- 1Yunnan Institute of Tropical Crops, Xishuangbanna, China
- 2Macadami Agricultural Engineering Research Center of Yunnan Province, Xishuangbanna, China
Macadamia nuts, scientifically designated as Macadamia integrifolia, are a highly valuable crop that originated in Australia. This study provides a comprehensive examination of the symbiotic relationships between various macadamia root systems and arbuscular mycorrhizal fungi (AMF). The four principal macadamia-producing regions in Yunnan Province were selected for investigation on the basis of meticulous criteria. To determine the AMF infection rate, the roots were stained. Furthermore, high-throughput sequencing was employed to conduct a comprehensive analysis of the fungal diversity in the rhizosphere soil. The findings were definitive, indicating that both normal and cluster roots are capable of establishing a symbiotic relationship with AMF. Secondary forests exhibited significantly elevated fungal diversity relative to normal roots, while cluster roots demonstrated the lowest diversity and notable regional variation, indicating that the environment exerts a considerable influence on inter-root fungi and AMF. The analysis of the fungal community composition revealed that the predominant groups were Ascomycetes and Basidiomycetes. The FUNGuild function prediction clearly indicated distinct differences in the fungal functions of secondary forests, cluster roots, and normal roots. This study provides a scientific foundation for the sustainable development of macadamia nuts and significantly contributes to a deeper comprehension of the intricate interactions between macadamia and AMF, thereby fostering the long-term stable and healthy growth of the macadamia nut industry.
1 Introduction
Macadamia nuts, also known as king of nuts, belong to the Proteaceae family. They are a crop domesticated from dicotyledons and are endemic to the subtropical rainforests of eastern Australia (Nock et al., 2019). Macadamia nuts are nutrient-rich foods that contain more than 70% fat, 9% protein, and 8 essential amino acids. They are also high in unsaturated fatty acids (Hardner et al., 2009), which provide significant nutritional and health benefits (Wall, 2010; Maguire et al., 2004). Studies have shown that long-term consumption of macadamia nuts may help prevent and treat cardiovascular diseases (Garg et al., 2003; Garg et al., 2007). Additionally, different parts of the macadamia nut, such as the kernel, husk, and oil meal, have various economic value. These parts can be processed into a range of products, including beverages (Topp et al., 2019), oils (Aradhya et al., 1998), activated charcoal, adsorbents, and feed. Furthermore, the macadamia tree is an ideal insect-resistant plant for landscaping due to its beautiful shape, dense branches and leaves, colorful and fragrant flowers, and solid and dense wood. In 2020, China accounted for 60.9% of the global total area of macadamia nut planting, with 266,133 hectares planted and 66,733 hectares harvested. In Yunnan Province, the planted and harvested areas were 235,333 hectares and 63,333 hectares, respectively, accounting for 88.43 and 94.91% of the total planted and harvested areas in the country. Yunnan is an ideal location for macadamia nut cultivation due to its low latitude, mild climate, abundant rainfall and fertile soil conditions. Currently, Yunnan has the largest macadamia nut planting area in the world, and its planting scale continues to increase annually.
However, the sustainable development of the macadamia nut industry still faces challenges due to differences in cultivation techniques and management levels between plantations in different regions, as well as changes in climate and environmental factors. To promote the long-term stable and healthy development of the macadamia nut industry, it is important to utilize plant–microbe interactions to improve resource utilization and reduce the need for pesticides and fertilizers (Avio et al., 2018). Arbuscular mycorrhizal fungus (AMF) represents a class of ancient and species - scarce symbiotic fungi associated with plant roots, predominantly found within approximately 27 genera across 4 orders of the Glomeromycota (Remy et al., 1994). Arbuscular mycorrhiza (AM) enters into a symbiotic relationship with roughly 80% of terrestrial plants (Redecker et al., 2000). They promote plant growth and development by increasing the plant’s ability to absorb water and nutrients (Ruiz-Lozano et al., 2016; Mo et al., 2016). Additionally, it exerts a broad influence on plant - plant interactions and community structures (Torrecillas et al., 2014; Savary et al., 2020), thereby playing a significant role in agricultural production as well as in ecosystem protection and restoration. Consequently, promoting the diversity of AMF communities constitutes an important avenue for enhancing the stability and productivity of plant communities within ecosystems. The diversity of mycorrhizal fungi can play an important role in improving the yield and quality of macadamia nuts. Research has shown that AMF can reduce the need for external inputs and help maintain the sustainability and stability of agroecosystems (Powell and Rillig, 2018). Maintaining stability and sustainability is crucial for the development of any crop ecosystem, including the emerging macadamia nut industry. AMF have been identified as key factors in addressing the sustainability of agricultural systems. However, there are differing views on whether AMF can prevent the colonization of macadamia root systems. It was previously believed that Proteaceae, including Macadamia nuts, are nonmycorrhizal (Lambers and Teste, 2013). Research has shown that flavonoids secreted by the root system during root exclusion in white lupin can inhibit mycorrhizal formation (Weisskopf et al., 2006a). However, it has also been suggested that these flavonoids may promote the germination of AMF spores or attract mycelia to roots (Pei et al., 2020).
Numerous studies have demonstrated the significant role of AMF in the success of exotic species invasion (Dawkins and Esiobu, 2017; Zhang et al., 2018; Paudel et al., 2014). The enhanced mutualisms hypothesis (EMH) proposes that invasive species can alter AMF communities and derive greater benefits from them than can symbiotic native plants. This is due to the ability of AMFs to enhance the survival and growth of invasive species, promoting their competitiveness at the introduced site (Lekberg et al., 2013; Zhang et al., 2017). However, the coordination process between AMF and the macadamia root system remains complex and unclear. Therefore, it is essential to understand the AMF composition of different types of macadamia root systems to determine the interactions between macadamia and AMF. Based on the available evidence, we hypothesize that the AMF communities differ among different root system types. In addition, it is important to note that the cooccurring AMFs with macadamia root systems may vary significantly depending on the region. To test our hypotheses, we collected interroot soils from different root systems (secondary forest, cluster roots, and normal roots) in the main macadamia-producing areas of Yunnan Province, including YingJiang, ChangNing, LanCang, and YunXian. Our investigation aimed to determine the impact of different root systems on the structure of AMF communities. The results of this study will aid in understanding the interrelationship between macadamia nuts and AMF, providing a crucial foundation for the sustainable development of macadamia nuts.
2 Materials and methods
2.1 Sample site overview
To prevent accidental errors, this study selected four main production areas of macadamia nuts in Yunnan Province as experimental sample plots. The areas selected were Ying Jiang (YJ), ChangNing (CN), LanCang (LC), and YunXian (YX) (Figure 1; Table 1). These areas have a highland monsoon climate, with an average altitude of 1,100 m above sea level. The average temperature is approximately 20°C in summer and 7°C in winter. The region experiences a rainy season from May to October each year, which contributes to more than 80% of the annual precipitation. Conversely, the dry season occurs from November to April and is characterized by lower precipitation levels. Furthermore, the region experiences temperate weather conditions, and the soil is deep, loose, and rich in organic matter, creating an optimal environment for cultivating macadamia nut trees.
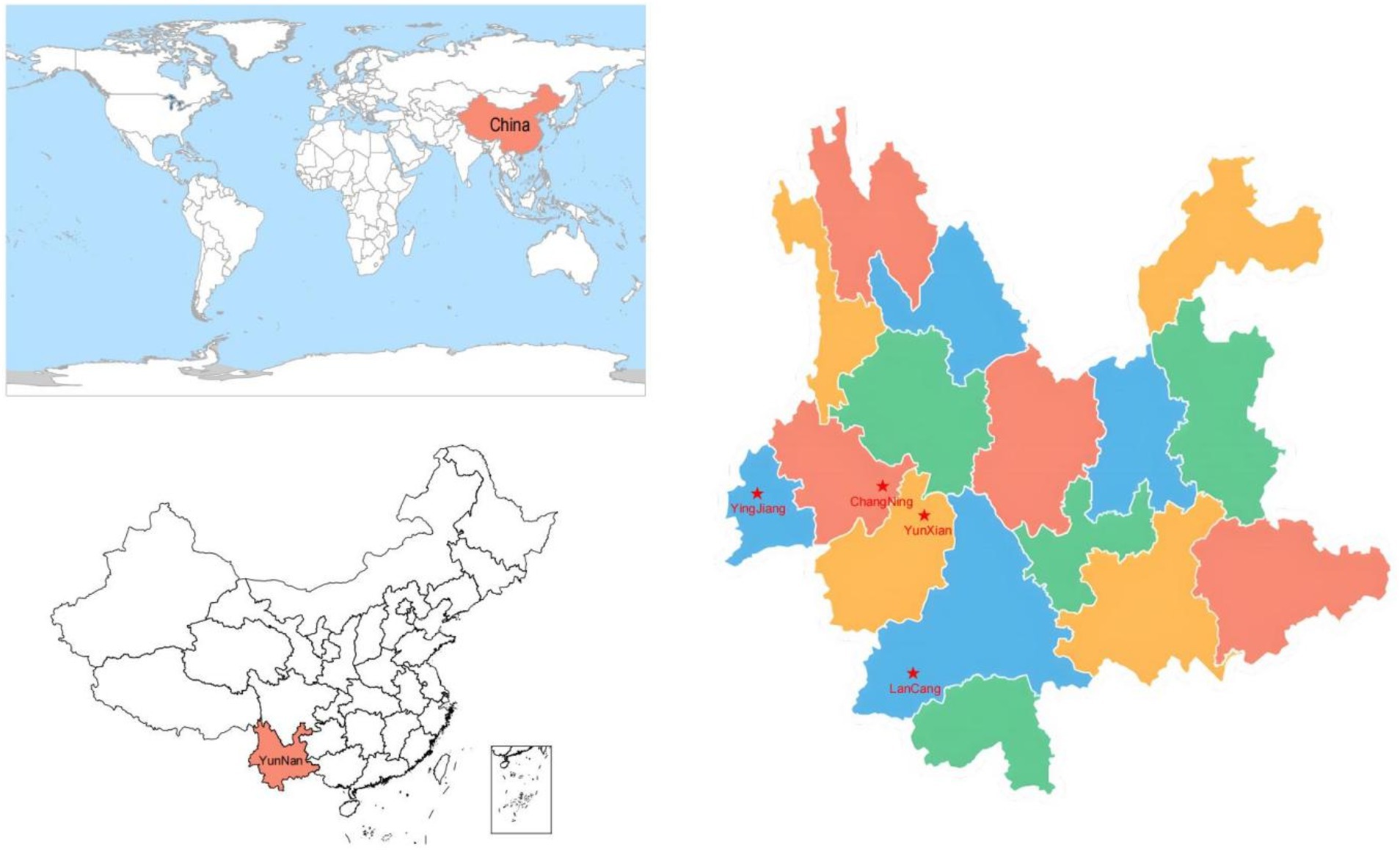
Figure 1. The asterisk indicates the study site in YunNan Province and China. YunXian (100°8′44″, 24°24′39″), YingJiang (97°54′30″, 24°44′46″), ChangNing (99°27′27″, 24°51′11″), LanCang (100°26′30″, 22°35′45″).
2.2 Soil sample collection and experimental method
Sampling was conducted in April 2022. Sample plots were selected within the macadamia nut plantation area in each region. The plots were located within 500 m × 500 m of flat terrain with uniform soil and consistent cultivation and management practices. Each sampling site was planted exclusively with similar-aged macadamia nut trees (Own Choice, O.C.) that were at least 8 years old. The study employed the five-point sampling method (Navarrete et al., 2015) to conduct random sampling. Each sampling point was spaced 20 meters apart. The macadamia nut root system was excavated using a soil auger within the soil layer of the sampled macadamia nut tree at a distance of 10–30 cm from the ground, which is the main distribution area of the macadamia nut root system. Normal and cluster roots were collected separately. Soil samples were collected within 2 cm of the root system of the same tree each time. Approximately 2 g per portion of soil sample was placed in 5 mL freezing tubes, snap frozen in liquid nitrogen for 30 min, and then transferred to a foam box filled with dry ice. The root system was rinsed with sterile water and stored in centrifuge tubes containing 60% ethanol fixative to determine the rate of mycorrhizal infestation from the branching mycorrhizae. The sampling methods for the root systems and interroot soils in the protection zones adjacent to the macadamia nut forest areas were consistent with the methods described above. Five biological replicates were taken from each group of interroot soils of normal and cluster roots in the O.C. In addition, three or four biological replicates were taken from each group in the adjacent protection zone, and DNA was extracted for sequencing analysis of fungal diversity. Sampling was kept consistent across the four areas.
2.3 Arbuscular mycorrhizal fungi staining and determination of infection rate
The method used to estimate the rate of AMF colonization in the macadamia roots was described in previous studies (Phillips and Hayman, 1970). In brief, the roots were rinsed multiple times with distilled water upon return to the laboratory and cut into approximately 1 cm segments. These segments were then placed in a 10% KOH solution and decolorized in a 90°C water bath for 60 min. After cooling, the segments were rinsed 3–5 times with distilled water and then acidified with a 2% HCL solution. Finally, the root segments were removed and rinsed for 30 min. Next, the sample was stained with a 0.12% trypan blue dye solution (250 mL of lactic acid, 250 mL of glycerol, 500 mL of water, and 1.2 mg of trypan blue) for 30 min at 90°C in a water bath. Subsequently, it was decolorized in a lactic acid-glycerol (lactic acid:glycerol = V:V = 1:1) solution. The decolorized root segments were then placed on slides, pressed, and counted under a light microscope using the grid-cross method (Giovannetti and Mosse, 1980). The mycorrhizal colonization rate was estimated according to previous studies (McGonigle et al., 1990).
2.4 DNA extraction and PCR amplification
The DNA from the samples was extracted using the CTAB/SDS method (Xia et al., 2019). DNA concentration and purity were monitored on 1% agarose gels. Based on the concentration, the DNA was diluted to 1 ng/μL with sterile water. The ITS regions were amplified using the primers ITS1F (5’-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5’-GCTGCGTTCTTCATCGATGC-3′) (White et al., 1990). The PCR mixtures were composed of 15 μL of Phusion® High-Fidelity PCR Master Mix (New England Biolabs), 0.2 μM of each primer, and 10 ng of target DNA. The cycling conditions consisted of an initial denaturation step at 98°C for 1 min, followed by 30 cycles at 98°C for 10 s, 50°C for 30 s, and 72°C for 30 s, and a final extension step of 5 min at 72°C. The PCR products were mixed with an equal volume of 1X loading buffer containing SYBR Green and subjected to electrophoresis on a 2% agarose gel for DNA detection. Subsequently, the PCR products were combined in equal proportions and purified using the Qiagen Gel Extraction Kit (Qiagen, Germany).
2.5 Library preparation and sequencing
The sequencing libraries were prepared using the NEBNext® UltraTM IIDNA Library Prep Kit (Cat No. E7645) following the manufacturer’s instructions. Library quality was evaluated using a Qubit@ 2.0 fluorometer (Thermo Scientific) and an Agilent Bioanalyzer 2,100 system. Subsequently, the library was sequenced on an Illumina NovaSeq platform, which produced 250 bp paired-end reads. The findings of this study were supported by data deposited in the CNGB Sequence Archive (CNSA) (Guo et al., 2020) of the China National GenBank DataBase (CNGBdb) (Chen et al., 2020) under the accession number CNP0005527.
2.6 Data processing
The samples were assigned paired-end reads based on their unique barcodes and were truncated by removing the barcodes and primer sequences. FLASH (version 1.2.11) (Magoč and Salzberg, 2011) was used to merge the paired-end reads. The raw tags were quality filtered using fastp (version 0.20.0) software to obtain high-quality clean tags. The clean tags were compared to the reference database1 using Vsearch (version 2.15.0) to detect chimeric sequences (Haas et al., 2011). The effective tags were denoised using the DADA2 module in QIIME2 software (version QIIME2-202006) to obtain initial amplicon sequence variants (ASVs). Species annotation was also performed using QIIME2 software. The absolute abundance of ASVs was normalized using a standard sequence number corresponding to the sample with the fewest sequences. Alpha diversity and beta diversity were analyzed based on the normalized data. To assess the diversity, richness, and uniformity of the communities in the samples, seven indices were used to calculate alpha diversity in QIIME2. The evaluated indices were the Observed_otus, Chao1, Shannon, Simpson, dominance, Good’s coverage, and Pielou_e indices. Beta diversity was calculated in QIIME2 to assess community composition complexity and compare differences between sample groups. Principal component analysis (PCA) was used for cluster analysis to reduce the dimensionality of the original variables. The R software (version 3.5.3) ggplot2 package was used. The fungal functions were annotated using the FUNGuild database (Nguyen et al., 2016) (accessed on 10 May 2022). Spearman’s correlation coefficient was calculated for the top 100 microbial genera, and only the connections with correlation coefficients greater than 0.6 or less than −0.6 and significant (p value less than 0.05) were considered valid for the Sankey diagram.
2.7 Statistical analysis
The data collected from the analyses of phenotypic and physiological parameters were subjected to one-way variance analysis (ANOVA) and Student’s t test using SPSS 22.0 software (IBM, New York, NY, United States). A p value less than 0.05 indicates a significant difference, while a p value less than 0.01 indicates a highly significant difference. The Kruskal–Wallis test in QIIME2 was used to analyze differences in alpha diversity between groups. Bioinformatic analysis was performed using the OmicStudio tools available at https://www.omicstudio.cn/tool (Lyu et al., 2023).
3 Results
3.1 Identification of AMF infestation in normal and cluster roots of macadamia nuts
To investigate the effects of AMF on various macadamia nut root systems, we stained both normal root systems and cluster roots with Tepan blue. The infection was then observed under a microscope (Figure 2). The AMF infestation rates were calculated using the cross-counting method. Both the normal root system (Figures 2A,B) and cluster roots (Figures 2C,D) were susceptible to AMF infection. The structures of AMF symbionts, such as mycelia and spores, were visible under an ordinary light microscope, indicating a symbiotic relationship between AMF and different types of macadamia root systems. Statistical analyses were conducted to confirm that AMF can infest both normal and cluster roots of macadamia nuts in the protection zone and the woodland zone. The results indicated that both types of root systems could infest both the protection zone and the forest zone, and the infestation rate of cluster roots was significantly greater than that of normal roots, with p values of 0.0002 (Figure 2E) and 0.0001 (Figure 2F), respectively.
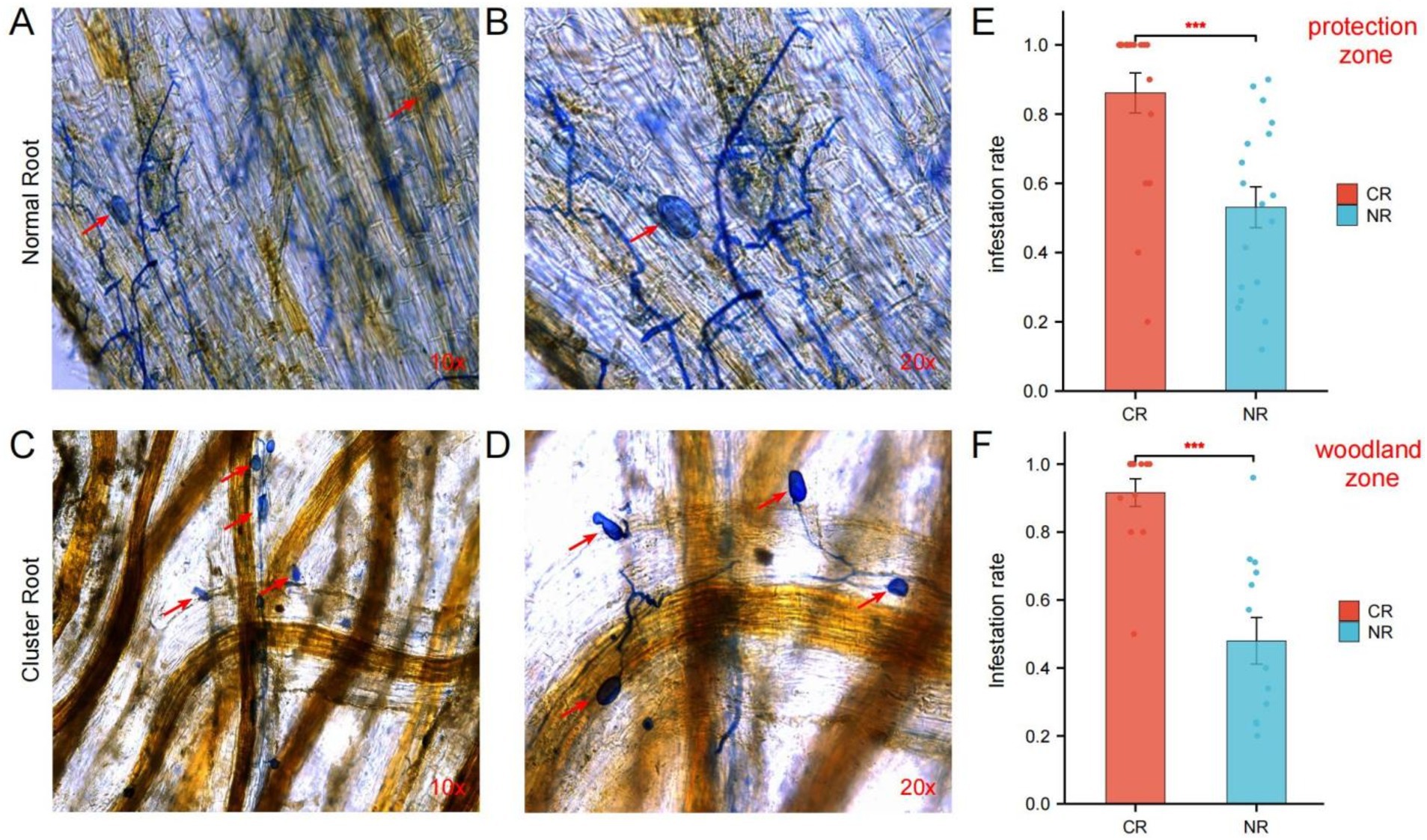
Figure 2. Infection of different types of root systems of macadamia nuts by AMF. The colonization of AMF in normal macadamia nut root systems was observed under a light microscope at 10× (A) and 20× (B) magnification, as well as in rows of roots (C,D). Red arrows indicate spores. The infection rates of AMF in normal and cluster roots of macadamia nuts were measured in the adjacent protection zone (E) and on the woodland zone (F). CR, cluster roots; NR, normal roots. ***p < 0.001 by one-way analysis of variance followed by Tukey’s post-hoc test.
3.2 Overview of soil fungal diversity at different locations
High-throughput sequencing was used to analyze fungal diversity in two types of macadamia nut root systems and in the interroot soils of adjacent secondary forests in four regional plantations. A total of 54 libraries were sequenced, with three or four replicates per group for secondary forests and five replicates per group for cluster and normal roots in each region. A total of 44,266,600 raw reads (PE250) were obtained, resulting in an average of approximately 81,974 raw reads per sample. After filtering, 42,568,660 qualified reads remained, resulting in an average of approximately 788,830 clean reads per sample. The average fragment length was 245.83 bp. The sequencing data in this study were of high quality and highly reliable, as indicated by the average values of Q20 and Q30, which are indicators of sequencing quality, being 98.67 and 95.30, respectively, and the average GC content of the sequenced fragments was 51.64% (Supplementary Table S1).
To identify the number of AMF communities in various regions and root types, we conducted statistical analyses based on the number of fungal species detected in each sampling area. The analysis included different root systems, such as secondary forests, clusters, and normal roots (Figure 3). The study revealed that among the four sampling areas, YunXian had the greatest number of detected fungal species (1429), while Changning had the least (937). Only 219 fungi were identified in all four regions, accounting for 7.23% of the total, indicating significant variation in the AMF community across regions (Figure 3A). Among the different root types, the greatest number of fungal species were identified in secondary forests (1784), followed by normal roots (1604), and the smallest number of fungal species were identified in cluster roots (1237). Of these, 495 fungal species were coidentified, representing 16.35% (Figure 3B). The AMF species identified in secondary forests, cluster roots, and normal roots were counted at the four sampling sites of ChangNing (Figure 3C), YingJiang (Figure 3D), LanCang (Figure 3E), and YunXian (Figure 3F). The percentage of mycorrhizal species identified across all four sites was approximately 10%.
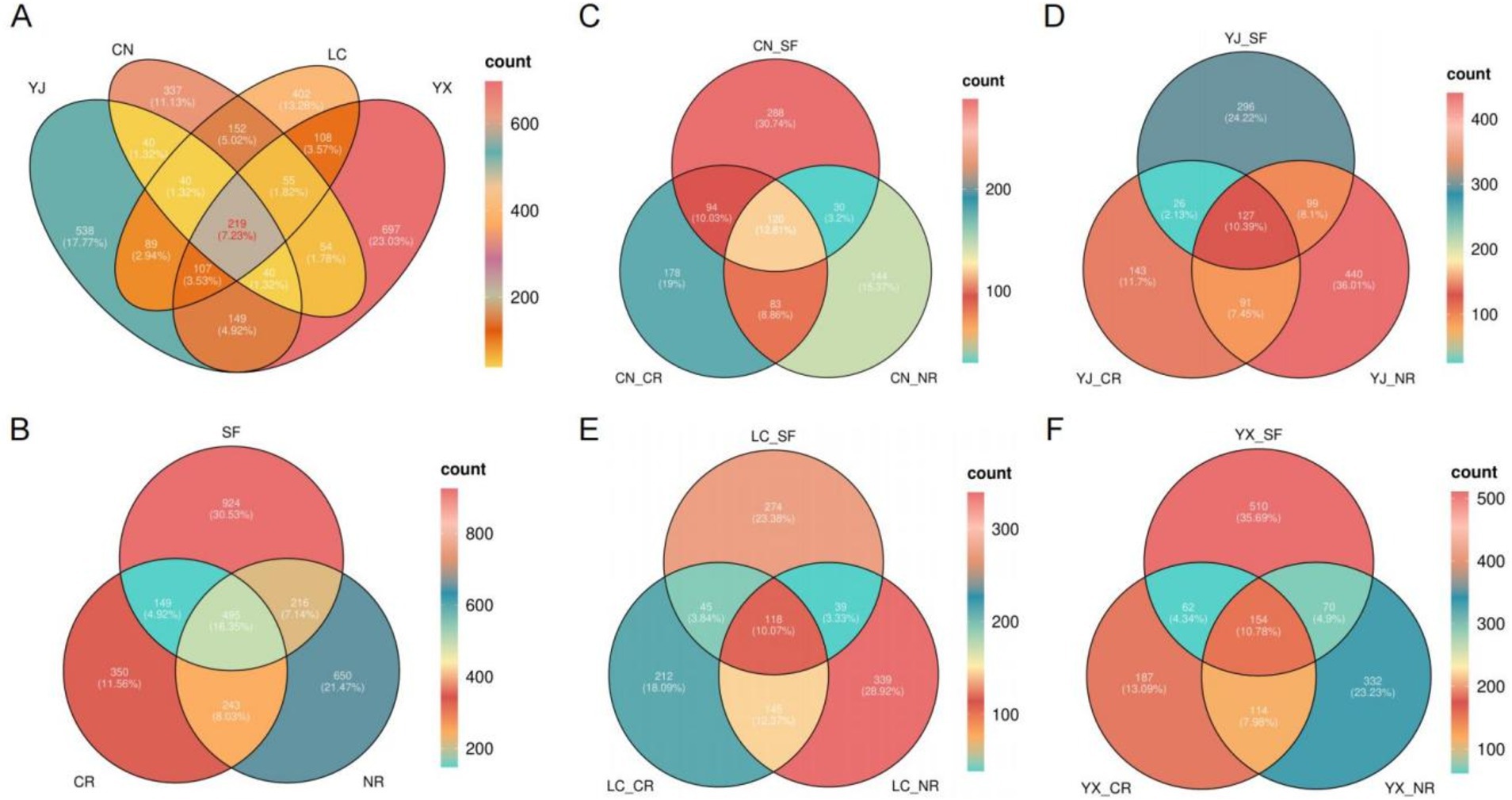
Figure 3. Identification of inter-root AMF fungal communities in macadamia nuts. (A) Venn diagrams of AMF communities in four different regions. (B) Venn diagram of AMF species identified in SF, CR, and NR. Venn diagrams of AMF species identified in SF, CR, and NR in different areas, CN (C), YJ (D), LC (E), and YX (F), respectively. Different colors indicate the number of fungi identified. SF, secondary forest; CR, cluster roots; NR, normal roots. CN, Changning; YJ, Yingjiang; LC, Lancang; YX, Yunxian.
3.3 Analysis of the diversity of mycorrhizal fungi in macadamia roots
To objectively assess the alpha diversity of the AMF, we used the Chao1 and Observed_otus indices for richness, the Shannon and Simpson indices for diversity, and the dominance, Pielou_e and Goods coverage indices for the alpha diversity of the AMF (Supplementary Table S2). We statistically analyzed the differences in the Chao1, Shannon, and Simpson indices (Figure 4). The Chao1 index was highest in SF, followed by NR and CR, which is consistent with the Venn diagram results. The Shannon and Simpson indices also followed the same trend as the Chao1 index, with the Simpson index showing a significant difference among the three groups (p = 0.031) (Figure 4A). At the ChangNing sampling site, the situation was different from that at the YingJiang site, with the NR site having the lowest Chao1 index and the CR site having the highest Shannon and Simpson indices (Figure 4B). The Chao1 index at the LanCang sampling site was lower than that at the other three sites. Within the sampling site, the index was lowest for NR and highest for SF. The Shannon and Simpson indices were highest for SF, followed by CR, and lowest for NR (Figure 4C). The YunXian sampling site showed a similar trend to that of YingJiang, possibly due to their geographical proximity (Figure 4D). In conclusion, the various measures of alpha diversity were significantly correlated with the presence of AMF fungi in macadamia nuts and their geographic environment.
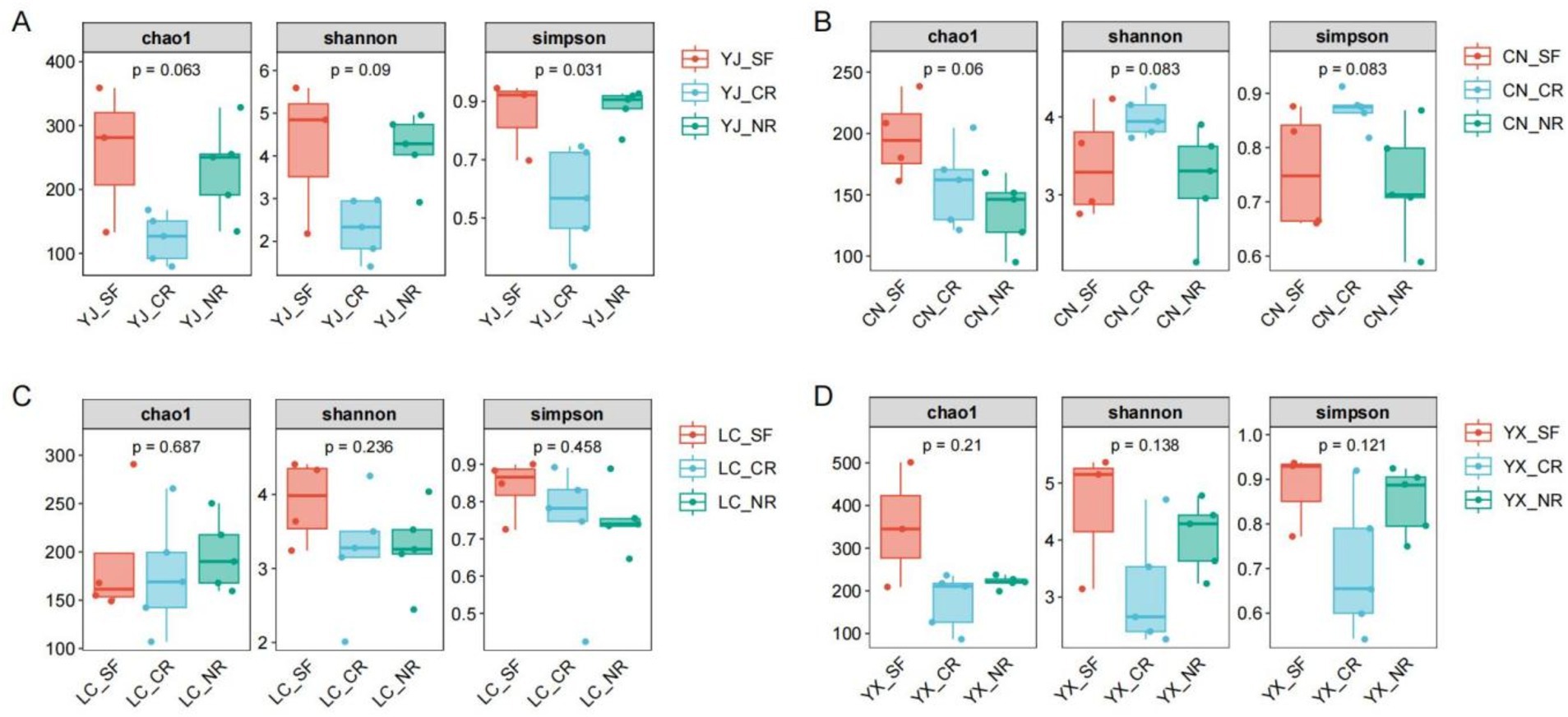
Figure 4. Analysis of alpha diversity of inter-root fungi of macadamia nuts in different regions. The box plots of Chao1, Shannon and Simpson indices of inter-root fungi in Yingjiang (A), Changning (B), Lancang (C) and Yunxian (D). Statistical differences determined using non-parametric testing Kruskal–Wallis test with 0.05 FDR Benjamini–Hochberg. SF, secondary forest; CR, cluster roots; NR, normal roots. CN, Changning; YJ, Yingjiang; LC, Lancang; YX, Yunxian.
To further explore the diversity of the AMF communities among the secondary forest, cluster, and normal root groups within each sampling site, we next conducted PCA analyses separately by region to distinguish between the groups (Figure 5). The PCA results revealed comparable trends for SF, CR, and NR across the four sampling sites. The interroot AMF communities in the secondary forests were significantly separated from those in the macadamia cluster and normal roots, while the sample distances between the cluster and normal roots were close. These results suggested that interroot AMF communities have a greater influence on host selection than interregional differences.
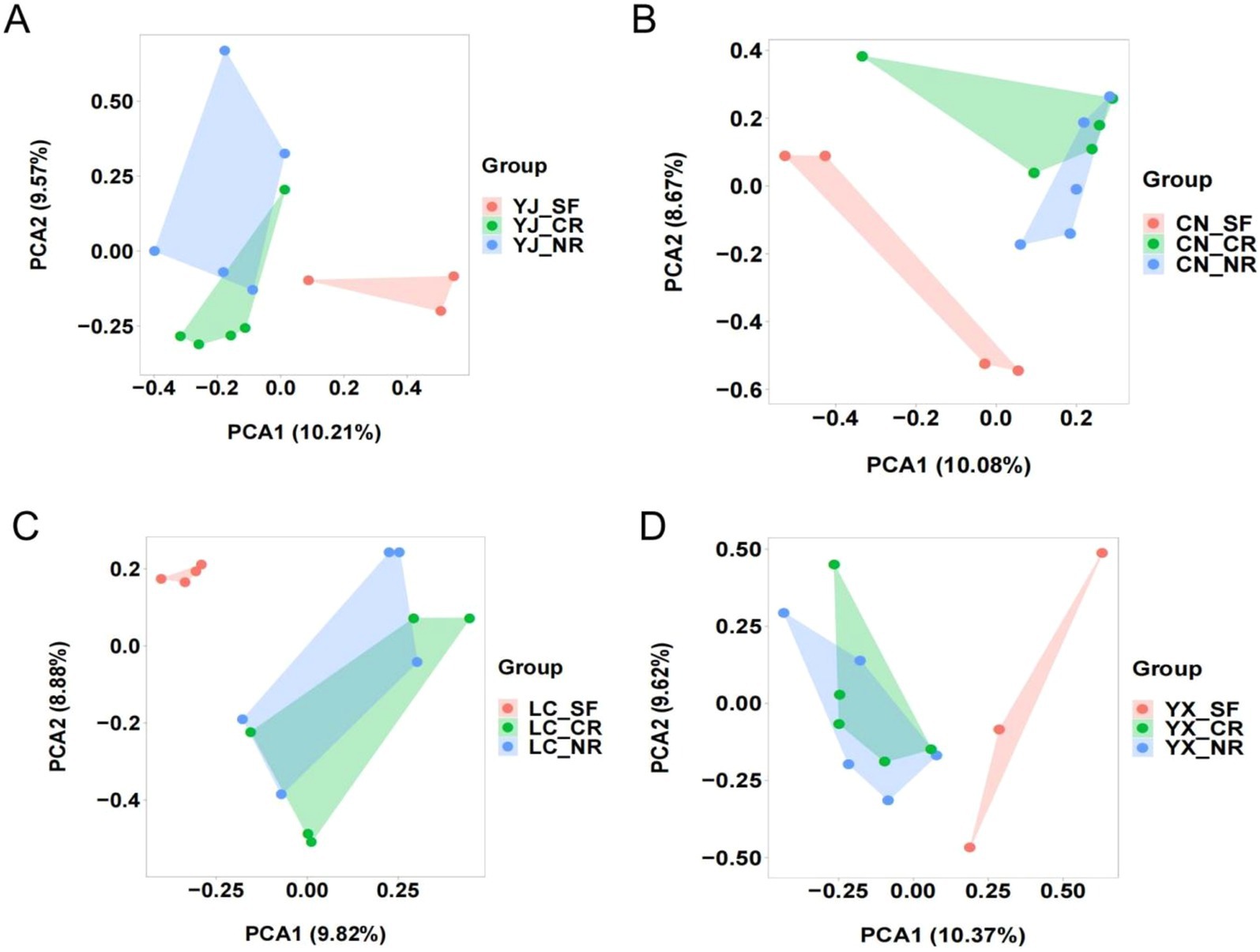
Figure 5. PCA analyses of inter-root fungi in secondary forests, cluster and normal roots in macadamia plantation areas in different regions. Each point represents a sample and different colours represent different groups. The four regions are Yingjiang (A), Changning (B), Lancang (C) and Yunxian (D). SF, secondary forest; CR, cluster roots; NR, normal roots. CN, Changning; YJ, Yingjiang; LC, Lancang; YX, Yunxian.
3.4 Analysis of the fungal community composition of macadamia roots
To analyze the most abundant AMF communities in secondary forests, cluster roots, and normal root systems, we examined the relative abundance of AMF at the phylum and genus levels (Figure 6). Figure 6A displays the top 30 fungi at the genus level, with Penicillium, Saitozyma, Talaromyces, Cladosporium, and Mycena being the five genera with the highest percentages of abundance. The 35 most abundant genera were distributed among four phyla: Ascomycota, Basidiomycota, Mortierellomycota, and Mucoromycota. Of these, 25 genera (71.43%) belonged to the phylum Ascomycota, while the Basidiomycota phylum contained 8 genera (22.86%). Moreover, Mortierellomycota and Mucoromycota had one genus each with a combined percentage of 5.71% (Figure 6B). Moreover, Ascomycota and Basidiomycota had the most significant impacts on the AMF in the interroots of the macadamia nuts. Additionally, the results suggested that the genera with the highest abundance differed among the groups, which also indicated a high degree of variability in the abundance of AMF among the different root types and environments.
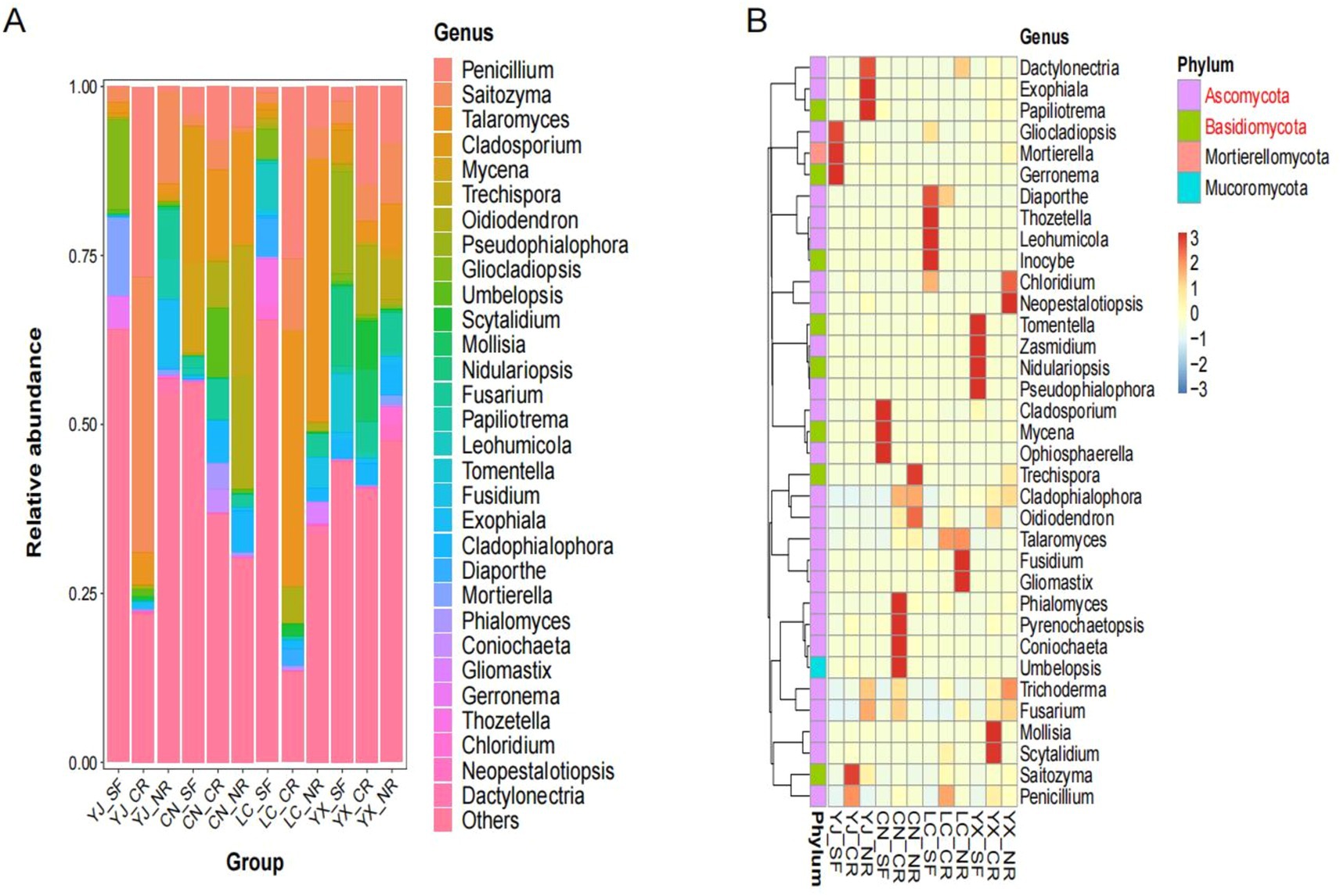
Figure 6. Abundance of fungal communities identified in different groups. (A) Species stacking diagram of fungi in genus level TOP30. (B) Heatmap of clustering of fungi in different groups in genus level TOP35 and the phylum to which they belong. SF, secondary forest; CR, cluster roots; NR, normal roots. CN, Changning; YJ, Yingjiang; LC, Lancang; YX, Yunxian.
3.5 Functional analysis of mycorrhizal fungal communities and correlation between different communities
We utilized the FunGuild database to obtain the ecological functions of the corresponding fungi based on the species classification of AMF. This research was performed to study the roles and functions these fungi play in ecology. We determined the functional abundance in the trophic mode and guild based on the annotation results of the database (Figure 7). The trophic mode categorizes organisms into three main groups: pathotrophs, symbiotrophs, and saprotrophs. These categories were interconnected, with saprotrophs having the highest percentage (Figure 7A). In contrast, according to the guild statistics, functional grouping types can be subdivided into animal pathogens, plantpathogens, foliar endophytes, etc. The most common functional type is Undefined_Saprotroph, followed by Endophyte−Plant_Pathogen. It can be clearly seen from the figure that there are obvious differences between the fungal functions of AMF in secondary forests and the functional classification of fungi in cluster roots and normal roots (Figure 7B).
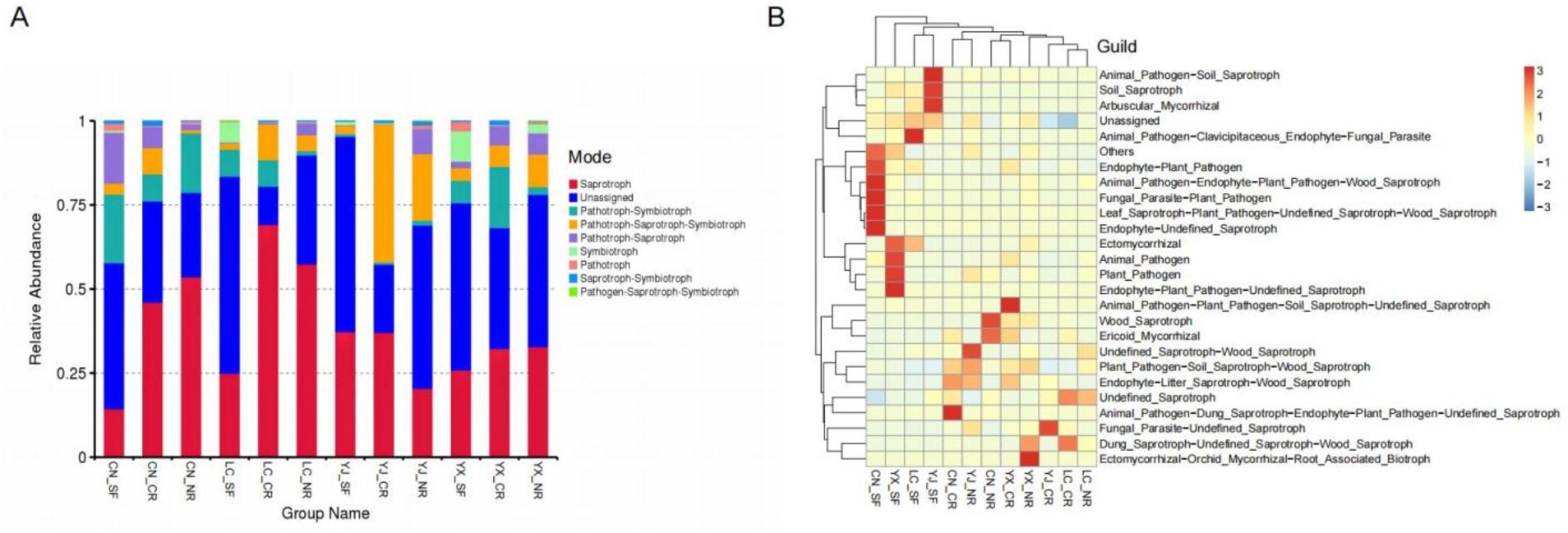
Figure 7. Variations in fungal functional groups compositions inferred by FUNGuild. The functional abundance of Trophic Mode (A) and Guild (B) were separately counted based on the database annotation results. The results were presented in bar stacked plots and clustered heatmaps. SF, secondary forest; CR, cluster roots; NR, normal roots. CN, Changning; YJ, Yingjiang; LC, Lancang; YX, Yunxian.
To better understand the relationships among the fungal genera associated with AMF, we analyzed the top 100 fungal genera by calculating the Spearman correlation coefficient (Figure 8). We then identified those with a correlation coefficient exceeding 0.6 or below −0.6, along with a statistically significant p value of less than 0.05. These significant connections were used to construct Sankey diagrams. This study revealed that genera associated with the fungal phyla Ascomycota and Basidiomycota had the greatest number of interactions, which was consistent with previous research. These interactions among various arbuscular mycorrhizal fungi highlight the complexity of the rhizosphere.
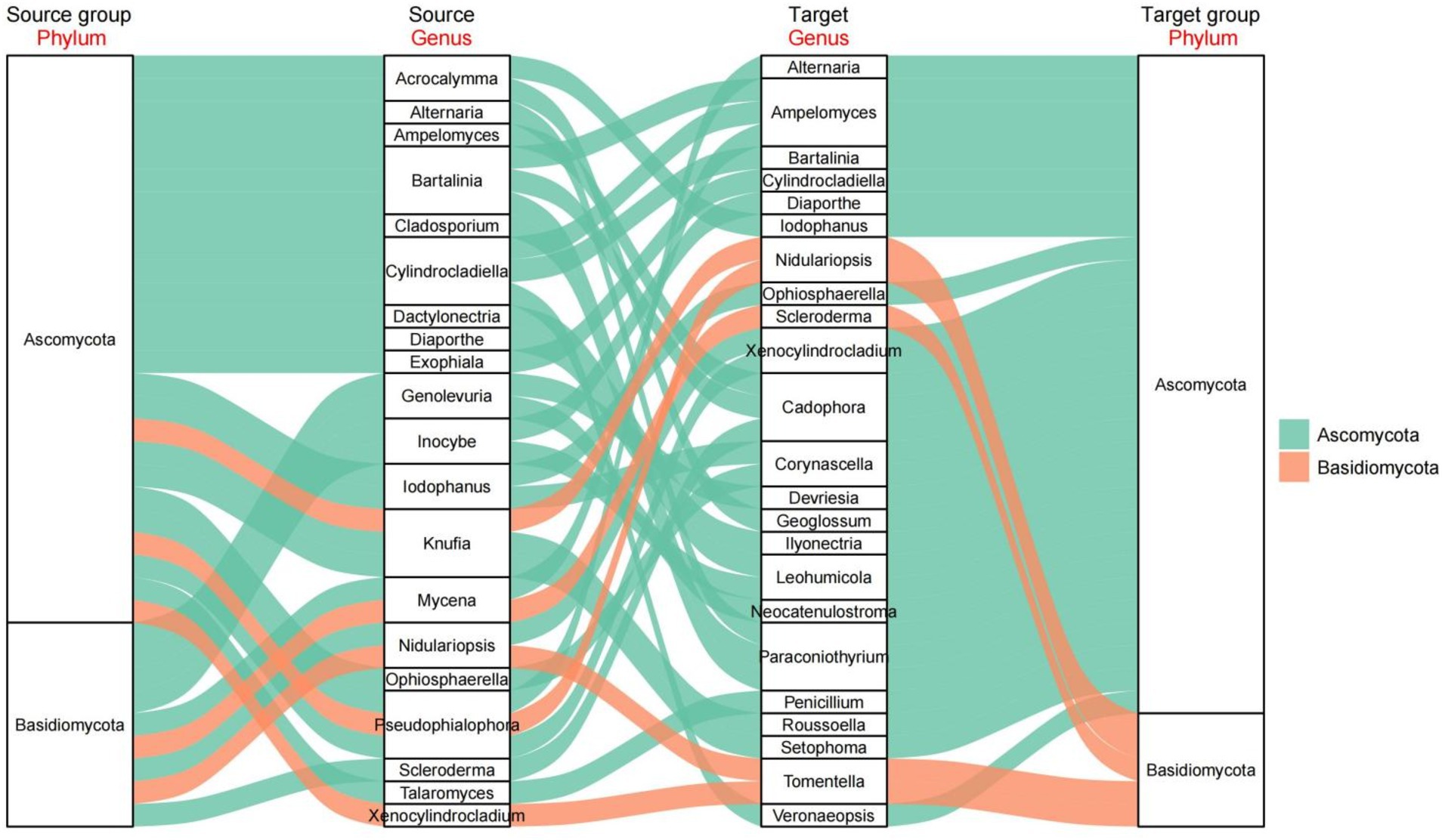
Figure 8. The Spearman correlation coefficients were calculated for the top 100 microbial genera. Valid connections for the drawing of Sankey diagrams were selected based on correlation coefficients greater than 0.6 or less than −0.6 and significant p-values (less than 0.05).
4 Discussion
4.1 The substantial potential value of AMF for macadamia nuts
Research has shown that AMF can reduce the need for external inputs and help maintain the sustainability of agroecosystems while stabilizing yields (Powell and Rillig, 2018). Arbuscular mycorrhizae (AMs) are symbiotic associations that can benefit more than 80% of terrestrial plants by expanding their root range and releasing organic acids and enzymes to improve nutrient uptake in the soil. This association can also increase plant resistance to biotic or abiotic stresses and improve ecosystems (Van Der Heijden, 2004; Bender et al., 2015; Jia et al., 2019; Gao et al., 2020). In nutrient-poor soils in the humid tropics, plants rely on AMF to form symbiotic relationships. AMF are believed to depend on the host plant for carbon fixation, while the plant receives various benefits, including increased uptake of nutrients, particularly immobile nutrients such as zinc and phosphorus (Sivakumar, 2013). However, there has been limited research conducted on the relationship between AMF and macadamia nuts. Previous studies have suggested that AMF may benefit macadamia nuts by increasing plant nutrient utilization efficiency. This is because AMF can enhance the ability of plant roots to absorb nutrients such as nitrogen, phosphorus, and potassium from the soil (Della Monica et al., 2015). AMF can improve the nutrient utilization efficiency of macadamia nut trees by facilitating the exchange of nutrients between the plant and the soil, which can increase nut yield and quality. Previous research has demonstrated that the inoculation of macadamia root systems with AMF can enhance seedling growth and increase the accumulation of proteins and sugars in the leaves. Furthermore, inoculation with AMF under water deficit conditions can regulate the content of soluble sugars and proline, thereby improving water availability in plant tissues, which can promote the growth of macadamia nuts (Yooyongwech et al., 2014). Second, they can enhance plant resistance to adverse conditions such as drought and salinity. For instance, under drought stress, AMF increase the uptake of trace elements, promote photosynthesis, and improve the drought tolerance of plants (Bowles et al., 2018). Additionally, salt stress can increase photosynthetic efficiency and water use by increasing K+/Na + ratios and the accumulation of compatible solutes such as proline and soluble carbohydrates (Porcel et al., 2012; Abdel Motaleb et al., 2020). Moreover, AMF can enhance microbial diversity in the soil surrounding roots. As a vital element of soil microorganisms, AMF can stimulate the reproduction and activity of other microorganisms in the soil (Qian et al., 2022). This can help maintain the ecological balance of the soil, improve soil fertility and biological activity, and provide a better environment for macadamia nut trees. The use of clumping fungi with biological control effects can effectively reduce the incidence of pests and diseases in macadamia nut trees, thereby improving both yield and nut quality. Reports have demonstrated that AMF activate the host to release relevant defense proteins, such as phenylalanine ammonia-lyase PAL, superoxide dismutase SOD, and PR proteins, during the process of establishing a symbiotic relationship with the host plant’s root system. This improves the host plant’s resistance to disease (Garmendia et al., 2006). Although research on the symbiotic relationship between AMF and macadamia nuts is still rather limited, and furthermore, the existence of a symbiotic relationship between them remains a matter of debate (Lambers and Teste, 2013), our results from AMF infection experiments on different types of roots in macadamia nut orchards across various regions have verified that AMF can indeed form a symbiotic relationship with macadamia nut roots (Figure 2). Consequently, we hypothesize that the enhancement in the yield and quality of macadamia nuts is attributable to these symbiotic AMF.
4.2 Diversity of the AMF communities in the different macadamia root systems
Macadamia nuts are a unique plant species with distinctive growth and root structures. One of these structures is root exclusion, which plays a significant role in plant growth. Root exclusion refers to the phenomenon of plant roots growing toward the surface or shallow soil layers, creating a distinct root arrangement. There is ongoing debate regarding whether AMF can colonize various types of macadamia nut root systems. In Australia, the country where macadamia nuts originate, the Solanum ophthalmicum plant, which includes macadamia nuts, was previously believed to be nonmycorrhizal (Lambers and Teste, 2013; Neumann and Martinoia, 2002). The formation of fungal spores is induced by the secretion of phenolic compounds, mainly isoflavones. This reduces fungal nutrient growth and the potential consumption of citrate, a carbon source (Weisskopf et al., 2006b). In addition, cluster roots secrete enzymes, such as chitinase and glucanase, which can degrade fungal cell walls and inhibit the growth of fungal communities (Cesco et al., 2010). However, not all plants with root-excluding systems have lost the ability to form symbiotic relationships with AMF. Recent studies have shown that some of these plants, such as Solanum ophthalmicum, are able to re-establish symbioses and interact with local AMFs when introduced to new environments (Tedersoo, 2017). For instance, macadamia nuts grown in fields in Bangladesh were found to have AMF colonization, which confirms the aforementioned findings (Bhuiyan et al., 2017). Our study confirmed that macadamia nut cluster roots can form a symbiotic relationship with AMF colonization, as was the case for both cluster roots and normal roots (Figure 2). High-throughput sequencing of interroot soils from secondary forests, macadamia cluster roots, and normal roots identified a total of 495 fungal species, representing 16.35% of the total. The highest number of fungal species was detected in secondary forests, followed by normal roots, and the lowest number was detected in cluster roots (Figure 3B). The alpha diversity of fungal communities in the macadamia cluster and normal roots from different regions varied (Figure 4). However, the PCA results consistently showed regional variability in microorganisms as well as host selectivity (Figure 5). Additionally, the functional classification provided by FUNGuild highlighted significant distinctions among the cluster roots, normal roots, and secondary forests of macadamia nuts. The fungal communities of the root cluster and normal roots displayed more closely related functions (Figure 7), which is consistent with the PCA findings.
4.3 Characteristics of the AMF fungal community composition and diversity of macadamia trees in different regions of Yunnan Province
This study analyzed the fungal community structure in various regions of macadamia plantations. The interroot soil AMFs in the four different habitats shared a total of 219 ASVs, which accounted for only 7.23% of all ASVs. These results suggest significant differences in the fungal community structure. The significant proportion of ASVs that are specific to particular regions (11.13–23.03%) indicates that soil fungi between roots exhibit a certain level of geographic specialization and host specificity (Figure 3A). The total number of microorganisms and the percentage of fungal populations in YunXian and ChangNing were similar. This similarity may be attributed to the geographic proximity of the two regions and the similarity of their soils, climates, and ecological environments (Figure 3). This study revealed that Ascomycetes and Basidiomycetes were the dominant groups of macadamia root fungi. Most of these fungi are terrestrial and capable of decomposing decayed plant and animal remains, as well as organic matter in the soil, releasing organic matter for plant uptake and utilization. The abundance of these fungi is related to soil fertility. Ascomycetes frequently form symbiotic mycorrhizae with plants, promoting their growth and development (Rosas-Medina et al., 2019). Ascomycetes and Basidiomycetes are also commonly found in soil (Bastias et al., 2006). Although the dominant fungal phyla of macadamia were similar across different regions, the composition and proportions of different geographic locations within the same rhizosphere type varied at the genus level. For example, in the cluster roots of ChangNing, the most abundant fungal genera were Trechispora, Cladophialophora, and Oidiodendron, while in YunXian, the most abundant genera were Chloridium, Neopestalotiopsis, and Trichoderma. The fungal genera that were dominant in LanCang were Talaromyces, Fusidium, and Gliomastix. In YingJiang, the dominant genera were Dactylonectria, Exophiala, and Papiliotrema. Overall, there was no overlap in the dominant fungal genera across these four areas (Figure 6B). The results for normal roots and secondary forests were similar to those for cluster roots. The findings of a previous study were consistent with the suggestion that the composition of interroot fungi varies considerably in different regions (Chen et al., 2022).
The results of the FUNGuild function prediction showed that the eight trophic types were unevenly distributed among the 12 groups, and the fungal functions of secondary forests were mainly concentrated in Pathotroph-Saprotroph, Symbiotroph, Pathotroph, and Pathogen-Saprotroph-Symbiotroph and showed regional specificity (Figure 7). The fungal functions of the cluster roots were primarily Saprotroph, Pathotroph-Saprotroph-Symbiotroph, and Saprotroph-Symbiotroph. In YunXian and ChangNing, the fungal function was mainly saprotrophic-symbiotrophic. The fungal trophic types of the normal and cluster macadamia roots were similar and consistent with the PCA results. Furthermore, this study revealed that the interroot fungal community functions of macadamia nuts in these four regions were mostly dominated by saprophytic trophic types, with a lower abundance of pathological trophic types. This suggested that the structural composition of the interroot fungal communities in the macadamia nut orchards in these regions was relatively healthy. Finally, the Spearman correlation coefficient calculation of the Sankey diagram clearly showed the complexity of the interroot AMF communities in macadamia nuts, which provides a reference for subsequent studies of the interactions among different AMF communities (Figure 8).
5 Conclusion
In this study, we employed high-throughput sequencing to examine the inter-root soils of four primary macadamia production zones in Yunnan Province. The results of the phenotypic characterization of root infection experiments indicated that both normal and cluster roots of macadamia nuts were capable of establishing symbiotic relationships with AMF. Furthermore, the diversity of fungi was found to be higher in secondary forests than in normal roots and lowest in cluster roots. There was a significant difference between regions, indicating that the environment exerts a considerable influence on inter-root fungi and AMF. The results of functional classification based on FUNGuild demonstrated that inter-root fungi exhibited notable differences among cluster roots, normal roots, and secondary forests of macadamia. The majority of the inter-root fungal communities of macadamia nuts in these regions were dominated by trophic types, indicating a relatively healthy structural composition. These findings contributed to a deeper understanding of the interactions between different types of macadamia nut root systems and AMF, and provided a valuable scientific basis for the sustainable development of the macadamia nut industry.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://db.cngb.org/, CNP0005527.
Author contributions
LG: Software, Validation, Writing – original draft. YN: Validation, Writing – review & editing. YC: Investigation, Writing – original draft. XH: Resources, Writing – original draft. ZW: Data curation, Writing – original draft. HY: Conceptualization, Visualization, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This research was funded by the National Natural Science Foundation of China (no. 32160743).
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1437577/full#supplementary-material
Footnotes
1. ^Unite database https://unite.ut.ee/.
References
Abdel Motaleb, N. A., Abd Elhady, S. A., and Ghoname, A. A. (2020). AMF and Bacillus megaterium neutralize the harmful effects of salt stress on bean plants. Gesunde Pflanz. 72, 29–39. doi: 10.1007/s10343-019-00480-8
Aradhya, M. K., Yee, L. K., Zee, F. T., and Manshardt, R. M. (1998). Genetic variability in macadamia. Genet. Resour. Crop. Evol. 45, 19–32. doi: 10.1023/A:1008634103954
Avio, L., Turrini, A., Giovannetti, M., and Sbrana, C. (2018). Designing the Ideotype mycorrhizal symbionts for the production of healthy food. Front. Plant Sci. 9:1089. doi: 10.3389/fpls.2018.01089
Bastias, B. A., Xu, Z., and Cairney, J. W. (2006). Influence of long-term repeated prescribed burning on mycelial communities of ectomycorrhizal fungi. New Phytol. 172, 149–158. doi: 10.1111/j.1469-8137.2006.01793.x
Bender, S. F., Conen, F., and Van der Heijden, M. G. A. (2015). Mycorrhizal effects on nutrient cycling, nutrient leaching and N2O production in experimental grassland. Soil Boil Biochem. 80, 283–292. doi: 10.1016/j.soilbio.2014.10.016
Bhuiyan, M. A. H., Banu, M. B., and Rahman, M. (2017). Assessment of arbuscular mycorrhizal association in some fruit and spice plants of rangamati hill district. Bangladesh J. Agril. Res. 42, 221–232. doi: 10.3329/bjar.v42i2.32810
Bowles, T. M., Jackson, L. E., and Cavagnaro, T. R. (2018). Mycorrhizal fungi enhance plant nutrient acquisition and modulate nitrogen loss with variable water regimes. Glob. Chang. Biol. 24, e171–e182. doi: 10.1111/gcb.13884
Cesco, S., Neumann, G., Tomasi, N., Pinton, R., and Weisskopf, L. (2010). Release of plant-borne flavonoids into the rhizosphere and their role in plant nutrition. Plant Soil 329, 1–25. doi: 10.1007/s11104-009-0266-9
Chen, Y., Chen, J., Chen, R., Xiao, L., Wu, X., Hu, L., et al. (2022). Comparison of the fungal community, chemical composition, antioxidant activity, and taste characteristics of Fu brick tea in different regions of China. Front. Nutr. 9:900138. doi: 10.3389/fnut.2022.900138
Chen, F. Z., You, L. J., Yang, F., Wang, L. N., Guo, X. Q., Gao, F., et al. (2020). CNGBdb: China national GeneBank DataBase. Yi Chuan 42, 799–809. doi: 10.16288/j.yczz.20-080
Dawkins, K., and Esiobu, N. (2017). Arbuscular and ectomycorrhizal Fungi associated with the invasive Brazilian pepper tree (Schinus terebinthifolius) and two native plants in South Florida. Front. Microbiol. 8:665. doi: 10.3389/fmicb.2017.00665
Della Monica, I. F., Saparrat, M. C. N., Godeas, A. M., and Scervino, J. M. (2015). The co-existence between DSE and AMF symbionts affects plant P pools through P mineralization and solubilization processes. Fungal Ecol. 17, 10–17. doi: 10.1016/j.funeco.2015.04.004
Gao, X., Guo, H., Zhang, Q., Guo, H., Zhang, L., Zhang, C., et al. (2020). Arbuscular mycorrhizal fungi (AMF) enhanced the growth, yield, fiber quality and phosphorus regulation in upland cotton (Gossypium hirsutum L.). Sci. Rep. 10:2084. doi: 10.1038/s41598-020-59180-3
Garg, M. L., Blake, R. J., and Wills, R. B. H. (2003). Macadamia nut consumption lowers plasma total and LDL cholesterol levels in hypercholesterolemic men. J. Nutr. 133, 1060–1063. doi: 10.1093/jn/133.4.1060
Garg, M. L., Blake, R. J., Wills, R. B., and Clayton, E. H. (2007). Macadamia nut consumption modulates favourably risk factors for coronary artery disease in hypercholesterolemic subjects. Lipids 42, 583–587. doi: 10.1007/s11745-007-3042-8
Garmendia, I., Aguirreolea, J., and Goicoechea, N. (2006). Defence-related enzymes in pepper roots during interactions with arbuscular mycorrhizal fungi and/or Verticillium dahliae. BioControl 51, 293–310. doi: 10.1007/s10526-005-4238-6
Giovannetti, M., and Mosse, B. (1980). An evaluation of techniques for measuring vesicular arbuscular mycorrhizal infection in roots. New Phytol. 84, 489–500. doi: 10.1111/j.1469-8137.1980.tb04556.x
Guo, X., Chen, F., Gao, F., Li, L., Liu, K., You, L., et al. (2020). CNSA: a data repository for archiving omics data. Database (Oxford) 2020:baaa055. doi: 10.1093/database/baaa055
Haas, B. J., Gevers, D., Earl, A. M., Feldgarden, M., Ward, D. V., Giannoukos, G., et al. (2011). Chimeric 16S rRNA sequence formation and detection in sanger and 454-pyrosequenced PCR amplicons. Genome Res. 21, 494–504. doi: 10.1101/gr.112730.110
Hardner, C. M., Peace, C., Lowe, A. J., Neal, J., and Williams, K. (2009). “Genetic resources and domestication of Macadamia” in Horticultural Reviews. ed. J. Janick (New York: John Wiley & Sons, Inc.), 1–125.
Jia, T., Wang, J., Chang, W., Fan, X., Sui, X., and Song, F. (2019). Proteomics analysis of E. angustifolia seedlings inoculated with arbuscular mycorrhizal Fungi under salt stress. Int. J. Mol. Sci. 20:788. doi: 10.3390/ijms20030788
Lambers, H., and Teste, F. P. (2013). Interactions between arbuscular mycorrhizal and non-mycorrhizal plants: do non-mycorrhizal species at both extremes of nutrient availability play the same game? Plant Cell Environ. 36, 1911–1915. doi: 10.1111/pce.12117
Lekberg, Y., Gibbons, S. M., Rosendahl, S., and Ramsey, P. W. (2013). Severe plant invasions can increase mycorrhizal fungal abundance and diversity. ISME J. 7, 1424–1433. doi: 10.1038/ismej.2013.41
Lyu, F. Y., Han, F. R., Ge, C. L., Mao, W. K., Chen, L., Hu, H. P., et al. (2023). OmicStudio: a composable bioinformatics cloud platform with real-time feedback that can generate high-quality graphs for publication. iMeta 2:e85. doi: 10.1002/imt2.85
Magoč, T., and Salzberg, S. L. (2011). FLASH: fast length adjustment of short reads to improve genome assemblies. Bioinformatics 27, 2957–2963. doi: 10.1093/bioinformatics/btr507
Maguire, L. S., O’Sullivan, S. M., Galvin, K., O’Connor, T. P., and O’Brien, N. M. (2004). Fatty acid profile, tocopherol, squalene and phytosterol content of walnuts, almonds, peanuts, hazelnuts and the macadamia nut. Int. J. Food Sci. Nutr. 55, 171–178. doi: 10.1080/09637480410001725175
McGonigle, T. P., Miller, M. H., Evans, D. G., Fairchild, G. L., and Swan, J. A. (1990). A new method which gives an objective measure of colonization of roots by vesicular–arbuscular mycorrhizal fungi. New Phytol. 115, 495–501. doi: 10.1111/j.1469-8137.1990.tb00476.x
Mo, Y., Wang, Y., Yang, R., Zheng, J., Liu, C., Li, H., et al. (2016). Regulation of plant growth, photosynthesis, Antioxidation and osmosis by an arbuscular mycorrhizal fungus in watermelon seedlings under well-watered and drought conditions. Front. Plant Sci. 7:644. doi: 10.3389/fpls.2016.00644
Navarrete, A. A., Tsai, S. M., Mendes, L. W., Faust, K., de Hollander, M., Cassman, N. A., et al. (2015). Soil microbiome responses to the short-term effects of Amazonian deforestation. Mol. Ecol. 24, 2433–2448. doi: 10.1111/mec.13172
Neumann, G., and Martinoia, E. (2002). Cluster roots-an underground adaptation for survival in extreme environments. Trends Plant Sci. 7, 162–167. doi: 10.1016/S1360-1385(02)02241-0
Nguyen, N. H., Song, Z., Bates, S. T., Branco, S., Tedersoo, L., Menke, J., et al. (2016). FUNGuild: an open annotation tool for parsing fungal community datasets by ecological guild. Fungal Ecol. 20, 241–248. doi: 10.1016/j.funeco.2015.06.006
Nock, C. J., Hardner, C. M., Montenegro, J. D., Ahmad Termizi, A. A., Hayashi, S., Playford, J., et al. (2019). Wild origins of Macadamia domestication identified through intraspecific chloroplast genome sequencing. Front. Plant Sci. 10:334. doi: 10.3389/fpls.2019.00334
Paudel, S., Baer, S. G., and Battaglia, L. L. (2014). Arbuscular mycorrhizal fungi (AMF) and success of Triadica sebifera invasion in coastal transition ecosystems along the northern Gulf of Mexico. Plant Soil 378, 337–349. doi: 10.1007/s11104-014-2026-8
Pei, Y., Siemann, E., Tian, B., and Ding, J. (2020). Root flavonoids are related to enhanced AMF colonization of an invasive tree. AoB Plants 12:plaa002. doi: 10.1093/aobpla/plaa002
Phillips, J. M., and Hayman, D. S. (1970). Improved procedures for clearing roots and staining parasitic and vesicular-arbuscular mycorrhizal fungi for rapid assessment of infection. Trans. Br. Mycol. Soc. 55, 158–IN18. doi: 10.1016/S0007-1536(70)80110-3
Porcel, R., Aroca, R., and Ruiz-Lozano, J. M. (2012). Salinity stress alleviation using arbuscular mycorrhizal fungi. A review. Agron Sustain Dev. 32, 181–200. doi: 10.1007/s13593-011-0029-x
Powell, J. R., and Rillig, M. C. (2018). Biodiversity of arbuscular mycorrhizal fungi and ecosystem function. New Phytol. 220, 1059–1075. doi: 10.1111/nph.15119
Qian, Y., Zhao, G., Zhou, J., Zhao, H., Mutter, T. Y., and Huang, X. (2022). Combined bioremediation of Bensulfuron-methyl contaminated soils with arbuscular mycorrhizal fungus and Hansschlegelia zhihuaiae S113. Front. Microbiol. 13:843525. doi: 10.3389/fmicb.2022.843525
Redecker, D., Kodner, R., and Graham, L. E. (2000). Glomalean fungi from the Ordovician. Science 289, 1920–1921. doi: 10.1126/science.289.5486.1920
Remy, W., Taylor, T. N., Hass, H., and Kerp, H. (1994). Four hundred–million–year–old vesicular arbuscular mycorrhizae. Proc. Natl. Acad. Sci. U. S. A. 91, 11841–11843. doi: 10.1073/pnas.91.25.11841
Rosas-Medina, M., Maciá-Vicente, J. G., and Piepenbring, M. (2019). Diversity of Fungi in soils with different degrees of degradation in Germany and Panama. Mycobiology 48, 20–28. doi: 10.1080/12298093.2019.1700658
Ruiz-Lozano, J. M., Aroca, R., Zamarreño, Á. M., Molina, S., Andreo-Jiménez, B., Porcel, R., et al. (2016). Arbuscular mycorrhizal symbiosis induces strigolactone biosynthesis under drought and improves drought tolerance in lettuce and tomato. Plant Cell Environ. 39, 441–452. doi: 10.1111/pce.12631
Savary, R., Dupuis, C., Masclaux, F. G., Mateus, I. D., Rojas, E. C., and Sanders, I. R. (2020). Genetic variation and evolutionary history of a mycorrhizal fungus regulate the currency of exchange in symbiosis with the food security crop cassava. ISME J. 14, 1333–1344. doi: 10.1038/s41396-020-0606-6
Sivakumar, N. (2013). Effect of edaphic factors and seasonal variation on spore density and root colonization of arbuscular mycorrhizal fungi in sugarcane fields. Ann. Microbiol. 63, 151–160. doi: 10.1007/s13213-012-0455-2
Tedersoo, L. (2017). “Global biogeography and invasions of ectomycorrhizal plants:past, present and future” in Biogeography of mycorrhizal symbiosis. ed. L. Tedersoo (Cham, Switzerland: Springer International), 469–531.
Topp, B. L., Nock, C. J., Hardner, C. M., Alam, M., and O’connor, K. M. (2019). “Macadamia (Macadamia spp.) breeding” in Advances in plant breeding strategies: Nut and beverage crops. eds. J. Al-Khayri, S. Jain, and D. Johnson (Cham: Springer), 221–251.
Torrecillas, E., del Mar Alguacil, M., Roldán, A., Díaz, G., Montesinos-Navarro, A., and Torres, M. P. (2014). Modularity reveals the tendency of arbuscular mycorrhizal fungi to interact differently with generalist and specialist plant species in gypsum soils. Appl. Environ. Microbiol. 80, 5457–5466. doi: 10.1128/AEM.01358-14
Van Der Heijden, M. G. A. (2004). Arbuscular mycorrhizal fungi as support systems for seedling establishment in grassland. Ecol. Lett. 7, 293–303. doi: 10.1111/j.1461-0248.2004.00577.x
Wall, M. M. (2010). Functional lipid characteristics, oxidative stability, and antioxidant activity of macadamia nut (Macadamia integrifolia) cultivars. Food Chem. 121, 1103–1108. doi: 10.1016/j.foodchem.2010.01.057
Weisskopf, L., Abou-Mansour, E., Fromin, N., Tomasi, N., Santelia, D., Edelkott, I., et al. (2006b). White lupin has developed a complex strategy to limit microbial degradation of secreted citrate required for phosphate acquisition. Plant Cell Environ. 29, 919–927. doi: 10.1111/j.1365-3040.2005.01473.x
Weisskopf, L., Tomasi, N., Santelia, D., Martinoia, E., Langlade, N. B., Tabacchi, R., et al. (2006a). Isoflavonoid exudation from white lupin roots is influenced by phosphate supply, root type and cluster-root stage. New Phytol. 171, 657–668. doi: 10.1111/j.1469-8137.2006.01776.x
White, T. J., Bruns, T. D., Lee, S. B., and Taylor, J. W. (1990). Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. Pcr Protocols 1, 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Xia, Y., Chen, F., Du, Y., Liu, C., Bu, G., Xin, Y., et al. (2019). A modified SDS-based DNA extraction method from raw soybean. Biosci. Rep. 39:BSR20182271. doi: 10.1042/BSR20182271
Yooyongwech, S., Phaukinsang, N., Cha-um, S., and Supaibulwatana, K. (2014). Arbuscular mycorrhiza improved growth performance in Macadamia tetraphylla L. grown under water deficit stress involves soluble sugar and proline accumulation. Plant Growth Regul. 69, 285–293. doi: 10.1007/s10725-012-9771-6
Zhang, F. J., Li, Q., Chen, F. X., Xu, H. Y., Inderjit, I., and Wan, F. H. (2017). Arbuscular mycorrhizal fungi facilitate growth and competitive ability of an exotic species Flaveria bidentis. Soil Biol. Biochem. 115, 275–284. doi: 10.1016/j.soilbio.2017.08.019
Keywords: Macadamia integrifolia, AMF, high-throughput sequencing, cluster root, FUNGuild
Citation: Gong L, Ning Y, Chen Y, He X, Wu Z and Yue H (2024) Combined cellular phenotyping and high-throughput sequencing analysis reveals the symbiotic relationships between different types of macadamia root systems and arbuscular mycorrhizal fungi. Front. Sustain. Food Syst. 8:1437577. doi: 10.3389/fsufs.2024.1437577
Edited by:
Duraisamy Saravanakumar, The University of the West Indies St. Augustine, Trinidad and TobagoReviewed by:
Debasis Mitra, Graphic Era University, IndiaJadson Belem De Moura, Evangelical School of Goianésia, Brazil
Copyright © 2024 Gong, Ning, Chen, He, Wu and Yue. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Hai Yue, aGFpeXVlMjAyNEAxMjYuY29t
 Lidan Gong1,2
Lidan Gong1,2 Hai Yue
Hai Yue