- 1Institute of Botany and Landscape Ecology, University of Greifswald, Greifswald, Germany
- 2Matthias Schleiden Institute, Plant Physiology, University of Jena, Jena, Germany
- 3Department of Environmental Science, Central University of Kerala, Kasaragod, India
Duckweeds include the world's smallest and fastest growing flowering plants that have the capacity to produce huge biomass with a broad range of potential applications like production of feed and food, biofuel and biogas. In order to achieve optimal and sustainable commercial system, it is necessary that suitable species and clones of duckweeds be identified and selected based on appropriate strategies. However, a high degree of reduction in their structural complexity poses serious problems in identification of closely related species of duckweeds, on a morphological basis. Use of molecular taxonomic tools is the present solution. The state of the art of molecular taxonomy of all the five genera of duckweeds (Spirodela, Landoltia, Lemna, Wolffiella, and Wolffia) is based mainly on the techniques of fingerprinting by amplified fragment length polymorphism (AFLP) and barcoding using sequences of plastidic DNA fragments. After more than 15 years of molecular taxonomic investigations, a certain viewpoint is now available demonstrating all five genera to be monophyletic. Also, the phenetic analyses had made huge progress in delineating the currently defined 36 species of duckweeds, although, all species cannot yet be defined with confidence. Wolffiella has turned out to be the most complicated genus as only 6 to 7 species out of the 10 can be reliably delineated. Further progress in the phylogenetic and phenetic analyses requires more advanced methods like next generation and/or whole genome sequencing. First results using the method genotyping-by-sequencing in the genus Lemna (in combination with metabolomic profiling by matrix-assisted laser desorption ionization time-of-flight mass-spectrometry (MALDI-TOF-MS) as well as AFLP and barcoding by plastidic sequences) are more promising: The species Lemna valdiviana and Lemna yungensis were united to one species, Lemna valdiviana. This reduced the total number of Lemnaceae species to 36.
Introduction
Lemnaceae (commonly known as water lens or duckweed) is a family of monocotyledonous water plants (Landolt, 1986; Landolt and Kandeler, 1987; Sree et al., 2016) characterized by the fastest growth rates among flowering plants (Sree et al., 2015b; Ziegler et al., 2015). As a consequence, the rate of biomass production is high, which provides the basis for different practical applications of duckweeds in the areas of food and feed sustainability (Landolt and Kandeler, 1987; Appenroth et al., 2017, 2018; Sonta et al., 2019) and biotechnology (Zhao et al., 2015; Chen et al., 2018; Kaur et al., 2018; Ren et al., 2018).
The biotechnological optimization as well as the commercial application of duckweeds require the knowledge of the taxa of these plants. Because of the miniaturization of the morphological units and extreme reduction of anatomical complexity, only a few experts worldwide can delineate duckweed species based on morphological markers. Considering these limitations, some decades back, Crawford and co-workers tried to delineate duckweed species based on chemical composition of flavonoids (Les et al., 1997) and analyzed isoforms of enzymes (allozymes), an advanced form of chemotaxonomy in that time. These authors wrote: “Allozymes were employed because they reflect discrete genetic differences and thus represent a potentially useful method for assessing whether one or two gene pools exist within what has been treated as two taxa” (Crawford et al., 1996). This method produced interesting results and had already offered an essential step ahead of the difficult morphological determination technique (Crawford and Landolt, 1995; Crawford et al., 1997, 2001). With the advent and development of molecular biology tools and techniques, analysis of taxa at the level of DNA offered deeper comprehension than chemotaxonomy. Therefore, the term molecular taxonomy is now almost exclusively used in connection with several methods of DNA analysis for taxonomic delineation.
At the level of genera, Les et al. (2002) already provided significant results demonstrating that all five genera are monophyletic taxa. Beside the delineation of species, identification of clones (Lemnaceae propagate mainly vegetatively resulting in genetically identical offsprings, called clones or strains) is also a requirement of the present day. Remarkably, the efficiency of physiological processes responsible for the commercial properties of these plants is not dependent on the genus or the species but is a characteristic of the clones of duckweeds. This clonal diversity has been proven e.g., for growth rate (Sree et al., 2015b; Ziegler et al., 2015), turion formation capacity (Kuehdorf et al., 2014), nutritional quality (Appenroth et al., 2018) as well as for starch accumulation capacity (Sree et al., 2015a; Ma et al., 2018). These exemplary investigations demonstrate that the physiological properties shown by a particular clone of a duckweed species is characteristic to that clone and cannot be extrapolated to the whole species or to the genus or to the family. For the screening and commercialization of clones with desirable characteristics and for maintaining these desirable genetic resources, it is, therefore, necessary to identify the particular clone of duckweed. However, clones of a given species, on a morphological basis, are highly similar and hence identification of clones requires the use of molecular tools. The progress in this approach is further supported by the increasing number of available whole genome sequences of duckweed species belonging to three of the five genera of the plant family, like Spirodela polyrhiza clone 9509 (Hoang et al., 2018, and references therein), Lemna minor 5500 (Van Hoeck et al., 2015) and Lemna minor 8627, Lemna gibba 7742a, and Wolffia australiana 8730 (www.lemna.org/; for a review see An et al., 2018). Further, 68 clones of Spirodela polyrhiza were recently sequenced by Xu et al. (2019) and 38 by Ho et al. (2019).
In the wake of the available resources, molecular taxonomic methods have attracted attention of the duckweed researchers. The aim of the present review is to address the questions: With what reliability can the presently available molecular methods be used to delineate, characterize and identify different taxa of Lemnaceae? The phylogenetic approach is of general interest as Lemnaceae occupy a unique basal position within monocot plants and we try to make it clear that the requirements for answering phenetic or phylogenetic questions are different.
Plant Family and Categorization Into Genera
Duckweeds constitute the family Lemnaceae Dumort. Although, it has been shown convincingly that Lemnaceae are very closely related to Araceae (Cusimano et al., 2011; Nauheimer et al., 2012; Choi et al., 2017), it has been further discussed that it is not necessary to integrate Lemnaceae into the family of Araceae (as subfamily Lemnoideae) and that this group can be treated in accordance with taxonomic rules as plant family, i.e., Lemnaceae Dumort (Appenroth et al., 2013, 2015; Bog et al., 2018, in press) (Figure 1). Based on the morphological markers, Landolt (1980, 1986) had categorized the family into two subfamilies, Wolffioideae (members devoid of roots) and Lemnoideae (members with varying numbers of roots). Normally, they would not be defined as taxa because the latter is paraphyletic. However, this categorization is very useful within a morphological key of determination (Landolt, 1980, 1986) of Lemnaceae species and therefore is used for practical reasons.
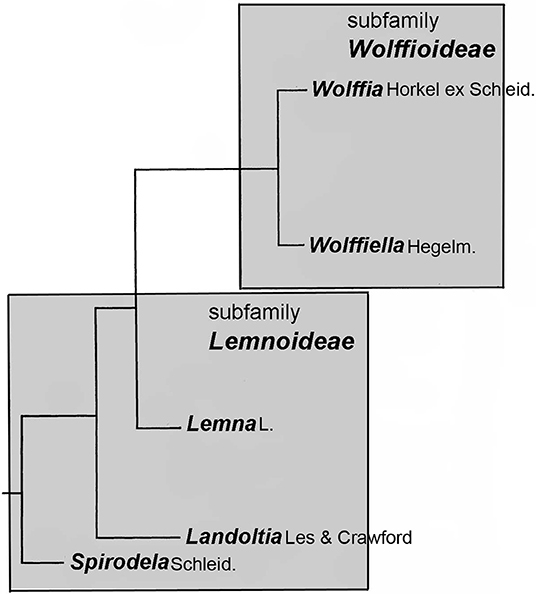
Figure 1. Structure of the family Lemnaceae in relation to genera. The presentation is related to the sequence data of Tippery et al. (2015).
After Les and Crawford (1999) defined the species Spirodela punctata as constituting a genus of its own, called Landoltia (which changed the nomenclature of the only species within this genus necessarily to Landoltia punctata (G.Mey.) Les & D.J. Crawford), the plant family now comprises of five genera (Figure 1). According to Les et al. (2002), all five genera are monophyletic. Already in 1986, Landolt suggested that evolution proceeded from the genus Spirodela to the genus Wolffia. However, the most evolved nature of the genus Wolffia could not yet be confirmed by molecular taxonomic tools. The phenetic tree structure of the plant family as reported by Les and Crawford (1999) and Les et al. (2002) was recently confirmed by Ding et al. (2017) using whole plastidic sequences of eight clones from all genera. However, the relative position of the genera Wolffiella vs. Wolffia in this tree cannot be defined because the branches representing these genera can be freely turned around the branching point. Remarkably, the evolutionary development in the family is correlated with a reduction of morphological and anatomical structures and increase in the number of derived characters, quantified as “degree of primitivity,” where most ancient/basal species have the highest “degree of primitivity” and derived species a lower degree (Landolt, 1986, p. 417–420). Interestingly, this “degree of primitivity” is negatively correlated with the DNA content per genome (Wang et al., 2011; Hoang et al., 2019). It is important to mention here that, this correlation exists at the level of genera but is hardly detectable at the level of species within the genera, as inferred from the data presented by Wang et al. (2011).
Delineation of Species
Although there were some preliminary attempts of use of molecular tools in the field of taxonomy of Lemnaceae (Jordan et al., 1996; Rothwell et al., 2004; Martirosyan et al., 2009), the era of molecular taxonomy of Lemnaceae kick-started with the hallmark paper of Les et al. (2002). For the first time, all species were included in the analysis and comprehensive trees were presented. The same group extended the analysis by investigating for the first time the nuclear fragments: internal and external transcribed spacer ITS and ETS (Tippery et al., 2015). With such procedure, it is possible to reveal the general structure of family and genera. However, as these studies included only one clone per species for the final evaluation, delineation of species is not possible, as the intraspecific genetic variations are not known. This problem was involuntarily demonstrated in their publication (Les et al., 2002) as a clone of the species named Lemna ecuadoriensis 8896 was included in the analysis of the whole family. Interestingly, a well separated branch was obtained for this clone indicating it as being a distinct species, Lemna ecuadoriensis. However, already in 2000, during a revision based on morphological markers, Landolt had merged clones of Lemna ecuadoriensis with those of Lemna obscura (Landolt, 2000) making the term Lemna ecuadoriensis a synonym. This revision was later confirmed by the AFLP analysis performed by Bog et al. (2010), where the clone 8896 (Lemna ecuadoriensis) was well integrated into the cluster of Lemna obscura. We conclude that the internal species structure of genera can be analyzed even when each species is represented only by a single clone. However, delineation of species as part of phenetic analysis indispensably requires the investigation of several clones per species in order to obtain reliable results.
The plant family Lemnaceae presently consists of 36 recognized species (Sree et al., 2016; Bog et al., in press, 2020). This present state has been achieved as a combinatorial result of use of molecular taxonomic tools on the clones of a species designated by using morphological markers. The molecular characterization of species until now has largely been based either on fingerprinting by AFLP or on barcoding using sequences of amplified plastidic fragments or nuclear fragments. Use of AFLP supposes that all individuals can be evaluated by the pattern of electrophoretic bands. However, we learnt that this is only possible within each of the genera of Lemnaceae but not for the whole plant family taken together. Therefore, the results of barcoding using AFLP were compiled separately for each genus (Bog et al., 2010, 2013, 2015, 2018), uniting only the small genera (in terms of the number of species) Spirodela and Landoltia (Bog et al., 2015). This problem does not exist in the barcoding procedure using plastidic fragments as markers. In the present paper, we will review the state of the art of molecular taxonomy of Lemnaceae with respect to each of the five genera.
Genera Spirodela and Landoltia
Historically, three species, Spirodela polyrhiza, Spirodela intermedia, and Spirodela punctata (synonym: Spirodela oligorrhiza) constituted the genus Spirodela (Landolt, 1986). The species Spirodela intermedia seems to be the most basal species of Lemnaceae as investigated on the basis of morphological markers. According to Landolt (1986), this species has the highest degree of primitivity i.e., it represents the highly differentiated plant body c.f. less differentiated plant body in Wolffia and Wolffiella as an adaptation to aquatic life style.
With the advent of molecular taxonomic tools to the family of Lemnaceae, these three species were separated into two genera, Spirodela and Landoltia (changing the nomenclature of the species, Spirodela punctata to Landoltia punctata), based mainly on the investigation of the plastidic sequence rbcL (Les and Crawford, 1999). These results were confirmed by investigating the whole plastidic sequence analysis of Spirodela polyrhiza and Landoltia punctata (Ding et al., 2017). Taken together, there is no doubt that these three species should be organized into the two different genera as mentioned above. In addition, the two species that presently constitute the genus Spirodela, Spirodela polyrhiza, and Spirodela intermedia, can be clearly delineated by all the plastidic markers used to date (Table 1). Moreover, the essential genome structure of the two species of Spirodela have been revealed recently (Hoang and Schubert, 2017).
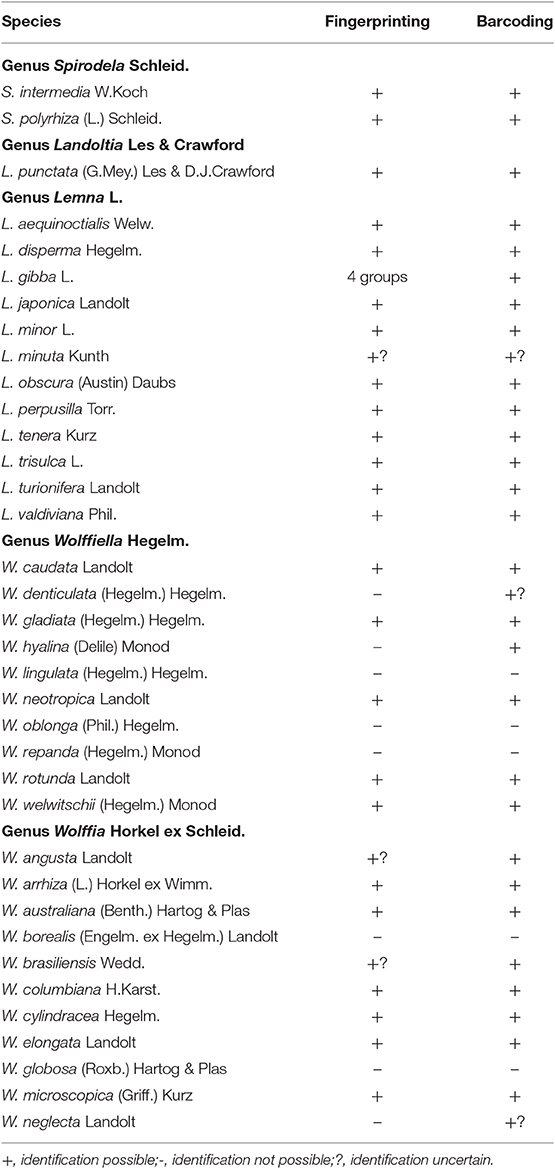
Table 1. Summary of the results of species identification by fingerprinting (AFLP) and barcoding by several plastidic fragments of 36 species of the family Lemnaceae Dumort.
Some of the initial molecular taxonomic investigations on these genera were carried out by Les et al. (2002) with only one clone per species included in the analysis. Wang et al. (2010) also separated all three species in an UPGMA tree based on atpF-atpH sequences. But, in this study the clone 9203 was misplaced as Spirodela polyrhiza although E. Landolt had identified this clone earlier as Spirodela intermedia (personal communication to KJA). This error was also pointed out by Borisjuk et al. (2015) who investigated 7 clones of Spirodela polyrhiza, 8 clones of Spirodela intermedia and 6 clones of Landoltia punctata. Eliminating this error, the combined cpDNA dataset based on atpF-atpH and psbK-psbI intergenic spacers clearly separated all of these three species with high bootstrap values. In a study focused on these two genera, Bog et al. (2015) investigated 24 clones of Spirodela polyrhiza, 14 clones of Spirodela intermedia and 17 clones of Landoltia punctata using the plastidic fragments rpl16, rps16, and atpF-atpH. In this study, all clones were categorized with high bootstrap values according to their morphological identification. However, the results showed that there was hardly any detectable intraspecific differentiation although a comparatively higher intraspecific genetic variation was observed in the species Spirodela intermedia than in the other two species.
Additionally, Bog et al. (2015) delineated these species using the method of AFLP. With this method, it was not only possible to delineate the two genera and three species clearly but it was also possible to separate (statistically significant) several genotypes within the species: 6 genotypes in Spirodela intermedia (out of 14 clones), 3 genotypes in Spirodela polyrhiza (24 clones) and 3 genotypes in Landoltia punctata (17 clones). However, this does not allow characterization of all investigated samples at the level of clones. Detection of a larger number of haplo-/genotypes in the species Spirodela polyrhiza and Landoltia punctata was possible by using plastidic inter-simple sequence repeat (ISSR) markers (Xue et al., 2012) or simple sequence repeat (SSR) markers of the plastidic genome (Feng et al., 2017) as well as nuclear SSR markers (Xu et al., 2018). Out of the three studies, the first identified 11 haplotypes of Spirodela polyrhiza and 11 haplotypes of Landoltia punctata in 97 investigated clones, resulting in the specific identification of ca. 22%, and the second identified 19 out of 68 clones (28%). An alternative method was recently presented by Chu et al. (2018) using the highly polymorphic regions of NB-ARC-related genes (nuclear-binding leucine-rich repeat protein) as markers in Spirodela polyrhiza. These authors could “uniquely” identify 20 out of 23 clones of Spirodela polyrhiza (ca. 87%), while the remaining 3 clones could not be distinguished from each other, but could be distinguished from the other 20. The extension of this method to clones belonging to other duckweed species requires the availability of whole genome sequences of those species. It should be mentioned here that from whole genome sequencing it is known that the intraspecific variation in Spirodela polyrhiza is extremely low (Michael et al., 2017; Hoang et al., 2018; Ho et al., 2019; Xu et al., 2019). Therefore, demonstrating differences between clones of this species is a kind of “toughness test” for the method employed. The present results of molecular investigations of the species in the genera Spirodela and Landoltia are summarized in the Table 1.
Genus Lemna
With the most recent results (Bog et al., in press) published in the journal “Taxon,” the currently accepted number of species in the genus Lemna is reduced to 12 from the previously accepted 13 species. These results will be discussed at a later part of this section.
The delineation of the species of the genus Lemna based on several plastidic sequences was published by Wang et al. (2010). Also, use of several plastidic and nuclear markers was investigated by Tippery et al. (2015) (e.g., ETS, ITS), and Borisjuk et al. (2015) (atpF-atpH and psbK-psbI). Presently, it is not yet possible to unify these results concerning the intrageneric structure. However, this problem is not specific to the genus Lemna but holds true also for all other genera of Lemnaceae. Landolt (1986) assigned Lemna gibba the highest morphological degree of primitivity and Lemna minuta and Lemna valdiviana the lowest degree. Perhaps the former represents the basal species and the latter ones the most derived species in the genus Lemna.
Using DNA-barcoding most of the species of Lemna could be easily defined in several projects. Wang et al. (2010) analyzed 8 of the Lemna species. In this study, Lemna japonica (only one clone investigated) was inserted in a clade of Lemna minor; Lemna minuta and Lemna valdiviana were not separated from each other and the bootstrap value of the branch to distinguish Lemna turionifera and Lemna trisulca was only 0.71. Borisjuk et al. (2015) investigated all Lemna species, in most cases represented by several clones. Lemna japonica was inserted in the clade of Lemna minor, and the separation of Lemna minuta, Lemna valdiviana, and Lemna yungensis was not completely clear. The fingerprinting method, AFLP used by Bog et al. (2010) was applied to all species of Lemna represented in total by 84 clones. With the exception of the group Lemna valdiviana/ Lemna yungensis, all species were delineated from each other including Lemna japonica. Clones of the species Lemna minuta were formally separated from those of the group of Lemna valdiviana/Lemna yungensis. However, with one exception, all clones of Lemna minuta were collected from Europe where they had invaded. Thus, the intraspecific similarity and dissimilarity to the other two species might have been overestimated by the selection of clones. Further results showed clearly that Lemna minuta is a species of its own but that there is no genetic difference between Lemna valdiviana and Lemna yungensis (Bog et al., in press, 2020). In order to unravel this issue, detailed investigation of these three species using AFLP, barcoding with atpF-atpH and psbK-psbI, metabolomic profiling by matrix-assisted laser desorption ionization time-of-flight mass-spectrometry (MALDI-TOF-MS) as well as genotyping-by-sequencing was performed (Bog et al., 2020). The results, all together, speak in favor of uniting Lemna valdiviana and Lemna yungensis to one single species, retaining Lemna valdiviana as a valid nomenclature for all the clones falling into these two species (with Lemna yungensis as a synonym). Thus, the number of currently accepted species of the genus Lemna was reduced from 13 to 12. In the Table 1 the results of molecular taxonomy for the genus Lemna are summarized.
We now move ahead from the root bearing members representing the subfamily of Lemnoideae to the rootless members, Wolffioideae, categorized into two genera, Wolffiella and Wolffia.
Genus Wolffiella
The genus Wolffiella represents the least investigated genus and comprises 10 species. Through the course of investigations, it turned out that this is the most difficult challenge for molecular taxonomy in the whole plant family (Bog et al., 2018). According to Landolt (1986), Wolffiella neotropica has the highest morphological degree of primitivity within this genus and should be the most basal species of the genus.
In general, application of AFLP or plastidic barcoding were only of limited success in this genus. Bog et al. (2018) investigated 67 clones representing all 10 species. AFLP, clearly distinguished the species Wolffiella caudata, Wolffiella gladiata, Wolffiella neotropica, Wolffiella rotunda, and Wolffiella welwitschii. Apart from confirming these five species, plastidic markers could delineate two additional species, Wolffiella hyalina and Wolffiella denticulata although the conclusion concerning the latter is restricted by the availability of only one clone. However, separation and identification of the three species Wolffiella lingulata, Wolffiella oblonga, and Wolffiella repanda appears to be a major problem within the genus. A Combination of the sequence data of Wang et al. (2010) and those of Bog et al. (2018) resulted in three groups: one constituting a mixture of clones from the species Wolffiella oblonga and Wolffiella gladiata, another small group exclusively containing Wolffiella oblonga clones and a larger mixed group of Wolffiella oblonga and Wolffiella lingulata. In conclusion the species Wolffiella lingulata, Wolffiella oblonga, and Wolffiella repanda could not be identified by any of the molecular methods but could be strictly defined on morphological bases. An interesting find from the Structure analysis of the AFLP data is the formation of hybrids between different species of Wolffiella (Bog et al., 2018). As an example, the existence of hybrids between Wolffiella welwitschii and Wolffiella lingulata were deduced by artificial simulation of F1 plants from the related parent clones and was tested by Principal Coordinate Analysis. The existence of such hybrids could be one of the reasons for the complications encountered in delineation of species of Wolffiella based on molecular methods. The results for the genus Wolffiella are summarized in the Table 1.
Genus Wolffia
The genus Wolffia includes 11 species of the simplest morphological structure. This genus not only contains the smallest flowering plants (Bog et al., 2013) but also the fastest growing angiosperms (Sree et al., 2015b). Interestingly, one of the members of this genus, Wolffia brasiliensis appears rather closer to Wolffiella species than to that of Wolffia. This is in agreement with the results of Wang et al. (2010) and Borisjuk et al. (2015) who both investigated three different clones of this species. Les et al. (2002) using one of these three clones obtained the same results from the analysis of cpDNA, however, not from the analysis based on nuclear DNA markers (ETS, 5.8S, 18S, 24S, ITS-1, and ITS2). Thus, analysis of the complete plastidic sequence, perhaps in combination with other nuclear markers would help to solve this mystery. Landolt characterized Wolffia brasiliensis and Wolffia borealis as those species with the highest degree of primitivity making them the candidates for the basal position within this genus. However, this prediction is neither supported by the trees shown in Bog et al. (2013) and Borisjuk et al. (2015) nor by the results of Tippery et al. (2015) based on either plastidic or nuclear sequences.
Bog et al. (2013) used the sequences of rps16 (54 clones) and rpl16 (55 clones), and identified the following species successfully: Wolffia angusta, Wolffia australiana, Wolffia brasiliensis, Wolffia cylindracea, Wolffia elongata, Wolffia microscopica, and Wolffia neglecta. Wolffia globosa has been separated into two groups by both AFLP and barcoding. One group consisted only of clones from this species, the other group of Wolffia globosa contained also clones of Wolffia neglecta and showed high similarity to Wolffia borealis. None of the methods recognized Wolffia borealis as a distinct species. Moreover, the difficulties in delineation of Wolffia borealis, Wolffia globosa and Wolffia neglecta were also observed by Borisjuk et al. (2015) who made use of the plastidic fragments, psbK-psbI and atpF-atpH. The presently available data are summarized in the Table 1. We have to conclude that besides the huge progress in the last decade in the field of molecular taxonomy, the results are not sufficient to identify all taxa of Wolffia.
Conclusions and Future Perspective
The present state of molecular taxonomy has reached a level that cannot be further improved by employing the same techniques. The limits of fingerprinting by AFLP and barcoding using plastidic sequences have been tested by using high number of clones covering as much as possible the species to be tested. In summary, the above mentioned in relation to the genus Wolffia is applicable to the whole family Lemnaceae: “The results are not sufficient to identify all taxa.” This is even more evident in the genus Wolffiella. The solution can only be the use of a molecular method that provides more markers per clone for both plastidic and nuclear genomes than the above mentioned methods of fingerprinting and barcoding. This can be reached by an approach close to whole genome sequencing and next generation sequencing. Alternatively, whole genome sequencing-based selection of highly polymorphic regions like the NB-ARC-related sequences could be used when the extension to a larger number of species will be possible in future. Further, the application of the method “genotyping-by-sequencing” to molecular taxonomy, as evidenced in the genus Lemna (Bog et al., in press) could be extended to the whole plant family which has the potential to solve some of the persistent problems in molecular taxonomy of Lemnaceae.
Author Contributions
All authors contributed to the writing and correcting of the article.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Abbreviations
AFLP, amplified fragment length polymorphism.
Note: The genera have not been abbreviated to the starting letter during repeated use of the species nomenclature in the text in order to avoid confusion (Genera Landoltia and Lemna start with “L” and genera Wolffiella and Wolffia start with “W”).
References
An, D., Li, C. S., Zhou, Y., Wu, Y. R., and Wang, W. Q. (2018). Genomes and transcriptomes of duckweeds. Front. Chem. 6:230. doi: 10.3389/fchem.2018.00230
Appenroth, K. J., Borisjuk, N., and Lam, E. (2013). Telling duckweed apart: genotyping technologies for Lemnaceae. Chin. J. Appl. Environ. Biol. 19, 1–10. doi: 10.3724/SP.J.1145.2013.00001
Appenroth, K. J., Crawford, D. J., and Les, D. H. (2015). After the genome sequencing of duckweed – how to proceed with research on the fastest growing angiosperm? Plant Biol. 17, 1–4. doi: 10.1111/plb.12248
Appenroth, K. J., Sree, K. S., Bog, M., Ecker, J., Seeliger, C., Böhm, V., et al. (2018). Nutritional value of the duckweed species of the genus Wolffia (Lemnaceae) as human food. Front. Chem. 6:483. doi: 10.3389/fchem.2018.00483
Appenroth, K. J., Sree, K. S., Böhm, V., Hammann, S., Vetter, W., Leiterer, M., et al. (2017). Nutritional value of duckweeds (Lemnaceae) as human food. Food Chem. 217, 266–273. doi: 10.1016/j.foodchem.2016.08.116
Bog, M., Baumbach, H., Schween, U., Hellwig, F., Landolt, E., and Appenroth, K. J. (2010). Genetic structure of the genus Lemna L. (Lemnaceae) as revealed by amplified fragment length polymorphism. Planta 232, 609–619. doi: 10.1007/s00425-010-1201-2
Bog, M., Himmelbach, A., Brandt, R., Wagner, F., and Appenroth, K. J. (2020). “Genotyping-By-Sequencing for species delimitation in Lemna section Uninerves Hegelm. (Lemnaceae),” in Compendium of Plant Genomes: The Duckweed Genomes, 1st Edn., eds X. H. Cao, P. Fourounjian, and W. Wang (Berlin; Heidelberg: Springer).
Bog, M., Landrock, M. F., Drefahl, D., Sree, K. S., and Appenroth, K. J. (2018). Fingerprinting by amplified fragment length polymorphism (AFLP) and barcoding by three plastidic markers in the genus Wolffiella Hegelm. Plant Syst. Evol. 304, 373–386. doi: 10.1007/s00606-017-1482-z
Bog, M., Lautenschlager, U., Landrock, M. F., Landolt, E., Fuchs, J., Sree, K. S., et al. (2015). Genetic characterization and barcoding of taxa in the genera Landoltia and Spirodela (Lemnaceae) by three plastidic markers and amplified fragment length polymorphism (AFLP). Hydrobiologia 749, 169–182. doi: 10.1007/s10750-014-2163-3
Bog, M., Schneider, P., Hellwig, F., Sachse, S., Kochieva, E. Z., Martyrosian, E., et al. (2013). Genetic characterization and barcoding of taxa in the genus Wolffia Horkel ex Schleid. (Lemnaceae) as revealed by two plastidic markers and amplified fragment length polymorphism (AFLP). Planta 237, 1–13. doi: 10.1007/s00425-012-1777-9
Bog, M., Sree, K. S., Fuchs, J., Hoang, P. T. N., Schubert, I., and Kuever, J. (in press). A taxonomic revision of Lemna sect. Uninerves (Lemnaceae). Taxon..
Borisjuk, N., Chu, P., Gutierrez, R., Zhang, H., Acosta, K., Friesen, N., et al. (2015). Assessment, validation and deployment strategy of a two barcode protocols for facile genotyping of duckweed species. Plant Biol. 17, 42–49. doi: 10.1111/plb.12229
Chen, G. K., Fang, Y., Huang, J., Zhao, Y. G., Li, Q., Lai, F., et al. (2018). Duckweed systems for eutrophic water purification through converting wastewater nutrients to high-starch biomass: comparative evaluation of three different genera (Spirodela polyrhiza, Lemna minor and Landoltia punctata) in monoculture or polyculture. RSC Adv. 8, 17927–17937. doi: 10.1039/C8RA01856A
Choi, K. S., Park, K. T., and Park, S. J. (2017). The chloroplast genome of Symplocarpus renifolius: a comparison of chloroplast genome structure in Araceae. Genes 8:324. doi: 10.3390/genes8110324
Chu, P., Wilson, G. M., Michael, T. P., Vaiciunas, J., Honig, J., and Lam, E. (2018). Sequence-guided approach to genotyping plant clones and species using polymorphic NB-ARC-related genes. Plant Molec. Biol. 98, 219–231. doi: 10.1007/s11103-018-0774-1
Crawford, D. J., and Landolt, E. (1995). Allozyme divergence among species of Wolffia (Lemnaceae). Plant Syst. Evol. 197, 59–69. doi: 10.1007/BF00984632
Crawford, D. J., Landolt, E., and Les, D. H. (1996). An allozyme study of two sibling species of Lemna (Lemnaceae) with comments on their morphology, ecology and distribution. Bull. Torrey Bot. Club 123, 1–6. doi: 10.2307/2996300
Crawford, D. J., Landolt, E., Les, D. H., and Kimball, R. T. (2001). Allozyme studies in Lemnaceae: variation and relationships in Lemna sections Alatae and Biformes. Taxon 50, 987–999. doi: 10.2307/1224716
Crawford, D. J., Landolt, E., Les, D. H., and Tepe, E. (1997). Allozyme variation and the taxonomy of Wolffiella. Aquat. Bot. 58, 43–54. doi: 10.1016/S0304-3770(97)00012-0
Cusimano, N., Bogner, J., Mayo, S. J., Boyce, P. C., Wong, S. A. Y., Hesse, M., et al. (2011). Relationships within the Araceae: Comparison of morphological patterns with molecular phylogeny. Am. J. Bot. 98, 654–668. doi: 10.3732/ajb.1000158
Ding, Y., Fang, Y., Guo, L., Li, Z., He, K., Thao, Y., et al. (2017). Phylogenetic study of Lemnoideae (duckweeds) through complete chloroplast genomes for eight accessions. PeerJ 5:e4186. doi: 10.7717/peerj.4186
Feng, B., Fang, A., Xu, Z., Xiang, C., Zhou, C. H., Jiang, F., et al. (2017). Development of a new marker system for identification of Spirodela polyrhiza and Landoltia punctata. Int. J. Genomics 2017:5196763. doi: 10.1155/2017/5196763
Ho, E. K. H., Bartkowska, M., Wright, S. I., and Agrawal, A. F. (2019). Population genomics of the facultatively asexual duckweed Spirodela polyrhiza. New Phytol. 224, 1361–1371. doi: 10.1111/nph.16056
Hoang, P. N. T., Michael, T. P., Gilbert, S., Chu, P., Motley, S. T., Appenroth, K. J., et al. (2018). Generating a high-confidence reference genome map of the greater duckweed by integration of cytogenomic, optical mapping, and Oxford Nanopore technologies. Plant J. 96, 670–684. doi: 10.1111/tpj.14049
Hoang, P. T. N., and Schubert, I. (2017). Reconstruction of chromosome rearrangements between the two most ancestral duckweed species Spirodela polyrhiza and S. intermedia. Chromosoma 126, 729–739. doi: 10.1007/s00412-017-0636-7
Hoang, P. T. N., Schubert, V., Meister, A., Fuchs, J., and Schubert, I. (2019). Variation in genome size, cell and nucleus volume, chromosome number and rDNA loci among duckweeds. Sci. Rep. 9:3234. doi: 10.1038/s41598-019-39332-w
Jordan, W. C., Courtney, M. W., and Neigel, J. E. (1996). Low levels of intraspecific genetic variation at a rapidly evolving chloroplast DNA locus in North American duckweeds (Lemnaceae). Am. J. Bot. 83, 430–439. doi: 10.1002/j.1537-2197.1996.tb12724.x
Kaur, M., Kumar, M., Sachdeva, S., and Puri, S. K. (2018). Aquatic weeds as the next generation feedstock for sustainable bioenergy production. Bioresour. Technol. 251, 390–402. doi: 10.1016/j.biortech.2017.11.082
Kuehdorf, K., Jetschke, G., Ballani, L., and Appenroth, K. J. (2014). The clonal dependence of turion formation in the duckweed Spirodela polyrhiza- an ecogeographical approach. Physiol. Plant. 150, 46–54. doi: 10.1111/ppl.12065
Landolt, E. (1980). Key to the Determination of Taxa Within the Family of Lemnaceae. Veröffentlichungen des Geobotanischen Institutes der Eidg. Technische Hochschule, Stiftung Rübel, Zurich, 13–21.
Landolt, E. (1986). The Family of Lemnaceae - A Monographic Study. Vol. 1. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae). Veröffentlichungen des Geobotanischen Institutes der Eidg. Technische Hochschule, Stiftung Rübel, Zurich.
Landolt, E. (2000). Contribution on the Lemnaceae of Ecuador. Fragmenta Floristica et Geobotanica 45, 221–237.
Landolt, E., and Kandeler, R. (1987). The Family of Lemnaceae - a Monographic Study. Vol. 2. Biosystematic Investigations in the Family of Duckweeds (Lemnaceae). Veröffentlichungen des Geobotanischen Institutes der ETH, Stiftung Ruebel, Zurich.
Les, D. H., and Crawford, D. J. (1999). Landoltia (Lemnaceae) a new genus of duckweeds. Novon 9, 530–533. doi: 10.2307/3392157
Les, D. H., Crawford, D. J., Landolt, E., Gabel, J. D., and Kimball, R. T. (2002). Phylogeny and systematics of Lemnaceae, the duckweed family. Syst. Bot. 27, 221–240. doi: 10.1043/0363-6445-27.2.221
Les, D. H., Landolt, E., and Crawford, D. J. (1997). Systematics of the Lemnaceae (duckweeds): inferences from micromolecular and morphological data. Plant Syst. Evol. 204, 161–177. doi: 10.1007/BF00989203
Ma, Y. B., Zhu, M., Yu, C. J., Wang, Y., Liu, Y., Li, M. L., et al. (2018). Large-scale screening and characterisation of Lemna aequinoctialis and Spirodela polyrhiza strains for starch production. Plant Biol. 20, 357–364. doi: 10.1111/plb.12679
Martirosyan, E. V., Ryzhova, N. N., Kochieva, E. Z., and Skryabin, K. G. (2009). Analysis of chloroplast rpS16 intron sequences in Lemnaceae. Mol. Biol. 43, 32–38. doi: 10.1134/S0026893309010051
Michael, T. P., Bryant, D., Gutierrez, R., Borisjuk, N., Chu, P., Zhang, H., et al. (2017). Comprehensive definition of genome features in Spirodela polyrhiza by high-depth physical mapping and short-read DNA sequencing strategies. Plant J. 89, 617–635. doi: 10.1111/tpj.13400
Nauheimer, L., Metzler, D., and Renner, S. S. (2012). Global history of the ancient monocot family Araceae inferred with models accounting for past continental positions and previous ranges based on fossils. New Phytol. 195, 938–950. doi: 10.1111/j.1469-8137.2012.04220.x
Ren, H. Y., Jiang, N., Wang, T., Omar, M. M., Ruan, W., and Ghafoor, A. (2018). Enhanced biogas production in the duckweed anaerobic digestion process. J. Energy Res. Technol. 140:041805. doi: 10.1115/1.4039782
Rothwell, G. W., van Atta, M. R., Ballard, H. E., and Stockey, R. A. (2004). Molecular phylogenetic relationships among Lemnaceae and Araceae using the chloroplast trnL-trnF intergenic spacer. Mol. Phylogenet. Evol. 30, 378–385. doi: 10.1016/S1055-7903(03)00205-7
Sonta, M., Rekiel, A., and Batorska, M. (2019). Use of duckweed (Lemna L.) in sustainable livestock production and aquaculture – a review. Ann. Anim. Sci. 19, 257–271. doi: 10.2478/aoas-2018-0048
Sree, K. S., Adelmann, K., Garcia, C., Lam, E., and Appenroth, K. J. (2015a). Natural variance in salt tolerance and induction of starch accumulation in duckweeds. Planta 241, 1395–1404. doi: 10.1007/s00425-015-2264-x
Sree, K. S., Bog, M., and Appenroth, K. J. (2016). Taxonomy of duckweeds (Lemnaceae), potential new crop plants. Emirate J. Food Agricult. 28, 291–302. doi: 10.9755/ejfa.2016-01-038
Sree, K. S., Sudakaran, S., and Appenroth, K. J. (2015b). How fast can angiosperms grow? Species and clonal diversity of growth rates in the genus Wolffia (Lemnaceae). Acta Physiol. Plant. 37:204. doi: 10.1007/s11738-015-1951-3
Tippery, N. P., Les, D. H., and Crawford, D. J. (2015). Evaluation of phylogenetic relationships in Lemnaceae using nuclear ribosomal data. Plant Biol. 17, 50–58. doi: 10.1111/plb.12203
Van Hoeck, A., Horemans, N., Monsieurs, P., Cao, H. X., Vandenhove, H., and Blust, R. (2015). The first draft genome of the aquatic model plant Lemna minor opens the route for future stress physiology research and biotechnological applications. Biotechnol. Biofuels 8:188. doi: 10.1186/s13068-015-0381-1
Wang, W., Kerstetter, R. A., and Michael, T. P. (2011). Evolution of genome size in duckweeds (Lemnaceae). J. Bot. 2011:570319. doi: 10.1155/2011/570319
Wang, W., Wu, Y., Yan, Y., Ermakova, M., Kerstetter, R., and Messing, J. (2010). DNA barcoding of the Lemnaceae, a family of aquatic monocots. BMC Plant Biol. 10:205. doi: 10.1186/1471-2229-10-205
Xu, N., Hu, F., Wu, J., Zhang, W., Wang, M., Zhu, M., et al. (2018). Characterization of 19 polymorphic SSR markers in Spirodela polyrhiza (Lemnaceae) and cross-amplification in Lemna perpusilla. Appl. Plant Sci. 6:e01153. doi: 10.1002/aps3.1153
Xu, S. Q., Stapley, J., Gablenz, S., Boyer, J., Appenroth, K. J., Sree, K. S., et al. (2019). Low genetic variation is associated with low mutation rate in the giant duckweed. Nat. Commun. 10:1243. doi: 10.1038/s41467-019-09235-5
Xue, H. L., Xiao, Y., Jin, Y. L., Li, X. B., Fang, Y., Zhao, H., et al. (2012). Genetic diversity and geographic differentiation analysis of duckweed using inter-simple sequence repeat markers. Mol. Biol. Rep. 39, 547–554. doi: 10.1007/s11033-011-0769-3
Zhao, X., Moates, G. K., Elliston, A., Wilson, D. R., Coleman, M. J., and Waldron, K. W. (2015). Simultaneous saccharification and fermentation of steam exploded duckweed: improvement of the ethanol yield by increasing yeast titre. Bioresour. Technol. 194, 263–269. doi: 10.1016/j.biortech.2015.06.131
Keywords: AFLP, DNA-barcoding, fingerprinting, Landoltia, Lemna, Spirodela, Wolffia, Wolffiella
Citation: Bog M, Appenroth K-J and Sree KS (2019) Duckweed (Lemnaceae): Its Molecular Taxonomy. Front. Sustain. Food Syst. 3:117. doi: 10.3389/fsufs.2019.00117
Received: 27 July 2019; Accepted: 02 December 2019;
Published: 17 December 2019.
Edited by:
Marvin Edelman, Weizmann Institute of Science, IsraelReviewed by:
Agnieszka Barbara Najda, University of Life Sciences of Lublin, PolandTahira Fatima, Purdue University, United States
Autar Krishen Mattoo, Beltsville Agricultural Research Center (USDA), United States
Copyright © 2019 Bog, Appenroth and Sree. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: K. Sowjanya Sree, a3Nvd3NyZWVAZ21haWwuY29t; a3Nvd3NyZWU5QGN1a2VyYWxhLmFjLmlu
 Manuela Bog
Manuela Bog Klaus-J. Appenroth
Klaus-J. Appenroth K. Sowjanya Sree
K. Sowjanya Sree