
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 12 July 2024
Sec. Crop Biology and Sustainability
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1432599
This article is part of the Research Topic Enhancing Soil and Crop Resilience: Strategies Against Climate-Driven Salinization and Degradation View all 7 articles
Phosphorus (P) is the second most important macro-element for plant growth, and its low availability in soil is a major obstacle to crop production. Inorganic phosphate (Pi) is the least available form in the soil, while organic phosphate (Po) is the most dominant one, up to 80% of which exists as inositol hexakisphosphate, also known as phytic acid (PA) that cannot be absorbed by plant roots unless hydrolyzed by microbial phytases. Similar to phosphate-solubilizing bacteria, many plant growth-promoting rhizobacteria (PGPR) can play a relevant role in phosphate turnover. In our study, we screened a series of PGPR strains for phytase activities using PA as a sole source of P. Three strains (named C2, N4, and S10) with relatively high phytase activities ranging from 42.84 to 100.55 Units g−1 were selected for barley growth assays. When barley plants grown in poor sandy soil and irrigated with a PA-containing solution were inoculated with each of these PGPR isolates, a significant growth enhancement was observed. This positive effect was well illustrated by an increase in root growth, plant height, and chlorophyll contents. In addition, the inoculated barley plants accumulated significantly higher Pi contents in leaves and roots compared to non-inoculated plants. Finally, the expression of a number of high-affinity Pi transporter genes (PHT1.1, PHT1.4, PHT1.8, and PHT1.6) in inoculated barley plants was downregulated especially in roots, compared to non-inoculated plants. This difference is most likely due to the bacterial phytases that change the P availability in the rhizosphere. In summary, these three strains can improve barley growth under phosphate-limited conditions and should be considered in developing eco-friendly biofertilizers as an alternative to conventional P fertilizers.
Phosphorus (P) is one of the most important nutrients needed for plant growth and development, and it has a direct effect on the yield and quality of crops. Two forms of P, namely, Pi and Po, can be found in the soil, but plants can take up only Pi, which is essential for major plant metabolic processes including photosynthesis, respiration, energy metabolism, and environmental stress response (Plaxton and Tran, 2011; Chien et al., 2018; Ham et al., 2018). However, Pi is very lowly available in the soil, while Po is the most dominant form of which up to 80% represents phytic acid (PA) that cannot be absorbed by plant roots. Low Pi availability is considered as one of the major limitations for agricultural production in many soils (Wang et al., 2010; Zhang et al., 2014). To cope with Pi deficiency, P fertilizers are used, but raw materials for their production are scarce and their effectiveness is severely limited as only 10–30% of the applied P is utilized by plants (Vance et al., 2003). The remainder portion is rendered unavailable by the formation of insoluble complexes of P with cations such as aluminum, iron, or calcium, as well as by P adsorption by mineral surfaces and organic matter (Kochian et al., 2004; López-Arredondo et al., 2017). Consequently, an excessive application of P is often required to meet the plants’ needs, hence leading to serious water and soil pollution (Cordell et al., 2009; Kochian, 2012; Pan et al., 2019).
Plants have developed a series of strategies to increase soil P availability. These strategies include morphological and biochemical changes at the soil–root interface. For example, increased root growth and branching, root hair proliferation, and the release of root exudates can increase the plant’s access to Pi from otherwise unavailable sources (Raghothama, 1999). In addition, plant roots exhibit phytase activities capable of hydrolyzing soil PA. However, all these strategies remain insufficient for the effective utilization of Po (Richardson et al., 2000; Brinch-Pedersen et al., 2002).
In this context, the use of phosphate-solubilizing bacteria (PSB) is gaining importance by increasing the availability of insoluble P to plants, while maintaining environmental sustainability. Indeed, it has been reported that PSB act through different mechanisms such as rhizosphere acidification and/or phytase excretion, resulting in an enhanced plant P uptake (Turner et al., 2003; Yu et al., 2011; Manzoor et al., 2017; Wan et al., 2020; Chungopast et al., 2021). Phytase-producing rhizobacteria have been reported to belong to Bacillus, Geobacillus, Burkholderia, Enterobacter, Pseudomonas, Serratia, Staphylococcus, and Acinetobacter genera (Shedova et al., 2008; Jorquera et al., 2018; Rix et al., 2022). It is worth noting that in the soil, many filamentous fungi are also known to produce active phytases capable of enhancing plant growth especially those produced by Aspergillus, Myceliophthora, Mucor, Penicillium, Rhizopus, and Trichoderma species (for review see Richardson et al., 2001; Dailin et al., 2019; Corrêa and Araújo, 2020).
Hordeum vulgare (barley) is one of the earliest domesticated cereal crops, derived from its wild relative Hordeum spontaneum, with domestication occurring approximately 11,000 years ago in the Fertile Crescent (Badr et al., 2000). Today, barley is the world’s fourth largest cereal crop after maize, rice, and wheat and contributes significantly as a food and feed resource, to global food security (Geng et al., 2022). In addition, barley is known to contain valuable genetic diversity toward adaptation to various environmental constraints (Tao et al., 2023). However, this does not preclude climate change, and its exacerbating effects on stress factors especially drought, high salinity, and mineral deficiencies in soil drastically limit global production (Sabagh et al., 2019). Moreover, drought results in an alteration of soil physiochemical properties and hence leads to reduced mobility and plant absorption of individual nutrients such as P (Amtmann and Blatt, 2009).
In the present study, a screening strategy was conducted to identify phytase-producing rhizospheric bacteria based on qualitative and quantitative assays. As a result, three strains exhibiting phytase activity and the ability to degrade PA were selected to assess their impact on barley seedlings grown in P-poor soil. Our physiological and molecular analyses reveal that these selected strains improve barley growth and P nutrition from PA as a sole phosphate source. Therefore, these phytase-producing strains could be part of a sustainable approach to address low soil P availability for enhancing crop production.
The seeds of barley (Hordeum vulgare L. cv. Rihane) were provided by the “Institut National des Grandes Cultures, Tunisie.” First, barley seeds were surface-sterilized for 15 min in a commercial bleach solution containing 3% sodium hypochlorite, washed with sterile distilled water three times, and sown on Petri dishes with sterile filter paper for germination. One week after germination, seedlings were transferred into pots containing an autoclaved soil mixture of sand and peat (10:1, v/v) and moved in a growth chamber (at 22–24°C) under long-day conditions (16-h light/8-h dark) where growth and inoculation assays took place.
Thirteen previously isolated bacterial strains (Sayahi et al., 2022) were tested for their phytase activity. The screening method consists of dropping 10 μL of bacterial suspension (OD = 0.1) from fresh colonies on the National Botanical Research Institute’s phosphate (NBRIP); glucose (10 g/L), Ca3(PO4)2 (5 g/L); MgCl2W6H2O (5 g/L), MgSO4W7H2O (0.25 g/L); KCl (0.2 g/L); and (NH4)2SO4 (0.1 g/L) solid medium (Nautiyal, 1999) containing 0.5 mM sodium phytate (Na-InsP6, P-8810, Sigma) as a sole P source instead of Ca3(PO4)2 and incubated for 72 h at 28°C. The strains displaying the ability to degrade phytate were discerned through the presence of a surrounding visible halo/zone on the agar plates. The solubilization efficiency (SE) can be calculated using the formula SE = (halo zone diameter/colony diameter) *100 (Nguyen et al., 1992). Subsequently, the surrounding halo zone was flooded with Murphy-Riley reagent which contains ammonium molybdate solution supplemented with ascorbic acid (10%) at room temperature for 5 min. The resulting blue staining corresponds to the Pi ions liberated as a result of phytate breakdown, and the colonies were considered according to Murphy and Riley (1962), as phytase-producing strains.
Among the 13 bacterial strains, 3 strains (named hereafter C2, N4, and S10) were selected to measure the phytase activity based on the determination of Pi released by the hydrolysis of sodium phytate as previously described by Engelen et al. (1994). In brief, 500 μL of cell suspensions (OD = 0.1) of the selected strains was incubated with a buffer containing 4.5 mM sodium phytate in 0.1 M Tris–HCl (pH 7.5 for S10 and pH5.5 for C2 and N4) supplemented with 2 mM CaC12 for 30 min at 30°C. The reaction was stopped by adding 750 μL of a 5% (w/v) trichloroacetic acid solution, and the released Pi was spectrophotometrically quantified at 700 nm by following the production of phosphomolybdate with the color reagent (four volumes of 1.5% ammonium molybdate solution in 5.5% sulfuric acid and one volume of 2.7% ferrous sulfate solution). One phytase unit (U) is defined as the amount of enzyme capable of releasing 1 mmol of Pi from sodium phytate per minute under the above reaction conditions.
To prepare the inoculum, a fresh colony of each strain was suspended in 50 mL LB medium and incubated at 30°C under constant shaking (180 rpm/min) for 18 h. Then, centrifugation at 6500 rpm for 20 min was performed, and the resulting pellets were washed three times with sterile distilled water and finally resuspended in sterilized water to reach a final concentration of 109 CFU/mL.
Barley seedlings were transferred into plastic pots containing a soil mixture of sterilized sand and peat (Potgrond H-Klasmann) with a 10:1 proportion (v/v). These pots, containing two plants each, were then arranged into four distinct treatment blocks: 1: non-inoculated seedlings and irrigated with tap water; 2: seedlings inoculated with one of the selected strains C2, N4, and S10 and watered with tap water; 3: non-inoculated seedlings watered with PA solution (0.33 mM); 4: bacterial- inoculated seedlings irrigated with PA solution. All treatments were performed in at least three individual replicates (n = 12). The inoculation was performed on the roots of each plantlets using 1 mL of cell suspension (109 CFU) as previously described (Sayahi et al., 2022). The irrigation with tap water or PA solution was repeated every 3 days for the first month and then once a week for the second month.
Primary and lateral roots were characterized by measuring their length and number using the Optimas 6.1 image analysis software, as described previously (Belgaroui et al., 2022). Leaf and root surface area has been analyzed using Fiji software (Schindelin et al., 2012).
Pi contents in roots and leaves were quantified using the molybdenum blue method (Murphy and Riley, 1962). In brief, ion extractions from weighed fresh leaves and roots (10–20 mg) were performed in water by incubation at 70°C for 30 min. Then, 300 μL of the diluted supernatant was mixed with 600 μL of ammonium molybdate solution and 100 μL of 10% ascorbic acid. In parallel, a standard curve with inorganic phosphate (K2HPO4) was prepared. The mixture was incubated at 45°C for 30 min, and absorbance at 820 nm was measured.
Chlorophyll a (Chl a), chlorophyll b (Chl b), and total chlorophyll (Chl a + Chl b) were examined with a modified method of Arnon (1949). The barley leaf samples (0.1 g) were dipped in 80% acetone and kept in the dark at 4°C for the extraction of chlorophyll pigment. The homogenates were collected and centrifuged at 12000 rpm for 10 min. The absorbance of the obtained supernatants was measured using a spectrophotometer at 646 nm and 663 nm, and total chlorophyll content was calculated according to the following formula.
.
.
.
The expression levels of Pi deficiency responsive genes including the PHT1 family of high-affinity Pi transporters (PHT1.1, PHT1.4, PHT1.6, PHT1.8, PHO2, and PHR1) were evaluated in barley roots and leaves. Two-month-old barley plants were grown and treated as described above, and total RNA was extracted using the TRIZOL reagent according to the manufacturer’s recommendations and then treated with DNase (Invitrogen) to remove any contaminating genomic DNA. cDNA was synthesized from 2 μg total RNA using an oligo (dT) primer and M-MLV reverse transcriptase (Invitrogen). Quantitative PCR (qRT-PCR) was performed on CFX 96 Touch Real-Time PCR System (Bio-Rad) using SYBR Green SYBR® Premix ExTaq (TAKARA). The housekeeping gene (HvEF-1α) was used as an endogenous reference for expression data normalization (Janská et al., 2013). The gene-specific primers (Supplementary Table S1) were designed from sequences of the respective genes downloaded from the National Center for Biotechnology Information (NCBI) using the Primer 3 software (version 0.4.0).1
The BLAST search for phytase in the genomic sequences of each strain was carried out by using two phytase genes from Serratia marcescens KS10 (accession number NZ_CP027798.1) and PSB-15 (accession number KR133277.1), as alignment queries. MaGe software2 was also used to browse the genomes of the strains C2, N4, and S10 for other phytase sequences. The Expasy Translate Tool3 was used to convert the genomic sequence into a protein sequence. Multiple sequence alignments of the deduced amino acid sequences with NZ_CP027798.1 or KR133277.1 were performed using the Clustal Omega program.4 HMMER5 analysis allowed the identification of the phytase-like domain.
Statistical analyses of the data were performed using a one-way analysis of variance (ANOVA) followed by Tukey’s test using GraphPad Prism6 software. Only differences with p-values <0.05 were considered significant.
From a collection of soil PGPR isolates that showed P solubilization (Sayahi et al., 2022), we have developed here a screening method for those exhibiting phytase activity. Among 13 bacterial strains, six strains showed clear halo zones around the colonies resulting from the solubilization of PA added as the sole P source on NBRIP-Pi agar (Supplementary Figure S1). Subsequent staining with the molybdate blue reagent revealed that all six strains were phytase-producing strains as the colonies turned blue, indicating the release of P ions (Figure 1). Three bacterial strains C2, N4, and S10 that showed intense blue color were selected. The P solubilization efficiency (SE) calculated by measuring the halo zones formed around phytase-producing colonies gave rise to the highest value of 70% for C2, followed by S10 (66.66%) and N4 (60%; Figure 1B).
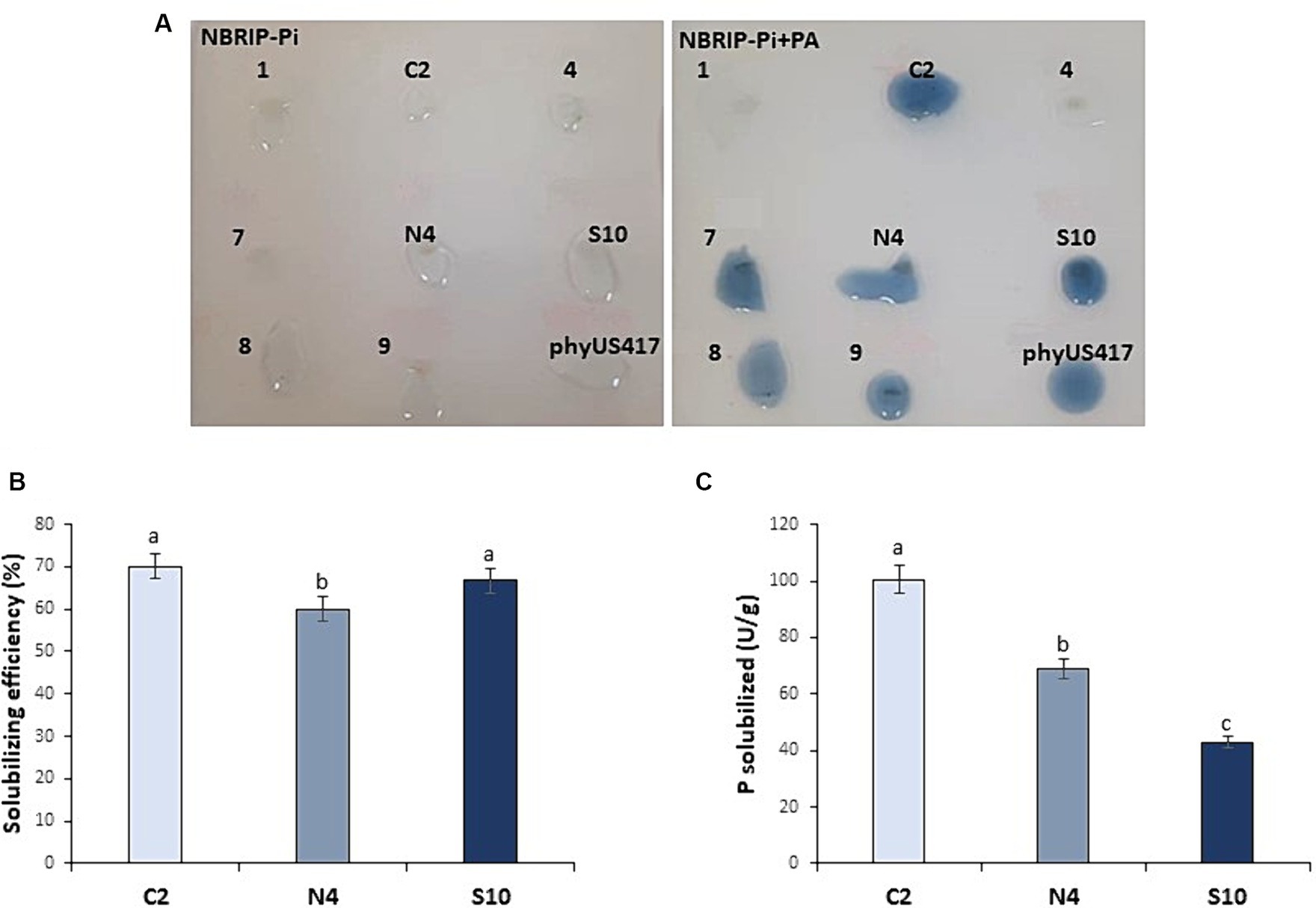
Figure 1. Screening for phytase-producing bacterial isolates. (A) Solubilization of phosphate by plant growth-promoting bacteria isolates grown on NBRIP-Pi containing PA as sole P source. The blue color around the colonies indicates the released phosphate (Pi) detected with the molybdate blue staining test. A purified phytase phyUS417 from Bacillus subtilis was used as a positive control. (B) P solubilization efficiency of the selected isolates (C2, N4, and S10) determined based on the calculation of the halo zones formed. (C) Quantitative assay to determine the phytase activity of the selected isolates. The data represent the means of three biological and experimental replicates. Different letters indicate that values are statistically different at a p-value of <0.05.
To evaluate the phytase activity on the three selected strains, LB overnight liquid cultures were used to measure the released Pi from the sodium phytate solution at 30°C in a reaction buffer containing 2 mM CaCl2 and at pH ranging from 5.5 (for C2 and N4) to 7.5 (for S10). The assay conditions allowed us to register variable phytase activities among the three bacterial isolates, consistent with the qualitative assay performed above. The highest level of phytase activity was observed in C2 (100.55 U/g) followed by N4 (69.10 U/g) and S10 (42.84 U/g; Figure 1C).
Genome sequence analysis revealed that the C2 strain belongs to Serratia sp. (BioSample SAMN38274870; Sayahi et al., 2024). The genome sequencing has been also initiated for the two other strains, and the retrieval of the corresponding 16S rDNA sequences from the draft genome allowed us to identify N4 (accession number PP621717.1) as Serratia marcescens and S10 as Citrobacter sp. species (accession number PP621716.1; Supplementary Figure S2). In addition, through genome browsing, we identified in each strain a gene sharing high homology with phytase-encoding sequences from Serratia marcescens KS10 (accession number NZ_CP027798.1) or PSB-15 (accession number KR133277.1) strains. The gene size ranges between 1,362 (C2 and N4) and 1,365 bp (S10) giving rise to proteins of 453 and 454 a.a. The two proteins from N4 and S10 share a strong homology (94% identity) and are closely related to KR133277.1 (Supplementary Figure S3A), confirmed previously as phytase-secreting bacteria (Zhang et al., 2011; Kalsi et al., 2016), while the one from C2 is highly homologous to NZ_CP027798.1 (Supplementary Figure S3B). In addition, the use of MaGe software allowed the identification in the genome of the N4 strain, of two other phytase sequences with open reading frames of 1,344 and 1,362 bp, giving rise to proteins of 447 and 453 a.a belonging to esterase-like activity of phytase family and phytase-like domain-containing proteins, respectively. Remarkably, the phytase-like domain-containing protein is 100% identical to the one identified in the C2 strain.
Moreover, protein analysis using HMMER software revealed the presence of a conserved phytase-like domain but only in the phytase sharing 100% identity between C2 and N4 strains. It should belong to the β_propeller class (according to Pfam Interpro) of phytases as the phytase-like domain (residues 34–371) is a repeated region that carries several highly conserved Glu and Asp residues that most likely incorporates the enzymatic activity of the PLC-like phosphodiesterase part of the proteins. In addition, the SignalP 6.0 server predicted the presence of signal peptides and the position of their cleavage sites in the phytase-like domain proteins of C2 and N4. The likelihood of the signal peptide was approximately 0.9986, and the location of the peptide cleavage site was predicted to be between positions 20 and 21, while the probability of the signal peptide for the esterase-like protein of the strain N4 was approximately 0.9989, and the location of the peptide cleavage site was predicted to be between positions 23 and 24.
To evaluate the capacity of the three phytase-producing strains (C2, N4, and S10) to promote barley growth under P-limited conditions, inoculation assays were performed on 1-week-old seedlings grown in pots containing poor soil (sand and peat) without any added P source. Barley seedlings were inoculated with one of the selected strains and then regularly irrigated either with water or with a PA solution.
Following 8 weeks of plant growth, we can see clearly that the three PGPR strains enhance barley growth in the presence of PA compared with the non-inoculated control plants. The positive effect of these strains on barley growth is best illustrated in root architecture, plant height, and leaf area (Figures 2, 3). When grown in the presence of PA, the inoculation with either of these three strains resulted in an increase of up to 32 and 190% in plant height and leaf area, respectively, compared to control plants. In addition, the inoculated barley exhibits more vigorous roots with a significant increase in primary root length ranging from 20 to 40% (Figure 3A), number of lateral roots (up to 45%; Figure 3B), lateral root length (up to 16%; Figure 3C), and most significantly in root surface area which was 3.4x higher (N4 strain) than that registered in non-inoculated plants (Figure 3D). All these observations strongly suggest that the selected phytase-producing strains can enhance barley growth in Pi-limited conditions.
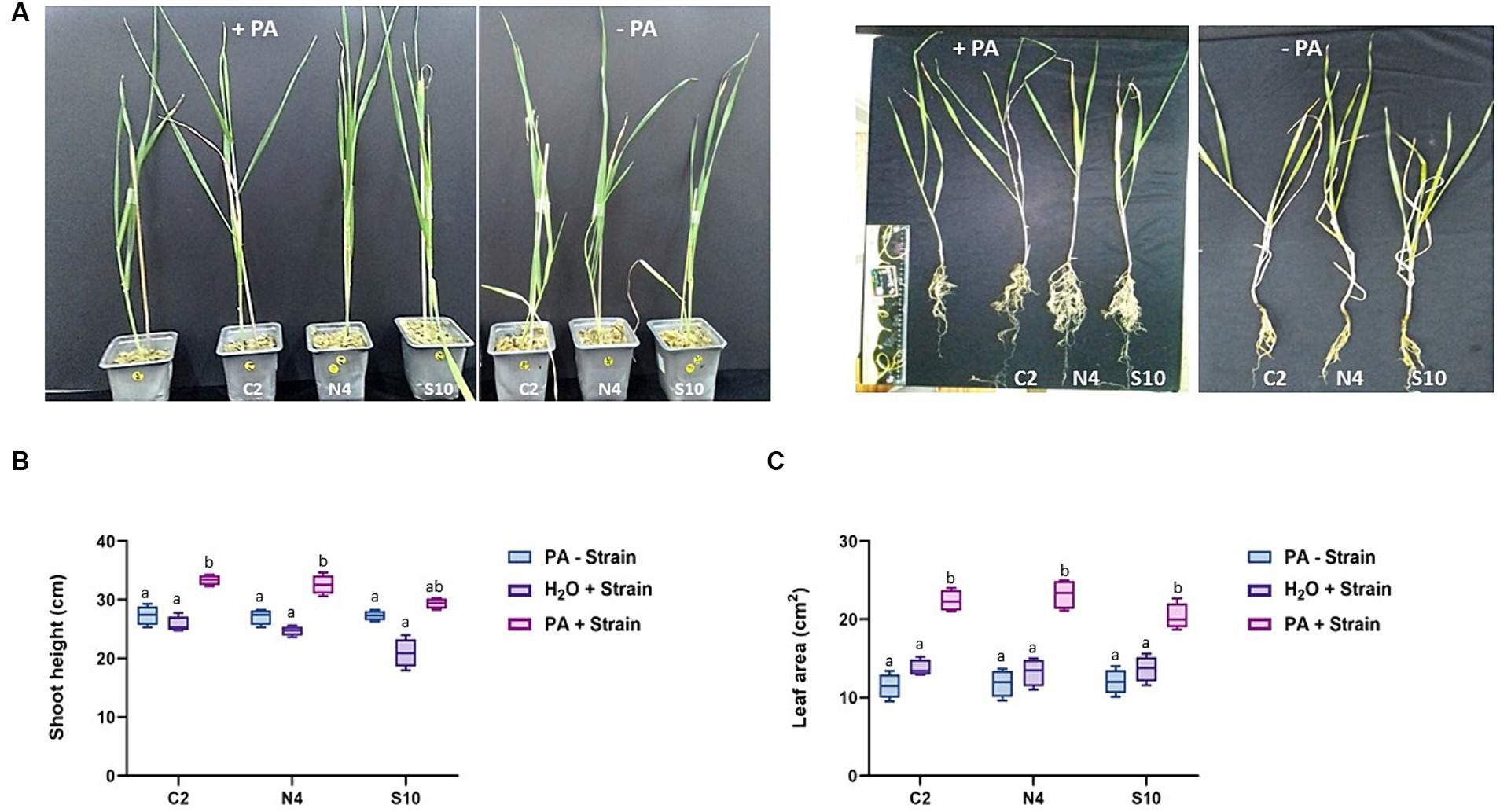
Figure 2. Effect of phytase-producing strains on barley growth under P-limited conditions. (A) Photographs of barley plants were taken after 8 weeks of post-inoculation with the phytase-producing strains (C2, N4, and S10) and irrigated either with water (-PA) or with a 0.33 mM PA solution (+PA). (B,C) Measurements of the shoot height and leaf area of barley plants grown as in A. The data are mean ± SD of three individual replicates (n = 12). Different letters indicate significantly different means at p < 0.05, as determined by one-way ANOVA followed by Tukey’s multiple comparison test.
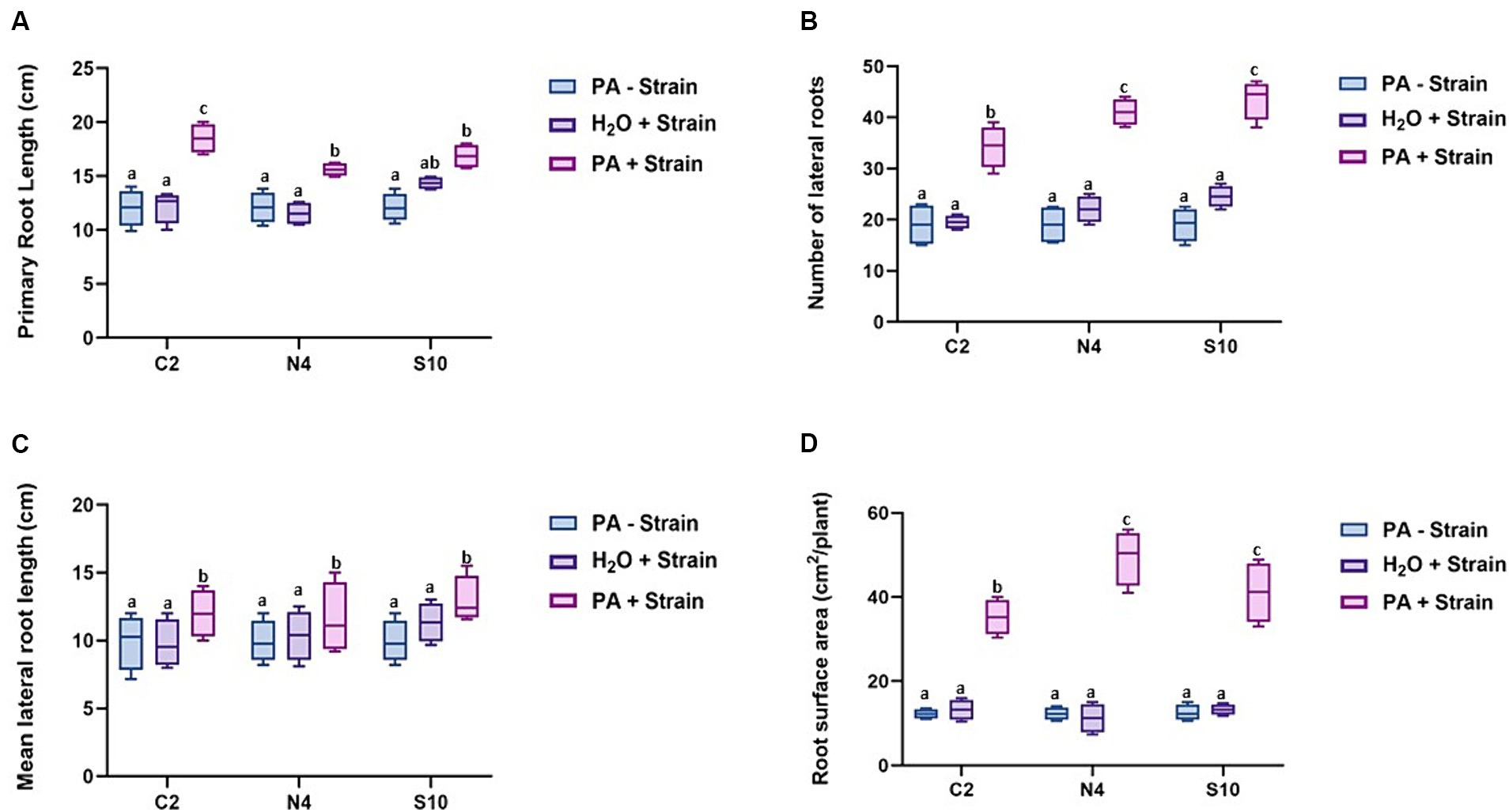
Figure 3. Effect of phytase-producing strains (C2, N4, and S10) on barley root architecture. (A) Primary root length. (B) Number of lateral roots. (C) Mean lateral root length. (D) Root surface area. All measurements were performed on barley plants grown in soil under P-limited conditions 8 weeks after post-inoculation with the selected strains (C2, N4, and S10). The barley plants were irrigated either with water (H2O) or with a 0.33 mM PA solution (PA). The data shown represent the mean ± SD of three independent experiments (n = 12). Different letters indicate significantly different means at p < 0.05, as determined by one-way ANOVA followed by Tukey’s multiple comparison test.
The difference observed at the phenotypic level prompted us to further investigate the effect of these three strains on the accumulation of Pi in the leaves and roots of barley plants grown under the inoculation conditions described above. A clear increase in Pi levels was observed in both roots and leaves of inoculated plants irrigated with PA compared to non-inoculated control plants or inoculated plants without PA supply (Figures 4A,B). For instance, the Pi content in the roots of plants inoculated with S10 and irrigated with PA was estimated to be 48.65 μmol g-1 FW which is 3.3x and 5x higher than those registered in the non-inoculated control barley (14.56 μmol g-1 FW) or barley inoculated without PA supply (9.48 μmol g-1 FW), respectively. Barley inoculated with N4 or C2 in the presence of PA also displayed a high root Pi accumulation reaching 47.25 μmol g-1 FW and 37.57 μmol g-1 FW, respectively (Figure 4A). Similarly, in leaves, barley plants irrigated with PA and inoculated with S10, N4, or C2 strains displayed a higher Pi accumulation (25.24, 25.03, and 22.30 μmol g-1 FW, respectively) compared to non-inoculated (12.64 μmol g-1 FW) or inoculated control plants without PA supply (14.87, 15.29, and 10.17 μmol g-1 FW; Figure 4B). These results confirm the potential of these three PGPR strains in improving Pi availability most likely through their phytase activity on PA substrate, hence increasing Pi contents in barley.
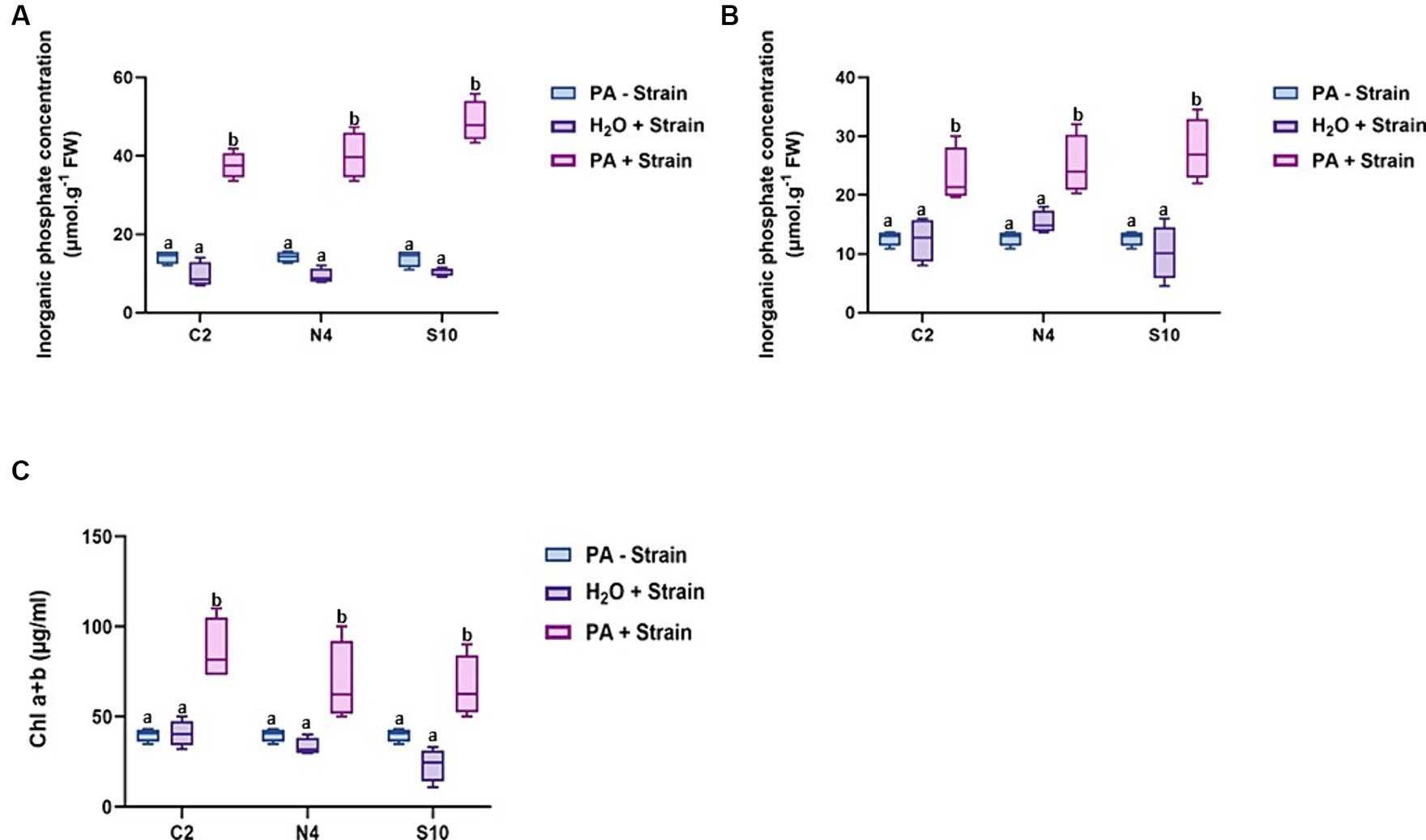
Figure 4. Inorganic phosphate content in roots and leaves and chlorophyll content in leaves of barley plants. (A) Pi levels in roots. (B) Pi levels in leaves. (C) Chlorophyll (Chla+b) content in leaves. Measurements were conducted on barley plants grown in soil under P-limited conditions 8 weeks after post-inoculation with the selected strains (C2, N4, and S10) and irrigated either with water (H2O) or with a PA solution (PA). Values are means ± SE of at least three biological replicates (n = 12). Different letters indicate significantly different means at p < 0.05, as determined by one-way ANOVA followed by Tukey’s multiple comparison test.
Knowing the relevance of Pi in photosynthesis, we expected that the increase in leaf Pi content of inoculated barley could enhance photosynthesis efficiency. To confirm this hypothesis, we compared the chlorophyll contents between non-inoculated and inoculated barley plants in the presence of PA. As shown in Figure 4C, the inoculation of barley with either of the three PGPR strains led to a significant increase in chlorophyll contents reaching up to 2x higher in the presence of the C2 strain.
We were then interested in checking whether the growth promotion and the change in Pi contents observed in barley plants following inoculation with the C2, N4, and S10 strains, and utilizing PA as the sole source of Pi, can be associated with a change in Pi transport and homeostasis. To this end, we performed expression analysis on a subset of barley genes involved in Pi transport and signaling. These genes include high-affinity Pi transporters belonging to the PHT1 family, acting in roots (PHT1.1, PHT1.4, and PHT1.8) or leaves (PHT1.1, PHT1.4, and PHT1.6), along with PHO2 (responsible for the root–shoot Pi translocation) and PHR1 transcription factor. Our results show that there is a significant difference in the expression of all Pi transporters (in roots and leaves) in inoculated barley plants grown on phosphate-poor soil compared with non-inoculated plants (Figures 5A,B). Notably, the expression of PHT1.1, PHT1.4, and PHT1.8 was significantly downregulated in barley roots grown in the presence of PA when inoculated with either of the three PGPR strains, compared to non-inoculated barley plants (Figure 5A). Similarly, the downregulation of the PHT1.1 and PHT1.4 genes, although less significant, was also recorded in the leaves of inoculated barley (Figure 5B). This downregulation of the high-affinity Pi transporters should result from the increased availability of Pi in the phosphate-poor soil, following the phytase-mediated hydrolysis of PA by the different PGPR strains C2, N4, and S10.
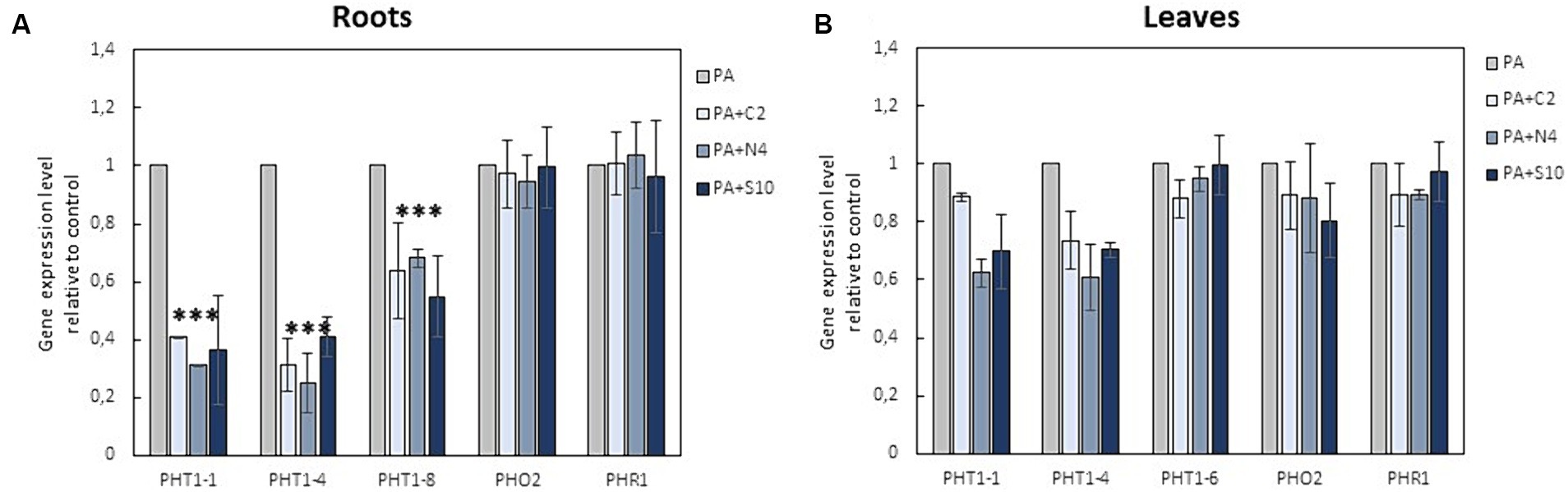
Figure 5. Expression profiles of phosphate transporters genes in shoots and roots of barley tissues after the application of the selected strains (C2, N4, and S10). qRT-PCR analysis of the HvPHT1.1, HvPHT1.4, HvPHT1.6, HvPHT1.8, PHO2, and PHR1 genes was carried out on roots (A) and shoots (B) collected from barley plants grown in soil under P-limited conditions 8 weeks after being post-inoculated with C2, N4, or S10 strain. The barley plants were irrigated either with water (H2O) or with a PA solution (0.33 mM PA). HvEF-1α was used as the reference gene for expression data normalization. Abundance of gene transcripts was normalized against their respective expression in control (non-inoculated plants). Data are mean ± SD of at least three biological replicates (n = 12). Asterisks indicate a statistically significant difference (p < 0.05), ANOVA.
Phosphorus is essential for plant growth and development. In many soils, the predominant phosphorus fraction is organic P, ranging from 50 to 95% (Gerke, 2022). This organic form poses a challenge for plant utilization since it cannot be absorbed directly by plant roots. The solubilization of PA, which constitutes up to 80% of soil organic P, has attracted significant attention for improving P-use efficiency since it can lead to higher yields while reducing the amounts of chemical fertilizers, thereby reducing environmental impact through minimized P runoff into surface water (Raboy, 2020). PA is dephosphorylated by a special class of phosphatases such as phytases and acidic/alkaline phosphatases, before being assimilated by plants (Tarafdar and Claassen, 1988; Tarafdar and Rao, 1996; Richardson et al., 2001; Yadav and Tarafdar, 2003; Richardson et al., 2009). In the rhizosphere, various fungi and bacteria play a crucial role in hydrolyzing this organic form of phosphorus (Rodriguez and Fraga, 1999; Singh and Satyanarayana, 2011).
Over the last decade, there has been an increased focus on isolating and identifying rhizobacteria beneficial to plants for use as inoculants (Saharan and Nehra, 2011; Karnwal, 2017; Lihan et al., 2021). The present study demonstrates that three PGPR strains exhibit relatively high phytase activity and can hydrolyze PA when used to inoculate with barley under P deficiency leading to improved P nutrition and plant growth. In an initial screening of a soil PGPR collection for phytase-producing strains, we employed PA as the sole source of P on the NBRIP agar medium. Our results indicate that solubilization halos surrounding bacterial colonies were observed in less than half of the tested strains. Additional staining with ammonium molybdate reagent confirmed that these bacteria have a phytase activity able to break down PA, releasing Pi into the growth medium (Figure 1A). Three strains (C2, N4, and S10) were selected for further analyses because of their relatively high phytase activity ranging from 42.8 to 100.55 U/g (Figure 1C).
Genome sequencing of the selected strains revealed that C2 and N4 should belong to the Serratia genus and S10 is affiliated with the Citrobacter genus. In previous studies, several Serratia and Citrobacter strains were reported as phytase-producing bacteria. The phytases found in Citrobacter (Kim et al., 2006; Patel et al., 2010; Tkachenko et al., 2021) harbor the conserved motifs (N-terminal RHGXRXP and the C-terminal HD) characteristic of the active center of phytases of the histidine acid phosphatase (HAP) family (Van Etten et al., 1991; Wodzinski and Ullah, 1996; Kumar et al., 2012). The majority of HAP members exhibit optimum catalytic efficiency at approximately pH 4.0–5.5, while most of the β-propeller phytases exhibit optimal activity within the neutral pH (6 to 8; Oh et al., 2001; Jang et al., 2018). In Serratia sp. TN49, two phytases were reported: One belongs to HAP and one to β-propeller phytase families with two β-propeller phytase domains (Zhang et al., 2011).
Based on phytase quantitative assays, the three PGPR strains exhibit optimal in vitro activities at pH 7.5 for S10 and pH 5.5 for C2 and N4 strains, in the presence of 2 mM Ca2+, which is required for the catalytic activity of all known β-propeller phytases (Fu et al., 2008; Jang et al., 2018). Therefore, our results confirm that the three strains have a phytase activity that effectively hydrolyzes PA under the tested conditions (Figure 1).
The genome analysis of the three PGPR strains studied here revealed the presence between one and three putative phytase genes that are highly homologous to phytase-encoding sequences from Serratia marcescens KS10 (accession number NZ_CP027798.1) and PSB-15 (accession number KR133277.1) strains. Among those, the same phytase sequence of 1,362 bp was found in C2 and N4 (the latter has two other phytase genes). The corresponding protein of 453 a.a. harbors a phytase-like domain (residues 34–371) carrying several highly conserved Glu and Asp residues typical of the β_propeller class of phytases that are known to have a preference for hydrolyzing every second phosphate from its PA substrate (Kerovuo et al., 2000). This phytase is most likely a secreted one as a peptide cleavage site (position 20–21) was predicted with a very high probability (0.9986).
On the other hand, numerous studies have revealed that transgenic plants secreting microbial-origin phytases can effectively utilize phytate when cultivated under laboratory conditions (Brinch-Pedersen et al., 2000; Ponstein et al., 2002; Hong et al., 2004; Lung et al., 2005; Bilyeu et al., 2008; Wang et al., 2013; Belgaroui et al., 2016). However, more sustainable approaches based on co-cultivating plants with bacteria that synthesize and secrete phytases or adding purified microbial phytases have shown their effectiveness in improving P plant nutrition and growth (Hayes et al., 2000; Li and Zhang, 2001; Idriss et al., 2002; Vessey, 2003; Fuentes-Ramirez and Caballero-Mellado, 2006). In this study, we investigated whether the action of phytase-producing strains, used to inoculate barley plants, influences growth. As far as we know, such an investigation was never reported on barley. Through inoculation assays, we aimed to mimic natural plant growth conditions by using poor soil where Pi is not or lowly available, and instead, we supplied PA as a Po source. The inoculation tests provided insights into the impact of each strain on plant morphology, physiology, and molecular processes. Our data indicated that the inoculation of barley plants with the three PGPR strains, when PA is present as the sole source of P, resulted in enhanced growth in the aerial part (Figure 2) and especially in terms of root development (Figures 2, 3). In a previous study, we have demonstrated that the addition of the purified phytase phyUS417 from Bacillus subtilis (used here as a positive control in our screening assays of phytase-producing bacterial isolates) enhanced the growth of Arabidopsis seedlings on P-low medium, with an increase of five times in shoot and root biomasses (Belgaroui et al., 2014). Therefore, one would expect that the plant inoculation with this phytase-producing strain may enhance growth in soil under P-limited conditions.
The modifications in the root architecture, including changes in primary root length, lateral root system, and root hair structure, are a major strategy adopted to enhance mineral acquisition in different plant species (Bates and Lynch, 2000; Péret et al., 2011; Smith and De Smet, 2012; Kong et al., 2019; Li et al., 2019; Sun et al., 2019). Here, the overall improvement in barley root growth (Figures 3A–D) can help in a better exploitation of low-fertility soils. In this regard, amending the soil with phytase-producing bacteria should be further investigated for setting sustainable cropping practices. Interestingly, the improvement of the root growth correlates with the presence of the phytase-producing strains and leads to a higher P accumulation in roots and leaves (Figures 4A,B). Consistently, the increase in P leaf contents was translated into higher chlorophyll contents indicating that the inoculated barley plants (especially with C2 strain) are able to maintain relatively higher photosynthetic capacity under Pi deficiency (Figure 4C). Previous studies have shown that Pi deficiency inhibits photosynthesis in barley, soybean, maize, tomato, and other crops (Giersch and Robinson, 1987; Jacob and Lawlor, 1992; Frydenvang et al., 2015; Chu et al., 2018). A recent study has demonstrated that Pi deficiency resulted in a decrease in phospholipid contents in leaf cell membranes, caused the peroxidation of membrane lipids, disrupted the ultrastructure of chloroplasts, and further inhibited photosynthesis (Li et al., 2022). Photosynthetic rate is of high relevance, especially in flag leaves of cereals, which are the main source of carbohydrates for grain filling and thus impact significantly the final yield (Fabre et al., 2016).
Taken together, our findings clearly demonstrate the ability of the three phytase-producing strains to increase the Pi availability (via PA hydrolysis enabled by bacterial enzymes) which results in higher P nutrition, photosynthetic capacity, and growth of barley under Pi-limited conditions. However, the exploitation of these strains under field conditions to enhance crop production remains a significant challenge as their effectiveness may be influenced by interactions with other microorganisms and environmental factors (Hameeda et al., 2008; Gontia-Mishra et al., 2017). Recently, it has been shown that applying biochar enhances crop productivity by increasing soil nutrient supply and microbial activity (Hussain et al., 2017). Moreover, biochar seems indeed to enhance soil quality and fertility and exert stimulating effects on the soil microbial biomass and respiration, which might result in higher grain yield (Nasiri et al., 2023). Remarkably, the application of Serratia odorifera, in combination with biochar, was shown to be more effective in increasing soil fertility and barley growth under drought stress, compared to the use of this PGPR strain alone (Gul et al., 2023). Therefore, a future soil amendment with the phytase-producing PGPR characterized here in combination with biochar can be an attractive approach to monitor barley growth not only in poor quality soil but also under drought conditions.
It is noteworthy that Pi uptake is particularly dependent on the high-affinity Pi/H+ symporters of the PHT1 (phosphate transporter 1) family (Shen et al., 2011). Members of this family play a critical role in Pi acquisition from soil solution and remobilization throughout plant development. Nine members of the PHT1 gene family have been found in the Arabidopsis genome (Mudge et al., 2002), 13 in rice (Oryza sativa; Paszkowski et al., 2002), and eight PHT1 genes in barley (Smith et al., 1999). Under P limitation, several PHT1 genes are rapidly induced as they all have a P1BS-like cis-element in their promoter, which is associated with the response to P-starvation (Rubio et al., 2001). In addition, they are under the control of a common transcriptional regulatory mechanism, governed by the MYB transcription factor PHR1 (for review, see Gojon et al., 2009). In this context, we decided to test the impact of barley inoculation with these three PGPR on the expression of a number of transporters of the PHT1 family. A significant downregulation of HvPHT1.1, PHT1.4, and PHT1.8 genes was registered in the roots of barley when inoculated with the three PGPR strains (Figure 5A), providing further evidence for the increase of Pi available in the soil, after PA hydrolysis by the bacterial phytases. A similar scenario was reported by Pacak et al. (2016) where HvIPS1, HvPHT1;1, HvPHT1;4, HvPHT1;6, and MIR399c exhibit a clear response to Pi deprivation. Following Pi replenishment, a reduction in the expression levels of these genes was observed. It has been demonstrated that PHO2 [PHOSPHATE2, ubiquitin-conjugating E2 enzyme (UBC24)] works with nitrogen limitation adaptation (NLA) protein, and they are both responsible for PHT1;4 degradation via the 26S proteasome (Park et al., 2014). PHO2 is involved in defining cellular Pi homeostasis and is responsible for the ubiquitin transfer to the protein substrate via the E3 enzyme (ubiquitin-protein ligase; Smalle and Vierstra, 2004). Furthermore, PHO2 is implicated in the degradation of the PHT1;1, PHT1;2, PHT1;3, PHT1;4, PHO1, and PHOSPHATE TRANSPORTER TRAFFIC FACILITATOR1 (PHF1; Liu et al., 2012; Huang et al., 2013). Therefore, it serves as a negative regulator of Pi uptake. However, we noticed that the expression of PHO2 was not significantly affected in the inoculated barley (Figure 5). It is known that Arabidopsis PHO2 exhibits ubiquitous expression, and it is only weakly affected by Pi status (Bari et al., 2006). Similarly, Pacak et al. (2016) showed that, in the later stages of barley development, Pi re-supply did not significantly alter the level of PHO2 expression, despite the expression of pri-miR399 and, consequently, mature miR399 was strongly decreased. PHR1 expression seems also to be unchanged in both barley roots and shoots (Figure 5) following inoculation with the three PGPR strains. This result is expected and agrees with a previous report where PHR1 expression appears to be constitutive, regardless of plant P status (Sega and Pacak, 2019). Overall, the observed growth promotion and downregulation of high-affinity Pi transporters in barley grown in the presence of PA demonstrate that Pi could be the end product of PA degradation by the bacterial phytases.
The present study reports the positive role of phytase-producing PGPRs in improving barley growth and phosphate nutrition. Our data clearly show that the phytase activity produced by the three PGPR strains is capable to solubilize PA and increase Pi availability for barley grown in Pi-limited conditions. This beneficial effect should be leveraged in using phytase-producing PGPR to improve soil P fertility and hence to reduce the use of conventional P fertilizers, especially in cereal production.
Publicly available datasets were analyzed in this study. This data can be found at: SAMN38274870; PP621716.1; PP621717.1.
WI: Validation, Writing – original draft, Writing – review & editing, Data curation, Investigation, Methodology, Visualization. NB: Validation, Writing – original draft, Writing – review & editing, Investigation, Methodology, Visualization. NS: Methodology, Writing – review & editing, Formal analysis. IG: Writing – review & editing, Data curation, Software. MH: Writing – review & editing, Conceptualization, Funding acquisition, Project administration, Supervision, Validation, Writing – original draft.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was supported by the European project INPLANTOMICS (Project number: 101078905) in the frame of the HORIZON-WIDERA-2021-ACCESS-03 Program.
We thank Abdelmalek Alioua and Valérie Cognat from the IBMP, Strasbourg, for their technical support with genome sequencing.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The handling editor MT declared a shared parent affiliation with the authors at the time of review.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1432599/full#supplementary-material
2. ^ https://mage.genoscope.cns.fr/microscope/metabolism/metabolicprofil.php?
3. ^ https://web.expasy.org/translate/
Amtmann, A., and Blatt, M. R. (2009). Regulation of macronutrient transport. New Phytol. 181, 35–52. doi: 10.1111/j.1469-8137.2008.02666.x
Arnon, D. I. (1949). Copper enzymes in isolated chloroplasts. Polyphenoloxidase in Beta vulgaris. Plant Physiol. 24, 1–15.
Badr, A., Sch, R., Rabey, H. E., Effgen, S., Ibrahim, H. H., Pozzi, C., et al. (2000). On the origin and domestication history of barley (Hordeum vulgare). Mol. Biol. Evol. 17, 499–510. doi: 10.1093/oxfordjournals.molbev.a026330
Bari, R., Datt Pant, B., Stitt, M., and Scheible, W. R. (2006). PHO2, microRNA399, and PHR1 define a phosphate-signaling pathway in plants. Plant Physiol. 141, 988–999. doi: 10.1104/pp.106.079707
Bates, T. R., and Lynch, J. P. (2000). The efficiency of Arabidopsis thaliana (Brassicaceae) root hairs in phosphorus acquisition. Am. J. Bot. 87, 964–970. doi: 10.2307/2656995
Belgaroui, N., Berthomieu, P., Rouached, H., and Hanin, M. (2016). The secretion of the bacterial phytase PHY-US 417 by Arabidopsis roots reveals its potential for increasing phosphate acquisition and biomass production during co-growth. Plant Biotechnol. J. 14, 1914–1924. doi: 10.1111/pbi.12552
Belgaroui, N., El Ifa, W., and Hanin, M. (2022). Phytic acid contributes to the phosphate-zinc signaling crosstalk in Arabidopsis. Plant Physiol. Biochem. 183, 1–8. doi: 10.1016/j.plaphy.2022.04.029
Belgaroui, N., Zaidi, I., Farhat, A., Chouayekh, H., Bouain, N., Chay, S., et al. (2014). Over-expression of the bacterial phytase US417 in Arabidopsis reduces the concentration of phytic acid and reveals its involvement in the regulation of sulfate and phosphate homeostasis and signaling. Plant Cell Physiol. 55, 1912–1924. doi: 10.1093/pcp/pcu122
Bilyeu, K. D., Zeng, P., Coello, P., Zhang, Z. J., Krishnan, H. B., Bailey, A., et al. (2008). Quantitative conversion of phytate to inorganic phosphorus in soybean seeds expressing a bacterial phytase. Plant Physiol. 146, 468–477. doi: 10.1104/pp.107.113480
Brinch-Pedersen, H., Olesen, A., Rasmussen, S. K., and Holm, P. B. (2000). Generation of transgenic wheat (Triticum aestivum L.) for constitutive accumulation of an aspergillus phytase. Mol. Breed. 6, 195–206. doi: 10.1023/A:1009690730620
Brinch-Pedersen, H., Sørensen, L. D., and Holm, P. B. (2002). Engineering crop plants: getting a handle on phosphate. Trends Plant Sci. 7, 118–125. doi: 10.1016/S1360-1385(01)02222-1
Chien, P. S., Chiang, C. P., Leong, S. J., and Chiou, T. J. (2018). Sensing and signaling of phosphate starvation: from local to long distance. Plant Cell Physiol. 59, 1714–1722. doi: 10.1093/pcp/pcy148
Chu, S., Li, H., Zhang, X., Yu, K., Chao, M., Han, S., et al. (2018). Physiological and proteomics analyses reveal low-phosphorus stress affected the regulation of photosynthesis in soybean. Int. J. Mol. Sci. 19:1688. doi: 10.3390/ijms19061688
Chungopast, S., Thongjoo, C., Islam, A. M., and Yeasmin, S. (2021). Efficiency of phosphate-solubilizing bacteria to address phosphorus fixation in Takhli soil series: a case of sugarcane cultivation, Thailand. Plant Soil 460, 347–357. doi: 10.1007/s11104-020-04812-w
Cordell, D., Drangert, J. O., and White, S. (2009). The story of phosphorus: global food security and food for thought. Glob. Environ. Chang. 19, 292–305. doi: 10.1016/j.gloenvcha.2008.10.009
Corrêa, T. L. R., and Araújo, D. (2020). Fungal phytases: from genes to applications. Braz. J. Microbiol. 51, 1009–1020. doi: 10.1007/s42770-020-00289-y
Dailin, D. J., Hanapi, S. Z., Elsayed, E., Sukmawati, D., Wan Azelee, N. I., Eyahmalay, J., et al. (2019). “Fungal phytases: biotechnological applications in food and feed industries” in Recent advancement in White biotechnology through Fungi. Fungal biology. eds. A. Yadav, S. Singh, S. Mishra, and A. Gupta (Cham: Springer), 65–99.
Engelen, A. J., Van Der Heeft, F. C., Randsdorp, P. H., and Smtt, E. L. (1994). Simple and rapid determination of phytase activity. J. AOAC Int. 77, 760–764. doi: 10.1093/jaoac/77.3.760
Fabre, D., Adriani, D. E., Dingkuhn, M., Ishimaru, T., Punzalan, B., Lafarge, T., et al. (2016). The qTSN4 effect on flag leaf size, photosynthesis and panicle size, benefits to plant grain production in rice, depending on light availability. Front. Plant Sci. 7:623. doi: 10.3389/fpls.2016.00623
Frydenvang, J., van Maarschalkerweerd, M., Carstensen, A., Mundus, S., Schmidt, S. B., Pedas, P. R., et al. (2015). Sensitive detection of phosphorus deficiency in plants using chlorophyll a fluorescence. Plant Physiol. 169, 353–361. doi: 10.1104/pp.15.00823
Fu, S., Sun, J., Qian, L., and Li, Z. (2008). Bacillus phytases: present scenario and future perspectives. Appl. Biochem. Biotechnol. 151, 1–8. doi: 10.1007/s12010-008-8158-7
Fuentes-Ramirez, L. E., and Caballero-Mellado, J. (2006). Bacterial biofertilizers. In: PGPR: Biocontrol and biofertilization. eds. A. Yadav (Dordrecht: Springer), 143–172.
Geng, L., Li, M., Zhang, G., and Ye, L. (2022). Barley: a potential cereal for producing healthy and functional foods. Food Quality and Safety 6:fyac012. doi: 10.1093/fqsafe/fyac012
Gerke, J. (2022). The central role of soil organic matter in soil fertility and carbon storage. Soil Systems 6:33. doi: 10.3390/soilsystems6020033
Giersch, C., and Robinson, S. P. (1987). Regulation of photosynthetic carbon metabolism during phosphate limitation of photosynthesis in isolated spinach chloroplasts. Photosynth. Res. 14, 211–227. doi: 10.1007/BF00032706
Gojon, A., Nacry, P., and Davidian, J. C. (2009). Root uptake regulation: a central process for NPS homeostasis in plants. Curr. Opin. Plant Biol. 12, 328–338. doi: 10.1016/j.pbi.2009.04.015
Gontia-Mishra, I., Swapnil, S., and Sharad,. (2017). Zinc solubilizing bacteria from the rhizosphere of rice as prospective modulator of zinc biofortification in rice. Rhizosphere 3, 185–190. doi: 10.1016/j.rhisph.2017.04.013
Gul, F., Khan, I. U., Rutherford, S., Dai, Z. C., Li, G., and Du, D. L. (2023). Plant growth promoting rhizobacteria and biochar production from Parthenium hysterophorus enhance seed germination and productivity in barley under drought stress. Front. Plant Sci. 14:1175097. doi: 10.3389/fpls.2023.1175097
Ham, B. K., Chen, J., Yan, Y., and Lucas, W. J. (2018). Insights into plant phosphate sensing and signaling. Curr. Opin. Biotechnol. 49, 1–9. doi: 10.1016/j.copbio.2017.07.005
Hameeda, B., Harini, G., Rupela, O. P., Wani, S. P., and Reddy, G. (2008). Growth promotion of maize by phosphate-solubilizing bacteria isolated from composts and macrofauna. Microbiol. Res. 163, 234–242. doi: 10.1016/j.micres.2006.05.009
Hayes, J. E., Richardson, A. E., and Simpson, R. J. (2000). Components of organic phosphorus in soil extracts that are hydrolysed by phytase and acid phosphatase. Biol. Fertil. Soils 32, 279–286. doi: 10.1007/s003740000249
Hong, C. Y., Cheng, K. J., Tseng, T. H., Wang, C. S., Liu, L. F., and Yu, S. M. (2004). Production of two highly active bacterial phytases with broad pH optima in germinated transgenic rice seeds. Transgenic Res. 13, 29–39. doi: 10.1023/B:TRAG.0000017158.96765.67
Huang, T. K., Han, C. L., Lin, S. I., Chen, Y. J., Tsai, Y. C., Chen, Y. R., et al. (2013). Identification of downstream components of ubiquitin-conjugating enzyme PHOSPHATE2 by quantitative membrane proteomics in Arabidopsis roots. Plant Cell 25, 4044–4060. doi: 10.1105/tpc.113.115998
Hussain, M., Farooq, M., Nawaz, A., Al-Sadi, A. M., Solaiman, Z. M., Alghamdi, S. S., et al. (2017). Biochar for crop production: potential benefits and risks. J. Soils Sediments 17, 685–716. doi: 10.1007/s11368-016-1360-2
Idriss, E. E., Makarewicz, O., Farouk, A., Rosner, K., Greiner, R., Bochow, H., et al. (2002). Extracellular phytase activity of Bacillus amyloliquefaciens FZB45 contributes to its plant-growth-promoting effect. Microbiology 148, 2097–2109. doi: 10.1099/00221287-148-7-2097
Jacob, J., and Lawlor, D. W. (1992). Dependence of photosynthesis of sunflower and maize leaves on phosphate supply, ribulose-1, 5-bisphosphate carboxylase/oxygenase activity, and ribulose-1, 5-bisphosphate pool size. Plant Physiol. 98, 801–807. doi: 10.1104/pp.98.3.801
Jang, W. J., Lee, J. M., Park, H. D., Choi, Y. B., and Kong, I. S. (2018). N-terminal domain of the beta-propeller phytase of Pseudomonas sp. FB15 plays a role for retention of low-temperature activity and catalytic efficiency. Enzym. Microb. Technol. 117, 84–90. doi: 10.1016/j.enzmictec.2018.06.008
Janská, A., Hodek, J., Svoboda, P., Zámečník, J., Prášil, I. T., Vlasáková, E., et al. (2013). The choice of reference gene set for assessing gene expression in barley (Hordeum vulgare L.) under low temperature and drought stress. Mol. Gen. Genomics. 288, 639–649. doi: 10.1007/s00438-013-0774-4
Jorquera, M. A., Gabler, S., Inostroza, N. G., Acuña, J. J., Campos, M. A., Menezes-Blackburn, D., et al. (2018). Screening and characterization of phytases from bacteria isolated from Chilean hydrothermal environments. Microb. Ecol. 75, 387–399. doi: 10.1007/s00248-017-1057-0
Kalsi, H. K., Singh, R., Dhaliwal, H. S., and Kumar, V. (2016). Phytases from Enterobacter and Serratia species with desirable characteristics for food and feed applications. 3 Biotech 6, 64–13. doi: 10.1007/s13205-016-0378-x
Karnwal, A. (2017). Isolation and identification of plant growth promoting rhizobacteria from maize (Zea mays L.) rhizosphere and their plant growth promoting effect on rice (Oryza sativa L.). J. Plant Protection Res. 57, 144–151. doi: 10.1515/jppr-2017-0020
Kerovuo, J., Rouvinen, J., and Hatzack, F. (2000). Analysis of myo-inositol hexakisphosphate hydrolysis by Bacillus phytase: indication of a novel reaction mechanism. Biochem. J. 352, 623–628. doi: 10.1042/bj3520623
Kim, Y. O., Kim, H. W., Lee, J. H., Kim, K. K., and Lee, S. J. (2006). Molecular cloning of the phytase gene from Citrobacter braakii and its expression in Saccharomyces cerevisiae. Biotechnol. Lett. 28, 33–38. doi: 10.1007/s10529-005-9684-9
Kochian, L. V., Hoekenga, O. A., and Pineros, M. A. (2004). How do crop plants tolerate acid soils? Mechanisms of aluminum tolerance and phosphorous efficiency. Annu. Rev. Plant Biol. 55, 459–493. doi: 10.1146/annurev.arplant.55.031903.141655
Kong, Y., Wang, B., Du, H., Li, W., Li, X., and Zhang, C. (2019). GmEXLB1, a soybean expansin-like B gene, alters root architecture to improve phosphorus acquisition in Arabidopsis. Front. Plant Sci. 10:808. doi: 10.3389/fpls.2019.00808
Kumar, V., Sinha, A. K., Makkar, H. P., De Boeck, G., and Becker, K. (2012). Phytate and phytase in fish nutrition. J. Anim. Physiol. Anim. Nutr. 96, 335–364. doi: 10.1111/j.1439-0396.2011.01169.x
Li, J., Chen, F., Li, Y., Li, P., Wang, Y., Mi, G., et al. (2019). ZmRAP2. 7, an AP2 transcription factor, is involved in maize brace roots development. Front. Plant Sci. 10:820. doi: 10.3389/fpls.2019.00820
Li, P., Yu, J., Feng, N., Weng, J., Rehman, A., Huang, J., et al. (2022). Physiological and transcriptomic analyses uncover the reason for the inhibition of photosynthesis by phosphate deficiency in Cucumis melo L. Int. J. Mol. Sci. 23:12073. doi: 10.3390/ijms232012073
Li, Z., and Zhang, H. (2001). Application of microbial fertilizers in sustainable agriculture. J. Crop. Prod. 3, 337–347. doi: 10.1300/J144v03n01_28
Lihan, S., Benet, F., Husaini, A. A. S. A., Apun, K., Roslan, H. A., and Hassan, H. (2021). Isolation and identification of plant growth promoting rhizobacteria from sago palm (Metroxylon sagu, Rottb.). Tropical Life Sciences Research 32, 39–51. doi: 10.21315/tlsr2021.32.3.3
Liu, T. Y., Huang, T. K., Tseng, C. Y., Lai, Y. S., Lin, S. I., Lin, W. Y., et al. (2012). PHO2-dependent degradation of PHO1 modulates phosphate homeostasis in Arabidopsis. Plant Cell 24, 2168–2183. doi: 10.1105/tpc.112.096636
López-Arredondo, D. L., Sánchez-Calderón, L., and Yong-Villalobos, L. (2017). Molecular and genetic basis of plant macronutrient use efficiency: concepts, opportunities, and challenges. Plant macronutrient use efficiency. 1–29. doi: 10.1016/B978-0-12-811308-0.00001-6
Lung, S. C., Chan, W. L., Yip, W., Wang, L., Yeung, E. C., and Lim, B. L. (2005). Secretion of beta-propeller phytase from tobacco and Arabidopsis roots enhances phosphorus utilization. Plant Sci. 169, 341–349. doi: 10.1016/j.plantsci.2005.03.006
Manzoor, M., Abbasi, M. K., and Sultan, T. (2017). Isolation of phosphate solubilizing bacteria from maize rhizosphere and their potential for rock phosphate solubilization–mineralization and plant growth promotion. Geomicrobiol J. 34, 81–95. doi: 10.1080/01490451.2016.1146373
Mudge, S. R., Rae, A. L., Diatloff, E., and Smith, F. W. (2002). Expression analysis suggests novel roles for members of the Pht1 family of phosphate transporters in Arabidopsis. Plant J. 31, 341–353. doi: 10.1046/j.1365-313X.2002.01356.x
Murphy, J. A. M. E. S., and Riley, J. P. (1962). A modified single solution method for the determination of phosphate in natural waters. Anal. Chim. Acta 27, 31–36. doi: 10.1016/S0003-2670(00)88444-5
Nasiri, S., Andalibi, B., Tavakoli, A., Delavar, M. A., El-Keblawy, A., Zwieten, L. V., et al. (2023). The mineral biochar alters the biochemical and microbial properties of the soil and the grain yield of Hordeum vulgare L. under drought stress. Landscape 12:559. doi: 10.3390/land12030559
Nautiyal, C. S. (1999). An efficient microbiological growth medium for screening phosphate solubilizing microorganisms. FEMS Microbiol. Lett. 170, 265–270. doi: 10.1111/j.1574-6968.1999.tb13383.x
Nguyen, C., Yan, W., Le Tacon, F., and Lapeyrie, F. (1992). Genetic variability of phosphate solubilizing activity by monocaryotic and dicaryotic mycelia of the ectomycorrhizal fungus Laccaria bicolor (Maire) PD Orton. Plant Soil 143, 193–199. doi: 10.1007/BF00007873
Oh, B. C., Chang, B. S., Park, K. H., Ha, N. C., Kim, H. K., Oh, B. H., et al. (2001). Calcium-dependent catalytic activity of a novel phytase from Bacillus amyloliquefaciens DS11. Biochemistry 40, 9669–9676. doi: 10.1021/bi010589u
Pacak, A., Barciszewska-Pacak, M., Swida-Barteczka, A., Kruszka, K., Sega, P., Milanowska, K., et al. (2016). Heat stress affects pi-related genes expression and inorganic phosphate deposition/accumulation in barley. Front. Plant Sci. 7:196647. doi: 10.3389/fpls.2016.00926
Pan, F., Guo, Z., Cai, Y., Liu, H., Wu, J., Fu, Y., et al. (2019). Kinetic exchange of remobilized phosphorus related to phosphorus-iron-sulfur biogeochemical coupling in coastal sediment. Water Resour. Res. 55, 10494–10517. doi: 10.1029/2019WR025941
Park, B. S., Seo, J. S., and Chua, N. H. (2014). NITROGEN LIMITATION ADAPTATION recruits PHOSPHATE2 to target the PHOSPHATE transporter PT2 for degradation during the regulation of Arabidopsis phosphate homeostasis. Plant Cell 26, 454–464. doi: 10.1105/tpc.113.120311
Paszkowski, U., Kroken, S., Roux, C., and Briggs, S. P. (2002). Rice phosphate transporters include an evolutionarily divergent gene specifically activated in arbuscular mycorrhizal symbiosis. Proceed. National Academy of Sci., USA 99, 13324–13329. doi: 10.1073/pnas.202474599
Patel, K. J., Vig, S., Nareshkumar, G., and Archana, G. (2010). Effect of transgenic rhizobacteria overexpressing Citrobacter braakii appA on phytate-P availability to mung bean plants. J. Microbiol. Biotechnol. 20, 1491–1499. doi: 10.4014/jmb.1006.06016
Péret, B., Clément, M., Nussaume, L., and Desnos, T. (2011). Root developmental adaptation to phosphate starvation: better safe than sorry. Trends Plant Sci. 16, 442–450. doi: 10.1016/j.tplants.2011.05.006
Plaxton, W. C., and Tran, H. T. (2011). Metabolic adaptations of phosphate-starved plants. Plant Physiol. 156, 1006–1015. doi: 10.1104/pp.111.175281
Ponstein, A. S., Bade, J. B., Verwoerd, T. C., Molendijk, L., Storms, J., Beudeker, R. F., et al. (2002). Stable expression of phytase (phyA) in canola (Brassica napus) seeds: towards a commercial product. Mol. Breed. 10, 31–44. doi: 10.1023/A:1020326219687
Raboy, V. (2020). Low phytic acid crops: observations based on four decades of research. Plan. Theory 9:140. doi: 10.3390/plants9020140
Raghothama, K. G. (1999). Phosphate acquisition. Annu. Rev. Plant Biol. 50, 665–693. doi: 10.1146/annurev.arplant.50.1.665
Richardson, A. E., Barea, J. M., and McNeill, A. M. (2009). Acquisition of phosphorus and nitrogen in the rhizosphere and plant growth promotion by microorganisms. Plant Soil 321, 305–339. doi: 10.1007/s11104-009-9895-2
Richardson, A. E., Hadobas, P. A., and Hayes, J. E. (2000). Acid phosphomonoesterase and phytase activities of wheat (Triticum aestivum L.) roots and utilization of organic phosphorus substrates by seedlings grown in sterile culture. Plant Cell Environ. 23, 397–405. doi: 10.1046/j.1365-3040.2000.00557.x
Richardson, A. E., Hadobas, P. A., and Hayes, J. E. (2001). Extracellular secretion of aspergillus phytase from Arabidopsis roots enables plants to obtain phosphorus from phytate. Plant J. 25, 641–649. doi: 10.1046/j.1365-313x.2001.00998.x
Rix, G. D., Sprigg, C., Whitfield, H., Hemmings, A. M., Todd, J. D., and Brearley, C. A. (2022). Characterisation of a soil MINPP phytase with remarkable long-term stability and activity from Acinetobacter sp. PLoS One 17:e0272015. doi: 10.1371/journal.pone.0272015
Rodriguez, H., and Fraga, R. (1999). Phosphate solubilizing bacteria and their role in plant growth promotion. Biotechnol. Adv. 17, 319–339. doi: 10.1016/S0734-9750(99)00014-2
Rubio, V., Linhares, F., Solano, R., Martín, A. C., Iglesias, J., Leyva, A., et al. (2001). A conserved MYB transcription factor involved in phosphate starvation signaling both in vascular plants and in unicellular algae. Genes Dev. 15, 2122–2133. doi: 10.1101/gad.204401
Sabagh, A. E., Hossain, A., Islam, M. S., Barutcular, C., Hussain, S., Hasanuzzaman, M., et al. (2019). Drought and salinity stresses in barley: consequences and mitigation strategies. Aust. J. Crop. Sci. 13, 810–820. doi: 10.3316/INFORMIT.580050100084352
Saharan, B. S., and Nehra, V. (2011). Plant growth promoting rhizobacteria: a critical review. Life Sci Med Res 6:30. doi: 10.4172/2157-7471.1000266
Sayahi, N., Abdelkefi, N., Ebel, C., Mechichi, T., and Hanin, M. (2024). Whole genome sequences of four plant growth promoting Rhizobacteria strains from different Tunisian rhizospheres. Zenodo. doi: 10.5281/zenodo.10558070
Sayahi, N., Djemal, R., Ben Merdes, K., Saidii, M. N., Yengui, M., Gdoura, R., et al. (2022). Characterization of Siccibacter sp. strain C2 a novel Rhizobacterium that enhances tolerance of barley to salt stress. Curr. Microbiol. 79:239. doi: 10.1007/s00284-022-02930-5
Schindelin, J., Arganda-Carreras, I., Frise, E., Kaynig, V., Longair, M., Pietzsch, T., et al. (2012). Fiji: an open-source platform for biological-image analysis. Nat. Methods 9, 676–682. doi: 10.1038/nmeth.2019
Sega, P., and Pacak, A. (2019). Plant PHR transcription factors: put on a map. Gene 10:1018. doi: 10.3390/genes10121018
Shedova, E., Lipasova, V., Velikodvorskaya, G., Ovadis, M., Chernin, L., and Khmel, I. (2008). Phytase activity and its regulation in a rhizospheric strain of Serratia plymuthica. Folia Microbiol. 53, 110–114. doi: 10.1007/s12223-008-0016-z
Shen, J., Yuan, L., Zhang, J., Li, H., Bai, Z., Chen, X., et al. (2011). Phosphorus dynamics: from soil to plant. Plant Physiol. 156, 997–1005. doi: 10.1104/pp.111.175232
Singh, B., and Satyanarayana, T. (2011). Microbial phytases in phosphorus acquisition and plant growth promotion. Physiol. Mol. Biol. Plants 17, 93–103. doi: 10.1007/s12298-011-0062-x
Smalle, J., and Vierstra, R. D. (2004). The ubiquitin 26S proteasome proteolytic pathway. Annu. Rev. Plant Biol. 55, 555–590. doi: 10.1146/annurev.arplant.55.031903.141801
Smith, F. W., Cybinski, D. H., and Rae, A. L. (1999). “Regulation of expression of genes encoding phosphate transporters in barley roots” in Plant nutrition—Molecular biology and genetics: Proceedings of the sixth international symposium on genetics and molecular biology of plant nutrition (Dordrecht: Springer Netherlands), 145–150.
Smith, S., and De Smet, I. (2012). Root system architecture: insights from Arabidopsis and cereal crops. Philosop. Trans. Royal Society B: Biolog. Sci. 367, 1441–1452. doi: 10.1098/rstb.2011.0234
Sun, H., Guo, X., Xu, F., Wu, D., Zhang, X., Lou, M., et al. (2019). Overexpression of OsPIN2 regulates root growth and formation in response to phosphate deficiency in rice. Int. J. Mol. Sci. 20:5144. doi: 10.3390/ijms20205144
Tao, W., Li, R., Li, T., Li, Z., Li, Y., and Cui, L. (2023). The evolutionary patterns, expression profiles, and genetic diversity of expanded genes in barley. Front. Plant Sci. 14:1168124. doi: 10.3389/fpls.2023.1168124
Tarafdar, J. C., and Claassen, N. (1988). Organic phosphorus compounds as a phosphorus source for higher plants through the activity of phosphatases produced by plant roots and microorganisms. Biol. Fertil. Soils 5, 308–312. doi: 10.1007/BF00262137
Tarafdar, J. C., and Rao, A. V. (1996). Contribution of aspergillus strains to acquisition of phosphorus by wheat (Triticum aestivum L.) and chick pea (Cicer arietinum Linn.) grown in a loamy sand soil. Appl. Soil Ecol. 3, 109–114. doi: 10.1016/0929-1393(95)00084-4
Tkachenko, A. A., Kalinina, A. N., Borshchevskaya, L. N., Sineoky, S. P., and Gordeeva, T. L. (2021). A novel phytase from Citrobacter gillenii: characterization and expression in Pichia pastoris (Komagataella pastoris). FEMS Microbiol. Lett. 368:fnaa217. doi: 10.1093/femsle/fnaa217
Turner, B. L., Mahieu, N., and Condron, L. M. (2003). Quantification of myo-inositol hexakisphosphate in alkaline soil extracts by solution 31P NMR spectroscopy and spectral deconvolution. Soil Sci. 168, 469–478. doi: 10.1097/01.ss.0000080332.10341.ed
Van Etten, R. L., Davidson, R., Stevis, P. E., MacArthur, H., and Moore, D. L. (1991). Covalent structure, disulfide bonding, and identification of reactive surface and active site residues of human prostatic acid phosphatase. J. Biol. Chem. 266, 2313–2319. doi: 10.1016/S0021-9258(18)52245-6
Vance, C. P., Uhde-Stone, C., and Allan, D. L. (2003). Phosphorus acquisition and use: critical adaptations by plants for securing a nonrenewable resource. New Phytol. 157, 423–447. doi: 10.1046/j.1469-8137.2003.00695.x
Vessey, J. K. (2003). Plant growth promoting rhizobacteria as biofertilizers. Plant Soil 255, 571–586. doi: 10.1023/A:1026037216893
Wan, W., Qin, Y., Wu, H., Zuo, W., He, H., Tan, J., et al. (2020). Isolation and characterization of phosphorus solubilizing bacteria with multiple phosphorus sources utilizing capability and their potential for lead immobilization in soil. Front. Microbiol. 11:752. doi: 10.3389/fmicb.2020.00752
Wang, X., Shen, J., and Liao, H. (2010). Acquisition or utilization, which is more critical for enhancing phosphorus efficiency in modern crops? Plant Sci. 179, 302–306. doi: 10.1016/j.plantsci.2010.06.007
Wang, Y., Ye, X., Ding, G., and Xu, F. (2013). Overexpression of phyA and appA genes improves soil organic phosphorus utilisation and seed phytase activity in Brassica napus. PLoS One 8:e60801. doi: 10.1371/journal.pone.0060801
Wodzinski, R. J., and Ullah, A. H. J. (1996). “Phytase” in Advances in applied microbiology. eds. S. L. Neidleman and A. I. Laskin vol. 42 (USA: Academic Press), 263–302.
Yadav, R. S., and Tarafdar, J. C. (2003). Phytase and phosphatase producing fungi in arid and semi-arid soils and their efficiency in hydrolyzing different organic P compounds. Soil Biol. Biochem. 35, 745–751. doi: 10.1016/S0038-0717(03)00089-0
Yu, X., Liu, X., Zhu, T. H., Liu, G. H., and Mao, C. (2011). Isolation and characterization of phosphate-solubilizing bacteria from walnut and their effect on growth and phosphorus mobilization. Biol. Fertil. Soils 47, 437–446. doi: 10.1007/s00374-011-0548-2
Zhang, L., Fan, J., Ding, X., He, X., Zhang, F., and Feng, G. (2014). Hyphosphere interactions between an arbuscular mycorrhizal fungus and a phosphate solubilizing bacterium promote phytate mineralization in soil. Soil Biol. Biochem. 74, 177–183. doi: 10.1016/j.soilbio.2014.03.004
Keywords: barley, plant growth promoting rhizobacteria, phytic acid, phytase, Pi
Citation: El Ifa W, Belgaroui N, Sayahi N, Ghazala I and Hanin M (2024) Phytase-producing rhizobacteria enhance barley growth and phosphate nutrition. Front. Sustain. Food Syst. 8:1432599. doi: 10.3389/fsufs.2024.1432599
Received: 14 May 2024; Accepted: 27 June 2024;
Published: 12 July 2024.
Edited by:
Mohamed Trigui, Institut Préparatoire aux Etudes d’Ingénieur de Sfax (IPEIS), TunisiaReviewed by:
Ajay Kumar Pandey, National Agri-Food Biotechnology Institute, IndiaCopyright © 2024 El Ifa, Belgaroui, Sayahi, Ghazala and Hanin. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moez Hanin, bW9lei5oYW5pbkBpc2JzLnVzZi50bg==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.