
94% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Sustain. Food Syst. , 21 February 2018
Sec. Agro-Food Safety
Volume 2 - 2018 | https://doi.org/10.3389/fsufs.2018.00005
Salmonella Enteritidis and Salmonella Typhimurium continue to be the most frequently identified serovars among confirmed cases of salmonellosis. In the current study, different genetic targets (safA, sdf I, STM4497, and typh) were compared, attending to their specificity and sensitivity in pure cultures and in spiked samples, in order to determine their capacity to accurately identify them by loop-mediated isothermal amplification (LAMP). For the genes selected to detect Enteritidis, both performed equally well regarding their specificity, but safA proved more sensitive than Sdf I; minor differences were observed among these genes when analyzing spiked food samples. Regarding the targets for Typhimurium, STM4497, and typh, the former demonstrated to be more specific and sensitive, both when analyzing pure cultures as well as spiked samples. These results highlight the importance of an adequate evaluation of the genetic targets selected, before their implementation for routine analyses.
Salmonella continues to be a major health issue in Europe, as well as in the rest of the world. This is supported by the figures reported by the European Food Safety Authority, who indicated that in 2015 a total of 94,625 salmonellosis cases were confirmed, representing a 1.9% increase compared to the previous year. In addition, it was also highlighted that the most prevalent serovar continues to be Salmonella Enteritidis (SE) and Salmonella Typhimurium (ST), representing 45.7 and 15.8% of all reported serovars confirmed in human cases (EFSA, 2015).
It has been extensively reported that classical microbiological methods, even though reliable, are lengthy and tedious. In this sense molecular applications have greatly allowed the reduction in the time needed to achieve the final result (Chapela et al., 2015; Law et al., 2015). One of the most popular methodologies, as evidenced by the increased number of publications in many different fields of microbiology, relies on the detection of specific genes by PCR/qPCR (Malorny et al., 2004; Park et al., 2013; Gianfranceschi et al., 2014; Garrido-Maestu et al., 2015; Maurischat et al., 2015). Novel technologies are continually being developed in order to further reduce costs and ease of implementation. Gaining increasing interest are the isothermal amplification approaches. Among them, we can find helicase-dependent amplification, nucleic acid sequence-based amplification or Recombinase Polymerase Amplification, among others (Fykse et al., 2007; Kuchta et al., 2014; Kim and Lee, 2016; Garrido-Maestu et al., 2018). One of the isothermal methods that has caught great attention in recent years is Loop-mediated isothermal AMPlification (LAMP), which was originally developed by Notomi et al. (2000). It relies on an auto-cycling strand displacement DNA synthesis performed by the Bst DNA polymerase large fragment. LAMP presents several advantages over PCR/qPCR, such as higher specificity due to the use of six primers instead of two, the reaction is performed at one single temperature, thus not needing a thermocycler; in addition, during the reaction, an insoluble product is formed (magnesium pyrophosphate) generating an increase in turbidity detectable with a naked eye, which can even be correlated with DNA concentration making the technique quantitative (Mori et al., 2001, 2004; Hara-Kudo et al., 2007). New approaches have been developed in recent years with the aim of improving naked eye detection of positive amplification, such as the use of gold nanoparticles (Arunrut et al., 2016; Kong et al., 2018). All these approaches are very attractive for the development of miniaturized devices due to their simplicity in temperature setup, and feasibility for in situ performance of the assays (Garrido-Maestu et al., 2017a).
Along with the increase in methodologies, many different genetic targets have been reported for different foodborne pathogen detection and characterization, including Salmonella (Zhang et al., 2011; Liu et al., 2012; Tang et al., 2012; Zhuang et al., 2014; Chen et al., 2015). With so many options, difficulties in selecting the most appropriate target have occurred. In the present study, two sets of genetic targets were tested for each serovar. For SE genes, safA and Sdf I, were selected. safA encodes for the major subunit of S. enterica atypical fimbriae and is involved in host-restricted colonization of the porcine ileum (Maurischat et al., 2015). Sdf I was reported to be specific for SE and is chromosomally encoded, but its function is unknown (Agron et al., 2001; Regan et al., 2008). Similarly, two gene targets specific for ST are represented by STM4497 and typh. STM4497 encodes for a putative cytoplasmic protein (Kim et al., 2006). The gene target typh, was described previously to be Typhimurium specific, but no specific biological function has been assigned (Olsen et al., 1995; Alvarez et al., 2004). The selection of these targets was based on the fact that LAMP assays have been previously published for most of them, and the inclusivity/exclusivity tests reported several target and non-target strains from the genus Salmonella and nearest phylogenetically related species. The aim of the present study was to evaluate these gene targets for the specific identification of SE and ST in foods, based on LAMP.
S1400 and CECT 4594 were considered the reference strains for SE and ST, respectively. These strains were selected for the sensitivity assays, as well as for all spiking experiments. In addition to these, a total of 34 strains were included in the evaluation of the specificity. Information regarding the strains tested is provided in Table 1.
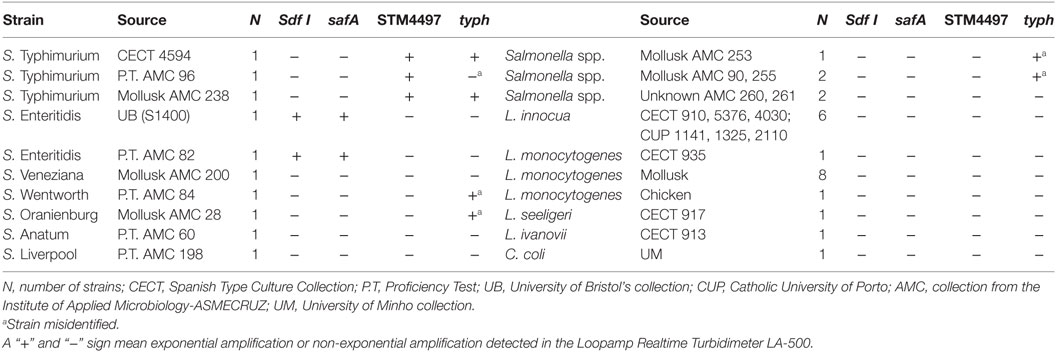
Table 1. Strain list selected for the evaluation of the specificity of the Loop-mediated isothermal AMPlification (LAMP) assays.
All pure bacterial cultures were performed by inoculating 1 single colony into 4 mL of buffered peptone water or tryptic soy broth (BPW, TSB, Biokar diagnostics S.A., France) and incubated at 37°C overnight. Whenever the cultures were used for food sample inoculation, 10-fold serial dilutions were performed in BPW, and then plated on TSB +15 g/L of agar. The plates were incubated as detailed previously. Finally, for food sample analysis, the enrichment broth selected was mTA10, as described by Garrido et al. (2013), and the incubation was performed at 37°C for 18–24 h.
One milliliter of an overnight enrichment culture was centrifuged at 4,000 × g for 5 min. The supernatant was discarded, and the pellet resuspended in 1 mL of Tris-EDTA 1× buffer (TE 1×, 10 mM Tris–HCl, 1 mM EDTA, Sigma–Aldrich, St. Louis, MS, USA) and centrifuged using the same conditions as described above. The pellet was resuspended in 300 µL of TE 1× and heated at 99°C for 15 min with constant agitation (1,000 rpm) in a shaker Thermomixer comfort heating block (Eppendorf AG, Germany). Finally, the bacterial cultures were centrifuged as detailed above, and the supernatant, containing bacterial DNA, was transferred to a clean tube and stored at −20°C. DNA concentration was measured with a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). DNA extracted from pure bacterial cultures of each reference strain was 10-fold serially diluted in TE 1×, this was performed for the determination of the lowest detectable concentration.
In order to avoid PCR inhibitors which may be associated with the food matrices, 1 mL of all food samples was centrifuged at 380 × g for 2 min to remove food debris, the supernatant was collected and centrifuged at 16,000 × g for 5 min. The pellet was resuspended in 1 mL of PBS and centrifuged again using the same conditions as previously described. After eliminating the PBS, the pellet was resuspended in 300 µL of 6% Chelex®100 (Bio-Rad Laboratories, Inc., USA), the samples were incubated at 56°C for 15 min with constant agitation (1,000 rpm). This step was followed by a thermal lysis at 99°C for 10 min. Finally, the samples were centrifuged at 16,000 × g for 5 min, and the supernatant was collected, and stored as described for pure cultures.
Two sets of genetic targets were tested for each serovar, safA and Sdf I for SE; and STM4497 and typh for ST. The evaluation of safA and STM4497 was based on the primers designed by Garrido-Maestu et al. (2017b) with the addition of newly designed loop primers, while for Sdf I and typh those designed by Yang et al. (2010) and Pavan Kumar et al. (2014), respectively, were chosen. The selection of the primers for safA and STM4497, was based on the fact that they exhibited excellent performance (accurate, sensitive, and specific detection) in food samples including chicken and turkey (Garrido-Maestu et al., 2017b). On the other hand, regarding those targeting Sdf I and typh, it was based on the fact that their specificity was tested against a reasonable panel of strains (5 SE and 8 non-SE, including 1 ST with Sdf I; and 28 ST, 22 non-ST and 6 non-Salmonella for typh). Further details regarding the sequences of all primers, are provided in Table 2.
All LAMP experiments were performed in a Loopamp Realtime Turbidimeter LA-500 (Eiken Chemical Co., Ltd., Japan). The reactions were prepared in a final volume of 25 µL, with 1 M Betaine (Sigma–Aldrich, St. Louis, MS, USA), 12.5 µL of Isothermal Master Mix (OptiGene Ltd., Horsham, UK) and the corresponding amount of primers (see Table 2), and 3 µL of template DNA. The amplification was accomplished at 65°C for 1 h. Even though the amplification was performed in 1 h, only those food samples reporting positive within the first 30 min were considered as such for safA, Sdf I, and STM4497, while for typh, due to the lack of loop primers, up to 40 min were considered acceptable.
A total of 87 food samples were spiked with different concentrations of both serovars, SE and ST. The types of foods selected covered those of high risk (raw chicken, turkey, raw, and cooked egg products). Twenty-five grams of each food sample were weighed and 225 mL of mTA10 broth were added and mixed for 30 s, then 1 mL of the appropriate bacterial dilution was added and incubated as detailed in the Section “Materials and Methods” “Bacterial Strains And Culture Media.” After enrichment, DNA was extracted as described in the Section “Materials and Methods” “Food Samples.”
The results obtained after the inoculation, and analysis of the different food samples were classified as Positive or Negative agreement (PA and NA) if matched those expected, while as Positive or Negative Deviations (PD and ND) if the results did not match. With these values, the following parameters were calculated: relative sensitivity, specificity and accuracy (SE/SP/AC), positive and negative predictive values (PPV/NPV), index kappa of concordance (κ), and the acceptability limit (AL); as described elsewhere (Malorny et al., 2003; Tomas et al., 2009; Anderson et al., 2011; Garrido et al., 2013; D’Agostino et al., 2016).
The evaluation of the specificity against a panel of 34 strains gave the expected results with both genes (safA and Sdf I) for SE. Both target strains were positive, while the 32 resulted negative. However, late amplification occurred with safA and we noticed that the threshold time (Tt) values higher than 35 min (positive strains reported Tt values below 30 min), occurred with certain strains, while this deviation in Tt values were not observed for Sdf I.
Regarding the identification of ST, all three target strains were detected with STM4497, without any interference due to non-target species. However, even the selection of typh gene, resulted in correct identification of all non-Salmonella strains, three Salmonella strains were misidentified as ST, AMC 28 (S. Oranienburg), 84 (S. Wentworth), and 253 (Salmonella non-Enteritidis/non-Typhimurium); while isolate AMC 96, previously confirmed as ST by qPCR with F3/B3 primers (Garrido-Maestu et al., 2017b) was not detected.
Regarding the LoD (lowest detectable concentration), for SE with safA it was possible to detect 0.00144 ng/µL over 5 consecutive 10-fold dilutions, while with Sdf I could only cover 3 dilutions reaching 0.144 ng/µL, as depicted in Figure 1A. Regarding ST, the lowest LoD was achieved with STM4497 reaching 0.00438 ng/µL, likewise safA over five consecutive dilutions; while with typh a higher value was detected as could only detect 4.38 ng/µL, see Figure 1B.
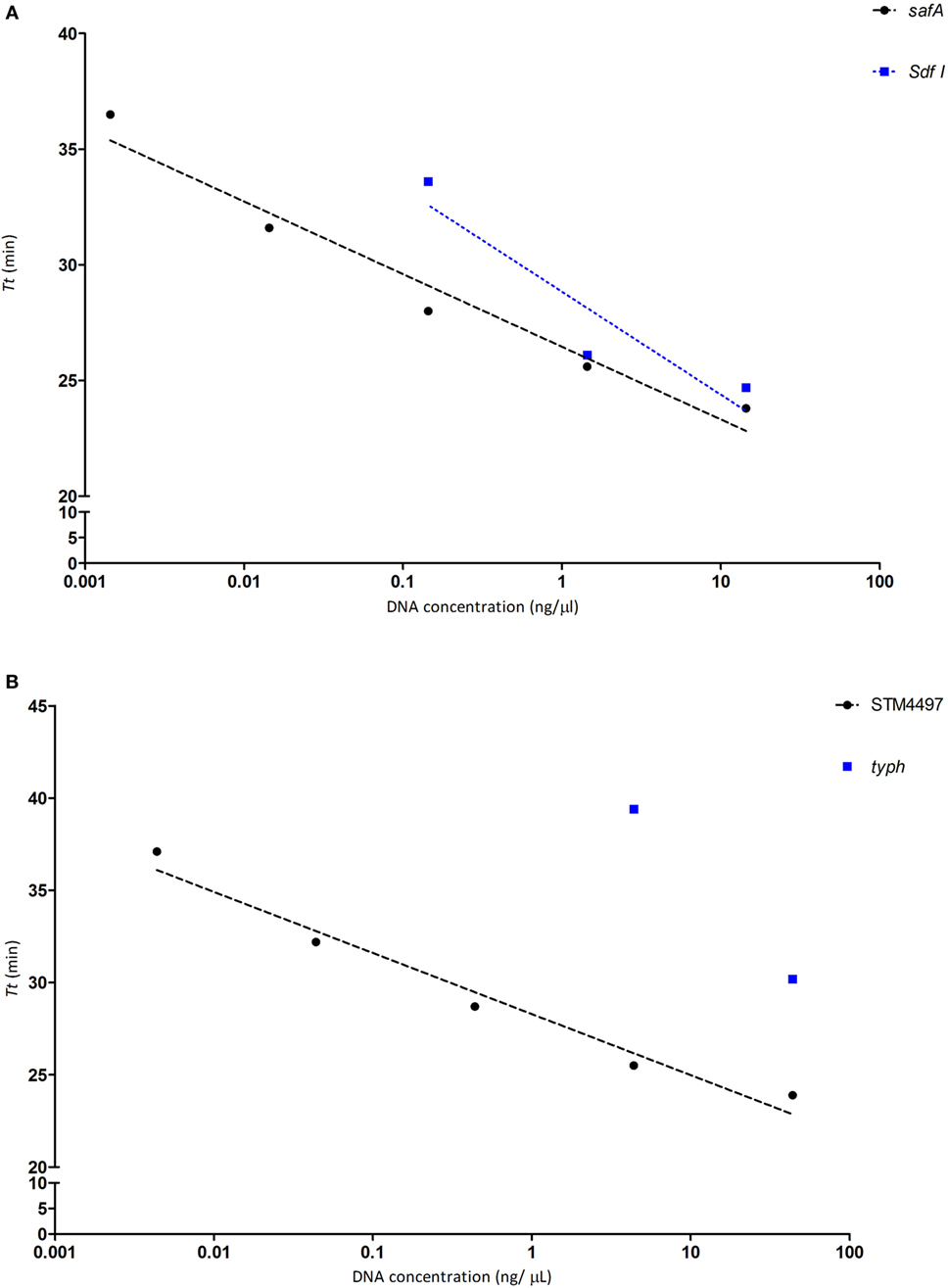
Figure 1. Determination of the limit of detection for each genetic target, with pure bacterial DNA for each target. The threshold time (Tt), obtained from 10-fold serial dilutions, plotted against the DNA concentration of strain S1400 for Salmonella Enteritidis (SE) and CECT 4594 for Salmonella Typhimurium (ST). (A) SE genes safA (black circles) and Sdf I (blue boxes). (B) ST genes STM4497 (black circles) and typh (blue boxes).
Different types of foods, i.e., chicken, turkey, eggs, and egg products, were spiked at different concentrations, and with different combination levels of each SE/ST strain. A total of 87 samples were analyzed. Both SE targets gave similar positive results where all but two samples were correctly identified. However, with the ST STM4497 gene target, again only two deviations were detected, while with the typh gene target, 16 of the 87 samples were misidentified. These results are summarized in Table 3.
Based on the results obtained from the spiked food samples, it was determined that both genetic targets for SE performed well, with minor differences among them. This was not the case for ST, as major differences were observed when targeting STM4497 and typh. The values obtained by the later were the worst of all the genes evaluated. It is worth highlighting that the k value obtained was of 0.62, which can be interpreted as “substantial agreement,” while for the rest, values higher than 0.9 were obtained. This is interpreted as “almost complete concordance.” The results are summarized in Table 4.
The increased acceptance of molecular methods for the detection of different bacterial pathogens in foods has led to the appearance of a great number of approaches for this purpose. In the current study, a set of four genes, two for SE and two for ST, were systematically compared in order to evaluate their adequacy to detect these, particularly problematic Salmonella serovars, in food samples.
Overall, it was observed that both genes evaluated targeting SE (safA and Sdf I) performed well, with minor differences. In the specificity test, all bacterial strains were correctly identified with both genes. Regarding the LoD with DNA extracted from pure bacterial cultures, safA was 100 times more sensitive than Sdf I (0.00144 ng/µL, compared to 0.144 ng/µL). The results obtained after the analysis of spiked samples were similar, being only detected one PD with safA and one ND associated with a sample co-inoculated with ST but with a concentration 107 times higher. This sample was also misidentified with Sdf I. With this second gene, no PD were detected but a second ND was obtained. This was associated with a sample with <10 cfu/25 g of SE. Overall, in spiked samples, both genes obtained values higher than 95% in all the parameters evaluated and the Acceptability Limit (AL) below 3 and 6, as recommended (D’Agostino et al., 2016).
Greater differences were observed in the evaluation of the genetic targets selected for ST. While STM4497 obtained very good results, typh proved more difficult. Under the conditions tested the lowest DNA concentration that was detectable with STM4497 was 0.00438 ng/µL, while with the typh target only two consecutive dilutions were positive and reached 4.38 ng/µL. In addition, while all pure culture strains were correctly identified with STM4497, a total of four strains were misidentified targeting typh gene (one ST was not detected, and three non-ST obtained positive results). It is worth considering that the specificity problems were only associated with Salmonella strains, as all the 19 non-Salmonella isolates were correctly identified as negative.
The differences in performance detected when testing pure cultures matched what was observed after sample inoculation. For STM4497, only two ND were obtained (these were associated with two samples co-inoculated with SE, thus it seems that this second strain grew more than ST). Regarding typh, a total of 16 deviations were detected. Out of these, six ND were associated with inoculation levels of <10 cfu/25 g, 1 ND was a sample co-inoculated with 107 times more SE, and one with an inoculum in the range of 102–103 cfu/25 g. As commented above, these results correlate with those obtained with DNA isolated from pure bacterial cultures, where STM4497 proved more sensitive than typh. Finally, eight PD were detected, five associated with non-spiked samples, and three more with samples inoculated with SE. All these erroneous results obtained in food samples, corroborated the specificity data obtained with pure bacterial cultures, and ended up in unacceptable AL values (0–16).
When the results obtained were compared with those previously published for each target, some differences were observed. For safA and STM4497 it was possible to reach a lower LoD, with DNA from pure cultures, than that reported by Garrido-Maestu et al. (2017b); but minor differences were observed with spiked samples. These discrepancies may be associated with small changes in the method, such as the inclusion of loop primers, which were not used in the original study.
The specificity results obtained with Sdf I matched those previously published by Yang et al., but the LoD was higher in the current study (Yang et al., 2010). Greater differences were observed for typh, with respect to the study of Pavan Kumar et al., who reported excellent specificity (Pavan Kumar et al., 2014). In our experiments, their primers were not able to correctly discriminate all the strains tested. This is in agreement with the fact that BLAST testing of these primers reported same results for ST as for other serovars. Regarding the LoD, once more, in the present study, the results were worse than those reported in the original paper, as we could only detect 4.3 ng/µL, while it was indicated that 0.002 ng/µL could be reached. As mentioned previously, the discrepancies found among this and the original studies may be related with slight differences in the methodology followed, i.e., small differences in the amplification temperature, end-point results with respect to real-time turbidity tracking, application of gel electrophoresis, among others.
The reasons behind the overall differences in performance obtained by these four genetic targets may be of diverse origin, from the quality of the selected sequences, the primer design process, to specific assay optimization. Thus, caution must be taken when attempting to directly implement previously published studies in routine laboratory testing.
The comparison of four genetic targets for the specific detection of SE and ST, in food samples, reported minor differences among all them, except for typh gene. In this sense either safA or Sdf I can be implemented for the detection of SE (the assays were equally specific, safA was 100 times more sensitive than sdf I, but after enrichment, both obtained optimal results in food samples). It is advised to select STM4497 over typh (more deviations were obtained during the evaluation of the specificity with typh, in addition to being 1,000 times less sensitive, and with the lowest quality parameters after evaluation in food samples).
AG-M designed the experiments and wrote the manuscript. SA and JC performed the experiments and helped writing the manuscript. MP helped in the design of the experiments and proofread the manuscript.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
This work was supported by a Marie Curie COFUND Action (Project No: 600375. NanoTRAINforGrowth - INL Fellowship programme in nanotechnologies for biomedical, environment and food applications), and by the project Nanotechnology Based Functional Solutions (NORTE-01-0145-FEDER-000019), supported by Norte Portugal Regional Operational Programme (NORTE2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF). Authors would like to thank Dr. Antonio Lozano-León, director of the Institute of Applied Microbiology-ASMECRUZ, the Center of Biological Engineering of the University of Minho, and Biomode® for kindly providing some of the bacterial strains used in the current study; and the Microbiology and Bioassays Laboratory of ANFACO-CECOPESCA for technical support.
Agron, P. G., Walker, R. L., Kinde, H., Sawyer, S. J., Hayes, D. C., Wollard, J., et al. (2001). Identification by subtractive hybridization of sequences specific for Salmonella enterica serovar enteritidis. Appl. Environ. Microbiol. 67, 4984–4991. doi: 10.1128/AEM.67.11.4984
Alvarez, J., Sota, M., Vivanco, A. B., Perales, I., Cisterna, R., Rementeria, A., et al. (2004). Development of a multiplex PCR technique for detection and epidemiological typing of Salmonella in human clinical samples. J. Clin. Microbiol. 42, 1734–1738. doi:10.1128/JCM.42.4.1734
Anderson, A., Pietsch, K., Zucker, R., Mayr, A., Muller-Hohe, E., Messelhausser, U., et al. (2011). Validation of a duplex real-time PCR for the detection of Salmonella spp. in different food products. Food Anal. Methods 4, 259–267. doi:10.1007/s12161-010-9142-8
Arunrut, N., Kampeera, J., Sirithammajak, S., Sanguanrut, P., Proespraiwong, P., Suebsing, R., et al. (2016). Sensitive visual detection of AHPND bacteria using loop-mediated isothermal amplification combined with DNA-functionalized gold nanoparticles as probes. PLoS ONE 11:e0151769. doi:10.1371/journal.pone.0151769
Chapela, M., Garrido-Maestu, A., and Cabado, A. G. (2015). Detection of foodborne pathogens by qPCR: a practical approach for food industry applications. Cogent Food Agric. 1, 1–19. doi:10.1080/23311932.2015.1013771
Chen, Z., Zhang, K., Yin, H., Li, Q., Wang, L., and Liu, Z. (2015). Detection of Salmonella and several common Salmonella serotypes in food by loop-mediated isothermal amplification method. Food Sci. Hum. Wellness 4, 75–79. doi:10.1016/j.fshw.2015.05.001
D’Agostino, M., Robles, S., Hansen, F., Ntafis, V., Ikonomopoulos, J., Kokkinos, P., et al. (2016). Validation of a loop-mediated amplification/ISO 6579-based method for analysing soya meal for the presence of Salmonella enterica. Food Anal. Methods 9, 2979–2985. doi:10.1007/s12161-016-0602-7
EFSA, E. (2015). The European union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2014. EFSA J. 13, 4329. doi:10.2903/j.efsa.2015.4329
Fykse, E. M., Skogan, G., Davies, W., Olsen, J. S., and Blatny, J. M. (2007). Detection of Vibrio cholerae by real-time nucleic acid sequence-based amplification. Appl. Environ. Microbiol. 73, 1457–1466. doi:10.1128/AEM.01635-06
Garrido, A., Chapela, M. J., Román, B., Fajardo, P., Lago, J., Vieites, J. M., et al. (2013). A new multiplex real-time PCR developed method for Salmonella spp. and Listeria monocytogenes detection in food and environmental samples. Food Control 30, 76–85. doi:10.1016/j.foodcont.2012.06.029
Garrido-Maestu, A., Azinheiro, S., Carvalho, J., Abalde-Cela, S., Carbó-Argibay, E., Diéguez, L., et al. (2017a). Combination of microfluidic loop-mediated isothermal amplification with gold nanoparticles for rapid detection of Salmonella spp. in food samples. Front. Microbiol. 8:2159. doi:10.3389/fmicb.2017.02159
Garrido-Maestu, A., Fuciños, P., Azinheiro, S., Carvalho, J., and Prado, M. (2017b). Systematic loop-mediated isothermal amplification assays for rapid detection and characterization of Salmonella spp., Enteritidis and Typhimurium in food samples. Food Control 80, 297–306. doi:10.1016/j.foodcont.2017.05.011
Garrido-Maestu, A., Azinheiro, S., Carvalho, J., Fuciños, P., and Prado, M. (2018). Development and evaluation of loop-mediated isothermal amplification, and recombinase polymerase amplification methodologies, for the detection of Listeria monocytogenes in ready-to-eat food samples. Food Control. 86:27–34. doi:10.1016/j.foodcont.2017.11.006
Garrido-Maestu, A., Chapela, M.-J., Peñaranda, E., and Cabado, A. G. (2015). Re-evaluation of enhanced qPCR prevalidated method for next-day detection of Salmonella spp., Shigella spp., Escherichia coli O157 and Listeria monocytogenes. Food Biotechnol. 29, 317–335. doi:10.1080/08905436.2015.1091977
Gianfranceschi, M. V., Rodriguez-Lazaro, D., Hernandez, M., Gonzalez-Garcia, P., Comin, D., Gattuso, A., et al. (2014). European validation of a real-time PCR-based method for detection of Listeria monocytogenes in soft cheese. Int. J. Food Microbiol. 184, 128–133. doi:10.1016/j.ijfoodmicro.2013.12.021
Hara-Kudo, Y., Nemoto, J., Ohtsuka, K., Segawa, Y., Takatori, K., Kojima, T., et al. (2007). Sensitive and rapid detection of vero toxin-producing Escherichia coli using loop-mediated isothermal amplification. J. Med. Microbiol. 56, 398–406. doi:10.1099/jmm.0.46819-0
Kim, H. J., Park, S. H., Lee, T. H., Nahm, B. H., Chung, Y. H., Seo, K. H., et al. (2006). Identification of Salmonella enterica serovar Typhimurium using specific PCR primers obtained by comparative genomics in salmonella serovars. J. Food Prot. 69, 1653–1661. doi:10.4315/0362-028X-69.7.1653
Kim, J. Y., and Lee, J. L. (2016). Rapid Detection of Salmonella enterica serovar enteritidis from eggs and chicken meat by real-time recombinase polymerase amplification in comparison with the two-step real-time PCR. J. Food Saf. 36, 402–411. doi:10.1111/jfs.12261
Kong, C., Wang, Y., Fodjo, E. K., Yang, G., Han, F., and Shen, X. (2018). Loop-mediated isothermal amplification for visual detection of Vibrio parahaemolyticus using gold nanoparticles. Microchimica Acta 185, 35. doi:10.1007/s00604-017-2594-4
Kuchta, T., Knutsson, R., Fiore, A., Kudirkiene, E., Höhl, A., Horvatek Tomic, D., et al. (2014). A decade with nucleic acid-based microbiological methods in safety control of foods. Lett. Appl. Microbiol. 59, 263–271. doi:10.1111/lam.12283
Law, J. W.-F., Ab Mutalib, N.-S., Chan, K.-G., and Lee, L.-H. (2015). Rapid methods for the detection of foodborne bacterial pathogens: principles, applications, advantages and limitations. Front. Microbiol. 5:770. doi:10.3389/fmicb.2014.00770
Liu, B., Zhou, X., Zhang, L., Liu, W., Dan, X., Shi, C., et al. (2012). Development of a novel multiplex PCR assay for the identification of Salmonella enterica Typhimurium and enteritidis. Food Control 27, 87–93. doi:10.1016/j.foodcont.2012.01.062
Malorny, B., Hoorfar, J., Bunge, C., and Helmuth, R. (2003). Multicenter validation of the analytical accuracy of Salmonella PCR: towards an international standard. Appl. Environ. Microbiol. 69, 290–296. doi:10.1128/AEM.69.1.290-296.2003
Malorny, B., Paccassoni, E., Fach, P., Bunge, C., Martin, A., and Helmuth, R. (2004). Diagnostic real-time PCR for detection of Salmonella in food. Appl. Environ. Microbiol. 70, 7046–7052. doi:10.1128/AEM.70.12.7046-7052.2004
Maurischat, S., Baumann, B., Martin, A., and Malorny, B. (2015). Rapid detection and specific differentiation of Salmonella enterica subsp. enterica Enteritidis, Typhimurium and its monophasic variant 4,[5],12:i:- by real-time multiplex PCR. Int. J. Food Microbiol. 193, 8–14. doi:10.1016/j.ijfoodmicro.2014.10.004
Mori, Y., Kitao, M., Tomita, N., and Notomi, T. (2004). Real-time turbidimetry of LAMP reaction for quantifying template DNA. J. Biochem. Biophys. Methods 59, 145–157. doi:10.1016/j.jbbm.2003.12.005
Mori, Y., Nagamine, K., Tomita, N., and Notomi, T. (2001). Detection of loop-mediated isothermal amplification reaction by turbidity derived from magnesium pyrophosphate formation. Biochem. Biophys. Res. Commun. 289, 150–154. doi:10.1006/bbrc.2001.5921
Notomi, T., Okayama, H., Masubuchi, H., Yonekawa, T., Watanabe, K., Amino, N., et al. (2000). Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 28, e63. doi:10.1093/nar/28.12.e63
Olsen, J. E., Aabo, S., Rasmussen, O. F., and Rossen, L. (1995). Oligonucleotide probes specific for the genus Salmonella and for Salm. Typhimurium. Lett. Appl. Microbiol. 20, 160–163. doi:10.1111/j.1472-765X.1995.tb00416.x
Park, M.-K., Weerakoon, K. A., Oh, J.-H., and Chin, B. A. (2013). The analytical comparison of phage-based magnetoelastic biosensor with TaqMan-based quantitative PCR method to detect Salmonella Typhimurium on cantaloupes. Food Control 33, 330–336. doi:10.1016/j.foodcont.2013.02.026
Pavan Kumar, P., Agarwal, R. K., Thomas, P., Sailo, B., Prasannavadhana, A., Kumar, A., et al. (2014). Rapid detection of Salmonella enterica subspecies enterica serovar Typhimurium by loop mediated isothermal amplification (LAMP) test from field chicken meat samples. Food Biotechnol. 28, 50–62. doi:10.1080/08905436.2013.870911
Regan, E. O., Mccabe, E., Burgess, C., Mcguinness, S., Barry, T., Duffy, G., et al. (2008). Development of a real-time multiplex PCR assay for the detection of multiple Salmonella serotypes in chicken samples. BMC Microbiol. 8:156. doi:10.1186/1471-2180-8-156
Tang, T., Cheng, A., Wang, M., Li, X., He, Q., Jia, R., et al. (2012). Development and clinical verification of a loop-mediated isothermal amplification method for detection of Salmonella species in suspect infected ducks. Poult. Sci. 91, 979–986. doi:10.3382/ps.2011-01992
Tomas, D., Rodrigo, A., Hernandez, M., and Ferrus, M. A. (2009). Validation of real-time PCR and enzyme-linked fluorescent assay-based methods for detection of Salmonella spp. in chicken feces samples. Food Anal. Methods 2, 180–189. doi:10.1007/s12161-009-9082-3
Yang, J.-L., Ma, G.-P., Yang, R., Yang, S.-Q., Fu, L.-Z., Cheng, A.-C., et al. (2010). Simple and rapid detection of Salmonella serovar Enteritidis under field conditions by loop-mediated isothermal amplification. J. Appl. Microbiol. 109, 1715–1723. doi:10.1111/j.1365-2672.2010.04800.x
Zhang, G., Brown, E. W., and González-Escalona, N. (2011). Comparison of real-time PCR, reverse transcriptase real-time PCR, loop-mediated isothermal amplification, and the FDA conventional microbiological method for the detection of Salmonella spp. in produce. Appl. Environ. Microbiol. 77, 6495–6501. doi:10.1128/AEM.00520-11
Keywords: Salmonella Enteritidis, Salmonella Typhimurium, LAMP, characterization, safA, Sdf I, typh, STM4497
Citation: Azinheiro S, Carvalho J, Prado M and Garrido-Maestu A (2018) Evaluation of Different Genetic Targets for Salmonella enterica Serovar Enteriditis and Typhimurium, Using Loop-Mediated Isothermal AMPlification for Detection in Food Samples. Front. Sustain. Food Syst. 2:5. doi: 10.3389/fsufs.2018.00005
Received: 14 December 2017; Accepted: 05 February 2018;
Published: 21 February 2018
Edited by:
Joshua B. Gurtler, Agricultural Research Service (USDA), United StatesReviewed by:
Tam L. Mai, IEH laboratories and Consultant Group, United StatesCopyright: © 2018 Azinheiro, Carvalho, Prado and Garrido-Maestu. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Marta Prado, bWFydGEucHJhZG9AaW5sLmludA==;
Alejandro Garrido-Maestu, YWxlamFuZHJvLmdhcnJpZG9AaW5sLmludA==
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.