- 1Food Science Department, College of Agriculture and Veterinary Medicine, United Arab Emirates University, Al-Ain, United Arab Emirates
- 2Analytical Biochemistry Research Center (ABrC), Universiti Sains Malaysia (USM), Penang, Malaysia
With hyperlipidemia posing a significant cardiovascular risk, innovative strategies are essential to unlock new therapeutic possibilities. Probiotic fermentation of milk proteins offers a natural and effective means to produce peptides with hypolipidemic properties, providing a promising approach to lowering lipid levels and reducing cardiovascular risk. In this study, fermented cattle milk (FCTM), fermented camel milk (FCM), fermented goat milk (FGM), and fermented sheep milk (FSM) were produced using a total of five probiotic bacterial strains to investigate the release of bioactive peptides (BAPs) with hypolipidemic potential via in vitro inhibitory activity toward pancreatic lipase (PL) during a 14-day refrigerated storage study. The PL inhibitory activities of these fermented milk (FM) varied according to the types of probiotic strains and milk types used. Overall, the Pediococcus pentosaceus MF000957 (PP-957) strain showed the highest PL inhibitory activity spanning across all milk types, and therefore, PP-957-derived fermented samples were analyzed for BAP identification by LCMS-QTOF. The identified BAPs were further analyzed using in silico and bioinformatics approaches for bioactivity prediction, molecular docking, and drug pharmacokinetic studies. Overall, four peptides derived from FCTM, one from FCM, and two peptides common in FGM and FSM were predicted as active PL inhibitors based on their binding energy and number of binding sites on the PL enzyme. All peptides were non-toxic, non-carcinogenic, and had appropriate drug-like properties. The outcomes of this study suggest that FM-derived peptides from animal milk are anticipated to be useful for combating hypercholesterolemia.
1 Introduction
The present world is contending with the escalating epidemic of obesity affecting millions of people, which, if not tackled, will have a long-term detrimental impact (Manzanarez-Quín et al., 2021). In general, obesity is strongly associated with hyperlipidemia, which acts as a risk factor for multiple health complications. Normally, dietary fats consumed in excess need to be hydrolyzed into smaller molecules by lipases in order to be absorbed. Over the past decades, Pancreatic lipase (PL) a carboxylesterase enzyme secreted by pancreatic acinar cells, has become the most important of these enzymes that play a vital digestive function for fat digestion and absorption in the small intestine (Liu et al., 2020; Kumar and Chauhan, 2021). PL is also one of the two key digestive enzymes responsible for triglyceride hydrolysis into glycerol and fatty acids, and the extreme accumulation of hydrolyzed fatty acids in the blood would lead to hyperlipidemia, hence increasing obesity occurrences. Inhibition of lipid metabolizing enzymes, specifically PL, is particularly prominent among the strategies for managing hyperlipidemia, as this could reduce dietary fat absorption and in turn the incidence of obesity (Birari and Bhutani, 2007; Liu et al., 2020; Kumar and Chauhan, 2021).
Using natural PL inhibitory compounds with no or little side effects has been encouraged because synthetic PL inhibitors have presented serious side effects (Birari and Bhutani, 2007; Mudgil et al., 2019). In response to the growing health concerns of consumers, functional foods or natural products are considered a promising approach to reducing obesity and its associated risks since they do not present any negative effects (Sridhar et al., 2019). The potential anti-hyperlipidemia influence of functional foods can act through (i) decreasing the bioavailability of nutrients or inhibition of lipase (Manzanarez-Quín et al., 2021), (ii) stimulation of energy expenditure, (iii) modulation of gut microbiota, or (iv) inhibition of adipocyte proliferation and differentiation (Trigueros et al., 2013; Mohamed et al., 2014). Thorough investigations on the anti-obesogenic effect of plant extracts, other legumes, and edible plants via PL inhibition are available. However, studies on the PL inhibitory potentials of food-derived BAPs are still in their infancy. Some reports do exist on the PL inhibitory activity of peptides from food proteins, including cow and camel casein hydrolysates (Mudgil et al., 2022), amaranth protein hydrolysates (Ajayi et al., 2021), camel whey hydrolysates (Baba et al., 2021), and brewer’s spent grain peptides (Garzón et al., 2020).
Fermented dairy products have become one of the most acclaimed raw materials for the development of functional foods due to their broad range of biologically active compounds (García-Burgos et al., 2020). BAPs with diverse activities have been generated during the fermentation of milk proteins with bacterial strains that produce physiologically active metabolites (Granato et al., 2010). Several authors have demonstrated the health-promoting benefits of FM besides their nutritional benefits, and thus they have been considered potential candidates for novel and functional foods to improve health (Khakhariya et al., 2023; Shukla et al., 2023; Pipaliya et al., 2024).
Previous studies have documented that FM contains anti-obesity, hyperlipidemia, and hypercholesterolemia properties (Cheng et al., 2015; Yoda et al., 2015; Pothuraju et al., 2016; Park et al., 2018; Tiss et al., 2020). However, in the literature, there is not enough information about the potential of using different probiotics for releasing peptides with PL inhibitory attributes. Only a few studies have demonstrated the lipase inhibitory activity of FM, which directly influences the management of obesity and increased body weight (Gil-Rodríguez and Beresford, 2019, 2020, 2021; Pipaliya et al., 2024); however, the identification of peptides that are released due to the fermentation of milk from different farm animals is still not being carried out. This research gap is crucial to be filled through the investigation of the fermentation of milk from different farm animals and their potential to release PL inhibitory peptides. To the best of our knowledge, there is no research available concerning the PL inhibitory potentials of FM sourced from different species using different probiotic species. Therefore, this present study aimed to investigate five different probiotic microorganisms, i.e., Lactiplantibacillus argentoratensis MF000943 (LA-943), Limosilactobacillus fermentum MF000944 (LF-944), Lactiplantibacillus pentosus MF000946 (LP-946), Pediococcus pentosaceus MF000957 (PP-957), and Enterococcus hirae MF000958 (EH-958), for their potential to produce FM from cattle, camel, goat, and sheep milk with high PL inhibitory activities and the impact of 14-day refrigerated storage. In the present study, the hypothesis was that proteins from the milk of individual animal species (cattle, camel, goat, and sheep) might influence their PL inhibitory properties due to their diverse structural, compositional, and genetic variability. Furthermore, different probiotic species can exert and produce a diverse range of BAPs that could demonstrate diverse biochemical properties (Ganatsios et al., 2021). The objective was to demonstrate the application potential of FM from different farm animals in the management of obesity and elucidate its relevance for the research community as well as the industry that is exploring novel functional foods.
2 Materials and methods
2.1 Chemicals, enzymes, and reagents
Enzyme PL (EC 3.4.23.1, source: porcine pancreas; L3126: 100–500 units/mg protein) was purchased from Sigma Aldrich (St. Louis, MO, United States). Chemicals and solvents of analytical grades, including o-phthaldialdehyde (OPA), p-nitrophenyl butyrate, formic acid, methanol (HPLC grade), acetonitrile (HPLC grade), β-mercaptoethanol, sodium tetra-borate, Trizma base, and SDS, were purchased from Sigma Aldrich (St. Louis, MO). The five probiotic bacterial strains, Lactiplantibacillus argentoratensis MF000943 (LA-943), Limosilactobacillus fermentum MF000944 (LF-944), Lactiplantibacillus pentosus MF000946 (LP-946), Pediococcus pentosaceus MF000957 (PP-957), and Enterococcus hirae MF000958 (EH-958), were previously isolated from raw camel milk in our laboratory (Ahmad et al., 2019). Culture media for microbial growth [De Man, Rogosa, and Sharpe (MRS)] and additional chemicals were purchased from BDH Middle East (Dubai, United Arab Emirates).
2.2 Milk samples collection
Raw cattle (Holstein Friesian), camel (Camelus dromedarius, local breed), goat, and SM used in this study were procured from local dairy farms located in the Al-Ain region of Abu Dhabi Emirate, UAE. The raw milk samples obtained were transferred to the laboratory under chilled conditions and immediately stored in the refrigerator.
2.3 Probiotic fermentation of milk samples
As explained in our previous publication (Mudgil et al., 2023), FM samples were obtained by inoculating pasteurized skim milk samples (cattle, camel, goat, and sheep) with each probiotic bacterial strain in its early stationary phase at an inoculum level of 5.0 log CFU/mL. The incubation was carried out at 37°C for 24 h under microaerophilic conditions. Thereafter, the batches of FM samples were cooled down to 15°C in ice water and then stored at 4°C for 14-day refrigerated storage with a sampling period of 0, 7, and 14 days. The FM produced from cattle, camels, goats, and sheep were referred to as FCTM, FCM, FGM, and FSM, respectively. Overall, at the end of this storage period experiment, a total of 60 FM were produced, and non-FM samples at each storage period and from each milk type were kept as controls.
2.4 Determination of degree of hydrolysis of milk proteins
The estimation of DH% of milk proteins upon fermentation by various probiotic bacteria was conducted as per the original method of Church et al. (1983) using OPA method with slight modifications as already described in our previous publication (Mudgil et al., 2023). The details of the method can be assessed in Supplementary material S1.1.
2.5 Pancreatic lipase inhibition assay
A method previously described by Baba et al. (2021) was used to analyze PL inhibitory activity. For detailed methodology, please refer to Supplementary material S1.2. The percent PL inhibition was calculated using the equation below:
Here, A is the absorbance of the control (E + Su-Sa), B is the absorbance of the control blank (only Su), C is the absorbance of the test reaction (E + Su + Sa), and D is the absorbance of the sample blank (Su + Sa), respectively. The IC50 values were calculated from the slope of the inhibition curve obtained by varying the peptide concentration of the sample.
2.6 Peptide sequencing and identification of bioactive peptides
The amino acid sequences of peptides from FM obtained through PP-957 strain fermentation were analyzed using liquid chromatography-mass spectrometry quadrupole time-of-flight (LCMS-QTOF) system (Agilent, CA, United States) according to the method of Sarah et al. (2016). The detailed methodology has been described in Supplementary material S1.3. The list of peptides was subsequently screened using the Peptide Ranker web server, available at http://distilldeep.ucd.ie/PeptideRanker/. Purposefully, average local confidence (ALC) was set above 80%, and peptides with a pepsite score above 0.80 were designated as potentially BAPs. Peptides novelty was checked against various databases such as BIOPEP-UWM,1 EROP-Moscow,2 PeptideAtlas,3 and PepBank.4 Additionally, a flexible peptide–protein docking server, HPEPDOCK 2.0, was used to calculate the docking score of selected peptides with PL enzyme (PBD ID:1ETH) (Zhou et al., 2018) and molecular interactions between each peptide and PL enzyme were explored using pepsite 2.0 web-based server available at http://pepsite2.russelllab.org/ (Trabuco et al., 2012).
2.7 Molecular docking
2.7.1 Preparation of peptides and enzyme structures
Triacylglycerol lipase, a human PL protein (PBD ID:1ETH), was downloaded in its three-dimensional (3D) form from the RCSB database.5 Any organic matter, attached ligands, and removal of water molecules from enzyme structure were performed by PyMOL software.6 De novo peptide structures were generated using PEPFOLD 3 software7 (Lamiable et al., 2016). The peptide structures were then optimized for geometrical stability as per the methodology described by Fadimu et al. (2023).
2.7.2 Peptide docking
The molecular interaction between selected FM-derived peptides and 1ETH was then generated using biomolecular interaction web portal high ambiguity-driven protein–protein docking (HADDOCK 2.4) (Honorato et al., 2021), an information-driven flexible docking approach freely available at https://wenmr.science.uu.nl/haddock2.4/. Upon docking, the binding affinity as kcal/mol, molecular interaction, and 2-D visualization were generated to understand peptide structure and activity relationship with lipase enzyme.
2.8 In silico analysis of selected peptides
An in silico evaluation of the various physicochemical properties such as molecular weight, charge, isoelectric point (Ip), and toxicity was predicted by the online web server ToxinPred, available at http://crdd.osdd.net/raghava/toxinpred/ (Gupta et al., 2013). Solubility predictions were made using http://pepcalc.com/ppc.php, carcinogenicity, and other drug-like properties such as parameters associated with absorption, distribution, metabolism, and excretion (ADME) were carried out using online tools ADMETlab 2.0 and ADMETlab 3.0 available at https://admetmesh.scbdd.com/ and https://admetlab3.scbdd.com/, respectively (Xiong et al., 2021; Fu et al., 2024). Furthermore, selected peptides underwent in silico simulated gastrointestinal digestion using enzyme activity for pepsin, trypsin, and chymotrypsin embedded inside BIOPEP-UWM: https://biochemia.uwm.edu.pl/biopep-uwm/ (Minkiewicz et al., 2019) to predict their stability to gastrointestinal digestion.
2.9 Statistical analysis
All FM prepared from five probiotic strains were produced in three batches, representing triplicates. Mean significant differences were calculated using multiple comparison test/two-way ANOVA, and mean significant letters were determined using the compact letter display feature that displays the results of multiple pairwise comparisons embedded within GraphPad prism 10.2.3 (Boston, MA, United States) at p < 0.05.
3 Results and discussion
3.1 Degree of hydrolysis
The DH of FM produced using different probiotic strains was investigated and presented in our previous publications (Mudgil et al., 2023, 2024). In summary, the DH and proteolytic activity of various probiotic bacterial strains in FM were investigated, demonstrating significant variations among strains and milk types (p < 0.05). Higher proteolytic activity was observed in FM compared to non-FM, attributed to the strains’ ability to effectively hydrolyze milk proteins and release small peptides (Pihlanto et al., 2010). The detailed discussion of these results can be assessed in our previous publications (Mudgil et al., 2023, 2024). In brief, the key findings summary indicated that over the storage period, increased proteolysis was observed, with its peak at 7 and 14 days, regardless of milk type or probiotic strains. Furthermore, among FCTM, LP-946 showed the highest activity after 14 days. In FCM, LF-944 was the most active. EH-958 and LA-943 exhibited higher proteolysis in FGM and FSM, respectively, suggesting substrate specificity in proteolytic behavior. Moreover, a comparative analysis between single strain activity across different milk types showed significant variability with more active hydrolysis of goat milk (GM) proteins. Overall, strain LA-943 was particularly active in GM and SM protein hydrolysis. Similarly, LF-944 also showed a preference for GM proteins over cattle milk (CTM) and SM proteins. In general, all probiotic strains remained active in hydrolyzing proteins throughout the 14 days of refrigerated storage. Interestingly, upon storage, FCTM exhibited higher proteolytic activity compared to FGM, FCM, and FSM. The reported findings are supported by the study of other researchers (Soleymanzadeh et al., 2016; Patel and Hati, 2018) indicating variability in proteolytic activity among lactic acid bacteria due to factors such as enzymatic conditions, cell envelope proteins, and genetic factors. Overall, LP-946 and EH-958 were the most effective strains in protein hydrolysis, particularly in FCTM and FGM, respectively. These insights are crucial for selecting specific probiotic strains for targeted milk fermentation processes.
3.2 Determination of pancreatic lipase inhibitory activities
In the current investigation, we assessed the inhibitory potencies of FM on PL enzymes associated with hyperlipidemia. The PL inhibitory activity of FM obtained using probiotic strains is reported as IC50 (protein concentration required to inhibit 50% of the original PL activity) in Figure 1. The IC50 inhibitory values demonstrating the FM potencies as PL inhibitors (i.e., lower IC50 value infers more potency at inhibiting PL) varied among the strains and milk types. The PL inhibitory activity of non-FM in this study for cattle, camel, goat, and sheep milk was 195.91, 114.82, 106.22, and 119.35 μg/mL, respectively. According to statistical results, the IC50 value for non-FM was higher in comparison to the values for FM inoculated with different probiotic strains for all four milk sources at 0, 7, and 14 days of fermentation. Consequently, increased PL inhibitory potency was recorded after the inoculation of probiotic strains, reaching an average of 5-fold increase for FCTM and FCM and a 2-fold increase for FGM and FSM, respectively, at the same fermentation period. This result confirmed the inhibitory potentials of FM against the PL enzymatic marker. The observations from this study were confirmed by a previous study, which already demonstrated that milk fermentation with Lactobacillus species maximized their potential to inhibit PL (Gil-Rodríguez and Beresford, 2019).
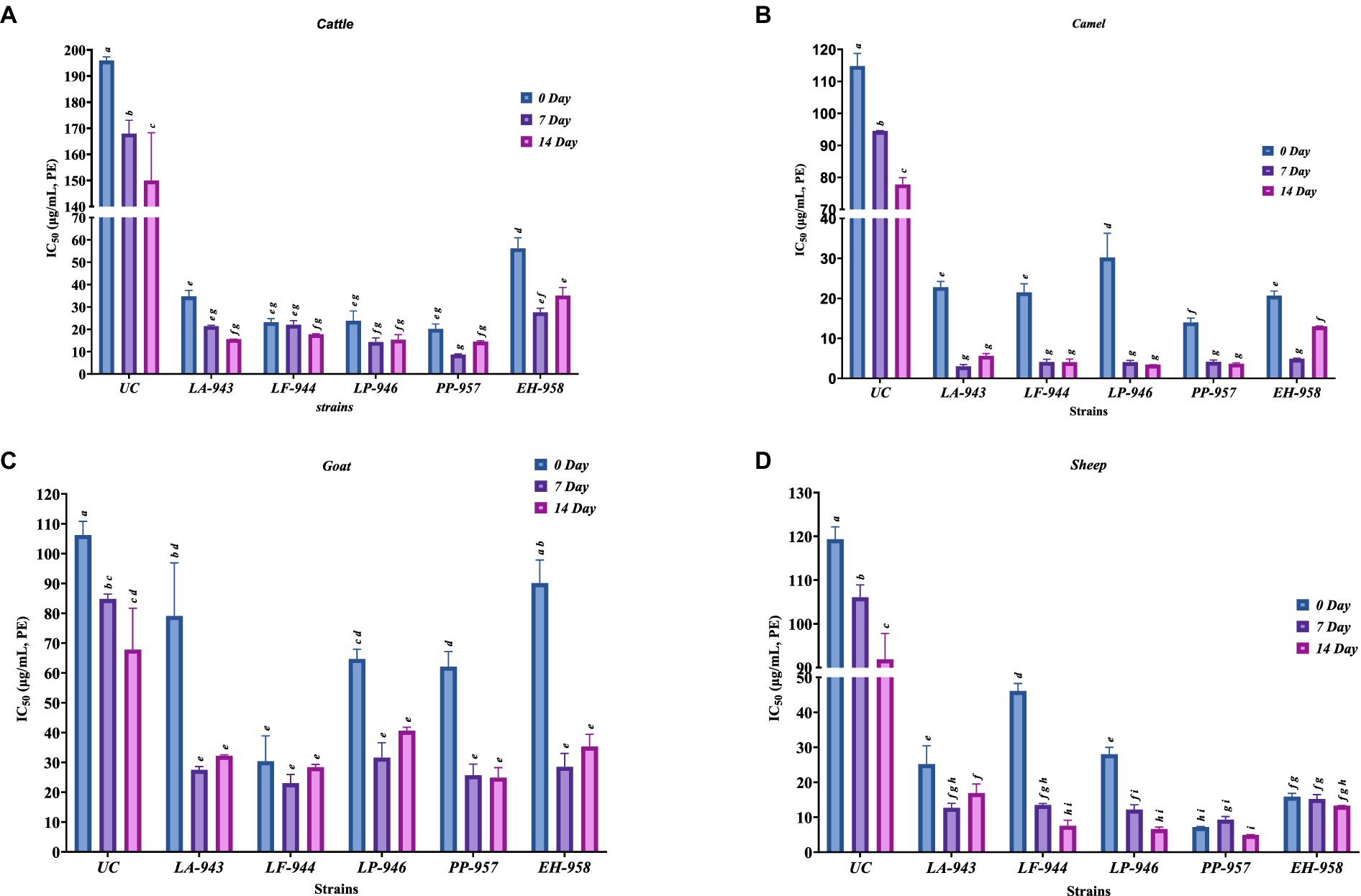
Figure 1. Pancreatic lipase IC50 inhibitory values (μg/mL) of fermented milk derived from cattle (A), camel (B), goat (C), and sheep (D) milk following fermentation with five distinct probiotic microorganisms during refrigerated storage for 14 days. Data represent mean ± SD. Different small alphabets on each bar represent significant differences between the samples. UC, Non-fermented milk; LA-943, Lactiplantibacillus argentoratensis MF000943; LF-944, Limosilactobacillus fermentum MF000944; LP-946, Lactiplantibacillus pentosus MF000946; PP-957, Pediococcus pentosaceus MF000957; and EH-958, Enterococcus hirae MF000958.
According to Figure 1, all the strains used in this study significantly decreased the IC50 value of FM, which varied at different fermentation periods of 0, 7, and 14 days. It is interesting to note that all the probiotic strains demonstrated high PL inhibitory activities at 0 days. These results suggest that the fermentation in the pre-culture has the potential to produce PL inhibitory peptides, as already demonstrated by Gonzalez-Gonzalez et al. (2011), after ACE-inhibition activity was performed on probiotic FM at 24 h. In FCTM, the PP-957 strain showed the most potent PL inhibitory activity (IC50: 20.2 μg/mL); however, no significant difference was observed in the IC50 values for samples fermented with LF-944 and LP-946 strains (p < 0.05), and the least potent strain was found to be EH-958 (IC50: 56.2 μg/mL). Similarly, CM and SM fermented by PP-957 strain showed significantly higher PL inhibitory activity with lower IC50 values of 14.2 and 7.21 μg/mL, respectively. Moreover, it is interesting to note that most of the milk fermented with the PP-957 strain developed the most potent PL inhibitory activity, except for FGM.
A further progression of the milk fermentation period to 7 days resulted in an imperative increase in PL inhibition (p < 0.05). As the fermentation duration increased, the PL inhibitory IC50 values of non-fermented and FM decreased, thus exhibiting higher PL inhibitory activity. The highest PL inhibitory activity was obtained in cattle milk fermented with PP-957 strain (IC50: 8.70 μg/mL at 7 days). On the other hand, camel milk fermented by the LA-943 strain produced the highest PL inhibitory activity (IC50: 3.01 μg/mL). The IC50 value for all GM fractions fermented with LA-943, LF-944, LP-946, and PP-957 strains showed no significant difference in the PL inhibitory activities (p > 0.05). Unexpectedly, FSM fermented by PP-957 strain also showed high PL inhibitory activity (IC50: 9.33 μg/mL). Moreover, the EH-958 probiotic strain produced FM with the least potent PL inhibitory activity among all the probiotic strains after 7 days of fermentation.
Furthermore, after 14 days of fermentation, the PL inhibitory activity of FM showed varied activities. The PP-957 strain displayed a similar tendency of higher PL inhibitory activity in all the FM when compared to other strains such as LF-944 and LP-946, while the FM inoculated with EH-958 exhibited the least potent PL inhibitory activities. It was clear that the PL inhibitory activity recorded among the different FM depends on the probiotic strain type used. From this study, all the probiotic strains exhibited a different ability to produce PL inhibitions during fermentation. This observation was confirmed by previous studies of Gil-Rodríguez and Beresford (2019) and Kinariwala et al. (2020), who reported that the PL inhibitory activity of FM had a greater dependence on the strain of starter cultures. Gonzalez-Gonzalez et al. (2011) reported that strain-dependent potentials might be due to different proteases and peptidase encryption in different bacteria displaying greater specificity for those peptide sequences in FM. Overall, the different proteolytic systems existing in specific bacteria influence their enzyme inhibitory activity.
Although some probiotic strains (e.g., LA-943 and LF-944) showed higher levels of PL inhibition during the fermentation periods, FM produced by PP-957 showed higher potencies to produce peptides from different milk proteins that could strongly inhibit PL. This could also indicate that this probiotic strain possesses a more potent proteolytic system than that of the other strains used in this study. Interestingly, P. pentosaceus strains have been previously reported to influence cholesterol metabolism (Jiang et al., 2021). Other studies have also reported the strong cholesterol-lowering ability of P. pentosaceus in both in vitro and in vivo experiments compared to other LABs (Damodharan et al., 2015; Lim et al., 2019). Taken together, FM produced by the PP-957 probiotic strain were confirmed to demonstrate more effective PL inhibitory properties, suggesting its superior anti-obesity potential.
Furthermore, here we reported for the first time that the source or nature of milk may also influence the PL inhibitory activity of FM despite being inoculated with the same probiotic strain. As shown in Figure 1, notable significant differences (p < 0.05) were observed in the PL inhibitory activity of FM sourced from different farm animals (cattle vs. camel vs. goat vs. SM). FGM exhibited much lower PL inhibitory capacities at 0, 7, and 14 days of fermentation, as indicated by high IC50 values. Contrarily, FGM using L. fermentum at 0 days showed similar PL inhibitory activity with FCTM and FCM counterparts (p > 0.05). Higher PL inhibitory activity was shown in FCM among all the FM samples throughout the fermentation periods. In line with these results, Elayan et al. (2010) have earlier reported the hypocholesterolemic validity of FCM in diet-induced hypercholesterolemic rats. Moreover, all FSM presented superior PL inhibitory activities than their FCTM counterpart. Therefore, according to these observations, inhibition of PL enzyme is not only governed by the strain type but also by the nature or composition of milk. The outcomes strongly suggested that both strains and milk sources are significant factors governing the PL inhibitory activities of peptides derived from FM. Overall, FCM was found to demonstrate higher PL inhibitory activity, irrespective of the type of probiotic strain used.
3.3 Characterization and identification of bioactive peptides with anti-hyperlipidemic activity from milk types
The FM (FCTM, FCM, FGM, and FSM) obtained after fermentation with PP-957 were subjected to LCMS-QTOF analysis for the characterization of peptide sequences. The interaction of the identified peptide sequences with the PL (1ETH) is presented based on their p value, potential binding sites, and docking scores, as shown in Table 1.
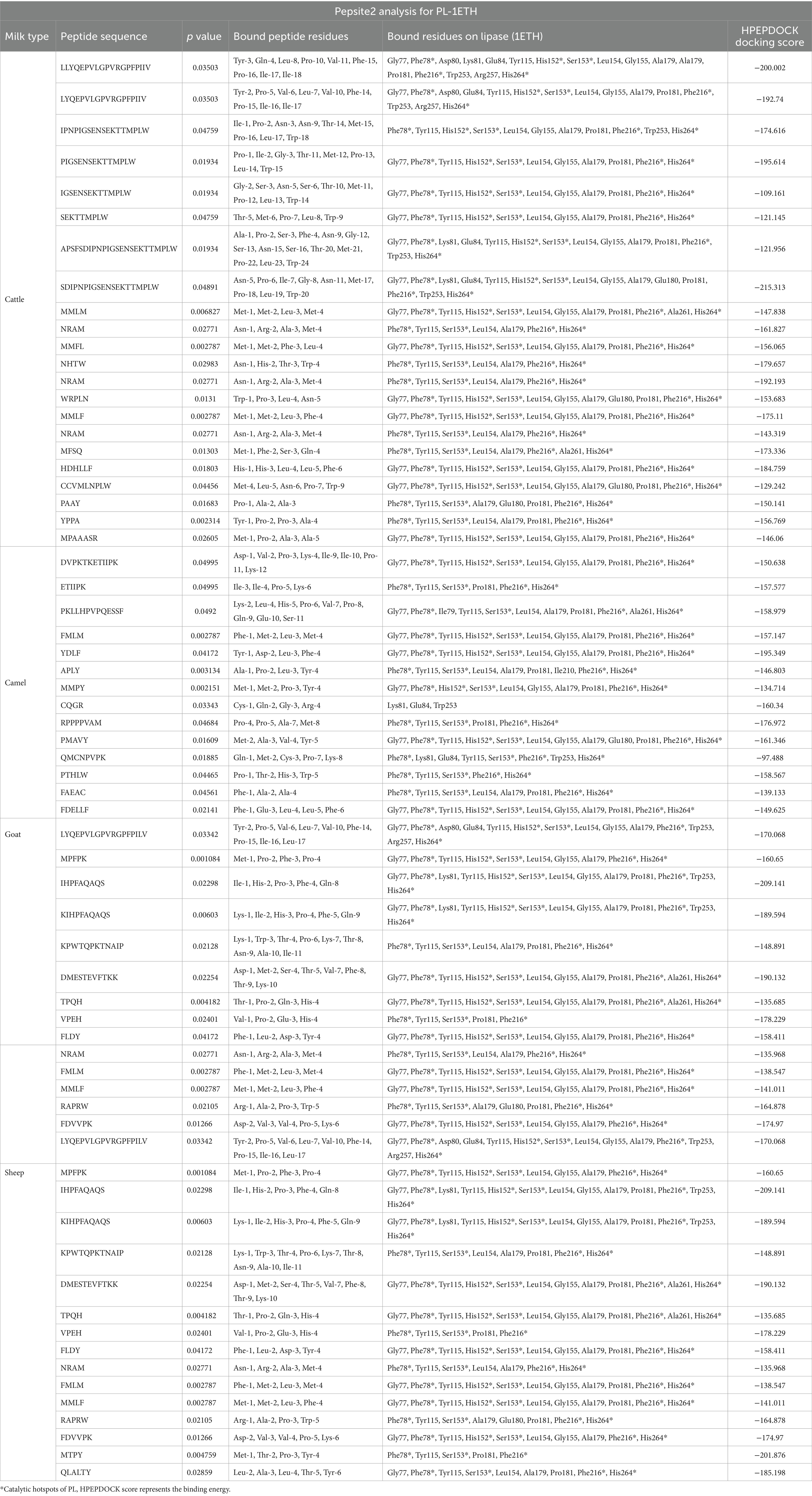
Table 1. Bioactive peptides derived from cattle, camel, goat, and sheep milk proteins and their interaction with pancreatic lipase (PL) (1ETH) as elucidated by pepsite 2.0.
According to the Peptide Ranker score (>0.80), a total of 47, 37, 44, and 45 peptides were identified as BAPs in PP-957-derived FCTM, FCM, FGM, and FSM, respectively (Supplementary Tables S1–S4). Among these, only 22 FCTM peptides, 14 FCM peptides, 16 FGM peptides, and 10 FSM peptides were identified as potent PL inhibitors with a significance level of <0.05, as presented in Table 1. It is well-known that PL plays a part in the hydrolysis and breakdown of dietary fats, thus yielding fatty acids as end products. The activation of PL occurs at the sites where residues Cys238 and Cys262 with the ability to form surface loops (lid) are removed, hence blocking substrate accessibility to active binding sites (Haque and Prabhu, 2016; Chia et al., 2023). PL has a catalytic triad consisting of six major amino acid residues Ser 153, Asp 177, and His 264, as part of the catalytic triad in the active site, and Phe78, His152, and Phe216 as part of the oxyanion hole that holds the substrate within the active site for hydrolysis. Therefore, binding of peptides with these six amino acids present in catalytic triad or oxyanion hole is of particular interest, as binding with any of these amino acids on PL enzyme could destabilize the transition state intermediates, causing inhibition of the enzyme’s hydrolytic action and eventually limiting the hydrolysis and absorption of lipids (Chia et al., 2023).
From the results obtained, all the peptides from all milk types were predicted to be potent PL inhibitors as they could bind to the important above-mentioned residues on the active site of PL. Regarding FCTM, all the identified peptides were found to bind strongly onto 4–5 potential residues of the PL enzyme catalytic triad (i.e., Phe78, His152, Ser153, Phe216, and His264), with docking scores ranging from −109.161 to −215.313 (Table 1). Interestingly, long peptides consisting of 9–24 amino acids (LLYQEPVLGPVRGPFPIIV, LYQEPVLGPVRGPFPIIV, IPNPIGSENSEKTTMPLW, PIGSENSEKTTMPLW, IGSENSEKTTMPLW, SEKTTMPLW, APSFSDIPNPIGSENSEKTTMPLW, and SDIPNPIGSENSEKTTMPLW) in FCTM appeared to be stronger PL inhibitors than short peptides as they significantly (p < 0.05) bind all five of the above-mentioned PL enzyme active sites. On the other hand, tetra-peptides (NRAM, NHTW, MMLF, MFSQ, PAAY, and YPPA) bind to four of these PL residues. Moreover, among the identified peptides LLYQEPVLGPVRGPFPIIV, PIGSENSEKTTMPLW, SDIPNPIGSENSEKTTMPLW, and NRAM were predicted to be the most potent, as they exhibited maximum docking energy with −200.002, −195.614, −215.313, and −192.193, respectively, against PL.
All the significant BAPs (p < 0.05) identified in FCM appear as potential PL inhibitors as they could bind 4–5 of these hot spots, except for peptide CQGR, which could not bind to any hot spot residues of PL (Table 1). Among FCM-derived peptides, YDLF was predicted to be a very potent PL inhibitor having the ability to bind onto five significant binding sites (i.e., His264, Phe78, and Ser 153, Asp 177, His152, and Phe216), as represented by a maximum docking score of −195.349. Baba et al. (2021) have also previously reported YDLF from camel whey protein hydrolysates as an important lipase-inhibiting peptide. Similarly, other peptides such as DVPKTKETIIPK, FMLM, MMPY, PMAVY, and FDELLF were also predicted to be potential PL inhibitors as they showed the ability to bind to five active site residues on PL. Concerning peptides derived from FGM, most of the BAPs identified were predicted to successfully bind to five active sites residues of PL. Five peptides from FGM (KPWTQPKTNAIP, NRAM, RAPRW, FDVVPK, and QLALTY) were able to bind to four active sites, and only two peptides (VPEH and MTPY) could bind three of the amino acid residues in the catalytic triad (Table 1). However, we presumed that peptides IHPFAQAQS and DMESTEVFTKK were qualified as the most effective lipase inhibitors due to their high docking scores of −209.141 and −190.132, respectively, from FGM and their ability to bind to five important PL active sites.
Moreover, it was found that seven peptides (LYQEPVLGPVRGPFPILV, IHPFAQAQS, MSQF, FMPY, FMLF, MMLM, and MMLF) generated from FSM could potentially bind to five hot spot residues on the active site of PL. Nonetheless, other peptides such as KTLVPQ, YTVAFE, and KASW were able to bind four and three of the reported sites, respectively, hence, considered potent lipase inhibitors (Table 1). In this study, peptides LYQEPVLGPVRGPFPILV and IHPFAQAQS, which were previously sequenced in FGM, were also identified in FSM, exhibiting similar binding patterns to the lipolytic sites of PL. Notably, the peptide HPFAQAQS also showed the highest docking scores in both FGM and FSM of −209.141 and −241.724, respectively, indicating satisfactory PL inhibitory activity. In addition, most of the peptides were also found to interact with bound residues of PL (i.e., Ala179, Pro181, Ile210, Ala261, and Try115) apart from binding with the characteristic catalytic triad, suggesting possible stearic inhibition of lipase enzyme (Mudgil et al., 2022). The above-discussed binding tendencies of potent peptides derived from different milk types to PL active sites were also previously reported by Mudgil et al. (2019, 2022) where they reported binding of PL-inhibiting peptides to various active hot spots on PL from enzyme-hydrolyzed cow and camel milk. It was also interesting to observe that despite higher inhibition (lower IC50 values) of PL by FCM, only one peptide (YDLF) met the necessary activity threshold for docking, and this deviation could be attributed to several factors such as synergistic effects, sequence specificity, or peptide length (Shao et al., 2023).
It is thought that the hydrophobicity and differences in the composition of protein’s primary structure could influence the variation in PL inhibition demonstrated by BAPs (Urbizo-Reyes et al., 2022). The majority of the selected peptides identified in all the milk types had an exceptionally high content of hydrophobic amino acids, such as proline, leucine, isoleucine, and phenylalanine, which might explain their higher PL inhibitory activity. In fact, the inhibition of lipase by hydrophobic amino acids could be seen in their capacity to bind easily to active sites of lipophilic enzymes. These results were similar to those reported by Baba et al. (2021), who reported the presence of hydrophobic amino acid residues, i.e., leucine and proline, in most of the lipase-inhibiting peptides from camel whey protein hydrolysates. More importantly, the presence of other hydrophobic amino acids such as Met (M) and Tyr (Y), whereas hydrophilic amino acids such as Ser (S) and His (H), is a common feature among lipid-lowering peptides that bind to the active site of lipase enzyme via electrostatic interactions and hydrogen bonding (Martinez-Villaluenga et al., 2010). Therefore, we assumed that both hydrophobic and hydrophilic amino acids present in the isolated peptides contributed to PL enzyme inhibition. Ajayi et al. (2021) reported peptides with excellent PL inhibitory activity from amaranth protein, which contained hydrophilic amino acid residues. Overall, we found out that the identified peptides exhibited higher binding ability to important catalytic residues and, thus, can be useful for combating hyperlipidemia.
3.4 Molecular docking analysis
The peptides with relatively higher HPEPDOCK docking scores i.e. LLYQEPVLGPVRGPFPIIV, LYQEPVLGPVRGPFPIIV, PIGSENSEKTTMPLW, and SDIPNPIGSENSEKTTMPLW from FCTM, YDLF from FCM, and IHPFAQAQS and DMESTEVFTKK from FGM and FSM, were screened and analyzed Using molecular docking techniques. On the whole, the weakest binding energy of −6.9 kcal/mol was observed for YDLF from FCM and the strongest binding energy of −11.4 kcal/mol for SDIPNPIGSENSEKTTMPLW from FCTM (Table 2). Peptides DMESTEVFTKK from FGM and FSM showed a binding energy of −10.6 kcal/mol, while another peptide from FCTM, i.e., PIGSENSEKTTMPLW, showed a binding energy of −10.3 kcal/mol, indicating strong binding force and, in turn, a robust binding effect. As indicated by Chen et al. (2024), binding energy lower than −7 kcal/mol suggests strong binding affinities. Hence, all these peptides suggest a stronger affinity of binding with PL enzyme, which in turn predict stronger inhibitions. These results indicated that the peptides have strong contacts with the receptor enzyme 1ETH. The visualization of binding poses in their 2-D form is presented in Figure 2, and their molecular interactions are presented in Table 2.
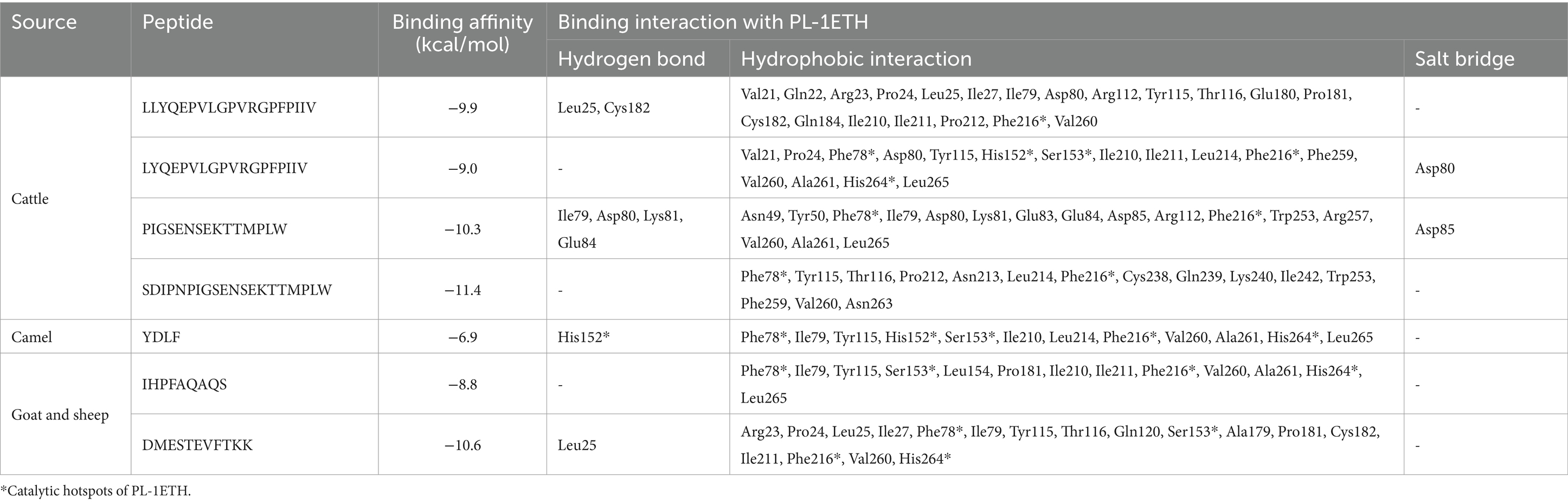
Table 2. Projected binding energies and molecular interactions as obtained by molecular docking of selected fermentation-derived peptides with PL-1ETH.
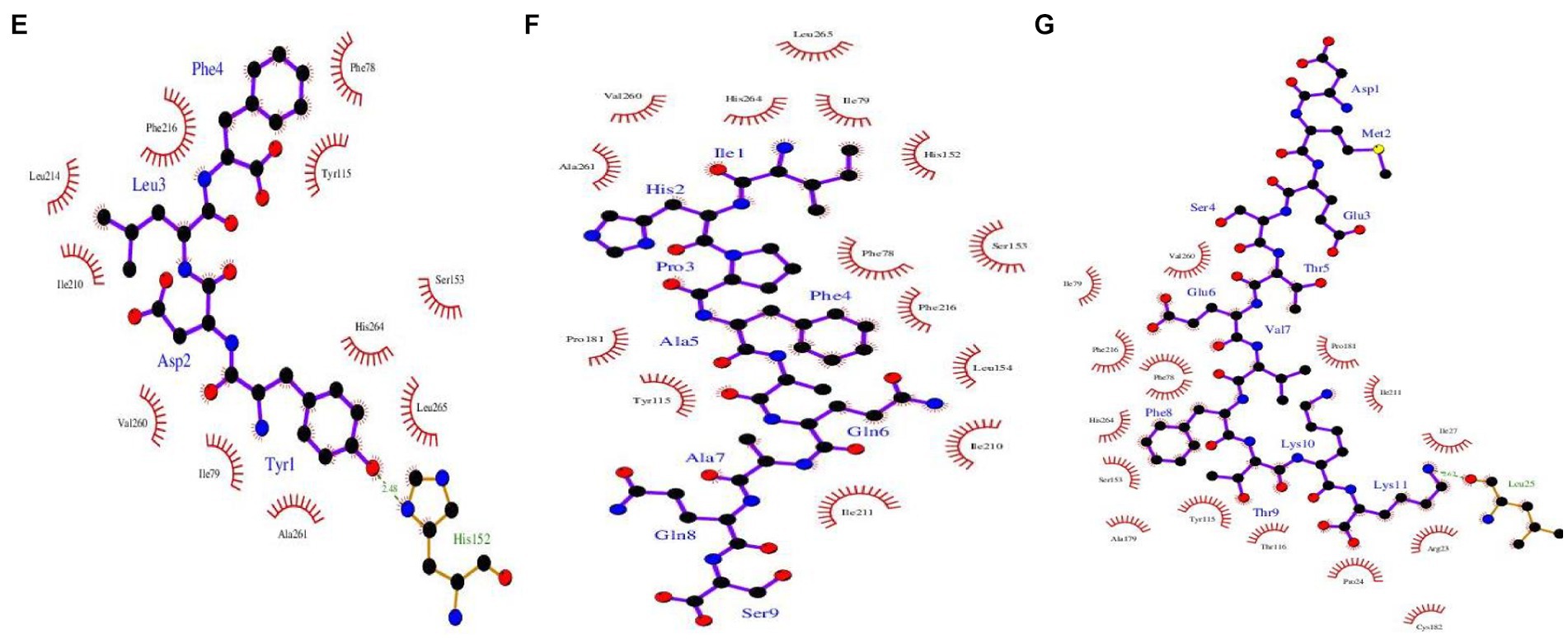
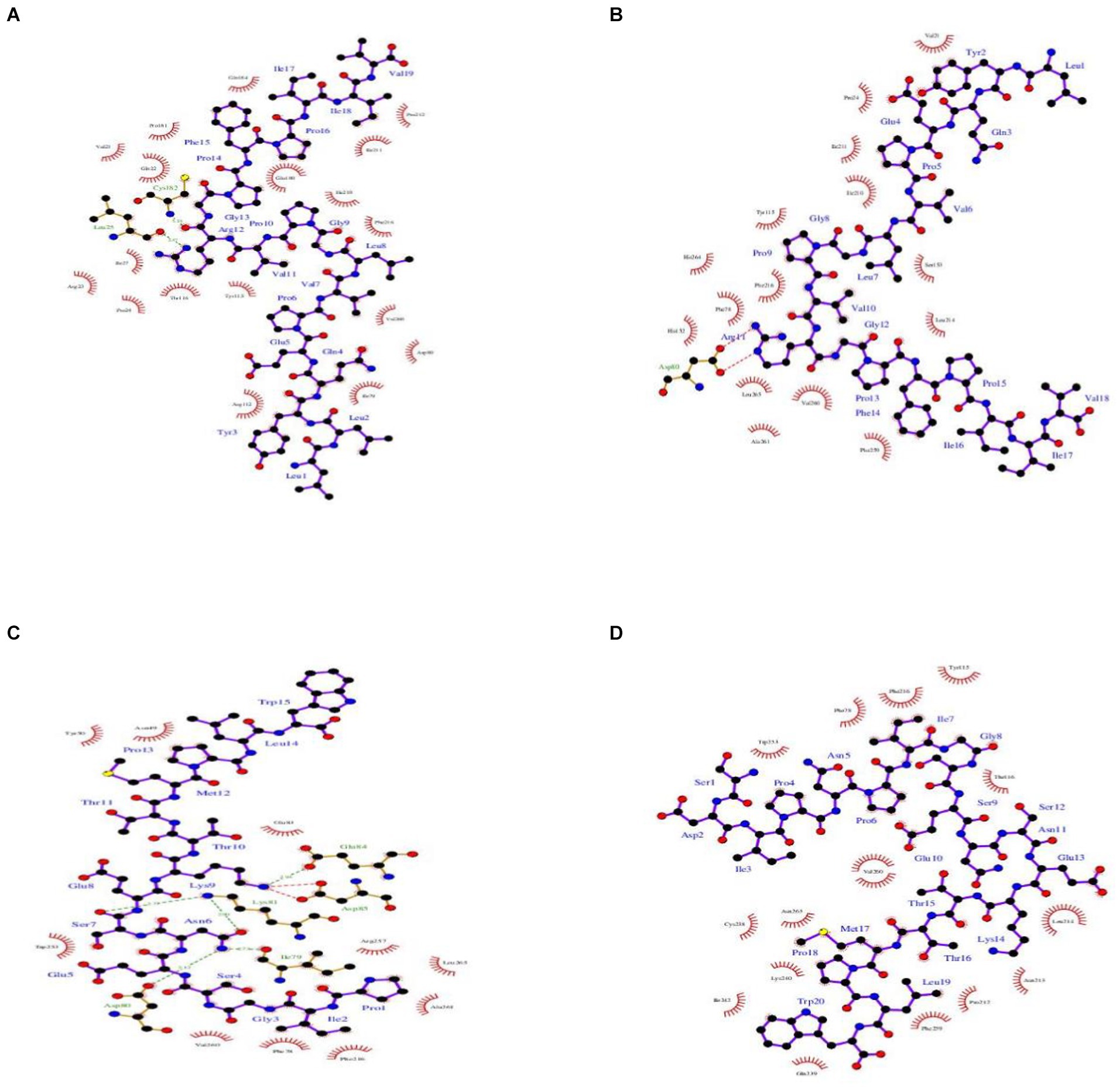
Figure 2. Binding poses and interactions between PL-1ETH with (A) LLYQEPVLGPVRGPFPIIV, (B) LYQEPVLGPVRGPFPIIV, (C) PIGSENSEKTTMPLW, (D) SDIPNPIGSENSEKTTMPLW, (E) YDLF, (F) IHPFAQAQS, and (G) DMESTEVFTKK. Peptide residues are joined through purple lines, whereas enzyme residues are joined through brown lines. The red, blue, black, and yellow dots indicate oxygen, nitrogen, carbon, and sulfur atoms, respectively. Green dotted line indicates a hydrogen bond, whereas the red dotted line indicates a salt bridge. Brick red eyelashes indicate hydrophobic interaction.
PIGSENSEKTTMPLW formed four hydrogen bonds with the amino acid residues Ile79, Asp80, Lys81, and Glu84 and a salt bridge with amino acid Asp85, whereas, hydrophobic interactions with 13 amino acids of 1ETH protein molecule were demonstrated. Peptides, YDLF showed hydrogen bonding with His152, a part of the oxyanion hole at the active site of PL. Whereas LLYQEPVLGPVRGPFPIIV showed two hydrogen bonds with Leu25 and Cys182, and peptide DMESTEVFTKK showed hydrogen bonding with Leu25. Another salt bridge was observed with Asp80 by LYQEPVLGPVRGPFPIIV peptide. Overall, it was observed that FCM-derived peptide YDLF and FCTM-derived peptide LYQEPVLGPVRGPFPIIV could bind to five of the six important active site residues (Ser 153, His 264, Phe78, His152, and Phe216). Similarly, FGM- and FSM-derived peptides IHPFAQAQS and DMESTEVFTKK could bind to four of these active site residues, indicating that these two peptides strongly interacted with the active cavity of PL, confirming their high hypolipidemic potential. Previous studies have also identified YDLF as a promising anti-obesity peptide; however, other peptides identified in this study have not been reported earlier for their PL inhibitory actions (Baba et al., 2021). Therefore, it can be suggested that probiotic fermentation can be used as an effective strategy for the production of novel anti-obesity peptides with stronger hypolipidemic potential.
3.5 In silico analysis of selected peptides
Absorption, distribution, metabolism, and excretion of various compounds, including BAPs, with the likelihood of being used as a nutraceutical and therapeutic agent have been very useful for the prediction of their drug-like characteristics. ADMETlab 2.0 and 3.0 are online prediction tools for pharmacokinetics assessment, including their medicinal chemistry (Xiong et al., 2021; Fu et al., 2024). These tools are an alternative to laborious and expensive experimental methods, along with being cost-effective (Ngo et al., 2023). Various physicochemical characteristics such as molecular weight (MWt); Ip, the logarithm of aqueous solubility value (LogS), the logarithm of the n-octanol/water distribution coefficient [Log P (Crippen method)], the number of hydrogen bond acceptors (HBAs), the number of hydrogen bond donors (HBDs), topological polar surface area (TPSA), human intestinal absorption (HIA), blood–brain barrier (BBB), metabolism as P450 CYP3A4 inhibitor (MB), clearance rate as a measure of excretion, mL/min/kg (CL), drug likeliness (Pfizer rule), toxicity, and carcinogenicity analysis were assessed, and the results are presented in Table 3. As seen in Table 3, all seven peptides that were selected qualified based on their drug likeliness as per the Pfizer rule. Solubility analysis as revealed by Log S and Log P indicated that four peptides, i.e., PIGSENSEKTTMPLW, SDIPNPIGSENSEKTTMPLW from FCTM; YDLF from FCM and DMESTEVFTKK from FGM and FSM, respectively, showed good solubility. Furthermore, as can be seen from LogS (water solubility) values, the values were well within the optimal range of −4 to 0.5 log mol/L described by the server as optimal. However, only two peptides, i.e., LLYQEPVLGPVRGPFPIIV (2.284), SDIPNPIGSENSEKTTMPLW (0.251), and DMESTEVFTKK (2.284), showed an optimal range of LogP values between 0 and 3 log mol/L. TPSA values were found to be higher than the optimal value of 0–140, based on the Veber rule. Similar to the results obtained by Wang et al. (2023), HIA and BBB values in all peptides indicated excellent values between 0 and 0.3. HIA rate is known to significantly influence the bioavailability of bioactive compounds across the intestine; similarly, BBB is crucial for maintaining the stability of the compound in the brain’s environment. Both systems permit the passage of selective nutrients and other macromolecules required while blocking harmful substances and eliminating metabolic waste. The possession of higher or recommended values for HIA and BBB indicates superior bioavailability of these peptides and is consistent with those obtained by Wang et al. (2023). Furthermore, based on the drug-like soft rule, all peptides except YDLF showed more than the optimal number of HBA and HBD. The bioavailability radar for these seven peptides, along with commercial drug orlistat, is shown in Supplementary Figure S1.
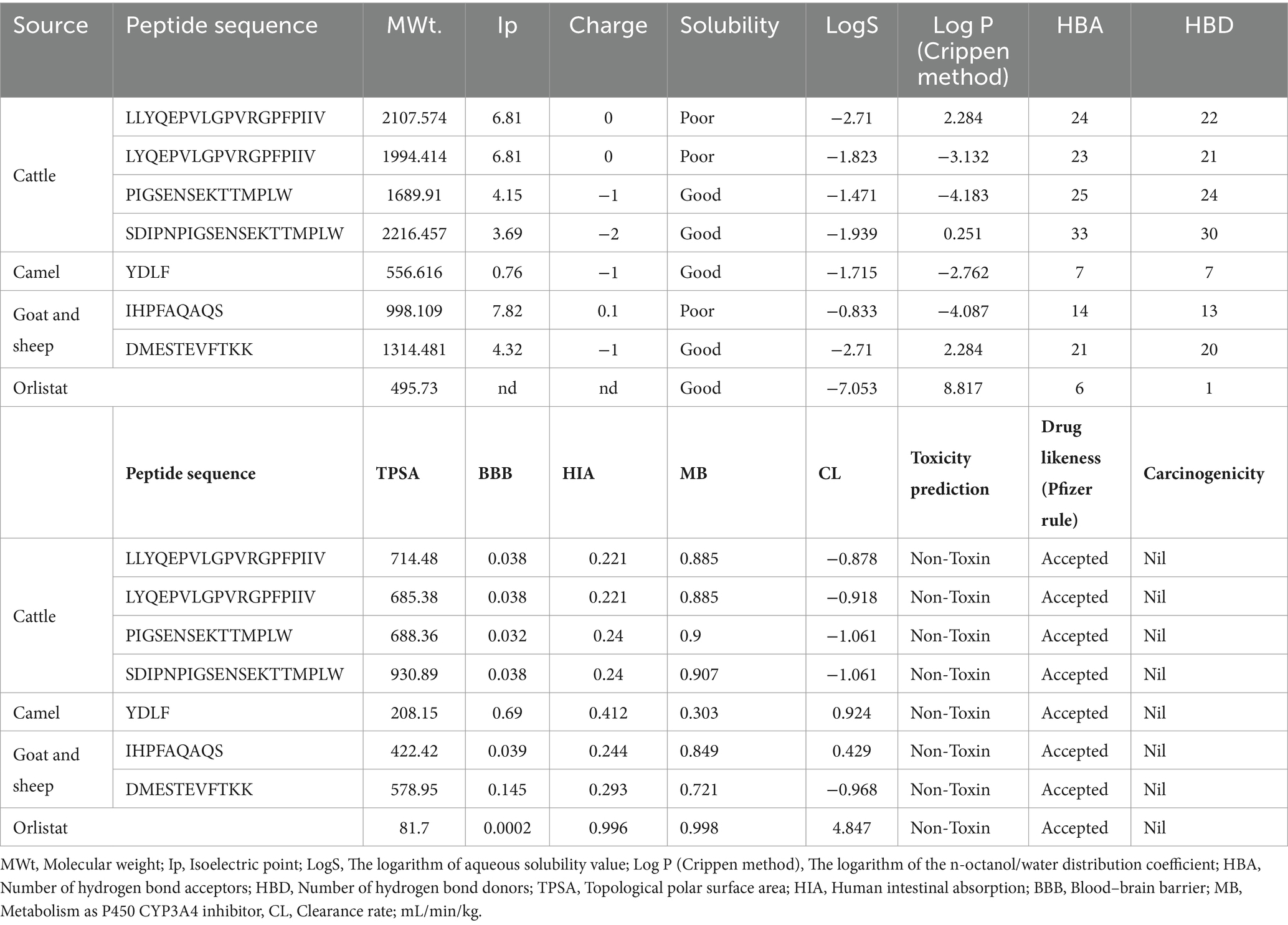
Table 3. In silico physicochemical properties and absorption, distribution, metabolism, excretion (ADME) profile of fermented milk-derived bioactive peptides from cattle, camel, goat, and sheep milk.
Further computational methods, such as BioPep UWM, were used, which provide insights into the stability of BAPs under a simulated gastrointestinal digestive environment, together with providing in-depth information into degradation sites, products generated, their sequence, and composition. The results obtained from in silico hydrolysis of selected seven peptides and their possible fragments upon gastric and gastrointestinal digestion are presented in Table 4. DHt values showed that peptide YDLF showed the highest value of 66.67%, followed by LLYQEPVLGPVRGPFPIIV with a DHt of 33.33%. The rest of the five peptides showed DHt values in the range of 26.32–30.00%. Overall, all peptides showed hydrolysis under optimum conditions of enzymatic action. The higher stability of peptide PIGSENSEKTTMPLW to digestion could be attributed to the presence of proline residues at the C-terminal and N-terminal, which is known to provide resistance to hydrolysis. The results are in accordance with those suggested by other researchers (Nielsen et al., 2017; Wang et al., 2023). Results as obtained from ADMET analysis for peptides obtained after in silico digestion are shown in the Supplementary material (Table 5). The bioactivity scoring for these hydrolyzed fragments using Peptide Ranker showed that only three peptides’ fractions (GPF, PL, and PF) showed higher values for bioactivity than the threshold of 0.8, with peptide ranking scores of 0.98, 0.81, and 0.99, respectively. These results indicated that these seven peptides, even after digestion, could produce BAPs that can further exert biological function even after digestion. Overall, the in-depth in silico analysis of the peptides provided valuable insights into the drug-like characteristics and bioavailability of the seven selected peptides, with several showing promising solubility and bioactivity post-digestion. These findings also underscore the potential of these peptides as effective nutraceuticals and therapeutic agents.
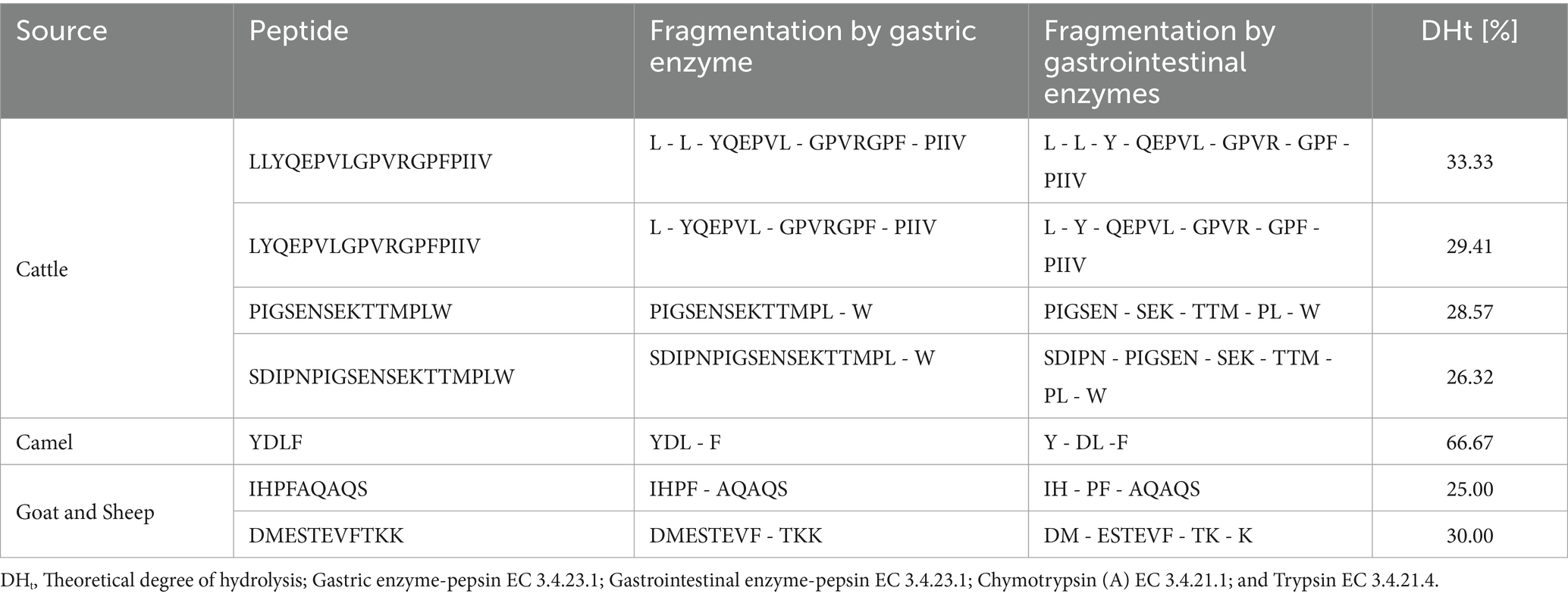
Table 4. Peptides stability upon in silico gastrointestinal digestion as predicted through BIOPEP-UWM.
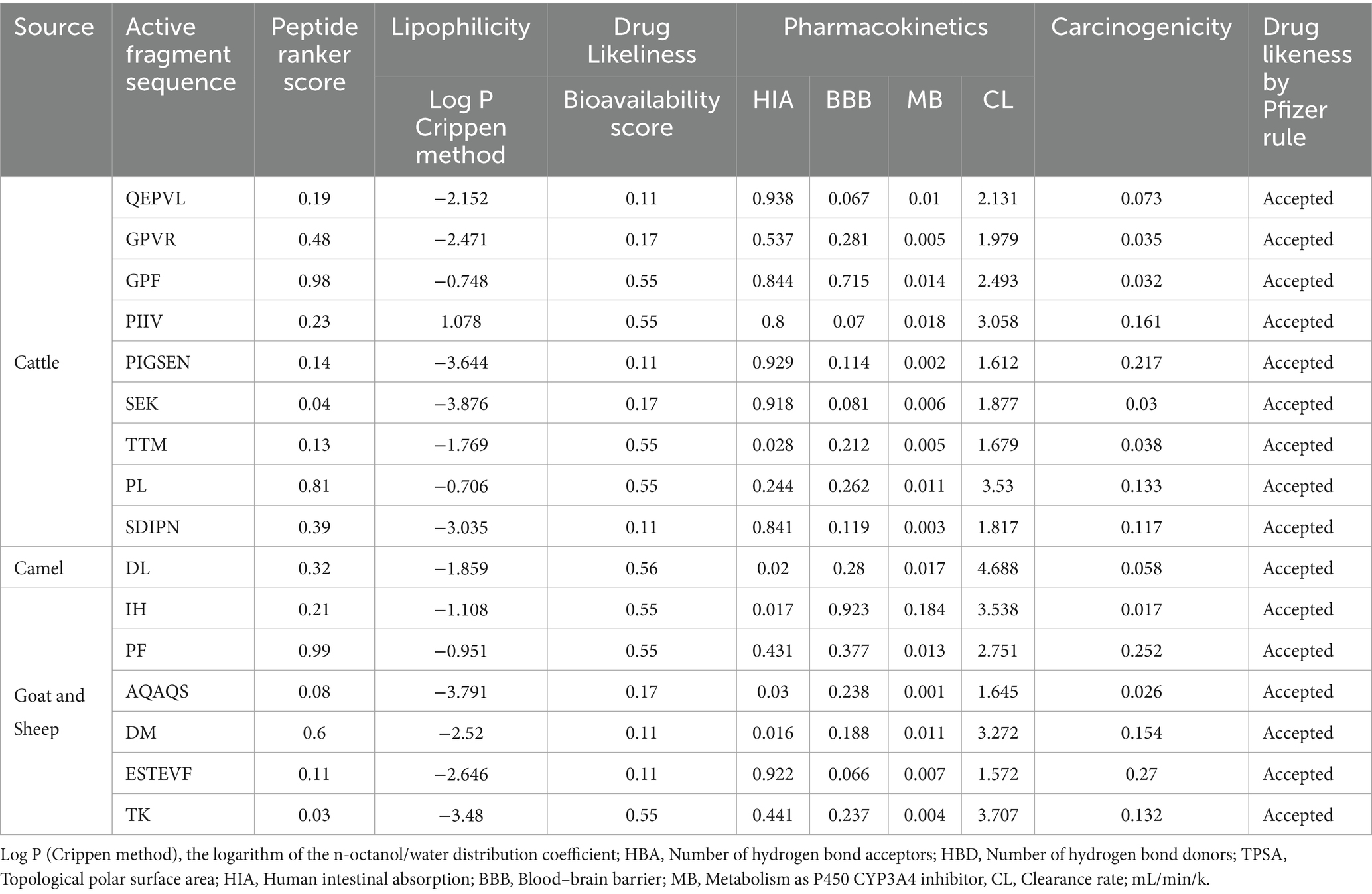
Table 5. Absorption, distribution, drug likeliness metabolism, excretion (ADME), and drug likeliness profile of after in silico gastrointestinal digestion through BIOPEP-UWM.
4 Conclusion
The findings from the present study suggested that the five different probiotic strains used for producing FM from cattle, camel, goat, and SM resulted in the generation of BAPs with hypercholesterolemic activity. FM produced by Pediococcus pentosaceus MF000957 (PP-957) probiotic strain was most effective in inhibiting PL, which appeared to be strain-dependent and substrate-dependent. Novel peptides identified from the different milk types have shown their ability to inhibit PL activities due to their structural features, hydrophobic amino acid compositions, interactions with catalytic residues, and their ability to bind to important PL active sites. However, more specific studies are needed to establish their mechanisms of action and efficacy. Importantly, most of these sequences have not yet been reported, suggesting that fermentation-derived peptides have the potential to produce new types of PL inhibitory peptides for preventing or reducing hyperlipidemia. Moreover, further studies can be undertaken for in-depth in vivo investigations into these selected peptides.
Data availability statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author contributions
PM: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Methodology, Investigation, Funding acquisition, Formal analysis, Data curation, Conceptualization. FA: Writing – review & editing, Writing – original draft, Methodology, Investigation. AA: Writing – original draft, Methodology, Investigation. P-GY: Writing – review & editing, Software, Methodology, Investigation. C-YG: Writing – review & editing, Supervision, Software, Methodology, Investigation, Formal analysis. SM: Writing – review & editing, Writing – original draft, Visualization, Validation, Supervision, Software, Resources, Project administration, Formal analysis, Data curation, Conceptualization.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. The authors are thankful to United Arab Emirates University for funding this research through a research grant (SURE Plus—G00002836) awarded to PI, PM.
Acknowledgments
I would like to acknowledge the use of ChatGPT for its invaluable assistance in enhancing the grammar and language of this research paper. Its support significantly contributed to the clarity and readability of the final manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declared that they were an editorial board member of Frontiers, at the time of submission. This had no impact on the peer review process and the final decision.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1443708/full#supplementary-material
Footnotes
1. ^https://biochemia.uwm.edu.pl/biopep-uwm/
4. ^http://pepbank.mgh.harvard.edu/
5. ^https://www.rcsb.org/structure/1ETH
7. ^https://mobyle.rpbs.univ-paris-diderot.fr/data/jobs/PEP-FOLD3/
References
Ahmad, M., Mudgil, P., and Maqsood, S. (2019). Camel whey protein microparticles for safe and efficient delivery of novel camel milk derived probiotics. LWT 108, 81–88. doi: 10.1016/j.lwt.2019.03.008
Ajayi, F. F., Mudgil, P., Gan, C.-Y., and Maqsood, S. (2021). Identification and characterization of cholesterol esterase and lipase inhibitory peptides from amaranth protein hydrolysates. Food Chem. X 12:100165. doi: 10.1016/j.fochx.2021.100165
Baba, W. N., Mudgil, P., Baby, B., Vijayan, R., Gan, C.-Y., and Maqsood, S. (2021). New insights into the cholesterol esterase-and lipase-inhibiting potential of bioactive peptides from camel whey hydrolysates: identification, characterization, and molecular interaction. J. Dairy Sci. 104, 7393–7405. doi: 10.3168/jds.2020-19868
Birari, R. B., and Bhutani, K. K. (2007). Pancreatic lipase inhibitors from natural sources: unexplored potential. Drug Discov. Today 12, 879–889. doi: 10.1016/j.drudis.2007.07.024
Chen, L., Chen, S., Rong, Y., Zeng, W., Hu, Z., Ma, X., et al. (2024). Identification and evaluation of antioxidant peptides from highland barley distiller's grains protein hydrolysate assisted by molecular docking. Food Chem. 434:137441. doi: 10.1016/j.foodchem.2023.137441
Cheng, M.-C., Tsai, T.-Y., and Pan, T.-M. (2015). Anti-obesity activity of the water extract of Lactobacillus paracasei subsp. paracasei NTU 101 fermented soy milk products. Food Funct. 6, 3522–3530. doi: 10.1039/C5FO00531K
Chia, T. Y., Gan, C.-Y., Shafie, M. H., Yap, P. G., Rodhi, A. M., Ahmad, A., et al. (2023). A comprehensive review on the pancreatic lipase inhibitory peptides: a future anti-obesity strategy. Electron. J. Gen. Med. 20, em470. doi: 10.29333/ejgm/12943
Church, F. C., Swaisgood, H. E., Porter, D. H., and Catignani, G. L. (1983). Spectrophotometric assay using o-phthaldialdehyde for determination of proteolysis in milk and isolated milk proteins. J. Dairy Sci. 66, 1219–1227. doi: 10.3168/jds.S0022-0302(83)81926-2
Damodharan, K., Lee, Y. S., Palaniyandi, S. A., Yang, S. H., and Suh, J.-W. (2015). Preliminary probiotic and technological characterization of Pediococcus pentosaceus strain KID7 and in vivo assessment of its cholesterol-lowering activity. Front. Microbiol. 6:768. doi: 10.3389/fmicb.2015.00768
Elayan, A. A., Suleiman, A., and Saleh, F. (2010). The hypocholesterolemic effect of gariss and gariss containing. Asian J. Biochem. 5, 205–209. doi: 10.3923/ajb.2010.205.209
Fadimu, G. J., Gan, C.-Y., Olalere, O. A., Farahnaky, A., Gill, H., and Truong, T. (2023). Novel antihypertensive peptides from lupin protein hydrolysate: an in-silico identification and molecular docking studies. Food Chem. 407:135082. doi: 10.1016/j.foodchem.2022.135082
Fu, L., Shi, S., Yi, J., Wang, N., He, Y., Wu, Z., et al. (2024). ADMETlab 3.0: an updated comprehensive online ADMET prediction platform enhanced with broader coverage, improved performance, API functionality and decision support. Nucleic Acids Res. 52. W422–W431. doi: 10.1093/nar/gkae236
Ganatsios, V., Nigam, P., Plessas, S., and Terpou, A. (2021). Kefir as a functional beverage gaining momentum towards its health promoting attributes. Beverages 7:48. doi: 10.3390/beverages7030048
García-Burgos, M., Moreno-Fernández, J., Alférez, M. J., Díaz-Castro, J., and López-Aliaga, I. (2020). New perspectives in fermented dairy products and their health relevance. J. Funct. Foods 72:104059. doi: 10.1016/j.jff.2020.104059
Garzón, A. G., Cian, R. E., Aquino, M. E., and Drago, S. R. (2020). Isolation and identification of cholesterol esterase and pancreatic lipase inhibitory peptides from brewer's spent grain by consecutive chromatography and mass spectrometry. Food Funct. 11, 4994–5003. doi: 10.1039/D0FO00880J
Gil-Rodríguez, A. M., and Beresford, T. P. (2019). Lipase inhibitory activity of skim milk fermented with different strains of lactic acid bacteria. J. Funct. Foods 60:103413. doi: 10.1016/j.jff.2019.06.015
Gil-Rodríguez, A. M., and Beresford, T. P. (2020). Lipase inhibitory activity assay for fermented milk. MethodsX 7:100999. doi: 10.1016/j.mex.2020.100999
Gil-Rodríguez, A. M., and Beresford, T. (2021). Bile salt hydrolase and lipase inhibitory activity in reconstituted skim milk fermented with lactic acid bacteria. J. Funct. Foods 77:104342. doi: 10.1016/j.jff.2020.104342
Gonzalez-Gonzalez, C. R., Tuohy, K. M., and Jauregi, P. (2011). Production of angiotensin-I-converting enzyme (ACE) inhibitory activity in milk fermented with probiotic strains: effects of calcium, pH and peptides on the ACE-inhibitory activity. Int. Dairy J. 21, 615–622. doi: 10.1016/j.idairyj.2011.04.001
Granato, D., Branco, G. F., Cruz, A. G., Faria, J. D. A. F., and Shah, N. P. (2010). Probiotic dairy products as functional foods. Compr. Rev. Food Sci. Food Saf. 9, 455–470. doi: 10.1111/j.1541-4337.2010.00120.x
Gupta, S., Kapoor, P., Chaudhary, K., Gautam, A., Kumar, R., and Raghava, G. P. S. (2013). In silico approach for predicting toxicity of peptides and proteins. PLoS One 8:e73957. doi: 10.1371/journal.pone.0073957
Haque, N., and Prabhu, N. P. (2016). Lid dynamics of porcine pancreatic lipase in non-aqueous solvents. Biochim. Biophys. Acta Gen. Sub. 1860, 2326–2334. doi: 10.1016/j.bbagen.2016.05.006
Honorato, R. V., Koukos, P. I., Jiménez-García, B., Tsaregorodtsev, A., Verlato, M., Giachetti, A., et al. (2021). Structural biology in the clouds: the WeNMR-EOSC ecosystem. Front. Mol. Biosci. 8:729513. doi: 10.3389/fmolb.2021.729513
Jiang, S., Cai, L., Lv, L., and Li, L. (2021). Pediococcus pentosaceus, a future additive or probiotic candidate. Microb. Cell Factories 20, 1–14. doi: 10.1186/s12934-021-01537-y
Khakhariya, R., Sakure, A. A., Maurya, R., Bishnoi, M., Kondepudi, K. K., Padhi, S., et al. (2023). A comparative study of fermented buffalo and camel milk with anti-inflammatory, ACE-inhibitory and anti-diabetic properties and release of bio active peptides with molecular interactions: in vitro, in silico and molecular study. Food Biosci. 52:102373. doi: 10.1016/j.fbio.2023.102373
Kinariwala, D., Panchal, G., Sakure, A., and Hati, S. (2020). Exploring the potentiality of lactobacillus cultures on the production of milk-derived bioactive peptides with antidiabetic activity. Int. J. Pept. Res. Ther. 26, 1613–1627. doi: 10.1007/s10989-019-09958-5
Kumar, A., and Chauhan, S. (2021). Pancreatic lipase inhibitors: the road voyaged and successes. Life Sci. 271:119115. doi: 10.1016/j.lfs.2021.119115
Lamiable, A., Thévenet, P., Rey, J., Vavrusa, M., Derreumaux, P., and Tufféry, P. (2016). PEP-FOLD3: faster de novo structure prediction for linear peptides in solution and in complex. Nucleic Acids Res. 44, W449–W454. doi: 10.1093/nar/gkw329
Lim, Y. H., Foo, H. L., Loh, T. C., Mohamad, R., and Abdullah, N. (2019). Comparative studies of versatile extracellular proteolytic activities of lactic acid bacteria and their potential for extracellular amino acid productions as feed supplements. J. Anim. Sci. Biotechnol. 10, 1–13. doi: 10.1186/s40104-019-0323-z
Liu, T.-T., Liu, X.-T., Chen, Q.-X., and Shi, Y. (2020). Lipase inhibitors for obesity: a review. Biomed. Pharmacother. 128:110314. doi: 10.1016/j.biopha.2020.110314
Manzanarez-Quín, C. G., Beltrán-Barrientos, L. M., Hernández-Mendoza, A., González-Córdova, A. F., and Vallejo-Cordoba, B. (2021). Invited review: potential antiobesity effect of fermented dairy products. J. Dairy Sci. 104, 3766–3778. doi: 10.3168/jds.2020-19256
Martinez-Villaluenga, C., Rupasinghe, S. G., Schuler, M. A., and Gonzalez De Mejia, E. (2010). Peptides from purified soybean β-conglycinin inhibit fatty acid synthase by interaction with the thioesterase catalytic domain. FEBS J. 277, 1481–1493. doi: 10.1111/j.1742-4658.2010.07577.x
Minkiewicz, P., Iwaniak, A., and Darewicz, M. (2019). BIOPEP-UWM database of bioactive peptides: current opportunities. Int. J. Mol. Sci. 20:5978. doi: 10.3390/ijms20235978
Mohamed, G. A., Ibrahim, S. R., Elkhayat, E. S., and El Dine, R. S. (2014). Natural anti-obesity agents. Bull. Faculty Pharm. Cairo Univ. 52, 269–284. doi: 10.1016/j.bfopcu.2014.05.001
Mudgil, P., Al Dhaheri, M. K. O., Alsubousi, M. S. M., Khan, H., Redha, A. A., Yap, P.-G., et al. (2024). Molecular docking studies on α-amylase inhibitory peptides from milk of different farm animals. J. Dairy Sci. 107, 2633–2652. doi: 10.3168/jds.2023-24118
Mudgil, P., Baba, W. N., Kamal, H., Fitzgerald, R. J., Hassan, H. M., Ayoub, M. A., et al. (2022). A comparative investigation into novel cholesterol esterase and pancreatic lipase inhibitory peptides from cow and camel casein hydrolysates generated upon enzymatic hydrolysis and in-vitro digestion. Food Chem. 367:130661. doi: 10.1016/j.foodchem.2021.130661
Mudgil, P., Baby, B., Ngoh, Y.-Y., Vijayan, R., Gan, C.-Y., and Maqsood, S. (2019). Identification and molecular docking study of novel cholesterol esterase inhibitory peptides from camel milk proteins. J. Dairy Sci. 102, 10748–10759. doi: 10.3168/jds.2019-16520
Mudgil, P., Gan, C.-Y., Affan Baig, M., Hamdi, M., Mohteshamuddin, K., Aguilar-Toalá, J. E., et al. (2023). In-depth peptidomic profile and molecular simulation studies on ACE-inhibitory peptides derived from probiotic fermented milk of different farm animals. Food Res. Int. 168:112706. doi: 10.1016/j.foodres.2023.112706
Ngo, N. T. T., Senadheera, T. R., and Shahidi, F. (2023). Antioxidant properties and prediction of bioactive peptides produced from Flixweed (sophia, Descurainis sophia L.) and Camelina (Camelina sativa (L.) Crantz) seed meal: integrated in vitro and in silico studies. Plan. Theory 12:3575. doi: 10.3390/plants12203575
Nielsen, S. D., Beverly, R. L., Qu, Y., and Dallas, D. C. (2017). Milk bioactive peptide database: a comprehensive database of milk protein-derived bioactive peptides and novel visualization. Food Chem. 232, 673–682. doi: 10.1016/j.foodchem.2017.04.056
Park, S., Kang, J., Choi, S., Park, H., Hwang, E., Kang, Y., et al. (2018). Cholesterol-lowering effect of Lactobacillus rhamnosus BFE5264 and its influence on the gut microbiome and propionate level in a murine model. PLoS One 13:e0203150. doi: 10.1371/journal.pone.0203150
Patel, R., and Hati, S. (2018). Production of antihypertensive (angiotensin I-converting enzyme inhibitory) peptides derived from fermented milk supplemented with WPC70 and calcium caseinate by Lactobacillus cultures. Rev. Res. Med. Microbiol. 29, 30–40. doi: 10.1097/MRM.0000000000000119
Pihlanto, A., Virtanen, T., and Korhonen, H. (2010). Angiotensin I converting enzyme (ACE) inhibitory activity and antihypertensive effect of fermented milk. Int. Dairy J. 20, 3–10. doi: 10.1016/j.idairyj.2009.07.003
Pipaliya, R., Basaiawmoit, B., Sakure, A. A., Maurya, R., Bishnoi, M., Kondepudi, K. K., et al. (2024). Peptidomics-based identification of antihypertensive and antidiabetic peptides from sheep milk fermented using Limosilactobacillus fermentum KGL4 MTCC 25515 with anti-inflammatory activity: in silico, in vitro, and molecular docking studies. Front. Chem. 12:1389846. doi: 10.3389/fchem.2024.1389846
Pothuraju, R., Sharma, R., Kavadi, P. K., Chagalamarri, J., Jangra, S., Bhakri, G., et al. (2016). Anti-obesity effect of milk fermented by Lactobacillus plantarum NCDC 625 alone and in combination with herbs on high fat diet fed C57BL/6J mice. Benefic. Microbes 7, 375–385. doi: 10.3920/BM2015.0083
Sarah, S., Faradalila, W., Salwani, M., Amin, I., Karsani, S., and Sazili, A. (2016). LC–QTOF-MS identification of porcine-specific peptide in heat treated pork identifies candidate markers for meat species determination. Food Chem. 199, 157–164. doi: 10.1016/j.foodchem.2015.11.121
Shao, B., Huang, X., Xu, M., Cheng, D., Li, X., and Li, M. (2023). Peptides isolated from black soybean synergistically inhibit the activity of angiotensin converting enzyme (ACE). J. Funct. Foods 106:105604. doi: 10.1016/j.jff.2023.105604
Shukla, P., Sakure, A., Maurya, R., Bishnoi, M., Kondepudi, K. K., Das, S., et al. (2023). Antidiabetic, angiotensin-converting enzyme inhibitory and anti-inflammatory activities of fermented camel milk and characterisation of novel bioactive peptides from lactic-fermented camel milk with molecular interaction study. Int. J. Dairy Technol. 76, 149–167. doi: 10.1111/1471-0307.12910
Soleymanzadeh, N., Mirdamadi, S., and Kianirad, M. (2016). Antioxidant activity of camel and bovine milk fermented by lactic acid bacteria isolated from traditional fermented camel milk (Chal). Dairy Sci. Technol. 96, 443–457. doi: 10.1007/s13594-016-0278-1
Sridhar, S., George, G., Verma, A., and Paul, A. T. (2019). Natural products-based pancreatic lipase inhibitors for obesity treatment. Nat. Bio-act. Comp. Product. Appl., 149–191. doi: 10.1007/978-981-13-7154-7_6
Tiss, M., Souiy, Z., Ben Abdeljelil, N., Njima, M., Achour, L., and Hamden, K. (2020). Fermented soy milk prepared using kefir grains prevents and ameliorates obesity, type 2 diabetes, hyperlipidemia and liver-kidney toxicities in HFFD-rats. J. Funct. Foods 67:103869. doi: 10.1016/j.jff.2020.103869
Trabuco, L. G., Lise, S., Petsalaki, E., and Russell, R. B. (2012). PepSite: prediction of peptide-binding sites from protein surfaces. Nucleic Acids Res. 40, W423–W427. doi: 10.1093/nar/gks398
Trigueros, L., Peña, S., Ugidos, A., Sayas-Barberá, E., Pérez-Álvarez, J., and Sendra, E. (2013). Food ingredients as anti-obesity agents: a review. Crit. Rev. Food Sci. Nutr. 53, 929–942. doi: 10.1080/10408398.2011.574215
Urbizo-Reyes, U., Liceaga, A. M., Reddivari, L., Kim, K.-H., and Anderson, J. M. (2022). Enzyme kinetics, molecular docking, and in silico characterization of canary seed (Phalaris canariensis L.) peptides with ACE and pancreatic lipase inhibitory activity. J. Funct. Foods 88:104892. doi: 10.1016/j.jff.2021.104892
Wang, J., Shao, B., Li, J., Wang, Z., Zhang, M., Jia, L., et al. (2023). Identification and in silico analysis of ACE-inhibitory peptides derived from milk fermented by Lacticaseibacillus paracasei. J. Agric. Food Chem. 71, 12462–12473. doi: 10.1021/acs.jafc.2c09148
Xiong, G., Wu, Z., Yi, J., Fu, L., Yang, Z., Hsieh, C., et al. (2021). ADMETlab 2.0: an integrated online platform for accurate and comprehensive predictions of ADMET properties. Nucleic Acids Res. 49, W5–W14. doi: 10.1093/nar/gkab255
Yoda, K., Sun, X., Kawase, M., Kubota, A., Miyazawa, K., Harata, G., et al. (2015). A combination of probiotics and whey proteins enhances anti-obesity effects of calcium and dairy products during nutritional energy restriction in aP2-agouti transgenic mice. Br. J. Nutr. 113, 1689–1696. doi: 10.1017/S0007114515000914
Keywords: probiotic fermentation, pancreatic lipase inhibition, milk proteins, dairy animals, bioactive peptides
Citation: Mudgil P, Ajayi FF, Alyafei AS, Yap P-G, Gan C-Y and Maqsood S (2024) Unlocking the hypolipidemic potential of bioactive peptides derived from probiotic fermented cattle, camel, goat, and sheep milk: a comprehensive investigation through in vitro,in silico, and molecular docking studies. Front. Sustain. Food Syst. 8:1443708. doi: 10.3389/fsufs.2024.1443708
Edited by:
Araceli Loredo, Autonomous University of Coahuila, MexicoReviewed by:
Qing-Feng Zhang, Jiangxi Agricultural University, ChinaTaner Sar, Bioextrax AB, Sweden
Heena Sharma, National Dairy Research Institute (ICAR), India
Copyright © 2024 Mudgil, Ajayi, Alyafei, Yap, Gan and Maqsood. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Priti Mudgil, cHJpdGkuZEB1YWV1LmFjLmFl; Sajid Maqsood, U2FqaWQubUB1YWV1LmFjLmFl
 Priti Mudgil
Priti Mudgil Feyisola Fisayo Ajayi
Feyisola Fisayo Ajayi Aysha Saleh Alyafei1
Aysha Saleh Alyafei1 Pei-Gee Yap
Pei-Gee Yap Chee-Yuen Gan
Chee-Yuen Gan Sajid Maqsood
Sajid Maqsood