- 1Department of Land Resources and Environmental Sciences, Montana State University, Bozeman, MT, United States
- 2Northern Agricultural Research Center, Montana State University, Havre, MT, United States
- 3Central Agricultural Research Center, Montana State University, Moccasin, MT, United States
- 4School of Food and Agriculture, University of Maine, Orono, ME, United States
Soil microbial communities are integral to highly complex soil environments, responding to changes in aboveground plant biodiversity, influencing physical soil structure, driving nutrient cycling, and promoting both plant growth and disease suppression. Cover crops can improve soil health, but little is known about their effects on soil microbial community composition in semiarid cropping systems, which are rapidly becoming warmer and drier due to climate change. This study focused on a wheat-cover crop rotation near Havre, Montana that tested two cover crop mixtures (five species planted early season and seven species planted mid-season) with three different termination methods (chemical, grazed, or hayed and baled) against a fallow control under ambient or induced warmer/drier conditions. Soil samples from the 2018 and 2019 cover crop/fallow phases were collected for bacterial community 16S rRNA gene sequencing. The presence and composition of cover crops affected evenness and community composition. Bacterial communities in the 2018 ambient mid-season cover crops, warmer/drier mid-season cover crops, and ambient early season cover crops had greater richness and diversity than those in the warmer/drier early season cover crops. Soil microbial communities from mid-season cover crops were distinct from the early season cover crops and fallow. No treatments affected bacterial alpha or beta diversity in 2019, which could be attributed to high rainfall. Results indicate that cover crop mixtures including species tolerant to warmer and drier conditions can foster diverse soil bacterial communities compared to fallow soils.
Introduction
Soils are indispensable and complex ecosystems vital to agricultural systems because of their roles in nutrient cycling and plant growth. Soil microbial communities are an important component of soil health, a relative measure of a soil's ability to function as a living ecosystem while supporting life aboveground and sustaining the socio-ecological functions of the land (Lehmann et al., 2020; Janzen et al., 2021). These belowground communities are involved in several key ecosystem functions interconnected with plant growth such as litter decomposition, nutrient cycling, and primary production (Philippot et al., 2013; Wagg et al., 2014; Delgado-Baquerizo et al., 2016).
While agricultural soil microbiota of semiarid climates have been understudied (Kim et al., 2013), over a third of the world's population depends on these systems (Delgado-Baquerizo et al., 2017). The importance of understanding the relationships between dryland agricultural systems and soil microbial communities magnifies as semiarid regions are predicted to become warmer and more widespread (Delgado-Baquerizo et al., 2017). Across the semiarid sections of the Northern Great Plains, the region where this study was conducted, temperatures and precipitation are projected to increase by the end of the century (Wienhold et al., 2018). However, reduced summer precipitation and higher evapotranspiration are likely to reduce effective soil moisture (Whitlock et al., 2017).
Soil bacterial communities in drylands are particularly sensitive to changes in aridity, and they decrease in abundance and diversity as soils become warmer and drier (Maestre et al., 2015; Naylor and Coleman-Derr, 2018). Increasing soil organic carbon is a possible solution to ameliorate the impact of climate change on soil microbiota (Whitmore et al., 2015), as this may bolster soil bacterial community resistance to drought (Moreno et al., 2019) and benefit agricultural production. One way to increase soil organic carbon is by reducing fallow periods and increasing the plant diversity of crop rotations (Bommarco et al., 2013).
Winter wheat—summer fallow rotations are commonly used in the semiarid sections of the Northern Great Plains as a method of soil moisture conservation (Kumar et al., 2020). However, this practice is only 10–40% efficient at storing water (Nielsen and Vigil, 2010) and fallow fields are susceptible to erosion, reductions in soil organic matter, salinization, and decreases in soil health (Carr et al., 2020). Replacing summer fallow with cover crops has the potential to increase soil organic matter and sequester soil organic carbon, even in semiarid regions (Blanco-Canqui et al., 2013a). This can aid in water retention over longer periods because soil organic carbon can increase potential water storage (Blanco-Canqui et al., 2013b), which is beneficial for both microbial communities and crops (Steenwerth and Belina, 2008).
Cover crops grown in semiarid regions are often terminated between late spring and early summer to preserve soil moisture (Carr et al., 2012; Nielsen et al., 2016) and contrasting termination methods may have different effects on soil microbial communities based on their inputs into soil and amount of physical disturbance (Liang et al., 2014; Castellano-Hinojosa and Strauss, 2020; Kim et al., 2020). Glyphosate is commonly used to terminate cover crops, but its effects on soil microbiota are not well understood (Schlatter et al., 2017; Kepler et al., 2020). It has been suggested that glyphosate may change soil microbial community composition by decreasing plant defenses and creating a favorable environment for pathogenic microbiota, or by suppressing taxa beneficial for plant growth (Van Bruggen et al., 2018, but see Kepler et al., 2020). Alternate methods of cover crop termination include haying and integrated crop-livestock management.
Soil bacterial diversity can increase under haying, possibly through disturbance-induced changes to root biomass, root exudates, and soil moisture (Foster et al., 2010), but studies are scarce. In contrast, the reintegration of livestock management with crop production is more thoroughly studied and has gained traction since the mid-2000s (Kumar et al., 2019), though the impacts on soil microbial communities are not fully understood and the ecological interactions of integrated crop-livestock production demand investigation. Grazing can influence microbial diversity and community structure directly through nutrient deposition (Yang et al., 2019b) and soil compaction (Zhou et al., 2010; Yang et al., 2019a) or indirectly by encouraging root production and enhancing rhizodeposition (Hamilton et al., 2008; Zhou et al., 2010; Yang et al., 2019a), by changing plant community composition, or by moderating soil nutrient availability (Yang et al., 2019a). For example, grazing can remove carbon from the system, suppressing the copiotrophic bacterial phyla Actinobacteria and supporting the oligotrophic Acidobacteria and Chloriflexi (Eldridge et al., 2017). Interactions with physicochemical soil characteristics such as pH and soil moisture may also explain grazing-induced changes in soil bacterial community structure (Yang et al., 2019a). In a dryland USDA-certified organic cropping system, sheep grazing with reduced tillage intensity has been shown to foster distinct soil bacterial communities in comparison with organic tilled and conventional no-till systems (Ouverson et al., 2021).
To our knowledge, no study has disentangled the relationships among cover crop diversity, agricultural management, and predicted climate conditions on soil microbiota. As many sections of northern Montana are expected to experience higher temperatures and decreased summer precipitation (Whitlock et al., 2017), understanding these relationships is imperative for the future agricultural stability of the region. This study assessed how soil bacterial communities respond to greater cover crop diversity under both ambient and induced warmer/drier conditions and the legacy effects of contrasting termination methods. Specifically, we tested the impacts of a winter wheat—cover crop rotation with cover crop mixtures varying in diversity (a chemical fallow control, an early season mixture of five species, and a mid-season mixture of seven species) terminated by three different methods (glyphosate application, cattle grazing, or cutting and baling). Open-top chambers and rain-out shelters were added to simulate climate change projections for the region by increasing temperature and decreasing precipitation compared to ambient conditions. We hypothesized that (1) the mid-season and early season treatments would have more diverse soil bacterial communities than fallow, (2) on average, ambient conditions would support higher soil bacterial diversity than warmer/drier conditions, and (3) the warmer/drier fallow plots terminated by glyphosate would have the lowest soil microbial community diversity while ambient cover crop plots terminated by grazing would have the highest soil microbial community diversity.
Materials and methods
Site and experimental design
This study was part of a crop rotation experiment that began in 2012–2013, conducted at the Montana State University Northern Agricultural Research Station, located southwest of Havre, Montana (48.49689 N, 109.8029 W). Data collected in 2018 and 2019 were from a subset of the cover crop phase which included a summer fallow treatment, an early season cover crop mixture of five species planted in early spring, and a mid-season cover crop mixture of seven species usually planted 10 days later. The site has an annual low and high temperature of 0 and 13.4°C, respectively, with an average annual precipitation of 305 mm.1 The soil underlying the study area is a mix of Joplin and Telstad clay loam (Fine-loamy, mixed, superactive, frigid Aridic Argiustolls)2 with an average pH of 7.34, 1.27% organic matter, and 46.5 ppm of total soil nitrogen. These deep soils (>150 cm) have similar surface textures of loam, parent material of till, and water holding capacities of over 18 centimeters.3
Details of the field trials, including experimental design, crop management, treatments, and field sampling procedures, are described in Bourgault et al. (2022), Wyffels et al. (2021), and DuPre et al. (2022). The experiment utilized two replicated field trials established as a restricted-randomized strip plot design (Figure 1). The two field trials, one planted with winter wheat (Triticum aestivum L.) and the other with either different cover crop mixtures or fallow, were rotated each year. The location of each treatment was randomized in the first year (2012–2013) of the experiment applied to 8 × 44 m strip plots and maintained throughout the duration of the study, allowing for the examination of legacy effects. Termination methods (herbicide, grazing, and haying and baling) were assigned perpendicular to the fallow and cover crop strip plots and applied in a strip plot design. We established two 0.56 m2 frames in each cover crop and fallow strip plots (8 × 13 m), then randomly assigned them to two different temperature and moisture conditions (“ambient” and “warmer/drier”, Figure 1).
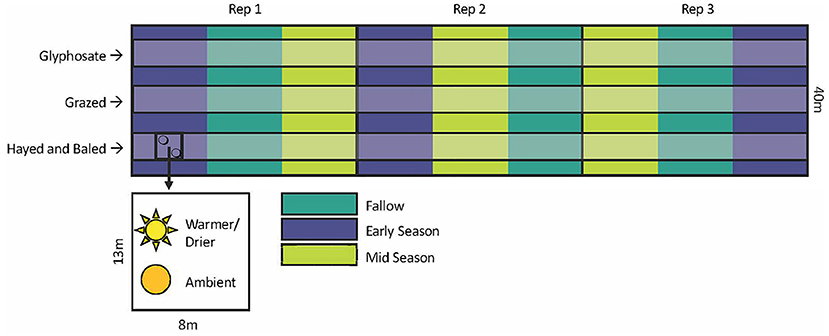
Figure 1. Experimental design showing 1 year of the winter wheat—cover crop rotation. Open-top chambers with rain-out shelters were located at each combination of the cover crop or fallow strip plots and termination treatments (9 per replication). Within a 13 × 8 m section of each crop and termination treatment, samples were taken from the 0.56 m2 frames placed within the open-top chamber, and from an equivalently sized marked area representing ambient conditions which was ~1 m away from the structure.
Soil samples were obtained from the summer fallow, early season cover crop mixture, and mid-season cover crop mixture treatments. Summer fallow began after the winter wheat harvest on July 12, 2017, and July 26, 2018, and ended on September 21, 2018, and September 13, 2019, respectively. All strip plots were sprayed with 2240 g/ha of ai RTIII glyphosate before planting. Summer fallow plots were also treated with 1120 g of ai RTIII glyphosate plus 340 g/ha of ai dicamba during the growing season to manage weed populations as needed.
Two cover crop mixtures with more legumes in proportion to other functional groups were selected from a total of 15 mixtures for this experiment (see Bourgault et al., 2022). All mixtures, categorized by planting date, were generally composed of legumes, brassicas, and cereals. The early season mixture was composed of radish (Raphanus raphanistrum L.), field pea (Pisum sativum L.), oat (Avena sativa L.), turnip (Brassica rapa L.), and hairy vetch (Vicia villosa Roth). The 2018 mid-season cover crop mixture was composed of oat, radish, field pea, turnip, lentil (Lens culinaris Medikus), and sorghum-sudangrass [Sorghum × drummondii (Steud.) Millsp. & Chase]. The 2019 mid-season cover crop mixture varied slightly due to logistics and was composed of oat, radish, turnip, soybean, hairy vetch, chickpea (Cicer arietinum L.), and millet (Setaria italica L.). Cover crops planting dates varied each year depending on local weather conditions and species phenology. The early season mixture was planted on May 4, 2018, and April 29, 2019, while the mid-season mixture was planted on May 14, 2018, and May 9, 2019. Cover crops were seeded at a depth of 2.54 cm at different seeding rates (Supplementary Table S1) using a 3.7-m Conservapak no-till disk planter and fertilized with 112 kg/ha N-P-K (20-20-20).
Termination of cover crops
Cover crops were terminated shortly after oat anthesis to preserve soil moisture for the next crop (O'Dea et al., 2013). Termination methods included herbicide (ai glyphosate applied at 2,500 g/ha), cutting and baling (henceforth referred to as “haying”), and grazing. Herbicide application and haying occurred on July 9, 2018, and July 8, 2019. Cattle grazing involved 8–10 yearling bulls fenced in the 12 × 360 m termination strip across the fallow and cover crop mixtures from July 11 to 13, 2018, and July 9 to 11, 2019. Four weeks after initial termination, regrowth of cover crops and weeds was terminated with 2,000 g/ha of ai RTIII glyphosate plus 340 g/ha of ai dicamba because haying and grazing did not sufficiently kill the cover crops.
Temperature and moisture conditions
Open-top chambers were combined with rain-out shelters during the growing season to mimic the warmer/drier climate projected for summers in Northern Montana. The open-top chambers, designed to increase the temperature by 1–2°C (Marion et al., 1997), had a basal diameter of 1.5 m and a top diameter of 1 m and were built out of low-opacity Sun-lite HP fiberglass material (1 mm thick; Solar Components Corporation, Manchester, New Hampshire). Following Yahdjian and Sala (2002), rain-out shelters decreased precipitation by up to 55% and were constructed out of wooden frames that supported gutters made of corrugated clear polycarbonate (Sunturf). The rain-out shelters were placed at an aspect of 220°-230° to account for the prevailing southwestern wind. GB-1 gypsum sensor blocks with a KS-D1 digital soil moisture tester (Delmhorst Instrument Company, Towaco, New Jersey) and iButtons (Maxim Integrated, San Jose) recorded soil moisture and temperature, respectively, 10 cm belowground in the ambient and warmer/drier treatments. The open-top chambers raised mean soil temperatures compared to ambient conditions by 0.8–0.9°C (DuPre et al., 2022).
Soil sampling
Soil samples from the cover crop/fallow phase were collected on July 1 of both years before crop termination, to ensure analysis of long-term termination treatment effects rather than short-term effects. Three 10 cm depth × 1.6 cm diameter soil cores were taken from a 0.56 m2 frame in the warmer/drier treatment and homogenized. The same procedure was repeated in the soil under ambient conditions. Instruments were cleaned with 70% isopropyl alcohol in between plots to minimize cross contamination. One gram of the homogenized soil was fixed in 9 mL of 70% isopropyl alcohol in the field and stored at ambient temperatures.
Laboratory methods
DNA extraction, amplification, and sequencing library preparation was performed as described in Ishaq et al. (2017, 2020). In 2018, a PowerLyzer PowerSoil DNA isolation kit was used to extract metagenomic DNA and in 2019, DNA extraction was done with the Qiagen equivalent (Mo-Bio Laboratories Inc., Carlsbad, California; Qiagen, Hilden, Germany). In both years, a negative control of molecular grade water was added to each extraction batch and an additional cleaning step was performed after extraction to precipitate DNA with ethanol and sodium acetate (Ishaq et al., 2020). The V3-V4 region of the 16S rRNA gene was amplified through 25 cycles of PCR with barcoded and indexed primers using the Q5 High-Fidelity Mastermix (New England BioLabs, Inc., Ipswitch, MA, USA) with 25 μL of mix, 6 μL molecular-grade water, 21 μL of forward and reverse barcoded primer mix at 1.09 uM concentration, and 4 μL sample DNA. The PCR protocol was as follows: 98°C for 30 s; 30 cycles of denaturation at 98°C for 10 s, annealing at 55°C for 30 s, elongation at 72°C for 30 s; then a final elongation at 72°C for 2 min. Primers included the MiSeq adaptors (A for forward, B for reverse), the sample index/barcodes, the two-nucleotide linker, and primers 341F (5′-ACTCCTACGGGAGGCAGCAG-3′) (Fadrosh et al., 2014) and 806R (5′-GGACTACHVGGGTWTCTAAT-3′) (Caporaso et al., 2011). Amplicons were purified using Omega Mag-Bind beads (Omega Bio-Tek, Norcross, Georgia) and quantified with a Qubit 4 Fluorometer (Invitrogen, Thermofisher Scientific). High-throughput sequencing with an Illumina MiSeq (Illumina, San Diego, CA) at the University of Oregon Genomics Core (Eugene, OR, USA) was performed using a 2 × 300-nucleotide V3 kit. PhiX at a 10% spike-in and molecular-grade sterilized water were used as positive and negative controls, respectively.
Bioinformatics
The DADA2 pipeline run in the software environment R 4.0.2 was used to filter paired end reads, remove chimeric sequences, and conduct taxonomic assignment (Callahan et al., 2016; R Development Core Team, 2020). The 2018 and 2019 samples were processed separately because they came from two different sequencing runs and reads per sample varied between years (Supplementary Figure S1). Due to low quality in the reverse reads, only the forward reads were used for this analysis. Sequences were trimmed by 10 bases at the start and 30 at the end positions during filtering, with no ambiguous bases allowed, a maximum expected error of two, and anything matching the phiX controls were removed. On average, the 2018 samples retained 75.1% of sequences with 88,413 reads in 35,082 unique sequences and the 2019 samples retained 70.5% sequences with 58,540 reads in 20,041 unique sequences. Error rates were learned on 2 × 106 randomly selected dereplicated reads and then used to identify Amplicon Sequence Variants (SVs). These are analogous to individuals as sequences have been grouped down to single-nucleotide polymorphism/single base differences by assessing overall error rate and probability of base error vs. polymorphism occurrence. Two-parent chimeras (bimeras) were removed, and the Silva NR version 138 database was used to assign taxonomy to sequence variants (SVs) (Yilmaz et al., 2014). Laboratory negative controls were used to remove identical SVs from experimental samples, using adapted code (Ishaq, 2017). Data were rarified to the size of the smallest sample library above 20,000 reads per sample, which was 33,572 reads per sample for 2018 and 21,423 reads per sample for 2019. This removed 10 samples and 284 absolute sequence variants from the 2018 dataset and 13 samples and 630 absolute sequence variants from the 2019 dataset.
Statistical procedures
All data analysis was performed in R 4.0.3–4.1.0 (R Development Core Team, 2021). The phyloseq and vegan packages were used for statistical analysis (McMurdie and Holmes, 2013; Okansen et al., 2020), while ggplot2 was used for data visualization (Wickham et al., 2016). Alpha or within-community diversity was assessed via observed sequence variants for bacterial community richness, Shannon's Diversity, and Shannon's Evenness. Linear regression with mixed effect models was used to explore the relationships between treatments and alpha diversity. Alpha diversity, beta diversity, and relative abundance were analyzed separately by year to account for environmental factors external to the experimental design. All regression analyses were performed using the lme4 package (Bates et al., 2015).
One sample in each year was a low outlier for Shannon's Diversity and Evenness; these two samples were excluded from the analysis of Shannon's Diversity and Evenness because they skewed the distribution of regression model residuals. A beta diversity heatmap revealed that the outlier from the 2019 samples had a high abundance of Enterobacteriaceae (Salmonella), which obscured the resolution of other taxa (Supplementary Figure S2).
Presence (fallow or cover crop) and composition (early season or mid-season) of cover crops, termination method, manipulated temperature and moisture conditions (ambient or warmer/drier), and the interactions among them were included as fixed effects with replication as a random effect in the multivariate explanatory models for alpha diversity. Type III One-Way Analysis of Variance determined which variables affected alpha diversity while Tukey's post-hoc comparisons evaluated differences in means among treatment levels. Correlations between environmental variables and bacterial richness, diversity, and evenness were explored using Pearson's correlation (Wei and Simko, 2021).
Bray-Curtis dissimilarities were calculated to assess between-community diversity with non-metric multidimensional scaling ordination (NMDS). Mean dissimilarities of soil bacterial communities grouped by treatment were compared using analysis of variance and Tukey's post-hoc tests. The betadisper function in the vegan package was used to assess the assumption of homogenous dispersion for performing a permutational analysis of variance (PERMANOVA), which was met. A PERMANOVA (Anderson, 2017) compared bacterial community dissimilarity among treatments, using the adonis function with 999 permutations. Data were stratified by replication to account for repeated measures. The pairwise.adonis2 function was used to examine post-hoc contrasts between significant factors (Martinez Arbizu, 2020).
Changes in relative bacterial abundance from 2018 to 2019 were represented with bar plots of the 10 most abundant phyla by the presence and composition of cover crops (Supplementary Figure S3). Phyla with a relative abundance sum of <0.9 were filtered out and ordered from most abundant to least. An analysis of variance and Tukey's post-hoc comparisons were performed on each remaining phyla to determine which variables affected relative abundance. Heatmaps displayed the 25 most abundant sequence variants labeled by family. Differential abundance analysis was performed using the DESeq2 package to determine which sequence variants differed by the presence and composition of cover crops (Love et al., 2014). The rfPermute package was used to conduct a random forest analysis to determine which taxa were significantly discriminatory between treatment states, highlighting the ones that were unique or defining features (Archer, 2021). Specifically, this procedure calculated which sequence variants could predict bacterial communities based on manipulated temperature and moisture conditions or the presence and composition of cover crops. For each comparison, 500 trees were made, with 100 permutations each. Out-of-bag estimates were checked as a form of model validation; these were the prediction errors for bootstrapped samples that did not contain elements of the original dataset.
Results
Alpha diversity
Of the 108 total soil samples collected, 85 passed through quality control: 44 samples with 16,994 unique sequence variants from 2018 and 41 samples with 16,989 unique sequence variants from 2019. When the 2018 and 2019 samples were analyzed together, year was the single highly significant predictor of bacterial richness. However, as the decrease from 2018 to 2019 in mean bacterial richness (Supplementary Figure S4) could not be explained by any treatments, the 2 years were analyzed separately. In 2018, bacterial richness was less variable in mid-season cover crops than in early season or fallow soils under both ambient and warmer/drier conditions (Figures 2A–F). Bacterial richness responded to the presence and composition of cover crops along with manipulated temperature and moisture conditions (Table 1). Additionally, there was an interaction between these variables (Table 1).
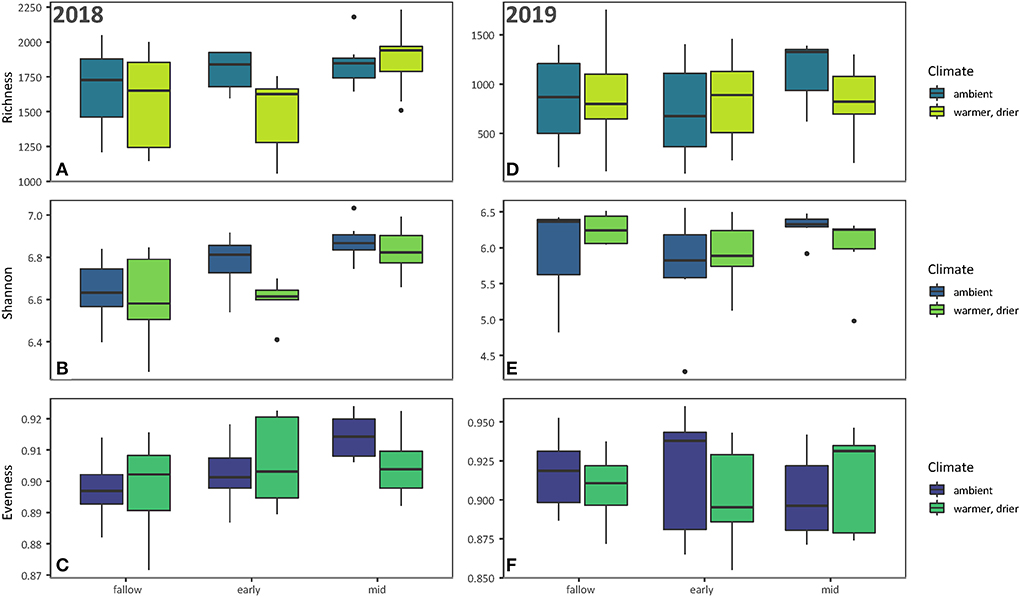
Figure 2. Alpha diversity metrics of soil bacterial communities associated with early season cover crops, fallow soils, or mid-season cover crops in both ambient and warmer/drier conditions for each year. From top to bottom, (A–C) display bacterial community richness, Shannon's Diversity, and Shannon's Evenness for 2018. (D–F) display the same for 2019.
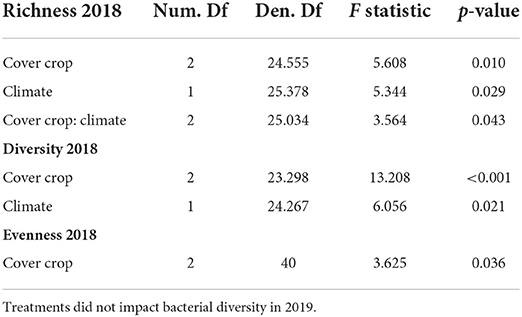
Table 1. Type III Analysis of Variance for mixed effect linear models of bacterial richness, Shannon's Diversity, and Shannon's Evenness for the 2018 samples.
There were, on average, 457 more sequence variants in the ambient early season cover crops than in the warmer/drier early season cover crops (Table 2). Bacterial richness was also higher in both the ambient and warmer/drier mid-season cover crops than in the warmer/drier early season cover crops, with an estimated difference of 475 and 465 sequence variants, respectively (Table 2).
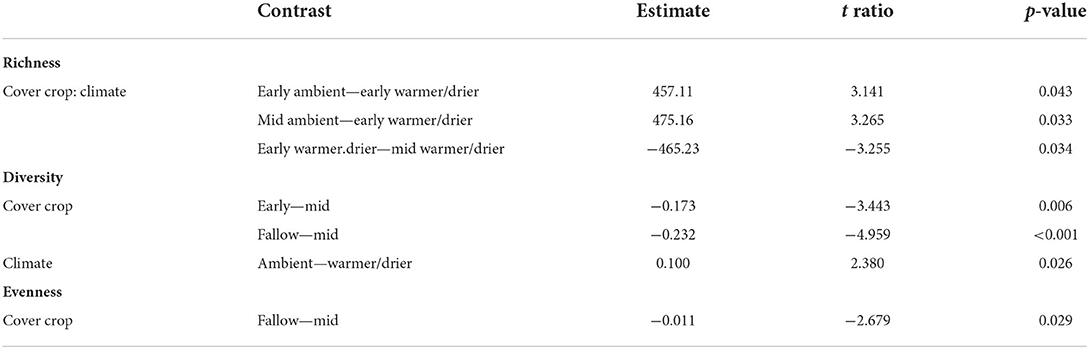
Table 2. Pairwise comparisons from Tukey's post-hoc analysis of alpha diversity models for the 2018 samples.
The diversity of bacterial communities also varied in response to the presence and composition of cover crops and temperature and moisture conditions (Table 1). Bacterial richness and diversity were more variable in the early season cover crops between ambient and warmer/drier conditions, while evenness varied more between ambient and warmer/drier conditions in the mid-season cover crops (Figures 2A–C). The mid-season cover crop soils had greater bacterial diversity than the early season and fallow soils, with an estimated difference in means of 0.173 and 0.232, respectively (Table 2). Mean Shannon's Diversity was 0.010 greater in ambient conditions than in warmer/drier conditions (Table 2). Bacterial evenness was only affected by the presence and composition of cover crops (Table 1), with mid-season cover crop soils 0.011 greater than fallow soils (Table 2). In contrast, no treatments were significant predictors of bacterial richness, diversity, or evenness in 2019 (Table 1).
Total aboveground plant biomass collected from a 0.56 m2 frame placed in the ambient and warmer/drier treatments of each strip plot (DuPre et al., 2022), average soil moisture the day of or prior to sampling, and the average soil temperature for one, three, and 7 days prior or up to the day of sampling were not correlated with bacterial richness or evenness across both years (Supplementary Figure S5). However, in 2018, bacterial diversity and plant biomass had a low positive correlation (0.34, p-value = 0.028) while plant biomass and soil moisture had a low negative correlation (−0.49, p-value = 0.002).
Beta diversity
The presence and composition of cover crops impacted bacterial community dissimilarity in 2018, accounting for 25.9% of the variation among communities (p-value = 0.001, Supplementary Table S2). The mid-season cover crop mixture had distinct soil bacterial communities from the early season mixture and fallow soils (Figure 3), confirmed with PERMANOVA pairwise contrasts (p-values = 0.001). Mid-season and early season communities had the highest mean dissimilarity of 76.2%, followed by mid-season and fallow communities at 76.0%. Communities in the mid-season cover crops had the lowest within-group mean dissimilarity of 53.8% on a weighted Bray-Curtis scale while those in fallow soils had the highest mean within-group dissimilarity of 64.7%.
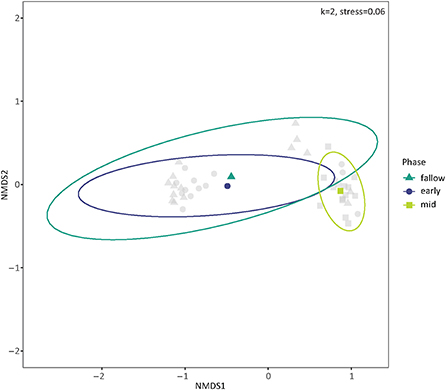
Figure 3. Ordination of soil bacterial communities in 2018 by the presence and composition of cover crops using non-metric multidimensional scaling (NMDS) on weighted Bray-Curtis dissimilarities. The centroids and multivariate t-distribution ellipses for bacterial communities in the cover crop/fallow phase are designated by color and shape (circle for early season, triangle for fallow, and square for mid-season). For the NMDS calculation, k = 2, and stress = 0.06.
The PERMANOVA on the 2019 samples indicated that none of the treatments or interactions impacted soil bacterial community dissimilarity. However, plant biomass was correlated with community dissimilarity (p-value = 0.037) and was included in the ordination as a vector. The average soil temperature for the 7 days prior to sample collection was also included (p-value = 0.052, Figure 4).
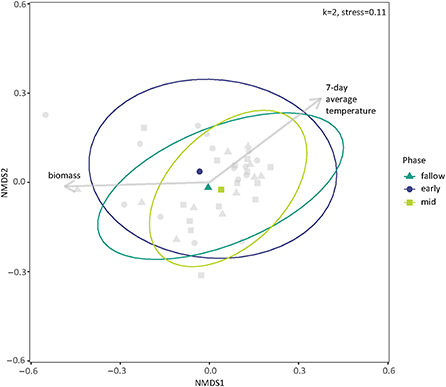
Figure 4. Ordination of soil bacterial communities in 2019 by presence and composition of cover crops using non-metric multidimensional scaling (NMDS) on weighted Bray-Curtis dissimilarities. The centroids and multivariate t-distribution ellipses for bacterial communities are designated by color and shape (circle for early season, triangle for fallow, and squared for mid-season). Plant biomass and average soil temperature for the 7 days preceding or including sampling were found to be potential vectors for the ordination. For the NMDS calculation, k = 2, and stress = 0.11.
Community composition: Relative abundance
Relative abundance of phyla differed by the presence and composition of cover crops but not by temperature and moisture conditions. Of the eight most abundant phyla in the 2018 samples, the relative abundances of Actinobacteria, Acidobacteria, Gemmatimonadota, and Planctomycetota were significantly different by the presence and composition of cover crops (Supplementary Table S3). Mid-season cover crop soils had a higher relative abundance of Gemmatimonadota and Planctomycetota than early season or fallow soils and a higher relative abundance of Actinobacteria than fallow soils (Supplementary Table S4). However, Acidobacteria was more abundant in early season and fallow soils. Community composition shifted from 2018 to 2019, marked by an increase in Actinobacteria and a decrease in Acidobacteria (Figure 5). The mean relative abundance of Actinobacteria increased from 28.3 to 49.8% while the mean relative abundance of Acidobacteria decreased from 23.9 to 6.5%. Of the six most abundant phyla in the 2019 samples, temperature and moisture conditions accounted for a significant change in the relative abundance of Gemmatimonadota (Supplementary Table S3), which had a higher estimated mean relative abundance in ambient soils than in warmer/drier soils (Supplementary Table S4). Heatmaps show that sequence variants from Pyrinomonadaceae and Rubrobacteriaceae were more abundant in 2018, while bacteria from the family designator JG30-KF-CM45 (order Thermomicrobiales) were more abundant in 2019 (Figure 6). Based on the heatmaps, several of the most common taxa found in the 2018 samples were not as abundant in the mid-season cover crop soils as the early season and fallow soils.
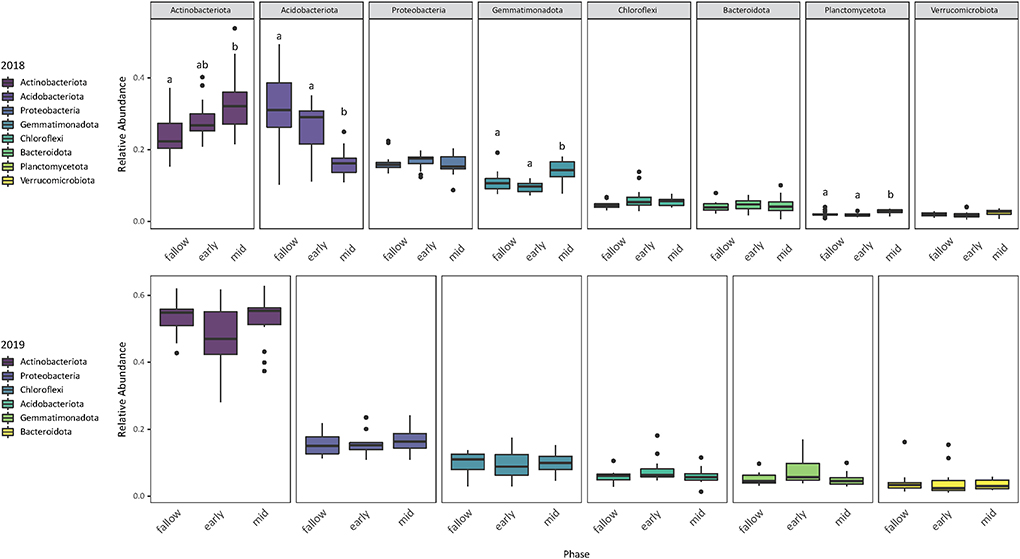
Figure 5. Relative abundance of the most common phyla (sum > 0.9) found in the 2018 and 2019 samples. Differences in relative abundance by the presence and composition of cover crops significant at the 0.05 alpha level are represented with lowercase letters.
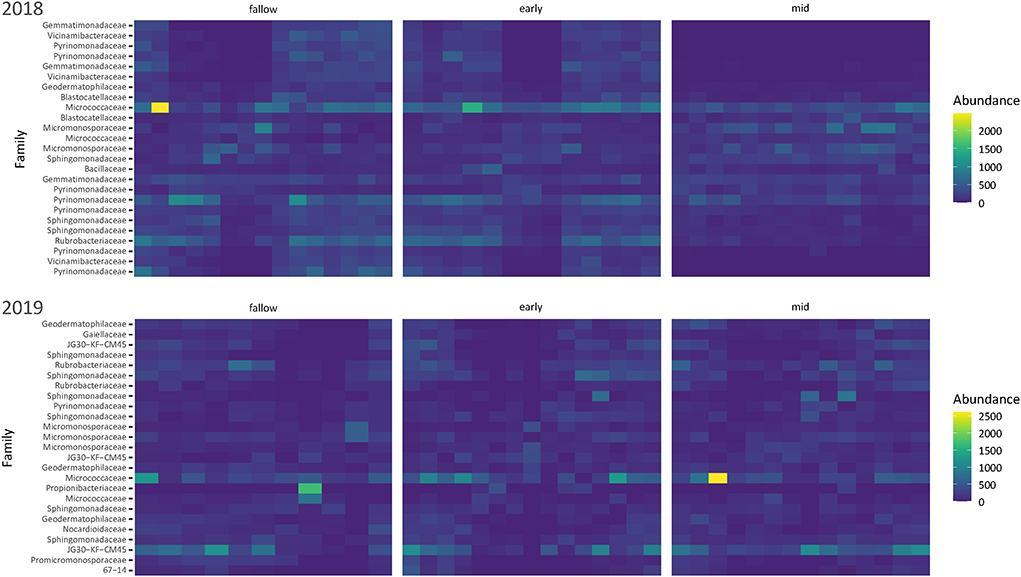
Figure 6. Heatmap of the most abundant sequence variants identified at the family level for each year. Taxa not present or at very low abundance in samples were set to display as the closest limit (0) for visual coherence.
DESeq2 was used on unrarified sample libraries to determine differential abundance of taxa among treatments (Weiss et al., 2017). Several bacterial genera relevant to soil health (Yang et al., 2019b) were differentially abundant between the 2018 mid-season and fallow soils as well as the mid-season and early season soils. Sequence variants from Nitrospira (family Nitrospiraceae) were more abundant in mid-season cover crop soils than in fallow soils (log2 fold change 6.40 and 2.16) or early season cover crop soils (log2 fold change 6.74). Sphingomonas (Sphingomonadaceae) were also more abundant in the mid-season cover crops than in fallow (log2 fold change 7.76) or early season cover crops (log2 fold change 7.70, 6.84). Nitrosomonadaceae and Nocardioidaceae were generally more abundant in the mid-season cover crops. However, Actinoplanes (Micromonosporaceae) were less abundant in mid-season soils than fallow (log2 fold change −6.09), but more abundant than in early season soils (log2 fold change 6.31). Three sequence variants from the phylum Actinobacteria were differentially expressed between the 2018 early season and fallow soils. One sequence variant belonging to the phylum Actinobacteria was differentially abundant between ambient and warmer/drier conditions (log2 fold change −23.96). In 2019, at most four sequence variants identifiable at the genus level were differentially abundant by the presence and composition of cover crops and one sequence variant of the genus Streptomyces (phylum Actinobacteria) was differentially abundant between ambient and warmer/drier soils (log2 fold change 24.45).
The random forest analysis could not predict bacterial communities for ambient or warmer/drier soils in both years (out-of-bag error 62.8 and 50.0%, respectively). Bacterial communities based on the presence and composition of cover crops could be successfully predicted in 2018 but not 2019, with a large variation in error (20.9 and 57.5%, respectively). The resulting top 50 sequence variants were ordered by calculated importance (p < 0.05) and faceted by manipulated temperature and moisture conditions. Pseudarthrobacter was the most abundant in both ambient and warmer/drier soils, except for the ambient mid-season cover crops, which had the highest abundance of Micromonosporaceae (Figure 7). Blastocatellaceaea were more abundant in warmer/drier fallow than in ambient fallow conditions. Sphingomonas remained abundant in the mid-season cover crops under both ambient and warmer/drier conditions but decreased in the early season cover crops and fallow (Figure 7).
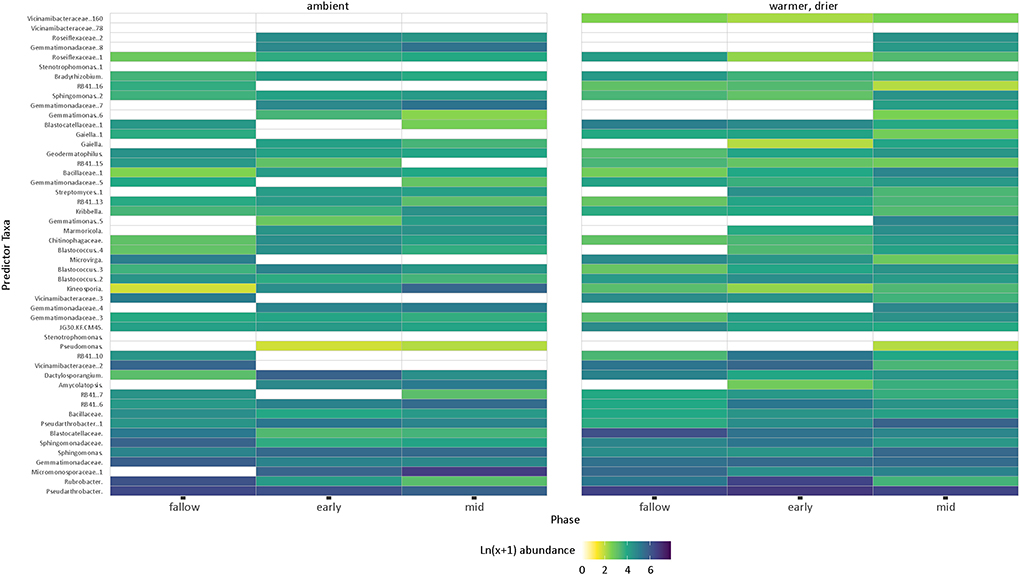
Figure 7. Random forest analysis of the 2018 samples showing the natural log(x+1) abundance of the top 50 predictor sequence variants under ambient and warmer/drier conditions. White spaces indicate taxa that are absent or at very low abundance. Sequence variants from unspecified genera are identified at the family level.
Discussion
Cover crop mixtures have been suggested as a replacement for summer fallow to increase soil organic carbon, soil health, and soil bacterial diversity (Blanco-Canqui et al., 2015). This study examined the effects of the presence and composition of cover crops, cover crop termination methods, and predicted temperature and moisture conditions on soil bacterial communities. While experimental treatments impacted bacterial communities sampled in 2018, no differences were observed in 2019. It is possible that high rainfall in 2019 played a dominant role in homogenizing soil bacterial communities. However, the 2018 results allowed us to disentangle some of the management and environmental factors conditioning the abundance and diversity of soil bacterial communities. In agreement with our first hypothesis, our results show that cover crop mixtures support higher bacterial richness and diversity than fallow, though this was moderated by manipulated temperature and moisture conditions. Confirming our second hypothesis, we observed that ambient conditions can foster more diverse soil bacterial communities than warmer and drier conditions. We did not find evidence to support our third hypothesis that chemically terminated fallow soils under warmer/drier conditions would have the lowest soil microbial community diversity and grazed cover crops in ambient conditions would have the highest soil microbial community diversity, probably because cover crop termination methods did not have an impact on soil bacterial communities.
Interaction between presence and composition of cover crops and predicted climate conditions
The effects of cover crops on soil bacterial communities range from marginal (Hartmann et al., 2006) to a strong response by particular taxa (Frasier et al., 2016; Finney et al., 2017; Martínez-García et al., 2018). Within the rhizosphere, cover crops may cause a “priming effect” by affecting soil organic matter through root turnover (Nivelle et al., 2016). Other studies have found that conservation tillage and arable weeds (Wortman et al., 2013) or irrigation (Calderón et al., 2016) can be more impactful on soil microbial communities than cover crops. In the studied systems, the 2018 mid-season cover crops fostered a soil bacterial community distinct from early season cover crops and fallow, reinforcing the selectivity of some plant species. However, the difference in planting dates cannot be separated from the cover crop treatments, limiting the interpretation of these results.
In 2018, an interaction between the presence and composition of cover crops and temperature and moisture conditions affected bacterial richness while only the former accounted for community dissimilarity. A related study at the same field site found that an interaction between these same variables affected plant biomass (DuPre et al., 2022). Cover crop species in the early and mid-season mixtures responded differently to changes in temperature and moisture. Specifically, oat biomass was higher in the ambient early season mixture than in warmer/drier conditions, while radish and turnip biomass was higher in the warmer/drier mid-season mixture than in ambient conditions (DuPre et al., 2022). Cereals exhibit greater stress under drier conditions compared to legumes (Daryanto et al., 2016), which may account for lower bacterial richness in the warmer/drier early season soils because microbial and plant community resilience are linked (de Vries et al., 2018). Additionally, brassicas select for unique microbial communities because of the compounds they produce (Rumberger and Marschner, 2003) and have been shown to suppress plant pathogens while increasing soil bacterial diversity when in rotation (Majchrzak et al., 2010; Li et al., 2017). Brassicas in both cover crop mixtures were seeded at a rate of <10% (kg/ha), but their mean biomass substantially increased in proportion to the legumes and grasses between the ambient and warmer/drier mid-season cover crops, compared to the ambient and warmer/drier early season cover crops (Supplementary Table S5). These factors may have driven the observed bacterial community dissimilarity between the early and mid-season cover crop mixtures.
Compared to the early season cover crops and fallow soils, the mid-season cover crops had higher differential abundance of several soil bacteria linked to aspects of soil health. For example, Sphingomonas is associated with plant health as an antagonist toward plant pathogens (Yang et al., 2019b). The mid-season cover crop mixture contained more legume species than the early season mixture, potentially increasing support for bacteria associated with nutrient cycling. Mesorhizobium fix nitrogen, Nitrospira participate in nitrification, and Nitrosomonadaceae and Nocardioidaceae are thought to play a role in carbon and nitrogen cycling (Yang et al., 2019b).
Effects of temperature and moisture conditions on soil bacterial communities
Environmental conditions strongly affect soil bacterial communities (Maestre et al., 2015; Naylor and Coleman-Derr, 2018; Ishaq et al., 2020; Ouverson et al., 2021). Precipitation in 2019 may have been a stronger influence on bacterial community alpha and beta diversity than the presence and composition of cover crops, termination methods, or simulated climate conditions. The study site received 188 mm more rainfall in June 2019 than in June 20184; there was 2.5 × as much precipitation in the 2 weeks preceding sample collection than during the same period the year before (Supplementary Figure S6), making it the 20th wettest June for Havre since 1970 (Menne et al., 2012a,b). It is likely that soils were saturated from cumulative precipitation, as evident by the higher soil moisture readings for 2019 (Supplementary Figure S7). Soils that normally undergo wet-dry cycles host distinct bacterial communities better adapted to these extremes (Fierer et al., 2003). In a study comparing bacterial communities under winter wheat in contrasting cropping systems, community dissimilarity increased toward the end of the growing season when conditions were hotter and drier (Ishaq et al., 2020). Without the selective pressure of decreased soil moisture, higher precipitation in 2019 may have homogenized soil bacterial communities instead.
In 2018, bacterial richness and diversity were lower in the warmer/drier treatment as hypothesized, though richness was not correlated with soil moisture and soil temperature. The early season cover crops, especially those in the warmer/drier conditions, may have driven the negative correlation between plant biomass and soil moisture and this relationship has been proposed as a mechanism for changes in soil microbial communities (de Vries et al., 2018). Bacterial richness and evenness have been observed to decrease in response to drier conditions and exhibit a lag between drought and recovery (de Vries et al., 2018). Soil bacterial community composition, however, does not have a consistent response to drought. Dominant taxa can either respond poorly to drought or not be affected compared to subordinate taxa (Sayer et al., 2017; de Vries et al., 2018). Also, altered precipitation can lead to shifts in vegetation community composition (Nielsen and Ball, 2015), resulting in changes in root structure and depth that in turn affect what root exudates and litter are available throughout the soil profile (van der Putten et al., 2013).
Cover crop termination and soil bacterial communities
Based on previous studies (Zhou et al., 2010; Eldridge et al., 2017; Ouverson et al., 2021), cattle grazed cover crop and fallow strip plots were expected to have higher bacterial diversity compared to glyphosate application and haying. However, no differences were found among soil bacterial communities based on cover crop termination method. In previous work, sheep grazing selected for dissimilar soil bacterial communities compared to tillage or chemical methods for cover crop termination and weed management (Ouverson et al., 2021). Long term exclusive cattle grazing and exclusive sheep grazing have different impacts on soil carbon storage, with no impact from cattle grazing observed in high diversity plant communities (Chang et al., 2018). The cropping system in this study is characterized by brief, targeted grazing, an approach that can negatively impact organic carbon storage without affecting microbial enzymatic activity (Tobin et al., 2020), which may account for the lack of soil bacterial response. Glyphosate application did not reduce bacterial diversity, which is consistent with previous work that found glyphosate application at recommended rates does not significantly change soil bacterial community structure (Rose et al., 2016; Kepler et al., 2020). In accordance with previous studies (Zhang et al., 2013; Carey et al., 2015), haying also had no effect.
In this study, the presence and composition of cover crops and interactions with soil temperature and moisture conditions had a greater effect on soil bacterial communities than cover crop termination methods. Our results suggest that in warmer and drier conditions, cover crop mixtures with similar composition to the mid-season mixture may encourage more diverse bacterial communities compared to cover crop mixtures composed of species from the early season mix. Future work could assess bacterial response to cover crop phases of single species and mixtures, testing those used in the early season and mid-season mixtures as well as other species commonly used as cover crops in the region. This would address the question of whether soil bacterial communities are influenced more by species or by the diversity of cover crops mixtures in a rotation. In either scenario, cover crops can be used to promote diverse soil bacterial communities as an important component of soil health.
Data availability statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found below: https://www.ncbi.nlm.nih.gov/, PRJNA782382.
Author contributions
TO performed laboratory protocols, data analysis, interpretation, and wrote the manuscript. SI performed laboratory protocols, facilitated sequencing, and contributed to code development. SI and JE contributed to data analysis. DB, FM, SI, JE, and TS developed the experimental design, methodology, analysis, and reviewed the manuscript. FM was the principal investigator for USDA funding. All authors contributed to the article and approved the submitted version.
Funding
The Montana Natural Resource and Conservation Service CIP Grant was the initial funding source for the project. Montana Wheat and Barley Committee funding was also instrumental to the initiation and early years of this long-term project. The Montana Research and Economic Development Initiative supported this project from 2015 to 2017, and the United States Department of Agriculture (USDA) National Institute of Food and Agriculture supported the project from 2018 to 2020 (USDA-NIFA; 2018-67013-27919). This publication was supported with funds provided by USDA-NIFA to Montana State University.
Acknowledgments
We would like to acknowledge the contributions of various Northern Agricultural Research Center staff who helped with various aspects of data collection over the years. Particularly, sincere thanks are due to Peggy Lamb, Maryse Bourgault, and Julia Dafoe for managing the experimental work over the years. Sincere thanks are also due to Roger Hybner for all aspects of the technical work related to this project, Tom Allen for planting and spraying operations, and Cory Parsons and the livestock crew for haying and putting up electrical fences and bringing cows and bulls in for grazing. Numerous short-term employees and student interns at NARC have also helped with various technical tasks and are also acknowledged. We would like to thank Chris Larson, Mary Ellyn DuPre, Shealyn Malone, Meiling Wong, James Schafer, Tony Leipicello, and Daniela Alambarrio for their assistance in installing the OTCs and with soil collection. We also thank Dr. Carl Yeoman at Montana State University and the Biology of the Built Environment (BioBE) lab at the University of Oregon for the use of their laboratory spaces and instruments, respectively. We are especially grateful for the assistance provided by Leslie Dietz of BioBE with laboratory procedures.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.911199/full#supplementary-material
Footnotes
1. ^https://wrcc.dri.edu/cgi-bin/cliMAIN.pl?mt3996.
2. ^https://casoilresource.lawr.ucdavis.edu/gmap/.
3. ^https://www.nrcs.usda.gov/.
4. ^https://agresearch.montana.edu/narc/weather_data/index.html.
References
Anderson, M. J. (2017). “Permutational multivariate analysis of variance (PERMANOVA),” in Wiley StatsRef: Statistics Reference Online (American Cancer Society), 1–15.
Archer, E. (2021). rfPermute: Estimate Permutation p-Values for Random Forest Importance Metrics. Available online at: https://CRAN.R-project.org/package=rfPermute (accessed March 2021).
Bates, D., Mächler, M., Bolker, B., and Walker, S. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Blanco-Canqui, H., Holman, J. D., Schlegel, A. J., Tatarko, J., and Shaver, T. M. (2013a). Replacing fallow with cover crops in a semiarid soil: effects on soil properties. Soil Sci. Soc. Am. J. 77, 1026–1034. doi: 10.2136/sssaj2013.01.0006
Blanco-Canqui, H., Shapiro, C. A., Wortmann, C. S., Drijber, R. A., Mamo, M., Shaver, T. M., et al. (2013b). Soil organic carbon: the value to soil properties. J. Soil Water Conserv. 68, 129A−134A. doi: 10.2489/jswc.68.5.129A
Blanco-Canqui, H., Shaver, T. M., Lindquist, J. L., Shapiro, C. A., Elmore, R. W., Francis, C. A., et al. (2015). Cover crops and ecosystem services: insights from studies in temperate soils. Agron. J. 107, 2449–2474. doi: 10.2134/agronj15.0086
Bommarco, R., Kleijn, D., and Potts, S. G. (2013). Ecological intensification: harnessing ecosystem services for food security. Trends Ecol. Evol. 28, 230–238. doi: 10.1016/j.tree.2012.10.012
Bourgault, M., Wyffels, S., Dafoe, J., Lamb, P., and Boss, D. (2022). Introducing cover crops as fallow replacement in the Northern Great Plains: II. Impact on following wheat crops. Renew. Agric. Food Syst. 37, 303–312. doi: 10.1017/S1742170521000508
Calderón, F. J., Nielsen, D., Acosta-Martínez, V., Vigil, M. F., and Lyon, D. (2016). Cover crop and irrigation effects on soil microbial communities and enzymes in semiarid agroecosystems of the central great plains of North America. Pedosphere 26, 192–205. doi: 10.1016/S1002-0160(15)60034-0
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: High-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carey, C. J., Beman, J. M., Eviner, V. T., Malmstrom, C. M., and Hart, S. C. (2015). Soil microbial community structure is unaltered by plant invasion, vegetation clipping, and nitrogen fertilization in experimental semi-arid grasslands. Front. Microbiol. 6, 466. doi: 10.3389/fmicb.2015.00466
Carr, P. M., Anderson, R. L., Lawley, Y. E., Miller, P. R., and Zwinger, S. F. (2012). Organic zero-till in the northern US Great Plains Region: opportunities and obstacles. Renew. Agric. Food Syst. 27, 12–20. doi: 10.1017/S174217051100041X
Carr, P. M., Bell, J. M., Boss, D. L., DeLaune, P., Eberly, J. O., Edwards, L., et al. (2020). Annual forage impacts on dryland wheat farming in the Great Plains. Agron. J. 113, 1–25. doi: 10.1002/agj2.20513
Castellano-Hinojosa, A., and Strauss, S. L. (2020). Impact of cover crops on the soil microbiome of tree crops. Microorganisms 8, 328. doi: 10.3390/microorganisms8030328
Chang, Q., Wang, L., Ding, S., Xu, T., Li, Z., Song, X., et al. (2018). Grazer effects on soil carbon storage vary by herbivore assemblage in a semi-arid grassland. J. Appl. Ecol. 55, 2517–2526. doi: 10.1111/1365-2664.13166
Daryanto, S., Wang, L., and Jacinthe, P.-A. (2016). Global synthesis of drought effects on maize and wheat production. PLoS ONE 11, e0156362. doi: 10.1371/journal.pone.0156362
de Vries, F. T., Griffiths, R. I., Bailey, M., Craig, H., Girlanda, M., Gweon, H. S., et al. (2018). Soil bacterial networks are less stable under drought than fungal networks. Nat. Commun. 9, 3033. doi: 10.1038/s41467-018-05516-7
Delgado-Baquerizo, M., Eldridge, D. J., Ochoa, V., Gozalo, B., Singh, B. K., and Maestre, F. T. (2017). Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 20, 1295–1305. doi: 10.1111/ele.12826
Delgado-Baquerizo, M., Maestre, F. T., Reich, P. B., Jeffries, T. C., Gaitan, J. J., Encinar, D., et al. (2016). Microbial diversity drives multifunctionality in terrestrial ecosystems. Nat. Commun. 7, 10541. doi: 10.1038/ncomms10541
DuPre, M. E., Seipel, T., Bourgault, M., Boss, D. L., and Menalled, F. D. (2022). Predicted climate conditions and cover crop composition modify weed communities in semiarid agroecosystems. Weed Res. 62, 38–48. doi: 10.1111/wre.12514
Eldridge, D. J., Delgado-Baquerizo, M., Travers, S. K., Val, J., Oliver, I., Hamonts, K., et al. (2017). Competition drives the response of soil microbial diversity to increased grazing by vertebrate herbivores. Ecology 98, 1922–1931. doi: 10.1002/ecy.1879
Fadrosh, D. W., Ma, B., Gajer, P., Sengamalay, N., Ott, S., Brotman, R. M., et al. (2014). An improved dual-indexing approach for multiplexed 16S rRNA gene sequencing on the Illumina MiSeq platform. Microbiome 2, 6. doi: 10.1186/2049-2618-2-6
Fierer, N., Schimel, J. P., and Holden, P. A. (2003). Influence of drying–rewetting frequency on soil bacterial community structure. Microb. Ecol. 45, 63–71. doi: 10.1007/s00248-002-1007-2
Finney, D. M., Buyer, J. S., and Kaye, J. P. (2017). Living cover crops have immediate impacts on soil microbial community structure and function. J. Soil Water Conserv. 72, 361–373. doi: 10.2489/jswc.72.4.361
Foster, B. L., Khavin, I. S., Murphy, C. A., Smith, V. H., Ramspott, M. E., Price, K. P., et al. (2010). Integrated responses of grassland biodiversity and ecosystem properties to hay management: a field experiment. Trans. Kans. Acad. Sci. 113, 103–119. doi: 10.1660/062.113.0208
Frasier, I., Noellemeyer, E., Figuerola, E., Erijman, L., Permingeat, H., and Quiroga, A. (2016). High quality residues from cover crops favor changes in microbial community and enhance C and N sequestration. Glob. Ecol. Conserv. 6, 242–256. doi: 10.1016/j.gecco.2016.03.009
Hamilton, I. I. I. E. W, Frank, D. A., Hinchey, P. M., and Murray, T. R. (2008). Defoliation induces root exudation and triggers positive rhizospheric feedbacks in a temperate grassland. Soil Biol. Biochem. 40, 2865–2873. doi: 10.1016/j.soilbio.2008.08.007
Hartmann, M., Fliessbach, A., Oberholzer, H.-R., and Widmer, F. (2006). Ranking the magnitude of crop and farming system effects on soil microbial biomass and genetic structure of bacterial communities: ranking the magnitude of crop and farming system effects. FEMS Microbiol. Ecol. 57, 378–388. doi: 10.1111/j.1574-6941.2006.00132.x
Ishaq, S. L. (2017). SueIshaq/Examples-DADA2-Phyloseq. Available online at: https://github.com/SueIshaq/Examples-DADA2-Phyloseq (accessed June 8, 2021).
Ishaq, S. L., Johnson, S. P., Miller, Z. J., Lehnhoff, E. A., Olivo, S., Yeoman, C. J., et al. (2017). Impact of cropping systems, soil inoculum, and plant species identity on soil bacterial community structure. Microb. Ecol. 73, 417–434. doi: 10.1007/s00248-016-0861-2
Ishaq, S. L., Seipel, T., Yeoman, C. J., and Menalled, F. D. (2020). Soil bacterial communities of wheat vary across the growing season and among dryland farming systems. Geoderma 358, 113989. doi: 10.1016/j.geoderma.2019.113989
Janzen, H. H., Janzen, D. W., and Gregorich, E. G. (2021). The ‘soil health' metaphor: illuminating or illusory? Soil Biol. Biochem. 159, 108167. doi: 10.1016/j.soilbio.2021.108167
Kepler, R. M., Epp Schmidt, D. J., Yarwood, S. A., Cavigelli, M. A., Reddy, K. N., Duke, S. O., et al. (2020). Soil microbial communities in diverse agroecosystems exposed to the herbicide glyphosate. Appl. Environ. Microbiol. 86, AEM.01744-19, e01744-19. doi: 10.1128/AEM.01744-19
Kim, M., Boldgiv, B., Singh, D., Chun, J., Lkhagva, A., and Adams, J. M. (2013). Structure of soil bacterial communities in relation to environmental variables in a semi-arid region of Mongolia. J. Arid Environ. 89, 38–44. doi: 10.1016/j.jaridenv.2012.09.014
Kim, N., Zabaloy, M. C., Guan, K., and Villamil, M. B. (2020). Do cover crops benefit soil microbiome? A meta-analysis of current research. Soil Biol. Biochem. 142, 107701. doi: 10.1016/j.soilbio.2019.107701
Kumar, S., Sieverding, H., Lai, L., Thandiwe, N., Wienhold, B., Redfearn, D., et al. (2019). Facilitating Crop–Livestock Reintegration in the Northern Great Plains. Agron. J. 111, 2141–2156. doi: 10.2134/agronj2018.07.0441
Kumar, V., Obour, A., Jha, P., Liu, R., Manuchehri, M. R., Dille, J. A., et al. (2020). Integrating cover crops for weed management in the semiarid U.S. Great Plains: opportunities and challenges. Weed Sci. 68, 311–323. doi: 10.1017/wsc.2020.29
Lehmann, J., Bossio, D. A., Kögel-Knabner, I., and Rillig, M. C. (2020). The concept and future prospects of soil health. Nat. Rev. Earth Environ. 1, 544–553. doi: 10.1038/s43017-020-0080-8
Li, T., Liu, T., Zheng, C., Kang, C., Yang, Z., Yao, X., et al. (2017). Changes in soil bacterial community structure as a result of incorporation of Brassica plants compared with continuous planting eggplant and chemical disinfection in greenhouses. PLoS ONE 12, e0173923. doi: 10.1371/journal.pone.0173923
Liang, S., Grossman, J., and Shi, W. (2014). Soil microbial responses to winter legume cover crop management during organic transition. Eur. J. Soil Biol. 65, 15–22. doi: 10.1016/j.ejsobi.2014.08.007
Love, M. I., Huber, W., and Anders, S. (2014). Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 15, 550. doi: 10.1186/s13059-014-0550-8
Maestre, F. T., Delgado-Baquerizo, M., Jeffries, T. C., Eldridge, D. J., Ochoa, V., Gozalo, B., et al. (2015). Increasing aridity reduces soil microbial diversity and abundance in global drylands. Proc. Natl. Acad. Sci. 112, 15684–15689. doi: 10.1073/pnas.1516684112
Majchrzak, B., Kurowski, T. P., Wachowska, U., and Jazwińska, E. (2010). Changes in soil microbial communities as a result of growing Brassicaceae crops. Acta Agrobot. 63, 161–169. doi: 10.5586/aa.2010.018
Marion, G. M., Henry, G. H. R., Freckman, D. W., Johnstone, J., Jones, G., Jones, M. H., et al. (1997). Open-top designs for manipulating field temperature in high-latitude ecosystems. Glob. Change Biol. 3, 20–32. doi: 10.1111/j.1365-2486.1997.gcb136.x
Martinez Arbizu, P. (2020). pairwiseAdonis: Pairwise Multilevel Comparison Using Adonis. Available online at: https://github.com/pmartinezarbizu/pairwiseAdonis (accessed March 2021).
Martínez-García, L. B., Korthals, G., Brussaard, L., Jørgensen, H. B., and De Deyn, G. B. (2018). Organic management and cover crop species steer soil microbial community structure and functionality along with soil organic matter properties. Agric. Ecosyst. Environ. 263, 7–17. doi: 10.1016/j.agee.2018.04.018
McMurdie, P. J., and Holmes, S. (2013). phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8, e61217. doi: 10.1371/journal.pone.0061217
Menne, M. J., Durre, I., Korzeniewski, B., McNeill, S., Thomas, K., Yin, X., et al. (2012a). Global Historical Climatology Network - Daily (GHCN-Daily), Version 3. [indicate subset used]. NOAA National Climatic Data Center. doi: 10.7289/V5D21VHZ
Menne, M. J., Durre, I., Vose, R. S., Gleason, B. E., and Houston, T. G. (2012b). An overview of the global historical climatology network-daily database. J. Atmospheric Ocean. Technol. 29, 897–910. doi: 10.1175/JTECH-D-11-00103.1
Moreno, J. L., Torres, I. F., García, C., López-Mondéjar, R., and Bastida, F. (2019). Land use shapes the resistance of the soil microbial community and the C cycling response to drought in a semi-arid area. Sci. Total Environ. 648, 1018–1030. doi: 10.1016/j.scitotenv.2018.08.214
Naylor, D., and Coleman-Derr, D. (2018). Drought stress and root-associated bacterial communities. Front. Plant Sci. 8, e02223. doi: 10.3389/fpls.2017.02223
Nielsen, D. C., Lyon, D. J., Higgins, R. K., Hergert, G. W., Holman, J. D., and Vigil, M. F. (2016). Cover crop effect on subsequent wheat yield in the central great plains. Agron. J. 108, 243–256. doi: 10.2134/agronj2015.0372
Nielsen, D. C., and Vigil, M. F. (2010). Precipitation storage efficiency during fallow in wheat-fallow systems. Agron. J. 102, 537–543. doi: 10.2134/agronj2009.0348
Nielsen, U. N., and Ball, B. A. (2015). Impacts of altered precipitation regimes on soil communities and biogeochemistry in arid and semi-arid ecosystems. Glob. Change Biol. 21, 1407–1421. doi: 10.1111/gcb.12789
Nivelle, E., Verzeaux, J., Habbib, H., Kuzyakov, Y., Decocq, G., Roger, D., et al. (2016). Functional response of soil microbial communities to tillage, cover crops and nitrogen fertilization. Appl. Soil Ecol. 108, 147–155. doi: 10.1016/j.apsoil.2016.08.004
O'Dea, J. K., Miller, P. R., and Jones, C. A. (2013). Greening summer fallow with legume green manures: on-farm assessment in north-central Montana. J. Soil Water Conserv. 68, 270–282. doi: 10.2489/jswc.68.4.270
Okansen, J., Blanchet, F. G., Friendly, M., Kindt, R., Legendre, P., McGlinn, D., et al. (2020). vegan: Community Ecology Package. Available online at: https://CRAN.R-project.org/package=vegan (accessed March 2021).
Ouverson, T., Eberly, J., Seipel, T., Menalled, F. D., and Ishaq, S. L. (2021). Temporal soil bacterial community responses to cropping systems and crop identity in dryland agroecosystems of the northern great plains. Front. Sustain. Food Syst. 5, 624242. doi: 10.3389/fsufs.2021.624242
Philippot, L., Spor, A., Hénault, C., Bru, D., Bizouard, F., Jones, C. M., et al. (2013). Loss in microbial diversity affects nitrogen cycling in soil. ISME J. 7, 1609–1619. doi: 10.1038/ismej,.2013.34
R Development Core Team (2020). R: A Language and Environment for Statistical Computing. Available online at: https://CRAN.R-project.org (accessed March 2021).
R Development Core Team (2021). R: A Language and Environment for Statistical Computing. R Development Core Team.
Rose, M. T., Cavagnaro, T. R., Scanlan, C. A., Rose, T. J., Vancov, T., Kimber, S., et al. (2016). “Impact of Herbicides on Soil Biology and Function,” in Advances in Agronomy, ed. D. L. Sparks (Academic Press), 133–220.
Rumberger, A., and Marschner, P. (2003). 2-Phenylethylisothiocyanate concentration and microbial community composition in the rhizosphere of canola. Soil Biol. Biochem. 35, 445–452. doi: 10.1016/S0038-0717(02)00296-1
Sayer, E. J., Oliver, A. E., Fridley, J. D., Askew, A. P., Mills, R. T. E., and Grime, J. P. (2017). Links between soil microbial communities and plant traits in a species-rich grassland under long-term climate change. Ecol. Evol. 7, 855–862. doi: 10.1002/ece3.2700
Schlatter, D. C., Yin, C., Hulbert, S., Burke, I., and Paulitz, T. (2017). Impacts of repeated glyphosate use on wheat-associated bacteria are small and depend on glyphosate use history. Appl. Environ. Microbiol. 83, e01354-17, e01354–e01317. doi: 10.1128/AEM.01354-17
Steenwerth, K., and Belina, K. M. (2008). Cover crops enhance soil organic matter, carbon dynamics and microbiological function in a vineyard agroecosystem. Appl. Soil Ecol. 40, 359–369. doi: 10.1016/j.apsoil.2008.06.006
Tobin, C., Singh, S., Kumar, S., Wang, T., and Sexton, P. (2020). Demonstrating short-term impacts of grazing and cover crops on soil health and economic benefits in an integrated crop-livestock system in South Dakota. Open J. Soil Sci. 10, 109. doi: 10.4236/ojss.2020.103006
Van Bruggen, A. H. C., He, M. M., Shin, K., Mai, V., Jeong, K. C., Finckh, M. R., et al. (2018). Environmental and health effects of the herbicide glyphosate. Sci. Total Environ. 616–617, 255–268. doi: 10.1016/j.scitotenv.2017.10.309
van der Putten, W. H., Bardgett, R. D., Bever, J. D., Bezemer, T. M., Casper, B. B., Fukami, T., et al. (2013). Plant-soil feedbacks: the past, the present and future challenges. J. Ecol. 101, 265–276. doi: 10.1111/1365-2745.12054
Wagg, C., Bender, S. F., Widmer, F., and van der Heijden, M. G. A. (2014). Soil biodiversity and soil community composition determine ecosystem multifunctionality. Proc. Natl. Acad. Sci. 111, 5266–5270. doi: 10.1073/pnas.1320054111
Wei, T., and Simko, V. (2021). R Package “Corrplot”: Visualization of a Correlation Matrix. Available online at: https://github.com/taiyun/corrplot (accessed March 2021).
Weiss, S., Xu, Z. Z., Peddada, S., Amir, A., Bittinger, K., Gonzalez, A., et al. (2017). Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 5, 27. doi: 10.1186/s40168-017-0237-y
Whitlock, C., Cross, W., Maxwell, B., Silverman, N., and Wade, A. A. (2017). 2017 Montana Climate Assessment. Bozeman and Missoula MT: Montana State University and University of Montana, Montana Institute on Ecosystems.
Whitmore, A. P., Kirk, G. J. D., and Rawlins, B. G. (2015). Technologies for increasing carbon storage in soil to mitigate climate change. Soil Use Manag. 31, 62–71. doi: 10.1111/sum.12115
Wickham, H., Navarro, D., and Pedersen, T. L. (2016). ggplot2: Elegant Graphics for Data Analysis. 2nd Edn. New York, NY: Springer-Verlag.
Wienhold, B. J., Vigil, M. F., Hendrickson, J. R., and Derner, J. D. (2018). Vulnerability of crops and croplands in the US Northern Plains to predicted climate change. Clim. Change 146, 219–230. doi: 10.1007/s10584-017-1989-x
Wortman, S. E., Drijber, R. A., Francis, C. A., and Lindquist, J. L. (2013). Arable weeds, cover crops, and tillage drive soil microbial community composition in organic cropping systems. Appl. Soil Ecol. 72, 232–241. doi: 10.1016/j.apsoil.2013.07.014
Wyffels, S. A., Bourgault, M., Dafoe, J. M., Lamb, P. F., and Boss, D. L. (2021). Introducing cover crops as a fallow replacement in the Northern Great Plains: I. Evaluation of cover crop mixes as a forage source for grazing cattle. Renew. Agric. Food Syst. 37, 1–11. doi: 10.1017/S1742170521000417
Yahdjian, L., and Sala, O. E. (2002). A rainout shelter design for intercepting different amounts of rainfall. Oecologia 133, 95–101. doi: 10.1007/s00442-002-1024-3
Yang, F., Niu, K., Collins, C. G., Yan, X., Ji, Y., Ling, N., et al. (2019a). Grazing practices affect the soil microbial community composition in a Tibetan alpine meadow. Land Degrad. Dev. 30, 49–59. doi: 10.1002/ldr.3189
Yang, J., Wang, Y., Cui, X., Xue, K., Zhang, Y., and Yu, Z. (2019b). Habitat filtering shapes the differential structure of microbial communities in the Xilingol grassland. Sci. Rep. 9, 19326. doi: 10.1038/s41598-019-55940-y
Yilmaz, P., Parfrey, L. W., Yarza, P., Gerken, J., Pruesse, E., Quast, C., et al. (2014). The SILVA and “All-species Living Tree Project (LTP)” taxonomic frameworks. Nucleic Acids Res. 42, D643–D648. doi: 10.1093/nar/gkt1209
Zhang, X., Chen, Q., and Han, X. (2013). Soil bacterial communities respond to mowing and nutrient addition in a steppe ecosystem. PLoS ONE 8, e84210. doi: 10.1371/journal.pone.0084210
Keywords: 16S rRNA gene, cover crop mixtures, cover crop termination, integrated crop-livestock management, climate change
Citation: Ouverson T, Boss D, Eberly J, Seipel T, Menalled FD and Ishaq SL (2022) Soil bacterial community response to cover crops, cover crop termination, and predicted climate conditions in a dryland cropping system. Front. Sustain. Food Syst. 6:911199. doi: 10.3389/fsufs.2022.911199
Received: 02 April 2022; Accepted: 26 September 2022;
Published: 13 October 2022.
Edited by:
Julio S. Bernal, Texas A&M University, United StatesReviewed by:
Hanna J. Poffenbarger, University of Kentucky, United StatesElena K. Perry, Genentech, Inc., United States
Copyright © 2022 Ouverson, Boss, Eberly, Seipel, Menalled and Ishaq. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Suzanne L. Ishaq, c3VlLmlzaGFxQG1haW5lLmVkdQ==
 Tindall Ouverson
Tindall Ouverson Darrin Boss2
Darrin Boss2 Jed Eberly
Jed Eberly Tim Seipel
Tim Seipel Fabian D. Menalled
Fabian D. Menalled Suzanne L. Ishaq
Suzanne L. Ishaq