- 1LSTM, Univ. Montpellier, IRD, CIRAD, INRAE, SupAgro, Montpellier, France
- 2LMI RICE-2, Univ. Montpellier, IRD, CIRAD, AGI, USTH, Hanoi, Vietnam
- 3IPME, Univ. Montpellier, CIRAD, IRD, Montpellier, France
- 4AMAP, Univ. Montpellier, IRD, CIRAD, INRAE, CNRS, Montpellier, France
- 5Sorbonne Université, UPEC, CNRS, IRD, INRA, Institut d'Écologie et des Sciences de l'Environnement, IESS, Bondy, France
- 6Eco&Sols, Univ. Montpellier, CIRAD, INRAE, IRD, SupAgro, Montpellier, France
Intensive coffee production is accompanied by several environmental issues, including soil degradation, biodiversity loss, and pollution due to the wide use of agrochemical inputs and wastes generated by processing. In addition, climate change is expected to decrease the suitability of cultivated areas while potentially increasing the distribution and impact of pests and diseases. In this context, the coffee microbiota has been increasingly studied over the past decades in order to improve the sustainability of the coffee production. Therefore, coffee associated microorganisms have been isolated and characterized in order to highlight their useful characteristics and study their potential use as sustainable alternatives to agrochemical inputs. Indeed, several microorganisms (including bacteria and fungi) are able to display plant growth-promoting capacities and/or biocontrol abilities toward coffee pests and diseases. Despite that numerous studies emphasized the potential of coffee-associated microorganisms under controlled environments, the present review highlights the lack of confirmation of such beneficial effects under field conditions. Nowadays, next-generation sequencing technologies allow to study coffee associated microorganisms with a metabarcoding/metagenomic approach. This strategy, which does not require cultivating microorganisms, now provides a deeper insight in the coffee-associated microbial communities and their implication not only in the coffee plant fitness but also in the quality of the final product. The present review aims at (i) providing an extensive description of coffee microbiota diversity both at the farming and processing levels, (ii) identifying the “coffee core microbiota,” (iii) making an overview of microbiota ability to promote coffee plant growth and to control its pests and diseases, and (iv) highlighting the microbiota potential to improve coffee quality and waste management sustainability.
Introduction
The coffee tree is a perennial plant belonging to the Rubiaceae family. The Coffea genus consists of c.a. one hundred species, but only Coffea arabica, C. canephora, and C. liberica are used for beverage production, the two formers representing around 70 and 30% of the world production, respectively (Davis et al., 2006; Vieira et al., 2006). Coffea arabica is native from the Ethiopian's highlands, between 1,300 and 2,000 m above the sea level, whereas the origin of C. canephora is more dispersed across the African tropical areas below 1,000 m (Wintgens, 2004). Coffee is the second most consumed beverage after water and the most traded tropical agricultural commodity (Mussatto et al., 2011; FAO, 2018). Around 25 million smallholder producers, especially in developing countries, rely on the coffee sector for their livelihood (FAO, 2018; ICO, 2019a). By order of importance, the main producing countries in 2018/2019 are Brazil, Vietnam, Colombia, Indonesia, Honduras, Mexico, Guatemala, and Ivory Coast (ICO, 2019b). Although the production and consumption have followed fairly parallel increasing trends over the past 50 years (FAO, 2015), the coffee market is periodically characterized by an oversupply in years of optimal environmental conditions due to innovation in cultivation techniques and planting material leading to a price decrease trend (Ponte, 2002; Bitzer et al., 2008). Farmers that rely on a perennial crop such coffee for their livelihood cannot either easily change their land use or anticipate their income. Therefore, it is difficult for them to adapt to fluctuations in market and environmental conditions (Amamo, 2014; Schroth and Ruf, 2014).
In order to increase yield, the modernization of the coffee production has heavily engaged in the use of new varieties, the reduction of shade, and the increase of plant density and agrochemical inputs (Perfecto et al., 1996). Nowadays, coffee management strategies fall along an intensity continuum ranging from natural or managed forests with coffee plants grown under tree canopy, to trees artificially planted to provide shade up to open sunlight plantation (Moguel and Toledo, 1999; Rice, 1999; Gobbi, 2000; Jezeer et al., 2018; Otero-Jiménez et al., 2018). Nevertheless, the intensive coffee systems result in serious environmental contamination due to excessive use of inputs (DaMatta, 2004), higher soil degradation (Ataroff and Monasterio, 1997) and are linked with a loss of biodiversity compared to traditional coffee systems (Perfecto et al., 1996; Guillemot et al., 2018). After harvest, coffee cherries undergo several processing steps aiming at removing all the external part of the fruit in order to reduce the water content to a level compatible with storage. To do so, three different processing techniques (dry, semi-wet, and wet) are implemented depending on the species, the country, and the farm size (Brando, 2004; Cleves, 2004; Schwan et al., 2012). However, coffee processing generates several by-products and wastes that can represent source of environmental pollution (Chanakya and De Alwis, 2004; Haddis and Devi, 2008; Beyene et al., 2012; Awoke et al., 2016). Finally, the coffee crop is already facing the climate change. This will decrease the suitability of the cultivated areas (Bunn et al., 2015; Ovalle-Rivera et al., 2015) and potentially increase the distribution and the impact of pests and diseases (Ghini et al., 2008; Jaramillo et al., 2011; Groenen, 2018).
The adverse effects of coffee cultivation and processing on the environment highlight the importance of developing sustainable solutions in order to maintain growers' livelihood while limiting the environmental impact in the climate change context. Thus, the benefit of smart agronomical systems, such as agroforestry, are increasingly highlighted (Méndez et al., 2010; De Beenhouwer et al., 2013; Vaast et al., 2016; Sauvadet et al., 2019; Gomes et al., 2020). Moreover, some high-technology microbial inputs (biofertilizers and biopesticides) are prone to increase the performances of such systems (reviewed in Singh et al., 2016a,b). Indeed, it is now well-documented that some of the microorganisms interacting with plants are directly beneficial by promoting their growth or indirectly by acting as antagonist of their pathogens (Compant et al., 2005; Olanrewaju et al., 2017). Therefore, to address the challenges associated with a sustainable crop management, research focused on the plant-associated microbes has been increasingly developed during the last decades (Berg et al., 2016; Compant et al., 2019; Arif et al., 2020). Nowadays, there is a shift in the ways of understanding the relationships between macroorganisms and microorganisms leading to the “holobiont” concept (Rosenberg and Zilber-Rosenberg, 2013; Bordenstein and Theis, 2015). According to this concept, the plants can be considered as superorganisms composed by the plant and associated microorganisms, the latter acting as an entire component of the host fitness by playing a role in the mineral nutrition, hormone balance, and adaptation capacity to biotic and abiotic stresses (Lemanceau et al., 2017; Simon et al., 2019). The development in this research area that describes the microbial communities has been accompanied with the use of specific terms such as “microbiome” and “microbiota” whose definitions are still debated (Marchesi and Ravel, 2015; Berg et al., 2020). In the present review, the term microbiota refers to all microorganisms interacting in a specific environment while the term microbiome encompasses their structural elements, molecules (e.g., DNA, metabolites) as well as the environmental conditions associated with the microbiota as initially described by Whipps et al. (1988) and clarified by Berg et al. (2020).
The use of microbes in the coffee farming and industry is still poorly exploited despite its potential capacity to reduce the amount of chemical inputs, improve coffee quality, and increase the farmer income through sustainable certifications (Mithöfer et al., 2017). Moreover, engineering the plant microbiome/microbiota and taking it into account in the new plant breeding strategies could represent some promising approaches to sustainably maintain the productivity (Nogales et al., 2016; Orozco-Mosqueda et al., 2018; Arif et al., 2020). The present review intends to (i) describe the diversity of the microorganisms that make up the coffee microbiota by focusing on archaea, bacteria, and fungi, (ii) summarize the current knowledge on the use of microorganisms to promote the coffee plant growth as well as to control the pests and diseases, and (iii) give an overview of their potential incorporation in the coffee processing and by-products management.
Strategies Used to Study the Coffee Microbiota
The first mention of the microorganisms associated with coffee plants dates from the nineteenth century and the description of the arbuscular mycorrhizal fungi (AMF) colonizing the roots of C. arabica and C. liberica (Janse, 1897). Since then, two major approaches have been used to describe the coffee microbiota diversity in combination with numerous identification strategies allowing to identify microorganisms at varying taxonomic levels from the highest (e.g., kingdom and phylum) to the lowest (e.g., genus and species).
The first one is the culture-dependent approach involving the isolation and the purification of the microorganisms. In that case, some basic morphological identifications using staining and microscopy were frequently employed to identify mycorrhizal species (Caldeira et al., 1983; Bertolini et al., 2020) or filamentous fungi (Mislivec et al., 1983; Casas-Junco et al., 2018). The morphology was often combined with standard biochemical tests such as those analyzing carbon sources utilization and enzymatic assays to identify bacteria (Pederson and Breed, 1946; Teshome et al., 2017) and fungi including yeasts (Agate and Bhat, 1966; Ranjini and Raja, 2019). Some more complex biochemical tests were sometimes applied to confirm the microorganisms' identity such as the multilocus enzyme electrophoresis (MLEE) in the case of nitrogen-fixing (N-fixing) bacteria (Jimenez-Salgado et al., 1997; Fuentes-Ramírez et al., 2001), the fatty acid methyl esters gas chromatography (FAME-GC) for bacterial isolates (Vega et al., 2005; Silva et al., 2012; Miguel et al., 2013), and the matrix-assisted laser desorption ionization–time of flight–mass spectrometry (MALDI-TOF-MS) for several bacteria and yeasts (Martins et al., 2020).
Regarding the molecular-based methods, the DNA–DNA reassociation study was one of the first molecular methods employed by bacterial taxonomists to describe the relatedness between bacterial species since the 1960s (Goris et al., 2007). Up to now, it is still the gold standard to identify new species as well as to discriminate bacterial isolates at the lowest taxonomic levels such as species and strain (Stackebrandt and Goebel, 1994; Janda and Abbott, 2007; Lagier et al., 2015). This method was successfully employed to describe some N-fixing bacterial species associated with coffee (Jimenez-Salgado et al., 1997; Estrada-De Los Santos et al., 2001). With the development of the first-generation sequencing technologies, DNA sequence comparisons contributed in an unprecedented manner to the number of identified microbial species (Rossi-Tamisier et al., 2015; Franco-Duarte et al., 2019).
The amplification and sequencing of simple genetic markers such as the rDNA gene repeats like the 16S rDNA of bacteria, as well as the 18S or 26S/28S rDNA and the ITS of fungi, have been extensively used (Sakiyama et al., 2001; Masoud et al., 2004; Oliveira et al., 2013; Prates Júnior et al., 2019; Martins et al., 2020). Sometimes, some housekeeping genes like those coding the β-tubulin (Samson et al., 2004; De Almeida et al., 2019) and TEF-1α factor (Mulaw et al., 2010, 2013) were sequenced for fungal identifications. The combination of several sequences in the multilocus sequence typing was also used to increase the reliability of the identification (Peterson et al., 2005). More recently, the whole-genome sequencing using the next-generation sequencing (NGS) technology was also employed to sequence the genome of a lactic acid bacterial strain of Pediococcus acidilactici isolated during the coffee fermentation (Muynarsk et al., 2019).
The second methodology does not require cultivation of the microorganisms. During its early development, it consisted in pooling DNA extractions, amplifying some DNA markers regions, and then sequencing them after some separation techniques such as the denaturing gradient gel electrophoresis (DGGE) or the cloning of single sequences. This procedure was used to study the endophytes in coffee cherries (Oliveira et al., 2013) and AMF (endophytic symbiotic fungi), colonizing the roots (Mahdhi et al., 2017) as well as the bacteria and fungi present during different coffee processing techniques (Vilela et al., 2010; Feng et al., 2016).
Nowadays, the culturable-independent strategy is increasingly used. The development of the NGS technologies also allows performing metabarcoding analyses involving the amplification and sequencing of specific marker genes to identify a whole community in an environmental DNA sample without the need of cloning or separation steps (Santos et al., 2020). For example, De Beenhouwer et al. (2015a,b) were among the first to use NGS in order to highlight the differences of AMF communities across a gradient of coffee management intensity. Then, two metabarcoding studies, describing the bacterial inhabitants of the coffee rhizosphere under organic or conventional cropping, were also performed (Caldwell et al., 2015; Rodríguez et al., 2020). In a recent work, Lamelas et al. (2020) examined the bacterial communities present in the C. arabica rhizosphere, in parasitic root-knot nematodes (females and eggs) as well as in healthy and nematode-infected coffee roots in order to determine the specific microbial assemblages correlated with the infection by Meloidogyne enterolobii and M. paranaensis. In another recent study, a metabarcoding analysis also highlighted the influence of edaphic and topographical factors on the bacterial and fungal communities associated with both rhizosphere and cherries of C. arabica (Veloso et al., 2020). Other authors also studied the fungi associated with C. arabica leaves infected by Hemileia vastatrix, the causal agent of the coffee leaf rust (CLR), with the aim to identify some potential mycoparasites (James et al., 2016). Recently, Fulthorpe et al. (2020) investigated both fungal and bacterial endophytes in C. arabica roots across a climatic gradient (temperature and humidity) in full sun and agroforestry cropping systems in Costa Rica and Nicaragua. Futhermore, the metabarcoding approach was also used to study the microbial (bacteria and fungi) communities linked with several postharvest processing steps and their impacts on coffee quality (De Bruyn et al., 2017; De Oliveira Junqueira et al., 2019; Zhang et al., 2019b; Elhalis et al., 2020a,b). Finally, the NGS approach involving the random sequencing of the fragmented DNA extract (shotgun sequencing) now allows to study the microbial diversity and to predict associated genes' function. This technique was used to perform a metagenomic analysis and to decipher the functional characteristics of the microbial communities found during C. arabica bean fermentation (Zhang et al., 2019a).
As usual, it is important to highlight that each approach displays its own strengths and weaknesses. On the one hand, the culture-dependent strategy allows isolating the microorganisms and further characterizing their biochemical and functional traits. However, it is laborious and time consuming with a limited capacity to cover the whole diversity of microorganisms because it is dependent of many parameters such as the culture media employed. Indeed, the concept of “unculturable microorganisms” was highlighted in the early twentieth century with the finding that there was far less colony able to grow on the medium than the number of cells observed by microscopy (Amann, 1911 in Ghosh and Bhadury, 2019). Nevertheless, this limit can now be bypassed with the use of various culture media leading to the development of the “culturomics” (Lagier et al., 2012).
On the other hand, the culture-independent approach is more labor/cost effective in studying the diversity of microorganisms as it allows identifying the uncultivable ones. This strategy can also picture the relative abundance of the microorganisms in metabarcoding studies and the potential function of associated genes when the metagenomic strategy is used. Despite that a bias can be introduced by the DNA extraction step when studying the microbial relative abundance, the introduction of an artificial community (mock) and the improvement of the DNA extraction protocols can help to standardize the results (Berg et al., 2020). Another constraint is the difficulty to reach the lowest taxonomic levels because of the limited amplicons length with the second-generation sequencers (Johnson et al., 2019; De Corato, 2020; Santos et al., 2020). Indeed, most of the metabarcoding studies related to coffee microorganisms' diversity were performed with second-generation sequencing platforms (Roche 454 and Illumina MiSeq) that allow sequencing only a part (usually hypervariable regions) of markers such as the 16S rDNA for bacteria and 18S rDNA for fungi or only smaller markers such as the ITS for fungi (Caldwell et al., 2015; De Beenhouwer et al., 2015a,b; De Bruyn et al., 2017; De Oliveira Junqueira et al., 2019; Zhang et al., 2019a,b; Elhalis et al., 2020a,b; Fulthorpe et al., 2020; Lamelas et al., 2020; Rodríguez et al., 2020; Veloso et al., 2020). Moreover, it has already been demonstrated for bacteria that the partial sequence does not achieve the taxonomic resolution obtained with the full-length 16S rDNA (Johnson et al., 2019). By contrast, the last technologies (third and fourth generations) now allow generating longer sequences compared to the advent of NGS, but this is done at the expense of the quality due to a higher sequencing error rate (Kulski, 2016; De Corato, 2020). Thus, James et al. (2016) were the only ones to study the coffee microbiota using a third-generation sequencing platform (PacBio) and concluded that the error rate remained very low. However, the full capacity of the platform remained unexploited as they sequenced only the ITS1-5.8S-ITS2 region of rDNA (<1 kb). Finally, it is important to have in mind that the data generated during metabarcoding/metagenomic analyses need a both statistical and bioinformatical treatment and the algorithms used still need to be improved (Ghosh and Bhadury, 2019).
To conclude, it is worth noting that both culture-dependent and independent approaches remain complementary. In other words, it is of a great interest to decipher the microbial diversity through metabarcoding/metagenomic analyses because this allows a better understanding of the interactions between coffee and microorganisms. Furthermore, the microbial diversity is a relevant indicator of environmental changes. However, it is necessary to isolate the microorganisms (e.g., to screen beneficial capacities and also to develop some biotechnological applications); hence, more efforts are certainly required to develop the culturomics approaches with the coffee microbiota as it has already been established to characterize the human (Lagier et al., 2018) and plant microbiota (Sarhan et al., 2019).
Coffee Microbiota Diversity
The main objective of the present review is to make an extensive survey of the literature describing the microbiota associated with coffee plants, including mainly the archaeal, bacterial, and fungal kingdoms. The keywords used for database search were Coffee, Coffea, microbiome, microbiota, archaea, bacteria, fungi, yeast, endophytes, epiphyte, rhizosphere, plant growth promotion, PGPR, PGPB, PGPF, and PGPM (plant growth promoting rhizobacteria, bacteria, fungi, and microorganisms, respectively), biocontrol, BCA (biocontrol agent), sustainable, biological, postharvest, processing, fermentation, wastes, and by-products. The databases screened were PubMed, Google Scholar, Web of Science, and SciELO. In total, 234 publications were found with identifications at least at the genus level and a well-defined origin of the microorganisms (rhizosphere, episphere, endosphere or associated with the cherries, beans, and wastes during the postharvest processes). The full detailed dataset describing the microorganisms' origin (continent, country, Coffea species, the type of colonization, the plant organs, the type of postharvest processing), the identification strategies, the thresholds used to filter the identifications, the accession numbers (when available), and all the other analyses described in the scientific articles and their potential applications are provided in the Supplementary Table 1.
Then, the microbiota was further divided into two principal components. The first one is the indigenous microbiota composed of the microorganisms living in close association with the coffee plants, in the soil at the vicinity of the roots (rhizosphere), at the surface (episphere), and inside the plant tissues (endosphere). The second component is related to the postharvest microbiota that encompasses all the microorganisms associated with the coffee cherries postharvest processing and its by-products including the fermentation, the drying steps, and the wastes (husks, pulps, and wastewater).
The indigenous and the postharvest coffee microbiota have been relatively equally studied with 115 and 127 publications, respectively (Table 1). To the best of our knowledge, the overall coffee microbiota is covering 22 phyla, encompassing 129 orders, 607 genera, and 923 species mainly belonging to the bacterial and fungal kingdoms. Indeed, only two archaeal phyla (including four orders and five genera without any species identification) have been described (Supplementary Table 1). This is not surprising since archaea were discovered relatively recently in the microbiology history (Woese et al., 1978) and remain quite difficult to cultivate (Song et al., 2019).
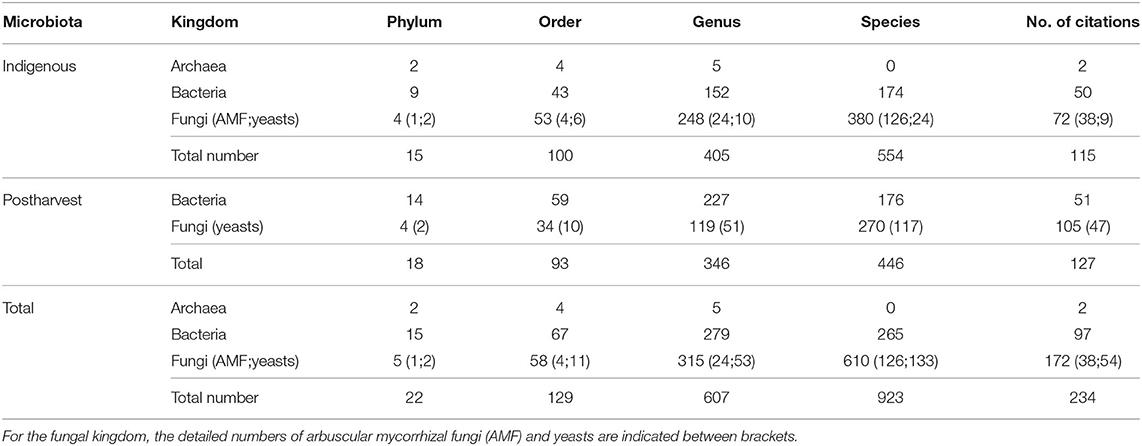
Table 1. Overview of the coffee microbiota with the numbers of phyla, orders, genera, species, and citations for the three kingdoms (archaea, bacteria, and fungi) constituting the coffee microbiota (indigenous and postharvest).
It is worth noting that our survey is a qualitative description of the coffee microbiota diversity. However, we tried to (i) give an insight into the relative abundances of the microorganisms based on their occurrence frequency in the literature and (ii) compare these results with the relative abundances identified through metabarcoding studies (when available).
The Indigenous Coffee Microbiota
Many biotic and abiotic factors influence the plant microbiota such as the soil physical–chemical characteristics (Fierer, 2017; Tkacz et al., 2020), the plant compartment (rhizosphere, episphere, and endosphere), and the organ studied (Compant et al., 2019; Berg et al., 2020; Tkacz et al., 2020). Moreover, the plant genotype is also believed to influence the microbial community (Patel et al., 2015; Mina et al., 2020). However, it is worth noting that most of the microorganisms associated with coffee have been described in C. arabica across 170 scientific articles while the remaining studies referred to C. canephora, C. liberica, and C. congensis or did not specified the Coffea species studied (Supplementary Table 1). Thereby, due to the multifactorial influence on the microbiota, we decided to split the indigenous microbiota in the following plant compartments, namely, the rhizosphere, the episphere, and the endosphere.
The Coffee Rhizospheric Microbiota
The soil represents an underestimated reservoir of microbial diversity for which a large part never been cultivated (Mendes et al., 2013). Plants are able to influence the diversity of microorganisms in their rhizosphere and to potentially select from the soil the beneficial ones through the production of root exudates (Hartmann et al., 2008; Mendes et al., 2013). As attested by the increasing use of the term “plant growth-promoting rhizobacteria” since its formulation by Kloepper and Schroth (1978), the rhizosphere microorganisms represent a well-known source of plant beneficial microorganisms able to improve the acquisition of nutrients as well as the resistance to biotic and abiotic stresses (Avis et al., 2008; De Zelicourt et al., 2013; Meena et al., 2017).
In the present review, we recorded 34 publications related to the coffee rhizospheric microbiota. Thirty-one used a culture-dependent approach, and only three studies employed a metabarcoding approach to describe the microorganisms in the coffee rhizosphere (Caldwell et al., 2015; Lamelas et al., 2020; Rodríguez et al., 2020). Based on this survey, we recorded that the rhizosphere microbiota diversity covers 12 phyla, 40 orders, 98 genera, and 58 species with as the most studied kingdoms the bacteria and fungi across 30 and 8 articles (Table 2).

Table 2. Rhizospheric archaea, bacteria, and fungi diversity including phyla, orders, and genera, as well as the numbers of species identified and citations.
The bacterial diversity is composed of eight phyla, 31 orders, 81 genera, and 42 species. The most encountered and diversified phylum is the Proteobacteria with 45 genera and 22 species described, with the Pseudomonas being the most diversified and common genus with 11 species identified across 14 publications. All studies that used the NGS to describe the coffee rhizosphere prokaryotic abundance and diversity also reported the dominance of the Proteobacteria phylum and the Pseudomonas genus (Caldwell et al., 2015; Lamelas et al., 2020; Rodríguez et al., 2020). The remaining diversity is distributed among the Acidobacteria, Actinobacteria, Bacteroidetes, Firmicutes, and Verrucomicrobia phyla.
The fungal diversity comprises three phyla, eight orders, 16 genera, and 16 species, the Ascomycota phylum being the most reported. Indeed, all the eight publications dealing with fungi in the coffee rhizosphere described some members of this phylum with 14 genera and 16 species identified. The most commonly identified genera belonging to this phylum are by number of citations Penicillium (6), Aspergillus (5), Fusarium (4), and Trichoderma (3). Even though there is no metabarcoding analysis of the fungal community in the coffee rhizosphere, it has already been reported for several other plant species that the rhizosphere is dominated by fungi from the Ascomycota phylum (Wang et al., 2017; Qiao et al., 2019; Schöps et al., 2020; Tkacz et al., 2020). By contrast, the other fungal phyla are underrepresented with only two isolates belonging to the Basidiomycota and Mucoromycota.
The Coffee Epiphytic Microbiota
In general, the plant epiphytic microbiota is composed of a high diversity of microorganisms able to attach and live on the surface of the above and belowground plant tissues (Vorholt, 2012; Newman and Cragg, 2020). This microhabitat requires a specific adaptation of the microorganisms to tolerate the particular environment at the surface of the plant tissues especially the leaves (Vorholt, 2012; Vandenkoornhuyse et al., 2015). Furthermore, the episphere is somehow considered as a boundary for the microorganisms limiting their establishment as endophytes (Vandenkoornhuyse et al., 2015).
Despite the fact that the episphere represents a source of plant beneficial microorganisms and bioactive compounds (Vandenkoornhuyse et al., 2015; Newman and Cragg, 2020), it is the least studied compartment of the indigenous coffee microbiota with only 19 articles (Table 3). All the reviewed studies used a culture-dependent approach to isolate the microorganisms at the surface of several coffee tissues, mainly from leaves (Vélez and Rosillo, 1995; Haddad et al., 2014) and cherries (Agate and Bhat, 1966; Compri et al., 2016), but also from roots (Velmourougane et al., 2000; Teshome et al., 2017) and stems (Velmourougane et al., 2000; Waller and Masaba, 2006). Two metabarcoding studies could have pictures the epiphytic communities at the surface of coffee leaves and cherries; however, the authors extracted DNA from the crushed organs making impossible to discriminate epiphytes from endophytes (James et al., 2016; Veloso et al., 2020) (Supplementary Table 1).

Table 3. Epiphytic bacteria and fungi diversity including phyla, orders, and genera, as well as the numbers of species identified and citations.
The coffee epiphytic diversity is composed of seven phyla, 25 orders, 52 genera, and 42 species (Table 3). The fungal kingdom is the most studied, encompassing 3 phyla, 15 orders, 34 genera, and 25 species. The most cited fungal phylum is the Ascomycota followed by the Basidiomycota and Mucoromycota and by citation number the genera Fusarium (6), Penicillium (5), and Aspergillus (4). Regarding the bacterial kingdom, it includes four phyla 10 orders, 18 genera, and 17 species, the Proteobacteria phylum being the most cited and diversified with 12 genera, 13 species and the Pseudomonas as the most commonly isolated genus.
The Coffee Endophytic Microbiota
Endophytic microorganisms are characterized by their capacity to colonize the internal part of the plant tissues without causing any negative symptoms to their host (Wilson, 1995; Hyde and Soytong, 2008). The endophytic lifestyle is therefore characterized by microorganisms spending only a part up to their entire life cycle within the plant tissues (Hardoim et al., 2015). It is worth noting that some endophytes can be vertically transmitted while other are characterized by diverse colonization patterns (Saikkonen et al., 2004; Hardoim et al., 2015; Frank et al., 2017). It is also believed that the internal colonization capacity allows endophytes to be less affected by soil condition fluctuations and by competition with other microorganisms (Santoyo et al., 2016). In addition, endophytes were reported to display plethora of activities that can be beneficial for the plants (Afzal et al., 2019; White et al., 2019; Yan et al., 2019).
It must be stressed that more than half of the studies related to coffee endophytes focused on AMF, which represent a particular class of endophytic symbiotic fungi belonging to the Mucoromycotina phylum and the Glomeromycotina sub-phylum (Spatafora et al., 2016). They are qualified of mutualistic obligate symbionts colonizing the plant roots while the external mycelium is foraging the soil to transfer some water and inorganic compounds (phosphorus, nitrogen, and other essential nutrients) to their host in exchange of a carbon source (Genre et al., 2020). Furthermore, their beneficial effects not only on the plant nutrition but also on their tolerance to biotic and abiotic stresses are now well-documented (Chen et al., 2018; Begum et al., 2019).
The endosphere is the most studied compartment of the indigenous coffee microbiota. We reviewed the content of 71 publications dealing with coffee endophytes, among which 65 employed a culture-dependent method to isolate bacterial and fungal endophytes from various C. arabica, C. canephora, and C. liberica tissues including cherries (Sakiyama et al., 2001; Vega et al., 2008; Miguel et al., 2013), leaves (Santamaría and Bayman, 2005; Bongiorno et al., 2016), roots (Raviraja et al., 1996; Jimenez-Salgado et al., 1997; Vega et al., 2006; Hoang et al., 2020; Duong et al., 2021), seeds (Vega et al., 2006; Duong et al., 2021), and stems (Vega et al., 2005, 2010). Basic culture-independent strategies were used in three studies to identify archaea, bacteria, and fungi including AMF inside the C. arabica roots and cherries (Oliveira et al., 2013; Mahdhi et al., 2017; Prates Júnior et al., 2019). Finally, three metabarcoding studies were also performed to study the AMF, endophytic bacteria, and fungi associated with C. arabica roots across some management and environmental gradients (De Beenhouwer et al., 2015a,b; Fulthorpe et al., 2020).
The coffee endophytic microbiota encompasses 12 phyla, 70 orders, 241 genera, and 350 species (Table 4). Fungi are the most studied microorganisms with a total 55 articles (38 related to AMF), followed by bacteria and archaea with 18 and 1 studies, respectively. The fungal kingdom is composed of four phyla, 39 orders, 149 genera, and 253 species. In terms of citations for the filamentous fungi and the yeasts, the Ascomycota phylum is by far the most studied with 18 citations, followed by the Basidiomycota (4), the Cryptomycota (1), and the Mucoromycota (1). This is also the case in terms of richness with 102 genera and 88 species belonging to the Ascomycota while the Basidiomycota phylum is represented by only 21 genera with two species identified. Finally, only one genus is reported for both the Cryptomycota and the Mucoromycota phyla with no species identified. In the Ascomycota phylum, the most cited genera by number of citations are Cladosporium (9), Colletotrichum (9), Aspergillus (6), Penicillium (6), Fusarium (6), and Trichoderma (6). It is noteworthy that Fulthorpe et al. (2020) also reported using a metabarcoding approach these genera in coffee roots from all the sites that they studied across a gradient of temperature and humidity. In the same study, these authors also highlighted that the genera Cladosporium and Penicillium represented more than 40 and 10% of all the fungal sequences, respectively. When focusing only on AMF, the most cited genera are Acaulospora (34), Gigaspora (30), Glomus (29), Claroideoglomus (18), Rhizophagus (18) Scutellospora (15), Ambispora (13), Funneliformis (12), Sclerocystis (11), Paraglomus (11), Archaeospora (10), and Entrophospora (10). The genera Glomus and Acaulospora are also the most diversified with 26 and 23 different species, respectively (see also Supplementary Table 2 for the complete list of species). Moreover, several metabarcoding studies also reported as dominant AMF associated with coffee the genera Glomus, Acaulospora, and Archaeospora under various management and environmental conditions (De Beenhouwer et al., 2015a,b; Fulthorpe et al., 2020).

Table 4. Endophytic archaea bacteria and fungi diversity including phyla, orders, and genera, as well as the numbers of species identified and citations.
Finally, the coffee endophytic bacteria diversity is composed of seven phyla, 28 orders, 88 genera, and 134 species. The most common bacterial phyla are the Firmicutes, Proteobacteria, and Actinobacteria with 15, 15, and 12 mentions in the literature, respectively. These phyla are also the most diversified with 42 genera and 61 species for the Proteobacteria, 28 genera and 39 species for the Actinobacteria, and 8 genera and 31 species for the Firmicutes. The most encountered genera in terms of number of citations are Bacillus (14), Enterobacter (8), Cedecea (6), Paenibacillus (6), Pseudomonas (6), and Pantoea (5). In addition, the authors of the only metabarcoding analysis dealing with endophytic bacteria also reported as dominant the genera Pantoea, Enterobacter, and Pseudomonas with a relative abundance of 17, 12, and 4% of all the bacterial sequences obtained from all the studied locations across a climatic gradient (Fulthorpe et al., 2020).
The Microbiota Associated With Coffee Postharvest Processes
Before discussing the microbiota associated with postharvest treatments, it is important to have in mind the various processes commonly used in coffee (reviewed in Brando, 2004; Cleves, 2004; Schwan et al., 2012). Independently of the Coffea species, all postharvest technics aim to remove all the external part of the cherries (exocarp, mesocarp, and endocarp) in order to produce green coffee beans to be commercialized. To do so, dry, semi-wet, and wet processes are implemented. The first one consists in drying the whole cherries and to mechanically remove the external parts (hulling) to obtain the green coffee beans. The two other methods involve the removal of the exocarp (skin) and a part of the mesocarp (pulp) of the fresh cherries leaving the beans with a remaining part of the mesocarp (mucilage). In the semidry process, the beans are then dried before the hulling (dry fermentation) while in the wet processing the mucilage layer is removed by a fermentation step in tanks (wet fermentation) before being dried and hulled. In the final steps, the coffee endocarp (“parchment” or “husk”) is removed to obtain the green coffee bean enveloped in its spermoderm (“silverskin”). Whatever the method used, it is well-known that the microorganisms play important roles during the postharvest processes, especially by degrading the mucilage layer during the fermentation (Agate and Bhat, 1966) but also by influencing either positively (Elhalis et al., 2020a) or negatively (Ndayambaje et al., 2019) the organoleptic quality of the final product as well as its safety with respect to the presence of mycotoxins (Urbano et al., 2001).
These are the reasons why the postharvest microbiota has been extensively studied in order to describe the diversity of microorganisms associated with the dry (Pasin et al., 2011; Evangelista et al., 2014) the semidry (Van Pee and Castelein, 1971; Silva et al., 2013) and the wet processes (Pederson and Breed, 1946; De Oliveira Junqueira et al., 2019). Furthermore, microorganisms associated with the beans during the storage (Mislivec et al., 1983; Ndayambaje et al., 2019) and with the wastes and by-products were also studied (Aquiahuatl et al., 1988; Pires et al., 2017; Oumer and Abate, 2018). These researches were conducted in order to better understand the role of the microbial component of the coffee processing and to develop some biotechnological applications to improve coffee quality as well as the wastes management sustainability.
Among the 127 publications related to the microbiota present after the harvest, most of them were performed using basic culture-dependent and independent methods. However, seven studies used NGS to perform some metabarcoding (one metagenomic) analyses of the bacterial and fungal communities associated with coffee fermentation (De Bruyn et al., 2017; De Carvalho Neto et al., 2018; De Oliveira Junqueira et al., 2019; Zhang et al., 2019a,b; Elhalis et al., 2020a,b).
Based on the survey of these studies, we can notice that the coffee postharvest microbiota is constituted by 18 phyla, 93 orders, 346 genera, and 446 species belonging to the bacterial and fungal kingdoms (Table 5). The fungi (including yeast) are the most studied microorganisms associated with the coffee postharvest steps with 105 references. This kingdom includes four phyla, 34 orders, 119 genera, and 270 species. In terms of citations, the most encountered fungal phyla are the Ascomycota (105), Mucoromycota (23), and Basidiomycota (19) and at the genus level the Aspergillus (67), Penicillium (45), Fusarium (37), Cladosporium (29), Mucor (16), and Rhizopus (16) focusing on filamentous fungi. Indeed, the presence of filamentous fungi has been extensively studied due to the presence of mycotoxins such the ochratoxins and aflatoxins (Joosten et al., 2001; Rezende et al., 2013). It is worth noting that all these toxigenic fungi belong to the Ascomycota phylum, the Eurotiales order, and the genera Aspergillus, Byssochlamys, and Penicillium with 23 toxigenic species identified to date (Supplementary Table 3). Regarding yeasts, the most cited genera are Pichia (28), Candida (26), Saccharomyces (21), Hanseniaspora (16), Torulaspora (15), Wickerhamomyces (13), Meyerozyma (12), and Debaryomyces (10). Furthermore, the yeast genera Pichia and Candida are also reported to be the dominant genera in term of relative abundance during the coffee fermentation across all the metabarcoding studies (De Bruyn et al., 2017; De Oliveira Junqueira et al., 2019; Zhang et al., 2019b; Elhalis et al., 2020a,b).

Table 5. Postharvest bacteria and fungi diversity including phyla, orders, and genera, as well as the numbers of species identified and citations.
Regarding the bacterial kingdom, it contains 14 phyla, 59 orders, 227 genera, and 176 species described across 51 publications. In matter of recurrence in the literature, the most cited are the Firmicutes (38), Proteobacteria (33), Actinobacteria (14), and the Bacteroidetes (7) at the phylum level and the Bacillus (21), Leuconostoc (20), Lactobacillus (18), Enterobacter (18), Pseudomonas (17), Klebsiella (16) Pantoea (12), Erwinia (11), Serratia (11), Lactococcus (10), and Acinetobacter (10) at the genus level. These findings are in accordance with the relative abundances registered using metabarcoding/metagenomic approach. Indeed, in such studies the lactic acid bacteria (LAB), especially from the genus Leuconostoc, and acetic acid bacteria (AAB) are often reported to be the dominant bacteria during the fermentation along with other bacterial genera (e.g., Bacillus, Erwinia, Pseudomonas) belonging to the Firmicutes and Proteobacteria phyla (De Bruyn et al., 2017; De Carvalho Neto et al., 2018; De Oliveira Junqueira et al., 2019; Zhang et al., 2019a,b; Elhalis et al., 2020a,b). Thus, a total of 22 species of AAB belonging to the genera Acetobacter, Gluconacetobacter, Gluconobacter, and Kozakia, as well as 23 species of LAB from the Bifidobacterium, Bacillus, Clostridium, Enterococcus, Lactobacillus, Lactococcus, Leuconostoc, Pediococcus, Streptococcus, and Weissella genera have been described during the coffee postharvest steps (see also Supplementary Table 4 for the complete list of species).
The Core Coffee Microbiota Related to Specific Functional Roles
The plant “core microbiota” can be described at different taxonomic, spatial, and temporal levels and is likely to contain highly competitive plant colonizers representing choice candidates for functional studies (Vandenkoornhuyse et al., 2015; Müller et al., 2016). Furthermore, it is hypothesized that the “core microbiota” is the result of a coevolution process in which the microbes have been selected to achieve essential functions for their hosts (Lemanceau et al., 2017; Compant et al., 2019).
In the framework of this review, we defined the “core coffee microbiota” corresponding to the microbial taxa (at genus and species levels) shared between (i) indigenous coffee microbiota plant compartments, namely, the rhizosphere, episphere, and endosphere (Figure 1; see also Supplementary Table 5 for the complete list of genera and species in each partitions), (ii) at least one indigenous coffee microbiota compartment and postharvest coffee microbiota (Figure 2; see also Supplementary Table 6 for the complete list of genera and species in each partitions), (iii) the continents (Figure 3; see also Supplementary Table 7 for the complete list of genera and species in each partitions), and (iv) all indigenous coffee microbiota plant compartments across all continents (Figure 4; see also Supplementary Table 8 for the complete list of genera and species in each partitions).
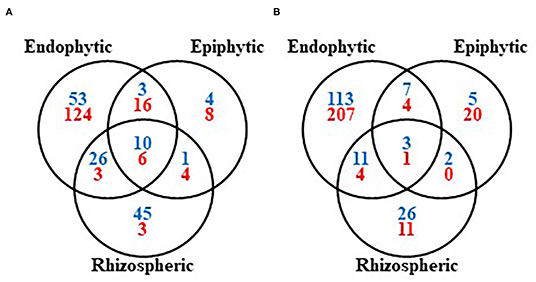
Figure 1. Venn diagrams of the archaea and bacteria (blue), and fungi (red) shared between endophytic, epiphytic, and rhizospheric compartments of the indigenous coffee microbiota. (A) At the genus level and (B) at the species level.
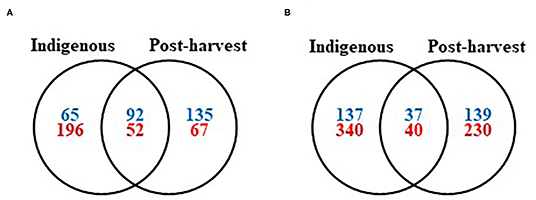
Figure 2. Venn diagrams of the archaea and bacteria (blue), and fungi (red) shared between at least one compartment of the indigenous coffee microbiota and the postharvest coffee microbiota. (A) At the genus level and (B) at the species level.
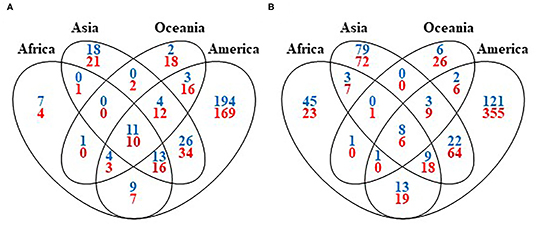
Figure 3. Venn diagrams of the archaea and bacteria (blue), and fungi (red) shared between continents. (A) At the genus level and (B) at the species level.
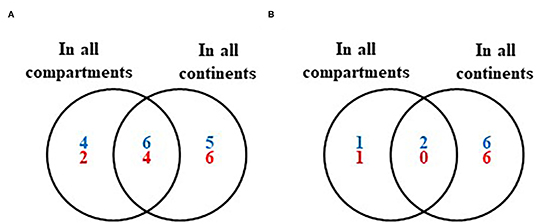
Figure 4. Venn diagrams of the bacteria (blue), and fungi (red) shared between all compartments of the indigenous coffee microbiota across all continents. (A) At the genus level and (B) at the species level.
Considering the taxa shared between the rhizosphere, endosphere, and episphere, a total of 10 bacterial genera (Bacillus, Burkholderia, Citrobacter, Enterobacter, Gluconacetobacter, Kocuria, Pseudomonas, Serratia, Stenotrophomonas, and Streptomyces) and three species (B. subtilis, P. putida, S. maltophilia), as well as six fungal genera (Aspergillus, Cladosporium, Cylindrocarpon, Fusarium, Penicillium, and Trichoderma), and one species (A. niger), are able to colonize all the coffee plant compartments (Figure 1; see also Supplementary Table 5). These microorganisms can therefore be considered as those presenting the highest competitiveness to colonize and survive in the coffee rhizosphere but also at the surface and inside coffee plant tissues/organs.
Regarding the microorganisms associated both to coffee plants in the field and during coffee processing, we highlighted 92 genera (32% of the total) and 37 species (12% of the total) for bacteria, as well as 52 genera (17% of the total) and 40 species (7% of the total) for the fungi, shared between the postharvest microbiota and at least one indigenous coffee microbiota compartment (Figure 2; see also Supplementary Table 6). Furthermore, 135 bacterial and 67 fungal genera, as well as 139 bacterial and 230 fungal species, were found only in the postharvest microbiota and are therefore introduced during the processing steps. These observations are particularly relevant regarding toxigenic fungi because among the 23 species found during postharvest processing, only 10 are member of indigenous microbiota. Indeed, the species A. flavus and A. ochraceus are also present as epiphytes while A. sclerotiorum, A. versicolor, A. westerdijkiae, P. crustosum, P. olsonii, and P. oxalicum as endophytes (Supplementary Table 3). Finally, A. niger and P. brevicompactum are present in all coffee plant compartments. Therefore, the presence of the other 13 toxigenic species could be avoided by taking stricter hygiene measures in the processing facilities and thus reducing the losses associated with fungal toxins contaminations.
Concerning the diversity between the coffee growing continents, 11 bacterial genera (Bacillus, Citrobacter, Enterobacter, Enterococcus, Erwinia, Klebsiella, Leuconostoc, Pantoea, Pseudomonas, Serratia, and Stenotrophomonas) and eighth species (B. megaterium, B. subtilis, E. cloacae, K. pneumoniae, L. mesenteroides, P. agglomerans, P. fluorescens, and S. maltophilia), together with 10 fungal genera (Aspergillus, Candida, Cladosporium, Fusarium, Hanseniaspora, Penicillium, Pichia, Rhodotorula, Torulaspora, and Wickerhamomyces) and six species (A. westerdijkiae, H. uvarum, P. brevicompactum, P. citrinum, R. mucilaginosa, and W. anomalus), are shared all across the coffee world (Figure 3; see also Supplementary Table 7). Thus, these microbial taxa, which are ubiquitous across continents, are able to adapt themselves to various environmental conditions.
Finally, a total of six bacterial genera (Bacillus, Citrobacter, Enterobacter, Pseudomonas, Serratia, and Stenotrophomonas) and two species (B. subtilis and S. maltophilia), as well as four fungal genera (Aspergillus, Cladosporium, Fusarium, and Penicillium), are present in all coffee plant compartments across all continents (Figure 4; see also Supplementary Table 8).
To summarize, the coffee microbiota contains some microorganisms that are either some competitive coffee plant colonizers or able to adapt themselves to various environmental conditions, and sometimes both. Therefore, these microorganisms should be further studied in order to exploit their capacities for the development of new biotechnological applications.
Biotechnological Applications of the Coffee Microbiota
Potential Uses as Plant Growth Promoting Agents
Coffee-associated microorganisms have been extensively studied for their potential use as plant-growth promoting agents including rhizospheric bacteria and fungi (Jimenez-Salgado et al., 1997; Posada et al., 2013; Kejela et al., 2016; Perea Rojas et al., 2019), epiphytic bacteria (Estrada-De Los Santos et al., 2001; Teshome et al., 2017), endophytic bacteria (Jimenez-Salgado et al., 1997; Silva et al., 2012) and AMF (Caldeira et al., 1983; Perea Rojas et al., 2019).
The microorganisms' capacity to promote the plant growth is often linked with the improvement of the plant mineral nutrition or the regulation of the plant hormonal balance (Egamberdieva et al., 2017; Kudoyarova et al., 2019; Aeron et al., 2020). Since nitrogen, phosphorus, and iron are among the most limiting nutrients for plants, the use of N-fixing, phosphorus-solubilizing (P-solubilizing), and siderophore-producing microorganisms could represent a sustainable strategy to reduce the reliance on chemical fertilizers (Vitousek et al., 2002; Scavino and Pedraza, 2013; Alori et al., 2017; Pahari et al., 2017; Prabhu et al., 2019; Smercina et al., 2019).
Thus, in vitro screenings were often used to highlight the microbiota potential capacity to improve the coffee plant nutrition. For example, the ability to fix the atmospheric nitrogen was demonstrated for some bacterial species associated with C. arabica roots belonging to the genera Gluconacetobacter (Jimenez-Salgado et al., 1997; Fuentes-Ramírez et al., 2001), Burkholderia (Estrada-De Los Santos et al., 2001), Azotobacter, Leifsonia, and Stenotrophomonas (Wedhastri et al., 2012). Another valuable microbial process is the improvement of nutrient availability. To this end, the capacity to solubilize the phosphorus has been demonstrated not only for numerous bacterial species associated with C. arabica, C. canephora, and C. liberica roots and seeds, belonging to the genera Acinetobacter, Aeromonas, Alcaligenes, Arthrobacter, Bacillus, Brachybacterium, Burkholderia, Caballeronia, Cellulomonas, Chromobacterium, Chryseobacterium, Chryseomonas, Citrobacter, Curtobacterium, Enterobacter, Gordonia, Kocuria, Luteibacter, Mycolicibacterium, Nocardia, Paenibacillus, Paraburkholderia, Pasteurella, Pseudomonas, Rhodococcus, Sphingomonas, Staphylococcus, Stenotrophomonas, and Vibrio (Baon et al., 2012; Muleta et al., 2013; Teshome et al., 2017; Duong et al., 2021) and also for some fungi from the genera Aspergillus, Chaetomium, Cladosporium, Cylindrocarpon, Fusarium, Humicola, Paecilomyces, and Penicillium (Posada et al., 2013; Perea Rojas et al., 2019). Another capacity is the iron mobilization through the production of siderophores as displayed by the bacterial genera Acinetobacter, Bacillus, Burkholderia, Caballeronia, Cellulomonas, Enterobacter, Escherichia, Luteibacter, Mycolicibacterium, Paraburkholderia, Lechevalieria, Mycobacterium, Pseudomonas, Nocardia, Paenibacillus, and Rhizobium associated with roots, leaves and seeds of C. arabica, C. canephora, and C. liberica (Silva et al., 2012; Kejela et al., 2016; Duong et al., 2021). Finally, the production of phytohormones (e.g., auxins) or regulators (e.g., the ACC deaminase enzyme able to lower ethylene level) was established for some members of the genera Bacillus, Brachybacterium, Burkholderia, Erwinia, Escherichia, Kocuria, Luteibacter, Methylobacterium, Mycobacterium, Mycolicibacterium, Nocardia, Ochrobactrum, Paenibacillus, Paracoccus, Pseudomonas, Rhizobium, Serratia, Sinomonas, and Sphingobium (Muleta et al., 2009; Baon et al., 2012; Silva et al., 2012; Kejela et al., 2016; Duong et al., 2021).
Even though highlighting some plant growth-promoting capacities in vitro represents a tool to select some potential beneficial microorganisms, the most important step to support the use of microorganisms in coffee production is the validation of the effects on the plants. Therefore, several in planta experiments under various controlled conditions (nursery, greenhouse, and phytotron) were performed to confirm the growth-promoting effect of the microorganisms. For example, Chattopadhyay et al. (2006) and Wedhastri et al. (2012) demonstrated the capacity of some N-fixing bacteria to promote the growth and the nutrient acquisition of C. canephora seedlings. Baon et al. (2012) and Cisneros-Rojas et al. (2017) also highlighted the increase in biomass of C. arabica and C. canephora seedlings after the inoculation of P-solubilizing bacteria. Furthermore, Medina et al. (2003) carried out some C. arabica seed inoculations with an N-fixing bacterial strain alone or in combination with some P-solubilizing bacteria and they were able to demonstrate a plant height increase of 33% with the co-inoculation. By testing numerous endophytic isolates (bacteria and fungi), Silva et al. (2012) showed that only 6 bacterial strains out of 234 isolates tested significantly promoted the growth of C. arabica seedlings while some isolates even had a deleterious effect. Trying to understand the mechanisms involved, the authors further characterized the most the efficient strains and demonstrated their ability to solubilize phosphorus, as well as to produce auxins and siderophores. This result underlines the interest to perform some preliminary beneficial capacity screenings before undertaking further larger experiments with the coffee plants.
The majority of other in planta experiments in controlled condition were focused on AMF. Several authors were able to demonstrate the positive effect of these symbiotic fungi on the growth of C. arabica seedlings and their nutrient acquisition including nitrogen, phosphorus, potassium, calcium, magnesium, and manganese (Caldeira et al., 1983; Vaast and Zasoski, 1992; Siqueira et al., 1995; Vaast et al., 1997b; Osorio et al., 2002). Other authors also emphasized the potential of the AMF co-inoculation with other plant growth-promoting microorganisms. Indeed, Pérez et al. (2002) observed an increased AMF colonization of C. canephora seedlings when combined with N-fixing bacteria. Moreover, González et al. (2004) underlined a consistent increase of the AMF effects on height and foliar area of C. arabica seedlings when N-fixing bacteria were applied simultaneously with the mycorrhizal fungi. Recently, Perea Rojas et al. (2019) confirmed the positive effect of AMF on C. arabica seedling growth and showed an improved efficiency by the addition of P-solubilizing fungi with a significant increase of the phosphorus content in the leaves.
The last step toward the development of sustainable alternatives to chemical fertilizers is the confirmation of the results obtained in controlled conditions in situ. Field experiments were exclusively conducted with AMF so far. Siqueira et al. (1993) were among the first authors to study the effect of mycorrhiza during 3 years in the field by inoculating different AMF strains (alone or in combination) on C. arabica seedlings under greenhouse conditions before to transfer the plants in the field. At the transplantation, the authors reported that most of the treatments were able to increase the plant biomasses and nutrient content (P and Cu) and after 6 months all the treatments increased the survival rate, stem diameter, and height of the plants. Finally, when superphosphate was applied some treatments also increased the yield with a mean increase of 74% with the most efficient one.
These beneficial effects of AMF were confirmed by several other studies. Indeed Colozzi-Filho et al. (1994) and Trejo et al. (2011) also observed an increase in C. arabica vegetative growth and nutrition (P, K, and Cu) under controlled conditions after the inoculation of different AMF isolates. These effects were maintained after the field transplantation along with an improvement of survival and first yield (up to 100% for some treatments). Siqueira et al. (1998) performed a 6-year field study during which they also confirmed the beneficial effect of AMF inoculation during the early development of coffee seedlings. However, after 26 months in the field the differences between the inoculated and control plants started to decrease and became insignificant during the following years. This finding was explained by the fact that the non-inoculated plants started to be colonized by indigenous mycorrhizal fungi after the transplantation. Therefore, the authors suggested that the AMF inoculation is really relevant only to increase the initial development of coffee seedlings and the first productions.
These findings also highlighted the need to further characterize the coffee-associated microorganisms especially under field conditions in order to develop some efficient microbiota-based alternatives to conventional fertilizers.
Potential Uses as Biocontrol Agents
Microorganisms colonizing several coffee plant compartments have been examined for their potential use as biocontrol agents including bacteria and fungi isolated from the rhizosphere (Mulaw et al., 2010; Kejela et al., 2016), the episphere (Haddad et al., 2014; Leong et al., 2014), and the endosphere (Silva et al., 2012; Hoang et al., 2020; Duong et al., 2021).
As for the plant growth-promoting agents, several in vitro screenings of potential biocontrol capacities were employed. For example, it was successfully demonstrated that numerous bacterial isolates (from the genera: Acinetobacter, Aeromonas, Alcaligenes, Bacillus, Brachybacterium, Brevibacillus, Burkholderia, Cedecea, Caballeronia, Cellulomonas, Chromobacterium, Chryseobacterium, Chryseomonas, Curtobacterium, Enterobacter, Erwinia, Escherichia, Flavobacterium, Herbaspirillum, Kitasatospora, Lechevalieria, Leifsonia, Luteibacter, Microbacterium, Micrococcus, Mycobacterium, Mycolicibacterium, Nocardia, Ochrobactrum, Paenibacillus, Paraburkholderia, Pasteurella, Pectobacterium, Pseudomonas, Rhizobium, Salmonella, Serratia, Sinomonas, Streptomyces, and Vibrio) were able to produce some enzymes including chitinases, gelatinases, lipases, and proteases (Muleta et al., 2009; Tiru et al., 2013; Kejela et al., 2016; Asyiah et al., 2018; Hoang et al., 2020; Duong et al., 2021) as well as some other active compounds like HCN and siderophores (Muleta et al., 2007; Silva et al., 2012; Tiru et al., 2013; Duong et al., 2021), already known to be involved in the biocontrol mechanisms (Compant et al., 2005; Saraf et al., 2014; Köhl et al., 2019).
Another approach that was extensively employed to highlight the biocontrol capacity of bacterial and fungal isolates involved the confrontation of the pathogen either directly with the antagonist (dual culture method) or with the compounds secreted in the culture medium (agar diffusion method). This strategy was successfully employed to demonstrate the ability of some biocontrol agents to inhibit the development of some of the major coffee diseases such as the CLR caused by the fungal pathogen H. vastatrix (Shiomi et al., 2006; Bettiol et al., 2007; Silva et al., 2008, 2012; Daivasikamani and Rajanaika, 2009; Haddad et al., 2013) or the coffee wilt disease (CWD) also known as tracheomycosis caused by the fungal pathogen Gibberella xylarioides (Muleta et al., 2007; Mulaw et al., 2010, 2013; Tiru et al., 2013). This methodology was also used to reveal the microorganisms biocontrol potential toward numerous other phytopathogens including Alternaria alternata, A. solani, Ambrosiella macrospora, Botrytis cinereal, Colletotrichum gloeosporioides, C. coffeicola, Fusarium oxysporum, F. solani, F. verticillioides, Glomerella sp., Macrophomina phaseolina, Myrothecium roridum, Pestalotia longisetula, Phoma sp., Phytophthora capsici, P. meadii, Pythium aphanidermatum, Rhizoctonia solani, and Sclerotinia sclerotiorum (Nair et al., 2002; Mulaw et al., 2013; Bongiorno et al., 2016; Kejela et al., 2016; Monteiro et al., 2017; Ranjini and Raja, 2019; Hoang et al., 2020; Duong et al., 2021) but also some pests such as the coffee berry borer Hypothenemus hampei (Vega et al., 2008), the root knot nematode Meloidogyne incognita (Mekete et al., 2009; Hoang et al., 2020), the burrowing nematode Radopholus duriophilus, and the root lesion nematode Pratylenchus coffeae (Duong et al., 2021), as well as some toxigenic fungi including Aspergillus carbonarius, A. flavus, A. niger, A. ochraceus, and A. westerdijkiae (Masoud and Kaltoft, 2006; Ramos et al., 2010; Djossou et al., 2011; Leong et al., 2014; De Melo Pereira et al., 2015a; De Almeida et al., 2019).
Several in planta experiments on C. arabica grown under controlled conditions (growth chamber and greenhouse) confirmed the capacity of several bacterial antagonists to decrease the spore germination, the disease severity, and the sporulation of H. vastatrix (Shiomi et al., 2006; Bettiol et al., 2007; Silva et al., 2008, 2012; Haddad et al., 2013, 2014; Culliao and Barcelo, 2015). Some authors underlined the fact that the treatment was more efficient when applied before the pathogen inoculation (Shiomi et al., 2006; Silva et al., 2012; Haddad et al., 2014), and other studies also demonstrated that the simultaneous applications of some bacterial isolates were as efficient as copper hydroxide in reducing CLR severity (Haddad et al., 2013).
In another greenhouse studies, Tiru et al. (2013) corroborated the in vitro results obtained with some G. xylarioides (CWD) antagonists. They showed that depending on the isolate and timing of application (before, after, or simultaneously), the inoculation of the biocontrol agents was able to significantly control the pathogen with a reduction of the disease severity comprised between 40 and 82.4%.
Nematode control capacity was also confirmed in planta. Indeed, Asyiah et al. (2018) demonstrated that a bacterial endophyte isolate was able to inhibit the penetration of the migratory endoparasitic nematode, P. coffeae, in the roots of C. arabica seedlings. Moreover, Vaast et al. (1997a) demonstrated that the inoculation of C. arabica seedlings with AMF 4 months before introducing P. coffeae significantly improved the tolerance to nematodes compared to the control without AMF. Finally, Mekete et al. (2009) confirmed the results obtained in vitro on tomato seedlings and showed that the inoculation of several bacterial endophytes significantly reduced the number of egg masses and galls caused by the root knot nematode M. incognita.
The only in situ experiments were conducted in order to test some microbial antagonists against H. vastatrix (CLR) under the field conditions. Vélez and Rosillo (1995) evaluated the efficiency of an isolate of the fungus Verticillium lecanii by spraying the antagonist 48 h before the inoculation of CLR spores. They noticed a delayed latent period (5 days), but the biocontrol agent failed to display a significant protecting effect although the number of lesions was reduced compared to the controls. More recently, Daivasikamani and Rajanaika (2009) tested some bacterial isolates as prevention treatment over a 2-year period and compared the results to those obtained with a copper-based (Bordeaux mixture) and a systemic fungicide (Triadimefon). By taking the average of the 2 years, the authors highlighted that the two best biocontrol agents were able to decrease the disease incidence by 36% with B. subtilis and 28% with P. fluorescens. However, the chemical fungicides were still more efficient with a mean disease incidence reduction of 44% with the Bordeaux mixture and 64% with Triadimefon. In another study, Haddad et al. (2009) assessed the efficiency of two bacterial isolates (Bacillus sp. and Pseudomonas sp.) compared to copper hydroxide in controlling CLR. Depending on the rate and time of application, the Bacillus isolate was as efficient as the copper-based fungicide in reducing the disease incidence and severity.
Another interesting way to control CLR was explored with the use fungal hyper-parasites. Indeed, the pathogenicity of fungal mycoparasites toward H. vastatrix has already been demonstrated on infected leaves (Carrion and Ruiz-Belin, 1988; Gómez-De La Cruz et al., 2017). The identification of CLR fungal hyper-parasites is therefore of prime interest and has already been done with a standard microbiological procedure (Carrion and Rico-Gray, 2002) as well as by a comparative metabarcoding analysis of the fungal communities associated with healthy and diseased coffee leaves (James et al., 2016).
Finally, the AMF abundance, diversity, and root colonization in C. arabica infested or not with H. vastatrix in the field were compared. Some authors reported a higher mycorrhizal colonization and spore density as well as the prevalence of some AMF genera in healthy plants, therefore highlighting the potential implication of mycorrhizal fungi in CLR tolerance (Monroy et al., 2019).
Together, these results emphasized the potential use of microbes as biocontrol agents that might be as efficient as the chemical pesticides and fungicides in controlling some of the major coffee pests and diseases.
Potential to Improve the Quality and the By-product Management
The quality of the coffee is not only influenced by several parameters during the production such as the plant genotype, environmental conditions, cultivation techniques and the associated microbiota (Toledo et al., 2016; Martins et al., 2020), but also by the type of postharvest processing (Gonzalez-Rios et al., 2007b; Lee et al., 2015; Poltronieri and Rossi, 2016), the storage conditions (Bucheli et al., 1998; Urbano et al., 2001; Geremew et al., 2016), and the roasting and brewing methods (Gonzalez-Rios et al., 2007a; Frost et al., 2020; Hu et al., 2020).
The involvement and potential use of microorganisms to improve the coffee quality is being increasingly studied. For example, the coffee undergoing the wet processing is often associated with a higher cup quality and it is now established that several groups of microorganisms especially the acetic acid bacteria (AAB), the lactic acid bacteria (LAB), and the yeasts are involved in the improvement of the quality by producing several metabolites, organic acids, and volatile compounds (De Bruyn et al., 2017; De Oliveira Junqueira et al., 2019; Zhang et al., 2019a,b; Elhalis et al., 2020b). In a recent study, Elhalis et al. (2020a) inhibited the yeast growth during the fermentation and compared the bean composition as well as the quality and sensory characteristics of coffee fermented with or without yeasts. Using this strategy, they were able to clearly demonstrate the implication of yeasts in the quality improvement of the wet-processed coffee. Finally, several authors also demonstrated the improvement of the quality after the inoculation of selected yeasts and lactic acid bacteria strains on coffee undergoing not only the wet (De Melo Pereira et al., 2015b, 2016; Da Silva Vale et al., 2019; Bressani et al., 2020) but also the semidry processing (Martinez et al., 2017).
Another explored aspect where microorganisms could have an impact on coffee quality is the potential control of toxigenic fungi. Indeed, it has been demonstrated that some yeasts and lactic acid bacteria are able to control the development and toxin production by toxigenic fungi during artificially contaminated coffee fermentation (Massawe and Lifa, 2010; De Melo Pereira et al., 2015a).
It is also important to mention that coffee processing generates several by-products including the fruit skin, pulp, parchment, silverskin, and used coffee grounds (Iriondo-DeHond et al., 2020). Several strategies have been explored to valorize these wastes by using them among others as animal feed, fertilizer, substrate for biogas/biodiesel production, compost for plants, and edible fungi and for earthworm production (Perraud-Gaime et al., 2000; Kondamudi et al., 2008; Ronga et al., 2016). However, these strategies are difficult to implement because of caffeine, tannin, and polyphenol contents that make the wastes toxic. For these reasons, microorganisms have also been studied for their potential utilization in detoxifying the coffee wastes and were therefore screened for their capacity to degrade the caffeine and tannins (Aquiahuatl et al., 1988; Roussos et al., 1995; Brand et al., 2000; Mazzafera, 2002; Nayak et al., 2012). Finally, microorganisms have also been explored for some industrial applications such as the production of enzymes with by-products such as amylases (Murthy et al., 2009), pectinases (Antier et al., 1993; Boccas et al., 1994; Sakiyama et al., 2001; Serrat et al., 2002; Masoud and Jespersen, 2006), proteases (Rodarte et al., 2011), and xylanases (Murthy and Naidu, 2012).
Consequently, the application of microbes in the coffee industry is of prime interest, not only to improve the quality of the final product but also to achieve a better management of the wastes and potentially add some value to by-products.
Conclusion and Future Trends
The coffee microbiota has been extensively studied and the potential applications of the microorganisms to improve the coffee plant fitness, either by directly promoting its growth or by acting as pathogen antagonists are now well-documented. Despite that a substantial work has been done in vitro and in planta under a controlled environment, there is a lack of assessment of the real potential of these microorganisms in situ under field conditions. This should be included as a research priority despite the difficulty of implementing this kind of experimental approach for a perennial crop such as coffee, which requires the monitoring of the plants for several years. Consumers, producers, industries, and governments are more and more concerned about the environmental and health issues associated with intensive agriculture and chemical inputs. Thus, microorganisms represent a promising alternative to improve the sustainability not only of the coffee production but also of the wastes' management as well as the quality of the final product. Despite the fact that metabarcoding studies now provide a more global understanding of the microbial communities associated with coffee, there are still some limitations regarding the taxonomic resolution. However, this issue should be resolved in the coming years with the improvement of both the sequencing technologies and the bioinformatics treatments. Nevertheless, this strategy remains complementary of the culture-dependent applied research that could be improved by some culturomics approaches, for example. The present review provides an extensive description of the diversity of microorganisms at both farming and processing levels and an overview of their potential uses. The present paper also highlights the need of further researches in this area.
Author Contributions
BD: writing—original draft. PM, J-LM, PV, ML, and RD: writing—review & editing. All authors contributed to the article and approved the submitted version.
Funding
This work was supported by the funding of the French Institute for the Research and Development (IRD) and the Laboratory of Mediterranean and Tropical Symbioses research unit (LSTM) in the framework of the France-Vietnam International Joint Laboratory (Rice, Interactions & Coffee in Environment-phase 2, RICE-2).
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to IRD for the funding and LMI RICE 2 for providing the necessary facilities. We thankfully acknowledge colleagues from IRD, CIRAD, and LMI RICE 2 for their critical editing.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2020.607935/full#supplementary-material
Supplementary Table 1. Coffee microbiota full description.
Supplementary Table 2. Arbuscular mycorrhizal fungi in association with coffee.
Supplementary Table 3. Toxigenic fungi producing Ochratoxin A (OTA), Ochratoxin B (OTB), and Aflatoxin B (AFB) in association with coffee.
Supplementary Table 4. Acetic acid bacteria (AAB) and lactic acid bacteria (LAB) in association with coffee.
Supplementary Table 5. Venn diagrams and partitions of the archaea, bacteria, and fungi shared between endophytic, epiphytic, and rhizospheric compartments of the indigenous coffee microbiota.
Supplementary Table 6. Venn diagrams and partitions of the archaea, bacteria, and fungi shared between at least one compartment of the indigenous coffee microbiota and the postharvest coffee microbiota.
Supplementary Table 7. Venn diagrams and partitions of the archaea, bacteria, and fungi shared between continents.
Supplementary Table 8. Venn diagrams and partitions of the bacteria and fungi shared between all compartments of the indigenous coffee microbiota across all continents.
References
Aeron, A., Khare, E., Jha, C. K., Meena, V. S., Aziz, S. M. A., Islam, M. T., et al. (2020). Revisiting the plant growth-promoting rhizobacteria: lessons from the past and objectives for the future. Arch. Microbiol. 202, 665–676. doi: 10.1007/s00203-019-01779-w
Afzal, I., Shinwari, Z. K., Sikandar, S., and Shahzad, S. (2019). Plant beneficial endophytic bacteria: mechanisms, diversity, host range and genetic determinants. Microbiol. Res. 221, 36–49. doi: 10.1016/j.micres.2019.02.001
Agate, A. D., and Bhat, J. V. (1966). Role of pectinolytic yeasts in the degradation of mucilage layer of Coffea robusta cherries. Appl. Microbiol. 14, 256–260.
Alori, E. T., Glick, B. R., and Babalola, O. O. (2017). Microbial phosphorus solubilization and its potential for use in sustainable agriculture. Front. Microbiol. 8:971. doi: 10.3389/fmicb.2017.00971
Amamo, A. A. (2014). Coffee production and marketing in Ethiopia. Eur. J. Bus. Manage. 6, 109–121. doi: 10.7176/EJBM
Amann, J. (1911). Die direkte zählung der wasserbakterien mittels des ultramikroskops. Centralblatt Bakteriol. 29, 381–384.
Antier, P., Minjares, A., Roussos, S., Raimbault, M., and Viniegra-Gonzalez, G. (1993). Pectinase-hyperproducing mutants of Aspergillus niger C28B25 for solid-state fermentation of coffee pulp. Enzyme Microb. Technol. 15, 254–260. doi: 10.1016/0141-0229(93)90146-S
Aquiahuatl, M. A., Raimbault, M., Roussos, S., and Trejo, M. R. (1988). “Coffee pulp detoxification by solid state fermentation: isolation, identification and physiological studies,” in Solid State Fermentation in Bioconversion of Agro-Industrial Raw Materials, ed M. Raimbault (Montpellier: ORSTOM), 13–26.
Arif, I., Batool, M., and Schenk, P. M. (2020). Plant microbiome engineering: expected benefits for improved crop growth and resilience. Trends Biotechnol. doi: 10.1016/j.tibtech.2020.04.015. [Epub ahead of print].
Asyiah, I. N., Husain, M., Iqbal, M., Hindersah, R., Narulita, E., and Mudakir, I. (2018). The endophytic bacteria isolation as biological control agent of Pratylenchus coffeae. Asian J. Microbiol. Biotechnol. Environ. Sci. 20,165–171.
Ataroff, M., and Monasterio, M. (1997). Soil erosion under different management of coffee plantations in the Venezuelan Andes. Soil Technol. 11, 95–108. doi: 10.1016/S0933-3630(96)00118-3
Avis, T. J., Gravel, V., Antoun, H., and Tweddell, R. J. (2008). Multifaceted beneficial effects of rhizosphere microorganisms on plant health and productivity. Soil Biol. Biochem. 40, 1733–1740. doi: 10.1016/j.soilbio.2008.02.013
Awoke, A., Beyene, A., Kloos, H., Goethals, P. L. M., and Triest, L. (2016). River water pollution status and water policy scenario in Ethiopia: raising awareness for better implementation in developing countries. Environ. Manage. 58, 694–706. doi: 10.1007/s00267-016-0734-y
Baon, J. B., Wedhastri, S., and Kurniawan, A. (2012). The ability of phosphate solubilizing bacteria isolated from coffee plant rhizosphere and their effects on Robusta coffee seedlings. J. Agric. Sci. Technol. A 2, 1064–1070. doi: 10.17265/2161-6256/2012.09A.005
Begum, N., Qin, C., Ahanger, M. A., Raza, S., Khan, M. I., Ashraf, M., et al. (2019). Role of arbuscular mycorrhizal fungi in plant growth regulation: implications in abiotic stress tolerance. Front. Plant Sci. 10:1068. doi: 10.3389/fpls.2019.01068
Berg, G., Rybakova, D., Fischer, D., Cernava, T., Vergès, M.-C. C., Charles, T., et al. (2020). Microbiome definition re-visited: old concepts and new challenges. Microbiome 8:103. doi: 10.1186/s40168-020-00875-0
Berg, G., Rybakova, D., Grube, M., and Köberl, M. (2016). The plant microbiome explored: implications for experimental botany. J. Exp. Bot. 67, 995–1002. doi: 10.1093/jxb/erv466
Bertolini, V., Montaño, N. M., Salazar-Ortuño, B. L., Chimal-Sánchez, E., and Varela, L. (2020). Diversidad de hongos micorrizógenos arbusculares en plantaciones de café (Coffea arabica) del volcán Tacaná, Chiapas, México. Acta Bot. Mexic. 127, 1–16. doi: 10.21829/abm127.2020.1602
Bettiol, W., Silva, H. S. A., de Melo, I. S., Terrasan, C. R. F., Tozzi, J. P. L., and Nunes, F. V. (2007). “Endophytic bacteria for biocontrol of coffee leaf rust (Hemileia vastatrix),” in Spa (Belgium: IOBC-WPRS, Bulletin OILB-SROP), 68.
Beyene, A., Kassahun, Y., Addis, T., Assefa, F., Amsalu, A., Legesse, W., et al. (2012). The impact of traditional coffee processing on river water quality in Ethiopia and the urgency of adopting sound environmental practices. Environm. Monit. Assess. 184, 7053–7063. doi: 10.1007/s10661-011-2479-7
Bitzer, V., Francken, M., and Glasbergen, P. (2008). Intersectoral partnerships for a sustainable coffee chain: really addressing sustainability or just picking (coffee) cherries? Global Environ. Change 18, 271–284. doi: 10.1016/j.gloenvcha.2008.01.002
Boccas, F., Roussos, S., Gutierrez, M., Serrano, L., and Viniegra-Gonzalez, G. (1994). Production of pectinase from coffee pulp in solid state fermentation system: selection of wild fungal isolate of high potency by a simple three-step screening technique. J. Food Sci. Technol. 31, 22–26.
Bongiorno, V. A., Rhoden, S. A., Garcia, A., Polonio, J. C., Azevedo, J. L., Pereira, J. O., et al. (2016). Genetic diversity of endophytic fungi from Coffea arabica cv. IAPAR-59 in organic crops. Ann. Microbiol. 66, 855–865. doi: 10.1007/s13213-015-1168-0
Bordenstein, S. R., and Theis, K. R. (2015). Host biology in light of the microbiome: ten principles of holobionts and hologenomes. PLoS Biol. 13:e1002226. doi: 10.1371/journal.pbio.1002226
Brand, D., Pandey, A., Roussos, S., and Soccol, C. R. (2000). Biological detoxification of coffee husk by filamentous fungi using a solid state fermentation system. Enzyme Microb. Technol. 27, 127–133. doi: 10.1016/S0141-0229(00)00186-1
Brando, C. H. J. (2004). “Harvesting and green coffee processing,” in Coffee: Growing, Processing, Sustainable Production, ed J. N. Wintgens (Weinheim: Wiley-VCH Verlag GmbH), 604–715. doi: 10.1002/9783527619627.ch24
Bressani, A. P. P., Martinez, S. J., Sarmento, A. B. I., Borém, F. M., and Schwan, R. F. (2020). Organic acids produced during fermentation and sensory perception in specialty coffee using yeast starter culture. Food Res. Int. 128:108773. doi: 10.1016/j.foodres.2019.108773
Bucheli, P., Meyer, I., Pittet, A., Vuataz, G., and Viani, R. (1998). Industrial storage of green Robusta coffee under tropical conditions and its impact on raw material quality and ochratoxin A content. J. Agric. Food Chem. 46, 4507–4511. doi: 10.1021/jf980468+
Bunn, C., Läderach, P., Ovalle Rivera, O., and Kirschke, D. (2015). A bitter cup: climate change profile of global production of Arabica and Robusta coffee. Clim. Change 129, 89–101. doi: 10.1007/s10584-014-1306-x
Caldeira, S. F., Chaves, G. M., and Zambolim, L. (1983). Associação de micorriza vesicular-arbuscular com café, limão-rosa e capim-gordura. Pesquisa Agropecuária Bras. 18, 223–228.
Caldwell, A. C., Silva, L. C. F., da Silva, C. C., and Ouverney, C. C. (2015). Prokaryotic diversity in the rhizosphere of organic, intensive, and transitional coffee farms in Brazil. PLoS ONE 10:e0106355. doi: 10.1371/journal.pone.0106355
Carrion, G., and Rico-Gray, V. (2002). Mycoparasites on the coffee rust in Mexico. Fungal Divers. 11, 49–60.
Carrion, G., and Ruiz-Belin, F. (1988). Laboratory inoculation of Verticillium lecanii on the coffee rust (Hemileia vastatrix). Rev. Mexic. Micol. 4, 317–321.
Casas-Junco, P. P., Ragazzo-Sánchez, J. A., de Jesus Ascencio-Valle, F., and Calderón-Santoyo, M. (2018). Determination of potentially mycotoxigenic fungi in coffee (Coffea arabica L.) from Nayarit. Food Sci. Biotechnol. 27, 891–898. doi: 10.1007/s10068-017-0288-7
Chanakya, H. N., and De Alwis, A. A. P. (2004). Environmental issues and management in primary coffee processing. Process Saf. Environ. Protect. 82, 291–300. doi: 10.1205/095758204323162319
Chattopadhyay, N., Swain, S., and Hore, J. K. (2006). Response of coffee seedlings to nitrogen fixing biofertilizers. Agric. Sci. Digest. 26, 103–106.
Chen, M., Arato, M., Borghi, L., Nouri, E., and Reinhardt, D. (2018). Beneficial services of arbuscular mycorrhizal fungi – from ecology to application. Front. Plant Sci. 9:1270. doi: 10.3389/fpls.2018.01270
Cisneros-Rojas, C. A., Sánchez-de Prager, M., and Menjivar-Flores, J. C. (2017). Efecto de bacterias solubilizadoras de fosfatos sobre el desarrollo de plántulas de café. Agron. Mesoamericana 28, 149–158. doi: 10.15517/am.v28i1.22021
Cleves, R. (2004). “Ecological processing of coffee and use of byproducts,” in Coffee: Growing, Processing, Sustainable Production, ed J. N. Wintgens (Weinheim: Wiley-VCH Verlag GmbH), 716–730. doi: 10.1002/9783527619627.ch25
Colozzi-Filho, A., Siqueira, J. O., Saggin Junior, O. J., Guimarães, P. T. G., and Oliveira, E. (1994). Efetividade de diferentes fungos micorrízicos arbusculares na formação de mudas, crescimento pós-transplante e produção do cafeeiro. Pesquisa Agropecuária Bras. 29, 1397–1406.
Compant, S., Duffy, B., Nowak, J., Clément, C., and Barka, E. A. (2005). Use of plant growth-promoting bacteria for biocontrol of plant diseases: principles, mechanisms of action, and future prospects. Appl. Environ. Microbiol. 71, 4951–4959. doi: 10.1128/AEM.71.9.4951-4959.2005
Compant, S., Samad, A., Faist, H., and Sessitsch, A. (2019). A review on the plant microbiome: ecology, functions, and emerging trends in microbial application. J. Adv. Res. 19, 29–37. doi: 10.1016/j.jare.2019.03.004
Compri, L., Florentino, L. A., Veiga, R. M., Da Silva, A. B., and Miranda, J. M. (2016). Diversidade de microrganismos em frutos do cafeeiro cultivado na proximidade do Lago de Furnas. Coffee Sci. Lavras 11, 285–289. doi: 10.25186/cs.v11i2.1094
Culliao, A. G. L., and Barcelo, J. M. (2015). Fungal and mycotoxin contamination of coffee beans in Benguet province, Philippines. Food Addit. Contamin. Part A 32, 250–260. doi: 10.1080/19440049.2014.1001796
Da Silva Vale, A., De Melo Pereira, G. V., De Carvalho Neto, D. P., Rodrigues, C., Pagnoncelli, M. G. B., and Soccol, C. R. (2019). Effect of co-inoculation with Pichia fermentans and Pediococcus acidilactici on metabolite produced during fermentation and volatile composition of coffee beans. Fermentation 5:67. doi: 10.3390/fermentation5030067
Daivasikamani, S., and Rajanaika (2009). Biological control of coffee leaf rust pathogen, Hemileia vastatrix Berkeley and Broome using Bacillus subtilis and Pseudomonas fluorescens. J. Biopest. 2, 94–98.
DaMatta, F. M. (2004). Ecophysiological constraints on the production of shaded and unshaded coffee: a review. Field Crops Res. 86, 99–114. doi: 10.1016/j.fcr.2003.09.001
Davis, A. P., Govaerts, R., Bridson, D. M., and Stoffelen, P. (2006). An annotated taxonomic conspectus of the genus Coffea (Rubiaceae). Bot. J. Linnean Soc. 152, 465–512. doi: 10.1111/j.1095-8339.2006.00584.x
De Almeida, Â. B., Corrêa, I. P., Furuie, J. L., De Farias Pires, T., Do Rocio Dalzoto, P., and Pimentel, I. C. (2019). Inhibition of growth and ochratoxin A production in Aspergillus species by fungi isolated from coffee beans. Braz. J. Microbiol. 50, 1091–1098. doi: 10.1007/s42770-019-00152-9
De Beenhouwer, M., Aerts, R., and Honnay, O. (2013). A global meta-analysis of the biodiversity and ecosystem service benefits of coffee and cacao agroforestry. Agric. Ecosyst. Environ. 175, 1–7. doi: 10.1016/j.agee.2013.05.003
De Beenhouwer, M., Muleta, D., Peeters, B., Van Geel, M., Lievens, B., and Honnay, O. (2015a). DNA pyrosequencing evidence for large diversity differences between natural and managed coffee mycorrhizal fungal communities. Agron. Sustain. Dev. 35, 241–249. doi: 10.1007/s13593-014-0231-8
De Beenhouwer, M., Van Geel, M., Ceulemans, T., Muleta, D., Lievens, B., and Honnay, O. (2015b). Changing soil characteristics alter the arbuscular mycorrhizal fungi communities of Arabica coffee (Coffea arabica) in Ethiopia across a management intensity gradient. Soil Biol. Biochem. 91, 133–139. doi: 10.1016/j.soilbio.2015.08.037
De Bruyn, F., Zhang, S. J., Pothakos, V., Torres, J., Lambot, C., Moroni, A. V., et al. (2017). Exploring the impacts of postharvest processing on the microbiota and metabolite profiles during green coffee bean production. Appl. Environ. Microbiol. 83:e02398–e02316. doi: 10.1128/AEM.02398-16
De Carvalho Neto, D. P., De Melo Pereira, G. V., De Carvalho, J. C., Soccol, V. T., and Soccol, C. R. (2018). High-throughput rRna gene sequencing reveals high and complex bacterial diversity associated with Brazilian coffee beans fermentation. Food Technol. Biotechnol. 56, 90–95. doi: 10.17113/ftb.56.01.18.5441
De Corato, U. (2020). Soil microbiota manipulation and its role in suppressing soil-borne plant pathogens in organic farming systems under the light of microbiome-assisted strategies. Chem. Biol. Technol. Agric. 7:17. doi: 10.1186/s40538-020-00183-7
De Melo Pereira, G. V., Beux, M., Pagnoncelli, M. G. B., Soccol, V. T., Rodrigues, C., and Soccol, C. R. (2015a). Isolation, selection and evaluation of antagonistic yeasts and lactic acid bacteria against ochratoxigenic fungus Aspergillus westerdijkiae on coffee beans. Lett. Appl. Microbiol. 62, 96–101. doi: 10.1111/lam.12520
De Melo Pereira, G. V., de Carvalho Neto, D. P., Medeiros, A. B. P., Soccol, V. T., Neto, E., Woiciechowski, A. L., et al. (2016). Potential of lactic acid bacteria to improve the fermentation and quality of coffee during on-farm processing. Int. J. Food Sci. Technol. 51, 1689–1695. doi: 10.1111/ijfs.13142
De Melo Pereira, G. V., Neto, E., Soccol, V. T., Medeiros, A. B. P., Woiciechowski, A. L., and Soccol, C. R. (2015b). Conducting starter culture-controlled fermentations of coffee beans during on-farm wet processing: growth, metabolic analyses and sensorial effects. Food Res. Int. 75, 348–356. doi: 10.1016/j.foodres.2015.06.027
De Oliveira Junqueira, A. C., de Melo Pereira, G. V., Coral Medina, J. D., Alvear, M. C. R., Rosero, R., de Carvalho Neto, D. P., et al. (2019). First description of bacterial and fungal communities in Colombian coffee beans fermentation analysed using Illumina-based amplicon sequencing. Sci. Rep. 9:8794. doi: 10.1038/s41598-019-45002-8
De Zelicourt, A., Al-Yousif, M., and Hirt, H. (2013). Rhizosphere microbes as essential partners for plant stress tolerance. Mol. Plant 6, 242–245. doi: 10.1093/mp/sst028
Djossou, O., Perraud-Gaime, I., Lakhal Mirleau, F., Rodriguez-Serrano, G., Karou, G., Niamke, S., et al. (2011). Robusta coffee beans post-harvest microflora: Lactobacillus plantarum sp. as potential antagonist of Aspergillus carbonarius. Anaerobe 17, 267–272. doi: 10.1016/j.anaerobe.2011.03.006
Duong, B., Nguyen, H. X., Phan, H. V., Colella, S., Trinh, P. Q., Hoang, G. T., et al. (2021). Identification and characterization of Vietnamese coffee bacterial endophytes displaying in vitro antifungal and nematicidal activities. Microbiol. Res. 242:126613. doi: 10.1016/j.micres.2020.126613
Egamberdieva, D., Wirth, S. J., Alqarawi, A. A., Abd_Allah, E. F., and Hashem, A. (2017). Phytohormones and beneficial microbes: essential components for plants to balance stress and fitness. Front. Microbiol. 8:2104. doi: 10.3389/fmicb.2017.02104
Elhalis, H., Cox, J., Frank, D., and Zhao, J. (2020a). The crucial role of yeasts in the wet fermentation of coffee beans and quality. Int. J. Food Microbiol. 333:108796. doi: 10.1016/j.ijfoodmicro.2020.108796
Elhalis, H., Cox, J., and Zhao, J. (2020b). Ecological diversity, evolution and metabolism of microbial communities in the wet fermentation of Australian coffee beans. Int. J. Food Microbiol. 321:108544. doi: 10.1016/j.ijfoodmicro.2020.108544
Estrada-De Los Santos, P., Bustillos-Cristales, R., and Caballero-Mellado, J. (2001). Burkholderia, a genus rich in plant-associated nitrogen fixers with wide environmental and geographic distribution. Appl. Environ. Microbiol. 67, 2790–2798. doi: 10.1128/AEM.67.6.2790-2798.2001
Evangelista, S. R., Silva, C. F., da Cruz Miguel, M. G. P., de Souza Cordeiro, C., Pinheiro, A. C. M., Duarte, W. F., et al. (2014). Improvement of coffee beverage quality by using selected yeasts strains during the fermentation in dry process. Food Res. Int. 61, 183–195. doi: 10.1016/j.foodres.2013.11.033
FAO (2015). FAO Statistical Pocketbook. Coffee. Food and Agriculture Organization of the United Nations (FAO).
FAO (2018). Food Outlook: Biannual Report on Global Food Markets, November 2018. Food and Agriculture Organization of the United Nations (FAO); Trade and Markets Division.
Feng, X., Dong, H., Yang, P., Yang, R., Lu, J., Lv, J., et al. (2016). Culture-dependent and -independent methods to investigate the predominant microorganisms associated with wet processed coffee. Curr. Microbiol. 73, 190–195. doi: 10.1007/s00284-016-1047-3
Fierer, N. (2017). Embracing the unknown: disentangling the complexities of the soil microbiome. Nat. Rev. Microbiol. 15, 579–590. doi: 10.1038/nrmicro.2017.87
Franco-Duarte, R., Cernáková, L., Kadam, S., Kaushik, K. S., Salehi, B., Bevilacqua, A., et al. (2019). Advances in chemical and biological methods to identify microorganisms-from past to present. Microorganisms 7:130. doi: 10.3390/microorganisms7050130
Frank, A., Saldierna Guzmán, J., and Shay, J. (2017). Transmission of bacterial endophytes. Microorganisms 5:70. doi: 10.3390/microorganisms5040070
Frost, S. C., Ristenpart, W. D., and Guinard, J. (2020). Effects of brew strength, brew yield, and roast on the sensory quality of drip brewed coffee. J. Food Sci. 85, 2530–2543. doi: 10.1111/1750-3841.15326
Fuentes-Ramírez, L. E., Bustillos-Cristales, R., Tapia-Hernández, A., Jiménez-Salgado, T., Wang, E. T., Martínez-Romero, E., et al. (2001). Novel nitrogen-fixing acetic acid bacteria, Gluconacetobacter johannae sp. nov. and Gluconacetobacter azotocaptans sp. nov., associated with coffee plants. Int. J. Syst. Evol. Microbiol. 51, 1305–1314. doi: 10.1099/00207713-51-4-1305
Fulthorpe, R., Martin, A. R., and Isaac, M. E. (2020). Root endophytes of coffee (Coffea arabica): variation across climatic gradients and relationships with functional traits. Phytobiomes J. 4, 27–39. doi: 10.1094/PBIOMES-04-19-0021-R
Genre, A., Lanfranco, L., Perotto, S., and Bonfante, P. (2020). Unique and common traits in mycorrhizal symbioses. Nat. Rev. Microbiol. 18, 649–660. doi: 10.1038/s41579-020-0402-3
Geremew, T., Abate, D., Landschoot, S., Haesaert, G., and Audenaert, K. (2016). Occurrence of toxigenic fungi and ochratoxin A in Ethiopian coffee for local consumption. Food Control 69, 65–73. doi: 10.1016/j.foodcont.2016.04.025
Ghini, R., Hamada, E., Pedro Júnior, M. J., Marengo, J. A., and do Valle Gonçalves, R. R. (2008). Risk analysis of climate change on coffee nematodes and leaf miner in Brazil. Pesquisa Agropecuária Bras. 43, 187–194. doi: 10.1590/S0100-204X2008000200005
Ghosh, A., and Bhadury, P. (2019). “Methods of assessment of microbial diversity in natural environments,” in Microbial Diversity in the Genomic Era, eds S. Das and H. Dash (London: Elsevier), 3–14. doi: 10.1016/B978-0-12-814849-5.00001-0
Gobbi, J. A. (2000). Is biodiversity-friendly coffee financially viable? An analysis of five different coffee production systems in western El Salvador. Ecol. Econ. 33, 267–281. doi: 10.1016/S0921-8009(99)00147-0
Gomes, L. C., Bianchi, F. J. J. A., Cardoso, I. M., Fernandes, R. B. A., Filho, E. I. F., and Schulte, R. P. O. (2020). Agroforestry systems can mitigate the impacts of climate change on coffee production: a spatially explicit assessment in Brazil. Agric. Ecosyst. Environ. 294:106858. doi: 10.1016/j.agee.2020.106858
Gómez-De La Cruz, I., Pérez-Portilla, E., Escamilla-Prado, E., Martínez-Bolaños, M., Carrión-Villarnovo, G. L. L., and Hernández-Leal, T. I. (2017). Selection in vitro of mycoparasites with potential for biological control on coffee leaf rust (Hemileia vastatrix). Mexican J. Phytopathol. 36, 172–183. doi: 10.18781/R.MEX.FIT.1708-1
González, C. B., Esmoris, C. S., and Rodríguez, M. I. (2004). “Utilización de Azotobacter en Coffea arabica L. y Coffea canephora Pierre ex Froehner. Resultados de 13 años de investigaciones,” in Convención Trópico 2004. II Congreso de Agricultura Tropical, Cuba (Havana), 15.
Gonzalez-Rios, O., Suarez-Quiroz, M. L., Boulanger, R., Barel, M., Guyot, B., Guiraud, J.-P., et al. (2007a). Impact of “ecological” post-harvest processing on coffee aroma: II. Roasted coffee. J. Food Compos. Anal. 20, 297–307. doi: 10.1016/j.jfca.2006.12.004
Gonzalez-Rios, O., Suarez-Quiroz, M. L., Boulanger, R., Barel, M., Guyot, B., Guiraud, J.-P., et al. (2007b). Impact of “ecological” post-harvest processing on the volatile fraction of coffee beans: I. Green coffee. J. Food Compos. Anal. 20, 289–296. doi: 10.1016/j.jfca.2006.07.009
Goris, J., Konstantinidis, K. T., Klappenbach, J. A., Coenye, T., Vandamme, P., and Tiedje, J. M. (2007). DNA–DNA hybridization values and their relationship to whole-genome sequence similarities. Int. J. Syst. Evol. Microbiol. 57, 81–91. doi: 10.1099/ijs.0.64483-0
Groenen, D. (2018). The effects of climate change on the pests and diseases of coffee crops in mesoamerica. J. Climatol. Weather Forecast. 6, 1–5. doi: 10.4172/2332-2594.1000239
Guillemot, J., le Maire, G., Munishamappa, M., Charbonnier, F., and Vaast, P. (2018). Native coffee agroforestry in the Western Ghats of India maintains higher carbon storage and tree diversity compared to exotic agroforestry. Agric. Ecosyst. Environ. 265, 461–469. doi: 10.1016/j.agee.2018.06.002
Haddad, F., Maffia, L. A., Mizubuti, E. S. G., and Teixeira, H. (2009). Biological control of coffee rust by antagonistic bacteria under field conditions in Brazil. Biol. Control 49, 114–119. doi: 10.1016/j.biocontrol.2009.02.004
Haddad, F., Saraiva, R. M., Mizubuti, E. S. G., Romeiro, R. S., and Maffia, L. A. (2013). Antifungal compounds as a mechanism to control Hemileia vastatrix by antagonistic bacteria. Trop. Plant Pathol. 38, 398–405. doi: 10.1590/S1982-56762013000500004
Haddad, F., Saraiva, R. M., Mizubuti, E. S. G., Romeiro, R. S., and Maffia, L. A. (2014). Isolation and selection of Hemileia Vastatrix antagonists. Eur. J. Plant Pathol. 139, 763–772. doi: 10.1007/s10658-014-0430-9
Haddis, A., and Devi, R. (2008). Effect of effluent generated from coffee processing plant on the water bodies and human health in its vicinity. J. Hazard. Mater. 152, 259–262. doi: 10.1016/j.jhazmat.2007.06.094
Hardoim, P. R., van Overbeek, L. S., Berg, G., Pirttilä, A. M., Compant, S., Campisano, A., et al. (2015). The hidden world within plants: ecological and evolutionary considerations for defining functioning of microbial endophytes. Microbiol. Mol. Biol. Rev. 79, 293–320. doi: 10.1128/MMBR.00050-14
Hartmann, A., Rothballer, M., and Schmid, M. (2008). Lorenz Hiltner, a pioneer in rhizosphere microbial ecology and soil bacteriology research. Plant Soil 312, 7–14. doi: 10.1007/s11104-007-9514-z
Hoang, H., Tran, L. H., Nguyen, T. H., Nguyen, D. A. T., Nguyen, H. H. T., Pham, N. B., et al. (2020). Occurrence of endophytic bacteria in Vietnamese Robusta coffee roots and their effects on plant parasitic nematodes. Symbiosis 80, 75–84. doi: 10.1007/s13199-019-00649-9
Hu, G., Peng, X., Gao, Y., Huang, Y., Li, X., Su, H., et al. (2020). Effect of roasting degree of coffee beans on sensory evaluation: research from the perspective of major chemical ingredients. Food Chem. 331:127329. doi: 10.1016/j.foodchem.2020.127329
ICO (2019a). Annual Review 2017/18. London: International Coffee Organization (ICO). Available online at: http://www.ico.org/
ICO (2019b). Coffee Market Report, March 2019. London: International Coffee Organization (ICO). Available online at: http://www.ico.org/
Iriondo-DeHond, A., Iriondo-DeHond, M., and del Castillo, M. D. (2020). Applications of compounds from coffee processing by-products. Biomolecules 10:1219. doi: 10.3390/biom10091219
James, T. Y., Marino, J. A., Perfecto, I., and Vandermeer, J. (2016). Identification of putative coffee rust mycoparasites via single-molecule dna sequencing of infected pustules. Appl. Environ. Microbiol. 82, 631–639. doi: 10.1128/AEM.02639-15
Janda, J. M., and Abbott, S. L. (2007). 16S rRna gene sequencing for bacterial identification in the diagnostic laboratory: pluses, perils, and pitfalls. J. Clin. Microbiol. 45, 2761–2764. doi: 10.1128/JCM.01228-07
Janse, J. M. (1897). Les endophytes radicaux de quelques plantes javanaises. Ann Jardin Bot. Buitenzorg 14, 53–212.
Jaramillo, J., Muchugu, E., Vega, F. E., Davis, A., Borgemeister, C., and Chabi-Olaye, A. (2011). Some like it hot: the influence and implications of climate change on coffee berry borer (Hypothenemus hampei) and coffee production in East Africa. PLoS ONE 6:e24528. doi: 10.1371/journal.pone.0024528
Jezeer, R. E., Santos, M. J., Boot, R. G. A., Junginger, M., and Verweij, P. A. (2018). Effects of shade and input management on economic performance of small-scale Peruvian coffee systems. Agric. Syst. 162, 179–190. doi: 10.1016/j.agsy.2018.01.014
Jimenez-Salgado, T., Fuentes-Ramirez, L. E., Tapia-Hernandez, A., Mascarua-Esparza, M. A., Martinez-Romero, E., and Caballero-Mellado, J. (1997). Coffea arabica L., a new host plant for Acetobacter diazotrophicus, and isolation of other nitrogen-fixing acetobacteria. Appl. Environ. Microbiol. 63, 3676–3683.
Johnson, J. S., Spakowicz, D. J., Hong, B.-Y., Petersen, L. M., Demkowicz, P., Chen, L., et al. (2019). Evaluation of 16S rRNA gene sequencing for species and strain-level microbiome analysis. Nat. Commun. 10:5029. doi: 10.1038/s41467-019-13036-1
Joosten, H. M. L. J., Goetz, J., Pittet, A., Schellenberg, M., and Bucheli, P. (2001). Production of ochratoxin A by Aspergillus carbonarius on coffee cherries. Int. J. Food Microbiol. 65, 39–44. doi: 10.1016/S0168-1605(00)00506-7
Kejela, T., Thakkar, V. R., and Thakor, P. (2016). Bacillus species (BT42) isolated from Coffea arabica L. rhizosphere antagonizes Colletotrichum gloeosporioides and Fusarium oxysporum and also exhibits multiple plant growth promoting activity. BMC Microbiol. 16:277. doi: 10.1186/s12866-016-0897-y
Kloepper, J. W., and Schroth, M. N. (1978). “Plant growth promoting rhizobacteria on radishes,” in Proc. 4th International Conference on Plant Pathogenic Bacteria, Vol. 2 (Angers), 879–882.
Köhl, J., Kolnaar, R., and Ravensberg, W. J. (2019). Mode of action of microbial biological control agents against plant diseases: relevance beyond efficacy. Front. Plant Sci. 10:845. doi: 10.3389/fpls.2019.00845
Kondamudi, N., Mohapatra, S. K., and Misra, M. (2008). Spent coffee grounds as a versatile source of green energy. J. Agric. Food Chem. 56, 11757–11760. doi: 10.1021/jf802487s
Kudoyarova, G., Arkhipova, T., Korshunova, T., Bakaeva, M., Loginov, O., and Dodd, I. C. (2019). Phytohormone mediation of interactions between plants and non-symbiotic growth promoting bacteria under edaphic stresses. Front. Plant Sci. 10:1368. doi: 10.3389/fpls.2019.01368
Kulski, J. K. (2016). “Next-generation sequencing - an overview of the history, tools, and ‘omic’ applications,” in Next Generation Sequencing - Advances, Applications and Challenges, ed J. K. Kulski (Rijeka: InTech), 3–60. doi: 10.5772/61964
Lagier, J.-C., Armougom, F., Million, M., Hugon, P., Pagnier, I., Robert, C., et al. (2012). Microbial culturomics: paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 18, 1185–1193. doi: 10.1111/1469-0691.12023
Lagier, J.-C., Dubourg, G., Million, M., Cadoret, F., Bilen, M., Fenollar, F., et al. (2018). Culturing the human microbiota and culturomics. Nat. Rev. Microbiol. 16, 540–550. doi: 10.1038/s41579-018-0041-0
Lagier, J.-C., Hugon, P., Khelaifia, S., Fournier, P.-E., La Scola, B., and Raoult, D. (2015). The rebirth of culture in microbiology through the example of culturomics to study human gut microbiota. Clin. Microbiol. Rev. 28, 237–264. doi: 10.1128/CMR.00014-14
Lamelas, A., Desgarennes, D., López-Lima, D., Villain, L., Alonso-Sánchez, A., Artacho, A., et al. (2020). The bacterial microbiome of Meloidogyne-based disease complex in coffee and tomato. Front. Plant Sci. 11:136. doi: 10.3389/fpls.2020.00136
Lee, L. W., Cheong, M. W., Curran, P., Yu, B., and Liu, S. Q. (2015). Coffee fermentation and flavor - an intricate and delicate relationship. Food Chem. 185, 182–191. doi: 10.1016/j.foodchem.2015.03.124
Lemanceau, P., Blouin, M., Muller, D., and Moënne-Loccoz, Y. (2017). Let the core microbiota be functional. Trends Plant Sci. 22, 583–595. doi: 10.1016/j.tplants.2017.04.008
Leong, K., Chen, Y., Pan, S., Chen, J., Wu, H., Chang, Y., et al. (2014). Diversity of lactic acid bacteria associated with fresh coffee cherries in Taiwan. Curr. Microbiol. 68, 440–447. doi: 10.1007/s00284-013-0495-2
Mahdhi, M., Tounekti, T., Al-Turki, T. A., and Khemira, H. (2017). Composition of the root mycorrhizal community associated with Coffea arabica in Fifa mountains (Jazan region, Saudi Arabia). J. Basic Microbiol. 57, 691–698. doi: 10.1002/jobm.201700075
Marchesi, J. R., and Ravel, J. (2015). The vocabulary of microbiome research: a proposal. Microbiome 3:31. doi: 10.1186/s40168-015-0094-5
Martinez, S. J., Bressani, A. P. P., da Cruz Pedrozo Miguel, M. G., Dias, D. R., and Schwan, R. F. (2017). Different inoculation methods for semi-dry processed coffee using yeasts as starter cultures. Food Res. Int. 102, 333–340. doi: 10.1016/j.foodres.2017.09.096
Martins, P. M. M., Batista, N. N., da Cruz Pedrozo Miguel, M. G., Simão, J. B. P., Soares, J. R., and Schwan, R. F. (2020). Coffee growing altitude influences the microbiota, chemical compounds and the quality of fermented coffees. Food Res. Int. 129:108872. doi: 10.1016/j.foodres.2019.108872
Masoud, W., Bjørg Cesar, L., Jespersen, L., and Jakobsen, M. (2004). Yeast involved in fermentation of Coffea arabica in East Africa determined by genotyping and by direct denaturating gradient gel electrophoresis. Yeast 21, 549–556. doi: 10.1002/yea.1124
Masoud, W., and Jespersen, L. (2006). Pectin degrading enzymes in yeasts involved in fermentation of Coffea arabica in East Africa. Int. J. Food Microbiol. 110, 291–296. doi: 10.1016/j.ijfoodmicro.2006.04.030
Masoud, W., and Kaltoft, C. H. (2006). The effects of yeasts involved in the fermentation of Coffea arabica in East Africa on growth and ochratoxin A (OTA) production by Aspergillus ochraceus. Int. J. Food Microbiol. 106, 229–234. doi: 10.1016/j.ijfoodmicro.2005.06.015
Massawe, G. A., and Lifa, S. J. (2010). Yeasts and lactic acid bacteria coffee fermentation starter cultures. Int. J. Postharvest Technol. Innov. 2, 41–82. doi: 10.1504/IJPTI.2010.038187
Mazzafera, P. (2002). Degradation of caffeine by microorganisms and potential use of decaffeinated coffee husk and pulp in animal feeding. Sci. Agric. 59, 815–821. doi: 10.1590/S0103-90162002000400030
Medina, A. D., López, J. A., Pérez, C. S., Albelo, N., and Reboso, A. G. (2003). Efecto de la aplicación de Azotobacter chroococcum y bacterias solubilizadoras de fósforo sobre el desarrollo de posturas de cafeto (Coffea arabica L.). Centro Agric. 30, 94–95.
Meena, V. S., Meena, S. K., Verma, J. P., Kumar, A., Aeron, A., Mishra, P. K., et al. (2017). Plant beneficial rhizospheric microorganism (PBRM) strategies to improve nutrients use efficiency: a review. Ecol. Eng 107, 8–32. doi: 10.1016/j.ecoleng.2017.06.058
Mekete, T., Hallmann, J., Kiewnick, S., and Sikora, R. (2009). Endophytic bacteria from Ethiopian coffee plants and their potential to antagonise Meloidogyne incognita. Nematology 11, 117–127. doi: 10.1163/156854108X398462
Mendes, R., Garbeva, P., and Raaijmakers, J. M. (2013). The rhizosphere microbiome: significance of plant beneficial, plant pathogenic, and human pathogenic microorganisms. FEMS Microbiol. Rev. 37, 634–663. doi: 10.1111/1574-6976.12028
Méndez, V. E., Bacon, C. M., Olson, M., Morris, K. S., and Shattuck, A. (2010). Agrobiodiversity and shade coffee smallholder livelihoods: a review and synthesis of ten years of research in Central America. Profess. Geogr. 62, 357–376. doi: 10.1080/00330124.2010.483638
Miguel, P. S. B., Delvaux, J. C., de Oliveira, M. N. V., Monteiro, L. C. P., de Souza Freitas, F., Tótola, M. D. C., et al. (2013). Diversity of endophytic bacteria in the fruits of Coffea canephora. Afr. J. Microbiol. Res. 7, 586–594. doi: 10.5897/AJMR12.2036
Mina, D., Pereira, J. A., Lino-Neto, T., and Baptista, P. (2020). Epiphytic and endophytic bacteria on olive tree phyllosphere: exploring tissue and cultivar effect. Microb. Ecol. 80, 145–157. doi: 10.1007/s00248-020-01488-8
Mislivec, P. B., Bruce, V. R., and Gibson, R. (1983). Incidence of toxigenic and other molds in green coffee beans. J. Food Prot. 46, 969–973. doi: 10.4315/0362-028X-46.11.969
Mithöfer, D., Méndez, V. E., Bose, A., and Vaast, P. (2017). Harnessing local strength for sustainable coffee value chains in India and Nicaragua: reevaluating certification to global sustainability standards. Int. J. Biodivers. Sc. Ecosyst. Serv. Manage. 13, 471–496. doi: 10.1080/21513732.2018.1460400
Moguel, P., and Toledo, V. M. (1999). Biodiversity conservation in traditional coffee systems of Mexico. Conserv. Biol. 13, 11–21. doi: 10.1046/j.1523-1739.1999.97153.x
Monroy, S. H., Brindis, R. C., Moreno, J. P., and Velarde, E. V. (2019). Endomycorrhizal diversity in coffee plants (Coffea arabica L.) infected with rust (Hemileia vastatrix). Nova Sci. 11, 102–123. doi: 10.21640/ns.v11i22.1642
Monteiro, M. C. P., Alves, N. M., Queiroz, M. V., de Pinho, D. B., Pereira, O. L., Souza, S. M. C., et al. (2017). Antimicrobial activity of endophytic fungi from coffee plants. Biosci. J. 33, 381–389. doi: 10.14393/BJ-v33n2-34494
Mulaw, T., Druzhinina, I., Kubicek, C., and Atanasova, L. (2013). Novel endophytic Trichoderma spp. isolated from healthy Coffea arabica roots are capable of controlling coffee tracheomycosis. Diversity 5, 750–766. doi: 10.3390/d5040750
Mulaw, T. B., Kubicek, C. P., and Druzhinina, I. S. (2010). The rhizosphere of Coffea arabica in its native highland forests of Ethiopia provides a niche for a distinguished diversity of Trichoderma. Diversity 2, 527–549. doi: 10.3390/d2040527
Muleta, D., Assefa, F., Börjesson, E., and Granhall, U. (2013). Phosphate-solubilising rhizobacteria associated with Coffea arabica L. in natural coffee forests of southwestern Ethiopia. J. Saudi Soc. Agric. Sci. 12, 73–84. doi: 10.1016/j.jssas.2012.07.002
Muleta, D., Assefa, F., and Granhall, U. (2007). In vitro antagonism of rhizobacteria isolated from Coffea arabica L. against emerging fungal coffee pathogens. Eng. Life Sci. 7, 577–586. doi: 10.1002/elsc.200700004
Muleta, D., Assefa, F., Hjort, K., Roos, S., and Granhall, U. (2009). Characterization of rhizobacteria isolated from wild Coffea arabica L. Eng. Life Sci. 9, 100–108. doi: 10.1002/elsc.200700031
Müller, D. B., Vogel, C., Bai, Y., and Vorholt, J. A. (2016). The plant microbiota: systems-level insights and perspectives. Annu. Rev. Genet. 50, 211–234. doi: 10.1146/annurev-genet-120215-034952
Murthy, P. S., and Naidu, M. M. (2012). Production and application of xylanase from Penicillium sp. utilizing coffee by-products. Food Bioprocess Technol. 5, 657–664. doi: 10.1007/s11947-010-0331-7
Murthy, P. S., Naidu, M. M., and Srinivas, P. (2009). Production of α-amylase under solid-state fermentation utilizing coffee waste. J. Chem. Technol. Biotechnol. 84, 1246–1249. doi: 10.1002/jctb.2142
Mussatto, S. I., Machado, E. M. S., Martins, S., and Teixeira, J. A. (2011). Production, composition, and application of coffee and its industrial residues. Food Bioprocess Technol. 4, 661–672. doi: 10.1007/s11947-011-0565-z
Muynarsk, E. S. M., de Melo Pereira, G. V., Mesa, D., Thomaz-Soccol, V., Carvalho, J. C., Pagnoncelli, M. G. B., et al. (2019). Draft genome sequence of Pediococcus acidilactici strain LPBC161, isolated from mature coffee cherries during natural fermentation. Microbiol. Resour. Announc. 8:e00332–e00319. doi: 10.1128/MRA.00332-19
Nair, J. R., Singh, G., and Sekar, V. (2002). Isolation and characterization of a novel Bacillus strain from coffee phyllosphere showing antifungal activity. J. Appl. Microbiol. 93, 772–780. doi: 10.1046/j.1365-2672.2002.01756.x
Nayak, S., Harshitha, M. J., Sampath, C., Anilkumar, H. S., and Rao, C. V. (2012). Isolation and characterization of caffeine degrading bacteria from coffee pulp. Indian J. Biotechnol. 11, 86–91. doi: 10.13057/biodiv/d200614
Ndayambaje, J. B., Nsabimana, A., Dushime, S., Ishimwe, F., Janvier, H., and Ongol, M. P. (2019). Microbial identification of potato taste defect from coffee beans. Food Sci. Nutr. 7, 287–292. doi: 10.1002/fsn3.887
Newman, D. J., and Cragg, G. M. (2020). Plant endophytes and epiphytes: burgeoning sources of known and “unknown” cytotoxic and antibiotic agents? Planta Med. 86, 891–905. doi: 10.1055/a-1095-1111
Nogales, A., Nobre, T., Valadas, V., Ragonezi, C., Döring, M., Polidoros, A., et al. (2016). Can functional hologenomics aid tackling current challenges in plant breeding? Brief. Funct. Genomics 15, 288–297. doi: 10.1093/bfgp/elv030
Olanrewaju, O. S., Glick, B. R., and Babalola, O. O. (2017). Mechanisms of action of plant growth promoting bacteria. World J. Microbiol. Biotechnol.33:197. doi: 10.1007/s11274-017-2364-9
Oliveira, M. N. V., Santos, T. M. A., Vale, H. M. M., Delvaux, J. C., Cordero, A. P., Ferreira, A. B., et al. (2013). Endophytic microbial diversity in coffee cherries of Coffea arabica from southeastern Brazil. Can. J. Microbiol. 59, 221–230. doi: 10.1139/cjm-2012-0674
Orozco-Mosqueda, M., del Rocha-Granados, M. C., Glick, B. R., and Santoyo, G. (2018). Microbiome engineering to improve biocontrol and plant growth-promoting mechanisms. Microbiol. Res. 208, 25–31. doi: 10.1016/j.micres.2018.01.005
Osorio, N. W., Alzate, J. M., and Ramírez, G. A. (2002). Coffee seedling growth as affected by mycorrhizal inoculation and organic amendment. Commun. Soil Sci. Plant Anal. 33, 1425–1434. doi: 10.1081/CSS-120004291
Otero-Jiménez, B., Vandermeer, J. H., and Tucker, P. K. (2018). Effect of coffee agriculture management on the population structure of a forest dwelling rodent (Heteromys desmarestianus goldmani). Conserv. Genet. 19, 495–499. doi: 10.1007/s10592-017-1016-9
Oumer, O. J., and Abate, D. (2018). Screening and molecular identification of pectinase producing microbes from coffee pulp. Biomed Res. Int. 2018:2961767. doi: 10.1155/2018/2961767
Ovalle-Rivera, O., Läderach, P., Bunn, C., Obersteiner, M., and Schroth, G. (2015). Projected shifts in Coffea arabica suitability among major global producing regions due to climate change. PLoS ONE 10:e0124155. doi: 10.1371/journal.pone.0124155
Pahari, A., Pradhan, A., Nayak, S. K., and Mishra, B. B. (2017). “Bacterial siderophore as a plant growth promoter,” in Microbial Biotechnology, eds J. K. Patra, C. N. Vishnuprasad, and G. Das (Singapore: Springer), 163–180. doi: 10.1007/978-981-10-6847-8_7
Pasin, L. A. A. P., Abreu, M. S., and de Souza, I. P. (2011). Influence of the fungi population on the physicochemical and chemical composition of coffee (Coffea arabica L.). Food Sci. Technol. 31, 681–687. doi: 10.1590/S0101-20612011000300020
Patel, J. S., Singh, A., Singh, H. B., and Sarma, B. K. (2015). Plant genotype, microbial recruitment and nutritional security. Front. Plant Sci. 6:608. doi: 10.3389/fpls.2015.00608
Pederson, C. S., and Breed, R. S. (1946). Fermentation of coffee. J. Food Sci. 11, 99–106. doi: 10.1111/j.1365-2621.1946.tb16331.x
Perea Rojas, Y., del, C., Arias, R. M., Medel Ortiz, R., Trejo Aguilar, D., Heredia, G., et al. (2019). Effects of native arbuscular mycorrhizal and phosphate-solubilizing fungi on coffee plants. Agrofor. Syst. 93, 961–972. doi: 10.1007/s10457-018-0190-1
Pérez, A., Bustamante, C., Rodríguez, R., Díaz, A., and Bertot, Y. (2002). Influencia de diferentes variantes de fertilización en el crecimiento y desarrollo de posturas de Coffea canephora Pierre. Cult. Trop. 23, 89–93.
Perfecto, I., Rice, R. A., Greenberg, R., and van der Voort, M. E. (1996). Shade coffee: a disappearing refuge for biodiversity. Bioscience 46, 598–608. doi: 10.2307/1312989
Perraud-Gaime, I., Saucedo-Castañeda, G., Augur, C., and Roussos, S. (2000). “Adding value to coffee solid by-products through biotechnology,” in Coffee Biotechnology and Quality, eds T. Sera, C. R. Soccol, A. Pandey, and S. Roussos (Dordrecht: Springer), 437–446. doi: 10.1007/978-94-017-1068-8_41
Peterson, S. W., Vega, F. E., Posada, F., and Nagai, C. (2005). Penicillium coffeae, a new endophytic species isolated from a coffee plant and its phylogenetic relationship to P. fellutanum, P. thiersii and P. brocae based on parsimony analysis of multilocus DNA sequences. Mycologia 97, 659–666. doi: 10.1080/15572536.2006.11832796
Pires, J. F., Cardoso, L., de, S., Schwan, R. F., and Silva, C. F. (2017). Diversity of microbiota found in coffee processing wastewater treatment plant. World J. Microbiol. Biotechnol. 33:211. doi: 10.1007/s11274-017-2372-9
Poltronieri, P., and Rossi, F. (2016). Challenges in specialty coffee processing and quality assurance. Challenges 7:19. doi: 10.3390/challe7020019
Ponte, S. (2002). The ‘latte revolution’? Regulation, markets and consumption in the global coffee chain. World Dev. 30, 1099–1122. doi: 10.1016/S0305-750X(02)00032-3
Posada, R. H., Heredia-Abarca, G., Sieverding, E., and Sánchez de Prager, M. (2013). Solubilization of iron and calcium phosphates by soil fungi isolated from coffee plantations. Arch. Agron. Soil Sci. 59, 185–196. doi: 10.1080/03650340.2011.610030
Prabhu, N., Borkar, S., and Garg, S. (2019). “Phosphate solubilization by microorganisms,” in Advances in Biological Science Research, eds S. N. Meena and M. M. Naik (London: Elsevier), 161–176. doi: 10.1016/B978-0-12-817497-5.00011-2
Prates Júnior, P., Moreira, B. C., da Silva, M., de, C. S., Veloso, T. G. R., Stürmer, S. L., et al. (2019). Agroecological coffee management increases arbuscular mycorrhizal fungi diversity. PLoS ONE 14:e0209093. doi: 10.1371/journal.pone.0209093
Qiao, Q., Zhang, J., Ma, C., Wang, F., Chen, Y., Zhang, C., et al. (2019). Characterization and variation of the rhizosphere fungal community structure of cultivated tetraploid cotton. PLoS ONE 14:e0207903. doi: 10.1371/journal.pone.0207903
Ramos, D. M. B., Silva, C. F., Batista, L. R., and Schwan, R. F. (2010). Inibição in vitro de fungos toxigênicos por Pichia sp. e Debaryomyces sp. isoladas de frutos de café (Coffea arabica). Acta Sci. Agron. 32, 397–402 doi: 10.4025/actasciagron.v32i3.3361
Ranjini, A. P., and Raja, N. (2019). Efficacy of biocontrol agents on Myrothecium roridum, the stem necrosis and leaf spot pathogen of coffee seedlings. J. Biopest. 12, 109–113.
Raviraja, N. S., Sridhar, K. R., and Bärlocher, F. (1996). Endophytic aquatic hyphomycetes of roots of plantation crops and ferns from India. Sydowia 48, 152–160.
Rezende, E., de, F., Borges, J. G., Cirillo, M. Â., Prado, G., Paiva, L. C., et al. (2013). Ochratoxigenic fungi associated with green coffee beans (Coffea arabica L.) in conventional and organic cultivation in Brazil. Braz. J. Microbiol. 44, 377–384. doi: 10.1590/S1517-83822013000200006
Rice, R. A. (1999). A place unbecoming: the coffee farm of Northern Latin America. Geogr. Rev. 89, 554–579. doi: 10.1111/j.1931-0846.1999.tb00234.x
Rodarte, M. P., Dias, D. R., Vilela, D. M., and Schwan, R. F. (2011). Proteolytic activities of bacteria, yeasts and filamentous fungi isolated from coffee fruit (Coffea arabica L.). Acta Sci. Agron. 33, 457–464. doi: 10.4025/actasciagron.v33i3.6734
Rodríguez, A. C., Calzada, R. T., Peña, C. G.-D., la, Ávila, J. G. A., Reyna, E. N., Paniagua, F. V., et al. (2020). A metagenomic approach in the evaluation of the soil microbiome in coffee plantations under organic and conventional production in tropical agroecosystems. Emirates J. Food Agric. 32, 263–270. doi: 10.9755/ejfa.2020.v32.i4.2092
Ronga, D., Pane, C., Zaccardelli, M., and Pecchioni, N. (2016). Use of spent coffee ground compost in peat-based growing media for the production of basil and tomato potting plants. Commun. Soil Sci. Plant Anal. 47, 356–368. doi: 10.1080/00103624.2015.1122803
Rosenberg, E., and Zilber-Rosenberg, I. (2013). The Hologenome Concept: Human, Animal and Plant Microbiota. Cham: Springer International Publishing. doi: 10.1007/978-3-319-04241-1
Rossi-Tamisier, M., Benamar, S., Raoult, D., and Fournier, P.-E. (2015). Cautionary tale of using 16S rRNA gene sequence similarity values in identification of human-associated bacterial species. Int. J. Syst. Evol. Microbiol. 65, 1929–1934. doi: 10.1099/ijs.0.000161
Roussos, S., Perraud-Gaime, I., and Denis, S. (1995). Biotechnological management of coffee pulp. Appl. Microbiol. Biotechnol. 42, 756–762. doi: 10.1007/BF00171958
Saikkonen, K., Wali, P., Helander, M., and Faeth, S. H. (2004). Evolution of endophyte-plant symbioses. Trends Plant Sci. 9, 275–280. doi: 10.1016/j.tplants.2004.04.005
Sakiyama, C. C. H., Paula, E. M., Pereira, P. C., Borges, A. C., and Silva, D. O. (2001). Characterization of pectin lyase produced by an endophytic strain isolated from coffee cherries. Lett. Appl. Microbiol. 33, 117–121. doi: 10.1046/j.1472-765x.2001.00961.x
Samson, R. A., Houbraken, J. A. M. P., Kuijpers, A. F. A., Frank, J. M., and Frisvad, J. C. (2004). New ochratoxin A or sclerotium producing species in Aspergillus section Nigri. Stud. Mycol. 50, 45–61.
Santamaría, J., and Bayman, P. (2005). Fungal epiphytes and endophytes of coffee leaves (Coffea arabica). Microb. Ecol. 50, 1–8. doi: 10.1007/s00248-004-0002-1
Santos, A., van Aerle, R., Barrientos, L., and Martinez-Urtaza, J. (2020). Computational methods for 16S metabarcoding studies using Nanopore sequencing data. Comput. Struct. Biotechnol. J. 18, 296–305. doi: 10.1016/j.csbj.2020.01.005
Santoyo, G., Moreno-Hagelsieb, G., del Carmen Orozco-Mosqueda, M. A., and Glick, B. R. (2016). Plant growth-promoting bacterial endophytes. Microbiol. Res. 183, 92–99. doi: 10.1016/j.micres.2015.11.008
Saraf, M., Pandya, U., and Thakkar, A. (2014). Role of allelochemicals in plant growth promoting rhizobacteria for biocontrol of phytopathogens. Microbiol. Res. 169, 18–29. doi: 10.1016/j.micres.2013.08.009
Sarhan, M. S., Hamza, M. A., Youssef, H. H., Patz, S., Becker, M., ElSawey, H., et al. (2019). Culturomics of the plant prokaryotic microbiome and the dawn of plant-based culture media - a review. J. Adv. Res. 19, 15–27. doi: 10.1016/j.jare.2019.04.002
Sauvadet, M., den Meersche, K. V., Allinne, C., Gay, F., de Melo Virginio Filho, E., Chauvat, M., et al. (2019). Shade trees have higher impact on soil nutrient availability and food web in organic than conventional coffee agroforestry. Sci. Tot. Environ. 649, 1065–1074. doi: 10.1016/j.scitotenv.2018.08.291
Scavino, A. F., and Pedraza, R. O. (2013). “The role of siderophores in plant growth-promoting bacteria,” in Bacteria in Agrobiology: Crop Productivity, eds D. K. Maheshwari, M. Saraf, and A. Aeron (Berlin; Heidelberg: Springer), 265–285. doi: 10.1007/978-3-642-37241-4_11
Schöps, R., Goldmann, K., Korell, L., Bruelheide, H., Wubet, T., and Buscot, F. (2020). Resident and phytometer plants host comparable rhizosphere fungal communities in managed grassland ecosystems. Sci. Rep. 10:919. doi: 10.1038/s41598-020-57760-x
Schroth, G., and Ruf, F. (2014). Farmer strategies for tree crop diversification in the humid tropics. A review. Agron. Sustain. Dev. 34, 139–154. doi: 10.1007/s13593-013-0175-4
Schwan, R., Silva, C., and Batista, L. (2012). “Coffee fermentation,” in Handbook of Plant-Based Fermented Food and Beverage Technology, 2nd Edn, eds Y. H. Hui and E. Ö. Evranuz (Boca Raton, FL: CRC Press), 677–690. doi: 10.1201/b12055-49
Serrat, M., Bermúdez, R. C., and Villa, T. G. (2002). Production, purification, and characterization of a polygalacturonase from a new strain of Kluyveromyces marxianus isolated from coffee wet-processing wastewater. Appl. Biochem. Biotechnol. 97, 193–208. doi: 10.1385/ABAB:97:3:193
Shiomi, H. F., Silva, H. S. A., Melo, I. S., de Nunes, F. V., and Bettiol, W. (2006). Bioprospecting endophytic bacteria for biological control of coffee leaf rust. Sci. Agric. 63, 32–39. doi: 10.1590/S0103-90162006000100006
Silva, C. F., Vilela, D. M., de Souza Cordeiro, C., Duarte, W. F., Dias, D. R., and Schwan, R. F. (2013). Evaluation of a potential starter culture for enhance quality of coffee fermentation. World J. Microbiol. Biotechnol. 29, 235–247. doi: 10.1007/s11274-012-1175-2
Silva, H. S. A., Terrasan, C. R. F., Tozzi, J. P. L., Melo, I. S., and Bettiol, W. (2008). Bacterias endofitas do cafeeiro e a inducao de enzimas relacionadas com o controle da ferrugem (Hemileia vastatrix). Trop. Plant Pathol. 33, 49–54. doi: 10.1590/S1982-56762008000100008
Silva, H. S. A., Tozzi, J. P. L., Terrasan, C. R. F., and Bettiol, W. (2012). Endophytic microorganisms from coffee tissues as plant growth promoters and biocontrol agents of coffee leaf rust. Biol. Control 63, 62–67. doi: 10.1016/j.biocontrol.2012.06.005
Simon, J.-C., Marchesi, J. R., Mougel, C., and Selosse, M.-A. (2019). Host-microbiota interactions: from holobiont theory to analysis. Microbiome 7:5. doi: 10.1186/s40168-019-0619-4
Singh, D. P., Singh, H. B., and Prabha, R., (eds.). (2016a). Microbial Inoculants in Sustainable Agricultural Productivity. Vol. 1: Research Perspectives. New Delhi: Springer. doi: 10.1007/978-81-322-2647-5
Singh, D. P., Singh, H. B., and Prabha, R., (eds.). (2016b). Microbial Inoculants in Sustainable Agricultural Productivity. Vol. 2: Functional Applications. New Delhi: Springer. doi: 10.1007/978-81-322-2644-4
Siqueira, J. O., Colozzi Filho, A., Saggin Júnior, O. J., Guimarães, P. T. G., and Oliveira, E. (1993). Crescimento de mudas e produção do cafeeiro sob influência de fungos micorrízicos e superfosfato. Rev. Bras. Ciênc. Solo 17, 53–60.
Siqueira, J. O., Saggin Junior, O. J., Colozzi-Filho, A., and De Oliveira, E. (1995). Influencia do substrato de formacao e da micorriza no crescimento de mudas de cafeeiro transplantadas. Pesquisa Agropecuária Bras. 30, 1417–1425.
Siqueira, J. O., Saggin-Júnior, O. J., Flores-Aylas, W. W., and Guimarães, P. T. G. (1998). Arbuscular mycorrhizal inoculation and superphosphate application influence plant development and yield of coffee in Brazil. Mycorrhiza 7, 293–300. doi: 10.1007/s005720050195
Smercina, D. N., Evans, S. E., Friesen, M. L., and Tiemann, L. K. (2019). To fix or not to fix: controls on free-living nitrogen fixation in the rhizosphere. Appl. Environ. Microbiol. 85:e02546–e02518. doi: 10.1128/AEM.02546-18
Song, G. C., Im, H., Jung, J., Lee, S., Jung, M., Rhee, S., et al. (2019). Plant growth-promoting archaea trigger induced systemic resistance in Arabidopsis thaliana against Pectobacterium carotovorum and Pseudomonas syringae. Environ. Microbiol. 21, 940–948. doi: 10.1111/1462-2920.14486
Spatafora, J. W., Chang, Y., Benny, G. L., Lazarus, K., Smith, M. E., Berbee, M. L., et al. (2016). A phylum-level phylogenetic classification of zygomycete fungi based on genome-scale data. Mycologia 108, 1028–1046. doi: 10.3852/16-042
Stackebrandt, E., and Goebel, B. M. (1994). Taxonomic note: a place for DNA-DNA reassociation and 16S rRna sequence analysis in the present species definition in bacteriology. Int. J. Syst. Evol. Microbiol. 44, 846–849. doi: 10.1099/00207713-44-4-846
Teshome, B., Wassie, M., and Abatneh, E. (2017). Isolation, screening and biochemical characterization of phosphate solubilizing rhizobacteria associated with Coffea arabica L. J. Fertil. Pest. 1, 1–7. doi: 10.4172/2471-2728.1000188
Tiru, M., Muleta, D., Berecha, G., and Adugna, G. (2013). Antagonistic effects of rhizobacteria against coffee wilt disease caused by Gibberella xylarioides. Asian J. Plant Pathol. 7, 109–122. doi: 10.3923/ajppaj.2013.109.122
Tkacz, A., Bestion, E., Bo, Z., Hortala, M., and Poole, P. S. (2020). Influence of plant fraction, soil, and plant species on microbiota: a multikingdom comparison. MBio 11:e02785. doi: 10.1128/mBio.02785-19
Toledo, P. R. A. B., Pezza, L., Pezza, H. R., and Toci, A. T. (2016). Relationship between the different aspects related to coffee quality and their volatile compounds. Compr. Rev. Food Sci. Food Saf. 15, 705–719. doi: 10.1111/1541-4337.12205
Trejo, D., Ferrera-Cerrato, R., García, R., Varela, L., Lara, L., and Alarcón, A. (2011). Efectividad de siete consorcios nativos de hongos micorrízicos arbusculares en plantas de café en condiciones de invernadero y campo. Rev. Chil. Hist. Nat. 84, 23–31. doi: 10.4067/S0716-078X2011000100002
Urbano, G. R., Taniwaki, M. H., Leitão, M. F., and Vicentini, M. C. (2001). Occurrence of ochratoxin A–producing fungi in raw Brazilian coffee. J. Food Prot. 64, 1226–1230. doi: 10.4315/0362-028X-64.8.1226
Vaast, P., Caswell-Chen, E. P., and Zasoski, R. J. (1997a). Influences of a root-lesion nematode, Pratylenchus coffeae, and two arbuscular mycorrhizal fungi, Acaulospora mellea and Glomus clarum on coffee (Coffea arabica L.). Biol. Fertil. Soils 26, 130–135. doi: 10.1007/s003740050355
Vaast, P., Harmand, J.-M., Rapidel, B., Jagoret, P., and Deheuvels, O. (2016). “Coffee and cocoa production in agroforestry—a climate-smart agriculture model,” in Climate Change and Agriculture Worldwide, ed E. Torquebiau (Dordrecht: Springer), 209–224. doi: 10.1007/978-94-017-7462-8_16
Vaast, P., Zasoski, R. J., and Bledsoe, C. S. (1997b). Effects of vesicular-arbuscular mycorrhizal inoculation at different soil P availabilities on growth and nutrient uptake of in vitro propagated coffee (Coffea arabica L.) plants. Mycorrhiza 6, 493–497. doi: 10.1007/s005720050153
Vaast, Ph., and Zasoski, R. J. (1992). Effects of VA-mycorrhizae and nitrogen sources on rhizosphere soil characteristics, growth and nutrient acquisition of coffee seedlings (Coffea arabica L.). Plant Soil 147, 31–39. doi: 10.1007/BF00009368
Van Pee, W., and Castelein, J. (1971). The yeast flora of fermenting Robusta coffee. East Afr. Agric. For. J. 36, 308–310. doi: 10.1080/00128325.1971.11662476
Vandenkoornhuyse, P., Quaiser, A., Duhamel, M., Le Van, A., and Dufresne, A. (2015). The importance of the microbiome of the plant holobiont. New Phytol. 206, 1196–1206. doi: 10.1111/nph.13312
Vega, F. E., Pava-Ripoll, M., Posada, F., and Buyer, J. S. (2005). Endophytic bacteria in Coffea arabica L. J. Basic Microbiol. 45, 371–380. doi: 10.1002/jobm.200410551
Vega, F. E., Posada, F., Catherine Aime, M., Pava-Ripoll, M., Infante, F., and Rehner, S. A. (2008). Entomopathogenic fungal endophytes. Biol. Control 46, 72–82. doi: 10.1016/j.biocontrol.2008.01.008
Vega, F. E., Posada, F., Peterson, S. W., Gianfagna, T. J., and Chaves, F. (2006). Penicillium species endophytic in coffee plants and ochratoxin A production. Mycologia 98, 31–42. doi: 10.3852/mycologia.98.1.31
Vega, F. E., Simpkins, A., Aime, M. C., Posada, F., Peterson, S. W., Rehner, S. A., et al. (2010). Fungal endophyte diversity in coffee plants from Colombia, Hawai'i, Mexico and Puerto Rico. Fungal Ecol. 3, 122–138. doi: 10.1016/j.funeco.2009.07.002
Vélez, P. E., and Rosillo, A. G. (1995). Evaluación del antagonismo del hongo Verticillium lecanii sobre Hemileia vastatrix en condiciones de invernadero y de campo. Cenicafé 46, 45–55.
Velmourougane, K., Panneerselvam, P., and Alwar, R. P. A. (2000). Qualitative and quantitative distribution of microflora associated with coffee plants and berries. in (Thirteenth plantation crops symposium 1998. Recent advances in plantation crops research, Coimbatore, India), 396–399.
Veloso, T. G. R., da Silva, M., de, C. S., Cardoso, W. S., Guarçoni, R. C., Kasuya, M. C. M., et al. (2020). Effects of environmental factors on microbiota of fruits and soil of Coffea arabica in Brazil. Sci. Rep. 10:14692. doi: 10.1038/s41598-020-71309-y
Vieira, L. G. E., Andrade, A. C., Colombo, C. A., Moraes, A. H., de, A., Metha, Â., et al. (2006). Brazilian coffee genome project: an EST-based genomic resource. Braz. J. Plant Physiol. 18, 95–108. doi: 10.1590/S1677-04202006000100008
Vilela, D. M., Pereira, G. V., de, M., Silva, C. F., Batista, L. R., and Schwan, R. F. (2010). Molecular ecology and polyphasic characterization of the microbiota associated with semi-dry processed coffee (Coffea arabica L.). Food Microbiol. 27, 1128–1135. doi: 10.1016/j.fm.2010.07.024
Vitousek, P. M., Cassman, K., Cleveland, C., Crews, T., Field, C. B., Grimm, N. B., et al. (2002). “Towards an ecological understanding of biological nitrogen fixation,” in The Nitrogen Cycle at Regional to Global Scales, eds E. W. Boyer and R. W. Howarth (Dordrecht: Springer), 1–45. doi: 10.1007/978-94-017-3405-9_1
Vorholt, J. A. (2012). Microbial life in the phyllosphere. Nat. Rev. Microbiol. 10, 828–840. doi: 10.1038/nrmicro2910
Waller, J. M., and Masaba, D. M. (2006). The microflora of coffee surfaces and relationships to coffee berry disease. Int. J. Pest Manage. 52, 89–96. doi: 10.1080/09670870600568311
Wang, Z., Li, T., Wen, X., Liu, Y., Han, J., Liao, Y., et al. (2017). Fungal communities in rhizosphere soil under conservation tillage shift in response to plant growth. Front. Microbiol. 8:1301. doi: 10.3389/fmicb.2017.01301
Wedhastri, S., Yudianti, N. F., Widada, J., and Baon, J. B. (2012). Ability of non symbiotic nitrogen-fixing bacteria isolated from coffee plant rhizosphere and their effects on Robusta coffee seedlings. J. Agric. Sci. Technol. A 2, 660–666. doi: 10.17265/2161-6256/2012.05A.009
Whipps, J., Lewis, K., and Cooke, R. (1988). “Mycoparasitism and plant disease control,” in Fungi in Biological Control Systems, ed M. Burge (Manchester: Manchester University Press), 161–187. doi: 10.1002/jctb.280480109
White, J. F., Kingsley, K. L., Zhang, Q., Verma, R., Obi, N., Dvinskikh, S., et al. (2019). Review: endophytic microbes and their potential applications in crop management. Pest Manag. Sci. 75, 2558–2565. doi: 10.1002/ps.5527
Wilson, D. (1995). Endophyte: the evolution of a term, and clarification of its use and definition. Oikos 73, 274–276. doi: 10.2307/3545919
Wintgens, J. N. (2004). “The coffee plant,” in Coffee: Growing, Processing, Sustainable Production, ed J. N. Wintgens (Weinheim: Wiley-VCH Verlag GmbH), 1–24. doi: 10.1002/9783527619627.ch1
Yan, L., Zhu, J., Zhao, X., Shi, J., Jiang, C., and Shao, D. (2019). Beneficial effects of endophytic fungi colonization on plants. Appl. Microbiol. Biotechnol. 103, 3327–3340. doi: 10.1007/s00253-019-09713-2
Zhang, S. J., De Bruyn, F., Pothakos, V., Contreras, G. F., Cai, Z., Moccand, C., et al. (2019a). Influence of various processing parameters on the microbial community dynamics, metabolomic profiles, and cup quality during wet coffee processing. Front. Microbiol. 10:2621. doi: 10.3389/fmicb.2019.02621
Keywords: wastes and by-products management, quality, biocontrol agent, plant growth promoting agents, core microbiota, coffee microbiota
Citation: Duong B, Marraccini P, Maeght J-L, Vaast P, Lebrun M and Duponnois R (2020) Coffee Microbiota and Its Potential Use in Sustainable Crop Management. A Review. Front. Sustain. Food Syst. 4:607935. doi: 10.3389/fsufs.2020.607935
Received: 18 September 2020; Accepted: 27 October 2020;
Published: 03 December 2020.
Edited by:
Everlon Cid Rigobelo, São Paulo State University, BrazilReviewed by:
Ugo De Corato, Energy and Sustainable Economic Development (ENEA), ItalyErica Lumini, Italian National Research Council, Italy
Copyright © 2020 Duong, Marraccini, Maeght, Vaast, Lebrun and Duponnois. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Robin Duponnois, cm9iaW4uZHVwb25ub2lzQGlyZC5mcg==
 Benoit Duong
Benoit Duong Pierre Marraccini
Pierre Marraccini Jean-Luc Maeght
Jean-Luc Maeght Philippe Vaast6
Philippe Vaast6 Robin Duponnois
Robin Duponnois