- 1Legumes Pathology, Integrated Crop Management, International Crops Research Institute for the Semi-arid Tropics, Patancheru, India
- 2Plant Pathology Division, Jawaharlal Nehru Krishi Vishwa Vidyalaya, Jabalpur, India
- 3Crop Protection Division, Indian Institute of Pulse Research, Kanpur, India
- 4Department of Plant Pathology, RAK College of Agriculture, Sehore, India
- 5Department of Plant Pathology, University of Agricultural Sciences, GKVK, Bangalore, India
- 6Pulse Research Station, Gujarat Agriculture University, Junagadh, India
- 7Agriculture Research Station, Badnapur, India
- 8Agriculture Research Station, Gulberga, India
- 9Department of Plant Pathology, Tirhut Collage of Agriculture, Samastipur, India
- 10Pulse Improvement Project, Mahatma Phule Krishi Vidyapeeth, Rahuri, India
Fusarium wilt (Fusarium oxysporum f. sp. ciceris) of chickpea is the major limitation to chickpea production worldwide. As the nature of the pathogen is soil borne, exploitation of host plant resistance is the most suitable and economical way to manage this disease. Present study was therefore conducted with an aim to find new, stable and durable sources of resistance of chickpea against Fusarium wilt through multi-environment and multi-year screening. During 2007/2008 crop season, 130 promising genotypes having <10% wilt incidence were selected from initial evaluation of 893 chickpea genotypes in wilt sick plot at ICRISAT, Patancheru. Of them 61 highly resistant lines were selected through further evaluation in 2008/2009 and 2009/2010 crop season. Finally, a set of 31 genotypes were selected to constitute a Chickpea Wilt Nursery (CWN) and tested at 10 locations in India for three cropping seasons (2010/2011, 2011/2012, and 2012/2013) coordinated through Indian Council of Agricultural Research (ICAR) and ICRISAT collaboration. The genotype and genotype × environment interaction (GGE) indicated significant variations (p ≤ 0.001) due to genotype × environment (G × E) interaction. Most of genotypes were resistant at two locations, ICRISAT (Patancheru) and Badnapur. On the contrary most of them were susceptible at Dholi and Kanpur indicating the variability in pathogen. GGE biplot analyses allowed the selection six genotypes ICCVs 98505, 07105, 07111, 07305, 08113, and 93706 with high resistance and stability across most of the locations and eight moderately resistant (<20% mean incidence) genotypes viz., ICCVs 08123, 08125, 96858, 07118, 08124, 04514, 08323, and 08117. As chickpea is grown in diverse agro-ecological zones and environments; these stable/durable sources can be used in future resistance breeding program to develop Fusarium wilt resistant cultivars.
Introduction
Among the legumes, chickpea (Cicer arietinum L.) has occupied a foremost place due to its high nutritional value. But the average global productivity of chickpea is hampered due to various biotic stresses (Reddy et al., 1990; Tarafdar et al., 2017, 2018). Among them Fusarium wilt caused by Fusarium oxysporum f. sp. ciceris (FOC) is one of the widely distributed diseases of chickpea and cause yield loss up to 10–100% depending on varietal susceptibility and climatic conditions (Jimenez-Diaz et al., 1989; Patil et al., 2015). The disease is more predominant in the Indian subcontinent, Spain, Ethiopia, Mexico, Tunisia, Turkey, and the United States (Westerlund et al., 1974; Halila and Strange, 1996; Ghosh et al., 2013). Since the disease is soil borne, chemical control is not effective and practical to implement (Sharma et al., 2017). Exploitation of host plant resistance is therefore the most trustworthy way to overcome the situation Numerous sources of resistance to Fusarium wilt in chickpea has been identified previously (Pande et al., 2006; Mirzapour et al., 2014; Chobe et al., 2016) and several are being utilized in resistance breeding program at ICRISAT and National Agricultural Research Stations (NARS) that has contributed in substantial increase of chickpea productivity in semi-arid regions of Africa and Asia (Sharma et al., 2012; Fikre et al., 2018). However, resistance sturdiness in these sources is affected due to G × E interaction and high genetic variability in the pathogen.
The pathogen was reported with eight races from over the world (Haware and Nene, 1982; Sharma et al., 2012). Races 0, 1A, 1B/C, 5, and 6, has been reported from United States and Spain and races 1A, 2, 3, and 4 from India. Although, gene for gene relationship of few FOC Avr gene and chickpea R gene has been proved, but chickpea and FOC interaction at molecular level is yet to be known (Sharma et al., 2016b). Present distribution of FOC races is not very clear owing to large exchange of germplasm and climate variability (Sharma et al., 2014) and existence of multiple races in one region. Therefore, in order to develop effective stratagems, for wilt management through host plant resistance, it is essential to obtain information on resistance stability of genotypes at multi-environment.
Several methods have been used to analyze the G × E interaction (Moore et al., 2019) and a number of multi-variate techniques such as GGE billet technique have been used by various researchers (Yan et al., 2000; Yan, 2001; Sharma et al., 2013, 2015). Biplot analysis of G × E data has been advanced such that many important questions, such as stability of genotypes, mean performance, discriminating ability, mega-environment investigation, representativeness of environment, and who-resistant-where pattern can be graphically addressed for better understanding.
In the above context, the present work was undertaken to identify stable, durable, and broad based sources of resistance to wilt under ICAR-ICRISAT collaboration in chickpea through multi-environment and multi-year field testing across the wilt hot spot locations in India.
Materials and Methods
Plant Materials
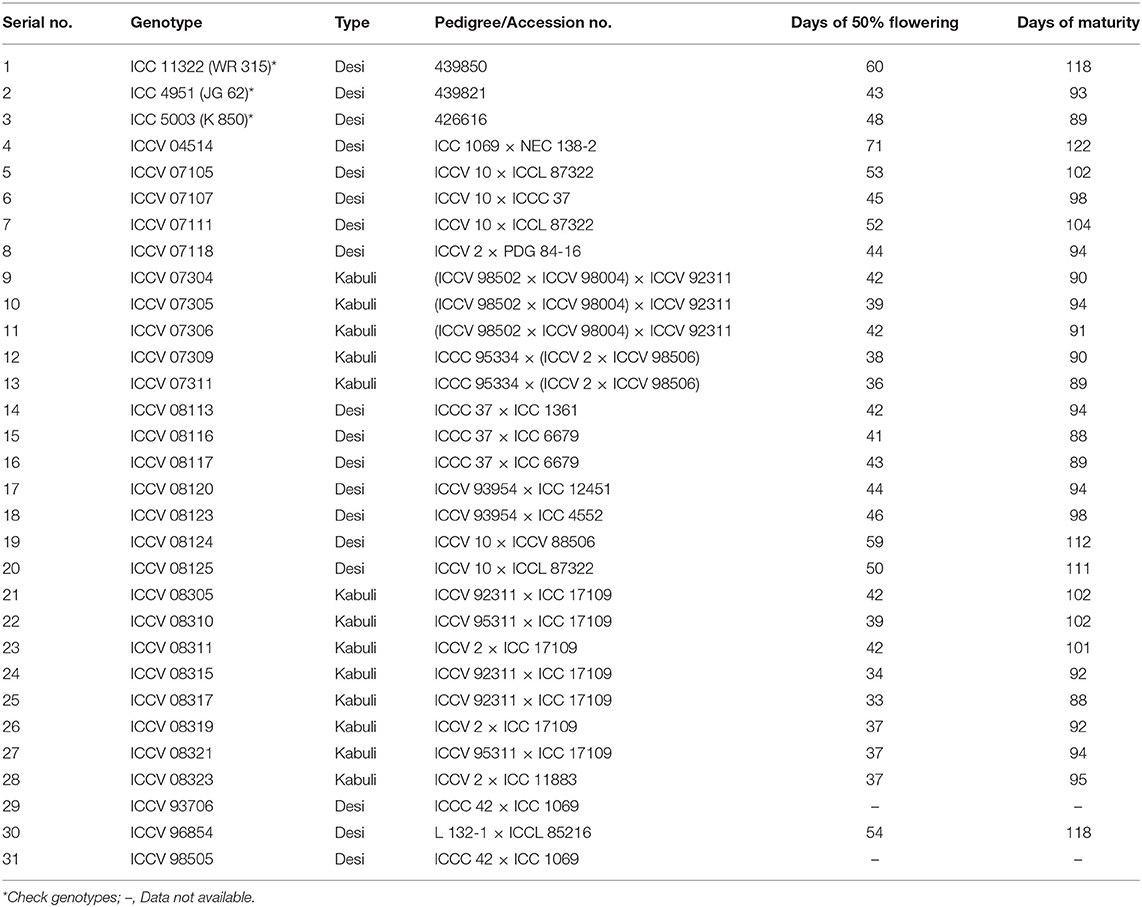
During 2007/2008 cropping season, a total of 893 chickpea genotypes including breeding lines and germplasm accessions were screened for Fusarium wilt resistant in a sick plot at ICRISAT, Patancheru under artificial epiphytotic conditions (Figure 1). Out of them, 130 promising lines (genotypes) having ≤10% wilt incidence was selected primarily for further evaluation. The selected genotypes were further evaluated in next two consecutive years, 2008/2009 and 2009/2010 through randomized complete block design (RCBD) in two replications. Each genotype was sown in a 4 m long row (2 rows/ replication) having 60 cm row to row distance. Susceptible check ICC 4951 (JG 62) and resistant check ICC 11322 (WR 315) were sown after every eight rows. Based on disease reaction in year of 2008/2009 and 2009/2010, a total of 61 highly wilt resistant genotypes were selected. Finally, a set of 31 genotypes (18 desi and 13 kabuli) including susceptible and resistant checks were selected based on consistent resistant reaction to constitute a Chickpea Wilt Nursery (CWN) for multi-environment and multi-year evaluation. The details of the 50% flowering, pod maturity duration and pedigree of the selected lines are described in Table 1.
Multi-environment Screening
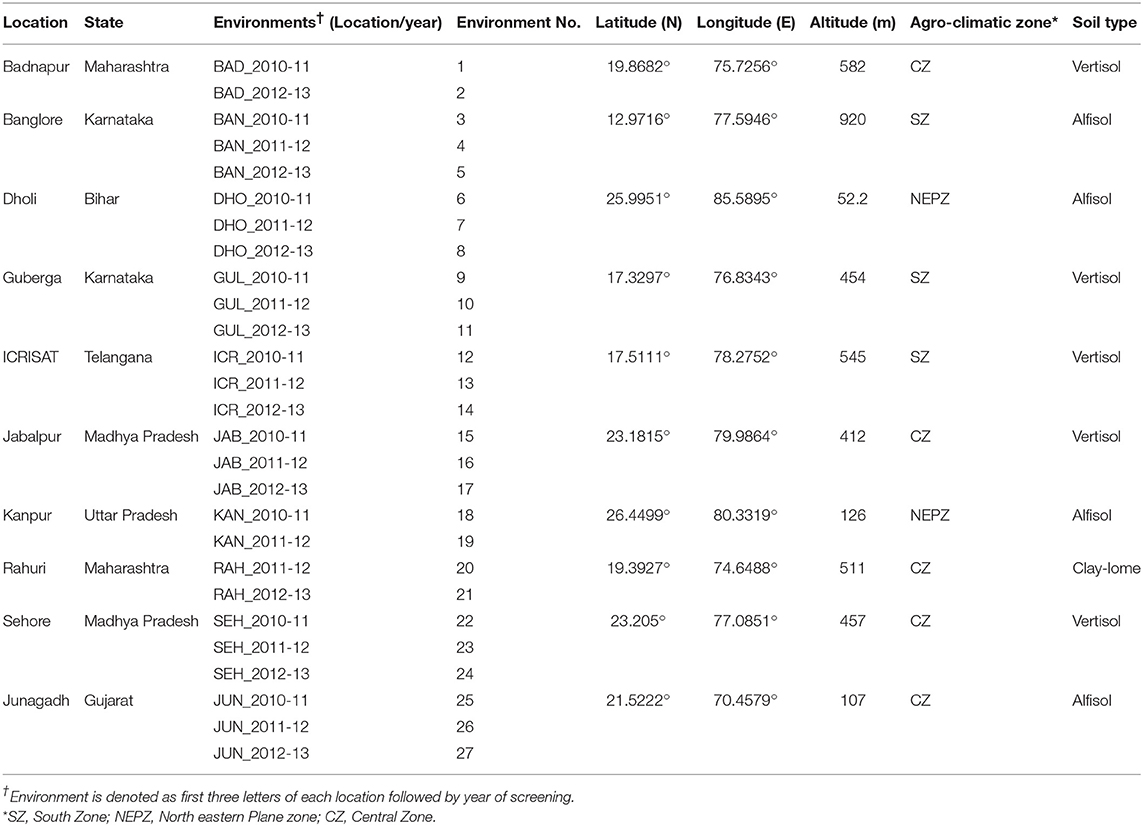
The CWN was evaluated for Fusarium wilt resistance at 10 different locations in India [Dholi, Banglore, ICRISAT (Patancheru), Rahuri, Sehore, Gulberga, Kanpur, Junagarh, Jabalpur, and Badnapur] for 3 years (2010/11, 2012/13, and 2013/14) in wilt sick plot. These locations encompass a wide diversity in agro-climatic zones with latitude from N 17.3297° at Gulberga to N 26.4499° at Kanpur, longitude from E 70.4579° at Junagadh to E 85.5895° at Dholi, and altitude from 52.2 m (Dholi) to 920 m (Banglore). The tested locations represent 27 environments and three soil types during three cropping seasons (Table 2 and Figure 2). Seeds of the test genotypes were supplied to the respective collaborators for multi-location testing against wilt. At each location, the nursery was evaluated in a RCBD with two replications as described above.
Data Collection and Statistical Analysis
Wilt incidence data was recorded replication across all locations during the years of evaluation. Per cent disease incidence in each test genotype was calculated using the following formula
Based on mortality of plants to Fusarium wilt, the test genotypes were divided into four categories, resistant (<10.0% plant mortality), moderately resistant (10.1–20.0% plant mortality), susceptible (20.1–40.0% plant mortality), and highly susceptible (>40.0% plant mortality).
To make residual normal the percentage data was arcsine transformed prior to analysis (Gomez and Gomez, 1984) and transformed data was used to test the significance of genotype (G), environment (E), and genotype × environment (G × E) interaction using MIXED procedure of SAS 9.4 (SAS Institute Inc, 2017) considering genotype, environment, and replication as random effects. Individual environment variances were modeled into combined analysis. BLUPs (Best Linear Unbiased Predictors) were estimated for G, E, and G × E interaction from combined analysis. Multiple comparisons were performed among test locations.
To identify relationship between environments, Spearman's rank correlation was performed using SAS PROC CORR procedure (SAS Institute Inc, 2017). The performance of all possible genotypes in two environments were compared and determined if the differences in performances is significantly “<0” in one environment and significantly “>0” in other environment (Yang et al., 2009; Ponnuswamy et al., 2018).
The GGE biplot, site regression model (Yan and Kang, 2002) was used to visualize the G × E interaction patterns and to distinguish (1) genotype performance and stable genotype across all environments, (2) environment effects, discriminating genotypes and (3) identify patterns where by specific genotypes can be recommended to a specific environment(s). Further, to understand the disease incidence and its distribution pattern among the test genotypes across the locations, boxplot analysis of environment × incidence, and genotype × incidence was carried out (Wiik and Rosenqvist, 2010).
Results
Identification of Resistant Genotypes Through Preliminary Screening
Preliminary screening of 893 genotypes at ICRISAT, Patancheru showed a broad range of response to wilt (resistant to susceptible). This allowed removal of susceptible materials and selection of most promising 61 genotypes having ≤10% wilt incidence. Subsequent screening of 61 genotypes enabled in constitution of a CWN consisting of 28 promising genotypes with <10% wilt incidence for multi-environment and multi-year screening in 10 diverse locations (Tables 1, 2).
Response of Genotypes to Wilt in Multi-environment
The wilt incidence in most of the chickpea genotypes varied across the locations and years. Among the genotypes, the variability in wilt incidence is evident from the frequency distribution of genotypes across locations over the years (Figure 3). For instance, most of the genotypes were resistant at ICRISAT, Patancheru (24 genotypes) followed by Badnapur (21 genotypes). On the contrary, none of the genotypes were found resistant in Kanpur and Sehore (Figure 3). Analysis of variance for wilt incidence showed that all the three sources of variation were highly significant (P < 0.0001); with a higher proportion of variation due to G × E (40.35 %) followed by G (35.6%) and E (24.05%) (Table 3).
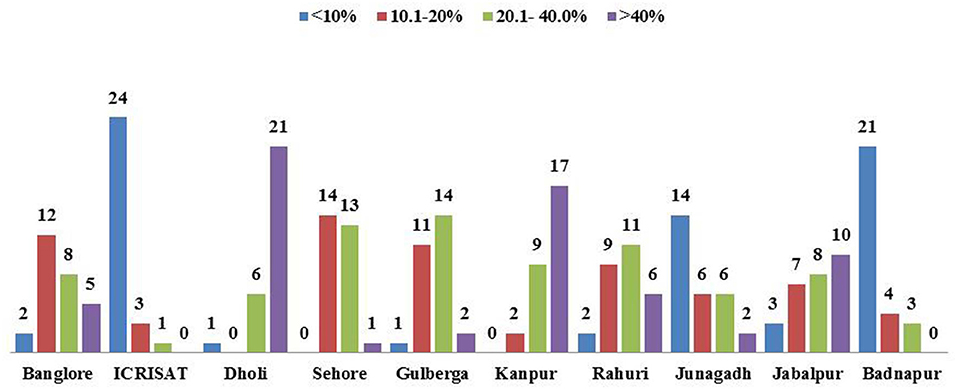
Figure 3. Frequency distribution of 28 test genotypes for levels of Fusarium wilt disease at 10 locations in India over 3 years. Rating of genotype reaction: resistant = 0–10% wilt incidence; moderately resistant = 10.1–20% wilt incidence; susceptible = 20.1–40% and highly susceptible = 40.1–100%.
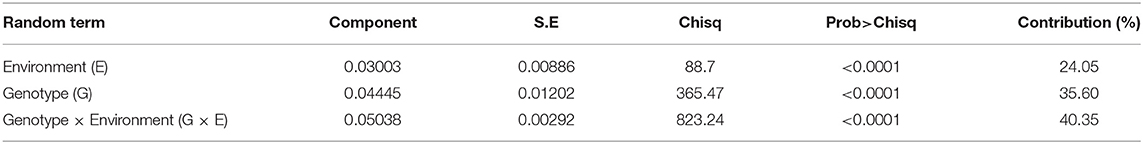
Table 3. Combined analysis of variance for wilt incidence of 31 genotypes across the locations during 2010/2011, 2011/2012, and 2012/2013 cropping seasons.
An adequate disease pressure was found in wilt sick plots at all the locations as evident from wilt incidence of susceptible check JG 62 (85.35–100%) (Table 4). Among the locations, the highest mean wilt incidence irrespective of genotypes (excluding check) over 3 years was observed in Kanpur (46.42%) followed by Dholi (45.75%), whereas the lowest mean wilt incidence was observed in Badnapur (8.45%) followed by ICRISAT (Patancheru) (9.52%) (Table 4). Significant differences in wilt resistant/ moderately resistant genotypes were observed in different locations (Tables S1, S2).
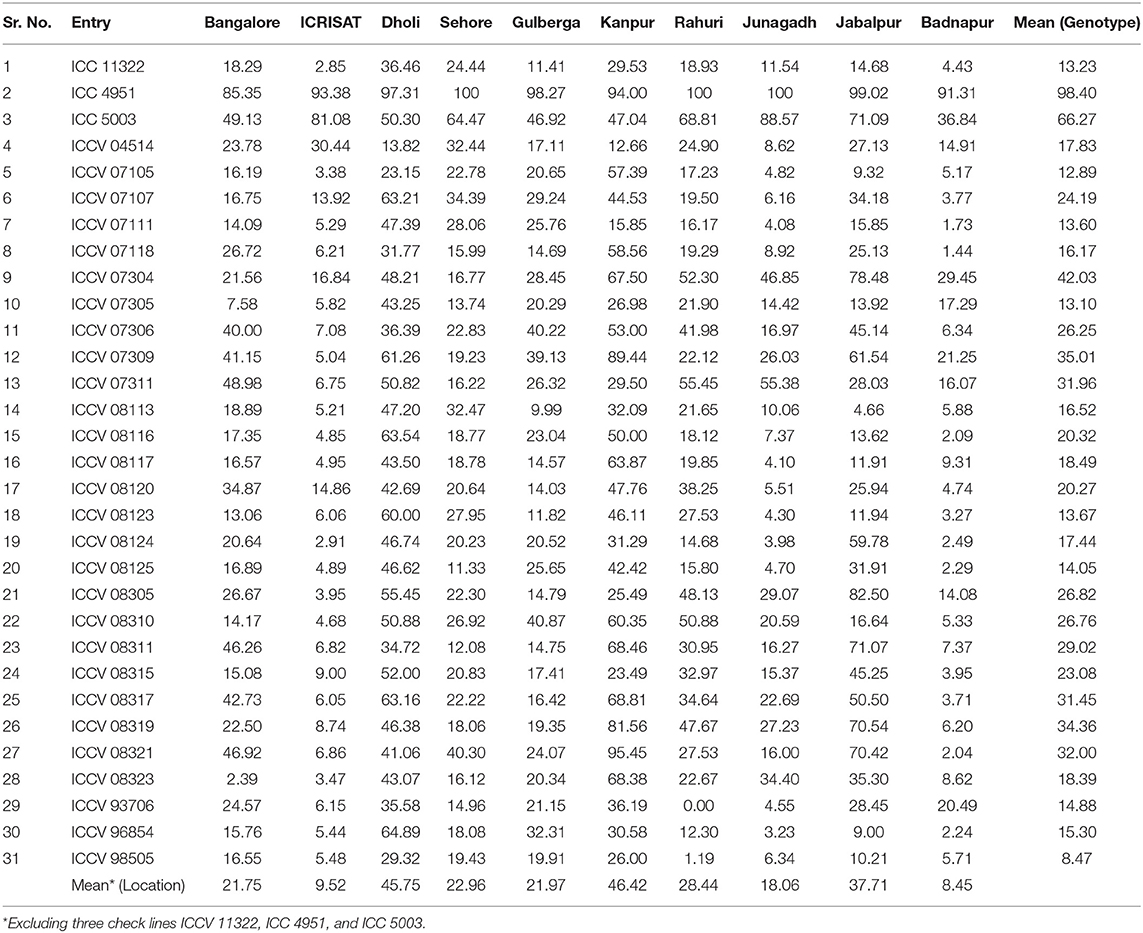
Table 4. Mean wilt incidence (%) of testing chickpea genotypes across the locations during 2010/2011, 2011/2012, and 2012/2013 cropping seasons.
Performance of genotypes differed over the years and locations as evident from the distribution pattern of disease incidence among the test genotypes across the locations (Figure 4). For example, three genotypes including susceptible check JG 62 and two genotypes ICCV 93706 and ICCV 96854 was stable throughout the environments, whereas the performance of four genotypes ICCV 07304, ICCV 08116, and ICCV 08311 and late wilt susceptible check ICC 5003 varied in different environments. Also, magnitude of wilt incidence varied depending on environmental variable at particular location. The analysis indicated that the location-wise performance of the genotypes was highly stable in Badnapur, followed by ICRISAT over the years, whereas the performance of genotypes varied highly in Jabalpur followed by Dholi and Banglore in same cropping seasons (Figure 5). Mean wilt incidence was highest at DHO_2012-13 (57.6%) followed by DHO_2010-11 (54.76%) and KAN_2011-12, whereas lowest average wilt incidence (4.86%) was observed in ICR_2012-13. Maximum range of average wilt incidence (2.44–100%) was recorded in JAB_2012-13.
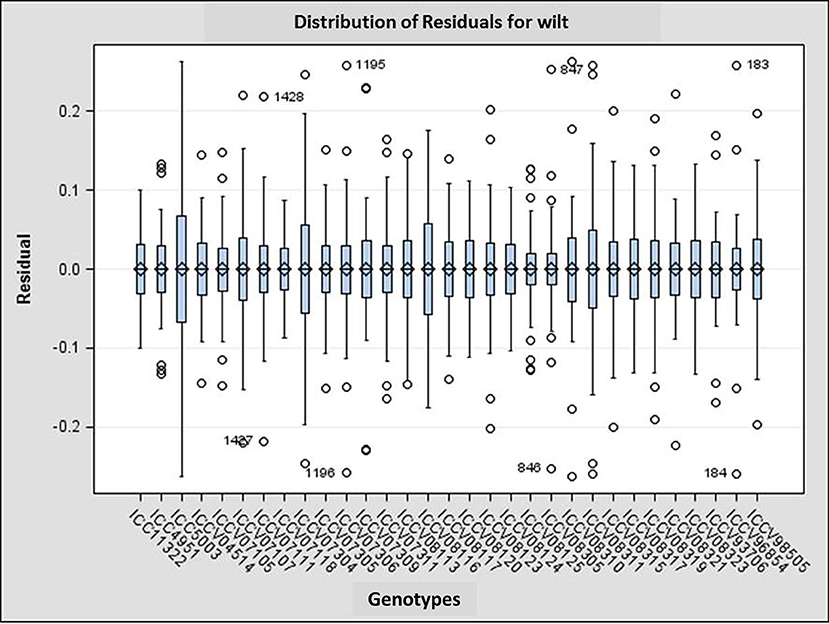
Figure 4. Distribution of residuals for wilt incidence of each genotype across 27 environments. Box edges represent the upper and lower quantile with median value shown in the middle of the box. Whiskers represented by “o” symbol. Individuals falling outside the range of whiskers shown as numbers.
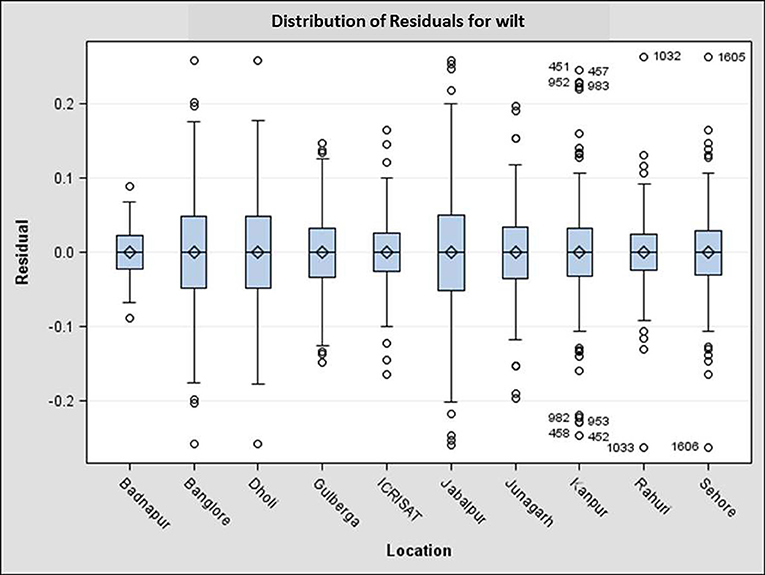
Figure 5. Distribution of residuals for genotypes across ten locations. Box edges represent the upper and lower quintile with median value shown in the middle of the box. Whiskers represented by “o” symbol. Individuals falling outside the range of whiskers shown as numbers.
During Fusarium wilt screening, the overall assessment through mean wilt incidence % indistinctly classified the tested genotypes in different resistant and susceptible phenotypic groups on the basis of their disease reaction, even though they are derived from the same parents. For example, the kabuli chickpea ICCVs 07304, 07305, and 07306 were derived from the multiple crosses of (ICCV 98502 × ICCV 98004) × ICCV 92311 (Table 1) showed considerable varying disease reaction from moderately resistant to highly susceptible with their mean wilt incidence range from 13.10 to 42.03%. Similarly, the desi chickpea ICCVs 93706 and 98505 were derived from cross of same parent lines ICCC 42 × ICC 1069, but the little variation was observed in their mean wilt incidence, 14.88 and 8.47%, respectively.
Correlation Between Environments
A significant positive correlation for the levels of wilt incidence was found in some of the test environments using Spearman's rank correlation analysis (P < 0.0001). For instance, the significant positive correlation (r = 0.60) was observed between environments of BAN_2010-11 (3) and SEH_2010-11 (22) with respect to wilt incidence, while in same cropping season, the negative correlation (r = −0.14) was found between environments of BAD_2010-11 (1) and GUL_2010-11 (9) (Table 5). Crossover interactions indicated diverse environments among the test locations. For instance, high entry rank value (24.09%) between the environments of JAB_2010-11 (15) and BAD_2010-11 (1) indicated the diverse environment between the locations. Conversely low entry rank value (3.84%) indicated less diversity between BAD_2010-11 (1) and GUL_2010-11 (9) (Table 5).
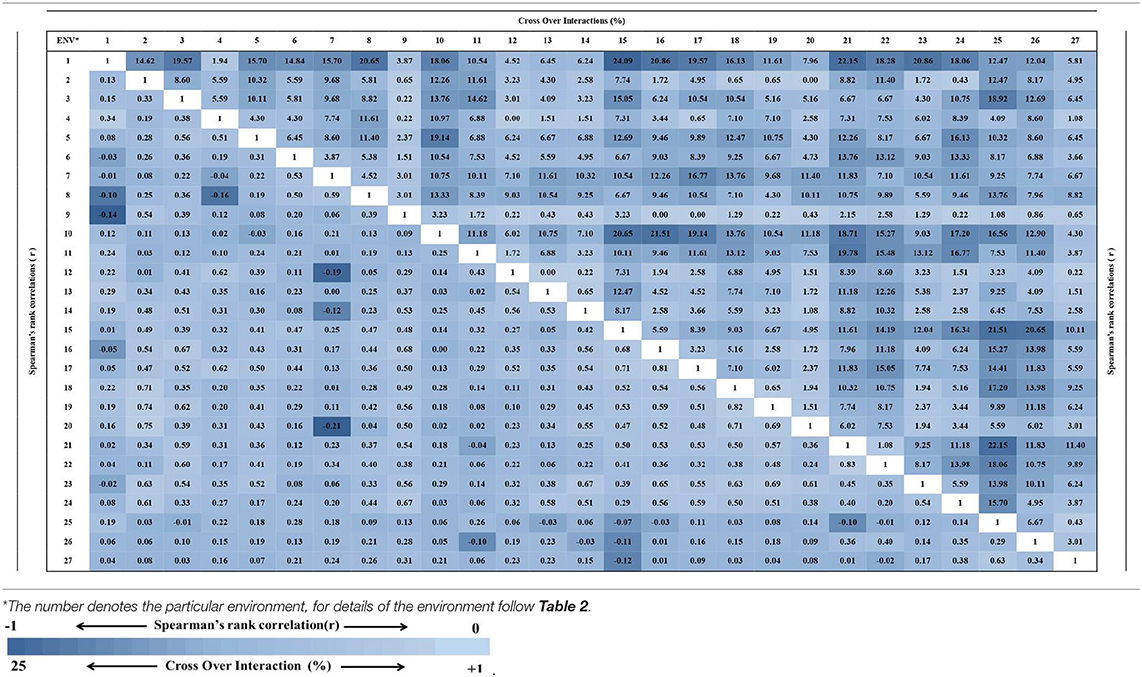
Table 5. Spearman's rank correlations (r) and cross over interactions (%) showing stability and comparison of the genotypes across the location.
Stability of Genotypes and Environments
GGE biplot analysis explained 66.11% of total variation, where PC1 (wilt incidence) and PC2 (resistance stability) accounted for 51.64 and 14.47% variation, respectively (Figure 6). It was found that the performance of test genotypes as indicated by proximity of vectors over the years within a specific location was nearly similar. For instance, the proximity of vectors for the environments of JAB_2010-11, JAB_2011-12, and JAB_2012-13 indicated that the performance of the tested genotypes to wilt in Jabalpur location was stable. Further, the longer vectors for the environments KAN_2010-11, KAN_2011-12, JAB_2011-12, and JAB_2012-13 indicated that those environments were most discriminating for genetic differentiation of genotypes. Conversely, the environments BAD_2010-11, BAN_2011-12, BAN_2012-13, DHO_2011-12, and DHO_2012-13 had smaller vectors indicating that tested genotypes were least discriminatory to those environments.
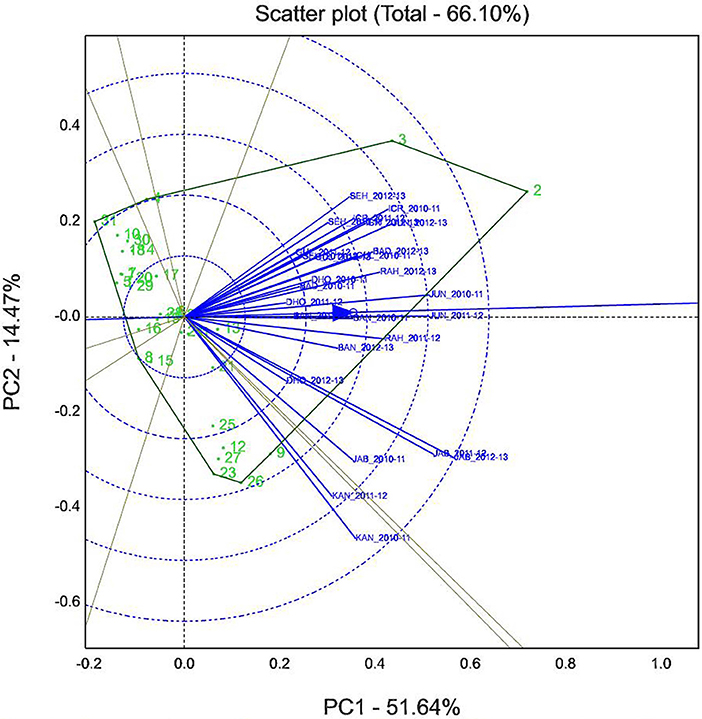
Figure 6. GGE biplot showing the relationship among 27 environments based on Fusarium wilt incidence of 31 chickpea genotypes.
An angle (nearly 90°) among the environments e.g., ICR_2010-11, ICR_2011-12, SEH_2010-11, and SEH_2012-13 with KAN_2010-11 and KAN_2011-12 indicated the moderate correlations within them. However, the higher PC1 scores and lower PC2 scores of the environments JAB_2010-11, JAB_2011-12, JAB_2012-13, KAN_2010-11, and KAN_2011-12 indicated better discriminating ability of these environments (Figure 6).
A six sided polygon on the biplot indicated that the genotypes positioned at the vertices of the polygon added most to the interaction (highest or lowest wilt incidence). The genotypes placed at the right side of the Y-axis indicated susceptibility to wilt irrespective of environments, whereas those genotypes placed at the left side are resistant to wilt across the environments. Out of 31, genotypes ICCV 07304 (9), ICCV 08319 (26), ICC 4951 (2), and ICC 5003 (3) were persistently more susceptible to wilt by being farthest from the point of source of biplot on the right side.
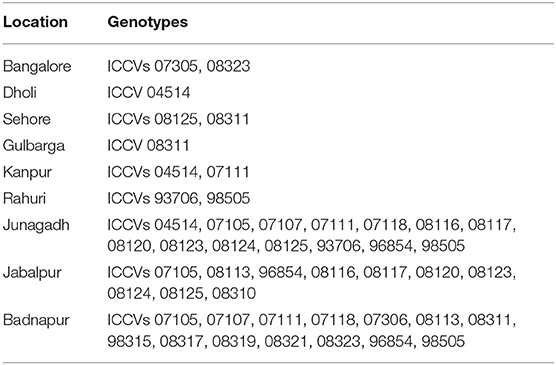
However, genotype ICCV 98505 (31) located farthest from the point of origin on the left side endorsed its resistance to wilt across the locations with high stability. Further, four genotypes namely genotype ICCV 07105 (5), ICCV 07111 (7), ICCV 07305 (10), and ICCV 93706 (29) placed on the right side of the origin-point revealed moderate level of stability to tested environment with low level of wilt incidence (Figure 6). Detail of the genotypes found suitable and adaptable for the location specific breeding program is provided in Table 6.
Discussion
Deploying wilt resistant chickpea cultivars is one of the sustainable strategies adopted by the breeders as a part of integrated disease management. The success of any breeding programme is generally dependent on the stable performance of any traits within the genotypes. Therefore, present study using G × E interaction on Fusarium wilt incidence, may play a crucial role to enhance chickpea productivity as this has enabled us to identify donors with stable and broad-based sources of resistance and existing variability in the pathogen population.
A multi-environment evaluation revealed significant differences (<0.0001) in G, E and G × E interaction for wilt. Differential reaction of the chickpea to Fusarium wilt in multi-environment can be attributed to variations in virulence in the pathogen population (Sharma et al., 2014). Presence of different virulence genes within the pathogen and their varied responses in different geographical locations may be responsible to varied wilt incidence across the environments, although the differential response of the genotypes in different environmental conditions cannot be let off (Kulkarni and Chopra, 1982).
High level of wilt incidence in susceptible genotypes at all locations indicated adequate disease pressure in sick plots. Average wilt incidence was found higher during the years of evaluation at Kanpur and Dholi than other test locations irrespective of genotypes. In contrast, average wilt incidence was lowest in ICRISAT (Patancheru) and Badnapur. Location-wise variation in wilt incidences might be attributed to differences in virulence of the pathogens or random distribution of the resistance gene(s) within the chickpea genotypes, or due to influence of both factors (Sharma et al., 2014). Presence of four pathogenic races (races 1A, 2, 3, 4) from India (Haware and Nene, 1982) has been reported where race 2 from Kanpur was found to be more virulent than race 1 from ICRISAT, Patancheru.
In our study, it was shown that the genotypes derived from same parents differed for their disease reaction e.g., kabuli chickpea ICCVs 07304, 07305, and 07306 indicating the segregation of the resistant genes. However, little variation in mean wilt incidence for the desi chickpea ICCVs 93706 and 98505 derived from same parent lines might be due to a tight link within the multiple genes contributing to the resistance response. Previous studies on genetic analysis reported that the resistance to FOC race 1 is governed either by one or two genes (Brinda and Ravikumar, 2005) or three genes (Singh et al., 1987) whereas the resistance of FOC race 2 is conferred by single recessive gene (Sharma and Muehlbauer, 2007) and resistance to FOC is also monogenic (Sharma et al., 2004). Therefore, it is clear that segregation of genes have a major role in controlling the disease. However, when such resistant genes are absent in the genotypes or the genotypes are challenged by a complex mixture of the different races of FOC in sick plot, the varied disease response of the genotype is expected.
Multi-environment evaluation of genotypes assisted in selection of stable and resistant genotypes [ICCV 98505 (31), ICCV 07105 (5), ICCV 07111 (7), ICCV 07305 (10), ICCV 08113 (14), and ICCV 93706 (29)]. These genotypes could be used as resistant donor for wilt in chickpea breeding programs at different locations. Identification of high stable genotypes with low disease incidence is the prime source of resistant breeding programs. The GGE biplot analysis has been widely used in resistant breeding program for selection of genotypes having high stability with low disease incidence such as spot blotch in wheat (Sharma and Duveiller, 2007), rust in soybean (Twizeyimana et al., 2008), Ascochyta blight in faba bean (Rubiales et al., 2012), sterility mosaic disease in pigeonpea (Sharma et al., 2015), Fusarium wilt in chickpea and pigeonpea (Sharma et al., 2012, 2016a), rust of field pea (Das et al., 2019). Further, the variance for G × E interaction was more in this study than the genotypic variance indicating that the disease incidence was affected by both genotypes and environments. Significant positive as well as negative correlation was found between test locations, however it was not in accordance with agro-ecological zones again indicating the interactive effects of genotypes and environment.
Multi-environment and multi-year screening against diseases possibly can be used as a model for future selection of genotypes and identified genotypes could be the prime source in resistance breeding programs for specific adaptation to a particular agro-climatic zone. Varying response of the genotypes throughout the environment reflected the influence of environment toward instability of wilt incidence. In this study we discriminated broad based and stable resistant chickpea genotypes for future resistance breeding programme. Badnapur, ICRISAT, and Rahuri found as ideal test locations for culling out superior and stable wilt resistant chickpea genotypes. For optimum resource utilization, elimination of unnecessary testing locations should be implemented.
Data Availability Statement
All datasets generated for this study are included in the manuscript/Supplementary Files.
Author Contributions
MSh conceived and planned the work with significant inputs from RG. MSh and RG coordinated the experiment. AT and DC compiled the data. AR and AK analyzed the data for statically. OG, NS, DS, MSa, MP, PHG, DM, JU, and PH contributed in the multi-location trials including data collection at their respective locations. PMG and SS provided the pedigree details and seeds for the trial. RG, AT, and DC drafted the manuscript. MSh finally edited the manuscript. All authors read the manuscript and agree with its content.
Funding
The funding support from CGIAR Research Program on Grain Legumes and Dryland Cereals (CRP–GLDC), ICRISAT, and SPLICE-Climate Change Program under Department of Science and Technology, Govt of India [DST/CCP/CoE/142/2018 (G)] is gratefully acknowledged.
Conflict of Interest
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We are thankful to the All India Coordinated Research Project (AICRP) on Chickpea for undertaking this activity under ICAR-ICRISAT collaboration. We also acknowledge the partnership with NARS pathologists of respective locations for evaluating the disease nursery over the years.
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fsufs.2019.00078/full#supplementary-material
References
Brinda, S., and Ravikumar, R. L. (2005). Inheritance of wilt resistance in chickpea—A molecular marker analysis. Curr. Sci. 88, 701–702.
Chobe, D. R., Gupta, O., and Pawar, M. (2016). Radiation induced mutation for resistance against races/pathotypes of Fusarium oxysporum f.sp ciceris in chickpea (Cicer arietinum L.). Indian Phytopath. 69, 260–265. Available online at: http://epubs.icar.org.in/ejournal/index.php/IPPJ/article/view/71426
Das, A., Gupta, S., Parihar, A. K., Saxena, D., Singh, D., Singha, K. D., et al. (2019).Deciphering genotype-by-environment interaction for targeting test environments and rust resistant genotypes in field pea (Pisum sativum L.). Front. Plant Sci. 10:825. doi: 10.3389/fpls.2019.00825
Fikre, A., Korbu, L., Eshete, M., Bekele, D., Girma, N., Mohamed, R., et al. (2018). A decade of research progress in chickpea and lentil breeding and genetics. Ethiop. J. Crop Sci. 6, 101–113. Available online at: http://oar.icrisat.org/11056/1/110-122.pdf
Ghosh, R., Sharma, M., Telangre, R., and Pande, S. (2013). Occurrence and distribution of chickpea diseases in central and southern parts of India. Am. J. Plant Sci. 4, 940–944. doi: 10.4236/ajps.2013.44116
Gomez, K. A., and Gomez, A. A. (1984). Statistical Procedure for Agricultural Research. New York, NY: Wiley Press.
Halila, M. H., and Strange, R. N. (1996). Identification of the causal agent of wilt of chickpea in Tunisia as Fusarium oxysporum f. sp. ciceris race 0. Phytopathol. Mediterr. 35, 67–74.
Haware, M. P., and Nene, Y. L. (1982). Races of Fusarium oxysporum f. sp. ciceri. Plant Dis. 66, 809–810. doi: 10.1094/PD-66-809
Jimenez-Diaz, R. M., Trapero-Casas, A., and de la Colina, J. C. (1989). “Races of Fusarium oxysporum f. sp. ciceris infecting chickpea in Southern Spain,” in Vascular Wilt Diseases of Plants, NATO ASI Series, eds E. C. Tjamos and C. H. Beckman (Berlin: Springer Verlag), 515–520. doi: 10.1007/978-3-642-73166-2_39
Kulkarni, R. N., and Chopra, V. L. (1982). Environment as the cause of differential interaction between host cultivars and pathogenic races. Phytopathology 72, 1384–1386. doi: 10.1094/Phyto-72-1384
Mirzapour, S., Darvishnia, M., Bazgir, E., and Goodarzi, D. (2014). Identification of resistant sources in chickpea against Fusarium wilt under greenhouse condition. Intl. J. Farm. Alli. Sci. 3, 772–776. Available online at: http://ijfas.com/wp-content/uploads/2014/08/772-776.pdf
Moore, R., Francesco, P. C., Marc, J. B., Danilo, H., Franke, L., Barroso, I., et al. (2019). A linear mixed-model approach to study multivariate gene–environment interactions. Nat. Genet. 51, 180–186. doi: 10.1038/s41588-018-0271-0
Pande, S., Kishore, G. K., Upadhyaya, H. D., and Rao, J. N. (2006). Identification of sources of multiple disease resistance in mini-core collection of chickpea. Plant Dis. 90, 1214–1218. doi: 10.1094/PD-90-1214
Patil, M., Om, G., Pawar, M., and Chobe, D. R. (2015). Effect of culture media, pH and temperature on the mycelial growth and sporulation of Fusarium oxysporum f. sp. ciceris isolates of chickpea from Central Zone of India. Res. J. Appl. Sci. 49, 54–60.
Ponnuswamy, R., Rathore, A., Vemula, A., Das, R., Singh, A., Balakrishnan, D., et al. (2018). Analysis of multi-location data of hybrid rice trials reveals complex genotype by environment interaction. Cereal Res. Communi. 46, 1–12. doi: 10.1556/0806.45.2017.065
Reddy, M. V., Nene, Y. L., Singh, G., and Bashir, M. (1990). “Strategies for management of foliar diseases of chickpea,” in Proceedings of the Second International Workshop on “Chickpea Improvement” (Patancheru: International Crops Research Institute for the Semi-Arid Tropics), 117–127.
Rubiales, D., Avila, C. M., Sillero, J. C., Hybl, M., Narits, L., Sass, O., et al. (2012). Identification and multi-environment validation of resistance to Ascochyta fabae in fababean (Vicia faba). Field Crops Res. 126,165–170. doi: 10.1016/j.fcr.2011.10.012
Sharma, K. D., and Muehlbauer, F. J. (2007). Fusarium wilt of chickpea: physiological specialization, genetics of resistance and resistance gene tagging. Euphytica 157, 1–14. doi: 10.1007/s10681-007-9401-y
Sharma, K. D., Winter, P., Kahl, G., and Muehlbauer, F. J. (2004). Molecular mapping of Fusarium oxysporum f. sp. ciceris race 3 resistance gene in chickpea. Theor. Appl. Genet. 108, 1243–1248. doi: 10.1007/s00122-003-1561-0
Sharma, M., Babu, K. T., Gaur, P. M., Ghosh, R., Rameshwar, T., Chaudhary, R. G., et al. (2012). Identification and multi-environment validation of resistance to Fusarium oxysporum f. sp. ciceris in chickpea. Field Crops Res. 135, 82–88. doi: 10.1016/j.fcr.2012.07.004
Sharma, M., Babu, T. K., Ghosh, R., Telangre, R., Rathore, A., Kaur, L., et al. (2013). Multi-environment field-testing for identification and validation of genetic resistance to Botrytis cinerea causing Botrytis grey mold in chickpea (Cicer arietinum L.). Crop Prot. 54, 106–113. doi: 10.1016/j.cropro.2013.07.020
Sharma, M., Ghosh, R., Rameshwar, T., and Pande, S. (2015). Multi-environment field testing to identify broad, stable resistance to sterility mosaic disease of pigeonpea. J. Gen. Plant Pathol. 81, 249–259. doi: 10.1007/s10327-015-0585-z
Sharma, M., Ghosh, R., Telangre, R., Rathore, A., Saifulla, M., Mahalinga, D. M., et al. (2016a). Environmental influences on pigeonpea-Fusarium udum interactions and stability of genotypes to Fusarium wilt. Front. Plant Sci. 7:253. doi: 10.3389/fpls.2016.00253
Sharma, M., Nagavardhini, A., Thudi, M., Ghosh, R., Pande, S., Varshney, R. K., et al. (2014). Development of DArT markers and assessment of diversity in Fusarium oxysporum f. sp. ciceris, wilt pathogen of chickpea (Cicer arietinum L.). BMC Genomics 15:454. doi: 10.1186/1471-2164-15-454
Sharma, M., Sengupta, A., Ghosh, R., Agarwal, G., Tarafdar, A., Nagavardhini, A., et al. (2016b). Genome wide transcriptome profiling of Fusarium oxysporum f sp. ciceris conidial germination reveals new insights into infection related genes. Sci. Rep. 6:37353. doi: 10.1038/srep37353
Sharma, M., Tarafdar, A., Ghosh, R., and Gopalakrishanan, S. (2017). “Biological control as a tool for eco-friendly management of plant pathogens,” in Advances in Soil Microbiology: Recent Trends and Future Prospects, eds T. Adhya, B. Mishra, K. Annapurna, D. Verma, and U. Kumar (Singapore: Springer Press), 153–188. doi: 10.1007/978-981-10-7380-9_8
Sharma, R. C., and Duveiller, E. (2007). Advancement toward new spot blotch resistant wheats in South Asia. Crop Sci. 47, 961–968. doi: 10.2135/cropsci2006.03.0201
Singh, H., Kumar, J., and Haware, M. P. (1987). “Genetics of resistance to Fusarium wilt in chickpeas,” in Genetics and Plant Pathogenesis, eds P. R. Day and G. J. Jellis (Oxford: Blackwell Scientific Publications, 339–342.
Tarafdar, A., Rani, T. S., Chandran, U. S. S., Ghosh, R., Chobe, D. R., and Sharma, M. (2018). Exploring combined effect of abiotic (soil moisture) and biotic (Sclerotium rolfsii Sacc.) stress on collar rot development in chickpea. Front. Plant Sci. 9:1154. doi: 10.3389/fpls.2018.01154
Tarafdar, A., Rani, T. S., Chandran, U. S. S., Ghosh, R., and Sharma, M. (2017). “Impact of moisture stress on collar rot (Sclerotium rolfsii Sacc.) development in chickpea (Cicer arietinum L.),” in Proceedings of ISMPP International Conference on “Plant Health for Human Welfare” (Jaipur: Indian Society of Mycology and Plant Pathology), 119.
Twizeyimana, M., Ojiambo, P., Ikotun, T., Ladipo, J. L., Hartman, G. L., and Bandyopadhyay, R. (2008). Evaluation of soybean germplasm for resistance to soybean rust (Phakopsora pachyrhizi) in Nigeria. Plant Dis. 92, 947–952. doi: 10.1094/PDIS-92-6-0947
Westerlund, F. V., Campbell, R. N., and Kimble, K. A. (1974). Fungal root rot and wilt of chickpea in California. Phytopathology 64, 632–635.
Wiik, L., and Rosenqvist, H. (2010). The economics of fungicide using winter wheat in sourthern Sweden. Crop Prot. 29,11–19. doi: 10.1016/j.cropro.2009.09.008
Yan, W. (2001). GGE biplot–a windows application for graphical analysis of multi-environment trial data and other types of two way data. Agronomy 93, 1111–1118. doi: 10.2134/agronj2001.9351111x
Yan, W., Hunt, L. A., Sheng, Q. L., and Szlavnics, Z. (2000). Cultivar evaluation and mega-environment investigation based on the GGE biplot. Crop Sci. 40, 597–605. doi: 10.2135/cropsci2000.403597x
Yan, W., and Kang, M. S. (2002). GGE Biplot Analysis: A Graphical Tool for Breeders, Geneticists, and Agronomists. New York: CRC Press. doi: 10.1201/9781420040371
Keywords: chickpea, GGE biplot, Fusarium wilt, multi-year, multi-environment
Citation: Sharma M, Ghosh R, Tarafdar A, Rathore A, Chobe DR, Kumar AV, Gaur PM, Samineni S, Gupta O, Singh NP, Saxena DR, Saifulla M, Pithia MS, Ghante PH, Mahalinga DM, Upadhyay JB and Harer PN (2019) Exploring the Genetic Cipher of Chickpea (Cicer arietinum L.) Through Identification and Multi-environment Validation of Resistant Sources Against Fusarium Wilt (Fusarium oxysporum f. sp. ciceris). Front. Sustain. Food Syst. 3:78. doi: 10.3389/fsufs.2019.00078
Received: 22 May 2019; Accepted: 06 September 2019;
Published: 20 September 2019.
Edited by:
Jose C. Jimenez-Lopez, Consejo Superior de Investigaciones Científicas (CSIC) Granada, SpainReviewed by:
Teresa Millan, Universidad de Córdoba, SpainAgnieszka Barbara Najda, University of Life Sciences of Lublin, Poland
Cengiz Toker, Akdeniz University, Turkey
Copyright © 2019 Sharma, Ghosh, Tarafdar, Rathore, Chobe, Kumar, Gaur, Samineni, Gupta, Singh, Saxena, Saifulla, Pithia, Ghante, Mahalinga, Upadhyay and Harer. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Mamta Sharma, bWFtdGEuc2hhcm1hQGNnaWFyLm9yZw==
 Mamta Sharma
Mamta Sharma Raju Ghosh
Raju Ghosh Avijit Tarafdar
Avijit Tarafdar Abhishek Rathore
Abhishek Rathore Devashish R. Chobe
Devashish R. Chobe Anil V. Kumar1
Anil V. Kumar1 Pooran M. Gaur
Pooran M. Gaur Srinivasan Samineni
Srinivasan Samineni D. R. Saxena
D. R. Saxena