
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 27 April 2023
Sec. Crop Biology and Sustainability
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1157575
 Lakshmidevi Paranthaman1†
Lakshmidevi Paranthaman1† Parthasarathy Seethapathy1,2†
Parthasarathy Seethapathy1,2† Deepu Pandita3*
Deepu Pandita3* Chellappan Gopalakrishnan1
Chellappan Gopalakrishnan1 Subbiah Sankaralingam4
Subbiah Sankaralingam4 Sakthivel Venkatesh4
Sakthivel Venkatesh4 Arunkumar Malaisamy5
Arunkumar Malaisamy5 Anu Pandita6
Anu Pandita6 Ryan Casini7
Ryan Casini7 Abed Alataway8
Abed Alataway8 Ahmed Z. Dewidar8
Ahmed Z. Dewidar8 Khalid F. Almutairi9
Khalid F. Almutairi9 Hosam O. Elansary9*
Hosam O. Elansary9*The tomato, one of the world’s most extensively cultivated and consumed vegetable crops is negatively impacted by various pathogens. This study aimed to observe the differentially expressed proteins in tomato samples in plant–pathogen-biocontrol interactions. The fungal pathogen associated with wilted plants were isolated and identified based on its morphological and molecular characteristics. Fourteen strains of Pseudomonas fluorescens from agricultural soils were identified and described using biochemical assays, molecular analyses, and screening for antagonistic ability against the Fusarium wilt pathogen. Results demonstrated that the potential of P. fluorescens (TPf12) positively influenced the expression of antagonism against tomato wilt disease. A total of 14 proteins expressed differently were revealed in the 2D-PAGE-MS investigation. Proteins such as nucleoside diphosphate kinase, phenylalanine ammonia-lyase, protein kinase family protein, Ser/Thr protein kinase-like are unchanged in FOL pathogen interaction, but up-regulated in FOL + TPf12 treated roots, and lipid transfer-like protein, and phenylalanine ammonia-lyase were down-regulated in FOL infested roots and upregulated in FOL + TPf12 treated tomato roots. Phenylalanine ammonia-lyase protein expression is commonly found in TPf12 bioenriched roots, and FOL + TPf12 treated roots, indicating its role in response to the application of TPf12 in tomato. A GC–MS analysis was performed to detect the bioactive metabolites synthesized by TPf12. Molecular docking investigations were conducted using the maestro’s GLIDE docking module of the Schrodinger Software program. Among the secondary metabolites, Cyclohexanepropanoic acid, 2-oxo-, methyl ester (CAS), and 3-o-(4-o-Beta-D-Galactopyranosyl-Beta-D-Galactopyraosyl)-2-Acetylamino-2-Deoxy-D-Galactose were shown to be top-ranked with a least docking score against each differently expressed proteins. The profiled molecules expressed differently due to plant-pathogen-biocontrol interactions may be directly or incidentally involved in the wilt disease resistance of tomato plants.
The tomato (Solanum lycopersicum L.), a member of the Solanaceae family, is one of the most economically important vegetable crops cultivated worldwide next to the potato (Quinet et al., 2019). The tomato is a dicotyledonous plant deeply studied as an ideal model for genetics, developmental biology, abiotic stress responses, disease resistance, and food science research (Rothan et al., 2019; Campos et al., 2022; Liu et al., 2022). The tomato is disproportionately more susceptible to biotic pests and pathogens than other cultivated crops (Zhang et al., 2022). Due to intense selection and severe genetic limitations during domestication and evolution, tomato cultivars have low genetic diversity. Owing to these factors, the tomato is susceptible to more than 200 diseases caused by diverse pathogens, including oomycetes, fungi, bacteria, phytoplasmas, viruses, viroids, and nematodes worldwide (Vitale et al., 2014; Panno et al., 2021). Among them, vascular wilt caused by Fusarium oxysporum (Schlectend. Fr.) f. sp. lycopersici (Sacc.) W.C. Snyder and H.N. Hansen (FOL) is the most economically destructive fungal disease due to its systemic infection, which severely affects all plant growth stages and is the foremost constraint in greenhouse and field-grown tomato production (Mousa et al., 2021). This is the most intensively studied plant disease described over 120 years ago (McGovern, 2015), widely disseminated, and ultimately reducing the crop yield by 45–55% and up to 80% in favorable conditions worldwide (Nirmaladevi et al., 2016; Srinivas et al., 2019; Panno et al., 2021). In the past, wilt incidence in various regions of India ranged from 25 to 55% due to FOL infection, posing a grave threat to its production in the country (Nirmaladevi et al., 2016). Warm weather (28°C), an acidic soil pH, and ammonium-based fertilizers exacerbate wilt symptoms in India (McGovern, 2015).
The germination of FOL soil-borne spores initiates the infection in response to chemosignals from the root exudates of the tomato. Infectious hyphae reach the rhizosphere and penetrate the epidermis. The mycelium then proliferates intra-or intercellularly in the cortex tissues till it penetrates the vascular tissues (Srinivas et al., 2019). At this stage, the fungus colonizes endophytically, remains exclusively within the xylem vessels by producing mycelium, spores, or polysaccharides, and induces tyloses as obstruction of xylem arteries in the host (Nirmaladevi et al., 2016). Further, FOL secretes tomatinase enzymes and toxins such as dehydrofusaric acid, fusaric acid, lycomarasmin, etc., in the colonized vessels, resulting in increased transpiration and decreased nutrient transfer, ultimately leading to vascular occlusion and wilting (Pareja-Jaime et al., 2008; Singh et al., 2017). The distinctive wilt symptoms are loss of turgidity, epinasty, flaccid and pale green to greenish-yellow leaf, drooping, browning, and wilting, which results in plant mortality (Ignjatov et al., 2012). At the warmest hours of the day, FOL wilt symptoms are often most apparent on mature tomato plants during flowering and fruit sets.
The Fusarium genus shows high-levels of genetic and functional diversity. It ranks among the top five most dynamic, versatile, ubiquitous, and necrotrophic plant pathogens in kingdom of fungi (Dean et al., 2012). Fusarium oxysporum (Fo) is a monocyclic, anamorphic species complex that embraces a variety of toxigenic pathogens of human, animal, and plant species in a host-specific manner and many non-pathogenic strains (Edel-Hermann and Lecomte, 2019). The fact that a pathogenic FOL isolate can infect both tomato seedlings, and immunocompromised mice, serves as a multi-host model for investigating inter-kingdom pathogenicity and virulence in fungi (Ortoneda et al., 2004). Over 106 well-characterized Formae speciales were recorded within F. oxysporum due to the virulence and pathogenic expansion of host ranges. Forma specialis of FOL is split into three physiological races, called races 1, 2, and 3, based on the different patterns of resistant and susceptible tomato varieties. Each race has a race-specific trait and a single dominant resistant gene (Edel-Hermann and Lecomte, 2019). Three parallel loci, I-1, I-2, and I-3, provide the pathogen-resistant genes that make the tomato resistant to the pathogen (McGovern, 2015). Even though there are ways to deal with these pathogenicity and virulence forms, they still limit tomato production.
Even though numerous management measures including soil solarization, crop rotation, and fungicides can prevent or minimize Fusarium (vascular) wilt, the vast majority are either environmentally damaging or unsuccessful. In intensive tomato farming, fungicides have been used to fight diseases that spread from the soil and increase crop yields. However, fungicides are transient and fail to protect plants during the entire growth period. In addition, fungicide-resistant plants can negatively affect the ecosystem and decrease the crop performance (Salman and Abuamsha, 2012). As a result, an alternative to these measures is to make and use effective plant-growth-promoting rhizobacteria (PGPR) preparations. Fluorescent pseudomonas species are ubiquitous in soils and on living roots, and they can use root exudates as food. The capacity of pseudomonads to bind to soils and the root surface, mobility and prototrophy, production of antibiotics, and hydrolytic enzymes are crucial for biocontrol fitness. In addition, they contain plant growth-promoting abilities, including nitrogen fixation, phosphate solubilization, iron chelation, and phytohormone synthesis. Due to their multifaceted functionality, fluorescent pseudomonads are the bioagent of interest in agriculture (Jayaraj and Radhakrishnan, 2008; Alzandi and Naguib, 2019; Suresh et al., 2022). In tomato, several fungal, oomycete, bacterial, viral, and nematode pests have been controlled using P. fluorescens strains (Jayaraj and Radhakrishnan, 2008; Mishra et al., 2014; Alzandi and Naguib, 2019; Dehghanian et al., 2020; Suresh et al., 2022). To be a product in the future, PGPR-mediated plant disease management may need to understand the possible interactions between the plant, pathogen, and biocontrol agent. Attempting to understand these interactions is a step forward for commercialization with ecological sustainability.
The primary hypothesis relies on the fact that most molecular investigations in tomato diseases have emphasized only two-way interactions (Mazzeo et al., 2014; Castillejo et al., 2015; Hu et al., 2019; Zhang et al., 2021), hence offering a preliminary understanding of disease suppression by chemical or biological agents. Furthermore, they lack information regarding the dynamical involvement of metabolites and expressed proteins during beneficial and detrimental interactions between microbes and plants. Recent advances in omics and bioinformatics tools have enhanced our understanding of triadic relationships and dependent mechanisms (Prabhukarthikeyan et al., 2017). Using methods that allow for comparative protein analysis, it is possible to find the vital functional proteins of the host in three-way interactions and understand the biomolecules that have built up during biocontrol interactions. Subsequently, molecular docking was explored to enlighten the possible protein interactions. Therefore, this insight is essential for successfully managing this disease and proving the efficiency of biocontrol agents.
To investigate this in terms of pathology, the identity and pathogenicity of F. oxysporum f. sp. lycoperisici were investigated, and the antagonism of P. fluorescens against the FOL was confirmed. The microbial bioactive compounds (MBCs) of P. fluorescens in vitro were documented using GC–MS. The interactive study attempted to outline the biocontrol mechanism with a complementary proteomic approach for investigating tomato defense mechanisms triggered by P. fluorescens against the infection of the FOL. In order to investigate differently expressed proteins during the triadic interactions, we employed two-dimensional electrophoresis (2-DE proteomics) with mass spectrometry (MALDI-TOF/TOF), followed by molecular docking investigations.
The wilted tomato plants were gathered from fields in Malumichampatti village, Tamil Nadu. The pathogen was isolated from discolored brown regions of root tissues, transferred aseptically to potato dextrose agar (PDA) media, and cultured for 7 days at 25 ± 2°C. The single-hyphal tip technique established a pure culture of the pathogen. A 9-mm disk of the fungus was transferred to 15 ml of sterile, solidified PDA from an 8-day-old culture. To analyze the pattern of mycelial development, phenotypic characteristics of hyphae, and spores, cultures were grown at room temperature (28 ± 2°C) for 7 days.
The FOL isolate was grown in sterile riverbed sand: corn granules (90:10 w/v with 20% distilled water) in a medium for 10 days at a temperature of 28 ± 2°C. The pathogenicity and virulence of the isolate were determined by artificially inoculating sand-corn granule inoculum in potting soil (autoclaved mixture of red soil, riverbed sand, and cow dung manure in a 1:1:1 weight ratio) at a concentration of 10% w/w, resulting in a spore concentration of 7 × 105. The potting mixture containing the fungus was used to fill pots with a diameter of 30 cm. Then, tomato cv. PKM-1 seeds were raised as three seedlings per pot with three replications in the greenhouse under 21–27°C and 60–70% R.H., with routine irrigation and proper drainage. Regularly, symptom manifestation was monitored for disease incidence.
The 10-day-old FOL culture was transferred to potato dextrose broth (PDB) 150 ml and cultured at ambient conditions for 1 week. Using the standard CTAB procedure, fungal mycelium was collected and processed for DNA extraction. In order to amplify the rDNA internal transcribed spacer (ITS) segment in the FOL genome, the whole DNA from a FOL isolate was utilized as the template in a Polymerase Chain Reaction (PCR) assay. In addition to the positive control, the universal ITS 1 and 4 primers were employed (Abd-Elsalam et al., 2003). Similarly, the F. oxysporum species-specific primers were used to amplify the ribosomal DNA intergenic spacer (IGS) regions of an isolated fungal genome (Balogun et al., 2008). Primers Sp13f and Sp13r confirmed the race (Hirano and Arie, 2006). Individual PCR reactions were conducted according to the cycle parameters stated in the literature. The PCR products were run on a 1.5% agarose gel and photographed with gel documentation equipment.
Fourteen isolates of P. fluorescens were obtained by serial dilution with King’s-B (KB) media from the roots of tomato crops cultivated in various regions of Tamil Nadu. The commercially available antagonistic strain P. fluorescens (Pf1) was obtained from the Plant Pathology-Culture Collection at Tamil Nadu Agricultural University in Coimbatore, India, for further investigations as a comparative strain. Using the dual culture technique in a PDA medium, the antifungal efficacy of P. fluorescens isolates was evaluated based on its potential to suppress mycelial proliferation. On one end of the plate, a 9-mm-diameter mycelial bit of the FOL was positioned, and on the opposite end, P. fluorescens strains were streaked. The absence of P. fluorescens on the plates served as a control. Five days were devoted to incubating the plates at 28 ± 2°C. Each experiment was performed three times. Following the incubation period, the FOL mycelial inhibition was assessed based on the growth inhibition equation: (C-T/C) × 100, where C is the measurement of FOL colony (mm) growth in the control plate, and T is the growth of FOL mycelium in the dual culture. Finally, the P. fluorescens isolate, where the FOL mycelial growth was largely inhibited, was selected as a potential isolate.
Biochemical reactions of P. fluorescens isolates were investigated using a biochemical test kit to characterize gram-negative rods [KB002 HiAssorted™ Biochemical Test Kit, HiMedia Laboratories Pvt. Ltd., Mumbai]. These test results were rated positive or negative and categorized using a deterministic scheme (Prabhukarthikeyan et al., 2015). The production of siderophore by certain P. fluorescens strains was determined using the chrome-azurol S (CAS) plate assay technique (Sasirekha and Srividya, 2016). In addition, selected isolates were streaked on tryptic soy agar medium (Himedia, Mumbai, and India) to test their ability to produce hydrogen cyanide (Devindrappa et al., 2022), compared to the Pf1 strain. Effective P. fluorescens were identified at the molecular level utilizing the 16S-23S rRNA intervening sequence primers and conditions (Ramesh Kumar et al., 2002), and the antibiotic gene of pyrrolnitrin was amplified using the primers (De Souza and Raaijmakers, 2003). DNA was extracted following the procedure described (Gobbin et al., 2007). Using the parameters specified in (Ramesh Kumar et al., 2002), DNA amplifications were conducted using a DNA thermal cycler (Eppendorf Master Cycler Gradient, New York). The amplicons were resolved on 1.5% agarose gel and stained with ethidium bromide (0.5 g ml−1) before being pictured and evaluated using a gel documentation system (Alpha Innotech Corporation, California).
Based on competitive screening, TPf12 was selected, and its fresh 24-h-old culture was inoculated into sterilized 300 ml of King’s B broth and incubated at 28°C for 5 days under constant agitation of 200 rpm. The culture supernatant was filtered with a bacteriological filter and centrifuged at 10,000 rpm for 30 min. With concentrated HCl, the pH of the cell-free supernatant was adjusted to 2.0, and the precipitate was incubated at 4°C overnight to form. The precipitate was collected and then re-suspended by centrifuging in 1 ml of 1 N NaOH to bring the pH to 7.0. The solution thus obtained was extracted twice with methanol. The methanolic extract was mixed with an equal volume of ethyl acetate and kept in a shaker at 120 rpm for 1 h. The ethyl acetate fraction was separated with a funnel and cooled in a rotary evaporator that turned at 80 rpm. The condensed form of the ethyl acetate fraction was dissolved in 1.5 ml of methanol: chloroform (1:1) and mixed thoroughly.
The agar diffusion method screened the collected microbial bioactive fraction of TPf12 against the FOL culture on the PDA medium. In the screening plates, four holes were bored 1 cm adjacent to the periphery at each 90° radius and filled with 1 μl of the collected fraction. Further solvent mixtures and sterile water were used for control plates, and inhibition zones were measured after 3 days of incubation at 28°C. After confirming the antifungal activity, the partially purified active fractions were analyzed and run on a PerkinElmer Clarus® 500 GC/MS with an AOC-20i auto-sampler system coupled with a column, Elite-1 (dimethyl polysiloxane 100%; 30 M × 0.25 mm × 0.25 μm) by following the flow rates of the carrier gas (1 ml/min), the split (10:1), and the injected load (2 μl). The column temperature was initially at 110°C for 2 min (hold), then increased to 200°C at a rate of 5°C/min for 9 min (hold). The injector temperature was 250°C, maintained for 36 min-the peak times of components in the chromatogram in GC and exposed to MS analysis. The MS electron impact (EI) energy was 70 eV, the Julet line was 200°C, and the source was at 200°C. EI mass scan (m/z) values between 45 and 450 aMU were recorded. The characters of compounds in the chromatogram were graphed in the TurboMass™ software platform and identified using database searches on the NIST Ver. 2.1 MS data library and a comparison of the spectra obtained through GC–MS.
The P. fluorescens liquid formulation (TPf12) was made according to the method (Manikandan et al., 2010). Thus, for the development of the liquid formulation of TPf12, 2% glycerol was added to the nutrient broth. The broth solution was cultured with 1 ml of TPf12 log-phase culture (3 × 101 CFU ml−1), and cells were cultured for 5 days at room temperature (25 ± 2°C). After that, the formulation was transferred into HDPE plastic jars and used in further tests. The tomato wilt pathogen FOL was placed in a Petri plate containing PDA. Four 6-mm sterile Whatman No. 40 filter paper disks were affixed 1 cm from the plate’s edge, encompassing the fungal disks on all four sides. Twenty-five microliters of the P. fluorescens (TPf12) formulation were dropped over filter paper disks. As a control, sterile distilled water was used instead of bacterial inoculum.
A glasshouse investigation was conducted at PL480, Tamil Nadu Agricultural University. In this investigation, the efficiency of TPf12 liquid formulation was compared to that of plants resulting from carbendazim seed treatment and soil drenching. The FOL strain was cultured in the sand: corn granule medium and inoculated 5% (w/w) with a sterilized potting mixture. Before sowing, the surface-sterilized seeds were treated with a TPf12 formulation at 1 ml/100 g for 10 min. The seeds were planted in pots with a diameter of 30 cm and filled with potting mix sterilized at 121°C at 15 psi for 2 h over 2 days, further soil drench at 5 ml/L/pot. Again, 20 days after the seeds were planted, 5 ml L−1 of the liquid formulation was added to the cropped soil. The inoculated control consisted of plants infected with FOL but not treated with TPf12 formulations and carbendazim. Three replications were performed. Each replication consisted of four pots, each of which contained three plants. Periodically, the plant growth, yield and incidence of wilt disease was observed, and the wilt incidence was recorded based on the percent disease incidence equation: number of infected plants/total number of plants × 100.
The following are treatment specifics: (1) healthy control (uninoculated), (2) soil inoculated with FOL pathogen alone, (3) seed treatment followed by soil drenching of (TPf12 liquid formulation) biocontrol alone, (4) pathogen and biocontrol agents subsequently (FOL + TPf12 liquid formulation). Each treatment was maintained with three replications. Observations about the symptom expression of wilt disease incidence were recorded in 20 days following the FOL pathogen alone. For proteome analysis, root samples were taken from both control and treated plants during the onset of symptoms in pathogen-inoculated control plants. The experiment was conducted twice with adequate repetitions.
Protein samples were extracted from each treatment (three times) separately and stored at −20°C. The frozen roots were ground to a powder with liquid nitrogen, then lysed in trichloroacetic acid (TCA), 10% in 2-propanone with 0.07% dithiothreitol (DTT) at −20°C for 1 h, followed by centrifugation at 12,000 rpm for 15 min. After 1 h in ice-cold 2-propanone containing 0.07% DTT at −20°C, pellets were centrifuged at 12,000 rpm per protocol (Kim and Swartz, 2004). This process was repeated three times until the supernatant became clear. The final deposited pellets were cooled and powdered in the freeze dryer to obtain the free residual fine powder. Further, sample buffer (200 μl; 6 M urea, 3 M thiourea, and 4% 3-[(3-cholamidopropyl) dimethylammonium]-1-propane sulfate (CHAPS), 1% DTT, 1% Bio-Rad ampholytes, and 35 mM Tris) was added to 10 mg of the powder. Protein was separated by incubating at 37°C with intermittent vortexing. After 1 h, the aqueous phase was separated by centrifuging at room temperature at 15000 rpm for 20 min, while the supernatant was frozen at −80°C for 2D-PAGE analysis. The protein quantitation was measured by Bradford protein assay.
The first-dimension separation was performed using linear immobiline® DryStrip gels as immobilized pH gradient (IPG) strips (17 cm, narrow range pH 4–7). The IPG dry strips were passively rehydrated in the reswelling tray for 12 h with 340 μl of rehydration buffer (8 M urea, 2% zwitter-ionic CHAPS, 0.2% DTT), and IPG buffer 0.5% (v/v) at pH 4.7–5.9, containing 150 μg of proteins. Isoelectric focusing (IEF) was conducted at 20°C using a GE Healthcare Multiphor II™ electrophoresis flatbed system. The operational conditions were 500 volts for 30 min, 1,000 volts for 30 min, and 3,500 volts for 11 h. The Immobiline DryStrip gel-focused strips were equilibrated twice for 15 min in each 10 ml of equilibration solution. Initial equilibration was performed in a solution containing 6 M urea, 30% (w/v) glycerol, 2% (w/v) SDS, 1% (w/v) DTT, and 50 mM Tris–HCl buffer with gentle shaking, and subsequently equilibrated in an equilibration solution where 4% (w/v) iodoacetamide (IAA) was used instead of DTT. Three replications were made to ensure reproducibility.
After IEF and equilibration, the proteins were separated by 30% SDS-PAGE gel in the second dimension. The equilibrated IPG strips were soaked with running buffer, then loaded on SDS-PAGE gel in the casted plates, encased with 2 ml of 0.5% (w/v) molten agarose solution at 5°C, and after solidification electrophoresed at an incessant current of 25 mA until the dye reached the bottom of the gel. Gels were removed from their gel cassettes and fixed in a 4:1:5 solution of methanol, acetic acid, and water. The gels were stained with silver using the technique described by Winkler et al. (Winkler et al., 2007). For each treatment, matching sets of replicate gels were produced and examined with similar equivalent sets from the other treatments. The entire match set consisted of 12 gels for a specific period.
A densitometric comparison of silver-stained gels was made using a flatbed scanner (ImageScanner III, GE Healthcare, USA). The data acquisition and protein spot analysis of replicate gel images were visualized and analyzed using ImageMaster™ 2D Platinum software 7.0 (GE Healthcare), a spot detection algorithm optimized to get relevant biological results. The abundance ratio is the volume of treated spots divided by the volume of control spots (treatments—FOL inoculated or FOL + P. fluorescens TPf12 interaction). The size and brightness of the protein spots were used to calculate the relative density. The molecular mass (Mr) of each protein was determined by co-electrophoresis of protein standard MW markers (14–94 kDa), and the isoelectric point (pI) was determined by the spot position along the IPG strip with a pH range of 4.7–5.8.
Over a pH range of 4–7, silver staining patterns were repeated reproducibly in 600–800 protein spots with molecular weights between 10 and 100 kDa. Among many dysregulated spots, 14 protein spots from gels that changed their abundance at least 2-fold according to a 0.05 level of significance (three replications) were excised using the procedure described by Prabhukarthikeyan et al. (Prabhukarthikeyan et al., 2022) and isolated peptides (1 μl) were treated with 1 μl of a matrix solution containing α-cyano-4-hydroxycinnamic acid in 50% acetonitrile, and 0.1% trifluoroacetic acid at a ratio of 1:2. Two microliters of the whole mixture were dropped onto a MALDI target plate and dried out. The sample plate was subjected to MALDI-TOF-MS analysis utilizing Bruker’s autoflex speed TOF/TOF MS/MS system (Bruker, US). After the mass-to-charge ratio analysis was done, the protein spectrum was retrieved using the Flexcontrol application (Shrimpex Biotech Pvt. Ltd., India).
MASCOT Distiller1 and Profound Expert System2 (Zhang and Chait, 2000) were used to searching the MALDI-TOF MS data against the databases of the Viridiplantae. The following parameters were permitted: taxonomy restrictions to Viridiplantae, trypsin with one missed cleavage, cleavage specificity, peptide acceptance of 100–300 ppm for fragment ions, permitted modifications, fixed Cys Carbamidomethyl (C), and variable oxidation of Met (M). The threshold Mowse score was calculated as -10Log(P), to evaluate the peptide mass fingerprinting. The functional categories of recognized proteins were given by linking their sequences to the universal database, NCBInr.
To provide preliminary information regarding the interaction between complexes. Molecular docking was used to analyze and screen compounds with multi-targeted proteins with higher binding affinity. The selected (S1–S14) differentially expressed proteins were chosen for molecular docking investigations. In the current investigation, the signal recognition particle receptor alpha subunit (SRP) was also used. Neither the crystal nor the NMR structure of the protein’s three-dimensional structure could be found. However, the Alphafold was used to get the model structures (predicted structures) based on the high-confidence model with more than 90 pLDDT values. The preprocessing steps were performed using the Maestro Platform’s Protein Preparation Wizard module, which assigned bond orders, added hydrogens, formed zero-order bonds to metals and di-sulfate bonds, changed seleno-methionines into methionines, and filled in any missing side chains. The OPLS4 force field was used to optimize the structure utilizing hydrogen bond optimization and energy minimization (Maestro, Schrödinger, LLC, New York, NY, USA).
The study focused on the secondary metabolites isolated from P. fluorescens, the chemicals represented in the three-dimensional structural data files (3D SDF) retrieved from the PubChem databases given in Supplementary Table S1. As part of the LigPrep module’s preparation, the structures were energy minimized using the OPLS4 force field, producing 32 distinct states of stereoisomeric and tautomeric entities. Schrödinger Release 2021–2: LigPrep, LigPrep, Schrödinger, LLC, New York, NY.
The Glide XP (extra precision) mode of Maestro’s Glide docking module was used to dock the target proteins and ligand molecules. The 2D interaction diagram of a ligand-protein complex molecule was generated using the ligand interaction module after the docked ligand, and protein interaction was analyzed using the XP pose viewer to find the optimistic pose. The resulting XP posture was further assessed to comprehend how ligand molecules interact with the target protein during the binding process (Leslie et al., 2021).
The statistical analyses were done using version v.92 of IRRISTAT, developed by the International Rice Research Institute in the Philippines. On the data, an Analysis of Variance (ANOVA) was performed. Prior to analysis, the percentages were arcsine transformed. The treatments were then compared using Duncan’s Multiple-Range Test (DMRT).
In the PDA, the FOL isolate produced a dense, white-to-pink mycelial growth, frequently with a purple hue. Microscopically, the FOL was identified based on macro-microconidia and chlamydospores characteristics. Microconidium is small, oval-shaped, single, or bicellular. Macroconidium is falcate or sickle-shaped, hyaline, multicellular, and contains four cells. The number of microconidia was greater than that of macroconidia. The macroconidia ranged in size from 25.0 μm to 38.7 μm and in width from 3.4 μm to 4.3 μm (Figures 1A–C). The FOL isolate inoculated on a tomato cv. PKM-1 developed typical wilt symptoms within 20 days after inoculation (Figures 2A, B). Infected vascular tissues exhibited a pinkish-reddish discoloration upon leaf drying, whereas similar symptoms did not appear in uninoculated pots. In order to confirm the FOL infection, the pathogen was reisolated and carefully examined. Primer pairs amplified the internal transcribed region (ITS) of the FOL isolate, ITS1/ITS4 at 560 bp, F. oxysporum-specific primers produced a 600 bp fragment, and race-specific primers of F. oxysporum amplified the predicted 450 bp fragment. The isolates belonged to FOL Race 1 (Figures 3A–C). In contrast, there was no amplification with the negative control.
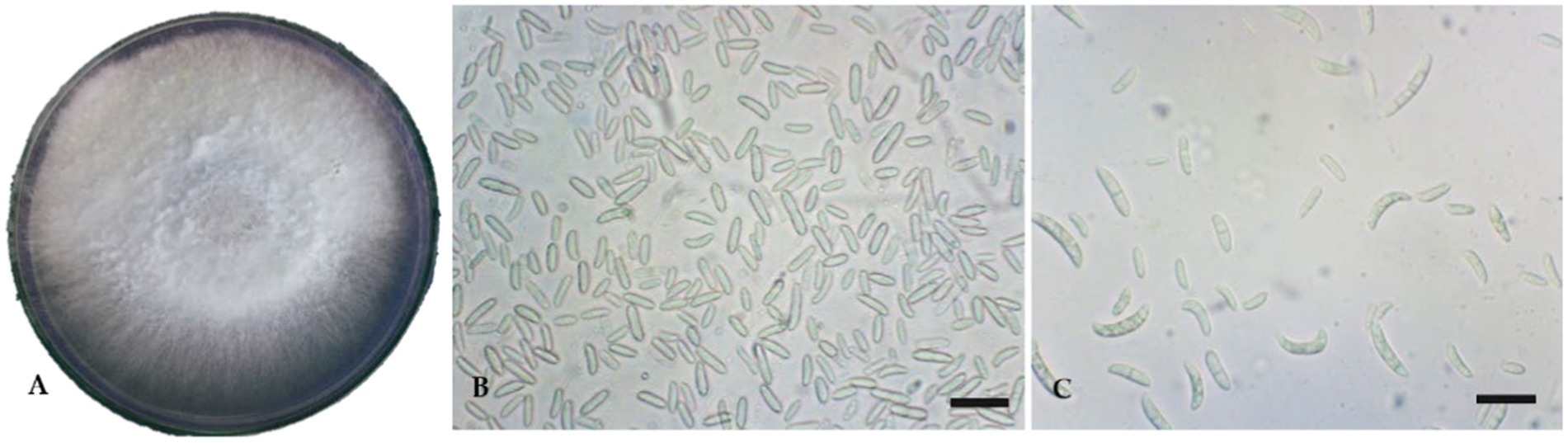
Figure 1. Cultural and micromorphological features of Fusarium oxysporum f. sp. lycopersici. Scale bars = 10 μm. (A) Culture plate; (B) Microconidia; (C) Macroconidia.
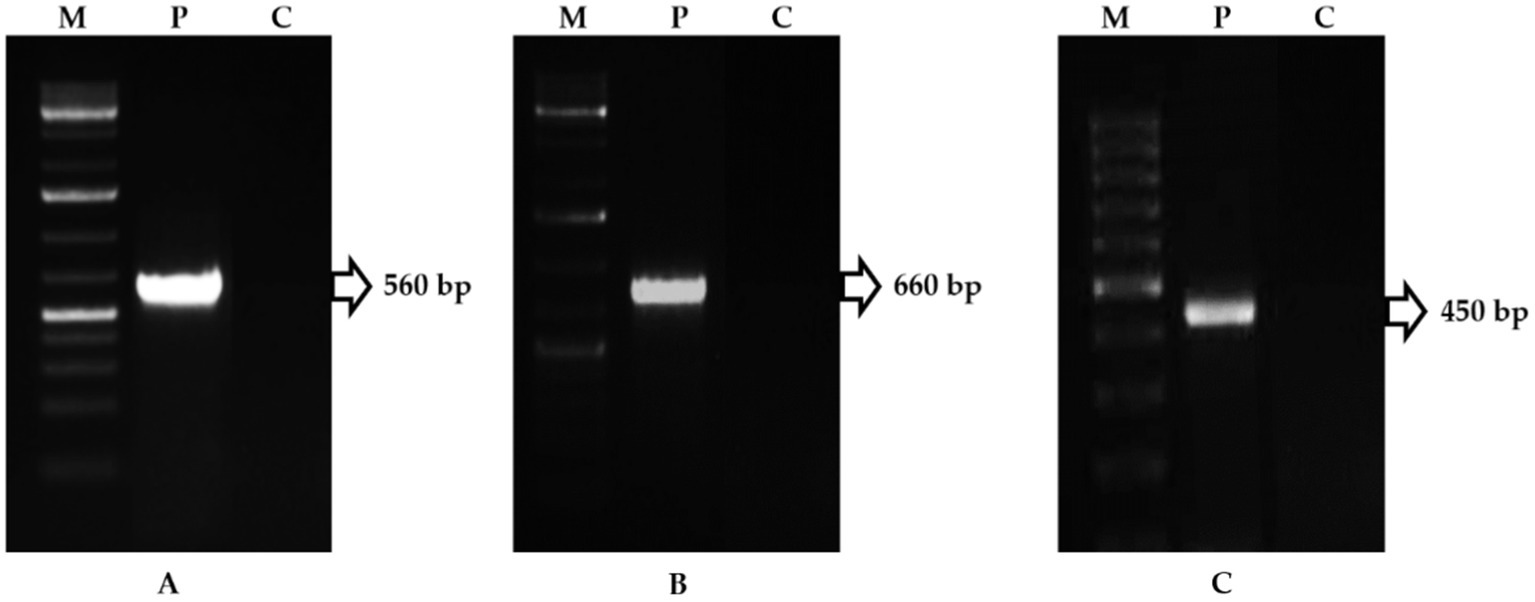
Figure 3. PCR amplification of Fusarium oxysporum f. sp. lycopersici genome (M-Marker, P-Pathogen, C-Control). (A) Amplicon of ITS1/ITS4 primers; (B) Amplicon of F. oxysporum-specific primers; (C) Amplicon of F. oxysporum-race-specific primers.
Pseudomonas fluorescens isolates were obtained from rhizosphere soils of tomato cultivated in Coimbatore, Villupuram, Pudukottai, and Tirupur districts. Among the P. fluorescens isolates tested for efficiency against FOL in vitro conditions, the isolate TPf12 inhibited the mycelia growth of FOL foremost (44.90%), followed by TPf8 (40.00%), TPf4 (35.90%), TPf10 (35.00%), and Pf1 (38.10%) was substantially more significant compared to control (Table 1).
The biochemical characters of isolates were screened using the following tests: citrate as a single source of carbon, urease to discern enzymatic activities, nitrate reduction to detect nitrate reductase, and the utilization of arabinose, glucose, lactose, lycine utilization, ornithine utilization, and phenylamine. Based on this, 10 tests were positive for the TPf12 isolate. All the isolates were determined to be P. fluorescens based on more than seven tests. On dark blue agar plates, P. fluorescens isolates TPf12, TPf8, TPf4, TPf10, and Pf1 exhibited a yellow halo around the streak, indicating siderophore synthesis (Figure 4A). According to the results, the intensity of siderophore synthesis was significant in the above-mentioned isolates. These five isolates were discovered to produce hydrogen cyanide (HCN), which induces plant resistance. The results demonstrated that TPf12 drastically altered the color of the filter paper from yellow to dark brown (Figure 4B). The ITS primers amplified a 560 bp portion from the 16S-23S rRNA intergenic region in 15 different Pseudomonas strains, confirming that these strains belonged to P. fluorescens (Figure 5A). Three of the 15 isolates shown for the presence of the pyrrolnitrin antibiotic gene, TPf11, TPf12, and TPf15, had an amplicon size of 786 bp (Figure 5B).
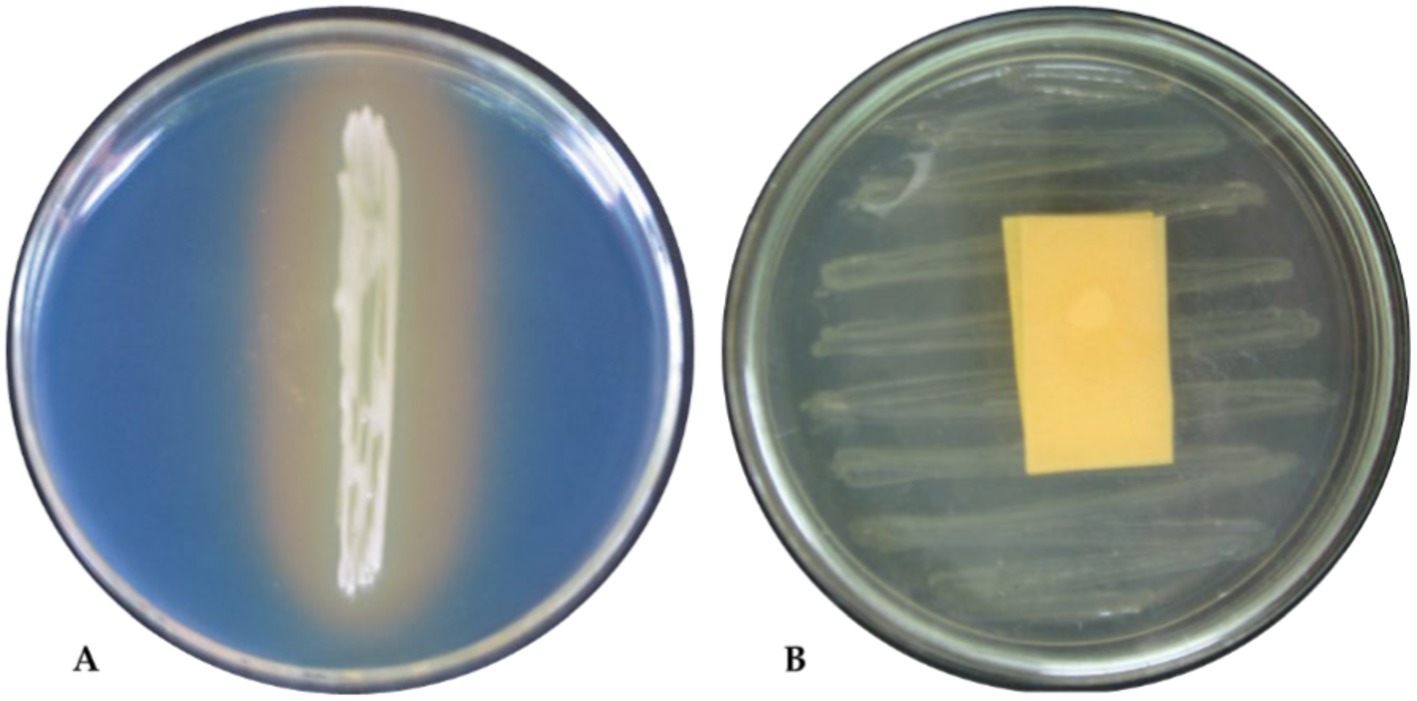
Figure 4. Biochemical tests of Pseudomonas fluorescens strain TPf12. (A) Siderophore test; (B) HCN test.
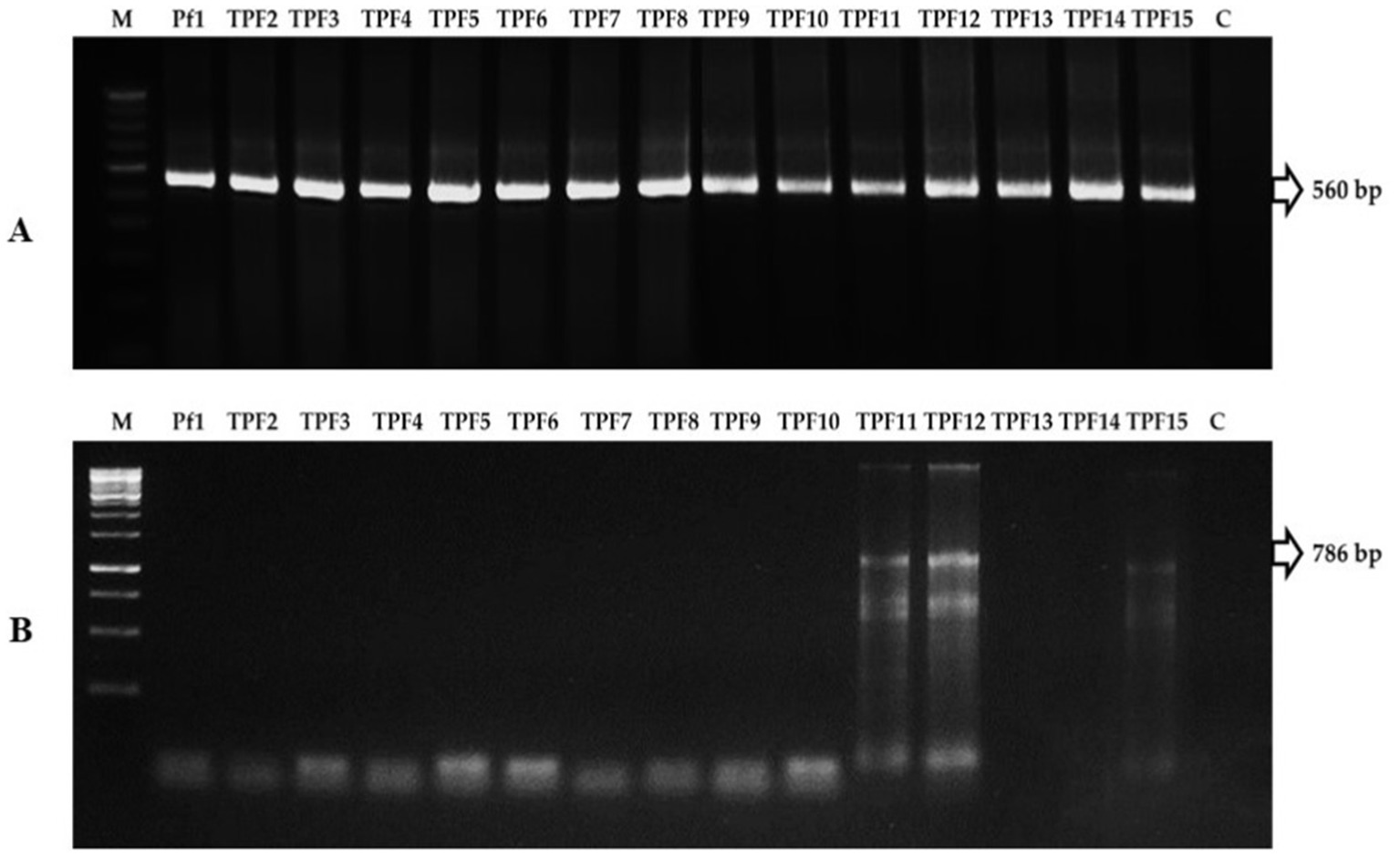
Figure 5. (A) PCR amplification of 16S-23S rRNA region in Pseudomonas fluorescens strains (M-Marker, Pf1-TPF15-strains, C-Control). (B) PCR amplification of pyrrolnitrin antibiotic gene region in P. fluorescens strains (M-Marker, Pf1-TPF15-strains, C-Control).
The efficacy of formulated TPf12 after 5 days was depicted as an inhibitory zone over the FOL fungal pathogen, and expansion of the bacterial colonies was recorded. In a greenhouse study, P. fluorescens TPf12 treatments exhibited no symptoms, whereas FOL pathogen-inoculated plants displayed wilt symptoms.
The purified crude antibiotic and extracellular antifungal compounds from P. fluorescens (TPf12) were analyzed through GC–MS and yielded 13 prominent peaks with a retention time of 3.63, 5.04, 8.91, 10.62, 10.59, 12.16, 13.65, 15.01, 18.56, 19.49, 20.21, 21.36, 21.74, 24.86, 26.69, 28.65, 29.32, 30.51, 31.30, 32.88, 33.72, 34.37, 36.60, 38.62, and 39.56 min, with peak area, and chemical compounds were predicted (Table 2).
The liquid formulation of TPf12 offered varying degrees of resistance against wilt disease. The formulation, applied three times from seed to pre-flowering, was considered superior but not better than carbendazim. Moreover, TPf12 liquid formulation treatments significantly reduced disease incidence compared to healthy plants. When the studied PGPR were applied as seed treatments and soil drenching (twice), strain TPf12 had the lowest disease incidence (18.6%), followed by seed treatments and pre-plant soil drenching, which had a disease incidence of 40%. Under greenhouse conditions, all of the TPf12 and carbendazim treatments led to more growth than the controls. Carbendazim increased seedling height marginally compared to control TPf12 by inhibiting disease more effectively. Carbendazim is effective in disease suppression, whereas TPf12 treatments are comparable to carbendazim in promoting plant productivity (Table 3).
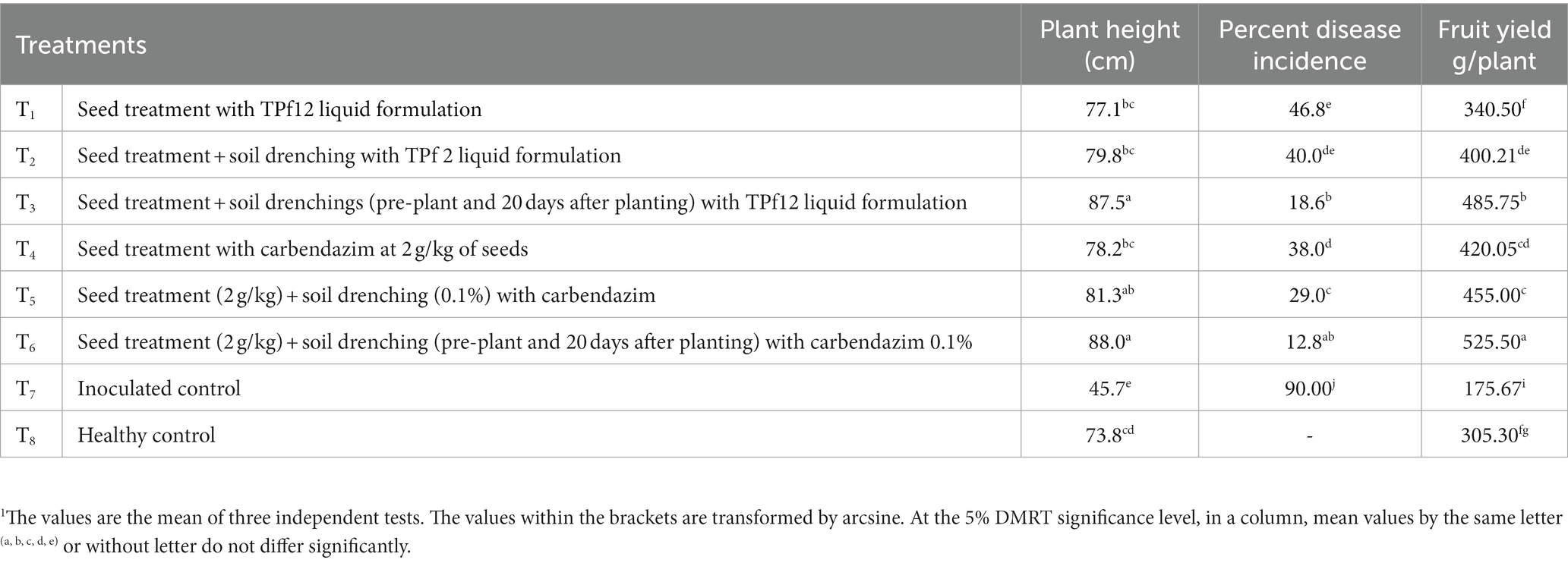
Table 3. Effect of TPf12 and carbendazim on wilt incidence and plant growth under greenhouse conditions.
The comparative root proteome analysis of tomato plants supplemented with a P. fluorescens (TPf12) liquid formulation and challenged with FOL revealed a substantial alteration in protein pattern, revealing the mechanism of resistance conferred by TPf12 against wilt disease. In all treatments, around 600–800 protein spots were expressed (Figures 6–9).
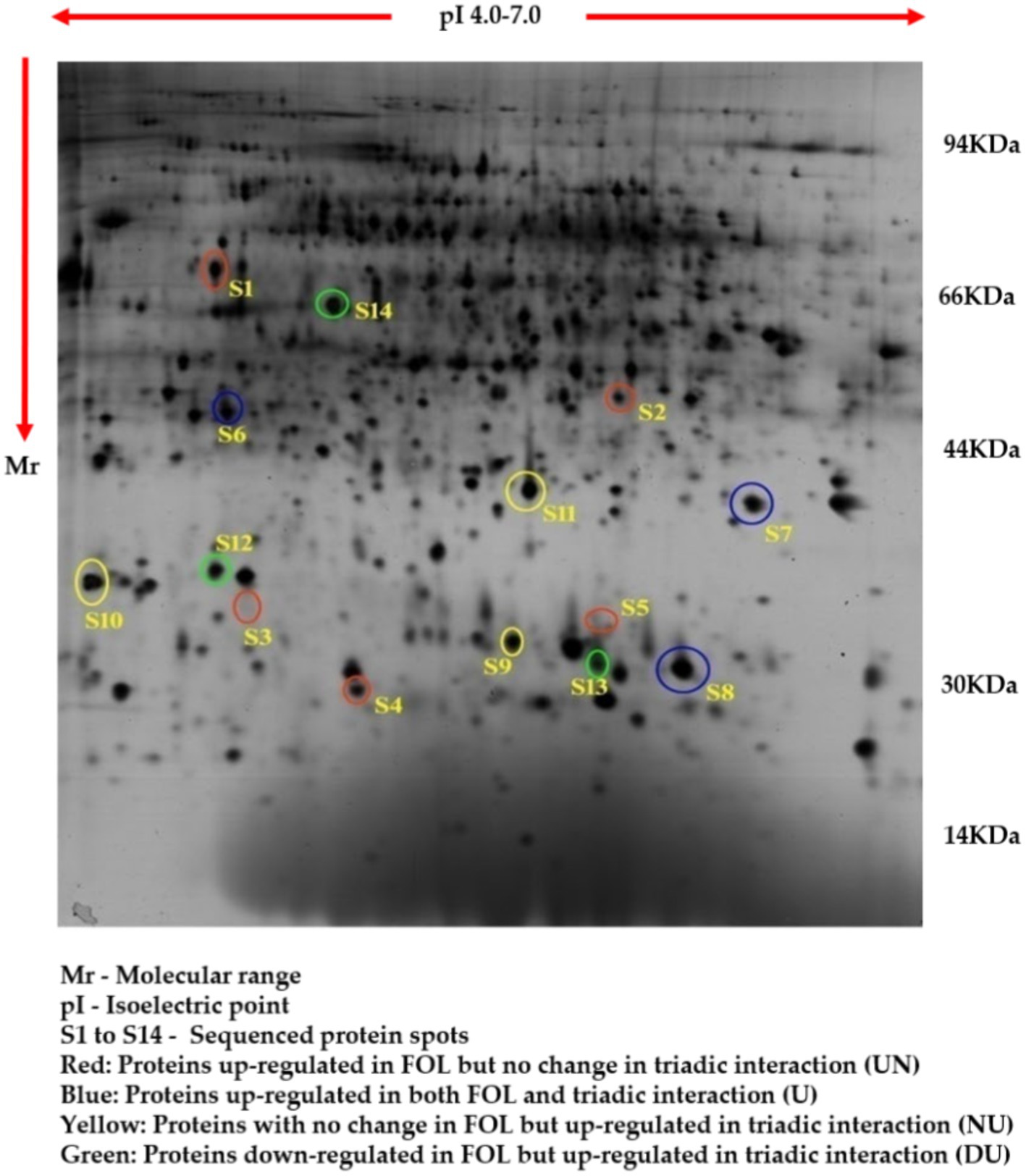
Figure 9. Proteome analysis of tomato roots during the triadic interactions of TPf1 biocontrol agents against FOL pathogen by 2D-PAGE.
Based on their differential regulation in terms of abundance ratio in the 2D-PAGE gels, 14 spots were classified into four classes; namely, Proteins up-regulated in FOL but no change in triadic interaction (UN), Proteins up-regulated in both FOL and triadic interaction (U), Proteins with no change in FOL but up-regulated in triadic interaction (NU), and Proteins down-regulated in FOL and but up-regulated in triadic interaction (DU). Five proteins (S1-S5) out of the 14 differentially expressed proteins were up-regulated in FOL-inoculated tomato roots but remained unchanged in interaction with FOL + TPf12 inoculated tomato roots. Three proteins (S6-S8) were significantly up-regulated in both pathogen inoculation and interaction treatments compared to the control. Three proteins, S9, S10, and S11, were up-regulated in interactions but not in the presence of the pathogen alone. Three proteins, S12, S13, and S14, were considerably downregulated in pathogen-treated plants and highly up-regulated in interaction treatments (Figures 10A–D).
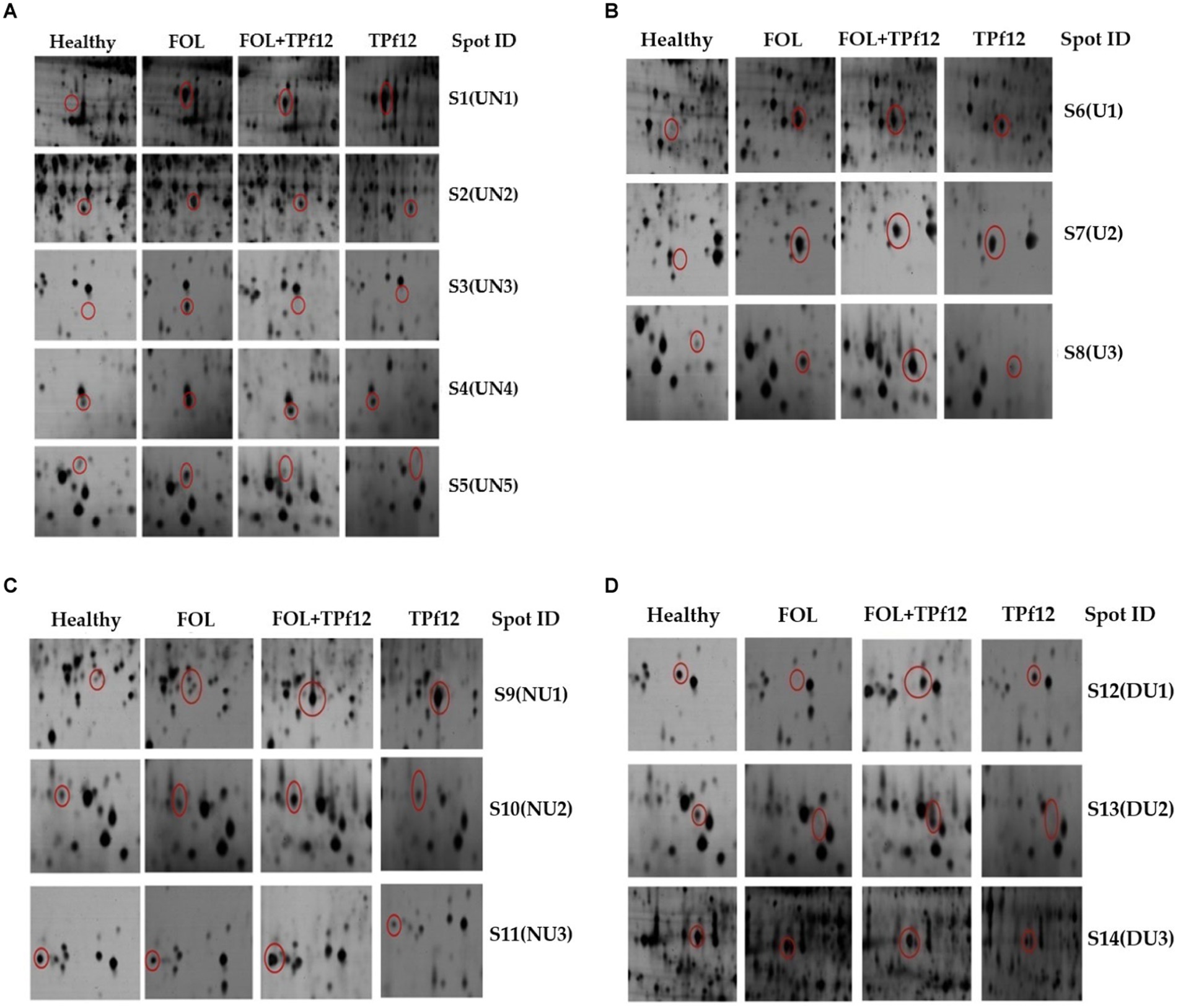
Figure 10. Protein expression patterns in tomato roots under different interactions by 2D-PAGE. (A) Upregulated in FOL and no change in triadic interaction (UN); (B) Upregulated in both FOL and triadic interaction. (C) No change in FOL and upregulated in triadic interaction (NU); (D) Downregulated in FOL and upregulated in triadic interaction.
The 14 differentially expressed functional proteins were extracted and recognized by MALDI-TOF/MS followed by a database examination using the Mascot software package and NCBInr. The 14 recognized proteins were TPA: jasmonate-induced protein, Delta DNA polymerase, Ribulose bisphosphate carboxylase, Chaperone protein, Cellulose synthase, a calcium-binding protein, GATA transcription factor, TPA: hypothetical protein, Nucleoside diphosphate kinase, Phenylalanine ammonia-lyase, Protein kinase, Ser/Thr protein kinase-like, lipid transfer-like protein, and Phenylalanine ammonia-lyase. The identities, NCBI accession numbers of homology proteins, scores, experimental and theoretical pI values, and molecular weights of all these recognized proteins are provided (Table 4).
The secondary metabolites from P. fluorescens were used in the molecular docking experiments against the target proteins chosen based on differentially expressed proteins. Differently expressed proteins (S1–S14) were used in the docking experiment as a GLIDE (ligand docking) module, and these proteins were normally prioritized in accordance with the docking score calculation in the form of G-Score. The lowest energy posture, or “binding confirmation,” indicates how well a docking method works. Multitargeted proteins’ G-score and GLIDE energy were noted. According to the docking complex, a hydrogen bond interaction typically occurs at a distance of roughly 3 Å.
The highest docking results were found in the protein of signal recognition particle receptor alpha subunit (SRP) against the compound of 3-o-(4-o-Beta-D-Galactopyranosyl-Beta-D-Galactopyraosyl)-2-Acetylamino-2-Deoxy-D-Galactose, and docking score is −9.704 kcal/mol. The amino acid interaction, bond length, and docking score of each differently expressed protein are listed in Table 5, further, 2-D and 3-D interaction map of metabolites are presented in Figures 11, 12.
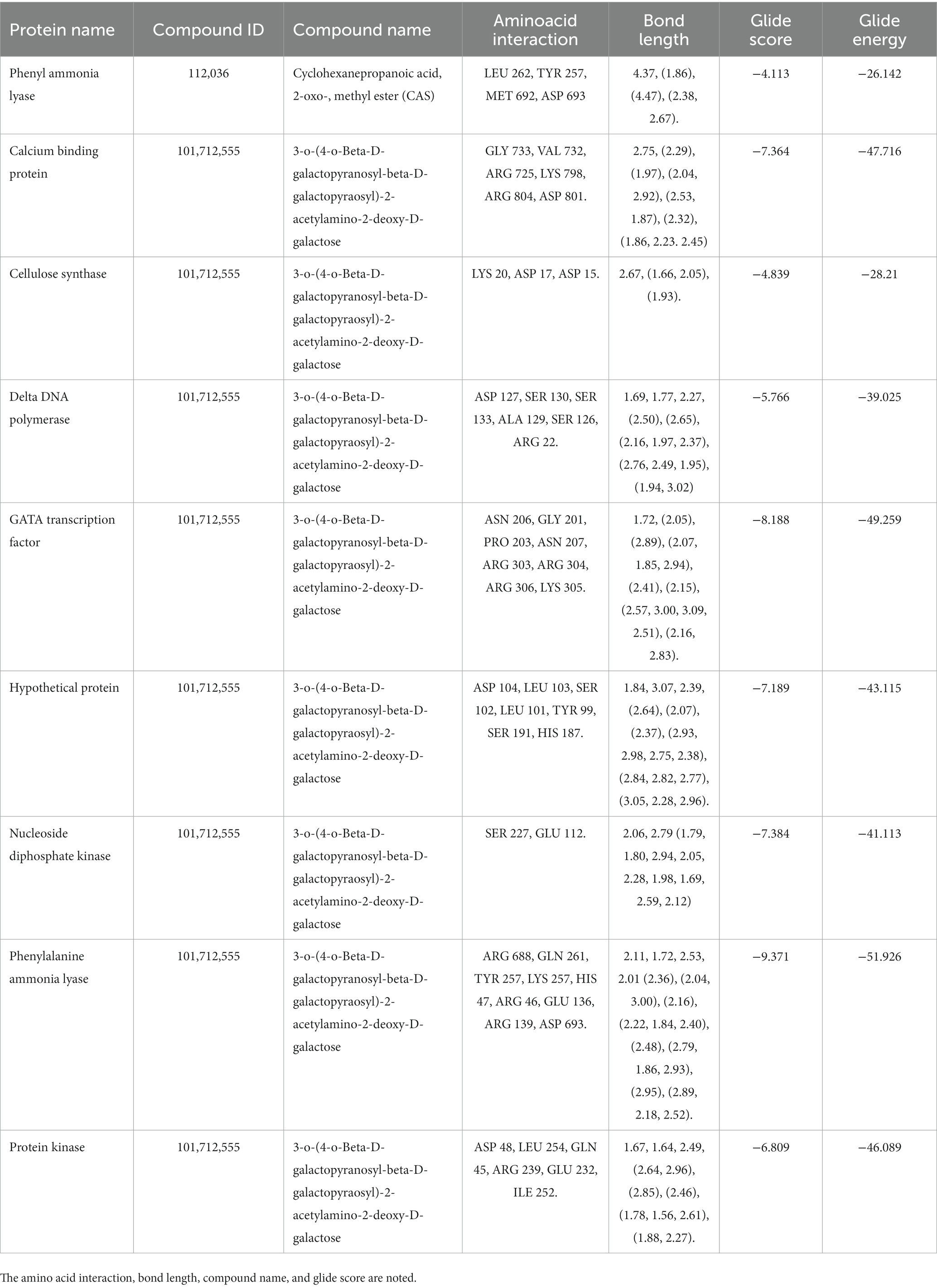
Table 5. The molecular docking experiment was carried out for the differently expressed proteins with secondary metabolite.
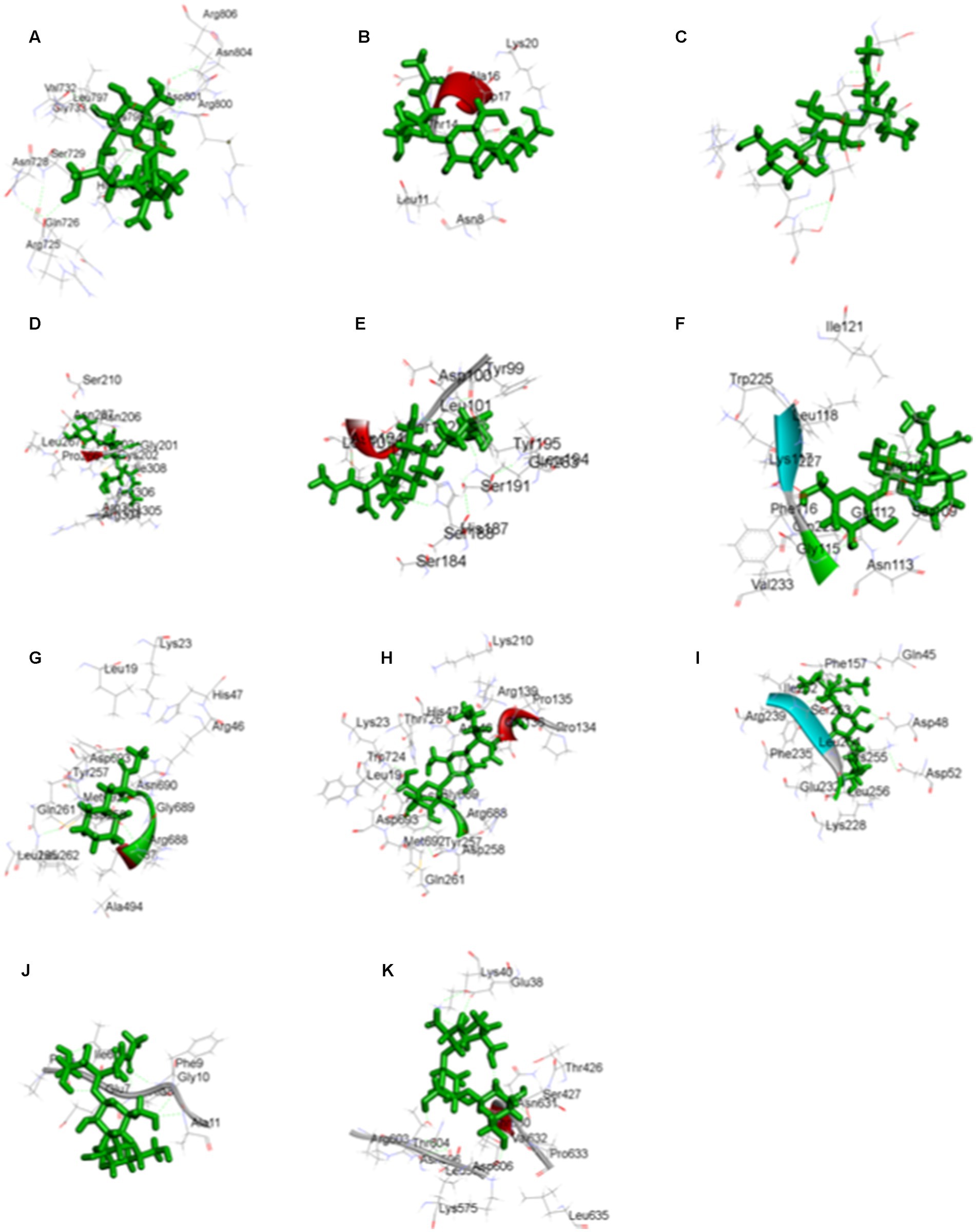
Figure 11. The three-dimensional interaction map of 3-o-(4-o-Beta-D-Galactopyranosyl-Beta-D-Galactopyraosyl) 2-Acetylamino-2-Deoxy-D-Galactose against (A) calcium binding protein, (B) Cellulose synthase, (C) Delta DNA Polymerase, (D) GATA Transcription factor, (E) hypothetical protein, (F) Nucleoside diphosphate kinase, (H) Phenylalanine ammonia lyase, (I) Protein kinase, (J) Ribulose biphosphate carboxylase, (K) SRP, and Cyclohexanepropanoic acid, 2-oxo-, methyl ester (CAS) with (G) Phenylalanine ammonia lyase.
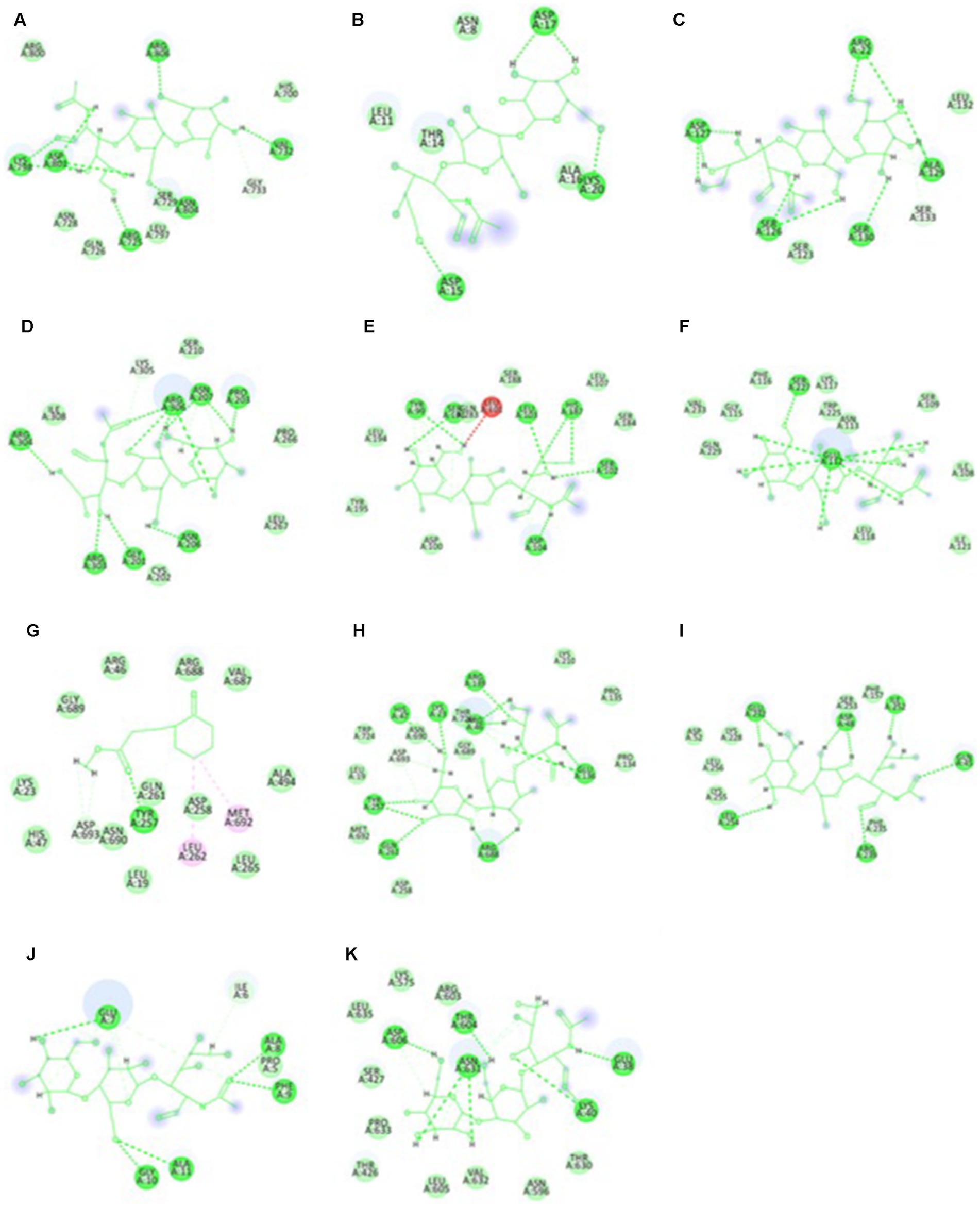
Figure 12. The two-dimensional interactional map of 3-o-(4-o-Beta-D-Galactopyranosyl-Beta-D-Galactopyraosyl) 2-Acetylamino-2-Deoxy-D-Galactose against (A) calcium-binding protein, (B) Cellulose synthase, (c) Delta DNA Polymerase, (D) GATA Transcription factor, (E) hypothetical protein, (F) Nucleoside diphosphate kinase, (H) Phenylalanine ammonia lyase, (I) Protein kinase, (J) Ribulose biphosphate carboxylase, (K) SRP, and Cyclohexanepropanoic acid, 2-oxo-, methyl ester (CAS) with (G) Phenylalanine ammonia lyase.
Since the late 1960s, the vascular wilt (fusariosis) of tomato has been observed in India. In 1988, maximum yield losses of 100 percent were recorded in the agro-climatic zones of Delhi, Maharashtra, and Tamil Nadu (Kapoor, 1988). Fusarium oxysporum f. sp. lycopersici causing vascular wilt disease, exists and is prevalent, wreaking havoc on tomato cultivation in Tamil Nadu (Manikandan et al., 2010). Early reports indicate that the recommended management strategies are less likely to be effective because the soil consists of numerous FOL pathotypes, and the infections are aggressive on the hosts (Manikandan et al., 2017). Based on the alarmingly widespread disease prevalence in the surveyed region, a wilted disease sample was collected from the Malumichampatti village of Tamil Nadu, observed the visible symptoms and vascular discoloration. The widespread incidence of wilt disease shows that it is a recurring issue, and all ruling varieties are prone to be vulnerable (Nirmaladevi et al., 2016). Due to the diversity and abundance of FOL in the cultivated soils in the study area, the pathogenicity assay confirmed the infectious nature of the collected sample by producing similar symptoms to those observed in the field as described and those reported in previous reports, compared to the control, which did not produce any symptoms. Effective management of plant diseases necessitates the timely detection and population diversity of pathogens in order to decide the suitable time, method and appropriate antagonists. Further, the phenotypic and genotypic characters in the study supported the confirmation of fungal species as FOL (Manikandan et al., 2018), specifically supported with the amplification of species-specific and race-specific primer pairs. The virulence profile of FOL isolates infecting tomato has been classified into three races based on their potential to infect a variety of cultivars with diverse resistance loci. Previously, race 1 type FOL was widely dispersed in India, necessitating the presence of the R gene in tomato plants for incompatibility reactions (Pritesh and Subramanian, 2013). In the present investigation, PCR amplification of FOL race 1 specific primers amplified a 450 bp fragment detected in all positive lanes, whereas no amplification was observed in control. Due to the variability in pathogenesis, it is customary for FOL to develop tactics to circumvent plant resistance (McGovern, 2015).
Chemical fungicides were examined extensively against FOL with inconclusive results (Amini and Dzhalilov, 2010); also, the pathogen may confer resistance to fungicidal molecules, which is a significant problem. Furthermore, using fungicides typically results in soil and tissue contamination hazards in tomato (Lopez-Lima et al., 2021). The inadequacy of management techniques for tomato wilt has resulted in substantial crop losses across the nation and significant economic losses for farmers (Nirmaladevi et al., 2016). Alternatively, biological control is an environmentally sustainable method for crop protection for suppressing pathogens in targeted areas, such as residential soil habitats, plant spheres, and infected host tissues, that disseminate, compete, and colonize. Despite this, interest in pathogen virulence, host response, and antagonistic effects of combating biocontrol agents has emerged due to increased pathogen populations and tomato farmers’ production concerns (Manikandan and Raguchander, 2014). It has been well-documented that P. fluorescens can be used in various formulations to manage crop diseases (Arya et al., 2018; Mohammed et al., 2019; Jayamohan et al., 2020; Singh et al., 2021). A bacterial biocontrol agent, P. fluorescens, has been chosen as a combating antagonist due to its size, rapid mobility towards the target, competitive saprophytic ability, siderophore, organic acids (Kamilova et al., 2006), HCN production in soil, plant spheres, and tissues, induced systemic resistance, plant growth promotion activity, and secreting antibiotics and enzymes for lysing the pathogen (de Lamo and Takken, 2020). In addition, the efficacy of the formulation and crop and soil-level adaptation, once applied, supported the study. Native P. fluorescens from tomato rhizosphere soils were isolated, screened against FOL isolate, and characterized based on biochemical tests and molecular methods, then comparatively screened with the ruling strain Pf1 against the FOL pathogen under lab conditions. In the dual plate screening assay, the native isolate TPf12 showed the highest inhibition to FOL, followed by TPf8, TPf4, and TPf10, compared to the commercialized strain Pf1. The biochemical characterization shows that the potential P. fluorescens isolates possess fundamental characteristics such as HCN, and siderophore synthesis. The potent isolates can produce antimicrobial compounds that enable them to fight plant diseases and stay alive in harsh environments. Also, biochemical properties and nutrition source consumption test results of various isolates positively indicated that these isolates belonged to P. fluorescens. It was further proven by amplifying ITS-1F and ITS-2R bacterial genome-specific primers with a 560 bp amplicon size, confirming it at the genus level. With its antagonistic action, three of the 15 P. fluorescens strains tested positive for pyrrolnitrin, including the strain TPf12, which showed promise in antagonist screening. The TPf12 liquid formulation worked well in the lab and greenhouse to suppress wilt disease by treating the seeds and drenching the soil. The antimicrobial secondary metabolites produced by plant-associated rhizobacteria are unknown to date. The present GC–MS study showed the presence of ketones, which have been reported as rich sources of bioactive volatiles from bacterial biocontrol agents and are effective against the growth and sporulation of Fusarium solani (Li et al., 2015) and F. oxysporum f. sp. cubense (Yuan et al., 2012), pathogens. These rhizobacterial pseudomonads produce secondary compounds that promote plant growth and increase systemic resistance by activating the cytokinin and ethylene signaling pathways (Li et al., 2015).
The latest developments in omics consists of improved methods for determining protein function and expression profiles. The functional proteins are necessary for pathogenicity in FOL during colonization and establishment for successful pathogen-host communication (Sun et al., 2004). Proteomic investigations characterized the expression of host leaf protein in response to fusaric acid interaction by FOL (Singh et al., 2017). Most proteomics studies on FOL have focused on two-way interactions, probably on FOL diversity in the soil (Manikandan et al., 2017), growth and reproduction, genomic integrity, and the infection rate potential (Manikandan et al., 2018), and virulence reactions, and mechanisms (Srinivas et al., 2019).
This study highlighted the differentially expressed proteins in tomato plants supplemented with a P. fluorescens TPf12 liquid formulation, and challenged inoculation with FOL, and untangled the resistance mechanism mediated by TPf12 in tomato against vascular wilt. The protein expression pattern of tomato roots subjected to four conditions (a healthy host, FOL inoculated, the interaction of FOL + TPf12 in the host, and the interaction of TPf12 with the host) were analyzed using a two-dimensional proteomic approach. The study revealed reproducible proteome differences between all possible host-pathogen and biocontrol interactions. During the triadic interaction, the proteome changed significantly in the tomato + FOL + TPf12 treatment compared to the tomato treated with FOL and the tomato treated with TPf12 liquid formulation. An earlier study on triadic interactions involving, bean plants, fungal pathogens (Botrytis cinerea, Rhizoctonia solani), and a biocontrol agent (Trichoderma atroviride), visualized the expressed a large number of protein factors associated with the three-player interactions (Marra et al., 2006). Examining the proteins directly responsible for cellular functions to comprehend the molecular biology and biochemistry of intrinsic plant-pathogen-biocontrol interactions is indispensable. Under field trials, instead of envisaging different dual interrelations (i.e., host-pathogen, host-biocontrol agent, or pathogen-biocontrol agent), a comprehensive framework of relationships involving multiple players is anticipated (Alfiky and Weisskopf, 2021). Thus, exploring and comprehending the triadic interactions between plants, antagonists, and pathogens is vital to understanding the pathway and developing a reliable product to accomplish field and market demands for bio-pesticides. Studying such triadic systems can reveal crucial crosstalk among divergent interacting partners that may not be evident in bipartite interactions.
The treatment with TPf12 alone and the combination treatment with FOL and TPf12-expressed proteins, which may have been involved in growth-promoting and defense-related activities, were evaluated. The pathogen-stimulated overexpression of proteins that may be involved in pathogenesis was observed in tomato plants. In the current study, 50% of proteins were abundant in TPf12 liquid formulation-mediated treatment, while only 25% were distinct in the pathogen-inoculated control. Among the 14 differentially expressed proteins, five proteins (S1–S5) were up-regulated in FOL inoculated, but no change was observed in the interaction of FOL + TPf12 inoculated in tomato roots. These five proteins were identified as TPA: jasmonate-induced protein (S1), Delta DNA polymerase (S2), Ribulose bisphosphate carboxylase (S3), Chaperone protein (S4), and Cellulose synthase (S4). The biosynthesis of JA from α-linolenic acid occurs via the octadecanoid pathway (Ninkovic et al., 2021) and is triggered by pathogen attack (Shen et al., 2018), wounding (Pandey et al., 2017), and osmotic stress (Tang et al., 2020). Pathogen-affected wheat showed expression of the non-host resistance-induced expressed sequence tag POLD2 during Blumeria graminis f. sp. hordei haustoria formation and HR. POLD2 encoding DNA polymerase delta subunit 2, is involved in DNA repair (Rezaei et al., 2022). Bipartite proteomics analyses of poplar inoculated with Botryosphaeria dothidea demonstrated upregulated expression of proteins in trees, specifically ribulose bisphosphate carboxylase small chain, which may be related to the constitutive mechanism of resistance.
Then, three spots (S6–S8) were discovered to be up-regulated in the roots of both FOL alone and FOL + TPf12-inoculated tomato plants. These three proteins exhibited similarities to the proteins listed below: Calcium-binding protein (S6), GATA transcription factor (S7), and TPA: hypothetical protein (S8). Calcium-binding protein, and GATA transcription factor are known to regulate the development of tissues by transcription in plants. It was discovered that the function of these proteins is involved in the regulation of intracellular Na + and K+ homeostasis and transcription factor activity. Calcium-binding secretion domain of Pseudomonas sp. is a potential pathway for nutrient uptake, bioactive metabolites, antimicrobial compounds, and ice recrystallization inhibition protein (Cid et al., 2018). The 2D-PAGE and mass spectrometry analyses of the chloroplast proteome in the diseased tomato leaves challenged with sulfated polysaccharide revealed the expression of GATA transcription factors under various physiological and pathological stress conditions for actively regulating chlorophyll biosynthesis (Mani et al., 2021).
Further, three proteins (S9–S11) were unchanged in FOL-inoculated but up-regulated in FOL + TPf12-treated tomato roots. These proteins have shown similarities with: Nucleoside diphosphate kinase (S9), phenylalanine ammonia-lyase (PAL; S10), and protein kinase family protein (S11). The functions of these proteins involve synthesizing nucleoside triphosphates other than ATP, activation of the phenylpropanoid pathway, and signaling for plant defense, development, growth, and hormone perception in response to pathogens. Nucleoside-diphosphate kinases (NDPKs, also known as NDP Kinases) are evolutionarily conserved biocatalysts ubiquitously found in living organisms ranging from bacteria to eukaryotes and are involved in the de novo syntheses of nucleotide triphosphates (Weng et al., 2021). Black scurf of potato by R. solani had a higher abundance of these enzymes during the virulence stage (Monazzah et al., 2022). A loss-of-function analysis revealed that NDK regulates sporulation and sclerotia production in Aspergillus flavus (Wang et al., 2019). Based on these findings, it is possible to conclude that a higher level of NDK expression in virulent isolates is responsible for the growth and virulence of the pathogen in host tissues. PAL is the initial enzyme in the phenylpropanoid biosynthesis pathway, which provides precursors for lignin, phenols, and salicylic acid. It is believed that the JA/ethylene signaling pathway of induced plant resistance activates the PAL gene. Induction of PAL by fluorescent pseudomonads was also reported in rice against blast (Magnaporthe oryzae; Suguna et al., 2020), tomato against wilt (F. oxysporum f. sp. lycopersici; Ramamoorthy et al., 2002), and mango against stem-end rot (Lasiodiplodia theobromae; Seethapathy et al., 2016). Plant-pathogen interactions in tomato may stimulate PAL activity (Vanitha et al., 2009; Song et al., 2011). The expression of protein kinase family proteins is widely studied during the interactions of biocontrol with several plant pathogens. The identified protein kinase family proteins may play a part in defense signal transmission from a local infection site to non-infected distal plant tissues and a persistent expression of acquired immunity (Hake and Romeis, 2019).
The last three proteins (S12-S14) were up-regulated in FOL + TPf12-treated tomato roots. This study identified three proteins with similarities to Ser/Thr protein kinase-like (S12), lipid transfer-like protein VAS-like (S13), and phenylalanine ammonia-lyase (S14). The function of these proteins was involved in resistance to biotic stress, defense response, disease resistance, and protein synthesis. A study demonstrated that the cotton cell wall-associated receptor-like kinases (WAKs) consisting of one or more Ser/Thr protein kinase-like are involved in defense against invading pathogens. However, their related signaling functions and regulatory mechanisms conversed with a DnaJ protein and were involved in defense against Verticillium dahliae (Feng et al., 2021). According to the review, the non-specific lipid transfer-like protein (LTP) is required for defense signaling by numerous chemical inducers of systemically acquired resistance in plants (Shine et al., 2019).
All of the differentially expressed proteins have diverse activities in plants, including plant growth promotion, nucleic acid replication, plant defense against stress, antioxidants, interacting with calcium ions and manganese, transcription, protein folding, and stabilization, involved in the pathway of plant cellulose biosynthesis, systemic resistance, involved in the regulatory mechanisms for restricting intracellular Na+ and K+ homeostasis, defense signaling, activation of the phenylpropanoid pathway, function in abiotic stress tolerance, and disease resistance. A comprehensive understanding or exposition of the mechanism of action is of great consequence since it may open new avenues for developing biological control agents against plant diseases. Therefore, molecular docking was carried out in the present study for the differently expressed proteins against secondary metabolites to understand the factors determining biocontrol activity. Among the secondary metabolites, the compounds Cyclohexanepropanoic acid, 2-oxo-, methyl ester (CAS), and 3-o-(4-o-Beta-D-Galactopyranosyl-Beta-D-Galactopyraosyl)-2-Acetylamino-2-Deoxy-D-Galactose were top-ranked with the least docking score against each differently expressed protein. These compounds were results above −4 kcal/mol of docking score. Proteomics, combined with other omics techniques, provides valuable information that can be used to comprehend the triadic interactions of plant-pathogen-biocontrol, the colonization process, the mechanism of suppression, and the upstream signaling of disease resistance.
So, even though genomic and transcriptomic studies of biopesticides have produced much data, the background scene still needs to be filled out. The proteomic approach compensates with a new perspective (Quirino et al., 2010). These insights, in turn, help select an effective biological agent and its mode of application for disease management strategies. They are possibilities for levering the induced host resistance and plant growth promotion to overcome the substantial crop losses.
The results indicated that P. fluorescens is an excellent producer of bioactive metabolites under in vitro conditions. Its application (seed treatment + soil drench) of the TPf12 liquid formulation effectively prevented vascular wilt disease in tomato. To our knowledge, this is the first attempt of triadic protein expression in tomato in response to F. oxysporum f. sp. solani infection under the antagonistic effects of P. fluorescens. We illustrate the importance of these studies for identifying and validating the biocontrol-mediated resistance in tomato and achieving sustainable plant disease management. This study dramatically impacts the ground level to reduce the long-term burden of wilt disease. It scientifically unveils the mechanism of disease resistance by the antagonist in the host. Proteomic analysis can provide broad and precise details on the “interaction proteomes” utilized by plants and microbes. Moreover, the enormity of the system, which necessitates the simultaneous actions of multiple players, indicates that omics tools and computational biology are required to visualize biocontrol agents’ interactions completely. Additional research is required to reveal the metabolomic interactions of TPf12 and FOL in tomato roots and comprehend the functional significance of bioactive metabolites in soil for pathogen suppression and plant growth promotion.
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding authors.
CG and PS: conceptualization. LP, PS, SS, DP, and AP: methodology. PS, AM, and DP: software. CG, SS, AP, DP, RC, AA, AD, KA, HE, and PS: validation. LP, SV, and PS: formal analysis. LP, PS, SV, and AM: investigation. PS, DP, and AP: resources. LP, PS, AP, DP, RC, AA, AD, KA, and HE: data curation. PS, SS, and DP: writing–original draft preparation. PS, RC, AA, AD, KA, HE, and DP: writing–review and editing. DP and PS: visualization. CG and SS: supervision. PS and DP: project administration. All authors contributed to the article and approved the submitted version.
This research was supported by King Saud University, Riyadh, Saudi Arabia (project number: RSP2023R118).
The authors would like to thank Researchers Supporting Project number (RSP2023R118), King Saud University, Riyadh, Saudi Arabia.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1157575/full#supplementary-material
Abd-Elsalam, K. A., Aly, I. N., Abdel-Satar, M. A., Khalil, M. S., and Verreet, J. A. (2003). PCR identification of fusarium genus based on nuclear ribosomal-DNA sequence data. Afr. J. Biotechnol. 2, 96–103. doi: 10.5897/AJB2003.000-1016
Alfiky, A., and Weisskopf, L. (2021). Deciphering trichoderma–plant–pathogen interactions for better development of biocontrol applications. J. Fungi 7, 1–18. doi: 10.3390/jof7010061
Alzandi, A. A., and Naguib, D. M. (2019). Pseudomonas fluorescens metabolites as biopriming agent for systemic resistance induction in tomato against fusarium wilt. Rhizosphere 11:100168. doi: 10.1016/j.rhisph.2019.100168
Amini, J., and Dzhalilov, F. S. (2010). The effects of fungicides on fusarium oxysporum F. SP. lycopersici associated with fusarium wilt of tomato. J. Plant Prot. Res. 50, 172–178. doi: 10.2478/v10045-010-0029-x
Arya, N., Rana, A., Rajwar, A., Sahgal, M., and Sharma, A. K. (2018). Biocontrol efficacy of Siderophore producing indigenous pseudomonas strains against fusarium wilt in tomato. Natl. Acad. Sci. Lett. 41, 133–136. doi: 10.1007/s40009-018-0630-5
Balogun, O. S., Hirano, Y., Teraoka, T., and Arie, T. (2008). PCR-based analysis of disease in tomato singly or mixed inoculated with fusarium oxysporum f. sp. lycopersid races 1 and 2. Phytopathol. Mediterr. 47, 50–60. doi: 10.14601/Phytopathol_Mediterr-2544
Campos, M. D., Félix, M. D. R., Patanita, M., Materatski, P., Albuquerque, A., Ribeiro, J. A., et al. (2022). Defense strategies: the role of transcription factors in tomato-pathogen interaction. Biol. (Basel) 11, 1–15. doi: 10.3390/biology11020235
Castillejo, M. Á., Bani, M., and Rubiales, D. (2015). Understanding pea resistance mechanisms in response to fusarium oxysporum through proteomic analysis. Phytochemistry 115, 44–58. doi: 10.1016/j.phytochem.2015.01.009
Cid, F. P., Maruyama, F., Murase, K., Graether, S. P., Larama, G., Bravo, L. A., et al. (2018). Draft genome sequences of bacteria isolated from the Deschampsia antarctica phyllosphere. Extremophiles 22, 537–552. doi: 10.1007/s00792-018-1015-x
de Lamo, F. J., and Takken, F. L. W. (2020). Biocontrol by fusarium oxysporum using endophyte-mediated resistance. Front. Plant Sci. 11, 1–15. doi: 10.3389/fpls.2020.00037
De Souza, J. T., and Raaijmakers, J. M. (2003). Polymorphisms within the prnD and pltC genes from pyrrolnitrin and pyoluteorin-producing pseudomonas and Burkholderia spp. FEMS Microbiol. Ecol. 43, 21–34. doi: 10.1016/S0168-6496(02)00414-2
Dean, R., Van Kan, J. A. L., Pretorius, Z. A., Hammond-Kosack, K. E., Di Pietro, A., Spanu, P. D., et al. (2012). The top 10 fungal pathogens in molecular plant pathology. Mol. Plant Pathol. 13, 414–430. doi: 10.1111/j.1364-3703.2011.00783.x
Dehghanian, S. Z., Abdollahi, M., Charehgani, H., and Niazi, A. (2020). Combined of salicylic acid and Pseudomonas fluorescens CHA0 on the expression of PR1 gene and control of Meloidogyne javanica in tomato. Biol. Control 141:104134. doi: 10.1016/j.biocontrol.2019.104134
Devindrappa, M., Kamra, A., Grover, M., and Gawade, B. (2022). Nematicidal rhizobacteria with plant growth-promoting traits associated with tomato in root-knot infested polyhouses. Egypt. J. Biol. Pest Control 32, 1–11. doi: 10.1186/s41938-022-00539-1
Edel-Hermann, V., and Lecomte, C. (2019). Current status of fusarium oxysporum formae speciales and races. Phytopathology 109, 512–530. doi: 10.1094/PHYTO-08-18-0320-RVW
Feng, H., Li, C., Zhou, J., Yuan, Y., Feng, Z., Shi, Y., et al. (2021). A cotton WAKL protein interacted with a DnaJ protein and was involved in defense against Verticillium dahliae. Int. J. Biol. Macromol. 167, 633–643. doi: 10.1016/j.ijbiomac.2020.11.191
Gobbin, D., Rezzonico, F., and Gessler, C. (2007). Quantification of the biocontrol agent Pseudomonas fluorescens Pf153 in soil using a quantitative competitive PCR assay unaffected by variability in cell lysis-and DNA-extraction efficiency. Soil Biol. Biochem. 39, 1609–1619. doi: 10.1016/j.soilbio.2007.01.015
Hake, K., and Romeis, T. (2019). Protein kinase-mediated signalling in priming: immune signal initiation, propagation, and establishment of long-term pathogen resistance in plants. Plant Cell Environ. 42, 904–917. doi: 10.1111/pce.13429
Hirano, Y., and Arie, T. (2006). PCR-based differentiation of fusarium oxysporum ff. Sp. lycopersici and radicis-lycopersici and races of F. oxysporum f. sp. lycopersici. J. Gen. Plant Pathol. 72, 273–283. doi: 10.1007/s10327-006-0287-7
Hu, X., Puri, K. D., Gurung, S., Klosterman, S. J., Wallis, C. M., Britton, M., et al. (2019). Proteome and metabolome analyses reveal differential responses in tomato-Verticillium dahliae-interactions. J. Proteome 207:103449. doi: 10.1016/j.jprot.2019.103449
Ignjatov, M., Milosevic, D., Nikolic, Z., Gvozdanovic-Varga, J., Jovicic, D., and Zdjelar, G. (2012). Fusarium oxysporum as causal agent of tomato wilt and fruit rot. Pestic. i fitomedicina 27, 25–31. doi: 10.2298/pif1201025i
Jayamohan, N. S., Patil, S. V., and Kumudini, B. S. (2020). Seed priming with pseudomonas putida isolated from rhizosphere triggers innate resistance against fusarium wilt in tomato through pathogenesis-related protein activation and phenylpropanoid pathway. Pedosphere 30, 651–660. doi: 10.1016/S1002-0160(20)60027-3
Jayaraj, J., and Radhakrishnan, N. V. (2008). Enhanced activity of introduced biocontrol agents in solarized soils and its implications on the integrated control of tomato damping-off caused by Pythium spp. Plant Soil 304, 189–197. doi: 10.1007/s11104-008-9539-y
Kamilova, F., Kravchenko, L. V., Shaposhnikov, A. I., Makarova, N., and Lugtenberg, B. (2006). Effects of the tomato pathogen fusarium oxysporum f. sp. radicis-lycopersici and of the biocontrol bacterium Pseudomonas fluorescens WCS365 on the composition of organic acids and sugars in tomato root exudate. Mol. Plant-Microbe Interact. 19, 1121–1126. doi: 10.1094/MPMI-19-1121
Kapoor, I. J. (1988). Fungi involved in tomato wilt syndrome in Delhi. Maharashtra Tamil Nadu. Indian Phytopathol. 41, 208–213.
Kim, D. M., and Swartz, J. R. (2004). Efficient production of a bioactive, multiple disulfide-bonded protein. Biotechnol. Bioeng. 85, 122–129. Available at. doi: 10.1002/bit.10865
Leslie, V. A., Mohammed Alarjani, K., Malaisamy, A., and Balasubramanian, B. (2021). Bacteriocin producing microbes with bactericidal activity against multidrug resistant pathogens. J. Infect. Publ. Health 14, 1802–1809. doi: 10.1016/j.jiph.2021.09.029
Li, X. Y., Mao, Z. C., Wu, Y. X., Ho, H. H., and He, Y. Q. (2015). Comprehensive volatile organic compounds profiling of bacillus species with biocontrol properties by head space solid phase microextraction with gas chromatography-mass spectrometry. Biocontrol Sci. Tech. 25, 132–143. doi: 10.1080/09583157.2014.960809
Liu, W., Liu, K., Chen, D., Zhang, Z., Li, B., El-Mogy, M. M., et al. (2022). Solanum lycopersicum, a model Plant for the Studies in developmental biology. Stress Biol. Food Sci. Foods 11:2402. doi: 10.3390/foods11162402
Lopez-Lima, D., Mtz-Enriquez, A. I., Carrión, G., Basurto-Cereceda, S., and Pariona, N. (2021). The bifunctional role of copper nanoparticles in tomato: effective treatment for fusarium wilt and plant growth promoter. Sci. Hortic. (Amsterdam). 277:109810. doi: 10.1016/j.scienta.2020.109810
Mani, S. D., Govindan, M., Muthamilarasan, M., and Nagarathnam, R. (2021). A sulfated polysaccharide κ-carrageenan induced antioxidant defense and proteomic changes in chloroplast against leaf spot disease of tomato. J. Appl. Phycol. 33, 2667–2681. doi: 10.1007/s10811-021-02432-0
Manikandan, R., Harish, S., Karthikeyan, G., and Raguchander, T. (2018). Comparative proteomic analysis of different isolates of fusarium oxysporum f.sp. lycopersici to exploit the differentially expressed proteins responsible for virulence on tomato plants. Front. Microbiol. 9, 1–13. doi: 10.3389/fmicb.2018.00420
Manikandan, R., Karthikeyan, G., and Raguchander, T. (2017). Soil proteomics for exploitation of microbial diversity in fusarium wilt infected and healthy rhizosphere soils of tomato. Physiol. Mol. Plant Pathol. 100, 185–193. doi: 10.1016/j.pmpp.2017.10.001
Manikandan, R., and Raguchander, T. (2014). Fusarium oxysporum f. sp. lycopersici retardation through induction of defensive response in tomato plants using a liquid formulation of Pseudomonas fluorescens (Pf1). Eur. J. Plant Pathol. 140, 469–480. doi: 10.1007/s10658-014-0481-y
Manikandan, R., Saravanakumar, D., Rajendran, L., Raguchander, T., and Samiyappan, R. (2010). Standardization of liquid formulation of Pseudomonas fluorescens Pf1 for its efficacy against fusarium wilt of tomato. Biol. Control 54, 83–89. doi: 10.1016/j.biocontrol.2010.04.004
Marra, R., Ambrosino, P., Carbone, V., Vinale, F., Woo, S. L., Ruocco, M., et al. (2006). Study of the three-way interaction between Trichoderma atroviride, plant and fungal pathogens by using a proteomic approach. Curr. Genet. 50, 307–321. doi: 10.1007/s00294-006-0091-0
Mazzeo, M. F., Cacace, G., Ferriello, F., Puopolo, G., Zoina, A., Ercolano, M. R., et al. (2014). Proteomic investigation of response to forl infection in tomato roots. Plant Physiol. Biochem. 74, 42–49. doi: 10.1016/j.plaphy.2013.10.031
McGovern, R. J. (2015). Management of tomato diseases caused by fusarium oxysporum. Crop Prot. 73, 78–92. doi: 10.1016/j.cropro.2015.02.021
Mishra, S., Jagadeesh, K. S., Krishnaraj, P. U., and Prem, S. (2014). Biocontrol of tomato leaf curl virus (ToLCV) in tomato with chitosan supplemented formulations of pseudomonas sp. under field conditions. Aust. J. Crop. Sci. 8, 347–355.
Mohammed, B. L., Hussein, R. A., and Toama, F. N. (2019). Biological control of fusarium wilt in tomato by endophytic rhizobactria. Energy Procedia 157, 171–179. doi: 10.1016/j.egypro.2018.11.178
Monazzah, M., Nasr Esfahani, M., and Tahmasebi Enferadi, S. (2022). Genetic structure and proteomic analysis associated in potato to Rhizoctonia solani AG-3PT-stem canker and black scurf. Physiol. Mol. Plant Pathol. 122:101905. doi: 10.1016/j.pmpp.2022.101905
Mousa, M. A. A., Abo-Elyousr, K. A. M., Abdel Alal, A. M. K., and Alshareef, N. O. (2021). Management fusarium wilt disease in tomato by combinations of bacillus amyloliquefaciens and peppermint oil. Agronomy 11, 1–13. doi: 10.3390/agronomy11122536
Ninkovic, V., Markovic, D., and Rensing, M. (2021). Plant volatiles as cues and signals in plant communication. Plant Cell Environ. 44, 1030–1043. doi: 10.1111/pce.13910
Nirmaladevi, D., Venkataramana, M., Srivastava, R. K., Uppalapati, S. R., Gupta, V. K., Yli-Mattila, T., et al. (2016). Molecular phylogeny, pathogenicity and toxigenicity of fusarium oxysporum f. sp. lycopersici. Sci. Rep. 6, 1–14. doi: 10.1038/srep21367
Ortoneda, M., Guarro, J., Madrid, M. P., Caracuel, Z., Roncero, M. I. G., Mayayo, E., et al. (2004). Fusarium oxysporum as a multihost model for the genetic dissection of fungal virulence in plants and mammals. Infect. Immun. 72, 1760–1766. doi: 10.1128/IAI.72.3.1760-1766.2004
Pandey, S. P., Srivastava, S., Goel, R., Lakhwani, D., Singh, P., Asif, M. H., et al. (2017). Simulated herbivory in chickpea causes rapid changes in defense pathways and hormonal transcription networks of JA/ethylene/GA/auxin within minutes of wounding. Sci. Rep. 7, 1–14. doi: 10.1038/srep44729
Panno, S., Davino, S., Caruso, A. G., Bertacca, S., Crnogorac, A., Mandić, A., et al. (2021). A review of the most common and economically important diseases that undermine the cultivation of tomato crop in the mediterranean basin. Agronomy 11, 1–45. doi: 10.3390/agronomy11112188
Pareja-Jaime, Y., Roncero, M. I. G., and Ruiz-Roldán, M. C. (2008). Tomatinase from fusarium oxysporum f. sp. lycopersici is required for full virulence on tomato plants. Mol. Plant-Microbe Interact. 21, 728–736. doi: 10.1094/MPMI-21-6-0728
Prabhukarthikeyan, S. R., Karthikeyan, G., and Raguchander, T. (2015). Biochemical characterization of fluorescent pseudomonads from turmeric rhizosphere. Biochem. Cell. Arch. 15, 299–303.
Prabhukarthikeyan, S. R., Manikandan, R., Durgadevi, D., Keerthana, U., Harish, S., Karthikeyan, G., et al. (2017). Bio-suppression of turmeric rhizome rot disease and understanding the molecular basis of tripartite interaction among Curcuma longa, Pythium aphanidermatum and Pseudomonas fluorescens. Biol. Control 111, 23–31. doi: 10.1016/j.biocontrol.2017.05.003
Prabhukarthikeyan, S. R., Parameswaran, C., Sawant, S. B., Naveenkumar, R., Mahanty, A., Keerthana, U., et al. (2022). Comparative proteomic analysis of Rhizoctonia solani isolates identifies the differentially expressed proteins with roles in virulence. J. Fungi 8, 1–18. doi: 10.3390/jof8040370
Pritesh, P., and Subramanian, R. B. (2013). Isolation of NBS-LRR class resistant gene (I2 gene) from tomato cultivar Heamsona. Afr. J. Biotechnol. 12, 6076–6078. doi: 10.5897/ajb2013.12194
Quinet, M., Angosto, T., Yuste-Lisbona, F. J., Blanchard-Gros, R., Bigot, S., Martinez, J. P., et al. (2019). Tomato fruit development and metabolism. Front. Plant Sci. 10, 1–23. doi: 10.3389/fpls.2019.01554
Quirino, B. F., Candido, E. S., Campos, P. F., Franco, O. L., and Krüger, R. H. (2010). Proteomic approaches to study plant-pathogen interactions. Phytochemistry 71, 351–362. doi: 10.1016/j.phytochem.2009.11.005
Ramamoorthy, V., Raguchander, T., and Samiyappan, R. (2002). Induction of defense-related proteins in tomato roots treated with Pseudomonas fluorescens Pf1 and fusarium oxysporum f. sp. lycopersici. Plant Soil 239, 55–68. doi: 10.1023/A:1014904815352
Ramesh Kumar, N., Thirumalai Arasu, V., and Gunasekaran, P. (2002). Genotyping of antifungal compounds producing plant growth-promoting rhizobacteria, Pseudomonas fluorescens. Curr. Sci. 82, 1463–1466.
Rezaei, A., Mahdian, S., Hashemi-Petroudi, S. H., Goodwin, P. H., Babaeizad, V., and Rahimian, H. (2022). Nonhost resistance EST profiling of wheat interacting with Blumeria graminis f. sp. hordei identifies genes for durable resistance to powdery mildew. Eur. J. Plant Pathol. 162, 793–806. doi: 10.1007/s10658-021-02416-3
Rothan, C., Diouf, I., and Causse, M. (2019). Trait discovery and editing in tomato. Plant J. 97, 73–90. doi: 10.1111/tpj.14152
Salman, M., and Abuamsha, R. (2012). Potential for integrated biological and chemical control of damping-off disease caused by Pythium ultimum in tomato. Biol. Control 57, 711–718. doi: 10.1007/s10526-012-9444-4
Sasirekha, B., and Srividya, S. (2016). Siderophore production by Pseudomonas aeruginosa FP6, a biocontrol strain for Rhizoctonia solani and Colletotrichum gloeosporioides causing diseases in chilli. Agric. Nat. Resour. 50, 250–256. doi: 10.1016/j.anres.2016.02.003
Seethapathy, P., Gurudevan, T., Subramanian, K. S., and Kuppusamy, P. (2016). Bacterial antagonists and hexanal-induced systemic resistance of mango fruits against lasiodiplodia theobromae causing stem-end rot. J. Plant Interact. 11, 158–166. doi: 10.1080/17429145.2016.1252068
Shen, Q., Liu, Y., and Naqvi, N. I. (2018). Fungal effectors at the crossroads of phytohormone signaling. Curr. Opin. Microbiol. 46, 1–6. doi: 10.1016/j.mib.2018.01.006
Shine, M. B., Xiao, X., Kachroo, P., and Kachroo, A. (2019). Signaling mechanisms underlying systemic acquired resistance to microbial pathogens. Plant Sci. 279, 81–86. doi: 10.1016/j.plantsci.2018.01.001
Singh, S., Balodi, R., Meena, P. N., and Singhal, S. (2021). Biocontrol activity of Trichoderma harzianum, Bacillus subtilis and Pseudomonas fluorescens against Meloidogyne incognita Fusarium oxysporum and Rhizoctonia solani. Indian Phytopathol. 74, 703–714. doi: 10.1007/s42360-021-00368-6
Singh, V. K., Singh, H. B., and Upadhyay, R. S. (2017). Role of fusaric acid in the development of ‘fusarium wilt’ symptoms in tomato: physiological, biochemical and proteomic perspectives. Plant Physiol. Biochem. 118, 320–332. doi: 10.1016/j.plaphy.2017.06.028
Song, W., Ma, X., Tan, H., and Zhou, J. (2011). Abscisic acid enhances resistance to Alternaria solani in tomato seedlings. Plant Physiol. Biochem. 49, 693–700. doi: 10.1016/j.plaphy.2011.03.018
Srinivas, C., Nirmala Devi, D., Narasimha Murthy, K., Mohan, C. D., Lakshmeesha, T. R., Singh, B. P., et al. (2019). Fusarium oxysporum f. sp. lycopersici causal agent of vascular wilt disease of tomato: biology to diversity– a review. Saudi J. Biol. Sci. 26, 1315–1324. doi: 10.1016/j.sjbs.2019.06.002
Suguna, S., Parthasarathy, S., and Karthikeyan, G. (2020). Induction of systemic resistant molecules in Phylloplane of Rice plants against Magnaporthe oryzae by Pseudomonas fluorescens. Int. Res. J. Pure Appl. Chem. 21, 25–36. doi: 10.9734/irjpac/2020/v21i330158
Sun, T. K., Sang, G. K., Du, H. H., Sun, Y. K., Han, J. K., Byung, H. L., et al. (2004). Proteomic analysis of pathogen-responsive proteins from rice leaves induced by rice blast fungus, Magnaporthe grisea. Proteomics 4, 3569–3578. doi: 10.1002/pmic.200400999
Suresh, P., Shanmugaiah, V., Rajagopal, R., Muthusamy, K., and Ramamoorthy, V. (2022). Pseudomonas fluorescens VSMKU3054 mediated induced systemic resistance in tomato against Ralstonia solanacearum. Physiol. Mol. Plant Pathol. 119:101836. doi: 10.1016/j.pmpp.2022.101836
Tang, G., Ma, J., Hause, B., Nick, P., and Riemann, M. (2020). Jasmonate is required for the response to osmotic stress in rice. Environ. Exp. Bot. 175:104047. doi: 10.1016/j.envexpbot.2020.104047
Vanitha, S. C., Niranjana, S. R., and Umesha, S. (2009). Role of phenylalanine ammonia lyase and polyphenol oxidase in host resistance to bacterial wilt of tomato. J. Phytopathol. 157, 552–557. doi: 10.1111/j.1439-0434.2008.01526.x
Vitale, A., Rocco, M., Arena, S., Giuffrida, F., Cassaniti, C., Scaloni, A., et al. (2014). Tomato susceptibility to fusarium crown and root rot: effect of grafting combination and proteomic analysis of tolerance expression in the rootstock. Plant Physiol. Biochem. 83, 207–216. doi: 10.1016/j.plaphy.2014.08.006
Wang, Y., Wang, S., Nie, X., Yang, K., Xu, P., Wang, X., et al. (2019). Molecular and structural basis of nucleoside diphosphate kinase–mediated regulation of spore and sclerotia development in the fungus aspergillus flavus. J. Biol. Chem. 294, 12415–12431. doi: 10.1074/jbc.RA119.007505
Weng, Q., Zhao, Y., Yanan, Z., Song, X., Yuan, J., and Liu, Y. (2021). Identification of salt stress-responsive proteins in maize (Zea may) seedlings using itraq-based proteomic technique. Iran. J. Biotechnol. 19, e2512–e2120. doi: 10.30498/IJB.2021.2512
Winkler, C., Denker, K., Wortelkamp, S., and Sickmann, A. (2007). Silver-and Coomassie-staining protocols: detection limits and compatibility with ESI MS. Electrophoresis 28, 2095–2099. doi: 10.1002/elps.200600670
Yuan, J., Raza, W., Shen, Q., and Huang, Q. (2012). Antifungal activity of bacillus amyloliquefaciens NJN-6 volatile compounds against fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 78, 5942–5944. doi: 10.1128/AEM.01357-12
Zhang, W., and Chait, B. T. (2000). ProFound: An Expert System for Protein. Anal. Chem. 72, 2482–2489. doi: 10.1021/ac991363o
Zhang, S., Griffiths, J. S., Marchand, G., Bernards, M. A., and Wang, A. (2022). Tomato brown rugose fruit virus: an emerging and rapidly spreading plant RNA virus that threatens tomato production worldwide. Mol. Plant Pathol. 23, 1262–1277. doi: 10.1111/mpp.13229
Keywords: proteomics, tomato, fusarium wilt disease, triadic-interaction, defense response, resistance mechanism, fluorescent pseudomonads
Citation: Paranthaman L, Seethapathy P, Pandita D, Gopalakrishnan C, Sankaralingam S, Venkatesh S, Malaisamy A, Pandita A, Casini R, Alataway A, Dewidar AZ, Almutairi KF and Elansary HO (2023) Levering proteomic analysis of Pseudomonas fluorescens mediated resistance responses in tomato during pathogenicity of Fusarium oxysporum f. sp. oxysporum. Front. Sustain. Food Syst. 7:1157575. doi: 10.3389/fsufs.2023.1157575
Received: 02 February 2023; Accepted: 07 April 2023;
Published: 27 April 2023.
Edited by:
Surendra K. Dara, Oregon State University, United StatesReviewed by:
Yazhong Jin, Heilongjiang Bayi Agricultural University, ChinaCopyright © 2023 Paranthaman, Seethapathy, Pandita, Gopalakrishnan, Sankaralingam, Venkatesh, Malaisamy, Pandita, Casini, Alataway, Dewidar, Almutairi and Elansary. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Deepu Pandita, ZGVlcHVwYW5kaXRhQGdtYWlsLmNvbQ==; Hosam O. Elansary, aGVsYW5zYXJ5QGtzdS5lZHUuc2E=
†These authors have contributed equally to this work and share first authorship
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.