- 1School of Marine Science, Ningbo University, Ningbo, China
- 2State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products, Ningbo University, Ningbo, China
Background: The fermented wax gourd, often referred to as “smelly wax gourd,” is a traditional food that undergoes natural fermentation. It’s a staple in eastern China and is recognized as Ningbo’s “city-fermented food.” Characterized by its distinct putrid flavor and soft texture, its safety, nutritional aspects, and sensory attributes have not been extensively studied.
Methods: In this research, the microbial community and flavor components of fermented wax gourd during its traditional fermentation were analyzed. The safety and impact on the gut microbiota were also assessed by administering it to healthy and pseudo-germ-free mice.
Results: The findings revealed that organic acids primarily contribute to the gourd’s flavor during fermentation. The aroma reminiscent of fruits is due to 2-methyl-butyric acid, while butyric, pentanoic, caproic, and octanoic acids are responsible for their characteristic smelly taste. In the fermentation of traditional foods, the abundance of substances and open fermentation contribute to the diversity of microorganisms in the system, and the reproduction and metabolism of microorganisms drive the fermentation of foods. From the results of this study, the flavor peaks on the 10th day of fermentation. Predominant microbes include Lactobacillus fermentum, Streptococcus equinus, Fusobacterium perfoetens, Weissella confusa, and Lactobacillus plantarum. Notably, Lactobacillus was the most abundant probiotic in the early fermentation stages. The “smelly” taste of smelly wax gourd was mainly derived from butyric acid, valeric acid, caproic acid, caprylic acid, p-methylphenol and other compounds, and the abundance of Caldicoprobacter algeriensis, Mariniphaga anaerophila, Streptococcus equi and Lactobacillus were significantly correlated with 4 of the above 5 acids. These four bacteria may contribute more to the “smelly” taste of smelly wax gourd. In the study, compared with the control group (CONT), the abundance of Helicobacter ganmani, H. Chanicola, Lactobacillus animalis, Lactobacillus gadi and Lactobacillus reuteri decreased in mice groups treated with anti-biological pretreatment followed by gavage of smelly wax gourd (A.SWG) and the smelly wax gourd (SWG) groups. Conversely, Muribaculum intestinale, Prevotellamassilia timonensis, Alistipes putredinis, Kineothrix alysoides and Clostridium indolis’ abundance of increases. Mice that underwent fecal microbiota transplantation (FMT) exhibited a higher abundance of probiotics like Bifidobacterium animalis, Bifidobacterium pseudolongum, Lactobacillus johnsonii, and others compared to the fecal culture microbiota transplantation (CMT) group. However, the CMT group had a higher presence of fermented and Royce lactobacilli.
Implication: Consuming fermented wax gourd can enhance the presence of beneficial probiotics and reduce pathogenic Helicobacter sp. in the mouse gut. Both Lactobacillus sp. and Bifidobacterium sp. showed increased abundance post fecal microbiota and fecal culture microbiota transplantation.
1 Introduction
The consumer demand for fermented vegetables with high nutritional value and probiotic microorganisms is increasing, also due to the consequence of the growing trend of vegetarianism and the increasing prevalence of cow’s milk allergy (Martins et al., 2013; Kumar et al., 2015; Flom and Sicherer, 2019; Okoye et al., 2023). Several famous traditional fermented vegetables are globally consumed and widely investigated, such as kimchi (Cheigh et al., 1994; Kim et al., 2021), Sichuan pickle (Rao et al., 2018) and sauerkraut (Beganović et al., 2015).
Fermented wax gourd (yan-dong-gua) also is a traditional fermented vegetable and famous cuisine in eastern China, dating back to the Song dynasty in Chinese history (A.D. 960–1279) (Wu et al., 2016b). Wax gourd, Benincasa hispida, consists of water 96.1 g, energy 54 kJ (13 kcal), protein 0.4 g, fat 0.2 g, carbohydrate 3.0 g (dietary fiber 2.9 g) per 100 g according to the data from the United States Department of Agriculture (Lim, 2012). The wax gourd has been consumed for many centuries for culinary and folk medical purposes as its health-promoting activities, such as alleviating cough and diabetes mellitus, stimulating urination, reducing body fat and blood triglycerides, and anti-obesity effects (Kang and Kwon, 2003; Ajuru and Nmom, 2017). Historically, fermented wax gourd served as a source of nutrients in winter when fresh vegetables were scarce due to its preserving a nutritive value of wax gourd, creating a desirable sensory and creamy texture property, and proving a particular safety food by proper homemade fermentation (Wu et al., 2016a). It is still popular and served in most restaurants in Ningbo, recognized as “the city fermented food” of Ningbo (one of the cities in east China). A fermented wax gourd is still homemade or restaurant-made naturally by adding salt (approximately 5–10%) and old fermented wax gourd solution as a starter at the beginning of the fermentation process and then through fermenting under anaerobic conditions for 10–20 days (Lan et al., 2012; Wu Z.-F. et al., 2015).
Unlike the famous and well-studied fermented vegetable (e.g., kimchi), the fermented wax gourd was only locally well-known, possibly due to the unique flavor and the lack of safety evaluation of this spontaneous fermentation, which is hard to bring modern consumer interests. Nonetheless, fermented gourds also offer the advantages of traditional vegetarian fermentation. On the one hand, fermented gourds prolong their shelf life, reduce their size and facilitate transportation (Ayed et al., 2020). On the other hand, during fermentation, the high enzymatic activity of lactic acid breaks down. It detoxifies many compounds (e.g., phenolic compounds, colorants, mycotoxins, pesticides, etc.) that are present in a variety of raw materials and, due to their high bioactivity, produces several biologically active substances (e.g., volatile fatty acids) (Park et al., 2014). It has been shown that lactic acid (LA) fermentation can reduce the risk of foodborne poisoning and health problems and that gram-negative bacteria’s growth is typically inhibited in the early stages of fermentation (Ogrodowczyk and Drabinska, 2021).
The microorganism plays an essential role in the flavor (Sidira et al., 2016; Xu et al., 2018), hygiene (Byakika et al., 2019), and functions of fermented foods during the process of fermentation (Choi et al., 2012; Ashraf and Shah, 2014; Wastyk et al., 2021; Diez-Ozaeta and Astiazaran, 2022). Lactic acid bacteria (LAB), such as Weissella spp. and Lactobacillus spp., were predominant bacteria in the ready-to-eat fermented wax gourd (Lan et al., 2009; Zhao et al., 2014).
In previous reports, the species of yeasts in pickled wax gourd, as well as the volatility compounds, were identified by 5.8 S rDNA-ITS (Internal Transcribed Spacer) sequencing and GC-MS (Gas Chromatography-Mass Spectrometry), respectively (Lin et al., 2023); and the microorganism of Pickled wax gourd was cultured with the medium in order to investigate the number and species of bacteria and yeast in pickled wax gourd (Wu Z.-F. et al., 2015). However, none of the previous studies can fully show the changes in microbial communities in the fermentation process of smelly wax gourd. Therefore, using 16S rDNA and ITS rDNA sequencing combination can investigate the succession of all microorganisms in the fermentation process of smelly wax gourd and subsequently clearly analyze the relationship between flavor components and microorganisms.
Numerous studies have revealed that gut microbiota plays a pivotal role in human health and disease. On the one hand, gut microbiota improves energy absorption and nutrients (Power et al., 2014). On the other hand, gut microbiota is closely associated with regulating human diseases, such as obesity, hypertension, metabolic syndrome, colitis, diabetes, etc. (Yoshimoto et al., 2013; Monk et al., 2016; Pevsner-Fischer et al., 2017; Chen et al., 2018). As previous reports, Lactobacillus isolated from kimchi could inhibit atopic dermatitis in NC/Nga mice, proving that Kimchi had an anti-obesity effect in diet-induced obese mice (Won et al., 2011; Park et al., 2012; Cui et al., 2015; Kim et al., 2020). However, studies of the interaction effect between fermented foods and gut microbiota are very limited so far, as well as smelly wax gourd. Therefore, it is necessary and feasible to investigate it.
In this study, high-throughput sequencing was used to investigate the link between the formation of volatiles and microorganisms during the fermentation of wax gourd, and also to explore the effects of these substances on the gut microbiota of mice, as well as the effects of FMT and cultured microbiota transplantation (CMT). It is expected to introduce a new perspective to the study of intestinal microbiota and provide a valuable theoretical basis for the study of fermented foods.
2 Materials and methods
2.1 Materials
The stinky winter melon was produced at the San Oxi Jia Cai market in Ningbo, Zhejiang Province, China. The standard methyl butyrate (0.898 g/mL, 25°C) was purchased from Shanghai Aladdin Biochemical Technology Co. of China. E.ZN.A. Fecal DNA kits were purchased from OMEGA (Norcross, GA, United States). In animal experiments, 25 specific pathogen-free (SPF)-grade 4 weeks-old male ICR mice (20 ± 2 g, Licence No. 1710300006) were obtained from Hangzhou Experimental Animal Center (Hangzhou, Zhejiang, China).
2.2 HS-SPME-GC-MS analysis
2.2.1 Internal standard, sample preparation and micro-extraction conditions
The winter melon salt embryos were immersed in the odour brine and sealed for fermentation at room temperature. The whole fermentation process lasted for 15 days. Samples were taken quickly from the center of the jar at the time points of 0 day (D0), 5 days (D5), 10 d (D10) and 15 days (D15) of fermentation, respectively, and mixed in the ratio of 1:1 (w:v), and homogenates were made in a high-speed tissue masher to be analyzed in the subsequent analysis. The standard of 0.898 g/mL methyl butyrate was prepared into a solution with a concentration of 89.8 mg/L, and stored in the refrigerator at −4°C for spare parts (Li et al., 2021).
Five grams of the homogenate sample was weighed into a 15 mL SPME headspace vial, 20 μL of methyl butyrate standard solution was added and the cap was quickly closed, and the extraction was carried out by inserting a 100 μm PDMS fibre tip in a water bath at 60°C for 45 min, and then inserting the extraction tip into the gas-phase injection port and resolving the sample for 3 min at 230°C after completion of the extraction (Yang et al., 2022).
2.2.2 GC-MS conditions
GC-MS analyzed the smelly wax gourd using GC-MS 7890A/5977A (Agilent Technologies Inc., Palo Alto, CA, United States), equipped with a silica capillary column (Vocol, 60 m × 0.32 mm × 1.8 μm thickness, Supelco, Inc., Bellefonte, PA, United States). The initial oven temperature was set at 32°C and kept warm for 5 min. The temperature was then increased to 45°C at a rate of 3°C/min and kept warm for 1 min for the first stage, then increased to 230°C at a rate of 5°C/min and kept warm for 20 min. The mass spectral ionization mode was EI, with a spectral scanning range of 45–550 m/z. The mass spectral transfer line temperature was 230°C, and the ion source temperature was 220°C. Volatile flavor compounds were analyzed semi-quantitatively by comparison to the chromatographic peak areas of internal standards.
2.3 Animal experimental design
2.3.1 Ethics statement
All experimental procedures and animal care adhered to the guidelines set by the “Experimental Animal Care and Use Guide” from the Experimental Animal Center of Ningbo University, which is affiliated with the Experimental Animal Sharing Service Platform of Zhejiang Province. The animal study received approval from the same center, under the approval number SYXK (Zhejiang 20080110).
2.3.2 Grouping and feeding
After 1 week of adaptation, the 15 4 weeks-old ICR male mice (weighing approximately 20 ± 2 g) were randomly divided into 3 groups, A.SWG group, control group (CONT) and SWG group (5 mice per group). To establish a pseudo-germ-free mouse model (Heimesaat et al., 2011; Brun et al., 2013), mice in A.SWG group first received 1 week of antibiotic pretreatment (comprising vancomycin 50 mg/kg, neomycin 100 mg/kg, metronidazole 100 mg/kg, and ampicillin 100 mg/kg; 100 μL per mouse per 12 h) by oral gavage and then they were given the smelly wax gourd mixture (20 g/kg/day) for 4 weeks (Haug et al., 2011). Mice in the SWG group were given saline for one week, then they were gavaged the smelly wax gourd mixture (20 g/kg/day) for 4 weeks. While CONT group mice were given the same amount of saline by gavage for 5 weeks.
Based on the established experiment (the smelly wax gourd treatment), the microbiota transplantation was carried out. After 4 weeks of intervention, daily collection of their feces under aseptic conditions was initiated, and a portion of the feces was used for gavage colonization into recipient mice, while another portion of the feces was cultured in anaerobic medium and then gavage colonized into recipient mice. The specific operation method is as follows:
Ten ICR mice were randomly divided into two groups with 5 mice in each group. One group received fecal microbiota transplantation (FMT) from the SWG group by gavage for 4 weeks (200 mg/day, 200 μL per mouse). The other group received culture microbiota transplantation (CMT) from the SWG group by gavage for 4 weeks (200 mg/day, 200 μL per mouse).
2.3.2.1 Fecal microbiota transplantation
The feces were resuspended in sterile saline at 100 mg/mL, and the solution was strongly mixed with Vortex-Genie for 10 s, and then centrifuged at 1,200 rpm for 3 min. The supernatant was collected and gavage into recipient mice. This process is guaranteed to be completed within 10 min to prevent changes in bacterial composition.
2.3.2.2 Fecal culture microbiota transplantation
The feces were cultured in the anaerobic medium at 100 mg/mL for 3 days and stored at −80°C. The feces were removed before each gavage and centrifuged at 1,200 rpm for 3 min. Then the culture medium was removed and an equal amount of normal saline was added. The solution was mixed vigorously for 10 s using a desktop vortex meter, and the supernatant was collected and gavage into recipient mice.
2.4 Total DNA extraction, PCR, and sequencing
Use the E.ZN.A. ® Fecal DNA kits extracted DNA from samples according to the manufacturer’s instructions, and the number and concentration of extracted DNA was measured by Thermo NanoDrop 2000 C (Thermo Fisher Scientific, MA, United States). The reagent, designed to uncover DNA from trace amounts of sample, is effective for the preparation of the DNA of most bacteria. Nuclear-free water was used for blank. Total DNA were eluted in 50 μL elution buffer, stored at −80°C, and subjected to PCR testing by LC Sciences Co., Ltd. in Zhejiang (Hangzhou China). The V3–V4 region of the 16S rRNA gene was amplified with slightly modified versions of primers 338F (5′-ACTCCTACGGGAGGCAGCA-3′) and 806R (5’-GGACTACHVGGGTWTCTAAT-3′). The 5′ ends of the primers were tagged with specific barcode per sample and universal sequencing primers. The total volume of PCR amplification was 25 μL reaction mixture, containing 25 ng template DNA, 12.5 μL PCR Premix Ex Taq TM Hot Start Version (12.5 μL) (Takara Biotechnology Co., Ltd., Dalian, Liaoning, China), each primer 0.1 μM, PCR-grade water adjusted volume. PCR conditions for amplified nucleus 16S fragment: 98°C initial denaturation 30 s, 98°C denaturation 10 s, 54°C annealing 30 s, 72°C stretching 45 s, final stretching 10 min, total 35 cycles (Zhang et al., 2018).
The PCR products were verified using 2% agarose gel electrophoresis and purified with a QIAquick PCR Purification Kit. Then purified PCR products were used to establish the amplicon library and the amplicon library sequenced by MiSeq Illumina 2×300 paired-end sequencing on an Illumina MiSeq platform (Illumina, San Diego, CA, United States) by LC Sciences Co., Ltd. (Hangzhou, Zhejiang, China).
2.5 Data analysis
As previously described (Lu et al., 2017), QIIME (version 1.8.0) (Caporaso et al., 2010), and PEAR (version 0.9.6) (Zhang et al., 2014) were used to filter the raw data as follows: (1) merging the reads that were overlapped by more than 10 bp, and removing the reads that could not be merged; (2) data belonging to each sample were identified through the barcode sequence; (3) when the average quality score on the 10 bp sliding window was below 20, the reads were truncated; and (4) the low-complexity sequences were also discarded by PRINDEQ (version 0.20.4) (Schmieder and Edwards, 2011). Then, Uchime (version 4.2.40) (Edgar et al., 2011) was used to identify and discard the chimera sequences. Uclust (version 1.1.579) assigns sequences with similarity ≥97% to the same Operational Taxonomic Unit (OTU) (Edgar, 2010). Representative sequences were chosen for each OTU, and taxonomic data were then assigned to each representative sequence using the RDP (Ribosomal Database Project) classifier (Wang et al., 2007). The OTUs abundance information is normalized using the sequence number criterion corresponding to the sample with the lowest sequence; the diversity complexity of a single sample species is analyzed using the four indices of Alpha diversity Chao 1, Observed species, Shannon, and Simpson, All indices in the sample were calculated using mothur (version 1.30.1) (Schloss et al., 2009). Beta diversity analysis was used to assess differences in sample species complexity. Calculation of beta diversity using Principal Coordinate (PCoA) and FastTree cluster analysis (2.1.3) (Price et al., 2010) Muscle (version 3.8.31) (Edgar, 2004), and the vegan package in R (version 3.2).
Values that align with a healthy distribution, determined using the standard deviation of the average, were analyzed using a one-way analysis of variance (ANOVA) followed by Tukey’s post hoc test. Data not fitting this distribution were analyzed using the Mann–Whitney test. A p-value of less than 0.05 indicates a statistically significant difference. Bar charts were created using GraphPad Prism8. To identify significant differences in the relative abundance of microbial groups between groups at the species level, we employed the Linear Discriminant Analysis Effect Size (LEfSe) method.
2.6 Accession numbers
The following sequences have been archived in the NCBI Sequence Read Archive database. Those associated with accession number PRJNA1030292 pertain to the microbial diversity observed during cucurbit fermentation. Meanwhile, data linked to accession number PRJNA1030303 are representative of the intestinal microorganisms in mice subjected to an odorous cucurbit-based diet.
3 Results
3.1 Changes of flavor substances and microbiota of smelly wax gourd
3.1.1 Volatile compounds of smelly wax gourd
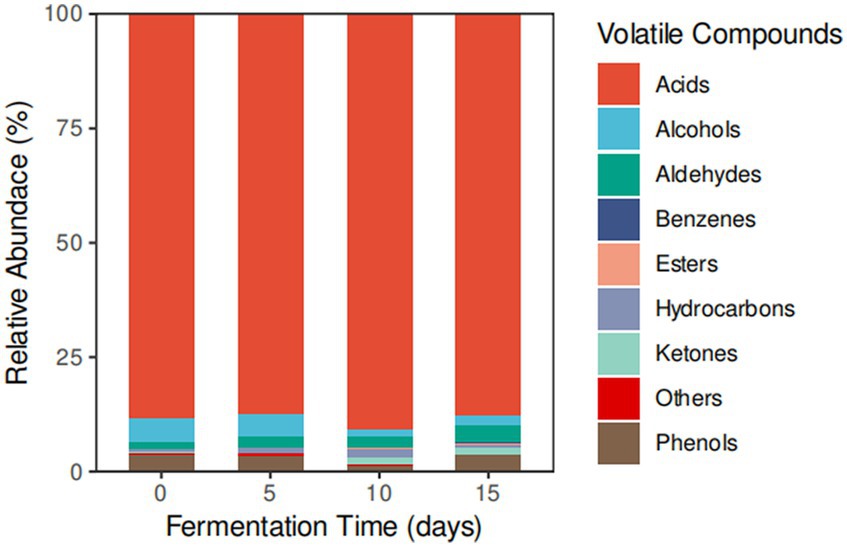
The volatile compounds present in the smelly wax gourd were analyzed using GC-MS, as detailed in Supplementary Table S1. This analysis identified a total of 86 volatile constituents across four different samples. These constituents comprised 15 acids, 20 alcohols, 8 aldehydes, 5 ketones, 7 phenols, 16 hydrocarbons, 4 esters, 2 benzenes, and 9 other compounds. Interestingly, the number of volatile compounds first decreased and then increased during the fermentation process.
Throughout fermentation, alcohol and phenol concentrations decreased until the fifth day. Conversely, the concentrations of acids, aldehydes, ketones, and hydrocarbons declined until the 10th day. Acids consistently had the highest concentration. During fermentation, the proportion of acid in the total flavor substances exhibited a parabolic trend, with percentages of 88.42 ± 0.35%, 87.21 ± 0.03%, 90.33 ± 0.02%, and 87.12 ± 0.28% at different stages. Butanoic acid was always the most prevalent acid. Alcohol content displayed an inverted parabolic trend with fermentation time, registering at 5.3 ± 0.34%, 5.11 ± 0.03%, 1.55 ± 0.02%, and 2.47 ± 0.28%. Among aldehydes, benzaldehyde, 2,5-dimethyl- consistently had the highest concentration at each fermentation stage, with values of 1.11 ± 0.19%, 1.92 ± 0.01%, 1.45 ± 0.09%, and 0.98 ± 0.09%. Notably, on the 10th day of fermentation, compounds like L-lactic acid (6.60 ± 0.01%), 2-methyl-2-Butanedioic acid (0.48 ± 0.01%), and 3-Ethylcyclopentanone (0.77 ± 0.05%) emerged. Benzene derivatives, specifically p-xylene and ethylbenzene, were only detected on the 10th and 15th days of fermentation. Further details can be found in Figure 1 and Supplementary Table S1.
3.1.2 Diversity of microbiota during the fermentation of wax gourd
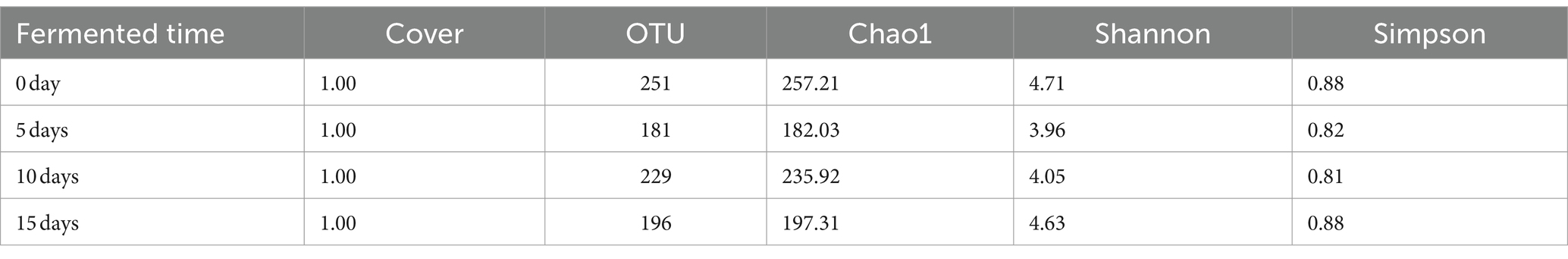
The alpha diversity of the smelly wax gourd index at different fermentation times was shown in Table 1. The coverage rate of each sample reached 1.00, indicating that the sequences in the sample library were measured. At the early stage of fermentation (0 day), the OTU number, Chao1, Shannon, and Simpson indices were highest, which were 251, 257.21, 4.71, and 0.88, respectively, indicating that the community richness and diversity of smelly wax gourd at 0d was the highest. On the contrary, 5 days was the lowest.
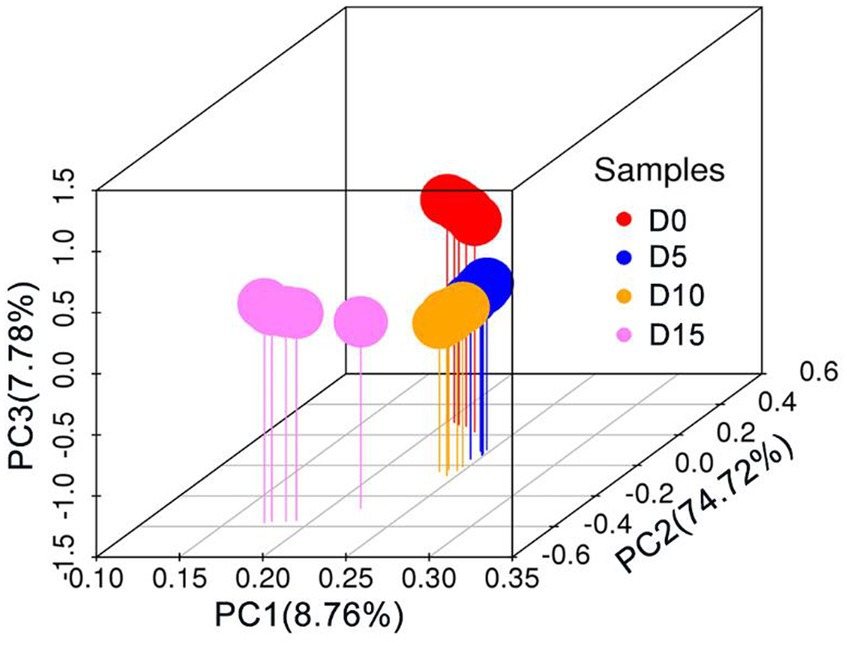
Beta diversity reflects diversity differences among samples. The overall structure of the microbiota at four fermentation periods of smelly wax gourd was analyzed by weighted UniFrac PCoA. As shown in Figure 2, the microbial structure of smelly wax gourd at 5 days and 10 days fermentation was close, while compared with fermentation at 0 day, fermentation at 15 days had the most significant difference in microflora structure, indicating the structure of the microbial community changes during the fermentation of smelly wax gourd.
3.1.3 Microbial shifts based on taxon-based analysis
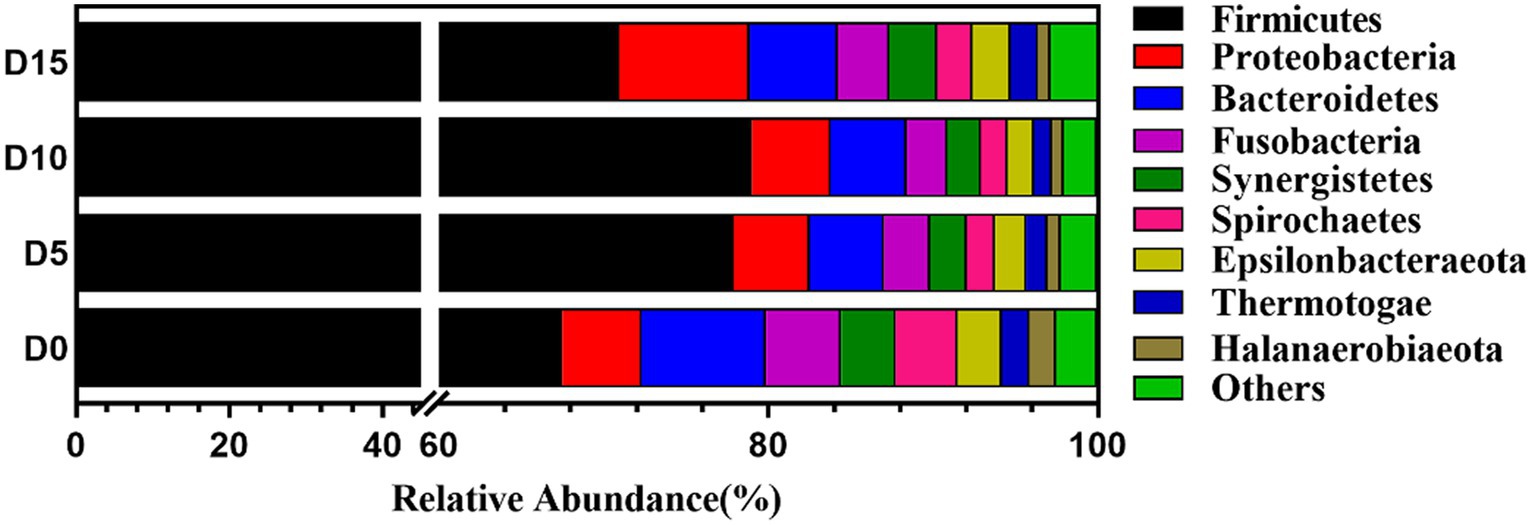
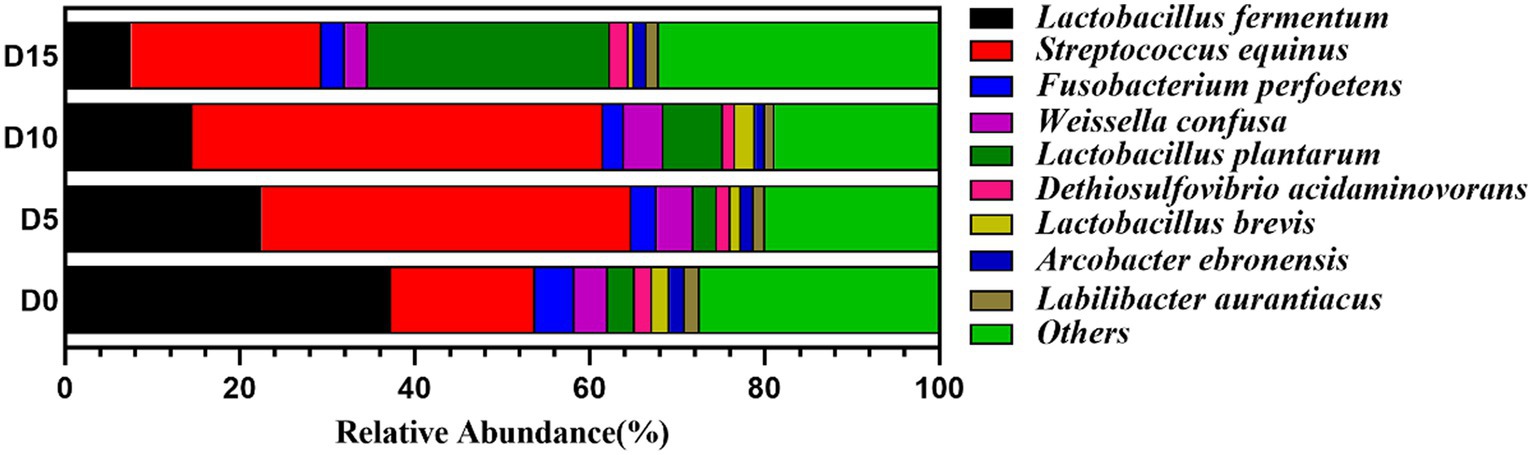
As shown in Figure 3, at the phylum level, the bacteria community of the smelly wax gourd was mainly composed of Firmicutes, Proteobacteria, Bacteroidetes, Fusobacteria, Synergistetes, and Spirochaetes. The six phyla accounted for 91.48, 93.78, 94.50, and 92.37% of the entire sequence of each group in four fermentation periods, respectively. Firmicutes were the most abundant phylum in all samples, accounting for more than 67% of the total sequence of each group. Their abundance increased with the prolonging of fermentation time, reaching the highest (79.02%) at D10 of fermentation, then decreased to 70.92%, while the abundance of Proteobacteria increased to 7.96% at D15.
At the species level, Lactobacillus fermentum, Streptococcus equinus, Fusobacterium perfoetens, Weissella confuse, and Lactobacillus plantarum were the dominant bacteria during the fermentation process of the wax gourd. The abundance of Lactobacillus fermentum was the highest (37.28%) at the D0 stage, then its abundance decreased to 22.46, 14.55, and 7.55% at D5, D10, and D15, respectively. The abundance of Streptococcus equinus increased significantly from 16.46% (D0) to 42.30% (D5) and then decreased from 46.98% (D10) to 21.76% (D15). The abundance of Lactobacillus plantarum was less than 7% in the first three fermentation stages but increased significantly to 27.68% in the D15 stage (Figure 4 and Supplementary Table S2).
3.1.4 The correlation between the bacteria and the flavor substance of smelly wax gourd
Based on the correlation analysis method, the correlation diagram of microbial species and volatile substance composition of smelly wax gourd was constructed (Figure 5). Among them, Streptococcus equinus and Weissella confusa were positively correlated with organic acid. In contrast, Lactococcus lactis, Pediococcus pentosaceus, Aminobacterium colombiense, M. anaerophila, C. algeriensis, Oceanimonas smirnovii, and Halocella cellulosilytica were positively correlated with organic acid, which was the dominant flavor substance in the smelly wax gourd.
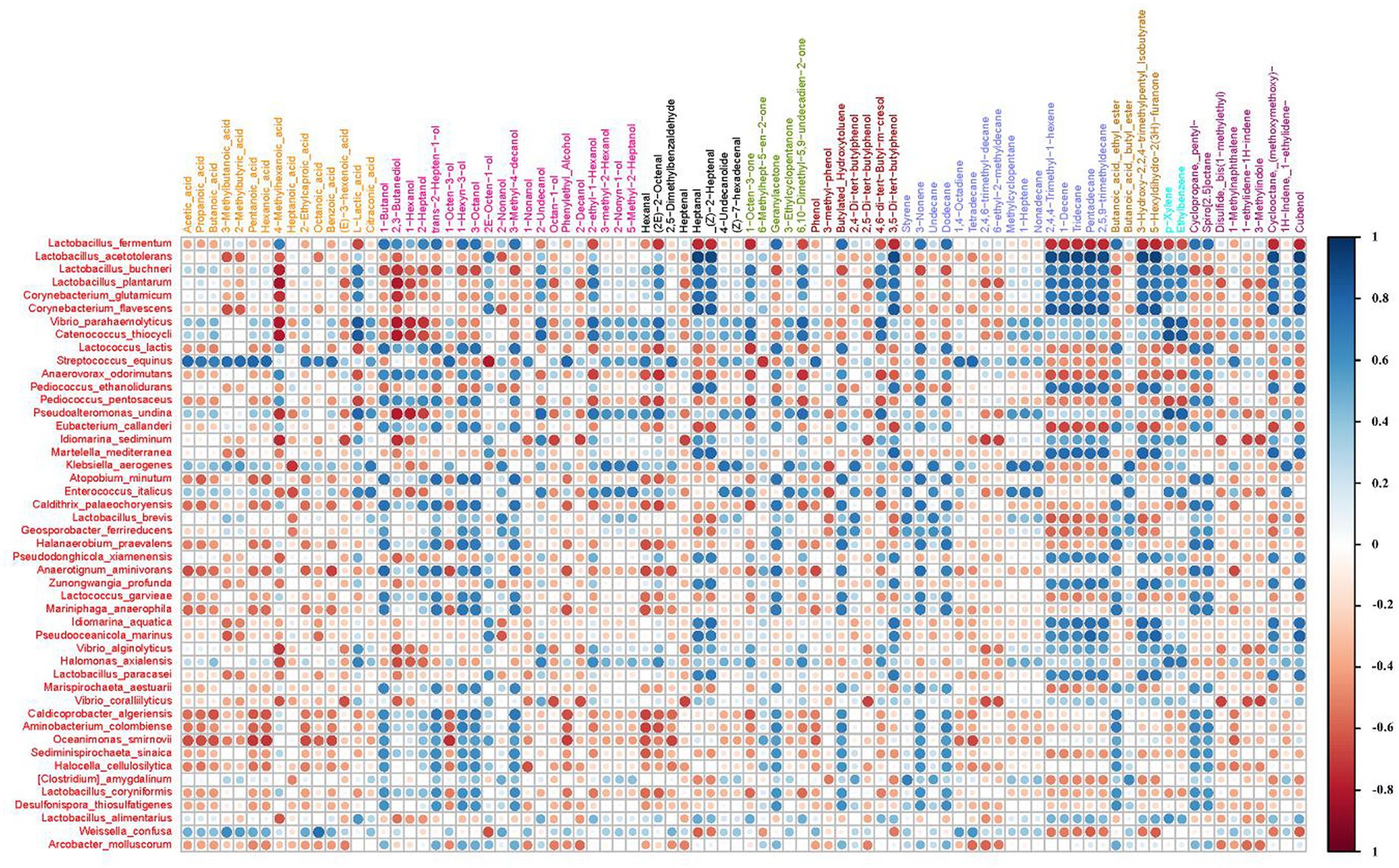
Figure 5. The corrplot of 47 species and 86 volatile substances. Line: species, column: volatile substances, blue is positive correlation, red is negative correlation. The larger the bubble, the greater the correlation, and vice versa.
Furthermore, significant correlations were observed between specific bacteria and various organic acids. Three bacteria showed significant positive correlations with acetic acid, while 11 exhibited negative correlations. Similarly, three bacteria displayed notable positive correlations with butyric acid, with 10 demonstrating negative associations. Additionally, 13 bacteria showed a significant positive correlation with lactic acid and seven showed a negative correlation. Similarly, two bacteria exhibited strong positive correlations with octanoic acid, whereas six had negative correlations. Two bacteria also displayed significant positive correlations with pentanoic acid, with 12 exhibiting negative associations, and two bacteria showed significant positive correlations with hexanoic acid, while 12 had negative correlations. Finally, five bacteria were notably positively correlated with 2,3-butanediol, while 18 showed negative correlations (Figure 5).
3.2 Effects of smelly wax gourd diet on gut microbiota in mice
3.2.1 Diversity of gut microbiota in mice
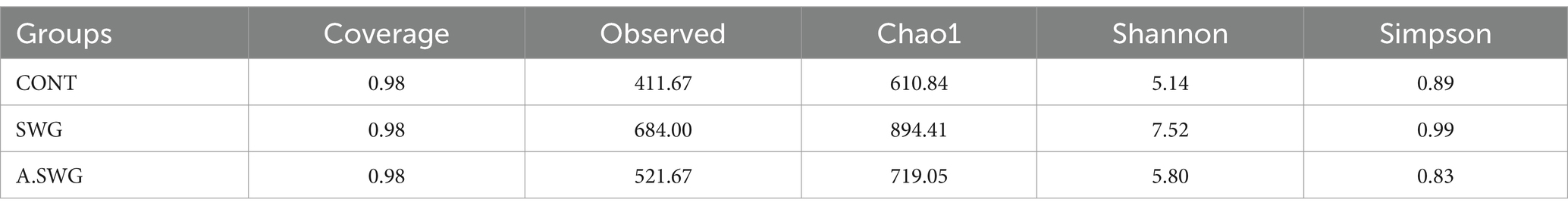
After 5 weeks of feeding, we sequenced fecal samples to elucidate the contribution of the smelly wax gourd treatment, fecal microbiota transplantation, and culture microbiota transplantation on the gut microbiota structure. The Chao1 observed species, Shannon, and Simpson indices were all alpha diversity indexes. Thereinto, the Chao1 and observed species indices represent community richness while others represent community diversity. In this study, the smelly wax gourd treatment increased community richness and diversity in mice, and the community richness and diversity in the SWG group were higher than that in A.SWG group (Table 2).
Beta diversity reflects diversity differences among samples. Weighted UniFrac PCoA analyzed the gut microbiota’s overall structure in three mice groups. As shown in Figure 6, the smelly wax gourd treatment caused the deviation of fecal microflora from the CONT group, and the deviation from the control group was furthest in the SWG group compared with the A.SWG group, indicating that diet has some effect on the gut microbiota compared to antibiotic treatment.
3.2.2 Microbial shifts based on taxon-based analysis
Based on the principle of relative abundance greater than 2%, a total of 10 dominant species were selected from the three groups, among which Helicobacter ganmani and H. canicola belong to Proteobacteria, Muribaculum intestinale, Prevotellamassilia timonensis, and Alistipes putredinis belong to Bacteroidetes, Lactobacillus animalis, Kineothrix alysoides, Lactobacillus reuteri, and Clostridium indolis belong to the Firmicutes, compared with the CONT group, the abundance of H. ganmani, H. canicola, L. animalis, Lactobacillus gasseri, and L. reuteri decreased in the SWG group. In contrast, the abundance of M. intestinale, P. timonensis, A. putredinis, K. alysoides, and C. indolis increased. The variation trend of the A.SWG group was consistent with that of the SWG group. In addition, the supplement of smelly wax gourd led to the disappearance of H. ganmani, and H. canicola (Figure 7 and Supplementary Table S3).
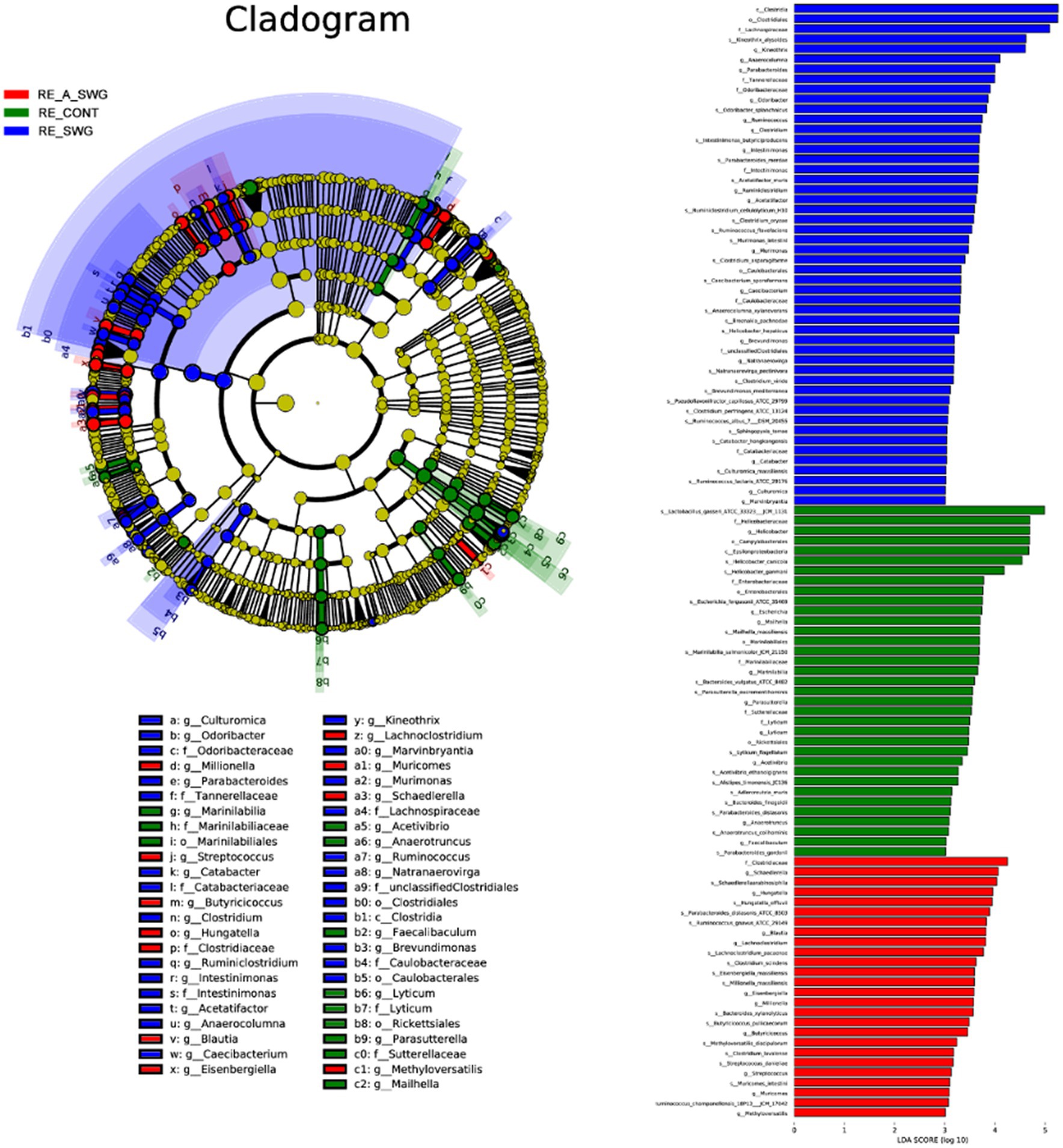
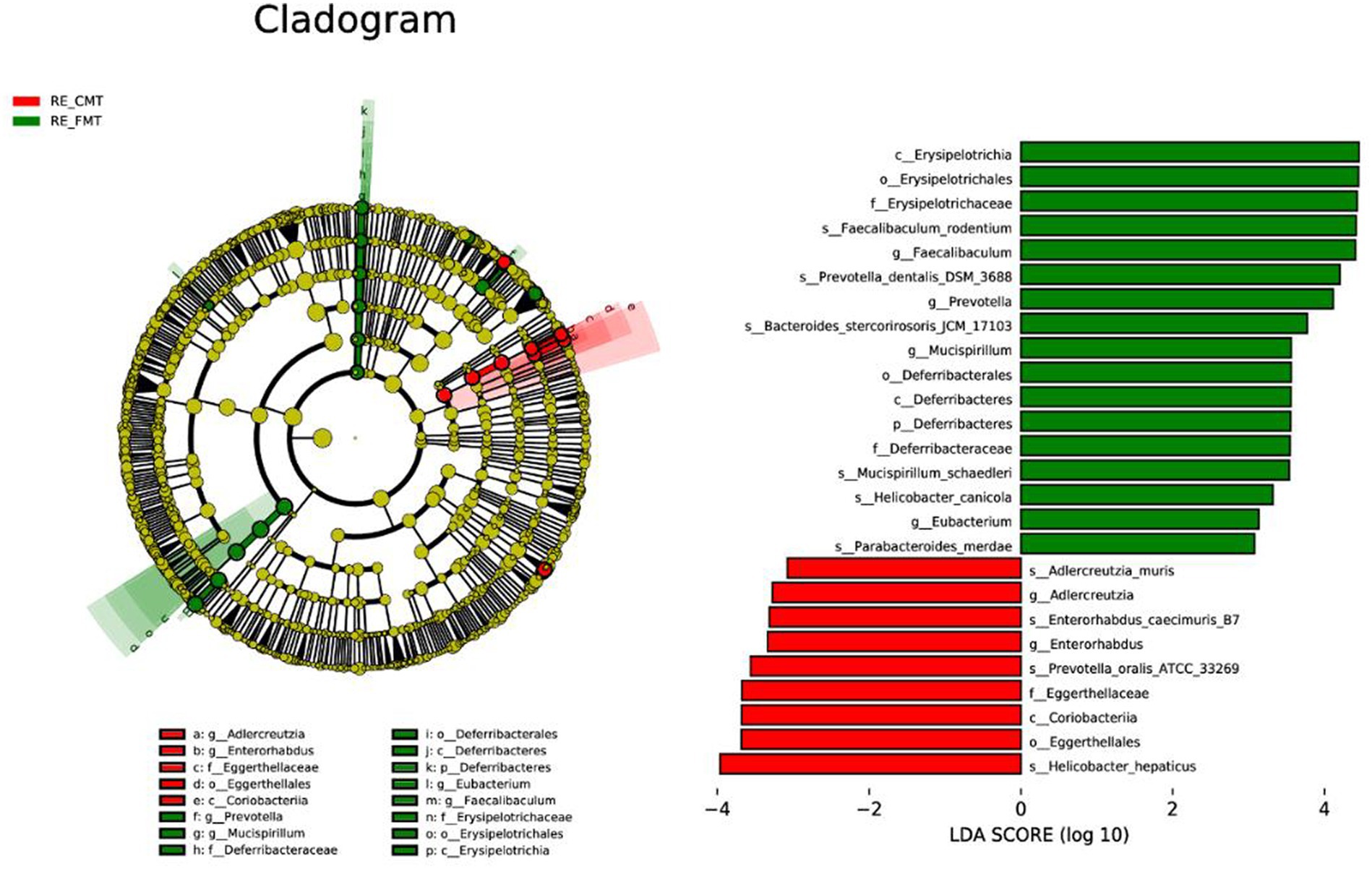
LEfSe analysis identified other representative species of each group as biomarkers to distinguish different groups. According to our LDA of the effective size, the abundances of many taxa were significantly different among the CONT, SWG, and A.SWG groups (LDA score >3.0; Figure 8). There were 55 biomarkers for species levels, including 16 species with high abundance in the CONT group, 24 species with high abundance in the SWG group, and 15 species with high abundance in the A.SWG group. Supplement of smelly wax gourd led to the disappearance of Bacteroides vulgatus, B. finegoldii, H. canicola, H. ganmani, Lyticum flagellatum and Mailhella massiliensis, but also adding some new species, including Clostridium perfringens, M. intestini, Caecibacterium sporoformans, C. oryzae, Culturomica massiliensis, H. hepaticus, P. merdae, Ruminococcus flavefaciens, R-lactaris, Sphingopyxis terrae, R. champanellensis and R. albus. In addition, a supplement of smelly wax gourd significantly decreased the abundance of L. gasseri (Supplementary Table S4).
3.3 Effects of FMT and CMT on gut microbiota in mice
3.3.1 Diversity of gut microbiota in mice
The Alpha diversity coverage of the two groups achieved 0.98, indicating that sample sequencing results can reflect the real situation of samples. The diversity results showed that the CMT group’s Observed index and Chao1 index were lower than the FMT group’s. In contrast, the Shannon index and Simpson index were similar, indicating that the difference between the two transplantation methods was reflected in the bacterial abundance, while that of the FMT group was higher (Table 3).
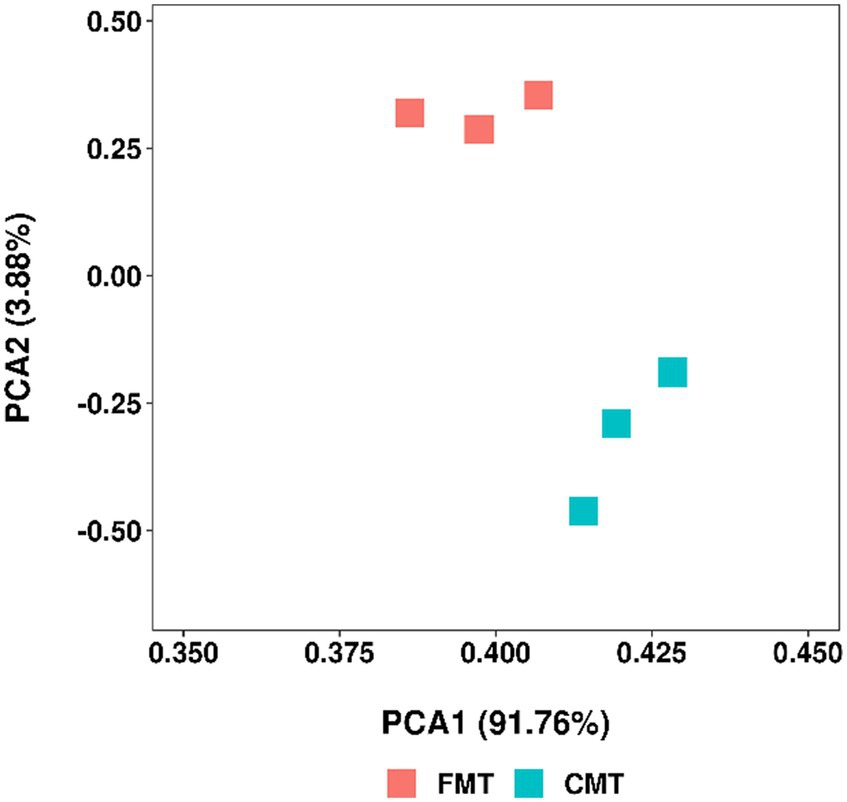
As shown in Figure 9, the gut microbiota of FMT and CMT was a deviation from each other.
3.3.2 Microbial shifts based on taxon-based analysis
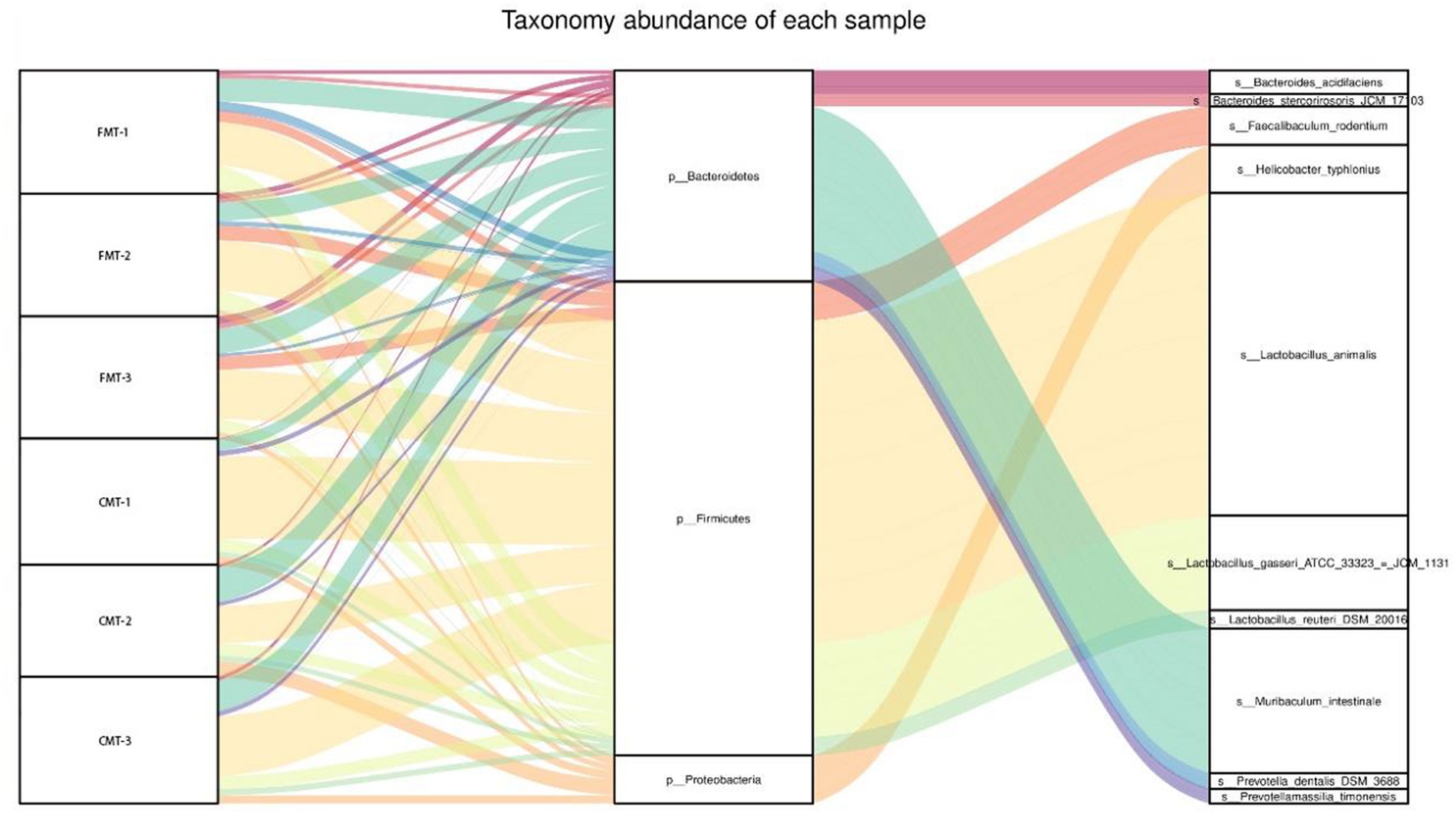
Based on the principle of relative abundance greater than 2%, a total of 10 dominant species were selected from the three groups, among which M. intestinale, Prevotellamassilia timonensis, Prevotella dentalis, Bacteroides acidifaciens, and Bacteroides stercorirosoris belong to Bacteroidetes, L. animalis, L. gasseri, Faecalibaculum rodentium, and L. reuteri belong to the Firmicutes, Helicobacter typhlonius belong to Proteobacteria. The abundance of L. animalis in the CMT group was 37.17%, was higher than that in the FMT group. The abundance of M. intestinale was higher in the CMT group (16.19%), while the abundance of L. gasseri was higher in the FMT group (11.68%). In addition, the abundance of P. dentalis, Bacteroides acidifaciens, F. rodentium, and Bacteroides stercorirosoris in the FMT group was higher than that in the CMT group. In comparison the abundance of L. reuteri, P. timonensis, and H. typhlonius in the FMT group was lower than that in the CMT group (Figure 10 and Supplementary Table S5).
LEfSe analysis (LDA score >3.0) based on the gut microbiota of the three groups of mice showed that there were 10 biomarkers at the species level, which showed that the abundance of P. dentalis, P. merdae, M. schaedleri, H. canicola, F. rodentium, and B. stercorirosoris was higher in the FMT group. However, the abundance of Prevotella oralis, H. hepatae, Enterorhabdus caecimuris, and Adlercreutzia muris was higher in the CMT group, among which P. dentalis, P. merdae, and B. stercorirosoris were not detected in the CMT group (Figure 11 and Supplementary Table S6).
4 Discussion
The flavor composition of fermented food is multifaceted. First of all, the flavor should derived from various flavoring substances contained in the raw vegetables themselves, which are still retained in the raw materials after fermentation, such as various intermediate products of the physiological metabolism of vegetables, as well as the trace volatile flavor substances contained in the vegetables themselves, which have important contributions to flavor. Secondly, the flavor should be derived from microbial fermentation. Various microorganisms in the system decompose the fermentable carbohydrates, proteins, and other substances in the raw materials to supply their growth and produce many substances with unique flavors. Thirdly, the formation of the flavor of kimchi is the result of the above factors. For example, organic acids contained in the raw materials may combine with alcohols produced by microorganisms to form aromatic esters (Kim et al., 2012).
Butyric acid, also known as caseic acid, has the odor of rotten butter, and other unpleasant odors, such as caproic acid and caprylic acid, form the odor of smelly wax gourd. In contrast 2-methyl butyric acid exists in the aroma components of various fruits, playing a particular role in enhancing aroma. The concentration of 2, 3-butanediol, hexanol, 1-nonyl alcohol, and octyl alcohol was the highest at the D5 phase. The concentration of acetaldehyde and anti-2-octenal was the highest at the D10 phase. These alcohols and aldehydes had high concentrations in pickled wax gourd and pickled amaranth stem (Wu Z.-F. et al., 2015; Wu et al., 2016b). Among the phenolic substances, p-methyl phenol has the highest concentration in the D5 phase, while phenol has the highest concentration in the D10 phase. P-methyl phenol has a unique odor, while phenol has the phenolic odor of plastic and rubber, which enhances the odor of brine. Phenols are also the main aroma components of Wang Zhihe stinky tofu (Liu et al., 2012). The peculiar smell of bamboo shoots during the fermentation of traditional bamboo shoots in Taiwan was caused by p-methyl phenol (Steinhaus and Schieberle, 2005; Selvaraj et al., 2013; Saito et al., 2018). Tyrosine is bamboo shoots’ primary free amino acid, which can be decomposed into p-methyl phenol (Tang et al., 2018). Since amaranth root and bamboo shoot are both high-fiber plants, it is speculated that p-methyl phenol may come from fermentation products of amaranth root. The structural formula of tyrosine contains phenol, so it is speculated that phenol may come from the decomposition of tyrosine, and there may be a specific correlation between phenol and p-methyl phenol (Doerner et al., 2009a,b; Franchi et al., 2018; Saito et al., 2018). In addition, volatile compounds such as methylcyclopentane, 1,4-octadiene, and butyl butyrate were also detected. To sum up, the flavoring substances in the D10 stage were the most abundant, and there were no harmful volatile substances, therefore, this may be considered the best time to consume it.
Lactobacillus is a gram-positive and anaerobic fermentation bacterium in various fermented foods (Holzapfel et al., 2001). In many cases, these Lactobacillus were also used as starter cultures for industrial and artisanal foods because they help maintain the flavor and texture of fermented foods. Their primary function is to convert the fermentation of sugars in raw materials into lactic acid. The production of antimicrobial peptides, exopolysaccharides, and a variety of other metabolites are their important properties isolated L. fermentum with potential probiotics from traditional dairy products (Ross et al., 2002; Tamime, 2002; Bao et al., 2010). The study found that L. fermentum has a high tolerance to simulated gastric intestinal fluid and bile salt, and broad antibacterial activity, with the most significant potential of probiotics. In addition, L. plantarum is versatile, widely found in the fermentation of dairy products, meat, and many vegetables, and is capable of surviving gastric transport and colonizing the intestines of humans and other mammals (de Vries et al., 2006). Of the hundreds of reports in the 20th century about the safe use of L. plantarum, only a few suggested that certain L. plantarum might be involved in infection.
Lactobacillus acetotolerans, Corynebacterium flavescens, and Pseudooceanicola marinus had high positive correlation coefficients with decanolactone, cubenol and pentadecane. Lactobacillus acidophilus and Corynebacterium flavus had high positive correlation coefficients with 2-Heptenaldehyde, decene, 2,4,4-trimethyl-1-hexene, 2,5,9-trimethyl-decane, 3, 5-tert-butylphenol, 2,2,4-trimethyl-1,3-pentanediol mono-isobutyrate, Cyclooctane, heptanal, and tridecane. The negative correlation coefficient was higher between L. plantarum and 2, 3-butanediol and 4-methyl-acetic acid. The “smelly” taste of the smelly wax gourd was mainly derived from the odor of butyric acid, valeric acid, caproic acid, octanoic acid, p-methyl phenol, and other compounds. At the same time, the abundance of C. algeriensis, M. anaerophila, S. equinus, and W. confuse was significantly correlated with 4 of the above 5 acids, indicating that these 4 bacteria might contribute more to the “smelly” taste of the smelly wax gourd.
It has been reported that L. gasseri in the upper intestine can play a role in blood glucose regulation by affecting the ACSL3-dependent fatty acid sensing pathway (Bauer et al., 2018), L. reuteri has been reported to exist naturally in the intestines of almost all vertebrates and mammals. It has a strong adhesion ability to the intestinal mucosa, which can improve the distribution of intestinal flora, antagonize the colonization of harmful bacteria, and avoid intestinal diseases (Yokota et al., 2018). In this study, the abundance of L. gasseri and L. reuteri were decreased in SWG and A.SWG groups. Supplementation with smelly wax gourd caused the disappearance of H. ganmani and H. canicola. In addition, the abundance of Ruminococcus was increased in SWG and A.SWG groups, which could produce short-chain fatty acids. Fecal microbiota transplantation increased the abundance of short-chain fatty acid bacteria such as B. acidoides, Bifidobacteria, and Faecalibaculum. In contrast some Helicobacter such as H. typhlonius, H. canicola, and H. ganmani, were present in both the FMT and CMT groups. In addition, compared with the CMT group, the abundance of L. gasseri, F. rodentium, B. acidoides, and B. stercorirosoris in the FMT group was higher, while the abundance of L. animalis, M. intestinale, L. reuteri, and H. typhlonius was lower.
5 Conclusion
In our study of stinky winter melon fermentation, it was found that the main flavor compounds were acids, with 2-methylbutyric acid providing the melon with an attractive fruity aroma. The peak abundance and best flavor were observed at 10 days of fermentation (D10), with key microorganisms including Lactobacillus fermentum, Streptococcus equinus, Clostridium perfringens, Mariniphaga anaerophila, and Lactobacillus brevis. Lactobacillus displayed potential probiotic effects and was most abundant at the initial stage (D0) of fermentation, while Streptococcus equinus, Mariniphaga anaerophila, and Caldicoprobacter algeriensis were linked to butyrate production. Caldicoprobacter algeriensis notably correlated with butyric, valeric, capric, and caproic acids. For optimal consumption, stinky winter melon fermented for 10 days preserved rich flavor compounds while minimizing the loss of lactic acid bacteria. Additionally, our investigation into fecal bacteria transplantation enhanced the diversity of intestinal flora in mice, particularly in the fecal transplantation group, showcasing the potential benefits of this approach and revealing distinct microbial genera such as Lactobacillus, Bifidobacterium, Mycobacterium, and Spirochetes compared to fecal culture transplantation. These findings offer insights into the culinary and probiotic potential of stinky winter melon and highlight the promise of fecal transplantation for promoting gut microbiota diversity.
Data availability statement
The data presented in the study are deposited in the NCBI repository, accession number PRJNA1030292 and PRJNA1030303.
Ethics statement
The animal study was approved by Laboratory Animal Center of Zhejiang Academy of Medical Sciences (Hangzhou, Zhejiang). The certificate number of MICE is SYXK Chol 2014-0001, No. 1710300006. The study was conducted in accordance with the local legislation and institutional requirements.
Author contributions
NW: Data curation, Investigation, Methodology, Visualization, Writing – original draft, Writing – review & editing. WB: Conceptualization, Data curation, Methodology, Writing – original draft. MG: Methodology, Writing – review & editing. JX: Conceptualization, Funding acquisition, Writing – review & editing. JH: Conceptualization, Methodology, Writing – review & editing. CL: Conceptualization, Methodology, Writing – review & editing. TM: Conceptualization, Methodology, Writing – review & editing. JZ: Conceptualization, Data curation, Writing – review & editing. WZ: Methodology, Writing – review & editing. XS: Conceptualization, Funding acquisition, Methodology, Project administration, Supervision, Writing – review & editing.
Funding
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work was sponsored by the Natural Science Foundation of Zhejiang Province (LQ22D060002 and LTGC23C190001), Fund of State Key Laboratory for Managing Biotic and Chemical Threats to the Quality and Safety of Agro-Products (ZS20190105), General Project of Zhejiang Provincial Department of Education (Y202146257), Ningbo International Collaboration Project (2023H023), and K. C. Wong Magna Fund of Ningbo University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2024.1314537/full#supplementary-material
References
Ajuru, M., and Nmom, F. (2017). A review on the economic uses of species of Cucurbitaceae and their sustainability in Nigeria. Am. J. Plant Biol. 2, 17–24.
Ashraf, R., and Shah, N. P. (2014). Immune system stimulation by probiotic microorganisms. Crit. Rev. Food Sci. Nutr. 54, 938–956. doi: 10.1080/10408398.2011.619671
Ayed, L., M'Hir, S., and Hamdi, M. (2020). Microbiological, biochemical, and functional aspects of fermented vegetable and fruit beverages. J. Chem. 2020:5790432. doi: 10.1155/2020/5790432
Bao, Y., Zhang, Y., Zhang, Y., Liu, Y., Wang, S., Dong, X., et al. (2010). Screening of potential probiotic properties of Lactobacillus fermentum isolated from traditional dairy products. Food Control 21, 695–701. doi: 10.1016/j.foodcont.2009.10.010
Bauer, P. V., Duca, F. A., Waise, T. Z., Dranse, H. J., Rasmussen, B. A., Puri, A., et al. (2018). Lactobacillus gasseri in the upper small intestine impacts an ACSL3-dependent fatty acid-sensing pathway regulating whole-body glucose homeostasis. Cell Metab. 27, 572–587.e6. doi: 10.1016/j.cmet.2018.01.013
Beganović, J., Pavunc, A. L., Gjuračić, K., Špoljarec, M., Šušković, J., and Kos, B. (2015). Improved sauerkraut production with probiotic strain Lactobacillus plantarum L4 and Leuconostoc mesenteroides LMG 7954. J. Food Sci. 76, M124–M129. doi: 10.1111/j.1750-3841.2010.02030.x
Brun, P., Giron, M. C., and Qesari, M. (2013). Toll-like receptor 2 regulates intestinal inflammation by controlling integrity of the enteric nervous system. Gastroenterology 145, 1323–1333. doi: 10.1053/j.gastro.2013.08.047
Byakika, S., Mukisa, I. M., Byaruhanga, Y. B., Male, D., and Muyanja, C. (2019). Influence of food safety knowledge, attitudes and practices of processors on microbiological quality of commercially produced traditional fermented cereal beverages, a case of Obushera in Kampala. Food Control 100, 212–219. doi: 10.1016/j.foodcont.2019.01.024
Caporaso, J. G., Kuczynski, J., Stombaugh, J., Bittinger, K., Bushman, F. D., Costello, E. K., et al. (2010). QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7, 335–336. doi: 10.1038/nmeth.f.303
Cheigh, H. S., Park, K. Y., and Lee, C. (1994). Biochemical, microbiological, and nutritional aspects of kimchi (Korean fermented vegetable products). Crit. Rev. Food Sci. Nutr. 34, 175–203. doi: 10.1080/10408399409527656
Chen, G., Xie, M., Wan, P., Chen, D., Dai, Z., Ye, H., et al. (2018). Fuzhuan brick tea polysaccharides attenuate metabolic syndrome in high-fat diet induced mice in association with modulation in the gut microbiota. J. Agric. Food Chem. 66, 2783–2795. doi: 10.1021/acs.jafc.8b00296
Choi, H., Kim, Y. W., Hwang, I., Kim, J., and Yoon, S. (2012). Evaluation of Leuconostoc citreum HO12 and Weissella koreensis HO20 isolated from kimchi as a starter culture for whole wheat sourdough. Food Chem. 134, 2208–2216. doi: 10.1016/j.foodchem.2012.04.047
Cui, M., Kim, H.-Y., Lee, K. H., Jeong, J.-K., Hwang, J.-H., Yeo, K.-Y., et al. (2015). Antiobesity effects of kimchi in diet-induced obese mice. J. Ethn. Foods 2, 137–144. doi: 10.1016/j.jef.2015.08.001
de Vries, M. C., Vaughan, E. E., Kleerebezem, M., and de Vos, W. M. (2006). Lactobacillus plantarum—survival, functional and potential probiotic properties in the human intestinal tract. Int. Dairy J. 16, 1018–1028. doi: 10.1016/j.idairyj.2005.09.003
Diez-Ozaeta, I., and Astiazaran, O. J. (2022). Fermented foods: an update on evidence-based health benefits and future perspectives. Food Res. Int. 156:111133. doi: 10.1016/j.foodres.2022.111133
Doerner, K., Cook, K., and Mason, B. (2009a). 3-methylindole production is regulated in Clostridium scatologenes ATCC 25775. Lett. Appl. Microbiol. 48, 125–132. doi: 10.1111/j.1472-765X.2008.02502.x
Doerner, K., Mason, B., Kridelbaugh, D., and Loughrin, J. (2009b). Fe (III) stimulates 3-methylindole and 4-methylphenol production in swine lagoon enrichments and Clostridium scatologenes ATCC 25775. Lett. Appl. Microbiol. 48, 118–124. doi: 10.1111/j.1472-765X.2008.02500.x
Edgar, R. C. (2004). MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 32, 1792–1797. doi: 10.1093/nar/gkh340
Edgar, R. C. (2010). Search and clustering orders of magnitude faster than BLAST. Bioinformatics 26, 2460–2461. doi: 10.1093/bioinformatics/btq461
Edgar, R. C., Haas, B. J., Clemente, J. C., Quince, C., and Knight, R. (2011). UCHIME improves sensitivity and speed of chimera detection. Bioinformatics 27, 2194–2200. doi: 10.1093/bioinformatics/btr381
Flom, J. D., and Sicherer, S. H. (2019). Epidemiology of cow’s Milk allergy. Nutrients 11:1051. doi: 10.3390/nu11051051
Franchi, O., Bovio, P., Ortega-Martínez, E., Rosenkranz, F., and Chamy, R. (2018). Active and total microbial community dynamics and the role of functional genes bamA and mcrA during anaerobic digestion of phenol and p-cresol. Bioresour. Technol. 264, 290–297. doi: 10.1016/j.biortech.2018.05.060
Haug, J. B., Myhr, R., and Reikvam, A. (2011). Pharmacy sales data versus ward stock accounting for the surveillance of broad-spectrum antibiotic use in hospitals. BMC Med. Res. Methodol. 11:166. doi: 10.1186/1471-2288-11-166
Heimesaat, M., Reikvam, D. H., and Erofeev, A. (2011). Depletion of murine intestinal microbiota: effects on gut mucosa and epithelial gene expression. PLoS One 6:e17996. doi: 10.1371/journal.pone.0017996
Holzapfel, W. H., Haberer, P., Geisen, R., Björkroth, J., and Schillinger, U. (2001). Taxonomy and important features of probiotic microorganisms in food and nutrition. Am. J. Clin. Nutr. 73, 365s–373s. doi: 10.1093/ajcn/73.2.365s
Kang, K.-J., and Kwon, S.-Y. (2003). Effects of wax gourd extracts on adipocyte differentiation and uncoupling protein genes (Ucps) expression in 3T3-Ll Preadipocytes. Nutr. Sci. 6, 148–154.
Kim, J., Bang, J., Beuchat, L. R., Kim, H., and Ryu, J.-H. (2012). Controlled fermentation of kimchi using naturally occurring antimicrobial agents. Food Microbiol. 32, 20–31. doi: 10.1016/j.fm.2012.05.007
Kim, E., Cho, E.-J., Yang, S.-M., Kim, M.-J., and Kim, H.-Y. (2021). Novel approaches for the identification of microbial communities in kimchi: MALDI-TOF MS analysis and high-throughput sequencing. Food Microbiol. 94:103641. doi: 10.1016/j.fm.2020.103641
Kim, W. K., Jang, Y. J., Han, D. H., Jeon, K., Lee, C., Han, H. S., et al. (2020). Lactobacillus paracasei KBL382 administration attenuates atopic dermatitis by modulating immune response and gut microbiota. Gut Microbes 12, 1–14. doi: 10.1080/19490976.2020.1819156
Kumar, B. V., Vijayendra, S. V. N., and Reddy, O. V. S. (2015). Trends in dairy and non-dairy probiotic products—a review. J. Food Sci. Technol. 52, 6112–6124. doi: 10.1007/s13197-015-1795-2
Lan, W.-T., Chen, Y.-S., Wu, H.-C., and Yanagida, F. (2012). Bio-protective potential of lactic acid bacteria isolated from fermented wax gourd. Folia Microbiol. 57, 99–105. doi: 10.1007/s12223-012-0101-1
Lan, W.-T., Chen, Y.-S., and Yanagida, F. (2009). Isolation and characterization of lactic acid bacteria from Yan-dong-gua (fermented wax gourd), a traditional fermented food in Taiwan. J. Biosci. Bioeng. 108, 484–487. doi: 10.1016/j.jbiosc.2009.06.009
Li, W., Chen, Y. P., Blank, I., Li, F., Li, C., and Liu, Y. (2021). GC × GC-ToF-MS and GC-IMS based volatile profile characterization of the Chinese dry-cured hams from different regions. Food Res. Int. 142:110222. doi: 10.1016/j.foodres.2021.110222
Lim, T. (2012). “Benincasa hispida cv-gr. wax gourd” in Edible medicinal and non-medicinal plants (Dordrecht: Springer), 167–178.
Lin, X. N., Bakyrbay, S., Liu, L. X., Tang, X. J., and Liu, Y. G. (2023). Microbiota succession and chemical composition involved in lactic acid bacteria-fermented pickles. Fermentation 9:330. doi: 10.3390/fermentation9040330
Liu, Y., Miao, Z., Guan, W., and Sun, B. (2012). Analysis of organic volatile flavor compounds in fermented stinky tofu using SPME with different fiber coatings. Molecules 17, 3708–3722. doi: 10.3390/molecules17043708
Lu, C., Zhou, J., Li, Y., Zhang, D., Wang, Z., Li, Y., et al. (2017). Structural modulation of gut microbiota in Bama minipigs in response to treatment with a “growth-promoting agent”, salbutamol. Appl. Microbiol. Biotechnol. 101, 5809–5818. doi: 10.1007/s00253-017-8329-y
Martins, E. M. F., Ramos, A. M., Vanzela, E. S. L., Stringheta, P. C., de Oliveira Pinto, C. L., and Martins, J. M. (2013). Products of vegetable origin: a new alternative for the consumption of probiotic bacteria. Food Res. Int. 51, 764–770. doi: 10.1016/j.foodres.2013.01.047
Monk, J. M., Lepp, D., Zhang, C. P., Wu, W., Zarepoor, L., Lu, J. T., et al. (2016). Diets enriched with cranberry beans alter the microbiota and mitigate colitis severity and associated inflammation. J. Nutr. Biochem. 28, 129–139. doi: 10.1016/j.jnutbio.2015.10.014
Ogrodowczyk, A. M., and Drabinska, N. (2021). Crossroad of tradition and innovation—the application of lactic acid fermentation to increase the nutritional and health-promoting potential of plant-based food products—a review. Pol. J. Food Nutr. Sci. 71, 107–134.
Okoye, C. O., Gao, L., Wu, Y., Li, X., Wang, Y., and Jiang, J. (2023). Identification, characterization and optimization of culture medium conditions for organic acid-producing lactic acid bacteria strains from Chinese fermented vegetables. Prep. Biochem. Biotechnol. 54, 49–60. doi: 10.1080/10826068.2023.2204507
Park, K. Y., Jeong, J. K., Lee, Y. E., and Daily, J. W. (2014). Health benefits of kimchi (Korean fermented vegetables) as a probiotic food. J. Med. Food 17, 6–20. doi: 10.1089/jmf.2013.3083
Park, J. A., Tirupathi Pichiah, P. B., Yu, J. J., Oh, S. H., Daily, J. W. 3rd, and Cha, Y. S. (2012). Anti-obesity effect of kimchi fermented with Weissella koreensis OK1-6 as starter in high-fat diet-induced obese C57BL/6J mice. J. Appl. Microbiol. 113, 1507–1516. doi: 10.1111/jam.12017
Pevsner-Fischer, M., Blacher, E., Tatirovsky, E., Ben-Dov, I. Z., and Elinav, E. (2017). The gut microbiome and hypertension. Curr. Opin. Nephrol. Hypertens. 26, 1–8. doi: 10.1097/MNH.0000000000000293
Power, S. E., O'Toole, P. W., Catherine, S., Paul, R., and Fitzgerald, G. F. (2014). Intestinal microbiota, diet and health. Br. J. Nutr. 111, 387–402. doi: 10.1017/S0007114513002560
Price, M. N., Dehal, P. S., and Arkin, A. P. (2010). FastTree 2—approximately maximum-likelihood trees for large alignments. PLoS One 5:e9490. doi: 10.1371/journal.pone.0009490
Rao, Y., Qian, Y., She, X., Yang, J., He, P., Jiang, Y., et al. (2018). Pellicle formation, microbial succession and lactic acid utilisation during the aerobic deteriorating process of Sichuan pickle. Int. J. Food Sci. Technol. 53, 767–775. doi: 10.1111/ijfs.13652
Ross, R. P., Morgan, S., and Hill, C. (2002). Preservation and fermentation: past, present and future. Int. J. Food Microbiol. 79, 3–16. doi: 10.1016/S0168-1605(02)00174-5
Saito, Y., Sato, T., Nomoto, K., and Tsuji, H. (2018). Identification of phenol-and p-cresol-producing intestinal bacteria by using media supplemented with tyrosine and its metabolites. FEMS Microbiol. Ecol. 94:fiy125. doi: 10.1093/femsec/fiy125
Schloss, P. D., Westcott, S. L., Ryabin, T., Hall, J. R., Hartmann, M., Hollister, E. B., et al. (2009). Introducing mothur: open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 75, 7537–7541. doi: 10.1128/AEM.01541-09
Schmieder, R., and Edwards, R. (2011). Quality control and preprocessing of metagenomic datasets. Bioinformatics 27, 863–864. doi: 10.1093/bioinformatics/btr026
Selvaraj, B., Pierik, A. J., Bill, E., and Martins, B. M. (2013). 4-Hydroxyphenylacetate decarboxylase activating enzyme catalyses a classical S-adenosylmethionine reductive cleavage reaction. J. Biol. Inorg. Chem. 18, 633–643. doi: 10.1007/s00775-013-1008-2
Sidira, M., Kandylis, P., Kanellaki, M., and Kourkoutas, Y. (2016). Effect of curing salts and probiotic cultures on the evolution of flavor compounds in dry-fermented sausages during ripening. Food Chem. 201, 334–338. doi: 10.1016/j.foodchem.2016.01.084
Steinhaus, M., and Schieberle, P. (2005). Characterization of odorants causing an atypical aroma in white pepper powder (Piper nigrum L.) based on quantitative measurements and orthonasal breakthrough thresholds. J. Agric. Food Chem. 53, 6049–6055. doi: 10.1021/jf0506030
Tamime, A. (2002). Fermented milks: a historical food with modern applications–a review. Eur. J. Clin. Nutr. 56, S2–S15. doi: 10.1038/sj.ejcn.1601657
Tang, H., Ma, J.-K., Chen, L., Jiang, L.-W., Xie, J., Li, P., et al. (2018). GC-MS characterization of volatile flavor compounds in stinky tofu brine by optimization of headspace solid-phase microextraction conditions. Molecules 23:3155. doi: 10.3390/molecules23123155
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naive Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wastyk, H. C., Fragiadakis, G. K., Perelman, D., Dahan, D., Merrill, B. D., Yu, F. B., et al. (2021). Gut-microbiota-targeted diets modulate human immune status. Cell 184, 4137–4153.e14. doi: 10.1016/j.cell.2021.06.019
Won, T. J., Kim, B., Lim, Y. T., Song, D. S., Park, S. Y., Park, E. S., et al. (2011). Oral administration of Lactobacillus strains from kimchi inhibits atopic dermatitis in NC/Nga mice. J. Appl. Microbiol. 110, 1195–1202. doi: 10.1111/j.1365-2672.2011.04981.x
Wu, Z., Mao, Y., Zhang, X., and Weng, P. (2016a). Symbolic metabolite analysis of pickled wax gourd in eastern China by 1H-NMR spectroscopy and multivariate data. Int. J. Food Prop. 19, 2052–2062. doi: 10.1080/10942912.2015.1099044
Wu, Z.-F., Sun, L., Zhang, X., Shen, X.-Q., and Weng, P.-F. (2015). Quantitative analysis of predominant yeasts and volatile compounds in the process of pickled wax gourd. CYTA - J. Food 14, 92–100. doi: 10.1080/19476337.2015.1052018
Wu, Z., Zhuang, B., Weng, P., and Zhang, X. (2016b). Fermentation quality characteristics and flavor formation changes during the process of pickled wax gourd in eastern Zhejiang. Int. J. Food Prop. 19, 409–419. doi: 10.1080/10942912.2015.1027775
Xu, Y., Li, L., Regenstein, J. M., Gao, P., Zang, J., Xia, W., et al. (2018). The contribution of autochthonous microflora on free fatty acids release and flavor development in low-salt fermented fish. Food Chem. 256, 259–267. doi: 10.1016/j.foodchem.2018.02.142
Yang, Y., Qian, M. C., Deng, Y., Yuan, H., and Jiang, Y. (2022). Insight into aroma dynamic changes during the whole manufacturing process of chestnut-like aroma green tea by combining GC-E-nose, GC-IMS, and GC × GC-TOFMS. Food Chem. 387:132813. doi: 10.1016/j.foodchem.2022.132813
Yokota, Y., Shikano, A., Kuda, T., Takei, M., Takahashi, H., and Kimura, B. (2018). Lactobacillus plantarum AN1 cells increase caecal L. reuteri in AN ICR mouse model of dextran sodium sulphate-induced inflammatory bowel disease. Int. Immunopharmacol. 56, 119–127. doi: 10.1016/j.intimp.2018.01.020
Yoshimoto, S., Loo, T. M., Atarashi, K., Kanda, H., Sato, S., Oyadomari, S., et al. (2013). Obesity-induced gut microbial metabolite promotes liver cancer through senescence secretome. Nature 499:97. doi: 10.1038/nature12347
Zhang, D., Han, J., Li, Y., Yuan, B., Zhou, J., Cheong, L., et al. (2018). Tuna oil alleviates d-galactose induced aging in mice accompanied by modulating gut microbiota and brain protein expression. J. Agric. Food Chem. 66, 5510–5520. doi: 10.1021/acs.jafc.8b00446
Zhang, J., Kobert, K., Flouri, T., and Stamatakis, A. (2014). PEAR: a fast and accurate Illumina Paired-End reAd mergeR. Bioinformatics 30, 614–620. doi: 10.1093/bioinformatics/btt593
Keywords: fermented wax gourd, microbial diversity, flavor substance, gut microbes, traditional food
Citation: Wang N, Bao W, Gouife M, Xu J, Han J, Lu C, Ming T, Zhou J, Zhang W and Su X (2024) Exploring the health benefits of traditionally fermented wax gourd: flavor substances, probiotics, and impact on gut microbiota. Front. Sustain. Food Syst. 8:1314537. doi: 10.3389/fsufs.2024.1314537
Edited by:
Huidi Wang, Southern Medical University, ChinaReviewed by:
Maria Simona Chi, University of Agricultural Sciences and Veterinary Medicine of Cluj-Napoca, RomaniaQingli Dong, University of Shanghai for Science and Technology, China
Copyright © 2024 Wang, Bao, Gouife, Xu, Han, Lu, Ming, Zhou, Zhang and Su. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jiajie Xu, dWppYWppZUBuYnUuZWR1LmNu; Xiurong Su, c3V4aXVyb25nX3B1YmxpY0AxNjMuY29t
†These authors have contributed equally to this work
 Nannan Wang
Nannan Wang Wei Bao1†
Wei Bao1† Jiajie Xu
Jiajie Xu Jiaojiao Han
Jiaojiao Han Chengyang Lu
Chengyang Lu Tinghong Ming
Tinghong Ming Xiurong Su
Xiurong Su