
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 02 February 2024
Sec. Crop Biology and Sustainability
Volume 8 - 2024 | https://doi.org/10.3389/fsufs.2024.1334421
This article is part of the Research TopicAbiotic Stresses in Field Crops: Response, Impacts and Management under Climate Change ScenarioView all 15 articles
 Muhammad Azrai1*
Muhammad Azrai1* Muhammad Aqil2
Muhammad Aqil2 N. N. Andayani2
N. N. Andayani2 Roy Efendi2
Roy Efendi2 Suarni2
Suarni2 Suwardi2
Suwardi2 Muhammad Jihad2
Muhammad Jihad2 Bunyamin Zainuddin2
Bunyamin Zainuddin2 Salim2
Salim2 Bahtiar3
Bahtiar3 Ahmad Muliadi2
Ahmad Muliadi2 Muhammad Yasin2
Muhammad Yasin2 Muhammad Fitrah Irawan Hannan2
Muhammad Fitrah Irawan Hannan2 Rahman2
Rahman2 Amiruddin Syam4
Amiruddin Syam4The frequent occurrence of drought, halting from unpredictable climate-induced weather patterns, presents significant challenges in breeding drought-tolerant maize to identify adaptable genotypes. The study explores the optimization of machine learning (ML) to predict both the grain yield and stress tolerance index (STI) of maize under normal and drought-induced stress. In total, 35 genotypes, comprising 31 hybrid candidates and four commercial varieties, were meticulously evaluated across three normal and drought-treated sites. Three popular ML were optimized using a genetic algorithm (GA) and ensemble ML to enhance data capture. Additionally, a Multi-trait Genotype-Ideotype Distance (MGIDI) was also involved to identify superior maize hybrids well-suited for drought conditions. The results highlight that the ensemble meta-models optimized by grid search exhibit robust performance with high accuracy across the testing datasets (R2 = 0.92 for grain yield and 0.82 for STI). The RF optimized by GA algorithm demonstrates slightly lower performance (R2 = 0.91 for grain yield and 0.79 for STI), surpassing the predictive performance of individual SVM-GA and KNN-GA models. Selection of the best-performing hybrids indicated that out of the six hybrids with the highest STI values, both the ensemble and MGIDI can accurately predict four hybrids, namely H06, H10, H13, and H35. Thus, combining ML with MGIDI enables researchers to discern traits for each genotype and holds promise for advancing the field of drought-tolerant maize breeding and expediting the development of resilient varieties.
The heightened concern relies on the impacts of global climate change on the intricate issue of plant water stress. The erratic shifts in precipitation patterns, coupled with increased temperatures, and the consequential elevation in evapotranspiration rates, collectively underscore a pivotal facet in the challenge of worsening drought conditions. These transformative alterations directly undermine the availability of water availability for plant growth, thereby magnifying the severity of drought scenarios. Consequently, this disruption results in a reduction in soil moisture content, while concurrently impeding the crucial uptake of water by the roots of plants. Furthermore, the heightened frequency and intensity of prolonged heatwaves, along with the changing climate, accelerate the evaporation process, which in turn intensifies the propensity for rapid water depletion, profoundly impacting the physiological well-being of plants.
The occurrence of drought in tropical regions, particularly in equatorial countries holds significant implications for agricultural productivity and necessitates adaptations in planting strategies. Bänziger et al. (2000) thoroughly investigated physical factors within the environment that exert stress on the cultivation of maize. Furthermore, Monneveux et al. (2008) underline a substantial reduction in maize productivity, ranging from 45% to 75%, due to water stress during the critical flowering period extending up to 2 weeks after silking. Moreover, in prolonged stress conditions, vulnerable genotypes exhibit an inability to produce viable seeds. The pursuit of drought-tolerant varieties demands a comprehensive plant breeding approach. It is noteworthy that the implementation of multi-location trials constitutes an essential prologue to more comprehensive field adaptation trials (Azrai et al., 2023).
As the dryland areas commonly exhibit limited water resources and low soil fertility, the process of genotype selection, usually aimed at identifying maize genotypes capable in efficient water uptake, holds significant importance within the realm of plant breeding, particularly for enhancing drought tolerance traits. The improvement of water use efficiency, achievable through the enhancement of plant water status, serves to facilitate the optimal distribution of assimilates and the improve kernel formation. Consequently, subjecting the maize genotypes assigned for development to rigorous pre-selection under water stress conditions before releasing as superior cultivars becomes imperative. Hence, the process of selecting for drought tolerance presents a multifaceted challenge, given the intricate interactions between genotypes and their environment.
Improving the selection efficiency of drought-tolerant maize genotypes involves directly observing their performance under water stress. Evaluating agronomic, morphological, and physiological traits linked to the plant’s drought tolerance greatly assists in enhancing adaptation (Bänziger et al., 2000). A thorough examination of agronomic, morphological, and physiological attributes across various hybrid maize genotypes grown under intense water stress has the potential to correlate with the resulting grain yield. Numerous techniques have been utilized to assess the superiority of genotype either based on single drought-tolerant traits, including the AMMI and GGE biplot methods. However, the imperative arises to account for multiple traits due to preferences expanding beyond a single factor. To tackle this challenge, Olivoto and Nardino (2021) introduced the MGIDI index for concurrent genotype selection grounded in multiple traits.
Cutting-edge computational technologies such as high-performance computing, specialized bioinformatics tools, and advancements in artificial intelligence (AI) and ML techniques are presently employed for the comprehensive analysis of intricate datasets. These innovative approaches empower breeders to derive significant and insightful conclusions from their data, thereby enabling the formulation of enhanced and streamlined breeding strategies. Furthermore, these techniques contribute to an enhanced comprehension of the underlying genetic foundations governing plant traits (Yoosefzadeh-Najafabadi et al., 2021b). Numerous strategies encompassing ML and deep learning, have emerged for exploring genotype stability under diverse abiotic and biotic stresses. These approaches extent various abiotic stress scenarios. Cheng et al. (2021) introduced a systematic feature reduction technique within ML, substantially boosting predictive precision in gene-to-trait models. Singh et al. (2023) incorporate modern image acquisition and ML to identify key determinants of biomass accumulation, encompassing both architectural and physiological traits. Meanwhile, the use of leaf reflectance has enabled the extraction of plant water status indicators via near-infrared and short-wave infrared canopy emissions. Other studies have capitalized on the potential of ensemble ML and deep learning models in various agricultural contexts, such as rapid maize parental line identification and classification using stacking ensemble ML (Aqil et al., 2022), accurate soybean yield and biomass estimation through hyperspectral vegetation indices (Yoosefzadeh-Najafabadi et al., 2021a), genotype classification under varying light conditions (Sakeef et al., 2023), leaf chlorophyll status assessment based on SPAD readings, encompassing different nitrogen levels (Zainuddin and Aqil, 2021), and successful prediction of grain yield by utilizing these techniques across a range of environmental and phenological data (Srivastava et al., 2022). These studies exemplify the adaptability of ML and deep learning models in evaluating the performance of genotypes and making yield adjustments under a wide range of abiotic stress conditions.
Biotic stress typically become apparent during the initial stages of plant growth, posing intricate challenges concerning their manual differentiation and necessitating a substantial investment of time. Consequently, the integration of artificial intelligence (AI) and ML has yielded significant advancements in the detection and management of agricultural diseases. Noteworthy studies encompass the utilization of a novel convolution model featuring modified rectified linear unit activation for the identification of diseases in cucumber plants (Agarwal et al., 2021), underscoring the considerable potential of AI in this domain. A deeper exploration of machine vision techniques to detect diseases in corn leaves (Austria et al., 2022), coupled with the use of MobileNet for maize seedling and weed detection (Cheng et al., 2021), as well as the innovative hybridization of ResNet and YOLO for paddy leaf disease recognition (Ganesan and Chinnappan, 2022), have provided invaluable insights. Roy and Bhaduri (2021) contributed a sophisticated deep learning model capable of multi-class disease detection. The efficacy of AI in managing crop diseases extends to maize diseases, as evidenced by the boosted framework (Gokulnath and Devi, 2020). Another noteworthy contribution by Sharma et al. (2020) involves a method that combines ML techniques with image preprocessing to classify plant diseases. Furthermore, the complex field scenarios of maize leaf blight detection have been effectively addressed through the utilization of deep learning techniques (Sun et al., 2020), and optimized neural network tailored for the identification of diseases in maize leaves (Waheed et al., 2020).
The objective of the research was to enhance the predictive accuracy of the grain yield and stress tolerance index of maize hybrids through fine-tuning via ensembles machine learning and genetic algorithms. Furthermore, the study compared the effectiveness of ML with MGIDI to select the best-performing hybrid candidates under normal and drought conditions.
The genetic material involved in the drought trials included a collection of 31 single cross hybrids, designated as H01 (DTH01) through H31 (DTH31), along with four commercial hybrids as checks, namely Bima14 (N51/MR 15), Bisi 18 (a commercial hybrid of BISI International), P 31 (hybrid of PT. Dupont Indonesia) and Pertiwi 3 (Supplementary Table S1). The N51 line was derived from Recombinant Inbred Lines (RILs) of the Syngenta genotype population through bulk selfing on a plant-to-plant basis, emphasizing drought tolerance. These hybridizations were undertaken with the intent of generating a wide spectrum of genotypes, exhibiting the potential for enhanced drought resilience. Meanwhile, the choice of the four commercial varieties was established in their proven ability to exhibit good resilience and achieve higher grain yields than other commercial varieties during cultivation in dry seasons in Indonesia.
For the assessment of potential drought-tolerant hybrid candidates during the cropping seasons of 2020–2021, three distinct locations were selected. These sites included the Maros experimental station/E1 (East longitude: 119°50; South longitude: −5°31), the Bajeng experimental station/E2 (East longitude: 119°57; South longitude: −5°33), and the Bontobili experimental station/E3 (East longitude: 119°58; South longitude: −5°01). These locations represented a diverse range of soil types, including Ultisols, Oxisols, and Inceptisols. Ultisols are rich in nutrients and suitable for farming while Oxisols are less fertile but offer good drainage and ecological importance. At the Maros site, the soil is classified as Inceptisols with varying fertility levels, ranging from low to moderate. The presence of these different soil types highlights the importance of considering soil factors when evaluating maize growth, as they significantly impact drought tolerance.
At each experimental site, there were two experimental plots: one with normal treatment involving regular plant watering and the other subjected to drought treatment, where irrigation halted at 40 DAP. The experimental plots followed a randomized complete block design (RCBD) with three replications. Plots were organized into four rows, each spanning 5 meters, with plant spacing set at 70 cm between rows and 20 cm within rows, and with 1 seed per hole. Fertilization was done twice, an initial application of 350 kg of NPK (15:15:15) and 150 kg of urea (46% N) per ha at 10 days after planting (DAP), followed by a second application of urea of 200 kg/ha at 30 DAP. Throughout the trial, negligible insect infestations were observed. Harvesting was done when plants reached physiological maturity (100–110 DAP). Maize cobs were harvested within a five-meter section from the middle rows of each replicate to ensure a representative sample for calculating yields. The collected ears from each plot were processed for yield component analysis. The moisture content of the maize kernels was measured using a digital grain moisture tester with 0.01 g resolution.
All tested maize genotypes under water stress conditions were subjected to the CIMMYT procedure (Bänziger et al., 2000), which involves imposing water stress on the plants from the flowering stage (50 DAP) until the milk-ripe stage (75 DAP). The water stress treatment was applied by withholding water supply when the plants were 40 DAP, causing the plants to experience water stress leading up to flowering at 50 DAP. This stress continued until the milky to kernel hardening stages (80 DAP), after which the plants were irrigated again. Observations encompassed a range of agronomic traits and yield components. From each plot, a random selection sample hybrid was gathered for analysis. The observed traits were: (1) plant height (PH), (2) ear height (EH), (3) stem diameter (SD), (4) leaf area (LA), (5) day to tasseling (DT), (6) day to silking (DS), (7) anthesis silking interval (ASI), (8) leaf angle (LAG), (9) SPAD, (10) husk cover (HC), (11) ear length (EL), (12) ear diameter (ED), (13) number of rows per cob (NR), (14) number of kernels per row (NKR), (15) 1,000 kernel weight (1,000 KW), (16) shelling percentage (SP), and (17) grain yield (GY). Both the stress tolerance index (STI) (Fernandez, 1992) and tolerance index (TOL) (Rosielle and Hamblin, 1981) were computed as metrics to quantify drought tolerance. These indices were calculated using the formulas (1) and (2) as follows:
Where Ys is the mean yield of each genotype under water stress, Yp is the mean yield of each genotype under normal condition, Ȳp is the grand mean of yield under normal conditions.
Feature selection (FS) was used to identify important attributes, eliminating irrelevant and redundant ones. This process allows for the creation of an optimized subset of features that remains unchanged by transformations, thereby improving the clarity and interpretability of learning models based on the selected feature subset. The underlying principle behind adopting feature selection includes the reduction of storage requirements and execution time, data dimensionality, and addressing concerns related to overfitting, thus promoting the refinement of model generalization. Consequently, feature selection improves the potential for enhancing model performance (Akhiat et al., 2019).
Three feature selection methods were utilized to assess the relationship between input variables and grain yield, namely Univariate Feature Selection (F-test), Recursive Feature Elimination (RFE), and LASSO (Least Absolute Shrinkage and Selection Operator) Regression. The study analyzed 17 agronomic parameters and yield components, evaluating model performance through R2 key metric. In the comparative analysis, LASSO and F-test demonstrated superior performance, boasting an R2 of 0.81, which outperformed the RFE. LASSO regression is additionally favored due to its utilization of fewer model inputs, thereby effectively reducing data dimensionality. The excellence of LASSO’s feature selection lay in its distinctive regularization methodology, adeptly penalizing substantial coefficients and compelling some to precisely zero, thereby eliminating irrelevant variables while maintaining model efficacy. LASSO model have also been applied as a powerful tools for reducing data dimensionality in crop yield prediction (Jhajharia et al., 2023). Feature selection using LASSO regression has selected 10 out of 16 agronomy and yield component parameters to predict grain yield and STI. The 10 selected variables as model inputs include ear height (EH), leaf area (LA), days to silking (DS), leaf angle (SD), SPAD, ear length (EL), ear diameter (ED), number of rows per cob (NR), number of kernels per row (NKR), and 1,000-kernel weight (1,000 KW). This formulation led to the refined model depicting grain yield and stress tolerance index, encapsulated by the function f (EH, LA, DS, SD, SPAD, EL, ED, NR, NKR, 1,000 KW). We examine the same model function to predict the stress tolerance index of each hybrid grown under normal and drought conditions.
The GA is a stochastic optimization method that operates independently of derivatives, derives its inspiration from natural selection and biological evolution. It showcases superior performance in comparison to other optimization approaches, spanning multiple dimensions. GA possesses a reduced susceptibility to being trapped in local minima. Notably, GA functions as a population-centric computational model, deeply rooted in principles of population genetics. It’s recognized primarily as a function optimizer, having displayed its efficacy as a robust global optimization technique, especially adept at handling multi-modal and non-continuous processes.
Figure 1 illustrates a representation of the novel hybrid algorithms, which are rooted in the SVM-GA, KNN-GA, and RF-GA frameworks. This conceptual model delineates an amalgamated approach to artificial intelligence, fusing together various ML techniques. These chromosomes experience selection, crossover (mixing), and mutation processes to generate new solution generations. The effectiveness of each solution is assessed using a pre-defined goal. As generations progress, the algorithm moves closer to optimal or nearly optimal parameter values, allowing effective parameter adjustment for different uses. The synergy is fortified by the optimization of network hyperparameters through the utilization of GA. The hyperparameters are meticulously fine-tuned by generating a population of candidate solutions (sets of hyperparameters), assessing their performance on a testing set and subsequently evolving the population across multiple generations to discover the optimal set of hyperparameters. Afterward, the ML technique, guided by GA’s processes, undertakes the training of the network.
Another predictive model that has gained popularity recently is ensemble ML. In this approach, datasets undergo a process of selecting base models followed by an ensuing meta-modelling step prior to generating predictions. The diagram of the ensemble ML process for yield prediction is shown in Figure 2. The diagram begins with thorough data preprocessing, including standardization. Afterward, the datasets are randomly divided into training and testing sets. The process of the training phase involves three specific ML models: SVM, KNN, and RF. To enhance parameter search efficiency and improve model predictions, a comprehensive search process fine-tunes the settings of each model using grid search. The algorithm generates three foundational base models, which are then input into the meta-model (SVM, KNN, and RF). This meta-model combines the separate predictions made by the SVM, KNN, and RF models mentioned earlier, resulting in an overall collective prediction. This combined prediction is applied to the testing dataset, generating yield predictions that are compared against actual yield values.
During the model running, 70% of the dataset was allocated for model training, while the remaining 30% was used for generating predictions. The accuracy of these predictions is evaluated using common metrics like R-squared (R2), Mean Absolute Error (MAE), Root Mean Square Error (RMSE), and Normalized Root Mean Square Error (n-RMSE), n-RMSE percentages falling within the 0%–10% range are considered as “excellent,” those ranging from 10%–20% are classified as “good,” values between 20%–30% are categorized as “fair,” and any percentages exceeding 30% are considered “poor” (Jamieson et al., 1991; Singh et al., 2023).
Nine combinations of ML and optimization models were subjected to training and prediction using cloud-based software services, GPU-based Google Colab. Nevertheless, the foundational structure of the models relies on comprehensive agronomy and yield component information for maize hybrids under both normal and drought treatment conditions. This data serves as a critical input, contributing to the accurate generation of predictions for grain yield and stress tolerance index. To construct predictive models for both grain yield and stress tolerance index, three classical ML algorithms, SVM, KNN and RF were employed, alongside traditional ML optimization using Genetic Algorithms (GA). These methods were coupled with ensemble techniques, where each SVM, KNN, and RF ML model was considered as a meta-model. In the search of enhancing predictive performance, the thoughtful adjustment of hyperparameters was integrated (Supplementary Table S2). Cross-validation was performed to some ML models to enable the training of higher-level ML models. To evaluate the effectiveness of all ML models, a comprehensive 10-fold cross-validation strategy was adopted.
The selection and manipulation of parameters for the SVM model involves a suite of hyperparameters, including the regularization parameter, kernel function and coefficient, as well as auxiliary parameters. Important parameters for KNN encompass leaf size, the number of neighbors, and the weighting scheme, while parameter configuration of RF model include n estimators for tree count, control over tree depth, and the quantity of features considered for optimal partitioning. The choice of a maximum of 50 iterations establishes a finite yet sufficient exploration horizon, balancing computational efficiency with the pursuit of optimal solutions. The modest population size of 10 individuals per generation, coupled with a 0.1 mutation probability, fosters exploration within the solution landscape. The elitism ratio of 0.01, along with the parents’ portion of 0.3 and a crossover probability of 0.5 was set to optimize the adjustment of hyperparameters (Supplementary Table S2).
The study also employed an MGIDI approach to discern the most appropriate genotypes, leveraging multi trait data (Olivoto and Nardino, 2021). Initially, the scaling process was implemented for each individual trait under consideration. Subsequently, a Factor Analysis (FA) was performed to facilitate the reduction of data dimensions and reveal underlying relationship structures. Finally, the computation of the MGIDI was done by quantifying the Euclidean distance between genotype scores and an ideotype defined. This index was evaluated through the application of the subsequent formula:
The score of the ith genotype in the jth factor (i = 1, 2, …, t; j = 1, 2, …, f) is represented by , where t and f denote the number of genotypes and factors, respectively. The score of the jth trait for the ideal genotype is represented by . The genotype with the lowest MGIDI value is considered to be more closely aligned with the ideal genotype, encapsulating the desired attributes for all evaluated traits. In the genotype selection procedure, selection discrepancies were computed for all traits while maintaining a selection intensity of 30%.
The ML script was executed in Google Colaboratory (Colab) environment, which is available at https://colab.research.google.com/?utm_source=scs-index. During the process of executing ML algorithms within Google Colab, pivotal libraries, i.e., scikit-learn KNN and RF, as well as SVM implementation from sklearn.svm, were employed. To expedite computations, Colab provides GPU acceleration, enhancing the speed of model training and tuning. Moreover, GridSearchCV from the scikit-learn library was employed to optimize parameters, enabling a thorough exploration of hyperparameters for each individual algorithm. This ensemble/meta-learning process adeptly harnesses Colab’s computational resources, libraries, and GPU acceleration to achieve refined predictions by amalgamating the strengths of KNN, RF, and SVM algorithms. In addition, genotype-versus-environment plots and MGIDI index calculations were generated using the “gamem” and “mgidi” functions of the “metan” package (Olivoto et al., 2019).
Based on visual inspection and the analysis of grain yield and other agronomic datasets, it has been determined that trial plots subjected to normal irrigation treatment produced a more favorable conditions for obtaining yield potency, as contrasting to plots subjected to drought treatments. The combined analysis of variance revealed the distinct influence of the environment on grain yield and agronomic parameters of maize genotypes (Supplementary Table S3). The analysis involved five environments (three normal and two drought), with the environment effect separated into drought effect (df = 1) and location within the drought effect (df = 4). The effect of the drought treatment was evident across all observed traits, excluding the husk cover trait. Similarly, the effect of location within the drought treatment was significant for all 17 observed traits, except husk cover, indicating that variations in trial location have an impact on genotype performance. The genotype’s effect was notable for all observed traits, except for SPAD, ED, and 1,000 KW traits, underscoring the phenotypic diversity of the hybrids in terms of agronomic, yield components, and grain yield. The interaction between genotype and location was significant for all observed traits. Additionally, the interaction between genotype and drought was significant for all traits, except husk cover and anthesis-to-silking interval. When examining the grain yields of hybrid maize, a notable finding exhibits significantly greater productivity in plots that receive regular irrigation. Figure 3 depicts the box plot graph of grain yield variation under normal and drought conditions (n = 525 for all environments).
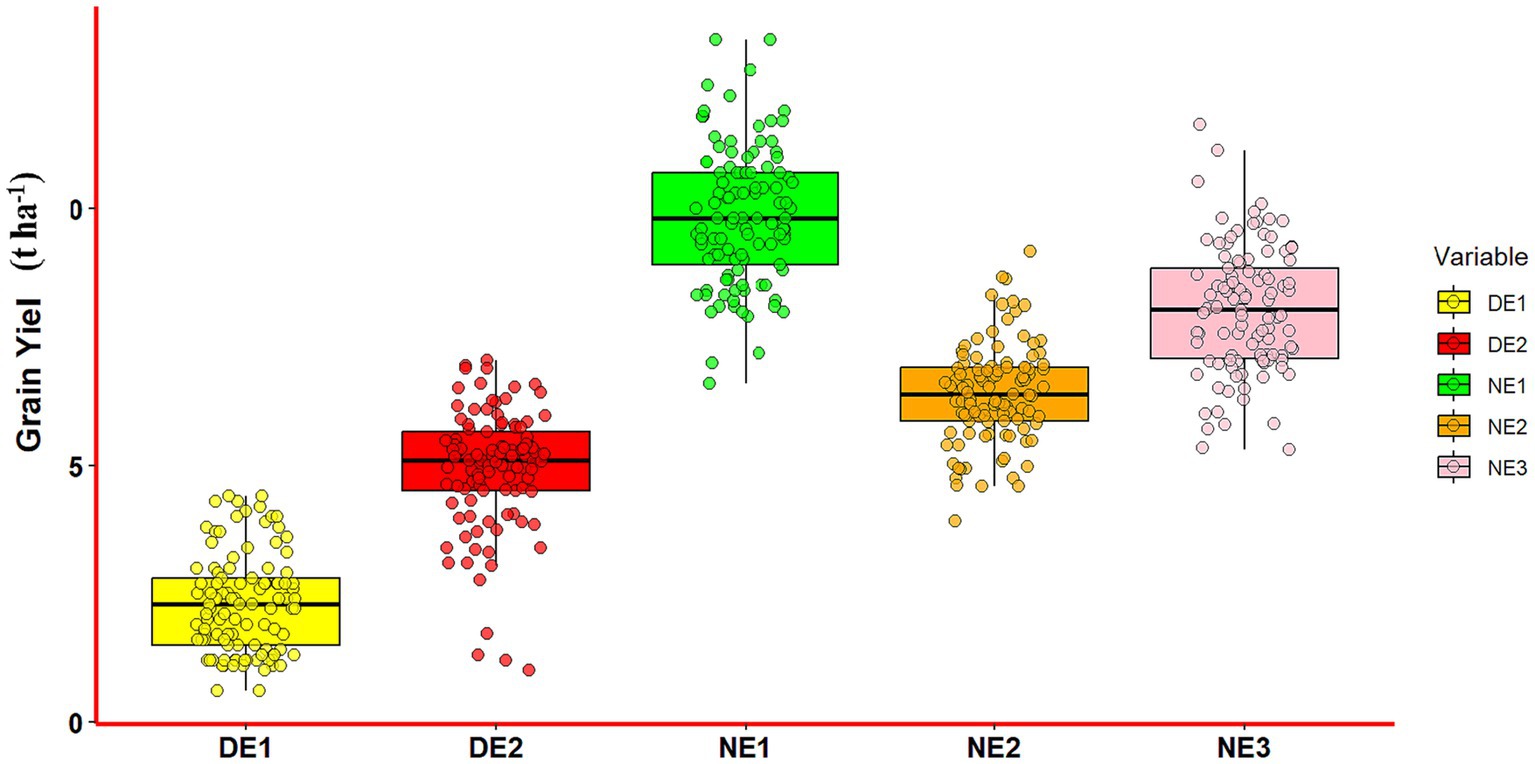
Figure 3. Box plots depicting the grain yield variation under normal and drought conditions (n = 105 for each environment, n = 525 for all environments, DE1 = drought E1, DE2 = drought E2, DE3 (data not available), NE1 = normal E1, NE2 = normal E2, NE3 = normal E3).
Figure 3 indicated a significant difference in maize yield across the entire plot for the normal/irrigated and drought plots. Several genotypes consistently produced grain yields comparable to the best of commercial variety (Bisi 18) under both irrigated and drought conditions. The PH and EH traits of maize genotypes exhibited a significant increase under normal conditions compared to drought conditions. Physiological traits, such as chlorophyll content (SPAD), showed significantly higher levels under normal conditions compared to drought conditions. This finding affirms that drought reduces chlorophyll concentration in maize leaves, leading to a reduction in nitrogen concentration (Kira et al., 2016; Széles et al., 2023). A significant reduction was also found in LA trait under drought condition, suggesting the plant’s response to water scarcity by adjusting its foliage area. Araus et al. (2021) found that the inability to maintain a larger area of green leaves under water stress conditions results in less sunlight being utilized for photosynthesis, thereby reducing yields. Several maize varieties exhibit leaf curling as a response to water stress, leading to a reduction in the photosynthetically active leaf extension of the plant. Additionally, it aids in reducing dehydration and lowering water consumption, especially during periods of high evaporation (Jordan, 1983). In addition to morphological aspect, water management such as the timing and intensity of water stress also have significant effects on maize growth (Çakir, 2004). Substantial reductions were also observed in number of kernel rows (NR), number of kernels per row (NKR), and 1,000 kernel weight (1,000 KW) under drought conditions, which might be indicative of the plant’s allocation of resources towards kernel development in the face of water scarcity.
A heatmap of correlation analyses was conducted among 17 traits to identify those related to grain yield, as illustrated in Figure 4. Particularly, a significant positive correlation emerged between yield plant height and ear height (r = 0.66***), SPAD reading with leaf area (r = −0.43**), including highly correlated between grain yield with EL (0.75***), ED (0.70***), NKR (0.71***), EH (0.48**), LA (−0.41**), NR (0.41**), and SP (0.39**). Plants subjected to drought stress exhibit the induction of reactive oxygen species (ROS) within cells, resulting in damage to chloroplasts and cell membranes. This leads to the rapid deterioration of chloroplasts, crucial sites for photosynthesis in leaves. The compromised chloroplasts diminish the maize plant’s capacity to produce energy and biomass (Sachdev et al., 2021). The impact of drought stress on chlorophyll is quantified by lower leaf chlorophyll meter (SPAD) values. Correlation analysis reveals a statistically significant correlation (r = 0.31**) between chlorophyll meter values and yield. This underscores the notion that maize plants capable of mitigating chloroplast damage under drought conditions yield higher grain quantities. Drought-tolerant plants adept at safeguarding chloroplast structures from damage enable leaves to sustain their greenness during drought stress, optimizing sunlight utilization for photosynthesis and consequently achieving elevated yields (Zahra et al., 2023).
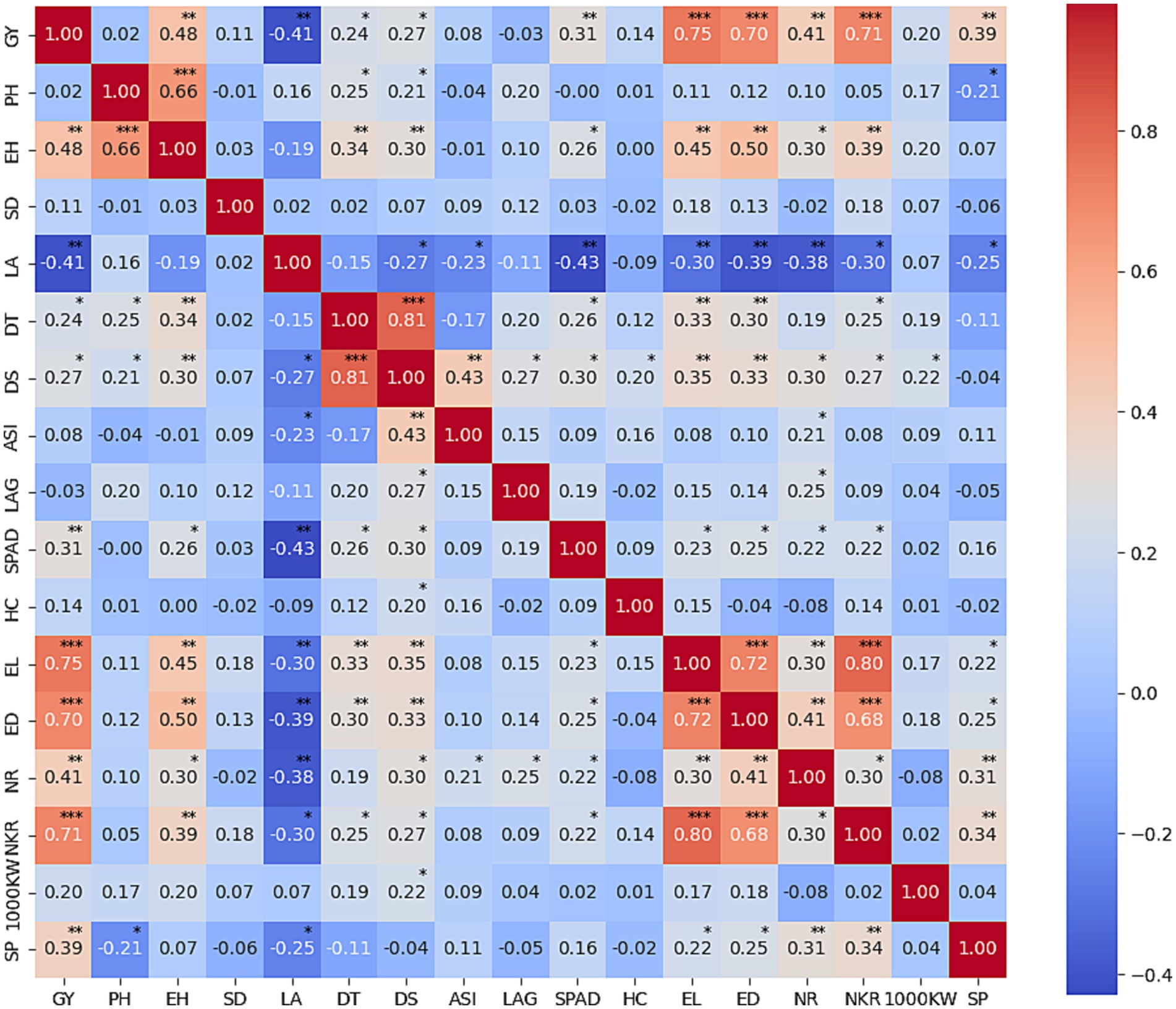
Figure 4. Heatmap of correlation analysis among agronomic and yield components under both normal and drought conditions.
The maize plant’s ability to shield leaf structures from drought-induced damage exhibits a significant correlation (r = −0.43**) with leaf angle. Leaf angle, an imperative agronomic trait in maize, influences the plant’s adaptability to drought conditions (Chotewutmontri and Barkan, 2016; Shao et al., 2016). A reduced leaf angle mitigates chloroplast exposure to intense light, preventing chloroplast damage (Zhang et al., 2022). Furthermore, a smaller leaf angle enhances planting density and maize plant biomass yield by facilitating greater light penetration through the canopy to reach lower leaves. Leaf angle demonstrates a noteworthy positive correlation with yield and yield components, with correlation coefficients ranging from 0.30 to 0.39. Therefore, manipulating leaf angles through selective breeding practices emerges as a pertinent strategy for augmenting the productivity and resilience of maize plants across diverse environmental conditions.
Yield components, such as cob length, cob diameter, seed row number, and seed number per row, manifest positive correlations with maize yield under drought stress conditions, featuring correlation coefficients ranging from 0.41** to 0.75***. These characteristics serve as reliable indicators of drought tolerance and valuable criteria for the selection of maize varieties to enhance yield under conditions of limited water availability.
The research integrates various foundational models to enhance grain yield predictive ability. The ensemble SVM, KNN, and RF models are structured with a meta-model at the helm, which employs SVM, KNN, and RF models as base models, respectively. Results of model performances during the training and prediction stages are shown in Table 1. The evaluation of the examined ML models centered around minimizing error. Moreover, model performance was assessed by minimizing the n-RMSE, which approaches below 10% for strong performance. R2 indicated perfect correlation at 1 and no correlation at 0.
Table 1 provides metric of the individual models, with both the SVM and KNN models delivering commendable results—each exhibiting a R2 value of 0.90 on the training set. However, in the case of SVM, there is a perceptible reduction in performance on the testing dataset (R2 = 0.69), potentially suggesting a challenge in generalization. On the other hand, the KNN model maintains a relatively high R2 of 0.77 on the testing set, indicating a robust balance between predictive accuracy and generalization. The RF model emerges as a standout performer, consistently achieving an R2 of 0.97 and 0.81 on both the training and testing datasets. This striking consistency underscores RF’s ability to generalize effectively, capturing complex relationships within the data. Sahu et al. (2017) reported that RF showed strong generalization capabilities for predicting grain yields in India.
The integration of RF with GA resulted in a significant enhancement in predicting grain yields, leading to an accuracy increase from 0.81 to 0.91 on testing datasets. Ensemble models, adeptly integrating predictions from multiple individual models, consistently present optimal results. These ensembles meta-model of SVM, KNN, and RF exhibit robustly high R2 values across both training and testing datasets. Utilizing an ensemble approach that incorporated three ML models as meta-models consistently resulted in high accuracy, with an R2 consistently surpassing 0.91 (Figure 5). The excellence of these ensemble models might be rooted in the diversity of underlying models within the ensemble itself, contributing to a more thorough exploration of the feature landscape.
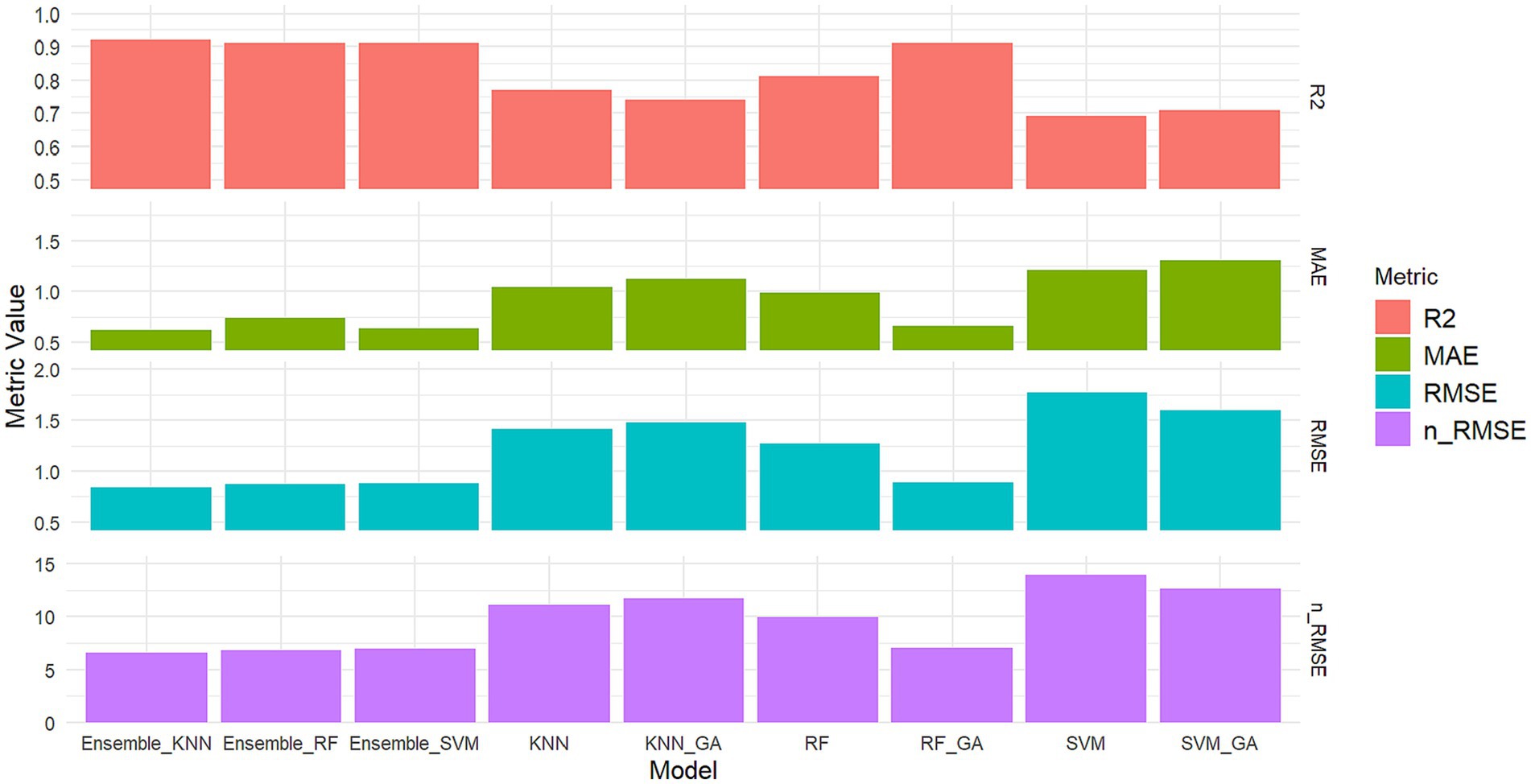
Figure 5. Comparative metrics for the nine examined ensembles and GA-ML models in grain yield prediction.
The analysis of the normalized n-RMSE values reveals distinctive performance characteristics among the considered crop yield prediction models. Ensemble modeling, combining predictions from KNN, RF, and SVM, stands out as a paradigm of excellence, showcasing an impressive n-RMSE range of 6.60% to 6.93%, classified as excellent prediction. This ensemble approach capitalizes on the diverse strengths of individual algorithms, resulting in a highly accurate and robust predictive model. Similarly, the standalone RF model and its optimized counterpart, RF + GA, exhibit notable excellence with n-RMSE values of 9.91% and 7.01%, respectively. These results underscore the significant impact of genetic algorithms in refining their predictive capabilities. In contrast, SVM emerges as the least favorable model for grain yield prediction, registering the highest n-RMSE value of 13.91%.
Comparative metrics for assessing model robustness in grain yield prediction in terms of R2, MAE, RMSE and n-RMSE are shown in Figure 5. Ensemble algorithms demonstrated superior performance in crop yield prediction (Ahmed, 2023), while RF is the optimized algorithm for accurately forecasting maize yields at the county level through the integration of diverse data sources (Pham and Olafsson, 2018). Aqil et al. (2022) reported that incorporating ensemble machine learning may enhance the accuracy of classifying maize plants. Figure 6 depicts a simple scatter plot presenting the correlation between observed and predicted grain yields across the nine analyzed models. These results underscore the significant impact of optimizing models via GA and ensemble to enhance their predictive capabilities.
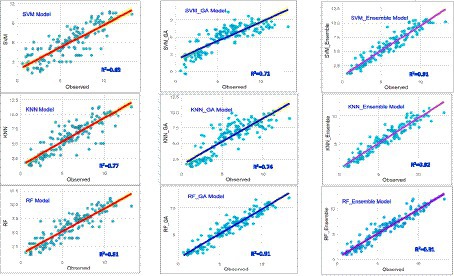
Figure 6. Scatter plot of the observed vs. predicted grain yield for nine examined ensemble models and GA-ML models.
The performance of the developed GA and ensemble ML models was also evaluated based on a quantitative assessment. Table 2 presents a similar assessment of model performance metrics for predicting the STI of genotypes cultivated in both irrigated and drought fields. Among the individual models, the RF model exhibited remarkable performance in both the training and testing phases, achieving high R2 values of 0.95 and 0.78, respectively. This indicates that the RF model captured a substantial portion of the variability in stress tolerance index predictions. The Ensemble KNN also demonstrated strong predictive accuracy, with R2 values of 0.96 and 0.82 for training and testing, respectively. This suggests that the ensemble approach effectively harnessed the collective strengths of individual KNN models to enhance predictive accuracy and generalization. Additionally, the ensemble SVM and RF models yielded competitive results with R2 values of 0.94 for both training and testing.
The analysis of the normalized n-RMSE values for STI indicated that none of the examined models had n-RMSE values falling below 10%, signifying reduced accuracy in predictive ability, although they remain within the “good” category with n-RMSE <20%. Ensemble KNN produced the lowest n-RMSE of 14.12% and is classified as a good prediction. The standalone SVM model produced the highest n-RMSE of 16.81%. The diminished accuracy in the predictive model is attributed to its failure to capture the underlying patterns or relationships in the data, resulting in larger prediction errors.
These ensemble methods leverage diverse models to improve predictive performance. The remaining models, including SVM, KNN, and their variants with GA optimization, displayed slightly lower R2 values in comparison. These results emphasize that ensemble techniques, particularly the Ensemble KNN and Ensemble RF models, stand out as robust approaches for predicting the stress tolerance index. The high R2 values achieved by these models indicate their capability to effectively capture and explain the variations in stress tolerance, making them valuable tools for accurate stress tolerance predictions. Comparative metrics for assessing model robustness in stress tolerance index prediction in terms of R2, MAE, RMSE and n-RMSE are shown in Figure 7. Aqil et al. (2022) reported that ensemble ML perform comparably with deep learning in the classification of maize tassels, reinforcing the efficacy of ensemble approaches in classification problems. The criteria for identifying a drought-tolerant maize genotype, as determined by STI value, involve a direct correlation: the greater the STI value of a maize genotype, the higher its productivity under stress conditions, including drought level. These findings highlight the potential of STI as a selective criterion for identifying maize genotypes capable of achieving high yields under stress conditions. Moradi et al. (2012) similarly reported that utilizing STI for selecting hybrid maize varieties can effectively identify tolerant genotypes with the potential for high yields in stress-prone environments.
The average yield in drought displayed a significantly positive correlation with yields in normal conditions, indicating that higher STI values corresponded to greater genotype tolerance. A three-dimensional plot was employed to visually represent the mean yield of hybrids under both irrigated and drought treatment (Figure 8). The correlation coefficient between Yp and TOL is 0.70, suggesting a moderately positive relationship, while the correlation between Ys and TOL is −0.51, indicating a moderate negative association. This implies that as Yp increases, there is a tendency for TOL to increase, while as Ys increases, TOL tends to decrease. These findings suggest that the STI and TOL could be incorporated into the selection criteria for identifying high-yielding genotypes under both normal and drought conditions. Among the test hybrids, 10 hybrids exhibited high STI values, including H33, H21, H17, H19, H14, H13, H10, H35, H30, and H06, with STI values ranging from 0.7 to 1.10. This suggests that these hybrids demonstrated greater tolerance to water stress. On the other hand, the two commercial checks, H32 and H34 displayed heightened vulnerability to drought and had lower grain yield than test varieties.
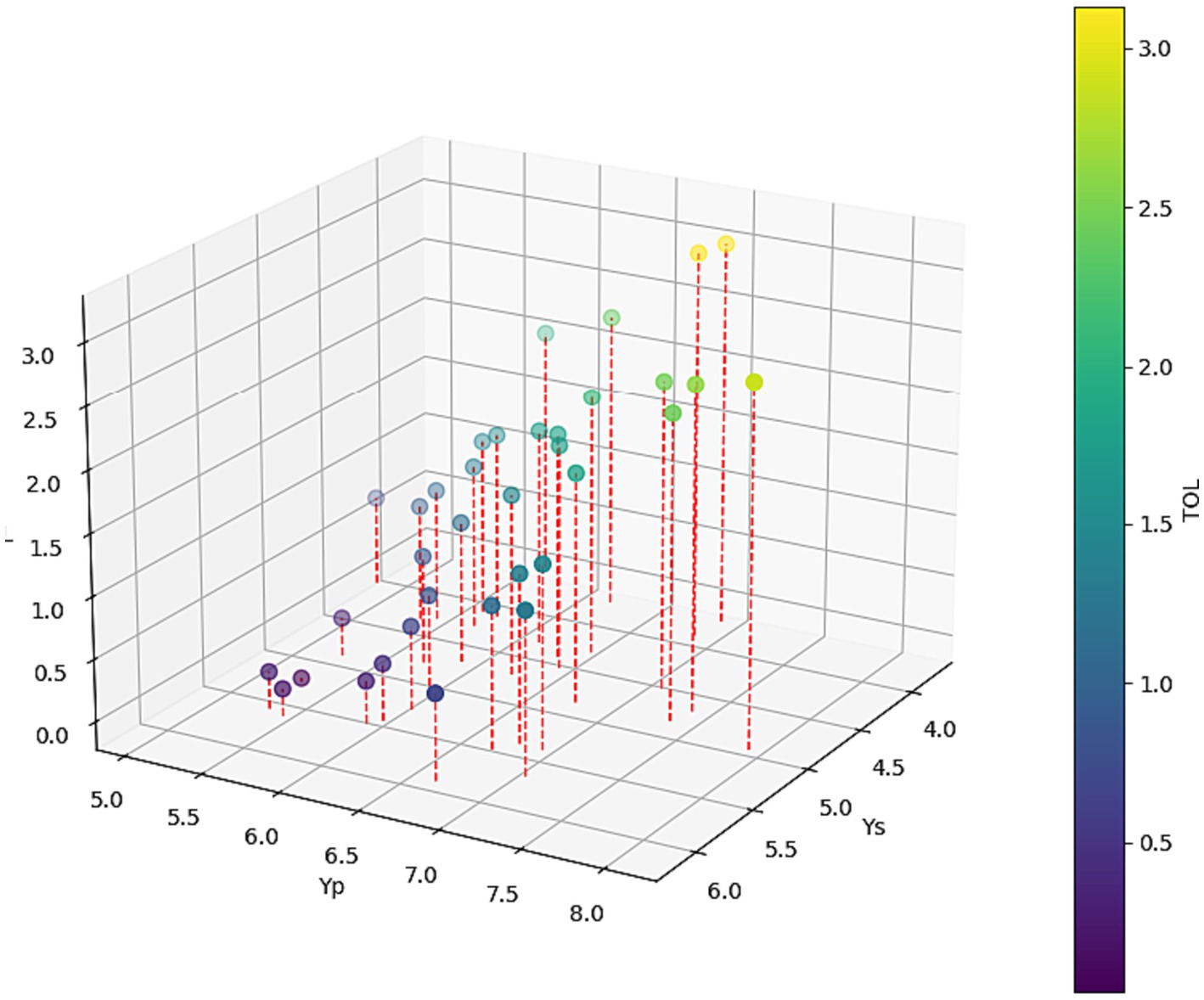
Figure 8. Three-dimensional plot depicting the mean yield for each hybrid under both drought and normal conditions, accompanied by their respective tolerance levels.
A comprehensive multi-trait methodology was implemented to ascertain the optimal hybrid pairings, with due consideration not only to maize yield but also covering various plant aspects and yield attributes. The genotype selection process included an initial evaluation of traits sensitive to multicollinearity. Restricted Maximum Likelihood (REM)/Best Linear Unbiased Prediction (BLUP) was used to calculate variance aspects within a mixed-effects framework, with genotype considered as a random effect and replication as a fixed effect. The likelihood ratio test results revealed that all assessed traits demonstrated a statistically significant genotype effect, surpassing the significance threshold of p < 0.05. Additionally, a more in-depth examination of these traits using the REML/BLUP approach revealed six principal components, which both accounted for 93.67% of the total variation.
This indicates that the six PC effectively captured a considerable degree of diversity within the characteristics. Similarly, the communalities of the variables ranged from 0.23 for the shelling percentage to 0.79 for trait plant height, with average uniqueness’s is 0.66. These figures imply that a substantial proportion of each variable’s variability was elucidated by those components. The assessment of precision in determining the mean trait value reveals significant genetic diversity among the hybrid genotypes, as evidenced by an accuracy level surpassing 0.86. This remarkable level of accuracy enables precise estimation of the genetic trait value. To maintain robust interpretative power and simplify data complexity, the nine analyzed traits were categorized into two factors, designated as FA. The factorial loadings and communalities resulting from factor analysis with varimax rotation are presented in Table 3. Communality refers to the shared characteristics or traits among different genotypes, while average uniqueness indicates the distinctive features or traits specific to each genotype. FA1 is associated with attributes such as plant height, ear height, days to tasseling, and shelling percentage, whereas FA2 is linked to characteristics including grain yield, leaf area, 1,000 kernel weight, ear length, and ear diameter. Factor values offer insights into the degree of each trait’s relationship with the underlying factor, with higher loadings indicating stronger associations.
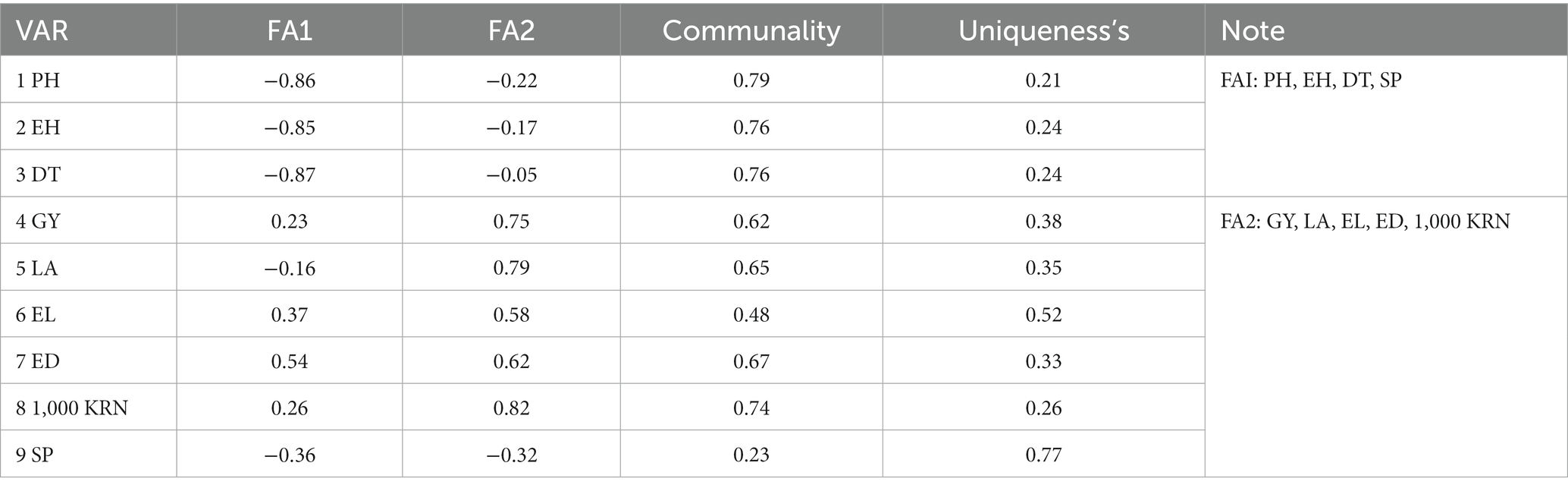
Table 3. The factorial loadings and communalities acquired through varimax rotation in the factor analysis.
FA1 prominently encapsulates traits relating to plant morphology and developmental attributes, exemplified by the involvement of variables such as plant height (PH), ear height (EH), days to tasseling (DT), and shelling percentage (SP). Notably, the distinct negative loadings exhibited by PH, EH, DT, and SP on FA1exhibit strong negative loadings (−0.86, −0.85, −0.87, and −0.36, respectively). This indicates that these variables are inversely related to FA1; when FA1 increases, these variables tend to decrease. Furthermore, FA1 markedly high communalities (0.79, 0.76, 0.76, and 0.23, respectively), underscore the robust interconnections among these traits, highlighting their mutual reliance on common underlying factors within the FA1 concept. FA2 predominantly encapsulates traits associated with crop yield and physical attributes, encompassing key variables such as grain yield (GY), leaf area (LA), ear length (EL), ear diameter (ED), and 1,000 kernel weight (1,000 KRN). FA2 display positive loadings (ranging from 0.58 to 0.82). This signifies a positive relationship between these variables and FA2, suggesting that when FA2 scores increase, these variables tend to increase as well. These strong loadings indicate that these variables are closely linked to FA2 and contribute substantially to its definition. These intricate findings offer a nuanced perspective on the intricate architecture of these plant traits, affording researchers valuable insights into potential trait groupings and interrelationships. This newfound understanding paves the way for more refined and targeted research endeavors, ultimately enhancing the efficacy and precision of agricultural practices and crop enhancement strategies.
The MGIDI index has been employed for the comprehensive evaluation of all measured traits. The procedure involves normalizing traits through BLUP to ascertain the genotype’s mean performance, followed by factor analysis and the computation of the genetic distance of hybrids from the ideotype. By utilizing a two-way table as the input dataset and applying a row-ranking approach based on desired outcomes, MGIDI provides an effective framework for evaluating the inherent strengths and weaknesses of the chosen genotypes. It offers an efficacious means of assessing the strengths and weaknesses inherent in the selected genotypes. The predicted genetic gains pertaining to the relevant traits within the MGIDI index are presented in Table 4. Specifically, an apparent reduction of −1.09% is anticipated in plant height, a characteristic considered desirable under certain circumstances due to its potential to mitigate lodging susceptibility and harvesting efficiency. Furthermore, ear height exhibits a more substantial decline of −2.02%, a phenomenon that is poised to enhance overall crop stability. This observation aligns with Andayani et al. (2018), which explain the negative implications of excessive plant height on stability and productivity, particularly in regions characterized by heavy precipitation and strong winds, such as equatorial zones. Days to tasseling exhibits a marginal decline of −0.0572%, indicative of relatively stable performance in this trait, while grain yield demonstrates a nearly imperceptible decrease of −0.0003%, underscoring the relative success of endeavors to maintain or augment yield within the context of FA1. Within the framework of FA2, leaf area is projected to experience an incremental increase of 0.00189%, thereby potentially augmenting photosynthetic efficiency and subsequent crop yields. Ear length is anticipated to extend by 0.0726%, signifying an increase in ear length, a factor poised to positively influence overall yield. Furthermore, ear diameter is expected to expand by 0.101%, likely leading to larger and potentially more productive ears. The 1,000 kernel weight is predicted to elevate by 0.128%, which bodes well for the improvement of seed size and quality. However, it is in the context of shelling percentage that the most substantial gain is observed, registering a notable increase of 4.8%. The heritability values within this study exhibit considerable variability, ranging from 0.30 for traits such as ear length and leaf angle to values exceeding 0.50 for traits including days to shelling percentage, ear diameter, and 1,000 kernel weight.
Based on Table 4, the overall gain achieved was 5.10% for traits targeted for improvement and a reduction of −3.16% for traits designated for minimization. Figure 9 presents a concise visual representation of genotype rankings based on their MGIDI index values, highlighting specific genotypes that align with the predefined selection criteria. Among the examined genotypes, H13, H31, H35, H06, H10, and H08 emerged as distinguished performers, denoted by their prominent highlighting, signifying their remarkable accomplishments. Additionally, four other genotypes, namely H11, H25, H32, and H30, also secured positions among the top 10 best-performing genotypes, showcasing their favorable characteristics across a spectrum of traits. These genotypes exhibit attributes that make them well-suited for the intended study or purpose. It is required to acknowledge, however, that while a robust correlation exists between genotype attributes and trait values, external environmental factors may impose limitations on the realization of heightened trait values. The MGIDI provides valuable insights into the strengths and weaknesses inherent in diverse genotypes, offering a convenient framework for discerning their advantages and limitations within the intricate context of multifaceted traits (Olivoto and Nardino, 2021).
The comprehensive evaluation of genotype attributes, effectively categorizing their influence on MGIDI divided into two distinct factors is shown in Figure 10. Particularly, attributes directing substantial influence located in a central position within the diagram, while those exerting a more marginal effect find their placement towards the periphery. The insights derived from this data regarding attribute contributions hold a potential role in the judicious selection of suitable parental contributors for the purpose of crossbreeding programs. It is evident that FA1 exerted a obviously greater influence on the MGIDI for genotypes H35, H25, and H31. This observation implies that these hybrids exhibited a relatively less favorable performance in terms of key attributes such as PH, EH, DT and SP traits. Conversely, FA1 had a notably diminished impact on the MGIDI of hybrids H08, H10, H08, H13, H30, H32, H25, and H35, thereby indicating their proficiency and excellence in manifesting the specified traits associated with FA1. FA2 had a more substantial influence on the MGIDI of genotypes H08, H10, H08, H13, H30, H32, while manifesting a relatively moderate effect on genotypes H25 and H35. Consequently, this distinction resulted in the latter two genotypes demonstrating strengths associated with FA2 within the framework. Optimal improvements are aspired for the traits GY, LA, EL, ED and 1,000 KRN within the domain of FA2. FA2 exhibits elevated values for these specific attributes to signify favorable outcomes.
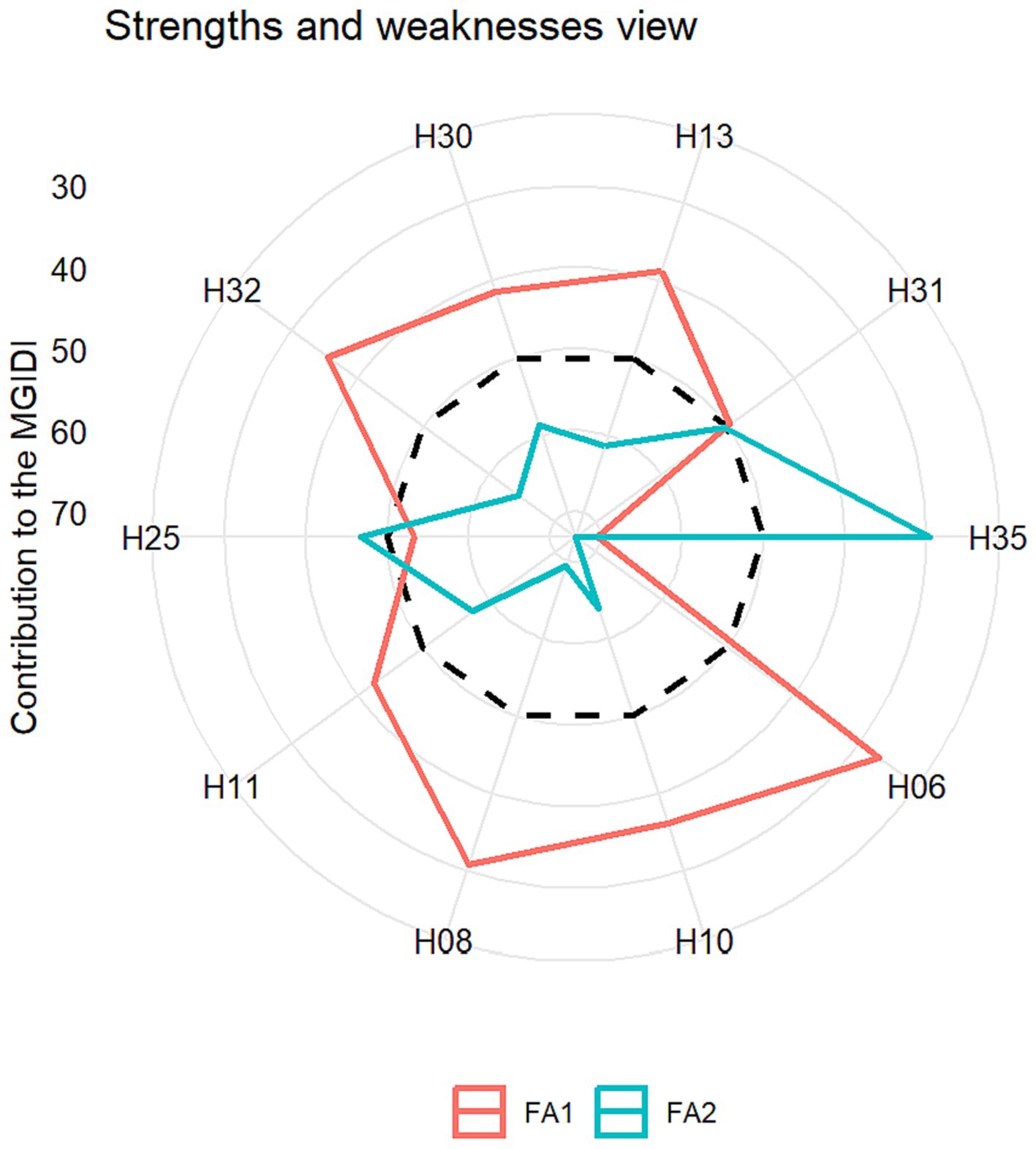
Figure 10. Comprehensive assessment of selected genotypes, highlighting their strengths and weaknesses weighed by MGIDI.
Through a comprehensive assessment of multiple traits, the process of genotype ranking has determined that among the chosen hybrids (H13, H31, H35, H06, H10, and H08), these six specifically exhibit the highest levels of performance. This ranking has been derived from their adeptness in seamlessly integrating grain yield with other target attributes. The practical application of the multiple trait combination index as an evaluative instrument has afforded researchers the ability to discern the traits that exert the most profound impact on each genotype. Yan and Frégeau-Reid (2018) reported that assessment of a genotype’s advantage must transcend the confines of isolated trait measurements. Instead, the evaluation should focus on the genotype’s proficiency in harmonizing grain yield with other meticulously chosen agronomic and yield components, thereby presenting a more holistic measure in assessment of screening or crossing program.
The study intricately investigated the interaction between ML techniques and a multi-trait selection model, resulting in the prediction of both grain yield and drought tolerance for maize crosses exposed to normal and drought-induced stress conditions. The results indicated that the optimized ML with GA and ensembles can substantially outperform the single ML model. Moreover, the incorporation of the Multi-trait Genotype-Ideotype Distance emerged as a crucial tool for identifying superior maize hybrids well-suited to drought-afflicted conditions, with its predictive performance benchmarked against that of the ML models. The empirical findings highlighted the elevated accuracy achieved by the RF-GA combination (R2 = 0.91 for grain yield and 0.79 for stress tolerance index), while the SVM-GA and KNN-GA models exhibited less favorable predictions. The remarkable consistency in achieving optimal outcomes was exemplified by the ensemble models, which harnessed the predictive capabilities of diverse individual models. These ensemble meta-models, incorporating SVM, KNN, and RF, consistently yielded R2 ≥ 0.92 for grain yield and 0.82 for STI across the testing datasets. The resounding effectiveness of these ensemble models can be attributed to the inherent diversity within the ensemble itself, enabling a comprehensive exploration of the intricate feature landscape. Among the six hybrids with the highest STI values, both the ML-based optimized model and MGIDI accurately predict four hybrids with high drought tolerance index, namely H06, H10, H13, and H35.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
MAz: Writing – original draft, Conceptualization. MAq: Methodology, Writing – original draft. NA: Writing – review & editing. RE: Writing – review & editing. Suarni: Writing – review & editing. Suwardi: Writing – review & editing. MJ: Writing – original draft. BZ: Writing – review & editing. Salim: Writing – original draft. Bahtiar: Writing – original draft. AM: Data analysis, Writing – original draft. MY: Writing – review & editing. MH: Writing – review & editing. Rahman: Data collection. AS: Writing – review & editing.
The author(s) declare financial support was received for the research, authorship, and/or publication of this article. We extend our gratitude to the Indonesia Endowment Fund for Education Agency (LPDP) through the Program for Funding Innovative Productive Research (Rispro Mandatori) under the National Research Priority (PRN), with reference number KEP-32/LPDP/2020. We also express our gratitude to Hasanuddin University for financing the publication of this research. Additionally, we acknowledge the IAARD, Ministry of Agriculture, Indonesia, for providing research facilities.
The research was conducted with the assistance of various genetic materials provided by CIMMYT Mexico. Thanks to the support provided by the technicians from Bajeng Experimental Station in Indonesia for this project.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1334421/full#supplementary-material
Agarwal, M., Gupta, S., and Biswas, K. K. (2021). A new Conv2D model with modified ReLU activation function for identification of disease type and severity in cucumber plant. Sustain. Comput. Informatics Syst. 30:100473. doi: 10.1016/j.suscom.2020.100473
Ahmed, S. (2023). A software framework for predicting the maize yield using modified multi-layer perceptron. Sustainability 15:3017. doi: 10.3390/su15043017
Akhiat, Y., Chahhou, M., and Zinedine, A. (2019). Ensemble feature selection algorithm. Int. J. Intell. Syst. Appl. 11, 24–31. doi: 10.5815/ijisa.2019.01.03
Andayani, N. N., Aqil, M., Efendi, R., and Azrai, M. (2018). Line × tester analysis across equatorial environments to study combining ability of Indonesian maize inbred. Asian J. Agric. Biol. 6, 213–220. Available at: https://www.asianjab.com/wp-content/uploads/2018/06/15.-OK_Line-_-tester-analysis-across-equatorial-environments-to-study-combining-ability-of-Indonesian-maize-inbred.pdf
Aqil, M., Azrai, M., Mejaya, M. J., Subekti, N. A., Tabri, F., Andayani, N. N., et al. (2022). Rapid detection of hybrid maize parental lines using stacking ensemble machine learning. Appl. Comput. Intell. Soft Comput. 2022, 1–15. doi: 10.1155/2022/6588949
Araus, J. L., Sanchez-Bragado, R., and Vicente, R. (2021). Improving crop yield and resilience through optimization of photosynthesis: panacea or pipe dream?. J. Exp. Bot. 72, 3936–3955.
Austria, Y. C., Mirabueno, M. C. A., Lopez, D. J. D., Cuaresma, D. J. L., Macalisang, J. R., and Casuat, C. D. (2022). EZM-AI: a Yolov5 machine vision inference approach of the Philippine corn leaf diseases detection system. 2022 IEEE International Conference on Artificial Intelligence in Engineering and Technology (IICAIET). 1–6.
Azrai, M., Aqil, M., Efendi, R., Andayani, N. N., Makkulawu, A. T., Iriany, R. N., et al. (2023). A comparative study on single and multiple trait selections of equatorial grown maize hybrids. Front. Sustain. Food Syst. 7:1185102. doi: 10.3389/fsufs.2023.1185102
Bänziger, M., Edmeades, G. O., Beck, D., and Bellon, M. (2000). Breeding for drought and nitrogen stress tolerance in maize: From theory to practice. CIMMYT. México-Veracruz. 68.
Çakir, R. (2004). Effect of water stress at different development stages on vegetative and reproductive growth of corn. Field Crops Res. 89, 1–16. doi: 10.1016/j.fcr.2004.01.005
Cheng, C. Y., Li, Y., Varala, K., Bubert, J., Huang, J., Kim, G. J., et al. (2021). Evolutionarily informed machine learning enhances the power of predictive gene-to-phenotype relationships. Nat. Commun. 12, 5627–5615. doi: 10.1038/s41467-021-25893-w
Chotewutmontri, P., and Barkan, A. (2016). Dynamics of chloroplast translation during chloroplast differentiation in maize. PLoS Genet. 12:e1006106. doi: 10.1371/journal.pgen.1006106
Fernandez, G. C. J. (1992). Effective selection criteria for assessing plant stress tolerance. Proceedings of the International Symposium on Adaptation of Vegetables and Other Food Crops in Temperature and Water Stress Chapter 25, Tainan Shanhua AVRDC Publications, 257–270.
Ganesan, G., and Chinnappan, J. (2022). Hybridization of ResNet with YOLO classifier for automated paddy leaf disease recognition: an optimized model. J. Field Robot. 39, 1085–1109. doi: 10.1002/rob.22089
Gokulnath, B. V., and Devi, G. U. (2020). Boosted-DEPICT: an effective maize disease categorization framework using deep clustering. Neural Comput. Applic. doi: 10.1007/s00521-020-05303-w
Jamieson, P. D., Porter, J. R., and Wilson, D. R. (1991). A test of the computer simulation model ARCWHEAT1 on wheat crops grown in New Zealand. Field Crops Res. 27, 337–350. doi: 10.1016/0378-4290(91)90040-3
Jhajharia, K., Mathur, P., Jain, S., and Nijhawan, S. (2023). Crop yield prediction using machine learning and deep learning techniques. Procedia Comput. Sci. 218, 406–417. doi: 10.1016/j.procs.2023.01.023
Jordan, W. R. (1983). “Whole plant response to water deficit: an overview” in Limitations to efficient water use in crop production. ed. H. M. Taylor (Madison, WI: American Society of Agronomy), 289–317.
Kira, O., Nguy-Robertson, A. L., Arkebauer, T. J., Linker, R., and Gitelson, A. A. (2016). Informative spectral bands for remote green LAI estimation in C3 and C4. Agric. For. Meteorol. 218–219, 243–249. doi: 10.1016/j.agrformet.2015.12.064
Monneveux, P., Sanchez, C., and Tiessen, A. (2008). Future progress in drought tolerance in maize needs new secondary traits and cross combinations. J. Agric. Sci. 146, 287–300. doi: 10.1017/S0021859608007818
Moradi, H., Akbari, G. A., Khorasani, S. K., and Ramshini, H. A. (2012). Evaluation of drought tolerance in corn (Zea mays L.) new hybrids with using stress tolerance indices. Eur. J. Sustain. Dev. 1:543. doi: 10.14207/ejsd.2012.v1n3p543
Olivoto, T., Lúcio, A. D. C., da Silva, J. A. G., Sari, B. G., and Diel, M. I. (2019). Mean performance and stability in multi-environment trials II: selection based on multiple traits. Agron. J. 111, 2961–2969. doi: 10.2134/agronj2019.03.0221
Olivoto, T., and Nardino, M. (2021). MGIDI: toward an effective multivariate selection in biological experiments. Bioinformatics 37, 1383–1389. doi: 10.1093/bioinformatics/btaa981
Pham, H., and Olafsson, S. (2018). Bagged ensembles with tunable parameters: Pham and Olafsson. Comput. Intell. 35, 184–203. doi: 10.1111/coin.12198
Rosielle, A. A., and Hamblin, J. (1981). Theoretical aspects of selection for yield in stress and non-stress environment. Crop Sci. 21, 943–946. doi: 10.2135/cropsci1981.0011183X002100060033x
Roy, A. M., and Bhaduri, J. (2021). A deep learning enabled multi-class plant disease detection model based on computer vision. AI 2, 413–428. doi: 10.3390/ai2030026
Sachdev, S., Ansari, S. A., Ansari, M. I., Fujita, M., and Hasanuzzaman, M. (2021). Abiotic stress and reactive oxygen species: generation, signaling, and defense mechanisms. Antioxidants 10:277. doi: 10.3390/antiox10020277
Sahu, S., Chawla, M., and Khare, N. (2017). An efficient analysis of crop yield prediction using Hadoop framework based on random forest approach. 2017 International Conference on Computing, Communication and Automation (ICCCA). 53–57.
Sakeef, N., Scandola, S., Kennedy, C., Lummer, C., Chang, J., Uhrig, R. G., et al. (2023). Machine learning classification of plant genotypes grown under different light conditions through the integration of multi-scale time-series data. Comput. Struct. Biotechnol. J. 21, 3183–3195. doi: 10.1016/j.csbj.2023.05.005
Shao, R. X., Xin, L. F., Zheng, H. F., Li, L. L., Ran, W. L., Mao, J., et al. (2016). Changes in chloroplast ultrastructure in leaves of drought-stressed maize inbred lines. Photosynthetica 54, 74–80. doi: 10.1007/s11099-015-0158-6
Sharma, P., Hans, P., and Gupta, S. C. (2020). Classification of plant leaf diseases using machine learning and image preprocessing techniques. 10th International Conference on Cloud Computing, Data Science & Engineering. 480–484. Available at: https://api.semanticscholar.org/CorpusID:215738169.
Singh, B., Kumar, S., Elangovan, A., Vasht, D., Arya, S., Duc, N. T., et al. (2023). Phenomics based prediction of plant biomass and leaf area in wheat using machine learning approaches. Front. Plant Sci. 14:1214801. doi: 10.3389/fpls.2023.1214801
Srivastava, A. K., Safaei, N., Khaki, S., Lopez, G., Zeng, W., Ewert, F., et al. (2022). Winter wheat yield prediction using convolutional neural networks from environmental and phenological data. Sci. Rep. 12, 3215–3214. doi: 10.1038/s41598-022-06249-w
Sun, J., Yang, Y., He, X., and Wu, X. (2020). Northern maize leaf blight detection under complex field environment based on deep learning. IEEE Access 8, 33679–33688. doi: 10.1109/ACCESS.2020.2973658
Széles, A., Horváth, É., Simon, K., Zagyi, P., and Huzsvai, L. (2023). Maize production under drought stress: nutrient supply, yield prediction. Plants 12:3301. doi: 10.3390/plants12183301
Waheed, A., Goyal, M., Gupta, D., Khanna, A., Hassanien, A. E., and Pandey, H. M. (2020). An optimized dense convolutional neural network model for disease recognition and classification in corn leaf. Comput. Electron. Agric. 175:105456. doi: 10.1016/j.compag.2020.105456
Yan, W., and Frégeau-Reid, J. (2018). Genotype by yield∗trait (GYT) biplot: a novel approach for genotype selection based on multiple traits. Sci. Rep. 8, 1–10. doi: 10.1038/s41598-018-26688-8
Yoosefzadeh-Najafabadi, M., Torabi, S., Tulpan, D., Rajcan, I., and Eskandari, M. (2021a). Genome-wide association studies of soybean yield-related hyperspectral reflectance bands using machine learning-mediated data integration methods. Front. Plant Sci. 12:777028. doi: 10.3389/fpls.2021.777028
Yoosefzadeh-Najafabadi, M., Tulpan, D., and Eskandari, M. (2021b). Using hybrid artificial intelligence and evolutionary optimization algorithms for estimating soybean yield and fresh biomass using hyperspectral vegetation indices. Remote Sens. 13:2555. doi: 10.3390/rs13132555
Zahra, N., Hafeez, M. B., Kausar, A., Al Zeidi, M., Asekova, S., Siddique, K., et al. (2023). Plant photosynthetic responses under drought stress: effects and management. J. Agron. Crop Sci. 209, 651–672. doi: 10.1111/jac.12652
Zainuddin, B., and Aqil, M. (2021). Analysis of the relationship between leaf color spectrum and soil plant analysis development. IOP Conf. Ser.: Earth Environ. Sci 911:012045. doi: 10.1088/1755-1315/911/1/012045
Keywords: drought, maize, machine learning, GA, ensemble
Citation: Azrai M, Aqil M, Andayani NN, Efendi R,,, Jihad M, Zainuddin B,,, Muliadi A, Yasin M, Hannan MFI, and Syam A (2024) Optimizing ensembles machine learning, genetic algorithms, and multivariate modeling for enhanced prediction of maize yield and stress tolerance index. Front. Sustain. Food Syst. 8:1334421. doi: 10.3389/fsufs.2024.1334421
Received: 07 November 2023; Accepted: 17 January 2024;
Published: 02 February 2024.
Edited by:
Mahesh Kumar, Indian Council of Agricultural Research (ICAR), IndiaReviewed by:
Sudhir Kumar, Indian Agricultural Research Institute (ICAR), IndiaCopyright © 2024 Azrai, Aqil, Andayani, Efendi, Suarni, Suwardi, Jihad, Zainuddin, Salim, Bahtiar, Muliadi, Yasin, Hannan, Rahman and Syam. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Muhammad Azrai, YXpyYWlAYWdyaS51bmhhcy5hYy5pZA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.