
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 30 July 2024
Sec. Crop Biology and Sustainability
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1413724
This article is part of the Research TopicRegulation of Ovule and Seed DevelopmentView all 5 articles
 Minwen Yan1†
Minwen Yan1† Hongyuan Xi1†
Hongyuan Xi1† Shiyin Hu2†
Shiyin Hu2† Jikun Song2
Jikun Song2 Bing Jia2
Bing Jia2 Pan Feng2
Pan Feng2 Liupeng Yang2
Liupeng Yang2 Jianjiang Ma2
Jianjiang Ma2 Li Wang2
Li Wang2 Wenfeng Pei2
Wenfeng Pei2 Bingbing Zhang2
Bingbing Zhang2 Jiwen Yu2*
Jiwen Yu2* Man Wu2*
Man Wu2* Shoulin Hu1*
Shoulin Hu1*Cotton (Gossypium spp.) not only serves as a primary textile fiber crop but also as a vital oilseed crop. It stands as the world’s fifth-largest oil crop and is rich in essential fatty acids. At present, the mechanisms underlying the biosynthesis of cottonseed oil have been extensively studied in cotton. 3-Hydroxyacyl-CoA dehydratase (HACD) is the third rate-limiting enzyme in the elongase complex, which plays a critical role in the biosynthesis of Very Long Chain Fatty Acids (VLCFA). However, the members of the HACD family and their roles in cottonseed oil remain uncharacterized in cotton. This study identified that G. arboreum and G. raimondii have two HACD genes, while four HACD genes exists in G. hirsutum, and G. barbadense. The phylogenetic relationships of the 12 HACD genes from the four cotton species further divided them into two subfamilies. Gene structure and conserved motif analysis revealed that members of the HACD family were relatively conserved during the evolution of cotton, but members within the same subfamily exhibited more similar structures. Homology and collinearity analysis suggest whole-genome duplication/segmental duplication may be a key factor in the amplification of the cotton HACD gene family. The qRT-PCR analysis of high-oil and low-oil genotype found significant differences in the expression levels of GhHACD1-4, which indicates GhHACD1-4 is expected to participate in the lipid oil biosynthesis process. Subcellular localization experiments confirmed the presence of the GhHACD2 inendoplasmic reticulum. The KEGG pathway enrichment analysis of co-expressed genes of GhHACD1 and GhHACD2 genes were conducted to confirm their potential involvement in fatty acid elongation and oil biosynthesis. Furthermore, transgenic overexpression analysis of GhHACD2 caused a 5.02% decrease in oil content compared with the control in yeast, while the levels of C28:0, C30:0, and VLCFAs were significantly improved. This study characterizes HACD gene family members in cotton and provides rich genetic resources for increasing cottonseed oil content and improving the nutritional value of cottonseed oil.
Cotton is an important economic crop worldwide, generally known for producing high-quality natural fiber. It is also the world’s fifth-largest oil crop after soybean, palm, rapeseed, and sunflower (Zia et al., 2021). Cotton seeds, as important by-products of cotton, also hold great potential and prospects as a valuable source for high-quality feed, plant oil, and biofuels (Wu et al., 2022). The composition of fatty acids in plant oil plays a decisive role in its nutritional value and uses (Napier et al., 2014). Cottonseed oil accounts for 17–27% of the seed weight (Wu et al., 2010), and is rich in fatty acids. Saturated fatty acids such as palmitic acid (C16:0) account for 26%, stearic acid (C18:0) accounts for 2%, and unsaturated fatty acids such as linoleic acid (C18:2) account for 58%, and oleic acid (C18:1) accounts for 13%. Cottonseed oil is a predominantly unsaturated plant oil with linoleic acid as its main component. Unsaturated fatty acids play significant roles in human health and nutrition, making cottonseed oil highly valuable for humans (Konukan et al., 2017; Zhao et al., 2021).
Fatty acids are primarily classified based on their carbon chain length. Short-chain fatty acids (SCFAs) contain fewer than 6 carbons; medium-chain fatty acids (MCFAs) have 6–12 carbons; long-chain fatty acids (LCFAs) contain 12–20 carbons; and very long-chain fatty acids (VLCFAs) contain more than 20 carbons. Almost all plant tissues have VLCFAs, which are essential for plant growth and play various critical roles in plant development (Haslam and Kunst, 2013). VLCFAs are important functional components of various lipid classes, including cuticular lipids in the higher plant epidermis and lipid-derived second messengers. The diversity of VLCFAs and their derivatives in the plant kingdom is profoundly attributed to the variable acyl-chain lengths, the degree of unsaturation, and the extent of hydroxylation each molecule undergoes (Batsale et al., 2021). VLCFAs in different locations in an organism have different forms of influence on various life activities. For example, they are essential for the synthesis of membrane lipids (e.g., phospholipids and sphingolipids) in plant cells (Devaiah et al., 2006). They also serve as precursors for the synthesis of cuticle waxes and suberins (Suh et al., 2005). Furthermore, VLCFAs can accumulate in the triacylglycerols (TAGs) of seeds, serving as stored lipids for reserve utilization (Kunst et al., 1992). Several distinguished VLCFAs, including, eicosapentaenoic acid (20:5), docosahexaenoic acid (22:6), erucic acid (C22:1), and nervonic acid (C24:1), constitute invaluable assets, offering substantial benefits to human health (Zhukov and Popov, 2022).
In plants, the biosynthesis of VLCFAs initiates with the de novo synthesis of C16/C18-CoA in the plastids, and then the C16/C18-CoA is catalytically elongated into VLC-acyl-CoAs with the help of fatty acid elongase complexes (FAE), which are located in the endoplasmic reticulum (Bates and Browse, 2012). The FAE complex consists of four enzymes, including 3-ketoacyl-CoA-synthase (KCS), 3-ketoacyl-CoA-reductase (KCR), 3-hydroxyacyl-CoA-dehydratase (HACD), and trans-2,3-enoyl-CoA-reductase (ECR). The elongation of VLCFAs proceeds in several steps; firstly, KCS catalyzes the condensation of acyl-CoA with malonyl-CoA to form3-ketoacyl-CoA. Then, the 3-ketoacyl-CoA is reduced to 3-hydroxyacyl-CoA by KCR enzymes. Subsequently, HACD dehydrates the 3-hydroxyacyl-CoA to trans-2,3-enoyl-CoA, which is eventually reduced by the fourth enzyme, ECR. This process results in the elongation of the acyl-CoA by two carbons. The reaction can be repeated, generating VLCFAs with various chain lengths ranging from C20 to more (Leonard et al., 2004; Haslam and Kunst, 2013).
The HACD gene is the third rate-limiting enzyme in the FAE complex and plays an indispensable role in the synthesis of VLCFAs. Initially discovered in yeast, the HACD gene was identified as a VLCFA dehydratase, crucial in the third step of very long-chain fatty acid synthesis (Denic and Weissman, 2007). Through yeast mutant phs1 complementation experiments, the homolog of HACD in Arabidopsis, known as PAS2/PTPLA, was discovered (Bach et al., 2008; Morineau et al., 2016). Assays of Arabidopsis mutants revealed that the loss of AtPAS2 function leads to embryo lethality, accompanied by a decrease in VLCFA content in the cuticle wax, seed oil, and sphingolipids (Bach et al., 2008). Another homolog of HACD in Arabidopsis, PTPLA, inhibits fatty acid elongation, and its loss of function in Arabidopsis increases VLCFA content (Morineau et al., 2016). In maize (Zea mays), ZmHCD was found to complement yeast mutants and complete VLCFA synthesis (Campbell et al., 2019). The GhPAS2 gene has been confirmed as a VLCFA dehydratase in fiber development of cotton, but its role in regulating fatty acid synthesis is not yet clearly understood (Wang et al., 2015). The very-long-chain (3R)-3-hydroxyacyl-CoA dehydratase (HACD) is involved in regulating the fatty acid elongation pathway in fiber development by analyzing a complex network of various RNA types, including mRNA, miRNA, lncRNA, and circRNA (Wang et al., 2024). However, HACD genes have not been reported to be involved in cottonseed oil and VLCFA biosynthesis.
This study undertook a whole-genome analysis of the HACD gene family members across four cotton species, revealing their phylogenetic relationships, gene structures, and expression patterns in cotton. The results highlighted the important regulatory role of the GhHACD2 gene in the synthesis of VLCFAs and lipids by heterologous expression of GhHACD2 in Saccharomyces cerevisiae. The research results provide new insights into the functional characterization of HACD genes in fatty acid biosynthesis in cotton.
Using G. hirsutum high-oil genotype (N0409, oil content 24.24%) and low-oil genotype (N6940, oil content 15.28%), the experiment was conducted in 2022 at the experimental station of the Cotton Research Institute of the Chinese Academy of Agricultural Sciences in Anyang, Henan, China. Samples were taken from the ovules at seven developmental stages (0, 5, 10, 15, 20, 25, and 30 days post-anthesis, DPA) after flowering in cotton. Each sample had three biological replicates. The freshly harvested samples were immediately placed in liquid nitrogen and stored at −80°C.
The gene annotation GFF3 and genome files of G. arboreum (HAU), G. raimondii (HAU), G. hirsutum (ZJU), and G. barbadense (ZJU) were retrieved from the Cotton Functional Genomics Database (CottonMD, http://yanglab.hzau.edu.cn/CottonMD/) (Yang et al., 2023c). The HACD protein sequences of Arabidopsis were obtained from the Arabidopsis Information Resource (TAIR, http://www.arabidopsis.org) database (Bano et al., 2021b). The HACD protein sequences of Glycine max and Brassica napus were obtained from the Soybean Genome Database (SoyBase, https://www.soybase.org/) and the Brassica Database (BRAD, http://www.brassicadb.cn/), respectively.
The protein sequences of the HACD genes AT5G10480 and AT5G59770 in Arabidopsis thaliana were used as reference sequences to search for HACD genes in cotton. The target files were aligned and searched using the local BLASTP (the e value≤ 10−5 and identity match ≥ 50%) and TBtools software, resulting in the identification of candidate HACD genes in four cotton species (Chen et al., 2020; Bano et al., 2023). The identified HACD genes were further confirmed by NCBI Batch-CDD1 search and SMART2 search for the presence of a conserved PTPLA protein structural domain (Letunic et al., 2021). The physicochemical properties of the HACD protein were analyzed using the online tool ExPASyProtParam (https://web.expasy.org/protparam/; Chattha et al., 2020).
The candidate HACD protein sequences from G. arboreum, G. raimondii, G. hirsutum, G. barbadense, Arabidopsis thaliana, Glycine max, and Brassica napus were aligned using the ClustalW tool (Kumar et al., 2016). The best phylogenetic tree of all HACD proteins was constructed using MEGA11, which using a neighbor-joining [NJ, bootstrap with 1,000 replicates and Jones-Taylor-Thornton (JTT) model] (Hall, 2013; Bano et al., 2021a). The constructed phylogenetic tree was visualized with the online iTOL tool.3
The MEME Suite4 was used to predict motifs in the HACD family members of four cotton varieties (Shafqat et al., 2021). The motif parameter was set to 10, while the other parameters were kept at their default values (Hall, 2013). TBtools was used to visualize the gene structure, protein motifs, and conserved domains of the HACD family members.
CottonMD genome annotation files and TBtools were used to visualize the chromosomal distribution of HACD genes in four cotton species. Multiple Collinearity Scan toolkit (MCScanX) was used to analyze gene duplication events and collinearity between four cotton species, as well as between diploid and tetraploid cotton species (Wang et al., 2012). Non-synonymous mutation rate (Ka), synonymous mutation rate (Ks), and Ka/Ks value of selection pressure were calculated using TBtools software. The haplotype distribution frequency of GhHACD in different cotton subgroups was obtained through CottonMD.5
Using the TBtools software, 2 kb promoter sequences upstream of the HACD gene transcription start sites in the genomes of four cotton species were extracted from the genomic database. Then, the retrieved sequences were analyzed for cis-elements using the PlantCARE website.6 After filtering and removing redundant information, the TBtools Simple BioSequence Viewer function was used to complete the visualization analysis.
The transcriptional data from different tissues, including epicalys, leaf, petal, pistil, root, sepal, stem, torus, fiber (25 DPA), and at different developmental stages of ovule (0, 1, 3, 5, 10, and 20 DPA), were obtained from the COTTONOMICS website7 for conducting expression analysis of GhHACD gene.
Following the grinding of the fresh samples, the FastPure Plant Total RNA Kit (Nanjing Vazyme Biotech. Co., Ltd.) was used to extract total RNA from cotton seeds and other tissues. The quality and quantity of RNA were then detected using a Nanodrop 2000 spectrophotometer (Thermo Fisher Scientific) (OD260/OD280). Next, the RNA was reverse transcribed into first-strand cDNA using the HiScript III Q RT SuperMix for qPCR kit (Vazyme Biotech Co., Ltd.). Specific primers for the gene were designed using SnapGene software (Supplementary Table S1). The ChamQ Universal SYBR qPCR Master Mix kit (Vazyme) was used for real-time fluorescence quantitative PCR, with the internal control reference being HISTONE3 (AF024716). The data were processed with the 2−ΔΔCT method, with three biological replicates set for each sample.
Performing Weighted Gene Co-expression Network Analysis (WGCNA) using transcriptomes from five developmental stages (0, 5, 10, 20, and 30 DPA) of embryo tissues from high-oil (3,008) and low-oil (3,012) genotype (Song et al., 2022). The co-expression network of GhHACD1 and GhHACD2 genes was analyzed using WGCNA with 49,661 genes. Among which, 16,828 genes were obtained and classified into MEturquoise modules (Song et al., 2022). Performing KEGG pathway analysis in MEturgugise modules genes using GENE DENOVO.8
The 35S-YFP subcellularly localized expression vector was constructed using the cDNA mixture from TM-1 as a template. The primer pairs GhHACD2-YFP-F (5′-GGGGACAAGTTTGTACAAAA AAGCAGGCTTAATGTCTCACCTGCTGAAGCT-3′) and GhHACD2-YFP-R(5′-GGGGACCACTTT GTACAAGAAAGCTG GG TAAATTTTCT TCTTCTTGTGGT-3′) were designed as per the reference genome TM-1. The recombinant plasmid p35S-GhHACD2-YFP was transformed into the Agrobacterium tumefaciens strain (GV3101). The bacterial liquid containing the target plasmid was then introduced into N. benthamiana leaves, and the YFP signal was observed through confocal laser scanning microscopy.
Using the cDNA mixture from TM-1 as a template, the primer pairs GhHACD2-HindIII-F (5′-TAATACGACT CACTATAGGG ATGTCTCACCTGCTGAAGCT-3′) and GhHACD2-BamHI-R (5′-GTGACATAACTAATTACATGATGTCAAATTTTCTTCTTCT TGT-3′) were designed as per the reference genome TM-1. The PCR product was inserted between the HindIII and BamHI sites of the pYES2 yeast expression vector. The recombinant plasmid pYES2-GhHACD2 was then transferred into the Saccharomyces cerevisiae (INVSc1) using the PEG/LiAc transformation technique (Ma et al., 2019). The INVSc1 strain containing the empty vector pYES2 served as a negative control (CK). Galactose and raffinose were added to the culture medium to induce transgenic yeast for gene expression. The obtained yeast cells were centrifuged and freeze-dried in a vacuum freeze dryer (Christ® Alpha I-5; Martin Christ).
The freeze-dried yeast cells were ground into powder, and each sample was arranged with three biological replicates. These samples were used for preparing fatty acid methyl esters (FAME), and the fatty acid composition was analyzed using a gas chromatograph Nexis GC-2030 (Shimadzu Corporation, Kyoto, Japan) (Xin et al., 2022). Quantitative analysis was performed based on the peak area of the internal standard C17:0 and the retention time of the fatty acid standards.
In a comprehensive analysis across four cotton species, 12 HACD genes were successfully identified. Specifically, two HACD genes were identified in the diploid cotton species G. arboreum and G. raimondii, respectively. The four HACD genes were renamed GaHACD1, GaHACD2, GrHACD1, and GrHACD2, respectively. In the tetraploid cotton species G. hirsutum and G. barbadense, four HACD genes were identified, respectively. The eight HACD genes were renamed GhHACD1, GhHACD2, GhHACD3, GhHACD4, GbHACD1, GbHACD2, GbHACD3, and GbHACD4, respectively. The physicochemical properties of the HACD genes in the four cotton species were analyzed, revealing that the protein length of cotton HACD family members ranged from 219 to 255 aa, with a molecular weight (MW) range of 25.24–29.55 kDa and an isoelectric point (pI) range of 9.32–9.79, indicating that they are all alkaline proteins. Among them, GaHACD1, GrHACD1, GhHACD1, GhHACD2, GbHACD1, and GbHACD2 showed similar protein lengths, MW, and pI, suggesting that they may have parallel functions in cotton (Table 1; Supplementary Table S2).
To systematically understand the evolutionary correlation and phylogenetic relationships of the cotton HACD gene family, protein sequences from Arabidopsis thaliana, Brassica napus, Glycine max, and four cotton HACDs were used to construct a phylogenetic tree (Figure 1). As shown in Figure 1, the HACD gene family is divided into two subgroups, A and B. AtHACD2 is grouped with GaHACD1, GrHACD1, GhHACD1, GhHACD2, GbHACD1, and GbHACD2 in the same branch, indicating homology between the HACD genes in this subgroup and ATHACD2. AtHACD1 is grouped with GaHACD2, GrHACD2, GhHACD3, GhHACD4, GbHACD3, and GbHACD4 in the same branch, suggesting homology between the HACD genes in this subgroup and AtHACD1. Each subgroup is further subdivided into two clusters, with a closer relationship observed between cotton and Glycine max HACD family members. Genes within the same branch demonstrate similar evolutionary levels and exhibit a higher degree of relatedness. Additionally, the number of genes varies among different subgroups, signifying significant divergence of HACD genes during the evolution of species.
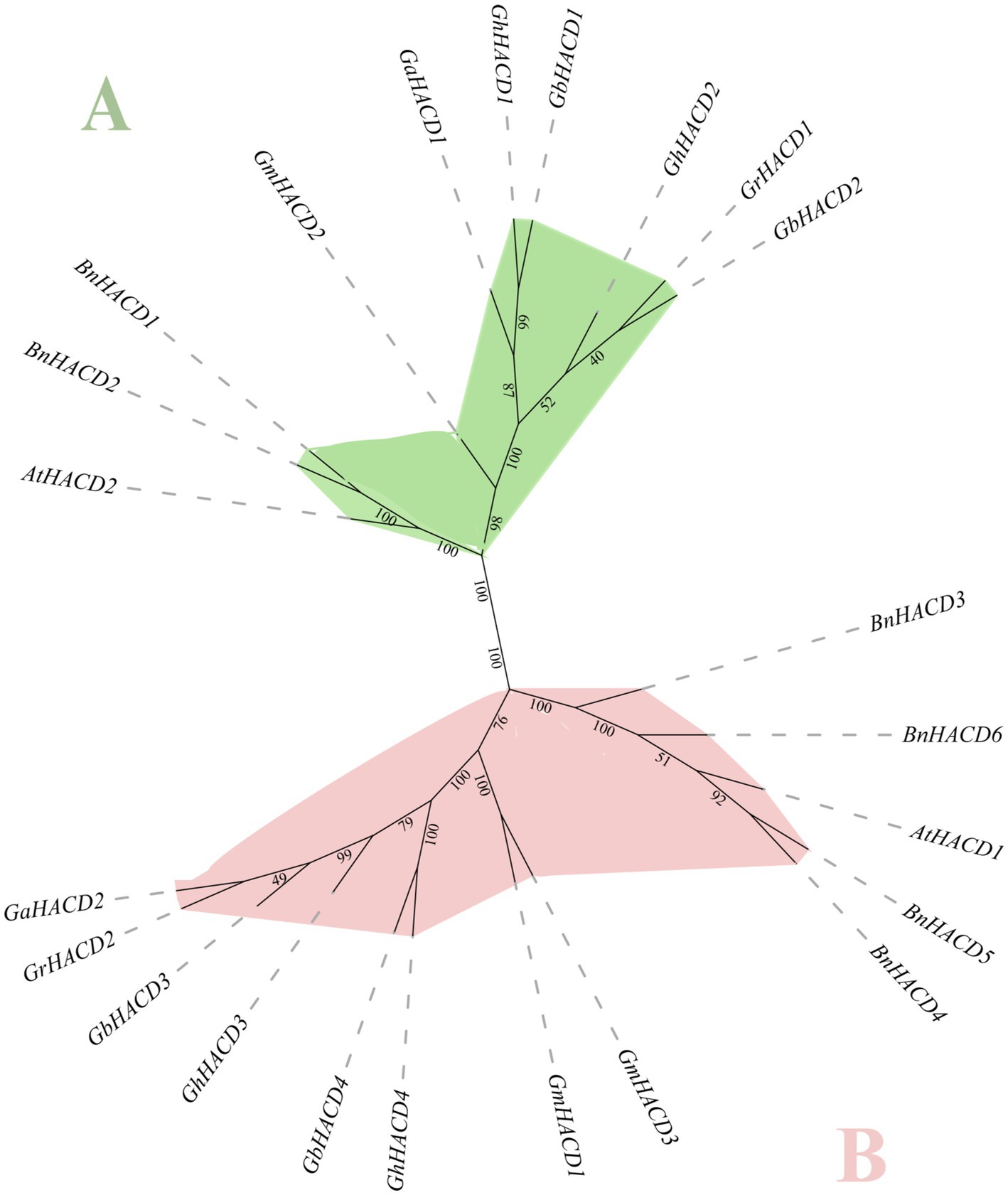
Figure 1. Phylogenetic analysis of the HACD genes in Arabidopsis, Glycine max, Brassica napus, and four cotton species. The phylogenetic tree was constructed using the neighbor-joining (NJ) method.
Analysis of the 10 conserved motifs of HACD proteins in four cotton species was done using the MEME online tool to study the characteristics of HACD genes (Figure 2). The conserved motifs within subgroups A and B exhibit substantial congruity. Intriguingly, Motif 10 was exclusively found in subgroup B. The distribution of motifs in HACD protein members within subgroup A is completely consistent, while differences in Motifs 9 and 7 were observed in GhHACD3, GhHACD4, GbHACD4, GaHACD2, GrHACD2, and GbHACD3 within subgroup B. It is worth noting that Motifs 1, 2, 3, 5, 6, 8, and 9 are identified in four cotton HACD proteins in both subgroup A and subgroup B. This observation underscores their highly conserved nature and signifies their role as conserved protein domains of HACD.
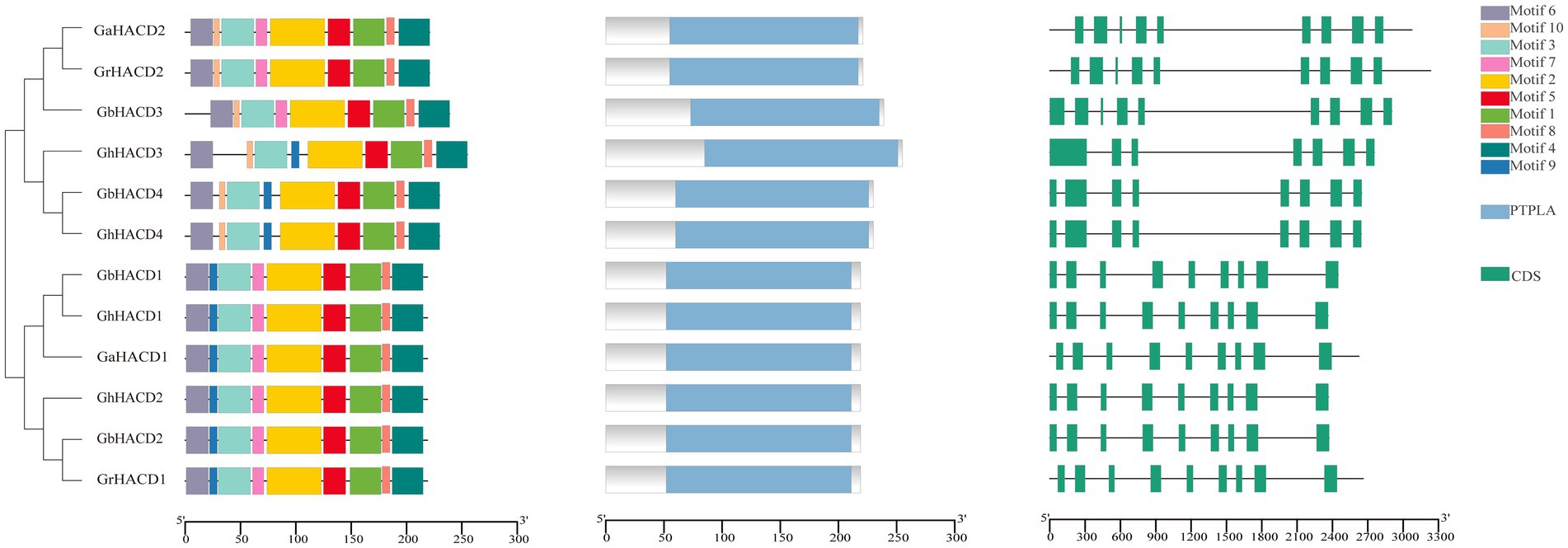
Figure 2. The conserved motifs, conserved domain, and gene structure (the exon–intron structure) of the HACD gene family in four cotton species.
To further explore the gene structure of HACD family members, we analyzed their intron/exon compositions (Figure 2). As shown in Figure 2, the exon-intron distribution pattern of HACD genes in subgroup A is consistent in that each HACD gene contains nine exons. The gene structures of HACD family members in subgroup B are relatively consistent, but GhHACD4 and GbHACD4 are devoid of a unique exon compared to the other members in subgroup B, and GhHACD3 lacks two exons. Conserved domain analysis of HACD indicates that all HACD proteins contain the conserved PTPLA domain. Overall, HACD genes exhibit high conservation during cotton evolution, but members within the same subgroup have more similar conserved protein regions and motif compositions. These results not only demonstrate the feasibility of phylogenetic evolution classification, but also suggest that they may have similar biological functions.
The chromosomal localization analysis of 12 HACD genes in four cotton species (Figure 3) was conducted. As shown in the Figure 3, in G. arboreum, HACD genes are located on chromosomes A01 and A11; in G. raimondii, HACD genes are located on chromosomes D02 and D11; in G. hirsutum and G. barbadense, HACD genes are distributed on chromosomes A03, A11, D03, and D11. The results indicate that during the course of evolution, HACD genes are evenly dispersed in the cotton genome (At and Dt subgenomes) without gene loss, which may be attributed to the relatively small number of HACD gene family members. Additionally, there is a high degree of similarity in the number, chromosomal distribution, and structure of HACD gene family members between the two tetraploid cotton species, G. hirsutum and G. barbadense, indicating a high level of conservation of HACD genes between them.
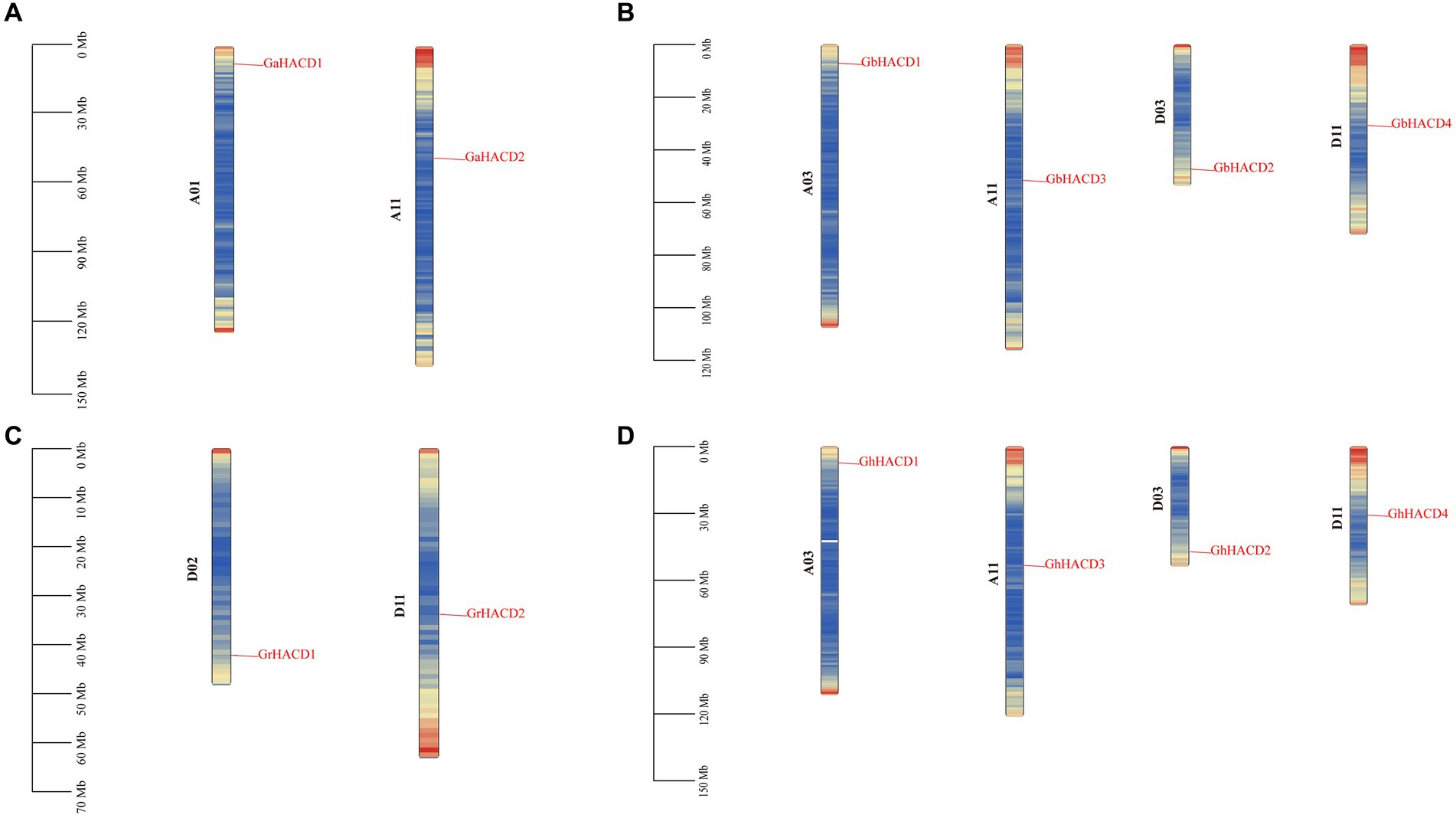
Figure 3. The distribution of the HACD gene on chromosomes of four cotton species. (A) Gossypium arboreum. (B) Gossypium barbadense. (C) Gossypium raimondii. (D) Gossypium hirsutum. The left scale indicates the chromosome length (Mb). Colors on the chromosomes indicate gene density (gene number/100 kb).
In order to clarify the duplication and evolutionary status of the cotton HACD genes, we conducted a gene duplication event analysis within four cotton species. Through collinearity analysis, we observed that in the two tetraploid cotton species, there are four pairs of collinear gene pairs (GhHACD1/GhHACD2, GhHACD3/GhHACD4, GbHACD1/GbHACD2, and GbHACD3/GbHACD4), and all eight HACD genes contain segmental repeat sequences (Figure 4A), without tandem repeats. Among them, there are no repeat relationships between HACD genes in the A subgenome and D subgenome of the diploid cotton species. In addition, the Ka/Ks ratios of all the duplicated genes were found less than 0.5 (Table 2).
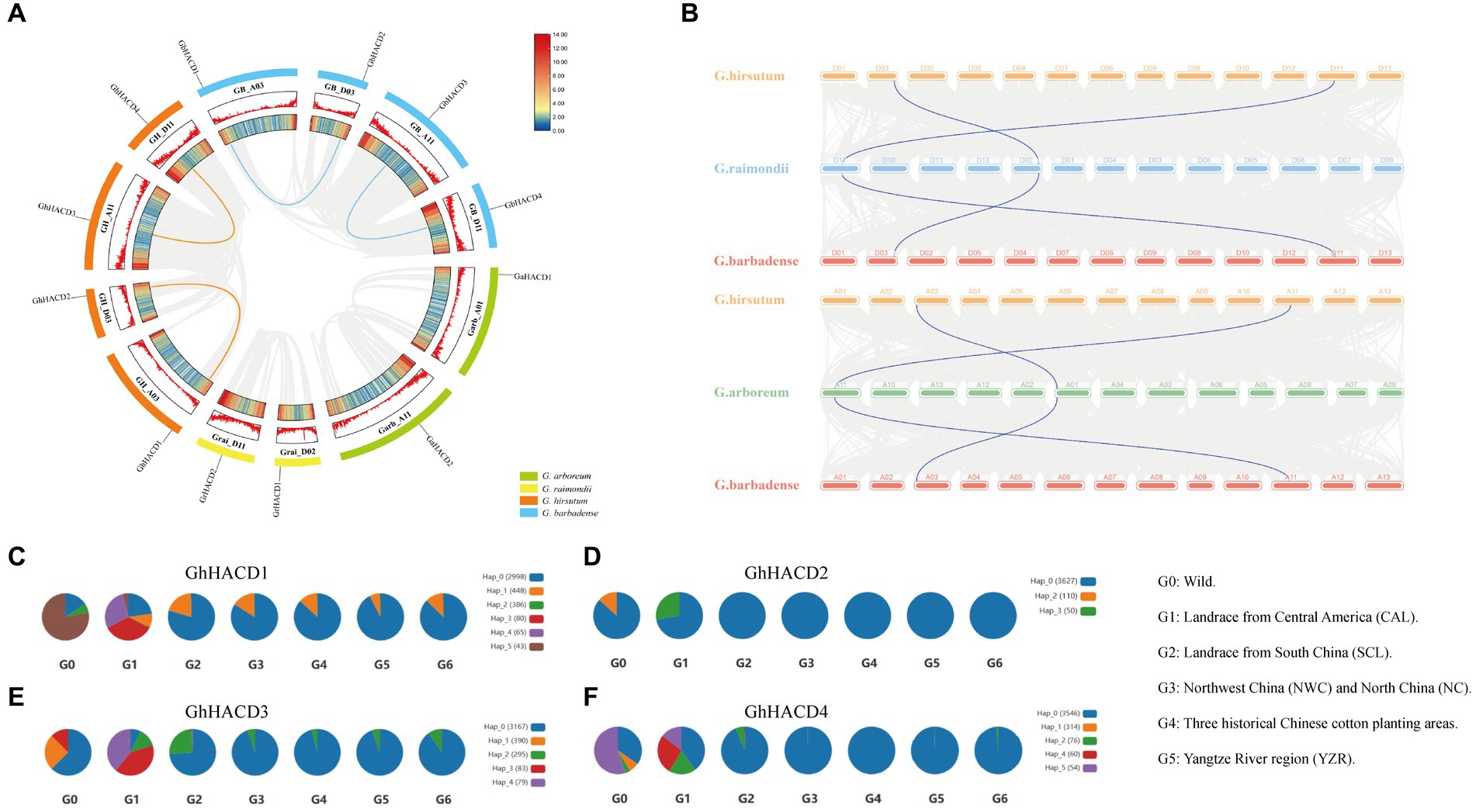
Figure 4. Analysis of HACD gene structure. (A) The distribution and collinearity of the HACD genes in four cotton species. Different color represents different cotton species, the red lines indicated the duplicated gene pairs of HACD. (B) Collinearity of HACD genes between different cotton species. The gray lines indicate colinear blocks, and the blue lines indicate the homozygous pairs of common Gossypium hirsutum or Gossypium barbadense with Gossypium arboreum and Gossypium raimondii, respectively. (C–F) The haplotype distribution frequency of GhHACD in different G. hirsutum Sub-population. (C) GhHACD1. (D) GhHACD2. (E) GhHACD3. (F) GhHACD4.
As is well known, the allotetraploid cotton species G. hirsutum and G. barbadense originated from the hybridization of the A subgenome (G. arboreum) and D subgenome (G. raimondii) (Hu et al., 2019). Subsequently, we investigated the collinearity relationship between tetraploid and diploid cotton species to further understand the duplication of the HACD gene family among different cotton species. In both the G. arboreum and G. raimondii genomes, we found two pairs of collinear homologous genes of HACD in the G. hirsutum (Figure 4B). Similarly, in the collinearity relationship between the HACD genes in the G. barbadense and the HACD genes in G. arboreum and G. raimondii, we also identified two homologous genes (Figure 4B). Thus, it can be inferred that the members of the HACD gene family are highly conserved in evolution. Additionally, we observed chromosomal translocation in the chromosomal segment containing the HACD genes during the process of polyploidization from diploid to tetraploid cotton species, with chromosome A01 from G. arboreum translocating to chromosome A03 in tetraploid cotton species, and chromosome D02 from G. raimondii translocating to chromosome D03 in tetraploid cotton species.
To explore the domestication selection of HACD genes in G. hirsutum, we analyzed the distribution of HACD genes in different G. hirsutum species and different regions (Figures 4C–F). The analysis revealed that the GhHACD1, GhHACD3, and GhHACD4 genes primarily had three or more haplotypes in G0 and G1. Among them, hap_0 accounted for 15.63, 62.5, and 34.48% in G0, and 22.91, 7.22, and 40.28% in G1 for GhHACD1, GhHACD3, and GhHACD4, respectively (Figures 4C,E,F). GhHACD2 had two haplotypes, with hap_0 occupying 86.67% in G0 and 72.22% in G1 (Figure 4D). With domestication progression, the proportion of hap_0 for GhHACD1, GhHACD3, and GhHACD4 increased to 80% or higher in G. hirsutum (G3, G4, G5, and G6), exhibiting only two haplotypes predominantly characterized by hap_0. GhHACD2 existed only in hap_0 in G3, G4, G5, and G6. These findings indicate that GhHACD have undergone significant purifying selection and share potential functional similarities.
To further understand the potential functions of the cotton HACD genes, we conducted a predictive analysis of cis-acting elements within the upstream 2,000 bp promoter sequence of the cotton HACD genes. As shown in Figure 5, the cotton HACD genes contain various cis-acting elements, mainly including MYBHv1 binding sites, growth and development response elements, hormone response elements, abiotic stress response elements, and light response elements. Hormone response elements include salicylic acid, jasmonic acid, gibberellins, auxins, and abscisic acid. Abiotic stress response-related elements include wound response, low temperature response, drought response, defense and stress response, and anaerobic induction. Growth and development response elements include diurnal rhythm control. The results suggest that the cotton HACD genes may play important roles in hormone and stress responses. It is noteworthy that the diurnal rhythm control element is exclusively present in GbHACD2, indicating that the GbHACD2 gene may be involved in plant growth and development.
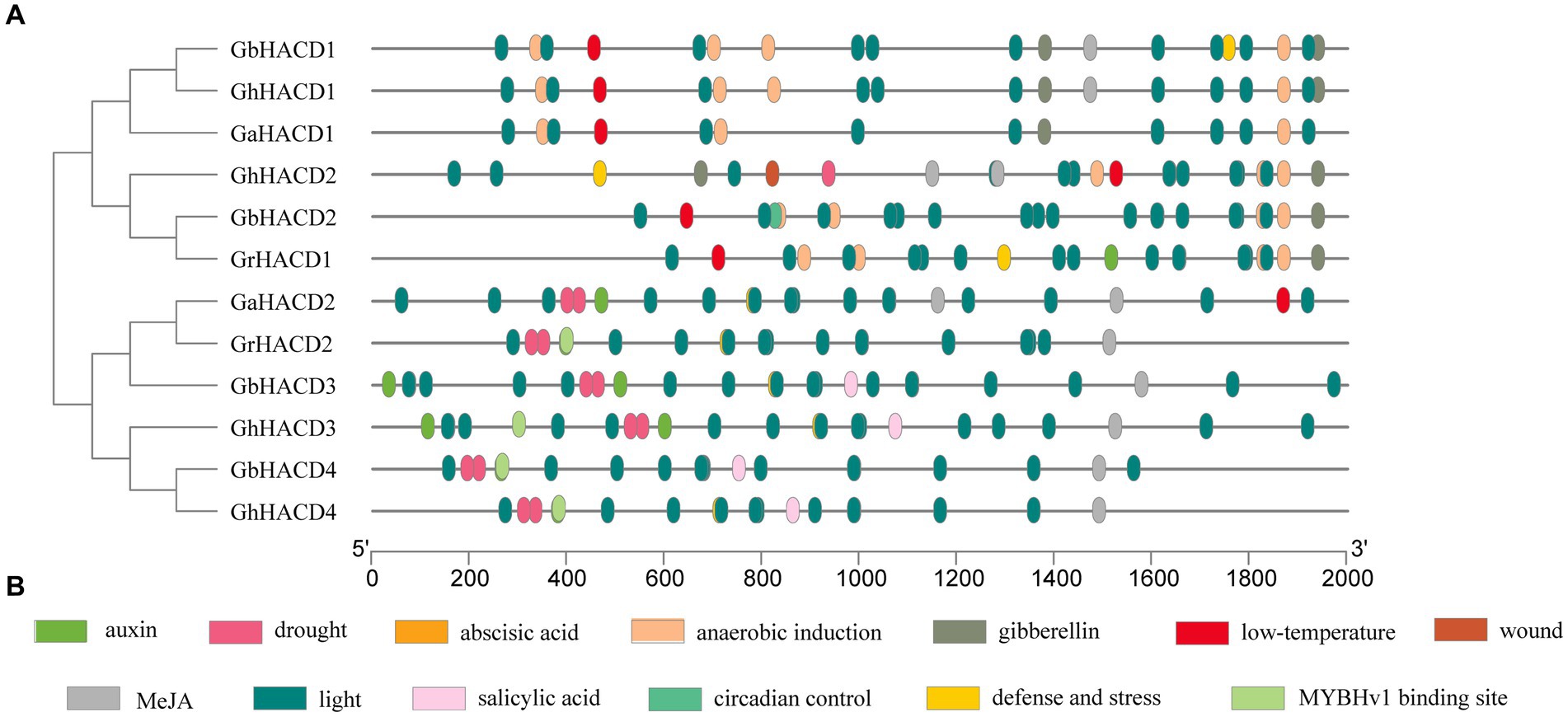
Figure 5. Promoter cis-element analysis of HACD gene family. (A) Positions of cis-acting elements on the HACD promoter sequences. (B) The rectangles of different colors represent distinct cis-acting elements. The black lines represent the length of the HACD promoters (in base pairs, bp).
To gain a deeper understanding of the expression patterns and potential biological roles of the HACD genes in G. hirsutum, we analyzed the expression patterns of GhHACD in various tissues using publicly available RNA-seq data. As shown in Figure 6A, 12 GhHACD genes are expressed in epicalys, leaf, root, sepal, stem, torus, ovule, and fiber of G. hirsutum, with lower expression levels in the petal and pistil. Compared to other tissues, GhHACD genes are expressed at higher levels during the development of cotton fiber and ovules, indicating a potential association with cotton fiber and ovule development.
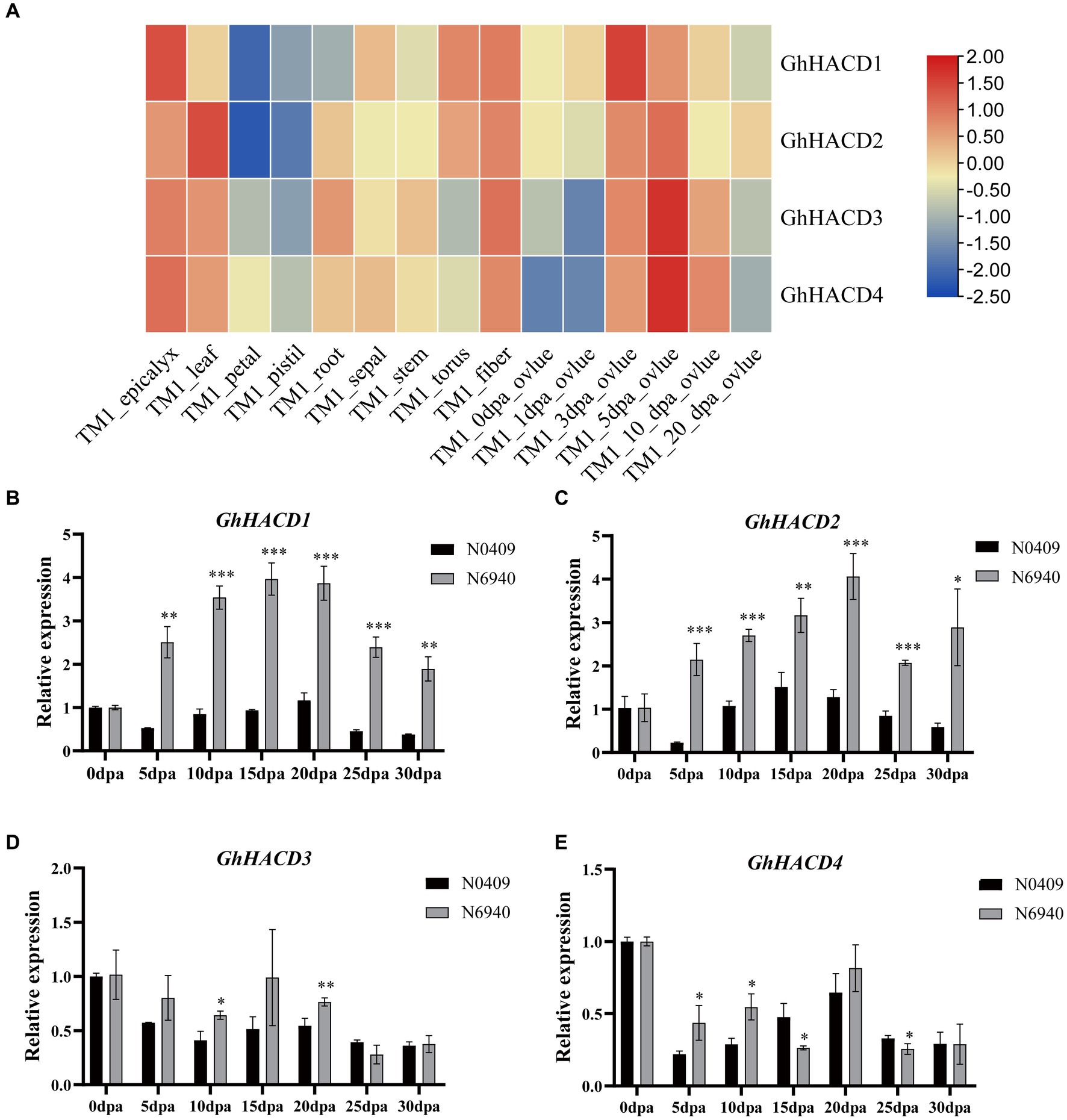
Figure 6. Expression profile of GhHACD. (A) Expression profile of GhHACD in different tissues. The heatmap demonstrates the expression level, the color gradient from blue to red presents increasing expression values. (B–E) Expression of GhHACD at 0 DPA, 5 DPA, 10 DPA, 15 DPA, 20 DPA, 25 DPA, and 30 DPA during cottonseed development of the two materials. N0409 represents the high-oil material and N6940 represents the low-oil material. The data were shown as mean ± standard error for three biological replicates.
Further analysis of GhHACD gene expression patterns during cotton seed development in high-oil genotype (N0409) and low-oil genotype (N6940) at 0, 5, 10, 15, 20, 25, and 30 DPA was conducted using qRT-PCR technology (Figure 6). As shown in the Figure 6, during embryonic development, the expression levels of GhHACD1 and GhHACD2 in the high-oil genotype were lower than those in the low-oil genotype, especially during the critical period of cottonseed oil accumulation at 20 and 25 DPA (Figures 6B,C). GhHACD3 and GhHACD4 genes showed significant differences in expression at 20 DPA, with low expression in N0409 and high expression in N6940 (Figures 6D,E). This research indicates that the GhHACD genes may play a role in negatively regulating the synthesis of cottonseed oil, particularly GhHACD1 and GhHACD2.
The significantly differentially expressed genes (GhHACD1 and GhHACD2) were further selected for analysis of co-expression networks and pathways. The co-expression network of GhHACD1 and GhHACD2 genes was analyzed using WGCNA with 49,661 genes in the transcriptome of the developing ovules from high-oil and low-oil genotype (Song et al., 2022). The analysis revealed that GhHACD1 and GhHACD2 were found in the MEturquoise module. To further elucidate the underlying mechanisms governing process of oil biosynthesis, the MEturquoise module was subjected to functional enrichment analyses through KEGG pathway annotations (Supplementary Figure S1). In the KEGG pathway enrichment analysis, 16,828 genes within the MEturquoise module were found to be associated with various metabolic pathways, with 251 genes specifically implicated in lipid metabolism. GhHACD1 and GhHACD2 were enriched in “Fatty acid elongation” metabolic pathways which including 10 genes. A total of three positively and six negatively co-expression with GhHACD1 and GhHACD2, respectively. Among which, including Protein-tyrosine phosphatase-like 2C PTPLA, 3-ketoacyl-CoA synthase, alpha/beta-Hydrolases superfamily protein, etc. (Supplementary Table S3). This analysis demonstrated that pathway enrichment analysis of co-expressed genes of GhHACD1 and GhHACD2 genes were conducted to confirm their potential involvement in fatty acid elongation and oil biosynthesis.
Based on the significant differences in expression patterns of GhHACD2 between high and low oil materials, as well as its conserved function during the domestication process of G. hirsutum, further research was conducted to investigate the expression site of GhHACD2 in plant cells. Transient expression of GhHACD2 was performed in N. benthamiana leaf epidermal cells. As shown in Figure 7, co-localization of the fusion expression construct 35S::GhHACD2::YFP with an endoplasmic reticulum marker was observed around the endoplasmic reticulum, indicating that GhHACD2 is located on the endoplasmic reticulum. This suggests that GhHACD2 might play a participatory role in the elongation mechanism of exceedingly long-chain fatty acids, an event that unfolds within the confines of the endoplasmic reticulum.
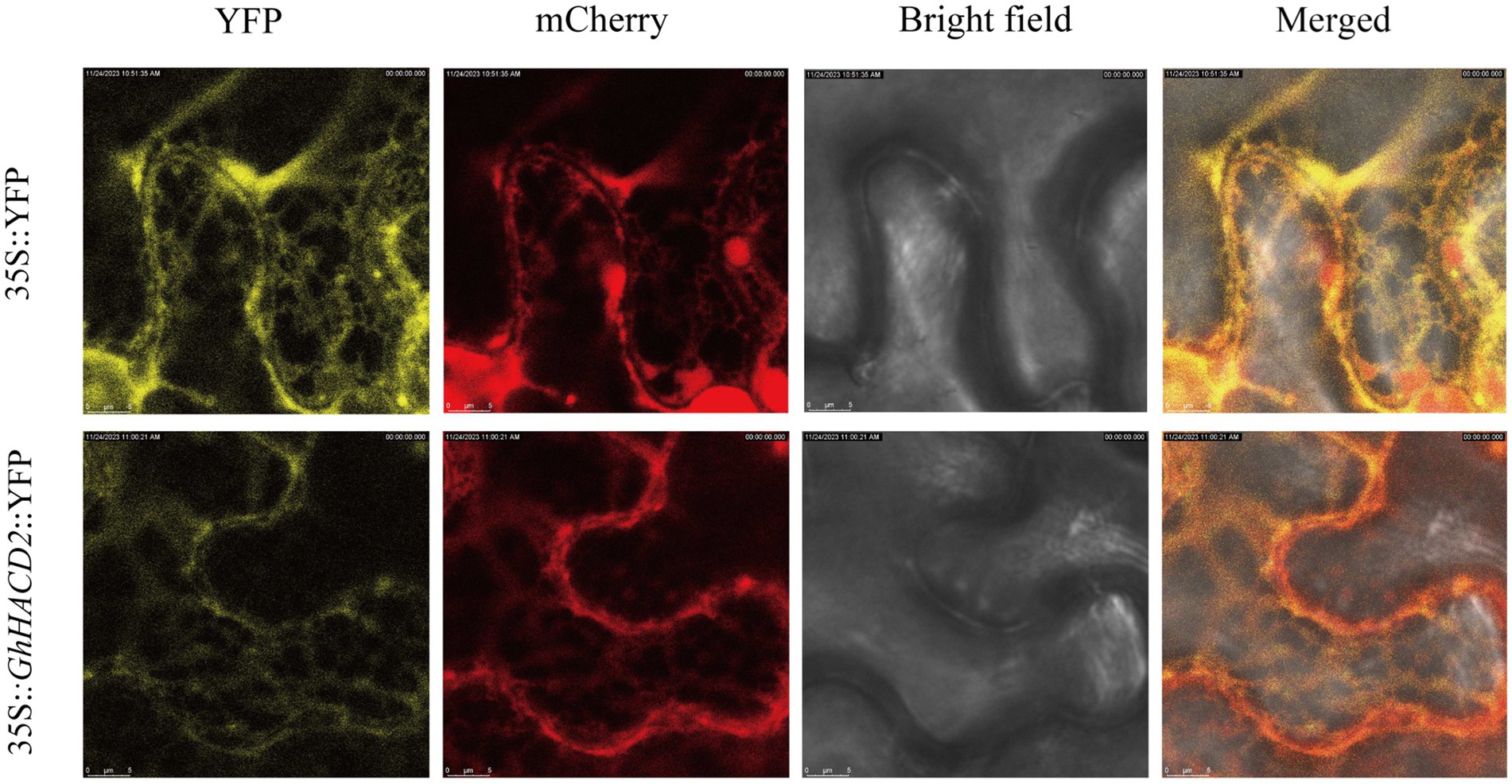
Figure 7. Subcellular localization analysis of GhHACD2. YFP empty vector was used as control. Scale bar = 5 μm.
To analyze the function of the protein encoded by GhHACD2, we heterologously overexpressed the GhHACD2 gene (pYES2-GhHACD2) in the S. cerevisiae strain INVScL1, using an empty vector (pYES2) as a control. The results, as shown in Figure 8, indicate that overexpression of GhHACD2 led to a significant decrease of 2.88 and 1.31% in C16:0 and C16:1, respectively, while C18:0, C18:1, and C18:2 increased significantly by 2.40, 2.06, and 11.77%, respectively, compared with the control. Long-chain fatty acids showed significant changes (Figure 8A). Seven VLCFAs were detected in yeast, including C20:0, C22:0, C22:2n6, C22:6n3, C24:0, C26:0, and C28:0 (Figure 8A). Overexpression of the GhHACD2 gene led to a 7.85 and 8.60% increase in the content of C26:0 and C28:0 in yeast, respectively, while C24:0 significantly decreased by 48.01%. Moreover, GhHACD2 overexpression increased the total VLCFAs in yeast by 3.88% (Figure 8B). Additionally, the VLCFA/LCFA ratio in GhHACD2 transgenic yeast significantly increased by 4.22% (Figure 8C). These results indicate that GhHACD2 promotes the synthesis of very long-chain fatty acids. Interestingly, overexpression of GhHACD led to a significant decrease of 5.02% in total oil content in yeast (Figure 8D).
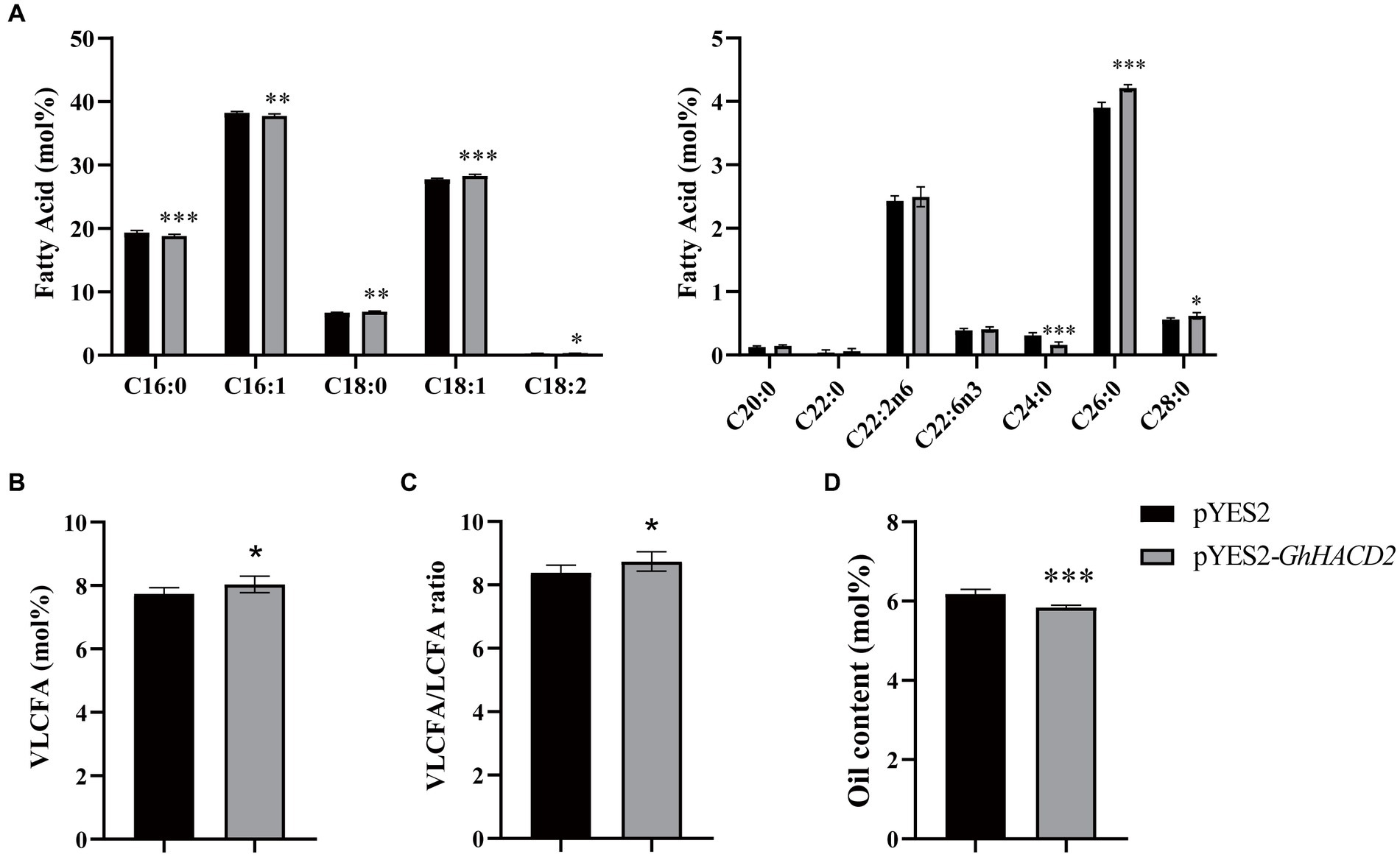
Figure 8. GC-2030 analysis of fatty acid components of yeast transformed with GhHACD2. (A) Individual fatty acid levels in GhHACD2-expressing yeast plants compared with the empty vector (pYES2), presented in the form relative mole percent. (B) Total VLCFA levels in GhHACD2-expressing yeast plants compared with the empty vector. (C) VLCFA/LCFA ratio in yeast transformed with empty vector and GhHACD2 genes. (D) Oil content (%) in yeast transformed with empty vector and GhHACD2 genes. Error bars represent the SD of three biological replicates (n ≥ 3). Significant differences were determined using the student’s t-test: *p < 0.05, **p < 0.01, ***p < 0.001.
Cotton is an important cash crop, serving as an excellent fiber crop as well as a significant source of plant oil and biofuels (Yang et al., 2023a). The demand for its seed oil can significantly increase with better seed oil quality contents, especially for human consumption. For high oil quality and contents, VLCFAs have higher importance in oilseed crops. VLCFAs play a crucial role in the biosynthesis of lipids such as phospholipids, sphingolipids, cuticular wax, suberin, and triglycerides. Therefore, VLCFAs are essential throughout the entire life process of plants, including cell membrane formation, protection against stress, and oil storage in seeds (Kim et al., 2023). The biosynthesis of VLCFAs is controlled by FAE complexes composed of four enzymes named KCS, KCR, HACD, and ECR. KCS has garnered considerable attention because it act as the rate-limiting enzyme for VLCFA synthesis (Huai et al., 2020). However, the functional studies of HACD genes in plants remain limited. So far, only two HACD genes have been identified in Arabidopsis thaliana, both confirmed to have dehydratase activity (Bach et al., 2008; Morineau et al., 2016). In G. hirsutum, GhPAS2 has been proven to encode 3-hydroxyacyl-CoA dehydratase, but its regulation of fatty acids remains unclear (Wang et al., 2015).
This study identified two HACD family members in diploid cotton species G. arboretum and G. raimondiiand four HACD family members in tetraploid cotton species G. hirsutum and G. barbadense through whole-genome screening. Studies have found that HACD genes are single genes in tomato and maize, with six genes in rice and three genes in sorghum (Campbell et al., 2019). Compared to other gene families, the HACD gene family is relatively smaller in size. Phylogenetic tree clustering revealed that these genes are divided into two branches, group A and B. Furthermore, several HACD proteins from the same species tend to cluster together, which may be a result of segmental duplication events of HACD genes in their genomes and also indicates the relative evolutionary independence of HACD genes in different species.
The tetraploid cotton species, G. hirsutum and G. barbadense belong to the AD genome and are allopolyploids that originated from hybridization between the A genome (G. arboreum) and the D genome (G. raimondii) millions of years ago (Hu et al., 2019). Cluster analysis reveals that HACD genes in two tetraploid cotton species, G. hirsutum and G. barbadense, clustered together in the same branch. This may stem from the relatively recent divergence time of HACD genes in G. hirsutum and G. barbadense and the conservation of HACD gene functions.
All members of the HACD gene family contain protein motif 1, 2, 3, 5, 6, 8, and 9, as well as the conserved PTPLA domain. In Arabidopsis thaliana, AtPTPLA has been shown to possess dehydratase activity (Morineau et al., 2016), suggesting that HACD genes may be involved in the third step of very long-chain fatty acid synthesis. The gene structures diversity plays a crucial role in gene family evolution, providing further support for phylogenetic clustering and aiding in protein function prediction (Bano et al., 2021b). The conserved motifs and gene structures of the cotton HACD members are preserved in most members, with some exceptions. Cotton HACD family members within the same subgroup have more similar gene structures and conserved motifs. For example, motif 10 is only present in the B subfamily, which may account for the differences between the two subfamilies. Overall, HACD genes exhibit high conservation during cotton evolution, but members within the same subfamily show closer conservation in protein regions and functions. In the previous study, the genomes of more advanced species contained fewer introns (Bano et al., 2021a). GhHACD4 contains a smaller number of introns (seven) and this gene has lost one of their introns during evolution, suggesting that intrafamilial specific genes may have specific functions (Bano et al., 2023). Furthermore, in the analysis of the differences between G. hirsutum and G. barbadense, we observed minimal differences in collinear genes. However, within different species, each has its own unique haplotypes that have been selectively retained. This may be attributed to the differential selection pressures faced by different cotton species. It also implies that the function of this gene is highly conserved and crucial for G. hirsutum.
Gene duplication plays a critical role in the duplication and expansion of genes. In the polyploidization process of cotton, genome expansion is mainly achieved through whole-genome duplication (WGD), tandem duplication, and segmental duplication (Shiraku et al., 2021). The HACD gene family in cotton, both in G. hirsutum and G. barbadense, underwent segmental duplication without tandem duplication. During the evolution of the cotton HACD gene family from diploid to tetraploid, they exhibit collinearity, indicating that segmental/WGD duplication may be the primary driving force for the evolution of the cotton HACD gene family. The Ka/Ks ratios of four HACD gene pairs of segmental duplications were all below 0.5. This further reveals the high conservation of the cotton HACD gene family members in evolution.
The expression patterns of genes can provide valuable clues for further exploring gene function (Zhu et al., 2021). In this study, using publicly available RNA-seq data (Hu et al., 2019), it was observed that the GhHACD gene is expressed in various tissues and organs during the development of G. hirsutum. Particularly, it shows higher expression during ovule development, indicating the potentially important role of GhHACD in cotton seed development. To further understand the role of GhHACD in cotton seed oil development, we analyzed the expression patterns of GhHACD during seed development in high-oil (N0409) and low-oil (N6940) genotypes. The results showed that the relative expression level of GhHACD is higher in low-oil genotype compared with high-oil genotype. Additionally, the expression levels of GhHACD1 and GhHACD2 genes significantly decreased during the key period of rapid oil accumulation in seed development at 25 and 30 DPA, compared to the early stages of 0–20 DPA, suggesting a negative correlation between the expression levels of the GhHACD1 and GhHACD2 and cotton seed oil content. To further elucidate the roles of GhHACD1 and GhHACD2 genes, their co-expression network was investigated. GhHACD1 and GhHACD2 were identified in the MEturgugise module by WGCNA analysis, and GhHACD1 and GhHACD2 were enriched in the fatty acid elongation pathway, further indicating their potential involvement in fatty acid elongation.
The synthesis of fatty acids is a complex process. First, they are synthesized de novo in the plastids and then elongated by the FAE complex on the endoplasmic reticulum to form VLCFA (Bates and Browse, 2012). In Arabidopsis thaliana, PTPLA interacts with several elongase subunits on the endoplasmic reticulum (Morineau et al., 2016). In order to confirm the subcellular localization of GhHACD2 and its functional site, this study conducted subcellular localization analysis, and the results revealed that GhHACD2 is localized in the endoplasmic reticulum, consistent with previous studies (Morineau et al., 2016).
Many studies have shown that the FAE complex regulates the production of VLCFAs. For example, the fe1 mutant of B. napus leads to a significant decrease in VLCFA content in rapeseed (Wang et al., 2010). Peanut KCS1 and KCS28 can regulate fatty acid elongation and promote the accumulation of VLCFAs in seeds, especially saturated VLCFAs, when overexpressed heterologously in Arabidopsis (Huai et al., 2020). The second member of the FAE complex, KCR, has been found in Arabidopsis to be involved in VLCFA elongation in the ER, and the inhibition of KCR1 activity leads to a decrease in total C20:0, C22:0, and VLCFA content in seeds (Beaudoin et al., 2009). Through heterologous expression of GhHACD2 in yeast, we observed an increase in VLCFA content, especially a significant increase in C26:0 and C28:0, confirming that GhHACD2 can regulate VLCFA synthesis. Partial loss of function of the HACD gene in Arabidopsis leads to changes in VLCFA content, which is reduced in the pas2 mutant and accumulates after overexpression of Arabidopsis with PTPLA (Bach et al., 2008; Morineau et al., 2016). Previous studies have shown that the elongase complex involved in limiting VLCFA synthesis only includes the KCS gene (Millar and Kunst, 1997; Huai et al., 2015). However, our findings demonstrate that the HACD gene (dehydratase) also limits VLCFA synthesis, consistent with earlier research (Bach et al., 2008).
Currently, research on VLCFAs in cotton primarily focuses on fiber development. For example, some plant hormones, such as brassinosteroid (BR) and strigolactones (SLs), regulate the cotton fiber elongation rate by regulating VLCFA biosynthesis (Tian et al., 2022; Yang et al., 2023b). However, this study revealed for the first time that overexpression of the GhHACD2 gene within yeast not only catalyzes the biosynthesis of VLCFAs but also impedes the overall synthesis of oil.
In this study, we identified 12 HACD genes from four species of cotton, classified them into two groups based on phylogenetic relationships, and discovered that their functions are substantially conserved in cotton. The cotton HACD gene family was analyzed in terms of phylogeny, chromosome distribution, gene structure, evolutionary pattern. Their gene expression patterns were confirmed during the oil formation stage in two contrasting cottonseed oil-producing geneotypes of cotton. Expression profile analysis indicated that differentially expressed GhHACD genes in high-oil and low-oil genotypemay negatively regulate cottonseed oil accumulation. Current research reveals that GhHACD2, of very long-chain carbon based fatty acids within yeast while concurrently impeding lipid deposition. These findings deepen our understanding of the role of the HACD family in VLCFA synthesis, providing insights into fatty acid synthesis-related genes and laying the platform for studying the regulatory network of cottonseed oil.
The original contributions presented in the study are included in the article/Supplementary material. Further inquiries can be directed to the corresponding authors.
MY: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft. HX: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft. ShiH: Conceptualization, Data curation, Investigation, Methodology, Visualization, Writing – original draft. JS: Conceptualization, Data curation, Methodology, Software, Writing – review & editing. BJ: Conceptualization, Data curation, Methodology, Software, Writing – review & editing. PF: Conceptualization, Data curation, Methodology, Software, Writing – review & editing. LY: Conceptualization, Data curation, Methodology, Software, Writing – review & editing. JM: Conceptualization, Data curation, Methodology, Supervision, Writing – review & editing. LW: Conceptualization, Data curation, Methodology, Supervision, Writing – review & editing. WP: Conceptualization, Data curation, Methodology, Supervision, Writing – review & editing. BZ: Data curation, Methodology, Supervision, Writing – review & editing. JY: Funding acquisition, Resources, Supervision, Writing – review & editing. MW: Funding acquisition, Resources, Supervision, Writing – review & editing. ShoH: Funding acquisition, Resources, Supervision, Writing – review & editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This study was funded by the National key R&D Program of China (2023YFD2301201 and 2023YFD2301200), the Scientific Research Project of Anyang city of China (2023C01NY003), the Scientific Research Project of Henan Province of China (222102110209), and the National Key Research and Development Program of China (2022YFD1200300 and 2022YFF1001400).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1413724/full#supplementary-material
SUPPLEMENTARY FIGURE S1 | KEGG pathway enrichment analysis of the genes in MEturgugise modules.
1. ^https://www.ncbi.nlm.nih.gov/cdd/
2. ^http://smart.embl-heidelberG.de/
4. ^https://meme-suite.org/meme/tools/meme/
5. ^http://yanglab.hzau.edu.cn/CottonMD/
6. ^http://bioinformatics.psb.ugent.be/webtools/plantcare/html/
Bach, L., Michaelson, L. V., Haslam, R., Bellec, Y., Gissot, L., Marion, J., et al. (2008). The very-long-chain hydroxy fatty acyl-CoA dehydratase PASTICCINO2 is essential and limiting for plant development. Proc. Natl. Acad. Sci. USA 105, 14727–14731. doi: 10.1073/pnas.0805089105
Bano, N., Fakhrah, S., Lone, R. A., Mohanty, C. S., and Bag, S. K. (2023). Genome-wide identification and expression analysis of the HD2 protein family and its response to drought and salt stress in Gossypium species. Front. Plant Sci. 14:1109031. doi: 10.3389/fpls.2023.1109031
Bano, N., Fakhrah, S., Mohanty, C. S., and Bag, S. K. (2021a). Genome-wide identification and evolutionary analysis of Gossypium tubby-like protein (TLP) gene family and expression analyses during salt and drought stress. Front. Plant Sci. 12:667929. doi: 10.3389/fpls.2021.667929
Bano, N., Patel, P., Chakrabarty, D., and Bag, S. K. (2021b). Genome-wide identification, phylogeny, and expression analysis of the bHLH gene family in tobacco (Nicotiana tabacum). Physiol. Mol. Biol. Plants 27, 1747–1764. doi: 10.1007/s12298-021-01042-x
Bates, P. D., and Browse, J. (2012). The significance of different diacylgycerol synthesis pathways on plant oil composition and bioengineering. Front. Plant Sci. 3:147. doi: 10.3389/fpls.2012.00147
Batsale, M., Bahammou, D., Fouillen, L., Mongrand, S., Joubès, J., and Domergue, F. (2021). Biosynthesis and functions of very-Long-chain fatty acids in the responses of plants to abiotic and biotic stresses. Cells 10:1284. doi: 10.3390/cells10061284
Beaudoin, F., Wu, X., Li, F., Haslam, R. P., Markham, J. E., Zheng, H., et al. (2009). Functional characterization of the Arabidopsis beta-ketoacyl-coenzyme a reductase candidates of the fatty acid elongase. Plant Physiol. 150, 1174–1191. doi: 10.1104/pp.109.137497
Campbell, A. A., Stenback, K. E., Flyckt, K., Hoang, T., Perera, M. A. D., and Nikolau, B. J. (2019). A single-cell platform for reconstituting and characterizing fatty acid elongase component enzymes. PLoS One 14:e0213620. doi: 10.1371/journal.pone.0213620
Chattha, W. S., Atif, R. M., Iqbal, M., Shafqat, W., Farooq, M. A., and Shakeel, A. (2020). Genome-wide identification and evolution of Dof transcription factor family in cultivated and ancestral cotton species. Genomics 112, 4155–4170. doi: 10.1016/j.ygeno.2020.07.006
Chen, C., Chen, H., Zhang, Y., Thomas, H. R., Frank, M. H., He, Y., et al. (2020). TBtools: an integrative toolkit developed for interactive analyses of big biological data. Mol. Plant 13, 1194–1202. doi: 10.1016/j.molp.2020.06.009
Denic, V., and Weissman, J. S. (2007). A molecular caliper mechanism for determining very long-chain fatty acid length. Cell 130, 663–677. doi: 10.1016/j.cell.2007.06.031
Devaiah, S. P., Roth, M. R., Baughman, E., Li, M., Tamura, P., Jeannotte, R., et al. (2006). Quantitative profiling of polar glycerolipid species from organs of wild-type Arabidopsis and a phospholipase Dalpha1 knockout mutant. Phytochemistry 67, 1907–1924. doi: 10.1016/j.phytochem.2006.06.005
Hall, B. G. (2013). Building phylogenetic trees from molecular data with MEGA. Mol. Biol. Evol. 30, 1229–1235. doi: 10.1093/molbev/mst012
Haslam, T. M., and Kunst, L. (2013). Extending the story of very-long-chain fatty acid elongation. Plant Sci. 210, 93–107. doi: 10.1016/j.plantsci.2013.05.008
Hu, Y., Chen, J., Fang, L., Zhang, Z., Ma, W., Niu, Y., et al. (2019). Gossypium barbadense and Gossypium hirsutum genomes provide insights into the origin and evolution of allotetraploid cotton. Nat. Genet. 51, 739–748. doi: 10.1038/s41588-019-0371-5
Huai, D., Xue, X., Li, Y., Wang, P., Li, J., Yan, L., et al. (2020). Genome-wide identification of Peanut KCS genes reveals that AhKCS1 and AhKCS28 are involved in regulating VLCFA contents in seeds. Front. Plant Sci. 11:406. doi: 10.3389/fpls.2020.00406
Huai, D., Zhang, Y., Zhang, C., Cahoon, E. B., and Zhou, Y. (2015). Combinatorial effects of fatty acid Elongase enzymes on Nervonic acid production in Camelina sativa. PLoS One 10:e0131755. doi: 10.1371/journal.pone.0131755
Kim, R. J., Han, S., Kim, H. J., Hur, J. H., and Suh, M. C. (2023). Arabidopsis 3-ketoacyl-CoA synthase 17 produces tetracosanoic acids required for synthesizing seed coat suberin. J. Exp. Bot. 75, 1767–1780. doi: 10.1093/jxb/erad381
Konukan, D. B., Yilmaztekin, M., Mert, M., Gener, O., and Dergisi, K. J. T. B. (2017). Tarm Bilimleri Dergisi Physico-chemical characteristic and fatty acids compositions of cottonseed oils. J. Agric. Sci. 23, 253–259.
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Kunst, L., Taylor, D. C., and Underhill, E. W. (1992). Fatty-acid elongation in developing seeds of Arabidopsis Thaliana. Plant Physiol. Biochem. 30, 425–434.
Leonard, A. E., Pereira, S. L., Sprecher, H., and Huang, Y. S. (2004). Elongation of long-chain fatty acids. Prog. Lipid Res. 43, 36–54. doi: 10.1016/s0163-7827(03)00040-7
Letunic, I., Khedkar, S., and Bork, P. (2021). SMART: recent updates, new developments and status in 2020. Nucleic Acids Res. 49, D458–D460. doi: 10.1093/nar/gkaa937
Ma, J., Liu, J., Pei, W., Ma, Q., Wang, N., Zhang, X., et al. (2019). Genome-wide association study of the oil content in upland cotton (Gossypium hirsutum L.) and identification of GhPRXR1, a candidate gene for a stable QTLqOC-Dt5-1. Plant Sci. 286, 89–97. doi: 10.1016/j.plantsci.2019.05.019
Millar, A. A., and Kunst, L. (1997). Very-long-chain fatty acid biosynthesis is controlled through the expression and specificity of the condensing enzyme. Plant J. 12, 121–131. doi: 10.1046/j.1365-313x.1997.12010121.x
Morineau, C., Gissot, L., Bellec, Y., Hematy, K., Tellier, F., Renne, C., et al. (2016). Dual fatty acid Elongase complex interactions in Arabidopsis. PLoS One 11:e0160631. doi: 10.1371/journal.pone.0160631
Napier, J. A., Haslam, R. P., Beaudoin, F., and Cahoon, E. B. (2014). Understanding and manipulating plant lipid composition: metabolic engineering leads the way. Curr. Opin. Plant Biol. 19, 68–75. doi: 10.1016/j.pbi.2014.04.001
Shafqat, W., Jaskani, M. J., Maqbool, R., Chattha, W. S., Ali, Z., Naqvi, S. A., et al. (2021). Heat shock protein and aquaporin expression enhance water conserving behavior of citrus under water deficits and high temperature conditions. Environ. Exp. Bot. 181:104270. doi: 10.1016/j.envexpbot.2020.104270
Shiraku, M. L., Magwanga, R. O., Cai, X., Kirungu, J. N., Xu, Y., Mehari, T. G., et al. (2021). Functional characterization of GhACX3 gene reveals its significant role in enhancing drought and salt stress tolerance in cotton. Front. Plant Sci. 12:658755. doi: 10.3389/fpls.2021.658755
Song, J., Pei, W., Wang, N., Ma, J., Xin, Y., Yang, S., et al. (2022). Transcriptome analysis and identification of genes associated with oil accumulation in upland cotton. Physiol. Plant. 174:e13701. doi: 10.1111/ppl.13701
Suh, M. C., Samuels, A. L., Jetter, R., Kunst, L., Pollard, M., Ohlrogge, J., et al. (2005). Cuticular lipid composition, surface structure, and gene expression in Arabidopsis stem epidermis. Plant Physiol. 139, 1649–1665. doi: 10.1104/pp.105.070805
Tian, Z., Zhang, Y., Zhu, L., Jiang, B., Wang, H., Gao, R., et al. (2022). Strigolactones act downstream of gibberellins to regulate fiber cell elongation and cell wall thickness in cotton (Gossypium hirsutum). Plant Cell 34, 4816–4839. doi: 10.1093/plcell/koac270
Wang, X. C., Li, Q., Jin, X., Xiao, G. H., Liu, G. J., Liu, N. J., et al. (2015). Quantitative proteomics and transcriptomics reveal key metabolic processes associated with cotton fiber initiation. J. Proteome 114, 16–27. doi: 10.1016/j.jprot.2014.10.022
Wang, N., Shi, L., Tian, F., Ning, H., Wu, X., Long, Y., et al. (2010). Assessment of FAE1 polymorphisms in three Brassica species using EcoTILLING and their association with differences in seed erucic acid contents. BMC Plant Biol. 10:137. doi: 10.1186/1471-2229-10-137
Wang, Y., Tang, H., Debarry, J. D., Tan, X., Li, J., Wang, X., et al. (2012). MCScanX: a toolkit for detection and evolutionary analysis of gene synteny and collinearity. Nucleic Acids Res. 40:e49. doi: 10.1093/nar/gkr1293
Wang, J., Wang, X., Wang, L., Nazir, M. F., Fu, G., Peng, Z., et al. (2024). Exploring the regulatory role of non-coding RNAs in fiber development and direct regulation of GhKCR2 in the fatty acid metabolic pathway in upland cotton. Int. J. Biol. Macromol. 266:131345. doi: 10.1016/j.ijbiomac.2024.131345
Wu, J., McCarty, J. C., and Jenkins, J. N. (2010). Cotton chromosome substitution lines crossed with cultivars: genetic model evaluation and seed trait analyses. Theor. Appl. Genet. 120, 1473–1483. doi: 10.1007/s00122-010-1269-x
Wu, M., Pei, W., Wedegaertner, T., Zhang, J., and Yu, J. (2022). Genetics, breeding and genetic engineering to improve cottonseed oil and protein: a review. Front. Plant Sci. 13:864850. doi: 10.3389/fpls.2022.864850
Xin, Y., Ma, J., Song, J., Jia, B., Yang, S., Wu, L., et al. (2022). Genome wide association study identiffes candidate genes related to fatty acid components in upland cotton (Gossypium hirsutum L.). Ind. Crop. Prod. 183:114999. doi: 10.1016/j.indcrop.2022.114999
Yang, Z., Gao, C., Zhang, Y., Yan, Q., Hu, W., Yang, L., et al. (2023a). Recent progression and future perspectives in cotton genomic breeding. J. Integr. Plant Biol. 65, 548–569. doi: 10.1111/jipb.13388
Yang, Z., Liu, Z., Ge, X., Lu, L., Qin, W., Qanmber, G., et al. (2023b). Brassinosteroids regulate cotton fiber elongation by modulating very-long-chain fatty acid biosynthesis. Plant Cell 35, 2114–2131. doi: 10.1093/plcell/koad060
Yang, Z., Wang, J., Huang, Y., Wang, S., Wei, L., Liu, D., et al. (2023c). CottonMD: a multi-omics database for cotton biological study. Nucleic Acids Res. 51, D1446–D1456. doi: 10.1093/nar/gkac863
Zhao, Y. P., Wu, N., Li, W. J., Shen, J. L., Chen, C., Li, F. G., et al. (2021). Evolution and characterization of acetyl coenzyme a: Diacylglycerol acyltransferase genes in cotton identify the roles of GhDGAT3D in oil biosynthesis and fatty acid composition. Genes (Basel) 12:1045. doi: 10.3390/genes12071045
Zhu, Y., Wang, Q., Gao, Z., Wang, Y., Liu, Y., Ma, Z., et al. (2021). Analysis of Phytohormone signal transduction in Sophora alopecuroides under salt stress. Int. J. Mol. Sci. 22:7313. doi: 10.3390/ijms22147313
Zhukov, A., and Popov, V. (2022). Synthesis of C(20-38) fatty acids in plant tissues. Int. J. Mol. Sci. 23:4731. doi: 10.3390/ijms23094731
Keywords: VLCFA, 3-hydroxyacyl-CoA dehydratase, phylogenetic analysis, co-expression, pathway, qRT-PCR
Citation: Yan M, Xi H, Hu S, Song J, Jia B, Feng P, Yang L, Ma J, Wang L, Pei W, Zhang B, Yu J, Wu M and Hu S (2024) Genome-wide analysis of HACD family genes and functional characterization of GhHACD2 for very long chain fatty acids biosynthesis in Gossypium hirsutum. Front. Sustain. Food Syst. 8:1413724. doi: 10.3389/fsufs.2024.1413724
Received: 07 April 2024; Accepted: 28 June 2024;
Published: 30 July 2024.
Edited by:
Pushp Sheel Shukla, Center for Cellular and Molecular Platforms, IndiaReviewed by:
Nasreen Bano, University of Pennsylvania, United StatesCopyright © 2024 Yan, Xi, Hu, Song, Jia, Feng, Yang, Ma, Wang, Pei, Zhang, Yu, Wu and Hu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiwen Yu, eXVqdzY2NkBob3RtYWlsLmNvbQ==; Man Wu, d3VtYW4yMDA0QDE2My5jb20=; Shoulin Hu, aHVzaG91bGluZ2h1QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.