
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
REVIEW article
Front. Sustain. Food Syst., 31 January 2022
Sec. Crop Biology and Sustainability
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.817932
This article is part of the Research TopicHeavy Metal(loid) Stress-alleviating and Phytostimulating Microorganisms: Dual-performing Warhorses in Soil-bioremediationView all 10 articles
Heavy metal (HM) contamination of soil and water resources is a global concern, which not only limits crop yield and quality, but also has serious environmental effects. Due to the non-biodegradable nature and toxicity, high concentration of HMs in food and environment is a serious threat to the entire ecosystem. Moreover, the target of supplying safe and quality food to the rising human population (expected to reach ~9–10 bn by the year 2050), necessitates effective treatment of the HM-contaminated soil. Various microbe-mediated bioremediation strategies such as biosorption, bioprecipiation, biostimulation, etc., have been found to be effective in uptake and conversion of HMs to less toxic forms. Further, in the past few years, the use of soil and plant-associated microbiome for HM stress alleviation is gaining attention among the scientific community. In general, microbes are spectacular in being dynamic and more responsive to environmental conditions in comparison to their host plants. Moreover, with the advancements in high throughput sequencing technologies, the focus is eventually shifting from just structural characterization to functional insights into the microbiome. The microbes inhabiting the HM-contaminated environments or associated with HM-tolerant plants are a source for exploring HM-tolerant microbial communities, which could be used for enhancing bioremediation efficiency and conferring HM tolerance in plants. This review discusses the application of omics techniques including metagenomics, metatranscriptomics, metaproteomics, and metabolomics, for rapid and robust identification of HM-tolerant microbial communities, mining novel HM resistance genes, and fabricating the HM resistome.
Heavy metals (HMs) constitute a class of non-biodegradable environmental pollutants, which have detrimental effects on both terrestrial and aquatic ecosystems (Banach et al., 2020). Heavy metals refer to the d-block elements with density >5 g/cm3 (Singh et al., 2016). Some HMs such as zinc (Zn), manganese (Mn), copper (Cu), iron (Fe), and nickel (Ni) act as enzyme cofactors (hence, are essential for physiological functions); most of them, however, disrupt normal cellular metabolism (Goyal et al., 2020). On the basis of their physiochemical characteristics, HMs could be divided into redox metals and non-redox metals. Redox active metals such as Mn, chromium (Cr), Cu, and Fe are responsible for causing oxidative damage to plant cells through production of reactive oxygen species (ROS) by Haber-Weiss and Fenton reactions (Valko and Cronin, 2005; Jozefczak et al., 2012). Such severe oxidative injury leads to disruption in cell homeostasis, degradation of DNA, proteins, and cell membrane components, and destruction of photosynthetic pigments, which may finally result in cell death (Schutzendubel and Polle, 2002; Flora, 2009). On the other hand, non-redox active metals such as aluminum (Al), cadmium (Cd), Zn, Ni, and mercury (Hg) generate oxidative stress indirectly by inhibiting the activity of antioxidants (through glutathione consumption or binding to sulfhydryl groups of proteins) or inducing the activity of ROS-producing enzymes (Emamverdian et al., 2015). Irrespective of the mode of action, consumption of HM-contaminated food and water leads to severe health complications such as kidney damage, cardiovascular diseases, neurological disorders, lung damage, and gastro-intestinal problems (Järup, 2003; Johri et al., 2010). HM-contamination of soil results in loss of fertility, primarily by altering the structure and composition of microbial communities and soil physicochemistry. For example, high concentration of Cd alters the soil microbial population and modifies the soil physiochemistry, thereby resulting in loss of some indigenous microbial species (that are unable to adapt Cd stress) (Salam et al., 2020). Therefore, considering the growing rate of organ-disorders in humans and environmental degradation, there is a need to employ effective measures to reduce HM contamination in environment, as well as to devise novel strategies for HM stress alleviation. This review article focuses on the application of omics technologies for identification of HM-tolerant microbiota, their genes/operons and mechanism of HM stress alleviation, for bioremediation and for imparting HM tolerance to crop plants.
The toxic concentrations of HMs in soil find its way through the vascular system and interfere with the function of biomolecules (such as protein, DNA, etc.) present in plant cells. However, as an adaptation to prolonged HM stress, some plant species have evolved morphological (formation of hairy roots, trichomes, thick wall, cuticle, etc.), and physiological (secretion of root exudates rich in organic acids, proline accumulation, and phytohormone production) adaptations (Rajkumar et al., 2012; Hauser, 2014; Boiteau et al., 2018; Tiwari and Lata, 2018). At the cellular level, tolerant plants use mechanisms including HM uptake and efflux, transport, sequestration, and chelation (Viehweger, 2014). Examples of some metal-tolerant plant species include Arabidopsis arenosa, Arabidopsis halleri, Deschampsia caespitosa, and Silene vulgaris (Borymski et al., 2018). The HM-tolerant plants could be divided into two groups, based on the tolerance mechanism: (i) species that avoid HM-uptake and hence, prevent accumulation in shoot system (ii) species that (hyper)accumulate and tolerate high concentrations of HMs. The hyperaccumulators include plant species such as Azolla filiculoides, Combretum erythrophyllum, A. halleri, etc., which employ measures such as over-expression of transport systems, high concentration of metal chelators, and greater ability to detoxify and accumulate HMs in their aerial organs (Viehweger, 2014).
In nature, plants do not exist as isolated entities; they are associated with microbial communities in the rhizosphere (soil around plant roots), phyllosphere (aerial parts of plant), and endosphere (inside the plant tissues), to form the holobiont. Research carried out in the past few decades have highlighted the role of microbiome in affecting overall plant health and responses, especially under abiotic and biotic stress conditions (Phurailatpam and Mishra, 2020; Mishra et al., 2021a,b). Moreover, since microbes are highly sensitive as well as adaptable to environmental factors (including HM contaminants), they act as powerful model systems to decipher mechanisms for alleviating HM stress in crop plants (Figure 1). Therefore, researchers have focused on analyzing the microbiota associated with metal tolerant plant species, to understand the contribution of microbiome in HM tolerance of the holobiont (Table 1). Omics techniques (such as metagenomics, metatranscriptomics, metaproteomics, and metabolomics) have provided important leads on the microbial structure and composition (species abundance and diversity), metabolic potential (HM-tolerant/detoxification genes and proteins), and plant-microbe crosstalk, in response to HM stress.
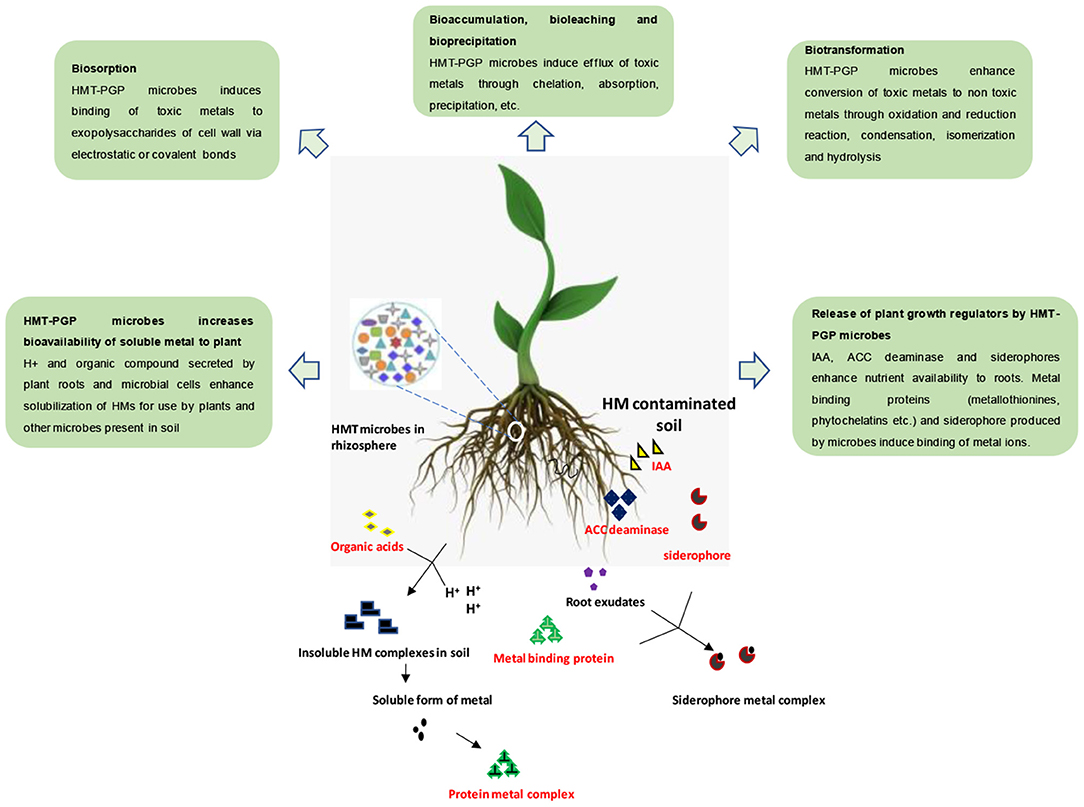
Figure 1. Mechanism of action of HM-tolerant (HMT) microbiome in bioremediation of HM-contaminated soil. The HMT microbiota present in the rhizosphere may exhibit plant growth promoting traits including secretion of phytohormones, siderophores, etc. (Adapted from Mishra et al., 2017).
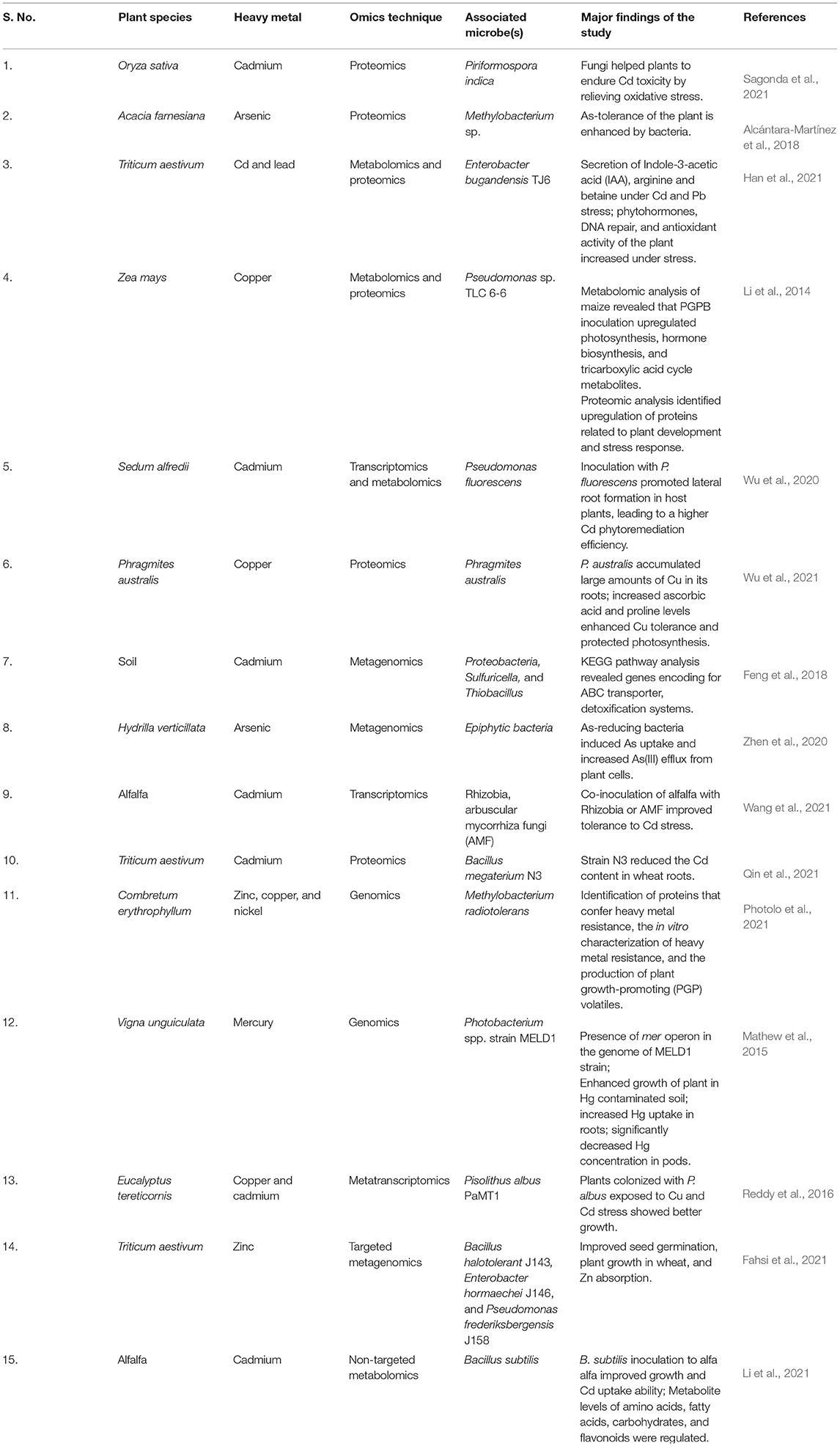
Table 1. Some recent studies involving omics technologies for unraveling the role of microbiota in HM-tolerance.
Until the past few decades, most of our knowledge on HM stress tolerance of plant-associated microbiota was based on culture-dependent approaches. For example, a culture-dependent study of two HM-contaminated sites in Portugal reported altered abundance and composition of bacterial communities, with most species belonging to Actinobacteria, Firmicutes, and Proteobacteria, and predominant genera belonging to Pseudomonas, Arthrobacter, and Bacillus (Pires et al., 2017). The HM-tolerant strains native to the contaminated sites, are promising candidates for their application in bioremediation. Though this approach enables isolation of microbial strains in tangible form; majority of the microbial communities being unculturable (under standard laboratory conditions) remains unnoticed and hence, unexplored (Staley and Konopka, 1985). Another approach involving PCR amplification of HM stress tolerance genes enables rapid screening of microbial strains; it however, suffers from the limitation of not being able to reveal novel candidates. Therefore, in order to circumvent such limitations, techniques based on functional analysis of metagenome have helped in identification and isolation of novel candidates (Handelsman et al., 1998; Majernik et al., 2001). The procedure involves extraction of total DNA from microbial communities of a particular environment, followed by screening for the desired “activity,” for example, screening for Na+(Li+)/H+antiporter activity in Escherichia coli (Majernik et al., 2001). This promising approach has been successfully used in detection and identification of genes inducing lithium-resistant phenotypes in E. coli.
The next generation sequencing (NGS) technologies have contributed toward increasing our understanding of various fields including plant microbiome. This is mainly attributed to the fact that majority of the microorganisms are unculturable under laboratory conditions, a phenomenon commonly known as “the great plate count anomaly” (Staley and Konopka, 1985). However, with recent technological advancements such as culturomics and FACS metagenomics, the percentage of 99% unculturability (Staley and Konopka, 1985) has been brought down to ~80% or even lesser (Bellali et al., 2021). Next generation sequencing offers an economical and rapid method for unraveling microbial diversity, which is otherwise inaccessible by conventional methods. The omics approaches have identified several genes from plants (Singh et al., 2016) and plant-associated microbiota, which could be utilized for imparting HM stress tolerance to the holobiont (Figure 2). For example, whole genome sequencing of Methylobacterium radiotolerans MAMP 4754, an endophytic strain associated with the hyperaccumulator plant Combretum erythrophyllum, has identified HM tolerance genes against Zn, Ni, and Cu (Photolo et al., 2021). Further, the knowledge of HM-tolerant microbiota (and their genes) could be incorporated in plant breeding programs aimed at generating HM-tolerant crops, in an approach known as holobiont breeding (Sahu and Mishra, 2021). The contribution of various omics approaches, namely metagenomics, metatranscriptomics, metaproteomics, and metabolomics in microbe-mediated HM tolerance, have been discussed below.
In metagenomics, microbial DNA sequencing is performed directly from the environmental sample, without involving the isolation of microbial communities (Akinsanya et al., 2015). Used for analyzing the structure and function of microbial communities, metagenomics could be divided into two types: high-throughput targeted amplicon sequencing and whole-genome shotgun metagenomics (Meena et al., 2017; Mishra et al., 2021b). The targeted amplicon sequencing involves specific amplification of ribosomal RNA genes (16S rRNA for bacteria and archaea and 18S rRNA or ITS for fungi) for determining the composition and diversity of microbial communities present in the samples. Being a cost-effective technique, 16S rRNA gene sequencing has been widely used for taxonomic profiling of microbial communities associated with HM-contaminated soil samples. For example, a study involving 16S rRNA gene sequencing of two nickel contaminated sites in southwest Slovakia revealed that phyla Euryarcheota followed by Crenarchaeota (both belonging to domain Archaea) were present in both regions, with same species richness at the genus level (Remenár et al., 2017). Likewise, a recent study involving targeted sequencing of V3–V4 region of 16S rRNA gene investigated the bacteriome composition of A. filiculoides (a metal hyperaccumulator) exposed to HM stress (Banach et al., 2020). It was found that Cyanobacteria and Proteobacteria comprised >97% of the sequencing reads. Apart from confirming the occurrence of known metal-tolerant genera, some (previously unidentified) potential metal tolerant genera were reported, which include Acinetobacter, Asticcacaulis, Anabaena, Bacillus, Brevundimonas, Burkholderia, Dyella, Methyloversatilis, Rhizobium, and Staphylococcus (Banach et al., 2020). It needs to be noted that although targeted amplicon sequencing does not directly provide information on the functional contribution of the associated microbial communities, some predictive metagenomics software such as PICRUSt and Tax4Fun predict the metabolic potential of bacterial communities (Douglas et al., 2020; Mishra et al., 2021b).
Shotgun metagenomics, on the other hand, provides information about both structural and functional attributes of the microbial communities. A recently conducted shotgun metagenomics study determined the HM resistome (collection of all the heavy metal resistance genes) of agricultural soil in Nigeria with (250 mg/kg) or without Cd contamination (Salam et al., 2020). The authors reported functional annotation of genes encoding for HM-translocating P-type ATPases, which are responsible for efflux and detoxification of Cd such as czcA, czcD, czrA, etc. In addition, resistance genes against other classes of HMs such as Co, Ni, Cu, Fe, Hg, etc. were also reported (Salam et al., 2020). Using multiple techniques namely comparative metagenomics, 16S rRNA gene sequencing and qPCR analysis, the diversity of bacterial microbiome in sediments of three HM-contaminated rivers of China were investigated (Chen et al., 2018). The core microbiota of contaminated sediments were represented by bacterial species belonging to the phyla Proteobacteria, Bacteroidetes, and Firmicutes, which were present in higher abundance at all the three sites. Besides, the functional annotation in shotgun metagenomics revealed genes mainly involved in DNA recombination and repair, and HM-resistance genes in the contaminated rivers (Chen et al., 2018). In a recent study, shotgun metagenomics approach was used to unravel the HM (and antibiotic) resistome of hydrocarbon-polluted soil samples collected from an automobile workshop at Taiwo, Nigeria (Salam, 2020). The functional annotation of the ORFs revealed several antibiotic (majority representing β-lactamase encoding genes) and HM tolerant genes, which constitute the resistome of the polluted area (Salam, 2020). The resistance genes against a wide range of HMs such as Cu, Ag, Ga, Zn, Fe, Cu, Cd, Ni, etc., were represented in the soil sample. Interestingly, most of the tolerant genes were found to reside on the mobile genetic elements, to promote their spread in the polluted soil (Salam, 2020).
Apart from identifying the HM-tolerant genera and HM-resistance genes in the environmental samples, metagenomic analyses have also provided insights into the role of microbial inoculation in enhancing the remediation capacity of some plant species. In one such study, the inoculation of a rhizobial bacterial species, Mesorhizobium loti HZ76, improved the phytoremediation ability of Robinia pseudoacacia (a deciduous tree, also known as black locust) growing in HM-contaminated soil (Fan et al., 2018). The 16S rRNA gene sequencing and shotgun metagenome sequencing of the rhizospheric microbes of R. pseudoacacia revealed that upon rhizobial inoculation, the genes encoding ATP-binding cassette transporters were upregulated, thereby highlighting the role of plant-microbiota interactions in enhancing the phytoremediation efficiency of the holobiont (Fan et al., 2018).
Metatranscriptomics is a high throughput method for detection of active microbial species and genes (that are actively transcribed in the sample) under the particular set of environmental conditions (Simon and Daniel, 2011; White et al., 2017). This technique has enabled researchers to explore changes in microbial mRNA pool of environmental samples, and to study the response mechanism of microbial communities to HM stress (Yu et al., 2021). Further, functional metatranscriptomics approach allows identification of genes from microbial communities responsible for adapting to extreme environmental conditions. A protocol for functional metatranscriptomics study would involve isolation of total RNA from an environmental sample, construction of cDNA library, screening for HM tolerance transcripts through bacterial or yeast complementation system, and finally sequencing for identification of transcripts of interest (Thakur et al., 2018; Mukherjee and Reddy, 2020). Thakur et al. (2018) used this approach to screen cDNA library prepared from Cd contaminated site. The study revealed a yeast transformant that exhibited significant tolerance against multiple stresses (2–4 mM Co, 150–300 μM Cu, and 10–12 mM Zn; Thakur et al., 2018).
Lehembre et al. (2013) used the functional meta-transcriptomics approach to decipher the functional contribution of microbiota toward HM resistance. The researchers performed functional screening of the soil eukaryotic metatranscriptome library (constituting total RNA from all the soil inhabiting microbes) for the ability to rescue (complement) the Cd or Zn sensitive phenotype of yeast mutants. The study identified some novel proteins which were previously uncharacterized with respect to HM resistance, such as BolA proteins and saccaropine dehydrogenase (for Zn tolerance), and C-terminal of aldehyde dehydrogenase (ADH) for Cd tolerance (Lehembre et al., 2013). In another study involving screening of eukaryotic cDNA library (prepared from HM contaminated soil), a clone (PLCc38) homologous to ADH, was found to be tolerant to Cu, Cd, Zn, and Co (Mukherjee et al., 2019). Aldehyde dehydrogenase enzymes eliminate the toxic aldehydes generated during various abiotic stresses (including HM stress).
In a recent published report, Yu et al. (2021) studied the response of soil microbiota to short term Cr6+ stress (1 mM Cr6+ for 30 min), using metatranscriptomics (and metagenomics) approach. The metagenomics study showed that 99% of the microbial communities (at genera level) were common between the control and stress groups. In contrast, the metatranscriptomics approach revealed that 83% of the microbes showed change in relative abundance (at RNA level) in response to stress. Among the upregulated genes were those involved in oxidative stress, and transport, resistance, and reduction of Cr6+. Further, ectopic expression of two unknown (upregulated) genes in Escherichia coli demonstrated their role in Cr6+ remediation. In a previous study, comparative metagenomic and metatranscriptomic study on Cr6+-contaminated (long term) riparian soil have been used to screen for genes involved in Cr6+ remediation (Pei et al., 2020). The omics analysis enabled the identification of six novel genes with Cr6+ tolerance property. Protein expression studies in E. coli with two genes, mcr and gsr, demonstrated reduction of ~50% Cr6+ in industrial wastewater contaminated with 200–600 μM of Cr6+ within 17 days (Pei et al., 2020).
Metaproteomics, also known as community proteomics or environmental proteomics, involves high-throughput study of all the proteins from microbial communities, extracted directly from the environment (Bastida et al., 2009; Gutleben et al., 2018; Li et al., 2019). A typical metaproteomics approach involves extraction of proteins from the environmental sample (soil, water, sediments, etc.), digestion of protein into peptides, followed by fractionation using 2D gel electrophoresis or liquid chromatography, and protein identification by mass spectrometry (against comparison to protein sequence database). It is a rapid method to identify and quantify the protein complement as well as protein–protein interactions in the community. Further, it provides a more authentic picture of the functional contribution of microbiota as often the DNA or RNA abundance does not co-relate well with protein abundance. Mattarozzi et al. (2017) investigated the rhizosphere of Biscutella laevigata (HM-tolerant) and Noccaea caerulescens (Ni hyperaccumulator) plants growing on serpentine soils by 16S rRNA gene sequencing and an LC-HRMS-based metaproteomics approach (Mattarozzi et al., 2017). Serpentine soils are characterized by high pH and HM concentration, and are low in nutrients and water holding capacity (Brady et al., 2005). The structural and functional characterization of microbial communities residing in soils contaminated with Ni, Co, and Cr revealed proteins involved in response to stimulus and metal transport. In addition, the taxonomic characterization revealed higher abundance of bacterial species namely Microbacterium oxidans, Pseudomonas oryzihabitans, Stenotrophomonas rhizophila, and Bacillus methylotrophicus, in the rhizosphere of these tolerant plants, in comparison to bulk soil (Mattarozzi et al., 2017). This study also highlighted the key interactions between bacterial communities and metal tolerant and hyperaccumulator plants in tackling HM stress. Moreover, another previous study examining the microbial diversity of Ni-contaminated serpentine soil has demonstrated that bacterial genera such as Pseudomonas and Streptomyces had over the years, become resistant to nickel ions by developing a highly potent nickel-resistant niche within the soil atmosphere (Mengoni et al., 2001).
The microbial transformation of Hg to MeHg, a potent neurotoxin that can bioaccumulate and biomagnify in food webs, is carried out by a group of bacteria known as mercury methylating bacteria. It is known that this transformation is an anaerobic process and depends upon the presence of hgcAB gene pair. Christensen et al. (2019) used a combination of shotgun metagenomics, 16S rRNA pyrosequencing, and metaproteomics approaches to determine the presence, distribution and diversity of mercury methylating bacteria in eight different sites in the USA. The metaproteomic (and metagenomic) analysis revealed that members belonging to Deltaproteobacteria phylum constituted the majority (~40–70%) of mercury methylators at all the sites. Further, there was poor co-relation between the Hg concentration in soil samples and abundance of mercury methylating bacteria (Christensen et al., 2019).
The shoot proteome analysis of A. halleri (a hyperaccumulator) grown under Cd and Zn stress, either in the presence or absence of Cd- and Zn-resistant bacterial strains, was investigated by Farinati et al. (2011). The proteomic analysis revealed that in the presence of Cd- and Zn-resistant microbes, there was an enhanced uptake of Cd and Zn in the shoots, upregulation of photosynthesis and stress-related proteins (for example, rubisco, malate dehydrogenase, and superoxide dismutase) and decreased abundance of plant defense-related proteins (Farinati et al., 2011).
Metabolomics is the large-scale study of total low-molecular weight compounds (<2 kDa) present at a particular developmental stage or environmental conditions (Grim et al., 2019). Metabolites are closer to the final phenotype of the organism, in comparison to transcripts and proteins. Besides, metabolomics studies are instrumental in bridging the gap between genotype and phenotype. Some plants known as metallophytes, have evolved mechanisms to tolerate high concentrations of HMs. Heavy Metal-tolerant plants could be obligate metallophytes that can survive only in high HM areas, or facultative metallophytes that are found in both normal and high HM-contaminated soils. Further, the recruitment of specific microbial communities in the rhizosphere could enhance the overall HM tolerance, and hence, remediation ability of plants. Plants produce a cocktail of small and large molecular weight, organic and inorganic compounds within their rhizosphere, to provide a nutrient rich environment for the microbial communities. The rhizosphere of metallophytes has been investigated to understand the exudates which could aid in the recruitment of specific metal-resistant microbial communities (Zhang et al., 2012; Borymski et al., 2018). Often the metal-resistant microbiota exhibit plant growth promoting properties such as nutrient solubilization, secretion of phytohormones, and enzymes such as 1-aminocyclopropane-1-carboxylate deaminase (Sessitsch et al., 2013; Sasse et al., 2018), which enable plants to survive in metalliferous soils that are nutrient poor.
Metabolomics of HM-contaminated soils and tolerant plants inhabiting HM-rich areas have been instrumental in identification of metal-resistant microbiota. Toyama et al. (2011) reported the production of plant metabolites such as sugar, short-chained organic acids, amino acids, and phenols, which act as source of nutrients for rhizospheric microbes, and accelerated the phytoremediation of pyrene and benzopyrene. The role of rhizospheric bacteria in biodegradation of these contaminants was indicated by their persistence in the control set (with autoclaved rhizosphere sediments of sterilized plants) (Toyama et al., 2011). For example, Phragmites australis, also known as common reed, is a tall wetland grass used for wetland phytoremediation. Further, a recent study used metabolomics approach to analyze 73 metabolomes associated with P. australis growing in different acid mine drainage sites. The researchers observed that the distinct parts of roots (endosphere vs. rhizosphere) secreted spatially defined metabolites, depending upon total dissolved solutes, pH, and the presence of different metals such as Fe, Cr, Cu, and Zn (Kalu et al., 2021). It also needs to be emphasized that the secreted metabolites did not significantly vary between the different acid mine drainage sites, indicating a conserved response to these contaminants.
Metabolomic studies of plants with or without microbial inoculation have been used to decipher the mechanism of HM tolerance. For instance, the Pb accumulation ability of Salix integra upon inoculation with some indigenous rhizospheric microbes was investigated by targeted metabolomics approach (Niu et al., 2021). Inoculation with Pb-resistant Bacillus sp. and Aspergillus niger, showed enhanced proline levels as well as increased superoxide dismutase and catalase (antioxidants) activity (Niu et al., 2021). This study also identified 410 metabolites, which mainly constituted organic acids, amino acids and carbohydrates, in response to microbial inoculation. Further, around half of the identified metabolites were associated with HM bioavailability (Niu et al., 2021), thereby corroborating the role of microbes in increasing the phytoremediation efficiency. In another recent report, metabolomics and proteomics approach was used to study the mechanism of enhanced tolerance of wheat to Cd and Pd stress on inoculation with Enterobacter bugandensis TJ6 strain, a HM-immobilizing bacterium (Han et al., 2021). The TJ6 bacterial strain employed multiple strategies, including ~50% reduced accumulation of Cd and Pd uptake, enhanced bioprecipitation and extracellular absorption, and secretion of arginine, betaine, and IAA. Further, the better tolerance of host plants was evident through improved DNA repair ability and antioxidant enzyme activity, and increased level of phytohormones in wheat roots (Han et al., 2021).
Phytoremediation is a cost-effective and environmental friendly approach. However, its large-scale application is limited by its lower efficiency. This issue could be addressed by inoculation of plants with microbes, in an approach known as plant-microbial remediation (Niu et al., 2021). For instance, the legume-Rhizobia interaction has been exploited to increase the phytoremediation ability of plants. The dual co-cultivation of Agrobacterium tumefaciens CCNWGS0286 (an IAA producing bacteria) and Sinorhizobium meliloti (nitrogen fixation) in alfa-alfa increases the zinc and copper accumulation ability, apart from promoting plant growth (Jian et al., 2019). The synergistic action of A. tumefaciens and S. meliloti enhanced the root nodulation (48% higher nodule number) and plant biomass (by >30% dry weight) under Cu and Zn stress, by promoting the antioxidant activity of plants (Jian et al., 2019). Similar mode of action of microbe-mediated HM stress alleviation was reported in a recently published study involving Cd stress in alfa alfa by Li et al. (2021). The growth and Cd uptake ability of alfa alfa was promoted upon inoculation with Bacillus subtilis due to enhanced antioxidant enzyme activity and reduced malondialdehyde (an indicator of oxidative lipid damage) levels in plants. In addition, the microbial inoculation increased Cd removal ability by >130%, and facilitated nutrient recycling as well (Li et al., 2021).
Natural environments, including HM-contaminated areas, contain many important microbial species that are unculturable (under standard lab conditions), and hence, inaccessible for further characterization. Omics approaches have been a boon in microbiome research, enabling identification and characterization of uncultured microbes, that constitute a major fraction (>90%) of the environmental samples. Further, metratranscriptomics and metaproteomics provide an insight into the “expressed” component of the community. Often two or more omics approaches are combined to corroborate the results and obtain an authentic picture of the microbiome. For instance, combining metaproteomics with metagenomics enables a co-relation between potential genetic diversity and actual “activity” occurring in the microbial communities.
However, the omics techniques are limited by technological challenges associated with extraction and fractionation procedures (high amounts of humic substances in soil), requirement of robust bioinformatic tools (huge computational load resulting from omics, normally from megabytes to terabytes of data) and sophisticated equipment and procedures. A major challenge associated with metabolomics study is the diverse chemical nature of metabolites, due to which no one protocol could be used to estimate all metabolites produced in a cell. Likewise, the metaproteome extracted from environmental samples are contaminated by proteins from other organisms such as protozoa, nematodes, etc. Therefore, a prior separation of microbial cells from the sample, followed by protein extraction and fractionation would be a better representation of protein fingerprint.
Investigation of HM-tolerant microbial communities from rhizospheric and endophytic microbiota associated with hyperaccumulator and facultative metallophytes (grown in high HM concentrations), is a powerful strategy of bioprospecting for bioremediation (Photolo et al., 2021). The higher abundance of some bacterial and fungal species in HM-contaminated sites represents the indigenous/native resistant population, which could be exploited for remediation of HMs. However, the isolation of microbial strains (in tangible/physical form) is a pre-requisite to explore the functional and ecological roles of potential microbial communities. Subsequently, carefully optimized media and culture conditions (mimicking the natural environmental conditions) could be used to isolate “unculturable” and novel species. Several such prokaryotic uncultivated but well-characterized species have been placed under the nomenclature Candidatus.
Another major challenge for plant-microbial remediation of HMs is the inability of inoculated microbes to maintain their number and sustain the remediation activity under field conditions. Therefore, it is important to consider the interaction between the added inoculants and microbial communities in nature (bulk soil, rhizosphere, and endosphere). Further, advancements in the field of protein and metabolite extraction, fractionation, and characterization methods as well as advances in instrumentation would boost the research on functional microbial ecology (focusing on the microbiome component of the plant holobiont), and open avenues for solving environmental issues, including HM contamination of environment. Further, the knowledge of HM-tolerant microbiota could be incorporated to assist the plant breeding programs aimed at generating HM-tolerant crops, in an approach known as holobiont breeding.
SM conceived the idea of the manuscript. LP, VD, NS, and SM prepared the manuscript and figures. All authors have read and approved the final version of the manuscript.
SM and VD would like to acknowledge the financial support provided by the Dayalbagh Educational Institute (Deemed-to-be-University), Agra, India, in the form of minor research project [Grant No. DEI/GBMF (1732020)/43].
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Akinsanya, M. A., Goh, J. K., Lim, S. P., and Ting, A. S. Y. (2015). Metagenomics study of endophytic bacteria in Aloe vera using next-generation technology. Genom. Data 6, 159–163. doi: 10.1016/j.gdata.2015.09.004
Alcántara-Martínez, N., Figueroa-Martínez, F., Rivera-Cabrera, F., Gutiérrez-Sánchez, G., and Volke-Sepúlveda, T. (2018). An endophytic strain of Methylobacterium sp. increases arsenate tolerance in Acacia farnesiana (L.) Willd: a proteomic approach. Sci. Total Environ. 625, 762–774. doi: 10.1016/j.scitotenv.2017.12.314
Banach, A. M., Kuzniar, A., Grzadziel, J., and Wolińska, A. (2020). Azolla filiculoides L. as a source of metal-tolerant microorganisms. PLoS ONE 15, e0232699. doi: 10.1371/journal.pone.0232699
Bastida, F., Moreno, J. L., Nicolas, C., Hernandez, T., and Garcia, C. (2009). Soil metaproteomics: a review of an emerging environmental science. Significance, methodology and perspectives. Eur. J. Soil Sci. 60, 845–859. doi: 10.1111/j.1365-2389.2009.01184.x
Bellali, S., Lagier, J.-C., Million, M., Anani, H., Haddad, G., Francis, R., et al. (2021). Running after ghosts: are dead bacteria the dark matter of the human gut microbiota? Gut Microbes 13, 1–12. doi: 10.1080/19490976.2021.1897208
Boiteau, R. M., Shaw, J. B., Pasa-Tolic, L., Koppenaal, D. W., and Jansson, J. K. (2018). Micronutrient metal speciation is controlled by competitive organic chelation in grassland soils. Soil Biol. Biochem. 120, 283–291. doi: 10.1016/j.soilbio.2018.02.018
Borymski, S., Cycoń, M., Beckmann, M., Mur, L. A., and Piotrowska-Seget, Z. (2018). Plant species and heavy metals affect biodiversity of microbial communities associated with metal-tolerant plants in metalliferous soils. Front. Microbiol. 9, 1425. doi: 10.3389/fmicb.2018.01425
Brady, K. U., Kruckeberg, A. R., and Bradshaw, H. D. Jr (2005). Evolutionary ecology of plant adaptation to serpentine soils. Annu. Rev. Ecol. Evol. Syst. 36, 243–266. doi: 10.1146/annurev.ecolsys.35.021103.105730
Chen, Y., Jiang, Y., Huang, H., Mou, L., Ru, J., Zhao, J., et al. (2018). Long-term and high-concentration heavy-metal contamination strongly influences the microbiome and functional genes in Yellow River sediments. Sci. Total Environ. 637, 1400–1412. doi: 10.1016/j.scitotenv.2018.05.109
Christensen, G. A., Gionfriddo, C. M., King, A. J., Moberly, J. G., Miller, C. L., Somenahally, A. C., et al. (2019). Determining the reliability of measuring mercury cycling gene abundance with correlations with mercury and methylmercury concentrations. Environ. Sci. Technol. 53, 8649–8663. doi: 10.1021/acs.est.8b06389
Douglas, G. M., Maffei, V. J., Zaneveld, J. R., Yurgel, S. N., Brown, J. R., Taylor, C. M., et al. (2020). PICRUSt2 for prediction of metagenome functions. Nat. Biotechnol. 38, 685–688. doi: 10.1038/s41587-020-0548-6
Emamverdian, A., Ding, Y., Mokhberdoran, F., and Xie, Y. (2015). Heavy metal stress and some mechanisms of plant defense response. ScientificWorldJournal. (2015) 2015, 756120. doi: 10.1155/2015/756120
Fahsi, N., Mahdi, I., Mesfioui, A., Biskri, L., and Allaoui, A. (2021). Plant growth-promoting Rhizobacteria isolated from the Jujube (Ziziphus lotus) plant enhance wheat growth, Zn uptake, and heavy metal tolerance. Agriculture 11, 316. doi: 10.3390/agriculture11040316
Fan, M., Xiao, X., Guo, Y., Zhang, J., Wang, E., Chen, W., et al. (2018). Enhanced phytoremdiation of Robinia pseudoacacia in heavy metal-contaminated soils with rhizobia and the associated bacterial community structure and function. Chemosphere 197, 729–740. doi: 10.1016/j.chemosphere.2018.01.102
Farinati, S., DalCorso, G., Panigati, M., and Furini, A. (2011). Interaction between selected bacterial strains and Arabidopsis halleri modulates shoot proteome and cadmium and zinc accumulation. J. Exp. Bot. 62, 3433–3447. doi: 10.1093/jxb/err015
Feng, G., Xie, T., Wang, X., Bai, J., Tang, L., Zhao, H., et al. (2018). Metagenomic analysis of microbial community and function involved in cd-contaminated soil. BMC Microbial. 18, 11. doi: 10.1186/s12866-018-1152-5
Flora, S. J. (2009). Structural, chemical and biological aspects of antioxidants for strategies against metal and metalloid exposure. Oxid. Med. Cell. Longev. 2, 191–206. doi: 10.4161/oxim.2.4.9112
Goyal, D., Yadav, A., Prasad, M., Singh, T. B., Shrivastav, P., Ali, A., et al. (2020). “Effect of heavy metals on plant growth: an overview,” in Contaminants in Agriculture, eds G. S. Naeem and A. Ansari (Cham: Springer), 79–101. doi: 10.1007/978-3-030-41552-5_4
Grim, C. M., Luu, G. T., and Sanchez, L. M. (2019). Staring into the void: demystifying microbial metabolomics. FEMS Microbiol. Lett. 366, fnz135. doi: 10.1093/femsle/fnz135
Gutleben, J., Chaib De Mares, M., Van Elsas, J. D., Smidt, H., Overmann, J., and Sipkema, D. (2018). The multi-omics promise in context: from sequence to microbial isolate. Crit. Rev. Microbiol. 44, 212–229. doi: 10.1080/1040841X.2017.1332003
Han, H., Zhang, H., Qin, S., Zhang, J., Yao, L., Chen, Z., et al. (2021). Mechanisms of Enterobacter bugandensis TJ6 immobilization of heavy metals and inhibition of Cd and Pb uptake by wheat based on metabolomics and proteomics. Chemosphere 276, 130157. doi: 10.1016/j.chemosphere.2021.130157
Handelsman, J., Rondon, M. R., Brady, S. F., Clardy, J., and Goodman, R. M. (1998). Molecular biological access to the chemistry of unknown soil microbes: a new frontier for natural products. Chem. Biol. 5, R245–R249. doi: 10.1016/S1074-5521(98)90108-9
Hauser, M. T. (2014). Molecular basis of natural variation and environmental control of trichome patterning. Front. Plant Sci. 5, 320. doi: 10.3389/fpls.2014.00320
Järup, L. (2003). Hazards of heavy metal contamination. Br. Med. Bull. 68, 167–182. doi: 10.1093/bmb/ldg032
Jian, L., Bai, X., Zhang, H., Song, X., and Li, Z. (2019). Promotion of growth and metal accumulation of alfalfa by coinoculation with Sinorhizobium and Agrobacterium under copper and zinc stress. PeerJ 7, e6875. doi: 10.7717/peerj.6875
Johri, N., Jacquillet, G., and Unwin, R. (2010). Heavy metal poisoning: the effects of cadmium on the kidney. Biometals 23, 783–792. doi: 10.1007/s10534-010-9328-y
Jozefczak, M., Remans, T., Vangronsveld, J., and Cuypers, A. (2012). Glutathione is a key player in metal-induced oxidative stress defenses. Int. J. Mol. Sci. 13, 3145–3175. doi: 10.3390/ijms13033145
Kalu, C. M., Ogola, H. J. O., Selvarajan, R., Tekere, M., and Ntushelo, K. (2021). Fungal and metabolome diversity of the rhizosphere and endosphere of Phragmites australis in an AMD-polluted environment. Heliyon 7, e06399. doi: 10.1016/j.heliyon.2021.e06399
Lehembre, F., Doillon, D., David, E., Perrotto, S., Baude, J., Foulon, J., et al. (2013). Soil metatranscriptomics for mining eukaryotic heavy metal resistance genes. Environ. Microbiol. 15, 2829–2840. doi: 10.1111/1462-2920.12143
Li, K., Pidatala, V. R., Shaik, R., Datta, R., and Ramakrishna, W. (2014). Integrated metabolomic and proteomic approaches dissect the effect of metal-resistant bacteria on maize biomass and copper uptake. Environ. Sci. Technol. 48, 1184–1193. doi: 10.1021/es4047395
Li, Q., Xing, Y., Fu, X., Ji, L., Li, T., Wang, J., et al. (2021). Biochemical mechanisms of rhizospheric Bacillus subtilis-facilitated phytoextraction by alfalfa under cadmium stress–Microbial diversity and metabolomics analyses. Ecotoxicol. Environ. Saf. 212, 112016. doi: 10.1016/j.ecoenv.2021.112016
Li, S., Hu, S., Shi, S., Ren, L., Yan, W., and Zhao, H. (2019). Microbial diversity and metaproteomic analysis of activated sludge responses to naphthalene and anthracene exposure. RSC Adv. 9, 22841–22852. doi: 10.1039/C9RA04674G
Majernik, A., Gottschalk, G., and Daniel, R. (2001). Screening of environmental DNA libraries for the presence of genes conferring Na+ (Li+)/H+ antiporter activity on Escherichia coli: characterization of the recovered genes and the corresponding gene products. J. Bacteriol. Res. 183, 6645–6653. doi: 10.1128/JB.183.22.6645-6653.2001
Mathew, D. C., Ho, Y. N., Gicana, R. G., Mathew, G. M., Chien, M. C., and Huang, C. C. (2015). A rhizosphere-associated symbiont, Photobacterium spp. strain MELD1, and its targeted synergistic activity for phytoprotection against mercury. PLoS ONE. 10, e0121178. doi: 10.1371/journal.pone.0121178
Mattarozzi, M., Manfredi, M., Montanini, B., Gosetti, F., Sanangelantoni, A. M., Marengo, E., et al. (2017). A metaproteomic approach dissecting major bacterial functions in the rhizosphere of plants living in serpentine soil. Anal. Bioanal. Chem. 409, 2327–2339. doi: 10.1007/s00216-016-0175-8
Meena, K. K., Sorty, A. M., Bitla, U. M., Choudhary, K., Gupta, P., Pareek, A., et al. (2017). Abiotic stress responses and microbe-mediated mitigation in plants: the omics strategies. Front. Plant Sci. 8, 172. doi: 10.3389/fpls.2017.00172
Mengoni, A., Barzanti, R., Gonnelli, C., Gabbrielli, R., and Bazzicalupo, M. (2001). Characterization of nickel-resistant bacteria isolated from serpentine soil. Environ. Microbiol. 3, 691–698. doi: 10.1046/j.1462-2920.2001.00243.x
Mishra, J., Singh, R., and Arora, N. K. (2017). Alleviation of heavy metal stress in plants and remediation of soil by rhizosphere microorganisms. Front. Microbiol. 8, 1706. doi: 10.3389/fmicb.2017.01706
Mishra, S., Bhattacharjee, A., and Sharma, S. (2021a). An ecological insight into the multifaceted world of plant-endophyte association. Crit. Rev. Plant Sci. 40, 127–146. doi: 10.1080/07352689.2021.1901044
Mishra, S., Goyal, D., and Phurailatpam, L. (2021b). Targeted 16S rRNA gene and ITS2 amplicon sequencing of leaf and spike tissues of Piper longum identifies new candidates for bioprospecting of bioactive compounds. Arch. Microbiol. 203, 3851–3867. doi: 10.1007/s00203-021-02356-w
Mukherjee, A., and Reddy, M. S. (2020). Metatranscriptomics: an approach for retrieving novel eukaryotic genes from polluted and related environments. 3 Biotech 10, 1–19. doi: 10.1007/s13205-020-2057-1
Mukherjee, A., Yadav, R., Marmeisse, R., Fraissinet-Tachet, L., and Reddy, M. S. (2019). Heavy metal hypertolerant eukaryotic aldehyde dehydrogenase isolated from metal contaminated soil by metatranscriptomics approach. Biochimie 160, 183–192. doi: 10.1016/j.biochi.2019.03.010
Niu, X. Y., Wang, S. K., Zhou, J., Di, D. L., Sun, P., and Huang, D. Z. (2021). Inoculation with indigenous rhizosphere microbes enhances aboveground accumulation of lead in Salix integra Thunb. by improving transport coefficients. Front. Microbiol. 12, 686812. doi: 10.3389/fmicb.2021.686812
Pei, Y., Tao, C., Ling, Z., Yu, Z., Ji, J., Khan, A., et al. (2020). Exploring novel Cr (VI) remediation genes for Cr (VI)-contaminated industrial wastewater treatment by comparative metatranscriptomics and metagenomics. Sci. Total Environ. 742, 140435. doi: 10.1016/j.scitotenv.2020.140435
Photolo, M. M., Sitole, L., Mavumengwana, V., and Tlou, M. G. (2021). Genomic and physiological investigation of heavy metal resistance from plant endophytic Methylobacterium radiotolerans MAMP 4754, Isolated from Combretum erythrophyllum. Int. J. Environ. Res. Public Health 18, 997. doi: 10.3390/ijerph18030997
Phurailatpam, L., and Mishra, S. (2020). “Role of plant endophytes in conferring abiotic stress tolerance,” in Plant Ecophysiology and Adaptation under Climate Change: Mechanisms and Perspectives II, ed M. Hasanuzzaman (Springer, Singapore), 603–628. doi: 10.1007/978-981-15-2172-0_22
Pires, C., Franco, A. R., Pereira, S. I., Henriques, I., Correia, A., Magan, N., et al. (2017). Metal (loid)-contaminated soils as a source of culturable heterotrophic aerobic bacteria for remediation applications. Geomicrobiol. J. 34, 760–768. doi: 10.1080/01490451.2016.1261968
Qin, S., Wu, X., Han, H., Pang, F., Zhang, J., and Chen, Z. (2021). Polyamine-producing bacterium Bacillus megaterium N3 reduced Cd accumulation in wheat and increased the expression of DNA repair-and plant hormone-related proteins in wheat roots. Environ. Exp. Bot. 189, 104563. doi: 10.1016/j.envexpbot.2021.104563
Rajkumar, M., Sandhya, S., Prasad, M. N. V., and Freitas, H. (2012). Perspectives of plant-associated microbes in heavy metal phytoremediation. Biotechnol. Adv. 30, 1562–1574. doi: 10.1016/j.biotechadv.2012.04.011
Reddy, M. S., Kour, M., Aggarwal, S., Ahuja, S., Marmeisse, R., and Fraissinet-Tachet, L. (2016). Metal induction of a Pisolithus albus metallothionein and its potential involvement in heavy metal tolerance during mycorrhizal symbiosis. Environ. Microbiol. 18, 2446–2454. doi: 10.1111/1462-2920.13149
Remenár, M., Harichová, J., Zámocký, M., Pangallo, D., Szemes, T., Budiš, J., et al. (2017). Metagenomics of a nickel-resistant bacterial community in an anthropogenic nickel-contaminated soil in southwest Slovakia. Biol. Plant. 72, 971–981. doi: 10.1515/biolog-2017-0117
Sagonda, T., Adil, M. F., Sehar, S., Rasheed, A., Joan, H. I., Ouyang, Y., et al. (2021). Physio-ultrastructural footprints and iTRAQ-based proteomic approach unravel the role of Piriformospora indica-colonization in counteracting cadmium toxicity in rice. Ecotoxicol. Environ. Saf. 220, 112390. doi: 10.1016/j.ecoenv.2021.112390
Sahu, P. K., and Mishra, S. (2021). Effect of hybridization on endophytes: the endo-microbiome dynamics. Symbiosis 84, 369–377. doi: 10.1007/s13199-021-00760-w
Salam, L. B. (2020). Unravelling the antibiotic and heavy metal resistome of a chronically polluted soil. 3 Biotech 10, 1–23. doi: 10.1007/s13205-020-02219-z
Salam, L. B., Obayori, O. S., Ilori, M. O., and Amund, O. O. (2020). Effects of cadmium perturbation on the microbial community structure and heavy metal resistome of a tropical agricultural soil. Bioresour. Bioprocess. 7, 1–19. doi: 10.1186/s40643-020-00314-w
Sasse, J., Martinoia, E., and Northen, T. (2018). Feed your friends: do plant exudates shape the root microbiome? Trends Plant Sci. 23, 25–41. doi: 10.1016/j.tplants.2017.09.003
Schutzendubel, A., and Polle, A. (2002). Plant responses to abiotic stresses: heavy metal-induced oxidative stress and protection by mycorrhization. J. Exp. Bot. 53, 1351–1365. doi: 10.1093/jexbot/53.372.1351
Sessitsch, A., Kuffner, M., Kidd, P., Vangronsveld, J., Wenzel, W. W., Fallmann, K., et al. (2013). The role of plant-associated bacteria in the mobilization and phytoextraction of trace elements in contaminated soils. Soil Biol. Biochem. 60, 182–194. doi: 10.1016/j.soilbio.2013.01.012
Simon, C., and Daniel, R. (2011). Metagenomic analyses: past and future trends. Appl. Environ. Microbiol. 77, 1153–1161. doi: 10.1128/AEM.02345-10
Singh, S., Parihar, P., Singh, R., Singh, V. P., and Prasad, S. M. (2016). Heavy metal tolerance in plants: role of transcriptomics, proteomics, metabolomics, and ionomics. Front. Plant Sci. 6, 1143. doi: 10.3389/fpls.2015.01143
Staley, J. T., and Konopka, A. (1985). Measurement of in situ activities of nonphotosynthetic microorganisms in aquatic and terrestrial habitats. Annu. Rev. Microbiol. 39, 321–346. doi: 10.1146/annurev.mi.39.100185.001541
Thakur, B., Yadav, R., Fraissinet-Tachet, L., Marmeisse, R., and Reddy, M. S. (2018). Isolation of multi-metal tolerant ubiquitin fusion protein from metal polluted soil by metatranscriptomic approach. J. Microbiol. Methods 152, 119–125. doi: 10.1016/j.mimet.2018.08.001
Tiwari, S., and Lata, C. (2018). Heavy metal stress, signaling, and tolerance due to plant-associated microbes: an overview. Front. Plant Sci. 9, 452. doi: 10.3389/fpls.2018.00452
Toyama, T., Furukawa, T., Maeda, N., Inoue, D., Sei, K., Mori, K., et al. (2011). Accelerated biodegradation of pyrene and benzo [a] pyrene in the Phragmites australis rhizosphere by bacteria–root exudate interactions. Water Res. 45, 1629–1638. doi: 10.1016/j.watres.2010.11.044
Valko, M., Morris, H., and Cronin, M. T. D. (2005). Metals, toxicity and oxidative stress. Curr. Med. Chem. 12, 1161–1208. doi: 10.2174/0929867053764635
Viehweger, K. (2014). How plants cope with heavy metals. Bot. Stud. 55, 1–12. doi: 10.1186/1999-3110-55-35
Wang, X., Fang, L., Beiyuan, J., Cui, Y., Peng, Q., Zhu, S., et al. (2021). Improvement of alfalfa resistance against Cd stress through rhizobia and arbuscular mycorrhiza fungi co-inoculation in Cd-contaminated soil. Environ. Pollut. 277, 116758. doi: 10.1016/j.envpol.2021.116758
White, R. A., Borkum, M. I., Rivas-Ubach, A., Bilbao, A., Wendler, J. P., Colby, S. M., et al. (2017). From data to knowledge: the future of multi-omics data analysis for the rhizosphere. Rhizosphere 3, 222–229. doi: 10.1016/j.rhisph.2017.05.001
Wu, J., Hu, J., Wang, L., Zhao, L., and Ma, F. (2021). Responses of Phragmites australis to copper stress: a combined analysis of plant morphology, physiology and proteomics. Plant Biol. 23, 351–362. doi: 10.1111/plb.13175
Wu, Y., Ma, L., Liu, Q., Vestergård, M., Topalovic, O., Wang, Q., et al. (2020). The plant-growth promoting bacteria promote cadmium uptake by inducing a hormonal crosstalk and lateral root formation in a hyperaccumulator plant Sedum alfredii. J. Hazard. Mater. 395, 122661. doi: 10.1016/j.jhazmat.2020.122661
Yu, Z., Pei, Y., and Zhao, S. (2021). Metatranscriptomic analysis reveals active microbes and genes responded to short-term Cr(VI) stress. Ecotoxicology 30, 1527–1537. doi: 10.1007/s10646-020-02290-5
Zhang, W. H., Huang, Z., He, L. Y., and Sheng, X. F. (2012). Assessment of bacterial communities and characterization of lead-resistant bacteria in the rhizosphere soils of metal-tolerant Chenopodium ambrosioides grown on lead–zinc mine tailings. Chemosphere 87, 1171–1178. doi: 10.1016/j.chemosphere.2012.02.036
Keywords: heavy metal, abiotic stress, bioremediation, omics, microbiome
Citation: Phurailatpam L, Dalal VK, Singh N and Mishra S (2022) Heavy Metal Stress Alleviation Through Omics Analysis of Soil and Plant Microbiome. Front. Sustain. Food Syst. 5:817932. doi: 10.3389/fsufs.2021.817932
Received: 18 November 2021; Accepted: 30 December 2021;
Published: 31 January 2022.
Edited by:
Krishnendu Pramanik, Visva-Bharati University, IndiaReviewed by:
Anirudha R. Dixit, Kennedy Space Center, United StatesCopyright © 2022 Phurailatpam, Dalal, Singh and Mishra. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Sushma Mishra, c3VzaG1hbWlzaHJhODdAZ21haWwuY29t; orcid.org/0000-0001-8012-3382
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.