
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 15 August 2022
Sec. Agroecology and Ecosystem Services
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.995894
This article is part of the Research TopicAdvances in Molecular Crop PathologyView all 9 articles
Bacterial fruit blotch (BFB) caused by Acidovorax citrulli is a worldwide threat to watermelon and melon production. The type III secretion system (T3SS) plays an important role in the virulence of A. citrulli in its host plants and to induce hypersensitive response (HR) in its non-host plants. Little is known, however, about the contribution of the T3SS to biofilm formation in A. citrulli. We mutated two T3SS-related genes hrcJ and hrpE, respectively, and compared the mutants with their wild-type strain Aac-5 of A. citrulli, and their complementation strains on virulence, HR, and biofilm formation. Both mutants significantly reduced virulence in watermelon and melon seedlings and their ability to induce HR in tobacco leaves. Such reduction phenotypes were significantly recovered to the wild-type level, when the mutant strains were complemented with the wild-type hrcJ and hrpE genes. Interestingly, the two T3SS-related gene mutants also displayed enhanced ability to form biofilm, suggesting a different role of biofilm in the virulence of the group II stains of A. citrulli.
Bacterial pathogens share common strategies to infect and colonize their hosts. One such strategy is to deliver type III effector proteins into host cells via the type III secretion system (T3SS) (Abramovitch and Martin, 2004) to overcome the defense response of their host. The T3SS is encoded by the hrp (HR and pathogenicity) cluster, which contains over 20 genes including hrp and hrc (HR and conserved) genes that encode a type III secretion tunnel, as well as avr (avirulence) and hop (Hrp-dependent out protein) genes that encode effector proteins (Staskawicz et al., 2001). In the hrp cluster, hrcJ is a key factor of virulence on a host and of HR on a non-host in Xanthomonas oryzae pv. oryzicola (Zhao et al., 2010). The HrcJ apparatus is located across the inner and outer membrane, likely held by its hydrophobic domain at the C-terminal and lipid moiety at the N-terminal. It is important for secretion of harpins, which are virulence factors that target the extracelluar space of plant tissues (Choi et al., 2013). The hrpE encodes a major component of a surface appendage named the Hrp pilus (Weber and Koebnik, 2005), which is involved in secretion of effector proteins. Bacterial pathogen X. oryzae pv. oryzae was unable to elicit HR on non-hosts and showed reduced virulence on its host when either hrcJ or hrpE was absent (Cho et al., 2008).
Bacterial fruit blotch is one of the most devastating diseases infecting watermelon and other melons, and poses a serious threat to cucurbit production worldwide (Yan et al., 2012). It is caused by the gram-negative bacterium Acidovorax citrulli that has been divided into two major groups: group I strains are more pathogenic to melon, while group II strains are highly aggressive on watermelon (Walcott et al., 2004; Yan et al., 2012). Additionally, group II strain W1 is reportedly unable to form biofilm, while group I strain M6 was observed to be able (Bahar et al., 2009). Recently, whole genome of representative strains belonging to Group II (AAC00-1, accession number NC_008752.1) and Group I [pslb65 (Wang et al., 2015a) and tw6 (Wang et al., 2015b)] were sequenced. Genomic analysis of the AAC00-1 strain isolated from the U. S. revealed that it contains a ~30 kb hrp cluster and genes coding for putative type III-secreted effectors.
Like other gram-negative plant-pathogenic bacteria, A. citrulli may also rely on the T3SS to translocate virulence proteins from the bacterial cell into the cytoplasm of the host plant cell (Johnson et al., 2011). Virulence of the A. citrulli strain AAC00-1 on watermelon was abolished when the hrcC gene was mutated (Johnson et al., 2011). The hrcV mutants generated in the background of group I strain M6 and group II strain W1 were unable to induce HR on non-host plant (tomato) and showed impaired virulence on their host plant (melon), suggesting the importance of T3SS in A. citrulli (Bahar and Burdman, 2010). Biofilm formation is required for causing disease symptoms of X. citri on lemon leaves (Rigano et al., 2007), and T3SS is necessary for the biofilm formation of X. citri (Zimaro et al., 2014). Biofilm formation is also critical for virulence in group I strain M6 of A. citrulli, since its mutants unable to form biofilm showed significantly impaired virulence on watermelon (Bahar et al., 2009). Little is known, however, about whether the T3SS affects biofilm formation in group II strains of A. citrulli. To investigate the role of the T3SS on biofilm formation in the group II strains of A. citrulli, we constructed the T3SS-related hrcJ and hrpE gene mutants and their complementation strains, and compared to their wild-type strain Aac-5, a group II strain isolated from watermelon in China.
Bacterial strains and plasmids used in this study are listed in Table 1. A. citrulli strains were grown in King's B broth (KB) (Walcott et al., 2000) or on KA plate (KB containing 15 g/L agar) with appropriate antibiotics at 28°C. Escherichia coli strains were grown in Luria Bertani (MacLean et al., 2006) broth or plate with appropriate antibiotics at 37°C. Antibiotics used in this study were rifampicin (Rif), ampicillin (Ap), gentamicin (Gm), chloramphenicol (Cm) 20 μg·ml−1 and kanamycin (Km) at concentrations of 100 μg ml−1 for ampicillin, 20 μg·ml−1 for chloramphenicol and 50 μg ml−1 for the other antibiotics.
The hrcJ and hrpE genes in the wild-type strain Aac-5 were inactivated by homologous integration as described by Windgassen et al. (2000), respectively, using the suicide vector pK18mobsacB (Schäfer et al., 1994). Primers for PCR amplification of the two genes were designed using the free online program Primer 3.0 (http://www.simgene.com/Primer3) (Table 2). Each reaction mixture contained 0.5 μl of DNA template, 6.25 μl of 2 × PCR Mix (TaKaRa, Dlian, China) and 0.5 μl of each primer for a total reaction volume of 12.5 μl. The PCR conditions were 94°C for 3 min, 30 cycles of 94°C for 30 s, 65°C for 30 s and 72°C for 90 s, followed by 72°C for 5 min. The 1,791-bp fragment of Aac-5 amplified by the hrcJ-up-F and hrcJ-dn-R primers (Table 2) contained an 864-bp coding region of the hrcJ gene, as well as 455- and 472-bp upstream and downstream sequences of the gene. The 1,962-bp fragment of Aac-5 amplified by the hrpE-up-F and hrpE-dn-R primers (Table 2) contained an 834-bp coding region of the hrpE gene, as well as 515- and 613-bp upstream and downstream sequences of the gene. After confirmation by sequencing, the fragments were digested by EcoRI and HindIII, as well as by EcoRI and SmaI, respectively, and cloned into pK18mobsacB to create plasmids pK18-hrcJ and pK18-hrpE (Table 1). The two plasmids were digested with BamHI and NdeI, and the hrcJ and hrpE gene regions were replaced with a Gm gene cassette (855 bp), respectively, to create plasmid pK18-hrcJGm and pK18-hrpEGm (Table 1). The pK18-hrcJGm and pK18-hrpEGm were introduced from E. coli DH5α into Aac-5, respectively, by triparental conjugation using pRK600 as a helper plasmid. Transconjugants were screened on KB supplemented with 10% sucrose and antibiotics (Rif, Ap and Gm) and confirmed by PCR using the hrcJ-F/hrcJ-R and hrpE-F/hrpE-R primers, respectively. To confirm the presence of the Gm cassette in the transconjugants, Southern blotting was performed with primers Gm-F/Gm-R using marker BM5000 (Biomed, 5,000 bp, 3,000 bp, 2,000 bp, 1,000 bp, 750 bp, 500 bp, 250 bp, 100 bp) as the probe. The confirmed hrcJ and hrpE mutant strains, ΔhrcJ and ΔhrpE (Table 1), was used for subsequent studies.
To generate complementation strains, the hrcJ and hrpE genes (864 bp and 834 bp, respectively) in Aac-5 were amplified using primers hrcJ-F/hrcJ-R and hrpE-F/hrpE-R, respectively (Table 2). The gene fragments were cloned separately into pBBR1MCS-2 to generate pBR1MCS-2-hrcJ and pBR1MCS-2-hrpE (Table 1), which were transferred into the mutant strains ΔhrcJ and ΔhrpE by triparental conjugation, respectively. Transconjugants named hrcJ-comp and hrpE-comp were identified through screening on KB [amended with Rif, Km and Gm (Table 1)]. All obtained plasmids and A. cirulli strains were confirmed by PCR and DNA sequencing.
To prepare bacterial inocula, A. citrulli strains were grown in KB for 24 h at 28°C, and their OD600 was adjusted to 0.3 (~108 CFU ml−1). Tobacco plants (Nicotiana benthamiana) were grown at room temperature (25°C) with a 12-h photoperiod and used when they were 6-week-old. HR assays were performed based on the method of Johnson et al. (2011) by infiltrating a fully expended tobacco leaf with 100 μl of the bacterial inoculum into the leaf area among the midrib and the lateral veins. Three leaves from different tobacco seedlings were infiltrated for each strain. Sterile water was used as a negative control. Infiltrated tobacco leaves were kept at 100% humidity at room temperature. The HR-associated cell death was recorded 24 h after inoculation.
The bacterial inocula was prepared as described above. For inoculation, two hundred milliliters of each inoculum were sprayed to all leaves of ten 3 to 4-week-old melons (Cucumis melo cultivar IVF, provided by Insitute of Vegetables and Flowers, Chinese Academy of Agricultural Sciences, Beijing, China) and watermelon (Citrullus lanatus cultivar Jingxin#6, provided by Beijing Academy of Agriculture and Forestry Sciences, Beijing, China) seedlings. Inoculated seedlings were kept in the greenhouse at 25–30°C and 80% relative humidity. Disease index (DI) was recorded 8 days after inoculation according to the method of Tian et al. (2015) with a modified disease severity scale. The disease severity in each seedling was rated as follows: 0, no symptoms; 1, 3, and 5, necrotic lesions on 25, 50 and 75% of the leaves, respectively; 7, necrotic lesions on ~100% of the leaves; and 9, complete death of the seedlings. The DI for each treatment was calculated based on the following formula:
where A is the disease severity rating (0, 1, 3, 5, 7, or 9), B is the number of seedlings associated with each disease severity rating, and C is the total number of seedlings used for each strain in an experiment. The experiment was repeated three times.
Biofilm formation was measured based on a previous method (Aschtgen et al., 2008). A. citrulli strains were grown overnight in KB and adjusted to OD600 of 1.0 with sterile water. Each cell suspension was diluted by a factor of 100 with KB (without any antibiotics) in a glass flask and incubated at 28°C for 7 days without agitation. The cell suspensions were then poured out slowly and the glass tubes were rinsed gently three times with sterile distilled water. The biofilm formed on the inner wall of the glass tubes was fixed by heating at 80°C in an oven (Memmert, Schwabach, Germany) for 50 min, stained with 1% crystal violet for 2 h, and washed three times with sterile distilled water. The stained biofilm was then dissolved in 3 ml of absolute ethanol for 12 h, and measured quantitatively at OD570 using a spectrophotometer (Biophotometer, Eppendorf, Hambug, Germany). The experiment was repeated three times.
Statistical analysis was performed using the Student's t-test in the Excel 2010 software (Microsoft Inc., Seattle, WA, USA). Differences were considered statistically significant if P < 0.05.
PCR amplification of the hrcJ and hrpE mutant strain ΔhrcJ and ΔhrpE with the hrcJ-up-F/hrcJ-dn-R and hrpE-up-F/hrpE-dn-R primers and the subsequent sequencing of the PCR products confirmed that strains ΔhrcJ and ΔhrpE contained truncated hrcJ and hrpE genes replaced by the Gm cassette, respectively (Figures 1A,B). The presence of the Gm cassette (855 bp) in ΔhrcJ and ΔhrpE was further confirmed by Southern blot and was absent from the wild-type strain Aac-5 (Figures 2A,B). The ΔhrcJ and ΔhrpE strains were stable after continuous culturing for 20 generations in KB medium. The fact that the hrcJ and hrpE complementation strain ΔhrcJ-comp and ΔhrpE-comp were KmR suggested the successful transfer of the plasmid pBR1MCS-2-hrcJ and pBR1MCS-2-hrpE into the ΔhrcJ and ΔhrpE strains, respectively. The presence of pBR1MCS-2-hrcJ and pBR1MCS-2-hrpE in ΔhrcJ-comp and ΔhrpE-comp was further confirmed by PCR using hrcJ-F/hrcJ-R and hrpE-F/hrpE-R primers, respectively, since only one PCR band was amplified as expected from each complementation strain. An 864-bp fragment from ΔhrcJ-comp and a 834-bp band from ΔhrpE-comp were amplified from pBR1MCS-2-hrcJ and pBR1MCS-2-hrpE, respectively, while no bands were amplified from the mutant strains ΔhrcJ and ΔhrpE (Figures 1A,B).
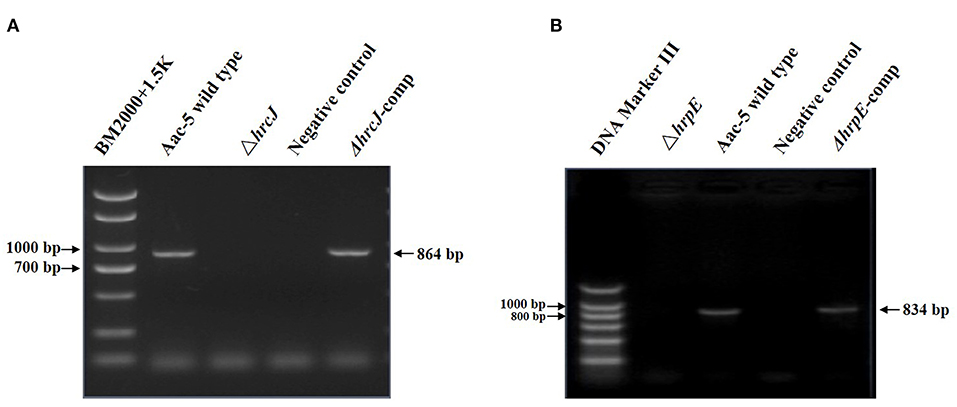
Figure 1. PCR results for confirmation of the hrcJ (A) and hrpE (B) mutants and complementation strains. (A) An 864-bp band of hrcJ gene was present in the lane of Aac-5 wild type and ΔhrcJ-comp, while was absent in the lane of ΔhrcJ and negative control (ddH2O as template). (B) An 834-bp band of hrpE gene was present in the lane of Aac-5 wild type and ΔhrpE-comp, while was absent in the lane of ΔhrpE and negative control.
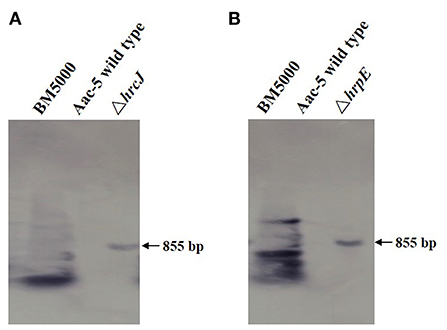
Figure 2. Southern blot for confirmation the presence of Gm in mutant strains of A. citrulli. The ΔhrcJ (A) and ΔhrpE (B) strain showed an 855-bp band on gel, while the wild type strain showed no band, indicating the mutant strains harbored a Gm cassette.
Twenty-four hours after infiltrating tobacco leaves with A. citrulli strains, typical and obvious HR necrotic lesions were observed where the infiltration was done with the wild-type strain Aac-5 (Figures 3A3,B3). Much like in the water control (Figures 3A1,B1), no such lesions were observed in areas of tobacco leaves infiltrated with the hrcJ and hrpE mutant strains ΔhrcJ and ΔhrpE, respectively (Figures 3A2,B2). When the ΔhrcJ and ΔhrpE mutant strains were complemented, however, similar necrotic HR lesions were observed in areas of tobacco leaves infiltrated with the complementation strains hrcJ-comp and hrpE-comp, as compared to those with the wild-type strain Aac-5 of A. citrulli (Figures 3A4,B4).

Figure 3. Effect of hrcJ (A) and hrpE (B) on hypersensitive response in tobacco (Nicotiana benthamiana) leaves infiltrated with water (1), hrcJ or hrpE mutant strain ΔhrcJ or ΔhrpE (2), wild-type strain Aac-5 (3), and the complementation strain ΔhrcJ-comp or ΔhrpE-comp (4).
Eight days after inoculation, more than 60% of leaves on watermelon seedlings inoculated with the wild-type strain Aac-5 developed necrotic lesions with a mean DI of 62.67% (Figures 4A,C). Even though this group II strain was not as virulent on melon as it was on watermelon, it still caused an average DI of 23.11% on melon (Figures 4B,D). When the hrcJ and hrpE genes were mutated, DIs caused by the mutant strains ΔhrpE and ΔhrcJ were 28.44 and 27.11% on watermelon (Figure 4C), and 9.04% and 10.52% on melon (Figure 4D), respectively. When the ΔhrpE and ΔhrcJ mutant strains were complemented, however, the complementation strains caused statistically similar DIs of 57.19 and 56.89% on watermelon, and 22.22 and 24.44% on melon, respectively, as compared to their wild-type strain Aac-5 (Figures 4C,D).
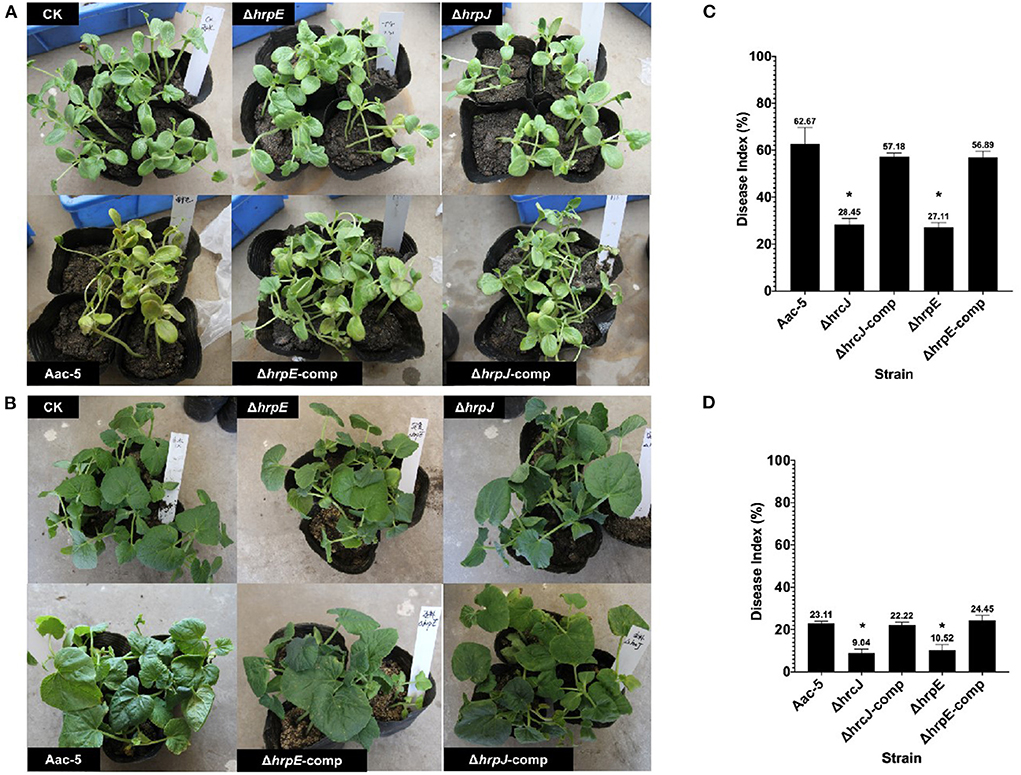
Figure 4. Virulence of A. citrulli strains on watermelon (A) and melon (B) seedlings 8 days after spray inoculation. Aac-5, wild-type strain; ΔhrcJ, hrcJ mutant strain; ΔhrcJ-comp, ΔhrcJ complementation strain; ΔhrpE, hrpE mutant strain; ΔhrpE-comp, ΔhrpE complementation strain. (C,D) Statistical analysis of the virulence of Aac-5 strains. Disease index for each A. citrulli strain was calculated from three experiments based on a 0 to 9 disease severity reading of each seedling in each experiment. Disease index with * indicates a significant difference as compared to that of the wild-type strain (P < 0.05).
Seven days after incubation, the absorbance measurement at OD570 for the stained biofilm dissolved with ethanol was 0.04 for the wild-type strain Aac-5, but significantly higher for the ΔhrcJ and ΔhrpE mutant strains at 0.123 and 0.093, respectively (P < 0.05) (Figure 5). When the mutant strains were complemented, however, OD570 was measured to be 0.048 and 0.046, similar to the wild-type level but significantly lower that the two mutant strains (P > 0.05) (Figure 5).
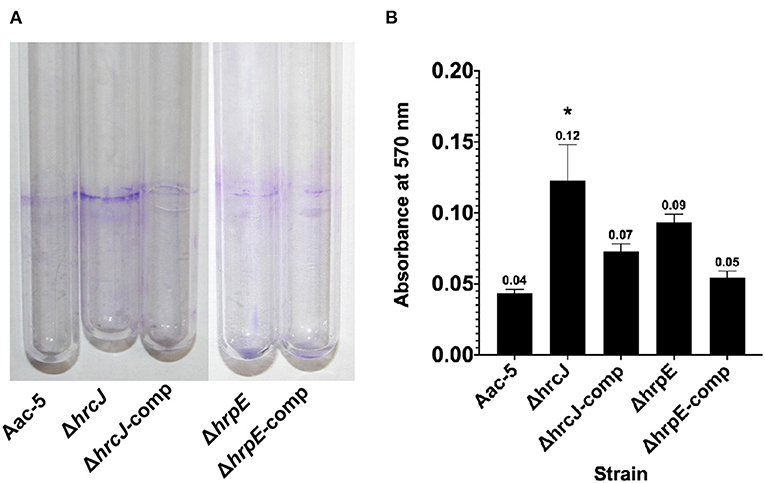
Figure 5. Effect of hrcJ and hrpE on biofilm formation of A. citrulli strains. (A) After 7 days incubation, the A. citrulli strains formed biofim on the inner wall of the cultural tubes, and then were visualized by staining with crystal violet. (B) Biofilm formation was quantified by measuring absorbance at 570 nm of the crystal violet stained biofilm dissolved with ethanol. The OD570 for each strain is the average of three experiments. The bars represent standard errors of the means. Asterisk indicates significant difference at P < 0.05 by the Student's t-test.
Pathogenicity assays on X. oryzae pv. oryzae have demonstrated that all hrp-hrc genes were very critical and their absence in the bacterium has led to loss of disease symptoms in the susceptible rice cultivar (Cho et al., 2008). Like X. oryzae, A. citrulli also requires a functional T3SS to cause disease on host plants and induce HR on non-host plants, since a hrcC deletion mutant of AAC00-1 abolished its pathogenicity on watermelon seedling tissues (Johnson et al., 2011), and the hrcV mutants of M6 (group I) and W1 (group II) strains failed to cause HR on tobacco and showed reduced virulence when inoculated to melon seeds (Bahar and Burdman, 2010). Genome sequencing of the group II strain AAC00-1 of A. citrulli revealed that it contains a hrp gene cluster coding for core proteins of the T3SS (Bahar et al., 2009). Besides the hrcC and hrcV genes, however, the function of other genes of T3SS in A. citrulli remains unknown. In our study, we investigated the biological functions of two of the T3SS genes, hrcJ and hrpE, in the ability of A. citrulli strain Aac-5, a group II strain from China, on biofilm formation, HR in tobacco and virulence in watermelon and melon seedlings. Aac-5 strain was isolated in Taiwan province of China, and was previously identified as a group II strain based on the Pulsed Field Gel Electrophoresis and Multilocus sequence typing (Yan et al., 2012). To achieve this goal, we therefore used Aac-5 as background to construct hrcJ and hrpE mutants and their complementation strains, and compared them to their wild-type strain Aac-5. The mutant strains ΔhrcJ and ΔhrpE were greatly reduced in virulence in watermelon and melon seedlings and lost ability to cause HR in tobacco leaves. Such reduction in virulence/loss of HR, however, was recovered/regained to the wild-type level, when the mutants were complemented. Our results revealed that in addition to the hrcC gene as reported previously (Johnson et al., 2011), both hrcJ and hrpE genes in the T3SS are required for virulence of A. citrulli in host plants and for HR in non-host plants.
The hrp clusters are most characterized in four plant pathogenic bacteria Erwinia amylovora, Pseudomonas syringae, Ralstonia solanacearum and X. campestris (Wengelnik and Bonas, 1996; Deng and Huang, 1999; Gijsegem et al., 2002; Rantakari et al., 2007). The clusters can be divided into two groups based on sequences, operon structures and regulation (Alfano and Collmer, 1997). Group I clusters include those of E. amylovora and P. syringae, and group II clusters include those of R. solanacearum and X. campestris. The most critical difference between the two cluster groups is that they are regulated by different regulators. Group I hrp operons are activated by HrpL and group II ones by HrpB (Alfano and Collmer, 1997). Based on the genome analysis of A. citrulli strain AAC00-1, the hrp operon contains a hrpB gene, suggesting that the hrp cluster in AAC00-1 belongs to group II hrp clustes. When the hrpB gene in X. citri was deleted, the bacterium greatly reduced its ability to form biofilm (Zimaro et al., 2014). In A. citrulli, the contribution of Type IV pili (TFP) to biofilm formation was revealed. The TFP, a form of surface appendage, is involved in many processes in bacteria including DNA transfer, twitching motility, and adherence to surfaces. The Type IV pili deficient mutants of the group I strain M6 of A. citrulli also greatly reduced biofilm formation as compared to its wild-type strain (Bahar et al., 2009). Whether the T3SS contributes to biofilm formation in A. citrulli strains, however, had been unknown before our study. The wild-type group II strain Aac-5 (Rantakari et al., 2007) we used in our study displayed low ability to form biofilm, compared to group I strain M6 of A. citrulli (Bahar et al., 2009). Interestingly, when the two T3SS-related genes hrcJ and hrpE were mutated, the mutant strains greatly increased their ability to form biofilm, but the increase was reduced to the wild-type level when the mutants were complemented, suggesting that hrcJ and hrpE negatively control biofilm formation in Aac-5. This is different from the positive role played by the Type IV pili in the group I strain M6 of A. citrulli, suggesting that different genes may control biofilm formation differently in A. citrulli, or that different strains of A. citrulli may use different strategies to attach to their in vitro environment or host plant surfaces.
In conclusion, our study showed that the T3SS genes hrcJ and hrpE positively control HR and virulence, but negatively contribute to biofilm formation in the group II strain Aac-5 of A. citrulli. This also suggests that biofilm formation in the group II strains of A. citrulli may not contribute, at least not directly, to virulence. Future research is needed to determine the biological function of biofilm, if any, in A. citrulli, especially in group II strains, and different functions biofilm may play in different groups of strains in A. citrulli.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author/s.
Conceptualization: TW, QH, and WG. Methodology: XA and YY. Writing—original draft preparation: WG. Writing—review and editing: QH. Funding acquisition: TZ. All authors have read and agreed to the published version of the manuscript.
This research was funded by National Natural Science Foundation of China, Grant Number 82104341; the Key Project at Central Government Level: the Ability Establishment of Sustainable Use for Valuable Chinese Medicine Resources, Grant Number 2060302; Scientific and Technological Innovation Project of China Academy of Chinese Medical Sciences, Grant Number CAAS-ASTIP-2016-IPP; China earmarked fund for Modern Agroindustry Technology Research System, Grant Number CARS-26.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abramovitch, R. B., and Martin, G. B. (2004). Strategies used by bacterial pathogens to suppress plant defenses. Curr. Opin. Plant Biol. 7, 356–364. doi: 10.1016/j.pbi.2004.05.002
Alfano, J. R., and Collmer, A. (1997). The type III (Hrp) secretion pathway of plant pathogenic bacteria: trafficking harpins, Avr proteins, and death. J. Bacteriol. 179, 5655. doi: 10.1128/jb.179.18.5655-5662.1997
Aschtgen, M.-S., Bernard, C. S., Bentzmann, S. D., Lloubès, R., and Cascales, E. (2008). SciN is an outer membrane lipoprotein required for type VI secretion in enteroaggregative Escherichia coli. J. Bacteriol. 190, 7523–7531. doi: 10.1128/JB.00945-08
Bahar, O., and Burdman, S. (2010). Bacterial fruit blotch: a threat to the cucurbit industry. Isr. J. Plant Sci. 58, 19–31. doi: 10.1560/IJPS.58.1.19
Bahar, O., Goffer, T., and Burdman, S. (2009). Type IV pili are required for virulence, twitching motility, and biofilm formation of Acidovorax avenae subsp. citrulli. Mol. Plant. Microbe Interact. 22, 909–920. doi: 10.1094/MPMI-22-8-0909
Cho, H.-J., Park, Y.-J., Noh, T.-H., Kim, Y.-T., Kim, J.-G., Song, E.-S., et al. (2008). Molecular analysis of the hrp gene cluster in Xanthomonas oryzae pathovar oryzae KACC10859. Microb. Pathog. 44, 473–483. doi: 10.1016/j.micpath.2007.12.002
Choi, M.-S., Kim, W., Lee, C., and Oh, C.-S. (2013). Harpins, multifunctional proteins secreted by gram-negative plant-pathogenic bacteria. Mol. Plant. Microbe Interact. 26, 1115–1122. doi: 10.1094/MPMI-02-13-0050-CR
Deng, W.-L., and Huang, H.-C. (1999). Cellular locations of Pseudomonas syringae pv. syringae HrcC and HrcJ proteins, required for harpin secretion via the type III pathway. J. Bacteriol. 181, 2298–2301. doi: 10.1128/JB.181.7.2298-2301.1999
Gijsegem, F. V., Vasse, J., Rycke, R. D., Castello, P., and Boucher, C. (2002). Genetic dissection of the Ralstonia solanacearum hrp gene cluster reveals that the HrpV and HrpX proteins are required for Hrp pilus assembly. Mol. Microbiol. 44, 935–946. doi: 10.1046/j.1365-2958.2002.02936.x
Johnson, K. L., Minsavage, G. V., Le, T., Jones, J. B., and Walcott, R. R. (2011). Efficacy of a nonpathogenic Acidovorax citrulli strain as a biocontrol seed treatment for bacterial fruit blotch of cucurbits. Plant Dis. 95, 697–704. doi: 10.1094/PDIS-09-10-0660
MacLean, A. M., MacPherson, G., Aneja, P., and Finan, T. M. (2006). Characterization of the β-Ketoadipate pathway in Sinorhizobium meliloti. Appl. Environ. Microbiol. 72, 5403–5413. doi: 10.1128/AEM.00580-06
Rantakari, A., Virtaharju, O., Vähämiko, S., Taira, S., Palva, E. T., Saarilahti, H. T., et al. (2007). Type III secretion contributes to the pathogenesis of the soft-rot pathogen Erwinia carotovora: partial characterization of the hrp gene cluster. Mol. Plant. Microbe Interact. 14, 962–968. doi: 10.1094/MPMI.2001.14.8.962
Rigano, L. A., Siciliano, F., Enrique, R., Sendín, L., Filippone, P., Torres, P. S., et al. (2007). Biofilm formation, epiphytic fitness, and canker development in Xanthomonas axonopodis pv. citri. Mol. Plant. Microbe Interact. 20, 1222–1230. doi: 10.1094/MPMI-20-10-1222
Schäfer, A., Tauch, A., Jäger, W., Kalinowski, J., Thierbach, G., and Pühler, A. (1994). Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene. 145, 69–73. doi: 10.1016/0378-1119(94)90324-7
Staskawicz, B. J., Mudgett, M. B., Dangl, J. L., and Galan, J. E. (2001). Common and contrasting themes of plant and animal diseases. Science. 292, 2285–2289. doi: 10.1126/science.1062013
Tian, Y., Zhao, Y., Wu, X., Liu, F., Hu, B., and Walcott, R. R. (2015). The type VI protein secretion system contributes to biofilm formation and seed-to-seedling transmission of Acidovorax citrulli on melon. Mol. Plant Pathol. 16, 38–47. doi: 10.1111/mpp.12159
Walcott, R. R., Fessehaie, A., and Castro, A. C. (2004). Differences in pathogenicity between two genetically distinct groups of Acidovorax avenae subsp. citrulli on cucurbit hosts. J. Phytopathol. 152, 277–285. doi: 10.1111/j.1439-0434.2004.00841.x
Walcott, R. R., Langston, D. B., Sanders, F. H., and Gitaitis, R. D. (2000). Investigating intraspecific variation of Acidovorax avenae subsp. citrulli using DNA fingerprinting and whole cell fatty acid analysis. Phytopathology. 90, 191–196. doi: 10.1094/PHYTO.2000.90.2.191
Wang, T., Sun, B., Yang, Y., and Zhao, T. (2015a). Genome sequence of Acidovorax citrulli group strain pslb65 causing bacterial fruit blotch of melons. Genome Announc. 3, e00327–e00315. doi: 10.1128/genomeA.00327-15
Wang, T., Yang, Y., and Zhao, T. (2015b). Genome sequence of a copper-resistant strain of Acidovorax citrulli causing bacterial fruit blotch of melons. Genome Announc. 3, e00310–e00315. doi: 10.1128/genomeA.00310-15
Weber, E., and Koebnik, R. (2005). Domain structure of HrpE, the hrp pilus subunit of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 187, 6175–6186. doi: 10.1128/JB.187.17.6175-6186.2005
Wengelnik, K., and Bonas, U. (1996). HrpXv, an AraC-type regulator, activates expression of five of the six loci in the hrp cluster of Xanthomonas campestris pv. vesicatoria. J. Bacteriol. 178, 3462–3469. doi: 10.1128/jb.178.12.3462-3469.1996
Windgassen, M., Urban, A., and Jaeger, K.-E. (2000). Rapid gene inactivation in Pseudomonas aeruginosa. FEMS Microbiol. Lett. 193, 201–205. doi: 10.1111/j.1574-6968.2000.tb09424.x
Yan, S., Yang, Y., Wang, T., Zhao, T., and Schaad, N. W. (2012). Genetic diversity analysis of Acidovorax citrulli in China. Eur. J. Plant Pathol. 136, 171–181. doi: 10.1007/s10658-012-0152-9
Zhao, W., Han, Y., Cui, Y., Zhao, M., Li, Y., Zou, L., et al. (2010). HrcJ is involved in Type-III apparatus formation of Xanthomonas oryzae pv. oryzicola for hypersensitive response in nonhost tobacco and pathogenicity in rice. Sci. Agric. Sin. 43, 79–86. doi: 10.3864/j.issn.0578-1752.2010.01.009
Keywords: Acidovorax citrulli, group II strains, type III secretion system, biofilm formation, virulence, hypersensitive response
Citation: Wang T, Huang Q, An X, Yang Y, Guan W and Zhao T (2022) Type III secretion system genes hrcJ and hrpE affect virulence, hypersensitive response and biofilm formation of group II strains of Acidovorax citrulli. Front. Sustain. Food Syst. 6:995894. doi: 10.3389/fsufs.2022.995894
Received: 16 July 2022; Accepted: 28 July 2022;
Published: 15 August 2022.
Edited by:
Zhengnan Li, Inner Mongolia Agricultural University, ChinaReviewed by:
Zhanbin Sun, Beijing Technology and Business University, ChinaCopyright © 2022 Wang, Huang, An, Yang, Guan and Zhao. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Wei Guan, d3luZ3dhbkB5YWhvby5jb20=; Tingchang Zhao, emhhb3RnY2dAMTYzLmNvbQ==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.