- 1Department of Plant Biotechnology, Center for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 2Department of Plant Molecular Biology and Bioinformatics, Center for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 3Department of Biotechnology, Agricultural College and Research Institute, Tamil Nadu Agricultural University, Madurai, Tamil Nadu, India
- 4Center for Plant Breeding and Genetics, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
- 5Center for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, Tamil Nadu, India
Sweet corn has dominated the urban market due to its sweetness, tenderness, and ease of digestibility. It's import and export values have dramatically increased during the past 10 years as a fresh, processed, and preserved commodity. However, the commercially available sweet corns are deficient in β-carotene. In our study, we introgressed the favorable allele of crtRB1 (responsible for high β-carotene) into the recurrent sweet corn inbred SC11-2 from maize donor parent UMI1230β1+ to develop the β-carotene-rich sweet corn genotype by marker aided breeding. The crtRB1 3′TE InDel marker was utilized for foreground selection of favorable genotype. A total of 103 polymorphic SSR markers were employed for background selection, resulting in a 96% recovery of recurrent parent genome (RPG). We recorded high β-carotene content (9.878–10.645 μg/g) in the introgressed lines compared to the recurrent parent, SC11-2 (0.989 μg/g). The sugar content ranged from 18 to 19.10% and was on par with the recurrent parent (20.40%). These biofortified inbreds can be used as a donor in maize breeding programs to develop sweet corn genotypes with high β-carotene content.
Introduction
Hidden hunger, caused by micronutrient malnutrition, is one of the prime focuses in this era. Around 30 vitamins and minerals cannot be synthesized in our body in sufficient quantity, but it has to be supplied through diet and are termed “essential micronutrient.” Micronutrient malnutrition has appeared to be a serious health issue worldwide (Bouis et al., 2019). Malnutrition affects all age groups irrespective of geographical location and economic status. Nearly two billion people suffer from micronutrient deficiency (Global Nutrition Report, 2018). Vitamin A deficiency (VAD) is a major cause of malnutrition, and it leads to several diseases in humans viz., measles, respiratory diseases, night blindness, xerophthalmia, growth retardation, generative disorders, and low immunity (Lozano-Alejo et al., 2007; Bouis and Saltzman, 2017).
Economic value of sweet corn increased due to its demand for fresh and processed vegetables. Sweet corn is preferred due to its sweet taste, tender nature, easy digestibility, and nutritional value (Mehta et al., 2017). Sweet corn demand is higher in urban cities where it is consumed as snacks, soups, and for preparing main courses. It is an integral part of the diet in the countries of South East Asia (Feng et al., 2015; Yang et al., 2018). Frozen sweet corn is estimated to have a global import of $433.60 million and an export of $426.25 million. Preserved sweet corn is estimated to have an import of $957.09 million and $908.31 million in export (FAOSTAT, 2022). Traditionally cultivated sweet corn varieties possess a significantly low level of provitamin A (Feng et al., 2015). Thus, traditional sweet corn does not make a significant contribution to the daily Provitamin A requirement in human beings.
Biofortification, medicinal supplementation, dietary diversity, and food fortification are widely used to combat malnutrition worldwide. Biofortification is an approach for increasing nutrient density in food crops by breeding. Biofortification is the preferred method to develop nutrition-rich staple crops that provide a sustainable solution to control malnutrition (Pfeiffer and McClafferty, 2007). The favorable allele related to β-carotene hydroxylase1 (crtRB1) increases the accumulation of proA in maize kernel endosperms by limiting the conversion of β-carotene into further downstream products. β-carotene acts as the precursor for vitamin A development (Yan et al., 2010). The proA biofortified sweet corn could resolve the problem of VAD, especially in urban areas. Hence, we need superior sweet corn donor plants with crtRB1 favorable allele to develop biofortified sweet corn hybrids.
Sweet corn is developed naturally through recessive mutation in genes related to the starch biosynthesis pathway. Mutation in the Shrunken2 (Sh2) allele, present in chromosome 3L of sweet corn, leads to the formation of recessive shrunken2 (sh2) (Baveja et al., 2021). Due to this sh2 loci, sweet corn accumulates 2–4-folds more sucrose and reducing sugar than maize (Nelson, 1980). This also determines the phenotypic appearance of the sweet corn kernel by shrinking it (Lertrat and Pulam, 2007). The development of commercial sweet corn lines predominantly uses the sh2 gene (Wong et al., 1994). Marker-assisted selection (MAS) has been successfully used for developing agronomically superior cultivars (Singh et al., 2011). MAS of the target allele shortens the breeding cycle and makes the breeding program more efficient and precise (Ribaut and Hoisington, 1998). Marker-assisted backcross breeding (MABB) is the most effective approach for transferring the gene of interest and improving the nutritional traits in maize (Singh et al., 2012; Babu et al., 2013; Chandran et al., 2019; Pukalenthy et al., 2019, 2020; Sagare et al., 2019; Sarankumar et al., 2019; Natesan et al., 2020; Das et al., 2021; Qutub et al., 2021; Talukder et al., 2022).
The current study describes the revival of sweetcorn-inbred SC11-2. We used a MABB strategy, introgressed the crtRB1 gene into sweetcorn inbred SC11-2, and developed agronomically superior lines rich in β-carotene. The newly developed lines will be valuable as a donor for the sweet corn biofortification program.
Materials and methods
Plant genetic materials and experimental site
A sh2 based sweet corn inbred SC11-2 developed at the Department of Millets, Tamil Nadu Agricultural University, Coimbatore, India was used as a recurrent parent. UMI1230β1+ was used as a donor parent from the Center for Plant Molecular Biology and Biotechnology, Tamil Nadu Agricultural University, Coimbatore, India. The donor parent was used to introduce the favorable target allele of crtRB1 into the recurrent parent SC11-2. All the field experiments were done at Eastern Block Farm, Tamil Nadu Agricultural University, Coimbatore, India, during 2019–2020.
Generation of backcross and self-progenies
The F1s were generated by a single cross between the recurrent parent, SC11-2, and the donor parent, UMI1230β1+, during Rabi, 2019 (November–March). The true F1s were selected using crtRB1 gene-specific markers. In Kharif 2020 (June–September), the BC1F1 population was developed by backcrossing the F1s with the recurrent parent. Only the shrunken seeds from the BC1F1 cobs were selected for forwarding into the next generation. These shrunken seeds were grown, and the second backcross was performed on the crtRB1 heterozygous plants in Rabi, 2020 (November–March) to develop the BC2F1. We obtained all the seeds as shrunken in this generation. The selected crtRB1 heterozygous plants in BC2F1 were selfed to produce the BC2F2 generation. The crtRB1 positive homozygous plants were selected from the segregating population of BC2F2 using the crtRB1 gene-specific marker and were selfed again to develop the stabilized BC2F3 population (Figure 1).
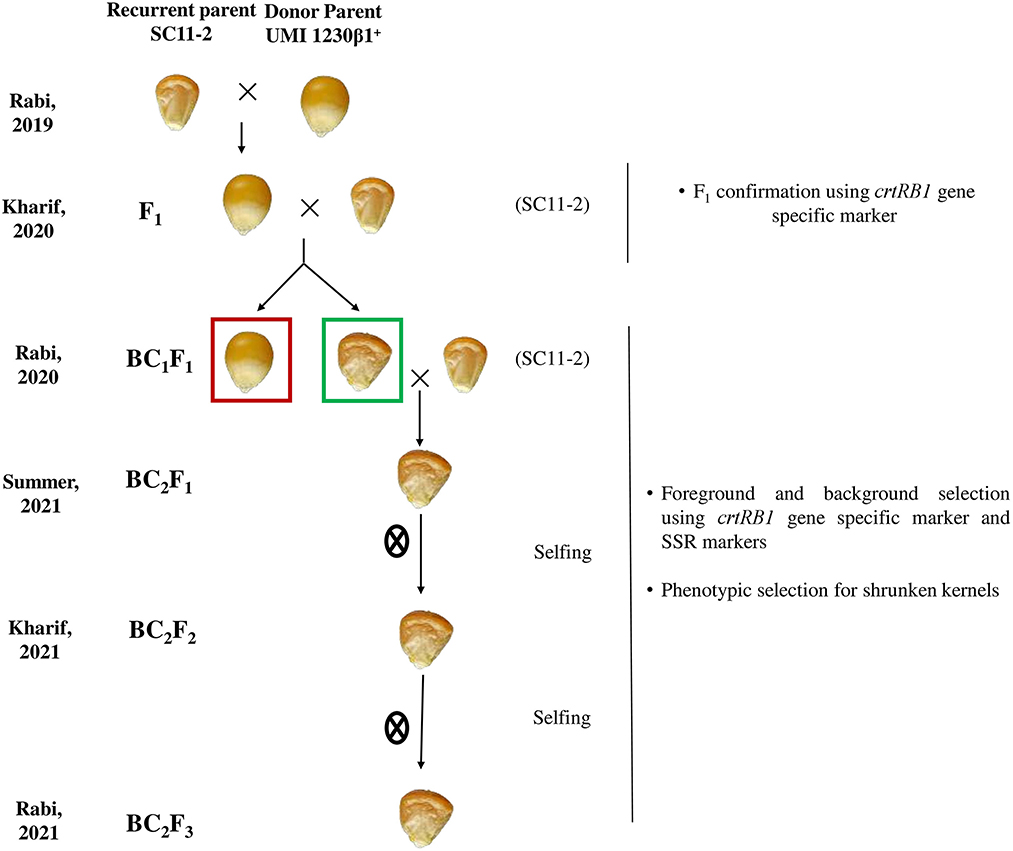
Figure 1. The selected flint kernel in the F1 generation is backcrossed with the recurrent parent (SC11-2). The shrunken kernel is selected (highlighted with green color box) in BC1F1 and these shrunken kernels are selected and selfed in the subsequent generations.
Foreground selection with gene-specific marker
Genomic DNA was extracted from 3 weeks old young seedlings of parents, backcrossed populations, and selfed BC2F2 population following the standard CTAB method (Murray and Thompson, 1980). crtRB1 gene-specific 3′TE InDel marker (Supplementary Table 1) was used for foreground selection (Yan et al., 2010). The crtRB1 gene exhibits three allelic forms due to crtRB1-3′TE polymorphism. Allele 1 (543 bp, without TE insertion), which is attributed to increase β-carotene by decreasing transcript expression of the crtRB1 gene, is the favorable allele. Allele 2 (296 + 875 bp; with 325 bp TE insertion) and allele 3 (296 + 1,221 + 1,880 bp; 1,250 bp TE insertion) are the unfavorable alleles. crtRB1 gene was amplified using polymerase chain reaction (PCR) (Eppendorf, Hamburg) following the procedure as described by Natesan et al. (2020). We screened for favorable allele at 543 bp and unfavorable allele at 296 bp. Under heterozygous conditions, both alleles were amplified. The PCR amplicons were resolved in 3% agarose gel at 120 V for 2 h. The resolved products in the agarose gel were visualized using a gel documentation system (BioRad, California, USA). This marker was used to select heterozygous plants in the BC1F1 and BC2F1 generations, and homozygotes in the BC2F2 generation.
Background selection with SSR markers
A total of 201 SSR markers covering 10 chromosomes of the maize genome were retrieved from the maize genome database (www.maizegdb.org). The SSR markers were custom synthesized from Eurofins Genomics India Pvt. Ltd., Bangalore, India. These SSR markers were used to screen the recurrent and donor parents to identify the parental polymorphism. The identified polymorphic markers were employed on foreground favorable plants of BC1F1, BC2F1, and BC2F2 to calculate the recovery of the recurrent parent genome (RPG). The polymerase chain reaction (PCR) and agarose gel electrophoresis were done following the method used by Chandrasekharan et al. (2022). Recovery of RPG for the selected plants was estimated using the formula:
“A” is the number of SSR markers homozygous to the donor plant, “B” is the number of SSR markers homozygous to the recurrent parent, and “H” is the number of SSR markers in the heterozygote state. Polymorphic information content (PIC) for each pair of SSR markers was estimated using the formula described by (Botstein et al., 1980; Supplementary Table 2).
Characterization of morphological traits
Morphological traits were recorded among the foreground positive plants with high recovery of RPG in BC1F1, BC2F1, and BC2F2 generation following the descriptors suggested by the International Board for Plant Genetic Resources (CTA, 1992). The traits recorded were 50% days to tasseling, 50% days to silking, plant height, ear height, tassel length, number of tassels, leaf length, leaf breadth, cob girth, number of kernel row per cob, number of kernel row, and single plant yield.
Sugar and β-carotene contents
Selfed cobs from BC2F3 generation were harvested 24 days after pollination (DAP) to check the total soluble solids (TSS) using the portable Brix meter (Neuffer and Sheridan, 1997). Total sugar was estimated following the Anthrone method (Hodge, 1962), and reducing sugars were estimated by the Nelson-Somogyi method (Nelson, 1944; Somogyi, 1952). Non-reducing sugar was estimated by the difference between the estimated value of total sugar and that of reducing sugar. To estimate the β-carotene, the matured BC2F3 cob kernels were dried in dark conditions before grinding into fine powder. The extraction solvents for the estimation were prepared according to the Harvest Plus protocol (Rodriguez-Amaya and Kimura, 2004). Quantification of β-carotene in the introgressed lines, recurrent, and donor parents were performed using Shimadzu HPLC Analytical C18G column 250 × 4.6 mm. Samples were eluted using YMC carotenoid C18 column. A photodiode array detector was used to detect the peaks. The mobile phase consisting of Acetonitrile: Methanol: Ethyl acetate at a ratio of 80:10:10 was made to pass through the column, under high pressure, at a flow rate of 1mL min−1. Five standards (0.1, 1, 10, 50, and 100 μg/g) (Kurilich and Juvik, 1999) of β-carotene (M/s. Sigma Aldrich India) were used to make the standard curve. β-carotene content was calculated in the inbreds and parents at 453 nm. The chromatograph was compared with the standard curve to calculate the β-carotene level in the kernels of the inbreds and the parents.
Statistical analysis
The Chi-square test was performed to test the goodness of fit in the segregation pattern of the crtRB1 alleles in BC1F1, BC2F1, and BC2F2 generations. A t-test was performed to assess the significance of the increase in β-carotene level in the introgressed lines over the recurrent parent.
Results
Marker polymorphism
The recurrent (SC11-2) and donor (UMI 1230β1+) parents were screened with a crtRB1 gene-specific marker which revealed an unfavorable allele (296 bp) in the recurrent parent and a favorable allele (543 bp) in the donor. Out of 201 SSR markers used for screening parental polymorphism, 103 markers showed polymorphism (51.24%), and the number of polymorphic markers in each chromosome ranged from 5 to 10. The polymorphic markers were used to screen the foreground favorable plants from BC1F1, BC2F1, and BC2F2 generation to trace the high RPG plants.
Accelerated development of SC11-2 with crtRB1 gene
F1 generation
The crtRB1 gene-specific marker was used to confirm heterozygosity in the F1s of SC11-2 × UMI 1230β1+. A total of 46 plants were screened in this generation, of which 42 were found to be heterozygous for crtRB1. The heterozygote F1 plants were backcrossed with the recurrent parent (SC11-2), and well-set BC1F1 cob designated as DBT 17-1 was selected for advancing it to the BC1F1 generation.
BC1F1 generation
A total of 96 plants from DBT 17-1 cob were screened in this generation using the crtRB1 gene-specific foreground marker. A total of 35 plants with unfavorable allele (296 bp) and 61 plants in heterozygous condition (296 + 543 bp) were confirmed (Figure 2). The segregation pattern for crtRB1 alleles deviated significantly from the expected Mendelian ratio (1:1) (Table 1). Recovery of RPG ranged from 72.30 to 76.60% in the crtRB1 heterozygous plants (Table 2). Three plants designated as DBT 17-1-1, DBT 17-1-6, and DBT 17-1-47, with higher RPG recovery, were selected and advanced to the next generation by backcrossing with the recurrent parent, SC11-2.
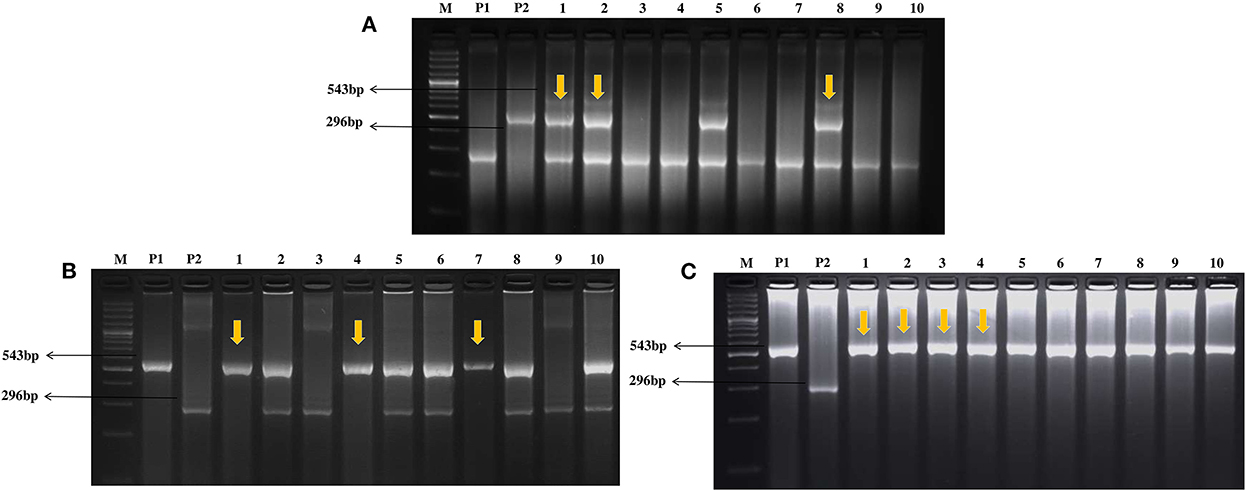
Figure 2. Foreground selection in backcross [(A) BC1F1, (B) BC2F2] and selfed generation [(C) BC2F3] of SC11-2 × UMI 1230β1+ using crtRB1 gene specific marker crtRB13′TE. (M) Ladder (100 bp), (P1) SC11-2, (P2) UMI 1230β1+, (1–10) progenies from BC1F1, BC2F2, and BC2F3 generation.
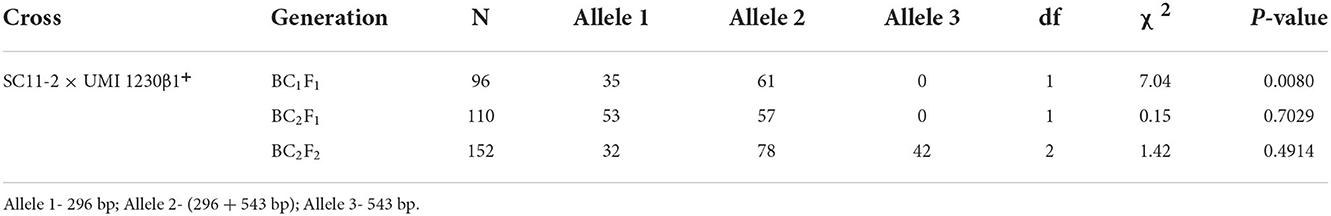
Table 1. Segregating pattern of crtRB1 alleles in the backcross and selfed populations derived from SC11-2 × UMI 1230β1+.

Table 2. Recovery percentage of recurrent parent genome in backcross and selfed populations derived from SC11-2 × UMI 1230β1+.
BC2F1 generation
In this generation, 110 plants were screened for the crtRB1 foreground marker, where 53 and 57 plants were detected as unfavorable and heterozygous, respectively (Figure 2). The segregation ratio of the alleles in this generation was as per the Mendelian ratio of 1:1 (Table 1). Recovery of RPG ranged from 84.30 to 89.36% (Table 2) in this generation. Three heterozygous plants, DBT 17-1-1-1, DBT 17-1-1-29, and DBT 17-1-1-71, with high recovery of RPG, were further selected and selfed to advance to BC2F2 generation.
BC2F2 generation
In BC2F2 generation, 152 segregating plants from DBT 17-1-1-1 cob were screened for crtRB1 alleles (Figure 2). The segregation of the crtRB1 alleles into 42 favorable positives (543 bp), 78 heterozygous (543+296 bp), and 32 unfavorable (296 bp) alleles were as per the Mendelian ratio of 1:2:1 (Table 1). Recovery of RPG in the foreground positive plants ranged from 91.20 to 94.68% (Table 2). Three positive plants with high recovery of RPG and phenotypic similarity with the recurrent parent were selected and selfed to forward to BC2F3 generation. The plants were designated as DBT 17-1-1-1-13, DBT 17-1-1-1-35, and DBT 17-1-1-1-101.
BC2F3 generation
In this population, a crtRB1 gene-specific marker was used to ascertain the presence of a positive allele (543bp) (Figure 2). Recovery of RPG was 94–96% in this generation. Biochemical analysis for sugar estimation and β-carotene was carried out in kernels of four positive plants with high recovery of RPG. The plants were designated as DBT 17-1-1-1-35-1, DBT 17-1-1-1-35-2, DBT 17-1-1-1-35-3 and DBT 17-1-1-1-35-4 (Table 1, Figure 3, Supplementary Figure 1).
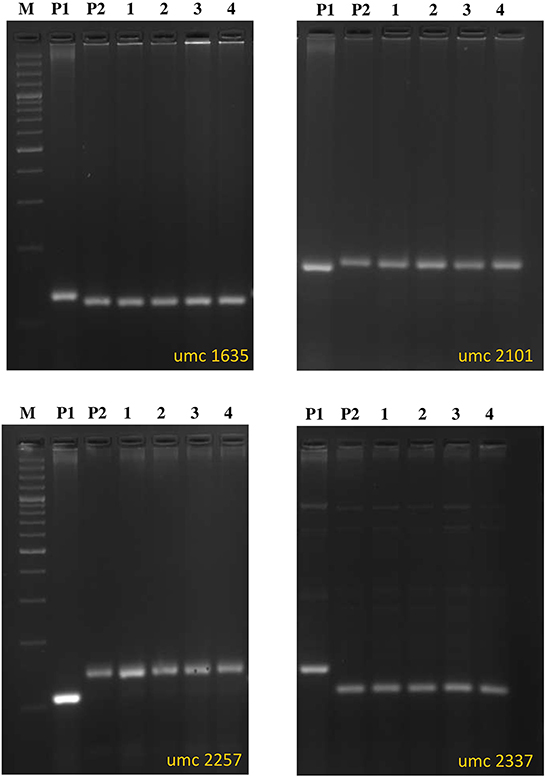
Figure 3. Background selection using SSR markers for selected improved lines from BC2F3 generation of SC11-2 × UMI 1230β1+. (M) Ladder (100 bp), (P1) UMI 1230β1+, (P2) SC11-2, (1) DBT 17-1-1-1-35-1, (2) DBT 17-1-1-1-35-2, (3) DBT 17-1-1-1-35-3, (4) DBT 17-1-1-1-35-4.
Morphological traits
A high degree of resemblance was found between the introgressed inbred and recurrent parent line concerning morphological characters and yield attributing traits. Four improved lines showed phenotypic resemblance from 87.63% (Cob girth) to 100% (Tassel length and number of kernel rows per cob). Among the four lines, DBT 17-1-1-1-35-3 showed maximum resemblance with the recurrent parent. For instance, days to tasselling, days to silking, plant height, ear height, number of tassel branches, lead length, cob length, and single plant yield showed more than 95% resemblance with SC11-2 (Figure 4, Table 3).

Figure 4. The morphological traits [(A) tassel, (B) silk, and (C) cob] in parents and improved line (DBT 17-1-1-1-35-1) from BC2F3 generation of SC11-2 × UMI 1230β1+.
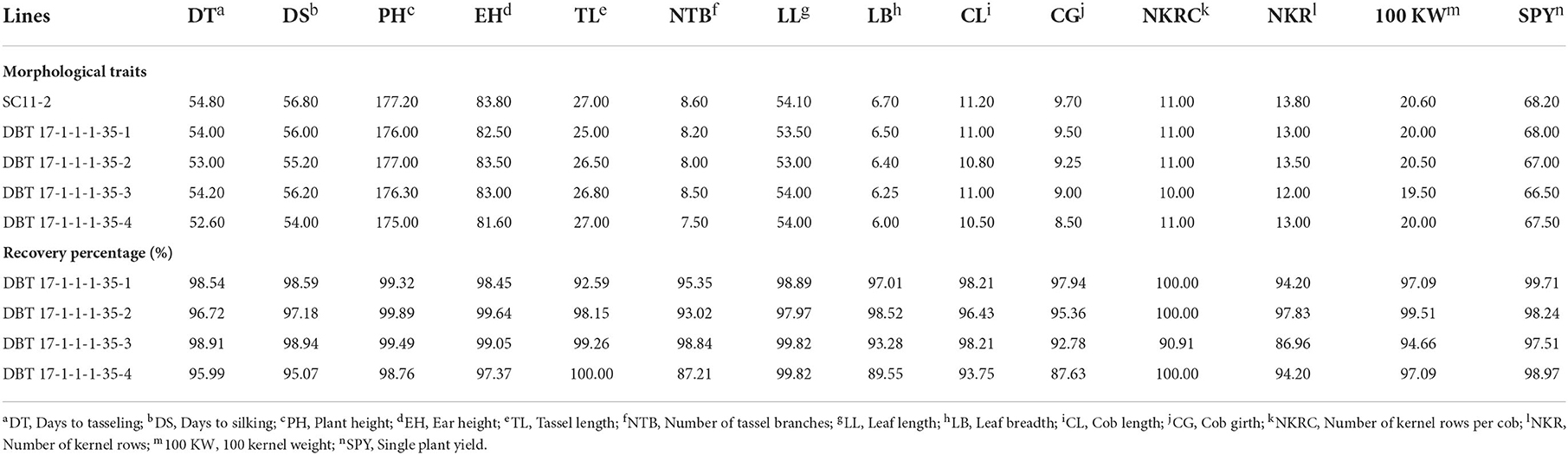
Table 3. Details of morphological traits and recovery percentage in the BC2F3 improved lines of SC11-2 × UMI 1230β1+ with respect to the recurrent parent SC11-2.
Sugar and β-carotene contents
TSS was estimated at 24DAP using the Brix meter and was found to be 19.00% in the recurrent parent kernels and 10.00% in the donor plant kernels. The introgressed inbred had an average TSS ranging from 16 to 18.5%. Total sugar (TS) in the recurrent parent was 20.40%, and in the donor parent, it was 5.25, whereas TS in the introgressed lines ranged from 18.35 to 19.65%. Reducing sugar (RS) in the recurrent parent and donor parent was estimated to be 3.5 and 2.1%, respectively, whereas RS in the introgressed lines was between 2.45 to 3.35%. Non-reducing sugar (NRS) was estimated to be 16.90% in the recurrent parent and 3.15% in the donor parent. NRS in the introgressed lines ranged from 15.30 to 17.20% (Figure 5, Table 4). The β-carotene level in the matured kernels of the recurrent parent was 0.989 and 11.608 ppm in the donor parent kernel. The β carotene level in the kernels of the introgressed lines ranged from 9.878 to 10.645 ppm. This result suggests that there was an average of 10.37-fold higher β-carotene levels in the introgressed lines compared to the recurrent line (Figure 5, Table 4).
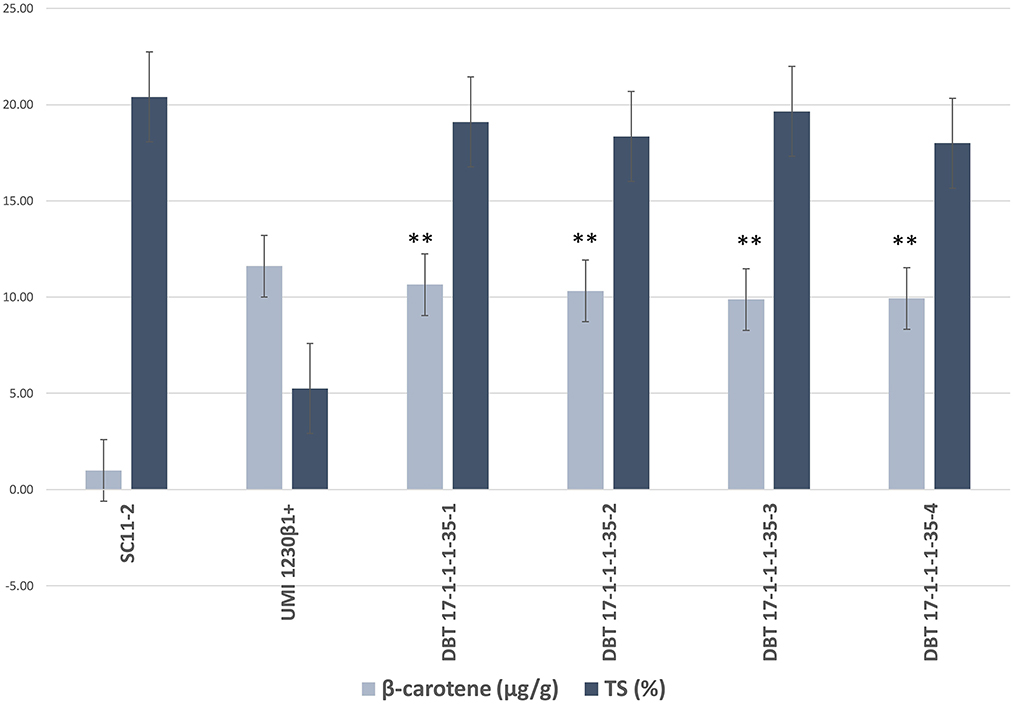
Figure 5. Total sugar (%) and β-carotene (μg/g) content in parents and improved lines from BC2F3 generation of SC11-2 × UMI 1230β1+. Error bars indicate means ± SE. Asterisks (**) indicate significant different from the recurrent parent β-carotene (μg/g) content using t-test (p < 0.01).
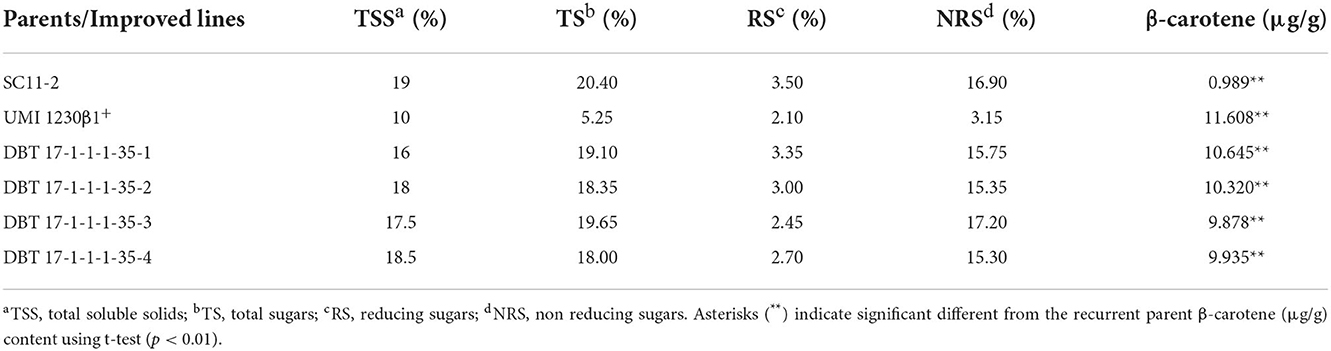
Table 4. Details of sugar and β-carotene contents in parents and BC2F3 improved lines of SC11-2 × UMI 1230β1+.
Discussion
Sweet corn demand has increased over the last decade and is preferred for its tenderness and sweet taste all over the world (Mehta et al., 2017). Sweet corn is eaten fresh in the milky stage which helps to retain the nutritional quality (Cabrera-Soto et al., 2018). Many commercially grown sweet corn is deficient in proA to meet the minimal daily requirement (Feng et al., 2015). Development of vitamin A biofortified sweet corn will provide essential vitamin A rich food (Hossain et al., 2019). In this study we developed vitamin A biofortified sweet corn inbred by marker assisted introgression of crtRB1 favorable allele in a promising sweet corn inbred, SC11-2.
Introgression of crtRB1 gene into sweet corn inbred SC11-2
The crtRB1 3′TE favorable allele is responsible for effecting a 2–10-fold increase in β-carotene content in maize (Babu et al., 2013). In this study, we developed sweet corn inbred with crtRB1 favorable allele using maize UMI1230β1+ as the donor parent and sweet corn inbred SC11-2 as a recurrent parent. crtRB1-3′TE-InDel markers located within the target genes facilitated selection of individual plants with favorable alleles of crtRB1 in the segregating populations (Gupta et al., 2013; Zunjare et al., 2018). Marker assisted selection of favorable progenies in the seedling stage before pollination resulted in saving time and resources (Sarika et al., 2018). The segregation pattern of the crtRB1 alleles in BC1F1 showed a significant segregation distortion (SD) due to the diverse backgrounds of the parents (Babu et al., 2013; Muthusamy et al., 2014). This could be due to the presence of naturally occurring gene mutants like dek (defective kernel) (Neuffer and Sheridan, 1997), gametophytic factors (ga) (Mangelsdorf and Jones, 1926), and embryo-specific mutation (emb) (Neuffer and Sheridan, 1997). In our study, MABB led to the high recovery of RPG in the introgressed progenies. The SSR markers used for background selection were distributed across the genome. This attributed to the selection of positive progenies with high recovery of RPG (Singh et al., 2012; Gupta et al., 2013). The recovery of RPG is responsible for the similarities in the agronomical traits of the introgressed lines and their recurrent parent (Muthusamy et al., 2014). Due to the fixation of various recurring parent allele proportions in the foreground positive plants, there is diversity in RPG recovery among the introgressed progenies (Muthusamy et al., 2014). Previous studies suggest a high recovery in RPG within two generations in the MABB program (Muthusamy et al., 2014; Liu et al., 2015; Hossain et al., 2018; Sarika et al., 2018). Background selection assisted in the indirect selection of loci corresponding to different phenotypic characteristics (Hossain et al., 2018).
Kernel sweetness and nutritional quality
The selection of shrunken kernels in BC1F1 helped to indirectly select the sh2 allele in the progenies (Khanduri et al., 2011; Mehta et al., 2017). As a result, the introgressed lines retained their sweetness and were on par with the recurrent parent. The Sh2 gene is located in chromosome 3 and is responsible for coding a large subunit of ADP-glucose pyrophosphorylase (AGPase), which synthesizes ADP-glucose from glucose-1-phosphate and adenosine triphosphate (Baveja et al., 2022). This conversion is blocked by the recessive sh2 allele, which restricts the starch biosynthesis of amylose and amylopectin. The sweetness was estimated and found to be maximum at 24 DAP (Chhabra et al., 2019). The variation in the sweetness across the introgressed lines may be due to the presence of loci influencing the sh2 gene (Qi et al., 2009; Mehta et al., 2020).
A significant increase of β-carotene in the introgressed inbred suggests that introgression of favorable allele (543 bp) alone has a major effect on the accumulation of β-carotene in the sweetcorn kernels. The crtRB1 gene is responsible for hydroxylation of β-carotene and β-cryptoxanthin into zeaxanthin. This reduces the proA concentration in the kernels. A natural variant of crtRB1 with no TE in the 3′ UTR reduces this conversion. This leads to more accumulation of proA carotenoids in the kernels (Yan et al., 2010). In China, four sh2 based sweet corn lines were developed by introgression of lcyE (Yang et al., 2018). But they reported a low increase in the proA (2.07–3.86 ppm) over traditional inbreds. In this study, we have reported a 10.7-fold increase in β-carotene content in the developed lines compared to the original inbred. This was achieved through the MAS of plants containing the favorable allele 1 of crtRB1.
The traditional sweet corn lines are low in proA. In our study, we introgressed crtRB1 gene encoding proA into traditional sweet corn inbred using marker-aided breeding. The crtRB1 gene-specific marker facilitated the precise identification of favorable genotypes from the segregating populations. SSR-based background selection expatiated the selection process, resulting in higher recovery of recurrent parent genome within two backcrosses. The marker-assisted breeding thus helped to develop biofortified sweet corn inbred with a 10.7-fold increase in β-carotene levels. This inbred can be used as a donor parent for crtRB1 favorable allele in developing vitamin A biofortified sweet corn hybrids. ProA biofortified sweet corn can help to mitigate the problem of hidden hunger in a sustainable manner in the urban areas of developing countries.
Data availability statement
The original contributions presented in the study are included in the article/Supplementary material, further inquiries can be directed to the corresponding author.
Author contributions
SN conceived and designed this study. IS and KR performed field and lab experiments. IS, KR, BM, and KA analyzed the data. VS, RR, and RM monitored the research and provided suggestions. IS, KR, KA, and SN wrote the manuscript. All authors contributed to the article and approved the submitted version.
Funding
Financial support from the Department of Biotechnology (DBT), Government of India (GOI), through the project entitled Incorporation of crtRB1 allele into sweet corn inbreds and north eastern landraces for development of biofortified sweet corn (BT/PR42333/NER/95/1866/2021 dt. 18.2.2022) was acknowledged. The funders had no role in the work design, data collection, and analysis, or decision and preparation of the manuscript.
Conflict of interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Supplementary material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2022.1004450/full#supplementary-material
References
Babu, R., Rojas, N. P., Gao, S., Yan, J., and Pixley, K. (2013). Validation of the effects of molecular marker polymorphisms in LcyE and CrtRB1 on provitamin A concentrations for 26 tropical maize populations. Theor. Appl. Genet. 126, 389–399. doi: 10.1007/s00122-012-1987-3
Baveja, A., Chhabra, R., Panda, K. K., Muthusamy, V., Mehta, B. K., Mishra, S. J., et al. (2022). Expression analysis of opaque2, crtRB1 and shrunken2 genes during different stages of kernel development in biofortified sweet corn. J. Cereal Sci. 105, 103466. doi: 10.1016/j.jcs.2022.103466
Baveja, A., Muthusamy, V., Panda, K. K., Zunjare, R. U., Das, A. K., Chhabra, R., et al. (2021). Development of multinutrient-rich biofortified sweet corn hybrids through genomics-assisted selection of shrunken2, opaque2, lcyE and crtRB1 genes. J. Appl. Genet. 62, 419–429. doi: 10.1007/s13353-021-00633-4
Botstein, D., White, R. L., Skolnick, M., and Davis, R. W. (1980). Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am. J. Hum. Genet. 32, 314.
Bouis, H. E., and Saltzman, A. (2017). Improving nutrition through biofortification: a review of evidence from HarvestPlus, 2003 through 2016. Glob. Food Secur. 12, 49–58 doi: 10.1016/j.gfs.2017.01.009
Bouis, H. E., Saltzman, A., and Birol, E. (2019). “Improving nutrition through biofortification,” in Agriculture for Improved Nutrition: Seizing the Momentum. p. 47–57.
Cabrera-Soto, L., Pixley, K. V., Rosales-Nolasco, A., Galicia-Flores, L. A., and Palacios-Rojas, N. (2018). Carotenoid and tocochromanol profiles during kernel development make consumption of biofortified “fresh” maize an option to improve micronutrient nutrition. J. Agric. Food Chem. 66, 9391–9398. doi: 10.1021/acs.jafc.8b01886
Chandran, S., Pukalenthy, B., Adhimoolam, K., Manickam, D., Sampathrajan, V., Chocklingam, V., et al. (2019). Marker-assisted selection to pyramid the opaque-2 (o2) and β-carotene (crtRB1) genes in maize. Front. Genet. 10, 859. doi: 10.3389/fgene.2019.00859
Chandrasekharan, N., Ramanathan, N., Pukalenthy, B., Chandran, S., Manickam, D., Adhimoolam, K., et al. (2022). Development of β-carotene, lysine, and tryptophan-rich maize (Zea mays) inbreds through marker-assisted gene pyramiding. Sci. Rep. 12, 8551. doi: 10.1038/s41598-022-11585-y
Chhabra, R., Hossain, F., Muthusamy, V., Baveja, A., Mehta, B. K., and Zunjare, R. U. (2019). Development and validation of breeder-friendly functional markers of sugary1 gene encoding starch-debranching enzyme affecting kernel sweetness in maize (Zea mays). Crop Pasture Sci. 70, 868–875. doi: 10.1071/CP19298
Das, A. K., Gowda, M. M., Muthusamy, V., Zunjare, R. U., Chauhan, H. S., Baveja, A., et al. (2021). Development of maize hybrids with enhanced vitamin-E, vitamin-A, lysine, and tryptophan through molecular breeding. Front. Plant Sci. 12, 659381. doi: 10.3389/fpls.2021.659381
FAOSTAT (2022). Food and Agriculture Data. Available online at: http://faostat.fao.org (accessed July 16, 2022).
Feng, F., Wang, Q., Liang, C., Yang, R., and Li, X. (2015). Enhancement of tocopherols in sweet corn by marker-assisted backcrossing of ZmVTE4. Euphytica 206, 513–521. doi: 10.1007/s10681-015-1519-8
Global Nutrition Report (2018). The 2022 Global Nutrition Report is Coming Soon. Available online at: https://globalnutritionreport.org/ (accessed July 1, 2022).
Gupta, H. S., Raman, B., Agrawal, P. K., Mahajan, V., Hossain, F., and Thirunavukkarasu, N. (2013). Accelerated development of quality protein maize hybrid through marker-assisted introgression of opaque-2 allele. Plant Breed. 132, 77–82. doi: 10.1111/pbr.12009
Hodge, J. E. (1962). Determination of reducing sugars and carbohydrates. Methods Carbohydr. Chem. 1, 380–394.
Hossain, F., Muthusamy, V., Pandey, N., Vishwakarma, A. K., Baveja, A., Zunjare, R. U., et al. (2018). Marker-assisted introgression of opaque2 allele for rapid conversion of elite hybrids into quality protein maize. J. Genet. 97, 287–298. doi: 10.1007/s12041-018-0914-z
Hossain, F., Sarika, K., Muthusamy, V., Zunjare, R. U., and Gupta, H. S. (2019). “Quality protein maize for nutritional security,” in Quality Breeding in Field Crops, eds A. M. I. Qureshi, Z. A. Dar, and S. H. Wani (Cham: Springer), 217–237.
Khanduri, A., Hossain, F., Lakhera, P. C., and Prasanna, B. M. (2011). Effect of harvest time on kernel sugar concentration in sweet corn. Indian J. Genet. Plant Breed. 71, 231. doi: 10.1007/978-3-030-04609-5_11
Kurilich, A. C., and Juvik, J. A. (1999). Quantification of carotenoid and tocopherol antioxidants in Zea mays. J. Agric. Food Chem. 47, 1948–1955. doi: 10.1021/jf981029d
Lertrat, K., and Pulam, T. (2007). Breeding for increased sweetness in sweet corn. Int. J. Plant Breed. 1, 27–30.
Liu, L., Jeffers, D., Zhang, Y., Ding, M., Chen, W., Kang, M. S., et al. (2015). Introgression of the crtRB1 gene into quality protein maize inbred lines using molecular markers. Mol. Breed. 35, 1–12. doi: 10.1007/s11032-015-0349-7
Lozano-Alejo, N., Carrillo, G. V., Pixley, K., and Palacios-Rojas, N. (2007). Physical properties and carotenoid content of maize kernels and its nixtamalized snacks. Innov. Food Sci. Emerg. Technol. 8, 385–389. doi: 10.1016/j.ifset.2007.03.015
Mangelsdorf, P. C., and Jones, D. (1926). The expression of Mendelian factors in the gametophyte of maize. Genetics 11, 423. doi: 10.1093/genetics/11.5.423
Mehta, B., Hossain, F., Muthusamy, V., Baveja, A., Zunjare, R., Jha, S. K., et al. (2017). Microsatellite-based genetic diversity analyses of sugary1-, shrunken2-and double mutant-sweet corn inbreds for their utilization in breeding programme. Physiol. Mol. Biol. Plants 23, 411–420. doi: 10.1007/s12298-017-0431-1
Mehta, B. K., Muthusamy, V., Zunjare, R. U., Baveja, A., Chauhan, H. S., Chhabra, R., et al. (2020). Biofortification of sweet corn hybrids for provitamin-A, lysine and tryptophan using molecular breeding. J. Cereal Sci. 96, 103093. doi: 10.1016/j.jcs.2020.103093
Murray, M. G., and Thompson, W. (1980). Rapid isolation of high molecular weight plant DNA. Nucleic Acids Res. 8, 4321–4326. doi: 10.1093/nar/8.19.4321
Muthusamy, V., Hossain, F., Thirunavukkarasu, N., Choudhary, M., Saha, S., Bhat, J. S., et al. (2014). Development of β-carotene rich maize hybrids through marker-assisted introgression of β-carotene hydroxylase allele. PLoS ONE 9, e113583. doi: 10.1371/journal.pone.0113583
Natesan, S., Duraisamy, T., Pukalenthy, B., Chandran, S., Nallathambi, J., Adhimoolam, K., et al. (2020). Enhancing β-carotene concentration in parental lines of CO6 maize hybrid through marker-assisted backcross breeding (MABB). Front. Nutr. 7, 134. doi: 10.3389/fnut.2020.00134
Nelson, N. (1944). A photometric adaptation of the somogyi method for the determination of glucose. J. Biol. Chem. 153, 375–380. doi: 10.1016/S0021-9258(18)71980-7
Nelson, O. E. (1980). Genetic control of polysaccharide and storage protein synthesis in the endosperms of barley, maize, and sorghum. Adv. Cereal Sci. Technol. 3, 41–71.
Neuffer, M. G., and Sheridan, W. F. (1997). Defective kernel mutants of maize. I. Genetic and lethality studies. Genetics 95, 929–944. doi: 10.1093/genetics/95.4.929
Pfeiffer, W. H., and McClafferty, B. (2007). “Biofortification: breeding micronutrient-dense crops,” in Breeding Major Food Staples, eds M. S. Kang and P. M. Priyadarshan (Ames, IA: Blackwell Publishing), 61–91. doi: 10.1002/9780470376447.ch3
Pukalenthy, B., Manickam, D., Adhimoolam, K., Mahesh, S. G., Ramanathan, N., Chandran, S., et al. (2020). Marker aided introgression of opaque 2 (o2) allele improving lysine and tryptophan in maize (Zea mays L.). Physiol. Mol. Biol. Plants 26, 1925–1930. doi: 10.1007/s12298-020-00857-4
Pukalenthy, B., Manickam, D., Chandran, S., Adhimoolam, K., Sampathrajan, V., Rajasekaran, R., et al. (2019). Incorporation of opaque-2 into ‘UMI 1200', an elite maize inbred line, through marker-assisted backcross breeding. Biotechnol. Biotechnol. Equip. 33, 144–153. doi: 10.1080/13102818.2018.1556121
Qi, X., Zhao, Y., Jiang, L., Cui, Y., Wang, Y., and Liu, B. (2009). QTL analysis of kernel soluble sugar content in supersweet corn. Afr. J. Biotechnol. 8, 6913–6917.
Qutub, M., Chandran, S., Rathinavel, K., Sampathrajan, V., Rajasekaran, R., Manickam, S., et al. (2021). Improvement of a yairipok chujak maize landrace from north eastern Himalayan region for β-carotene content through molecular marker-assisted backcross breeding. Genes 12, 762. doi: 10.3390/genes12050762
Ribaut, J. M., and Hoisington, D. (1998). Marker-assisted selection: new tools and strategies. Trends Plant Sci. 3, 236–239. doi: 10.1016/S1360-1385(98)01240-0
Rodriguez-Amaya, D. B., and Kimura, M. (2004). HarvestPlus Handbook for Carotenoid Analysis, Vol. 2. Washington, DC: International Food Policy Research Institute (IFPRI).
Sagare, D. B., Shetti, P., Surender, M., and Reddy, S. S. (2019). Marker-assisted backcross breeding for enhancing β-carotene of QPM inbreds. Mol. Breed. 39, 31. doi: 10.1007/s11032-019-0939-x
Sarankumar, C., Karthikeyan, A., Vanniarajan, C., Kokiladevi, E., Ravikesavan, R., and Senthil, N. (2019). Frequency distribution of yield related traits in BC1F1, BC2F1 and BC2F2 backcross population of maize. Electro. J. Plant Breed. 10, 592–600. doi: 10.5958/0975-928X.2019.00075.9
Sarika, K., Hossain, F., Muthusamy, V., Zunjare, R. U., Baveja, A., Goswami, R., et al. (2018). Marker-assisted pyramiding of opaque2 and novel opaque16 genes for further enrichment of lysine and tryptophan in sub-tropical maize. Plant Sci. 272, 142–152. doi: 10.1016/j.plantsci.2018.04.014
Singh, A., Singh, V. K., Singh, S. P., Pandian, R. T. P., Ellur, R. K., Singh, D., et al. (2012). Molecular breeding for the development of multiple disease resistance in Basmati rice. AoB Plants 2012, pls029. doi: 10.1093/aobpla/pls029
Singh, A. K., Gopalakrishnan, S., Singh, V. P., Prabhu, K. V., Mohapatra, T., Singh, N. K., et al. (2011). Marker assisted selection: a paradigm shift in basmati breeding. Indian J. Genet. Plant Breed. 71, 120.
Somogyi, M. (1952). Notes on sugar determination. J. Biol. Chem. 195, 19–23. doi: 10.1016/S0021-9258(19)50870-5
Talukder, Z. A., Muthusamy, V., Chhabra, R., Gain, N., Reddappa, S. B., Mishra, S. J., et al. (2022). Combining higher accumulation of amylopectin, lysine and tryptophan in maize hybrids through genomics-assisted stacking of waxy1 and opaque2 genes. Sci. Rep. 12, 706. doi: 10.1038/s41598-021-04698-3
Wong, A. D., Juvik, J. A., Breeden, D. C., and Swiader, J. M. (1994). Shrunken2 sweet corn yield and the chemical components of quality. J. Am. Soc. Hortic. Sci. 119, 747–755. doi: 10.21273/JASHS.119.4.747
Yan, J., Kandianis, C. B., Harjes, C. E., Bai, L., Kim, E. H., Yang, X., et al. (2010). Rare genetic variation at Zea mays crtRB1 increases β-carotene in maize grain. Nat. Genet. 42, 322–327. doi: 10.1038/ng.551
Yang, R., Yan, Z., Wang, Q., Li, X., and Feng, F. (2018). Marker-assisted backcrossing of lcyE for enhancement of proA in sweet corn. Euphytica 214, 1–12. doi: 10.1007/s10681-018-2212-5
Keywords: crtRB1, genomics aided breeding, maize, sweet corn, vitamin A deficiency
Citation: Saha I, Rathinavel K, Manoharan B, Adhimoolam K, Sampathrajan V, Rajasekaran R, Muthurajan R and Natesan S (2022) The resurrection of sweet corn inbred SC11-2 using marker aided breeding for β-carotene. Front. Sustain. Food Syst. 6:1004450. doi: 10.3389/fsufs.2022.1004450
Received: 27 July 2022; Accepted: 30 November 2022;
Published: 22 December 2022.
Edited by:
Abdul Wakeel, University of Agriculture, Faisalabad, PakistanReviewed by:
Yuanda Lv, Jiangsu Academy of Agricultural Sciences (JAAS), ChinaLakshmi Narayana Vemireddy, Acharya N. G. Ranga Agricultural University, India
Tayyaba Shaheen, Government College University, Faisalabad, Pakistan
Copyright © 2022 Saha, Rathinavel, Manoharan, Adhimoolam, Sampathrajan, Rajasekaran, Muthurajan and Natesan. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Senthil Natesan, c2VudGhpbF9uYXRlc2FuQHRuYXUuYWMuaW4=
 Iman Saha
Iman Saha Krishnakumar Rathinavel
Krishnakumar Rathinavel Bharani Manoharan
Bharani Manoharan Karthikeyan Adhimoolam
Karthikeyan Adhimoolam Vellaikumar Sampathrajan
Vellaikumar Sampathrajan Ravikesavan Rajasekaran
Ravikesavan Rajasekaran Raveendran Muthurajan
Raveendran Muthurajan Senthil Natesan
Senthil Natesan