
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 30 January 2023
Sec. Agro-Food Safety
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.957099
This article is part of the Research Topic Plant-Based Antimicrobials and Their Role in Food Safety View all 5 articles
 Quail Das1
Quail Das1 Teri-Lyn Hasted1,2
Teri-Lyn Hasted1,2 Dion Lepp1
Dion Lepp1 Xianhua Yin1
Xianhua Yin1 Joshua Tang1
Joshua Tang1 Gabhan Chalmers2
Gabhan Chalmers2 Kelly Ross3
Kelly Ross3 Patrick Boerlin2
Patrick Boerlin2 Moussa S. Diarra1*
Moussa S. Diarra1*Extraintestinal pathogenic Escherichia coli (ExPEC) includes several serotypes that have been associated with colibacillosis in poultry, as well as urinary tract infections and newborn meningitis in humans. This study investigated the antimicrobial activities of ceftriaxone (AXO) and cranberry pomace extracts (CRAN) alone or in combination (CC) against multidrug-resistant (MDR) ExPEC from broiler. The growth-inhibitory activity of CRAN and synergy tests by a checkerboard method were determined in cation-adjusted Mueller–Hinton broth (CAMHB). The transcriptomic profile of the MDR E. coli O7:H18 (ST38) grown in CAMHB supplemented with sub-inhibitory concertation of CRAN and AXO alone or in combination was obtained by RNA-seq. The MIC of CRAN for all isolates was 16 mg/mL. An additive activity was observed between 4 mg/mL of CRAN and 4 μg/mL of AXO. Compared to the control, the transcriptomic analysis revealed that 4 mg/ml of (1/4MIC) CRAN and its combination with 4 μg/mL of (1/8MIC) AXO (CC) exposures resulted in 727 and 712 differentially expressed genes, respectively (false discovery rate < 0.001 and log2-fold change > 2), in the studied E. coli. Major virulence genes including adhesins (fim, flg, csg, and yad), protectins (omp, tra, waa, and hly), secretion systems (hof, pho, and vir), and quorum sensing (lsr), which are energetically expensive for bacteria, were downregulated. Most importantly, 1/4MIC of CRAN or CC downregulated the β-lactamase blaCMY-2 and efflux pump including tolC, mdtEIJ, gadEW, and their regulator gene evgS, while upregulating the cysteine biosynthesis and oxidative stress-related regulatory genes including cys, dmlA, sbp, nrdGHI, soxSR, and rpoH. Downregulation of multiple enzymes involved in TCA cycles and upregulation of Fe–S cluster coordinated by Cys and Isc proteins reflect the regulation of energy metabolism of the studied E. coli upon CRAN or CC exposure. The downregulation of outer membrane protein genes that control permeability barriers, along with different antimicrobial resistance genes, demonstrates that CRAN may have the unique potential to enhance the antimicrobial activities of third-generation cephalosporins such as AXO against MDR E. coli.
The use of antibiotics in poultry over many decades contributed to the rise of antimicrobial resistance (AMR) in several bacterial species, some of which could be zoonotic. Prophylactic use of antibiotics in livestock production plays an important role in the rise of AMR in bacteria (Ching and Zaman, 2021). A 2-year prospective study of small poultry flocks in Ontario, Canada, showed that 46.65% of the chicken (167/358) were pan-susceptible, while a high proportion of E. coli isolates (≥40% of isolates) were resistant to tetracycline, (15–39% of isolates) streptomycin, sulfisoxazole, and ampicillin, and a small proportion of isolates (5–14% of isolates) were resistant to trimethoprim–sulfamethoxazole and gentamicin (Varga et al., 2019). Among animal reservoirs, fluoroquinolone and β-lactam-resistant E. coli were most frequent in poultry, where tetA, blaTEM − 1B, sul2, aadA1, and aph(6)-Id were the most prevalent AMR genes (Leekitcharoenphon et al., 2021). Multidrug-resistant (MDR) bacteria, largely extended-spectrum β-lactamase (ESBL) producing Gram-negative bacteria including E. coli harboring the ampC-like blaCMY − 2 gene, were more common in chicken-derived isolates than other retail meat products in Canada (Awosile et al., 2021). Antimicrobial resistance genes (ARGs) are frequently located on mobile genetic elements (MGEs) such as plasmids, transposons, and integrons, which allow their dissemination among bacterial populations (Chalmers et al., 2017). Interestingly, MGEs have been increasingly associated with the rise and distribution of both virulence functions and ARGs (Juhas et al., 2009). Clear associations between resistance and virulence traits in avian pathogenic bacterial strains have been reported (Cepas et al., 2019).
Based on the location of pathologies (diseases), E. coli strains can be classified as intestinal pathogenic E. coli (InPEC) and extraintestinal pathogenic E. coli (ExPEC). In avian pathogenic E. coli (APEC), the etiological agent of colibacillosis in poultry is an important subset of ExPEC presenting a zoonotic concern (Aleksandrowicz et al., 2021). Strains of APEC share virulence traits and resistance patterns with human ExPEC such as those causing urinary tract infection (UTI). Often APEC of serotypes O1, O2, O65, and O78 are associated with colibacillosis (da Silva and Mendonça, 2012). Virulence attributes involved in the pathogenesis of APEC include adhesins, fimbriae, flagella, lipopolysaccharide, and other factors to overcome hosts' defense mechanisms (Aleksandrowicz et al., 2021). In addition, Gram-negative bacteria employ numerous (I through IX) secretion systems (SSs) for communication, virulence functions, adhesion to surfaces, nutrient acquisition, or growth inhibition of competing bacteria (Meuskens et al., 2019). Among them, type III (T3SS), type IV (T4SS), and type VI (T6SS) secretion systems transfer proteins and inject virulence factors into eukaryotic cells (Souza et al., 2015).
In order to combat AMR and reduce the spread of AGRs, antibiotic use has been restricted in animal production in Canada for several years [Government of Canada, 2017; https://canadagazette.gc.ca/rp-pr/p2/2017/2017-05-17/html/sor-dors76-eng.html]. For example, Canadian poultry farmers require a veterinary prescription and have chosen to end the use of Category I, II, and III antibiotics [Chicken Farmers of Canada, 2020; https://www.chickenfarmers.ca/antibiotics/]. Unfortunately, these measures could lead to an increase in bacterial diseases such as colibacillosis incidence in poultry. Hence, farmers/food producers are looking for alternatives to protect their chickens while increasing or maintaining food safety. Cranberry extracts could be an alternative solution on their own or a contributing tool, due to their high contents of polyphenolic compounds known to have antimicrobial activities. Cranberry extracts have been reported to exhibit in vitro antimicrobial effects through pleiotropic mechanisms which include the alteration of bacterial cell surface structure and integrity in S. aureus (Diarra et al., 2013); dose-dependent reduction of salt tolerance, bile-salt hydrolase activity, and biofilm formation in Listeria monocytogenes (Diarra et al., 2020); reduction of virulence genes expression and cell membrane biogenesis of Salmonella, nutrient chelation through affecting the citrate metabolism, enterobactin synthesis, and transport system in E. coli (Harmidy and Tufenkji, 2011) and Salmonella (Das et al., 2019); decreasing the adhesion genes related to flagellar motility (Das et al., 2019); and reduction of the production of quorum sensing-regulated virulence determinants of Pseudomonas aeruginosa (Maisuria et al., 2016).
This study investigated the antibacterial activities of an extract from cranberry pomace alone or in combination with a third-generation cephalosporin (ceftriaxone) against different E. coli from the broiler. The signatures of tested products were determined on the transcriptional profile in an MDR broiler ExPEC isolate of serotype O7:H18 (ST38) recently reported to cluster with human UTI isolates and virulence in day-old chicks (Rehman et al., 2022), to unveil overall patterns of gene expression. This work could support the development of cranberry extracts as cost-effective alternatives to combat potential zoonotic MDR pathogens from broilers.
The extracts of cranberry pomace were prepared from organic American cranberry (Vaccinium macrocarpon) pomace as previously described (Ross et al., 2017). In brief, phenolic-rich compounds were extracted with 80% ethanol from the cranberry pomace. After extraction, ethanol was removed by rotary evaporation and the remaining extracts were freeze-dried at −30°C for 10–11 days to generate crude cranberry pomace extracts (CRAN) and stored at −20°C. A stock solution was prepared by solubilizing 300 mg/mL of dry powder of CRAN in 80% ethanol, filter sterilized through a sterile cellulose acetate filter assembly (pore size, 0.2 mm), and stored at −20°C until used in antimicrobial assays.
Eight E. coli isolates from broiler chickens were included in this study based on their antimicrobial susceptibility profiles (Rehman et al., 2022) (Appendix). The extraintestinal pathogenic E. coli O7:H18 (ST38) isolate was selected for RNA-seq. This E. coli was obtained from chicken cloacae. It was resistant to several antimicrobial classes and classified as MDR (Appendix). The isolates were grown on cation-balanced Mueller–Hinton Agar (CAMHA, Becton Dickinson, Mississauga, ON). Colony forming units per ml (CFU/ml) were determined by viable bacterial counts on Tryptic Soy Agar (TSA, Becton Dickinson). For growth studies, a single colony of each strain was inoculated into 5 ml of CAMHB, and the cultures were incubated at 37°C with agitation (200 rpm) for 20 h.
The minimum inhibitory concentration (MIC) of CRAN against all E. coli was determined following the broth microdilution method according to the Clinical Laboratory Standard Institute's guidelines. To determine the MICs, concentrations of CRAN ranging from 0.5 to 16 mg/mL were prepared in a honeycomb of 100-well microtiter plates (Oy Growth Curves Ab Ltd, Helsinki, Finland). Plates were loaded in a Bioscreen (Growth Curves USA, Piscataway, NJ), and growth readings were taken every 15 min at 37°C for 24 h to automatically record the optical densities at 600 nm (OD600nm). Media without inoculum, but with the studied concentrations of tested products, were included as blanks. The OD of blanks was subtracted from the OD of inoculated wells containing equivalent concentrations of studied products. The OD data were used to produce OD/time plots using Microsoft Excel. Kanamycin A (Sigma Aldrich) which MICs were known against all isolates, was used as a control on each plate to ensure the reproducibility/validity of the results.
Checkerboard assay was performed by a broth microdilution method similar to that used for standard MIC determinations to evaluate the effect of varying concentrations of CRAN with antimicrobials in at least three independent replicates. Antimicrobials investigated include ceftiofur, ceftriaxone (AXO), ampicillin, kanamycin, streptomycin, tetracycline, florfenicol, and sulfisoxazole. The fractional inhibitory concentration (FIC) indices were calculated as follows: FIC index = FICA + FICB = A/MICA + B/MICB, where A and B are MICs of compounds A and B in combination, MICA and MICB are the individual MICs of compounds A and B, and FICA and FICB are the FICs of compounds A and B. Indifference for drug interactions or an additive effect is demonstrated if the FIC index is in the range of 0.5–4, a synergy if the FIC index is ≤0.5, whereas an antagonistic effect is represented by an FIC index >4 (Diarra et al., 2013). Controls without CRAN were performed for each E. coli isolate combination to confirm the efficacy of CRAN only, and not related to solvent.
The transcriptional profile of E. coli O7:H18 (ST38) grown in CAMHB supplemented with sub-inhibitory concertation of CRAN and AXO alone or in combination (CC) was compared by RNA-seq. Bacterial cells in all culture conditions were incubated with agitation at 37°C for 3 h followed by centrifugation at 6,000×g for 10 min at room temperature. The pellets were then kept with RNAlater Stabilization Solution (Thermo Fisher Scientific) for 24 h at 4°C, washed with ice-cold 1x PBS, and re-centrifuged again at 6,000×g for 10 min. Total RNA was extracted using the PureLink RNA Mini Kit (Ambion, Invitrogen Fisher Scientific, Carlsbad, CA, USA) according to the manufacturer's instructions. The RNA concentration and purity were determined using a NanoDrop 2,000c (Thermo Scientific) and the integrity of the extracted RNA was examined using Agilent Bioanalyzer RNA Nano assay (Agilent Technologies, CA, USA). Samples with RNA integrity number (RIN) ≥8 were used for library construction. RNA (2 μg) was treated with 2U Turbo DNase (Invitrogen) at 37°C for 30 min, and four biological replicates from each treatment group were used for RNA-seq analysis.
To investigate the transcriptional response of E. coli O7:H18 exposure to antimicrobial agents, a comparative transcriptomic analysis was performed among CRAN, CC, and AXO exposed to E.coli cells with control. A total of 2 μg of each RNA sample was used as the input material for RNA sample preparations, and five biological replicates were analyzed for a total of 20 (5 × 4) samples. Bacterial-stranded mRNA sequencing libraries were constructed with the TruSeq Stranded mRNA Library Prep Kit (Illumina, RS-122-9004DOC) according to the manufacturer's instructions. The filtered reads were mapped to the E. coli O7:H18 (ST38) using bowtie v1.1.1, and the counts per feature were determined with the HTSeq-count tool within HTSeq v0.6.1 (Das et al., 2019). Differential gene expression (DGE) analysis was performed in R v3.2.5 with the edgeR package v3.6.8 (Das et al., 2019). Genes with an absolute log2-fold change of > 2 and false discovery rate (FDR) < 0.001 in negative control vs. (A) CRAN; (B) AXO; (C) CC were considered differentially expressed. Over-representation analysis (ORA) and gene-set enrichment analysis (GSEA) were performed on KEGG pathways and GO terms (GSEA on KEGG pathways only). ORA tests if certain terms (KEGG pathways or GO annotations) are more frequently associated with DEGs, compared to background genes (i.e., whole genome). In GSEA, a list of all genes is sorted by log2FC value, and terms are identified that are enriched in either the top (upregulated genes) or bottom (downregulated genes) of the list.
To further confirm/validate RNA-seq results, quantitative reverse transcriptase PCR (qRT-PCR) was carried out for 13 genes that were determined in our previous study in the same isolate (Rehman et al., 2022). These genes were selected based on their modulation pattern in RNA-seq analysis. Gene names and primer sequences used for RT-qPCR are presented in Table 1. All primers were designed with Primer 3 software with an amplicon size of ~82–128 bp. The efficiency of primers was checked by melting curve and standard curve analysis. The reaction mixture contained 20 μL final volume with 1 μL of cDNA (1:10 dilution), 3 μL of each primer (1 mM), 3 μL of nuclease-free water, and 10 μL of Universal SYBR Green Supermix (Bio-Rad, Canada). The RT-qPCR assay was performed on the 7,500 Real-time PCR System (Applied Biosystems, Foster, USA). The amplification program was as follows: one cycle at 95°C for 3:20 min, and 40 cycles at 95°C for 30 s, 58°C for 30 s, and 72°C for 30 s. The 2−ΔΔCt method was used to analyze the relative gene expression data, using rpoD and 16S as reference genes for normalization. All RT-qPCR assays were conducted using three biological and three technical replicates.
All experiments were performed in four separate assays, each including five technical replicates. The RNA-seq differential expression between the control group and treatments with CRAN, AXO, and CC was analyzed using the Bioconductor package edgeR, which is analogous to t- and F-statistics tools using a generalized linear model (GLM) and the likelihood ratio test. A log2-fold change of > 2 and false discovery rate (FDR) cutoffs ≤0.001 were used. The false discovery rates were calculated using the Benjamini–Hochberg method (Das et al., 2019). The qPCR results are represented as means ± standard deviations, and significant differences (P < 0.05) were determined by the Global Mixed Model Analysis of Variance procedure of Statistical Analysis System (SAS) version 9.4.
In order to verify the antibacterial effects of ethanolic extracts of cranberry pomace (CRAN), the growth curve of different multidrug-resistant E. coli strains was generated (Figure 1). The strains were treated with 0 to 16 mg/mL of CRAN. As shown in Figure 1, the MIC of CRAN for all strains was 16 mg/mL, although 8 mg/mL displayed an extended lag phase duration and a lower maximum growth rate. No significant differences were observed between the MICs of strains isolated from chicken and those isolated from human sources.
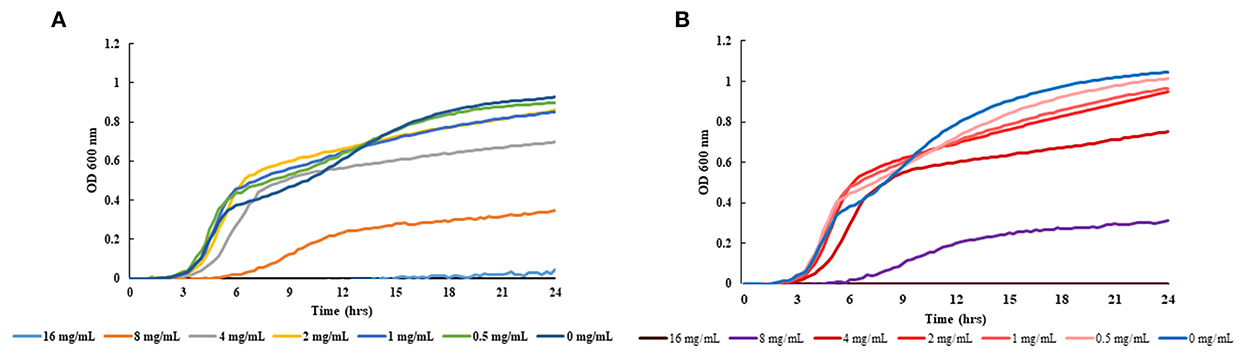
Figure 1. Growth curve obtained using the Bioscreen C analyzer representing optical density at 600 nm for (A) multidrug-resistant isolate 2532 E. coli O7:H18 (ST38) and (B) pan susceptible isolate 2440 E. coli O7:H6 (ST362). Three replicates for each concentration (0 to 16 mg/mL) of ethanolic extracts of cranberry pomace tested are represented in the legend.
Checkerboard assays were performed to investigate the relationship between CRAN and certain antibiotics to which E. coli O7:H18 (ST38) showed resistance. The results revealed that the best reduction in the MICs of both CRAN and ceftriaxone (MIC 128 μg/ml) was achieved (Figure 2). Combination of 0.125, 4, or 8 mg/mL of CRAN with ceftriaxone yielded FIC indices of 0.50, 0.37, and 0.50, respectively, against all tested strains. Sub-MIC (1/4) of CRAN potentiated the inhibitory activity of ceftriaxone against E. coli O7:H18 (ST38) by lowering its MIC by 8X time (Table 2). The potentiated activity of CRAN against other antibiotics was not consistent. No indifference or antagonistic effects were observed.
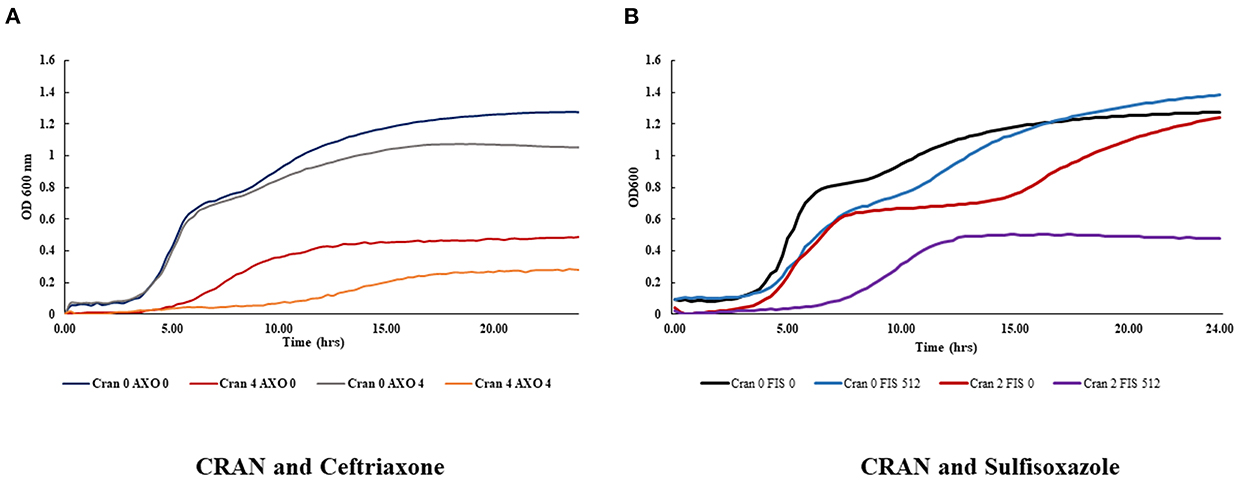
Figure 2. Growth kinetics of E. coli O7:H18 (ST38) was monitored by the Bioscreen C in a checkerboard assay in the combination of sub-lethal concentrations of cranberry pomace extracts (CRAN) and (A) ceftriaxone (AXO) or (B) sulfisoxazole (FIS).
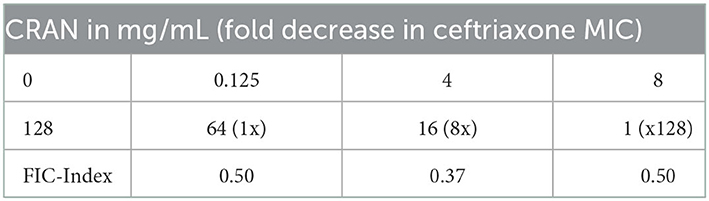
Table 2. Minimal inhibitory concentration (MIC) of ceftriaxone (μg/mL) in the presence of an increasing concentration of the cranberry extract (CRAN) in mg/mL.
The molecular responses of E. coli O7:H18 (ST38) to 1/4MIC of CRAN and its combination with <1/8MIC of AXO (CC) were investigated using RNA-seq after a 3-h exposure period. No bactericidal effect was observed during this period. Exposure to 1/4 MIC CRAN markedly affected the transcriptomic profile of the studied E. coli and appears to be greater than the combined effect of CC (Figure 3A). In pairwise comparisons, overall CRAN induced the highest number of significantly differentially expressed genes (DEGs) compared to that of the untreated control. Exposure to CRAN resulted in 727 DEGs of 4,800 coding sequences, with 232 and 495 genes being upregulated and downregulated, respectively [false discovery rate (FDR) <0.001 and log2-fold change of >2]. Almost a similar number of genes were differentially expressed (712 of 4,800 coding genes) with CC exposure, among which 213 genes were upregulated, while 499 genes were downregulated. No significant difference was observed between CRAN and CC treatments except for citrate lyase synthetase genes (Figure 3B). Treatment with CRAN and CC resulted in the DEGs of citrate lyase synthetase (citC), citrate lyase acyl carrier gamma subunit (citD), citrate lyase citryl-ACP lyase beta subunit (citE), and putative ribosomal protein (ykgMO) which were upregulated with CC compared to CRAN exposure. As expected, exposure to <1/8 MIC AXO (4 μg/mL) did not result in any DEGs at the same threshold limit [false discovery rate (FDR) <0.001, and log2-fold change of >2]. This indicated the potentiation of AXO by CRAN in the CC treatment. Principal component analysis (PCA) also confirmed the significant effect of CRAN and CC on E. coli O7:H18 (ST38) relative to the untreated control and AXO.
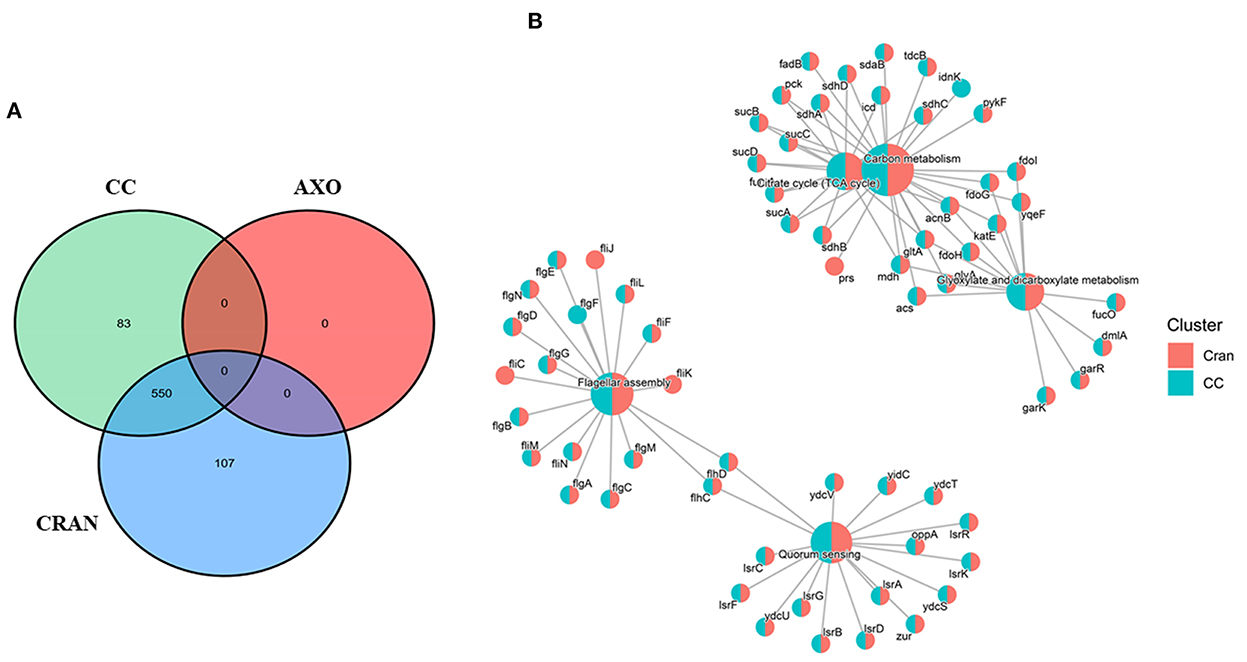
Figure 3. (A) Venn diagram showing overlap of differentially expressed genes following exposure to 4 mg/mL of cranberry pomace extracts (CRAN), 4 μg/mL of AXO, and (4 μg/mL AXO + 4 mg/mL CRAN) CC for 3 h. (B) Gene set enrichment analysis (GSEA) grouped by KEGG pathway showing the clustering of a similar set of differential gene expressions due to CRAN and CC exposures compared to control. A log2-fold change of >2 and false discovery rate (FDR) cutoffs ≤0.001 are used.
Differential expression of genes involved in particular cellular functions or their assignment to the KEGG pathways database reveals that the majority of genes were involved in “microbial metabolism in diverse environments”. Almost 13 genes involved in this category responsible for sulfate/thiosulfate ATP-binding cassette (ABC) transport were differentially expressed and upregulated (log2FC ≥4). Genes from this pathway are interconnected with pathways related to the metabolism of carbon, glyoxylate and dicarboxylate, tryptophan, and in the tricarboxylic acid (TCA) cycle as well (Figures 4, 7). Other categories downregulated by CRAN and CC exposures were those having major effects on flagellar assembly, quorum sensing, and ABC transport (Figure 4) associated with virulence factors of bacteria.
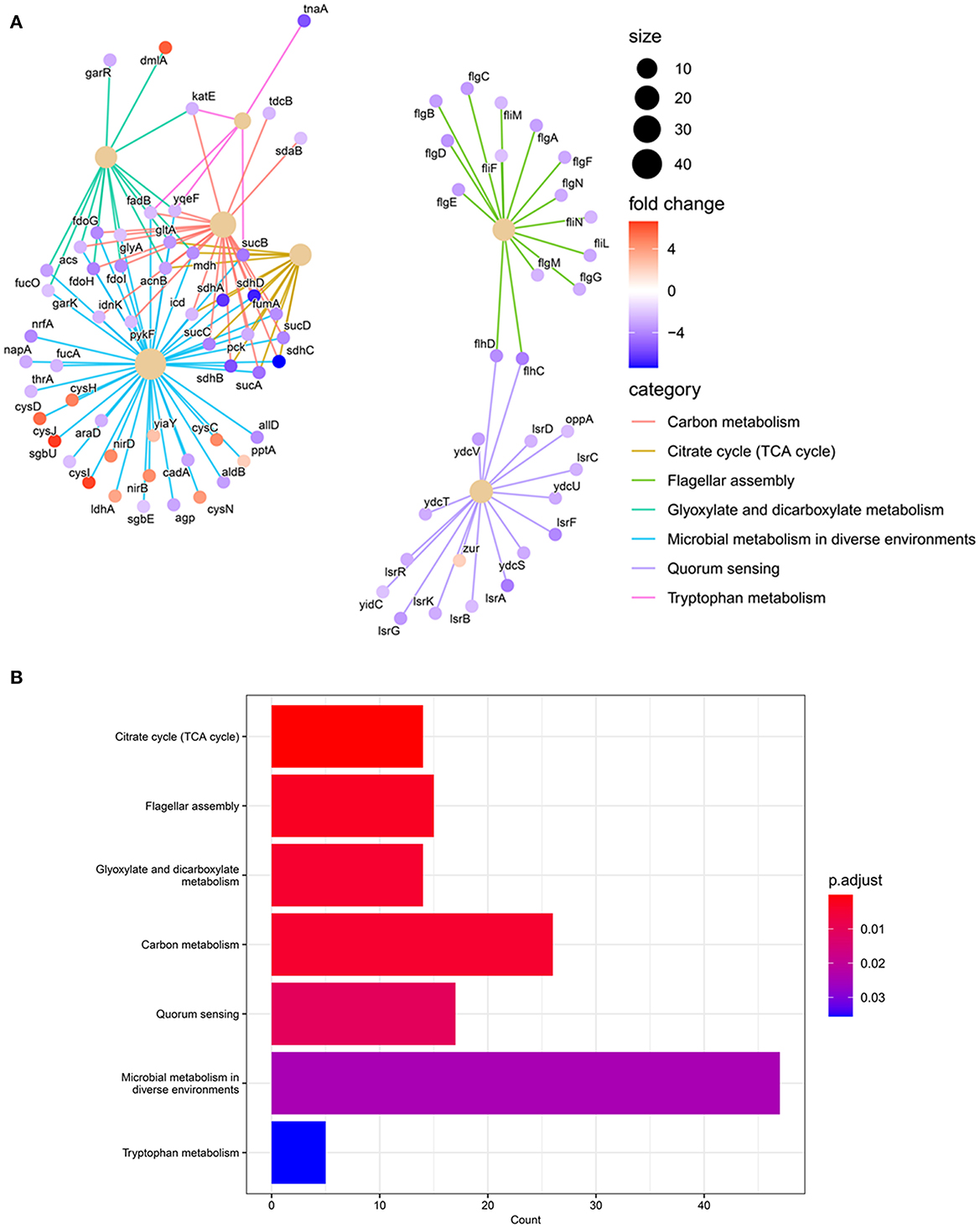
Figure 4. Gene annotation enrichment analysis of KEGG pathway. (A) Differentially expressed genes (DEGs) and KEGG pathway enrichment analysis in 4 μg/mL AXO + 4 mg/mL CRAN (CC) compared to control. The blue circle indicates downregulation, while the red circle indicates upregulation. The darker the color, the stronger the effect. These are combined from 10 different pathways, with none of the pathways providing more than three interactions. (B) Count of affected genes in each KEGG pathway. A log2-fold change of > 2 and false discovery rate (FDR) cutoffs ≤0.001 are used.
Further clarification of the gene functions by GO functional analysis based on DEGs has found about nine important functional categories with transcriptional changes due to CRAN and CC exposures (Figure 5). These categories include genes for proteins related mostly to the “generation of precursor metabolites and energy”; “energy derivation by the oxidation of organic compounds”; “cellular respiration;” and “electron transport chain”. Significant predominant DEGs involved networks among “iron–sulfur cluster binding”,“carbohydrate transmembrane transporter activity”, “monosaccharide transmembrane transporter activity”, “sugar transmembrane transporter activity”, and “heme binding”. Major downregulated genes (nrfABCDEFG) were grouped into the TCA cycle, heme binding, while upregulated DEGs (nirBDC) were found particularly with cellular respiration (Figure 5).
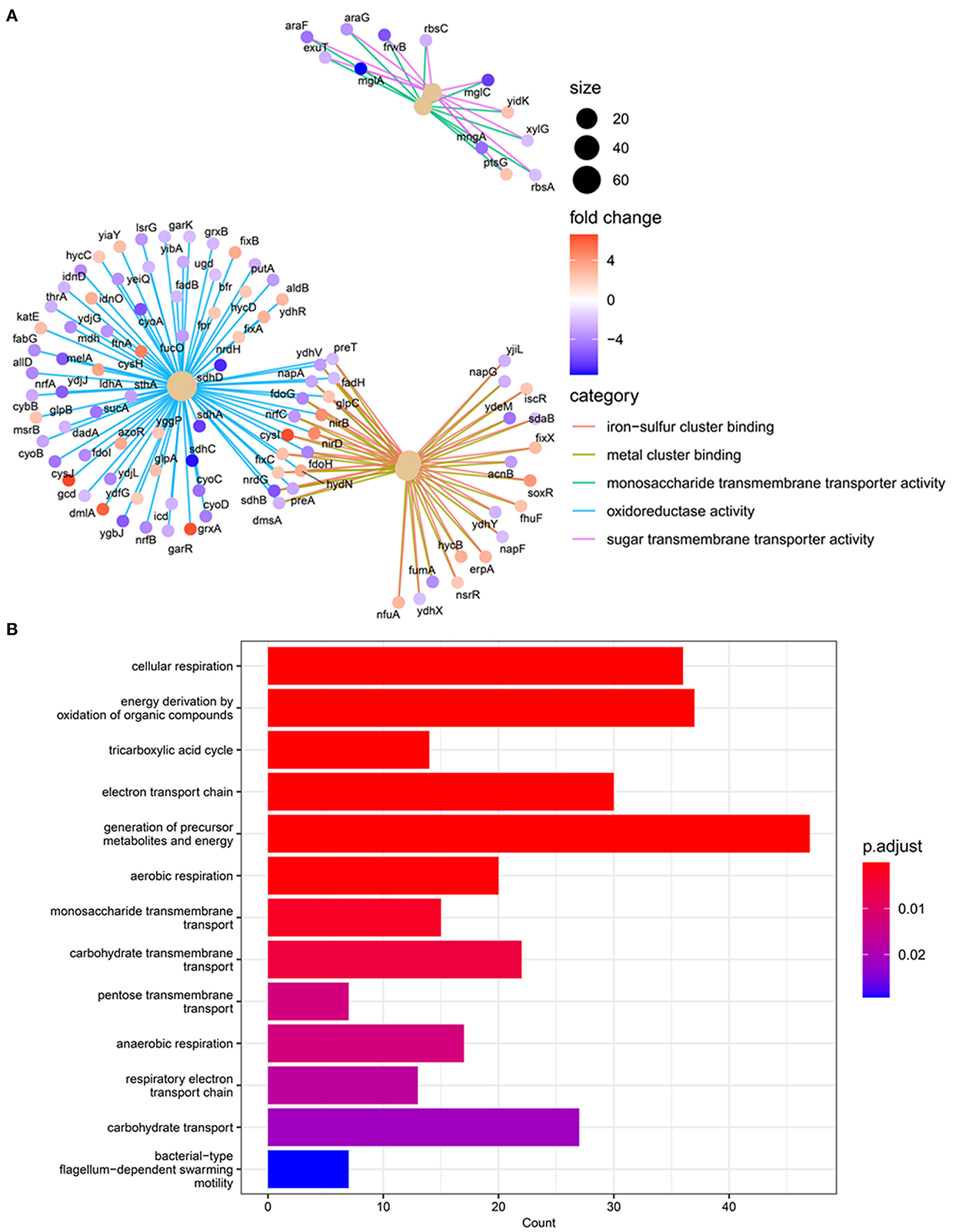
Figure 5. Gene annotation enrichment analysis of GO pathway networking. (A) Differentially expressed genes (DEGs) and GO pathway enrichment analysis of the DEGs in the different treatments of 4 mg/mL of CRAN, and (4 μg/mL AXO + 4 mg/mL CRAN) CC compared to control. The blue circle indicates downregulation, while the red circle indicates upregulation. The darker the color, the stronger the effect. These were combined from 10 different pathways, with none of the pathways providing more than three interactions. (B) Count of affected genes in each GO pathway. A log2-fold change of >2 and false discovery rate (FDR) cutoffs ≤0.001 are used.
To understand the physiological adaptations resulting in multidrug resistance in E. coli O7:H18 (ST38), changes in DEG of resistance-related genes were determined. Almost similar patterns of DEG were exhibited by CRAN and CC exposures (Figure 3B). DEG of 49 genes was observed, with downregulation of tolC, acrAB, emrK, mdtEFG, gadEW, evgS, yjcS, blaCMY − 2, and upregulation of multiple antimicrobial resistance operon (marRAB), tetracycline resistance repressor gene (tetR), quaternary ammonium compound resistance gene (sugE), and regulatory genes for bacterial oxidative defense (soxSR), compared to control. These findings show that CRAN perturbs major multidrug resistance mechanisms in E. coli (Figure 6). The TolC-dependent Acr is a well-known efflux pump system responsible for multidrug expelling and also signaling molecules that coordinates quorum sensing (QS) in Gram-negative bacteria. Exposure to CRAN and CC downregulated the expression of tolC (log2-fold change of >2) along with acrABDEF (although DEGs are <log2FC) and 18 QS-related genes (Figures 4, 7). Significant downregulation of non-specific E. coli cell surface porin genes involved in both antibiotic transport and membrane integrity (ompCFW) was observed.
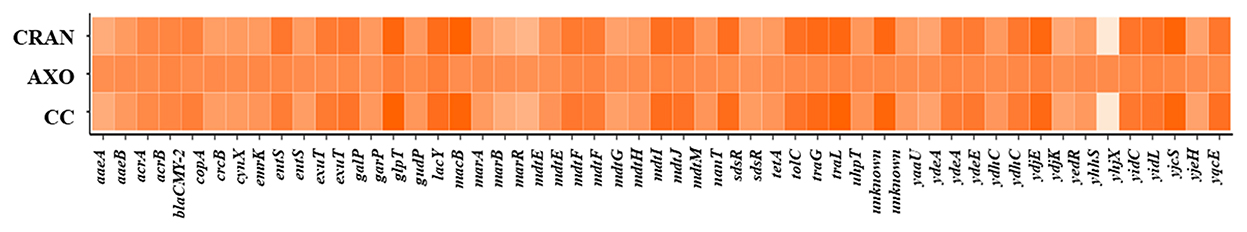
Figure 6. Cluster analysis of differential gene expression (DEG). Heat maps showing log2FC DEGs of antimicrobial resistance genes of E. coli O7:H18 (ST38) treated with 4 mg/mL of CRAN, 4 μg/mL of AXO, and 4 μg/mL AXO + 4 mg/mL CRAN (CC) compared to control for 3 h.
Expression of six major families of efflux pumps of prokaryotes including (i) ATP-binding cassette (ABC), (ii) multidrug and toxin extrusion (MATE) family, (iii) major facilitator superfamily (MFS), and (iv) resistance nodulation cell division (RND) family were largely affected in studied E. coli after exposure to CRAN and CC. Significant DEGs of numerous (65) genes related to efflux pumps have been observed where MFS transporter_DHA3 family_macrolide efflux protein (gene unknown), outer membrane protein_heavy metal efflux system (tolC), and membrane fusion protein_multidrug efflux system (emrK, mdtE) were also downregulated. In contrast, the MFS transporter_DHA1 family: quinolone resistance protein (yhjX, entS), inner membrane transport protein (yedR), OPA family, and glycerol-3-phosphate transporter (mdtG) were upregulated from 2- to 11-folds [false discovery rate (FDR) < 0.001 and log2-fold change of > 2]. Interestingly, the downregulation of some of the MFS transporter from the DHA1 family associated with quinolone resistance (ydhC, exuT, lacY, and gudP), ACS family regulate hexuronate transport (ydjE and yqcE), dicarboxylic acid transport (nanT), proline/betaine transporter (ydjK), and 4-hydroxybenzoate transport (garP) were observed after CRAN and CC exposures.
Different virulence factors for adhesion, invasion, colonization, proliferation, and systemic dissemination are deployed by E. coli during infections. Significant transcriptional downregulation of important genes related to these virulence factors of the studied E. coli have been revealed by RNA-seq (Table 3, Figure 4). Both CRAN and CC upregulated genes in the cys operon (cysACDEHKIJNPW) by 2- to 7-fold. In addition, genes encoding other proteins in the nrd system, such as nrdGH and soxSR, were significantly upregulated as well. Genes regulated by marRAB overlapping with those in the soxRS regulon were also upregulated by CRAN/CC exposure.
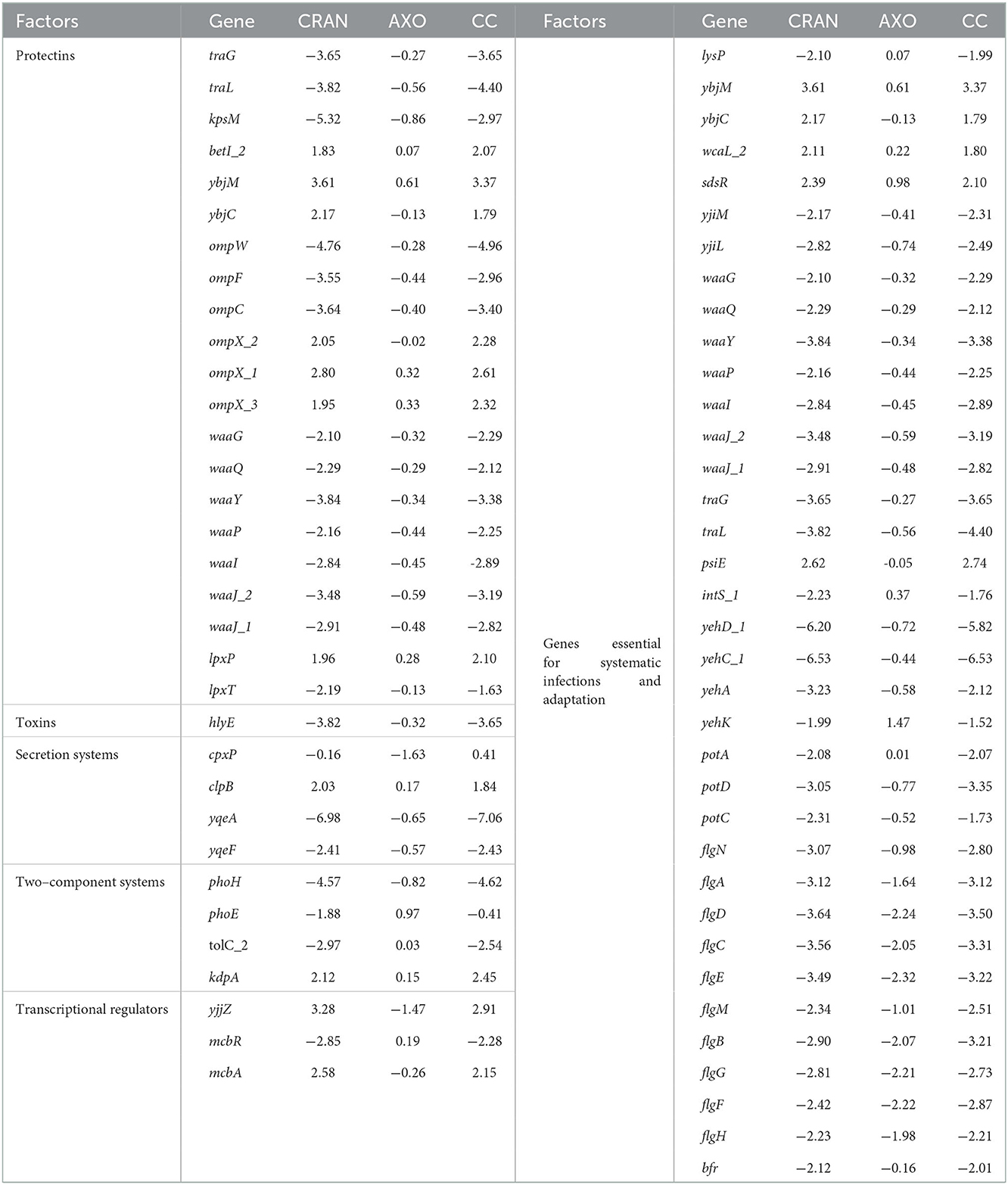
Table 3. Differentially expressed virulence and pathogenesis genes in E. coli O7:H18 (ST38) exposed to 4 mg/mL of CRAN, 4 μg/mL of AXO, and 4 μg/mL AXO + 4 mg/mL CRAN (CC).
Sub-MIC of CRAN alone or its combination with sub-|MIC with AXO caused downregulation of genes such as the type 1 fimbriae (fimC), flagellar hook (flgABCDEFGHMN), curli (csgABDEF), and yadCMK that promotes intracellular survival and motility.
The iron transport system genes modulated by CRAN and CC included ferric enterobactin transporter (fepE), bacterioferritin (bfr), enterobactin synthesis, and transport gene (entS). In addition, the manganese transporter (mntS) gene was downregulated as well.
In adverse conditions, E. coli deploys different protecting genes such as those encoding the complement resistance protein (traGL), capsular polysaccharides (kpsM), outer membrane protein (ompCFW), and major core oligosaccharides operon proteins (waaGJLPQY). Exposure to CRAN and CC significantly downregulated (>2 log2FC) these protectin-related genes. The toxin hlyE (putative avian hemolysis) gene was (>3.5 log2FC) downregulated as well.
The QS, an auto-inducer-based cell communication system, plays an important role in APEC pathogenesis, and biofilm formation genes (lsrABCDFGK) associated with the regulation of QS were significantly downregulated after CRAN and CC exposure compared to control (Figure 7).
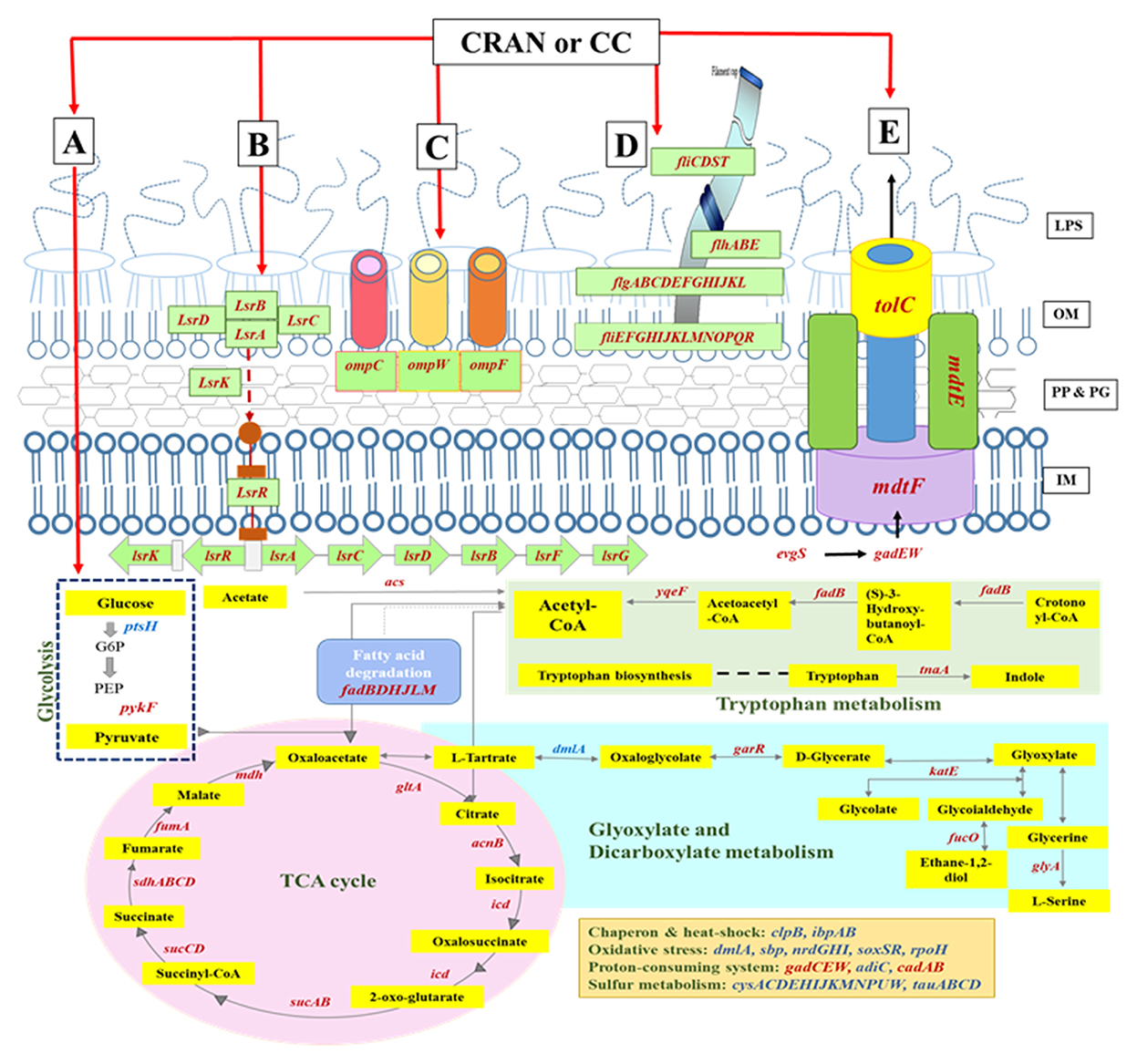
Figure 7. Schematic representation of the differentially expressed genes involved in the E. coli O7:H18 (ST38) upon exposure to CRAN (4 mg/mL of CRAN) or CC (4 μg/mL AXO + 4 mg/mL CRAN) compared to control. The main routes of metabolism (A), quorum sensing (B), outer membrane porins (C), flagella (D), and efflux pump (E)-related transcribed genes are indicated (italics); upregulated genes are marked in blue, and downregulated genes are marked in red. Metabolites are highlighted in yellow rectangles. A log2-fold change of >2 and false discovery rate (FDR) cutoffs ≤0.001 are used.
The Type IV secretion system (hofBC and virB) and their regulator yqeAF encoding factors on the bacterial membrane to enhance attachment were downregulated (>6 log2FC) in presence of CRAN and CC.
In adverse conditions, bacteria enable by downregulating the expression of TCs genes such as phoEH which is important for biofilm formation, and pathogenesis were downregulated in E. coli. Several TCs along with the transcription regulators and McbR (control the colanic acid biofilm formation) gene were downregulated following CRAN and CC exposures. However, an uncharacterized McbA protein gene was upregulated under the same condition.
Mixed expression patterns were observed with metabolism-associated genes. The operons acs-yjcS (encoding acetate assimilation, proliferation, and colonization), potACD (involved in putrescine transport), sucABCD, and sdhABCD (involved in the TCA cycle) were downregulated. However, nirBCD was involved in nitrite transport; and cysABCDEHIJKMNPUW, tauC of sulfur metabolism, was upregulated after exposure to CRAN and CC. Genes involved in maintaining cell envelope, integrity, metabolism, transport systems, and virulence (ybjMC, wcaL, and sdsR) were upregulated, while yjiML and lysP were downregulated. Moreover, genes for LPS synthesis (waaGJLPQY), plasmid function (traGL and psiE), phage-related (intS), adhesion, invasion, and motility (yehACDK, potACD, flgACDN, and bfr) were significantly downregulated by CRAN and CC treatments (Table 3). The schematic representation of sophisticated E. coli O7:H18 (ST38) responses to the CRAN and CC exposure is illustrated in Figure 7.
Upregulation of major heat shock and extra-cytoplasmic stress responses (cysACDEHIJKMNPUW, dmlA, sbp, nrdGHI, soxSR, rpoH, clpB, ibpAB), DNA damage repair (uvrA, uvrB, ruvA, mfd), and chaperones (clpB, ibpAB, spy, hscA, stpA) genes was observed, while the downregulation of major virulence genes was observed by CRAN and CC treatments (Figure 7).
Downregulation of outer membrane porins, OmpF, and OmpC; hypothetical proteins-YicS (play a role in motility, biofilm formation) and cpdB (20,30-cyclic phosphodiesterase)-mediated colonization were observed by CRAN and CC exposures.
Thirteen genes were randomly selected for RT-qPCR (Table 1) to validate the RNA-seq results. Seven of the selected gene showed significantly different transcription levels in response to CRAN or CC compared to the control culture. RNA-seq and RT-qPCR results were in agreement for all tested genes although the values of fold-change were different. In RNA-seq, fimB and fimC were slightly downregulated, while upregulation was observed in RT-qPCR with AXO exposure. Regarding exposure to CRAN and CC, downregulation of fimB, fimC, tolC, and blaCMY − 2 and upregulation of entC, fliG, and floR were observed both in RNA-seq and RT-qPCR compared to control, which indicated the reliability of the RNA-seq expression analysis (Figure 8).
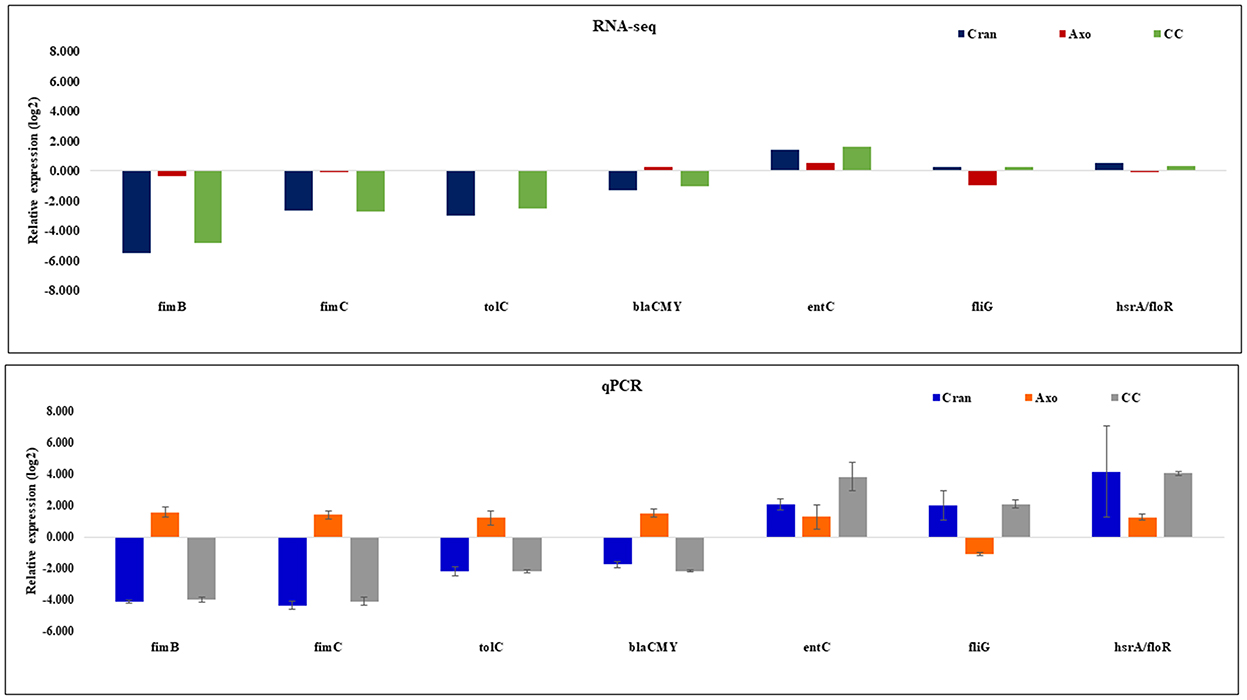
Figure 8. Validation of the RNA-seq data for selected genes by RT-qPCR. RT-qPCR data confirmed the expression trends observed in the RNA-seq data for seven genes in 4 mg/mL of CRAN, 4 μg/mL of AXO, and 4 μg/mL AXO + 4 mg/mL CRAN (CC) compared to control, where rpoD was used as a reference gene.
It is well recognized that AMR is one of the top global public health threats, and the lack of new/effective antibiotics makes the situation even more complicated. The emergence of MDR E. coli, most importantly from food-producing animals, will require the development of antimicrobial agents not only to improve animal health and productivity but also to decrease food safety and public health risks. Avian pathogenic E. coli is a causative agent of septicemia, pericarditis, and mortality in birds causing important economic loss to the poultry industry. Ampicillin, streptomycin, sulfonamide, and tetracycline are some of the antibiotics commonly used to treat E. coli infections (Poirel et al., 2018). In the present study, we have selected eight pan-susceptible and MDR E. coli, among which an E. coli O7:H18 (ST38) isolates, previously reported as an ExPEC, was used for RNA-seq. This isolate presented a resistance spectrum of amoxicillin–clavulanic-acid–ceftiofur–ceftriaxone–ampicillin–cefoxitin–streptomycin–sulfisoxazole–tetracycline–chloramphenicol resistance (Rehman et al., 2022). This isolate induced mortality in day-old chick while clustering with human UTI strains and resistant to ceftriaxone an important third-generation cephalosporin. Urinary tract infections due to Enterobacteriaceae including E. coli in children are commonly treated with a second- or third-generation cephalosporin such as ceftriaxone (Tratselas et al., 2014). Ceftriaxone exhibits time-dependent bactericidal activity and concentrations may be sub-therapeutic in patients. Cranberry fruit compounds have been reported to exert anti-adhesion and antimicrobial activities against several pathogenic bacteria and have been suggested to prevent urinary tract infections caused by uropathogenic P-type E. coli (Foo et al., 2000; Cimolai and Cimolai, 2007; Howell, 2007). Thus, the earlier description justifies the use of cranberry and ceftriaxone. The cranberry pomace extracts used in this study induced similar concentration-dependent growth-inhibitory actions against all selected isolates. Varying MIC ranges of cranberry products having different compositions have been reported (Das et al., 2017). Highly soluble proanthocyanidins (PAC) from cranberry juice extracts showed higher antimicrobial activities compared to whole berry pomace-derived products (Howell et al., 2022), which could be the reason for the higher MIC value (16 mg/mL) found in the present study. However, from an economic perspective, pomace-derived cranberry extracts can be a potential natural antimicrobial agent which can be used against antibiotic-resistant bacteria in livestock.
Extracts of American cranberry (Vaccinium macrocarpon) are well-known for their antimicrobial activity against urinary tract infections caused by E. coli. Although the anti-adhesive effects of cranberry extracts on P-fimbriae-mediated adhesion of uropathogenic E. coli are the most reported mechanism (Howell et al., 2022), the underlying mode of antimicrobial-actions of cranberry products is still the subject of investigations. Using a DNA array-based approach, treatment of E. coli with cranberry products showed the downregulation of ompF, tolQ, and gad involved in membrane functions, maintenance of ionic balance, and protection against high-proton-concentration environments (González-Lamothe et al., 2009). The emergence of MDR E. coli along with their association with virulence in poultry production has driven us to look for potentiating activity of a cranberry pomace extract (CRAN) of various antibiotics among which ceftriaxone showed an additive inhibitory activity with CRAN against ceftriaxone-resistant E. coli isolates (MIC >32 μg/mL). Ceftriaxone is a third-generation cephalosporin used to treat E. coli infections. The World Health Organization (WHO) has declared ceftriaxone-resistant Enterobacteriaceae as priority pathogens of critical importance (Chua and Stewardson, 2019). Combination of sub-MIC of CRAN with sub-MICs of ceftriaxone (AXO) or sulfisoxazole (MIC>256 μg/mL) induced strong growth-inhibitory activity against an MDR E. coli O7:H18 (ST38); however, the potentiation of sulfisoxazole activity was moderate (data not shown). These results indicated that the potentiation level of CRAN is antimicrobial class-dependent. The findings in the present study are in agreement with those reported by Gallique et al. (2021) and provide evidence of the β-lactam-potentiating activity of cranberry compounds against MDR bacteria (Diarra et al., 2013).
In Gram-negative bacteria, T3SS acts as a channel that allows the chaperon-regulated translocation of effector proteins directly into the host cell (Slater et al., 2018), while T6SS influences the expression of type 1 fimbriae and flagella, two extracellular structures required for adhesion and biofilm formation (Navarro-Garcia et al., 2019). Thus, all studies on the antimicrobial activity of cranberry extracts, particularly against Gram-negative bacteria, have shown that its PAC content is responsible for affecting genes involved in bacterial virulence (Ranfaing et al., 2018; Maisuria et al., 2019; Howell et al., 2022). The affected genes are involved in bacterial adhesion, invasion, flagellar biosynthesis, motility pathway, iron metabolism, stress response, and biofilm development. The biofilm-forming capacity has not been evaluated in this study. However, results showed differential expression of biofilm-associated genes in the study isolates exposed to CRAN or CC. These findings in regard to polyphenols' ability to affect biofilm formation are in agreement with our previous study (Diarra et al., 2020). Our transcriptomic analysis revealed a similar differential expression of those virulence genes in the sub-MIC of CRAN and its combination with the sub-MIC of AXO (CC)-treated E. coli cells. While exposure to only sub-MIC of AXO did not result in any significant DEGs, CC exposure displayed adjuvant properties of CRAN by interfering with virulence, quorum sensing, and metabolism mechanisms of E. coli, indicating that CRAN may have the potential to enhance the effectiveness of existing third-generation cephalosporins such as AXO.
Tetracycline is among the antibiotics frequently used to treat colibacillosis in chickens (Kathayat et al., 2021). As a result, E. coli developed resistance against tetracycline as a consequence of the selective pressure (Poirel et al., 2018). Although, nine tetracycline efflux genes have been identified in the studied E. coli O7:H18 (ST38), the upregulation of the tetracycline resistance repressor protein TetR was observed in our RNA-seq assay. Moreover, the yhjX MFS transporter, which has been reported to regulate the growth of E. coli in the presence of a sub-MIC of gentamicin, was upregulated more than 11-fold in the presence of CRAN and CC. The production of ESBL and AmpC β-lactamases is the most common cause of ceftriaxone resistance in E. coli. In addition to these acquired resistance mechanisms, E. coli also develop intrinsic resistance mechanisms, such as drug efflux and low membrane permeability against β-lactam antibiotics (Gallique et al., 2021). We have found the downregulation of the ampC-like blaCMY − 2 gene and the upregulation of sugE (quaternary ammonium compound resistance) often associated with β-lactamase class C upon CRAN and CC exposures. β-lactams cross the outer membrane by diffusion through the OmpC and OmpF porins. The outer membrane of Gram-negative bacteria is composed of lipopolysaccharides (LPS) and glycerophospholipid bilayers which restrict the penetration of polar antibiotics mostly by porins (Xiao et al., 2016; Choi, 2019). Single point mutations can reduce the channel size or alter electrostatic interactions in the channel, decreasing permeability and reducing antibiotic susceptibility (Bajaj et al., 2016). Reduction of expression of these porins (OmpCFW) along with blaCMY − 2 in CRAN- and CC-treated E. coli compared to control could explain in part the potentiation of AXO by CRAN. These findings could lead to the development of cranberry products in combination with third-generation cephalosporin such as AXO against resistant E. coli.
The autoinducer-2 (AI-2) quorum sensing (QS) system mediates interspecies communication by controlling genes involved in flagella, motility, and chemotaxis (Yu et al., 2021). Our data showed that CRAN affected QS via the downregulation of the lsrABCDFGK operon which could be due to its PACs content previously reported to inhibit QS in P. aeruginosa in a fruit fly animal model (Maisuria et al., 2016). The used CRAN in the present study contained 89% phenolic compounds where 88% accounted for total tannins (Ross et al., 2017). Moreover, UPLC-MS analysis measurement showed that anthocyanin (peonidin-3-O-galactoside and cyanidin-3-O-galactoside) and flavonols (quercetin-3-O-galactoside and myricetin-3-O-galactoside) were the principal components of CRAN along with their phenolic components (protocatechuic acid, catechin, epicatechin, chlorogenic acid, and trans-cinnamic acid) (Das et al., 2019; Diarra et al., 2020). Catechin gallates such as epicatechin gallate and epigallocatechin gallate have been reported to enhance the multidrug NorA efflux system in Staphylococcus aureus at low concentrations (Sharma and Gupta, 2019).
Intrinsic multidrug resistance (MDR) in bacterial species occurs by mutation to facilitate the upregulation of broad-spectrum efflux pumps, including acrAB and tolC (Rosner, 2013). The synergistic action of efflux pumps and the outer permeability membrane regulates intracellular antibiotics penetration (Krishnamoorthy et al., 2017). The outer membrane pore-forming protein TolC of E. coli interacts with nine inner membrane efflux pumps to create networking among cell envelope, metabolism, and efflux pumps. We observed the downregulation of tolC and other associated regulons (phoH and evgAS), while the upregulation of paralogous activators of tolC (marAB and soxSR) was noted. Inactivation of all these genes may lead to a significant increase in susceptibility to antibiotics while impacting bacterial physiology and virulence (Zgurskaya et al., 2011). Moreover, the observed downregulation of tolC and other associated membrane fusion protein genes (acrAB, emrK, and mdtE) belonged to the multidrug efflux system and biofilm formation, which indicates that CRAN may act against biofilm formation in agreement with our previous research (Diarra et al., 2020).
Three types of transport systems including ABC-type, RND-type, and MFS-type involve TolC (Garmory, 2004). In the present study, ABC transporters that help in the uptake of various nutrients (metal ions, such as iron, zinc, and manganese) were differentially expressed in presence of CRAN or CC. Accordingly, more than 50 genes associated with ABC transportation were largely downregulated with CRAN and CC exposures compared to control. As an example, potABCDF involved in the polyamine transportation of E. coli was downregulated with CRAN and CC exposures.
Under adverse conditions, bacteria often use a metabolic cue to regulate virulence and metabolic functions (Rohmer and Hocquet, 2011). The iron chelation capability of cranberry extracts has been described in many studies. Exposure of E. coli to cranberry extracts downregulated the expression of several iron uptake-related genes including ent, feo, fep, and exb, which are known to be negatively regulated by the transcriptional repressor Fur to limit the accumulation of intracellular iron and to prevent iron-derived oxidative stresses (González-Lamothe et al., 2009). However, upregulation of iron cofactors such as iron–sulfur (Fe–S) clustering has not been previously observed following E. coli treatment with cranberry products. In the present study, multiple Fe–S cluster assembly pathways predominantly coordinated by Cys and Isc proteins (Kertesz, 2001) were strongly upregulated in E. coli treated with CRAN and CC. Furthermore, acid stress also causes sulfate reduction that may activate cysteine synthesis which is important for enzyme catalysis, protein folding, and assembly (Yu et al., 2021). Cysteine is the most common amino acid residue in the Fe–S clusters upregulated by the CRAN and CC treatments. Moreover, oxidative stress may induce marAB along with the transcriptional regulator soxRS as was observed in this study (Wang et al., 2009). The induction of sulfur metabolism in response to cysteine biosynthesis could be a cell protection strategy induced by oxidative stress due to exposure to CRAN and CC treatments.
The final enzyme in glycolysis, pyruvate kinase, generates ATP at the substrate level and it was downregulated 2.5-fold under the CRAN and CC treatments. The absence of oxygen may cause downregulation of the citric acid cycle which leads to incomplete oxidation of sugars (Förster, 2014). We have seen the downregulation of multiple enzymes involved in TCA cycles which reflects the regulation of energy metabolism of E.coli under CRAN or CC stress. This response could be a cellular energy conservation procedure to produce less ATP. Moreover, the downregulation of methionine-(R)-sulfoxide reductase protein (MsrB) and hyperosmotically inducible periplasmic protein (OsmY), which are crucial for the amelioration of cellular damage from reactive oxygen species, and upregulation of Fe–S cluster proteins (Cys, NfuA, TusA, YtfEGH, and IscAR) increased from 2- to 7-fold. This could lead to a repair mechanism of cell protection from oxidative stress or metal chelation activities of CRAN pomace extracts.
Findings in the present study confirmed the antibacterial activities of cranberry extract against pathogenic E. coli isolates which could be associated with the downregulation of several metabolism pathways involved in their survival and virulence. The increased and decreased potentiation of β-lactam antibiotics (third-generation cephalosporin) and biofilm formation capacity, respectively, need to be established in an in vivo challenge model in future studies to investigate its potential to enhance the effectiveness of existing third-generation cephalosporins such as AXO in controlling colibacillosis and other infections caused by multi-resistant ExPEC of poultry and human.
Growth assays and transcriptomic analysis revealed an additive inhibitory activity between extracts of cranberry pomace and ceftriaxone that may help to develop strategies against cephalosporin-resistant and virulent E. coli. This additive activity translates to reducing the MIC values of both cranberry products and ceftriaxone. Downregulation of major cell envelope or membrane components revealed that exposure to our investigated cranberry extracts and their combination with ceftriaxone affects E. coli cell structure integrity. The most important findings indicated that CRAN alone or in combination with AXO (1) downregulated the ampC-like blaCMY − 2, (2) enhanced cysteine biosynthesis and several regulatory genes responsive to oxidative stress, and (3) hindered the efflux systems pump by downregulating the expression of their major determinants. Inactivation of efflux pump activity has been previously reported to negatively impact virulence and biofilm formation in Gram-negative bacteria. Most importantly, the alteration of the outer membrane proteins that control the permeability barriers along with expression reduction of different AGRs demonstrates the ability of our cranberry pomace extract to potentiate the activities of third-generation cephalosporins against resistant E. coli.
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/geo/, GSE205240.
MD (principal investigator) conceptualized the study and its design with PB. QD, T-LH, XY, DL, GC, JT, and MD performed experiments and data analysis. QD wrote the manuscript with major contributions of KR, MD, and PB. MD provided overall guidance, mentorship, and resources throughout the scope of this study. All authors approved this work for publication.
This study was supported by the Agriculture and Agri-Food Canada to MD through the A-Base (PSS #2574, J-001788), the Genomics Research and Development Initiative (PSS #1858 J-001262), and projects on Mitigating Antimicrobial Resistance.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Aleksandrowicz, A., Khan, M. M., Sidorczuk, K., and Noszka, M. (2021). Whatever makes them stick–Adhesins of avian pathogenic Escherichia coli. Vet. Microbiol. 257, 109095. doi: 10.1016/j.vetmic.2021.109095
Awosile, B., Eisnor, J., Saab, M. E., and Heider, L. (2021). Occurrence of extended-spectrum β-lactamase and AmpC-producing Escherichia coli in retail meat products from the Maritime Provinces, Canada. Canad. J. Microbiol. 67, 537–547. doi: 10.1139/cjm-2020-0442
Bajaj, H., Scorciapino, M. A., Moyni,é, L., Page, M. G., Naismith, J. H., Ceccarelli, M., et al. (2016). Molecular basis of filtering carbapenems by porins from β-lactam-resistant clinical strains of Escherichia coli. J. Biol. Chem. 291, 2837–2847. doi: 10.1074/jbc.M115.690156
Cepas, V., López, Y., Muñoz, E., Rolo, D., Ardanuy, C., Mart,í, S., et al. (2019). Relationship between biofilm formation and antimicrobial resistance in gram-negative bacteria. Microbial Drug Resist. 25, 72–79. doi: 10.1089/mdr.2018.0027
Chalmers, G., Cormier, A. C., Nadeau, M., Côt,é, G., and Reid-Smith, R. J. (2017). Determinants of virulence and of resistance to ceftiofur, gentamicin, and spectinomycin in clinical Escherichia coli from broiler chickens in Québec, Canada. Vet. Microbiol. 203, 149–157. doi: 10.1016/j.vetmic.2017.02.005
Chicken Farmers of Canada (2020). Antibiotics. Available online at: https://www.chickenfarmers.ca/antibiotics/ (accessed Feb 28, 2022).
Ching, C., and Zaman, M. H. (2021). Identification of multiple low-level resistance determinants and coselection of motility impairment upon Sub-MIC ceftriaxone exposure in Escherichia coli. mSphere. 6, e00778–21. doi: 10.1128/mSphere.00778-21
Choi, U. (2019). Antimicrobial Agents that Inhibit the Outer Membrane Assembly Machines of Gram-Negative Bacteria. doi: 10.4014/jmb.1804.03051
Chua, K. Y. L., and Stewardson, A. J. (2019). Individual and community predictors of urinary ceftriaxone-resistant Escherichia coli isolates, Victoria, Australia. Antimicrob. Resist. Infect. Control 8, 36. doi: 10.1186/s13756-019-0492-8
Cimolai, N., and Cimolai, T. (2007). The cranberry and the urinary tract. Eur. J. Clin. Microbiol. Infect. Dis. 26, 767–776. doi: 10.1007/s10096-007-0379-0
da Silva, G. J., and Mendonça, N. (2012). Association between antimicrobial resistance and virulence in Escherichia coli. Virulence 3, 18–28. doi: 10.4161/viru.3.1.18382
Das, Q., Islam, M. R., Marcone, M. F., and Warriner, K. (2017). Potential of berry extracts to control foodborne pathogens. Food Control 73, 650–662. doi: 10.1016/j.foodcont.2016.09.019
Das, Q., Lepp, D., Yin, X., Ross, K., McCallum, J. L., Warriner, K., et al. (2019). Transcriptional profiling of Salmonella enterica serovar Enteritidis exposed to ethanolic extract of organic cranberry pomace. PLoS ONE 14, e0219163. doi: 10.1371/journal.pone.0219163
Diarra, M. S., Block, G., Rempel, H., Oomah, B. D., Harrison, J., McCallum, J., et al. (2013). In vitro and in vivo antibacterial activities of cranberry press cake extracts alone or in combination with β-lactams against Staphylococcus aureus. BMC Complem. Altern. Med. 13, 1–14. doi: 10.1186/1472-6882-13-90
Diarra, M. S., Hassan, Y. I., Block, G. S., Drover, J. C., and Delaquis, P. (2020). Antibacterial activities of a polyphenolic-rich extract prepared from American cranberry (Vaccinium macrocarpon) fruit pomace against Listeria spp. LWT 123, 109056. doi: 10.1016/j.lwt.2020.109056
Foo, L. Y., Lu, Y., and Howell, A. B. (2000). A-Type proanthocyanidin trimers from cranberry that inhibit adherence of uropathogenic P-Fimbriated escherichia coli. J. Nat. Prod. 63, 1225–1228. doi: 10.1021/np000128u
Förster, A. H. (2014). Metabolic engineering of Escherichia coli for production of mixed-acid fermentation end products. Front. Bioeng. Biotechnol. 2, 16. doi: 10.3389/fbioe.2014.00016
Gallique, M., Wei, K., Maisuria, V. B., Okshevsky, M., McKay, G., Nguyen, D., et al. (2021). Cranberry-derived proanthocyanidins potentiate β-lactam antibiotics against resistant bacteria. Appl. Environ. Microbiol. 87, e00127–e00121. doi: 10.1128/AEM.00127-21
Garmory, H. S. (2004). ATP-binding cassette transporters are targets for the development of antibacterial vaccines and therapies. Infect. Immun. 72, 6757–6763. doi: 10.1128/IAI.72.12.6757-6763.2004
González-Lamothe, R., Mitchell, G., Gattuso, M., Diarra, M. S., and Malouin, F. (2009). Plant antimicrobial agents and their effects on plant and human pathogens. Int. J. Mol. Sci. 10, 3400–3419. doi: 10.3390/ijms10083400
Government of Canada (2017). Food and Drugs Act. Regulations Amending the Food and Drug Regulations (Veterinary Drugs-Antimicrobial Resistance). Available online at: http://gazette.gc.ca/rp-pr/p2/2017/2017-05-17/html/sor-dors76-eng.html (accessed Feb 28, 2022).
Harmidy, K., and Tufenkji, N. (2011). Perturbation of host cell cytoskeleton by cranberry proanthocyanidins and their effect on enteric infections. PLoS ONE 6, e27267. doi: 10.1371/journal.pone.0027267
Howell, A. B. (2007). Bioactive compounds in cranberries and their role in prevention of urinary tract infections. Mol. Nutr. Food Res. 51, 732–737. doi: 10.1002/mnfr.200700038
Howell, A. B., Dreyfus, J. F., and Chughtai, B. (2022). Differences in urinary bacterial anti-adhesion activity after intake of Cranberry dietary supplements with soluble vs. insoluble proanthocyanidins. J. Diet. Suppl. 19, 621–639. doi: 10.1080/19390211.2021.1908480
Juhas, M., Van Der Meer, J. R., Gaillard, M., Harding, R. M., and Hood, D. W. (2009). Genomic islands: tools of bacterial horizontal gene transfer and evolution. FEMS Microbiol. Rev. 33, 376–393. doi: 10.1111/j.1574-6976.2008.00136.x
Kathayat, D., Lokesh, D., and Ranjit, S. (2021). Avian pathogenic Escherichia coli (APEC): an overview of virulence and pathogenesis factors, zoonotic potential, and control strategies. Pathogens 10, 467. doi: 10.3390/pathogens10040467
Kertesz, M. A. (2001). Bacterial transporters for sulfate and organosulfur compounds. Res. Microbiol. 152, 279–290. doi: 10.1016/S0923-2508(01)01199-8
Krishnamoorthy, G., Leus, I. V., Weeks, J. W., Wolloscheck, D., and Rybenkov, V. V. (2017). Synergy between active efflux and outer membrane diffusion defines rules of antibiotic permeation into Gram-negative bacteria. MBio 8, e01172–e01117. doi: 10.1128/mBio.01172-17
Leekitcharoenphon, P., Johansson, M. H. K., Munk, P., Malorny, B., Skarzyńska, M., Wadepohl, K., et al. (2021). Genomic evolution of antimicrobial resistance in Escherichia coli. Sci. Rep. 11, 1–12. doi: 10.1038/s41598-021-93970-7
Maisuria, V. B., Los Santos, Y. L. D., and Tufenkji, N. (2016). Cranberry-derived proanthocyanidins impair virulence and inhibit quorum sensing of Pseudomonas aeruginosa. Sci. Rep. 6, 1–12. doi: 10.1038/srep30169
Maisuria, V. B., Okshevsky, M., and Déziel, E. (2019). Proanthocyanidin interferes with intrinsic antibiotic resistance mechanisms of gram-negative bacteria. Adv. Sci. 6, 1802333. doi: 10.1002/advs.201802333
Meuskens, I., Saragliadis, A., and Leo, J. C. (2019). Type V secretion systems: an overview of passenger domain functions. Front. Microbiol. 10, 1163. doi: 10.3389/fmicb.2019.01163
Navarro-Garcia, F., Ruiz-Perez, F., and Cataldi, Á. (2019). Type VI secretion system in pathogenic Escherichia coli: structure, role in virulence, and acquisition. Front. Microbiol. 1965. doi: 10.3389/fmicb.2019.01965
Poirel, L., Madec, J. Y., Lupo, A., Schink, A. K., Kieffer, N., Nordmann, P., et al. (2018). Antimicrobial resistance in Escherichia coli. Microbiol. Spectrum 6, 6–4. doi: 10.1128/microbiolspec.ARBA-0026-2017
Ranfaing, J., Dunyach-Remy, C., Louis, L., and Lavigne, J. P. (2018). Propolis potentiates the effect of cranberry (Vaccinium macrocarpon) against the virulence of uropathogenic Escherichia coli. Sci. Rep. 8, 1–11. doi: 10.1038/s41598-018-29082-6
Rehman, M. A., Rempel, H., Carrillo, C. D., Ziebell, K., Allen, K., Manges, A., et al. (2022). Virulence genotype and phenotype of multiple antimicrobial-resistant escherichia coli isolates from broilers assessed from a “One-Health” perspective. J. Food Prot. 85, 336–354. doi: 10.4315/JFP-21-273
Rohmer, L., and Hocquet, D. (2011). Are pathogenic bacteria just looking for food? Metabolism and microbial pathogenesis. Trends Microbiol. 19, 341–348. doi: 10.1016/j.tim.2011.04.003
Rosner, J. L. (2013). Reduction of cellular stress by TolC-dependent efflux pumps in Escherichia coli indicated by BaeSR and CpxARP activation of spy in efflux mutants. J. Bacteriol. 195, 1042–1050. doi: 10.1128/JB.01996-12
Ross, K., Ehret, D., Godfrey, D., Fukumoto, L., and Diarra, M. (2017). Characterization of pilot scale processed Canadian organic cranberry (Vaccinium macrocarpon) and blueberry (Vaccinium angustifolium) juice pressing residues and phenolic-enriched extractives. Int. J. Fruit Sci. 17, 202–232. doi: 10.1080/15538362.2017.1285264
Sharma, A., and Gupta, V. K. (2019). Efflux pump inhibitors for bacterial pathogens: from bench to bedside. Indian J. Med. Res. 149, 129. doi: 10.4103/ijmr.IJMR_2079_17
Slater, S. L., Sågfors, A. M., Pollard, D. J., Ruano-Gallego, D., and Frankel, G. (2018). “The type III secretion system of pathogenic Escherichia coli,” in Frankel G., Ron E., eds Escherichia coli, a Versatile Pathogen. Current Topics in Microbiology and Immunology, vol 416 (Cham: Springer). doi: 10.1007/82_2018_116
Souza, D., Oka, G., Alvarez-Martinez, C., et al. (2015). Bacterial killing via a type IV secretion system. Nat. Commun. 6, 6453. doi: 10.1038/ncomms7453
Tratselas, A., Simitsopoulou, M., Giannakopoulou, A., Dori, I., Saoulidis, S., Kollios, K., et al. (2014). Effect of Ceftriaxone on the Outcome of Murine Pyelonephritis Caused by Extended-Spectrum-β-Lactamase-Producing Escherichia coli. doi: 10.1128/AAC.03974-14
Varga, C., Guerin, M. T., Brash, M. L., Slavic, D., and Boerlin, P. (2019). Antimicrobial resistance in fecal Escherichia coli and Salmonella enterica isolates: a two-year prospective study of small poultry flocks in Ontario, Canada. BMC Vet. Res. 15, 1–10. doi: 10.1186/s12917-019-2187-z
Wang, S., Deng, K., Zaremba, S., Deng, X., Lin, C., Wang, Q., et al. (2009). Transcriptomic response of Escherichia coli O157, H7 to oxidative stress. Appl. Environ. Microbiol. 75, 6110–6123. doi: 10.1128/AEM.00914-09
Xiao, M., Lai, Y., Sun, J., and Chen, G. (2016). Transcriptional regulation of the outer membrane porin gene ompW reveals its physiological role during the transition from the aerobic to the anaerobic lifestyle of Escherichia coli. Front. Microbiol. 7, 799. doi: 10.3389/fmicb.2016.00799
Yu, L., Zhang, S., Xu, Y., Mi, X., Xing, T., Li, J., et al. (2021). Acid resistance of E. coli O157, H7 O26: H11 exposure to lactic acid revealed by transcriptomic analysis. LWT 136, 110352. doi: 10.1016/j.lwt.2020.110352
Zgurskaya, H. I., Krishnamoorthy, G., and Ntreh, A. (2011). Mechanism and function of the outer membrane channel tolc in multidrug resistance and physiology of enterobacteria. Front. Microbiol. 2, 189. doi: 10.3389/fmicb.2011.00189
Keywords: cranberry, multi-drug resistant E. coli, 3rd generation cephalosporin, β-lactamase, efflux pump
Citation: Das Q, Hasted T-L, Lepp D, Yin X, Tang J, Chalmers G, Ross K, Boerlin P and Diarra MS (2023) Transcriptional profiling of extraintestinal Escherichia coli exposed to cranberry pomace extracts alone or in combination with ceftriaxone. Front. Sustain. Food Syst. 6:957099. doi: 10.3389/fsufs.2022.957099
Received: 30 May 2022; Accepted: 09 December 2022;
Published: 30 January 2023.
Edited by:
Swati Vishwas Pund, Biobay, IndiaReviewed by:
Priti Desai, Institute of Advanced Research (IAR), IndiaCopyright © 2023 Gabhan Chalmers, Patrick Boerlin, and His Majesty the King in Right of Canada, as represented by the Minister of Agriculture and Agri-Food Canada for the contribution of Quail Das, Teri-Lyn Hasted, Dion Lepp, Xianhua Yin, Joshua Tang, Kelly Ross and Moussa S. Diarra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Moussa S. Diarra,  bW91c3NhLmRpYXJyYUBhZ3IuZ2MuY2E=
bW91c3NhLmRpYXJyYUBhZ3IuZ2MuY2E=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.