- Crop Cultivation System, Agronomy College, Northeast Agricultural University, Harbin, China
Introduction: Drought stress has gradually become a limiting factor for plant growth. Soybean, a crop with a higher water demand than other plants, is particularly sensitive to water deficits.
Method: To explore the effect of drought stress on soybean protein expression, we used drought-tolerant soybean genotype HeiNong44 (HN44) and sensitive soybean genotype HeiNong65 (HN65) as experimental materials, PEG-6000 as an osmotic regulator, and tandem mass tag (TMT) technology to identify the differential expression of soybean proteins.
Results: We identified that 48 upregulated and 38 downregulated proteins in HN44 and 55 upregulated and 13 downregulated proteins in HN65.
Discussion: KEGG analysis showed that these differentially expressed proteins were involved in carbohydrate metabolism, signal transduction, amino acid metabolism, lipid metabolism, and programmed cell death. Drought stress usually caused an increase in the activity of plant antioxidant enzymes, the obstruction of photosynthetic synthesis, and a significant change in the content of plant hormones. We found that HN44 responds to drought stress mainly through sugar decomposition, increased antioxidant enzyme activity and lipid metabolism. HN65 responds to drought stress mainly through ABA synthesis, increased antioxidant enzyme activity and amino acid metabolism. Our study also found that differential proteins in the starch and sucrose metabolic pathway play a key role in supplying energy and regulating osmotic potential.
1. Introduction
Owing to climate and environmental changes, long-term insufficient rainfall and limited water supply in many areas result in severe drought stress on crops. Currently, drought stress is one of the most unfavorable abiotic stresses for crop growth (Gupta et al., 2020). Water deficit induces plants to produce large amounts of reactive oxygen species (ROS), which destroy biological macromolecules such as DNA, proteins, and lipids in plant cells, change cell structure, and affect the plant metabolic processes of plants (Silva et al., 2010; Zhou et al., 2022a). Carbohydrate regulation is one of the most important physiological processes under drought stress (Baslam et al., 2020). Under drought stress, plant water potential decreases (Hu et al., 2022). The adverse effects of drought stress on plants are mainly manifest in its interference with many physiological processes, such as plant nitrogen-assimilates production and metabolism (Du et al., 2020), weakened photosynthetic capacity (Sudhir et al., 2005), signal transduction (Li et al., 2020), and endogenous hormone metabolism (Demirkol, 2021), and can even cause plant death in severe cases (Jangir et al., 2021).
To adapt and reduce the damage caused by drought stress, plants regulate their metabolism and growth according to the water conditions, resulting in morphological, physiological, and molecular changes (Jupa et al., 2016; Gupta et al., 2020). Among them, the molecular changes are most complex. Many genes and metabolites related to plant drought resistance have been identified through genomic, transcriptomic, and metabolomic studies (Jaiswal et al., 2019). Proteomics has also been widely used in crop drought resistance research to complement other omics (Wang et al., 2021). For example, studies have shown that the overexpression of calmodulin-binding protein 60 (CBP60) in soybean under drought conditions can stimulate the regulation of calcium signaling pathways, increase the content of osmotic adjustment substances (such as proline), reduce the water loss rate and ROS content of soybean roots, and effectively enhance soybean drought tolerance (Yu et al., 2021). In addition, various proteins are involved in carbohydrate metabolism, protein processing, ROS detoxification, secondary metabolism, ion and water transport, and other drought resistance pathways (Zhang and Shi, 2018).
Soybean (Glycine max (L). Merr.) originates in the Yellow River Basin in China and is an annual herbaceous plant (Gonyane and Sebetha, 2021). Soybeans are widely used in many industries, such as food and animal husbandry. Among these, 29% of edible oils and 70% of plant proteins are derived from soybeans (Rahman et al., 2022). However, soybeans require more water and are highly sensitive to water deficits. Drought stress can reduce soybean yield by approximately 40% (Ingwers et al., 2022). A full physiological study confirmed that soybeans can reduce the damage caused by drought stress through osmotic adjustment, ROS scavenging, and endogenous hormone changes (Zhou et al., 2022a). Some studies have also investigated drought-resistance regulatory genes in soybeans (Jin et al., 2022). With continuous technological development, plant studies have become increasingly extensive. Several studies have widely used proteomic technology as an important technique for studying plants under various abiotic stresses, such as salt, drought, flooding, high temperature, and heavy metals (Wang et al., 2016).
The selection of varieties with different stress resistances can make the results more evident and better reveal the drought resistance mechanism of soybeans (Zhou et al., 2022a). Therefore, we used soybean varieties HeiNong44 (HN44) and HeiNong65 (HN65) with different drought resistance as test materials, PEG-6000 as an osmotic adjustment substance to simulate drought stress, and the tandem mass tag (TMT) quantitative proteomics method to determine the differential proteins under drought stress and normal treatment conditions, as well as functional analysis. Our aim was to clarify the underlying mechanisms of soybean resistance to drought stress by comparing at the protein level the drought response of genotypes differing in drought tolerance, and to provide a theoretical basis for selection and breeding of soybean varieties with improved drought-resistance.
2. Materials and methods
2.1. Plant materials and drought treatment
The drought stress experiment was conducted at the Northeast Agricultural University (N45° 44′ 43.87, E126° 43′ 50.42). The experimental materials were HeiNong44 (HN44) and HeiNong 65 (HN65), which were developed by the Heilongjiang Academy of Agricultural Sciences. Previous studies have shown that the drought resistance of HN44 was stronger than that of HN65 (Wang et al., 2022a). A potting experiment was conducted. First, the river sand was washed, dried, and placed in a flowerpot with a height of 35 cm and an inner diameter of 30 cm. After watering with 2 L per pot, eight plump and uniform seeds were sown. Then, 500 mL distilled water was added at one-time every day until the true leave of all seedlings was fully expanded. Three soybean seedlings with consistent growth were retained in each pot, and the other seedlings were cut with scissors. After thinning, 500 mL of Hoagland nutrient solution (Waheed et al., 2019) was added daily until all seedlings grew into three compound leaves. The drought treatment was then performed. Control plants were supplied daily with 500 mL nutrient solution per pot, and drought treated plants were supplied daily with 500 mL nutrient solution containing 15% PEG-6000 for 4 days until a moderate drought stress has been reached (Wang et al., 2012, 2022a; GB/T 32136-2015, 2015). The water potential of nutrient solution containing 15% PEG-6000 was −0.33 MPa (Chen et al., 2015). Each treatment consisted of three replicates, with five pots per replicate. The samples comprised a mixture of the second leaves of all plants per replicate. The sampling time was 8:00–9:00 a.m. on the day after treatment. After sampling, the replicate was quickly frozen in liquid nitrogen, then stored in a −80°C ultra-low temperature refrigerator (Haier, DW-86L828J, Qingdao, Shandong Province, China).
2.2. Determination of stress response parameters
2.2.1. Determination of relative water content of leaves
Determination of Relative Water Content (RWC) of leaves was made by the BADR Method (Badr and Bruggemann, 2020) using the equation RWC [%] = [(FM – DM) / (TM – DM)] × 100, where FM, TM, and DM are the fresh, turgid, and dry masses, respectively. Three leaf discs for each accession plants exposed to drought and corresponding control plants were cut and immediately weighted (FM), then saturated to turgidity by immersing in cold water over night, briefly dried, weighted (TM), oven-dried at 80°C for 24 h, and weighted (DM).
2.2.2. Determination of relative electrical conductivity
The electrical conductivity (EC) of leaves was measured to assess cell membrane damage, as described by Song et al. (2020). In brief, sampled leaves were washed using deionized water, cut into 0.5-cm pieces, aspirating 15 min used vacuum pressure pumping (SCJ-10, Huaou Precision, Xian, China), and immersed in deionized water for 30 min. Then, EC of the solution was measured using a conductivity meter (DDS-307, Electric Scientific Instruments, Shanghai, China) and recorded as S1. After boiling the leaf samples for 15 min, EC of the solution at room temperature was measured again and recorded as S2. A relative EC (REC) was calculated as S1/S2 × 100%.
2.2.3. Determination of malondialdehyde (MDA): thiobarbituric acid method
According to the experimental method of Wang et al. (2022b). Frozen sample (0.1 g) was weighted into cold mortar, 1 mL of 10% trichloroacetic acid (TCA) was added, and the homogenate was ground in an ice bath, and centrifuged at 12,000 rpm for 10 min. Then, 0.2 mL of 0.67% thiobarbituric acid (TBA) was added to 0.4 mL of each supernatant, mixed well and kept in a wather bath at 100°C for 30 min; after cooling, the samples were centrifuged again. The absorbance values of the supernatants were measured at 450, 532, and 600 nm, and the malondialdehyde (MDA) content was calculated according to the following formula and expressed on a moist weight basis. CMDA = 6.45 (A532 − A600) − 0.56A450 (μmol/g).
2.2.4. Determination of proline (Pro): ninhydrin method
According to the experimental method of Bates et al. (1973) and Wang et al. (2022b). Frozen sample (0.1 g) was weighted into cold mortar, 1 mL of 3% sulfosalicylic acid was added and samples were homogenised. Then the homogenate was transferred into a centrifuge tube in a boiling water bath for 10 min. After cooling to 25°C, and centrifuged at 12,000 rpm for 10 min; then, 0.4 mL of glacial acetic acid and 0.6 mL of chromogenic solution were added to 0.4 mL of the supernatant. This mixture was heated in a boiling water bath for 40 min and then cooled before 1 mL of toluene was added. After stratification, the toluene layer was withdrawn and absorbance was measured at 520 nm using a spectrophotometer. Pro content in the sample was calculated according to the following formula:
where C is the proline content (μg) in the extract as calculated from the standard curve; V is the total volume of extract (mL); A is the volume (mL) absorbed during measurement; and W is the sample weight (g).
2.2.5. Determination of soluble sugars (SSs): anthrone method
According to the experimental method of Yemm and Willis (1954) and Wang et al. (2022b). Frozen sample (0.1 g) was weighted into mortar, 1 mL distilled water was added, homogenized and centrifuged for 10 min at 12,000 rpm. Then, 0.2 mL of sample extracts or distilled water for the control were added to 1 mL anthrone reagent, shaken well, boiled in a boiling water bath for 10 min, removed, and cooled, and absorbance was measured at 620 nm using a spectrophotometer. Soluble sugar content (mg/g) = sugar content (mg) × dilution (10 − fold)/ {sample weight (g) × 106} × 100.
2.2.6. Determination of soluble proteins (SPs): coomassie brilliant blue G-250 method
According to the experimental method of Bradford (1976) and Wang et al. (2022b). Frozen sample (0.1 g) was weighted into mortar, 1 mL distilled water was added, homogenized and centrifuged for 10 min at 12,000 rpm. The sample extracts (0.2 mL) were placed in test tubes and added with 1 mL Coomassie brilliant blue G-250 reagent; the mixture was shaken vigorously, and after 2 min, absorbance was measured at 595 nm. Soluble protein content was calculated using the corresponding standard curve obtained using bovine serum albumin (BSA).
2.3. Preparation and concentration determination of protein
The Tandem Mass Tag (TMT) technology was used for sample preparation, extraction analysis, protein identification, and quantification according to the standard procedures of Metware Biotechnology Co., Ltd. (www.metware.cn, accessed on April 1, 2022) (Wuhan, China). The lysate (1.5% SDS/100 mM Tris-Cl) was mixed with the sample, homogenized, and centrifuged to obtain the supernatant. The proteins in the solution were precipitated using acetone. Subsequently, iodoacetamide (IAA) was added, and the alkylation reaction was carried out in the dark at 20°C. The protein concentration was determined using the Bradford method.
Then the samples were labeled with TMT (The amount of protein used for TMT labeling was equal to that used for Bradford method). Marking was performed according to the TMT instructions (Thermo Scientific™, Massachusetts, United States). After labeling, Sep-PakC18 was used for desalination. After vacuum drying, the mixed samples were separated by the high pH reverse chromatographic separation method, vacuum dried, and stored in a refrigerator at −80°C for testing.
2.4. Liquid chromatographic-mass pectrometric (LC/MS)
Mass spectrometry data were collected using an Orbitrap Exploris 480 mass spectrometer (Thermo Scientific™, Massachusetts, United States) in series with an EASY-nLC1200 liquid-phase liquid chromatography-mass spectrometry system (Thermo Scientific™, Massachusetts, United States). The peptide sample was dissolved in the loading buffer and separated using an analytical column (Metware Biotechnology, Wuhan, China). The parameters of the analytical column are 75 μm × 25 cm, C18, 2 μm, 100 Å. Two mobile phases were used to establish the analytical gradient. The liquid phase flow rate was 300 nL/min. Mass spectrometry data were collected in the DDA mode. Each scan cycle consisted of one full MS scan and 20 subsequent MS/MS scans. The Turbo TMT function was turned on.
2.5. Qualitative and quantitative analysis of differential proteins
Mass spectrometry data were retrieved using MaxQuant (V1.6.6), and the Andromeda database retrieval algorithm was used. The Uniprot.Glycine_max.20190126 Fast Proteome Reference Database was used for retrieval. In addition, 20 ppm was set for the primary and secondary mass spectrometry matching tolerance and 4.5 ppm for the main search. The search results were screened at the protein and peptide levels using a 1% FDR for subsequent analysis. Proteins with fold-change ≥1.2 and p-value ≤0.05 were defined as upregulated proteins and proteins with fold-change ≤0.83 and p-value ≤0.05 were defined as down-regulated proteins.
BLAST alignment (blastp, evalue ≤ le-5) was used for GO, KEGG, and KOG database annotation. Alignment results with the highest scores were selected as the annotation results. In addition, BLAST software (Springer Nature, Berlin, Germany) combined the UniProt and InterPro databases for domain annotation, and MultiLoc2 was used for subcellular localization prediction.
All our experiments contain three replicates biological to ensure the stability of the experimental data.
2.6. Data analysis
Physiological and biochemical data were analyzed using IBM SPSS (version 21.0: IBM Corporation, Armonk, NY, United States) for T-test, Microsoft Office Excel 2021 (USA) for histogram and pie charts, and OriginPro2021 (Origin Lab Corp, Northampton, MA, United States) for principal component analysis and cluster analysis and draw Venn and bubble diagram. KEGG enrichment pathway was drawn by Adobe Illustrator (version 2022 v26.0.0.730: Adobe Systems Incorporated, State of California, United States). Protein interaction analyses were performed using the String DB protein interaction database and visualized using Cytoscape (version 3.9.1: Netscape, State of Illinois, United States).
3. Results
3.1. Stress response parameters
We measured the effects of drought stress on the physiological indices of soybeans (Figure 1). Under drought stress, the relative water content of plant leaves decreased significantly, HN44 decreased by 15.49%, HN65 decreased by 24.67%. The relative water content of HN44 leaves was 5.55% and 18.41% higher than that of HN65 under control and drought conditions, respectively. Under drought stress, the relative electrical conductivity and MDA content of HN44 increased by 76.98% and 44.57%, respectively, and HN65 increased by 140.15% and 58.97%, respectively. The relative conductivity and MDA of HN44 leaves were 21.60% and 10.24% lower than those of HN65 under control. Under drought conditions, HN44 was 42.22% and 18.37% lower than HN65.
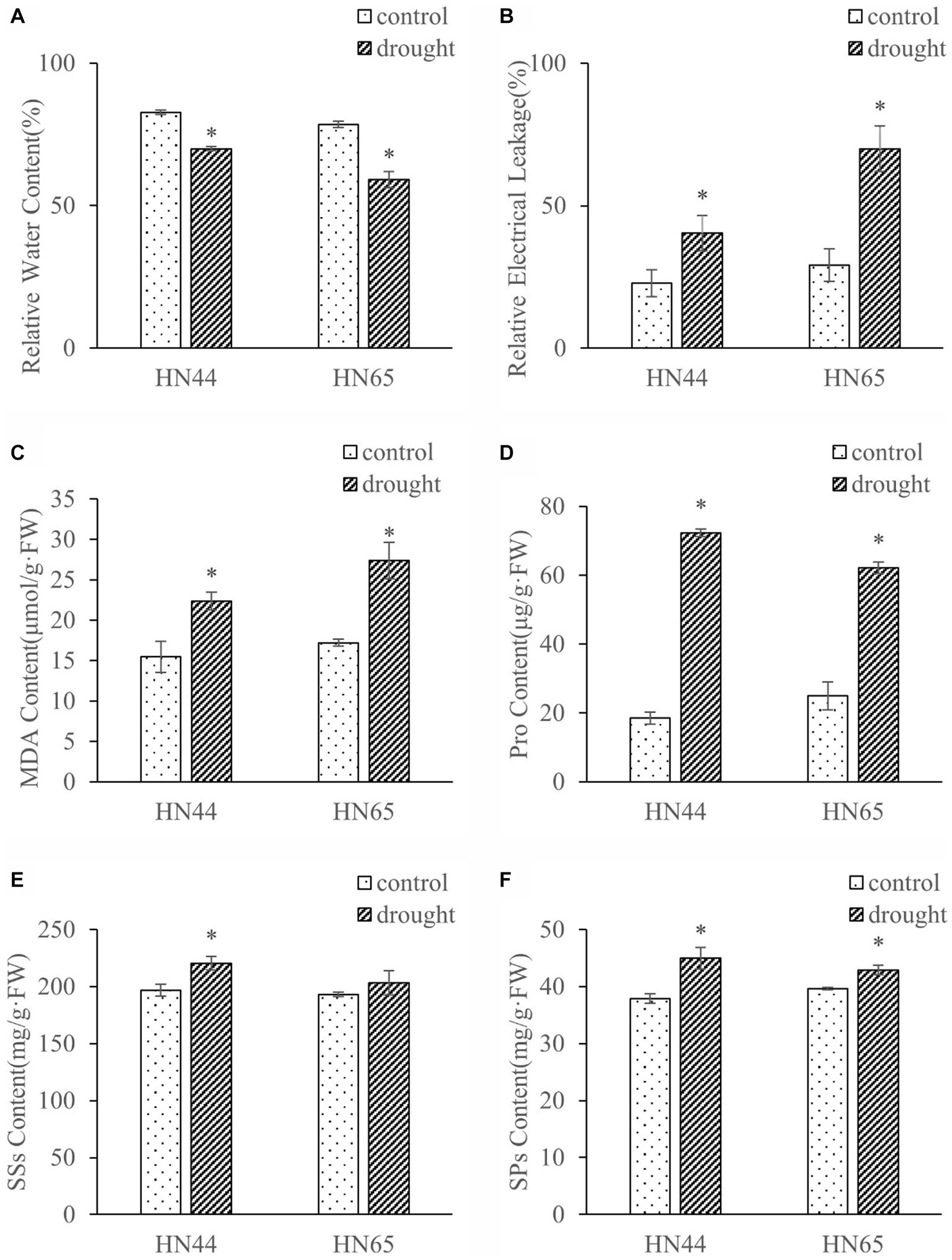
Figure 1. Effects of drought stress on relative water content (A), relative electrical conductivity (B), MDA (C), Pro (D), SSs (E), and SPs (F) contents in leaves of HN44 and HN65. *indicate significant differences between control and drought according to T test at the 5% level, and data are presented as mean ± SE (standard error) (n = 3). HN44, Hei Nong 44; HN65, Hei Nong 65; MDA, malonaldehyde; Pro, proline; SSs, soluble sugars; SPs, soluble proteins.
Drought stress increased the content of Pro, SSs, and SPs in HN44 leaves by 291.98%, 12.03%, and 18.81%, respectively, and in HN65 leaves by, 149.58%, 5.33%, and 8.23%, respectively. Under drought stress, Pro, SSs and SPs of HN44 were 16.30%, 8.33%, and 4.90% higher than those of HN65, respectively. In addition, the rate of increase in Pro, SSs, and SPs content in HN65 was lower than that in HN44, and the SSs content of HN65 did not significantly change under drought stress. Overall, the results showed that drought stress significantly affected the soybean physiology and that the drought resistance of HN44 was stronger than that of HN65.
3.2. Quantification of differential proteins
A total of 86 differentially expressed proteins were identified in HN44 and 68 in HN65, of which 29 were shared by HN44 and HN65 (Figure 2A). There were 48 upregulated and 38 downregulated proteins in HN44 and 55 upregulated and 13 downregulated proteins in HN65 (Figure 2B). Principal component analysis (PCA) showed that there was a significant difference between the control and drought treatments of HN44 and HN65 (Figure 2C), indicating that drought stress had a significant impact on soybean growth.
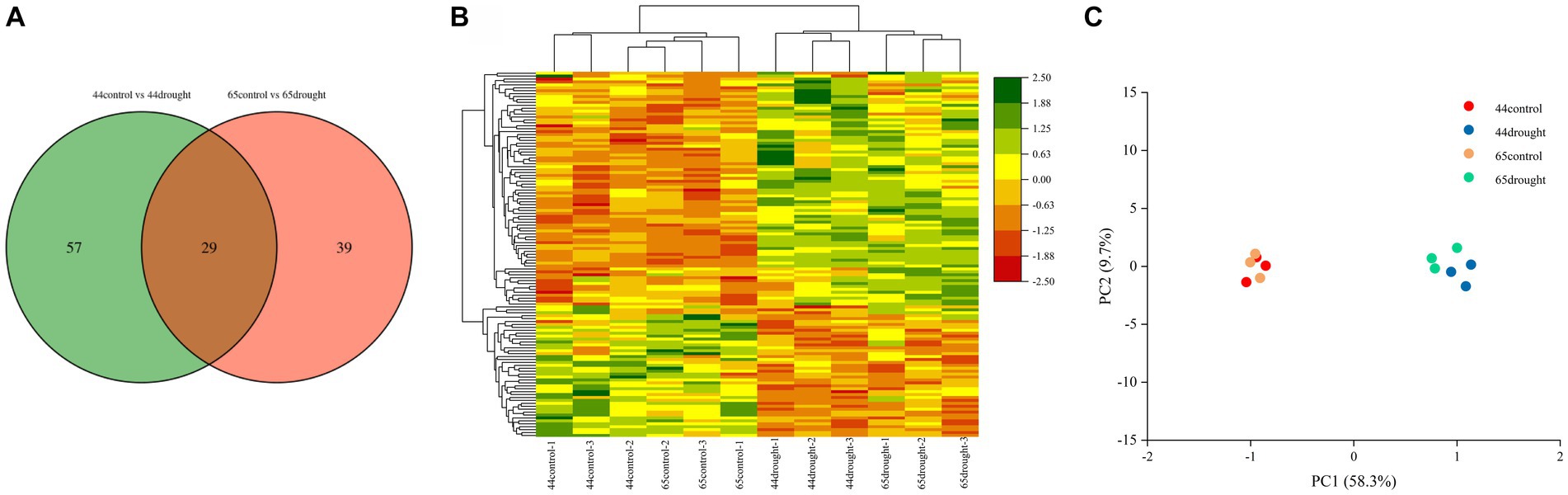
Figure 2. Differential protein number and principal component analysis under drought stress. (A) Venn diagram of differential proteins. (B) Cluster analysis of differential proteins. (C) Principal component analysis of differential proteins.
GO analysis showed that the differentially expressed proteins in HN44 were mainly involved in the cell membrane and cytoplasm composition, purine ribonucleoside triphosphate binding, metal ion binding, and protein binding (Figure 3A). The differentially expressed proteins in HN65 were mainly involved in metal ion binding, hydrolase and transferase activity, membrane function, and response to stress (Figure 3B). KEGG analysis showed that the differentially expressed proteins in HN44 and HN65 cells were mainly involved in protein turnover, post-translational modification, chaperones, transport and catabolism, signal transduction mechanisms, lipid transport and metabolism, and carbohydrate transport and metabolism (Figures 3C,D). To alleviate the damage caused by stress, energy and osmotic adjustment substances are provided to plants by increasing the content of soluble sugars, free amino acids, and soluble proteins. Subcellular localization showed that the differentially expressed proteins in HN44 were mainly located in the chloroplast, cytoplasm, and extracellular regions, accounting for 34.88%, 27.91%, and 18.60% of the total differentially expressed proteins, respectively (Figure 3E). The differentially expressed proteins of HN65 were mainly located in the cytoplasm, chloroplasts, and nucleus, accounting for 33.82%, 32.35%, and 16.18% of the total differentially expressed proteins, respectively (Figure 3F).
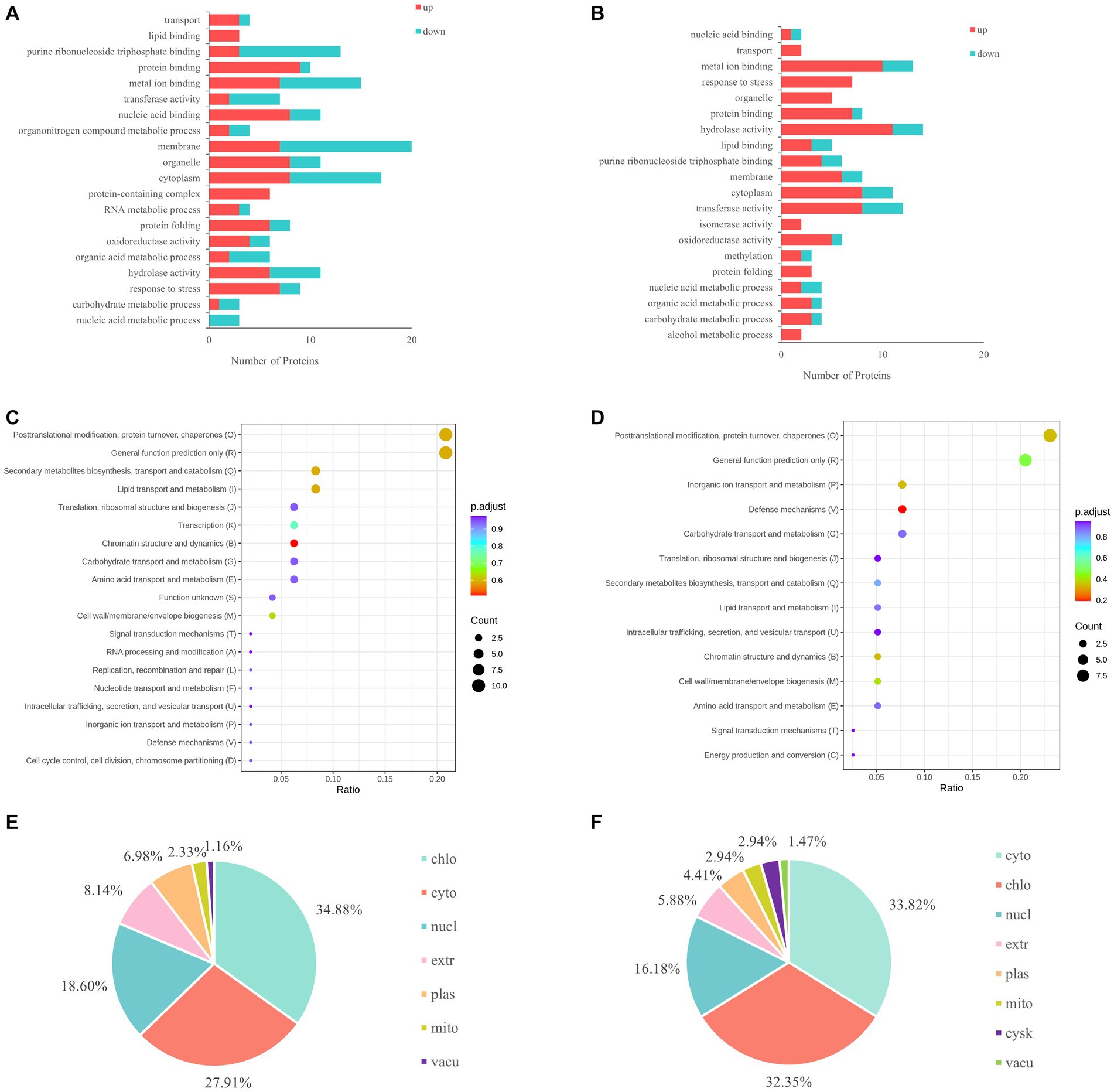
Figure 3. Differential function and enrichment analysis of proteins under drought stress. (A) GO analysis of HN44. (B) GO analysis of HN65. (C) KEGG analysis of HN44. (D) KEGG analysis of HN65. (E) Subcellular localization of HN44. (F) Subcellular localization of HN65. chlo, chloroplast; cyto, cytoplasm; nucl, nucleus; extr, extra cell; plas, plasma membrane; mito, mitochondrion; vacu, vacuolus; cysk, cytoskeleton.
3.3. The important role of carbon metabolism in plant drought resistance
Non-structural carbon, represented by soluble sugars in plants, is an important product of plant carbohydrate metabolism. It reflects the physiological status of plants as well as plays a staged buffering role in environmental stress and effectively reduces the damage caused by adversity to plants (Baslam et al., 2020). A total of 35 related proteins were detected in starch and sucrose metabolic pathways. And found the original sugar metabolism pathway was changed. In HN44, catabolism was enhanced and anabolism was weakened. Specific performance in the proteins related to endoglucanase-1,3-β-D-glucosidase in HN44 were significantly increased. In contrast, the expression of granule-bound starch synthase, α-amylase, glycogen phosphorylase, and endoglucanase-related proteins significantly decreased, indicating that the synthesis of starch in HN44 leaves was reduced under drought conditions. In HN65, the sucrose synthase-related proteins decreased significantly under drought stress, which promoted the accumulation in soluble sugar content, such as glucose and fructose, consequently promoting a decrease in cell water potential and reduced cell water loss. The responses of HN44 and HN65 to drought differed, and the method of regulating drought was also significantly different (Figure 4).
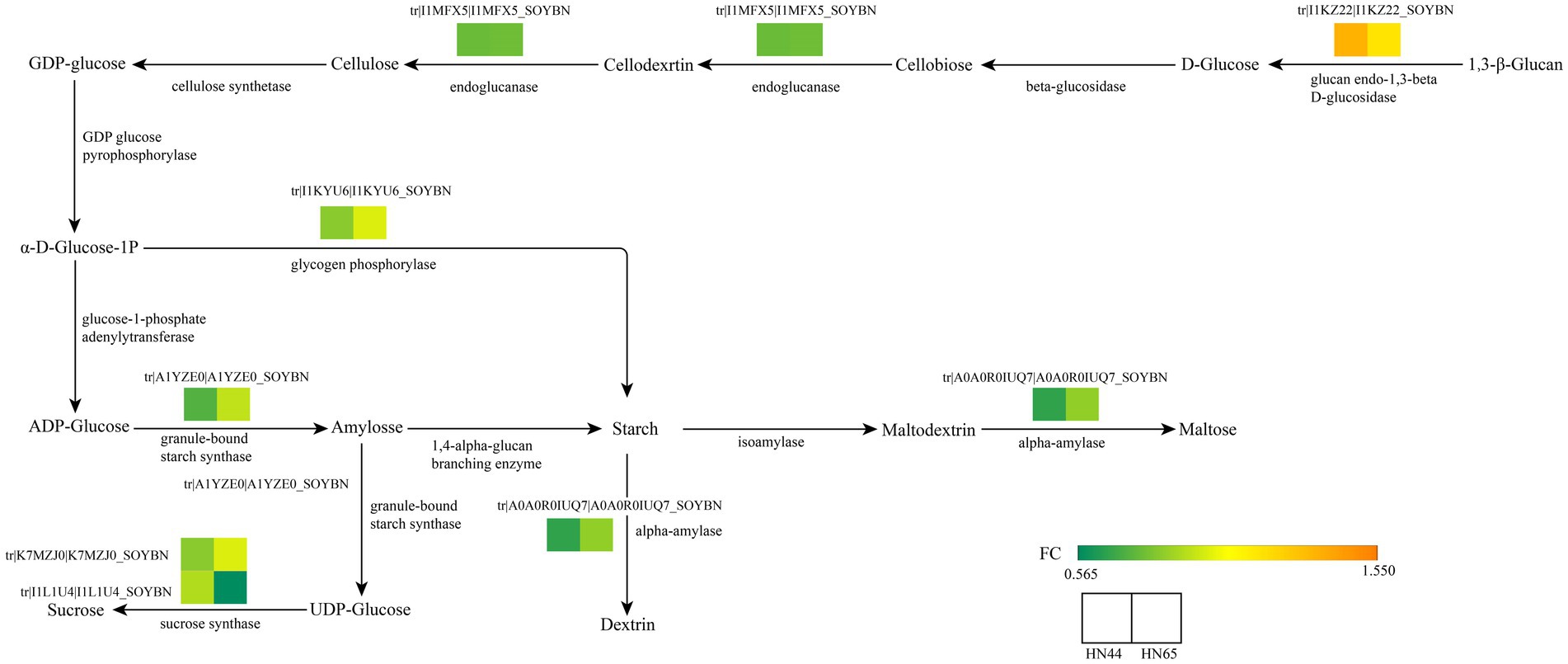
Figure 4. Key genes of starch and sucrose metabolism based on the KEGG enrichment pathway. From green to orange indicates a FC value from 0.565 to 1.550.
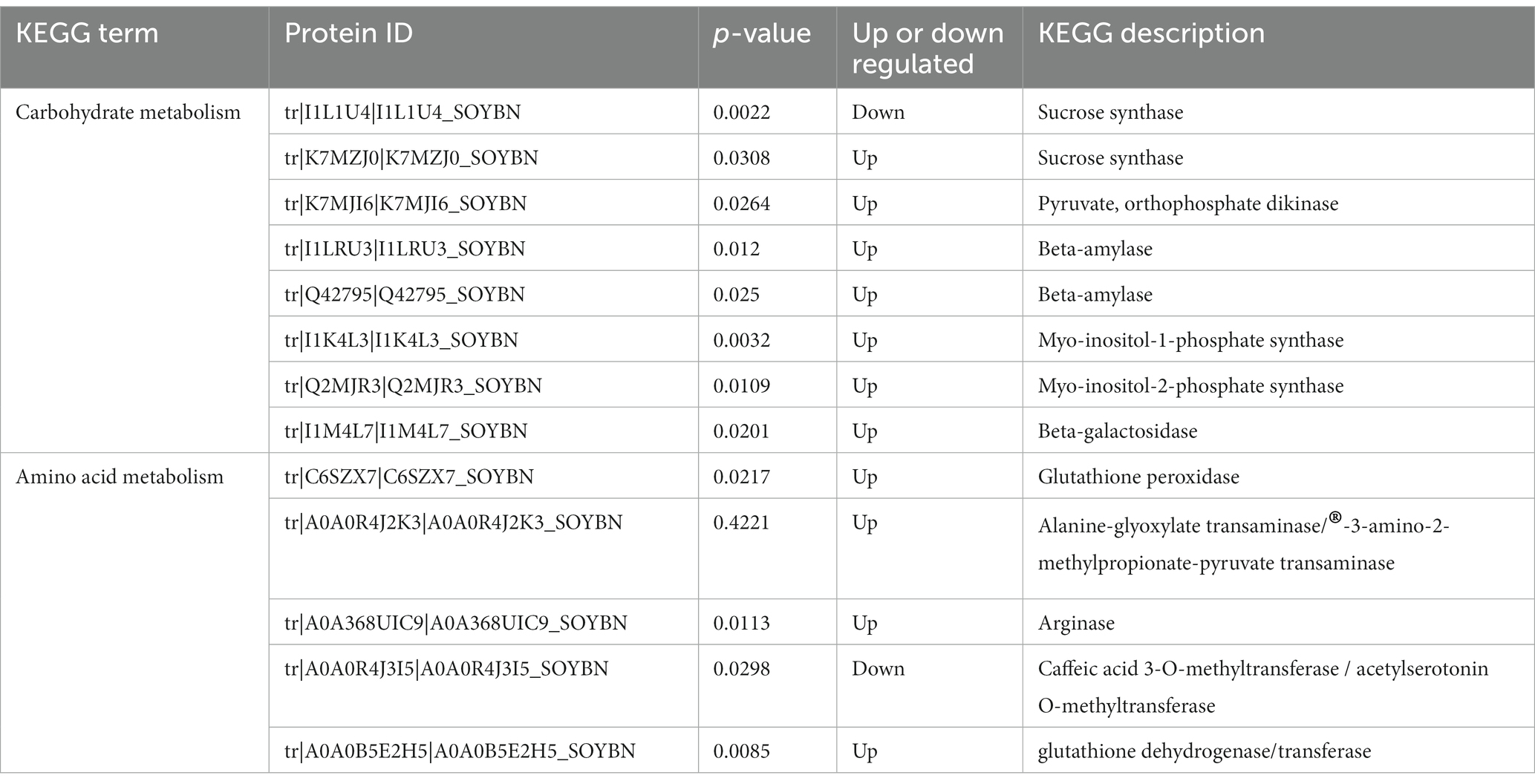
Carbohydrate metabolism is an important process that provides energy to plants (Yu et al., 2023). KEGG analysis showed that carbohydrates played an important role in the resistance of HN44 and HN65 to drought stress. In the KEGG pathway enrichment analysis of HN44 cells, we identified 13 differentially expressed proteins related to carbohydrate metabolism, of which six differentially expressed proteins were upregulated. One protein was related to the expression of l-ascorbate oxidase, and one protein was related to the expression of phospholipid: diacylglycerol acyltransferase. Three proteins were associated with the expression of myo-inositol-1-phosphate synthase, and one protein was associated with the expression of glucan endo-1,3-beta-glucosidase. Seven downregulated proteins were related to the expression of glycogen phosphorylase, alpha-amylase, endoglucanase, 3-hydroxyisobutyryl-CoA hydrolase, UDP-glucuronate 4-epimerase, UDP-apiose/xylose synthase, and granule-bound starch synthase (Table 1).
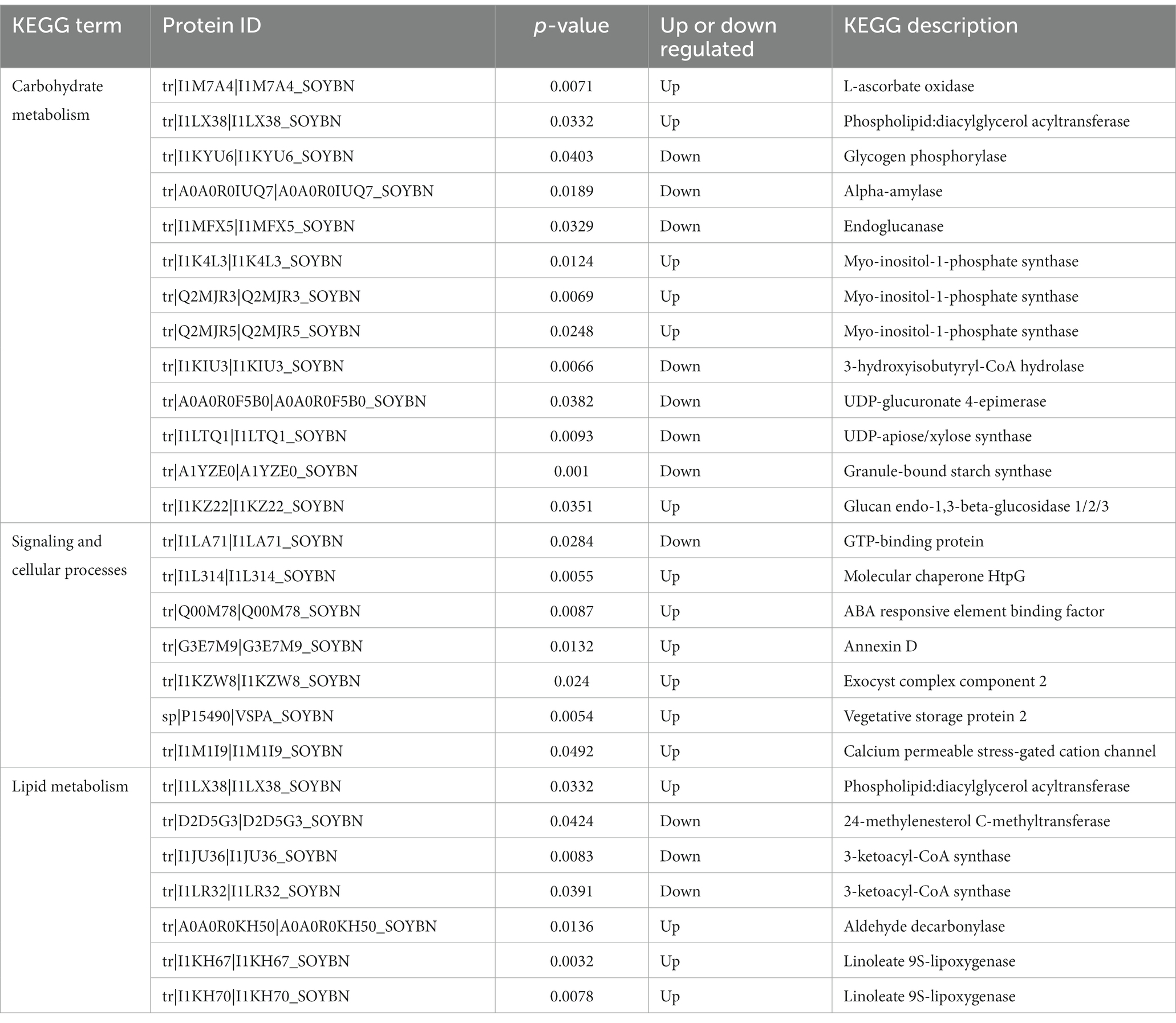
Table 1. HN44 differential proteins are involved in signal transduction and cellular processes as well as carbohydrate and lipid metabolism.
Under drought stress, eight differentially expressed proteins were related to carbohydrate metabolism, of which seven were upregulated. One protein was related to sucrose synthase, and another was related to pyruvate and orthophosphate dikinase. Two proteins were associated with beta-amylase, two proteins were associated with phosphate synthase, and one protein was associated with beta-galactosidase. The expression level of one protein related to sucrose synthase decreased (Table 2). Plants must adapt to adversity by changing the different forms of carbon under drought conditions. In addition, drought stress considerably impacts important life processes, such as signal transduction and lipid and amino acid metabolism.
3.4. Important role of amino acids in plant drought resistance
KEGG result showed that drought stress significantly affects the metabolism of sucrose and starch as well as affects the metabolism of many amino acids. Our results showed that drought stress had a significant effect on the content of free amino acids. Two proteins involved in amino acid metabolism under drought conditions were upregulated in HN44, which related to primary amine oxidase and alanine-glyoxylic acid transamina®(R)-3-amino-2-methylpropionate pyruvate transaminase. They are located in chloroplast and mitochondrion respectively, and play a vital role in redox reactions and amino acid metabolism. Four proteins were downregulated, including, which were mainly associated with aspartate kinase, cystathionine gamma synthase, 3-hydroxyisobutyryl-CoA hydrolase, and aminocyclopropanecarboxylate oxidase. They are located in chloroplast, chloroplast, nucleus and cytoplasm, respectively, and play a crucial role in regulating carbon flow, cysteine metabolism, valine catabolism and ethylene synthesis. Among the proteins involved in amino acid synthesis under drought conditions in HN65, four were upregulated and one was downregulated. The upregulated proteins were related to glutathione peroxidase, alanine-glyoxylate transamina®(R)-3-amino-2-methylpropionate-pyruvate transaminase, and arginase. They are located in chloroplasts, mitochondria, chloroplasts and chloroplasts, respectively, and play an important role in antioxidant response, glyoxylic acid detoxification and immune response. The downregulated protein mainly related to caffeic acid 3-O-methyltransferase/acetylserotonin O-methyltransferase. It is located in the cytoskeleton, involved in the biosynthesis of lignin, is an important component of stable cell structure.
3.5. Important roles of other proteins in drought resistance
After GO enrichment analysis, we identified eight proteins related to the stress response in HN44 cells, and seven were upregulated. Among these, three proteins were involved in the response to reactive oxygen species, and one protein was associated with antioxidant responses. One protein was involved in systemic acquired resistance, and two proteins were involved in response to heat and drought stress. One protein related to the response to heat stress was downregulated, also related to physiological processes such as ATP activity and protein renaturation.
Seven upregulated proteins related to stress response were identified in HN65, of which two were involved in response to reactive oxygen species and heat stress, one was involved in the biosynthesis of abscisic acid, and two were involved in response to heat stress and drought stress and two were involved in antioxidant enzyme activities (including peroxidase and glutathione peroxidase activities and superoxide dismutase activity), and no downregulated proteins related to stress response were identified. In addition, drought stress increased the oxidoreductase activity of HN44 cells and promoted organic acid and alcohol metabolism in HN65 cells.
3.6. Differential protein–protein interaction (PPI) networks
Proteins are important carriers of life activities, and interactions between proteins are involved in many life processes. PPI analysis can identify key regulatory factors and effectively reveal interactions between proteins. We used a STRING database to construct a protein–protein interaction (PPI) network to better understand the relationship between differential proteins. The PPI networks of HN44 and HN65 cells included 47 and 28 nodes, respectively (Figure 5). In the PPI network of HN44 and HN65, we screened three proteins (A0A6N2BR27, A0A445IS56 and A0A445F5B9) expressed by SGT1-1, RAR1-1, and RAR1-2 gene as hub proteins (degree ≥ 20), and the molecular functions of them were protein-folding chaperone binding, metal in binding and metal ion binding. Fu et al. (2009) considered that SGT1-1, RAR1-1 and RAR1-2 are important components of soybean triggering immune effects. The three hub proteins were closely related to the HSP20 protein family of HN44, nucleolin, calcium ion permeability stress-gated cation channel proteins, UDPG glucuronide epimerase, and other differentially expressed proteins. In HN65 cells, the three hub proteins were closely related to differentially expressed proteins, such as GTP-binding protein, molecular chaperone G, calcium ion permeability stress-gated cation channel protein, pyruvate, and orthophosphate dikinase. Thus, these three proteins, which are related to plant immunity and disease resistance, play vital roles under drought stress.
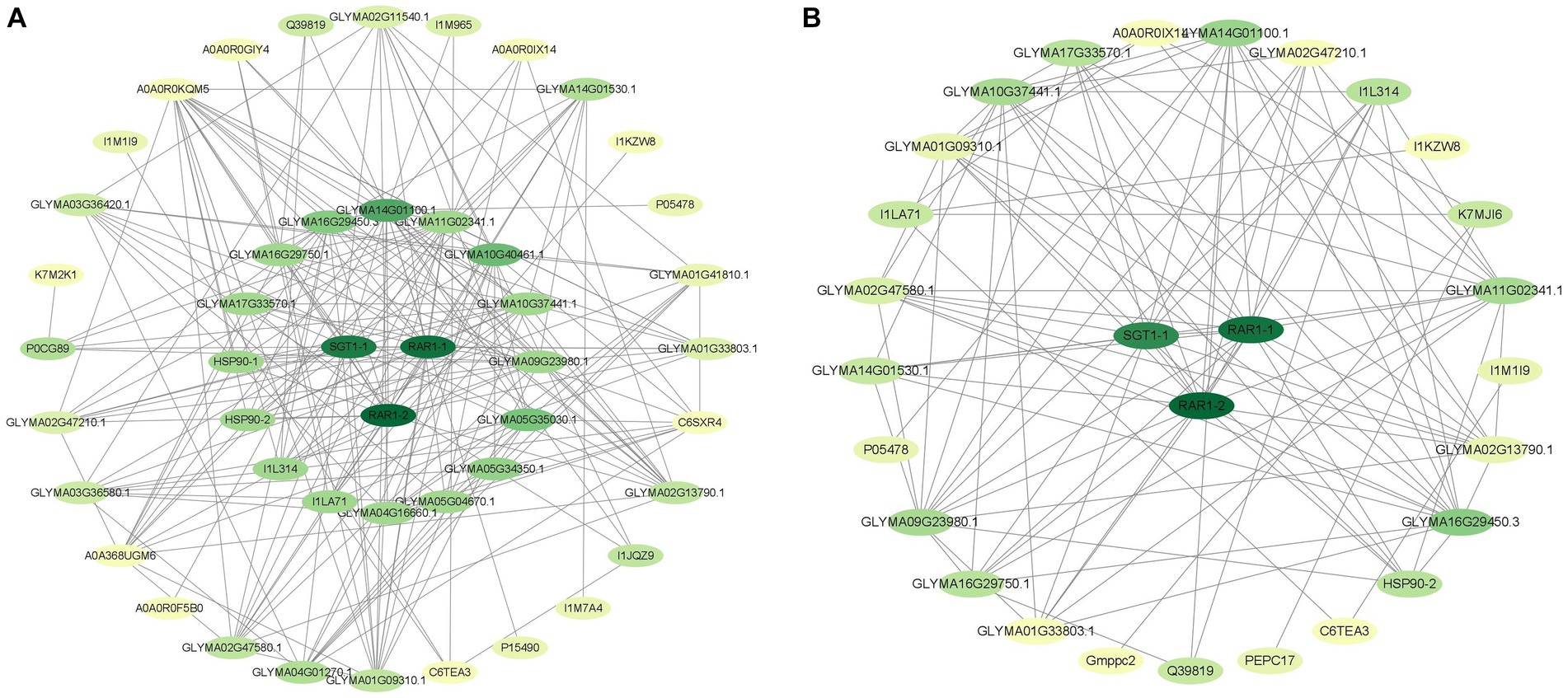
Figure 5. (A) The interaction network of differential proteins in HN44. (B) Differential protein interaction network of HN65. Each node represents a protein, and the connection between nodes indicates the interaction between proteins. The darker the color of a node, the more connections it has.
4. Discussion
Under normal water conditions, the water potential of plant cells was in a state of dynamic equilibrium. When drought occurs, osmotic water loss is greater than osmotic water absorption of plant cells, the original dynamic balance was broken, and the osmotic potential in plant cells increases rapidly to cope with osmotic water loss caused by drought (Guo et al., 2012; Lv et al., 2019). As an excellent osmotic regulator, PEG-6000 is easily soluble in water and has a large molecular weight and cannot enter plant cell wall, resulting in a high extracellular osmotic potential. It can also break the dynamic balance of the original water potential of the cells, causing cell water loss and achieving the purpose of simulating drought (Ahmed et al., 2022). The nutrient solution was added with 15% PEG-6000 for 4 days to achieve moderate drought stress equivalent to natural drought conditions (Wang et al., 2012, 2022a; GB/T 32136-2015, 2015). Drought stress causes slow plant growth rate, visible morphological changes such as leaf yellowing, accumulation of ROS, membrane lipid peroxidation, and an increase in osmotic regulators, seriously affecting the growth and development of plants (Vahideh et al., 2021; Zhou et al., 2022b). Previous proteomic studies have shown that drought stress significantly affects soybean protein metabolism. Drought stress considerably impacts energy metabolism, followed by defense-related proteins (programmed death, signal transduction, and osmotic regulation) (Yu et al., 2016). Our research also proves this point. Drought stress also significantly affects plant secondary metabolite synthesis, amino acid metabolism, protein folding, calcium homeostasis, and signal transduction (Wang and Komatsu, 2018). Our study screened many differentially expressed proteins, including energy metabolism, nucleic acid metabolism, carbohydrate metabolism, and secondary metabolism.
Carbon metabolism is an important part of energy metabolism, and the accumulation of soluble sugars in plants under drought stress is an important way to resist drought (Guo et al., 2021; Wang et al., 2022b). Our study showed that the expression levels of starch synthase, and other key enzymes in the synthesis of polysaccharides in drought-resistant soybean HN44 decreased under drought stress, and the expression levels of endoglucanases such as glucan endo-1,3-β-D-glucosidase increased, causing an increase in soluble sugar content and effectively alleviating drought stress (Zhou et al., 2022a). However, the expression levels of glucan endo-1,3-β-D-glucosidase and other endonucleases in soybean HN65 with weak drought resistance did not change significantly. The sucrose synthase-related protein of HN65 changed significantly. The expression of the sucrose synthase protein in HN65 soybeans with weak drought resistance increased or decreased under drought stress, indicating that drought stress changed the carbon metabolism of HN65, which may be an important way to endow it with new drought stress tolerance. Halder et al. (2022) demonstrated a similar conclusion. Drought stress causes a significant change in the activity of starch synthase in wheat, which blocks the synthesis of starch, changes energy metabolism, and enhances drought resistance. Simultaneously, it also led to a decrease in wheat grain weight and thinner stems. Ali et al. (2023) suggested that the main mechanism by which drought stress changes sucrose synthase activity in maize and enhances drought resistance is by promoting sucrose decomposition into sugar and glucose, increasing soluble sugar concentrations, and alleviating drought stress. Sucrose synthase is a key factor in plant abiotic stress tolerance; however, its mechanism under abiotic stress requires further investigation (Halder et al., 2022).
Carbohydrate metabolism in soybeans under drought stress is strictly regulated by calcium homeostasis. Calcium ions change the carbon metabolism process of plants under stress by regulating pyruvate carboxylase, causing an increase in the content of fructose and sucrose in plant leaves and increasing their ability to resist drought stress (Wang and Komatsu, 2018). Calcium homeostasis is an important signal transduction component involved in almost all eukaryote physiological regulations (Yu et al., 2021). Although the mechanism of calcium homeostasis in biological stress processes has not been fully elucidated, many studies have shown that the expression of calcium-related proteins and the level of calcium ions in plants increase sharply after various abiotic stress stimuli (Medvedev, 2005; Zhang et al., 2022). Our study also attained a similar conclusion; the expression levels of calcium homeostasis-related proteins (tr|I1M1I9|I1M1I9_SOYBN) and signal transduction-related proteins in HN65 and HN44 leaves increased significantly after drought stress. Calcium homeostasis is involved in carbon metabolism to enhance plant drought resistance as well as activating ABA signal transduction, synthesizing secondary metabolites, increasing intracellular ion concentration, energy metabolism, and other drought resistance pathways (Kudla et al., 2018).
Through PPI network analysis, the three hub proteins at the center of the differential protein network were related to plant immunity and stress resistance. This indicates that defense-related proteins play a crucial role in the adaptation of soybeans to drought stress (Yu et al., 2016; Wang and Komatsu, 2018). Our study found that drought stress caused the response of antioxidant enzyme system of soybean, such as catalase, antioxidant enzymes and other stress-related protein expression levels, to reduce or remove oxidative stress caused by reactive oxygen species (ROS) and protect growth and metabolism of plants (Silva et al., 2010; Koh et al., 2015). In addition, glutathione and ascorbic acid also can achieve the purpose of scavenging ROS by binding to toxic substances (Zhu B. et al., 2023). And drought stress causes a significant increase in necroptosis-related proteins, achieving tissue homeostasis and stabilizing the development of organisms by self-cleaning some aging and death cells. There are also an important way for plants to adapt to drought stress (Yamada and Yoshida, 2019).
The metabolism and content of free amino acids are highly susceptible to drought stress, resulting in their large accumulation (Zhu et al., 2020). Free amino acids act as osmotic regulators to reduce water loss and have strong antioxidant activity, which is essential for maintaining the redox homeostasis of cells (Yu et al., 2017; Li et al., 2022). Our study demonstrated that the activities of aspartate kinase, cystathionine gamma synthase, and aminocyclopropanecarboxylate oxidase, which are involved in the metabolism of cysteine and methionine, were significantly decreased under drought stress. In contrast, the activity of alanine-glyoxylate transamina®(R)-3-amino-2-methylpropionate-pyruvate transaminase was significantly upregulated. Changes in amino acid metabolic pathways, leading to changes in the proportion of amino acids that maintain oxidative homeostasis, are also important for plants to resist adversity (Li et al., 2022). We also found that primary amine oxidase and alanine-glyoxylate transam®se/(R)-3-amino-2-methylpropionate pyruvate transaminase, which synthesize neutral amino acids, such as serine, glycine, alanine, and threonine, were significantly upregulated. The significant upregulation of neutral amino acid expression does not cause changes in intracellular charge while maintaining osmotic potential and is an excellent contributor to osmotic regulation (Zhu et al., 2020).
At present, some studies have used omics technology to demonstrate the drought resistance mechanism of soybean, but most of them are limited to a single variety or yield formation period, and there are few studies on multiple varieties and seedling stage (Zhao et al., 2021). We believe that the use of varieties with different stress resistance at the same time can make the results more convincing (Farzad et al., 2023). Therefore, we choose two varieties with great difference in stress resistance for comparison. It was found that there were large differences between drought-resistant varieties and sensitive varieties, which may be an important reason for the large differences in drought resistance among different varieties. Proteomics technology was widely used in the study of plant stress resistance as a new technology. However, in order to fully understand the mechanism of plant stress resistance, it was not enough to rely solely on proteomics technology (Zhu W. C. et al., 2023). In addition, we have previously measured transcriptomics and metabolomics. Similar to proteomics, drought stress seriously affected carbohydrate metabolism, amino acid metabolism, hormone metabolism and other pathways in soybean. Varieties with different drought resistance showed great differences. In addition, the results of transcriptomics also showed that drought stress seriously affected plant photosynthesis. Metabolomics showed that flavonoids are one of the important substances for soybean to resist drought stress (Wang et al., 2022c).
5. Conclusion
We identified 86 differentially expressed proteins (DEPs) in HN44 cells. These differentially expressed proteins are involved in signal transduction, energy metabolism, stress responses, and other biological processes. A total of 68 differential proteins were identified in the leaves of HN65 cells. These differentially expressed proteins are involved in stress response, material hydrolysis, redox and energy metabolism, and other biological processes. HN44 responds to drought stress mainly through sugar decomposition, increased antioxidant enzyme activity and lipid metabolism. HN65 responds to drought stress mainly through ABA synthesis, increased antioxidant enzyme activity and amino acid metabolism. Proteomics, as a powerful research method, is of great significance for revealing plant adversity. However, it is not enough for us to study the mechanism of drought resistance difference between the two genotypes of soybean. In the future, we will study the drought resistance of soybean from more perspectives, aiming to contribute to the study of soybean drought resistance mechanism.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.proteomexchange.org/, PXD040486.
Author contributions
SD and QZ made conceptualization. SD offered the methodology. QZ processed data. QZ, SS, XinW, ZW, and XiyW did the investigation. SD offered resources. QZ wrote—original draft preparation. SD, CY, and CM monitored experiments and guided manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by Heilongjiang Postdoctoral Research Start-up Fund, China, grant number LBH-Q21078 and Heilongjiang Provincial Natural Science Foundation, China, Grant number LH2021C023.
Acknowledgments
Thanks to the seed materials had been provided by Heilongjiang Academy of Agricultural Sciences and the experimental platform had been provided by Northeast Agricultural University.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher’s note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Ahmed, M., Ahmed, M. S. K., Mehmood, M. Z., Ahmad, S., and Hasanuzzaman, M. (2022). Changes in germination and seedling traits of sesame under simulated drought. Phyton 91, 713–726. doi: 10.32604/PHYTON.2022.018552
Ali, A. E. E., Husselmann, L. H., Tabb, D. L., and Ludidi, N. (2023). Comparative proteomics analysis between maize and Sorghum uncovers important proteins and metabolic pathways mediating drought tolerance. Life 13:170. doi: 10.3390/life13010170
Badr, A., and Bruggemann, W. (2020). Special issue in honor of prof. Reto J. Strasser—comparative analysis of drought stress response of maize genotype using chlorophyll fluorescence measurements and leaf relative water content. Photosynthetica 58, 638–645. doi: 10.32615/ps.2020.014
Baslam, M., Mitsui, T., Sueyoshi, K., and Ohyama, T. (2020). Recent advances in carbon and nitrogen metabolism in C3 plants. IJMS 22:318. doi: 10.3390/ijms22010318
Bates, L. S., Waldren, R. P., and Teare, I. D. (1973). Rapid determination of free proline for water-stress studies. Plant Soil 39, 205–207. doi: 10.1007/BF00018060
Bradford, M. M. (1976). A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal. Biochem. 72, 248–254. doi: 10.1016/0003-2697(76)90527-3
Chen, F., Wei, M., Ye, Y., and Yuan, W. (2015). Calculation of chemical potential in dilute solution and derivation of osmotic pressure formula. Univ. Chem. 30, 63–67. doi: 10.3866/pku.DXHX20150463
Demirkol, G. (2021). PopW enhances drought stress tolerance of alfalfa via activating antioxidative enzymes, endogenous hormones, drought related genes and inhibiting senescence genes. Plant Physiol. Biochem. 166, 540–548. doi: 10.1016/j.plaphy.2021.06.036
Du, Y., Zhao, Q., Chen, L., Yao, X. D., and Xie, F. T. (2020). Effect of drought stress at reproductive stages on growth and nitrogen metabolism in soybean. Agronomy 10:302. doi: 10.3390/agronomy10020302
Farzad, F., Behzad, S., Payam, M., Gholamreza, A., and Hamid, M. (2023). Nutritional value and agronomic traits of forage sorghum under drought stress. Biocatal. Agric. Biotechnol. 48:102624. doi: 10.1016/J.BCAB.2023.102624
Fu, D., Ghabrial, S., and Kachroo, A. (2009). GmRAR1 and GmSGT1 are required for basal, R gene–mediated and systemic acquired resistance in soybean. Mol. Plant-Microbe Interact. 22, 86–95. doi: 10.1094/MPMI-22-1-0086
GB/T 32136-2015 . Grade of agricultural drought. Beijing, China: China Meteorological Administration (2015).
Gonyane, M. B., and Sebetha, E. T. (2021). The effect of plant density, zinc added to phosphorus fertilizer sources and location on selected yield parameters of soybean. LR 45, 196–202. doi: 10.18805/LRF-647
Guo, R., Hao, W. P., and Gong, D. Z. (2012). Effects of PEG-6000 simulated water stress on physiological and ecological indexes of winter wheat with different drought resistance. J. Crops. 5, 43–47. doi: 10.16035/j.issn.1001-7283.2012.05.014
Guo, J., Qu, L., Hu, Y., Lu, W., and Lu, D. (2021). Proteomics reveals the effects of drought stress on the kernel development and starch formation of waxy maize. BMC Plant Biol. 21:434. doi: 10.1186/s12870-021-03214-z
Gupta, A., Rico-Medina, A., and Caño-Delgado, A. I. (2020). The physiology of plant responses to drought. Science 368, 266–269. doi: 10.1126/science.aaz7614
Halder, T., Choudhary, M., Liu, H., Chen, Y., Yan, G., and Siddique, K. H. M. (2022). Wheat proteomics for abiotic stress tolerance and root system architecture: current status and future prospects. Proteomes 10:17. doi: 10.3390/proteomes10020017
Hu, W., Zhang, J., Wu, Z., Loka, D. A., Zhao, W., Chen, B., et al. (2022). Effects of single and combined exogenous application of abscisic acid and melatonin on cotton carbohydrate metabolism and yield under drought stress. Ind. Crop. Prod. 176:114302. doi: 10.1016/j.indcrop.2021.114302
Ingwers, M. W., Steketee, C. J., Yadav, S. K., and Li, Z. (2022). Relationships among carbon isotope composition, growth, and foliar nitrogen in soybean. J. Crop Improv. 36, 90–107. doi: 10.1080/15427528.2021.1910092
Jaiswal, S., Gautam, R. K., Singh, R. K., Krishnamurthy, S. L., Ali, S., Sakthivel, K., et al. (2019). Harmonizing technological advances in phenomics and genomics for enhanced salt tolerance in rice from a practical perspective. Rice 12:89. doi: 10.1186/s12284-019-0347-1
Jangir, P., Shekhawat, P. K., Bishnoi, A., Ram, H., and Soni, P. (2021). Role of serendipita indica in enhancing drought tolerance in crops. Physiol. Mol. Plant Pathol. 116:101691. doi: 10.1016/j.pmpp.2021.101691
Jin, T., Shan, Z., Zhou, S., Yang, Q., Gai, J., and Li, Y. (2022). GmDNAJC7 from soybean is involved in plant tolerance to alkaline-salt, salt, and drought stresses. Agronomy 12:1419. doi: 10.3390/agronomy12061419
Jupa, R., Plavcová, L., Flamiková, B., and Gloser, V. (2016). Effects of limited water availability on xylem transport in liana Humulus lupulus L. Environ. Exp. Bot. 130, 22–32. doi: 10.1016/j.envexpbot.2016.05.008
Koh, J., Chen, G., Yoo, M. J., Zhu, N., Dufresne, D., Erickson, J. E., et al. (2015). Comparative proteomic analysis of Brassica napus in response to drought stress. J. Proteome Res. 14, 3068–3081. doi: 10.1021/pr501323d
Kudla, J., Becker, D., Grill, E., Hedrich, R., Hippler, M., Kummer, U., et al. (2018). Advances and current challenges in calcium signaling. New Phytol. 218, 414–431. doi: 10.1111/nph.14966
Li, B., Feng, Y., Zong, Y., Zhang, D., Hao, X., and Li, P. (2020). Elevated CO2-induced changes in photosynthesis, antioxidant enzymes and signal transduction enzyme of soybean under drought stress. Plant Physiol. Biochem. 154, 105–114. doi: 10.1016/j.plaphy.2020.05.039
Li, Y., Zou, J., Zhu, H., He, J., Setter, T. L., Wang, Y., et al. (2022). Drought deteriorated the nutritional quality of cottonseed by altering fatty acids and amino acids compositions in cultivars with contrasting drought sensitivity. Environ. Exp. Bot. 194:104747. doi: 10.1016/j.envexpbot.2021.104747
Lv, X. P., Gao, H. J., Zhang, L., Wang, Y. P., Shao, K. Z., Zhao, Q., et al. (2019). Dynamic responses of Haloxylon ammodendron to various degrees of simulated drought stress. Plant Physiol. Biochem. 139, 121–131. doi: 10.1016/j.plaphy.2019.03.019
Medvedev, S. S. (2005). Calcium signaling system in plants. Russ. J. Plant Physiol. 52, 249–270. doi: 10.1007/s11183-005-0038-1
Rahman, M. D. M., Mostofa, M. G., das, A. K., Anik, T. R., Keya, S. S., Ahsan, S. M., et al. (2022). Ethanol positively modulates photosynthetic traits, antioxidant defense and osmoprotectant levels to enhance drought acclimatization in soybean. Antioxidants 11:516. doi: 10.3390/antiox11030516
Silva, E. N., Ferreira-Silva, S. L., Fontenele, A. d. V., Ribeiro, R. V., Viégas, R. A., and Silveira, J. A. G. (2010). Photosynthetic changes and protective mechanisms against oxidative damage subjected to isolated and combined drought and heat stresses in Jatropha curcas plants. J. Plant Physiol. 167, 1157–1164. doi: 10.1016/j.jplph.2010.03.005
Song, H., Huang, Y., and Gu, B. (2020). QTL-Seq identifies quantitative trait loci of relative electrical conductivity associated with heat tolerance in bottle gourd (Lagenaria siceraria). PLoS One 15:e0227663. doi: 10.1371/journal.pone.0227663
Sudhir, P. R., Pogoryelov, D., Kovacs, L., Garab, G., and Murthy, S. D. S. (2005). The effects of salt stress on photosynthetic electron transport and thylakoid membrane proteins in the cyanobacterium Spirulina platensis. BMB Rep. 38, 481–485. doi: 10.5483/BMBRep.2005.38.4.481
Vahideh, K., Ali, R. Y., Afshin, T., Bahman, F., and Andrea, M. (2021). Concentrations-dependent effect of exogenous abscisic acid on photosynthesis, growth and phenolic content of Dracocephalum moldavica L. under drought stress. Planta 253:127. doi: 10.1007/s00425-021-03648-7
Waheed, H., Javaid, M. M., Shahid, A., Ali, H. H., Nargis, J., and Mehmood, A. (2019). Impact of foliar-applied Hoagland’s nutrient solution on growth and yield of mash bean (Vigna mungo L.) under different growth stages. J. Plant Nutr. 42, 1133–1141. doi: 10.1080/01904167.2019.1607380
Wang, X., and Komatsu, S. (2018). Proteomic approaches to uncover the flooding and drought stress response mechanisms in soybean. J. Proteome 172, 201–215. doi: 10.1016/j.jprot.2017.11.006
Wang, X., Li, X., and Dong, S. (2022a). Screening and identification of drought tolerance of spring soybean at seedling stage under climate change. Front. Sustain. Food Syst. 6:988319. doi: 10.3389/fsufs.2022.988319
Wang, L., Liu, L., Pei, Y., Dong, S., Sun, C., Zu, W., et al. (2012). Drought resistance identification of soybean germplasm resources at germination stage. J. Northeast Agric. Univ. 2012 43, 36–43. doi: 10.19720/j.cnki.issn.1005-9369.2012.01.007
Wang, A., Ma, C., Ma, H., Qiu, Z., and Wen, X. (2021). Physiological and proteomic responses of pitaya to PEG-induced drought stress. Agriculture 11:632. doi: 10.3390/agriculture11070632
Wang, X., Oh, M., Sakata, K., and Komatsu, S. (2016). Gel-free/label-free proteomic analysis of root tip of soybean over time under flooding and drought stresses. J. Proteome 130, 42–55. doi: 10.1016/j.jprot.2015.09.007
Wang, X., Song, S., Wang, X., Liu, J., and Dong, S. (2022c). Transcriptomic and metabolomic analysis of seedling-stage soybean responses to PEG-simulated drought stress. Int. J. Mol. Sci. 23:6869. doi: 10.3390/ijms23126869
Wang, X., Wu, Z., Zhou, Q., Wang, X., Song, S., and Dong, S. (2022b). Physiological response of soybean plants to water deficit. Front. Plant Sci. 12:809692. doi: 10.3389/fpls.2021.809692
Yamada, K., and Yoshida, K. (2019). Mechanical insights into the regulation of programmed cell death by p53 via mitochondria. Biochim Biophys Acta Mol Cell Res 1866, 839–848. doi: 10.1016/j.bbamcr.2019.02.009
Yemm, E. W., and Willis, A. J. (1954). The estimation of carbohydrates in plant extracts by anthrone. Biochem. J. 57, 508–514. doi: 10.1042/bj0570508
Yu, H., Cao, Y., Wang, Z., Zhang, J., Yang, L., Zhao, Z., et al. (2023). Identification of the most sensitive stage of cotton microspore development to water deficit and analysis of carbohydrate metabolism related to pollen viability. Environ. Exp. Bot. 206:105168. doi: 10.1016/j.envexpbot.2022.105168
Yu, X., James, A. T., Yang, A., Jones, A., Mendoza-Porras, O., Bétrix, C. A., et al. (2016). A comparative proteomic study of drought-tolerant and drought-sensitive soybean seedlings under drought stress. Crop Pasture Sci. 67:528. doi: 10.1071/CP15314
Yu, Q., Liu, Y. L., Sun, G. Z., Liu, Y. X., Chen, J., Zhou, Y. B., et al. (2021). Genome-wide analysis of the soybean calmodulin-binding protein 60 family and identification of GmCBP60A-1 responses to drought and salt stresses. Int. J. Mol. Sci. 22:13501. doi: 10.3390/ijms222413501
Yu, X., Yang, A., and James, A. T. (2017). Comparative proteomic analysis of drought response in roots of two soybean genotypes. Crop Pasture Sci. 68:609. doi: 10.1071/CP17209
Zhang, X., Feng, Y., Khan, A., Ullah, N., Li, Z., Zaheer, S., et al. (2022). Quantitative proteomics-based analysis reveals molecular mechanisms of chilling tolerance in grafted cotton seedlings. Agronomy 12:1152. doi: 10.3390/agronomy12051152
Zhang, C., and Shi, S. (2018). Physiological and proteomic responses of contrasting alfalfa (Medicago sativa L.) varieties to PEG-induced osmotic stress. Front. Plant Sci. 9:242. doi: 10.3389/fpls.2018.00242
Zhao, B., Zhang, S., Yang, W., Li, B., Lan, C., Zhang, J., et al. (2021). Multi-omic dissection of the drought resistance traits of soybean landrace LX. Plant Cell Environ. 44, 1379–1398. doi: 10.1111/pce.14025
Zhou, Q., Li, Y., Wang, X., Yan, C., Ma, C., Liu, J., et al. (2022a). Effects of different drought degrees on physiological characteristics and endogenous hormones of soybean. Plan. Theory 11:2282. doi: 10.3390/plants11172282
Zhou, Q., Song, S., Wang, X., Yan, C., Ma, C., and Dong, S. (2022b). Effects of drought stress on flowering soybean physiology under different soil conditions. Plant Soil Environ. 68, 487–498. doi: 10.17221/237/2022-PSE
Zhu, W. C., Miao, X. X., Qian, J., Chen, S. J., Jin, Q. X., Li, M. Z., et al. (2023). A translatome-transcriptome multi-omics gene regulatory network reveals the complicated functional landscape of maize. Genome Biol. 24:60. doi: 10.1186/S13059-023-02890-4
Zhu, B., Xu, H., Guo, X., Lu, J., Liu, X., and Zhang, T. (2023). Comparative analysis of drought responsive transcriptome in Brassica napus genotypes with contrasting drought tolerance under different potassium levels. Euphytica 219:25. doi: 10.1007/s10681-023-03156-7
Keywords: drought stress, soybean protein expression, tandem mass tag technology, osmotic adjustment, proteomics
Citation: Dong S, Zhou Q, Yan C, Song S, Wang X, Wu Z, Wang X and Ma C (2023) Comparative protein profiling of two soybean genotypes with different stress tolerance reveals major components in drought tolerance. Front. Sustain. Food Syst. 7:1200608. doi: 10.3389/fsufs.2023.1200608
Edited by:
Duraisamy Saravanakumar, The University of the West Indies St. Augustine, Trinidad and TobagoReviewed by:
Irina Ivanova Vaseva, Bulgarian Academy of Sciences (BAS), BulgariaLyudmila Petrova Simova-Stoilova, Bulgarian Academy of Sciences (BAS), Bulgaria
Copyright © 2023 Dong, Zhou, Yan, Song, Wang, Wu, Wang and Ma. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Chunmei Ma, Y2h1bm1tMTk3NEAxNjMuY29t
 Shoukun Dong
Shoukun Dong Qi Zhou
Qi Zhou Chao Yan
Chao Yan Shuang Song
Shuang Song Xin Wang
Xin Wang Zihao Wu
Zihao Wu Xiyue Wang
Xiyue Wang