- 1Food Microbiology Laboratory, Department of Food Technology, University of North Bengal, Siliguri, West Bengal, India
- 2Food Microbiology Laboratory, Department of Botany, Kurseong College, Darjeeling, West Bengal, India
- 3Department of Bioinformatics, Kalinga University, Atal Nagar-Nava Raipur, Chhattisgarh, India
- 4Molecular Cytogenetics Laboratory, Department of Botany, University of North Bengal, Siliguri, India
Endemic fermented vegetables play a crucial role in local cuisine and culture, with traditionally fermented raw green vegetables such as gundruk being a popular practice in North Bengal and Sikkim, India. However, there is a gap in the microbial profiling of these vegetables. This study aimed to explore the diversity of dominant bacterial populations in fermented leafy vegetables using both culture-dependent and culture-independent methods. In addition to isolating bacteria using conventional methods, we conducted phenotypic and biochemical characterization, community DNA isolation, and amplicon sequencing. We also introduced a new approach in bioinformatics analysis: reverse ecology, which analyzes complementation and competition among participant microbes. In conventional culture-dependent techniques, LAB genera such as Lactobacillus, Enterococcus, Leuconostoc, and Pediococcus have been identified as predominant consortia, whereas metagenomic analysis revealed that the microbiome of fermented dried leafy vegetables was mainly composed of Firmicutes, Proteobacteria, Actinobacteria, Bacteroides, and Planctomycetes at the phylum level. Within the Lactobacillaceae family, predominant types included Lactobacillus, Lactococcus, Pediococcus, Leuconostoc, Enterococcus, Vagococcus, Weissella, and Carnobacterium. The microbial metabolism revealed key pathways, such as carbon metabolism, glycolysis, gluconeogenesis, and glyoxylate. Aromatic amino acid degradation, fatty acid metabolism, amino sugar metabolism, nucleotide sugar metabolism, and biosynthesis of nucleotide sugar pathways were also active. The competition index among microbes and human metabolic data was low (0.32–0.44), indicating minimal competition for nutrition. Complementation indices between bacteria and humans were high (0.76–0.88), suggesting a beneficial impact of gundruk microbial populations on human health.
Introduction
Fermentation of vegetables was started centuries ago to enhance the stability of fresh ones, to make them safer to eat in the absence of refrigeration, and to increase their flavor. The primary vegetables that are fermented in the world are cucumbers, mangoes, carrots, radishes, cabbage, green leaf-based vegetables (sauerkraut, kimchi, etc.), and olives. One such leafy vegetable-based fermented food, an ethnic food of the Nepalese community residing in the Himalayan regions of Nepal, Bhutan, and India, is gundruk (Ghimire et al., 2020). The description of gundruk preparation and its significance within Nepalese culture is fascinating. Gundruk’s fermentation process, akin to kimchi and sauerkraut, not only preserves vegetables but also enhances their nutritional value and flavor. The involvement of lactic acid bacteria (LAB) in the fermentation process contributes to the unique taste and brings potential health benefits, similar to those found in probiotic foods.
The utilization of LAB, particularly strains such as Lactobacillus plantarum and Pediococcus pentosaceus, underscores the traditional wisdom in food preservation methods. These bacteria aid in fermentation, in addition to offering probiotic properties, potentially aiding in digestion and immuno-modulation.
Furthermore, the nutritional richness of gundruk, including its high content of vitamins, minerals, and therapeutic compounds, makes it a valuable dietary addition, particularly for lactating mothers. Its antimicrobial properties, along with the ability to degrade anti-nutritional factors, further highlight its potential health benefits.
The genesis of gundruk as a method to preserve leafy vegetables, susceptible to spoilage, is evident (Karki et al., 1983). This fermented product, lacking salt and dried, is derived from local greens such as rayo-saag, cauliflower, mustard, cabbage, and radish leaves (Tamang and Tamang, 2010; Das and Deka, 2012; Gautam and Sharma, 2015). Gundruk production typically occurs in the winter months when leafy vegetables are abundant and the weather less humid (Dahal et al., 2005; Tamang and Tamang, 2010). Analogous to kimchi, sauerkraut, sunki, and suan-cai from Korea, Germany, Japan, and China, respectively, gundruk shares commonalities in its fermentation process (Tamang et al., 2005; Tamang and Tamang, 2010; Ray and Swain, 2013).
Overall, gundruk stands as a prime example of traditional food preservation techniques merging with modern scientific methods, resulting in a culturally significant, nutritious, and potentially health-promoting food product. Nevertheless, microbial makeup involved in gundruk is for the most part very illusive. The initiation of fermentation in gundruk is attributable to LAB; Lactobacillus cellobiosus, followed by P. pentosaceus, L. plantarum, Lactobacillus casei, and L. casei spp. pseudoplantarum (Ghimire et al., 2020). The other microorganisms associated with gundruk are Lactobacillus fermentum, Lactobacillus paracasei, Leuconostoc fallax, and Pediococcus acidilactici (Sharma and Sarkar, 2015; Senapati et al., 2016). According to Karki et al. (1983), Lactiplantibacillus plantarum and P. pentosaceus are the dominant microflora involved in the natural fermentation of gundruk (Karki et al., 1983).
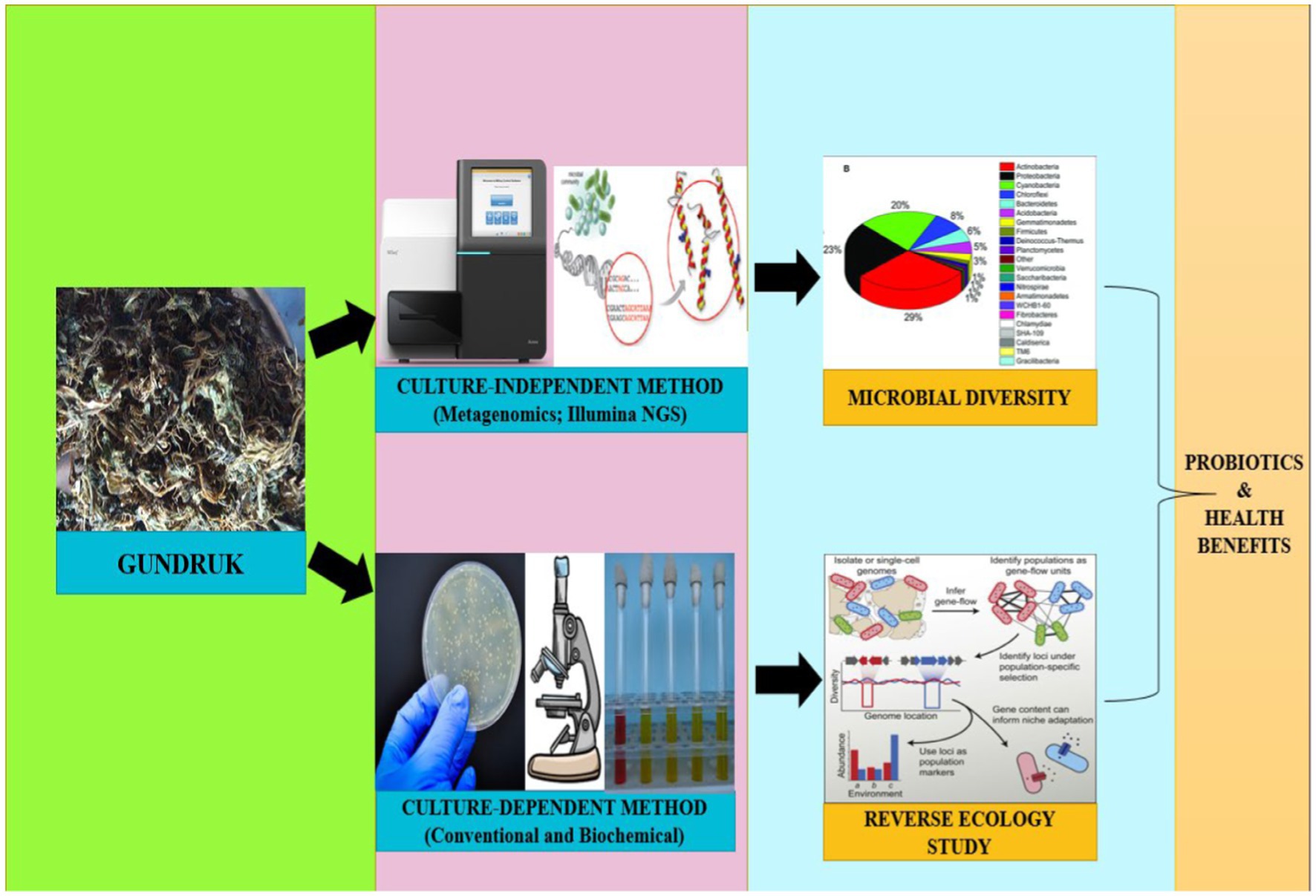
This study therefore aimed to obtain a profile of bacterial diversity associated with gundruk samples based on phenotypic and biochemical tests. We intended to involve both culture-dependent and culture-independent approaches for these purposes. Besides using traditional microbiological and biochemistry methods, we used the next-generation sequencing system to analyze bacterial diversity in gundruk samples, which we believe represents a commendable advancement. This method facilitates a thorough evaluation of microbial composition, enabling a comprehensive assessment of probiotic potential and associated health benefits of gundruk. We also employed various statistical methods like reverse ecology analysis to study the complementation of beneficial microbiomes and human health. Reverse environmental bacterial ecology, often referred to as ‘reverse ecology’, is a bioinformatics approach used to infer the ecological roles of microbial taxa based on their genomic information. Rather than starting with environmental samples and attempting to identify the microorganisms present, reverse ecology begins with the genomes of known microbial species and seeks to predict their ecological functions and interactions within ecosystems.
Methods and materials
Survey and sample collection
A thorough survey was carried out across villages, local markets, and haats in Darjeeling, Kalimpong, Sikkim, Mirik, and Kurseong to gather comprehensive information on the traditional knowledge of gundruk preparation. The survey aimed to observe any variations between modern and traditional preparation methods. To aid this, a questionnaire was used, which was designed to collect specific data including the head of the family’s name, age, address, ethnicity, and academic qualifications. These details helped track which ethnic groups are involved in fermented food preparation and whether age and education influence the transmission of fermentation knowledge. The questionnaire also assessed the knowledge, awareness, and experience of individuals involved in fermentation, shedding light on hygiene standards and the use of starters. Furthermore, it documented whether fermented foods were for personal consumption or marketed, potentially indicating local business opportunities. Additionally, the questionnaire recorded the names and ages of those inheriting fermentation knowledge for future reference. This survey aimed to understand local beliefs about the health benefits of fermented foods, potentially informing future research. Finally, the survey studied and compared traditional and modern fermentation methods to gain insights into evolving practices.
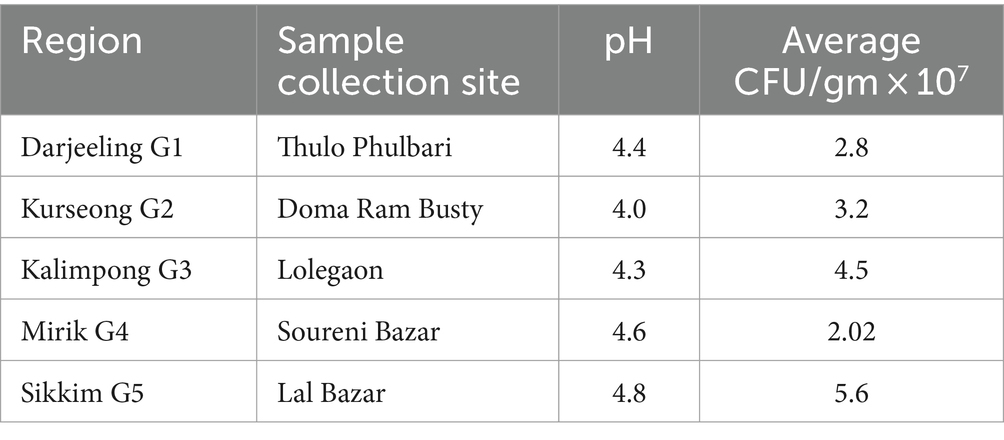
Five samples (G1, G2, G3, G4, and G5) of traditionally fermented gundruk were collected aseptically from the villages and markets of the surveyed regions and coded as gundruk G1 from Darjeeling, gundruk G2 from Kurseong, gundruk G3 from Kalimpong, gundruk G4 from Mirik, and gundruk G5 from Sikkim. The samples were packed in gamma-irradiated sterile bottles, sealed, and then transported to the laboratory for further analysis. Traditionally prepared gundruk can be kept for over a year or more in a dry condition at room temperature; therefore, samples were kept in the laboratory in desiccators at room temperature (Sha et al., 2017, 2018).
Isolation of bacteria
The gundruk samples (10 g) were crushed with mortar and pestle and mixed with 90 mL of sterile physiological saline (0.85% w/v) and homogenized in a stomacher laboratory blender (Seward, United Kingdom) for 1 min; 9 mL of serial dilution blanks was prepared using saline to determine the total viable count (TVC) in nutrient agar medium, and isolation of LAB was done on MRS (M641, HiMedia) agar plates supplemented with 1% CaCO3. The plates were incubated at 30°C in a gas jar (LE002, HiMedia) for 48–72 h under anaerobic conditions (Ghatani and Tamang, 2017). After incubation, distinct colony morphology was noted and single colonies were picked randomly from the plates were streaked for purifying and sub-culturing on fresh MRS agar plates.
Phenotypic and biochemical characterization
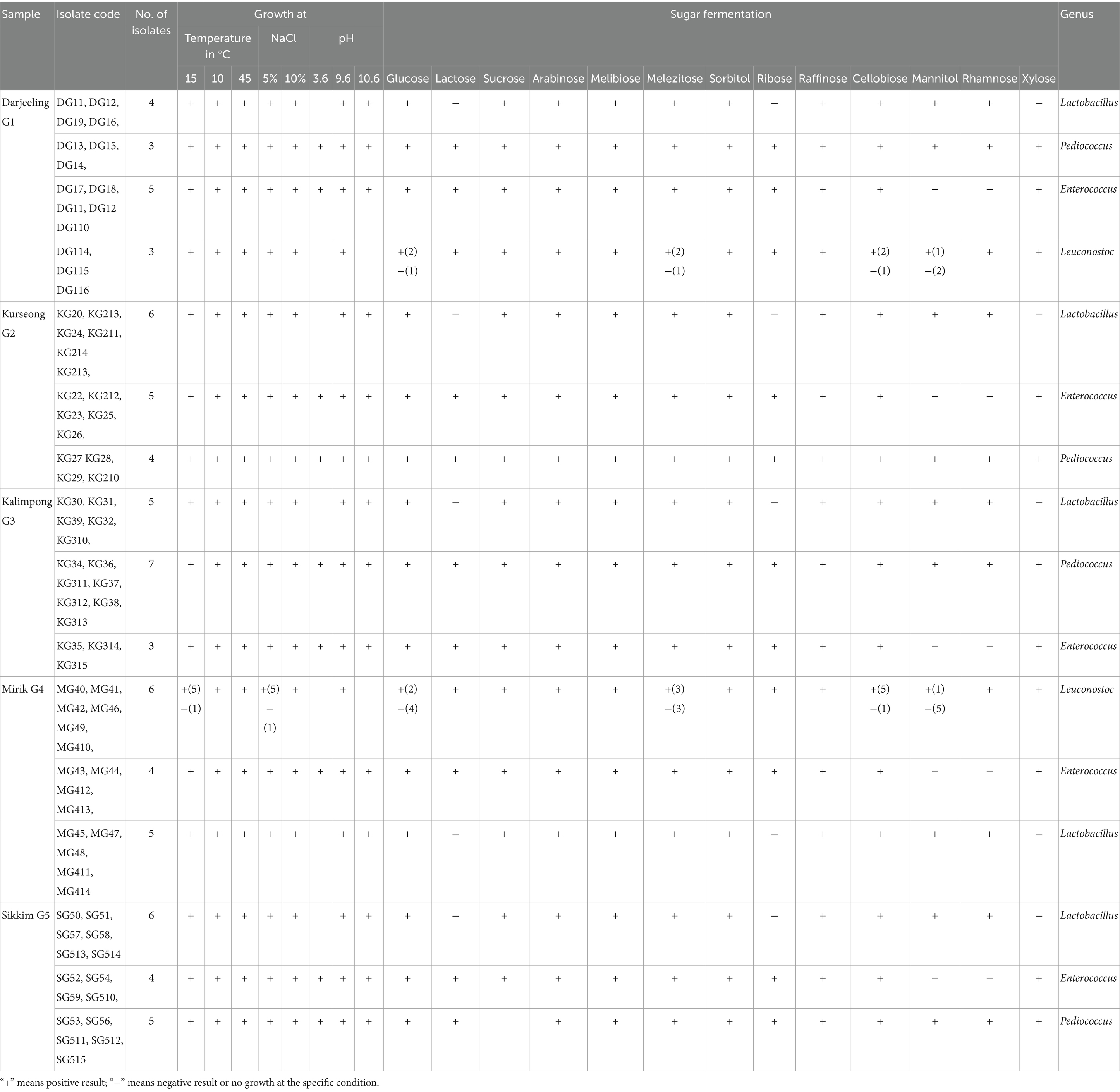
Phenotypic characterization of the bacterial isolates was performed based on cell morphology, Gram’s reaction, colony morphology, and arginine hydrolysis (Holt et al., 1994). The isolates were then grouped tentatively. Other tests included growth at different temperatures (10°C, 15°C, and 45°C), pH (3.6, 9.6, and 10.6), and salt tolerance (5 and 10%) (Ghatani and Tamang, 2017). Sugar or carbohydrate fermentation tests with 13 sugars, namely, glucose, sucrose, lactose, cellobiose, raffinose, rhamnose, sorbitol, arabinose, melibiose, ribose, melezitose, mannitol, and xylose, as well as nitrate reduction tests, and urease enzyme tests, were also performed (Hammes and Hertel, 2003).
Community DNA isolation
DNA was extracted from gundruk samples using the DNeasy PowerSoil Pro Kit. The quality of genomic DNA (gDNA) was assessed by loading 5 μL onto a 1% agarose gel to confirm the presence of a single intact band. Gel electrophoresis was conducted at 110 V for 30 min. PCR products were quantified using the Qubit dsDNA HS Assay kit (Life Tech), with 1 μL of each sample measured for concentration using a Qubit® 2.0 Fluorometer (Kumbhare et al., 2015). The DNA was then stored at −20°C until further use (Sha et al., 2017, 2019).
Preparation of libraries and amplicon sequencing
The hypervariable region V3–V4 of the bacterial 16S rRNA gene was targeted using universal primer sets i5 Forward (CCTACGGGNBGCASCAG-) and i7 Reverse (GACTACNVGGGT ATCTAATCC-), as described by Caporaso et al. (2010). PCR amplification was performed with the following reaction conditions: initial denaturation at 98°C for 5 min, followed by 28 cycles of denaturation at 98°C for 15 s, annealing at 55°C for 30 s, and elongation at 72°C for 30 s. A final extension step was carried out at 72°C for 10 min (Sha et al., 2017). The resulting amplicon libraries were purified using 1× AMpure XP beads, checked for quality on an Agilent DNA1000 chip using a Bioanalyzer 2100, and quantified with a Qubit Fluorometer 2.0 using the Qubit dsDNA HS Assay Kit (Life Technologies). Sequencing was performed following Illumina MiSeq Sequencer protocols (Illumina, San Diego, CA, USA). Raw sequences generated from MiSeq were processed using the QIIME 1.7 suite (Caporaso et al., 2010). Specifically, sequences were aligned with FLASH and quality trimmed using mothur, retaining only sequences with quality scores above 30 and a length greater than 150 bp for further analysis (Sha et al., 2017, 2019).
Cluster generation and sequencing
After determining the Qubit concentration for the libraries and the mean peak size from the Bioanalyzer profile, the libraries were loaded onto the Illumina platform at an appropriate concentration (10–20 pM) for cluster generation and sequencing. Paired-end sequencing was utilized, allowing the template fragments to be sequenced in both the forward and reverse directions on the Illumina platform. Kit reagents were employed to bind samples to complementary adapter oligos on the paired-end flow cell. These adapters were designed to facilitate selective cleavage of the forward strands after re-synthesis of the reverse strand during sequencing. The copied reverse strand was then used to sequence from the opposite end of the fragment following the protocols of the Illumina MiSeq Sequencer (Caporaso et al., 2010).
Bioinformatics analysis
Sequence reads were assigned to operational taxonomic units (OTUs) in QIIME v1.7 using a closed reference-based OTU picking approach with the SILVA database. OTU picking was conducted using the UCLUST method with a similarity threshold of 97%. Taxonomic assignments were performed using the RDP naïve Bayesian classifier in mothur v1.25, with taxonomic classification determined by the Ribosomal Database Project (RDP) Classifier at a confidence level of 80%. Rarefaction curves were generated via MEGAN 5. Alpha diversity indices, including Chao, Shannon, and Simpson biodiversity indices, were calculated using QIIME after rarefying all samples to the same sequencing depth. Additionally, metabolic pathway analysis was performed considering the type strains of genera with read counts exceeding 100, utilizing the KEGG database. A heatmap was generated using the R program. Furthermore, reverse ecology analysis was conducted on the considered type strains using the RevEcoR package in R, with metabolic pathway information gathered from the KEGG database (Sarkar et al., 2022).
Results
Survey
The preparation method of gundruk as observed in one of the households under the survey is outlined in Figures 1, 2.
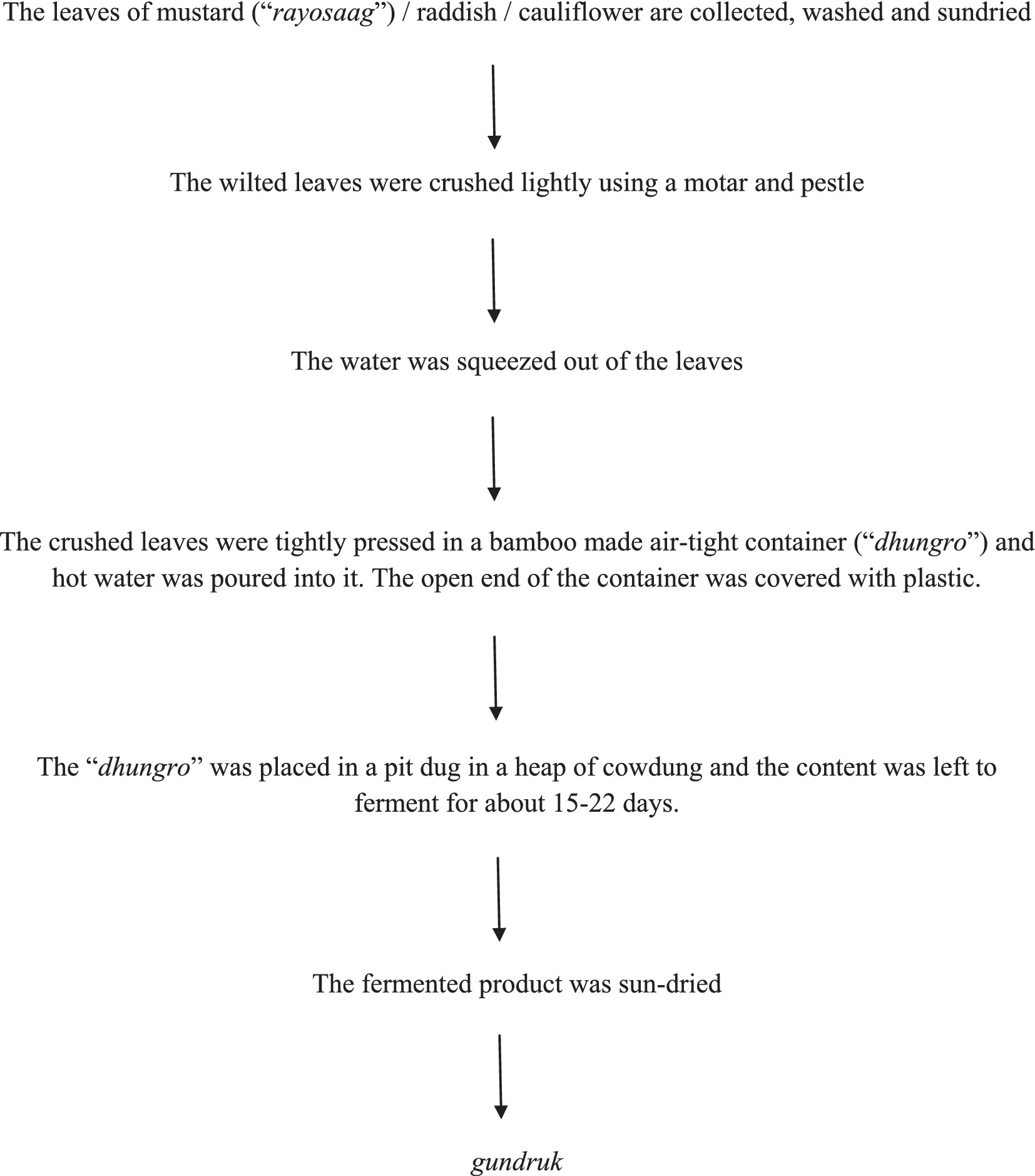
Figure 1. Preparation process of gundruk as observed during the survey in a household at Thulo Phulbari, Darjeeling.

Figure 2. (A) Washing of the radish leaves. (B) Sun-drying of the leaves. (C) Crushing of the leaves using mortar and pestle. (D) Squeezing water from the crushed leaves. (E) Packing the crushed and squeezed leaves inside a “dhungro.” (F) Pouring hot water into the content of the “dhungro.” (G) Covering the open end of the “dhungro” with plastic to make it air-tight. (H) Placing the “dhungro” in a pit dug in a heap of cow dung to allow fermentation of the raw material. (I) Sun-dried gundruk.
Isolation of bacteria
The total viable count (TVC) in nutrient agar medium is given in Table 1. The average CFU/gm × 107 in gundruk samples ranged from 2.02 to 5.6, and the pH ranged from 4 to 4.8. From the gundruk samples of Darjeeling, Kurseong, Kalimpong, Mirik, and Sikkim, a total of 75 isolates were obtained.
Phenotypic and biochemical characterization
The isolated colonies showed a circular shape, with an entire margin, low convex, and white color surrounded by a clear transparent region on MRS agar with 1% CaCO3. The isolates were gram-positive and catalase-negative. The phenotypic and biochemical characterization suggested that the LAB belonging to Lactobacillus (26), Enterococcus (21), Leuconostoc (9), and Pediococcus (19) genera were present in the gundruk samples. The data of the phenotypic and biochemical characterization are given in Table 2.
Metabolic pathway analysis
The metabolic pathways of microbial populations in fermented foods like gundruk play a critical role in determining food quality and potential health impacts on consumers. Our comprehensive investigation revealed key metabolic pathways present in gundruk microbial populations.
Carbon metabolism, aromatic compound degradation, glycolysis, gluconeogenesis, glyoxylate, and dicarboxylate metabolism were identified as the main metabolic pathways in gundruk microbial populations. Carbon metabolism, being a major energy source in living organisms, along with glycolysis and gluconeogenesis, were the most utilized pathways. This is consistent with the anaerobic conditions typical of fermentation, where fermenting microbes primarily utilize glucose for energy. The balance between glycolysis and gluconeogenesis helps regulate glucose levels within bacterial cells (Figure 2).
Furthermore, significant usage of aromatic amino acid degradation pathways indicates the microbial population’s ability to utilize complex aromatic compounds. Additionally, fatty acid metabolism, amino sugar metabolism, nucleotide sugar metabolism, and biosynthesis of nucleotide sugar pathways were found to be active in gundruk microbial populations.
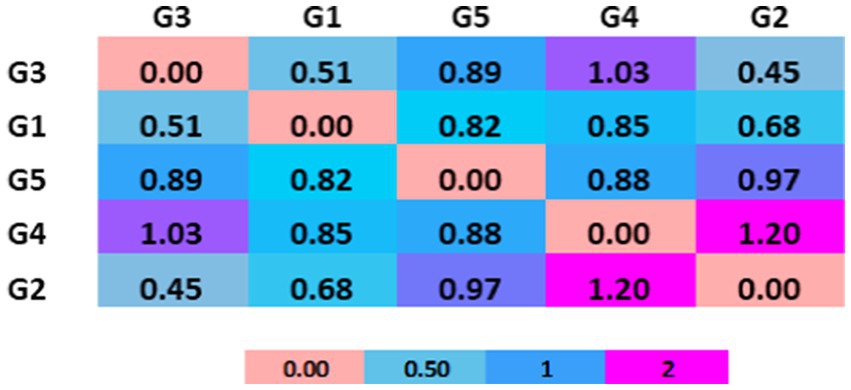
To assess microbial diversity, we conducted a comparative analysis using Shannon and Beta diversity indices, as well as a Venn diagram representation. These analyses provide insights into the richness and distribution of microbial species across gundruk samples (Figure 3).
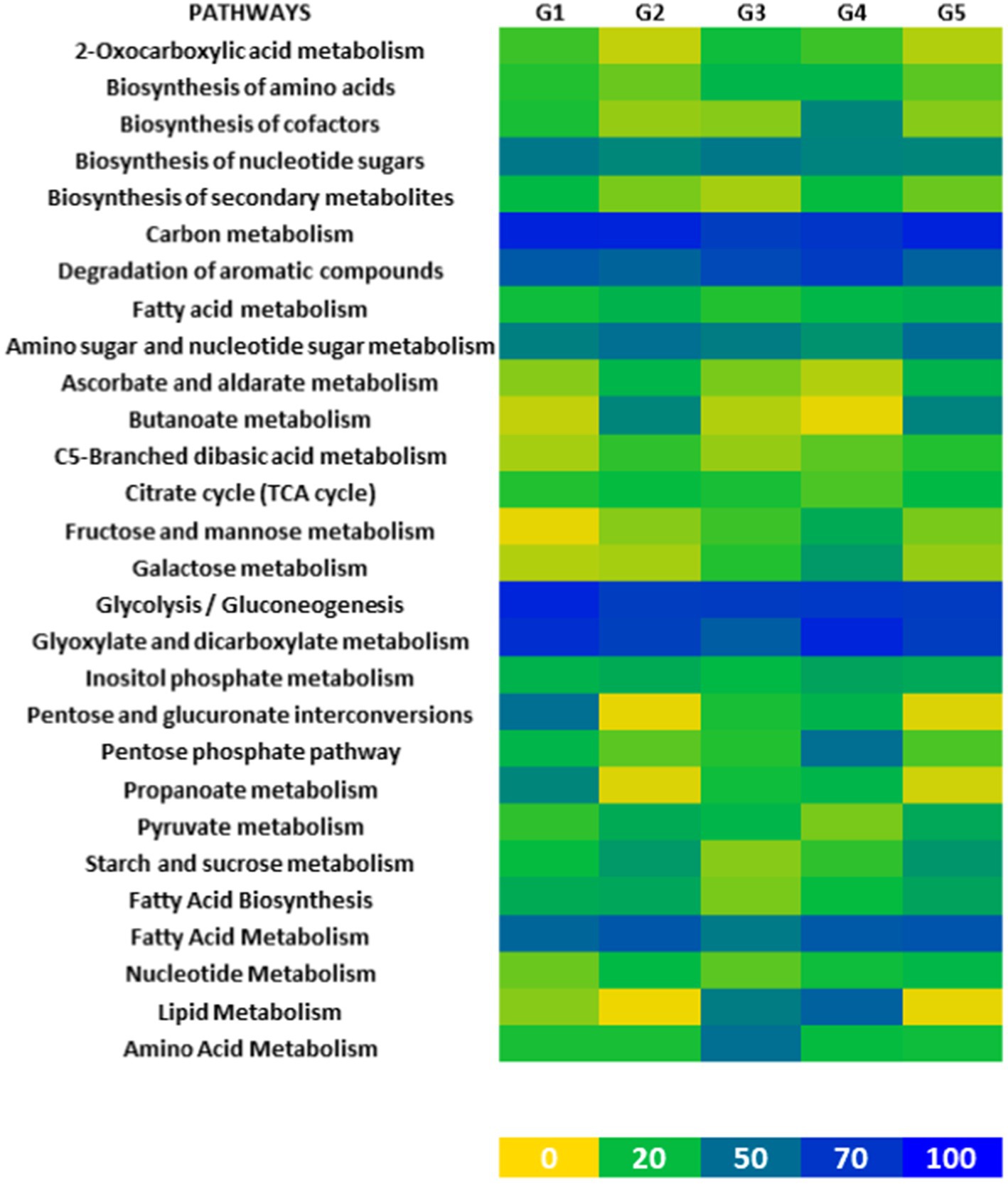
Figure 3. Heatmap based on the major metabolic pathways found among the microbial population of five different gundruk samples. MetaCyc server was used for this analysis. The total numbers of genes associated with different metabolic pathways of considered microbes were taken into account, and then, an average was done for each gundruk sample. The color code has been indicated in this heatmap. The color ranges from yellow to greenish blue to deep blue as the count increases from 0 to 100.
Overall, understanding the metabolic pathways of gundruk microbial populations is essential for ensuring food quality and safety as well as for exploring potential health benefits associated with fermented foods (Figures 4–8).
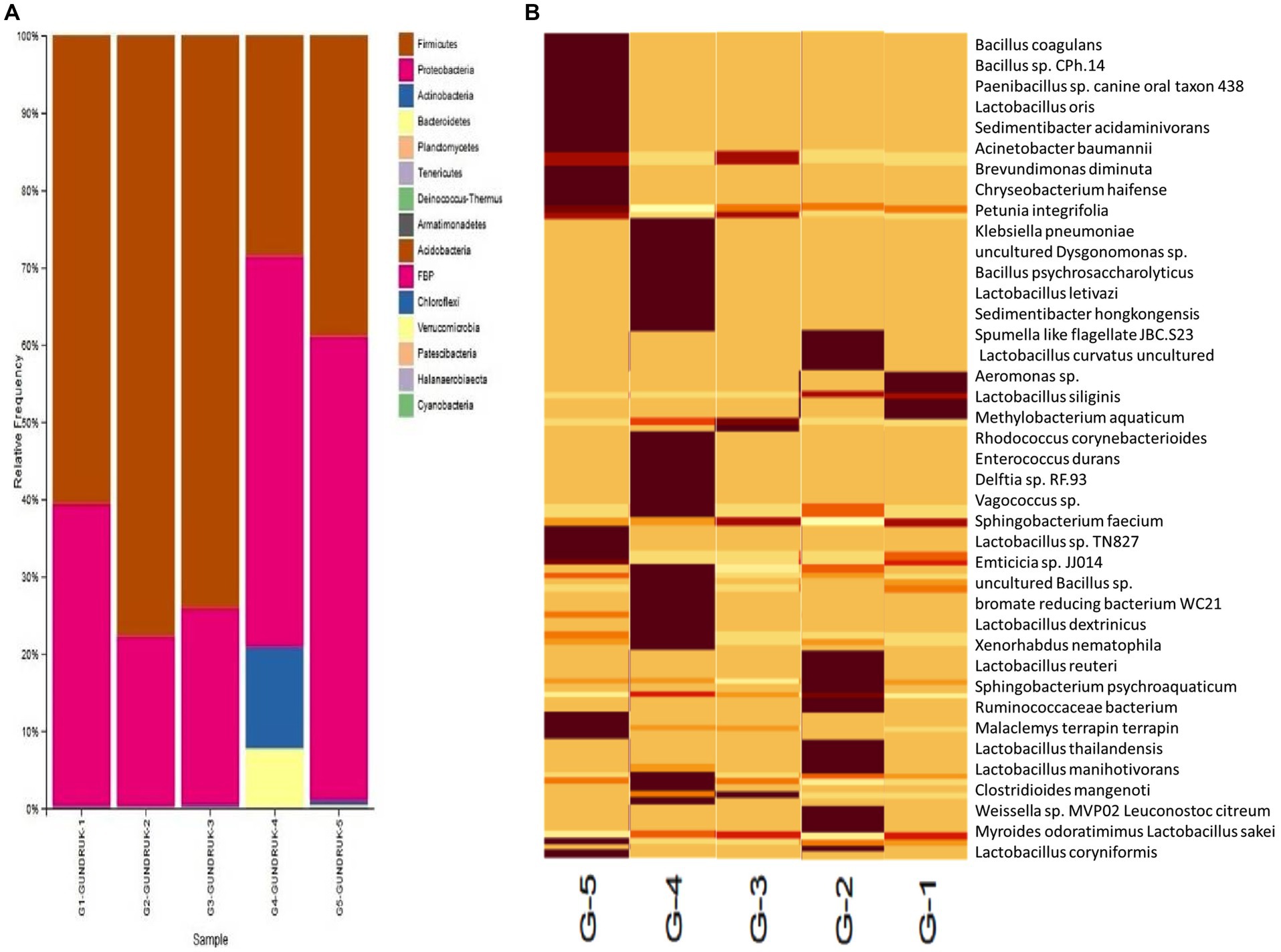
Figure 4. (A) Bar diagram for representing the relative frequency of different microbial genera of five different gundruk samples. (B) Heatmap for representing the relative frequency of different microbial species present in five different gundruk samples. The bright brown color represents the higher population of a specific species. The lighter the yellow color, the lower the species population in each of the five gundruk samples.
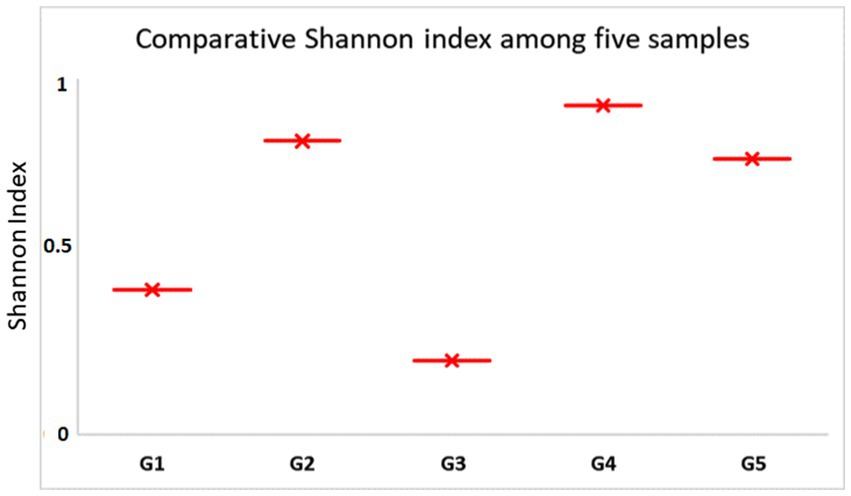
Figure 5. Comparative Shannon index among five different gundruk samples. The G4 sample has the highest Shannon index, indicating high microbial biodiversity followed by G5 and G2. G1 and G3 show comparatively less biodiversity.
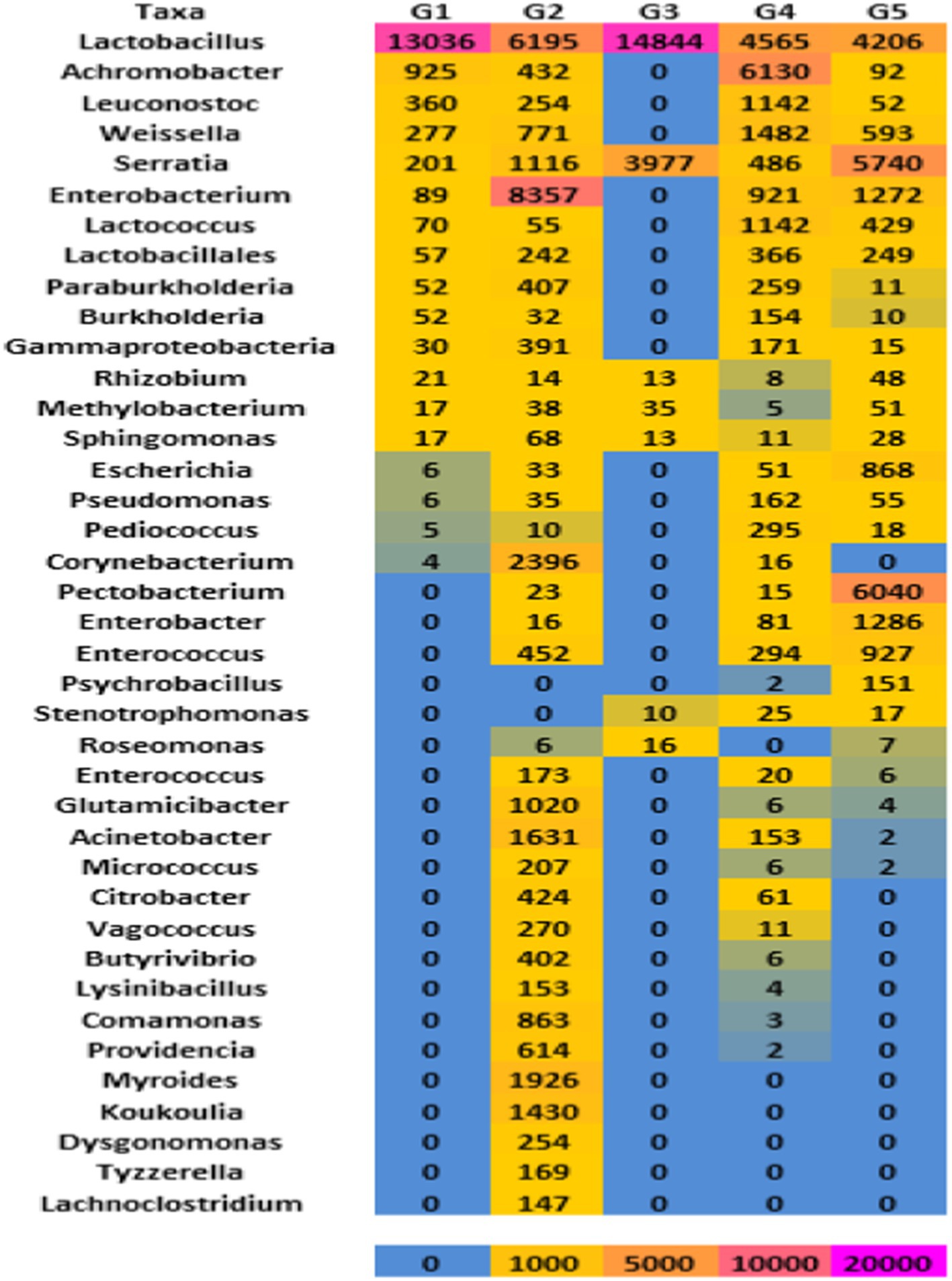
Figure 6. Heatmap based on total read counts of different microbial genera present among five gundruk samples. The color code has been indicated.
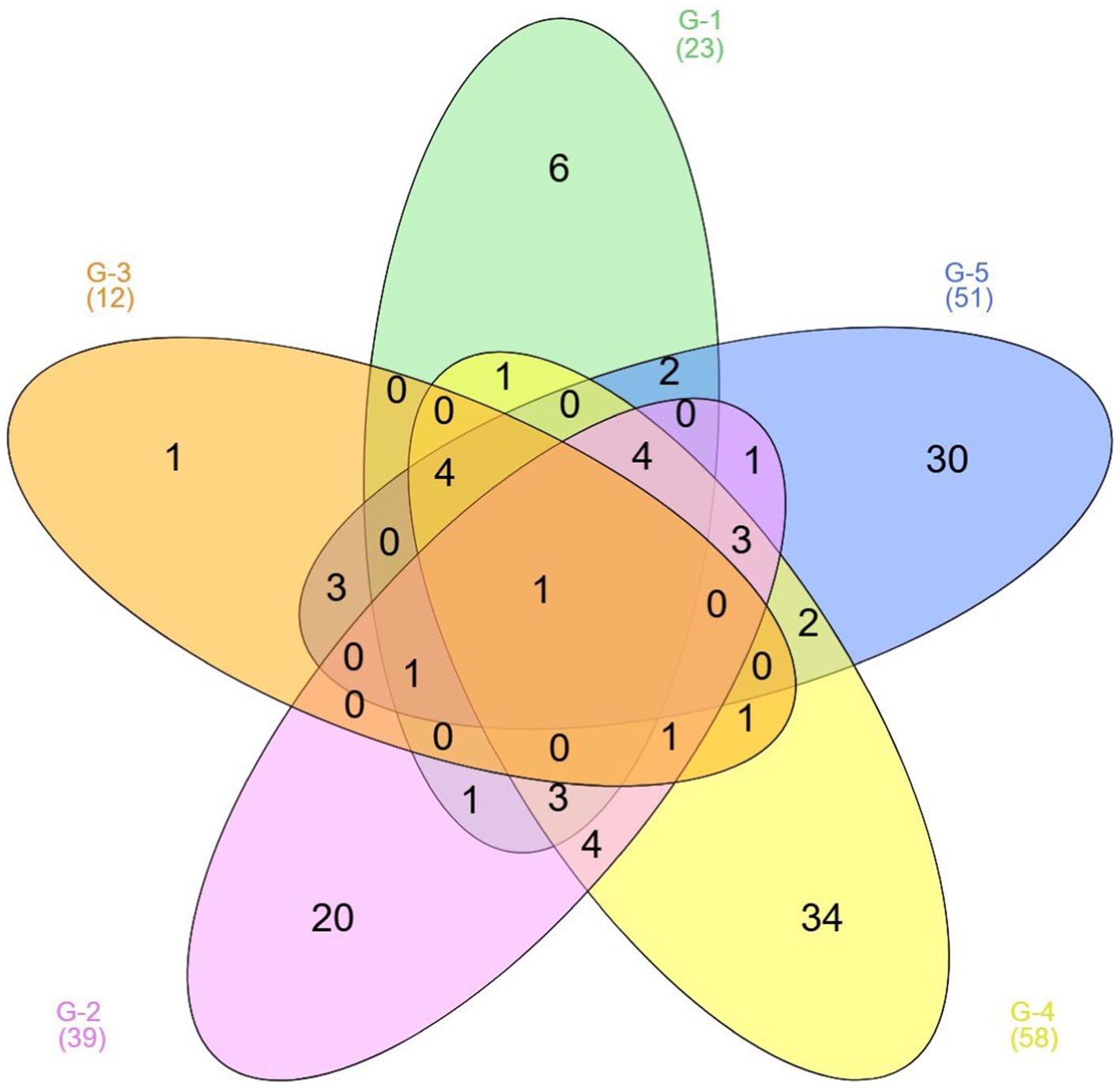
Figure 7. Venn diagram analysis based on unique and shared microbial genera among five gundruk samples.
Reverse ecology analysis
The reverse ecology analysis was performed based on the competition and complementation index. Both these indices range from 0 to 1. The competition index among the considered microbes was found to be very low (0.32–0.44). Moreover, their competition with human metabolic data was also low (0.34–0.43). The complementation index among those microbes ranged from 0.6 to 0.8. This is a clear representation of stable interaction and more specifically cooperation among the bacterial population of gundruk. Moreover, their competition with human was also 0.34–0.43, which is lower than their complementation index of 0.76–0.88. This indicated that the major microbial population in the gundruk samples was not competing with humans for their nutrition. Moreover, this result also suggested that the gundruk microbial population may have a positive or complementing effect on the human system, which means after the consumption of gundruk, the population of good gut microbes may increase in the human gut. This increase in good gut bacteria population may not only help in the betterment of the human digestive system but also enhance the effectivity of our endocrine and nervous system through the gut–pituitary axis and gut–brain axis (Mayer et al., 2015; Sudo, 2016; Figures 5–8).
Discussion
Lactobacillaceae are the most ubiquitous group of food-related bacteria that are widely used in the food and beverage industries. They have GRAS status, and this makes them useful for such applications. These beneficial bacteria have probiotic attributes and are used for treating gastric diseases, inflammatory bowel diseases, colon cancer, allergies, lowering cholesterol, diabetes, cancer, immune modulation, preventing the colonization of pathogenic bacteria, and improving lactose tolerance (Lebeer et al., 2008; Van Baarlen et al., 2011).
The conventional phenotypic and biochemical characterization of LAB isolated from the five samples of gundruk identified LAB belonging to Lactobacillus (26), Enterococcus (21), Leuconostoc (9), and Pediococcus (19) genera. A study conducted by Tamang, 2022 has also reported that the predominant bacterial genera observed in all the gundruk samples are Lactobacillus, Serratia, Weissella, Lactococcus, Leuconostoc, Enterococcus, Pediococcus, Burkholderia, unclassified (derived from Bacteria), Cronobacter, Achromobacter, and Bacillus. In addition, this result is on par with those reported earlier by Karki et al. (1983), which revealed gundruk fermentation to be started by Lactobacillus cellobiose and P. pentosaceus and finally by L. casei subspecies pseudoplantarum and L. plantarum in gundruk pickle of Nepal. The dominance of Lactobacillus in all five samples of gundruk has been observed. Leuconostoc fallax, Lactobacillus brevis, P. pentosaceus, L. plantarum, and P. acidilactici were also reported in a study where traditional fermented vegetables in Eastern Himalayas were studied by Tamang et al. (2005) and Sha et al. (2012, 2013).
The major bacterial phyla identified in the gundruk samples include Firmicutes, Actinobacteria, Proteobacteria, and Bacteroidetes. We found differences in the microbial population among the five different gundruk samples using the Shannon diversity index. This difference is probably due to the geographic locations from where the samples were collected and the preparation strategy the native people used for gundruk preparation. Some people use only “Rayo saag” or mustard leaves, and some use mustard leaves along with cabbage and other vegetables. Based on the constituents, the microbial diversity differs, and as a result, the Shannon index also differs.
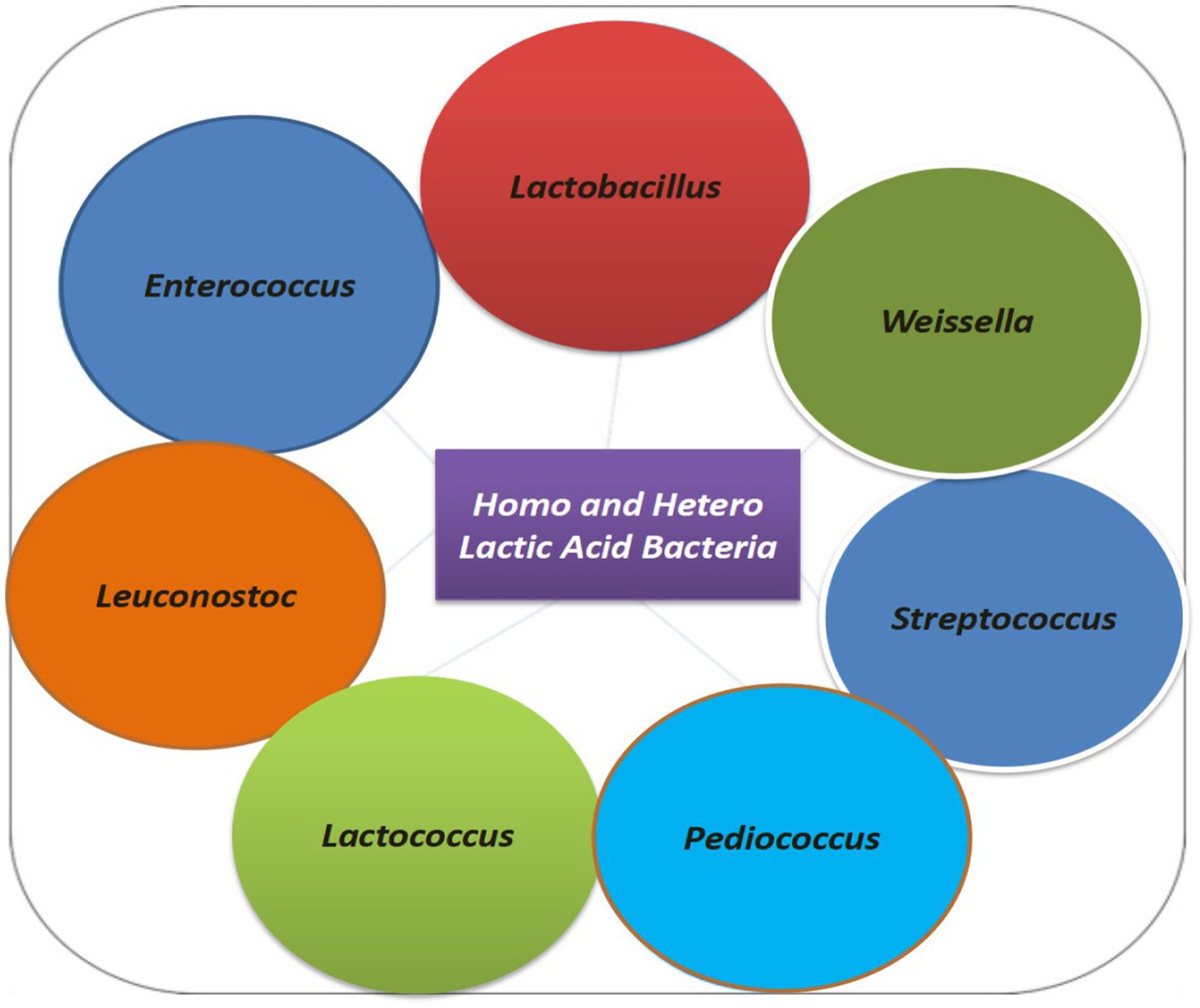
This is the first study of its kind where the total bacterial flora from the gundruk sample from parts of Sikkim and North Bengal is considered by next-generation sequencing (NGS). Our NGS investigation provides complete bacterial diversity analysis using an in-depth sequencing approach of the traditional ethnic fermented gundruk samples of the Himalayan regions of India. Quantitative differences were observed in the presence of diverse bacterial species among various samples of gundruk, which could be the consequence of differences in the preparation, period of incubation method, and most importantly the type of particular food preservation (Tamang, 2016; Sha et al., 2017, 2018). Alpha diversity estimation of different geographically diverse samples using microbial species richness and Shannon index (non-parametric) suggested higher bacterial diversity with the dominance of Lactobacillus followed by the Lactococcus, Pediococcus, and Leuconostoc. Metagenomic investigation showed that phylum Firmicutes, Proteobacteria, Actinobacteria, Bacteroides, and Planctomycetes were predominant members of the bacterial communities in the end product; similar results also reported by Tamang et al. (2016) and Sha et al. (2018). Our metagenomics results showed that lactic acid bacteria are categorized into the homo-fermentative group and the hetero-fermentative group based on secondary metabolism and their synthesis during fermentation (Surve et al., 2022). LABs have also been classified according to their acid production due to sugar utilization or sugar fermentation and their growth at a particular temperature range (Parvez et al., 2006). Lactic acid bacteria can ferment carbohydrates by utilizing the ED metabolic pathways and can be grouped into two types: the homo-fermentative LAB group and the hetero-fermentative LAB group based on their capacity to ferment or utilize various carbon sugars (Kuipers et al., 2000). The homo-fermentative LAB such as Lactococcus and Streptococcus sp. yields two molecules of lactic acids from one glucose molecule, whereas the hetero-fermentative LAB, such as Leuconostoc, Weissella produces lactate, ethanol, CO2, and lactate, from one molecule of glucose (Salminen et al., 1998). Our study reveals both the homo-fermentative and hetero-fermentative LAB groups, which are responsible for the aroma and flavor development in fermented gundruk (Figure 9).
The lactic acid synthesized by the LAB groups is responsible for the sourness of the gundruk and biopreservation (Tamang et al., 2008; Tamang, 2016; Surve et al., 2022). Leuconostoc strains, which are the hetero-fermentative LAB observed in our analysis, probably play an important role in biopreservation and flavor development (Kumari et al., 2022; Surve et al., 2022). LAB isolated from Indian vegetable-based fermented foods showed antimicrobial properties, acidification, anti-nutritive factors, and probiotic attributes (Tamang et al., 2009).
Pediococcus, Leuconostoc, and Bifidobacteria with enzyme α-galactosidase hydrolyze sugar raffinose and use them as a growing substrate for fermenting to lactate (Kumari et al., 2022). A similar genus of LAB has also been found in presently investigated gundruk samples. Bacteriocin-producing Lactobacillus isolated from gundruk, reported by Gautam and Sharma (2015), plays an important role as a natural preservative or biopreservative for the preservation of gundruk for 1–2 years in normal environmental conditions. Similar strains of lactic acid bacteria have also been observed in our metagenomic analysis. The reverse ecology analysis and other bioinformatics analyses revealed strong complementing features of the gundruk microbial population with the human system, which validates the probiotic nature of the gundruk and its beneficial role on humans. These results were in correlation with the earlier reports of culture-dependent analysis showing the dominance of Lactobacillus, Lactococcus, and Pediococcus genera in gundruk (Tamang et al., 2008). Interestingly, no Enterobacterium, Rhizobium, Pseudomonas, and Escherichia were observed in our phenotypic or biochemical results, which could be due to a lower abundance of these microbial consortia, age of the sample, limited sample size, and finally also due to inadequate cell lysis that may prevent the release of nucleases (Dolci et al., 2015). In accordance with the previous results, the exposure of food samples to various external environmental conditions, geographical regions, and manufacturing processes has various impacts on the microbial composition of the final products (Nam et al., 2012). Ethnic communities have traditional knowledge of preparations and the natural preservation of excess leafy vegetables, which are harvested in large quantities during winter seasons without cold storage and modern freezing facilities. This is a good example of the ‘biological preservation’ of perishable vegetables or leafy vegetables by using lactic acid bacterial fermentations and LAB-produced antimicrobial compounds, mostly bacteriocins, which preserve the food products naturally by preventing the growth of enteric bacteria and other pathogenic microbes (Sha et al., 2018; Das et al., 2020). LAB produces a sufficient amount of lactic and acetic acid for the prevention of the growth of pathogenic microorganisms in foods (Adams and Nicolaides, 1997). NGS raw data investigation showed the presence of a few Staphylococcus, Enterobacteriaceae, and Microbacteriaceae also observed in our gundruk samples that may be suppressed by lactic acid and bacteriocins produced by LAB, and it ultimately improves the self-life of gundruk (Gautam and Sharma, 2015).
This is the first next-generation sequencing result presented in this study that has helped to explore the diversity of fermentative hetero- and homo-fermentative LAB microflora, especially unique genera of Lactobacillus, Lactococcus, Pediococcus, Leuconostoc, Enterococcus, Vagococcus, Weissella, and Carnobacterium that were not reported before. LAB in gundruk is found to possess fermentative and functional properties, which can be used as an ideal starter culture(s) for the desirable production of foods. Hence, these LAB genera, with their probiotic attributes when used as starter cultures during the fermentation process of gundruk, can impart potential health benefits to the consumers.
Our study reveals the dominance of the genera Streptococcus and Lactobacillus, which can be used for the production of dairy and non-dairy fermented foods as a starter culture (Kumar et al., 2022). In the present investigations, Lactobacillus sp. is a major group in gundruk, it synthesizes the enzyme β-galactosidase, an enzyme that converts lactose to glucose and galactose and has various beneficial roles in fermentation in the dairy industry and overcomes lactose-intolerant phenomena (Saqib et al., 2017; Ayivi and Ibrahim, 2022). Currently, the high prevalence of lactose indigestion or lactose intolerance globally is a major problem for human populations. This is due to the absence of the enzyme lactase, making it incapable of metabolizing or utilizing lactose in food products (Saqib et al., 2017). The absence or decreased levels of the enzyme beta-lactase in a large portion of the human population creates an opportunity for the potential application of β-galactosidase enzyme in providing long-lasting remedies through its addition and incorporation in dairy and non-dairy food products (Movahedpour et al., 2022). The lactose-intolerant human population, therefore, stands a great benefit from assimilating foods with the help of this enzyme. Due to their great health applications, this enzyme can be explored technologically for many applications at the dairy and non-dairy fermented food industries level (Jurado et al., 2002; Saqib et al., 2017). The beta-lactase enzyme can also be used industrially to enhance the sensorial characteristics and aroma of food products by increasing their solubility rates and sweetness and also enhancing the formation of monosaccharide sugar (Jurado et al., 2002). Thus, gundruk microbiota can play an important role in the biosynthesis of β-galactosidase enzymes, used to overcome the lactose intolerance phenomena.
Lactobacillus is the most important bacterial genera among the various LAB, which is used for the production of postbiotics (Aggarwal et al., 2022). Our investigation revealed that the Lactobacillus genera is one of the predominant consortia of gundruk and thus can be used as postbiotics. Postbiotics are metabolic products or byproducts secreted or synthesized by viable or non-viable bacterial cells (lactic acid bacteria) or released after bacterial lysis, providing physiological benefits to the host organisms. A wide range of antimicrobial compounds or peptides are synthesized by viable and non-viable lactic acid bacteria, which fall into two categories: low molecular weight compounds (such as diacetylene, hydrogen peroxide, and carbon dioxide) and high molecular weight compounds (such as bacteriocins and bacteriocins-like byproducts). These compounds are known as postbiotics (Šuškovic et al., 2010; de Almeida Júnior et al., 2015). Enterococcus and Leuconostoc, which have been observed in our samples, find application as a biopreservative and anti-fungal agent for their antagonistic activities against many filamentous fungi producing diverse bioactive compounds and metabolites, such as ⍺-hydroxyisobutyric acid, β-phenyllactic acid, 1,3-butanediol, and phenethylamine (Fugaban et al., 2023).
Postbiotics development may create a new arena as preventives and therapeutic agents for diseases, such as diabetes, wound healing, mellitus, and adjunctive therapeutic agents and as biopreservation of food, packaging of food, food supplements, functional food, pharmaceuticals products, and biofilm control. Our study can be correlated with the study done by Aggarwal et al. (2022), it can be concluded that gundruk samples can also be a good source of postbiotics or metabiotics having various health attributes to impart (Figure 10). Fermented gundruk that contains consortia of viable and non-viable microbes (LAB) can synthesize primary and secondary metabolites, and therapeutics and nutraceuticals can play an important role in the sustenance of human health (Aggarwal et al., 2022).
Our study mainly focused on microbial diversity and ecology of gundruk of North Bengal. Incidentally, this is the first study of its kind where the total bacterial flora from gundruk sample from parts of Sikkim and North Bengal is considered by next-generation sequencing (NGS) followed by detailed reverse ecology analysis. However, its metabolomics and functionality studies can be conducted in the future for the development and commercialization of the products as well as profiling of secondary metabolites produced by LAB.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/bioproject/PRJNA910670/.
Ethics statement
Written informed consent was obtained from the individual(s) for the publication of any potentially identifiable images or data included in this article.
Author contributions
KG: Conceptualization, Data curation, Funding acquisition, Validation, Writing – original draft. SS: Conceptualization, Data curation, Validation, Writing – original draft. ST: Data curation, Methodology and Writing – original draft. IS: Conceptualization, Data curation, Formal analysis, Resources, Software, Writing – original draft. GS: Data curation, Formal analysis, Methodology, Resources, Writing – original draft. AS: Conceptualization, Formal analysis, Resources, Supervision, Writing – original draft.
Funding
The author(s) declare that no financial support was received for the research, authorship, and/or publication of this article.
Acknowledgments
The authors gratefully acknowledge the support of the Department of Science and Technology—SERB, New Delhi, and the Department of Food Technology, University of North Bengal.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Adams, M. R., and Nicolaides, L. (1997). Review of the sensitivity of different foodborne pathogens to fermentation. Food Control 8, 227–239. doi: 10.1016/S0956-7135(97)00016-9
Aggarwal, S., Sabharwal, V., Kaushik, P., Joshi, A., Aayushi, A., and Suri, M. (2022). Postbiotics: from emerging concept to application. Front. Sustain. Food Syst. 6:887642. doi: 10.3389/fsufs.2022.887642
Ayivi, R. D., and Ibrahim, S. A. (2022). Lactic acid bacteria: an essential probiotic and starter culture for the production of yoghurt. Int. J. Food Sci. Technol. 57, 7008–7025. doi: 10.1111/ijfs.16076
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods. 7, 335–336. doi: 10.1038/nmeth.f.303
Dahal, N. R., Karki, T. N., Swamylingappa, B., Li, Q., and Gu, G. (2005). Traditional foods and beverages of Nepal—a review. Food Rev. Int. 21, 1–25. doi: 10.1081/FRI-200040579
Das, A., and Deka, S. (2012). Mini review fermented foods and beverages of the North-East India. Int. Food Res. J. 19, 377–392.
Das, S., Mishra, B. K., and Hati, S. (2020). Techno-functional characterization of indigenous Lactobacillus isolates from the traditional fermented foods of Meghalaya, India. Curr. Res. Food Sci 3, 9–18. doi: 10.1016/j.crfs.2020.01.002
de Almeida Júnior, W. L. G., da Silva Ferrari, Í., de Souza, J. V., da Silva, C. D. A., da Costa, M. M., and Dias, F. S. (2015). Characterization and evaluation of lactic acid bacteria isolated from goat milk. Food Control 53, 96–103. doi: 10.1016/j.foodcont.2015.01.013
Dolci, P., Alessandria, V., Rantsiou, K., and Cocolin, L. (2015). “Advanced methods for the identification, enumeration, and characterization of microorganisms in fermented foods” in Advances in fermented foods and beverages. ed. W. Holzapfel (Sawston, Cambridge: Woodhead Publishing), 157–176.
Fugaban, J. I. I., Jung, E. S., Todorov, S. D., and Holzapfel, W. H. (2023). Evaluation of antifungal metabolites produced by lactic acid bacteria. Probiotics Antimicrob. Proteins 15, 1447–1463. doi: 10.1007/s12602-022-09995-5
Gautam, N., and Sharma, N. (2015). A study on characterization of new bacteriocin produced from a novel strain of Lactobacillus spicheri G2 isolated from Gundruk-a fermented vegetable product of north East India. J. Food Sci. Technol. 52, 5808–5816. doi: 10.1007/s13197-015-1710-x
Ghatani, K., and Tamang, B. (2017). Assessment of probiotic characteristics of lactic acid bacteria isolated from fermented yak milk products of Sikkim, India: chhurpi, shyow, and khachu. Food Biotechnol. 31, 210–232. doi: 10.1080/08905436.2017.1335212
Ghimire, A., Kumar Sah, A., and Poudel, R. (2020). Kinetics and modeling of growth and lactic acid production in Gundruk, a Himalayan fermented vegetable dish. Food Sci. Nutr. 8, 5591–5600. doi: 10.1002/fsn3.1854
Hammes, W. P., and Hertel, C. (2003). “The genus Lactobacillus” in The prokaryotes an electronic resource for the microbiological community (New York, NY: Springer-Verlag), 19–54.
Holt, J. G., Krieg, N. R., Sneath, P. H. A., Staley, J. T., and Williams, S. T. (1994). Bergey’s manual of determinative bacteriology. 9th Edn. Baltimore, MD: Williams & Wilkins, 965–1599.
Jurado, E., Camacho, F., Luzon, G., and Vicaria, J. M. (2002). A new kinetic model proposed for enzymatic hydrolysis of lactose by a bgalactosidase from Kluyveromyces fragilis. Enzym. Microb. Technol. 31, 300–309. doi: 10.1016/S0141-0229(02)00107-2
Karki, T., Okada, S., Baba, T., Itoh, H., and Kozaki, M. (1983). Studies on the microflora of Nepalese pickles Gundruk studies on “microorganisms and their role in Gundruk fermentation” part I. Nippon Shokuhin Kogyo Gakkaishi 30, 357–367. doi: 10.3136/nskkk1962.30.357
Kuipers, O. P., Buist, G., and Kok, J. (2000). Current strategies for improving food bacteria. Res. Microbiol. 151, 815–822. doi: 10.1016/S0923-2508(00)01147-5
Kumar, D., Lal, M. K., Dutt, S., Raigond, P., Changan, S. S., Tiwari, R. K., et al. (2022). Functional fermented probiotics, prebiotics, and synbiotics from non-dairy products: a perspective from nutraceutical. Mol. Nutr. Food Res. 66:2101059. doi: 10.1002/mnfr.202101059
Kumari, M., Kokkiligadda, A., Dasriya, V., and Naithani, H. (2022). Functional relevance and health benefits of soymilk fermented by lactic acid bacteria. J. Appl. Microbiol. 133, 104–119. doi: 10.1111/jam.15342
Kumbhare, S. V., Dhotre, D. P., Dhar, S. K., Jani, K., Apte, D. A., Shouche, Y. S., et al. (2015). Insights into diversity and imputed metabolic potential of bacterial communities in the continental shelf of Agatti Island. PLoS One. 10:e0129864. doi: 10.1371/journal.pone.0129864
Lebeer, S., Vanderleyden, J., and De Keersmaecker, S. C. (2008). Genes and molecules of lactobacilli supporting probiotic action. Microbiol. Mol. Bio. Rev. 72, 728–764. doi: 10.1128/MMBR.00017-08
Mayer, E. A., Tillisch, K., and Gupta, A. (2015). Gut/brain axis and the microbiota. J. Clin. Invest. 125, 926–938. doi: 10.1172/JCI76304
Movahedpour, A., Ahmadi, N., Ghalamfarsa, F., Ghesmati, Z., Khalifeh, M., Maleksabet, A., et al. (2022). β-Galactosidase: from its source and applications to its recombinant form. Biotechnol. Appl. Biochem. 69, 612–628. doi: 10.1002/bab.2137
Nam, Y. D., Lee, S. Y., and Lim, S. I. (2012). Microbial community analysis of Korean soy- bean pastes by next-generation sequencing. Int. J. Food Microbiol. 155, 36–42. doi: 10.1016/j.ijfoodmicro.2012.01.013
Parvez, S., Malik, K. A., Ah Kang, S., and Kim, H. Y. (2006). Probiotics and their fermented food products are benefificial for health. J. Appl.Microbiol. 100, 1171–1185. doi: 10.1111/j.1365-2672.2006.02963.x
Ray, R. C., and Swain, M. R. (2013). “Indigenous fermented foods and beverages of Odisha, India: an overview” in Indigenous fermented foods of South Asia (Boca Raton: CRC Press), 1–6.
Salminen, S., Bouley, C., Boutron, M. C., Cummings, J. H., Franck, A., Gibson, G. R., et al. (1998). Functional food science and gastrointestinal physiology and function. Br. J. Nutr. 80, S147–S171. doi: 10.1079/BJN19980108
Saqib, S., Akram, A., Halim, S. A., and Tassaduq, R. (2017). Sources of β-galactosidase and its applications in food industry. 3 Biotech. 7, 1–7. doi: 10.1007/s13205-017-0645-5
Sarkar, I., Kar, P., Sen, G., Chhetri, S., Bhattacharya, M., Bhattacharyya, S., et al. (2022). Metagenomic outlooks of microbial dynamics influenced by organic manure in tea garden soils of North Bengal, India. Arch.Microbiol 204, 1–9. doi: 10.1007/s00203-021-02635-6
Senapati, A. K., Ann, A., Raj, A., Gupta, A., Sharma, A., Neopany, B., et al. (2016). “Diversity of indigenous fermented foods and beverages of South Asia” in Indigenous fermented foods of South Asia. 1st ed (Boca Raton: CRC Press), 69–106.
Sha, S. P., Ghatani, K., and Tamang, J. P. (2013). Dalbari, a traditional pulse based fermented food of West Bangal. Int. J. Agric. Food Sci.Technol. 4, 6–10.
Sha, S. P., Jani, K., Sharma, A., Anupma, A., Pradhan, P., Shouche, Y., et al. (2017). Analysis of bacterial and fungal communities in Marcha and Thiat, traditionally prepared amylolytic starters of India. Sci. Rep. 7, 1–7. doi: 10.1038/s41598-017-11609-y
Sha, S. P., Suryavanshi, M. V., Jani, K., Sharma, A., Shouche, Y., and Tamang, J. P. (2018). Diversity of yeasts and molds by culture-dependent and culture-independent methods for mycobiome surveillance of traditionally prepared dried starters for the production of Indian alcoholic beverages. Front. Microbiol. 9, 1–15. doi: 10.3389/fmicb.2018.02237
Sha, S. P., Suryavanshi, M. V., and Tamang, J. P. (2019). Mycobiome diversity in traditionally prepared starters for alcoholic beverages in India by high-throughput sequencing method. Front.Microbiol. 10, 1–11. doi: 10.3389/fmicb.2019.00348
Sha, S. P., Thakur, N., Tamang, B., and Tamang, J. P. (2012). Haria, a traditional rice fermented alcoholic beverage of West Bengal. Int. J. Agric. Food Sci. Technol. 3, 157–160.
Sharma, A., and Sarkar, P. K. (2015). Microbial diversity in ethno-fermented foods of Indian Himalayan region. ENVIS Bull. Hima. Ecol. 23, 85–91.
Sudo, N. (2016). “The hypothalamic-pituitary-adrenal axis and gut microbiota: a target for dietary intervention?” in The gut-brain axis. (Cambridge, Massachusetts: Academic Press), 293–304.
Surve, S., Shinde, D. B., and Kulkarni, R. (2022). Isolation, characterization and comparative genomics of potentially probiotic Lactiplantibacillus plantarum strains from Indian foods. Sci. Rep. 12, 1–16. doi: 10.1038/s41598-022-05850-3
Šuškovic, J., Kos, B., Beganovi, C. J., LebošPavunc, A., Habjani, C. K., and Matoši, C. S. (2010). Antimicrobial activity – the most important property of probiotic and starter lactic acid bacteria. Food Technol. Biotechnol. 48, 296–307.
Tamang, J. P. (2016). Ethnic fermented foods and alcoholic beverages of Asia. 409. Delhi, India: Springer.
Tamang, J. P. (2022). “Ethno-microbiology” of ethnic Indian fermented foods and alcoholic beverages. J. Appl. Microbiol. 133, 145–161. doi: 10.1111/jam.15382
Tamang, B., and Tamang, J. P. (2010). In situ fermentation dynamics during production of gundruk and khalpi, ethnic fermented vegetable products of the Himalayas. Indian J. Microbiol. 50, 93–98. doi: 10.1007/s12088-010-0058-1
Tamang, B., and Tamang, J. (2014). Traditional knowledge of biopreservation of perishable vegetable and bamboo shoots in Northeast India as food resources. Indian J. Tradit. Knowl. 8, 89–95.
Tamang, J. P., Watanabe, K., and Holzapfel, W. H. (2016). Diversity of microorganisms in global fermented foods and beverages. Front. Microbiol. 7:377. doi: 10.3389/fmicb.2016.00377
Tamang, J. P., Tamang, B., Schillinger, U., Franz, C. M., Gores, M., and Holzapfel, W. H. (2005). Identification of predominant lactic acid bacteria isolated from traditionally fermented vegetable products of the eastern Himalayas. Int. J. Food Micrtobiol. 105, 347–356. doi: 10.1016/j.ijfoodmicro.2005.04.024
Tamang, B., Tamang, J. P., Schillinger, U., Franz, C. M., Gores, M., Holzapfel, W. H., et al. (2008). Phenotypic and genotypic identification of lactic acid bacteria isolated from ethnic fermented bamboo tender shoots of North East India. Int. J. Food Microbiol. 121, 35–40. doi: 10.1016/j.ijfoodmicro.2007.10.009
Tamang, J. P., Tamang, B., Schillinger, U., Guigas, C., and Holzapfel, W. H. (2009). Functional properties of lactic acid bacteria isolated from ethnic fermented vegetables of the Himalayas. Int. J. Food Microbiol. 135, 28–33. doi: 10.1016/j.ijfoodmicro.2009.07.016
van Baarlen, P., Troost, F., van der Meer, C., Hooiveld, G., Boekschoten, M., Brummer, R. J., et al. (2011). Human mucosal in vivo transcriptome responses to three lactobacilli indicate how probiotics may modulate human cellular pathways. Proc. Natl. Acad. Sci. U. S. A. 108, 4562–4569. doi: 10.1073/pnas.1000079107
Keywords: vegetable-based fermented foods, Lactobacillaceae family, NGS, reverse ecology, metabolic pathway
Citation: Ghatani K, Sha SP, Thapa S, Sarkar I, Sen G and Sen A (2024) Investigating bacterial diversity involved in the production of vegetable-based ethnic fermented food of North Bengal and their metabolic pathways with reverse ecology approach. Front. Sustain. Food Syst. 8:1322192. doi: 10.3389/fsufs.2024.1322192
Edited by:
Araceli Loredo, Autonomous University of Coahuila, MexicoReviewed by:
Adhitya Pitara Sanjaya, Sebelas Maret University, IndonesiaArmando Robledo-Olivo, Universidad Autónoma Agraria Antonio Narro, Mexico
Francisco Hernandez Centeno, Universidad Autónoma Agraria Antonio Narro, Mexico
Copyright © 2024 Ghatani, Sha, Thapa, Sarkar, Sen and Sen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Kriti Ghatani, Z2hhdGFuaWtAbmJ1LmFjLmlu; Shankar Prasad Sha, c2hhbmthcnByYXNhZHNoYUBnbWFpbC5jb20=; Arnab Sen, YXJuYWIuc2VuQG5idS5hYy5pbg==
†These authors share first authorship
 Kriti Ghatani
Kriti Ghatani Shankar Prasad Sha
Shankar Prasad Sha Subarna Thapa1
Subarna Thapa1 Indrani Sarkar
Indrani Sarkar Gargi Sen
Gargi Sen