
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst. , 27 June 2022
Sec. Agroecology and Ecosystem Services
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.943080
This article is part of the Research Topic Advances in Molecular Crop Pathology View all 9 articles
Phytoplasmas are important prokaryotic pathogenic bacteria without cell walls, which were formerly known as mycoplasma-like organisms, and belong to the Mollicutes class, Candidatus Phytoplasma genus. They are widely distributed in plants and insects, and can cause serious diseases in important food crops, vegetables, fruit trees, ornamental plants and trees, resulting in huge economic losses. To date, more than 100 phytoplasma diseases have been reported in China, which are distributed throughout the country. Jujube witches'-broom, paulownia witches'-broom, wheat blue dwarf, banana bunchy top, sugarcane white leaf, rice orange leaf and mulberry dwarf represent the phytoplasma diseases causing the most serious damage in China. New phytoplasma diseases and their strains are being reported continuously, indicating that phytoplasmas are more diverse than previously thought. Phytoplasmas are mainly transmitted by insect vectors, such as leafhopper and planthopper, and can also be spread by grafting or Cuscuta australis (known as dodder). Mixed infections of phytoplasmas and viruses, bacteria, and spiroplasmas have also become a serious problem in several crops and are responsible for more synergistic losses. With the continuous development and improvement of technology, molecular biological detection has become the main technique for phytoplasma detection and identification. Currently, research on phytoplasma diseases in China mainly focuses on pathogen identification and classification, and insect vector and host diversity; however, there is less focus on pathogenicity, comparative genomics, and effect factors. More research attention has been paid to wheat blue dwarf phytoplasma, paulownia witches'-broom phytoplasma, jujube witches'-broom phytoplasma, and sugarcane white leaf phytoplasma. Other phytoplasma diseases have been reported; however, there have been no in-depth studies. In this paper, the history and present situation of phytoplasma research, and the status, distribution, and diversity of phytoplasma diseases are summarized, and some possible research directions of phytoplasma in the future in China are proposed.
Phytoplasma was first discovered by Doi et al. in Japan in 1967, and was initially termed a mycoplasma-like organism (MLO) (Doi et al., 1967). In 1992, at the 9th International Organization of Mollicutes (IOM) conference, it was first proposed to replace MLO with “phytoplasma” (Yang et al., 2020). Phytoplasma belongs to Mollicutes, Candidatus Phytoplasma genus. It is a pleiomorphic procaryotic organism with no cell wall that inhabits the phloem in infected plants and has a volume of 50–1,000 nm (Lai et al., 2008; Yang et al., 2020). Based on the 16S rRNA gene sequence, phytoplasma has been identified and classified into 52 Candidatus species, comprising 34 groups and more than 100 subgroups presently (Yang et al., 2020).
Phytoplasmas cause more than 1,000 kinds of plant disease worldwide, resulting in large economic losses to agricultural and forestry production (Che et al., 2009). The typical symptoms of phytoplasma infection include witches'-broom, yellow leaves, floral metamorphosis, plant dwarfing, and phloem tissue necrosis (Yang et al., 2014a; Geng et al., 2015). Phytoplasmas can be transmitted by vegetative propagation of host plants, parasitic plants, and piercing-sucking insect vectors, such as leafhopper and planthopper (Weintraub and Beanland, 2006; Sugio et al., 2011). Although a few phytoplasmas can be cultured in vitro (Contaldo et al., 2012), their culture conditions are strict and their artificial cultivation is difficult, making the study of phytoplasma pathogenesis challenging. The first complete sequence of a phytoplasma genome was produced in 2014, allowing a preliminary understanding of the pathogenic effectors of phytoplasma (Oshima et al., 2004). To date, 7 complete phytoplasma genome sequences (Oshima et al., 2004; Bai et al., 2006; Kube et al., 2008; Tran-Nguyen et al., 2008; Andersen et al., 2013; Wang et al., 2018; Liang et al., 2020) and 19 phytoplasma genome sketches and incomplete genome series have been reported (Saccardo et al., 2012; Chung et al., 2013; Chen et al., 2014; Kakizawa et al., 2014; Mitrovic et al., 2014; Chang et al., 2015; Lee et al., 2015; Quaglino et al., 2015; Fischer et al., 2016; Zamorano and Fiore, 2016; Zhu et al., 2017; Sparks et al., 2018; Town et al., 2018; Music et al., 2019). The phytoplasma genome is between 530 kb and 1,350 kb, with a G+C content of 24–35% (Yang et al., 2014b). Based on the whole genome sequence of phytoplasmas, many phytoplasma effector proteins have been identified. However, mechanistically, only Secreted AY-WB Protein 11 (SAP11), Secreted AY-WB Protein 54 (SAP54), Secreted AY-WB Protein 05 (SAP05) and Tengu have been studied in depth, which are multifunctional effector proteins (MacLean et al., 2011; Sugio et al., 2011; Minato et al., 2014; Tan et al., 2016; Huang et al., 2021).
Before the application of molecular techniques, the early detection and identification of phytoplasma mainly depends on ultrathin section electron microscope observation, fluorescence microscope, symptomatology, host reaction, relationship with transmitted vectors and antibiotic experiment. Since the 1980's, with the development of molecular biology detection technology, especially PCR-based techniques are now used to detect various phytoplasmas associated with plants and insects and study in various fields of phytoplasmas diseases research (Smart et al., 1996; Lee et al., 1998; Lu et al., 2016). To date, research on phytoplasma diseases has made some important progress in China. Research has mainly focused on the detection and identification of phytoplasmas, insect vector identification, and host diversity. In the future, in-depth research should be carried out in the fields of interaction and transmission mechanisms between phytoplasmas and insect vectors, the pathogenic mechanism of phytoplasmas, and the control of diseases. In this paper, the history and present situation of phytoplasma research, and the status, distribution, and diversity of phytoplasma diseases are summarized, and some possible research directions of phytoplasma in the future in China are proposed.
Phytoplasma disease has a long history in China. As early as 1640 CE, there was a record of mulberry dwarf disease (Yang et al., 2020). In 1965, Kuai et al. (2000) reported the existence of mulberry dwarf disease in east, south, and north China. In 1958, sugarcane white leaf (SCWL) was reported in Taiwan, China (Ling, 1962). In 1987, Zhou et al. (1987) used electron microscopy to show that SCWL also occurred in some cultivars in sugarcane producing areas such as Fujian, Guangxi Nanning, and Yunnan Kaiyuan. In 2013, Li et al. (2013b) first confirmed the existence of SCWL in Baoshan, Yunnan, China using nested PCR. Subsequently, the disease was reported in Lincang and Puer, Yunnan (Zhang et al., 2019, 2020). Rice orange leaf disease caused by a phytoplasma was first found in Xishuangbanna, Yunnan Province in 1978 (Wu et al., 1980). Then, it was historically outbreak in parts of southern China during the late 1980's to the early 1990's (Zhang et al., 1995). In 1982, tomato big bud disease was first discovered in Hainan Island, China (Tang et al., 1986). In 1984, rubber witches' -broom disease was reported in China, and then phytoplasma was also observed in rubber tree tissues with witches' -broom symptoms using electron microscopy (Chen et al., 1991). In 1985, areca nut yellow leaf disease was reported in Wanning and Tunchang, Hainan, China (Yu et al., 1986). In the same year, rice yellow dwarf was found in Fujian, China (Lin and Xie, 1985). In the 21st century, further phytoplasma diseases have been reported in China. Cactus witches' broom disease was first reported in Yunnan, China in 2002 (Cai et al., 2002). In 2004, transmission electron microscopy was used to observe the ultrastructure of diseased apricot trees with chlorotic leaf curling in Xinjiang, which confirmed that a phytoplasma was the causative pathogen (He et al., 2012). In 2006, spiraea witches' -broom disease was reported in Qingzhou, Shandong (Gao et al., 2007). In 2009, locust trees witches' -broom disease was found in Taian, Shandong (Yu et al., 2012). In July 2010, the cactus witches' -broom disease caused by the 16SrII-C subgroup was first reported in Yangling, Shanxi (Li et al., 2012). Tomato yellowing disease caused by a 16SrII-A phytoplasma was first reported in Yunnan Province in 2011 (Dong et al., 2013). In 2012, the camellia yellow flower disease caused by a 16SrV-B subgroup phytoplasma was first reported in Taian, Shandong, China (Gao et al., 2015). Reed witches' -broom disease was first reported in China in 2013 (Li et al., 2013a). In 2020, Eclipta and lilac witches' broom caused by a phytoplasma were reported in Haikou and Beijing, China, respectively (Chen et al., 2020; Yang et al., 2020). To date, more than 100 kinds of phytoplasma diseases have been reported in China. Among them, jujube witches'- broom, mulberry dwarf, paulownia witches' -broom, wheat blue dwarf are widely distributed and have caused serious damaged in China. SCWL disease, rice orange leaf disease, cherry lethal yellow, groundnut witches' broom, and areca nut yellow leaf disease, are distributed in some areas in China (Zhang et al., 1995; Mou et al., 2011; Yu et al., 2016; Song et al., 2018) (Table 1).
Although compared with plant fungi, bacteria and virus diseases, may be due to the application of pesticides, the losses caused by phytoplasmas may less than fungi, bacteria and virus diseases, they also can caused significant economic losses to agricultural and forestry production in China. For example, wheat stripe rust, a fungus disease of wheat, has been prevalent three times in 1950, 1964 and 1990, causing 6 billion kg, 3 billion kg and 2.6 billion kg of yield loss respectively in China (Wan, 2000). In 2006, wheat yellow mosaic virus damaged 2 million hectares of wheat in 9 provinces of China, causing losses of 1.5 billion kg (Yue et al., 2008a). Since the 1960's, outbreaks of wheat blue dwarf have occurred more than 10 times in Shaanxi, China, resulting in an annual loss of ~50 million kg of wheat yield (Chen et al., 2014). According to the statistics in 2006, Paulownia witches' -broom disease has affected 880,000 hm2 of wood production and caused billions of dollars in economic losses (Yue et al., 2008b; Geng et al., 2015). Mulberry dwarf disease has occurred in various silkworm breeding areas in China, with a relatively high incidence rate, causing serious economic losses to sericulture production (Zhao et al., 2017). Since the discovery of SCWL in Baoshan, Yunnan, China in 2012, SCWL has spread rapidly in many sugarcane areas, such as Lincang, Baoshan and Puer in Yunnan, causing marked economic losses (Zhang et al., 2019, 2020). The occurrence area of SCWL in Yunnan has expanded to more than 6,000 hm2, and the loss of sugarcane yield has reached 100% in severely affected fields (Huang et al., 2018). Currently, areca nut yellow leaf disease is spreading rapidly in Hainan, with an area of about 53,333 hm2 and is increasing year by year. The yield reduction has ranged from 10 to 60%, and the annual loss caused by areca nut yellow leaf disease was more than 2 billion RMB (Wang, 2019). Jujube witches' broom is a devastating phytoplasma disease for jujube trees, which has occurred in all jujube production areas in China. The incidence rate in severely affected areas reached 60–80%, and the mortality rate exceed 30%. The annual loss caused by jujube witches' broom reached hundreds of millions of RMB (Zhao et al., 2006; Wang et al., 2018). Phytoplasma disease has become an important disease in potato production in southwest China, severely restricting the farmers' income and the quality of the potatoes (Liang et al., 2020). Phytoplasma diseases have become one of the most important diseases to threaten the development of China's cherry industry in recent years. They mainly cause symptoms of cherry phyllody and yellowing, and eventually lead to rapid plant death within 2–5 years, which has caused significant losses to the Chinese cherry industry (Mu et al., 2019).
Doi et al. (1967) first observed phytoplasma in plant tissues using electron microscopy. Since then, Chinese researchers have used electron microscopy to observe phytoplasmas on various plants such as Zephyranthes candida, sugarcane, jujube, amorpha, and mulberry (Shi et al., 1984; Zhou et al., 1987; Chen and Li, 1994; Jin and Wang, 1994; Xu and Feng, 1998). Since the 1980's, serology has gradually been developed and applied, and many phytoplasmic detection methods based on serology have been established (Lu et al., 2016). Chinese scholars have successfully detected phytoplasma of paulownia witches' -broom, areca nut yellow leaf disease, and wheat blue dwarf using serological methods (Mou et al., 2011; Yang et al., 2014a,b). In the early 1990's, PCR technology was first used to detect phytoplasma diseases (Deng and Hiruki, 1991). Their high detection sensitivity has meant that PCR-based methods have become the most important techniques to detect and identify phytoplasmas. In addition to conventional PCR techniques, nested PCR, real-time PCR, immune-capture PCR (IC-PCR), and competitive quantitative PCR are currently used to detect phytoplasmas. In recent years, real-time PCR has been widely used to detect phytoplasma diseases in China. Liao et al. (2002) successfully applied real-time PCR to detect phytoplasmas of coconut lethal yellow, apple proliferation, and elm yellow disease. Ren et al. (2015) used real-time PCR to determine the phytoplasma concentration in different jujube witches' -broom disease-resistant varieties. Che et al. (2017) established a real-time PCR rapid detection method for phytoplasma from areca nut yellow leaf disease in Hainan. Loop-mediated isothermal amplification (LAMP) is a nucleic acid amplification technology established by Notomi et al. (2000). To date, this technology has been used widely in phytoplasma detection. For example, Han et al. (2015) established a visual LAMP detection method to detect phytoplasma of Jujube witches' broom; Wang et al. (2017) established a LAMP detection method to detect five species of 16 SrI group phytoplasmas using the tuf gene as a target. Han et al. (2020) established a LAMP method for the rapid detection of phytoplasma of apricot chlorotic leaf curl according to the tuf gene conserved sequence of a Xinjiang isolate.
There are more than 100 phytoplasma diseases reported in China and mainly caused by 11 phytoplasma groups, including 16SrI, 16SrII, 16SrIII, 16SrV, 16SrVI, 16SrVII, 16SrVIII, 16SrX, 16SrXI, 16SrXII, and 16SrXIV, which are distributed throughout China (Gong et al., 1990; Cai, 2007; Li et al., 2007, 2013a,b; Chang et al., 2012; Li, 2015) (Figure 1). Among them, in addition to the phytoplasma diseases that seriously endanger production, such as paulownia witches' broom, mulberry dwarf, jujube witches'-broom and Eucalyptus witches'-broom, new phytoplasma diseases have been reported continuously, and some have caused serious economic losses (Lin et al., 2007; Xu et al., 2009). For example, areca nut yellowing disease, which only occurred in India and Hainan Province in China, has spread rapidly in Hainan, resulting in an infected areca nut area of more than 2,667 hm2. The areca nut orchard with early infection was completely destroyed, and the incidence rate reached 90%. The yield was reduced by 78–80%, and new areca nut orchards have been constantly harmed (Che, 2010). The phytoplasma disease that endangers wheat production was named wheat blue dwarf (WBD), and was first reported in China by An et al. (2006). Since 2000, this disease has been prevalent in Shaanxi and Shanxi, resulting in large losses to wheat production and posing a serious threat to the high and stable yield of wheat. In Xichang, Sichuan Province, the phytoplasma of cherry lethal yellowing disease almost destroyed a hundred-year-old cherry orchard (Li, 2004). In 2012, Li et al. (2013a,b, 2014) first detected a phytoplasma disease named sugarcane white leaf disease (SCWL) in sugarcane planting regions of Baoshan and Lincang in Southwest Yunnan, China. SCWL has strong transmissibility and spreads rapidly. The diseased plant rate in seriously affected fields was as high as 100%, and the loss of sugar yield was serious, which was disastrous for the safe production of sugarcane.
Phytoplasmas in China are widely distributed and diverse. Currently, the phytoplasmas found in China can be divided into 11 groups, including 16SrI, 16SrII, 16SrIII, 16SrV, 16SrVI, 16SrVII, 16SrVIII, 16SrX, 16SrXI, 16SrXII, and 16SrXIV, among which 16SrI is having wide host range, followed by 16SrII and 16SrV groups (Lai et al., 2008). The representative phytoplasmas of each group were selected to construct a phylogenetic tree. The results showed that the 10 phytoplasma groups could be divided into three major clades. 16SrV, 16SrVI, 16SrVIII, 16SrXIV, 16SrXI, 16SrIII, and 16SrII were clustered into one clade; 16SrI and 16SrXII were clustered into the 2nd clade; and 16SrX formed a separate clade (Figure 2). Qiu et al. (1998) confirmed that among 20 plants infected with phytoplasma, 13 harbored phytoplasmas belonging to 16SrI, two harbored phytoplasmas belonging to 16SrVII, three harbored phytoplasmas belonging to 16SrII, and two harbored phytoplasmas belonging to 16SrVI. Cai (2007) studied the diversity of phytoplasma strains and related diseases in Yunnan, and identified 15 phytoplasma diseases belonging to the 16SrI-B, 16SrII-C, 16SrIII-B, and 16SrV-B subgroups. Among them, cactus witches' broom caused by phytoplasmas of the 16SrI-B subgroup and 16SrII groups was widely distributed in Yunnan Province. Che et al. (2009) first discovered the existence of 16SrV phytoplasma in naturally infected periwinkle. Zhang et al. (2016) analyzed SCWL phytoplasmas in Yunnan, which showed that SCWL is caused by phytoplasmas from group 16SrXI, including subgroup 16SrXI-B and a new subgroup, 16SrXI-D.
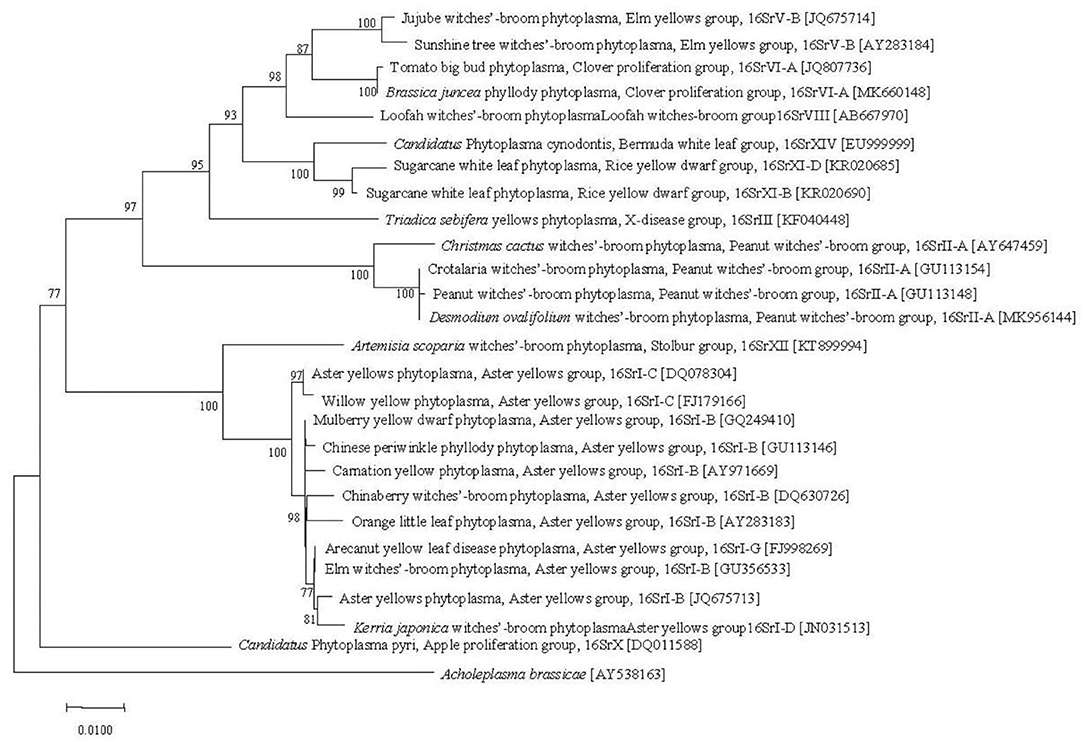
Figure 2. Phylogenetic tree constructed with 16S rDNA sequences of main phytoplasmas in China. Numbers at each node are the bootstrap values of the Neighbor-Joining tree (1,000 replications). The homologous sequence of Acholeplasma brassicae was used as the outgroup.
Mixed infections of phytoplasmas with viruses, bacteria, and spiroplasmas have been reported in many plants in China. A mixed infection of jujube witches' broom phytoplasma and paulownia witches' broom phytoplasmas in jujube witches' broom plants has been confirmed (Li et al., 2011). Two phytoplasmas belonging to the 16SrI-D and 16SrII-A subgroups were detected in lilac cluster leaf samples, which suggested that the disease might be caused by the combined infection of the two phytoplasmas (Wang, 2008). Momordica grosvenori (monkfruit) blister leaf witches' -broom disease is caused by the mixed infection of Luohanguo mosaic virus and a phytoplasma (Li, 2005). Beefwood witches'-broom disease is caused by the mixed infection of Rick-like bacteria and a phytoplasma (Zhang et al., 1983). Rubber witches'- broom disease is caused by the mixed infection of a phytoplasma and bacteria (Chen et al., 1998). Kelp leaf roll disease is caused by the mixed infection of a phytoplasma and a spiroplasma (Wang et al., 1983). Compared with the losses caused by a single pathogen, the losses caused by these mixed infections are more serious.
Compared with other plant pathogens, such as bacteria and viruses, the study of phytoplasma have been focused on its identification and classification, diversity, vector insects, effect factors, comparative genome and gene function in some developed countries such as Italy, the United States, Britain, France and Germany, and large agricultural countries such as India and Brazil. As far as China is concerned, phytoplasma research is concentrated in the identification of new phytoplasma and host diversity. More research attention has been paid to wheat blue dwarf phytoplasma, paulownia witches'-broom phytoplasma, jujube witches'-broom phytoplasma, rice orange leaf phytoplasm and sugarcane white leaf phytoplasm. Genome research has made some progress, for example, the complete genome sequence of jujube witches'-broom Phytoplasma (jwb-nky) was determined in China (Wang et al., 2018). Then, four out of 19 phytoplasma genome sketches and incomplete genome series were determined by Chinese scientists (Chung et al., 2013; Chen et al., 2014; Chang et al., 2015; Zhu et al., 2017). However, the pathogenicity research is very little. Other phytoplasma diseases have been reported, but there is no in-depth study (Ma et al., 2020). Since the discovery of phytoplasmas in 1640, research on phytoplasma diseases in China has made a great progress. More and more phytoplasma diseases and phytoplasma species have been found and identified, and the molecular mechanisms of phytoplasma disease transmission and epidemics have been gradually revealed. To date, more than 100 kinds of phytoplasma diseases have been found and identified, and the response of host plants to phytoplasma infection has been analyzed. However, in view of the spreading and damaging of phytoplasma, and its precise control, the following research directions should be emphasized in the future: (1) To establish the preservation and transmission methods of phytoplasmas, and a high-throughput system of phytoplasma race screening and host resource evaluation. (2) Digging up molecular markers closely linked to resistance, fine locating and cloning of phytoplasma resistance-related genes. (3) Mining of the variable and invariant regions of the genome to infer the genetic relationship and population structure characteristics of phytoplasma. (4) Analysis of the genomic characteristics of phytoplasma from different hosts, and mining of important functional genes and secreted proteins. (5) The development of rapid detection methods for phytoplasmas in the field, the generation of a prediction model for the occurrence and loss of disease in the short and medium term, and clarification of the transmission characteristics and epidemiological laws governing the disease. (6) To accelerate the breeding of excellent resistant varieties and developing economic and practical control products and technologies. (7) Analyze and study the influence of transmission vector on the epidemic of phytoplasma diseases, and to effectively cut off insect vector transmission and control strategies.
X-YW and R-YZ completed the writing of the first draft of the paper. JL, Y-HL, and H-LS literature and W-FL conducted the literature references searching and writing some part of the paper. Y-KH is the initiator and leader of the project and paper modification. All authors contributed to the article and approved the submitted version.
This work was supported by China Agriculture Research System of MOF and MARA (CARS-170303), the National Natural Science Foundation of China (31760504),Yunling Industry and Technology Leading Talent Training Program Prevention and Control of Sugarcane Pests (2018LJRC56), and Yunnan Province Agriculture Research System (YNGZTX-4-92).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
An, F. Q., Wu, Y. F., Sun, X. Q., Gu, P. W., and Yang, Y. (2006). Homologic analysis of tuf Gene for elongation factor Tu of phytoplasma from wheat blue dwarf. Sci. Agric. Sin. 39, 74–80. doi: 10.3321/j.issn:0578-1752.2006.01.011
Andersen, M. T., Liefting, L. W., Havukkala, I., and Beever, R. E. (2013). Comparison of the complete genome sequence of two closely related isolates of ‘Candidatus Phytoplasma australiense' reveals genome plasticity. BMC Genomics. 14, 529. doi: 10.1186/1471-2164-14-529
Bai, X., Zhang, J., Ewing, A., Miller, S. A., Radek, A. J., Shevchenko, D. V., et al. (2006). Living with genome instability: the adaptation of phytoplasmas to diverse environments of their insect and plant hosts. J. Bacteriol. 188:3682–3696. doi: 10.1128/JB.188.10.3682-3696.2006
Cai, H (2007). Diversity of Phytoplasma Strains and Associated With Diseases in Yunnan Province. (PhD dissertation). Kunming: Yunnan Agricultural University.
Cai, H., Chen, H. R., Li, F., and Kong, B. H. (2002). First report of a phytoplasma associated with cactus witches'-broom in Yunnan (China). Plant Pathol. 51, 394. doi: 10.1046/j.1365-3059.2002.00707.x
Chang, S. H., Cho, S. T., Chen, C. L., Yang, J. Y., and Kuo, C. H. (2015). Draft genome sequence of a 16SrII-A subgroup phytoplasma associated with purple coneflower (Echinacea purpurea) witches' broom disease in Taiwan. Genome Announc. 3, e01398. doi: 10.1128/genomeA.01398-15
Chang, W. C., Li, X. D., Shao, Y. H., XU, J. L., and Zhu, X. P. (2012). Molecular identification of the phytoplasma associated with kerria witches'-broom. Acta Phytopathol. Sin. 42, 541–545. doi: 10.13926/j.cnki.apps.2012.05.013
Che, H. Y (2010). Diversity of Phytoplasma Disease and Molecular Detection of Phytoplasma Associated With Arecanut Yellow Leaf in Hainan Province. (PhD dissertation). Xianyang: Northwest A&F University.
Che, H. Y., Cao, X. R., and Luo, D. Q. (2017). Research advances in pathogenic detection technique for arecanut yellow leaf disease. Chin. J. Trop. Agric. 37, 67–72. doi: 10.12008/j.issn.1009-2196.2017.02.016
Che, H. Y., Luo, D. Q., Fu, R. Y., Ye, S. B., and Wu, Y. F. (2009). Sequence analysis of 16S ribosomal DNA of phytoplasma associated with periwinkle yellows disease in Hainan. Acta Phytopathol. Sin. 39, 212–216. doi: 10.3321/j.issn:0412-0914.2009.02.015
Chen, J. S., and Li, D. B. (1994). A study on mycoplasma-like organisms causing yellow disease on autumn Zephyrlily. Acta Microbiol. Sin. 34, 245–247. CNKI:SUN:WSXB.0.1984-02-008
Chen, M. R., Luo, D. Q., Huang, Q. C., and Ye, S. B. (1998). Preparation and application of antiserium of causative agent of witches' broom of hevea brasiliensis. Chin. J. Trop. Crop. 4, 34–38.
Chen, M. R., Yang, S. H., Zheng, G. B., Chen, Z. Y., and Shen, J. Y. (1991). Identification of causative agent of rubber witches' broom disease and its relation with brown bast. Chin. J. Trop. Crop. 12, 65–73.
Chen, W., Li, Y., and Fang, X. P. (2020). Detection and molecular characterization of a phytoplasma in Ecliptaprostrata in China. J. Gen. Plant Pathol. 86, 60–64. doi: 10.1007/s10327-019-00871-9
Chen, W., Li, Y., Wang, Q., Wang, N., and Wu, Y. F. (2014). Comparative genome analysis of wheat blue dwarf phytoplasma, an obligate pathogen that causes wheat blue dwarf disease in China. PLoS ONE. 9, e96436. doi: 10.1371/journal.pone.0096436
Chung, W. C., Chen, L. L., Lo, W. S., Lin, C. P., and Kuo, C. H. (2013). Comparative analysis of the peanut witches'-broom phytoplasma genome reveals horizontal transfer of potential mobile units and effectors. PLoS ONE. 8, e62770. doi: 10.1371/journal.pone.0062770
Contaldo, N., Bertaccini, A., Paltrinieri, S., Windsor, H., and Windsor, D. (2012). Axenic culture of plant pathogenic phytoplasmas. Phytopathol. Mediterr. 51, 607–617. doi: 10.2298/GENSR1203701Z
Deng, S., and Hiruki, C. (1991). Genetic relatedness between two nonculturable mycoplasmalike organisms revealed by nucleic acid hybridization and polymerase chain reaction. Phytopathology. 81, 1475–1479. doi: 10.1094/Phyto-81-1475
Doi, Y., Teranaka, M., and Yora, K. (1967). Mycoplasma- or PLT group-likemicroorganisms found in the phloem elements of plants infectedwith mulberry dwarf,potato witches' broom, asteryellows, or paulownia witches' broom. Ann. Phytopathol. Soc. Jpn. 33, 259–266. doi: 10.3186/jjphytopath.33.259
Dong, J. H., Zhang, L., Li, W. H., McBeath, J. H., and Zhang, Z. K. (2013). ‘Candidatus Phytoplasma aurantifolia'-related strain associated with tomato yellows disease in China. J. Gen. Plant Pathol. 79, 366–369. doi: 10.1007/s10327-013-0463-5
Fischer, A., Santana-Cruz, I., Wambua, L., Olds, C., Midega, C., Dickinson, M., et al. (2016). Draft genome sequence of “Candidatus Phytoplasma oryzae” strain Mbita1, the causative agent of napier grass stunt disease in Kenya. Genome Announc. 4, e00297–e00216. doi: 10.1128/genomeA.00297-16
Gao, R., Wang, J., Li, X. D., Zhu, X. P., and Tian, J. Z. (2007). First report of spirea witches'-broom disease in China. Plant Dis. 91, 635. doi: 10.1094/PDIS-91-5-0635C
Gao, Y., Dong, Y. Z., Tan, W. P., Sun, G. Z., Zhu, Y. R., and Zhu, X. P. (2015). Detection and identification of an elm yellows group phytoplasma associated with camellia in China. J Phytopathol. 163, 560–566. doi: 10.1111/jph.12354
Geng, X. S., Shu, J. P., Wang, H. J., and Zhang, W. (2015). Research advance on transmission, epidemic and control of phytoplasmal disease. Chin. Agric. Sci. Bulletin. 31, 164–170. CNKI:SUN:ZNTB.0.2015-25-032
Gong, Z. X., Chen, Z. Y., and Shen, J. Y. (1990). Atlas of Mycoplasma-Like Organism in Chinese Plants. Beijing: Science Press
Han, J., Chen, K. W., Ji, W. B., Chen, H., and Tang, Z. H. (2020). Establishment of visual LAMP detection method for phytoplasma isolated from apricot chlorotic leaf roll (ACLR) in Xinjiang. Guizhou Agric. Sci. 48, 131–135. CNKI:SUN:GATE.0.2020-02-028
Han, J., Luo, M., He, G. L., Zhang, X. L., and Xiang, X. F. (2015). Development of a loop-mediated isothermal amplification assay for visual detection of jujube witches' broom phytoplasma. Acta Agric. Boreali Occident. Sin. 24, 125–131. doi: 10.7606/j.issn.1004-1389.2015.06.020
He, T. M., Han, B., Wu, Y. X., Li, W. H., and He, F. J. (2012). Advance and prospective of apricot chlorotic leaf roll. Biotechnol. Bull. 20–26.
Huang, W. J., MacLean, A. M., Sugio, A., Maqbool, A., Busscher, M., Cho, A. T., et al. (2021). Parasitic modulation of host development by ubiquitin-independent protein degradation. Cell. 184, 5201-5214.e12. doi: 10.1016/j.cell.2021.08.029
Huang, Y. K., Li, W. F., Zhang, R. Y., and Wang, X. Y. (2018). Color Illustration of Diagnosis and Control for Modern Sugarcane Diseases, Pests, and Weeds. Singapore: Springer Nature Singapore.
Jin, K. X., and Wang, Y. (1994). A new fasciation disease of amorpha fruticose associated with mycoplasma-like organisms. Acta Phytopathol. Sin. 24, 96. CNKI:SUN:ZWBL.0.1994-01-019
Kakizawa, S., Makino, A., Ishii, Y., Tamaki, H., and Kamagata, Y. (2014). Draft genome sequence of “Candidatus phytoplasma asteris” strain OY-V, an unculturable plant-pathogenic bacterium. Genome Announc. 2, e00944. doi: 10.1128/genomeA.00944-14
Kuai, Y. Z., Zhang, Z. K., and Chen, H. R. (2000). Kinds of plant mycoplasma like organisms in China. J. Yunnan Agric. Univ. 15, 153–160. doi: 10.3969/j.issn.1004-390X.2000.02.019
Kube, M., Schneider, B., Kuhl, H., Dandekar, T., Heitmann, K., Migdoll, A. M., et al. (2008). The linear chromosome of the plant-pathogenic mycoplasma ‘Candidatus Phytoplasma mali'. BMC Genomics. 9, 306. doi: 10.1186/1471-2164-9-306
Lai, F., Li, Y., Xu, Q. C., and Tian, G. Z. (2008). The present status on classification of phytoplasmas. Microbiol. China. 35, 291–295. doi: 10.3969/j.issn.0253-2654.2008.02.025
Lee, I. M., Gundersen-Rindal, D. E., Davis, R. E., and Bartoszyk, I. M. (1998). Revised classification scheme of phytoplasma based on RFLP analyses of 16S rDNA and ribosomal protein gene sequences. Int. J. Syst. Bacteriol. 48, 1153–1169. doi: 10.1099/00207713-48-4-1153
Lee, I. M., Shao, J., Bottner-Parker, K. D., Gundersen-Rindal, D. E., and Davis, R. E. (2015). Draft genome sequence of “Candidatus phytoplasma pruni” strain CX, a plant-pathogenic bacterium. Genome Announc. 3, e01117. doi: 10.1128/genomeA.01117-15
Li, C. L., Du, Y. J., Xiang, B. C., and Zhang, P. (2013a). First report of the association of a 'Candidatus Phytoplasma ulmi' isolate with a witches' broom disease of reed in China. New Dis. Rep. 28, 13. doi: 10.5197/j.2044-0588.2013.028.013
Li, T. T., Wu, G. X., Sun, X. C., Qing, L., and Yang, S. Y. (2011). Phytoplasma compound infection was found on jujube trees in China. Innovation of Plant Protection Science and Technology and Specialization of Disease and Pest Control-2011 Annual Conference of China Society of Plant Protection. Beijing: China Agricultural Science and Technology Press, 728.
Li, W. F., Wang, X. Y., Huang, Y. K., Shan, H. L., Shen, K., Luo, Z. M., et al. (2014). A quarantining sugarcane white leaf disease caused by phytoplasma found in sugarcane field in Yunnan. Acta Phytopathol. Sin. 44, 556–560. doi: 10.13926/j.cnki.pps.2014.05.016
Li, W. F., Wang, X. Y., Huang, Y. K., Shen, K., Shan, H. L., Luo, Z. M., et al. (2013b). First report of sugarcane white leaf phytoplasma in Yunnan province, China. Can. J. Plant Pathol. 35, 407–410. doi: 10.1080/07060661.2013.825815
Li, W. H., Xu, L., He, T. M., Zhang, D. H., Tang, Z. H., and Fan, G. Q. (2007). Preliminary identification on apricot chlorotic leafroll in Xinjiang. Acta Agricul. Boreali Occidentalis Sin. 16, 207–209. doi: 10.3969/j.issn.1004-1389.2007.06.046
Li, X. X (2005). A brief discussion on occurrence regularity and prevention countermeasures of Momordica blister leaf witches'-broom. Soc. Hortic. 16, 31–32. doi: 10.3969/j.issn.1674-5868.2005.03.018
Li, Y (2004). Molecular Detection and Identification of Several Different Phytoplasmas From Woody Plants in China. (PhD dissertation). Nanjing: Chinese Academy of Forestry.
Li, Z. N (2015). Molecular Diagnosis of Phytoplasma Diseases on Common Plants in Northwest China and Diversity Study of the Associated Phytoplasmas. (PhD dissertation). Yangling: Northwest A&F University.
Li, Z. N., Zhang, L., Liu, P., Bai, Y. B., Yang, X. G., and Wu, Y. F. (2012). Detection and molecular characterization of cactus witches'-broom disease associated with a group 16SrII phytoplasma in northern areas of China. Trop. Plant Pathol. 37, 210–214. doi: 10.1590/S1982-56762012000300008
Liang, J. S., Zhu, J. L., Xu, P., Zhang, P., Li, C. H., and Tang, W. (2020). Research progress of phytoplasma and potato related diseases. Acta Horticulturae Sinica. 47, 1777–1792. doi: 10.16420/j.issn.0513-353x.2020-0475
Liao, X. L., Zhu, S. F., Chen, H. Y., Huang, W. S., Luo, K., Zhao, W. J., et al. (2002). Establishment of real-time fluorescent PCR method with TaqMan probe for phytoplasma detection and identification. Acta Phytopathol. Sin. 32, 361–367. doi: 10.3321/j.issn:0412-0914.2002.04.013
Lin, C. L., Li, H. F., and Fan, Z. F. (2007). “Two novel Plasmids from Paulo wniawitches' broom Phytoplasma and its relatednss to other phytoplasmas,” in Proceedings of the Third Asian Conference on Plant Pathology. Yogyakarta, Indonesia, 149–150.
Lin, Q. Y., and Xie, L. H. (1985). The Pathogen of rice yellow dwarf disease. J. Fujian Agric. Coll. 14, 103–108.
Lu, H. Y., Wei, H., and Yang, G. (2016). Research progress on phytoplasma disease. Fujian J. Agric. Sci. 31, 326–332. doi: 10.19303/j.issn.1008-0384.2016.03.022
Ma, Q., Yuan, Z. L., Zhang, L., Sun, P. P., Li, X. Y., and Li, Z. N. (2020). Bibliometric analysis of scientific research of phytoplasma. J. Inner Mong. Agric. Univ. 41, 92–100. doi: 10.16853/j.cnki.1009-3575.2020.05.016
MacLean, A., Sugio, A., Makarova, O., Findlay, K., Grieve, V., Toth, R., et al. (2011). Phytoplasma effector SAP54 inducesindeterminate leaf-like flower development in Arabidopsis plants. Plant Physiol. 154, 831–841. doi: 10.1104/pp.111.181586
Minato, N., Himeno, M., Hoshi, A., Maejima, K., Komatsu, K., Takebayashi, Y., et al. (2014). The phytoplasmal virulence factor TENGU causes plant sterility by down regulating of the jasmonic acid and auxin pathways. Sci. Rep. 4, 7399. doi: 10.1038/srep07399
Mitrovic, J., Siewert, C., Duduk, B., Hecht, J., Molling, K., Broecker, F., et al. (2014). Generation and analysis of draft sequences of ‘Stolbur' phytoplasma from multiple displacement amplification templates. J. Mol. Microbiol. Biotechnol. 24, 1–11. doi: 10.1159/000353904
Mou, H. Q., Zhou, T., Zhao, W. J., Zhu, S. F., Lin, C. L., Li, H. F., et al. (2011). Preparation and application of antiserum against antigenic membrane protein of Paulownia witches' broom phytoplasma. Acta Phytopathol. Sin. 41, 161–170. CNKI:SUN:ZWBL.0.2011-02-009
Mu, X., Zhao, Y., Li, C. Y., and Wang, W. X. (2019). International advances of the research on the phytoplasma diseases incherry. J. Fruit Sci. 36, 1754–1762. CNKI:SUN:GSKK.0.2019-12-016
Music, M. S., Samarzija, I., Hogenhout, S. A., Haryono, M., Cho, S. T., and Kuo, C. H. (2019). The genome of ‘Candidatus Phytoplasma solani' strain SA-1 is highly dynamic and prone to adopting foreign sequences. Syst. Appl. Microbiol. 42, 117–127. doi: 10.1016/j.syapm.2018.10.008
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, 1–7. doi: 10.1093/nar/28.12.e63
Oshima, K., Kakizawa, S., Nishigawa, H., Jung, H. Y., Wei, W., Suzuki, S., et al. (2004). Reductive evolution suggested from the complete genome sequence of a plant-pathogenic phytoplasma. Nat. Genet. 36, 27–29. doi: 10.1038/ng1277
Qiu, B. S., Li, H. H., Shi, C. L., and Jin, K. X. (1998). Amplification of phytoplasma 16s rDNA from 20 infected plants in China and their RFLP analysis. Sci. Silvae Sin. 34, 67–74.
Quaglino, F., Kube, M., Jawhari, M., Abou-Jawdah, Y., Siewert, C., Choueiri, E., et al. (2015). ‘Candidatus phytoplasma phoenicium' associated with almond witches'-broom disease: from draft genome to genetic diversity among strain populations. BMC Microbiol. 15, 148. doi: 10.1186/s12866-015-0487-4
Ren, Z. G., Wang, H., Lin, C. L., Liu, X., Song, C. S., Feng, S. K., et al. (2015). A real-time (SYBR Green I) PCR assay for detection and quantification of jujube witches'-broom phytoplasma in the grafted jujube cultivar scions with different resistance. Acta Phytopathol. Sin. 45, 520–529.
Saccardo, F., Martini, M., Palmano, S., Ermacora, P., Scortichini, M., Loi, N., et al. (2012). Genome drafts of four phytoplasma strains of the ribosomal group 16SrIII. Microbiology. 158, 2805–2814. doi: 10.1099/mic.0.061432-0
Shi, C. L., Zhang, F. W., and Chen, Z. W. (1984). Scanning electron microscopic observation on mlo in freeze-fractured phloem tissues of witche's-broom diseased jujube. Acta Microbiol. Sin. 24, 139–141. CNKI:SUN:WSXB.0.1984-02-008
Smart, C. D., Schneider, B., Blomquist, C. L., Guerra, L. J., Harrison, N. A., Ahrens, U., et al. (1996). Phytoplasma-specific PCR primers based on sequences of the 16S-23S rRNA spacer region. Appl. Environ. Microbiol. 62, 2988–2993. doi: 10.1128/aem.62.8.2988-2993.1996
Song, Y. S., Zhu, N. B., Cui, Z. Q., Dong, Y. Q., Cui, D. Y., and Yue, F. Z. (2018). Species analysis of forest pest in China III. Mites, nematodes, bacteria, phytoplasmas, viruses. Forest Pest and Disease. 37, 34–37. doi: 10.19688/j.cnki.issn1671-0886.20180016
Sparks, M. E., Bottner-Parker, K. D., Gundersen-Rindal, D. E., and Lee, I. M. (2018). Draft genome sequence of the new jersey aster yellows strain of ‘Candidatus Phytoplasma asteris'. PLos ONE. 13, e0192379. doi: 10.1371/journal.pone.0192379
Sugio, A., Kingdom, H., MacLean, A., Grieve, V., and Hogenhout, S. (2011). Phytoplasma protein effector SAP11 enhances insect vector reproduction by manipulating plant development and defense hormone biosynthesis. Proc. Natl Acad. Sci. U. S. A. 108, 1254–1263. doi: 10.1073/pnas.1105664108
Tan, C., Li, C., Tsao, N., Su, L., Lu, Y., Chang, S., et al. (2016). Phytoplasma SAP11 alter 3-isobutyl- 2-methoxyprazine biosynthesis in Nicotainabenthamiana by suppressing NbOMT1. J. Exp. Bot. 67, 4415–4425. doi: 10.1093/jxb/erw225
Tang, W. W., Luo, X. H., Zhang, S. G., Kao, C. W., and Fan, H. C. (1986). Preliminary identification of tomato big-bud and tomato rosette discovered in Hainan island. Acta Phytopathol. Sin. 16, 105–108.
Town, J. R., Wist, A. T., Perez-Lopez, A. E., Olivier, B., and Dumonceauxa, A. (2018). Genome sequence of a plant-pathogenic bacterium, “Candidatus Phytoplasma asteris” strain TW1. Microbiol. Resour. Announce. 7, e01109. doi: 10.1128/MRA.01109-18
Tran-Nguyen, L. T. T., Kube, M., Schneider, B., Reinhardt, R., and Gibb, K. S. (2008). Comparative genome analysis of ‘Candidatus Phytoplasma australiense' (subgroup tuf-Australia I; rp-A) and ‘Ca. Phytoplasma asteris' strains OY-M and AY-WB. J. Bacteriol. 190, 3979–3991. doi: 10.1128/JB.01301-07
Wang, C. Y (2019). Comprehensive prevention and control measures of areca nut yellow leaf disease in Hainan. China Trop. Agric. 5, 29–31. doi: 10.3969/j.issn.1673-0658.2019.05.008
Wang, J (2008). The Analyses of Paulownia Witches' Broom Phytoplasma Elongation Factor tuf Gene and Investigation of Other Host Ptants of PaWB Phytoplasma. (Phd dissertation). Taian: Shandong Agricultural University.
Wang, J., Song, L. Q., Jiao, Q. Q., Yang, S. K., Gao, R., Lu, X. B., et al. (2018). Comparative genome analysis of jujube witches'-broom Phytoplasma, an obligate pathogen that causes jujube witches'-broom disease. BMC Genomics. 19, 689. doi: 10.1186/s12864-018-5075-1
Wang, Q. K., Shi, C. L., and Ma, J. C. (1983). Isolation and cultivation of MLO associated with coiling-stunt disease of sea tangle. Acta microbial. Sin. 23, 73–74.
Wang, S. J., Wang, S. K., Lin, C. L., Yu, S. S., Wang, L. F., Pu, C. G., et al. (2017). Loop-mediated isothermal amplification assay for detection of five phytoplasmas belonging to 16SrIgroup based on target tuf Gene. Sci. Silvae Sin. 53, 54–63. doi: 10.11707/j.1001-7488.20170807
Weintraub, P. G., and Beanland, L. (2006). Insect vectors of phytoplasmas. Annu. Rev. Entomol. 51, 91–111. doi: 10.1146/annurev.ento.51.110104.151039
Wu, Z. Q., He, Y. K., Xu, S. R., and Chang, S. C. (1980). The occurrence of rice orange leaf disease in Yunnan Province. Acta Phytopathol. Sin. 10, 55–58.
Xu, J. H., and Feng, M. G. (1998). Ultrastructures of a mycoplasma-like organism causing mulberry dwarf disease. Acta Microbiol. Sin. 38, 386–389. doi: 10.1088/0256-307X/15/12/024
Xu, Q. C., Tian, G. Z., Wang, Z. L., Kong, F. H., Li, Y., and Wang, H. (2009). Molecular detection and variability of jujube witches'-broom phytoplasmas from different cultivars in various regions of China. Acta Microbiol. Sin. 49, 1510–1519. doi: 10.13343/j.cnki.wsxb.2009.11.017
Yang, J., Liao, Y. J., Ning, J. H., Wang, J. Z., Wang, H., and Ren, Z. G. (2020). Identification of a phytoplasma associated with Syringa reticulata witches' broom disease in China. Forest Pathol. 50, e12592. doi: 10.1111/efp.12592
Yang, W. J., Yu, N. T., Zhang, Y. L., Wang, J. H., and Liu, Z. X. (2014a). Cloning and prokaryotic expression of areca phytoplasma membrane protein gene and preparation of its polclonal antiserum. Chin. J. Trop. Crops. 35, 2243–2248. doi: 10.3969/j.issn.1000-2561.2014.11.024
Yang, Y., Che, H. Y., Cao, X. R., and Luo, D. Q. (2014b). Research progress in phytoplasma genomes. Plant Prot. 40, 1–6. doi: 10.3969/j.issn.0529-1542.2014.06.001
Yu, H., Feng, S. F., and Zheng, J. H. (1986). Investigation report on yellow disease of areca palm in Hainan. Trop. Agricul. Sci. 3, 45–49.
Yu, S. S., Xu, Q. C., Lin, C. L., Wang, S. J., and Tian, G. Z. (2016). Genetic diversity of phytoplasmas: research status and prospects. Biodiversity Sci. 24, 205–215. doi: 10.17520/biods.2015127
Yu, Z. C., Cao, Y., Zhang, Q., Deng, D. F., and Liu, Z. Y. (2012). 'Candidatus Phytoplasma ziziphi' associated with Sophora japonica witches' broom disease in China. J. Gen. Plant Pathol. 78, 298–300. doi: 10.1007/s10327-012-0385-7
Yue, H. N., Wu, Y. F., Li, Y. R., Wei, T., Hou, W., and Wu, K. K. (2008a). Simultaneous detection of three wheat virus BSMV, BYDV-PAV, WYMV and WBD phytoplasma by multiplex PCR. Sci. Agric. Sin. 41, 2663–2669. doi: 10.3864/j.issn.0578-1752.2008.09.013
Yue, H. N., Wu, Y. F., Shi, Y. Z., Wu, K. K., and Li, Y. R. (2008b). First report of paulownia witches' broom phytoplasma in China. Plant Dis. 92, 1134. doi: 10.1094/PDIS-92-7-1134A
Zamorano, A., and Fiore, N. (2016). Draft genome sequence of 16SrIII-J phytoplasma, a plant pathogenic bacterium with a broad spectrum of hosts. Genome Announc. 4, e00602–e00616. doi: 10.1128/genomeA.00602-16
Zhang, J. N., Xu, D., Liu, Z. J., and Deng, L. Z. (1983). Study on pathogen of witche's broom of casuarina. Acta Phytopathol. Sin. 13, 37–41.
Zhang, R. Y., Li, W. F., Huang, Y. K., Wang, X. Y., Shan, H. L., Li, J., et al. (2019). Molecular identification of sugarcane white leaf in Puer, Yunnan Province, China. Sugar Tech. 21, 734–736. doi: 10.1007/s12355-019-00721-0
Zhang, R. Y., Li, W. F., Huang, Y. K., Wang, X. Y., Shan, H. L., Luo, Z. M., et al. (2016). Group 16SrXI phytoplasma strains, including subgroup 16SrXI-B and a new subgroup, 16SrXI-D, are associated with sugar cane white leaf. Int. J. Syst. Evol. Microbiol. 66, 487–491. doi: 10.1099/ijsem.0.000712
Zhang, R. Y., Shan, H. L., Huang, Y. K., Wang, X. Y., Li, J., Li, W. F., et al. (2020). Survey of incidence and nested PCR detection of sugarcane white leaf in different varieties. Plant Dis. 104, 2665–2668. doi: 10.1094/PDIS-11-19-2482-RE
Zhang, S. G., Fan, H. Z., Xiao, H. G., Xie, S. D., Zhou, X. M., Cai, H. X., et al. (1995). On the identification of rice orange leaf occurring in Guangdong Province. Acta Phytopathol. Sin. 25, 233–237. CNKI:SUN:ZWBL.0.1995-03-009
Zhao, J., Liu, M. J., Dai, L., and Zhou, J. Y. (2006). The Variations of endogenous hormones in Chinese jujube infected with witches' broom disease. Sci. Agric. Sin. 39, 2255–2260. doi: 10.3321/j.issn:0578-1752.2006.11.014
Zhao, X. J., Zhang, N., Li, T. T., Zhao, C. G., and Sun, X. C. (2017). Molecular detection of phytoplasma associated with mulberry dwarf disease in Beibei of Chongqing. J. Southwest Univ. 39, 16–20. doi: 10.13718/j.cnki.xdzk.2017.01.003
Zhou, Z. J., Lin, Q. Y., and Xie, L. H. (1987). Sugarcane white leaf disease occurrence and observation of its pathogens by electron microscopic. J. Fujian Agric. Coll. 16, 165–168. CNKI:SUN:FJND.0.1987-02-013
Keywords: China, phytoplasma disease, status, distribution, diversity
Citation: Wang X-Y, Zhang R-Y, Li J, Li Y-H, Shan H-L, Li W-F and Huang Y-K (2022) The Diversity, Distribution and Status of Phytoplasma Diseases in China. Front. Sustain. Food Syst. 6:943080. doi: 10.3389/fsufs.2022.943080
Received: 13 May 2022; Accepted: 06 June 2022;
Published: 27 June 2022.
Edited by:
Zhen He, Yangzhou University, ChinaReviewed by:
Yongqiang Li, Beijing University of Agriculture, ChinaCopyright © 2022 Wang, Zhang, Li, Li, Shan, Li and Huang. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Ying-Kun Huang, aHVhbmd5azY0QDE2My5jb20=
†These authors have contributed equally to this work
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.