- Department of Food and Animal Science, Tennessee State University, Nashville, TN, United States
This study explores into the levels of coliform contamination, prevalence of antibiotic-resistant bacteria, and the hygienic practices in goat meat retail stores. Goat meat from 10 retail stores was analyzed for E. coli, Salmonella, and S. aureus using serological and PCR methods. Antimicrobial susceptibility testing was performed using the Kirby-Bauer disk diffusion method. Data on hygienic practices were collected through a structured observational questionnaire. Pearson’s correlation analysis was also employed to establish the relationship between hygienic practices and coliform loads. The average coliform loads on goat meat ranged between 0.88–5.04 log10 cfu/g. Our results revealed that 52% of examined goat meat was deemed unacceptable (>3.30 log10 CFU/g). The overall level of good meat handling practices among meat handlers in our study was 45.75%. Further, the study establishes a significant correlation between the level of food safety practices and coliform load. Hence, stores with higher hygienic practice scores exhibited lower coliform loads. The prevalence of S. aureus (44%) in goat meat was significantly higher (p < 0.05) as compared to E. coli (29%), and Salmonella spp. (20%). E. coli isolates displayed the highest resistance to penicillin (31.2%), Salmonella spp. to oxytetracycline (13.9%), and S. aureus to ampicillin (29.0%). Resistance was observed across selected antibiotic classes, particularly in beta-lactams and tetracyclines, with penicillin (78.5%) and oxytetracycline (64.5%) exhibiting notable resistance. Cephalosporin resistance was noted, with 48.4 and 33.3% of isolates showing resistance to cephalothin and cefpodoxime, respectively. Bacterial isolates also demonstrated resistance to phenicol antibiotics, including chloramphenicol (9.7%) and florfenicol (16.1%), respectively. Approximately 44.1% of bacterial isolates displayed multidrug resistance and MAR index ranged from 0.25 to 0.75. The study’s findings reveal heightened levels of coliform contamination, the presence of pathogenic and multidrug-resistant bacteria in goat meat, and suboptimal meat handling practices in retail stores. The significance of improving food safety practices in retail settings is emphasized to ensure the safety of goat meat, a matter of increasing importance due to its growing demand globally.
Introduction
Meat serves as a crucial source of high-quality protein and bioavailable vitamins, along with essential minerals like iron, zinc, and phosphorus (Adesokan et al., 2015). Despite these nutritional benefits, meat poses a potential risk for spreading foodborne illnesses due to its elevated protein content and nearly neutral pH, creating an environment conducive to bacterial growth and survival (Ahmed and Shimamoto, 2014). Coliforms, common in meats, serve as reliable indicators of fecal contamination and the potential presence of pathogens (Seo et al., 2019; Some et al., 2021). In goat and sheep production, notable pathogenic bacteria, including Staphylococcus aureus, E. coli, and Salmonella spp., pose a significant health risk (Hanlon et al., 2018; Ariffin et al., 2019). The contamination of meat by E. coli is closely associated with fecal contamination. According to Stein and Katz (2017), small ruminant meat is a source of E. coli infection in humans. E. coli being a common commensal intestinal bacterium in both animals and humans, serves as a significant zoonotic agent linked to infectious diseases in both domains (Getaneh, 2019; Sarowska et al., 2019). The prevalence of E. coli O157:H7 in goat feces at slaughter has been reported at 11.1%, posing a potential spill-over to the carcass during slaughtering and subsequently contaminating fresh meat. Salmonella and S. aureus are also highlighted as pathogens of concern in goat and sheep production (Hanlon et al., 2018; Ariffin et al., 2019). Salmonella spp. is a common pathogen reported to cause illnesses in both animals and humans (Chlebicz and Śliżewska, 2018), and numerous outbreaks have been linked to the consumption of contaminated goat products (Robinson et al., 2020). Li et al. (2021) suggests that animal-derived foods, such as meat, can serve as a mode of transmission for Salmonella to humans. Additionally, the presence of S. aureus in goat products has been documented (Angelidis et al., 2020), posing a potential threat to public health, especially given its association with udder infections in goat farming (Nelli et al., 2022).
The global escalation of multidrug resistance (MDR) poses a significant public health threat (Catalano et al., 2022). To mitigate the development and transmission of antimicrobial resistance between animals and humans, the judicious use of antimicrobial agents in food-producing animals is imperative (Lekshmi et al., 2017). Excessive antibiotic use in food-producing farms is identified as a primary contributor to the increasing prevalence of antimicrobial resistance (Ma et al., 2021). The overuse and misuse of antibiotics in both medical and agricultural settings are expected to contribute to the proliferation of antibiotic-resistant bacteria (Mancuso et al., 2021). Particularly worrisome is the antimicrobial resistance observed in Escherichia coli, the most common gram-negative pathogen in humans (Jans et al., 2018). Widespread resistance in Escherichia coli to fluoroquinolones, aminoglycosides, and cephalosporins; some of the most widely used antibiotics, adds to the gravity of the situation (Jans et al., 2018). Additionally, contaminated raw meat stands out as a primary source of antibiotic-resistant S. enterica infections in humans (Jaja et al., 2019). During slaughtering, antimicrobial-resistant bacteria may leak and contaminate meat, posing a potential route for transmission to humans through food (Mcewen and Fedorka-Cray, 2017). Meats are identified as both a cradle and vehicle for the dispersion of antibiotic-resistant bacteria to humans, emphasizing the need for urgent attention and intervention (Bosilevac et al., 2015).
During production, processing, transportation, and retailing, meats are frequently contaminated with pathogenic organisms (Koutsoumanis et al., 2021). Several factors may contribute to the contamination of carcasses during slaughter such as animals’ skin and dung, equipment including cutting tools, an unhygienic environment, non-compliance with proper slaughter processes, and a lack of personal hygiene (Wardhana, 2019). According to Kamala and Kumar (2018) and Sánchez-Aldana et al. (2020), microbiological quality of meat depends mostly on the slaughter process, sanitation during processing and packaging, maintenance of adequate cold chain storage from the processing to the retail level, and finally, sanitation during handling at the retail stores.
In recent years, there has been a notable surge in the demand for goat meat in the U.S., as evidenced by increased imports (Nakai, 2018) and a rise in meat goat inventory from 591,543 in 1990 to 2,075,000 in 2018 (Ibrahim et al., 2020; Mazhangara et al., 2019). This heightened demand can be attributed to factors such as the growing immigrant population, which traditionally favors goat meat, and the shifting consumer preferences towards healthier food choices like low-fat, low-cholesterol, and low-carb diets (Degala et al., 2018). The inclination of immigrants towards goat meat, highlighted by Ryan et al. (2021), is coupled with its perceived nutritional benefits compared to other red meats (Silva et al., 2022). This has positioned goat and sheep meat as the fourth most consumed meat in the U.S., following beef, pork, and poultry (Aravani et al., 2022). Specifically in Tennessee, the demand for goat meat has experienced a significant upswing over the past two decades, driven by a growing ethnic population that increased from 159,004 in 2000 to 304,801 in 2013 (Ekanem et al., 2016). This surge in demand has propelled the state of Tennessee to become the second-largest producer of goat meat in the country, trailing only behind Texas (Sang, 2016). The increasing demand for goat meat in Nashville, emphasizes the importance of addressing food safety practices in retail stores to safeguard consumer well-being. The main goal of this study was to assess coliform contamination levels, antibiotic resistance patterns of bacteria, and the relationship between coliform contamination and hygienic practices in retail establishments selling goat meat throughout the city. This research is vital to ensure the safety and quality of goat meat, particularly considering its growing popularity and production.
Materials and methods
Selection of ethnic retail stores, sample collection, and preparation
The study was approved by the Tennessee State University Institutional Review Board (HS-2021-4597). A total of one hundred (n = 100) goat meat samples were acquired from ten (n = 10) ethnic retail stores in Nashville, Davidson County, spanning a period of 7 months, from March 2022 to September 2022. Fresh meat was procured in duplicate from each selected store and discreetly labelled with letters A to J to ensure confidentiality. To prevent multiple samplings from the same shipments, meat samples were obtained at a biweekly interval from various stores. The quantity of meat samples obtained on each collection day depended on the availability of meat in the stores. All collected meat samples were labelled with store identification letters and collection dates. Promptly after collection, the meat samples were transported to the Tennessee State University Food Safety and Microbiology Laboratory in a cooler filled with ice. Microbial analysis was conducted within 2 h upon arrival at the laboratory. Each meat sample yielded three sub-samples, each weighing fifty grams (50 g), which were aseptically placed in sterile stomacher bags (Fisher Scientific, Pittsburgh, PA, United States). Subsequently, 450 mL of 0.1% sterile buffered peptone water (BPW) (Oxoid, Solon, OH, United States) was added, and the samples were homogenized for 2 min at 230 rpm using a 400 Circulator Stomacher® (Seaward, Norfolk, United Kingdom). The investigation of each homogenized sample included determining the total coliform count and prevalence of generic E. coli, Salmonella spp., and S. aureus in retail goat meat. Coliforms and E. coli levels were determined using the plating method, and the prevalence of bacteria was evaluated based on the number of positive samples.
Determination of coliform load and Escherichia coli
The homogenized meat samples underwent serial dilution with sterile peptone water (Oxoid, Basingstoke, United Kingdom). Using the serially diluted homogenate, 1 mL from each sample was pipetted from 10−1 to 10−5 dilutions, and each was spread-plated onto two petri dishes containing selective media Harlequin E. coli /Coliform Agar (Neogen, Lansing, Michigan, United States) which enables simultaneous enumeration of E. coli and coliforms in food and environmental samples. The plated samples were then incubated at 35°C for 24 h (Georgopoulou et al., 2020). After incubation, plates displaying 25–250 colonies were counted. The total coliform count per gram of meat was calculated as:
where N represents the number of colonies per mL/g of product, ∑C is the sum of all colonies on all counted plates, n1 is the number of plates in the first dilution counted, n2 is the number of plates in the second dilution counted, and d is the dilution from which the initial counts were obtained.
The number of coliform colony forming units per gram (CFU/g) of goat meat samples was categorized as acceptable (≤100 CFU/g or ≤2 log10 CFU/g), marginally acceptable (100–2000 CFU/g or 2 log10–3.30 log10 CFU/g), or unacceptable (≥2000 CFU/g or ≥3.30 log10 CFU/g) based on the International Commission on Microbiological Specifications for Food (ICMSF) guidelines. For the isolation of E. coli, characteristic colony morphology on Harlequin E. coli/Coliform Agar, such as dark blue, blue-green, or purple colonies, was considered presumptive E. coli. These colonies underwent further identification using E. coli latex agglutination test kits (Prolex, Round Rock, TX, United States), with results interpreted following the manufacturer’s instructions. Presumptive colonies were then inoculated into tryptic soy broth (TSB; Oxoid, Basingstoke, United Kingdom), incubated overnight at 37°C, and preserved in 50% glycerol at −80°C for subsequent confirmation through polymerase chain reaction (PCR). Coliforms, which possess the ß-galactosidase enzyme produced rose-pink colonies.
Isolation of Salmonella spp
To isolate Salmonella spp., a 25-gram portion of each meat sample was placed in a sterile stomacher bag. Subsequently, 225 mL of sterile lactose broth was added, and the mixture was homogenized for 2 min at 230 rpm using a 400 Circulator Stomacher® (Seaward, Norfolk, United Kingdom). The homogenized sample was then transferred into a sterile screw-cap jar, securely capped, and allowed to stand for 60 min at room temperature. Then, the pH of each sample was adjusted to 6.8, thoroughly mixed, and incubated overnight at 35°C. After overnight incubation, 0.1 mL of the mixture was transferred to 10 mL Rappaport-Vassiliadis (RV) medium and incubated at 42°C for 24 h. Subsequently, ten microliters of RV broth were streaked in duplicates on Xylose Lysine Deoxycholate (XLD) agar plates, which were then incubated at 35°C for 24 h. Post-incubation, three colonies exhibiting typical Salmonella spp. characteristics on XLD agar, appearing as red or red with a black center, were selected as presumptive colonies. These presumptive Salmonella spp. colonies underwent further identification using latex agglutination kits (Wellcolex, Santa Fe, KS, United States), and the results were interpreted following the manufacturer’s instructions. The identified colonies were inoculated into tryptic soy broth (TSB; Oxoid, Basingstoke, United Kingdom), incubated overnight at 37°C, and preserved in 50% glycerol at −80°C for subsequent confirmation.
Isolation of Staphylococcus aureus
To isolate, S. aureus, a 1 mL aliquot from each homogenate was transferred to 9 mL of brain heart infusion broth (BHI) (Oxoid, Basingstoke, United Kingdom) and incubated at 37°C for 24 h. Subsequently, 1 mL aliquots from BHI were transferred to 9 mL of sterile peptone water (PW) and incubated as previously described. Loops of broth containing ten microliters of the organism were then streaked in duplicates on mannitol salt agar and incubated aerobically at 35°C for 24 h. Yellowish colonies observed on mannitol salt agar were chosen as presumptive S. aureus (Raji et al., 2016). These presumptive S. aureus colonies underwent further identification using latex agglutination kits (Bacistaph, Santa Fe, KS, United States), and the results were interpreted following the manufacturer’s instructions. Presumptive colonies were subsequently inoculated into tryptic soy broth (TSB; Oxoid, Basingstoke, United Kingdom), incubated overnight at 37°C, and preserved in 50% glycerol at −80°C for further confirmation.
Bacterial DNA extraction and PCR confirmation of presumptive isolates
Presumptive isolates of E. coli, Salmonella spp., and S. aureus were cultured overnight at 37°C in TSB. DNA extraction from the overnight cultures (>5 × 106 cells) was performed using the DNeasy® UltraClean® Microbial Kit (QIAGEN, Hilden, Germany) following the manufacturer’s instructions. DNA concentrations and integrity were determined by using a NanoDrop 2000 (Thermo Scientific, Pittsburgh, PA, United States) and agarose gel electrophoresis, respectively. Confirmation of presumptive isolates of E. coli, Salmonella spp., and S. aureus was carried out through PCR. The Hotstar Taq Plus Master Mix (Qiagen, Hilden, Germany) was employed in this study. The PCR mixture (20 μL) consisted of 0.4 μL each of forward and reverse primers, 2 μL of coral load concentrate 10× (dye), 5.2 μL of Rnase-free water (RH20), 2 μL of DNA template, and a 10 μL solution of Taq PCR Master Mix polymerase. The working concentrations for the primers were 10 ng/μL and 100 ng/μL for the DNA template. The sequences of primer pair used for E. coli target gene (16Sr RNA) was 5′-AGAGTTTGATCATGGCTCAG-3′ and 5′-GGACTACCAGGGTATCTAAT-3′ (Mamun et al., 2016), whereas the primer pair used for targeting Salmonella spp. (sdiA) was 5′-CGGTGGTTTTAAGCGTACTCTT-3′ and 5′-CGAATATGCTCCACAAGGTTA-3′ (Paião et al., 2013). S. aureus (16Sr RNA) primer pair was 5′-CCTATAAGACTGGGATAACTTCGGG-3′ and 5′-CTTTGAGTTTCAACCTTGCGGTCG-3′ (Mason et al., 2001). PCR amplification products (20 μL aliquots) were subjected to electrophoresis on a 1.5% agarose gel (FMC Bioproducts, Rockland, Maine) in 1X TAE buffer (Tris-acetate-EDTA). Ethidium bromide (Fisher, Fair Lawn, United States) was used for enhanced visualization. Gel electrophoresis was conducted for 1 h and 30 min at 70 volts in 1X TAE buffer, and the results were visualized using a Bio-Rad Gel Doc Imager (735 BR EZ).
Antimicrobial susceptibility and multiple antibiotic resistance
The Kirby Bauer disk diffusion method was used for antimicrobial susceptibility test. E. coli, Salmonella spp., and S. aureus isolates were subjected twelve antibiotics (Oxoid, Hampshire, United Kingdom) categorized into six classes with corresponding strength in parentheses: (class 1) β-lactam: ampicillin (AMP; 10 μg), and penicillin (P; 10iu); (class 2) β-lactam/β-lactamase inhibitor combinations: amoxicillin/clavulanic acid (AMC; 20/10 μg); (class 3) cephalosporins: cefpodoxime (CPD; 10 μg), cephalothin (KF; 30 μg), and ceftiofur (XNL; 30 μg); (4 class) phenicol: chloramphenicol (C; 30 μg), and florfenicol (FFC; 30 μg); (class 5) tetracyclines: doxycycline (DO; 30 μg), tetracycline (TE; 30 μg), and oxytetracycline (T; 30 μg); and (class 6) aminoglycosides: neomycin (N; 30 μg). Bacterial colonies were inoculated in TSB and incubated at 37°C for 18 h. The turbidity of the broth was adjusted to 0.5 McFarland and spread-plated on Müeller Hinton Agar. On each Müeller Hinton Agar plate (Åhman et al., 2019), four antibiotic disks were positioned to avoid overlapping of inhibition zones. The plates were then incubated for 24 h at 37°C. Following incubation, the zones of inhibition were measured, and results were categorized as resistant, intermediate, or susceptible using the Clinical Laboratory Standard Institute (CLSI Clinical and Laboratory Standards Institute, 2020) guidelines. The diameter of inhibition zone (DIZ) was considered as a measure of the antibacterial activity. To identify multidrug-resistant (MDR) strains, the number of antibiotics to which each bacterium exhibited resistance was noted. MDR isolates were defined as those showing intermediate susceptibility or resistance to drugs in three or more antibiotic classes (Wang et al., 2021).
The multiple antibiotic resistance (MAR) indexes were calculated using the formula:
where “a” represents the number of antibiotics to which an isolate was resistant, and “b” represents the overall number of antibiotics tested, according to Titilawo et al. (2015) and interpreted as per Igere et al. (2020).
Investigation of food safety practices among retail goat meat handlers
The food safety practices of goat meat handlers were examined while they were involved in the handling and sale of goat meat to consumers in ten retail stores in Nashville, Tennessee. Each store had approximately 2 to 3 employed workers on-site at the time of the study. The study took place between March and September 2022, and data collection involved direct personal observation using a structured questionnaire survey, which was designed based on previous studies (Yenealem et al., 2020; Al Banna et al., 2021; Sarma et al., 2022). The study focused on food safety practices outlined in Table 1, with each correctly executed practice earning one (1) point. For evaluation purposes, a score of ≥70% indicated that meat handlers implemented seven or more out of the 10 hygienic practices specified in the observational questions. Stores achieving this score were categorized as demonstrating “good” food hygiene practices, while those scoring below 70% were classified as having “poor” food hygiene practices, following the criteria established by Akabanda et al. (2017). The meat handling practice score from each store was then compared with its average total coliform colony-forming units per gram (cfu/g) of goat meat to explore potential correlations between hygienic practices and coliform contamination levels.
Statistical analysis
Descriptive statistics were utilized to present the mean log and standard deviation (SD) of coliform counts in goat meat from various stores. Coliform counts were further categorized into percentages of acceptable, marginally acceptable, and unacceptable, following the guidelines outlined by ICMSF. Plate counts between 25 and 250 cfu/plate were considered countable and log transformed. Microsoft Excel version 2020 (Microsoft, Redmond, WA, United States) was employed for data analysis. Variations in antimicrobial resistance patterns among bacteria isolated from goat meat in retail stores were determined using the chi-square test. To explore potential correlations, Pearson’s correlation was applied to assess the relationship between hygienic practices and coliform contamination levels. A p-value of <0.05 was considered statistically significant in all analyses.
Results
Coliform levels in retail goat meat
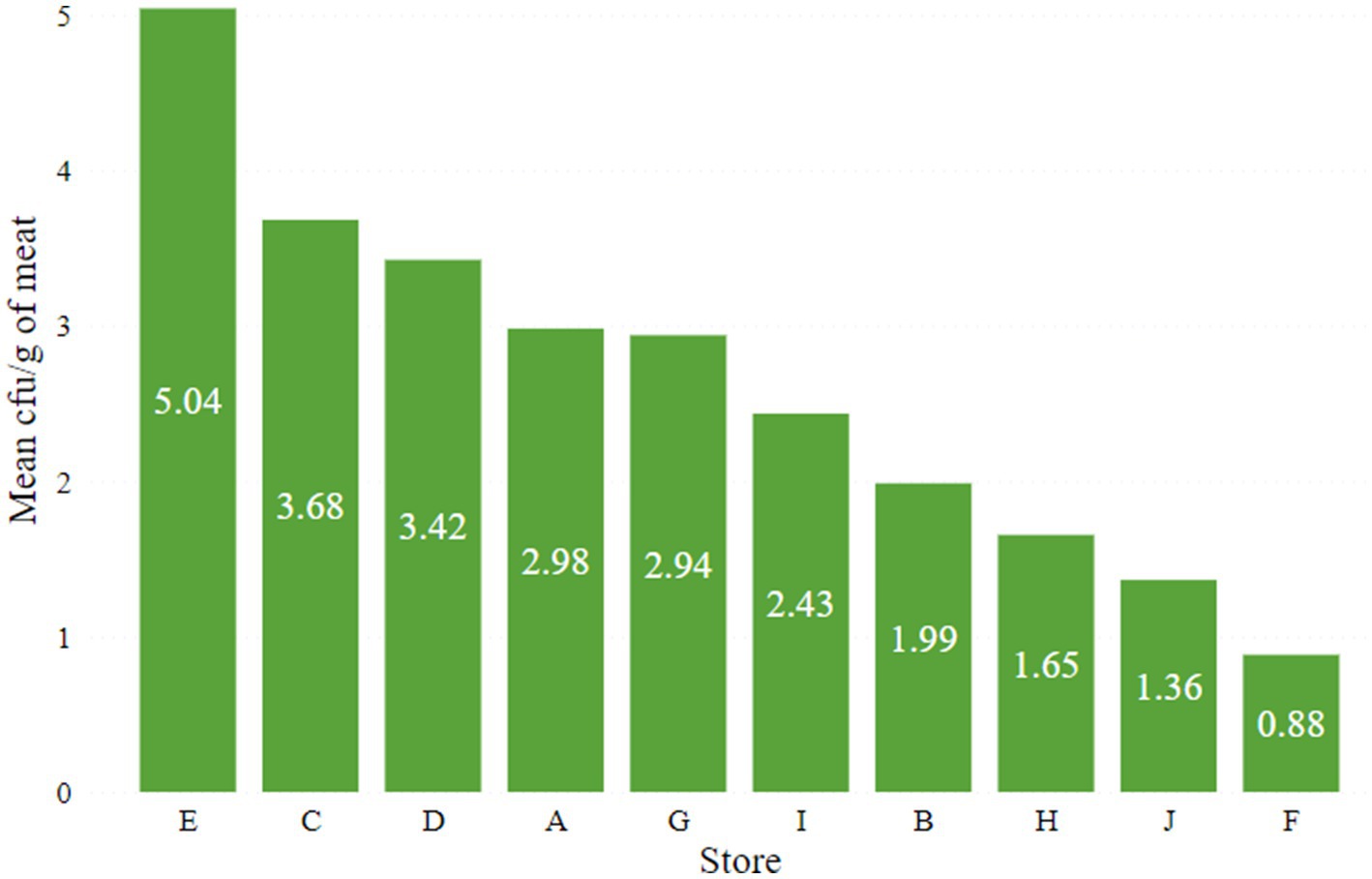
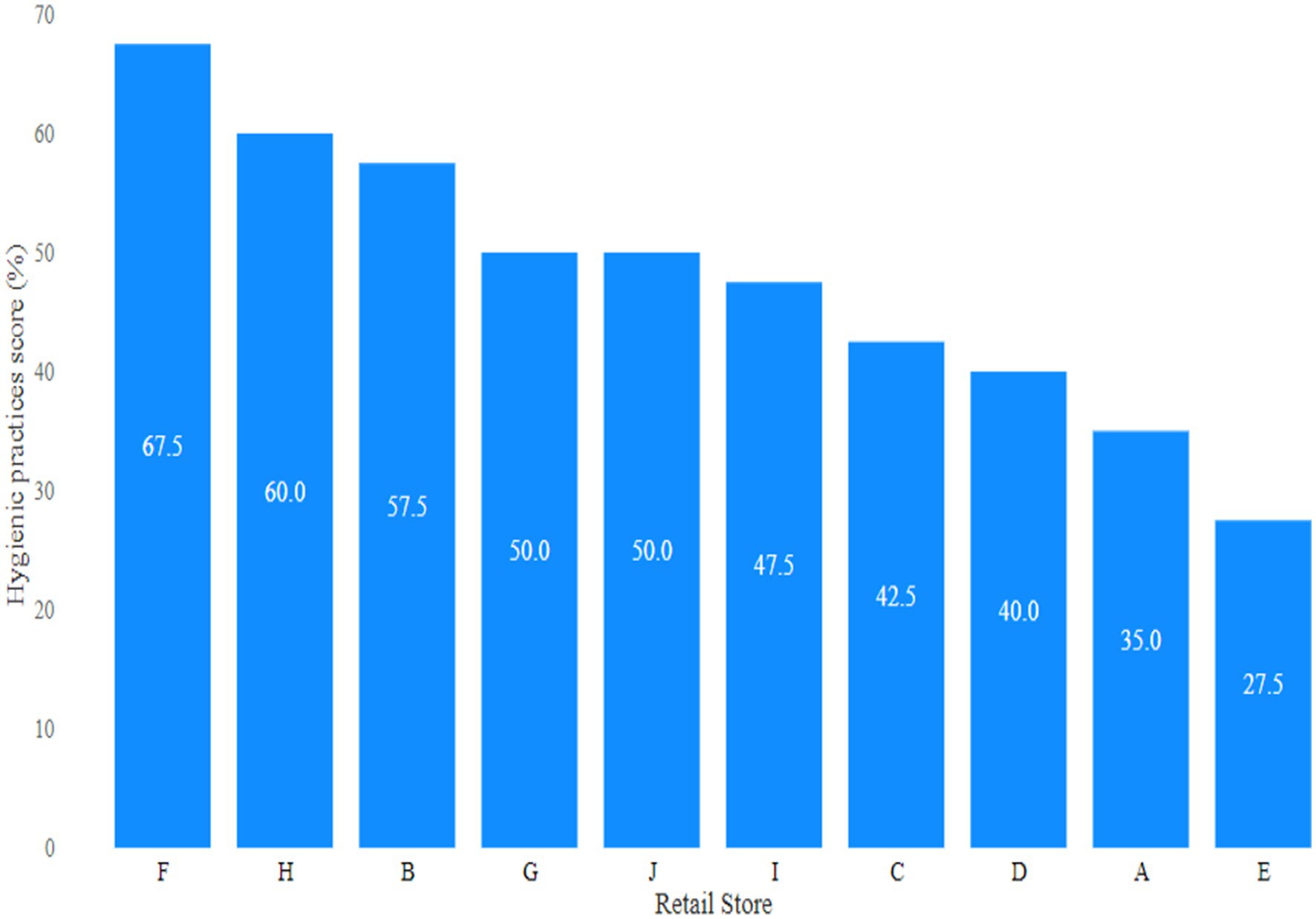
The lowest (0.88 log10 cfu/g) and highest (5.04 log10 cfu/g) average coliform loads in meat were observed in retail stores F and E, respectively (Figure 1). The mean coliform load in meat from store E was significantly (p < 0.05) higher compared to all other stores. Following the guidelines established by ICMSF, the distribution of coliform levels in the goat meat samples revealed that 52% were unacceptable (>2000 cfu/g or 3.30 log10 cfu/g), 21% marginally acceptable (100–2000 cfu/g or 2–3.30 log10 cfu/g), and 27% acceptable (≤100 cfu/g or ≤2 log10 cfu/g) as indicated in Figure 2.
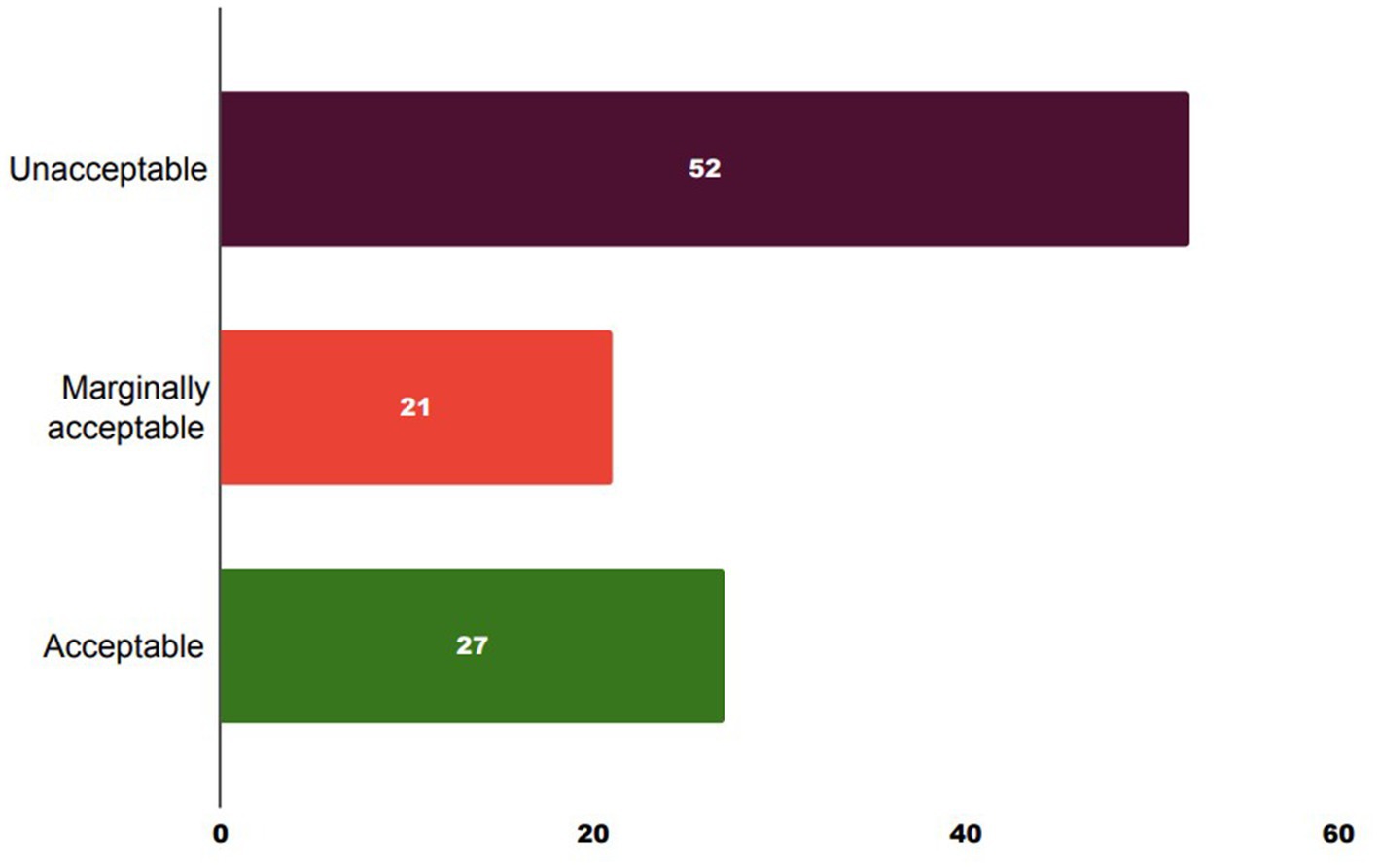
Figure 2. Total coliform load in retail goat meat. Unacceptable: >2000 cfu/g or 3.30 log10 cfu/g, marginally acceptable: 100–2000 cfu/g or 2–3.30 log10 cfu/g, and acceptable: ≤100 cfu/g or ≤2 log10 cfu/g.
Recovery of E. coli and Salmonella spp., and S. aureus in fresh goat meat
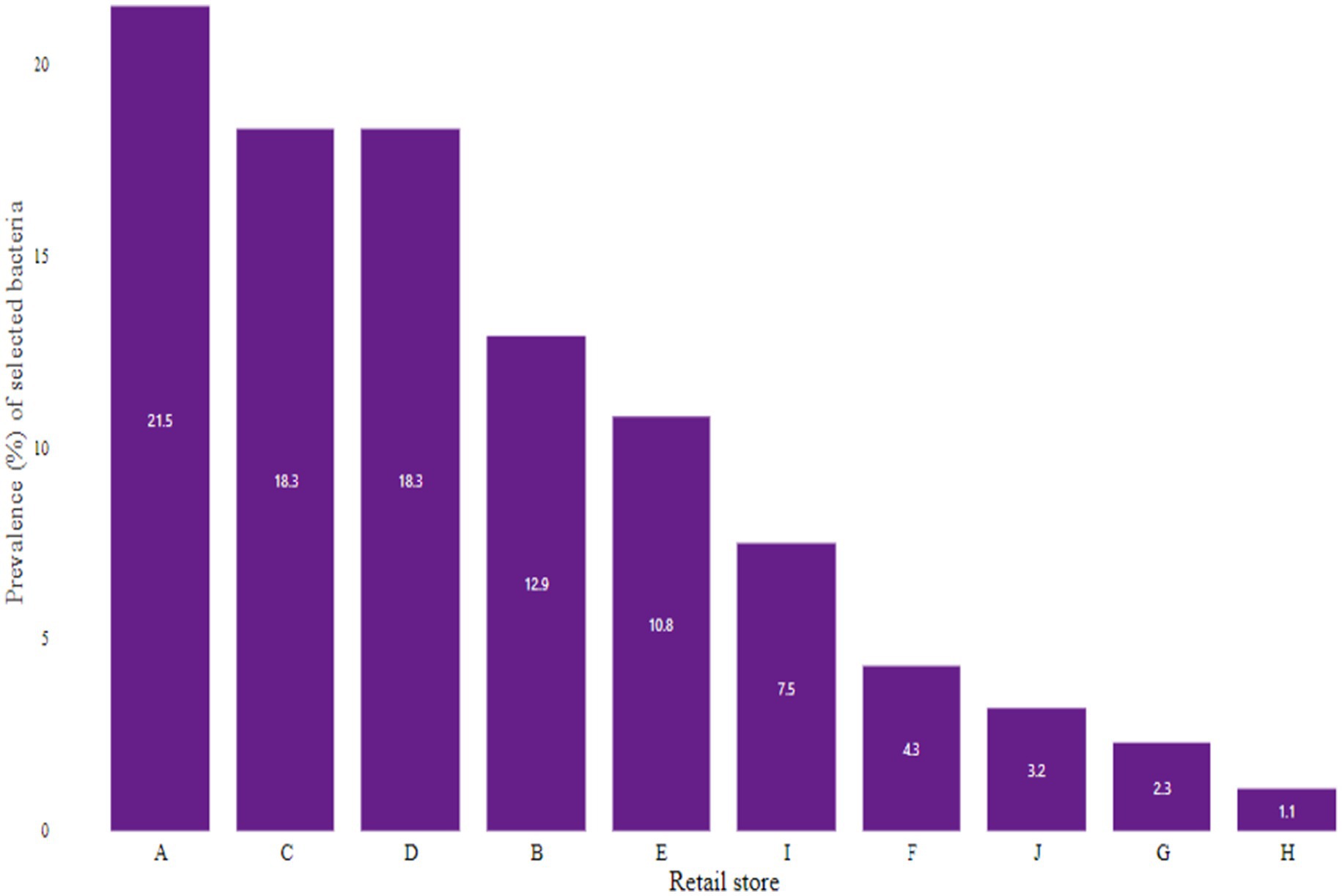
From the presumptive isolates identified by cultural methods and latex agglutination tests, bacteria were confirmed by the amplification of specific target genes 16S rRNA, sdiA, and 16S rRNA for E. coli, Salmonella spp., and S. aureus, respectively (Figures 3–5). The prevalence of E. coli and Salmonella spp., and S. aureus, both combined are summarized as indicated in Figure 6. The highest prevalence of bacteria isolates was recorded from meats in store A at 21.5% (20/93), followed by C and D at 18.3% (17/93), B at 12.9% (12/93), while G and H samples recorded the least at a rate of 2.3% (2/93), and 1.1% (1/93), respectively.
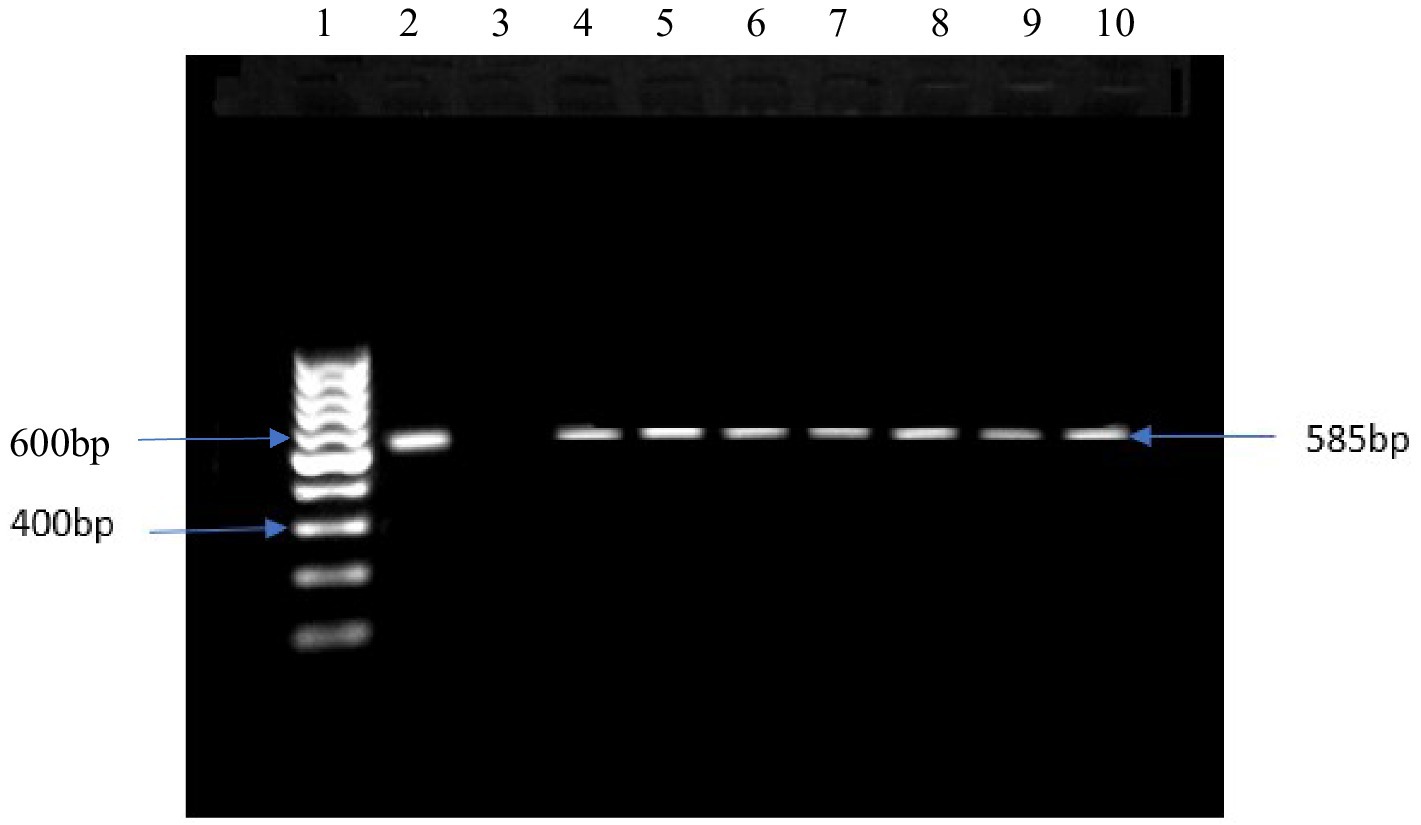
Figure 3. DNA amplification of 16srRNA gene in E. coli Lane 1: 1 kb ladder, lane 2: positive control: E. coli ATCC 11775, lane 3: negative control, lane 4–10: positive samples.
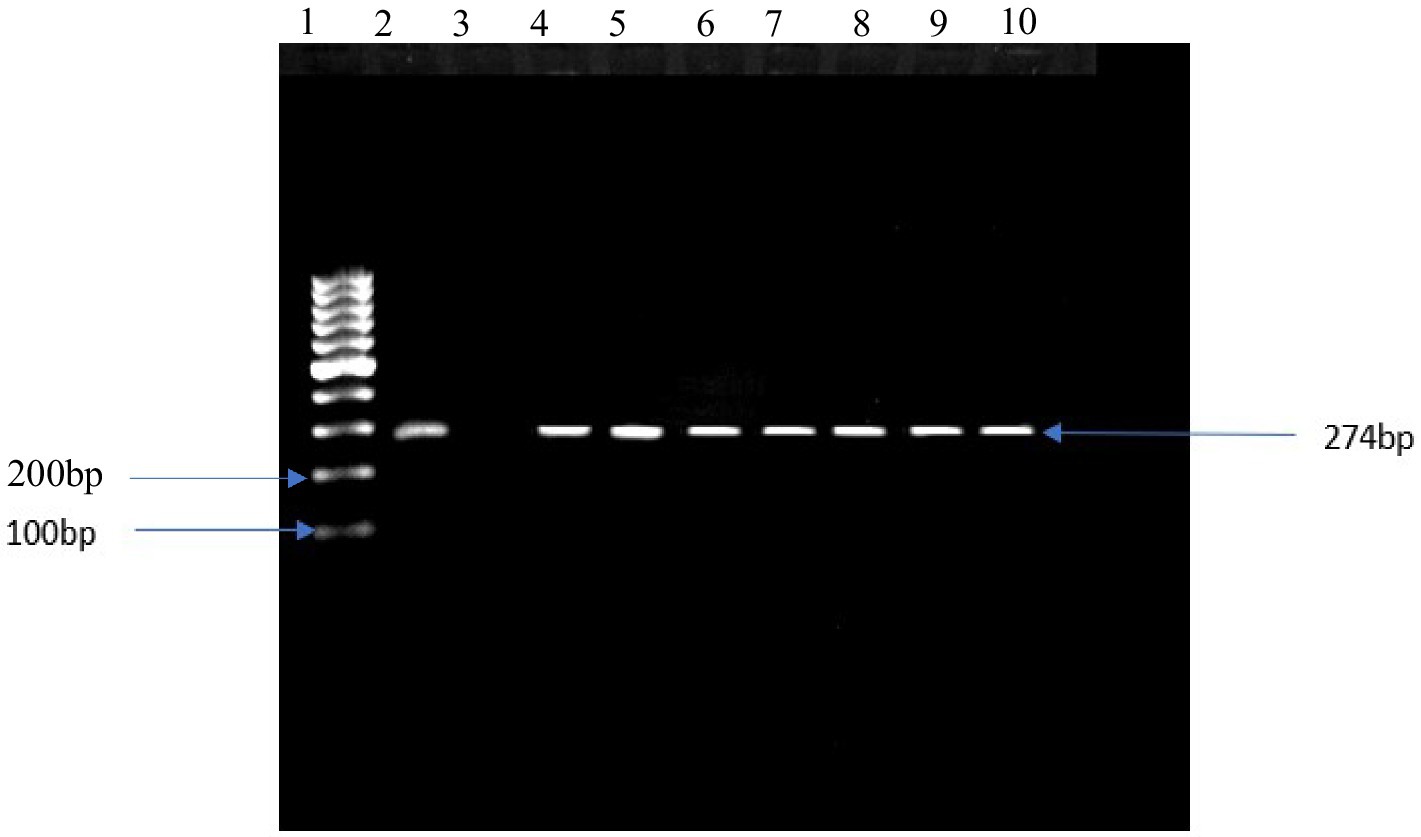
Figure 4. DNA amplification of the sdiA gene in Salmonella spp. Lane 1: 1 kb ladder, lane 2: positive control salmonella typyimurium ATTC 13311, lane 3: negative control, lane 4–10: positive samples.
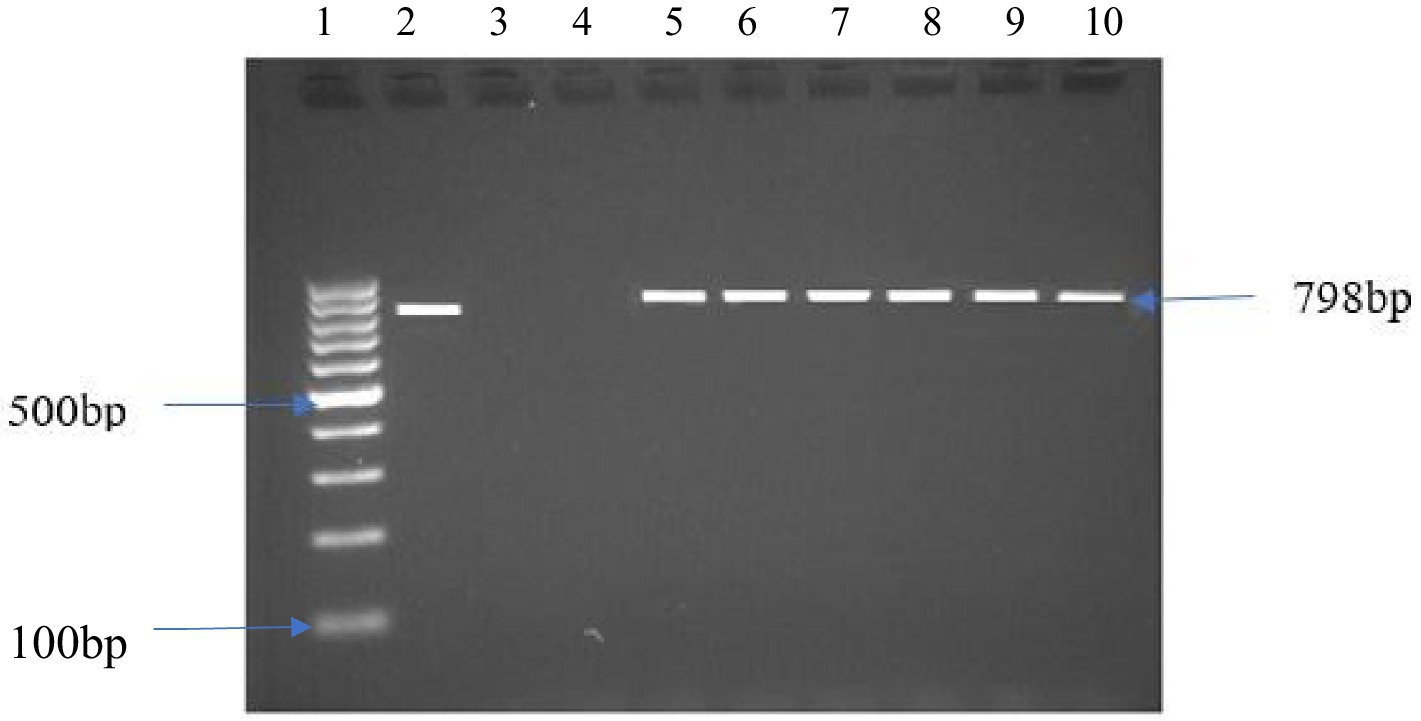
Figure 5. DNA amplification of the 16srRNA gene in S. aureus Lane 1: 1 kb ladder, lane 2: positive control S. aureus ATCC 11775, lane 3: negative control, lane 4: negative sample, lane 5–10: positive samples.
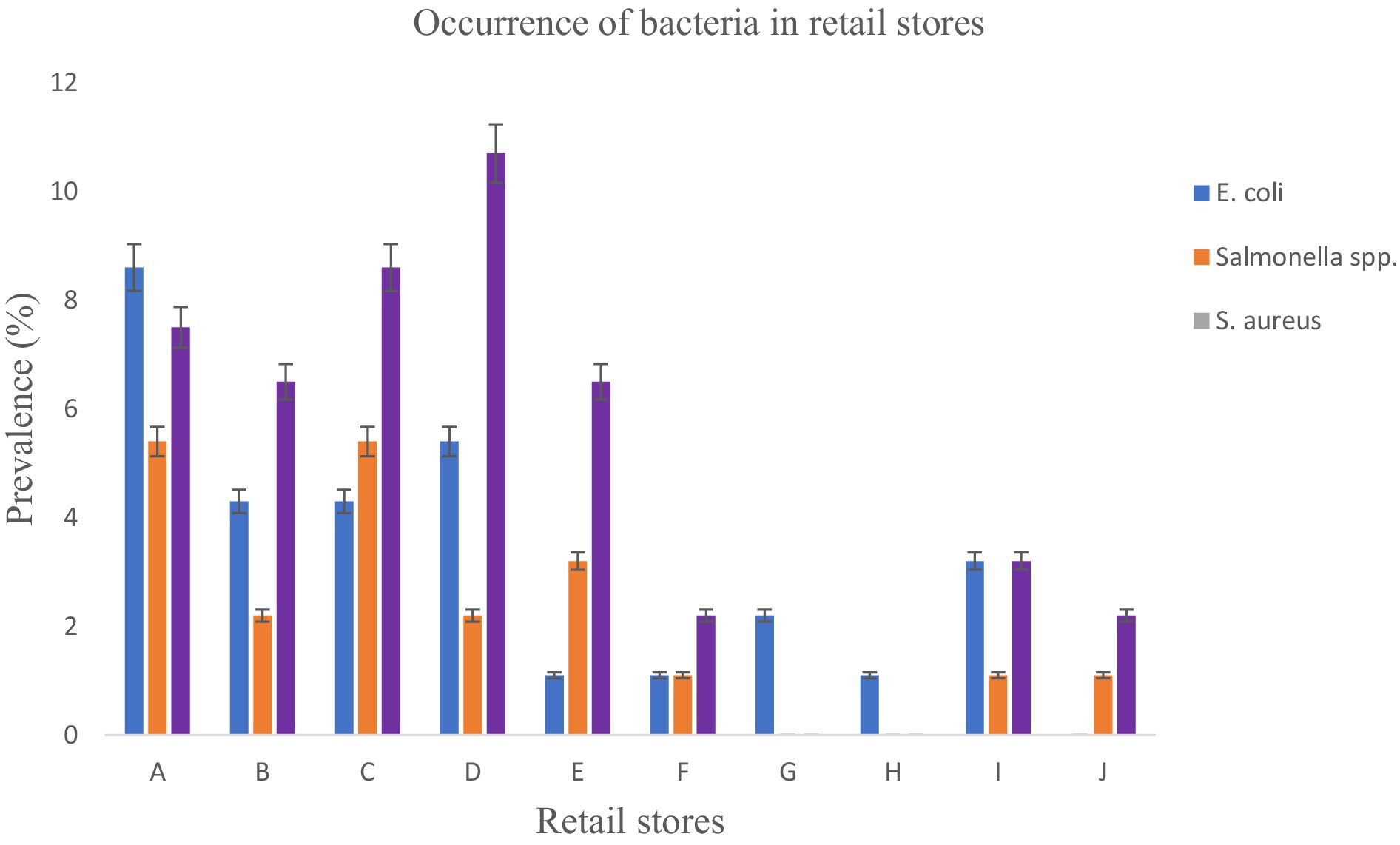
Figure 7 shows the levels of specific bacteria found in each store. Of the total 93 bacteria isolates analyzed, S. aureus showed the highest prevalence at 47.3%, significantly surpassing E. coli (31.2%) and Salmonella spp. (21.5%) with a p-value <0.05. Store D exhibited the highest occurrence of S. aureus at 10.7%, followed by C at 8.6%, and B and E both at 6.5%. Stores F and J had a prevalence of 2.2%, and stores G and H showed no presence of S. aureus. On the other hand, Salmonella spp. isolates were found in varying levels in different stores as: 5.4% for A and C, 3.2% for E, 2.2% for B and D, 1.1% for F, I and J and H recorded no Salmonella. Regarding E. coli isolates recovered from goat meat, store A exhibited the highest prevalence at 8.6%, followed by D (5.4%), B (4.3%), and C (4.3%). E. coli in goat meat from store A was significantly higher (p < 0.05) compared to stores F, H, and J. Notably, E. coli was not recovered from store E.
Resistance of E. coli, Salmonella spp., and S. aureus to specific antibiotics
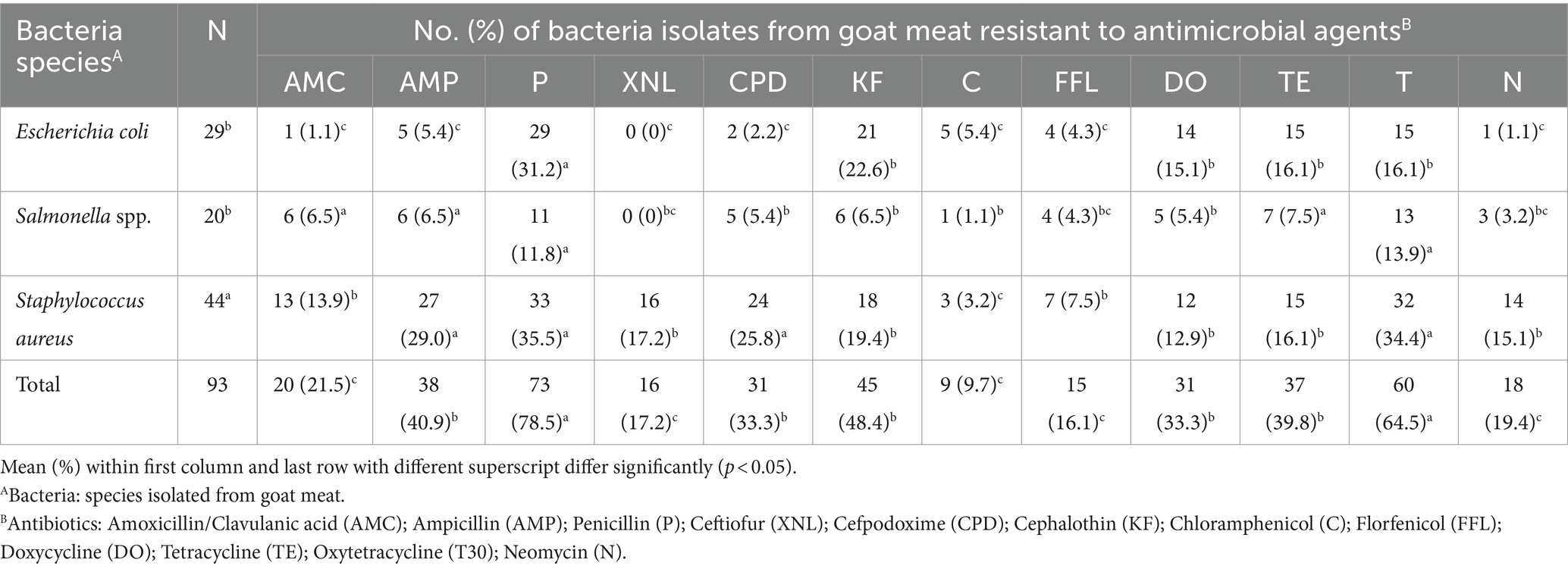
The increasing trend of foodborne bacterial pathogens developing antibiotic resistance is adding to the rising challenge of foodborne infections, leading to infections that are challenging to treat and pose greater complications for consumers. Table 1 displays antibiotic resistance to selected antibiotics. AMR analysis demonstrated that the S. aureus isolates were resistant to all evaluated antibiotics. Particularly, noteworthy is its significant resistance (p < 0.05) to penicillin (35.5%; 33/93), oxytetracycline (34.4%; 32/93), ampicillin (29.0%; 27/93), and cefpodoxime (25.8%; 24/93) compared to other antibiotics. Salmonella spp. displayed notable resistance, with the highest rates observed for oxytetracycline (13.9%; 13/93) and penicillin (11.8%; 11/93), significantly differing (p < 0.05) from ceftiofur (0%; 0/93) and chloramphenicol (1.1%; 1/93). Notably, Salmonella spp. exhibited resistance to at least one of the twelve antibiotics, except for ceftiofur. E. coli isolates demonstrated significant resistance (p < 0.05) to penicillin (31.2%; 29/93) when compared to cephalothin (22.6%; 21/93), tetracycline and oxytetracycline (16.1%; 15/93), and doxycycline (15.1%; 14/93). All E. coli and Salmonella spp. isolates were sensitive to ceftiofur (100%).
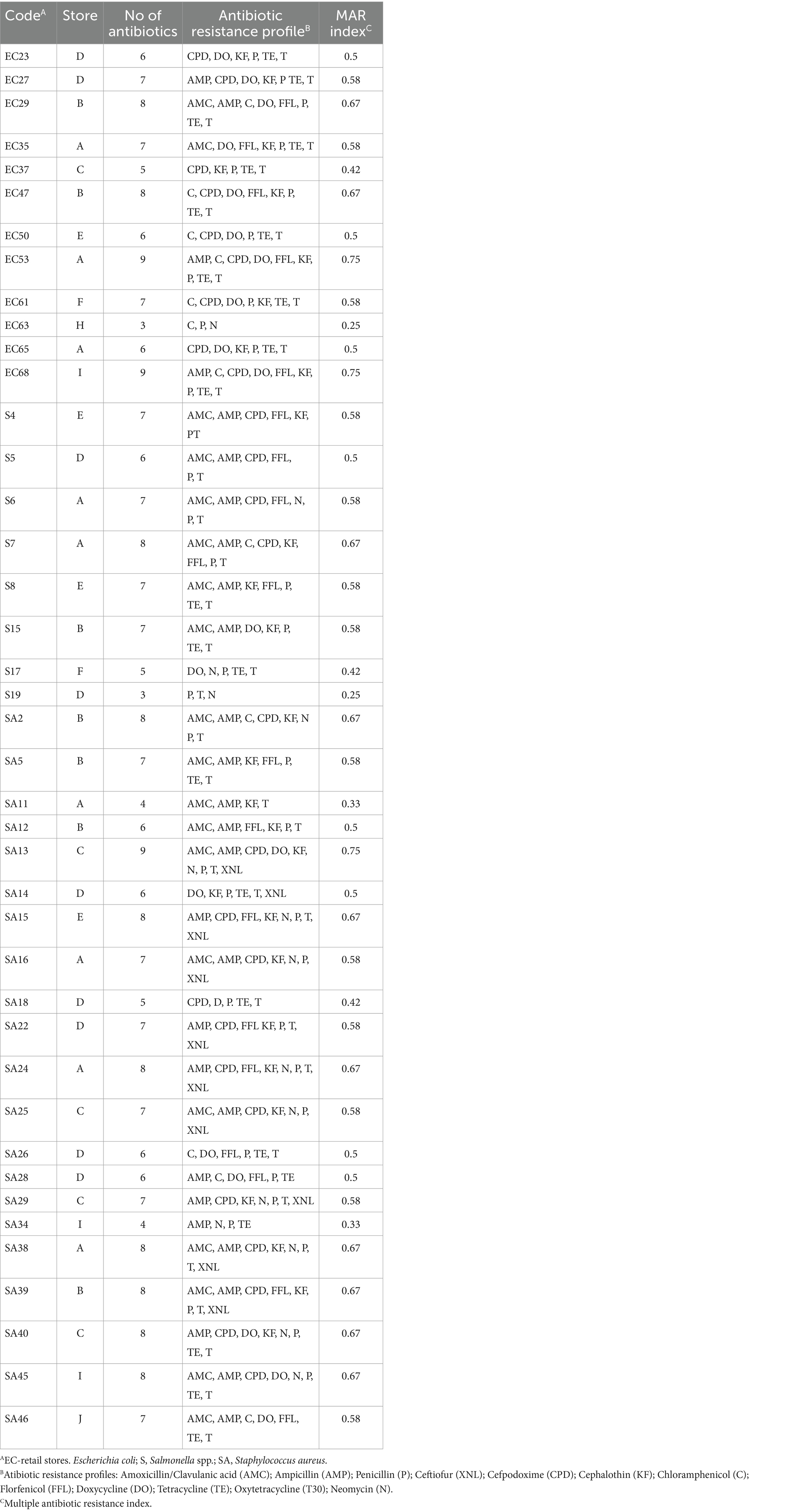
Multidrug-resistant patterns and MAR index
In this study, multidrug-resistant (MDR) of E. coli, Salmonella spp., and S. aureus was identified. The antimicrobial susceptibility test indicated that 29.3% of E. coli, 19.5% of Salmonella spp., and 51.2% of S. aureus isolates displayed multidrug resistance, as detailed in Table 2. MAR index ranged from 0.25 to 0.75. Resistance patterns of AMP-C-CPD-DO-FFL-KF-P-TE-T, AMP-C-CPD-DO-FFL-KF-P-TE-T, and AMC-AMP-CPD-DO-KF-N-P-T-XNL were observed in EC53, EC68, and SA13 isolates, respectively (Table 3). E. coli isolates from various stores exhibited diverse drug resistance patterns, such as AMC-DO-FFL-KF-P-TE-T, AMP-C-CPD-DO-FFL-KF-P-TE-T, and AMC-AMP-CPD-FFL-N-P-T in store A, AMC-AMP-C-DO-FFL-P-TE-T and C-CPD-DO-FFL-KF-P-TE-T in store B, and CPD-DO-KF-P-TE-T and AMP-CPD-DO-KF-P-TE-T in store D. Despite being the least recovered in this study, Salmonella spp. isolates demonstrated a diverse array of multiple drug resistance patterns. Distinct resistance patterns observed among Salmonella spp. isolates from different stores, included AMC-AMP-CPD-FFL-N-P-T and AMC-AMP-C-CPD-KF-FFL-P-T in store A, AMC-AMP-CPD-FFL-P-T and P-T-N in store D, and AMC-AMP-CPD-FFL-KF-PT, and AMC-AMP-KF-FFL-P-TE-T in store E. S. aureus exhibited a noteworthy display of twenty-one multi-drug resistance patterns, emphasizing the complexity of drug resistance (Table 4).
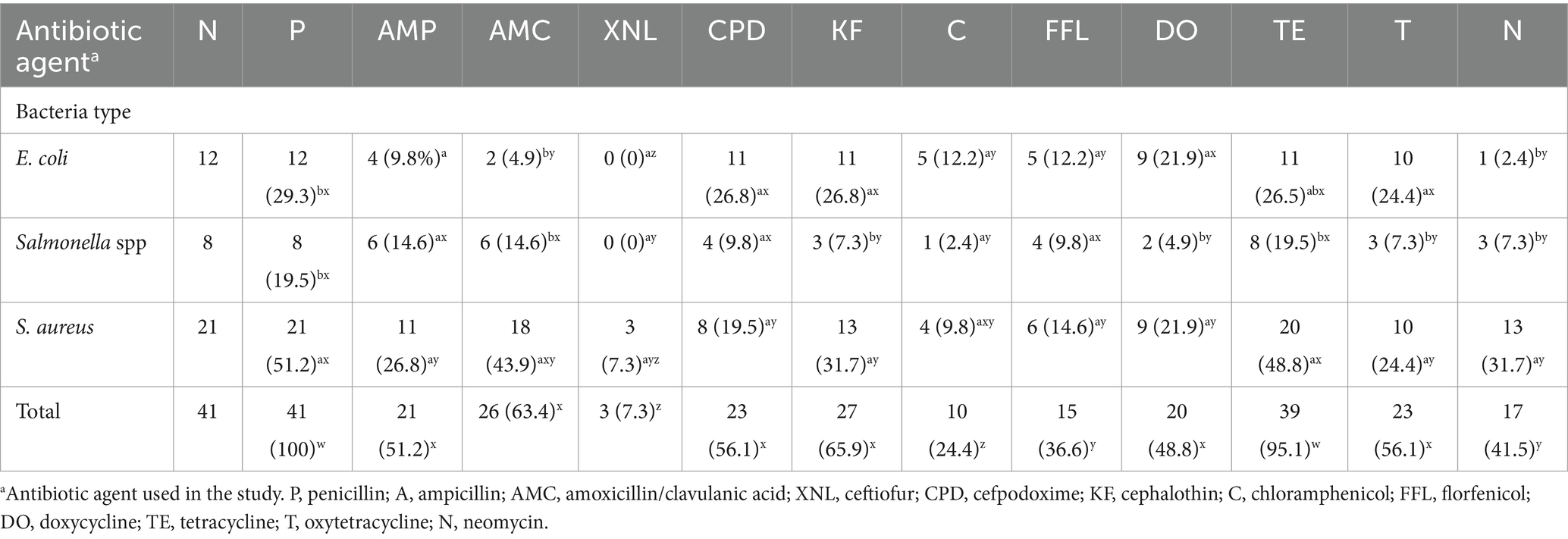
Table 2. Multi-drug resistance of E. coli spp., Salmonella spp. and S. aureus to various antimicrobial agents in goat meat.
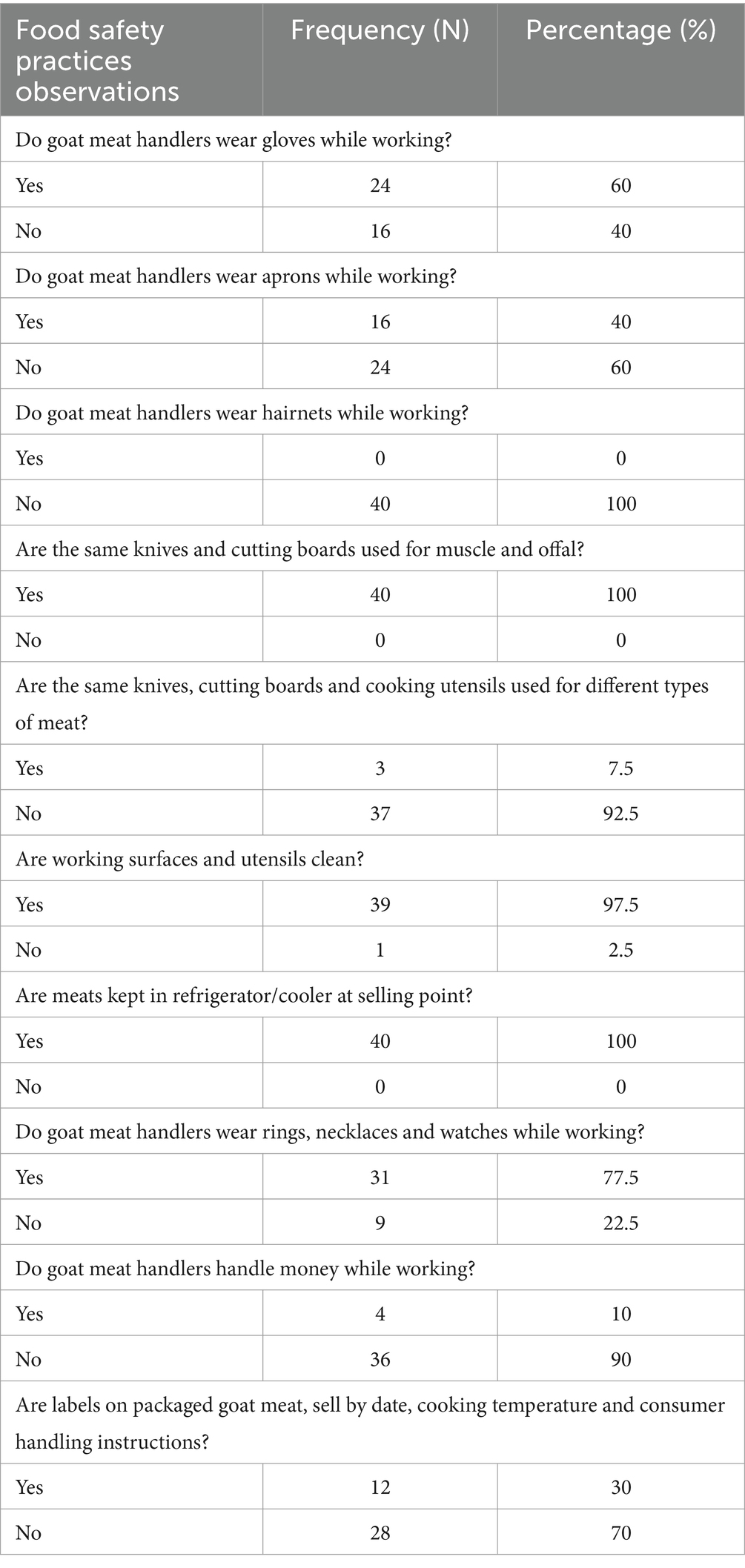
Table 3. Observational questions and answers for assessment of food safety practices of meat handlers in retail stores.
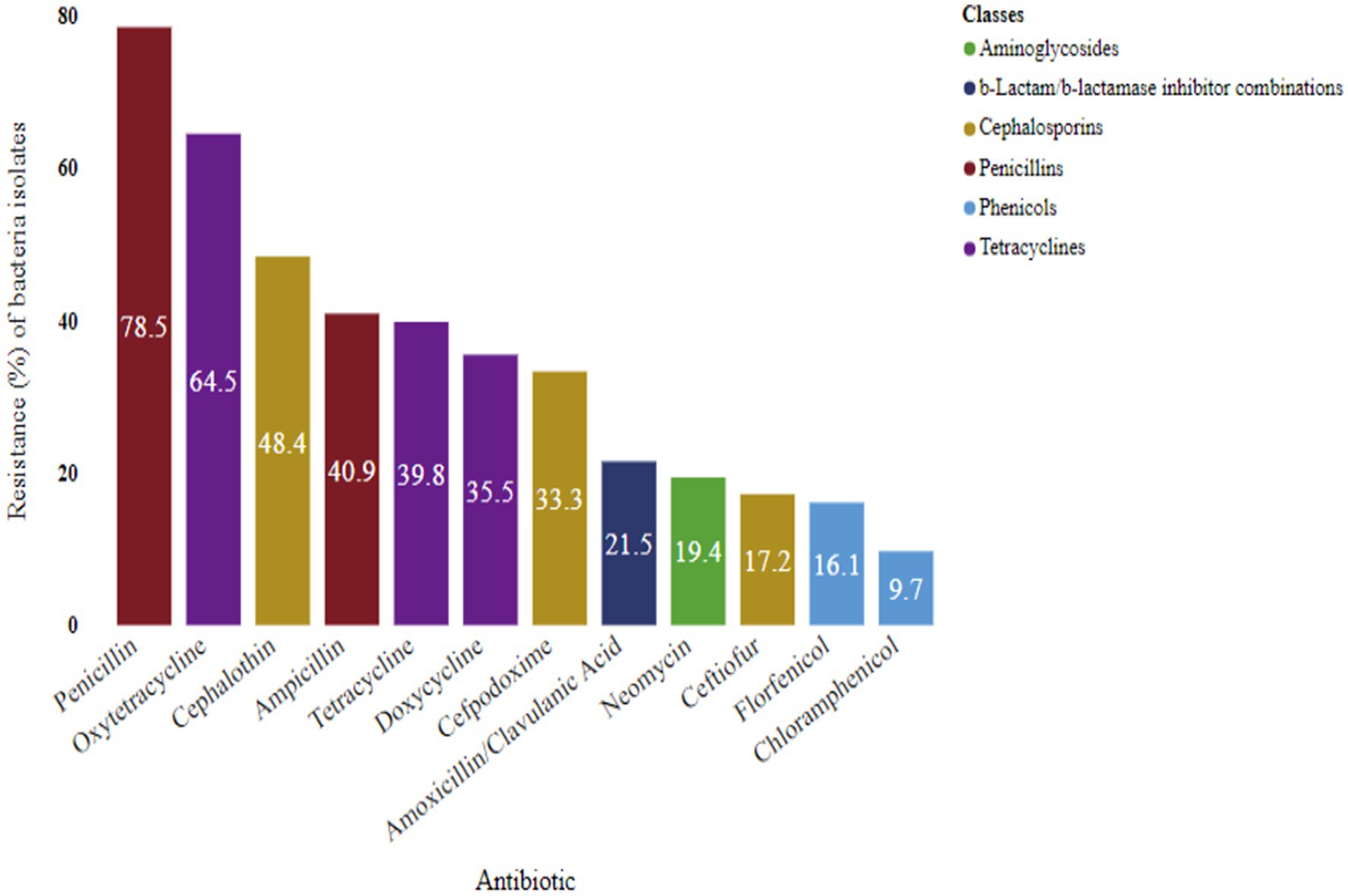
Resistance of bacteria to antibiotic classes
Notably, beta-lactams, such as penicillin (78.5%) and ampicillin (40.9%), exhibited significant resistance. Tetracyclines also displayed resistance, with oxytetracycline (64.5%), tetracycline (39.8%), and doxycycline (35.5%) showing notable levels. Cephalosporin resistance was observed at rates of 48.4, 33.3, and 17.2% for cephalothin, cefpodoxime, and ceftiofur, respectively. Neomycin, an aminoglycoside, demonstrated a resistance of 19.4%. Among the antibiotic classes, phenicol showed the least resistance, with chloramphenicol (9.7%) and florfenicol (16.1%) exhibiting lower resistance to bacterial isolates (Figure 8).
Retail goat meat handlers’ food safety practices
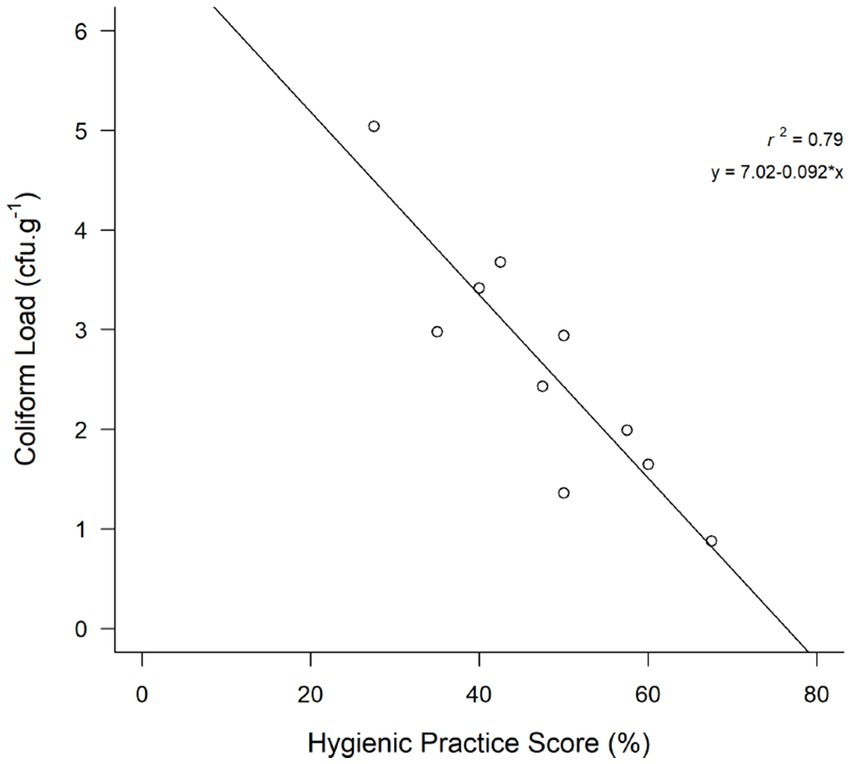
The data presented pertains to the food safety practices observed among goat meat handlers in retail stores (Figure 9). Hygienic practices score of goat meat handlers in retail stores are presented in Table 3. The findings revealed that none of the stores met the criteria for “good hygienic practices,” with none scoring 70% or more on the hygienic practices scale, as depicted in the study. The overall frequency of glove use was 60% and apron use was 40%. Strikingly, the absence of hairnet use was noted in all retail stores. A significant proportion of participants (92.5%) did not use the same knives, cutting boards, and utensils for different types of meats. However, 100% of retail stores used the same knife and cutting board for both muscle meat and offal (organ meat). Meat handlers (10%) handled money while working. Approximately 70% of retail stores lacked proper labels on packaged goat meat, including important information such as the sell-by date, cooking temperature, and consumer handling instructions. Pearson’s correlation analysis showed that the coefficient for hygienic practices and the coliform count was −0.8913968 (p < 0.05) suggesting that hygienic practices affected coliform contamination in the retail stores (see Figure 10).
Discussion
The aim of this study was to evaluate the prevalence of coliforms, antimicrobial resistance (AMR) of E. coli, Salmonella spp., and S. aureus on goat meat, along with assessing meat handling practices in retail stores. The results indicated that 96% of the tested meat samples were positive for coliform, surpassing the 75.6% reported. The mean coliform counts observed in our study was exceeded the findings of Thapa (2016). Considering specifications, 52% of goat meat was deemed unacceptable (>2000 cfu/g). This aligns with the results of Al-Mahmood (2020), reporting that 51.5% of retail meats from North Carolina, South Carolina, and Georgia exceeded total coliform counts of 3 log10 cfu/g. In our study, 21% of goat meat was marginally acceptable (100–2000 cfu/g), and 27% was acceptable (≤100 cfu/g). The presence of coliforms suggests potential unhygienic conditions during goat slaughter and meat portioning into different parts (Laban et al., 2021), indicating fecal contamination or unhygienic meat handling (Abuzaid et al., 2020). Studies have indicated that individuals involved in slaughtering and meat selling, lacking stringent hygienic practices, significantly contribute to coliform contamination of meat (Rani et al., 2017; Abayneh et al., 2019; Sebsibe and Asfaw, 2020). Frequent handling, unhygienic practices, meat exposure to contaminated environment, equipment, and utensils increase the microbial contamination (Augustin et al., 2020; Mallhi et al., 2019; Das et al., 2019; Alimi et al., 2022). The microbial safety of retail meat products is crucial, given that raw meats are often associated with cases of foodborne diseases (Bantawa et al., 2018). Implementing effective measures, such as providing regular hands-on meat safety training for meat handlers, is essential to prevent coliform contamination. Coliforms serve as indicators of pathogenic bacteria, making proactive training crucial in maintaining food safety standards in the meat handling process.
Our study observed a prevalence of E. coli at 31.2%, contrary to the findings of Wilson-Smith et al. (2021), who reported a higher prevalence (60%) in retail goat meat. Conversely, Kim et al. (2020) reported a lower prevalence (16.7%) compared to our study. Also, our findings align with Ajulo et al. (2020), supporting their report on contamination of goat meat with E. coli. Commensal bacteria such as E. coli inhabit the intestinal tract of food animals. During slaughter, processing, and meat handling, deficits in hygiene cause bacteria contamination of fresh meat (Ramos et al., 2020). According to Stein and Katz (2017), small ruminant meat serves as a reservoir for E. coli. Certain strains of E. coli, including E. coli O157:H7, have the potential to cause severe illnesses in both animals and humans (Yakubu et al., 2020). In the current study, the prevalence of Salmonella spp. (21.5%) exceeded the 9% contamination reported by Naik et al. (2015) in goat meat but was lower than the higher prevalence of 65% reported by Sangeetha et al. (2020). Additionally, Salmonella has been identified in 12% of goat meat samples in India (Das et al., 1990) and 3.3% of goat meat in Kathmandu, Nepal (Maharjan et al., 2006). This presents a noteworthy risk of zoonotic transmission through the consumption of contaminated goat meat. In our study, the observed prevalence of S. aureus at 47.3% aligns with Latha et al. (2017) findings of 40% but differs from Torki Baghbaderani et al. (2020) lower prevalence of 20.4% in retail goat meat. The prevalence of S. aureus (non-MRSA) in raw, unprocessed red meat has been reported in Egypt (Al-Amery et al., 2019). The growth of S. aureus and the production of enterotoxins in food are attributed to improper handling and inadequate storage conditions that support the proliferation of this pathogen (Umeda et al., 2017). Contaminated equipment can also serve as a means of transferring S. aureus to meats; this pathogen has been identified on all equipment used in abattoirs (Adugna et al., 2018). Following appropriate slaughter and food handling protocols is crucial to minimize the risk of contamination with pathogenic microorganisms. The occurrence of E. coli, Salmonella spp., and S. aureus in goat meat is concerning, as they are the leading causes of foodborne diseases. Therefore, understanding and adhering to safety measures along the entire meat supply chain is crucial to safeguard consumers’ health.
This study outlines the antimicrobial resistance profiles of E. coli spp., Salmonella spp., and S. aureus to commonly used antibiotics. The prevalence and potential for pathogenicity and antibiotic resistance gene acquisition in E. coli, Salmonella spp., and S. aureus is becoming a growing threat to human health (Výrostková et al., 2021). According to this study, E. coli isolates were significantly (p < 0.05) resistant to penicillin as compared to cephalothin, tetracycline and oxytetracycline, and doxycycline. Mwanyika et al. (2016) reported a notable level of antimicrobial resistance in E. coli isolated from goat meat. Our findings align with those of Momtaz et al. (2013), who reported similar resistance patterns in E. coli isolates from goat meat, emphasizing the need for ongoing monitoring due to E. coli’s role as an indicator for antibiotic resistance in foods (EFSA Panel on Biological Hazards (BIOHAZ) et al., 2021). Considering the potential health threat posed by antibiotic-resistant E. coli to consumers, it is imperative to implement measures aimed at reducing their occurrence in fresh goat meat (Rega et al., 2022).
Our study reveals that Salmonella spp. exhibited resistance to all antibiotics except for ceftiofur. Notably, Salmonella significantly showed higher (p < 0.05) resistance to oxytetracycline and penicillin, compared to ceftiofur and chloramphenicol. The resistance of Salmonella to oxytetracycline may be linked to its extensive use in prophylaxis and disease treatment in food animal production, including goats, since its approval by the USFDA (Mog et al., 2020). Our findings underscore the potential of goat meat as a source of antibiotic-resistant salmonella spp. S. aureus demonstrated resistance to all antibiotics evaluated in our study, with the highest resistance observed against penicillin, followed by oxytetracycline, ampicillin and cefpodoxime, significantly differing (p < 0.05) from other antibiotics. These findings are consistent with those reported by Mechesso et al. (2021), highlighting the extensive resistance of S. aureus isolates from goat carcasses to penicillin. The predominant resistance trait in goat meat isolates was displayed to penicillin, maybe attributed to its widespread use in animal production in the U.S., particularly in sheep and goat farming (Susan, 2020). The resistance of S. aureus to multiple clinically important antibiotics raises concerns about potential dissemination to humans through frequent contact with infected animals and the food chain (Chang et al., 2015). Antimicrobials are commonly used in livestock for prevention and control of diseases (Vanderhaeghen and Dewulf, 2017), as well as for sustainable production. However, it is documented that antibiotics use in food animal production is a foremost cause of the evolving AMR in humans (Manyi-Loh et al., 2018).
This study identified multidrug-resistant (MDR) bacterial isolates, defined as resistance to three or more antibiotic classes (Wang et al., 2021). The observed MAR index ranged from 0.25 to 0.75, with prevalent values at 0.58 and 0.67, indicating a high-risk source of contamination often associated with frequent antibiotic usage (Kahn et al., 2019). Bacteria with a multiple antibiotic resistance index (MARI) greater than 0.2 are indicative of a high-risk source of contamination, where the use of multiple antibiotics is to a great degree (Mthembu et al., 2019). E. coli, Salmonella spp. and S. aureus in our results was MDR. E. coli and Salmonella spp. exhibited resistance to eleven out of the twelve antimicrobials tested, raising serious concerns. Multidrug-resistant Enterobacteriaceae have been linked to higher mortality rates compared to other bacteria (Alqasim et al., 2018; Scheich et al., 2018). S. aureus displayed resistance to all twelve antibiotics, posing a significant public health concern due to its association with high mortality rates from systemic infections and food poisoning (Hong et al., 2018; Vanamala et al., 2021). The distinct drug resistance patterns identified in this study raise concerns for public health, considering the widespread use of these drugs for infection treatment and prevention in both animals and humans (Geletu et al., 2022).
This study assessed the susceptibility of isolated bacteria from goat meat to various major classes of antibiotics. Notably, a high resistance to beta-lactams (penicillin) was shown for isolated bacteria. The presence of beta-lactam-resistant bacteria in goat meat is alarming due to the potential transmission to humans through the food chain, leading to treatment failures for infections caused by these bacteria (Chishimba et al., 2016). To mitigate this risk, it is imperative to reduce the use of beta-lactam antibiotics in farm animals to curb the development and spread of resistance through the food chain (World Health Organization, 2017). Continuous surveillance of beta-lactam-resistant bacteria in meat is essential for detecting emerging patterns and guiding appropriate interventions. Additionally, the study found resistance among bacterial isolates to tetracyclines, including oxytetracycline and doxycycline. Tetracyclines are commonly employed in food-animal production, their use has been linked to the emergence of tetracycline resistance in bacteria (Roberts and Schwarz, 2016). Furthermore, bacteria isolated from goat meat displayed resistance to cephalosporins, such as cephalothin, cefpodoxime, and ceftiofur. Given the frequent use of cephalosporins in animals, their association with cephalosporin resistance in bacteria is noteworthy (Cameron-Veas et al., 2015). The lower resistance levels to ceftiofur may be attributed to differences in the mechanism of action between 3rd and 1st generation cephalosporins (Collineau et al., 2020). Moreover, neomycin, classified as an aminoglycoside, exhibited a lower resistance rate. Neomycin is commonly used to treat susceptible bacterial infections in goats and sheep. Resistance to phenicol, including chloramphenicol and florfenicol, was also noted in the study. The relatively lower resistance levels may also be explained by the infrequent use of phenicol in livestock production.
Based on the outcomes of our study, it is evident that most of the retail stores investigated exhibited inadequate practices in handling meat. None of the stores met the criteria for “good hygienic practices,” as none achieved hygienic practices score of 70% or higher. The overall level of good meat handling practices among meat handlers in our study was found to be 45.75%. However, previous studies have reported higher percentages of good meat handling practices, such as 66.4% (Yenealem et al., 2020) and 86.8% (Tegegne and Phyo, 2017). Examining specific practices, the overall frequency of glove and apron use was 60 and 40%, respectively, with no observed use of hairnets. This contrasts with findings from other studies, where hairnet usage was reported at 96.6% in Saudi Arabia (Al-Shabib et al., 2016), apron usage at 95.9% in Iran (Ansari-Lari et al., 2010), and glove usage at 89.7% in Bangladesh (Al Banna et al., 2021). Notably, a significant proportion of participants in this study did not use the same knives, cutting boards, and utensils for different types of meats. Nevertheless, all retail stores (100%) employed the same knife and cutting board for both muscle meat and offal, which is not a recommended practice. This approach poses the risk of transferring foodborne pathogens from offal meats to muscle meats. According to Abdalrahman et al. (2015), edible offal is frequently contaminated with pathogenic bacteria. In terms of personal accessories, meat handlers at eight stores (77.5%) in our study wore rings, necklaces, and watches, surpassing the 70% reported by Kanko et al. (2023). The frequency of handling money while working was 10% in our study, lower than the 45.5% reported in Ethiopia (Birhanu et al., 2017) and 87% in Kenya (Chepkemoi et al., 2015). All stores had refrigerated meat, with only three stores providing labels, sell-by dates, and consumer handling instructions. Consumer handling instructions provide guidance on storage conditions, cooking temperatures, and other essential information. This empowers consumers to handle and prepare food safely, minimizing the risk of contamination and foodborne illnesses.
Pearson’s correlation analysis revealed a significant negative correlation (coefficient: −0.8913968, p < 0.05) between hygienic practices and coliform count, suggesting that hygienic practices influenced coliform contamination in fresh goat meat. Stores with higher hygienic practices scores demonstrated lower coliform loads than those with lower scores. These findings align with Mahato’s (2019) observation that meat from stores with unhygienic meat handling practices and poor sanitary conditions exhibited high microbial counts compared to those with hygienic handling practices and good sanitation. In our study, the presence of E. coli in goat meat serves as an indicator of fecal contamination (Anihouvi et al., 2020), suggesting potential exposure to human or animal feces due to inadequate hygienic and handling practices in slaughterhouses or retail outlets. E. coli is a good indicator of fecal contamination (Anihouvi et al., 2020) and therefore, their detection in of goat meat samples suggested a contamination by human or animal feces through inappropriate hygienic and handling practices in slaughterhouses or at retail outlets. The prevalence of multidrug-resistant (MDR) E. coli, Salmonella spp., and S. aureus underscore the potential safety concerns associated with goat meat purchased from retailers, emphasizing the necessity for enhanced hygienic practices. Furthermore, this study indicates that goat meat may pose a risk as a potential source of MDR bacteria. Unhygienic meat handling has specifically been associated with meat-borne diseases (Sulleyman et al., 2018). Other studies also attribute 97% of foodborne illness outbreaks to food handler behaviors or mistakes (Tauxe, 2019; Lin and Roberts, 2020). Promoting hygienic behaviors in food handling is considered a plausible and potentially effective strategy to safeguard consumers from foodborne illnesses and deaths (Wambui et al., 2017).
In summary, this study establishes a significant correlation between hygienic practices and coliform load in retail stores selling fresh goat meat in Nashville, Tennessee. Stores with higher hygienic practice scores demonstrated lower coliform loads compared to those with lower scores. Our findings highlight the potential threat to consumer health posed by the presence of multidrug-resistant E. coli, Salmonella spp., and S. aureus in goat meat, particularly concerning the growing demand for meat products. The observed resistance in bacteria from goat meat spans major antibiotic classes, including beta-lactam, tetracycline, and cephalosporins. It is crucial to exercise caution during the slaughter process to prevent fecal contamination of goat meat, as this could introduce pathogenic bacteria, including multi-resistant strains. The results strongly advocate for the development of educational materials to enhance the food safety knowledge of goat meat handlers, addressing both meat safety and broader food safety concerns. Furthermore, it is advisable for consumers to cook meats at recommended temperatures to effectively destroy foodborne pathogens.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The studies involving humans were approved by the Tennessee State University Institutional Review Board (HS-2021-4597). The studies were conducted in accordance with the local legislation and institutional requirements. The participants provided their written informed consent to participate in this study.
Author contributions
MO: Writing – review & editing, Writing – original draft, Validation, Methodology, Investigation, Formal analysis, Data curation, Conceptualization. AK-N: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. AB: Methodology, Data analysis, Data curation, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by USDA-National Institute of Food and Agriculture Award #2021-68006-33836.
Acknowledgments
The authors acknowledge the USDA—National Institute of Food and Agriculture for funding this study. The authors are also grateful to the College of Agriculture at Tennessee State University for facilitating this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abayneh, M., Tesfaw, G., Woldemichael, K., Yohannis, M., and Abdissa, A. (2019). Assessment of extended-spectrum β-lactamase (ESBLs)–producing Escherichia coli from minced meat of cattle and swab samples and hygienic status of meat retailer shops in Jimma town, Southwest Ethiopia. BMC Infect. Diseases 19, 1–8. doi: 10.1186/s12879-019-4554-6
Abdalrahman, L. S., Wells, H., and Fakhr, M. K. (2015). Staphylococcus aureus is more prevalent in retail beef livers than in pork and other beef cuts. Pathogens 4, 182–198. doi: 10.3390/pathogens4020182
Abuzaid, K. E. A., Shaltout, F., Salem, R., and El-Diasty, E. M. (2020). Microbial aspect of some processed meat products with special reference to aflatoxins. Benha Vet. Med. J. 39, 24–28. doi: 10.21608/bvmj.2020.44886.1274
Adesokan, H. K., Akinseye, V. O., and Adesokan, G. A. (2015). Food safety training is associated with improved knowledge and behaviours among foodservice establishments’ workers. Int. J. Food Sci. 2015:328761. doi: 10.1155/2015/328761
Adjei, V. Y., Mensah, G. I., Kunadu, A. P. H., Tano-Debrah, K., Ayi, I., and Addo, K. K. (2022). Microbial safety of beef along beef value chains in the Ashaiman municipality of Ghana. Front. Vet. Sci. 9:813422. doi: 10.3389/fvets.2022.813422
Adugna, F., Pal, M., and Girmay, G. (2018). Prevalence and Antibiogram assessment of Staphylococcus aureus in beef at municipal abattoir and butcher shops in Addis Ababa, Ethiopia. Biomed. Res. Int. 2018:5017685. doi: 10.1155/2018/5017685
Åhman, J., Matuschek, E., and Kahlmeter, G. (2019). The quality of antimicrobial discs from nine manufacturers EUCAST evaluations in 2014 and 2017. Clin. Microbiol. Infect. 25, 346–352. doi: 10.1016/j.cmi.2018.05.021
Ahmed, A. M., and Shimamoto, T. (2014). Isolation and molecular characterization of Salmonella enterica, Escherichia coli O157: H7 and Shigella spp. from meat and dairy products in Egypt. Int. J. Food Microbiol. 168, 57–62. doi: 10.1016/j.ijfoodmicro.2013.10.014
Ajulo, H. O., Ajulo, M. O., and Ekereumoh, N. S. E. (2020). Isolation and identification of Salmonella and Escherichia coli from raw goat meat in Uyo Metropolis. Asian Food Sci. J. 14, 50–60. doi: 10.9734/afsj/2020/v14i430138
Akabanda, F., Hlortsi, E. H., and Owusu-Kwarteng, J. (2017). Food safety knowledge, attitudes and practices of institutional food-handlers in Ghana. BMC Public Health 17, 1–9. doi: 10.1186/s12889-016-3986-9
Al Banna, M., Disu, T. R., Kundu, S., Ahinkorah, B. O., Brazendale, K., Seidu, A. A., et al. (2021). Factors associated with food safety knowledge and practices among meat handlers in Bangladesh: a cross-sectional study. Environ. Health Prev. Med. 26, 1–12. doi: 10.1186/s12199-021-01004-5
Al-Amery, K., Elhariri, M., Elsayed, A., El-Moghazy, G., Elhelw, R., El-Mahallawy, H., et al. (2019). Vancomycin-resistant Staphylococcus aureus isolated from camel meat and slaughterhouse workers in Egypt. Antimicrob. Resist. Infect. Control 8:129. doi: 10.1186/s13756-019-0585-4
Alimi, B. A., Lawal, R., and Odetunde, O. N. (2022). Food safety and microbiological hazards associated with retail meat at butchery outlets in north-Central Nigeria. Food Control 139:109061. doi: 10.1016/j.foodcont.2022.109061
Alqasim, A., Abu Jaffal, A., and Alyousef, A. A. (2018). Prevalence of multidrug resistance and extended-spectrum β-lactamase carriage of clinical uropathogenic Escherichia coli isolates in Riyadh, Saudi Arabia. Int. J. Microbiol. 2018:1. doi: 10.1155/2018/3026851
Al-Shabib, N. A., Mosilhey, S. H., and Husain, F. M. (2016). Cross-sectional study on food safety knowledge, attitude and practices of male food handlers employed in restaurants of King Saud University, Saudi Arabia. Food Control 59, 212–217. doi: 10.1016/j.foodcont.2015.05.002
Angelidis, A. S., Komodromos, D., Giannakou, R., Arsenos, G., Gelasakis, A. I., Kyritsi, M., et al. (2020). Isolation and characterization of Staphylococcus aureus and methicillin-resistant Staphylococcus aureus (MRSA) from milk of dairy goats under low-input farm management in Greece. Vet. Microbiol. 247:108749. doi: 10.1016/j.vetmic.2020.108749
Anihouvi, D. G., Kpoclou, Y. E., Assogba, M. F., Iko Afé, O. H., Lègba, G., Scippo, M. L., et al. (2020). Microbial contamination associated with the processing of grilled pork, a ready-to-eat street food in Benin. J. Food Saf. 40:e12731. doi: 10.1111/jfs.12731
Ansari-Lari, M., Soodbakhsh, S., and Lakzadeh, L. (2010). Knowledge, attitudes and practices of workers on food hygienic practices in meat processing plants in Fars, Iran. Food Control 21, 260–263. doi: 10.1016/j.foodcont.2009.06.003
Aqib, A. I., Nighat, S., Ahmed, R., Sana, S., Jamal, M. A., Kulyar, M. F. E. A., et al. (2019). Drug susceptibility profile of Staphylococcus aureus isolated from Mastitic Milk of goats and risk factors associated with goat mastitis in Pakistan. Pak. J. Zool. 22, 1–450.
Araby, E., Nada, H. G., El-Nour, A., Salwa, A., and Hammad, A. (2020). Detection of tetracycline and streptomycin in beef tissues using charm II, isolation of relevant resistant bacteria and control their resistance by gamma radiation. BMC Microbiol. 20, 1–11. doi: 10.1186/s12866-020-01868-7
Aravani, V. P., Sun, H., Yang, Z., Liu, G., Wang, W., Anagnostopoulos, G., et al. (2022). Agricultural and livestock sector’s residues in Greece & China: comparative qualitative and quantitative characterization for assessing their potential for biogas production. Renew. Sust. Energ. Rev. 154:111821. doi: 10.1016/j.rser.2021.111821
Ariffin, S. M. Z., Hasmadi, N., Syawari, N. M., Sukiman, M. Z., Ariffin, M. F. T., Chai, M. H., et al. (2019). Prevalence and antibiotic susceptibility pattern of Staphylococcus aureus, Streptococcus agalactiae and Escherichia coli in dairy goats with clinical and subclinical mastitis. J. Anim. Health Prod. 7, 32–37. doi: 10.17582/journal.jahp/2019/7.1.32.37
Augustin, J. C., Kooh, P., Bayeux, T., Guillier, L., Meyer, T., Jourdan-Da Silva, N., et al. (2020). Contribution of foods and poor food-handling practices to the burden of foodborne infectious diseases in France. Food Secur. 9:1644. doi: 10.3390/foods9111644
Bantawa, K., Rai, K., Subba Limbu, D., and Khanal, H. (2018). Food-borne bacterial pathogens in marketed raw meat of Dharan, eastern Nepal. BMC. Res. Notes 11, 1–5. doi: 10.1186/s13104-018-3722-x
Birhanu, W., Weldegebriel, S., Bassazin, G., Mitku, F., Birku, L., and Tadesse, M. (2017). Assessment of microbiological quality and meat handling practices in butcher shops and abattoir found in Gondar town, Ethiopia. Int. J. Microbiol. Res. 8, 59–68.
Bosilevac, J. M., Gassem, M. A., Al Sheddy, I. A., Almaiman, S. A., Al-Mohizea, I. S., Alowaimer, A., et al. (2015). Prevalence of Escherichia coli O157: H7 and Salmonella in camels, cattle, goats, and sheep harvested for meat in Riyadh. J. Food Prot. 78, 89–96. doi: 10.4315/0362-028X.JFP-14-176
Cameron-Veas, K., Solà-Ginés, M., Moreno, M. A., Fraile, L., and Migura-Garcia, L. (2015). Impact of the use of β-lactam antimicrobials on the emergence of Escherichia coli isolates resistant to cephalosporins under standard pig-rearing conditions. Appl. Environ. Microbiol. 81, 1782–1787. doi: 10.1128/AEM.03916-14
Catalano, A., Iacopetta, D., Ceramella, J., Scumaci, D., Giuzio, F., Saturnino, C., et al. (2022). Multidrug resistance (MDR): a widespread phenomenon in pharmacological therapies. Molecules 27:616. doi: 10.3390/molecules27030616
Chang, Q., Wang, W., Regev-Yochay, G., Lipsitch, M., and Hanage, W. P. (2015). Antibiotics in agriculture and the risk to human health: how worried should we be? Evol. Appl. 8, 240–247. doi: 10.1111/eva.12185
Chepkemoi, S., Lamuka, P. O., Abong, G. O., and Matofari, J. (2015). Sanitation and hygiene meat handling practicesin small and medium enterprises butcheries in kenya: case study of nairobi and isiolo counties. J Food Saf. 15, 64–74.
Chishimba, K., Hang’Ombe, B. M., Muzandu, K., Mshana, S. E., Matee, M. I., Nakajima, C., et al. (2016). Detection of extended-spectrum beta-lactamase-producing Escherichia coli in marketready chickens in Zambia. Int. J. Microbiol. 2016:5275724. doi: 10.1155/2016/5275724
Chlebicz, A., and Śliżewska, K. (2018). Campylobacteriosis, salmonellosis, yersiniosis, and listeriosis as zoonotic foodborne diseases: a review. Int. J. Environ. Res. Public Health 15:863. doi: 10.3390/ijerph15050863
CLSI Clinical and Laboratory Standards Institute (2020). Performance standards for antimicrobial susceptibility testing. 30th Edn. Wayne, PA, USA: CLSI Supplement M100; CLSI.
Collineau, L., Chapman, B., Bao, X., Sivapathasundaram, B., Carson, C. A., Fazil, A., et al. (2020). A farm-to-fork quantitative risk assessment model for Salmonella Heidelberg resistant to third-generation cephalosporins in broiler chickens in Canada. Int. J. Food Microbiol. 330:108559. doi: 10.1016/j.ijfoodmicro.2020.108559
Das, A. K., Nanda, P. K., Das, A., and Biswas, S. (2019). Hazards and safety issues of meat and meat products. Food safety and human health, 145–168. Academic Press
Das, M. S., Roy, D. K., and Das, S. (1990). Occurrence of salmonellae in slaughtered pigs, goat meat, meat handlers and slaughtered-house workers. J. Commun. Dis. 22, 39–42.
Degala, H. L., Mahapatra, A. K., Demirci, A., and Kannan, G. (2018). Evaluation of non-thermal hurdle technology for ultraviolet light to inactivate Escherichia coli K12 on goat meat surfaces. Food Control 90, 113–120. doi: 10.1016/j.foodcont.2018.02.042
EFSA Panel on Biological Hazards (BIOHAZ)Koutsoumanis, K., Allende, A., Álvarez-Ordóñez, A., Bolton, D., Bover-Cid, S., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19:e06651. doi: 10.2903/j.efsa.2021.6651
Ekanem, E., Mafuyai, M., Tegegne, F., and Bhavsar, H. (2016). What is driving the demand for goat meat in Tennessee. J. Food Distrib. Res. 47, 1–2.
Feng, P., Weagant, S. D., Grant, M. A., Burkhardt, W., Shellfish, M., and Water, B. (2017). Bacteriological analytical manual (BAM), enumeration of Escherichia coli and the coliform bacteria.
Geletu, U. S., Usmael, M. A., and Ibrahim, A. M. (2022). Isolation, identification, and susceptibility profile of E. coli, Salmonella, and S. aureus in dairy farm and their public health implication in Central Ethiopia. Vet. Med. Int. 2:1887977.
Georgopoulou, M. P., Syngouna, V. I., and Chrysikopoulos, C. V. (2020). Influence of graphene oxide nanoparticles on the transport and cotransport of biocolloids in saturated porous media. Colloids Surf. B: Biointerfaces 189:110841. doi: 10.1016/j.colsurfb.2020.110841
Getaneh, T. (2019). Bacteriological quality and safety of fresh meat from some butcher houses in Feresbet town. Northern Ethiopia: Amhara Regional State.
Ghougal, K., Moreno Roldán, E., and Espigares Rodríguez, E. (2021). Risk factors related to bacterial contamination by Enterobacteriaceae and fecal coliforms and the prevalence of Salmonella spp. in Algerian farms, slaughterhouses, and butcheries: A two-year follow-up study. AIMS 6, 768–785. doi: 10.3934/agrfood.2021046
Hanlon, K. E., Miller, M. F., Guillen, L. M., Echeverry, A., Dormedy, E., Cemo, B., et al. (2018). Presence of Salmonella and Escherichia coli O157 on the hide, and presence of Salmonella, Escherichia coli O157 and Campylobacter in feces from small-ruminant (goat and lamb) samples collected in the United States, Bahamas, and Mexico. Meat Sci. 135, 1–5. doi: 10.1016/j.meatsci.2017.08.003
Hassan, M. K., Jahan, L., Sultana, P., Hasan, A., and Siddique, M. P. (2021). Detection and Antibiogram of different bacterial agents from market goat meat. Res. Agric. Livestock Fisheries 8, 135–143. doi: 10.3329/ralf.v8i1.53275
Hong, J., Kim, J., Quan, L. H., Heu, S., and Roh, E. (2018). Purification and characterization of pasteuricin produced by Staphylococcus pasteuri RSP-1 and active against multidrug-resistant Staphylococcus aureus. J. Food Prot. 81, 1768–1775. doi: 10.4315/0362-028X.JFP-18-111
Ibrahim, M., Pattanaik, N., Onyango, B., Walker, E., Bledsoe, M., and Sudbrock, C. (2020). Factors affecting goat meat demand and willingness to pay a premium price for domestically produced goat meat in the southern United States. J. Food Distrib. Res. 51, 57–61.
Igere, B. E., Okoh, A. I., and Nwodo, U. U. (2020). Antibiotic susceptibility testing (AST) reports: a basis for environmental/epidemiological surveillance and infection control amongst environmental vibrio cholerae. Int. J. Environ. Res. Public Health 17:5685. doi: 10.3390/ijerph17165685
Jaja, I. F., Bhembe, N. L., Green, E., Oguttu, J., and Muchenje, V. (2019). Molecular characterization of antibiotic-resistant Salmonella enterica isolates recovered from meat in South Africa. Acta Trop. 190, 129–136. doi: 10.1016/j.actatropica.2018.11.003
Jans, C., Sarno, E., Collineau, L., Meile, L., Stärk, K. D., and Stephan, R. (2018). Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front. Microbiol. 9:362. doi: 10.3389/fmicb.2018.00362
Kahn, L. H., Bergeron, G., Bourassa, M. W., De Vegt, B., Gill, J., Gomes, F., et al. (2019). From farm management to bacteriophage therapy: strategies to reduce antibiotic use in animal agriculture. Ann. N. Y. Acad. Sci. 1441, 31–39. doi: 10.1111/nyas.14034
Kamala, K., and Kumar, V. P. (2018). Food products and food contamination. Microbial contamination and food degradation, 1–19. Academic Press
Kanko, T., Seid, M., and Alemu, M. (2023). Evaluation of bacteriological profile of meat contact surfaces, handling practices of raw meat and its associated factors in butcher shops of Arba Minch town, southern Ethiopia-a facility based cross sectional study. Int. J. Food Contam. 10, 1–13.
Kim, S., Kim, H., Kim, Y., Kim, M., Kwak, H., and Ryu, S. (2020). Antimicrobial resistance of Escherichia Coli from retail poultry meats in Korea. J. Food Prot. 83, 1673–1678. doi: 10.4315/JFP-20-150
Laban, S. E., Mashaly, M. M., Aly, A. M., Maher, N. E., and Zaki, M. M. (2021). Evaluation of different hygienic practices applied in slaughterhouses and its effect on beef quality. Adv. Animal Vet. Sci. 9, 429–437.
Larry, M, and James, T. Peeler (2021). Bacteriological Analytical Manual (BAM), Chapter 3: Aerobic Plate Count, 2001, Available at: https://www.fda.gov/food/laboratory-methods-food/bam-chapter-3-aerobic-plate-count (Accessed May 6, 2024)
Latha, C., Anu, C. J., Ajaykumar, V. J., and Sunil, B. (2017). Prevalence of Listeria monocytogenes, Yersinia enterocolitica, Staphylococcus aureus, and Salmonella enterica typhimurium in meat and meat products using multiplex polymerase chain reaction. Veterinary world 10, 927–931. doi: 10.14202/vetworld.2017.927-931
Lekshmi, M., Ammini, P., Kumar, S., and Varela, M. F. (2017). The food production environment and the development of antimicrobial resistance in human pathogens of animal origin. Microorganisms 5:11. doi: 10.3390/microorganisms5010011
Li, S., He, Y., Mann, D. A., and Deng, X. (2021). Global spread of Salmonella enteritidis via centralized sourcing and international trade of poultry breeding stocks. Nat. Commun. 12:5109. doi: 10.1038/s41467-021-25319-7
Lin, N., and Roberts, K. R. (2020). Using the theory of planned behavior to predict food safety behavioral intention: a systematic review and meta-analysis. Int. J. Hosp. Manag. 90:102612. doi: 10.1016/j.ijhm.2020.102612
Ma, F., Xu, S., Tang, Z., Li, Z., and Zhang, L. (2021). Use of antimicrobials in food animals and impact of transmission of antimicrobial resistance on humans. Biosafety Health 3, 32–38. doi: 10.1016/j.bsheal.2020.09.004
Maharjan, M., Joshi, V., Joshi, D. D., and Manandhar, P. (2006). Prevalence of Salmonella species in various raw meat samples of a local market in Kathmandu. Ann. N. Y. Acad. Sci. 1081, 249–256. doi: 10.1196/annals.1373.031
Mahato, S. (2019). Relationship of sanitation parameters with microbial diversity and load in raw meat from the outlets of the metropolitan city Biratnagar, Nepal. Int. J. Microbiol. 2019, 1–17. doi: 10.1155/2019/3547072
Mallhi, I. Y., Sohaib, M., Khan, A. U., and Nawaz, M. (2019). Evaluating food safety knowledge, practices, and microbial profile of meat in abattoirs and butchery shops in Lahore, Pakistan. J. Food Saf. 39:e12612. doi: 10.1111/jfs.12612
Mamun, M. M., Parvej, M. S., Ahamed, S., Hassan, J., Nazir, K. H. M. N. H., Nishikawa, Y., et al. (2016). Prevalence and characterization of shigatoxigenic Escherichia coli in broiler birds in Mymensingh. Bangladesh J. Vet. Med. 14, 5–8. doi: 10.3329/bjvm.v14i1.28809
Mancuso, G., Midiri, A., Gerace, E., and Biondo, C. (2021). Bacterial antibiotic resistance: the most critical pathogens. Pathogens 10:1310. doi: 10.3390/pathogens10101310
Manyi-Loh, C., Mamphweli, S., Meyer, E., and Okoh, A. (2018). Antibiotic use in agriculture and its consequential resistance in environmental sources: potential public health implications. Molecules 23:795. doi: 10.3390/molecules23040795
Mason, W. J., Blevins, J. O. N. S., Beenken, K., Wibowo, N., Ojha, N., and Smeltzer, M. S. (2001). Multiplex PCR protocol for the diagnosis of staphylococcal infection. JCM 39, 3332–3338. doi: 10.1128/JCM.39.9.3332
Mazhangara, I. R., Chivandi, E., Mupangwa, J. F., and Muchenje, V. (2019). The potential of goat meat in the red meat industry. Sustain. For. 11:3671. doi: 10.3390/su11133671
McEwen, S. A., and Fedorka-Cray, P. J. (2017). Antimicrobial use and resistance in animals’, clinical infectious diseases: an official publication of the infectious diseases. Soc. Am. 34, S93–S106. doi: 10.1086/340246
Mechesso, A. F., Moon, D. C., Ryoo, G. S., Song, H. J., Chung, H. Y., Kim, S. U., et al. (2021). Resistance profiling and molecular characterization of Staphylococcus aureus isolated from goats in Korea. Int. J. Food Microbiol. 336:108901. doi: 10.1016/j.ijfoodmicro.2020.108901
Mog, M., Ngasotter, S., Tesia, S., Waikhom, D., Panda, P., Sharma, S., et al. (2020). Problems of antibiotic resistance associated with oxytetracycline use in aquaculture: a review. J. Entomol. Zool. Stud 8, 1075–1082.
Momtaz, H., Dehkordi, F. S., Rahimi, E., Ezadi, H., and Arab, R. (2013). Incidence of Shiga toxinproducing Escherichia coli serogroups in ruminant’s meat. Meat Sci. 95, 381–388. doi: 10.1016/j.meatsci.2013.04.051
Mthembu, T. P., Zishiri, O. T., and El Zowalaty, M. E. (2019). Molecular detection of multidrug-resistant Salmonella isolated from livestock production systems in South Africa. Infect. Drug Resist. 12, 3537–3548. doi: 10.2147/IDR.S211618
Mwanyika, G., Call, D. R., Rugumisa, B., Luanda, C., Murutu, R., Subbiah, M., et al. (2016). Load and prevalence of antimicrobial-resistant Escherichia coli from fresh goat meat in Arusha. J. Food Protect. 79, 1635–1641. doi: 10.4315/0362-028X.JFP-15-573
Naik, V. K., Shakya, S., Patyal, A., and Gade, N. E. (2015). Isolation and molecular characterization of Salmonella spp. from chevon and chicken meat collected from different districts of Chhattisgarh, India. Veterinary World 8, 702–706. doi: 10.14202/vetworld.2015.702-706
Nakai, J. (2018). Food and agriculture Organization of the United Nations and the sustainable development goals. Sustain. Dev. 22, 1–450.
Nedelkova, M., Delova, A., Petreska, T., Ivanovska, Z. Z., Petrushevska-Tozi, L., and Bull, M. P. (2019). Assessment of microbial contamination of drinking water with total coliform bacteria and Escherichia coli in the Bitola region. Macedonian Pharmaceutical Bull. 65, 23–32. doi: 10.33320/maced.pharm.bull.2019.65.02.003
Nelli, A., Voidarou, C., Venardou, B., Fotou, K., Tsinas, A., Bonos, E., et al. (2022). Antimicrobial and methicillin resistance pattern of potential mastitis-inducing Staphylococcus aureus and coagulase-negative staphylococci isolates from the mammary secretion of dairy goats. Biology 11:1591. doi: 10.3390/biology11111591
Paião, F. G., Arisitides, L. G. A., Murate, L. S., Vilas-Bôas, G. T., Vilas-Boas, L. A., and Shimokomaki, M. (2013). Detection of Salmonella spp, Salmonella Enteritidis and typhimurium in naturally infected broiler chickens by a multiplex PCR-based assay. Braz. J. Microbiol. 44, 37–42. doi: 10.1590/S1517-83822013005000002
Pathare, N. A., Asogan, H., Tejani, S., Al Mahruqi, G., Al Fakhri, S., Zafarulla, R., et al. (2016). Prevalence of methicillin resistant Staphylococcus aureus (MRSA) colonization or carriage among health-care workers. J. Infect. Public Health 9, 571–576. doi: 10.1016/j.jiph.2015.12.004
Raji, M. A., Garaween, G., Ehricht, R., Monecke, S., Shibl, A. M., and Senok, A. (2016). Genetic characterization of Staphylococcus aureus isolated from retail meat in Riyadh, Saudi Arabia. Front. Microbiol. 7:911. doi: 10.3389/fmicb.2016.00911
Ramos, S., Silva, V., Dapkevicius, M. D. L. E., Caniça, M., Tejedor-Junco, M. T., Igrejas, G., et al. (2020). Escherichia coli as commensal and pathogenic bacteria among food-producing animals: health implications of extended spectrum β-lactamase (ESBL) production. Animals 10:2239. doi: 10.3390/ani10122239
Rani, Z. T., Hugo, A., Hugo, C. J., Vimiso, P., and Muchenje, V. (2017). Effect of post-slaughter handling during distribution on microbiological quality and safety of meat in the formal and informal sectors of South Africa: a review. South Afr. J. Animal Sci. 47, 255–267. doi: 10.4314/sajas.v47i3.2
Rega, M., Andriani, L., Cavallo, S., Bonilauri, P., Bonardi, S., Conter, M., et al. (2022). Antimicrobial Resistant E. coli in pork and wild boar meat: a risk to consumers. Food Secur. 11:3662. doi: 10.3390/foods11223662
Rehman, R., and Gohar, M. (2022). Evaluation of microbial contamination in meat and its control using preservatives. Int. J. Innov. Sci. Technol. Eval 4, 404–415. doi: 10.33411/IJIST/2022040211
Riffiandi, N., Maradon, G. G., and Pertiwi, V. R., (2022), The detection of microbial contamination on goat meat from traditional market in Bandar Lampung City. IOP conference series: Earth and environmental science, 1012, 12004, IOP Publishing
Roberts, M. C., and Schwarz, S. (2016). Tetracycline and phenicol resistance genes and mechanisms: importance for agriculture, the environment, and humans. J. Environ. Qual. 45, 576–592. doi: 10.2134/jeq2015.04.0207
Robinson, E., Travanut, M., Fabre, L., Larréché, S., Ramelli, L., Pascal, L., et al. (2020). Outbreak of Salmonella Newport associated with internationally distributed raw goats’ milk cheese, France, 2018. Epidemiol. Infect. 148:e180. doi: 10.1017/S0950268820000904
Ryan, N., Gurung, N., Byrant, J., Abrahamsen, F., and McElhenney, W. (2021). Sustainable winter-feeding practices for meat goats within the southeastern United States. Professional Agric. Workers J. 7, 52–65.
Sánchez-Aldana, D., Galicia-García, T., and Leal-Ramos, M. Y. (2020). Microbiological quality and food safety challenges in the meat industry. Food Microbiol. Biotechnol., 85–102. doi: 10.1201/9780429322341-5
Sang, F. C. (2016). An evaluation of goat meat consumption in Tennessee. 01. ETD Collection for Tennessee State University. Paper AAI10243606. Available at: https://digitalscholarship.tnstate.edu/dissertations/AAI10243606
Sangeetha, A., Balakrishnan, S., Kowsalya, P., Manimaran, K., Dhanalakshmi, M., and Sivakumar, T. (2020). Microbial safety of meat sold in Orathanadu region, Thanjavur. J. Entomol. Zool. Stud. 8, 811–814.
Sarma, P. K., Alam, M. J., and Begum, I. A. (2022). Red meat handlers’ food safety knowledge, attitudes, and practices in the Dhaka megacity of Bangladesh. Int. J. Food Prop. 25, 1417–1431. doi: 10.1080/10942912.2022.2083638
Sarowska, J., Futoma-Koloch, B., Jama-Kmiecik, A., Frej-Madrzak, M., Ksiazczyk, M., BuglaPloskonska, G., et al. (2019). Virulence factors, prevalence, and potential transmission of extraintestinal pathogenic Escherichia coli isolated from different sources: recent reports. Gut pathogens 11, 1–16. doi: 10.1186/s13099-019-0290-0
Scheich, S., Weber, S., Reinheimer, C., Wichelhaus, T. A., Hogardt, M., Kempf, V. A., et al. (2018). Bloodstream infections with gram-negative organisms and the impact of multidrug resistance in patients with hematological malignancies. Ann. Hematol. 97, 2225–2234. doi: 10.1007/s00277-018-3423-5
Sebsibe, M. A., and Asfaw, E. T. (2020). Occurrence of multi-drug resistant Escherichia coli and Escherichia coli O157: H7 in meat and swab samples of various contact surfaces at abattoir and butcher shops in Jimma town, southwest district of Ethiopia. Infection and Drug Resistance 13, 3853–3862. doi: 10.2147/IDR.S277890
Seo, M., Lee, H., and Kim, Y. (2019). Relationship between coliform bacteria and water quality factors at weir stations in the Nakdong River, South Korea. Water 11:1171. doi: 10.3390/w11061171
Silva, N. D., Whittaker, W., and Chidmi, B. (2022). Potential factors influencing goat meat consumption in Oklahoma: An Econometric Approach
Some, S., Mondal, R., Mitra, D., Jain, D., Verma, D., and Das, S. (2021). Microbial pollution of water with special reference to coliform bacteria and their nexus with environment. Energy Nexus 1:100008. doi: 10.1016/j.nexus.2021.100008
Stein, R. A., and Katz, D. E. (2017). Escherichia coli, cattle, and the propagation of disease. FEMS Microbiol. Lett. 364:e50. doi: 10.1093/femsle/fnx050
Sulleyman, K. W., Adzitey, F., and Boateng, E. F. (2018). Knowledge and practices of meat safety by meat sellers in the Accra metropolis of Ghana. Int. J. Vet. Sci. 7, 167–171.
Susan, Schoenian (2020). Healthy animals produce healthy food. Antibiotic use in sheep, goat, and livestock industry. Available at: www.sheepandgoat.com/healthylivestock
Tanih, N. F., Sekwadi, E., Ndip, R. N., and Bessong, P. O. (2015). Detection of pathogenic Escherichia coli and Staphylococcus aureus from cattle and pigs slaughtered in abattoirs in Vhembe District, South Africa. Sci. World J. 2015:195972. doi: 10.1155/2015/195972
Tauxe, R. (2019). Outbreak: foodborne illness and the struggle for food safety. Emerg. Infect. Dis. 25:2342. doi: 10.3201/eid2512.191192
Tegegne, H. A., and Phyo, H. W. W. (2017). Food safety knowledge, attitude and practices of meat handler in abattoir and retail meat shops of Jigjiga town, Ethiopia. J. Prev. Med. Hyg. 58, E320–E327. doi: 10.15167/2421-4248/jpmh2017.58.4.737
Titilawo, Y., Sibanda, T., Obi, L., and Okoh, A. (2015). Multiple antibiotic resistance indexing of Escherichia coli to identify high-risk sources of faecal contamination of water. Environ. Sci. Pollut. Res. 22, 10969–10980. doi: 10.1007/s11356-014-3887-3
Torki Baghbaderani, Z., Shakerian, A., and Rahimi, E. (2020). Phenotypic and genotypic assessment of antibiotic resistance of Staphylococcus aureus bacteria isolated from retail meat. Infection and Drug Resistance 13, 1339–1349. doi: 10.2147/IDR.S241189
U.S. Department of Agriculture, Food Safety and Inspection Service . (2019). Microbiology laboratory guidebook. Available at: https://www.fsis.usda.gov/wps/portal/fsis/topics/science/laboratoriesandprocedures/guidebooks-andmethods/microbiology-laboratoryguidebook/microbiologylaboratory-guidebook (Accessed May 14, 2024).
Umeda, K., Nakamura, H., Yamamoto, K., Nishina, N., Yasufuku, K., Hirai, Y., et al. (2017). Molecular and epidemiological characterization of staphylococcal foodborne outbreak of Staphylococcus aureus harboring seg, sei, sem, sen, seo, and selu genes without production of classical enterotoxins. Int. J. Food Microbiol. 256, 30–35. doi: 10.1016/j.ijfoodmicro.2017.05.023
Vanamala, K., Tatiparti, K., Bhise, K., Sau, S., Scheetz, M. H., Rybak, M. J., et al. (2021). Novel approaches for the treatment of methicillin-resistant Staphylococcus aureus: using nanoparticles to overcome multidrug resistance. Drug Discov. Today 26, 31–43. doi: 10.1016/j.drudis.2020.10.011
Vanderhaeghen, W., and Dewulf, J. (2017). Antimicrobial use and resistance in animals and human beings. Lancet Planet. Health. 1, e307–e308. doi: 10.1016/S2542-5196(17)30142-0
Výrostková, J., Regecová, I., Dudriková, E., Marcinčák, S., Vargová, M., Kováčová, M., et al. (2021). Antimicrobial resistance of Enterococcus sp. isolated from sheep and goat cheeses. Food Secur. 10:1844. doi: 10.3390/foods10081844
Wambui, J., Karuri, E., Lamuka, P., and Matofari, J. (2017). Good hygiene practices among meat handlers in small and medium enterprise slaughterhouses in Kenya. Food Control 81, 34–39. doi: 10.1016/j.foodcont.2017.05.036
Wang, W. Y., Chiu, C. F., Lee, Y. T., Hsueh, P. R., and Tsao, S. M. (2021). Molecular epidemiology and phenotypes of invasive methicillin-resistant vancomycin-intermediate Staphylococcus aureus in Taiwan. J. Microbiol. Immunol. Infect. 55:1203. doi: 10.1016/j.jmii.2021.09.003
Wardhana, D. K. (2019). Risk factors for bacterial contamination of bovine meat during slaughter in ten Indonesian abattoirs. Vet. Med. Int. 2019:e7064. doi: 10.1155/2019/2707064
Wilson-Smith, J., Pollard, D., and Marshall, R. (2021). The prevalence of Escherichia coli O157: H7 and Salmonella in frozen and fresh goat meat. The demand for goat meat is on the rise in the US a common concern in the production and harvest of animals for human consumption is the presence of foodborne pathog. J. Stud. Res. 10:e1424. doi: 10.47611/jsr.v10i4.1424
World Health Organization (2017). Critically important antimicrobials for human medicine. World Health Organization. Available at: https://www.who.int/foodsafety/publications/antimicrobialssixth/en/
Yakubu, R. O., Lawan, M. K., Kwaga, J. K. P., and Kabir, J. (2020). Isolation, molecular detection and antimicrobial susceptibility profile of Escherichia coli O157: H7 in household-reared small ruminants in Zaria Metropolis, Kaduna state, Nigeria. Sahel J. Vet. Sci. 17, 16–23. doi: 10.54058/saheljvs.v17i4.196
Yenealem, D. G., Yallew, W. W., and Abdulmajid, S. (2020). Food safety practice and associated factors among meat handlers in Gondar town: a cross-sectional study. J. Environ. Public Health 2020:1. doi: 10.1155/2020/7421745
Keywords: antibiotic resistance, Salmonella, S. aureus , E. coli , goat meat, retail stores, hygienic practices
Citation: Owusu M, Basnet A and Kilonzo-Nthenge A (2024) Antibiotic resistant bacteria in goat meat and hygienic practices among retail stores in Nashville, Tennessee. Front. Sustain. Food Syst. 8:1460350. doi: 10.3389/fsufs.2024.1460350
Edited by:
Fatima Zahra Jawhari, Higher Institute of Nursing and Health Techniques, Fes Branch, MoroccoReviewed by:
Lizyben Chidamba, Vaal University of Technology, South AfricaHajar Belhassan, Sidi Mohamed Ben Abdellah University, Morocco
Copyright © 2024 Owusu, Basnet and Kilonzo-Nthenge. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Agnes Kilonzo-Nthenge, YWtpbG9uem9udGhlbmdAdG5zdGF0ZS5lZHU=
†ORCID: Ashesh Basnet, orcid.org/0000-0002-2920-6542
Agnes Kilonzo-Nthenge, orcid.org/0000-0002-3000-2002
 Moses Owusu
Moses Owusu Agnes Kilonzo-Nthenge
Agnes Kilonzo-Nthenge