- 1Applied Phycology and Biotechnology Division, Council of Scientific and Industrial Research (CSIR)-Central Salt and Marine Chemicals Research Institute, Bhavnagar, India
- 2Academy of Scientific and Innovative Research (AcSIR), Ghaziabad, India
Kappaphycus alvarezii seaweed extract (KSWE) has been known for its plant biostimulant and stress alleviation activities on various crops. However, very few reports are available depicting its impact at the molecular level, which is crucial in identifying the mechanism of action of KSWE on plants. Here, maize leaf tissue of control and KSWE-treated plants were analyzed for their transcriptional changes under drought stress. KSWE was applied foliarly at the V5 stage of maize crop under drought, and leaf transcriptome analysis was performed. It was found that a total of 380 and 631 genes were up- and downregulated, respectively, due to the application of KSWE. Genes involved in nitrate transportation, signal transmission, photosynthesis, transmembrane transport of various ions, glycogen, and starch biosynthetic processes were found upregulated in KSWE-treated plants, while genes involved in the catabolism of polysaccharide molecules such as starch as well as cell wall macromolecules like chitin and protein degradation were found downregulated. An overview of differentially expressed genes involved in metabolic as well as regulatory processes in KSWE-treated plants was also analyzed via Mapman tool. Phytohormone signaling genes such as cytokinin-independent 1 (involved in cytokine signal transduction), Ent-kaurene synthase and GA20 oxidase (involved in gibberellin synthesis), and gene of 2-oxoglutarate-dependent dioxygenase enzyme activity (involved in ethylene synthesis) were found upregulated while 9-cis-epoxycarotenoid dioxygenase (a gene involved in abscisic acid synthesis) was found downregulated due to the application of KSWE. Modulation of gene expression in maize leaf tissue in response to KSWE treatment elucidates mechanisms to ward off drought stress, which can be extended to understand similar phenomenon in other crops as well. This molecular knowledge can be utilized to make the use of KSWE more efficient and sustainable.
Introduction
Sustainable agriculture is the need of the hour to reduce the burden of climate change and global warming on the environment. According to the Global Sustainable Development Report, the current food system is contributing 19–29% of total greenhouse gas emissions (Messerli et al., 2019). Present agriculture practices are also unsustainable, leading to eutrophication, groundwater contamination, atmospheric pollution, and soil acidification (Messerli et al., 2019). Furthermore, due to ever-increasing atmospheric temperature and changes in global rainfall patterns, abiotic stressors have become an immense challenge for global agriculture. Without scientific interventions in a sustainable way, one is less likely to achieve global food targets and sustainable development goals. Biostimulants in the form of humic or fulvic acids, seaweed extracts, protein hydrolysates, chitosan or biopolymers, and beneficial microorganisms (bacteria or fungi) are frequently applied to plants to increase their nutrient efficiency, productivity, and stress tolerance capacity in an environment-friendly way (Du Jardin, 2015). Kappaphycus alvarezii seaweed extract (KSWE) is one such type of biostimulant. The potential of KSWE for its plant-growth-promoting (Rathore et al., 2009; Pramanick et al., 2014; Raverkar et al., 2016) and stress tolerance activities on various crops has been well-documented in the literature (Patel et al., 2018; Trivedi et al., 2018a). It was also reported to lessen the carbon footprint by saving ~2.06 kg CO2 equivalent per ton of cane production (Singh et al., 2018) and 9.5 kg CO2 equivalent per ton of rice production (Sharma et al., 2017). The same authors also successfully substituted 50% recommended dose of chemical fertilizers with Kappaphycus seaweed extract without compromising the yield, thus reducing the intensive use of chemical fertilizers. Despite many reported beneficial effects, studies on its mechanism of action are still scarce. Alterations in endogenous phytohormone levels (Agarwal et al., 2016), enzymatic and non-enzymatic antioxidants (Trivedi et al., 2018a), reactive oxygen species (ROS) (Trivedi et al., 2018b), and various components of photosynthetic machinery (Trivedi et al., 2018a) have been reported earlier for the application of KSWE. However, many of these physiological changes are the result of biochemical alterations governed by molecular mechanisms. Very little transcriptomic work has been performed to understand the effects of KSWE at molecular level under drought stress. Identifying the mechanism of action of seaweed extract on plants is crucial to further make its use more efficient and sustainable. Agarwal et al. (2016) reported changes in plant defense-responsive genes upon application of KSWE in tomato seedlings, but limited to a few genes only. We earlier reported complete transcriptomic analysis of maize root tissue under drought after drench application of KSWE, while in the present study, we are reporting a complete transcriptomic picture of maize leaf tissue treated with foliar application of KSWE under drought. Drought stress severely affects many physiological processes in plants. Due to decrease in water availability to the root cells, plants experience modulation in root growth, decrease above ground biomass, damage to photosynthetic pigments leading to impaired photosynthesis and transpiration, accumulation of compatible solutes and ROS, and decreased reproduction (Mansori et al., 2016; Bhusal et al., 2021).
Maize is an efficient water user with a transpiration ratio of 388, as compared to wheat and soybean, which have a transpiration ratio of 613 and 704, respectively (Jensen, 1973). Although maize uses water relatively efficiently, drought stress severely affects all growth stages and especially hampers yield, if drought strikes at the reproductive stage (NeSmith and Ritchie, 1992; Earl and Davis, 2003; Li-Ping et al., 2006). Maize has very unique water requirement and needs ~350–450 mm of water to complete its entire life cycle. Single maize plant consumes ~250 L of water up to its maturity. Ten to 16 kg of kernels will be produced from each consumed millimeter of water (Du Plessis, 2003). According to Rana and Rao (2000), 368 kg of water is required by maize to generate 1 kg of dry matter. Due to reduced water potential and decreased relative water content under drought, leaf folding and stomata closure occur, which decrease leaf transpiration rate, leading to increase in leaf temperature (Siddique et al., 2001), which negatively affects yield. Drought reduces the growth and development of silk, and speeds up the pollen shedding, which causes an increase in the anthesis-silking time interval (ASI). Productivity decreases as ASI increases. Buitink et al. (1996) reported that viability and specific gravity of pollens greatly abridged if moisture level decreased below 0.4 g per gram of pollen. Improper silk development and reduced pollen viability negatively affect kernel rows and number of kernels per row, which drastically reduces the grain yield of maize (Anjum et al., 2011).
Thus, our aim in the present study is to elucidate the drought alleviatory mechanism of KSWE, in maize, up to molecular level by transcriptomic analysis and to test our hypothesis that KSWE positively modulates number of genes in plants. Hence, an experiment was designed in maize leaf tissue treated with foliar application of KSWE at the V5 growth stage under drought conditions. The V5 growth stage of the maize plant is the stage when five fully developed collars were visible. It was selected for foliar application in the present study as it has a crucial role in deciding the ultimate yield of the plant (Trivedi et al., 2017).
Materials and Methods
Preparation of K. alvarezii Seaweed Extract
KSWE was prepared as per the procedure described in Eswaran et al. (2005). It was prepared in a bulk quantity and stored at 4°C with preservatives [propylparaben (0.02%), methyl paraben (0.2%) and potassium benzoate (0.1%)] (Singh et al., 2016) added and used as required. Liquid seaweed extract prepared by this method was considered as 100%, and from that, 10% dilution was made as per the requirement of the treatments. Detailed composition of 100% concentrated KSWE (presented here in Table 1) was the same as in Singh et al. (2016) as the extract from the same batch was also used in the present study.
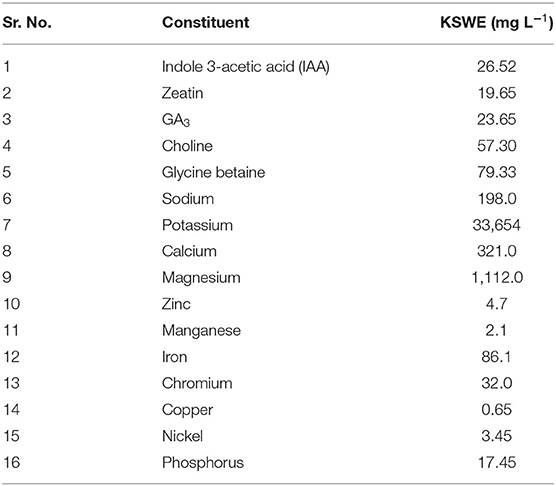
Table 1. Constituents of KSWE (Singh et al., 2016).
Study Area and Experimental Design
A pot experiment was conducted using maize (variety Sugar 75, Syngenta) at the net house facility (21°44′57.6″N latitude, 72°08′39.3″E longitude) of CSMCRI, Bhavnagar, Gujarat, India during November–March 2014. The experiment was laid out with two treatments subjected to foliar application of water with preservatives (control) (denoted as KT2) and 10% KSWE (denoted as KT6) at the V5 stage (24 DAS) of the crop under drought stress. Application stage and concentration of KSWE were selected based on our previously reported studies (Layek et al., 2015; Trivedi et al., 2017). Moisture stress was applied by withholding water up to 7 days at the V5 growth stage. Each treatment was replicated six times. Each pot having single plant represented one replicate. Transcriptome study was carried out from two samples prepared by pooling leaf samples of three replicates from each treatment. Two technical replicates were also taken from each sample during transcriptomic analysis. Pots were filled with 32 kg of soil. Chemical fertilizers were given at a rate of 120:60:40 kg ha−1 of N:P2O5:K2O in the form of urea, single super phosphate, and sulfate of potash, respectively.
Growth, Yield, and Photosynthetic Attributes
Plant height, leaf length, and leaf width were measured at 55 DAS, while chlorophyll index and photosynthesis rate were measured at 31 DAS. All attributes were measured in six replications except photosynthetic rate, which was measured in three replications. Plant height was recorded from the surface to the last leaf collar developed. Leaf length and width were recorded from each leaf and presented as the average leaf length and width per plant. Green cobs were harvested at 104 DAS from each plant, and cob attributes were measured from them. Full cob length, highest cob girth, and weight were recorded from each cob immediately after harvest. Green cob weight in g plant−1 was represented as green cob yield.
Chlorophyll index and photosynthetic rate were recorded from the last fully matured leaf and measured using the chlorophyll content meter (Model CCM-200, Opti-Sciences Inc., USA) and infrared gas analyzer system (IRGA; Model Li-6400XT, LI-COR, USA), respectively. During measurement of photosynthetic rate, photosynthetic photon flux density (PPFD) was kept at 1,000 μmol m−2 s−1 while CO2 in the leaf chamber was retained at a fixed concentration by continuously allowing air to pass through the leaf chamber from an open end.
RNA-Sequencing and de novo Assembly
Leaf tissue was collected from control and KSWE-treated plants of V5 stage and stored at −80°C for further RNA isolation procedure. Total RNA was isolated from leaves by using NucleoSpin RNA Plant kit following the manufacturer's protocol. The quantity and integrity of total RNA were assessed by an Epoch microplate reader (BioTek, USA) and Agilent 2100 Bioanalyzer (USA), respectively. Library construction was carried out in those samples which had an RNA integrity number (RIN) value of ≥7. Library construction was carried out with 2 μg of total RNA. The RNA-Seq library was prepared by RNA TruSeq Stranded mRNA LT sample prep kit (Illumina, San Diego) following the manufacturer's protocols. On each library, qubit quantitation assay and the NGS Fragment Analysis (Agilent 2100 Bioanalyzer, Santa Clara, CA) were conducted (Song et al., 2016). The Illumina HiSeq 2500 (SciGenom Labs Private Limited, Cochin, Kerala, India) was used to generate paired end reads (read length 2 × 100 base).
The fastq file summary, trimming summary, and overall alignment summary are given in Supplementary Material 1. An average of 87.2% of total reads passed ≥30 Phred score. The average base composition and quality of overall samples were also determined (figures not given). Sequence reads were trimmed based on quality parameters. Only high-quality sequences were used for further analysis, and the low-quality bases were trimmed out. Cutadapt tool (version 1.8.1) was used in the pre-processing step for adapter sequence removal. The trimming summary was provided in Supplementary Material 1. Unwanted sequences like non-polyA-tailed RNAs, ribosomal RNAs, transfer RNAs, adapter sequences, and others were removed by using bowtie2 (version 2.2.6), perl scripts, and picard tools (version 1.115).
The Zea mays genome and gene model downloaded from MaizeSequence.org (http://ftp.maizesequence.org/release-5b/assembly/ZmB73_RefGen_v2.tar.gz) was used to align pre-processed reads by Tophat program (version 2.1.0). The tape station profiles of the KT2 and KT6 sample and library were performed (figures not given). The pre-processed reads were used for reference-based pairwise alignment. The overall alignment summary is also given in Supplementary Material 1.
Expression Estimation and Differential Gene Expression Analysis
The aligned reads were used for estimating expression of the genes and transcripts using cufflinks program (version 2.2.1) after its aligning with the reference gene model. The differential expression analysis for the sample comparison was performed using cuffdiff program of cufflinks package (Trapnell et al., 2009, 2010).
Transcript abundance was calculated using the FPKM method (Fragments Per Kilobase of transcript per Million mapped reads) (Trapnell et al., 2010) using the formula given below:
Where C is the number of mappable reads that fell onto the gene's exons, N is the total number of mappable reads that uniquely aligned to all genes, and L is the number of bases pairs in the exon.
FPKM ≥ 1 for at least one of the two samples being compared and p-values 0.01 and 0.05 separately were used as cutoffs for up- and downregulated genes and isoforms in the analysis (Kumar et al., 2020). The differential expression FPKM plot of the sample comparison was also plotted and given as Supplementary Material 2.
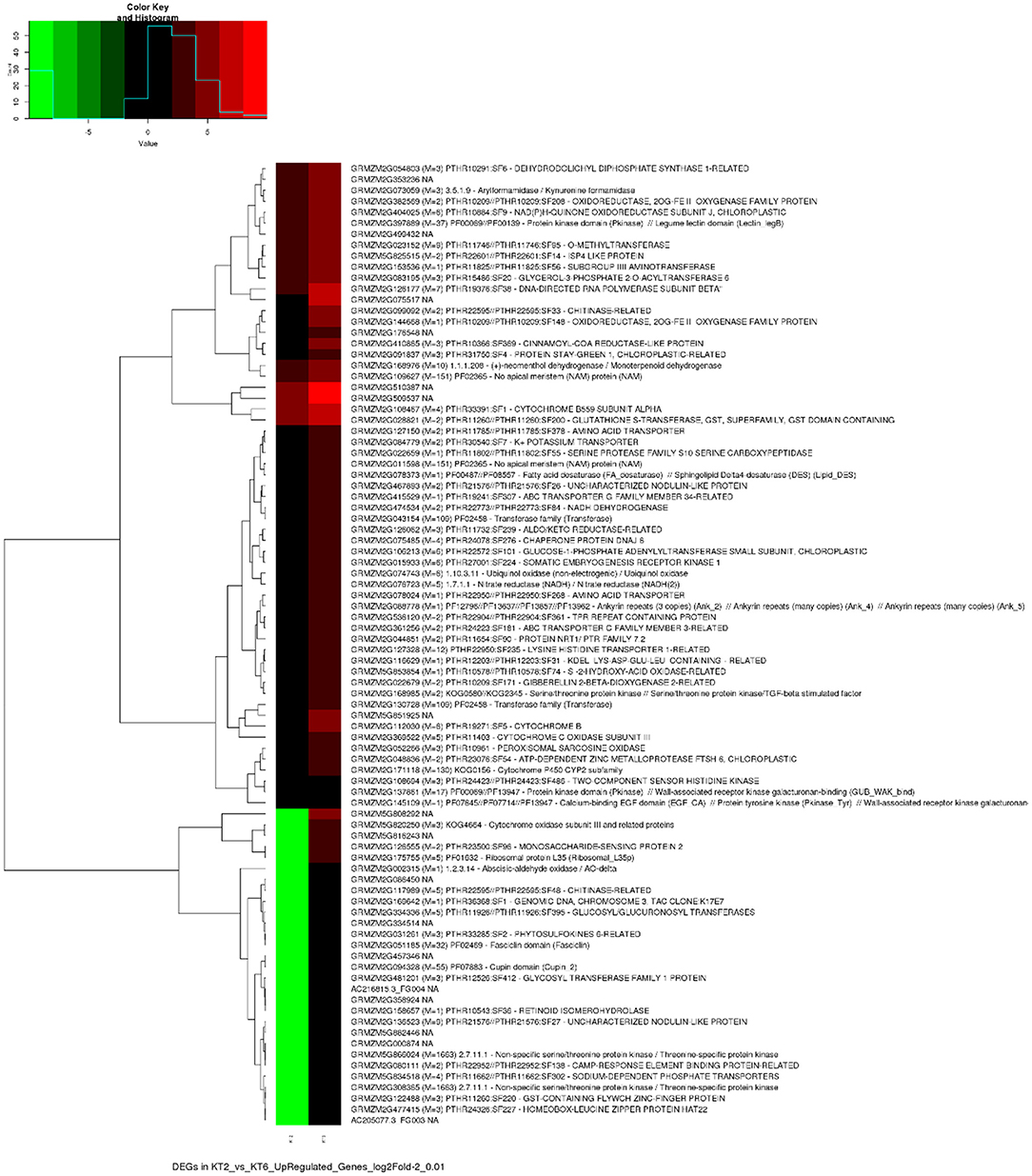
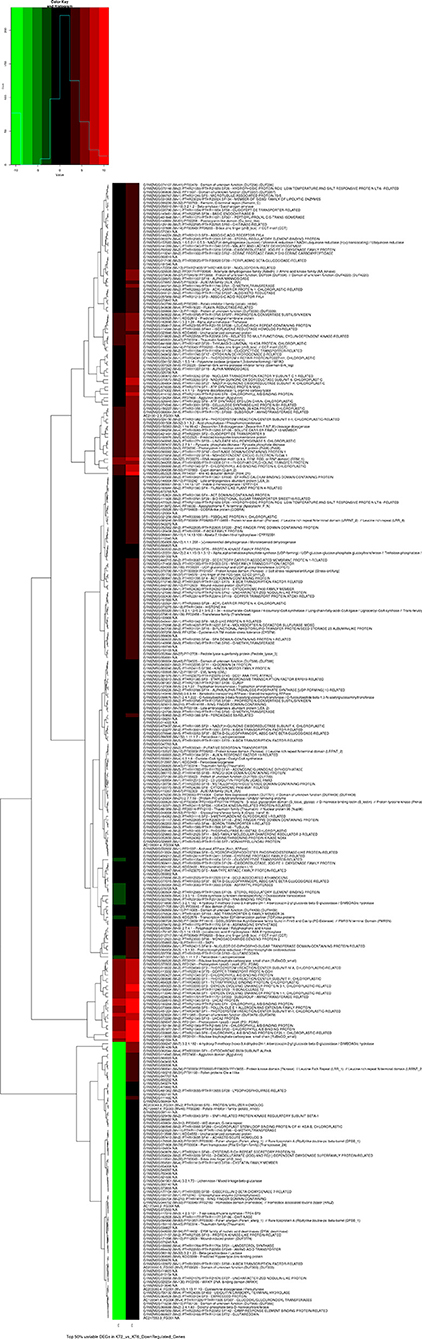
Clustered heatmaps of the top 50% highly variable upregulated and downregulated genes of the sample comparison with p-value cutoff 0.01 and log2Fold-2 are given as Figures 1, 2, respectively.
Gene Ontology (GO) Enrichment Analyses and Pathway Identification
GO annotations were attained to understand the functional classification of differential gene expression data (Ashburner et al., 2000; Kakumanu et al., 2012). It includes the molecular functions, biological processes, and cellular components with which the differentially expressed genes were associated. The pathway of differentially expressed genes were plotted by MapMan (Thimm et al., 2004). Different samples comparison were annotated for biochemical pathway analysis with Plant MetGenMAP (Joung et al., 2009).
Statistical Analysis
Statistical analyses of growth, yield, and photosynthetic data were carried out using Microsoft Excel computer software. The significance was calculated by paired t-test at p < 0.05.
Results
Growth, Yield, and Photosynthetic Attributes
Figures 3A–F display the changes recorded in growth and yield attributes along with chlorophyll index and photosynthetic rate.
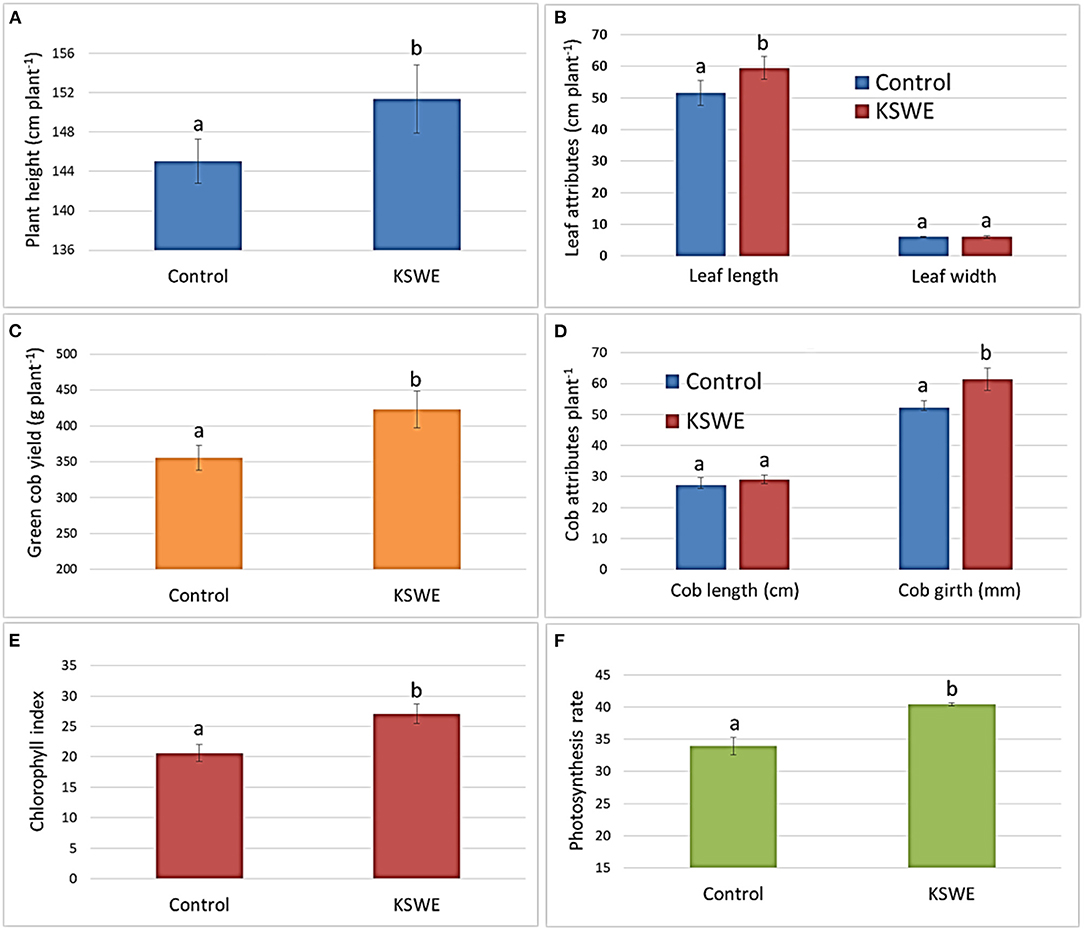
Figure 3. (A–F) Growth, yield, and photosynthetic attributes of KSWE and control plants. Values represented are mean of six replicates except photosynthetic rate, which is the mean of three replicates. Bars followed by different alphabets are significantly different using t-test (p < 0.05).
KSWE-treated plants showed significantly higher plant height (151 ± 3.5 cm) and leaf length (59 ± 3.6 cm) compared to control plants (145 ± 2.2 and 51 ± 3.9 cm, respectively). No difference was recorded in case of leaf width at 55 DAS. Green cob yield and cob girth recorded at harvest showed significant changes. KSWE-treated plants recorded 19% higher green cob yield (423 ± 26 g plant−1) compared to control plant, which was 356 ± 17 g plant−1 only. Cob girth was also found significantly higher in KSWE-treated plants (61 ± 3.7 mm) compared to that in control (52 ± 2.2 mm). Additionally, chlorophyll index and photosynthetic rate recorded after 7 days of treatment showed significant changes in both of the parameters. Leaves of KSWE-treated plants showed significantly higher chlorophyll index (27 ± 1.6) and photosynthetic rate (40.4 ± 0.2) compared to control plants (21 ± 1.4 and 34 ± 1.4, respectively).
Transcriptomic Analysis
Transcriptomic analysis of control (water sprayed) and KSWE-treated plants, both subjected to soil moisture stress at the V5 stage, generated 65,271,409 (6.5 Gb) of the total number of reads, among which 87.2% reads had Phred score ≥ 30. Table 2 gives the number of upregulated and downregulated genes and isoforms, obtained with a p-value cutoff of 0.01 and 0.05 and FPKM ≥ 1. Compared to control, it was found that 380 and 631 genes (p ≤ 0.05) were upregulated and downregulated, respectively, by KSWE application at the V5 stage under soil moisture stress conditions. A comprehensive list of the expressed up- and downregulated genes has been given in Supplementary Material 3, including its normalized expression values, fold changes, and GO annotation.
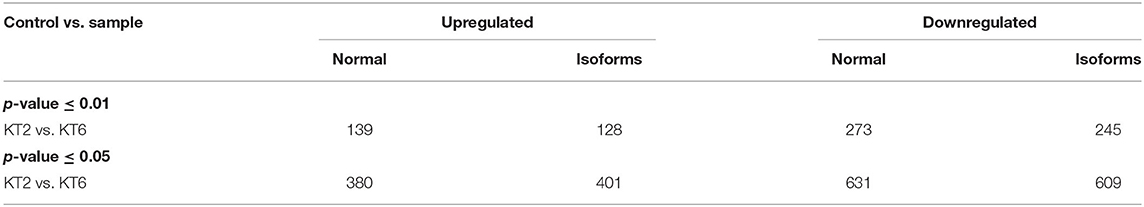
Table 2. Total up- and downregulated genes along with isoforms (at p-value ≤ 0.01 and 0.05; FPKM ≥ 1) obtained using cuffdiff analysis.
Gene Ontology (GO) and Function Analysis of Differentially Expressed Genes (DEGs)
GO annotations for the functional classification of DEGs (biological processes, molecular functions, and cellular components) were carried out separately for the relevant comparisons and for their GO enrichment plots for upregulated and downregulated genes which are shown in Figure 4. Compared to control, a total of 17, 41, and 27 DEGs were upregulated and 67, 86, and 66 were downregulated at log2FC ≥ 2 and p-value cutoff ≤ 0.01, for biological processes, molecular functions, and cellular components, respectively, by KSWE treatment (Figure 4).
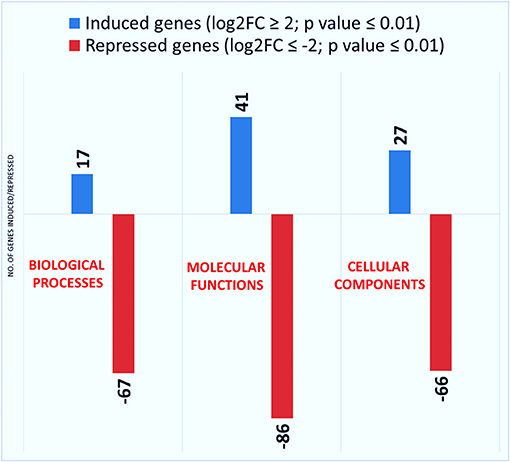
Figure 4. Number of DEGs in control vs. KSWE in relation to biological processes, molecular functions, and cellular functions, Log2FC ≥ 2; p ≤ 0.01.
Some of the induced biological processes by KSWE included nitrate assimilation (nitrate transportation, GRMZM2G076723), signal transmission (sphingolipid biosynthesis, GRMZM2G078373), photosynthesis (electron transport, GRMZM2G108467 and GRMZM2G112030), transmembrane transport of various ions (like zinc, potassium, and molybdenum, GRMZM2G161310 and GRMZM2G084779), and glycogen and starch biosynthetic process (GRMZM2G106213). Furthermore, some of the repressed biological processes included catabolism of polysaccharide molecules such as starch (GRMZM2G058310) as well as cell wall macromolecules like chitin (GRMZM2G129189), ubiquitin-dependent protein degradation (GRMZM2G059865), and transmembrane transport of Na+ ion (GRMZM2G086258) (Supplementary Material 3).
Functional Characterization of Genes
Pathway identification for DEGs was carried out using Mapman tool based on sufficient similarity to any protein of known function encoded by the genes (Figure 5). An overview of DEGs involved in metabolic processes in KSWE-treated sample compared to control was generated through Mapman and represented in Figure 5A.
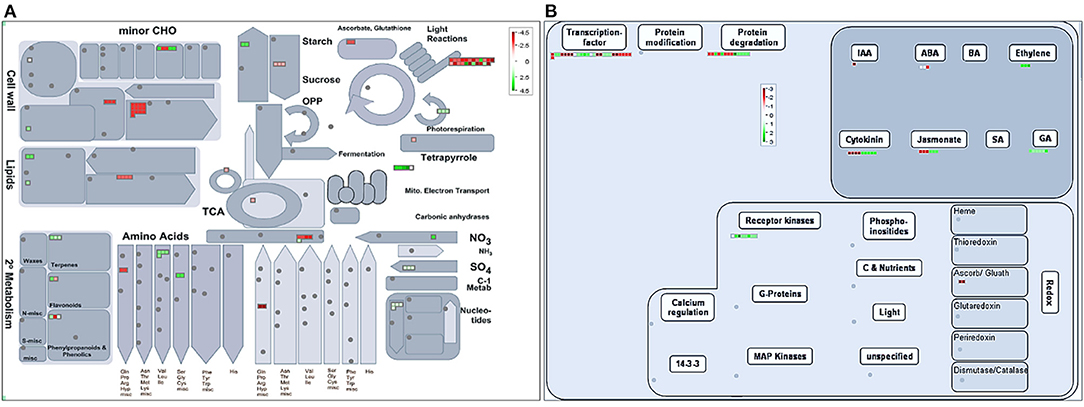
Figure 5. Mapman diagrams representing up- and downregulated genes due to KSWE application at V5 stage under drought. (A) Metabolism overview. (B) Regulation overview. Fold changes are indicated by the color scale (red and green represent up- and downregulated genes, respectively).
Genes involved in cell wall degradation [cellulases and beta−1,4-glucanases (GRMZM2G017186)], lipid degradation [glycerophosphodiesterase (GRMZM2G018820)], and amino acid degradation [arginine decarboxylase (GRMZM2G374302)] were downregulated, while those involved in phospholipid synthesis [Glycerol-3-phosphate acyltransferase (GRMZM2G083195)], terpenoid synthesis [dehydrodolichyl diphosphate synthase (GRMZM2G054803) and ent-kaurene synthase (GRMZM2G049538)], and amino acid synthesis [branched-chain-amino-acid transaminase (GRMZM2G153536)] were upregulated due to KSWE application at the V5 stage under drought stress. Genes involved in N metabolism [Nitrate reductase 1 (GRMZM2G076723)], S assimilation [sulfate adenylyltransferase (GRMZM2G051270)], and pyrimidine nucleotide synthesis [orotate phosphoribosyltransferase (GRMZM2G367026)] were also found upregulated due to KSWE treatment when compared to control. Among the genes involved in the light reaction of photosynthesis, upregulation of NADH dehydrogenase and PSII cytochrome b559 was noted, while downregulation of genes encoding Chlorophyll a-b binding protein 2 and PSI polypeptide subunits was noted by KSWE application compared to control.
Similarly, an overview of DEGs involved in regulatory processes in the KSWE-treated sample compared to control was represented as Figure 5B. MYB-related transcription factor, NAC domain transcription factor, and WRKY domain transcription factor were found upregulated, while Constans-like zinc finger and CCAAT box binding factor were found downregulated in leaves of KSWE-treated plants compared to water-sprayed control, under stress. Wall associated kinases and serine/threonine kinase, which are from signaling receptor kinase category, were found upregulated. Furthermore, there was a significant modulation in genes involved in hormonal regulation as well. Genes involved in cytokinin signal transduction like cytokinin-independent 1 (GRMZM2G108694) was found upregulated while cytokinin oxidase 7 (GRMZM2G348452) was found downregulated. Additionally, Ent-kaurene synthase (GRMZM2G049538) and GA20 oxidase (GRMZM2G060940) involved in gibberellin synthesis were found upregulated in KSWE-treated maize leaf tissue. 9-cis-epoxycarotenoid dioxygenase (GRMZM2G150363), which is an enzyme having a role in abscisic acid synthesis, was downregulated due to KSWE application. Moreover, there was upregulation in 2-oxoglutarate-dependent dioxygenase enzyme activity, which was involved in ethylene synthesis (Supplementary Material 3).
A list of some important differentially expressed genes at log2FC ≥ 2 and p-value ≤ 0.001 (either up- or downregulated) with their biological process, molecular function, and cellular component along with gene ids are given in Table 3.
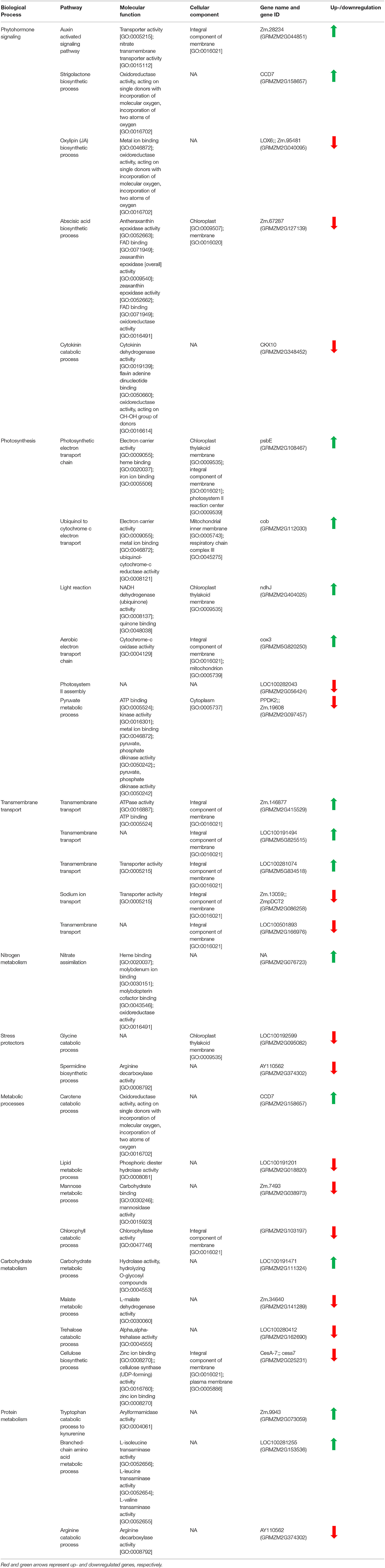
Table 3. Classification of upregulated (log2FC ≥ 2) and downregulated (log2FC ≤ −2) genes (at p-value ≤ 0.001) based on biological process and pathway in which they are involved.
Discussion
Drought is one of the major environmental factors reducing the productivity of crops worldwide. We earlier reported that KSWE increased the growth and productivity of maize crop under normal as well as soil moisture stress conditions (Trivedi et al., 2018a,b). Concerning biochemical changes, we have found that KSWE increased the rate of photosynthesis, and levels of antioxidant enzymes simultaneously decreased different ROS and lipid peroxidation in the foliarly treated maize plants compared to its control (Trivedi et al., 2018a). Additionally, proline, soluble sugars, and amino acids, which act as osmoprotectants during the stress conditions, were also reported higher in KSWE-treated wheat plants by Patel et al. (2018). These biochemical changes brought positive influence on growth and yield attributes and increased the productivity of treated maize or wheat plants (Patel et al., 2018; Trivedi et al., 2018a). In the present study, we have also noticed improvement in growth and yield attributes, which is in accordance to previously published reports (Patel et al., 2018; Trivedi et al., 2018a,b). Leaves of KSWE-treated plants showed higher photosynthetic rate and chlorophyll index, which might have resulted in higher growth and green cob yield compared to control plants, which were water sprayed only. There are also reports available that KSWE can influence plants up to the molecular level. For example, Agarwal et al. (2016) reported a higher transcript level of pathogenesis-related genes due to KSWE application under biotic stress in tomato seedlings. MPK2 (mitogen-activated kinase pathway gene) and Pti4 (transcription factor) were found elevated in Macrophomina-affected tomato plants treated with KSWE, resulting in increased stress tolerance compared to its control (Agarwal et al., 2016). In another study, Patel et al. (2018) reported increased expression of antioxidant genes, MAP kinase gene, and stress-responsive WRKY transcription factor in KSWE-treated wheat plants. In the present study, we tried to show the complete change in the transcriptional profile of maize leaf tissue when treated with KSWE compared to its control.
Gibberellins (GAs) are an important group of phytohormones. They play crucial roles in seed germination, dormancy, leaf expansion, stem elongation, root growth, flower development, and fruit senescence (Qin et al., 2013; Ghosh et al., 2017). The level of GAs decrease when plants are under salt, drought, or cold stress as a defensive strategy to reduce the growth and fight against the stress (Ghosh et al., 2017). Litvin et al. (2016) reported downregulation of GA20 oxidase transcript level (involved in GAs biosynthesis) in tomato plants in response to drought stress. Our study revealed that KSWE upregulated genes involved in the biosynthesis of GAs such as ent-kaurene synthase and GA20 oxidase. Ent-kaurene synthase is the key enzyme involved in GA biosynthetic pathway. Margis-Pinheiro et al. (2005) showed that silencing of that resulted in semidwarf rice plants. GA20 oxidase is involved in the final step of the biologically active GAs synthesis. Higher levels of bioactive GAs were found in mutants overexpressing the GA20 oxidase gene (Qin et al., 2013). The increased transcript level of GA biosynthetic genes even under drought stress compared to its control indicates that KSWE-treated plants successfully mitigated the stress and were able to continue their growth even under harsh conditions.
Other transcript analyses also supported this hypothesis as NCED4, an abscisic acid (ABA) biosynthesis gene, was found to be downregulated in KSWE-treated plants compared to its control. ABA is a major stress hormone. In response to drought, cold, or salt stress, plant increases the ABA concentration, which plays a crucial role in stomatal closure, reducing water loss, delays germination and synthesis of various storage proteins and lipids, and helps the plant maintain the water balance (Tuteja, 2007). However, downregulation in ABA biosynthesis gene found in KSWE-treated plants under drought stress suggests successful stress mitigation achieved in the treated plants. KSWE-treated plants showed higher growth and yield response as well-compared to its control in the present study. Genes involved in cytokinin signal transduction like cytokinin-independent 1 (CKI 1) was found to be upregulated while cytokinin oxidase 7 (CKX 7) was found to be downregulated. The upregulation of CKI 1 and downregulation of cytokinin degradation related to CKX 7 and cytokinin oxidase 1 (COX 1) suggest an increase in cytokinin signal transduction due to KSWE application at the V5 stage in maize. In addition, the gene involved in auxin activated signaling pathway was also found upregulated, suggesting higher auxin signaling in treated plants as compared to control. Auxin is also a well-known phytohormone that regulates many plant growth processes under drought stress (Shi et al., 2014). Increased auxin signaling suggests improved plant growth even under stress conditions. Agarwal et al. (2016) also reported higher endogenous IAA levels in tomato seedlings treated with KSWE.
Furthermore, the gene of arginine decarboxylase (ADC; a key enzyme involved in polyamine biosynthesis) was found downregulated in KSWE-treated plants. Polyamines are low-molecular-weight compounds, which generally increase in plant cells in response to environmental stress conditions (Chen et al., 2019). Downregulation of the ADC gene indicates lower polyamine biosynthesis in KSWE-treated maize leaves. This might have been due to the reason stated earlier that KSWE-treated plants were not experiencing as much stress as control plants. There are also reports that seaweed extracts can also contain polyamines (Papenfus et al., 2012), which may be the reason plant decreased their endogenous synthesis as they got it from exogenous seaweed application.
Moreover, nitrogen supply in plants is important for maintaining proper photosynthesis. It has been reported that a high amount of assimilated nitrogen is used only in the photosynthesis process in plants (Zhong et al., 2017). Due to drought, photosynthesis is hampered, which resulted in a reduction in nitrogen metabolism as well (Zhong et al., 2017). However, in our study, we have found that gene involved in nitrogen metabolism such as Nitrate reductase 1 was upregulated, suggesting improved nitrogen metabolism that helped plants to efficiently perform photosynthesis even under drought conditions. In accordance with this, we earlier recorded higher photosynthesis also in maize plants when subjected to foliar KSWE application under drought (Trivedi et al., 2018a). This molecular information regarding hormonal regulation, nitrogen or sulfur metabolism, photosynthesis, and signal transduction put us forward in the way of finding the mechanism of KSWE on plants. Further validation by taking individual genes is necessary for the future to confirm the results obtained in the present study.
Conclusion
K. alvarezii seaweed extract is known for its bio-stimulatory and stress alleviation activities on plants, and this study adds more information to substantiate these reports. This transcriptomic analysis also unravels missing gene expression patterns that KSWE imparts on drought stress in maize. Principally, KSWE upregulated the genes involved in IAA, GA, and cytokinin signal transduction and downregulated the gene involved in ABA biosynthesis. Other key genes that were up- and downregulated are those involved in photosynthesis and nitrogen metabolism, polysaccharide synthesis, and signal transduction. These genes collectively portray the drought alleviatory mechanism of Kappaphycus seaweed extract in maize.
Data Availability Statement
The data presented in the study are deposited in the NCBI's Gene Expression Omnibus repository and are accessible through GEO Series accession number GSE188626 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE188626).
Author Contributions
KT: data curation, formal analysis, investigation, methodology, and writing-original draft. VG: supervision, validation, and writing-review and editing. RK: data curation, formal analysis, and software. AG: conceptualization, funding acquisition, resources, supervision, visualization, and writing-review and editing. All authors contributed to the article and approved the submitted version.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Publisher's Note
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Acknowledgments
The authors wish to thank the Council of Scientific and Industrial Research (CSIR) and Central Salt and Marine Chemicals Research Institute (CSMCRI) for supporting the work under the project MLP-0016. KT and RK thank the Academy of Scientific and Innovative Research (AcSIR) for enrolment in a Ph.D. program. KT's work was financially supported by CSIR in the form of CSIR-SRF. Dr. K Eswaran and Dr. Vaibhav Mantri from CSMCRI MARS Mandapam Camp, Tamil Nadu, India were specially acknowledged for providing Kappaphycus alvarezii seaweed extract. SciGenom Labs Private Limited, Cochin, Kerala was duly acknowledged for help in sequencing analysis. This manuscript bears CSIR-CSMCRI communication no. 177/2021.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.774978/full#supplementary-material
References
Agarwal, P., Patel, K., Das, A. K., Ghosh, A., and Agarwal, P. K. (2016). Insights into the role of seaweed Kappaphycus alvarezii sap towards phytohormone signalling and regulating defence responsive genes in Lycopersicon esculentum. J. Appl. Phycol. 28, 2529–2537. doi: 10.1007/s10811-015-0784-1
Anjum, S. A., Wang, L. C., Farooq, M., Hussain, M., Xue, L. L., and Zou, C. M. (2011). Brassinolide application improves the drought tolerance in maize through modulation of enzymatic antioxidants and leaf gas exchange. J. Agron. Crop Sci. 197, 177–185. doi: 10.1111/j.1439-037X.2010.00459.x
Ashburner, M., Ball, C. A., Blake, J. A., Botstein, D., Butler, H., Cherry, J. M., et al. (2000). Gene ontology: tool for the unification of biology. Nat. Genet. 25, 25–29. doi: 10.1038/75556
Bhusal, B., Poudel, M. R., Rishav, P., Regmi, R., Neupane, P., Bhattarai, K., et al. (2021). A review on abiotic stress resistance in maize (Zea mays L.): effects, resistance mechanisms and management. J. Biol. Today's World. 10, 1–3.
Buitink, J., Walters-Vertucci, C., Hoekstra, F. A., and Leprince, O. (1996). Calorimetric properties of dehydrating pollen; analysis of desiccation tolerant and an intolerant species. Plant Physiol. 111, 235–242. doi: 10.1104/pp.111.1.235
Chen, D., Shao, Q., Yin, L., Younis, A., and Zheng, B. (2019). Polyamine function in plants: metabolism, regulation on development, and roles in abiotic stress responses. Front. Plant Sci. 9:1945. doi: 10.3389/fpls.2018.01945
Du Jardin, P. (2015). Plant biostimulants: definition, concept, main categories and regulation. Scientia Horticulturae 196, 3–14. doi: 10.1016/j.scienta.2015.09.021
Du Plessis, J. (2003). Maize Production. Potchefstroom: Directorate Agricultural Information Services, Department of Agriculture in Cooperation With ARC-Grain Crops Institute.
Earl, H. J., and Davis, R. F. (2003). Effect of drought stress on leaf and whole canopy radiation use efficiency and yield of maize. Agron. J. 95, 688–696. doi: 10.2134/agronj2003.6880
Eswaran, K., Ghosh, P. K., Siddhanta, A. K., Shambhubhai, P. J., Chellaiah, P., Shantibhai, M. A., et al. (2005). Integrated method for production of carrageenan and liquid fertilizer from fresh seaweeds. US. Pat. No. 6,893,479.
Ghosh, A., Trivedi, K., Kumar, R., Kubavat, D., and Agarwal, P. K. (2017). “Gibberellic acid signalling and abiotic stress tolerance: past and present,” in Mechanisms Behind Phytohormonal Signalling and Crop Abiotic Stress Tolerance, eds V. P. Singh, S. Singh, and S. M. Prasad (Hauppauge, NY: Nova Science Publishers), 189–207.
Jensen, M. E. (1973). Consumptive Use of Water and Irrigation Water Requirements. New York, NY: American Society of Civil Engineers.
Joung, J. G., Corbett, A. M., Fellman, S. M., Tieman, D. M., Klee, H. J., Giovannoni, J. J., et al. (2009). Plant MetGenMAP: an integrative analysis system for plant systems biology. Plant Physiol. 151, 1758–1768. doi: 10.1104/pp.109.145169
Kakumanu, A., Ambavaram, M. M. R., Klumas, C., Krishnan, A., Batlang, U., Myers, E., et al. (2012). Effects of drought on gene expression in maize reproductive and leaf meristem tissue revealed by RNA-Seq. Plant Physiol. 160, 846–867. doi: 10.1104/pp.112.200444
Kumar, R., Trivedi, K., Anand, K. V., and Ghosh, A. (2020). Science behind biostimulant action of seaweed extract on growth and crop yield: insights into transcriptional changes in roots of maize treated with Kappaphycus alvarezii seaweed extract under soil moisture stressed conditions. J. Appl. Phycol. 32, 599–613. doi: 10.1007/s10811-019-01938-y
Layek, J., Das, A., Ramkrushna, G. I., Trivedi, K., Yesuraj, D., Chandramohan, M., et al. (2015). Seaweed sap: a sustainable way to improve productivity of maize in North-East India. Int. J. Environ. Stud. 72, 305–315. doi: 10.1080/00207233.2015.1010855
Li-Ping, B. A. I., Fang-Gong, S. U. I., Ti-Da, G. E., Zhao-Hui, S. U. N., Yin-Yan, L. U., and Guang-Sheng, Z. H. O. U. (2006). Effect of soil drought stress on leaf water status, membrane permeability and enzymatic antioxidant system of maize. Pedosphere 16, 326–332. doi: 10.1016/S1002-0160(06)60059-3
Litvin, A. G., van Iersel, M. W., and Malladi, A. (2016). Drought stress reduces stem elongation and alters gibberellin-related gene expression during vegetative growth of tomato. J. Am. Soc. Horticult. Sci. 141, 591–597. doi: 10.21273/JASHS03913-16
Mansori, M., Chernane, H., Latique, S., Benaliat, A., Hsissou, D., and El Kaoua, M. (2016). Effect of seaweed extract (Ulva rigida) on the water deficit tolerance of Salvia officinalis L. J. Appl. Phycol. 28, 1363–1370. doi: 10.1007/s10811-015-0671-9
Margis-Pinheiro, M., Zhou, X. R., Zhu, Q. H., Dennis, E. S., and Upadhyaya, N. M. (2005). Isolation and characterization of a Ds-tagged rice (Oryza sativa L.) GA-responsive dwarf mutant defective in an early step of the gibberellin biosynthesis pathway. Plant Cell Rep. 23, 819–833. doi: 10.1007/s00299-004-0896-6
Messerli, P., Murniningtyas, E., Eloundou-Enyegue, P., Foli, E. G., Furman, E., Glassman, A., et al. (2019). Global Sustainable Development Report 2019: The Future Is Now–Science for Achieving Sustainable Development. New York.
NeSmith, D. S., and Ritchie, J. T. (1992). Maize (Zea mays L.) response to a severe soil water-deficit during grain-filling. Field Crops Res. 29, 23–35. doi: 10.1016/0378-4290(92)90073-I
Papenfus, H. B., Stirk, W. A., Finnie, J. F., and Van Staden, J. (2012). Seasonal variation in the polyamines of Ecklonia maxima. Bot. Mar. 55, 539–546. doi: 10.1515/bot-2012-0150
Patel, K., Agarwal, P., and Agarwal, P. K. (2018). Kappaphycus alvarezii sap mitigates abiotic-induced stress in Triticum durum by modulating metabolic coordination and improves growth and yield. J. Appl. Phycol. 30, 2659–2673. doi: 10.1007/s10811-018-1423-4
Pramanick, B., Brahmachari, K., Ghosh, A., and Zodape, S. T. (2014). Effect of seaweed saps on growth and yield improvement of transplanted rice in old alluvial soil of West Bengal. Bangladesh J. Bot. 43, 53–58. doi: 10.3329/bjb.v43i1.19746
Qin, X., Liu, J. H., Zhao, W. S., Chen, X. J., Guo, Z. J., and Peng, Y. L. (2013). Gibberellin 20-oxidase gene OsGA20ox3 regulates plant stature and disease development in rice. Mol. Plant Microbe Interact. 26, 227–239. doi: 10.1094/MPMI-05-12-0138-R
Rana, B. S., and Rao, M. H. (2000). Technology for Increasing Sorghum Production and Value Addition. Hyderabad: National Research Center for Sorghum, Indian Council of Agricultural Research.
Rathore, S. S., Chaudhary, D. R., Boricha, G. N., Ghosh, A., Bhatt, B. P., Zodape, S. T., et al. (2009). Effect of seaweed extract on the growth, yield and nutrient uptake of soybean (Glycine max) under rainfed conditions. South African J. Bot. 75, 351–355. doi: 10.1016/j.sajb.2008.10.009
Raverkar, K. P., Pareek, N., Chandra, R., Chauhan, S., Zodape, S. T., and Ghosh, A. (2016). Impact of foliar application of seaweed saps on yield, nodulation and nutritional quality in green gram (Vigna radiata L). Legume Res. Int. J. 39, 315–318. doi: 10.18805/lr.v39i2.9535
Sharma, L., Banerjee, M., Malik, G. C., Gopalakrishnan, V. A. K., Zodape, S. T., and Ghosh, A. (2017). Sustainable agro-technology for enhancement of rice production in the red and lateritic soils using seaweed based biostimulants. J. Clean. Prod. 149, 968–975. doi: 10.1016/j.jclepro.2017.02.153
Shi, H., Chen, L., Ye, T., Liu, X., Ding, K., and Chan, Z. (2014). Modulation of auxin content in Arabidopsis confers improved drought stress resistance. Plant Physiol. Biochem. 82, 209–217. doi: 10.1016/j.plaphy.2014.06.008
Siddique, M. R. B., Hamid, A., and Islam, M. S. (2001). Drought stress effects on water relations of wheat. Bot. Bull. Acad. Sin. 41, 35–39.
Singh, I., Anand, K. V., Solomon, S., Shukla, S. K., Rai, R., Zodape, S. T., et al. (2018). Can we not mitigate climate change using seaweed based biostimulant: a case study with sugarcane cultivation in India. J. Clean. Prod. 204, 992–1003. doi: 10.1016/j.jclepro.2018.09.070
Singh, S., Singh, M. K., Pal, S. K., Trivedi, K., Yesuraj, D., Singh, C. S., et al. (2016). Sustainable enhancement in yield and quality of rain-fed maize through Gracilaria edulis and Kappaphycus alvarezii seaweed sap. Journal of applied Phycology. 28, 2099–2112. doi: 10.1007/s10811-015-0680-8
Song, L., Prince, S., Valliyodan, B., Joshi, T., dos Santos, J. V. M., Wang, J., et al. (2016). Genome-wide transcriptome analysis of soybean primary root under varying water-deficit conditions. BMC Genom. 17, 1–17. doi: 10.1186/s12864-016-2378-y
Thimm, O., Bläsing, O., Gibon, Y., Nagel, A., Meyer, S., Krüger, P., et al. (2004). MAPMAN: a user-driven tool to display genomics data sets onto diagrams of metabolic pathways and other biological processes. Plant J. 37, 914–939. doi: 10.1111/j.1365-313X.2004.02016.x
Trapnell, C., Pachter, L., and Salzberg, S. L. (2009). TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25, 1105–1111. doi: 10.1093/bioinformatics/btp120
Trapnell, C., Williams, B. A., Pertea, G., Mortazavi, A., Kwan, G., Van Baren, M. J., et al. (2010). Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515. doi: 10.1038/nbt.1621
Trivedi, K., Anand, K. V., Kubavat, D., Kumar, R., Vaghela, P., and Ghosh, A. (2017). Crop stage selection is vital to elicit optimal response of maize to seaweed bio-stimulant application. J. Appl. Phycol. 29, 2135–2144. doi: 10.1007/s10811-017-1118-2
Trivedi, K., Anand, K. V., Kubavat, D., Patidar, R., and Ghosh, A. (2018a). Drought alleviatory potential of Kappaphycus seaweed extract and the role of the quaternary ammonium compounds as its constituents towards imparting drought tolerance in Zea mays L. J. Appl. Phycol. 30, 2001–2015. doi: 10.1007/s10811-017-1375-0
Trivedi, K., Anand, K. V., Vaghela, P., and Ghosh, A. (2018b). Differential growth, yield and biochemical responses of maize to the exogenous application of Kappaphycus alvarezii seaweed extract, at grain-filling stage under normal and drought conditions. Algal Res. 35, 236–244. doi: 10.1016/j.algal.2018.08.027
Tuteja, N. (2007). Abscisic Acid and abiotic stress signaling. Plant Signal. Behav. 2, 135–138. doi: 10.4161/psb.2.3.4156
Keywords: Kappaphycus alvarezii, abiotic stress, biostimulant, maize, transcriptome analysis, drought
Citation: Trivedi K, Gopalakrishnan VAK, Kumar R and Ghosh A (2021) Transcriptional Analysis of Maize Leaf Tissue Treated With Seaweed Extract Under Drought Stress. Front. Sustain. Food Syst. 5:774978. doi: 10.3389/fsufs.2021.774978
Received: 13 September 2021; Accepted: 04 November 2021;
Published: 02 December 2021.
Edited by:
Pushp Sheel Shukla, Dalhousie University, CanadaReviewed by:
Serenella Nardi, University of Padua, ItalySowmyalakshmi Subramanian, McGill University, Canada
Copyright © 2021 Trivedi, Gopalakrishnan, Kumar and Ghosh. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Arup Ghosh, YXJ1cGdob3NoQGNzbWNyaS5yZXMuaW4=
 Khanjan Trivedi
Khanjan Trivedi Vijay Anand K. Gopalakrishnan
Vijay Anand K. Gopalakrishnan Ranjeet Kumar1,2
Ranjeet Kumar1,2 Arup Ghosh
Arup Ghosh