- 1Research Base of Zhengzhou University, State Key Laboratory of Cotton Bio-Breeding and Integrated Utilization, School of Agricultural Sciences, Zhengzhou University, Zhengzhou, China
- 2State Key Laboratory of Cotton Bio-Breeding and Integrated Utilization, Institute of Cotton Research, Chinese Academy of Agricultural Sciences, Anyang, China
- 3Western Agricultural Research Center, Chinese Academy of Agricultural Sciences, Changji, China
Aphis gossypii Glover (Hemiptera: Aphididae) causes considerable damage to crop yields globally. Solanum nigrum (Solanales: Solanaceae), an annual malignant weed, serves as a crucial weed host for A. gossypii. However, the potential mechanisms by which A. gossypii adapts to different hosts during the transition between crops, such as Gossypium hirsutum (Malvales: Malvaceae) and S. nigrum remain elusive. We calculated the life table parameters of A. gossypii after rearing on S. nigrum for ten generations. The fifth generation of A. gossypii (T5) exhibited the strongest adaptability to S. nigrum, demonstrating notably higher values of r (intrinsic rate of increase), λ (finite rate of increase), and fecundity compared to the first generation of A. gossypii (T1). Upon retransferring T1, T5, and the tenth generation of A. gossypii (T10) were retransferred to G. hirsutum (designated as T1-M, T5-M, and T10-M, respectively), the T5-M showed superior r, λ, and fecundity compared to both T1-M and T10-M. 16S rRNA sequencing and qPCR analyses indicated a significant decrease in the diversity of the symbiotic bacterial community in both T5 and T10. Notably, Buchnera and Arsenophonus were two dominant symbiotic bacteria related to metabolism and host adaptability in A. gossypii. The relative abundance of Buchnera in T5 and T10 significantly increased compared to M and T, while the relative abundance of Arsenophonus decreased markedly. KEGG (Kyoto Encyclopedia of Genes and Genomes) function prediction analysis suggested that the roles of symbiotic bacteria in A. gossypii are primarily linked to metabolic processes. Therefore, the adaptation of A. gossypii to S. nigrum enhances its population expansion on G. hirsutum, potentially involving the metabolic functions of Buchnera and Arsenophonus. These findings provide a theoretical foundation for the scientific management of A. gossypii and S. nigrum in the fields.
1 Introduction
Aphis gossypii Glover (Hemiptera: Aphididae) is a polyphagous pest renowned for causing substantial losses to both crop yields and quality across the globe (Morando et al., 2021). Additionally, non-crop plants (such as weeds) might contribute to the host transfer process of A. gossypii. These weeds, located within or adjacent to agricultural fields, can serve as alternative hosts, supporting the growth and reproduction of aphid populations (Caballero-López et al., 2011). And the secondary metabolites of different plants are different, and aphids that feed on different host plants for a long time may have population differentiation (Razmjou et al., 2010; Vorburger et al., 2017), thus increasing the biodiversity of farmland pests. However, there are several aspects remain elusive: the specific selection of plant hosts during aphid’s transfer from overwintering hosts to crops, the precise role of weeds in the aphid’s adaptation to host crops, and the underlying adaptive mechanisms that facilitate aphid’s transition between various hosts.
Microbial symbiosis is widely present in insects, with bacteria and insects co-evolving over hundreds of millions of years. This symbiotic relationship is vital for the survival and reproduction of insects. In the case of aphids, bacteria offer numerous benefits, including nutrition supply (Douglas et al., 2001; Rabatel et al., 2013) and protection from natural predators (Frago et al., 2017; Hrček et al., 2016). The symbiotic microorganisms harbored by aphids play a pivotal role in the process of host transfer. Typically, a shift in hosts alters the bacterial community structure within A. gossypii (Zhao et al., 2016). Furthermore, the bacterial symbionts present in aphids are influenced by the types of plants the aphids consume (Chandler et al., 2008). Notable differences exist in populations of Acyrthosiphon pisum on peach trees, Medicago sativa, and clover, and these variations are closely linked to their symbiotic microflora (Simon et al., 2003). Similarly, Aphis craccivora demonstrated distinct microbial communities when feeding on M. sativa and Robinia pseudoacacia, indicating a broad association between bacterial symbionts and host plants (Brady and White, 2013). Studies indicates that when the same aphid species feeds on different host plants, the prevalence of their symbionts often varies (Henry et al., 2015). Specifically, Hamiltonella was only detected in A. craccivora feeding on M. sativa, whereas Arsenophonus was exclusively present in A. craccivora feeding on R. pseudoacacia (Brady and White, 2013). However, there is a dearth of studies exploring the microbial changes of A. gossypii upon its transfer from cotton to alternative host plants. In consequence, it remains unclear how host adaptation alters the diversity of microbes within A. gossypii.
Solanum nigrum (Solanales: Solanaceae) is an annual broadleaf weed that grows rapidly in the field, distinguished by its vigorous growth, high fecundity, brief growth cycle, and persistent fruitfulness (Zhao et al., 2017). When fully mature, S. nigrum can occupy significant space, impeding light penetration and competing for nutrients with crops. These competitions lead to slower growth of crops, ultimately reducing both yield and quality. S. nigrum serves as an alternative host for aphids, especially in environments where cultivated crops are temporarily unavailable. Therefore, A. gossypii (Perng, 2002), Myzus persicae (Almohamad et al., 2006), Aulacorthum solani (Milbrath and Biazzo, 2012) and Brevicoryne brassicae (Coutts et al., 2006) often utilize S. nigrum as a host for growth and reproduction. In addition, S. nigrum stands as a important weed in corn and soybean fields (Dalley et al., 2004), with the potential to be parasitized by aphids. This host adaptability can pose a threat to crops, yields and ultimately food security. Nevertheless, the mechanisms by which A. gossypii adapts to S. nigrum, as well as the subsequent effects on its population expansion capacity upon returning to the original host crop, remains undefined.
Numerous studies have investigated the adaptability of A. gossypii to various host plants and the alterations in their internal microorganisms. However, the shifts in the symbiotic bacterial community of A. gossypii following host transfer, as well as their adaptive mechanism to plant hosts during this transition is not clear. Therefore, this paper initially constructed a life table for multi-generational growth of A. gossypii on S. nigrum, thereby assessing its adaptability to this host. Subsequently, utilizing 16S rRNA sequencing technology, we analyzed the dynamics of symbiotic microbial communities during A. gossypii adaptation to S. nigrum.
The results of this study offer insights into the adaptive mechanism of A. gossypii to the weed host S. nigrum, which is instrumental in unveiling its broader adaptive strategies following host transfers. Ultimately, this research provides theoretical guidance for the scientific prevention and control of A. gossypii.
2 Materials and methods
2.1 Insects and plants
A. gossypii used in this study were collected from the cotton seedlings of the experimental farm of the Institute of Cotton Research of CAAS (Anyang City, Henan Province). These collected aphid were confirmed by examination of the mtDNA COI gene sequences as A. gossypii (Wang et al., 2016). A. gossypii were subsequently reared in laboratory for multiple generations without exposure to insecticides. Additionally, seeds of Gossypium hirsutum (Malvales: Malvaceae) (CCRI49) and S. nigrum sourced from the same experimental farm. A. gossypii, G. hirsutum, and S. nigrum were maintained under controlled environmental conditions (temperature 25 ± 1°C, relative humidity 65 ± 5%, and photoperiod 14 h: 10 h photoperiod).
2.2 Construction of life table for A. gossypii
Fresh leaf of G. hirsutum or S. nigrum were positioned in a plastic petri dish (9 cm diameter, with a 1 cm layer of 1.8% agar). A wingless adult aphid was placed onto the leaf. The nymph of A. gossypii was transferred on a new leaf of G. hirsutum (labeled M) and S. nigrum (labeled T1). Subsequently, the nymph borning from T1 adult was moved to fresh S. nigrum leaf (labeled T2), and this process continued until T10. Concurrently, nymph from T1, T5, and T10 was transferred to G. hirsutum leaves (designated as T1-M, T5-M, and T10-M, respectively). Aphid growth was monitored daily, noting their lifespan and fecundity until their demise. Each aphid served as an individual replicate, with 60 replicates per generation. The leaves are replaced every 2 to 3 days, and the feeding conditions of A. gossypii are the same as mentioned above.
2.3 16S rRNA sequencing
Thirty adults of M, T1, T5, and T10 were gathered within 48 h after the final molt. These samples underwent sterilization with 75% alcohol for 5 min (Chang et al., 2023), followed by quick-frozen with liquid nitrogen, and were then stored at −80°C. From each generation, four samples were collected. The total DNA of A. gossypii samples was extracted using the FastDNA Spin kit for soil (MP Biomedicals, United States). The concentration and purity of the extracted DNA were determined using NanoDrop 2000C (Thermo Scientific, United States), and the integrity of DNA was assessed by 1.2% agarose gel electrophoresis.
The genes in the V3 ~ V4 region of bacterial 16S rRNA were amplified with universal primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′) (Xu et al., 2016), with the extracted DNA as template for 16S rRNA sequencing. PCR amplification, quantification, identification, and purification PCR product, and library preparation as described previously (Zhao et al., 2016). Sequencing was then performed on an Illunina Miseq PE 300 platform of Majorbio Bio-Pharm Technology Co. Ltd. (Shanghai, China). Details of the analysis process can be found in Supplementary Method S1. Sequencing data have been uploaded to NCBI (BioProject ID PRJNA1155176).
2.4 Absolute quantification of bacterial community
Based on the results of 16S rRNA sequencing, three symbiotic dominant bacteria (bacteria with relative abundance greater than 10% were defined as dominant bacteria) were chosen for absolute quantitative analysis. The copy numbers across various generations were calculated. The bacterial target sequence was cloned into a pUC19 cloning vector (Zhili Zhongte Wuhan Biotechnology Co., Ltd., China), which subsequently served as a standard vector (confirmed through sequencing). The copy numbers calculation process and method of the sample target bacteria refer to previous studies (Chang et al., 2023). Primer sequences (Supplementary Method S2) and PCR amplification efficiency were shown in Supplementary Table S1. It was ensured that the amplification efficiency (E value) fell within the 0.9 and 1.1, with a correlation (R2) is above 0.99. The quantitative polymerase chain reaction (qPCR) were set up according to the MonAmp™ SYBR® Green qPCR Mix manual (Wuhan Mona Biological Co., Ltd.) and executed using the quantitative fluorescence PCR instrument (CFX Opus 96, Bio-rad, Singapore).
2.5 Statistical analysis
Utilizing the theory of age-stage, sex life table theory, the raw life table data of all individuals were analyzed through TWOSEX-MSChart program (http://140.120.197.173/Ecology/, Ver. 24/01/2023) (Chi, 1988; Chi and Liu, 1985). Bootstrap technique was used to estimate the variance and standard deviation of population parameters. Paired bootstrap test in TWOSEX-MSChart program was used to test the difference significance of the data. Additionally, GraphPad Prism (9.0.0) (GraphPad Software Corporation) was used to plot. Student’s t-test and Log-rank (Mantel-Cox) test were executed via IBM SPSS Statistics 20 (International Business Machines Corporation), with p < 0.05 indicating statistical difference.
3 Results
3.1 Effects of S. nigrum on multigenerational life table parameters of A. gossypii
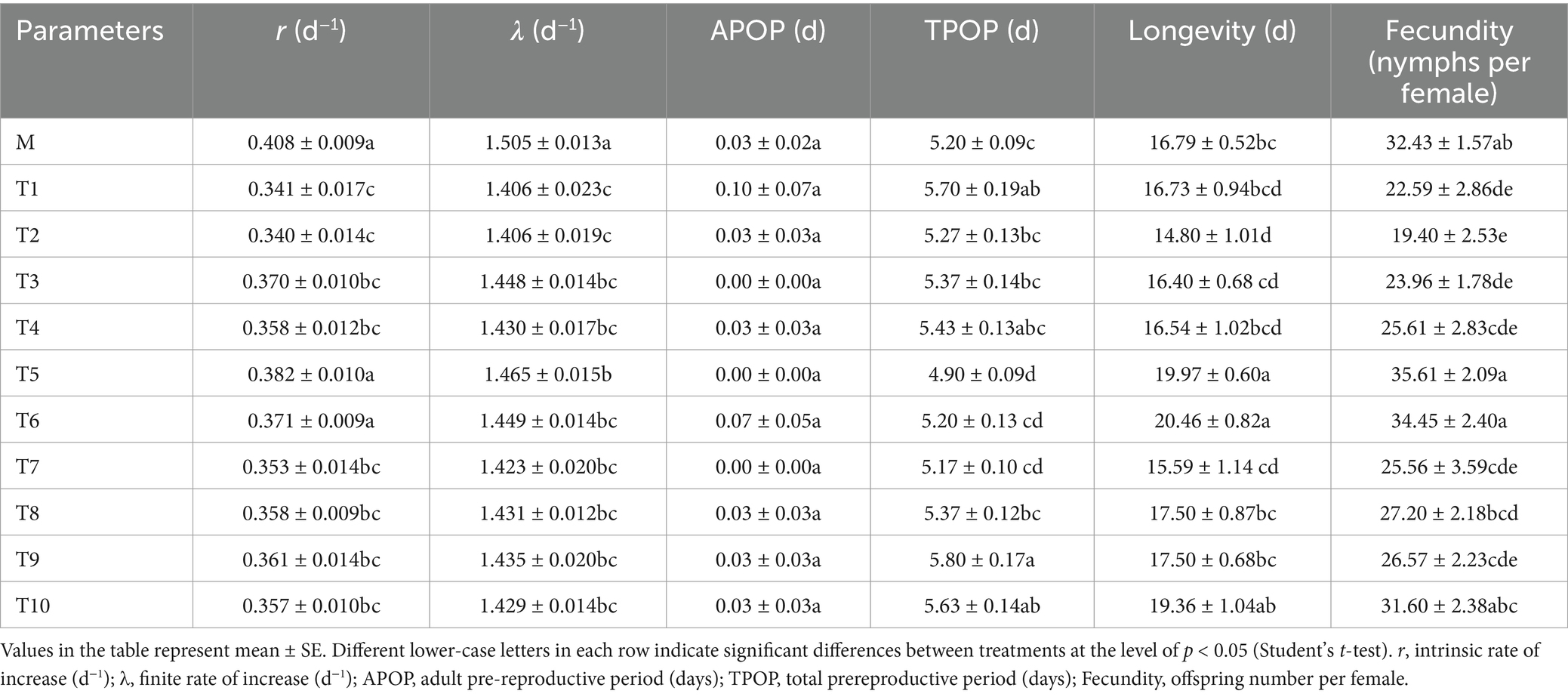
The life table parameters of different generations of A. gossypii reared on S. nigrum are shown in Table 1. The results showed that, excluding T5 (0.382 d−1) and T6 (0.371 d−1), the r (intrinsic rate of increase) value of M (0.408 d−1) was significantly greater than that of other generations. The λ (finite rate of increase) of each generation was significantly lower than that of M (1.505 d−1), yet T5 (1.465 d−1) exhibited the highest λ among all generations. In comparison to M (5.20 d), the TPOP (total prereproductive period) of T1 (5.70 d), T9 (5.80 d), and T10 (5.63 d) increased significantly, whereas the TPOP of T5 (4.90 d) decreased notably. The longevity of T5 (19.97 d) and T6 (20.46 d) was surpassed that of M (16.79 d) significantly, and their fecundity also proved higher across all generations. Moreover, the lx (age-specific survival rate) of T5 was elevated compared to other generations between 15 to 20 days (Figure 1A), the mx (age-specific fecundity) of M was superior to other generations (from 3 to 10 days), followed by an alternating leadership between T5 and T10 in mx (Figure 1B).
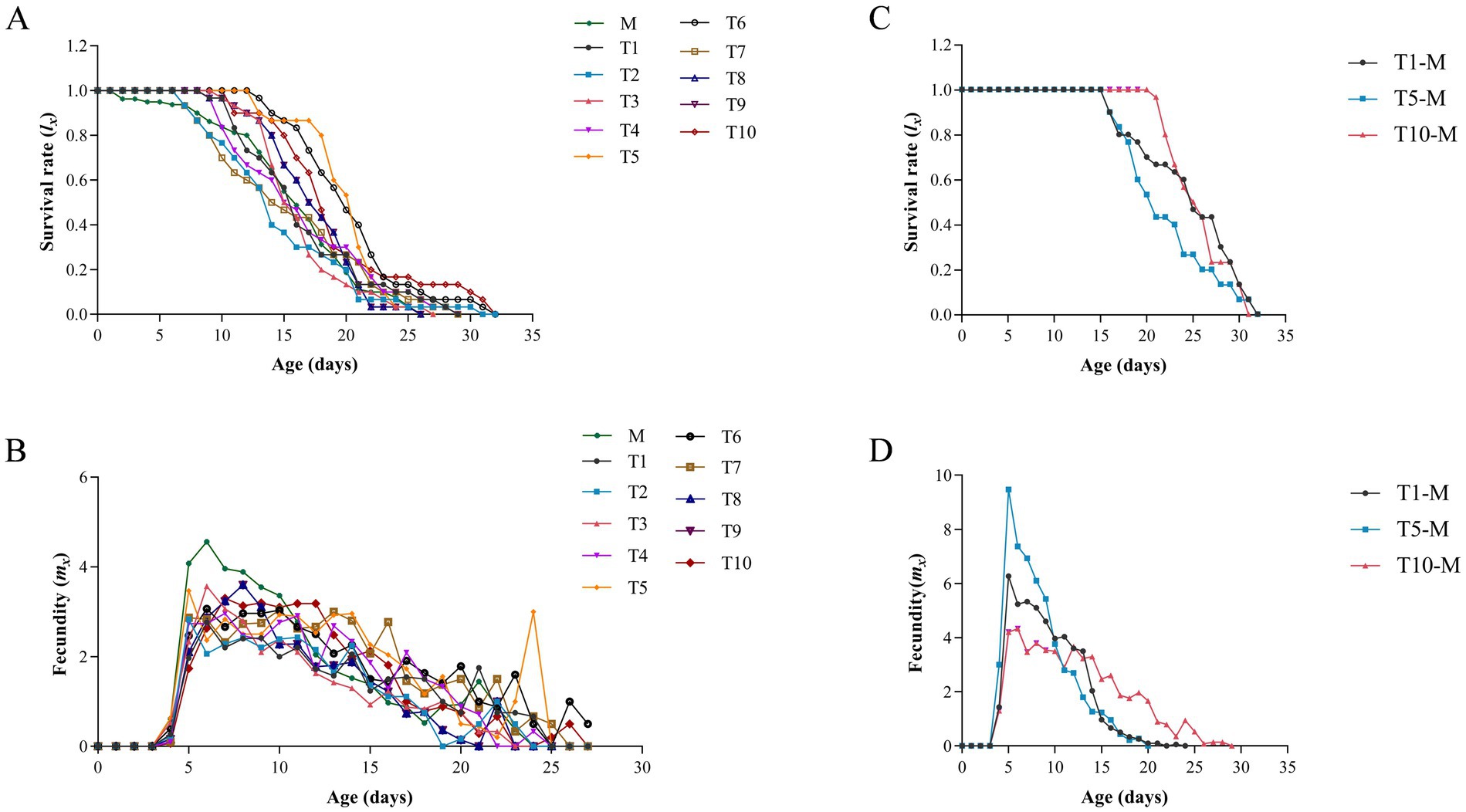
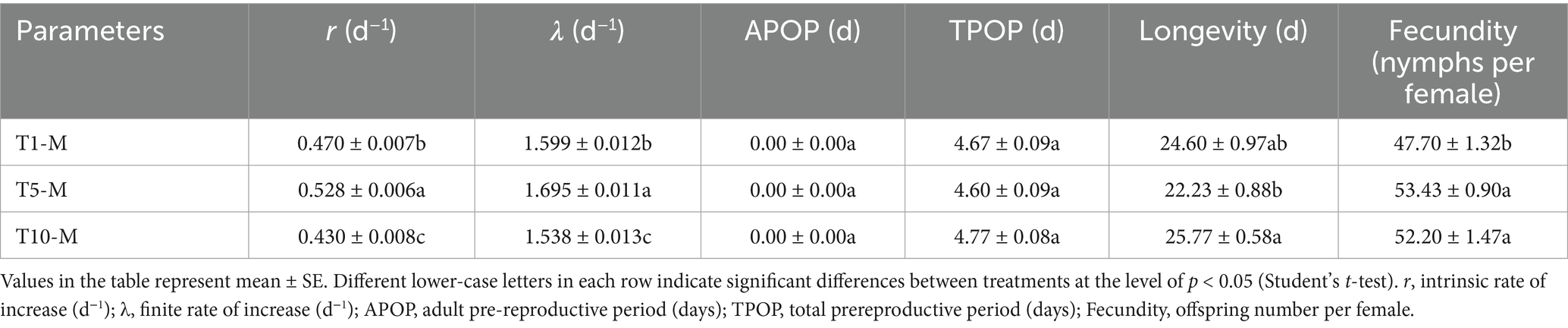
Figure 1. Age specific survival rate (lx) (A) and fecundity (mx) (B) of A. gossypii from S. nigrum ten generation and on G. hirsutum. Age specific survival rate (lx) (C) and fecundity (mx) (D) of A. gossypii on S. nigrum retransferring to G. hirsutum.
The life table parameters of A. gossypii after transferred from S. nigrum to G. hirsutum are shown in Table 2. According to the population parameter listed in Table 1, the T1, T5, and T10 of A. gossypii were retransferred to their original host (G. hirsutum). The results revealed that the r and λ values of T5-M (r = 0.528 d−1, λ = 1.695 d−1) were significantly higher compared to those of T1-M (r = 0.470 d−1, λ = 1.599 d−1) and T10-M (r = 0.430 d−1, λ = 1.538 d−1). Furthermore, the fecundity of T5-M (53.43) and T10-M (52.20) proved to be significantly greater than that of T1-M (47.70). However, the longevity of T5-M (22.23 d) was considerably shorter than that of T10-M (25.77 d). No substantial disparities were detected in APOP (adult pre-reproductive period) and TPOP across the three generations. The value of lx of T5-M was higher than that of T1-M and T10-M (from 16 to 23 days) (Figure 1C). Additionally, the mx of T5-M prevailed over other two generations (from 5 to 10 days), while the mx of T10-M emerged as the highest after 13 days (Figure 1D).
3.2 Summary of the 16S rRNA sequencing
To clarify the differences in microbial community diversity of A. gossypii on S. nigrum and G. hirsutum, the M, T1, T5, and T10 of A. gossypii were selected for sequencing, based on distinct biological data. A total of 799,239 high-quality, optimized sequences, representing 343,078,429 bases. The average sequencing coverage exceeded 99% across all groups, demonstrating adequate sequencing quantity and reasonable depth (Supplementary Table S2).
Rarefaction curves based on Sobs index (Supplementary Figure S1A) and Shannon index (Supplementary Figure S1B) showed that increasing the sample volume would not produce more OTUs (operational taxonomic units), suggesting that the data accurately reflects the composition of the majority of bacterial communities present in the samples. In comparison to M and T1, the Shannon index for T5 and T10 decreased, while the Simpson index increased (Supplementary Table S2). Furthermore, both Chao 1 (decreased 18.11%) and Ace (decreased 22.64%) indices decreased relative to M after continuously feed A. gossypii with S. nigrum.
3.3 Influence of bacterial community of A. gossypii feeding with S. nigrum
M, T1, T5, and T10 exhibited unique OTUs with counts of 33, 22, 16, and 76, respectively, while 119 OTUs were shared by the four groups (Figure 2A). Principal Co-ordinates Analysis (PCoA) revealed significant disparities in genus level composition of A. gossypii among M, T5, and T10 (Figure 2B).
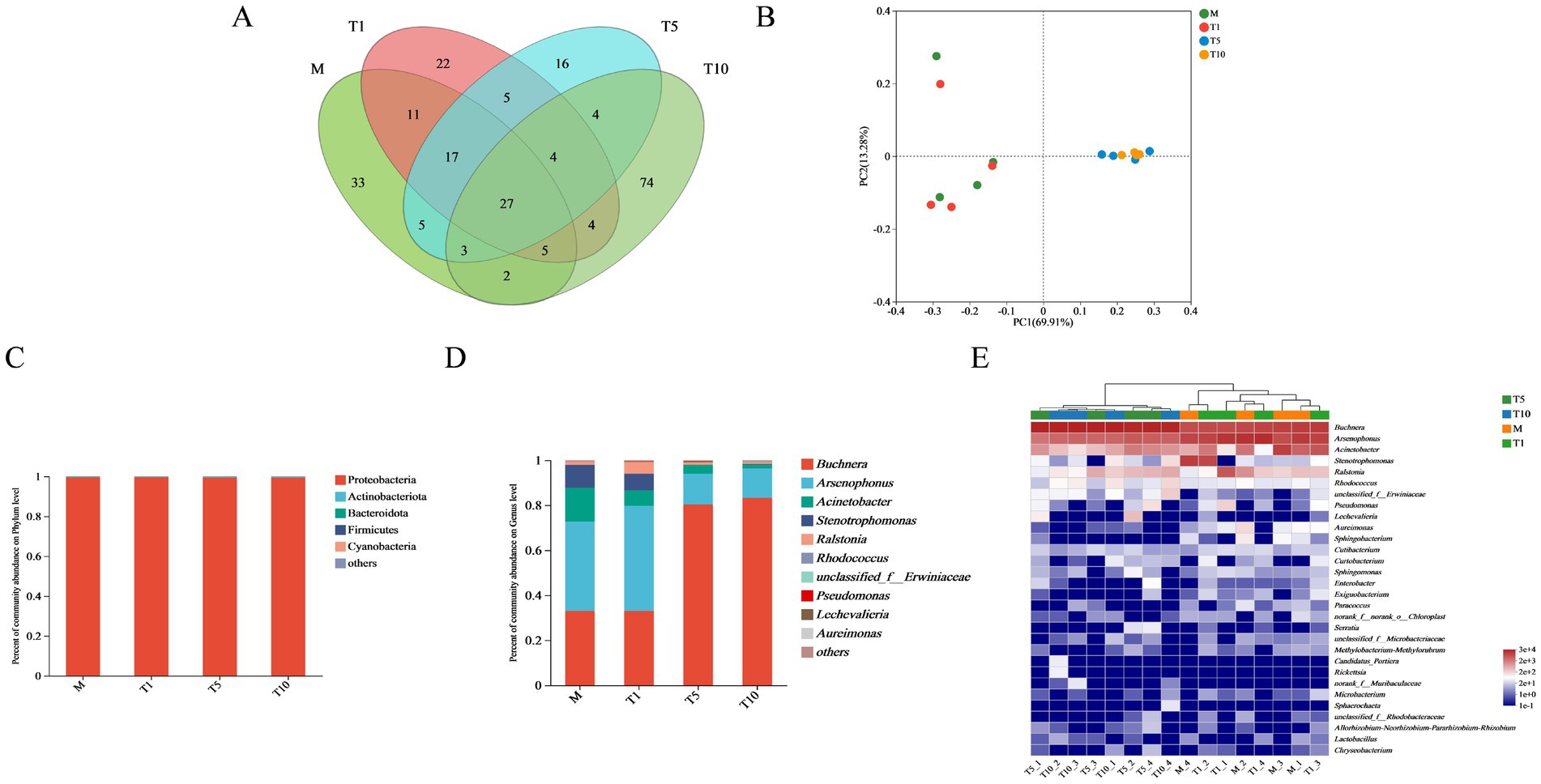
Figure 2. Influence of continuous feeding of S. nigrum on bacterial community of A. gossypii. The Venn diagram (A) illustrates the number of shared and unique bacteria in the four treatments. (B) Difference of OTUs types in different treatments based on Principal co-ordinates analysis (PCoA). The relative abundance of dominant bacterial communities of A. gossypii at the phylums (C) and genus (D) levels after feeding on S. nigrum. (E) Heatmap shows the relative abundance of the dominant bacterial community at the genus level for each sample.
The histograms of bacterial phylums and genus in four groups of A. gossypii were mapped to visualize the composition and distribution of bacterial communities. Then, the difference of symbiotic bacteria abundance among each group was compared at each taxonomic level by averaging the samples within each group. At the phylum level, the five most abundant bacteria were identified as Proteobacteria, Actinobacteriota, Bacteroidota, Firmicutes, and Cyanobacteria (Figure 2C). Notably, Proteobacteria was the highest relative abundance, accounting for 99.58% in M, and similarly high percentages in T1 (99.62%), T5 (99.48%), and T10 (99.41%).
At the genus level, Buchnera and Arsenophonus were the dominant bacteria all four groups (Figure 2D). Specifically, the superiority bacteria in M were Arsenophonus (39.59%), Buchnera (33.02%), Acinetobacter (15.14%), and Stenotrophomonas (10.15%) (Supplementary Table S3). In T1, the relative abundance of Buchnera, Acinetobacter, and Stenotrophomonas decreased to 33.01, 6.89, and 7.37%, respectively. In compared to M, the relative abundance of Buchnera in T5 and T10 increased to 80.30 and 83.28%, respectively. Conversely, the relative abundance of Arsenophonus decreased to 13.68 and 13.11%, respectively, while those of Acinetobacter decreased to 3.95 and 1.66%, respectively.
The aggregation of bacterial community at the genus level in A. gossypii was visualized through a heat map (Figure 2E). In general, the relative abundance of Arsenophonus, Buchnera, Acinetobacter, and Stenotrophomonas in A. gossypii was higher in all groups. The relative abundance of Buchnera in T5 and T10 was higher than that in M and T1, whereas the relative abundance of Arsenophonus, Acinetobacter, and Stenotrophomonas was comparatively lower in T5 and T10 than in M and T1.
The significant changes in bacterial abundance were compared among the four groups (Figure 3). There were no significant alterations in the abundance of the four prevalent bacteria (Arsenophonus, Buchnera, Acinetobacter, and Stenotrophomonas) in T1 compared with M (Figure 3A). However, compared with M, the abundance of Buchnera in T5 increased significantly by 58.88%, while the abundance of Arsenophonus decreased significantly by 65.44% (Figure 3B). Similarly, the abundance of Buchnera in T10 rose significantly by 60.35%, and the abundance of Arsenophonus dropped significantly by 66.89% (Figure 3C). Nevertheless, the abundance of Acinetobacter and Stenotrophomonas remained largely unchanged in both T5 and T10.
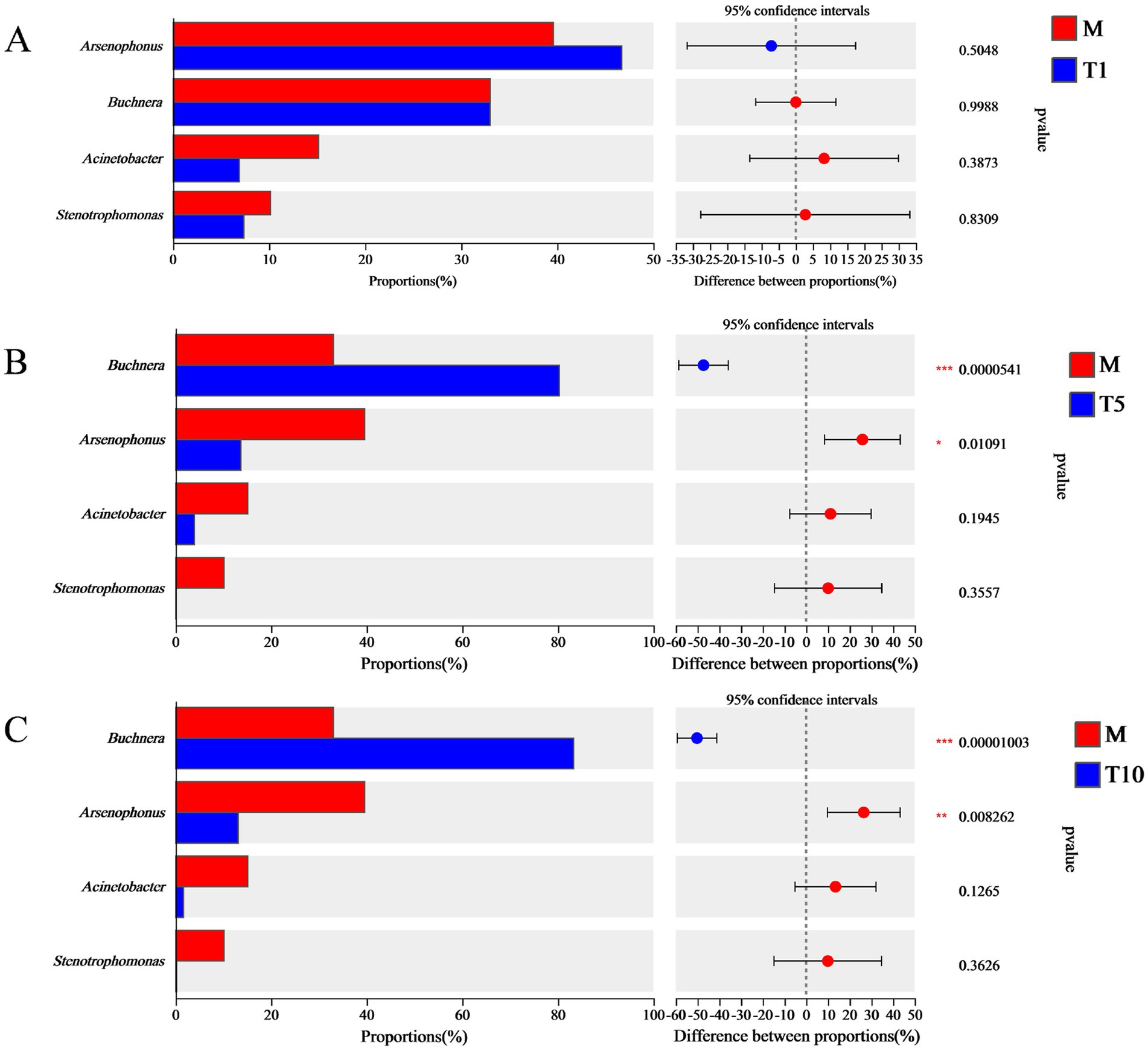
Figure 3. Difference analyses of multiple generations of S. nigrum feeding on bacterial community in A. gossypii. Analysis of bacterial community differences between M and T1 (A), M and T5 (B), M and T10 (C). Student’s t test is used for significance analysis (*p < 0.05, **p < 0.01, and ***p < 0.001).
3.4 Absolute quantitative analysis of dominant bacteria
Analysis of dominant bacteria (Buchnera, Arsenophonus, and Acinetobacter) in M, T1, T5, and T10 of A. gossypii by qPCR found that, the abundance of Buchnera was higher in all groups, with a significant increase in copy number observed in T5 and T10 compared to T1 and M. Conversely, the copy number of Arsenophonus was significantly reduced in T5 and T10 compared to T1 and M. No notable difference was detected in the copy number of Acinetobacter among the four groups (Figure 4).
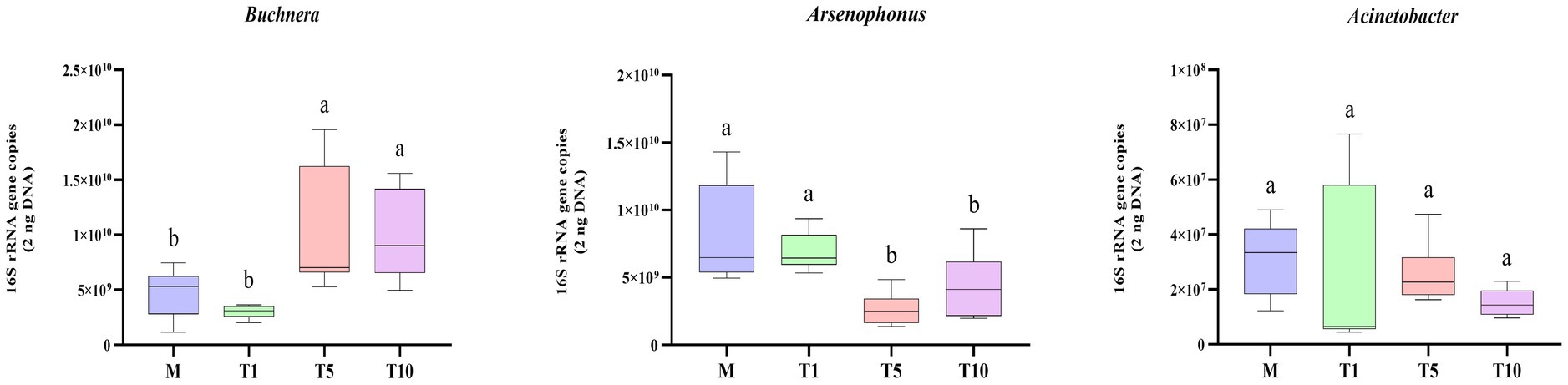
Figure 4. The 16S rRNA gene copy number of bacterial community in A. gossypii fed on G. hirsutum and different generations of S. nigrum. The lower-case letters in each figure indicated that there was a significant difference between the control group and the treatment group at the p < 0.05 level (Student’s t-test).
3.5 Functional prediction
To investigate the role of different microbial communities in A. gossypii after host shift, PICRUSt2, combined with KEGG database, was used to predict and analyze the function of these microbial community. The analysis revealed that 73.41% of the microorganisms in A. gossypii were involve in metabolism (Supplementary Figure S2). According to the KEGG secondary classification function prediction, the top five predicted functions fall under “global and overview maps,” “carbohydrate metabolism,” “amino acid metabolism,” “metabolism of cofactors and vitamins,” and “energy metabolism” (Figure 5). Furthermore, microorganisms in T5 and T10 displayed a notable concentration in “carbohydrate metabolism,” “membrane transport,” “cellular community-prokaryotes,” “signal transduction,” and other pathways, compared with those in T1 and M. While the symbiotic bacteria in T1 and M showed a significant focus on “metabolism of cofactors and vitamins,” “energy metabolism,” “nucleotide metabolism,” and other functions (Supplementary Figure S3).
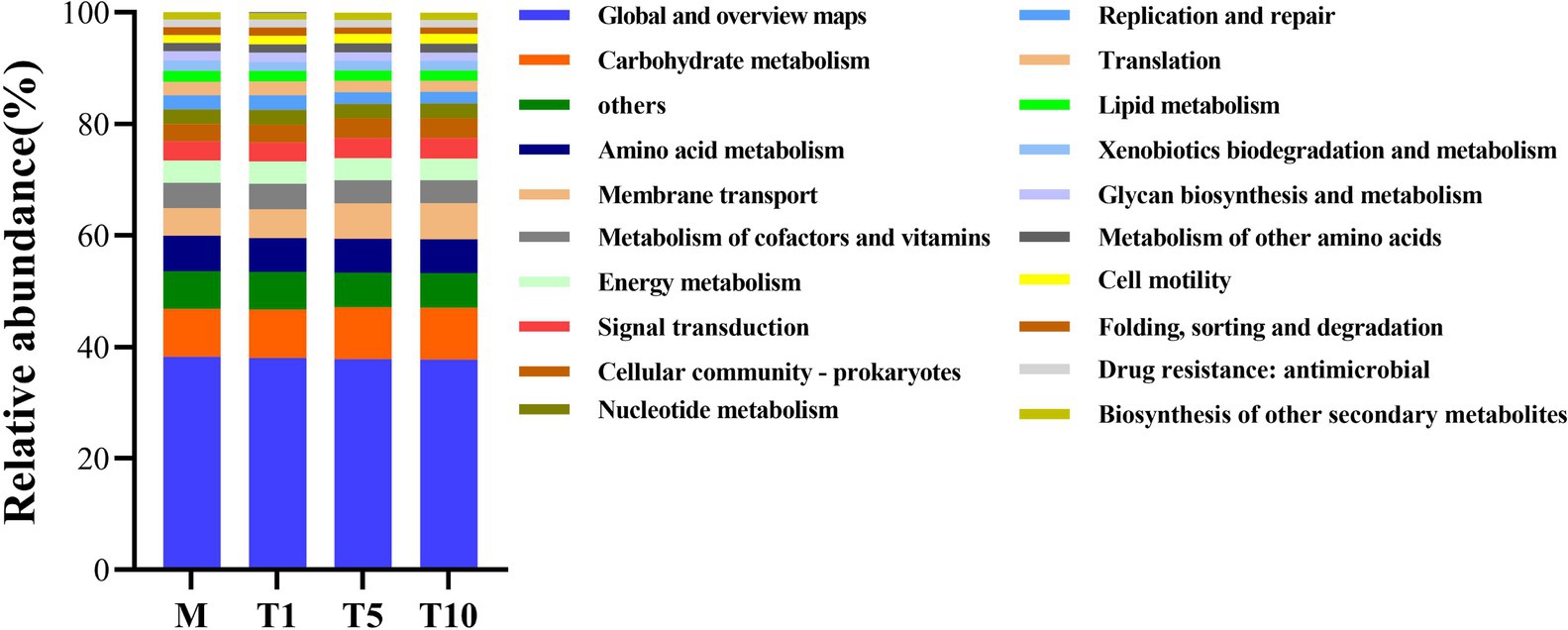
Figure 5. Functional prediction of KEGG secondary classification of microbes in A. gossypii under different groups.
4 Discussion
Although A. gossypii is hosted by numerous plants, research on the role of weeds in its host shift remains limited. S. nigrum stands as a prominent weed in agricultural fields. Nevertheless, the adaptability mechanism of A. gossypii to S. nigrum, and the subsequent effects on its population expansion capacity upon returning to original host crop, are still not fully understood. S. nigrum serves as an alternative host for A. gossypii, particularly in environments where cultivated crops are temporarily unavailable. Our research emphasizes the multi-generational adaptation of S. nigrum and its role in enhancing the reproduction rates of A. gossypii. This adaptation of the weed may increase habitat suitability for the aphid, thereby facilitating its population growth. Elevated aphid populations can significantly impact crop yields through direct feeding damage (Ramalho et al., 2012) and the transmission of plant viruses (Smith and Chuang, 2014). In regions such as the North China Plain, where crops like cotton, corn, and vegetables are essential, the increased reproduction of A. gossypii could result in considerable economic losses (Zhang et al., 2018). A. gossypii can diminish plant vigor, stunt growth, and reduce both the quality and quantity of agricultural produce (Zhong et al., 2022), directly affecting farmers’ livelihoods and regional food supplies.
Host adaptation is a long process. The research revealed that no significant difference in r between A. gossypii initially reared on Hibiscus syriacus and those on Cucumis sativus after three generations (Zheng et al., 2007). When Tetranychus viennensis transfers from apple trees to other fruit trees, the first generation experiences negative effects on life table parameters, but these effects dissipate in succeeding generation (Li et al., 2006). A. gossypii develops faster on the original host compared to non-original host, and its r is higher on the original host (Satar et al., 2013), indicating inhibited development on the non-original hosts. Transferring A. gossypii from cotton to C. sativus and Abelmoschus manihot for four consecutive generations did not enhance its development (Satar et al., 2013). The first-generation life table parameters of Amphitetranychus viennensis suffered when transferred from Prunus serotine, which is conducive to its growth, to Malus domestica (Kafil et al., 2007). Likewise, when A. viennensis was transferred to Prunus avium, its first-generation life table parameters fared better than the third generation, showing no gradual adaptation to P. avium (Kafil et al., 2007). However, the difference of life table of a pest on different host crops may be caused by the difference of secondary metabolites (Abdelsalam et al., 2016), more experiments are essential to uncover the exact mechanism.
Our results demonstrated that A. gossypii gradually adapted to S. nigrum, with T5 demonstrating significant progress. The shape of lx and mx curves illustrate the interplay of life history traits and determine r, a common indicator for evaluating the adaptability of pest to plants (Bethke et al., 1998; Sandström and Pettersson, 1994). A later decline in the lx curve and higher peak in the mx indicate a the greater r and, consequently, higher aphid population growth potential. The r of A. gossypii on S. nigrum peaked at T5, signifying the highest population growth potential for this generation on S. nigrum. In the absence of alternative crops like G. hirsutum, A. gossypii can rapidly multiply on S. nigrum, maintaining a dense population.
Microbial symbionts play a pivotal role in enhancing the adaptability of pests to their hosts. In this study, significant changes were observed in the bacterial community of T5 and T10 of A. gossypii feeding on S. nigrum. At the phylum level, the abundance and species of bacterial communities remained stable across all groups, and the highest abundance of bacteria was Proteobacteria. Proteobacteria are recognized as a vital component of the intestinal flora in numerous herbivorous insects and fostering a beneficial symbiotic relationship with their hosts (Show et al., 2022). These insects provide a stable environment for Proteobacteria, which, in turn, endow their hosts with novel metabolic capabilities (Latorre et al., 2003).
Buchnera is a member of the Proteobacteria, which is the dominant bacterium in aphids growing on various host plants (Ma et al., 2021). Its roles extend to DNA replication, transcription, protein translation, secretion, energy metabolism, and amino acid biosynthesis for aphids (Baumann et al., 1995; Richards et al., 2010; Tian et al., 2023). In our study, a notable increase in the relative abundance of Buchnera was observed in T5 and T10, while no significant alteration was detected in the M and T1. Similarly, Buchnera content was significantly higher in A. gossypii reared on C. sativus for over 10 years compared to those on cotton (Zhang et al., 2016). The relative abundance of Buchnera in A. gossypii reared on Cucurbita pepo was also significantly higher than those on cotton (Xu et al., 2023). Collectively, the elevated abundance of Buchnera is related to the adaptation of A. gossypii to new host plants.
Arsenophonus (Proteobacteria) is an intracellular symbiotic bacteria of insects, generally belonging to the secondary symbiotic bacteria, which has a wide range of hosts and rich biodiversity (Chen et al., 2014). This bacterium plays a crucial part in enhancing the growth performance of aphids during amino acid deficiency (Tian et al., 2023; Wang et al., 2024). Furthermore, it mitigates the harmful effects of parasitic wasps to aphids (Heidari Latibari et al., 2023) and mediates host specialization of aphids (Tian et al., 2019). Notably, Arsenophonus is also the dominant bacteria in A. gossypii (Xu et al., 2020; Zhang et al., 2021). In our study, the relative abundance of Arsenophonus was significantly lower in T5 and T10 compared to M and T1. This finding suggests a decline in Arsenophonus as A. gossypii adapts to S. nigrum. Similar study has showed that a reduction in Arsenophonus abundance of plutella xylostella after treatment with the insecticide sulfoxaflor (Shang et al., 2021).
Based on the changes in the abundance of symbiotic bacteria and predictions from the KEGG function database, Buchnera and Arsenophonus may regulated metabolic processes, enabling A. gossypii to gradually adapt to the S. nigrum. Across various life stages of Adelphocoris suturalis, the function of symbiotic bacteria predominantly focuses on metabolic pathway (Xue et al., 2021). The signal transduction function of intestinal bacteria increased after 17 generations of P. xylostella shifted from Raphanus sativus to Amygdalus persica (Yang et al., 2020). In comparison to the findings of this study, KEGG function prediction analysis suggests that Buchnera and Arsenophonus contribute to the adaptation of A. gossypii to S. nigrum by engaging in metabolic functions, particularly in “carbohydrate metabolism,” “membrane transport,” “metabolism of cofactors and vitamins,” “energy metabolism,” and so on. To gain a comprehensive understanding of these disparities, further investigation is necessary, possibly including metagenomic analyses to explore the functional significance of these symbiotic bacteria and to unravel the causal relationships.
In this study, the growth and reproduction parameters of A. gossypii demonstrated a gradual adaptation process as its generations progressed on S. nigrum. Notably, the T5 generation of A. gossypii exhibited the highest level of adaptation to S. nigrum. Upon transferring A. gossypii, which had been domesticated by S. nigrum, back to G. hirsutum, T5-M displayed the greatest values for r, λ, and fecundity. Furthermore, significant changes were observed in the symbiotic bacteria of A. gossypii after several generations on S. nigrum compared to those on G. hirsutum. The abundance of Buchnera in T5 and T10 of A. gossypii reared on S. nigrum increased significantly, whereas the abundance of Arsenophonus decreased significantly.
The adaptability of A. gossypii to transition between distinct host species, for instance, from S. nigrum to G. hirsutum, underscores the intricate interactions within agricultural ecosystems. These interactions can impact biodiversity and crop yields, as the presence of adaptable pests like A. gossypii has the potential to exacerbate pest infestation on economically crucial crops such as cotton, corn or soybean. By comprehending the mechanisms of host adaptation and the symbiotic relationships involved, more effective strategies could be devised to reduce the negative influence of A. gossypii on food production.
5 Conclusion
In conclusion, this article unveils the consequences of adaptation of A. gossypii to S. nigrum on their capacity for population growth and the abundance of dominant bacteria, while clarifying the function of S. nigrum in the host transition process of A. gossypii. The findings of this study offer valuable insights for the development of an innovative control technology system targeting A. gossypii, as well as advancing scientific and comprehensive methods for managing pests and weeds in fields.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/, PRJNA1155176.
Ethics statement
The manuscript presents research on animals that do not require ethical approval for their study.
Author contributions
PW: Formal analysis, Investigation, Writing – original draft. Y-XJ: Formal analysis, Investigation, Writing – original draft. Y-JM: Conceptualization, Supervision, Writing – original draft. DW: Investigation, Resources, Writing – original draft. Y-PS: Formal analysis, Writing – original draft. HH: Investigation, Software, Writing – original draft. CW: Data curation, Investigation, Writing – original draft. X-PS: Methodology, Supervision, Writing – original draft. XR: Methodology, Validation, Writing – original draft. YM: Project administration, Writing – review & editing. XM: Funding acquisition, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was supported by National Key R&D Program of China (2022YFD1400300), the earmarked fund for CARS (CARS-15-21), the Agricultural Science and Technology Innovation Program (ASTIP) (CAAS-ZDRW202412), and Central Public-interest Scientific Institution Basal Research Fund (No. 1610162023025).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Generative AI statement
The authors declare that no Generative AI was used in the creation of this manuscript.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1523123/full#supplementary-material
References
Abdelsalam, S. A., Awad, A. M. A., Abdelrahman, M. A. A., Nasser, M. A. K., and Abdelhamid, N. M. R. (2016). Antioxidant defense response of the green peach aphid, Myzus persicae against secondary metabolites of the host plants cumin, anise, and coriander. J. Agric. Sci. Technol. 18, 1583–1592.
Almohamad, R., Verheggen, F., Francis, F., and Haubruge, E. (2006). Evaluation of hoverfly Episyrphus balteatus De Geer (Diptera: Syrphidae) oviposition behaviour toward aphid-infested plants using a leaf disc system. Commun. Agric. Appl. Biol. Sci. 71, 403–412
Baumann, P., Baumann, L., Lai, C. Y., Rouhbakhsh, D., Moran, N. A., and Clark, M. A. (1995). Genetics, physiology, and evolutionary relationships of the genus Buchnera: intracellular symbionts of aphids. Ann. Rev. Microbiol. 49, 55–94. doi: 10.1146/annurev.mi.49.100195.000415
Bethke, J. A., Redak, R. A., and Schuch, U. K. (1998). Melon aphid performance on chrysanthemum as mediated by cultivar, and differential levels of fertilization and irrigation. Entomol. Exp. Appl. 88, 41–47. doi: 10.1046/j.1570-7458.1998.00344.x
Brady, C. M., and White, J. A. (2013). Cowpea aphid (Aphis craccivora) associated with different host plants has different facultative endosymbionts. Ecol. Entomol. 38, 433–437. doi: 10.1111/een.12020
Caballero-López, B., Blanco-Moreno, J. M., Pérez-Hidalgo, N., Michelena-Saval, J. M., Pujade-Villar, J., Guerrieri, E., et al. (2011). Weeds, aphids, and specialist parasitoids and predators benefit differently from organic and conventional cropping of winter cereals. J. Pest. Sci. 85, 81–88. doi: 10.1007/s10340-011-0409-7
Chandler, S. M., Wilkinson, T. L., and Douglas, A. E. (2008). Impact of plant nutrients on the relationship between a herbivorous insect and its symbiotic bacteria. Proc. R. Soc. B-Biol. Sci. 275, 565–570. doi: 10.1098/rspb.2007.1478
Chang, G., Xue, H., Ji, J., Wang, L., Zhu, X., Zhang, K., et al. (2023). Risk assessment of predatory lady beetle Propylea japonica's multi-generational exposure to three non-insecticidal agrochemicals. Sci. Total Environ. 886:163931. doi: 10.1016/j.scitotenv.2023.163931
Chen, Y., Wang, W. W., Chen, Y., and Fu, Q. (2014). Research progress on the bacterial symbiont Arsenophonus of insects. Acta Agric. Zhejiangensis 26, 530–536. doi: 10.3969/j.issn.1004-1524.2014.02.49
Chi, H. (1988). Life-table analysis incorporating both sexes and variable development rates among individuals. Environ. Entomol. 17, 26–34. doi: 10.1093/ee/17.1.26
Chi, H., and Liu, H. (1985). Two new methods for the study of insect population ecology. Bull. Inst. Zool. Acad. Sin. 24, 225–240.
Coutts, B. A., Hawkes, J. R., and Jones, R. A. C. (2006). Occurrence of beet western yellows virus and its aphid vectors in over-summering broad-leafed weeds and volunteer crop plants in the grainbelt region of South-Western Australia. Aust. J. Agric. Res. 57, 975–982. doi: 10.1071/AR05407
Dalley, C. D., Kells, J. J., and Renner, K. A. (2004). Effect of glyphosate application timing and row spacing on corn (Zea mays) and soybean (Glycine max) yields. Weed Technol. 18, 165–176. doi: 10.1614/02-150A
Douglas, A. E., Minto, L. B., and Wilkinson, T. L. (2001). Quantifying nutrient production by the microbial symbionts in an aphid. J. Exp. Biol. 204, 349–358. doi: 10.1242/jeb.204.2.349
Frago, E., Mala, M., Weldegergis, B. T., Yang, C., McLean, A., and Godfray, H. C. J. (2017). Symbionts protect aphids from parasitic wasps by attenuating herbivore-induced plant volatiles. Nat. Commun. 8:1860. doi: 10.1038/s41467-017-01935-0
Heidari Latibari, M., Moravvej, G., Rakhshani, E., Karimi, J., Arias-Penna, D. C., and Butcher, B. (2023). Arsenophonus: a double-edged sword of aphid defense against parasitoids. Insects. 14:763. doi: 10.3390/insects14090763
Henry, L. M., Maiden, M. C. J., Ferrari, J., Godfray, H. C. J., and Bourke, A. (2015). Insect life history and the evolution of bacterial mutualism. Ecol. Lett. 18, 516–525. doi: 10.1111/ele.12425
Hrček, J., McLean, A. H. C., and Godfray, H. C. J. (2016). Symbionts modify interactions between insects and natural enemies in the field. J. Anim. Ecol. 85, 1605–1612. doi: 10.1111/1365-2656.12586
Kafil, M., Allahyari, H., and Saboori, A. (2007). Effect of host plants on developmental time and life table parameters of Amphitetranychus viennensis (Acari: Tetranychidae). Exp. Appl. Acarol. 42, 273–281. doi: 10.1007/s10493-007-9095-9
Latorre, A., Gil, R., Silva, F. J., Martínez-Torres, D., and Moya, A. (2003). Tempo and mode of genomic evolution in endosymbiotic bacteria of insects: the case of Buchnera aphidicola. Symbiosis 34, 301–316.
Li, D., Hou, Y., and Shen, Z. (2006). Influence of host plant species on the development and reproduction of hawthorn spider mite Tetranychus viennensis Zacher. Acta Ecol. Sin. 1, 182–189. doi: 10.1007/s11461-006-0010-z
Ma, Y. J., He, H. P., Zhao, H. M., Xian, Y. D., Guo, H., Liu, B., et al. (2021). Microbiome diversity of cotton aphids (Aphis gossypii) is associated with host alternation. Sci. Rep. 11:5260. doi: 10.1038/s41598-021-83675-2
Milbrath, L. R., and Biazzo, J. (2012). Development and reproduction of the foxglove aphid (hemiptera: Aphididae) on invasive swallow-worts (vincetoxicum spp.). Environ. Entomol. 41, 665–668. doi: 10.1603/EN11239
Morando, R., Da Silva, I. F., Da Silva Santana, A., Sampaio, G. S. L., Lourenção, A. L., and Baldin, E. L. L. (2021). Assessing cotton genotypes for resistance to Aphis gossypii (Hemiptera: Aphididae). J. Econ. Entomol. 114, 387–396. doi: 10.1093/jee/toaa303
Perng, J. J. (2002). Life history traits of Aphis gossypii glover (Hom., Aphididae) reared on four widely distributed weeds. J. Appl. Entomol. 126, 97–100. doi: 10.1046/j.1439-0418.2002.00613.x
Rabatel, A., Febvay, G., Gaget, K., Duport, G., Baa-Puyoulet, P., Sapountzis, P., et al. (2013). Tyrosine pathway regulation is host-mediated in the pea aphid symbiosis during late embryonic and early larval development. BMC Genomics 14:235. doi: 10.1186/1471-2164-14-235
Ramalho, F. S., Fernandes, F. S., Nascimento, A. R. B., Nascimento, J. L., Malaquias, J. B., and Silva, C. A. D. (2012). Feeding damage from cotton aphids, Aphis gossypii glover (Hemiptera: Heteroptera: Aphididae), in cotton with colored fiber intercropped with fennel. Ann. Entomol. Soc. Am. 105, 20–27. doi: 10.1603/AN11122
Razmjou, J., Vorburger, C., Moharramipour, S., Mirhoseini, S. Z., and Fathipour, Y. (2010). Host-associated differentiation and evidence for sexual reproduction in Iranian populations of the cotton aphid, Aphis gossypii. Entomol. Exp. Appl. 134, 191–199. doi: 10.1111/j.1570-7458.2009.00951.x
Richards, S., Gibbs, R. A., Gerardo, N. M., Moran, N., Nakabachi, A., Stern, D., et al. (2010). Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8:e1000313. doi: 10.1371/journal.pbio.1000313
Sandström, J., and Pettersson, J. (1994). Amino acid composition of phloem sap and the relation to intraspecific variation in pea aphid (Acyrthosiphon pisum) performance. J. Insect Physiol. 40, 947–955. doi: 10.1016/0022-1910(94)90133-3
Satar, S., Kersting, U., and Yokomi, R. (2013). Presence of two host races of Aphis gossypii glover (Hemiptera: Aphididae) collected in Turkey. Ann. Appl. Biol. 162, 41–49. doi: 10.1111/j.1744-7348.2012.00578.x
Shang, J., Yao, Y. S., Zhu, X. Z., Wang, L., Li, D. Y., Zhang, K. X., et al. (2021). Evaluation of sublethal and transgenerational effects of sulfoxaflor on Aphis gossypii via life table parameters and 16S rRNA sequencing. Pest Manag. Sci. 77, 3406–3418. doi: 10.1002/ps.6385
Show, B. K., Banerjee, S., Banerjee, A., GhoshThakur, R., Hazra, A. K., Mandal, N. C., et al. (2022). Insect gut bacteria: a promising tool for enhanced biogas production. Rev. Environ. Sci. Biotechnol. 21, 1–25. doi: 10.1007/s11157-021-09607-8
Simon, J. C., Carré, S., Boutin, M., Prunier-Leterme, N., Sabater-Muñoz, B., and Latorre, A. (2003). Host-based divergence in populations of the pea aphid: insights from nuclear markers and the prevalence of facultative symbionts. Proc. R. Soc. B-Biol. Sci. 270, 1703–1712. doi: 10.1098/rspb.2003.2430
Smith, C. M., and Chuang, W. P. (2014). Plant resistance to aphid feeding: behavioral, physiological, genetic and molecular cues regulate aphid host selection and feeding. Pest Manag. Sci. 70, 528–540. doi: 10.1002/ps.3689
Tian, P. P., Chang, C. Y., Miao, N. H., Li, M. Y., and Liu, X. D. (2019). Infections with Arsenophonus facultative endosymbionts Alter performance of aphids (Aphis gossypii) on an amino-acid-deficient diet. Appl. Environ. Microbiol. 85, e01407–e01419. doi: 10.1128/aem.01407-12
Tian, P. P., Zhang, Y. L., Huang, J. L., Li, W. Y., and Liu, X. D. (2023). Arsenophonus interacts with Buchnera to improve growth performance of aphids under amino acid stress. Appl. Environ. Microbiol. 11:e01792-23. doi: 10.1128/spectrum.01792-23
Vorburger, C., Herzog, J., and Rouchet, R. (2017). Aphid specialization on different summer hosts is associated with strong genetic differentiation and unequal symbiont communities despite a common mating habitat. J. Evol. Biol. 30, 762–772. doi: 10.1111/jeb.13040
Wang, Y. H., Mikaelyan, A., Coates, B. S., and Lorenzen, M. (2024). The genome of Arsenophonus sp. and its potential contribution in the corn Planthopper, Peregrinus maidis. Insects. 15:113. doi: 10.3390/insects15020113
Wang, L., Zhang, S., Luo, J. Y., Wang, C. Y., Lv, L. M., Zhu, X. Z., et al. (2016). Identification of Aphis gossypii glover (Hemiptera: Aphididae) biotypes from different host plants in North China. PLoS One 11:e0146345. doi: 10.1371/journal.pone.0146345
Xu, S., Jiang, L., Qiao, G., and Chen, J. (2020). The bacterial Flora associated with the polyphagous aphid Aphis gossypii glover (Hemiptera: Aphididae) is strongly affected by host plants. Microb. Ecol. 79, 971–984. doi: 10.1007/s00248-019-01435-2
Xu, W., Liu, W., Li, J., Zhu, X., Wang, L., Li, D., et al. (2023). Buchnera breaks the specialization of the cotton-specialized aphid (Aphis gossypii) by providing nutrition through zucchini. Front. Nutr. 10:1128272. doi: 10.3389/fnut.2023.1128272
Xu, N., Tan, G., Wang, H., and Gai, X. (2016). Effect of biochar additions to soil on nitrogen leaching, microbial biomass and bacterial community structure. Eur. J. Soil Biol. 74, 1–8. doi: 10.1016/j.ejsobi.2016.02.004
Xue, H., Zhu, X., Wang, L., Zhang, K., Li, D., Ji, J., et al. (2021). Gut bacterial diversity in different life cycle stages of Adelphocoris suturalis (Hemiptera: Miridae). Front. Microbiol. 12:670383. doi: 10.3389/fmicb.2021.670383
Yang, F. Y., Saqib, H. S. A., Chen, J. H., Ruan, Q. Q., Vasseur, L., He, W. Y., et al. (2020). Differential profiles of gut microbiota and metabolites associated with host shift of plutella xylostella. Int. J. Mol. Sci. 21, 1–15. doi: 10.3390/ijms21176283
Zhang, Y. C., Cao, W. J., Zhong, L. R., Godfray, H. C. J., and Liu, X. D. (2016). Host plant determines the population size of an obligate symbiont (Buchnera aphidicola) in aphids. Appl. Environ. Microbiol. 82, 2336–2346. doi: 10.1128/AEM.04131-15
Zhang, Q., Lu, Z., Li, L., Guan, X., Ouyang, F., Zhang, Y., et al. (2018). Ecological effects of farmland landscape patterns on the populations of cotton aphids, Aphis gossypii glover, in North China. Shengtai Xuebao 38, 1366–1374. doi: 10.5846/stxb201701040031
Zhang, S., Su, H., Jiang, W., Hu, D., Ali, I., Jin, T., et al. (2021). Symbiotic microbial studies in diverse populations of Aphis gossypii, existing on altered host plants in different localities during different times. Ecol. Evol. 11, 13948–13960. doi: 10.1002/ece3.8100
Zhao, Y., Zhang, S., Luo, J. Y., Wang, C. Y., Lv, L. M., and Cui, J. J. (2016). Bacterial communities of the cotton aphid Aphis gossypii associated with Bt cotton in northern China. Sci. Rep. 6:22958. doi: 10.1038/srep22958
Zhao, B. M., Zhu, Y. Y., and Zhang, Q. (2017). Efficacy of Oxyfluorfen and Pendimethalin on Solanum nigrum in film covered cotton field. Agrochemicals. 56, 612–620. doi: 10.16820/j.cnki.1006-0413.2017.08.021
Zheng, C. L., Liu, X. D., and Zhai, B. P. (2007). The fitness of host biotypes of the cotton aphid (Aphis gossypii glover) to host plants and their reciprocal transfer pathways. Acta Ecol. Sin. 27, 1879–1886.
Keywords: cotton aphid, 16S rRNA, life table, host shift, symbiotic bacterium
Citation: Wang P, Jing Y-X, Ma Y-J, Wang D, Shan Y-P, Hu H, Wu C, Song X-P, Ren X, Ma Y and Ma X (2025) Multi-generational adaptation to Solanum nigrum increases reproduction and decreases microbial diversity of Aphis gossypii. Front. Sustain. Food Syst. 8:1523123. doi: 10.3389/fsufs.2024.1523123
Edited by:
Liming Ye, Ghent University, BelgiumReviewed by:
Sarah Nanyiti, National Crops Resources Research Institute (NaCRRI), Uganda: Kun Xue, Minzu University of China, ChinaCopyright © 2025 Wang, Jing, Ma, Wang, Shan, Hu, Wu, Song, Ren, Ma and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xiangliang Ren, cmVueGlhbmdsaWFuZ0BjYWFzLmNu; Yan Ma, YXltYXlhbkAxMjYuY29t; Xiaoyan Ma, bWF4eV9jYWFzQDEyNi5jb20=
†These authors share first authorship
 Peng Wang1,2†
Peng Wang1,2† Hongyan Hu
Hongyan Hu Changcai Wu
Changcai Wu Xiangliang Ren
Xiangliang Ren Yan Ma
Yan Ma Xiaoyan Ma
Xiaoyan Ma