
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 12 March 2021
Sec. Crop Biology and Sustainability
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.621689
This article is part of the Research Topic Plant Growth-Promoting Microorganisms for Sustainable Agricultural Production View all 50 articles
 Sire Diedhiou-Sall1
Sire Diedhiou-Sall1 Komi B. Assigbetsee2
Komi B. Assigbetsee2 Aminata N. Badiane3
Aminata N. Badiane3 Ibrahima Diedhiou4
Ibrahima Diedhiou4 M. Khouma5†
M. Khouma5† Richard P. Dick6*
Richard P. Dick6*The Sahel is an ecologically vulnerable region where increasing populations with a concurrent increase in agricultural intensity has degraded soils. Agroforestry offers an approach to remediate these landscapes. A largely unrecognized agroforestry resource in the Sahel are the native shrubs, Piliostigma reticulatum, and Guiera senegalensis that to varying degrees already coexist with row crops. These shrubs improve soil quality, redistribute water from the deep soil to the surface (hydraulic lift), and can improve crop growth. However, little information is available on whether these shrubs affect spatial and temporal dynamics of microbial communities. Therefore, the objective of this study was to determine microbial composition and activity in the wet and dry seasons of soil in the: shrub rhizosphere (RhizS), inter-root zone (IntrS), and outside the influence of shrub soil (OutS) for both G. senegalensis and P. reticulatum in Senegal. A 3 × 2 factorial field experiment was imposed at two locations (490 and 700 mm annual rainfall with G. senegalensis and P. reticulatum, respectively), that had the soil sampling treatments of three locations (RhizS, IntrS, and OutS) and two seasons (wet and dry). Soils were analyzed for: microbial diversity (DGGE with bacterial 16S or fungal 28S rRNA gene sequences phospholipids fatty acid, PLFA); enzyme activities; microbial biomass carbon (MBC); and nitrogen (N) mineralization potential. For the DGGE profiling, the bacterial community responded more to the rhizosphere effect, whereas, the fungal community was more sensitive to season. PLFA, MBC, enzyme activities and inorganic N were significantly higher in both seasons for the RhizS. The presence of shrubs maintained rhizosphere microbial communities and activity during the dry season. This represents a paradigm shift for semi-arid environments where logically it would be expected to have no microbial activity in the extended dry season. In contrast this study has shown this is not the case that rather the presence of shrub roots maintained the microbial community in the dry season most likely due to hydraulic lift and root exudates. This has implications when these shrubs are in cropped fields in that decomposition and mineralization of nutrients can proceed in the dry season. Thus, enabling accumulation of plant available nutrients during the dry season for uptake by crops in the rainy season.
The Sahel is an ecologically vulnerable region where increasing populations with a concurrent increase in agricultural intensity has degraded soils. Agroforestry offers an approach to remediate these landscapes. A largely unrecognized agroforestry resource in the Sahel are the native shrubs, Piliostigma reticulatum, and Guiera senegalensis that to varying degrees already coexist with row crops. These two species dominate over any other shrub species and are coppiced in the spring prior to the cropping/rainy season. Subsequently, the shrubs regrow during the long dry season.
Previous recommendations were to remove shrubs from cropped fields because of perceived competition of shrubs with crops for nutrients and water, and reduced crop yield (Somarriba, 1988; Kater et al., 1992). However, a preliminary study in Niger in farmers' fields showed millet growing within the influence of G. senegalensis rhizospheres had greater growth than when grown outside the influence of the shrub (Wezel et al., 2000). Long-term statistically valid field studies have shown that G. senegalensis (Dossa et al., 2012) and P. reticulatum (Dossa et al., 2013; Bright et al., 2017) in Senegal can increase yields for groundnut (Arachis hypogaea L.) and millet [Pennisetum glaucum (L.) R. Br.] with or with inorganic fertilizer.
These pedo-ecological benefits by woody species have been shown in natural, semi-arid desert environments. This phenomenon has been characterized as soil fertility and hydrologic resource islands that develop beneath woody species and influence the biogeochemistry and decomposition of uncultivated desert environments (Schlesinger et al., 1996; Schlesinger and Pilmanis, 1998; Rango et al., 2006; Schade and Hobbie, 2006). Brooker et al. (2016) proposed that plant-plant interactions hold potential to facilitate sustainable crop production and address food insecurity in developing countries where subsistence farmers use limited external inputs.
Rhizospheres produce organic root exudates and affect soil moisture, which changes the numbers and activities of microbial communities living under the influence of plant roots (Lynch and Whipps, 1990; Diedhiou et al., 2009). A large proportion of the C released by roots is in the form of water-soluble substances such as sugars, organic acids and amino acids. The differences in type and quantity of C available in different root zones thereby select for distinct rhizospheric community structures (Young, 1998; Yang and Crowley, 2000). In addition, these communities may vary with respect to plant species (Westover et al., 1997).
An important finding for G. senegalensis and P. reticulatum is that their roots redistribute water from the subsoil to the surface, a phenomenon known as hydraulic redistribution (Kizito et al., 2007, 2012). Hydraulic redistribution is the movement of water from regions of higher soil water potential (subsoil) to regions of lower soil water potential (surface soil) via plant roots, and is typically characteristic of semi-arid to arid environments (Richards and Caldwell, 1987) as well as mesic environment during drought (Dawson, 1993; Caldwell et al., 1998). The amount of water redistributed can go as high as 0.1 mm day−1 for P. reticulatum and 0.2 mm day−1 for G. senegalensis (Kizito et al., 2007). Hydraulic redistribution could be important to maintain microbial communities and drive biogeochemical processes in shrub rhizosphere soil during the long dry season of semi-arid or arid environments (Saul-Tcherkas and Steinberger, 2011). Previous research has shown that soil beneath canopies of P. reticulatum or G. senegalensis increased decomposition, microbial biomass C, and enzyme activities over soil outside the influence of these shrubs. Also, Debenport et al. (2015) found that root zone soil of pearl millet [Pennisetum glaucum (L.) R. Br.] grown within the influence of P. reticulatum or G. senegalensis had increased diversity and abundance with some genera in both bacterial and fungal communities over soil outside the influence of shrubs. However, little information is available on whether shifts in microbial communities remain during the dry season due to hydraulic lift.
Therefore, the hypothesis was that the presence of shrubs will shift soil microbial communities with the objective to determine microbial composition and activity in the wet and dry seasons of shrub (RhizS), inter-root soil (IntrS), and outside the influence of shrub soil (OutS).
The study was conducted in two agro-ecological zones of Senegal within the major cropping region known as the Peanut Basin. The first site, Keur Matar Arame (KMA) is located at Thies (N14° 46 W16° 51) where G. senegalensis is the main shrub species found in farmers' fields. The soil is a sandy, ferruginous Oxisol (FAO, 1998). The KMA site has low rainfall with ~300 mm per annum with a water table depth of ~ 10 m. The second site is located at Nioro (N13° 45 W15° 47), predominantly occupied by P. reticulatum shrubs on a sandy, lateritic area classified as an Oxisol (FAO, 1998). The area has unimodal rainfall, 700 mm per annum and a mean annual temperature of 32°C. The water table is at ~8 m. The sites were both in farmers' fields that had been under long-term cropping of millet [Pennisetum glaucum (L.) R. Br.] and groundnuts (Arachis hypogaea L.) that included coppicing of aboveground biomass each spring. The coppiced residues were normally burned or occasionally some portion was removed and taken back for household use. Both sites in the rainy season were under millet cultivation which were planted in a 1 × 1 m pattern.
Soil samples from rhizosphere soil (RhizS), inter-root soil (IntrS), and outside the influence of shrub soil (OutS) were collected in relation for both shrub species (0–15 cm depth) during the dry season (March 2005), and the wet season (August 2005). At each experimental site, soils were sampled randomly under six plants at three locations: (1) shrub rhizosphere soil RhizS (2) soil beneath shrub canopy but between roots or inter-root soil (IntrS); and (3) samples collected two meters away from the shrubs which was outside the influence of shrub (OutS). The OutS were taken between millet plants at a distance of 0.5 m from millet plants. The rhizosphere soils was obtained by peripheral excavation of surface soil followed by careful excising the roots from the larger crown or tap roots. The excised roots were gently shaken and then the soil retained on the roots was collected for analysis which is defined as rhizosphere soil. The soil samples of three pairs of shrubs for each species were combined (resulted in 3 field replicates for each shrub species), homogenized and then crushed to pass through a 2 mm mesh screen. The soil samples were maintained at field moisture and stored at 4°C until analysis. Soil extractions for nucleic acid and fatty acid analyses outlined below to characterize soil microbial communities were done within 2 days and other analyses were done within a week after sampling. Soils from old and new roots were pooled for all analyses except the for the DGGE analysis.
Microbial biomass was determined by chloroform–fumigation extraction (CFE) following the method of Amato and Ladd (1988) with some modifications. Briefly, 10 g of the moist soil was fumigated with chloroform (ethanol free) and then incubated for 10 days. Fumigated samples and unfumigated control samples were extracted with 2 M KCl solution for 60 min on a rotator shaker. After filtration, 2 ml of the filtrates were mixed with 0.5 ml of 0.4 M sodium citrate solution. Ninhydrin-reactive N was determined colorimetrically at 750 nm (Schinner et al., 1996) (Evolution II, Alliance-Instrument, France). Microbial biomass C (MBC) was estimated by multiplying by 21 the gain in ninhydrin-reactive N after fumigation (Amato and Ladd, 1988). Results were expressed as μg C g−1 of dry soil.
Soil inorganic-N species were quantified in the same extract, colorimetrically in KCl extracts (2 M KCl) using the method of Bremner (1965). Ammonium was quantified using the reaction of Berthelot modified with the indophenol blue at 660 nm. For the nitrate, nitrite is mixed with the sulfanilamide to form a diazo complex and the absorbance was read at 525 nm. Inorganic nitrogen was assessed as the sum of ammonium and nitrate, and the results were expressed as μg N g−1 of dry soil.
All soil enzyme analyses were performed on field moist soil. The β-glucosidase and chitinase activities were measured using a modified method originally described by Hayano (1973) and Ndour et al. (2001). Fresh soil samples (100 mg) were incubated for 2 h at 37 °C, with 100 μl of 5 mM para-nitrophenyl β-d-glucopyranoside (pNP) for β-glucosidase and 5 mM para-nitrophenyl N-acetyl glucosaminide for chitinase as the substrates. The pNP released was measured 15 min after stopping the reaction at 400 nm for both enzymes. The results were expressed as μg pNP released g−1 h−1. Phosphatase activity was determined following the method described by Tabatabai and Bremner (1969). The method by Kandeler and Gerber (1988) was used to determine the urease activity. Two analytical replicates and one control were analyzed for each soil sample.
Phospholipids fatty acids (PLFA) were extracted from the soil by Bligh and Dyer (1959) method as modified by Schutter and Dick (2000). Briefly, lipids were extracted from 3 g of soil samples using a chloroform-methanol-phosphate buffer solvent. PLFA were separated from neutral lipids and glycolipids using silicic acid columns (Supelco, Bellefonte, PA, USA). The phospholipid fractions were then converted to methyl-esters by alkaline methanolysis. PLFAs from all samples were extracted in triplicates. PLFAs were analyzed by gas chromatography (GC) (Agilent Ultra 2 column; temperature ramping from 120 to 260°C at a rate of 5°C per min) using helium as the carrier gas, and peaks were detected by flame ionization detector (Frostegård and Bååth, 1996).
Individual fatty acid methyl esters (FAME) were identified and quantified using the MIDI Sherlock Microbial Identification System (MIDI, Newark, Delaware, USA) and with the mixture of 37 FAME (FAME 37 47885-4; Supelco, Inc), 24 bacterial FAME mixture (P-BAME 24 47080-U; Supelco, Inc.). Each individual fatty acid was expressed as a percentage of the total amount of fatty acids (mol%) found in a given sample. PLFA data with <0.5% of the total relative abundance were not included in the data set. PLFA biomass was estimated by adding the amount of all fatty acids detected and was expressed in nano moles of PLFA per g of dry weight of soil (nmol g−1 dw) (White et al., 1979; Frostegård et al., 1991; Bossio et al., 1998). For each microbial group and total PLFA (PLFAtot), the total amount of PLFA identified was calculated to represent their contribution to the total microbial biomass.
A total of 23 PLFA markers out of 27 identified (91%) were used for the multivariate analyses for G. senegalensis, while a total of 23 PLFA out of 29 identified (94%) were used for multivariate analysis for P. reticulatum. The combined masses of FAMEs reported as typical of fungi (18:2ω6c, 18:1ω9c) (Frostegård and Bååth, 1996; Olsson, 1999), Gram-negative (GN) bacteria (18:1ω7c; 17:0cy; 19:0cy) (Wilkinson, 1988), Gram-positive (GP) bacteria (15:0i; 15:0a; 16:0i; 17:0i; 17:0a) (O'Leary and Wilkinson, 1988) and actinomycetes (ACT) (16:0 10-Me; 17:0 10-Me; 18:0 10-Me) (Kroppenstedt, 1992) were used as signatures for these microbial groups. The summation of individual fungal or bacterial fatty acids were used to calculate fungal to bacterial ratios (FUN/BACT) as an indicator of the general change in the soil microbial community structure (Bardgett et al., 1998; Olsson, 1999; Zelles, 1999; Fierer et al., 2002). The ratio of 19:0cy to18:1ω7c was calculated and is primarily an indicator to water stress (Guckert et al., 1986; Lundquist et al., 1999). The ratio of saturated to monounsaturated fatty acids (SAT/MONO) was calculated and is an indicator nutrient deprivation (Bossio and Scow, 1998; Larkin, 2003).
Changes in soil microbial community structure were analyzed by DGGE profile of 16S rRNA and 28S rRNA gene sequences for bacteria and fungi, respectively. The method by Porteous et al. (1997) was used to extract soil DNA from 0.5 g of soil samples in triplicates.
Bacterial 16S rRNA was amplified using universal bacterial DGGE primers 338f-GC clamp and 518r primers (Muyzer et al., 1993). Amplification was performed by 5 min of denaturation at 94°C, 30 cycles of 45 s each at 94°C, 45 s at 52°C and 1 min at 72°C, followed by a final extension at 72°C for 10 min using a PTC-100 thermal cycler. The primers 403f-GC (U1) and 662r (U2) were used to amplify the 28S rRNA of the fungal population (Sandhu et al., 1995). The following cycle was used to amplify fungal 28S rRNA: 5 min of denaturation at 94°C, 35 cycles of 30 s at 94°C, 1 min at 52°C and 2 min at 72°C, followed by a final extension at 72°C for 10 min using the same thermal cycler. The PCR tag “Ready- to-go” (Amersham Biosciences, USA) was used to amplify both the 16S and the 28S rRNA. PCR products were confirmed on 1.0% agarose gels after staining with ethidium bromide solution (0.25 μg ml−1). PCR products were then quantified using a spectrophotometer to measure the optical density.
Equivalent quantities of PCR products were resolved in an 8% polyacrylamide gel (37.5:1 acrylamide:bisacrylamide) in 0.5 × TAE buffer (20 mM Tris-HCl, 10 mM acetate, 0.5 mM EDTA) and denaturants (100% denaturant contains 7 M urea and 40% deionized formamide). A gradient of denaturants ranged from 40 to 70% for bacterial communities and 35–65% for fungal communities. Electrophoresis was performed on an Ingeny apparatus (Ingeny phorU, Netherlands) at a constant voltage of 75 for 16 h. The software Bio-profil Biogene program (Vilber Lourmat) was used to analyze the DGGE profile. The detected bands were used to construct a matrix indicating presence or absence of bands in each sample.
Spatial and temporal shifts in the composition of PLFA profiles were analyzed by principal components analysis (PCA) using the PC-ORD package (MjM Software Design, Gleneden Beach, OR) (McCune and Grace, 2002) after converting the PLFA to mol% of peak totals. Scores of samples on the first two axes were then analyzed by ANOVA. For the DGGE analysis, matrices of presence and absence of bands was then subject to ordination analyses to identify groups of similar samples, or structure in the dataset (Fromin et al., 2002). A second matrix containing summed values for PLFA or functional measurements were used to construct joint plots that overlaid on the PCA plots. The joint plots were used to visualize the relationship between a set of variables (in this case, taxonomic groups, enzyme activities, and stress indicators) and PCA scores. The angle and length of a line indicate the direction and strength of the relationship (McCune and Grace, 2002). The enzyme activities of the samples were analyzed using ANOVA analysis on SAS software (SAS Institute, 1996) to assess spatial and temporal effects. In addition, correlations between PLFA and enzyme activities were investigated using the Mantel test (McCune and Grace, 2002).
Diversity indices were calculated as below: the Shannon-Wiener index, H′ = -∑(ni/N) ln(ni/N) (Shannon, 1948); the Simpson dominance index, λ = ∑(ni/N)2 (Simpson, 1949); and the evenness index, e = H′/lnS (Pielou, 1966); where S is the total number of PLFA peaks, ni is the area of ith PLFA peak, and N is the sum of the area of all PLFA peaks for each sample.
Inorganic N consistently for both shrub species and seasons, the OutS had the lowest levels but it was not significant at P < 0.05.
PLFAtot and MBC are indexes of the total microbial biomass and did not show the same treatment effects, except that they both had the RhizoS consistently being highest within a season and shrub species (Table 1). MBC consistently showed a stepwise decrease of: RhizoS>IntS>OutS within a season and shrub species. It also showed consistently higher levels in the wet season over the dry season at all sampling locations and for both species (Table 1).
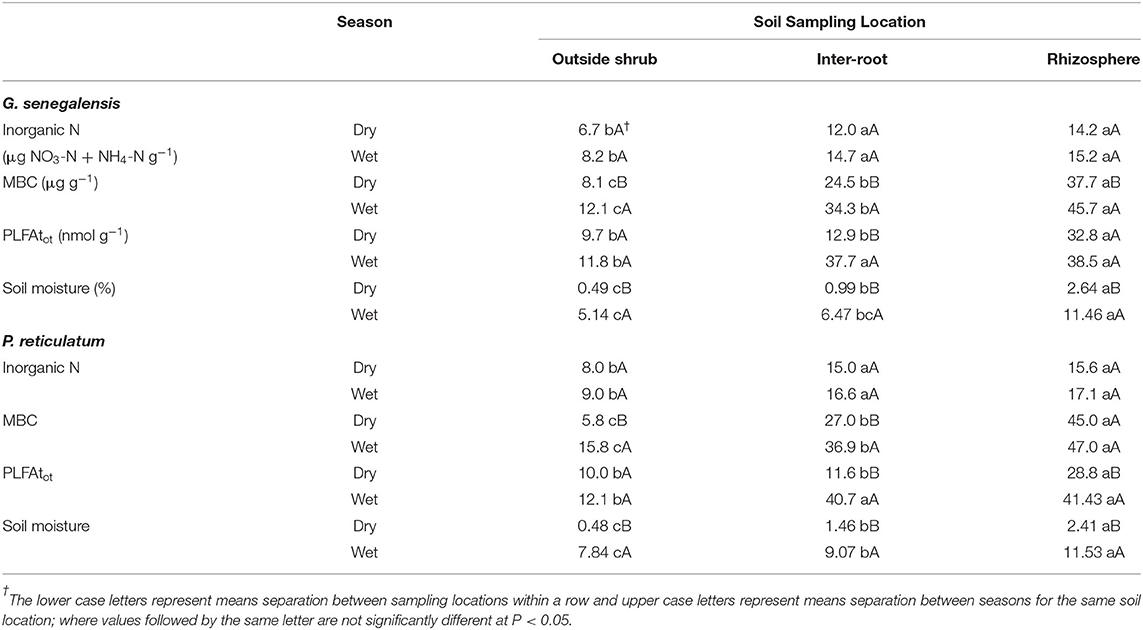
Table 1. Soil inorganic N, MBC, PLFAtot and moisture of soil inside and outside the influence of G. senegalensis or P. reticulatum during the wet and dry season.
Overall, both shrub species showed differences in PLFAtot between RhizS and OutS (Table 1) in both seasons with InterR being similar to RhizS in the wet season but intermediate in the dry season. During the dry season, PLFAtot was significantly higher (P < 0.05) for RhizS than IntrS and OutS of both shrub (Table 1).
Acid phosphatase, β-glucosidase and chitinase activities were higher for the rhizosphere soil than the bulk soil (data not shown); the IntrS soil had significantly higher activities than did the non-rhizosphere soil. This is true for both seasons for those enzymes except the chitinase activity which during the dry season was the same whether it was a bulk soil or a non-rhizosphere soil. For urease, activity was significantly higher in rhizosphere soil than in bulk and non-rhizosphere soil only during the dry season. During the wet season, this activity was the same in the bulk and rhizosphere soil but significantly different from the non-rhizosphere soil.
Soil moisture was greatly reduce in the dry season. However, the RhizS soil had significantly higher levels than the other two sampling locations in the dry season for both shrub species.
Consistently, across all the PLFA functional groups in the dry season, RhizS was significantly higher than IntrS and OutS (Figure 1). On average for the dry season, fungal PLFA was 80% greater in the RhizS compared to IntrS for both shrub species (Figure 1). Conversely, in the wet season the fungal PLFAs in RhizS were only about 20% higher than the IntrS and often statistically showing no differences. Similar levels of increase were observed in the RhizS soil for the Gram+ bacterial PLFAs (65%) when compared to IntrS during the dry season. The wet season resulted in a significant increase of all microbial PLFAs in both shrubs, which was more prominent in the case of IntrS compared to those in dry season.
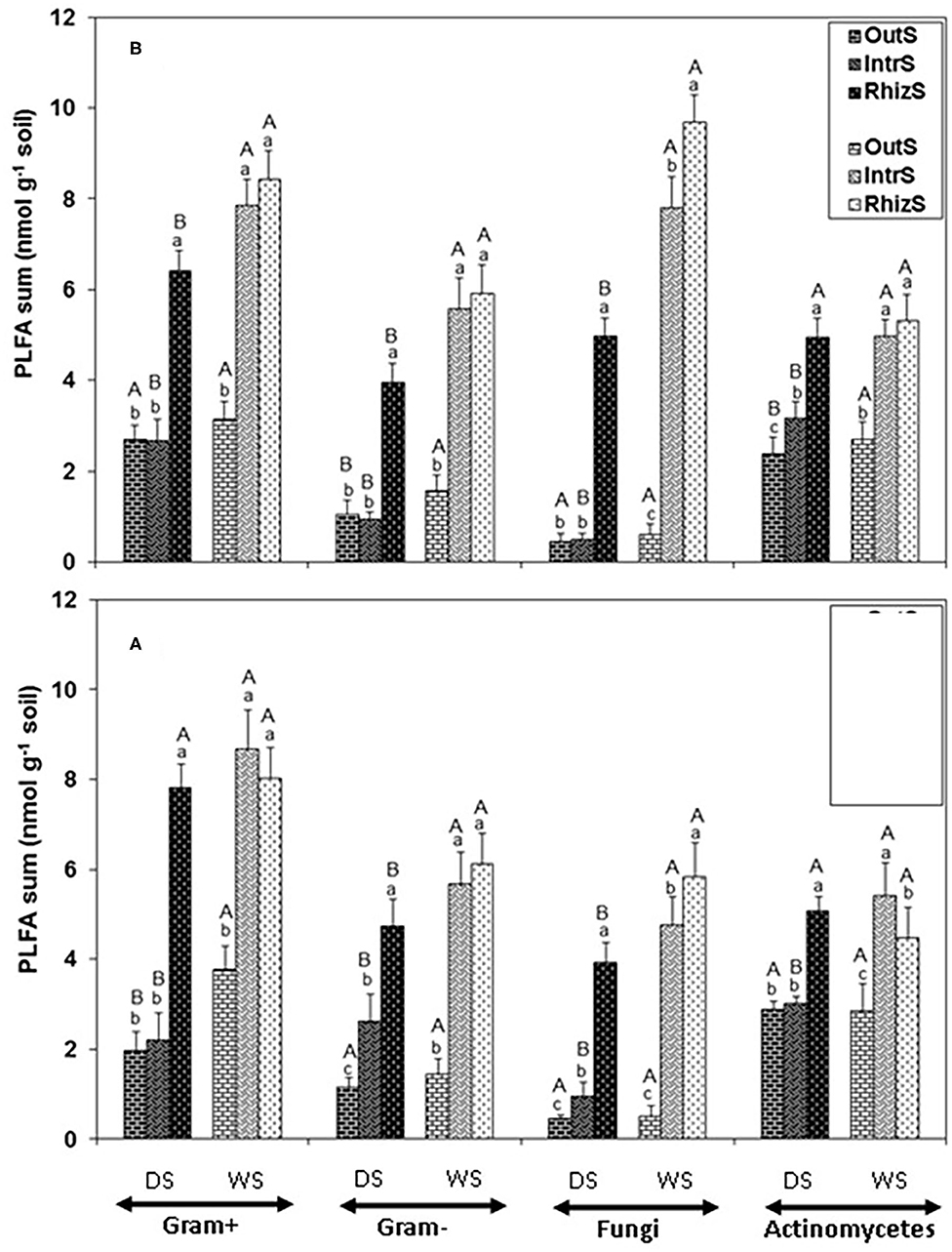
Figure 1. Summation of PLFAs representing Gram+, Gram-, fungi and actinomycetes microorganisms extracted from soil associated with (A) G. senegalensis, and (B) P. reticulatum. Soil samples (RhizS, shrub rhizosphere soil; IntrS, inter-root soil; and OutS, outside the influence of shrub soil) are from the dry season (DS) and wet season (WS). Bars are standard deviations. The lower case letters represent means separation between sampling locations and upper case letters represent mean separation between seasons for the same soil location.
Comparison of the major functional groups measured by PLFA (Gram+, Gram-, fungi, and actinomycetes) showed no major differences among these relative to soil sampling location. That is, for Gram+, Gram-, fungi, and actinomycetes, patterns of each of the sampling locations within a season were quite similar for both shrub species—which was that RhizS remained high between seasons, OutS was always low, and IntrS tended to be not significantly different from the RhizS in the rainy season, with IntrS dropping significantly in the dry season. The most striking effect was on the PLFAs of fungi at the G. senegalensis site, which for OutS and IntrS became extremely low in the dry season but then for IntrS showed a dramatic increase in the wet season (1,495% increase). The other general outcome was that averaging across all sampling sites and seasons for both shrub species showed that, on a nmol basis, Gram+ was higher than Gram- bacteria.
Principal component analysis of soil microbial community had a total variance of 65% explained by the first two axes, with the first axis explaining 47% of the total variance in PLFA community composition (Figure 2). Plant species did not show marked differences for the microbial community structures. The strongest factor in structuring the communities was the location of the soil sample; OutS vs. RhizS (p < 0.001). PCA showed that the communities from the IntrS during the wet season were similar to those of the RhizS. Samples from the OutS clustered, and within this cluster they were separated primarily based on the season and then by the shrub species.
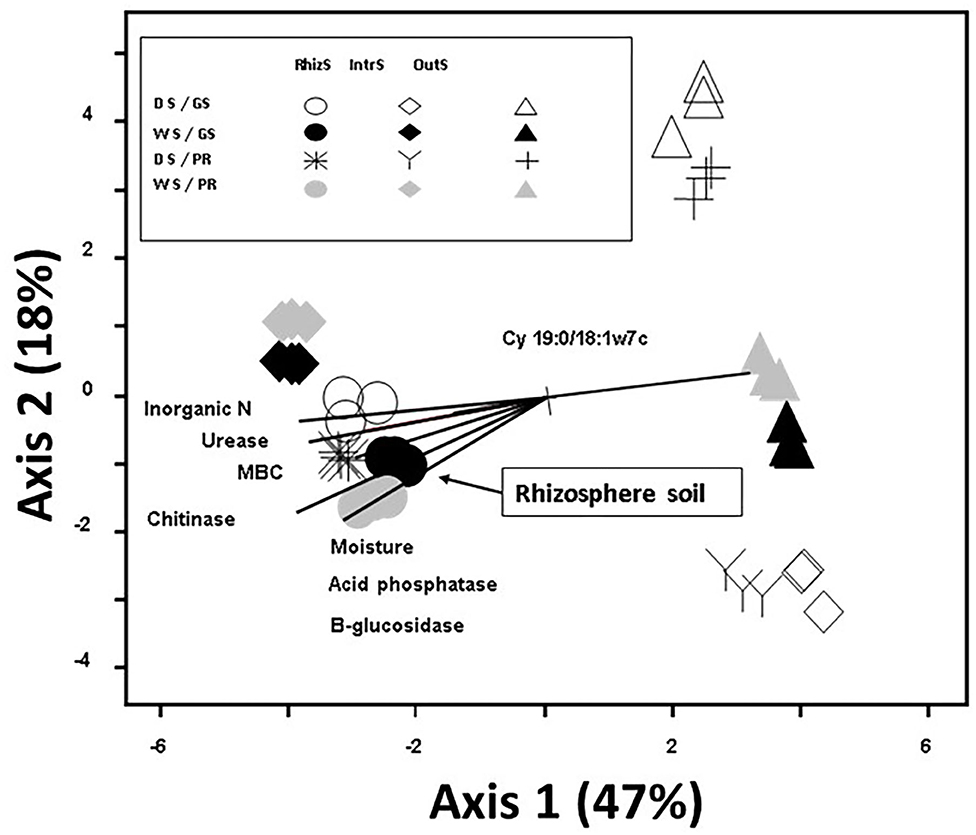
Figure 2. Principle Component Analysis based on 23 phospholipid microbial markers from soil associated with G. senegalensis (GS) and P. reticulatum (PR) which are shrubs that co-exist with row crops in the Sahel. The vectors are joint plots of enzyme activities. Soil samples (RhizS, shrub rhizosphere soil; IntrS, inter-root soil; and OutS, outside the influence of shrub soil) are from the dry season (DS) and wet season (WS).
The FUN/BACT PLFA ratio was also affected by the presence of shrubs, which was highest in RhizS, followed by IntrS and OutS for both shrubs during both wet and dry seasons (Figure 3), except for P. reticulatum in dry season, where it was similar between IntrS and OutS. The FUN/BACT ratio was significantly higher in all soil locations of both shrubs during the wet season, except for OutS of G. senegalensis, where the ratio was higher in the dry season.
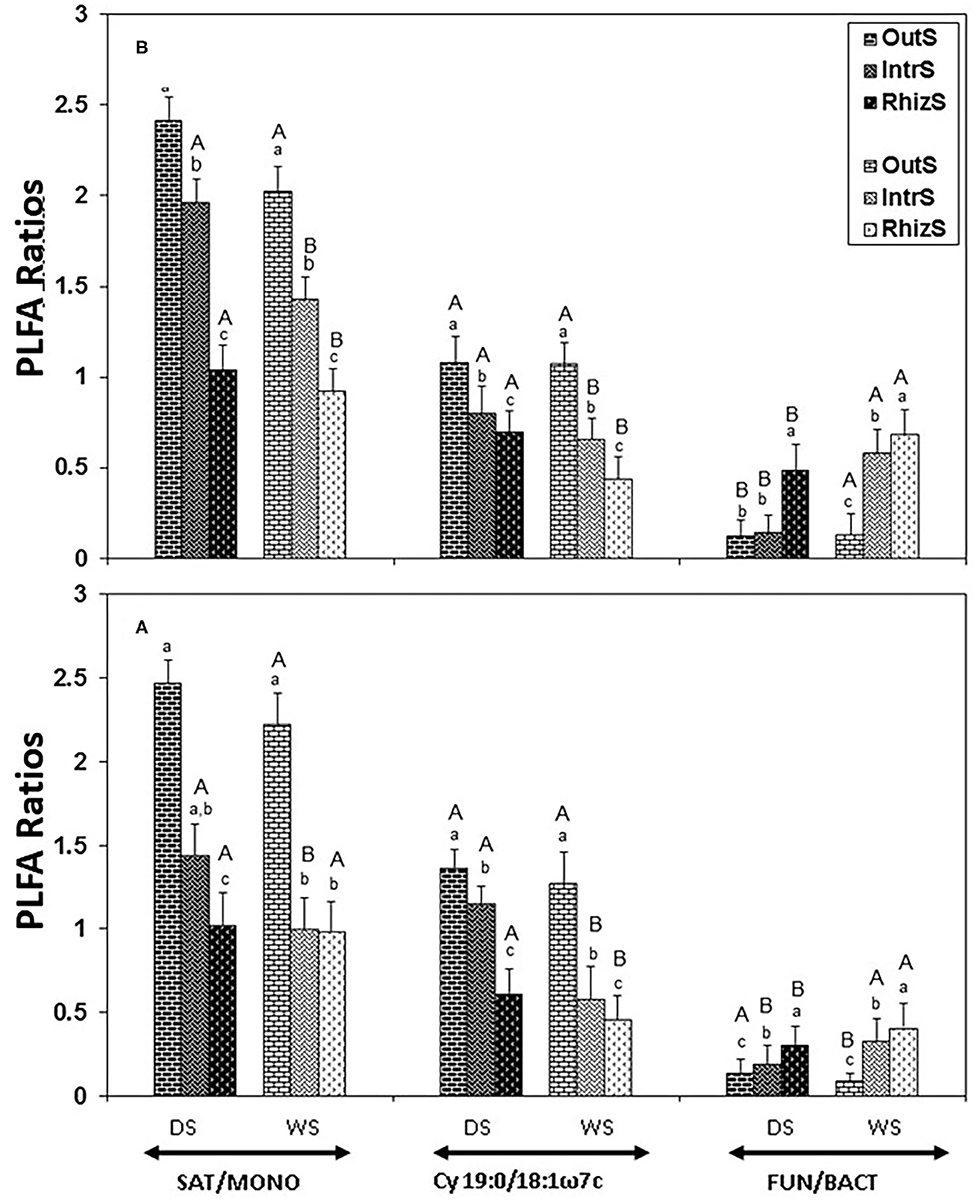
Figure 3. Stress indicators (19:0cy/18:1ω7c or SAT/MONO ratios) and FUN/BACT PLFA ratio extracted from soil associated with (A) G. senegalensis, and (B) P. reticulatum. Soil samples (RhizS, shrub rhizosphere soil; IntrS, inter-root soil; and OutS, outside the influence of shrub soil) are from the dry season (DS) and wet season (WS). Bars are standard deviations. The lower case letters represent mean separation between sampling locations and upper case letters represent mean separation between seasons for the same soil location.
Analysis of DGGE banding data from 16S DNA bacteria profile (Figure 4A) using PCA (Figure 4B) explained a total of 33% for the first two axes. Samples from the RhizS clustered and separated from the IntS and OutS (ANOVA p < 0.01), whereas there was minimal separation due to season for the DGGE bacterial profile (Figure 4B). The comparison of the DGGE bacterial banding between new roots and old roots for both species (Figure 4A) showed that the number of bands were higher and more intense in soil associated with new roots than soil associated with old roots during the wet season. During the dry season, intensity of bands was similar, and there was a small but non-significant difference for the number of bands between those two communities.
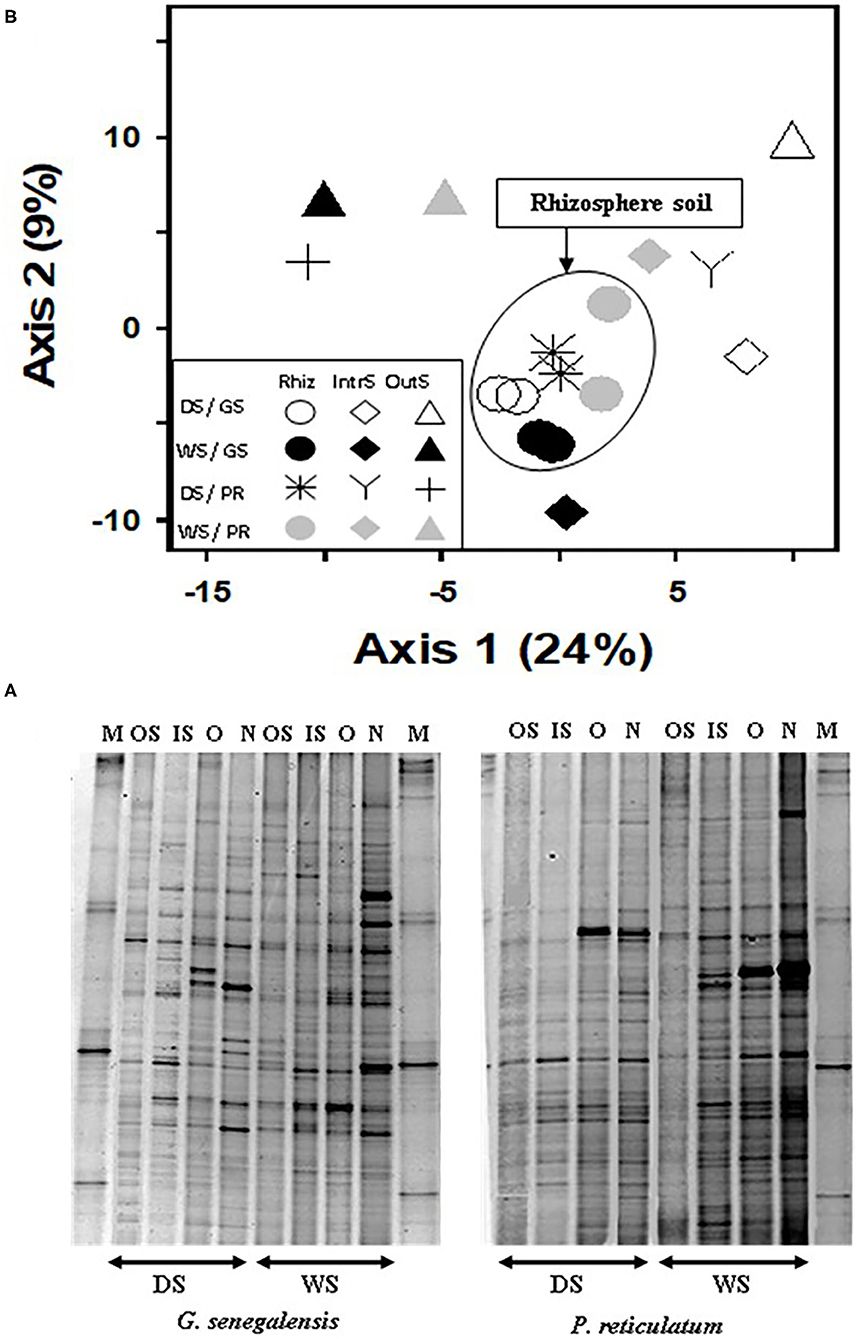
Figure 4. (A) Bacterial DGGE profile of 16S rDNA PCR product extracted from soil, and (B) PCA ordination analysis of DGGE banding profiles. Samples are from the wet (WS) and dry season (DS) in two sites with G. senegalensis (GS) and P. reticulatum (PR) shrubs. M, marker; OS, outside the influence of shrub soil; IS, inter-root soil; O, old roots; and N, new roots. In DGGE gel image M, marker; OS, outside the influence of shrub soil; IS, inter-root soil; O, old roots; and N, new roots. In PCA plot RhizS, shrub rhizosphere soil; IntrS, inter-root soil; and OutS, outside the influence of shrub soil.
For the fungal community, the first two axes of PCA of bands explained 33% of the data (Figure 5). Samples clustered for the dry season but not based on sampling location. This was the opposite of bacterial samples where there was a weak seasonal effect but strong soil sampling location effect. During the dry season, fungal community in the RhizS was similar to IntS. The opposite was observed during the wet season where there was a separation between RhizS and IntS.
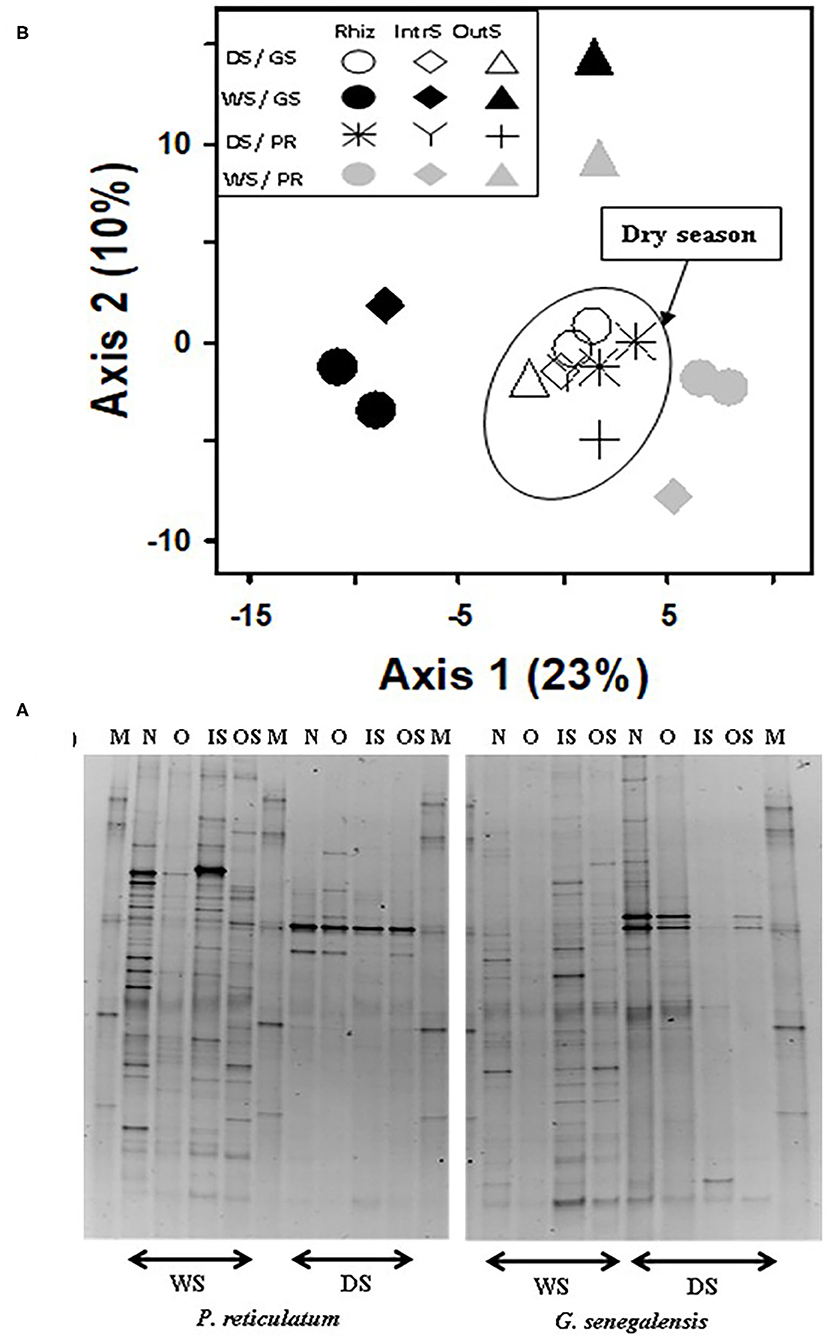
Figure 5. (A) Fungal DGGE profile of 28S rDNA PCR product extracted from soil and (B) PCA ordination analysis of DGGE banding profiles. Samples are from the wet (WS) and dry season (DS) in two sites with G. senegalensis (GS) and P. reticulatum (PR) shrubs. In DGGE gel image M, marker; OS, outside the influence of shrub soil; IS, inter-root soil; O, old roots; and N, new roots. In PCA plot RhizS, shrub rhizosphere soil; IntrS, inter-root soil; and OutS, outside the influence of shrub soil.
The stress indicators, Cy 19:0/18:1ω7c and SAT/MONO, were lowest in RhizS followed by IntrS and OutS during both the dry and wet season in the case of both shrubs (Figure 3). The only exception was that SAT/MONO during the wet season for G. senegalensis was similar between RhizS and IntrS.
Both Cy19:0/18:1ω7c and SAT/MONO in OutS did not differ significantly between dry and wet seasons. However, Cy19:0/18:1ω7c and SAT/MONO in RhizS and IntrS were significantly higher in dry season than in the wet season of both shrubs, except for SAT/MONO in RhizS of G. senegalensis, which was similar between dry and wet season.
The Shannon-Wiener diversity index (H') of PLFA was highest in RhizS for both shrubs in both seasons (Table 2). In the dry season H' index of IntrS and OutS were statistically similar, whereas, in wet season H' index of Intr S was statistically similar to Rhiz S in both shrubs. The evenness scores, although lower than H' index, demonstrated the same pattern as H' index for both shrubs in both seasons. The dominance index (D), in contrast, was highest for OutS and lowest for RhizS in dry season of both shrubs. In the wet season, the D index in G. senegalensis was highest for RhizS, with OutS showing no statistically significant differences between RhizS and IntrS. Whereas, in the case of P. reticulatum, IntrS demonstrated the highest dominance index, followed by RhizS and OutS.
The microbial diversity in OutS did not change between seasons based on diversity indices analysis, as H', E and D indices remained statistically similar between dry and wet seasons in both shrubs (Table 2). The Shannon and evenness index in RhizS also stayed the same between the dry and wet seasons, whereas the dominance index in RhizS was higher in the wet season than in the dry season in both shrubs. The IntrS exhibited higher H', E and D indices values in the wet season than in the dry season in both shrubs, except for dominance in G. senegalensis, which was higher in the dry season.
Both PLFAtot and MBC showed that RhizS maintained the highest microbial biomass over soil of the IntS and OutS locations in both wet and dry season (Table 1). Similarly, diversity analysis using PLFAs showed that RhizS had greater microbial diversity than these other sampling locations, again in both seasons (Table 2). Similar to our findings, several studies on crop systems have shown that rhizospheric soil has higher population densities of bacteria and fungi compared to inter-root soil (Curl and Truelove, 1986; Norton and Firestone, 1991; Maloney et al., 1997; Semenov et al., 1999; Steer and Harris, 2000; Marschner et al., 2002; Butler et al., 2003). Furthermore, it is consistent with Debenport et al. (2015), who showed greater diversity in soil beneath canopies of P. reticulatum and G. senegalensis than soil outside the influence of these shrubs using phylogenetic biomarkers. However, this study was only done on samples collected in the rainy season. Our current study shows that this diversity and increased microbial biomass in rhizosphere soil of these two shrub species is maintained in the dry season (~6 months after rains had stopped).
Summing PLFAs into the functional groups (Gr+, Gr-, fungi, and actinomycetes) indicated that across sampling locations, the pattern of response was quite similar for all these functional groups (in dry season RhizS >> IntrS ≥ OutS; in wet season RhizS similar to IntrS > OutS). This reflects the fact that PLFAs represent broad microbial functional groups. Nonetheless, diversity analysis of PLFAs and DGGE analysis for bacteria do suggest the RhizS is maintaining a more diverse community in both seasons and; on the other extreme, OutS has the lowest diversity. Based on PLFA profiling, the G. senegalensis site showed a major shift in fungal biomass between seasons in the IntrS compared to the OutS that was the lowest compared to all other samplings. The soil at the G. senegalensis site is very sandy and has low organic matter (as is typical of where G. senegalensis dominates in the northern/drier region of Senegal) (Dossa et al., 2010).
The most notable effects of the RhizS based on PLFA profiling was for the fungal and Gr+ bacteria which were significantly greater than OutS in both seasons and shrub species. This is in accordance with the study by Priha et al. (2001) who found that fungal markers 18:2ω6,9c and branched fatty acids, which have commonly been found in Gr+ bacteria (O'Leary and Wilkinson, 1988) were dominant in birch rhizosphere (Betula pendula Roth). They attributed this difference to a higher dominance of Bacillus species in birch rhizosphere. Butler et al. (2003) also showed a greater amount of the PLFA marker i15:0, which is generally attributed to Gr+ bacteria, in the rhizospheric than soil outside the shrub canopy. The same authors found that 18:2ω6,9c was the most highly enriched when 13C-labeled glucose or acetate was added to soils. Analysis of rhizosphere DNA indicated that Gram+ bacteria can be more dominant in the rhizosphere (Smalla et al., 2001).
When water is widely available in the rainy season, this RhizS microbial response in the rhizosphere is largely due to elevated organic inputs that are released and available around roots. Roots produce soluble organic compounds such as carbohydrates, proteins, ad amino acids, sloughed off root cells, and mucilage. This complex mixture of organic compounds provides a source of reduced carbon, nitrogen, and other nutrients to support larger and more diverse microbial communities. This is supported by the higher inorganic N level we found beneath the canopy of both shrubs and by Diedhiou-Sall et al. (2013) who, at the same site as our study, found 45 and 33% greater total C, and 10 and 11% greater total N soil beneath than outside the canopies of P. reticulatum and G. senegalensis, respectively.
However, in the dry season it would be expected that microbial biomass and diversity would be diminished, even with the organic inputs in the RhizS, because of a lack of water. This dry season response did happen for the soil outside the influence of both shrub species and the IntrS location. However, microbial biomass and the diversity based on PLFA markers in the RhizS were maintained in the dry season and significantly higher than IntS. In contrast, there was no difference in the rainy season between IntS and RhizS for these measures, but outside the influence of the shrub, these microbial properties were lower at that time. This would suggest that there is significant diffusion of C and nutrient-rich exudates from the shrub roots and/or that long-term root turnover beneath the shrubs has built up labile organic resources in the IntrS. So, in the summer when water is not limiting, it has very similar microbial properties to the RhizS.
Soil moisture varied widely in the IntrS and particularly for the OutS locations because of the extended dry season from November to June when there is virtually no rainfall. This is likely the major reason for the large changes in microbial communities we noted between seasons for these soil locations based on PLFA analysis. In contrast, PLFA profiling of the RhizoS microbial communities was much more similar and stable between seasons. This can be attributed to hydraulic lift by shrubs that release water year around (Kizito et al., 2012; Bogie et al., 2018). Bogie et al. (2018) confirmed, using isotopically labeled water, that hydraulically lifted water by G. senegalensis is transferred to adjacent millet plants.
The soil moisture data in our study supports the role of hydraulic lift by these shrubs in providing water to microorganisms during the dry season. Indeed, this is reinforced with RhizS having significantly higher levels than the other sampling locations at both sites in the dry season (Table 1). Extrapolating from Bogie et al. (2018) on the same soils, the Kuer Matar soil has a wilting point of 1.87%, whereas the soil moisture in the current study in the dry season was 2.64% for the RhizS soil which is estimated to be 47% of field capacity. Although a similar relationship of elevated RhizS at 2.41% soil moisture for the Nioro site it was only slightly higher than wilting point of 2.33% [based on estimate using data from Bogie et al. (2018)]. None-the-less, this was over 5-fold higher than soil outside the influence of P. reticulatum during the dry season.
Another effect is microclimate shading of the shrubs (Young, 1995; Stark and Firestone, 1996; Eviner, 2001; Myers et al., 2001). Indeed, Kizito (2006) showed that shrubs buffer diurnal temperature by a 5°C difference. This could affect the microorganisms directly by the lower temperature and by reducing moisture losses below the canopy that is present in the dry season. However, this effect would only occur in the extended dry season for our study site because, following farming practices, the shrubs were coppiced in late spring in preparation for the cropping season.
Litter inputs provide organic inputs and nutrient sources for the near-surface soil for both the RhizS and IntrS microorganisms (Ben-David et al., 2011). Since this is a cropped field, there is some shallow tillage which presumably could incorporate litter to a depth of 5–10 cm. This would be an additional factor for the lower levels of microbial properties outside the influence of the shrubs, that receives limited or only crop residue.
Diversity analysis followed the conclusions reported above for microbial biomass and PLFA profiling of the microbial community. The Shannon (H') and Evenness (E) indices were higher for RhizS than IntrS and OutS during the dry season, but in the wet season, H' and E indices of RhizS and IntrS were similar—again showing that rhizosphere soil and soil between roots but below the canopy become similar from a diversity perspective when there is adequate water. The higher H' index of RhizS in the dry season is perhaps due to hydraulic lift by shrubs (Kizito et al., 2012), whereas IntrS moisture fluctuates between dry and wet season causing lower diversity in dry season. Over the year, soil beneath the canopy receives litter inputs and has root turnover—thus, when there is adequate water in the wet season, IntrS supports greater diversity of microbial community.
The OutS showed a higher Simpson's dominance than RhizS and IntrS during the wet season due to low soil moisture and nutrient availability outside the canopy in the dry season. In the wet season, an increase in dominance was noticed in both RhizS and IntrS compared to the dry season, whereas the Simpson index in OutS remained unaffected between seasons. Enhancement of relative abundance of several populations in RhizS and IntrS in the wet season could be due to the influence of the shrubs on soil microorganisms under optimal moisture and nutrient conditions.
Enzyme activities provide information on the potential of the soil to carry out discrete reactions, which for out study were ones involved in degradation of organic inputs and nutrient mineralization. Of the sampling locations, RhizS consistently had the highest levels of activity, and joint plots of enzyme activities showed a strong correlation to the rhizosphere community composition. Thus, this would suggest that the greater diversity that increases from the OutS, to the IntrS, to the RhizS results in a soil community that has more potential to decompose litter and root inputs and to release nutrients. This follows Dossa et al. (2009) who found that soil from beneath these same shrub species had greater nitrogen and phosphorus mineralization potential.
Correlation analysis of PLFA biomarker functional groups showed high r-values among these groups and with PLFAtot (Supplementary Table 1). This would suggest that although all groups were different on an abundance basis this difference was similar or consistent across groups as evidenced by the high correlations.
During the dry season, the lack of water probably limited fungal growth and activity, more so than bacteria. This may explain the lack of difference between the OutS and the inter-root soil for chitinase activity, which is largely associated with the presence of fungi (Sinsabaugh et al., 1993; Sinsabaugh and Moorhead, 1994; Bandick and Dick, 1999). Urease activity was closely related to PLFA biomass which may be due to the fact that ammonium (the product of urease activity) is tightly linked to microbial assimilation (Sylvia et al., 2005). Since organic C and total P is higher in the soil beneath these shrubs (Dossa et al., 2009), increases in acid phosphatase and β-glucosidase activities in the rhizosphere would be expected, as the microbial community would stimulate these enzymes in response to elevated levels of litter substrates and root exudates into the RhizS over OutS that would lack these organic inputs. This is supported by inorganic N levels that were highest in the RhizS soil, which results from the activity of hydrolytic enzymes involved in releasing N from organic matter. Overall, there was a good correlation between enzyme activities and PLFAtot (r = 0.70, p < 0.01). Fernandes (2006) also found significant correlation between enzyme activities and PLFA analysis. These results and that of the joint plots indicate that RhizS of these shrubs has greater potential to decompose organic matter and mineralize nutrients than the IntrS and OutS.
The PLFA profiling showed a consistent seasonal and sampling location effect on microbial community structure in both univariate microbial PLFA functional group data and based on diversity/clustering using multivariate PCA analysis. A somewhat similar effect for bacterial DGGE diversity analysis was found but not for fungal DGGE PCA analysis across both shrub species.
These divergent effects between PLFA and DGGE are because they measure different parameters. The PLFA method is a direct and quantitative chemical extraction of a cellular biomarker of living cells and is representative of major microbial groups and the overall community. Conversely, the PCR-DGGE analysis depends on amplifying DNA which, for one, may not have come from living material. Furthermore, amplification can be biased, while detecting only 1–2% of the microbial population representing the dominant species present in an environmental sample (MacNaughton et al., 1999). Thus, the DGGE analysis is likely representing a sub-population of what is represented by PLFA biomarkers. Smalla et al. (2001) reported a seasonal difference in the composition of the bacteria community using DGGE analysis. But in their study, they compared the microbial communities over many years. In the present study, the dry season was contrasted with the wet season in a 6-month period.
For the DGGE fungal community, the seasonal effect was more evident in separating the communities than the rhizospheric effect. Buyer et al. (2002), using PLFA methods, also reported the weak separation of the fungal community primarily based on the rhizosphere influence, but they found a clear rhizospheric effect for the bacterial community.
In the present study, the stress indicators 19:0cy/18:1ω7c and SAT/MONO were higher in the IntrS than the RhizS during both dry and wet seasons, with greater differences noticed during the dry season, where higher levels means greater stress (Willers et al., 2015). The lower stress for the RhizoS location in dry season can be attributed to greater water availability (Table 1) that can only be due to hydraulic lift by the shrubs. In all seasons the RhizoS would be under lower nutrient stress due to the release of root exudates and root tissue turnover (Zelles et al., 1992; Kieft et al., 1994, Zelles et al., 1995; Bossio and Scow, 1998).
The FUN/BACT ratio was highest in RhizS during both seasons, with less difference, but still significantly different between IntrS and RhizS during the wet season. This ratio has been suggested as an indicator of soil microbial community stability (Bardgett and McAlister, 1999; Zelles, 1999). In the present study, water is available in the RhizS that not only directly supports the microbial community but also enables decomposition of litter to proceed in the dry season, providing C and nutrient resources for microorganisms. This could explain fewer differences in FUN/BACT ratio between IntrS and RhizS in the wet season than in the dry season.
The two shrub species were on two different soil types. Therefore, it was not possible to compare shrub species. The degree to which rhizospheric communities are controlled by plant species, rather than soil type, is not clear, as there are studies in which plant species growing in the same soil had similar rhizospheric microbial communities, indicating that the influence of the soil may be greater than that of the plant (Buyer et al., 1999; Latour et al., 1999, Blackwood and Paul, 2003). Overall, results of DGGE analysis should be analyzed carefully, because PCR-DGGE can detect only part of the dominant species present in an environmental sample (MacNaughton et al., 1999). The sensitivity of analysis is chiefly limited by the resolving power of the DGGE gel and the imaging system used (Jongman et al., 1995; Fromin et al., 2002). We found PLFA profiling and enzyme activities were more sensitive than DGGE in reflecting the microbial differences across the various soil sampling locations.
The presence of shrubs resulted in more active and diverse communities compared to soil outside the influence of the shrubs. Moreover, during the dry season, shrubs maintained a wetter soil environment, which can be attributed to hydraulic redistribution of water from the wet subsoil to the drier surface soil and was likely important in stimulating the microbial communities. The RhizS had higher enzyme activities in soil in the dry season that was quite similar to the wet season data. PLFA microbial profiling and enzyme activities were significantly inter-correlated and clearly showed strong rhizospheric and seasonal effects.
The results of the present study have implications for plant-microbial interactions relative to biogeochemical processes and organic matter decomposition. A priori it would be expected that in these semi-arid agro-ecosystems, any biologically-driven soil processes would largely stop during the dry season. The results of this study suggest otherwise: indeed, that diversity and activity of microbial communities are maintained in the shrub rhizosphere soil during the dry season, presumably due to hydraulic distribution. This is important because these shrubs co-exist with crops throughout the Sahel and therefore, during the dry season, shrub rhizospheres should be able to promote organic matter decomposition and nutrient mineralization and allow for release of plant available N, P, and S. The lack of rain during the dry season would allow for the accumulation of inorganic nutrients that are plant available, because there is no rainfall to cause leaching of nutrients beneath the rooting zone. This follows what farmers report: that if a crop can be established and maintained with the first rains, they get their highest yields on millet, because they can presumably take advantage of nutrients that have accumulated over the dry season. In contrast, if they have to replant when the early rains do not sustain crop growth of the first planting, yields are lower because the accumulated nutrients were leached out of the rooting zone during the early rains (personal communication, Ibrahim; Diedhiou, 2020). Indeed, the elevated levels of inorganic N in both seasons at the RhizS site would support the importance of shrubs combined with hydraulic lift in driving biological nutrient mineralization that would result in the accumulation of nutrients over the dry season that would subsequently be available to crops in the early growing season.
Clearly, our results show that shrubs promote microbial communities and their activity, but further studies are required to determine if these shrub rhizospheres also harbor beneficial microorganisms that could assist crops during the growing season and whether shrub rhizospheres influence the crop rhizosphere microbial communities that grow within the influence of these shrubs. If shrubs harbor microorganisms that can fix N, solubilize P, produce plant hormones, and/or suppress diseases, this would be very helpful for subsistence farmers to reduce or eliminate external, purchased inputs for crop production. It offers the opportunity to develop shrub-crop systems that are optimized to take advantage of local biological interactions to promote food productivity and reduce risk in this semi-arid environment.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
All authors listed have made a substantial, direct and intellectual contribution to the work, and approved it for publication.
This research was supported by National Science Foundation under Grant No. 0120732.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The authors are grateful to the Senegalese National Research Institute (ISRA) and the French Research Institute for Development (IRD). Special appreciation goes to the ISRA team in Dakar, Nioro and Bambey, for their committed service through the entire work. The authors are indebted to Joan Sandeno and Nazya Lakhani-Vogelsang for editing this manuscript.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.621689/full#supplementary-material
Amato, M., and Ladd, J. N. (1988). Assay for microbial biomass based on ninhydrin-reactive nitrogen in extracts of fumigated soils. Soil Biol. Biochem. 20, 107–114. doi: 10.1016/0038-0717(88)90134-4
Bandick, A. K., and Dick, R. P. (1999). Field management effects on soil enzyme activities. Soil Biol. Biochem. 31, 1471–1479. doi: 10.1016/S0038-0717(99)00051-6
Bardgett, R. D., and McAlister, M. (1999). The measurement of soil fungal: bacterial biomass ratios as an indicator of ecosystem self-regulation in temperate meadow grasslands. Biol. Fertil. Soils 29, 282–290. doi: 10.1007/s003740050554
Bardgett, R. D., Wardle, D. A., and Yeates, G. W. (1998). Linking above-ground and below-ground interactions: how plant responses to foliar herbivory influence soil organisms. Soil Biol. Biochem. 30, 1867–1878. doi: 10.1016/S0038-0717(98)00069-8
Ben-David, E. A., Zaady, E., Sher, Y., and Nejidat, A. (2011). Assessment of the spatial distribution of soil microbial communities in patchy arid and semi-arid landscapes of the Negev Desert using combined PLFA and DGGE analyses. FEMS Microbiol. Ecol. 76, 492–503. doi: 10.1111/j.1574-6941.2011.01075.x
Blackwood, C. B., and Paul, E. A. (2003). Eubacterial community structure and population size within the soil light fraction, rhizosphere, and heavy fraction of several agricultural systems. Soil Biol. Biochem. 35, 1245–1455. doi: 10.1016/S0038-0717(03)00188-3
Bligh, E. G., and Dyer, W. J. (1959). A rapid method for total lipid extraction and purification. Canad. J. Biochem. Physiol. 37, 911–917. doi: 10.1139/o59-099
Bogie, N., Bayala, R., Diedhiou, I., Dick, R. P., and Ghezzehei, T. A. (2018). Alteration of soil physical properties and processes after ten years of intercropping with native shrubs in the Sahel. Soil Tillage Res. 182, 153–163. doi: 10.1016/j.still.2018.05.010
Bossio, D. A., and Scow, K. M. (1998). Impacts of carbon and flooding on soil microbial communities: phospholipid fatty acid profiles and substrate utilization patterns. Microb. Ecol. 35, 265–278. doi: 10.1007/s002489900082
Bossio, D. A., Scow, K. M., Gunapala, N., and Graham, K. J. (1998). Determinants of soil microbial communities: effects of agricultural management, season, and soil type on phospholipid fatty acid profiles. Microb. Ecol. 36, 1–12. doi: 10.1007/s002489900087
Bremner, J. M. (1965). Nitrogen availability indexes. Agronomy 9, 1324–1345. doi: 10.2134/agronmonogr9.2.c37
Bright, M. B. H., Diedhiou, I., Bayala, R., Assigbetse, K., Chapuis-Lardy, L., Ndour, Y., et al. (2017). Long-term Piliostigma reticulatum intercropping in the Sahel: crop productivity, carbon sequestration, nutrient cycling, and soil quality. Agric. Ecosyst. Environ. 242, 9–22. doi: 10.1016/j.agee.2017.03.007
Brooker, R. B., Karley, A. J., Adrian, C., Newton, A. C., Pakeman, R. J., and Schob, C. (2016). Facilitation and sustainable agriculture: a mechanistic approach to reconciling crop production and conservation. Funct. Ecol. 30, 98–107. doi: 10.1111/1365-2435.12496
Butler, J. L., Williams, M. A., Bottomley, P. J., and Myrold, D. D. (2003). Microbial community dynamics associated with rhizosphere carbon flow. Appl. Environ. Microbiol. 69, 6793–6800. doi: 10.1128/AEM.69.11.6793-6800.2003
Buyer, J. S., Roberts, D. P., and Russek-Cohen, E. (1999). Microbial community structure and function in the spermosphere as affected by soil and seed type. Can. J. Microbiol. 45, 138–144. doi: 10.1139/w98-227
Buyer, J. S., Roberts, D. P., and Russek-Cohen, E. (2002). Soil and plant effects on microbial community structure. Can. J. Microbiol. 48:955. doi: 10.1139/w02-095
Caldwell, M. M., Dawson, T. E, and Richards, J. H. (1998). Hydraulic lift: consequences of water efflux from the roots of plants. Oecologia 113, 151–161. doi: 10.1007/s004420050363
Curl, E. A., and Truelove, D. (1986). The Rhizosphere. New York, NY: Springer-Verlag. doi: 10.1007/978-3-642-70722-3
Dawson, T. E. (1993). Hydraulic lift and water-use by plants: implications for water-balance, performance and plant interactions. Oecologia 95, 565–574. doi: 10.1007/BF00317442
Debenport, S., Assigbetse, K., Bayala, R., Chapuis-Lardy, L., Dick, R. P., and McSpadden Gardener, B. B. (2015). Shifting populations in the root-zone microbiome of millet associated with enhanced crop productivity in the Sahel. Appl. Environ. Microbiol. 81, 2841–2851. doi: 10.1128/AEM.04122-14
Diedhiou, S., Assigbetsee, K. B., Goudiaby, A. O. K., Diedhiou, I., Badiane, A. N., and Sène, M. (2020). Arid agroecosystem shrubs enhance enzyme activities during the dry season. Am. J. Plant Sci. 11, 180–188. doi: 10.4236/ajps.2020.112014
Diedhiou, S., Dossa, E. L., Badiane, A. N., Diedhiou, I., Sene, M., and Dick, R. P. (2009). Decomposition and spatial microbial heterogeneity associated with native shrubs in soils of agroecosystems in semi-arid Senegal. Pedobiologia 52, 273–286. doi: 10.1016/j.pedobi.2008.11.002
Diedhiou-Sall, S., Dossa, E. L., Badiane, A. N., Assigbetsee, K. B., Diedhiou, I., Ndiaye, N. A. S., et al. (2013). Microbiology and macrofaunal activity in soil beneath shrub canopies during residue decomposition in agroecosystems of the Sahel. Soil Science Soc. Am. J. 77, 501–511. doi: 10.2136/sssaj2012.0284
Dossa, E. L., Diedhiou, I., Khouma, M., Sene, M., Badiane, A. N., Samba, S. A. N., et al. (2013). Crop productivity and nutrient dynamics in a shrub-based farming system of the Sahel. Agronomy J. 105, 1237–1246. doi: 10.2134/agronj2012.0432
Dossa, E. L., Diedhiou, I., Khouma, M., Sene, M., Lufafa, A., Kizito, F., et al. (2012). Crop productivity and nutrient dynamics in a Shrub (Guiera senegalensis)–based farming system of the Sahel. Agronomy Soils Environ. Q. 104, 1255–1264. doi: 10.2134/agronj2011.0399
Dossa, E. L., Diedhiou, S., Compton, J. E., Assigbetse, K. B., and Dick, R. P. (2010). Spatial patterns of P fractions and chemical properties in soils of two native shrub communities in Senegal. Plant Soil. 327, 185–198. doi: 10.1007/s11104-009-0044-8
Dossa, E. L., Khouma, M., Diedhiou, I., Sene, M., Kizito, F., Badiane, A. N., et al. (2009). Carbon, nitrogen and phosphorus mineralization potential of semiarid Sahelian soils amended with native shrub residues. Geoderma 148, 251–260. doi: 10.1016/j.geoderma.2008.10.009
Eviner, V. T. (2001). Linking Plant Community Composition and Ecosystem Dynamics: Interactions of Plant Traits Determine the Ecosystem Effects of Plant Species and Plant Species Mixtures. PhD thesis. Univ. California, Berkeley.
FAO (1998). World Reference Base for Soil Resources. Rome: Food and Agriculture Organization of the United Nations.
Fernandes, M. F. (2006). Fatty Acids Profiling of Soil Microbial Communities: A Comparison of Extraction Methods and Temporal Dynamics in Plant Residue Amended Soils. PhD thesis, Oregon State University, Corvallis.
Fierer, N., Schimel, J. P., and Holden, P. A. (2002). Variation in microbial community composition through two soil depth profiles. Soil Biol. Biochem. 35, 167–176. doi: 10.1016/S0038-0717(02)00251-1
Fromin, N., Hamelin, J., Tarnawski, S., Roesti, D., Jourdain-Miserez, K., et al. (2002). Statistical analysis of denaturing gel electrophoresis (DGE) fingerprinting patterns. Environ. Microbiol. 4, 634–643. doi: 10.1046/j.1462-2920.2002.00358.x
Frostegård, A., and Bååth, E. (1996). The use of phospholipid fatty acid analysis to estimate bacterial and fungal biomass in soil. Biol. Fertil. Soils 22, 59–65. doi: 10.1007/BF00384433
Frostegård, A., Tunlid, A., and Bååth, E. (1991). Microbial biomass measured as total phosphate in soils of different organic content. J. Microbiol. Methods 14, 151–163. doi: 10.1016/0167-7012(91)90018-L
Guckert, J. B., Hood, M. A., and White, D. C. (1986). Phospholipid ester-linked fatty acid profile changes during nutrient deprivation of Vibrio cholerae: increases in the trans/cis ratio and proportions of cyclopropyl fatty acids. Appl. Environ. Microbiol. 52, 794–801. doi: 10.1128/AEM.52.4.794-801.1986
Hayano, K. (1973). A method for determination of b-glucosidase activity in soil. Soil Sci. Pl. Nutr. 19, 103–108. doi: 10.1080/00380768.1973.10432524
Jongman, R. H. G., Ter Braak, C. J. F., and Van Tongeren, O. F. R. (1995). Data Analysis in Community and Landscape Ecology. New York, NY: Cambridge University Press. doi: 10.1017/CBO9780511525575
Kandeler, E., and Gerber, H. (1988). Short-term assay of soil urease activity using colorimetric determination of ammonium. Biol. Fertil. Soils 6, 68–72. doi: 10.1007/BF00257924
Kater, L. J., Kante, S., and Budelman, A. (1992). Karité (Vitellaria paradoxa) and néré (Parkia biglobosa) associated with crops in south Mali. Agroforestry Syst. 18, 89–105. doi: 10.1007/BF00115407
Kieft, T. L., Ringelberg, D. B., and White, D. C. (1994). Changes in ester-linked phospholipid fatty acid profiles of subsurface bacteria during starvation and desiccation in a porous medium. Appl. Environ. Microbiol. 60, 3292–3299. doi: 10.1128/AEM.60.9.3292-3299.1994
Kizito, F. (2006). Hydrological Consequences of Two Native Shrubs in Semi-Arid Senegal: Patterns, Processes, Concerts and Methods. PhD thesis. Oregon State Univ, Corvallis.
Kizito, F., Dragila, M. I., Sen,è, M., Brooks, J. R., Meinzer, F. C., Diedhiou, I., et al. (2012). Hydraulic redistribution by two semi-arid shrub species: implications for Sahelian agro-ecosystems. J. Arid Environ. 83, 69–77. doi: 10.1016/j.jaridenv.2012.03.010
Kizito, F., Sène, M., Dragila, M. I., Lufafa, A., Diedhiou, I., Dossa, E., et al. (2007). Soil water balance of annual crop–native shrub systems in Senegal's Peanut Basin: the missing link. Agricult. Water Manage. 90, 137–148. doi: 10.1016/j.agwat.2007.02.015
Kroppenstedt, R. M. (1992). “The genus nocardiopsis,” in The Prokaryotes. A Handbook on the Biology of Bacteria: Ecophysiology, Isolation, Identification, Applications, eds A. Balows, H. G. Tru$per, W. Dworkin, W. Harder, and K. H. Schleifer (New York, NY: Springer), 1139–1156.
Larkin, R. P. (2003). Characterization of soil microbial communities under different potato cropping systems by microbial population dynamics, substrate utilization, and fatty acid profiles. Soil Biol. Biochem. 35, 1451–1466. doi: 10.1016/S0038-0717(03)00240-2
Latour, X., Philippot, L., Corberand, T., and Lemanceau, P. (1999). The establishment an introduced community of fluorescent pseudomonads in the soil and the rhizosphere is affected by the soil type. FEMS Microb. Ecol. 30, 163–170. doi: 10.1111/j.1574-6941.1999.tb00645.x
Lundquist, E. J., Scow, K. M., Jackson, L. E., Uesugi, S. L., and Johnson, C. R. (1999). Rapid response of soil microbial communities from conventional, low input, and organic farming systems to a wet/dry cycle. Soil Biol. Biochem. 31, 1661–1675. doi: 10.1016/S0038-0717(99)00080-2
Lynch, J. M., and Whipps, J. M. (1990). Substrate flow in the rhizosphere. Plant Soil 129, 1–10. doi: 10.1007/BF00011685
MacNaughton, S. J., Stephen, J. R., Venosa, A. D., Davis, G. A., Chang, Y. J., et al. (1999). Microbial population changes during bioremediation of an experimental oil spill. Appl. Environ. Microbiol. 65, 3566–3574. doi: 10.1128/AEM.65.8.3566-3574.1999
Maloney, P. E., van Bruggen, A. H. C., and Hu, S. (1997). Bacterial community structure in relation to the carbon environments in lettuce and tomato rhizospheres and in bulk soil. Microb. Ecol. 34, 109–117. doi: 10.1007/s002489900040
Marschner, P., Neumann, G., Kania, A., Weisskopf, L., and Lieberei, R. (2002). Spatial and temporal dynamics of bacterial community composition in the rhizosphere of cluster roots of white lupin (Lupinus albus L.). Plant Soil 246, 167–174. doi: 10.1023/A:1020663909890
McCune, B., and Grace, J. B. (2002). Analysis of Ecological Communities. Gleneden Beach, OR: MJM software design.
Muyzer, G., de Waal, E. C., and Uitterlinden, A. G. (1993). Profiling of complex microbial populations by DGGE analysis of PCR-amplified genes encoding for 16S rRNA. Appl. Environ. Microbiol. 59, 695–700. doi: 10.1128/AEM.59.3.695-700.1993
Myers, R. T., Zak, D. R., White, D. C., and Peacock, A. (2001). Landscape-level patterns of microbial community composition and substrate use in upland forest ecosystems. Soil Sci. Soc. Am. J. 65, 359–367. doi: 10.2136/sssaj2001.652359x
Ndour, N. Y. B., Chotte, J. L., Pate, E., Masse, D., and Rouland, C. (2001). Use of soil enzyme activities to monitor soil quality in natural and improved fallows in semi-arid tropical regions. Appl. Soil Ecol. 18, 229–238. doi: 10.1016/S0929-1393(01)00159-7
Norton, J. M., and Firestone, M. K. (1991). Carbon flow in the rhizosphere of ponderosa pine seedlings. Soil Biol. Biochem. 22, 449–455. doi: 10.1016/0038-0717(90)90177-2
O'Leary, W. M., and Wilkinson, S. G. (1988). “Gram-positive bacteria,” in Microbial Lipids, Vol. 1, eds C. Ratledge and S. G. Wilkinson (San Diego, CA: Academic Press), 117–201.
Olsson, P. A. (1999). Signature fatty acids provide tools for determination of the distribution and interactions of mycorrhizal fungi in soil. FEMS Microbiol. Ecol. 29, 303–310. doi: 10.1111/j.1574-6941.1999.tb00621.x
Pielou, E. C. (1966). The measurement of diversity in different types of biological collections. J. Theor. Biol. 13, 131–144. doi: 10.1016/0022-5193(66)90013-0
Porteous, L. A., Seidler, R. J., and Watrud, L. S. (1997). An improved method for purifying DNA from soil for polymerase chain reaction amplification and molecular ecology applications. Mol. Ecol. 6, 787–791. doi: 10.1046/j.1365-294X.1997.00241.x
Priha, O., Grayston, S. J., Hiukka, R., Pennanen, T., and Smolander, A. (2001). Microbial community structure and characteristics of the organic matter in soils under Pinus sylvestris, Picea abies and Betula pendula seedlings at two forest sites. Biol Fertil. Soils 33, 17–24. doi: 10.1007/s003740000281
Rango, A., Tartowskia, S. L., Laliberte, A., Wainwright, J., and Parsons, A. (2006). Islands of hydrologicallyenhanced biotic productivity in natural and managed arid ecosystems. J. Arid Environ. 65, 235–252. doi: 10.1016/j.jaridenv.2005.09.002
Richards, J. H., and Caldwell, M. M. (1987). Hydraulic lift, substantial water transport between soil layers by Artemisia tridentata roots. Oecologia 73, 486–489. doi: 10.1007/BF00379405
Sandhu, G. S., Kline, B. C., Stockman, L., and Roberts, G. D. (1995). Molecular probes for diagnosis of fungal infections. J Clin. Microbiol. 33, 2913–2919. doi: 10.1128/JCM.33.11.2913-2919.1995
Saul-Tcherkas, V., and Steinberger, Y. (2011). Soil microbial diversity in the vicinity of a negev desert Shrub—Reaumuria negevensis. Microbial. Ecol. 61, 64–81. doi: 10.1007/s00248-010-9763-x
Schade, J. D., and Hobbie, S. E. (2006). Spatial and temporal variation in islands of fertility in the Sonoran Desert. Biogeochemistry 73, 541–553. doi: 10.1007/s10533-004-1718-1
Schinner, F. R., Öhlinger, R., Kandeler, E., and Margesin, R. (1996). Methods in Soil Biology. Berlin: Springer Verlag. doi: 10.1007/978-3-642-60966-4
Schlesinger, W. H., and Pilmanis, A. M. (1998). Plant–soil interactions in deserts. Biogeochemistry. 42, 169–187. doi: 10.1007/978-94-017-2691-7_9
Schlesinger, W. H., Raikes, J. A., Hartley, A. E., and Cross, A. F. (1996). On the spatial pattern of soil nutrients in desert ecosystems. Ecology 77, 364–374. doi: 10.2307/2265615
Schutter, M. E., and Dick, R. P. (2000). Comparison of fatty acid methyl ester (FAME) methods for characterizing microbial communities. Soil Sci. Soc. Am. J. 64, 1659–1668. doi: 10.2136/sssaj2000.6451659x
Semenov, A. M., Bruggen, A. H. C., and Zelenev, V. V. (1999). Moving waves of bacterial populations and total organic carbon along roots of wheat. Microbial. Ecol. 37, 116–128. doi: 10.1007/s002489900136
Shannon, C. E. (1948). The mathematical theory of communication. Bell Syst. Tech. J. 27, 379–423. doi: 10.1002/j.1538-7305.1948.tb00917.x
Sinsabaugh, R. L., Antibus, R. K., Linkins, A. E., McClaugherty, C. A., Rayburn, L., Repert, D., et al. (1993). Wood decomposition: nitrogen and phosphorus dynamics in relation to extracellular anzyme activity. Ecology 74, 1586–1593. doi: 10.2307/1940086
Sinsabaugh, R. L., and Moorhead, D. L. (1994). Resource allocation to extracellular enzyme production: a model for nitrogen and phosphorous control of litter decomposition. Soil Biol. Biochem. 26, 1305–1311. doi: 10.1016/0038-0717(94)90211-9
Smalla, K., Wieland, G., Buchner, A., Zock, A., Parzy, J., Kaiser, S., et al. (2001). Bulk and rhizosphere soil bacterial communities studied by denaturing gradient gel electrophoresis: plant-dependent enrichment and seasonal shifts revealed. Appl. Environ. Microbiol. 67, 4742–4751. doi: 10.1128/AEM.67.10.4742-4751.2001
Somarriba, E. (1988). Pasture growth and floristic composition under the shade of guava (Psidium gujava L.) trees in Costa Rica. Agroforest. Syst. 6, 153–162. doi: 10.1007/BF02344752
Stark, J. M., and Firestone, M. K. (1996). Kinetic characteristics of ammonium-oxidizer communities in a California oak woodland–annual grassland. Soil Biol. Biochem. 28, 1307–1317. doi: 10.1016/S0038-0717(96)00133-2
Steer, J., and Harris, J. A. (2000). Shifts in the microbial community in rhizosphere and non-rhizosphere soils during the growth of Agrostis stolonifera. Soil Biol. Biochem. 32, 869–878. doi: 10.1016/S0038-0717(99)00219-9
Sylvia, D. M., Fuhrmann, J. J., Hartel, P. G., and Zoberer, D. A. (2005). Principles and Applications of Soil Microbiology. Upper Saddle River, NJ: Pearson/Prentice Hall.
Tabatabai, M. A., and Bremner, J. M. (1969). Use of p-nitrophenyl phosphate for assay of soil phosphatase activity. Soil Biol. Biochem 1, 301–307. doi: 10.1016/0038-0717(69)90012-1
Westover, K. M., Kennedy, A. C., and Kelley, S. E. (1997). Patterns of rhizosphere microbial community structure associated with co-occurring plant species. J. Ecol. 85, 863–873. doi: 10.2307/2960607
Wezel, A., Rajot, J. L., and Herbrig, C. (2000). Influence of shrubs on soil characteristics and their function in Sahelian agro-ecosystems in semi-arid Niger. J. Arid Environ. 44, 383–398. doi: 10.1006/jare.1999.0609
White, D. C., Davis, W. M., Nickels, J. S., King, J. D., and Bobbie, R. J. (1979). Determination of the sedimentary microbial biomass by extractable lipid phosphate. Oecologia 40, 51–62. doi: 10.1007/BF00388810
Wilkinson, S. G. (1988). “Gram-negative bacteria.” in Microbial lipids, Vol. 1, eds C. Ratledge and S. G. Wilkinson (San Diego, CA: Academic Press).
Willers, C., Jansen van Rensburg, P. J., and Claassens, S. (2015). Phospholipid fatty acid profiling of microbial communities–a review of interpretations and recent applications. J. Appl. Microb. 119, 1207–1218. doi: 10.1111/jam.12902
Yang, C. H., and Crowley, D. E. (2000). Rhizosphere microbial community structure in relation to root location and plant iron nutritional status. Appl. Environ. Microbiol. 66, 345–351. doi: 10.1128/AEM.66.1.345-351.2000
Young, I. M. (1995). Variation in moisture contents between bulk soil and the rhizosheath of wheat (Triticum aestivum L. cv. Wembley). New Phytol. 130, 135–139. doi: 10.1111/j.1469-8137.1995.tb01823.x
Young, I. M. (1998). Biophysical interactions at the root: soil interface: a review. J. Agric. Sci. 130, 1–7. doi: 10.1017/S002185969700498X
Zelles, L. (1999). Fatty acid patterns of phospholipids and lipopolysaccharides in the characterisation of microbial communities in soil: a review. Biol. Fertil. Soils 29, 111–129. doi: 10.1007/s003740050533
Zelles, L., Bai, Q. Y., Beck, T., and Beese, F. (1992). Signature fatty acids in phospholipids and lipopolysaccharides as indicators of microbial biomass and community structure in agricultural soils. Soil Biol. Biochem. 24, 317–323. doi: 10.1016/0038-0717(92)90191-Y
Keywords: microbial diversity, soil, shrub-intercropping, DGGE, PLFA
Citation: Diedhiou-Sall S, Assigbetsee KB, Badiane AN, Diedhiou I, Khouma M and Dick RP (2021) Spatial and Temporal Distribution of Soil Microbial Properties in Two Shrub Intercrop Systems of the Sahel. Front. Sustain. Food Syst. 5:621689. doi: 10.3389/fsufs.2021.621689
Received: 26 October 2020; Accepted: 04 February 2021;
Published: 12 March 2021.
Edited by:
Everlon Cid Rigobelo, Universidade Estadual Paulista, BrazilReviewed by:
Xingang Zhou, Northeast Agricultural University, ChinaCopyright © 2021 Diedhiou-Sall, Assigbetsee, Badiane, Diedhiou, Khouma and Dick. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Richard P. Dick, ZGljay43OEBvc3UuZWR1
†Deceased
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.