- 1Institute of Microbiology, Government College University, Faisalabad, Pakistan
- 2Department of Zoology, Government College University, Faisalabad, Pakistan
- 3Department of Biochemistry, Government College University, Faisalabad, Pakistan
Probiotic properties of isolated lactic acid bacteria (LAB) from sustainable foods including camel milk are the potential research domains. For this purpose, camel milk samples (n = 20), from four different herds of Camelus dromedarius, were processed for the identification of LAB strains based on biochemical profiles followed by amplification and sequence analysis of the 16S rRNA gene. The probiotic characteristics, i.e., acids and bile salts tolerance, antimicrobial susceptibility profiles, hemolytic and antimicrobial activities, auto-aggregation assay, and adhesion to HT-29 epithelial cells were determined. Thirteen out of 20 milk samples were initially found positive for the growth of probiotics or LAB which were further confirmed as Lacticaseibacillus casei (5) and Pediococcus pentosaceus (3). The probiotics/LAB strains showed maximum survival (%) = 92.06 ± 1.82 and 81.35 ± 3.64 against acids and bile salts, respectively. The LAB strains were found sensitive to amoxicillin, ceftazidime, imipenem, linezolid, ofloxacin, tetracycline, tobramycin, and vancomycin. None of the LAB strains showed hemolytic activity. L. casei-04 strain showed a maximum zone of inhibition (15.33 ± 0.58) against multidrug-resistant E. coli AZ1 strain whereas, L. casei-05 showed a maximum zone of inhibition (16.33 ± 1.15) against methicillin-resistant S. aureus Saba-1 strain. L. casei-03 showed maximum percentage auto-aggregation (28.65 ± 1.96) at 4 h while L. casei-01 showed (41.10 ± 3.03) at 24 h of incubation. Maximum adhesion was shown by P. pentosaceus-01 (11.14%) followed by L. casei-02 (9.73%). Altogether, the current findings suggested that camel milk has significant potential of providing probiotics/LAB strains into human food chain and enabling camel milk as potential sustainable food.
1 Introduction
The term “Probiotics” is defined as “viable, non-pathogenic microorganisms which when administered in adequate amounts confer a health benefit on the host” (Khurshid and Akash, 2020; Anwar et al., 2021). Dairy products including milk, yogurt, and cheese are considered conventional and sustainable sources of different probiotics and lactic acid bacteria (LAB). Camel milk or its fermented products are potential research areas for the isolation and identification of probiotics/LAB strains (Shori, 2017) that could be considered as classical example of sustainable foods. Historically, the camels have a pivotal role in the cultural and economic developments of several communities, i.e., the Arabian and Middle East regions of the world (Burger et al., 2019). Camel milk is also useful as biomedicine against different clinical conditions including generalized edema, asthma, jaundice, diabetes, anemia, and piles (El-Fakharany et al., 2017; Behrouz et al., 2022). The bacteriocin-producing LAB strains from camel milk are recently discussed (Rahmeh et al., 2019). The proteins and peptide molecules from camel milk contribute to different biological pathways including digestion, intestinal absorption, gut immunity, and generalized growth of the individual (Rahmeh et al., 2019; Swelum et al., 2021). For example, lactoferrin is one of the defense proteins of camel milk which regulates iron metabolism along with induction and modulation of the immune system (Mahala et al., 2022). Some of the previous studies reported the antimicrobial properties of camel milk against different pathogens including parasites, fungi, and bacteria (Swelum et al., 2021).
Further, camel milk is also an enriched source of probiotics which can ferment carbohydrates and produce lactic acid. The probiotics/LAB strains of camel milk consisted of different species of Lacticaseibacillus, Leuconostoc, Lactococcus, Pediococcus, and Bifidobacterium which have a beneficial impact on human health (El-Zahar et al., 2021; Mahala et al., 2022). The probiotics/LAB strains are resistant to gastric pH and bile salts and are generally recognized as safe (GRAS) microorganisms that are safe to use in human or veterinary medicine (Rahmeh et al., 2019; Afzal et al., 2020). The LAB strains from camel milk can inhibit the growth of different bacterial pathogens by producing different antimicrobial compounds, i.e., hydrogen peroxide, organic acids, and bacteriocins. The LAB strains adjust the intestinal microbial balance and inhibit the adhesion of pathogenic bacteria to the intestinal epithelium, promote digestion, boost immune function and confer resistance to different infections (Azizi et al., 2017; Rahmeh et al., 2019). This study was designed to address the gap regarding the scarcity of global data on the isolation and in vitro assessment of potential probiotic or LAB from camel milk that represents a promising component of sustainable food systems. In vitro assessment, that was conducted in the current study includes molecular analysis, acid and bile salt tolerance assays, antibiotic susceptibility profiles, hemolytic activity, antimicrobial activity, auto-aggregation assay and adhesion to human cell line of the isolated LAB strains.
2 Methods
2.1 Sample collection and initial isolation
The milk samples (n = 20) were collected in duplicates from four different camel herds (Camelus dromedarius) using 15 mL falcon tubes (MTC-Bio, San Diego, USA) from District Faisalabad-Pakistan. The samples were immediately transported to the research laboratory under temperature-controlled conditions using ice-chest and were processed within 24 h of collection. 100 μL of each sample was inoculated on de Man Rogosa and Sharpe agar (MRS agar) (Oxoid-UK) and incubated in the anaerobic chamber (Oxoid-UK) at 37°C. The bacterial growth was recorded after 72–96 h and morphological characteristics were recorded. Each type of bacterial growth was processed separately. Pure bacterial cultures were stored using MRS broth (Oxoid-UK) supplemented with 20–30% glycerol (Oxoid-UK) at −80°C (Kabir et al., 2020).
2.2 Morphological and biochemical characteristics
The bacterial growth was initially identified based on cultural, morphological and biochemical characteristics of each sample. For this purpose, the stained smears were examined under a light microscope (IRMECO, Germany) at 100X. The isolates were processed for biochemical characteristics according to the standard protocols established by the American Society for Microbiology (ASM), i.e., catalase, indole, oxidase, methyl red, triple sugar iron, and Voges-Proskauer test (Kabir et al., 2020; Waheed et al., 2021).
2.3 Molecular analysis of the isolates
Initially identified isolates were further screened on molecular basis by extracting the DNA from purified cultures using the commercially available DNA extraction kit (GeneJET Genomic DNA Purification Kit, Thermo Scientific, United Kingdom) as described by the manufacturer. The quantification of bacterial DNA was performed with Colibri Micro volume Spectrophotometer (Titertek-Berthold, Germany). 16S rRNA gene was amplified using the universal primers: 27F (5´-AGAGTTTGATCMTGGCTCAG-3′) and 1492R (5´-TACGGYTACCTTGTTACGACTT-3′). The amplification reaction was performed in total 25 μL reaction volume [containing 1 μL of each forward and reverse primers, 12.5 μL of Master Mix (TaqMan ™, ThermoFisher Scientific), and 1 μL of genomic DNA] for 40 cycles using a Thermal cycler (BIO-RAD, T100™ Thermal Cycler, California). The amplified product was subjected to electrophoresis using 1.5% agarose with 1X Tris-EDTA buffer and stained with ethidium bromide (Kabir et al., 2020; Waheed et al., 2021). The purified PCR product was dispatched to Macrogen™, Korea for sequencing, and the sequences were analyzed and compared with the existing GenBank database.1 Molecular Evolutionary Genetic Analysis software (Mega-X) was used for analysis as described (Swelum et al., 2021; Waheed et al., 2021).
2.4 Tolerance to acids and bile salts
The bacterial isolates were processed for the evaluation of the tolerance to acids and bile salts according to the standard protocols. Briefly, in the acid tolerance test, bacterial cultures were first incubated overnight in 100 mL of MRS broth (Oxoid-UK) at 37°C. The pH of fresh MRS broth was adjusted to 1.5 using 0.2 N HCl and inoculated with bacterial cultures and incubated for 5–7 h at 37°C. The pH of the control MRS broth was adjusted to 6.5 which was also inoculated with bacterial cultures. Afterward, each inoculation was spread on MRS agar plates (Oxoid-UK) and incubated at 37°C for 48–72 h. For the bile tolerance test, MRS agar (Oxoid-UK) was prepared and supplemented with 1.5% (W/V) bile salts (Oxoid-UK) along with control MRS agar (without bile salts). After solidification, the bacterial cultures were streaked and incubated with similar conditions (Tambekar and Bhutada, 2010). Survival rates were measured by counting the Log CFU/mL by the given formula:
where Log CFUT = Log CFU/mL at time and Log CFUI = Log CFU/mL at initial time.
2.5 Antibiotic susceptibility testing
The bacterial isolates were examined for the antibiotic susceptibility profile by the Kirby-Bauer disc diffusion method (Hudzicki, 2009) on Muller Hinton Agar (Oxoid-UK). Following antibiotic discs (Oxoid-UK) were used to determine the antibiotic susceptibility profiles, i.e., Amoxicillin (AMC-30 μg), Ceftazidime (CAZ-30 μg), Imipenem (IPM-10 μg), Linezolid (LZD-10 μg), Ofloxacin (OFX-5 μg), Tetracycline (TET-30 μg), Tobramycin (TOB-10 μg), and Vancomycin (VA-5 μg). The discs were placed onto the agar and incubated overnight at 37°C. Later, the diameters of clear zones around the discs were measured and the results were demonstrated in terms of sensitive/intermediate/resistant according to the guidelines of the Clinical Laboratory Standards Institute (CLSI-2016).
2.6 Hemolytic activity
For the determination of hemolytic activity, Columbia agar (Oxoid, UK) was prepared followed by supplementation with 5% sheep blood. The bacterial isolates were streaked on agar plates and incubated at 37°C for 24 h. Hemolysis was noted as a greenish zone (α-hemolysis), clear zone (β-hemolysis), or no clear zone (γ-hemolysis) as described (Jang et al., 2019). Staphylococcus aureus Saba-1 (NCBI GenBank Number = MN453615.1) was used as the positive control.
2.7 Antimicrobial activity of LAB
The bacterial isolates were analyzed for the antimicrobial activity against multidrug-resistant E. coli AZ1 strain (NCBI GenBank Number = MF185146.1) and methicillin-resistant S. aureus Saba-1 strain (NCBI GenBank Number = MN453615.1) according to the recently described protocol with a slight modification of Tryptic Soy agar (TSA) (Oxoid, UK) preparation (Jang et al., 2019). Briefly, 3 μL of the overnight incubated LAB cultures were spotted on freshly prepared MRS agar plates and incubated anaerobically for 24 h at 37°C. In the next step, 100 μL overnight incubated cultures of described E. coli and S. aureus were inoculated into TSA soft agar, and the soft agar was overlaid. The plates were incubated for 24 h at 37°C. The zones of inhibition (mean ± SD) were recorded.
2.8 Auto-aggregation assay
The percentage auto-aggregation was measured (with some modifications) as described (Jang et al., 2019). Briefly, overnight cultured bacteria were centrifuged at 14000 g and the pellet was washed twice with PBS. The initial absorbance (A0) was adjusted to 0.3 ± 0.05 and 5 mL of bacterial suspension was incubated at 37\u00B0C. The time lapsed absorbance (AT) was measured at 0, 4, and 24 h, and the percentage auto-aggregation was estimated as:
where A0 = initial absorbance and AT = absorbance at a specific time.
2.9 Adhesion to HT-29 cells
The adhesion ability of LAB strains was estimated according to the recently described protocol using the HT-29 cell line (Jang et al., 2019) with a slight modification of initial bacterial count (Log CFU/mL = 6.7 ± 0.1) followed by 2 h of incubation at 37°C.
2.10 Statistical analysis
The mean ± SD was calculated using a Microsoft Excel spreadsheet, further all the procedures were conducted in triplicates.
3 Results
3.1 Initial identification of isolates
A total of 13 camel milk samples were found positive for characteristics LAB growth, whereas 7 samples did not show any characteristics LAB growth up to 6 days of incubation or characteristics biochemical profiles (hence, excluded from the study). The bacterial colonies were observed as round, smooth and creamy white with raised entire margins. Further, microscopically, all the isolates were observed as Gram-positive. The isolates were initially identified as Lacticaseibacillus (n = 5), Pediococcus (n = 3), Enterococcus (n = 3), and Bacillus (n = 2) based on microscopic/biochemical profiles. Enterococcus and Bacillus were also excluded from the current study.
3.2 Molecular identification of the isolates
The amplification of 16S rRNA of each isolate produced a single band of about ~1,400–1,500 bp product which corresponds to the size of the 16S rRNA gene. Further, the sequence analysis of the bacterial isolates resulted in the identification of LAB strains, i.e., Lacticaseibacillus casei (n = 5) and Pediococcus pentosaceus (n = 3), whereas Enterococcus faecium (n = 3) and Bacillus aerophilus (n = 2) were excluded from the study.
3.3 Acid and bile tolerance test
The result of acid tolerance (%) data showed that isolated LAB strains tolerated pH 1.5. The L. casei-01 showed a maximum (92.06 ± 1.82) while L. casei-02 showed the least survival rate (77.38 ± 1.19), as shown in Figure 1. The bile tolerance (%) data showed that isolated LAB strains tolerated the 1.5% (W/V) bile salts. The L. casei-01 showed a maximum (81.35 ± 3.64) while L. casei-04 showed the least survival rate (77.78 ± 0.69), as shown in Figure 2.
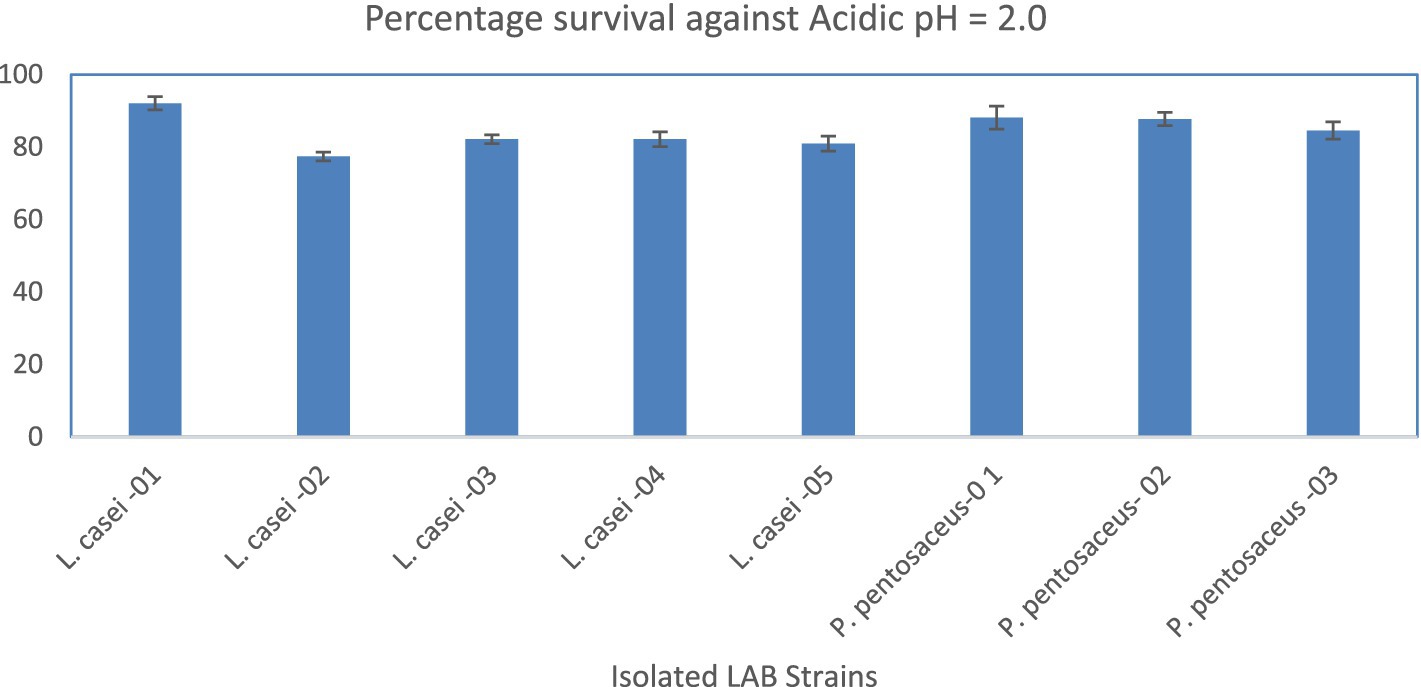
Figure 1. Percentage survival of the LAB strains against acidic pH (1.5). X-axis = Isolates, Y-axis = Percentage Survival.
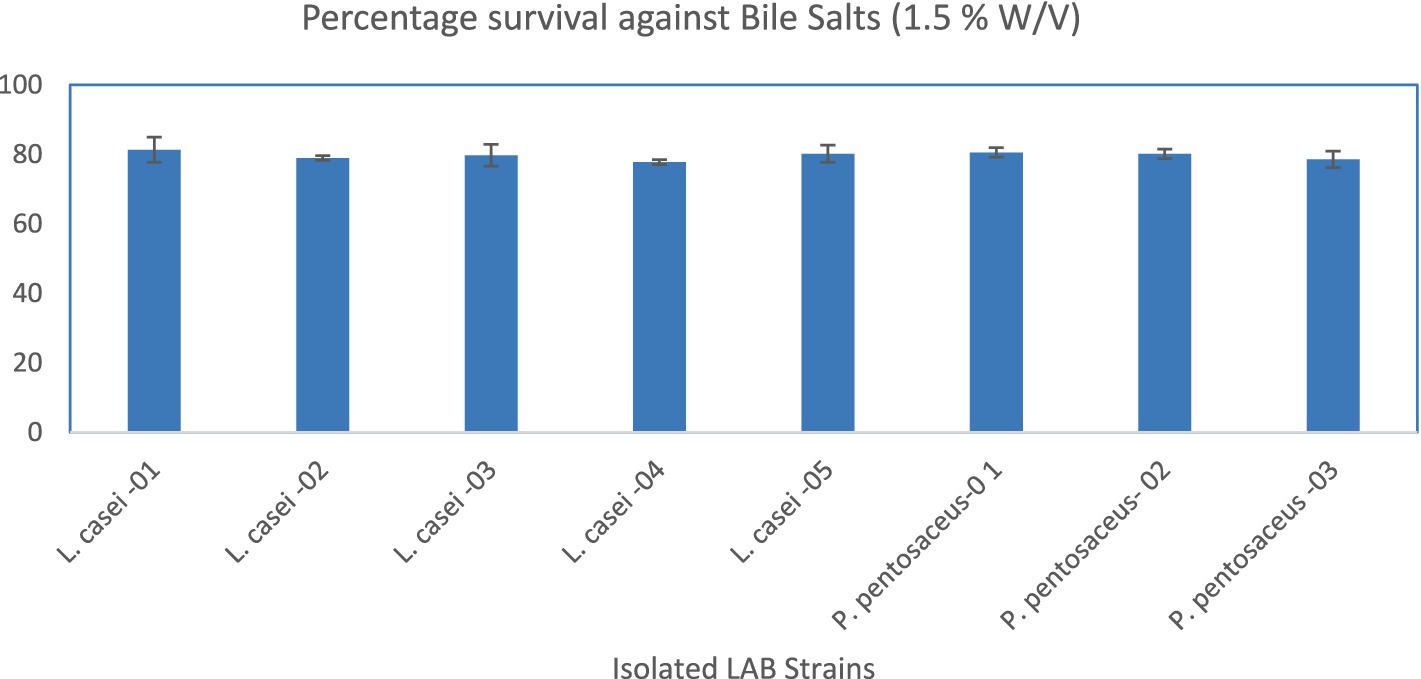
Figure 2. Percentage survival of the LAB strains against bile salts (1.5% W/V). X-axis = Isolates, Y-axis = Percentage Survival.
3.4 Antibiotic susceptibility testing
The isolated LAB strains were sensitive to the antibiotics which were used in the current study.
3.5 Hemolytic activity
None of the isolated LAB strains showed hemolytic activity.
3.6 Antimicrobial activity
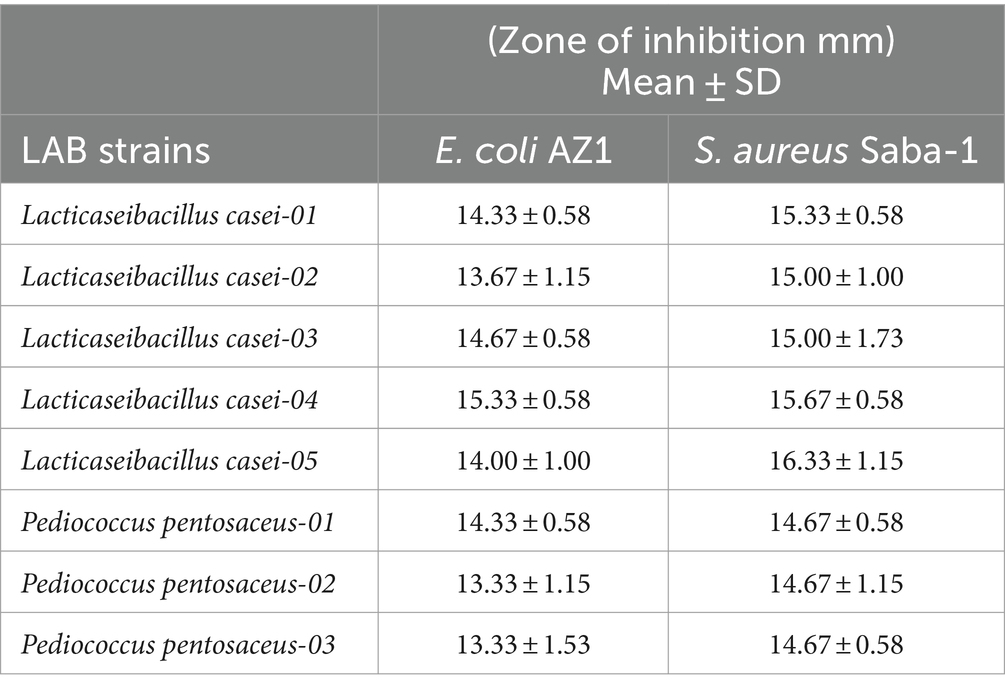
The L. casei-04 showed the maximum zone of inhibition (15.33 ± 0.58) against E. coli AZ1 strain whereas, L. casei-05 showed the maximum zone of inhibition (16.33 ± 1.15) against S. aureus Saba-1, as described in Table 1.
3.7 Auto-aggregation assay
The L. casei-03 showed maximum percentage auto-aggregation (28.65 ± 1.96) at 4 h of incubation whereas, the maximum percentage auto-aggregation was measured for L. casei-01 (41.10 ± 3.03) at 24 h of incubation, as shown in Figure 2.
3.8 Adhesion to HT-29 cells
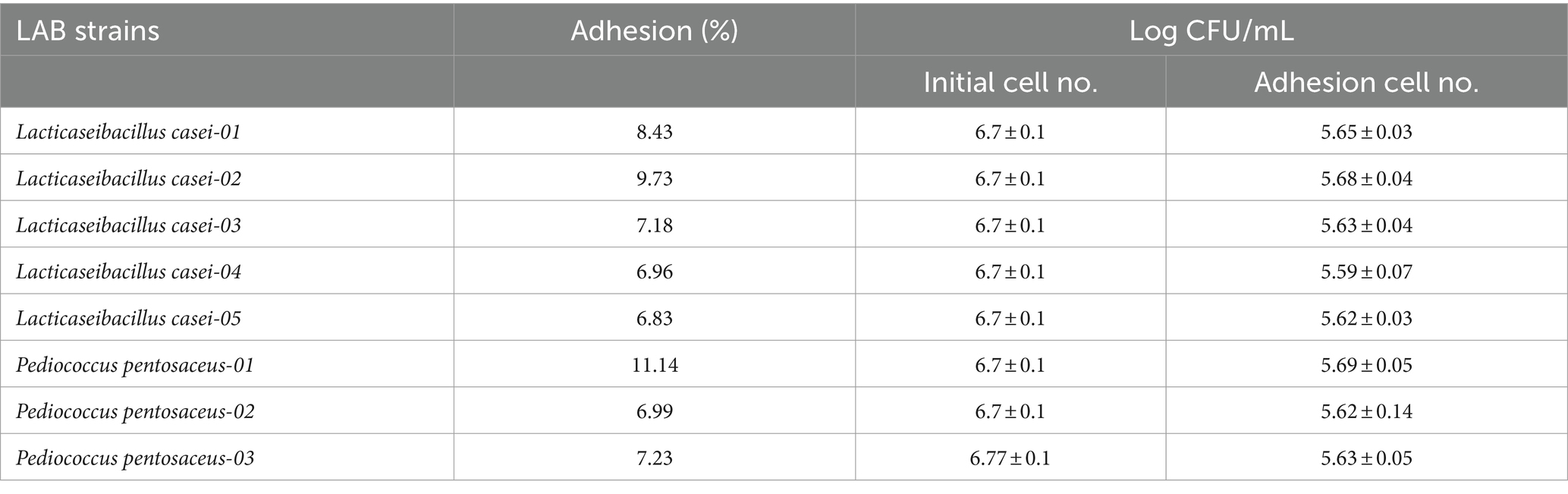
Maximum adhesion was shown by P. pentosaceus-01 (11.14%, Log CFU/mL = 5.69 ± 0.05) followed by L. casei-02 (9.73%, Log CFU/mL = 5.68 ± 0.04) as described in Table 2.
4 Discussion
Milk and other dairy products have been the befitting sources of probiotics that are included in human food for thousands of years (Kariyawasam et al., 2021). Among different milk-producing animals, camels are the best livestock that can efficiently survive in tropical and sub-tropical regions and are a good source of milk, meat, and leather (Hawaz et al., 2016). Camel milk is a good source of probiotic bacteria, i.e., Lacticaseibacillus, Streptococcus, and Bifidobacterium as described in some of the previous studies (Kadri et al., 2015; El-Zahar et al., 2021). Therefore, in the current study, we targeted the isolation and molecular identification of probiotics/LAB strains from camel milk. For this purpose, camel milk samples (n = 20) were collected from four different herds (Camelus dromedarius) from District Faisalabad-Pakistan (Figure 3) (Hussain et al., 2022; Aslam et al., 2023).

Figure 3. Percentage auto-aggregation of the LAB strains at 4 and 24 h of incubation. X-axis = Isolates, Y-axis = Percentage Auto-aggregation.
After the primary isolation and biochemical identification, the bacterial isolates were confirmed by molecular identification by amplification and sequence analysis of the 16S rRNA gene. The cultural characteristics were observed according to the previously described findings regarding different strains of probiotics from camel milk (Benmechernene et al., 2013). The results of cultural or biochemical identification showed that 13 out of 20 milk samples were positive for LAB strains, however 8 isolates were confirmed as LAB strains based on 16S rRNA gene amplification and sequence analysis. The PCR amplification resulted in a single band of about 1,400–1,500 bp product. The sequence data resulted in identification as probiotics/LAB strains, i.e., Lacticaseibacillus casei (n = 5) and Pediococcus pentosaceus (n = 3). The 16S rRNA identification and sequencing were described as a confirmatory identification tool for different bacterial isolates (Kabir et al., 2020; Waheed et al., 2021).
The isolated LAB strains showed acid tolerance and the survival rates (%) ranged from 77.38 ± 1.19 to 92.06 ± 1.82, while bile salts tolerance and the survival rates (%) ranged from 77.78 ± 0.69 to 81.35 ± 3.64. The survival rate against acids or bile salts was found comparable to the previous data (Jang et al., 2019). However, they calculated the acid tolerance in the presence of 0.3% pepsin, but we demonstrated the exposure to acid and bile salt followed by survival rates separately. Another study described that probiotics/LAB strains from camel milk can tolerate acidic pH, and increased bile salt concentrations (Sharma et al., 2021). Previously it was reported as 74 ± 04 survival rate (%) under gastrointestinal conditions (Vimont et al., 2017). Tolerance to acids or bile salts are considered as core potentials of the LAB strains, as orally administered probiotics or probiotic containing products/foods should survive the gastric or intestinal environment to exert their health benefits that include improved digestion, immune modulation, and pathogen inhibition etc.
The isolated LAB strains were found sensitive to different antibiotics, i.e., amoxicillin, ceftazidime, imipenem, linezolid, ofloxacin, tetracycline, tobramycin, and vancomycin. Some of the previous studies reported similar findings regarding susceptibility profiles (Benmechernene et al., 2013). However, a few studies described resistance to tetracycline by Lactobacillus paracasei (Comunian et al., 2010). Hence, a critical analysis of antibiotic susceptible profiles is required to investigate the isolates. Potential probiotics/LAB strains should be susceptible to all classes of antibiotics before the consideration as “probiotics/LAB strains to control the possible spread of different classes of antimicrobial resistance genes among humans, animals or environment. None of the LAB strains showed hemolytic activity as shown in one of the previous studies (Hamed and Elattar, 2013). This is a fact that probiotics/LAB strains must not show any type of hemolytic activity and do not have negative impact on ecosystem of other individuals. Similar findings were recently described regarding the probiotic’s characterization of Lactobacillus brevis KU15153 (Jang et al., 2019).
In the current study, the LAB strains inhibited the growth of pathogenic bacteria, i.e., multidrug-resistant E. coli AZ1 (maximum zone of inhibition = 15.33 ± 0.58) and S. aureus Saba-1 (maximum zone of inhibition = 16.33 ± 1.15). These findings were in accordance with the previous studies. A study reported the antimicrobial activity of LAB strains against Salmonella Typhimurium and Bacillus cereus (Šalomskienė et al., 2015). Another study has reported the antimicrobial activity of the LAB strain against S. typhimurium and S. aureus (Jang et al., 2019). However, we demonstrated this activity against MDR E. coli and methicillin-resistant S. aureus. The literature described that the mechanism of this sort of inhibition involves the metabolic products of LAB strains including bacteriocins, hydrogen peroxide, lactic acid and acetic acid (Anastasiadou et al., 2008). The current findings showed maximum auto-aggregation (%) 28.65 ± 1.96 and 41.10 ± 3.03 at 4 and 24 h of incubation. The studies have also demonstrated the au-to-aggregation abilities of the LAB strains from camel milk (Abushelaibi et al., 2017; Jang et al., 2019). This ability of the LAB strains is to prevent in vivo colonization of the intestinal epithelial cells. The antimicrobial potential is contributed by different metabolites of probiotics/LAB strains, i.e., hydrogen peroxide, organic acids or bacteriocins. Further, probiotics/LAB strains have potential to ferment milk to obtain wide range of products, i.e., yogurt, cheese, and kefir etc. (Shori, 2017; Kariyawasam et al., 2021). Similarly, probiotics/LAB strains could also be utilized for enhancing the bioavailability of different nutrients including vitamins and minerals.
In the current study, HT-29 epithelial cells adhesion ranged from 6.96 to 11.14%. These findings were partially in accordance with a previous study that used the HT-29 cells and Caco-2 cells and reported increased adhesion percentage (Vimont et al., 2017). Adhesion to cell membrane receptor is beneficial to combat the intestinal colonization by pathogenic bacteria using competitive exclusion. In conclusion, the current study has demonstrated that probiotics/LAB strains from camel milk have potential probiotic characteristics which can be further evaluated using suitable animal models.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
Ethical approval was not required for the studies on humans in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
ZN: Conceptualization, Writing – original draft, Writing – review & editing, Data curation. MKZ: Writing – original draft, Writing – review & editing, Formal analysis. MS: Formal analysis, Writing – original draft, Writing – review & editing, Data curation. RA: Formal analysis, Writing – original draft, Writing – review & editing. AY: Formal analysis, Writing – original draft, Writing – review & editing. MAZ: Writing – original draft, Writing – review & editing, Conceptualization, Supervision, Validation.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors acknowledge the Institute of Microbiology, Government College University, Faisalabad-Pakistan for utilizing the Labs and available resources.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Footnotes
References
Abushelaibi, A., Al-Mahadin, S., El-Tarabily, K., Shah, N. P., and Ayyash, M. (2017). Characterization of potential probiotic lactic acid bacteria isolated from camel milk. LWT-Food Sci Technol 79, 316–325. doi: 10.1016/j.lwt.2017.01.041
Afzal, M., Mazhar, S. F., Sana, S., Naeem, M., Rasool, M. H., Saqalein, M., et al. (2020). Neurological and cognitive significance of probiotics: a holy grail deciding individual personality. Future Microbiol. 15, 1059–1074. doi: 10.2217/fmb-2019-0143
Anastasiadou, S., Papagianni, M., Filiousis, G., Ambrosiadis, I., and Koidis, P. (2008). Growth and metabolism of a meat isolated strain of Pediococcus pentosaceus in submerged fermentation: purification, characterization and properties of the produced pediocin SM-1. Enzym. Microb. Technol. 43, 448–454. doi: 10.1016/j.enzmictec.2008.05.007
Anwar, H., Iftikhar, A., Muzaffar, H., Almatroudi, A., Allemailem, K. S., Navaid, S., et al. (2021). Biodiversity of gut microbiota: impact of various host and environmental factors. Biomed. Res. Int. 2021:5575245. doi: 10.1155/2021/5575245
Aslam, F., Rehman, M., Saleem, G., Ashraf, K., Hafeez, M. A., and Saqib, M. (2023). Identification and molecular characterization of theileria annulata with associated risk factors in naturally infected camels from selected districts in Punjab, Pakistan. Pakistan Vet J. 43, 79–84. doi: 10.29261/pakvetj/2022.084
Azizi, F., Habibi Najafi, M. B., and Edalatian Dovom, M. R. (2017). The biodiversity of Lactobacillus spp. from Iranian raw milk Motal cheese and antibacterial evaluation based on bacteriocin-encoding genes. AMB Express 7:176. doi: 10.1186/s13568-017-0474-2
Behrouz, S., Saadat, S., Memarzia, A., Sarir, H., Folkerts, G., and Boskabady, M. H. (2022). The antioxidant, anti-inflammatory and immunomodulatory effects of camel Milk. Front. Immunol. 13:855342. doi: 10.3389/fimmu.2022.855342
Benmechernene, Z., Chentouf, H. F., Yahia, B., Fatima, G., Quintela-Baluja, M., Calo-Mata, P., et al. (2013). Technological aptitude and applications of Leuconostoc mesenteroides bioactive strains isolated from Algerian raw camel milk. Biomed. Res. Int. 2013:418132. doi: 10.1155/2013/418132
Burger, P. A., Ciani, E., and Faye, B. (2019). Old World camels in a modern world - a balancing act between conservation and genetic improvement. Anim. Genet. 50, 598–612. doi: 10.1111/age.12858
Comunian, R., Daga, E., Dupré, I., Paba, A., Devirgiliis, C., Piccioni, V., et al. (2010). Susceptibility to tetracycline and erythromycin of Lactobacillus paracasei strains isolated from traditional Italian fermented foods. Int. J. Food Microbiol. 138, 151–156. doi: 10.1016/j.ijfoodmicro.2009.11.018
El-Fakharany, E. M., El-Baky, N. A., Linjawi, M. H., Aljaddawi, A. A., Saleem, T. H., Nassar, A. Y., et al. (2017). Influence of camel milk on the hepatitis C virus burden of infected patients. Exp. Ther. Med. 13, 1313–1320. doi: 10.3892/etm.2017.4159
El-Zahar, K. M., Hassan, M. F. Y., and Al-Qaba, S. F. (2021). Protective effect of fermented camel Milk containing Bifidobacterium longum BB536 on blood lipid profile in Hypercholesterolemic rats. J. Nutr. Metab. 2021:1557945. doi: 10.1155/2021/1557945
Hamed, E., and Elattar, A. (2013). Identification and some probiotic potential of lactic acid bacteria isolated from Egyptian camels milk. Life Sci. J. 10, 1952–1961.
Hawaz, E., Guesh, T., Kebede, A., and Menkir, S. (2016). Characterization of lactic acid bacteria from camel milk and their technological properties to use as a starter culture. East Afr. J. Sci. 10, 49–60.
Hudzicki, J. (2009). Kirby-Bauer disk diffusion susceptibility test protocol. Washington, DC: American Society for Microbiology, 55–63.
Hussain, S., Saqib, M., Ashfaq, K., and Sindhu, Z. D. (2022). First molecular evidence of coxiella burnetii in ticks collected from dromedary camels in Punjab, Pakistan. Pakistan Vet. J. 42, 276–280. doi: 10.29261/pakvetj/2021.073
Jang, H. J., Lee, N.-K., and Paik, H.-D. (2019). Probiotic characterization of Lactobacillus brevis KU15153 showing antimicrobial and antioxidant effect isolated from kimchi. Food Sci. Biotechnol. 28, 1521–1528. doi: 10.1007/s10068-019-00576-x
Kabir, S., Shahid, M., Waseem, M., Muzammil, S., Nawaz, Z., Rasool, M., et al. (2020). Dairy origin lactobacilli: functional analyses and antagonistic potential against multidrug-resistant foodborne pathogens. Int. Food Res. J. 27, 131–140.
Kadri, Z., Spitaels, F., Cnockaert, M., Praet, J., El Farricha, O., Swings, J., et al. (2015). Enterococcus bulliens sp. nov., a novel lactic acid bacterium isolated from camel milk. Antonie Van Leeuwenhoek 108, 1257–1265. doi: 10.1007/s10482-015-0579-z
Kariyawasam, K., Lee, N. K., and Paik, H. D. (2021). Fermented dairy products as delivery vehicles of novel probiotic strains isolated from traditional fermented Asian foods. J. Food Sci. Technol. 58, 2467–2478. doi: 10.1007/s13197-020-04857-w
Khurshid, M., and Akash, M. S. (2020). Probiotic preparations for infantile gastroenteritis: the clinical and economic perspective. Future Microbiol. 15, 567–569. doi: 10.2217/fmb-2019-0111
Mahala, N., Mittal, A., Lal, M., and Dubey, U. S. (2022). Isolation and characterization of bioactive lactoferrin from camel milk by novel pH-dependent method for large scale production. Biotechnol. Rep. 36:e00765. doi: 10.1016/j.btre.2022.e00765
Rahmeh, R., Akbar, A., Kishk, M., Al-Onaizi, T., Al-Azmi, A., Al-Shatti, A., et al. (2019). Distribution and antimicrobial activity of lactic acid bacteria from raw camel milk. New Microbes New Infect. 30:100560. doi: 10.1016/j.nmni.2019.100560
Šalomskienė, J., Abraitienė, A., Jonkuvienė, D., Mačionienė, I., and Repečkienė, J. (2015). Selection of enhanced antimicrobial activity posing lactic acid bacteria characterised by (GTG)5-PCR fingerprinting. J. Food Sci. Technol. 52, 4124–4134. doi: 10.1007/s13197-014-1512-6
Sharma, A., Lavania, M., Singh, R., and Lal, B. (2021). Identification and probiotic potential of lactic acid bacteria from camel milk. Saudi J. Biol. Sci. 28, 1622–1632. doi: 10.1016/j.sjbs.2020.11.062
Shori, A. B. (2017). Camel milk and its fermented products as a source of potential probiotic strains and novel food cultures: a mini review. Pharma Nutr. 5, 84–88. doi: 10.1016/j.phanu.2017.06.003
Swelum, A. A., El-Saadony, M. T., Abdo, M., Ombarak, R. A., Hussein, E. O. S., Suliman, G., et al. (2021). Nutritional, antimicrobial and medicinal properties of Camel's milk: a review. Saudi J. Biol. Sci. 28, 3126–3136. doi: 10.1016/j.sjbs.2021.02.057
Tambekar, D., and Bhutada, S. (2010). An evaluation of probiotic potential of Lactobacillus sp. from milk of domestic animals and commercial available probiotic preparations in prevention of enteric bacterial infections. Recent Res. Sci. Technol. 2, 82–88.
Vimont, A., Fernandez, B., Hammami, R., Ababsa, A., Daba, H., and Fliss, I. (2017). Bacteriocin-producing Enterococcus faecium LCW 44: a high potential probiotic candidate from raw camel milk. Front. Microbiol. 8:865. doi: 10.3389/fmicb.2017.00865
Keywords: antimicrobial susceptibility, Camelus dromedarius, camel milk, sustainable foods, lactic acid bacteria
Citation: Nawaz Z, Zahoor MK, Shafique M, Athar R, Yasmin A and Zahoor MA (2024) In vitro assessment of probiotic properties of lactic acid bacteria isolated from camel milk: enhancing sustainable foods. Front. Sustain. Food Syst. 8:1437201. doi: 10.3389/fsufs.2024.1437201
Edited by:
Kathleen L. Hefferon, Cornell University, United StatesReviewed by:
Vignesh Sivanandham, Indian Institute of Food Processing Technology, IndiaŁukasz Łopusiewicz, West Pomeranian University of Technology, Poland
Copyright © 2024 Nawaz, Zahoor, Shafique, Athar, Yasmin and Zahoor. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Asif Zahoor, ZHJhc2lmemFob29yQGdjdWYuZWR1LnBr
 Zeeshan Nawaz
Zeeshan Nawaz Muhammad Kashif Zahoor
Muhammad Kashif Zahoor Muhammad Shafique
Muhammad Shafique Rasham Athar
Rasham Athar Aysha Yasmin
Aysha Yasmin Muhammad Asif Zahoor
Muhammad Asif Zahoor