- 1Tim Taylor Department of Chemical Engineering, Kansas State University, Manhattan, KS, United States
- 2Department of Ecology and Evolutionary Biology, University of Kansas, Lawrence, KS, United States
- 3Department of Agronomy, Kansas State University, Manhattan, KS, United States
- 4Division of Biology, Kansas State University, Manhattan, KS, United States
Plant growth-promoting bacteria (PGPB) are valuable for supporting sustainable food production and may alleviate the negative impacts of chemical fertilizers on human health and the environment. While single-strain inoculations have proven unreliable due to poor survival and colonization in the rhizosphere, application of PGPB in multispecies consortia has the potential to improve these outcomes. Here, we describe a new approach for screening and identifying bacterial consortia that improve the growth of corn relative to plants inoculated with a single strain. The method uses the microwell recovery array (MRA), a microfabricated high-throughput screening device, to rapidly explore the maize (Zea mays L.) rhizobiome for higher-order combinations of bacteria that promote the growth and colonization of the nitrogen-fixing PGPB, Azospirillum brasilense. The device simultaneously generates thousands of random, unique combinations of bacteria that include A. brasilense and members of the maize rhizobiome, then tracks A. brasilense growth in each combination during co-culture. Bacteria that show the highest levels of A. brasilense growth promotion are then recovered from the device using a patterned light extraction technique and are identified. With this approach, the screen uncovered growth-promoting consortia consisting primarily of bacteria from the Acinetobacter-Enterobacter-Serratia genera, which were then co-inoculated with A. brasilense on axenic maize seedlings that were monitored inside a plant growth chamber. Compared to maize plants inoculated with A. brasilense alone, plants that were co-inoculated with these consortia showed accelerated growth after 15 days. Follow-up root colonization assays revealed that A. brasilense colonized at higher levels on roots from the co-inoculated seedlings. These findings demonstrate a new method for rapid bioprospecting of root and soil communities for complementary PGPB and for developing multispecies consortia with potential use as next-generation biofertilizers.
1. Introduction
Increasing crop production and enhancing plant health in a sustainable manner is critical in the face of climate change (Farooq et al., 2022), increasing population (Komarek and Msangi, 2019), reduction of cultivable lands (Maranguit et al., 2017), and pest or pathogen mediated diseases in crops (Leisner et al., 2022). While chemical fertilizers and pesticides remain necessary for enhancing food production, their indiscriminate use has proven unsustainable and negatively impacts human and environmental health (Chaudhary et al., 2022). Much effort has focused on exploiting plant growth-promoting bacteria (PGPB) as bioinoculants, i.e., biofertilizers, to enhance plant growth and address the limitations of chemical approaches (Mitter et al., 2021; Bhat et al., 2022). Biofertilizers offer a low-energy, environmentally friendly, and sustainable approach to promoting plant health and increasing biomass production. PGPB can fix nitrogen (Aasfar et al., 2021), produce phytohormones (Egamberdieva et al., 2017), stimulate root development (Gowtham et al., 2022), provide pathogen defense (Zhu et al., 2022), and alleviate environmental and human-induced stresses (Ma et al., 2019; Dodds et al., 2020). However, PGPB are often unreliable and pose a high economic risk amongst agricultural producers, limiting their broad adoption (Nakkeeran et al., 2006; Tabassum et al., 2017).
Currently, the biofertilizer market is dominated by diazotrophic PGPB, such as Rhizobium spp., Bradyrhizobium spp., Actinorhizobium spp., Azotobacter spp., and Azospirillum spp. (Stamenković et al., 2018; Pedrosa et al., 2020). Azospirillum brasilense is one of the most widely adopted diazotrophs, and it displays versatile C- and N-metabolism. It also promotes plant growth through additional mechanisms, including phytohormone production (Alzate Zuluaga et al., 2022), development of stress tolerance (Pedrosa et al., 2020), siderophore production (Ferreira et al., 2019; Timofeeva et al., 2022), phosphate solubilization (Bargaz et al., 2021), and phytopathogen inhibition (Viejobueno et al., 2021). With this array of benefits, significant effort has been given to understanding the microbial interactions that influence the association and colonization of A. brasilense and other diazotrophic bacteria with non-leguminous maize (Zea mays) crops (Dent and Cocking, 2017; Oliveira et al., 2018; Van Deynze et al., 2018). Prior studies have also shown that co-inoculation of A. brasilense with complementary organisms such as Bradyrhizobium (Wang et al., 2018; dos Santos Lima Fagotti et al., 2019) and cyanobacteria isolates (Hungria et al., 2010; Verma et al., 2011; Ferreira et al., 2013; Bashan et al., 2014) improves maize yield. This suggests that A. brasilense applied in a symbiotic, multi-species consortium may be key for its improvement as a biofertilizer. However, this requires isolating and identify new collections of complementary bacteria that are also well-adapted to the root environment.
As crop roots harbor diverse collections of beneficial bacteria (Philippot et al., 2013), recent bioprospecting efforts have targeted the rhizosphere and endosphere to uncover new PGPB strains (Mitter et al., 2021). For example, Aloo et al. (2021) recently prospected the potato rhizosphere to uncover isolates with multiple plant growth promoting traits, including bacteria from the Klebsiella, Serratia, Enterobacter and Citrobacter genus (2021). Jochum et al. (2019) screened the rhizosphere of perennial grass in semi-arid regions to uncover Bacillus and Enterobacter strains that improved the drought tolerance of wheat by modifying root architecture (2019). With respect to maize, Ikeda et al. (2020) uncovered Cellulosimicrobium, Bacillus, Stenotrophomonas, and Enterobacter strains from the maize root which were found to promote maize growth in greenhouse studies, albeit in a genotype-specific manner (2020). These studies demonstrate that crop roots are a rich resource of bacteria that have potential application as biofertilizers, and motivate screening the maize rhizosphere for bacteria that are interactive and complementary toward A. brasilense.
Uncovering favorable PGPB interactions is a daunting task due to the high degree of species diversity in root communities, which depends on soil type/conditioning, irrigation, and climate at different agricultural sites (Lee et al., 2020). Such a task necessitates the application of novel high-throughput screening technologies that can rapidly explore interactions in a microbiome and accelerate the pace of discovery. Here, we apply the microwell recovery array (MRA) (Barua et al., 2021) to rapidly search the maize (Zea mays L.) rhizobiome for collections of root-associated bacteria that improve A. brasilense growth. Subsequent experiments investigate the impact of these consortia on A. brasilense's root colonization and the resulting plant growth. The results demonstrate the unique ability of the MRA as a bioprospecting tool that generates combinations of microbes from root or soil samples that have positive effects on maize growth. The technique has the potential be extended to other plants and offers a new approach to screening plant rhizosphere for multispecies consortia that are potentially useful as biofertilizers.
2. Materials and methods
2.1. Preparation of Azosprillium brasilense strain Sp7-GFP and of Zea mays rhizobacteria
All bacteria strains and plasmids used in this research are listed in Supplementary Table 1. A. brasilense Sp7-GFP, herein referred to as Sp7, was received from the Alexandre lab (University of Tennessee, Knoxville) and was stored in 25% glycerol at −80 °C. Sterile inoculation loops were used to pick cells from the frozen stocks for culture in R2A broth media (pH: 7.2 ± 0.2, Teknova) in sterile test tubes supplemented with ampicillin (100 μg/mL) and tetracycline (5 μg/mL) for 24 hrs (28°C, 215 rpm). For collection of rhizosphere-associated maize (Zea mays) microbes, soil and root samples were first collected from a conservation tillage maize field located near Glen Eder, Kansas (Field coordinate: 39.3533616 and −98.362541, 1147 m above sea level). The growth stage at the time of sampling was VT (beginning of flowering). Soil samples from the site were sent to K-State Soil Testing Lab for a complete analysis of carbon, nitrogen, phosphorous content, and soil pH. The comprehensive analysis of the soil samples is listed in Supplementary Table 2. A total of 4 sets of soil and root samples were collected from maize plants, immediately stored on ice, transferred to the laboratory, and stored overnight at 4°C. 200 g of each root sample were washed with 200 mL of sterile, ice-cold 1X phosphate buffer saline (PBS buffer: 8 g/L NaCl, 0.2 g/L KCL, 0.2 g/L KH2PO4,1.15 g/L Na2HPO4, pH 7) for 20 min (Machado and Colli Mull, 2017). The resulting suspensions were filtered using 0.8 μm sterile filters and centrifuged at 4,400 rpm for 20 min to collect pellets composed of bacteria harvested from maize roots. Extracted cells from each root sample were combined, resuspended in 25% glycerol, and stored at −80°C.
2.2. Media selection
Microbes extracted from maize roots were inoculated from glycerol stocks and incubated in TY Media (10 g/L Bacto-tryptone, 10 g/L NaCl, 5 g/L Yeast Extract, pH: 7 ± 0.2), R2A media (0.50 g/L Yeast extract, 0.50 g/L Proteose Peptone, 0.50 g/L Casamino acids, 0.50 g/L Glucose, 0.50 g/L Soluble starch, 0.30 g/L Na-pyruvate, 0.30 g/L K2HPO4, 0.05 g/L MgSO4.7H2O, pH: 7 ± 0.2), or LB media (10 g/L Tryptone, 10 g/L NaCl, 5 g/L Yeast Extract, pH: 7 ± 0.2), in sterile test tubes for 24 h (28°C, 215 rpm). After incubation, cells grown in each culture media were stored at −80°C in 25% glycerol before the 16S amplicon sequencing used to identify the media that recovered the highest diversity of maize root-associated bacteria (described in Section 2.3).
2.3. 16S rRNA gene community sequencing of maize root isolates
Purified gDNA samples of cells directly extracted from maize roots (Section 2.1) or from cells after culture in TY, R2A, and LB media (Section 2.2) were obtained from the respective glycerol stocks. E.Z.N.A soil DNA kit (Omega Bio-Tek, Norcross, GA) and DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) were used for DNA extraction. gDNA was diluted to 20 ng/μL in 100 μL aliquots and sent to Integrated Genomics Facility (Department of Plant Pathology, Kansas State University, Manhattan, KS) for library preparation using Nextera XT index Kit v2 (Illumina, Inc., San Diego, CA) and Illumina MiSeq sequencing (2 x 300 bp PE) of the hypervariable V4 region of the 16S rRNA gene (MiSeq platform, PE-300bp format). The following Earth Microbiome Project primers were used to generate amplicons: 515F: 5'-GTGCCAGCMGCCGCGGTAA-3' and 806R: 5'- GGACTACHVGGGTWTCTAAT-3' (Caporaso et al., 2011). Cutadapt was used to remove adapter and primer sequences from raw reads (Martin, 2011), and reads with any ambiguous bases or more than 0.15 expected errors were discarded. The trimmed 16S-v4 reads were further quality-filtered, truncated, de-noised, and merged using DADA2 software (Callahan et al., 2016) to generate an amplicon sequence variant (ASV) table. Bimeric ASVs were identified using the consensus method and removed. ASVs were identified using the Naïve Bayesian Classifier (Wang et al., 2007) trained on the RDP trainset v16. The ASV table and taxonomic assignments were imported into R v.4.1.2 and analyzed using the tidyverse and phyloseq packages (McMurdie and Holmes, 2013; Wickham et al., 2019). After discarding ASVs that could not be confidently identified as either bacteria or archaea, the dataset included 499 ASVs with a mean of 58,962 reads per sample (standard deviation = 12,835).
2.4. MRA design, fabrication, and preparation
MRAs were fabricated by following the protocols described by Barua et al. (2021). Each array was divided into a 7 × 7 grid of sub-arrays, consisting of a 15 × 15 microwell arrays of 10 μm diameter, 20 μm depth, and 30 μm pitch. A total of 11,025 microwells were etched on a single 3-inch diameter N-type silicon wafer (University Wafers), coated with 1 μm thick layer of Parylene N (PDS 2010 Labcoater, Specialty Coating Systems), using standard photolithography techniques described in previous publications (Masigol et al., 2017, 2018). Cells were next seeded into the MRA devices. R2A was selected as the media for use in MRA operation, as this media was found to recover the most diverse collections of bacteria (Section 2.3, Supplementary Figures 1, 2). A. brasilense Sp7 and microbes extracted from maize roots were first cultured in R2A media to mid-log phase then resuspended in R2A to an OD600 of 0.2. Next, bacteria cells were inoculated in microwell arrays using protocols described previously (Barua et al., 2021). Based on prior MRA screens, we estimate this generated inoculum densities of ~40 cells per well, where wells are inoculated with a unique combinations of bacteria species (Hansen et al., 2016; Barua et al., 2021). For co-culture studies, cultures of Sp7 (OD600 = 0.2) and root-extracted microbes were mixed at a ratio of 1:1 to reach a final OD600 of 0.2. 700 μL of each cell suspension was seeded on top of microwell arrays for 1 h at room temperature. Then, the substrates were dried, followed by the parylene lift-off process to remove cells attached to the array's background regions (Hansen et al., 2016). A polyethylene glycol (PEG) photodegradable membrane was attached on top of the MRA seeded with cells. This membrane attachment step has been previously described in detail (van der Vlies et al., 2019; Fattahi et al., 2021), a summary of the protocol is also provided in the Supplementary material.
2.5. Time-lapse fluorescent microscopy and image analysis
Time-lapse fluorescent microscopy (TLFM) images were acquired with a Nikon Eclipse Ti-U inverted microscope equipped with a 20× objective, a motorized XYZ stage, a humidified live-cell incubation chamber (Tokai Hit), a DS-QiMc monochromatic digital camera, and NIS Elements image acquisition software. The prepared microwell arrays were placed in a custom 3D printed scaffold in order to keep them submerged under liquid media while imaging. The design of the scaffold was described previously (Barua et al., 2021). The scaffold was then placed inside a humidified live-cell incubation chamber at 28°C for co-culture and imaging. A FITC filter was used to image Sp7 (20×, 200 ms, 17.1× gain) with a neutral density filter and with 25% standard light intensity to ensure imaging without photobleaching. Brightfield images were also taken at each section of the array after fluorescent imaging. Images of the microwell arrays (3,600 microwells total) were taken every 60 min during culture. GFP fluorescent images from Sp7 were analyzed following the protocol described by Timm et al. (2017). The Protein Array Analyzer tool in ImageJ was used to generate growth profiles for each microwell to identify the top 2 wells with the highest growth levels for extraction. The time-lapse fluorescent images were imported as image sequences corrected by subtracting darkfield images from illumination field images with the image calculator plugin. Then image backgrounds were removed by selecting a 125-radius sliding paraboloid, and illumination correction was performed using the calculator plus plugin. Finally, the growth of each strain in the microwells was calculated using the ImageJ “Micro Array” plugin (Timm et al., 2017).
2.6. Recovery and storage of isolates from microwell arrays
Cells were extracted from microwell arrays using a Polygon400 patterned illumination tool (Mightex Systems, Pleasanton, CA) following previously described procedures (van der Vlies et al., 2019). The two promoter wells (A, B) showing the greatest final Sp7 growth levels after 24 h of co-culture were opened by exposing the well perimeter to 365 nm UV light in a ring pattern area with a 10 μm inner diameter and a 20 μm outer diameter as described in Fattahi et al. (2021). The extraction process was done sequentially for two promoter wells, the opened wells were then washed from the well using wash buffer (0.05% Tween in R2A media) into a ~ 200 mL volume, and cell suspensions were plated onto agar-R2A for overnight culture (28°C) and recovery of individual colonies. Five colonies from the two wells with unique colony morphologies were sampled from the plates, these colonies were cultured in R2A media and finally stored in 25% glycerol stocks. These consortia were labeled as promoting consortia 1-5 (PC1, PC2, PC3, PC4, and PC5).
2.7. Validation using 96-well plates
It was previously demonstrated that microbe-microbe interactions observed in the MRA microwells can be recapitulated in a 96-well plate validation assay (Barua et al., 2021). This approach was applied here to validate the growth enhancement of A. brasilense in co-culture with consortia PC1-PC5. To obtain cell-free culture fluid (CFCF), R2A liquid media was inoculated with each individual consortium directly from glycerol stocks using a sterile loop, and each sample was cultured (28°C, 300 rpm) in 2mL of media overnight. Cells were removed from the media by centrifugation (2000g, 10 min). To obtain conditioned media, the CFCF was then mixed with Sp7 in R2A media at a 1:1 volumetric ratio to reach an initial OD600 value of 0.2 (final volume = 100 μL), at which point growth was quantified with a Biotek Epoch 2 Multi-Mode Microplate Reader (28°C, 300 rpm). Unconditioned media (UCM), which represents the Sp7 growth in monoculture, was obtained following the same procedure, except 1X PBS was added to Sp7 in R2A media instead of CFCF. To verify that the OD600 measurement was due to Sp7 growth, CFCF from selected isolates without inoculation of Sp7 was also measured. A total of n=3 independent replicates were calculated for each culture condition. Growth rates and carrying capacities for each condition were quantified using Growthcurver (Sprouffske and Wagner, 2016). Statistical tests were performed using Statistical Analysis System (SAS) analytical software. Differences were identified by comparing Sp7 growth metrics in UCM with Sp7 growth metrics in CFCF from each separate consortium (PC1-PC5) using the Wilcoxon two-sample test to demonstrate Sp7 growth promotion with the consortia.
2.8. 16S rRNA gene community sequencing of microwell extracts
Consortia (PC1-PC5) were first cultured in R2A media by direct inoculation from glycerol stocks into R2A media and then incubated overnight (28°C, 215 rpm). Genomic DNA from each culture was then extracted using DNeasy Blood & Tissue Kit (Qiagen, Germantown, MD) following manufacture's protocol and DNA quality and quantity were measured using a NanoDrop spectrophotometer. DNA quantities (Ads260) of 39–47 ng/mL and DNA qualities (Ads260/Ads280) between 1.74 and 1.88 were measured. Genomic DNA samples were temporarily stored at 4°C in TE buffer, then sent to MR DNA (Shallowater, TX, U.S.A.) for 16S diversity analysis using an Illumina MiSeq sequencing (PE-300bp format). Updated Earth Microbiome primers (515F: 5′-GTGYCAGCMGCCGCGGTAA-3′ and 806R: 5′- GGACTACNVGGGTWTCTAAT-3′) were used to amplify the V4 region of the 16S rRNA gene (Apprill et al., 2015; Parada et al., 2016). Raw sequences were processed into ASV tables using the bioinformatic pipeline described above (Section 2.3). After removing reads that were unclassifiable at the kingdom level or classified as mitochondria or chloroplasts, the dataset included 153 ASVs. Rare ASVs (<0.5% relative abundance) were then removed, leaving 10 ASVs that together accounted for 99.1% of the original data. In this final dataset, the number of reads per consortium ranged from 401,854 to 592,830.
2.9. Plant growth studies to validate survival and colonization of A. brasilense Sp7
Plant growth studies were conducted to validate the enhancement of maize growth, as well as survival and colonization of Sp7 in maize rhizosphere when Sp7 was combined with five of the promoter consortia using the protocol described by Niu and Kolter (Niu and Kolter, 2018). First, Sp7 and each of one of the five consortia were inoculated in 2 mL of R2A broth and cultured separately overnight (28°C, 215 rpm). 50 μL of each culture was then transferred to 2 mL of fresh R2A broth and cultured for another 8 h at 28°C. Cells were then centrifuged at 4,400 rpm for 10 min and resuspended in 1X PBS. Cell suspensions from each consortium (PC1-PC5) as well as Sp7 were diluted to ~108 cells per milliliter. Each consortium was then mixed with Sp7 in 50 mL Falcon tubes in equal volume to prepare the final formulations (Sp7+PC1, Sp7+PC2, Sp7+PC3, Sp7+PC4, Sp7+PC5). Combining Sp7 with the promoting consortia in this way ensured that Sp7 was present at comparable levels for each of the five formulations. 80 surface-sterilized and germinated maize seedlings (genotype B73) were soaked in each formulation. Sp7 monoculture cell suspension and formulations without Sp7 (PC1, PC2, PC3, PC4, PC5), and a sterile 1X PBS control were included as controls. Seedlings were incubated without shaking at room temperature for 1 hr. Inoculated maize seedlings were then transferred onto 20 ml ½ Murashige and Skoog (MS) agar (0.8%) in 16 x 150 mm glass tubes with sterile forceps. Sterile empty glass tubes of the same size were attached to the tubes containing seeds in a mouth-to-mouth way using air porous tape (Niu and Kolter, 2018). Sealed glass tubes were then placed in test tube racks and were transferred to a plant growth chamber (Carolina Plant Environmental chamber, Carolina Biological Supply Company) under diurnal light under the following conditions: 8 h of light (day) at 27°C and 9,500 lux of light intensity, 16 h of dark (night) at 23°C, and relative humidity of 54%, and the growth of the maize seedlings was measured every 5 days for 15 days to measure plant height and Sp7 rhizosphere colonization in each sample. Plant growth was measured across n=6 replicate plants for each condition. The Wilcoxon two-sample test (SAS software) was used to identify differences in plant growth by comparing plant height after inoculation with Sp7 only to plant height after inoculation with Sp7 plus each separate consortium (PC1 + Sp7 to PC5 + Sp7).
2.10. Quantification of A. brasilense Sp7 abundance on maize roots
Roots generated from maize seedlings that were inoculated with Sp7 or with Sp7 and one of the five consortia (PC1-PC5) were sampled on day 15 after inoculation. Roots were then cut and washed with sterile 1x PBS to remove agar adhered to the root surface. A 1-cm long primary root fragment below the maize kernel was then cut, weighed (~20 mg each), and transferred into 1.5 mL centrifuge tubes containing six glass beads (diameter: 3 mm, Propper) and 1 mL sterile 1x PBS buffer. The bacterial cells colonized on the root surfaces were dislodged by sonication (amplitude: 30%; pulse: on 01 sec, off 01 s; time: 30 s) for 1 m, then vortexing for another 1 min. This step was repeated twice, then the tube was put on ice for 1 min. The cell suspensions were collected and sequentially diluted by a dilution factor of 108. 10 μL of the diluted suspensions were then spread on R2A agar supplemented with 200 μg/mL of ampicillin and 10 μg/mL of tetracycline to selectively recover Sp7. The plates were air-dried before incubating the plates at 28 °C in the dark for 16 to 60 hrs. The number of the colony-forming units (CFUs) was counted. Sp7 abundance, defined as CFU per mg root, was then calculated using the equation below (Niu and Kolter, 2018):
Sp7 abundance measurements were performed for n=3 replicates for each condition. The Wilcoxon two-sample test (SAS software) was used to identify differences in Sp7 abundance by comparing roots inoculated with Sp7 only to roots inoculated with Sp7 and each separate consortium (PC1+Sp7 to PC5+Sp7).
3. Results and discussion
3.1. Overview of the bioprospecting workflow and MRA device operation
The bioprospecting approach (Figure 1A) involves sampling maize roots to isolate the rhizobiome, then selection of a culture media that recovers rhizobiome isolates at highest taxonomic diversity. The MRA device then uses a co-culture screening process to select a combination of bacteria from the rhizobiome that best promotes the growth of Sp7. The promoting interaction between Sp7 and the consortia found in the MRA is validated in independent 96-well plate assays and the consortia are genomically characterized. After this, Sp7 and the promoting consortia are inoculated onto seedlings to investigate the impact on plant growth. A key step in this workflow is the MRA screening process (Figure 1B), which uses a stochastic cell seeding process characterized previously (Halsted et al., 2016; Hansen et al., 2016, 2021) to generate random collections of rhizobiome isolates and Sp7 cells in small (10 μm diameter) microwells which are compositionally unique from each other. Cells are then sealed into wells with a photodegradable membrane and co-cultured in the selected media. Sp7 growth is monitored in parallel across 3600 microwells using TLFM to reveal the location of individual wells containing consortia that best promote Sp7 growth rate and population size. These wells are tagged as promoter wells. Using a patterned light source, promoting consortia are extracted out of promoter wells (van der Vlies et al., 2019; Fattahi et al., 2020, 2021) and plated for recovery.
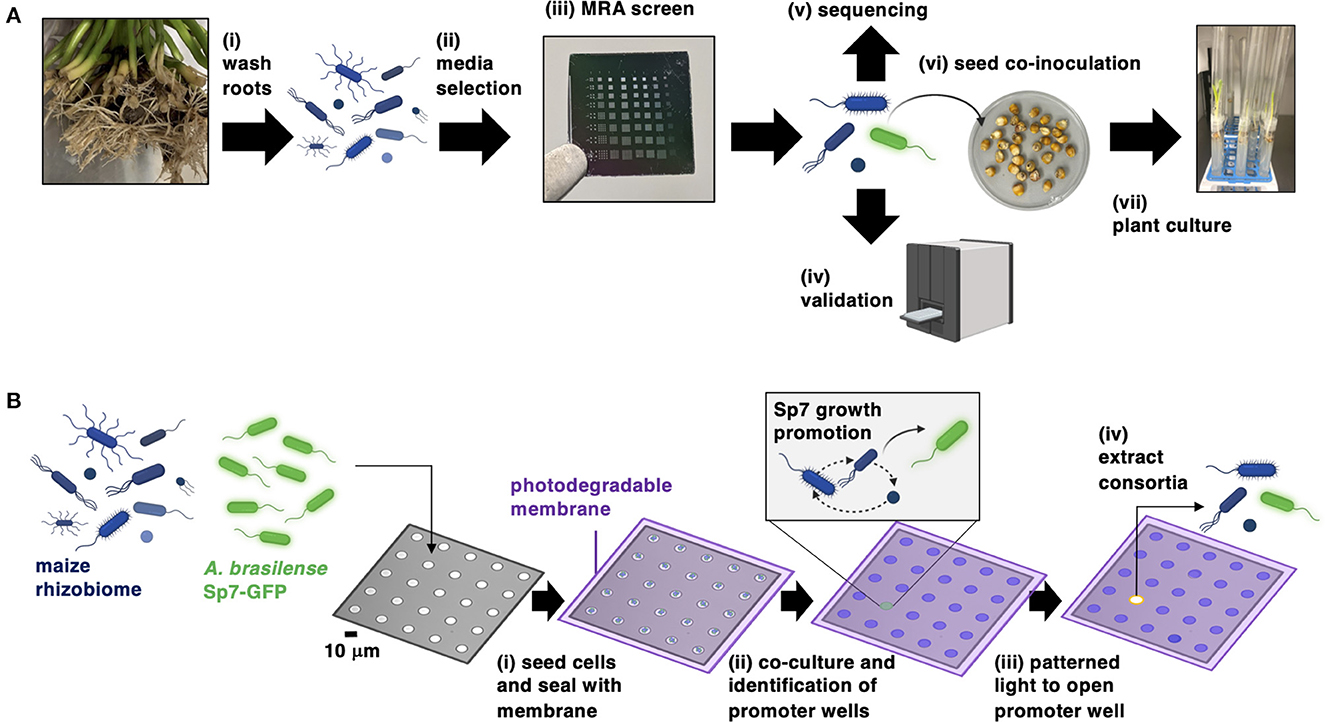
Figure 1. (A) Workflow for bioprospecting the maize rhizosphere for isolates (blue cells) that improve the colonization of Sp7 (green cells) in maize roots. (B) Operational steps of the MRA device for screening and isolating consortia from the maize rhizosphere that promote the growth of Sp7. Figure was prepared with the assistance of BioRender software.
3.2. Media selection and MRA screening results
The goal of the media selection was to identify the media that would enable observation of a maximum number of interactions during MRA co-culture screen and that would recover the highest diversity of bacteria from the device. Here, LB, TY, and R2A media were considered and used to culture bacteria from maize rhizosphere, cultures were then characterized with 16S-V4 rRNA gene sequencing. Community analysis for the rhizosphere microbiome showed members of four bacterial phyla (Proteobacteria, Firmicutes, Bacteroidetes, and Actinobacteria), with the genera Pseudomonas, Serratia, Acinetobacter, Enterobacter, and Stenotrophomonas at high relative abundances (Supplementary Figure 1). As expected, diversity of the original root washes was much higher than the diversity of any of the cultured communities. Of the three media, R2A supported a slightly higher diversity of bacterial ASVs (Supplementary Figure 2) and was selected as the culture media for the MRA screen and for all further cultures.
After media selection, the MRA workflow (Figure 1B) was followed to screen for unknown interactions between the microbes extracted from maize rhizosphere and Sp7. Monoculture control arrays seeded with Sp7 only were first performed (Figure 2A) to generate baseline growth curves. Then co-culture arrays seeded with Sp7 and the root isolates in a 1:1 ratio were performed (Figure 2B). Compared to the monoculture control, significant changes in Sp7 growth metrics were observed with co-culture. Averaged endpoint fluorescence levels of Sp7 monoculture across the measured wells showed relatively low variance (s2 = 67) compared to co-culture (s2 = 553), indicating a wide range of impact due to the addition of the various combinations of rhizobacteria in wells across the array. In the co-culture wells, a noticeable lag time was observed for Sp7 growth compared to the monoculture wells, and low levels of Sp7 growth were most frequently observed, which could be due to competitive and/or antagonistic interactions with many of the rhizobacteria. In fact, relative to the monoculture wells, Sp7 exhibited enhanced population growth in only 16.5% of the co-culture array. Wells with highest Sp7 growth and end-point fluorescence signals were identified in the co-culture array using Grubb's outlier testing, and these “promoter wells” were targeted for selective recovery of the putative promoter isolates.
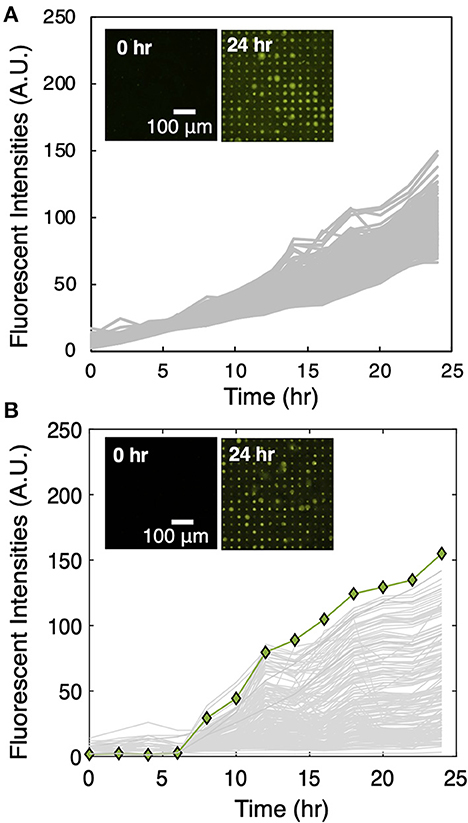
Figure 2. (A) Sp7 growth trajectories across a 225 microwell subset in the MRA device during monoculture. (Inset) Corresponding TLFM images of a sample 15 × 15 array of microwells during Sp7 monoculture. (B) Sp7 growth trajectories of a sample 15 × 15 array containing a Sp7 promoter well during Sp7 co-culture with microbes recovered from maize rhizosphere. Growth trajectories that result in the highest end-point fluorescent signal (green line) indicate a well where highest levels of Sp7 growth promotion occurred. (Inset) Corresponding TLFM images of the sample 15 × 15 array of microwells during Sp7 co-culture.
3.3. Extraction, identification, and validation of promoter consortia
Consortia from the top two promoter wells on the co-culture MRA were extracted for characterization using 16S-V4 rRNA gene sequencing and then off-chip validation and application. Cells from the promoter wells became visible with brightfield microscopy after culture, as the cells protruded out of the well boundary due to the high levels of growth (Figure 3A, i). Promoter wells were exposed to ring patterned light (Figure 3A, ii) to remove the membrane from the top of the wells while minimizing cellular exposure to UV light, releasing cells into solution. After exposure, the cell mass could be observed moving out of an opened well (Figure 3A, iii), at which point it was retrieved by washing the arrays with R2A media. After this process, visible cells appeared removed from the promoter wells, and an opening in the membrane around the well was clearly visible (Figure 3A, iv), verifying the successful extraction. During previous development of this extraction protocol, it was demonstrated that minimal cross-contamination from other wells or from the outside environment occurred (van der Vlies et al., 2019), which ensured that the isolates recovered here originated from the promoter well. The extraction process was done sequentially for two promoter wells, and the extract from each individual well was then plated on R2A-agar to recover individual colonies.
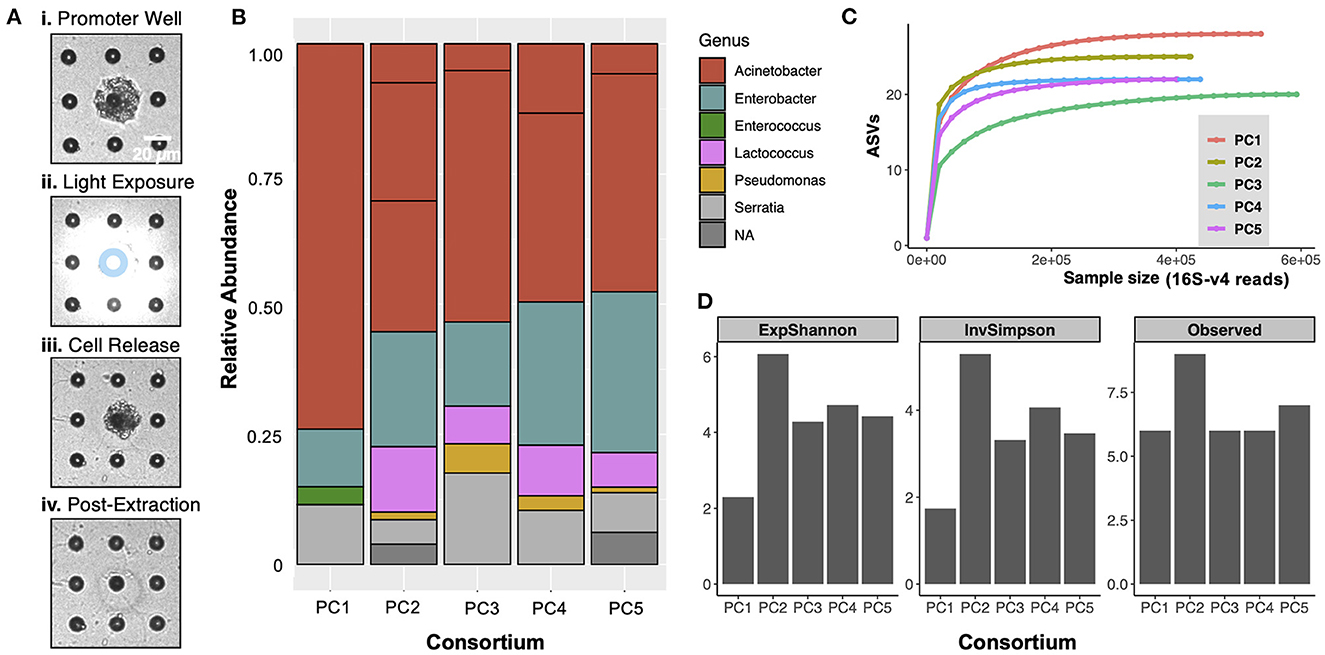
Figure 3. (A) Removal of communities from promoter wells. The perimeter of the promoter well was exposed to UV light in a 10 μm inner diameter and 20 μm outer diameter ring pattern (denoted by blue ring in (ii)) to open the photodegradable membrane above the well while also minimizing direct UV exposure to cells. After membrane degradation, cells were released from only the opened well. (B) Taxonomic bar plots of the composition of the 5 promoter consortia (PC1-PC5), extracted from promoter wells. ‘NA' indicates reads that were not assigned to one of the dominant genera. (C) Rarefaction curves generated prior to removal of rare ASVs indicate that the sequencing effort was sufficient to capture the full diversity of each promoter consortium. (D) Alpha diversity metrics are presented for the 5 promoter consortia. “ExpShannon” = Shannon diversity, calculated as e[Shannon index] . “InvSimpson” = Inverse Simpson diversity. “Observed” = number of ASVs.
After extraction and recovery, 16S-V4 rRNA gene of PC1-PC5 was performed to characterize the taxa present in the promoting consortia extracted from the promoter wells (Figures 3B–D). The analysis revealed that each consortium was dominated with cells from the Acinetobacter, Enterobacter, and Serratia genera, as well as smaller amounts of Lactococcus cells (<10%) and some Pseudomonas cells (<2%). Measures of alpha diversity for the consortia (Figure 3D) were lower than those of the input (Supplementary Figure 2), demonstrating that the device was able to refine the rhizosphere communities that were input into the device toward interacting members. It should be noted that the compositions of all consortia were similar, with only minor differences in composition at the genus level. This was expected because these consortia originated from one of two wells, which were the top two wells on the array for promoting Sp7 growth. Because of these similarities, it was hypothesized that the five promoting consortia would function similarly to one other. However, with the compositional differences noted, each consortium was still included as independent seedling inoculum in the following studies. It is also notable that some of the genera dominant within these consortia could potentially have inherent plant growth promoting function, independent of Sp7. For example, several species in the Serratia genus have been reported to fix nitrogen [e.g., Serratia nematodiphila, Serratia ureilytica (Tang et al., 2020)] and produce indole-3-acetic acid and siderophores [e.g., Serratia marcescens (Khan et al., 2017; Matteoli et al., 2018)].
To verify the on-chip observations, interactions between each PC and Sp7 were recapitulated in an independent off-chip co-culture, following our previous validation assay procedure (Barua et al., 2021). Culture in CFCF accounts for the PC's diffusive effects on Sp7 in a scaled-up solution volume (from a 1.6 pL microwell volume to a 100 mL volume on the 96 well plate). As a control, a monoculture of Sp7 (OD600 = 0.2) in unconditioned media (UCM), which was R2A culture media instead supplemented with a blank solution (1X PBS) at a 1:1 ratio, was also performed and used for comparison. As a negative control, conditioned media from a PC was inoculated without Sp7 to verify that the measured growth in the other experiments was solely due to Sp7 and not due to contaminating microbes. Growth trajectories of each culture reveled that, congruent with microwell observations, higher levels of Sp7 growth occurred when cultured when in CFCF from each promoting consortium (Figure 4A). The corresponding growth metrics show significantly increased the growth rate and carrying capacity of Sp7 compared to the UCM monoculture control (Figures 4B, C, raw data provided in Supplementary Figure 3). This demonstrates that the Sp7 promotion effect from each consortium that was initially identified on the MRA device could be independently recapitulated, and motivated further investigation of the complementary formulations in the context of a root host.
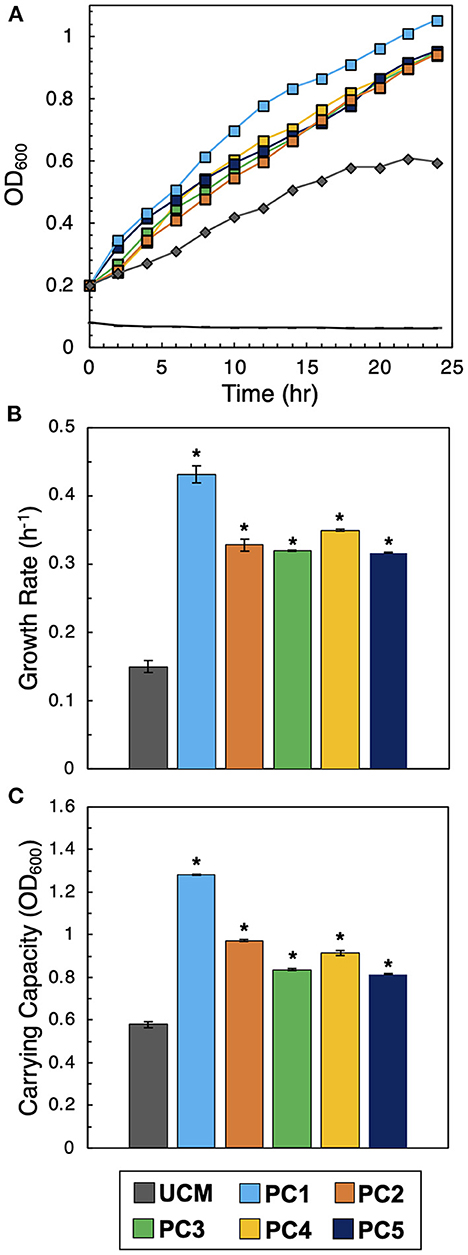
Figure 4. Validation of interactions. (A) Sp7 growth curves after inoculation into unconditioned media (UCM) or conditioned media from each of the five consortia (PC1-PC5). The black control line represents conditioned media without Sp7. (B) Corresponding growth rates and (C) carrying capacities of Sp7 in UCM or media conditioned with the promoter consortia (PC1-PC5). Statistical differences were identified by comparing Sp7 growth metrics in consortia-conditioned media with Sp7 monoculture growth metrics in UCM (Wilcoxon two-sample test, *P < 0.01, n = 3 independent experiments).
3.4. Effect of A. brasilense- isolate co-inoculation on maize growth
With a panel of five consortia with verified growth promotion on Sp7 in liquid co-culture, the effect of co-inoculation with Sp7 and each individual consortium on the growth of axenic maize seedlings was studied in a plant growth chamber environment. After inoculating the surface-sterilized and germinated seeds with a cell suspension containing both Sp7 and each consortium at a Sp7:consortium cellular ratio of 1:1, the axenic seedlings were transferred to a glass tube growth chamber and allowed to grow in ½ MS agar for 15 days (Figure 5A). Two control treatments were included for comparison, these were sterilized seedlings with no inoculation and sterilized seedlings inoculated with Sp7 only. Because several genera of bacteria found in the consortia samples also had known plant growth promoting properties, surface-sterilized and germinated seedlings inoculated only with individual consortia but no Sp7 were added as a final control study. This enabled a distinction between consortia-mediated plant growth due to Sp7 promotion and potential plant growth enhancement due to the promoting consortia alone.
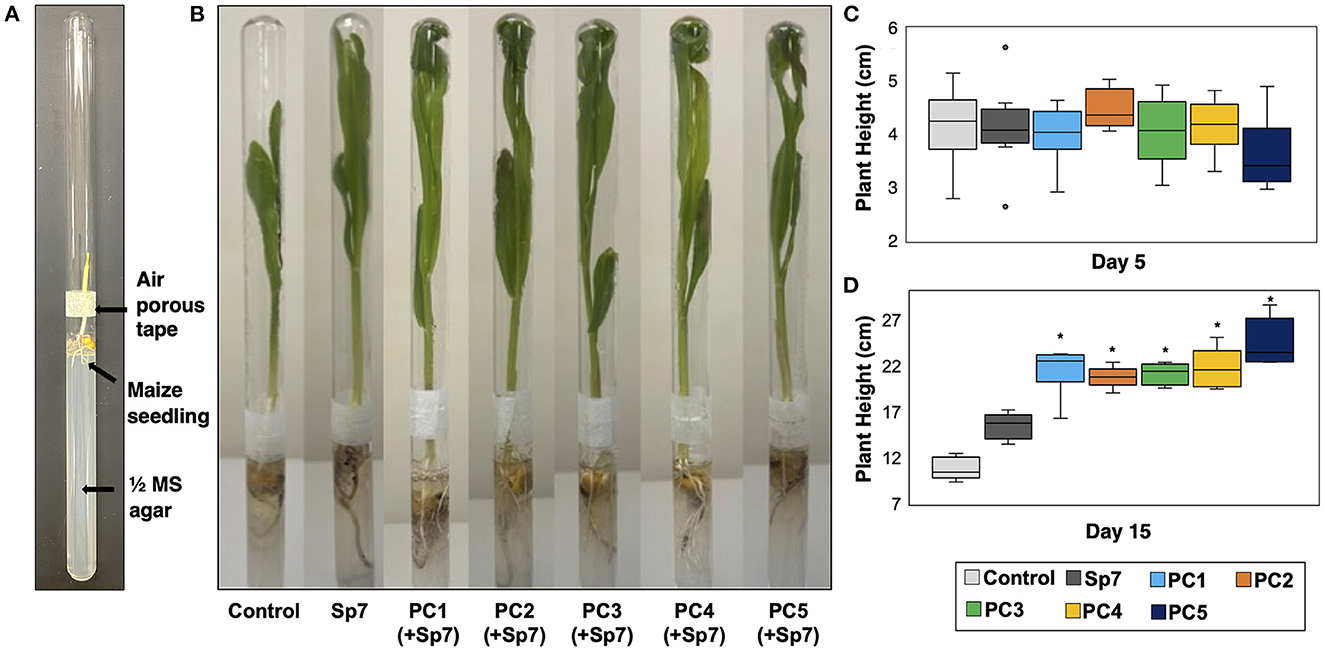
Figure 5. Growth of axenic maize seedlings in a growth chamber environment. (A) A double-tube growth chamber was used for accomodating surface-sterilized and germinated maize seedlings after inoculation with an ultrapure water blank (control), Sp7, or Sp7 with a promoter consortium (PC1-PC5). Two test tubes were attached in a mouth-to-mouth fashion with air-porous tape. (B) Growth of the maize seedlings in the double-tube growth chamber at day 15. (C) Comparison of plant heights for each treatment at day 5 and (D) at day 15 (*Wilcoxon Rank test: p-value < 0.01). n = 6 replicate trials.
In all the treatments involving Sp7, leaf emergence from all maize seedlings was observed on Day 2. It was therefore concluded that the promoting consortia combined with Sp7 did not have an observed impact on leaf emergence compared to single strain inoculation with Sp7. Co-inoculations also did not significantly impact plant height at Day 5 (Figure 5C). However, significant differences were observed at Day 15 (Figures 5B, D). At this time, seedlings grown after inoculating with Sp7 only showed significantly greater plant height compared to the axenic maize seedlings grown in no-inoculum conditions, demonstrating the positive impact of A. brasilense alone. Additionally, axenic seedlings soaked with Sp7 and each promoting consortium showed significantly greater plant height at Day 15 (p < 0.01) compared to both control treatments of Sp7 only and no inoculum. No significant differences in plant height were found between seedlings inoculated with Sp7 plus any one of the five consortia. This was not surprising given the similar composition of each promoting consortium (Figure 3B). Finally, 15-day plant height for the seedlings inoculated only with a promoting consortium but without Sp7 showed diminished growth compared to seedlings inoculated with Sp7 only, and minimal or no improvement relative to uninoculated seedlings (Figure 6). This demonstrated that the promoter consortia (PC1–PC5) must be combined with Sp7 to provide improvement in plant growth.
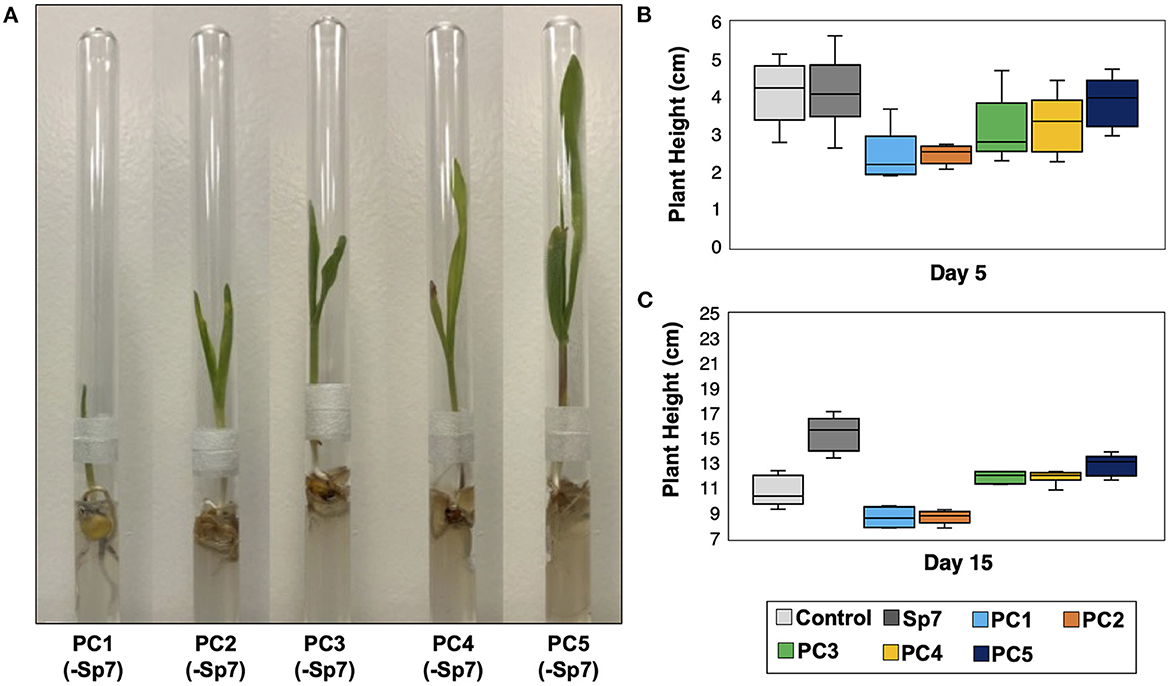
Figure 6. Growth of axenic maize seedlings after innoculation with each promoter consortium (PC1-PC5) but without Sp7. (A) Plant growth in growth chambers after 15 days. Comparision of plant heights for each consortium at (B) day 5 and (C) day 15 with sterile seedlings (control) and with seedlings inoculated with Sp7 only. n = 6 replicate trials.
3.5. Promoter consortia increase A. brasilense population size on maize roots
Given the A. brasilense Sp7 growth promotion observed in MRA and 96-well plate co-cultures, combined with improved maize growth outcomes with seedling co-inoculations, we hypothesized that the promoter consortia enhanced plant growth by increasing Sp7 population sizes on root hosts during the plant growth studies. To test this hypothesis, we measured Sp7 abundances on the maize roots used in the plant growth studies by plating on antibiotic-supplemented R2A agar. As a control to verify that only Sp7 would be recovered from these plating experiments, the five consortia (PC1-PC5) were also plated on R2A agar plates supplemented with the Sp7 resistant antibiotics. No cell colonies were observed here (Supplementary Figure 4), verifying that the colonies observed in the plating experiment were Sp7. Quantification of Sp7 abundance from each sample revealed that three of the consortia (PC2, PC3, and PC5) generated the most significant increases in Sp7 root colonization levels (p < 0.01), while the other two (PC1, PC4) still generated significantly elevated levels of Sp7 (p < 0.05) relative to seedlings inoculated with Sp7 only (Figure 7). The differences in quantified Sp7 abundance levels between the different promoting consortia were mostly likely due to the differences in the consortia composition noted previously (Figure 3B). However, all formulations resulted in elevated Sp7 root colonization to some degree and all led to improved and equivalent plant growth after 15 days (Figure 5D).
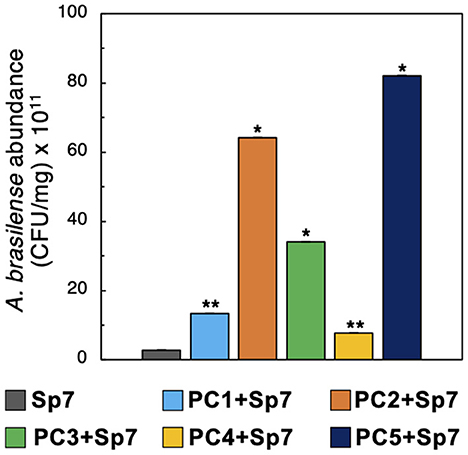
Figure 7. Sp7 abundance in maize roots. Sp7 abundance (CFU/mg root) on maize roots inoculated with Sp7 only or co-inoculated with Sp7 and promoter consortia (PC1-PC5). (*Wilcoxon Rank test: p-value < 0.01; **Wilcoxon Rank test: p-value < 0.05). n = 3 replicate trials.
4. Discussion
Taken together, the findings show that by using promoting consortia to elevate the level of Sp7 population on the root host, the growth of maize could be accelerated. This suggests that the approach of pairing a well-known PGPB, such as Sp7, with collections of rhizosphere isolates that have positive interactions with the PGPB could be a viable strategy for advancing the formulation of next-generation, consortia-based biofertilizers for improved food production. These findings support a growing body of literature suggesting that applying PGPB as complementary, multi-species consortia instead of single-strain inoculations can improve crop health and development (Backer et al., 2018; Trivedi et al., 2020).
While the much of effort in developing PGPB formulations has focused on identifying and using complementary pair-wise interactions, a key advantage of the MRA platform is the capability to assemble cells at a consortium level and screen multi-membered interactions. By using small wells (10 μm diameter) that only partition a low number of cells, random consortia are generated across the array space, where cells are confined together at a length scale that facilitates intercellular interactions. While small wells are critical to platform design, this feature also presents challenges related to removal of consortia from the wells. Although an optimized, light-based extraction system is used (van der Vlies et al., 2019), cellular recovery from wells may still be incomplete, which may result in a recovered composition that is modified from the well composition. Likewise, culture-based recovery steps after extraction could shift community compositions from what it was in the promoter well. However, these drawbacks are mitigated by performing 96-well plate co-culturing validation assays that evaluate weather interactions present on the chip recapitulate in vitro. This is a critical step in this workflow. If the interaction can be successfully recapitulated, as done here, one can proceed with using the promoting consortium in plant growth studies.
Finally, it should be noted that the platform is reliant on co-culture during the screening and recovery steps, and is therefore only able to screen and recover consortia that consist of bacteria that are culturable in the selected media. This underscores the need to carefully select the media most appropriate to the specific screen. For example, depending on the specific goals of the study, it may be desirable to target the screen toward a specific group of bacteria (e.g., Rhizobium), in which case it would be appropriate to select a media that supports the growth of a wide range of isolates of that group. The approach taken here was to consider common culture media and select the one that recovered the highest diversity of bacteria. While this inevitably introduces bias that limits the interactions that can be found and the composition of consortia that can be recovered, from an application standpoint, culturability is a pre-requisite for generating enough inoculant biomass for seed coating. For improvement, one may consider performing the co-cultures in the presence of root exudates, which may contain natural growth factors that fit the metabolic requirements of more diverse collections of isolates.
5. Conclusions
Poor survival and colonization of PGPB after single strain inoculations highlight the need to exploit microbe-microbe interactions to improve the growth, survival, and function of beneficial microbes in the rhizosphere. Here, we use a new technology platform for high-throughput screening of multispecies consortia that included A. brasilense, a well-known diazotrophic PGPB, and thousands of combinations of maize rhizobiome isolates taken from the field. Utilizing the unique capability of the platform to screen microbial interactions at a consortium level, consortia consisting of bacteria primarily of the Acinetobacter, Enterobacter, and Serratia genera that promoted the growth of A. brasilense were uncovered from the maize rhizosphere. Furthermore, for the first time, we demonstrated that interactions initially observed in the screening platform can also be observed in the context of root hosts, and that application of these MRA-assembled consortia can be used to elicit improved plant development. By coupling a well-known PGPB with bacteria native to roots that were observationally found to promote its growth and colonization, we present a new bioprospecting approach to discover multispecies consortia that have potential application as next-generation biofertilizers for food production. Future studies involve investigating the effect of these consortia on maize growth in more complex, soil-based systems in larger scale greenhouse studies and expanding the MRA platform capabilities to screening consortia that include fungi, which may further enhance plant responses. Moreover, the workflow developed here—consisting of field sampling, media selection, MRA screening, cross validation, and application—lays a foundation for future bioprospecting endeavors aimed at developing formulations of plant-associated microbes that can improve seedling vigor, crop yield, and tolerance to drought, salinity, and heat stress.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found in the article/Supplementary material.
Author contributions
NB, KC, MW, TP, and RH conceived and designed experiments. NB and KC performed experiments. MW and DR developed or contributed analysis tools, samples, reagents, and materials. NB, TP, and RH wrote the paper. All authors contributed to the article and approved the submitted version.
Funding
We acknowledge support from the Global Food Systems Seed Program at Kansas State University and from the National Science Foundation Award 1828571. KC and MW were supported by the National Science Foundation under Award No. IOS-2033621 to MW as well as No. OIA-1656006 to Kristin Bowman-James and matching support from the State of Kansas through the Kansas Board of Regents.
Acknowledgments
We thank Gladys Alexandre and Elena Ganusova (Biochemistry and Cellular and Molecular Biology, University of Tennessee Knoxville) for kindly providing A. brasilense Sp7-GFP strains. We also acknowledge the Nebraska Nanoscale Facility at the University of Nebraska Lincoln for assistance in microwell fabrication. A portion of this manuscript has been included in a Ph.D. dissertation that is available online at the Kansas State Research Exchange (K-REx) repository (Barua, 2021).
Conflict of interest
RH and TP currently have a U.S. patent on this technology (Hansen et al., 2022).
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2023.1106528/full#supplementary-material
References
Aasfar, A., Bargaz, A., Yaakoubi, K., Hilali, A., Bennis, I., Zeroual, Y., et al. (2021). Nitrogen fixing azotobacter species as potential soil biological enhancers for crop nutrition and yield stability. Front. Microbiol. 12, 8739. doi: 10.3389/fmicb.2021.628379
Aloo, B. N., Mbega, E. R., Makumba, B. A., Hertel, R., and Daniel, R. (2021). Molecular identification and in vitro plant growth-promoting activities of culturable potato (Solanum tuberosum L.) Rhizobacteria in Tanzania. Potato Res. 64, 67–95. doi: 10.1007/s11540-020-09465-x
Alzate Zuluaga, M. Y., Miras-Moreno, B., Monterisi, S., Rouphael, Y., Colla, G., Lucini, L., et al. (2022). Integrated metabolomics and morpho-biochemical analyses reveal a better performance of azospirillum brasilense over plant-derived biostimulants in counteracting salt stress in tomato. Int. J. Mol. Sci. 23, 14216. doi: 10.3390/ijms232214216
Apprill, A., McNally, S. P., Parsons, R. J., and Weber, L. (2015). Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquatic Microb. Ecol. 75, 129–137. doi: 10.3354/ame01753
Backer, R., Rokem, J. S., Ilangumaran, G., Lamont, J., Praslickova, D., Ricci, E., et al. (2018). Plant growth-promoting rhizobacteria: context, mechanisms of action, and roadmap to commercialization of biostimulants for sustainable agriculture. Front. Plant Sci. 9, 1473. doi: 10.3389/fpls.2018.01473
Bargaz, A., Elhaissoufi, W., Khourchi, S., Benmrid, B., Borden, K. A., Rchiad, Z., et al. (2021). Benefits of phosphate solubilizing bacteria on belowground crop performance for improved crop acquisition of phosphorus. Microbiol. Res. 252, 110. doi: 10.16./j.micres.2021.126842
Barua, N. (2021). Screening and Discovery of Symbiotic and Antagonistic Microbial Networks Using Microwell Recovery Arrays. PhD dissertation, Chemical Engineering, Kansas State University.
Barua, N., Herken, A. M., Stern, K. R., Reese, S., Powers, R. L., Morrell-Falvey, J. L., et al. (2021). Simultaneous discovery of positive and negative interactions among rhizosphere bacteria using microwell recovery arrays. Front. Microbiol. 11, 6010. doi: 10.3389/fmicb.2020.601788
Bashan, Y. de-Bashan, L.E., Prabhu, S.R., and Hernandez, J. P. (2014). Advances in plant growth-promoting bacterial inoculant technology: formulations and practical perspectives (1998–2013). Plant Soil 378, 1–33. doi: 10.1007/s11104-013-1956-x
Bhat, B. A., Tariq, L., Nissar, S., Islam, S. T., Islam, S. U., Mangral, Z., et al. (2022). The role of plant-associated rhizobacteria in plant growth, biocontrol and abiotic stress management. J. Appl. Microbiol. 133, 2717–2810. doi: 10.1111/jam.15796
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., Holmes, S. P., et al. (2016). DADA2: high-resolution sample inference from illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proceed. Nat. Acad. Sci. 108(supplement_1), 4516–4522. doi: 10.1073/pnas.1000080107
Chaudhary, P., Singh, S., Chaudhary, A., Sharma, A., and Kumar, G. (2022). Overview of biofertilizers in crop production and stress management for sustainable agriculture. Front. Plant Sci. 13, 340. doi: 10.3389/fpls.2022.930340
Dent, D., and Cocking, E. (2017). Establishing symbiotic nitrogen fixation in cereals and other non-legume crops: the greener nitrogen revolution. Agricult. Food Sec. 6, 7. doi: 10.1186/s40066-016-0084-2
Dodds, W. K., Zeglin, L. H., Ramos, R. J., Platt, T. G., Pandey, A., Michaels, T., et al. (2020). Connections and feedback: aquatic, plant, and soil microbiomes in heterogeneous and changing environments. BioScience 70, 548–562. doi: 10.1093/biosci/biaa046
dos Santos Lima Fagotti, D., Abrantes, J. L. F., Cerezini, P., Fukami, J., Nogueira, M. A., del Cerro, P., et al. (2019). Quorum sensing communication: bradyrhizobium-Azospirillum interaction via N-acyl-homoserine lactones in the promotion of soybean symbiosis. J. Basic Microb. 59, 38–53. doi: 10.1002/jobm.201800324
Egamberdieva, D., Wirth, S. J., Alqarawi, A. A., Abd_Allah, E.F., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8, 2104. doi: 10.3389/fmicb.2017.02104
Farooq, M. S., Uzair, M., Raza, A., Habib, M., Xu, Y., Yousuf, M., et al. (2022). Uncovering the research gaps to alleviate the negative impacts of climate change on food security: a review. Front. Plant Sci. 13, 7535. doi: 10.3389/fpls.2022.927535
Fattahi, N., Barua, N., van der Vlies, A. J., and Hansen, R. R. (2021). Photodegradable hydrogel interfaces for bacteria screening, selection, and isolation. JoVE 177, e63048. doi: 10.3791/63048
Fattahi, N., Nieves-Otero, P. A., Masigol, M., van der Vlies, A. J., Jensen, R. S., Hansen, R. R., et al. (2020). Photodegradable hydrogels for rapid screening, isolation, and genetic characterization of bacteria with rare phenotypes. Biomacromolecules 21, 3140–3151. doi: 10.1021/acs.biomac.0c00543
Ferreira, A. S., Pires, R. R., Rabelo, P. G., Oliveira, R. C., Luz, J. M. Q., Brito, C. H., et al. (2013). Implications of Azospirillum brasilense inoculation and nutrient addition on maize in soils of the Brazilian Cerrado under greenhouse and field conditions. Appl. Soil Ecol. 72, 103–108. doi: 10.1016/j.apsoil.2013.05.020
Ferreira, C. M. H., Soares, H. M. V. M., and Soares, E. V. (2019). Promising bacterial genera for agricultural practices: an insight on plant growth-promoting properties and microbial safety aspects. Sci. Total Environ. 682, 779–799. doi: 10.1016/j.scitotenv.2019.04.225
Gowtham, H. G., Singh, S. B., Shilpa, N., Aiyaz, M., Nataraj, K., Udayashankar, A. C., et al. (2022). Insight into recent progress and perspectives in improvement of antioxidant machinery upon PGPR augmentation in plants under drought stress: a review. Antioxidants 11, 1763. doi: 10.3390/antiox11091763
Halsted, M., Wilmoth, J. L., Briggs, P. A., Hansen, R. R., Briggs, D. P., Timm, A. C., et al. (2016). Development of transparent microwell arrays for optical monitoring and dissection of microbial communities. J. Vac. Sci. Technol. B 34, 06KI03. doi: 10.1116/1.4962739
Hansen, R. R., Anderson, A. C., Barua, N., and McGinley, L. M. (2021). Using ongoing laboratory problems as active learning research projects in transport phenomena. Chem. Engin. Edu. 55, 51–60. doi: 10.18260/2-1-370.660-120873
Hansen, R. R., Platt, T. G., and van der Vlies, A. J. (2022). Hydrogel Membrane and Methods for Selective Retrieval of Microbial Targets. USA Patent 11, 235.330B2.
Hansen, R. R., Timm, A. C., Timm, C. M., Bible, A. N., Morrell-Falvey, J. L., Pelletier, D. A., et al. (2016). Stochastic assembly of bacteria in microwell arrays reveals the importance of confinement in community development. PLoS ONE 11, e0155080. doi: 10.1371/journal.pone.0155080
Hungria, M., Campo, R. J., Souza, E. M., and Pedrosa, F. O. (2010). Inoculation with selected strains of Azospirillum brasilense and A. lipoferum improves yields of maize and wheat in Brazil. Plant Soil 331, 413–425. doi: 10.1007/s11104-009-0262-0
Ikeda, A. C., Savi, D. C., Hungria, M., Kava, V., Glienke, C., Galli-Terasawa, L. V., et al. (2020). Bioprospecting of elite plant growth-promoting bacteria for the maize crop. Acta Scientiarum. Agronomy 42, 364. doi: 10.4025/actasciagron.v42i1.44364
Jochum, M. D., McWilliams, K. L., Borrego, E. J., Kolomiets, M. V., Niu, G., Pierson, E. A., et al. (2019). Bioprospecting plant growth-promoting rhizobacteria that mitigate drought stress in grasses. Front. Microbiol. 10, 2106. doi: 10.3389/fmicb.2019.02106
Khan, A. R., Park, G. S., Asaf, S., Hong, S. J., Jung, B. K., Shin, J. H., et al. (2017). Complete genome analysis of Serratia marcescens RSC-14: a plant growth-promoting bacterium that alleviates cadmium stress in host plants. PLoS ONE 12, e01710. doi: 10.1371/journal.pone.0171534
Komarek, A. M., and Msangi, S. (2019). Effect of changes in population density and crop productivity on farm households in Malawi. Agricult. Econ. 50, 615–628. doi: 10.1111/agec.12513
Lee, S. A., Kim, J. M., Kim, Y., Joa, J-. H., Kang, S-. S., Ahn, J-. H., et al. (2020). Different types of agricultural land use drive distinct soil bacterial communities. Sci Rep-Uk 10, 8. doi: 10.1038/s41598-020-74193-8
Leisner, C. P., Potnis, N., and Sanz-Saez, A. (2022). Crosstalk and trade-offs: plant responses to climate change-associated abiotic and biotic stresses. Plant Cell Environ. 4, 1. doi: 10.22541/au.166462027.71310652/v1
Ma, Y., Vosátka, M., and Freitas, H. (2019). Editorial: beneficial microbes alleviate climatic stresses in plants. Front. Plant Sci. 10, 595. doi: 10.3389/fpls.2019.00595
Machado, G. P., Colli Mull, P. M. G. JG, Aguero Chapin, G, Riva de la Riva, GA, and de la Vargas Samano, C. D. (2017). Plant growth promoting bacteria isolated from a mexican natural ecosystem induce water stress resistance in maize and sorghum plants. J. Microb. Biochem. Technol. 9, 209–219. doi: 10.4172/1948-5948.1000367
Maranguit, D., Guillaume, T., and Kuzyakov, Y. (2017). Land-use change affects phosphorus fractions in highly weathered tropical soils. CATENA 149, 385-393. doi: 10.1016/j.catena.2016.10.010
Martin, M. (2011). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 3. doi: 10.14806/ej.17.1.200
Masigol, M., Barua, N., Lokitz, B. S., and Hansen, R. R. (2018). Fabricating reactive surfaces with brush-like and crosslinked films of azlactone-functionalized block co-polymers. Jove-J Vis Exp, ARTN 5, e57562. doi: 10.3791/57562
Masigol, M., Barua, N., Retterer, S. T., Lokitz, B. S., and Hansen, R. R. (2017). Chemical copatterning strategies using azlactone-based block copolymers. J. Vac. Sci. Technol. B 35, Artn 06gj01. doi: 10.1116/1.4991881
Matteoli, F. P., Passarelli-Araujo, H., Reis, R. J. A., Rocha, da., de Souza, L. O., Aravind, E. M., et al. (2018). Genome sequencing and assessment of plant growth-promoting properties of a Serratia marcescens strain isolated from vermicompost. BMC Genom. 19, 750. doi: 10.1186/s12864-018-5130-y
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e610. doi: 10.1371/journal.pone.0061217
Mitter, E. K., Tosi, M., Obregón, D., Dunfield, K. E., and Germida, J. J. (2021). Rethinking crop nutrition in times of modern microbiology: innovative biofertilizer technologies. Front. Sustain. Food Sys. 5. doi: 10.3389/fsufs.2021.606815
Nakkeeran, S., Fernando, W. G. D., and Siddiqui, Z. A. (2006). “Plant growth promoting rhizobacteria formulations and its scope in commercialization for the management of pests and diseases,” in PGPR: Biocontrol and Biofertilization, Z.A. Siddiqui, ed. (Springer Netherlands), pp. 257-296. doi: 10.1007/1-4020-4152-7_10
Niu, B., and Kolter, R. (2018). Quantification of the composition dynamics of a maize root-associated simplified bacterial community and evaluation of its biological control effect. Bio. Protoc. 8, 2885. doi: 10.21769/BioProtoc.2885
Oliveira, I. J., Fontes, J. R. A., Pereira, B. F. F., and Muniz, A. W. (2018). Inoculation with Azospirillum brasiliense increases maize yield. Chem. Biol. Technol. Agricult. 5, 6. doi: 10.1186/s40538-018-0118-z
Parada, A. E., Needham, D. M., and Fuhrman, J. A. (2016). Every base matters: assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 18, 1403-10. doi: 10.1111/1462-2920.13023
Pedrosa, F. O., Oliveira, A. L. M., Guimarães, V. F., Etto, R. M., Souza, E. M., Furmam, F. G., et al. (2020). The ammonium excreting Azospirillum brasilense strain HM053: a new alternative inoculant for maize. Plant Soil 451, 45–56. doi: 10.1007/s11104-019-04124-8
Philippot, L., Raaijmakers, J. M., Lemanceau, P., and van der Putten, W. H. (2013). Going back to the roots: the microbial ecology of the rhizosphere. Nat. Rev. Microbiol. 11, 789–799. 10.1038/nrmicro3109. doi: 10.1038/nrmicro3109
Sprouffske, K., and Wagner, A. (2016). Growthcurver: an R package for obtaining interpretable metrics from microbial growth curves. BMC Bioinform. 17, 172. doi: 10.1186/s12859-016-1016-7
Stamenković, S., Beškoski, V., Karabegović, I., Lazić, M., and Nikolić, N. (2018). Microbial fertilizers: a comprehensive review of current findings and future perspectives. Span. J. Agric. Res. 16, e09R.01. doi: 10.5424/sjar/2018161-12117
Tabassum, B., Khan, A., Tariq, M., Ramzan, M., Iqbal Khan, M. S., Shahid, N., et al. (2017). Bottlenecks in commercialisation and future prospects of PGPR. Appl. Soil Ecol. 121, 102–117. doi: 10.1016/j.apsoil.2017.09.030
Tang, A., Haruna, A. O., Majid, N. M. A., and Jalloh, M. B. (2020). Effects of selected functional bacteria on maize growth and nutrient use efficiency. Microorganisms 8, 854. doi: 10.3390/microorganisms8060854
Timm, A. C., Halsted, M. C., Wilmoth, J. L., and Retterer, S. T. (2017). Assembly and tracking of microbial community development within a microwell array platform. J. Vis. Exp. 124, 55701. doi: 10.3791/55701
Timofeeva, A. M., Galyamova, M. R., and Sedykh, S. E. (2022). Bacterial siderophores: classification, biosynthesis, perspectives of use in agriculture. Plants 11, 3065. doi: 10.3390/plants11223065
Trivedi, P., Leach, J. E., Tringe, S. G., Sa, T., and Singh, B. K. (2020). Plant–microbiome interactions: from community assembly to plant health. Nat. Rev. Microbiol. 18, 607–621. doi: 10.1038/s41579-020-0412-1
van der Vlies, A. J., Barua, N., Nieves-Otero, P. A., Platt, T. G., and Hansen, R. R. (2019). On demand release and retrieval of bacteria from microwell arrays using photodegradable hydrogel membranes. Acs. Appl. Bio. Mater. 2, 266–276. doi: 10.1021/acsabm.8b00592
Van Deynze, A., Zamora, P., Delaux, P-. M., Heitmann, C., Jayaraman, D., Rajasekar, S., et al. (2018). Nitrogen fixation in a landrace of maize is supported by a mucilage-associated diazotrophic microbiota. PLOS Biol. 16, e2006352. doi: 10.1371/journal.pbio.2006352
Verma, R., Chourasia, S. K., and Jha, M. N. (2011). Population dynamics and identification of efficient strains of Azospirillum in maize ecosystems of Bihar (India). 3 Biotech 1, 247–253. doi: 10.1007/s13205-011-0031-7
Viejobueno, J., Albornoz, P. L., Camacho, M., de los Santos, B., Martínez-Zamora, M.G., and Salazar, S.M. (2021). Protection of strawberry plants against charcoal rot disease (Macrophomina phaseolina) induced by Azospirillum brasilense. Agronomy 11, 195. doi: 10.3390/agronomy11020195
Wang, J., Li, Q., Xu, S., Zhao, W., Lei, Y., Song, C., et al. (2018). Traits-based integration of multi-species inoculants facilitates shifts of indigenous soil bacterial community. Front. Microbiol. 9, 1692. doi: 10.3389/fmicb.2018.01692
Wang, Q., Garrity, G. M., Tiedje, J. M., and Cole, J. R. (2007). Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 73, 5261–5267. doi: 10.1128/AEM.00062-07
Wickham, H., Averick, M., Bryan, J., Chang, W., D'Agostino McGowan, L., Francois, R., et al. (2019). Welcome to the tidyverse. J. Open Source Software 4, 1686. doi: 10.21105/joss.01686
Keywords: biofertilizers, consortia, Azospirillum, plant growth promoting bacteria, microbial interactions, high throughput screening, microwell arrays
Citation: Barua N, Clouse KM, Ruiz Diaz DA, Wagner MR, Platt TG and Hansen RR (2023) Screening the maize rhizobiome for consortia that improve Azospirillum brasilense root colonization and plant growth outcomes. Front. Sustain. Food Syst. 7:1106528. doi: 10.3389/fsufs.2023.1106528
Received: 23 November 2022; Accepted: 27 March 2023;
Published: 18 April 2023.
Edited by:
Micaela Tosi, University of Guelph, CanadaReviewed by:
Expedito Olimi, Graz University of Technology, AustriaBecky Nancy Aloo, University of Eldoret, Kenya
Oluwaseyi Samuel Olanrewaju, North-West University, South Africa
Copyright © 2023 Barua, Clouse, Ruiz Diaz, Wagner, Platt and Hansen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ryan R. Hansen, cnJoYW5zZW5Aa3N1LmVkdQ==
 Niloy Barua
Niloy Barua Kayla M. Clouse
Kayla M. Clouse Dorivar A. Ruiz Diaz
Dorivar A. Ruiz Diaz Maggie R. Wagner
Maggie R. Wagner Thomas G. Platt
Thomas G. Platt Ryan R. Hansen
Ryan R. Hansen