min normalization method. The prediction phase used an ANN method for predicting soil nutrient properties. A recommendation system called the GeaGrow mobile application was then developed based on the results obtained from the prediction stage on the available nutrients in the soil for the specific crops being considered in this paper. The amount of fertilizer needed to put the soil in the perfect condition for optimal crop yield is recommended for that location using the developed recommendation system. The results showed that by combining soil texture with its water retention properties, NPK values, and Organic Carbon content, the model predicts the pH level of the soil. Upon testing, the Neural Network model achieved an MAE of 2.1388 and an accuracy of 97.86% in the first stage, an MAE of 4.1354 and an accuracy of 95.86% in the second stage, an MAE of 0.0478 and an accuracy of 99.95% in the third stage, and MAE of 0.2785 and accuracy of 99.72% in the fourth stage. This study makes substantial contributions to the field of agricultural science by integrating machine learning into soil nutrient mapping and optimizing fertilizer application for specific crops, marking a significant leap in the use of advanced technologies in agriculture. The developed method allows smallholder farmers to solve soil nutrient mapping and optimal fertilizer application problems; making it a broader application for precision agriculture. The limitations of the study include reliance on static datasets and challenges with real-time data integration for effective model validations. As a next step in the SmartSoil team project, the study suggests integrating real-time weather data and expanding crop coverage.
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
OF: Writing – original draft, Writing – review & editing. OOj: Writing – original draft, Writing – review & editing. MB: Writing – original draft, Writing – review & editing. MA: Writing – original draft, Writing – review & editing. JA: Writing – original draft, Writing – review & editing. DF: Writing – original draft, Writing – review & editing. FA: Writing – original draft, Writing – review & editing. OA: Writing – original draft, Writing – review & editing. OOl: Writing – original draft, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the European Union Under the Accelerating Inclusive Green Growth through Agri-Based Digital Innovation in West Africa (AGriDI), which is an ACP Innovation Fund (grant ref. FED/2020/420–491) led by the International Centre for Insect Physiology and Ecology (ICIPE), Nairobi Kenya.
We would like to thank the reviewers for their valuable comments and feedback, which have greatly improved the quality and clarity of our manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Gen AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Ahmad, F., Javed, K., Tahir, A., Khan, M. U. G., Abbas, M., Rabbani, M., et al. (2024). Identifying key soil characteristics for Francisella tularensis classification with optimized machine learning models. Sci. Rep. 14:1743. doi: 10.1038/s41598-024-51502-z
Akhter, R., and Sofi, S. A. (2022). Precision agriculture using IoT data analytics and machine learning. J. King Saud Univ. Comput. Inf. Sci. 34, 5602–5618. doi: 10.1016/j.jksuci.2021.05.013
Araujo-Carrillo, G. A., Varón-Ramírez, V. M., Jaramillo-Barrios, C. I., Estupiñan-Casallas, J. M., Silva-Arero, E. A., Gómez-Latorre, D. A., et al. (2021). IRAKA: the first Colombian soil information system with digital soil mapping products. Catena 196:104940. doi: 10.1016/j.catena.2020.104940
Barathkumar, S., Sellamuthu, K. M., Sathyabama, K., Malathi, P., Kumaraperumal, R., and Devagi, P. (2024). Advancements in soil quality assessment: a comprehensive review of machine learning and AI-driven approaches for nutrient deficiency analysis. Commun. Soil Sci. Plant Anal. 56, 251–276. doi: 10.1080/00103624.2024.2406484
Bouyoucos, C. J. (1962). Hydrometer method improved for making particle size analysis of soils. Agron. J. 54, 464–465. doi: 10.2134/agronj1962.00021962005400050028x
Bremmer, J. M., and Mulvaney, C. (1982). “Nitrogen-Total. Methods of soil analysis. Part 2” in Chemical and biological properties. eds. A. L. Page, R. H. Miller, and D. R. Keeney (Madison, WI: American Society of Agronomy, Soil Science Society of America), 526–595.
Buthelezi, M. N. M., Lottering, R., Peerbhay, K., and Mutanga, O. (2024). Assessing the extent of land degradation in the eThekwini municipality using land cover change and soil organic carbon. Int. J. Remote Sens. 45, 1339–1367. doi: 10.1080/01431161.2024.2307945
Esmaeilizad, A., Shokri, R., Davatgar, N., and Kari Dolatabad, H. (2024). Exploring the driving forces and digital mapping of soil biological properties in semi-arid regions. Comput. Electron. Agric. 220:108831. doi: 10.1016/j.compag.2024.108831
Garosi, Y., Sheklabadi, M., Conoscenti, C., Pourghasemi, H. R., and van Oost, K. (2019). Assessing the performance of GIS-based machine learning models with different accuracy measures for determining susceptibility to gully erosion. Sci. Total Environ. 664, 1117–1132. doi: 10.1016/j.scitotenv.2019.02.093
Guo, Y., Yang, Y., Li, R., Liao, X., and Li, Y. (2024). Cadmium accumulation in tropical island paddy soils: from environment and health risk assessment to model prediction. J. Hazard. Mater. 465:133212. doi: 10.1016/j.jhazmat.2023.133212
Hudson, B. D. (1992). The soil survey as paradigm-based science. Soil Sci. Soc. Am. J. 56, 836–841. doi: 10.2136/sssaj1992.03615995005600030027x
Ibrahim, M., Haider, A., Lim, J. W., Mainali, B., Aslam, M., Kumar, M., et al. (2024). Artificial neural network modelling for the prediction, estimation, and treatment of diverse wastewaters: a comprehensive review and future perspective. Chemosphere 362:142860. doi: 10.1016/j.chemosphere.2024.142860
Júnior, M. R. B., Moreira, B. R. D. A., Carreira, V. D. S., Filho, A. L. D. B., Trentin, C., Souza, F. L. P. D., et al. (2024). Precision agriculture in the United States: a comprehensive meta-review inspiring further research, innovation, and adoption. Comput. Electron. Agric. 221:108993. doi: 10.1016/j.compag.2024.108993
Khanna, A., Jain, S., Choudhury, T., Kotecha, K., and Sar, A. (2024). “AI-driven prediction of medicinal property presence in plants cultivated with nutrient-enriched manure,” in International Conference on Emerging Trends in Expert Applications & Security (Singapore: Springer Nature Singapore), 73–82.
Kumar, J., Chawla, R., Katiyar, D., Chouriya, A., Nath, D., Sahoo, S., et al. (2023). Optimizing irrigation and nutrient Management in Agriculture through artificial intelligence implementation. Int. J. Environ. Clim. Change 13, 4016–4022. doi: 10.9734/ijecc/2023/v13i103077
Kurani, A., Doshi, P., Vakharia, A., and Shah, M. (2023). A comprehensive comparative study of artificial neural network (ANN) and support vector machines (SVM) on stock forecasting. Ann. Data Sci. 10, 183–208. doi: 10.1007/s40745-021-00344-x
Lamichhane, S., Kumar, L., and Wilson, B. (2019). Digital soil mapping algorithms and covariates for soil organic carbon mapping and their implications: a review. Geoderma 352, 395–413. doi: 10.1016/j.geoderma.2019.05.031
Landeta-Escamilla, O., Alvarado-Lassman, A., Sandoval-González, O. O., Flores-Cuautle, J. J. A., Rosas-Mendoza, E. S., Martínez-Sibaja, A., et al. (2023). Determination of soil agricultural aptitude for sugar cane production in Vertisols with machine learning. PRO 11:1985. doi: 10.3390/pr11071985
Makungwe, M., Chabala, L. M., Chishala, B. H., and Lark, R. M. (2021). Performance of linear mixed models and random forests for spatial prediction of soil pH. Geoderma 397:115079. doi: 10.1016/j.geoderma.2021.115079
Martinho, V. J. P. D. (2024). “Predictive machine learning approaches to agricultural output” in Machine learning approaches for evaluating statistical information in the agricultural sector. eds. P. Jarvis, A. Rey, and C. Petsikos (Cham: Springer), 1–17.
Mkuhlani, S., Kephe, P. N., Rusere, F., and Ayisi, K. (2024). Editorial: modelling approaches for climate variability and change mitigation and adaptation in resource constrained farming systems. Front. Sust. Food Syst. 8:1510162. doi: 10.3389/fsufs.2024.1510162
Nelson, D. W., and Sommers, L. E. (1982). “Total carbon, organic carbon, and organic matter” in Methods of soil analysis. Part 2. eds. A. L. Page, R. H. Miller, and D. R. Keeney (Madison, WI: ASA and SSSA), 539–579.
Padhiary, M., Roy, P., Dey, P., and Sahu, B. (2025). “Harnessing AI for automated decision-making in farm machinery and operations: optimizing agriculture,” in Enhancing Automated Decision-Making Through AI. London: IGI Global Scientific Publishing, 249–282.
Purohit, K., Kumar Singh, A., and Chatterjee, S. (2024). Enhancing agriculture production through smart assessment of soil nutrients. J. Crop Improv. 38, 1–18. doi: 10.1080/15427528.2024.2355249
Qiu, Z., Ma, F., Li, Z., Xu, X., Ge, H., and du, C. (2021). Estimation of nitrogen nutrition index in rice from UAV RGB images coupled with machine learning algorithms. Comput. Electron. Agric. 189:106421. doi: 10.1016/j.compag.2021.106421
Reddy, T. V., Reddy, R. A., Prasanna, K. S., Shiva, S. S., Meghana, S., Reddy, T. S. S., et al. (2024). “Design and developing AI-driven agro-sage for optimal precision agriculture,” in 2024 5th International Conference on Smart Electronics and Communication (ICOSEC), IEEE, 1538–1542.
Sarkar, T., and Mishra, M. (2018). Soil erosion susceptibility mapping with the application of logistic regression and artificial neural network. J. Geovis. Spat. Anal. 2:8. doi: 10.1007/s41651-018-0015-9
Tziachris, P., Aschonitis, V., Chatzistathis, T., Papadopoulou, M., and Doukas, I. D. (2020). Comparing machine learning models and hybrid geostatistical methods using environmental and soil covariates for soil pH prediction. ISPRS Int. J. Geo Inf. 9:276. doi: 10.3390/ijgi9040276
Wadoux, A. M.-C., Minasny, B., and McBratney, A. B. (2020). Machine learning for digital soil mapping: applications, challenges and suggested solutions. Earth Sci. Rev. 210:103359. doi: 10.1016/j.earscirev.2020.103359
Xing, Y., Zhang, X., and Wang, X. (2024). Enhancing soil health and crop yields through water-fertilizer coupling technology. Front. Sust. Food Syst. 8:1494819. doi: 10.3389/fsufs.2024.1494819
Zeraatpisheh, M., Garosi, Y., Reza Owliaie, H., Ayoubi, S., Taghizadeh-Mehrjardi, R., Scholten, T., et al. (2022). Improving the spatial prediction of soil organic carbon using environmental covariates selection: a comparison of a group of environmental covariates. Catena 208:105723. doi: 10.1016/j.catena.2021.105723
Keywords: precision farming, machine learning, soil data analytics, fertilizer optimization, GeaGrow mobile application
Citation: Folorunso O, Ojo O, Busari M, Adebayo M, Adejumobi J, Folorunso D, Ayo F, Alabi O and Olabanjo O (2025) GeaGrow: a mobile tool for soil nutrient prediction and fertilizer optimization using artificial neural networks. Front. Sustain. Food Syst. 9:1533423. doi: 10.3389/fsufs.2025.1533423
Received: 23 November 2024; Accepted: 24 February 2025;
Published: 06 March 2025.
Edited by:
Elsayed Said Mohamed, National Authority for Remote Sensing and Space Sciences, EgyptReviewed by:
Abdelraouf M. Ali, National Authority for Remote Sensing and Space Sciences, EgyptCopyright © 2025 Folorunso, Ojo, Busari, Adebayo, Adejumobi, Folorunso, Ayo, Alabi and Olabanjo. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olusegun Folorunso, Zm9sb3J1bnNvb0BmdW5hYWIuZWR1Lm5n
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.