
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst., 09 February 2021
Sec. Agroecology and Ecosystem Services
Volume 5 - 2021 | https://doi.org/10.3389/fsufs.2021.596316
This article is part of the Research TopicMicrobiome Research in AgroecosystemsView all 8 articles
 Ju-Chun Chang1
Ju-Chun Chang1 Shin-Shan Wu1
Shin-Shan Wu1 Yao-Chia Liu1
Yao-Chia Liu1 Yu-Hong Yang1
Yu-Hong Yang1 Yi-Fang Tsai1
Yi-Fang Tsai1 Yi-Hsuan Li1
Yi-Hsuan Li1 Ching-Tzu Tseng2
Ching-Tzu Tseng2 Li-Cheng Tang1
Li-Cheng Tang1 Yu-Shin Nai1*
Yu-Shin Nai1*Chemical pesticides have been used for pest control for many decades, but they cause serious problems, including insecticide resistance, secondary pest resurgence, and negative environmental impacts. Therefore, sustainable alternatives to chemical pesticides are necessary for pest control. Entomopathogenic fungi (EPF) are natural epizootic pathogens of insects, and some of them have been used as microbial biocontrol agents. Herein, we attempted to construct an entomopathogenic fungal library (EFLib) via the “Tenebrio molitor pathogenicity-based fungal collection method (TmPC)” to select EPF for control of the serious agricultural pest Spodoptera litura. A total of 172 soil samples were collected in northern and central Taiwan for the EFLib construction. The isolation efficiency of TmPC was 64.02%. The EFLib consisted of 101 isolates and was designated as the National Chung Hsing University (NCHU) EFLib. Among these isolates, 26 showed high virulence (mortality = 100%) to T. molitor larvae. Based on the results of molecular identification, the highly virulent isolates belonged to seven genera, including Beauveria, Clonostachys, Fusarium, Cordyceps, Penicillium, Purpureocillium, and Metarhizium. To evaluate the potential of these isolates for Spodoptera litura control, 12 isolates were selected for pathogenicity screening against S. litura larvae. A total of six EPF isolates belonging to the genera Beauveria and Metarhizium showed rapid eradication of the S. litura larvae. To rank the potential of these fungal strains for pest management, the six isolates were subjected to thermotolerance and conidial production assays, and a novel effective conidia number (ECN) formula was applied. The results indicated that the ECN index of Beauveria australis (NCHU-113) was much higher than that of Metarhizium isolates. However, among the Metarhizium isolates, NCHU-95 showed the highest ECN index. Altogether, NCHU-69 and NCHU-113 should be further tested in field trials. To our knowledge, this is the first attempt to integrate pathogenicity or virulence and ECN data into EPF screening and ranking, providing a baseline for mass selection of potential EPF strains for further applications.
Entomopathogenic fungi (EPF) are a group of epizootic insect pathogens in the natural environment that serve as natural regulators of insect populations, including those of pests (Gardner et al., 2005; Gürlek et al., 2018). In general, the mode of action of EPF infection includes adhesion of conidia onto the cuticle of target insects or another portal of entry such as the trachea (Holder and Keyhani, 2005; Gürlek et al., 2018). During the fungal infection process, enzymes such as lipases, proteases, and chitinases are produced and secreted for penetration of the host cuticle by formation of an appressorium (Yang et al., 2016; Kim et al., 2018; Lee et al., 2019).
In terms of microbial control, EPF can act efficiently under a wide range of humidity and temperature conditions; therefore, EPF have high potential to be developed as microbial control agents for insect pests in agriculture, horticulture, and forestry (Wraight and Carruthers, 1999, Lacey et al., 2015). To our knowledge, a high proportion of EPF species belong to the phyla Ascomycota and Zygomycota. Among these fungi, the genera Beauveria (i.e., B. bassiana), Hirsutella, Cordyceps, Akantomyces, Metarhizium (i.e., M. anisopliae), and Purpureocillium belonging to the phylum Ascomycota, and Entomophaga, Entomophthora, Pandora, and Furia belonging to the phylum Zygomycota are the most studied EPF, and some of these organisms have also been used in commercial products for integrated pest management (IPM) (Meyling and Eilenberg, 2007, Gilbert and Gill, 2010).
Before the development of biopesticides based on EPF, it is necessary to collect and establish a library of EPF to facilitate mass screening of virulence against target insect pests. Several methods could be used for EPF collection. EPF could be collected from soil directly or from soil with insect bait as well as from insect cadavers showing mycosis found in the field (Rangel et al., 2010, Barra et al., 2013, Kim et al., 2018). Among the methods mentioned above, the collection of EPF from soil with insect bait showed more efficiency than those directly from soil (Kim et al., 2018). In addition, indigenous EPF isolated from their native locations may be more efficient for controlling the local pests of those areas (Jackson et al., 2000, Gulsar et al., 2004, Kim et al., 2018). Therefore, for the purpose of microbial pest management in local areas, the isolation of indigenous EPF from the soil with insect bait is an ideal strategy.
Many efforts have been made to isolate EPF from soil samples by antibiotic-based methods; however, the efficiencies of isolation and pathogenicity screening with these methods were lower than those with insect bait-based methods (i.e., Galleria mellonella larvae) (Meyling and Eilenberg, 2007, Tuininga et al., 2009, Gürlek et al., 2018). Previous studies have shown that the larvae of yellow mealworms, Tenebrio molitor (Coleoptera: Tenebrionidae), have also been used as bait for the isolation of EPF from soil, and the isolation efficiency was much higher than that of the antibiotic-based method from soil samples; therefore, “T. molitor-mediated EPF library construction” provides a new strategy for EPF isolation and screening (Vänninen et al., 2000, Kim et al., 2016).
The tobacco cutworm, Spodoptera litura Fabricius (Lepidoptera: Noctuidae), is an important polyphagous pest species. S. litura is considered an important pest worldwide which causes serious economic damage to important crop plants, i.e., fruits, soybean, and vegetables (Otuka et al., 2020). It has been reported that the larvae of S. litura feed on more than 380 plant species (Ghumare and Mukherjee, 2003, Xue et al., 2010, Wu et al., 2019). However, the chemical control of S. litura causes problems such as insecticide resistance and environmental residues (Ahmad and Mehmood, 2015, Wu et al., 2019). Therefore, alternative control strategies are needed to control this pest.
In this study, we mainly followed a method described previously (Kim et al., 2018) to isolate EPF from soil using “T. molitor-mediated EPF library construction” in Taiwan. After isolation, molecular identification and phylogenetic analysis were performed to clarify the taxonomic position of these EPF from soil samples, and pathogenicity screening against T. molitor larvae and Spodoptera litura larvae was performed. To determine the potential of EPF for field applications, commercialization, and mass production, their thermotolerance and conidial production were also determined. Moreover, based on thermotolerance analysis and conidial production data, a formula, first reported in this study, for the effective conidia number (ECN) was used to estimate the potential of EPF under environmental stress. This procedure could be applied as a standard operating procedure (Supplementary Figure 1) for the selection of EPF for either laboratory screening and bioassays or the development of fungal biopesticides and their applications in pest management approaches.
To isolate EPF, soil samples were collected from six different locations in the cities of Yilan and Taichung in Taiwan (Figure 1). The soil samples were collected from October to November 2018 in Yilan and from December 2018 to March 2019 in Taichung. Detailed information on each sampling site is summarized in Table 1. Soil samples were collected following the method described by Kim et al. (2018). The samples were collected at a depth of approximately 10 cm from randomly selected sites in mountainous areas or campuses in Yilan city and Taichung city. Approximately 100 g of soil samples were collected and stored in sealed plastic bags at room temperature for ca. 1 h (transport time) and then immediately subjected to a fungal isolation protocol in the laboratory.
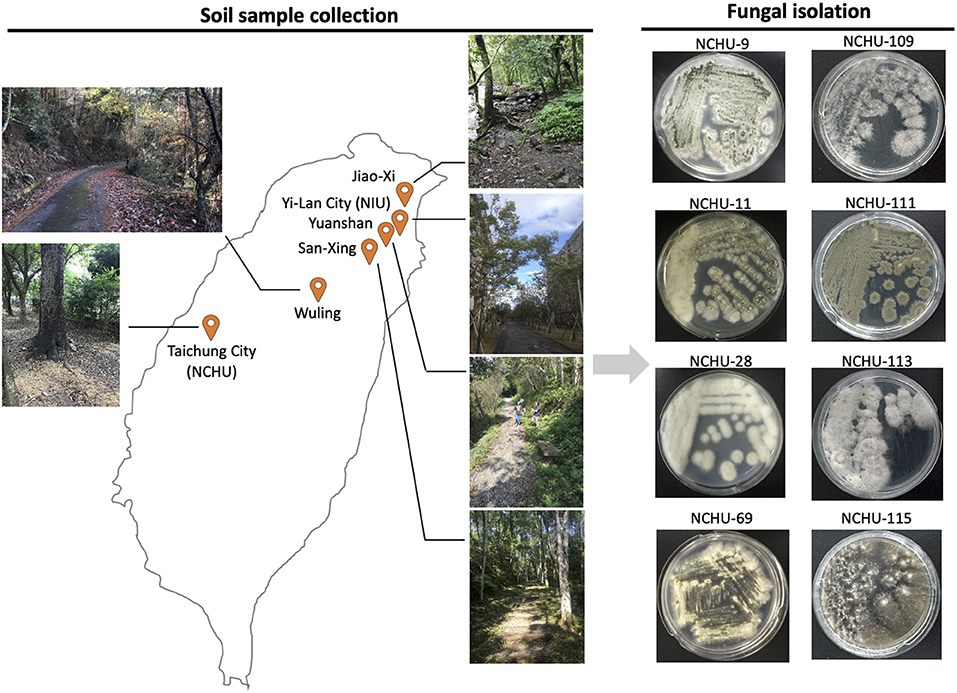
Figure 1. Locations of soil sample collections and illustration of fungal isolation and pathogenicity screening using mealworms. The entomopathogenic fungal isolates were designated the National Chung Hsing University Entomopathogenic Fungi (NCHU), and the fungal collection library was established.
The protocol for EPF isolation followed a method described previously (Kim et al., 2018). Entomopathogenic fungi were isolated from soil samples using 4th instar Tenebrio molitor larvae. Each soil sample was placed in a plastic cup, and five T. molitor larvae were placed on the surface of the soil for exposure to the soil samples. A lid was placed on the plastic cup to maintain the optimal relative humidity (~70%), and the plastic cup, which contained T. molitor larvae, was incubated at 25°C in the laboratory for 10 days. All the samples were observed daily for mycosis of the cadavers, and the percentage of mycosis was recorded. The cadavers exhibiting mycosis were transferred to a clean bench, and the conidia were collected using sterile tooth picks and transferred to quarter strength Sabouraud dextrose agar medium (¼ SDA) by the streak-plate method. The cultured plates were incubated at 25°C for 7 d. The cultured fungi were reisolated on ¼ SDA plates for several rounds to obtain pure fungal cultures (Figure 1).
To select EPF, fungal isolates obtained from the above procedure were used for the first round of insect pathogenicity screening using five T. molitor larvae in one plate (Kim et al., 2018). Briefly, a total of five 4th instar larvae of T. molitor were exposed to 10-d cultured fungi on ¼ SDA at 25°C in a 60 mm plate. The plate was sealed with parafilm and incubated at 25°C in the dark. Control larvae were placed on ¼ SDA plates without fungi. The mortality of the T. molitor larvae was observed daily until 10 d after inoculation. The surface of cadavers with mycosis confirmed the presence of fungal infection, and the fungal isolates were further subjected to a second round of pathogenicity screening to confirm their virulence to T. molitor larvae in addition to molecular identification.
To confirm the pathogenicity indicated in the first screening, 26 selected fungal isolates were subjected to a second round of pathogenicity screening. The conidia of each fungal isolate were harvested with 0.03% Silwet solution, and the number of conidia was counted using a hemocytometer. The conidial suspension was then adjusted to 1 × 107 conidia/mL for the following experiment. A volume of 10 μL of each conidial suspension was spread on a 60 mm ¼ SDA plate and incubated at 25°C for 10 d for the second round of pathogenicity screening using five T. molitor larvae. Briefly, 4th instar larvae of T. molitor were exposed to the fungal culture plates. The plates were sealed with parafilm and incubated at 25°C in the dark. Control larvae were placed on ¼ SDA plates without fungi. The mortality of the T. molitor larvae was observed and recorded daily for 10 d. The second round of pathogenicity screening of each fungal isolate was repeated in triplicate.
Fungal genomic DNA was extracted from the hyphae of 10-days cultures grown on ¼ SDA using a Yeast Genomic DNA Kit (Geneaid, TW). The molecular identification at the genus level was based on the internal transcribed spacer (ITS) region. To amplify the ITS region (ITS1-5.8S-ITS2), the primer set ITS1F (5′- TCCGTAGGTGAACCTGCGG-3′)/ITS4R (5′-TCCTCCGCTTATTGATATGC-3′) was used for PCR (White et al., 1990). A total of 12 selected entomopathogenic fungal isolates were subjected to second-round molecular identification at the species level based on the transcriptional elongation factor (tef ) region using tef-983F (5′-GCYCCYGGHCAYCGTGAYTTYAT-3′) and tef-2218R (5′- ATGACACCRACRGCRACRGTYTG-3′) (Rehner and Buckley, 2005, Quandt et al., 2014, Kepler et al., 2017) for PCR. The PCR mixture consisted of 10 μl of 2 X SuperRed PCR Master Mix (Tools, TW), 1 μl of fungal genomic DNA (ca. 100 ng/μl), 1 μl of each primer (10 μM), and 7 μl of ddH2O in a 20 μl reaction volume. PCR was performed using a thermal cycler (ABI Perkin Elmer 9700, USA). The ITS region was amplified with the following reaction parameters: an initial cycle of denaturation at 94°C for 1 min, followed by 35 cycles of denaturation at 94°C for 30 sec, annealing at 55°C for 30 s, and extension at 72°C for 30 s. An additional cycle comprising a 7 min extension at 75°C was included as the final step. Transcription elongation factor 1 alpha (Tef ) amplification was performed by the following reaction parameters: an initial cycle of denaturation at 94°C for 2 min, followed by 35 cycles of denaturation at 94°C for 1 min, annealing at 56°C for 30 s, and extension at 72°C for 1 min. An additional cycle comprising a 10 min extension at 72°C was included as the final step. The PCR products were analyzed by electrophoresis using a 1% agarose gel in TAE buffer and purified using a DNA Cleanup Kit (Geneaid, TW). The purified PCR products were sent to a sequencing service company (Genomics Co., TW) for direct sequencing. The obtained fungal sequences were assembled and cleaned/edited using SnapGene Viewer and blasted through the NCBI GenBank database for identification and determination of similarity with already identified organisms.
The ITS sequences of 52 entomopathogenic fungi were subjected to phylogenetic analysis, including 26 entomopathogenic fungal isolates from this study and 26 strains/isolates from the NCBI GenBank (Supplementary Tables 1, 3). In addition, the tef sequences of 11 Metarhizium spp. and one Beauveria sp. promising entomopathogenic fungal isolates from this study were also subjected to phylogenetic analysis, respectively, with those of 60 other entomopathogenic fungi (32 Beauveria spp and 28 Metarhizium spp) (Supplementary Table 1). The multiple sequences were aligned by the ClustalX 2.1 software (Thompson et al., 1997), and the conserved sequences were manually trimmed with GeneDoc (Nicholas and Nicholas, 1997) for phylogenetic analysis. The ITS phylogenetic tree was constructed by the MEGA7 software based on the neighbor-joining (NJ) criteria and the evolutionary distances were computed using the Kimura 2-parameter method (Kumar et al., 2016). Bootstrap analyses were performed to evaluate the robustness of the phylogenies using 1000 replicates for NJ analyses. The phylogenetic analysis of the 11 Metarhizium spp. and one Beauveria sp. promising entomopathogenic fungal isolates with those of other EPF based on tef nucleotide sequences were performed based on the Maximum Likelihood (ML), Minimum Evolution (ME), and NJ analyses using the Kimura 2-parameter method. Bootstrap analyses were performed to evaluate the robustness of the phylogenies using 1000 replicates for the three models.
Based on the pathogenicity screening data obtained with T. molitor larvae, 12 isolates belonging to the genera Metarhizium and Beauveria were selected for pathogenicity screening against the tobacco cutworm S. litura (Lepidoptera: Noctuidae). The artificial rearing of S. litura followed a method described previously (Ou-Yang and Chu, 1988). A colony of S. litura was maintained and fed with artificial diets under laboratory conditions. Larvae were kept in screened cages at 25 ± 1°C with 70% relative humidity (RH) and a 12:12 (L:D) photoperiod. Each fungal isolate was cultured at 25°C on ¼ SDA medium for 10 d for pathogenicity screening. Five 4th instar larvae were exposed to the 10-days cultures of each fungal isolate on ¼ SDA medium for 24 h at 25 ± 1°C. After exposure, each larva was transferred to an individual well of a new 30-well plastic plate and fed with artificial diets. The numbers of live S. litura larvae were recorded daily for 10 d. A total of three replicates were performed to evaluate the potential of the EPF for pest control. According to the results, a total of six fungal isolates, including five Metarhizium isolates (NCHU-9 (Metarhizium lepidiotae), NCHU-11 and NCHU-64 (Metarhizium pinghaense), NCHU-69 (Metarhizium anisopliae), NCHU-95 (M. anisopliae), and one Beauveria isolate [NCHU-113 (Beauveria australis)], were selected for evaluation in the field.
Each selected fungal isolate was cultured on 1/4 SDA medium in a 55 mm Petri dish at 25 ± 1°C in the dark for 7 d for morphological observation. The observation of conidia and conidiogenesis was performed with a Nikon CI-L optical microscope at 400 × magnification and the images were captured by a SGHD-3.6C camera. The length and width of conidia and phialides were measured by the SAGE VISION Image software. In total 20 conidia and phialides were measured for each fungal isolate.
The thermotolerance assay was conducted following a previous method (Kim et al., 2018). Each of the fungal isolates was cultured on ¼ SDA medium at 25 ± 1°C in darkness for 10 d. Conidial suspensions of each isolate were prepared as mentioned above. The conidial suspensions were then adjusted to 1 × 107 conidia/mL. A total of 1 mL of conidial suspension was vortexed for 1 min and placed in a 45°C incubator for 0, 30, 60, 90, and 120 min. Then, 5 μL of each heat-exposed isolate was dropped on ¼ SDA medium and incubated at 25 ± 1°C for 18 h. The germination percentage of the isolates was determined by randomly counting the number of germinated and ungerminated conidia in five fields at 200 × under light microscopy (Kim et al., 2018). Each treatment was replicated three times.
The conidial production assay was conducted following a previous method (Kim et al., 2018), and the conidial suspensions of each isolate were prepared as described above and adjusted to 1 × 107 conidia/mL. Ten microliters of sample was inoculated (dropped) on ¼ SDA and incubated at 25°C for 7, 10, and 14 d. At each time point, a 5-mm agar block was detached from the center of the colony and placed in 1 mL of 0.03% Silwet solution in a 1.5 ml tube. The sample was vortexed at 3,000 rpm at room temperature for 15 min, and the conidial density was calculated by a hemocytometer. The conidial production assay was repeated three times for each isolate.
To further evaluate the effective conidia number (ECN) under a simulated environment, the conidial production and thermotolerance data were combined, and the ECN was calculated using the following steps:
1. The fold-change in conidial production (FC) at each time point was calculated as follows:
where x = the time point for data collection; ncp = the number of conidia after each day of growth; and I = the initial number of seeded conidia (105 conidia in this study).
2. The ECN was calculated at each time point under treatment using the following formula:
where y = the ECN of the time point under treatment; TT0 = the germination rate of conidia not undergoing heat stress (= germination rate of 0 min heat treatment); and the stress coefficient = the conidia germination rate at different times of heat treatment (z).
To simulate the application under field conditions, the accumulation of heat stress was added to the ECN calculation as the “stress coefficient.” In this study, the “stress coefficient” is the conidial germination rate under different times (z) of heat treatment; thus, when y = 0, TTz is equal to 100%. Therefore, the formula for calculating the total ECN is listed below:
In this study, the ECNsum was also equal to the following for each fungal strain:
In addition, the conidial production and thermotolerance data were further analyzed with principal component analysis (PCA) to evaluate the similarity between each selected potential EPF isolate.
The data of insect mortality in the bioassays were analyzed using a T-test and the LT50 of each fungal strain also included generalized linear models (GLMs) with a quasibinomial error distribution and a log link function to account for overdispersion. The above analyses were conducted using SPSS ver. 12.1 (SPSS Inc., 2009) at the 0.05 (α) level of significance. The length and width of conidia and phialides were analyzed based on the Welch's ANOVA test using the R studio.
There were 101 fungal isolates obtained from 172 soil samples using the “T. molitor-mediated EPF library construction” method (Table 1). The fungal isolation efficiency was 64.02%. Based on the pathogenicity screening with T. molitor larvae, 26 fungal isolates showed high virulence against T. molitor larvae (100% mortality rate), and 19 fungal isolates showed medium virulence against T. molitor larvae (60~80% mortality rate), while the other fungal isolates showed lower virulence to T. molitor larvae, suggesting that ~50% of the 101 fungal isolates might have insect-killing activity and belong to the EPF group (Supplementary Table 2). Herein, the EPF library was established, and this collection was named the National Chung Hsing University (NCHU) EPF library.
Of 101 fungal isolates, 26 fungal isolates were selected for the second round of virulence tests against T. molitor larvae based on the primary mealworm pathogenicity screening. The mortality rate of T. molitor larvae after fungal treatment showed a similar correlation to that obtained with the primary pathogenicity screening (Figure 2; Supplementary Table 2). All 26 fungal isolates showed significantly high virulence to T. molitor larvae (df = 9, P < 0.01). There were a total of 17 fungal isolates that showed the highest virulence to T. molitor larvae (100% mortality rate) at 10 d post inoculation (d.p.i.), and three fungal isolates caused a 93.3% mortality rate in T. molitor larvae, while the other fungal isolates resulted in a lower mortality rate (20–80%) in T. molitor larvae at 10 d.p.i. and were considered lower virulence isolates (Figure 2). Among the 17 fungal isolates, 12 isolates, NCHU-9, 10, 11, 27, 29, 30, 32, 64, 69, 95, 102, and 113, caused 100% mortality in T. molitor larvae at 5 d.p.i., which revealed the fast insect-killing activity of these strains. Thus, the strains were selected for further virulence testing against S. litura.
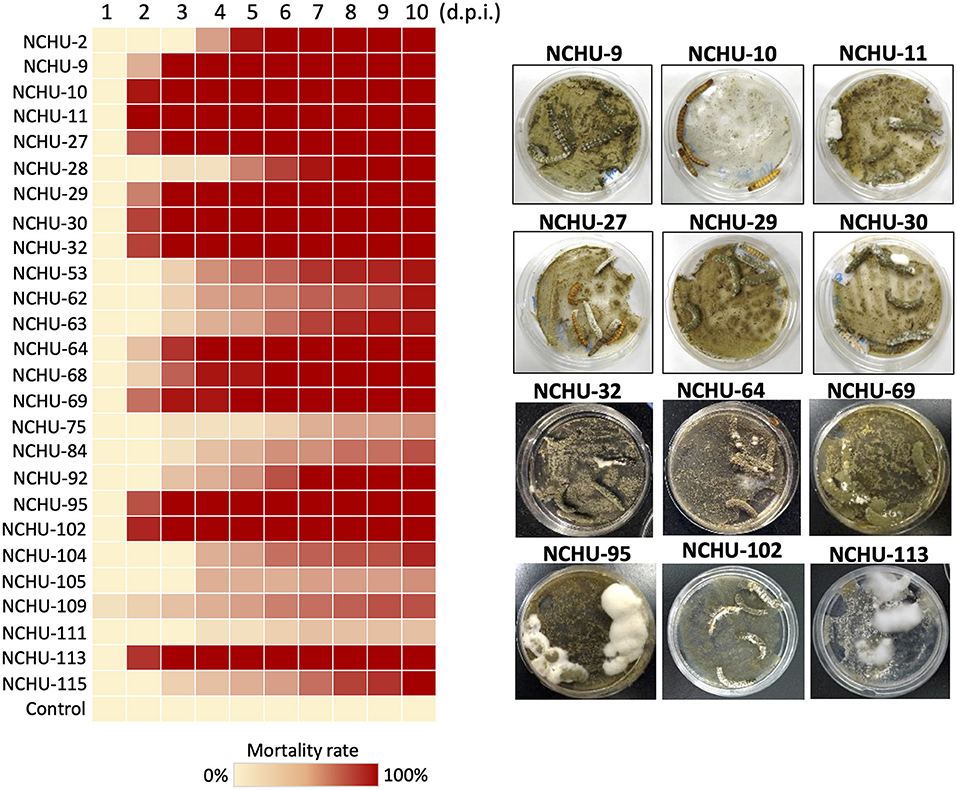
Figure 2. Mortality heat map of 26 fungal isolates against mealworms. Based on the primary mealworm pathogenicity screening, 26 fungal isolates were selected based on their rapid killing and high virulence against mealworms. The mycoses of 12 rapid killing and high virulence fungal isolates are shown in parallel. Each fungal strain was repeated three times. d.p.i., days post inoculation.
A total of 26 isolates were subjected to molecular identification and phylogenetic analysis. Based on the molecular characterization of the ITS region, 26 fungal isolates could be classified into seven genera, including Beauveria, Clonostachys, Fusarium, Cordyceps, Penicillium, Purpureocillium, and Metarhizium (Figure 3; Supplementary Table 3). Most of the fungal isolates belonged to the genus Metarhizium. All the taxonomic positions of the 26 fungal isolates at the genus level were confirmed by phylogenetic analysis based on the sequences of their ITS1-5.8S-ITS2 region (Figure 3). All 26 fungal isolates were grouped into well-defined clades, revealing their high similarity to reported isolates from GenBank.
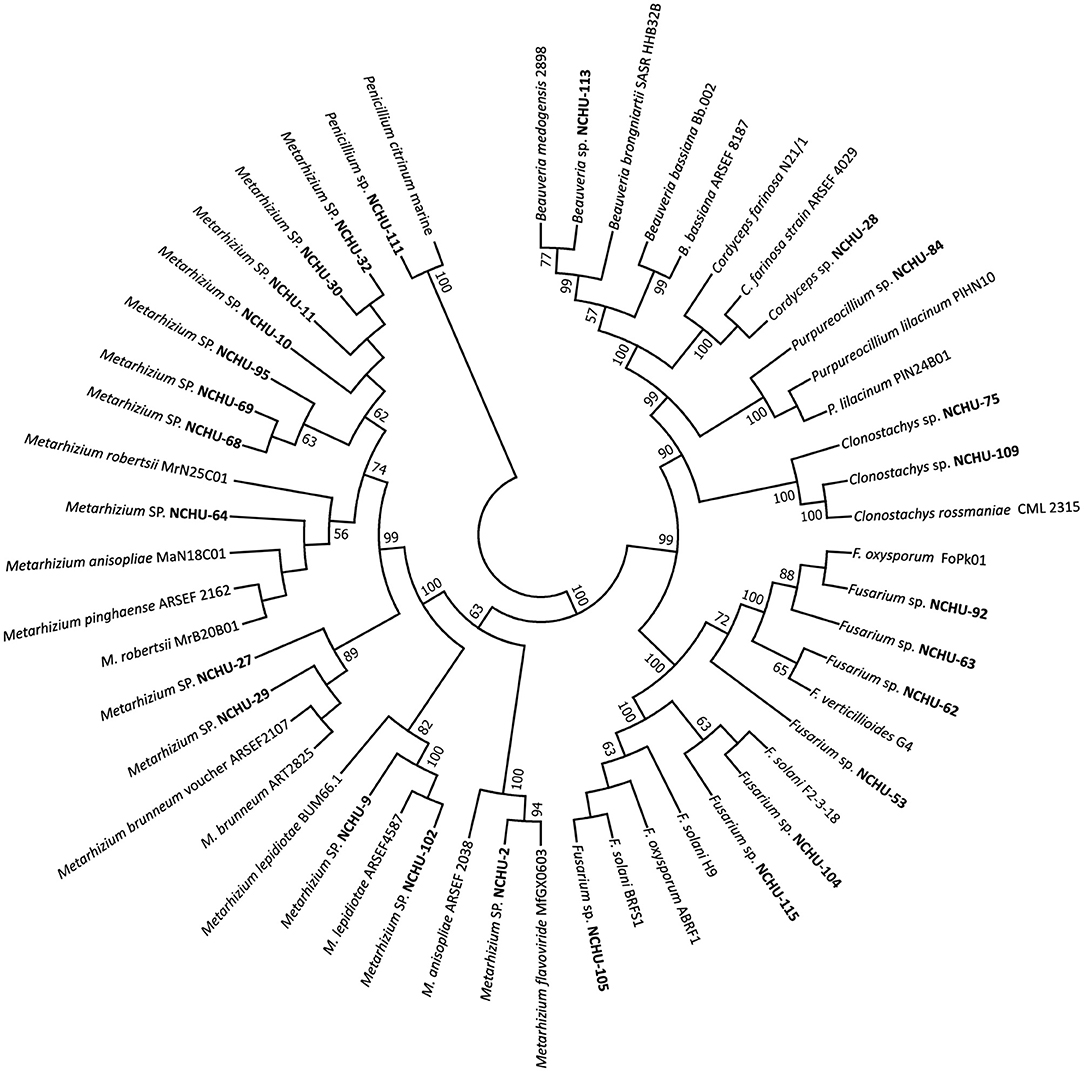
Figure 3. A phylogenetic tree was constructed by using the neighbor-joining (NJ) method based on the fungal ITS for 26 potential entomopathogenic fungal isolates in MEGA7. Bootstrap analyses were performed to evaluate the robustness of the phylogenies using 1000 replicates.
The tef region was also sequenced for the species-level identification of 12 selected fungal isolates. The tef sequences of the 12 selected fungal isolates were subjected to phylogenetic analysis, and the sequences were also submitted to the NCBI Genbank (Supplementary Table 3). Of these isolates, 11 belonged to the genus Metarhizium (Figure 4A); among these isolates, four species of Metarhizium spp. were identified, including Metarhizium lepidiotae (NCHU-9, NCHU-102), Metarhizium pinghaense (NCHU-10, NCHU-11, NCHU-30, NCHU-32, NCHU-64), Metarhizium brunneum (NCHU-27, NCHU-29), and Metarhizium anisopliae (NCHU-69, NCHU-95) (Figure 4A), and one isolate was identified as Beauveria australis (NCHU-113) (Figure 4B).
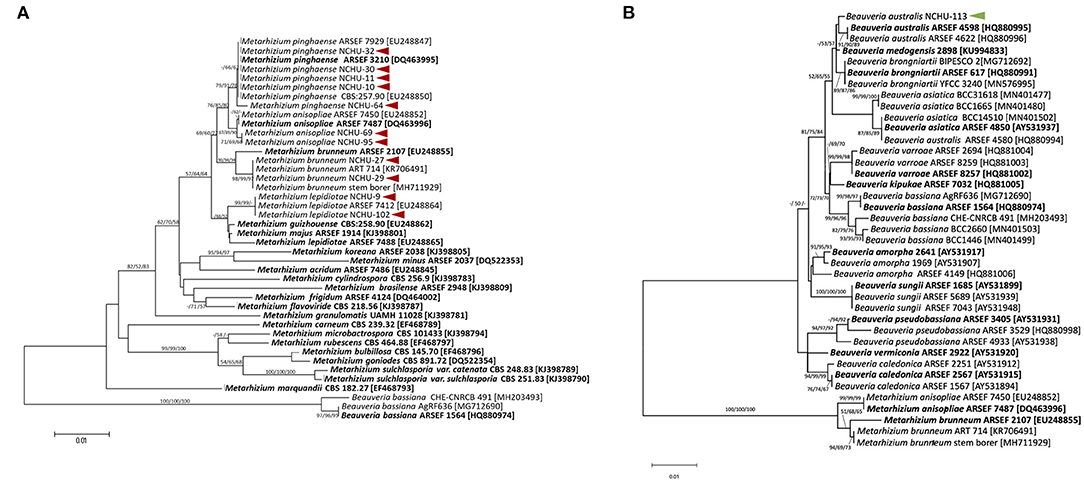
Figure 4. The phylogenetic trees were constructed using the ML, ME, and NJ analyses method based on the fungal transcriptional elongation factor 1 alpha (tef ) for (A) 11 promising Metarhizium spp. and (B) one Beauveria sp. in MEGA7. Bootstrap analyses were performed to evaluate the robustness of the phylogenies using 1000 replicates and the bootstrap values > 50% for ML, ME, and NJ trees at each node (ML/ME/NJ). The arrows indicated the promising fungal isolates from this study.
A total of 12 fungal isolates showing pathogenicity and rapid insect-killing activity against T. molitor larvae were selected for further pathogenicity screening against S. litura larvae (LT50 = 2.49, df = 9, P < 0.01). Based on the results, seven of the selected fungal isolates showed 100% mortality, and the other five isolates showed 80% mortality to S. litura larvae after 8 days of treatment, indicating that the fungi that were isolated based on the T. molitor-mediated EPF library construction method showed virulence against lepidopteran insects (Figure 5). Although the seven fungal isolates showed 100% mortality against S. litura larvae at 8 d.p.i., the length of time required to reach 100% mortality was different, suggesting that different biological characteristics of fungal isolates might influence the infection process based on our bioassay data (Figure 5). Of the seven fungal isolates, five isolates belonging to the genus Metarhizium, including M. lepidiotae (NCHU-9), M. pinghaense (NCHU-11, NCHU-64), and M. anisopliae (NCHU-69 and 95), revealed rapid killing activity (100% mortality at 6 d.p.i., LT50 = 2.94, 2.22, 2.84, 2.57 and 2.96) for Metarhizium spp. and 80% mortality at 3 d.p.i. for B. australis (NCHU-113) (LT50 = 1.13) in S. litura larvae, and were subjected to further evaluation in terms of their conidial production and thermotolerance.
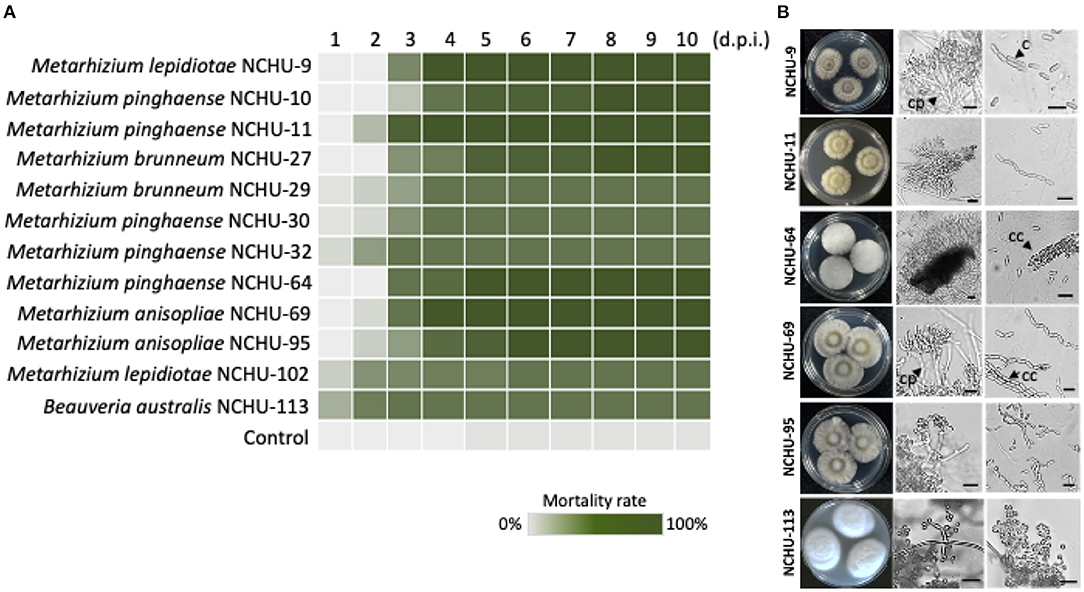
Figure 5. Mortality heat map of 12 fungal isolates against S. litura. (A) Each fungal strain was repeated three times. d.p.i., days post inoculation. Six entomopathogenic fungal isolates were selected for morphological observation. (B) The fungal colony growth characteristics are shown in parallel. Cp, conidiophores; c, conidium; cc, cylindrical conidia. Scale bar = 10 μm.
Based on morphological observations, the fungal cultures showed different colors on the 1/4 SDA medium (Figure 5B). Branched conidiophores and short, blocky conidiogenous cells were clearly observed in M. lepidiotae (NCHU-9), M. pinghaense (NCHU-11, and 64), M. anisopliae (NCHU-69 and 95) (Figure 5B). The phialides and conidia sizes of six selected fungal isolates are similar to those of close-type fungal strains (Table 2; Supplementary Table 4). The conidiogenous cells of B. australis (NCHU-113) were solitary and usually grew in dense and ampulliform in base (Figure 5B). In addition, both short and long conidial types were found on the hymenial surfaces, and cylindrical conidia were also observed in all isolates of M. lepidiotae (NCHU-9: 6.5 ± 0.45 μm × 2.7 ± 0.13 μm), M. pinghaense (NCHU-11: 6.3 ± 0.41 μm × 2.7 ± 0.16 μm; NCHU-64: 6.8 ± 0.53 μm × 2.9 ± 0.31 μm), and M. anisopliae (NCHU-69: 6.3 ± 0.34 μm × 2.6 ± 0.19 μm; NCHU-95: 6.0 ± 0.82 μm × 2.9 ± 0.29 μm) (Table 2; Figure 5B). B. australis produced globose conidia (NCHU-113: 2.3 ± 0.24 μm × 2.3 ± 0.24 μm), and conidia aggregated to form spherical clusters among the aerial hyphae (Table 2; Figure 5B).
From the conidia production assay, the number of conidia produced was counted at different times after 7, 10, and 14 days of cultivation (Figure 6). In general, the production of conidia was dramatically increased from 7 to 10 days of cultivation in both Metarhizium spp. and B. australis, peaking at 14 days of cultivation (Figure 6). For conidial production after 14 days of cultivation, B. australis (NCHU-113) produced a 1.3 × 108 conidia/per 5 mm block, while the production of conidia was much lower in the five Metarhizium isolates (Figure 6). Among the five Metarhizium isolates, M. pinghaense (NCHU-11) showed the highest conidial production, which was 2.1 × 106 after 14 d of cultivation, followed by M. lepidiotae (NCHU-9) and M. pinghaense (NCHU-64) (Figure 6).
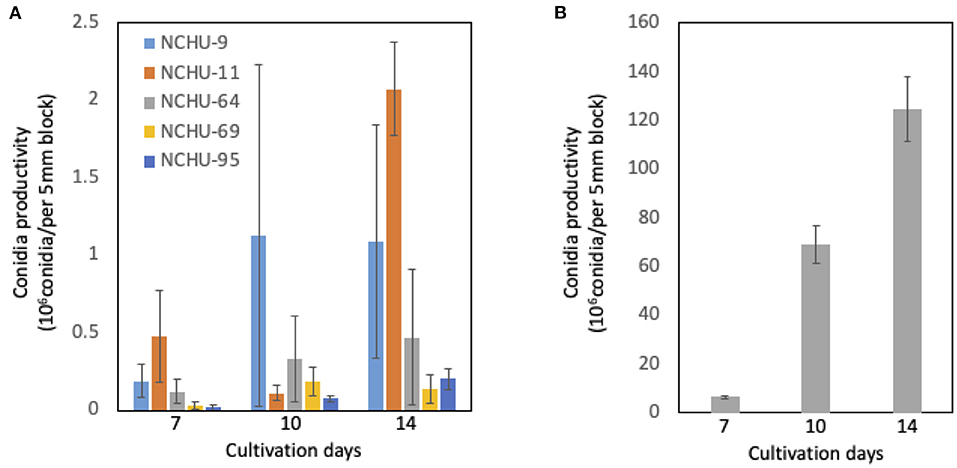
Figure 6. (A-B) Conidial production of six fungal isolates. The conidial production data were collected after 7, 10, and 14 d of culture. The conidia of three blocks (5 mm in diameter) were calculated for each fungal isolate. The experiments were replicated three times for each fungal isolate.
From the thermotolerance assay, the conidia of six isolates were exposed to 45°C for different treatment times, including 0, 30, 60, 90, and 120 min, and the germination rates were further evaluated (Figure 7). The highest original germination rates (0 min heat treatment) were found in B. australis (NCHU-113) (98.67%), followed by M. anisopliae (NCHU-95) (98.52%) and M. pinghaense (NCHU-11) (97.94%), while M. lepidiotae (NCHU-9) showed the lowest original germination rates (77.25%) (Figure 7). After exposure for 30 min, the conidial germination rate of M. pinghaense (NCHU-64) was reduced to 0% and the germination rate of B. australis (NCHU-113), M. pinghaense (NCHU-11), and M. lepidiotae (NCHU-9) also dropped to 3.48 ~16.3% after exposure for 30 min, while M. anisopliae, NCHU-95 and NCHU-69, showed 89.1 and 85.8% germination rates, respectively (Figure 7). After 60 min of treatment, thermotolerance was significantly decreased in the Metarhizium isolates (NCHU-9, 69, and 95) and B. australis (NCHU-113), ranging from 0.8% (NCHU-9) to 36.7% (NCHU-95) (Figure 6). After exposure for 90 min and 120 min, the Metarhizium isolates (NCHU-9, 69, and 95) and B. australis (NCHU-113) showed conidia germination rates of only 0.4–5.2% and 0.4–2%, respectively (Figure 7).
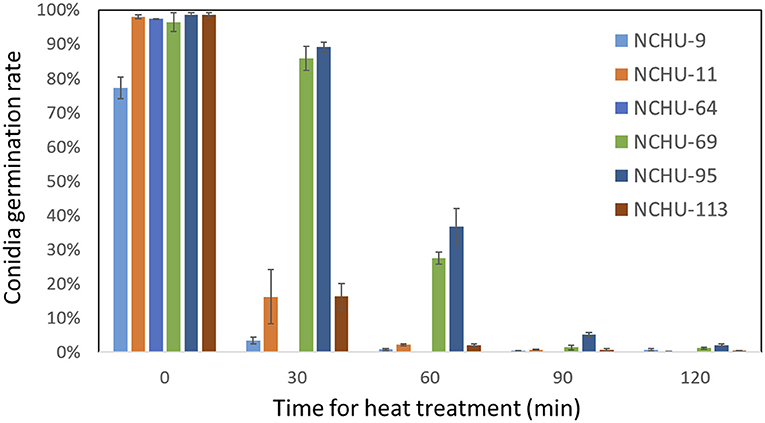
Figure 7. Thermotolerance of six fungal isolates under different durations of 45°C heat treatment. The experiments were replicated three times for each fungal isolate.
The conidial production and thermotolerance data was combined based on the ECN formula, which was first proposed in this study (Figure 8). The integration of conidial production and the stress coefficient (in the current study, an indicator of thermotolerance) could help us select a high-efficiency EPF for field application and further commercialization. For the five Metarhizium isolates, the ECN results indicated that M. anisopliae (NCHU-95) has the highest potential to be developed as a biopesticide, followed by M. anisopliae (NCHU-69) and M. pinghaense (NCHU-11). The isolate of B. australis (NCHU-113) showed a higher ECN than the five Metarhizium isolates, indicating that different species exhibit high variation in either conidial production or tolerance to environmental stresses (Figure 8). The PCA results revealed high coordination with the ECN results (Supplementary Figure 2), suggesting that the ECN formula could be used to evaluate the hierarchy of the viability-related parameters of fungi in the natural environment.
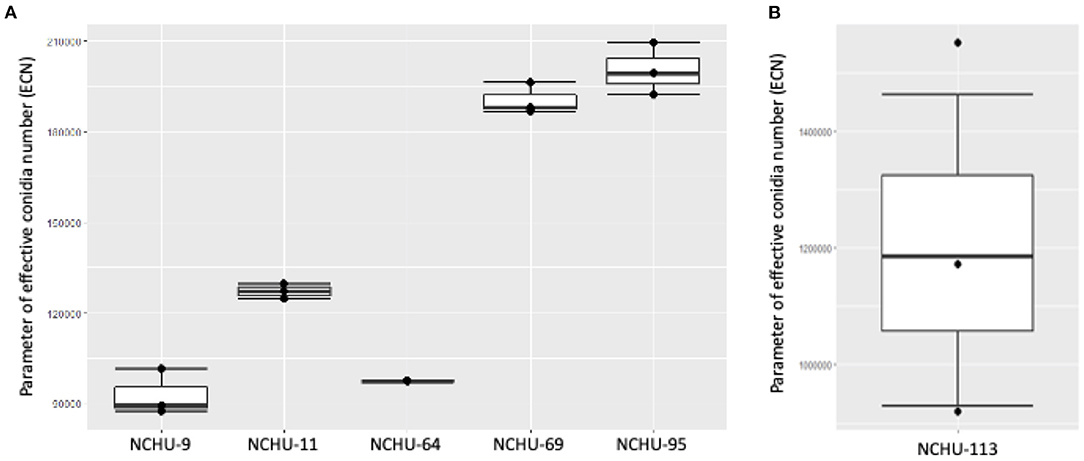
Figure 8. The effective conidia numbers (ECNs) of (A) Metarhizium spp. and (B) Beauveria australis six fungal isolates showed variation in thermotolerance and conidial production among these isolates.
In this study, an EPF library was established in Taiwan, and this collection was named the National Chung Hsing University (NCHU) EPF library. According to our data, most fungal isolates belonging to the genus Metarhizium showed high virulence against T. molitor larvae, while the virulence of some fungal species, such as Fusarium spp. (NCHU-53, 62, 63, 104, 105, and 115), varied (Figure 3). Although the genus Fusarium is usually considered an important plant pathogen, many studies have reported pathogenicity tests of Fusarium isolates from insects, soil, and vegetables against one or more species of insects. Mortality varies (5 to 100%) between different isolates against different insects (Santos et al., 2020). Therefore, the pathogenicity of Fusarium lineages to insect species has been noted in recent years; however, more data should be provided to prove the specificity of these Fusarium for host insects and the side effects and safety with regard to other non-target organisms before fully exploiting Fusarium for controlling insect pests (Santos et al., 2020). In fact, EPF infect their hosts mainly through adhesion to the insect cuticle. The penetration of EPF is related to several enzymes, such as lipases, proteases, and chitinases (Yang et al., 2016, Lee et al., 2019). It has been reported that Metarhizium species showed high activities of cuticle penetration-related enzymes and had high virulence against different insect hosts under laboratory conditions (Golo et al., 2015, Kim et al., 2018), indicating that the amounts of cuticle penetration-related enzymes among different isolates of the same species or different species of EPF showed a positive correlation with the pathogenesis of the EPF. Although most EPF show a broad host range, virulence factors for specific hosts still need to be further confirmed.
By following the previously reported “T. molitor-mediated EPF library construction method” (Kim et al., 2018), we established an EPF library from soil in Taiwan. This method allowed for high efficiency of fungal isolation, and the insect bait revealed the specificity of the EPF isolates. It was reported that dodine, which is a common antibiotic used for EPF isolation, could suppress the growth and sporulation of fungal colonies at high concentrations (Rangel et al., 2010), while other antibiotic-based fungal isolate platforms might also result in the overgrowth of opportunistic soil fungi on selective media supplemented with antibiotics (Lacey, 1997, Kim et al., 2018). Compared to the “T. molitor-mediated EPF library construction method,” the antibiotic-based fungal isolate platform showed lower fungal isolation efficiency. Another concern for antibiotic-based fungal isolate platforms is that fungi isolated by these platforms show lower insect host pathogenicity (Kim et al., 2018). Therefore, several studies have also reported the alteration of the EPF isolation method to the insect-bait method, such as applying G. mellonella (Lepidoptera: Pyralidae) larvae and T. molitor (Coleoptera: Tenebrionidae) for screening indigenous EPF in native soil samples (Vänninen et al., 2000, Tuininga et al., 2009, Barra et al., 2013, Kim et al., 2018). Indeed, the use of yellow mealworm larvae was proven to yield EPF from soil samples in the current study and could be a highly efficient method for the establishment of an EPF library under laboratory conditions.
By using a similar method for isolating EPF from Taiwan and Korea, the majority of EPF isolates from both countries were M. anisopliae. According to previous reports, the most frequently isolated EPF from Korean soil samples were B. bassiana, I. fumosorosea, M. anisopliae, I. javanicus, and Pochonia bulbillosa (Shin et al., 2013), while based on our molecular identification results, most of the fungal isolates belonged to the genus Metarhizium, followed by the genera Fusarium, Beauveria, and Clonostachys, indicating that the soil of the various locations and environments had different microbiota diversity and richness (Shin et al., 2013, Kim et al., 2016) (Figure 2). As the sample sites in this study shared similar environmental characteristics, the different diversity and richness of the microbiota in different locations might be linked to the different populations of insect hosts in these natural environments. In addition, it has also been suggested that the frequent isolation of certain EPF may be due to their high fitness in the environment (i.e., high tolerance to temperatures, dryness, nutritional limitation, etc.) (Kim et al., 2018). This fungal diversity in soil should be further investigated with regard to microorganism resource investigations in the future.
A total of six promising fungal isolates, which belonged to the genera Metarhizium (NCHU-9, 11, 64, 69, and 95) and B. australis (NCHU-113), revealed rapid killing activity against S. litura larvae and were subject to further evaluation in terms of their conidial production and thermotolerance. Coordinately, the five Metarhizium isolates also showed high virulence against T. molitor, indicating that Metarhizium isolates shared virulence against coleopteran and lepidopteran insects. Therefore, the spectrum of EPF virulence against their hosts could extend between different orders of insects. As mentioned above, different EPF isolates or species showed variations in the activities of cuticle penetration-related enzymes, which might influence the pathogenesis of EPF. The different times required to kill insect hosts might also be correlated with the timing or level of cuticle penetration-related enzyme expression. Additionally, one B. australis (NCHU-113) also showed fast-killing activity (LT50= 1.23) against S. litura larvae (Figure 4), suggesting that the species of EPF expressed these enzymes at earlier times and higher levels during the fungal infection process than other fungal isolates or species. On the other hand, insect immune mechanisms are also one of the key factors blocking EPF infection (i.e., the Toll pathway), and this issue is worthy of investigation in the future.
With the increasing need for biopesticides, EPF will play a critical role in IPM worldwide. Conidia play a major role in the infectious stage against crop pests and require special consideration in terms of the current market for biopesticides (Muniz-Paredes et al., 2017, Zulfiana et al., 2020). However, there is a serious bottleneck in the mass production of conidia; thus, successful mass production of fungal conidia is important for commercial purposes and field applications. According to our data, conidial production varied among the different isolates of Metarhizium. Therefore, it is possible to select a high conidia production EPF strain under laboratory conditions to facilitate mass production. It was interesting that the conidia production varied among the different isolates of Metarhizium. It has been reported that genes involved in conidiogenesis, such as a regulator of G protein signaling (cag8), may have different strain- or species-specific expression levels, which could influence conidial production (Fang and Bidochka, 2006). For commercialization purposes, the shelf-life of EPF products is important for field applications (Kim et al., 2011). Moreover, EPF applied in the biological control of agricultural pests may suffer from environmental stresses, such as elevated temperatures, especially in tropical or subtropical areas (Alali et al., 2019). Therefore, the selection of an EPF with high thermotolerance is the first step in the development of biocontrol agents. Based on our thermotolerance assay, four fungal isolates showed higher thermotolerance than others; among them, three Metarhizium isolates (NCHU-9, 69, and 95) showed strong thermotolerance, even after exposure to 45°C for up to 60 min. Most of the fungal isolates in this study were isolated from northern Taiwan, which is located in a subtropical area, and the strong thermotolerance of Metarhizium isolates might also be a consequence of environmental domestication. It was also noted that these four fungal isolates (NCHU-9, 69, 95, and 113) also showed high virulence against either T. molitor or S. litura larvae; therefore, fungal isolation in Taiwan should be continued to discover more valuable EPF for biological control.
The conidial production and thermotolerance data were combined based on the ECN formula, which was first proposed in this study. To our knowledge, this is the first report that attempted to combine two sets of parameters to generate the ECN under laboratory conditions. As mentioned above, there are multiple circumstances to consider for successful biocontrol in the natural environment; therefore, based on the ECN formula, the stress coefficient could be extended (i.e., the addition of UV stress) to better estimate field conditions.
In this study, we constructed an entomopathogenic fungal library (EFLib) via the “Tenebrio molitor pathogenicity-based fungal collection method (TmPC)” in the Insect Pathology Laboratory of National Chung Hsing University. The results showed that this procedure provided a useful and efficient strategy for either the development of fungal insecticides for pest management or for further research purposes. The characteristics of EPF, such as their conidial production and thermotolerance, should be considered before commercialization; however, some EPF strains might have high conidial production but lower tolerance to environmental stresses. Herein, based on the ECN formula first proposed in this study, potential fungal strains for use in the natural environmental could be selected. Moreover, the flexibility of the “stress coefficient” was demonstrated, showing that more stresses could be accounted for depending on the natural conditions (i.e., addition of UV stress). To our knowledge, this is the first attempt to combine two kinds of parameters to generate an ECN under laboratory conditions to better estimate field conditions.
The original contributions presented in the study are included in the article/Supplementary Material, further inquiries can be directed to the corresponding author/s.
J-CC collected samples, isolated fungi, performed the molecular identification, and bioassay. S-SW insect rearing and performed the bioassay. Y-HY insect rearing and performed the bioassay. Y-FT performed the thermotolerance and conidia production assay. Y-HL analyzed the data. C-TT collected samples, isolated fungi, and reviewed the manuscript. L-CT insect rearing and reviewed the manuscript. Y-SN designed the experiments and wrote the manuscript. All authors contributed to the article and approved the submitted version.
This research was supported by grant 109-2313-B-005−048 -MY3 from the Ministry of Science and Technology (MOST).
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2021.596316/full#supplementary-material
Supplementary Figure 1. The experimental flowchart in this study. The numbers in parentheses indicate the amount of fungal isolates for the experiments.
Supplementary Figure 2. Principal component analysis (PCA) of six fungal isolates based on thermotolerance and conidial production data among these isolates. The numbers in parentheses in the axis labels indicate the percentage of variance explained.
Supplementary Table 1. The sequences of entomopathogenic fungi from the NCBI Genbank.* Ex-type strains are in bold.
Supplementary Table 2. The mortality rates obtained with primary pathogenicity screening of fungal isolates.
Supplementary Table 3. Genbank accession numbers of entomopathogenic fungi in this study.
Supplementary Table 4. Morphological data of four entomopathogenic fungal type strains.
Ahmad, M., and Mehmood, R. (2015). Monitoring of resistance to new chemistry insecticides in Spodoptera litura (Lepidoptera: Noctuidae) in Pakistan. J. Econ. Entomol. 108, 1279–1288. doi: 10.1093/jee/tov085
Alali, S., Mereghetti, V., Faoro, F., Bocchi, S., Al Azmeh, F., and Montagna, M. (2019). Thermotolerant isolates of Beauveria bassiana as potential control agent of insect pest in subtropical climates. PLoS ONE 14:e0211457. doi: 10.1371/journal.pone.0211457
Barra, P., Rosso, L., Nesci, A., and Etcheverry, M. (2013). Isolation and identification of entomopathogenic fungi and their evaluation against Tribolium confusum, Sitophilus zeamais, and Rhyzopertha dominica in stored maize. J. Pest. Sci. 86, 217–226. doi: 10.1007/s10340-012-0460-z
Fang, W., and Bidochka, M. J. (2006). Expression of genes involved in germination, conidiogenesis and pathogenesis in Metarhizium anisopliae using quantitative real-time RT-PCR. Mycol. Res. 110, 1165–1171. doi: 10.1016/j.mycres.2006.04.014
Gardner, W. A., Eilenberg, J., and Glare, T. (2005). “Entomopathogenic fungi and their role in regulation of insect populations,” in Insect Control, Biological and Synthetic Agents, eds by L. I. Gilbert and S. S. Gill (London).
Ghumare, S. S., and Mukherjee, S. N. (2003). Performance of Spodoptera litura Fabricius on different host plants: influence of nitrogen and total phenolics of plants and mid-gut esterase activity of the insect. Indian J. Exp. Biol. 41, 895–899.
Gilbert, L. I., and Gill, S. S. (2010). Insect Control: Biological and Synthetic Agents. San Diego: Academic Press.
Golo, P. S., Santos, H. A., Perinotto, W. M., Quinelato, S., Angelo, I. C., Camargo, M. G., et al. (2015). The influence of conidial Pr1 protease on pathogenicity potential of Metarhizium anisopliae senso latu to ticks. Parasitol. Res. 114, 2309–2315. doi: 10.1007/s00436-015-4426-y
Gulsar, B. J., Subahasan, K., and Lyer, R. (2004). Occurrence and distribution of entomopathogenic nematodes in white grub endemic areas of Kerala. J. Plant Crops 32: 333–334. Available online at: http://scholar.google.com/scholar_lookup?&title=Occurrence%20and%20distribution%20of%20entomopathogenic%20nematodes%20in%20white%20grub%20endemic%20areas%20of%20Kerala&journal=J%20Plant%20Crops&volume=32&pages=333-334&publication_year=2004&author=Gulsar%20Banu%2CJ&author=Subahasan%2CK&author=Iyer%2CR
Gürlek, S., Sevim, A., Sezgin, F. M., and Sevim, E. (2018). Isolation and characterization of Beauveria and Metarhizium spp. from walnut fields and their pathogenicity against the codling moth, Cydia pomonella (L.) (Lepidoptera: Tortricidae). Egypt. J. Biol. Pest Control 28:50. doi: 10.1186/s41938-018-0055-y
Holder, D. J., and Keyhani, N. O. (2005). Adhesion of the entomopathogenic fungus Beauveria (Cordyceps) bassiana to substrata. Appl. Environ. Microbiol. 71, 5260–5266. doi: 10.1128/AEM.71.9.5260-5266.2005
Jackson, T. A., Alves, S. B., and Pereira, R. M. (2000). Success in Biological Control of Soildwelling Insects by Pathogens and Nematodes. London: Springer. doi: 10.1007/978-94-011-4014-0_9
Kepler, R. M., Luangsa-Ard, J. J., Hywel-Jones, N. L., Quandt, C. A., Sung, G. H., Rehner, S. A., et al. (2017). A phylogenetically-based nomenclature for Cordycipitaceae (Hypocreales). IMA Fungus 8, 335–353. doi: 10.5598/imafungus.2017.08.02.08
Kim, J. C., Lee, M. R., Kim, S., Lee, S. J., Park, S. E., Nai, Y. S., et al. (2018). Tenebrio molitor-mediated entomopathogenic fungal library construction for pest management. J. Asia-Pacific Entomol. 21, 196–204. doi: 10.1016/j.aspen.2017.11.018
Kim, J. S., Kassa, A., Skinner, M., Hata, T., and Parker, B. L. (2011). Production of thermotolerant entomopathogenic fungal conidia on millet grain. J. Ind. Microbiol. Biotechnol. 38, 697–704. doi: 10.1007/s10295-010-0850-2
Kim, S., Lee, S. J., Nai, Y. S., Yu, J. S., Lee, M. R., Yang, Y. T., et al. (2016). Characterization of T-DNA insertion mutants with decreased virulence in the entomopathogenic fungus Beauveria bassiana JEF-007. Appl. Microbiol. Biotechnol. 100, 8889–8900. doi: 10.1007/s00253-016-7734-y
Kumar, S., Stecher, G., and Tamura, K. (2016). MEGA7: Molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. doi: 10.1093/molbev/msw054
Lacey, L. A., Grzywacz, D., Shapiro-Ilan, D. I., Frutos, R., Brownbridge, M., and Goettel, M. S. (2015). Insect pathogens as biological control agents: back to the future. J. Invertebr. Pathol. 132, 1–41. doi: 10.1016/j.jip.2015.07.009
Lee, S. J., Yang, Y. T., Kim, S., Lee, M. R., Kim, J. C., Park, S. E., et al. (2019). Transcriptional response of bean bug (Riptortus pedestris) upon infection with entomopathogenic fungus, Beauveria bassiana JEF-007. Pest Manag. Sci. 75, 333–345. doi: 10.1002/ps.5117
Meyling, N. V., and Eilenberg, J. (2007). Ecology of the entomopathogenic fungi Beauveria bassiana and Metarhizium anisopliae in temperate agro-ecosystems: potential for conservation biological control. Biol. Control 43, 145–155. doi: 10.1016/j.biocontrol.2007.07.007
Muniz-Paredes, F., Miranda-Hernandez, F., and Loera, O. (2017). Production of conidia by entomopathogenic fungi: from inoculants to final quality tests. World J. Microbiol. Biotechnol. 33:57. doi: 10.1007/s11274-017-2229-2
Nicholas, K. B., and Nicholas, H. B. J. (1997). Genedoc: A Tool for Editing and Annoting Multiple Sequence Alignments. Embnet.news.
Otuka, A., Matsumura, M., and Tokuda, M. (2020). Dispersal of the common cutworm, spodoptera litura, monitored by searchlight trap and relationship with occurrence of soybean leaf damage. Insects 11:427. doi: 10.3390/insects11070427
Ou-Yang, S. C., and Chu, Y. I. (1988). The comparison of the development of the tobacoo cutworm (Spodoptera litura (F.)) reared with natural and artifical diets. Formosan Entomol. 8, 143–150.
Quandt, C. A., Kepler, R. M., Gams, W., Araujo, J. P., Ban, S., Evans, H. C., et al. (2014). Phylogenetic-based nomenclatural proposals for Ophiocordycipitaceae (Hypocreales) with new combinations in Tolypocladium. IMA Fungus 5, 121–134. doi: 10.5598/imafungus.2014.05.01.12
Rangel, D. E. N., Dettenmaier, S. J., Fernandes, E. K. K., and Roberts, D. W. (2010). Susceptibility of Metarhizium spp. and other entomopathogenic fungi to dodine-based selective media. Biocontr. Sci. Technol. 20, 375–389. doi: 10.1080/09583150903518370
Rehner, S. A., and Buckley, E. (2005). A Beauveria phylogeny inferred from nuclear ITS and EF1-α sequences: evidence for cryptic diversification and links to Cordyceps teleomorphs. Mycologia 97, 84–98. doi: 10.3852/mycologia.97.1.84
Santos, A. C. S., Diniz, A. G., Tiago, P. V., and Oliveira, N. T. (2020). Entomopathogenic Fusarium species: a review of their potential for the biological control of insects, implications and prospects. Fungal Biol. Rev. 34, 41–57. doi: 10.1016/j.fbr.2019.12.002
Shin, T. Y., Lee, W. W., Ko, S. H., Choi, J. B., Bae, S. M., Choi, J. Y., et al. (2013). Distribution and characterization of entomopathogenic fungi from Korean soils. Biocont. Sci. Technol. 23, 288–304. doi: 10.1080/09583157.2012.756853
Thompson, J. D., Gibson, T. J., Plewniak, F., Jeanmougin, F., and Higgins, D. G. (1997). The CLUSTAL_X windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res. 25, 4876–4882. doi: 10.1093/nar/25.24.4876
Tuininga, A. R., Miller, J. L., Morath, S. U., Daniels, T. J., Falco, R. C., Marchese, M., et al. (2009). Isolation of entomopathogenic fungi from soils and Ixodes scapularis (Acari: Ixodidae) ticks: prevalence and methods. J. Med. Entomol. 46, 557–565. doi: 10.1603/033.046.0321
Vänninen, I., Tyni-Juslin, J., and Hokkanen, H. (2000). Persistence of augmented Metarhizium anisopliae and Beauveria bassiana in Finnish agricultural soils. Biocontr. Sci. Technol. 45, 201–222. doi: 10.1023/A:1009998919531
White, T. J., Bruns, T., Lee, S. J. W. T, and Taylor, J. W. (1990). “Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics,” in PCR Protocols: A Guide to Methods and Applications, Vol. 18, eds M. A. Innis, D. H. Gelfand, J. J. Sninsky, and T. J. White (London: Academic Press), 315–322. doi: 10.1016/B978-0-12-372180-8.50042-1
Wraight, S. P., and Carruthers, R. I. (1999). Biopesticides: Use and Delivery, Vol. 5. New Jersey, NY: Humana Press.
Wu, J., Yu, X., Wang, X., Tang, L., and Ali, S. (2019). Matrine enhances the pathogenicity of Beauveria brongniartii against Spodoptera litura (Lepidoptera: Noctuidae). Front. Microbiol. 10:1812. doi: 10.3389/fmicb.2019.01812
Xue, M., Pang, Y. H., Wang, H. T., Li, Q. L., and Liu, T. X. (2010). Effects of four host plants on biology and food utilization of the cutworm, Spodoptera litura. J. Insect. Sci. 10:22. doi: 10.1673/031.010.2201
Yang, Y. T., Lee, S. J., Nai, Y. S., Kim, S., and Kim, J. S. (2016). Up-regulation of carbon metabolism-related glyoxylate cycle and toxin production in Beauveria bassiana JEF-007 during infection of bean bug, Riptortus pedestris (Hemiptera: Alydidae). Fungal Biol. 120, 1236–1248. doi: 10.1016/j.funbio.2016.07.008
Keywords: entomopathogenic fungi, Tenebrio molitor, Spodoptera litura, Beauveria, Metarhizium, effective conidia number
Citation: Chang J-C, Wu S-S, Liu Y-C, Yang Y-H, Tsai Y-F, Li Y-H, Tseng C-T, Tang L-C and Nai Y-S (2021) Construction and Selection of an Entomopathogenic Fungal Library From Soil Samples for Controlling Spodoptera litura. Front. Sustain. Food Syst. 5:596316. doi: 10.3389/fsufs.2021.596316
Received: 19 August 2020; Accepted: 11 January 2021;
Published: 09 February 2021.
Edited by:
Adam C. N. Wong, University of Florida, United StatesReviewed by:
Komivi Senyo Akutse, International Centre of Insect Physiology and Ecology (ICIPE), KenyaCopyright © 2021 Chang, Wu, Liu, Yang, Tsai, Li, Tseng, Tang and Nai. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Yu-Shin Nai, eXNuYWlAbmNodS5lZHUudHc=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.