- 1School of Horticulture and Landscape, Yangzhou University, Yangzhou, China
- 2State Key Laboratory of Vegetable Germplasm Innovation, Tianjin, China
Horticultural crops are susceptible to various biotic stressors including fungi, oomycetes, bacteria, viruses, and root-knot nematodes. These pathogens limit the growth, development, yield, and quality of horticultural crops, and also limit their adaptability and geographic distribution. The continuous cropping model in horticultural facilities exacerbates soil-borne diseases, and severely restricts yield, quality, and productivity. Recent progress in the understanding of mechanisms that confer tolerance to different diseases through innovative strategies including host-induced gene silencing (HIGS), targeting susceptibility genes, and rootstocks grafting applications are reviewed to systematically explore the resistance mechanisms against horticultural plant diseases. Future work should successfully breed resistant varieties using these strategies combined with molecular biologic methods.
Introduction
Horticultural crops, including vegetables, fruits, and ornamentals, provide nutrients, biologically active substances, and aesthetic values. Additionally, they are an integral part of economies and contribute significantly to the agricultural production (Shipman et al., 2021). However, there are various challenges to the growth, production, and processing of horticultural crops. Diseases caused by fungi, oomycetes, bacteria, viruses, and root-knot nematodes often lead to crops yield reductions, quality deterioration, and post-harvest loss (Zhang et al., 2014; Wang et al., 2015; Zhang M. et al., 2021). Huge efforts have been made to create disease-resistant cultivars by the traditional breeding, however, this method is limited because of unavailable natural resistance sources. Consequently, using conventional approaches to breed resistant cultivars is still a big challenge. To reduce the negative effects of diseases in horticultural crops, alternative strategies for generating disease-resistant varieties and searching environmentally-friendly control of plant diseases are urgent requirements.
In this review, we summarize the progress in developing plant tolerance to disease with a focus on three aspects: host-induced gene silencing (HIGS) technology, targeting susceptibility (S) genes, and rootstocks grafting applications. Each approach is based on the in-depth study of mechanisms underlying plant defense responses. Finally, we discuss the current challenges and future research directions, aiming to offer a reference and recommendations for further research on horticultural crops' diseases resistance.
Host-induced gene silencing (HIGS) technology
HIGS mechanisms
HIGS is an RNA interference (RNAi)-based technology that silences virulence genes in pathogens, virus, or insects by expressing double-stranded RNAs or hairpin RNAs in host plants that are complementary to essential genes, thereby conferring protection to engineered plants from infection (Govindarajulu et al., 2015; Cai et al., 2018; Koch and Wassenegger, 2021; Zand Karimi and Innes, 2022). In detail, RNAi is a conserved part of post-transcriptional gene-regulation, which has served as a available and powerful genetic tool to accelerate the research in plant biotechnology and to develop the potentially useful agronomical traits (Huang et al., 2006; Baum et al., 2007; Mao et al., 2007). Dicer, an RNaseIII-like enzyme, which can split a precursor dsRNA into small-interfering RNA or microRNA duplexes to make the gene post-transcriptional silencing (Tinoco et al., 2010; Govindarajulu et al., 2015). The double-stranded siRNAs generate from the an RNA-induced silencing complex, which contains an argonaute protein including an endonucleolytic activity for cleavage of target RNAs and a small RNA-binding domain (Ketting, 2011). Subsequently, the activated RNA-induced silencing complex loosens the siRNAs in an ATP-dependent reaction, and produces an anti-sense strand targeting complementary mRNA transcripts through base-pairing interactions for degradation of the targeted mRNA to inhibit protein translation (Hamilton and Baulcombe, 1999; Baulcombe, 2004). This technology has been proven to be a useful tool for investigating the functions of candidate pathogenic genes in pathogens and creating transgenic crops to better control diseases (Nunes and Dean, 2012).
HIGS target pathogenic genes for different diseases resistance
Recently, an increasing number of HIGS products or studies have been developed and become an effective strategy to control pathogen infections (Table 1). Cotton Verticillium wilt serves as a seriously soil-borne disease and is caused by the genus Verticillium, which makes negative impacts for a wide range of plants and is also a constant threat to agriculture worldwide. HIGS targeting pathogenic genes in Virticillium dahliae such as hygrophobins1 (VdH1) (Zhang et al., 2016a), G protein signaling (RGS1) (Xu et al., 2018), and acetolactate synthases (VdILV2 and VdILV6) (Wei et al., 2020) effectively control Verticillium wilt. Also, a independent study has shown that cotton plants can transfer microRNAs (miR166 and miR159) into V. dahliae pathogens to target corresponding virulence genes Clp-1 (Ca2+-dependent cysteine protease) and HiC-15 (isotrichodermin C-15 hydroxylase) to confer disease resistance (Zhang et al., 2016b). Furthermore, targeting pathogenic genes (Ave1, Sge1, and NLP1) of the invaded V. dahliae in tomato and Arabidopsis thaliana can be used to generate Verticillium wilt resistance (Song and Thomma, 2018). Powdery mildew (PM) fungi (Blumeria graminis) are obligate biotrophic pathogens that cause damage in thousands of plant species including wheat (Triticum aestivum) and barley (Hordeum vulgare). HIGS has been exploited to silence effector genes in B. graminis, which results in reduced fungal development and enhances resistance to PM (Nowara et al., 2010; Pliego et al., 2013). Rust fungi are caused by the Puccinia striiformis f. sp. tritici or P. graminis f. sp. Tritici, which leads to the devastating diseases in wheat or other cereal species globally. HIGS of essential pathogenic genes in the invaded fungi has also shown promise for the engineering of resistance in many host plants (Yin et al., 2011; Zhang et al., 2012; Panwar et al., 2013). A independent result shows that CRISPR-Cas9 disruption of TaPsIPK1, a wheat receptor-like cytoplasmic kinase gene, leads to immune priming without constitutive activation of defense responses and confers durable and broad-spectrum resistance against Pst without affecting important agronomic traits (Wang et al., 2022). The lettuce downy mildew (DM) is caused by a biotrophic oomycete (Bremia lactucae), which is the most important disease of lettuce worldwide. HIGS of virulence genes HAM34 and CES1 results in greatly reduced growth and sporulation of B. lactucae, and effective control of DM in lettuce (Govindarajulu et al., 2015). In addition, transgenic rice targeting MoAP1 via expressing RNA hairpins can highly enhance resistance to 11 tested M. oryzae strains (Guo et al., 2019).
Fusarium serves as a genus of filamentous fungi containing many different plant pathogens resulted in various devastating diseases including Fusarium wilt. HIGS technology has also been shown to be used for preventing Fusarium species pathogens in different crops (Koch et al., 2013; Chauhan and Rajam, 2022). The fungal CYP51 encodes the cytochrome P450 lanosterol C-14α-demethylase and plays essential functions for fungal growth, ergosterol biosynthesis and pathogenicity. Targeting of CYP51 genes obtain high efficiency of fungal inhibition to different Fusarium species pathogens in vitro and in planta through spray applications (spray-induced gene silencing, SIGS) or HIGS methods (Koch et al., 2013, 2019; He et al., 2019). Also, Fusarium head blight (FHB) is a serious disease in wheat, barley, and maize, which is caused by the F. graminearum and F. culmorum species and results in annual yield losses from high disease pressure. HIGS of the β-1, 3-glucan synthase gene FcGls1 in the invaded fungi highly enhance FHB resistance in the plant spike and leaf inoculation assays (Chen et al., 2016).
Tomato Fusarium wilt serves as primarily vascular disease, which is caused by the Fusarium oxysporum f. sp. lycopersici (Fol) and leads to the high disease pressure and yield losses. The fasciclin-like proteins (FLPs) in the Fol pathogen play important roles in the cell-to-cell adhesions and signaling cascade, and HIGS targeting the pathogenic genes (FoFLP1, FoFLP3, FoFLP4, and FoFLP5) shows the reduction of spore count and germination frequency, and disease symptoms in the infected plants (Chauhan and Rajam, 2022). Currently, banana cultivation is also seriously threatened by Fusarium wilt caused by F. oxysporum f. sp. cubense (Foc), and HIGS of two ergosterol biosynthetic genes ERG6/ERG11 in the invaded Foc results in highly resistance to Fusarium wilt in banana (Dou et al., 2020).
The above studies using HIGS technology not only provide reference for horticultural crops to generate disease resistance, but also obtain a theoretical foundation for developing double stranded RNA fungicides to control crop fungal diseases. Accordingly, the molecular mechanisms that underlie HIGS technologies have been defined the specificity, stability, and durability required for future field applications. HIGS strategies will serve as a genetic protection against pathogens applicable to highly disease-susceptible horticultural crops.
Susceptibility genes and application
Susceptibility genes and CRISPR-Cas9 technology
Pathogens cause a huge threat to crop quality and productivity, and it is worse that most of plants or crops don't pose resistance to diseases. S genes exist in susceptible crop varieties and are required for successful pathogens infection. Typically, pathogens use the host plants' S genes to accelerate their invading and proliferation. On the basis of the plant-pathogen interactions, S genes mainly have three molecular mechanisms. Firstly, the basic compatibility assists the pathogens recognition and penetration in hosts; secondly, the sustained compatibility promotes pathogens proliferation and spread; and thirdly, these genes can negatively regulate immune signals (van Schie and Takken, 2014). S-gene-mediated defense responses involves the underlying target S genes to confer typically broad-spectrum and durable disease resistance to the invaded pathogens (Zaidi et al., 2018). Current reviews have described the different technologies to analyze molecular mechanisms or applications of S genes (Lapin and Van den Ackerveken, 2013).
Though developed recently, many new breeding techniques associated with genetic engineering are used to successfully generate and commercialize high-yield and durable disease-resistant crop varieties. Recently, several plant breeding technologies have been developed and exploit the high-throughput genotyping and phenotyping methods to establish gene editing and speed-breeding platforms (Li et al., 2018; Watson et al., 2018), including the latest tools, such as CRISPR–Cas9 system. This technology can easily target interesting genes in different plant species containing Arabidopsis, tobacco and rice (Jiang et al., 2013), wheat (Wang et al., 2014), and maize (Char et al., 2017), to introduce agronomically important traits including disease resistance (Wang et al., 2014; Zaidi et al., 2016a). Targeting S genes via CRISPR system has been successfully and widely applied because of its high specificity, greater efficacy, and also it can be designed and implemented easily.
Targeting S genes for plant disease resistance
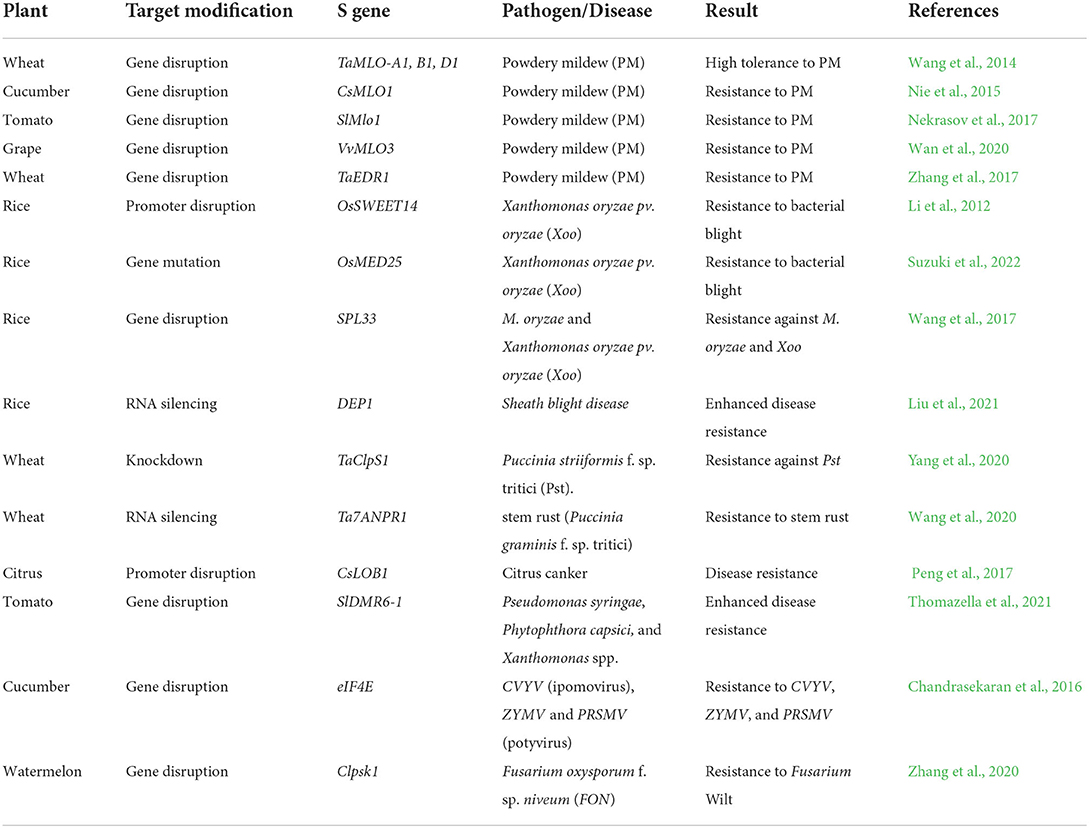
Recent studies have demonstrated S genes to be effective strategies to obtain resistance against various pathogens including viruses (Baltes et al., 2015; Chandrasekaran et al., 2016; Zaidi et al., 2016b; Aman et al., 2018), bacteria (Peng et al., 2017), and fungi (Shan et al., 2013; Wang et al., 2014). Disruption of S genes can confer broad-spectrum disease resistance and this technology has been applied in many economically important plant species (Table 2). Mildew resistance locus O (Mlo) encodes a transmembrane protein containing seven trans-membrane domains. Mlo is a well-known S gene and is conserved in monocots and dicots plants, which always serves as a typical and prominent example in durable pathogen-resistance programs. Targeting Mlo with CRISPR–Cas9 system has continued to confer resistance of powdery mildew (PM) in wheat (Wang et al., 2014), cucumber (Nie et al., 2015), tomato (Nekrasov et al., 2017), and grapevine (Wan et al., 2020). Also, wheat Tamlo-R32 mutant with a 304-kilobase pair targeted deletion in MLO-B1 locus that confers broad-spectrum and durable robust powdery mildew resistance and retains crop growth and yields (Li et al., 2022). Another PM-susceptibility locus is named as EDR1 encoding a Raf-like mitogen-activated protein, which significantly enhances the PM resistance via targeted the S gene by CRISPR–Cas9 technology in wheat (Zhang et al., 2017). Watermelon (Citrullus lanatus) wilt is one of the most devastating diseases, which is caused by Fusarium oxysporum f. sp. niveum (FON) and affects watermelon quality and yields in the world. Clpsk1 gene, encodes the phytosulfokine (PSK) precursor attenuated plant immune response, and knockout of Clpsk1 by CRISPR/Cas9 system confers highly resistance to FON in watermelon (Zhang et al., 2020).
Many S genes have also been identified and applied in disease resistance in the rice. The bacterial blight caused by the Xanthomonas oryzae pv. oryzae (Xoo), which can transfer sugars from the plant cell to the apoplast meeting pathogen's nutritional needs through several endogenous transactivator-like effectors (TALEs). A sugar transporter, OsSWEET14, is a typical S gene of Xoo strains targeted by four different transcription activator-like (TAL) effectors. Utilized genome editing technology can successfully target OsSWEET14 disruption and significantly enhance resistance against Xoo (Li et al., 2012). A subunit of the mediator multiprotein complex, OsMED25, which serves as a important adaptor between transcription factors (TFs) and RNA polymerase II, and plays crucial roles in lateral root development mediated by jasmonic acid (JA) in rice. Loss of function OsMED25 mutants exhibit high resistance to Xoo through regulating JA- and auxin-signaling (Suzuki et al., 2022). Another Xoo-susceptibility locus, SPL33, encoding a eEF1A-like protein, which is used to trigger broad-spectrum and durable resistance against different pathogens including Xoo and M. oryzae (Wang et al., 2017). Additionally, knockdown of G-protein γ subunit DEP1, which is a key player in the transmission of extracellular signals via membrane-spanning G-protein-coupled receptors to intracellular effectors, highly enhance resistance against sheath blight disease in rice (Liu et al., 2021).
Furthermore, wheat stem rust serves as a devastating disease and is caused by Puccinia graminis f. sp. tritici, which can lead to the estimated annual losses of US $1 billion to wheat production worldwide (http://www.usda.gov/nass). Two independent results have shown that targeting TaClpS1 or Ta7ANPR1 can trigger resistance to stem rust in wheat (Wang et al., 2020; Yang et al., 2020). Citrus canker is one of the most destructive diseases caused by Xanthomonas citri subsp.citri (Xcc), which affects citrus crop production and results in severe yield losses (Stover et al., 2014). LATERAL ORGAN BOUNDARIES 1 (CsLOB1), a typical S gene for citrus canker, which can promote pathogen growth and erumpent pustule formation. And loss-functions of CsLOB1 with CRISPR–Cas9 technology highly enhance resistance against citrus canker (Peng et al., 2017). Another susceptibility locus, DMR6, has also been used to confer broad-spectrum and durable resistance against fungal, oomycete, and bacterial pathogens in tomato (Thomazella et al., 2021). Moreover, apart from fungal and bacterial pathogens, the S genes have also been used to trigger immunity against different viral pathogens. A cap-binding protein, eukaryotic translation initiation factor 4E (eIF4E), serves as essential roles in the cellular infection cycle of different viruses, and a study has shown that targeting eIF4Es through the CRISPR–Cas9 method highly increase viruses resistance in cucumber (Chandrasekaran et al., 2016).
As most of horticultural crops resources are susceptible to different diseases, identifying and targeting S genes to develop resistant crop varieties is a continuous strategy to meet urgent requirements. The genome-edited horticultural crops in various studies have also been achieved transgene free, and this protection approach can be applicable to highly susceptible varieties.
Grafting in disease resistance
Grafting technology
Grafting is a traditional technique and widely implemented in modern agriculture to control soil-borne diseases caused by bacteria, fungi, oomycetes, viruses or root-knot nematodes of different crops in a sustainable and environmentally friendly approach (Nawaz et al., 2016; Li and Chen, 2017; Li and Zhao, 2021; Thies, 2021). Briefly, this technology is commonly accomplished by connecting two plant segments, the upper one named the scion and the lower part known as the rootstock. Successful grafts depend on an anatomical connector that surgically joins the rootstock and scion, and creates a dual plant system expressing superior traits on either half of the junction. Grafting technology acts as a disease management tactic that has been rapidly expanded to the horticultural crops including solanaceous and cucurbit fruiting vegetables. A good rootstock/scion combination forms a robust root system to guarantee nutrient transport and resistance to deal with different stresses (Louws et al., 2010). Additionally, grafting has also been associated with crop performance, yield, fruit quality, and nutritional value required by farmers and consumers (Kyriacou and Rouphael, 2018), and provides advances to control abiotic stresses, to reduce the using of chemical and fertilizer (Rouphael et al., 2008; Proietti et al., 2010). Currently, grafting has been used to explore research into the disease-resistant mechanisms by studying the functions of transmissible signals including genes, RNAs, proteins, hormones, and metabolites between rootstock and scions (Harada, 2010; Goldschmidt, 2014; Xu et al., 2022).
Applications of rootstocks grafting to manage soil-borne diseases
Plant grafting appears to be the effective and sustainable methods to control the soil-borne pathogens including fungi, bacteria, viruses, and nematodes. The resistant rootstocks mainly contain intra-specific (within the same species), inter-specific (different species), and inter-generic (different genera). These selective rootstocks always have resistant genes or with non-host resistance mechanisms, seem to be an important and typical weapon to manage against soil-borne diseases (Louws et al., 2010). For example, root-knot nematodes (RKN) is one of the most important limiting soil-borne diseases for vegetable production (Hallmann and Meressa, 2018), which is caused by the most damaging species Meloidogyne spp., and have a wide range of host plants (Greco and DiVito, 2009; Jones et al., 2013).
RKN are obligate endoparasitic nematodes, which move among the soil particles, and subsequently, penetrate near the elongation zone of the host roots. Root parenchyma cells can form hypertrophy, hyperplasia, and affect the water and nutrients uptake in the infected host plants, and results in pathogenetic symptoms including dwarfism, wilting, nutrient deficiency, and plant death. Many vegetable and fruit crops containing tomato (Solanum lycopersicum), eggplant (Solanum melongena), cucumber (Cucumis sativus), and watermelon (Citrullus lanatus) have potential for improving pathogen resistance through grafting technology (Kokalis-Burelle and Rosskopf, 2011). In tomato, the family Solanaceae or hybrids have typically been used to select the nematode resistant rootstocks for grafting. The muskmelons (Cucumis melo) has not yet identified RKN resistance. However, the hybrids of C. melo and C. metuliferus have shown potential as rootstocks posing the root-knot nematode resistant, which are used to graft onto commercial musk melon cultivars (Sigüenza et al., 2005; Kubota et al., 2008). In addition, the hybrids C. ficifolius × C. myriocarpus and C. ficifolius × C. anguria are tolerant to RKN (M. cuscannonballus) and resistant to Fusarium oxysporum f. sp. melon. When grafted, they preserve the different quality in the melon fruit compared to self-grafted or non-grafted plants (Cáceres et al., 2017). The wild watermelon (Citrullus lanatus var. citroides) germplasm and commercial watermelon rootstock (C. lanatus) have significantly less galling than the diploid seeded watermelon “Fiesta,” bottle gourd rootstocks and the Cucurbita moschata × C. maxima squash hybrid rootstock (Thies et al., 2008), which may be useful as candidate rootstocks for overcoming the watermelon RKN (Thies et al., 2010). These results will provide a number of alternative rootstocks that are available for different growers in the near future.
Fusarium species are common soil-borne pathogens, which can persist in soils, straw, and seeds for decades by colonizing alternate hosts leading to long-term disease cycles (Martínez et al., 2003). Fusarium, is well-studied and rootstocks resistant to Fusarium are widely available. Therefore, Fusarium can be managed through grafting. In general, the Fusarium causes wilt through recognizing host roots, penetrating, and colonizing the vascular tissue slowly, which provides an advantage for severe rootstocks or scion/rootstock combinations that reduces water stress. Therefore, the grafting practices with available resistance of rootstocks have provided successes to effectively manage Fusarium pathogens in multiple crops.
Many studies have shown that use of rootstocks with resistance to Fusarium wilt have been successfully obtained in vegetable annual crops through grafting. Globally, watermelon Fusarium wilt is the most production-limiting disease, which is caused by the pathogen Fusarium oxysporum f. sp. niveum (FON) and contains four races, designated 0, 1, 2, and 3 (Zhang et al., 2015). Watermelon seedlings grafted onto the bottle gourd rootstocks are highly resistant to FON compared with self-grafted watermelon crops (Huh et al., 2002; Zhang Z. Q. et al., 2021), and also facilitate to increase fruit quality and total yield (Davis et al., 2008). Additionally, bottle gourd [Lagenaria siceraria (Molina) Standl.] and inter-specific hybrid squash (Cucurbita maxima Duch. ex Lam. × C. moschata Duch. ex Poir) rootstocks-grafted plants are highly resistant to Fusarium wilt caused by FON races 1 and 2 (Davis et al., 2008; Keinath and Hassell, 2014a,b). Furthermore, in Turkey, grafted a diploid watermelon cultivar “Crimson Tide” is highly resistant to FON race 1, however, bottle gourd grafted watermelon can control the unidentified FON races including FON race 2 in soil and increase yields (Yetisir et al., 2003). In Spain, the interspecific hybrid squash “Shintoza” rootstock grafted triploid watermelon can overcome against unidentified FON races and increase yields by over 3-fold (Miguel et al., 2004). Thus, grafting is an effective strategy to overcome different FON races present or predominates in a field.
Melon (Cucumis melo L.) Fusarium wilt is a devastating soil-borne disease caused by the Fusarium oxysporum f. sp. melonis (FOM), which heavily affects melon cultivation and production. A study finds among a panel of 65 melon germplasm lines, “K134068,” “K133069,” “Wondae,” and “PI414723,” shows increased resistance to FOM. The resistant rootstocks grafted “Earl's elite” (Muskmelon) found that the yield, quality, and FOM-resistance are better than those of non-grafted melons (Dong et al., 2013). Additional rootstock species are available for melon grafting include wax gourd (Benincasa hispida) and pumpkin (Cucurbita spp.) (Traka-Mavrona et al., 2000). It is reported that the Cucurbita rootstock grafted melon can affect the plant growth (Ruiz and Romero, 1999), fruit quality, yield, and Fusarium wilt phenotype (Ruiz et al., 1997; Traka-Mavrona et al., 2000; Nisini et al., 2002).
Cucumber FW is also one of most destructive soil-borne disease, and the continuous cropping of cucumber in the horticultural facilities causes the FW to occur frequently, and severely restricts the high-yield, high-quality and high efficient cultivation of cucumber. Currently, grafting cucumber onto rootstocks is the most effective and sustainable technique to prevent FW (Reddy, 2016; Shi et al., 2016). Five different rootstocks of cucurbits including Super Shintoza, Bottle gourd, VSS-61F1, Cobalt, and Ferro have been used evaluate the resistance to Fusarium wilt under high-temperature stress conditions. Among them, the VSS-61 F1 rootstock has a high grafting efficiency, high compatibility between the rootstock and the scion, and also provides high resistance against Fusarium wilt (Shalaby et al., 2022). Similar results have been confirmed that grafted cucumber can form carbohydrates and lignin deposites as protective substances such as necrotic layer to prevent Fusarium invasion (Sabry et al., 2022). The inter-specific F1 hyprid of Cucurbita maxima × C. moschata has been widely untilized as rootstock species for grafting cucumbers around the world (Lee and Oda, 2003). To date, although the vast range of disease-susceptible cucumber scion–rootstock interactions have been analyzed, the response of grafted seedlings to pathogens or disease-resistant mechanisms are still difficult to investigate (Leonardi and Romano, 2004; Al-Debei et al., 2012). Also, many studies have shown that the pumpkin rootstocks have been used to identify candidate resistant genes to defend Fusarium wilt for increasing cucumber yields and profits (Xu et al., 2022).
Currently, more and more horticultural plants have been grafted onto the compatible disease-resistant rootstocks to manage against soil-borne diseases or abiotic stress (Huang et al., 2015). The available rootstocks, grafting compatibilies, and methods are important factors for increasing grafting success rates, influence the corp quality and yield, and overcoming soilborne pathogens. The present results indicated that disease-susceptible horticultural plants grafting onto suitable rootstocks can significantly reduce the disease incidence and increase crop yield and quality.
Challenges and perspectives
Although large efforts have also been made to generate disease-resistant horticultural cultivars via traditional breeding, many pathogens have still not been effectively and sustainably controlled. The lack of disease-resistant germplasm and continuous cropping practices used in most crop producing areas highly limits pathogens control. At present, an increasing number of studies highlight the use of alternative techniques including HIGS, S gene targeting, and rootstocks grafting to effectively and sustainably improve plant resistance against different pathogens. Thereafter, practical problems in the diseases of horticultural plants defense may be solved by creating novel cultivars with above approaches. HIGS silences the virulence genes of invaded pathogens to suppress pathogenic symptoms. Finding susceptibility genes from disease-susceptible horticultural cultivars and targeting these genes through different methods including gene editing and speed-breeding platforms may confer durable and broad-spectrum resistance against pathogens infections. Additionally, grafting is generally effective against diseases reported and used as environmentally friendly technique in modern agriculture to overcome soil-borne diseases such as RKN and FW. Many independent studies better investigate that rootstocks pose different resistant genes to defense soil-borne diseases, and furthermore, the resistant genes will be used to innovate varieties against these diseases. In this sense, additional efforts must be made in control pathogens invasion to increase crop quality and productivity. Accordingly, these new strategies summarized in this review are available for the management of different diseases invaded in the horticultural crops.
Author contributions
XC designed, structured, and prepared manuscript initially. JX searched and collated references and wrote the complete manuscript. All authors contributed to the manuscript version and publication. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the Natural Science Foundation of Jiangsu Province (BK20190887) and the Yangzhou City's Green and Golden Phoenix Program.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
References
Al-Debei, H. S., Makhadmeh, I., Abu-AlRuz, I., Al-Abdallat, A. M., Ayad, J. Y., and AlAmin, N. (2012). Influence of different rootstocks on growth and yield of cucumber (Cucumis sativus L.) under the impact of soil-borne pathogens in Jordan. J. Food Agric. Environ. 10, 343–349. doi: 10.1016/j.jff.2012.03.002
Aman, R., Ali, Z., Butt, H., Mahas, A., Aljedaani, F., Khan, M. Z., et al. (2018). RNA virus interference via CRISPR/Cas13a system in plants. Genome Biol. 19, 1. doi: 10.1186/s13059-017-1381-1
Baltes, N. J., Hummel, A. W., Konecna, E., Cegan, R., Bruns, A. N., Bisaro, D. M., et al. (2015). Conferring resistance to geminiviruses with the CRISPR-Cas prokaryotic immune system. Nat. Plants 1, 15145. doi: 10.1038/nplants.2015.145
Baum, J. A., Bogaert, T., Clinton, W., Heck, G. R., Feldmann, P., Ilagan, O., et al. (2007). Control of coleopteran insect pests through RNA interference. Nat. Biotechnol. 25, 1322–1326. doi: 10.1038/nbt1359
Cáceres, A., Perpiña, G., Ferriol, M., Picó, B., and Gisbert, C. (2017). New Cucumis rootstocks for melon: ‘UPV-FA' and ‘UPV-FMy'. Hort. Sci. 52, 792–797. doi: 10.21273/HORTSCI11791-17
Cai, Q., Qiao, L., Wang, M., He, B., Lin, F. M., Palmquist, J., et al. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126–1129. doi: 10.1126/science.aar4142
Chandrasekaran, J., Brumin, M., Wolf, D., Leibman, D., Klap, C., Pearlsman, M., et al. (2016). Development of broad virus resistance in non-transgenic cucumber using CRISPR/Cas9 technology. Mol. Plant Pathol. 17, 1140–1153. doi: 10.1111/mpp.12375
Char, S. N., Neelakandan, A. K., Nahampun, H., Frame, B., Main, M., Spalding, M. H., et al. (2017). An Agrobacterium-delivered CRISPR/Cas9 system for high-frequency targeted mutagenesis in maize. Plant Biotechnol. J. 15, 257–268. doi: 10.1111/pbi.12611
Chauhan, S., and Rajam, M. V. (2022). RNAi-mediated down-regulation of fasciclin-like proteins (FoFLPs) in Fusarium oxysporum f. sp. lycopersici results in reduced pathogenicity and virulence. Microbiol. Res. 260, 127033. doi: 10.1016/j.micres.2022.127033
Chen, W., Kastner, C., Nowara, D., Oliveira-Garcia, E., Rutten, T., Zhao, Y., et al. (2016). Host-induced silencing of Fusarium culmorum genes protects wheat from infection. J. Exp. Bot. 67, 4979–4991. doi: 10.1093/jxb/erw263
Davis, A. R., Perkins-Veazie, P., Sakata, Y., Lopez-Galarza, S., Maroto, J. V., Lee, S. G., et al. (2008). Cucurbit grafting. Crit. Rev. Plant. Sci. 27, 50–74. doi: 10.1080/07352680802053940
Dong, K. P., Son, S. H., Su, K., Lee, W. M., and Huh, Y. C. (2013). Selection of melon genotypes with resistance to Fusarium wilt and monosporascus root rot for rootstocks. Plant Breed. Biotechnol. 1, 277–282. doi: 10.9787/PBB.2013.1.3.277
Dou, T., Shao, X., Hu, C., Liu, S., Sheng, O., Bi, F., et al. (2020). Host-induced gene silencing of Foc TR4 ERG6/11 genes exhibits superior resistance to Fusarium wilt of banana. Plant Biotechnol. J. 18, 11–13. doi: 10.1111/pbi.13204
Goldschmidt, E. E. (2014). Plant grafting: new mechanisms, evolutionary implications. Front. Plant Sci. 5, 727. doi: 10.3389/fpls.2014.00727
Govindarajulu, M., Epstein, L., Wroblewski, T., and Michelmore, R. W. (2015). Host-induced gene silencing inhibits the biotrophic pathogen causing downy mildew of lettuce. Plant Biotechnol. J. 13, 875–883. doi: 10.1111/pbi.12307
Greco, N., and DiVito, M. (2009). “Population dynamics and damage level,” in Root-knot Nematodes, Eds R. N. Perry, M. Moens, and J. L. Starr (Wallingford: CABI International), 246–274.
Guo, X. Y., Li, Y., Fan, J., Xiong, H., Xu, F. X., Shi, J., et al. (2019). Host-induced gene silencing of MoAP1 confers broad-spectrum resistance to Magnaporthe oryzae. Front Plant Sci. 10, 433. doi: 10.3389/fpls.2019.00433
Hallmann, J., and Meressa, B. H. (2018).“Nematode parasites of vegetables,” in Plant Parasitic Nematodes in Subtropical and Tropical Agriculture, eds R. A. Sikora, D. Coyne, J. Hallmann, and P. Timper (Wallingford: CABI International), 346–410.
Hamilton, A. J., and Baulcombe, D. C. (1999). A species of small antisense RNA in post-transcriptional gene silencing in plants. Science 286, 950–952. doi: 10.1126/science.286.5441.950
Harada, T. (2010). Grafting and RNA transport via phloem tissue in horticultural plants. Sci. Hortic. 125, 545–550. doi: 10.1016/j.scienta.2010.05.013
He, F., Zhang, R., Zhao, J., Qi, T., Kang, Z., and Guo, J. (2019). Host-induced silencing of Fusarium graminearum genes enhances the resistance of Brachypodium distachyon to Fusarium head blight. Front. Plant Sci. 10, 1362. doi: 10.3389/fpls.2019.01362
Huang, G., Allen, R., Davis, E. L., Baum, T. J., and Hussey, R. S. (2006). Engineering broad root-knot resistance in transgenic plants by RNAi silencing of a conserved and essential root-knot nematode parasitism gene. Proc. Natl. Acad. Sci. U.S.A. 103, 14302–14306. doi: 10.1073/pnas.0604698103
Huang, Y., Kong, Q. S., Chen, F., and Bie, Z. L. (2015). History, current status and future prospects of vegetable grafting in China. Acta Hortic. 1086, 31–39. doi: 10.17660/ActaHortic.2015.1086.2
Huh, Y. C., Om, Y. H., and Lee, J. M. (2002). Utilization of Citrullus germplasm with resistance to Fusarium wilt (Fusarium oxysporum f. sp. niveum) for watermelon rootstocks. Acta Hort. 588, 127–132. doi: 10.17660/ActaHortic.2002.588.18
Jiang, W., Zhou, H., Bi, H., Fromm, M., Yang, B., and Weeks, D. P. (2013). Demonstration of CRISPR/Cas9/sgRNA mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res. 41, e188. doi: 10.1093/nar/gkt780
Jones, J. T., Haegeman, A., Danchin, E. G. J., Gaur, H. S., Helder, J., Jones, M. G. K., et al. (2013). Top10 plant-parasitic nematodes in molecular plant pathology. Mol. Plant Pathol. 14, 946–961. doi: 10.1111/mpp.12057
Keinath, A. P., and Hassell, R. L. (2014a). Control of Fusarium wilt of watermelon by grafting onto bottlegourd or interspecific hybrid squash despite colonization of rootstocks by Fusarium. Plant Dis. 98, 255–266. doi: 10.1094/PDIS-01-13-0100-RE
Keinath, A. P., and Hassell, R. L. (2014b). Suppression of Fusarium wilt caused by Fusarium oxysporum f. sp. niveum race 2 on grafted triploid watermelon. Plant Dis. 98, 1326–1332. doi: 10.1094/PDIS-01-14-0005-RE
Ketting, R. F. (2011). The many faces of RNAi. Dev. Cell 20, 148–161. doi: 10.1016/j.devcel.2011.01.012
Koch, A., Höfle, L., Werner, B. T., Imani, J., Schmidt, A., Jelonek, L., et al. (2019). SIGS vs HIGS: a study on the efficacy of two dsRNA delivery strategies to silence Fusarium FgCYP51 genes in infected host and non-host plants. Mol. Plant Pathol. 20, 1636–1644. doi: 10.1111/mpp.12866
Koch, A., Kumar, N., Weber, L., Keller, H., Imani, J., and Kogel, K. H. (2013). Host-induced gene silencing of cytochrome P450 lanosterol C14α-demethylase-encoding genes confers strong resistance to Fusarium species. Proc. Natl. Acad. Sci. U.S.A. 110, 19324–19329. doi: 10.1073/pnas.1306373110
Koch, A., and Wassenegger, M. (2021). Host-induced gene silencing-mechanisms and applications. New Phytol. 231, 54–59. doi: 10.1111/nph.17364
Kokalis-Burelle, N., and Rosskopf, E. N. (2011). Microplot evaluation of rootstocks for control of meloidogyne incognita on grafted tomato, muskmelon, and watermelon. J. Nematol. 43, 166–171. doi: 10.1002/jez.b.21423
Kubota, C., McClure, M. A., Kokalis-Burelle, N., Bausher, M. G., and Rosskopf, E. N. (2008). Vegetable grafting: history, use and current technology status in North America. HortScience 43, 1664–1669. doi: 10.21273/HORTSCI.43.6.1664
Kyriacou, M. C., and Rouphael, Y. (2018). Towards a new definition of quality for fresh fruits and vegetables. Sci. Hortic. 234, 463–469. doi: 10.1016/j.scienta.2017.09.046
Lapin, D., and Van den Ackerveken, G. (2013). Susceptibility to plant disease: more than a failure of host immunity. Trends Plant Sci. 18, 546–554. doi: 10.1016/j.tplants.2013.05.005
Lee, J. M., and Oda, M. (2003). Grafting of herbaceous vegetable and ornamental crops. Hortic. Rev. 28, 124. doi.org/10.1002/9780470650851.ch2.
Leonardi, C., and Romano, D. (2004). Recent issues on vegetable grafting. Acta Hortic. 631, 163–174. doi: 10.17660/ActaHortic.2004.631.21
Li, H., Rasheed, A., Hickey, L. T., and He, Z. (2018). Fast-forwarding genetic gain. Trends Plant Sci. 23, 184–186. doi: 10.1016/j.tplants.2018.01.007
Li, S., Lin, D., Zhang, Y., Deng, M., Chen, Y., Lv, B., et al. (2022). Genome-edited powdery mildew resistance in wheat without growth penalties. Nature 602, 455–460. doi: 10.1038/s41586-022-04395-9
Li, T., Liu, B., Spalding, M. H., Weeks, D. P., and Yang, B. (2012). High-efficiency TALEN-based gene editing produces disease-resistant rice. Nat. Biotechnol. 30, 390–392. doi: 10.1038/nbt.2199
Li, X. Z., and Chen, S. X. (2017). Screening and identification of cucumber germplasm and rootstock resistance against the root-knot nematode (Meloidogyne incognita). Genet. Mol. Res. 16, gmr16029383. doi: 10.4238/gmr16029383
Li, Y., and Zhao, D. (2021).Transcriptome analysis of scions grafted to potato rootstock for improving late blight resistance. BMC Plant Biol. 21, 272. doi: 10.1186/s12870-021-03039-w
Liu, M. J., Mei, Q., Xue, C. Y., Wang, Z. Y., Li, D. P., Zhang, Y. X., et al. (2021). Mutation of G-protein γ subunit DEP1 increases planting density and resistance to sheath blight disease in rice. Plant Biotechnol. J. 19, 418–420. doi: 10.1111/pbi.13500
Louws, F. J., Rivard, C. L., and Kubota, C. (2010). Grafting fruiting vegetables to manage soilborne pathogens, foliar pathogens, arthropods and weeds. Sci. Hortic. 127, 127–146. doi: 10.1016/j.scienta.2010.09.023
Mao, Y. B., Cai, W. J., Wang, J. W., Hong, G. J., Tao, X. Y., Wang, L. J., et al. (2007). Silencing a cotton bollworm P450 monooxygenase gene by plant-mediated RNAi impairs larval tolerance of gossypol. Nat. Biotechnol. 25, 1307–1313. doi: 10.1038/nbt1352
Martínez, R., Aguilar, M. I., Guirado, M. L., Alvarez, A., and Gomez, J. (2003). First report of Fusarium wilt of cucumber caused by Fusarium oxysporum in Spain. Plant Pathol. 52, 410. doi: 10.1046/j.1365-3059.2003.00832.x
Miguel, A., Varoto, J. V., San Bautista, A., Baizauli, C., Cebolla, V., Pascual, B., et al. (2004). The grafting of triploid watermelonis an advantageous alternative to soil fumigation by methyl bromide for control of Fusarium wilt. Sci. Hortic. 103, 9–17. doi: 10.1016/j.scienta.2004.04.007
Nawaz, M. A., Imtiaz, M., Kong, Q., Cheng, F., Ahmed, W., Huang, Y., et al. (2016). Grafting: a technique to modify ion accumulation in horticultural crops. Front. Plant Sci. 7, 1457. doi: 10.3389/fpls.2016.01457
Nekrasov, V., Wang, C., Win, J., Lanz, C., Weigel, D., and Kamoun, S. (2017). Rapid generation of a transgene-free powdery mildew resistant tomato by genome deletion. Sci. Rep. 7, 482. doi: 10.1038/s41598-017-00578-x
Nie, J., Wang, Y., He, H., Guo, C., Zhu, W., Pan, J., et al. (2015). Loss-of-function mutations in CsMLO1 confer durable powdery mildew resistance in cucumber (Cucumis sativus L.). Front. Plant Sci. 6, 1155. doi: 10.3389/fpls.2015.01155
Nisini, P. T., Colla, G., Granati, E., Temperini, O., Crino, P., and Saccardo, F. (2002). Rootstock resistance to Fusarium wilt and effect on fruit yield and quality of two muskmelon cultivars. Sci. Hortic. 93, 281–288. doi: 10.1016/S0304-4238(01)00335-1
Nowara, D., Gay, A., Lacomme, C., Shaw, J., Ridout, C., Douchkov, D., et al. (2010). HIGS: host induced gene silencing in the obligate biotrophic fungal pathogen. Plant Cell 22, 3130–3141. doi: 10.1105/tpc.110.077040
Nunes, C. C., and Dean, R. A. (2012). Host-induced gene silencing: a tool for understanding fungal host interaction and for developing novel disease control strategies. Mol. Plant Pathol. 13, 519–529. doi: 10.1111/j.1364-3703.2011.00766.x
Panwar, V., McCallum, B., and Bakkeren, G. (2013). Host-induced gene silencing of wheat leaf rust fungus Puccinia triticina pathogenicity genes mediated by the barley stripe mosaic virus. Plant Mol. Biol. 81, 595–608. doi: 10.1007/s11103-013-0022-7
Peng, A., Chen, S., Lei, T., Xu, L., He, Y., Wu, L., et al. (2017). Engineering canker-resistant plants through CRISPR/Cas9-targeted editing of the susceptibility gene CsLOB1 promoter in citrus. Plant Biotechnol. J. 15, 1509–1519. doi: 10.1111/pbi.12733
Pliego, C., Nowara, D., Bonciani, G., Gheorghe, D. M., Xu, R., Surana, P., et al. (2013). Host-induced gene silencing in barley powdery mildew reveals a class of ribonuclease-like effectors. Mol. Plant Microbe Interact. 26, 633–642. doi: 10.1094/MPMI-01-13-0005-R
Proietti, S., Rouphael, Y., Colla, G., Cardarelli, M., De Agazio, M., Zacchini, M., et al. (2010). Fruit quality of mini-watermelon as affected by grafting and irrigation regimes. J. Sci. Food Agric. 88, 1107–1114. doi: 10.1002/jsfa.3207
Reddy, P. P. (Ed.). (2016). “Grafted vegetables for management of soilborne pathogens,” in Sustainable Crop Protection Under Protected Cultivation, Ed P. P. Reddy (Singapore: Springer Science + Business Media), pp. 83–97.
Rouphael, Y., Cardarelli, M., Rea, E., and Colla, G. (2008). Grafting of cucumber as a means to minimize copper toxicity. Environ. Exp. Bot. 63, 49–58. doi: 10.1016/j.envexpbot.2007.10.015
Ruiz, J. M., Belakbir, A., Lopez-Cantarero, I., and Romero, L. (1997). Leaf-macronutrient content and yield in grafted melon plants, a model to evaluate the influence of rootstock genotype. Sci. Hortic. 71, 227–234. doi: 10.1016/S0304-4238(97)00106-4
Ruiz, J. M., and Romero, I. (1999). Nitrogen efficiency and metabolism in grafted melon plants. Sci. Hortic. 81,113–123. doi: 10.1016/S0304-4238(98)00200-3
Sabry, S., Ali, A. Z., Abdel-Kader, D. A., and Abou-Zaid, M. I. (2022). Histopathological and biochemical aspects of grafted and non-grafted cucumber infected with stem rot caused by Fusarium spp. Saudi. J. Biol. Sci. 29, 1770–1780. doi: 10.1016/j.sjbs.2021.10.053
Shalaby, T. A., Taha, N. A., Rakha, M. T., El-Beltagi, H. S., Shehata, W. F., Ramadan, K. M. A., et al. (2022). Can grafting manage Fusarium wilt disease of cucumber and increase productivity under heat stress? Plants 11, 1147. doi: 10.3390/plants11091147
Shan, Q., Wang, Y, Li, J., Zhang, Y., Chen, K., Liang, Z., et al. (2013). Targeted genome modification of crop plants using a CRISPR–Cas system. Nat. Biotechnol. 31, 686–688. doi: 10.1038/nbt.2650
Shi, L., Du, N., Yuan, Y., Shu, S., Sun, J., and Guo, S. (2016). Vinegar residue compost as a growth substrate enhances cucumber resistance against the Fusarium wilt pathogen Fusarium oxysporum by regulating physiological and biochemical responses. Environ. Sci. Pollut. Res. 23, 18277–18287. doi: 10.1007/s11356-016-6798-7
Shipman, E. N., Yu, J., Zhou, J., Albornoz, K., and Beckles, D. M. (2021). Can gene editing reduce post harvest waste and loss of fruit, vegetables, and ornamentals? Hortic. Res. 8, 1–21. doi: 10.1038/s41438-020-00428-4
Sigüenza, C., Schochow, M., Turini, T., and Ploeg, A. (2005). Use of Cucumis metuliferus as a rootstock for melon to manage Meloidogyne incognita. J. Nematol. 37, 276–280.
Song, Y., and Thomma, B. P. H. J. (2018). Host-induced gene silencing compromises Verticillium wilt in tomato and Arabidopsis. Mol. Plant Pathol. 19, 77–89. doi: 10.1111/mpp.12500
Stover, E., Driggers, R., Richardson, M. L., Hall, D. G., and Lee, R. F. (2014). Incidence and severity of asiatic citrus canker on diverse citrus and citrus-related germplasm in a florida field planting. Hortscience 49, 4–9. doi: 10.21273/HORTSCI.49.1.4
Suzuki, G., Fukuda, M., Lucob-Agustin, N., Inukai, Y., and Gomi, K. (2022). The mutation of rice MEDIATOR25, OsMED25, induces rice bacterial blight resistance through altering jasmonate- and auxin-signaling. Plants 11, 1601. doi: 10.3390/plants11121601
Thies, J. A. (2021). Grafting for managing vegetable crop pests. Pest Manag. Sci. 77, 4825–4835. doi: 10.1002/ps.6512
Thies, J. A., Ariss, J., Hassell, R., Kousik, C. S., Olson, S., and Levi, A. (2010). Grafting for managing southern root-knot nematode, Meloidogyne incognita, in watermelon. Plant Dis. 94, 1195–1199. doi: 10.1094/PDIS-09-09-0640
Thies, J. A., Ariss, J., Kousik, C. S., and Hassell, R. (2008). Grafting–a tool for managing root-knot nematodes in watermelon? Phytopathology 98, S156.
Thomazella, D. P. T., Seong, K., Mackelprang, R., Dahlbeck, D., Geng, Y., Gill, U. S., et al. (2021). Loss of function of a DMR6 ortholog in tomato confers broad-spectrum disease resistance. Proc. Natl. Acad. Sci. U.S.A. 118, e2026152118. doi: 10.1073/pnas.2026152118
Tinoco, M. L., Dias, B. B., Dall'Astta, R. C., Pamphile, J. A., and Aragão, F. J. (2010). In vivo trans-specific gene silencing in fungal cells by in planta expression of a double-stranded RNA. BMC Biol. 8, 27. doi: 10.1186/1741-7007-8-27
Traka-Mavrona, E., Koutsika-Sotiriou, M., and Pritsa, T. (2000). Response of squash (Cucurbita spp.) as rootstock for melon (Cucumis melo L.). Sci. Hortic. 83, 353–362. doi: 10.1016/S0304-4238(99)00088-6
van Schie, C. C., and Takken, F. L. (2014). Susceptibility genes 101: how to be a good host. Annu. Rev. Phytopathol. 52, 551–581. doi: 10.1146/annurev-phyto-102313-045854
Wan, D. Y., Guo, Y., Cheng, Y., Hu, Y., Xiao, S., Wang, Y., et al. (2020). CRISPR/Cas9-mediated mutagenesis of VvMLO3 results in enhanced resistance to powdery mildew in grapevine (Vitis vinifera). Hortic Res. 7, 116. doi: 10.1038/s41438-020-0339-8
Wang, M., Sun, Y. M., Sun, G. M., Liu, X., Zhai, L., Shen, Q., et al. (2015). Water balance altered in cucumber plants infected with Fusarium oxysporum f. sp. cucumerinum. Sci. Rep. 5, 7722. doi: 10.1038/srep07722
Wang, N., Tang, C., Fan, X., He, M., Gan, P., Zhang, S., et al. (2022). Inactivation of a wheat protein kinase gene confers broad-spectrum resistance to rust fungi. Cell 185, 2961–2974. e19. doi: 10.1016/j.cell.2022.06.027
Wang, S., Lei, C., Wang, J., Ma, J., Tang, S., Wang, C., et al. (2017). SPL33, encoding an eEF1A-like protein, negatively regulates cell death and defense responses in rice. J. Exp. Bot. 68, 899–913. doi: 10.1093/jxb/erx001
Wang, X., Zhang, H., Nyamesorto, B., Luo, Y., Mu, X., Wang, F., et al. (2020). A new mode of NPR1 action via an NB-ARC-NPR1 fusion protein negatively regulates the defence response in wheat to stem rust pathogen. New Phytol. 228, 959–972. doi: 10.1111/nph.16748
Wang, Y., Cheng, X., Shan, Q., Zhang, Y., Liu, J., Gao, C., et al. (2014). Simultaneous editing of three homoeoalleles in hexaploid bread wheat confers heritable resistance to powdery mildew. Nat. Biotechnol. 32, 947–951. doi: 10.1038/nbt.2969
Watson, A., Ghosh, S., Williams, M. J., Cuddy, W. S., Simmonds, J., Rey, M. D., et al. (2018). Speed breeding is a powerful tool to accelerate crop research and breeding. Nat. Plants 4, 23–29. doi: 10.1038/s41477-017-0083-8
Wei, C., Qin, T., Li, Y., Wang, W., Dong, T., and Wang, Q. (2020). Host-induced gene silencing of the acetolactate synthases VdILV2 and VdILV6 confers resistance to Verticillium wilt in cotton (Gossypium hirsutum L.). Biochem. Biophys. Res. Commun. 524, 392–397. doi: 10.1016/j.bbrc.2020.01.126
Xu, J., Wang, X., Li, Y., Zeng, J., Wang, G., Deng, C., et al. (2018). Host-induced gene silencing of a regulator of G protein signalling gene (VdRGS1) confers resistance to Verticillium wilt in cotton. Plant Biotechnol. J. 16, 1629–1643. doi: 10.1111/pbi.12900
Xu, J., Xian, Q. Q., Wang, K., Dong, J. P., Zhang, C. Y., Du, S. L., et al. (2022). Screening and identification of candidate Fusarium wilt-resistance genes from pumpkin. Hortic. Plant J. 8, 583–592. doi: 10.1016/j.hpj.2021.11.011
Yang, Q., Islam, M. A., Cai, K., Tian, S., Liu, Y., Kang, Z., et al. (2020). TaClpS1, negatively regulates wheat resistance against Puccinia striiformis f. sp. tritici. BMC Plant Biol. 20, 555. doi: 10.1186/s12870-020-02762-0
Yetisir, H., Sari, N., and Yücel, S. (2003). Rootstock resistance to Fusarium wilt and effect on watermelon fruit yield and quality. Phytoparasitica 31, 163–169. doi: 10.1007/BF02980786
Yin, C., Jurgenson, J., and Hulbert, S. H. (2011). Development of a host induced RNAi system in the wheat stripe rust fungus Puccinia striiformis f. sp. tritici. Mol. Plant Microbe Interact. 24, 554–561. doi: 10.1094/MPMI-10-10-0229
Zaidi, S. S., Mansoor, S., Ali, Z., Tashkandi, M., and Mahfouz, M. M. (2016b). Engineering plants for geminivirus resistance with CRISPR/Cas9 system. Trends Plant Sci. 21, 279–281. doi: 10.1016/j.tplants.2016.01.023
Zaidi, S. S., Mukhtar, M. S., and Mansoor, S. (2018). Genome editing: targeting susceptibility genes for plant disease resistance. Trends Biotechnol. 36, 898–906. doi: 10.1016/j.tibtech.2018.04.005
Zaidi, S. S., Tashkandi, M., Mansoor, S., and Mahfouz, M. M. (2016a). Engineering plant immunity: using CRISPR/Cas9 to generate virus resistance. Front. Plant Sci. 7, 1673. doi: 10.3389/fpls.2016.01673
Zand Karimi, H., and Innes, R. W. (2022). Molecular mechanisms underlying host-induced gene silencing. Plant Cell 34, 3183–3199. doi: 10.1093/plcell/koac165
Zhang, H., Guo, J., Voegele, R. T., and Kang, Z. S. (2012). Functional characterization of calcineurin homologs PsCNA1/PsCNB1 in Puccinia striiformis f. sp.tritici using a host-induced RNAi system. PLoS ONE 7, e49262. doi: 10.1371/journal.pone.0049262
Zhang, M., Liu, Q., Yang, X., Xu, J., Liu, G., Yao, X., et al. (2020). CRISPR/Cas9-mediated mutagenesis of Clpsk1 in watermelon to confer resistance to Fusarium oxysporum f. sp. niveum. Plant Cell Rep. 39, 589–595. doi: 10.1007/s00299-020-02516-0
Zhang, M., Xu, J., Ren, R., Liu, G., Yao, X., Lou, L., et al. (2021). Proteomic analysis of Fusarium oxysporum-induced mechanism in grafted watermelon seedlings. Front. Plant Sci. 12, 632758. doi: 10.3389/fpls.2021.632758
Zhang, M., Yang, X. P., Xu, J. H., Liu, G., Yao, X. F., and Li, P. F. (2015). Physiological responses of watermelon grafted onto bottle gourd to Fusarium oxysporum f. sp. niveum infection. Acta Hort. 1086, 107–111. doi: 10.17660/ActaHortic.2015.1086.12
Zhang, S. P., Miao, H., Yang, Y. H., Xie, B. Y., Wang, Y., and Gu, X. F. (2014). A major quantitative trait locus conferring resistance to Fusarium wilt was detected in cucumber by using recombinant inbred lines. Mol. Breed. 34, 1805–1815. doi: 10.1007/s11032-014-0140-1
Zhang, T., Jin, Y., Zhao, J. H., Gao, F., Zhou, B. J., Fang, Y. Y., et al. (2016a). Host-induced gene silencing of the target gene in fungal cells confers effective resistance to the cotton wilt disease pathogen Verticillium dahliae. Mol. Plant 9, 939–942. doi: 10.1016/j.molp.2016.02.008
Zhang, T., Zhao, Y. L., Zhao, J. H., Wang, S., Jin, Y., Chen, Z. Q., et al. (2016b). Cotton plants export microRNAs to inhibit virulence gene expression in a fungal pathogen. Nat. Plants 2, 16153. doi: 10.1038/nplants.2016.153
Zhang, Y., Bai, Y., Wu, G., Zou, S., Chen, Y., Gao, C., et al. (2017). Simultaneous modification of three homoeologs of TaEDR1 by genome editing enhances powdery mildew resistance in wheat. Plant J. 91, 714–724. doi: 10.1111/tpj.13599
Keywords: horticultural crops, diseases, HIGS, CRISPR-Cas9 technology, graft
Citation: Xu J, Zhang N, Wang K, Xian Q, Dong J and Chen X (2022) Exploring new strategies in diseases resistance of horticultural crops. Front. Sustain. Food Syst. 6:1021350. doi: 10.3389/fsufs.2022.1021350
Received: 17 August 2022; Accepted: 17 October 2022;
Published: 08 November 2022.
Edited by:
Lei Jiang, Anhui Agricultural University, ChinaReviewed by:
Ziyi Yin, Shandong Agricultural University, ChinaMeixiang Zhang, Shaanxi Normal University, China
Muxing Liu, Nanjing Agricultural University, China
Copyright © 2022 Xu, Zhang, Wang, Xian, Dong and Chen. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Xuehao Chen, eGhjaGVuQHl6dS5lZHUuY24=
 Jun Xu1
Jun Xu1 Xuehao Chen
Xuehao Chen