
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst. , 27 June 2023
Sec. Crop Biology and Sustainability
Volume 7 - 2023 | https://doi.org/10.3389/fsufs.2023.1086660
This article is part of the Research Topic UN International Day for Plant Health: a Focus on Cassava View all 4 articles
 Padinjarepulikkiyil Sankaran Hareesh†
Padinjarepulikkiyil Sankaran Hareesh† Thulasi Raveendrannair Resmi†
Thulasi Raveendrannair Resmi† Madhavi N. Sheela
Madhavi N. Sheela Thangaraj Makeshkumar*
Thangaraj Makeshkumar*Cassava is one of the most important food and industrial crops in Asia, Africa, and South America. Cassava mosaic disease (CMD), caused by cassava mosaic geminivirus, is one of the major constraints to cassava cultivation. In Asia, the disease is caused by the Indian cassava mosaic virus and Sri Lankan cassava mosaic virus. Phytosanitation, vector control, breeding, and genetic engineering strategies have been widely adopted to manage the disease. This study provides a comprehensive review of the disease spread, the development of diagnostic methods, and various approaches employed for the management of CMD in South and Southeast Asia.
Cassava (Manihot esculenta Crantz) originated in the south edge of the Brazilian Amazon and was domesticated by indigenous South Americans ~12,000–17,000 years ago (Allem, 1994; Oslen and Schaal, 1999). The starchy root of this woody perennial shrub serves as a staple food for more than 800 million people around the world and is also an important industrial commodity. Its adaptability to poor soil and relatively dry weather conditions makes it attractive to smallholder farmers. Cassava has gained global attention as a “famine crop” with production increasing from ~124 to 278 million tons over the period from 1980 to 2018. It is the fourth most important staple food in the world after rice, wheat, and maize, with a global caloric intake of 2.6% (Rey and Vanderschuren, 2017; Tafesse et al., 2021).
Cassava cultivation has severely been affected by several insect pests and viruses. Cassava brown streak virus and cassava mosaic geminivirus (CMG) affect cassava cultivation to a large extent (Bock et al., 1978; Lennon et al., 1986). Cassava mosaic disease (CMD) caused by CMG (family: Geminiviridae; genus: Begomovirus) is the major constraint to cassava cultivation with an estimated crop loss of USD 1.9–2.7 billion annually (Patil and Fauquet, 2009). The cassava plants infected with CMG exhibit symptoms of leaf curling and distortion, mosaic patterns, yellowing, and stunting, which result in decreased photosynthesis and reduced yield (Figure 1). The virus is transmitted by whitefly (Fauquet and Stanley, 2003). In Africa, the major viruses associated with cassava mosaic disease are African cassava mosaic virus (ACMV; Stanley and Gay, 1983), East African cassava mosaic Malawi virus (EACMMV; Zhou et al., 1998), East African cassava mosaic Cameroon virus (EACMCMV; Fondong et al., 2000), East African cassava mosaic virus (EACMV; Pita et al., 2001), South African cassava mosaic virus (SACMV; Berrie et al., 2001), East African cassava mosaic Zanzibar virus (EACMZV; Maruthi et al., 2004), East African cassava mosaic Kenya virus (EACMKV; Bull et al., 2006), Cassava mosaic Madagascar virus (CMMGV; Harimalala et al., 2012), and African cassava mosaic Burkina Faso virus (ACMBFV; Tiendrébéogo et al., 2012). In Asia, the disease is caused by the Indian cassava mosaic virus (ICMV) and the Sri Lankan cassava mosaic virus (SLCMV; Hong et al., 1993; Saunders et al., 2002).
The genome of CMG is bipartite, designated as DNA A and DNA B, encapsidated in twinned icosahedral particles with a molecular size of ~2.7 kb (Stanley and Gay, 1983; Zhang et al., 2001). DNA A consists of genes, AC1, AC2, AC3, AC4, AVI, and AV2, which are important in replication, transcription, defense, and encapsidation. The two genes in DNA B, BC1, and BV1, are involved in cell-to-cell and systemic movement of the virus (Hanley-Bowdoin et al., 2013). DNA A and DNA B components share a common region or intergenic region of ~200 nucleotides where the regulatory elements required for replication and transcription are situated (Hong et al., 1993; Saunders et al., 2002).
Based on genetic diversity and geographical origin, begomoviruses are classified into “Old World” and “New World” viruses. Members of Old World viruses are monopartite as well as bipartite, while members of New World viruses have bipartite genomes (Nawaz-ul-Rehman and Fauquet, 2009; Zhou, 2013). In addition to the major encapsidated genomic components, most of the Old World viruses and some New World begomoviruses are found associated with sub-genomic DNA components such as betasatellites, alphasatellites, delta satellites, and defective interfering DNAs (Briddon and Stanley, 2006; Patil and Dasgupta, 2006; Fiallo-Olive et al., 2012). These are circular single-stranded DNA components having genome sizes of ~½ of their helper viruses. Betasatellites contain a βC1 ORF and a non-anucleotide sequence. βC1 is essential for the pathogenesis and suppression of gene silencing (Cui et al., 2004; Eini et al., 2012), and they depend on a helper virus for replication. Alpha satellites encode an ORF alpha-Rep and are self-replicating. Both alphasatellites and betasatellites depend on their helper viruses for encapsidation and movement (Nawaz-ul-Rehman and Fauquet, 2009). The only natural association of an alphasatellite with a cassava mosaic infection has been reported from cassava fields in Madagascar infected with EACMKV. These alphasatellites, designated as “cassava mosaic alphasatellites,” were closely related to Cotton leaf curl Gezira virus alphasatellite (Harimalala et al., 2013). Alphasatellites or betasatellites associated with SLCMV or ICMV infection have not yet been reported, although it has been shown that SLCMV DNA A can interact with Ageratum yellow vein virus betasatellite and that interaction expands its host range (Saunders et al., 2002).
Geminiviruses often produce deleted versions of their genomes and give rise to defective interfering DNAs (DI DNAs) by recombination, deletion, and rearrangement of the parent genome. This was first reported in association with ACMV infection (Stanley and Townsend, 1985). Defective DNAs depend on the parent virus for replication, movement, and transmission although they contain an intergenic region with all cis-acting elements (Patil and Dasgupta, 2006). They ameliorate the symptoms and cause a reduction in helper DNA levels (Bach and Jeske, 2014). This could be due to their competition with the helper virus for essential host and viral factors and thereby inhibiting the replication of the helper virus. DI DNAs are generally derived from DNA B but a type of DI-DNA derived from the DNA-A of EACMV has also been reported (Ndunguru et al., 2006). Though they usually form in relation to infections in experimental hosts, the natural association of DI DNA-B with SACMV-infected cassava plants in South Africa was reported by Rey et al. (2012). Upon infection with SLCMV infectious clones, DI DNAs were produced from both DNA A and DNA B components. The DNA-B-derived DI DNAs are found associated with a decrease in levels of DNA-B with a change in the symptoms from downward leaf curling in the older leaves to upward leaf-rolling in newly emerging leaves (Patil et al., 2007).
Two novel episomal begomovirus-associated sequences, namely sequences enhancing geminivirus symptoms (SEGS-1 and SEGS-2), were identified in the cassava field affected by CMD. SEGS-2 contains sequences highly similar to the cassava genome and a 26-bp sequence identical to alphasatellite replication origin. This supports the hypothesis of its possible evolution by the recombination between a cassava genomic sequence and an alphasatellite. Arabidopsis thaliana plants co-infected with ACMV and SEGS-2 developed severe CMD symptoms. The satellite molecule when co-inoculated with Cabbage leaf curl virus could infect an ecotype of A. thaliana, recalcitrant to geminivirus infection. This confirms its status as a satellite molecule capable of enhancing CMD symptoms (Ndunguru et al., 2016; Aimone et al., 2021b).
Recombinations, mutations, and pseudorecombinations or reassortment of the viral components occurring in mixed infections are the major causes of the emergence of new viruses (Fondong et al., 2000; Seal et al., 2006). Recombinant viruses have been associated with severe outbreaks of CMD in Africa. The intergenic region and the 5′ part of the Rep gene were identified as hot spots of recombination in begomoviruses (Lefeuvre et al., 2007). EACMV-like viruses are highly prone to recombination compared with ACMV. The highly virulent strain, EACMV-UG, was a recombinant virus that resulted from the exchange of a portion of a coat protein (CP) gene of an EACMV genome with a homologous CP gene fragment of EACMV (Zhou et al., 1997).
The Sri Lankan cassava mosaic virus DNA A has many characteristics of a monopartite begomovirus. Sequence analysis and pseudorecombination studies revealed that SLCMV evolved into a bipartite virus by recombination with ICMV DNA B. High-sequence similarity of SLCMV DNA B and ICMV DNA B outside their common regions reinforces this hypothesis. In the phylogenetic analysis of viruses causing CMD in Africa, and South and Southeast Asia, SLCMV and ICMV formed a separate clade, revealing their distinct evolutionary lineage compared with their African counterparts (Figure 2 and Table 1). SLCMV DNA A alone produces typical monopartite symptoms of upward leaf curling and stunting in Nicotiana benthamiana, proving its evolutionary intermediate position between bipartite and monopartite viruses. Pseudorecombination studies of SLCMV DNA A and ICMV DNA B suggest that SLCMV can trans-replicate ICMV DNA B in spite of the difference in their Rep binding sequences, revealing its complex infectious patterns. Moreover, its ability to interact with the DNAβ component of Ageratum yellow vein virus (AYVV) to produce systemic infection in Ageratum conyzoides also indicates that a monopartite ancestor of SLCMV might have existed in association with a betasatellite component (Saunders et al., 2002). SLCMV is highly recombinogenic. Studies by Crespo-Bellido et al. (2021) also showed the evolution of SLCMV by interspecific recombination, which was further supported by PCR-RFLP analyses (Borah and Dasgupta, 2012). SLCMV Rep is even shown to be triggering transposon activity in Agrobacterium tumefaciens (Resmi et al., 2014).
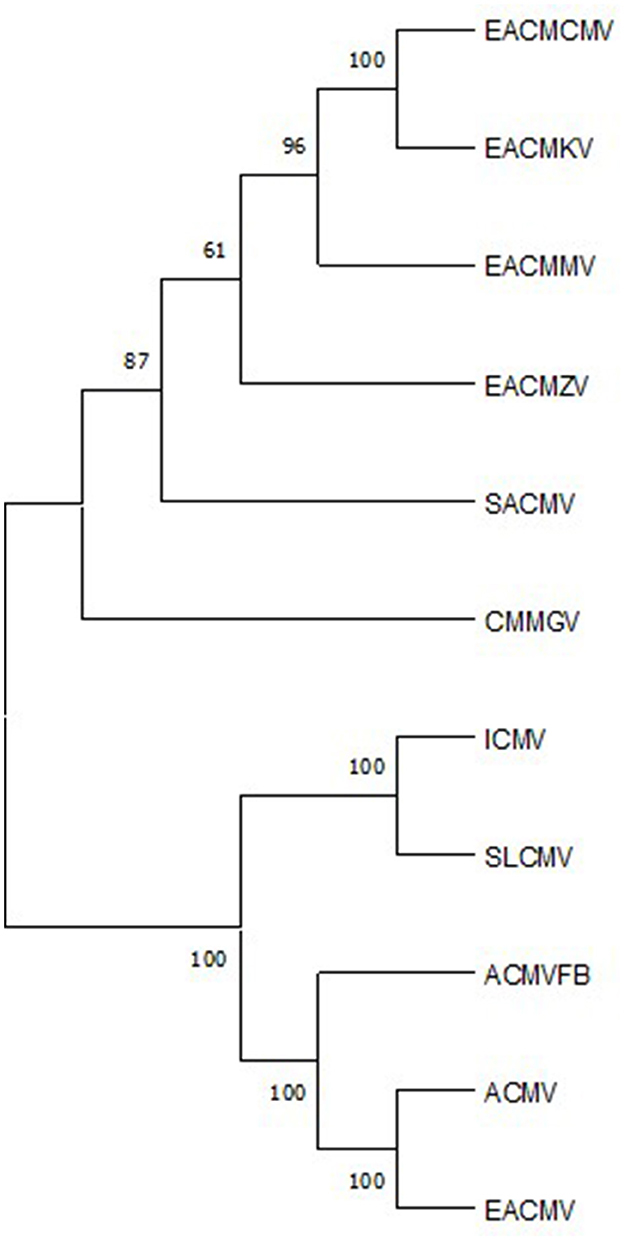
Figure 2. Maximum-likelihood phylogenetic tree obtained from the alignment of full-length nucleotide sequences of the cassava mosaic viruses given in Table 1. The sequences were aligned with CLUSTAL W, and the tree was constructed using MEGA11 software with 1,000 bootstrap replications.
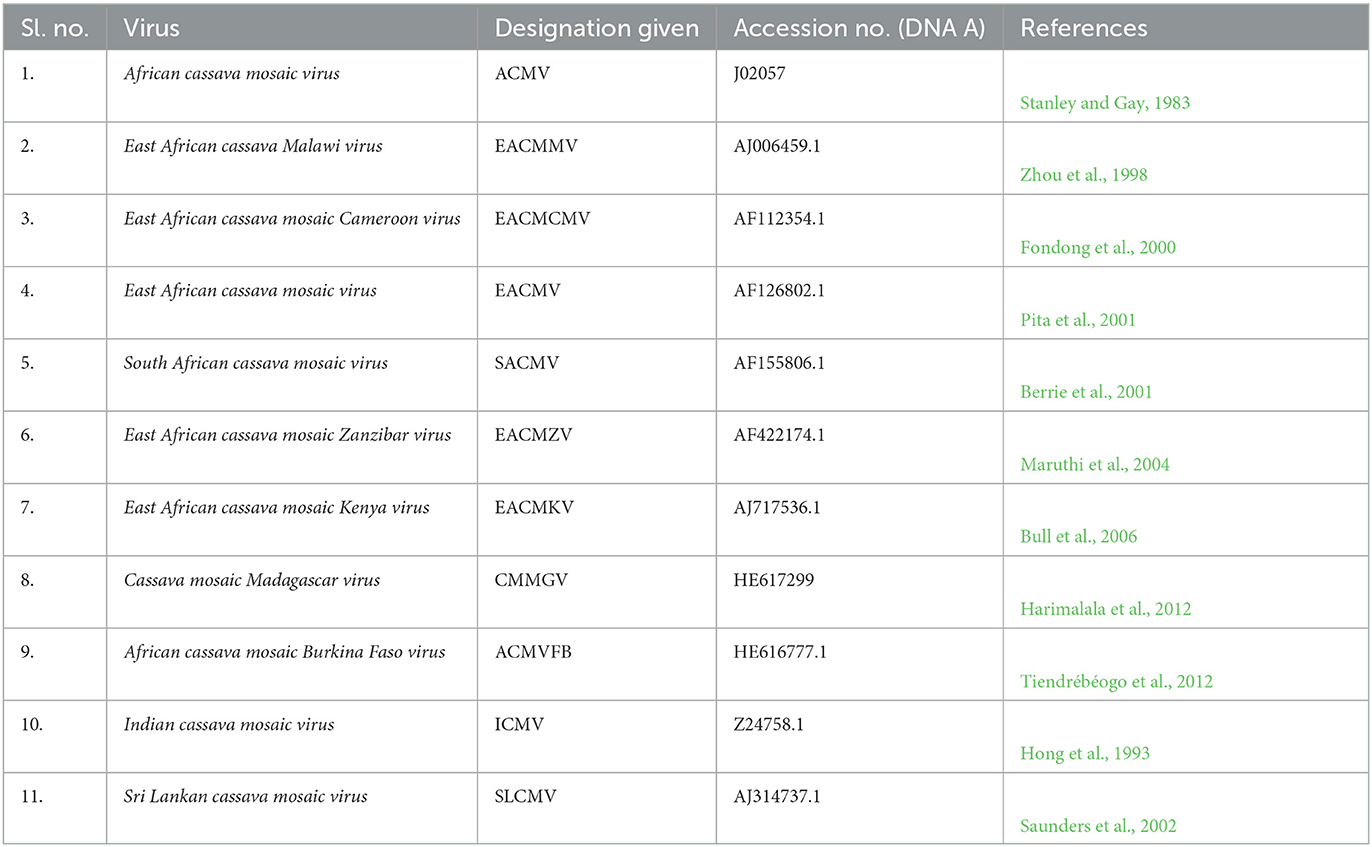
Table 1. List of virus species used in the phylogenetic analysis with accession numbers and designations used.
Weeds from the family Euphorbiaceae are shown to be potential reservoirs of cassava mosaic geminiviruses. Various ICMV and ACMV strains were found to be associated with Jatropha curcas mosaic disease in South India (ICMV-Dharwad) and Southeast Asia (ICMV-Singapore; Gao et al., 2010; Wang et al., 2014). Many SLCMV strains isolated from Southeast Asia exhibit a seven amino acid deletion at the carboxy-terminal of the replication-associated protein (Rep). This deletion is associated with decreased virulence of the virus compared with the virus harboring the full-length Rep protein, hence identified as the virulence determinant. To date, only one isolate identified from South Asia contains this truncated Rep, which shows a similarity to the SLCMV-Col strain isolated from Sri Lanka. However, the SLCMV-Col strain cloned from China did not show this mutation and exhibited hypervirulence. Further studies are needed to understand whether this is an adaptation of the virus to exist in the plant in the dormant stage without triggering host defense mechanisms (Siriwan et al., 2020; Wang et al., 2020).
Ecological conditions and farming practices influence the evolution of plant viruses. Evaluating nucleotide diversity in ACMV and EACMCV using high-throughput sequencing on a susceptible landrace Kibandameno, Aimone et al. (2021a) have shown that cropping practices can affect viral evolution and disease progression. Vegetative propagation over several generations has a significant impact on diversity at the nucleotide level and was unique to each of the viruses studied. Such changes were represented by biased base substitution toward C → T and G → A and the highest nucleotide diversity in the non-coding regions of DNA A and B, except the promoter regions. In addition to this, increased variations were also observed in coding regions and predicted to have deleterious effects on protein structure, owing to relaxed selection pressure over half a dozen vegetative passages. The effect of temperature changes or the presence of SEGS did not have a significant impact on the viruses (Aimone et al., 2021b).
Cassava mosaic geminivirus is vectored by the whitefly, Bemisia tabaci (Fauquet and Stanley, 2003). The species complex consists of ~35 genetically distinct cryptic species (Liu et al., 2012). In Africa, the virus transmission by whiteflies plays a major role in the spread of CMD (Legg et al., 2014). Field studies in India and Vietnam confirmed that white fly-mediated infections occurred only in younger leaves and accounted for a very less percentage compared with cutting-borne infections. The infections borne from diseased planting material occurred both in younger and older leaves (Jose et al., 2011; Minato et al., 2019). Transmission efficiencies of SLCMV by three whitefly species in Asia, namely Asia II 1, Mediterranean (MED), and Middle East-Asia Minor (MEAM1), vary significantly, and only Asia II 1 whiteflies transmit this virus efficiently (Chi et al., 2020). Temperature, rainfall, and wind affect the whitefly population (Fauquet and Fargette, 1990). The infection begins from the plants situated at the edge of a plot and gradually spreads inward to the newly planted cassava stands (Saokham et al., 2021). Monitoring the plants at regular intervals helps in preventing further spread of the disease in fields. Even though whiteflies contribute only to the secondary source of infection (Minato et al., 2019), their control is important to eradicate CMG infection in cassava and its spread to alternate hosts.
Cassava mosaic disease was first reported in Tanzania (Warburg, 1894), and it was proposed as a viral disease by Zimmermann (1906; reviewed in Patil and Fauquet, 2009). The virus was initially designated as Cassava latent virus, and it was sequenced and renamed as African cassava mosaic virus (Bock et al., 1981; Bock and Woods, 1983; Stanley and Gay, 1983). The incidence of CMD in India was initially reported by Abraham (1956) and Malathi et al. (1985) from Kerala. The virus was first cloned and sequenced from the samples collected from Kerala and named as Indian cassava mosaic virus (Hong et al., 1993). A second virus species, Sri Lankan cassava mosaic virus was identified from infected cassava samples from Sri Lanka (Saunders et al., 2002). Later, SLCMV was also identified from infected cassava material collected from Kerala (Dutt et al., 2005). CMD is also prevalent in major cassava-cultivating districts of Tamil Nadu with a disease incidence of more than 90% (Rajinimala et al., 2011). A survey conducted across 80 locations in nine districts of Tamil Nadu revealed that the majority of the samples were infected with SLCMV. Mixed infections of SLCMV and ICMV were also observed in a few samples. Sequence analysis displayed only a low level of genetic variability in these samples, observed as scattered single nucleotide changes (Kushawaha et al., 2018). In a recent survey conducted in West Bengal, India, an incidence of 61–92% was recorded with varying symptoms from mild chlorotic pattern to severe mosaic and leaf distortion. Sequence analysis showed that the isolates are similar to SLCMV from India (Adhikary et al., 2018). Unlike in Africa, where CMD causes remarkable crop losses, CMD has not yet turned out as a serious threat in India as the yield loss ranges only between 10 and 15% (Rothenstein et al., 2006).
Ever since the advent of CMD at Ratanakiri, Cambodia, in the year 2015 (Wang et al., 2016), SLCMV has been spreading rapidly in Southeast Asian countries. CMD was confirmed in farmers' fields in Vietnam and China in 2017 and Thailand in 2019 (Uke et al., 2018; Minato et al., 2019; Wang et al., 2020). During the field surveys, CMD has also been identified in Laos in 2020 (Chittarath et al., 2021). All the strains isolated so far from the South and Southeast Asia are closely related to previously characterized SLCMV isolates.
Minato et al. (2019) investigated the geographical distribution of SLCMV in Vietnam and Cambodia in the year 2016. In the systematic nationwide survey in Vietnam, 15 districts with the largest cultivating area were selected and a total of 2,640 samples were collected from 179 fields. Similarly, in Cambodia, 16 districts were included in the survey, collecting 3,840 samples from 240 fields. PCR-based diagnostics identified 49 plants representing nine fields, accounting for 2% of the total field samples collected. Among these positive plants, 14% did not show any visible symptoms, and the majority of positive samples belonged to two provinces in Eastern Cambodia-Ratanakiri and Stung Treng. However, no infection was detected in other samples collected from either country. The incidence of within-field level in two fields in Ratanakiri province was 25 and 37.5%, and other fields showed SLCMV incidence. The within-field level incidence in two fields in Ratanakiri province was 25% and 37.5%. In Stung Treng province it ranged from 6.3 to 56.3% among various fields, and, interestingly, it was much higher compared to Ratanakiri province where SLCMV was detected initially (Minato et al., 2019).
In Thailand, Cassava plantations in five provinces, along the Thailand–Cambodia border were surveyed during 2018–2019 in order to understand the incidence of CMD caused by SLCMV. The survey showed an overall incidence of 40% with the highest incidence of 80% in the Prachinburi area, followed by 43% in Sakaeo (Saokham et al., 2021).
During field surveys in Laos in the year 2020, eight fields were studied, of which the fields from the Kong district showed a considerable incidence of SLCMV. Only 6.7% of plants showed visible symptoms, but 18.3% of the plants were positive when tested by PCR. The visible symptoms were confined to older leaves, owing to the use of contaminated stakes, as the majority of cassava grown in Laos is from neighboring countries, where CMD was already reported (Chittarath et al., 2021).
The severity of the disease depends on various factors such as the age of the plant, cultivar, virus strain, and climatic conditions. The effect of SLCMV infection on the root yield of cassava was attributed to 16–33% when infected cuttings were used as a planting material (Uke et al., 2021). When 1–3-month-old plants showed moderate to severe mosaic symptoms, 5–7-month-old plants displayed only mild chlorosis. Whitefly-borne infections were less severe than infected-cutting-borne infections. In many cases, the infected plants remained asymptomatic. It was also found that genotypes, such as CMR-89 and Ryong 11, were susceptible to CMD (Saokham et al., 2021).
The studies conducted so far confirm that SLCMV is the major virus causing cassava mosaic disease in South and Southeast Asia, and it is more successful as a pathogen than ICMV. Though ICMV and SLCMV co-exist in cassava fields in the Indian sub-continent, in mixed infections, SLCMV suppresses ICMV and the latent ICMV can re-emerge when the recovery type cassava genotype recuperates from the SLCMV infection. This dominance of SLCMV might be the reason for the prevalence of SLCMV over ICMV in cassava fields (Karthikeyan et al., 2016). In Asia, no new cassava-infecting geminivirus species have been reported other than SLCMV and ICMV. The overlapping occurrence of SLCMV and ICMV in the cassava fields of South India and their presence in other host plants, which serve as reservoirs of various geminiviruses, demand immediate attention due to their recombinogenic properties. The long-distance spread of CMD mainly occurs due to the movement of contaminated plant materials, and the local spread can be affected by the whitefly vectors. Though ACMV has not yet been reported from Asia except for an odd incident in Oman (Khan et al., 2013), the recovery of ACMV from cotton plants in Pakistan hints at the risk of interspecific recombination. If Asian and African CMGs get an opportunity to co-exist in the same host, the emergence of highly virulent CMG could happen. Thus, proper surveillance measures are needed and should be imposed to curtail the spread of the disease.
Effective and rapid diagnostic methods need to be developed and implemented to detect CMGs and control their spread effectively. Molecular and serological methods are commonly used to detect the viruses. The serological techniques, such as double antibody sandwich ELISA (DAS-ELISA) and triple antibody sandwich ELISA (TAS-ELISA), have been successfully used to detect and distinguish between CMGs and to produce the first CMG distribution map for Africa (Sequeira and Harrison, 1982; Swanson and Harrison, 1994; Charoenvilaisiri et al., 2021). Malathi et al. (1989) produced ICMV antiserum and tested the field-infected plants by the DAS-ELISA method. Using TAS-ELISA, dot immunobinding assay (DIBA), and tissue blot immunoassay (TBIA), ICMV infection was detected both in symptomatic and asymptomatic cassava leaf samples (Makeshkumar and Nair, 2001; Hegde et al., 2010). ELISA is cost-effective and simple and hence adapted for the large-scale screening of the virus, but ELISA sometimes fails to detect asymptomatic infections.
Different types of PCRs such as immunocapture PCR (IC-PCR), spot capture PCR (SC-PCR), print capture PCR (PC-PCR), were tested and found that all are equally effective in detecting ICMV. The nucleic acid spot hybridization technique was also demonstrated to be more effective in the detection of ICMV using CP gene non-radioactive probe (Makeshkumar et al., 2005). Different sets of primers targeting the conserved AC1 and AV1 regions have been used by various groups in India and Southeast Asia to detect ICMV and SLCMV from field-infected samples (Dutt et al., 2005; Anitha et al., 2019; Uke et al., 2022). Multiplex PCR is useful in detecting mixed infections (Patil et al., 2005), and real-time PCR also has been developed for the detection and quantification of viruses from asymptomatic plants (Makeshkumar et al., 2005; Deepthi et al., 2012).
For the specific detection of SLCMV in Southeast Asia, most research groups have used primer sets that specifically target AV1 (CP). Coat protein-targeting primers, reported by Dutt et al. (2005) were used to detect SLCMV in cassava samples collected from Cambodia (Wang et al., 2016). On the other hand, Minato et al. (2019) reported the use of a PCR-primer set targeting the whole ORF of the AC1 gene (Duraisamy et al., 2013) to screen for SLCMV in Cambodia and Vietnam, while Uke et al. (2018) used the rolling circle amplification (RCA) process to characterize the virus detected in Vietnam.
Loop-mediated isothermal amplification (LAMP) can detect viruses even from stem tissues and it does not require a thermal cycler. The sensitivity of the LAMP assay is 10,000 times higher than the conventional PCR. A commercial LAMP kit containing dried reagents is stable at room temperature and can be used for the on-site detection of CMGs (Uke et al., 2022).
The advent of next-generation sequencing (NGS) and third-generation sequencing technologies has taken the virus diagnosis far ahead. For example, shotgun metagenomic analysis does not require any previous knowledge of viral sequences. Millions of RNA/DNA fragments can be sequenced simultaneously, allowing the diagnosis of mixed infections of all viruses. But this method is associated with high costs. Recently, Leiva et al. published a nanopore-based full genome sequence of SLCMV from Thailand (Leiva et al., 2020). Further research on affordable next-generation platforms could help in the early diagnosis of the virus and thereby reduce crop losses. The multispectral imaging system (MSIS) consists of an electron multiplying charge-coupled device (EMCCD) camera that can detect changes in the red:far-red fluorescence image ratio due to CMD stress at an early stage. The technique is highly useful for monitoring the health of crops from proximal sensing platforms (Raji et al., 2015). An active multispectral imaging (A-MSI) device combined with machine learning and CIDER sequencing was also used for the early detection of cassava-infecting viruses in a cost-effective way (Mehta et al., 2020; Peng et al., 2022).
Conventional breeding is considered the main egress from the problems faced by cassava growers worldwide (Lynam and Byerlee, 2017). Cassava breeding faces several challenges such as long breeding cycles as long as 8 years, low multiplication rate, and most importantly, the use of heterozygous progenitors, which detains conventional back-crossing (Ceballos et al., 2004, 2020).
Identifying cassava mosaic geminivirus-resistant genetic resources for breeding has been a major focus in the battle against CMD (Malik et al., 2020). Over the past few decades, several conventional breeding programs were carried out, in order to impart resistance against CMD, both in Africa and Asia. To date, three sources of resistance, named CMD1, CMD2, and CMD3, have been identified and partially characterized and are also being used for the development of CMD-resistant cassava varieties (Rey and Vanderschuren, 2017).
CMD1 is a recessive polygenic locus initially reported in a wild relative of cassava, Manihot glaziovii Mull. Arg. (Nichols, 1947; Fregene et al., 2001). Conversely, CMD2 resistance, a dominant single genetic locus on chromosome 12, was identified in tropical M. esculenta (TME) populations cultivated as landraces across West Africa (Akano et al., 2002; Okogbenin et al., 2012). The varieties carrying CMD2 resistance locus have been the main source for CMD resistance breeding programs in Africa, Latin America, and Asia, as they exhibited a high level of resistance to almost all cassava mosaic begomoviruses. The loss of CMD2 type of resistance was reported (LCR phenotypes) upon regeneration during de novo morphogenesis (Beyene et al., 2016). However, it was not observed in the varieties developed through conventional breeding programs. Recently, Lim et al. (2022) demonstrated that CMD2 and LCR phenotypes have a genomic basis and co-localize on the cassava genome. An independently evolved non-synonymous single-nucleotide polymorphism (SNP) variant in the delta subunit 1 of polymerase (MePOLD1) was found segregating with CMD resistance, which is considered a novel type of resistance protein in plants. MePOLD might have been selected as a chimeric clonal variant under several selections by African farmers, and its monogenic dominant nature enabled their use in breeding programs in Africa, India, and Southeast Asia (Rabbi et al., 2014). This is important information for breeders who can use the SNPs as markers when introgressing CMD2 into non-resistant varieties. CMD3 is produced by the crossing between TME and TMS genotypes (Lokko et al., 2005). However, the genetic analysis of a cultivar named TMS 97/2205 and its offspring revealed that CMD3 is a new quantitative trait locus (QTL). It provides putative resistance to CMD and is not linked to CMD2 (Okogbenin et al., 2012).
The pioneering work in molecular breeding is the identification of SSR and RFLP markers associated with putative single dominant resistance gene CMD2 (Akano et al., 2002; Fregene et al., 2006). A high-density map of cassava sequencing revealed a single putative chromosomal location of CMD2-associated dominant resistance (Rabbi et al., 2014; Wolfe et al., 2016).
Cassava breeding programs in India were initiated by Central Tuber Crop Research Institute (CTCRI) with its rich cassava germplasm, holding several indigenous and exotic landraces and breeding lines including wild relatives of cassava (Malik et al., 2020). The collaboration of CTCRI with CIAT since 1996, accepting different combinations of F1 seeds, has been instrumental in extensive breeding programs in the country (Abraham et al., 2000; Legg et al., 2015; Malik et al., 2020).
Subsequent to several evaluation trials of MNga-1 (a breeding line from IITA) in the field conditions of India, CTCRI has released a variety named Sree Padmanabha in the state of Tamil Nadu, which showed high-level resistance to ICMV and SLCMV, and has been used in developing resistant clones through intervarietal hybridization (Nair et al., 1998; Unnikrishnan et al., 2002; Abraham et al., 2006; George et al., 2012). In addition, evaluation of the MNga-1 seedling population yielded CMR1 and CMR129 with CMD resistance and high starch content (Unnikrishnan et al., 2011). The West African landraces with CMD2 resistance, introduced in India, are used to generate heterotic hybrids through inbreeding and are being evaluated (Sheela et al., 2012). Interspecific breeding of cassava with M. caerulescens and other species resulted in the prebreeding population of CMD-resistant clones with a high level of resistance to CMD and are being used as donor parents in the breeding programs (Sheela et al., 2002, 2004; Unnikrishnan et al., 2002; Malik et al., 2020). Over the past two decades, ICAR-CTCRI has evaluated several resistant sources of CMD, including interspecific back-cross lines, inbred lines, and introductions from CIAT, Cali, and Columbia. ICAR-CTCRI and AICRP(TC) have released five CMD-resistant varieties, namely Sree Reksha, Sree Sakthi, Sree Suvarna, PDP-CMR-1, and Sree Kaveri, which are very popular among farmers (Malik et al., 2020).
The long breeding cycle of cassava is a major limiting factor for developing and implementing breeding programs. It has been found that the over-expression of the Arabidopsis FLOWERING LOCUS T (FT) gene encoding florigen improves floral development in cassava (Adeyemo et al., 2017). Hence, the utilization of FT gene orthologs of cassava in the breeding programs could efficiently lessen the long breeding cycle, and efforts are underway.
Various control strategies have been exploited in order to control the disease, and to some extent, planting resistant varieties had a promising effect. The development of disease-free planting material was another focus area in tackling CMD in Southeast Asia. In 2016, the Science and Technology Research Partnership for Sustainable Development (SATREPS) project funded by the Japanese Government was launched for the overall management of cassava diseases. It is a collaborative research program between the Agricultural Genetics Institute (AGI), Plant Protection Research Institute (PPRI), Hung Loc Agricultural Research Center (HLARC), Nong Lam University (NLU), Vietnam; University of Battambang (UBB), Cambodia, and the Rayong Field Crops Research Center (RFCRC), Thailand. This project was instrumental in the development of diagnostic tools, pest management technologies, and novel molecular breeding strategies. Production and dissemination of healthy planting materials (Tokunaga et al., 2018) were another major focus area of the project.
The International Center for Tropical Agriculture (CIAT) and The International Institute of Tropical Agriculture (IITA) were involved in the development and introduction of CMD-resistant materials in Thailand and Vietnam subsequent to the outbreak of CMD in these countries and Thailand. In the SATREPS project, a breeding line (C-33) that contained CMD2 loci was introduced in the field of prevailing CMD to serve as a potential source against SLCMV infection. This line showed normal growth and remained asymptomatic under the high-SLCMV pressure in the fields of Tay Ninh province, Vietnam, while the Asian cultivars exhibited CMD symptoms 3 months after planting. In addition to this, it was also shown that SLCMV was not transmitted when C-33 plants were used as a rootstock in grafting experiments (Vu et al., 2020), suggesting that the CMD2 as a resistant source for breeding cassava, which is suitable for cultivation in Southeast Asian countries (Uke et al., 2021). These approaches were useful to address the CMD problem in Southeast Asia and to generate new cassava varieties with high resistance to CMD. These collaborative projects could strengthen the efforts to minimize CMD and offer significant contributions to food security in Southeast Asian countries.
Advancements in the field of plant genetic engineering have unveiled a new arena in the management of plant diseases. Various strategies have been successfully employed in engineering disease resistance in plants. The initial trials in the management of plant viruses were based on the concept of pathogen-derived resistance (PDR; Hamilton, 1980; Sanford and Johnston, 1985; Dong and Ronald, 2019), and several models have been shown to be effective against a broad range of viruses. This is based on the fact that when transgenic plants express viral genes or viral genomic regions, it may lead to improved resistance against invading viruses (Lindbo and Falk, 2017; Rosa et al., 2018). Pioneer studies based on the concept of PDR were focusing on protein-mediated resistance, involving the use of full-length functional protein coding sequences that provide a moderate level of protection against a wide range of related viruses. PDR generated by transgene expression largely depends on the sequence homology of the viruses to be targeted (Ratcliff et al., 1997). However, the PDR was not restricted to such fully functional protein alone, but the presence of the non-translatable sequence could also attenuate the virus infection to a certain level and was later identified as an RNA-mediated silencing mechanism (Lindbo and Dougherty, 1992). The discovery of RNA interference has explained the mechanism of antiviral defense both in protein/non-translated sequence-mediated resistance (Lindbo and Falk, 2017; Rosa et al., 2018). Small RNAs (sRNAs) regulate the gene expression of target genes by mRNA degradation or translational repression. Similarly, the small interfering RNAs (siRNAs) formed from the viral genes/genomic regions through their expression in the host can target the invading viral genes as they have extensive base-pair complementarity with virus genes. These sequences can either be in sense or antisense orientation (Lin et al., 2019).
RNAi strategies have been effectively employed in conferring resistance against many viruses, including cassava mosaic geminiviruses. The capability of the ACMV-derived AC1 gene to impart strong resistance against CMD is a classic example of utilizing RNAi technology in Cassava (Chellappan et al., 2004). The AC1 gene from ACMV, when expressed constitutively in the host, conferred resistance to ACMV and related species EACMV and also to SLCMV, although SLCMV is considered a far distant species as per the species demarcation threshold described by Brown et al. (2015). Cassava plants with increased ACMV resistance using improved antisense RNA technology by targeting the viral mRNAs of Rep (AC1), TrAP (AC2), and REn (AC3) were developed by Zhang and co-workers (Zhang et al., 2005). The transgenic ACMV-resistant plants showed reduced viral DNA accumulation in their infected leaves, proving the high efficacy of antisense RNAs against viral mRNAs encoding essential non-structural proteins.
Vanderschuren et al. (2007) developed transgenic cassava plants producing siRNAs from an intron-containing dsRNA homologous to the common region containing a bidirectional promoter of DNA-A of ACMV. Symptom manifestations were attenuated in two of the transgenic lines, with reduced accumulation of viral DNA in their leaves, compared to wild plants. It was concluded that in transgenic cassava plants expressing hairpin RNA, the natural RNA silencing mechanism against the virus was triggered by the formation of virus-derived dsRNAs prior to infection. In another study, the hairpin RNA turnover was correlated with the level of resistance in transgenic lines. The significance of the load of hairpin-derived small RNAs in geminivirus resistance was demonstrated in CMD-susceptible cassava cultivar (TMS60444) expressing a hairpin dsRNA homologous to a conserved region of the replication-associated protein coding sequence (Rep/AC1; Vanderschuren et al., 2009). Transgenic cassava lines, with a high turnover of AC1-homologous small RNAs, showed a corresponding increase in immunity when the viral load was elevated.
Resistance to SLCMV was imparted through RNA silencing in the KU50 line, an important cultivar used for non-food purposes (Ntui et al., 2015). Transgenic KU50 lines expressing dsRNA homologous to the region between AV2 and AV1 in the DNA A of SLCMV displayed high levels of resistance to SLCMV compared to the wild-type plants without any growth impairment. The use of the coat protein gene in imparting resistance against SLCMV was demonstrated by Gogoi et al. (2019), in N. benthamiana. The coat protein gene (AV1) was cloned both in sense and antisense orientation under the control of the CaMV 35S promoter, and the transgenic N. benthamiana plants were developed. Upon challenging agroinfectious clones of SLCMV, the plants showed delayed symptom expression and a high level of resistance against SLCMV.
RNAi technology is currently being used extensively in engineering virus resistance. However, its efficacy is limited when targeting a virus species with <90% identity to the transgene-derived dsRNA. Hence, profiling virus sequence diversity has become a prerequisite to the development of RNAi constructs. A new full-length circular DNA sequencing method, termed circular DNA enrichment sequencing (CIDER-seq) has been developed and effectively demonstrated for the assessment of virus sequence diversity in transgenic plants. Intriguingly, it showed a considerable shift of the virus species toward a population with less homology to the transgene-derived dsRNA (Mehta et al., 2019a).
The importance of cassava improvement through genetic engineering is that it helps to overcome the high heterozygosity of cassava plants, the trait separation occurring in traditional breeding, and the faster achievement of improved target traits (Liu et al., 2011). Hence, genetic engineering is considered the most important tool for the improvement of cassava. The challenges in delivering new products to farmers using this system are either biological or associated with regulation, approval, and perception of crops enhanced through genetic engineering tools (Bart and Taylor, 2017).
In order to subvert the host immune response, pathogens utilize their virulence factors in association with certain host processes to complete pathogenesis (Mandadi and Scholthof, 2013; Toruño et al., 2016). In instances where the pathogen depends on a singular host-encoded susceptibility gene or small gene families, editing of such candidate genes could stall the disease progression. Genome editing refers to the process of inducing nucleotide modifications at targeted loci in the genome of organisms (Hsu et al., 2014). This technique utilizes nuclease-mediated double-stranded DNA breaks in the target sequence coupling with cellular DNA repair mechanism, homology-directed repair (HDR), and non-homologous end joining (NHEJ) breaks that introduce modification at the target site (Puchta et al., 1996; Gorbunova and Levy, 1997). Current nuclease-based genome editing tools focus on techniques based on zinc-finger nucleases (ZFNs), transcription activator-like effector nucleases (TALENs), and clustered regularly interspaced short palindromic repeats/CRISPR-associated (CRISPR/Cas) (Kim et al., 1996; Christian et al., 2010; Cong et al., 2013; Mali et al., 2013; Gaj et al., 2016).
CRISPR-/Cas9-based genome editing was first reported in A. thaliana (Li et al., 2013) and N. benthamiana (Nekrasov et al., 2013) and can be used in managing CMD as well. In cassava, CRISPR/Cas9-mediated editing of the phytoene desaturase (MePDS) gene in the genome of TME 204 and cv TM60444 cassava cultivars were reported (Odipio et al., 2017). Although several trials have been carried out with various genes including the disease-susceptibility gene, MeSWEET10a, using the CRISPR/Cas9 system, the homozygous mutations in the T0 generation were insignificant. However, in A. thaliana, when Cas9 was expressed under YAO promoter (pYAO:hSp Cas9 binary vector), the number of targeted and homozygous mutations was increased compared to the cauliflower mosaic virus (CaMV) 35S promoter. Similarly, the use of the pYAO: hSpCas9 binary vector to knock out the MePDS gene in cassava improved the efficiency of homozygous mutations. This system is highly promising for use in cassava gene editing (Wang et al., 2022).
Conversely, being DNA viruses, the CRISPR/Cas system can be used to target the CMG genome itself, as demonstrated successfully in N. benthamiana and A. thaliana, against geminiviruses (Ali et al., 2015, 2016; Baltes et al., 2015; Ji et al., 2015). A likely problem associated with this system is the evolution of editing resistance of viral genomes and the formation of truncated proteins which may be functional, hence the system becomes unrewarding (Mehta et al., 2019b). An alternative to this is the targeting of non-coding motifs located in the intergenic regions of the viruses that are crucial for replication and has been demonstrated with the LIR motif, wherein reduced geminiviral accumulation was observed (Ali et al., 2016). In addition to this, targeting multiple loci of the same virus was also more beneficial (Ali et al., 2015; Baltes et al., 2015).
The adaptability of cassava in poorly fertile soils, its drought tolerance, and its relaxed harvesting period make the crop very attractive in tropical regions. Cassava cultivation has remarkably increased in South and Southeast Asia in recent years owing to its demand in use as a source for biofuel production apart from starch and its derivatives. The expansion of crop area along with a large demand for planting materials imposes threats of the virus being transported across the region and its further spread. Transport of infected materials among different countries in Southeast Asia makes it nearly impossible to find cassava fields devoid of CMD. In South and Southeast Asia, most of the cassava cultivating fields are mainly affected by SLCMV; ICMV is also detected in a few fields of South India. The cassava-infecting begomoviruses have evolved rapidly due to their aggressiveness in recombining with related viruses and infect alternate hosts. Primarily, training farmers for the visual identification of the symptoms and creating awareness among them to use disease-free stem cutting for planting could help to restrict the disease's spread. Deployment of cultivars resistant to SLCMV developed through conventional breeding has been a major focus area in the management of CMD. Several genetic engineering strategies were proven to be effective against SLCMV, but the concerns of possible environmental and health hazards are lessening their acceptance. However, novel marker-free genome editing technologies appear to be more promising, and the development of farmer-preferred Asian cultivars resistant to SLCMV is underway. Successful deployment of efficient, and economical diagnostic methods for the early detection of viruses and routine surveillance will certainly help in devising proper strategies to contain the disease. This will provide clues for detecting the emergence of any new strain. There is an urgent need for a coordinated effort by all countries affected by this disease to monitor the disease's progression and enforce strict prohibitions on the movement of infected planting materials to areas that are currently free from CMD. As an immediate solution, large-scale multiplication and distribution of resistant cultivars will help growers in reducing the inoculum source and overcoming the present crisis to a considerable extent.
PH and TR wrote the manuscript. TM and MS evaluated and edited the manuscript. All authors contributed to the article and approved the submitted version.
The Director, ICAR-Central Tuber Crops Research Institute, is thankfully acknowledged for extending all the facilities. Kerala Higher Education Council, Govt of Kerala, India has granted postdoctoral fellowship to PH and TR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abraham, A. (1956). Tapioca cultivation in India. Indian Council of Agricultural Research. New Delhi, India. Farm Bull. 17, 20.
Abraham, K., Edison, S., Unnikrishnan, M., Sheela, M. N., Vimala, B., Sreekumari, M. T., et al. (2006). Tuber crops varieties released by CTCRI, Thiruvananthapuram: Technical Bulletin No. 24. CTCRI. p. 64.
Abraham, K., Nair, S. G., and Naskar, S. K. (2000). “Cassava breeding and varietal dissemination in India-Major achievements during past 25-30 years. Cassava's Potentials in the 21st Century: Present Situation and Future Research and Development Needs,” in Sixth Asian Cassava Research Workshop (Ho Chi Minh City), 174–184.
Adeyemo, O. S, Chavarriaga, P., Tohme, J., Fregene, M., and Davis, S.J. (2017). Overexpression of Arabidopsis FLOWERING LOCUS T (FT) gene improves floral development in cassava (Manihot esculenta, Crantz). PLoS ONE. 12, e0181460. doi: 10.1371/journal.pone.0181460
Adhikary, N. K., Kumar, M., and Tarafdar, J. (2018). Occurrence of Sri Lankan Cassava Mosaic Virus (SLCMV) and its characterization in West Bengal. Int. J. Curr. Microbiol. App. Sci. 7, 2887–2893. doi: 10.20546/ijcmas.2018.701.344
Aimone, C. D., De León, L., Dallas, M. M., Ndunguru, J., Ascencio-Ibáñez, J. T., and Hanley-Bowdoin, L. (2021b). A new type of satellite associated with cassava mosaic begomoviruses. J. Virol. 13, e0043221. doi: 10.1101/2021.03.11.434950
Aimone, C. D., Lavington, E., Hoyer, J. S., Deppong, D. O., Mickelson-Young, L., and Jacobson, A. (2021a). Population diversity of cassava mosaic begomoviruses increases over the course of serial vegetative propagation. J. Gen. Virol. 102, e001622. doi: 10.1099/jgv.0.001622
Akano, O., Dixon, O., Mba, C., Barrera, E., and Fregene, M. (2002). Genetic mapping of a dominant gene conferring resistance to cassava mosaic disease. Theor. Appl. Genet. 105, 521–525. doi: 10.1007/s00122-002-0891-7
Ali, Z., Abul-faraj, A., Li, L., Ghosh, N., Piatek, M., Mahjoub, A., et al. (2015). Efficient virus-mediated genome editing in plants using the CRISPR/Cas9 system. Mol. Plant 8, 1288–1291. doi: 10.1016/j.molp.2015.02.011
Ali, Z., Ali, S., Tashkandi, M., Zaidi, S. S., and Mahfouz, M. M. (2016). CRISPR/Cas9-mediated immunity to geminiviruses: differential interference and evasion. Sci. Rep. 6, 26912. doi: 10.1038/srep26912
Allem, A. C. (1994). The origin of Manihot esculenta Crantz (Euphorbiaceae). Genet. Resour. Crop Evol. 41, 133–150. doi: 10.1007/BF00051630
Anitha, J., Edison, S., and Makeshkumar, T. (2019). Occurrence and distribution of Sri Lankan cassava mosaic virus in the stems of cassava. J. Root Crops 45, 32–40. Available online at: https://journal.isrc.in/index.php/jrc/article/view/552
Bach, J., and Jeske, H. (2014). Defective DNAs of Beet curly top virus from long-term survivor sugar beet plants. Virus Res. 183, 89–94. doi: 10.1016/j.virusres.2014.01.028
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1, 15145. doi: 10.1038/nplants.2015.145
Bart, R. S., and Taylor, N. J. (2017). New opportunities and challenges to engineer disease resistance in cassava, a staple food of African small-holder farmers. PLoS Pathog. 11, e1006287. doi: 10.1371/journal.ppat.1006287
Berrie, L. C., Rybicki, E. P., and Rey, M. E. (2001). Complete nucleotide sequence and host range of South African cassava mosaic virus: further evidence for recombination amongst begomoviruses. J. Gen. Virol. 82, 53–58. doi: 10.1099/0022-1317-82-1-53
Beyene, G., Chauhan, R. D., Wagaba, H., Moll, T., Alicai, T., Miano, D., et al. (2016). Loss of CMD2-mediated resistance to cassava mosaic disease in plants regenerated through somatic embryogenesis. Mol. Plant Pathol. 17, 1095–1110. doi: 10.1111/mpp.12353
Bock, K. R., Guthrie, E. J., and Figueiredo, G. (1981). A strain of Cassava latent virus occurring in coastal districts of Kenya. Ann. Appl. Biol. 99, 151–159. doi: 10.1111/j.1744-7348.1981.tb05142.x
Bock, K. R., Guthrie, E. J., and Meredith, G. (1978). Distribution, host range, properties and purification of Cassava latent virus, a geminivirus. Ann. Appl. Biol. 90, 361–367. doi: 10.1111/j.1744-7348.1978.tb02644.x
Bock, K. R., and Woods, R. D. (1983), The etiology of African cassava mosaic disease. Plant Dis. 67, 994–995. doi: 10.1094/PD-67-994
Borah, B. K., and Dasgupta, I. (2012). PCR-RFLP analysis indicates that recombination might be a common occurrence among the cassava infecting begomoviruses in India. Virus Genes 45, 327–332. doi: 10.1007/s11262-012-0770-5
Briddon, R. W., and Stanley, J. (2006). Subviral agents associated with plant single stranded DNA viruses. Virology 344, 198–210. doi: 10.1016/j.virol.2005.09.042
Brown, J. K., Zerbini, F. M., Navas-Castillo, J., Moriones, E., Ramos-Sobrinho, R., Silva, J. C., et al. (2015). Revision of begomovirus taxonomy based on pairwise sequence comparisons. Arch. Virol. 160, 1593–1619. doi: 10.1007/s00705-015-2398-y
Bull, S. E., Briddon, R. W., Sserubombwe, W. S., Ngugi, K., Markham, P. G., and Stanley, J. (2006). Genetic diversity and phylogeography of cassava mosaic viruses in Kenya. J. Gen. Virol. 87, 3053–3065. doi: 10.1099/vir.0.82013-0
Ceballos, H., Iglesias, C. A., Pérez, J. C., and Dixon, A. G. (2004). Cassava breeding: opportunities and challenges. Plant Mol. Biol. 56, 503–516. doi: 10.1007/s11103-004-5010-5
Ceballos, H., Rojanaridpiched, C., Phumichai, C., Becerra, L. A., Kittipadakul, P., and Iglesias, C. (2020). Excellence in cassava breeding: perspectives for the future. Crop Breed Genet. Genom. 2, e200008. doi: 10.20900/cbgg20200008
Charoenvilaisiri, S., Seepiban, C., Kumpoosiri, M., Rukpratanporn, S., Warin, N., Phuangrat, B., et al. (2021). Development of a triple antibody sandwich enzyme-linked immunosorbent assay for cassava mosaic disease detection using a monoclonal antibody to Sri Lankan cassava mosaic virus. Virol. J. 18, 100. doi: 10.1186/s12985-021-01572-6
Chellappan, P., Masona, M. V., Vanitharani, R., Taylor, N. J., and Fauquet, C. M. (2004). Broad spectrum resistance to ssDNA viruses associated with transgene induced gene silencing in cassava. Plant Mol. Biol. 56, 601–611. doi: 10.1007/s11103-004-0147-9
Chi, Y., Pan, L. L., Bouvaine, S., Fan, Y. Y., Liu, Y. Q., Liu, S. S., et al. (2020). Differential transmission of Sri Lankan cassava mosaic virus by three cryptic species of the whitefly Bemisia tabaci complex. Virology 15, 141–149. doi: 10.1016/j.virol.2019.11.013
Chittarath, K., Jimenez, J., Vongphachanh, P., Leiva, A. M., Sengsay, S., Lopez-Alvarez, D., et al. (2021). First report of Sri Lankan cassava mosaic virus and cassava mosaic disease in Laos. Plant Dis. 20, 1868. doi: 10.1094/PDIS-09-20-1868-PDN
Christian, M., Cermak, T., Doyle, E. L., Schmidt, C., Zhang, F., Hummel, A., et al. (2010). Targeting DNA double-strand breaks with TAL effector nucleases. Genetics 186, 757–761. doi: 10.1534/genetics.110.120717
Cong, L., Ran, F. A., Cox, D., Lin, S., Barretto, R., Habib, N., et al. (2013). Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823. doi: 10.1126/science.1231143
Crespo-Bellido, A., Hoyer, J. S., Dubey, D., Jeannot, R. B., and Duffy, S. (2021). Interspecies recombination has driven the macroevolution of cassava mosaic begomoviruses. J. Virol. 21, e0054121. doi: 10.1128/JVI.00541-21
Cui, X., Tao, X., Xie, Y., Fauquet, C. M., and Zhou, X. (2004). A DNAβ associated with Tomato yellow leaf curl China virus is required for symptom induction. J. Virol. 78, 13966–13974. doi: 10.1128/JVI.78.24.13966-13974.2004
Deepthi, D. C., Makesh kumar, T., Unnikrishnan, M., and Winter, S. (2012). “Quantitative analysis of geminiviruses in recovery phenotypes of cassava,” in Second Scientific Conference of the Global Cassava Partnership for the 21st Century (GCP-21-II), Kampala, Uganda, June 18–22 2012. Kampala.
Dong, O. X., and Ronald, P. C. (2019). Genetic engineering for disease resistance in plants: recent progress and future perspectives. Plant Physiol. 180, 26–38. doi: 10.1104/pp.18.01224
Duraisamy, R., Natesan, S., Muthurajan, R., Gandhi, K., Lakshmanan, P., Karuppusamy, N., et al. (2013). Molecular studies on the transmission of Indian cassava mosaic virus (ICMV) and Sri Lankan cassava mosaic virus (SLCMV) in cassava by Bemisia tabaci and cloning of ICMV and SLCMV replicase gene from cassava. Mol. Biotechnol. 53, 150–158. doi: 10.1007/s12033-012-9503-1
Dutt, N., Briddon, R. W., and Dasgupta, I. (2005). Identification of a second begomovirus, Sri Lankan cassava mosaic virus causing cassava mosaic disease in India. Arch. Virol. 150, 2101–2108. doi: 10.1007/s00705-005-0579-9
Eini, O., Dogra, S. C., Dry, I. B., and Randles, J. W. (2012). Silencing suppressor activity of a begomovirus DNA β encoded protein and its effect on heterologous helper virus replication. Virus Res. 167, 97–101. doi: 10.1016/j.virusres.2012.03.012
Fauquet, C., and Fargette, D. (1990). African cassava mosaic virus: etiology, epidemiology, and control. Plant Dis. 74, 404–411. doi: 10.1094/PD-74-0404
Fauquet, C. M., and Stanley, J. (2003). Geminivirus classification and nomenclature: progress and problems. Ann. Appl. Biol. 142, 165–189. doi: 10.1111/j.1744-7348.2003.tb00241.x
Fiallo-Olive, E., Martinez-Zubiaur, Y., Moriones, E., and Navas-Castillo, J. (2012). A novel class of DNA satellites associated with New World begomoviruses. Virology 426, 1–6. doi: 10.1016/j.virol.2012.01.024
Fondong, V. N., Pita, J. S., Rey, M. E. C., De Kochko, A., Beachy, R. N., and Fauquet, C. M. (2000). Evidence of synergism between African cassava mosaic virus and a new double-recombinant geminivirus infecting cassava in Cameroon. J. Gen. Virol. 81, 287–297. doi: 10.1099/0022-1317-81-1-287
Fregene, M., Morante, N., Sanchez, T., Marin, J., Ospina, C., Barrera, E., et al. (2006). Molecular markers for introgression of useful traits from wild Manihot relatives of cassava, marker-assisted selection (MAS) of disease and root quality traits. J. Root Crops 32, 1–31.
Fregene, M., Okogbenin, E., Mba, C., Angel, F., Suarez, M. C., Janneth, G., et al. (2001). Genome mapping in cassava improvement: challenges, achievements and opportunities. Euphytica 120, 159–165. doi: 10.1023/A:1017565317940
Gaj, T., Sirk, S. J., Shui, S. L., and Liu, J. (2016). Genome-editing technologies: principles and applications. Cold Spring Harb. Perspect. Biol. 8, a023754. doi: 10.1101/cshperspect.a023754
Gao, S., Qu, J., Chua, N. H., and Ye, J. (2010). A new strain of Indian cassava mosaic virus causes a mosaic disease in the bio-diesel crop Jatropha curcas. Arc. Virol. 155, 607–612. doi: 10.1007/s00705-010-0625-0
George, J., Suresh Kumar, P., and Unnikrishnan, M. (2012). Descriptions of recommended released varieties under AICRP on tuber crops 1975-2011: Technical Bulletin No. 51. Trivandrum: CTCRI, 122.
Gogoi, A., Kaldis, A., Dasgupta, I., Borah, B., and Voloudakis, A. (2019). Sense- and antisense-mediated resistance against Sri Lankan cassava mosaic virus SLCMV) in Nicotiana benthamiana. Biol. Plant. 2019, 79. doi: 10.32615/bp.2019.079
Gorbunova, V., and Levy, A. A. (1997). Non-homologous DNA end joining in plant cells is associated with deletions and filler DNA insertions. Nucl. Acids Res. 25, 4650–4657. doi: 10.1093/nar/25.22.4650
Hamilton, R. (1980). “Defenses triggered by previous invaders: viruses,” in Plant Disease: An Advance Treatise, eds J. G. Horsfall and E. B. Cowling (New York, NY: Academic Press), 279–230.
Hanley-Bowdoin, L., Bejarano, E. R., Robertson, D., and Mansoor, S. (2013). Geminiviruses: masters at redirecting and reprogramming plant processes. Nat. Rev. Microbiol. 11, 777–788. doi: 10.1038/nrmicro3117
Harimalala, M., De Bruyn, A., Hoareau, M., Andrianjaka, A., Ranomenjanahary, S., Reynaud, B., et al. (2013). Molecular characterization of a new alphasatellite associated with a cassava mosaic geminivirus in Madagascar. Arch. Virol. 158, 1829–1832. doi: 10.1007/s00705-013-1664-0
Harimalala, M., Lefeuvre, P., De Bruyn, A., Tiendrébéogo, F., Hoareau, M., Villemot, J., et al. (2012). A novel cassava-infecting begomovirus from Madagascar: Cassava mosaic Madagascar virus. Arch. Virol. 157, 2027–2030. doi: 10.1007/s00705-012-1399-3
Hegde, V., Jeeva, M. L., Makeshkumar, T., Misra, R. S., and Veena, S. S. (2010). Diagnostic Techniques for Diseases of Tropical Tuber Crops. Technical Bulletin Series 50. (Thiruvananthapuram: Central Tuber Crops Research Institute), 56.
Hong, Y. G., Robinson, D. J., and Harrison, B. D. (1993). Nucleotide sequence evidence for the occurrence of three distinct whitefly transmitted geminiviruses in cassava. J. Gen. Virol. 74, 2437–2443. doi: 10.1099/0022-1317-74-11-2437
Hsu, P. D., Lander, E. S., and Zhang, F. (2014). Development and applications of CRISPR-Cas9 for genome engineering. Cell 157, 1262–1278. doi: 10.1016/j.cell.2014.05.010
Ji, X., Zhang, H., Zhang, Y., Wang, Y., and Gao, C. (2015). Establishing a CRISPR-Cas-like immune system conferring DNA virus resistance in plants. Nat. Plants 1, 15144. doi: 10.1038/nplants.2015.144
Jose, A., Makeshkumar, T., and Edison, S. (2011). Survey of cassava mosaic disease in Kerala. J. Root Crops 37, 41–47. Available online at: https://journal.isrc.in/index.php/jrc/article/view/8
Karthikeyan, C., Patil, B. L., Borah, B. K., Resmi, T. R., Turco, S., Pooggin, M. M., et al. (2016). Emergence of a latent Indian cassava mosaic virus from cassava which recovered from infection by a non-persistent Sri Lankan cassava mosaic virus. Viruses 8, 264. doi: 10.3390/v8100264
Khan, A. J., Akhtar, S., Al-Matrushi, A. M., Fauquet, C. M., and Briddon, R. W. (2013). Introduction of East African cassava mosaic Zanzibar virus to Oman harks back to “Zanzibar, the capital of Oman”. Virus Genes 46, 195–198. doi: 10.1007/s11262-012-0838-2
Kim, Y. G., Cha, J., and Chandrasegaran, S. (1996). Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc. Natl. Acad. Sci. U. S. A. 93, 1156–1160. doi: 10.1073/pnas.93.3.1156
Kushawaha, A. K., Rabindran, R., and Dasgupta, I. (2018). Rolling circle amplification-based analysis of Sri Lankan cassava mosaic virus isolates from Tamil Nadu, India, suggests a low level of genetic variability. Virus Dis. 29, 61–67. doi: 10.1007/s13337-018-0432-x
Lefeuvre, P., Martin, D. P., Hoareau, M., Naze, F., Delatte, H., Thierry, M., et al. (2007). Begomovirus ‘melting pot' in the south-west Indian Ocean islands: Molecular diversity and evolution through recombination. J. Gen. Virol. 88, 3458–3468. doi: 10.1099/vir.0.83252-0
Legg, J. P., Lava Kumar, P., Makesh kumar, T., Tripathi, L., Ferguson, M., Kanju, E., et al. (2015). Cassava virus diseases: biology, epidemiology, and management. Adv. Virus Res. 91, 85–142. doi: 10.1016/bs.aivir.2014.10.001
Legg, J. P., Shirima, R., Tajebe, L. S., Guastella, D., Boniface, S., and Jeremiah, S. (2014). Biology and management of Bemisia whitefly vectors of cassava virus pandemics in Africa. Pest Manag. Sci. 70, 1446–1453. doi: 10.1002/ps.3793
Leiva, A. M., Siriwan, W., Lopez-Alvarez, D., Barrantes, I., Hemniam, N., Saokham, K., et al. (2020). Nanopore-based complete genome sequence of a Sri Lankan cassava mosaic virus (geminivirus) strain from Thailand. Microbiol. Resour. Announc. 2020, 19. doi: 10.1128/MRA.01274-19
Lennon, A. M., Aiton, M. M., and Harrison, B. D. (1986). “Cassava viruses from South America,” in Annual Report 1985 Rep (Dundee: Scottish Crop Research. Institute), 167.
Li, W., Teng, F., Li, T., and Zhou, Q. (2013). Simultaneous generation and germline transmission of multiple gene mutations in rat using CRISPR-Cas systems. Nat. Biotechnol. 31, 684–686. doi: 10.1038/nbt.2652
Lim, Y. W., Mansfeld, B. N., Schläpfer, P., Ben, N., Gilbert, K. B., Narayanan, N. N., et al. (2022). Mutations in DNA polymerase δ subunit 1 co-segregate with CMD2-type resistance to cassava mosaic geminiviruses. Nat. Commun. 13, 3933. doi: 10.1038/s41467-022-31414-0
Lin, Z. J. D., Taylor, N. J., and Bart, R. (2019). Engineering disease-resistant cassava. Cold Spring Harb. Perspect. Biol. 1, a034595. doi: 10.1101/cshperspect.a034595
Lindbo, J. A., and Dougherty, W. G. (1992). Untranslatable transcripts of the Tobacco etch virus coat protein gene sequence can interfere with Tobacco etch virus replication in transgenic plants and protoplasts. Virology 189,725–733. doi: 10.1016/0042-6822(92)90595-G
Lindbo, J. A., and Falk, B. W. (2017). The Impact of “coat protein-mediated virus resistance” in applied plant pathology and basic research. Phytopathology 107, 624–634. doi: 10.1094/PHYTO-12-16-0442-RVW
Liu, J., Zheng, Q., Ma, Q., Gadidasu, K. K., and Zhang, P. (2011). Cassava genetic transformation and its application in breeding. J. Integr. Plant Biol. 53, 552–569. doi: 10.1111/j.1744-7909.2011.01048.x
Liu, S. S., Colvin, J., and De Barro, P. J. (2012). Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there? J. Integr. Agr. 11, 176–186. doi: 10.1016/S2095-3119(12)60002-1
Lokko, Y, Danquah, E., Offei, S., Dixon, A., and Gedil, M. (2005). Molecular markers associated with a new source of resistance to the cassava mosaic disease. Afr. J. Biotech. 4, 873–881. doi: 10.4314/ajb.v4i9.71131
Lynam, J., and Byerlee, D. (2017). Forever Pioneers—CIAT: 50 Years Contributing to a Sustainable Food Future… and Counting. CIAT Publication No. 444. Cali: International Center for Tropical Agriculture (CIAT).
Makeshkumar, T., Anoopsankar Nair, R. R., and Edison, S. (2005). Detection of Indian cassava mosaic virus through polymerase chain reaction and nucleic acid hybridization techniques. J. Root Crops 31, 1–6.
Makeshkumar, T., and Nair, R. R. (2001). “Serological and nucleic acid based detection of ICMD,” in 5th Intl. Scientific Meeting of Cassava Biotechnol. Network (CBN-V), held at St. Louis, USA during 4-9 Nov. 2001. St. Louis.
Malathi, V. G., Nair, N. G., and Shantha, P. (1985). Cassava Mosaic Disease. Technical Bulletin Series: Vol. 5. Trivandrum: Central Tuber Crops Research Institute.
Malathi, V. G., Varma, A., and Nambisan, B. (1989). Detection of Indian cassava mosaic virus by ELISA. Curr. Sci. 58, 149–150.
Mali, P., Yang, L., Esvelt, K. M., Aach, J., Guell, M., Di Carlo, J. E., et al. (2013). RNA-guided human genome engineering via Cas9. Science 339, 823–826. doi: 10.1126/science.1232033
Malik, A. I., Kongsil, P., Nguy?n, V. A., Ou, W., Sholihin, A., Srean, P., et al. (2020). Cassava breeding and agronomy in Asia: 50 years of history and future directions. Breed. Sci. 70, 145–166. doi: 10.1270/jsbbs.18180
Mandadi, K. K., and Scholthof, K. B. G. (2013). Plant immune responses against viruses: how does a virus cause disease? Plant Cell 25, 1489–1505. doi: 10.1105/tpc.113.111658
Maruthi, M. N., Seal, S., Colvin, J., Briddon, R. W., and Bull, S. E. (2004). East African cassava mosaic Zanzibar virus-a recombinant begomovirus species with a mild phenotype. Arch. Virol. 149, 2365–2377. doi: 10.1007/s00705-004-0380-1
Mehta, D., Cornet, L., Hirsch-Hoffmann, M., Zaidi, S. S., and Vanderschuren, H. (2020). Full-length sequencing of circular DNA viruses and extrachromosomal circular DNA using CIDER-seq. Nat. Protoc. 15, 1673–1689. doi: 10.1038/s41596-020-0301-0
Mehta, D., Hirsch-Hoffmann, M., Were, M., Patrignani, A., Zaidi, S. S., Were, H., et al. (2019a). A new full-length circular DNA sequencing method for viral-sized genomes reveals that RNAi transgenic plants provoke a shift in geminivirus populations in the field. Nucleic Acids Res. 47, e9. doi: 10.1093/nar/gky914
Mehta, D., Stürchler, A., Anjanappa, R. B., Zaidi, S. S., Hirsch-Hoffmann, M., Gruissem, W., et al. (2019b). Linking CRISPR-Cas9 interference in cassava to the evolution of editing-resistant geminiviruses. Genome Biol. 20, 3. doi: 10.1186/s13059-019-1678-3
Minato, N., Sok, S., Chen, S., Delaquis, E., Phirun, I., Le, V. X., et al. (2019). Surveillance for Sri Lankan cassava mosaic virus (SLCMV) in Cambodia and Vietnam one year after its initial detection in a single plantation in 2015. PLoS ONE 22, e0212780. doi: 10.1371/journal.pone.0212780
Nair, S. G., Unnikrishnan, M., Sheela, M. N., Abraham, K., Vimala, B., Sreekumari, M. T., et al. (1998). Tuber Crops Varieties Released by CTCRI Techmical Bulletin Series: Vol. 24, 6–14. Trivandrum: CTCRI.
Nawaz-ul-Rehman, M. S., and Fauquet, C. M. (2009). Evolution of geminiviruses and their satellites. FEBS Lett. 583, 1825–1832. doi: 10.1016/j.febslet.2009.05.045
Ndunguru, J., De León, L., Doyle, C. D., Sseruwagi, P., Plata, G., Legg, J. P., et al. (2016). Two novel DNAs that enhance symptoms and overcome CMD2 resistance to cassava mosaic disease. J. Virol. 28, 4160–4173. doi: 10.1128/JVI.02834-15
Ndunguru, J., Legg, J. P., Fofana, I. B. F., Aveling, T. A. S., Thompson, G., and Faquet, C. M. (2006). Identification of a defective molecule derived from DNA-A of the bipartite begomovirus of East African cassava mosaic virus. Plant Pathol. 55, 2–10. doi: 10.1111/j.1365-3059.2005.01289.x
Nekrasov, V., Staskawicz, D., Weigel, J. D., Jones, B., and Kamoun, S. (2013). Targeted mutagenesis in the model plant Nicotiana benthamiana using Cas9 RNA-guided endonuclease. Nat. Biotechnol. 31, 691–693. doi: 10.1038/nbt.2655
Nichols, R. F. W. (1947). Breeding cassava for resistance. East Afr. Agr. J. 12, 184–194. doi: 10.1080/03670074.1947.11664554
Ntui, V. O., Kong, K., Khan, R. S., Igawa, T., Janavi, G. J., Rabindran, R., et al. (2015). Resistance to Sri Lankan cassava mosaic virus (SLCMV) in genetically engineered cassava cv. KU50 through RNA silencing. PLoS ONE 10, e0120551. doi: 10.1371/journal.pone.0120551
Odipio, J., Alicai, T., Ingelbrecht, I., Nusinow, D. A., Bart, R., and Taylor, N. J. (2017). Efficient CRISPR/Cas9 genome editing of phytoene desaturase in cassava. Front. Plant Sci. 18, 780. doi: 10.3389/fpls.2017.01780
Okogbenin, E., Egesi, C. N., Olasanmi, B., Ogundapo, O., Kahya, S., Hurtado, P., et al. (2012). Molecular marker analysis and validation of resistance to cassava mosaic disease in elite cassava genotypes in Nigeria. Crop Sci. 52, 2576–2586. doi: 10.2135/cropsci2011.11.0586
Oslen, K. M., and Schaal, B. A. (1999). Evidence on the origin of cassava: phylogeography of Manihot esculenta. Proc. Natl. Acad. Sci. U. S. A. 96, 5586–5591. doi: 10.1073/pnas.96.10.5586
Patil, B. L., and Dasgupta, I. (2006). Defective interfering DNAs of plant viruses. Crit. Rev. Plant Sci. 25, 47–64. doi: 10.1080/07352680500391295
Patil, B. L., Dutt, N., Briddon, R. W., Bull, S. E., Rothenstein, D., Borah, B. K., et al. (2007). Deletion and recombination events between the DNA-A and DNA-B components of Indian cassava-infecting geminiviruses generate defective molecules in Nicotiana benthamiana. Virus Res. 124, 59–67. doi: 10.1016/j.virusres.2006.10.003
Patil, B. L., and Fauquet, C. M. (2009). Cassava mosaic geminiviruses: actual knowledge and perspectives. Mol. Plant Pathol. 10, 685–701. doi: 10.1111/j.1364-3703.2009.00559.x
Patil, B. L., Rajasubramaniam, S., Bagchi, C., and Dasgupta, I. (2005). Both Indian cassava mosaic virus and Sri Lankan cassava mosaic virus are found in India and exhibit high variability as assessed by PCR-RFLP. Arch. Virol. 150, 389–397. doi: 10.1007/s00705-004-0399-3
Peng, Y., Dallas, M. M., Ascencio-Ibáñez, J. T., Hoyer, J. S., Legg, J., Hanley-Bowdoin, L., et al. (2022). Early detection of plant virus infection using multispectral imaging and spatial-spectral machine learning. Sci. Rep. 24, 3113. doi: 10.1038/s41598-022-06372-8
Pita, J. S., Fondong, V. N., Sangare, A., Otim-Nape, G. W., Ogwal, S, and Fauquet, C. M. (2001). Recombination, pseudo recombination and synergism of geminiviruses are determinant keys to the epidemic of severe cassava mosaic disease in Uganda. J. Gen. Virol. 82, 655–665. doi: 10.1099/0022-1317-82-3-655
Puchta, H., Dujon, B., and Hohn, B. (1996). Two different but related mechanisms are used in plants for the repair of genomic double-strand breaks by homologous recombination. Proc. Natl. Acad. Sci. U. S. A. 93, 5055–5060. doi: 10.1073/pnas.93.10.5055
Rabbi, I. Y., Hamblin, M. T., Lava Kumar, P., Gedil, M. A., Ikpan, A. S., Jannink, J. L., et al. (2014). High-resolution mapping of resistance to cassava mosaic geminiviruses in cassava using genotyping-by-sequencing and its implications for breeding. Virus Res. 186, 87–96. doi: 10.1016/j.virusres.2013.12.028
Raji, S. N., Subhash, N., Ravi, R., Saravanan, R., Mohanan, C. N., Nita, N., et al. (2015). Detection of mosaic virus disease in cassava plants by sunlight-induced fluorescence imaging: a pilot study for proximal sensing. Int. J. Remote Sens. 36, 2880–2897. doi: 10.1080/01431161.2015.1049382
Rajinimala, R., Rabindran, R., Mohan, R., and Sethuraman, K. (2011). Survey for the occurrence of cassava mosaic disease in Tamil Nadu. J. Root Crops 37, 197–199. Available online at: https://journal.isrc.in/index.php/jrc/article/view/49
Ratcliff, F., Harrison, B. D., and Baulcombe, D. C. (1997). A similarity between viral defense and gene silencing in plants. Science 276, 1558–1560 doi: 10.1126/science.276.5318.1558
Resmi, T. R., Nivedhitha, S., Karthikeyan, C., and Veluthambi, K. (2014). Sri Lankan cassava mosaic virus replication associated protein (Rep) triggers transposition of IS426 in Agrobacterium. FEMS Microbiol. Lett. 360, 42–50. doi: 10.1111/1574-6968.12584
Rey, C., and Vanderschuren, H. (2017). Cassava mosaic and brown streak diseases: current perspectives and beyond. Annu. Rev. Virol. 29, 429–452. doi: 10.1146/annurev-virology-101416-041913
Rey, M. E., Ndunguru, J., Berrie, L. C., Paximadis, M., Berry, S., Cossa, N., et al. (2012). Diversity of dicotyledenous-infecting geminiviruses and their associated DNA molecules in southern Africa, including the south-west Indian Ocean islands. Viruses 4,1753–1791. doi: 10.3390/v4091753
Rosa, C., Kuo, Y. W., Wuriyanghan, H., and Falk, B. W. (2018). RNA interference mechanisms and applications in plant pathology. Annu. Rev. Phytopathol. 56, 581–610. doi: 10.1146/annurev-phyto-080417-050044
Rothenstein, D., Haible, D., Dasgupta, I., Dutt, N., Patil, B. L., and Jeske, H. (2006). Biodiversity and recombination of cassava-infecting begomoviruses from southern India. Arch. Virol. 151, 55–69. doi: 10.1007/s00705-005-0624-8
Sanford, J. C., and Johnston, S. A. (1985). The concept of parasite-derived resistance -deriving resistance genes from the parasite's own genome. J. Theor. Biol. 113, 395–405. doi: 10.1016/S0022-5193(85)80234-4
Saokham, K., Hemniam, N., Roekwan, S., Hunsawattanakul, S., Thawinampan, J., Siriwan, W., et al. (2021). Survey and molecular detection of Sri Lankan cassava mosaic virus in Thailand. PLoS ONE 16, e0252846. doi: 10.1371/journal.pone.0252846
Saunders, K., Salim, N., Mali, V. R., Malathi, V. G., Briddon, R, Markham, P. G., et al. (2002). Characterisation of Sri Lankan cassava mosaic virus and Indian cassava mosaic virus: evidence for acquisition of a DNA B component by a monopartite begomovirus. Virology 293, 63–74. doi: 10.1006/viro.2001.1251
Seal, S., Vanden Bosch, F., and Jeger, M. (2006). Factors influencing begomovirus evolution and their increasing global significance: implications for sustainable control. Crit. Rev. Plant Sci. 25, 23–46. doi: 10.1080/07352680500365257
Sequeira, J. C., and Harrison, B. D. (1982). Serological studies on cassava latent virus. Ann. Appl. Biol. 101, 33–42. doi: 10.1111/j.1744-7348.1982.tb00798.x
Sheela, M. N., Abraham, K., and Moorthy, S. N. (2012). Development of Elite Inbreds of Cassava and Their HSE in Heterosis Breeding: Annual Report 2011–2012. Thiruvananthapuram: CTCRI 25.
Sheela, M. N., Easwari Amma, C. S., Unnikrishnan, M., and Nair, S. G. (2002). “Expression of cassava mosaic tolerance in inter-specific backcross progeny of cassava (Manihot esculenta Crantz),” in Intemational Conference on Vegetables, November 1, 1–14, 2002. Bangalore.
Sheela, M. N., Unnikrishnan, M., Edison, S., and Easwari Amma, C. S. (2004). “Manihot caerulescens - A new source of resistance to cassava mosaic disease (ICMD),” in Sixth inter- national scientific meeting of the Cassava Biotechnology Network (Cali: CIAT). p. 48.
Siriwan, W., Jimenez, J., Hemniam, N., Saokham, K., Lopez-Alvarez, D., Leiva, A. M., et al. (2020). Surveillance and diagnostics of the emergent Sri Lankan cassava mosaic virus (Fam. Geminiviridae) in Southeast Asia. Virus Res. 285, 197959. doi: 10.1016/j.virusres.2020.197959
Stanley, J., and Gay, M. (1983). Nucleotide sequence of Cassava latent virus DNA. Nature 301, 2660–2662. doi: 10.1038/301260a0
Stanley, J., and Townsend, R. (1985). Characterisation of DNA forms associated with Cassava latent virus infection. Nucleic Acids Res. 13, 2189–2206. doi: 10.1093/nar/13.7.2189
Swanson, M. M., and Harrison, B. D. (1994). Properties, relationships and distribution of cassava mosaic geminiviruses. Trop. Sci. 34, 15–25.
Tafesse, A., Mena, B., Belay, A., Aynekulu, E., Recha, J. W., Osano, P. M., et al. (2021). Cassava production efficiency in southern Ethiopia: the parametric model analysis. Front. Sustain. Food Syst. 5, 758951. doi: 10.3389/fsufs.2021.758951
Tiendrébéogo, F., Lefeuvre, P., Hoareaum, M., Harimalala, M. A., De Bruyn, A., Villemot, J., et al. (2012). Evolution of African cassava mosaic virus by recombination between bipartite and monopartite begomoviruses. Virol. J. 9, 67–73. doi: 10.1186/1743-422X-9-67
Tokunaga, H., Baba, T., Ishitani, M., Ito, K., Kim, O. K., Ham, L. H., et al. (2018). “Sustainable management of invasive cassava pests in Vietnam, Cambodia, and Thailand,” in Crop Production under Stressful Conditions, eds M. Kokubun and S. Asanuma (Springer: Singapore), 131–157
Toruño, T. Y., Stergiopoulos, I., and Coaker, G. (2016). Plant–pathogen effectors: cellular probes interfering with plant defenses in spatial and temporal manners. Annu. Rev. Phytopathol. 54, 419–441. doi: 10.1146/annurev-phyto-080615-100204
Uke, A., Hoat, T. X., Quan, M. V., Liem, N. V., Ugaki, M., Natsuaki, K. T., et al. (2018). First report of Sri Lankan cassava mosaic virus infecting cassava in Vietnam. Plant Dis. 18, 805. doi: 10.1094/PDIS-05-18-0805-PDN
Uke, A., Khin, S., Kobayashi, K., Satou, T., Kim, O. K., Hoat, A., et al. (2022). Detection of Sri Lankan cassava mosaic virus by loop-mediated isothermal amplification using dried reagents. J. Virol. Methods. J299, 114336. doi: 10.1016/j.jviromet.2021.114336
Uke, A., Tokunaga, H., Utsumi, Y., Vu, N. A., Nhan, P. T., Srean, P., et al. (2021). Cassava mosaic disease and its management in Southeast Asia. Plant Mol. Biol. 109, 301–311. doi: 10.1007/s11103-021-01168-2
Unnikrishnan, M., EaswariAmma, C. S., Sreekumari, M. T., Sheela, M. N., and Mohan, C. (2002). “Cassava germplasm conservation and improvement in India,” in CIAT-VII Asian Cassava Research Workshop (Bangkok: CIAT). p. 4.
Unnikrishnan, M., Mohan, C., Santha, V., Ramanathan Pillai, S., and Anantharaman, M. (2011). Varietal Improvenent in Cassava for Mosaic Disease Resistance, Yield and Quality: Annual Report 2010-2011 (Thiruvananthapuram: CTCRI), 26–29.
Vanderschuren, H., Akbergenov, R., Pooggin, M. M., Hohn, T., Gruissem, W., and Zhang, P. (2007). Transgenic cassava resistance to African cassava mosaic virus is enhanced by viral DNA-A bidirectional promoter-derived siRNAs. Plant Mol. Biol. 64, 549–557. doi: 10.1007/s11103-007-9175-6
Vanderschuren, H., Alder, A., Zhang, P., and Gruissem, W. (2009). Dose-dependent RNAi-mediated geminivirus resistance in the tropical root crop cassava. Plant Mol. Biol. 70, 265–272. doi: 10.1007/s11103-009-9472-3
Vu, N. A., Tuan, L. N., Hung, N., Quynh, D. T. N., Tokunaga, H., Seki, M., et al. (2020). Study on quick testing method of resistant ability of cassava plants to mosaic disease. J. Vietnam Agric. Sci. Technol. 6, 85–90.
Wang, D., Zhang, X., Yao, X., Zhang, P., Fang, R., and Ye, J. (2020). A 7-amino-acid motif of rep protein essential for virulence is critical for triggering host defense against Sri Lankan cassava mosaic virus. Mol. Plant-Microbe Interact. 33, 78–86. doi: 10.1094/MPMI-06-19-0163-FI
Wang, G., Sun, Y. W., Xu, R. R., Qu, J., Tee, C. S., Jiang, X. Y., et al. (2014). DNA-A of a highly pathogenic Indian cassava mosaic virus isolated from Jatropha curcas causes symptoms in Nicotiana benthamiana. Virus Genes 48, 402–405. doi: 10.1007/s11262-014-1034-3
Wang, H. L., Cui, X. Y., Wang, X. W., Liu, S. S., Zhang, Z. H., Zhou, X. P., et al. (2016). First report of Sri Lankan cassava mosaic virus infecting cassava in Cambodia. Plant Dis. 100, 1029–1029. doi: 10.1094/PDIS-10-15-1228-PDN
Wang, Y. J., Lu, X. H., Zhen, X. H., Yang, H., Che, Y. N., Hou, J. Y., et al. (2022). Transformation and genome editing system for cassava cultivar SC8. Genes 14, 1650. doi: 10.3390/genes13091650
Wolfe, M. D., Rabbi, I. Y., Egesi, C., Hamblin, M., Kawuki, R., Kulakow, P., et al. (2016). Genome-wide association and prediction reveals genetic architecture of cassava mosaic disease resistance and prospects for rapid genetic improvement. Plant Genome 9, 1–13. doi: 10.3835/plantgenome2015.11.0118
Zhang, P., Vanderschuren, H., Fütterer, J., and Gruissem, W. (2005). Resistance to cassava mosaic disease in transgenic cassava expressing antisense RNAs targeting virus replication genes. Plant Biotechnol J. 3, 385–397. doi: 10.1111/j.1467-7652.2005.00132.x
Zhang, W., Olson, N. H., Baker, T. S., Faulkner, L., Agbandje-McKenna, M., Boulton, M. I., et al. (2001). Structure of the Maize streak virus geminate particle. Virology 279, 471–477. doi: 10.1006/viro.2000.0739
Zhou, X. (2013). Advances in understanding begomovirus satellites. Annu. Rev. Phytopathol. 51, 357–381. doi: 10.1146/annurev-phyto-082712-102234
Zhou, X., Liu, Y., Calvert, L., Munoz, C., Otim-Nape, G. W., Robinson, D. J., et al. (1997). Evidence that DNA-A of a geminivirus associated with severe cassava mosaic disease in Uganda has arisen by interspecific recombination. J. Gen. Virol. 78, 2101–2111. doi: 10.1099/0022-1317-78-8-2101
Zhou, X., Robinson, D. J., and Harrison, B. D. (1998). Types of variation in DNA-A among isolates of East African cassava mosaic virus from Kenya, Malawi and Tanzania. J. Gen. Virol. 79, 2835–2840. doi: 10.1099/0022-1317-79-11-2835
Keywords: cassava, CMD, breeding, genetic engineering, recombination
Citation: Hareesh PS, Resmi TR, Sheela MN and Makeshkumar T (2023) Cassava mosaic disease in South and Southeast Asia: current status and prospects. Front. Sustain. Food Syst. 7:1086660. doi: 10.3389/fsufs.2023.1086660
Received: 01 November 2022; Accepted: 01 June 2023;
Published: 27 June 2023.
Edited by:
Titus Alicai, National Agricultural Research Organisation, UgandaReviewed by:
Wilhelm Gruissem, ETH Zürich, SwitzerlandCopyright © 2023 Hareesh, Resmi, Sheela and Makeshkumar. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Thangaraj Makeshkumar, bWFrZXNoY3RjcmlAZ21haWwuY29t
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.