- Food Security and Safety Focus Area, Faculty of Natural and Agricultural Sciences, North-West University, Mmabatho, South Africa
The microorganisms inhabiting soil perform unique functions in the growth and development of plants. However, little is known about how plant health status affects their potential functions. We examined the functional diversity of the microbiome inhabiting the rhizosphere of powdery mildew diseased and healthy tomato plants alongside the bulk soils in South Africa's Northwest Province employing a shotgun metagenomics approach. We envisaged that the functional categories would be abundant in the healthy rhizosphere (HR) of the tomato plant. We collected soil from the rhizosphere of healthy, powdery mildew diseased tomato plants (DR), and bulk soil (BR). After that, their DNA was extracted. The extracted DNA was subjected to shotgun metagenomic sequencing. Our result using the SEED subsystem revealed that a total of fifteen (15) functional categories dominated the healthy rhizosphere, seven (7) functional categories dominated the diseased rhizosphere. At the same time, six (6) functions dominated the bulk soil. Alpha (α) diversity assessment did not reveal a significant difference (p > 0.05) in all the soil samples, but a considerable difference was observed for beta (β) diversity (P = 0.01). The functional categories obtained in this research were highly abundant in HR. Therefore, this study shows that the functions groups of the rhizosphere microbiomes were more abundant in HR samples as compared to others. The high prevalence of functions groups associated with rhizobiomes in the tomato rhizosphere indicates the need for more research to establish the functional genes associated with these rhizosphere microbiomes.
Introduction
The microbial community present in the soil plays important functions like the acquisition of nutrients like calcium, carbon, iron, magnesium, nitrogen, potassium, and sulfur, among many others, by plants and the transformation of exudates produced from the plant root into organic materials (Fadiji et al., 2021b). The microbial communities in the rhizosphere are abundant and diverse. They enhance plants' resistance to biotic and abiotic stresses (Kumar et al., 2019; Wu et al., 2020; Monohon et al., 2021). Microbial communities inhabiting the rhizosphere soil of tomato plants can act against the activity of spoilage organisms that can introduce diseases to tomato plants (Ahmad et al., 2012). These microbes' efforts improve endurance to non-living conditions, which eventually leads to improvement of plant growth (Ayangbenro and Babalola, 2021).
Tomato plant roots have a close relationship with microbiota found in its rhizosphere soil under natural systems (Lee et al., 2019). These microbiota are a part of a special microbial community that contributes to the growth and development of the tomato plant, thereby improving its health status (Ajilogba et al., 2013). Knowledge of the characteristics of microbiota potentiality aids the safety of plants' health, necessary for the functionality of the microbiome in the rhizosphere (French et al., 2021b).
The functions of the microbial community in the rhizosphere of tomato plants are manifold. They include the disintegration of metabolic material or products that boost the production of nutrients and other substances required for plants' growth (Korenblum et al., 2020; Omotayo et al., 2021). Sulfur cycling, phosphorus, potassium, and protein metabolism are among the various metabolic processes in the rhizosphere of plant crops, which improves the health condition of the plants (Dixon et al., 2020; Teklić et al., 2021). Tomato diseases and pests are known to reduce productivity (Ajilogba and Babalola, 2013). Among these diseases are early blight (Alternaria solani) reported by Akhtar et al. (2019), powdery mildew used as a case study in this research (Oidium neolycopersici) as reported by Sun et al. (2019), tomato late blight (Phytophthora infestans) reported by Mazumdar et al. (2021), bacterial wilt (Ralstonia solanacearum) reported by Zhang et al. (2020b), and soft rot (Xanthomonas species and Fusarium oxysporum) (Khalil Bagy and Abo-Elyousr, 2019; Oh et al., 2019; Safari et al., 2021) among others.
Plant roots provide mechanical support and the transporting of water and mineral salts via xylem and phloem tissues transports sugar and other organic substrates to various parts of the plant including its stem, branches, and leaves. They can participate in definitive purposes like chemical substance and exudation (Jacobsen et al., 2018). The region between microbial communities and roots of plants is the rhizosphere, where carbohydrates and organic acid are examples of the metabolites produced as rhizodeposits secreted from the roots (Hassan et al., 2019; Adedeji and Babalola, 2020; Tian et al., 2020). The exudates have molecules that draw microbes to the rhizosphere (Liu et al., 2020). The rhizosphere's primary habitat for microbial communities contains distinct, active, and abundant microorganisms unlike bulk soils (Molefe et al., 2021; Wahdan et al., 2021).
Functional diversity is defined as a component recognized by microbial ecologists as the important link between biodiversity that is generally concerned with what microbial communities do in their ecosystem. It helps to determine the trophic relationships, associations between microbes, their involvement in the process of biogeochemical cycles, and their responses to environmental change. The functions of the microorganisms found in the rhizosphere soil are abundant and for this reason, promote the quality and fertility of the soil (Zhu et al., 2020). Various studies have examined the rhizosphere microbiome associated with Solanum lycopersicum (Nuzzo et al., 2020; Wen et al., 2020; French et al., 2021a) but the functional potential of the microbial community in its rhizosphere employing a shotgun approach is understudied. There are limited functional features of root-associated microbiomes in sufficient rhizosphere soil of Solanum lycopersicum, even though recent research used a whole-genome sequencing approach to analyze the functional potential of the rhizosphere microbiome of tomatoes (Guerrieri et al., 2021; Zhou et al., 2021). However, the principle limiting these procedures is that they cannot reveal sufficient perception of the functions investigated from soil microbial communities. The introduction of culture-independent procedures like high-throughput sequencing, especially shotgun metagenomics, has helped to subdue this limitation. Shotgun metagenomics made unraveling of the functional potentials of microbiomes in a particular environment possible. To understand functional soil microbial diversity, we present one of the foremost attempts at examining its interaction with powdery mildew diseased, healthy tomato plants, and bulk soils. We hypothesized that the functional diversity of microbial communities in the tomato rhizosphere would be more abundant in the healthy rhizosphere of tomatoes compared to the diseased and bulk soils. This will further help examine the impact of plants' health status on the functional potentials of the rhizosphere microbiome.
Materials and Methods
Soil Sampling and Processing
The study was carried out at the North-West University farm at this coordinate (25°47′19.1″S, 25°37′05.1″E; 25°47′17.0″S, 25°37′03.2″E; 26°019′36.9″S, 26°053′19.0″E altitude, 159 km). Slight rain (300–600 mm) that comes with thunderstorms is experienced annually in this region between August to March in summer and the temperature ranges between 20 and 40°C. During the winter between April to July, the temperature falls to 8°C and lower. The Roma tomato cultivar is cultivated on this farm, where it has been produced for more than 7 years.
Rhizosphere soil samples were obtained from three plots, each with three replicates. The first plot contained a healthy tomato plantation. The second contained a powdery mildew diseased tomato plantation characterized by the bright yellow spots and powdery appearance on the leaves and stems, and the last plot was the bulk soil of a natural grassland with no tomato plantation. The rhizosphere soils were obtained from the root region of the tomato plant. With the aid of a sterilized soil auger, the soil was dug at the range of 4–15 cm deep and the soil samples were collected into sterile zip-lock plastic bags. Soils adhering to the root of the tomato plant was shaken and collected in the bag. The plots were separated by 40 m (healthy, and diseased). The bulk plot was separated by 50 m from the other plots. Replicates were obtained from the rhizosphere of 5 tomato plants (healthy and diseased) and the collected rhizosphere soils were pooled together. Therefore, 15 tomato plant rhizospheres were pooled together to form 3 replicates. The healthy rhizosphere soil was denoted “HR,” diseased rhizosphere “DR,” and bulk soil “BR.” The soil samples were collected in mid-March 2021, just before the tomato harvest. The rhizosphere and bulk soils were dug from a depth of 5–15 cm. The soil samples were collected and placed into sterile plastic bags (Ziploc), kept in cold boxes containing ice at −4°C, and transported to the laboratory (Babalola et al., 2022). Upon reaching the laboratory, the samples were stored in a cold room at −20°C.
DNA Extraction
According to the manufacturer's instructions, total soil DNA was extracted from 0.5 g of obtained soil with the NucleoSpin Soil kit (Macherey-Nagel, Germany). Fifty microliter of elution buffer was used to dissolve the extracted DNA and then sent out to Molecular Research DNA Lab for shotgun sequencing in the United States of America. With the aid of nanodrop, the quality of the DNA was assessed. The DNA quantity and concentration were quantified using a Qubit® dsDNA HS Assay Kit with a Qubit® fluorometer (Life Technologies, Carlsbad, CA, USA). The DNA libraries were organized after being passed through fragmentation and adapter sequence removal with Nextera DNA Flex library provision materials manufactured by Illumina. Qiagen DNEasy PowerClean Pro Cleanup Kit was employed to determine the purity of the DNA. The DNA samples were made to pass through the fragmentation process and adapter sequences. After the final concentration of DNA had been determined, the size of the libraries produced was quantified with the aid of the Agilent 2100 Bioanalyzer manufactured by Agilent Technologies, USA. Furthermore, the libraries acquired were pooled in 0.7 nM, the equimolar ratio, after which the Illumina NovaSeq 6000 system was employed for 300 cycles of pair-end sequencing.
Data Analyses
The acquired metagenome sequences were uploaded onto the online server MG-RAST (Hong et al., 2019). The quality control of the raw data was executed on the online server. This procedure entails cutting off the low reads sequences, filtering ambiguous bases, artificial sequences, and other adapter sequences from data already sequenced with the aid of the Trimmomatic version 0.33 (Bolger et al., 2014). The annotation of sequences already processed was conducted with the aid of BLAT according to Kent (2002) after the quality control procedure against the M5NR database, as explained by Wilke et al. (2012). This allowed for the incorporation of non-redundant databases. Following the method of Mitra et al. (2011) that explained how the SEED Subsystem was employed to classify the microbiomes as observed in Supplementary Figure S1. Likewise, functional profiling was carried out using the SEED Subsystem databases 1, 2, 3 levels with certain quantities like 10−5 e-value deduction together with at least 60% sequence similarity to a subsystem. On other sequences that could be annotated no analysis was further considered. Normalization of data sequences was examined to inhibit the various errors of the experiments on MG-RAST. The functional levels were obtained after functional tables were summed up and sequence data that were not categorized were maintained for statistical analysis. However, the functional diversity abundance was calculated in percentages. Thereafter, independent examination of the 9 sequences was conducted employing MG-RAST. The mean obtained from the 3 replicates of the rhizosphere and bulk soil (HR, DR, and BR) were used for statistical analysis. On the NCBI SRA data set, the raw sequences data were deposited with the bioproject number PRJNA766489 and the following Sequence Read Archive (SRA) with accession numbers SRX12366062, SRX12366063, SRX12366064 (Healthy); SRX12366065, SRX12366066, SRX12366067 (Diseased); SRX12366068, SRX12366069, SRX12366070 (Bulk).
Statistical Analysis
The heatmap through Z-score was employed to produce representing heatmaps with the functional groups of relative abundance. The assessments of functional diversity were conducted using Shannon and Pielou indices for the microbiome in rhizospheric soil of tomato plants and bulk soil samples. Furthermore, a Kruskal–Wallis test was conducted to compare the indices, and these analyses were carried out on the PAST 3.20 version (Hammer et al., 2001). To determine the functional differences and β-diversity in the soil samples. Principal Coordinate Analysis was employed. Euclidean-based Principal Component Analysis (PCA) was employed to distribute different functional groups from rhizosphere soil samples of the tomato plant. CANOCO 5.0 version was employed to plot the graphs PCA and PCoA.
Results
The Metagenome Sequencing, Quality Control, and Protein Annotation
The sum of sequences uploaded was 5,038,729,957 bp. This was recorded for rhizosphere soil samples of the tomato plant and bulk soil samples. The mean sequence data for the healthy soil (HR) was 2152004650.3 bp, diseased rhizosphere soil (DR) 1,409,528,303 bp, and bulk soil 1477197003bp. After the quality control had been executed, the mean of the sequences and mean G + C content were obtained for HR (765,041,235 bp, 63%), DR (1,352,124,415 bp, 64%), and BR (138,521,8693 bp, 64%) (Table 1). The identified protein sequences in rhizosphere soil of tomato plants and bulk soil samples were 3,985,526 for HR, 4,178,206 for DR, and 2,515,439 for BR (Table 1).
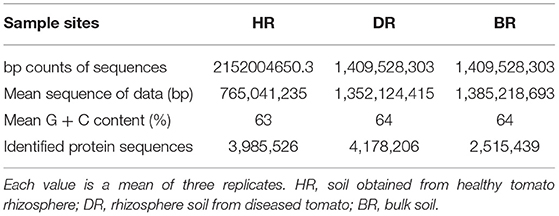
Table 1. Analysis of sequenced data and diversity evaluation of the shotgun metagenomes of the tomato plant rhizosphere.
Analysis of Functional Diversity of Microbial Biomes Associated With the Rhizosphere of the Diseased and Healthy Tomatoes and Bulk Soil
Our results on SEED Subsystem level 1, revealed 28 functional categories from the microbiome inhabiting the rhizosphere soil of the tomato plants and bulk soil. The HR soil was dominated by 15 functional categories namely: Carbohydrates (C), Miscellaneous (Mis), Cofactors, Vitamins, Prosthetic Groups, Pigments (CVPgP), Cell Wall and Capsule (CwC), Nucleosides and Nucleotides (NN), Fatty Acids, Lipids, Isoprenoids (FacLI), Clustering-based subsystems (CbS), Stress Response (SR), Nitrogen Metabolism (NM), Regulation and Cell signaling (RCs), Motility and Chemotaxis (MC), Cell Division and Cell Cycle (CdCc), Potassium metabolism (KM), Dormancy and Sporulation (DS), and Photosynthesis (P) (Figure 1). In the rhizosphere of diseased tomato plants seven (7) functional categories were identified including Phosphorus Metabolism (PhM), Amino Acids and Derivatives (AaD), DNA Metabolism (DNAM), Sulfur Metabolism (SM), Virulence, Disease and Defense (VDD), Metabolism of Aromatic Compounds (MAC), and Iron acquisition and metabolism (FeAM) (Figure 1). In bulk soil six (6) functional categories were identified: Membrane Transport (MemT), Protein Metabolism (ProM), Respiration, RNA Metabolism, Phages, Prophages, Transposable elements, and Plasmids (PPTeP) (Figure 1). The differences identified between the functional groups of the rhizosphere soil and bulk soil did not differ significantly (p > 0.05) (Supplementary Table S2). The PCA graph in this research enabled us to observe how the functional categories were distributed in the soil samples. This is illustrated in Figure 2 which shows the abundance of 15 functional categories in the healthy rhizosphere of tomatoes, 7 in the diseased rhizosphere of tomatoes, and 6 in bulk soil.
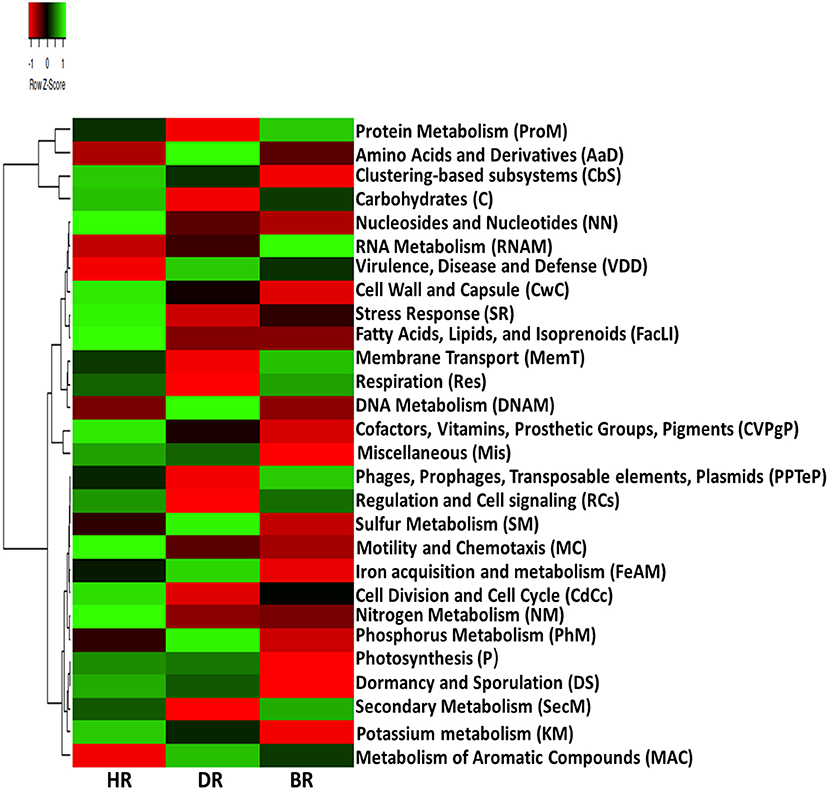
Figure 1. Heatmap data of primary metabolism of the microbiomes in the healthy, diseased rhizosphere soil of tomato plants and bulk soil samples at Level 1. Each plot situated on the relative abundances expresses the gradient color with a Z-score. HR, soil obtained from healthy tomato rhizosphere; DR, rhizosphere soil from diseased tomato; BR, bulk soil.
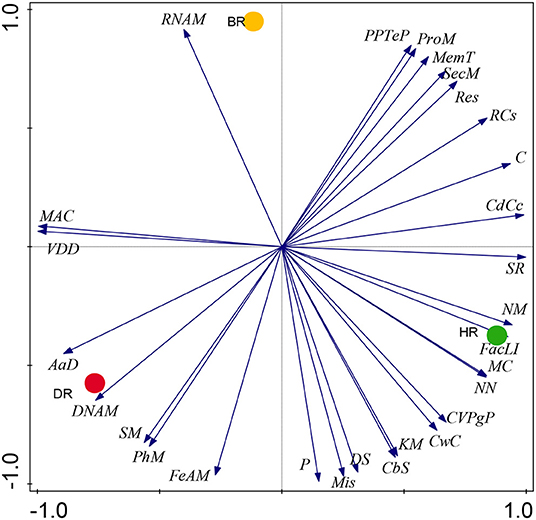
Figure 2. The PCA graph showing prominent metabolism of the microbial biomes in the healthy, diseased rhizosphere soil of tomato plants and bulk soil samples at Level 1. Axis 1 (59%) and Axis 2 (41%) revealed the variations based on Euclidean differences. HR=soil obtained from healthy tomato rhizosphere, DR, rhizosphere soil from diseased tomato; BR, bulk soil.
At subsystem level 2 functional categories, the unknown functions dominated with 21%, followed by Plant-prokaryote DOE project (6%), Protein biosynthesis (5%), Central carbohydrate metabolism (5%), DNA repair (3%), Branched-chain amino acid (2%), Monosaccharides (2%), Purines (2%), Arginine, urea cycle, polyamines (2%), ABC transporter (2%), and fatty acids (1%). Seven functions were obtained in DR: Resistance to antibiotics and toxic compounds (3%), Lysine, threonine, methionine, and cysteine (3%), RNA processing and modification (2%), Folate and pterins (2%), Protein degradation (2%), Aromatic amino acids and derivatives (1%), and DNA replication (1%). While 3 Electron donating reactions (2%), di and oligosaccharides (2%), and One-carbon Metabolism (1%) dominated the BR samples (Figure 3).
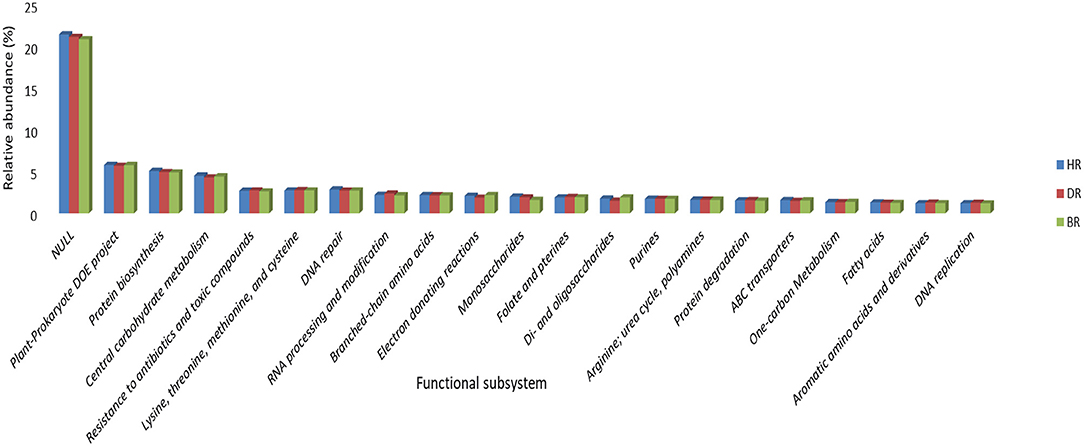
Figure 3. Level 2 subsystem arrangement of functional diversity annotation in the rhizosphere and bulk soil. HR, soil obtained from healthy tomato rhizosphere; DR, rhizosphere soil from diseased tomato; BR, bulk soil.
α and β Diversity Assessment of the Functional Groups in the Rhizosphere Soil of Tomato Plant and Bulk Soil
The functional group diversity at level 1 statistically explained the Simpson, Shannon, and evenness index of the SEED subsystem. The Kruskal–Wallis test showed that the functional categories were not significantly different (p > 0.05) (Table 2). However, the PCoA plot unveiled differences between the 28 functional groups present in the HR, DR, and BR at level 1 (Figure 4). The similarity among the soil samples was observed with one-way ANOSIM (R = 0.67 and p = 0.01).
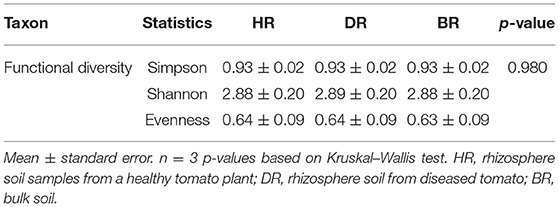
Table 2. The functional group diversity and evenness assessment of the rhizobiome at level 1 of the SEED subsystem of the soil samples.
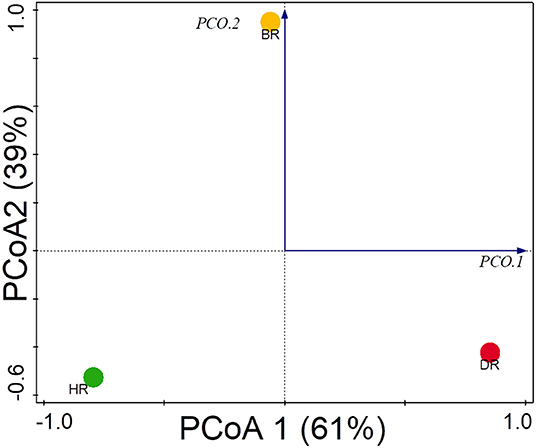
Figure 4. PCoA graph for the functional groups identified at the SEED subsystem Level 1 of rhizosphere microorganisms from each site based on Euclidean dissimilarities. HR, soil obtained from healthy Tomato Rhizosphere; DR, Rhizosphere soil from diseased tomato; BR, Bulk soil.
Discussion
This study examined the functional diversity of the microbiome in the rhizosphere soils of powdery mildew diseased tomato plants and bulk soil obtained from North to West University Farm, South Africa. Our results show that there is an eventual shift in the microbiome with functional group diversity present in the healthy rhizosphere of tomato plants compared to their presence in diseased rhizosphere soil and bulk soil. The microbial communities move from bulk soil to the healthy and diseased rhizosphere of tomatoes as the result of the exudates produced by the tomato plant root embedded in rhizosphere soil (Berendsen et al., 2012; Li et al., 2014). The DNA samples extracted from the soil samples were sequenced with shotgun metagenomics. The data obtained after the sequencing were uploaded to the MG-RAST database to conduct the analysis. The prevalent phyla obtained after the analysis were archaea, eubacteria, and fungi.
The functions of the soil samples' microbiomes are illustrated with the aid of SEED subsystem analysis. This subsystem assists in gathering and categorizing functional groups into different categories. The categories fall into 5 different subsystem levels. The highest level is Level 1, which is composed of different metabolisms in the subsystems namely anabolism and catabolism. Other levels display circumstantial pathways or specific genes necessary in the metabolism processes (Overbeek et al., 2014; Brettin et al., 2015).
In this study, the abundance of the functional groups exhibited by the microbiomes was determined in the healthy rhizosphere soil of tomatoes. At subsystems level 1, they do not reveal significant differences across the soil samples i.e., both rhizosphere and bulk soils (p > 0.05) (Supplementary Table S2). The functional categories obtained in the healthy tomato rhizosphere soil were most abundant compared to diseased rhizosphere and bulk soil as reported in this study as the result of the healthy nature of plants. This supports the hypothesis that rhizospheric microbes may be grouped based on their functional categories and not strictly their taxonomical diversity (Ramírez-Flandes et al., 2019). The functional groups were abundant in the rhizosphere due to the presence of microbial communities that contribute to the production of root exudate (Huang et al., 2014).
Our results agree with that of Tian et al. (2015). They reported how the functional categories, including carbohydrate, and protein metabolism, biological nitrogen fixation, and degradation of plant polysaccharides among other functions were harbored and enriched bacterial populations with the root-knot nematode, Meloidogyne incognita, in tomato roots. Castellano-Hinojosa et al. (2021) revealed the functional categories that include amino acid transport and metabolism, carbohydrate transport and metabolism, and carbon and nitrogen fixation are present in the rhizosphere soil of tomatoes. These contribute to the growth of tomatoes which corresponds with our results. Fadiji et al. (2021a) further explained that the functional diversity of carbohydrates, cell wall and capsule, membrane transport, clustering-based subsystems, cofactors, vitamins, prosthetic groups, pigments, lipids, isoprenoids, iron acquisition and metabolism, DNA metabolism, dormancy and sporulation, cell division and cell cycle, fatty acids, metabolism of aromatic compounds, motility and chemotaxis, nitrogen metabolism, phages, prophages, transposable elements, plasmids, and miscellaneous were abundant in the root rhizosphere of maize plants obtained from different farmlands. This indicated the abundant activity of the different microbial communities present in the root of the maize plants, thereby improving the plants' health.
The results obtained from the α-diversity investigation based on the categories of functions identified in the soil metagenomes reached the theoretical limit of 2.81, thereby reducing the subsystems found in the soil samples (Dinsdale et al., 2008). The evenness value of the functions obtained in the soil sample was low as obtained in Table 2. We were able to show the prevalent metabolisms of different types and various features of functional groups in the studied rhizosphere and bulk soil. The results obtained revealed that all the functional groups related to the soil samples do not differ significantly (p > 0.05) (Table 1).
The PCoA plot revealed a special separation between the rhizosphere soil (healthy and diseased) and bulk soil (Figure 4). The ANOSIM indicated significant differences between the functional categories of the rhizosphere and bulk soil (p-values = 0.01). The PCA revealed the abundancy of the level metabolisms of the microbiomes in the HR as hypothesized compared to DR and BR. Our result was in accordance with the study of Mendes et al. (2019) that reported on the abundance of functional categories in the rhizosphere soil of the plant. The variance obtained from the soil sample signals that the microbial metagenomes ascertain the specific functions in the rhizosphere soil. On the PCA plot, each metagenome point shows the composition of sequences joined to each subsystem together with the vector arrows that explain the degree of a metabolic pathway to the point at which distribution is influenced.
Healthy tomato rhizosphere soil possessed some predominant functional categories (Supplementary Table S2, Figure 1). The result was expected because the rhizosphere soil has been reported to contain exudates that are volatile organic compounds (VOC) produced by tomato roots (Gulati et al., 2020; Korenblum et al., 2020). These exudates are released from the root hairs of plants and consist of amino acids, carbon-rich compounds, organic acids, proteins, sugars, and secondary metabolites, which are consumed by the microbial community inhabiting the rhizosphere soil (Emmanuel and Babalola, 2020). Bardgett and Van Der Putten (2014) reported that the microbial community diversity inhabiting the soil substantially shapes biological diversity at the aboveground level. Microbiomes in the rhizosphere soil carry out important functions in evolutionary and ecological responses. Madany et al. (2020) reported how salicylic acid-induced resistance in tomatoes against Orobanche (broomrape), and observed its effect on carbon (fatty acids, organic acids, and sugars) and nitrogen (amino acids and polyamines) metabolism of the tomato plant.
At level 3 functional categories, the prominent metabolic route with carbohydrate metabolism is more pronounced in HR (Supplementary Figure S1A). This result is in line with earlier study by Mendes et al. (2019) which revealed how microorganisms needed a high prevalence of carbohydrate as an energy source for rhizosphere microbiome composition and functioning. Nitrogen is an important macronutrient required for the sustainability of life in both plants and animals (Zhang et al., 2020a). This gaseous element “N2” is acquirable in the atmosphere at an uncombined state >75% (Bolinches et al., 2020). Nitrogen-fixing bacteria in the soil fix N2 into plant-accessible amino acids, ammonia, nitrate, and urea. Nitrogen metabolism, as illustrated in Supplementary Figure S1B, indicates metabolic pathways that include ammonia assimilation, allantoin utilization, nitrate and nitrite ammonification, nitric oxide synthetization, and nitrogen fixation. According to Zheng et al. (2020), nitrogen metabolic pathways in the rhizosphere of tomato plants improve its growth and tomato fruit yield in greenhouse plantations. Our results were in accordance with that of Castellano-Hinojosa et al. (2021) namely that nitrogen metabolism was abundant in the rhizosphere of tomato plants upon inorganic nitrogen fertilization.
Sulfur metabolism was also found to dominate the HR at subsystem level 3 functional categories. The principal functions in HR are inorganic sulfur assimilation, sulfatase and sulfatase modifying factors, sulfur oxidation, galactosylceramide and sulfatide metabolism, alkanesulfonates utilization as shown in Supplementary Figure S1C. Sulfur is among the major macroelements required by tomato and potato plants to prevent the invasion of plant diseases by producing sulfur-containing defense compounds (SDCs) like glutathione, glutathione-S-transferase, phytochelatins, and other sulfur-containing proteins (Rausch and Wachter, 2005). Sulfur metabolism was conducive in the root of the tomato plants, thereby improving their wellbeing (Wang et al., 2017). Another report revealed how the microbial community plays a significant role in the process of enzymatic sulfur metabolism. The genes that encode for prominent enzymes' production for microbial sulfur oxidation, nitrogen fixation, and carbon fixation in T2 (slightly acidic tailings) were prominently obtained from Thiobacillus, Acidithiobacillus, Methylococcus capsulatus, and Thiobacillus denitrificans (Chen et al., 2013). Phosphorus metabolism was also prevalent in HR. The functional categories in this subsystem include phosphate metabolism, the pentose phosphate pathway, phosphorus uptake, high-affinity phosphate transporter, alkylphosphonate utilization, nucleoside triphosphate pyrophosphohydrolase MazG, polyprenyl diphosphate biosynthesis, and phosphonate metabolism among others (Supplementary Figure S1D). In the rhizospheres of soybean, alkylphosphonate and phosphorous uptake were relatively abundant as reported by Mendes et al. (2014) which correspond with to the results of this study.
Secondary metabolism is included among the subsystem level 3 functional groups. These functions, including biosynthesis of methionine, auxins, tannins, heme, siroheme, threonine, and homoserine are abundant in the HR (Supplementary Figure 1E). It has been reported that the abundance of the microbial communities in the healthy rhizosphere of tomatoes assists in the production of hormones by the tomato plants (Nuzzo et al., 2020). The biosynthesis of hormones that include auxin, gibberellin, cytokine, abscisic acid, jasmonic acid, and salicylic acid participate in the control of the mineral nutrient conditions (Rubio et al., 2009; Patel et al., 2016, 2021). The syntheses support a close interrelation between hormonal stimuli and nutritional homeostasis. These hormones are known to improve the growth of the tomato plant root and shoot. They are responsible for producing new cells that further promote cell elongation and apical dominance of the plant part (Chowdappa et al., 2013).
Conclusion
In this study, we present one of the first reports to prove that plant health status affects the functional diversity of microbial communities that inhabit powdery mildew diseased, healthy tomato rhizosphere, and bulk soils. We report that the general functional categories were abundant in the healthy rhizosphere of tomato plants with some of the prominent ones including the metabolism of carbohydrates, nitrogen, phosphorus, sulfur, and secondary metabolites. The Alpha diversity of the study showed no significant differences in functional groups of the rhizosphere functional groups, but significant different was obtained in the beta diversity of the functional groups of the rhizosphere microorganisms. Due to the existence of various functions explaining how the beneficial rhizosphere microorganisms express important genes, we propose further research should be conducted on rhizosphere microbiomes' ability to produce functional genes which contribute to production of various crops in of agricultural practices. Also, this study expresses those functions associated with plant-growth promotion were abundant in the microbial communities present in the healthy rhizosphere of tomato plants. Therefore, our study is fundamental for the improvement of tomato production by investigating the important features of the rhizosphere organisms to improve sustainable agriculture.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and Bioproject number can be found below: NCBI [Bioproject number: PRJNA766489].
Author Contributions
AA did the laboratory work, analyzed and interpreted the results with the help of AF, and wrote the first draft of the manuscript. OB supervised all the listed coauthors, helped structure the research, verified the analytical methods, and edited or commented on the document at all stages. All authors have read and agreed to the published version of the manuscript.
Funding
This work was funded by the National Research Foundation of South Africa grants (UID123634 and UID132595).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
We acknowledge the National Research Foundation, South Africa grants (UID123634 and UID132595) that support research in our laboratory.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.894312/full#supplementary-material
References
Adedeji, A. A., and Babalola, O. O. (2020). Secondary metabolites as plant defensive strategy: a large role for small molecules in the near root region. Planta 252, 61. doi: 10.1007/s00425-020-03468-1
Ahmad, F., Babalola, O. O., and Tak, H. I. (2012). Potential of MALDI-TOF mass spectrometry as a rapid detection technique in plant pathology: identification of plant-associated microorganisms. Anal. Bioanal. Chem. Res. 404, 1247–1255. doi: 10.1007/s00216-012-6091-7
Ajilogba, C. F., and Babalola, O. O. (2013). Integrated management strategies for tomato fusarium wilt. Biocontrol Sci. 18, 117–127. doi: 10.4265/bio.18.117
Ajilogba, C. F., Babalola, O. O., and Ahmad, F. (2013). Antagonistic effects of bacillus species in biocontrol of tomato fusarium wilt. Stud. Ethno Med. 7, 205–216. doi: 10.1080/09735070.2013.11886462
Akhtar, K. P., Ullah, N., Saleem, M. Y., Iqbal, Q., Asghar, M., and Khan, A. R. (2019). Evaluation of tomato genotypes for early blight disease resistance caused by Alternaria solani in Pakistan. Plant Pathol. 101, 1159–1170. doi: 10.1007/s42161-019-00304-8
Ayangbenro, A. S., and Babalola, O. O. (2021). Reclamation of arid and semi-arid soils: the role of plant growth-promoting archaea and bacteria. Curr. Plant Biol. 25, 100173. doi: 10.1016/j.cpb.2020.100173
Babalola, O. O., Adedayo, A. A., and Fadiji, A. E. (2022). Metagenomic survey of tomato rhizosphere microbiome using the shotgun approach. Microbiol. Resour. Announc. 11, e01131–e01121. doi: 10.1128/mra.01131-21
Bardgett, R. D., and Van Der Putten, W. H. (2014). Belowground biodiversity and ecosystem functioning. Nature 515, 505–511. doi: 10.1038/nature13855
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. H.M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformation 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Bolinches, A., Paredes, A. J., Garrido, A., and De Stefano, L. (2020). A comparative analysis of the application of water quality exemptions in the European Union: the case of nitrogen. Sci. Total Environ. 739, 139891. doi: 10.1016/j.scitotenv.2020.139891
Brettin, T., Davis, J. J., Disz, T., Edwards, R. A., Gerdes, S., Olsen, G. J., et al. (2015). RASTtk: a modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 5, 8365. doi: 10.1038/srep08365
Castellano-Hinojosa, A., Strauss, S. L., González-López, J., and Bedmar, E. J. (2021). Changes in the diversity and predicted functional composition of the bulk and rhizosphere soil bacterial microbiomes of tomato and common bean after inorganic N-fertilization. Rhizosphere 18, 100362. doi: 10.1016/j.rhisph.2021.100362
Chen, L. X., Li, J. T., Chen, Y. T., Huang, L. N., Hua, Z. S., Hu, M., et al. (2013). Shifts in microbial community composition and function in the acidification of a lead/zinc mine tailings. Environ. Microbiol. 15, 2431–2444. doi: 10.1111/1462-2920.12114
Chowdappa, P., Mohan Kumar, S. P., Jyothi Lakshmi, M., and Upreti, K. K. (2013). Growth stimulation and induction of systemic resistance in tomato against early and late blight by Bacillus subtilis OTPB1 or Trichoderma harzianum OTPB3. Biol. Control. 65, 109–117. doi: 10.1016/j.biocontrol.2012.11.009
Dinsdale, E. A., Edwards, R. A., Hall, D., Angly, F., Breitbart, M., Brulc, J. M., et al. (2008). Functional metagenomic profiling of nine biomes. Nature 452, 629–632. doi: 10.1038/nature06810
Dixon, M., Simonne, E., Obreza, T., and Liu, G. (2020). Crop response to low phosphorus bioavailability with a focus on tomato. Agron 10, 617. doi: 10.3390/agronomy10050617
Emmanuel, O. C., and Babalola, O. O. (2020). Productivity and quality of horticultural crops through co-inoculation of arbuscular mycorrhizal fungi and plant growth promoting bacteria. Microbiol. Res. 239, 126569. doi: 10.1016/j.micres.2020.126569
Fadiji, A. E., Ayangbenro, A. S., and Babalola, O. O. (2021a). Shotgun metagenomics reveals the functional diversity of root-associated endophytic microbiomes in maize plant. Curr. Plant Biol. 25, 100195. doi: 10.1016/j.cpb.2021.100195
Fadiji, A. E., Kanu, J. O., and Babalola, O. O. (2021b). Impact of cropping systems on the functional diversity of rhizosphere microbial communities associated with maize plant: a shotgun approach. Arch. Microbiol. 203, 3605–3613. doi: 10.1007/s00203-021-02354-y
French, E., Kaplan, I., and Enders, L. (2021a). Foliar aphid herbivory alters the tomato rhizosphere microbiome, but initial soil community determines the legacy effects. Front. Sustain. Food Syst. 5, 96. doi: 10.3389/fsufs.2021.629684
French, E., Kaplan, I., Iyer-Pascuzzi, A., Nakatsu, C. H., and Enders, L. (2021b). Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 7, 256–267. doi: 10.1038/s41477-020-00830-9
Guerrieri, M. C, Fiorini, A., Fanfoni, E., Tabaglio, V., Cocconcelli, P. S., et al. (2021). Integrated genomic and greenhouse assessment of a novel plant growth-promoting rhizobacterium for tomato plant. Front. Plant Sci. 12, 500. doi: 10.3389/fpls.2021.660620
Gulati, S., Ballhausen, M.-B., Kulkarni, P., Grosch, R., and Garbeva, P. (2020). A non-invasive soil-based setup to study tomato root volatiles released by healthy and infected roots. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-69468-z
Hammer, Ø., Harper, D., and Ryan, P. D. (2001). PAST: paleontological statistics software package for education and data analysis. palaeontol. Electron. 4, 1–9. Available online at: http://palaeo-electronica.org
Hassan, M., Mcinroy, J., and Kloepper, J. (2019). The interactions of rhizodeposits with plant growth-promoting rhizobacteria in the rhizosphere: a review. Agriculture 9, 142. doi: 10.3390/agriculture9070142
Hong, C. E., Kim, J. U., Lee, J. W., Bang, K. H., and Jo, I. H. (2019). Metagenomic analysis of bacterial endophyte community structure and functions in Panax ginseng at different ages. 3 Biotech 9, 1–8. doi: 10.1007/s13205-019-1838-x
Huang, X.-F., Chaparro, J. M., Reardon, K. F., Zhang, R., Shen, Q., and Vivanco, J. M. (2014). Rhizosphere interactions: root exudates, microbes, and microbial communities. Botany 92, 267–275. doi: 10.1139/cjb-2013-0225
Jacobsen, A. L., Valdovinos-Ayala, J., Rodriguez-Zaccaro, F. D., Hill-Crim, M. A., Percolla, M. I., and Venturas, M. D. (2018). Intra-organismal variation in the structure of plant vascular transport tissues in poplar trees. Trees 32, 1335–1346. doi: 10.1007/s00468-018-1714-z
Kent, W. J. (2002). BLAT—the BLAST-like alignment tool. Genome Res. 12, 656–664. doi: 10.1101/gr.229202
Khalil Bagy, H. M. M., and Abo-Elyousr, K. M. (2019). Antibacterial activity of some essential oils on bacterial spot disease of tomato plant caused by Xanthomonas axonopodis pv. vesicatoria. Int. J. Plant Pathol. 8, 53–61. doi: 10.33687/phytopath.008.02.2967
Korenblum, E., Dong, Y., Szymanski, J., Panda, S., Jozwiak, A., Massalha, H., et al. (2020). Rhizosphere microbiome mediates systemic root metabolite exudation by root-to-root signaling. Proc. Natl. Acad. Sci. U. S. A. 117, 3874–3883. doi: 10.1073/pnas.1912130117
Kumar, M., Kour, D., Yadav, A. N., Saxena, R., Rai, P. K., Jyoti, A., et al. (2019). Biodiversity of methylotrophic microbial communities and their potential role in mitigation of abiotic stresses in plants. Biologia 74, 287–308. doi: 10.2478/s11756-019-00190-6
Lee, S. A., Kim, Y., Kim, J. M., Chu, B., Joa, J.-H., Sang, M. K., et al. (2019). A preliminary examination of bacterial, archaeal, and fungal communities inhabiting different rhizocompartments of tomato plants under real-world environments. Sci. Rep. 9, 1–15. doi: 10.1038/s41598-019-45660-8
Li, J.-G., Ren, G.-D., Jia, Z.-J., and Dong, Y.-H. (2014). Composition and activity of rhizosphere microbial communities associated with healthy and diseased greenhouse tomatoes. Plant Soil. 380, 337–347. doi: 10.1007/s11104-014-2097-6
Liu, W., Zhao, Q., Zhang, Z., Li, Y., Xu, N., Qu, Q., et al. (2020). Enantioselective effects of imazethapyr on Arabidopsis thaliana root exudates and rhizosphere microbes. Sci. Total Environ. 716, 137121. doi: 10.1016/j.scitotenv.2020.137121
Madany, M. M. Y., Obaid, W. A., Hozien, W., Abdelgawad, H., Hamed, B. A., and Saleh, A. M. (2020). Salicylic acid confers resistance against broomrape in tomato through modulation of C and N metabolism. Plant Physiol. Biochem. 147, 322–335. doi: 10.1016/j.plaphy.2019.12.028
Mazumdar, P., Singh, P., Kethiravan, D., Ramathani, I., and Ramakrishnan, N. (2021). Late blight in tomato: insights into the pathogenesis of the aggressive pathogen Phytophthora infestans and future research priorities. Planta 253, 1–24. doi: 10.1007/s00425-021-03636-x
Mendes, L. W., De Chaves, M. G., Fonseca, M. D. C., Mendes, R., Raaijmakers, J. M., and Tsai, S. M. (2019). Resistance breeding of common bean shapes the physiology of the rhizosphere microbiome. Front. Microbiol. 10, 2252. doi: 10.3389/fmicb.2019.02252
Mendes, L. W., Kuramae, E. E., Navarrete, A. A., Van Veen, J. A., and Tsai, S. M. (2014). Taxonomical and functional microbial community selection in soybean rhizosphere. ISME J. 8, 1577–1587. doi: 10.1038/ismej.2014.17
Mitra, S., Rupek, P., Richter, D. C., Urich, T., Gilbert, J. A., Meyer, F., et al. (2011). Functional analysis of metagenomes and metatranscriptomes using SEED and KEGG. BMC Bioinform. 12, S21. doi: 10.1186/1471-2105-12-S1-S21
Molefe, R. R., Amoo, A. E., and Babalola, O. O. (2021). Metagenomic insights into the bacterial community structure and functional potentials in the rhizosphere soil of maize plants. J. Plant Interact. 16, 258–269. doi: 10.1080/17429145.2021.1936228
Monohon, S. J., Manter, D. K., and Vivanco, J. M. (2021). Conditioned soils reveal plant-selected microbial communities that impact plant drought response. Sci. Rep. 11, 1–11. doi: 10.1038/s41598-021-00593-z
Nuzzo, A., Satpute, A., Albrecht, U., and Strauss, S. L. (2020). Impact of soil microbial amendments on tomato rhizosphere microbiome and plant growth in field soil. Microb. Ecol. 80, 398–409. doi: 10.1007/s00248-020-01497-7
Oh, J.-W., Chun, S. C., and Chandrasekaran, M. (2019). Preparation and in vitro characterization of chitosan nanoparticles and their broad-spectrum antifungal action compared to antibacterial activities against phytopathogens of tomato. Agron 9, 21. doi: 10.3390/agronomy9010021
Omotayo, O., Igiehon, O., and Babalola, O. (2021). Metagenomic study of the community structure and functional potentials in maize rhizosphere microbiome: elucidation of mechanisms behind the improvement in plants under normal and stress conditions. Sustainability 13, 8079. doi: 10.3390/su13148079
Overbeek, R., Olson, R., Pusch, G. D., Olsen, G. J., Davis, J. J., Disz, T., et al. (2014). The SEED and the rapid annotation of microbial genomes using subsystems technology (RAST). Nucleic Acids Res. 42, D206–D214. doi: 10.1093/nar/gkt1226
Patel, R. R., Patel, D. D., Bhatt, J., Thakor, P., Triplett, L. R., and Thakkar, V. R. (2021). Induction of pre-chorismate, jasmonate and salicylate pathways by Burkholderia sp. RR18 in peanut seedlings. J. Appl. Microbiol. 131, 1417–1430. doi: 10.1111/jam.15019
Patel, R. R., Thakkar, V. R., and Subramanian, R. B. (2016). Simultaneous detection and quantification of phytohormones by a sensitive method of separation in culture of Pseudomonas sp. Curr. Microbiol. 72, 744–751. doi: 10.1007/s00284-016-1012-1
Ramírez-Flandes, S., González, B., and Ulloa, O. (2019). Redox traits characterize the organization of global microbial communities. Proc. Natl. Acad. Sci. U. S. A. 116, 3630–3635. doi: 10.1073/pnas.1817554116
Rausch, T., and Wachter, A. (2005). Sulfur metabolism: a versatile platform for launching defence operations. Trends Plant Sci. 10, 503–509. doi: 10.1016/j.tplants.2005.08.006
Rubio, V., Bustos, R., Irigoyen, M. L, Cardona-López, X., Rojas-Triana, M., et al. (2009). Plant hormones and nutrient signaling. Plant Mol. Biol. 69, 361–373. doi: 10.1007/s11103-008-9380-y
Safari, Z. S., Ding, P., Nakasha, J. J., and Yusoff, S. F. (2021). Controlling Fusarium oxysporum tomato fruit rot under tropical condition using both Chitosan and Vanillin. Coatings 11, 367. doi: 10.3390/coatings11030367
Sun, G., Feng, C., Guo, J., Zhang, A., Xu, Y., Wang, Y., et al. (2019). The tomato Arp2/3 complex is required for resistance to the powdery mildew fungus Oidium neolycopersici. Plant Cell Environ. 42, 2664–2680. doi: 10.1111/pce.13569
Teklić, T., Paradiković, N., Špoljarević, M., Zeljković, S., Lončarić, Z., and Lisjak, M. (2021). Linking abiotic stress, plant metabolites, biostimulants and functional food. Ann. Appl. Biol. 178, 169–191. doi: 10.1111/aab.12651
Tian, B.-Y., Cao, Y., and Zhang, K.-Q. (2015). Metagenomic insights into communities, functions of endophytes and their associates with infection by root-knot nematode, Meloidogyne incognita, in tomato roots. Sci. Rep. 5, 17087. doi: 10.1038/srep17087
Tian, T., Reverdy, A., She, Q., Sun, B., and Chai, Y. (2020). The role of rhizodeposits in shaping rhizomicrobiome. Environ. Microbiol. Rep 12, 160–172. doi: 10.1111/1758-2229.12816
Wahdan, S. F. M., Heintz-Buschart, A., Sansupa, C., Tanunchai, B., Wu, Y.-T., Schädler, M., et al. (2021). Targeting the active rhizosphere microbiome of Trifolium pratense in grassland evidences a stronger-than-expected belowground biodiversity-ecosystem functioning link. Front. Microbiol. 12, 73. doi: 10.3389/fmicb.2021.629169
Wang, J., Niu, W., Zhang, M., and Li, Y. (2017). Effect of alternate partial root-zone drip irrigation on soil bacterial communities and tomato yield. Appl. Soil Ecol. 119, 250–259. doi: 10.1016/j.apsoil.2017.06.032
Wen, T., Zhao, M., Liu, T., Huang, Q., Yuan, J., and Shen, Q. (2020). High abundance of Ralstonia solanacearum changed tomato rhizosphere microbiome and metabolome. BMC Plant Biol. 20, 166. doi: 10.1186/s12870-020-02365-9
Wilke, A., Harrison, T., Wilkening, J., Field, D., Glass, E. M., Kyrpides, N., et al. (2012). The M5nr: a novel non-redundant database containing protein sequences and annotations from multiple sources and associated tools. BMC Bioinform. 13, 141. doi: 10.1186/1471-2105-13-141
Wu, Y., Qu, M., Pu, X., Lin, J., and Shu, B. (2020). Distinct microbial communities among different tissues of citrus tree Citrus reticulata cv. Chachiensis. Sci. Rep. 10, 1–9. doi: 10.1038/s41598-020-62991-z
Zhang, X., Davidson, E. A., Zou, T., Lassaletta, L., Quan, Z., Li, T., et al. (2020a). Quantifying nutrient budgets for sustainable nutrient management. Glob. Biogeochem. Cycles 34, 1–25. doi: 10.1029/2018GB006060
Zhang, Y., Hu, A., Zhou, J., Zhang, W., and Li, P. (2020b). Comparison of bacterial communities in soil samples with and without tomato bacterial wilt caused by Ralstonia solanacearum species complex. BMC Microbiol. 20, 89. doi: 10.1186/s12866-020-01774-y
Zheng, X., Wang, Z., Zhu, Y., Wang, J., and Liu, B. (2020). Effects of a microbial restoration substrate on plant growth and rhizosphere bacterial community in a continuous tomato cropping greenhouse. Sci. Rep. 10, 1–11. doi: 10.1038/s41598-020-70737-0
Zhou, L., Song, C., Li, Z., and Kuipers, O. P. (2021). Antimicrobial activity screening of rhizosphere soil bacteria from tomato and genome-based analysis of their antimicrobial biosynthetic potential. BMC Genet. 22, 29. doi: 10.1186/s12864-020-07346-8
Keywords: microbial diversity, root exudate, SEED subsystem, tomato, shotgun metagenomics, powdery mildew
Citation: Adedayo AA, Fadiji AE and Babalola OO (2022) Plant Health Status Affects the Functional Diversity of the Rhizosphere Microbiome Associated With Solanum lycopersicum. Front. Sustain. Food Syst. 6:894312. doi: 10.3389/fsufs.2022.894312
Received: 11 March 2022; Accepted: 11 April 2022;
Published: 11 May 2022.
Edited by:
Everlon Cid Rigobelo, São Paulo State University, BrazilReviewed by:
Gabriel I. Ballesteros, University of Talca, ChileSalma Mukhtar, Connecticut Agricultural Experiment Station, United States
Copyright © 2022 Adedayo, Fadiji and Babalola. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Olubukola Oluranti Babalola, b2x1YnVrb2xhLmJhYmFsb2xhQG53dS5hYy56YQ==
 Afeez Adesina Adedayo
Afeez Adesina Adedayo Ayomide Emmanuel Fadiji
Ayomide Emmanuel Fadiji Olubukola Oluranti Babalola
Olubukola Oluranti Babalola