- 1Guangdong Vtr Bio-Tech Co., Ltd., Zhuhai, Guangdong, China
- 2Guangdong Feed Additive Research and Development Center, Zhuhai, Guangdong, China
Introduction: The demand for gluconic acid (GA) has risen recently, driven by its extensive applications in the food, healthcare, and construction industries. The biocatalysis of gluconic acid, facilitated by glucose oxidase and catalase, hinges on enzyme stability, significantly influencing catalytic efficiency. Nonetheless, catalase requires enhancements in thermal stability and activity to meet the requirements of practical applications.
Methods: We evaluated ten catalases expressed in Aspergillus niger, ultimately selecting the catalase from the thermophilic fungus Thermoascus aurantiacus, labeled as TaCat, for its superior thermal stability and operational performance. We further characterized the enzymatic properties of the recombinant catalase, focusing on its thermostability. Simultaneously, we used AlphaFold2 for structural predictions and conducted in-depth analyses via accelerated molecular dynamics simulations.
Results and discussion: We successfully obtained a strain with the highest catalase activity by optimizing signal peptides and overexpressing the crucial heme synthesis enzyme. Enzyme production reached an impressive 321,779.5 U/mL in a 50-L fermenter. Our application studies confirmed the considerable advantages of TaCat in terms of GA production. In conclusion, TaCat, distinguished by its remarkable thermal stability and high activity, holds substantial potential for GA production.
1 Introduction
Gluconic acid (GA), an oxidation product of glucose, is a mild, noncorrosive, nontoxic organic acid with excellent thermal stability (Anastassiadis and Morgunov, 2007). Due to these unique physiological and chemical properties, GA and its derivatives are widely applied across various industries, including food, feed, pharmaceuticals, textiles, and construction (Cañete-Rodríguez et al., 2016). The annual production is estimated at 100,000 tons (Mu et al., 2019). The demand for GA and its salts (especially sodium, calcium, iron, copper, and zinc gluconates) is rapidly increasing at a rate of 9% per year (Banerjee et al., 2018), indicating notable growth potential in its manufacturing industry. Currently, the production of GA mainly relies on microbial fermentation and the dual-enzyme catalytic method, the latter of which uses glucose oxidase (GOD) and catalase (CAT). Compared with traditional microbial fermentation, the dual-enzyme method is highly efficient in catalyzing GA production ex vivo, achieving near 100% conversion, requiring less time, and yielding impurity-free results (Wong et al., 2008; Han et al., 2018). During this process, GOD catalyzes the oxidation of glucose to GA while producing hydrogen peroxide, but hydrogen peroxide inhibits the activity of GOD. Thus, CAT is introduced to break down hydrogen peroxide, removing its inhibitory impact on GOD and supplying the oxygen required for GOD (Han et al., 2018). Throughout the entire reaction process, the role of CAT is crucial: the addition of CAT reduces the exposure of GOD to peroxides, enhances the stability of GOD, and ensures the continuation of the whole reaction (Singh and McShane, 2010).
Catalase is extensively used in various sectors, such as medicine, food, and industry, owing to its superior antioxidative properties (Glorieux and Calderon, 2017; Kaushal et al., 2018; Sepasi Tehrani and Moosavi-Movahedi, 2018; Galasso et al., 2021). Commercially, catalases are obtained primarily from animal liver or produced through fermentation using A. niger. Extraction from animal liver involves a complex process and incurs high costs, whereas catalases derived from A. niger have poor enzyme stability (Yang et al., 1988; Meyer et al., 1997; Eberhardt et al., 2004). The stability of enzyme preparations during reactions is critical for industrial applications (Malik and Javed, 2021; Luan and Duan, 2022), particularly since gluconic acid reactions occur at 42–45°C for 20–30 hours. However, catalases from different sources, such as Bacillus subtilis (Philibert et al., 2016), Serratia marcescens (Jia et al., 2017), Penicillium rubens (Koleva et al., 2024), and Thermus thermophilus (Erçin, 2008), generally exhibit low activity or stability. Improving the thermostability and catalytic activity of catalases is essential to meet market demands for highly active and stable enzymes. This enhancement also helps expand their use in biocatalysis.
A. niger stands out for its strong protein secretion abilities and unique food safety features. It is widely used in various fields, including food processing and biomedicine (Ntana et al., 2020; Cairns et al., 2021). In A. niger, the fermentation yield of glucoamylase (GlaA) can reach 30 g/L (Xu et al., 2018). Although Aspergillus species are widely used for recombinant protein production, high-level expression of proteins from nonmodel organisms remains challenging (Erçin, 2008; Gressler et al., 2015). To increase the secretion and expression of heterologous proteins in Aspergillus, researchers have used several strategies, with signal peptide optimization proven to be particularly effective in increasing target protein levels (Tsang et al., 2009; Madhavan et al., 2017; Ntana et al., 2020). Additionally, heme is an essential cofactor for catalase, which is typically synthesized in most organisms through a highly conserved biosynthetic pathway (Layer et al., 2010). However, heme addition is not suitable for mass production processes because of its high hydrophobicity and cost. The best approach to overcome the limiting effect of heme is to modulate heme metabolism to increase the intracellular heme content. 5-Aminolevulinic acid synthases (HemeA) and 5-aminolevulinic acid dehydratase (HemeB) are crucial enzymes in the synthesis of heme (Nilsson, 2009; Su et al., 2019; Ye et al., 2022). Kwon et al. overexpressed HemeA, resulting in nearly a 40-fold increase in the amount of porphyrin produced (Kwon et al., 2003). This study aimed to enhance heme content by overexpressing both HemeA and HemeB, thereby increasing catalase production.
In this study, we evaluated 10 catalases expressed in A. niger. Through fermentation enzyme activity and thermostability analysis, we ultimately selected the catalase from the thermophilic fungus Thermoascus aurantiacus (TaCat) due to its excellent thermostability and operational performance. To further increase the production of TaCat in A. niger, we utilized signal peptide and heme metabolism optimization strategies, successfully creating a high-producing catalase strain with an enzyme activity of 321779.5 U/mL in a 50 L fermenter, the highest level recorded thus far. Additionally, the suitability of TaCat for GA production was further confirmed through performance evaluations in a 20 L bioreactor. Given these characteristics, TaCat is anticipated to be an effective biocatalyst for producing GA under conditions feasible for industrial applications.
2 Materials and methods
2.1 Strains, plasmids, and materials
Aspergillus niger VT-2 (CICC2422), obtained from Hongying Biotechnology Co., Ltd. (Hunan, China), was cultured on potato dextrose agar (PDA) at 30°C. Escherichia coli TOP10 was used for all the molecular cloning experiments. DNA polymerase (PrimeSTAR™ HS), restriction enzymes, and T4 DNA ligase were purchased from Takara Bio, Inc. (Dalian, China). Oligonucleotides were synthesized by Sangon Biotech (Shanghai, China), and multiple DNA fragments were assembled via NEBuilder® HiFi DNA Assembly Master Mix (Catalog No. E2621S). Reagents were obtained from Sigma-Aldrich (Sigma-Aldrich Co.). Unless otherwise specified, all chemicals were of analytical grade. Details of the strains, plasmids, and primers used in this study are available in Supplementary Tables S1–S3.
2.2 Construction of recombinant vectors for catalase activity, transformation, and selection of engineered recombinant strains
Ten catalases were identified from thermophilic or mesophilic fungi, including AnCat from Aspergillus niger (NCBI: AAA68206.1), TtCat from Thermothelomyces thermophilus (NCBI: XP_003663032.1), MtCat from Mycothermus thermophilus (NCBI: PP951965), TaCat from Thermoascus aurantiacus (NCBI: PP951966), PvCat from Paecilomyces variotii (NCBI: GAD95766.1), AfCat from Aspergillus fumigatus (NCBI: XP_748550.1), PmCat from Penicillium marneffei (NCBI: EEA19216.1), NcCat from Neurospora crassa (NCBI: XP_957826.1), ReCat from Rasamsonia emersonii (NCBI: XP_013328798.1), and TrCat from Talaromyces rugulosus (NCBI: XP_035343757.1). The nucleotide sequences of AnCat, TtCat, MtCat, TaCat, PvCat, AfCat, PmCat, NcCat, ReCat, and TrCat were obtained through gene synthesis by Shanghai Sangon Biotech, with synthetic gene fragments flanking 30 bp homology arms on both sides to the expression vector pANEXP. Using the NEBuilder® HiFi DNA Assembly method, we then seamlessly connected the XbaI-linearized pANEXP vector with the synthesized fragments, forming the corresponding ten pANEXP-CAT expression vectors (pANEXP-ANCAT, pANEXP-TTCAT, pANEXP-MTCAT, pANEXP-TACAT, pANEXP-PVCAT, pANEXP-AFCAT, pANEXP-PMCAT, pANEXP-NCCAT, pANEXP-RECAT, and pANEXP-TRCAT).
Following the protocol established by Wernars et al. (1986), we prepared and transformed the protoplasts of the A. niger strain VT-002. We linearized the ten plasmids via Bgl II (Thermo Scientific, Waltham, MA, USA) and then transformed them into A. niger VT-2 through protoplast-mediated transformation. After transformation, we spread the transformed protoplasts on potato dextrose agar (PDA) plates supplemented with 1 M sorbitol and hygromycin B (300 μg/mL).
From the transformants of each catalase, 200 colonies were selected and transferred to 24-well deep-well plates, with each well containing 2 mL potato dextrose broth (PDB), and incubated for four days. After initial screening, the top six strains with the highest enzyme activity from each catalase transformant were chosen for further screening in a 24-well plate. The strain with the highest activity from each catalase group was subsequently selected for shake flask fermentation, which was conducted in three flasks. The strains with the highest enzyme activity were identified using the primers for the Pgla-fw/Tter-rw PCR. The harvested supernatants were centrifuged and filtered to measure catalase activity, and protein expression in the fermentation broth was analyzed via SDS–PAGE. Stability analyses under application conditions (45°C, pH 5.5) were conducted for each sample, with enzyme activity tested every 8 hours to assess stability under these conditions.
2.3 Strategies for enhancing the enzyme activity of catalase in A. niger
In this study, the pANEXP-TACAT vector was digested with SacI and XbaI, and the dual enzyme-cutting vector was recombined with synthesized long primers (S2, S3, S4, and S5 signal peptide-fw in Supplementary Table S3) via homologous recombination to replace the original S1 signal peptide. The S2, S3, S4, and S5 signal peptides were then inserted into the pANEXP-TACAT vector, transformed into A. niger for selection, and subjected to enzyme activity assays and SDS–PAGE analysis. Details of the high-enzyme activity screening process are described in Section 2.2.
The assembly of the heme-critical enzyme overexpression vector proceeded as follows: expression cassettes for Pamy+HemeA+TrpC and Ptef1+HemeB+TrpC were constructed via fusion PCR. These were then seamlessly linked with the amdS (GenBank: M16371.1) segment and the PUC57 vector fragment via the NEBuilder® HiFi DNA Assembly method and transformed into Escherichia coli TOP10 competent cells, yielding the final construct, the pAN-amdS-hemeA+hemeB vector. The correct vector was identified via PCR. Following NotI digestion and transformation with the strain with the highest enzyme activity, AN-S5-TaCat, high-enzyme-activity strains were selected and analyzed for enzyme activity and heme content. The details of this process are described in Section 2.2.
2.4 Catalase production in a 50-L fermenter
The inoculum was spread onto 9 cm petri dishes containing 20 mL of PDA agar medium and incubated at 30°C for 4–5 days. Subsequently, 6–8 A. niger colonies were selected and inoculated into 250 mL Erlenmeyer flasks containing 100 mL of CD medium (sucrose 30 g/L, NaNO3 2 g/L, K2HPO4 1 g/L, MgSO4 0.5 g/L, KCl 0.5 g/L, FeSO4 0.01 g/L, pH 7.3), which were then cultured statically at 30°C for 72 hours. Afterward, 30 mL of the mycelial culture grown in CD medium was transferred into five separate 500 mL Erlenmeyer flasks, each containing 200 mL of liquefied starch medium. The cultures were incubated at 30°C with shaking at 200 rpm for 3 days. The liquefied starch medium consisted of 45 g/L liquefied corn starch, 0.04 g/L calcium chloride, 28 g/L corn steep liquor, and 10.0 g/L soybean meal. Pre-liquefaction of the corn starch was achieved using high-temperature amylase.
Following the shake flask cultivation, 1000 mL of primary seed culture was inoculated into a 20-liter fermenter containing 10 L of liquefied starch medium. During fermentation, the temperature was maintained at 30–32°C, and the pH was controlled between 5.0 and 5.5 with the addition of 25% ammonia solution. Stirring speed was set to 500 rpm, and the aeration rate was 20 L/min. The residual sugar concentration in the fermentation broth was kept between 15 and 30 g/L. After 64 hours of culture, 4 L of seed culture from the 20 L fermenter was transferred into a 50 L fermenter containing 25 L of liquefied starch medium. Fermentation conditions were similar, with temperature controlled at 30–32°C and pH maintained at 5.0–5.5 using ammonia solution. The sugar concentration in the feed solution was approximately 450 g/L, and the residual sugar concentration in the fermentation broth was kept between 5 and 20 g/L by manual control. Stirring speed remained at 500 rpm, and the aeration rate was increased to 40 L/min. The sugar feeding began after 24 hours of fermentation when the residual sugar concentration dropped below 10 g/L. A constant feeding rate of 2–3 g/L/h was maintained to keep the residual sugar level between 5 and 25 g/L. Between 85 and 110 hours, the feeding rate was lowered to 1–2 g/L/h due to a decline in sugar consumption. After 110 hours, the feeding rate was increased to 2–3.5 g/L/h as sugar consumption accelerated. At 140 h, the feeding rate was again reduced to 1–2 g/L/h. The fermentation process was completed after 210 to 220 hours. Throughout the cultivation, catalase activity, wet weight, and residual sugar concentration were continuously monitored.
2.5 Biochemical characterization of recombinant rTaCat
Catalase activity was measured following the method described by Thorup (Thorup et al., 1961). To determine the optimal temperature and pH, we performed enzyme assays across temperatures ranging from 25°C to 90°C and pH values ranging from 2.5 to 8.0. During pH stability tests, we mixed diluted enzyme solutions with citrate-phosphate buffers at various pH values. The pH stability was determined post-incubation at 25°C for 24 hours. For the pH stability tests, we mixed the diluted enzyme solutions with citrate–phosphate buffers at various pH values. Additionally, we evaluated the tolerance of recombinant catalase to different metal ions by testing concentrations of 5 mM for Ca2+, Mg2+, Na+, K+, Ni2+, Zn2+, Al3+, Co2+, Fe3+, Ag+, Fe2+, and Cu2+.
2.6 Thermal stability analysis and glycosylation analysis of rTaCat
Glycosylation analysis was validated via sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS–PAGE). The protein concentration in the supernatant was diluted to between 0.1 g/L and 0.5 g/L, combined with glycoprotein denaturing buffer (NEB, B1704S), boiled at 100°C for 10 min, and then incubated with Endo H (NEB, P0702S) at 37°C for 1 hour, followed by SDS–PAGE alongside untreated samples.
NetNGlyc (https://services.healthtech.dtu.dk/services/NetNGlyc-1.0/) was used to predict glycosylation sites. Thermostability analysis was conducted by measuring the heat tolerance of Endo H-treated and untreated rTaCat at 75°C, 80°C, and 90°C for 5 min. Additionally, the thermostabilities of rTaCat and rAnCat were compared after incubation for 1 h at 60°C, 65°C, and 70°C.
2.7 Molecular dynamics simulation analysis
The protein structure modeling and molecular dynamics simulation analyses are as follows: initially, TaCat and AnCat were modeled, and side-chain structures were optimized via Alphafold2 (Jumper et al., 2021) and Rosetta relax (Nivón et al., 2013), followed by molecular dynamics simulations via the optimized structure models. All molecular dynamics simulations in this study were performed via the AMBER99SB-ILDN force field in Gromacs 2022.5 (Singh et al., 2018). The detailed simulation process is provided in Supplementary material S1.
2.8 Sodium gluconate production application test
The production of GA involves catalytic generation from glucose in a 20-liter reactor. A total of 10 L of 36% glucose solution was added to a 20 L bioreactor, along with approximately 1 mL of antifoaming agent, catalase, and glucose oxidase. For sodium gluconate production, catalase and glucose oxidase were added to the reaction mixture at final concentrations of 300000 U/mL (added amount 20 g/kg) and 15,000 U/mL (added amount 5 g/kg), respectively. The reaction proceeded at 45°C with a pH of 5.5 at 500 rpm under a pressure of 0.1 MPa. During this process, the dissolved oxygen and alkali addition were monitored, and samples were taken to test the residual sugar content.
3 Results
3.1 Exploration of catalase with thermal stability
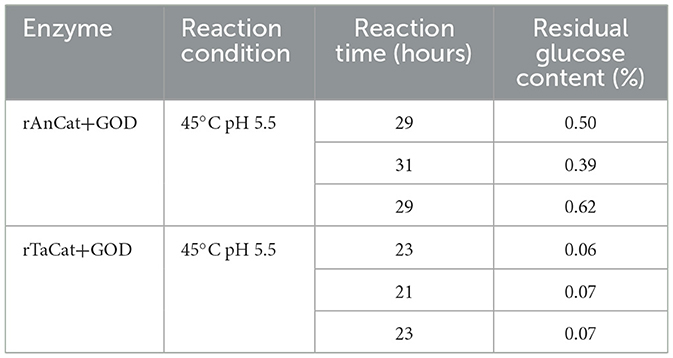
In search of catalases with thermal stability, we identified 10 catalases from thermophilic or mesophilic fungi. Through sequence homology analysis, we constructed a phylogenetic tree for these 10 catalases via MEGA software (Figure 1A). We subsequently cloned these catalase genes into the A. niger expression vector pANEXP and transformed them into A. niger host protoplasts for integration into the A. niger genome (Figure 1B). SDS–PAGE analysis revealed that 7 of the catalases were successfully expressed in A. niger (Figure 1C), including catalase from Thermoascus aurantiacus (TaCat), catalase from Paecilomyces variotii (PvCat), catalase from Aspergillus fumigatus (AfCat), catalase from Penicillium marneffei (PmCat), catalase from Neurospora crassa (NcCat), catalase from Rasamsonia emersonii (ReCat), and catalase from Aspergillus niger (AnCat). Among them, the TaCat and PmCat transformants presented the highest enzyme activities in shake-flask fermentation, with 9240 U/mL and 12433 U/mL, respectively (Figure 1D). The enzyme activities of the other catalases ranged from 5000 to 8000 U/mL, while TtCat from Thermothelomyces thermophilus, MtCat from Mycothermus thermophilus, and TrCat from Talaromyces rugulosus showed little to no enzyme activity. Under simulated conditions of a sodium gluconate reaction (45°C, pH 5.0), TaCat exhibited superior thermal stability, maintaining 90.6% activity, whereas the stability of the other transformants remained below 72% (Figure 1E). TaCat was selected as the most promising candidate catalase due to the data showing its excellent enzyme activity and thermal stability.
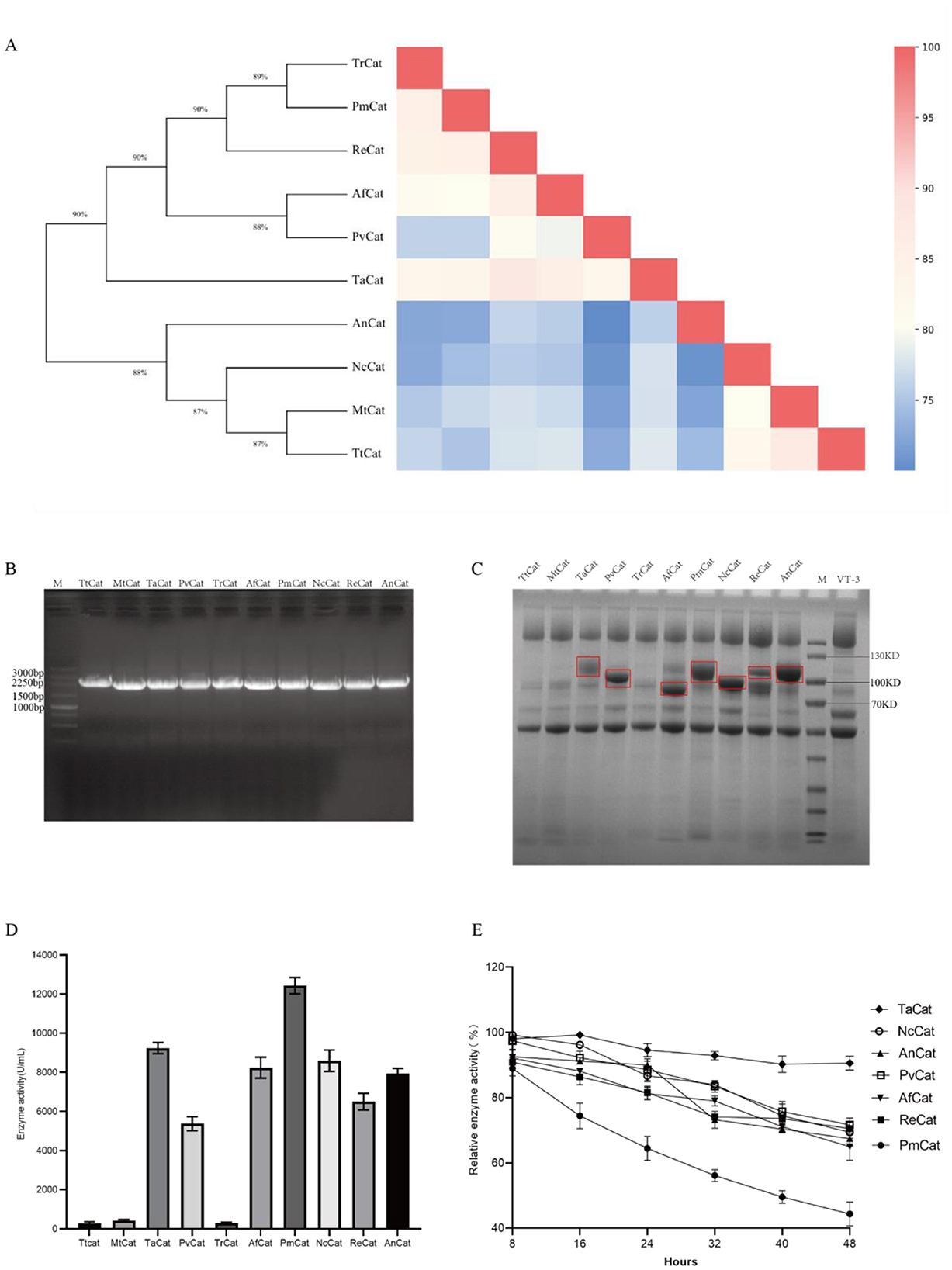
Figure 1. Recombinant expression and characterization of catalases from ten different sources in A. niger. (A) Maximum likelihood phylogenetic tree and percentage identity matrix of the 10 selected catalases. The color bar indicates the percent identity of the enzymes, with detailed values in Supplementary Table S2. (B) PCR-based identification of the catalase gene in various transformants. (C) SDS–PAGE analysis of ten catalases in A. niger (TtCat, MtCat, TaCat, PvCat, TrCat, AfCat, PmCat, NcCat, ReCat, and AnCat); M: protein molecular weight marker; VT-3: wild-type host. (D) Enzyme activity assay of ten different catalases in A. niger. (E) Stability analysis of seven catalases (TaCat, PvCat, AfCat, PmCat, NcCat, ReCat and AnCat).
3.2 Strategies for enhancing the expression of catalase in A. niger
To further increase the enzymatic activity of the catalase TaCat in A. niger, we increased production through signal peptide optimization and enhancement of the production of the catalase cofactor heme. The secretion levels of the recombinant protein are markedly influenced by the type of signal peptide used. For our research, we chose four frequently used signal peptides that exhibit high expression: S2, S3, S4, and S5. Specifically, S2 is the glucoamylase signal peptide of A. niger, S3 is the glucoamylase of Rasamsonia emersonii, S4 is the lipase signal peptide from A. niger, and S5 is the amylase signal peptide of A. niger. In this study, we replaced the original S1 signal peptide with a homologous recombination sequence, constructing S2, S3, S4, and S5 signal peptides into the pANEXP-TACAT vector for expression in A. niger. As illustrated in Figure 2A, the enzyme activities of recombinant strains modified with the S2, S4, and S5 signal peptides were significantly higher than those of the strains modified with the wild-type S1 signal peptide. Notably, the activities of the S2 and S5 peptides were 1.15 times and 1.29 times greater than those of the wild-type strains, respectively, and they substantially exceeded those of the control. Figure 2B demonstrates that the protein level of catalase expressed with the S5 signal peptide was higher than that expressed with the wild-type S1 signal peptide. These findings suggest that signal peptides derived from A. niger are effective for the expression of exogenous proteins in A. niger. Based on these results, AN-S5-TaCat was selected as the candidate for further investigation.
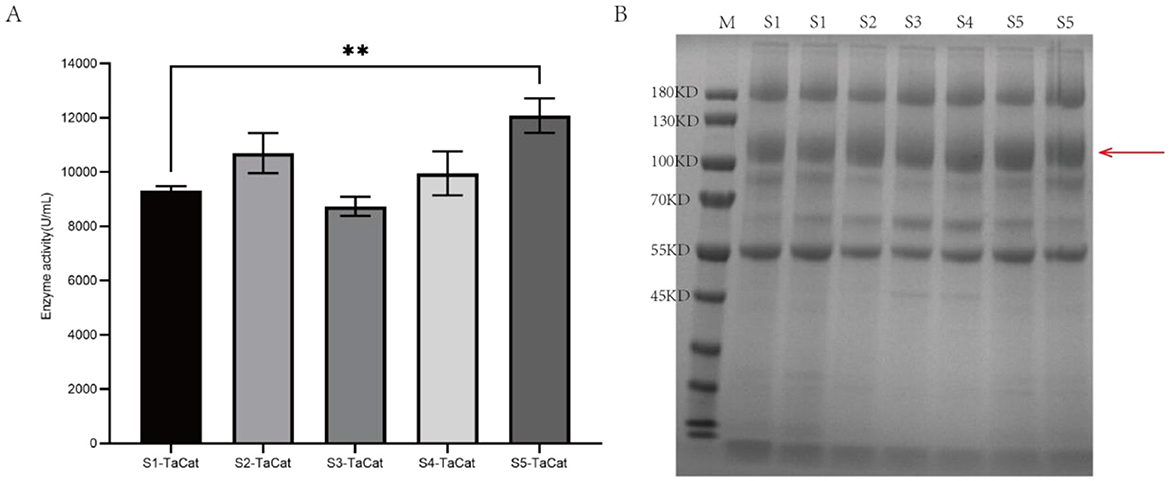
Figure 2. Effects of signal peptide optimization on catalase secretion in A. niger. (A) Enzymatic activity of recombinant strains (AN-S1-TaCat, AN-S2-TaCat, AN-S3-TaCat, AN-S4-TaCat, and AN-S5-TaCat) with five different signal peptides (** denotes p < 0.01, indicating significant differences compared with the control. The experiments were conducted with three biological replicates and three technical replicates). (B) SDS–PAGE analysis of recombinant strains with five different signal peptides (S1, S2, S3, S4, and S5).
Heme is an essential cofactor in heme-dependent catalase (Pedersen et al., 2008; Maresca et al., 2018), and the conversion of apoprotein to heme protein depends on the use of heme provided by the heme biosynthesis pathway. As shown in Figure 3A, eight specific enzymes are involved in the biosynthesis of fungal heme; we focused on two key enzymes, HemeA and HemeB. Initially, by employing NEBuilder® HiFi DNA Assembly, we assembled the expression cassettes for HemeA, HemeB, amdS, and fragments of the PUC57 vector into the complete vector pAN-amdS-HemeA+HemeB (shown in Figure 3B). Following linearization, the vector was transformed into the S5-TaCat protoplast strain of A. niger via protoplast transformation and plated onto CD+ acetamide plates. When the transformants were streaked onto PDA plates, the recombinant strain was reddish, whereas the background strain was grayish white (Figure 3C). As shown in Figure 3D, the enzymatic activity of the transformant AN-S5-TaCat-HemeA+HemeB substantially increased to 17761.3 U/mL, a 50.5% increase over that of AN-S5-TaCat. We also assessed the intracellular heme levels and detected an 88.1% increase in AN-S5-TaCat-HemeA+HemeB compared with AN-S5-TaCat.
Figure 3. Effects of heme synthesis regulation on catalase activity. (A) Heme biosynthesis pathway. (B) Construction process of the pAN-amdS-HemeA+HemeB vector. (C) The color of the strains on PDA plates: the heme-regulated recombinant strain AN-S5-TaCat-HemeA+HemeB is light red, whereas AN-S5-TaCat is off-white. (D) Enzyme activity assay and intracellular heme measurement of the heme-regulated recombinant strain and AN-S5-TaCat.
3.3 Catalase production in a 50-L fermenter
To explore the industrial applicability of the newly constructed catalase strain AN-S5-TaCat-HemeAB, the production capacity of the engineered strain was determined by culturing it in a 50-L fermenter. As depicted in Figure 4, the enzymatic activity of the strain's catalase continuously increased during the entire 213-h fermentation process, finally reaching a high level of 321,779.5 U/mL. The increase in enzymatic activity followed an approximate S-shaped curve, with slow growth occurring between 0–93 hours and 164–213 hours and logarithmic growth occurring during the 93–164-hour period. In the fermenter, the engineered cells grew rapidly, reaching a peak wet cell weight of 610 mg-CWW/mL at 68 hours. Afterward, the wet weight began to decrease and fluctuated, ending at 490 mg-CWW/mL. Additionally, we observed that the mycelia gradually separated into smaller filaments at ~80 h (in Supplementary Figure S1). The high level of catalase activity and rapid growth indicate that the improved strain has substantial industrial application potential.
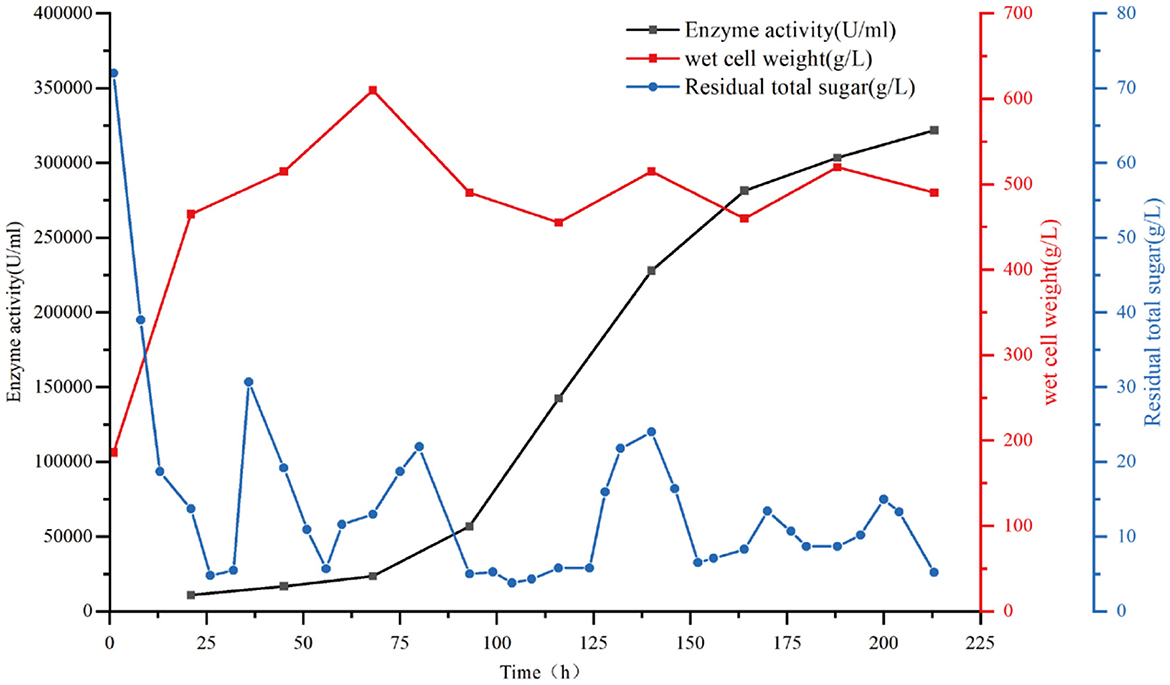
Figure 4. Production of catalase from the recombinant strain AN-S5-TaCat-hemeA+ hemeB via fed-batch fermentation in a 50-L fermenter. The black squares represent catalase activity; the red squares indicate cell growth; the blue line represents residual total sugar.
3.4 Biochemical characterization of recombinant rTaCat
The enzymatic characteristics of catalase produced by the recombinant strain AN-S5-TaCat-hemeAB were studied. The catalase activity was measured within the range of 30–90°C, and as shown in Figure 5A, the optimal reaction temperature for rTaCat was 40°C, with the relative activity maintained above 75% between 25 and 75°C. The optimal pH for rTaCat is 6.0, with the relative activity remaining above 70% within a pH range of 4.0–9.0 (Figure 5B). Research on pH stability (Figure 5C) revealed that rTaCat remains relatively stable from pH 4.0 to 10.0, preserving more than 80% of its enzymatic activity after 8-h incubation. This property of the catalase is advantageous for its use in the sodium gluconate conversion process, which generally occurs under acidic conditions (pH 5.0–6.0). Additionally, most of the residual chemicals and ions detected in this study had a negative impact on the enzymatic activity of rTaCat (Figure 5D). Na+, Ca2+, and K+ had little inhibitory effect on the activity of the enzyme. The enzyme retained only approximately 30% and 40% of its activity in the presence of Cu2+ and Co2+, respectively. It is speculated that the metal ions form coordination bonds with key amino acids of the rTaCat catalase, altering its conformation and thus affecting enzyme activity.
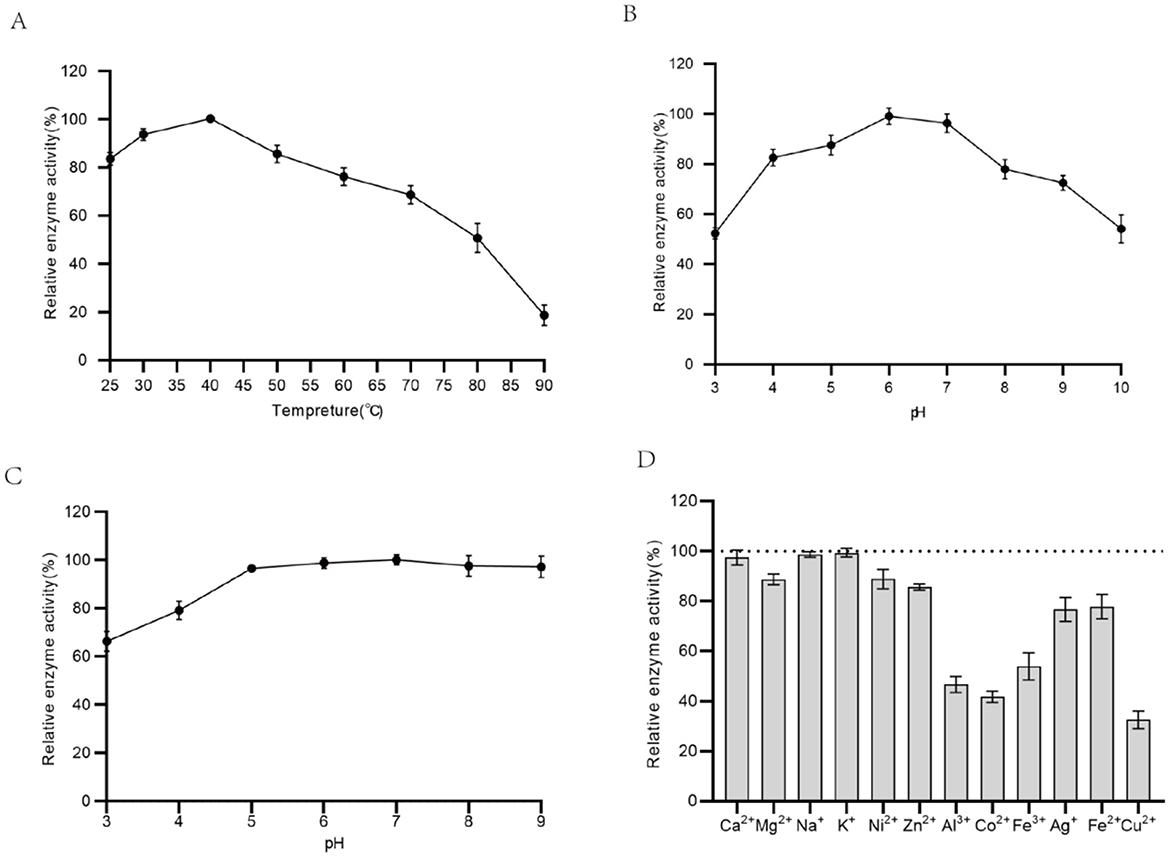
Figure 5. Biochemical characterization of the recombinant catalase rTaCat. (A) Effect of temperature; (B) effect of pH; (C) pH stability; (D) effects of metal ions.
3.5 Thermal stability analysis and glycosylation analysis of rTaCat
The theoretical molecular weight of rTaCat is presumed to be ~90 kDa. Nonetheless, SDS–PAGE (Figure 6A) demonstrated that the molecular weight of rTaCat expressed in A. niger exceeds 100 kDa, surpassing the theoretical estimate. Given the glycosylation system in A. niger, we speculate that this band may represent the glycosylated state. To verify this hypothesis, rTaCat was digested with the endoglycosidase Endo H, which removes nearly all N-linked oligosaccharides from the glycoprotein. We observed that the band for the rTaCat protein decreased in size after Endo H treatment, with a molecular weight of ~90 kDa (Figure 6A), which is consistent with the theoretical molecular weight. Glycosylation prediction revealed 7 potential glycosylation sites (Figure 6B). Among these potential glycosylation sites, N62, N131, and N562 exhibit high signal N-glycosylation. Furthermore, the thermal stability of both rTaCat and its deglycosylated form was assessed. As shown in Figure 6C, the deglycosylated catalase slightly decreased in thermal stability at 75°C and 85°C, but markedly decreased after heating at 90°C, with retention ranging from 38% to 15%.
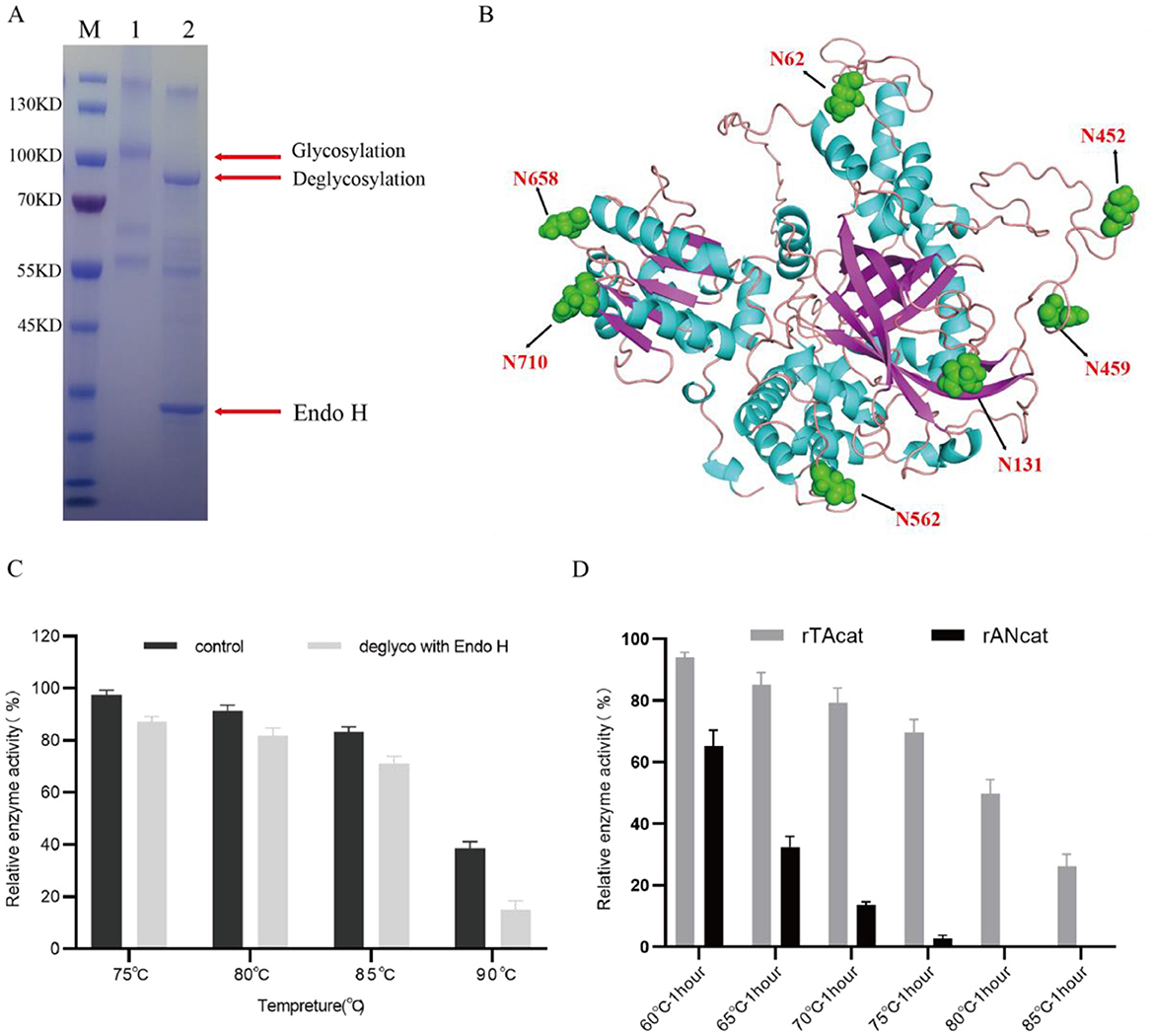
Figure 6. Thermal stability analysis and glycosylation analysis of rTaCat. (A) SDS–PAGE analysis of catalase glycosylation. Lane M: Molecular weight markers; Lane 1: Supernatant of recombinant AN-S5-TaCat-hemeA+hemeB culture; Lane 2: Supernatant of recombinant AN-S5-TaCat-hemeA+hemeB culture treated with Endo H. (B) The protein structure of catalase TaCat predicted by ALPHAFOLD, with the seven predicted glycosylation sites marked. (C) The thermostability of rTaCat before and after Endo H treatment at 75°C, 80°C, 85°C, and 90°C for 5 min. The activity of the untreated enzyme was set at 100%. (D) Enzyme activity retention rates of rTaCat and rAnCat treated for 1 h at 60°C, 65°C, 70°C, 75°C, 85°C, and 90°C. The activity of the untreated enzyme was set at 100%.
We compared the high temperature tolerances of rTaCat and rAnCat produced from the engineered strains. The catalase rAnCat, derived from A. niger, is a commonly used commercial product. Figure 6D shows that, compared with rAnCat, rTaCat has notably greater heat resistance. Throughout the temperature range tested, rTaCat demonstrated superior thermal stability, maintaining near or exceeding 80% relative activity between 60°C and 70°C and retaining 49.7% activity even after one hour at 80°C. In contrast, rAnCat has virtually no activity above 70°C, highlighting its extreme instability at high temperatures.
3.6 Molecular dynamics simulation analysis
We predicted the three-dimensional structures of TaCat and AnCat via AlphaFold2 and performed accelerated molecular dynamics simulations on the optimal model structures. Using AlphaFold2, we predicted the three-dimensional protein structures of the TaCat and AnCat catalases, obtaining highly accurate pLDDT scores of 93.13 and 93.02, respectively. Selecting the optimal model structures as initial conformations, we conducted 50 ns of accelerated molecular dynamics simulations on these two enzymes. We explored the theoretical differences in the stability and influencing factors of the two types of catalases, as shown in Figures 7A, B. Over three simulations, the root mean square deviation (RMSD) indicated protein system stability, with TaCat consistently showing a lower RMSD than AnCat with more stable fluctuations, suggesting superior structural stability for TaCat. Moreover, by measuring the radius of gyration (Rg), we found that the TaCat structure was more compact than the AnCat structure was, further implying its greater stability (Figures 7A, B). TaCat also displayed a more extensive hydrogen bond network than AnCat across the simulations, which was often linked to improved thermostability (Figures 7E, F). The root mean square fluctuation (RMSF) represents the average change in the atomic position over time, indicating the flexibility and intensity of protein amino acid movements throughout the simulation. Supplementary Figure S2 shows notable fluctuations in the TAcat protein between the regions of 1–100 aa and 400–490 aa. If further improvements in thermal stability are needed, focusing on studying and mutating this region will be our priority.
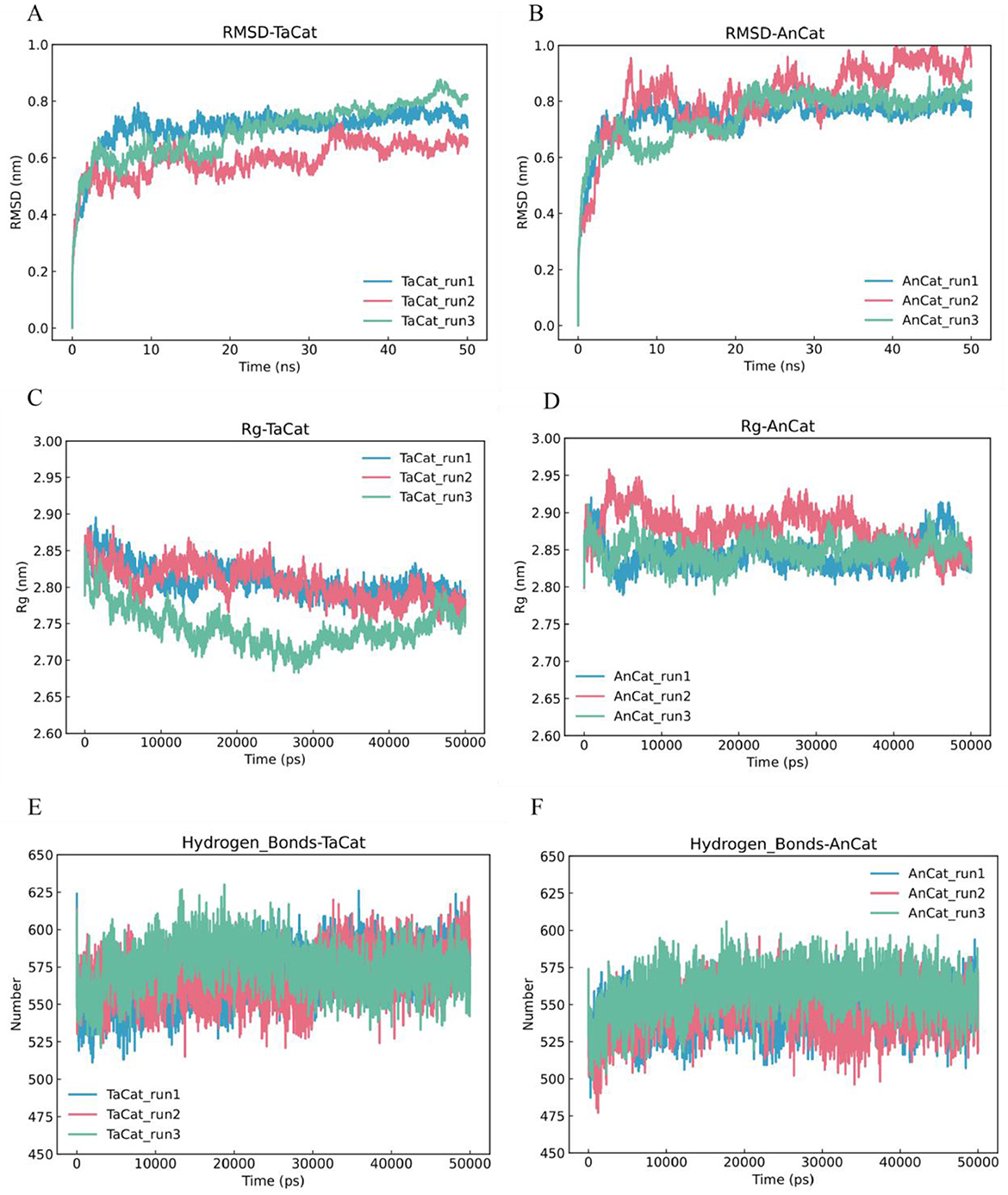
Figure 7. Molecular dynamics simulation analysis of TaCat and AnCat. (A) Root mean square deviation (rmsd) of TaCat. (B) Root mean square deviation (rmsd) of AnCat. (C) Radius of gyration (Rg) of TaCat. (D) Radius of gyration (Rg) of AnCat. (E) Hydrogen bond simulation of TaCat. (F) Hydrogen bond simulation of AnCat. The blue, red, and green lines represent the three parallel simulations.
3.7 Sodium gluconate production application test
To demonstrate the practicality of rTaCat catalase in the biocatalytic production of gluconic acid (GA), a sodium gluconate application test was conducted in a 20 L bioreactor. The reaction conditions were maintained at 45°C and pH 5.5, with a stirring speed of 500 rpm and a pressure of 0.1 MPa. The results, detailed in Table 1, highlight the performance differences between TaCat and AnCat. Under identical conditions with the same quantity of glucose oxidase (GOD) added, TaCat demonstrated rapid kinetics, achieving complete conversion of glucose to GA in an average of 22 hours across three trials. In contrast, AnCat exhibited slower reaction kinetics, with an incomplete reaction observed after an average of 30 hours in three trials. Collectively, these findings underscore the superior efficacy of TaCat in sodium gluconate production compared with that of the conventional catalase AnCat.
4 Discussion
Gluconic acid (GA) and its alkaline salts are extensively utilized in the food, feed, chemical, pharmaceutical, and construction sectors (Yan et al., 2022). The demand for gluconic acid in the global market has substantially risen in recent years (Raj et al., 2024). Nonetheless, GA production faces challenges in terms of cost efficiency and eco-friendly manufacturing. Currently, the main method for producing gluconic acid is the dual-enzyme catalytic method, which uses glucose oxidase (GOD) and catalase (CAT). The efficiency of glucose conversion to GA in this process is highly contingent upon the stability of these two enzymes (Mafra et al., 2015). For example, Mu et al. (2019) enhanced its stability via computer-aided design, doubling GA production. Ning et al. (2018) increased the thermostability of the S100A and D408W mutants, markedly extending their half-lives over those of the wild type. Conversely, research on the use of catalase in GA production is relatively rare. The currently used A. niger catalase still does not fully meet the production needs in terms of stability and thermal stability. In this study, we selected 10 catalase genes from thermophilic or mesophilic fungi and expressed them recombinantly in A. niger to search for highly stable CAT. We report a thermostable catalase, TaCat, which is more stable than AnCat under gluconic acid application conditions and has remarkable thermal stability and application effects. Through the optimization of signal peptide and heme metabolism, fermentation enhancement is remarkable, with fermentation enzyme activity reaching 321,779.5 U/mL, which is the highest level reported thus far. Considering the above research, we believe that the newly discovered TaCat may be a promising alternative for addressing the poor stability of catalase and the long transformation time in the production process of gluconic acid.
In recent years, researchers have increasingly been interested in using enzymes from the thermophilic fungus Thermoascus aurantiacus, which typically exhibit higher reaction rates and thermal stability than the fungal enzymes currently in use (McClendon et al., 2012; Gabriel et al., 2020; Battisti et al., 2024). In this study, we verified that TaCat sourced from T. aurantiacus presented the highest thermal resistance and stability among the 10 catalases screened. Given that TaCat has enhanced stability at 45°C and pH 5.5 and can completely convert D-glucose to gluconic acid within ~22 hours, this enzyme is particularly effective in addressing the low conversion efficiency of catalases in high-temperature environments. Thus, TaCat is highly suitable for the efficient production of gluconic acid at high temperatures. Identifying thermostable enzymes from T. aurantiacus or other thermophilic fungi could be a quick approach. According to Juliane Almeida Battisti et al., the multifunctional polygalacturonase PI3S3 from T. aurantiacus shows increased activity at elevated temperatures (Battisti et al., 2024). Bhaumik R. Dave et al. Also found that the endoglucanase from T. aurantiacus exhibits very high stability at 50°C−80°C (Dave et al., 2015).
Furthermore, we predicted the 3D structures of TaCat and the commonly used AnCat using AlphaFold2 and analyzed accelerated molecular dynamics simulations of the best model structures. The results of these simulations of the RMSD, RMSF, Rg, and hydrogen bond network are consistent with our thermostability tests (Figure 7). The simulations also preliminarily revealed key elements involved in the internal structural differences in protein stability between the two proteins and provided directions for further targeted modification of catalases.
Signal peptide optimization is a key strategy for enhancing protein expression and secretion efficiency (Tsang et al., 2009). To increase the secretory ability of TaCat, we validated five highly efficient signal peptides. According to Figure 2B, transformants with the S2 (glucoamylase) and S5 (amylase) signal peptides achieved 15% and 29% greater yields than those with the wild-type S1 signal peptide, respectively. This result is consistent with observations by Li et al. (2023), where signal peptide optimization significantly increased secretion and enzyme activity in studies of A. niger xylanase. Similarly, Si-Yuan et al. (2020) studied the effects of cbhI, ANL, and glaA signal peptides on lipase expression. They reported that cbhI increased yields by 1.90–3.13 times compared with ANL and glaA, highlighting the critical role of signal peptide choice in increasing specific protein production. Hence, signal peptides derived from A. niger aid in the secretion and expression of proteins in A. niger, especially the signal peptides for glucoamylase and amylase.
Heme has been identified as a bottleneck in the synthesis of catalases by A. niger (Franken et al., 2011). We addressed this challenge by overexpressing the key enzymes HemeA and HemeB. The results showed that this regulation not only increased heme levels by 88.1% but also increased the fermentative enzyme activity of catalase by 50.5%. Research by Agris Pentjuss et al. demonstrated that overexpression of HemA, HEM2, HEMB, and HEM12 resulted in a 41% increase in intracellular heme yield (Pentjuss et al., 2023). In related studies, Rekdal et al. (2024) successfully quadrupled the heme content by regulating the expression of four key heme biosynthetic enzymes in Aspergillus oryzae and noted that the recombinant strain colonies also presented red characteristics. Similarly, Elrod and Cherry (1999) markedly enhanced the enzyme activity of peroxidases by overexpressing 5-aminolevulinic acid synthases (hemA) in A. oryzae, increasing it by 1.9 times. Based on these results, we speculate that by modulating the increase in heme content, other heme-containing enzymes may also be able to increase the activity of fermentation enzymes.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PP951965; https://www.ncbi.nlm.nih.gov/, PP951966.
Author contributions
JHu: Conceptualization, Data curation, Formal analysis, Investigation, Methodology, Writing – original draft. JW: Conceptualization, Project administration, Resources, Supervision, Writing – original draft. JHe: Formal analysis, Investigation, Methodology, Writing – original draft. YW: Methodology, Software, Visualization, Writing – original draft. LC: Investigation, Methodology, Writing – original draft. SZ: Formal analysis, Methodology, Writing – original draft. YB: Methodology, Validation, Writing – original draft. YL: Conceptualization, Project administration, Resources, Supervision, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This study was supported by the National Key Research and Development Program of China (2022YFC2805105).
Acknowledgments
Many thanks to Dr. Jin Xing and Dr. Qiang Wang for their help with the experimental analysis and data analysis.
Conflict of interest
JHu, JW, JHe, YW, LC, SZ, YB, and LL were employed by Guangdong Vtr Bio-Tech Co., Ltd.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1465445/full#supplementary-material
References
Anastassiadis, S., and Morgunov, I. G. (2007). Gluconic acid production. Recent Pat. Biotechnol. 1, 167–180. doi: 10.2174/187220807780809472
Banerjee, S., Kumar, R., and Pal, P. (2018). Fermentative production of gluconic acid: a membrane-integrated Green process. J. Taiwan Instit. Chem. Eng. 84, 76–84. doi: 10.1016/j.jtice.2018.01.030
Battisti, J. A., Rocha, G. B., Rasbold, L. M., Delai, V. M., Costa, M. S. S., Kadowaki, M. K., et al. (2024). Purification, biochemical characterization, and biotechnological applications of a multifunctional enzyme from the Thermoascus aurantiacus PI3S3 strain. Sci. Rep. 14:5037. doi: 10.1038/s41598-024-55665-7
Cairns, T. C., Barthel, L., and Meyer, V. (2021). Something old, something new: challenges and developments in Aspergillus niger biotechnology. Essays Biochem. 65, 213–224. doi: 10.1042/EBC20200139
Cañete-Rodríguez, A. M., Santos-Dueñas, I. M., Jiménez-Hornero, J. E., Ehrenreich, A., Liebl, W., and García-García, I. (2016). Gluconic acid: properties, production methods and applications—An excellent opportunity for agro-industrial by-products and waste bio-valorization. Proc. Biochemist. 51, 1891–1903. doi: 10.1016/j.procbio.2016.08.028
Dave, B. R., Sudhir, A. P., and Subramanian, R. B. (2015). Purification and properties of an endoglucanase from Thermoascus aurantiacus. Biotechnology Reports. 6, 85–90. doi: 10.1016/j.btre.2014.11.004
Eberhardt, A. M., Pedroni, V., Volpe, M., and Ferreira, M. L. (2004). Immobilization of catalase from Aspergillus niger on inorganic and biopolymeric supports for H2O2 decomposition. Appl. Catalysis B: Environm. 47, 153–163. doi: 10.1016/j.apcatb.2003.08.007
Elrod, S. L., and Cherry, J. R. (1999). Aspergillus oryzae 5-Aminolevulinic Acid Synthases and Nucleic Acids Encoding Same. US Patent No. 5,871,991. United States Patent and Trademark Office (USPTO). Assignee: Novozymes Inc.
Erçin, H. Ö. (2008). Cloning of the Scytalidium Thermophilum Bifunctional Catalase/Phenol Oxidase Gene and Expression in Aspergillus sojae (Master Thesis) Ankara: Middle East Technical University, Available: https://open.metu.edu.tr/handle/11511/17539 (accessed December 31, 2023).
Franken, A. C. W., Lokman, B. C., Ram, A. F. J., Punt PJvan den Hondel, C.A. M. J, and de Weert, S. (2011). Heme biosynthesis and its regulation: towards understanding and improvement of heme biosynthesis in filamentous fungi. Appl. Microbiol. Biotechnol. 91, 447–460. doi: 10.1007/s00253-011-3391-3
Gabriel, R., Prinz, J., Jecmenica, M., Romero-Vazquez, C., Chou, P., Harth, S., et al. (2020). Development of genetic tools for the thermophilic filamentous fungus Thermoascus aurantiacus. Biotechnol. Biofuels. 13:167. doi: 10.1186/s13068-020-01804-x
Galasso, M., Gambino, S., Romanelli, M. G., Donadelli, M., and Scupoli, M. T. (2021). Browsing the oldest antioxidant enzyme: catalase and its multiple regulation in cancer. Free Radical Biol. Med. 172, 264–272. doi: 10.1016/j.freeradbiomed.2021.06.010
Glorieux, C., and Calderon, P. B. (2017). Catalase, a remarkable enzyme: targeting the oldest antioxidant enzyme to find a new cancer treatment approach. Biol. Chem. 398, 1095–1108. doi: 10.1515/hsz-2017-0131
Gressler, M., Hortschansky, P., Geib, E., and Brock, M. (2015). A new high-performance heterologous fungal expression system based on regulatory elements from the Aspergillus terreus terrein gene cluster. Front. Microbiol. 6:184. doi: 10.3389/fmicb.2015.00184
Han, X., Liu, G., Song, W., and Qu, Y. (2018). Production of sodium gluconate from delignified corn cob residue by on-site produced cellulase and co-immobilized glucose oxidase and catalase. Bioresour. Technol. 248, 248–257. doi: 10.1016/j.biortech.2017.06.109
Jia, X., Lin, X., Lin, C., Lin, L., and Chen, J. (2017). Enhanced alkaline catalase production by Serratia marcescens FZSF01: enzyme purification, characterization, and recombinant expression. Elect. J. Biotechnol. 30, 110–117. doi: 10.1016/j.ejbt.2017.10.001
Jumper, J., Evans, R., Pritzel, A., Green, T., Figurnov, M., Ronneberger, O., et al. (2021). Highly accurate protein structure prediction with AlphaFold. Nature. 596, 583–589. doi: 10.1038/s41586-021-03819-2
Kaushal, J., Mehandia, S., Singh, G., Raina, A., and Arya, S. K. (2018). Catalase enzyme: Application in bioremediation and food industry. Biocatal. Agric. Biotechnol. 16, 192–199. doi: 10.1016/j.bcab.2018.07.035
Koleva, Z., Abrashev, R., Angelova, M., Stoyancheva, G., Spassova, B., Yovchevska, L., et al. (2024). A novel extracellular catalase produced by the antarctic filamentous fungus penicillium rubens III11-2. Fermentation. 10:1. doi: 10.3390/fermentation10010058
Kwon, S. J., de Boer, A. L., Petri, R., and Schmidt-Dannert, C. (2003). High-level production of porphyrins in metabolically engineered escherichia coli: systematic extension of a pathway assembled from overexpressed genes involved in heme biosynthesis. Appl. Environ. Microbiol. 69, 4875–4883. doi: 10.1128/AEM.69.8.4875-4883.2003
Layer, G., Reichelt, J., Jahn, D., and Heinz, D. W. (2010). Structure and function of enzymes in heme biosynthesis. Protein Sci. 19, 1137–1161. doi: 10.1002/pro.405
Li, Y., Li, C., Aqeel, S. M., Wang, Y., Zhang, Q., Ma, J., et al. (2023). Enhanced expression of xylanase in Aspergillus niger enabling a two-step enzymatic pathway for extracting β-glucan from oat bran. Bioresour. Technol. 377:128962. doi: 10.1016/j.biortech.2023.128962
Luan, S., and Duan, X. (2022). A Novel thermal-activated β-galactosidase from Bacillus aryabhattai GEL-09 for lactose hydrolysis in milk. Foods 11:372. doi: 10.3390/foods11030372
Madhavan, A., Pandey, A., and Sukumaran, R. K. (2017). Expression system for heterologous protein expression in the filamentous fungus Aspergillus unguis. Bioresour. Technol. 245, 1334–1342. doi: 10.1016/j.biortech.2017.05.140
Mafra, A. C. O., Furlan, F. F., Badino, A. C., and Tardioli, P. W. (2015). Gluconic acid production from sucrose in an airlift reactor using a multi-enzyme system. Bioprocess Biosyst. Eng. 38, 671–680. doi: 10.1007/s00449-014-1306-2
Malik, W. A., and Javed, S. (2021). Biochemical characterization of cellulase from bacillus subtilis strain and its effect on digestibility and structural modifications of Lignocellulose rich biomass. Front Bioeng Biotechnol. 9:800265. doi: 10.3389/fbioe.2021.800265
Maresca, D., Zotta, T., and Mauriello, G. (2018). Adaptation to aerobic environment of Lactobacillus johnsonii/gasseri strains. Front Microbiol. 9:157. doi: 10.3389/fmicb.2018.00157
McClendon, S. D., Batth, T., Petzold, C. J., Adams, P. D., Simmons, B. A., and Singer, S. W. (2012). Thermoascus aurantiacus is a promising source of enzymes for biomass deconstruction under thermophilic conditions. Biotechnol. Biofuels. 5:54. doi: 10.1186/1754-6834-5-54
Meyer, A. S., Pedersen, L. H., and Isaksen, A. (1997). The effect of various food parameters on the activity and stability of catalase from Aspergillus niger and catalase from bovine liver. Food Chem. 60, 137–142. doi: 10.1016/S0308-8146(95)00252-9
Mu, Q., Cui, Y., Tian, Y., Hu, M., Tao, Y., and Wu, B. (2019). Thermostability improvement of the glucose oxidase from Aspergillus niger for efficient gluconic acid production via computational design. Int. J. Biol. Macromol. 136, 1060–1068. doi: 10.1016/j.ijbiomac.2019.06.094
Nilsson, R, Schultz, I. J, Pierce, E. L, Soltis, K. A, Naranuntarat, A, and Ward, D. M. (2009). Discovery of genes essential for heme biosynthesis through large-scale gene expression analysis. Cell Metab. 10, 119–130. doi: 10.1016/j.cmet.2009.06.012
Ning, X., Zhang, Y., Yuan, T., Li, Q., Tian, J., Guan, W., et al. (2018). Enhanced thermostability of glucose oxidase through computer-aided molecular design. Int. J. Mol. Sci. 19:2. doi: 10.3390/ijms19020425
Nivón, L. G., Moretti, R., and Baker, D. (2013). A pareto-optimal refinement method for protein design scaffolds. PLoS ONE. 8:e59004. doi: 10.1371/journal.pone.0059004
Ntana, F., Mortensen, U. H., Sarazin, C., and Figge, R. (2020). Aspergillus: a powerful protein production platform. Catalysts. 10:9. doi: 10.3390/catal10091064
Pedersen, M. B., Garrigues, C., Tuphile, K., Brun, C., Vido, K., Bennedsen, M., et al. (2008). Impact of aeration and heme-activated respiration on Lactococcus lactis gene expression: identification of a heme-responsive operon. J. Bacteriol. 190, 4903–4911. doi: 10.1128/JB.00447-08
Pentjuss, A., Bolmanis, E., Suleiko, A., Didrihsone, E., Suleiko, A., Dubencovs, K., et al. (2023). Pichia pastoris growth—coupled heme biosynthesis analysis using metabolic modelling. Sci. Rep. 13:15816. doi: 10.1038/s41598-023-42865-w
Philibert, T., Rao, Z., Yang, T., Zhou, J., Huang, G., Irene, K., et al. (2016). Heterologous expression and characterization of a new heme-catalase in Bacillus subtilis 168. J. Indust. Microbiol. Biotechnol. 43, 729–740. doi: 10.1007/s10295-016-1758-2
Raj, M., Singh, M., Kumar, V., Devi, T., Upadhyay, S. K., Mishra, M., et al. (2024). Gluconic acid: strategies for microbial production using organic waste and applications. Physical Sciences Reviews. 9, 2371–2383. doi: 10.1515/psr-2022-0163
Rekdal, V. M., van der Luijt, C. R. B., Chen, Y., Kakumanu, R., Baidoo, E. E. K., Petzold, C. J., et al. (2024). Edible mycelium bioengineered for enhanced nutritional value and sensory appeal using a modular synthetic biology toolkit. Nat. Commun. 15:2099. doi: 10.1038/s41467-024-46314-8
Sepasi Tehrani, H., and Moosavi-Movahedi, A. A. (2018). Catalase and its mysteries. Prog. Biophys. Mol. Biol. 140, 5–12. doi: 10.1016/j.pbiomolbio.2018.03.001
Singh, A., Singh, A., Grover, S., Pandey, B., Kumari, A., and Grover, A. (2018). Wild-type catalase peroxidase vs G279D mutant type: Molecular basis of Isoniazid drug resistance in Mycobacterium tuberculosis. Gene. 641, 226–234. doi: 10.1016/j.gene.2017.10.047
Singh, S., and McShane, M. (2010). Enhancing the longevity of microparticle-based glucose sensors towards one month continuous operation. Biosens. Bioelectron. 25:1075. doi: 10.1016/j.bios.2009.09.026
Si-Yuan, Z., Yan, X., and Xiao-Wei, Y. (2020). Improved homologous expression of the acidic lipase from Aspergillus niger. 30, 196–205. doi: 10.4014/jmb.1906.06028
Su, T., Guo, Q., Zheng, Y., Liang, Q., Wang, Q., and Qi, Q. (2019). Fine-Tuning of hemB using CRISPRi for increasing 5-aminolevulinic acid production in Escherichia coli. Front. Microbiol. 10, 1731. doi: 10.3389/fmicb.2019.01731
Thorup, O. A., Strole, W. B., and Leavell, B. S. (1961). A method for the localization of catalase on starch gels. J. Lab. Clin. Med. 58, 122–128.
Tsang, A., Butler, G., Powlowski, J., Panisko, E. A., and Baker, S. E. (2009). Analytical and computational approaches to define the Aspergillus niger secretome. Fungal Genet. Biol. 46, S153–S160. doi: 10.1016/j.fgb.2008.07.014
Wernars, K., Goosen, T., Swart, K., and van den Broek, J. H. W. (1986). Genetic analysis of Aspergillus nidulans AmdS+ transformants. Molec. Gen. Genet. 205, 312–317. doi: 10.1007/BF00430444
Wong, C. M., Wong, K. H., and Chen, X. D. (2008). Glucose oxidase: natural occurrence, function, properties and industrial applications. Appl. Microbiol. Biotechnol. 78, 927–938. doi: 10.1007/s00253-008-1407-4
Xu, Y., Wang, Y.-H., Liu, T.-Q., Zhang, H., Zhang, H., and Li, J. (2018). The GlaA signal peptide substantially increases the expression and secretion of α-galactosidase in Aspergillus niger. Biotechnol. Lett. 40, 949–955. doi: 10.1007/s10529-018-2540-5
Yan, Y., Liu, X., Jiang, X., Zhang, W., Wang, Y., Zhang, Y., et al. (2022). Surface charge modifications modulate glucose oxidase pH-activity profiles for efficient gluconic acid production. J. Clean. Prod. 372:133817. doi: 10.1016/j.jclepro.2022.133817
Yang, H.-S., Yang, H.-C., and Tani, Y. (1988). Catalase from Aspergillus niger KUF-04. Microbiol. Biotechnol. Letters. 16, 193–198,
Keywords: catalase, Aspergillus niger, gluconic acid, thermal stability, signal peptide, heme
Citation: Huang J, Wang J, He J, Wu Y, Chen L, Zhou S, Bian Y and Li Y (2024) Enhanced production of thermostable catalase for efficient gluconic acid biocatalysis. Front. Sustain. Food Syst. 8:1465445. doi: 10.3389/fsufs.2024.1465445
Received: 16 July 2024; Accepted: 14 November 2024;
Published: 04 December 2024.
Edited by:
Shenglin Sun, Shandong Agricultural University, ChinaReviewed by:
Nicolás Oscar Soto-Cruz, TecNM-Instituto Tecnológico de Durango, MexicoP. Sankarganesh, Hindustan Institute of Technology and Science, India
Copyright © 2024 Huang, Wang, He, Wu, Chen, Zhou, Bian and Li. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yangyuan Li, bGl5YW5neXVhbkB2dHJiaW8uY29t
 Jiang Huang
Jiang Huang Jun Wang1,2
Jun Wang1,2 Yeyu Bian
Yeyu Bian Yangyuan Li
Yangyuan Li