- 1Plataforma de Bioinsumos, Instituto Nacional de Investigación Agropecuaria, Montevideo, Uruguay
- 2Unité Mixte de Recherche en Écologie Fonctionnelle & Biogéochimie des Sols et Agroécosystemes, INRA-CIRAD-IRD-SupAgro, Montpellier, France
Phosphorus deficiency can be a major limitation to legume growth when plant nitrogen nutrition depends on symbiotic nitrogen fixation. One possible approach to overcome this constraint is the selection of plant and rhizobial genotypes capable of metabolizing complex forms of phosphorus in the nodules. The aim of this research was to study the rhizobial phytase transcript abundance in nodules of two soybean cultivars (Glycine max (L.) Merr.) grown under two different phosphorus conditions in hydroaeroponic conditions. An in situ RT-PCR of a rhizobial phytase was performed in microtome sections of soybean nodules of two cultivars growing under phosphorus sufficiency vs. phosphorus deficiency. The results showed that the plant cultivar may influence the level of transcript abundance of the bacterial phytase and in consequence affect the phosphorus use efficiency of nitrogen-dependent Bradyrhizobium spp.-soybean symbioses. Thus, the selection of a good combination of plant and rhizobial genotypes should be a priority when breeding for phosphorus deficiency is performed.
Introduction
Phosphorus (P) is the second limiting element for plant growth after nitrogen (N), thus being an essential nutrient for agricultural production. Total soil P, in both inorganic and organic forms, is abundant in most soils (Richardson, 2001). However, only 1–5% of the total P is dissolved in the soil solution as orthophosphate ions, or H2, and absorbed by plants (Richardson and Simpson, 2011; Khan et al., 2013). Despite being often overlooked, soil organic P (Po) comprises 30–50% of soil total P (Hinsinger et al., 2015), whether as part of the biomass or associated to the soil organic matter (Maougal et al., 2014a). Phytates, in particular, may represent up to 80% of the total Po (Quiquampoix and Mousain, 2005). However, organic forms of P are unavailable to plants unless mineralization takes place (Lazali et al., 2013). Phytases are enzymes responsible for the mineralization of phytates. They are classified according to their hydrolytic mechanism, the substrate specificity, their protein structure and optimal pH: histidine acid phosphatase (HAP) (EC 3.1.3.8), β-propeller phytase (BPP) (EC 3.1.3.8), cystein phytase (CPhy) and purple acid phytase (PAP) (EC 3.1.3.2) (Mullaney and Ullah, 2003; Jorquera et al., 2008a). There are a lot of rhizospheric bacterial genera that can mineralize phytate such as Enterobacter, Pseudomonas, Burkholderia, Bacillus and Klebsiella, among others (Yoon et al., 1996; Richardson and Hadobas, 1997; Richardson, 2001; Hill et al., 2007; Jorquera et al., 2008b; Maougal et al., 2014a; Ramesh et al., 2014). Several plant species have the ability to increase the activity of phytases in the rhizosphere in response to P deficiency (Li et al., 1997).
P has a crucial role for symbiotic nitrogen fixation (SNF) and the subsequent ammonium assimilation, which take place in legume nodules; both processes are highly energy-consuming and depend on the energy status of nodules (Araújo et al., 2008; Lazali et al., 2016). P deficiency comprises a limitation for SNF in legumes (Graham et al., 2003), resulting in a decrease in SNF and, consequently, in slow plant growth (Israel, 1987). Previous reports showed that mutations on three Sinorhizobium meliloti P transport systems, OrfA-Pit, PstSCAB, and phoCDET, failure to obtain sufficient P for growth during the infection process, forming “empty nodules,” which contain very few infected cells, in the alfalfa (Medicago sativa) symbiont and fail to fix N2 (Bardin et al., 1996; Yuan et al., 2006). Atlhough, the expression of one of these transports could be necessary and sufficient for SNF in alfalfa (Yuan et al., 2006). The content of P is greater in nodules than in other plant organs under conditions of either P sufficiency or deficiency (Tang et al., 2001; Schulze et al., 2006). Legumes allocate different type of P sources in nodules, and leaves start to deplete before nodules, when they grow under P deficiency (Sulieman et al., 2010). It has been reported for different legumes, such as M. sativa, Medicago truncatula, soybean (Glycine max), common bean (Phaseolus vulgaris), pea (Pisum sativum), and Sophora flavescens, that nodules become a P sink under P deficient growing conditions (Sa and Israel, 1991; Kouas et al., 2005; Sulieman et al., 2013a; Hu et al., 2018).
For the above-stated reasons, P deficiency is a major limitation to legume production in which N nutrition depends on SNF (Araújo et al., 2008). Effective approaches to P deficiency include: (1) increasing P acquisition by mechanisms such as root exudation and morphology (Gahoonia et al., 2007; Ao et al., 2010; Cheng et al., 2011); (2) inoculation with mycorrhizal fungi or with P-solubilizing and/or mineralizing bacteria or fungi (Rosas et al., 2006; Bucher, 2007; Elkoca et al., 2007; Harvey et al., 2009); (3) selecting plant genotypes with greater enzymatic activity, capable of metabolizing phosphate compounds in the nodules (Araújo et al., 2008; Lopez-Arredondo et al., 2014; Drevon et al., 2015; Castro-Guerrero et al., 2016); (4) selecting specific rhizobia with P-mineralizing abilities, capable of metabolizing complex forms of P in the nodules, such as phytate (Lazali et al., 2016).
The present study analyzes this last approach, aiming to test if one of the two strains used to inoculate Uruguayan soybean (SEMIA 587) could mineralize the phytate present in soybean nodules, increasing P content and therefore the efficiency of the SNF. In Uruguay, SNF has been upheld as a long-term public policy since the 1960's (Altier et al., 2013). As a result of research and outreach policies, 100% of the farmers have adopted the use of inoculation technology (Lindrström et al., 2010). In Uruguay, there are two officially recommended strains for soybean inoculation, Bradyrhizobium elkanii strain SEMIA 587 and Bradyrhizobium elkanii strain SEMIA 5019 (syn. 29w) (Olivera et al., 2016). Specific references of these two strains can be found in Hungria et al. (1998). First, we tested the ability of the strain SEMIA 587 to mineralize phytate in vitro and afterwards, we studied the rhizobial phytase transcript abundance in soybean nodules for a better understanding of the role of this enzyme in the acquisition and cycling of phytate in soybean. We also addressed whether the transcript abundance levels significantly vary between two cultivars.
Materials and Methods
Growth Curves
The ability of the strain Bradyrhizobium elkanii SEMIA 587 to use phytate was tested at pH 6 and 7 by inoculating the following Angle medium (Angle et al., 1991) modified by Richardson and Hadobas (1997): 1 mM KNO3, 2 mM MgSO47H2O, 4 mM CaSO4, 55 mM glucose, 50 mg L−1 thiamine-HCl, 0.5 mL L−1 Fe-citrate 1%, 0.2 mL L−1 micronutrient solution containing per liter (2.82 g H3BO3, 98 mg CuSO45H2O, 3.08 g MnSO4H2O, 0.29 g NaMoO42H2O, 4.41 g ZnSO47H2O) and 6 mM Na-phytate (Sigma, ref P0109). The optical density at 600 nm was measured every 24 h during seven days (168 h). The above-mentioned medium supplemented with 1.5 mM KH2PO4, instead of Na-phytate, was used as control. Three replicates for both growing conditions were used.
Rhizobial HAP Phytase Gene Primers Design
The design of the rhizobial HAP phytase primers was performed online at the National Center of Biotechnology Information (NCBI, http://blast.ncbi.nlm.nih.gov/Blast.cgi). The highest hit by BLASTp to rhizobiaceae group of a known amino acid sequence of the Escherichia coli HAP phytase (GenBank accession number: AAN28334.1) was obtained with a conserved hypothetical HAP-type phytase of Azorhizobium caulinodans (GenBank accession number: 5692057 AZC_4319). The gene sequence of this hypothetical HAP-type phytase was obtained from A. caulinodans full genome (GenBank accession number: AP009384.1). Thereafter, the following pair of primers were designed with the primer-BLAST tool from this HAP gene-sequence: forward primer (HAPfor), 5′CAGTTCACGCCAAAGATGCC3′; reverse primer (HAPrev), 5′CGCGTATGGTCCATCCTGAA3′. Reverse primer containing three consecutive mismatched bases near its 3′ terminus can hybridize to the target RNA at a temperature (in this study: 42°C) which is not high enough for DNA denaturation. Furthermore, due to the lower affinity between this mismatched primer and genomic DNA (gDNA), it is a very inefficient primer for gDNA amplification at high annealing temperatures (in this study: 62°C). Consequently, the mismatched primer can be used to selectively amplify the newly synthesized complementary DNA (cDNA), even in the presence of high quantities of gDNA (Koo and Jaykus, 2000).
In vitro RT-PCR of Rhizobial Phytase Transcripts
HAP designed primer pair along with a primer pair that amplifies BBP described by Farhat et al. (2008) (BPPfor, 5′GATGCAGCTGATGATCCTGCG3′; BPPrev, 5′ATTTTCTCCGTCCTGTGCGAC3′), were used to amplified B. elkanii SEMIA 587 phytases expressed in presence of phytate as unique P source. B. elkanii SEMIA 587 RNA and gDNA were extracted with the RNeasy Plus and DNeasy Plant Mini Kits (QIAGEN), respectively. RNA was extracted from B. elkanii SEMIA 587 cultivated in Angle medium supplemented with Na-phytate and KH2PO4.
Retrotranscription reactions were carried out in 25 μL, but first, 1 μg RNA and 3 μL of 10 μM HAP or BPP reverse primers were incubated at 70°C during 5 min and at 4°C for 2 min. Then, 5 X Buffer (Promega), 10 mM of each dNTP, 0.1% bovine serum albumin (BSA), and 200 U Moloney murine leukemia virus (M-MLV) reverse transcriptase H- (Promega), were added and incubated at 42°C during 60 min.
A touchdown PCR was performed with HAP and BPP primers and with cDNA and gDNA as templates, to test if at different temperatures it was able to amplify a phytase. PCR reactions were carried out in 25 μL volumes containing 1 μL target, 10 X Buffer (Invitrogen), 50 mM MgCl2, 10 mM of each dNTP, 10 μM forward primer (HAP or BPP), 20 μM reverse primer (HAP or BPP), 0.1% bovine serum albumin (BSA), 5 U Taq Polymerase (Invitrogen), under the following thermal conditions: 95°C for 2 min, 30 cycles of 95°C for 30 s, 70°C for 45 s, and gradually reduced 1°C till reaching 60°C, 72°C for 45 s, with a final extension at 72°C for 3 min.
When PCR products were obtained in the touchdown PCR, a gradient PCR was performed with cDNA and gDNA as templates, to obtain the optimal hybridization temperature of primers. PCR reactions were carried out in 25 μL volumes containing 1 μL target, 10 X Buffer (Invitrogen), 50 mM MgCl2, 10 mM of each dNTP, 10 μM forward primer, 20 μM reverse primer, 0.1% BSA, 5 U Taq Polymerase (Invitrogen), under the following thermal conditions: 94°C for 3 min, 35 cycles of 94°C for 30 s, 64-59°C for 45 s, 72°C for 45 s, with a final extension at 72°C for 3 min. Consequently, the optimal hybridization temperature of primers HAP was 62°C.
All amplification products were checked by electrophoresis in 1% agarose gel.
Hydroaeroponic Culture of Soybean
Two commercial soybean cultivars, namely N5909 (Nidera) and SR532 (Santa Rosa), were used in this study. SR532 and N5909 have a short and long production cycle, respectively. B. elkanii SEMIA 587 was used to inoculate these two cultivars. Before sowing, the soybean seeds were surface sterilized, first with 95% ethanol for 3 min and afterwards, with 3% NaOCl for 3 min more. Then, seeds were rinsed five times with sterile distilled water. Later, seeds were imbibed in sterile distilled water for 4 h, and then transferred for germination on sterile 15 g L−1 agar-water plates (Beyhaut et al., 2006). After germination, inoculation was performed by 30 min soaking 10-day-old seedlings in a suspension of B. elkanii SEMIA 587 containing ~108 cfu mL−1. The inoculum was prepared by growing the strain SEMIA 587 in yeast extract mannitol broth during four days at 28°C at 120 rpm.
Twenty inoculated seedlings were transferred into each container, with base dimensions 0.2 × 0.4 m and height 0.2 m, refilled weekly with 40 L of Vadez et al. (1996) nutrient solution. In order to avoid initial N deficiency and ensure optimal nodulation, urea was supplied at 1 mmol plant−1 during the initial 15 days of growth. Thereafter, the plants were grown in N-free solution. They were grown under both deficient and sufficient P supply in hydroaeroponic conditions with a permanent flow of 400 mL min−1 of compressed air to ensure the oxygenation of the culture solution (Hernandez and Drevon, 1991). P was supplied weekly in the form of KH2PO4, at 75 vs. 250 μmol plant−1 as P deficiency vs. P sufficiency, respectively (Vadez et al., 1996). The pH was adjusted to 6.8 with 0.2 g L−1 CaCO3 (Hernandez and Drevon, 1991). The experiment was carried out in a greenhouse, 28/20°C under 16/8 h day/night cycle, with an additional light supply of 400 μmol photons m−2 s−1 and 70% relative humidity during the day.
Shoot, Root and Nodule Biomass
Forty two days after transfer (DAT), soybean plants were harvested and shoot, roots and nodules were separated. Dry weights (DW) were determined after drying at 70°C for three days. Shoot accumulated P was determined by the vanado-molybdate method in Laboratorio Oriental (http://labo.com.uy/).
The efficiency in use of the rhizobial symbiosis (EURS) was estimated by the slope of the regression model of shoot biomass as a function of nodule biomass (y = ax + b), where a corresponds to the EURS and b corresponds to the plant biomass production without nodules (Zaman-Allah et al., 2007).
In situ RT-PCR of Rhizobial Phytase Transcripts
The in situ RT-PCR was made according to Lazali et al. (2013) and Maougal et al. (2014b). Three nodules of 3 mm diameter of three plants of each cultivar and P treatment were carefully detached from roots at 42 DAT. They were thoroughly washed with diethyl pyrocarbonate (DEPC) treated water, then fixed in 4% paraformaldehyde, 45% ethanol and 5% acetic acid, kept for 2 h under vacuum, and stored overnight at 4°C. Fixed nodules were extensively rinsed with two baths of DEPC treated water during 5 min each, two baths of 1 X phosphate buffered saline (PBS, 5 mM Na2HPO4, 300 mM NaCl, pH 7.5) during 10 min each, one bath of 1 X PBS plus 0.2% glycine during 10 min and one last bath with 1 X PBS during 10 min.
Thereafter, the fixed nodules were dehydrated in baths of increasing concentrations of ethanol and butanol: one bath of 50% ethanol during 30 min; two baths of 70% ethanol during 30 min and 1 h; one bath of 70% of ethanol overnight at 4°C; two baths with 100% ethanol for 1 h; and in the end, at least, three baths with 100% butanol during one week. Then, the nodules were included in paraffin and cut into transversal 12 μm thick sections using a microtome (Micro-cut H1200 Vibrating Microtome). The resulting sections were placed on slides treated with silane to increase cell adhesion. Thereafter, the sections were deparaffinated through three baths of Safesolv (Q Path®) for 10 min each, one bath of Safesolv during 5 min; three baths of 100% ethanol during 10 min each; one bath of 70% ethanol during 5 min; one bath of 50% ethanol during 5 min. This was continued by section rehydration with one bath of 1 X PBS during 10 min, one bath of 1 X PBS plus glycine 0.2% during 2 min, one bath of 1 X PBS during 10 min and one bath of 1X PBS for 5 min at 65°C.
The first cDNA strand was synthesized from total RNA of nodular sections, that were incubated at 42°C during 1 h in 100 μL reverse transcriptase mix containing 5 X Buffer (Promega), 10 mM of each dNTP, 10 μM HAPrev, 0.1% BSA, and 200 U M-MLV reverse transcriptase H- (Promega). Negative controls (NRT) were prepared by omitting the reverse transcriptase. Afterwards, the reverse transcriptase mix was removed, by adding two times 1 mL of 1 X PBS and incubating the slides two times more in 50 mL of 1 X PBS during 10 min. PCR reactions were carried out in 100 μL volumes containing 10 X Buffer (Invitrogen), 50 mM MgCl2, 10 mM of each dNTP, 1 mM digoxigenin-11-uridine-triphosphate (Dig-11-dUTP), 10 μM HAPfor, 20 μM HAPrev, and 5 U Taq polymerase (Invitrogen), under the following thermal conditions: 35 cycles of 94°C for 30 s, 62°C for 45 s, and 72°C for 45 s, with a final extension at 72°C for 3 min.
For detection of the amplified cDNA in the nodules, the PCR mix was removed, and samples were washed in 100 μl of blocking solution under gentle agitation in the dark at 37°C. The blocking solution was replaced by 100 μl of alkaline phosphatase conjugated anti-dioxygenin Fab fragment (Roche Diagnostics) diluted 1:1000 in 2% BSA. The samples were incubated at room temperature for 90 min, then washed three times to remove excess antibody. Detection of alkaline phosphatase was carried out using an ELF-97® endogenous phosphatase detection kit (Molecular Probes). The ELF substrate was diluted 1:40 in the alkaline detection buffer (Molecular Probes), vigorously shaken, and then filtered through a 0.22 μm filter (Millex®-GV, Millipore) to remove any aggregates of the substrate that may have formed during storage. Samples were incubated in 20 μL ELF substrate–buffer solution for 20 min in the dark and transferred to washing buffer (1 X PBS, 25 mM EDTA, and 5 mM levamisole, pH 8.0). Three washings of 1 min were performed. Samples were mounted for observations with a BX61® microscope (Olympus) equipped with an epifluorescence condenser, a Hoechst/DAPI filter set configured with an excitation filter of 360–370 nm, a dichroic mirror of 400 nm and an emission of 420 nm, and a gray View II® camera (ORCA AG). Image analysis was performed using ImageJ software (Schneider et al., 2012) as an image analysis program. The intensity of the fluorescent signal emitted by the rhizobial phytase transcript was measured as number of green pixels per image.
Statistical Analysis
Shoot, nodules and root DW, shoot accumulated P and intensity of the fluorescent signal emitted by the rhizobial phytase transcript were analyzed through analysis of variance (ANOVA) and tested for differences between cultivars and P growing conditions using a post-hoc Tukey's HSD test (p < 0.05) in R3.1.3 software (https://www.r-project.org/) using the “agricolae” package (de Mendiburu, 2020). The relationship between nodule and shoot biomass was tested by regression analysis.
Results
Phytate Mineralization by B. elkanii SEMIA 587
B. elkanii SEMIA 587 showed similar growth curves when it was cultivated in Angle medium supplemented with Na-phytate or KH2PO4, as P sources, at both pH evaluated (6 and 7) (Figures 1A,B). Therefore, this strain was able to mineralize phytate and to use it as a P source.
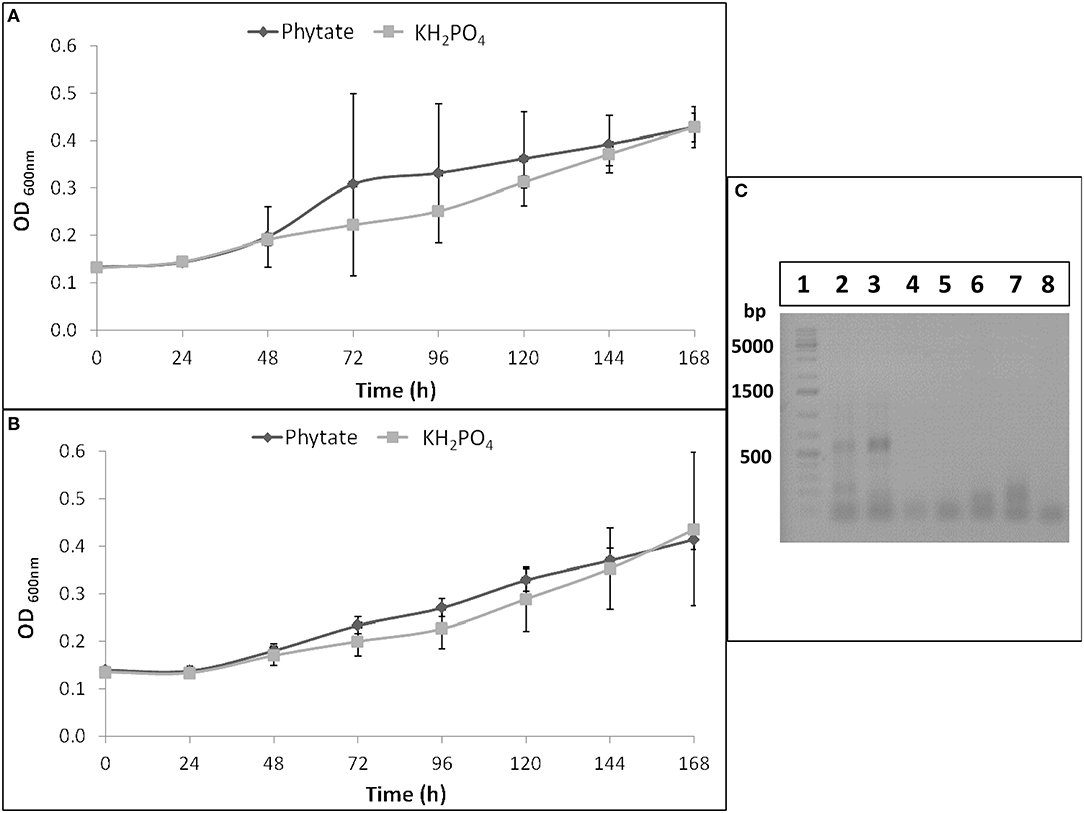
Figure 1. B. elkanii SEMIA 587 growth curves in Angle medium at pH 6 (A) or at pH 7 (B) supplemented with 1.5 mM KH2PO4 (light gray) or with 6 mM Na-phytate (dark gray). Data represent average and standard deviation of three replicates. (C) RT-PCR or PCR products of B. elkanii SEMIA 587 grown in Angle medium supplemented with Na-phytate or with KH2PO4 as P source in 1% agarose gel. Lane 1, GeneRuler 1 kb Plus DNA Ladder (Thermo Scientific); Lane 2, RT-PCR performed with HAP primers and cDNA as template obtained from SEMIA 587 cultivated with phytate as P source; Lane 3, RT-PCR performed with HAP primers (and BSA) and cDNA as template obtained from SEMIA 587 cultivated with phytate as P source; Lane 4, RT-PCR performed with BPP primers and cDNA as template obtained from SEMIA 587 cultivated with phytate as P source; Lane 5, PCR performed with HAP primers (and BSA) and gDNA as template obtained from SEMIA 587 cultivated with phytate as P source; Lane 6, PCR performed with BPP primers and gDNA as template obtained from SEMIA 587 cultivated with phytate as P source; Lane 7, RT-PCR performed with HAP primers (and BSA) and cDNA as template obtained from SEMIA 587 cultivated with KH2PO4 as P source; Lane 8, RT-PCR performed with BPP primers and cDNA as template obtained from SEMIA 587 cultivated with KH2PO4 as P source.
Two primers pairs, that amplify two types of phytases, HAP and BPP, were tested through a retro-transcription and a subsequent touchdown PCR in B. elkanii SEMIA 587. An amplification product was obtained when SEMIA 587 was cultivated with Na-phytate as P source and when HAP primers were used, but not when BPP primers were used (Figure 1C). When SEMIA 587 was cultivated with KH2PO4 as P source, no amplification products were obtained either when HAP or BPP primers were used (Figure 1C). Additionally, we demonstrated that HAP gene was not amplified from gDNA with these HAP primers (Figure 1C), due to the lower affinity between the mismatched primer and the gDNA at the used annealing temperature (62°C).
Efficiency in Use of the Rhizobial Symbiosis for Plant Growth
Cultivar N5909 cultivated under P sufficiency showed more shoot, nodules and root DW than under P deficiency (p = 0.0001, p = 0.0007, p = 0.0300, respectively), whereas no significant differences of shoot, nodules and root DW under P sufficiency and P deficiency in cultivar SR532 were found (Figure 2).
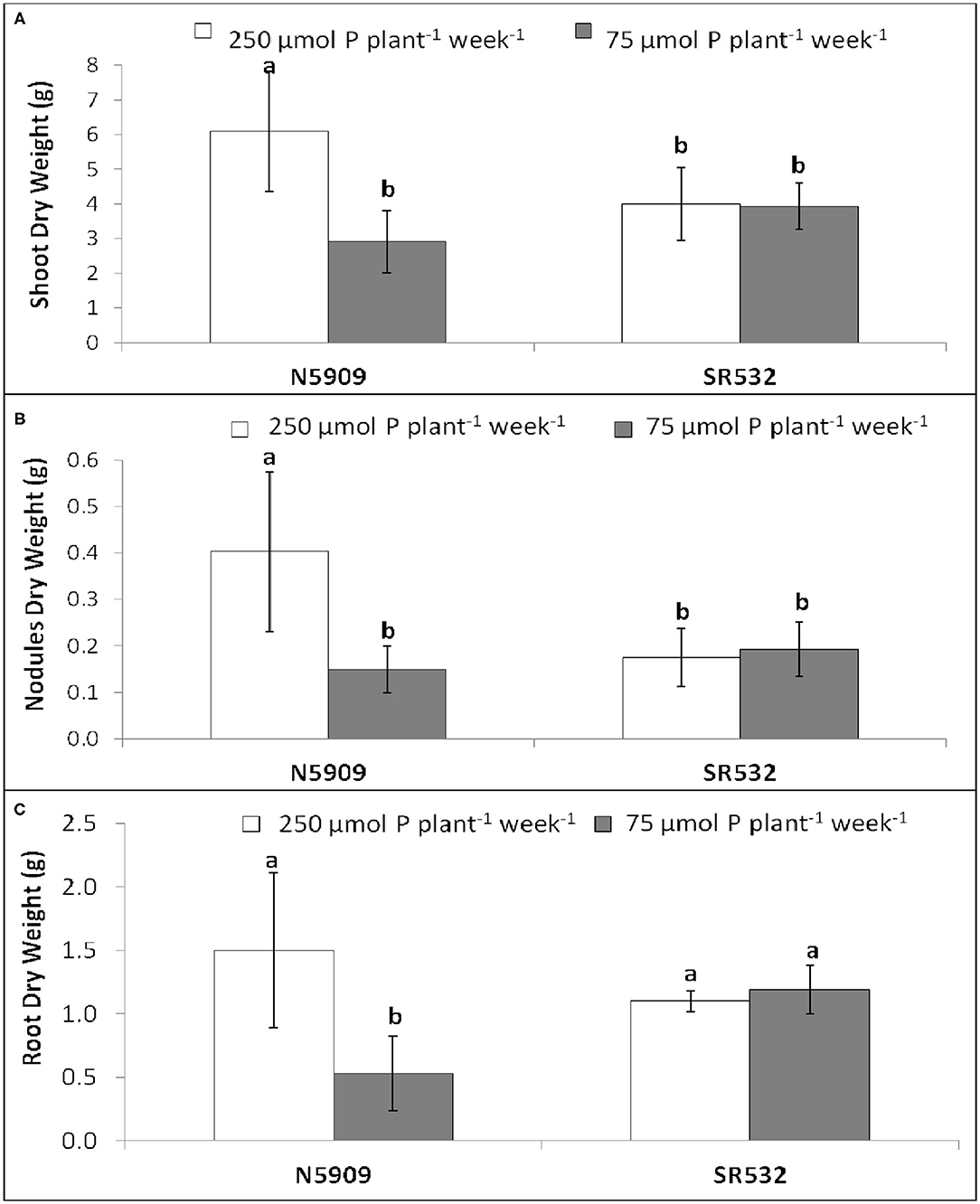
Figure 2. (A) Shoot dry weight, (B) nodules dry weight, and (C) root dry weight of G. max cultivars N5909 and SR532 inoculated with B. elkanii SEMIA 587 under P sufficiency (250 μmol P plant−1 week−1) (white) vs. P deficiency (75 μmol P plant−1 week−1) (gray). Data represent average and standard deviation of ten replicates harvested at 42 DAT. Treatments with a common letter are not significantly different (p > 0.05).
The EURS was estimated as the regression slope of shoot biomass as a function of nodule biomass, whenever the correlation between both parameters was significant (up to R2 = 0.719). Under P sufficiency, SR532 was the most efficient, and showed similar EURS under P deficiency, whereas N5909 increased its EURS under P deficiency (Figure 3).
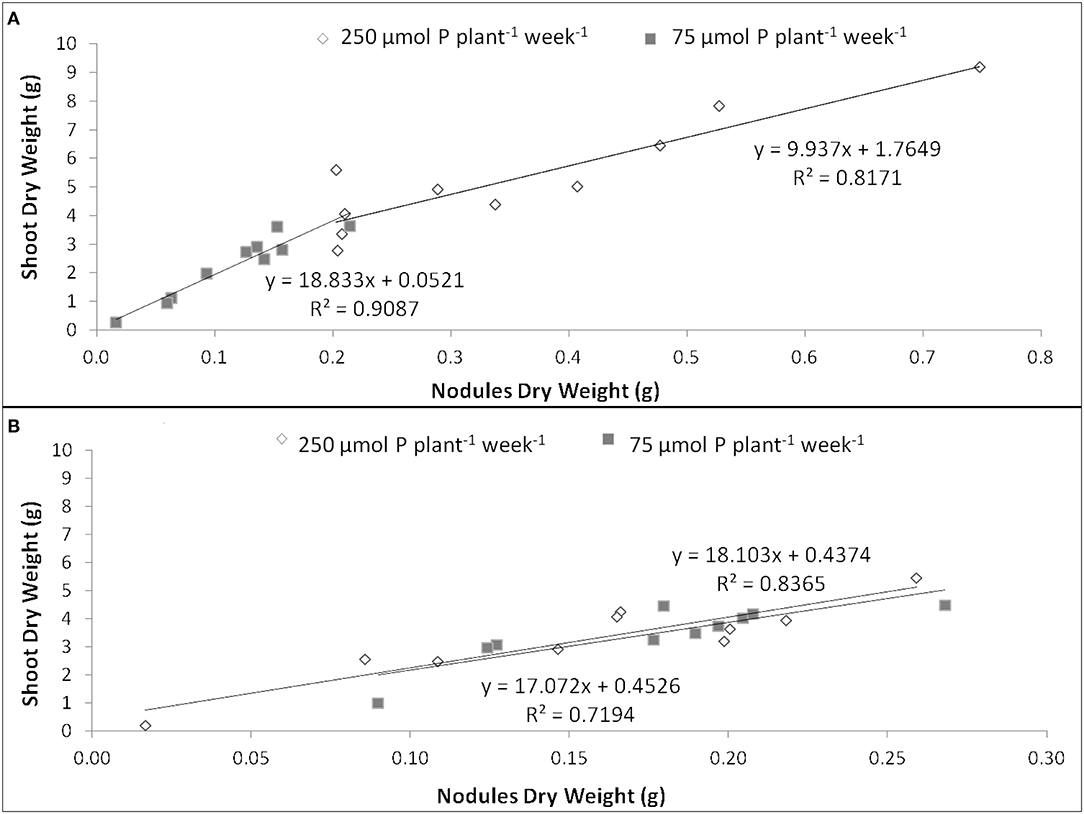
Figure 3. Efficiency in use of the rhizobial symbiosis (EURS) in G. max cultivars N5909 (A) and SR532 (B) under P sufficiency (250 μmol P plant−1 week−1) (white) vs. P deficiency (75 μmol P plant−1 week−1) (gray). Data are individual values harvested at 42 DAT.
Cultivar N5909 accumulated more P in its shoots under P sufficiency than under P deficiency (p = 0.0008), whereas cultivar SR532 did not show any significant difference in shoot accumulated P between P treatments (Figure 4).
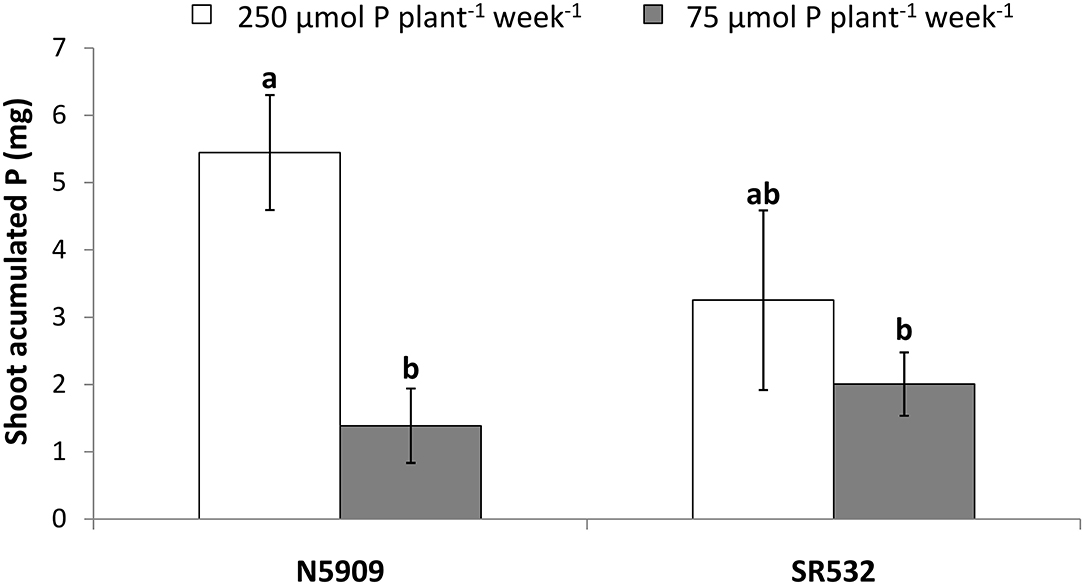
Figure 4. Shoot accumulated phosphorus in G. max cultivars N5909 and SR532 inoculated with B. elkanii SEMIA 587 under P sufficiency (250 μmol P plant−1 week−1) (white) vs. P deficiency (75 μmol P plant−1 week−1) (gray). Data represent average and standard deviation of ten replicates harvested at 42 DAT. Treatments with a common letter are not significantly different (p > 0.05).
Rhizobial Phytase Transcript Abundance in Soybean Nodules
Using the In situ RT-PCR technique we were able to show the transcript abundance of the rhizobial phytase in nodular sections of soybean cultivars N5909 and SR532 inoculated with B. elkanii SEMIA 587 cultivated under P sufficiency and P deficiency (Figures 5A–D). The NRT nodules not subjected to reverse transcription did not display any fluorescent signal (Figures 5E,F), confirming that fluorescence was not due to methodological artifacts. The rhizobial phytase was transcripted in all bacteroidal symbiosomes in the infected zone, with a high transcript abundance rate since the fluorescent signal, as assessment of number of transcripts, was intense for all the treatments (Figures 5A–D).
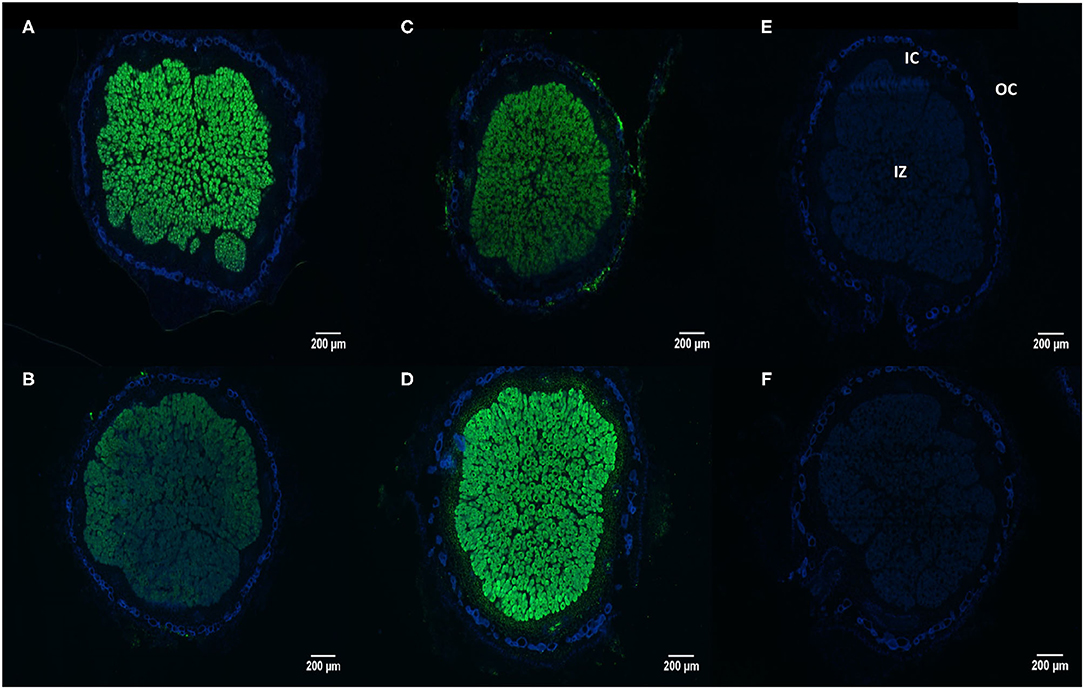
Figure 5. In situ RT-PCR localization of rhizobial phytase transcripts in nodule sections of G. max-B. elkanii SEMIA 587 symbiosis, under P sufficiency (250 μmol P plant−1 week−1), (A) cultivar N5909; (B) cultivar SR532; and under P deficiency (75 μmol P plant−1week−1), (C) cultivar N5909; (D) cultivar SR532. Negative controls (without reverse transcriptase) for: (E) cultivar N5909; (F) cultivar SR532. Transcripts are colored in green and nodular tissues in blue. IZ (infected zone), IC (inner cortex), OC (outer cortex).
In the case of cultivar SR532, the fluorescent signal was more intense under P deficiency in comparison to P sufficiency (p < 0.0001), whereas in the case of cultivar N5909, there were no significant differences between the intensity of the fluorescent signal under P deficiency or sufficiency. The intensity of the fluorescent signal in SR532 under P deficiency was equal to the intensity of this signal in N5909 under both P conditions (Figure 6). The ANOVA showed a significant interaction between the factors P supply and cultivar for the variable intensity of fluorescent signal (p = 0.0010).
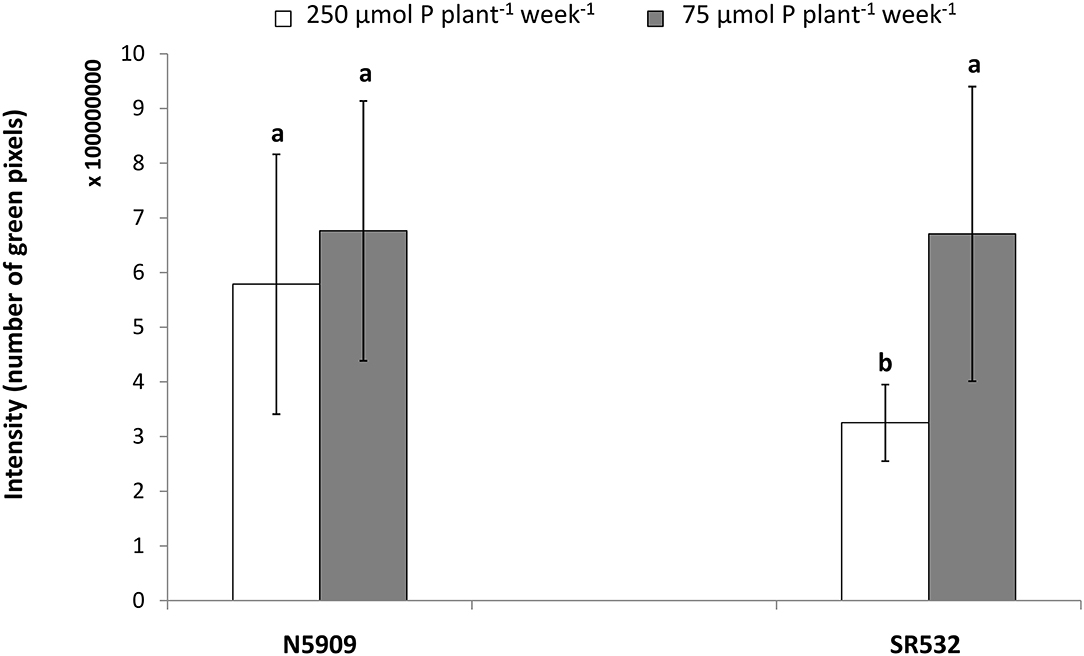
Figure 6. Intensity of the fluorescent signal emitted by the rhizobial phytase transcripts in nodules sections of G. max cultivars N5909 and SR532 inoculated with B. elkanii SEMIA 587 under P sufficiency (250 μmol P plant−1 week−1) (white) vs. P deficiency (75 μmol P plant−1 week−1) (gray). Data represent average and standard deviation of three nodules of three plants. The y-axis intensity of fluorescence is expressed in number of green pixels and must by 108. Treatments with a common letter are not significantly different (p > 0.05).
Discussion
The present study shows for the first time that a rhizobial strain could mineralize phytate and use it as a P source and that we could locate and observe the distribution of the transcripts of a rhizobial phytase within nodules of N2-fixing soybean plants, thanks to the in situ RT-PCR methodology carried out. This method has proven suitable to quantify the level of transcripts in prokaryote (Maougal et al., 2014b) and in eukaryotes cells (van Aarle et al., 2007; Bargaz et al., 2012; Lazali et al., 2013), and in the case of the present study, we were able to quantify the expression of rhizobial phytase transcripts with this method. The rhizobial phytase transcripts were localized only in the bacteroidal cells. It has been shown that P in G. max nodules was higher in the infected zone in comparison to the cortical region (revised by Hu et al., 2018). Thus, the high level of transcripts in the infection zone may respond to bacteroidal requirement for metabolism and survival, as well as bacteria multiplication (Bargaz et al., 2012; Lazali et al., 2013).
Legumes nodules are plant tissues with high concentration of P and when plants are under P-depletion, they follow a strategy to maintain SNF and viable leaf tissue as long as possible, expressing several P cycling genes in nodules (Cabeza et al., 2014). Plant phosphatase transcripts have been previously shown as affected by plant available P (Araújo et al., 2008; Bargaz et al., 2012; Lazali et al., 2013). Previously, the transcript abundance and activity of the PAP phytase of common beans was shown to increase when plants were grown under P deficiency, independently of the plant genotype (Araújo et al., 2008; Lazali et al., 2013). In addition, Bargaz et al. (2012) demonstrated that the transcript abundance and activity of the plant phosphoenol pyruvate phosphatase (PEPase) increased under P deficiency for several common bean genotypes. Contrary, the two soybean cultivars performed contrastingly when grown under low- and high-P supply. While cultivar N5909 did not show differences in intensity of the fluorescent signal under low- and high-P, cultivar SR532 showed a high transcript abundance of the rhizobial phytase in the nodules when grown under P deficiency. Moreover, the transcript abundance of the rhizobial phytase in the nodules of SR532 under P sufficiency seemed to be repressed compared to the transcript abundance in nodules of N5909 under the same conditions. These facts suggest that the transcript abundance rate of the rhizobial phytase depends on the plant genotype and P level, although further testing of other soybean genotypes is required. Additionally, the high transcript abundance level of the rhizobial phytase in the nodules of the cultivar SR532 under P deficiency, suggests that this enzyme contributes to the adaptation of this cultivar to P deficiency. Cultivar SR532 under P deficiency shows a correlation of high bacterial phytase transcripts with high EURS, suggesting a high regulation between EURS and the P content. Overall, our results suggest that the high tolerance of SR532 to P deficiency, as compared to N5909, is associated with better capacity to maintain SNF under low P supply based on the ability to increase the rhizobial phytase transcript abundance under such condition. Phytases might play a major role for internal plant metabolism rather than for obtaining P from the soil phytate (Tang et al., 2006). Phytate seems to act as a phosphate storage (Raboy, 2003) and rhizobial phytases can mineralize the phytate present in the soybean nodules, increasing the concentration of orthophosphates ions necessary for the SNF (Lazali et al., 2013).
Although further studies are required to identify new rhizobial strains with phytase activity and to test their interaction with various soybean genotypes, the differential transcript abundance shown in this research suggests that this phenomenon can contribute to the external and internal use efficiency of phytate for SNF. Sulieman et al. (2013b) found that different strains of S. meliloti showed different symbiotic efficiency with M. truncatula growing under different P levels. Thus, the identification and selection of an efficient combination of rhizobia and soybean genotype might play an important role in soybean breeding and production, aiming to reduce synthetic P fertilizer inputs and to improve sustainability in soils with low available P. We propose to consider the ability of a strain to mineralize phytate at the moment of evaluate new N2 fixing rhizobial strains, and to evaluate how the plant genotype influences this process, looking for combinations of rhizobia and plant that enhances P metabolism, thus SNF, under low available P soils, to select the best adapted symbiosis to P deficient soils.
Data Availability Statement
The raw data supporting the conclusions of this article will be made available by the authors, without undue reservation.
Author Contributions
VC performed the experimental work, analyzed data, and wrote the manuscript. LA and CT coordinated the experimental work. NA, EB, and J-JD conceived, planned the study, and helped to write the manuscript. All authors read and approved the final manuscript.
Funding
This research was funded by the Uruguayan Agencia Nacional de Investigación e Innovación (ANII) (grants MOV_CA_2016_1_127378 and POS_NAC_2014_1_102525) and by Agropolis Foundation, Montpellier, France, through Fabatropimed Project under the reference of ID 100-009.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
The authors thank Mustapha Teffahi, Luis Rojas, and Billal Kirdi for assistance with nodule collecting. We acknowledge Silvia Garaycochea for reviewing this manuscript.
References
Altier, N., Beyhaut, E., and Pérez, C. (2013). “Root nodule and rhizosphere bacteria for forage legume growth promotion and disease management,” in Bacteria in Agrobiology: Crop Productivity, eds D. K. Maheshwari, M. Saraf, and A. Aeron (Berlin, Heidelberg: Springer), 167–183. doi: 10.1007/978-3-642-37241-4_7
Angle, J. S., McGrath, S. P., and Chaney, R. L. (1991). New culture medium containing ionic concentrations of nutrients similar to concentrations found in the soil solution. Appl. Environ. Microbiol. 57, 3674–3676. doi: 10.1128/AEM.57.12.3674-3676.1991
Ao, J., Tian, J., Fu, J., Yan, X., and Liao, H. (2010). Genetic variability for root morph-architecture traits and root growth dynamics as related to phosphorus efficiency in soybean. Funct. Plant Biol. 37, 304–312. doi: 10.1071/FP09215
Araújo, A. P., Plassard, C., and Drevon, J. J. (2008). Phosphatase and phytase activities in nodules of common bean genotypes at different levels of phosphorus supply. Plant Soil 312, 129–138. doi: 10.1007/s11104-008-9595-3
Bardin, S., Dan, S., Osteras, M., and Finan, T. M. (1996). A phosphate transport system is required for symbiotic nitrogen fixation by Rhizobium meliloti. J. Bacteriol. 178, 4540–4547. doi: 10.1128/JB.178.15.4540-4547.1996
Bargaz, A., Ghoulam, C., Amenc, L., Lazali, M., Faghire, M., Abadie, J., et al. (2012). A phosphoenol pyruvate phosphatase transcript is induced in the root nodule cortex of Phaseolus vulgaris under conditions of phosphorus deficiency. J. Exp. Bot. 63, 4723–4730. doi: 10.1093/jxb/ers151
Beyhaut, E., DeHaan, L. R., Byun, J. L., Sheaffer, C. C., and Graham, P. H. (2006). Response to inoculation in Illinois bundleflower. Can. J. Plant Sci. 86, 919–926. doi: 10.4141/P05-097
Bucher, M. (2007). Functional biology of plant phosphate uptake at root and mycorrhizal interfaces. New Phytol. 173, 11–26. doi: 10.1111/j.1469-8137.2006.01935.x
Cabeza, R. A., Liese, R., Lingner, A., von Stieglitz, I., Neumann, J., Salinas-Riester, G., et al. (2014). RNA-seq transcriptome profiling reveals that Medicago truncatula nodules acclimate N2 fixing before emerging P deficiency reaches the nodules. J. Exp. Bot. 65, 6035–6048. doi: 10.1093/jxb/eru341
Castro-Guerrero, N. A., Isidra-Arellano, M. C., Mendoza-Cozatl, D. G., and Valdés-López, O. (2016). Common bean: a legume model on the rise for unraveling responses and adaptations to iron, zinc, and phosphate deficiencies. Front. Plant Sci. 7:600. doi: 10.3389/fpls.2016.00600
Cheng, L., Bucciarelli, B., Shen, J., Allan, D., and Vance, C. P. (2011). Update on white lupin cluster root acclimation to phosphorus deficiency update on lupin cluster roots. Plant Physiol. 156, 1025–1032. doi: 10.1104/pp.111.175174
de Mendiburu, F. (2020). Agricolae: Statistical Procedures for Agricultural Research. R package version 1.3-2. Available online at: https://CRAN.R-project.org/package=agricolae
Drevon, J. J., Abadie, J., Alkama, N., Andriamananjara, A., Amenc, L., Bargaz, A., et al. (2015). “Phosphorus use efficiency for N2 fixation in the rhizobial simbiosis with legumes,” in Biological Nitrogen Fixation, ed F. J. de Bruijn (Hoboken, NJ: John Wiley & Sons, Inc.), 455–464. doi: 10.1002/9781119053095.ch46
Elkoca, E., Kantar, F., and Sahin, F. (2007). Influence of nitrogen fixing and phosphorus solubilizing bacteria on the nodulation, plant growth, and yield of chickpea. J. Plant Nutr. 31, 157–171. doi: 10.1080/01904160701742097
Farhat, A., Chouayekh, H., Ben Farhat, M., Bouchaala, K., and Bejar, S. (2008). Gene cloning and characterization of a thermostable phytase from Bacillus subtilis US417 and assessment of its potential as a feed additive in comparison with a commercial enzyme. Mol. Biotechnol. 40, 127–135. doi: 10.1007/s12033-008-9068-1
Gahoonia, T. S., Ali, R., Malhotra, R. S., Jahoor, A., and Rahman, M. M. (2007). Variation in root morphological and physiological traits and nutrient uptake of chickpea genotypes. J. Plant Nutr. 30, 829–841. doi: 10.1080/15226510701373213
Graham, P. H., Rosas, J. C., Estevez de Jensen, C., Peralta, E., Tlusty, B., Acosta-Gallegos, J., et al. (2003). Addressing edaphic constraints to bean production: the bean/cowpea CRSP project in perspective. Field Crops Res. 82, 179–192. doi: 10.1016/S0378-4290(03)00037-6
Harvey, P. R., Warren, R. A., and Wakelin, S. (2009). Potential to improve root access to phosphorus the role of non-symbiotic microbial inoculants in the rhizosphere. Crop Pasture Sci. 60, 144–151. doi: 10.1071/CP08084
Hernandez, G., and Drevon, J. J. (1991). Influence of oxygen and acetylene during in situ open flow assays of nitrogenase activity (C2H2 reduction) in Phaseolus vulgaris root nodules. J. Plant Physiol. 138, 587–590. doi: 10.1016/S0176-1617(11)80246-4
Hill, J. E., Kysela, D., and Elimelech, M. (2007). Isolation and assessment 313 of phytate-hydrolysing bacteria from the DelMarVa Peninsula. Environ. Microbiol. 9, 3100–3107. doi: 10.1111/j.1462-2920.2007.01420.x
Hinsinger, P., Herrmann, L., Lesueur, D., Robin, A., Trap, J., Waithaisong, K., et al. (2015) “Impact of roots, microorganisms and microfauna on the fate of soil phosphorus in therhizosphere,” in Phosphorus Metabolism in Plants, eds W. C. Plaxton H. Lambers (Hoboken, NJ: JohnWiley & Sons, Ltd), 377–408. doi: 10.1002/9781119312994.apr0528.
Hu, Y., Jiao, J., Liu, L. X., Sun, Y. W., Chen, W. F., Sui, X. H., et al. (2018). Evidence for phosphate starvation of rhizobia without terminal differentiation in legume nodules. Mol. Plant Microbe Interact. 31, 1060–1068. doi: 10.1094/MPMI-02-18-0031-R
Hungria, M., Boddey, L. H., Santos, M. A., and Vargas, M. A. T. (1998). Nitrogen fixation capacity and nodule occupancy by Bradyrhizobium japonicum and B. elkanii strains. Biol. Fertil. Soils. 27, 393–399. doi: 10.1007/s003740050449
Israel, D. W. (1987). Investigation of the role of phosphorus in symbiotic dinitrogen fixation. Plant Physiol. 84, 835–840. doi: 10.1104/pp.84.3.835
Jorquera, M., Hernández, M., Rengel, Z., Marschner, P., and Mora, M., and de la, L. (2008b). Isolation of culturable phosphobacteria with both phytate-mineralization and phosphate-solubilization activity from the rhizosphere of plants grown in a volcanic soil. Biol. Fertil. Soils. 44, 1025–1034. doi: 10.1007/s00374-008-0288-0
Jorquera, M., Martínez, O., Maruyama, F., Marschner, P., and de la Luz Mora, M. (2008a). Current and future biotechnological applications of bacterial phytases and phytase-producing bacteria. Microbes Environ. 23, 182–191. doi: 10.1264/jsme2.23.182
Khan, M. S., Ahmad, E., Zaidi, A., and Oves, M. (2013). “Functional aspect of phosphate solubilizing bacteria: importance in crop production,” in Bacteria in Agrobiology: Crop Productivity, eds D. K. Maheshwari, M. Saraf, and A. Aeron (Berlin, Heidelberg: Springer), 237–332.263. doi: 10.1007/978-3-642-37241-4_10
Koo, K., and Jaykus, L. A. (2000). Selective amplification of bacterial RNA: use of a DNA primer containing mismatched bases near its 3' terminus to reduce false-positive signals. Lett. Appl. Microbiol. 31, 187–192. doi: 10.1046/j.1365-2672.2000.00798.x
Kouas, S., Labidi, N., Debez, A., Abdelly, C., Kouas, S., and Labidi, N. (2005). Effect of P on nodule formation and N fixation in bean. Agron. Sustain. Dev. 25, 389–393. doi: 10.1051/agro:2005034
Lazali, M., Brahimi, S., Merabet, C., Latati, M., Benadis, C., Maougal, R. T., et al. (2016). Nodular diagnosis of contrasting recombinant inbred lines of Phaseolus vulgaris in multi-local field tests under Mediterranean climate. Eur. J. Soil Biol. 79, 100–107. doi: 10.1016/j.ejsobi.2016.02.002
Lazali, M., Zaman-Allah, M., Amenc, L., Ounane, G., Abadie, J., and Drevon, J. J. (2013). A phytase gene is over expressed in root nodules cortex of Phaseolus vulgaris–rhizobia symbiosis under phosphorus deficiency. Planta 238, 317–324. doi: 10.1007/s00425-013-1893-1
Li, M., Osaki, M., Rao, I. M., and Tadano, T. (1997). Secretion of phytase from the roots of several plant species under phosphorus-deficient conditions. Plant Soil 195, 161–169. doi: 10.1023/A:1004264002524
Lindrström, K., Murwira, M., Willems, A., and Altier, N. (2010). The biodiversity of beneficial microbe-host mutualism: the case of rhizobia. Res. Microbiol. 161, 453–463. doi: 10.1016/j.resmic.2010.05.005
Lopez-Arredondo, D. L., Leyva-Gonzalez, M. A., Gonzalez-Morales, S. I., Lopez-Bucio, J., and Herrera-Estrella, L. (2014). Phosphate nutrition: improving low-phosphate tolerance in crops. Annu. Rev. Plant Biol. 65, 95–123. doi: 10.1146/annurev-arplant-050213-035949
Maougal, R. T., Bargaz, A., Sahel, C., Amenc, L., Djekoun, A., Plassard, C., et al. (2014b). Localization of the Bacillus subtilis beta-propeller phytase transcripts in nodulated roots of Phaseolus vulgaris supplied with phytate. Planta 239, 901–908. doi: 10.1007/s00425-013-2023-9
Maougal, R. T., Brauman, A., Plassard, C., Abadie, J., Djekoun, A., and Drevon, J. J. (2014a). Bacterial capacities to mineralize phytate increase in the rhizosphere of nodulated common bean (Phaseolus vulgaris) under P deficiency. Eur. J. Soil Biol. 62, 8–14. doi: 10.1016/j.ejsobi.2014.02.006
Mullaney, E. J., and Ullah, A. H. J. (2003). The term phytase comprises several different classes of enzymes. Biochem. Biophys. Res. Commun. 312, 179–184. doi: 10.1016/j.bbrc.2003.09.176
Olivera, L., Rodriguez, E., Ceretta, S., and Beyhaut, E. (2016). Repelentes de aves aplicados a la semilla de soja: compatibilidad con el inoculante y residualidad en cotiledones. Agrociencia Uruguay. 20, 51–60.
Quiquampoix, H., and Mousain, D. (2005). “Enzymatic hydrolysis of organic-phosphorus,” in Organic-Phosphorus in the Environment, eds B. L. Turner, E. Frossard, and D. S. Baldwin (Wallingford: CAB International), 89–112. doi: 10.1079/9780851998220.0089
Raboy, V. (2003). Myo-inositol-1,2,3,4,5,6-hexakisphosphate. Phytochemistry 64, 1033–1043. doi: 10.1016/S0031-9422(03)00446-1
Ramesh, A., Sharma, S. K., and Yadav, N. (2014). Phosphorus mobilization from native soil P-pool upon inoculation with phytate-mineralizing and phosphate-solubilizing Bacillus aryabhattai isolates for improved P-acquisition and growth of soybean and wheat crops in microcosm conditions. Agric. Res. 3, 118–127. doi: 10.1007/s40003-014-0105-y
Richardson, A. E. (2001). Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Funct. Plant Biol. 28, 897–906. doi: 10.1071/PP01093
Richardson, A. E., and Hadobas, P. A. (1997). Soil isolates of Pseudomonas spp. that utilize inositol phosphates. Can. J. Microbiol. 43, 509–516. doi: 10.1139/m97-073
Richardson, A. E., and Simpson, R. J. (2011). Soil microorganisms mediating phosphorus availability. Plant Physiol. 156, 989–996. doi: 10.1104/pp.111.175448
Rosas, B. S., Andrés, J. A., Rovera, M., and Correa, N. S. (2006). Phosphate solubilizing Pseudomonas putida can influence the rhizobia legume simbiosis. Soil Biol. Biochem. 38, 3502–3505. doi: 10.1016/j.soilbio.2006.05.008
Sa, T. M., and Israel, D. W. (1991). Energy status and functioning of phosphorus-deficient soybean nodules. Plant Physiol. 97, 928–935. doi: 10.1104/pp.97.3.928
Schneider, C. A., Rasband, W. S., and Eliceiri, K. W. (2012). NIH Image to ImageJ: 25 years of image analysis. Nat. Methods. 9, 671–675. doi: 10.1038/nmeth.2089
Schulze, J., Temple, G., Temple, S. J., Beschow, H., and Vance, C. P. (2006). Nitrogen fixation by white lupin under phosphorus deficiency. Ann. Bot. 98, 731–740. doi: 10.1093/aob/mcl154
Sulieman, S., Fischinger, S. A., Gresshoff, P. M., and Schulze, J. (2010). Asparagine as a major factor in the N-feedback regulation of N2 fixation in Medicago truncatula. Physiol. Plant. 140, 21–31. doi: 10.1111/j.1399-3054.2010.01380.x
Sulieman, S., Schulze, J., and Tran, L. S. P. (2013a). Comparative analysis of the symbiotic efficiency of Medicago truncatula and Medicago sativa under phosphorus deficiency. Int. J. Mol. Sci. 14, 5198–5213. doi: 10.3390/ijms14035198
Sulieman, S., van Ha, C., Schulze, J., and Tran, L. S. P. (2013b). Growth and nodulation of symbiotic Medicago truncatula at different levels of phosphorus availability. J. Exp. Bot. 64, 2701–2712. doi: 10.1093/jxb/ert122
Tang, C., Hinsinger, P., Jaillard, B., Rengel, Z., and Drevon, J. J. (2001). Effect of phosphorus deficiency on the growth, symbiotic N2 fixation and proton release by two bean (Phaseolus vulgaris) genotypes. Agronomie 21, 683–689. doi: 10.1051/agro:2001161
Tang, J., Leung, A., Leung, C., and Lim, B. L. (2006). Hydrolysis of precipitated phytate by three distinct families of phytases. Soil Biol. Biochem. 38, 1316–1324. doi: 10.1016/j.soilbio.2005.08.021
Vadez, V., Rodier, F., Payre, H., and Drevon, J. J. (1996). Nodule permeability to O2 and nitrogenase-linked respiration in bean genotypes varying in the tolerance of N2 fixation to P deficiency. Plant Physiol. Biochem. 34, 871–878.
van Aarle, I. M., Viennois, G., Amenc, L. K., Tatry, M. V., Luu, D. T., and Plassard, C. (2007). Fluorescent in situ RT -PCR to visualize the expression of a phosphate transporter gene from an ectomycorrhizal fungus. Mycorrhiza 17, 487–494. doi: 10.1007/s00572-007-0127-4
Yoon, S. J., Choi, Y. J., Min, H., Cho, K. K., Kim, J. W., Lee, S. C., et al. (1996). Isolation and identification of phytase-producing bacterium, Enterobacter sp. 4, and enzymatic properties of phytase enzyme. Enzyme Microb. Technol. 18, 449–454. doi: 10.1016/0141-0229(95)00131-X
Yuan, Z. C., Zaheer, R., and Finan, T. M. (2006). Regulation and properties of PstSCAB, a high-affinity, high-velocity phosphate transport system of Sinorhizobium meliloti. J. Bacteriol. 188, 1089–1102. doi: 10.1128/JB.188.3.1089-1102.2006
Keywords: nodule RT-PCR in situ, phytase, phytate mineralization, rhizobia, soybean, symbiotic nitrogen fixation, sustainability
Citation: Cerecetto V, Beyhaut E, Amenc L, Trives C, Altier N and Drevon J-J (2021) Contrasting Expression of Rhizobial Phytase in Nodules of Two Soybean Cultivars Grown Under Low Phosphorus Availability. Front. Sustain. Food Syst. 4:607678. doi: 10.3389/fsufs.2020.607678
Received: 17 September 2020; Accepted: 15 December 2020;
Published: 15 January 2021.
Edited by:
Everlon Cid Rigobelo, Universidade Estadual Paulista, BrazilReviewed by:
Peter Gresshoff, The University of Queensland, AustraliaNoemi Carla Baron Cozentino, Universidade Estadual Paulista, Brazil
Copyright © 2021 Cerecetto, Beyhaut, Amenc, Trives, Altier and Drevon. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Elena Beyhaut, ZWJleWhhdXRAaW5pYS5vcmcudXk=; Jean-Jacques Drevon, ZHJldm9uampAeWFob28uZnI=
 Victoria Cerecetto
Victoria Cerecetto Elena Beyhaut
Elena Beyhaut Laurie Amenc2
Laurie Amenc2 Nora Altier
Nora Altier