- 1Department of Food Science and Technology, Virginia Tech, Blacksburg, VA, United States
- 2Department of Food Science and Technology, Texas A&M University, College Station, TX, United States
- 3Department of Biomedical Engineering, Tufts University, Medford, MA, United States
- 4Department of Food Science and Technology, University of California, Davis, Davis, CA, United States
- 5Department of Biological and Agricultural Engineering, University of California, Davis, Davis, CA, United States
Cellular agriculture holds the potential to address sustainability, food security, and agricultural resilience. Within the cell-based meat supply chain, one of the key steps is developing sustainable scaffolding. In this study, we evaluated the impact of decellularized banana leaves, various coating materials including soy protein and gelatine, and different cell seeding strategies on cell viability, cell growth, cell alignment, and the response of the materials to thermal processing. Kinetics of the quality degradation of the scaffolds with and without cells were determined through kinetics equations. The efficiency of decellularization was verified through DNA quantification, which decreased from 445 ng/mg in fresh banana leaves to non-detectable levels in the decellularized samples. The alignment of cells on gelatin-coated samples was the highest among the samples, with a dominant orientation of 65.8°, compared to soy-coated and uncoated samples, with dominant orientations of 9.2° and −6.3°, respectively. The kinetics of shrinkage indicated that coating with soy and the presence of cells increased the activation energy due to the higher energy required for protein denaturation. The kinetics of area changes in plain scaffolds without cells followed a first-order pattern, while with seeded cells a second-order pattern was followed. Overall, the results showed that decellularized banana leaves provide sustainable scaffoldings for cellular agriculture applications. In addition, soy coating provided many benefits for decellularized samples by supporting cell adhesion and cell proliferation.
1 Introduction
Projections indicate that the world's population is expected to reach 10 billion people by the year 2050 from the current level of eight billion amplifying pressure on existing agricultural systems (Röös et al., 2017; Eibl et al., 2021). While this necessitates a corresponding increase of food and agricultural commodities, there are sustainability concerns over food security due to highly limited arable land and the looming challenges posed by climate change. In order to attain this objective, it is imperative to make significant advancements in agricultural production within the foreseeable future (Rischer et al., 2020). One potential alternative to current food production norms involves the adoption of cellular agriculture, which presents a more sustainable and ecologically conscious approach to long-term agricultural production.
Cellular agriculture offers numerous advantages for sustainable and ethical food production. The proposed approaches reduce environmental impact when scaled up, primarily by requiring fewer resources and emitting fewer greenhouse gases (Soice and Johnston, 2021), reduce foodborne pathogens risks and improve animal welfare (McNamara and Bomkamp, 2022).
A fundamental feature in cultivated meat research and production is the development of biomaterials used for scaffolding (Xiang et al., 2022). This feature involves seeding cells onto or into a biomaterial tissue-like platform known as a scaffold, which plays a crucial role in supporting the growth, differentiation, and proliferation of cell lines relevant to skeletal muscle formation (Bomkamp et al., 2022). There are two general approaches for scaffolding in tissue engineering, bottom-up and top-down. In the first approach, the scaffold is fabricated from the bottom-up using natural biomaterials or synthetic polymers, and the top-down approach uses modifications of preexisting or naturally occurring structures, such as plant tissues modified to increase porosity, to generate the scaffolds (Nichol and Khademhosseini, 2009). Plant cellular materials can be chemically removed from whole plant tissue, the remaining extracellular matrix (ECM) can be isolated without excessive damage to the structure and biochemical characteristics. Decellularized tissue from common plants can be considered a potential option for cultivated meat scaffolding, since these are edible, safe to consume, and biocompatible, while also offering important attributes such as texture and porosity (Jones et al., 2023). To allow animal cells to attach, proliferate, and ultimately differentiate into muscle fibers, it is critical to identify plant matrices with 50–200 μm anisotropically aligned pore spaces (Duffy et al., 2016). Our initial experiments revealed that, among the nine plant sources tested, banana leaves emerged as the optimal choice for generating a decellularized scaffold. The banana leaf scaffold exhibited favorable mechanical properties, maintaining structural integrity in media, and possessing an ideal pore size conducive to cell adhesion.
Using chemical solutions such as acids and bases or ionic solutions, and detergents are efficient strategies for decellularization. Choosing a chemical that successfully removes cells from the tissue with minimal damage to the ECM is a crucial step in the decellularization process. Since the final products will be edible, it is essential for the solutions used to be generally recognized as safe (GRAS) and regulated for use in the food industry. Sodium dodecyl sulfate (SDS) and Polysorbate-20 that are used in the present study are examples of detergents that are regulated by the US Food and Drug administration (FDA) and are employed in different steps in food processing (Jones et al., 2023).
In previous studies, the efficiency of using decellularized spinach leaves as scaffolds for growing bovine satellite cells and producing cultured meat was assessed (Jones et al., 2021). Cheng et al. (2020) assessed a range of decellularized fruit and vegetables for scaffolding, such as carrot, celery, broccoli, leek, cucumber, potato, apple, asparagus, and green onion. The results of quantitative analysis of C2C12 and human skeletal muscle cells alignment and differentiation on these structures revealed that decellularized green onion scaffold was a cost-effective substrate for cultivated meat production. In another study, Perreault et al. (2023) investigated agricultural byproducts, such as corn husks and jackfruit rinds for scaffolds via decellularization, and suggested using these wastes as sustainable substrate sources. However, identifying a sustainable anisotropic decellularized structure and optimizing the cell seeding process in a low cost, scalable method has not yet been explored for a cultivated meat application. In addition to the raw materials selection for these decellularized scaffolds, the process of cell seeding and the downstream development of food products, including changes that occur during cooking (Ovissipour et al., 2017), should also be considered.
The primary objective of the present study was to assess the suitability of decellularized banana leaves as scaffolds for cultivated meat production. Various seeding techniques and coating materials were explored to optimize the process. Additionally, the quality of the materials was assessed using standard protocols after subjecting the products to pasteurization time and temperature schedules. The findings from this study will offer comprehensive insights, as well as widely applicable techniques, into the utilization of decellularized banana leaves as an environmentally sustainable scaffold to produce cell-based meat.
2 Materials and methods
2.1 Decellularization process
Fresh banana leaves (Musa spp.) were obtained from the local grocery store, cleaned to remove dirt and debris, and disinfected using 70% ethanol across the surface of each sample. Banana leaves were sampled at a central point between the midrib and edge of each leaf using a biopsy punch. Two punches with two sizes including 6 and 8 mm were used. For DNA and imaging, a 6 mm punch, and for thermal processing, an 8 mm punch were applied. Surface area exposure to decellularizing solutions was increased via an additional 1 mm biopsy punch in the center of each disk. Samples were immediately placed in a 1% sodium dodecyl sulfate (SDS; Thermo Scientific) solution and agitated in an orbital shaker at 100 RPM at ambient temperature for 3 days. This was followed by three washing steps with deionized water, allowing 5 min of shaking between washes. Disks were submerged in a solution of 10% bleach (Chlorox, 7.5% Sodium hypochlorite) and 1% Tween 20 (Thermo Fisher), which was used as a detergent to remove remaining cellular material from the cell wall matrix of the plant tissue. The samples were then returned to the orbital shaker and agitated for 24–48 h, until the samples were colorless, as an indicator of complete cell removal. Samples were then washed with deionized water, dried, and lyophilized before storage at −80°C until characterization and use.
2.2 Scaffold preparation
Decellularized scaffolds were thawed, sterilized in 70% ethanol solution for 1 h, and then washed vigorously in phosphate-buffered saline (PBS). Scaffolds in each coating group (0.1% porcine gelatine, and 0.1% soy protein isolate) were suspended in the coating solution for an additional 2 h, while the uncoated (control) scaffolds were suspended in growth media until cell seeding.
2.3 Cell culture
Zebrafish fibroblast stem cells (ZEM2S, CRL-2147™) were acquired from the American Type Cell Culture (ATCC), thawed, and maintained to passage 5–10 before use in scaffold seeding. The thawing method consisted of allowing the ampoules to reach 28°C, at which point the cells were resuspended in growth media and centrifuged at 500 g for 7 min. The supernatant was removed and the cells were immediately resuspended in growth media. Cells were seeded at 4,000 cells/cm2 in T-75 flasks. The subcultures were initiated when each flask reached 70–85% confluency, and consisted of the removal of spent cell culture media, a single wash with PBS, and cell detachment via trypsinization. Trypsin-EDTA (1 mL, 0.025%; Thermo Fisher) was distributed onto the cells and incubated at 28°C for 3–5 min until cells visibly detached. Trypsinized cells were transferred to a 15 mL falcon tube with 6 mL additional media, and subsequently centrifuged at 500 g for 7 min. The supernatant was removed and the remaining white pellet was resuspended in growth media. At this point, cells were counted using the Countess Automated Cell Counter (Thermo Fisher) and were then used in seeding experiments or seeded in T-75 flasks for further passage. For counting the cells, a 10 μL cell suspension was combined with 10 μL of Trypan blue dye and loaded onto each side of a cell counting chamber slide. Both live counts were taken and averaged to maximize consistency and accuracy for cell seeding. To prepare growth media, Leibovitz's L15 (Gibco™, Grand Island, NY, USA) with phenol red indicator was first dissolved in 985 mL of sterile distilled water, and the pH of the media was adjusted to 7.8 via addition of 1 M HCl solution. The L15 media was filtered using a 0.22 μm polyether sulfone (PES) filter (Millipore Sigma-Aldrich). The growth media consisted of 50 mL aliquots prepared every 2 weeks of sterile 89% Leibovitz L-15 Medium, 9% Fetal Bovine Serum (FBS), and 2% antibiotic-antimycotic.
2.4 Cell seeding
Seeding density was maintained at 300,000–500,000 cells suspended in 15–20 μL of FBS per each scaffold in both control and vacuum seeded groups. In the control seeding process, each scaffold was seeded by directly pipetting the seeding solution onto each scaffold and allowing an incubation phase of 30 min for the attachment of the cells before gently addition of growth media. The vacuum seeding process was carried out in accordance with the method described by Lúquez-Caravaca et al. (2023), with minor modification. The scaffolds were individually placed in 1.5 mL Eppendorf tubes before adding the seeding solution directly onto the surface of each scaffold. The tubes were briefly centrifuged at low speed to ensure that the scaffolds were in full contact with the seeding solution and no large air bubbles were visible in the solution. The Eppendorf tubes were placed with the cap open in a vacuum bag, and the opening of each bag was folded over at least twice to ensure sterility while transferring tubes to the vacuum sealer. Each bag was vacuum sealed at 95% vacuum with a 5 s hold time, and then removed from the vacuum bag in sterile conditions. The scaffolds were incubated at 28°C in complete growth media, and characterized at three time points: immediately after seeding (0 h), 24 h, and at 48 h post cell seeding.
2.5 Imaging
2.5.1 Scanning electron microscopy
Lyophilized scaffolds were sputter-coated with a 10 nm layer of Pt/Pd on a Leica ACE600 Sputter (Deerfield, IL, USA). Scaffolds were then imaged at an accelerating voltage of 5 kV on a FEI Quanta 600 FEG scanning electron microscope (Thermo Fisher). Pore size was measured using ImageJ software Version 1.47a (National Institutes of Health (NIH), Rockville, MD, USA). The image scale (500 μm) was used to the define the scale, and the diameter of more than 30 pores were measured.
2.5.2 F-actin cytoskeleton staining
To fix the scaffolds, 4% paraformaldehyde (PFA) solution in PBS was added to each well and scaffolds were incubated for 30 min at room temperature. Cell seeding distribution was screened in the control and vacuum seeded scaffolds via DAPI (4′,6-diamidino-2-phenylindole), which was used as a blue fluorescent nuclear stain. DAPI was diluted to 1 ng/mL and added to each well. The plates were incubated for 15 min, and DAPI containing solution was removed. The scaffolds were washed in PBS three times before being transferred to a glass slide for viewing.
2.5.3 Brightfield and confocal microscopy
An Olympus Inverted Microscope CKX53 with a phase contrast attachment was used to monitor cell growth and attachment. Fluorescence imaging was performed using a fluorescence imaging attachment at respective wavelengths for each stain.
2.5.4 Immunofluorescent staining
Scaffolds were fixed with 4% paraformaldehyde for 30 min at room temperature. After fixation, blocking was performed with 3% Bovine serum albumin (Millipore Sigma, USA) for 30 min. The samples were incubated overnight with Paired Box 7 (PAX 7) antibodies (Invitrogen, USA) in blocking solution at 4°C. Afterwards, the samples were incubated in DAPI (1:1000, Thermo Fisher), AlexaFluor TM 488 phalloidin (1:400, Invitrogen) and Goat anti rabbit AlexaFluor 594 (1:400, Invitrogen) for 1 h at room temperature. Imaging by confocal laser scanning microscopy (CLSM) with 3D z-stack reconstruction was performed on the TCS SP8 microscope (Leica, Wetzlar, Germany).
2.6 DNA quantification
The Quant-iTTM PicogreenVR (PG) dsDNA Kit (Invitrogen, Orlando, USA) was used to determine the effectiveness of the decellularization process as well as the DNA content of the scaffolds at each time point, adapted from Dikici et al. (2019). Fresh banana leaf samples were cut using a 6 mm biopsy punch and 1 mm biopsy punch in the center of each sample. All other samples were decellularized and vacuum seeded as described above, then removed from growth media at the time of characterization. Samples were washed once with PBS and transferred to microtubes, snap frozen in liquid nitrogen, and stored at −80°C for later quantification. Immediately upon removal from the −80°C freezer, 100 μL of cell digestion buffer was added to each microtube. Cell digestion buffer consisted of 10 mM Tris-HCl, 1 mM ZnCl2, and 1% Triton-X100 in distilled water (dH2O). Each scaffold was manually ground into a fibrous pulp using microtube pestles until homogenous (minimal fiber visibly remaining), about 30 s per sample, and vortexed for 60 s, then allowed to digest overnight at 4°C. Picogreen (PG) working solution was prepared by diluting 20 Tris-EDTA (TE) 1:20 in dH2O (1 TE), then diluting the PG reagent 1:200 in 1 TE. Samples were subjected to 3 freeze-thaw (FT) cycles (10 min at −80°C and 20 min at 37°C, with 15 s vortexing between each cycle). The microtubes were centrifuged at 11,000 g for 5 min to separate fiber from lysate. Then, 100 μL of the lysate was removed and combined thoroughly via pipetting with 100 mL PG working solution in Qubit tubes. Samples were protected from light and incubated at room temperature for 10 min. Fluorescence reading was conducted with the Qubit™ 4 Fluorometer (Thermo Fisher) at an excitation wavelength of 485 nm and an emission wavelength of 528 nm. The efficiency of decellularization was determined by comparing the DNA content (%) in fresh and decellularized leaves.
2.7 Fourier-transform infrared-red
The spectra of both fresh and decellularized banana leaves were collected using a Fourier-transform infrared spectrometer (FTIR; PerkinElmer Universal ATR, PerkinElmer, Waltham, MA, USA). The spectrometer was coupled with a sampling accessory that included a Frontier Universal Diamond/ZnSe ATR crystal. The spectra were collected from 4,000 to 600 cm−1 with a 32-scan rate and a resolution of 4 cm−1. The samples were analyzed in triplicates and means of the spectra were used for further analysis. Principal Component Analysis (PCA) was used via a loading plot constructed for PC1, which accounted for over 85% of the variables. This plot aimed to identify the most crucial variable responsible for the distinctions between the fresh (control) and decellularized banana leaves.
2.8 Cell seeding efficiency
The efficiency of cell seeding was determined via DNA quantification using Equation 1.
2.9 Cell alignment
Cell alignment was measured 48 h after cell seeding on the different scaffolds. After staining with DAPI, AlexaFluor 488 for Actin, and AlexaFluor 594 for the PAX7 antibody, the images were obtained using confocal laser microscopy as described above. The merged images were utilized for cell alignment measurements using the FIJI-Image 1.8.0_322 image process program by using an ellipse to each cell nucleus and cytoskeleton and measuring the angle of the maximum diameter relative to the horizontal axis of the image. The OrientationJ Plug-in was downloaded from www.epfl.ch/demo/orientation/ and added to the software to measure the orientation (Rezakhaniha et al., 2012; Jones et al., 2021). Color survey and 3D surface survey were created as well to visualize the orientation of cell cytoskeleton and the surface topography properties of each cell-seeded scaffold.
2.10 Thermal treatment
Different sets of scaffolds, including uncoated and soy-coated vacuum-seeded samples were utilized to assess the kinetics of area shrinkage after cooking at different pasteurization temperatures (60, 65, 70, and 75°C) for varying durations, in accordance with the methods outlined previously in Ovissipour et al. (2017) (N = 4). These times and temperatures were selected based on target bacteria and industry requirements to provide equivalent lethality and sufficient heating to inactivate Listeria monocytogenes (Ovissipour et al., 2017). Each scaffold was carefully positioned within an individually custom-built cylindrical aluminum test cell. These test cells were originally developed for kinetic studies of muscle foods (Ovissipour et al., 2017). Subsequently, the test cells were immersed in a water bath at each designated temperature point and retrieved at the specified time intervals, as detailed in Table 1. Following removal from the water bath, the aluminum sample cells were immediately submerged in an ice-water mixture to rapidly cool the samples and arrest the cooking process.
2.10.1 Area shrinkage
Area changes (shrinkage or swelling) was calculated based on the area of a sample prior and after heating, using ImageJ software Version 1.47a [National Institutes of Health (NIH), Rockville, MD, USA]. The shrinkage ratio was calculated using Equation 2:
2.10.2 Kinetics analysis
Quality degradation was calculated based on the area shrinkage. Reaction rates for quality degradation (area shrinkage) (C) in isothermal conditions are expressed in the following Equation 3 (Kong et al., 2007a):
where k is the rate constant, C is the quality parameter (area shrinkage) at time t, and n is the order of reaction. To find the best empirical relationship, data was analyzed using zero-, first-, and second-order kinetic models in as Equations 4–6:
zero-order:
first-order:
second-order:
where C0 is the initial amount of the quality at time zero, Ct is the value at time t, k is the rate constant. Then, Arrhenius equation was applied to determine the degradation rate constant (k) on temperature, which is described as follows:
where Ea is the activation energy of the reaction (kJ/mol), R is universal gas constant (8.3145 J/mol/K), T is the absolute temperature (K) and k0 is the frequency factor (per min). By applying the Equation 7 to a reaction, a plot of the rate constant on semi-logarithmic scale as a function of reciprocal absolute temperature (1/T) should yield a straight line. The activation energy can be determined as the slope of the line multiplied by the gas constant R.
2.11 Statistical analysis
The data normality and homoscedasticity were tested to confirm normal distribution using the Shapiro–Wilk test and normal quantile plots (NQP), and the even distribution of variability was tested using Levene's test. The data are presented as means for each treatment along with their respective standard deviations. A one-way ANOVA was conducted to assess the cell viability, cell adhesion and DNA content. To pinpoint any statistically significant distinctions among the treatment means, Tukey's multiple comparison test was employed.
3 Results and discussion
3.1 Decellularization, FTIR spectra, and DNA quantification
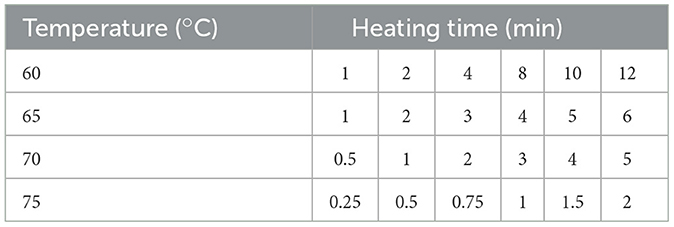
Banana leaf disks browned within 3 days after exposure to the SDS solution, and subsequently became translucent after 24 h exposure to the bleach solution (Figure 1A). To verify the efficiency of the decellularization process, DNA analysis was employed and indicated that the decellularization protocol reduced the DNA levels from 445 ng/mg in the fresh banana leaves, to non-detectable level in the decellularized samples (Figure 1B; n = 9). The average pore size for the decellularized banana leaves was 147 ± 31 μm (n = 30 pores; Figure 1C) which potentially provides a suitable platform for cell seeding and cell attachment and was in the range of pore sizes in decellularized plant tissues previously studied, which ranging from 50 to 300 μm (Jones et al., 2021; Thyden et al., 2022). While <50 ng/mg DNA is deemed sufficient for plant tissue decellularization for developing tissue in biomedical field (Crapo et al., 2011), for large-scale cultivated meat production and to reduce the environmental footprint and minimize chemical applications, adhering to this standard may not be required. This threshold was originally established to mitigate adverse host reactions to xenogenic DNA from implanted decellularized tissue in patients. The efficiency of decellularization can vary depending on the type of plant tissue and the specific decellularization process employed. For instance, decellularizing spinach leaves required a 9-day process (Jones et al., 2021), while in the case of broccoli florets, only 48 h were needed for complete decellularization (Thyden et al., 2022). In the present study, we were able to achieve complete decellularization in 4 days by implementing process modifications. Specifically, by introducing the small hole in the center of the banana leaf disks, the duration of the decellularization process was reduced from the original 8 to 4 days. A more pressing concern for the development of a food safe material is the safety of decellularizing agents. While SDS and Tween 20 at designated concentrations are considered to be GRAS by the FDA, a suitable replacement for bleach in the final step of the decellularization process must be considered.
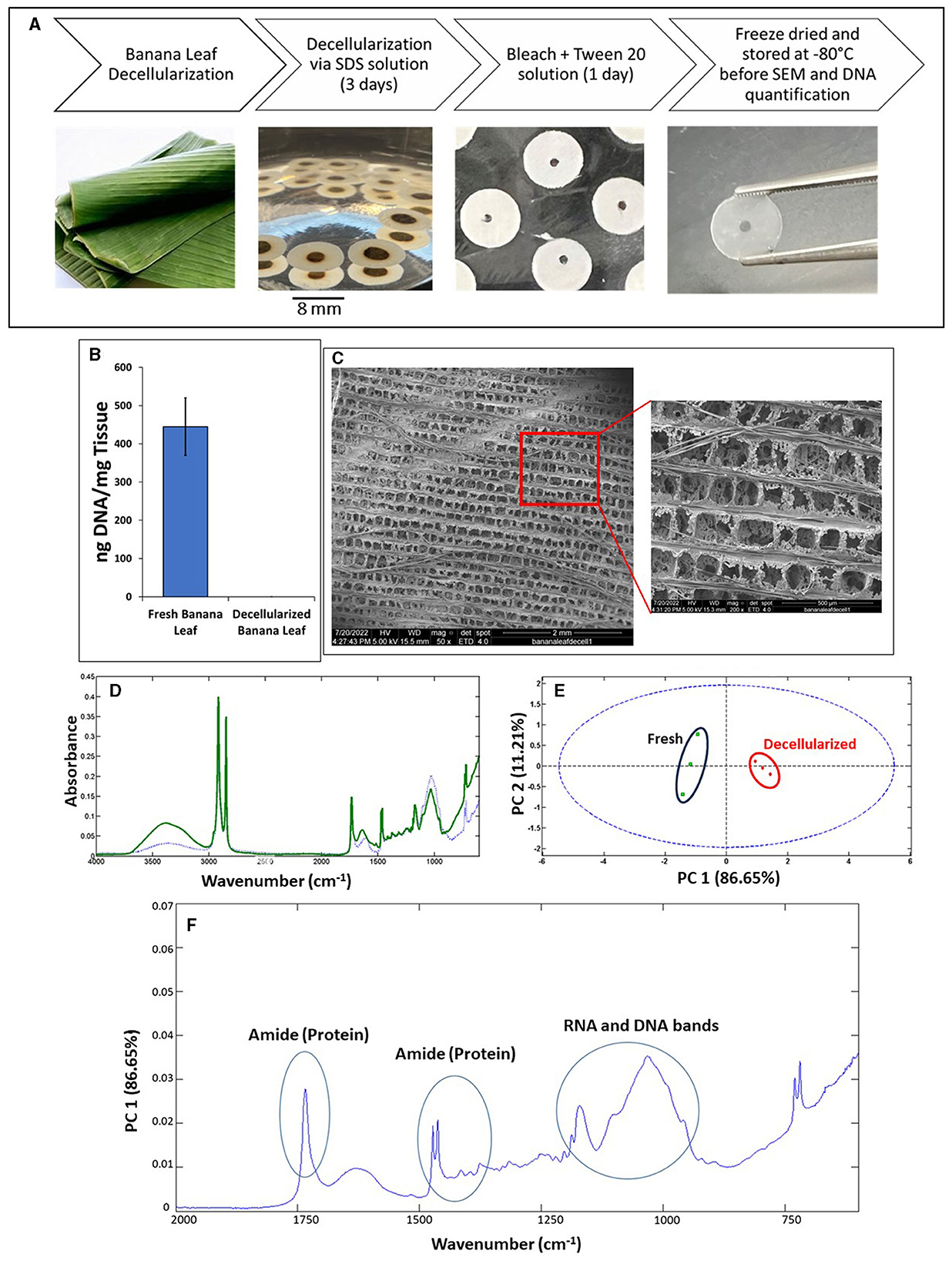
Figure 1. (A) Decellularization process description from fresh banana leaf to completely decellularized translucent banana leaf; (B) Total DNA of fresh and decellularized banana leaves (n = 9); (C) SEM pictures of decellularized banana leaves with mean pores size of 147 μm; (D) FTIR spectra of fresh (green) and decellularized (blue) banana leaves; (E) PCA model for fresh and decellularized banana leaves; (F) Loading plot for PC1, indicating the main peaks for Amides (protein), RNA and DNA.
The spectra of the banana leaf (green line) and the decellularized banana leaf (blue dots) and principal component analysis (PCA) are illustrated in Figures 1D and 1E. The results indicated a decrease in peak intensity and the elimination of certain peaks within the spectral range 1,350 to 1,220 cm−1 in the decellularized banana leaf, which corresponds to the presence of DNA and RNA peaks (Movasaghi et al., 2008). Furthermore, the peak observed at ~1,520 cm−1, commonly associated with cytosine, guanine or amide II, is absent in the decellularized banana leaf. The absence of a peak corresponding to the amide or adenine group was noted at 1,650 cm−1 in the decellularized banana leaf. Furthermore, a significant decrease in peak intensity was observed at ~2,950 cm−1 in the decellularized banana leaf sample, which primarily corresponds to the stretching of protein CH bonds (Mello and Vidal, 2012). The results from loading plots also confirmed that the protein and DNA peak intensities accounted for the difference in fresh and decellularized banana leaves as seen by DNA quantification (Figure 1F).
3.2 Cell seeding efficiency
Two distinct methods for cell seeding were employed, including natural or control seeding by directly pipetting the seeding solution onto each scaffold and allowing an incubation phase of 30 min, and vacuum infusion for 5 s. The imaging data suggested that vacuum infusion was more efficient for cell seeding in comparison to naturally seeding (30 min; Figure 2). Although natural seeding produced very high cell density around the exposed perimeter of each scaffold, cells were not able to penetrate or migrate toward the center of the scaffold over 24 or 48 h. Thus, cell seeding through vacuum infusion was selected for the rest of the experiments.
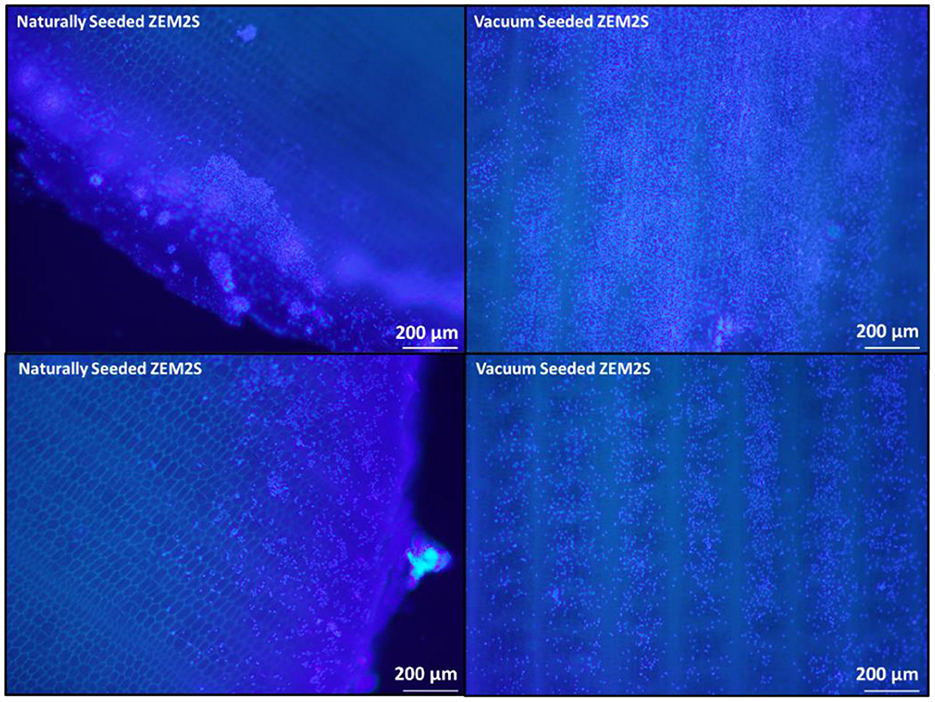
Figure 2. DAPI image from naturally and vacuum seeded cells on the surface of the banana leaves, immediately after seeding. These images indicate high cell biomass around the edges of the decellularized banana leaves, in naturally seeded samples, compared to the uniform cell distribution through the scaffold surface in vacuum seeded cells. Both scaffolds used were uncoated with the thickness of 2,000 μm.
The cell seeding efficiency for the vacuum seeding method across all scaffold groups including uncoated, soy-coated, and gelatin-coated (n = 27) were 35, 29, and 33% of the total cells in the seeding solution. Following cell seeding, the DNA content in the decellularized banana leaves without coatings, as well as those coated with soy and gelatin, were 107 ± 33.5, 99 ± 18, and 88 ± 31.2 ng/mg tissue, respectively. These results indicate that the decellularized banana leaf had the capacity to absorb cells up to ~25% of its original (pre-decellularization capacity; 445 ng/mg).
3.3 Short-term cell growth indicated by DNA content
DNA quantification indicated no significant difference in cell seeding among all groups immediately after seeding (Figure 3). After 24 h, the soy coated scaffolds (262.5 ± 97 ng/mL) had a higher DNA content than the gelatin coated group (173.63 ± 48.6 ng/mL). DNA quantification indicated that cell growth was reduced between 24 and 48 h. In some previous studies, a reduction in cell numbers after 7 days of incubation was not observed (Jones et al., 2021). Vacuum infusion method in this study induced deeper penetration of cells, which may have restricted access to media and nutrient uptake due to the high cell density and thickness of the decellularized banana leaves (2,000 μm), leading to hypoxic conditions after 24 h. Further functionalization, such as increasing surface area exposure of the pores to media, may be necessary to maintain long term cell culture necessary for cell differentiation studies. However, the technique employed still represents a viable method for effectively seeding cell biomass onto the scaffolds. Infusing cells through a vacuum process not only expedites the cell seeding stage but also eliminates the need for a distinct bioreactor dedicated to cell seeding, proliferation, and differentiation on the scaffold, making the cell-based production more cost effective.
Figure 3. DNA Content over 48 h in uncoated, soy- and gelatin-coated scaffolds as compared to the initial seeding solution distributed onto each scaffold. Minimum outlier indicates that one of the replicates in soy coated samples had non-detectable level of DNA; Maximum outlier in gelatin-coated samples illustrates that one of the replicates showed 230 ng/mL DNA.
3.4 Cell differentiation alignment
When examining cell adhesion, the soy-coated samples exhibited higher levels of cell adhesion, as well as cell alignment and spreading along the existing pore structure. Elongated, spindle like morphology was observed, which is an indicator of transition from a fibroblast-like to myoblast-like phenotype (Benarroch et al., 2023). Gelatin coated scaffolds were not able to retain cell attachment through the cell immunostaining procedure, indicating that the gelatin coating prevented cells from entering the matrix. Cell differentiation efficiency using PAX7 indicated that the cells on uncoated and gelatin-coated banana leaves had begun to show differentiation potential compared to the soy-coated samples. Other researchers also reported delayed differentiation in cells seeded on plant scaffold (Jones et al., 2021) (Figures 4A–C). Delayed differentiation potentially suggests increased proliferation and self-renewal. This would lead to an increase in the number of cells and their colonization of a much larger area within the scaffold in the first few days (short term). Additionally, a higher number of the cells could differentiate into myofibers (Riederer et al., 2012).
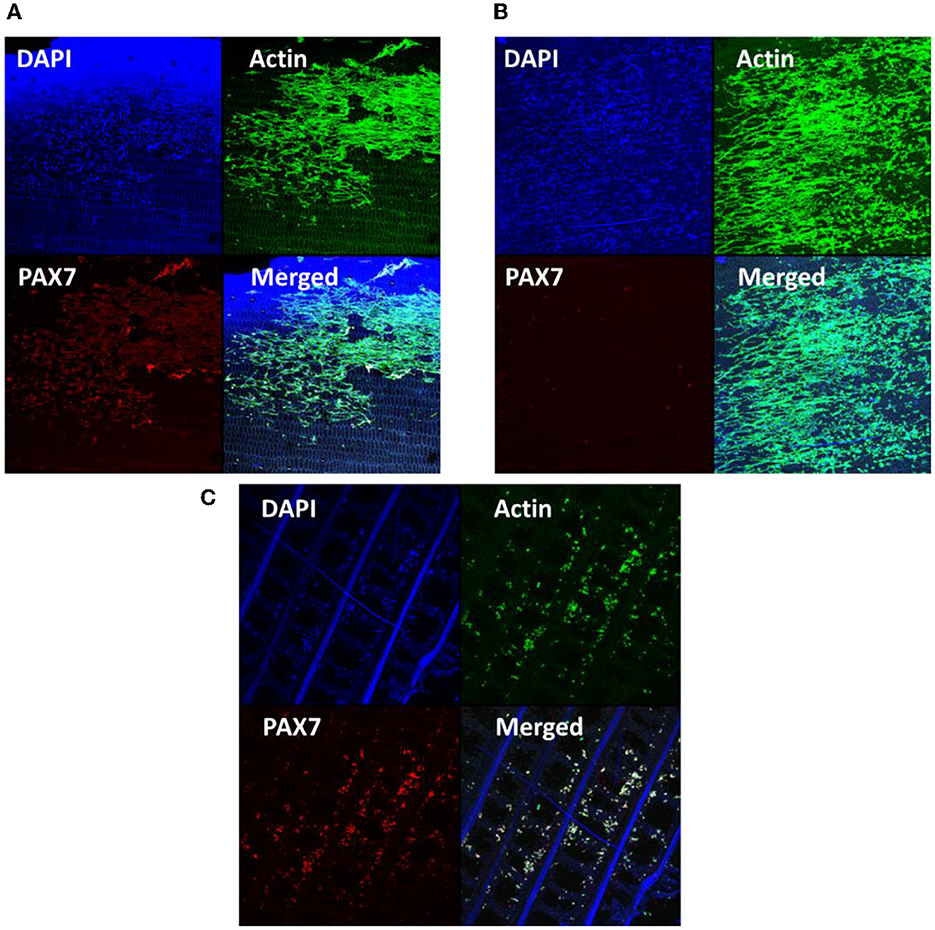
Figure 4. DAPI, Action, PAX7, and merged images for Zebrafish cells vacuum seeded on: (A) uncoated decellularized banana leaves; (B) soy-coated decellularized banana leaves; (C) gelatin-coated decellularized banana leaves.
The results of alignment indicated that the cells did not align on the uncoated scaffolds (Figure 5), similar to the findings of other researchers (Jones et al., 2021). However, the addition of soy and gelatin as coating materials enhanced cell alignment. Specifically, the dominant orientations based on the image processing results, were −6.3°, 9.2°, and 65.8° for uncoated, soy-coated, and gelatin-coated samples, respectively (Figure 5C). The organized alignment of cells within various tissues is vital for maintaining specific functions including cell growth and cell differentiation (Yin et al., 2010; Subramony et al., 2013).
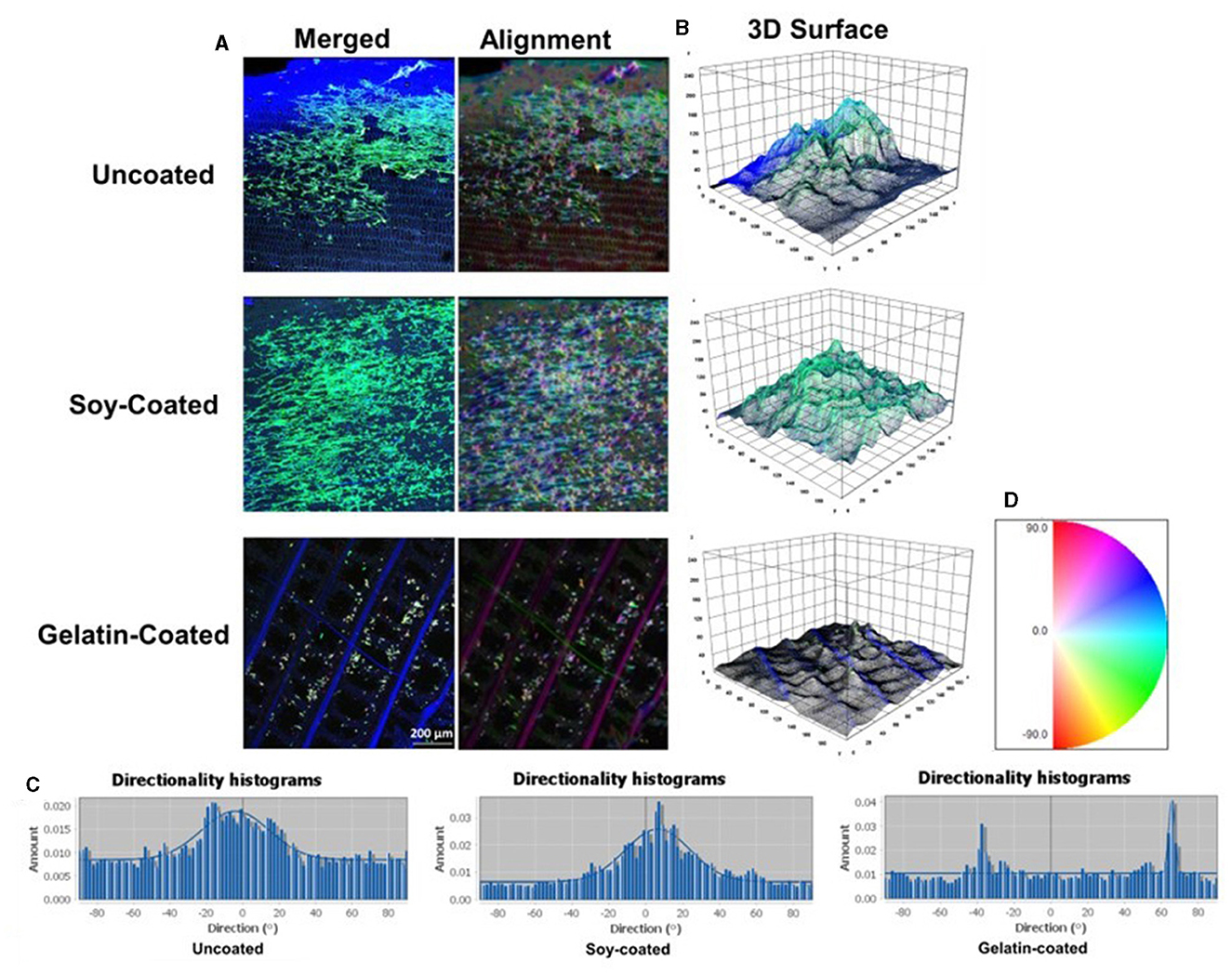
Figure 5. (A) Cell alignment color survey indicating the direction of Zebrafish cytoskeleton on uncoated, soy-coated and gelatin-coated scaffolds; (B) 3D surface analysis of the cytoskeleton filaments on uncoated, soy-coated and gelatin-coated scaffolds; (C) Cytoskeleton microfilament orientation histogram of Zebrafish cells cultured on uncoated, soy-coated, and gelatin-coated banana leaves; (D) The orientation degree ranges from −90 to +90, defining the histograms (C). Any number above 0 indicates that the orientation is positive toward 90, and any degree below 0 is considered as negative toward −90.
For example, no cell differentiation (Subramony et al., 2013) and less differentiated cells (Yin et al., 2010) were observed on non-aligned scaffolds compared to aligned scaffolds. The role of alignment in cell differentiation is crucial to the extent that even after applying mechanical stimulation, no fibroblastic differentiation was observed on the non-aligned scaffolds. This underscores the significant impact of scaffold topography on stem cell differentiation (Subramony et al., 2013). To induce such alignment, ideal scaffolds should replicate the characteristics and morphologies of natural tissues (Yin et al., 2010; Subramony et al., 2013). Aligned structures that guide cell orientation are employed to facilitate tissue regeneration and repair.
3D surface plots revealed that the uncoated banana leaves were rougher than the soy- and gelatin-coated samples due to the porous structure. Cell alignment on plant-based materials is influenced by various factors, including surface topography of the decellularized plant tissues, interconnectivity and vascularization of the matrix, the tissue source, and the type of cells (Jones et al., 2021). These findings underscore the complexity of cell behavior on plant-based materials and the need for further investigation. This insight would provide valuable insights into optimizing scaffold designs and coating strategies for aligned muscle development in cultivated seafood production, ultimately advancing the field of cellular agriculture.
3.5 Area shrinkage and activation energy
Area shrinkage is considered one of the most important quality attributes in meat and seafood products in response to heat (Ovissipour et al., 2013, 2017). Area shrinkage is the result of cook loss which is defined as loss of moisture and protein denaturation (Ovissipour et al., 2013, 2017). The results of fresh, cooked and fried scaffolds with the cells are illustrated in Figures 6A–E. The samples from cooking were used to assess tissue shrinkage (Figures 6I–IV, Table 2). In plain uncoated and soy-coated decellularized banana leaves, swelling or increase in the samples area was observed. Soy-coated samples swelled significantly less (Figure 6III) compared to uncoated samples (Figure 6I). Seeding the cells onto the scaffold resulted in different findings. The area shrinkage in cell seeded samples was similar to previous findings in conventional seafood products in which by increasing the temperature, the area shrinkage was increased (Ovissipour et al., 2013, 2017). Area shrinkage in the samples without cells followed the first order with activation energy of 130 and 118 kJ/mol for uncoated, and soy-coated scaffolds, respectively. However, after cell seeding into the scaffolds, the kinetics of area shrinkage shifted to second order, with significantly higher activation energy, reaching 195 and 198 kJ/mol for uncoated and soy-coated scaffolds, respectively. Coating the scaffold with soy protein did not significantly change the activation energy. The results indicated that the activation energy for decellularized banana leaf scaffold, without cell seeding, was relatively high compared to area shrinkage of conventional livestock meat activation energy and was significantly increased after seeding cells on to the scaffolds. This is primarily due to the unseeded scaffold being composed of long chain polysaccharides such as lignin and hemicellulose, and the addition of intracellular protein and water content affecting thermal resistance in seeded samples. Activation energy for area shrinkage in blue mussel (Mytilus edulis; Ovissipour et al., 2013) and Atlantic salmon (Salmo salar; Ovissipour et al., 2017) were 92 and 107 kJ/mol, respectively. Area shrinkage is the result of cook loss due to the protein deformation (denaturation and aggregation) in meat products (Ovissipour et al., 2013, 2017).
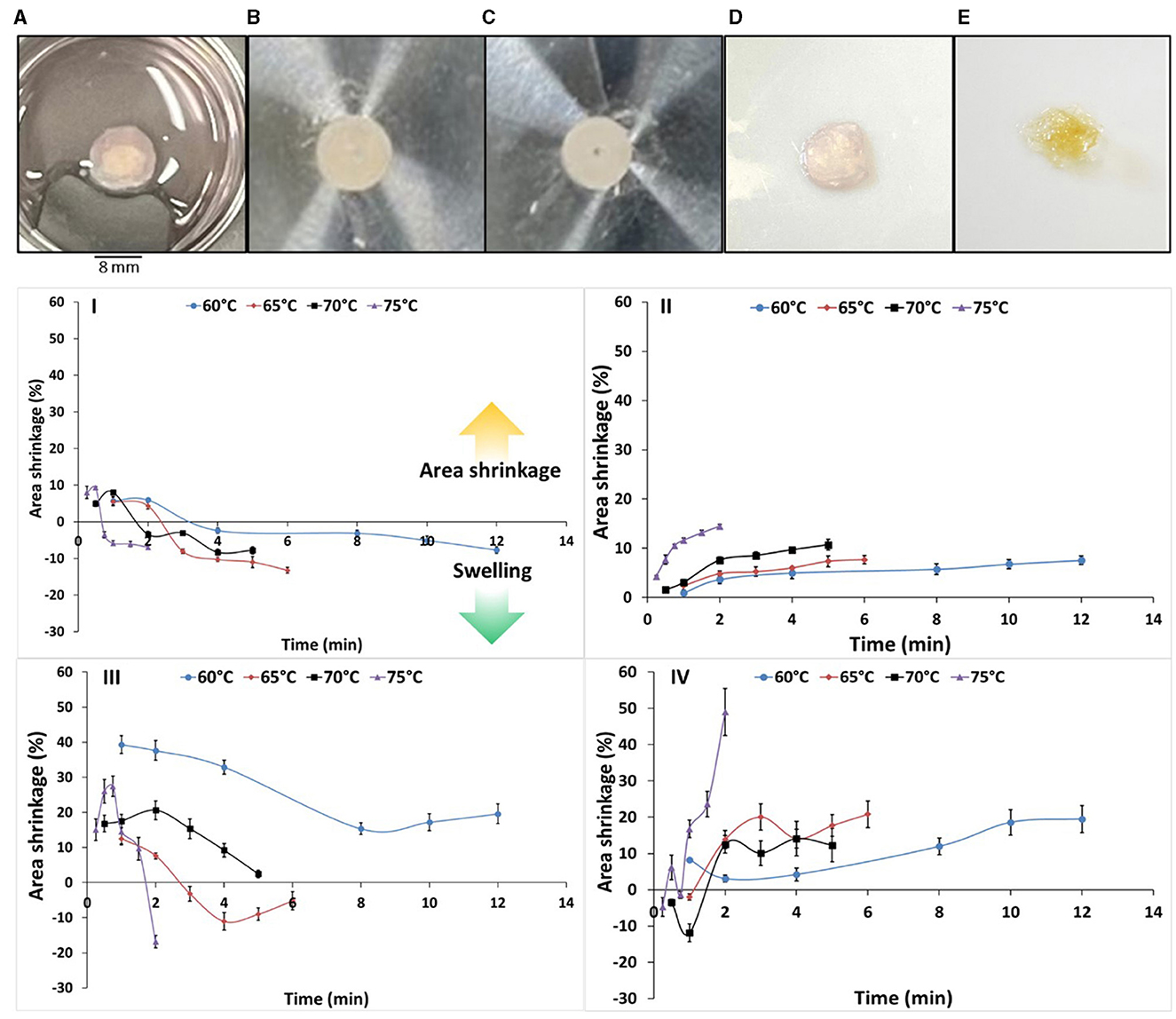
Figure 6. Decellularized banana leaf scaffolds seeded with cells: (A) fresh; (B) before cooking; (C) after cooking at 75°C; (D) fresh sample before frying; (E) after frying; and area shrinkage (%) of (I) uncoated decellularized banana leaf; (II) cell seeded uncoated decellularized banana leaf; (III) soy-coated decellularized banana leaf; (IV) cell seeded soy-coated decellularized banana leaf. The positive areas illustrate the area shrinkage, and the negative areas indicate the swelling and increase in the area due to the impact of heat on the scaffold and possible gelation.
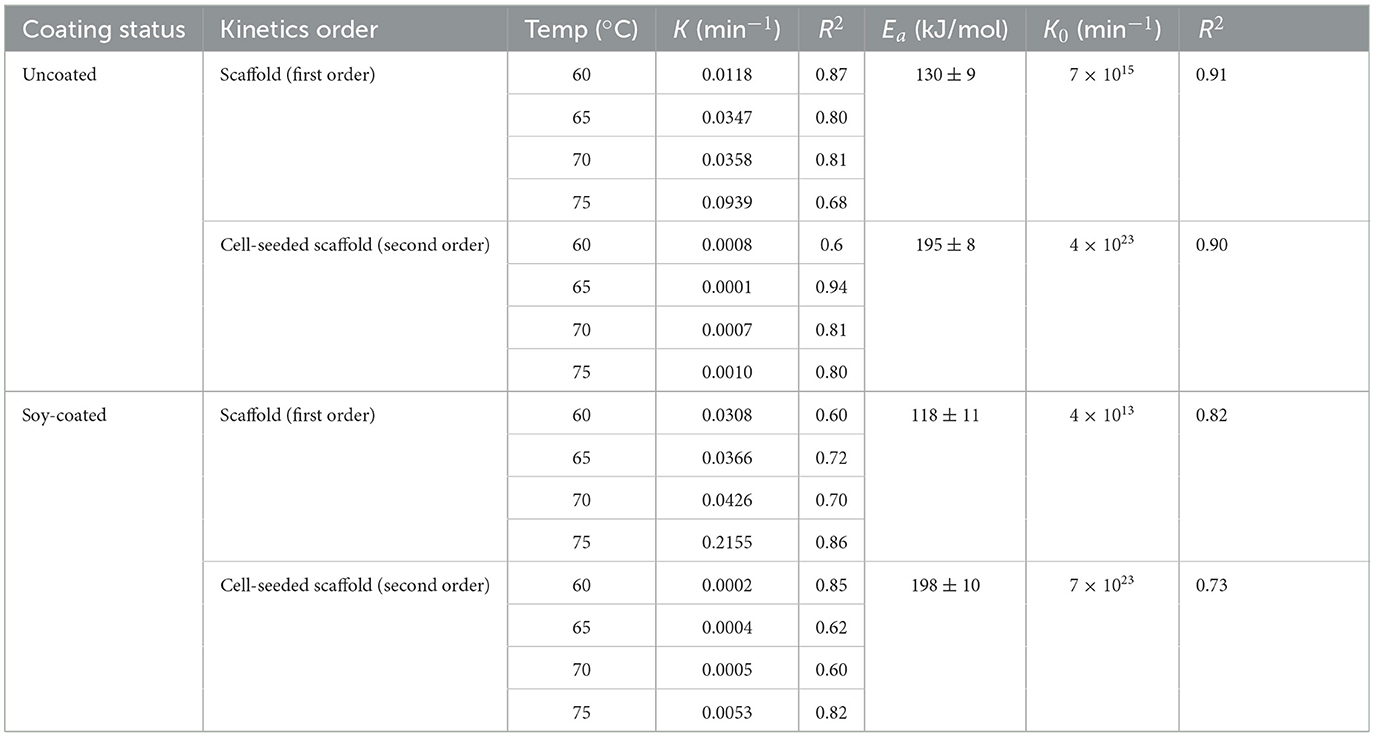
Table 2. Kinetic parameters of area shrinkage of zebrafish cells seeded on uncoated and soy-coated banana leaves.
After decellularization, the protein content of the scaffold is significantly reduced. This can result in the swelling of carbohydrates in decellularized plants during thermal processing. However, in cell-seeded samples, the protein and moisture contributed significantly to protein denaturation and moisture loss, resulting in high area shrinkage in thermally processed samples. Additionally, the area shrinkage was higher in soy-coated samples due to the availability of more protein for denaturation and the higher cell density on soy-coated samples compared to the uncoated samples.
In conventional livestock meat products, heating results in denaturation of myosin and shrinkage of myofibrils, expanding extracellular spaces and resulting in the expulsion of water (Ofstad et al., 1993). In these conventional meat products, muscle fiber diameters and sarcomere length become shorter due to protein denaturation, resulting in water soluble proteins and fats being expelled from the tissue (Kong et al., 2007a,b; Ovissipour et al., 2017). Fish muscle tissue is distinct from mammalian muscle due to low thermal stability of collagen (Yunoki et al., 2003) and shortened myosepta, contributing to tender and flaky cooked texture (Listrat et al., 2016). In previous studies on area shrinkage in aquatic food products, significant area shrinkage was observed due to thermal processing in salmon (Oncorhynchus gorbuscha; Kong et al., 2007a,b), cod (Gadus morhua; Skipnes et al., 2007, 2011), Atlantic salmon (S. salar; Ofstad et al., 1993; Ovissipour et al., 2017), and blue mussel (M. edulis; Ovissipour et al., 2013). Conventional aquatic food products contain lower collagen contents (Skipnes et al., 2011), resulting in higher susceptibility to heat and higher area shrinkage compared to conventional livestock meat products (Ovissipour et al., 2017). The magnitude of area shrinkage and activation energy vary for different fish. Aquatic organisms with higher muscle fat and collagen content, and stronger connective tissues such as in salmon, exhibit greater heat resistance with higher activating energy (107 kJ/mol; Ovissipour et al., 2017) compared to tissues with significantly less fat and collagen content and poor connective tissues such as in mussel (92 kJ/mol; Ovissipour et al., 2013). The findings in the present study present a promising outcome for the use of plant-based scaffolds enriched with protein in cultivated meat production. The observed high activation energy in decellularized banana leaves further supports the potential of such scaffolds to enhance thermal stability and overall cooking characteristics of cell-based seafood products. This could potentially lead to the development of cell-based fish filets that maintain quality, nutritional value, and structural integrity perceived as texture during cooking. Further research and development in this area could impact the cultivated meat industry by providing consumers with sustainable, high-quality alternatives to traditional seafood products.
4 Conclusions
Banana leaves offer a cost-effective and sustainable material for scaffold development for cultured meat production. Their potential for decellularized plant tissues for scaffolds in cultivated meat products is an emerging technology. Traditional cell seeding on scaffolds often suffers from inefficiency, necessitating separate bioreactor setups for cell proliferation and differentiation. The present results demonstrate that the use of pressure differential seeding in a highly interconnected, porous scaffold improves cell infusion and significantly enhanced cell seeding efficiency. This may obviate the need for separate bioreactor setups, and has potential to thereby reduce production costs. Furthermore, the kinetics of quality degradation after thermal processing (e.g., cooking) reveal that the thermal behavior kinetics of banana leaves with cells closely resemble that of traditional seafood products, suggesting that banana leaves can serve as a scaffold source for cell-based seafood and meat production.
Data availability statement
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding authors.
Ethics statement
Ethical approval was not required for the studies on animals in accordance with the local legislation and institutional requirements because only commercially available established cell lines were used.
Author contributions
RS: Investigation, Methodology, Writing—original draft, Writing—review & editing. AB: Data curation, Investigation, Methodology, Writing—original draft. AA: Data curation, Investigation, Methodology, Software, Writing—review & editing. XL: Investigation, Methodology, Writing—review & editing. CN: Investigation, Methodology, Writing—review & editing. DK: Funding acquisition, Methodology, Resources, Supervision, Writing—review & editing. NN: Conceptualization, Funding acquisition, Investigation, Methodology, Supervision, Writing—review & editing. RO: Conceptualization, Data curation, Formal analysis, Funding acquisition, Investigation, Methodology, Project administration, Resources, Supervision, Validation, Visualization, Writing—original draft, Writing—review & editing.
Funding
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. This research was financially supported by the Agriculture and Food Research Initiative (AFRI) Sustainable Agricultural Systems program, grant no. 2021-699012-35978 from the USDA National Institute of Food and Agriculture, and Texas A&M AgriLife Research.
Acknowledgments
We would like to extend our gratitude to Joseph Awika and Youjun Deng from Texas A&M University for their invaluable support throughout the course of this study.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Benarroch, L., Madsen-Østerbye, J., Abdelhalim, M., Mamchaoui, K., Ohana, J., Bigot, A., et al. (2023). Cellular and genomic features of muscle differentiation from isogenic fibroblasts and myoblasts. Cells 12:1995. doi: 10.3390/cells12151995
Bomkamp, C., Skaalure, S. C., Fernando, G. F., Ben-Arye, T., Swartz, E. W., and Specht, E. A. (2022). Scaffolding biomaterials for 3D cultivated meat: prospects and challenges. Adv. Sci. 9:2102908. doi: 10.1002/advs.202102908
Cheng, Y.-W., Shiwarski, D. J., Ball, R. L., Whitehead, K. A., and Feinberg, A. W. (2020). Engineering aligned skeletal muscle tissue using decellularized plant-derived scaffolds. ACS Biomater. Sci. Eng. 6, 3046–3054. doi: 10.1021/acsbiomaterials.0c00058
Crapo, P. M., Gilbert, T. W., and Badylak, S. F. (2011). An overview of tissue and whole organ decellularization processes. Biomaterials 32, 3233–3243. doi: 10.1016/j.biomaterials.2011.01.057
Dikici, S., Claeyssens, F., and MacNeil, S. (2019). Decellularised baby spinach leaves and their potential use in tissue engineering applications: studying and promoting neovascularisation. J. Biomater. Appl. 34, 546–559. doi: 10.1177/0885328219863115
Duffy, R. M., Sun, Y., and Feinberg, A. W. (2016). Understanding the role of ECM protein composition and geometric micropatterning for engineering human skeletal muscle. Ann. Biomed. Eng. 44, 2076–2089. doi: 10.1007/s10439-016-1592-8
Eibl, R., Senn, Y., Gubser, G., Jossen, V., van den Bos, C., and Eibl, D. (2021). Cellular agriculture: opportunities and challenges. Ann. Rev. Food Sci. Technol. 12, 51–73. doi: 10.1146/annurev-food-063020-123940
Jones, J. D., Rebello, A. S., and Gaudette, G. R. (2021). Decellularized spinach: an edible scaffold for laboratory-grown meat. Food Biosci. 41:100986. doi: 10.1016/j.fbio.2021.100986
Jones, J. D., Thyden, R., Perreault, L. R., Varieur, B. M., Patmanidis, A. A., Daley, L., et al. (2023). Decellularization: leveraging a tissue engineering technique for food production. ACS Biomater. Sci. Eng. 9, 2292–2300. doi: 10.1021/acsbiomaterials.2c01421
Kong, F., Tang, J., Rasco, B., and Crapo, C. (2007a). Kinetics of salmon quality changes during thermal processing. J. Food Eng. 83, 510–520. doi: 10.1016/j.jfoodeng.2007.04.002
Kong, F., Tang, J., Rasco, B., Crapo, C., and Smiley, S. (2007b). Quality changes of salmon (Oncorhynchus gorbuscha) muscle during thermal processing. J. Food Sci. 72, S103–S111. doi: 10.1111/j.1750-3841.2006.00246.x
Listrat, A., Lebret, B., Louveau, I., Astruc, T., Bonnet, M., Lefaucheur, L., et al. (2016). How muscle structure and composition influence meat and flesh quality. Sci. World J. 2016:3182746. doi: 10.1155/2016/3182746
Lúquez-Caravaca, L., Ogawa, M., Rai, R., Nitin, N., Moreno, J., García-Martínez, T., et al. (2023). Yeast cell vacuum infusion into fungal pellets as a novel cell encapsulation methodology. Appl. Microbiol. Biotechnol. 107, 5715–5726. doi: 10.1007/s00253-023-12681-3
McNamara, E., and Bomkamp, C. (2022). Cultivated meat as a tool for fighting antimicrobial resistance. Nat. Food 3, 791–794. doi: 10.1038/s43016-022-00602-y
Mello, M. L. S., and Vidal, B. (2012). Changes in the infrared microspectroscopic characteristics of DNA caused by cationic elements, different base richness and single-stranded form. PLoS ONE 2012:43169. doi: 10.1371/journal.pone.0043169
Movasaghi, Z., Rehman, S., and ur Rehman, D. I. (2008). Fourier transform infrared (FTIR) spectroscopy of biological tissues. Appl. Spectr. Revi. 43, 134–179. doi: 10.1080/05704920701829043
Nichol, J. W., and Khademhosseini, A. (2009). Modular tissue engineering: engineering biological tissues from the bottom up. Soft Matter 5, 1312–1319. doi: 10.1039/b814285h
Ofstad, R., Kidman, S., Myklebust, R., and Hermansson, A.-M. (1993). Liquid holding capacity and structural changes during heating of fish muscle: cod (Gadus morhua L.) and salmon (Salmo salar). Food Struct. 12:4.
Ovissipour, M., Rasco, B., Tang, J., and Sablani, S. (2017). Kinetics of protein degradation and physical changes in thermally processed Atlantic salmon (Salmo salar). Food Bioprocess Technol. 10, 1865–1882. doi: 10.1007/s11947-017-1958-4
Ovissipour, M., Rasco, B., Tang, J., and Sablani, S. S. (2013). Kinetics of quality changes in whole blue mussel (Mytilus edulis) during pasteurization. Food Res. Int. 53, 141–148. doi: 10.1016/j.foodres.2013.04.029
Perreault, L. R., Thyden, R., Kloster, J., Jones, J. D., Nunes, J., Patmanidis, A. A., et al. (2023). Repurposing agricultural waste as low-cost cultured meat scaffolds. Front. Food Sci. Technol. 3:1208298. doi: 10.3389/frfst.2023.1208298
Rezakhaniha, R., Agianniotis, A., Schrauwen, J. T. C., Griffa, A., Sage, D., Bouten, C., et al. (2012). Experimental investigation of collagen waviness and orientation in the arterial adventitia using confocal laser scanning microscopy. Biomech. Model. Mechanobiol. 11, 461–473. doi: 10.1007/s10237-011-0325-z
Riederer, I., Negroni, E., Bencze, M., Wolff, A., Aamiri, A., Di Santo, J. P., et al. (2012). Slowing down differentiation of engrafted human myoblasts into immunodeficient mice correlates with increased proliferation and migration. Mol. Ther. 20, 146–154. doi: 10.1038/mt.2011.193
Rischer, H., Szilvay, G. R., and Oksman-Caldentey, K.-M. (2020). Cellular agriculture-industrial biotechnology for food and materials. Curr. Opin. Biotechnol. 61, 128–134. doi: 10.1016/j.copbio.2019.12.003
Röös, E., Bajželj, B., Smith, P., Patel, M., Little, D., and Garnett, T. (2017). Greedy or needy? Land use and climate impacts of food in 2050 under different livestock futures. Glob. Environ. Change 47, 1–12. doi: 10.1016/j.gloenvcha.2017.09.001
Skipnes, D., Johnsen, S., Skåra, T., Sivertsvik, M., and Lekang, O. (2011). Optimization of heat processing of farmed Atlantic cod (Gadus morhua) muscle with respect to cook loss, water holding capacity, color, and texture. J. Aquat. Food Prod. Technol. 20, 331–340. doi: 10.1080/10498850.2011.571808
Skipnes, D., Østby, M. L., and Hendrickx, M. E. (2007). A method for characterising cook loss and water holding capacity in heat treated cod (Gadus morhua) muscle. J. Food Eng. 80, 1078–1085. doi: 10.1016/j.jfoodeng.2006.08.015
Soice, E., and Johnston, J. (2021). How cellular agriculture systems can promote food security. Front. Sustain. Food Syst. 5:753996. doi: 10.3389/fsufs.2021.753996
Subramony, S. D., Dargis, B. R., Castillo, M., Azeloglu, E. U., Tracey, M. S., Su, A., et al. (2013). The guidance of stem cell differentiation by substrate alignment and mechanical stimulation. Biomaterials 34, 1942–1953. doi: 10.1016/j.biomaterials.2012.11.012
Thyden, R., Perreault, L. R., Jones, J. D., Notman, H., Varieur, B. M., Patmanidis, A. A., et al. (2022). An edible, decellularized plant derived cell carrier for lab grown meat. Appl. Sci. 12:5155. doi: 10.3390/app12105155
Xiang, N., Yuen, J. S., Stout, A. J., Rubio, N. R., Chen, Y., and Kaplan, D. L. (2022). 3D porous scaffolds from wheat glutenin for cultured meat applications. Biomaterials 285:121543. doi: 10.1016/j.biomaterials.2022.121543
Yin, Z., Chen, X., Chen, J. L., Shen, W. L., Nguyen, T. M. H., Gao, L., et al. (2010). The regulation of tendon stem cell differentiation by the alignment of nanofibers. Biomaterials 31, 2163–2175. doi: 10.1016/j.biomaterials.2009.11.083
Keywords: banana leaves, decellularization, cell seeding, quality, seafood, cellular agriculture
Citation: Banavar A, Sarkarat R, Amirvaresi A, Li X, Nguyen C, Kaplan DL, Nitin N and Ovissipour R (2024) Decellularized banana leaves: eco-friendly scaffolds for cell-based seafood. Front. Sustain. Food Syst. 8:1341151. doi: 10.3389/fsufs.2024.1341151
Received: 19 November 2023; Accepted: 15 January 2024;
Published: 02 February 2024.
Edited by:
Yan Ping Chen, Shanghai Jiao Tong University, ChinaReviewed by:
Tyler John Barzee, University of Kentucky, United StatesSayantani Dutta, Central Food Technological Research Institute (CSIR), India
Copyright © 2024 Banavar, Sarkarat, Amirvaresi, Li, Nguyen, Kaplan, Nitin and Ovissipour. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Reza Ovissipour, b3Zpc3NpQHRhbXUuZWR1; Nitin Nitin, bm5pdGluQHVjZGF2aXMuZWR1
†These authors have contributed equally to this work
 Amiti Banavar1†
Amiti Banavar1† Reyhaneh Sarkarat
Reyhaneh Sarkarat Xinxin Li
Xinxin Li David L. Kaplan
David L. Kaplan Nitin Nitin
Nitin Nitin Reza Ovissipour
Reza Ovissipour