- 1Southern Plains Agricultural Research Center, United States Department of Agriculture, Agricultural Research Service, College Station, TX, United States
- 2Department of Food Science, Center for Food Safety, University of Arkansas, Fayetteville, AR, United States
- 3Jones-Hamilton Co., Walbridge, OH, United States
- 4Department of Computer Science and Computer Engineering, University of Arkansas, Fayetteville, AR, United States
Due to global climate change, water is becoming increasingly scarce. The poultry industry is a major consumer of fresh water; therefore, in order to reduce their environmental footprint, companies are in the beginning stages of evaluating water for re-use. Re-use water is classified as water from the poultry plant that has been collected, sanitized, and used again during poultry processing. Necessarily, this water must be potable as it otherwise produces a significant risk to the poultry industry. One of the most commonly used sanitizers in processing water is peracetic acid (PAA). At high concentrations, PAA is corrosive to the metal equipment in the plant and introduces a significant occupational hazard to plant workers. Inorganic sanitizers, such as sodium bisulfate (SBS), have documented antimicrobial and sanitation effects on a variety of surfaces. In this study, SBS and PAA were compared for their ability to sanitize re-use water collected from a local poultry plant. Fresh, commercial poultry processing plant re-use water was collected at the end of a processing shift and used within 1 h of the collection. Microcosms were created to simulate the sanitation environment and a time course collection of live microorganisms were collected and evaluated for aerobic plate counts, total Enterobacteriaceae, and Salmonella load. The microcosms contained either SBS (1, 2, or 3%) or 200 ppm PAA throughout the course of the study. Water samples were collected at 0, 15, 30, and 60 min post-sanitation. The water was evaluated for total Enterobacteriaceae and microbial load using traditional plating methods and the total DNA was extracted and sequenced using the 16S rDNA Illumina MiSeq v3 Platform targeting the V4 region of the prokaryote rDNA molecule. All sanitation methods resulted in nearly undetectable XLD and APC counts by 60 m (P < 0.05), with 1 and 2% SBS having a slight rebounding of APC counts by 60 m. The sequences were analyzed using the QIIME2.2018.08 pipeline. Changes in alpha diversity and beta diversity were indicated across time and by treatment group, which may reflect the cumulative effects of the sanitation treatment (killing, delayed killing, and the emergence of resistant populations). Overall, based on the ANCOM results, SBS groups had less Pseudmonas and more Bacteroides while PAA groups had greater Actiobacter and Gallinobacterium (Q < 0.05). The results of this study indicate that SBS is as efficacious as PAA for decreasing bacterial loads in poultry re-use water, but it may favor different populations of bacteria in that water. Therefore, while more studies are needed to evaluate the changes in the microbiome when re-use water is applied to carcasses, evidence presented herein suggest that the choice of sanitizer in water re-use sanitation may impact downstream microbial loads.
Introduction
When factoring in seasonal variation, two-thirds of the world's population face severe water scarcity during at least 1 month of the year (Mekonnen and Hoekstra, 2016). As the human population continues to increase and consume more water, scarcity of this natural resource will become more profound (Arnell, 2004; Alcamo et al., 2007; Schewe et al., 2014). In the U.S., due to the geography, there are large portions of the country subject to water restrictions to protect this natural resource, including the great plains and southwestern United States (Averyt et al., 2011). Unfortunately, agricultural activity has a large impact on water use, attributed to 80% of the current water usage in order to support horticulture, livestock, and energy demands (Shannon et al., 2008). With increased scrutiny from the public sector, the agricultural industry is facing significant pressure to conserve water use.
The poultry industry has a significant water use foot print. In poultry processing alone, an estimated 26.5 L of water are used to process each carcass (Avula et al., 2009). Reducing the total water consumption of a poultry processing facility per bird will likely lead to unintended food safety risks (Lillard, 1980; Walsh et al., 2018). Importantly, water is a major component of the Hazard Analysis and Critical Control Point (HACCP) plans implemented industry-wide; and, even without sanitizers water can be effective to dilute pathogens and reduce biofilm forming bacteria (Srey et al., 2013; United States Department of Agriculture Food Safety Inspection Service, 2016). Water reuse in processing is covered by 9 CFR 416.2 and is enforced by the United States Department of Agriculture (USDA). This regulation states that water re-use is allowed if the water is equivalent to potable standards (United States Department of Agriculture Food Safety Inspection Service, 2016). While reducing the total amount of water per bird is unlikely, the ability to re-use water collected from the plant is promising. This approach would lessen the burden of the plants on the municipal supply while satisfying federal and industry-wide regulations. Current restrictions require that re-use water can be used on carcasses at the same stage or later on the processing line where it is collected prior to sanitation (United States Department of Agriculture Food Safety Inspection Service, 2016).
The foundations for improving water re-use and ensuring food safety have already been implemented throughout the processing industry, though validation and additional research is needed (Micciche et al., 2019). To reduce the total microbial burden and load on carcasses, sanitizers, such as chlorine or peracetic acid (PAA) are added to water and used during the wash steps, dips, pre-chillers, and chillers throughout poultry processing (Tsai et al., 1992; Kitis, 2004; Bauermeister et al., 2008). Free chlorine is regulated in re-use water to no more than 5 ppm (United States Department of Agriculture Food Safety Inspection Service, 2003). As re-use water has to be potable, this can be concerning as studies have shown that low levels of chlorine (5 ppm) may reduce total microbial burdens without preventing biofilm formation (Bertelli et al., 2018; Fish and Boxall, 2018). Peracetic acid (PAA) may be utilized in higher concentrations (2,000 ppm) and has been demonstrated to have potentially bactericidal effects on bacteria and the allowable inclusion rate is increasing (21 CFR 173.370; Fatemi and Frank, 1999). Specifically, PAA is effective against significant foodborne bacteria like Campylobacter and Salmonella (Nagel et al., 2013). While efficacious, currently there is pressure from federal agencies as well as the poultry plants to reduce the use of PAA. PAA can be corrosive toward equipment, pose an environmental hazard toward plant workers, and is unstable which requires a large inclusion rate of the chemical throughout processing (Casani et al., 2005; National Research Council, 2010; Walsh et al., 2018). While bactericidal, resistance is mounting and other sanitation methods need to be investigated in order to find safer, more effective avenues to improve food safety.
One likely alternative to PAA and chlorine would be inorganic acids. Sodium bisulfate (SBS) has shown efficacy against Campylobacter in poultry litter and Salmonella on chicken drumsticks (Line, 2002; Dittoe et al., 2019). Previously our group set out to determine if SBS may be an effective antimicrobial for Salmonella in water intended for re-use using an in vitro microcosm study. Data indicated that SBS at 1, 2, and 3% w/v was effective against Salmonella as compared to PAA and that these effects may be bactericidal (Micciche et al., 2018). However, the previous study did not document how these concentrations impact other important microorganisms, such as total Enterobacteriaceae or APC load as well as the overall microbial population dynamics. Changes in microbial population ecology may impact spoilage and food safety risks, therefore are necessary to document before recommendations can be made (Kim et al., 2017).
Water potability necessitates a total reduction in microbial load. Water re-use in poultry plants is required to make the water clean enough to use in processes upstream from where it was collected. Therefore, the goal of this study was to evaluate whether or not sanitation with SBS (1, 2, and 3%) was effective at reducing specific microbial groups and how those sanitizers impact microbial ecology within 1 h of exposure to the sanitizer. One hour of sanitation was chosen as that is the likely time for the water to be processed prior to re-circulation. This study was conducted without neutralization as it more likely represents the real-world risk that re-use water poses in a poultry plant. By evaluating the changes of the microbial ecology and recoverable pathogens from 0 to 60 min post-sanitation, the investigation helps determine if SBS is a viable option for the sanitation of water re-use as compared to PAA.
Materials and Methods
Water Collection
Processing water (n = 5) was collected from a Northwest Arkansas commercial poultry processing facility at the intake of the re-use water sanitization system as described in Micciche et al. (2018). The intake water was chlorinated with 20–50 ppm of total chlorine at the facility. The collected, pre-sanitation re-use water at this processing facility was intended to be re-circulated post-sanitation to the inside-outside bird wash station post-evisceration. The water was transported to the laboratory at room temperature and used within 1 h of collection.
Microcosms and Assessment of Antimicrobial Effects
Upon arrival, an aliquot (100 μL) of each water sample was plated onto Tryptic Soy Agar (TSA) and Xylose Lysine Deoxycholate (XLD) to determine the total microbial load prior to sanitation and then incubated at 37°C for 24 h (Difco Laboratories, Detroit, MI, USA). The initial microbial load and population was assessed by aliquoting 1 mL of the pre-sanitized re-use water, centrifuged at 12,000 rpm for 10 min, the supernatant was removed and the pellet was frozen at −20°C until DNA extractions were performed.
At the start of the study, 0 m, 20 mL of re-use water was aliquoted to an Erlenmeyer flask that individually contained one of the following acid sanitizers to generate the following concentrations: 0.02% PAA (v/v) (Sigma-Aldrich, St. Louis, MO, USA), and 1, 2, and 3% SBS (w/v) (Jones-Hamilton Co, Walbridge, Ohio, USA). A no treatment control was also included.
Throughout the course of the study, the microcosms were incubated in an oscillating incubator (150 rpm) at 25°C to simulate the turbulent flow of water through plant piping. At 30 and 60 m post-sanitation, 100 μL aliquots were direct plated onto TSA or XLD in duplicate, and 1 mL was pelleted and frozen as previously described for DNA extractions in singlets. The TSA and XLD were incubated and enumerated as per the manufacturer's instructions. The terminal time point of 60 m was chosen as water re-use within a processing facility re-enters circulation within 1 h of sanitation (United States Environmental Protection Agency, 2010).
DNA Extractions
The DNA was extracted from frozen pelleted re-use water using the Gram-positive protocol for the QIAGEN DNeasy Blood & Tissue Kit (Qiagen, Valencia, CA, USA), with the final elution from the spin column utilizing 50 μL nuclease free deionized water instead of the elution buffer ATE. The DNA concentration and purity were estimated using a NanoDrop 1000 Spectrophotometer (Thermo Fisher Scientific, Waltham, MA, USA) and then diluted to 10 ng/μL. The extracted diluted DNA was stored at −20°C until library preparations commenced.
Library Preparation and Sequencing
A library targeting the V4 region of 16S rRNA gene was created as detailed in Kozich et al. (2013) and Park et al. (2017) with the only change being an increase in PhiX in order to control for the low GC content in the water microbiome (10%). Each library was diluted to 20 nM and was mixed with HT1 Buffer along with a PhiX control v3 (20 nM) and 0.2 N NaOH, to generate a final concentration of the library at 12 pM.
Sequencing Analysis
The sequencing reads files were downloaded from the Illumina BaseSpace website, and the QIIME2 pipeline (version 2018.11) was used (Bolyen et al., 2018). Paired end sequences were demultiplexed, trimmed, and denoised through DADA2 (Callahan et al., 2016). The assembled sequences were utilized to construct operational taxonomic units (OTUs) with 97% identity. Representative sequences were classified based on the Greengenes 16S rRNA gene database from phylum to genus levels (DeSantis et al., 2006). The α-diversity (Observed OTUs, Shannon Diversity, Pielous Evenness, Faith PD) and β-diversity indices (Bray-Curtis, Jaccard Distance, Weighted and Unweighted UniFrac) were generated using the standard pipeline. Principal coordinate analysis (PCoA) of β-diversity was also performed and plotted with QIIME2, with the continuous variable of time. The Analysis of Compositions of Microbiomes (ANCOM) was performed using the standard pipeline (Mandal et al., 2015).
Statistical Analysis of Count Data and Microbiome Compositional Changes Due to Treatment
The CFU counts were log10 transformed prior to statistical analyses. The prevalence counts were considered positive if enumerable colonies were detectable on the plate. Analyses for prevalence were through the Generalized Regression for Prevalence MP SAS (Mandal et al., 2015).
Proc GLM in JMP SAS 14.0 (SAS; Cary, North Carolina, United States) with linear contrasts were used to evaluate the main effects and the interaction thereof (time, treatment, time · treatment). If the main effects and the interaction were significant, pairwise linear contrasts were performed to determine the leverage effects. The statistical analyses regarding diversity and compositional abundance was contained within the QIIME2 platform. The initial statistical evaluation of the overall effect was considered significant with P < 0.05. Kruskal-Wallis pairwise comparisons of α diversity evaluating the differences of each of the levels within the main effect were considered significant at Q < 0.05, with the q-value representing the incorporation of the false discovery rate (Kruskal-Wallis; Siegel and Castellan, 1988). In order to evaluate the initial differences in similarity between treatment groups with beta diversity, permutation multivariate analysis of variance (PERMANOVA) was used to determine if β-diversity indices were significant, using 999 permutations in total for each pair-wise contrast. The main effect was considered significance at P < 0.05, and the individual pair-wise contrasts considered significant at Q < 0.05.
Indiscriminate OTU picking based on interest and arbitrary focus ultimately leads to significant false discovery rates, therefore in order to ascertain statistically significant differences in microbial composition, of significantly different OTUs between the interaction of time and treatment were performed by ANCOM (Analysis of Microbial Composition) through the QIIME2 q2-composition plugin at the genus level (Mandal et al., 2015). ANCOM is appropriate given the low number of samples as ANCOM increases statistical power as well as incorporating a stringent and effective discrete false discovery rate. QIIME2 q2-composition plugin was used for the final stage of the analysis (Mandal et al., 2015). The main effect with the most divergent and significant β-diversity effects were selected for the final analyses, with the genus focused on as it provided the greatest resolution for identifying indicator and pathogenic organisms present in re-use water. The q-value was used for significance (Q < 0.05). Data was exported from QIIME2 then imported into Excel (Microsoft; Redmond, WA, USA). Composite bar graphs reflecting the compositional changes between the treatment groups were created from significant results.
Results and Discussion
Impact of Acid Sanitizers on Microbial Load
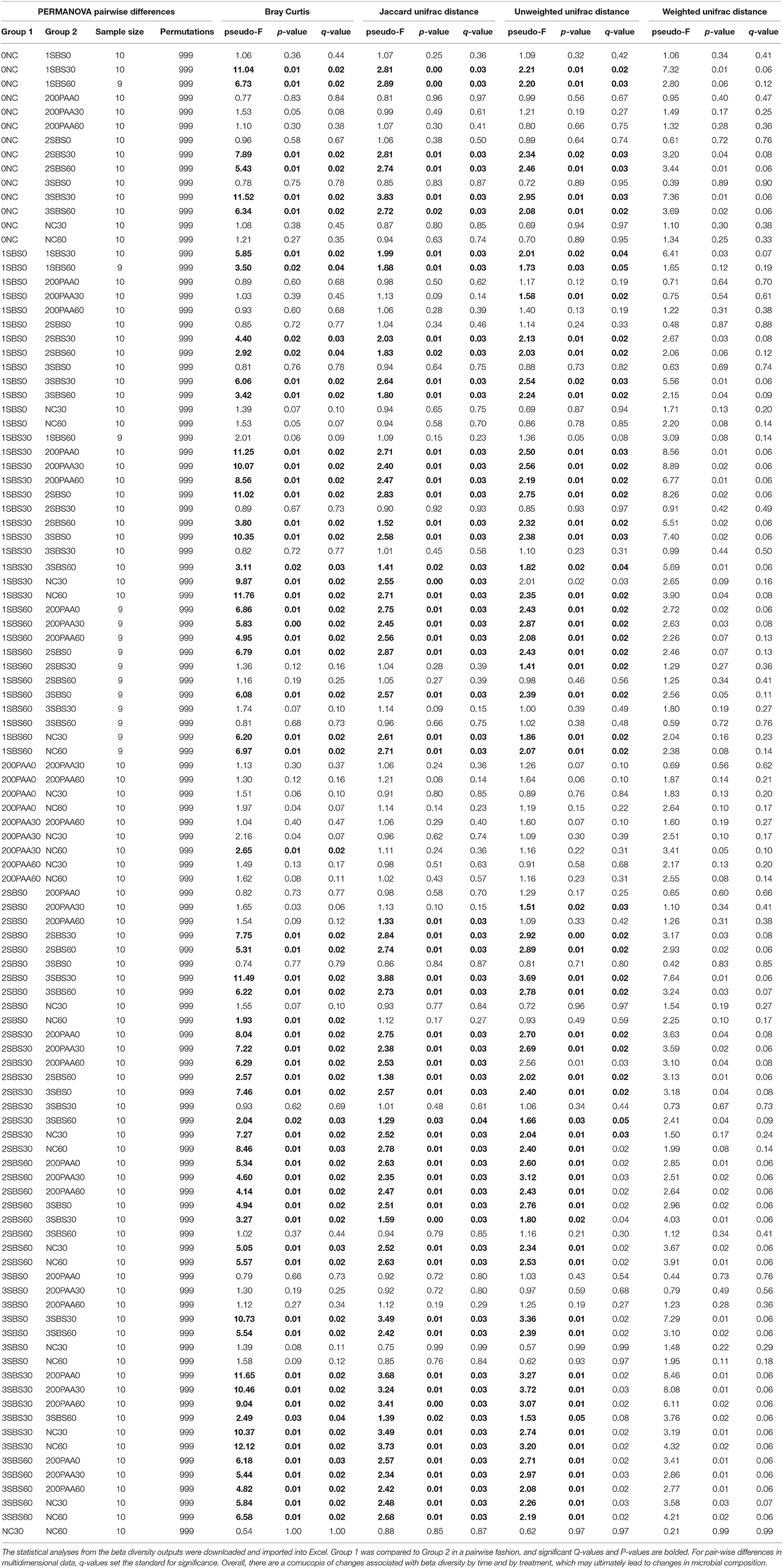
Figure 1A documents the effects of the sanitizers on total APC load, where the main effects of time, treatment, and the interaction thereof were statistically significant for load (P < 0.05). For prevalence (Figure 1B), only treatment and treatment by time interactions were significant for prevalence (P < 0.05). For all treatments, there were significant reductions in recoverable APC counts and prevalence as compared to the NTC group (P < 0.05 Treatment; P < 0.05 Time; P < 0.05 Interaction). The non-treatment control (NTC) group was not significantly different over time and there was also no difference in load or prevalence for the treatment groups at 0 m.
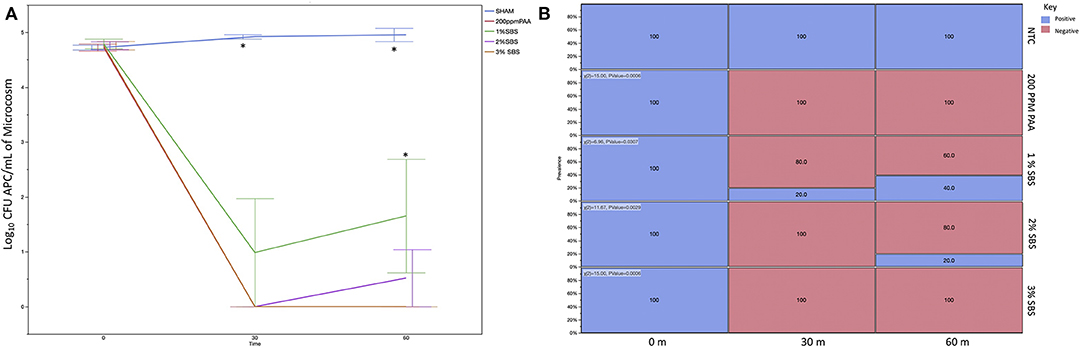
Figure 1. (A) Log10 APC CFU/mL Re-Use Water. Microcosms were created with either PAA or SBS (1, 2, and 3%). Samples were collected at 0, 30, and 60 m post-sanitation and were plated on APC media and incubated aerobically for 24 h. The plates were enumerated and statistically analyzed, with significance defined at P < 0.05 (asterisk indicates significant differences). There were no differences between the groups at 0 m. However, at 30 m, the treatment groups had all exhibited a 4-log10 reduction in APC. PAA and 3% PAA had no detectable APC after 30 m. 1 and 2% SBS had a slight increase in APC load by 60 m. (B) Prevalence of APC Counts Isolated from Re-Use Water. The recoverable APC were considered positive plates for prevalence. The individual trends were analyzed via χ2 distribution and their pairwise differences were evaluated statistically. The NTC (Sham) experiment had no change in prevalence throughout the experiment. The 200 PPM and 3% SBS groups had a complete loss of recoverable APC by 30 m. The 2 and 3% SBS groups both exhibited a rebound in their APC prevalence.
However, starting at 30 m, the NTC control has over a 4 log10 load greater APC load as compared to the sanitizer treatment groups, with all sanitation groups having a significant reduction in APC CFU (P < 0.0001). There was no statistically significant difference between 30 and 60 m (P = 0.277) when comparing the effect of time alone for APC load. However, at 60 m, the APC log CFU counts rebounded in the 1% SBS and 2% SBS group. Both the 1% SBS and 2% sanitation treatment are significantly different at 60 m, with an average difference of 1.13 log10 CFU between the groups (SEM = 0.50; P = 0.026). However, both the 1% SBS sanitation treatment and the 2% sanitation treatment had significantly greater recoverable CFUs than the PAA and 3% SBS group (both effects: P < 0.01). The 200 ppm PAA and 3% SBS group were not statistically different from one another, both effectively reducing the total recoverable APC load below the limit of detection. In terms of prevalence, both the 1 and 2% SBS groups had positive plates at 60 m, with 40% of the plates positive for 1% SBS and 20% for 2% SBS (Figure 1B). Both prevalence rates were significantly different than the other treatments and controls at 60 m.
Total changes in microbial load recovered from XLD plates changed throughout the experiment. For microbial load, the effect of time, treatments and the interaction thereof were statistically significant (Figure 2A; P < 0.001). Prevalence effects of treatment and time by treatment effects were also statistically significant, however time was not (Figure 2B; P < 0.001). As expected, no significant differences between the plate counts were observed at the start of the experiment (0 m; P > 0.05). Additionally, the sanitation treatments were all able to reduce the recoverable XLD plate counts as compared to the NTC (P < 0.001). At 30 m, 2% SBS was greater than the other treatments (P < 0.05). The 2% SBS group also had 20% of the plates positive for recoverable CFUs from XLD media (P < 0.05). At 60 m, none of treatment groups yielded recoverable bacteria in any of the microcosm samples, with the exception of the NTC.
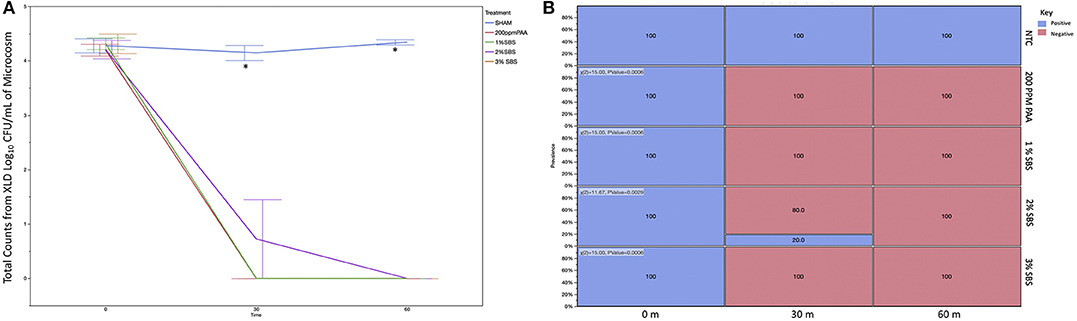
Figure 2. (A). Log10 CFU/mL of CFUs on XLD from Re-Use Water. Microcosms were created with either PAA or SBS (1, 2, and 3%). Samples were collected at 0, 30, and 60 m post-sanitation and were plated on APC media and incubated aerobically for 24 h. The plates were enumerated and statistically analyzed, with significance defined at P < 0.05 (asterisk indicates significant differences). There were no differences between the groups at 0 m and there was a complete reduction in recoverable counts by 30 m, which was sustained throughout the course of the experiment. (B) Prevalence of CFU Counts from XLD Isolated from Re-Use Water. The recoverable CFUs from XLD were considered positive plates for prevalence analysis. The individual trends were analyzed via χ2 distribution and their pairwise differences were evaluated statistically. The NTC and 1% SBS had positive plates at 30 m, with the remainder of the treatment groups not having positive plate counts from 30 m onward. At 60 m, only the NTC group was both not different than 0 m and had recoverable counts. All other treatment groups had no positive plates at 60 m.
Alpha and Beta-Diversity of the Acid Sanitized Microcosms
In order to determine if differences in composition were due to differences in unique OTUs, α-diversity was analyzed and pairwise contrasts of the significant effects were produced for the metrics: Observed OTUs, Shannon Diversity, Pielou's Evenness, Faith PD (Figure 3; Table 1). There were no significant effects for the OTU counts between treatments or measured effects (data not shown). However, Pielou's Evenness, Shannon Diversity, and Faith PD indices all had significant effects of time. Using the Faith PD diversity metric, there was a decrease in diversity between 0 m (Q < 0.0–2) and an increase in α-diversity from 30 and 60 m (Q < 0.02). There was no difference in alpha diversity from 0 to 60 m. Pilou's Evenness and the Shannon Diversity index showed similar significant differences over time. Both indices demonstrated a decrease in either diversity or read evenness from 0 to 30 m and 0 to 60 m (Q < 0.01; Figure 3; Table 1). Therefore, the effects of time had quantitative effects on community richness and evenness.
Figure 3. Alpha diversity significance by time throughout the course of sanitation (A) Faith's diversity index; (B) Pielou's evenness index; (C) Shannon diversity index. When comparing pairwise differences, the same letter within graph is not significantly different whereas different letters are statistically significantly different (Q < 0.05). Overall, diversity deceases with time.
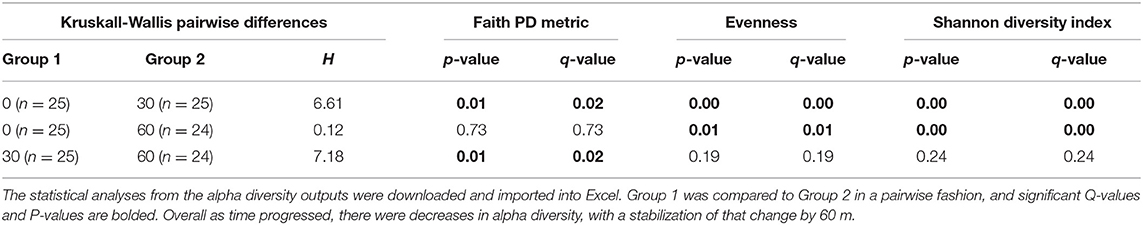
Table 1. Significant Kruskall-Wallis pairwise differences in alpha diversity indices whose main effects were significant.
β-diversity analysis yielded significant differences in the interaction effects over time. With α-diversity not statistically significant for the effect of time nor the interaction of the treatment by time, the community dissimilarity arose due to the effects of the sanitizers over time alone. PERMANOVA was used for the main effects, with full factorial pair-wise contrasts documenting the significant effects of the sanitizers on microbial community composition (significant relationships bolded; Table 2). The weighted unifrac distance matrix was not significant for any main effects nor the interaction. The effect of SBS vs. PAA was continually reinforced with all of the other β-diversity metrics as the contrasts between SBS and PAA at each time point were significant (Q < 0.05). The visualization of this separation is evident in Figure 4, with the blue line showing the separation between the SBS versus the PAA treatment. PAA and the NTC were not statistically significantly different from one another throughout the trial, while all SBS treatment groups had differences in β-diversity indices. Therefore, the effect of the sanitizer on the microbial community is different, with PAA being more similar to the NTC than SBS.
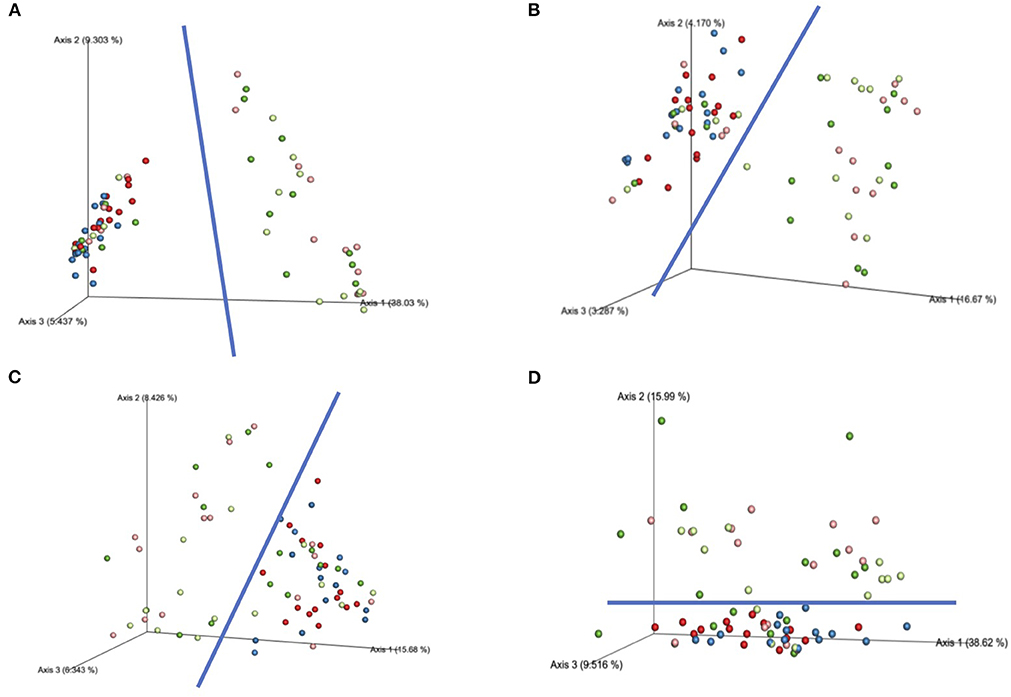
Figure 4. Significant beta diversity indices by time and treatment throughout the course of sanitation. (A) Bray-Curtis Matrix, (B) Jaccard Matrix, (C) Unweighted Unifrac Distance Matrix, (D) Weighted Unifrac Distance Matrix. The colors: Red: 200 PAA, Blue: NTC, Light Green 1% SBS, Dark Green 2% SBS, Pink, 3% SBS, Blue overreaching circle represents a majority of the SBS treatments. Dark blue line represents the separation between the SBS treatments and the controls (average).
ANCOM Analysis of Compositional Diversity
The use of ANCOM to discern quantitative compositional differences between the treatment groups over time indicate significant shifts by treatment and time. The populations present in the ANCOM analysis are all statistically significantly different than the background populations (Q < 0.05) and each other. The composite diagram is shown on Figure 5. Compared to treatment or time alone, the interaction of time by treatment was specifically focused on due to the differences in β-diversity indices. The NTC group had relative differences in Microvirgula, Barnsiella, and Halomonas over time. The SBS treatment groups had a greater percentage of Stentrophomonas, Schwanella, and Bacteroides. The 3% SBS group had the greatest abundance in Bacteroides overall, showing a stepwise increase over time in this population. Likewise, the Aeromonadaceae, Actinobacter, Gallinobacterium, Schwanella, Halomonas, and Pseudomonadaceae populations decreased over time in the 3% SBS group. The PAA group also has more Gallinobacterium, which was not detectable in the 30 or 60 m 3% SBS group. Comparing the 3% SBS group with the 200 ppm PAA group, there were increases in Bacteroides, Schwanella and a decrease in Neisseriaceae in the SBS group. Consistently in the SBS groups, there were increases over time in Schwanella, with fluctuations in Halomonas between each group.
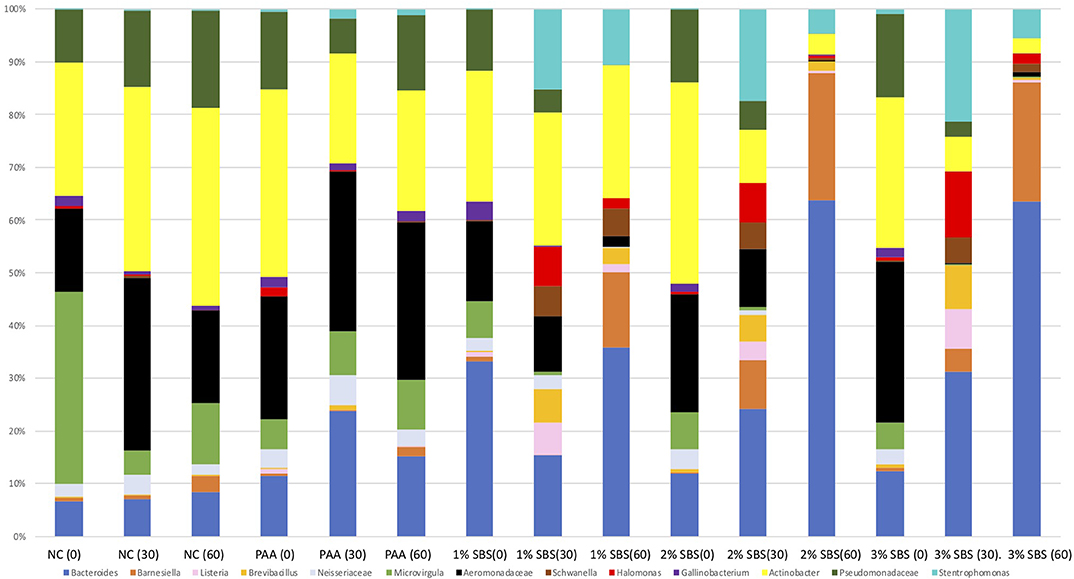
Figure 5. The Analysis of Composition of Microbiomes (ANCOM) throughout sanitation. Relative stack bar plots were created from the top 11 OTUs with a Q < 0.05 and a significant leverage effect that were statistically associated with the time by treatment effect. Ultimately, significant variations in microbial composition occur over time and by treatment.
Discussion
While microbiome data is an important snapshot into the microbial ecology of re-use water as it goes through the sanitation process, it is not necessarily indicative of the species that are recoverable. Unlike rinsate studies where the microbial ecology drives shelf-life and food safety risks to the customer, re-use water likely will go through additional, and unintended, sanitation by the continuous infusion of antimicrobials throughout processing. Therefore, injured bacterial populations are not necessarily important as their continued biological weakening will occur as their application in the plant will include the use of additional antimicrobials, such as chlorine or PAA. Rinsate bacteria recovered from the carcass will have time to recover and pose a significant risk, whereas colonies isolated prior to re-use will not likely have the time to recover. However, live and healthy bacteria quantified directly without enrichment or recovery media will absolutely pose a threat, and may increase the total microbial load on the carcass. Increased microbial loads may be more difficult to reduce if re-use water is implemented and added to that load. Assessing sanitized re-use water for recovered bacteria is necessary as recovered bacteria would likely point to a lack of a bactericidal effect.
Therefore, total aerobic plate counts (APC) and total counts from XLD media were used for this study. Aerobic plate counts throughout poultry processing are an important indicator of total microbial load and the risk of foodborne pathogens (Kim et al., 2017; Samapundo et al., 2019). The initial goal of using XLD was to recover any Salmonella present in re-use water that may be resistant to sanitation. However, there was no recoverable Salmonella. While not traditionally used in this manner, XLD can serve as a general screen for total Enterobacteriaceae and Pseudomonas. Accordingly, the plate counts were instead used to determine if the treatments reduced the overall populations recoverable on XLD. The use of XLD is interesting as it yields some basic indicator species for poor sanitation, like E. coli, Klebsiella, and Citrobacter, as well as Pseudomonas and Proteus which are known spoilage organisms (Restaino et al., 1976; Toule and Murphy, 1978; Guntzel, 1991; De Boer, 1998; Fielding et al., 2012; Raposo et al., 2017).
The efficacy of PAA and 3% SBS for the sanitation of re-use water in microcosms is indistinguishable at 30 m post-sanitation; however, their effects on microbial ecology are drastically different. The 1 and 2% SBS group are also effective at 60 m, but as enumerable APC counts at 60 m occurred, the concentration may not be high enough to result in bactericidal effects. However, all treatments ultimately reduced the recoverable XLD counts. It is unknown as to the effect of downstream antimicrobials on the water nor the effects on bacterial loads on carcasses. Additional studies need to be conducted to evaluate the use of re-use water on the composition of carcass microbial ecology.
Changes to microbial ecology may be driven by a number of factors. First, we assume the bacterial OTUs present at the end of sanitation are alive and actively contributing to the ecology of the water. Therefore, the changes in microbial ecology may be an artifact of bactericidal effects on the cell and the resulting DNA degradation. Consequently, the OTUs present at the end of sanitation may be recovered from hardier cells or even dead cells that have DNA still intact.
As there are no differences in alpha diversity due to treatment nor the interaction of time and treatment, the ecological changes of the water may be reflective of a population shifts over time. Theoretically, if the sanitation process is killing a microbial population, resistant populations may be blooming, populations may be dying, and everything may actively be occurring. Therefore, data presented herein may be a cumulative effect resulting from the changes in microbial ecology and composition due to treatment. More research is needed to determine if the microbial ecology of the re-use water due to the treatment effects ultimately impacts the carcass. Changes in microbial load and carcass populations can affect food safety and spoilage. A rise in Pseudomonas as indicated by the PAA data and the 1% SBS data is indicative of a potential increase risk of reducing the shelf life. More studies are required to determine if those effects are a genuine risk. It does appear that the relative abundance of spoilage bacteria decreases over time when the microcosms are treated with SBS.
Conclusions
Water is already a significant cost to the poultry industry. Compound this cost with the rise in water scarcity, it will become important to re-use water in the future and conserve high-quality, fresh water for consumption or critical processing points. The study presented herein indicates that SBS at 3% may be a good alternative to PAA for sanitizing re-use water for the reduction of live microorganisms; however, the changes in microbial ecology are much more difficult to interpret. The efficacy of SBS (3%) is similar to that of PAA and it may favor a different population that has the potential to impact spoilage risks. However, more research is needed to determine if water that has been sanitized with SBS has differences in spoilage risk as well as to its contribution to carcass rinses.
Data Availability Statement
This article contains previously unpublished data. The name of the repository and accession number(s) are not available.
Author Contributions
KF, CK, DT, and SR conceived the experiment. KF, AM, PR, and DT conducted the experiment. PR provided sequencing support and training to AM. KF analyzed the data (bioinformatics and statistics), wrote the paper, produced the figures, and submitted the final manuscript. AM provided a first attempted draft of the paper. MK was required for authorship by ARS as KF was a federal employee at the time of work under the supervision of MK.
Conflict of Interest
CK is employed by the company Jones-Hamilton.
The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
Alcamo, J., Flörke, M., and Märker, M. (2007). Future long-term changes in global water resources driven by socio-economic and climatic changes. Hydrol. Sci. J. 52, 247–275. doi: 10.1623/hysj.52.2.247
Arnell, N. W. (2004). Climate change and global water resources: SRES emissions and socio-economic scenarios. Glob. Environ. Change 14, 31–52. doi: 10.1016/j.gloenvcha.2003.10.006
Averyt, K., Fisher, J., Huber-Lee, A., Lewis, A., Macknick, J., Madden, N., et al. (2011). Freshwater Use by U.S. Power Plants: Electricity's Thirst for a Precious Resource, A Report of the Energy and Water in a Warming World Initiative. Cambridge, MA: Union of Concerned Scientists.
Avula, R. Y., Nelson, H. M., and Singh, R. K. (2009). Recycling of poultry process wastewater by ultrafiltration. Innov. Food Sci. Emerg. Technol. 10, 1–8. doi: 10.1016/j.ifset.2008.08.005
Bauermeister, L. J., Bowers, J. W., Townsend, J. C., and McKee, S. R. (2008). Validating the efficacy of peracetic acid mixture as an antimicrobial in poultry chillers. J. Food Protec. 71, 1119–1122. doi: 10.4315/0362-028X-71.6.1119
Bertelli, C., Courtois, S., Rosikiewicz, M., Piriou, P., Aeby, S., Robert, S., et al. (2018). Reduced chlorine in drinking water distribution systems impacts bacterial biodiversity in biofilms. Front. Microbiol. 9:2520. doi: 10.3389/fmicb.2018.02520
Bolyen, E., Rideout, J. R., Dillon, M. R., Bokulich, N. A., Abnet, C., Al-Ghalith, G. A., et al. (2018). QIIME 2: reproducible, interactive, scalable, and extensible microbiome data science. PeerJ Preprints 6:e27295v2. doi: 10.7287/peerj.preprints.27295v2
Callahan, B. J., McMurdie, P. J., Rosen, M. J., Han, A. W., Johnson, A. J. A., and Holmes, S. P. (2016). DADA2: high-resolution sample inference from Illumina amplicon data. Nat. Methods 13:581. doi: 10.1038/nmeth.3869
Casani, S., Rouhany, M., and Knøchel, S. (2005). A discussion paper on challenges and limitations to water reuse and hygiene in the food industry. Water Res. 39, 1134–1146. doi: 10.1016/j.watres.2004.12.015
De Boer, E. (1998). Update on media for isolation of Enterobacteriaceae from foods. 45, 45–53. doi: 10.1016/S0168-1605(98)00146-9
DeSantis, T. Z., Hugenholtz, P., Larsen, N., Rojas, M., Brodie, E. L., Keller, K., et al. (2006). Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72, 5069–5072. doi: 10.1128/AEM.03006-05
Dittoe, D. K., Atchley, J. A., Feye, K. M., Lee, J., Knueven, C. J., and Ricke, S. C. (2019). The efficacy of sodium bisulfate salt (SBS) as an antimicrobial on whole chicken drumsticks artificially inoculated with Salmonella Enteritidis. Front. Vet. Sci. 6:6. doi: 10.3389/fvets.2019.00006
Fatemi, P., and Frank, J. F. (1999). Inactivation of Listeria monocytogenes/Pseudomonas biofilms by peracid sanitizers. J. Food Protec. 62, 761–765. doi: 10.4315/0362-028X-62.7.761
Fielding, B. C., Mnabisa, A., Gouws, P. A., and Morris, T. (2012). Antimicrobial-resistant Klebsiella species isolated from free-range chicken samples in an informal settlement. Arch. Med. Sci. 8, 39–42. doi: 10.5114/aoms.2012.27278
Fish, K., and Boxall, J. (2018). Biofilm microbiome (Re) growth dynamics in drinking water distribution systems are impacted by chlorine concentration. Front. Microbiol. 9:2519. doi: 10.3389/fmicb.2018.02519
Guntzel, M. N. (1991). “Escherichia, Klebsiella, Enterobacter, Serratia, Citrobacter, and Proteus,” in Medical Microbiology, ed S. Baron (New York, NY: Churchill Livingstone), 377–387.
Kim, B. R., Shin, J., Guevarra, R., Lee, J. H., Kim, D. W., Seol, K. H., et al. (2017). Deciphering diversity indices for better understanding of the microbial communities. J. Microbiol. Biotechnol. 27, 2089–2093. doi: 10.4014/jmb.1709.09027
Kitis, M. (2004). Disinfection of wastewater with peracetic acid: a review. Environ. Int. 30, 47–55. doi: 10.1016/S0160-4120(03)00147-8
Kozich, J. J., Westcott, S. L., Baxter, N. T., Highlander, S. K., and Schloss, P. D. (2013). Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 79, 5112–5120. doi: 10.1128/AEM.01043-13
Lillard, H. S. (1980). Effect on broiler carcasses and water of treating chiller water with chlorine or chlorine dioxide. J. Poultry Sci. 59, 1761–1766. doi: 10.3382/ps.0591761
Line, J. E. (2002). Campylobacter and Salmonella populations associated with chickens raised on acidified litter. Poult. Sci. 81, 1473–1477. doi: 10.1093/ps/81.10.1473
Mandal, S., Van Treuren, W., White, R. A., Eggesbø, M., Knight, R., and Peddada, S. D. (2015). Analysis of composition of microbiomes: a novel method for studying microbial composition. Microb. Ecol. Health Dis. 26:27663. doi: 10.3402/mehd.v26.27663
Mekonnen, M. M., and Hoekstra, A. Y. (2016). Four billion people facing severe water scarcity. Sci. Adv. 2:e1500323. doi: 10.1126/sciadv.1500323
Micciche, A. C., Feye, K. M., Rubinelli, P. M., Lee, J. A., Knueven, C. J., and Ricke, S. C. (2019). Comparison of acid sanitizers on Salmonella Typhimurium inoculated commercial poultry processing reuse water. Front. Sustain. Food Syst. 2:90. doi: 10.3389/fsufs.2018.00090
Micciche, A. C., Feye, K. M., Rubinelli, P. M., Wages, J. A., Knueven, C., and Ricke, S. C. (2018). The implementation and food safety issues associated with poultry processing reuse water for conventional poultry production systems in the United States. Front. Sustain. Food Syst. 2:70. doi: 10.3389/fsufs.2018.00070
Nagel, G. M., Bauermeister, L. J., Bratcher, C. L., Singh, M., and McKee, S. R. (2013). Salmonella and Campylobacter reduction and quality characteristics of poultry carcasses treated with various antimicrobials in a post-chill immersion tank. Int. J. Food Microbiol. 165, 281–286. doi: 10.1016/j.ijfoodmicro.2013.05.016
National Research Council (2010). Committee on Acute Exposure Guideline Levels. Acute Exposure Guideline Levels for Selected Airborne Chemicals: Volume 8. Washington, DC: National Academies Press (US); 7, Peracetic Acid Acute Exposure Guideline Levels. Available online at: https://www.ncbi.nlm.nih.gov/books/NBK220001/
Park, S. H., Kim, S. A., Rubinelli, P. M., Roto, S. M., and Ricke, S. C. (2017). Microbial compositional changes in broiler chicken cecal contents from birds challenged with different Salmonella vaccine candidate strains. Vaccine 35, 3204–3208. doi: 10.1016/j.vaccine.2017.04.073
Raposo, A., Pérez, E., de Faria, C. T., Ferrús, M. A., and Carrascosa, C. (2017). “Food spoilage by pseudomonas spp.—an overview,” in Food Borne Pathogens and Antibiotic Resistance, ed O. V. Singh (Hoboken, NJ: John C. Wiley & Sons), 41–58. doi: 10.1002/9781119139188.ch3
Restaino, L., Grauman, G. S., McCall, W. A., and Hill, W. M. (1976). Effects of varying concentrations of novobiocin incorporated into two Salmonella plating media on the recovery of four Enterobacteriaceae. Appl. Environ. Microbiol. 33, 585–589. doi: 10.1128/AEM.33.3.585-589.1977
Samapundo, S., Baenst, I. D., Aerts, M., Cnockaert, M., Devlieghere, F., and Van Damme, P. (2019). Tracking the sources of psychotrophic bacteria contaminating chicken cuts during processing. Food Microbiol. 81, 40–50. doi: 10.1016/j.fm.2018.06.003
Schewe, J., Heinke, J., Gerten, D., Haddeland, I., Arnell, N. W., Clark, D. B., et al. (2014). Multimodel assessment of water scarcity under climate change. Proc. Natl. Acad. Sci. U.S.A. 111, 3245–3250. doi: 10.1073/pnas.1222460110
Shannon, M. A., Bohn, P. W., Elimelech, M., Georgiadis, J. G., Marinas, B. J., and Mayes, A. M. (2008). Science and technology for water purification in the coming decades. Nature 452:301. doi: 10.1038/nature06599
Siegel and Castellan (1988). Non-parametric Statistics for the Behavioral Sciences. New York, NY: MacGraw Hill Int.
Srey, S., Jahid, I. K., and Ha, S. D. (2013). Biofilm formation in food industries: a food safety concern. Food Control 31, 572–585. doi: 10.1016/j.foodcont.2012.12.001
Toule, G., and Murphy, O. (1978). A study of bacteria contaminating refrigerated cooked chicken; their spoilage potential and possible origin. J. Hyg. 81, 161–169. doi: 10.1017/S0022172400024980
Tsai, L. S., Schade, J. E., and Molyneux, B. T. (1992). Chlorination of poultry chiller water: chlorine demand and disinfection efficiency. Poult. Sci. 71, 188–196. doi: 10.3382/ps.0710188
United States Department of Agriculture Food Safety Inspection Service (2003). Use of Chlorine to Treat Poultry Chiller Water. FSIS Notice 45-03. Available online at: http://www.fsis.usda.gov/OPPDE/rdad/FSISNotices/45-03.htm (accessed November 30, 2018).
United States Department of Agriculture Food Safety Inspection Service (2016). Sanitation Performance Standards Compliance Guide. Available online at: https://www.fsis.usda.gov/wps/portal/fsis/topics/regulatory-compliance/compliance-guides-index/sanitation-performance-standards/sanitation-compliance-guide (accessed November 30, 2018).
United States Environmental Protection Agency (2010). Sampling and Consideration of Variability (Temporal and Spatial) For Monitoring of Recreational Waters. Available online at: https://www.epa.gov/sites/production/files/2015-11/documents/sampling-consideration-recreational-waters.pdf (accessed May 18, 2019).
Keywords: sodium bisulfate, microbiome, re-use water, peracetic acid, poultry processing
Citation: Feye KM, Micchichi AC, Rubinelli PM, Knueven CJ, Thompson DR, Kogut MH and Ricke SC (2020) The Effect of Acid Sanitizers on the Microbiome of Re-use Water. Front. Sustain. Food Syst. 4:85. doi: 10.3389/fsufs.2020.00085
Received: 11 June 2019; Accepted: 12 May 2020;
Published: 03 July 2020.
Edited by:
Paula Teixeira, Catholic University of Portugal, PortugalReviewed by:
Guadalupe Virginia Nevárez-Moorillón, Autonomous University of Chihuahua, MexicoJosé Antonio Teixeira, University of Minho, Portugal
Copyright © 2020 Feye, Micchichi, Rubinelli, Knueven, Thompson, Kogut and Ricke. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Steven C. Ricke, c3JpY2tlQHVhcmsuZWR1
 Kristina M. Feye
Kristina M. Feye Andrew C. Micchichi
Andrew C. Micchichi Peter M. Rubinelli
Peter M. Rubinelli Carl J. Knueven
Carl J. Knueven Dale R. Thompson
Dale R. Thompson Michael H. Kogut
Michael H. Kogut Steven C. Ricke
Steven C. Ricke