- Department of Entomology, Purdue University, West Lafayette, IN, United States
Aboveground herbivory can impact the root-associated microbiome, while simultaneously different soil microbial communities influence herbivore performance. It is currently unclear how these reciprocal top-down and bottom-up interactions between plants, insects and microbes vary across different soils and over successive plant generations. In this study, we examined top-down impacts of above-ground herbivory on the rhizosphere microbiome across different soils, assessed bottom-up impacts of soil microbial community variation on herbivore performance, and evaluated their respective contributions to soil legacy effects on herbivore performance. We used Macrosiphum euphorbiae (potato aphid) and Solanum pimpinellifolium (wild tomato) to capture pre-domestication microbiome interactions with a specialist pest. First, using 16S rRNA sequencing we compared bacterial communities associated with rhizospheres of aphid-infested and uninfested control plants grown in three different soils over three time points. High aphid infestation impacted rhizosphere bacterial diversity in a soil-dependent manner, ranging from a 22% decrease to a 21% increase relative to uninfested plants and explained 6–7% of community composition differences in two of three soils. We next investigated bottom-up and soil legacy effects of aphid herbivory by growing wild tomatoes in each of the three soils and a sterilized “no microbiome” soil, infesting with aphids (phase one), then planting a second generation (phase two) of plants in the soil conditioned with aphid-infested or uninfested control plants. In the first phase, aphid performance varied across plants grown in different soil sources, ranging from a 20 to 50% increase in aphid performance compared to the “no microbiome” control soil, demonstrating a bottom-up role for soil microbial community. In the second phase, initial soil community, but not previous aphid infestation, impacted aphid performance on plants. Thus, while herbivory altered the rhizosphere microbiome in a soil community-dependent manner, the bottom-up interaction between the microbial community and the plant, not top-down effects of prior herbivore infestation, affected herbivore performance in the following plant generation. These findings suggest that the bottom-up effects of the soil microbial community play an overriding role in herbivore performance in both current and future plant generations and thus are an important target for sustainable control of herbivory in agroecosystems.
Introduction
Development of sustainable agricultural practices relies on understanding and managing the impacts of soil microbial communities on plant health (Finkel et al., 2017; Toju et al., 2018; French et al., 2021). In particular, microbes that develop around plant roots (e.g., the rhizosphere), play critical roles in nutrient acquisition and protection against a range of environmental stressors (Berendsen et al., 2012; Liu et al., 2020). For example, the contribution of rhizosphere-associated microbial communities to increasing pathogen protection is well-characterized (Rudrappa et al., 2008; Berendsen et al., 2018; Kwak et al., 2018; Yuan et al., 2018; Wei et al., 2019) and can even extend over multiple plant generations due to enrichment of protective microbial community members in soils (Berendsen et al., 2018; Yuan et al., 2018). However, the role of the rhizosphere microbiome in protection against herbivore damage is poorly understood, though evidence increasingly suggests plant-associated microbes influence plant-herbivore interactions (Badri et al., 2013; Pineda et al., 2017; Hu et al., 2018).
Two reciprocal processes are hypothesized to influence plant-insect-microbiome interactions (Figure 1) (Friman et al., 2020). “Top-down” effects of herbivory on plant physiology (e.g., induced defenses or root exudation) can lead to changes in below-ground microbial communities (Hamilton et al., 2008; Gosset et al., 2009; Lee et al., 2012; Lebeis et al., 2015; Kong et al., 2016; Hu et al., 2018; Wilkinson et al., 2019) (Figure 1A), while “bottom-up” effects of soil or root-associated microbiomes on plant nutrition or defense can impact herbivore performance (Pineda et al., 2012; Badri et al., 2013; Benítez et al., 2017; Hubbard et al., 2019; Blundell et al., 2020) (Figure 1B). Both top-down and bottom-up processes are inherently interconnected, making quantification of plant-insect-microbiome interactions complicated. When aboveground herbivory alters the microbial community surrounding plant roots, these top-down effects can then influence bottom-up effects on herbivore performance over time. Likewise, if soil microbial communities impact plant-herbivore interactions, then bottom-up effects could also lead to variation in responsiveness of the rhizosphere microbiome to top-down effects of herbivore feeding. Studies have yet to tease apart these two reciprocal processes and it is unclear if both shape plant-insect interactions to a similar degree.
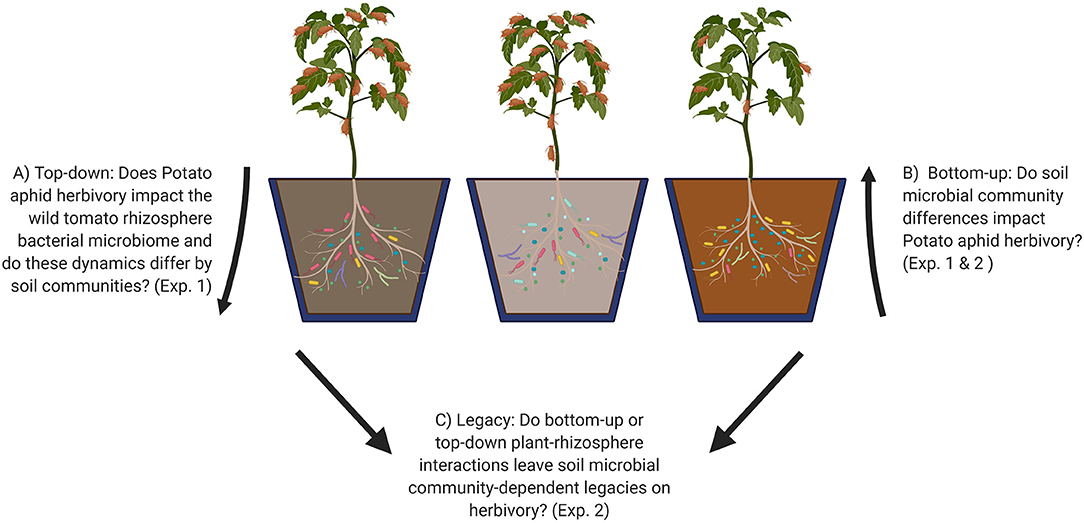
Figure 1. Graphical representation of experimental questions (A–C) addressed in this study. Numbers in parentheses refer to the experiments that address each question. Experiment 1: Top-down and bottom-up effects of aphid-tomato-microbiome interactions. Experiment 2: Soil microbial legacy effects of aphid infestation. Made in Biorender.
Top-down and bottom-up effects can occur through a number of mechanisms. Herbivory may generate top-down effects on soil microbial communities surrounding plant roots by triggering induced systemic plant defenses (Gosset et al., 2009; Lebeis et al., 2015), altering carbon flux and modulation of root exudates (Hamilton et al., 2008), or even through deposition of nutrient-rich excrement (i.e., honeydew or frass) (Seeger and Filser, 2008; Milcu et al., 2015). In some cases, top-down effects alter plant stress response and therefore herbivory is hypothesized to elicit a “cry for help” to recruit soil microbes to the rhizosphere that provide protection (Yi et al., 2011; Liu et al., 2020). Thus, far a few studies have shown enrichment of potentially beneficial microbes after insect infestation in pepper (Yang et al., 2011; Lee et al., 2012); ragwort (Kostenko et al., 2012; Bezemer et al., 2013) and maize (Holland, 1995); though direct evidence for the “cry for help” hypothesis is still lacking. In addition to top-down effects, “bottom-up” effects of the soil microbiome on herbivore performance can occur through induction of chemical defenses or alteration of plant nutritional quality (Pineda et al., 2017). For example, single microbial strains that induce systemic plant defenses can alter insect performance either positively or negatively (Valenzuela-Soto et al., 2010; Pineda et al., 2012; Coppola et al., 2019). At the microbial community level, Badri et al. (2013) showed a positive correlation between amino acid content and Trichoplusia ni larval weight when Arabidopsis plants were grown in microbiomes derived from different soils, suggesting a bottom-up role for the microbiome in modulating leaf nutrient content with cascading effects on insect performance.
Although the top-down and bottom-up effects of microbial communities are increasingly recognized as important in shaping plant-insect interactions, it is unclear what role they together play in determining soil microbial legacy effects that impact plant health and herbivore performance when plants are grown in the same soil over multiple generations. Soil microbial legacies, defined as the effects of previous planting and environmental conditions on the microbial community that feedback onto plant health in successive plantings, likely play an important role in the outcome of insect-plant interactions (Figure 1C) (Kaplan et al., 2018), similar to evidence from plant-pathogen interactions (Berendsen et al., 2018; Yuan et al., 2018). Several studies show different plant species leave legacies that impact herbivory in successive generations of plants grown in the same soil (Kos et al., 2015; Zhu et al., 2018; Carrillo et al., 2019; Pineda et al., 2019). For example, top-down effects resulting from root exudation of benzoxazinoids, a class of plant defensive chemicals specific to cereal species, are linked to alterations in the soil microbiome that affect both plant growth and insect pest resistance in subsequent generations (Hu et al., 2018). Microbial community differences between soils that have bottom-up impacts on plant health may also lead to different legacy impacts, though this has been poorly studied thus far (Bezemer et al., 2006; Harrison and Bardgett, 2010; Pizano et al., 2019). In general, it remains unclear if and how herbivory generates microbial soil legacies through top-down effects on the soil microbiome (Kostenko et al., 2012; Bezemer et al., 2013) and to what extent variation across soil types that differ in physico-chemical properties and native microbial communities contributes to legacy impacts. Thus, there is a need for research that investigates top-down and bottom-up plant-microbiome-herbivore interactions and how these together contribute to soil legacies impacting plant health in different soil types.
To address these knowledge gaps in plant-insect-microbiome interactions, we investigated top-down and bottom-up interactions of foliar herbivores infesting plants grown in three different soil microbiomes and their subsequent legacy effects (Figure 1). We used potato aphid (Macrosiphum euphorbiae), a common North American pest of solanaceous crops that causes leaf curling and chlorosis (Walgenbach, 1997). Potato aphids also transmit viruses, promote mold growth on leaves, and attract other damage-causing insects (Lange and Bronson, 1981; Walgenbach, 1997). Together, these effects can result in yield losses and reduced fruit quality (Walgenbach, 1997). We chose S. pimpinellifolium, a close wild relative of tomato, as the host plant to minimize effects of crop domestication, such as reduced ability to interact positively with the microbiome (Pérez-Jaramillo et al., 2015). We asked the following questions: (1) Does herbivory have similar top-down effects on rhizosphere communities across soils? (2) Do bottom-up effects on herbivore performance vary across soils? (3) How do top-down and bottom-up effects impact soil legacies on herbivory in the following generation of plants? This study is an important step in the development of effective microbiome management strategies for sustainable insect control in agro-ecosystems.
Methods
To examine the relationship between aphid herbivory and the plant-root associated microbiome (rhizosphere), we performed two experiments to test bottom-up, top-down, and legacy effects of wild tomato-rhizosphere microbiome interactions under aphid herbivory. First, we infested wild tomato plants grown in three different soil microbial communities with potato aphids and used 16S sequencing to assess top-down impacts of herbivory on rhizosphere bacterial communities (Figure 1A) as well as measures of aphid performance to assess bottom-up effects of the soil microbial community on herbivory (Figure 1B). In a second experiment, we first conditioned the three different soils with aphid-infested or control plants, then grew a new set of plants in the conditioned soils and measured aphid performance to investigate how bottom-up and top-down plant-microbiome interactions lead to legacy effects on herbivory (Figure 1C).
Plant and Insect Materials
Seeds for both experiments derived from five S. pimpinellifolium plants grown from seeds obtained from the Tomato Genetic Resource Center (University of California Davis, USA) under greenhouse conditions in Spring 2019. For both experiments, seeds were sterilized in 50% bleach (Chlorox brand sodium hypochlorite mixed with ultrapure water) for 30 min, rinsed five times with sterile ultrapure water, and stored overnight at 4C before planting.
Wingless, adult M. euphorbiae were used to infest herbivore-treated plants for both experiments. The aphid colony was maintained on the same genotype of S. pimpinellifolium in a growth chamber at 100 μm/s light 16 h day/8 h night cycle at 24°C day/20°C night. The colony was started from a potato aphid infestation on tomato plants in a high tunnel in Indiana in October 2018 and has been continuously maintained under laboratory growth chamber conditions. For both experiments, aphids were age synchronized by placing ten adult females from the colony on 3-week-old tomato plants and allowing them to reproduce for 48 h. After 48 h the adult aphids were removed, and their offspring were allowed to mature for an additional 5 days (for a total of 7 days from initial adult infestation). These age-synchronized aphids were then used for both experiments.
Soil Collection
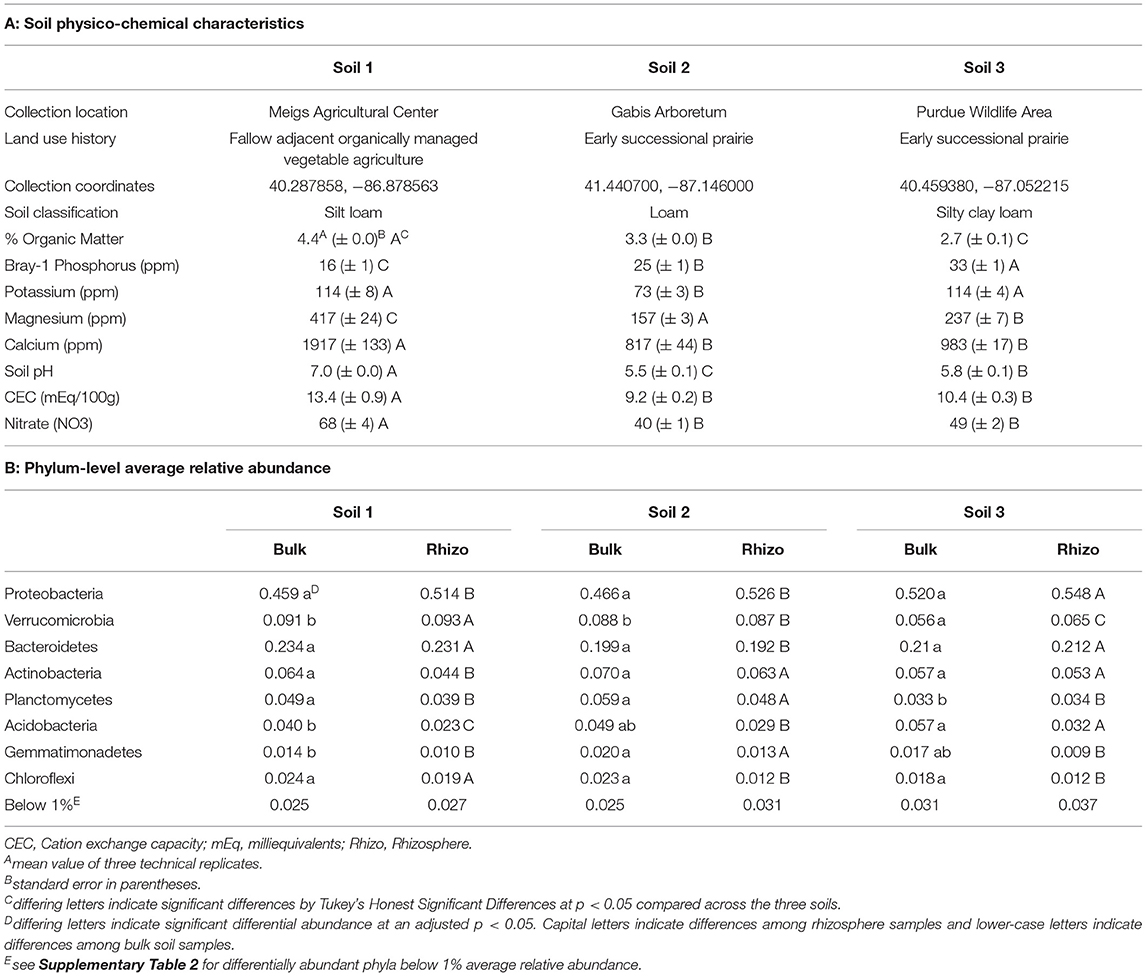
Soils for both experiments were collected from Purdue Agriculture Centers and Research Areas (Table 1A) from the top 10 cm of the soil horizon at each location and stored at 4C until use. The soils were chosen from undisturbed or minimally disturbed soils in Indiana to represent high microbial diversity. Soil 1 was collected from a fallow area adjacent to a plot that has been organically managed since 2012 with a combination of cover crops and vegetable plots at the Meigs Horticultural Research Farm. Soil 2 was collected from a preserved prairie site established in 1998 located at the Gabis Arboretum in Northwest Indiana. Soil 3 was collected from a site within the Purdue Wildlife Area cropped with corn and soybeans until 1999 and established as a prairie site in 2002. Since 2008, it has been managed with prescribed fire to encourage plant diversity. Soils were homogenized by hand and visible insects or invertebrates were removed. Soil classification, percent organic matter, Bray-1 phosphorus, potassium (K), magnesium (Mg), calcium (Ca), soil pH, cation exchange capacity (CEC), and total nitrate (NO3) were determined by a commercial laboratory (A&L Great Lakes Laboratories; Fort Wayne, Indiana, USA).
Experiment 1: Bottom-Up and Top-Down Effects of Tomato-Aphid-Microbiome Interactions
To test bottom-up effects of different soil microbial communities on aphid herbivory as well as top-down effects of herbivory on the tomato rhizosphere bacterial communities over time, a greenhouse experiment was designed that included three soil sources, three aphid infestation levels (0, 2, 10 aphids) and four time points (pre-infestation, 2, 7, and 14 days post-infestation) for a total of 30 pots per replicate block (3 soils × 3 aphid treatments × 3 time points + 3 pre-infestation samples, one for each soil). As each pot was sampled destructively, a different set of individual pots was included for each time point. Pots were arranged in a randomized complete block design with one pot per treatment in each of four blocks to account for environmental variation in the greenhouse. Each block represents one biological replicate of each treatment, leading to a total of four replicates per treatment and 120 pots for the experiment.
Soil and Seed Preparation
Three soils (labeled 1, 2, and 3) were used for this experiment (Table 1). Potting mix (Metro-mix 510, Sungro Horticulture, Massachusetts, USA) was autoclaved for 2.5 h at 235°C to sterilize. Soils were mixed with autoclaved potting mix 1:1 by volume. This was done to prevent excess soil compaction in pots that severely reduces tomato seedling germination and growth. Mixed soils were then transferred into four-inch square pots that were seeded with two sterile seeds and arranged by block in a temperature-controlled greenhouse (24–30°C) with a 16:8 h day/night cycle and high pressure sodium lighting. Pots were watered as needed with drip stakes and thinned to one plant after germination.
Aphid Infestation and Sample Collection
After ~2.5 weeks (leaf stage 4–5), plants were infested with either 0, 2 or 10 aphids and placed in mesh cages with a frame for support. Rhizospheres were harvested from plants at Day 0 (pre-infestation), and post-infestation on Days 2, 7, and 14 to examine top-down effects of aphid infestation on the rhizosphere bacterial community. Four replicate pots were harvested for each aphid treatment and soil on each date. For each sampling date, fresh shoots were collected and weighed as a measure of plant performance, and the total number of aphids was counted on each plant as a measure of aphid performance. These two measures were used as a proxy for bottom-up effects of soil type. We acknowledge that due to the reciprocal nature of top-down and bottom-up processes, aphid and plant performance may also result from top-down effects on rhizosphere communities that then feedback onto aphid performance via bottom-up effects on plant physiology, but we are unable to separate these effects in this experimental design.
Rhizosphere samples were collected by gently removing the root system from the pot and removing soil until only the rhizosphere remained (~1 mm from root surface). Then roots + rhizosphere were placed in sterile 250 mL flasks containing 50 mL of 1 × PBS buffer and agitated to dislodge rhizosphere soil. Samples were transferred to 50 mL conical tubes and centrifuged at 3000 RPM for 15 min. The supernatant was decanted, ~250 mg of rhizosphere soil was weighed into a 1.5 mL Eppendorf tube, and then flash frozen in liquid nitrogen. Bulk soil samples were collected from unplanted pots. Soil samples were then stored at −80°C until needed for DNA extraction.
DNA Extraction and Sequencing
To analyze bacterial community composition through next generation sequencing, DNA was first extracted with Qiagen DNeasy PowerSoil kits as per kit instructions. One negative water control and one mock community DNA control (ZymoBIOMICS Microbial Community Standard, Zymo Research, Irvine, California, USA) were included on each plate. Samples were sent to the University of Minnesota Genomics Center for sequencing of the V4 region of the 16S ribosomal gene following the optimized method described in Gohl et al. (2016). Sequencing was performed on the Illumina MiSeq with V2 chemistry and paired end 250 bp sequencing. Primers used were standard V4 region primers 515F GTGCCAGCMGCCGCGGTAA and 806R GGACTACHVGGGTWTCTAAT (Caporaso et al., 2011). Sample demultiplexing was performed by the University of Minnesota Genomics Center with Illumina software.
Sequence Pre-processing
16S rRNA sequencing of the V4 region resulted in a total of 11.5 M reads. Adapter removal and primer clipping was performed with Trimmomatic (v 0.36) (Bolger et al., 2014) and Cutadapt (v 1.13) (Martin, 2015). Reads were then processed through the dada2 (v 1.14.1) pipeline by filtering and trimming based on read quality, inferring error rates, merging paired end reads, removing chimeras, and assigning taxonomy with the Silva reference database v. 132 (Callahan et al., 2016). Likely contaminant sequences inferred from negative controls, as well as archaeal, mitochondrial and plastid 16S sequences and low abundance sequences (fewer than 2 reads across 10% of samples) were filtered out, leaving 2,149 amplicon sequence variants (ASVs) in the final dataset (Davis et al., 2018). After preprocessing and removing samples with fewer than 2,000 reads, the dataset contained ~3 million reads in 123 samples, averaging 24,164 reads per sample (see Supplementary Table 1 for sequencing summary).
Statistical Analyses
To examine the bottom-up effects of soil community and aphid infestation on plant performance, a linear mixed model was developed in R (v. 3.6.3) (Team, 2018) to test the effects of soil type, aphid level, and their interactions on fresh shoot weight with block included as a random factor. One model was developed for each time point as the model including time point as a variable did not meet the assumption of normality (Shapiro-Wilks test p < 0.05). To examine the bottom-up effects of soil community on aphid performance, a linear mixed model was developedto test the effects of soil type, aphid level, time, and their interactions on total aphid counts with block included as a random factor.
To compare bacterial diversity measures across samples, richness and Shannon diversity were calculated with the Phyloseq (v 1.30.0) package (McMurdie and Holmes, 2013) after subsampling to the smallest library size (2,223 reads) 100 times and averaging the results. To evaluate the effects of aphid infestation on community diversity over time in the three soils, linear mixed models were developed. Initial models included soil source, time point (post-infestation Days 2, 7, 14), aphid infestation level (0, 2 or 10) and all interactions as fixed factors with block included as a random factor, and each diversity measure (richness and Shannon diversity) was modeled separately. Statistical models tested a complete crossed design (Soil Source × Time × Aphid Level), therefore the pre-infestation time point was not included. Significant interactions occurred in these initial complete models (e.g., Soil × Time, Soil × Aphid Level), therefore we developed separate linear mixed models for each soil source to test the effects of time point, aphid infestation level, and their interaction as fixed factors and block as a random factor for each diversity measure. Separate models were developed for high (10 initial aphids) and low (2 initial aphids) infestation level comparisons to the non-infested control. Because the data set failed to meet the assumptions of homogeneity of variance and normality, non-parametric, rank-based two-way ANOVA was also performed using raov() in the Rfit (v 0.24.2) package to test the fixed effects of time, aphid level and their interaction on both diversity measures. Results from these models were consistent with the linear mixed model results, so we report the results of the linear mixed models. Rank-based results can be found in the R code uploaded to github. All linear mixed models were tested with the lmer() function in the lme4 package (v. 1.1–23) and lmerTest package (v 3.1–3) (Bates et al., 2015; Kuznetsova et al., 2017).
Then, to examine community compositional differences across samples, β diversity distance calculations and constrained analysis of principle coordinates (CAP) were performed with Phyloseq and vegan (v 2.5–6) (Dixon, 2003) packages with reads proportionally scaled to the smallest library size (code courtesy of Denef lab tutorial—http://deneflab.github.io/MicrobeMiseq/). Bray-Curtis distance, which includes both number of taxa and their relative abundance in its distance calculations, and Jaccard distance, which includes only presence/absence information on taxa, were used for distance measures. CAP analysis allows for comparison of multivariate data points according to a priori hypotheses (e.g., effects of time and aphid infestation on bacterial community composition) (Anderson and Willis, 2003). DESeq2 (v 1.26.0) (Love et al., 2014) was used to identify differentially abundant taxa between soils at the phylum level and between aphid-infested or control plants at the family and ASV levels. To identify over- or under-represented taxa exclusive to herbivore-infested or control plants in each soil, over-representation analysis was performed using Fisher's Exact Test on the number of ASVs in each phyla exclusive to the aphid-infested or control rhizospheres compared to the total number of ASVs in that phylum across the dataset. The Bonferroni test was used to correct for multiple comparisons (Abdi, 2007). All code for analysis and figure generation can be found in the Purdue University Github at https://github.itap.purdue.edu/LaramyEndersGroup/Aphid-Rhizosphere-Microbiome. All sequences have been deposited to NCBI's Short Read Archive (SRA) under the BioProject number PRJNA676117.
Experiment 2: Soil Microbial Legacy Effects of Aphid Infestation
We next designed a plant-soil feedback experiment to test (1) bottom-up effects of the soil microbial community and (2) legacy effects of both aphid herbivory and differing initial soil microbial communities on aphid herbivory in a subsequent generation of plants grown in the same soil. See Supplementary Figure 1 for a graphical representation of the feedback experiment.
Phase 1: Conditioning
For the initial conditioning phase of the experiment, sterile soil substrate was created by mixing potting mix (Metro-mix 510, Sungro Horticulture, Massachusetts, USA) and top soil (Wrede's Rocks, Lafayette, IN, USA) 1:1 by volume and steam sterilizing for 2 h. The sterile mix was then mixed with 10% live soil by volume for each of the three soils. This technique permitted the introduction of the microbial community from each soil source while minimizing soil physico-chemical differences. For the “no microbiome” control, 100% sterile soil mix was used. Mixed soils were then transferred into 36 pot flats. Pots were seeded with 2 sterile seeds each and caged in tents (Bioquip, Rancho Dominguez, California, USA) in the greenhouse with supplemental high pressure sodium lighting 16 h a day. Twenty pots were planted for each soil mixture. Flats were bottom-watered as needed to keep soil moist. When plants reached the 3 leaf stage (about 2 weeks), between 12 and 16 plants (depending on germination) were infested with 10 adult aphids for a total of 54 infested plants. An additional four un-infested plants grown in each soil type served as “no aphid” control plants, resulting in 66 total plants for the experiment. Plants were then individually caged with micro-perforated bags sealed with elastic. After 1 week, aphid performance was measured by counting the number of aphids on each plant to quantify the “bottom-up” effect of different soil microbial communities on aphid performance. Shoot weights were not measured for this experiment.
Phase 2: Response
Then, to investigate legacy effects, three pots with and without aphid infestation were selected randomly from each of three soil treatments from the conditioning phase to be carried forward for the response phase of the experiment. The “no microbiome” control was not carried forward to the response phase. The entire soil sample from each pot was collected and used to inoculate sterile soil mix, made as above, at a 10% rate by volume. After mixing thoroughly with aseptic technique, each soil mix was placed into ten pots that were then seeded with two sterile S. pimpinellifolium seeds each and thinned to one plant after germination. The flats from each replicate were then arranged randomly in three blocks in the greenhouse. After 2 weeks, eight plants from each treatment were infested with ten adult aphids. One week after infestation, aphid performance was measured by counting the total number of aphids on each plant to quantify the effects of both different soil types and herbivory in the previous generation on herbivory in the response phase.
Statistical analysis was performed in R with lm(), anova(), and TukeyHSD() functions. Two-way ANOVA with post-hoc Tukey Honest Significant Differences were used to test (1) the effects of phase 1 (conditioning) aphid infestation on phase 2 (response) aphid performance in each soil source with prior aphid infestation, soil, and their interaction as fixed factors and (2) the overall effects of phase (phase 1 conditioning vs. phase 2 response) and soil on aphid performance with phase, soil, and their interaction as fixed factors. These models did not meet the assumptions of homogeneity of variance and normality. However, the results of the rank-based test raov() gave the same results. Thus, the results of the two-way ANOVA are reported. All plots were made with the ggplot2 (v. 3.3.0) package (Wickham, 2017) and arranged in Inkscape (v. 0.92.3).
Results
Overall, Soil Types Differed in Physico-Chemical Properties, and Bacterial Community Composition
The three soils used in our experiments varied in both physical and chemical characteristics (Table 1A). Soil 1 had higher organic matter, magnesium, calcium, cation exchange capacity (CEC), soil pH, and nitrate, but lower Bray-1 (available) phosphorus than the other two soils. Soils 2 and 3 also differed from one another in organic matter, phosphorus, potassium, magnesium, and pH. All three soils were classified as loamy soils, though Soil 1 was a silt loam and Soil 3 was a silty clay loam.
Bacterial 16S rRNA sequencing of rhizosphere samples collected in Experiment 1 was used to compare the structure and composition of communities across the different soils (Table 1, Figure 2, Supplementary Tables 1–3). Principal coordinate analysis revealed that bacterial community composition varied primarily by soil type (Figure 2, Supplementary Table 3), which alone explained about 42% of the variation in the entire dataset when all experimental samples were considered. In contrast, the difference between bulk and rhizosphere samples explained only about 2% of variation in bacterial communities. To characterize the taxonomic differences among the soils, we then examined the relative abundances of taxa at the phylum level across bulk and rhizosphere samples from the three soil types (Table 1B). Overall, Proteobacteria (52.6%), Bacteroidetes (21.2%), and Verrucomicrobia (8.1%) were the most highly abundant taxa across all the soils (Table 1, Supplementary Table 2). However, several phyla were significantly differentially abundant across rhizospheres of the three soil sources, including Verrucomicrobia, Acidobacteria, and Proteobacteria. For example, Proteobacteria, the most highly abundant phylum, was significantly more abundant in Soil 3 (54.8%) than in Soil 1 (51.4%) or Soil 2 (52.6%) rhizospheres. Soil 2 samples also had higher relative abundance of Gemmatimonadetes, but lower abundance of Bacteroidetes than the other two soils, while Soil 1 had higher Chloroflexi and lower Actinobacteria. Fewer differences were observed among bulk soil samples, likely due to lower sample sizes, though relative abundances of Verrucomicrobia and Planctomycetes were lower in Soil 3 compared to the other two soils. No differences in overall bacterial species diversity were observed across bulk soil samples of the three different soils (Supplementary Table 4), indicating that differences in soil physico-chemical characteristics did not impact overall taxonomic diversity, though community composition varied.
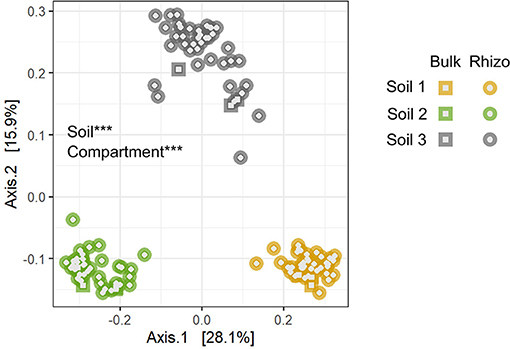
Figure 2. The three soil types differed significantly in bacterial community composition. Asterisks indicate main factors that contribute significantly to compositional variation among samples (***p < 0.001) by Bray-Curtis distance, tested by PERMANOVA. Bulk, soil samples not associated with plants; Rhizo, soil samples collected from rhizospheres. Compartment refers to bulk vs. rhizosphere samples.
Top-Down Effects: Does Aphid Herbivory Impact the Tomato Rhizosphere Microbiome Across Different Soil Communities? (Experiment 1)
High Initial Aphid Infestation Had Time and Soil-Dependent “Top-Down” Effects on Rhizosphere Bacterial Community Composition and Diversity
In contrast to initial predictions that top-down effects of aphid herbivory on the rhizosphere would be similar across soils due to a common “cry for help” response, we observed soil-dependent effects of herbivory on both the diversity and structure of rhizosphere bacterial communities. We first characterized the top-down effects of aphid herbivory on bacterial taxonomic diversity between aphid-infested and un-infested control plant rhizospheres across the three soils over time. Interestingly, though no main effects were significant, the interaction between soil and aphid level was significant for both richness (p = 0.018) and Shannon diversity (p = 0.024) when comparing high initial aphid infestation (10 aphids) and uninfested controls (Figure 3, Supplementary Table 4). This significant interaction suggested that high initial aphid infestation resulted in soil-dependent differences in diversity. Therefore, we then tested the effects of aphid infestation and time in each soil individually to characterize these differences. High initial aphid infestation resulted in significantly reduced bacterial species richness in Soil 3, where the number of amplicon sequence variants (ASVs) per sample in the rhizospheres of aphid-infested plants decreased by 22.2% compared to un-infested control plants (Figure 3; statistical results in Supplementary Table 4). Interestingly, the opposite pattern appeared in Soil 1, with bacterial richness increasing by 21.2% in rhizospheres of aphid-infested plants compared to uninfested controls; however, this trend was only marginally significant (p = 0.096). Similar trends were observed for Shannon diversity, which takes both the number of taxa and their relative abundance into account (Supplementary Figure 2, Supplementary Table 4). In contrast, rhizospheres of plants infested with low initial aphid density showed no variation in bacterial richness or Shannon diversity compared to un-infested plants in any soil over the time course (Supplementary Figure 3, Supplementary Table 4). We also found a significant interaction between soil and time in the initial overall models for high initial aphid infestation (Richness, p = 0.042; Shannon diversity, p = 0.029). When looking at the soil-specific models, we found a marginally significant effect of time for both diversity measures in Soil 1 (Richness, p = 0.076; Shannon diversity, p = 0.083) and for Shannon diversity in Soil 2 (p = 0.091). Bacterial community diversity trended upward overall in Soil 1 over time, while it trended downward in Soil 2.
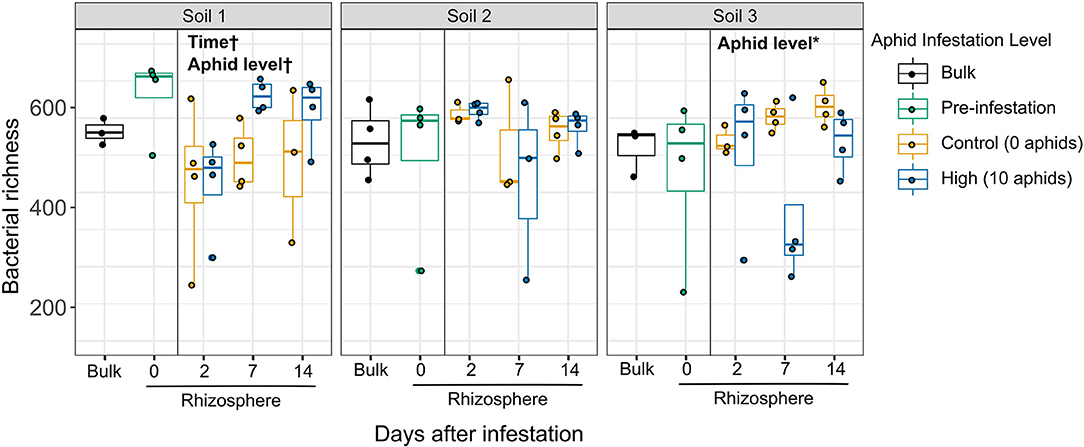
Figure 3. Top-down effects of high levels of aphid herbivory on rhizosphere bacterial taxonomic diversity vary across soils. Boxplots showing average bacterial richness (number ASVs/sample) of individual samples, comparing high aphid-infestation and no aphid infestation in each soil type (soils analyzed separately). Statistically different factors (*p < 0.05, †p < 0.1) are labeled on the panel to which they correspond.
Next, we used constrained analysis of principal coordinates (CAP) to investigate the effects of aphid infestation over multiple time points on bacterial community composition in each soil (Figure 4). High levels of aphid infestation significantly affected rhizosphere bacterial community composition in Soil 1 (Figure 4A) and Soil 3 (Figure 4B), explaining 6% of overall bacterial community variation, using Bray-Curtis distance. However, low aphid infestation did not impact community composition measured by Bray-Curtis distance in any soil (Supplementary Figure 4). For all three soils, rhizosphere bacterial community composition varied significantly over the 2 week experiment for both high and low infestation densities, with time explaining 14–23% of variation between rhizosphere samples (Figure 4, Supplementary Figure 4, Supplementary Table 5).
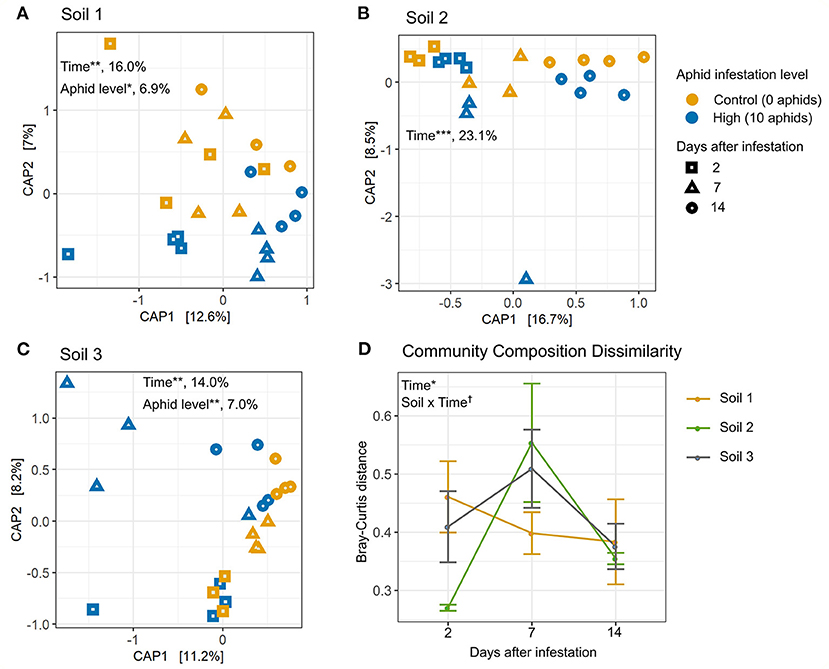
Figure 4. Top-down effects of aphid herbivory alter rhizosphere bacterial community composition in two soils, and impacts vary over time and soil type. Canonical analysis of principle component (CAP) plots showing Bray-Curtis distance between sample community compositions in (A) Soil 1, (B) Soil 2, and (C) Soil 3. Bray-Curtis distance represents variation among samples due to both taxa presence and relative abundance. Points on plots represent individual samples. Color represents aphid infestation level and shape represents the number of days after infestation. Percent variance explained is listed after significant CAP factors. Distance between aphid-infested and control samples within each block were plotted over time for (D) Bray-Curtis distance. Statistically different factors (*p < 0.05, **p < 0.01, ***p < 0.001, †p < 0.1) are labeled on the panel to which they correspond.
Next, we compared patterns of dissimilarity in community composition (i.e., Bray-Curtis distance) between aphid-infested and control plants over time and between soil types. The dissimilarity in taxonomic composition between aphid-infested and control samples varied significantly over the time points evaluated, peaking at 7 days (Figure 4D). Additionally, the interaction between soil and time was marginally significant (p = 0.07; Supplementary Table 6), suggesting that the effects of aphid infestation on rhizosphere community composition over time differed by soil type. In particular, rhizosphere communities from aphid-infested plants differed from their controls to a similar extent over time in Soil 1, while dissimilarity between aphid-infested and control communities peaked at 7 days for Soils 2 and 3, suggesting a transient effect of aphid infestation on the rhizosphere bacterial community in these two soils.
Firmicutes, Particularly Bacillaceae, Are More Diverse and Abundant in Aphid-Infested Rhizospheres of Soil 1 Compared to Un-infested Controls
We next investigated the presence/absence of taxa between aphid-infested and control plants in each soil type to identify underlying sources of variation that could contribute to observed differences in bacterial diversity and composition (Figures 3, 4A–C). Because the differences between aphid-infested and control rhizospheres were highest at 7 days after infestation, we focused only on this time point for our analysis. Consistent with differences in bacterial species richness, Soil 1 had a higher proportion of taxa that were found exclusively (i.e., unique to a treatment) in the aphid-treated rhizospheres (25.3%) compared to the un-infested controls (6.9%) at Day 7 (Figure 5A). The opposite pattern was observed in Soil 3, where a larger proportion of taxa were found exclusively in the control plants (29.1%) compared to the aphid-infested plants (11.2%) (Figure 5C). Soil 2 had similar proportions of taxa found exclusively in rhizospheres of either control or infested plants (20.0 vs. 14.0%) compared to the other two soils (Figure 5B). Next, we performed an over-representation analysis at the phylum level to determine if any taxa exclusive to aphid-infested or control rhizospheres in a given soil were significantly over or under-represented. We found that the phylum Firmicutes was significantly overrepresented in aphid-infested plants in Soil 1 (Fisher's exact test, adjusted p < 0.05) at 7 days after infestation (Figure 5D, Supplementary Table 7), suggesting an increased diversity of Firmicutes present in aphid-infested rhizospheres of Soil 1-grown plants.
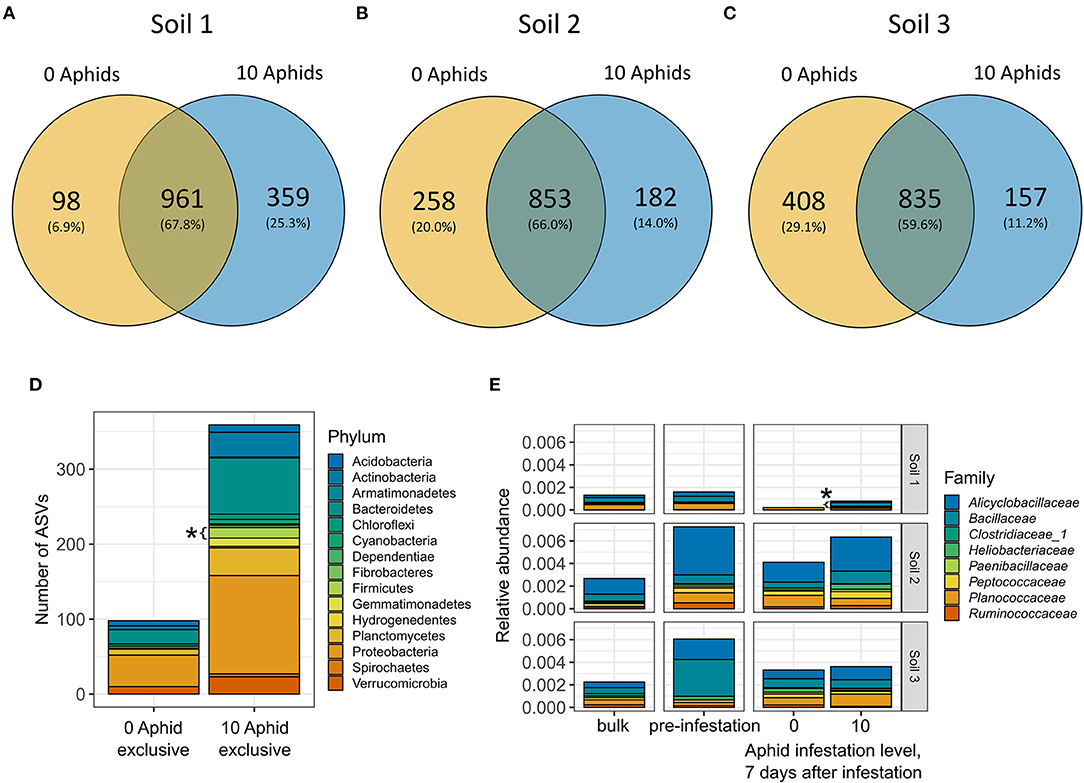
Figure 5. Differential abundance and presence/absence analysis show an enrichment of Firmicutes in the aphid-infested plants grown in Soil 1 at 7 days after infestation. Venn Diagrams showing numbers and proportions of ASVs found exclusively in the rhizospheres of aphid-infested, control plants, or both in (A) Soil 1, (B) Soil 2, and (C) Soil 3. (D) Stacked barplot showing the taxonomic composition at the phylum level of ASVs found exclusively in control or aphid-infested rhizospheres of Soil 1. Significantly over-represented phyla (adjusted p < 0.05) are marked with an asterisk. (E) Relative abundance of Firmicutes, colored by family, across the three soils (rows). Columns represent bulk soil, pre-infestation rhizosphere, and control and aphid-infested rhizospheres at 7 days after infestation. Significantly differentially abundant families (adjusted p < 0.05) are marked with an asterisk.
We next examined differential abundance of taxa between aphid-infested and control rhizosphere communities at 7 days after infestation as CAP analysis showed differences between aphid-infested and control rhizosphere bacterial communities in Soils 1 and 3 (Figure 4). Only a few low abundance taxa were differentially abundant between aphid-infested and control plants at the ASV level, with ten differentially abundant taxa in Soil 1, and one each in Soils 2 and 3 (Supplementary Table 8). No bacterial families were differentially abundant between aphid-infested and control rhizospheres in either prairie soil treatment. Three families were however differentially abundant in Soil 1, with Bacillaceae (Phylum: Firmicutes) (Figure 5E) and Azospirillaceae significantly enriched in aphid-infested rhizospheres, and Pedosphaeraceae depleted in aphid-infested compared to control rhizospheres (Supplementary Table 8). Though Bacillaceae were enriched in aphid-infested Soil 1 plants, Soil 1 had the lowest overall relative abundance of Firmicutes compared to the other two soils (Figure 5E, Supplementary Table 2).
Bottom-Up Effects: Do Soil Microbial Community Differences Impact Aphid Herbivory (Experiments 1 and 2)?
Soil Microbial Community Source Impacted Bottom-Up Effects on Plant and Aphid Performance
We next asked whether fresh shoot weight and aphid population growth were impacted by different soil microbial communities in a bottom-up manner (Figure 1B). In Experiment 1, fresh shoot weight was significantly impacted by soil source at each time point with plants grown in Soil 1 generally having the lowest shoot weight. By 14 days after infestation, all three soils were significantly different, with plants grown in Soil 2 having the highest shoot weights and Soil 1 the lowest (Supplementary Figure 5, Supplementary Table 9) Aphid infestation did not significantly impact shoot weight at any time point. Total aphid population size per plant changed significantly over time and with initial aphid infestation level, as expected (Supplementary Figure 6, Supplementary Table 10). However, soil source was not a significant factor, though the small number of replicates and wide variation in aphid population growth in Soils 1 and 2 suggested that a higher-powered experiment may reveal differential impacts of soil microbial community source on aphid performance. Thus, in Experiment 2 we incorporated higher replication (12–16 replicates per soil), focused on only the high initial aphid infestation level, and counted aphid populations on each plant 7 days after infestation. Additionally, in order to minimize the effects of soil physico-chemical differences we mixed each of the soils at a 10% rate with a 1:1 sterile mix of potting soil and topsoil. We included a “no microbiome” control to compare and to what extent the magnitude of bottom-up effects would differ across soils in comparison to an initially sterile no-microbiome control. In Experiment 2, soil microbial community source significantly affected aphid population growth in phase 1 (Figure 6, Supplementary Table 10). Surprisingly, aphid performance was significantly lower on the “no microbiome” control plants than on plants with an intact soil microbiome (i.e., no evidence of a soil microbiome induced herbivore suppression). However, we did observe significant differences in bottom-up effects of the microbiome on aphid performance across soils compared to the “no microbiome” control. Soil 1 had the lowest performance with only a 19% increase over the “no microbiome” control, while Soil 3 had the highest aphid performance with an ~52% increase over the control. Aphid performance in plants grown in Soil 2 (43% increase compared to “no microbiome” control) was not significantly different from either of the other two soils.
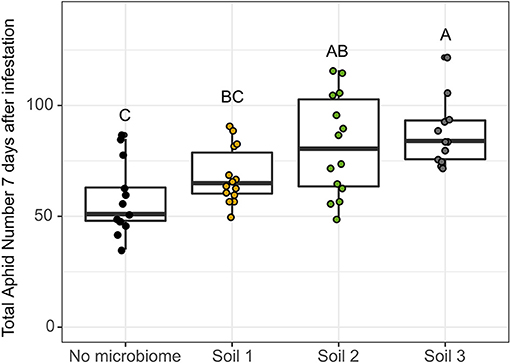
Figure 6. Native soil microbial community has a “bottom-up” impact on aphid performance after 7 days of infestation. Aphid performance was measured as the total number of aphids present on the plant 7 days after infestation with ten adult, wingless aphids. “No microbiome” control refers to pre-sterilized soil with no introduced native microbial community. Differing letters indicate significant differences between groups at p < 0.05 with Tukey's Honest Significant Differences test.
Legacy Effects: Do Bottom-Up or Top-Down Plant-Rhizosphere Interactions Leave Soil Microbial Community-Dependent Legacies on Herbivory? (Experiment 2)
Soil Microbial Community Source, but Not Herbivory, Impacted Soil Legacy Effects on Aphid Performance
Because we observed both top-down impacts of herbivory on the rhizosphere as well as bottom-up differences in aphid performance across soils (Figures 3–6), we then investigated whether aphid infestation would result in a feedback effect on herbivore performance in a following generation of plants grown in that soil (Figure 1C). We compared aphid performance between phase one (conditioning) and two (response) across the different soil sources to investigate whether soil legacy effects differed. Overall aphid performance was unchanged from phase one to phase two (Figure 7). Conditioning with either aphid-infested or uninfested control plants in phase one had no effect on aphid performance in phase two plants (Supplementary Figure 7, Supplementary Table 11), though soil type did have a significant effect, thus suggesting top-down effects of herbivory play minimal to no role in determining legacy effects on herbivory over multiple generations in this system. However, there was a significant interaction between phase and soil, where aphid performance trended upwards for Soil 1, while it trended downward in Soils 2 and 3 (Figure 7, Supplementary Table 11), suggesting a role for initial soil microbial community (e.g., bottom-up effects) in determining soil legacy effects on aphid performance.
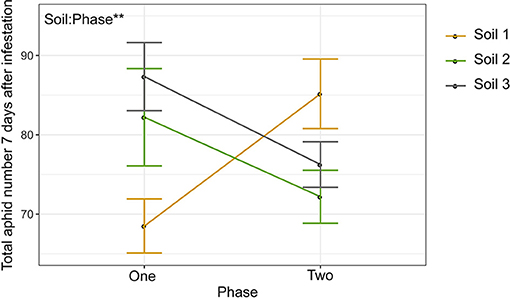
Figure 7. Soil microbial community source affects legacy impacts on herbivore performance. Interaction plot showing mean aphid performance between Phase 1 and 2. Error bars represent standard error of the mean. Significant factors are shown (**p < 0.01).
Discussion
In this study, we found that high levels of aphid herbivory have a top-down impact on the diversity and composition of the rhizosphere bacterial microbiome of wild tomato (S. pimpinellifolium), but this effect depended on the soil microbial communities in which the plants were grown (Figures 2–5). Our study also provides evidence for a bottom-up effect of the rhizosphere microbiome on aphids as soil source impacted herbivore performance (Figure 6). Additionally, soil microbial community, but not aphid infestation, impacted legacy effects on herbivory, suggesting that bottom-up plant-rhizosphere interactions are important for determining soil legacies over multiple generations (Figure 7).
Top-Down Effects of Herbivory on the Rhizosphere Microbiome Are Soil-Dependent
Few other studies thus far have examined the impacts of above-ground herbivory on the below-ground soil microbiome, and most have focused only on culturable bacteria (Yang et al., 2011; Lee et al., 2012) or cultivation-independent measures of fungal taxa (Kostenko et al., 2012; Bezemer et al., 2013; Schaeffer et al., 2017; Wilkinson et al., 2019). In this study, top down effects of aphid herbivory on rhizosphere bacterial community diversity varied according to the soil in which the plants were grown, ranging from a 20% decrease to a 20% increase, with no effects in one soil (Figure 3). When considering community composition, the effects of herbivory accounted for ~6–7% of variation in the rhizosphere bacterial community in two of the soils (Figure 4). Similar to our study, herbivory on ragwort (Jacobaea vulgaris) altered rhizosphere community composition by ~10%, though the herbivore tested was a chewing insect rather than a sap-feeding insect, and the focus was on the fungal community (Bezemer et al., 2013). In contrast, O'Brien et al. (2018) observed no effect of green peach aphid (Myzus persicae) herbivory on cabbage rhizosphere bacterial community diversity or composition when plants were grown under controlled conditions in an agricultural field soil.
Several factors may have played a role in the soil-type dependent differences we observed in top-down microbiome responses to herbivory. For example, some soils may contain strains of bacteria that are more responsive to plant defensive chemistries activated in response to herbivore feeding. Plants in the Solanaceae family generally activate the salicylic acid pathway in response to piercing/sucking insects like aphids (Digilio et al., 2010; Coppola et al., 2019), and salicylic acid has been implicated in structuring root bacterial communities (Lebeis et al., 2015), although these effects were observed in the endosphere rather than the rhizosphere. In addition, differences in root exudation in response to both herbivory (Hamilton et al., 2008) and soil chemistry (Zhang et al., 1997; Carvalhais et al., 2013; Neumann et al., 2014) is another possible route to herbivore-triggered microbiome modulation that could lead to top-down effects dependent on soil properties and native microbial communities. Interestingly, the top-down effects we observed on the rhizosphere appeared to be transient, peaking at 7 days after infestation. These transient shifts in rhizosphere communities may in part be connected to the timing of induction of plant defensive hormone signaling or root exudation, which can be a dynamic process (Weidenhamer et al., 2014; Coppola et al., 2019) that only temporarily alters microbial communities. Further research will be required to elucidate mechanisms and temporal dynamics of soil microbial community-dependent responses to herbivory.
Though we observed significant, but transient, top down effects of herbivory on rhizosphere community composition and diversity, we found few significant shifts in taxonomic abundance or presence/absence of specific groups that explain differences in aphid performance across the soils. We observed changes in three bacterial families in the rhizosphere in aphid-infested plants grown in Soil 1, which had the lowest overall aphid performance, with Pedosphaeraceae (phylum: Verrucomicrobia) being depleted and Bacillaceae (phylum: Firmicutes) and Azospirillaceae (phylum: Proteobacteria) being enriched. Bacterial strains belonging to the Pedosphaeraceae family are known to break down complex carbon molecules (Senechkin et al., 2010), suggesting a shift in root exudate composition under aphid infestation. Azospirillaceae are free-living nitrogen fixers, often having plant-growth promoting capabilities (Cassán et al., 2020), though impacts on biotic stress have not been documented. Some strains of Bacillaceae have been shown to induce systemic resistance against insects (Valenzuela-Soto et al., 2010; Gadhave and Gange, 2016), though further work is needed to determine if the strains present in this soil are protective against insects. However, Soil 1 had an overall higher abundance of Bacillaceae before aphid infestation, suggesting that infested Soil 1 plants are maintaining their populations of Bacillaceae rather than recruiting them to the rhizosphere in a “cry for help” in response to herbivore attack, as has been observed when pepper plants were challenged with green peach aphid after inoculation with a strain of Bacillus subtilis (Lee et al., 2012). Additionally, Soil 1 had the lowest abundance of Bacillaceae compared to the other two soils. In the other two soils, Bacillaceae relative abundance also decreased over time but was not significantly different between aphid-infested and control plants. Thus, it is unclear whether maintenance of Bacillaceae relative abundance or overall increase in presence of Firmicutes in response to aphid infestation played a role in the reduced aphid performance on plants grown in Soil 1.
Overall, our data provides little evidence for the “cry for help” hypothesis (Yi et al., 2011; Liu et al., 2020), suggesting that this mechanism is less important for plant defenses against herbivory or depends heavily on the soil community in which affected plants are growing. Instead, the overall soil-dependent shifts in community diversity in response to aphid infestation may have played a role in aphid performance as diversity increased in Soil 1, which had the lowest aphid performance, and decreased in Soil 3, which had the highest aphid performance (Figures 3, 6), suggesting a negative correlation between bacterial diversity response to infestation and aphid performance, though more soil microbial communities will need to be tested to confirm this phenomenon. In chrysanthemum, soil bacterial community diversity differences were similarly negatively correlated with thrips performance, though the differences in community diversity resulted from previous soil conditioning with a variety of plant species (Pineda et al., 2019), not by the thrips themselves. Increased diversity has been positively linked with soil and plant health (Hartmann et al., 2015; Bender et al., 2016; El Mujtar et al., 2019; Saleem et al., 2019), though it is currently unclear whether this relationship is causal and in some cases higher diversity does not equate to a healthier or more beneficial microbiome (Shade, 2017). Future work will be needed to further understand the mechanistic relationship between rhizosphere bacterial community diversity and plant protection from herbivory.
Bottom-Up Effects of the Soil Microbiome on Aphid Performance Differ Among Soils, but Consistently Increase Plant Susceptibility to Aphids Compared to a “No Microbiome” Control
Consistent with other studies that observed bottom-up effects of soil inoculum on insect performance (Badri et al., 2013; Benítez et al., 2017; Hubbard et al., 2019; Blundell et al., 2020), we found that differences in aphid performance depended on the source of the soil microbial community (Figures 6, 7). While we did not measure effects of microbiome differences on plant nutritional quality or defense, we observed differences in shoot weight between plants grown in the different soils consistent with aphid performance (e.g., Soil 1 had both the lowest shoot weights and aphid population growth) (Supplementary Figure 5), suggesting that the effects of the soil microbiome on plant growth and aphid performance in this experiment were nutritionally linked. Previous studies have correlated compositional microbiome differences with plant nutritional content (Badri et al., 2013) and secondary chemistry (Benítez et al., 2017; Blundell et al., 2020) that resulted in differences in insect performance.
Surprisingly, we observed a positive impact overall of an intact microbiome on aphid performance compared to the pre-sterilized, “no microbiome” control. This is in contrast with several other studies that observed that intact microbiomes resulted in decreased insect performance or damage due to green peach aphids, flea beetles (Hubbard et al., 2019), cabbage looper (Badri et al., 2013), and thrips (Pineda et al., 2019) compared to a disrupted or “no microbiome” control. This discrepancy could be due to differences in soil microbial community sources and methods of introduction, methods of creating a “no microbiome” control, or microbiome effect differences across feeding guilds. The contrasting results with green peach aphid (Hubbard et al., 2019) may also be related to assay differences. That study measured aphid populations due to natural infestation, which is a result of both aphid performance as well as attraction to the plant, while our study measured only aphid performance after experimental infestation on caged plants. Our observation of increased aphid performance in intact microbiome treatments compared to the “no microbiome” control may be the result of triggered susceptibility or increased plant nutritional quality due to the presence of the microbiome, consistent with studies that have shown that some bacterial strains trigger susceptibility to aphids (Pineda et al., 2012) and alter nutritional quality (Badri et al., 2013).
Bottom-Up, but Not Top-Down, Microbiome-Plant-Herbivore Interactions Contribute to Legacy Impacts on Aphid Performance
Though bottom-up and top-down microbiome-plant-herbivore interactions are both thought to impact herbivore performance (Friman et al., 2020), their respective roles in legacy impacts on herbivory are poorly characterized. Unlike previous studies in ragwort (Kostenko et al., 2012; Bezemer et al., 2013), we did not observe a legacy impact of above-ground herbivory on herbivore performance on wild tomato in the subsequent planting. This discrepancy may result from differences in legacy impacts between feeding guilds as the previous studies examined legacy impacts of a chewing insect. Piercing/sucking insects like aphids may have little impact on soil legacy. Alternatively, given that we did not observe a negative impact of aphid herbivory on plant performance, it is possible that the plants in our study were not exposed to the aphids long enough to condition the soil sufficiently to observe legacy effects (Lepinay et al., 2018). However, we did see an impact of soil microbial community source on herbivore legacy with two of the soils resulting in negative feedbacks and one soil resulting in a positive feedback on aphid performance in the following generation. Interestingly, the soil with the positive feedback had the lowest aphid performance in the first phase of the experiment, providing further evidence against the “cry for help” hypothesis.
Multiple mechanisms could have been involved in the bottom-up effects of soil on aphid performance in both the initial and following generation of plants. First, observed differences in aphid performance and soil legacies may have been due directly to differences in soil physico-chemical characteristics. For example, phosphorus (P) and nitrogen (N) levels have been positively linked to aphid performance measures (Jansson and Ekbom, 2002). Aphid performance correlated with Bray-1 P measurements, with Soil 3 having the highest aphid performance in Phase 1 and highest Bray-1 P levels, though the same correlation was not seen with N levels. Additionally, soil type and N levels have been shown to impact plant-soil feedbacks on plant performance (Bezemer et al., 2006; Manning et al., 2008), though feedback effects on herbivory have not been studied. However, since we observed differences in aphid performance when minimizing soil physico-chemical differences through 10% soil inoculation into the same sterile soil substrate, it is more likely that microbial community differences impacted aphid performance, possibly through effects on plant growth, though direct effects of soil chemistry cannot be ruled out. While direct effects of soil characteristics were likely not the major drivers of the differences observed in aphid performance and legacy impacts, differences in soil physico-chemical characteristics such as pH, organic matter content, and nutrient levels have been linked to differences in soil microbial communities (Fierer, 2017). The three soils that we tested all had major differences in these parameters, which likely contributed to the differences we observed in their bacterial communities, thus leading to indirect impacts of soil chemistry on herbivore performance and legacy impacts.
Conclusions and Implications for Agriculture
In conclusion, we have shown that both top-down and bottom-up plant-herbivore interactions depend on the soil microbial community in which the plant is grown, with bottom-up effects in particular impacting herbivore performance in both current and following plant generations. Therefore, the impact of variation in soil microbiomes on plant responses to herbivory has important, and thus far under-appreciated, implications for insect pest management in agroecosystems. Not all soil communities may have the potential to provide benefits to crop health and protection from pests. Our work highlights the need to screen plant-herbivore-microbiome interactions across many soil microbiomes, which will be essential for identifying soil microbial community characteristics important in determining plant protection from insects. Additionally, agricultural management practices (i.e., nutrient management, rotation, inter-cropping, irrigation) as well as crop species and cultivar are all known to impact the soil microbial community (French et al., 2021). Thus, compositional and functional alteration of native soil communities that result from chosen agricultural management practices will likely impact top-down, bottom-up and legacy effects on herbivore performance. Further work is needed investigating plant-herbivore-microbiome interactions under field conditions, especially different agroecosystem management regimes (i.e., conventional, organic, low-input). Developing tools to predict how a given native microbial community will interact with a plant experiencing herbivory stress will be critical to creating strategies for managing microbiomes for crop health in agro-ecosystems, whether through interventions with microbial inoculations, predictive monitoring to assess potential impacts of a given microbial community on herbivore-crop interactions, or implementation of agricultural practices that positively direct the microbiome.
Data Availability Statement
The datasets presented in this study can be found in online repositories. The names of the repository/repositories and accession number(s) can be found at: http://www.ncbi.nlm.nih.gov/bioproject/676117, PRJNA676117.
Author Contributions
EF and LE developed research questions and designed experiments. EF conducted experiments, completed data analysis, created figures and tables, and wrote the manuscript. IK and LE provided conceptual guidance and edited the manuscript. All authors contributed to the article and approved the submitted version.
Funding
This research was funded by USDA-NIFA Grant #2019-67013-29635 awarded to PD Enders. USDA-NIFA postdoctoral fellowship #2019-07257 awarded to E. French as a funding source.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank members of Enders and Kaplan labs for help with rhizosphere collection and aphid counting and Wadih Ghanem for assistance in aphid rearing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.629684/full#supplementary-material
References
Abdi, H. (2007). “The Bonferonni and Sˇidák Corrections for Multiple Comparisons,” in Encyclopedia of Measurement and Statistics, ed. N. Salkind (Thousand Oaks, CA: Sage), 103–107. doi: 10.4135/9781412952644
Anderson, M. J., and Willis, T. J. (2003). Canonical analysis of principal coordinates: a useful method of constrained ordination for ecology. Ecology 84, 511–525. doi: 10.1890/0012-9658(2003)084[0511:CAOPCA]2.0.CO;2
Badri, D. V., Zolla, G., Bakker, M. G., Manter, D. K., and Vivanco, J. M. (2013). Potential impact of soil microbiomes on the leaf metabolome and on herbivore feeding behavior. New Phytol. 198, 264–273. doi: 10.1111/nph.12124
Bates, D., Mächler, M., Bolker, B. M., and Walker, S. C. (2015). Fitting linear mixed-effects models using lme4. J. Stat. Softw. 67, 1–48. doi: 10.18637/jss.v067.i01
Bender, S. F., Wagg, C., and van der Heijden, M. G. A. (2016). An underground revolution: biodiversity and soil ecological engineering for agricultural sustainability. Trends Ecol. Evol. 31, 440–452. doi: 10.1016/j.tree.2016.02.016
Benítez, E., Paredes, D., Rodríguez, E., Aldana, D., González, M., Nogales, R., et al. (2017). Bottom-up effects on herbivore-induced plant defences: a case study based on compositional patterns of rhizosphere microbial communities. Sci. Rep. 7:6251. doi: 10.1038/s41598-017-06714-x
Berendsen, R. L., Pieterse, C. M. J., and Bakker, P. a H. M. (2012). The rhizosphere microbiome and plant health. Trends Plant Sci. 17, 478–486. doi: 10.1016/j.tplants.2012.04.001
Berendsen, R. L., Vismans, G., Yu, K., Song, Y., De Jonge, R., Burgman, W. P., et al. (2018). Disease-induced assemblage of a plant-beneficial bacterial consortium. ISME J. 12, 1496–1507. doi: 10.1038/s41396-018-0093-1
Bezemer, T. M., Lawson, C. S., Hedlund, K., Edwards, A. R., Brook, A. J., Igual, J. M., et al. (2006). Plant species and functional group effects on abiotic and microbial soil properties and plant-soil feedback responses in two grasslands. J. Ecol. 94, 893–904. doi: 10.1111/j.1365-2745.2006.01158.x
Bezemer, T. M., van der Putten, W. H., Martens, H., van de Voorde, T. F. J., Mulder, P. P. J., and Kostenko, O. (2013). Above- and below-ground herbivory effects on below-ground plant-fungus interactions and plant-soil feedback responses. J. Ecol. 101, 325–333. doi: 10.1111/1365-2745.12045
Blundell, R., Schmidt, J. E., Igwe, A., Cheung, A. L., Vannette, R. L., Gaudin, A. C. M., et al. (2020). Organic management promotes natural pest control through altered plant resistance to insects. Nat. Plants 6, 483–491. doi: 10.1038/s41477-020-0656-9
Bolger, A. M., Lohse, M., and Usadel, B. (2014). Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. doi: 10.1093/bioinformatics/btu170
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13, 581–583. doi: 10.1038/nmeth.3869
Caporaso, J. G., Lauber, C. L., Walters, W. A., Berg-Lyons, D., Lozupone, C. A., Turnbaugh, P. J., et al. (2011). Global patterns of 16S rRNA diversity at a depth of millions of sequences per sample. Proc. Natl. Acad. Sci U.S.A. 108, 4516–4522. doi: 10.1073/pnas.1000080107
Carrillo, J., Ingwell, L. L., Li, X., and Kaplan, I. (2019). Domesticated tomatoes are more vulnerable to negative plant-soil feedbacks than their wild relatives. J. Ecol. 107, 1365–2745. doi: 10.1111/1365-2745.13157
Carvalhais, L. C., Dennis, P. G., Fan, B., Fedoseyenko, D., Kierul, K., Becker, A., et al. (2013). Linking plant nutritional status to plant-microbe interactions. PLoS ONE 8:e68555. doi: 10.1371/journal.pone.0068555
Cassán, F., Coniglio, A., López, G., Molina, R., Nievas, S., de Carlan, C. L. N., et al. (2020). Everything you must know about Azospirillum and its impact on agriculture and beyond. Biol. Fertil. Soils 56, 461–479. doi: 10.1007/s00374-020-01463-y
Coppola, M., Diretto, G., Digilio, M. C., Woo, S. L., Giuliano, G., Molisso, D., et al. (2019). Transcriptome and metabolome reprogramming in tomato plants by Trichoderma harzianum strain T22 primes and enhances defense responses against aphids. Front. Physiol. 10:745. doi: 10.3389/fphys.2019.00745
Davis, N. M., Proctor Di, M., Holmes, S. P., Relman, D. A., and Callahan, B. J. (2018). Simple statistical identification and removal of contaminant sequences in marker-gene and metagenomics data. Microbiome 6, 1–14. doi: 10.1186/s40168-018-0605-2
Digilio, M. C., Corrado, G., Sasso, R., Coppola, V., Iodice, L., Pasquariello, M., et al. (2010). Molecular and chemical mechanisms involved in aphid resistance in cultivated tomato. New Phytol. 187, 1089–1101. doi: 10.1111/j.1469-8137.2010.03314.x
Dixon, P. (2003). VEGAN, a package of R functions for community ecology. J. Veg. Sci. 14, 927–930. doi: 10.1111/j.1654-1103.2003.tb02228.x
El Mujtar, V., Muñoz, N., Prack Mc Cormick, B., Pulleman, M., and Tittonell, P. (2019). Role and management of soil biodiversity for food security and nutrition; where do we stand? Glob. Food Sec. 20, 132–144. doi: 10.1016/j.gfs.2019.01.007
Fierer, N. (2017). Embracing the unknown: Disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Finkel, O. M., Castrillo, G., Herrera Paredes, S., Salas González, I., and Dangl, J. L. (2017). Understanding and exploiting plant beneficial microbes. Curr. Opin. Plant Biol. 38, 155–163. doi: 10.1016/j.pbi.2017.04.018
French, E., Kaplan, I., Iyer-Pascuzzi, A., Nakatsu, C. H., and Enders, L. (2021). Emerging strategies for precision microbiome management in diverse agroecosystems. Nat. Plants 7, 256–267. doi: 10.1038/s41477-020-00830-9
Friman, J., Pineda, A., van Loon, J. J. A., and Dicke, M. (2020). Bidirectional plant-mediated interactions between rhizobacteria and shoot-feeding herbivorous insects: a community ecology perspective. Ecol. Entomol. 46, 1–10. doi: 10.1111/een.12966
Gadhave, K. R., and Gange, A. C. (2016). Plant-associated Bacillus spp. alter life-history traits of the specialist insect Brevicoryne brassicae L. Agric. For. Entomol. 18, 35–42. doi: 10.1111/afe.12131
Gohl, D. M., Vangay, P., Garbe, J., MacLean, A., Hauge, A., Becker, A., et al. (2016). Systematic improvement of amplicon marker gene methods for increased accuracy in microbiome studies. Nat. Biotechnol. 34, 942–949. doi: 10.1038/nbt.3601
Gosset, V., Harmel, N., Göbel, C., Francis, F., Haubruge, E., Wathelet, J. P., et al. (2009). Attacks by a piercing-sucking insect (Myzus persicae Sultzer) or a chewing insect (Leptinotarsa decemlineata Say) on potato plants (Solanum tuberosum L.) induce differential changes in volatile compound release and oxylipin synthesis. J. Exp. Bot. 60, 1231–1240. doi: 10.1093/jxb/erp015
Hamilton, E. W., Frank, D. A., Hinchey, P. M., and Murray, T. R. (2008). Defoliation induces root exudation and triggers positive rhizospheric feedbacks in a temperate grassland. Soil Biol. Biochem. 40, 2865–2873. doi: 10.1016/j.soilbio.2008.08.007
Harrison, K. A., and Bardgett, R. D. (2010). Influence of plant species and soil conditions on plant-soil feedback in mixed grassland communities. J. Ecol. 98, 384–395. doi: 10.1111/j.1365-2745.2009.01614.x
Hartmann, M., Frey, B., Mayer, J., Mäder, P., and Widmer, F. (2015). Distinct soil microbial diversity under long-term organic and conventional farming. ISME J. 9, 1177–1194. doi: 10.1038/ismej.2014.210
Holland, J. N. (1995). Effects of above-ground herbivory on soil microbial biomass in conventional and no-tillage agroecosystems. Appl. Soil Ecol. 2, 275–279. doi: 10.1016/0929-1393(95)00055-2
Hu, L., Robert, C. A. M., Cadot, S., Zhang, X., Ye, M., Li, B., et al. (2018). Root exudate metabolites drive plant-soil feedbacks on growth and defense by shaping the rhizosphere microbiota. Nat. Commun. 9:2738. doi: 10.1038/s41467-018-05122-7
Hubbard, C. J., Li, B., McMinn, R., Brock, M. T., Maignien, L., Ewers, B. E., et al. (2019). The effect of rhizosphere microbes outweighs host plant genetics in reducing insect herbivory. Mol. Ecol. 28, 1801–1811. doi: 10.1111/mec.14989
Jansson, J., and Ekbom, B. (2002). The effect of different plant nutrient regimes on the aphid Macrosiphum euphorbiae growing on petunia. Entomol. Exp. Appl. 104, 109–116. doi: 10.1046/j.1570-7458.2002.00997.x
Kaplan, I., Pineda, A., and Bezemer, M. (2018). Application and theory of plant–soil feedbacks on aboveground Herbivores 319–343. doi: 10.1007/978-3-319-91614-9_14
Kong, H. G., Kim, B. K., Song, G. C., Lee, S., and Ryu, C. M. (2016). Aboveground whitefly infestation-mediated reshaping of the root microbiota. Front. Microbiol. 7:1314. doi: 10.3389/fmicb.2016.01314
Kos, M., Tuijl, M. A. B., de Roo, J., Mulder, P. P. J., and Bezemer, T. M. (2015). Species-specific plant-soil feedback effects on above-ground plant-insect interactions. J. Ecol. 103, 904–914. doi: 10.1111/1365-2745.12402
Kostenko, O., van de Voorde, T. F. J., Mulder, P. P. J., van der Putten, W. H., and Bezemer, T. M. (2012). Legacy effects of aboveground – belowground interactions. Ecol. Lett. 15, 813–821. doi: 10.1111/j.1461-0248.2012.01801.x
Kuznetsova, A., Brockhoff, P. B., and Christensen, R. H. B. (2017). lmerTest Package: tests in linear mixed effects models. J. Stat. Softw. 82. doi: 10.18637/jss.v082.i13
Kwak, M. J., Kong, H. G., Choi, K., Kwon, S. K., Song, J. Y., Lee, J., et al. (2018). Rhizosphere microbiome structure alters to enable wilt resistance in tomato. Nat. Biotechnol. 36, 1100–1116. doi: 10.1038/nbt.4232
Lange, W. H., and Bronson, L. (1981). Insect pests of tomatoes. Annu. Rev. Entomol. 26, 345–371. doi: 10.1146/annurev.en.26.010181.002021
Lebeis, S. L., Paredes, S. H., Lundberg, D. S., Breakfield, N., Gehring, J., McDonald, M., et al. (2015). Salicylic acid modulates colonization of the root microbiome by specific bacterial taxa. Science 349, 860–864. doi: 10.1126/science.aaa8764
Lee, B., Lee, S., and Ryu, C. M. (2012). Foliar aphid feeding recruits rhizosphere bacteria and primes plant immunity against pathogenic and non-pathogenic bacteria in pepper. Ann. Bot. 110, 281–290. doi: 10.1093/aob/mcs055
Lepinay, C., Vondráková, Z., Dostálek, T., and Münzbergová, Z. (2018). Duration of the conditioning phase affects the results of plant-soil feedback experiments via soil chemical properties. Oecologia 186, 459–470. doi: 10.1007/s00442-017-4033-y
Liu, H., Brettell, L. E., Qiu, Z., and Singh, B. K. (2020). Microbiome-mediated stress resistance in plants. Trends Plant Sci. 25, 733–743. doi: 10.1016/j.tplants.2020.03.014
Love, M. I., Anders, S., and Huber, W. (2014). Differential analysis of count data - the DESeq2 package. Genome Biol. 15:550. doi: 10.1186/s13059-014-0550-8
Manning, P., Morrison, S. A., Bonkowski, M., and Bardgett, R. D. (2008). Nitrogen enrichment modifies plant community structure via changes to plant-soil feedback. Oecologia 157, 661–673. doi: 10.1007/s00442-008-1104-0
Martin, M. (2015). Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J. 17, 1–3. doi: 10.14806/ej.17.1.200
McMurdie, P. J., and Holmes, S. (2013). Phyloseq: an R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 8:e61217. doi: 10.1371/journal.pone.0061217
Milcu, A., Bonkowski, M., Collins, C. M., and Crawley, M. J. (2015). Aphid honeydew-induced changes in soil biota can cascade up to tree crown architecture. Pedobiologia 58, 119–127. doi: 10.1016/j.pedobi.2015.07.002
Neumann, G., Bott, S., Ohler, M. A., Mock, H. P., Lippmann, R., Grosch, R., et al. (2014). Root exudation and root development of lettuce (Lactuca sativa L. cv. Tizian) as affected by different soils. Front. Microbiol. 5:2. doi: 10.3389/fmicb.2014.00002
O'Brien, F. J. M., Dumont, M. G., Webb, J. S., and Poppy, G. M. (2018). Rhizosphere bacterial communities differ according to fertilizer regimes and cabbage (Brassica oleracea var. Capitata L.) harvest time, but not aphid herbivory. Front. Microbiol. 9:1620. doi: 10.3389/fmicb.2018.01620
Pérez-Jaramillo, J. E., Mendes, R., and Raaijmakers, J. M. (2015). Impact of plant domestication on rhizosphere microbiome assembly and functions. Plant Mol. Biol. 90, 635–644. doi: 10.1007/s11103-015-0337-7
Pineda, A., Kaplan, I., and Bezemer, T. M. (2017). Steering soil microbiomes to suppress aboveground insect pests. Trends Plant Sci. 22, 770–778. doi: 10.1016/j.tplants.2017.07.002
Pineda, A., Kaplan, I., Hannula, S. E., Ghanem, W., and Bezemer, T. M. (2019). Conditioning the soil microbiome through plant-soil feedbacks suppresses an aboveground insect pest. New Phytol. 226:595–608. doi: 10.1111/nph.16385
Pineda, A., Zheng, S. J., van Loon, J. J. A., and Dicke, M. (2012). Rhizobacteria modify plant-aphid interactions: a case of induced systemic susceptibility. Plant Biol. 14, 83–90. doi: 10.1111/j.1438-8677.2011.00549.x
Pizano, C., Kitajima, K., Graham, J. H., and Mangan, S. A. (2019). Negative plant–soil feedbacks are stronger in agricultural habitats than in forest fragments in the tropical Andes. Ecology 100, 1–12. doi: 10.1002/ecy.2850
Rudrappa, T., Czymmek, K. J., Paré, P. W., and Bais, H. P. (2008). Root-secreted malic acid recruits beneficial soil bacteria. Plant Physiol. 148, 1547–1556. doi: 10.1104/pp.108.127613
Saleem, M., Hu, J., and Jousset, A. (2019). More than the sum of its parts: microbiome biodiversity as a driver of plant growth and soil health. Annu. Rev. Ecol. Evol. Syst. 50, 145–168. doi: 10.1146/annurev-ecolsys-110617-062605
Schaeffer, R. N., Wilson, C. M., Radville, L., Barrett, M., Whitney, E., Roitman, S., et al. (2017). Individual and non-additive effects of exotic sap-feeders on root functional and mycorrhizal traits of a shared conifer host. Funct. Ecol. 31, 2024–2033. doi: 10.1111/1365-2435.12910
Seeger, J., and Filser, J. (2008). Bottom-up down from the top: Honeydew as a carbon source for soil organisms. Eur. J. Soil Biol. 44, 483–490. doi: 10.1016/j.ejsobi.2008.07.008
Senechkin, I. V., Speksnijder, A. G. C. L., Semenov, A. M., van Bruggen, A. H. C., and van Overbeek, L. S. (2010). Isolation and partial characterization of bacterial strains on low organic carbon medium from soils fertilized with different organic amendments. Microb. Ecol. 60, 829–839. doi: 10.1007/s00248-010-9670-1
Shade, A. (2017). Diversity is the question, not the answer. ISME J. 11, 1–6. doi: 10.1038/ismej.2016.118
Team, R. C. (2018). R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing. Vienna. Available online at: https://www.R-project.org/
Toju, H., Peay, K. G., Yamamichi, M., Narisawa, K., Hiruma, K., Naito, K., et al. (2018). Core microbiomes for sustainable agroecosystems. Nat. Plant 4, 247–257. doi: 10.1038/s41477-018-0139-4
Valenzuela-Soto, J. H., Estrada-Hernández, M. G., Ibarra-Laclette, E., and Délano-Frier, J. P. (2010). Inoculation of tomato plants (Solanum lycopersicum) with growth-promoting Bacillus subtilis retards whitefly Bemisia tabaci development. Planta 231, 397–410. doi: 10.1007/s00425-009-1061-9
Walgenbach, J. F. (1997). Effect of Potato Aphid (Homoptera: Aphididae) on yield, quality, and economics of staked-tomato production. J. Econ. Entomol. 90, 996–1004. doi: 10.1093/jee/90.4.996
Wei, Z., Gu, Y., Friman, V. P., Kowalchuk, G. A., Xu, Y., Shen, Q., et al. (2019). Initial soil microbiome composition and functioning predetermine future plant health. Sci. Adv. 5:eaaw0759. doi: 10.1126/sciadv.aaw0759
Weidenhamer, J. D., Mohney, B. K., Shihada, N., and Rupasinghe, M. (2014). Spatial and temporal dynamics of root exudation: how important is heterogeneity in allelopathic interactions? J. Chem. Ecol. 40, 940–952. doi: 10.1007/s10886-014-0483-4
Wickham, H. (2017). ggplot2: Elegant Graphics for Data Analysis. New York, NY: Springer- Verlag. doi: 10.1007/978-0-387-98141-3
Wilkinson, T. D. J., Miranda, J. P., Ferrari, J., Hartley, S. E., and Hodge, A. (2019). Aphids influence soil fungal communities in conventional agricultural systems. Front. Plant Sci. 10, 1–13. doi: 10.3389/fpls.2019.00895
Yang, J. W., Yi, H. S., Kim, H., Lee, B., Lee, S., Ghim, S. Y., et al. (2011). Whitefly infestation of pepper plants elicits defence responses against bacterial pathogens in leaves and roots and changes the below-ground microflora. J. Ecol. 99, 46–56. doi: 10.1111/j.1365-2745.2010.01756.x
Yi, H. S., Yang, J. W., Ghim, S. Y., and Ryu, C. M. (2011). A cry for help from leaf to root: aboveground insect feeding leads to the recruitment of rhizosphere microbes for plant self-protection against subsequent diverse attacks. Plant Signal. Behav. 6, 1192–1194. doi: 10.4161/psb.6.8.15780
Yuan, J., Zhao, J., Wen, T., Zhao, M., Li, R., Goossens, P., et al. (2018). Root exudates drive the soil-borne legacy of aboveground pathogen infection. Microbiome 6, 1–12. doi: 10.1186/s40168-018-0537-x
Zhang, F. S., Ma, J., and Cao, Y. P. (1997). Phosphorus deficiency enhances root exudation of low-molecular weight organic acids and utilization of sparingly soluble inorganic phosphates by radish (Raghanus sativus L.) and rape (Brassica napus L.) plants. Plant Soil 196, 261–264. doi: 10.1023/A:1004214410785
Keywords: aphid, tomato, rhizosphere, microbiome, plant-soil (above-ground-below-ground) interactions
Citation: French E, Kaplan I and Enders L (2021) Foliar Aphid Herbivory Alters the Tomato Rhizosphere Microbiome, but Initial Soil Community Determines the Legacy Effects. Front. Sustain. Food Syst. 5:629684. doi: 10.3389/fsufs.2021.629684
Received: 15 November 2020; Accepted: 10 March 2021;
Published: 08 April 2021.
Edited by:
Mariana Benítez, National Autonomous University of Mexico, MexicoReviewed by:
Ek Del-Val, National Autonomous University of Mexico, MexicoNeelendra K. Joshi, University of Arkansas, United States
Copyright © 2021 French, Kaplan and Enders. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Laramy Enders, bGVuZGVyc0BwdXJkdWUuZWR1
 Elizabeth French
Elizabeth French Ian Kaplan
Ian Kaplan Laramy Enders*
Laramy Enders*