
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 28 October 2022
Sec. Crop Biology and Sustainability
Volume 6 - 2022 | https://doi.org/10.3389/fsufs.2022.1020634
Volatile organic compounds (VOCs) are involved in microbial interspecies communication and in the mode of action of various antagonistic interactions. They are important for balancing host-microbe interactions and provide the basis for developing biological control strategies to control plant pathogens. We studied the interactions between the bacterial antagonist Serratia plymuthica HRO-C48 and three fungal plant pathogens Rhizoctonia solani, Leptosphaeria maculans and Verticillium longisporum. Significant differences in fungal growth inhibition by the Serratia-emitted VOCs in pairwise dual culture assays and changes in the transcriptome of the bacterium and in the volatilomes of both interacting partners were observed. Even though the rate of fungal growth inhibition by Serratia was variable, the confrontation of the bacterium with the VOCs of all three fungi changed the levels of expression of the genes involved in stress response, biofilm formation, and the production of antimicrobial VOCs. Pairwise interacting microorganisms switched between defense (downregulation of gene expression) and attack (upregulation of gene expression and metabolism followed by growth inhibition of the interacting partner) modes, subject to the combinations of microorganisms that were interacting. In the attack mode HRO-C48 significantly inhibited the growth of R. solani while simultaneously boosting its own metabolism; by contrast, its metabolism was downregulated when HRO-C48 went into a defense mode that was induced by the L. maculans and V. longisporum VOCs. L. maculans growth was slightly reduced by the one bacterial VOC methyl acetate that induced a strong downregulation of expression of genes involved in almost all metabolic functions in S. plymuthica. Similarly, the interaction between S. plymuthica and V. longisporum resulted in an insignificant growth reduction of the fungus and repressed the rate of bacterial metabolism on the transcriptional level, accompanied by an intense volatile dialogue. Overall, our results indicate that VOCs substantially contribute to the highly break species-specific interactions between pathogens and their natural antagonists and thus deserving of increased consideration for pathogen control.
Microbiome-based crop management is facing enormous challenges due to the complexity of interactions between plant and diverse microbiota associated with it (Bakker et al., 2020). In addition to harmless saproptrophs or biotrophs, nutrient rich root exudates also attract plant pathogens that may cause diverse plant diseases (Whipps, 2001). In a healthy plant there is a steady balance between pathogens and beneficial/harmless microorganisms, yet many factors can unbalance this dynamic, creating a predisposition for the accumulation of pathogenic or antagonistic microbes within the host microbiome (Berg, 2019). Agricultural intensification is a significant unbalancing factor as it has a negative influence on the fungal networks associated with plant roots, and on the abundance of keystone taxa, thus reducing the indigenous antagonistic potential of the root microbiome to combat plant pathogens (Banerjee et al., 2018). Soil borne plant pathogens such as Verticillium longisporum (C. Stark) Karapapa et al. (1997), Rhizoctonia solani Kühn (teleomorph Thanatephorus cucumeris [Frank] Donk) and Leptosphaeria maculans (anamorph Phoma lingam) are responsible for substantial yield losses amongst diverse crops (Gugel and Petrie, 1992; Karapapa et al., 1997; Sneh et al., 2013). These pathogens are very difficult to control by means of conventional methods due to the complexity of their life cycles, the persistence of their survival structures (microsclerotia) in soil for a very long time and the breadth of their host ranges. The implementation of biological control strategies has therefore been recommended as a viable environmentally friendly alternative to the use of pesticides in the practice of sustainable agriculture (Köhl et al., 2019).
In order to develop effective microbiome-based biological control strategies of phytopathogens we need to be able to understand the complexity of interactions between the pathogenic and potentially beneficial microorganisms. One of many such interaction pathways involves volatile organic compounds (VOCs) produced by microorganisms (Kai et al., 2007; Weisskopf et al., 2021). VOCs are small odorous compounds that typically have less than 15 C-atoms and low molecular mass. They play an important role in interactions between members of natural microbiota and can induce various phenotypical and biochemical responses in the interacting partners (Kai et al., 2007; Schulz et al., 2010; Schmidt et al., 2015). Recently they became the focus of significant attention due to their antimicrobial properties and ability to influence the growth of microbial pathogens over large distances (Schmidt et al., 2016, 2017; Schulz-Bohm et al., 2017). Understanding VOCs-based communication between microorganisms and revealing their roles in biocontrol mechanisms is an important step in developing novel, environmentally friendly solutions for combating fungal plant pathogens.
S. plymuthica spp. are known for their antifungal activities against plant pathogenic fungi such as Verticillium spp., R. solani Kühn and L. maculans (Grosch et al., 2005; Müller and Berg, 2008; Abuamsha et al., 2011). In particular, the biocontrol agent S. plymuthica HRO-C48 has been shown to be active against V. dahliae Kleb, R. solani Kühn, and Sclerotinia sclerotiorum (Kurze et al., 2001; Kai et al., 2007; Abuamsha et al., 2011). Moreover, it has been successfully used for controlling Verticillium wilt and other diseases caused by soil borne fungi when used as a soil amendment in strawberry fields (RhizoStar® Kurze et al., 2001). The application of S. plymuthica HRO-C48 to the seeds of the oilseed rape reduced the degree of Verticillium wilt in plants grown under greenhouse conditions (Müller and Berg, 2008). The same treatment showed a significant improvement of the germination rate of plants grown in a field with a history of natural Verticillium wilt infection (Rybakova et al., 2017a, 2020).
Substantial knowledge has been generated for the past decade on soluble metabolites of S. plymuthica HRO-C48 involved in biological control (Frankowski et al., 2001; Liu et al., 2007; Müller et al., 2009; Pang et al., 2009). Various Serratia spp. are described as being producers of both antifungal antibiotics pyrrolnitrin and sodorifen (Müller et al., 2009; Neupane et al., 2015; Domik et al., 2016; Schmidt et al., 2017; Pawar and Chaudhari, 2020). The ability of S. plymuthica HRO-C48 to produce the broad spectrum antibiotic pyrrolnitrin has been shown to be regulated by quorum sensing mediated by N-acyl homoserine lactone signals (Liu et al., 2007; Müller et al., 2009; Pang et al., 2009). In contrast to the large amount of literature available on the topic of Serratia's soluble metabolites, very little information is available about its VOCs-mediated interactions with fungal pathogens (Kai et al., 2007; Weise et al., 2014). Previous studies have indicated that S. plymuthica HRO-C48 emits a broad spectrum of VOCs that are involved in its antifungal activity (Kai et al., 2007; Müller et al., 2009). In the study conducted by Kai et al., only two volatile compounds produced by HRO-C48 out of 14 detected VOCs were identified (Kai et al., 2007). Recent advances in microbial VOCs identification, as well as the availability of S. plymuthica HRO-C48 genomic information (BioSample: SAMN04515841) allowed us to further examine the mode of action of HRO-C48 against pathogenic fungi via its volatiles.
We combined three complementary methods in our study. We applied (1) in vitro volatile dual plate assays to assess fungal growth inhibition by the biocontrol agent, (2) gas chromatography–mass spectrometry (GC-MS) headspace Solid Phase Microextraction (SPME) assays to detect and identify VOCs involved in bacterial-fungal interactions, and (3) transcriptomics assays to assess changes in gene expression of S. plymuthica HRO-C48 when exposed to fungal volatiles. Particular emphasis was placed on the crosstalk between the bacterial and fungal partners that facilitated deepening of our insight into microbial interactions at the molecular level. This novel approach allowed for a significant contribution to understanding the mode of action of the biological S. plymuthica HRO-C48 over long distances and embraced the complexity of its VOCs-mediated interactions with soil borne plant pathogens - another important step toward attaining sustainable agroecosystems.
S. plymuthica HRO-C48 was isolated from the rhizosphere of oilseed rape and characterized as antagonists toward fungal pathogens (Kalbe et al., 1996). The fungal plant pathogens V. longisporum (C. Stark) Karapapa et al. (1997) strain EVL43 (Messner et al., 1996), and R. solani Kühn (teleomorph Thanatephorus cucumeris [Frank] Donk) AG2-2IIIB isolate (in this study further called R. solani AG2) were provided by the strain collection of the TU Graz, institute of environmental biotechnology. L. maculans MB 158 (anamorph: Phoma lingam) isolate (Stachowiak et al., 2006; Kaczmarek et al., 2014) was provided by Malgorzata Jedryczka, Poznan. All microorganisms in this study were grown on potato dextrose agar (PDA; Carl-Roth, Karlsruhe, Germany) at 25°C, unless otherwise indicated.
Antagonism of S. plymuthica HRO-C48 toward fungal plant pathogens V. longisporum ELV 43, L. maculans MB 158, R. solani AG2 and R. solani AG2 was tested with the Two Clamp VOCs Assay (Cernava et al., 2015). Fungal strains were pre-cultured at 22°C from 4 to 7 days plate under natural day-night cycle and were inoculated as a plug with a diameter of 0.6 cm onto PDA six-well plates (Greiner Bio-One, Frickenhausen, Germany). A colony of S. plymuthica HRO-C48 was taken from a two-day old culture on a solid nutrient agar (NA; Sifin, Berlin, Germany) plate incubated at 30°C and spread out in four lines horizontal and four vertical resulting in a grid pattern. Bacterial and fungal inocula were co-incubated at 20°C keeping the two six-well plates facing each other with the fungal strain on the bottom. The wells were separated by a perforated (0.5 cm diameter) sterile 2 mm silicone foil allowing exchange of VOCs without direct contact of microorganisms. The whole construct was fixed by four clamps, one at each side of the plate. The assay was performed in three replicates and three repetitions of each antagonistic assay were performed. The fungal growth was determined by measuring the colony diameter after 3, 6, and 9 days of co-incubation. Values significantly different from control group values (P < 0.05) were defined using pairwise t-test or non-parametric Mann–Whitney U-test, depending on the distribution of the samples as described by Rybakova et al. (2016).
GC-MS headspace Solid Phase Micro Extraction (SPME) experiments were carried out as described by Cernava et al. (2015). V. longisporum ELV43 and S. plymuthica HRO-C48, separately inoculated on PDA slope agar in headspace vials were co-incubated for 3 days at 20°C in three replicates. The following dual combinations of samples were prepared: (a) each fungus and an empty vial with a PDA slope agar (negative control for fungi); (b) each fungus with S. plymuthica HRO-C48; (c) S. plymuthica HRO-C48 and an empty vial with a PDA slope agar (negative control for bacteria); and (d) empty vial with a PDA slope agar without microorganisms' contact (PDA control). Each jar contained three replicates of each sample. During the cultivation, the jars were completely sealed. After 3 days of coincubation the jars were re-opened under sterile conditions and vials were incubated for further 2 h before they were sealed with crimp seals.
The SPME was conducted with an automated sampler with a 50/30 μm Divinylbenzene/Carboxen/Polydimethylsiloxane (DVB/CAR/PDMS) StableFlex fiber with a length of 2 cm. Compounds present in the headspace were enriched for 30 min at 35°C. The instruments that were used for separation and detection of the compounds, respectively, were the GC7890B together with a MS5977A (Agilent Technologies, Waldbronn, Germany). The separation column which was used was a HP-5ms column (5%-phenyl)-methylpolysiloxane, 30 m × 250 μm 0.25 μm (Length × Inner Diameter × Coating, 250°C) followed by electron ionization and detection within a mass range of 35–450 AU. First the GC column was kept at 40°C for 2 min, then the temperature was increased to 110°C (rate = 5°C/min), then again increased to 280°C (rate = 10°C/min) and maintained there for 3 min. The helium flow rate was 1.2 mL/min. Identification of the volatile compounds was performed with NIST MS Search 2.2 included in the Software-Package of the NIST 2014 database (MLA, 1997). The ratio of each peak was calculated the following way: the area of the peak divided through the total area of all peaks in the chromatogram, multiplied by 100%. The compound suggestion with the best relative spectrum match (RMatch) from the NIST14 database was manually checked. Together with the RI-Match the identification suggestion was accepted or deleted. From this resulting peaks values data, the values of the respective controls (PDA incubated with respective microorganism) were subtracted. The substances detected in the PDA control were not included in the final volatile list as it was assumed that they were contaminants from the PDA medium itself. The raw data of GC-MS analyses is provided in Supplementary Table S1.
Fungal strains V. longisporum ELV 43, L. maculans MB 158 and R. solani AG2 were pre-cultivated at 22°C for 4 to 7 days on PDA plates under natural day-night cycle. The plugs with a diameter of 0.6 cm were transferred to a fresh PDA plate and co-incubated with another plate containing S. plymuthica HRO-C48 pre-cultured on NA for 2 days at 30°C. HRO-C48 and one of the fungal isolates were exposed to each other's VOCs in the dark for 3 days at 20°C in two replicates. Control sample consists of pre-cultured S. plymuthica HRO-C48 co-incubated with a non-inoculated PDA plate. The plates were fixed with parafilm facing each other together so that only VOCs could interchange between bacterial and fungal (or control) cultures. After the co-incubation the bacterial cultures were harvested using RNAse away (Sigma-Aldrich) – treated Drigalsky spatula in an RNase free environment. 0.2 g of cell material was transferred to RNase free tubes containing RNA later (Thermo Fisher Scientific, Massachusetts, USA). The ratio of RNA later to sample was 1:5 as suggested by GATC Biotech (Konstanz, Germany). The tubes were inverted for about 30 sec. and sent in duplicates to GATC Biotech (Konstanz, Germany) where the samples were processed according to company's proprietary protocols including RNA extraction, depletion of ribosomal RNA, fragmentation of mRNA, random-primed synthesis of cDNA, double strand synthesis and library preparation. Sequencing was performed using Illumina HiSeq 2000 and 50 bp single read mode resulting in 41,345,272–51,940,177 raw reads per sample. Raw reads were subjected to quality-based trimming using Trimmomatic v0.39 (Bolger et al., 2014). At the 3' end, reads were clipped at position 11, whereas the position for clipping reads at the 5' end was set through Phred quality score cut-off at 28. Finally, only reads ≥ 40 bp passed filtering step representing 71–92% of the input. Quality reads were mapped to the genome assembly of S. plymuthica HRO-C48 available at NCBI (RefSeq assembly accession: GCF_001590765.1) using Bowtie 2 with default parameter (Langmead and Salzberg, 2012). Resulting mapping files were provided to StringTie (Pertea et al., 2015) to exclusively count reads that were mapped within coding regions of reference transcripts generated by NCBI's automated annotation pipeline. All read numbers along the processing workflow from raw reads to reads finally mapped to the reference genome are provided in Supplementary Table S2.
For exploratory gene expression analysis, read count tables were further processed in R by employing the RNA-Seq analysis platform Degust v4.1.1 (Powell, 2019). Analysis steps included (i) normalization of raw counts using CPM (counts per million mapped reads) method, (ii) library size normalization using TMM (trimmed mean of M-value) method and (iii) testing for differentially expressed genes (DEGs) using glmQLFit function implemented in edgeR (Lun et al., 2016). Genes were defined as differentially expressed at fold-change cut-off ±1.5 and a FDR of 0.01. Putative gene products and corresponding COG categories were considered for analyzing differentially expressed genes (DEGs) in a functional context. For predicting gene-encoded products and their functional role, protein sequences translated from CDS features were mapped to the eggNOG protein database v5.0 (Huerta-Cepas et al., 2019) using eggNOG-mapper web v2.1.7 (Cantalapiedra et al., 2021). The complete list of the DEGs including functional annotations as well as data from expression analysis (normalized counts, fold-changes and significance levels) for three conditions are provided in Supplementary Table S2. Venn diagrams were produced using online tool Venny 2.1.0 (Oliveros, 2007).
In order to assess antifungal effects of S. plymuthica HRO-C48 via its volatiles, we performed specific, pairwise VOCs assays with HRO-C48 and three fungal plant pathogens, R. solani AG2, L. maculans MB 158 and V. longisporum ELV 43 on solid PDA. PDA was chosen as a growth medium for both interacting partners as it facilitated a stronger inhibition of fungal growth by bacterial volatiles compared to the fungal growths on both Waksman and R2A agars in a previous experiment (data not shown). After 3, 6, and 9 days of co-inoculation with bacterial VOCs, the mycelial growth of the selected fungi was measured and compared to the untreated control. Only R. solani AG2 was significantly inhibited by the VOCs of S. plymuthica HRO-C48 after 3 and 6 days of co-incubation. The growth of L. maculans MB 158 and V. longisporum ELV 43 was reduced under all tested conditions, compared to the untreated control, however this reduction was not statistically significant (Figure 1). The strongest inhibition of mycelial growth of all three tested fungi by S. plymuthica HRO-C48 VOCs, when compared to the untreated control, was observed when both microorganisms were incubated for 3 days. Therefore, this condition was selected for further experiments.
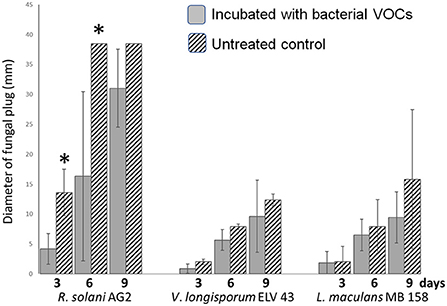
Figure 1. VOCs assay of three fungal plant pathogens (R. solani AG2, L. maculans MB 158 and V. longisporum ELV 43) and S. plymuthica HRO-C48 on solid PDA for 3, 6, and 9 days. Dashed columns show growth of fungi in the presence of S. plymuthica HRO-C48 VOCs, while gray columns illustrate the growth of the selected fungi without bacterial VOCs (untreated control). The assay was performed in three replicates. The bars represent standard errors, while an asterisk *denotes values that varied significantly from control group values (P < 0.05) defined using pairwise t-test or non-parametric Mann–Whitney U-test, depending on the distribution of the samples.
VOCs produced by S. plymuthica HRO-C48 after 3 days of co-incubation with the VOCs of the three selected fungi were analyzed using GC-MS headspace SPME and compared to database entries (MLA, 1997). In total, 28 different substances were detected in S. plymuthica HRO-C48 samples. Twenty-two of the detected VOCs are mentioned in current literature as having an antagonistic effect on other microorganisms (Supplementary Table S3). Some of them were up- and some downregulated depending on the interacting fungus. A half of the identified VOCs (15 out of 28) produced by S. plymuthica HRO-C48 belonged to the core volatilome under the conditions tested, while the other half varied depending on the fungus that the bacterium interacted with (Supplementary Table S3 and Figure 2A). Acetic acid and methylthiolacetate were the only two substances that were detected in all samples treated with fungal VOCs, while not being detected in the control. These two substances that were emitted in all treatments are known for their antimicrobial and, in particular, antifungal properties (Pimenta et al., 2012; Ossowicki et al., 2017). Contact of S. plymuthica HRO-C48 with V. longisporum ELV 43 VOCs resulted in a significantly stronger upregulation of VOCs production in the bacterium compared to the other fungi tested: 22 upregulated VOCs (14 substances with putative antimicrobial function) vs. two downregulated VOCs with unknown function. VOCs produced by R. solani AG2 had an intermediate effect on the VOCs production in Serratia (Figure 2A). Eight VOCs (six had a predicted antimicrobial function) were upregulated, and five were downregulated due to the contact with fungal volatiles. L. maculans MB 158 VOCs had the weakest effect on the bacterial VOCs production. Only five substances (three of them predicted to be antimicrobial) were upregulated in the bacterium. No downregulation of VOCs production in Serratia due to L. maculans volatiles was observed (Figure 2A). The detailed information on the specific volatiles produced by S. plymuthica HRO-C48 as a response to treatments with fungal VOCs is provided in the Supplementary File S1.
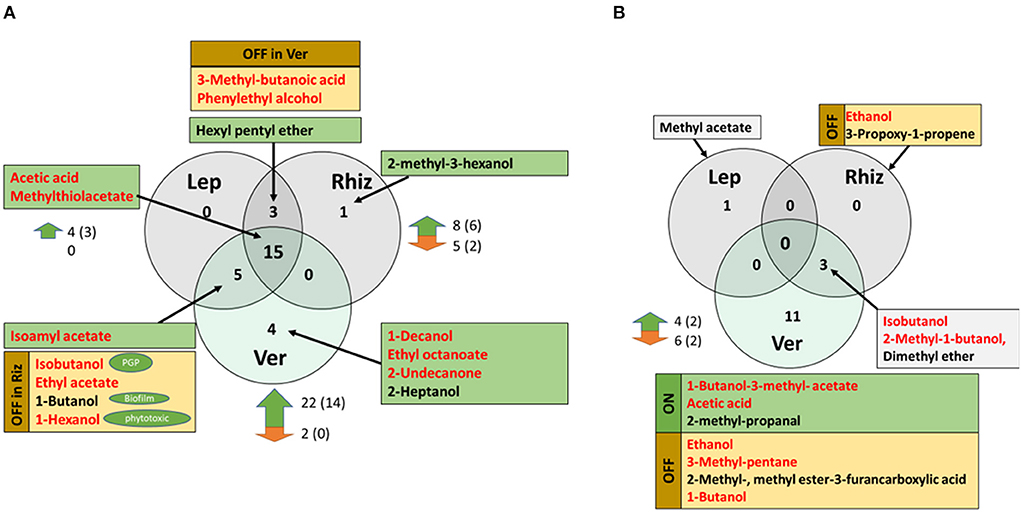
Figure 2. (A) Common and unique VOCs which were produced by S. plymuthica HRO-C48 when incubated with Rhizoctonia solani AG2 (Rhiz), Leptospheria maculans MB 158 (Lep) and Verticillium longisporum ELV 43 (Ver), respectively. The substances that were not found in the untreated control, thus were “switched on” in any of the treatments are highlighted in green. The substances that were switched off in any of the treatments but found in other treatment(s) and in the control, are highlighted in yellow and indicated with the “OFF” sign for a respective treatment. Red font indicates that the substances were mentioned in the scientific literature as antimicrobial, while other predicted functions are indicated in green ovals (“PGP” = plant growth promoting, “Biofilm” = effect on the biofilm formation; “phytotoxic” – predicted toxic effect on plants). Black arrows point to the respective set these substances belong to. The numbers to the right of the green and red arrows mean the number of strongly (significant or over 100%) up- or downregulated volatiles due to the treatment with fungal volatiles, respectively. The number of substances that may have antimicrobial function are shown in brackets. For references please see Supplementary Table S3. (B) Common and unique VOCs that were produced by R. solani AG2 (Rhiz), L. maculans MB 158 (Lep) and V. longisporum ELV 43 (Ver), when incubated with S. plymuthica HRO-C48 VOCs. The substances that were not found in the untreated control (“switched on” in any of the treatments) are highlighted in green and indicated with “ON”. The substances that were switched off in any of the treatments but found in other treatment(s) and in the control, are highlighted in yellow and indicated with the “OFF” sign for the respective treatments. Gray boxes indicate that the respective substances were also detected in the untreated control. Red font indicates that the substances were mentioned in the scientific literature as antimicrobial or inducing some responses in microorganisms. The arrows point to the respective set these substances belong to. The numbers to the right of the green and red arrows indicate the number of strongly (significant or over 100%) up- or downregulated volatiles due to the treatment with bacterial volatiles, respectively. The amount of the substances that may have antimicrobial function are shown in brackets. For references please see Supplementary Table S4.
Altogether, V. longisporum ELV 43 VOCs showed the strongest effect on VOC production in S. plymuthica HRO-C48 and L. maculans MB 158 the weakest.
There were no common VOCs produced by all three fungi. This was mostly due to the fact, that in the headspace of L. maculans MB 158 samples only one volatile, methyl acetate, was identified (Figure 2B and Supplementary Table S4). It was found in both treated and the untreated control of the fungus. V. longisporum ELV 43 produced the highest number of volatiles among the three tested fungi. Out of 14 VOCs that were detected in the volatilome of V. longisporum four were strongly upregulated and six downregulated due to the contact with S. plymuthica HRO-C48 volatiles (Figure 2B). Three substances were identified in the volatilome of R. solani AG2: isobutanol, 2-methyl-1-butanol, and dimethyl ether (a substance with unknown function). All three substances were shared with those produced by V. longisporum ELV 43 (Figure 2B). Two further substances were completely downregulated in the volatilome of R. solani AG2 due to the contact with S. plymuthica HRO-C48. Six VOCs were either completely switched off due to the contact with bacterial volatiles in V. longisporum, or significantly downregulated (Figure 2B). The detailed information on the specific volatiles produced by the three tested fungi as a response to treatment with bacterial VOCs and their predicted effects on the microorganisms is provided in the Supplementary File S1.
To investigate the response of S. plymuthica HRO-C48 to the volatiles emitted by V. longisporum ELV 43, R. solani AG2 and L. maculans MB 158, the changes at the transcriptome level after 3 days of exposure to fungal VOCs were compared. We identified 339 differentially expressed genes (DEGs, significance based on foldchange ≥ (±1, 5) and with p-adjusted values ≤ 0.01, which corresponds to 3% of the total detected transcripts (4990). From these 339 DEGs, 149 were up- and 190 were downregulated (Figure 3A). The strongest difference in the regulation of genes was found between the treatments of S. plymuthica HRO-C48 with R. solani and L. maculans volatiles (Figure 3B). The total number of upregulated genes due to the contact with the R. solani VOCs was significantly higher than that of the downregulated genes (230 vs. 88 genes, respectively). The co-incubation of the S. plymuthica HRO-C48 with L. maculans VOCs, on the other hand, resulted in a much stronger downregulation, rather than upregulation of the genes (276 vs. 90 genes, respectively). The interaction of S. plymuthica HRO-C48 with V. longisporum VOCs resulted in a significant gene downregulation as compared to upregulation (187 vs. 133, respectively). V. longisporum and R. solani shared most of the upregulated genes (Figure 3B), while V. longisporum and L. maculans share most of the downregulated genes (Figure 3C). Thirty-nine DEGs were common among the genes upregulated, while 63 genes were significantly downregulated in S. plymuthica HRO-C48 due to the contact with the VOCs of all three fungi (Figure 3B). Some of the upregulated genes were predicted to code for phage shock proteins, or proteins involved in stress response and virulence, such as for example, Type III secretion system lipoprotein chaperone (Tosi et al., 2011; Horstman and Darwin, 2012; Dunstan et al., 2013). Among the downregulated genes those coding for membrane proteins, multiple stress resistance proteins, murein hydrolase proteins and putative Ferritin-like protein were identified. Downregulation of these proteins is generally associated with cellular stress, decrease of cellular movement, enhancement of biofilm formation processes and protection of DNA against enzymatic or oxidative attack (Smith, 2004; Zhang et al., 2007; Johansen et al., 2008; Vollmer et al., 2008). A list of all genes which were up- or downregulated in response to confrontation with volatiles from the pathogenic fungi: R. solani AG2, L. maculans MB 158 and V. longisporum ELV 43 and detailed information on these genes have been provided in Supplementary Table S2 and Supplementary File S1.
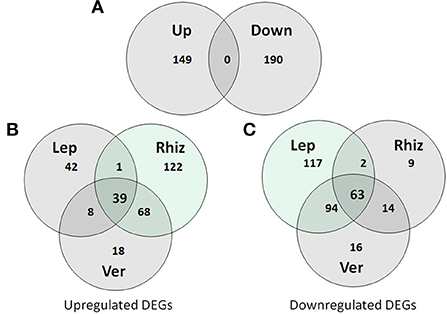
Figure 3. (A) Total distribution of up- and downregulated genes (DEGs) in the treatments of S. plymuthica HRO-C48 with fungal VOCs. Up- (B) and down- (C) regulation of the genes in S. plymuthica HRO-C48 due to the contact with the VOCs of R. solani AG2 (Rhiz), L. maculans MB 158 (Lep) and V. longisporum ELV 43 (Ver). The numbers in the circles indicate the number of DEGs due to the contact with the volatiles of each fungal strain. The light green color indicates the treatment resulting in the highest amount of the up- or downregulated transcripts among the three conditions tested.
A strong downregulation of gene expression assigned to almost all functions in S. plymuthica HRO-C48 that was subjected to VOCs from V. longisporum ELV43 or L. maculans MB158 was observed, while treatments with R. solani AG2 VOCs resulted rather in upregulation of genes associated with these functions (Figure 4). This difference was especially strong in the DEGs assigned to the processing of genetic information. Among those, functions of 130 DEGs were assigned to the expression of ribosomal RNA (rRNA). Thirty-nine of the rRNA genes were upregulated due to exposure to R. solani VOCs, while 81 were downregulated due to exposure of S. plymuthica HRO-C48 to V. longisporum and L. maculans VOCs (Supplementary Table S2). Among the DEGs, functions involving cellular metabolism, cell wall structure biogenesis, cellular motility, defense mechanisms, and intra and extracellular processes were also mainly downregulated due to the contact with V. longisporum and L. maculans VOCs and upregulated upon R. solani VOCs treatments. The detailed descriptions of all DEGs, their differential expression due to the treatments as well as their predicted functions are summarized in the Supplementary Table S2 for upregulated and downregulated genes, respectively.
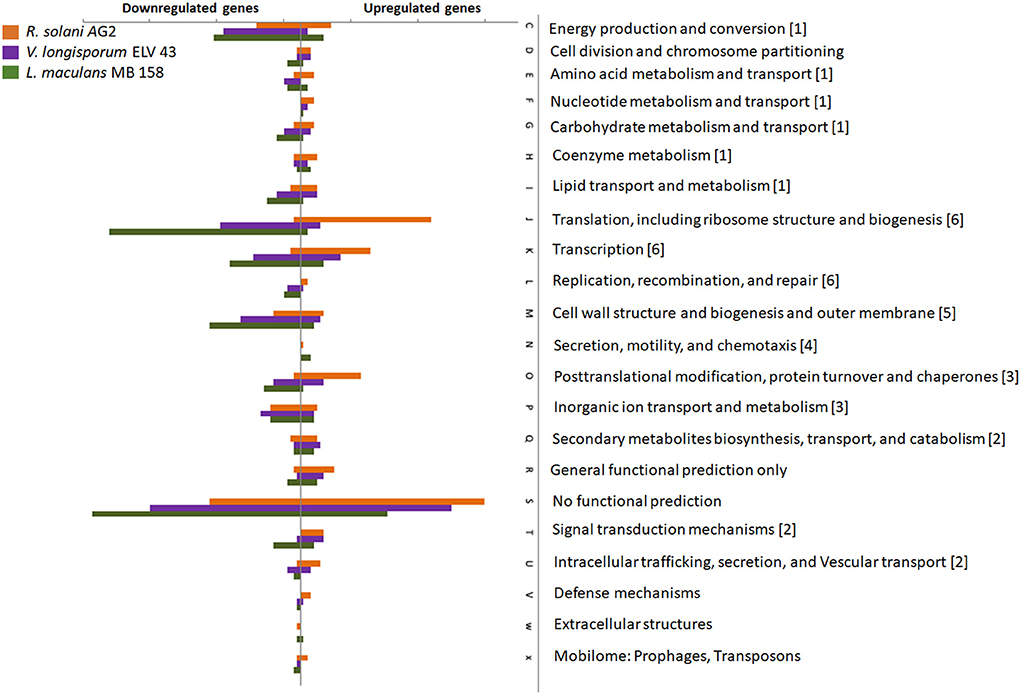
Figure 4. Impact of fungal volatiles on gene expression in S. plymuthica HRO-C48. The number of genes with at least 1.5-fold change in relation to the untreated control is depicted. If a transcript was assigned to multiple classes, it was included in each of the assigned classes' counts. Orange, violet, and green bars indicate genes with significantly changed expression due to the contact with R. solani AG2, V. longisporum ELV43 or L. maculans MB158 VOCs, respectively. The letters represent different COG functions, while numeric figures appended at the end of each COG function represent the functional classification of differentially expressed genes (DEGs). The genes in the diagram correspond to the genes listed in the Supplementary Table S2. The numbers in brackets correspond to the following functional annotations: 1: metabolism, 2: Extracellular communication, 3: Intracellular processes, 4: secretion, motility, and chemotaxis, 5: cellular structure, 6: genetic information processing.
We concluded a manual search for DEGs involved in selected functions with the focus on virulence, stress response, biofilm formation and cellular motility and identified 13 genes that are mentioned in the literature as being associated with virulence. R. solani VOCs resulted in a much stronger upregulation of predicted virulence-associated genes than downregulation. In the samples treated with V. longisporum and L. maculans VOCs, the expression of virulence-associated genes was on the other hand often downregulated (Supplementary Table S5). Only one DEG putatively involved in the production of secondary metabolites was found (SOD10_RS02040: acyl carrier protein putatively involved in fatty acid biosynthesis), while two other identified genes were neither downregulated nor upregulated (Supplementary Table S5). Additionally, we specifically searched the complete transcriptome of S. plymuthica HRO-C48 for the known antibiotic pyrrolnitrin and sodorifen biosynthesis clusters. One gene, SOD10_RS11220 (coding for a monodechloroaminopyrrolnitrin synthase PrnB) related to the pyrrolnitrin biosynthetic cluster (Pawar and Chaudhari, 2020) was insignificantly upregulated in all three treatments (Supplementary Table S5). Two further genes, SOD10_RS18500 and SOD10_RS19890 (both coding for isopentenyl-diphosphate isomerase) putatively belong to the sodorifen biosynthetic gene cluster (Domik et al., 2016; Schmidt et al., 2017). The expression of only one of them, SOD10_RS19890, was significantly upregulated due to R. solani AG2 volatiles.
Eight DEGs were identified that are putatively involved in cell motility and biofilm formation (Supplementary Table S5). As previously mentioned, the VOCs of all three fungal pathogens enhanced processes involved in biofilm formation the decrease in cell motility of S. plymuthica HRO-C48. Additional search for motility-associated DEGs indicated an increase of cellular motility in the bacteria due to contact with L. maculans MB158 and V. longisporum ELV 43 VOCs as shown by downregulation of genes involved in the Cpx response (Vogt and Raivio, 2012). Same was true for R. solani AG2 VOCs that resulted in upregulation of a gene coding for the Sec-independent protein secretion pathway components putatively involved in secretion of pili and flagella (Kostakioti et al., 2005). Such an upregulation may facilitate an increase in cellular motility.
Twenty-five genes involved in stress response were significantly up or downregulated in S. plymuthica HRO-C48 due to the treatment with VOCs of R. solani AG2, L. maculans MB158 or V. longisporum ELV 43 (Supplementary Table S5). In addition to the downregulation of the predicted multiple stress resistance proteins BhsA, outer membrane lipoprotein, Ferritin-like protein and upregulation of phage shock proteins B in all three treatments with fungal volatiles which is associated with cellular stress, several other stress-associated changes in S. plymuthica HRO-C48 transcriptome were found. For example, a significant downregulation of four genes involved in protection against oxidative stress was observed.
A significant upregulation of five genes involved in the TA system only in samples treated with R. solani AG2 VOCs was found. Both, toxin (CcdB and pndA protein) and the post-segregation antitoxin were upregulated in S. plymuthica HRO-C48 (Supplementary Table S5). The TA system serves to stabilize the plasmid within a bacterial population. TA system-associated genes are activated by antibiotic or heat stress. The plasmid maintenance protein CcdB causes growth arrest when expressed at low levels, while its high levels cause cell death (Tripathi et al., 2012).
A detailed analysis of the DEGs involved in virulence, stress response, biofilm formation and cellular motility is provided in the Supplementary File 1 and Supplementary Table S5.
In the present study, we showed how the VOCs of the biocontrol agent S. plymuthica HRO-C48 are influenced by volatiles produced by the plant pathogenic fungi R. solani AG2, L. maculans MB 158 and V. longisporum ELV 43, and vice versa. We discovered a strong species-specific dialogue on both volatilome and transcriptome levels that resulted in various degrees of fungal growth repression.
Our study showed that the core volatilome of S. plymuthica HRO-C48 consists of 15 volatiles under the tested conditions, much more than described previously (Kai et al., 2007). Only six of them were identified in the volatilomes of five other Serratia spp., while many others were present in other species but not detected in our experiments (Weise et al., 2014). Interestingly, all VOCs that were found to be common among six Serratia species possess strong antimicrobial properties. Recently, the volatile antibiotic sodorifen was identified as an important contributor to the mode of action of S. plymuthica spp. (Domik et al., 2016; Schmidt et al., 2017), however, no sodorifen was detected in the S. plymuthica HRO-C48 volatilome under any testing conditions. Transcriptomic analysis showed a low, non-significant upregulation of two genes coding for isopentenyl-diphosphate that may be attributed to the sodorifen cluster. We therefore conclude that S. plymuthica HRO-C48 is not an active sodorifen producer under the conditions that were implemented.
The growth of all three fungi tested was inhibited by S. plymuthica HRO-C48 volatiles, while only R. solani AG2 growth inhibition was significant. In addition to the fact that three thirds of the VOCs produced by S. plymuthica HRO-C48 are mentioned in the literature as antimicrobial substances (Supplementary Table S3), these results provide further evidence that VOCs significantly contribute to the biocontrol effect of S. plymuthica. The production of acetic acid and methylthiolacetate was induced by the VOCs of all three fungi tested. These strong antimicrobial substances (Pimenta et al., 2012; Ossowicki et al., 2017) were not previously detected in Serratia volatilomes (Weise et al., 2014) and were not present in the untreated S. plymuthica HRO-C48 samples. Activation of acetic acid and methylthiolacetate production by the volatiles of all three fungi suggests that they belong to the common defense mechanism of S. plymuthica HRO-C48 against volatile-mediated fungal attacks.
We found that S. plymuthica HRO-C48 influenced the growth of the fungi, and that it was strongly influenced by the fungal VOCs that were perceived as signals by the biocontrol agent. Fungal VOCs influenced the expression of genes encoding proteins of various cellular processes in HRO-C48. The common response of S. plymuthica HRO-C48 to the VOCs of all three fungi was a change in expression of genes involved in response to stress, such as envelope damage, biofilm formation and motility-associated genes. A general stress response in microorganisms is an evolutionary tool acting on a global level to provide protection against various stresses (Guan et al., 2017). A transition from single cells to a biofilm and loss of flagellar motility in bacteria is also associated with environmental stress (Guttenplan and Kearns, 2013). Four genes coding for phage-shock proteins were upregulated in at least one of the treatments with fungal volatiles. They belong to the stress response system that prevents lethal cytoplasmic membrane permeability (Darwin, 2005; Horstman and Darwin, 2012). Decreased permeability of the cell membrane in its turn prevents the influx of antimicrobials into the cell, thus protecting it from even greater harm. Interestingly, a foregoing study found that VOCs emitted by F. culmorum made the cell wall of S. plymuthica PRI-2C more permeable for VOCs to enter and be taken up by the cell (Schmidt et al., 2017). Increase in cell wall permeability in PRI-2C was also accompanied by a significant increase in bacterial growth when exposed to VOCs emitted by the fungal pathogen F. culmorum (Schmidt et al., 2017). This indicates a rather positive effect of F. culmorum VOCs on Serratia spp. compared to the stress-inducing effects that were observed with the fungi selected for our study. Based on these observations, we suggest that stress response, biofilm formation, decrease of cellular motility and production of antifungal substances acetic acid and methylthiolacetate belong to the general response of S. plymuthica HRO-C48 upon contact with potentially damaging fungal volatiles.
We observed that S. plymuthica HRO-C48 was strongly influenced by the VOCs of the fungal pathogens with significant changes in gene expression levels under the conditions chosen. A significant up or downregulation of 3% of all detected transcripts due to the contact with fungal volatiles was found. The overall pattern of how gene expression was influenced by fungal volatiles was very much dependent on the fungal volatiles to which the bacterium was exposed. While the expression of most of the genes associated with energy metabolism, stress response, motility, biofilm formation and virulence was upregulated in S. plymuthica HRO-C48 due to contact with R. solani VOCs, genes associated with the same functions were mainly downregulated when treated with V. longisporum and L. maculans VOCs (Figure 5). Three VOCs, two of them with known antimicrobial properties, were detected in the volatilome of R. solani AG2 exposed to S. plymuthica HRO-C48 volatiles. These VOCs induced the strongest differential gene expression levels (230 up vs. 88 downregulated DEGs), in comparison to the other two fungi studied. R. solani AG2 VOCs activated genes responsible for stress defense, virulence, and biofilm formation, but also those involved in a general metabolism, cellular motility, and signal transduction (especially rRNA coding genes and those involved in TA system) in the bacterium. Schmidt et al. (2017) also observed an upregulation of the expression of DEGs involved in the same functions in S. plymuthica PRI-2C following the exposure to F. culmorum VOCs (Schmidt et al., 2017). A significant upregulation of genes whose products were involved in general metabolism, including carbon, energy, amino acids and nucleic acids, and their transporters was also observed by Neupane et al. (2015) for Serratia proteamaculans S4 upon a direct contact with R. solani (Neupane et al., 2015). Genes coding for the antibiotic pyrrolnitrin biosynthesis and transporters were 8 to 14-fold upregulated in S. proteamaculans S4 and S. plymuthica AS13 (Neupane et al., 2015). The involvement of pyrrolnitrin produced by Serratia spp. and Pseudomonas spp. in the inhibition of various fungal plant pathogens has been shown on several occasions (Müller et al., 2009; Neupane et al., 2015; Pawar and Chaudhari, 2020). In our study in which S. plymuthica HRO-C48 was exposed to R. solani AG2 volatiles, only an insignificant increase (a log fold change of 0.15) of the gene expression coding for monodechloroaminopyrrolnitrin synthase PrnB was detected (Supplementary Table S2). On one hand this observation may be due to isolate-specific differences in the mode of action between the AS13 and HRO-C48 strains. On the other hand, it may reflect the ability of Serratia spp. to sense whether the enemy is confronted via direct contact or via its volatiles and thus upregulate the synthesis of soluble antibiotics only when the direct contact is anticipated.
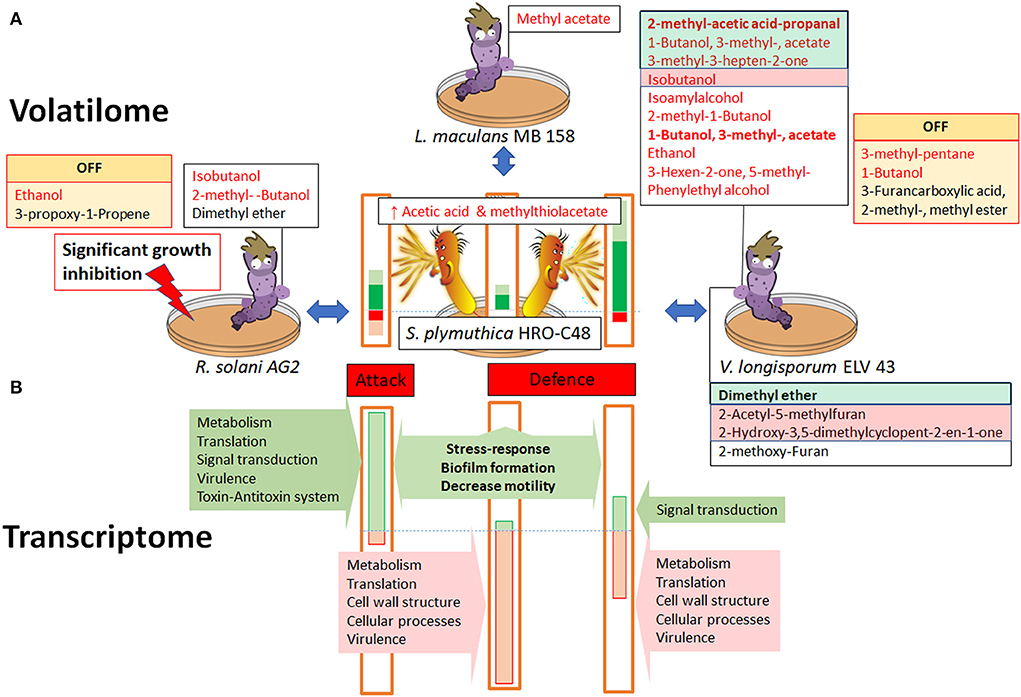
Figure 5. Volatile in vitro interactions between S. plymuthica HRO-C48 and the fungal plant pathogens R. solani AG2, L. maculans MB 158 and V. longisporum ELV 43. Panel (A) shows the interactions on the level of the volatilome and growth inhibition. Significantly up- or downregulated volatiles produced by fungi are highlighted in green and red, respectively. Those volatiles that were “switched off” in the respective volatilome compared to the untreated control, are labeled with “OFF” sign. Changes in the bacterial volatilome are shown schematically as colored bars (green upregulated, red downregulated). The heights of the bars correspond with the proportion of up- (green) or down (red) regulated VOCs, while VOCs with predicted antimicrobial effects are shown in dark green or red, respectively. Panel (B) shows the proportion of up- and downregulated genes involved in different metabolic functions in S. plymuthica HRO-C48 due to exposure to volatiles of the three selected fungi as detected by the transcriptomic analysis. The dashed lines indicate the borders between up and downregulated substances. Please see corresponding Supplementary Tables S2–S4 for the exact substance names and references. The correspondence of the gene and VOCs expression changes to the “defense” and “attack” modes was extrapolated based on the available literature.
On the volatilome level, the VOCs of R. solani AG2 induced a rather moderate response in the bacterium, compared to the changes due to exposure to L. maculans MB 158 and V. longisporum ELV 43 VOCs (Figures 2A, 5). Bacterial volatiles not only significantly inhibited the growth of R. solani AG2, they also induced a complete downregulation of two out of five volatiles produced by R. solani grown alone. Based on those observations we speculate that R. solani AG2 VOCs boost the cellular processes in the biocontrol agent, especially those involved in attacking other microbes. Such active bacterial attacks result in the significant inhibition of fungal growth by the biocontrol agent. Because many of the microbial VOCs are by-products of primary metabolism, the significant downregulation of the volatilome in AG2 may also be regarded as the consequence of stress induced by bacterial volatiles. Literature sources describe some microbial VOCs as important carbon sources for other microorganisms or as substances with positive effects on the growth of neighboring bacteria in the rhizosphere (Schulz-Bohm et al., 2017). A boosting effect on the S. plymuthica PRI-2C metabolism due to exposure to the F. culmorum VOCs was also reported by Schmidt et al. (2017). The authors observed, among other findings, that PRI-2C exposed to fungal VOCs induced the production of sodorifen, an antimicrobial substance most probably used by the bacterium to attack other microorganisms (Schmidt et al., 2017). It is therefore also possible to speculate that some of the R. solani AG2 VOCs are attack signals for S. plymuthica HRO-C48, and serve additionally as a source of nutrition that boost its metabolism.
Naznin et al. (2013) indicated an emission of two substances, 2-methyl-propanol and 3-methyl-butanol by a plant growth promoting Leptosphaeria sp. GS8-3 (Naznin et al., 2013). Our study identified only one volatile substance, methyl acetate, with a sufficient certainty in the volatilome of L. maculans MB 158. It was not detected in the volatilomes of the two other tested fungi, however it is mentioned in literature as a typical fungal VOC (Plaszkó et al., 2020). Methyl acetate is usually metabolized by various pro- and eukaryotes into its toxic compounds methanol and acetic acid by esterases and has a half maximal effective concentration of 6 g l−1 for microorganisms (Echa, 2022). Exposure to L. maculans MB 158 VOCs (most probably, to methyl acetate) resulted in the strongest downregulation of genes involved in metabolism and cellular functioning in HRO-C48 compared to the other tested conditions (276 downregulated vs. 90 upregulated DEGs). The functions of the downregulated genes included those involved in cellular metabolism, translation (especially, rRNA genes expression), cell wall structure and virulence. A similar transcriptional response pattern where highly translated transcripts with high copy numbers (e.g. those of rRNA genes) were significantly downregulated was also observed in the transcriptome of Escherichia coli exposed to osmotic upshift (Bartholomäus et al., 2016). It was suggested by the authors that such a relocation of a transcriptional focus allows the cell to reallocate translation resources to non-degraded and newly synthesized mRNAs. Despite strong repression of most of the DEGs by L. maculans MB 158 VOCs, the volatile production in the bacterium was enhanced with more than half of upregulated VOCs possessing antimicrobial properties. In comparison to the effects of the three fungi tested, L. maculans induced the weakest response on the bacterial volatilome. We therefore conclude that methyl acetate produced by L. maculans was the most deleterious VOC among all detected fungal volatiles in our study, while the fungus itself was only mildly affected by the bacterial VOCs. We speculate that the exposure to the MB 158 volatilome shifted the metabolism of S. plymuthica HRO-C48 into the defense mode via downregulation of genes involved in cellular functioning, while the defense mechanism on the volatile level was upregulated.
The strongest change in the volatilome of S. plymuthica was induced by V. longisporum ELV 43 VOCs. Of the total 26 detected VOCs, 14 were strongly upregulated or even switched on in S. plymuthica HRO-C48 upon contact with V. longisporum VOCs, while only two substances were downregulated (Figure 2A). In addition, the V. longisporum ELV 43 volatilome was affected the strongest by bacterial volatiles, compared to the other two fungi tested (Figure 2B). Both interacting microorganisms significantly changed the numbers of the produced antimicrobial VOCs as a reaction to one another, indicating an active dialogue on the volatile level. A comparison to a foregoing study where V. longisporum ELV 43 and another biocontrol agent P. polymyxa Sb3-1 interacted with each other via their volatiles (Rybakova et al., 2017b) suggests that the volatile interaction strongly depends on the interacting bacterial partner. While V. longisporum was inhibited in growth by the volatiles of both biocontrol agents, this inhibition was only significant when ELV 43 and Sb3-1 interacted via their VOCs for 9 days (Rybakova et al., 2017b). Only three identified VOCs produced by ELV 43 were common between the studies. Among them were the antimicrobial VOCs isobutanol and isoamyl alcohol (Supplementary Table S4). Their production was enhanced when the fungus was confronted with S. plymuthica VOCs, while P. polymyxa volatiles did not significantly influence their production. The production of 1-butanol was switched off in V. longisporum due to the contact with S. plymuthica VOCs and upregulated upon contact with P. polymyxa volatiles. Only isobutanol was produced under all conditions tested: 3 and 6 days of co-incubation with Sb3-1 and 3 days incubation with HRO-C48 as well as in all non-incubated controls. When we compared and contrasted our own results with those of previous studies, we observed that V. longisporum ELV 43 volatilome is substantially influenced by bacterial volatiles as well as by environmental conditions. The ability of V. longisporum to react to various stresses via adaptation of its volatilome appears to be one of the possible factors that make the development of successful biological measures against Verticillium wilt so challenging (Rybakova et al., 2020).
The changes on the transcriptional level in S. plymuthica HRO-C48 due to the contact of V. longisporum ELV 43 VOCs involved higher gene downregulation than upregulation (187 vs. 133, respectively). We found that the expression patterns of S. plymuthica HRO-C48 exposed to V. longisporum ELV 43 VOCs were similar to those exposed to L. maculans MB 158 volatiles, with 94 repressed DEGs shared between the transcriptomes of the two fungal pathogens. For example, a large number of genes coding for highly translated transcripts, such as rRNAs were downregulated in HRO-C48 due to the contact with both V. longisporum ELV 43 and L. maculans MB 158 VOCs. Genes involved in virulence were also downregulated in S. plymuthica HRO-C48 exposed to V. longisporum ELV 43 VOCs. The majority of the upregulated DEGs shared between V. longisporum ELV 43 and R. solani AG2 VOCs treatments (15 out of 68 upregulated DEGs) were either associated with stress response or had no known functions assigned to them. We therefore suggest that the volatile-mediated interaction between S. plymuthica HRO-C48 and V. longisporum ELV 43 in our experimental approach may be best described as an active competition. This competition involves the mobilization of all resources by the bacterium and an active production of potentially antifungal volatile substances by both microorganisms.
Our data suggests that VOCs-mediated interactions of S. plymuthica HRO-C48 with R. solani AG2 induced attack mode, while L. maculans MB 158 and V. longisporum ELV 4 VOCs induced defense mode in the bilateral dialogues. We are aware that growth stages of the bacteria and fungi may have an effect on the interaction patterns and that our data only reflects the in vitro interactions between the implemented microbial species. Our results may therefore not reveal the true ecological roles of the detected VOCs in rhizosphere soil or in the host plant. Under natural conditions the VOCs of the surrounding microbiota, as well as plant derived volatiles and a variety of VOCs distribution patterns through the soil particles influence the inter-species dialogues. Nevertheless, we propose that deciphering the peculiarities of the volatile dialogues between the biological agents and their plant pathogenic counteragents in vitro will support future research and contribute to improved understanding of the complexity of the mode of action of biocontrol agents on the molecular level. These novel insights will ultimately assist in the development of next generation biological control solutions for sustainable agriculture.
he datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: https://www.ncbi.nlm.nih.gov/bioproject/?term=PRJNA874900.
GB, HM, and DR designed the study. HM analyzed transcriptomic raw data and assisted with data interpretation. EO assisted with data analysis and figures. AS analyzed GC-MS data. TC assisted with the manuscript writing and data interpretation. DR wrote the manuscript with input from the other authors. All authors read and approved the final manuscript.
This project was partly funded by the European Union in frame of FP7-KBBE-2013-7-single-stage (BIOCOMES; No. 612713), the European Union Horizon 2020 Research and Innovation (EXCALIBUR; No.81794), and by the Austrian Research Promotion Agency (FFG; No. 836466). TC's involvement has been supported by the Federal Ministry of Science, Research and Economy (BMWFW), the Federal Ministry of Traffic, Innovation and Technology (bmvit), the Styrian Business Promotion Agency SFG, the Standortagentur Tirol, the Government of Lower Austria and ZIT - Technology Agency of the City of Vienna through the COMET-Funding Program managed by the 16FFG (grant number 282482).
The authors gratefully acknowledge support from NAWI Graz. We are also grateful to Riccardo Mancinelli (Graz, Austria) and Isabell M. Trinkl (Graz, Austria) for their assistance and Jürgen Köhl (Wageningen, Netherlands) for discussions in frame of BIOCOMES.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.1020634/full#supplementary-material
Abuamsha, R., Salman, M., and Ehlers, R. U. (2011). Effect of seed priming with Serratia plymuthica and Pseudomonas chlororaphis to control Leptosphaeria maculans in different oilseed rape cultivars. Eur. J. Plant Pathol. 130, 287–295. doi: 10.1007/s10658-011-9753-y
Bakker, P. A. H. M., Berendsen, R. L., Van Pelt, J. A., Vismans, G., Yu, K., Li, E., et al. (2020). (2020). The soil-borne identity and microbiome-assisted agriculture: looking back to the future. Mol. Plant 13, 1394–1401. doi: 10.1016/j.molp.2020.09.017
Banerjee, S., Schlaeppi, K., and van der Heijden, M. G. A. (2018). Keystone taxa as drivers of microbiome structure and functioning. Nat. Rev. Microbiol. 16, 567–576. doi: 10.1038/s41579-018-0024-1
Bartholomäus, A., Fedyunin, I., Feist, P., Sin, C., Zhang, G., Valleriani, A., et al. (2016). Bacteria differently regulate mRNA abundance to specifically respond to various stresses. Philos. Trans. R. Soc. Math. Phys. Eng. Sci. 374, 20150069. doi: 10.1098/rsta.2015.0069
Berg, G. (2019). Microbiome definition re-visited: old concepts and new recommendations. Microbiome. 8, 103. doi: 10.1186/s40168-020-00875-0
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Cantalapiedra, C., Hernández-Plaza, A., Letunic, I., Bork, P., and Huerta-Cepas, J. (2021). eggNOG-mapper v2: functional annotation, orthology assignments, and domain prediction at the metagenomic scale. Mol. Biol. Evol. 1, 38. doi: 10.1101/2021.06.03.446934
Cernava, T., Aschenbrenner, I. A., Grube, M., Liebminger, S., and Berg, G. (2015). A novel assay for the detection of bioactive volatiles evaluated by screening of lichen-associated bacteria. Front. Microbiol. 6, 398. doi: 10.3389/fmicb.2015.00398
Darwin, A. J. (2005). The phage-shock-protein response. Mol. Microbiol. 57, 621–628. doi: 10.1111/j.1365-2958.2005.04694.x
Domik, D., Magnus, N., and Piechulla, B. (2016). Analysis of a new cluster of genes involved in the synthesis of the unique volatile organic compound sodorifen of Serratia plymuthica 4Rx13. FEMS Microbiol. Lett. 7, 363. doi: 10.1093/femsle/fnw139
Dunstan, R. A., Heinz, E., Wijeyewickrema, L. C., Pike, R. N., Purcell, A. W., Evans, T. J., et al. (2013). Assembly of the type ii secretion system such as found in Vibrio cholerae depends on the novel pilotin AspS. PLOS Pathog. 9, e1003117. doi: 10.1371/journal.ppat.1003117
Echa, R. D. (2022). Methyl acetate. Available online at: https://echa.europa.eu/registration-dossier/-/registered-dossier/15222/6/2/8 (accessed February 18, 2021).
Frankowski, J., Lorito, M., Scala, F., Schmid, R., Berg, G., Bahl, H., et al. (2001). Purification and properties of two chitinolytic enzymes of Serratia plymuthica HRO-C48. Arch. Microbiol. 176, 421–426. doi: 10.1007/s002030100347
Grosch, R., Faltin, F., Lottmann, J., Kofoet, A., and Berg, G. (2005). Effectiveness of 3 antagonistic bacterial isolates to control Rhizoctonia solani Kühn on lettuce and potato. Can. J. Microbiol. 51, 345–353. doi: 10.1139/w05-002
Guan, N., Li, J., Shin, H., Du, G., Chen, J., Liu, L., et al. (2017). Microbial response to environmental stresses: from fundamental mechanisms to practical applications. Appl. Microbiol. Biotechnol. 101, 3991–4008. doi: 10.1007/s00253-017-8264-y
Gugel, R., and Petrie, G. (1992). History, occurrence, impact, and control of blackleg of rapeseed. Can. J. Plant Pathol. 14, 36–45. doi: 10.1080/07060669209500904
Guttenplan, S. B., and Kearns, D. B. (2013). Regulation of flagellar motility during biofilm formation. FEMS Microbiol. Rev. 37, 849–871. doi: 10.1111/1574-6976.12018
Horstman, N. K., and Darwin, A. J. (2012). Phage shock proteins B and C prevent lethal cytoplasmic membrane permeability in Yersinia enterocolitica. Mol. Microbiol. 85, 445–460. doi: 10.1111/j.1365-2958.2012.08120.x
Huerta-Cepas, J., Szklarczyk, D., Heller, D., Hernández-Plaza, A., Forslund, S. K., Cook, H., et al. (2019). eggNOG 5.0: a hierarchical, functionally and phylogenetically annotated orthology resource based on 5090 organisms and 2502 viruses. Nucleic Acids Res. 47, D309–D314. doi: 10.1093/nar/gky1085
Johansen, J., Eriksen, M., Kallipolitis, B., and Valentin-Hansen, P. (2008). Down-regulation of outer membrane proteins by noncoding RNAs: unraveling the cAMP–CRP-and σE-dependent CyaR–ompX regulatory case. J. Mol. Biol. 383, 1–9. doi: 10.1016/j.jmb.2008.06.058
Kaczmarek, J., Latunde-Dada, A. O., Irzykowski, W., Cools, H. J., Stonard, J. F., Brachaczek, A., et al. (2014). Molecular screening for avirulence alleles AvrLm1 and AvrLm6 in airborne inoculum of Leptosphaeria maculans and winter oilseed rape (Brassica napus) plants from Poland and the UK. J. Appl. Genet. 55, 529–539. doi: 10.1007/s13353-014-0235-8
Kai, M., Effmert, U., Berg, G., and Piechulla, B. (2007). Volatiles of bacterial antagonists inhibit mycelial growth of the plant pathogen Rhizoctonia solani. Arch. Microbiol. 187, 351–360. doi: 10.1007/s00203-006-0199-0
Kalbe, C., Marten, P., and Berg, G. (1996). Strains of the genus Serratia as beneficial rhizobacteria of oilseed rape with antifungal properties. Microbiol. Res. 151, 433–439. doi: 10.1016/S0944-5013(96)80014-0
Karapapa, V., Bainbridge, B., and Heale, J. (1997). Morphological and molecular characterization of Verticillium longisporum comb. nov., pathogenic to oilseed rape. Mycol. Res. 101, 1281–1294. doi: 10.1017/S0953756297003985
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10, 845. doi: 10.3389/fpls.2019.00845
Kostakioti, M., Newman, C. L., Thanassi, D. G., and Stathopoulos, C. (2005). Mechanisms of protein export across the bacterial outer membrane. J. Bacteriol. 187, 4306–4314. doi: 10.1128/JB.187.13.4306-4314.2005
Kurze, S., Bahl, H., Dahl, R., and Berg, G. (2001). (2001). Biological Control of Fungal Strawberry Diseases by Serratia plymuthica HRO-C48. Plant Dis. 85, 529–534. , 5.529. doi: 10.1094/PDIS.2001.85.5.529
Langmead, B., and Salzberg, S. L. (2012). Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359. doi: 10.1038/nmeth.1923
Liu, X., Bimerew, M., Ma, Y., Müller, H., Ovadis, M., Eberl, L., et al. (2007). Quorum-sensing signaling is required for production of the antibiotic pyrrolnitrin in a rhizospheric biocontrol strain of Serratia plymuthica. FEMS Microbiol. Lett. 270, 299–305. doi: 10.1111/j.1574-6968.2007.00681.x
Lun, A. T. L., Chen, Y., and Smyth, G. K. (2016). It's DE-licious: A Recipe for Differential Expression Analyses of RNA-seq Experiments Using Quasi-Likelihood Methods in edgeR. Methods Mol. Biol. Clifton NJ 1418, 391–416. doi: 10.1007/978-1-4939-3578-9_19
Messner, R., Schweigrofler, W., Ibl, M., Berg, G., and Prillinger, H. (1996). Molecular characterization of the plant pathogen Verticillium dahliae Kleb. using RAPD-PCR and sequencing of the 18SrRNA-gene. J. Phytopathol. 144, 347–354. doi: 10.1111/j.1439-0434.1996.tb00305.x
MLA (1997). NIST Chemistry WebBook. 7th Edn. Washington, DC: National Institute of Standards and Technology.
Müller, H., and Berg, G. (2008). Impact of formulation procedures on the effect of the biocontrol agent Serratia plymuthica HRO-C48 on Verticillium wilt in oilseed rape. BioControl 53, 905–916. doi: 10.1007/s10526-007-9111-3
Müller, H., Westendorf, C., Leitner, E., Chernin, L., Riedel, K., Schmidt, S., et al. (2009). Quorum-sensing effects in the antagonistic rhizosphere bacterium Serratia plymuthica HRO-C48. FEMS Microbiol. Ecol. 67, 468–478. doi: 10.1111/j.1574-6941.2008.00635.x
Naznin, H. A., Kimura, M., Miyazawa, M., and Hyakumachi, M. (2013). Analysis of volatile organic compounds emitted by plant growth-promoting fungus Phoma sp. GS8-3 for growth promotion effects on tobacco. Microbes Environ. 28, 42–49. doi: 10.1264/jsme2.ME12085
Neupane, S., Finlay, R. D., Alström, S., Elfstrand, M., and Högberg, N. (2015). Transcriptional responses of the bacterial antagonist Serratia plymuthica to the fungal phytopathogen R hizoctonia solani. Environ. Microbiol. Rep. 7, 123–127. doi: 10.1111/1758-2229.12203
Oliveros, J. C. (2007). Venny. An interactive tool for comparing lists with Venn's diagrams. Available online at: https://bioinfogp.cnb.csic.es/tools/venny/index.html (accessed April 7, 2021).
Ossowicki, A., Jafra, S., and Garbeva, P. (2017). The antimicrobial volatile power of the rhizospheric isolate Pseudomonas donghuensis P482. PLoS ONE 12. doi: 10.1371/journal.pone.0174362
Pang, Y., Liu, X., Ma, Y., Chernin, L., Berg, G., Gao, K., et al. (2009). Induction of systemic resistance, root colonization and biocontrol activities of the rhizospheric strain Serratia plynmuthica are dependent on N-acyl homoserine lactones. Eur. J. Plant Pathol. 124, 261–268. doi: 10.1007/s10658-008-9411-1
Pawar, S. P., and Chaudhari, A. B. (2020). Pyrrolnitrin biosynthesis from rhizospheric Serratia spp. with antifungal activity and binding interactions of PrnF with ligands. doi: 10.21203/rs.3.rs-56362/v1
Pertea, M., Pertea, G. M., Antonescu, C. M., Chang, T.-.C, Mendell, J. T., et al. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat. Biotechnol. 33, 290–295. doi: 10.1038/nbt.3122
Pimenta, R. S., Moreira Da Silva, J. F., Buyer, J. S., and Janisiewicz, W. J. (2012). Endophytic fungi from plums (Prunus domestica) and their antifungal activity against Monilinia fructicola. J. Food Prot. 75, 1883–1889. doi: 10.4315/0362-028X.JFP-12-156
Plaszkó, T., Szucs, Z., Kállai, Z., Csoma, H., Vasas, G., Gonda, S., et al. (2020). Volatile Organic Compounds (VOCs) of Endophytic Fungi Growing on Extracts of the Host, Horseradish (Armoracia rusticana). Metabolites 10, 451. doi: 10.3390/metabo10110451
Rybakova, D., Mancinelli, R., Wikström, M., Birch-Jensen, A. S., and Postma, J. (2017a). The structure of the Brassica napus seed microbiome is cultivar-dependent and affects the interactions of symbionts and pathogens. Microbiome 5, 104. doi: 10.1186/s40168-017-0310-6
Rybakova, D., Rack-Wetzlinger, U., Cernava, T., Schaefer, A., Schmuck, M., Berg, G., et al. (2017b). Aerial warfare: a volatile dialogue between the plant pathogen verticillium longisporum and its antagonist Paenibacillus polymyxa. Front. Plant Sci. 8, 1294. doi: 10.3389/fpls.2017.01294
Rybakova, D., Schmuck, M., Wetzlinger, U., Varo-Suarez, A., Murgu, O., Müller, H., et al. (2016). Kill or cure? The interaction between endophytic Paenibacillus and Serratia strains and the host plant is shaped by plant growth conditions. Plant Soil 405, 65–79. doi: 10.1007/s11104-015-2572-8
Rybakova, D., Wikström, M., Birch-Jensen, F., Postma, J., Ehlers, R. U., Schmuck, M., et al. (2020). Verticillium wilt in oilseed rape—the microbiome is crucial for disease outbreaks as well as for efficient suppression. Plants 9, 866. doi: 10.3390/plants9070866
Schmidt, R., Cordovez, V., Boer, D. E., Raaijmakers, W. J., and Garbeva, P. (2015). Volatile affairs in microbial interactions. ISME J. 9, 2329–2335. doi: 10.1038/ismej.2015.42
Schmidt, R., Etalo, D. W., Jager de, W, Gerards, V., Zweers, S., De Boer, H., et al. (2016). Microbial small talk: volatiles in fungal–bacterial interactions. Front. Microbiol. 6, 1495. doi: 10.3389/fmicb.2015.01495
Schmidt, R., Jager, V. D., Zühlke, D., Wolff, C., Bernhardt, J., Cankar, K., et al. (2017). Fungal volatile compounds induce production of the secondary metabolite Sodorifen in Serratia plymuthica PRI-2C. Sci. Rep. 7, 862. doi: 10.1038/s41598-017-00893-3
Schulz, S., Dickschat, J. S., Kunze, B., Wagner-Dobler, I., Diestel, R., Sasse, F., et al. (2010). Biological activity of volatiles from marine and terrestrial bacteria. Mar. Drugs 8, 2976–2987. doi: 10.3390/md8122976
Schulz-Bohm, K., Martín-Sánchez, L., and Garbeva, P. (2017). Microbial volatiles: small molecules with an important role in intra- and inter-kingdom interactions. Front. Microbiol. 8, 2484. doi: 10.3389/fmicb.2017.02484
Smith, J. L. (2004). The physiological role of ferritin-like compounds in bacteria. Crit. Rev. Microbiol. 30, 173–185. doi: 10.1080/10408410490435151
Sneh, B., Jabaji-Hare, S., Neate, S., and Dijst, G. (2013). Rhizoctonia Species: Taxonomy, Molecular Biology, Ecology, Pathology and Disease Control. Berlin: Springer Science and Business Media.
Stachowiak, A., Olechnowicz, J., Jedryczka, M., Rouxel, T., Balesdent, M. H., et al. (2006). Frequency of avirulence alleles in field populations of Leptosphaeria maculans in Europe. Eur. J. Plant Pathol. 114, 67–75. doi: 10.1007/s10658-005-2931-z
Tosi, T., Nickerson, N. N., Mollica, L., Jensen, M. R., Blackledge, M., Baron, B., et al. (2011). Pilotin–secretin recognition in the type II secretion system of Klebsiella oxytoca. Mol. Microbiol. 82, 1422–1432. doi: 10.1111/j.1365-2958.2011.07896.x
Tripathi, A., Dewan, P. C., Barua, B., and Varadarajan, R. (2012). Additional role for the ccd operon of F-plasmid as a transmissible persistence factor. Proc. Natl. Acad. Sci. 109, 12497–12502. doi: 10.1073/pnas.1121217109
Vogt, S. L., and Raivio, T. L. (2012). Just scratching the surface: an expanding view of the Cpx envelope stress response. FEMS Microbiol. Lett. 326, 2–11. doi: 10.1111/j.1574-6968.2011.02406.x
Vollmer, W., Joris, B., Charlier, P., and Foster, S. (2008). Bacterial peptidoglycan (murein) hydrolases. FEMS Microbiol. Rev. 32, 259–286. doi: 10.1111/j.1574-6976.2007.00099.x
Weise, T., Thürmer, A., Brady, S., Kai, M., Daniel, R., Gottschalk, G., et al. (2014). VOC emission of various Serratia species and isolates and genome analysis of Serratia plymuthica 4Rx13. FEMS Microbiol. Lett. 352, 45–53. doi: 10.1111/1574-6968.12359
Weisskopf, L., Schulz, S., and Garbeva, P. (2021). Microbial volatile organic compounds in intra-kingdom and inter-kingdom interactions. Nat. Rev. Microbiol. 19, 391–404. doi: 10.1038/s41579-020-00508-1
Whipps, J. M. (2001). Microbial interactions and biocontrol in the rhizosphere. J. Exp. Bot. 52, 487–511. doi: 10.1093/jxb/52.suppl_1.487
Keywords: microbiome, transcriptome, volatilome, biocontrol, Serratia, Verticillium, Rhizoctonia, Leptospheria
Citation: Rybakova D, Müller H, Olimi E, Schaefer A, Cernava T and Berg G (2022) To defend or to attack? Antagonistic interactions between Serratia plymuthica and fungal plant pathogens, a species-specific volatile dialogue. Front. Sustain. Food Syst. 6:1020634. doi: 10.3389/fsufs.2022.1020634
Received: 16 August 2022; Accepted: 10 October 2022;
Published: 28 October 2022.
Edited by:
Micaela Tosi, University of Guelph, CanadaReviewed by:
Sowmyalakshmi Subramanian, McGill University, CanadaCopyright © 2022 Rybakova, Müller, Olimi, Schaefer, Cernava and Berg. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Daria Rybakova, ZGFyaWFyeWJAZ21haWwuY29t
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.