
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
SYSTEMATIC REVIEW article
Front. Sustain. Food Syst. , 03 February 2022
Sec. Agro-Food Safety
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.824714
 Ariel Brunn1
Ariel Brunn1 Zaharat Kadri-Alabi1
Zaharat Kadri-Alabi1 Arshnee Moodley2
Arshnee Moodley2 Luca Guardabassi3,4
Luca Guardabassi3,4 Phil Taylor5
Phil Taylor5 Ana Mateus4
Ana Mateus4 Jeff Waage1*
Jeff Waage1*Background: The role of the crop environment as a conduit for antimicrobial resistance (AMR) through soil, water, and plants has received less attention than other sectors. Food crops may provide a link between the agro-environmental reservoir of AMR and acquisition by humans, adding to existing food safety hazards associated with microbial contamination of food crops.
Objectives: The objectives of this review were: (1) to use a systematic methodology to characterize AMR in food crop value chains globally, and (2) to identify knowledge gaps in understanding exposure risks to humans.
Methods: Four bibliographic databases were searched using synonyms of AMR in food crop value chains. Following two-stage screening, phenotypic results were extracted and categorized into primary and secondary combinations of acquired resistance in microbes of concern based on established prioritization. Occurrence of these pathogen-AMR phenotype combinations were summarized by sample group, value chain stage, and world region. Sub-analyses on antimicrobial resistance genes (ARG) focused on extended-spectrum beta-lactamase and tetracycline resistance genes.
Results: Screening of 4,455 citations yielded 196 studies originating from 49 countries, predominantly in Asia (89 studies) and Africa (38). Observations of pathogen-phenotype combinations of interest were reported in a subset of 133 studies (68%). Primary combinations, which include resistance to antimicrobials of critical importance to human medicine varied from 3% (carbapenem resistance) to 13% (fluoroquinolones), whereas secondary combinations, which include resistance to antimicrobials also used in agriculture ranged from 14% (aminoglycoside resistance) to 20% (aminopenicillins). Salad crops, vegetables, and culinary herbs were the most sampled crops with almost twice as many studies testing post-harvest samples. Sub-analysis of ARG found similar patterns corresponding to phenotypic results.
Discussion: These results suggest that acquired AMR in opportunistic and obligate human pathogens is disseminated throughout food crop value chains in multiple world regions. However, few longitudinal studies exist and substantial heterogeneity in sampling methods currently limit quantification of exposure risks to consumers. This review highlights the need to include agriculturally-derived AMR in monitoring food safety risks from plant-based foods, and the challenges facing its surveillance.
Recognition of waning antimicrobial efficacy and the rise of untreatable bacterial infections in hospitals has driven the issue of antimicrobial resistance (AMR) to the forefront of the global health agenda. Initial efforts to address antimicrobial misuse and overuse have prioritized human health care systems and livestock production, however other sources of AMR, such as discharges from wastewater treatment facilities, generate a reservoir of antimicrobial resistant microbes in the environment that may also be pertinent to human health (O'Neill, 2016; Singer et al., 2016). Within the agricultural sector, particular attention has been paid to food animal production due to large volume use of antimicrobials and the risk of transmission of zoonotic pathogens to humans through animal-source foods. Antimicrobial use in livestock is estimated to comprise 73% of total global consumption and is likely to rise with anticipated growth in demand for animal protein and further intensification of livestock production systems in low- and middle-income countries (LMICs) (Van Boeckel et al., 2019). However, recent modeling suggests reductions in antimicrobial use in animal agriculture may have a limited impact on decreasing AMR infections in human populations (Van Bunnik and Woolhouse, 2017; Van Boeckel et al., 2019; Booton et al., 2021). Thus, a focus on AMR risks from livestock alone may be too narrow.
Comparatively little attention has been paid to the wider agricultural environment as a starting point and a conduit for AMR transmission to people through soil, water, and plants (Chatterjee et al., 2018; FAO and WHO, 2019; Wellcome, 2020). Crops grown for human food and animal feed are of particular interest as they may provide a direct link between the agro-environmental reservoir of resistant microbes and humans and animals. Antimicrobial resistant bacteria (ARB) harboring antimicrobial resistance genes (ARG) can be introduced to the crop environment through a range of inputs, including manure used as a crop fertilizer, contaminated irrigation water, crop field contaminated with raw sewage due to flooding or illegal discharges, or residues from antimicrobials used to counter crop pests. A consequence of antimicrobial drug metabolism in livestock is the excretion of active drug compounds in manure and urine, as well as ARB, which risk introduction to crop fields after application (You and Silbergeld, 2014; Lima et al., 2020). Similarly, open water sources used for crop irrigation may be contaminated with antimicrobial residues and ARB after pollution with industrial waste, hospital sewage, or agricultural effluents, contaminating it with antimicrobial chemicals and ARB (Fahrenfeld et al., 2013). Direct application of antimicrobials for control of bacterial phytopathogens, such as streptomycin use against Erwinia amylovora (fire blight) in apple and pear orchards, may leave residues that contaminate food crops and leach into local soil and water environments (Stockwell and Duffy, 2012; McManus, 2014). Other antimicrobial compounds such as biocides, heavy metals, and disinfectants have been shown to co-select resistance in soil bacteria in the crop environment (Seiler and Berendonk, 2012). Given that edible components of many crops undergo minimal or no processing prior to ingestion by humans or animals, antimicrobial residues, ARB, and ARG in crops could represent an emerging foodborne hazard.
The presence of active antimicrobial metabolites and residues in crop environments generates selection pressure for the emergence of AMR. At a sufficient concentration, susceptible microbes adapt to the presence of these residues, either through de novo mutations or by sharing mobile genetic elements (MGE) through horizontal gene transfer (HGT) to gain a survival advantage (You and Silbergeld, 2014; Martínez et al., 2015). Microbial proliferation of resistant strains can occur in the soil, on the plant surface, or within the plant tissue itself and contributes to further spread of MGE to other microbes within the local environment (Zhang H. et al., 2017).
While various methods of transmission and emergence of AMR on crops have been described, the human risk from exposure associated with food crops remains unclear. Ingested enteropathogenic bacteria harboring ARG are an obvious concern, however, the ability of bacteria to share ARG through HGT implies that even non-pathogenic species may be important vectors in the spread of ARG to the human microbiome, potentially raising the risk of future therapeutic failure (Manaia, 2017; Hölzel et al., 2018).
Human commensal bacteria might also acquire ARG from environmental microbes and plant pathogens present in crops (Sundin, 2000; Gołȩbiewski et al., 2007). In the human gut, multiplication of these commensal bacterial “vectors” could facilitate accumulation of ARG, with each exposure to contaminated food potentially also increasing the diversity of ARGs (Manaia, 2017). As such, acquisition of AMR determinants through food consumption could compromise human health not only through exposure to foodborne pathogens with acquired resistance toward therapeutic antimicrobials, but also through ingestion of non-pathogenic microbial occupants of plant-based foods carrying potentially hazardous ARG (Lammie and Hughes, 2016).
Within the food crop value chain, both pre-harvest and post-harvest factors can contribute to ARB contamination of plant-based foods. For example, cross-contamination by equipment or value chain actors can occur during harvest and processing (Richter et al., 2020); washing (O'Flaherty et al., 2019); or handling in markets (Hiko et al., 2016). Since contamination of food can occur at any point in the value chain, identifying patterns of introduction and spread may help to define critical control points where interventions could be applied to improve production of safe plant-based foods, particularly as the human health risks of foodborne AMR remain underexplored. Until recently, risk analyses of foodborne AMR (based on Codex Alimentarius Guidelines, FAO/WHO., 2015) has focused almost exclusively on animal-sourced foods (Caffrey et al., 2019; Collineau et al., 2019). Moreover, AMR surveillance programs do not generally sample plant-based foods, thereby constraining baseline data available for quantitative microbial risk assessments.
Food crop production in LMICs may be particularly susceptible to AMR contamination arising from close integration of crop and livestock production, agricultural and industrial contamination of irrigation water resources, insufficient local sanitation and hygiene measures alongside wastewater re-use, dependency on antimicrobials to compensate for poor animal husbandry, and inadequate biosecurity practices with limited surveillance and enforcement of animal health and phytosanitary regulations (Van Boeckel et al., 2015; FAO, 2016; Berthe et al., 2019). In addition, poor regulation in the antimicrobial supply chain and fewer barriers to over-the-counter purchase of antimicrobials by farmers, combined with limited access to agricultural extension services, facilitate opportunities for misapplication of antimicrobials (Rodríguez et al., 2006; McCubbin et al., 2021). For these reasons, crop production systems in LMICs may be an underestimated source of ARB to humans in those regions, and potentially other regions, through international trade in plant foods.
In this scoping review, we employed a systematic approach to explore broad patterns of AMR characteristics in crops grown for human consumption and to consolidate the current state of knowledge, identifying critical knowledge gaps while minimizing potential bias. The objectives of this review were: (1) to summarize and compare baseline descriptive data on AMR detected in crops and crop inputs globally, and (2) to identify knowledge gaps and opportunities to improve the understanding of potential human exposure risks. We had a particular interest in examining the literature from LMICs, where research in foodborne hazards is still limited.
The review was conducted from May 2020 to July 2021 using the guidelines from the published extension to the Preferred Reporting Items for Systematic Reviews and Meta-Analysis for scoping reviews (Tricco et al., 2018). An advisory group made up of experts specializing in AMR-related aspects of veterinary medicine, plant pathology, microbiology, agronomy, water management, and anthropology were invited to provide insight on the research methods, descriptive results synthesis, and summaries of evidence.
An a priori protocol was written, setting out the objectives and methods of the review using the “Population, Concept, and Context” framework (Peters et al., 2015) to identify four initial research questions: (1) In pre- and post-harvest production stages, what is the current evidence that ARB and ARG contamination in human opportunistic or obligate pathogenic microbes and selected phytopathogens (plant pathogenic bacteria) occurs in food crops?; (2) What is the evidence that contamination of food crops with antimicrobial residues leads to emergence of ARB important to human health?; (3) What is currently understood about transmission dynamics of ARB- and ARG-contaminated food crops to consumers?; (4) What is known about the human health risks of AMR in the food crop value chain with particular regard to LMICs?
Inclusion criteria comprised studies from all geographic origins. Where imported produce was sampled and the country of sample origin was given, this was cited as the source of the sample rather than where the authors were based. Given the diversity of microbes in the agro-environment and the broad range of antimicrobials available for study, we selected a subset of microbes of particular interest to human and plant health and chose antimicrobial classes categorized by the World Health Organization (WHO) as Highest Priority—Critically Important Antimicrobials (Glycopeptides; Polymixins; Cephalosporins; Quinolones), High Priority—Critically Important Antimicrobials (Carbapenems; Aminoglycosides; Aminopenicillins), and Highly Important Antimicrobials (Amphenicols; Tetracyclines) (WHO, 2019). The microbes considered are listed in Table 1 and include human opportunistic and obligate pathogens, and zoonotic species, as well as two representative bacterial phytopathogens common to crops (Erwinia amylovora, Xanthomonas species). Since treatment for these phytopathogens with clinical antimicrobials is approved in some countries and reported in the literature (Stockwell and Duffy, 2012; McManus, 2014; Zhang T. et al., 2017), they were included in this review to capture reports on the presence of clinically relevant ARG which might be transferable to human microbes. Additionally, we were interested in understanding if transferable resistance traits were reported from antimicrobials exclusively used in food crops, such as kasugamycin and zhongshenmycin (Zhang T. et al., 2017), and from agricultural azole use on Aspergillus fumigatus (Gisi, 2014). A full list of antimicrobials selected and their prioritization in medical, veterinary and plant sectors (reported use) is included in Supplementary Table 1.
Piloting of search terms was carried out in the MEDLINE database and validated against a selection of ten studies that had been gathered during the initial question-forming process. The search string in MEDLINE used Medical Subject Headings and keywords and all other databases were searched with database-specific variations on this string: ((antimicrobial or antibacterial or antibiotic or antifungal) adj3 (resistant or resistance)) AND ((fruit* or citrus or vegetable* or salad* or grain* or cereal*) OR (cultivar* or orchard*)) AND ((human* or patient* or “community acquired” or “public health” or “one health”)); the exact search string in MEDLINE is available as Supplementary Figure 1. The full set of eligibility criteria are summarized in Table 1.
Four bibliographic databases were searched on July 21st, 2020: MEDLINE; CAB Direct, Embase, and Scopus and citations transferred to Microsoft Excel (Version 2102, Microsoft Corporation, Redmond, WA, USA) for removal of duplicates. Citations then underwent screening at the title and abstract level, in which three questions answered in the affirmative permitted a study to undergo full text screening: (1) Is the study a primary research article or systematic review about ARB, ARG or antibiotic or antifungal residues? (2) Does the research discuss or describe AMR emergence, contamination and/or transmission of antimicrobial residues, ARB or ARG relevant to human health through the food crop value chain? (3) Does the study include Escherichia coli, Staphylococcus aureus, Salmonella enterica, Klebsiella pneumonia, Shigella spp., Acinetobacter baumannii, Listeria monocytogenes, Enterococcus faecium, Enterococcus faecalis, Bacillus spp., Erwinia amylovora, Xanthomonas spp. or Aspergillus fumigatus?
If any one of the questions did not apply to the citation, it was excluded from the review. A second reviewer (JW) screened 100 of the excluded studies using the same questions to validate the primary screening process; disagreements were discussed and resolved by consensus with the primary reviewer (AB). Hand-searching reference lists from 22 narrative reviews added 12 additional citations to the literature search.
A data extraction template in Microsoft Excel was developed with input from members of the advisory group. Each study underwent data extraction by a single person and was validated using a cross-over method to identify missed information or disagreements, which were subsequently resolved by consensus or a third person (AB). The data extraction table contained data points including study characteristics, study methods and samples, antimicrobial susceptibility test results and antimicrobial resistance genes through the food crop value chain. Data extraction results were stratified according to sample type, microbial species, and phenotypic antimicrobial resistance results.
The initial descriptive analysis of the literature includes all studies that underwent data extraction as one of the objectives was to characterize the body of published works on the topic; subsequent analysis however focused on a subset of studies that reported phenotypic and/or genotypic results in the following microbes, selected for their relevance to human health: E. coli, K. pneumonia, S. enterica, Shigella spp., S. aureus, L. monocytogenes, E. faecium, E. faecalis, and A. baumanii. Studies on the impact of antimicrobial residues on the microbes of interest were few and primarily focused on environmental and human toxicity rather than AMR. As these outcomes were deemed too broad to be included in this review, they are summarized in a separate report available from the authors. No studies in the data set isolated E. amylovora or contained information on antibiotics used exclusively in crops, namely kasugamycin or zhongshenmycin, so no analyses were undertaken on these variables.
To date, there is no recognized standard of qualifying the human risk of exposure to microbes with AMR traits occurring in plant-based foods, though some authors have recently proposed broad frameworks to conceptualize environmental AMR risks (Martínez et al., 2015; EFSA BIOHAZ Panel et al., 2021). To facilitate our analysis on AMR risks from food crops, we grouped human pathogens with acquired AMR phenotypes (Table 2). These pathogen-AMR phenotype combinations were adapted from the WHO Global Priority Pathogens List (PPL) (Tacconelli et al., 2018) and termed primary pathogen-phenotype combinations. They combine opportunistic and obligate human pathogens of Critical, High, and Medium importance on the PPL with phenotypic resistance to antimicrobials that have a narrow spectrum of activity and to which there are fewer alternatives should resistance develop. Our categorization largely corresponds to a recently published ranking of highest priority ARB occurring in the food value chain by the European Food Safety Authority (EFSA BIOHAZ Panel et al., 2021).
Given the global scope of our review, we were also interested in an additional category of pathogen-AMR phenotype combinations (termed secondary pathogen-phenotype combinations) focused on phenotypic resistance toward antimicrobials that are frequently used in veterinary medicine and crop protection but may not rank as high risk in a public health context. These secondary pathogen-phenotype combinations were included because these antimicrobials are used by crop producers and may become dispersed in the environment after agricultural application (EFSA BIOHAZ Panel et al., 2021). Secondary pathogen-phenotype combinations comprised PPL pathogens as well as Listeria monocytogenes and Aspergillus fumigatus, the former a contaminant of food processing establishments and the latter a common soil inhabitant in the crop environment. Resistance to chloramphenicol, which is no longer authorized for use in livestock production in many countries due to the risk of aplastic anemia in humans from residue exposure, was included due to its historical use and as an indirect indicator of unregulated use of antimicrobials in agriculture (Li et al., 2013; Lu et al., 2020; McCubbin et al., 2021). A full list of the primary and secondary pathogen-phenotype combinations is provided in Table 2.
Samples were categorized into ten groups comprising seven different kinds of crops and three groups from the crop environment: soils, irrigation water and manure fertilizer. Crops were grouped according to whether the edible component is grown above or below ground and whether crops are consumed fresh (salad crops, fruit), consumed either cooked or fresh (vegetables, culinary herbs), or require cooking (root crops, pulses and grains). Ready-to-eat (RTE) plant-based foods (defined as value-added foods intended to be consumed immediately with no further preparation or cooking needed, such as a prepared salad) were also included. Full categorization of sample groups can be found in Supplementary Table 2. Food samples were characterized as pre-harvest if they were collected in the field prior to harvest, or post-harvest if they were collected at harvest, during processing, or in retail markets. Among sample groups, irrigation water, manure fertilizer, and soil inputs to crop production were always categorized as pre-harvest samples and RTE plant-based foods were always categorized as post-harvest samples. Patterns and trends were explored in the subset of studies (n = 133) that provided phenotypic results according to sample group, stage of value chain sampling, and country of origin.
The data synthesis process enumerated observations of acquired phenotypic resistance of interest reported in sample groups within each study as presence or absence, for example a study reporting ampicillin resistance detected in retail carrots of any quantity was counted as one observation of ampicillin resistance in root vegetables. This descriptive method was used on account of substantial heterogeneity among both study sampling and study data reporting methods. The full list of citations reporting phenotypic and sample observations are found in Supplementary Table 3.
Studies providing ARG results were analyzed separately and stratified by antimicrobial class and sample group. Two families of ARG, extended-spectrum beta-lactamase (ESBL) and tetracycline resistance genes, were selected for further study because they have known functional resistance genes that correspond to phenotypic resistance with a high degree of sensitivity (Neuert et al., 2018; Collineau et al., 2019) and represent resistance to antimicrobials of critical medical application and extensive agricultural usage, respectively.
Keyword searches in four bibliographic databases yielded 4,455 studies after duplicates removal (Figure 1). Studies that measured antimicrobial residues in the crop environment are reviewed in another report (n = 17). The study set after relevance screening of full texts yielded 196 studies for the final review. Reasons for exclusion of 70 studies were as follows: six studies represented an opinion or narrative review; nine studies sampled plant-foods that were cooked or fermented; seven studies focused on non-eligible animal or environmental samples; 25 studies investigated non-AMR aspects of microbial contamination such as virulence traits or focused on microbe species not included in this review, and 23 studies reported AMR results of food crop samples aggregated with non-crop samples, such as meat or wildlife feces.
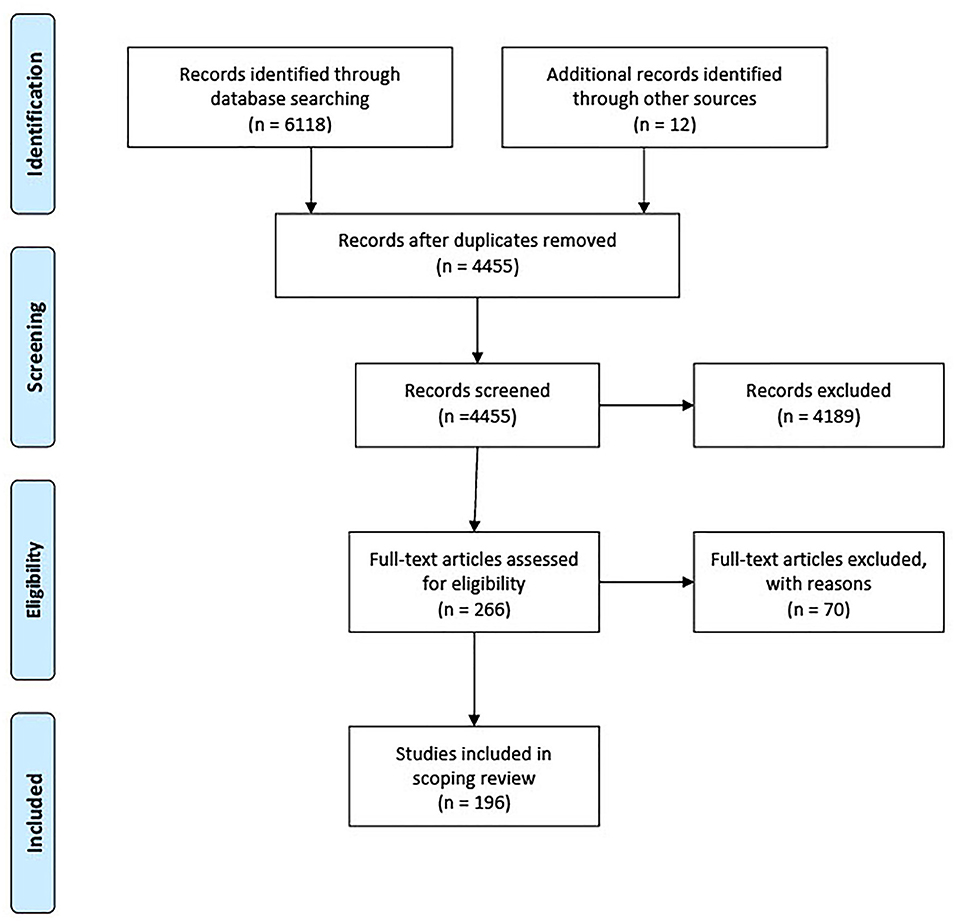
Figure 1. PRISMA flow chart of study selection process for scoping review. Studies published between January 1st, 2000, to July 21st, 2020, were included.
One hundred and ninety-six studies analyzed samples from the food crop value chains in 49 countries, 63% (n = 31) of which are classed as LMICs (World Bank, 2020). Sample observations from LMICs comprised 69% of studies evaluated (n = 136). While most studies investigated AMR in domestic samples, five studies explicitly collected imported plant foods (Veldman et al., 2014; Zurfuh et al., 2016; Chanseyha et al., 2018; Jans et al., 2018; Jung and Rubin, 2020). Studies from Asia were highly represented (46%, 90/196), with half of these being conducted in China (45). Other regions were also represented to a lesser extent: Africa (38), the Americas (33), Europe (33) and Oceania (2).
Publications on the topic have increased recently, with almost two-thirds of studies published after 2016 (128/196). Most studies were observational studies (n = 158, 80.6%), in which researchers surveyed crop samples for the presence of AMR. Relatively few studies involved an experimental field trial (n = 33; 16.9%), in which researchers investigated the effects of an exposure, usually contaminated agricultural inputs, on AMR transmission to crops. Of the experimental trials, 14 studies examined exposure of crops or crop soil to manure and nine studies did the same with irrigation water, to measure transfer of ARB and ARG to crops.
Studies conducted in LMICs were somewhat more likely to use an observational study design (85% of LMIC studies) than in high income countries (HICs, 70% of HIC studies). Observational study designs consisted of 147 cross-sectional; 2 case control; and 9 descriptive reports; probability sampling techniques were rarely used or reported in observational studies (n = 26; 13.3%). One systematic review and four quantitative studies were also included, two of which were quantitative human exposure assessments.
Most studies collected and tested multiple sample types such that the data set comprised 1,008 sample observations from the food crop value chain. Almost half of all samples were from crops in which the consumed part is grown above ground (479 observations) including: salad crops (232); vegetables (136); culinary herbs (59); fruit crops (29); and pulses and grains (23). Root crops and RTE plant-based foods comprised 86 and 32 records, respectively. Some samples were only reported as an aggregate of mixed vegetables or fruit and these were classified as “mixed produce” in our review (n = 68). Several studies also collected samples of crop-related agricultural inputs, including field soil (n = 196); irrigation water (n = 83); and manure fertilizer (n = 64).
Figure 2 shows the number of observations of microbial isolates recovered summarized by sample group. The four most frequently recorded species of interest to this review were E. coli (35% of isolate observations), S. enterica (19%), and E. faecium or E. faecalis (15%). When observations from these four species were combined, they comprised over two-thirds of culture-positive sample records in vegetables (72% of positive samples), salad crops (71%), as well as most of the microbial observations in irrigation water and manure samples (94 and 90%, respectively). The most reported species in studies that identified culture-positive root crop observations were E. faecium and E. faecalis (52% of observations); whereas L. monocytogenes were the most frequently reported microbe isolated in positive samples of pulses and grains (45%). Other microbes were rarely reported in the study set, together comprising <10% of total microbial observations across all sample groups, these were A. baumanii, Bacillus spp., Shigella spp., and Xanthomonas spp.
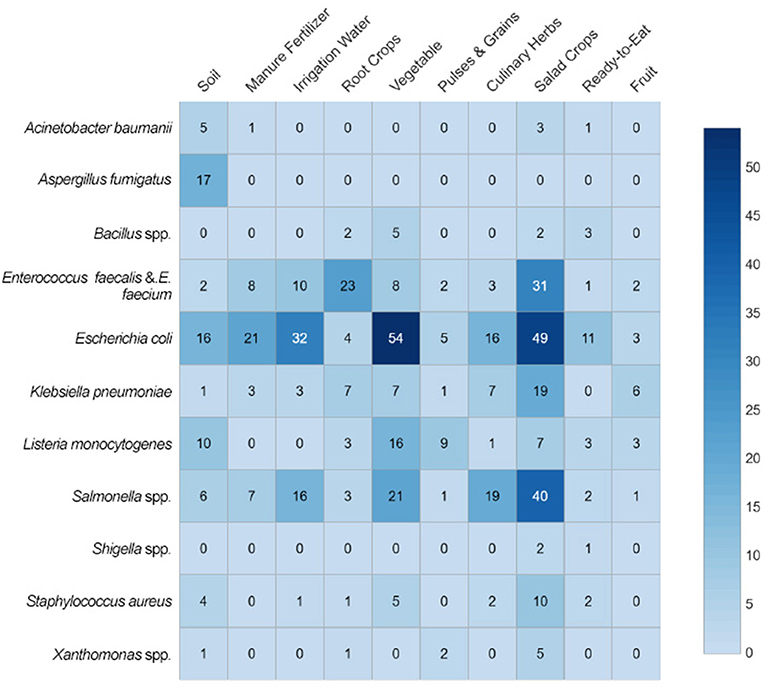
Figure 2. Microbial isolates of sample groups reported in studies published between 2000 and 2020. In this heat map, shaded cells indicate the distribution of microbes of interest isolated from sample groups reported in 196 studies, with the number of observations of microbe isolates per sample group indicated in the cell. Studies could report multiple isolates from multiple sample groups, so the number of observations is not equal to the number of studies.
Studies employed a range of methods for detecting AMR. Most common were culture-based methods using broth dilution (n = 29) or agar diffusion (such as disk diffusion and e-tests) (n = 106) in comparison to non-culture-based methods (n = 105), which included six metagenomic studies. Culture-based methods mainly relied on interpretive criteria of the Clinical and Laboratory Standards Institute (CLSI, including NCCLS, n = 108), followed by the European Committee of Antimicrobial Susceptibility Testing (EUCAST, n = 25), standards of the Deutsches Institut für Normung (DIN, n = 2), and the Comité Antibiogramme - Société Française de Microbiologie (CA-FSM, n = 3).
Studies with samples containing positive microbial isolates were stratified by phenotypic resistance observations and selected if they met definitions for primary and secondary pathogen-phenotype combinations (see Table 2). These observations formed a subset of 133 studies conducted in 38 countries that underwent further analysis by sample group as illustrated in Figure 3. More than twice as many resistance observations and just over 50% more sample observations were recorded for secondary than for primary pathogen-phenotype combinations. Observations of each primary pathogen-phenotype combination as a proportion of total resistance observations rose from 3% (carbapenem resistance: IMI-R microbes) to 10 and 13% (third generation cephalosporin resistance: CAZ-R and CTX-R microbes; fluoroquinolone resistance: CIP-R microbes, respectively), whereas the proportion of detected secondary combinations were greater overall, ranging from 14% (aminoglycoside resistance: GEN-R and STR-R microbes) to 20% (aminopenicillin resistance: AMP-R and AMO-R microbes) of resistance observations. Observations of secondary pathogen-phenotype combinations were detected in every sample group other than RTE foods and fruit crops, where results were variable, whereas observations of primary pathogen-phenotype combinations were not reported at all in RTE foods and pulses and grains (and inconsistently in fruit crops). Certain sample groups had resistance observed in <50% of sample observations, these included pulses and grains, RTE foods, and fruit crops; these three sample groups had the fewest sample observations overall. Resistance to vancomycin in S. aureus, E. faecium and E. faecalis and resistance to azoles in Aspergillus fumigatus were rarely observed and not included in Figure 3; vancomycin resistance in enterococci was reported in two studies on vegetable samples, and azole resistance in A. fumigatus was reported in four studies that tested soil samples.
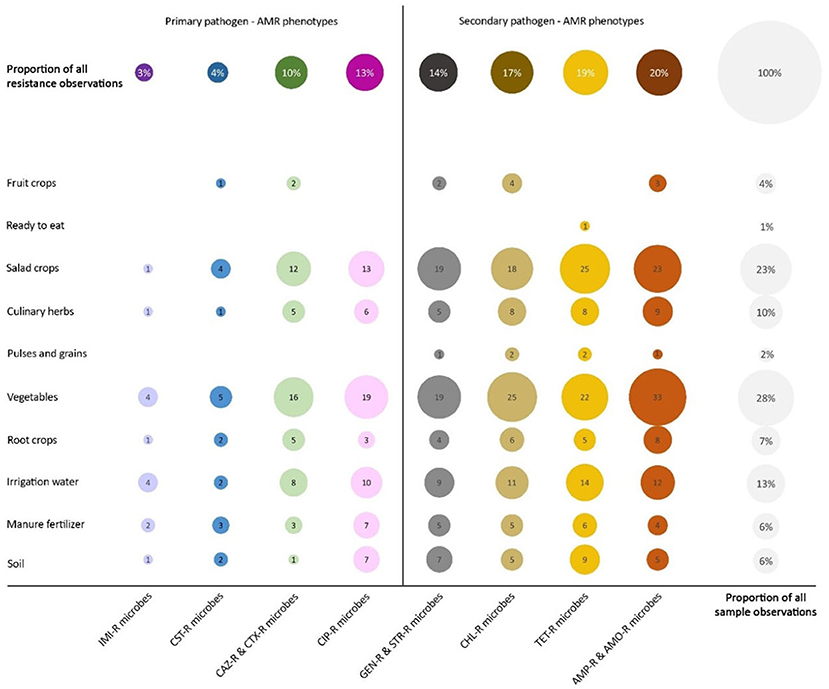
Figure 3. Selected pathogen-AMR phenotype combinations detected in sample groups reported in studies published between 2000 and 2020. The number of AMR observations of primary and secondary pathogen-phenotype combinations are shown. IMI-R microbes: A. baumanii, E. coli, K. pneumoniae, Salmonella and Shigella spp.; CST-R microbes: E. coli, K. pneumoniae, Salmonella and Shigella spp.; CAZ-R and CTX-R microbes: E. coli, K. pneumoniae, Salmonella and Shigella spp.; CIP-R microbes: Salmonella spp., E. coli and K. pneumoniae; GEN-R and STR-R microbes: E. coli, K. pneumoniae and S. aureus; CHL-R microbes: E. coli, K. pneumoniae, Salmonella spp., Shigella spp., S. aureus, E. faecium and E. faecalis; TET-R microbes: E. coli, K. pneumoniae, Salmonella spp., Shigella spp., S. aureus and L. monocytogenes; AMP-R and AMO-R microbes: E. coli, K. pneumoniae, Salmonella spp., Shigella spp., L. monocytogenes, E. faecium and E. faecalis. Circle size and central number indicate the aggregate number of observations corresponding to each pathogen-phenotype combination distributed across sample groups per study. The uppermost row indicates the relative proportion of all AMR observations represented by each pathogen-phenotype category in the column directly beneath. The right-most column (uncoloured) represents the relative distribution of sample groups, irrespective of susceptibility testing result.
Figure 4 shows the distribution of studies reporting primary and secondary pathogen-phenotype combinations across world regions, sub-grouped by whether they involved pre- or post-harvest samples. More than twice as many studies reporting primary pathogen-phenotype combinations were conducted in Asia and Africa (n = 57) than in other regions combined (n = 22), and studies from Asia were also three times more likely to collect samples at the post-harvest stage of the food value chain. Of the AMR phenotype combinations, the distribution of reported chloramphenicol resistance in food value chain samples favored LMICs in Asia and Africa. In contrast, vancomycin resistance (including intermediate susceptibility) in enterococci was exclusively reported in food crops from HICs[-108mm] Q16 (Johnston and Jaykus, 2004; Kim and Woo, 2017).
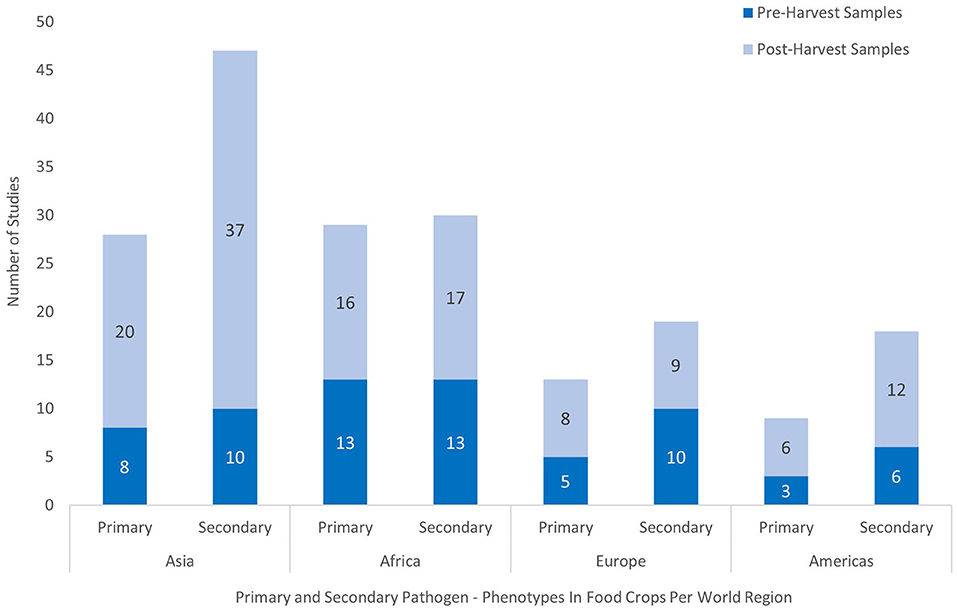
Figure 4. Analysis of the world region origin of occurrence of pathogen-phenotype combinations in studies conducted on the food crop value chain. Selected studies published between 2000 and 2020 are grouped by the stage of sampling at the pre- or post-harvest level of the value chain and by priority level of the pathogen-phenotype combination.
Over half of all studies used genotypic methods solely or in addition to phenotypic methods to characterize AMR (105 studies). A minority of these studies (21/105) employed advanced sequencing methods such as whole genome and metagenomic sequencing to characterize resistant isolates and to evaluate related clones for source attribution analysis. ARG conferring resistance to all antimicrobial classes of interest were observed. Figure 5 shows the distribution of ARG belonging to two of these antimicrobial classes, ESBLs (A) and tetracycline resistance genes (B), reported per sample group. ESBL genes, including CTX-M, SHV, OXA, and TEM beta-lactamases, were reported in a range of sample groups with both SHV and CTX-M genes (and its variants) reported in every group, from soil and salad crops to fruit crops. However, these ESBLs were most frequently detected in vegetables, irrigation water, salad crops, and culinary herb samples and mirrored patterns of phenotypic third-generation cephalosporin resistance. In studies that identified CTX-M variants, one of the most globally disseminated variants of the CTX-M beta-lactamase family, CTX-M-15 (Bevan et al., 2017), was more frequently reported than other variants and detected in food crops often eaten raw, including culinary herbs, salad, and fruit crops, as well as in vegetables and root crops.
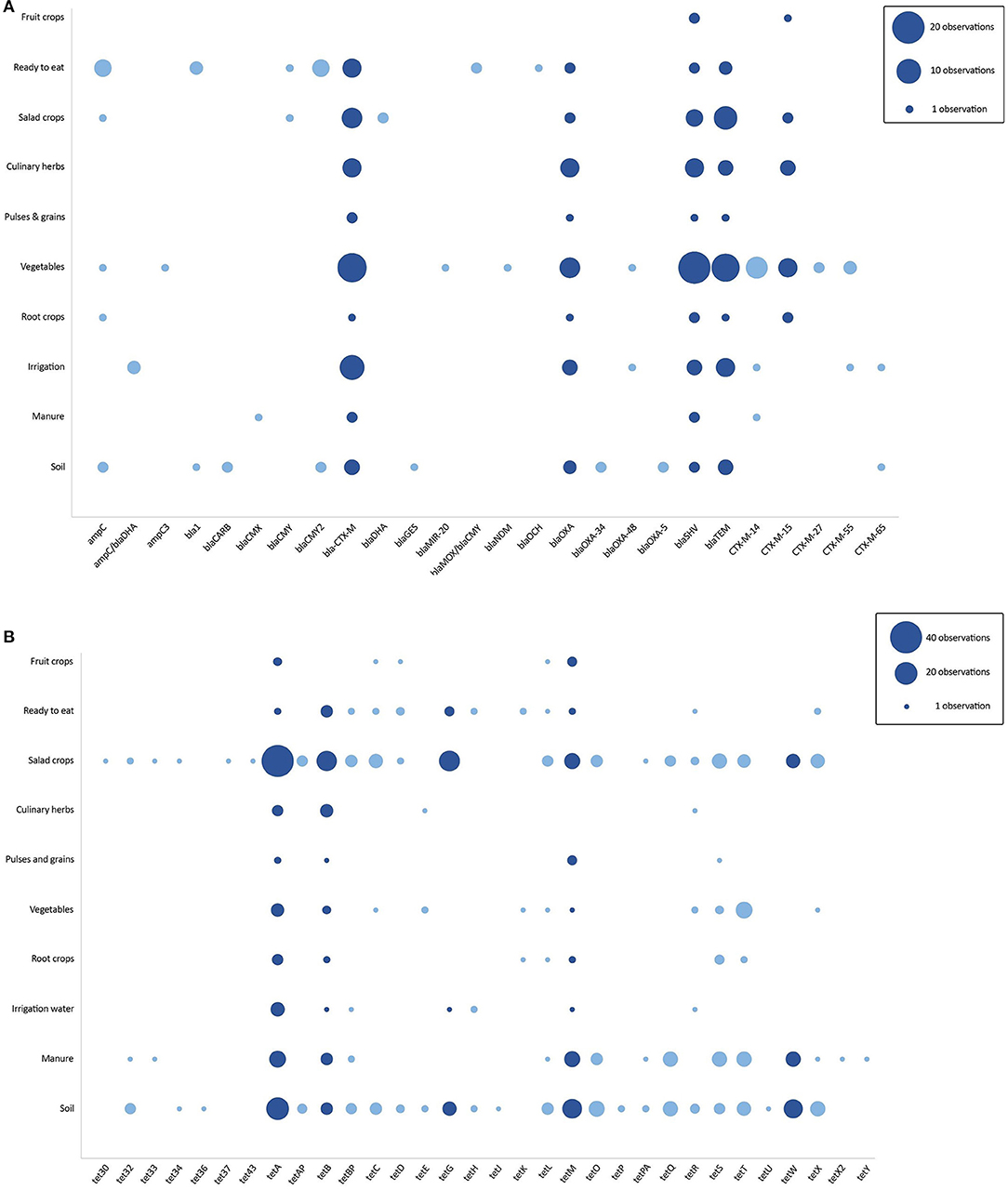
Figure 5. (A,B) Distribution of extended-spectrum beta-lactamase (A) and tetracycline resistance (B) genes across sample groups. Each filled circle corresponds to observations of specific ARG (x-axis) recorded in each sample group (y-axis). The size of the circle is proportional to the number of observations recorded from the study set (studies published between 2000 and 2020). Dark blue shading corresponds to major ARG.
Tetracycline resistance genes, particularly tetA, tetB, and tetM, were observed to occur in a diversity of sample groups with a greater frequency than ESBLs. Sample groups most highly represented included salad crops, soil, and manure samples. This pattern was partly reflected in the corresponding phenotypic profile in which salad crops, vegetables, irrigation water and soil were most common. Soil and manure samples were more likely to contain observations of tetA, tetM, and tetW genes, while tetA, tetB, and tetG were particularly frequent in salad crops.
Numerous studies have been published recently on AMR in food crop value chains worldwide. This review confirms the widespread reporting of resistance to antimicrobials of medical importance in human pathogenic microbes isolated from crops, both in the field and marketplace. We applied a systematic approach to evaluate studies reporting both phenotypic and genotypic results and from this we describe characteristics of AMR in the food crop value chain. We stress that firm conclusions cannot be drawn on the prevalence and relative importance of different kinds of resistance and AMR transmission pathways because of the substantial heterogeneity between study methods and conditions, nonetheless, some broad, indicative patterns emerge from our analysis.
We report observations of resistance to antimicrobials, particularly aminopenicillins, and tetracyclines, in microbes on food crops including those routinely eaten raw or with minimal preparation primarily in Asia and Africa, with the fewest studies from the Americas. This pattern is similar to results from recent mapping of AMR surveys on meat samples from LMICs which identified “hotspots” of resistance in several locations in Asia and certain areas in Africa and found the highest rates of resistance included tetracyclines and penicillins (Van Boeckel et al., 2019). Tetracyclines and penicillins have also been identified as the two most frequently used antimicrobials in livestock production (OIE, 2020) suggesting that AMR trends in crops may partly reflect those occurring in other agricultural sectors with substantial antimicrobial usage. Of great concern, however, are multiple reports of resistance to antimicrobials considered critically important to medical therapy detected on samples taken throughout the food crop value chain. This includes resistance to third generation cephalosporins in Gram negative enteropathogenic bacteria, which comprised 10% of phenotypic resistance observations detected in crops and associated inputs, particularly vegetables, salad crops and irrigation water. This pattern was largely replicated in the analysis of ESBL genes. Resistance to imipenem, a carbapenem antimicrobial used for the treatment of infections resistant to third and fourth generation cephalosporins and multi-drug resistant Gram-negative infections (Zemelman et al., 1993; Park, 2014), was also observed. Despite its classification as a High Priority Critically Important Antimicrobial (WHO, 2020), imipenem resistance was detected in samples from vegetables, salad crops, root crops, culinary herbs, irrigation water, manure fertilizer, and soil. These results concur with findings from a recent review summarizing carbapenem resistance in environmental reservoirs that confirmed presence of plasmid-mediated carbapenemase genes in environmental water sources and agricultural settings and suggested wastewater treatment plant effluents as a primary contaminating source (Mills and Lee, 2019). Furthermore, colistin resistance in Enterobacteriaceae was reported in multiple crops including fruit, salad, root, culinary herbs, and vegetables. Colistin (polymyxin E) is an antimicrobial primarily used in veterinary medicine that has in recent decades been re-embraced by the medical community as a last-resort option to combat carbapenem-resistant Enterobactericeae and Acinetobacter spp. infections, including K. pneumoniae (Skov and Monnet, 2016; Pragasam et al., 2017). Although in this review colistin resistance was primarily described in E. coli, two reports described resistance in K. pneumoniae in post-harvest fruit and vegetable crops from China and India, respectively (Ghafur et al., 2019; Yang et al., 2019). Since colistin resistance in Klebsiella has previously been reported primarily in clinical isolates from hospitals (Liu et al., 2016; Li et al., 2018), this may imply a human, rather than animal, origin and indicates the complexity of intersecting transmission pathways for AMR in crops.
The subset of studies reporting culture results showed that of the bacterial species of interest to this review, E. coli and S. enterica species were the most studied in food crop samples. Although virulence characteristics were not consistently identified among authors, the association of these microbes causing foodborne illness originating from food crops is well-documented. Source attribution analysis by Sivapalasingam et al. (2004) found that Salmonella-contaminated spinach and lettuce were the largest contributors to produce-associated gastrointestinal illness in the USA. Similarly, one of the largest outbreaks of enterohemorrhagic E. coli O104:H4 to occur in Germany caused 3,801 cases and 46 deaths and was linked to contaminated sprouts (Mellmann et al., 2011). A more recent meta-analyses of pathogen contamination of retail produce in Europe reported higher pooled prevalence estimates of Salmonella (1.7%) and of Shiga toxin-producing E. coli (1.4%) in leafy greens, with slightly lower estimates in lettuce (1.2 and 0.7%, respectively) (Silva et al., 2017). While our review appears to support findings that S. enterica and E. coli are important food crop contaminants, the frequency of their isolation may reflect selective sampling at the expense of other bacterial genera, particularly those that are of less consequence to human health or that are more difficult to isolate in the laboratory.
In our review, we found two studies that reported resistance in bacterial pathogens of plants, specifically Xanthomonas spp. Rodríguez et al. (2006) reported phenotypic and genotypic resistance, with evidence of plasmid carriage, to gentamycin and oxytetracycline in lettuce. A second study in our review reported a prevalence of 0.75% of streptomycin resistance in leaf blight (Xanthomonas oryzae pv. oryzae) and bacterial blight (Xanthomonas oryzae pv. oryzicola) of rice crops (Xu et al., 2010). The authors of this study suggested that other control methods such as use of resistant cultivars and alternating antibacterial chemicals may have reduced selection pressure for resistance development, contributing to the low prevalence (Xu et al., 2010). Plasmid-borne AMR in phytopathogens could nonetheless constitute a risk of AMR dissemination to human commensal or pathogenic bacteria through horizontal gene transfer. Earlier reports have demonstrated transferability of plasmid-borne antimicrobial resistant genes from the plant pathogen Erwinia amylovora to human pathogens Corynebacterium striatum, Pseudomonas aeruginosa, and Pasteurella multocida (Sundin, 2000; Tauch et al., 2000).
Research on antifungal resistance in Aspergillus fumigatus in crops was also uncommon in the studies included in this review. While A. fumigatus is not a plant pathogen, it is a regular inhabitant of soils and decaying vegetations, and an opportunistic pathogen of immunosuppressed patients (Gisi, 2014). Since the vast majority of phytopathogens are fungal (Berger et al., 2017), A. fumigatus represent a good sentinel for monitoring azole resistance due to fungicide application in the agricultural environment. Two studies included in our review linked the use of triazole fungicides in crops to azole-resistant A. fumigatus collected from vegetable crop soils (Ren et al., 2017; Alvarez-Moreno et al., 2019). However, two studies from Italy suggested that although resistance occurs in crop environments, fungal clonal expansion is unlikely due to presence of genetically distinct strains (Prigitano et al., 2014, 2019) further supporting the hypothesis that wind-driven dispersal may be an important mechanism of spread (Fisher et al., 2018). Taken together, these studies suggest that direct application of fungicides for crop protection purposes may result in antifungal resistance that could theoretically constitute a threat to food safety especially in exposed immunocompromised individuals. While A. fumigatus is rarely implicated in fungal foodborne disease outbreaks, there have been reported cases of gastrointestinal aspergillosis due to ingestion of the pathogen in humans (CDC, 2014; Dias et al., 2018). Nevertheless, the paucity of studies and low prevalence reported are insufficient to confirm this use as an important threat to consumers.
Fewer studies used genotypic methods than phenotypic methods and advanced genomic analysis was uncommon in studies included in this review. Despite this, ARG associated with all antimicrobial classes of interest were observed in food crops and their value chains. Our data synthesis focused on the distribution of two types of ARG, ESBLs and tetracycline resistance genes, reported in agricultural environments (Bevan et al., 2017; Ye et al., 2018). Both types were detected in a diversity of sample groups, however a greater number of tetracycline resistance genes observations were reported than ESBLs. The ESBL variant, CTX-M-15, dominates human, animal, and environmental settings globally (Cantón et al., 2012; Bevan et al., 2017) and was observed along with other ESBL genes including SHV, OXA, and TEM, in vegetables, salad crops, culinary herbs and irrigation water samples, similar to phenotypic findings (Figure 3).
A recent literature review of ARG distribution patterns suggested that tetracycline ARG are overrepresented in non-clinical settings such as farms, in comparison to hospitals (Zhuang et al., 2021). In food crop environments, tetracycline resistance genes are highly associated with agricultural determinants (Lugo-Melchor et al., 2010; Campos et al., 2013; Jones-Dias et al., 2017; Blau et al., 2019; Xiong et al., 2019; Gao et al., 2020) and we observed a predominance of tetA, tetB, tetG, tetM, and tetW in soil and manure samples. The accumulation of tetracycline resistance genes in agricultural soil is influenced by a strong sorption affinity of tetracycline residues to soil (Pan and Chu, 2016) and tetracycline antimicrobial use in livestock production and possibly crop protection. Moreover, the diversity of microbes and nutrient density of manure and sewage-sludge fertilized soils, may amplify HGT to human and zoonotic pathogens (Lima et al., 2020) and contribute to uptake and contamination of surface grown food crops, such as vegetables and lettuce (Holvoet et al., 2013; Rahube et al., 2014; Ye et al., 2018). ESBL and tetracycline resistance gene accumulation in food crops may be linked to contaminated irrigation water, manure, and fertilized soils (Huang et al., 2021; Zhuang et al., 2021), however the risk to human health after exposure remains unclear (Martínez et al., 2015; Collineau et al., 2019).
We were interested in understanding transmission and dissemination of AMR through the food value chain and concomitant food safety risks. Experimental trials are a useful method to explore AMR exposure and transmission, and of the 33 studies using this design and included in our review, 14 focused on exposure to manure and nine studies on irrigation water exposure. Soil fertilization with manure remains a common and cost-effective source of nutrient recycling for crop production and is a useful method to manage rising volumes of manure production due to increasing demand for animal protein. However, manure can have a high concentration of active antimicrobial metabolites, ARB, and ARG as a result of veterinary antimicrobial usage in livestock (Zhang H. et al., 2017; Lima et al., 2020). Few countries set drug residue limits or monitor other contaminants prior to use of manure as fertilizer, and our study indicated a clear interest in crop contamination from manure sources, with more experimental field trials studying this transmission pathway than any other routes of pre-harvest exposure. Mitigation of AMR exposure in animal and human waste may be feasible. For example, a series of longitudinal studies conducted in Canada found that increasing the number of months between soil application of sewage sludge and crop planting lessens ARG subsequently detected in crops (Rahube et al., 2014; Tien et al., 2017).
Unlike manure, irrigation water may have multiple sources of contamination: industrial, livestock and human sewage (Gekenidis et al., 2018). This may explain the higher number of observations of primary and secondary pathogen-phenotype combinations reported in irrigation water samples in our data synthesis. Both the irrigation delivery method and the level of microbial pollution of irrigation water sources can affect crop contamination and consumer risk (Carey et al., 2016; Jongman and Korsten, 2016; Guzman-Otazo et al., 2019). For example, sprinkle irrigation, which applies drops of water from an overhead sprinkler, poses a greater risk of microbial adulteration of edible plant components that are grown above ground due to direct surface exposure, than subsurface irrigation methods do (Jung et al., 2014). Contamination of water sources is also a problem where there is little oversight of disposal of untreated animal and human wastes, as illustrated in a recent study in rural eastern China that investigated E. coli harboring CTX-M genes isolated from environmental and agricultural samples. In this study, genetic relatedness analysis of isolates suggested vegetable contamination was attributable to irrigation water polluted with effluents from nearby swine farms (Zou et al., 2019).
Over half of reported food crop samples in studies included in this review were foods consumed raw or with minimal preparation (salad crops, culinary herbs, and fruit). Unlike animal-based foods, there is no required cooking process for these foods that would destroy potential pathogens and ARG. Reducing microbial contamination of such fresh produce remains an important food safety challenge (Murray et al., 2017). While washing and peeling may reduce the risk of surface contamination, resistance determinants can also be internalized from the surrounding soil through the roots of the plant via microbiota associated with plant tissues (endophytes) or enter plant tissues through the stem or open wounds (Yang et al., 2016; Zhang H. et al., 2017; Wei et al., 2020). In some cases, water used for washing may contribute to surface contamination when untreated or re-used wash water is applied to produce in the post-harvest stage (Richter et al., 2020). Understanding the effect of processing and preparation on ARB and ARG in plant-based foods would be valuable given indications that antimicrobial use may be greater than expected in certain crops such as rice (Taylor and Reeder, 2020).
Quantitative microbial risk assessment measures human exposure to microbial hazards and estimates risks associated with consumption of such hazards using dose-response data (FAO/WHO., 2015). Two quantitative human exposure assessments were included in the study set. Both studies evaluated consumer exposure to resistant E. coli through lettuce consumption and the influence of irrigation and post-harvest washing and processing measures on exposure risk (Njage and Buys, 2017; O'Flaherty et al., 2019). In both studies, consumer exposure was most influenced by microbial loads in irrigation water. This might be influenced by the limited effective post-harvest processing methods available to reduce microbial contamination of produce, particularly when contamination occurs as biofilms or through internalization of microbes in plant cells (Frankel et al., 2009). In addition, certain post-harvest steps in the value chain such as non-refrigerated storage conditions and unhygienic handling by value chain actors may lead to colony expansion of pre-harvest microbial contaminants and cross-contamination (Njage and Buys, 2017). Although data on dose-response is still missing, these exposure assessment models are a first step toward conducting quantitative microbial risk assessment.
In this review, 70% (136/196) of studies were conducted in LMICs. This greater degree of research intensity could reflect concerns that human health risks from AMR in the food crop value chain may be greatest in LMICs (Berthe et al., 2019; FAO and WHO, 2019). Most studies occurred in Asia and Africa and more than twice as many of these studies detected primary pathogen-phenotype combinations in food crop value chains relative to studies in other regions combined. Seven of the ten countries that contributed the largest number of studies to this review were LMICs. Notably, four of these – China, India, Nigeria, and Mexico are among the top ten most populous countries (UN, 2019), necessitating huge production of food crops. It is evident that China has invested in research on this topic with almost one quarter of all included studies conducted there. Overall, a greater number of studies sampled food crops in the post-harvest, or retail stages of the value chain, and this trend was particularly clear in studies from Asia. As suggested in a review on consumer perceptions of food safety in LMICs, this may be due to increasing consumer concern for retail food quality as well as interest in control options to reduce exposure applied post-harvest (Grace, 2015). In countries where environmental regulations and WASH standards are less enforced, mitigating consumer risks through post-exposure interventions may be an easier task than pre-harvest measures.
Studies in LMICs were five times more likely to report resistance to chloramphenicol than those in HICs. Chloramphenicol is an antimicrobial banned from food production in most countries due to non-dose related and potentially fatal adverse effects from residue exposure (EFSA CONTAM Panel, 2018). While resistance to this antimicrobial may result from use of other phenicol antimicrobials in animal agriculture or be caused by co-selection due to the presence of chloramphenicol resistance genes in transferable genetic elements carrying other resistance genes (Lu et al., 2020), some evidence also exists of continuing veterinary use of chloramphenicol in LMICs (McCubbin et al., 2021).
The evidence in this review points to contamination of food crops with medically important AMR and an attendant risk of exposure for consumers. However, a full understanding of the magnitude and impact of this risk is hampered by the lack of source attribution studies and an incomplete picture of the health consequences of single and continuous exposure events to resistance determinants through food crop consumption. In addition, there is limited and inconsistent data on practices that influence local exposure to ARBs, ranging from crop irrigation methods and regional veterinary antimicrobial usage in livestock and crops, through to food type, consumption quantities, and preparation preferences (e.g., raw or cooked). Nonetheless, recently shifting ideological and nutritional preferences globally have raised the profile of food crops and plant-food diets as a contributor to human AMR exposures. We identified four key knowledge gaps related for future research: (1) understanding the prevalence of AMR on food crops; (2) untangling the web of transmission of AMR determinants through the agro-environment; (3) defining and quantifying antimicrobial usage in the agro-environment; and (4) quantifying the burden of AMR infections in humans due to exposure to contaminated food crops.
Understanding the characteristics and prevalence of AMR in food crops is an important first step toward defining risk factors that bring the agro-environmental reservoir of AMR into contact with the human microbiome. While independently conducted cross-sectional studies can provide point prevalence estimates, various methodological biases may overrepresent the risk (discussed further in section Limitations) and surveys provide little information on temporal patterns. The Global Antimicrobial Resistance Surveillance System (GLASS) introduced in 2015 is an example of a cooperative global surveillance program for AMR in hospital infections (WHO, 2020), however, no such program for standardized data collection on AMR in food crop value chains yet exist, despite an increasingly interconnected global food industry. While recent revisions to the Codex Alimentarius Guidelines on Integrated Monitoring and Surveillance of Foodborne Antimicrobial Resistance recommend a risk-based approach to crop sampling at all levels of the production, detailed guidelines from international plant regulatory bodies such as the International Plant Protection Convention (IPPC) with respect to crop types, processing stage, and reporting of antimicrobial use and resistance data are still lacking (FAO/WHO, 2021).
Unexpected pathogen-AMR phenotype combinations in food crops such as colistin resistant K. pneumoniae suggest a need for a clearer understanding of sources and transmission pathways of AMR in human pathogens through food crop value chains. However, few studies used methods that permit source attribution analysis in the food crop value chain. Studies using advanced sequencing methods remained rare (n = 21) and just over half of all studies included in the study set (n = 105) used any genotypic methods (either solely or in addition to reporting phenotypic results). Whole Genome Sequencing (WGS) is increasingly being employed in food safety studies and as part of surveillance programs for foodborne diseases as it provides a highly discriminating method of identifying sequence characteristics of pathogens to enable their origins to be traced in the food value chain. For data on AMR occurrence in microbe populations, metagenomic sequencing provides a higher degree of confidence than conventional phenotypic and PCR-based genotypic methods and can be applied to monitor population changes through the food value chain, which may be useful for human exposure assessments (Oniciuc et al., 2018). For these reasons, various authors have proposed applying WGS and metagenomic sequencing into quantitative microbial risk assessments (Greig et al., 2015; Oniciuc et al., 2018; Collineau et al., 2019). Quantifying the burden of AMR in food crops may be a more achievable objective with a concerted effort to incorporate advanced sequencing into AMR studies on foods, however many LMICs lack the laboratory capacity and in-country expertise to implement these methods.
This review sought evidence of AMR through the food crop value chain to better understand the exposure hazard to consumers. Very few studies investigated or reported AMR development as a result of antimicrobial use against phytopathogens. Antimicrobial stewardship programs aimed at human and animal sectors may be less effective if indiscriminate use in crops is left unaddressed. There is limited data on use of antimicrobials in the crop sector and new approaches to gathering this information are needed as existing regulatory programs may be difficult and time-consuming to set up, particularly in LMICs. In this regard, an informal extension database called PlantWise provided a recent example of how data from an advisory program for crop producers in LMICs might be obtained and used as an indirect record of antimicrobial usage in this sector (Taylor and Reeder, 2020).
Finally, quantifying the burden of AMR infections in humans due to exposure to contaminated crops is a nascent field of research with just two quantitative microbial risk assessments included in the search results of this scoping review of the published literature. Challenges of risk modeling of AMR exposure through crops include various assumptions about survival of microbes harboring AMR on food crops; preparation and consumption habits in individuals and populations; and production methods for different crops in different regions (Njage and Buys, 2017). The knowledge gaps listed here include suggestions for improved data capture to underpin and refine assumptions currently used in exposure assessment and hazard characterization to improve quantitative microbial risk assessments and inform population-level mitigation options.
This review used a systematic approach to assess the published literature on ARB and ARG in food crops. Nevertheless, the methods of this review preclude us from validating specific transmission pathways of AMR from the crop environment to humans or comparing the levels of resistance present in different sources. For this reason, it is not possible to identify key risks of exposure of humans to AMR from food crops or to identify practices that could best mitigate these risks. Previous reviews on environmental sources of AMR concluded that research on the topic was predisposed to problematic study design, substantial bias, and a lack of standardization in methods, which challenges comparability and understanding (Greig et al., 2015; Bueno et al., 2017).
Some studies included in this review measured the proportion of samples in which resistance was found. However, we have been cautious not to present prevalence of AMR in our analysis because of evident selection bias, reporting bias, and potential publication bias. While most studies were observational in design, fewer than 15% of studies reported the use of probability-based sampling methods, which would enable proportions to be extrapolated beyond the sample population. Where random sampling methods were employed, prevalence estimates of resistant organisms in samples were rarely reported, with most authors preferring to report prevalence of resistance among isolates instead. Aggregation of phenotypic results in mixed groups of samples (e.g., meat and vegetables) excluded 23 studies from this review and several studies did not include information on the total amount of samples tested or provided full susceptibility results meaning that our analysis could only report on the presence of resistance, rather than prevalence. In addition, we cannot exclude the effect of publication bias, in which studies with resistance results are prioritized for publication and lead to overstated conclusions of presence of resistant organisms.
Finally, attention to AMR in crops as a risk to human health has focused on antibiotics and resistant bacteria, but fungal pathogens present a larger burden of disease to plants and the crop sector. Widespread control efforts make use of fungicides, contributing to selective pressure and resistance development to available antifungal therapy for plant protection, and also reducing the efficacy of antifungal drugs used in the treatment of human fungal infections, called the medical azoles (Gisi, 2014; Fisher et al., 2018). The low number of studies included in this review on antifungal resistance related to crop application of fungicides may be a result of search terms that focused on human health risks, which tend to favor bacterial pathogens.
This review contributes to the growing understanding on AMR of medical significance occurring in food crop value chains posing a potential food safety risk to consumers. Our results indicate that human pathogenic microbes on food crops harbor resistance traits that appear to reflect antimicrobial usage trends in animal agriculture and are particularly evident on food crops that are consumed raw or with minimal preparation. In contrast, bacteria resistant to antimicrobials of critical importance in human medicine, such as carbapenems and glycopeptides, appear more rarely in food crops in the published literature but point to the complexity of contamination pathways involving human and agricultural sources. The results from this review support the biological plausibility of medically important AMR exposure risks for consumers and workers from the food crop value chain and highlight the need to include food crops in national and global food safety surveillance programs related to AMR. However, as we have shown, incorporating AMR surveillance on food crops represents a considerable challenge because of the lack of standardized microbial indicators and types of AMR, inconsistent methods of measuring resistance and resistance determinants, diverse agricultural production systems with variable inputs and complex value chains with differing processing and distribution methods, and a vast number of edible crops produced and provided to consumers around the world. Finding comparable indicators to monitor AMR in food crops will require considerable and sustained input from various stakeholders but it is a necessary step for antimicrobial stewardship efforts.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
AB and JW conceptualized the study. AB, AMo, LG, PT, and AMa contributed to development of the study protocol. AB and ZK-A performed data extraction and data analysis. AB prepared the draft the manuscript, with support from ZK-A and JW. JW supervised the study. All authors reviewed the results and contributed to editing of the final manuscript.
This work was partially supported by the CGIAR Research Program on Agriculture for Nutrition and Health (A4NH).
The opinions expressed here belong to the authors, and do not necessarily reflect those of A4NH or CGIAR.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The authors wish to thank the advisors to this review for their insights and feedback on the protocol, analysis, and manuscript, including Clare Chandler (London School of Hygiene and Tropical Medicine); Angkana Legakul (London School of Hygiene and Tropical Medicine); Javier Mateo-Sagasta, and Mahesh Jampani (International Water Management Institute). In addition, we thank Manon Roustit for her assistance in data extraction and contributions to analysis.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.824714/full#supplementary-material
Alvarez-Moreno, C., Lavergne, R. A., Hagen, F., Morio, F., Meis, J. F., and Le Pape, P. (2019). Fungicide-driven alterations in azole-resistant Aspergillus fumigatus are related to vegetable crops in Colombia, South America. Mycologia 111, 217–224. doi: 10.1080/00275514.2018.1557796
Berger, S., El Chazli, Y., Babu, A. F., and Coste, A. T. (2017). Azole resistance in Aspergillus fumigatus: A consequence of antifungal use in agriculture? Front. Microbiol. 8, 1024. doi: 10.3389/fmicb.2017.01024
Berthe, F. C. J., Wadsworth, J., Thiebaud, A., Marquez, P. V., and Baris, E. (2019). Pulling Together to Beat Superbugs: Knowledge and Implementation Gaps in Addressing Antimicrobial Resistance. Washington D.C. Available online at: https://www.worldbank.org/en/topic/agriculture/publication/pulling-together-to-beat-superbugs-knowledge-and-implementation-gaps-in-addressing-antimicrobial-resistance (accessed June 30, 2021).
Bevan, E. R., Jones, A. M., and Hawkey, P. M. (2017). Global epidemiology of CTX-M β-lactamases: temporal and geographical shifts in genotype. J. Antimicrob. Chemother. 72, 2145–2155. doi: 10.1093/jac/dkx146
Blau, K., Jacquiod, S., Sørensen, S. J., Su, J.-Q., Zhu, Y.-G., Smalla, K., et al. (2019). Manure and doxycycline affect the bacterial community and its resistome in lettuce rhizosphere and bulk soil. Front. Microbiol. 10:725. doi: 10.3389/fmicb.2019.00725
Booton, R. D., Meeyai, A., Alhusein, N., Buller, H., Feil, E., Lambert, H., et al. (2021). One Health drivers of antibacterial resistance: quantifying the relative impacts of human, animal and environmental use and transmission. One Health 12:100220. doi: 10.1016/j.onehlt.2021.100220
Bueno, I., Williams-Nguyen, J., Hwang, H., Sargeant, J. M., Nault, A. J., and Singer, R. S. (2017). Impact of point sources on antibiotic resistance genes in the natural environment: a systematic review of the evidence. Anim. Health Res. Rev. 18, 112–127. doi: 10.1017/S146625231700007X
Caffrey, N., Invik, J., Waldner, C. L., Ramsay, D., and Checkley, S. L. (2019). Risk assessments evaluating foodborne antimicrobial resistance in humans: a scoping review. Microb. Risk Anal. 11, 31–46. doi: 10.1016/j.mran.2018.08.002
Campos, J., Mourão, J., Pestana, N., Peixe, L., Novais, C., and Antunes, P. (2013). Microbiological quality of ready-to-eat salads: an underestimated vehicle of bacteria and clinically relevant antibiotic resistance genes. Int. J. Food Microbiol. 166, 464–470. doi: 10.1016/j.ijfoodmicro.2013.08.005
Cantón, R., González-Alba, J. M., and Galán, J. C. (2012). CTX-M enzymes: origin and diffusion. Front. Microbiol. 3:110. doi: 10.3389/fmicb.2012.00110
Carey, S. A., Goldstein, R. E. R., Gibbs, S. G., Claye, E., Xin, H., and Sapkota, A. R. (2016). Occurrence of vancomycin-resistant and -susceptible Enterococcus spp. in reclaimed water used for spray irrigation. Environ. Res. 147, 350–355. doi: 10.1016/j.envres.2016.02.030
CDC (2014). Clinical Features of Aspergillosis. Available online at: https://www.cdc.gov/fungal/diseases/aspergillosis/symptoms/clinical-features.html (accessed October 30, 2021).
Chanseyha, C., Sadiq, M. B., Cho, T. Z. A., and Anal, A. K. (2018). Prevalence and analysis of antibiotic resistant genes in Escherichia coli and salmonella isolates from green leaf lettuce. Chiang Mai J. Sci. 45, 1274–1286. Available online at: https://www.thaiscience.info/Journals/Article/CMJS/10989379.pdf (accessed July 21, 2020).
Chatterjee, A., Modarai, M., Naylor, N. R., Boyd, S. E., Atun, R., Barlow, J., et al. (2018). Quantifying drivers of antibiotic resistance in humans: a systematic review. Lancet Infect. Dis. 18, e368–e378. doi: 10.1016/S1473-3099(18)30296-2
Collineau, L., Boerlin, P., Carson, C. A., Chapman, B., Fazil, A., Hetman, B., et al. (2019). Integrating whole-genome sequencing data into quantitative risk assessment of foodborne antimicrobial resistance: a review of opportunities and challenges. Front. Microbiol. 10:1107. doi: 10.3389/fmicb.2019.01107
Dias, C., Duarte-Ribeiro, F., Pipa, S., and Mota, M. (2018). A Rare and potentially catastrophic infection: primary intestinal aspergillosis—case report in an HIV patient. Case Rep. Infect. Dis. 2018:1–3. doi: 10.1155/2018/3269847
EFSA BIOHAZ Panel (European Food Safety Panel on Biological Hazards), Koutsoumanis, K., Allende, A., Alvarez-Ordonez, A., Bolton, D., Bover-Cid, S., et al. (2021). Role played by the environment in the emergence and spread of antimicrobial resistance (AMR) through the food chain. EFSA J. 19:e06651. doi: 10.2903/j.efsa.2021.6651
EFSA CONTAM Panel (2018). Scientific Opinion on Chloramphenicol in Food and Feed. Available online at: https://www.efsa.europa.eu/en/efsajournal/pub/3907 (accessed June 30, 2021).
Fahrenfeld, N., Ma, Y., O'Brien, M., and Pruden, A. (2013). Reclaimed water as a reservoir of antibiotic resistance genes: distribution system and irrigation implications. Front. Microbiol. 4:130. doi: 10.3389/fmicb.2013.00130
FAO (2016). Drivers, Dynamics, and Epidemiology of Antimicrobial Resistance in Animal Production. Available online at: https://doi.org/10.1016/S0305-417900134-X (accessed June 30, 2021).
FAO WHO (2019). Joint FAO/WHO Expert Meeting in collaboration with OIE on Foodborne Antimicrobial Resistance: Role of the Environment, Crops and Biocides – Meeting Report. Microbiological Risk Assessment Series no. 34. Rome. Available online at: https://www.who.int/publications/i/item/9789241516907 (accessed June 30, 2021).
FAO/WHO (2015). Codex Texts on Foodborne Antimicrobial Resistance. Available online at: http://www.fao.org/3/a-i4296t.pdf (accessed October 25, 2021).
FAO/WHO (2021). “Joint FAO/WHOFood Standards Programme: Guidelines on Integrated Monitoring and Surveillance of Foodborne Antimicrobial Resistance.” Agenda Item 6. Eighth Virtual Session. Available online at: https://www.fao.org/fao-who-codexalimentarius/meetings/detail/en/?meeting=TFAMR&session=8 (accessed October 25, 2021).
Fisher, M. C., Hawkins, N. J., Sanglard, D., and Gurr, S. J. (2018). Worldwide emergence of resistance to antifungal drugs challenges human health and food security. Science 360, 739–742. doi: 10.1126/science.aap7999
Frankel, G., Robert, K. S., David, P., Cedric, N. B., and Paul, H. (2009). Fresh Produce as a potential vector for bacterial human pathogens. Microbial Biotechnol. 2, 595–597. doi: 10.1111/j.1751-7915.2009.00137.x
Gao, F. Z., He, L. Y., He, L. X., Zou, H. Y., Zhang, M., Wu, D. L., et al. (2020). Untreated swine wastes changed antibiotic resistance and microbial community in the soils and impacted abundances of antibiotic resistance genes in the vegetables. Sci. Total Environ. 741:140482. doi: 10.1016/j.scitotenv.2020.140482
Gekenidis, M. T., Qi, W., Hummerjohann, J., Zbinden, R., Walsh, F., and Drissner, D. (2018). Antibiotic-resistant indicator bacteria in irrigation water: high prevalence of extended-spectrum beta-lactamase (ESBL)-producing Escherichia coli. PLoS ONE 13:e0207857. doi: 10.1371/journal.pone.0207857
Ghafur, A., Shankar, C., Gnanasoundari, P., Venkatesan, M., Mani, D., Thirunarayanan, M. A., et al. (2019). Detection of chromosomal and plasmid-mediated mechanisms of colistin resistance in Escherichia coli and Klebsiella pneumoniae from Indian food samples. J. Glob. Antimicrobial Resist. 16, 48–52. doi: 10.1016/j.jgar.2018.09.005
Gisi, U.. (2014). Assessment of selection and resistance risk for demethylation inhibitor fungicides in Aspergillus fumigatus in agriculture and medicine: a critical review. Pest Manag. Sci. 70, 352–364. doi: 10.1002/ps.3664
Gołȩbiewski, M., Kern-Zdanowicz, I., Zienkiewicz, M., Adamczyk, M., Zyliǹska, J., Baraniak, A., et al. (2007). Complete nucleotide sequence of the pCTX-M3 plasmid and its involvement in spread of the extended-spectrum β-lactamase gene blaCTX-M-3. Antimicrobial Agents Chemother. 51, 3789–3795. doi: 10.1128/AAC.00457-07
Grace, D.. (2015). Food safety in low and middle income countries. Int. J. Environ. Res. Public Health 12, 10490–10507. doi: 10.3390/ijerph120910490
Greig, J., Rajić, A., Young, I., Mascarenhas, M., Waddell, L., and Lejeune, J. (2015). A scoping review of the role of wildlife in the transmission of bacterial pathogens and antimicrobial resistance to the food chain. Zoonoses Public Health 62, 269–284. doi: 10.1111/zph.12147
Guzman-Otazo, J., Gonzales-Siles, L., Poma, V., Bengtsson-Palme, J., Thorell, K., Flach, C. F., et al. (2019). Diarrheal bacterial pathogens and multi-resistant enterobacteria in the Choqueyapu River in La Paz, Bolivia. PLoS ONE 14:e0210735. doi: 10.1371/journal.pone.0210735
Hiko, A., Abdata, K., Muktar, Y., Woyesa, M., and Mohammed, A. (2016). Contamination of Ethiopian paper currency notes from various food handlers with E. coli and antimicrobial resistance test. SpringerPlus 5:1065. doi: 10.1186/s40064-016-2742-z
Holvoet, K., Sampers, I., Callens, B., Dewulf, J., and Uyttendaele, M. (2013). Moderate prevalence of antimicrobial resistance in Escherichia coli isolates from lettuce, irrigation water, and soil. Appl. Environ. Microbiol. 79, 6677–6683. doi: 10.1128/AEM.01995-13
Hölzel, C. S., Tetens, J. L., and Schwaiger, K. (2018). Unraveling the role of vegetables in spreading antimicrobial-resistant bacteria: a need for quantitative risk assessment. Foodborne Pathog. Dis. 15, 671–688. doi: 10.1089/fpd.2018.2501
Huang, J., Mi, J., Yan, Q., Wen, X., Zhou, S., Wang, Y., et al. (2021). Animal manures application increases the abundances of antibiotic resistance genes in soil-lettuce system associated with shared bacterial distributions. Sci. Total Environ. 787:147667. doi: 10.1016/j.scitotenv.2021.147667
Jans, C., Sarno, E., Collineau, L., Meile, L., Stärk, K. D. C., and Stephan, R. (2018). Consumer exposure to antimicrobial resistant bacteria from food at Swiss retail level. Front. Microbiol. 9:362. doi: 10.3389/fmicb.2018.00362
Johnston, L. M., and Jaykus, L. A. (2004). Antimicrobial resistance of Enterococcus species isolated from produce. Appl. Environ. Microbiol. 70, 3133–3137. doi: 10.1128/AEM.70.5.3133-3137.2004
Jones-Dias, D., Carvalho, A. S., Moura, I. B., Manageiro, V., Igrejas, G., Caniça, M., et al. (2017). Quantitative proteome analysis of an antibiotic resistant Escherichia coli exposed to tetracycline reveals multiple affected metabolic and peptidoglycan processes. J. Proteomics 156, 20–28. doi: 10.1016/j.jprot.2016.12.017
Jongman, M., and Korsten, L. (2016). Genetic diversity and antibiotic resistance of Escherichia coli isolates from different leafy green production systems. J. Food Prot. 79, 1846–1853. doi: 10.4315/0362-028X.JFP-16-117
Jung, D., and Rubin, J. E. (2020). Identification of antimicrobial resistant bacteria from plant-based food products imported into Canada. Int. J. Food Microbiol. 319:108509. doi: 10.1016/j.ijfoodmicro.2020.108509
Jung, Y., Jang, H., and Matthew, K. R. (2014). Effect of the food production chain from farm practices to vegetable processing on outbreak incidence. Microb. Biotechnol. 7, 517–527. doi: 10.1111/1751-7915.12178
Kim, M. C., and Woo, G. J. (2017). Characterization of antimicrobial resistance and quinolone resistance factors in high-level ciprofloxacin-resistant Enterococcus faecalis and Enterococcus faecium isolates obtained from fresh produce and fecal samples of patients. J. Sci. Food Agric. 97, 2858–2864. doi: 10.1002/jsfa.8115
Lammie, S. L., and Hughes, J. M. (2016). Antimicrobial resistance, food safety, and one health: the need for convergence. Annu. Rev. Food Sci. Technol. 7, 287–312. doi: 10.1146/annurev-food-041715-033251
Li, J., Shao, B., Shen, J., Wang, S., and Wu, Y. (2013). Occurrence of chloramphenicol-resistance genes as environmental pollutants from swine feedlots. Environ. Sci. Technol. 47, 2892–2897. doi: 10.1021/es304616c
Li, Y., Sun, Q., Shen, Y., Zhang, Y., Yang, J., Shu, L., et al. (2018). Rapid increase in prevalence of carbapenem-resistant enterobacteriaceae (CRE) and Emergence of colistin resistance gene mcr-1 in CRE in a hospital in Henan, China. J. Clin. Microbiol. 56, e01932–e01917. doi: 10.1128/JCM.01932-17
Lima, T., Domingues, S., and Silva, G. J. D. (2020). Manure as a potential hotspot for antibiotic resistance dissemination by horizontal gene transfer events. Vet. Sci. 7:110. doi: 10.3390/vetsci7030110
Liu, Y. Y., Wang, Y., Walsh, T. R., Yi, L. X., Zhang, R., Spencer, J., et al. (2016). Emergence of plasmid-mediated colistin resistance mechanism MCR-1 in animals and human beings in China: a microbiological and molecular biological study. Lancet Infect. Dis. 16, 161–168. doi: 10.1016/S1473-3099(15)00424-7
Lu, W., Wang, M., Wu, J., Jiang, Q., Jin, J., Jin, Q., et al. (2020). Spread of chloramphenicol and tetracycline resistance genes by plasmid mobilization in agricultural soil. Environ. Pollut. 260:113998. doi: 10.1016/j.envpol.2020.113998
Lugo-Melchor, Y., Quiñones, B., Amézquita-López, B. A, León-Félix, J., García-Estrada, R., et al. (2010). Characterization of tetracycline resistance in Salmonella enterica strains recovered from irrigation water in the Culiacan Valley, Mexico. Microbial Drug Resist. 16, 185–190. doi: 10.1089/mdr.2010.0022
Manaia, C. M.. (2017). Assessing the risk of antibiotic resistance transmission from the environment to humans: non-direct proportionality between abundance and risk. Trends Microbiol. 25, 173–181. doi: 10.1016/j.tim.2016.11.014
Martínez, J. L., Coque, T. M, and Baquero, F. (2015). What is a resistance gene? Ranking risk in resistomes. Nat. Rev. Microbiol. 13, 116–123. doi: 10.1038/nrmicro3399
McCubbin, K. D, Ramatowski, J. W., Buregyeya, E., Hutchinson, E., Kaur, H., Mbonye, A. K, et al. (2021). Unsafe “crossover-use” of chloramphenicol in Uganda: importance of a One Health approach in antimicrobial resistance policy and regulatory action. J. Antibiotics. 74, 417–420. doi: 10.1038/s41429-021-00416-3
McManus, P. S.. (2014). Does a drop in the bucket make a splash? Assessing the impact of antibiotic use on plants. Curr. Opin. Microbiol. 19, 76–82. doi: 10.1016/j.mib.2014.05.013
Mellmann, A., Harmsen, D., Cummings, C. A., Zentz, E. B., Leopold, S. R, Rico, A., et al. (2011). Prospective genomic characterization of the german enterohemorrhagic Escherichia coli O104: H4 outbreak by rapid next generation sequencing technology. PLoS ONE 6:e22752. doi: 10.1371/journal.pone.0022751
Mills, M. C., and Lee, J. (2019). The threat of carbapenem-resistant bacteria in the environment: evidence of widespread contamination of reservoirs at a global scale. Environ. Pollut. 255:113143. doi: 10.1016/j.envpol.2019.113143
Murray, K., Wu, F., Shi, J., Jun Xue, S., and Warriner, K. (2017). Challenges in the microbiological food safety of fresh produce: limitations of post-harvest washing and the need for alternative interventions. Food Qual. Saf. 1, 289–301. doi: 10.1093/fqsafe/fyx027
Neuert, S., Nair, S., Day, M. R., Doumith, M., Ashton, P. M, Mellor, K. C., et al. (2018). Prediction of phenotypic antimicrobial resistance profiles from whole genome sequences of non-typhoidal Salmonella enterica. Front. Microbiol. 9, 1–11. doi: 10.3389/fmicb.2018.00592
Njage, P. M. K., and Buys, E. M. (2017). Quantitative assessment of human exposure to extended spectrum and AmpC β-lactamases bearing E. coli in lettuce attributable to irrigation water and subsequent horizontal gene transfer. Int. J. Food Microbiol. 240, 141–151. doi: 10.1016/j.ijfoodmicro.2016.10.011
O'Flaherty, E., Solimini, A. G., Pantanella, F., De Giusti, M., and Cummins, E. (2019). Human exposure to antibiotic resistant-Escherichia coli through irrigated lettuce. Environ. Int. 122, 270–280. doi: 10.1016/j.envint.2018.11.022
OIE (2020). Annual Report on Antimicrobial Agents Intended for Use in Animals. Fourth Report. Available online at: https://www.oie.int/app/uploads/2021/03/a-fourth-annual-report-amr.pdf (accessed July 10, 2021).
O'Neill, J.. (2016). Tackling Drug-Resistant Infections Globally: Final Report and Recommendations. London, UK: The Review on Antimicrobial Resistance. Available online at: https://amr-review.org/Publications.html (accessed June 30, 2021).
Oniciuc, E. A, Likotrafiti, E., Alvarez-Molina, A., Prieto, M., Santos, J. A, and Alvarez-Ordóñez, A. (2018). The present and future of whole genome sequencing (WGS) and whole metagenome sequencing (WMS) for surveillance of antimicrobial resistant microorganisms and antimicrobial resistance genes across the food chain. Genes 9, 1–28. doi: 10.3390/genes9050268
Pan, M., and Chu, L. M. (2016). Phytotoxicity of veterinary antibiotics to seed germination and root elongation of crops. Ecotoxicol. Environ. Saf. 126, 228–237. doi: 10.1016/j.ecoenv.2015.12.027
Park, S. H.. (2014). Third-generation cephalosporin resistance in gram-negative bacteria in the community: a growing public health concern. Korean J. Internal Med. 29, 27–30. doi: 10.3904/kjim.2014.29.1.27
Peters, M. D. J., Godfrey, C. M., Khalil, H., McInerney, P., Parker, D., and Soares, C. B. (2015). Guidance for conducting systematic scoping reviews. Int. J. Evid. Based Healthc. 13, 141–146. doi: 10.1097/XEB.0000000000000050
Pragasam, A. K., Shankar, C., Veeraraghavan, B., Biswas, I., Nabarro, L. E. B., Inbanathan, F. Y., et al. (2017). Molecular mechanisms of colistin resistance in klebsiella pneumoniae causing bacteremia from India -A first report. Front. Microbiol. 7:2135. doi: 10.3389/fmicb.2016.02135
Prigitano, A., Esposto, M. C., Roman, òL., Auxilia, F., and Tortorano, A. M. (2019). Azole-resistant Aspergillus fumigatus in the Italian environment. J. Global Antimicrob. Resist. 16, 220–224. doi: 10.1016/j.jgar.2018.10.017
Prigitano, A., Venier, V., Cogliati, M., De Lorenzis, G., Esposto, M. C, and Tortorano, A. M. (2014). Azole-resistant Aspergillus fumigatus in the environment of northern Italy, May 2011 to June 2012. Eurosurveillance 19:20747. doi: 10.2807/1560-7917.ES2014.19.12.20747
Rahube, T. O., Marti, R., Scott, A., Tien, Y. C., Murray, R., Sabourin, L., et al. (2014). Impact of fertilizing with raw or anaerobically digested sewage sludge on the abundance of antibiotic-resistant coliforms, antibiotic resistance genes, and pathogenic bacteria in soil and on vegetables at harvest. Appl. Environ. Microbiol. 80, 6898–6907. doi: 10.1128/AEM.02389-14
Ren, J., Jin, X., Zhang, Q., Zheng, Y., Lin, D., and Yu, Y. (2017). Fungicides induced triazole-resistance in Aspergillus fumigatus associated with mutations of TR46/Y121F/T289A and its appearance in agricultural fields. J. Hazard. Mater. 326, 54–60. doi: 10.1016/j.jhazmat.2016.12.013
Richter, L., du Plessis, E. M., Duvenage, S., and Korsten, L. (2020). Occurrence, phenotypic and molecular characterization of extended-spectrum- and AmpC- beta-lactamase producing Enterobacteriaceae isolated from selected commercial spinach supply chains in South Africa. Front. Microbiol. 11:638. doi: 10.3389/fmicb.2020.00638
Rodríguez, C., Lang, L., Wang, A,., Altendorf, K., García, F., Lipski, A., et al. (2006). Lettuce for human consumption collected in Costa Rica contains complex communities of culturable oxytetracycline- and gentamicin-resistant bacteria. Appl. Environ. Microbiol. 72, 5870–5876. doi: 10.1128/AEM.00963-06
Seiler, C., and Berendonk, T. U. (2012). Heavy metal driven co-selection of antibiotic resistance in soil and water bodies impacted by agriculture and aquaculture. Front. Microbiol. 3:399. doi: 10.3389/fmicb.2012.00399
Silva, B. N., Cadave, V., Teixeira, J. A., and Gonzales-Barron, U. (2017). Meta-analysis of the incidence of foodborne pathogens in vegetables and fruits from retail establishments in Europe. Curr. Opin. Food Sci. 18, 21–28. doi: 10.1016/j.cofs.2017.10.001
Singer, A. C., Shaw, H. V. R., and Hart, A. (2016). Review of antimicrobial resistance in the environment and its relevance to environmental regulators. Front. Microbiol. 7:1728. doi: 10.3389/fmicb.2016.01728
Sivapalasingam, S., Friedman, C. R., Cohen, L., and Tauxe, R. V. (2004). Fresh produce: a growing cause of outbreaks of foodborne illness in the United States, 1973 through 1997. J. Food Prot. 67, 2342–2353. doi: 10.4315/0362-028X-67.10.2342
Skov, R. L., and Monnet, D. L. (2016). Plasmid-mediated colistin resistance (mcr-1 gene): three months later, the story unfolds. Eurosurveillance 21, 1–6. doi: 10.2807/1560-7917.ES.2016.21.9.30155
Stockwell, V. O., and Duffy, B. (2012). Use of antibiotics in plant agriculture. OIE Revue Scientifique et Technique 31, 199–210. doi: 10.20506/rst.31.1.2104
Sundin, G. W.. (2000). Examination of base pair variants of the strA-strB streptomycin resistance genes from bacterial pathogens of humans, animals and plants. J. Antibiotics 46, 848–849. doi: 10.1093/jac/46.5.848
Tacconelli, E., Carrara, E., Savoldi, A., Harbarth, S., Mendelson, M., Monnet, D. L., et al. (2018). Discovery, research, and development of new antibiotics: the WHO priority list of antibiotic-resistant bacteria and tuberculosis. Lancet Infect. Dis. 18, 318–327. doi: 10.1016/S1473-3099(17)30753-3
Tauch, A., Krieft, S., Kalinowski, J., and Pühler, A. (2000). The 51,409-bp R-plasmid pTP10 from the multiresistant clinical isolate Corynebacterium striatum M82B is composed of DNA segments initially identified in soil bacteria and in plant, animal, and human pathogens. Mol. Gen. Genet. 263, 1–11. doi: 10.1007/PL00008668
Taylor, P., and Reeder, R. (2020). Antibiotic use on crops in low and middle-income countries based on recommendations made by agricultural advisors. CABI Agricul. Biosci. 1:1. doi: 10.1186/s43170-020-00001-y
Tien, Y. C., Li, B., Zhang, T., Scott, A., Murray, R., Sabourin, L., et al. (2017). Impact of dairy manure pre-application treatment on manure composition, soil dynamics of antibiotic resistance genes, and abundance of antibiotic-resistance genes on vegetables at harvest. Sci. Total Environ. 581–582, 32–39. doi: 10.1016/j.scitotenv.2016.12.138
Tricco, A. C., Lillie, E., Zarin, W., O'Brien, K. K., Colquhoun, H., Levac, D., et al. (2018). PRISMA extension for scoping reviews (PRISMA-ScR): checklist and explanation. Ann. Intern. Med. 169, 467–473. doi: 10.7326/M18-0850
UN (2019). UN(United Nations) Department of Economic and Social Affairs, Population Division. World Population Prospects 2019: Highlights (ST/ESA/SER.A/423). Available online at: https://population.un.org/wpp/Publications/Files/WPP2019_Highlights.pdf (accessed June 30, 2021).
Van Boeckel, T. P., Brower, C., Gilbert, M., Grenfell, B. T., Levin, S. A., Robinson, T. P., et al. (2015). Global trends in antimicrobial use in food animals. Proc. Natl. Acad. Sci. U. S. A.. 112, 5649–54. doi: 10.1073/pnas.1503141112
Van Boeckel, T. P., Pires, J., Silvester, R., Zhao, C., Song, J., Criscuolo, N. G., et al. (2019). Global trends in antimicrobial resistance in animals in low- and middle-income countries. Science 365:eaaw1944. doi: 10.1126/science.aaw1944
Van Bunnik, B. A. D., and Woolhouse, M. E. J. (2017). Modelling the impact of curtailing antibiotic usage in food animals on antibiotic resistance in humans. R. Soc. Open Sci. 4:161067. doi: 10.1098/rsos.161067
Veldman, K., Kant, A., Dierikx, C., van Essen-Zandbergen, A., Wit, B., and Mevius, D. (2014). Enterobacteriaceae resistant to third-generation cephalosporins and quinolones in fresh culinary herbs imported from Southeast Asia. Int. J. Food Microbiol. 177, 72–77. doi: 10.1016/j.ijfoodmicro.2014.02.014
Wei, H., Ding, S., Qiao, Z., Su, Y., and Xie, B. (2020). Insights into factors driving the transmission of antibiotic resistance from sludge compost-amended soil to vegetables under cadmium stress. Sci. Total Environ. 729:138990. doi: 10.1016/j.scitotenv.2020.138990
Wellcome (2020). The Global Response to AMR: Momentum, Success and Critical Gaps. Available online at: https://wellcome.org/reports/global-response-amr-momentum-success-and-critical-gaps (accessed June 30, 2021).
WHO (2019). Critically Important Antimicrobials for Human Medicine, 6th revision. Geneva: World Health Organization. Available online at: https://www.who.int/publications/i/item/9789241515528 (accessed March 24, 2021).
WHO (2020). Global Antimicrobial Resistance and Use Surveillance System (GLASS) Report: Early Implementation 2020. Available online at: http://www.who.int/glass/resources/publications/early-implementation-report-2020/en/ (accessed March 24, 2021).
World Bank (2020). Country and Lending Groups, The World Bank Website. Available online at: https://datahelpdesk.worldbank.org/knowledgebase/articles/906519-world-bank-country-and-lending-groups (accessed March 24, 2021).
Xiong, L., Sun, Y., Shi, L., and Yan, H. (2019). Characterization of antimicrobial resistance genes and class 1 integrase gene in raw meat and aquatic product, fresh vegetable and fruit, and swine manure in southern China. Food Control 104, 240–246. doi: 10.1016/j.foodcont.2019.05.004
Xu, Y., Zhu, X. F., Zhou, M. G., Kuang, J., Zhang, Y., Shang, Y., et al. (2010). Status of streptomycin resistance development in xanthomonas oryzae pv. oryzae and xanthomonas oryzae pv. oryzicola in China and their resistance characters. J. Phytopathol. 158, 601–608. doi: 10.1111/j.1439-0434.2009.01657.x
Yang, F., Shen, C., Zheng, X., Liu, Y., Ahmed, M., Zhao, Z., et al. (2019). Plasmid-mediated colistin resistance gene mcr-1 in Escherichia coli and Klebsiella pneumoniae isolated from market retail fruits in Guangzhou, China. Infect. Drug Resist. 12, 385–389. doi: 10.2147/IDR.S194635
Yang, Q., Zhang, H., Guo, Y., and Tian, T. (2016). Influence of chicken manure fertilization on antibiotic-resistant bacteria in soil and the endophytic bacteria of pakchoi. Int. J. Environ. Res. Public Health 13:662. doi: 10.3390/ijerph13070662
Ye, Q., Wu, Q., Zhang, S., Zhang, J., Yang, G., Wang, J., et al. (2018). Characterization of extended-spectrum β-lactamase-producing Enterobacteriaceae from retail food in China. Front. Microbiol. 9:1709. doi: 10.3389/fmicb.2018.01709
You, Y., and Silbergeld, E. K. (2014). Learning from agriculture: understanding low-dose antimicrobials as drivers of resistome expansion. Front. Microbiol. 5:284. doi: 10.3389/fmicb.2014.00284
Zemelman, R., Bello, H., Dominguez, M., Gonzalez, G., Mella, S., and Garcia, A. (1993). Activity of imipenem, third-generation cephalosporins, aztreonam and ciprofloxacin against multi-resistant gram-negative bacilli isolated from chilean hospitals. J. Antimicrob. Chemother. 32, 413–419. doi: 10.1093/jac/32.3.413
Zhang, H., Li, X., Yang, Q., Sun, L., Yang, X., Zhou, M., et al. (2017). Plant growth, antibiotic uptake, and prevalence of antibiotic resistance in an endophytic system of pakchoi under antibiotic exposure. Int. J. Environ. Res. Public Health 14:1336. doi: 10.3390/ijerph14111336
Zhang, T., Toepfer, S., Wang, B., Peng, H, Luo, H., Wan, X., et al. (2017). Is business linkage affecting agricultural advisory services? Int. J. Agric. Extension 05, 59–77. Available online at: https://www.plantwise.org/wp-content/uploads/sites/4/2019/03/Zhang-Et-Al-2016-Business-Linkage-Agriextensio-Intjagriext.pdf (accessed August 15, 2021).
Zhuang, M., Achmon, Y., Cao, Y., Liang, X., Chen, L., Wang, H., et al. (2021). Distribution of antibiotic resistance genes in the environment. Environ. Pollut. 285:117402. doi: 10.1016/j.envpol.2021.117402
Zou, H., Zheng, B., Sun, M., Ottoson, J., Li, Y., Berglund, B., et al. (2019). Evaluating dissemination mechanisms of antibiotic-resistant bacteria in rural environments in China by using CTX-M-producing Escherichia coli as an indicator. Microb. Drug Resist. 25, 975–984. doi: 10.1089/mdr.2018.0431
Zurfuh, K., Poirel, L., Nordmann, P., Nüesch-Inderbinen, M., Hächler, H., and Stephan, R. (2016). Occurrence of the plasmid-borne mcr-1 colistin resistance gene in extended-spectrum-β-lactamase-producing enterobacteriaceae in river water and imported vegetable samples in Switzerland. Antimicrob. Agents Chemother. 60, 2594–2595. doi: 10.1128/AAC.00066-16
Keywords: food safety, antimicrobial resistance, food crops, Asia, Africa
Citation: Brunn A, Kadri-Alabi Z, Moodley A, Guardabassi L, Taylor P, Mateus A and Waage J (2022) Characteristics and Global Occurrence of Human Pathogens Harboring Antimicrobial Resistance in Food Crops: A Scoping Review. Front. Sustain. Food Syst. 6:824714. doi: 10.3389/fsufs.2022.824714
Received: 29 November 2021; Accepted: 03 January 2022;
Published: 03 February 2022.
Edited by:
Arun K. Bhunia, Purdue University, United StatesReviewed by:
Yanhong Liu, United States Department of Agriculture (USDA), United StatesCopyright © 2022 Brunn, Kadri-Alabi, Moodley, Guardabassi, Taylor, Mateus and Waage. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Jeff Waage, amVmZi53YWFnZUBsc2h0bS5hYy51aw==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.