
95% of researchers rate our articles as excellent or good
Learn more about the work of our research integrity team to safeguard the quality of each article we publish.
Find out more
ORIGINAL RESEARCH article
Front. Robot. AI , 25 February 2025
Sec. Field Robotics
Volume 12 - 2025 | https://doi.org/10.3389/frobt.2025.1548143
This article is part of the Research Topic Revolutionizing Agriculture: The Role of Robotics and AI in Smart Farming View all articles
Robust perception systems allow farm robots to recognize weeds and vegetation, enabling the selective application of fertilizers and herbicides to mitigate the environmental impact of traditional agricultural practices. Today’s perception systems typically rely on deep learning to interpret sensor data for tasks such as distinguishing soil, crops, and weeds. These approaches usually require substantial amounts of manually labeled training data, which is often time-consuming and requires domain expertise. This paper aims to reduce this limitation and propose an automated labeling pipeline for crop-weed semantic image segmentation in managed agricultural fields. It allows the training of deep learning models without or with only limited manual labeling of images. Our system uses RGB images recorded with unmanned aerial or ground robots operating in the field to produce semantic labels exploiting the field row structure for spatially consistent labeling. We use the rows previously detected to identify multiple crop rows, reducing labeling errors and improving consistency. We further reduce labeling errors by assigning an “unknown” class to challenging-to-segment vegetation. We use evidential deep learning because it provides predictions uncertainty estimates that we use to refine and improve our predictions. In this way, the evidential deep learning assigns high uncertainty to the weed class, as it is often less represented in the training data, allowing us to use the uncertainty to correct the semantic predictions. Experimental results suggest that our approach outperforms general-purpose labeling methods applied to crop fields by a large margin and domain-specific approaches on multiple fields and crop species. Using our generated labels to train deep learning models boosts our prediction performance on previously unseen fields with respect to unseen crop species, growth stages, or different lighting conditions. We obtain an IoU of 88.6% on crops, and 22.7% on weeds for a managed field of sugarbeets, where fully supervised methods have 83.4% on crops and 33.5% on weeds and other unsupervised domain-specific methods get 54.6% on crops and 11.2% on weeds. Finally, our method allows fine-tuning models trained in a fully supervised fashion to improve their performance in unseen field conditions up to +17.6% in mean IoU without additional manual labeling.
The demand for food is constantly increasing due to the growing world population, requiring new methods to optimize crop production (Horrigan et al., 2002; Ewert et al., 2023; Storm et al., 2024; Walter et al., 2017). The use of robotic systems in agriculture has the promise to make processes, such as monitoring fields (Ahmadi et al., 2020; Boatswain Jacques et al., 2021), phenotyping (Weyler et al., 2022b), and weed spraying (Wu et al., 2020), more efficient and sustainable (Cheng et al., 2023). Commonly, robotic platforms perceive their environment using deep learning methods to semantically interpret complex data collected with onboard sensors (Dainelli et al., 2024). However, these approaches usually require large amounts of human-labeled data to achieve satisfactory performance for real-world deployment and often fall short in unseen field conditions (Wang et al., 2022; Magistri et al., 2023).
In this paper, we examine the problem of automated semantic crop-weed segmentation in RGB images, enabling robots to perform tasks, such as automated weeding (Balabantaray et al., 2024; Saqib et al., 2023), controlled usage of pesticide (Murugan et al., 2020), harvesting (Pan et al., 2023), or phenotyping (Weyler et al., 2022a). We aim to maximize a semantic segmentation neural network’s performance in various field deployment conditions, e.g., different growth stages, crop species, or lighting conditions, without human-labeled training data. This is crucial to ensure a robust crop-weed segmentation in new unseen fields to enable robots to perform weeding and harvesting. Our approach automatically labels onboard RGB images based on the robot’s pose and the current map of the field semantically segmented into crops and weeds. In this way, semantic labels are generated using the robot’s spatial information and the field arrangement’s crop row structure.
Previous heuristic-based methods for unsupervised semantic segmentation in agriculture proposed by Lottes et al. (2017) and Winterhalter et al. (2018) rely on poorly generalizing assumptions about field arrangements, e.g., absence of weeds in the crop row (Lottes et al., 2017), constant distance between plants’ rows (Lottes and Stachniss, 2017; Winterhalter et al., 2018), or non-overlapping vegetation components (Lottes et al., 2017). Although fully supervised deep learning-based approaches do not rely on geometric assumptions, they rely on in-domain human-labeled training data. The performance of such approaches is satisfactory when being deployed in conditions similar to those they were trained on. However, their performance usually rapidly deteriorates in novel deployment conditions, e.g., different crop species, weeds pressure, lighting conditions, or growth stage, requiring new human-labeled training data. This is costly and makes these approaches impractical for application when there is not enough time, money, or data to re-train the approach on new field conditions.
The main contribution of this paper is a novel heuristic approach for unsupervised soil-weed-crop segmentation in managed agricultural fields addressing these limitations. Our method automatically generates labels used to train deep semantic segmentation networks. The overview of our pipeline is shown in Figure 1. Our pipeline takes the current RGB image and the camera pose of the robotic platform as input to compute a semantic map of the field. As a key novelty, we use the semantic map to enforce the spatial consistency of labels. To this end, we propagate the information about the crop rows in the map, leading to better crop segmentation across different growth stages. Additionally, we do not assign labels to vegetation components that are close to the crop rows but are not classified as crops. This reduces possible labeling errors and thus improves model predictions after training on our generated labels. We use the generated image-label pairs to train an uncertainty-aware evidential semantic segmentation network (Sensoy et al., 2018). At inference, as a post-processing step, we exploit the predicted uncertainties to refine the final semantic predictions.
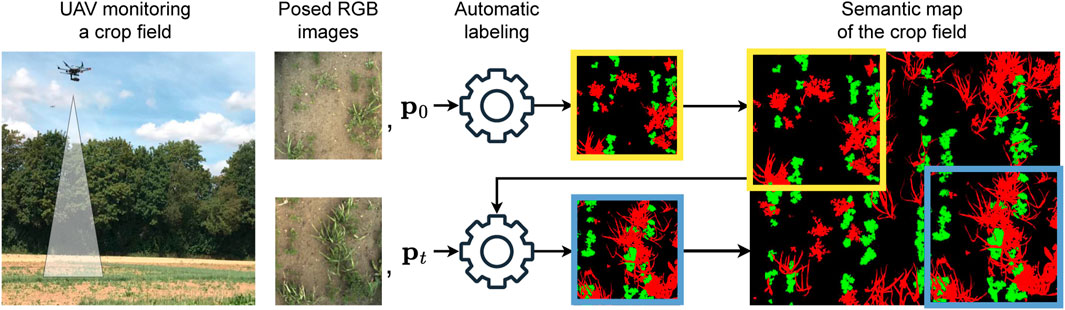
Figure 1. The overview of our pipeline to generate semantic labels for images of crop fields. We use a robotic platform equipped with an RGB camera to collect posed images of the field. Each image gets processed by our automatic labeling method, generating a semantic segmentation of the image to fuse into the semantic map. At each time step, we use the current semantic map to generate the image’s semantic label and update the map accordingly.
In sum, we make three key claims: our approach (i) generates more accurate semantic labels than previous unsupervised label generation approaches on multiple crop species, growth stages, and lighting conditions; (ii) we outperform previous unsupervised semantic segmentation approaches by combining our spatially consistent generated labels and uncertainty-aware semantic neural networks; and (iii) improve performance of fully supervised models on previously unseen crops, growth stages, or soil conditions after fine-tuning using our automatically generated labels. These claims are backed up by our experimental evaluation. We open-source our code upon paper acceptance.
Our work uses heuristic-based computer vision techniques for semantic segmentation of RGB images to automatically generate weed-crop segmentation labels of agricultural fields for training a semantic segmentation network. We train the network in an uncertainty-aware fashion using evidential deep learning (Sensoy et al., 2018) to post-process predictions at inference time based on their uncertainty.
Otsu (1979) proposed using gray-level histograms for binary image segmentation based on an automatic threshold assuming a bimodal distribution for fore- and background pixels. Pong et al. (1984) propose the region-growing algorithm segmenting images in multiple regions after providing initial seeds for each region. Similarly, the Watershed algorithm (Najman and Schmitt, 1996) requires user-defined markers to segment objects using a distance function. To overcome the need for initial seeds, Canny (1986) used edge detectors to distinguish regions. To incorporate statistical image features for segmentation, Loyd (1982) adopted the K-means algorithm. To allow automatic robotic intervention in the fields, Riehle et al. (2020) and Gao et al. (2020) applied semantic segmentation techniques to the agricultural domain. Lottes et al. (2017) further advance these general-purpose approaches by exploiting the field arrangement and deploying their method on a ground field robot. Similarly, our approach also exploits the field arrangement to automatically segment images. In contrast, we additionally enhance spatial label consistency using robotic semantic mapping. Further, we do not assign labels to image parts likely to include labeling errors. In this way, we reduce the number of erroneous crop and weed instances, which is essential to achieve high prediction performance and consistent uncertainty estimation of the trained deep neural network.
Recent approaches mainly use neural networks to extract latent image features for semantic segmentation. Various convolutional neural network architectures (Romera et al., 2018; He et al., 2017), and more recently, Vision transformers (Strudel et al., 2021; Cao et al., 2023) have been applied to semantic segmentation. A large portion of these approaches have also been evaluated or adapted to the agricultural domain. Cui et al. (2024) propose an improvement to the U-net architecture by Ronneberger et al. (2015) to segment corns and weeds while Zenkl et al. (2022) use the DeepLabV3 architecture by Chen et al. (2017) to segment wheat. These approaches usually require vast amounts of per-pixel human-labeled training data, covering all the desired crop species, growth stages, lighting conditions, and other deployment conditions to ensure promising test-time performance. Hence, many works have investigated how to reduce the labeling effort of deep learning-based approaches. One popular method is pre-training the network on different easy-to-label tasks, e.g., image classification (Deng et al., 2009) or using self-supervision (Chen et al., 2020), and fine-tuning the pre-trained network using few human-labeled per-pixel annotations specific to the target application. Other works propose to train networks on sparse labels instead of dense per-pixel labels (Lee et al., 2022), so called weakly supervised semantic segmentation. In the agricultural domain, Zhao et al. (2023) reduce the need for per-pixel labels using scrawl annotations, i.e., manually drawn lines, to weakly supervise a semantic segmentation model. Chen et al. (2024) remove per-pixel annotations completely, only exploiting reference images to localize disease symptoms in plants, using an innovative class activation mapping method. In contrast to Chen et al. (2024), we propose a new unsupervised approach to automatically generate per-pixel semantic segmentation labels exploiting domain knowledge of the field arrangement. Our semantic labels can be directly used for network training without the need for human labels or for fine-tuning pre-trained networks on unseen fields.
Classical neural networks are known to often provide overconfident wrong point estimate predictions (Abdar et al., 2021). Several works, including the one by Lakshminarayanan et al. (2017), use ensembles of multiple independently initialized and trained neural networks to quantify predictive uncertainty. Although ensembles improve prediction performance and model calibration, they induce high computational costs during training. Gal and Ghahramani (2016) propose Monte Carlo dropout to approximate predictive uncertainty with a single network trained with dropout. At inference, multiple forward passes with independently sampled dropout masks are performed to compute predictive uncertainty. Although more compute-efficient at train time, Monte Carlo dropout produces overconfident predictions compared to ensembles (Beluch et al., 2018b). More recently, Sensoy et al. (2018) proposed evidential deep learning for image classification to predict uncertainty using a single forward pass. As evidential deep learning performs on par with ensembles while drastically reducing online compute requirements, we adapt the evidential deep learning framework to our semantic segmentation task using the predictive uncertainties for label post-processing, facilitating deployment on compute-constrained robots. We use the network’s uncertainty to correct its prediction about the weeds, which is the most under-represented class and, thus, the most uncertain for the model.
Our approach combines a heuristic-based method to automatically generate partial but consistent per-pixel semantic labels. In contrast to learning-based approaches, our approach does not require human-labeled data and, at the same time, improves label consistency and, thus, the network’s prediction performance over previous heuristic-based approaches.
We propose a heuristic-based approach to automatically semantically segment RGB images of agricultural fields collected using unmanned ground vehicles (UGVs) or unmanned aerial vehicles (UAVs) in three classes: soil, crop, and weed. Based on the robot’s pose, we fuse each generated semantic image label in an online-built global semantic field map. A key aspect of our approach is that we enforce spatial label consistency based on the global semantic field map. To reduce the possibility of labeling errors, we only label the detected rows as crops and the vegetation components that are far away from the rows as weeds. In this way, we trade off label quality with quantity to improve prediction performance after training our uncertainty-aware semantic segmentation network (Sensoy et al., 2018) on labels extracted from the global semantic field map. At inference time, we post-process the network’s predictions using their associated uncertainty to refine uncertain vegetation predictions.
We perform semantic mapping to enforce spatial consistency across automatically generated semantic labels. Furthermore, the semantic map allows us to extract image-label pairs from the map with different rotations, positions, and scales. We assume that our robotic system is equipped with a downwards-facing RGB camera. At each time step
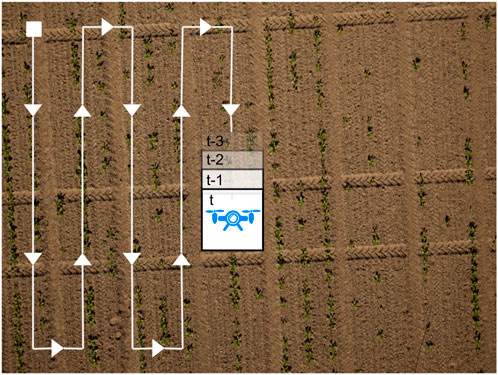
Figure 2. Example of a typical UAV mission. The coverage path along which we fuse semantic image labels is depicted in white, the square is the initial pose, and the arrows indicate the direction of movement. The images can overlap, but it is not required. This path maximizes the crop field coverage and is typically used in aerial data collection missions.
At each time step
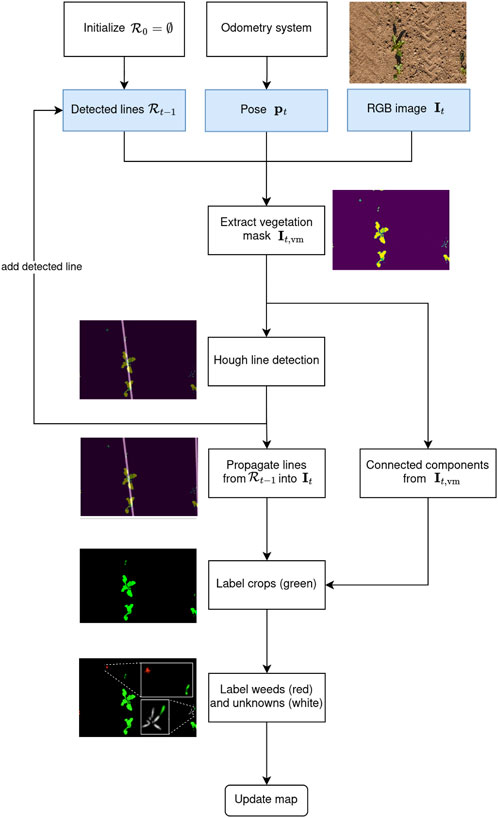
Figure 3. Flowchart of our automatic labeling approach with an example image. At time step
We compute a binary vegetation mask
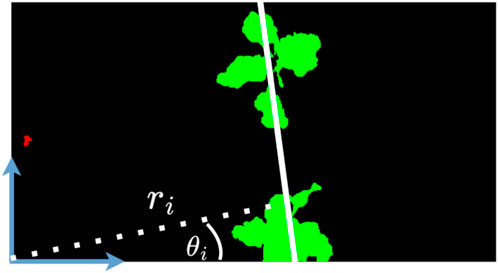
Figure 4. Given the vegetation mask, we visualize the line detected by the Hough transform (in white). Considering the origin as the bottom left corner, we show the parameters
We use our semantic map
For each line
Where
As we propagate our line predictions from previously recorded images into the current image, we use an eroded version of the vegetation mask
Naively classifying any vegetation component in
We aim to estimate crop sizes along the detected rows using these distances
where
Once we finish our mapping mission as described in Section 3.2, we can extract any number of image-label pairs with any size, rotation, and aspect ratio. We use the extracted labels to train a semantic segmentation network. We follow the evidential deep learning framework by Sensoy et al. (2018) to predict semantic segmentation and the network’s prediction uncertainty at the same time. Estimating the prediction uncertainty allows us to account for the “unknown” class by refining the network’s semantic predictions in a post-processing step described in Section 3.4.
The key idea behind evidential deep learning is to predict a Dirichlet distribution over all possible class probabilities instead of a single point estimate as in deterministic deep neural networks. In this way, the evidential network minimizes the prediction error while maximizing the prediction uncertainty for ambiguous image parts. We use evidential deep learning instead of Bayesian deep learning approaches (Gal and Ghahramani, 2016; Beluch et al., 2018a) as it is empirically shown to produce similarly or better-calibrated prediction uncertainties (Sensoy et al., 2018) while being computationally more efficient during training than ensemble methods and during inference than Monte Carlo dropout.
We train the network to minimize the Bayes risk cross-entropy for a pixel
where
for all
which is the average over all image pixels, iterating over all training images. At inference time, the network predicts the semantic class and an uncertainty for each pixel, that we use for our label refinement.
We use the network’s predicted Dirichlet distribution
where
We compute a binary mask
This threshold helps us avoid detecting as weeds a lot of small spikes of uncertainty that could arise because of shadows, reflections, or insects. In this way, we only act upon vegetation components where there is a large uncertain area. If the network is uncertain about the prediction of crop component
If crop component
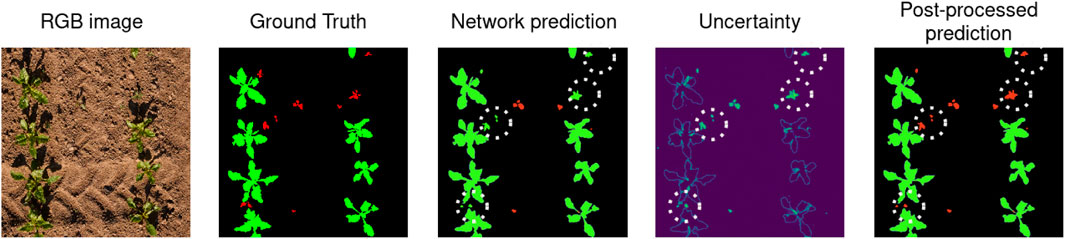
Figure 5. For the RGB input image on the left, we show the semantic ground truth labels, where crops are represented in green, weeds in red, and the soil in black. Then, we show our network’s prediction and we highlights some mistakes using white dotted circles, where weeds are mislabeled as crops. The fourth image shows our network’s uncertainty. As expected, the network is mostly uncertain about the boundaries of the plants and about the weeds, we see that even the weeds labeled as crops in our prediction have high uncertainty. The last image shows our post-processed prediction, after we label as weeds the highly uncertain vegetation components. We can see that this corrects many of the network’s errors.
The main focus of this work is an automatic labeling pipeline for semantic soil-weed-crop segmentation of RGB images. The results of our experiments support our key claims: our approach (i) generates more accurate semantic labels than previous unsupervised label generation approaches on multiple datasets; (ii) we outperform previous unsupervised semantic segmentation approaches by combing our spatially consistent generated labels and uncertainty-aware semantic neural networks; and (iii) we improve the performance of fully supervised models on previously unseen crops, growth stages, and soil conditions after fine-tuning the network using our automatically generated labels.
We use four datasets, three of which are publicly available: PhenoBench (Weyler et al., 2024), as well as the Carrots and Onions from Lincoln University (Bosilj et al., 2020), and a Sugar Beets dataset introduced by Weyler et al. (2022b). The Carrots dataset was recorded in Lincolnshire, United Kingdom, in June. The field is under substantial weed pressure and contains weeds with a similar appearance to the crop. Furthermore, several regions of vegetation contain crops and weeds in close proximity. The Onions dataset was also recorded in Lincolnshire, United Kingdom, but in April. The weed pressure is lower compared to the Carrots dataset. The PhenoBench dataset was recorded in Meckenheim, Germany, on different dates between May and June to capture different growth stages. The field contains two varieties of sugar beets and six different weed varieties. The weed pressure varies as the dataset contains images from fields that were fully, partially, or not treated at all with herbicides. The Sugar Beets dataset was also recorded in Meckenheim, Germany, over five different weekly sessions. The field is arranged with small spacing between plants and shows high weed pressure, inducing challenging conditions. We refer to Table 1 for information about the camera, image resolution, and ground sampling distance of the datasets.

Table 1. Details for the datasets used in the paper: name, reference paper, camera sensor, image resolution and GSD.
We use ERFNet (Romera et al., 2018) as our network trained using the Adam (Kingma and Ba, 2015) optimizer, a learning rate of 0.01, and a batch size of 32. We set
We use the intersection over union (IoU) (Everingham et al., 2010) as a metric for all of our experiments. For the automatic labeling pipeline, we also report the boundary IoU (Cheng et al., 2021) to have a better understanding of the approaches’ limitations. The reported mean IoU (mIoU) values are the macro-averages over all classes.
We use three baselines: two are general-purpose unsupervised semantic segmentation networks not specifically developed for the agricultural domain, while one is an automatic labeling method specifically developed for the agricultural domain. The first baseline is STEGO by Hamilton et al. (2022), which provides an official implementation for the evaluation alongside their models. We use the model trained on MS COCO (Lin et al., 2014) with the vision transformer architecture (Dosovitskiy et al., 2021). STEGO predicts different per-pixel features and then clusters them using self and cross attention mechanisms (Vaswani et al., 2017). Our second baseline is U2Seg by Niu et al. (2024), which builds on top of STEGO and uses instance information to overcome some of the limitations of the previous work; they also open-source their code and provide their models. U2Seg proposes a universal segmentation, coupling instances and semantic classes at training time, to predict clusters at inference time for which they recover class and instance labels. We use the model trained on Imagenet (Deng et al., 2009) and MS COCO with 800 clusters. Lottes et al. (2016) propose a domain-specific method for generating per-pixel crop and weed labels. They use a vegetation mask to detect the main crop row and then label all other vegetation components as weeds. We use their official implementation, removing the NIR image channels. We evaluate their automatically generated labels (base) and the performance of ERFNet trained on their labels (learned). We train the same network with the same training hyperparameters on their and our generated label to ensure a fair comparison. We report the results of ERFNet trained on the manually annotated training set of PhenoBench and evaluated on the validation set as an upper performance bound.
In the first experiment, we show that our automatic labeling pipeline generates more accurate semantic soil-weed-crop labels than other methods on multiple datasets. We compare against two general-purpose unsupervised semantic segmentation networks and the domain-specific approach by Lottes et al. (2016).
We show the results on all four datasets in Table 3. The general-purpose approaches perform worse than the domain-specific methods across all datasets, except for U2Seg on the Onions dataset. As Onions have thin leaves, they are hard to detect with common color histogram thresholding methods, such as the one by Lottes et al. (2016). Furthermore, the weeds in this dataset are the same size as the crops, leading to bias in crop row detection and introducing a higher risk of confusing weeds and crops. Our approach for label generation, referred to as Ours (base), shows higher crop label quality than Lottes (base) while performing on par or better in terms of weed label quality. Particularly, Lottes (base) confuses substantially more weeds with crops, while our approach, by design, does not assign labels to hard-to-label vegetation components, as described in Section 3.2. The Carrots dataset is the only one where U2Seg outperforms the domain-specific approaches, which suffer from the weed pressure when estimating the crop rows. Our method consistently outperforms all other baselines across all datasets with different crop species, weed pressure, growth stages and lighting conditions. Most approaches fail on the Onions dataset due to brighter illumination and thin crops. In contrast, our approach improves by approx.
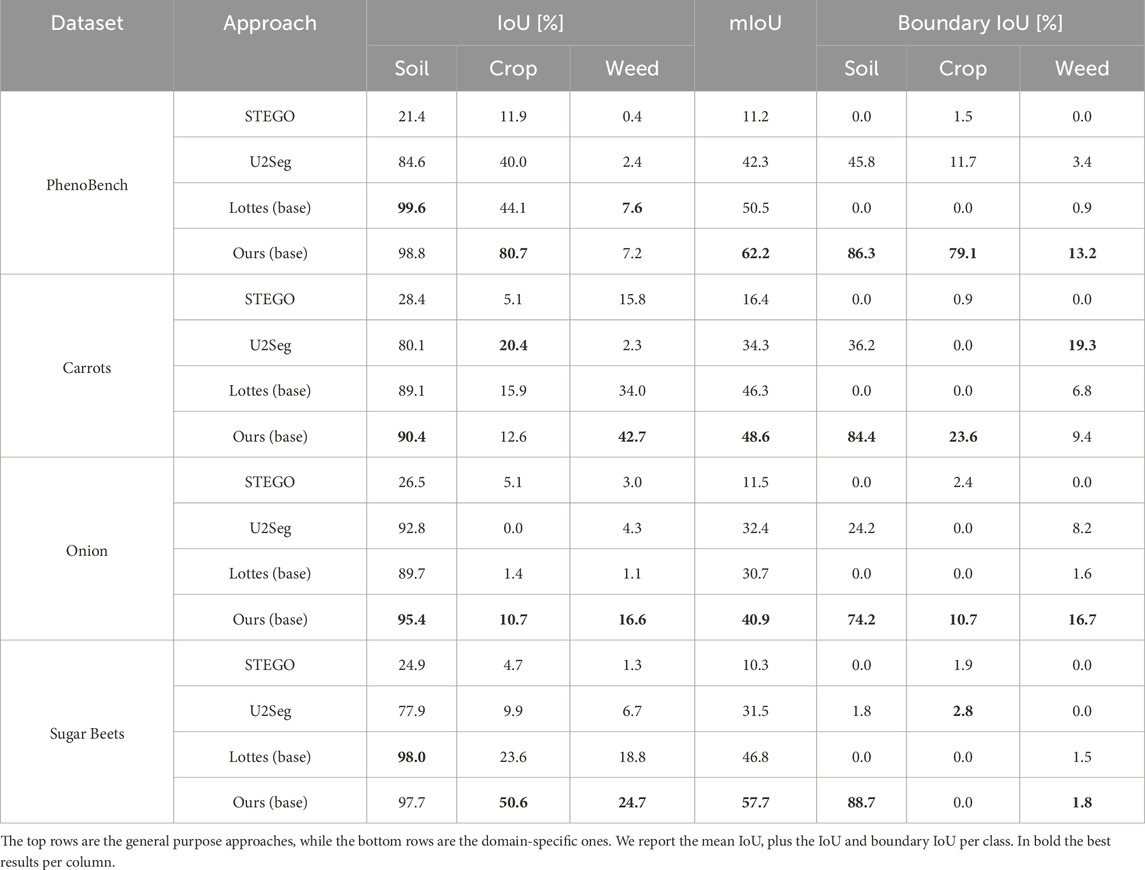
Table 3. Performance of all the baselines on the PhenoBench dataset, Carrots dataset, Onion dataset, and Sugar Beets dataset.
The boundary IoU confirms the result of the standard IoU metric. As shown in Table 3, the approach by Lottes et al. (2016) poorly segments boundaries on most of the datasets. This might be due to wrongly segmented vegetation masks. Aiming to include the boundary of weeds more accurately may worsen the overall performance since soil could be wrongly considered as vegetation. We hypothesise that our approach might suffer from the same problem on the Carrots dataset. The difference between IoU and boundary IoU per class suggests that we underestimate the size of weeds, i.e., high IoU but low boundary IoU for weeds, and overestimate crop size, i.e., low IoU but high boundary IoU for crops. On the Carrots dataset U2Seg outperforms the other methods on the weeds boundary IoU. The weed IoU suggests that U2Seg overestimates weeds, thus obtaining a boost as the total number of pixels in the IoU computation is low. On the Onions dataset, our method’s IoU and boundary IoU are almost the same irrespective of the semantic class since the crops and weeds are thin. Thus, the boundary area covers the whole vegetation instance. The other approaches fail to correctly assign weed and crop boundaries on the Onion dataset, which follows from the low weed and crop IoU. On the Sugar Beets dataset, all approaches fail to predict boundaries, most likely due to unusually high weed pressure. Our method accurately segments soil boundaries, suggesting that it at least successfully differentiates between soil and vegetation. Overall, the results suggest that most approaches underestimate the size of vegetation, both crops and weeds. Instead, our automatic labeling method shows the strongest boundary segmentation performance across all methods and classes on most datasets, often by a large margin compared to the second-best method. This further verifies our claim that our automatic labeling pipeline generates more accurate semantic soil-weed-crop labels than previous methods. We show qualitative results of Lottes et al. (2016) and our approach in Figure 6.
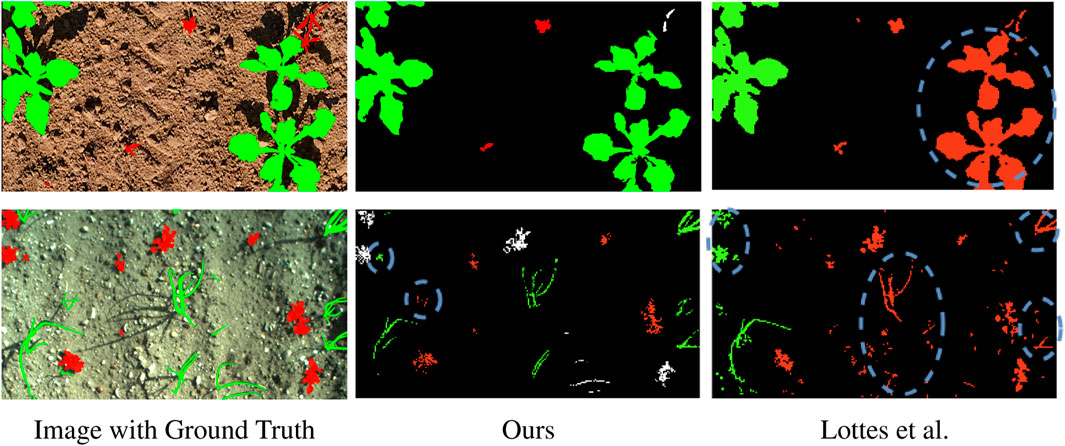
Figure 6. Qualitative results of our and Lottes et al. methods on PhenoBench (top row) and Onions (bottom row). Soil is black, crops are green, weeds are red, vegetation that we leave unlabeled is white. In the dashed blue circles, we highlight segmentation errors.
The second experiment evaluates the performance of our automatic label generation combined with network training and uncertainty post-processing on the PhenoBench dataset. We show that training the evidential ERFNet using our automatically generated labels outperforms other unsupervised semantic segmentation models. The general-purpose learning-based approaches have not been fine-tuned on human-labeled field images to ensure a fair comparison. Our approach and Lottes et al. (2016) generate labels on the PhenoBench training set. We use the public training set of images to have a fair comparison with the fully suprvised ERFNet model, trained on the manual labels. Trained models are evaluated on the official PhenoBench validation set.
Table 4 summarizes the results. We use (learned) to refer to the results obtained by ERFNet after being trained on the labels generated by the approach, and we use (+uncertainty) to refer to the previous results once we post-process them using the uncertainty estimated by the model. The approach by Lottes et al. (2016) confuses more crops with weeds since it naively assigns all vegetation components that are not on the main crop row to the weed class. Hence, Lottes et al. (2016) introduce inconsistent labels in the model’s training data. Thus, training the ERFNet on Lottes’ labels does not yield uncertainty estimations that are useful for improving the predictions during post-processing. Additionally, this leads to smaller performance improvements after training on their labels than after training on our labels. Using our generated labels to train the ERFNet substantially improves the weed and crop predictions over directly using our generated labels. We further improve mIoU and weed predictions by exploiting the estimated uncertainties in Ours (uncertainty) for post-processing. Most importantly, Ours (uncertainty) noticeably closes the performance gap between fully supervised and state-of-the-art unsupervised approaches. However, the ERFNet trained on human-labeled training images still predicts weeds more accurately. As the fully supervised model predicts more weeds, it also confuses weeds with crops more often. Hence, our approach performs better on both the crop and soil classes. This experiment confirms that our method’s conservative approach to labeling, ignoring vegetation components likely to introduce labeling errors combined with evidential deep learning, is a viable solution to largely reduce the need for manually annotated images.
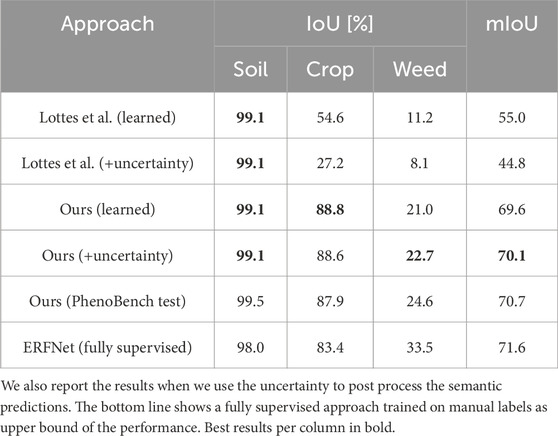
Table 4. Performance of ERFNet trained on the labels generated by ours and the approach by Lottes et al.
In the third experiment, we show that our approach enhances the performance of networks trained in a fully supervised fashion by fine-tuning on unseen fields using our automatically generated labels. We do not use our evidential network but train an ERFNet using the standard cross-entropy loss to seamlessly fine-tune existing networks pre-trained in a fully supervised fashion. We train two ERFNets, one on each of the human-labeled training sets of PhenoBench and Sugar Beets. We deploy the two models on all four datasets without fine-tuning. Then, we fine-tune the two models leveraging our automatically generated labels for the Sugar Beets and PhenoBench datasets. Each model is fine-tuned on the dataset that it was not trained on.
In Table 5, we show the performance of the two models. In brackets, we provide the performance difference after fine-tuning, where blue and red indicate performance improvements or degradations, respectively. The gray rows show the models’ performances on the dataset they were trained on. Due to the domain gap between datasets, the models that were not fine-tuned have a lower performance when evaluated on unseen data. Fine-tuning the models makes the performance over the original training data worse as they aim to learn features that are common to both datasets. Our results suggest that using our automatically generated labels helps to close the performance gap on previously unseen datasets with different crops, soil types, lighting conditions, and sensor setups. Generally, our fine-tuned models perform better on all classes and datasets, even on the Onions and Carrots datasets, the model was not pre-trained nor fine-tuned on. Only the model that is fine-tuned on the Sugar Beets dataset does not improve performance on the Carrots dataset. We hypothesize this is because the PhenoBench dataset is approx.
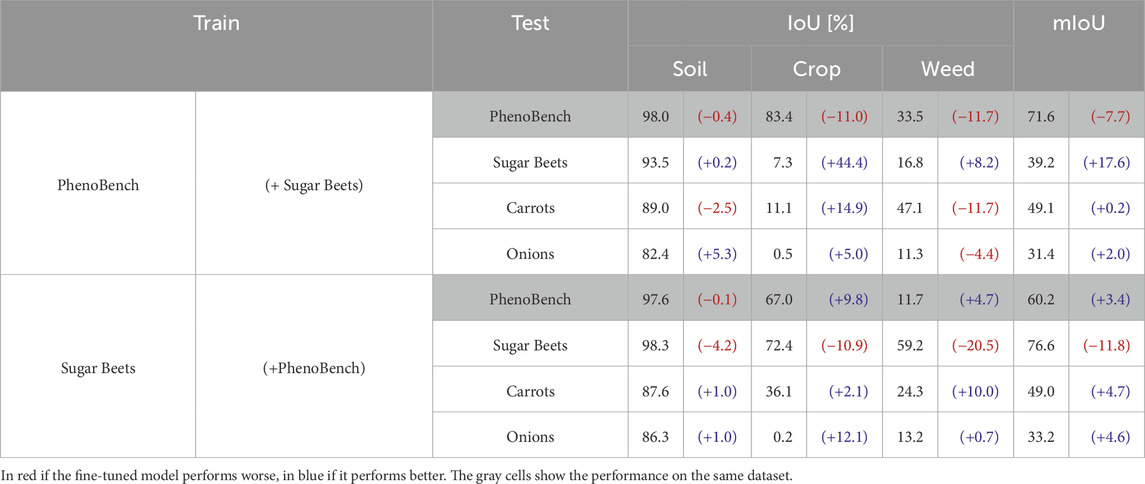
Table 5. Performance of fully supervised models trained on manually annotated data, and in brackets the difference with respect to the model after fine-tuning.
A robust perception system is crucial for the successful deployment of robotic platforms in arable fields. Most perception systems rely on data-driven machine learning approaches to train vision models that automatically interpret the data collected with onboard sensors, such as RGB cameras. Thus, reliable and accurate learning-based perception systems are crucial to providing valuable information to farmers or autonomous robots. Most learning-based semantic segmentation approaches assume access to large amounts of human-labeled data required to train the vision model. However, their performance rapidly decreases in field conditions they were not trained on, i.e., different crop species, growth stages, weed pressure, and lighting conditions.
To address this issue, we proposed an automatic labeling approach to obtain semantic information from RGB images of agricultural fields. Our method shows semantic segmentation performance close to the performance of a model trained on large amounts of human-labeled data in a fully supervised fashion. This significantly reduces the need for manually annotated data, reducing costs and relaxing the need for domain experts. The arable field dataset works considered in our experimental evaluation report an average of 2 h per image for labeling the Onions dataset, 3–4 h per image for the Carrots dataset, and 1–3.5 h per image for the PhenoBench dataset. All of the datasets went through at least two labeling rounds, doubling the costs. This highlights the need for new labeling paradigms beyond fully supervised model training while maintaining strong prediction performance. Our method is a crucial step towards closing the performance gap between models trained in an unsupervised fashion and fully supervised models without adding additional labeling costs.
In our experiments, we show that the fully supervised approach has a lower performance in segmenting crops compared to our unsupervised method, as it is trained on more weed instances. Nevertheless, the fully supervised method still shows the highest mIoU. The unsupervised methods are not exposed to enough weed labels, making them assign the crop class more often. Since the number of crop pixels is generally higher, these errors have a smaller impact on the crop than on the weed segmentation. We also investigate how to use our automatic labeling in combination with supervised methods to improve the overall performance in challenging scenarios, i.e., in unseen fields with new crop species and different weed pressure. Fine-tuning comes at the cost of performing worse on the pre-training dataset, as shown in Table 5. The degradation largely depends on the size and similarity of the pre-training and automatically labeled dataset used for fine-tuning. Future work could investigate continuous learning methods to train on the newly automatically labeled images without catastrophically forgetting what has already been learned.
The need for posed images can be a limitation of our method as it cannot be applied to a dataset of unposed images. However, most of the agricultural datasets are recorded using aerial or ground vehicles that, by default, provide spatial information while recording images in the field, often using GNSS systems such as GPS. Furthermore, we assume deployment in a managed agricultural field. If this assumption does not hold and the weeds are larger than the crops, our crop row detection fails and leads to degraded results. Our results, as well as those by Lottes et al. (2016), show that we could make use of a better vegetation mask to improve unsupervised methods. One possible solution would be to use NIR images, which are less dependent on the lighting conditions compared to RGB images. NIR images are already commonly used for crop segmentation in agriculture (Sahin et al., 2023; Colorado et al., 2020). Moreover, our approach leverages uncertainty estimates to post-process semantic predictions. Current state-of-the-art methods are known to produce partially miscalibrated uncertainty estimates (Beluch et al., 2018a). Thus, our post-processing could benefit from improvements in uncertainty-aware deep learning. Finally, we plan to deploy our approach on a real robot to perform field trials.
In this paper, we presented a novel approach to automatically generate semantic soil-crop-weed labels of images from agricultural fields. We evaluated our approach on four datasets recorded with different robotic platforms and in various fields. Our approach outperforms previous domain-agnostic and domain-specific unsupervised labeling approaches. Furthermore, we showed that our generated labels allow fine-tuning networks trained in a fully supervised fashion on one dataset to other agricultural fields, e.g., different species, growth stages, and field conditions. In this way, our approach increases the semantic segmentation generalization capabilities of existing networks for soil-weed-crop segmentation without additional human labeling effort.
The original contributions presented in the study are included in the article/supplementary material, further inquiries can be directed to the corresponding author.
GR: Conceptualization, Formal Analysis, Investigation, Methodology, Software, Validation, Visualization, Writing–original draft, Writing–review and editing. JR: Conceptualization, Investigation, Methodology, Writing–original draft, Writing–review and editing. MP: Conceptualization, Supervision, Validation, Writing–original draft, Writing–review and editing. JB: Conceptualization, Formal Analysis, Supervision, Validation, Writing–original draft, Writing–review and editing. CS: Conceptualization, Funding acquisition, Project administration, Resources, Supervision, Validation, Writing–original draft, Writing–review and editing.
The author(s) declare that financial support was received for the research, authorship, and/or publication of this article. This work has been funded by the Deutsche Forschungsgemeinschaft (DFG, German Research Foundation) under Germany’s Excellence Strategy, EXC2070-390732324-PhenoRob.
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
The author(s) declare that no Generative AI was used in the creation of this manuscript.
All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article, or claim that may be made by its manufacturer, is not guaranteed or endorsed by the publisher.
Abdar, M., Pourpanah, F., Hussain, S., Rezazadegan, D., Liu, L., Ghavamzadeh, M., et al. (2021). A review of uncertainty quantification in deep learning: techniques, applications and challenges. Inf. fusion 76, 243–297. doi:10.1016/j.inffus.2021.05.008
Ahmadi, A., Nardi, L., Chebrolu, N., and Stachniss, C. (2020). “Visual servoing-based navigation for monitoring row-crop fields,” in Proc. Of the IEEE intl. Conf. On robotics and automation (ICRA).
Balabantaray, A., Behera, S., Liew, C., Chamara, N., Singh, M., Jhala, A. J., et al. (2024). Targeted weed management of palmer amaranth using robotics and deep learning (yolov7). Front. Robotics AI 11, 1441371. doi:10.3389/frobt.2024.1441371
Beluch, W. H., Genewein, T., Nurnberger, A., and Kohler, J. M. (2018a). “The power of ensembles for active learning in image classification,” in Proc. Of the IEEE/CVF conf. On computer vision and pattern recognition (CVPR), 9368–9377.
Beluch, W. H., Genewein, T., Nürnberger, A., and Köhler, J. M. (2018b). “The power of ensembles for active learning in image classification,” in Proc. Of the IEEE/CVF conf. On computer vision and pattern recognition (CVPR).
Boatswain Jacques, A. A., Adamchuk, V. I., Park, J., Cloutier, G., Clark, J. J., and Miller, C. (2021). Towards a machine vision-based yield monitor for the counting and quality mapping of shallots. Front. Robotics AI 8, 627067. doi:10.3389/frobt.2021.627067
Bosilj, P., Aptoula, E., Duckett, T., and Cielniak, G. (2020). Transfer learning between crop types for semantic segmentation of crops versus weeds in precision agriculture. J. Field Robotics (JFR) 37, 7–19. doi:10.1002/rob.21869
Canny, J. F. (1986). A computational approach to edge detection. IEEE Trans. Pattern Analysis Mach. Intell. (TPAMI) 8, 679–698. doi:10.1109/TPAMI.1986.4767851
Cao, H., Wang, Y., Chen, J., Jiang, D., Zhang, X., Tian, Q., et al. (2023). “Swin-unet: unet-like pure transformer for medical image segmentation,” in Proc. Of the europ. Conf. On computer vision (ECCV).
Chen, J., Guo, J., Zhang, H., Liang, Z., and Wang, S. (2024). Weakly supervised localization model for plant disease based on siamese networks. Front. Plant Sci. 15, 1418201. doi:10.3389/fpls.2024.1418201
Chen, L., Papandreou, G., Schroff, F., and Adam, H. (2017). Rethinking atrous convolution for semantic image segmentation. arXiv preprint arXiv:1706.05587.
Chen, T., Kornblith, S., Norouzi, M., and Hinton, G. (2020). “A simple framework for contrastive learning of visual representations,” in Proc. Of the intl. Conf. On machine learning (ICML).
Cheng, B., Girshick, R., Dollár, P., Berg, A. C., and Kirillov, A. (2021). “Boundary iou: improving object-centric image segmentation evaluation,” in Proc. Of the IEEE/CVF conf. On computer vision and pattern recognition (CVPR), 15334–15342.
Cheng, C., Fu, J., Su, H., and Ren, L. (2023). Recent advancements in agriculture robots: benefits and challenges. Machines 11, 48. doi:10.3390/machines11010048
Colorado, J. D., Calderon, F. C., Mendez, D., Petro, E., Rojas, J. P., Correa, E. S., et al. (2020). A novel nir-image segmentation method for the precise estimation of above-ground biomass in rice crops. PLOS ONE 15, e0239591. doi:10.1371/journal.pone.0239591
Cui, J., Tan, F., Bai, N., and Fu, Y. (2024). Improving u-net network for semantic segmentation of corns and weeds during corn seedling stage in field. Front. Plant Sci. 15, 1344958. doi:10.3389/fpls.2024.1344958
Dainelli, R., Bruno, A., Martinelli, M., Moroni, D., Rocchi, L., Morelli, S., et al. (2024). Granoscan: an ai-powered mobile app for in-field identification of biotic threats of wheat. Front. Plant Sci. 15, 1298791. doi:10.3389/fpls.2024.1298791
Deng, J., Dong, W., Socher, R., Li, L., Li, K., and Fei-Fei, L. (2009). “Imagenet: a large-scale hierarchical image database,” in Proc. Of the IEEE conf. On computer vision and pattern recognition (CVPR).
Dosovitskiy, A., Beyer, L., Kolesnikov, A., Weissenborn, D., Zhai, X., Unterthiner, T., et al. (2021). “An image is worth 16x16 words: transformers for image recognition at scale,” in Proc. Of the intl. Conf. On learning representations (ICLR).
Everingham, M., Van Gool, L., Williams, C., Winn, J., and Zisserman, A. (2010). The pascal visual object classes (VOC) challenge. Intl. J. Comput. Vis. (IJCV) 88, 303–338. doi:10.1007/s11263-009-0275-4
Ewert, F., Baatz, R., and Finger, R. (2023). Agroecology for a sustainable agriculture and food system: from local solutions to large-scale adoption. Annu. Rev. Resour. Econ. 15, 351–381. doi:10.1146/annurev-resource-102422-090105
Felzenszwalb, P. F., and Huttenlocher, D. P. (2004). Efficient graph-based image segmentation. Intl. J. Comput. Vis. (IJCV) 59, 167–181. doi:10.1023/b:visi.0000022288.19776.77
Gal, Y., and Ghahramani, Z. (2016). “Dropout as a bayesian approximation: representing model uncertainty in deep learning,” in Proc. Of the intl. Conf. On machine learning (ICML).
Gao, J., Wang, B., Wang, Z., Wang, Y., and Kong, F. (2020). A wavelet transform-based image segmentation method. Intl. J. Light Electron Opt. 208, 164123. doi:10.1016/j.ijleo.2019.164123
Hamilton, M., Zhang, Z., Hariharan, B., Snavely, N., and Freeman, W. T. (2022). “Unsupervised semantic segmentation by distilling feature correspondences,” in Proc. Of the intl. Conf. On learning representations (ICLR).
He, K., Gkioxari, G., Dollár, P., and Girshick, R. (2017). “Mask R-CNN,” in Proc. Of the IEEE intl. Conf. On computer vision (ICCV).
Höffmann, M., Patel, S., and Büskens, C. (2023). Optimal coverage path planning for agricultural vehicles with curvature constraints. Agriculture 13, 2112. doi:10.3390/agriculture13112112
Horrigan, L., Lawrence, R. S., and Walker, P. (2002). How sustainable agriculture can address the environmental and human health harms of industrial agriculture. Environ. health Perspect. 110, 445–456. doi:10.1289/ehp.02110445
Hough, P. V. C. (1959). “Machine analysis of bubble chamber pictures,” in Proc. Of the intl. Conf. On high-energy accelerators and instrumentation.
Kingma, D., and Ba, J. (2015). “Adam: a method for stochastic optimization,” in Proc. Of the intl. Conf. On learning representations (ICLR).
Lakshminarayanan, B., Pritzel, A., and Blundell, C. (2017). “Simple and scalable predictive uncertainty estimation using deep ensembles,” in Proc. Of the conf. Neural information processing systems (NIPS).
Lee, J., Oh, S. J., Yun, S., Choe, J., Kim, E., and Yoon, S. (2022). “Weakly supervised semantic segmentation using out-of-distribution data,” in Proc. Of the IEEE/CVF conf. On computer vision and pattern recognition (CVPR).
Lin, T., Maire, M., Belongie, S., Hays, J., Perona, P., Ramanan, D., et al. (2014). “Microsoft COCO: common objects in context,” in Proc. Of the europ. Conf. On computer vision (ECCV).
Lottes, P., Höferlin, M., Sander, S., Müter, M., Schulze-Lammers, P., and Stachniss, C. (2016). “An effective classification system for separating sugar beets and weeds for precision farming applications,” in Proc. Of the IEEE intl. Conf. On robotics and automation (ICRA).
Lottes, P., Höferlin, M., Sander, S., and Stachniss, C. (2017). Effective vision-based classification for separating sugar beets and weeds for precision farming. J. Field Robotics (JFR) 34, 1160–1178. doi:10.1002/rob.21675
Lottes, P., and Stachniss, C. (2017). “Semi-supervised online visual crop and weed classification in precision farming exploiting plant arrangement,” in Proc. Of the IEEE/RSJ intl. Conf. On intelligent robots and systems (IROS).
Loyd, S. P. (1982). Least squares quantization in pcm. IEEE Trans. Inf. Theory 28, 129–137. doi:10.1109/TIT.1982.1056489
Magistri, F., Weyler, J., Gogoll, D., Lottes, P., Behley, J., Petrinic, N., et al. (2023). From one field to another – unsupervised domain adaptation for semantic segmentation in agricultural robotics. Comput. Electron. Agric. 212, 108114. doi:10.1016/j.compag.2023.108114
Murugan, K., Shankar, B. J., Sumanth, A., Sudharshan, C. V., and Reddy, G. V. (2020). “Smart automated pesticide spraying bot,” in Proc. Of the intl. Conf. On intelligent sustainable systems (ICISS).
Najman, L., and Schmitt, M. (1996). Geodesic saliency of watershed contours and hierarchical segmentation. IEEE Trans. Pattern Analysis Mach. Intell. (TPAMI) 18, 1163–1173. doi:10.1109/34.546254
Niu, D., Wang, X., Han, X., Lian, L., Herzig, R., and Darrell, T. (2024). “Unsupervised universal image segmentation,” in Proc. Of the IEEE/CVF conf. On computer vision and pattern recognition (CVPR).
Otsu, N. (1979). A threshold selection method from gray-level histograms. IEEE Trans. Syst. Man, Cybern. 9, 62–66. doi:10.1109/tsmc.1979.4310076
Pan, Y., Magistri, F., Läbe, T., Marks, E., Smitt, C., McCool, C., et al. (2023). “Panoptic mapping with fruit completion and pose estimation for horticultural robots,” in Proc. Of the IEEE/RSJ intl. Conf. On intelligent robots and systems (IROS).
Pong, T.-C., Shapiro, L. G., Watson, L. T., and Haralick, R. M. (1984). Experiments in segmentation using a facet model region grower. Comput. Vis. Graph. Image Process. 25, 1–23. doi:10.1016/0734-189x(84)90046-x
Riehle, D., Reiser, D., and Griepentrog, H. W. (2020). Robust index-based semantic plant/background segmentation for rgb-images. Comput. Electron. Agric. 169, 105201–201. doi:10.1016/j.compag.2019.105201
Romera, E., Alvarez, J. M., Bergasa, L. M., and Arroyo, R. (2018). ERFNet: efficient residual factorized ConvNet for real-time semantic segmentation. IEEE Trans. Intelligent Transp. Syst. (TITS) 19, 263–272. doi:10.1109/tits.2017.2750080
Ronneberger, O., Fischer, P., and Brox, T. (2015). “U-net: convolutional networks for biomedical image segmentation,” in Proc. Of the medical image computing and computer-assisted intervention (MICCAI).
Sahin, H. M., Miftahushudur, T., Grieve, B., and Yin, H. (2023). Segmentation of weeds and crops using multispectral imaging and crf-enhanced u-net. Comput. Electron. Agric. 211, 107956. doi:10.1016/j.compag.2023.107956
Saqib, M. A., Aqib, M., Tahir, M. N., and Hafeez, Y. (2023). Towards deep learning based smart farming for intelligent weeds management in crops. Front. Plant Sci. 14, 1211235. doi:10.3389/fpls.2023.1211235
Sensoy, M., Kaplan, L., and Kandemir, M. (2018). “Evidential deep learning to quantify classification uncertainty,” in Proc. Of the conf. On neural information processing systems (NeurIPS).
Storm, H., Seidel, S., Klingbeil, L., Ewert, F., Vereecken, H., Amelung, W., et al. (2024). Research priorities to leverage smart digital technologies for sustainable crop production. Eur. J. Agron. 156, 127178. doi:10.1016/j.eja.2024.127178
Strudel, R., Garcia, R., Laptev, I., and Schmid, C. (2021). “Segmenter: transformer for semantic segmentation,” in Proc. Of the IEEE/CVF intl. Conf. On computer vision (ICCV).
Vaswani, A., Shazeer, N., Parmar, N., Uszkoreit, J., Jones, L., Gomez, A. N., et al. (2017). “Attention is all you need,” in Proc. Of the conf. On neural information processing systems (NeurIPS).
Walter, A., Finger, R., Huber, R., and Buchmann, N. (2017). Smart farming is key to developing sustainable agriculture. Proc. Natl. Acad. Sci. 114, 6148–6150. doi:10.1073/pnas.1707462114
Wang, J., Lan, C., Liu, C., Ouyang, Y., Qin, T., Lu, W., et al. (2022). Generalizing to unseen domains: a survey on domain generalization. IEEE Trans. Knowl. Data Eng. 35, 1–8072. doi:10.1109/TKDE.2022.3178128
Weyler, J., Magistri, F., Marks, E., Chong, Y. L., Sodano, M., Roggiolani, G., et al. (2024). Phenobench: a large dataset and benchmarks for semantic image interpretation in the agricultural domain. IEEE Trans. Pattern Analysis Mach. Intell. (TPAMI) 46, 9583–9594. doi:10.1109/TPAMI.2024.3419548
Weyler, J., Magistri, F., Seitz, P., Behley, J., and Stachniss, C. (2022a). “In-field phenotyping based on crop leaf and plant instance segmentation,” in Proc. Of the IEEE winter conf. On applications of computer vision (WACV).
Weyler, J., Quakernack, J., Lottes, P., Behley, J., and Stachniss, C. (2022b). Joint plant and leaf instance segmentation on field-scale uav imagery. IEEE Robotics Automation Lett. (RA-L) 7, 3787–3794. doi:10.1109/lra.2022.3147462
Winterhalter, W., Fleckenstein, F. V., Dornhege, C., and Burgard, W. (2018). Crop row detection on tiny plants with the pattern Hough transform. IEEE Robotics Automation Lett. (RA-L) 3, 3394–3401. doi:10.1109/lra.2018.2852841
Wu, X., Aravecchia, S., Lottes, P., Stachniss, C., and Pradalier, C. (2020). Robotic weed control using automated weed and crop classification. J. Field Robotics (JFR) 37, 322–340. doi:10.1002/rob.21938
Zenkl, R., Timofte, R., Kirchgessner, N., Roth, L., Hund, A., Van Gool, L., et al. (2022). Outdoor plant segmentation with deep learning for high-throughput field phenotyping on a diverse wheat dataset. Front. Plant Sci. 12, 774068. doi:10.3389/fpls.2021.774068
Keywords: agricultural automation, robotic crop monitoring, deep learning for agricultural robots, semantic scene understanding, automatic labeling, unsupervised learning
Citation: Roggiolani G, Rückin J, Popović M, Behley J and Stachniss C (2025) Unsupervised semantic label generation in agricultural fields. Front. Robot. AI 12:1548143. doi: 10.3389/frobt.2025.1548143
Received: 19 December 2024; Accepted: 31 January 2025;
Published: 25 February 2025.
Edited by:
Dario Calogero Guastella, University of Catania, ItalyReviewed by:
Nataliya Strokina, Tampere University, FinlandCopyright © 2025 Roggiolani, Rückin, Popović, Behley and Stachniss. This is an open-access article distributed under the terms of the Creative Commons Attribution License (CC BY). The use, distribution or reproduction in other forums is permitted, provided the original author(s) and the copyright owner(s) are credited and that the original publication in this journal is cited, in accordance with accepted academic practice. No use, distribution or reproduction is permitted which does not comply with these terms.
*Correspondence: Gianmarco Roggiolani, Z3JvZ2dpb2xAdW5pLWJvbm4uZGU=
Disclaimer: All claims expressed in this article are solely those of the authors and do not necessarily represent those of their affiliated organizations, or those of the publisher, the editors and the reviewers. Any product that may be evaluated in this article or claim that may be made by its manufacturer is not guaranteed or endorsed by the publisher.
Research integrity at Frontiers

Learn more about the work of our research integrity team to safeguard the quality of each article we publish.