Metagenomic probing toward an atlas of the taxonomic and metabolic foundations of the global ocean genome
Explainer
Front Sci, 16 January 2024
This explainer is part of an article hub, related to lead article https://doi.org/10.3389/fsci.2023.1038696
Creating a comprehensive catalog of ocean life
The world’s oceans brim with microscopic life—most of which is unknown to science. These tiny organisms play a critical role in marine health and the Earth’s climate. Moreover, their genes are a major resource for science and society, with potential applications in biotechnology, energy, medicine, and food. Finding out which microbes live where, and what they do, is extremely challenging. But Laiolo et al. present a breakthrough: the “KMAP Global Ocean Gene Catalog 1.0”. Built using powerful new technologies, this open-source database is the most extensive analysis of ocean microbes to date. It lays the foundations for compiling a global ocean genome—a long-standing goal of microbial oceanography.
This explainer describes how the catalog was developed, its main findings, and potential applications.
What is the global ocean genome?
The global ocean genome is the complete set of genes from every marine organism, and the information these genes encode. It comprises genes across the tree of life—from bacteria and archaea to fungi, plants, and animals—which code for everything these organisms need to survive, reproduce, and interact with their environment. As such, the global ocean genome is the foundation of marine biodiversity, marine ecosystems, and all biogeochemical processes taking place in the ocean.
What is the KMAP Global Ocean Gene Catalog 1.0?
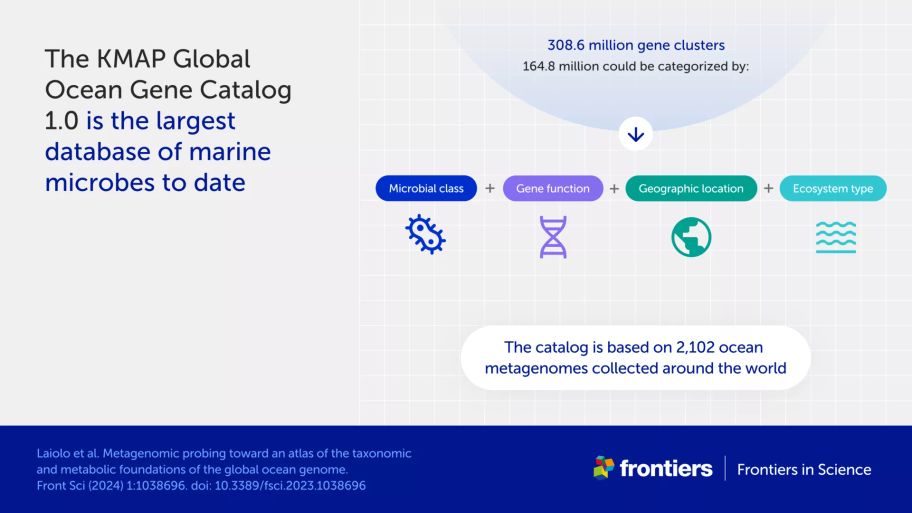
The KMAP Global Ocean Gene Catalog 1.0 is the largest, open-source database of gene sequences from marine organisms to date—and so an important step toward a global ocean genome. Focusing on marine microbes, it consists of 308.6 million gene groups from over 2,100 ocean samples, of which just over half could be categorized according to:
taxonomy: which organism type, or even species, the genes belong to
function: what the protein encoded by each gene does
geographic location: where the sample containing the gene was collected
zone/ecosystem: what depth the sample was collected from, and whether it was a water or ocean floor sample.
How was the KMAP Global Ocean Gene Catalog 1.0 created?
Building a catalog of genetic material present in ocean samples is a complex process requiring powerful technologies that have only recently become available.
The first step is to sequence all the DNA in each sample. This was once a lengthy and expensive process, but a sophisticated technique called shotgun sequencing can do the task quickly and efficiently. The method compiles a “metagenome”: the pooled set of genetic information from every organism within a sample. The authors obtained 2,102 such ocean metagenomes from the European Nucleotide Archive, a repository of genetic data.
The next step is to identify individual genes within the millions of DNA sequences present in the metagenomes. The authors did this using the recently developed “King Abdullah University of Science and Technology (KAUST) Metagenomic Analysis Platform” (KMAP)—whose sophisticated algorithms and massive computational power identified 308.6 million gene groups present in the ocean metagenomes.
The final step is to pinpoint what the genes do and which organism they came from. The platform did this by comparing the identified gene sequences, and the predicted proteins they encode, with existing databases. While many were new to science, 52% matched known sequences and could be categorized as described above.
What geographic locations and ocean zones are represented in the catalog?
The majority of samples used to create the KMAP Global Ocean Gene Catalog 1.0 come from the Pacific (40.6%) and Atlantic (28.0%) oceans. The rest were collected from the Indian Ocean (12.9%), the Mediterranean Sea (11.6%), the Arctic Ocean (5.4%), and the Southern Ocean (1.5%).
Most (78.5%) samples are from the upper ocean zone (depth of up to 200 meters). Of the remaining, 7.2% are from the mesopelagic zone (depths of 200-1,000 meters), 10.2% are from the dark ocean (below 1,000 meters), and about 4.1% are sediments from the benthic zone (ocean floor).
What are the applications of the catalog?
The KMAP Ocean Gene Catalog 1.0 has important applications in various fields of research and industry. These include:
biotechnology and pharmaceutical innovation: researchers can search the catalog for genes encoding compounds or proteins that could be useful for drug development, agriculture, and other industries
understanding ocean biodiversity: the analysis extends our understanding of where different microbes are found and their role in biogeochemical processes that shape marine and planetary health—including processes relevant to climate change
tracking the impact of human activities on marine life: the catalog serves as a baseline for monitoring the effect of global warming, pollution, and other human-induced changes to marine environments
guiding future research: scientists can use the catalog to form research questions about specific habitats, microbial classes, and other areas of marine biology.

What are the early findings of the KMAP Ocean Gene Catalog 1.0?
The catalog highlights the diversity of marine life and the role of different microbes in different marine habitats. Key findings include:
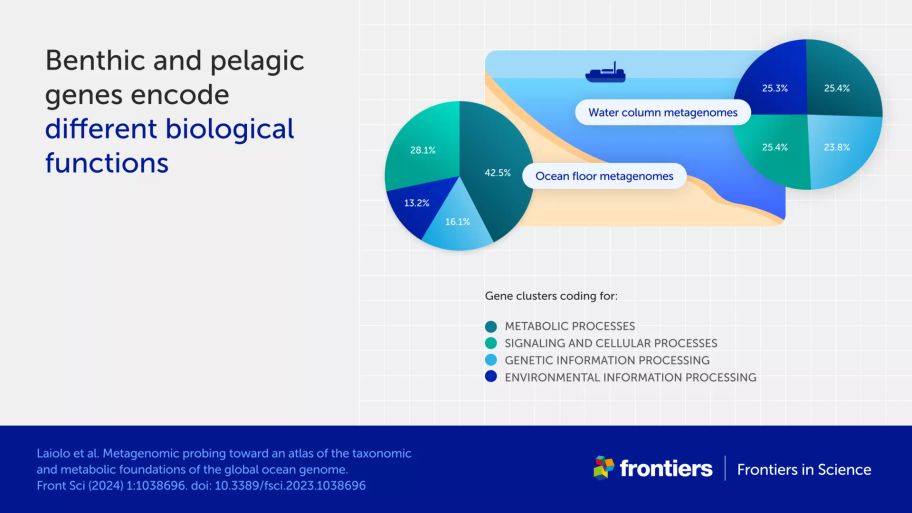
microbial activity on the ocean floor is different from that in the water column, including differing and highly specialized pathways for metabolizing the main climate-active molecules (carbon, nitrogen, and sulfur)
bacteria are the largest contributors to unique genes in the ocean
viral genomes contain far more innovation than previously realized
fungi account for over 50% of gene groups identified in the mesopelagic zone, implying these microbes play a more important role in ocean processes than previously considered.
What are the next steps for building a global ocean genome?
The authors identify several areas of research for fully exploring the global ocean genome. One priority is to increase the number of deep sea and ocean floor samples. In particular, the ocean floor is a very diverse environment and so is likely to contain many undiscovered genes and functions. Another priority is to extend the analysis to include RNA viruses. More comprehensive sample data—like location details, depth, temperature, salinity, and nutrient concentration—could also provide valuable context for future analyses.
The authors additionally mention several technological challenges to overcome. These include increased computational power to analyze metagenomes as new genes are added to existing repositories, and improvements to gene prediction frameworks. This could help categorize the 48% of gene clusters that the authors could not characterize, as well as the metagenome sequences where no genes were identified.
Even once complete, the global ocean genome will need be updated regularly as the ocean is always changing. This means continuous collaborative efforts are needed on a global scale to fully understand, harness, and monitor the complexity of ocean biodiversity across all marine environments.
The authors stress that these efforts must be supported by the development of open access platforms—as well as an intellectual property framework that simultaneously ensures adequate investment in future expeditions and the just and equitable sharing of benefits derived from genes identified in international waters.