- School of Veterinary Medicine, Wollo University, Dessie, Ethiopia
Food-producing animals including bovine species are major reservoirs for different food-borne pathogens. Staphylococcus aureus is among the causes of food-borne diseases globally that can be transmitted mainly through consumption of contaminated foods of animal origin and emergence of multidrug resistant bacteria like methicillin resistant S. aureus (MRSA) become a significant public health concern. A cross-sectional study was conducted from October 2019 to July 2021 to estimate the prevalence, identify associated risk factors and determine antibiogram profiles of S. aureus and MRSA from foods of bovine origin in Dessie and Kombolcha towns. A total of 384 foods of bovine origin samples were collected using random sampling techniques. Isolation and characterization of S. aureus were done according to the standard bacteriological protocols. Agar disc diffusion method was employed to determine the in vitro antimicrobial resistance pattern of S. aureus and MRSA isolates. The collected data were analyzed using descriptive and inferential statistics. The overall prevalence of S. aureus and MRSA were found to be the equal (39.3%). The prevalence of S. aureus was 55.6, 44.0, 41.1, 36.4, 16.7, and 0.0% in yogurt, beef swab, udder milk, carcass swab, tank milk, and cheese samples, respectively. A statistically significant difference was observed in the prevalence of S. aureus among the different sample types (P < 0.05). The prevalence of S. aureus in milk samples from cows with and without treatment history was 47.1 and 26.0%, respectively. The difference in the prevalence of S. aureus among treatment history categories was statistically significant (P < 0.05). Higher prevalence of S. aureus was recorded in carcass swab samples collected from Dessie town (50.0%), municipal abattoirs (46.7%), slaughtering process with poor hygiene (57.1%); and carcasses slaughtered by butchers with poor hygiene (62.1%). 100.0, 97.4, 90.1, and 74.8% of S. aureus isolates were resistant to Cefoxitin, Penicillin G, Ampicillin, and Nalidixic acid, respectively. 97.3% of S. aureus isolates showed multidrug resistance to three and more than three drugs. To reduce the high magnitude of S. aureus contamination of foods of bovine origin, improvement of cattle health and good hygienic procedures along the production chain should be implemented in the study areas.
1 Introduction
Food-borne diseases are globally important public health problems that can be transmitted to humans through consumption of food and water contaminated with microorganisms or chemicals (Mehrnaz et al., 2021). These diseases can occur in sporadic or outbreak forms and are frequently associated with morbidity, mortality, and economic losses around the world (Hemalata and Virupakshaiah, 2016; Khater et al., 2021; Borena et al., 2022; Pal et al., 2022).
Food-borne pathogens are responsible for a wide range of illnesses with serious consequences for both human health and the economy (Bintsis, 2017). Globally, food-borne disease outbreaks are becoming more frequent and more than 250 pathogens of food-borne illnesses have been identified (Liu et al., 2019; Al-Mohaithef, 2021). Due to the emergence of food-borne pathogens, microbial contamination of food is a significant public health concern (Al-Mohaithef, 2021). Food-producing animals including bovine species are the major reservoirs of different food-borne pathogens and foods of animal origin like meat, egg, milk and their products are the major sources and vehicles of food-borne infections (Oliver et al., 2005; Zakary et al., 2011; Heredia and Garcia, 2018; Kandil et al., 2018; Abebe et al., 2020).
Even though various microbial pathogens can contaminate food products of humans (Chao et al., 2007), both sporadic cases and outbreaks of food-borne illness are most commonly caused by bacteria and viruses (Truchado and Randazzo, 2022). Due to the high magnitude of occurrence and seriousness, some bacterial food-borne pathogens are significantly important (Loir et al., 2003). The leading bacterial causes of food-borne diseases include Staphylococcus aureus (S. aureus), Listeria monocytogenes (L. monocytogenes), Salmonella enterica, Shiga-toxin producing and other Escherichia coli strains, Campylobacter jejuni, Vibrio species, and Bacillus cereus (Zhao et al., 2014).
Among the major food-borne bacterial pathogens, S. aureus is the leading and most common cause of food-borne infections around the globe (Tsepo et al., 2016; Ayele et al., 2017; Fetsch and Johler, 2018; Hiko, 2018; Gebremedhin et al., 2022) and is responsible for food poisoning through production of enterotoxins (SEs; Denayer et al., 2017). S. aureus is a well-known, ubiquitous, commensal and opportunistic pathogen which can cause a wide spectrum of diseases in both humans and animals (Daka et al., 2012; Elemo et al., 2017; Adugna et al., 2018; Grace and Fetsch, 2018; Mphahlele et al., 2020; Mehrnaz et al., 2021; Alembo and Torka, 2023). Contaminated animal-source foodstuff mainly milk, dairy products and meat are common vehicles that are frequently implicated in Staphylococcal Food Poisoning (Abunna et al., 2017; Abebe et al., 2020; Feyissa et al., 2023).
In addition to the high prevalence and toxic effect of S. aureus, the emergence and increasing prevalence of antimicrobial resistant bacteria, in particular of methicillin resistant S. aureus (MRSA), due to the inappropriate and widespread use of antibiotics is a major concern to human and veterinary medicine worldwide (Chao et al., 2007; Tsepo et al., 2016; Abraha et al., 2018; Fetsch and Johler, 2018). The magnitude of MRSA in food producing animals is increasing and the emergence of livestock-associated MRSA has become a growing concern for public health as it can be transmitted to humans via food chain (Lim et al., 2010; Mekuria et al., 2013; Ndahi et al., 2014; Rodríguez-Lázaro et al., 2017; Chai et al., 2020; Keyvan et al., 2020; Khater et al., 2021; Gajewska et al., 2022).
In many developing countries, the magnitude of food-borne diseases is relatively high. In order to reduce this high prevalence, the detection of food-borne pathogens in food is very important (Zhao et al., 2014). However, the true magnitude of food-borne diseases in these countries is not accurately estimated due to the poor surveillance systems (Savariraj et al., 2018). In Ethiopia, there is limited published information on the occurrence and distribution of S. aureus and its antibiotic resistance pattern in food animals and foods of animal origin (Mekuria et al., 2013; Abebe et al., 2020). Foods of animal origin are produced under unsanitary conditions and there is the habit of consumption of raw or under cooked cow milk and beef in most parts of Ethiopia (Demme and Abegaz, 2015). Moreover, there was no accessible published report on the magnitude and antimicrobial resistance pattern of S. aureus from foods of bovine origin in Dessie and Kombolcha towns. Detection of this bacterial pathogen in foods of bovine origin is crucial to implement different strategies which help to reduce bacterial contamination along the production chain to improve the safety of these food products. Hence, this study was conducted to estimate the prevalence, identify associated risk factors and determine antimicrobial resistance pattern of S. aureus and methicillin resistant S. aureus (MRSA) from foods of bovine origin in Dessie and Kombolcha towns.
2 Materials and methods
2.1 Study area
The study was conducted in Dessie and Kombolcha towns of South Wollo Zone, Eastern Amhara Region, Ethiopia. Kombolcha and Dessie towns are located at a distance of 376 km and 401 killometers from Addis Ababa, respectively. Dessie is situated at 39°38′E-41°13′East longitude and 11°8′N-11°46′ North latitude whereas Kombolcha town is located at 39°45′E longitude and 11°6′ N latitude. The topography of Dessie is a highland type with an elevation between 2,470 and 2,550 meters above sea level. On the other hand, Kombolcha town is located at a range of altitudes between 1, 500 and 1, 840 meter above sea level with mean annual rainfall of 1,046 mm. The mean annual rainfall of Dessie town ranges from 1,100 to 1,200 mm and the annual minimum and maximum temperatures of the town are 9 and 23.7°C, respectively. With regard to Kombolcha town, the annual minimum and maximum temperatures are 12.9 and 28.1°C, respectively (Eskinder et al., 2010; Abebe et al., 2023).
2.2 Study population
Milking dairy cows from dairy farms, carcasses from municipal and ELFORA abattoirs, cheese and yogurt from milk product retail shops and beef from butcher shops and restaurants in the study sites were considered as study population. In Kombolcha town, 164 registered dairy farms were found during the time of sample collection. The number of milking cows in these farms was 586 and daily average milk yield in the town was 6,261 liters (Kombolcha Town Animal Production Health Office, 2019). In Dessie town, the number of dairy farms and milking cows included in the study were 28 and 196, respectively. Around 30 to 35 oxen were slaughtered on Friday of each week at Dessie municipal abattoir and 5 oxen were slaughtered per day in average at the abattoir. The samples were collected during holidays at Kombolcha municipal abattoir since small numbers of oxen were slaughtered per day due to expansion of illegal field slaughtering. The number of oxen slaughtered at Kombolcha ELFORA abattoir ranged from 80 to 140 (120 in average).
2.3 Study design
A cross sectional study was conducted from October 2019 to July 2021 to estimate the prevalence, identify associated risk factors and determine antibiotic resistance pattern of S. aureus from foods of bovine origin in Dessie and Kombolcha towns.
2.4 Sample size determination
The sample size (n) was determined based on the formula recommended by Thrusfield (2005).
Where, n = Sample size, Z = Statistic for a level of confidence,
d = Required absolute precision, Pexp = Expected prevalence
Since there was no previous accessible and published report on the same title in the study sites, 50% of prevalence of S. aureus contamination was expected. Moreover, the sample size was calculated at 95% confidence interval with 5% desired absolute precision. Based on the above given formula and numbers, the sample size was calculated to be 384.
2.5 Sampling technique
Probability sampling methods were employed to select foods of bovine origin samples from different sources in the study sites. Milking cows were selected using simple random sampling technique from the sampling frame of total milking cows in dairy farms of the two study sites. Systematic random sampling method was employed to select carcass samples among cattle slaughtered at abattoirs of study sites and every 3rd carcasses were selected. Moreover, samples from tank milk and milk product shops, and beef swab samples from butcher shops and restaurants were selected using random sampling technique.
2.6 Sample collection
After the milkers prepared the cows for milking through usual practice, about 25 ml of milk sample was collected aseptically from all quarters of the selected individual cow's udder during the middle of milking procedure using sterile labeled screw cupped glass bottles. Using sterile screw cupped glass bottles, around 25 ml of sample from well-mixed tank milk and 25 ml/g of yogurt and cheese samples from milk product shops were collected aseptically. From the surface and deep part of the selected carcasses (neck, thorax, abdomen, breast, and crutch), carcass swab samples were collected at abattoirs using sterile cotton swabs and the beef swab samples were collected from different sites of individual beef at butcher shops and restaurants, and the swab samples were transferred into labeled test tubes containing 5 ml of sterile 0.85% NaCl solution. Moreover, different variables of each sample type were recorded on predesigned data recording format. The collected samples were transported in ice box containing ice packs to Veterinary Microbiology Laboratory of School of Veterinary Medicine, Wollo University on the day of collection, stored aseptically in refrigerator and analyzed within 24 h.
2.7 Standard organisms for quality control
A standard strain of S. aureus, which was obtained from Amhara Public Health Institute (APHI) Dessie branch, was used as quality control standard organisms.
2.8 Isolation and identification of S. aureus and methicillin-resistant Staphylococcus aureus
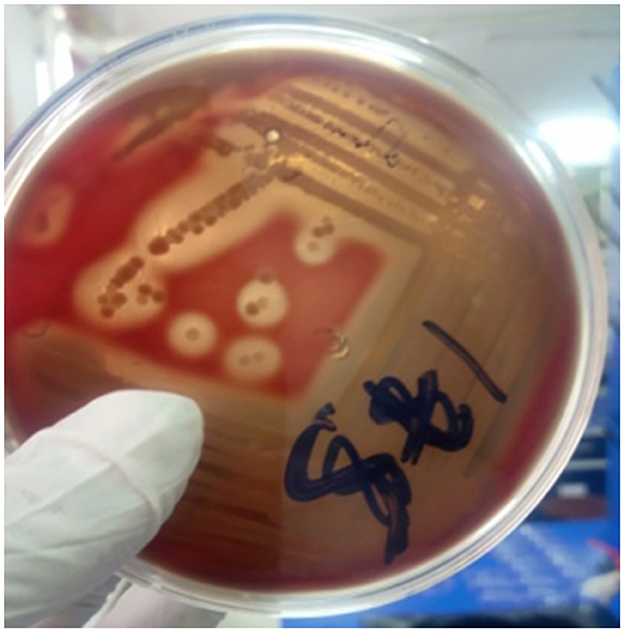
Standard microbiological protocols described by Quinn et al. (1999) and Quinn et al. (2002) were followed for the isolation and identification of S. aureus from different samples of foods of bovine origin. Briefly, from each well-mixed original sample, 1 ml was transferred into 9 ml of sterile peptone water (Micromaster, India) and incubated aerobically at 37°C for 24 h. A loopful of the enriched sample was streaked aseptically on blood agar base (HiMedia Laboratories Pvt. Ltd., India) enriched with 5% sheep blood plates and incubated aerobically at 37°C for 24–48 h. Then, the plates were examined for growth of bacterial colonies, colonial morphology (round, smooth, and white or yellow colonies) and presence or absence of hemolysis. Those isolates that were suspected as Staphylococcus species on the basis of their morphological aspects and β-hemolytic pattern on the surface of blood agar plates (Figure 1) were observed and recorded (Bannerman, 2003).
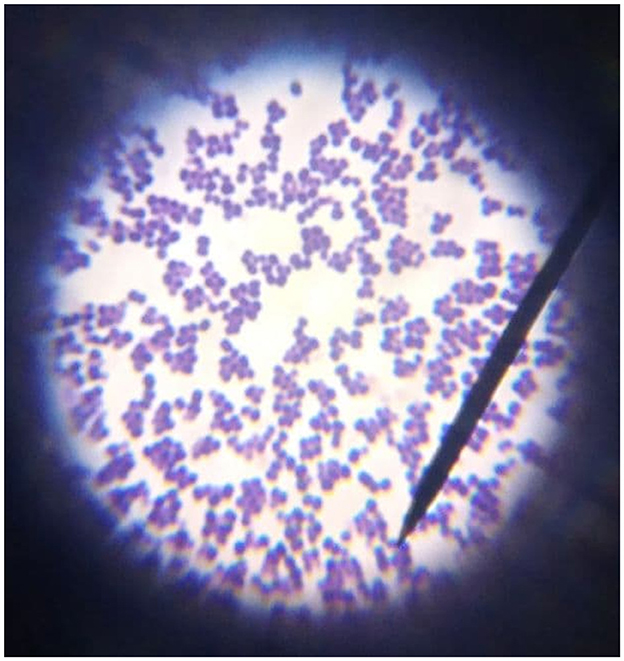
Pure presumed colonies were subcultured on nutrient agar plates (HiMedia Laboratories Pvt. Ltd., India) and subjected to Gram's stain and catalase test. Colonies that showed Gram-positive cocci occurring in bunched, grapelike irregular clusters (Figure 2) and positive for catalase test (forming bubbles) were presumptively identified as Staphylococcus.
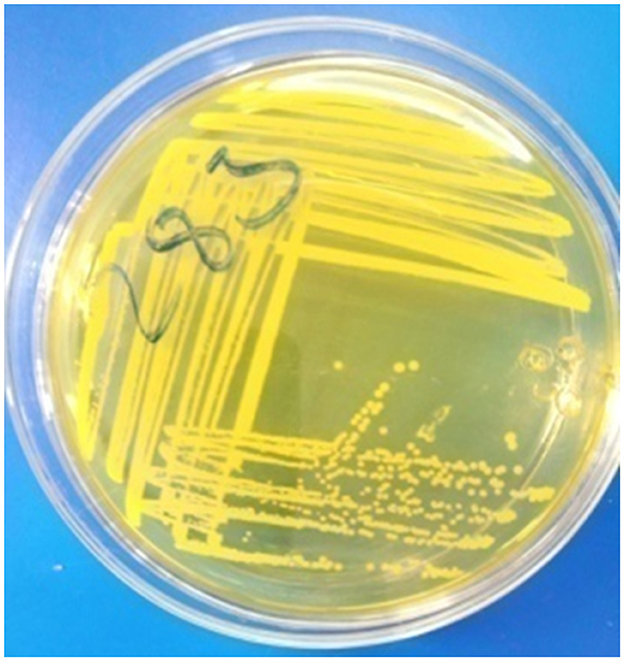
The presumptive colonies were taken from nutrient agar plates and subcultured onto nutrient broth (Sisco Research Laboratories Pvt. Ltd., India) and incubated aerobically at 37°C for 24 h. A loopful of the subcultured isolates was streaked aseptically onto Mannitol Salt Agar (MSA; Microxpress, India; Figure 3) and incubated aerobically at 37°C for 24–48 h. Presumed colonies were then subcultured on nutrient agar plates and incubated aerobically at 37°C for 24 h for conducting different biochemical tests.
The purified S. aureus isolates were identified through different biochemical tests including Catalase test, Oxidation-Fermentation (OF) test, detection of hemolysis, DNase test, and both slide and tube Coagulase tests. Isolates which were hemolysis positive (complete hemolysis around the colonies; Figure 1), Gram-positive (blue color cocci with bunch of grapes arrangement; Figure 2), Catalase positive (forming bubbles), yellowish color colonies with yellow color of MSA (Figure 3), slide coagulase positive (clump formation), tube coagulase positive (clot formation), OF positive (both open and sealed media changed to yellow color), and DNase positive (distinct pink clear zone around the streak line) were considered as S. aureus (Quinn et al., 2002).
In this study, the Kirby-Bauer disc diffusion method using Cefoxitin (30 μg) and Oxacillin (1 μg) discs (Mast Group Ltd., Merseyside, U.K) was done for identifying MRSA strains from S. aureus isolates. However, S. aureus isolates resistant for Cefoxitin disc diffusion method were confirmed as MRSA since CLSI (2020) suggested that Oxacillin disc diffusion method is not reliable for S. aureus and recommended the use of the Cefoxitin for MRSA detection as it is a better inducer of PBP-2a encoding mecA gene. Moreover, Cefoxitin disc diffusion has 100% sensitivity and 100% specificity using PCR for mecA gene as gold standard comparison test (Ahmad et al., 2013; Fernandes et al., 2005; Pramodhini et al., 2011).
2.9 Antimicrobial susceptibility testing of S. aureus
All the identified S. aureus isolates were subjected to in vitro antimicrobial susceptibility test using the standard Kirby-Bauer disc diffusion method on the Muller-Hinton Agar (HiMedia Laboratories Pvt. Ltd., India; Bauer et al., 1966). Along with Cefoxitin (30 mg) and Oxacillin (1 μg), other ten antibiotics discs (Mast Group Ltd., Merseyside, U.K) were used: Amoxicillin/clavulanic acid (30 mg), Ampicillin (25 mg), Kanamycin (30 mg), Nalidixic acid (30 mg), Norfloxacin (2 μg), Penicillin G (10 IU), Streptomycin (10 mg), Sulphamethoxazole-trimethoprim (25 μg), Tetracycline (10 mg), and Vancomycin (5 mg).
The subcultured colonies were transferred into test tubes containing 5 ml of sterile 0.85% saline solution and homogenized until the turbidity of the bacterial suspension is comparable to the 0.5 McFarland turbidity standards. The sterile cotton swab was immersed into the adjusted suspension and the inoculum was then spread uniformly over the entire surface of the Mueller-Hinton Agar plate (HiMedia Laboratories Pvt. Ltd., India). Using sterile thumb forceps, antibiotic discs were placed at a minimum distance of 24 mm on the agar surface after the inoculated plates dried for 3–5 min and gently pressed. The plates were inverted within 15 min of the antibiotic discs placement and incubated aerobically at 37°C for 24 h. Then, the diameters of growth inhibition zones of each antibiotic disc were measured using digital caliper and the S. aureus isolates were grouped as Sensitive (S), Intermediate (I), and Resistant (R) based on the Clinical and Laboratory Standard Institute interpretation tables (CLSI, 2020; Figure 4). Bacterial isolates resistant to one or more antibiotic types in three or more antibiotic classes were considered multidrug-resistant (Magiorakos et al., 2012).
2.10 Data management and analysis
All collected raw data were compiled, entered, and coded in Microsoft Excel 2007 spread sheet and transferred to STATA Version 12 software for statistical analysis. Descriptive and inferential statistical techniques were used to analyze the collected data. Descriptive statistics such as mean, frequency, and/or percentage were calculated. Besides to proportion, chi-square test (χ2), P-value, and logistic regression were computed to see the association of risk factors with that of the occurrence of the isolates and the degree of association was determined using Odds ratio (OR) with 95% confidence interval (CI). P-values < 0.05 were considered as statistically significant.
3 Results
3.1 Overall prevalence
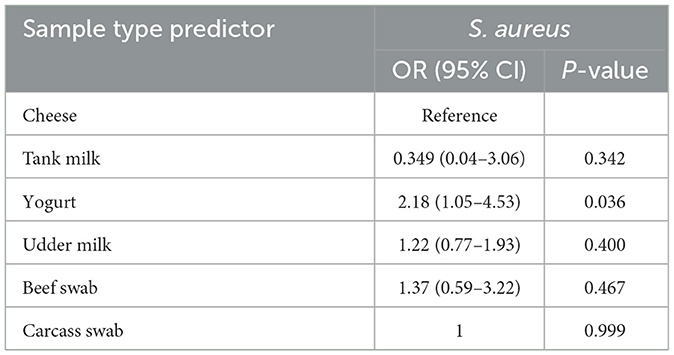
Out of the total of 384 examined samples of foods of bovine origin, 151 (39.3%) were found to be contaminated with S. aureus. The site-based prevalence of S. aureus was 40.9 and 37.9% in Dessie and Kombolcha towns, respectively. The difference in the prevalence of S. aureus among the study sites was not statistically significant (P > 0.05). The prevalence of S. aureus was 55.6, 44.0, 41.1, 36.4, 16.7, and 0.0% in yogurt, beef swab, udder milk, carcass swab, tank milk, and cheese samples, respectively. A statistically significant difference in the S. aureus prevalence (P < 0.05) was observed among the different sample types of foods of bovine origin (Table 1).
The contamination of yogurt with S. aureus was 2.18 times more likely to occur compared to cheese and it was statistically significant (P < 0.05; Table 2).
3.2 Prevalence of S. aureus among variables of different sample types
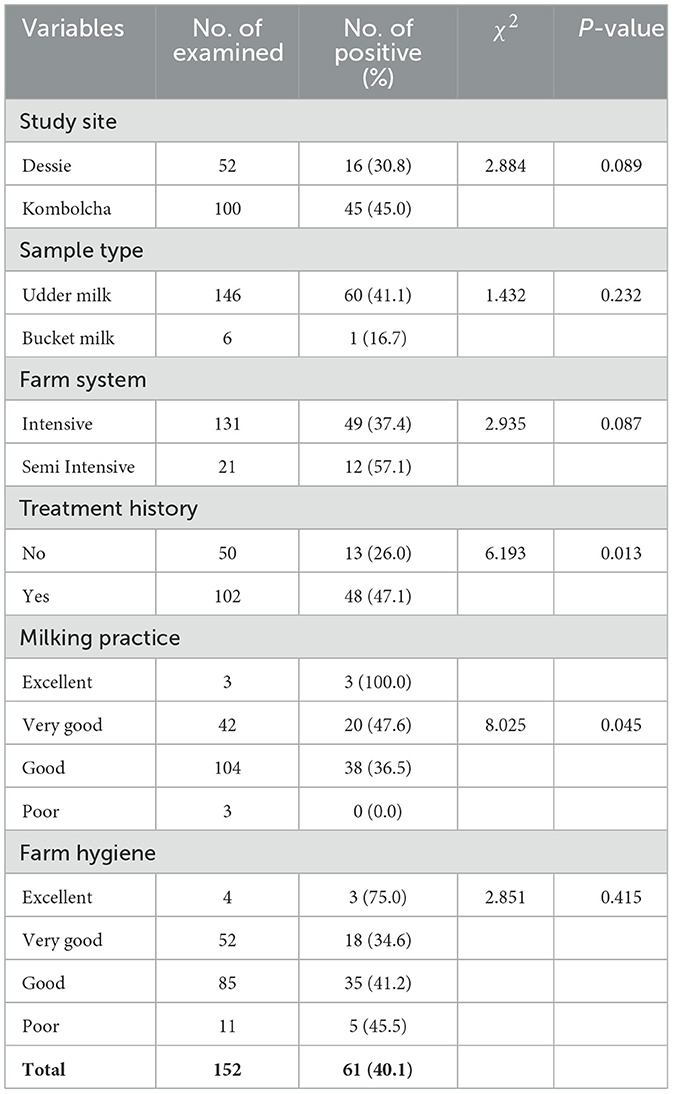
The recorded prevalence of S. aureus in milk samples from cows with treatment history was 47.1%. The difference in the prevalence of S. aureus among treatment history categories was statistically significant (P < 0.05; Table 3).
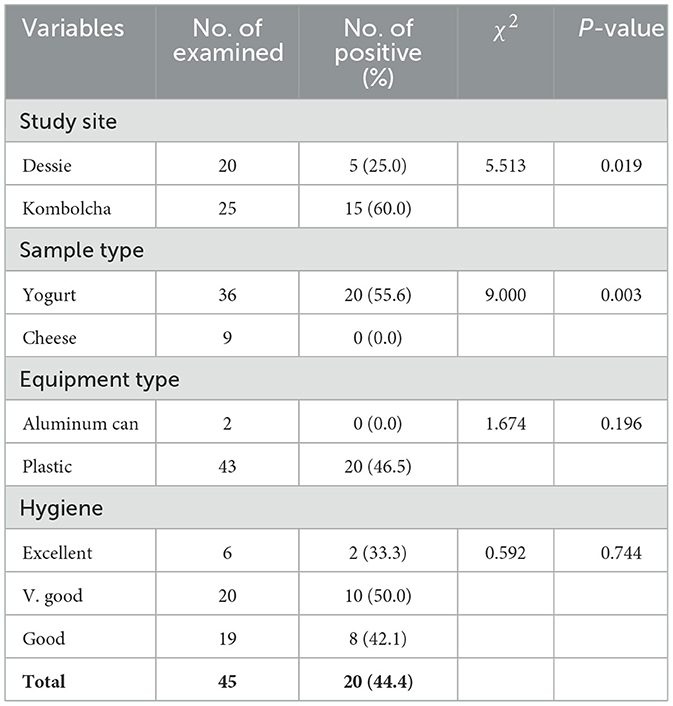
Though S. aureus was not detected from cheese, it was isolated from 55.6% of yogurt samples and the difference was statistically significant (P < 0.05). The proportion of S. aureus from milk products was higher in Kombolcha town (60.0%) than Dessie town (25.0%). Thus, there was a statistically significant difference in the prevalence of S. aureus from milk products among the two study sites (P < 0.05; Table 4).
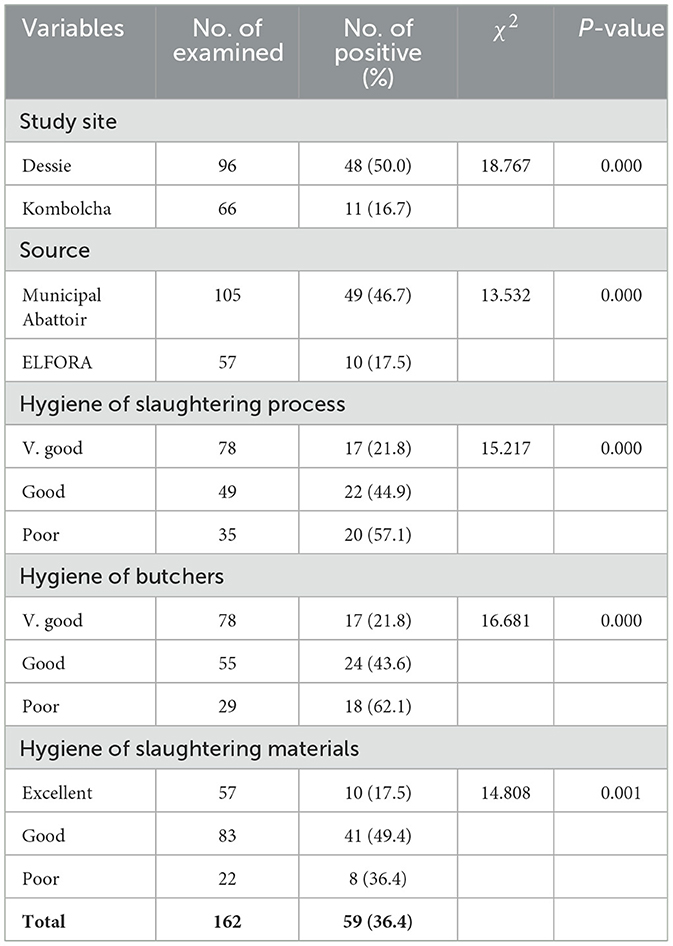
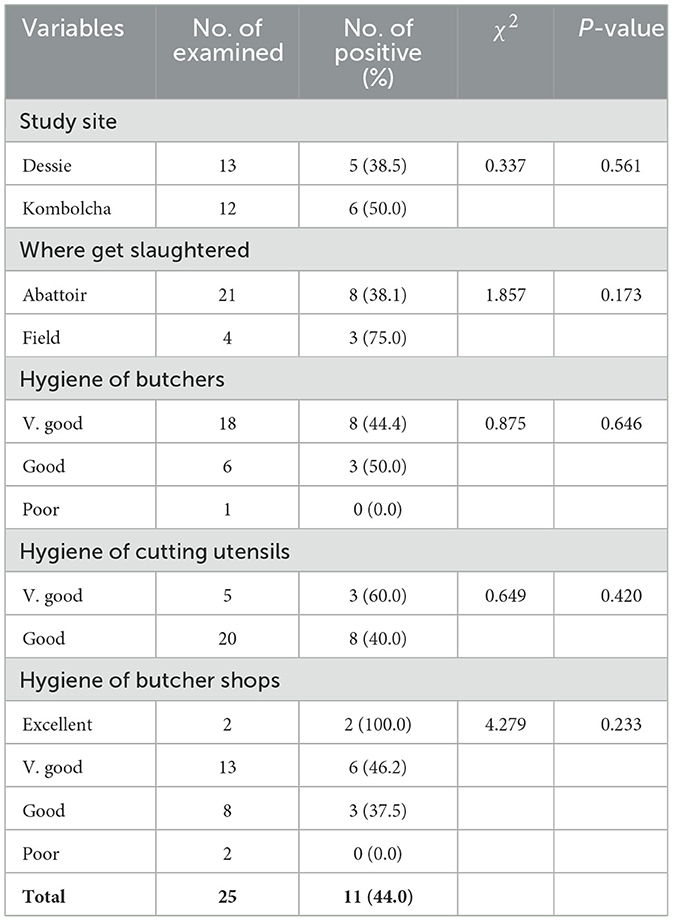
A statistically significant difference in the S. aureus prevalence (P < 0.05) was observed among all hypothesized variables of carcass swab samples as shown in Table 5. Higher prevalence rates of S. aureus were recorded in carcass swab samples collected from Dessie town (50.0%), municipal abattoirs (46.7%), slaughtering process with poor hygiene (57.1%); and carcasses slaughtered by butchers with poor hygiene (62.1%) as presented in Table 5.
The difference in the prevalence of S. aureus among all hypothesized variables of beef swab samples was not statistically significant (P > 0.05) as shown in Table 6.
3.3 In vitro antimicrobial susceptibility pattern of S. aureus isolates
Phenotypical antibiotic sensitivity test of 151 S. aureus isolates against 12 antimicrobial agents revealed that 100.0, 97.4, 90.1, and 74.8% of the isolates were resistant to Cefoxitin, Penicillin G, Ampicillin, and Nalidixic acid, respectively. Most of the isolates (93.4%) were susceptible to Norfloxacin as illustrated in Table 7. Moreover, 100.0% of resistant to Cefoxitin indicated that all of the S. aureus isolates were classified as MRSA. Hence, the overall prevalence of MRSA in the present study was 39.3%.

Table 7. In vitro antimicrobial sensitivity pattern of S. aureus isolated from different sample types of foods of bovine origin.
The result shown in Figure 5 indicated that 3 (2.0%), 1 (0.7%), 14 (9.3%), 21 (13.9%), 42 (27.8%), 47 (31.1%), 11 (7.3%), 9 (6.0%), and 3 (2.0%) of S. aureus isolates showed resistance to one, two, three, four, five, six, seven, eight, and nine antibiotics, respectively. In general, 147 isolates of S. aureus (97.3%) showed multidrug resistance to three and more than three antimicrobial drugs.
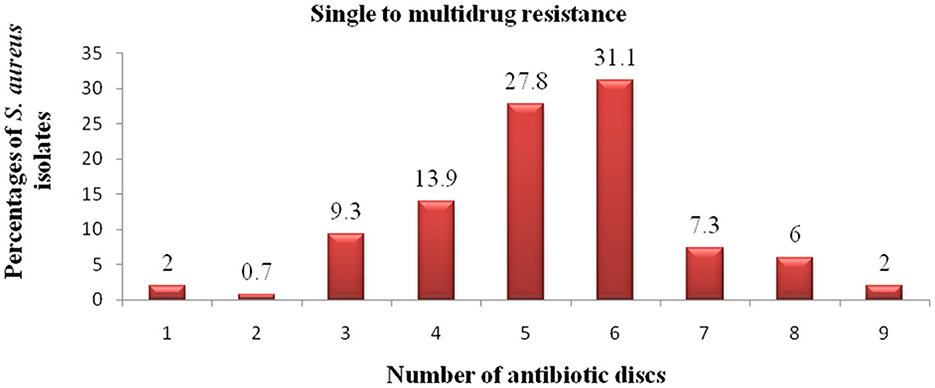
Figure 5. Antibiotic resistance pattern of S. aureus isolates. The numbers (1–9) in the x-axis indicate the total number of antibiotics to which the S. aureus isolates showed resistance. The exact percentage of resistant S. aureus isolates to each number of antibiotics is indicated above each column.
4 Discussion
In the present study, the overall prevalence of S. aureus was 39.3%, which was nearly comparable with the findings of Weldeselassie et al. (2020) (39.1%), Addis et al. (2011) (39.5%), Sudhanthiramani et al. (2015) (39.09%), Enquebaher et al. (2015) (38.7%), Zerabruk et al. (2019) (37.5%), Derib et al. (2017) (37.14%), Awad et al. (2017) (42.0%), and Abo-Shama (2014) (36.7%) in Mekelle town, Debre-Zeit town, Tirupathi (India), Tigray region, Addis Ababa, Wolaita Sodo, Dakahliya and Damietta Governorates (Egypt), and Sohag Governorate (Egypt), respectively. The high proportion of S. aureus in foods of bovine origin is indicative of poor hygienic measures during production, handling, storage, and distribution (Jahan et al., 2015). S. aureus is an obvious contributor to milk contamination since it is the most common cause of bovine mastitis (Matallah et al., 2019).
However, the current prevalence of S. aureus was higher than the previous reports of -, Hiwot et al. (2016) in Arsi and East Shewa Zones (3.2%), Arafa et al. (2016) in Great Cairo zone (Egypt) (4.67%), Argaw et al. (2018) in Selected Districts of Jimma Zone (5.0%), Riva et al. (2015) in Northern Italy (9.1%), Kalayu et al. (2020) in Mekelle town (12.5%), Basanisi et al. (2017) in South Italy (12.9%), Beyene et al. (2017) in Addis Ababa (14.67%), Ektik et al. (2017) in Balikesir (Turkey) (14.85%), Massawe et al. (2019) in Mbeya Region (Tanzania; 15.0%), Shrestha et al. (2021) in Chitwan (Nepal; 15.2%), Mekuria et al. (2013) in Addis Ababa (16.2%), Abunna et al. (2017) in and around Asella town (16.24%), Asiimwe et al. (2017) in South-West Uganda (17.15%), Lemma et al. (2021) in Addis Ababa (20.5%), Hamid et al. (2017) in Jammu (India; 22.5%), Ayele et al. (2017) in Sebeta town (23.4%), Reta et al. (2016) in Jigjiga City (24.2%), Jahan et al. (2015) in Bangladesh (25.53%), Tsepo et al. (2016) in North West Province (South Africa; 26.5%), Tessema and Tsegaye (2017) in Alage (Ziway; 28.2%), Girmay et al. (2020) in Shire (Tigray; 29.09%), Tesfaye et al. (2021) in and around Adama town (30.6%), Olatoye et al. (2018) in Oyo State (Nigeria; 31.5%), Matallah et al. (2019) in Algeria (31.56%), and Hassan et al. (2018) in Asella (34.3%).
On the contrary, prevalence rates of S. aureus higher than the present study were reported by Salauddin et al. (2020) in Rangpur division (Bangladesh; 100.0%), Ateba et al. (2010) in North West province (South Africa; 100.0%), De-Oliveira et al. (2011) in Bahia (Brazil; 68.0%), Lingathurai and Vellathurai (2011) in Madurai (South India; 61.7%), Gundogan and Avci (2014) in Turkey (56.0%), Megersa et al. (2012) in Hawassa town (53.5%), Wu et al. (2018) in China (50.4%), Daka et al. (2012) in Hawassa town (48.75%), Mohanta and Mazumder (2015) in Southern Assam (India; 47.86%), Limbu et al. (2020) in Dharan (Nepal; 45.0%), and Elemo et al. (2017) in Asella town (44.62%). This variation in the prevalence of S. aureus between the present study and previous reports could arise from differences in dairy and beef cattle production and health, milking and slaughtering procedures and hygiene, cleanliness of equipments, and hygienic conditions during handling, transportation, storage, and distribution up to consumption. Additionally, differences in the research methods used by the investigators, such as study design, sample source, sampling method, sample size, sample type, and methods of detection in laboratories could also contribute to the variations of the results in the different reports.
In the current study, the occurrence of S. aureus was highest in yogurt (55.6%), followed by beef swab (44.0%), udder milk (41.1%), carcass swab (36.4%), tank milk (16.7%), and cheese (0.0%). A statistically significant difference in the S. aureus prevalence (P < 0.05) was observed among different sample types of foods of bovine origin. The contamination of yogurt with S. aureus was 2.18 times more likely to occur compared to cheese and it was statistically significant (P < 0.05). The higher prevalence of S. aureus in yogurt might be associated with the initial contamination of the milk due to unsatisfactory hygienic practices during milking and poor dairy cow health status or further contamination of the milk during collection and transportation as well as storage for long time under ambient temperature.
The prevalence of S. aureus was higher in milk samples from cows with teat treatment history (47.1%) than cows without treatment history (26.0%) and the difference was statistically significant (P < 0.05). In dairy cattle, S. aureus is the most common mastitis-causing bacterium (Kotzamanidis et al., 2021). Thus, the higher occurrence of S. aureus in milk samples from cows with teat treatment history might be due to mastitis since cows with previous mastitis history are more prone to re-infection.
Though S. aureus was not detected from cheese, it was isolated from 55.6% of yogurt samples and the difference was statistically significant (P < 0.05). According to Pazakova et al. (1997), the antibacterial action of yogurt is insufficient to prevent food poisoning in the case of high S. aureus contamination of milk. Moreover, Soliman and Ahmed (2019) stated that S. aureus could contaminate yogurt and survived fermentation process for 8–10 days.
The proportion of S. aureus from milk products was higher in Kombolcha town (60.0%) than Dessie town (25.0%). Thus, there was a statistically significant difference in the prevalence of S. aureus from milk products among the two study sites (P < 0.05). The difference might be associated with the variation in hygienic practices at dairy environment, herd health status of dairy farms and variation in sanitation during collection, transportation and storage of milk.
A statistically significant difference in S. aureus prevalence (P < 0.05) was observed among all hypothesized variables of carcass swab samples. The carcass swab samples collected from Dessie town were more contaminated by S. aureus (50.0%) than those collected from Kombolcha town (16.7%). This could be associated with the difference in slaughtering operations and hygienic practices at abattoirs. Higher prevalence rate of S. aureus was recorded in carcass swab samples collected from municipal abattoirs (46.7%) than ELFORA (17.5%). The higher prevalence of S. aureus in carcass swab samples collected from municipal abattoirs could arise from the slaughtering operation. In municipal abattoirs, slaughtering process was conducted on the floor which could increases the risk of contamination from the ground, outer integument, slaughter men and other sources. The higher prevalence of S. aureus from carcasses slaughtered under poor hygienic operation (57.1%) and carcasses slaughtered by butchers with poor hygiene (62.1%) were not astonishing since S. aureus are commonly associated with poor hygiene and sanitation (Bagumire and Karumuna, 2017) and contamination of beef at the slaughterhouse in particular occurs because of inadequate hygienic conditions and poor handling practice (Fasanmi et al., 2018). The difference in the prevalence of S. aureus among different hypothesized variables of beef swab samples was not statistically significant (P > 0.05).
S. aureus is a notorious bacterium which rapidly develops resistance to many antibiotics of different classes (Parvin et al., 2021) and the acquisition of resistance to several antibiotics worsen the risk and severity of its infection. Moreover, the emergence and spread of livestock-associated MRSA is a great concern for public health (Tamendjari et al., 2021). In the present study, the in vitro antibiotic sensitivity testing revealed that 97.3% of the S. aureus isolates showed multidrug resistance to three and more than three drugs which was consistent with previous reports of Girmay et al. (2020) in Shire town, Beyene et al. (2017) in Addis Ababa, Weldeselassie et al. (2020) in Mekelle town who reported 100.0, 100.0, and 93.75% of multidrug resistant isolates, respectively.
In the current study, the large proportions of the S. aureus isolates were phenotypically resistant to Cefoxitin (100.0%), Penicillin G (97.4%), Ampicillin (90.1%), and Nalidixic acid (74.8%). Thus, all isolates were classified as MRSA and the overall prevalence of MRSA in the present study was 39.3%. The 100% resistance of S. aureus to Cefoxitin was consistent with the previous findings of Ayele et al. (2017) in Sebeta town, Aliyu et al. (2019) in Nasarawa State (Nigeria) and Yakubu et al. (2020) in Nasarawa State (Nigeria) who reported similar 100.0% resistance to Cefoxitin. However, Mekonnen et al. (2018) reported 100% sensitive S. aureus isolates to Cefoxitin in North-Western Ethiopia and Sudhanthiramani et al. (2015) reported low magnitude of Cefoxitin resistance (4.65%) in Tirupathi, India.
The high resistance pattern of the isolates to Penicillin G (97.4%) was in agreement with the previous studies reported by Ayele et al. (2017) (98.85%) in Sebeta town, Pati and Mukherjee (2016) (96.0%) in Northern Plains of India, Daka et al. (2012) (100.0%) in Hawassa area, Girmay et al. (2020) (100.0%) in Shire town, Derib et al. (2017) (100%) in Wolaita Sodo town, Ayana et al. (2017) (100%) in Bishoftu town, and Mathenge et al. (2017) (99.6%) in Nairobi, Kenya. The higher resistance to Ampicillin (90.1%) was consistent with the reports of Pati and Mukherjee (2016) (93.0%), Mathenge et al. (2017) (93.1%), and Lemma et al. (2021) (94.2%) in Northern Plains of India, Nairobi (Kenya), and Addis Ababa, respectively. However, 100% Ampicillin susceptible isolates were reported by Adugna et al. (2018) in Addis Ababa. According to Girmay et al. (2020), the production of β-lactamase enzyme by S. aureus could be responsible for its resistance to penicillin and other related antibiotics. The proportion resistance to Nalidixic acid obtained in the present study was consistent with the report of Anueyiagu and Isiyaku (2015) who reported 71.4% resistance to Nalidixic acid in Nigeria.
The antimicrobial sensitivity testing result indicated that 52.3% of the isolates showed resistance to Tetracycline which was consistent with Mekonnen et al. (2018) who reported 54.0% resistance to Tetracycline in North-Western Ethiopia. However, 100.0% susceptibility to Tetracycline was reported by Sori et al. (2011) in Jimma town. In the current study, 38.4% of the isolates were also resistant to Amoxicillin/Clavulanic acid which was consistent with the report of Pati and Mukherjee (2016) (37.0%) in Northern Plains of India. Furthermore, high proportions of S. aureus isolates (90.7%) showed intermediate susceptibility to Vancomycin. However, Massawe et al. (2019) and Pati and Mukherjee (2016) reported S. aureus isolates which were 100.0% susceptible to Vancomycin in Mbeya Region (Tanzania) and Northern Plains of India, respectively. Most of the S. aureus isolates (93.4%) were susceptible to Norfloxacin which was in accordance with the finding of Sori et al. (2011) who reported 100.0% sensitivity to Norfloxacin in Jimma town.
In human and veterinary medicine, the extensive, indiscriminate, and injudicious use of antibiotics may lead to development of drug resistance and the magnitude of drug resistance among pathogens is increasing (Mokgophi et al., 2021; Qamar et al., 2020). Thus, the high resistance pattern of S. aureus to readily available and relatively inexpensive antibiotics might be associated with these factors.
5 Conclusion and recommendations
The present study revealed that the prevalence of S. aureus from foods of bovine origin was very high. The treatment history of milking cows was the risk factor for the high prevalence of milk contamination with S. aureus. The sources of secondary contamination including municipal abattoirs with slaughtering operation on the floor, poor hygiene of the slaughtering process and butchers with poor hygiene were the determinants of high magnitude of detection of S. aureus in carcass samples. Nearly all of the S. aureus isolates showed multidrug resistance to readily and commonly used antimicrobial agents. The gold standard techniques for identifying MRSA, specifically detection of mecA gene and identifying PBP2a through latex agglutination were not done in this study due to limitations in budget and their unavailability in nearby laboratories. Hence, improvement of cattle health and good hygienic procedures in farms, abattoirs, milk product and butcher shops, and restaurants should be implemented to reduce primary and secondary bacterial contamination of foods of bovine origin. Moreover, rational use of antibiotics should be practiced in animal and public health and regular assessment of antibiotic resistance among pathogenic bacteria in food animals and their products should be done. Further studies on molecular characterization of S. aureus and their resistant genes should be carried out in the study areas.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
Ethics statement
The animal studies were approved by Research and Publication Director, Wollo University. The studies were conducted in accordance with the local legislation and institutional requirements. Written informed consent was obtained from the owners for the participation of their animals in this study.
Author contributions
EA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. GG: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. MA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. NA: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Software, Supervision, Validation, Visualization, Writing – original draft, Writing – review & editing. YT: Conceptualization, Funding acquisition, Writing – review & editing. SA: Conceptualization, Funding acquisition, Writing – review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research work was funded by Wollo University Research and Community Service Vice President Office and Wollo University Post Graduate Directorate Office. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Acknowledgments
The authors would like to thank Research and Community Service Vice President and Post Graduate Directorate Offices of Wollo University for the financial funding. The authors are also extremely grateful to animal health and production officers, veterinarians, farm attendants, dairy farm owners and/or managers, milk product shops owners and/or workers, restaurant managers and owners, butchers and other abattoir workers, and other inhabitants in the study sites for their voluntariness and assistance during sample and data collection.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Abebe, E., Gugsa, G., and Ahmed, M. (2020). Review on major food-borne zoonotic bacterial pathogens. J. Trop. Med. 2020, 1–19. doi: 10.1155/2020/4674235
Abebe, E., Gugsa, G., Ahmed, M., Awol, N., Tefera, Y., Abegaz, S., et al. (2023). Occurrence and antimicrobial resistance pattern of E. coli O157: H7 isolated from foods of Bovine origin in Dessie and Kombolcha towns, Ethiopia. PLoS Negl. Trop. Dis. 17:e0010706. doi: 10.1371/journal.pntd.0010706
Abo-Shama, U. H. (2014). Prevalence and antimicrobial susceptibility of Staphylococcus aureus isolated from cattle, buffalo, sheep and goat's raws milk in Sohag Governorate, Egypt. Assiut. Vet. Med. J. 60, 63–72. doi: 10.21608/avmj.2014.170753
Abraha, H., Hadish, G., Aligaz, B., Eyas, G., and Workelule, K. (2018). Antimicrobial resistance profile of Staphylococcus aureus isolated from raw cow milk and fresh fruit juice in Mekelle, Tigray, Ethiopia. J. Vet. Med. Anim. Health 10, 106–113. doi: 10.5897/JVMAH2017.0664
Abunna, F., Ashenafi, D., Beyene, T., Ayana, D., Mamo, B., and Duguma, R. (2017). Isolation, identification and antimicrobial susceptibility profiles of Salmonella isolates from dairy farms in and around Modjo Town, Ethiopia. Ethiop. Vet. J. 21, 92–108. doi: 10.4314/evj.v21i2.7
Addis, M., Pal, M., and Kyule, M. N. (2011). Isolation and identification of Staphylococcus species from raw bovine milk in DebreZeit, Ethiopia. Vet. Res. 4, 45–49. doi: 10.3923/vr.2011.45.49
Adugna, F., Pal, M., and Girmay, G. (2018). Prevalence and antibiogram assessment of Staphylococcus aureus in beef at municipal abattoir and butcher shops in Addis Ababa, Ethiopia. Biomed. Res. Int. 1, 1–7. doi: 10.1155/2018/5017685
Ahmad, B., Jamil, J., and Ahmed, J. (2013). On of clinical methods for the phenotypic detection of Methicillin resistant Staphylococcus aureus: disc diffusion methods with BrillianceTM MRSA. Afr. J. Microbiol. Res. 7, 3096–3100. doi: 10.5897/AJMR12.2177
Alembo, E. A., and Torka, T. T. (2023). Prevalence, contamination level, and associated factors of methicillin-resistant Staphylococcus aureus in raw cow milk at selected Districts of Gamo Zone, Southern Ethiopia. Vet. Med. Int. 2023, 1–11. doi: 10.1155/2023/6238754
Aliyu, Y., Abdullahi, I. O., Whong, C. M. Z., and Olayinka, B. O. (2019). Antibiotic resistant phenotypes of Staphylococcus aureus isolated from fresh and fermented milk in parts of Nasarawa State, Nigeria. Afr. J. Microbiol. Res. 13, 446–456. doi: 10.5897/AJMR2019.9175
Al-Mohaithef, M. (2021). Awareness of foodborne pathogens among students: a cross-sectional study in the Kingdom of Saudi Arabia. Int. J. Food Sci. 2021, 1–6. doi: 10.1155/2021/9971748
Anueyiagu, K. N., and Isiyaku, A. W. (2015). Isolation, identification of Staphylococcus aureus from bovine milk and its antibiotics susceptibility. Int. J. Livest. Prod. 6, 74–77. doi: 10.5897/IJLP2015.0248
Arafa, A. A., Ibrahim, E. S., Fouad, E. A., and Gaber, E. S. (2016). Antibiotic resistance of Staphylococci concerning strains included in food industry in Egypt. Int. J. Pharm. Clin. Res. 8, 1583–1589.
Argaw, S., Addis, M., and Degefu, H. (2018). Identification and antimicrobial resistance pattern of Staphylococci isolated from cottage cheese (Ayib) and yoghurt (Ergo) in selected districts of Jimma Zone, Ethiopia. Health Sci. J. 12, 1–8. doi: 10.21767/1791-809X.1000549
Asiimwe, B. B., Baldan, R., Trovato, A., and Cirillo, M. D. (2017). Prevalence and molecular characteristics of Staphylococcus aureus, including methicillin resistant strains, isolated from bulk can milk and raw milk products in Pastoral communities of South-West Uganda. BMC Infect. Dis. 17, 1–8. doi: 10.1186/s12879-017-2524-4
Ateba, C. N., Mbewe, M., Moneoang, M. S., and Bezuidenhout, C. C. (2010). Antibiotic-resistant Staphylococcus aureus isolated from milk in the Mafikeng Area, North West province, south Africa. S. Afr. J. Sci. 106,1–6. doi: 10.4102/sajs.v106i11/12.243
Awad, A., Ramadan, H., Nasr, S., Ateya, A., and Atwa, S. (2017). Genetic characterization, antimicrobial resistance patterns and virulence determinants of Staphylococcus aureus isolated form bovine mastitis. Pak. J. Biol. Sci. 20, 298–305. doi: 10.3923/pjbs.2017.298.305
Ayana, H. W., Mekonnen, B. T., Bulle, A. S., and Berecha, M. S. (2017). Isolation and identification of methicillin-resistant Staphylococcus aureus from mastitic dairy cows in Bishoftu town, Ethiopia. Afr. J. Microbiol. Res. 11, 1606–1613. doi: 10.5897/AJMR2017.7088
Ayele, Y., Gutema, D. F., Edao, M. B., Girma, R., Tufa, B. T., Beyene, J. T., et al. (2017). Assessment of Staphylococcus aureus along milk value chain and its public health importance in Sebeta, Central Oromia, Ethiopia. BMC Microbiol. 7, 1–7. doi: 10.1186/s12866-017-1048-9
Bagumire, A., and Karumuna, R. (2017). Bacterial contamination of ready-to-eat meats vended in highway markets in Uganda. Afr. J. Food Sci. 11, 160–170. doi: 10.5897/AJFS2016.1550
Bannerman, T. L. (2003). “Staphylococcus, Micrococcus, and other catalase positive cocci that grow aerobically,” in Manual of Clinical Microbiology, 8th Edn, eds. P. R. Murray, E. J. Baron, J. H. Jorgensen, M. A. Pfaller and R. H. Yolken (Washington, DC: ASM Press), 384–404.
Basanisi, M. G., La Bella, G., Nobili, G., Franconieri, I., and La Salandra, G. (2017). Genotyping of methicillin-resistant Staphylococcus aureus (MRSA) isolated from milk and dairy products in South Italy. Food Microbiol. 62, 141–146. doi: 10.1016/j.fm.2016.10.020
Bauer, A. W., Kirby, M., Sheris, J. D., and Turch, M. (1966). Antibiotic susceptibility testing by standard single disc method. Am. J. Clin. Pathol. 45,493–496. doi: 10.1093/ajcp/45.4_ts.493
Beyene, T., Hayishe, H., Gizaw, F., Beyi, F. A., Abunna, F., Mammo, B., et al. (2017). Prevalence and antimicrobial resistance profile of Staphylococcus in dairy farms, abattoir and humans in Addis Ababa, Ethiopia. BMC Res. Notes. 10, 1–9. doi: 10.1186/s13104-017-2487-y
Borena, B. M., Dilgasa, L., Zewdu, E. G., Sarba, E. J., Marami, L. M., Kelbesa, K. A., et al. (2022). Listeria species occurrence and associated risk factors and antibiogram of Listeria monocytogenes in milk and milk products in Ambo, Holeta, and Bako towns, Oromia Regional State, Ethiopia. Vet. Med. Int. 2022, 1–11. doi: 10.1155/2022/5643478
Chai, M. H., Faiq, T. A. M., Ariffin, S. M. Z., Suhaili, Z., Sukiman, M. Z., and Ghazali, M. F. (2020). Prevalence of methicillin resistant Staphylococcus aureus in raw goat milks from selected farms in Terengganu, Malaysia. Trop. Anim. Sci. J. 43, 64–69. doi: 10.5398/tasj.2020.43.1.64
Chao, G., Zhou, X., Jiao, X., Qian, X., and Xu, L. (2007). Prevalence and antimicrobial resistance of foodborne pathogens isolated from food products in China. Foodborne Pathog. Dis. 4, 277–284. doi: 10.1089/fpd.2007.0088
CLSI (2020). Performance Standards for Antimicrobial Susceptibility Testing; M100, 30th Edn. A CLSI Supplement for Global Application (Wayne, PA), 1–294.
Daka, D., G/silassie, S., and Yihdego, D. (2012). Antibiotic-resistance Staphylococcus aureus isolated from cow's milk in the Hawassa area, South Ethiopia. Ann. Clin. Microbiol. Antimicrob. 11, 1–6. doi: 10.1186/1476-0711-11-26
Demme, B., and Abegaz, S. (2015). Isolation and identification of major bacterial pathogen from clinical mastitis cow raw milk in Addis Ababa, Ethiopia. A. J. A. D. 4, 44–51. doi: 10.5829/idosi.ajad.2015.4.1.9156
Denayer, S., Delbrassinne, L., Nia, Y., and Botteldoorn, N. (2017). Food-borne outbreak investigation and molecular typing: high diversity of Staphylococcus aureus strains and importance of toxin detection. Toxins 9, 1–13. doi: 10.3390/toxins9120407
De-Oliveira, L. P., Barros, L. S. S., Silva, V. C., and Cirqueira, M. G. (2011). Study of Staphylococcus aureus in raw and pasteurized milk consumed in the Reconcavo area of the State of Bahia. Brazil. J. Food Process. Technol. 2, 1–5. doi: 10.4172/2157-7110.1000128
Derib, B. T., Birhanu, B. T., Tessema, T. S., and Wubshet, A. K. (2017). Isolation and identification of methicillin resistant Staphylococcus aureus (MRSA) from bovine mastitic milk in and around Wolaita Sodo, Southern Ethiopia. J. Vet. Sci. Res. 2, 1–11. doi: 10.23880/oajvsr-16000136
Ektik, N., Gökmen, M., and Çibik, R. (2017). The prevalence and antibiotic resistance of methicillin-resistant Staphylococcus aureus (MRSA) in milk and dairy products in Balikesir, Turkey. J. Hellenic. Vet. Med. Soc. 68, 613–620. doi: 10.12681/jhvms.16062
Elemo, K. K., Sisay, T., Shiferaw, A., and Fato, M. A. (2017). Prevalence, risk factors and multidrug resistance profile of Staphylococcus aureus isolated from bovine mastitis in selected dairy farms in and around Asella town, Arsi Zone, South Eastern Ethiopia. Afr. J. Microbiol. Res. 11, 1632–1642. doi: 10.5897/AJMR2017.8529
Enquebaher, T., Siv, S., Knut, R., Taran, S., and Judith, A. N. (2015). Staphylococcus aureus and other Staphylococcus species in milk and milk products from Tigray Region, Northern Ethiopia. Afr. J. Food Sci. 9, 567–576. doi: 10.5897/AJFS2015.1373
Eskinder, Z., Eyassu, Y., and Mitiku, H. (2010). “Assessment of the impact of industrial effluents on the quality of irrigation water and changes on soil characteristics (a case of Kombolcha town),” in Fourteenth International Water Technology Conference, IWTC 14, 2010 (Cairo), 711–727.
Fasanmi, O. G., Makinde, G. E. O., Popoola, M. A., Fasina, O. F., Matere, J., Kehinde, O. O., et al. (2018). Potential risk factors associated with carcass contamination in slaughterhouse operations and hygiene in Oyo state, Nigeria. Int. J. Livest. Prod. 9, 211–220. doi: 10.5897/IJLP2018.0491
Fernandes, C. J., Fernandes, L. A., and Collignon, P. (2005). Cefoxitin resistance as a surrogate marker for the detection of methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 55, 506–510. doi: 10.1093/jac/dki052
Fetsch, A., and Johler, S. (2018). Staphylococcus aureus as a foodborne pathogen. Curr. Clin. Microbiol. Rep. 5, 88–96. doi: 10.1007/s40588-018-0094-x
Feyissa, N., Alemu, T., Birri, D. J., and Dessalegn, A. (2023). Isolation, identification, and determination of antibiogram characteristics of Staphylococcus aureus in cow milk and milk products (yoghurt and cheese) in West Showa Zone, Ethiopia. Int. Dairy J. 137:105503. doi: 10.1016/j.idairyj.2022.105503
Gajewska, J., Chajecka-Wierzchowska, W., and Zadernowska, A. (2022). Occurrence and characteristics of Staphylococcus aureus strains along the production chain of raw milk cheeses in Poland. Molecules 27:6569. doi: 10.3390/molecules27196569
Gebremedhin, E. Z., Ararso, A. B., Borana, B. M., Kelbesa, K. A., Tadese, N. D., Marami, L. M., et al. (2022). Isolation and identification of Staphylococcus aureus from milk and milk products, associated factors for contamination, and their antibiogram in Holeta, Central Ethiopia. Vet. Med. Int. 2022, 1–13. doi: 10.1155/2022/6544705
Girmay, W., Gugsa, G., Taddele, H., Tsegaye, Y., Awol, N., Ahmed, M., et al. (2020). Isolation and identification of methicillin-resistant Staphylococcus aureus (MRSA) from milk in Shire dairy farms, Tigray, Ethiopia. Vet. Med. Int. 2020, 1–7. doi: 10.1155/2020/8833973
Grace, D., and Fetsch, A. (2018). Staphylococcus aureus-a Foodborne Pathogen: Epidemiology, Detection, Characterization, Prevention, and Control: An Overview (Nairobi: International Livestock Research Institute), 3–10.
Gundogan, N., and Avci, E. (2014). Occurrence and antibiotic resistance of Escherichia coli, Staphylococcus aureus and Bacillus cereus in raw milk and dairy products in Turkey. Int. J. Dairy Technol. 67, 562–569. doi: 10.1111/1471-0307.12149
Hamid, S., Bhat, M. A., Mir, I. A., Taku, A., Badroo, G. A., Nazki, S., et al. (2017). Phenotypic and genotypic characterization of methicillin-resistant Staphylococcus aureus from Bovine mastitis. Vet. World 10, 363–367. doi: 10.14202/vetworld.2017.363-367
Hassan, A., Hiko, A., Bogale, K., Abera, B., and Tsegaye, B. (2018). Antimicrobial resistance profiles of Staphylococcus aureus isolates along Asella Municipal Beef Abattoir Line, South Eastern Ethiopia. J. Vet. Sci. Technol. 9, 1–5. doi: 10.4172/2157-7579.1000539
Hemalata, V. B., and Virupakshaiah, D. B. M. (2016). Isolation and identification of food borne pathogens from spoiled food samples. Int. J. Curr. Microbiol. Appl. Sci. 5, 1017–1025. doi: 10.20546/ijcmas.2016.506.108
Heredia, N., and Garcia, S. (2018). Animals as sources of food-borne pathogens: a review. Anim. Nutri. 4, 250–255. doi: 10.1016/j.aninu.2018.04.006
Hiko, A. (2018). Staphylococcus aureus and methicillin resistant S. aureus in commercial soft drink with antimicrobial resistance test on isolates in Ethiopia. J. Microb. Biochem. Technol. 10, 40–45. doi: 10.4172/1948-5948.1000393
Hiwot, D., Savoinni, G., Cattaneo, D., Gabriella, S., and Martino, P. (2016). Bacteriological quality of milk in raw bovine bulk milk in the selected milk collection centers: smallholder dairy processing Ethiopia. J. Ve.t Sci. Ani. Husb. 4, 1–5. doi: 10.15744/2348-9790.4.201
Jahan, M., Rahman, M., Parvej, S., Chowdhury, S. Z. H., Haque, E., Talukder, A. K., et al. (2015). Isolation and characterization of Staphylococcus aureus from raw cow milk in Bangladesh. J. Adv. Vet. Anim. Res. 2, 49–55. doi: 10.5455/javar.2015.b47
Kalayu, A. A., Woldetsadik, D. A., Woldeamanuel, Y., Wang, S., Gebreyes, W. A., and Teferi, T. (2020). Burden and antimicrobial resistance of S. aureus in dairy farms in Mekelle, Northern Ethiopia. BMC Vet. Res. 16, 1–8. doi: 10.1186/s12917-020-2235-8
Kandil, A. A., Elhadidy, M., El-Gamal, A., and Al-Ashmawy, M. A. (2018). Identification of S. aureus and E. coli from dairy products intended for human consumption. Adv. Anim. Vet. Sci. 6, 509–513. doi: 10.17582/journal.aavs/2018/6.11.509.513
Keyvan, E., Yurdakul, O., Demirtas, A., Yalcin, H., and Bilgen, N. (2020). Identification of methicillin-resistant Staphylococcus aureus in bulk tank milk. Food Sci. Technol. 40, 150–156. doi: 10.1590/fst.35818
Khater, D. F., Lela, R. A., El-Diasty, M., Moustafa, S. A., and Wareth, G. (2021). Detection of harmful foodborne pathogens in food samples at the points of sale by MALDT-TOF MS in Egypt. BMC Res. Notes 14, 1–6. doi: 10.1186/s13104-021-05533-8
Kombolcha Town Animal Production and Health Office (2019). Annual Report of Dairy Cattle Production in Kombolcha Town (Amhara).
Kotzamanidis, C., Vafeas, G., Giantzi, V., Anastasiadou, S., Mygdalias, S., Malousi, A., et al. (2021). Staphylococcus aureus isolated from ruminants with mastitis in Northern Greece Dairy Herds: genetic relatedness and phenotypic and genotypic characterization. Toxins 13, 1–16. doi: 10.3390/toxins13030176
Lemma, F., Alemayehu, H., Stringer, A., and Eguale, T. (2021). Prevalence and antimicrobial susceptibility profile of Staphylococcus aureus in milk and traditionally processed dairy products in Addis Ababa, Ethiopia. Biomed Res. Int. 2021, 1–7. doi: 10.1155/2021/5576873
Lim, S. K., Nam, H. M., Park, H. J., Lee, H. S., Choi, M. J., Jung, S. C., et al. (2010). Prevalence and characterization of methicillin-resistant Staphylococcus aureus in raw meat in Korea. J. Microbiol. Biotechnol. 20, 775–778. doi: 10.4014/jmb.0912.12022
Limbu, D. S., Bantawa, K., Limbu, D. K., Devkota, M., and Ghimire, M. (2020). Microbiological quality and adulteration of pasteurized and raw milk marketed in Dharan, Nepal. Hi. J. O. S. T. 3, 37–44. doi: 10.3126/hijost.v4i0.33864
Lingathurai, S., and Vellathurai, P. (2011). Bacteriological quality and safety of raw cow milk in Madurai, South India. Webmed. Cent. Microbiol. 1, 1–10. doi: 10.3329/bjsir.v48i2.15741
Liu, Y., Cao, Y., Wang, T., Dong, Q., Li, J., and Niu, C. (2019). Detection of 12 common food-borne bacterial pathogens by TaqMan real-time PCR using a single set of reaction conditions. Front. Microbiol. 10, 1–9. doi: 10.3389/fmicb.2019.00222
Loir, L. Y., Baron, F., and Gautier, M. (2003). Staphylococcus aureus and food poisoning. Genet. Mol. Res. 2, 63–76.
Magiorakos, A. P., Srinivasan, A., Carey, R. B., Carmeli, Y., Falagas, M. E., Giske, C. G., et al. (2012). Multidrug-resistant, extensively drug-resistant and pandrug-resistant bacteria: an international expert proposal for interim standard definitions for acquired resistance. Clin. Microbiol. Infect. 18, 268–281. doi: 10.1111/j.1469-0691.2011.03570.x
Massawe, H. F., Mdegela, R. H., and Kurwijila, R. L. (2019). Antibiotic resistance of Staphylococcus aureus isolates from milk produced by smallholder dairy farmers in Mbeya Region, Tanzania. Int. J. One Health 5, 31–37. doi: 10.14202/IJOH.2019.31-37
Matallah, A. M., Bouayad, L., Boudjellaba, S., Mebkhout, F., Hamdi, T. M., and Ramdani-Bouguessa, N. (2019). Staphylococcus aureus isolated from selected dairies of Algeria: prevalence and susceptibility to antibiotics. Vet. World 12, 205–210. doi: 10.14202/vetworld.2019.205-210
Mathenge, J. M., Mbaria, J. M., Okemo, P. O., Ng′ang′a, P. M., and Gicheru, M. M. (2017). Multiple drug resistance Staphylococcus aureus isolated in foods of animal origin in Nairobi, Kenya. East Afr. Med. J. 94, 180–185.
Megersa, B., Manedo, A., Abera, M., Regassa, A., and Abunna, F. (2012). Mastitis in lactating cows at Hawassa Town: prevalence, risk factors, major bacterial causes and treatment response to routinely used antibiotics. Am-Euras. J. Sci. Res. 7, 86–91. doi: 10.5829/idosi.aejsr.2012.7.2.6391
Mehrnaz, M., Mohammad, M. S. D., Ramin, M. N. F., Hossein, M. A., and Mahdieh, P. (2021). Prevalence of SEA and SEB producing Staphylococcus aureus isolated from foodborne-outbreaks in Iran. Afr. J. Microbiol. Res. 15, 535–542. doi: 10.5897/AJMR2021.9525
Mekonnen, S. A., Lam, T. J. G. M., Hoekstra, J., Rutten, V. P. M. G., Tessema, T. S., Broens, E. M., et al. (2018). Characterization of Staphylococcus aureus isolated from milk samples of dairy cows in small holder farms of North-Western Ethiopia. BMC Vet. Res. 14, 1–8. doi: 10.1186/s12917-018-1558-1
Mekuria, A., Asrat, D., Woldeamanuel, Y., and Tefera, G. (2013). Identification and antimicrobial susceptibility of Staphylococcus aureus isolated from milk samples of dairy cows and nasal swabs of farm workers in selected dairy farms around Addis Ababa, Ethiopia. Afr. J. Microbiol. Res. 7, 3501–3510. doi: 10.5897/AJMR12.2060
Mohanta, A., and Mazumder, P. B. (2015). Detection of Staphylococci in raw milk and milk products and evaluation of their antibiotic sensitivity: a report from Southern Assam, India. IOSR J. Environ. Sci. Toxicol. Food Technol. 9, 17–22.
Mokgophi, T. M., Gcebe, N., Fasina, F., and Adesiyun, A. A. (2021). Antimicrobial resistance profiles of Salmonella isolates on chickens processed and retailed at outlets of the informal market in Gauteng Province, South Africa. Pathogens 10, 1–15. doi: 10.3390/pathogens10030273
Mphahlele, M. P., Oguttu, J. W., Petzer, I. M., and Qekwana, D. N. (2020). Prevalence and antimicrobial drug resistance of Staphylococcus aureus isolated from cow milk samples. Vet. World 13, 2736–2742. doi: 10.14202/vetworld.2020.2736-2742
Ndahi, M. D., Kwaga, J. K. P., Bello, M., Kabir, J., Umoh, V. J., Yakubu, S. E., et al. (2014). Prevalence and antimicrobial susceptibility of Listeria monocytogenes and methicillin-resistant Staphylococcus aureus strains from raw meat and meat products in Zaria, Nigeria. Lett. Appl. Microbiol. 58, 262–269. doi: 10.1111/lam.12183
Olatoye, I. O., Uba, U. C., and Akintunde, S. O. (2018). Prevalence of Staphylococcus aureus contamination in bovine milk from local dairy herd in Oyo State, Nigeria. Trop. Vet. 36, 87–93.
Oliver, S. P., Jayarao, B. M., and Almeida, R. A. (2005). Foodborne pathogens in milk and the dairy farm environment: food safety and public health implications. Foodborne Pathog. Dis. 2, 115–129. doi: 10.1089/fpd.2005.2.115
Pal, M., Ketchakmadze, D., Durglishvili, N., and Ketchakmadze, K. (2022). Staphylococcus aureus: a major pathogen of food poisoning: a rare research report. Nutr. Food Process 5, 1–3. doi: 10.31579/2637-8914/082
Parvin, S., Ali, Y., Talukder, S., Nahar, A., Chowdhury, E. H., Rahman, T., et al. (2021). Prevalence and multidrug resistance pattern of methicillin resistant S. aureus isolated from frozen chicken meat in Bangladesh. Microorganisms 9, 1–16. doi: 10.3390/microorganisms9030636
Pati, B. K., and Mukherjee, R. (2016). Characterization of Staphylococcus aureus isolates of bovine mastitis origin and antibiotic sensitivity pattern from Northern Plains of India. J. Vet. Res. Ani. Husb. 1, 1–5.
Pazakova, J., Turek, P., and Laciakova, A. (1997). The survival of Staphylococcus aureus during the fermentation and storage of yoghurt. J. Appl. Microbiol. 82, 659–662. doi: 10.1111/j.1365-2672.1997.tb02877.x
Pramodhini, S., Thenmozhivalli, P. R., Selvi, R., Dillirani, V., Vasumathi, A., and Agatha, D. (2011). Comparison of various phenotypic methods and mecA based PCR for the detection of MRSA. J. Clin. Diagn. Res. 5, 1359–1362.
Qamar, A., Ismail, T., and Akhtar, S. (2020). Prevalence and antibiotic resistance of Salmonella spp. in South Punjab-Pakistan. PLoS ONE 15, 1–15. doi: 10.1371/journal.pone.0232382
Quinn, P. J., Carter, M. E., Markey, B., and Carter, G. R., (eds.). (1999). “Enterobacteriaceace,” in Clinical Veterinary Microbiology (London: Mosby International Limited), 226–460.
Quinn, P. J., Markey, B. K., Carter, M. E., Donelly, W. J., and Leonard, F. C. (2002). Veterinary Microbiology and Microbial Disease (Hoboken, NJ: Blackwell Publishing Company), 465–475.
Reta, M. A., Bereda, T. W., and Alemu, A. N. (2016). Bacterial contaminations of raw cow's milk consumed at Jigjiga City of Somali Regional State, Eastern Ethiopia. Int. J. Food Contam. 3, 1–9. doi: 10.1186/s40550-016-0027-5
Riva, A., Borghi, E., Cirasola, D., Colmegna, S., Borgo, F., Amato, E., et al. (2015). Methicillin-resistant Staphylococcus aureus in raw milk: prevalence, SCCmec typing, enterotoxin characterization, and antimicrobial resistance patterns. J. Food Prot. 78, 1142–1146. doi: 10.4315/0362-028X.JFP-14-531
Rodríguez-Lázaro, D., Oniciuc, E., García, G. P., Gallego, D., Fernández-Natal, I., Dominguez-Gil, M., et al. (2017). Detection and characterization of Staphylococcus aureus and methicillin-resistant S. aureus in foods Confiscated in EU Borders. Front. Microbiol. 8, 1–10. doi: 10.3389/fmicb.2017.01344
Salauddin, M., Rowshan, A. M., Hossain, K. M., Nazir, K. H. M. N.H., Noreddin, A., and El Zowalaty, M. E. (2020). Molecular detection of multidrug resistant Staphylococcus aureus isolated from bovine mastitis milk in Bangladesh. Vet. Sci. 7, 1–10. doi: 10.3390/vetsci7020036
Savariraj, W. R., Ravindran, N. B., Kannan, P., Paramasivam, R., Senthilkumar, T. M. A., Kumarasamy, P., et al. (2018). Prevalence, antimicrobial susceptibility and virulence genes of Staphylococcus aureus isolated from pork meat in retail outlets in India. Food Saf. 39, 1–8. doi: 10.1111/jfs.12589
Shrestha, A., Bhattarai, R. K., Luitel, H., Karki, S., and Basnet, H. B. (2021). Prevalence of methicillin-resistant Staphylococcus aureus and pattern of antimicrobial resistance in mastitis milk of cattle in Chitwan, Nepal. BMC Vet. Res. 17, 1–7. doi: 10.1186/s12917-021-02942-6
Soliman, N. S. M., and Ahmed, L. I. (2019). Survival of Staphylococcus aureus in Bio-Yoghurt. Open J. Appl. Sci. 9, 564–572. doi: 10.4236/ojapps.2019.97044
Sori, T., Hussien, J., and Bitew, M. (2011). Prevalence and susceptibility assay of Staphylococcus aureus isolated from bovine mastitis in dairy farms of Jimma town, South West Ethiopia. J. Anim. Vet. Adv. 10, 745–749. doi: 10.3923/javaa.2011.745.749
Sudhanthiramani, S., Swetha, C. S., and Bharathy, S. (2015). Prevalence of antibiotic resistant Staphylococcus aureus from raw milk samples collected from the local vendors in the region of Tirupathi, India. Vet. World. 8, 478–481. doi: 10.14202/vetworld.2015.478-481
Tamendjari, S., Bouzebda, F. A., Chaib, L., Aggad, H., Ramdani, M., and Bouzebda, Z. (2021). Antibiotic resistance of Staphylococcus aureus isolated from raw cow and goat milk produced in the Tiaret and Souk Ahras areas of Algeria. Vet. World. 14, 1929–1934. doi: 10.14202/vetworld.2021.1929-1934
Tesfaye, K., Gizaw, Z., and Haile, A. F. (2021). Prevalence of mastitis and phenotypic characterization of methicillin-resistant Staphylococcus aureus in lactating dairy cows of selected dairy farms in and around Adama Town, Central Ethiopia. Environ. Health Insights 15, 1–8. doi: 10.1177/11786302211021297
Tessema, D., and Tsegaye, S. (2017). Study on the prevalence and distribution of Staphylococcus aureus in raw cow milk originated from Alage Atvet College Dairy Farm, Ethiopia. J. Nutr. Food Sci. 7, 1–5. doi: 10.4172/2155-9600.1000586
Thrusfield, M. (2005). Veterinary Epidemiology, 3rd Edn (London: Black Well Science Ltd., A Blackwell Publishing Company), 230–234.
Truchado, P., and Randazzo, W. (2022). New challenges for detection and control of foodborne pathogens: from tools to people. Foods 11:1788. doi: 10.3390/foods11121788
Tsepo, R., Ngoma, L., Mwanza, M., and Ndou, R. (2016). Prevalence and antibiotic resistance of Staphylococcus aureus isolated from beef carcasses at abattoirs in North West Province. J. Hum. Ecol. 56, 188–195. doi: 10.1080/09709274.2016.11907055
Weldeselassie, M., Gugsa, G., Kumar, A., Tsegaye, Y., Awol, N., Ahmed, M., et al. (2020). Isolation and characterization of Staphylococcus aureus from food of bovine origin in Mekelle, Tigray, Ethiopia. Open Microbiol. J. 14, 234–241. doi: 10.2174/1874285802014010234
Wu, S., Huang, J., Wu, Q., Zhang, J., Zhang, F., Yang, X., et al. (2018). Staphylococcus aureus isolated from retail meat and meat products in China: incidence, antibiotic resistance and genetic diversity. Front. Microbiol. 9, 1–14. doi: 10.3389/fmicb.2018.02767
Yakubu, A., Abdullahi, I. O., Whong, C. Z., and Olayinka, B. (2020). Prevalence and antibiotic susceptibility profile of Staphylococcus aureus from milk and milk products in Nasarawa State, Nigeria. Sokoto J. Vet. Sci. 18, 1–12. doi: 10.4314/sokjvs.v18i1.1
Zakary, E. M., Nassif, M. Z., and Mohammed, G. M. (2011). Detection of Staphylococcus aureus in bovine milk and its product by real time PCR assay. Glob. J. Biotechnol. Biochem. 6, 171–177.
Zerabruk, K., Retta, N., Muleta, D., and Tefera, A. T. (2019). Assessment of microbiological safety and quality of minced meat and meat contact surfaces in selected butcher shops of Addis Ababa. J. Food Qual. 2019, 1–8. doi: 10.1155/2019/3902690
Keywords: antimicrobial resistance, Dessie, Kombolcha, MRSA, prevalence, risk factors, S. aureus
Citation: Abebe E, Gugsa G, Ahmed M, Awol N, Tefera Y and Abegaz S (2024) Occurrence, associated risk factors and antimicrobial resistance patterns of Staphylococcus aureus and methicillin resistant S. aureus from foods of bovine origin in Dessie and Kombolcha towns, Ethiopia. Front. Sustain. Food Syst. 8:1422850. doi: 10.3389/fsufs.2024.1422850
Received: 11 June 2024; Accepted: 04 November 2024;
Published: 26 November 2024.
Edited by:
M. Leonor Faleiro, University of Algarve, PortugalReviewed by:
Jitendra Patel, Agricultural Research Service (USDA), United StatesGuodong Zhang, United States Food and Drug Administration, United States
Copyright © 2024 Abebe, Gugsa, Ahmed, Awol, Tefera and Abegaz. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Getachew Gugsa, Z3Vnc2FnQHlhaG9vLmNvbQ==
 Engidaw Abebe
Engidaw Abebe Getachew Gugsa
Getachew Gugsa